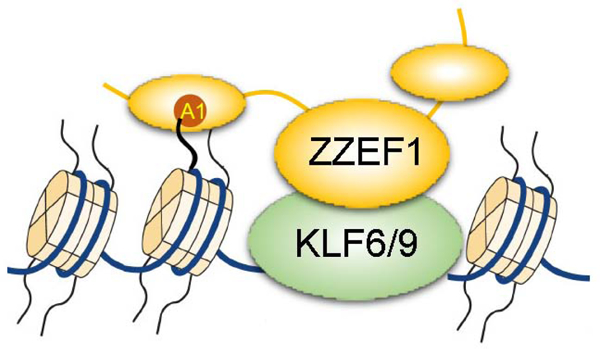
SUMMARY
The ZZ-type zinc finger and EF-hand domain protein 1 (ZZEF1) is a multidomain-containing protein. Mutations of ZZEF1 has been implicated in several kinds of human diseases such as diabetes and cancers. However, the function of the ZZEF protein remains largely unknown. Here we show that ZZEF1 functions as a histone H3 reader. The second ZZ domain of ZZEF1 (ZZEF1ZZ2) binds to the N-terminus of histone H3 and is capable of accommodating common epigenetic marks on the H3 tail. The N-terminal amino acids, especially Ala1, of H3 and an acidic cavity of ZZEF1ZZ2 are critical for the ZZ-H3 interaction. RNA-seq analysis in human lung cancer cell line H1299 reveals that downregulated genes upon ZZEF1 depletion are specifically enriched in genes regulated by Krüppel-like factors. Indeed, ZZEF1 physically interacts with KLF9 and KLF6, and regulates a common set of target genes of these transcription factors. Together, our findings suggest a model in which ZZEF1 binds to histone H3 tail and promotes KLF9/6-mediated gene regulation.
Keywords: ZZEF1, ZZ domain, H3, Krüppel-like factors, KLF9, KLF6
Graphical Abstract
The ZZ domain is an evolutionarily conserved protein domain present in ~20 eukaryotic proteins involved in fundamental cytosolic and nuclear cellular processes 1. The ZZ domain is comprised of ~50 amino acids and topologically belongs to a large family of RING fingers that also includes other zinc fingers, such as the PHD finger and the FYVE domain. Although relatively small in size, the ZZ domain is a versatile signaling module that has been implicated in a diverse array of cellular events. For instance, the ZZ domain of p62 recognizes arginylated substrates promoting selective autophagy 2, whereas the ZZ domains of p300 and ZZZ3 are capable of binding to either unmodified or acetylated histone H3 tails 3,4. In addition, a set of the ZZ domain-containing proteins has been suggested to recognize proteins or RNA 5,6, however biological roles of many ZZ domains remain uncharacterized.
The zinc finger ZZ-type and EF-hand domain-containing protein 1 (ZZEF1) gene is conserved in vertebrates. A number of high-throughput sequencing studies indicate that coding region single-nucleotide polymorphisms or copy number alterations are associated with type 2 diabetes and several types of human cancer, such as breast, pancreatic, gastric and esophageal cancers 7,8. However, until now there have been no studies delineating the role of ZZEF1 or its ZZ domains (Fig. 1a). To determine the function of the ZZEF1 ZZ domains, we first performed sequence alignment with the ZZ domains from p300 and ZZZ3, which we recently identified as novel histone H3 readers 3,4. Sequence alignment revealed a high degree conservation of the residues necessary for zinc-coordinating and H3 binding activities (Supplementary Fig. S1), suggesting that the ZZ domains of ZZEF1 may bind to histones. To test this hypothesis, we performed in vitro histone peptide pull-downs experiments with both ZZ domains of ZZEF1. We found that the second ZZ domain (ZZEF1ZZ2) specifically binds to the H3(1–22) peptide, whereas the first ZZ domain does not bind to any histone peptide tested (Supplementary Fig. S2). Unlike the PHD1 finger of KDM5A, whose binding to H3 tail was abolished by methylation on H3R2 or H3K4, ZZEF1ZZ2 binding to H3 was not affected by posttranslational modifications (PTMs) commonly present in histone H3 tail (Fig. 1b). The interaction between ZZEF1 and H3 was further validated by Flag immunoprecipitation (IP) in HEK 293T cells expressing Flag tagged-H3 and a short isoform of ZZEF1, ZZEF1ISO5, that contains the ZZ domains (Fig. 1c and Supplementary Fig. S3). Together, these data suggest that ZZEF1ZZ2 binds to histone H3 both in vitro and in cells.
