Abstract
Age-dependent changes in reactive oxygen species (ROS) levels are critical in leaf senescence. While H2O2-reducing enzymes such as catalases and cytosolic ASCORBATE PEROXIDASE1 (APX1) tightly control the oxidative load during senescence, their regulation and function are not specific to senescence. Previously, we identified the role of ASCORBATE PEROXIDASE6 (APX6) during seed maturation in Arabidopsis (Arabidopsis thaliana). Here, we show that APX6 is a bona fide senescence-associated gene. APX6 expression is specifically induced in aging leaves and in response to senescence-promoting stimuli such as abscisic acid (ABA), extended darkness, and osmotic stress. apx6 mutants showed early developmental senescence and increased sensitivity to dark stress. Reduced APX activity, increased H2O2 level, and altered redox state of the ascorbate pool in mature pre-senescing green leaves of the apx6 mutants correlated with the early onset of senescence. Using transient expression assays in Nicotiana benthamiana leaves, we unraveled the age-dependent post-transcriptional regulation of APX6. We then identified the coding sequence of APX6 as a potential target of miR398, which is a key regulator of copper redistribution. Furthermore, we showed that mutants of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7), the master regulator of copper homeostasis and miR398 expression, have a higher APX6 level compared with the wild type, which further increased under copper deficiency. Our study suggests that APX6 is a modulator of ROS/redox homeostasis and signaling in aging leaves that plays an important role in developmental- and stress-induced senescence programs.
The involvement of ASCORBATE PEROXIDASE6 in developmental and stress-induced leaf senescence is tightly regulated at the transcriptional and post-transcriptional levels.
Introduction
Senescence marks the last step in the development of annual plants, culminating in the death of tissues, and finally, the entire organism (Kim et al., 2016).
Leaf senescence is developmentally controlled by several internal cues that include the age of the plant, the age of the leaf, and the onset of the reproductive phase. It involves hormone-associated pathways mediated by ethylene, jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA). Interactions with environmental stimuli such as changes in light conditions, photoperiodicity, temperature, reduced water availability, prolonged darkness, oxidative stress, etc. often affect or interact with the internal cues, which can induce early senescence or enhance its degree. Many of these and other unfavorable environmental conditions, such as drought and salinity, trigger premature senescence via promoting the accumulation of reactive oxygen species (ROS; Allu et al., 2014; Kim et al., 2016).
Fluctuations in the H2O2 level that accompany plants during their life cycle play an important role in the initiation and progression of age-induced senescence. High resolution temporal transcriptomic profiling of leaf development in Arabidopsis (Arabidopsis thaliana) showed that the ROS-responsive gene cluster is activated in 3-week-old plants, well before leaves have reached their full size (Breeze et al., 2011). The initiation of senescence typically coincides with the decline in the growth rate, which in Arabidopsis is concomitant with the transition to flowering (Thomas, 2013). During this developmental phase in Arabidopsis, the increase in intracellular H2O2 concentrations is coupled with the temporal decrease in the activity of two key H2O2-scavenging enzymes, CATALASE2 (CAT2) and ASCORBATE PEROXIDASE1 (APX1; Ye et al., 2000; Zimmermann et al., 2006; Bresson et al., 2017).
During the “reorganization” phase, the second stage of senescence, the loss of chlorophyll (Chl) and chloroplast degradation slowly progress while still supporting the energy requirements of the cells. During this phase, the bulk of photoassimilates and available nutrient are being exported to sink tissues before the cytoplasm and nucleus degrade. The loss of Chl and other macromolecules can lead to the accumulation of ROS, further promoting senescence (Foyer and Noctor, 2009; Chen et al., 2012; Khanna-Chopra, 2012; Jajic et al., 2015; Bresson et al., 2017). Thus, the regulation of the oxidative load during senescence is critical to allow a timely and efficient sink to source nutrient mobilization under favorable conditions, and for reducing the risk of excessive oxidative damage and loss of organic material under stress conditions (Foyer and Noctor, 2005). For instance, the expression and activity of the Arabidopsis CATALASE3 (CAT3) and APX1 enzymes gradually increase from the time of inflorescence bolting late into senescence, when much of the Chl has degraded (Zimmermann et al., 2006).
