Abstract
Sepsis is a leading cause of mortality in the intensive care unit. Early prediction of sepsis can reduce the overall mortality rate and cost of sepsis treatment. Some studies have predicted mortality and development of sepsis using machine learning models. However, there is a gap between the creation of different machine learning algorithms and their implementation in clinical practice.
This study utilized data from the Medical Information Mart for Intensive Care III. We established and compared the gradient boosting decision tree (GBDT), logistic regression (LR), k-nearest neighbor (KNN), random forest (RF), and support vector machine (SVM).
A total of 3937 sepsis patients were included, with 34.3% mortality in the Medical Information Mart for Intensive Care III group. In our comparison of 5 machine learning models (GBDT, LR, KNN, RF, and SVM), the GBDT model showed the best performance with the highest area under the receiver operating characteristic curve (0.992), recall (94.8%), accuracy (95.4%), and F1 score (0.933). The RF, SVM, and KNN models showed better performance (area under the receiver operating characteristic curve: 0.980, 0.898, and 0.877, respectively) than the LR (0.876).
The GBDT model showed better performance than other machine learning models (LR, KNN, RF, and SVM) in predicting the mortality of patients with sepsis in the intensive care unit. This could be used to develop a clinical decision support system in the future.
Keywords: intensive care unit, machine learning, mortality prediction, sepsis
1. Introduction
Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection.[1] Sepsis is not only a common and potentially life-threatening condition, but is also a major global health issue.[2] An estimated more than 30 million people develop sepsis every year worldwide, potentially leading to 6 million deaths.[3] Sepsis is one of the most burdensome diseases worldwide because of high treatment costs and excessively lengthy hospital stays.[4] However, early diagnosis and accurate identification of the risk factors, as well as the appropriate treatment, reduce the overall mortality rate, and improve patient outcomes.[5] It is difficult to diagnose sepsis early due to various sources of infection in patients with sepsis and the difference in host response. Early and timely detection of sepsis has always been the focus of research.[6]
Some studies have shown that machine learning can be used to build prognostic models for both mortality and sepsis development.[7–10] To predict septic mortality, previous studies have employed big data and machine learning models such as stochastic gradient boosting, support vector machine (SVM), naive Bayes, logistic regression (LR), and random forest (RF).[11–13] Machine learning helps to analyze complex data automatically and produces significant results. Machine learning-based approaches have the potential for increased sensitivity and specificity by training sepsis patient data.[14–16] However, the possibility of some techniques, including ensemble algorithms, has not yet been addressed in improving the prediction outcomes. It is also necessary to find methods for generating accurate predictions.
To address these issues, we conducted an exploratory study to evaluate the efficiency of different classification algorithms in predicting death in adult patients with sepsis. In this study, we compared the gradient boosting decision tree (GBDT) model with other machine learning approaches (LR, k-nearest neighbor, RF, and SVM) using the prediction of sepsis in-hospital mortality as the use case.
2. Methods
2.1. Dataset
This study used the Medical Information Mart for Intensive Care III (MIMIC-III) V1.4. MIMIC-III is a large, freely available database comprising anonymous health-related data associated with over 53,423 adult patients admitted to critical care units at the Beth Israel Deaconess Medical Center in Boston between 2001 and 2012.[17]
2.2. Ethics statement
Because our study used an open-access database, no further local institutional review board approval was required. Data analysis and model development procedures followed the MIMIC-III guidelines and regulations.
2.3. Sepsis definition
Septic patients were identified using the International Classification of Diseases-9th Revision, Clinical Modification (ICD-9-CM) code for sepsis from records in the database. The codes included: 003.1 (salmonella septicemia); 022.3 (anthrax septicemia); 038.0 to 038.9 (subcodes of septicemia); 054.5 (herpetic septicemia). In October 2002, new diagnostic codes came into effect. They were included in the study: 995.91 (systemic inflammatory response syndrome caused by the infectious process without organ dysfunction) and 995.92 (systemic inflammatory response syndrome caused by the infectious process with organ dysfunction).[18]
2.4. Data extraction and imputation
We developed SQL scripts that contain a large number of SQL statements to query the MIMIC-III database for all adult patients (≥ 18 years). We used the indicators when the patients entered the ICU for the first time to predict the in-hospital mortality rate. Researchers extracted all data of sepsis patients 48 hours after ICU admission. Data extracted included patient age, sex, ethnicity, length of hospital stay, Glasgow coma scale, percutaneous oxygen saturation, vital signs, and laboratory values. Sepsis was defined as an ICD-9. Researchers restricted our search to patients aged 18 years or older, and we excluded patients with data missing more than 30%.[19] For each variable with less than 30% missing values, we replaced the missing values by means in each group.
