Abstract
The coronavirus disease-19 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is rampant in the world and is a serious threat to global health. The SARS-CoV-2 RNA has been detected in various environmental media, which speeds up the pace of the virus becoming a global biological pollutant. Because many engineered nanomaterials (ENMs) are capable of inducing anti-microbial activity, ENMs provide excellent solutions to overcome the virus pandemic, for instance by application as protective coatings, biosensors, or nano-agents. To tackle some mechanistic issues related to the impact of ENMs on SARS-CoV-2, we investigated the molecular interactions between carbon nanoparticles (CNPs) and a SARS-CoV-2 RNA fragment (i.e., a model molecule of frameshift stimulation element from the SARS-CoV-2 RNA genome) using molecular mechanics simulations. The interaction affinity between the CNPs and the SARS-CoV-2 RNA fragment increased in the order of fullerenes < graphenes < carbon nanotubes. Furthermore, we developed quantitative structure–activity relationship (QSAR) models to describe the interactions of 17 different types of CNPs from three dimensions with the SARS-CoV-2 RNA fragment. The QSAR models on the interaction energies of CNPs with the SARS-CoV-2 RNA fragment show high goodness-of-fit and robustness. Molecular weight, surface area, and the sum of degrees of every carbon atom were found to be the primary structural descriptors of CNPs determining the interactions. Our research not only offers a theoretical insight into the adsorption/separation and inactivation of SARS-CoV-2, but also allows to design novel ENMs which act efficiently on the genetic material RNA of SARS-CoV-2. This contributes to minimizing the challenge of time-consuming and labor-intensive virus experiments under high risk of infection, whilst meeting our precautionary demand for options to handle any new versions of the coronavirus that might emerge in the future.
Keywords: COVID-19, Coronavirus, Genetic material, Nanomaterials, Interaction
Graphical Abstract
1. Introduction
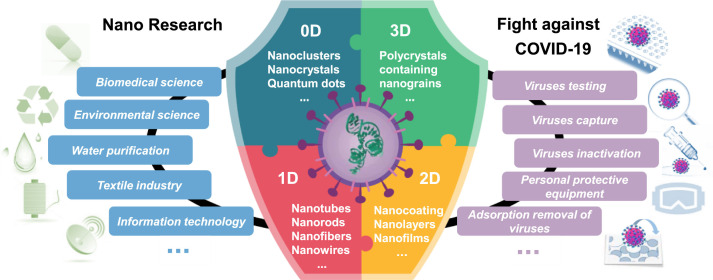
The outbreak of a new coronavirus which by now has lasted for more than a year, has brought great disaster to human beings (Diffenbaugh et al., 2020, Slot et al., 2020, Snape and Viner, 2020). To deal with the most serious global public health emergency in recent decades, the governments of various countries have employed a number of measures to control the epidemic situation (Cheng et al., 2020). Scientists from various fields are stimulated to either improve existing or to develop new “weapons” against the coronavirus (Allawadhi et al., 2020, Florindo et al., 2020, Talebian et al., 2020). In response to the virus pandemic, nanoscience and nanotechnology are offering opportunities and challenges ( Fig. 1).
Fig. 1.
Use of compositional ENM libraries, including zero-dimension (0D), one-dimension (1D), two-dimension (2D), and three-dimension (3D) nanomaterials, to link nanotechnology-based application to virus fighting experience.
Viruses such as the avian influenza virus (H5N1), the severe acute respiratory syndrome coronavirus (SARS), the swine influenza virus (H1N1), and the Middle East respiratory syndrome coronavirus (MERS-CoV) are nature’s nanostructures (Kostarelos, 2020) which easily enter biological entities because of their special nanostructure-related advantages. Similarly, engineered nanomaterials (ENMs) also have such special properties due to the small size and hence relative large surface to volume ratio, which is in part why ENMs have been widely used for a variety of biomedical applications. In this respect there is a growing need for design of ENMs that are highly specific and efficiently taken up into target cells. The controllable physicochemical properties (e.g., size, shape, and surface) of ENMs facilitate their direct interactions with viral particles (e.g., interacting with viral envelope proteins and nucleic acids) or with host cell surface receptors to inhibit virus-cell interactions (Chen et al., 2020). Hence, there is an urgent need to realize whether ENMs as anti-viral nano-agents can offer effective therapeutic strategies to combat the emergence of the coronavirus disease-19 (COVID-19). Although there has been criticism about the negative impact of nanotechnology, the time for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has now come to highlight the knowledge and previous experience of nanotechnologists in vaccine and drug development, delivery, and distribution (Anonymous, 2020). At the same time, the presence of the SARS-CoV-2 RNA in the environment such as in air (Mohan et al., 2021, Morawska and Cao, 2020) and water (Kitajima et al., 2020, Mohan et al., 2021) requires us to confirm whether ENMs because of their strong tendency to adsorb to any biotic or abiotic moiety, can efficiently remove the novel coronavirus. Understanding the interaction mechanisms between ENMs and macromolecules that are part of SARS-CoV-2 is an important foundation for the application of ENMs versus COVID-19.
