Abstract
Indole-3-butyric acid (IBA) is an endogenous storage auxin important for maintaining appropriate indole-3-acetic acid (IAA) levels, thereby influencingprimary root elongation and lateral root development. IBA is metabolized into free IAA in peroxisomes in a multistep process similar to fatty acid β-oxidation. We identified LONG CHAIN ACYL-COA SYNTHETASE 4 (LACS4) in a screen for enhanced IBA resistance in primary root elongation in Arabidopsis thaliana. LACSs activate substrates by catalyzing the addition of CoA, the necessary first step for fatty acids to participate in β-oxidation or other metabolic pathways. Here, we describe the novel role of LACS4 in hormone metabolism and postulate that LACS4 catalyzes the addition of CoA onto IBA, the first step in its β-oxidation. lacs4 is resistant to the effects of IBA in primary root elongation and dark-grown hypocotyl elongation, and has reduced lateral root density. lacs6 also is resistant to IBA, although both lacs4 and lacs6 remain sensitive to IAA in primary root elongation, demonstrating that auxin responses are intact. LACS4 has in vitro enzymatic activity on IBA, but not IAA or IAA conjugates, and disruption of LACS4 activity reduces the amount of IBA-derived IAA in planta. We conclude that, in addition to activity on fatty acids, LACS4 and LACS6 also catalyze the addition of CoA onto IBA, the first step in IBA metabolism and a necessary step in generating IBA-derived IAA.
An enhancer mutant revealed an acyl-CoA synthetase that catalyzes CoA addition to indole-3-butryic acid, required for the β-oxidation steps necessary to generate indole-3-butryic acid-derived IAA.
Introduction
Auxin controls plant cell elongation, division, and differentiation and is, therefore, crucial for development throughout the life span of a plant. Because of its diverse and extensive effects, proper levels of indole-3-acetic acid (IAA), the primary signaling auxin, are tightly controlled through synthesis, degradation, transport, and sequestration (Korasick et al., 2013). Auxin can be sequestered in inactive forms by conjugation to amino acids or side chain elongation to form indole-3-butyric acid (IBA). IAA conjugates are converted back to IAA with a single hydrolysis reaction (Korasick et al., 2013). IBA is structurally similar to IAA but the side chain is two carbons longer; IBA is metabolized to IAA in the peroxisome in a multistep process (Hu et al., 2012). These unique input pathways vary between individual cells, organs, and developmental stages but all function to ensure proper levels of this crucial hormone.
IBA-derived auxin influences cotyledon expansion, apical hook formation, and several aspects of root architecture, including primary root elongation, lateral root development, root hair elongation, and adventitious root development (Korasick et al., 2013; Frick and Strader, 2018). Evidence suggests that IBA to IAA metabolism in the root tip is the primary source of auxin to initiate lateral root development (De Rybel et al., 2012; Xuan et al., 2015) and reinforces the auxin gradient to elongate lateral roots (Strader and Bartel, 2011). Recent evidence suggests IBA β-oxidation is required for root hair elongation under low-phosphate conditions, connecting IBA effects to changing environmental conditions (Trujillo-Hernandez et al., 2020). Such developmental roles are hypothesized to be due to the actions of IBA-derived IAA instead of direct effects of IBA. Analysis of IBA-response (ibr) mutants suggests IBA is β-oxidized into IAA in a stepwise process paralleling fatty acid β-oxidation, the process in which two carbons are removed from fatty acids (Hu et al., 2012).
In plants, the peroxisome is the sole location of fatty acid β-oxidation. Peroxisomes house enzymes for three crucial β-oxidation pathways: fatty acid β-oxidation, jasmonic acid (JA) synthesis, and IBA conversion to IAA (Hu et al., 2012; Kao et al., 2018). Peroxisomes must actively import necessary enzymes and substrates required for these pathways. Substrates for β-oxidation are imported into peroxisomes through PEROXISOMAL ABC TRANSPORTER PXA1/PED3/CTS (Zolman et al., 2001; Hayashi et al., 2002). These molecules are broken down into two carbons per cycle.
Following fatty acid import, fatty acid β-oxidation begins with CoA addition by acyl-CoA synthetases LACS6 and LACS7 (Fulda, 2004). CoA addition is necessary to activate the fatty acid for recognition before metabolism proceeds. Fatty acyl-CoA esters are oxidized by acyl-CoA oxidases, hydrated and then dehydrogenated by the multifunctional proteins ABNORMAL INFLORESCENCE MERISTEM 1 (AIM1) and MULTIFUNCTIONAL PROTEIN 2 (MFP2), and cleaved by the 3-ketoacyl-CoA thiolases KAT2/PED1 and KAT5 to release acetyl-CoA and a shortened fatty acid (Graham, 2008; Hu et al., 2012). This process can be repeated as needed. For instance, very long-chain fatty acids, as found in seed storage lipids, enter the pathway multiple times to release many molecules of acetyl CoA. Acetyl-CoA can then enter the citric acid cycle to ultimately be oxidized for energy production (Hu et al., 2012). Peroxisomes are, therefore, crucial for early development and seedling establishment because plants rely on the energy produced by the breakdown of fatty acids stored as triacylglycerol before they are photosynthetically active (Graham, 2008). Plants with severe defects in fatty acid β-oxidation are compromised in their ability to generate the energy necessary for germination and growth for early seedling establishment. Defects in fatty acid β-oxidation can be exposed by examining growth on media without sucrose, which requires metabolism of seed storage lipids. Addition of sucrose to the media bypasses this requirement, allowing development to proceed. Mutants that require exogenous sucrose for normal growth are designated as sucrose dependent.
In a similar process, JA is the product of three rounds of β-oxidation of 12-oxo-phytodienoic acid, by the actions of ACX1, ACX5, AIM1, and PED1/KAT2 (Hu et al., 2012; Wasternack and Hause, 2013). The role of peroxisomes in JA synthesis makes them important for JA effects including fertility and response to abiotic and biotic stressors (Wasternack and Hause, 2013).
Peroxisomes also contribute to the pool of free IAA by β-oxidizing IBA. IBA is hypothesized to undergo one round of β-oxidation to remove the two-carbon elongation, producing IAA. Forward genetic screens have revealed proteins that are acting in IBA metabolism. ibr3, ibr10, and ibr1 were identified in Arabidopsis thaliana as resistant to the inhibitory effects of IBA on root elongation. The predicted protein functions of these peroxisomal enzymes suggest that the process of IBA β-oxidation is mechanistically related to fatty acid metabolism, with the action of IBR3, a predicted oxidase, IBR10, a predicted hydratase, and IBR1, a predicted dehydrogenase, to release IAA (Zolman et al., 2007, 2008). IBR3, IBR10, and IBR1 appear specific to IBA metabolism as mutants do not have phenotypes associated with defects in fatty acid β-oxidation, JA synthesis, or peroxisomal import (Zolman et al., 2007, 2008). Mutations to known β-oxidation enzymes, including aim1 and ped1, have IBA-response phenotypes as well (Hayashi et al., 1998; Zolman et al., 2000); these phenotypes could indicate redundant enzymatic activity, with multiple enzymes acting on IBA at each step, or enzyme activity on multiple substrates affecting both pathways. Alternatively, IBA-resistant responses in the mutants acx1, acx3, chy1, and ech2 suggest that indirect mechanisms, such as CoA limitations (Adham et al., 2005) or accumulation of toxic intermediates (Zolman, Monroe-Augustus, et al., 2001; Li et al., 2019), also influence peroxisomal metabolic pathways.
IBA metabolism requires that the peroxisome must actively import functional proteins, substrates, and cofactors necessary for the contained metabolic processes (Hu et al., 2012; Kao et al., 2018). Enzymes destined for the peroxisome matrix typically contain one of two peroxisome targeting signals (PTSs): PTS1, a C-terminal tripeptide with the consensus amino acids [S][RK][LM], or PTS2, a R[LI]X5HL near the N-terminus (Reumann, 2004; Lanyon-Hogg et al., 2010). IBA-resistant phenotypes can be caused by mutations specific to IBA metabolism, such as defects in IBA-specific enzymes or by disrupting function of the organelle as a whole. For instance, mutations in PEX5 and PEX7, receptors required for import of enzymes into the peroxisome matrix (Zolman et al., 2000; Woodward and Bartel, 2005; Khan and Zolman, 2010) and the PXA1/CTS/PED3 (Zolman et al., 2001) peroxisomal substrate transporter each disrupt IBA responses.
Although many components specific to IBA metabolism and peroxisome function have been identified, our understanding is incomplete. For instance, the CoA synthetase initiating IBA β-oxidation and a peroxisomal IAA efflux carrier have not been identified (Hu et al., 2012; Damodaran and Strader, 2019). We sought to identify additional components of IBA metabolism by continuing the forward genetic screen for IBA-resistant plants. In this study, we describe our identification of long-chain CoA synthetase 4 (LACS4) from an enhancer screen using an ibr3 mutant background. The mutant phenotypes and enzymatic activity of LACS4 suggest that it functions as the initial step in IBA metabolism, charging IBA with CoA allowing entry into the β-oxidation process. This discovery completes the enzymatic components that metabolize IBA to IAA.
Results
Isolation of mutants with enhanced IBA resistance
We conducted a screen to identify Arabidopsis mutants with altered responses to IBA in root development. To increase our chances of finding novel components, an enhancer screen was performed. ibr3 is resistant to both the inhibition of IBA on root elongation and the stimulatory effect of IBA on lateral root initiation (Zolman et al., 2007). ibr3 was chosen as the background because it displays weaker IBA resistance than other ibr mutants (Zolman et al., 2008), facilitating the potential to capture a range of enhancement phenotypes.
ibr3-1 seeds were treated with ethylmethane sulfonate (EMS) to induce point mutations. Mutagenized populations were screened for seedlings exhibiting enhanced resistance to IBA in root length. This phenotype parallels the original screen (Zolman et al. 2000) and is facile relative to lateral root or root hair phenotypes. We hypothesized we could identify factors involved in the function or regulation of IBR3, the conversion of IBA to IAA, and/or general peroxisome function in Arabidopsis.
