Abstract
Biomolecule-based nanostructures are inherently multifunctional and harbour diverse biological activities, which can be explored for cancer nanomedicine. The supramolecular properties of biomolecules can be precisely programmed for the design of smart drug delivery vehicles, enabling efficient transport in vivo, targeted drug delivery and combinatorial therapy within a single design. In this Review, we discuss biomolecule-based nanostructures, including polysaccharides, nucleic acids, peptides and proteins, and highlight their enormous design space for multifunctional nanomedicines. We identify key challenges in cancer nanomedicine that can be addressed by biomolecule-based nanostructures and survey the distinct biological activities, programmability and in vivo behaviour of biomolecule-based nanostructures. Finally, we discuss challenges in the rational design, characterization and fabrication of biomolecule-based nanostructures, and identify obstacles that need to be overcome to enable clinical translation.
Subject terms: Nanotechnology in cancer, Cancer therapy
Multifunctional nanostructures are explored for smart cancer nanomedicine. This Review discusses nanostructures made of biomolecules, including polysaccharides, nucleic acids, peptides and proteins, which are inherently multifunctional and allow the targeting and delivery of cancer drugs to tumour tissue.
Introduction
A major advantage of nanostructures in biomedical applications is their multifunctional capacity. The flexibility of nanomaterial design and modification allows us to exploit different nano–bio interactions with a single system. Such multifunctionality is particularly important for the treatment of complex and heterogeneous diseases, such as cancer. Numerous anticancer functionalities can be combined in a nanostructure-based therapy approach, including targeted delivery, sequential targeting, stimuli-responsiveness, theranostics, combination treatment and built-in logic gates, to realize refined therapeutic actions1–4. However, the application of nanostructures in cancer therapy also faces several challenges; for example, a series of biological barriers interfere with the transport of nanostructures to target tissues, limiting delivery efficiency5,6. In addition, nanoformulations are subject to reticuloendothelial clearance and often show limited tumour penetration, as compared with free drugs. Therefore, effort has been dedicated to the development of functionalities that specifically overcome or circumvent biological barriers, clearance and limited penetration into tissues. The great variety of nanomaterials further leads to diverse and complicated in vivo behaviours, and the introduction of multiple components can increase the risk of unpredicted loss of function or adverse effects. It is therefore important to apply multifunctional nanomaterial design judiciously, taking into consideration the particular properties of the nanomaterial or functional units and the specific medical requirements.
In cancer nanomedicine, natural and engineered biomolecules (that is, polysaccharides, nucleic acids, peptides and proteins) have been explored for nanostructure construction7–11. These biomolecules are generally biocompatible and biodegradable, and often intrinsically bioactive as targeting and/or therapeutic agents. The protein-based nanoformulation Abraxane (albumin-bound paclitaxel) is already used in the clinic12, and several biomolecule-derived nanostructures are currently in clinical trials (Table 1). Although liposomes and polymeric particles remain the nanoplatforms with the most translational success thus far4,13, biomolecule-based building blocks offer specific properties that cannot be facilely reproduced in these synthetic materials. For example, the supramolecular interactions of peptide and nucleic acid materials can be precisely manipulated, and natural-protein-based nanosystems can mimic proteins in terms of molecular recognition or enzymatic activities. Importantly, multifunctional strategies can be designed using biomolecule-derived elements, each with distinct characteristics and limitations. However, a holistic view of biomolecule-based nanostructures in the broad context of cancer nanomedicine remains elusive. In this Review, we discuss biomolecule-based nanomaterials, highlighting specific multifunctional features in the context of cancer targeting and therapy.
Table 1.
Biomolecule-based nanostructures approved or in clinical trials for cancer therapy
| Biomolecule | Nanostructure | Name | Therapeutic modality | Active pharmaceutical ingredient(s) | Indication(s) | Status | ClinicalTrials.gov identifier |
|---|---|---|---|---|---|---|---|
| Polysaccharide | Cyclodextrin–PEG copolymer nanoparticle | NLG207, formerly CRLX101 | Targeted molecular therapy | Camptothecin (topoisomerase I inhibitor) | Rectal cancer | Phase I/II | NCT02010567 |
| Relapsed/refractory small-cell lung cancer; urothelial carcinoma | Phase I/II | NCT02769962 | |||||
| Metastatic castration-resistant prostate cancer | Phase II | NCT03531827 | |||||
| Protein | Albumin nanoparticle | Nab-paclitaxel (Abraxane) | Chemotherapy | Paclitaxel | Breast cancer; non-small-cell lung cancer; pancreatic cancer | Approved by FDA (2005; 2012; 2013) | – |
| Nab-rapamycin (ABI‑009) | Chemotherapy | Rapamycin | PEComa | Phase II | NCT02494570 | ||
| Non-muscle-invasive bladder cancer | Phase I/II | NCT02009332 | |||||
| Solid tumours | Phase I | NCT00635284 | |||||
| Advanced PEComa; malignancy with relevant genetic mutations or mTOR pathway activation | Expanded Access | NCT03817515 | |||||
| Aldoxorubicin (DOXO-EMCH, INNO-206) | Chemotherapy | Doxorubicin | Advanced solid tumour | Phase I | NCT01673438 | ||
| Glioblastoma | Phase II | NCT02014844 | |||||
| Metastatic, locally advanced or unresectable soft tissue sarcoma | Phase III | NCT02049905 | |||||
| Pancreatic ductal adenocarcinoma | Phase II | NCT01580397 | |||||
| Nab-docetaxel (ABI-008) | Chemotherapy | Docetaxel | Hormone-refractory prostate cancer | Phase I/II | NCT00477529 | ||
| Nucleic acid | Spherical nucleic acid | NU-0129 | Targeted molecular therapy | Bcl2L12 targeted RNA | Glioblastoma; recurrent glioblastoma | Early phase I | NCT03020017 |
| Cavrotolimod (AST-008) | Immunotherapy | AST-008 (toll-like receptor 9 agonist oligonucleotide) | Healthy volunteer study | Phase I | NCT03086278 | ||
| Immunotherapy in combination with chemotherapy | AST-008, Pembrolizumab, Cemiplimab | Solid tumours; melanoma; head and neck squamous cell carcinoma; cutaneous squamous cell carcinoma; Merkel cell carcinoma | Phase Ib/II | NCT03684785 |
mTOR, mechanistic target of rapamycin; PEComa, perivascular epithelioid cell tumours; PEG, polyethylene glycol.
Challenges in cancer nanomedicine
The fate of a nanosystem administered to an organism is influenced by a large network of factors. In particular, the efficiency of antitumour nanostructures is limited by difficulties in targeting, dynamic in vivo changes of the materials and multiple biological barriers. These complex and interconnected obstacles require multilayered countermeasures that can only be achieved with multifunctionality.
Cancer progression is a collective result of numerous pathological events14, which are often not tumour-specific but involve normal physiological processes distorted by the disease. Moreover, tumour cells are highly heterogeneous and have high mutation rates, resulting in pathological differences between different types of cancers, individuals and intratumoural regions. To address this heterogeneity, nanostructure-based combinatorial or multiple-target therapies are being explored. For example, coupling tumour-cell-targeted methods with strategies that modulate the tumour microenvironment (TME) (Box 1) enables the therapy to be tailored to distinct tumour characteristics. In highly fibrotic tumours, cancer-associated fibroblasts (CAFs) or stellate cells can be co-targeted, in addition to cancer cells, to inhibit the secretion of growth factors and pro-inflammatory cytokines and to distribute nanostructure-delivered therapeutics across the dense stroma15–17. Moreover, the recurrence of tumours after vasculature occlusion by targeted delivery of thrombin can be considerably decreased by co-delivery of doxorubicin, probably because the chemotherapeutic helps to eradicate tumour regions that are less populated by blood vessels18.
Although the development of a tumour-specific drug is extremely difficult, optimizing the pharmacokinetics and biodistribution of a less specific drug can substantially increase its efficacy. Indeed, the capacity of tumour-targeted delivery, either through passive targeting based on the enhanced permeability and retention (EPR) effect, active targeting or stimuli-responsive drug release, has been considered a key advantage of nanomedicines in cancer treatment2. However, compared with conventional drug formulations, the in vivo behaviour and fate of multifunctional nanostructures are more complex, which can make them particularly prone to inconsistencies across patients and between preclinical models and clinical applications19.
The properties of nanostructures are instantly affected when entering the body by interactions at the nano–bio interface. These interactions can lead to instabilities, such as particle aggregation, decomposition, loss of functional units or release of hazardous species, which greatly affect delivery efficiency and biosafety20. Contact with physiological fluids also leads to cloaking of the nanostructure with serum proteins (the protein corona), potentially shielding surface functionalities (such as targeting ligands) and interfering with drug release21. Antifouling modifications, such as coating with poly(ethylene glycol) (PEG), are often applied to reduce nanoparticle‒protein interactions. Alternatives based on biomolecules, especially proteins and peptides, have also been reported22,23. In addition, interference of the protein corona with surface functionalities depends on the functionalization technique; for example, antibodies pre-adsorbed onto polymeric nanoparticles retain their targeting ability better than chemically conjugated molecules, perhaps owing to more robust surface coverage24. By tailoring nanosurface modifications, the protein corona composition can be manipulated and exploited to increase circulation, reduce toxicity or improve targeting21,25. For example, Onpattro, a commercially available small interfering RNA (siRNA) formulation, recruits the liver-targeting protein, apolipoprotein E, onto the cholesterol-containing lipid nanoparticle surface in vivo26. Short peptides have also been used to manipulate the binding modes of serum apolipoproteins on the nanoparticle surface to improve delivery across the blood–brain barrier27. Although interference from non-specific adsorption needs to be carefully characterized28, this approach suggested the possibility of modulating corona proteins more precisely and using them as additional functionality.
In the bloodstream, phagocytes readily adhere to protein corona-coated nanostructures through specific receptors, which leads to removal of the nanostructures from the circulation by the reticuloendothelial system (RES)29. Thus, antifouling coatings, such as PEG, are commonly used to prolong the circulation time of nanostructures. Nonetheless, PEG modification only partly reduces immunogenicity, and the stimulation of PEG-binding antibodies has raised safety concerns30,31. More active antifouling strategies include surface decoration with moieties based on CD4732,33, a ‘self-marker’ protein that inhibits phagocytosis, or with extracted blood cell membranes (displaying CD47 on their surface)31,34, to help nanomaterials to evade clearance by phagocytes. In mouse models, these methods have demonstrated superior stealth properties compared with PEGylation31,32,34. However, corona-mediated clearance can also benefit macrophage- or RES organ-targeted delivery. For example, by precisely alternating the charge of cationic liposomes, they can be selectively directed to the liver and spleen to deliver genes35.
To reach tumour tissue, nanosystems first need to extravasate from the circulation1,36, which has long been thought to be enabled by leaky tumour blood vasculature. However, the EPR effect greatly varies between tumours and even between different regions of the same tumour, and its clinical significance remains controversial37–39. Multifunctional nanoparticles can actively increase intratumoural extravasation by creating local hypertension, causing vascular relaxation or inducing additional damage in tumour blood vessels. Such strategies may partly address the problem of EPR heterogeneity38,40, but stratification of patients and imaging-guided personalized medicine will prove essential to tailor nanomaterial-based therapeutic strategies1,2,41.
In addition, extravasation efficiency is affected by the nanomaterial properties; for example, in flowing blood, spherical nanoparticles tend to stay close to the middle of the vessel, whereas rod- or plate-shaped structures are more likely to drift towards the vessel walls and exhibit strong interactions with endothelial cells, owing to the larger contact area36,42. The surface softness of extracellular vesicles substantially impacts extravasation and cellular uptake in vivo43. Endothelial adhesion, and thus extravasation, can be enhanced by coating nanoparticles with cell membranes derived from leucocytes44. Extravasation is generally thought to occur by paracellular leaking; however, other mechanisms, such as active transport by endothelial cells39,45, also play a role, and are receiving attention in terms of nanosystem design and targeting46,47.
Most anticancer nanoformulations are designed for intravenous use. Of note, challenges posed by biological barriers during nanoparticle transport are related to the administration route; for example, multifunctional strategies have not yet addressed the problem of intravenously injected nanomaterial accumulation in the liver, which can help to reduce the toxicity of delivered drugs to other tissues and improve the overall tolerance12, but remains a problem for efficient drug delivery to the tumour. The administration route determines the biodistribution pattern of drugs, and routes alternative to intravenous injection with corresponding targeting strategies are explored for specific cancers (Box 2). Intradermal and subcutaneous injection, for example, may be particularly useful to target the lymphatic system for tumour immunization48, whereas orally delivered nanoparticles can target colon cancer49.
The tumour and the TME also provide a barrier to nanostructure-based drug delivery. High interstitial fluid pressure and desmoplastic stroma limit the penetration of nanostructures6,36. Moreover, deeper tumour regions are less populated by blood vessels. In addition, nanoparticles can be captured by tumour-associated macrophages (TAMs) instead of tumour cells following extravasation41. These intratumoural complications have motivated the development of TME-targeting strategies (Box 1), exploiting potential targets that are more accessible than tumour cells. Indeed, TME-related targets, such as the fibroblast activation protein on CAFs16, fibrin–fibronectin complex in tumour blood vessels18 or TAMs50,51, have already been applied for nanoparticle delivery. For therapies that require intracellular delivery (for example, gene delivery and antigen delivery), cellular uptake and endosomal escape are needed. However, unlike hydrophobic small drugs, nanoparticles cannot passively diffuse across the plasma membrane. Therefore, membrane-protein-targeted ligands, cell-penetrating peptides or fusogenic materials are used to promote cellular uptake5,52.
Box 1 Tumour microenvironment targets for multifunctional nanostructures.
Stroma and fibroblasts
In the tumour microenvironment (TME), quiescent stromal cells become proliferative and highly secretory, resulting in a dense extracellular matrix (ECM), which acts as an intratumoural barrier against drug penetration. Moreover, ECM components, cytokines and chemokines secreted by activated cancer-associated fibroblasts and mesenchymal stromal cells promote tumour cell proliferation, invasion and acquisition of drug resistance15. Stromal cell proteins (such as fibroblast activation protein α) and ECM components (especially ECM enzymes) can be used as targets for delivery or selective drug release16. Drugs targeting stromal abnormalities can be coupled with TME-targeting strategies to reverse the activation of stromal cells, inhibit protumoural signalling, or reduce the synthesis and secretion of fibrotic ECM components such as hyaluronan and collagen15,17.