Molecular simulation methods could be effective tools in exploring the interactions between ENMs and biomacromolecules with key biological functions (Ge et al., 2011), which would facilitate the design of novel nano-agents and improve the development and application of new therapeutic techniques. Quantitative structure–activity relationships for nanomaterials (nano-QSARs) are guided by the classical QSAR model and combine impacts of (non-tested) ENMs with their specific physicochemical properties (Chen et al., 2017, Puzyn et al., 2011). This provides a new way for rapid screening and priority testing of those ENMs that are predicted to be the most effective anti-viral agents. Combined with molecular simulation, nano-QSAR will play an increasingly important role in the fight against COVID-19 and future virus pandemics.
Many ENMs are known to exhibit high anti-microbial activity; either via induction of oxidative stress by for instance metal-based ENMs from which ions dissolve having the ability to generate oxidative stress (Sánchez-López et al., 2020), or via photothermal/photocatalytic effects, lipid extraction, inhibition of bacterial metabolism, isolation by wrapping the microbes (Maleki Dizaj et al., 2015) with a nano-layer due to the high adsorption and high mechanical strength of ENMs. Also carbon nanoparticles (CNPs) are recognized as a promising nanomaterial for the detection, filtering, and inactivation of viruses. According to recent studies the physical interaction of carbon-based nanomaterials with bacteria, rather than oxidative stress, is the primary antimicrobial activity of these nanostructures (Maleki Dizaj et al., 2015). An additional benefit of carbon-based nanoparticles is that their high bio-safety and high biological compatibility, which are the characteristics used to comply with various medical drugs (Aasi et al., 2020, Łoczechin et al., 2019, Palmieri and Papi, 2020, Vermisoglou et al., 2020). Therefore, this work is devoted to assessing and quantifying the interactions of the key fragment of the SARS-CoV-2 RNA (Zhang et al., 2020) with CNPs of different type and dimension. The fragment is a model molecule of frameshift stimulation element (FSE) from the SARS-CoV-2 RNA genome (Zhang et al., 2020). What is more, the FSE plays an important role in the virus replication cycle and has emerged as a major drug target (Lan et al., 2021). Subsequently, a predictive model is developed which quantifies the relationship between the structural properties of CNPs and these interactions.
2. Computational methods
2.1. Annealing simulations
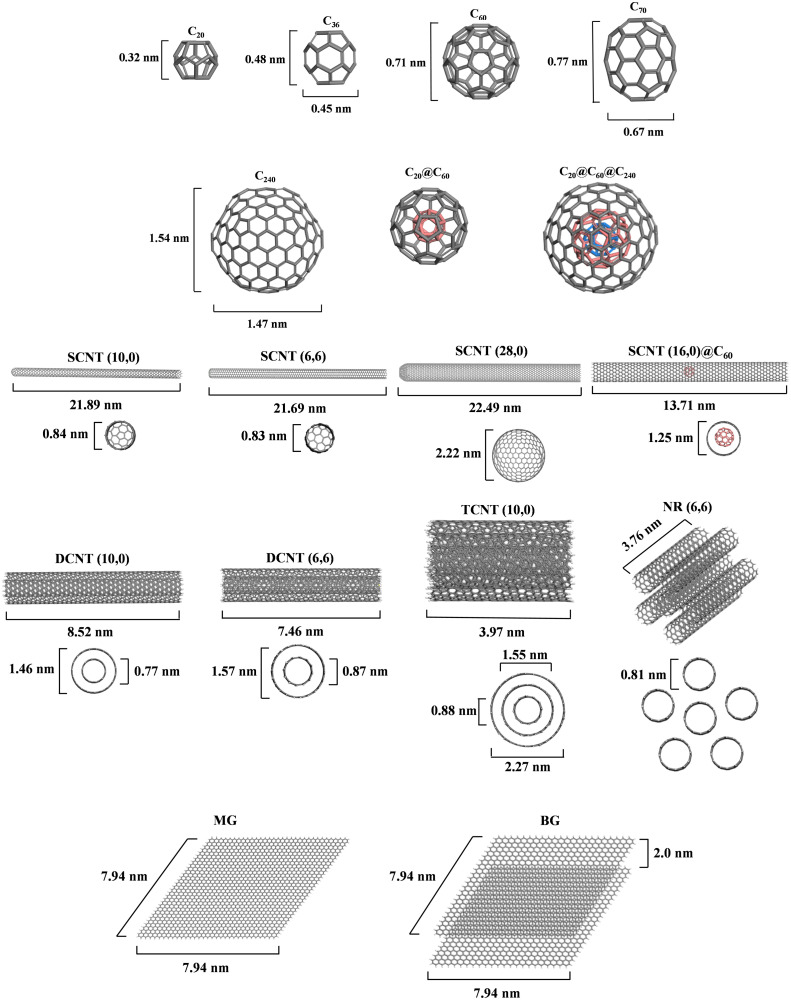
The three-dimensional structure of the SARS-CoV-2 fragment determined by Zhang et al. (2020) was used for the simulations. Seventeen CNPs from three families of fullerenes, carbon nanotubes (CNTs), and graphene were selected as model molecules ( Fig. 2). The constructed fullerene molecular models include C20, C36, C70, C240, carbon nanoballs C60 and C20@C60, as well as the carbon nano-onion C20@C60@C240. The constructed carbon nanotube molecular models include single-walled carbon nanotube (SCNT) (10,0), SCNT (6,6), SCNT (28,0), double-walled carbon nanotube (DCNT) (10,0), DCNT (6,6), triple-walled carbon nanotube (TCNT) (10,0), nanorope (NR) (6,6), and the complexes SCNT (16,0) with C60 named SCNT (16,0)@C60. The constructed graphene molecular models include mono-layer graphene (MG) and bilayer graphene (BG). To search for the best geometry with various forms of energy for each complex of the CNPs with the SARS-CoV-2 RNA fragment, a classical annealing simulation was performing with the Materials Studio software package (Ver. 8.0). The universal force field was adopted to perform this simulation. The cutoff radius was chosen to be 18.5 Å. The annealing simulation was performed as