2.5. Prediction model
In this study, 5 prediction models of the GBDT, LR, K-nearest neighbor (KNN), RF, and SVM models were established and compared.
GBDT is a new algorithm that combines decision trees and holistic learning techniques.[20] Its basic idea is to combine a series of weak base classifiers into a strong base classifier.[21] In the learning process, a new regression tree is constructed by fitting residuals to reduce the loss function until the residuals are less than a certain threshold, or the number of regression trees reaches a certain threshold. The advantages of GBDT are good training effect, less overfitting, and flexible handling of various data types, including continuous and discrete values.[22]
LR is a statistical method for analyzing datasets, and it is also a supervised machine-learning algorithm developed for learning classification problems.[23] It is one of the most widely used methods in health sciences research, especially in epidemiology.[24] Some studies have shown that LR is effective in analyzing mortality factors and predicting mortality.[25,26]
The KNN is an algorithm that stores all available instances and classifies new instances based on a similarity measure (such as distance functions).[27] It has been widely used in classification and regression prediction problems owing to its simple implementation and outstanding performance.[28]
SVM was derived from the statistical learning theory proposed by Cortes and Vapnik.[29] SVM maps the original datasets from the input space to the high-dimensional feature space, thus simplifying the classification problems in the feature space. Its main advantage is that it uses kernel tricks to build expert knowledge about the problem, minimizing both model complexity and prediction error.[30]
RF is an ensemble supervised machine learning algorithm. It uses a decision tree as the base classifier. RF produces many classifiers and combines their results through majority voting.[31]
In this study, we used the Scikit-learn toolkit to train and test the model. We conducted a 5-fold cross-validation using only the encounters allocated to the training set. For the cross-validation results, a paired t test was used to measure the significant difference between the models.
3. Results
A total of 3937 patients were included, with 34.3% in-hospital mortality in the MIMIC-III v1.4 database (Fig. 1). The main characteristics of the patients with sepsis are shown in Table 1. Compared with those who survived, patients who died were older (68.9 ± 14.9) versus 65.5 ± 16.7 years (P < .01).
Figure 1.
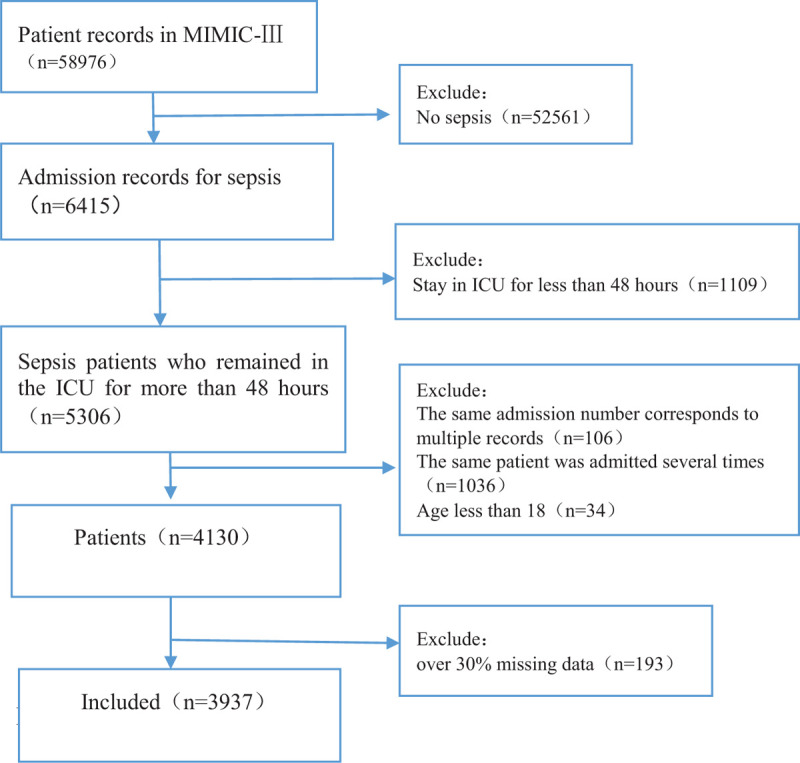
The flowchart for including patients in the study.
Table 1.
