Abstract
Although the main cellular target of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is thought to be alveolar cells, the absence of their tractable culture system precludes the development of a clinically relevant SARS-CoV-2 infection model. Here, we establish an efficient human alveolosphere culture method and sphere-based drug testing platform for SARS-CoV-2. Alveolospheres exhibit indolent growth in a Wnt- and R-spondin-dependent manner. Gene expression, immunofluorescence, and electron microscopy analyses reveal the presence of alveolar cells in alveolospheres. Alveolospheres express ACE2 and allow SARS-CoV-2 to propagate nearly 100,000-fold in 3 days of infection. Whereas lopinavir and nelfinavir, protease inhibitors used for the treatment of human immunodeficiency virus (HIV) infection, have a modest anti-viral effect on SARS-CoV-2, remdesivir, a nucleotide prodrug, shows an anti-viral effect at the concentration comparable with the circulating drug level. These results demonstrate the validity of the alveolosphere culture system for the development of therapeutic agents to combat SARS-CoV-2.
Keywords: alveolosphere, lung, stem cell niche, drug screening, COVID-19, organoid
Graphical abstract
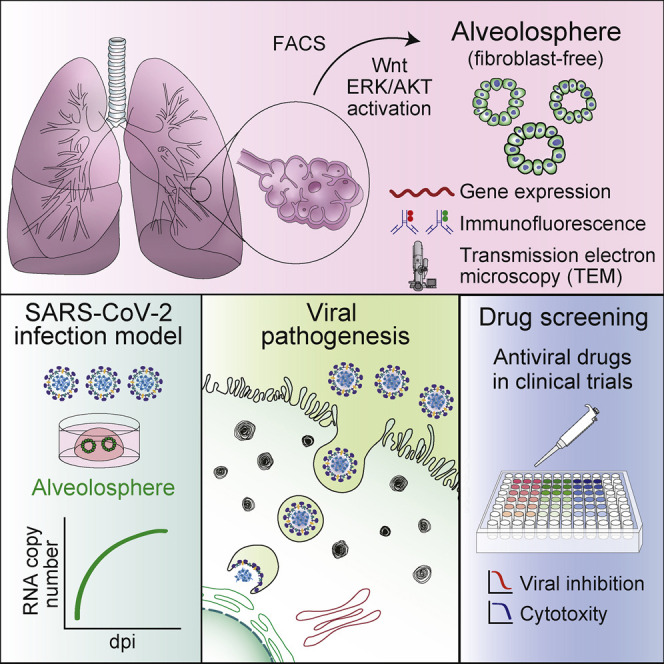
Ebisudani et al. establish a clinically relevant culture platform for human alveolus. They apply this sphere-based system to SARS-CoV-2 infection. The reproduction of SARS-CoV-2 in alveolospheres enables evaluation of anti-viral drug inhibitory effects, such as with remdesivir. This model will contribute to the accurate evaluation of candidate drugs against COVID-19.
Introduction
The pandemic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is currently posing a major public health issue. Due to the extremely rapid worldwide spread of the disease, the treatment of COVID-19 currently relies on supportive therapy, such as the use of dexamethasone (RECOVERY Collaborative Group et al., 2021) and the compassionate use of anti-viral drugs that have been developed and clinically used for the treatment of other viral infections (Beigel et al., 2020). In vitro validation of the anti-SARS-CoV-2 effect of the repurposed drugs has been performed on infection-competent yet clinically irrelevant cell lines, such as Vero cells (Zhou et al., 2020b). It remains unknown to what extent such experimental cell lines recapitulate the therapeutic effect of the drugs on COVID-19, given that species-unique genetic variants in viral receptors, such as ACE2, can potentially affect viral entry into target cells (Hoffmann et al., 2020). For instance, genetic variants in mouse and hamster Ace2 account for the species difference of disease severity during SARS-CoV-2 infection (Sia et al., 2020). Vero cells, which are derived from African green monkey kidney, require artificial overexpression of TMPRSS2 for optimal SARS-CoV-2 infection (Takayama, 2020). Therefore, there has been an urgent need for human cell-based infection models that are amenable for the screening of anti-SARS-CoV-2 drugs (Elbadawi and Efferth, 2020).
Recent advances in organoid technology have enabled modeling of SARS-CoV-2 infection using human intestinal epithelial organoids and airway epithelium (Lamers et al., 2020; Zhou et al., 2020a). However, an organoid or sphere culture system for the most clinically relevant target cells, human alveolar cells (Xu et al., 2020), has been lacking. Here, we established a sphere culture system for human alveolus and found that SARS-CoV-2 infects alveolospheres with a high infectivity. This human-relevant platform recapitulates the anti-viral drug responses of alveolar cells and will contribute to the accurate pre-clinical testing of candidate drugs for the treatment of COVID-19.
Results
Long-term expansion of alveolospheres without a fibroblast niche
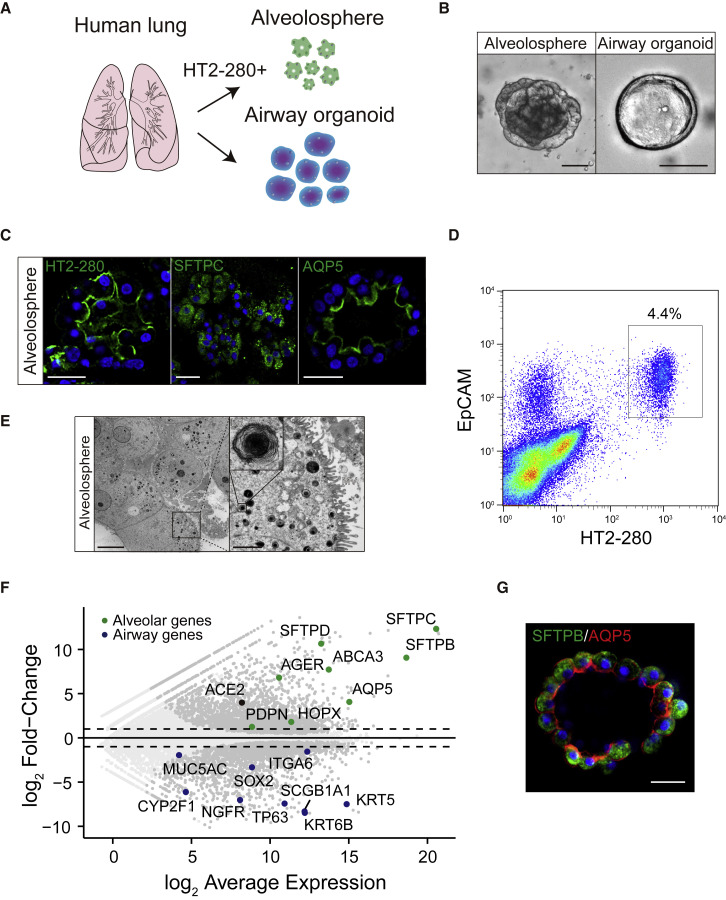
We isolated epithelium from the alveolar region of the human lung and cultured them with a previously reported culture condition for human airway organoids (Hui et al., 2018; Sachs et al., 2019). The established organoids showed a cystic morphology that resembles previously reported human airway organoids but lacked cells expressing alveolar markers (Sachs et al., 2019). Based on the culture condition for adult stem cell-derived organoids (Fujii et al., 2018; Fujii and Sato, 2021), we added the following generic niche factors to the airway organoid medium: Afamin-Wnt-3A, epidermal growth factor (EGF), Noggin, R-spondin, A83-01, insulin-like growth factor (IGF)-1, fibroblast growth factor (FGF)-2, FGF-7, and FGF-10 (WENRAIFs). This culture condition gave rise to not only airway organoids but also spheres with alveolar marker expression (Figures 1 A–1C). The spheres, hereafter referred to as alveolospheres, showed slow growth and were outcompeted by airway organoids over time. To selectively establish alveolospheres, we employed fluorescence-activated cell sorting (FACS) and sorted cells that expressed EpCAM (pan-epithelium marker) and HT2-280 (alveolar type 2 epithelium marker) (Gonzalez et al., 2010) (Figure 1D). The sorted cells formed pure alveolospheres. Electron microscopy depicted lamellar bodies and secretory organelles for the storage of surfactant proteins, and demonstrated the conservation of AT2 cells in alveolospheres at the ultrastructure level (Figure 1E). RNA sequencing (RNA-seq) analysis revealed that airway organoids expressed airway epithelium markers, such as KRT5, TP63, and SCGB1A1, whereas the alveolospheres expressed both AT2 (SFTPB, SFTPC, and ABCA3) and AT1 markers (AQP5 and HOPX) (Figure 1F). Immunostaining depicted the expression of both AT1 and AT2 markers in most alveolosphere cells. The dual-positive cells were reminiscent of intermediate AT2-AT1 cells, which are observed during alveolar regeneration in mice (Choi et al., 2020) (Figure 1G) and persisted following removal of niche factors or treatment with 10% serum (Figures S1A and S1B).