follows: a total of 200 annealing cycles which represented an optimal balance of total energy (Fig. S1, Supplementary Material) were simulated with an initial temperature of 200 K, a midcycle temperature of 300 K, and 50 heating ramps per cycle, with 100 dynamic steps per ramp. The canonical ensemble (NVT ensemble, in which the number of molecules [N], volume [V], and temperature [T] of the system are kept constant) was used and the molecular dynamic simulations were performed with a time step of 1.0 fs and a Nosé thermostat. After each cycle, the lowest energy configuration was optimized. The ‘van der Waals’ energies, electrostatic and total potential energies of the studied systems were calculated using the annealing simulation.
Fig. 2.
Structure, morphology, and character of the studied models of carbon nanoparticles.
For the interaction systems, interaction energy (E int) is used to evaluate the stability of the complexes of the CNPs with the SARS-CoV-2 RNA fragment. The magnitude of E int is an indication of the magnitude of the driving force towards complexation. A negative value reflects a stable adsorption on the CNPs. E int was calculated by
| (1) |
where E CNP-covRNA, E CNP, and E covRNA represent the energies (van der Waals, electrostatic, or total potential energies) of the complex, the isolated CNPs, and the individual SARS-CoV-2 RNA fragment, respectively.
2.2. Development of a predictive nano-QSAR model for Eint
Based on the interactions between the CNPs and the SARS-CoV-2 RNA fragment implied by the annealing simulations, several constitutional geometric and topological descriptors ( Table 1) such as molecular weight (M W), overall surface area (OSA), volume (Vol), specific surface area (SSA), and sum of degrees (SDeg), were selected to correlate with E int so as to construct predictive models. OSA and Vol were calculated using Multiwfn 3.8 software (Lu and Chen, 2012a, Lu and Chen, 2012b). The SSA values were obtained directly from the derivation of OSA and Vol. SDeg was calculated using Chem3D Ultra (Ver. 19.0). Orthogonal partial least squares (OPLS) regression was performed with Simca (Ver. 14.1 Umetri AB & Erisoft AB) to select variables and to develop models. Randomization tests proposed for testing the rationality of the models were performed using the RAND () function to generate the pseudo-random numbers of the E int derived from the total potential energy.
Table 1.
Molecular parameters of the carbon nanoparticles.
| CNPsa | Chemical formula | Mol weight (g/mol) | Overall surface area (nm2) | Volume (nm3) | Specific surface area (m2/g) | Sum of degrees |
|---|---|---|---|---|---|---|
| C20 | C20 | 240.220 | 1.859 | 0.234 | 4659.147 | 60 |
| C36 | C36 | 432.396 | 2.678 | 0.404 | 3729.094 | 108 |
| C60 | C60 | 720.660 | 3.812 | 0.645 | 3185.747 | 180 |
| C70 | C70 | 840.770 | 4.325 | 0.750 | 3098.086 | 210 |
| C240 | C240 | 2882.640 | 13.127 | 2.538 | 2742.369 | 720 |
| C20@C60 | C80 | 960.880 | 4.340 | 0.824 | 2720.323 | 240 |
| C20@C60@C240 | C320 | 3843.520 | 10.283 | 3.047 | 1611.094 | 960 |
| SCNT (10,0) | C2010H22 | 24,164.286 | 108.576 | 21.858 | 2705.906 | 6010 |
| SCNT (6,6) | C2100 | 25,223.100 | 113.148 | 22.822 | 2701.464 | 6288 |
| SCNT (28,0) | C5846 | 70,291.912 | 314.016 | 62.889 | 2690.280 | 1098 |
| DCNT (10,0) | C2282H58 | 29,767.822 | 65.384 | 22.949 | 1322.735 | 1495 |
| DCNT (6,6) | C2080H68 | 27,148.064 | 61.193 | 21.367 | 1357.418 | 1454 |
| TCNT (10,0) | C2146H130 | 28,069.814 | 50.583 | 21.944 | 1085.212 | 2173 |
| NR (6,6) | C2160H144 | 28,266.192 | 92.495 | 23.584 | 1970.605 | 3204 |
| SCNT (16,0)@C60 | C2108H32 | 27,415.828 | 109.455 | 22.416 | 2404.270 | 1091 |
| MG | C2046H126 | 26,763.882 | 124.221 | 23.031 | 2795.087 | 6012 |
| BG | C2112H180 | 27,677.568 | 77.180 | 22.648 | 1679.300 | 6156 |
= more details in Fig. 2.