Blood vessels
In many (but not all)241 solid tumours, angiogenesis is shifted towards the pro-angiogenic state, leading to a tortuous, interrupted vasculature network, which limits drug perfusion. Vascular endothelial cells conditioned by the TME actively participate in T-cell exclusion or metastasis, and the pro-coagulative state of tumours leads to the activation of platelets, which can promote tumour invasion242–244. Endothelial receptors and tumour-associated microthrombi can be targeted to localize nanostructures to the tumour vasculature40. Drugs can be released into the TME to modulate intratumour angiogenic signalling (for example, the vascular endothelial growth factor (VEGF) pathway) or to neutralize excessive pro-angiogenic factors, which are generated in response to cytotoxic therapies245.
Immune cells
Most immune-cell-targeted nanotherapies aim to enhance the anticancer cytotoxic T-cell response. Treatments that directly target T cells using immune checkpoint (cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) or programmed cell death protein 1 (PD-1)) inhibitors have shown promise against hot tumours, in which active T cells are able to infiltrate but are functionally blocked by immune checkpoint signalling246. In cold tumours, T cells are not activated, owing to the presence of immunosuppressive immune cells, which can be targeted to enhance T-cell recruitment. Nanovaccines that deliver tumour antigens and immunostimulatory agents to dendritic cells can improve their maturation and antigen presentation247,248. Reprogramming of protumoural macrophages to antitumoural phenotypes can revive their antitumour cytotoxic activity and boost T-cell responses50,51. Natural killer (NK) cells spontaneously attack cancer cells without requiring antigen sensitization. Targeting NK cells with cytokines (for example, IL-15) or antibodies that induce stimulation and expansion provides an alternative to T-cell-based immunotherapy249.
Lymphatic vessels
Solid tumours lack functional lymphatic drainage; however, lymphatic vessels are important highways for metastasizing cancer cells. Indeed, high levels of lymphangiogenesis in tumours correlate with poor outcome250,251. Tumour-associated lymphatic endothelial cells overexpress immunosuppressive molecules, such as PD-L1, but also promote dendritic cell migration into lymph nodes, associated with high infiltration of naive T cells250. Tumoural lymphangiogenesis can be modulated by blocking VEGFR3, the receptor of VEGF-C, a tumour-cell-secreted lymphangiogenic factor251. However, targeting ligands for lymphatic vessels remain limited (with the example of the peptide LyP-1252).
Box 2 Administration routes for cancer-targeting nanostructures.
Intravenous administration is characterized by a rapid therapeutic response, high drug bioavailability and high administration control. Structurally complex and biologically unstable nanosystems are often not suitable for gastrointestinal absorption or intramuscular injection, and thus intravenous injection is often the method of choice. In addition, anti-metastasis therapies require systemic drug administration. However, intravenous administration suffers from rapid drug clearance, undesired tissue distribution and accumulation, and subsequent adverse side effects.
Oral delivery benefits from good patient compliance and may provide easy access for gastrointestinal tract targeting. However, the harsh and complex environment in the gastrointestinal tract (high pH variation, metabolic enzymes, mucus-mediated clearance) provides challenges for nanostructures. Mucus-adhering and endothelium-penetrating elements are usually required to increase intestinal adsorption and penetration of nanomaterials.
Inhalation and intranasal administration allow access to the respiratory tract (for example, for the treatment of lung cancer) and enable sustained and less invasive targeting, as compared with intravenous injection. Nanostructures also require mucus-adhering functionalities to enhance mucus adhesion and penetration (as for other mucosal routes, such as vaginal delivery)253. However, inhaled or intranasally delivered drugs can also be systemically distributed; indeed, intranasally delivered drugs have been found to accumulate in the central nervous system253.
Direct injection and infusion into tissues adjacent to the tumour site is an option if tumours are readily accessible. Local administration routes bypass many biological barriers (for example, drugs administered into the brain bypass the blood–brain barrier), but tissue penetration is still required. Approaches to enhance nanomaterial penetration and retention in the dermis and subcutaneous tissue remain limited254. Locally injected small particles are prone to rapid lymphatic drainage, and thus subcutaneous administration can deliver nanoparticles to lymph nodes and immune cells that reside in the lymphatic system.
Each route of administration leads to a distinct drug distribution and distinct patterns of adverse effects. Safety issues related to non-intravenous routes, such as inhalation or skin contact, need to be considered255. Systematic studies of route-specific toxicities in the context of cancer nanomedicine will be essential.
Multifunctional nanostructures
Multifunctional nanoformulations enable multitargeted and/or combinatorial therapy, theranostics, tumour-specific drug delivery, bypassing of biological barriers and improved pharmaceutical properties (for example, pharmacokinetics, stability, biodegradation and biocompatibility), as compared with free drugs (Table 2). A multifunctional nanoplatform for cancer therapy typically consists of a structural framework, functional units and loaded cargo(es), which may also be considered a functional element (Fig. 1). To simplify fabrication and reduce unwanted side effects, the framework can also serve as functional element. For example, peptide drugs can be modified and self-assembled into nanosized particles to improve their pharmacokinetic profile. Biomolecules have particularly useful properties as backbone building blocks and can act as versatile substrates for multifunctionalization.
Table 2.
Anticancer functionalities of biomolecule-based nanostructures
| Anticancer functions | Functional components | Aims and advantages | Refs |
|---|---|---|---|
| Bypassing biological barriers | |||
| Reduced immune recognition and RES clearance | Low-protein-adsorption peptides or polysaccharides; anti-phagocytosis peptides; blood cell membranes; albumin; DNA tetrahedron; ferritin | Prolonged circulation time and evasion of liver and spleen capture | 34,90,175,177,233,234 |
| Enhanced extravasation | Albumin; leucocyte membranes; rod-like nanostructures | High intratumoural delivery efficiency; high exposure of functional elements to molecular targets; microenvironmental trigger | 44,146,175 |
| Enhanced tumour tissue penetration | Albumin; stroma-targeted ligands; cell-penetrating peptides | Treatment of less accessible tumour cells deep within tumour tissue | 16,177 |
| Delivery across the blood–brain barrier | Albumin; DNA tetrahedron; brain-targeted peptides | Treatment of brain tumours with intravenously delivered formulations | 27,175 |
| Delivery across the air–blood barrier | Albumin | Facilitated pulmonary delivery of agents to treat lung cancer | 178 |
| Delivery across the intestinal endothelium | Chitosan surface coating; endothelium-penetrating ligands | Treatment of intestinal gut tumours; orally administered chemotherapy | 49,61,65,235 |
| Tumour-targeted delivery | |||
| Ligand-receptor recognition | Aptamers; albumin; peptides; folic acid; ferritin | Selective drug delivery to and accumulation in specific tissues, cells or subcellular organelles | 18,58,67,88,90,91,99,150,177,179,233,234 |
| Protein corona manipulation | Adsorbed serum proteins | 27 | |
| Stimuli responsiveness | |||
| Stimuli-triggered decomposition of nanostructures | Protonable polysaccharides and peptides (pH); enzyme-cleavable peptides (tumour-specific enzymes); temperature-sensitive peptides (heat); disulfide bonds (GSH); irradiation-sensitive bonds (external light or radioactive irradiation); DNA nanoflower (H2O2); DNA nanohydrogel (pH); spherical nucleic acid (GSH); DNA origami (GSH); quantum dot DNA hydrogels (DNase); DNA coordination polymer (pH); albumin (H2O2); ferritin (pH); GroEL (ATP) | Rapid and complete drug release from otherwise stable nanostructures; selective drug release in desired location | 16,59,63,93,157,216,234 |
| Stimuli-triggered (in situ) self-assembly (including gel formation) of nanostructures | Albumin; stimuli-responsive peptides | Delivery of nanostructures not suitable for administration in the assembled form owing to size, shape or mechanical properties | 148,150,179 |
| Stimuli-triggered shape transformation of nanostructures | Peptide assembly; DNA origami (protein); DNA nanosuitcase (mRNA); DNA clamp (antibody); GroEL (ATP) | Rapid and complete drug release from otherwise stable nanostructures; selective drug release in desired location | 3,95,96,99,150,216,217 |
| Stimuli-triggered activation and shedding of targeting, therapeutic or imaging functionality of nanostructures | Aptamer; ferritin; enzyme-cleavable peptides; breakable bonds | Minimization of non-specific nano–bio interactions; imaging signal detection during delivery; selective drug release in desired location | 55,88,191 |
| Stimuli-triggered surface transformation (for example, charge reverse) of nanostructures | Protonable peptides and polysaccharides | Selective alteration of surface charge or functional groups at the tumour site to facilitate cell penetration, lysosome escape or other biological effects | 57 |
| Anticancer drug building blocks | |||
| Targeting ligand | Antibodies; therapeutic peptides and aptamers | Low metabolic burden and minimal adverse effects | 127,157,196,197,202 |
| Self-assembly material | Therapeutic peptides and aptamers, CpG DNA nanococoon, DNA–RNA nanocapsules, plant viral nanoparticles, protozoa protein, E2 subunit of pyruvate dehydrogenase | ||
| Combinatorial therapy | |||
| Multidrug loading | Albumin; DNA origami; amphiphilic polysaccharides; amphiphilic peptides | Combined advantages of multiple drugs or therapeutic modalities; mechanism-based cooperation to improve tumour sensitivity to other treatment and to overcome resistance or to prevent recurrence | 18,57,63,175,178 |
| Cooperation with immunotherapy | Immune cell-targeting ligands; DNA nanohydrogel; DNA–RNA nanocapsules; ferritin; hepatitis B core protein | 127,185,192 | |
| Cooperation with phototherapy or radiotherapy | DNA coordination polymer; albumin; nanostructures containing sensitizer or anti-angiogenetic drug | 67,93,176 | |
| Theranostics | |||
| Simultaneous imaging and treatment of tumour tissue | Nanostructures containing fluorophores; magnetic nanoparticles or other imaging agents | Simultaneous diagnosis of hidden tumours or small metastasis | 91,148 |
| Real-time monitoring of drug release and activity | Separable fluorophore–quencher pairs; fluorescent molecules with stimuli-dependent emission properties | Monitoring of in vivo activity of nanostructures or tumoural response to delivered drug | 148 |
CpG, cytosine-phosphate–guanine; GSH, glutathione; RES, reticuloendothelial system.
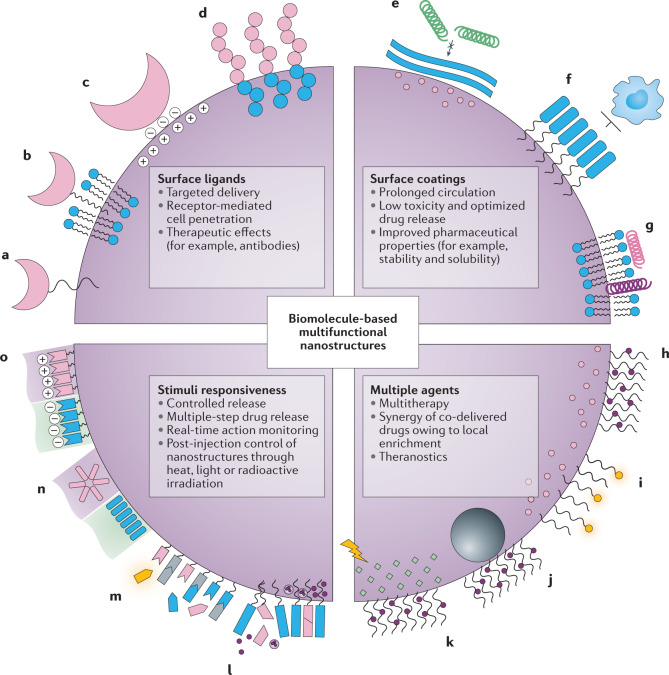
Fig. 1. Functional elements in antitumour multifunctional nanomedicines.
a–d | Surface ligands recognize tumour-associated molecules and epitopes for tumour targeting, penetration and therapy. Ligands can be attached through chemical conjunction (part a), hydrophobic interaction (part b) or electrostatic adsorption onto the nanoparticle surface (part c), or integrated as a functional motif into a peptide- or nucleic-acid-based molecular design (part d). e–g | Surface coatings shield the internal structure from non-specific nano–bio interactions and prevent early clearance during transport and distribution. e | Low protein adsorption on bioinert materials. f | Anti-phagocytosis molecules. g | Blood-cell-derived membrane camouflage. h–k | Combination therapy or theranostic designs may be coupled with antitumour drugs. h | Multiple drug loading. i | Real-time imaging. j | Magnetic resonance steering. k | Phototherapy. l–o | Stimuli-responsive elements. l | Degradable building blocks trigger the disassembly of the nanocarrier to release its contents. m | Surface functionalities for selective activation by a stimulus. Bioactive components may be released or exposed on the nanostructure surface, which can be further monitored in real time by using quenchable or activable imaging agents. n | Environmental stimuli may induce structural transformation of nanostructures, often through stimuli-triggered re-assembly of the building units. o | Nanosurfaces may reverse their overall charge in specific microenvironmental niches.
Polysaccharide-based nanostructures
Polysaccharides are biopolymers consisting of monosaccharide units linked together by glycosidic bonds. Polysaccharide-based materials provide a range of functional groups and show good biodegradability, stability, long-term durability and, in general, low immunogenicity8,53. Many polysaccharides, such as cellulose, starch, alginate and chitosan, are abundant in nature and can be extracted from renewable sources54,55. These characteristics make polysaccharides interesting alternatives to synthetic polymers in nanomedicine. Almost all known types of polysaccharides have been explored for cancer-targeted drug delivery or theranostics8,54,56.
Long-chained polysaccharides can be fabricated into diverse nanostructures for drug loading. Polysaccharide-based building blocks are commonly assembled into micelles, and anticancer drugs can then either be encapsulated or linked to the saccharide chain. Grafting with polymers, hydrophobic tails or protonable groups allows tuning of solubility and amphilicity. Saccharides containing primary amines (for example, chitosan), amides (for example, chitosan, heparin and hyaluronan), carboxyls (for example, heparin and hyaluronan) or primary hydroxyl groups (for example, hyaluronan and cyclodextrin) can be covalently modified, enabling the formation of amide bonds or ester bonds, and the glucosidic ring can also be opened by oxidation8. Targeting ligands57,58, stimuli-sensitive elements (most commonly redox-responsive disulfide bonds)55,59,60 or other biologically active entities can thus be integrated into the biopolymer backbone61. Certain saccharides also allow additional modes for material‒drug interaction; for example, negatively charged drugs, such as nucleic acids, can be electrostatically attached to protonable amine groups of chitosan, enabling gene delivery. NLG207, a topoisomerase inhibitor nanoformulation currently in phase I/II trials (Table 1), consists of β-cyclodextrin–PEG copolymer chains that self-assemble into nanoparticles through guest‒host recognition between cyclodextrin and the conjugated active component camptothecin, which improves the solubility and tumour exposure of the otherwise hydrophobic drug62.