Figure 1.
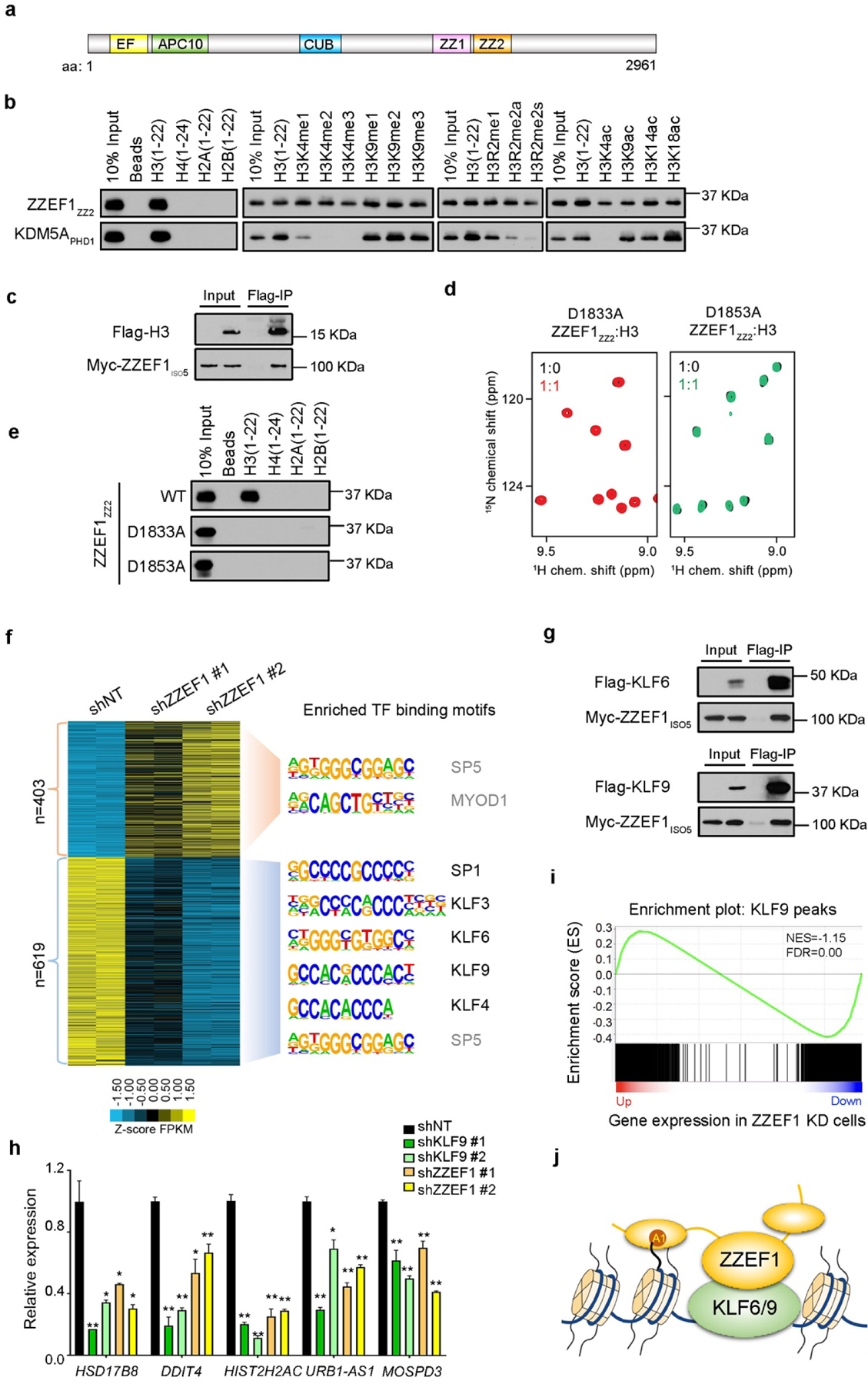
ZZEF1 binds to histone H3 and the KLF6/9 transcription factors to stimulate gene expression. a, Schematic representation of ZZEF1 protein structure. EF: EF-hand motif; APC10: APC10/DOC1-like domain; CUB; CUB domain; ZZ1: the first ZZ domain; ZZ2: the second ZZ domain. b, Western blot analysis of peptide pull-downs of GST-tagged ZZEF1ZZ2 and KDM5APHD1 with the indicated histone peptides. c, Western blot analysis of Flag co-IP in cells expressing H3-Flag and Myc- ZZEF1iso5. d, Superimposed 1H, 15N HSQC spectra of the indicated ZZEF1ZZ2 mutants titrated with the H3(1–12) peptide. e, Western blot analysis of peptides pull-downs of WT ZZEF1ZZ2 and the indicated mutants with histone peptides. f, Heatmap of upregulated (n=403) and downregulated (n=619) genes in ZZEF1 shRNA knockdown (KD) cells compared with control (shNT) cells. Enriched consensus motifs and transcription factors (TFs) are shown on the right. TFs that are not expressed (RPKM<1) in H1299 cells are shown in gray. g, Western blot analysis of Flag co-IP in cells expressing Flag-KLF6 or KLF9 and Myc-ZZEF1iso5. h, qRT-PCR analysis of gene expressions in control (shNT) and KLF9 or ZZEF1 KD H1299 cells. Error bars indicate SD of three replicates. *: p<0.05; **: p<0.01 (Student’s t-test) i, GSEA analysis showing that down-regulated genes in ZZEF1 KD cells are enriched for KLF9 occupancy. NES: normalized enrichment score, FDR: false discovery rate. j, Working model of ZZEF1 binding to H3 and KLF6/9 to promote gene expression. A1: Ala 1 of histone H3.
To assess binding of ZZEF1ZZ2 to histone H3 in detail, we expressed 15N-labeled ZZEF1ZZ2 and tested it in 1H, 15N heteronuclear single quantum coherence (1H,15N HSQC) experiments. Large resonance changes in intermediate exchange regime were induced by the unmodified H3 (aa 1–12), H3K4me3 and H3K4ac peptides, indicating direct and tight binding and confirming the pull-down data (Supplementary Fig. S4). In order to measure binding affinities by tryptophan fluorescence, we