Figure 1.
Establishment of human alveolospheres
(A) Schematics of sorting-based isolation and establishment of alveolospheres from human lung tissues.
(B) Representative bright-field images of an alveolosphere (left) and airway organoid (right).
(C) Immunostaining of HT2-280 (green, left), SFTPC (green, middle), and AQP5 (green, right) in alveolospheres.
(D) Representative gating strategy for sorting HT2-280+ EPCAM+ human AT2 cells.
(E) Transmission electron microscopy images of alveolospheres. Microvilli (MVs) were observed in spheres at the apical lumen. The inset (right) shows a lamellar body (LB).
(F) An MA plot comparing the transcriptomes between alveolospheres (n = 4) and airway organoids (n = 2) (passages 4–6). Genes with false discovery rate (FDR) < 0.01 are shown in dark gray. Representative genes that are upregulated in alveolospheres (green) and airway organoids (blue) are highlighted. Dotted lines show log2 (fold change) values of 1 and −1.
(G) Co-staining of SFTPB (green) and AQP5 (red) in alveolospheres. Nuclear counterstaining, Hoechst 33342 (C and G). Scale bar: 2 μm (E, right), 10 μm (E, left), 25 μm (B, right, C, and G), and 100 μm (B, left).
See also Figure S1.
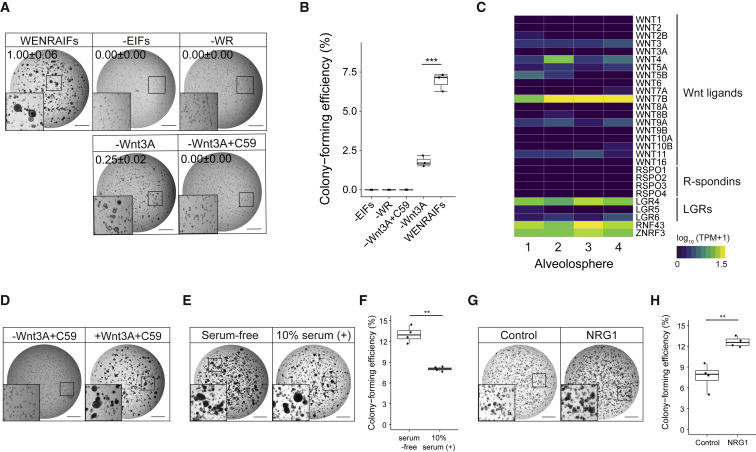
To define essential niche factors for alveolospheres, we analyzed the growth of spheres under various growth factor conditions. Activation of Wnt and ERK/AKT pathways was indispensable for the growth of alveolospheres, because removal of Wnt-3A/R-spondin or EIFs (EGF/IGF-1/FGF-2/FGF-7/FGF-10) significantly reduced sphere formation (Figures 2A and 2B). In contrast, alveolospheres showed a partial growth when Wnt-3A alone was removed, suggesting the contribution of autocrine/paracrine secretion of Wnt ligands (Figures 2A and 2B). Indeed, RNA-seq analysis identified robust expression of WNT7B that was previously shown to activate canonical Wnt signaling (Seino et al., 2018) in alveolospheres (Figure 2C). R-spondin-dependent sphere growth was abolished upon treatment with a porcupine inhibitor that abrogates the production of active Wnt ligands. Supplementation of Wnt-3A rescued this growth-suppressive effect, reinforcing the dependence of alveolospheres on autocrine/paracrine Wnt signaling (Figure 2D). Additionally, the expression of R-spondin receptor, LGR4, in alveolospheres was consistent with the requirement of R-spondin for efficient Wnt activation (de Lau et al., 2011) (Figure 2C). In addition, alveolospheres exhibited better growth when supplied with serum-free Afamin-Wnt-3A medium (Mihara et al., 2016) compared with the standard serum-stabilized Wnt-3A-conditioned medium. The difference between the effects of these Wnt conditioned media implied growth inhibition by serum components. In support of this, treatment with serum blunted the growth-promoting effect of Afamin-stabilized Wnt-3A medium (Figures 2E and 2F). Recombinant Wnt3A and Wnt7B failed to propagate alveolospheres, presumably because of their lower Wnt-activating potency than that of afamin-stabilized Wnt3A (Mihara et al., 2016) (Figures S2A and S2B).
Figure 2.
Refined culture condition of human alveolospheres
(A) Dependency of alveolospheres on Wnt, EGF, and FGF pathway activation. Images of alveolosphere (passage 7, day 18) cultured with Afamin-Wnt-3A, EGF, Noggin, R-spondin-1, A83-01, IGF-1, FGF-2, FGF-7, and FGF-10 (WENRAIFs, top left), WNRA (−EIFs, top right), ENAIFs (−WR, top middle), ENRAIFs (−Wnt3A, bottom left), or ENRAIFs and Porcupine inhibitor (C59) (−Wnt3A+C59, bottom right). Inset shows higher magnification. Organoid area relative to the WENRAIFs condition (mean ± SEM) was shown on each image.
(B) Colony-forming efficiency of alveolospheres (passage 7) cultured with WENRAIFs, −EIFs, −WR, −Wnt3A, or −Wnt3A+C59. Inset shows higher magnification. Data are demonstrated as mean ± SEM. ∗∗∗p < 0.001, Welch’s unpaired t test.
(C) mRNA expression (log10 (TPM+1)) of the indicated Wnt pathway-related genes in alveolospheres derived from four independent patients (n = 4).
(D) Representative images of alveolospheres (passage 7) cultured without Wnt-3A and C59 (left) and with Wnt-3A and C59 (right).
(E) Representative bright-field images of alveolospheres expanded from single cells cultured with WENRAIFs with (right) or without (serum-free, left) 10% serum. 5,000 cells were plated per well.
(F) Colony-forming efficiency of alveolospheres (passage 4) in the presence (right) or absence (serum-free, left) of 10% serum. Data are demonstrated as mean ± SEM. ∗∗p < 0.01, Welch’s unpaired t test.
(G) Representative bright-field images of alveolospheres cultured with WENRAIFs with (right) or without (control, left) neureglin-1 (NRG1).
(H) Colony-forming efficiency of alveolospheres (passage 8) cultured with (right) or without (control, left) NRG1. Data are demonstrated as mean ± SEM. ∗∗p < 0.01, Welch’s unpaired t test.
Scale bars: 1 mm (A, D, E, and G). See also Figure S2.