Some polysaccharides have been exploited for their intrinsic targeting and therapeutic functions. Hyaluronic acid is a major component of the extracellular matrix and non-immunogenic. Its specific receptor CD44 is overexpressed in many types of cancer8,56, and thus hyaluronic-acid-based nanostructures can be applied for CD44-mediated targeting and cell uptake60,63,64. The protonatable amine groups of chitosan facilitate intestinal mucus adhesion and penetration, and chitosan nanoparticles can therefore be used for oral chemotherapy61,65. The biological properties of other polysaccharide nanomaterials, however, have been less explored. For example, fucoidan, a fucose-rich sulfated polysaccharide derived from brown seaweed, specifically binds to P-selectin, an adhesion protein that facilitates metastasis by mediating the interactions between tumour cells and platelets or blood vessel endothelia66. Fucoidan-containing nanoparticles have been shown to be effective P-selectin-targeted drug carriers67,68; however, fucoidan is difficult to purify, and its monosaccharide composition, molecular weight and structural characteristics vary depending on the source and purification method, which may limit rigorous study and clinical translation66.
The clinical translation of polysaccharide nanostructures remains difficult, because many polysaccharides are derived from natural products. Depending on the source material and processing method, material parameters such as monosaccharide composition, glycosidic bond type, charge density, molecular weight and biomacromolecule contamination can differ, which can cause variations in biological effects8,66. For example, alginate, a seaweed-derived carbohydrate, is usually regarded as biosafe; by contrast, alginate with high mannuronate, rather than guluronate, content can induce the production of inflammatory cytokines in vitro69. For the majority of polysaccharide-based materials, systematic in vivo data are limited, and it is not known how the abovementioned parameters affect biomedical performance (for example, immunogenicity, pharmacokinetics, cellular uptake and biodegradation profile), impeding clinical translation. Moreover, the impacts of material characteristics on delivery efficiency are complex to decipher, leading to often inconsistent results. For example, the correlation of molecular weight and efficiency of chitosan as gene carrier has been controversial; it has been suggested that nanoparticles fabricated with long chitosan chains have a disadvantage in terms of RNA release, but promote cellular uptake and endosomal escape64. However, superiority of long-chain chitosan was observed only for mRNA transfection and not for siRNA silencing.
The potential advantage of polysaccharides as economic and renewable biomass remains underexplored in cancer nanomedicine. Polysaccharides in multifunctional nanostructures have mostly served as building blocks thus far and not as effectors of biological actions. Identifying their potential to trigger biological actions, in comparison with mainstream platforms such as liposomes and polymeric particles, would be of value. In any case, clinical translation of polysaccharide-based nanostructures will require systematic in vivo characterization and the development of methods for scalable fabrication and quality control.
Nucleic-acid-based nanostructures
Nucleic acids (that is, DNA and RNA) are natural building blocks that permit a bottom-up approach of molecular self-assembly enabled by highly stringent base pairing. They offer programmable molecular interactions and provide an almost unlimited number of sequences that can be used for the fabrication of defined nanostructures11,70. They can be chemically fabricated in high yield and with low batch-to-batch variation in quality. In addition, negatively charged phosphate backbones or nucleobases that can chelate metal ions and form π-stacking can be modified to optimize pharmacokinetics and function71. By introducing biological tags and stimuli-responsive units into DNA or RNA architectures, the features of nucleic acids can be extended far beyond their natural genetic roles. These attributes can be exploited for cancer targeting and therapy.
DNA and RNA nanotechnology has enabled the design of multidimensional nanostructures with high spatial accuracy for a range of applications72,73. DNA has high thermodynamic stability and pH tolerance, allowing DNA nanofabrication by precise molecular self-assembly71. Chemical modification strategies can also increase the stability of RNA, enabling RNA self-assembly into nanostructures74,75. For example, the 2′-OH of the ribose sugar moiety can be substituted with 2′-fluorine, 2′-O-methyl, 2′-O-methoxyethyl or 2′-amine to overcome the thermal and enzymatic susceptibility of RNA76. Moreover, the linkage between bases can be modified by phosphorothioate, boranophosphate, peptide nucleic acids and nucleic acid analogues, as well as by direct uracil or adenine modifications77,78. Complex nanoarchitectures based on nucleic acids can be engineered by various methods, including tile assembly79, origami techniques80, single-stranded tile methods81, rolling circle amplification82,83 and in silico design. Software packages, such as caDNAno, CanDo, oxDNA and ENERG-MD, have been widely used to design DNA origamis84. However, accurate computational methods remain limited for RNA nanostructure design, although several computer programs have been developed, including NanoTiler, Mfold, RNA designer, Sfold and NUPACK76. Natural and custom-made nucleic acids can be engineered into 2D and 3D DNA, RNA, DNA–RNA and heterologous hybrid nanostructures, such as dynamic machines and supramolecular assemblies11,85. After decades of development, such nanostructures have become powerful tools in biomedical research (Fig. 2).
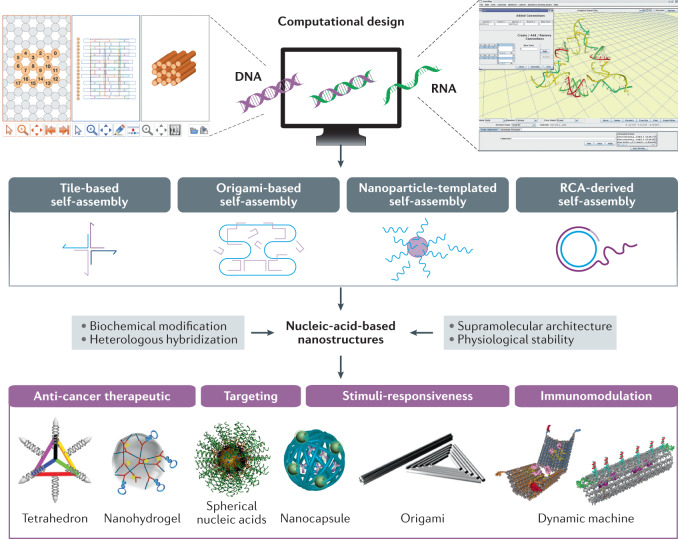
Fig. 2. Nucleic-acid-based nanostructures for cancer therapy.
Nucleic-acid-based nanostructures, including DNA, RNA, DNA–RNA and heterologous hybrid nanostructures, can be constructed with high spatial accuracy in different dimensions, using tile assembly, origami techniques, rolling circle amplification (RCA), nanoparticle-templated assembly and in silico design (for example, caDNAno, a software package for DNA origami design; NanoTiler, a computer program for RNA nanostructure design). These nanostructures, such as tetrahedrons91, nanohydrogels236, spherical structures237, nanocapsules83, origamis238 and dynamic machines3,99, can be applied as multifunctional anticancer systems. CaDNAno interface reprinted with permission from ref.239, OUP. NanoTiler program reprinted with permission from ref.240, Elsevier. Tetrahedron reprinted from ref.91, Springer Nature Limited. Spherical nucleic acids reprinted with permission from ref.237, ACS. Nanocapsule reprinted with permission from ref.83, Wiley. Origami reprinted with permission from ref.238, ACS. Dynamic machine reprinted with permission from ref.3, AAAS.
The most striking feature of nucleic-acid-based nanostructures is their spatial addressability, which can be achieved by modifying the sequence. Thus, functional molecules can be accurately integrated at specific locations in the nucleic acid structure to tailor the formulation for targeted delivery, smart cargo loading and release, simultaneous detection and visualization, and/or combinatorial therapeutic regimes11,84. Nucleic-acid-based assemblies can exploit the targeting modalities of DNA and RNA to engage cellular components. For example, aptamers, which are single-stranded DNA or RNA molecules (20–100 nucleotides in length) with defined secondary and tertiary structures, can interface with molecular targets with high specificity and affinity, comparable to antibodies86,87. By simply hybridizing or integrating aptamers with nucleic acid nanostructures, they can direct the specific recognition and binding of therapeutic agents by receptors expressed on malignant cells or to TME-specific biomarkers. The incorporation or self-assembly of microRNAs (miRNAs) or siRNAs into nucleic acid nanostructures enables gene delivery, and by adding cancer-cell-specific aptamers, non-specific adsorption and off-target effects can be prevented88. Therapeutic aptamers (for example, anti-human epidermal growth factor receptor 2 (HER2) aptamers) possess anticancer activity and enable tumour cell targeting, which improves efficacy and decreases the payload of nucleic acid nanostructures89.
Nanostructures based on nucleic acids can also be modified with other targeting ligands, such as antibodies, peptides and small molecules, through covalent or non-covalent linkage90,91. Additionally, nucleic acid nanostructures with certain hydrodynamic sizes (10–200 nm) and geometries (for example, triangular DNA origami) passively accumulate within tumour tissue, which improves targeting75,92. Efficient cargo delivery can also be achieved by incorporating environmentally responsive molecular switches in the nanostructures. For example, targetable properties of the intracellular environment and the TME (Box 2) or external physical stimuli can be exploited for cancer targeting and therapy93,94.
Controllable drug release is a central aspect of targeted delivery in anticancer therapeutics, which can be accomplished by stimulus-responsive disassembly of nanocarriers. The computer-aided, controlled design of smart nanomachines or nanorobots based on nucleic acids allows the engineering of logic-gated therapeutics84. Cargo release kinetics at a specified target site can be customized by reconfiguration of nanostructures in response to dynamic changes in specific molecular switches or environmental cues. One of the earliest logic-gated DNA nanorobots is a hollow origami barrel that contains antibody Fab fragments against human leucocyte antigens3. A logical AND gate is programmed through a two-input system based on aptamer encoding; two different DNA aptamer ‘locks’ can only be opened by simultaneous recognition of both ‘keys’ on the target cell surface, which leads to opening of the nanostructures and release of an antibody cargo that inhibits cell growth. Dual lock-and-key strategies can also be incorporated into DNA nanodevices for conditional siRNA delivery and release in specific cellular environments, yielding high gene silencing efficiency and low off-target toxicity88,95.
Reversible loading and release of nucleic acid cargo can also be achieved by a conformational change of an antibody-driven DNA-based nanomachine96. In particular, DNA origami nanorobots are explored for the precise delivery of therapeutics in vivo97,98. For example, a DNA origami nanorobot can precisely deliver a protease, which has strong adverse effects when administered in its free form, to tumour-associated blood vessels in tumour-bearing mice, with a high biosafety profile in both mice and Bama miniature pigs99. In this case, mechanical opening of the tubular DNA nanorobots is triggered by the specific binding of tumour endothelium-targeted DNA aptamers, leading to the release of thrombin from the inner cavity of the nanorobot, which causes intravascular thrombosis at the tumour site. Recently, a DNA machine communication network was developed that mimics the cascade reaction in the vertebrate adaptive immune system by stimulating immune cells upon the invasion of pathogens, demonstrating the great promise of DNA-based nanotherapeutics responding to sophisticated targets100. Taking advantage of the metastability of co-transcriptionally generated nucleic acid structures, a self-driven RNA nanomachine has been designed as an miRNA sensor, exploiting a mechanism beyond simple structural transformation101. Although most proof-of-principle nucleic-acid-based nanomachines have not yet been thoroughly validated in vivo, they highlight the opportunities for dynamic nucleic-acid-based machines and nanorobots for targeted smart cancer treatment. Various other construction methods and applications of DNA machines have been explored84.
Although referred to as machines or robots, dynamic nucleic-acid-based nanostructures mainly rely on blood circulation for delivery and do not provide autonomous movement, as demonstrated in other micro- or nanostructures driven by exogenous power, such as magnetic fields, acoustic fields, electric fields and/or light energy, or by endogenous chemical reaction energy102. Such active transport mechanisms face several technical challenges; they require self-propelled motion; transport needs to be sustained by chemical or biological fuels; and navigation has to be controlled in the complex body environment, which shows irregular Brownian motion and can lead to interactions between the nanostructure and biological molecules103. The implementation of feedback mechanisms, such as automatic selection and optimal response chosen from a menu of available programs in response to a signal, will facilitate smart theranostics executed by nucleic-acid-based nanomachines.
DNA and RNA molecules are indispensable to essential immunological events, and thus have the ability to target and/or modulate immune cells104. Indeed, recombinant plasmid DNA and mRNA are promising vaccine platforms105,106, as demonstrated in COVID-19 vaccines. Currently, there are 22 mRNA-based and 15 DNA-based COVID-19 vaccine candidates in or entering clinical trials, including three mRNA–lipid nanoparticle vaccines encoding the SARS-CoV-2 spike protein or its receptor-binding domain107. The immunogenicity of a nucleic acid sequence can be completely reversed by a single nucleotide or base change, implying that precise control of the immune activity of nucleic acid nanoarchitectures is feasible70. Thus, the initial design of nucleic acid nanoarchitectures should take into account whether immune responses should be exploited (for example, for vaccines and immunotherapies) or mitigated (for example, for immunologically quiescent drug delivery).
The composition, structure and immunosuppressive properties of nucleic acid sequences need to be considered in the design of delivery vehicles based on nucleic acids to ensure suppression of immunogenicity during theranostic cargo delivery108–110. DNA nanostructures evoke weaker immune responses than DNA–RNA hybrids, which in turn are less immunostimulatory than their RNA counterparts104,111. Therefore, more DNA- than RNA-based nanostructures have been constructed as immunologically inert drug carriers for cancer treatment despite the fact that some RNA structures, such as the RNA four-way junction (4WJ-X), do not elicit detectable immune stimulation in vivo112. As an alternative to decreasing the number of RNA strands, RNA strands can be modified with a 5′ end cap or certain RNA motifs can be embedded in the nanostructure in a certain orientation to attenuate immunogenicity113,114. Of note, drug-loading triangular DNA origami, a self-assembled DNA tetrahedral framework and 3D DNA nanotubes do not elicit detectable immunogenic responses in tumour-bearing mouse models91,99,115. The in vivo stability of nanostructures, which is typically enhanced by surface modifications, also affects the immune response, for example by altering the accessibility of vulnerable sites to nucleases116. Immunorecognition can be assessed by quantitative structure–activity relationship modelling, and preliminary results reveal that molecular weight, melting temperature and half-life largely dictate the immunomodulatory activity of nucleic acid nanostructures104,117,118. Therefore, a design road map is crucial for the precise tailoring of the immune processing of nucleic acid nanostructures in cancer therapy.