Z377 is defective in several IBA responses
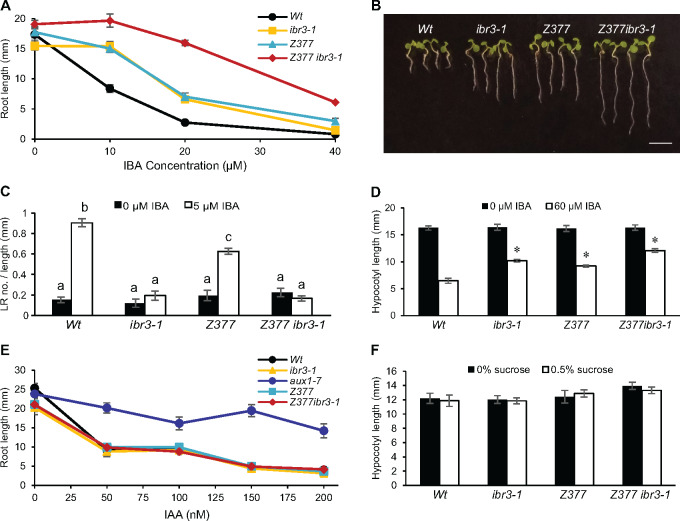
Exogenous IBA inhibits root elongation of wild-type (Wt) plants and ibr3-1 at higher concentrations. One enhancer mutant, Z377 ibr3-1, was selected for further characterization based on its increased resistance to IBA in root elongation (Figure 1A). Wild-type plants grown on 20 µM IBA have a primary root length <20% of that when grown without hormone supplementation (Figure 1, A and B). ibr3-1 is double that with a primary root length of 40% the root length without hormone. In stark contrast, Z377 ibr3-1 retains a root length of >80% on the same IBA concentration.
Figure 1.
Z377 has phenotypes specific to disruptions in IBA metabolism. (A) Primary root elongation of 7-d-old seedlings grown on increasing concentrations of IBA (± se, n ≥10). (B) Images of 7-d-old seedlings grown on 20 μM IBA. Scale bar = 5mm. (C) Lateral root density of 8-d-old seedlings was quantified by dividing the number of lateral roots by the primary root length. Statistical significance was determined by two-way ANOVA with post hoc Tukey HSD (± se, n ≥8, P < 0.001). Common letters indicate no significant difference. (D) Length of dark-grown hypocotyls. Seedlings were grown for 1 d in light and 5 d in darkness. Statistical significance was determined by two-tailed t test versus Wt on the same condition (± se, n ≥ 15, *P < 0.001). (E) Primary root length of 7-d-old seedlings grown on increasing concentrations of IAA (± se, n ≥ 8). (F) Hypocotyl length of seedlings grown on media with and without sucrose for 1 d in light and 5 d in darkness. Statistical significance determined by one-tailed t test between sucrose treatments (± se, n ≥ 13, P <0.05).
Following its confirmation as an ibr3 enhancer in root elongation, Z377 ibr3-1 was examined for defects in lateral root initiation. IBA is effective at inducing lateral rooting; a wild-type plant will develop branched root architecture when grown in the presence of IBA compared to an elongated, less branched root in the absence of hormone (Zolman et al., 2000). Decreased induction of secondary roots is a prominent phenotype in many IBA-resistant mutants, including ibr3-1 (Zolman et al., 2000, 2007). Lateral root development can be quantified by counting the number of lateral roots that develop per millimeter along the primary root. Z377 ibr3-1 is defective in lateral root initiation when stimulated with IBA, resembling wild-type grown in the absence of hormone (Figure 1C). This phenotype is similar to the ibr3-1 parent line.
IBA has prominent effects on root development but also affects shoot development (Strader et al., 2011; Frick and Strader, 2018). To determine if the IBA-response defects of Z377 ibr3-1 are specific to roots or affected throughout the plant, hypocotyl elongation was measured as an indicator. Wild-type plants have reduced hypocotyl length when grown on IBA than media without hormone (Figure 1D). The enhanced resistance of Z377 ibr3-1 to IBA extends to dark-grown hypocotyl elongation, indicating the IBA-response defects are not limited to specific tissues.
To understand the mutation leading to these enhanced responses, the Z377 single mutant was separated from ibr3-1 following a backcross to wild-type. F2 progeny were characterized at a genotypic and phenotypic level. IBA-resistant individuals were selected, then tested to determine their genotype at the IBR3 locus. A subset of lines was identified that showed elongated roots on IBA but a wild-type IBR3 genotype. These lines were hypothesized to represent the effects of the Z377 mutation alone. These mutants had IBA resistance in root elongation assays intermediate between wild-type seedlings and the Z377 ibr3-1 enhancer and were comparable to the single ibr3 mutation (Figure 1, A and B). Surprisingly, Z377 remained sensitive to IBA-induced lateral rooting (Figure 1C). Z377 develops a similar mean number of lateral roots to wild-type, with somewhat reduced lateral root density because of its longer primary root. This phenotype is distinct from ibr3, which has statistically similar lateral root density when grown with or without IBA (Figure 1C), and other ibr mutants (Zolman et al., 2008). Additional backcrosses revealed that the Z377 mutation segregates in a recessive manner.
Z377 phenotypes are specific to IBA
To further understand the range of Z377-related phenotypes, we tested other common responses related to IBA metabolism using the single mutant and enhancer line. First, Z377 and Z377 ibr3-1 were tested for responses to IAA to determine if these mutants had IBA-specific defects or a generalized resistance to auxin. Z377 and Z377 ibr3-1 remained sensitive to inhibition of primary root elongation by IAA (Figure 1E).
Enzymatic components of the IBA metabolic pathway discovered to date are contained within the peroxisome and several mutants with fatty acid β-oxidation defects also have IBA-resistant phenotypes (Hayashi et al., 1998; Zolman et al., 2000; Adham et al., 2005). To examine if Z377 is involved in fatty acid β-oxidation or peroxisomal functions, Z377 was tested for sucrose-dependent growth. Both Z377 and Z377 ibr3-1 have comparable elongation of dark-grown hypocotyls in media lacking or containing sucrose (Figure 1F).
These initial experiments suggest that the causative mutation in Z377 specifically disrupts IBA metabolism, but not fatty acid β-oxidation, and the mutants have normally functioning peroxisomes.
Z377 is defective in LACS4
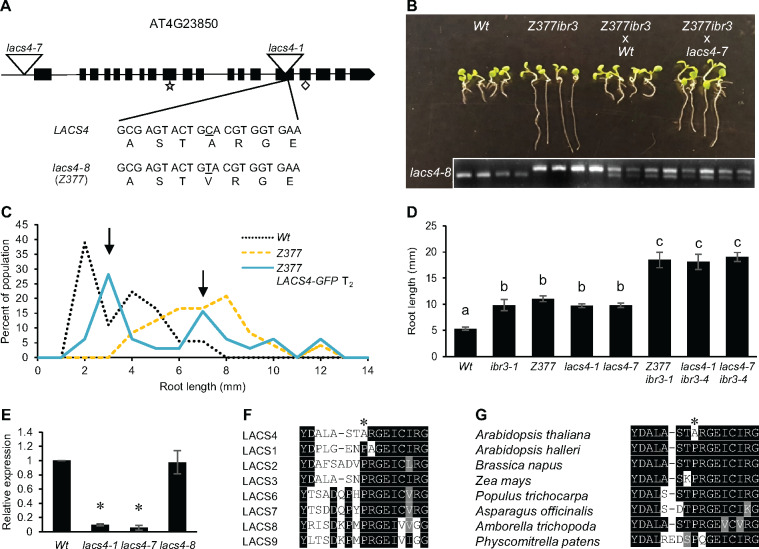
Whole-genome sequencing was used to identify the causative mutation in Z377. DNA extracted from 10 independently isolated Z377 ibr3-1 lines from a backcross to ibr3-1 was pooled for sequencing. Parallel experiments were done for wild-type and ibr3-1. Gene candidates were first narrowed by identifying homozygous single nucleotide polymorphisms consistent with EMS mutagenesis in Z377 ibr3-1. Mutations common between Z377 ibr3-1 and wild-type or Z377 ibr3-1 and ibr3-1 were eliminated, leaving 10 mutations unique to Z377 ibr3-1. Nine candidate genes were clustered on chromosome 4 (Supplementary Table S1). AT4G23850, which encodes LACS4, showed an alanine to valine substitution in the Z377 mutant (Figure 2A) and became the prime candidate because LACS6 and LACS7 are known peroxisomal enzymes responsible for priming fatty acids with CoA in fatty acid β-oxidation (Fulda, 2004). We hypothesized that LACS4 may be the CoA synthetase responsible for catalyzing the addition of CoA onto IBA as this enzyme has not yet been identified (Hu et al., 2012). Other candidate genes did not have a clear link to peroxisomes, auxin responses, IBA metabolism, or lipid metabolism, further supporting the hypothesis that the mutation in LACS4 might be the causative mutation in Z377.
Figure 2.
Z377 is defective in lacs4. (A) Schematic of LACS4 gene structure. Exons are represented as black boxes whereas introns are lines. Locations of T-DNA insertion mutants used in this study are depicted as inverted triangles. The mutation in Z377 changes nucleotide 1,400 from C to T, resulting in an alanine to valine substitution at residue 467. As a result, Z377 was renamed as lacs4-8. Predicted AMP and fatty acid binding motifs (UniProt Consortium, 2019) are indicated by a star and diamond, respectively. (B) Z377 ibr3-1 was crossed to Wt and lacs4-7 in a test of non-complementation. F1 seedlings were grown on 20 µM IBA for 7 d. Seedlings were genotyped for the point mutation in LACS4 in Z377 to confirm the cross was successful. The Wt LACS4 allele is the smaller product whereas the mutant lacs4 is the larger product. (C) Z377 seedlings segregating a Wt copy of LACS4 tagged with a C-terminal GFP were grown on 15 µM IBA for 7 d. Length of primary root was measured for each individual. Data are represented as percent of the population with indicated primary root length (n ≥ 18 Wt and Z377, n = 32 Z377 35S: LACS4-GFP). (D) Primary root elongation of 7-d-old seedlings grown on 20 µM IBA. Statistical significance was determined by one-way ANOVA with post hoc Tukey HSD test (±SE, n ≥ 11, P < 0.05). Common letters indicate no significant difference. (E) Expression of LACS4 relative to Wt in 5-d-old seedlings grown under white light on filter paper. LACS4 expression is normalized against UBQ10. Statistical significance was determined by a two-tailed t test (± se, n = 5, *P < 0.001). (F) and (G) T-Coffee alignment of LACS proteins of A. thaliana (F) and LACS4 orthologs in divergent plant species (G). Residues depicted are 460–475 of LACS4 from A. thaliana. The amino acid residue mutated in lacs4-8 is indicated with an asterisk. Black shading indicates the same amino acid. Gray shading indicates similar amino acids.