Alveolospheres exhibited steady yet slow growth and enabled bi- or tri-weekly passaging at a split ratio of 1:2. To improve scalability, we further screened other candidate niche factors that were nominated based on previous literature (Cao et al., 2016; Katsura et al., 2019; Liang et al., 2016; Nabhan et al., 2018; Zepp et al., 2017). One of the candidates, neuregulin-1 (NRG1), which is a ligand for tyrosine kinase receptors of the ERBB family, promoted the growth of alveolospheres (Figures 2G, 2H, S2C, and S2D). With NRG1 supplementation, we successfully derived nine lines of alveolospheres from surgically resected lung tissues. The NRG1-added culture condition allowed alveolospheres to expand for over 8 months, albeit with a gradual decrease of colony formation efficiency in the late phase (Figures S2E–S2G). NRG1 supplementation did not alter AT1-AT2 differentiation status (Figure S1B). Alveolospheres tolerated GFP transduction by electroporation (Figure S2H) and could potentially be applied to further experiments. These results together demonstrated robust propagation of alveolar cells in a fibroblast-free sphere culture system.
Robust SARS-CoV-2 infection modeling using human alveolospheres
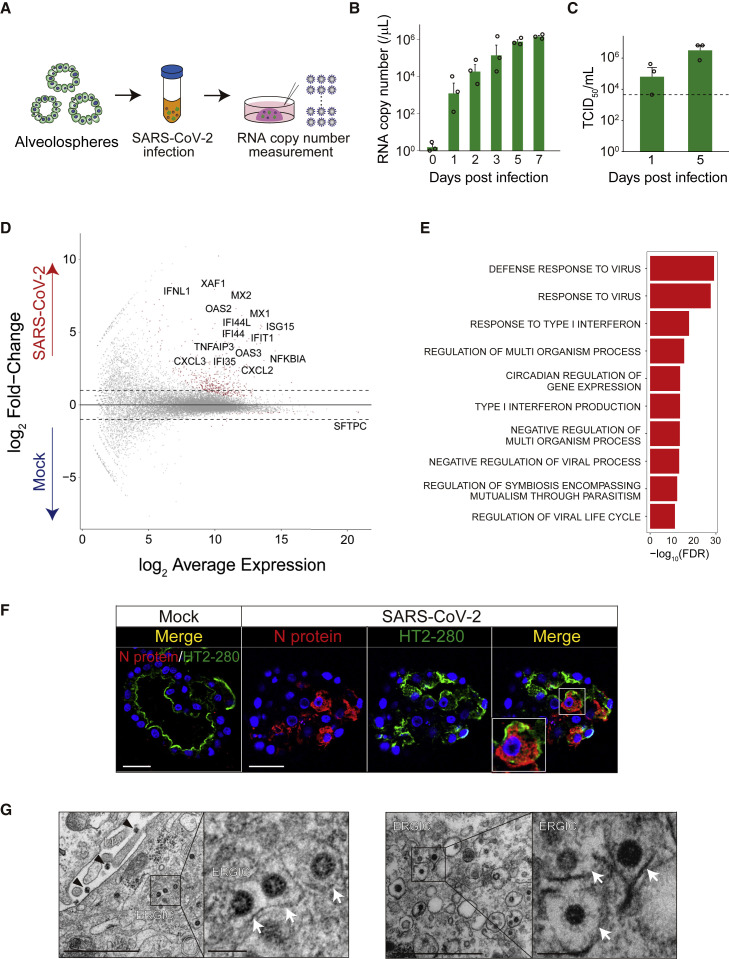
We next sought to develop a SARS-CoV-2 infection model using human alveolospheres. ACE2, a receptor for SARS-CoV-2, was robustly expressed in alveolospheres at RNA and protein levels (Figures 1F and S3A). We obtained viral particles from a patient diagnosed with COVID-19 in Japan. Following removal of Matrigel with a non-enzymatic matrix depolymerizer, the spheres were exposed to SARS-CoV-2 and re-embedded in Matrigel (Figure 3 A). Viral RNA copies in the medium increased nearly 1,000,000-fold on day 7 of infection, indicating sustained SARS-CoV-2 replication in alveolosphere culture (Figure 3B). A TCID50 assay further revealed steady replication and release of active SARS-CoV-2 (Figure 3C). In support of the robust host-virus responses in SARS-CoV-2-infected alveolospheres, their RNA-seq analysis revealed the enrichment of genes that are related to interferon response and apoptosis compared with the mock-infected alveolospheres (Figures 3D, 3E, S3B, and S3C). Around 40% of the RNA-seq reads in alveolospheres were mapped onto the viral genome (Figure S3D), demonstrating active SARS-CoV-2 infection. A long-term culture of alveolospheres did not affect the infection efficiency (Figure S3E). These results indicated efficient SARS-CoV-2 infection in human alveolospheres.
Figure 3.
A human alveolosphere-based platform for SARS-CoV-2 infection
(A) Schematics of SARS-CoV-2 infection to alveolospheres and measurement of viral RNA copy number in the supernatant.
(B) Analysis of SARS-CoV-2 RNA copy number in the culture medium of alveolospheres (passage 3) (multiplicity of infection [MOI] = 10). Copy numbers are shown as RNA copies per 1 μL of culture medium.
(C) TCID50 assay using the culture medium of alveolospheres (passage 3, MOI = 10). Dotted line indicates detection limit. The data of (B) and (C) are representative of at least three independent experiments.
(D) An MA plot comparing the transcriptomes between SARS-CoV-2-infected (n = 2) and mock-infected alveolospheres (n = 2) (MOI = 2). Genes with FDR < 0.01 are shown in red. Dotted lines show log2 (fold change) values of 1 and −1.
(E) Gene Ontology analysis of genes with FDR < 0.01 in SARS-CoV-2-infected alveolospheres.
(F) Co-staining of SARS-CoV-2 nucleocapsid (N) protein (red) and HT2-280 (top, green) or AQP5 (bottom, green) in mock-infected (left) or SARS-CoV-2-infected (right) alveolospheres (passage 5) (MOI = 5).
(G) Transmission electron microscopy images of human alveolospheres (passage 3) infected with SARS-CoV-2 (MOI = 5). Black arrowhead (top left) indicates secreted virions. White arrow indicates budding viral particles in ERGIC. Inset shows higher magnification.
Data are demonstrated as mean ± SEM (B and C). Scale bars, 100 nm (G, right), 1 μm (G, left), and 25 μm (F). Nuclear counterstaining, Hoechst 33342 (F). See also Figure S3.
Immunohistological analysis visualized the expression of SARS-CoV-2 nucleocapsid proteins in approximately 30% of infected alveolospheres (Figures 3F and S3F). Interestingly, most cells infected with SARS-CoV-2 expressed HT2-280+ but were devoid of SFTPC (Figures S3G and S3H). Electron microscopic imaging and immunogold staining confirmed intracellular and secreted SARS-CoV-2 particles with the structural nucleocapsid protein and membrane spike protein (Figures 3G and S3I). Of note, electron microscopy captured budding of SARS-CoV-2 from the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), suggesting that the sphere model can simulate the release of viral particles from ERGIC during SARS-CoV-2 infection (Stertz et al., 2007).
