Supplemental Digital Content is available in the text.
Keywords: artificial intelligence, big data, database, data anonymization, data science, General Data Protection Regulation, Health Insurance Portability and Accountability Act, machine learning
Abstract
OBJECTIVES:
Critical care medicine is a natural environment for machine learning approaches to improve outcomes for critically ill patients as admissions to ICUs generate vast amounts of data. However, technical, legal, ethical, and privacy concerns have so far limited the critical care medicine community from making these data readily available. The Society of Critical Care Medicine and the European Society of Intensive Care Medicine have identified ICU patient data sharing as one of the priorities under their Joint Data Science Collaboration. To encourage ICUs worldwide to share their patient data responsibly, we now describe the development and release of Amsterdam University Medical Centers Database (AmsterdamUMCdb), the first freely available critical care database in full compliance with privacy laws from both the United States and Europe, as an example of the feasibility of sharing complex critical care data.
SETTING:
University hospital ICU.
SUBJECTS:
Data from ICU patients admitted between 2003 and 2016.
INTERVENTIONS:
We used a risk-based deidentification strategy to maintain data utility while preserving privacy. In addition, we implemented contractual and governance processes, and a communication strategy. Patient organizations, supporting hospitals, and experts on ethics and privacy audited these processes and the database.
MEASUREMENTS AND MAIN RESULTS:
AmsterdamUMCdb contains approximately 1 billion clinical data points from 23,106 admissions of 20,109 patients. The privacy audit concluded that reidentification is not reasonably likely, and AmsterdamUMCdb can therefore be considered as anonymous information, both in the context of the U.S. Health Insurance Portability and Accountability Act and the European General Data Protection Regulation. The ethics audit concluded that responsible data sharing imposes minimal burden, whereas the potential benefit is tremendous.
CONCLUSIONS:
Technical, legal, ethical, and privacy challenges related to responsible data sharing can be addressed using a multidisciplinary approach. A risk-based deidentification strategy, that complies with both U.S. and European privacy regulations, should be the preferred approach to releasing ICU patient data. This supports the shared Society of Critical Care Medicine and European Society of Intensive Care Medicine vision to improve critical care outcomes through scientific inquiry of vast and combined ICU datasets.
The adoption of machine learning is progressing at a revolutionary pace (1), even in the traditionally conservative medical domain (2). The promise of creating models that can diagnose, classify, predict, or optimize treatment using the wealth of routinely collected clinical information to individualize future treatments is exciting (3). However, this requires large amounts of patient data, ideally from multiple hospitals. Fortunately, electronic health records (EHRs) are widely adopted amongst ICUs (4), and ICU admissions generate vast amounts of data from patient monitors and life support devices (3).
Broad-scale data sharing with the critical care community implies that knowledge generation and validation could be accelerated. In addition, it may aid in addressing the problem of irreproducibility in research as analyses can easily be replicated, especially if code-sharing is enforced (5, 6). However, technical, legal, ethical, and privacy concerns have so far prevented large-scale data sharing both in general and by the critical care medicine community specifically (7). Currently, only two datasets containing comprehensive deidentified data from critical care patients are freely and openly accessible, the Medical Information Mart for Intensive Care (MIMIC) and eICU databases (8–10). However, they contain data collected exclusively in the United States, which limit generalizability to other healthcare systems such as those found in Europe which are differently organized and resourced. Specifically, model and knowledge transferability is hampered by substantive differences in ICU case mix, treatment strategies, and organization between continents, thus severely hindering the transition from bytes to bedside (11).
Consequently, the Society of Critical Care Medicine (SCCM) and the European Society of Intensive Care Medicine (ESICM) have identified ICU patient data sharing as one of the priorities under their Joint Data Science Collaboration (12). With the ultimate goal of harmonizing data across different sites and databases, they seek to encourage ICUs to make their data available by providing expert technical, legal, and ethical advice as well as recommendations on best practices (13). In collaboration with Amsterdam University Medical Centers (Amsterdam UMC), The Netherlands, known for their work in bringing data science to the bedside (14, 15), we therefore now report on the development and release of Amsterdam University Medical Centers database (AmsterdamUMCdb), the first freely accessible comprehensive and high-resolution European critical care database. The project is also the first to specifically address technical, legal, and ethical challenges in full compliance with both U.S. and European legislation, including the U.S. Health Insurance Portability and Accountability Act as well as the European General Data Protection Regulation (GDPR) (16, 17). The specific aim of this article is to describe the steps taken to develop AmsterdamUMCdb and to guide ICUs worldwide to responsibly share their data to benefit future critically ill patients.
MATERIALS AND METHODS
The department sourcing AmsterdamUMCdb is a mixed surgical-medical tertiary referral center for critical care medicine at an academic medical center in Amsterdam, The Netherlands, with up to 32 critical care beds and up to 12 high-dependency beds. Data from multiple clinical information systems were combined into the so-called “data lake” (Fig. 1A) (eMethods, Supplemental Digital Content 1, http://links.lww.com/CCM/G194). The table structure and underlying data model were designed with relevance for critical care, utility for machine learning, and human readability in mind. Fields that are typically needed together to understand the content were joined, for example, measurements with both identifiers and understandable names (Data Format, Supplemental Digital Content 2, http://links.lww.com/CCM/G195).
Figure 1.