3. Results and discussion
3.1. Modeling the interaction of CNPs with the SARS-CoV-2 RNA fragment
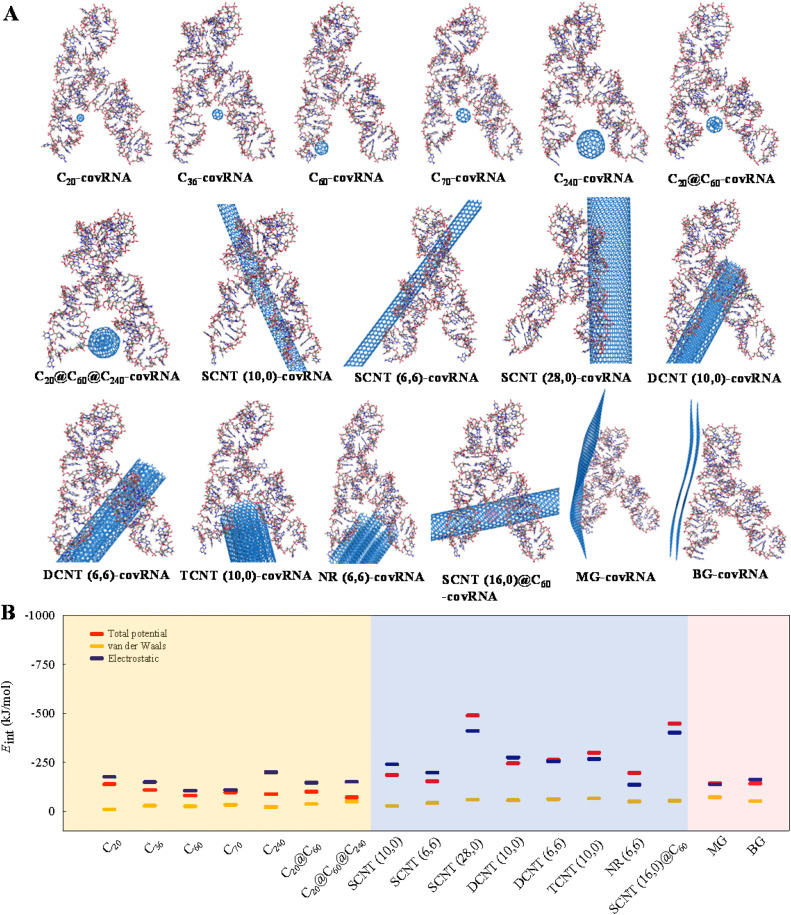
In order to reveal the mechanisms of the interactions of CNPs with the SARS-CoV-2 RNA fragment, the E int derived from the total potential energies, the ‘van der Waals’ energies, and electrostatic energies are summarized in Fig. 3 and Table S1 (Supplementary Material). The optimized conformations obtained after the annealing simulations are shown in Fig. 3A. The computed values are negative, indicating that the CNPs can form stable complexes with the SARS-CoV-2 RNA fragment. Fig. 3B shows that CNTs have the highest absolute energy of interaction with the SARS-CoV-2 RNA fragment among the studied CNPs, as derived from the total potential energies. This suggests a strong interaction between the CNTs and the SARS-CoV-2 RNA fragment.
Fig. 3.
Optimized structures of the complexes of the CNPs with the SARS-CoV-2 RNA fragment (abbreviated as covRNA) obtained after the annealing/geometry optimization procedure (A) and the calculated total potential energy interaction energies (Eint), ‘van der Waals’ interaction energies, and electrostatic interaction energies between the CNPs and the SARS-CoV-2 RNA fragment using the simulated annealing method. The first 7 pictures (A) and the orange block (B) represent the fullerenes, the middle 8 pictures (A) and blue block (B) represent the nanotubes, the last 2 pictures (A) and the pink block (B) represent the graphenes.