Clinically relevant anti-viral drug validation against SARS-CoV-2
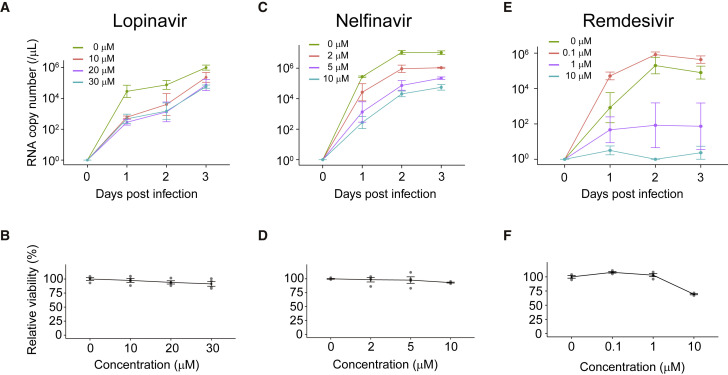
We next applied human alveolospheres to drug testing. A number of clinical trials are running to repurpose or reposition several drugs for the treatment of COVID-19. The budding of SARS-CoV-2 from ERGIC, which was known to be protease dependent, prompted us to select lopinavir and nelfinavir, protease inhibitors used for the treatment of human immunodeficiency virus (HIV) infection, from drug candidates (Cao et al., 2020; Chu et al., 2004; Musarrat et al., 2020). A recent study using Vero E6 cells demonstrated that lopinavir has an anti-viral effect on SARS-CoV-2 with IC50 and CC50 values of 9.12 and >50 μM, respectively (Jeon et al., 2020). Lopinavir induced a cytotoxic response in alveolospheres at a concentration of 40 μM (data not shown), which is lower than the CC50 value in Vero E6 cells, and we thus tested lopinavir within the range of 0–30 μM. Importantly, treatment with 10–30 μM lopinavir led to a significant decrease in SARS-CoV-2 viral RNA copy numbers on 1 day postinfection (dpi) (Figure 4 A). The treatment did not induce apparent cytotoxicity (Figure 4B), and alveolospheres tolerated a 3-day treatment with 20 μM lopinavir. Of note, a comparable level of drug concentration was observed in the sera of lopinavir-treated patients (Cao et al., 2020). Despite the therapeutic effect on 1 dpi, the viral titer increased over time (Figure 4A). A similar trend was observed for nelfinavir (Figures 4C and 4D), which is known to have an anti-viral effect at a lower concentration (Musarrat et al., 2020), and therefore these protease inhibitors may have a modest therapeutic potential. We next tested remdesivir, a viral RNA-dependent RNA polymerase inhibitor, given its efficacy in the treatment of COVID-19 (Beigel et al., 2020; Grein et al., 2020; Wang et al., 2020). Strikingly, remdesivir dramatically inhibited the replication of SARS-CoV-2 in alveolospheres over a 3-day period (Figures 4E and 4F). Taken together, these results demonstrated the utility of alveolospheres for clinically relevant in vitro validation of anti-viral drugs.
Figure 4.
Testing of anti-viral drugs using SARS-CoV-2-infected human alveolospheres
(A, C, and E) The time course of SARS-CoV-2 RNA copy number in the culture medium of alveolospheres (passage 4) treated with lopinavir (0–30 μM) (A), nelfinavir (0–10 μM) (C), and remdesivir (0–10 μM) (E) at indicated concentrations (MOI = 5).
(B, D, and F) The viability of alveolospheres after treatment with lopinavir (B), nelfinavir (D), or remdesivir (F) was analyzed by an ATP activity assay. The viability is shown as the relative value to the untreated control. Copy numbers are shown as RNA copies per 1 μL of culture medium (A, C, and E). Data are demonstrated as mean ± SEM (A–F).
Discussion
The pandemic of COVID-19 infection has been casting a devastating impact on the global society, and the development of its preventative and therapeutic measures is of utmost urgency for worldwide healthcare. To promptly respond to this crisis, the scientific community has established a number of in vitro SARS-CoV-2 infection models that facilitate our understanding of the disease pathophysiology and the development of therapeutic strategies. These models mainly employ classic cell lines, as well as human tissue-based systems, including colon and airway organoids (Lamers et al., 2020) and ex vivo-cultured respiratory tract epithelium (Hui et al., 2020). Human tissue-based SARS-CoV-2 infection models have not been applied to drug screening to date. A tractable culture system for adult alveolar epithelium, which is considered to be the critical site of SARS-CoV-2 infection, has also been lacking. Therefore, there has been an urgent demand for the development of a tissue-relevant culture system for human alveolar cells that enables robust propagation of SARS-CoV-2 and drug testing.
The culture of alveolar cells has classically adopted a 2D-culture format, in which AT2 cells preferentially differentiate into AT1 cells and lose self-renewing capacity within a few days (Wang et al., 2007). Due to this drawback, a previous study has failed to efficiently propagate SARS-CoV-2 in the 2D-cultured alveolar epithelium (Hou et al., 2020). Recently, researchers succeeded in the short-term propagation of alveolar epithelium using a 3D culture platform (Barkauskas et al., 2013; Evans and Lee, 2020). The growth of 3D-cultured alveoli in this system was strictly dependent on co-cultured fibroblasts, suggesting the essential role of fibroblast-derived niche factors in the maintenance of alveolar epithelium (Nabhan et al., 2018; Zepp et al., 2017). In the current study, we defined niche factors that enable the long-term self-renewal of the human alveolar epithelium. Similar niche factors, including Wnt, FGF, and EGF ligand families, are expressed in lung fibroblasts (Nabhan et al., 2018; Zepp et al., 2017), and such molecules may be key fibroblast-secreted factors that support alveolar growth in the previous co-culture system. In contrast to our culture condition, the co-culture format nevertheless did not allow long-term expansion of alveolospheres. This shortened lifespan may be attributed to undefined substances in the co-culture medium, including those included in the serum, which had a detrimental effect on alveolospheres. The standard Wnt-3A conditioned medium contains serum to stabilize the lipophilic Wnt protein, and its use has also complicated the establishment of alveolospheres. In the course of optimizing the culture condition for alveolospheres, we contingently replaced standard Wnt-3A conditioned medium with Afamin-stabilized serum-free Wnt-3A conditioned medium (Mihara et al., 2016), which enabled robust propagation of alveolospheres. During the preparation of our manuscript, reports on the propagation of the human alveolar epithelium have appeared (Katsura et al., 2020; Salahudeen et al., 2020; Youk et al., 2020). These studies also underscored the importance of activating Wnt signal without serum in expanding the alveolar epithelium (Katsura et al., 2020; Youk et al., 2020). Consistent with predominant AT2-AT1 intermediate cells in our alveolospheres, a single-cell RNA-seq (scRNA-seq) analysis of alveolospheres revealed the co-expression of AT1 and AT2 markers (Youk et al., 2020). Although other studies showed that human serum (Katsura et al., 2020) or culture on glass (Salahudeen et al., 2020) induced AT1 differentiation, further investigation will be required to derive a robust condition for efficient maturation of AT1 and AT2 cells. In the current study, we established alveolospheres using non-cancer lung tissues from patients with lung cancers or a lung hamartoma. Future studies will reveal to what extent non-tumor alveolar epithelium from cancer-bearing patients harbor (epi)genetic lesions.
As a step toward clinical translation of human alveolospheres, we established a clinically relevant model of SARS-CoV-2 infection. Consistent with the previous scRNA-seq data analysis (Sungnak et al., 2020), human alveolospheres expressed ACE2 and were highly susceptible to SARS-CoV-2 infection. Upon infection, we observed an increase in the viral titer in the supernatant by approximately six orders of magnitude. Electron microscopy confirmed viral replication at the ultrastructure level. These results indicate that our alveolosphere culture condition maintains the host molecular machinery required for the replication of SARS-CoV-2. One of the advantages of using alveolospheres in infection models is that, in contrast with the previous sphere-based models, alveolospheres allow for straightforward expansion of the cell types that SARS-CoV-2 target in the human body. Future studies may capitalize on the clinically relevant tissue origin to shed light on the pathogenesis of COVID-19. For instance, although accumulating evidence demonstrated strong associations between COVID-19 severity and underlying respiratory diseases, as well as smoking history (Leung et al., 2020), it remains unknown whether disease exacerbation in this scheme is directly driven by the disease-compromised epithelium or occurs secondary to the dysregulation of the immune system. Extensive studies using alveolospheres derived from patients with various respiratory diseases may address such as-yet-unanswered questions in the future.