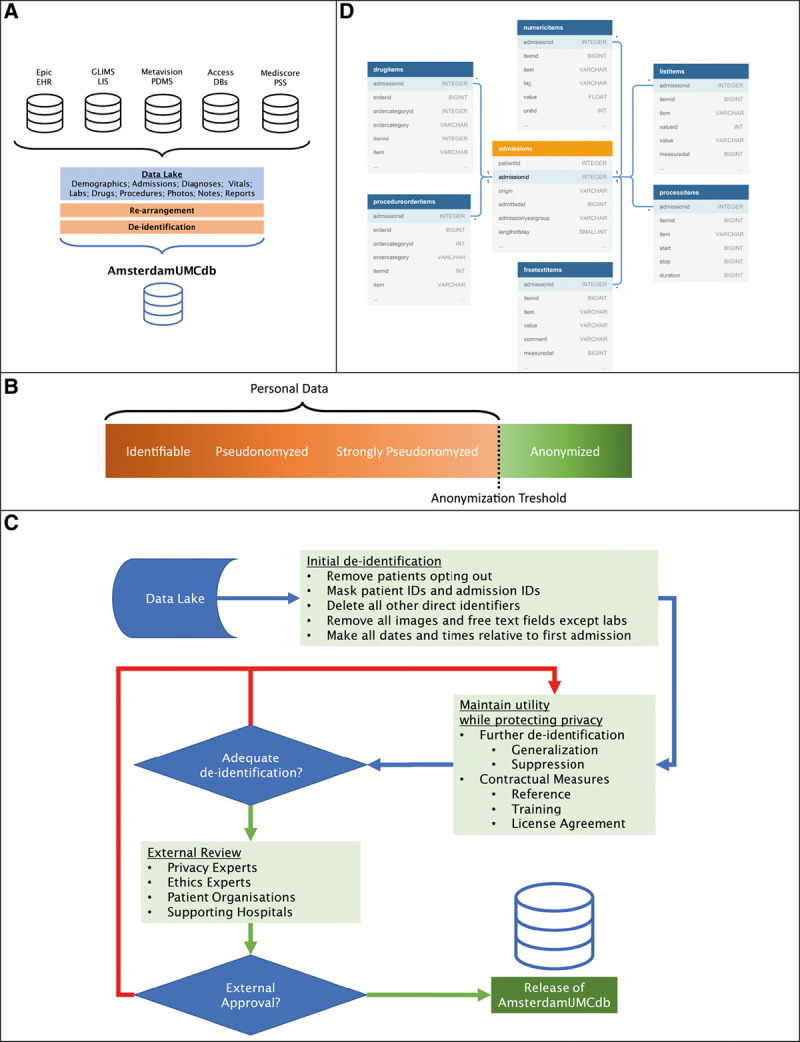
An overview of the process to create AmsterdamUMCdb from different source databases (A). The anonymization threshold separates personal data from anonymous data (B). The applied risk-based deidentification strategy demonstrating the iterative nature of performing deidentification (C). Final table structure depicting the relations with the admissions table (D). Capitalized words in the tables refer to data types used: INTEGER (whole number), SMALINT (small-range integer), BIGINT (large-range integer), FLOAT (floating-point number) or VARCHAR (variable size character data). DBs = databases, EHR = electronic health record, GLIMS = General Laboratory Information Management System, ID = identifier, LIS = Laboratory Information System, PDMS = Patient Data Management System, PSS = patient scoring system.
Risk-Based Deidentification Strategy
We used a risk-based iterative deidentification strategy to create a dataset that would qualify as anonymous data for the purposes of HIPAA and GDPR, maintaining utility while preserving privacy (Fig. 1, B and C) (18–20). This approach considers the possibility of attacks on the database by linking public or private information to reidentify patients. To mitigate attacks, we used different deidentification techniques to generate a database that is safe by design (eMethods and eTables 1 and 2, Supplemental Digital Content 1, http://links.lww.com/CCM/G194). In addition, we set up contractual and governance processes (21, 22). These processes include an end-user license agreement (EULA, Supplemental Digital Content 3, http://links.lww.com/CCM/G196) that, in contrast to EULA’s from other databases, require the signature of a practicing intensivist, the need for end users to complete a training on scientific integrity, and audits by representatives from patient organizations as well as experts on privacy and ethics. In addition, an access protocol for source data and an opt out procedure, as well as education and delegation logs were installed. Our deidentification strategy is subject to a continuous quality control cycle. This includes regular assessment of reidentification risk, when new data will be added.
Assessment of Reidentification Risk
We assessed reidentification risk using hypothetical adversaries representing different attack types. These include the “friendly researcher,” who might inadvertently reidentify an acquaintance; the “rogue researcher,” who might deliberately reidentify someone using public information; and the “rogue insurance company,” who might seek, illegally, to reidentify someone using corporate data (18, 21).
First, we determined the risk of reidentification should AmsterdamUMCdb be completely public, that is, without considering contractual and governance processes. To this end, we determined which items adversaries could have knowledge about the so-called “quasi-identifiers.” We applied statistical principles to determine reidentification risk of individual patient data (23), in particular k-anonymity and l-diversity with specific consideration for missing values (24). A database is said to have k-anonymity, when individuals, selected using quasi-identifiers, cannot be distinguished from at least k-1 other individuals in the database. The data has l-diversity if there are at least l distinct values for a sensitive attribute (e.g., diagnosis) within a group of persons with k-anonymity. We determined k-anonymity and l-diversity for all possible combinations of “quasi-identifiers” and calculated reidentification risk (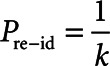 ) for each group (18, 21, 22).
) for each group (18, 21, 22).
Second, we determined the likelihood for adversaries to attempt reidentification and their likelihood of having access to AmsterdamUMCdb. For the “friendly researcher,” we used the Dunbar estimate of average number of friends (25), that is, 150, and the prevalence of ICU admissions (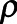 ), that is, 20,109 patients in the database divided by 13.7 million adult inhabitants in the Netherlands (26), to calculate the likelihood of knowing someone in the database using the formula
), that is, 20,109 patients in the database divided by 13.7 million adult inhabitants in the Netherlands (26), to calculate the likelihood of knowing someone in the database using the formula 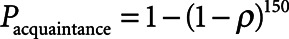 . This is a very conservative estimate, especially for researchers outside of the Netherlands. The likelihood for both researchers of having access to the data is 1. In contrast, for the “rogue insurance” company, access to the data requires a data breach, which is estimated to occur in up to 27% of databases maintained under HIPAA (18). This estimate is reasonable for a conservative risk assessment as HIPAA’s deidentification requirements are considered less strict than GDPR requirements (27).
. This is a very conservative estimate, especially for researchers outside of the Netherlands. The likelihood for both researchers of having access to the data is 1. In contrast, for the “rogue insurance” company, access to the data requires a data breach, which is estimated to occur in up to 27% of databases maintained under HIPAA (18). This estimate is reasonable for a conservative risk assessment as HIPAA’s deidentification requirements are considered less strict than GDPR requirements (27).