Nucleic acid nanostructures also offer the possibility of activating an immune response against tumours. Anticancer immune responses can be boosted by pathogen-associated molecular patterns, such as single-stranded DNA or double-stranded RNA, which are recognized by toll-like receptors (TLRs) on immune cells119. Unmethylated cytosine-phosphate–guanine oligodeoxynucleotides (CpG ODNs), for example, are processed by TLR9 and have thus been used as efficient adjuvants to induce strong immune responses in cancer immunotherapy120,121. Sequence-specific immunostimulation permits streamlined manufacturing of nucleic-acid-based nanostructures, without the need for complex multifunctional formulation. To maximize immune responses, an antigen can be formulated with a nucleic acid adjuvant through tailored engineering. For example, a DNA tetrahedron activates long-lasting immune responses in a TLR9-dependent manner in vertebrates, when co-assembled with CpG ODN adjuvants122,123. Spherical nucleic acids are also clinically promising nanostructures; here, high-density oligonucleotide nanoparticles precisely incorporate CpG ODNs for the co-delivery of tumour-associated antigens124,125. Using a high-throughput method, ~1,000 spherical nucleic acid architectures have been synthesized at the picomolar scale, and activation of the TLR9 pathway could be quantitatively screened by mass spectrometry118. Nucleic acid nanovaccines can create immunity in situations in which antitumour immune responses to other therapies are limited126. For example, a self-assembled hybrid DNA–RNA-peptide nanovaccine can deliver tandem CpG ODNs and signal transducer and activator of transcription 3 (STAT3) short hairpin RNA adjuvants, as well as tumour-specific peptide neoantigens, into antigen-presenting cells (APCs) in lymph nodes for synergistic cancer immunotherapy127. DNA and RNA aptamers have been used as antagonists of cytotoxic T-lymphocyte-associated protein 4 or programmed cell death protein 1 receptors to improve T-cell responses for cancer immunotherapy128,129. Thus, precisely incorporating aptamers into nucleic-acid-based nanostructures is a viable cancer immunotherapy strategy, enabling homogeneous synthesis of the nanostructures and modulation of various APCs. In addition, intracellular trafficking of nanostructures can be leveraged to trigger sensor-specific immune recognition in specific compartments by functional nucleic acid elements, facilitating cross-presentation of an antigen payload.
The clinical translation of nucleic-acid-based nanostructures requires the development of economically viable and robust methods for mass production, as well as stable modification of the functional nanostructures. The scalability challenge has been partially addressed by technological breakthroughs in producing highly purified nucleic acid strands and nanostructures in vivo. Macroscopic amounts of DNA origami can be produced by the simultaneous generation of long single-stranded DNA (ssDNA) scaffold strands and short staple strands in a litre-scale bioreactor culture of M13 bacteriophages. This approach reduces the production cost of DNA origami from ~US$200 per milligram to around 20 cents130. To improve chemical synthesis through adding of 2′-protecting groups, single-stranded RNA can also be transcribed from gene-encoded DNA and folded into designed nanostructures inside Escherichia coli cells, similar to the mass production of recombinant proteins131. Although large-scale synthesis at the kilogram level is expected to reduce the price of pharmaceutical oligonucleotides to a few dollars per milligram, the requirements of chemistry, manufacturing and controls. and of good manufacturing practices. will certainly increase production costs108.
Structural integrity is essential for the nanostructures to function in vivo76,132. DNA nanostructures can be stabilized through covalent crosslinking between strands, without the need for additional chemical modification or cofactors133. In addition, polymer-based (such as PEGylated oligolysines and dendritic oligonucleotides) or lipid-based coatings can be applied134–136. Certain structural elements, such as single-duplex edges, make 3D DNA origami more stable in physiological environments, as compared to close-packed bundles of helices137–139. To rigorously determine the structural integrity and pharmacokinetics of nucleic acid architectures in the highly heterogeneous TME, robust techniques, surpassing currently used fluorescent or radioactive labelling in terms of stability, spatiotemporal resolution and sensitivity, are needed at the molecular and in vivo levels. The interaction between nucleic-acid-based nanostructures and cellular DNA or RNA must also be kept at a minimum. Therefore, standards for the predictive and rational design of nanostructures are indispensable to achieve clinical translation.
Peptide-based nanostructures
Peptides are perhaps the most extensively studied materials for self-assembly-based nanosystems9,140–142. Compared with base-pairing-guided assembly of nucleic acids, the self-assembling behaviour of peptide molecules is less programmable. However, with 20 different natural amino acids, peptides offer a broader range of customizable physicochemical properties by sequence-specific engineering, as compared with nucleic acids. Peptides can be easily synthesized and functionalized, and are usually more stable in storage than proteins and nucleic acids. Moreover, many peptide motifs bear distinct biological roles, particularly for molecular recognition and signal transduction143.
Peptide self-assembly is a synergistic result of non-covalent intermolecular forces, including hydrophobic, electrostatic, hydrogen, van der Waals and π–π interactions. By manipulating these forces, either through sequence design or through additional functionalization, the peptides can be assembled into particles, fibres, tubes, sheets or 3D gels with various dimensions, morphologies and surface chemistries140. The assembly processes can also be co-mediated or assisted by other agents; for example, hydrophobic small molecule drugs can transform amphiphilic peptide nanofibres into spherical structures, owing to the formation of tight cores driven by drug‒peptide hydrophobic interactions16. Inspired by the assisted folding process of proteins, folding-assisting enzymes have been applied to form α-helical peptide assemblies with very short peptides, which is not feasible without the help of enzymes144.
A thorough understanding of peptide supramolecular chemistry is important for the accurate tailoring of their biological performance. For example, a modular system of nanoscale peptide cages modified with short charged peptides can promote endocytosis in vitro by controlling the charge of the cages145. A ‘patched’ hollow peptide tube can be engineered by mixing two types of peptides, which individually self-assemble into tubular and spherical structures, respectively. The hollow tube is sealed on both ends by half-spheres146, and can be applied as a drug-encapsulating vehicle, enabling extravasation into tumour tissue, comparable to other rod-like nanocarriers.
Minor changes in the forces governing peptide assembly can lead to substantial structural transformations140, rendering peptide-based materials particularly versatile as stimuli-sensitive devices. Peptides can also alter their functionality in response to tumour-related stimuli; for example, pH-sensitive cell-penetrating peptides147 are mainly internalized in the slightly acidic TME and not in healthy tissue. In a ‘reverse’ stimuli-responsive strategy, disassembled ordered structures, free peptides or small nanoassemblies aggregate at the tumour site to form larger nanostructures for sustained drug release, real-time imaging or to induce nanomaterial-mediated cytotoxicity. This approach reduces potential adverse side effects owing to rapid clearance of off-target free peptides148,149. For example, a hydrophobic photosensitizer (Purpurin18)-linked tumour-homing peptide is specifically cleaved by gelatinase, an enzyme overexpressed in the TME148. Once the hydrophilic ‘head’ of the peptide conjugate is removed, the residues assemble into nanofibres mediated by the photosensitizer ‘tail’. This assembly-induced retention effect increases the intratumoural half-life of the photosensitizer, enhancing both the photoacoustic imaging signal and the photothermal therapy efficacy. Peptide structures assembled in situ can remain in tumours for up to a month, much longer than intravenously administered peptide nanoparticles143. Moreover, post-administration self-assembly offers a way to deliver peptide nanostructures that are unsuitable for intravenous transportation, such as long fibres or gel-like networks. For example, a short peptide-based amphiphile selectively assembles into pericellular fibres in response to hypoxia-induced TME acidity. The fibres formed in situ readily bind to the tumour cell surface and can co-deliver a molecule-targeted drug. In addition, the fibres limit cell motility and contribute to cell death through shape-related lysosomal injury after receptor-mediated internalization, thereby enhancing the therapeutic effect150.
In contrast to proteins, bioactive peptides can be synthetically engineered, allowing easy multifunctional design and facile discovery by in vitro and in vivo screening techniques127,151. Specific peptide ligands have been developed for a range of targets, including cancer cells, TME cells, extracellular components, tissues susceptible to metastasis and biological barriers143,152,153. Furthermore, motifs can be incorporated that enable penetration through lipid bilayers to localize peptide nanoparticles in distinct subcellular compartments, by targeting membranes of the cell, mitochondria or nucleus152,154. Importantly, the immunogenicity of peptide assemblies varies considerably with the molecular design. Many peptide assemblies are non-immunogenic in animals, whereas some peptide structures elicit potent antibody responses, which can last for more than a year142,155. Therefore, peptides can be designed with different immunogenicity for vaccination or delivery applications.
Therapeutic peptides are a major class of anticancer drugs156. However, free peptides suffer from rapid clearance, limited stability in vivo and poor membrane permeability. These limitations can be overcome by fabricating them into nanoparticles. Effector peptides can be released from nanostructures in response to specific stimuli, minimizing the loss of peptide activity owing to the addition of additional moieties, such as targeting motifs. For example, the short peptide T4 (NLLMAAS) is a potent angiogenesis inhibitor, but highly hydrophobic and rapidly cleared from the bloodstream. Using an enzyme-cleavable peptide linker, the sequence can be integrated into pH-responsive self-assembled nanoparticles, which become loose in the weakly acidic TME, leading to the exposure of cleavage sites and eventual release of the small bioactive peptide157. Maintaining the nanostructure of apposed peptides at the tumour site may also elicit multivalent or multimeric effects and enhance the potency of therapeutic peptides158. For example, in mice, certain immunological effects are elicited only by nanoparticles coated with peptide–major histocompatibility complexes (pMHC) and not by pMHC alone. Here, the high ligand density on the nanoparticle surface accelerates the kinetics of interactions between pMHC and T-cell receptor, and promotes local clustering of receptor molecules on T-cell membranes, amplifying the magnitude of signalling159. Although not directly applicable to cancer treatment, this study confirms that a multifunctional nanostructure can elicit greater biological responses than the sum of its individual components, which may be particularly beneficial for peptide and protein nanostructures that can modulate intratumoural signal transduction.
With advanced simulation and fabrication tools, new functional peptides and nanostructures can be created. For example, temperature-sensitive peptide hydrogels have been widely used for long-term drug delivery; however, their function usually depends on complex motifs, making it difficult to further tune their functions on the sequence level. Alternatively, computation-assisted peptide design allows the generation of a library of peptides with accurately tuned thermal hysteresis properties, by alternating the periodical combination of two simple modules160. Microfluidics can be used to aid the formation of thermodynamically unfavourable self-assembly products of aromatic dipeptides by maintaining them in a non-equilibrium state, highlighting the possibility of accurately manipulating the formation of peptide assemblies on a kinetic level161. Coupled with in vitro and in vivo screening techniques, these methods to precisely predict and control the supramolecular behaviours of peptides will become crucial for the development of multifunctional nanostructures capable of complex actions (Fig. 3).
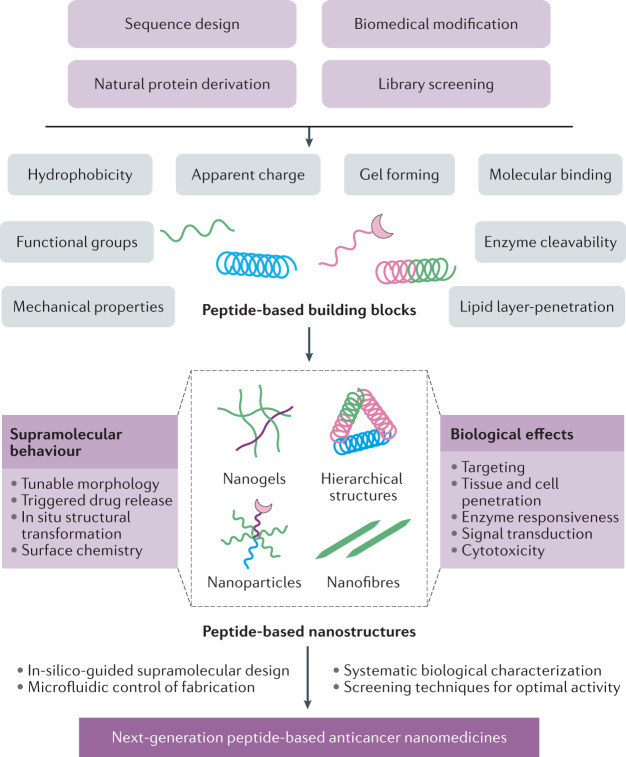
Fig. 3. Development of peptide-based multifunctional nanostructures.
Peptides and peptide-based building blocks are designed or selected according to their physicochemical properties and biological functions. By optimizing these characteristics, the self-assembly and supramolecular behaviour (for example, size, shape, surface functional groups, stimuli responsiveness) and biological activities (for example, targeting, cell penetration, therapeutic effects) can be manipulated to achieve optimal performance. The next-generation peptide-based multifunctional nanomedicines will benefit from computationally assisted design, precisely controlled fabrication (for example, microfluidic platforms) and systematic biological data, as well as advanced screening techniques.
Natural-protein-derived nanostructures
Proteins have remarkable structural and functional diversity, can be genetically programmed and are involved in essentially all cellular processes. Proteins and their assemblies — in particular, caged or virus-like protein structures — have long been explored for drug delivery, especially for immunotherapy, owing to their excellent biocompatibility, biodegradability, mechano-chemical features, wide variety of possible modifications, unique molecular recognition and responsive features at precise locations162–165. Various nanostructures derived from natural plant and animal proteins (for example, gliadin, legumin, sericin, protamine, collagen, gelatin, serum albumin and ferritin) have been applied for cancer theranostics166–168. The preparation method, site-selective modification and recombinant engineering of natural-protein-derived nanostructures, such as emulsion and solvent extraction, desolvation, salt precipitation, polyelectrolyte complexation and electrospraying, are selected based on the physicochemical properties and composition of the proteins and therapeutic agents169. For anticancer nanomedicines, the intrinsic biological activities of proteins can be exploited, including drug encapsulation, specific targeting, stimulus-dependent conformational changes, synergistic theranostics and enzymatic activities, epitomizing the multifunctionality of biomolecule-based nanostructures170,171. Indeed, nanostructures derived from natural proteins have already found their way into the clinic — for example, albumin nanoparticles172.