To test our findings in an unbiased manner, T-DNA mutants disrupting each candidate gene were acquired and screened for IBA resistance in primary root elongation (Supplementary Table S1). Only Salk_120357 showed an IBA-resistant response in root elongation assays like Z377 (Supplemental Figure S1A). Salk_120357 is inserted in the 5′–untranslated region of LACS4 (Figure 2A). Because several T-DNA alleles of lacs4 were recently named (Zhao et al., 2019), we named this previously uncharacterized Salk line lacs4-7. In addition to resistance in primary root elongation, lacs4-7 shows partial sensitivity to induction of lateral root formation, another phenotype consistent with Z377 (Supplementary Figure S1B).
To confirm causality, we completed non-complementation testing for Z377 ibr3-1. F1 seedlings of Z377 ibr3-1 backcrossed to wild-type were sensitive to IBA (Figure 2B), indicating that the mutation is recessive. We then crossed Z377 ibr3-1 with lacs4-7. F1 progeny of this cross had intermediate resistance to IBA in primary root elongation, similar to Z377 alone, supporting that the mutation in LACS4 is causative.
Separately, we generated a complementation line of the Z377 mutant transformed with the LACS4 coding sequence. Z377 lines segregating a 35S:LACS4 transgene containing a green fluorescent protein (GFP) tag were analyzed to observe if IBA responses were recovered by the presence of a wild-type copy of LACS4. Primary root length of segregating lines grown on 15 µM IBA was measured and scored for rescue relative to the IBA-resistant phenotype of Z377. The primary root IBA-resistant phenotype of Z377 is successfully complemented by a LACS4 construct, as indicated by the dual peaks representing the populations of rescued and nonrescued phenotypes within this segregating line (Figure 2C). The large peak with IBA-sensitive plants has root lengths less than the average root length of Z377, which suggests the addition of a wild-type LACS4 copy restores wild-type to IBA in primary root elongation. The smaller peak of the segregating population shows plants with elongated roots, with growth consistent to the Z377 mutant.
Finally, we demonstrated causation by generating novel allele combinations of lacs4 and ibr3 and looking for similar enhanced IBA-resistant responses. For this assay, we used lacs4-7 and also expanded our analysis to include lacs4-1, a T-DNA allele found in an exon (Figure 2A) that has been previously described (Jessen et al., 2011, 2015; Zhao et al., 2019). We crossed lacs4-1 and lacs4-7 separately with a T-DNA line disrupting IBR3 (Zolman et al., 2007). lacs4-1 ibr3-4 and lacs4-7 ibr3-4 have enhanced resistance to IBA in primary root elongation and indeed recapitulated the phenotype seen in the original Z377 ibr3-1 line (Figure 2D). Following these tests, we concluded that the mutation in LACS4 is causative for the IBA-response defects of Z377. The Z377 mutant is now named lacs4-8 (Figure 2A).
Missense mutation at residue 467 disrupts LACS4 function in IBA metabolism
The lacs4 mutation in Z377 is a substitution of an alanine to a valine at residue 467 (Figure 2A). Despite being what is traditionally defined as a weak mutation, lacs4-8 yields IBA-resistant phenotypes as strong as each T-DNA allele (Figure 2D). To determine if lacs4-8 disrupts function by altered expression or protein activity, expression of LACS4 was tested in all three mutants using gene-specific primers. We found that LACS4 is expressed in lacs4-8 at wild-type levels, whereas lacs4-1 and lacs4-7 have dramatically reduced expression (Figure 2E), as previously noted for lacs4-1 (Jessen et al., 2015). This result suggests the mutation in lacs4-8 results in translation of a nonfunctional LACS4 protein, which is as compromised in IBA metabolism as much as plants lacking LACS4 completely.
In A. thaliana LACS4, residue 467 encodes an alanine. Interestingly, alignment of the LACS4 protein sequence with the Arabidopsis LACS proteins revealed that residue 467 is a conserved proline in all other family members (Figure 2F). We expanded our inquiry to LACS4 orthologs in diverse species and discovered that this residue again is a completely conserved proline, even in the close relative Arabidopsis halleri (Figure 2G). This strong conservation suggests this alanine residue is specific for A. thaliana LACS4 function and may be a site sensitive to perturbations.
lacs4 and lacs6 are resistant to IBA
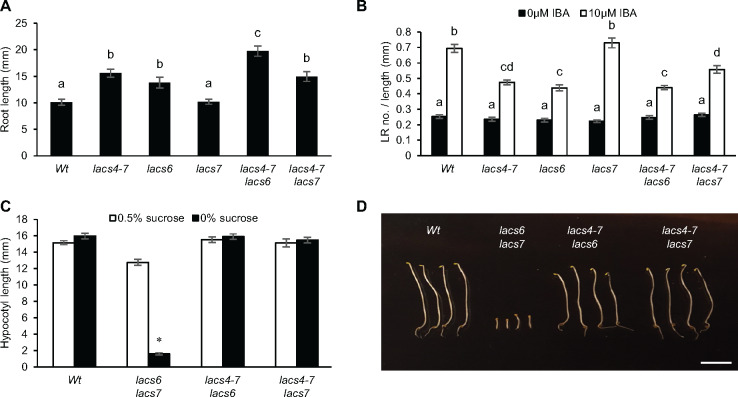
Nine LACS genes are encoded in the Arabidopsis genome (Shockey et al., 2002). As several LACS proteins have overlapping roles (Jessen et al., 2011, 2015; Zhao et al., 2019), we examined all lacs mutants for IBA resistance. Available T-DNA lines were screened for root elongation on IBA. We found lacs6 is the only other lacs mutant resistant to IBA in primary root elongation (Supplemental Figure S2). The finding that lacs6, but not lacs7, is resistant to IBA in primary root elongation (Figure 3A) was surprising as LACS6 and LACS7 are closely related, both localized to the peroxisome, and have overlapping activity in the first step of fatty acid β-oxidation (Shockey et al., 2002; Fulda, 2004). lacs6 is comparable to lacs4 in that it also has slightly reduced lateral root density when treated with IBA (Figure 3B).
Figure 3.
LACS4 and LACS6 have overlapping activity in IBA metabolism but not fatty acid β-oxidation. (A) Primary root elongation of 7-d-old seedlings on 10 µM IBA. Statistical significance was determined by one-way ANOVA with post hoc Tukey HSD test (± se, n ≥ 12, P < 0.05). Common letters indicate no significant difference. (B) Lateral root density of 8-d-old seedlings was quantified by dividing the number of lateral roots by the primary root length. Statistical significance determined by two-way ANOVA with post hoc Tukey HSD (± se, n ≥ 14, P < 0.05). Common letters indicate no significant difference. (C) Hypocotyl elongation of seedlings grown with and without sucrose for 1 d in light and 5 d in darkness. Statistical significance determined by two-tailed t test between sucrose treatments (± se, n ≥ 17, *P < 0.001). (D) Representative hypocotyls of seedlings grown for 1 d in light and 5 d in darkness without sucrose. Scale bar = 5mm.
To further explore the relationships between LACS4, LACS6, and LACS7, double mutants of lacs4 with lacs6 or lacs7 were generated. lacs4-7 lacs6, but not lacs4-7 lacs7, displayed enhanced resistance to IBA in primary root elongation when compared with single mutants (Figure 3A). This enhancement was not seen in the lateral root phenotype; lacs4-7 lacs6 lateral root density was not statistically different from the single mutants (Figure 3B). This evidence supports that lacs4 and lacs6 are not defective in lateral root initiation and that their IBA-defective responses are predominately seen in the primary root. The finding that lacs6 is resistant to IBA and enhances the IBA resistance in primary root elongation of lacs4 suggests that LACS4 and LACS6 may work in combination in IBA to IAA conversion. These experiments also demonstrate that LACS7 does not have a role in IBA to IAA conversion.
lacs4 is not sucrose dependent, even in combination with lacs6 or lacs7
Redundancy is common with β-oxidation enzymes (Hu et al., 2012). Single mutants may not have sucrose-dependent phenotypes even when a biochemical defect is present. For example, lacs6 and lacs7 appear wild-type but growth is severely compromised in a lacs6 lacs7 double mutant (Fulda, 2004). lacs4 did not display sucrose-dependent phenotypes (Figure 1F). The overlapping role of LACS4 and LACS6 in IBA responses led us to analyze if lacs4 has a sucrose-dependent phenotype in higher order mutants. lacs4-7 lacs6 and lacs4-7 lacs7 were evaluated for sucrose dependence. Neither double mutant was defective in hypocotyl elongation on media lacking sucrose, indicating lacs4 does not display the sucrose-dependent phenotype consistent with fatty acid β-oxidation defects (Figure 3, C and D). We conclude that fatty acid β-oxidation is not detectably compromised when LACS4 is mutated.
lacs4 and lacs6 responses are specific to auxins that are β-oxidized
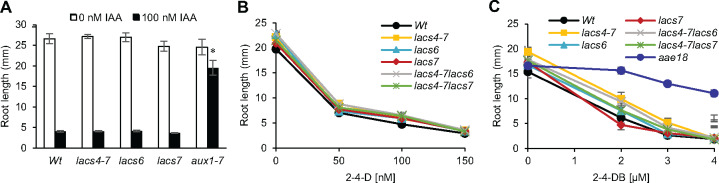
Our evidence demonstrates that lacs4 and lacs6 are resistant to IBA. We previously showed that lacs4 responses were specific to IBA; to explore if lacs6 responses are specific to IBA, primary root elongation was tested on IAA and found to be inhibited similarly to wild-type (Figure 4A). In addition, we tested these lacs mutants on the synthetic auxin 2,4-dichlorophenoxyacetic acid (2-4-D). lacs4, lacs6, lacs4-7 lacs6, and lacs4-7 lacs7 are sensitive to 2-4-D-based inhibition of primary root elongation to the same extent as wild-type (Figure 4B). This indicates that neither lacs4 nor lacs6 are disrupting generalized auxin responses.