As the reorganization phase progress, there is a steady decrease in the antioxidative capacity that accompanies the increase in ROS accumulation, which persists during the termination of senescence (Leshem, 1988; Orendi et al., 2001; Navabpour et al., 2003; Zimmermann and Zentgraf, 2005; Foyer and Noctor, 2009; Smykowski et al., 2010). Interestingly, in the very last stages of senescence termination, when most of the Chl is degraded, low expression and activity of the seed-specific CAT1 become detectable in leaves (Dat et al., 2000; Zimmermann et al., 2006), indicating that the tight control of ROS level persists until the very end.
In Arabidopsis and maize, it was shown that while the level of ROS increases with the progression of senescence in aging leaves and plants, the extent of irreversible protein oxidation (i.e. protein carbonylation) drastically decrease rather than increase (Johansson et al., 2004; Foyer and Noctor, 2009). This evidence supports the paradigm that ROS exert their effects through specific-signaling pathways rather than acting as indiscriminating oxidants, including those activating senescence and programmed cell death (Foyer and Noctor, 2009).
This senescence-associated redox program may involve the H2O2-responsive senescence key regulators WRKY53, which is considered a master regulator of age-induced leaf senescence, ORS1 (ANAC052), and the redox-sensitive REVOLTA (Zentgraf et al., 2010; Balazadeh et al., 2011; Xie et al., 2014). Many key components of the ROS network that function in this program, including ROS-scavenging enzymes and ROS-signaling factors, are yet unknown.
In this study, we show that the expression of cytosolic APX6 increases with the age of the leaf and the plant, indicating it is a bona fide senescence-associated gene. We further show that high osmotic conditions, dark stress, and ABA activate APX6 expression in an age-dependent manner. Concomitantly, APX6-deficient mutants prematurely induced senescence programs triggered by the transition to flowering, extended darkness, and ethylene. The earlier onset of senescence in the mutants was accompanied by higher levels of H2O2 compared with the wild-type and reduced ability to adjust the leaves’ redox state. Using transient expression assays in N. benthamiana leaves, we discovered that the constitutive expression of APX6 was restricted to old and dying cells and absent in younger tissues. Using small RNA target analysis, we identified miR398 as a potential post-transcriptional regulator of APX6. Correspondingly, mutants of SQUAMOSA promoter binding protein-like 7 (SPL7), the master regulator of miR398, showed increased levels of APX6 in as yet flowering or senescing plants. Our results unraveled the distinct role of APX6 in senescence control, providing additional evidence for the importance of tightly regulating ROS homeostasis during this developmental phase.
Results
Age-dependent expression of cytosolic APXs
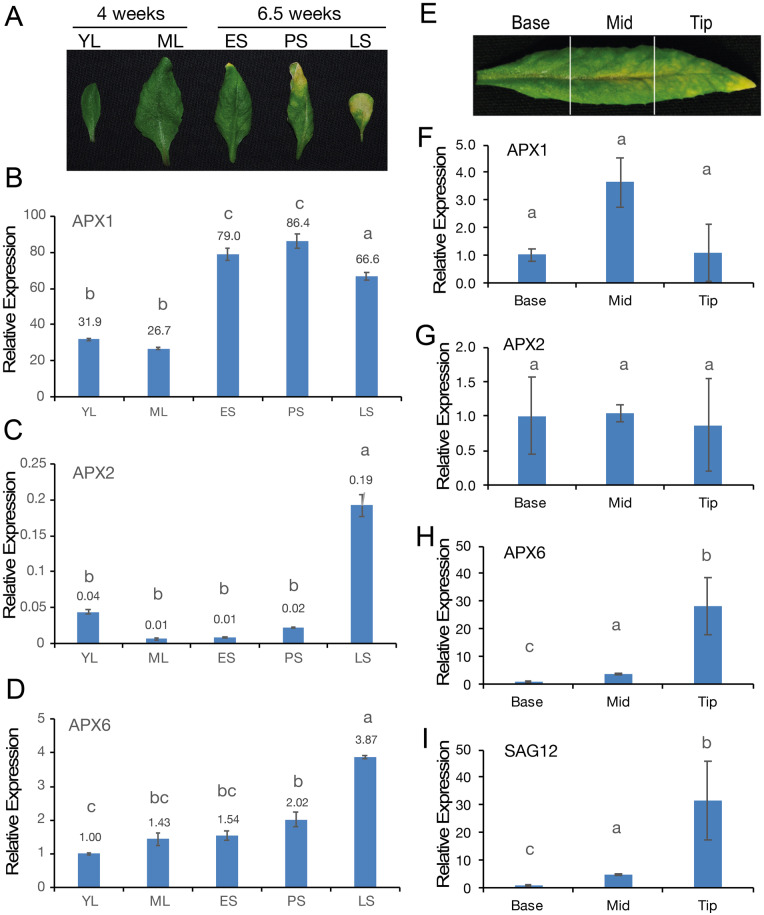