Serum albumin is the most abundant protein in blood. The long half-life of albumin of 19 days makes it an excellent candidate for drug delivery. Serum albumin naturally crosses vascular endothelia through GP60-receptor-aided transcytosis, and concentrates inside tumours by binding to secreted protein acidic and rich in cysteine (SPARC)173,174. Biomimetic penetration of the blood–brain barrier has been achieved using a dual drug-loaded bovine serum albumin nanostructure for glioma targeting and chemotherapy175. Proteins possess a variety of functional surface groups, such as thiol, amino and carboxylic groups, that allow drug binding. Indeed, modified serum albumin-based nanostructures have been applied as targeted and responsive cancer nanomedicines in vivo176,177. In addition to intravenous administration, an inhalable human serum albumin conjugate can be applied for the treatment of drug-resistant lung cancer in a mouse model178. A nanovaccine self-assembled in vivo from endogenous human serum albumin and Evans blue-conjugated CpG or tumour-specific neoantigens has been shown to potentiate the innate and adaptive immunity in multiple syngeneic tumours in mice179. Abraxane is the first albumin-based nanomedicine approved by the FDA for the treatment of advanced breast, lung and pancreatic cancer172,180. Other albumin-derived anticancer formulations, such as aldoxorubicin, nab-docetaxel and nab-rapamycin, have also been investigated in clinical trials13,181,182, highlighting the great promise of albumin-based cancer nanomedicines (Table 1).
Ferritin is a ubiquitous, well-characterized protein with a rigid octahedral cage comprising 24 subunits166. The cavity of ferritin reversibly self-assembles under acidic conditions, making it an excellent natural nanostructure for therapeutic cargo encapsulation and delivery170. The ferritin nanocage binds to transferrin receptor 1, which is overexpressed in a range of cancer cells, particularly in hypoxic tumour tissue183,184. Foreign recognition ligands, such as the peptide arginyl-glycyl-aspartic acid (RDG) and CD47-binding Sirpα variants, introduced by genetic engineering or self-assembling, further expand ferritin functions185,186. Ferritin can also function as a bioreactor for the templated synthesis of biomineral nanoparticles, which can be used for cancer imaging and therapy187–190. Cancer-targeted catalytic therapy has been explored by engaging the pH-responsive multienzyme-like activities of engineered ferritin–carbon nanosphere conjugates191. However, the full potential of ferritin-derived nanostructures has yet to be unleashed.
Nanostructures derived from caged proteins are well suited for cancer immunotherapy owing to their geometries and surface properties, which are similar to viruses192–194. As vaccine platforms and natural adjuvants, these nanostructures promote passive localization of tumour antigens in the lymphatic system, endocytosis of antigens by APCs and antigen presentation to adaptive immune cells162,163. Virus-like nanoparticles (VLPs) are caged protein structures isolated from viruses. VLPs lack infectious genetic material, but feature repetitive surface structures and pathogen-associated molecular patterns, which activate innate immunity and thus can be exploited for cancer vaccine delivery195. VLPs from plant viruses (such as cowpea mosaic virus and tobacco mosaic virus) have been thoroughly evaluated for antigen delivery in different cancer models, and vaccine formulations incorporating bacteriophage Qβ have shown promise in clinical studies196–200.
With their highly ordered structures and symmetry, non-viral protein cages have been engineered to elicit anticancer immune responses201–204. For example, by virtue of passive targeting to lymph nodes, antigen-loaded ferritin nanocages can trigger specific cytotoxic CD8 T-cell induction and prolong the survival of colon-tumour-bearing mice185,205. Vaccination with tumour antigens caged in heat-shock proteins has proven effective in clinical studies in different cancer types206,207. Some cage protein-based adjuvants, such as natural vaults, a ribonucleic protein complex in eukaryotic organisms with a hollow, barrel-shaped structure, favour cell-mediated over humoral immune responses when incorporated into cancer vaccines208. Various strategies have been developed for protein-cage-based nanoplatform cancer vaccines to evade neutralization and rapid clearance209. However, the exact mechanisms underlying the potential antitumour immunological effects of many natural-protein complexes remain to be explored.
Cells possess many sophisticated functional proteins that perform complex mechanical tasks through stimulus-responsive mechanical motion210. The engineering of such proteins into intelligent nanomachines has long been investigated for anticancer therapies, but remains challenging211. For example, molecular motor enzymes fuelled by chemical energy show open-closed conformational changes of interlocked movable subunits, triggered by hydrolysis of adenosine-5′-triphosphate (ATP)212. The bacterial chaperonin GroEL has such ATPase activity, and its hydrophobic double-ring cylinder has excellent structural stability as well as heat and acid resistance213. These dynamic structural features can be used to release entrapped guest molecules. Indeed, the ATP-switchable cavity of GroEL has been used as smart nanocarrier for the intracellular delivery and release of drugs, membrane proteins and nanoparticles214,215. Nanotubes assembled by multiple barrel-shaped GroEL units have been surface functionalized with a boronic acid derivative, which enables cancer cell uptake216. Intracellular drug release is triggered through intracellular ATP-responsive disassembly and esterase cleavage. GroEL also binds to plectin1, which is a structural protein highly expressed on some types of cancer cells, and thus can be used to release drugs in response to elevated ATP in the TME217.
Natural-protein-based drug delivery vehicles offer many benefits for cancer therapy; however, clinical translation remains challenging. For example, the structural heterogeneity of proteins leads to chemical and mechanical variability. This problem can be addressed through chemical modification or integration of synthetic materials that do not interfere with the desired in vivo activity7. Large-scale synthesis and recombinant protein production at reasonable cost will be required for clinical applications. In addition, formulation stability and long shelf-life are often difficult to achieve owing to the rapid degradation of proteins. Moreover, nanostructure absorption, in vivo distribution, metabolism and excretion must be further clarified by sensitive in vivo detection techniques. The functionalization of nanostructures is restricted by the structures and characteristics of the original proteins. This limitation can be addressed by de novo design and engineering of proteins with tailored therapeutic potential218.
Perspectives
Biomolecule-based nanostructures integrate the material and biological advantages of natural building blocks, enabling the multifunctional design of drug carriers. However, the more complex the materials, the more challenging the clinical translation. In addition, a thorough understanding of nanomaterial‒tumour interactions will be required to fully reveal the potential of biomolecule-based anticancer nanostructures.
Rational design and fabrication
Biomolecule-derived nanomaterials are highly variable in terms of molecular and supramolecular structures, and can interact and interfere with biological pathways in vivo. Therefore, the efficacy of biomolecule-based nanostructures can be particularly sensitive to the molecular composition of their building blocks, which makes system optimization challenging. Computational simulation and statistical models enable the rational design of biomolecule-based nanostructures; for example, molecular dynamics simulations have been applied to screen and identify hydrogel-forming tripeptides219. In silico tools can also be used to simulate protein adsorption, extravasation, cell uptake and drug release220,221, or to develop structure–activity relationships to analyse safety222. However, predicting the in vivo behaviour of biomolecule-based systems is difficult, compared with inorganic or polymeric nanostructures, owing to the complexity of biomolecule-based nanostructures and a lack of systematic experimental in vivo data.
To explore the multifunctional advantage of nanomaterials, systems with highly sophisticated spatial structures and responsive mechanisms have been designed100,101,146. However, accurate and reproducible fabrication methods for such complex structures remain limited. Nucleic acids and peptides, whose supramolecular behaviour and thus structure formation can be reproducibly programmed, may provide an advantage, compared with proteins. Microfluidic and high-throughput techniques further allow automation and monitoring of nanomaterial synthesis and characterization161,223,224.
Delivery evaluation
A major challenge in the development of tumour-targeted nanomedicines is the lack of tools to estimate and compare in vivo performance. Pharmacokinetic parameters are not always sufficient for describing the mechanism-of-action of nanostructures, such as multimeric and aggregation-induced effects. Current methodologies (for example, chromatography-based small-molecule drug quantification) often cannot discriminate between different nanostructure-associated drug forms, in particular if therapeutic effects are only activated following a multistep mechanism. In addition, the incorporation of active components in biomolecule-based nanostructures often requires complicated strategies. By contrast, most nanoformulations in clinical trials are drug-encapsulating liposomes or polymeric micelles with simple structures, facilitating standardization of in vivo evaluation and comparison with free drug formulations. Therefore, theoretical and statistical models need to be specifically developed for biomolecule-based nanostructures, to pre-clinically assess their action and behaviour in vivo.
Accurate delivery assessment can further be achieved by using probes and contrast agents for high-resolution, real-time in vivo imaging of nanostructures. These probes should ideally differentiate between different nanostructure forms. In particular, owing to differences in the impact of biological barriers on nanostructure transport between individuals, nanomedicines could be coupled with imaging agents (nanotheranostics) to enable real-time delivery evaluation2,12. High-content, animal-free evaluation of nanomaterial delivery (for example, using organs-on-a-chip and tumour-on-a-chip devices) may allow nanomedicine screening and drug response prediction224,225. However, these models cannot yet accurately predict intratumoural delivery efficiency for individual patients. Machine learning approaches can help to analyse large and heterogeneous datasets and thus may allow the identification of new imaging markers. Several deep learning algorithms based on computed tomography and magnetic resonance imaging have been developed to predict important prognostic factors, such as tumour grade, molecular expression pattern or immune phenotype226,227. The use of artificial intelligence may enable not only patient stratification, but also the design of personalized nanostructure delivery strategies228.
Combating metastasis
Metastasis of tumour cells remains the main cause of cancer-related deaths. Metastasized cancer cells do not necessarily have the same targetable genetic or biological characteristics as primary tumour cells229,230. In particular, early metastases are difficult to target by nanoparticle-based strategies owing to the lack of targetable TME characteristics, such as fully developed vasculatures. Nanomedical therapies are being explored for the treatment of metastasis, including early metastases, pre-metastatic tissues and circulating tumour cells1,231. For example, nanoparticles coated with biological membranes extracted from pro-metastatic cells, such as platelets232 and neutrophils34, have been investigated for anti-metastasis drug delivery. Identifying metastatic sites for surgical removal or localized treatments remains difficult, and therefore applying nanostructures that can specifically bind to and target metastatic cancer cells may prove valuable. Multifunctional nanostructures could also incorporate imaging modalities, which is particularly important for the development of anti-metastatic nanomedicines12.
Multifunctional nanomedicine is actively explored in preclinical and clinical research. Biomolecule-based nanomaterials are inherently multifunctional, and a thorough understanding of their building blocks, in concert with advances in rational design and in vivo evaluation, will make these nanostructures promising candidates for clinical cancer treatments.
Acknowledgements
We sincerely acknowledge all researchers and their work in our field that we were unable to cite because of space limitations. This work was supported by the National Basic Research Plan of China (2018YFA0208900), the National Key R&D Program of China (2018YFE0205300), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB36000000), the Key Research Program of Frontier Sciences CAS (ZDBS-LY-SLH039) and the K. C. Wong Education Foundation (GJTD-2018-03).
Author contributions
J.W. and Y.L. contributed equally to researching the data for the Review, discussing the content, and reviewing and editing the manuscript. G.N. conceived the structure and focus and was responsible for writing the Review. All authors approved the contents of the Review.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
caDNAno: https://cadnano.org/index.html
CanDo: https://cando-dna-origami.org/
ENERG-MD: http://bionano.physics.illinois.edu/origami-structure
Mfold: http://www.unafold.org/
NUPACK: http://www.nupack.org/
oxDNA: https://oxdna.org/
RNA designer: http://www.rnasoft.ca/cgi-bin/RNAsoft/RNAdesigner/rnadesign.pl
These authors contributed equally: Jing Wang, Yiye Li.