Patient demographic information.
| Variable | Death (n = 1352) | Survival (n = 2585) | P value |
| Gender | |||
| Female | 578 (42.8%) | 1147 (44.4%) | .344 |
| Male | 774 (57.2%) | 1438 (55.6%) | .344 |
| Age (y) (mean, SD) | 68.9 ± 14.9 | 65.5 ± 16.7 | <.01 |
| Ethnicity | |||
| Caucasian | 950 (70.3%) | 1894 (73.3%) | .047 |
| Hispanic | 37 (2.7%) | 90 (3.5%) | .218 |
| African American | 109 (8.1%) | 246 (9.5%) | .143 |
| Other | 256 (18.9%) | 355 (13.7%) | <.01 |
| ICU days (mean, SD) | 17.4 ± 18.1 | 18.2 ± 16.5 | .176 |
Death = death of septic patients during hospitalization.
The results of the 5 machine learning methods found in 5-fold cross-validation are shown in Tables 2 and 3. It included the area under the receiver operating characteristic curve (AUROC), precision, recall, accuracy, and F1 score. In our study, accuracy was defined by dividing the number of correctly predicted observations by the total number of observations. Precision is calculated by dividing the number of correctly predicted positive observations by the number of predicted positive observations. Recall is the proportion of correctly predicted positive observations to all observations in the actual class. The F1 score is a weighted average of the accuracy and recall, representing the balance between these 2 values.[32]
Table 2.
Comparison of performance of the 5 models.
| LR | KNN | SVM | RF | GBDT | |
| AUC | 0.876 | 0.877 | 0.898 | 0.980 | 0.992 |
| Precision | 0.723 | 0.806 | 0.828 | 0.931 | 0.948 |
| Recall | 0.776 | 0.624 | 0.749 | 0.885 | 0.917 |
| Accuracy | 0.821 | 0.819 | 0.860 | 0.938 | 0.954 |
| F1 score | 0.715 | 0.702 | 0.780 | 0.907 | 0.933 |
Table 3.
Comparison of AUROC and F1 among the different models.
| AUROC | F1 | P(AUROC) | P(F1) | |
| GBDT | 0.992 (0.989–0.994) | 0.933 (0.929–0.938) | <0.01 vs LR<0.01 vs KNN<0.01 vs RF<0.01 vs SVM | <0.01 vs LR<0.01 vs KNN<0.01 vs RF<0.01 vs SVM |
| LR | 0.876 (0.864–0.885) | 0.715 (0.704–0.723) | 0.774 vs KNN<0.01 vs RF0.012 vs SVM | 0.354 vs KNN<0.01 vs RF<0.01 vs SVM |
| KNN | 0.877 (0.871–0.885) | 0.702 (0.665–0.730) | <0.01 vs RF0.010 vs SVM | <0.01 vs RF<0.01 vs SVM |
| RF | 0.980 (0.978–0.984) | 0.907 (0.896–0.930) | <0.01 vs SVM | <0.01 vs. SVM |
| SVM | 0.898 (0.880–0.914) | 0.780 (0.771–0.801) |
The AUROC ranged from 0.876 to 0.992 for the 5 predictive models. GBDT showed the largest AUROC (0.992), highest precision (94.8%), recall (91.7%), accuracy (95.4%), and F1 score (0.933). LR showed the lowest AUC (0.876), precision (0.723), and recall (0.776). The receiver operating characteristic curves of these predictive models are shown in Figure 2. GBDT ranks the individual variables based on their relative influence, and the top 10 variables are presented in Figure 3.
Figure 2.
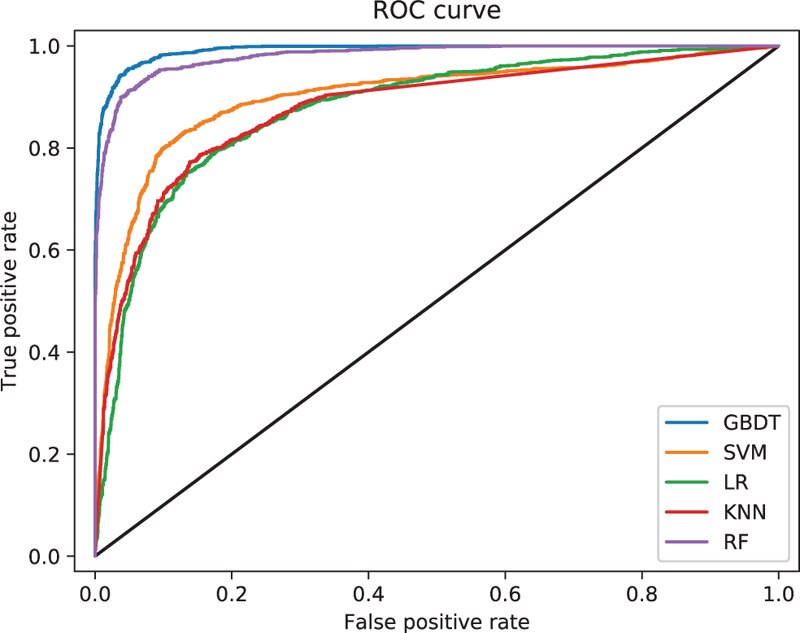
Comparison of the ROC curve of the 5 models. ROC = receiver operating characteristic curve.