To investigate the effect of leaf age on the expression of the three cytosolic (c)APXs, APX1, APX2, and APX6, we compared their relative transcript abundance using RT-qPCR. To this end, we sampled young and mature leaves (YL and ML, respectively) of 4-weeks old plants and leaves at different senescence stages of 6.5-week-old plants (early-, progressive-, and late- senescing [ES, PS, and LS, respectively] leaves; Figure 1, A). As expected, APX1 was the dominant cAPX (Figure 1, B–D). The level of APX1 was between two- and three-fold higher in week 7, in the ES and PS leaves, compared with week 4 in YL and ML. However, the expression of APX1 did not increase with the age of the leaf within the plant, as the expression was comparable between the ES and PS leaves and then decreased by 25% in LS leaves (Figure 1, B). These results indicated that the increase in APX1 expression was in a plant age-dependent but not leaf developmental stage-dependent manner. Also, as expected, the expression of the exclusively stress-activated APX2 (Suzuki et al., 2013) remained negligible throughout the plant and leaf development. However, it was significantly induced in late senescing leaves (Figure 1, C). In contrast, APX6 showed a gradual increase with the age of the leaf in both the 4- and 6.5-weeks old plant, indicating its expression is upregulated in a leaf-age-dependent manner (Figure 1, D). Yet, although APX6 was nearly four-fold higher in late senescing leaves of 6.5-week-old plants compared with young green leaves of 4-week-old plants, it was 17-fold lower than the level of APX1 (Figure 1, B and D). We further examined the expression of the cAPXs in the base, middle, and tip of early senescing leaves in 6.5-week-old plants (Figure 1, E–I). Only APX6 displayed a gradual increase in expression along the leaf blade with about a 4- and a 30-fold higher level in the mid and tip sections, respectively, compared with the base (Figure 1, H). The changes in the level of APX6 correlated with the changes in the expression of the senescence marker gene SAG12 (Figure 1, I).
Figure 1.
Expression of cytosolic APXs during leaf aging. (A) A representative image of leaves used for the qPCRs in B–D. Young leaf (YL) and fully expanded mature green leaf (ML) from 4-week-old plants, and leaves from 6.5-week-old plants at different stages of senescence; early senescence (ES; yellowing leaf area <25%), progressive senescence (PS; yellowing leaf area ∼25%–50%), and late senescence (LS; yellowing leaf area >50%). (B–D) Quantitative real-time PCR analysis of the APX1, APX2, and APX6 transcript levels at different stages of leaf development, as presented in A. The relative fold change expression for all three APXs in the different leaves is shown relative to the abundance of APX6 in the YL in D. Values above the bars indicate the fold change expression level. Letters above each bar indicate statistical significance at the level of P < 0.05 as indicated by one-way ANOVA Tukey’s test. (E) A representative image of ES leaf divided into three segments (base, mid, and tip) used for the qPCRs in F–I. (F–I) The relative expression of the cAPXs and SAG12 in the different leaf sections. Results of expression in the mid and tip sections are presented relative to the level of each gene at the base segment. SE indicates an average of n = 3 replicates. Letters above each bar indicate statistical significance at the level of P < 0.05 as indicated by one-way ANOVA Tukey’s test.
Taken together, these results indicated that APX6 expression is upregulated in leaves with an age- and senescence-dependent manner.