References
- 1.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer. 2016;17:20. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Zhang W, Zhu G, Xie J, Chen X. Rethinking cancer nanotheranostics. Nat. Rev. Mater. 2017;2:17024. doi: 10.1038/natrevmats.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 4.Anselmo AC, Mitragotri S. Nanoparticles in the clinic: an update. Bioeng. Transl. Med. 2019;4:e10143. doi: 10.1002/btm2.10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Z, Ukidve A, Kim J, Mitragotri S. Targeting strategies for tissue-specific drug delivery. Cell. 2020;181:151–167. doi: 10.1016/j.cell.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A. Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science. 2012;338:903. doi: 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta LP, Manchineella S, Govindaraju T. Biomolecules-derived biomaterials. Biomaterials. 2020;230:119633. doi: 10.1016/j.biomaterials.2019.119633. [DOI] [PubMed] [Google Scholar]
- 8.Liu K, Jiang X, Hunziker P. Carbohydrate-based amphiphilic nano delivery systems for cancer therapy. Nanoscale. 2016;8:16091–16156. doi: 10.1039/C6NR04489A. [DOI] [PubMed] [Google Scholar]
- 9.Habibi N, Kamaly N, Memic A, Shafiee H. Self-assembled peptide-based nanostructures: smart nanomaterials toward targeted drug delivery. Nano Today. 2016;11:41–60. doi: 10.1016/j.nantod.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakkar A, Traverso G, Farokhzad OC, Weissleder R, Langer R. Evolution of macromolecular complexity in drug delivery systems. Nat. Rev. Chem. 2017;1:63. doi: 10.1038/s41570-017-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeman NC, Sleiman HF. DNA nanotechnology. Nat. Rev. Mater. 2018;3:17068. doi: 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- 12.Lammers T, et al. Cancer nanomedicine: is targeting our target? Nat. Rev. Mater. 2016;1:16069. doi: 10.1038/natrevmats.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res. 2016;33:2373–2387. doi: 10.1007/s11095-016-1958-5. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Valkenburg KC, de Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018;15:366–381. doi: 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji T, et al. Transformable peptide nanocarriers for expeditious drug release and effective cancer therapy via cancer-associated fibroblast activation. Angew. Chem. Int. Ed. 2016;55:1050–1055. doi: 10.1002/anie.201506262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, et al. Reversal of pancreatic desmoplasia by re-educating stellate cells with a tumour microenvironment-activated nanosystem. Nat. Commun. 2018;9:3390. doi: 10.1038/s41467-018-05906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, et al. Combination of tumour-infarction therapy and chemotherapy via the co-delivery of doxorubicin and thrombin encapsulated in tumour-targeted nanoparticles. Nat. Biomed. Eng. 2020;4:732–742. doi: 10.1038/s41551-020-0573-2. [DOI] [PubMed] [Google Scholar]
- 19.Metselaar JM, Lammers T. Challenges in nanomedicine clinical translation. Drug Deliv. Transl. Res. 2020;10:721–725. doi: 10.1007/s13346-020-00740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nel AE, et al. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 21.Cai R, Chen C. The crown and the scepter: roles of the protein corona in nanomedicine. Adv. Mater. 2019;31:1805740. doi: 10.1002/adma.201805740. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee I, Pangule RC, Kane RS. Antifouling coatings: recent developments in the design of surfaces that prevent fouling by proteins, bacteria, and marine organisms. Adv. Mater. 2011;23:690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 23.Chelmowski R, et al. Peptide-based SAMs that resist the adsorption of proteins. J. Am. Chem. Soc. 2008;130:14952–14953. doi: 10.1021/ja8065754. [DOI] [PubMed] [Google Scholar]
- 24.Tonigold M, et al. Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat. Nanotechnol. 2018;13:862–869. doi: 10.1038/s41565-018-0171-6. [DOI] [PubMed] [Google Scholar]
- 25.Hamad-Schifferli K. Exploiting the novel properties of protein coronas: emerging applications in nanomedicine. Nanomedicine. 2015;10:1663–1674. doi: 10.2217/nnm.15.6. [DOI] [PubMed] [Google Scholar]
- 26.Setten RL, Rossi JJ, Han S-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. Brain-targeted drug delivery by manipulating protein corona functions. Nat. Commun. 2019;10:3561. doi: 10.1038/s41467-019-11593-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirshafiee V, Kim R, Park S, Mahmoudi M, Kraft ML. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials. 2016;75:295–304. doi: 10.1016/j.biomaterials.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Sun F, Liu S, Jiang S. Anti-PEG antibodies in the clinic: current issues and beyond PEGylation. J. Control. Rel. 2016;244:184–193. doi: 10.1016/j.jconrel.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu C-MJ, et al. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl Acad. Sci. USA. 2011;108:10980. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez PL, et al. Minimal ‘self’ peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science. 2013;339:971. doi: 10.1126/science.1229568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, et al. Co-coating of receptor-targeted drug nanocarriers with anti-phagocytic moieties enhances specific tissue uptake versus non-specific phagocytic clearance. Biomaterials. 2017;147:14–25. doi: 10.1016/j.biomaterials.2017.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang T, et al. Nanoparticles coated with neutrophil membranes can effectively treat cancer metastasis. ACS Nano. 2017;11:1397–1411. doi: 10.1021/acsnano.6b06477. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Q, et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015;33:941. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols JW, Bae YH. EPR: evidence and fallacy. J. Control. Rel. 2014;190:451–464. doi: 10.1016/j.jconrel.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura H, Jun F, Maeda H. Development of next-generation macromolecular drugs based on the EPR effect: challenges and pitfalls. Expert Opin. Drug Deliv. 2015;12:53–64. doi: 10.1517/17425247.2014.955011. [DOI] [PubMed] [Google Scholar]
- 39.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016;1:16014. doi: 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- 40.Li S, et al. Nanoparticle-mediated local depletion of tumour-associated platelets disrupts vascular barriers and augments drug accumulation in tumours. Nat. Biomed. Eng. 2017;1:667–679. doi: 10.1038/s41551-017-0115-8. [DOI] [PubMed] [Google Scholar]
- 41.Miller MA, et al. Predicting therapeutic nanomedicine efficacy using a companion magnetic resonance imaging nanoparticle. Sci. Transl. Med. 2015;7:314ra183. doi: 10.1126/scitranslmed.aac6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah S, Liu Y, Hu W, Gao J. Modeling particle shape-dependent dynamics in nanomedicine. J. Nanosci. Nanotechnol. 2011;11:919–928. doi: 10.1166/jnn.2011.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang Q, et al. The softness of tumour-cell-derived microparticles regulates their drug-delivery efficiency. Nat. Biomed. Eng. 2019;3:729–740. doi: 10.1038/s41551-019-0405-4. [DOI] [PubMed] [Google Scholar]
- 44.Palomba R, et al. Biomimetic carriers mimicking leukocyte plasma membrane to increase tumor vasculature permeability. Sci. Rep. 2016;6:34422. doi: 10.1038/srep34422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sindhwani S, et al. The entry of nanoparticles into solid tumours. Nat. Mater. 2020;19:566–575. doi: 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Jiang J, Meng H. Transcytosis — an effective targeting strategy that is complementary to ‘EPR effect’ for pancreatic cancer nano drug delivery. Theranostics. 2019;9:8018–8025. doi: 10.7150/thno.38587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandit S, Dutta D, Nie S. Active transcytosis and new opportunities for cancer nanomedicine. Nat. Mater. 2020;19:478–480. doi: 10.1038/s41563-020-0672-1. [DOI] [PubMed] [Google Scholar]
- 48.Guo Y, et al. Erythrocyte membrane-enveloped polymeric nanoparticles as nanovaccine for induction of antitumor immunity against melanoma. ACS Nano. 2015;9:6918–6933. doi: 10.1021/acsnano.5b01042. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, et al. Reversing tumor stemness via orally targeted nanoparticles achieves efficient colon cancer treatment. Biomaterials. 2019;216:119247. doi: 10.1016/j.biomaterials.2019.119247. [DOI] [PubMed] [Google Scholar]
- 50.Shields CW, et al. Cellular backpacks for macrophage immunotherapy. Sci. Adv. 2020;6:eaaz6579. doi: 10.1126/sciadv.aaz6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv. Drug Deliv. Rev. 2017;114:206–221. doi: 10.1016/j.addr.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 53.Thambi T, Phan VHG, Lee DS. Stimuli-sensitive injectable hydrogels based on polysaccharides and their biomedical applications. Macromol. Rapid Commun. 2016;37:1881–1896. doi: 10.1002/marc.201600371. [DOI] [PubMed] [Google Scholar]
- 54.Lin N, Huang J, Dufresne A. Preparation, properties and applications of polysaccharide nanocrystals in advanced functional nanomaterials: a review. Nanoscale. 2012;4:3274–3294. doi: 10.1039/c2nr30260h. [DOI] [PubMed] [Google Scholar]
- 55.Hu H, et al. Redox-responsive polycation-functionalized cotton cellulose nanocrystals for effective cancer treatment. ACS Appl. Mater. Interfaces. 2015;7:8942–8951. doi: 10.1021/acsami.5b02432. [DOI] [PubMed] [Google Scholar]
- 56.Swierczewska M, Han HS, Kim K, Park JH, Lee S. Polysaccharide-based nanoparticles for theranostic nanomedicine. Adv. Drug Deliv. Rev. 2016;99:70–84. doi: 10.1016/j.addr.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butt AM, et al. Doxorubicin and siRNA codelivery via chitosan-coated pH-responsive mixed micellar polyplexes for enhanced cancer therapy in multidrug-resistant tumors. Mol. Pharm. 2016;13:4179–4190. doi: 10.1021/acs.molpharmaceut.6b00776. [DOI] [PubMed] [Google Scholar]
- 58.Shi G-N, et al. Enhanced antitumor immunity by targeting dendritic cells with tumor cell lysate-loaded chitosan nanoparticles vaccine. Biomaterials. 2017;113:191–202. doi: 10.1016/j.biomaterials.2016.10.047. [DOI] [PubMed] [Google Scholar]
- 59.Thambi T, et al. Bioreducible carboxymethyl dextran nanoparticles for tumor-targeted drug delivery. Adv. Healthc. Mater. 2014;3:1829–1838. doi: 10.1002/adhm.201300691. [DOI] [PubMed] [Google Scholar]
- 60.Han HS, et al. Bioreducible shell-cross-linked hyaluronic acid nanoparticles for tumor-targeted drug delivery. Biomacromolecules. 2015;16:447–456. doi: 10.1021/bm5017755. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, et al. Amphiphilic carboxymethyl chitosan–quercetin conjugate with P-gp inhibitory properties for oral delivery of paclitaxel. Biomaterials. 2014;35:7654–7665. doi: 10.1016/j.biomaterials.2014.05.053. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt KT, et al. Measurement of NLG207 (formerly CRLX101) nanoparticle-bound and released camptothecin in human plasma. J. Pharm. Biomed. Anal. 2020;181:113073. doi: 10.1016/j.jpba.2019.113073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi KY, et al. Versatile RNA interference nanoplatform for systemic delivery of RNAs. ACS Nano. 2014;8:4559–4570. doi: 10.1021/nn500085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lallana E, et al. Chitosan/hyaluronic acid nanoparticles: rational design revisited for RNA delivery. Mol. Pharm. 2017;14:2422–2436. doi: 10.1021/acs.molpharmaceut.7b00320. [DOI] [PubMed] [Google Scholar]
- 65.Kim T, et al. Nanoparticle-patterned multicompartmental chitosan capsules for oral delivery of oligonucleotides. ACS Biomater. Sci. Eng. 2018;4:4163–4173. doi: 10.1021/acsbiomaterials.8b00806. [DOI] [PubMed] [Google Scholar]
- 66.Wu L, et al. A review about the development of fucoidan in antitumor activity: progress and challenges. Carbohydr. Polym. 2016;154:96–111. doi: 10.1016/j.carbpol.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Shamay Y, et al. P-selectin is a nanotherapeutic delivery target in the tumor microenvironment. Sci. Transl. Med. 2016;8:345ra87. doi: 10.1126/scitranslmed.aaf7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Juenet M, et al. Thrombolytic therapy based on fucoidan-functionalized polymer nanoparticles targeting P-selectin. Biomaterials. 2018;156:204–216. doi: 10.1016/j.biomaterials.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 69.Otterlei M, et al. Induction of cytokine production from human monocytes stimulated with alginate. J. Immunother. 1991;10:286–291. doi: 10.1097/00002371-199108000-00007. [DOI] [PubMed] [Google Scholar]
- 70.Jani MS, Veetil AT, Krishnan Y. Precision immunomodulation with synthetic nucleic acid technologies. Nat. Rev. Mater. 2019;4:451–458. doi: 10.1038/s41578-019-0105-4. [DOI] [Google Scholar]
- 71.Zhao Y, et al. Nanofabrication based on DNA nanotechnology. Nano Today. 2019;26:123–148. doi: 10.1016/j.nantod.2019.03.004. [DOI] [Google Scholar]
- 72.Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 73.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/S1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 74.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu C, et al. Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Lett. 2018;414:57–70. doi: 10.1016/j.canlet.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jasinski D, Haque F, Binzel DW, Guo P. Advancement of the emerging field of RNA nanotechnology. ACS Nano. 2017;11:1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 78.Piao X, Xia X, Mao J, Bong D. Peptide ligation and RNA cleavage via an abiotic template interface. J. Am. Chem. Soc. 2015;137:3751–3754. doi: 10.1021/jacs.5b00236. [DOI] [PubMed] [Google Scholar]
- 79.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 80.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 81.Wei B, Dai M, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ali MM, et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- 83.Wang C, Sun W, Wright G, Wang AZ, Gu Z. Inflammation-triggered cancer immunotherapy by programmed delivery of CpG and anti-PD1 antibody. Adv. Mater. 2016;28:8912–8920. doi: 10.1002/adma.201506312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramezani H, Dietz H. Building machines with DNA molecules. Nat. Rev. Genet. 2020;21:5–26. doi: 10.1038/s41576-019-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang P, Ko SH, Tian C, Hao C, Mao C. RNA–DNA hybrid origami: folding of a long RNA single strand into complex nanostructures using short DNA helper strands. Chem. Commun. 2013;49:5462–5464. doi: 10.1039/c3cc41707g. [DOI] [PubMed] [Google Scholar]
- 86.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li, L. et al. Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angew. Chem. Int. Ed. 10.1002/anie.202003563 (2020). [DOI] [PubMed]
- 88.Ren K, et al. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat. Commun. 2016;7:13580. doi: 10.1038/ncomms13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma W, et al. An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2. Nano Lett. 2019;19:4505–4517. doi: 10.1021/acs.nanolett.9b01320. [DOI] [PubMed] [Google Scholar]
- 90.Jiang D, et al. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl. Mater. Interfaces. 2016;8:4378–4384. doi: 10.1021/acsami.5b10792. [DOI] [PubMed] [Google Scholar]
- 91.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Q, et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8:6633–6643. doi: 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- 93.Yang Y, et al. G-quadruplex-based nanoscale coordination polymers to modulate tumor hypoxia and achieve nuclear-targeted drug delivery for enhanced photodynamic therapy. Nano Lett. 2018;18:6867–6875. doi: 10.1021/acs.nanolett.8b02732. [DOI] [PubMed] [Google Scholar]