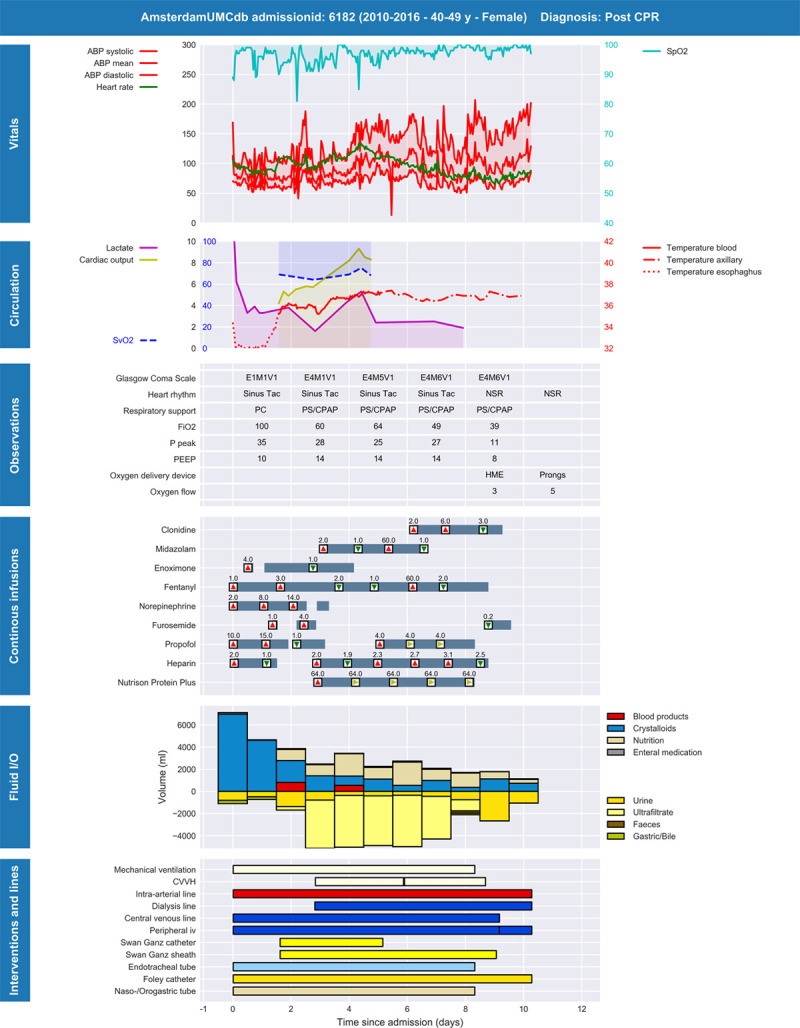
Finally, we chose an average risk approach for the “friendly researcher” and “rogue insurance company” and a maximum risk approach for the “rogue researcher.” The maximum risk approach assumes that the “rogue researcher” will attempt to reidentify persons with the highest chance of reidentification (i.e., the outliers). However, the “friendly researcher” and “rogue insurance company” will only aim to identify specific persons, not just anyone, and the risk of a successful reidentification attack of a specific person, not merely an outlier, is determined by the average risk. However, by applying further field and record suppression, we established at least k-2 anonymity. This conservative approach is called “strict average risk” (21). We ensured that the final risks of reidentification (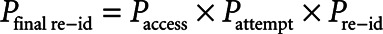 ) by these adversaries did not exceed commonly accepted thresholds (e.g., 0.05–0.10) (18, 21, 23).
) by these adversaries did not exceed commonly accepted thresholds (e.g., 0.05–0.10) (18, 21, 23).
Legal and Ethical Considerations
Our deidentification and governance strategies were externally audited by privacy experts to ensure anonymization. To prevent risk of bias, an internal team carried out a data privacy impact assessment and an external team performed an audit. The external team was led by a member of the privacy expert group at the Netherlands Federation of UMCs. The Regional Medical Ethics Committee confirmed that the creation of AmsterdamUMCdb was not eligible for their assessment as no specific research question was involved. The Ethics in Intensive Care Medicine group provided external ethics review and appraisal.
Communication and Engagements
Since sharing patient data is a sensitive subject, we developed a media and communication strategy based on full disclosure and multiangled views. This included a joint press release aiming to reach a large audience to assess public perception. In addition, we obtained explicit approval from our executive board and involved all stakeholders early on. These included the Dutch patient organization IC Connect and the Dutch Foundation of Family and Patient Centered Care, which both confirmed their perception of added value in releasing patient data, when privacy protection has been carried out appropriately. The Dutch Society of Intensive Care Medicine, and its Research Network, also made data sharing a priority. This led to multiple representatives of other ICUs expressing their intent to share their patient data responsibly.
RESULTS
Version 1.0.2 of AmsterdamUMCdb was released in March 2020 as “comma separated values” files. Access may be requested from Amsterdam Medical Data Science (https:/amsterdammedicaldatascience.nl/). Following approval, files can be downloaded from the Dutch Data Archiving and Networked Services (https://doi.org/10.17026/dans-22u-f8vd). Codes used for the analyses of this article, as well as detailed descriptions of the data, are available online (https://github.com/AmsterdamUMC/AmsterdamUMCdb). As shown in Figure 1D, the data model and table structure combine usability for machine learning with human readability (AmsterdamUMCdb Tables, Supplemental Digital Content 2, http://links.lww.com/CCM/G195).
Summary of Available Data
Table 1 shows patient demographics and data characteristics and Figures 2 and 3 illustrate the diversity of available data. The database contains approximately 1 billion clinical data points related to 23,106 admissions of 20,109 unique patients between 2003 and 2016. The median ICU length of stay was 1.08 days (interquartile range, 0.83–3.76 d). The released dataset includes patient monitor and life support device data (up to one value every minute), laboratory measurements, clinical observations and scores, medical procedures and tasks, medication, fluid balance, diagnosis groups, and clinical outcomes.
TABLE 1.