Generally, the interaction affinity between the CNPs and the SARS-CoV-2 RNA fragment increased in the order of fullerenes < graphenes < CNTs. In addition, the computed electrostatic interaction energies between the CNPs and the SARS-CoV-2 RNA fragment are similar to the E int values derived from the total potential energies (Fig. 3B). This implies that the electrostatic interaction contributes mainly to the mechanism of interaction. Wang et al. (2017) also concluded that electrostatic interactions contribute to the gaseous adsorption energies of organic molecules onto carbon-based nanomaterials by means of polyparameter linear free energy relationships. As the SARS-CoV-2 has a positive charge (Zhang et al., 2020), whereas the studied CNPs are neutral, the electrostatic interactions are mainly ion-induced dipole interactions.
3.2. Nano-QSAR prediction of the interaction of CNPs with the SARS-CoV-2 RNA fragment
The OPLS regression technique was used to find the most suited descriptors (Table 1) for developing models to quantify the E int derived from the total potential energies ((2), (3), (4)). SSA is the parameter that most significantly correlates with the E int values of fullerenes, and there is a positive correlation between SSA and the absolute value of E int. For CNTs and graphenes, OSA and SDeg are the parameters that correlate most significantly with the E int values. At the same time, OSA showed a positive correlation with the absolute E int, whereas SDeg displayed a negative correlation with the absolute E int. For the whole set of fullerenes, CNTs, and graphenes, M W and SDeg are the parameters correlating most significantly with the E int values. Moreover, M W presented a positive correlation with the absolute value of E int, while SDeg had a negative correlation with the absolute value of E int.
Among the selected descriptors, M W usually describes the size of a molecule. SDeg as a molecular descriptor of topology is the sum of degrees of every atom, and an atom’s degree is the number of non-hydrogen atoms to which it is bonded. Moreover, SSA and OSA are known to be associated with the steric structures of NPs. Note that surface properties such as surface area generally dominate the behavior (Yang and Xing, 2010) and effects (Mottier et al., 2016) of CNPs. Taken together, the selected nano-specific descriptors not only are easy to obtain, but also can explain the interaction mechanism.
Fullerenes:
| (2) |
n = 7, R 2 = 0.804, RMSE = 0.485, Q2 CUM = 0.737.
CNTs and graphenes:
| (3) |
n = 10, R 2 = 0.849, RMSE = 0.440, Q2 CUM = 0.681.
Fullerenes, CNTs, and graphenes:
| (4) |
n = 17, R 2 = 0.804, RMSE = 0.473, Q2 CUM = 0.710.
where n stands for the number of CNPs, R 2 is squared regression coefficient, RMSE is root mean squared error, and Q2 CUM is the cumulative percentage of variance explained for extracted components. The values of Q2 CUM of the models are higher than 0.5, suggesting the good robustness and internal predictability of the models and the models thus have high goodness-of-fit.
To further ensure the reliability of the obtained models, randomization tests were carried out by generating a fake pool of data for the E int values derived from the total potential energies (Table S2, Supplementary Material). The E int values were scrambled in two ways, namely a single sample one and a full one, to generate the pseudo-random numbers. As shown in Table S2 (Supplementary Material), all the OPLS models obtained with the scrambled data exhibited non-competitive R 2 and Q2 CUM values, as comparison to the three models provided in (2), (3), (4). Thus, it is clear that the developed models are reliable and grasp the most significant information used to interpret the interactions of CNPs with the SARS-CoV-2 RNA fragment. The outcome of the randomization testing also shows that the nano-specific descriptors are relevant.
3.3. Implications of nano-QSAR based approaches in battling coronaviruses
A virus can be regarded as a nanoscale particle consisting of outer-capsid proteins and inner-core nucleic acids (RNA or DNA). ENMs can not only directly interact with viral particles including the envelope protein and the nucleic acids, but they can also competitively bind with the cell receptors. As aforementioned, CNPs can interact with the SARS-CoV-2 RNA fragment and stabilize it. Knowing the interaction affinity between ENMs and virus particles is important for accurately inferring the efficacy of antiviral nano-agents, which can be applied to disrupt the viral replication cycle (Chen et al., 2020) and even directly to destroy its structure.
The SARS-CoV-2 RNA has been detected in environmental media (Al Huraimel et al., 2020, Kitajima et al., 2020, Mohan et al., 2021, Morawska and Cao, 2020), which causes the novel coronavirus to become an environmental pollutant especially in air, in sewage (e.g. via the stool of contaminated patients) and in watersheds. It is known that ENMs, especially carbon-based nanomaterials, are widely utilized in the adsorption and separation of environmental pollutants because of their strong adsorption capacity and high adsorption efficiency (Ji et al., 2013, Pan and Xing, 2008, Wang et al., 2017, Yang and Xing, 2010). It is reported that the SARS-CoV-2 RNA is likely to persist for a long time in untreated wastewater (Ahmed et al., 2020). Hence, it is important to elucidate the interactions of the CNPs with the SARS-CoV-2 RNA fragment. Besides, the knowledge of these interactions can deepen and expand related research in other nanotechnology-based applications, e.g., disinfectants for personal protective equipment and sensors for SARS-CoV-2 detection.