- 94.Veetil AT, et al. Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol. 2017;12:1183–1189. doi: 10.1038/nnano.2017.159. [DOI] [PubMed] [Google Scholar]
- 95.Bujold KE, Hsu JCC, Sleiman HF. Optimized DNA ‘nanosuitcases’ for encapsulation and conditional release of siRNA. J. Am. Chem. Soc. 2016;138:14030–14038. doi: 10.1021/jacs.6b08369. [DOI] [PubMed] [Google Scholar]
- 96.Ranallo S, Prevost-Tremblay C, Idili A, Vallee-Belisle A, Ricci F. Antibody-powered nucleic acid release using a DNA-based nanomachine. Nat. Commun. 2017;8:15150. doi: 10.1038/ncomms15150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Amir Y, et al. Universal computing by DNA origami robots in a living animal. Nat. Nanotechnol. 2014;9:353–357. doi: 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arnon S, et al. Thought-controlled nanoscale robots in a living host. PLoS ONE. 2016;11:e0161227. doi: 10.1371/journal.pone.0161227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li S, et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018;36:258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 100.Ma PQ, Huang Q, Li HD, Yin BC, Ye BC. Multimachine communication network that mimics the adaptive immune response. J. Am. Chem. Soc. 2020;142:3851–3861. doi: 10.1021/jacs.9b11545. [DOI] [PubMed] [Google Scholar]
- 101.Kobori S, Nomura Y, Yokobayashi Y. Self-powered RNA nanomachine driven by metastable structure. Nucleic Acids Res. 2019;47:6007–6014. doi: 10.1093/nar/gkz364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Agrahari V, et al. Intelligent micro-/nanorobots as drug and cell carrier devices for biomedical therapeutic advancement: promising development opportunities and translational challenges. Biomaterials. 2020;260:120163. doi: 10.1016/j.biomaterials.2020.120163. [DOI] [PubMed] [Google Scholar]
- 103.Wu Z, Chen Y, Mukasa D, Pak OS, Gao W. Medical micro/nanorobots in complex media. Chem. Soc. Rev. 2020;49:8088–8112. doi: 10.1039/D0CS00309C. [DOI] [PubMed] [Google Scholar]
- 104.Hong E, et al. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 2018;18:4309–4321. doi: 10.1021/acs.nanolett.8b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jackson NAC, Kester KE, Casimiro D, Gurunathan S, DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jeyanathan M, et al. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Keller A, Linko V. Challenges and perspectives of DNA nanostructures in biomedicine. Angew. Chem. Int. Ed. 2020;59:15818–15833. doi: 10.1002/anie.201916390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Afonin KA, et al. Triggering of RNA interference with RNA–RNA, RNA–DNA, and DNA–RNA nanoparticles. ACS Nano. 2015;9:251–259. doi: 10.1021/nn504508s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo S, et al. Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles. Mol. Ther. Nucleic Acids. 2017;9:399–408. doi: 10.1016/j.omtn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Halman JR, et al. Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res. 2017;45:2210–2220. doi: 10.1093/nar/gkx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo S, et al. Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy. Nat. Commun. 2020;11:972. doi: 10.1038/s41467-020-14780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hyde JL, et al. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343:783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rackley L, et al. RNA fibers as optimized nanoscaffolds for siRNA coordination and reduced immunological recognition. Adv. Funct. Mater. 2018;28:1805959. doi: 10.1002/adfm.201805959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu J, et al. A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew. Chem. Int. Ed. 2018;57:15486–15490. doi: 10.1002/anie.201809452. [DOI] [PubMed] [Google Scholar]
- 116.Surana S, Shenoy AR, Krishnan Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 2015;10:741–747. doi: 10.1038/nnano.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson MB, et al. Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling. Small. 2017;13:1701255. doi: 10.1002/smll.201701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yamankurt G, et al. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat. Biomed. Eng. 2019;3:318–327. doi: 10.1038/s41551-019-0351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kroll AV, et al. Biomimetic nanoparticle vaccines for cancer therapy. Adv. Biosyst. 2019;3:1800219. doi: 10.1002/adbi.201800219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Klinman DM. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 2004;4:249–258. doi: 10.1038/nri1329. [DOI] [PubMed] [Google Scholar]
- 121.Khisamutdinov EF, et al. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res. 2014;42:9996–10004. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu X, et al. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li J, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 124.Goldberg MS. Improving cancer immunotherapy through nanotechnology. Nat. Rev. Cancer. 2019;19:587–602. doi: 10.1038/s41568-019-0186-9. [DOI] [PubMed] [Google Scholar]
- 125.Wang S, et al. Rational vaccinology with spherical nucleic acids. Proc. Natl Acad. Sci. USA. 2019;116:10473–10481. doi: 10.1073/pnas.1902805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Umeki Y, et al. Induction of ptent antitumor immunity by sustained release of cationic antigen from a DNA-based hydrogel with adjuvant activity. Adv. Funct. Mater. 2015;25:5758–5767. doi: 10.1002/adfm.201502139. [DOI] [Google Scholar]
- 127.Zhu G, et al. Intertwining DNA–RNA nanocapsules loaded with tumor neoantigens as synergistic nanovaccines for cancer immunotherapy. Nat. Commun. 2017;8:1482. doi: 10.1038/s41467-017-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- 129.Devaraj SGT, Rao LGL, Zu Y, Chang J, Iyer SP. DNA aptamer against anti-programmed cell death-1 (anti-PD1-apt) induces robust anti-leukemic activity in vitro and in vivo humanized NSG mice with myeloid leukemia xenografts. Blood. 2017;130:1373. [Google Scholar]
- 130.Praetorius F, et al. Biotechnological mass production of DNA origami. Nature. 2017;552:84–87. doi: 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- 131.Li M, et al. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat. Commun. 2018;9:2196. doi: 10.1038/s41467-018-04652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bae W, Kocabey S, Liedl T. DNA nanostructures in vitro, in vivo and on membranes. Nano Today. 2019;26:98–107. doi: 10.1016/j.nantod.2019.03.001. [DOI] [Google Scholar]
- 133.Gerling T, Kube M, Kick B, Dietz H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 2018;4:eaau1157. doi: 10.1126/sciadv.aau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anastassacos FM, Zhao Z, Zeng Y, Shih WM. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 2020;142:3311–3315. doi: 10.1021/jacs.9b11698. [DOI] [PubMed] [Google Scholar]
- 135.Kim Y, Yin P. Enhancing biocompatible stability of DNA nanostructures using dendritic oligonucleotides and brick motifs. Angew. Chem. Int. Ed. 2020;59:700–703. doi: 10.1002/anie.201911664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perrault SD, Shih WM. Lipid membrane encapsulation of a 3D DNA nano octahedron. Methods Mol. Biol. 2017;1500:165–184. doi: 10.1007/978-1-4939-6454-3_12. [DOI] [PubMed] [Google Scholar]
- 137.Wiraja C, et al. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat. Commun. 2019;10:1147. doi: 10.1038/s41467-019-09029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Benson E, et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 139.Veneziano R, et al. Designer nanoscale DNA assemblies programmed from the top down. Science. 2016;352:1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Santis E, Ryadnov MG. Peptide self-assembly for nanomaterials: the old new kid on the block. Chem. Soc. Rev. 2015;44:8288–8300. doi: 10.1039/C5CS00470E. [DOI] [PubMed] [Google Scholar]
- 141.Qin H, Ding Y, Mujeeb A, Zhao Y, Nie G. Tumor microenvironment targeting and responsive peptide-based nanoformulations for improved tumor therapy. Mol. Pharm. 2017;92:219–231. doi: 10.1124/mol.116.108084. [DOI] [PubMed] [Google Scholar]
- 142.Eskandari S, Guerin T, Toth I, Stephenson RJ. Recent advances in self-assembled peptides: implications for targeted drug delivery and vaccine engineering. Adv. Drug Deliv. Rev. 2017;110-111:169–187. doi: 10.1016/j.addr.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 143.Qi G-B, Gao Y-J, Wang L, Wang H. Self-assembled peptide-based nanomaterials for biomedical imaging and therapy. Adv. Mater. 2018;30:1703444. doi: 10.1002/adma.201703444. [DOI] [PubMed] [Google Scholar]
- 144.Liang C, et al. Enzyme-assisted peptide folding, assembly and anti-cancer properties. Nanoscale. 2017;9:11987–11993. doi: 10.1039/C7NR04370H. [DOI] [PubMed] [Google Scholar]
- 145.Beesley JL, et al. Modifying self-assembled peptide cages to control internalization into mammalian cells. Nano Lett. 2018;18:5933–5937. doi: 10.1021/acs.nanolett.8b02633. [DOI] [PubMed] [Google Scholar]
- 146.Ueda M, et al. End-sealed high aspect ratio hollow nanotubes encapsulating an anticancer drug: torpedo-shaped peptidic nanocapsules. ACS Nano. 2019;13:305–312. doi: 10.1021/acsnano.8b06189. [DOI] [PubMed] [Google Scholar]
- 147.Wyatt LC, et al. Peptides of pHLIP family for targeted intracellular and extracellular delivery of cargo molecules to tumors. Proc. Natl Acad. Sci. USA. 2018;115:E2811–E2818. doi: 10.1073/pnas.1715350115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang D, et al. In situ formation of nanofibers from purpurin18–peptide conjugates and the assembly induced retention effect in tumor sites. Adv. Mater. 2015;27:6125–6130. doi: 10.1002/adma.201502598. [DOI] [PubMed] [Google Scholar]
- 149.Dimatteo R, Darling NJ, Segura T. In situ forming injectable hydrogels for drug delivery and wound repair. Adv. Mater. 2018;127:167–184. doi: 10.1016/j.addr.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li J, et al. New power of self-assembling carbonic anhydrase inhibitor: short peptide-constructed nanofibers inspire hypoxic cancer therapy. Sci. Adv. 2019;5:eaax0937. doi: 10.1126/sciadv.aax0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mann AP, et al. A peptide for targeted, systemic delivery of imaging and therapeutic compounds into acute brain injuries. Nat. Commun. 2016;7:11980. doi: 10.1038/ncomms11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang C, et al. Peptide-based multifunctional nanomaterials for tumor imaging and therapy. Adv. Funct. Mater. 2018;28:1804492. doi: 10.1002/adfm.201804492. [DOI] [Google Scholar]
- 153.Zhang G, et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat. Med. 2012;18:307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 154.Field LD, Delehanty JB, Chen Y, Medintz IL. Peptides for specifically targeting nanoparticles to cellular organelles: quo vadis? Acc. Chem. Res. 2015;48:1380–1390. doi: 10.1021/ar500449v. [DOI] [PubMed] [Google Scholar]
- 155.Rudra JS, et al. Modulating adaptive immune responses to peptide self-assemblies. ACS Nano. 2012;6:1557–1564. doi: 10.1021/nn204530r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lau JL, Dunn MK. Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 157.Zhang L, et al. Cooperatively responsive peptide nanotherapeutic that regulates angiopoietin receptor Tie2 activity in tumor microenvironment to prevent breast tumor relapse after chemotherapy. ACS Nano. 2019;13:5091–5102. doi: 10.1021/acsnano.8b08142. [DOI] [PubMed] [Google Scholar]
- 158.Gray BP, Li S, Brown KC. From phage display to nanoparticle delivery: functionalizing liposomes with multivalent peptides improves targeting to a cancer biomarker. Bioconjug. Chem. 2013;24:85–96. doi: 10.1021/bc300498d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Singha S, et al. Peptide-MHC-based nanomedicines for autoimmunity function as T-cell receptor microclustering devices. Nat. Nanotechnol. 2017;12:701–710. doi: 10.1038/nnano.2017.56. [DOI] [PubMed] [Google Scholar]
- 160.Roberts S, et al. Injectable tissue integrating networks from recombinant polypeptides with tunable order. Nat. Mater. 2018;17:1154–1163. doi: 10.1038/s41563-018-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mason TO, et al. Synthesis of nonequilibrium supramolecular peptide polymers on a microfluidic platform. J. Am. Chem. Soc. 2016;138:9589–9596. doi: 10.1021/jacs.6b04136. [DOI] [PubMed] [Google Scholar]
- 162.Molino NM, Wang SW. Caged protein nanoparticles for drug delivery. Curr. Opin. Biotechnol. 2014;28:75–82. doi: 10.1016/j.copbio.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Neek M, Kim TI, Wang S-W. Protein-based nanoparticles in cancer vaccine development. Nanomedicine. 2019;15:164–174. doi: 10.1016/j.nano.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhuang J, et al. Nanoparticle delivery of immunostimulatory agents for cancer immunotherapy. Theranostics. 2019;9:7826–7848. doi: 10.7150/thno.37216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Edwardson TGW, Hilvert D. Virus-inspired function in engineered protein cages. J. Am. Chem. Soc. 2019;141:9432–9443. doi: 10.1021/jacs.9b03705. [DOI] [PubMed] [Google Scholar]
- 166.Cannon KA, Ochoa JM, Yeates TO. High-symmetry protein assemblies: patterns and emerging applications. Curr. Opin. Struct. Biol. 2019;55:77–84. doi: 10.1016/j.sbi.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 167.Jain A, Singh SK, Arya SK, Kundu SC, Kapoor S. Protein nanoparticles: promising platforms for drug delivery applications. ACS Biomater. Sci. Eng. 2018;4:3939–3961. doi: 10.1021/acsbiomaterials.8b01098. [DOI] [PubMed] [Google Scholar]
- 168.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 169.Kuan SL, Bergamini FRG, Weil T. Functional protein nanostructures: a chemical toolbox. Chem. Soc. Rev. 2018;47:9069–9105. doi: 10.1039/C8CS00590G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.He J, Fan K, Yan X. Ferritin drug carrier (FDC) for tumor targeting therapy. J. Control. Rel. 2019;311:288–300. doi: 10.1016/j.jconrel.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 171.Jiang B, Fang L, Wu K, Yan X, Fan K. Ferritins as natural and artificial nanozymes for theranostics. Theranostics. 2020;10:687–706. doi: 10.7150/thno.39827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kratz F. A clinical update of using albumin as a drug vehicle — a commentary. J. Control. Rel. 2014;190:331–336. doi: 10.1016/j.jconrel.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 173.Desai N, Trieu V, Damascelli B, Soon-Shiong P. SPARC expression correlates with tumor response to albumin-bound paclitaxel in head and neck cancer patients. Transl. Oncol. 2009;2:59–64. doi: 10.1593/tlo.09109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hidalgo M, et al. SPARC expression did not predict efficacy of nab-paclitaxel plus gemcitabine or gemcitabine alone for metastatic pancreatic cancer in an exploratory analysis of the phase III MPACT trial. Clin. Cancer Res. 2015;21:4811–4818. doi: 10.1158/1078-0432.CCR-14-3222. [DOI] [PubMed] [Google Scholar]
- 175.Lin T, et al. Blood–brain-barrier-penetrating albumin nanoparticles for biomimetic drug delivery via albumin-binding protein pathways for antiglioma therapy. ACS Nano. 2016;10:9999–10012. doi: 10.1021/acsnano.6b04268. [DOI] [PubMed] [Google Scholar]