Figure 3.
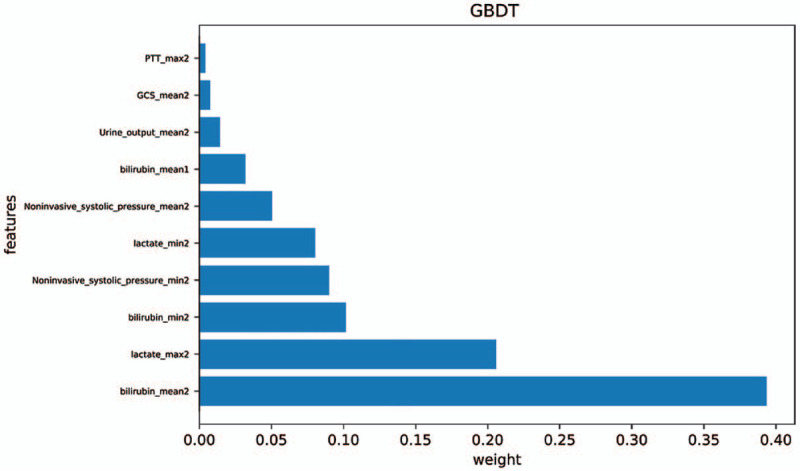
Top-10 variable importance of GBDT. GBDT = gradient boosting decision tree, GCS = Glasgow coma scale, max2 = parameter maximum in 48 hours of admission, Mean1 = average of parameters within 24 hours of admission, mean2 = average of parameters within 48 hours of admission, min2 = parameter minimum in 48 hours of admission, PTT = partial thromboplastin time.
Figure 2 shows the comparison of AUROC for predicting death in patients with sepsis according to the 5 predicted models. The AUROC and F1 score of GBDT were higher than those of the other models. The AUROC of the RF, SVM, and KNN showed better performance than LR but worse than GBDT. GBDT was significantly different from the other models (P < .01).
4. Discussion
In this study, there was no significant difference between the sexes of patients with sepsis who died and those who survived (P = .344). There was a significant difference in age between the death and survival groups (P < .01). There was no significant difference in the number of days in the hospital between patients with sepsis who died and those who survived (P = .176).
The results showed that GBDT had the largest AUROC (0.992) and highest precision (0.948), recall (0.917), accuracy (0.954), and F1 score (0.933) for predicting death in patients with sepsis. The results were better than those of the other models. This is because GBDT is based on the tree model and inherits the advantages of the tree model: it is robust to outliers and has little noise interference; its uncorrelated features have low interference and can deal with missing values well. A tree model is a decision support tool that uses a tree-like diagram or model to represent a decision and its possible consequences, including chance event outcomes, costs of resources, and utility.[33]
This study has some limitations. First, the study was performed at a single institution; the performance of machine learning techniques might be different when applied to a sample of different institutions with a different distribution of covariates. This study mainly involved Caucasians (72.2%), African Americans (9.0%), and Hispanics (3.2%). The results of this study need to be further verified in other ethnic groups due to ethnic differences.