replaced H1877 at the C-terminus of ZZEF1ZZ2 with a tryptophan and verified that this replacement does not affect the ZZEF1ZZ2 fold (Supplementary Fig. S5). The tryptophan fluorescence measurements revealed that ZZEF1ZZ2 binds to the unmodified H3 peptide with affinity of 4.3 ± 0.1 μM, and comparable binding affinities were measured for the interactions with H3K4me3 (4.8 ± 0.1 μM) and H3K4ac (11 ± 1 μM) (Supplementary Fig. S4 and S6). Thus, in agreement with peptide pull-down experiments, the NMR and tryptophan fluorescence results indicate that ZZEF1ZZ2 binds to histone H3 tail well and irrespectively to post-translational modifications present on H3K4. Structural modeling suggests that similar to other known H3-binding ZZ domains 3,9, ZZEF1ZZ2 recognizes Ala1 of H3 by two conserved aspartic residues, D1833 and D1853 (Supplementary Fig. S7). Indeed, binding of ZZEF1ZZ2 to H3 was abolished when either residue was mutated to alanine, as observed in NMR titration and peptide pulldown experiments (Fig. 1d and 1e).
To determine the role of ZZEF1 in regulation of gene expression, we knocked down (KD) ZZEF1 gene expression in H1299, a human non-small cell lung carcinoma cell line, using two independent shRNAs. Both shRNAs efficiently depleted endogenous ZZEF1 proteins (Supplementary Fig. S8). RNA-seq analysis identified total 1022 common differentially expressed genes (DEGs) in both ZZEF1 KD cells compared with cells expressing a control non-targeting shRNA, with 619 genes down-regulated and 403 genes upregulated (Fig. 1f and Supplementary Table S1). Gene ontology (GO) analysis revealed that down-regulated genes upon ZZEF1 depletion are mainly enriched in pathways involving exosome secreting, mitochondrial functions, and metabolic processes, whereas up-regulated genes are genes involved in transcription, cell proliferation, and the Notch signaling pathway (Supplementary Fig. S9). Motif analysis of the promoters the 1022 DEGs revealed enrichment for SP1, SP5, MYOD1, and the Krüppel-like factor (KLF) family members (Fig. 1f).