All human beta-coronaviruses share a certain degree of genetic and structural homology (Shin et al., 2020). As reported, the SARS-CoV-2 genome sequence homology with SARS-CoV and MERS-CoV is 77% and 50%, respectively (Kim et al., 2020). Hence, the nano-QSAR models developed for SARS-CoV-2 in the present study are likely to be suitable for forecasting the interactions between the CNPs and other beta-coronaviruses. Furthermore, we advocate to keep the modeling as simple as it can be, and to filter those molecular descriptors which are easy to obtain and are related to antimicrobial (physical) properties. In the face of the urgency of the COVID-19 pandemic, nano-QSAR is a useful tool to investigate the impacts of nanotechnology on the novel coronavirus, and it has the advantages of preliminary screening of effective ENMs that will save valuable research time - the step towards validating the models by means of experimental research can be started and done faster in a justified way. Eventually this may lead to saving efforts and preventing infection during experimental testing.
Nanomaterials entered the consumer market around 2000, meaning that by now nanoparticles are used in many products. For instance, within toothpaste titanium dioxide nanoparticles can be found as well as in creams to whiten products (Braakhuis et al., 2021, Rompelberg et al., 2016), and silver nanoparticles are used within many cosmetics like anti-aging creams (Kaul et al., 2018). The use of carbon-based nanomaterials for antiviral purposes is not so far off (Patel et al., 2019), and the virus killing activity of differently shaped carbon-based nanomaterials is intensively discussed (Innocenzi and Stagi, 2020, Serrano-Aroca et al., 2021). Serrano-Aroca et al. (2021) concluded that carbon-based nanomaterials had antiviral activity against 13 enveloped positive-sense single stranded RNA viruses, including SARS-CoV-2. It has been shown that the toxicity of nanomaterials is difficult to unravel, the antimicrobial activity of the nanoparticles depends on their composition, surface modification, intrinsic properties (Innocenzi and Stagi, 2020). Especially for unwanted toxicity, rigid high aspect ratio carbon fibers might be an issue in this respect that need to be dived furth into. Nonetheless, the use of carbon-based biocompatible nanomaterials as antivirals is still an almost unexplored field, while the published results show promising prospects.
4. Conclusions
To sum up, through molecular mechanics simulations, we have mainly addressed the molecular interactions between CNPs and the SARS-CoV-2 RNA fragment. The estimated E int suggests that the electrostatic interaction could be the predominant driving force for the interactions. The models on E int developed by OPLS show high goodness-of-fit and robustness. Four nanostructural descriptors (M W, SSA, OSA, and SDeg) were found to be the decisive factors controlling E int.
CRediT authorship contribution statement
Fan Zhang: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Writing - original draft preparation, Visualization, Funding acquisition. Zhuang Wang: Conceptualization, Formal analysis, Resources, Writing - review & editing, Funding acquisition, Project administration. Martina G. Vijver: Conceptualization, Formal analysis, Writing - review & editing, Supervision, Funding acquisition, Project administration. Willie J. G. M. Peijnenburg: Conceptualization, Writing - review & editing, Supervision, Funding acquisition, Project administration.
Declaration of Competing Interest
The authors declare no competing financial interest.
Acknowledgments
This article pays tribute to those who are fighting against COVID-19. This work was supported by the National Natural Science Foundation of China (31971522) to Zhuang Wang and the European Union’s Horizon 2020 research and innovation program “NanoinformaTIX” (814426) that supported Willie J.G.M. Peijnenburg and Martina G. Vijver. Fan Zhang greatly acknowledges the support from the China Scholarship Council (202008320308). We also thank the reviewers for their valuable comments on the manuscript.
Supplementary material
Variation of total energy of the complexes of the carbon nanoparticles with SARS-CoV-2 RNA fragment during Forcite Anneal optimization; Calculated total potential energy interaction energies (E int), ‘van der Waals’ interaction energies, and electrostatic interaction energies between the carbon nanoparticles (CNPs) and the SARS-CoV-2 RNA fragment (covRNA); OPLS regression models obtained from the fake pool data of the interaction energies derived from the total potential energy.
Edited by: Professor Bing Yan
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ecoenv.2021.112357.
Appendix A. Supplementary material
Supplementary material
.