Characteristics of Patients and Data in Amsterdam University Medical Centers Database (AmsterdamUMCdb)
| Characteristics | Total | ICU | Medium Care Unit (High-Dependency Unit) |
|---|---|---|---|
| Distinct patients, n | 20,109 | 16,518 | 4,295 |
| ICU admissions, n | 23,106 | 18,386 | 4,720 |
| ICU length of stay, d, median (IQR) | 1.08 (0.83–3.67) | 1.25 (0.92–4.71) | 0.83 (0.71–1.62) |
| Gender | |||
| Male, n (%) | 12,799 (63.65) | 10,565 (63.96) | 2,234 (52.01) |
| Age, yr, n (%) | |||
| 18–39 | 2,202 (10.95) | 1,538 (9.31) | 743 (17.30) |
| 40–49 | 1,897 (9.43) | 1,356 (8.21) | 613 (14.27) |
| 50–59 | 3,405 (16.93) | 2,740 (16.59) | 800 (18.63) |
| 60–69 | 5,272 (26.22) | 4,518 (27.35) | 954 (22.21) |
| 70–79 | 5,293 (26.32) | 4,635 (28.06) | 824 (19.19) |
| 80+ | 2,040 (10.14) | 1,731 (10.48) | 361 (8.41) |
| Admission year, n (%) | |||
| 2003–2009 | 8,556 (42.55) | 7,940 (48.07) | 809 (18.84) |
| 2010–2016 | 11,553 (57.45) | 8,578 (51.93) | 3,486 (81.16) |
| Admission type, n (%) | |||
| Surgical admissions | 11,294 (48.88) | 8,942 (48.63) | 2,352 (49.83) |
| Urgent admissions | 6,246 (27.03) | 4,985 (27.11) | 1,261 (26.72) |
| Reason for admission, n (%) | |||
| Cardiothoracic surgery | 5,935 (25.69) | 5,759 (31.32) | 176 (3.73) |
| Sepsis | 3,136 (13.57) | 2,751 (14.96) | 385 (8.16) |
| Respiratory failure | 1,568 (6.79) | 1,402 (7.63) | 166 (3.52) |
| Neurosurgery | 1,619 (7.01) | 739 (4.02) | 880 (18.64) |
| Trauma | 902 (3.90) | 613 (3.33) | 289 (6.12) |
| Gastrointestinal surgery | 1,149 (4.97) | 800 (4.35) | 349 (7.39) |
| Vascular surgery | 1,037 (4.49) | 791 (4.30) | 246 (5.21) |
| Cardiac arrest | 959 (4.15) | 958 (5.21) | 1 (0.02) |
| Neurologic disorders (nontraumatic) | 628 (2.72) | 475 (2.58) | 153 (3.24) |
| Cardiac disorders (including cardiogenic shock) | 538 (2.33) | 485 (2.64) | 53 (1.12) |
| Supportive therapies, n (%) | |||
| Vasopressors and/or inotropes | 13,575 (58.75) | 12,809 (69.67) | 766 (16.23) |
| Mechanical ventilation | 16,680 (72.19) | 16,305 (88.68) | 375 (7.94)a |
| Renal replacement therapy | 1,140 (4.93) | 1,136 (6.18) | 4 (0.08) |
| Outcome, n (%) | |||
| Death at unit discharge | 2,288 (9.90) | 2,216 (12.05) | 72 (1.53) |
| Death < 1 yr after discharge | 4,730 (20.47) | 4,002 (21.77) | 728 (15.42) |
| Severity scores | |||
| Urgent patients | |||
| APACHE II score, median (IQR) | 19 (13–26) | 21 (16–27) | 12 (8–16) |
| SOFA score (day 1), median (IQR) | 7 (4–10) | 8 (5–10) | 2 (1–4) |
| Elective patients | |||
| APACHE II score, median (IQR) | 16 (12–20) | 17 (14–21) | 11 (8–15) |
| SOFA score (day 1), median (IQR) | 6 (4–8) | 6 (4–8) | 2 (1–4) |
APACHE = Acute Physiology And Chronic Health Evaluation, IQR = interquartile range, SOFA = Sequential Organ Failure Assessment.
aNoninvasive ventilation.
For conciseness, only major categories of reason for admission are shown. Reasons for admission documented without diagnostic codes (i.e., full text only) were excluded from the analysis. APACHE II score ranges from 0 to 71; higher ranges indicate greater severity of illness. SOFA score ranges from 0 to 24; higher ranges indicate greater severity of illness.
Figure 2.
Overview of the diversity of data in Amsterdam University Medical Centers Database (AmsterdamUMCdb). The plots show a selection of the most common data shown as percentage of availability for all admissions: device data that have been automatically filed (A), observations and scores that were entered manually (B), laboratory measurements (C), and administered drugs (D). ALAT = alanine aminotransferase, APACHE = Acute Physiology and Chronic Health Evaluation, ASAT = aspartate aminotransferase, CO = cardiac output, DNAR = do not attempt resuscitation, PEEP = positive end-expiratory pressure, SDD = selective decontamination of the digestive tract, Spo2 = peripheral oxygen saturation, TSH = thyroid-stimulating hormone.
Figure 3.
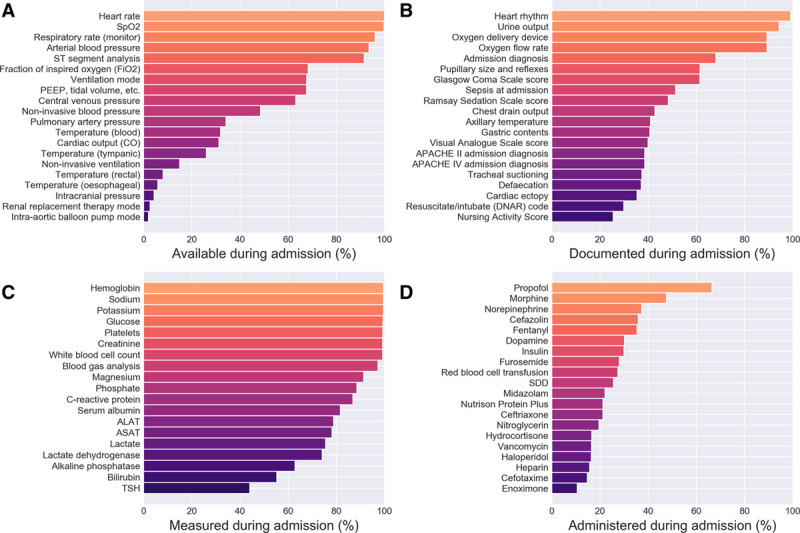
Example of time series data from an ICU admission in Amsterdam University Medical Centers Database (AmsterdamUMCdb) displayed as a graphical timeline. The data are from a patient admitted after cardiopulmonary resuscitation who received mild therapeutic hypothermia and developed shock and acute kidney injury with initiation of renal replacement therapy. The series show a selection of data documented throughout the admission: vital variables, clinical observations, infusions of medication, fluid input and output, supportive care, and inserted catheters, drains, and catheters. Data have been downsampled for readability and translated to English from the original Dutch variables and values. ABP = arterial blood pressure, CPAP = continuous positive airway pressure, CPR = cardiopulmonary resuscitation, CVVH = continuous veno-venous hemofiltration, I/O = input/output, NSR = normal sinus rhythm, PC = pressure control ventilation, PEEP = positive end-expiratory pressure, PS = pressure support ventilation, Spo2 = peripheral oxygen saturation, Svo2 = venous oxygen saturation.
Deidentification and Assessment of Reidentification Risk
Compared with the source data, our iterative deidentification strategy resulted in complete deletion of 265 admissions (1.13%) of 60 patients (0.30%) and suppression of weight, height, and/or diagnosis fields of 415 patients (1.80%). Table 2 summarizes the assumed adversary background knowledge and the formal risk analysis. The most prominent risk was presented by the “rogue researcher” (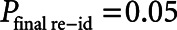 ). Compared with our risk-based strategy, the HIPAA Safe Harbor method would have led to unacceptably high reidentification risks (
). Compared with our risk-based strategy, the HIPAA Safe Harbor method would have led to unacceptably high reidentification risks (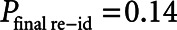 ) (eTable 2, Supplemental Digital Content 1, http://links.lww.com/CCM/G194) with the potential of reidentification of over half of the patients (n = 12,153) in the database by the “rogue researcher.”