- 176.Chung SW, et al. Albumin-binding caspase-cleavable prodrug that is selectively activated in radiation exposed local tumor. Biomaterials. 2016;94:1–8. doi: 10.1016/j.biomaterials.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 177.Sarett SM, et al. Lipophilic siRNA targets albumin in situ and promotes bioavailability, tumor penetration, and carrier-free gene silencing. Proc. Natl Acad. Sci. USA. 2017;114:E6490–E6497. doi: 10.1073/pnas.1621240114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Choi SH, et al. Inhalable self-assembled albumin nanoparticles for treating drug-resistant lung cancer. J. Control. Rel. 2015;197:199–207. doi: 10.1016/j.jconrel.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 179.Zhu G, et al. Albumin/vaccine nanocomplexes that assemble in vivo for combination cancer immunotherapy. Nat. Commun. 2017;8:1954. doi: 10.1038/s41467-017-02191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gradishar WJ. Albumin-bound paclitaxel: a next-generation taxane. Expert Opin. Pharmacother. 2006;7:1041–1053. doi: 10.1517/14656566.7.8.1041. [DOI] [PubMed] [Google Scholar]
- 181.Chawla SP, et al. First-line aldoxorubicin vs doxorubicin in metastatic or locally advanced unresectable soft-tissue sarcoma: a phase 2b randomized clinical trial. JAMA Oncol. 2015;1:1272–1280. doi: 10.1001/jamaoncol.2015.3101. [DOI] [PubMed] [Google Scholar]
- 182.Cirstea D, et al. Dual inhibition of akt/mammalian target of rapamycin pathway by nanoparticle albumin-bound-rapamycin and perifosine induces antitumor activity in multiple myeloma. Mol. Cancer Ther. 2010;9:963–975. doi: 10.1158/1535-7163.MCT-09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huang XL, et al. Hypoxia-tropic protein nanocages for modulation of tumor- and chemotherapy-associated hypoxia. ACS Nano. 2019;13:236–247. doi: 10.1021/acsnano.8b05399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fan K, et al. Ferritin nanocarrier traverses the blood brain barrier and kills glioma. ACS Nano. 2018;12:4105–4115. doi: 10.1021/acsnano.7b06969. [DOI] [PubMed] [Google Scholar]
- 185.Lee EJ, et al. Nanocage-therapeutics prevailing phagocytosis and immunogenic cell death awakens immunity against cancer. Adv. Mater. 2018;30:11. doi: 10.1002/adma.201705581. [DOI] [PubMed] [Google Scholar]
- 186.Zhen Z, et al. RGD-modified apoferritin nanoparticles for efficient drug delivery to tumors. ACS Nano. 2013;7:4830–4837. doi: 10.1021/nn305791q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang Z, et al. Biomineralization-inspired synthesis of copper sulfide–ferritin nanocages as cancer theranostics. ACS Nano. 2016;10:3453–3460. doi: 10.1021/acsnano.5b07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sun C, et al. Controlling assembly of paired gold clusters within apoferritin nanoreactor for in vivo kidney targeting and biomedical imaging. J. Am. Chem. Soc. 2011;133:8617–8624. doi: 10.1021/ja200746p. [DOI] [PubMed] [Google Scholar]
- 189.Fan J, et al. Direct evidence for catalase and peroxidase activities of ferritin–platinum nanoparticles. Biomaterials. 2011;32:1611–1618. doi: 10.1016/j.biomaterials.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 190.Sun C, et al. Fine-tuned h-ferritin nanocage with multiple gold clusters as near-infrared kidney specific targeting nanoprobe. Bioconjug. Chem. 2015;26:193–196. doi: 10.1021/bc5005284. [DOI] [PubMed] [Google Scholar]
- 191.Fan K, et al. In vivo guiding nitrogen-doped carbon nanozyme for tumor catalytic therapy. Nat. Commun. 2018;9:1440. doi: 10.1038/s41467-018-03903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shan WJ, et al. Bioengineered nanocage from HBc protein for combination cancer immunotherapy. Nano Lett. 2019;19:1719–1727. doi: 10.1021/acs.nanolett.8b04722. [DOI] [PubMed] [Google Scholar]
- 193.Chowdhury S, et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019;25:1057–1063. doi: 10.1038/s41591-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tang L, et al. Enhancing T cell therapy through TCR-signaling-responsive nanoparticle drug delivery. Nat. Biotechnol. 2018;36:707–716. doi: 10.1038/nbt.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mohsen MO, Zha L, Cabral-Miranda G, Bachmann MF. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 196.Shukla S, et al. Plant viral nanoparticles-based HER2 vaccine: immune response influenced by differential transport, localization and cellular interactions of particulate carriers. Biomaterials. 2017;121:15–27. doi: 10.1016/j.biomaterials.2016.12.030. [DOI] [PubMed] [Google Scholar]
- 197.Lizotte PH, et al. In situ vaccination with cowpea mosaic virus nanoparticles suppresses metastatic cancer. Nat. Nanotechnol. 2016;11:295–303. doi: 10.1038/nnano.2015.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kemnade JO, et al. Tobacco mosaic virus efficiently targets DC uptake, activation and antigen-specific T cell responses in vivo. Vaccine. 2014;32:4228–4233. doi: 10.1016/j.vaccine.2014.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Goldinger SM, et al. Nano-particle vaccination combined with TLR-7 and -9 ligands triggers memory and effector CD8(+) T-cell responses in melanoma patients. Eur. J. Immunol. 2012;42:3049–3061. doi: 10.1002/eji.201142361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Serradell MC, et al. Efficient oral vaccination by bioengineering virus-like particles with protozoan surface proteins. Nat. Commun. 2019;10:361. doi: 10.1038/s41467-018-08265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Molino NM, Neek M, Tucker JA, Nelson EL, Wang SW. Viral-mimicking protein nanoparticle vaccine for eliciting anti-tumor responses. Biomaterials. 2016;86:83–91. doi: 10.1016/j.biomaterials.2016.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Neek M, et al. Co-delivery of human cancer-testis antigens with adjuvant in protein nanoparticles induces higher cell-mediated immune responses. Biomaterials. 2018;156:194–203. doi: 10.1016/j.biomaterials.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Molino NM, Neek M, Tucker JA, Nelson EL, Wang SW. Display of DNA on nanoparticles for targeting antigen presenting cells. ACS Biomater. Sci. Eng. 2017;3:496–501. doi: 10.1021/acsbiomaterials.7b00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Molino NM, Anderson AK, Nelson EL, Wang SW. Biomimetic protein nanoparticles facilitate enhanced dendritic cell activation and cross-presentation. ACS Nano. 2013;7:9743–9752. doi: 10.1021/nn403085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Han JA, et al. Ferritin protein cage nanoparticles as versatile antigen delivery nanoplatforms for dendritic cell (DC)-based vaccine development. Nanomedicine. 2014;10:561–569. doi: 10.1016/j.nano.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 206.Specht HM, et al. Heat shock protein 70 (Hsp70) peptide activated natural killer (NK) cells for the treatment of patients with non-small cell lung cancer (NSCLC) after radiochemotherapy (RCTx) — from preclinical studies to a clinical phase II trial. Front. Immunol. 2015;6:162. doi: 10.3389/fimmu.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Crane CA, et al. Individual patient-specific immunity against high-grade glioma after vaccination with autologous tumor derived peptides bound to the 96 KD chaperone protein. Clin. Cancer Res. 2013;19:205–214. doi: 10.1158/1078-0432.CCR-11-3358. [DOI] [PubMed] [Google Scholar]
- 208.Kar UK, et al. Vault nanocapsules as adjuvants favor cell-mediated over antibody-mediated immune responses following immunization of mice. PLoS ONE. 2012;7:e38553. doi: 10.1371/journal.pone.0038553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schwarz B, et al. Symmetry controlled, genetic presentation of bioactive proteins on the P22 virus-like particle using an external decoration protein. ACS Nano. 2015;9:9134–9147. doi: 10.1021/acsnano.5b03360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.van den Heuvel MGL, Dekker C. Motor proteins at work for nanotechnology. Science. 2007;317:333–336. doi: 10.1126/science.1139570. [DOI] [PubMed] [Google Scholar]
- 211.Chen HB, et al. Precise nanomedicine for intelligent therapy of cancer. Sci. China Chem. 2018;61:1503–1552. doi: 10.1007/s11426-018-9397-5. [DOI] [Google Scholar]
- 212.Kinbara K, Aida T. Toward intelligent molecular machines: directed motions of biological and artificial molecules and assemblies. Chem. Rev. 2005;105:1377–1400. doi: 10.1021/cr030071r. [DOI] [PubMed] [Google Scholar]
- 213.Hayer-Hartl M, Bracher A, Hartl FU. The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem. Sci. 2016;41:62–76. doi: 10.1016/j.tibs.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 214.Ishii D, et al. Chaperonin-mediated stabilization and ATP-triggered release of semiconductor nanoparticles. Nature. 2003;423:628–632. doi: 10.1038/nature01663. [DOI] [PubMed] [Google Scholar]
- 215.Deaton J, et al. Functional bacteriorhodopsin is efficiently solubilized and delivered to membranes by the chaperonin GroEL. Proc. Natl Acad. Sci. USA. 2004;101:2281–2286. doi: 10.1073/pnas.0307759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Biswas S, et al. Biomolecular robotics for chemomechanically driven guest delivery fuelled by intracellular ATP. Nat. Chem. 2013;5:613–620. doi: 10.1038/nchem.1681. [DOI] [PubMed] [Google Scholar]
- 217.Yuan Y, et al. Chaperonin-GroEL as a smart hydrophobic drug delivery and tumor targeting molecular machine for tumor therapy. Nano Lett. 2018;18:921–928. doi: 10.1021/acs.nanolett.7b04307. [DOI] [PubMed] [Google Scholar]
- 218.Kuhlman B, Bradley P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019;20:681–697. doi: 10.1038/s41580-019-0163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Frederix PWJM, et al. Exploring the sequence space for (tri-)peptide self-assembly to design and discover new hydrogels. Nat. Chem. 2015;7:30–37. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- 220.Sacha GM, Varona P. Artificial intelligence in nanotechnology. Nanotechnology. 2013;24:452002. doi: 10.1088/0957-4484/24/45/452002. [DOI] [PubMed] [Google Scholar]
- 221.Pietro M, Bernhard Aribo S. In silico models for nanomedicine: recent developments. Curr. Med. 2018;25:4192–4207. doi: 10.2174/0929867324666170417120725. [DOI] [PubMed] [Google Scholar]
- 222.Puzyn T, et al. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 2011;6:175–178. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]
- 223.Ahn J, et al. Microfluidics in nanoparticle drug delivery; from synthesis to pre-clinical screening. Adv. Drug Deliv. Rev. 2018;128:29–53. doi: 10.1016/j.addr.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 224.He Z, Ranganathan N, Li P. Evaluating nanomedicine with microfluidics. Nanotechnology. 2018;29:492001. doi: 10.1088/1361-6528/aae18a. [DOI] [PubMed] [Google Scholar]
- 225.Carvalho MR, et al. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci. Adv. 2019;5:eaaw1317. doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dana J, Agnus V, Ouhmich F, Gallix B. Multimodality imaging and artificial intelligence for tumor characterization: current status and future perspective. Semin. Nucl. Med. 2020;50:541–548. doi: 10.1053/j.semnuclmed.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 227.Sun R, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 228.Adir O, et al. Integrating artificial intelligence and nanotechnology for precision cancer medicine. Adv. Mater. 2020;32:1901989. doi: 10.1002/adma.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hong MKH, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 2015;6:6605. doi: 10.1038/ncomms7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Vignot S, Besse B, André F, Spano J-P, Soria J-C. Discrepancies between primary tumor and metastasis: a literature review on clinically established biomarkers. Crit. Rev. Oncol. Hematol. 2012;84:301–313. doi: 10.1016/j.critrevonc.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 231.Schroeder A, et al. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer. 2012;12:39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 232.Hu Q, et al. Anticancer platelet-mimicking nanovehicles. Adv. Mater. 2015;27:7043–7050. doi: 10.1002/adma.201503323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liang M, et al. H-ferritin-nanocaged doxorubicin nanoparticles specifically target and kill tumors with a single-dose injection. Proc. Natl Acad. Sci. USA. 2014;111:14900–14905. doi: 10.1073/pnas.1407808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ahn B, et al. Four-fold channel-nicked human ferritin nanocages for active drug loading and pH-responsive drug release. Angew. Chem. Int. Ed. 2018;57:2909–2913. doi: 10.1002/anie.201800516. [DOI] [PubMed] [Google Scholar]
- 235.Khatun Z, Nurunnabi M, Reeck GR, Cho KJ, Lee Y-K. Oral delivery of taurocholic acid linked heparin–docetaxel conjugates for cancer therapy. J. Control. Rel. 2013;170:74–82. doi: 10.1016/j.jconrel.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 236.Li J, et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 2015;137:1412–1415. doi: 10.1021/ja512293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J. Am. Chem. Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 238.Jiang Q, et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012;134:13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 239.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bindewald E, Grunewald C, Boyle B, O’Connor M, Shapiro BA. Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler. J. Mol. Graph. Model. 2008;27:299–308. doi: 10.1016/j.jmgm.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Donnem T, et al. Non-angiogenic tumours and their influence on cancer biology. Nat. Rev. Cancer. 2018;18:323–336. doi: 10.1038/nrc.2018.14. [DOI] [PubMed] [Google Scholar]
- 242.Hida K, Maishi N, Torii C, Hida Y. Tumor angiogenesis — characteristics of tumor endothelial cells. Int. J. Clin. Oncol. 2016;21:206–212. doi: 10.1007/s10147-016-0957-1. [DOI] [PubMed] [Google Scholar]
- 243.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 244.Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat. Rev. Cancer. 2011;11:123–134. doi: 10.1038/nrc3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tozer GM, Kanthou C, Baguley BC. Disrupting tumour blood vessels. Nat. Rev. Cancer. 2005;5:423–435. doi: 10.1038/nrc1628. [DOI] [PubMed] [Google Scholar]
- 246.Nam J, et al. Cancer nanomedicine for combination cancer immunotherapy. Nat. Rev. Mater. 2019;4:398–414. doi: 10.1038/s41578-019-0108-1. [DOI] [Google Scholar]
- 247.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Joshi MD, Unger WJ, Storm G, van Kooyk Y, Mastrobattista E. Targeting tumor antigens to dendritic cells using particulate carriers. J. Control. Rel. 2012;161:25–37. doi: 10.1016/j.jconrel.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 249.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 250.Ma Q, Dieterich LC, Detmar M. Multiple roles of lymphatic vessels in tumor progression. Curr. Opin. Immunol. 2018;53:7–12. doi: 10.1016/j.coi.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 251.Garnier L, Gkountidi A-O, Hugues S. Tumor-associated lymphatic vessel features and immunomodulatory functions. Front. Immunol. 2019;10:720. doi: 10.3389/fimmu.2019.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat. Med. 2002;8:751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- 253.Cheng CJ, Tietjen GT, Saucier-Sawyer JK, Saltzman WM. A holistic approach to targeting disease with polymeric nanoparticles. Nat. Rev. Drug Discov. 2015;14:239. doi: 10.1038/nrd4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yang Y, et al. Effect of size, surface charge, and hydrophobicity of poly(amidoamine) dendrimers on their skin penetration. Biomacromolecules. 2012;13:2154–2162. doi: 10.1021/bm300545b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.De Matteis V. Exposure to inorganic nanoparticles: routes of entry, immune response, biodistribution and in vitro/in vivo toxicity evaluation. Toxics. 2017;5:29. doi: 10.3390/toxics5040029. [DOI] [PMC free article] [PubMed] [Google Scholar]