Figure 4.
LACS4 and LACS6 are specific to auxins that must be β-oxidized. (A) Primary root elongation of 7-d-old seedlings grown with and without IAA. Statistical significance determined by two-tailed t test to Wt of the same treatment (± se, n ≥ 12, aux1-7 n = 8, *P < 0.001). (B) Primary root elongation of 7-d-old seedlings grown on increasing concentrations of 2-4-D (± se, n ≥ 16). (C) Primary root elongation of 7-d-old seedlings grown on increasing concentrations of 2-4-DB (± se, n ≥ 10).
lacs4 and lacs6 were also tested on (2,4-dichlorophenoxy)butyric acid (2-4-DB), a synthetic auxin that has a side chain that is two carbons longer than 2-4-D and may be β-oxidized in a similar mechanism to that of IBA (Wain and Wightman, 1954). We used the peroxisomal acyl-activating enzyme, aae18, as a control as it is specific for activating 2-4-DB for β-oxidation and remains sensitive to IBA (Wiszniewski et al., 2009). lacs4 is resistant to 2-4-DB only at low concentrations and not to the same extent as aae18 (Figure 4C). lacs6 remained sensitive to 2-4-DB (Figure 4C), consistent with previously published results (Fulda, 2004). Resistance to 2-4-DB was not enhanced in the lacs4-7 lacs6 double mutant, supporting that lacs6 is sensitive to this auxin. The differential effect of LACS4 and LACS6 on 2-4-DB may indicate that LACS4 preferentially participates in auxin β-oxidation compared to LACS6 or that LACS4 may be capable of accepting a broader range of substrates than LACS6.
LACS4 localization
All proteins identified in IBA to IAA conversion are in the peroxisome matrix or have roles in general peroxisome function (Hu et al., 2012). However, a previous study (Jessen et al., 2015) reported LACS4 localizes to the endoplasmic reticulum (ER). Based on the phenotypes described above and the potential connection of LACS4 with IBA activation, we hypothesized LACS4 also would associate with peroxisomes.
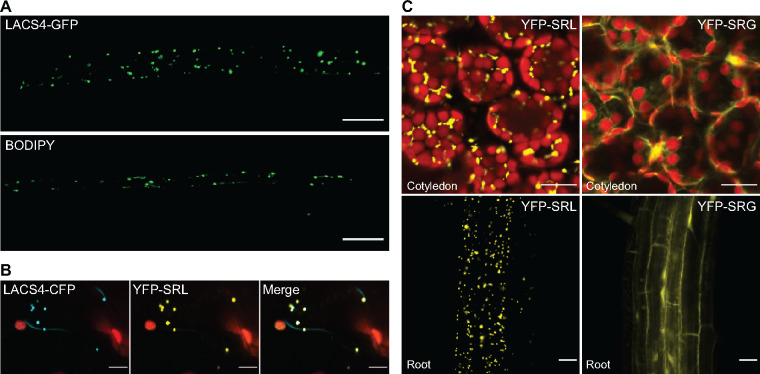
A full-length LACS4 cDNA was used to generate a C-terminal GFP fusion with a flexible linker between LACS4 and the fluorescent tag. This construct was transformed into wild-type and lacs4-8 plants. This construct rescued the IBA-resistant phenotype of lacs4-8, demonstrating that the construct is functional (Figure 2C). LACS4-GFP could be viewed as distinct puncta. These puncta looked comparable in size and number to peroxisomes stained with the peroxisome-specific dye BODIPY (Landrum et al., 2010) in wild-type seedlings (Figure 5A).
Figure 5.
LACS4 localizes to puncta reminiscent of peroxisomes. (A) Visualization of LACS4-GFP and BODIPY signals in peroxisomes of 7-d-old Arabidopsis. Scale bar = 20 µm. (B) Localization of transiently expressed LACS4-CFP and YFP-SRL in Nicotiana benthamiana. Scale bar = 10 µm. (C) Representative confocal images of YFP-SRL and YFP-SRG signals in cotyledons and roots of 7-d-old transgenic Arabidopsis. Scale bar = 20 µm.
To determine if these puncta were peroxisomes, Nicotiana benthamiana was co-infiltrated with Agrobacterium tumefaciens containing a 35S:LACS4-CFP, a cyan fluorescent protein attached to the LACS4 coding sequence with a flexible linker, and UBQ10:YFP-SRL, a yellow fluorescent protein ending with a canonical PTS1. Transient expression of LACS4-CFP could be seen as discrete puncta that colocalized with the peroxisome marker (Figure 5B). This result supports our hypothesis that LACS4 can associate with peroxisomes.
Proteins internalized into the peroxisome typically contain a PTS. LACS4 does not have a canonical PTS1 or PTS2. However, the PTS1 C-terminal tripeptide (typically [S] [RK] [LM]), can accept degeneracy at these residues (Chowdhary et al., 2012). The LACS4 C-terminal amino acids are SRG, one amino acid different from a canonical PTS1. To test if the C-terminal tripeptide of LACS4 functions as a noncanical PTS, a UBQ10:YFP construct was mutated to end either with SRG or SRL terminal amino acids. YFP-SRL localized as discrete puncta (Figure 5C), demonstrating peroxisome localization. In contrast, YFP-SRG was seen as a diffuse fluorescent signal (Figure 5C). This experiment demonstrates that if LACS4 is entering into the peroxisome, it is doing so by a different mechanism than a C-terminal tripeptide PTS or that additional upstream residues are required beyond the consensus amino acids. Alternatively, LACS4 may associate with the peroxisome without being internalized.
LACS4 directly acts on IBA
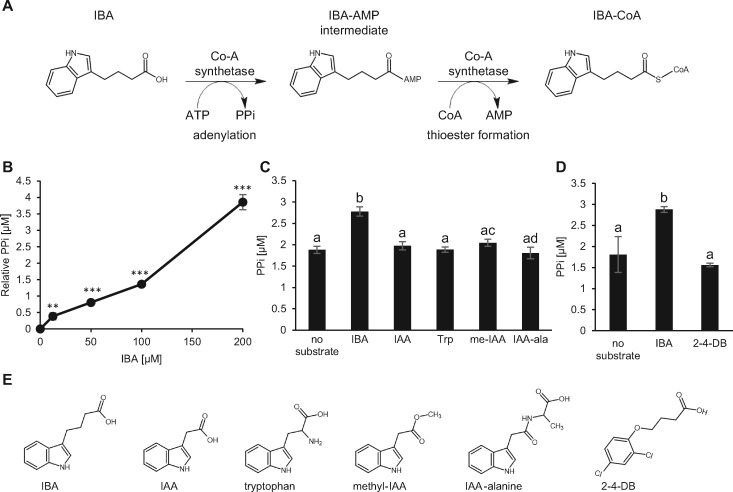
LACS4 CoA synthetase activity on fatty acids has been demonstrated (Shockey et al., 2002). The IBA-resistant phenotype of lacs4 suggests that LACS4 also is involved in IBA metabolism. To determine if the mechanism of IBA resistance of lacs4 is due to a wild-type role for CoA synthetase activity on IBA, in vitro enzyme activity assays were performed with recombinant LACS4. A CoA synthesis reaction consumes ATP to yield pyrophosphate (PPi) and AMP (Figure 6A). We examined enzyme activity by measuring the production of PPi with a fluorometric assay. After 10 min of incubation with IBA, LACS4 produced significantly more PPi than when incubated with no substrate, and PPi production increased with increasing concentrations of IBA (Figure 6, B and C).
Figure 6.
LACS4 has activity on IBA substrate in vitro. (A) CoA synthetase reaction on IBA depicting intermediates and byproducts. (B) Recombinant LACS4 was incubated with increasing concentrations of IBA dissolved in water for 10 min in vitro. Production of PPi was measured and represented relative to µM PPi detected without substrate. Statistical significance was determined by a two-tailed t test (±SE, n = 6, **P<0.01, ***P<0.001). (C) Recombinant LACS4 was incubated with 100 μM of IBA, IAA, tryptophan (Trp), methyl-IAA (me-IAA), and IAA-alanine (IAA-Ala) for 10 min in vitro before PPi was measured. Statistical significance was determined by one-way ANOVA with post hoc Tukey HSD test (± se, n = 6, P < 0.01). Common letters indicate no significant difference. (D) Recombinant LACS4 was incubated with 100 μM of IBA, 2-4-DB, or no substrate for 10 min in vitro before PPi was measured. Statistical significance was determined by one-way ANOVA with post hoc Tukey HSD test (± se, n = 6, P < 0.05). Common letters indicate no significant difference. (E) Chemical structures of IBA, IAA, tryptophan, methyl-IAA, IAA-alanine, and 2-4DB.
To evaluate substrate specificity, LACS4 was tested on IAA, IAA-alanine, methyl-IAA, and tryptophan, an auxin precursor. These substrates were chosen because each are endogenous, relevant to auxin metabolism, and structurally similar to IBA. Each possess an indole ring identical to that of IBA but have side chains of varying lengths and modifications (Figure 6E). Chain length is of interest because LACS4 has activity on fatty acids ranging in length from 14 to 18 carbons (Shockey et al., 2002). Methyl-IAA was included because it has a side chain as the same length of IBA, but does not have a terminal carboxyl group to participate in adenylation. Despite significant LACS4 activity on IBA, activity was not detected for IAA, IAA-alanine, methyl-IAA, or tryptophan (Figure 6C). This result demonstrates that LACS4 has specific activity on IBA, but not other structurally similar auxins or carboxylic acids. This biochemical data are consistent with lacs4 being resistant specifically to the effects of IBA.
We expanded our in vitro analysis of LACS4 substrates to include 2-4-DB. 2-4-DB is a synthetic auxin and not endogenous to plants. lacs4 is weakly resistant to 2-4-DB at low levels. However, LACS4 did not have activity on 2-4-DB (Figure 6D), suggesting that it is not a bona fide substrate.