) (eTable 2, Supplemental Digital Content 1, http://links.lww.com/CCM/G194) with the potential of reidentification of over half of the patients (n = 12,153) in the database by the “rogue researcher.”
TABLE 2.
Assumed Background Knowledge and Assessment of Reidentification Risk After Risk-Based Deidentification
| Hypothetical Adversary | |||
|---|---|---|---|
| Friendly Researcher | Rogue Researcher | Rogue Insurance Company | |
| Assumed background knowledge | |||
| Gender | X | X | X |
| Age | X | X | X |
| Weight | X | X | |
| Height | X | X | |
| Admission date | X | X | X |
| Survival at discharge | X | X | X |
| Number of ICU admissions | X | ||
| Assessment of reidentification risk | |||
| P(access) | 1.00 | 1.00 | 0.27 |
| P(intention) | 0.20 | 0.10 | 0.10 |
| Average risk | |||
| P(reidentification) | 0.047 | 0.047 | 0.009 |
| k-anonymity | 89 | 89 | 682 |
| l-diversity | 26 | 26 | 65 |
| Maximum risk | |||
| P(reidentification) | 0.50 | 0.50 | 0.50 |
| k-anonymity | 2 | 2 | 2 |
| l-diversity | 2 | 2 | 2 |
| P final risk | 0.01 | 0.05 | 0.0002 |
Strict average risk is used for determining final risk for the “friendly researcher” and the “rogue insurance company,” whereas maximum risk was used for the “rogue researcher.” For the “friendly researcher,” P(intention) is acquaintance risk, the risk of knowing somebody in the database.
Legal Considerations
Legal analyses focused on the GDPR as the data are from European patients. Recital 26 states that “the principles of data protection should not apply to personal data rendered anonymous in such a manner that the data subject is not or no longer identifiable. To determine whether a natural person is identifiable, account should be taken of all the means reasonably likely to be used, considering all objective factors, such as the costs of and the amount of time required for identification, the available technology at the time of the processing and technological developments” (17).
Two independent teams auditing the project reported that “the design, database management and governance are state-of-the-art.” Based on our reidentification risk assessment, they concluded that “taking into account all means reasonably likely to be used for reidentification, reidentification is not reasonably likely for AmsterdamUMCdb which can therefore be considered as anonymous information in the context of the GDPR.” In addition, Amsterdam UMC is NEN7510 (ISO 27001) certified, which ensures that strict information governance protocols are in place for the protection of source data.
The teams also audited the legal aspects of the data processing to develop the database from the source data, which arguably “is” subject to the GDPR. Article 9 of the GDPR identifies health data as one of the special categories of personal data for which data processing is prohibited without a specific lawful basis. In our case, data processing was performed on the lawful basis of scientific research as specified in article 89 of the GDPR, since it is not realistically feasible to operate on the basis of explicit consent given the large numbers of subjects included in the database.
Ethical Considerations
From an ethical perspective, medical confidentiality precludes sharing of potentially identifiable information to others without explicit consent of the patient unless overwhelmingly in the public interest. Although obtaining informed consent would clearly allow ethically acceptable secondary use of health-related data, obtaining consent from critical care patients may be extremely difficult to achieve in practice (28). This is especially the case if large numbers of patients are involved, where there is high mortality or where patients may have impaired levels of consciousness, all of which are true in the ICU. In addition, refusals and untraceable patients will reduce data quality and introduce selection bias that may lead to biased results and algorithms that can contribute to further health disparities. Thus, obtaining individual patient informed consent for sharing critical care databases is neither desirable nor feasible. The ethical principle “duty of easy rescue” applies here (29). When the burden of performing an action is small and the benefit is putatively large, we ought to act. For the case of data sharing using proper risk-based deidentification strategies, the risk is minimal. But, the benefit for other and future generations of patients as well as society is potentially large.
Communication
AmsterdamUMCdb received extensive coverage by international media including prime time Dutch television news. Overall public sentiment was positive, with only a small minority of reactions related to privacy issues. Among millions of people reached, only one request for data removal was received. This is in line with the results of a recent extensive poll among over 7,000 patients by the Dutch Patient Federation showing that over 97% of patients, asked to share their data, were comfortable with data sharing for medical research to help future patients (30). Only a very small number of patients (1%) refused out of fear of inadequate protection of their data.
DISCUSSION
The SCCM/ESICM Joint Data Science Collaboration results from the shared vision of these societies in exploring how common data definitions, data science, and data sharing may impact clinical care, quality improvement, and scientific inquiry in critical care. With the ultimate goal of harmonizing data across different sites and databases, responsible data sharing is an important first step. Wide availability of critical care databases is of paramount importance to ensure model performance, validity, generalizability, and transferability. Since AmsterdamUMCdb is already being used by over 40 research institutions worldwide to develop new (machine learning) models and validate published ones, it demonstrates the need for wide availability of anonymized medical data.
The GDPR provides a framework to support and regulate the sharing of sensitive data. However, member states are free to impose additional local statutory requirements and regulations. A similar example exists in the United States where the California Consumer Privacy Act is generally perceived to be more restrictive than HIPAA. Since the risk assessment for proper deidentification is pivotal in our approach and not a mere academic exercise, one of the challenges will be to gain insight into the potential adversaries. The background information that potential adversaries may possess will differ from country to country depending on the health information routinely exchanged with third parties. Correctly identifying these sources of information that could be used to reidentify patients requires consulting clinical lead, local hospital administration, or business intelligence departments.
Our risk-based deidentification strategy operationalizes the definition of anonymity by using thresholds (Fig. 1B). This resonates well with recital 26 of the GDPR which calls for consideration of all means reasonably likely to be used for reidentification to determine whether natural persons are identifiable. However, different definitions have also been proposed, focusing on the prevention of singling out, linkability, or inference, even if only of theoretical concern (31). This is a problematic position to take for releasing critical care databases as these are great examples of extensive time series data, given the high resolution of data from monitors and life support devices. All patient records will be unique if enough data are released: the “curse of dimensionality.” And indeed, some may argue that no deidentification strategy is able to guarantee zero risk of reidentification and that individual critical care patient data could never be released (23, 32). Nevertheless, although many previously released public databases suffered from incorrect deidentification strategies or were only deidentified by following HIPAA Safe Harbor methods, and thus were not able to withstand reidentification attacks (33), recent evidence suggests that when a dataset is properly deidentified, even when releasing redacted narratives, this may only lead to low confidence matches even after investing a significant amount of time (34). In addition, generating synthetic data from real patient data may further reduce reidentification risks, and AmsterdamUMCdb has been selected to feature in the prestigious Conference on Neural Information Processing Systems competition of December 2020 to evaluate the feasibility of this technique (35, 36).