We are currently facing a major challenge in developing effective anti-viral drugs against SARS-CoV-2, and the establishment of efficient drug screening systems has been warranted. However, recent studies on alveolosphere culture did not perform drug screening (Katsura et al., 2020; Salahudeen et al., 2020; Youk et al., 2020). Using the alveolosphere-based SARS-CoV-2 infection model, we determined the anti-viral effect of US Food and Drug Administration (FDA)-approved drugs that have been used for the treatment of COVID-19. Viral proteinase is an essential molecule that allows SARS-CoV-2 to be released from ERGIC and is a possible anti-viral drug target. Although two protease inhibitors, lopinavir and nelfinavir, significantly decreased viral titers at 1 dpi, both drugs could not terminate viral replication. In contrast with this result but consistent with human clinical data (Beigel et al., 2020; Grein et al., 2020; Wang et al., 2020), remdesivir showed a potent anti-viral effect at concentrations within the range of serum drug levels in patients. Current screening of anti-SARS-CoV-2 drugs mainly utilizes Vero cells as host cells. Immortalized cell lines are, however, less relevant to native human tissues than primary cells, and their routine use may lead to the underestimation of drug-induced cytotoxicity and potential adverse effects. Vero cells indeed tolerate an exposure to high-concentration lopinavir, whereas alveolospheres do not. Nevertheless, the slow growth of alveolospheres poses a limitation in applying our infection model to large-scale drug screening. Thus, the alveolosphere-based infection model is considered to be rather suitable for the validation of drugs selected by scalable cell-line-based drug screening. Such combinatory approaches will provide an excellent strategy to discover potent drugs against SARS-CoV-2 and other viruses that infect the lower respiratory system.
Due to the difficulty in conducting large-scale clinical trials at the early phase of the disease pandemic, the therapeutic effect of anti-viral drugs on COVID-19 largely remains controversial (Beigel et al., 2020; Wang et al., 2020). Bridging the gap between conventional in vitro drug testing systems and clinical practice using appropriate models for SARS-CoV-2 infection, including those using human alveolospheres, will contribute to the accurate evaluation of candidate drugs for the treatment of COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa fluor 647-mouse monoclonal anti-human CD326 (EpCAM) (clone 9C4) | BioLegend | Cat#324212, RRID: AB_756086 |
| Mouse IgG2b, κ isotype control antibody | BioLegend | Cat#400301 |
| Mouse monoclonal anti-HT2-280 | Terrace Biotech | Cat#TB-27AHT2-280, RRID: AB_2832931 |
| Alexa fluor 488-donkey anti-mouse IgM, μ chain specific | Jackson ImmunoResearch Laboratories | Cat#715-545-020, RRID: AB_2340844 |
| Mouse monoclonal anti-SARS-CoV/SARS-CoV-2 nucleocapsid | Sino Biological | Cat#40143-MM05, RRID: AB_2827977 |
| Mouse monoclonal anti-SARS-CoV-2 (COVID-19) spike (clone 1A9) | GeneTex | Cat#GTX632604, RRID: AB_2864418 |
| Mouse monoclonal anti-SFTPB | Santa Cruz Biotechnology | Cat#sc133143, RRID: AB_2285686 |
| Rabbit polyclonal anti-SFTPC | Millipore | Cat#AB3786, RRID: AB_91588 |
| Rabbit monoclonal anti-AQP5 | Abcam | Cat#ab92320, RRID: AB_2049171 |
| Goat polyclonal anti-ACE2 | R&D Systems | Cat#AF933, RRID: AB_355722 |
| Donkey polyclonal anti-mouse IgG (H+L), Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-21202, RRID: AB_141607 |
| Donkey polyclonal anti-mouse IgG (H+L), Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-31571, RRID: AB_162542 |
| Donkey polyclonal anti-rabbit IgG (H+L), Alexa Fluor 568 | Thermo Fisher Scientific | Cat#A10042, RRID: AB_2534017 |
| Donkey polyclonal anti-goat IgG (H+L), Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21447, RRID: AB_141844 |
| Goat anti-mouse IgG, colloidal gold (5 nm) conjugated | EY Laboratories | Cat#GAF-011-5 |
| Bacterial and virus strains | ||
| SARS-CoV-2 | This paper | JPN/Kanagawa/KUH003, DDBJ: LC630936 |
| Chemicals, peptides, and recombinant proteins | ||
| Advanced DMEM/F12 | Thermo Fisher Scientific | Cat#12634010 |
| HEPES | Thermo Fisher Scientific | Cat#15630080 |
| Penicillin-streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| GlutaMAX supplement | Thermo Fisher Scientific | Cat#35050061 |
| Matrigel | Corning | Cat#356231 |
| B-27 supplement | Thermo Fisher Scientific | Cat#17504044 |
| N-Acetyl-L-cysteine | Sigma-Aldrich | Cat#A9165 |
| [Leu15]-gastrin I human | Sigma-Aldrich | Cat#G9145 |
| Afamin-Wnt-3A serum-free conditioned medium | Mihara et al., 2016 | N/A |
| Recombinant human Wnt3A | R&D Systems | Cat#5036-WN |
| Recombinant human Wnt7B | Abnova | Cat#H00007477-P01 |
| Recombinant mouse EGF | Thermo Fisher Scientific | Cat#PMG8043 |
| Recombinant human IGF-1 | BioLegend | Cat#590904 |
| Recombinant human FGF-basic | Peprotech | Cat#100-18B |
| Recombinant human FGF10 | Peprotech | Cat#100-26 |
| Recombinant human KGF (FGF-7) | Peprotech | Cat#100-19 |
| Recombinant human Epiregulin | BioLegend | Cat#550202 |
| Recombinant human HB-EGF | Peprotech | Cat#100-47 |
| Recombinant human neuregulin-1 (Heregulinβ-1) | Peprotech | Cat#100-03 |
| Recombinant human HGF | Peprotech | Cat#100-39 |
| Recombinant human IL-6 | Peprotech | Cat#AF-200-06 |
| Recombinant human SDF-1 | Peprotech | Cat#300-28A |
| Recombinant human IL1-β | Peprotech | Cat#200-01B |
| Recombinant human TNF-α | Peprotech | Cat#300-01A |
| Wnt-C59 | ShangHai Biochempartner | Cat#1243243-89-1 |
| Recombinant mouse noggin | Peprotech | Cat#250-38 |
| R-spondin-1 conditioned medium | Ootani et al., 2009 | N/A |
| A83-01 | Tocris | Cat#2939 |
| SB202190 | Sigma-Aldrich | Cat#S7067 |
| Y-27632 | FUJIFILM Wako Pure Chemical | Cat#253-00513 |
| Fetal bovine serum | BioWest | Cat#S1820 |
| Cell recovery solution | Corning | Cat#354253 |
| Liberase TH research grade | Roche | Cat#05401151001 |
| TrypLE Express | Thermo Fisher Scientific | Cat#12605010 |
| Red Blood Cell Lysis Buffer | Roche | Cat#11814389001 |
| Propidium Iodide | FUJIFILM Wako Pure Chemical | Cat# 169-26281 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#H3570 |
| Puromycin | Thermo Fisher Scientific | Cat#A1113803 |
| G418, Geneticin | Thermo Fisher Scientific | Cat#10131035 |
| Lopinavir | Sigma-Aldrich | Cat#SML1222 |
| Nelfinavir | Selleck | Cat#S4282 |
| Remdesivir | Selleck | Cat#S8932 |
| ProLong Diamond Antifade Mountant | Thermo Fisher Scientific | Cat#P36961 |
| Power Block Universal Blocking Reagent | BioGenex | Cat#HK083-50K |
| BTXpress Solution | BTX | Cat#45-0805 |
| Opti-MEM I Reduced Serum Medium | Thermo Fisher Scientific | Cat#31985062 |
| Critical commercial assays | ||
| RNeasy Plus Mini Kit | QIAGEN | Cat#74134 |
| Direct-zol RNA MicroPrep Kit | Zymo Research | Cat#R2061 |
| TruSeq RNA Library Prep Kit v2 | Ilumina | Cat#RS-122-2001 |
| SARS-CoV-2 Detection Kit -N2 set- | TOYOBO | Cat#NCV-302 |
| CellTiter-Glo | Promega | Cat#G9681 |