Evidence that LACS4 has increasing activity with increasing IBA substrate and exhibits specificity for IBA over related auxins strongly supports that LACS4 directly acts on IBA and can charge IBA with the necessary CoA before entering the β-oxidation pathway.
lacs4 has reduced auxin signaling in root tip
IBA conversion to IAA is an important source of auxin for root development (Strader and Bartel, 2011; De Rybel et al., 2012; Xuan et al., 2015) and contributes to establishing and maintaining the total auxin pool (Spiess et al., 2014). To examine if LACS4 can influence overall auxin signaling, lacs4-8 and lacs4-8 ibr3-1 were crossed to DR5:GUS, a reporter line that contains the synthetic auxin-responsive reporter DR5 driving expression of the β-glucoronidase (GUS) enzyme (Ulmasov et al., 1997). Seedlings were evaluated for relative strength of the reporter in root caps, the hypothesized location of IBA conversion into IAA (De Rybel et al., 2012; Xuan et al., 2015). In media without hormone, staining in wild-type, ibr3, and lacs4-8 was comparable. DR5:GUS induction was slightly reduced in lacs4-8 ibr3-1. We hypothesize this to be due to reduced metabolism of endogenous IBA into IAA in the root cap. Upon treatment with IBA, wild-type plants showed increased and expanded induction of the DR5:GUS construct. However, lacs4-8 and lacs4-8 ibr3-1 were not induced to the same levels as wild-type (Figure 7), suggestive of reduced auxin signaling (Ulmasov et al., 1997). Staining was similar in all genotypes incubated with IAA (Figure 7), demonstrating that the seedlings still retain the ability to sense and respond to IAA; only the IAA generated from the breakdown of IBA is compromised. The reduced GUS induction in lacs4 supports a hypothesis that LACS4 is functionally contributing to the total auxin pool available in the root cap and is doing so through the mechanism of IBA to IAA conversion.
Figure 7.
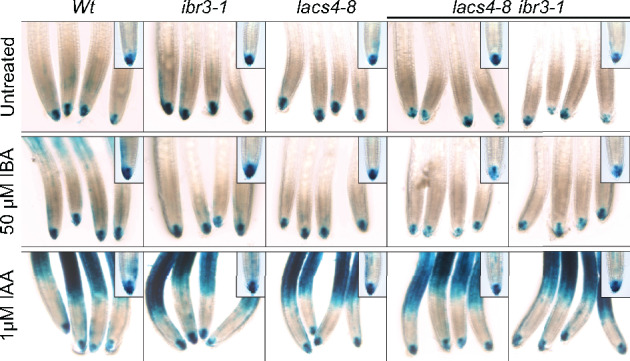
lacs4 and lacs4 ibr3 display reduced IBA-responsive DR5 activation. Staining of DR5:GUS in root tips of 5-d-old seedlings. Plants were incubated in liquid PN with or without hormone gently rocking for 2 h. Plants were stained for GUS for 3 h at 37°C and washed with 50, 75, and 95% EtOH before imaging. Groups were imaged with a 20× objective, inset is 40× objective.
Discussion
IBA metabolism to IAA parallels fatty acid β-oxidation; enzymes with similar activities likely will be required for the analogous steps. It is hypothesized that IBA is charged with CoA by a CoA synthetase before entering the pathway. Our identification of LACS4 in an unbiased screen for IBA resistance was intriguing, as a CoA synthetase functioning on IBA had not yet been reported. Separation of the new allele from the previously characterized ibr3 IBA-response mutant background (Zolman et al., 2000; Zolman et al., 2007) revealed that lacs4 has a notable IBA-resistance profile on its own. Mutations in LACS4 make Arabidopsis resistant to the inhibition of IBA in primary root elongation and dark-grown hypocotyl elongation (Figure 1, A and D), consistent with previously characterized IBA-response mutants (Zolman et al., 2000; Zolman et al., 2007, 2008; Strader et al., 2011).
LACS4 CoA synthetase activity is specific to IBA in vitro. LACS4 was not active against other auxin substrates and lacs4 remains sensitive to IAA in primary root elongation (Figures 4, 6). Our data support a model in which LACS4 catalyzes CoA addition onto IBA, the first step in IBA to IAA conversion, and is required for normal contribution of IBA-derived IAA in the root.
LACS4 function
LACSs in all organisms, from prokaryotes to humans, primarily activate fatty acids with CoA making LACS enzymes critical for utilization of fatty acids (Groot et al., 1976; Watkins, 1997). The Arabidopsis genome contains nine LACS genes with some overlapping activity, but which vary in substrate specificities, localization, and expression (Shockey et al., 2002; Zhao et al., 2019). Teasing apart the role of LACSs has been difficult as most single mutants have no obvious lipid phenotypes. Higher-order mutants have revealed specific roles of some Arabidopsis LACS proteins.
LACS4 has overlapping activity with LACS1, LACS2, LACS8, and LACS9 with roles in a diverse array of lipid biosynthesis. lacs4 appears wild-type in vegetative growth, fertility, and seed oil content (Jessen et al., 2011, 2015; Zhao et al., 2019), but phenotypes began to emerge upon combining lacs4 with other mutants. LACS4 and LACS9 act in glycerolipid synthesis and work together to transfer lipids from the ER to the plastid. lacs4 lacs9 had reduced stature, reduced seed weight, altered seed oil content, and impaired seed development (Jessen et al., 2015; Zhao et al., 2019). LACS8 also may have overlapping activity as a lacs4 lacs8 lacs9 triple mutant is embryo lethal (Jessen et al., 2015; Zhao et al., 2019). LACS4 and LACS1 together are required for synthesis of pollen coat lipids, as lacs1 lacs4 shows male sterility (Jessen et al., 2011). lacs1 lacs2 lacs4 has altered seed oil content and dramatically reduced cuticle waxes (Zhao et al., 2019). These studies link LACS4 activity to vegetative growth, seed oil accumulation, and fertility, highlighting that LACS4 has a wide impact on growth and fecundity.
Our results implicating LACS4 activity in IBA metabolism expands the roles for LACS4 to span lipid and hormone metabolism. Our work reveals LACS4 also influences early seedling development, particularly root systems, through IBA to IAA conversion and contributes to the overall pool of free IAA, as indicated by reduced induction of DR5:GUS in lacs4-8 ibr3-1 in the root cap (Figure 7).
Mirroring the overlapping roles of LACS family enzymes described above, our study also implicated LACS6 as another CoA synthetase that can act on IBA. LACS6 is localized in the peroxisome matrix (Fulda, 2004; Jessen et al., 2015), consistent with our initial hypothesis that the acyl-CoA synthetase acting on IBA would also be localized where all other IBA metabolizing enzymes are located (Hu et al., 2012). lacs6 is resistant to IBA in primary root elongation (Figure 3) at levels comparable to lacs4, supporting the hypothesis that LACS6 acts directly on IBA. Alternatively, lacs6 could indirectly disrupt IBA metabolism. Disruption to LACS6 could be indirectly influencing IBA metabolism via a mechanism such as CoA limitation, slowed peroxisome metabolism due to accumulation of fatty acid intermediates, or accepting exogenous IBA as substrate because it is in excess. Such a model might suggest lacs7 also would display IBA resistance, as LACS6 and LACS7 are both highly expressed in the same tissues and have overlapping activity on fatty acid substrates (Shockey et al., 2002). However, whereas lacs6 is resistant to IBA, lacs7 has wild-type responses to IBA in all phenotypes tested and no additional enhancement was noted in the lacs4 lacs7 double mutant in primary root elongation or lateral root initiation assays. Future work characterizing the substrate profile of LACS6 will be necessary to establish a direct or indirect role of LACS6 in IBA metabolism.
lacs4 and lacs6 single mutants have IBA-resistant phenotypes (Figure 3), whereas defects in fatty acid β-oxidation, seed oil accumulation, and glycolipid synthesis are only seen in higher-order mutants with other lacs mutants (Figure 3 and Shockey et al., 2002; Fulda, 2004; Jessen et al., 2015).
Together these findings illustrate that activity of LACS4 and LACS6 on IBA is unique, although expanded studies of this enzyme family will reveal additional details of enzyme activity and pathway interactions. For instance, lacs8-2 shows a slight hypersensitivity to IBA (Supplemental Figure S2); lacs8 is of interest as other studies report overlapping activity of LACS4 and LACS8 (Jessen et al., 2015; Zhao et al., 2019). lacs3 also has a slight hypersensitivity to IBA but has reduced primary root length without hormone treatment, potentially indicating a general growth defect instead of an IBA-specific effect. Other LACS enzymes have not been thoroughly investigated yet, but future experiments will reveal their roles on fatty acids and potentially other related substrates.
Additional interactions may be required for full IBA responses in Arabidopsis
lacs4 develops lateral roots in response to IBA stimulation, although not to the same extent as wild type (Figures 1, 3); this differential pattern of resistance across developmental stages is rare in IBA-response mutants. For instance, acx3 also develops lateral roots in the presence of IBA but is resistant in primary root elongation (Eastmond et al., 2000; Adham et al., 2005). The distinct primary and lateral root IBA-responses of acx3 is attributed to its expression pattern. ACX3 expression in roots is seen in the root tip and tips of elongated lateral roots but not lateral root primordia, even upon IBA treatment (Eastmond et al., 2000; Adham et al., 2005).
The mechanism for LACS4 differential responses remains an open question. LACS4 and LACS6 are highly expressed in root tissue compared to other LACSs (Shockey et al., 2002) and analysis of a LACS4 promoter:GUS construct demonstrated that LACS4 expression is detected in the root tip but is absent from the elongation zone (Zhao et al., 2019). Analysis of LACS4 expression in lateral root primordia is required to determine if the different IBA response phenotypes of lacs4 can be attributed to cell-specific expression or activity.
Interestingly, lacs6 has been previously characterized and shows sensitivity to the synthetic IBA analog 2,4-DB (Fulda, 2004), similar to our results (Figure 4C). AAE18 is an acyl-activating enzyme found in peroxisomes with predicted overlapping activity. Notably, aae18 mutants are sensitive to IBA, but resistant to 2,4-DB (Figure 3C; Wiszniewski et al., 2009). This unique pattern of responses represents another area for future investigation, as connections and overlap between LACS4, LACS6, and AAE18 could be revealing for different tissues or conditions as suggested by their unique but overlapping phenotypes.