Both SP5 and MYOD1 are expressed at very low levels (RPKM<1) in H1299 cells (Supplementary Table S1), thus we speculated that ZZEF1 may associate with SP1 and/or the KLF family of transcription factors to promote gene expression. To test this hypothesis, we first determined the interactions between ZZEF1 and these transcription factors by co-IP experiments in cells. Among the few KLF family proteins tested, we found that ZZEF1 physically interacted with KLF6 and KLF9, but not SP1 or other KLF proteins (Fig. 1g and Supplementary Fig. S10). Furthermore, quantitative reverse-transcription PCR (qRT-PCR) showed that both KLF9 KD and ZZEF1 KD led to target gene repression (Fig. 1h and Supplementary Fig. S11). Finally, to determine whether ZZEF1 functions as a transcription coregulator of KLF9 and KLF6 across different cell types, we performed Gene Set Enrichment Analysis (GSEA) of ZZEF1-regulated genes on the set of overlapped genes that have KLF9 occupancy in HEK293, HepG2 and MCF-7 cells (Supplementary Table S2) or KLF6 ChIP-seq peaks in HepG2 cells (Supplementary Table S3). GSEA results revealed that the down-regulated genes in ZZEF1 KD cells are enriched in both KLF9 and KLF6 occupancies (Fig. 1i and Supplementary Fig. S12), suggesting that ZZEF1 likely function as a transcription coactivator of KLF9 and KLF6 across different cell types.
ZZEF1 is a large protein comprised of five conserved domains with unknown function. Our results suggest that ZZEF1 binds to histone H3 tail and facilitates binding of KLF9 and KLF6 at target gene promoters to stimulate active transcription (Fig. 1j). The KLF family members regulate various cellular functions, such as proliferation, differentiation, apoptosis, as well as the development and homeostasis of several types of tissue 10. Dysregulation of the KLF family members, including KLF6 and KLF9, are observed in a broad range of human diseases, including cancers. Our finding that ZZEF1 binds both KLF6/9 and histone H3 suggests that ZZEF1 may play an important role in these processes in diverse cell types. The capability of the ZZ2 domain to accommodate different PTMs on histone H3 tail enables ZZEF1 to function as a versatile co-regulator that facilitates the association of KLF6 and KLF9 with chromatin at specific epigenetic states. However, since we were unable to perform a rescue experiment with H3-binding deficient point mutants of ZZEF1 (because of the large gene size), the precise role of the ZZ domain in vivo needs to be further validated. Nevertheless, to the best of our knowledge, our study is the first report of the function of ZZEF1. Our findings in vitro suggest a role of the ZZEF1 ZZ2 domain in cells in targeting the KLF family proteins to chromatin for transcriptional regulation.
Supplementary Material
HIGLIGHTS.
The second ZZ domain of ZZEF1 (ZZEF1ZZ2) is a reader of histone H3
Recognition of Ala1 of H3 by aspartic residues of ZZEF1ZZ is essential
ZZEF1 is a transcriptional coregulators of Krüppel-like factors
Acknowledgements
We thank the Van Andel Institute Genomics Core for Next Generation Sequencing. This work was supported in part by grants from NCI CA204020 to X.S. and NIH HL151334, CA252707 and AG067664 to T.G.K. X.S. is a Leukaemia & Lymphoma Society Career Development Program Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
X.S. is a SAB member of EpiCypher. All other authors declare no competing interests.
References
- 1.Zhang Y, Mi W, Xue Y, Shi X & Kutateladze TG The ZZ domain as a new epigenetic reader and a degradation signal sensor. Crit Rev Biochem Mol Biol 54, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y et al. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat Commun 9, 4373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y et al. The ZZ domain of p300 mediates specificity of the adjacent HAT domain for histone H3. Nat Struct Mol Biol 25, 841–849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mi W et al. The ZZ-type zinc finger of ZZZ3 modulates the ATAC complex-mediated histone acetylation and gene activation. Nat Commun 9, 3759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J et al. Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2. Proc Natl Acad Sci U S A 115, 10010–10015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afroz T et al. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev 28, 1498–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahajan A et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50, 559–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joo JE et al. Heritable DNA methylation marks associated with susceptibility to breast cancer. Nat Commun 9, 867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J et al. Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1. Structure (2020). [DOI] [PMC free article] [PubMed]
- 10.Bieker JJ Kruppel-like factors: three fingers in many pies. J Biol Chem 276, 34355–8 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