| iPGell | GenoStaff | Cat#PG20-1 |
| Deposited data | ||
| Raw RNA sequencing count data of alveolospheres and airway organoids | This paper | GEO: GSE171430 |
| Raw RNA sequencing count data of mock-infected and SARS-CoV-2 infected alveolospheres | This paper | GEO: GSE171430 |
| Experimental models: Cell lines | ||
| Human: alveolospheres: see Table S2 | This paper | N/A |
| Human: airway organoids: see Table S2 | This paper | N/A |
| Vero E6/TMPRSS2 | Matsuyama et al., 2020 | JCRB: JCRB1819 |
| Recombinant DNA | ||
| PB-CMV-MCS-EF1a-GFP-Puro vector | System Biosciences | Cat#PB513B-1 |
| Super PiggyBac Transposase Expression Vector | System Biosciences | Cat#PB200PA-1 |
| Software and algorithms | ||
| R (version 3.6.0) | Comprehensive R Archive Network | https://cran.r-project.org/ |
| cutadapt (version 1.18) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/installation.html |
| STAR (version 2.6.1b) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| RSEM (version 1.3.3) | Li and Dewey, 2011 | https://github.com/deweylab/RSEM |
| R Bioconductor package DESeq2 | Bioconductor, Love et al., 2014 | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R Bioconductor package fgsea | Bioconductor, Korotkevich et al., 2019 (bioRxiv) | https://www.bioconductor.org/packages/release/bioc/html/fgsea.html |
| MSigDB v7.0 | Broad Institute | https://www.gsea-msigdb.org/gsea/msigdb/index.jsp |
| LAS X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/ |
| BZ-X analyzer | Keyence | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Toshiro Sato (t.sato@keio.jp).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The datasets generated during this study is deposited to Gene Expression Omnibus (GEO) under accession codes GSE171430. This study did not generate/analyze computer codes or algorithms.
Experimental model and subject details
Isolation and culture of human alveolospheres
Human normal lung samples were obtained from patients between Sep 19, 2018 and Jul 20, 2020, with written informed consent after an approval by the ethical committee of Keio University School of Medicine. All spheres were established and maintained in a CO2 incubator (5% CO2, 37°C) as previously described with modifications (Fujii et al., 2018; Sachs et al., 2019). Briefly, normal lung tissues of patients were sampled from macroscopically normal peripheral regions within 1–2 cm from the edge with gross margins of at least 10 cm from the tumors. The tissues were washed with ice-cold PBS and cut into small pieces. The fragments were digested with Liberase TH (Roche) at 37°C for 30 min. Prior to plating, collected epithelia were washed with PBS supplemented with 10% fetal bovine serum (FBS) to inactivate digestive enzymes. For isolation of alveolar type 2 (AT2) cells from the bulk population, the cells were stained and sorted by FACS using Alexa Fluor 647-conjugated anti-human EpCAM (9C4) (324212, BioLegend, 1:50) and anti-HT2-280 (TB-27AHT2-280, Terrace Biotech, 1:50) antibodies. Alexa Fluour 488-conjugated anti-mouse IgM antibody (715-545-020, Jackson ImmunoResearch, 1:100) was used as a secondary antibody. The cells were washed with cold PBS and stained with Propidium Iodide (FUJIFILM Wako Pure Chemical) to exclude dead cells. Single viable cells were gated and sorted. Control staining was performed with anti-mouse IgG2b, κ isotype control (400301, Biolegend, 1:50) and Alexa Fluour 647-conjugated anti-mouse IgG antibody (A-31571, Thermo Fisher Scientific, 1:50), and Alexa Fluour 488-conjugated anti-mouse IgM antibody without anti-HT2-280 antibody. Isolated AT2 cells, or non-AT2 where indicated, were embedded in Matrigel (Corning) and cultured with the medium for alveolospheres (Table S1). We seeded 30,000 cells per well (48-well plate, Corning), and typically observed around 10–30 organoid colonies per well. The screening of growth factors including recombinant human neuregulin-1 (Heregulinβ-1) is described below. Airway organoids were established as previously reported (Sachs et al., 2019). Spheres were passaged every 1–4 weeks using TrypLE express (Thermo Fisher Scientific) with gentle pipetting. We added 10 μM Y-27632 (FUJIFILM Wako Pure Chemical) for two days after passaging. Clinical characteristics of the tissue donors are available in Table S2.
Isolation and expansion of SARS-CoV-2
SARS-CoV-2 (JPN/Kanagawa/KUH003) was isolated in April 2020 from of a patient who had complained of sore throat. The patient’s nasopharyngeal swab sample tested positive for SARS-CoV-2 by PCR with a viral titer of 2.6 × 105/mL, and the patient was referred to Kitasato University Hospital in Japan. On admission, the patient did not complain symptoms except for a low-grade fever and showed a normal blood oxygen partial pressure. The serum was negative for SARS-CoV-2 S1 antibody. Viral particles were prepared by culturing virus-infected Vero E6/TMPRSS2 cells (JCRB1819) (Matsuyama et al., 2020) with DMEM with 2% FBS, 1 mg/mL G418 and 100 U/mL penicillin/100 μg/mL streptomycin for 48 h at 37°C in 5% CO2. The supernatant was cleared of cell debris and was stored at –80°C. The total nucleotide sequence data of this viral strain is deposited in DNA Data Bank of Japan (DDBJ) under accession number LC630936.
Method details
Growth factor screening for efficient expansion of human alveolospheres
To screen growth factors that promote the growth of human alveolospheres, we FACS-sorted and expanded HT2-280+ EpCAM+ alveolospheres as described above for three passages. Alveolospheres were dissociated into single cells with TrypLE Express and filtered using a 20-μm pore cell strainer (Sysmex), and 5,000 cells were plated onto each well of a 48-well plate. The cells were overlaid with the WENRAIFs medium supplemented with the following growth factors: recombinant human Epiregulin (BioLegend), recombinant human HB-EGF (PeproTech), recombinant human neuregulin-1 (Heregulinβ-1) (PeproTech), recombinant human HGF (PeproTech), recombinant human IL-6 (PeproTech), recombinant human SDF-1 (PeproTech), recombinant human IL1-β (PeproTech) and recombinant human TNF-α (PeproTech). The concentration of the growth factors was set to 100 ng/mL. Alveolospheres were also treated with 100 nM porcupine inhibitor (Wnt-C59; ShangHai Biochempartner) to block autocrine/paracrine Wnt signaling, or stimulated with 300 ng/mL recombinant human WNT3A (R&D Systems), 300 ng/mL recombinant human WNT7B (Abnova) or afamin-Wnt3A where indicated. Images of each well were captured using a BZ-X800 digital microscope (Keyence). Sphere area was automatically calculated using BZ-X800 Analyzer (Keyence). Colonies with areas > 2,500 μm2 were used for area measurement.
Electroporation of alveolospheres
GFP labeling of alveolospheres was performed by co-electroporating a piggyBac GFP-Puro expression vector (PB513B-1, System Biosciences) and a PBase vector (System Biosciences) into human normal alveolospheres as previously described (Fujii et al., 2015). Following selection with puromycin, GFP-positive clones were manually isolated and expanded.