Unique aspects of lacs4-8 mutation
Our lacs4 allele has a weak missense mutation of an alanine to valine. Residue 467 is not located in a known substrate or cofactor binding pocket or active site (Figure 2). Our structural analysis and functional prediction using Phyre2 (Kelley et al., 2015) suggest this residue is not in a functionally important domain or region of secondary structure. Our genetic evidence that a substitution at residue 467 disrupts function unveils questions about which regions in LACS4 are important for function and if they are equally important for lipid and IBA metabolism.
We also completed mutational analysis using the SuSPect method (Yates et al. 2014). Substitution to any amino acid at residue 467 is predicted to have near neutral effects. However, despite this prediction, there is no amino acid degeneracy at this residue; this residue is a completely conserved alanine in all A. thaliana accessions and a proline in all other Arabidopsis LACS proteins and LACS4 orthologs in other plant species, including closely related Arabidopsis species (Figure 2). Such conservation suggests an evolutionary pressure to maintain the amino acids at this site. It is striking that LACS4 of A. thaliana encodes a divergent alanine at this site. The PAM250 matrix (Schwarz and Dayhoff, 1979) predicts that proline is most often and equally likely to be substituted for alanine or serine. Although the recent divergence of A. thaliana at this amino acid is surprising, a substitution for alanine may be favorable and necessary for function. This biological evidence of strong conservation increases our confidence that this residue is sensitive to perturbations, despite bioinformatics predictive analysis.
Our genetic evidence supports that residue 467 is important for protein function, folding, or stability, given that a point mutation disrupts plant physiology to the same extent as null alleles (Figure 2E). Continued analysis of the LACS4 domain structure generally and this region specifically is required to determine if this site is required for binding to a substrate, interacting protein, or required to give the protein a precise structure.
LACS4 localization remains an open question
In our analysis of localization, LACS4-GFP could be viewed as distinct puncta in Arabidopsis (Figure 5). These puncta looked comparable to BODIPY-stained peroxisomes. The punctate localization was also seen when LACS4-CFP was transiently expressed in N. benthamiana and colocalized with YFP:SRL-labeled peroxisomes. This evidence that LACS4 associates with peroxisomes is consistent with localization of other IBA to IAA enzymatic components (Hu et al., 2012). Subcellular localization prediction tools yield disparate predictions. WoLF-PSORT (Horton et al., 2007) and Suba4 (Hooper et al., 2017) most often predict cytosol localization followed by peroxisome localization, but also predict localization to all other cellular compartments.
If LACS4 is localized to the peroxisome matrix, it is unclear how import occurs as LACS4 does not possess a canonical PTS1 or PTS2. The LACS4 C-terminal amino acids SRG are not a sufficient PTS for entry into the peroxisome (Figure 5), although additional upstream amino acids not tested could facilitate import (Brocard and Hartig, 2006; Chowdhary et al., 2012). Some peroxisomal proteins without a PTS piggyback onto PTS containing proteins (Thoms, 2015) and protein oligomers can be imported even when subunits lack a PTS (Brown and Baker, 2008). This piggybacking mechanism has been reported for protein phosphatase 2A in Arabidopsis (Kataya et al., 2015) and even proteins that are not part of a complex, such as Nicotinamidase I which is co-imported into the peroxisome with glycerol-3-phosphate dehydrogenase 1 in Saccharomyces cerevisiae (Saryi et al., 2017).
LACS4 previously has been described as ER localized in Arabidopsis (Jessen et al., 2015) and Brassica napus (Tan et al., 2014). However, the mammalian ortholog, namely ACS4, localizes to the mitochondria, ER, and peroxisomes (Lewin et al., 2001, 2002; Milger et al., 2006; Grevengoed et al., 2014). This finding opens the question of LACS4 localization and the possibility of dual localization. A detailed study of LACS4 throughout development and under changing conditions, as well as identification of an interacting protein that could serve as a vehicle into the peroxisome, is necessary to elucidate the localization of LACS4 and understand the implications of potential dual localization over space and time.
Potential mechanisms of LACS4 activation of IBA
Our genetic and biochemical data support that LACS4 charges IBA with CoA. Genetic evidence suggests LACS6 does this as well. We hypothesize these two enzymes work together at this step via one of three possible mechanisms (Figure 8).
Figure 8.
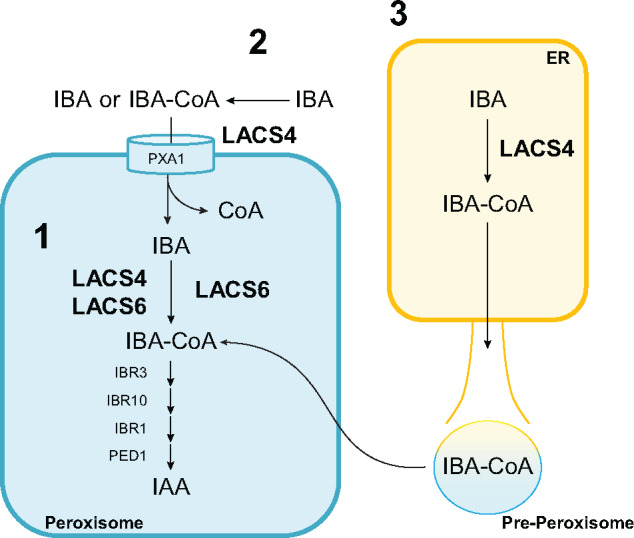
Three possible models of LACS4 activation of IBA, which affects IBA metabolism to IAA in the peroxisome. (1) LACS4 and LACS6 are both located inside the peroxisome where together they charge IBA with CoA. LACS4 may localize solely to the peroxisome or also localize to the ER and/or cytoplasm. (2) LACS4 may exist in the cytoplasm and loosely associated with the peroxisome without being internalized by the organelle to generate a cytoplasmic pool of IBA-CoA. IBA-CoA is imported into the peroxisome where LACS6 can reattach the CoA onto IBA. (3) ER-localized LACS4 may generate a pool of IBA-CoA that is actively imported into the peroxisome or is contained within pre-peroxisome structures that become mature peroxisomes. CoA cleaved off during import is reattached by the peroxisomally contained LACS6.
All enzymatic components of IBA to IAA conversion discovered to date are contained within the peroxisome. Our LACS4 localization experiments demonstrate LACS4 could be localized in the peroxisome matrix as well. This localization supports the hypothesis that LACS4 and LACS6 together charge IBA with CoA inside the peroxisome.
Alternatively, LACS4 may reside outside of the peroxisome, associated with the outer membrane. LACS4 generates a cytosolic pool of IBA-CoA, which is imported into the peroxisome. Active import by PXA1 is coupled with CoA hydrolysis (Nyathi et al., 2010; Lousa et al., 2013) and in this model, IBA could be reactivated by peroxisomal LACS6 in the matrix.
However, previous work also has reported LACS4 localization in the ER (Jessen et al., 2015). LACS4 may generate a pool of IBA-CoA in the ER for import into the peroxisome, which could be reactivated by LACS6.
Finally, there could be two pools of LACS4 with distinct functions. One pool may be ER localized and function primarily in fatty acid metabolism, whereas a peroxisomal pool may be primarily associated with hormone metabolism. Such a model is consistent with our mutant analysis suggesting loss of LACS4 affects IBA responses, but does not affect peroxisomal lipid metabolism during early seedling development.
Each of these proposed mechanisms have a novel aspect that is intriguing. If LACS4 is peroxisomal, then it is unclear how it is imported without a canonical PTS. LACS4 could enter via the piggyback mechanism, an extended signal sequence, or a noncanonical PTS. If LACS4 is solely localized to the ER, it is the first enzyme with a role in IBA metabolism not linked to the peroxisome. However, peroxisomes have a close relationship with the ER. Peroxisomes bud off the ER membrane to become independent entities, as described in a model of de novo biogenesis, or the ER may serve as a source of membranes and membrane proteins, as described in a semi-autonomous model (Mullen and Trelease, 2006; Hu et al., 2012; Agrawal and Subramani, 2016). IBA-CoA generated from LACS4 in the ER may be moved into the peroxisome via these membrane fusion models rather than active import. If LACS4 is only localized to the ER, this furthers the growing evidence that implicates the ER in auxin metabolism. IBA can be formed via acetylation of IAA and is associated with the ER membrane in maize (Zea mays; Ludwig-Müller and Epstein, 1992; Ludwig-muller and Hilgenberg, 1995). Auxin-conjugate hydrolases reside here, IAA actively is transported into the ER, and the ER may regulate the amount of auxin transported to the nucleus for signaling (Bartel and Fink, 1995; Davies et al., 1999; Friml and Jones, 2010; Middleton et al., 2018).
Finally, LACS4 may exist in both the ER and the peroxisome. Multiple pools of LACS4 would accommodate the pleiotropic activity of LACS4 in lipid and IBA metabolism. Localization to multiple organelles is seen in the mammalian isoform ACS4, which is localized to the ER, peroxisome, and mitochondrial membrane (Grevengoed et al., 2014). In mammals, superoxide dismutase and lactate dehydrogenase are localized primarily to the cytosol with a smaller peroxisomal pool that enter via piggybacking mechanisms (Wanders and Waterham, 2006; Islinger et al., 2009). The import of these proteins into the peroxisome is dependent on the amount of available carrier protein, offering an explanation for the comparatively reduced amount of protein in the peroxisome, which is often masked in in vitro studies (Thoms, 2015). Dual localization of ACS4 and the ability of superoxide dismutase and lactate dehydrogenase to enter the peroxisome without PTSs while maintaining primary localization elsewhere substantiates the possibility that LACS4 could have also dual localization in Arabidopsis and suggests a rationale for why LACS4 does not have a PTS, why peroxisomal localization has not been previously detected, and why a significant role in fatty acid β-oxidation is not observed.
Summary
The roles of IBA-derived IAA, and auxin storage forms more broadly, have been investigated in detail but continue to be studied to determine the activity and relative importance of each throughout plant growth, development, and responses to changing environmental conditions. Identification of LACS4 as an activator of IBA provides a more complete view of the IBA metabolic pathway, facilitating additional studies to increase our understanding of alternative auxin input pathways during plant development. In addition, this work gives new light to the role of LACS4 within a cell and the importance of LACS4 in plant development. LACS4 stands at the crossroads of hormone and lipid metabolism, two molecular pathways required to balance growth, development, and responses to the environment throughout the lifespan of a plant.