References
- Aasi A., Aghaei S.M., Moore M.D., Panchapakesan B. Pt-, Rh-, Ru-, and Cu-single-wall carbon nanotubes are exceptional candidates for design of anti-viral surfaces: a theoretical study. Int. J. Mol. Sci. 2020;21:5211. doi: 10.3390/ijms21155211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Huraimel K., Alhosani M., Kunhabdulla S., Stietiya M.H. SARS-CoV-2 in the environment: modes of transmission, early detection and potential role of pollutions. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allawadhi P., Khurana A., Allwadhi S., Joshi K., Packirisamy G., Bharani K.K. Nanoceria as a possible agent for the management of COVID-19. Nano Today. 2020;35 doi: 10.1016/j.nantod.2020.100982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous Nanotechnology versus coronavirus. Nat. Nanotechnol. 2020;15:617. doi: 10.1038/s41565-020-0757-7. [DOI] [PubMed] [Google Scholar]
- Braakhuis H.M., Gosens I., Heringa M.B., Oomen A.G., Vandebriel R.J., Groenewold M., Cassee F.R. Mechanism of action of TiO2: recommendations to reduce uncertainties related to carcinogenic potential. Annu. Rev. Pharmacol. Toxicol. 2021;61:203–223. doi: 10.1146/annurev-pharmtox-101419-100049. [DOI] [PubMed] [Google Scholar]
- Chen G., Peijnenburg W.J.G.M., Xiao Y., Vijver M.G. Current knowledge on the use of computational toxicology in hazard assessment of metallic engineered nanomaterials. Int. J. Mol. Sci. 2017;18:1504. doi: 10.3390/ijms18071504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ma J., Xu M., Liu S. Antiviral nanoagents: more attention and effort needed? Nano Today. 2020;35 doi: 10.1016/j.nantod.2020.100976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Barceló J., Hartnett A.S., Kubinec R., Messerschmidt L. COVID-19 government response event dataset (CoronaNet v.1.0) Nat. Hum. Behav. 2020;4:756–768. doi: 10.1038/s41562-020-0909-7. [DOI] [PubMed] [Google Scholar]
- Diffenbaugh N.S., Field C.B., Appel E.A., Azevedo I.L., Baldocchi D.D., Burke M., Burney J.A., Ciais P., Davis S.J., Fiore A.M., Fletcher S.M., Hertel T.W., Horton D.E., Hsiang S.M., Jackson R.B., Jin X., Levi M., Lobell D.B., McKinley G.A., Moore F.C., Montgomery A., Nadeau K.C., Pataki D.E., Randerson J.T., Reichstein M., Schnell J.L., Seneviratne S.I., Singh D., Steiner A.L., Wong-Parodi G. The COVID-19 lockdowns: a window into the earth system. Nat. Rev. Earth Environ. 2020;1:470–481. [Google Scholar]
- Florindo H.F., Kleiner R., Vaskovich-Koubi D., Acúrcio R.C., Carreira B., Yeini E., Tiram G., Liubomirski Y., Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C., Du J., Zhao L., Wang L., Liu Y., Li D., Yang Y., Zhou R., Zhao Y., Chai Z., Chen C. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. 2011;108:16968–16973. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenzi P., Stagi L. Carbon-based antiviral nanomaterials: graphene, C-dots, and fullerenes. A perspective. Chem. Sci. 2020;11:6606–6622. doi: 10.1039/d0sc02658a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Chen W., Xu Z., Zheng S., Zhu D. Graphene nanosheets and graphite oxide as promising adsorbents for removal of organic contaminants from aqueous solution. J. Environ. Qual. 2013;42:191–198. doi: 10.2134/jeq2012.0172. [DOI] [PubMed] [Google Scholar]
- Kaul S., Gulati N., Verma D., Mukherjee S., Nagaich U. Role of nanotechnology in cosmeceuticals: a review of recent advances. J. Pharm. 2018;2018 doi: 10.1155/2018/3420204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-M., Chung Y.-S., Jo H.J., Lee N.-J., Kim M.S., Woo S.H., Park S., Kim J.W., Kim H.M., Han M.-G. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostarelos K. Nanoscale nights of COVID-19. Nat. Nanotechnol. 2020;15:343–344. doi: 10.1038/s41565-020-0687-4. [DOI] [PubMed] [Google Scholar]
- Lan T.C.T., Allan M.F., Malsick L.E., Khandwala S., Nyeo S.S.Y., Sun Y., Guo J.U., Bathe M., Griffiths A., Rouskin S. Insights into the secondary structural ensembles of the full SARS-CoV-2 RNA genome in infected cells. bioRxiv 2020.06.29.178343 [Preprint]. 19 February 2021. 2021 doi: 10.1101/2020.06.29.178343. [DOI] [Google Scholar]