RNA isolation
Human alveolospheres and airway organoids were cultured from single cells for 7–14 days in the identical culture medium containing Afamin-Wnt-3A, R-spondin-1, EGF, Noggin and A83-01. RNA was extracted from spheres using the RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions. Total RNA from mock-infected or SARS-CoV-2 infected (2 dpi) human alveolospheres cultured with optimized condition was extracted using the Direct-zol RNA MicroPrep Kit (Zymo Research) according to the manufacturer’s instructions.
RNA sequencing for normal alveolospheres and airway organoids
RNA quality was evaluated with an Agilent 2100 bioanalyzer (Agilent). Sequence library was prepared with TruSeq RNA Library Prep Kit v2 (Illumina) and sequenced with HiSeq X or NovaSeq 6000 (Illumina). Adaptors were removed from raw fastq files with cutadapt (version 1.18) (Martin, 2011) and the reads were aligned to human genome (hg38) using STAR (version 2.6.1b) (Dobin et al., 2013). The expression levels of the human genes in Ensembl release 81 were estimated with RSEM (version 1.3.3) (Li and Dewey, 2011). Differentially expressed gene analysis was performed using nbinomLRT in the R Bioconductor package DESeq2 (Love et al., 2014).
RNA sequencing for mock-infected and SARS-CoV-2-infected alveolospheres
RNA quality was evaluated with an Agilent 2100 bioanalyzer (Agilent). Sequence library was prepared with TruSeq RNA Library Prep Kit v2 (Illumina) and sequenced with HiSeq X Ten (Illumina). Adaptor sequences were removed from raw fastq files using cutadapt (version 1.18). Then, the reads were aligned to a combined reference genome comprising the hg38 and the SARS-CoV2 genome (GenBank: NC_045512.2) using STAR (version 2.6.1b). The expression levels of the human genes in Ensembl release 81 were estimated with RSEM (version 1.3.3). Differentially expressed gene analysis was performed using nbinomLRT function in R bioconductor package DESeq2. Gene ontology (GO) enrichment was tested with Fisher’s exact test and P values were adjusted with the Benjamini-Hochberg method. The GO biological process ontology database was obtained from MSigDB v7.0, and gene sets with size less than 15 or larger than 500 were discarded in the analysis. For gene set enrichment analysis (GSEA), read count data was normalized with size factors using DESeq2. GSEA was performed using the R Bioconductor package fgsea with 10,000 permutations (Korotkevich et al., 2019). Gene sets used in GSEA analysis (HALLMARK_INTERFERON_ALPHA_RESPONSE, GOBP_RESPONSE_TO_TYPE_I_INTERFERON, HALLMARK_APOPTOSIS and GOBP_EXTRINSIC_APOPTOTIC_SIGNALING_PATHWAY) were downloaded from Molecular Signatures Database (MSigDB v7.3)
SARS-CoV-2 infection to alveolospheres
Before viral infection, alveolospheres were released from Matrigel by treating Matrigel-embedded spheres with Cell Recovery Solution (Corning) for 30 min on ice. Released alveolospheres were suspended in the culture medium and were infected with SARS-CoV-2 (JPN/Kanagawa/KUH003) at multiplicity of infection (MOI) of 2–10 for 1 h at 37°C in 5% CO2. After viral infection, the alveolospheres were washed twice with the basal medium: Advanced DMEM/F-12 supplemented with 10 mM HEPES, 2 mM GlutaMAX and 100 U/mL penicillin/100 μg/mL streptomycin. Infected alveolospheres were suspended with Matrigel and dispensed onto 96-well plates as 20 μL droplets and overlaid with 200 μL of medium per well. Each well contained up to 10,000 cells. Three-μL of supernatant per well was harvested at indicated time points. The samples were stored at –80°C until measurement of the viral RNA copy numbers and/or TCID50. Alveolospheres treated with heat-inactivated virus (65°C for 10 min) served as mock-infected negative control. All experiments with live SARS-CoV-2 were performed in biosafety level 3 (P3) facilities.
Measurement of viral RNA copies
The Viral RNA copy number of SARS-CoV-2 was determined by measuring the amount of viral genomic RNA using quantitative real-time RT-PCR (qRT-PCR). Viral genomic RNA copies in cell culture supernatants were directly measured using a SARS-CoV-2 Detection Kit -N2 set- (TOYOBO) according to the manufacturer’s protocol. qRT-PCR was performed using a LightCycler 96 device (Roche).
Viral titration by TCID50 assay
Vero E6/TMPRSS2 cells were seeded on a 96-well tissue culture plate one day before titration assay. Confluent Vero E6/TMPRSS2 cells were washed once and overlaid with DMEM medium supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin and 2% FBS. Virus supernatants were serially diluted from 150 to 3 × 107 on a dilution plate. Diluted supernatants were added onto the Vero E6/TMPRSS2 plates in duplicate. After 4 days from infection, the end-point of viral dilution leading to cytopathic effect (CPE) in 50% of inoculated wells was estimated using the Reed and Muench method.
Drug testing for SARS-CoV-2 infection
SARS-CoV-2-infected alveolospheres were treated with lopinavir, nelfinavir and remdesivir at indicated concentrations immediately after re-embedding. Three-μL of the supernatant was harvested at indicated time point and kept at –80°C until measurement of viral RNA copy numbers. Drugs were added on 0 dpi, and the viral RNA copy numbers was measured as described above. To assess cytotoxicity of drugs, we plated single cell-dissociated alveolospheres at a density of 1,000 cells per well and treated them with drugs at indicated concentrations. Sphere viability assays were performed using CellTiter-Glo (Promega) on day 1 of treatment.
Transmission electron microscopy
Human alveolospheres with (2 dpi) or without infection of SARS-CoV-2 were immersed in 2% glutaraldehyde in 0.1 M PBS overnight at 4°C. Subsequently, samples were post-fixed with 2% osmium tetroxide for 2 h at 4°C, dehydrated in a series of ethanol and propylene oxide, and embedded in Epon 812. Ultrathin (80–90 nm) sections were prepared using an ultramicrotome, and the sections were double-stained with uranyl acetate and lead citrate. Images were captured with a transmission electron microscope (H-7600, Hitachi) at 100kV.
Immunoelectron microscopy
Human alveolospheres with infection of SARS-CoV-2 (2 dpi) were fixed with 4% PFA and 0.1% glutaraldehyde in 0.1 M PBS for 1h, and immersed in 70% ethanol for 30 min. Subsequently, samples were post-fixed with 1% osmium tetroxide, dehydrated, substituted with propylene oxide, and embedded in Epon 812 (64°C for 3 days). Ultrathin (100 nm) sections were collected on Collodion-coated nickel grids. The sections were treated with 10% sodium periodate for 30 min, 0.2 M Glycine/PBS for 30 min, 2% BSA/PBS for 20 min, and incubated with mouse anti-SARS-CoV-2 Spike (1A9) (GTX632604, GeneTex, 1:10 or 1:50) antibodies at room temperature overnight. After treatment with goat serum for 20 min, the sections were then incubated with Colloidal gold (5 nm) conjugated goat anti-mouse antibodies (GAF-011-5, EY Laboratories, 1:50) for 3 h at room temperature. Subsequently, the sections were post-fixed with 1% glutaraldehyde/PBS for 5 min and double-stained with uranyl acetate and lead citrate. Images were captured with a transmission electron microscope (H-7600, Hitachi) at 100kV of accelerating voltage.