Materials and methods
Plant materials
All plants are A. thaliana of the Columbia (Col-0) background. ibr3-1 has a point mutation generated by EMS (Zolman et al., 2007). ibr3-4 (SALK_004657), lacs1-2 (SALK_138782), lacs2-3 (GABI_368C02), lacs3-1 (SALK_ 027707), lacs4-1 (SALK_101543), lacs4-7 (SALK_120357), lacs6 (SALK_069510c), and lacs7 (SALK_146444), lacs8-2 (SALK_105118), and lacs9-4 (SALK_124615) are T-DNA lines obtained from the Arabidopsis Biological Resource Center. All mutants were genotyped to confirm homozygous mutations (Supplementary Tables S2 and S3).
lacs4-1, lacs4-7, lacs6, and lacs7 were backcrossed to wild type, reisolated by genotyping and confirmed homozygous. All double mutants were obtained from directed outcrossings.
Mutagenesis and mutant isolation
ibr3-1 seeds were soaked in 0.24% EMS (v/v) for 18 h in the dark, washed extensively, and transferred to soil. M2 seedlings were grown on 25 µM IBA and screened for individuals with elongated roots compared to the ibr3-1 background. M3 progeny were retested to validate the mutant phenotype.
Z377 ibr3-1 was backcrossed to ibr3-1 to generate lines for sequencing. Z377 was isolated from segregating generations of the Z377 ibr3-1 backcross to wild type by screening for seedlings with an intermediate IBA-resistant root phenotype that genotyped wild-type for ibr3-1. Z377 ibr3-1 and Z377 were backcrossed to wild-type twice to generate lines for phenotypic analysis.
Growth conditions for phenotypic assays
Seeds were sterilized with 30% bleach (v/v) and 0.1% triton (v/v), rinsed, suspended in 0.1% agar (w/v), and imbibed at 4°C for 3–5 d. Seeds were plated on plant nutrient media (PN; Haughn and Somerville, 1986) supplemented as indicated. Plates were incubated at 22°C under continuous yellow light for the specified number of days.
Primary root length was measured after 7 d of growth on indicated media. Lateral root density was determined by growing seeds on PN for 4 d, transferring seedlings to PN or PN + 5 µM IBA plates and growing for an additional 4 d. Lateral roots were counted with a Leica Zoom 2000.
Sucrose dependence and effects of auxins on hypocotyl length were performed by growing seeds on indicated media for 1 d. Plates were then incubated in darkness for an additional 5 d before hypocotyls were measured.
Whole-genome sequencing and mutant genotyping
Ten independently isolated Z377 ibr3-1 lines from a backcross to ibr3-1 were selected and pooled for sequencing in parallel with wild type and ibr3-1. DNA was extracted from approximately 2,000 seedlings grown on filter paper for 10 d. Sequencing was performed at the Genome Technology Access Center at Washington University in St Louis on an Illumina-HiSeq2000. Gene candidates were first narrowed down by identifying homozygous single nucleotide polymorphisms consistent with EMS mutagenesis in coding regions. Mutations common between Z377 ibr3-1 and wild type or ibr3-1 were eliminated, leaving mutations unique to Z377 ibr3-1. dCAPS primers (Neff et al., 2002) were designed to genotype the point mutation in lacs4-8 (Supplemental Table 2).
Generation of constructs
LACS4 cDNA was amplified with LACS4Topo-F and LACS4Topo-R from cDNA synthesized from wild-type RNA. LACS4 was cloned into the Gateway entry vector pcr8/Topo. The stop codon of LACS4 was replaced with a 6-amino-acid, glycine-serine flexible linker with the Q5 Site Directed Mutagenesis Kit (NEB) using the primers LACS4-cterm linker F and LACS4 c-term linker R. This LACS4 construct was moved into plant gateway expression vectors pEarlyGate102 and pEarlyGate103 (Earley et al., 2006) with LR clonase (Thermo Fisher).
For recombinant expression, LACS4 cDNA was amplified with primers LACS4S and LACS4B, digested with Sal1-HF and BamHI-HF, and ligated into the pZA31 expression vector which adds an N-terminal HN tag and controls expression through the TetR system (Lutz and Bujard, 1997). pZA31:LACS4 was transformed into Escherichia coli DH5ɑZ1, a strain modified to constitutively express TetR from the chromosome for bacterial expression (Lutz and Bujard, 1997).
UBQ10:YFP-GW (Michniewicz et al., 2015) was mutated by site-directed mutagenesis to end with SRL and SRG C-terminal tripeptides with the primers YFP-SRG F, YFP-SRG R, YFP-SRL F, and YFP-SRL R using the Quikchange XL site-directed mutagenesis kit (Agilent Technologies).
Constructs were confirmed by sequencing. Primer sequences are listed in Supplementary Table S3.
Plant transformation
Wild-type and Z377 were transformed with the pEarlyGate103:LACS4 fluorescent fusion protein construct. UBQ10:YFP-SRL and UBQ10:YFP-SRG were independently transformed into wild-type. Arabidopsis transformation was done via the floral-dip method with Agrobacterium tumefaciens (Clough and Bent, 1998). Transformants were selected by growing seedlings on 10 µg mL−1 Basta for 14 d. Homozygous lines were identified by screening T3s on Basta.
Transient LACS4 expression
LACS4 localization was observed through transient expression in N. benthamiana (Sparkes et al., 2006). pEarlyGate102:LACS4 and UBQ10:YFP-SRL in A. tumefaciens were grown overnight at 30°C. Cultures were incubated in infiltration media (10 mM MgCl2, 10 mM MES, and 200 µM acetosyringone) and gently rocked at room temperature for 4 h. The cells were resuspended in fresh infiltration media to an OD600 of 1. Cultures of each construct were mixed at a ratio of 1:1 and infiltrated into leaves of young N. benthamiana plants using a 1-mL needleless syringe. Leaves were imaged 48 h after infiltration.
Confocal microscopy
Confocal images were obtained with a Zeiss LSM 700 laser scanning confocal microscope using the 20X lens. pEarlyGate103:LACS4 was imaged in 7-d-old Arabidopsis seedlings of T2s showing rescued IBA-resistant phenotypes when grown on 15 µM IBA. Seven-day-old wild-type Arabidopsis seedlings were stained with 5 µM 8-(4-Nitrophenyl) BODIPY (Toronto Research Chemicals) for 10 min and excited with a 488-nm laser.
RNA extraction and expression quantification
Seedlings were grown on filter paper under white light for 5 d. RNA was extracted by grinding seedlings in liquid nitrogen and using the IBI Total RNA Mini Kit (Plant; MidSci). cDNA was synthesized using ProtoScript II Reverse Transcriptase (NEB). LACS4 expression was determined by RT-qPCR using Bullseye EvaGreen qPCR Mix (MidSci) on a BioRad CFX96 Real Time PCR System. LACS4 expression was normalized to UBIQUITIN 10 and calculated with the ΔΔCT method. Primers were designed with QuantPrime (Arvidsson et al., 2008) and are listed in Supplemental Table S3. No reverse transcriptase reactions were used as a control. Experiments were performed with both biological and technical triplicates.
Gus staining
The DR5:GUS reporter line (Ulmasov et al., 1997) was crossed to ibr3-1 and lacs4-8 ibr3-1. Five-day-old seedlings were then incubated in liquid PN with no hormone, 50 µM IBA, or 1 µM IAA and gently rocked at room temperature for 2 h. GUS was visualized by incubating seedlings in 0.5 mg mL−1 X-Gluc for 3 h at 37°C, followed by ethanol (EtOH) washes (Bartel and Fink, 1994). Plants were stored in 50% glycerol (v/v) at 4°C and imaged with Evos XL Core (Thermo Fisher) on 20× and 40× objectives.
LACS4 purification from Escherichia coli DH5ɑZ1
DH5ɑZ1 containing the pZA31:LACS4 construct was grown with 25 µg mL−1 chloramphenicol at 37°C to an OD600 of 0.5. LACS4 expression was induced with 200 µg mL−1 anhydrotetracycline, then grown overnight at 16°C. Cells were pelleted, resuspended in 50 mM Tris pH 7.5, 300 mM NaCl, 0.1% Triton X-100 (v/v), and Pierce Protease Inhibitor Mini Tablets (Thermo Fisher). Cells were lysed by incubating with 2 mg/mL lysozyme at 4°C for 1 h followed by sonication with 10 30-s pulses. Cell debris was removed by centrifuging for 30 min at 20,000g at 4°C. Supernatant was then passed through a 0.22-µM filter to remove remaining debris.
The filtered supernatant was passed through an equilibrated 5-mL cobalt column on an ÄKTA prime FPLC (GE Healthcare) to capture HN-tagged LACS4. LACS4 recombinant protein was eluted in 1-mL fractions with 50 mM Tris pH 7.5, 300 mM NaCl, with increasing imidazole gradient up to 200 mM. Eluted fractions containing LACS4 were pooled and dialysed at 4°C in 10 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA, and 40% glycerol (v/v). Protein concentration was determined by Bradford assay. LACS4 was aliquoted and stored at –80°C.
LACS4 activity
LACS4 CoA synthetase activity was assayed by measuring PPi production. A 1 µg of LACS4 was incubated in a 50-µL reaction of 0.5 mM CoA, 45 mM ATP, 0.1% Triton X-100 (v/v), 1 mM dithiothreitol, 100 mM Tris pH 7.5, and the indicated substrate for 10 min at 24°C. 10-mM stocks of IBA potassium salt (Chem Cruz), IAA sodium salt (Cayman Chemical Co), IAA-alanine (Milipore-Sigma), methyl-IAA (Milipore-Sigma), tryptophan, and 2-4-DB dissolved in 50% EtOH (v/v) were tested as substrates. LACS4 incubated with 5% EtOH (v/v) served as the negative control. PPi was measured with a Pyrophosphate Assay Kit (Millipore-Sigma) according to instructions by adding 50 µL of reaction buffer to each CoA synthetase reaction and incubating 15 min at 24°C. PPi fluorescence was measured with λex=316 nm, λem= 456 nm on a Biotek Cytation 3.