A strength of our approach is that it resulted in the removal of only a relatively small number of records and fields. This should ensure adequate usability for data science. However, a significant limitation of our strategy is that in an earlier step, all free-text fields and imaging have been removed. In addition, the main limitation of AmsterdamUMCdb is that it has been sourced from a single Dutch ICU, thereby limiting generalizability. A further limitation of our risk-based strategy is that it does not directly take into account the risk of prospective collection, since adding new records to a deidentified database may unintentionally reveal information (e.g., approximate discharge dates). Also, given that access to the data is one of the determinants of reidentification risk, violating the EULA by distributing the database to third parties, which cannot reasonably be monitored, could increase risk. Finally, no risk-based deidentification strategy will fully prevent against malignant intent such as criminal or terrorist activity, since in those cases, adversaries will have gained access to additional sensitive resources. However, in these rare circumstances, not reidentification but disclosure of those other resources should most likely be the main concern.
In contrast to other openly accessible ICU databases (8–10), our table structure sacrifices disk and memory space for the sake of interpretability using denormalization (Data Format, Supplemental Digital Content 2, http://links.lww.com/CCM/G195). We believe a simplified data model will expedite the process from data to bedside.
Although many of the challenges for responsible data sharing have been addressed, several other problems should not be overlooked. First, some investigators perceive the data as their property and believe to have the exclusive right to publish based on these data (37). Second, hospitals may see data as revenue or means to attract industry experts, such as Google, although this has shown to generate negative publicity (38). Third, several EHR vendors, at least in the United States, are actively lobbying against data sharing based on privacy arguments yet are also actively developing revenue streams based on smart analytics (39). Finally, hospitals may fear that cases where quality of care was arguably suboptimal would be uncovered by competitors, lawyers, or the public.
Although previous efforts by individual critical care departments in Belgium and Spain have been prevented by local ethics committees (7), there have been some notable initiatives by consortia of ICUs to promote data sharing. These include the Critical Care Health Informatics Collaborative and M@tric initiatives, collecting large amounts of data from six U.K. and three Belgian ICUs, respectively (40, 41). However, their current focus is on sharing data between founding ICUs only.
It could be argued that reliance on big data from EHR’s to improve patient outcomes might lead to a greater disparity between higher and lower resource countries, as the latter often lack implementation of an EHR. However, we see an opportunity for using large databases from high resource countries to better than before determine the added benefit of (costly) medical interventions.
Costs of data governance, including the costs of a risk-based approach, were not specifically addressed. Although the actual data extraction with iterative deidentification including manual checks has been estimated to have taken two full time equivalents of combined clinical and nonclinical staff for a month, the legal and ethical process, by the nature of the sensitivity of releasing healthcare data, required between 9 and 12 months in our case. We imagine by having created a framework for sharing ICU data, future releases of AmsterdamUMCdb but more importantly datasets from other institutions will require a more reasonable timeframe.
As mentioned, currently all freely available critical care datasets come from the United States (9, 10, 42). This may be partially explained by differences between the privacy laws of the United States and Europe. The HIPAA Privacy Rule provides two methods by which health information can be designated as deidentified. One method requires expert determination of very low risk of reidentification, comparable with our approach. The other is arguably easier to adhere to by requiring the removal of 18 specific elements (eTable 2, Supplemental Digital Content 1, http://links.lww.com/CCM/G194) and absence of actual knowledge that the remaining data are reidentifiable. It may be argued that this pathway was not designed for the current digital age with the everincreasing availability of public and private datasets (27). Our approach to data governance complies with both U.S. and European regulations and should be considered the model for accretion for future deidentified healthcare datasets. However, because risk-based deidentification is not without its caveats or costs, we expect all major medical societies, including SCCM and ESICM, to play an import role in disseminating the state-of-the-art of responsible disclosure of healthcare data.
Connecting ICUs that consider sharing their patient data is important for creating a worldwide network as well as a knowledge base to address challenges related to privacy and ethics. Besides sharing data, ICUs should require end users to share their queries and code to ensure reproducible research. This would greatly contribute to the broader goals of the SCCM/ESICM Joint Data Science Collaboration that include advancing critical care by harmonizing datasets across sites and continents to facilitate a common language spoken across a global collaborative (12).
CONCLUSIONS
The SCCM/ESICM Joint Data Science Collaboration envisions global ICU data sharing and harmonization. The development and release of AmsterdamUMCdb serves as an example on how to address the many technical, legal, ethical, and privacy challenges related to responsible data sharing using a multidisciplinary approach. A risk-based deidentification strategy, that complies with both U.S. and European privacy regulations, should be the preferred approach to releasing ICU patient data. To accelerate delivering on the promise of data driven approaches, we encourage other ICUs to follow this example.