- Łoczechin A., Séron K., Barras A., Giovanelli E., Belouzard S., Chen Y.-T., Metzler-Nolte N., Boukherroub R., Dubuisson J., Szunerits S. Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Appl. Mater. Interfaces. 2019;11:42964–42974. doi: 10.1021/acsami.9b15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Chen F. Quantitative analysis of molecular surface based on improved Marching Tetrahedra algorithm. J. Mol. Graph. Model. 2012;38:314–323. doi: 10.1016/j.jmgm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- Maleki Dizaj S., Mennati A., Jafari S., Khezri K., Adibkia K. Antimicrobial activity of carbon-based nanoparticles. Adv. Pharm. Bull. 2015;5:19–23. doi: 10.5681/apb.2015.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S.V., Hemalatha M., Kopperi H., Ranjith I., Kumar A.K. SARS-CoV-2 in environmental perspective: occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 2021;405 doi: 10.1016/j.cej.2020.126893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier A., Mouchet F., Laplanche C., Cadarsi S., Lagier L., Arnault J.-C., Girard H.A., León V., Vázquez E., Sarrieu C., Pinelli É., Gauthier L., Flahaut E. Surface area of carbon nanoparticles: a dose metric for a more realistic ecotoxicological assessment. Nano Lett. 2016;16:3514–3518. doi: 10.1021/acs.nanolett.6b00348. [DOI] [PubMed] [Google Scholar]
- Palmieri V., Papi M. Can graphene take part in the fight against COVID-19? Nano Today. 2020;33 doi: 10.1016/j.nantod.2020.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B., Xing B. Adsorption mechanisms of organic chemicals on carbon nanotubes. Environ. Sci. Technol. 2008;42:9005–9013. doi: 10.1021/es801777n. [DOI] [PubMed] [Google Scholar]
- Patel K.D., Singh R.K., Kim H.-W. Carbon-based nanomaterials as an emerging platform for theranostics. Mater. Horiz. 2019;6:434–469. [Google Scholar]
- Puzyn T., Rasulev B., Gajewicz A., Hu X., Dasari T.P., Michalkova A., Hwang H.-M., Toropov A., Leszczynska D., Leszczynski J. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 2011;6:175–178. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]
- Rompelberg C., Heringa M.B., van Donkersgoed G., Drijvers J., Roos A., Westenbrink S., Peters R., van Bemmel G., Brand W., Oomen A.G. Oral intake of added titanium dioxide and its nanofraction from food products, food supplements and toothpaste by the Dutch population. Nanotoxicology. 2016;10:1404–1414. doi: 10.1080/17435390.2016.1222457. [DOI] [PubMed] [Google Scholar]
- Sánchez-López E., Gomes D., Esteruelas G., Bonilla L., Lopez-Machado A.L., Galindo R., Cano A., Espina M., Ettcheto M., Camins A., Silva A.M., Durazzo A., Santini A., Garcia M.L., Souto E.B. Metal-based nanoparticles as antimicrobial agents: an overview. Nanomaterials. 2020;10:292. doi: 10.3390/nano10020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Aroca Á., Takayama K., Tuñón-Molina A., Seyran M., Hassan S.S., Pal Choudhury P., Uversky V.N., Lundstrom K., Adadi P., Palù G., Aljabali A.A.A., Chauhan G., Kandimalla R., Tambuwala M.M., Lal A., Abd El-Aziz T.M., Sherchan S., Barh D., Redwan E.M., Bazan N.G., Mishra Y.K., Uhal B.D., Brufsky A. Carbon-based nanomaterials: promising antiviral agents to combat COVID-19 in the microbial-resistant era. ACS Nano. 2021 doi: 10.1021/acsnano.1c00629. doi: 10.1021/acsnano.1c00629. [DOI] [PubMed] [Google Scholar]
- Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., Steinmetz N.F. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- Slot E., Hogema B.M., Reusken C.B.E.M., Reimerink J.H., Molier M., Karregat J.H.M., IJlst J., Novotný V.M.J., van Lier R.A.W., Zaaijer H.L. Low SARS-CoV-2 seroprevalence in blood donors in the early COVID-19 epidemic in the Netherlands. Nat. Commun. 2020;11:5744. doi: 10.1038/s41467-020-19481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape M.D., Viner R.M. COVID-19 in children and young people. Science. 2020;370:286–288. doi: 10.1126/science.abd6165. [DOI] [PubMed] [Google Scholar]
- Talebian S., Wallace G.G., Schroeder A., Stellacci F., Conde J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020;15:618–621. doi: 10.1038/s41565-020-0751-0. [DOI] [PubMed] [Google Scholar]
- Vermisoglou E., Panáček D., Jayaramulu K., Pykal M., Frébort I., Kolář M., Hajdúch M., Zbořil R., Otyepka M. Human virus detection with graphene-based materials. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen J., Wei X., Hernandez Maldonado A.J., Chen Z. Unveiling adsorption mechanisms of organic pollutants onto carbon nanomaterials by density functional theory computations and linear free energy relationship modeling. Environ. Sci. Technol. 2017;51:11820–11828. doi: 10.1021/acs.est.7b02707. [DOI] [PubMed] [Google Scholar]
- Yang K., Xing B. Adsorption of organic compounds by carbon nanomaterials in aqueous phase: polanyi theory and its application. Chem. Rev. 2010;110:5989–6008. doi: 10.1021/cr100059s. [DOI] [PubMed] [Google Scholar]
- Zhang K., Zheludev I.N., Hagey R.J., Wu M.T.-P., Haslecker R., Hou Y.J., Kretsch R., Pintilie G.D., Rangan R., Kladwang W., Li S., Pham E.A., Bernardin-Souibgui C., Baric G.D., Sheahan T.P., D′Souza V., Glenn J.S., Chiu W., Das R. Cryo-electron microscopy and exploratory antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. bioRxiv 2020.07.18.209270 [Preprint]. 20 July 2020. 2020 doi: 10.1101/2020.07.18.209270. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material