Accession numbers
Gene sequences in this study can be found in Arabidopsis Genome Initiative database under the following accession numbers: IBR3, AT3G06810; LACS4, AT4G23850; LACS6, AT3G05970; LACS7, AT5G27600; LACS1, AT2G47240; LACS2, AT1G49430; LACS3, AT1G64400; LACS8, AT2G04350; LACS9, AT1G77590; AUX1, AT2G38120; AAE18, AT1G55320; UBQ10, AT4G05320. Sequences for long chain Co-A synthetase 4 orthologs can be accessed through Genbank under the following accession numbers: Arabidopsis halleri, ACC91252.1; Brassica napus, NP_001302548.1; Zea mays, XP_008652385.1, Populus trichocarpa, XP_024453969.1, Amborella trichopoda, XP_006850521.1, Physcomitrella patens, XP_024370732.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. lacs4-7 is comparable to Z377 in primary root length and lateral root density when grown on IBA.
Supplemental Figure S2. lacs4 and lacs6 are resistant to IBA in primary root elongation.
Supplemental Table S1 . Descriptions of candidate genes mutated in Z377 and the mutant allele tested for resistance to IBA.
Supplementary Table S2. Primer pairs and enzymes required for genotyping each mutant used in this study.
Supplementary Table S3. Sequences for new primers used in this study.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center at Ohio State University for seed stocks and the Salk Institute Genomic Analysis Laboratory for generating the Arabidopsis T-DNA insertion mutants. We thank Lon Chubiz for thoughtful discussions, donation of pZA31 and Escherichia coli DH5ɑZ1, and assistance purifying LACS4, Lucia Strader for assistance in whole-genome sequencing of Z377, and Ying Li for regular discussions and encouragement.
Funding
This research was supported by the National Institutes of Health (R15GM116090) and the University of Missouri Research Board.
V.J. and B.K.Z. designed the project. B.K.Z. conducted the initial screen and V.J. designed and performed further experiments. V.J. and B.K.Z. wrote the article. B.K.Z. agrees to serve as the author responsible for contact and ensures communication (zolmanb@umsl.edu).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Bethany Karlin Zolman (zolmanb@umsl.edu).
References
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Agrawal G, Subramani S (2016) De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim Biophys Acta 1863: 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR (1994) Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA 91: 6649–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748 [DOI] [PubMed] [Google Scholar]
- Brocard C, Hartig A (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim Biophys Acta 1763: 1565–1573 [DOI] [PubMed] [Google Scholar]
- Brown L-A, Baker A (2008) Shuttles and cycles: transport of proteins into the peroxisome matrix (review). Mol Membr Biol 25: 363–375 [DOI] [PubMed] [Google Scholar]
- Chowdhary G, Kataya ARA, Lingner T, Reumann S (2012) Non-canonical peroxisome targeting signals: identification of novel PTS1 tripeptides and characterization of enhancer elements by computational permutation analysis. BMC Plant Biol 12: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Damodaran S, Strader LC (2019) Indole 3-butyric acid metabolism and transport in Arabidopsis thaliana. Front Plant Sci 10: 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Xuan W, Overvoorde P, Strader LC, Kepinski S, Hoye R, Brisbois R, Parizot B, Vanneste S, et al. (2012) A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat Chem Biol 8: 798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Hooks MA, Williams D, Lange P, Bechtold N, Sarrobert C, Nussaume L, Graham IA (2000) Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J Biol Chem 275: 34375–34381 [DOI] [PubMed] [Google Scholar]
- Frick EM, Strader LC (2018) Roles for IBA-derived auxin in plant development. J Exp Bot 69: 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Jones AR (2010) Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiol 154: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M (2004) Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Grevengoed TJ, Klett EL, Coleman RA (2014) Acyl-CoA metabolism and partitioning. Annu Rev Nutr 34: 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot PHE, Scholte HR, Hülsmann WC (1976) Fatty acid activation: specificity, localization, and function. Adv Lipid Res 14: 75–126 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M (2002) Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol 43: 1–11 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid–resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45: D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islinger M, Li KW, Seitz J, Völkl A, Lüers GH (2009) Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes—evidence for a natural piggyback import mechanism in mammals. Traffic 10: 1711–1721 [DOI] [PubMed] [Google Scholar]
- Jessen D, Olbrich A, Knüfer J, Krüger A, Hoppert M, Polle A, Fulda M (2011) Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J 68: 715–726 [DOI] [PubMed] [Google Scholar]
- Jessen D, Roth C, Wiermer M, Fulda M (2015) Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol 167: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y-T, Gonzalez KL, Bartel B (2018) peroxisome function, biogenesis, and dynamics in plants. Plant Physiol 176: 162–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataya ARA, Heidari B, Hagen L, Kommedal R, Slupphaug G, Lillo C (2015) Protein phosphatase 2A holoenzyme is targeted to peroxisomes by piggybacking and positively affects peroxisomal β-oxidation. Plant Physiol 167: 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan BR, Zolman BK (2010) pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol 154: 1602–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC (2013) Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M, Smertenko A, Edwards R, Hussey PJ, Steel PG (2010) BODIPY probes to study peroxisome dynamics in vivo. Plant J 62: 529–538 [DOI] [PubMed] [Google Scholar]
- Lanyon-Hogg T, Warriner SL, Baker A (2010) Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biol Cell 102: 245–263 [DOI] [PubMed] [Google Scholar]
- Lewin TM, Kim J-H, Granger DA, Vance JE, Coleman RA (2001) Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J Biol Chem 276: 24674–24679 [DOI] [PubMed] [Google Scholar]
- Lewin TM, Van Horn CG, Krisans SK, Coleman RA (2002) Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Arch Biochem Biophys 404: 263–270 [DOI] [PubMed] [Google Scholar]
- Li Y, Liu Y, Zolman BK (2019) Metabolic alterations in the enoyl-CoA hydratase 2 mutant disrupt peroxisomal pathways in seedlings. Plant Physiol 180: 1860–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lousa CDM, De Marcos Lousa C, van Roermund CWT, Postis VLG, Dietrich D, Kerr ID, Wanders RJA, Baldwin SA, Baker A, Theodoulou FL (2013) Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc Natl Acad Sci USA 110: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J, Epstein E (1992) Indole-3-acetic acid is converted to indole-3-butyric acid by seedlings of Zea mays L. Prog Plant Growth Regul 13: 188–193 [Google Scholar]
- Ludwig-muller J, Hilgenberg W (1995) Characterization and partial purification of indole-3-butyric acid synthetase from maize (Zea mays). Physiol Plant 94: 651–660 [Google Scholar]
- Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Frick EM, Strader LC (2015) Gateway-compatible tissue-specific vectors for plant transformation. BMC Res Notes 8: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton AM, Dal Bosco C, Chlap P, Bensch R, Harz H, Ren F, Bergmann S, Wend S, Weber W, Hayashi K-I, et al. (2018) Data-driven modeling of intracellular auxin fluxes indicates a dominant role of the ER in controlling nuclear auxin uptake. Cell Rep 22: 3044–3057 [DOI] [PubMed] [Google Scholar]
- Milger K, Herrmann T, Becker C, Gotthardt D, Zickwolf J, Ehehalt R, Watkins PA, Stremmel W, Fullekrug J (2006) Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J Cell Sci 119: 4678–4688 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN (2006) The ER-peroxisome connection in plants: development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim Biophys Acta Mol Cell Res 1763: 1655–1668 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Nyathi Y, De Marcos Lousa C, van Roermund CW, Wanders RJA, Johnson B, Baldwin SA, Theodoulou FL, Baker A (2010) The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2Delta mutant for metabolism of long-chain fatty acids and exhibits fatty acyl-CoA-stimulated ATPase activity. J Biol Chem 285: 29892–29902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saryi NAA, Hutchinson JD, Al-Hejjaj MY, Sedelnikova S, Baker P, Hettema EH (2017). Pnc1 piggy-back import into peroxisomes relies on Gpd1 homodimerisation. Sci Rep 7: 42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Dayhoff M (1979) Matrices for Detecting Distant Relationships. In: Dayhoff M, editor, Atlas of Protein Sequences. National Biomedical Research Foundation, Silver Springs, Maryland USA, pp. 353–358. [Google Scholar]
- Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Spiess GM, Hausman A, Yu P, Cohen JD, Rampey RA, Zolman BK (2014) Auxin input pathway disruptions are mitigated by changes in auxin biosynthetic gene expression in Arabidopsis. Plant Physiol 165: 1092–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B (. 2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X-L, Xiao-li TAN, Zheng X-F, Zhang Z-Y, Wang Z, Heng-chuan XIA, Changming LU, GU Shou-lai (2014) Long chain acyl-coenzyme A synthetase 4 (BnLACS4) gene from Brassica napus enhances the yeast lipid contents. J Integr Agric 13: 54–62 [Google Scholar]
- Thoms S (2015) Import of proteins into peroxisomes: piggybacking to a new home away from home. Open Biol 5: 150148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Hernandez JA, Bariat L, Strader LC, Reichheld J-P, Belin C (2020) A glutathione-dependent control of the indole butyric acid pathway supports Arabidopsis root system adaptation to phosphate deprivation. J Exp Bot 71: 4843–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47: D506–D515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain RL, Wightman F (1954) The growth regulating activity of certain omega-substituted alkyl carboxylic acids in relation to their beta-oxidation within the plant. Proc R Soc Lond B Biol Sci 142: 525–536 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75: 295–332 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA (1997) Fatty acid activation. Prog Lipid Res 36: 55–83 [DOI] [PubMed] [Google Scholar]
- Wiszniewski AAG, Zhou W, Smith SM, Bussell JD (2009) Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol Biol 69: 503–515 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, Möller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mähönen AP, Vanneste S, et al. (2015) Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol 25: 1381–1388 [DOI] [PubMed] [Google Scholar]
- Yates CM, Filippis I, Kelley LA, Sternberg MJE (2014) SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J Mol Biol 426: 2692–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Haslam TM, Sonntag A, Molina I, Kunst L (2019) Functional overlap of long-chain acyl-CoA synthetases in Arabidopsis. Plant Cell Physiol 60: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SP, Bartel B (2001) chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal beta-hydroxyisobutyryl-CoA hydrolase. J Biol Chem 276: 31037–31046 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.