Immunocytochemistry
Sections with a 4-μm thickness were prepared from archived paraffin-embedded spheres. Whole-mount immunofluorescent staining of spheres was performed as previously described (Fujii et al., 2018). Briefly, spheres were isolated from Matrigel using Cell Recovery Solution (Corning) and fixed with 4% paraformaldehyde for 20 min at room temperature. The spheres were washed three times with PBS and permeabilized with 0.2% Triton X-100 in PBS for 20 min at room temperature. The spheres were subsequently blocked with Power Block Universal Blocking Reagent (BioGenex) for 10 min at room temperature. The spheres were reacted with primary anti-bodies at 4°C overnight with gentle rocking. The spheres were then washed three times with PBS and stained with secondary antibodies for 30 min at room temperature with rocking and light protection. Primary antibodies used for imaging analysis were mouse anti-SARS-CoV/SARS-CoV-2 nucleocapsid (40143-MM05, Sino Biological, 1:200), mouse anti-HT2-280 (TB-27AHT2-280, Terrace Biotech, 1:200), mouse anti-SFTPB (sc133143, Santa Cruz Biotechnology, 1:200), rabbit anti-SFTPC (AB3786, Millipore, 1:200), rabbit anti-AQP5 (ab92320, Abcam, 1:200), and goat anti-ACE2 (AF933, R&D, 1:200) antibodies. Secondary antibodies were Alexa Fluor 488-, 568- or 647- conjugated anti-goat, -rabbit or -mouse antibodies (Thermo Fisher Scientific, 1:250). For double immunofluorescence with the primary antibodies from the same spices, we utilized microwaving between the first and second staining cycles. Nuclei were counterstained with Hoechst 33342 (Thermo Fisher Scientific, 1:1000). Images were captured using a confocal microscope (SP8, Leica).
Quantification and statistical analyses
Difference between means from separate groups was determined using Welch’s unpaired t tests. The level of significance is indicated as the P value in each experiment. Asterisks in figures indicate the following: ∗, P value < 0.05; ∗∗, P value < 0.01; ∗∗∗, P value < 0.001; n.s., P value > 0.05. The data are demonstrated as mean ± SEM. Graphs with statistical analysis were made with the R software. For further statistical details, refer to each figure legend.
Acknowledgments
We thank Chinatsu Yonekawa for technical assistance. This work was supported by the Japan Agency for Medical Research and Development (AMED) (grant numbers JP20nk0101612, JP21fk0108415, JP21cm0106576, and JP21ck0106471) and JSPS KAKENHI (grant number JP19K08610). This work was also supported by COVID-19 Kitasato project. A.M., and K.T. were supported by the Japan Society for the Promotion of Science Research Fellowships for Young Scientists.
Author contributions
Conceptualization, S.S., H.Y., K.K., and T.S.; methodology, T.E., S.S., K.K., and T.S.; investigation, T.E., S.S., K.H., A.M., R.T.-T., M.F., K.T., J.H., K.S., and K.K.; formal analysis, T.E. and K.T.; resources, T.H., H.A., K.F., H.Y., K.K., and T.S.; writing, T.E., S.S., M.F., and T.S.; visualization, T.E., S.S., and J.H.; funding acquisition, H.Y., K.K., and T.S.; project administration, K.K. and T.S.
Declaration of interests
T.S. is an inventor on several patents related to organoid culture.
Published: May 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109218.
Supplemental information
References
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., Hogan B.L. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. ACTT-1 Study Group Members Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Lis R., Ginsberg M., Chavez D., Shido K., Rabbany S.Y., Fong G.H., Sakmar T.P., Rafii S., Ding B.S. Targeting of the pulmonary capillary vascular niche promotes lung alveolar repair and ameliorates fibrosis. Nat. Med. 2016;22:154–162. doi: 10.1038/nm.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Park J.E., Tsagkogeorga G., Yanagita M., Koo B.K., Han N., Lee J.H. Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell. 2020;27:366–382.e7. doi: 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., Kao R.Y., Poon L.L., Wong C.L., Guan Y., et al. HKU/UCH SARS Study Group Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbadawi M., Efferth T. Organoids of human airways to study infectivity and cytopathy of SARS-CoV-2. Lancet Respir. Med. 2020;8:e55–e56. doi: 10.1016/S2213-2600(20)30238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K.V., Lee J.H. Alveolar wars: The rise of in vitro models to understand human lung alveolar maintenance, regeneration, and disease. Stem Cells Transl. Med. 2020;9:867–881. doi: 10.1002/sctm.19-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Sato T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 2021;20:156–169. doi: 10.1038/s41563-020-0754-0. [DOI] [PubMed] [Google Scholar]
- Fujii M., Matano M., Nanki K., Sato T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 2015;10:1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- Fujii M., Matano M., Toshimitsu K., Takano A., Mikami Y., Nishikori S., Sugimoto S., Sato T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell. 2018;23:787–793.e6. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Gonzalez R.F., Allen L., Gonzales L., Ballard P.L., Dobbs L.G. HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J. Histochem. Cytochem. 2010;58:891–901. doi: 10.1369/jhc.2010.956433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Ching R.H.H., Chan S.K.H., Nicholls J.M., Sachs N., Clevers H., Peiris J.S.M., Chan M.C.W. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 2018;6:846–854. doi: 10.1016/S2213-2600(18)30236-4. [DOI] [PubMed] [Google Scholar]
- Hui K.P.Y., Cheung M.C., Perera R.A.P.M., Ng K.C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.W., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. e00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Kobayashi Y., Tata P.R., Hogan B.L.M. IL-1 and TNFα Contribute to the Inflammatory Niche to Enhance Alveolar Regeneration. Stem Cell Reports. 2019;12:657–666. doi: 10.1016/j.stemcr.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J., et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019 doi: 10.1101/060012. [DOI] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J.M., Niikura M., Yang C.W.T., Sin D.D. COVID-19 and COPD. Eur. Respir. J. 2020;56:2002108. doi: 10.1183/13993003.02108-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Zhang Y., Xie T., Liu N., Chen H., Geng Y., Kurkciyan A., Mena J.M., Stripp B.R., Jiang D., Noble P.W. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat. Med. 2016;22:1285–1293. doi: 10.1038/nm.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet. 2011;17:10–12. [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara E., Hirai H., Yamamoto H., Tamura-Kawakami K., Matano M., Kikuchi A., Sato T., Takagi J. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/α-albumin. eLife. 2016;5:e11621. doi: 10.7554/eLife.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarrat F., Chouljenko V., Dahal A., Nabi R., Chouljenko T., Jois S.D., Kousoulas K.G. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J. Med. Virol. 2020;92:2087–2095. doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan A.N., Brownfield D.G., Harbury P.B., Krasnow M.A., Desai T.J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., Kuo C.J. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Böttinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen A.A., Choi S.S., Rustagi A., Zhu J., van Unen V., de la O S.M., Flynn R.A., Margalef-Català M., Santos A.J.M., Ju J., et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino T., Kawasaki S., Shimokawa M., Tamagawa H., Toshimitsu K., Fujii M., Ohta Y., Matano M., Nanki K., Kawasaki K., et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell. 2018;22:454–467.e6. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. In Vitro and Animal Models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020;41:513–517. doi: 10.1016/j.tips.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Edeen K., Manzer R., Chang Y., Wang S., Chen X., Funk C.J., Cosgrove G.P., Fang X., Mason R.J. Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am. J. Respir. Cell Mol. Biol. 2007;36:661–668. doi: 10.1165/rcmb.2006-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J., Kim T., Evans K.V., Jeong Y.I., Hur Y., Hong S.P., Kim J.H., Yi K., Kim S.Y., Na K.J., et al. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–919.e10. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp J.A., Zacharias W.J., Frank D.B., Cavanaugh C.A., Zhou S., Morley M.P., Morrisey E.E. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell. 2017;170:1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study is deposited to Gene Expression Omnibus (GEO) under accession codes GSE171430. This study did not generate/analyze computer codes or algorithms.