Although the Sequential Organ Failure Assessment (SOFA) criterion is the latest definition of sepsis, the use of SOFA as a criterion for sepsis may lead to some bias due to missing data in the MIMIC III database. If the event death occurs during the assessment period, data from some patients, many of whom have high scores, will be missing, leading to survival bias.[34] SOFA criteria may lead to delayed diagnosis and intervention in cases of severe infection.[35] Some authors have reported that the use of SOFA criteria requires further exploration.[1] The sepsis standard (ICD-9) used in this study is an imperfect characterization of sepsis. Nevertheless, we believe it is useful in developing sepsis prediction tools, as evidenced by the improvements in sepsis-related clinical outcomes using a sepsis prediction algorithm trained on the same standard.[6]
5. Conclusions
In this study, researchers established and evaluated a GBDT prediction model for death in patients with sepsis in the ICU. The GBDT model showed better performance than other machine learning models in predicting death in patients with sepsis in the ICU. Among these models, the GBDT model showed the best performance with the highest AUROC and F1 scores. The evaluation results demonstrated that GBDT is an effective algorithm that offers the best predictive performance for predicting death in patients with sepsis. In future studies, we intend to verify the performance of the GBDT model in hospitals with different demographic and clinical characteristics, as well as in nonintensive care units. It can also be used to develop a clinical decision support system.
Author contributions
Jialin Liu and Ke Li conceived the study. Qinwen Shi, Siru Liu, Jialin Liu, Ke Li, and Yilin Xie performed the analysis, interpreted the results, and drafted the manuscript. All authors have revised the manuscript accordingly. All authors read and approved the final manuscript.
Conceptualization: Jialin Liu, Ke Li.
Data curation: Ke Li, Jialin Liu, Qinwen Shi, Siru Liu, Yilin Xie.
Formal analysis: Jialin Liu, Ke Li, Qinwen Shi.
Funding acquisition: Ke Li, Jialin Liu.
Investigation: Ke Li, Jialin Liu, Qinwen Shi, Siru Liu, Yilin Xie.
Methodology: Jialin Liu, Ke Li, Qinwen Shi, Siru Liu.
Project administration: Jialin Liu.
Resources: Jialin Liu, Ke Li.
Supervision: Jialin Liu, Ke Li.
Validation: Jialin Liu.
Writing – original draft: Jialin Liu, Ke Li, Qinwen Shi, Siru Liu, Yilin Xie.
Writing – review & editing: Jialin Liu, Ke Li, Qinwen Shi, Siru Liu, Yilin Xie.
Footnotes
Abbreviations: AUROC = area under the receiver operating characteristic curve, GBDT = gradient boosting decision tree, ICD-9-CM = International Classification of Diseases-9th Revision, Clinical Modification, ICU = intensive care unit, IRB = institutional review board, KNN = k-nearest neighbor, LR = logistic regression, MIMIC-III = medical information mart for intensive care III, RF = random forest, SOFA = sequential organ failure assessment, SVM = support vector machine.
How to cite this article: Li K, Shi Q, Liu S, Xie Y, Liu J. Predicting in-hospital mortality in ICU patients with sepsis using gradient boosting decision tree. Medicine. 2021;100:19(e25813).
The data that support the findings of this study are available from the Medical Information Mart for Intensive Care (MIMIC-III), but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. However, data are available from the authors upon reasonable request and with permission from MIMIC-III.
The datasets generated during and/or analyzed during the current study are publicly available.
This work was supported by a special project of the central government guiding local science and technology development under Grant No. 2020ZYD001, Sichuan Science and Technology Program under Grant Nos. 2020YFS0162 and 2019JDPT0008.
The authors have no conflicts of interest to disclose.
References
- [1].Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet 2020;395:200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med 2016;193:259–72. [DOI] [PubMed] [Google Scholar]
- [4].Novosad SA. Vital signs: epidemiology of sepsis: prevalence of health care factors and opportunities for prevention. MMWR Morb Mortal Wkly Rep 2016;65:864–9. [DOI] [PubMed] [Google Scholar]
- [5].Grudzinska FS, Aldridge K, Hughes S, et al. Early identification of severe community-acquired pneumonia: a retrospective observational study. BMJ Open Respir Res 2019;6:e000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Barton C, Chettipally U, Zhou Y, et al. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput Biol Med 2019;109:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Calvert J, Mao Q, Hoffman JL, et al. Using electronic health record collected clinical variables to predict medical intensive care unit mortality. Ann Med Surg 2016;11:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Raita Y, Goto T, Faridi MK, et al. Emergency department triage prediction of clinical outcomes using machine learning models. Crit Care 2019;23:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hu, YF, Lee VCS, Tan K. Prediction of Clinicians’ Treatment in Preterm Infants with Suspected Late-onset Sepsis-An ML Approach. 13th IEEE Conference on Industrial Electronics and Applications (ICIEA); May 31–Jun 02, 2018; Wuhan, China. [Google Scholar]