ACKNOWLEDGMENTS
Amsterdam University Medical Centers (Amsterdam UMC) is grateful to the Executive Committee and the Data Science Section of the Society of Critical Care Medicine and the European Society of Intensive Care Medicine for initiating their visionary Data Science Collaboration; to our colleagues at the Massachusetts Institute of Technology Laboratory for Computational Physiology for extensive discussions on many aspects related to the creation, deidentification and distribution of critical care databases and for producing the MIMIC and eICU databases that were a great inspiration; to our Privacy Officers Marcel van der Haagen and Michel Paardekooper for expert advice; to our Communications department, in particular Nicole de Haan and Jan Hol for help with and execution of our communication strategy; and to the Executive Board of Amsterdam UMC, in particular Dr. Mark Kramer, MD, PhD for strongly supporting this initiative. The AmsterdamUMCdb Collaborators consist of the authors and Luca F. Roggeveen, MD, Lucas M. Fleuren, MD, Tingjie Guo, MSc, Department of Intensive Care Medicine, Amsterdam Medical Data Science (AMDS), Amsterdam Cardiovascular Sciences (ACS), Amsterdam Infection and Immunity Institute (AI&II), Amsterdam UMC, Vrije Universiteit, Universiteit van Amsterdam, Amsterdam, The Netherlands; Diederik A. M. P. J. Gommers, MD, PhD, Department of Intensive Care Medicine, Erasmus MC, Rotterdam, The Netherlands and Executive Committee, Dutch Society of Intensive Care (NVIC), Utrecht, The Netherlands; Lilian C. M. Vloet, PhD, Department of Emergency and Critical Care, HAN University of Applied Sciences, Nijmegen, The Netherlands and Foundation Family and Patient Centered Care, Alkmaar, The Netherlands and IQ Healthcare, Radboud Institute of Health Sciences, Scientific Center for Quality of Healthcare, Nijmegen, the Netherlands; Bas C. T. van Bussel, MD, PhD, Iwan C. C. van der Horst, MD, PhD, Department of Intensive Care Medicine, Maastricht University Medical Center+, Maastricht University, Maastricht, The Netherlands; Olaf L. Cremer, MD, PhD, Department of Intensive Care Medicine, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands; Sander Rigter, MD, Department of Anesthesia, Intensive Care Medicine and Pain Medicine, St. Antonius Hospital, Nieuwegein, The Netherlands; Tim Frenzel, MD, PhD, J. G. (Hans) van der Hoeven, MD, PhD, Department of Intensive Care Medicine, Radboudumc Nijmegen, Nijmegen, The Netherlands; Rob J. Bosman, MD, Department of Intensive Care, OLVG, Amsterdam, The Netherlands; R. P. (Peter) Pickkers, MD, PhD, Department of Intensive Care Medicine, Radboudumc Nijmegen, Nijmegen, The Netherlands and Research Collaboration on Critical Care in The Netherlands (RCCNet), Dutch Society of Intensive Care Medicine (NVIC), Utrecht, The Netherlands; Leo M. A. Heunks, MD, PhD, Department of Intensive Care Medicine, Amsterdam Medical Data Science (AMDS), Amsterdam Cardiovascular Sciences (ACS), Amsterdam Infection and Immunity Institute (AI&II), Amsterdam UMC, Vrije Universiteit, Universiteit van Amsterdam, Amsterdam, The Netherland and Research Collaboration on Critical Care in The Netherlands (RCCNet), Dutch Society of Intensive Care Medicine (NVIC), Utrecht, The Netherlands; Arjen J. C. Slooter, MD, PhD, Department of Intensive Care Medicine, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands and Research Collaboration on Critical Care in The Netherlands (RCCNet), Dutch Society of Intensive Care Medicine (NVIC), Utrecht, The Netherlands; Nicole P. Juffermans, MD, PhD, Department of Intensive Care Medicine, Amsterdam Medical Data Science (AMDS), Amsterdam Cardiovascular Sciences (ACS), Amsterdam Infection and Immunity Institute (AI&II), Amsterdam UMC, Vrije Universiteit, Universiteit van Amsterdam, Amsterdam, The Netherlands and Department of Intensive Care, OLVG, Amsterdam, The Netherlands and Research Collaboration on Critical Care in The Netherlands (RCCNet), Dutch Society of Intensive Care Medicine (NVIC), Utrecht, The Netherlands; Leo A. Celi, MD, MS, MPH, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA and Division of Pulmonary, Critical Care and Sleep Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA. The SCCM/ESICM Joint Data Science Task Force consists of these authors: Matthew Churpek, MD, MPH, PhD, Department of Medicine, University of Wisconsin, Madison, WI; Gilles Clermont, MD, Department of Critical Care Medicine, CRISMA Laboratory, University of Pittsburgh, Pittsburgh, PA; Ari Ercole, MD, PhD, Division of Anaesthesia, University of Cambridge, Cambridge, United Kingdom and Data Science Section, European Society of Intensive Care Medicine, Brussels, Belgium; Paul W. G. Elbers, MD, PhD, Department of Intensive Care Medicine, Amsterdam Medical Data Science (AMDS), Amsterdam Cardiovascular Sciences (ACS), Amsterdam Infection and Immunity Institute (AI&II), Amsterdam UMC, Vrije Universiteit, Universiteit van Amsterdam, Amsterdam, The Netherlands and Data Science Section, European Society of Intensive Care Medicine, Brussels, Belgium.
Supplementary Material
Footnotes
*See also p. 1003.
The Regional Medical Ethics Committee deemed that the creation of Amsterdam University Medical Centers Database (AmsterdamUMCdb) was not eligible for their assessment as no specific research question was involved. The process of developing the database was audited by an external team led by a member of the privacy expert group at the Netherlands Federation of University Medical Centers. The Ethics in Intensive Care Medicine group provided external ethics review and appraisal.
Drs. Thoral and Elbers wrote the deidentification process draft and drafted the article. Mr. Peppink and Mr. Driessen designed the data extraction process. Dr. Sijbrands led the external privacy auditing process. Dr. Kompanje lead the ethics review. Dr. Thoral, Mr. Peppink, Mr. Driessen, and Dr. Elbers analyzed and interpreted the data and performed the iterative deidentification. Drs. Kaplan, Bailey, Kesecioglu, Cecconi, Ercole, Churpek, Clermont, and Girbes were involved in starting the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration, participated in extensive discussions related to data deidentification and sharing, and audited these processes. Drs. van der Schaar, Ercole, and Elbers evaluated machine learning applicability. All authors read, commented on, and approved the final article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccmjournal).