- [10].Khojandi A, Tansakul V, Li X, et al. Prediction of sepsis and in-hospital mortality using electronic health records. Methods Inf Med 2018;57:185–93. [DOI] [PubMed] [Google Scholar]
- [11].García-Gallo JE, Fonseca-Ruiz NJ, Celi LA, et al. A machine learning-based model for 1-year mortality prediction in patients admitted to an Intensive Care Unit with a diagnosis of sepsis. Med Intensiva 2020;44:160–70. [DOI] [PubMed] [Google Scholar]
- [12].Gultepe E, Green JP, Nguyen H, et al. From vital signs to clinical outcomes for patients with sepsis: a machine learning basis for a clinical decision support system. J Am Med Inform Assoc 2014;21:315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad Emerg Med 2016;23:269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Calvert JS, Price DA, Chettipally UK, et al. A computational approach to early sepsis detection. Comput Biol Med 2016;74:69–73. [DOI] [PubMed] [Google Scholar]
- [15].Calvert J, Desautels T, Chettipally U, et al. High-performance detection and early prediction of septic shock for alcohol-use disorder patients. Ann Med Surg (Lond) 2016;8:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform 2016;4:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016;3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thomas BS, Jafarzadeh SR, Warren DK, et al. Temporal trends in the systemic inflammatory response syndrome, sepsis, and medical coding of sepsis. BMC Anesthesiol 2015;15:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin K, Hu Y, Kong G. Predicting in-hospital mortality of patients with acute kidney injury in the ICU using random forest model. Int J Med Inform 2019;125:55–61. [DOI] [PubMed] [Google Scholar]
- [20].Chen T, Zhu L, Niu R, et al. Mapping landslide susceptibility at the Three Gorges Reservoir, China, using gradient boosting decision tree, random forest and information value models. J Mt Sci 2020;17:670–85. [Google Scholar]
- [21].Rao H, Shi X, Rodrigue AK, et al. Feature selection based on artificial bee colony and gradient boosting decision tree. Appl Soft Comput J 2019;74:634–42. [Google Scholar]
- [22].Zhang Y, Zhang R, Ma Q, et al. A feature selection and multi-model fusion-based approach of predicting air quality. ISA Transactions 2020;100:210–20. [DOI] [PubMed] [Google Scholar]
- [23].Bisong E. Logistic Regression. In: Building Machine Learning and Deep Learning Models on Google Cloud Platform. Berkeley, CA: Apress, 2019. [Google Scholar]
- [24].Zhang Z. Model building strategy for logistic regression: purposeful selection. Ann Transl Med 2016;4:111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cheng TH, Sie YD, Hsu KH, et al. Shock index: a simple and effective clinical adjunct in predicting 60-day mortality in advanced cancer patients at the emergency department. Int J Environ Res Public Health 2020;17:4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ekmekcigil E, Ünalp Ö, Uğuz A, et al. Management of iatrogenic bile duct injuries: multiple logistic regression analysis of predictive factors affecting morbidity and mortality. Turk J Surg 2018;34:264–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Havaei M, Jodoin PM, Larochelle AH. Efficient interactive brain tumor segmentation as within-brain kNN classification. International Conference on Pattern Recognition 2014;556–61. [Google Scholar]
- [28].Shichao Z, Xuelong L, Ming Z, et al. Learning k for kNN Classification. ACM Trans Intell Syst Technol 2017;8:43. [Google Scholar]
- [29].Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273–97. [Google Scholar]
- [30].Raghavendra NS, Deka PC. Support vector machine applications in the field of hydrology: a review. Appl Soft Comput 2014;19:372–86. [Google Scholar]
- [31].Kulkarni V, Sinha P, Petare M. Weighted hybrid decision tree model for random forest classifier. J Instit Engineers Series B 2016;97:209–17. [Google Scholar]
- [32].Powers DMW. Evaluation: from precision, recall and f-measure to roc, informedness, markedness & correlation. J Mach Learning Technol 2011;2:37–63. [Google Scholar]
- [33].Das S, Dahiya S, Bharadwaj A. An online software for decision tree classification and visualization using c4.5 algorithm (ODTC). 8th International Conference on Computing for Sustainable Global Development (INDIACom) 2014;1:962–5. [Google Scholar]
- [34].Lambden S, Laterre PF, Levy MM, et al. The SOFA score-development, utility and challenges of accurate assessment in clinical trials. Crit Care 2019;23:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Simpson SQ. New Sepsis criteria: a change we should not make. Chest 2016;149:1117–8. [DOI] [PubMed] [Google Scholar]