Dr. Sijbrands’ institution received funding from European Institute of Innovation and Technology (EIT) Health and Amgen. Drs. Kaplan and Bailey received funding from Society of Critical Care Medicine. Dr. Cecconi received funding from Directed Systems, Edwards Lifesciences, and Cheetah Medical. Dr. Churpek’s institution received funding from an EarlySense research grant; he is supported by National Institutes of Health (NIH) R01 (GM123193), and he has a patent pending for risk stratification algorithm for hospitalized patients (money from royalties from the University of Chicago). Dr. Clermont received funding from the NIH, Department of Defense, National Science Foundation, and NOMA AI. The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019; 380:1347–1358 [DOI] [PubMed] [Google Scholar]
- 2.Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018; 319:1317–1318 [DOI] [PubMed] [Google Scholar]
- 3.Bailly S, Meyfroidt G, Timsit J-F. What’s new in ICU in 2050: Big data and machine learning. Intensive Care Med. 2017; 44:1524–1527 [DOI] [PubMed] [Google Scholar]
- 4.Cosgriff CV, Celi LA, Stone DJ. Critical care, critical data. Biomed Eng Comput Biol. 2019; 10:1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupple A, Singerman D, Celi LA. The reproducibility crisis in the age of digital medicine. npj Digit Med. 2019; 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruns SB, Ioannidis JPA. p-curve and p-hacking in observational research. PLoS One. 2016; 11:e0149144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLennan S, Shaw D, Celi LA. The challenge of local consent requirements for global critical care databases. Intensive Care Med. 2019; 45:246–248 [DOI] [PubMed] [Google Scholar]
- 8.Mark R. The story of MIMIC. In: Secondary Analysis of Electronic Health Records. MIT Critical Data (Eds). Cham, CH: Springer; 2016Available at: https://www.ncbi.nlm.nih.gov/books/NBK543630/. Accessed September 22, 2020 [PubMed] [Google Scholar]
- 9.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard TJ, Johnson AEW, Raffa JD, et al. The eICU collaborative research database, a freely available multi-center database for critical care research. Sci Data. 2018; 5:180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anesi GL, Admon AJ, Halpern SD, et al. Understanding irresponsible use of intensive care unit resources in the USA. Lancet Respir Med. 2019; 7:605–612 [DOI] [PubMed] [Google Scholar]
- 12.Kaplan LJ, Cecconi M, Bailey H, et al. Imagine…(a common language for ICU data inquiry and analysis). Crit Care Med. 2020; 48:273–275 [DOI] [PubMed] [Google Scholar]
- 13.European Society of Intensive Care Medicine - Data Science Section. Data Science – ESICM. Available at: https://www.esicm.org/groups/data-science/. Accessed January 20, 2020
- 14.Roggeveen LF, Fleuren LM, Guo T, et al. Right dose right now: Bedside data-driven personalized antibiotic dosing in severe sepsis and septic shock - rationale and design of a multicenter randomized controlled superiority trial. Trials. 2019; 20:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbers PW, Girbes A, Malbrain ML, et al. Right dose, right now: Using big data to optimize antibiotic dosing in the critically ill. Anaesthesiol Intensive Ther. 2015; 47:457–463 [DOI] [PubMed] [Google Scholar]
- 16.govinfo. Health Insurance Portability and Accountability Act (1996). Public Law 104–191. Available at: https://www.govinfo.gov/app/details/PLAW-104publ191. Accessed September 22, 2020 [PubMed]
- 17.EUR-Lex: General Data Protection Regulation (2016).Regulation (EU) 2016/679. Available at: https://eur-lex.europa.eu/eli/reg/2016/679/oj. Accessed September 22, 2020
- 18.El Emam K: Guide to the De-Identification of Personal Health Information. Boca Raton: Auerbach Publications; 2013. Available at: https://www.taylorfrancis.com/books/9780429100659. Accessed September 22, 2020 [Google Scholar]
- 19.Arbuckle L, Ritchie F. The five safes of risk-based anonymization. IEEE Secur Priv. 2019; 17:84–89 [Google Scholar]
- 20.Prasser F, Kohlmayer F, Kuhn KA. The importance of context: Risk-based de-identification of biomedical data. Methods Inf Med. 2016; 55:347–355 [DOI] [PubMed] [Google Scholar]
- 21.El Emam K, Arbuckle L: Anonymizing Health Data: Case Studies and Methods to Get You Started. Sebastopol, O’Reilly Media, Inc., 2013 [Google Scholar]
- 22.El Emam K, Rodgers S, Malin B, et al. Anonymising and sharing individual patient data. BMJ. 2015; 350:h1139–h1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandt M, Franconi L, Guerke C, et al. : Guidelines for the Checking of Output Based on Microdata Research. ESSNet SD, 2010. Available at: https://research.cbs.nl/casc/ESSnet/guidelines_on_outputchecking.pdf. Accessed February 16, 2020
- 24.Ciglic M, Eder J, Koncilia C. k-Anonymity of microdata with NULL values. In: Database and Expert Systems Applications. Decker H, Lhotská L, Link S, et al. (Eds). DEXA 2014. Lecture Notes in Computer Science. Springer Verlag; 2014, pp 328–342 [Google Scholar]
- 25.Gonçalves B, Perra N, Vespignani A. Modeling users’ activity on twitter networks: Validation of Dunbar’s number. PLoS One. 2011; 6:e22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centraal Bureau voor de Statistiek (CBS). CBS statline. Available at: https://opendata.cbs.nl/statline/#/CBS/nl/. Accessed January 20, 2020
- 27.Cohen IG, Mello MM. Big data, big tech, and protecting patient privacy. J Am Med Assoc. 2019; 322:1141–1142 [DOI] [PubMed] [Google Scholar]
- 28.Mentzelopoulos SD, Slowther AM, Fritz Z, et al. Ethical challenges in resuscitation. Intensive Care Med. 2018; 44:703–716 [DOI] [PubMed] [Google Scholar]
- 29.Porsdam Mann S, Savulescu J, Sahakian BJ. Facilitating the ethical use of health data for the benefit of society: Electronic health records, consent and the duty of easy rescue. Philos Trans A Math Phys Eng Sci. 2016; 374:20160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patiëntenfederatie Nederland. Delen medische data voor meeste patiënten geen punt | Nieuws, 2019. Available at: https://www.patientenfederatie.nl/nieuws/delen-medische-data-voor-meeste-patienten-geen-punt. Accessed December 27, 2019
- 31.El Emam K, Alvarez C. A critical appraisal of the Article 29 Working Party Opinion 05/2014 on data anonymization techniques. Int Data Priv Law. 2015; 5:73–87 [Google Scholar]
- 32.Rocher L, Hendrickx JM, de Montjoye YA. Estimating the success of re-identifications in incomplete datasets using generative models. Nat Commun. 2019; 10:3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Emam K, Jonker E, Arbuckle L, et al. A systematic review of re-identification attacks on health data. PLoS One. 2011; 6:e28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branson J, Good N, Chen JW, et al. Evaluating the re-identification risk of a clinical study report anonymized under EMA policy 0070 and health canada regulations. Trials. 2020; 21:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foraker R, Mann DL, Payne PRO. Are synthetic data derivatives the future of translational medicine? JACC Basic Transl Sci. 2018; 3:716–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jordon J, Jarrett D, Yoon J, et al. Hide-and-seek privacy challenge. arXiv 2020; arXiv 2007.12087
- 37.Figueiredo AS. Data sharing: Convert challenges into opportunities. Front Public Health. 2017; 5:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachter RM, Cassel CK. Sharing health care data with digital giants: Overcoming obstacles and reaping benefits while protecting patients. JAMA. 2020; 323:507–508 [DOI] [PubMed] [Google Scholar]
- 39.Farr C. Epic and About 60 Hospitals Sign Letter Opposing HHS Proposed Data Rules. 2020. Available at: https://www.cnbc.com/2020/02/05/epic-about-60-hospitals-sign-letter-opposing-hhs-proposed-data-rules.html. Accessed February 23, 2020
- 40.Harris S, Shi S, Brealey D, et al. Critical Care Health Informatics Collaborative (CCHIC): Data, tools and methods for reproducible research: A multi-centre UK intensive care database. Int J Med Inform. 2018; 112:82–89 [DOI] [PubMed] [Google Scholar]
- 41.M@tric Project. M@tric, 2019. Available at: https://www.matric.be/. Accessed December 22, 2019
- 42.Shillan D, Sterne JAC, Champneys A, et al. Use of machine learning to analyse routinely collected intensive care unit data: A systematic review. Crit Care. 2019; 23:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