Spatial–temporal age-dependent expression of APX6
We further compared the age-dependent expression of APX6, using transgenic plants expressing an APX6pro::GUS construct. Within the first 2 weeks of the plant’s development, GUS activity was below the visible detection threshold (Figures 2, A, 8, A–F). Between weeks 3 and 4, GUS activity was moderate in cotyledons and was very weak at the tip of the first true leaves (Figure 2, B). By weeks 5 and 6, GUS expression was seen at the tip of fully developed leaves and spreading throughout the blade as the leaves got older leaves, and yet restricted in younger leaves (Figure 2, C). The appearance of APX6pro::GUS expression, starting from the tip and outer edge of a rosette leaf, is consistent with the initiation pattern of leaf senescence and the directionality of Chl degradation (Guo and Gan, 2006; Farage-Barhom et al., 2008). In 7-week-old plants, the age-dependent increase in GUS activity was most evident with the strongest expression in yellowing leaves (Figure 2, D, E). In contrast, GUS expression under the APX1 promoter (APX1pro::GUS) resulted in a strong and relatively uniform spatial GUS staining in leaves from the early seedling stage to aging plants (Supplemental Figure S1). By 9 weeks, APX6pro::GUS expression became detectable in young leaves, and the intensity grew even stronger in mature and senescing leaves (Supplemental Figure S2, A). Interestingly, a strong APX6pro::GUS expression was detected in sepals of mature post-anthesis flowers showing a further increase with the aging of the tissue (Supplemental Figure S2, B). Collectively, these results indicate that APX6 expression is dependent on the age of the plant as well as the age of the tissue and support our conclusion that APX6 has a distinct role in senescence.
Figure 2.
Age-dependent activation of the APX6 promoter. APX6pro::GUS expression in leaves at different ages of the plants. (A) 1-week old, (B) 3.5-weeks old, (C) 5-weeks old, and (D, E) 7-week old plant. Photograph of the leaves in E was taken prior to GUS staining. Scale bar = 5 mm. The arrowheads in B point to inserts showing enlargements of the indicated first true leaves having weak blue staining at the very tip of the blade. Scale bar in the inserts = 1 mm.
Early senescence in apx6 mutants
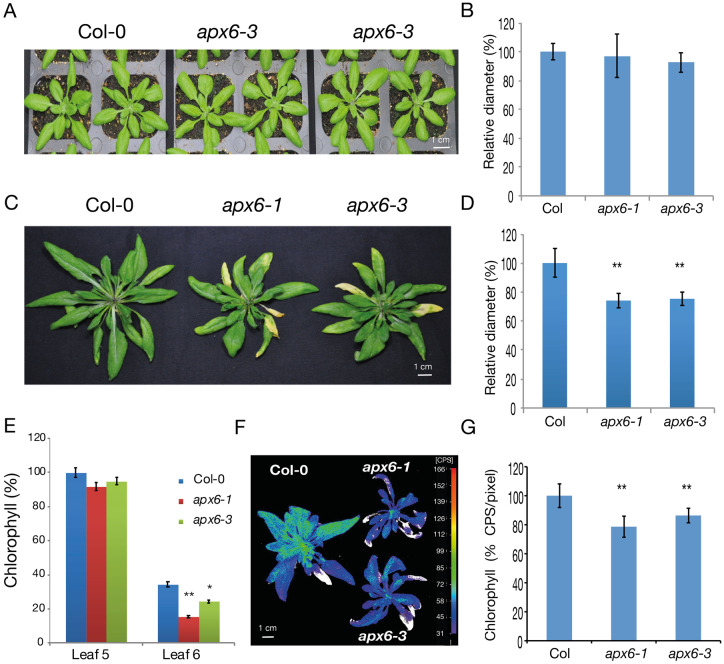
To evaluate the function of APX6 during senescence, we compared the growth and development of two independent Arabidopsis T-DNA knockout lines of APX6, apx6-1 and apx6-3 (Chen et al., 2014a) with the wild-type plant. From the early stages of the vegetative growth to maturity, 4 weeks after germination, no remarkable difference was observed between the wild-type and apx6 mutants (Figure 3, A and B). However, in 6–7-week-old flowering apx6 plants, early signs of senescence were apparent in the older leaves, and the diameter of the rosette was reduced compared with the wild type (Figure 3, C–F and Supplemental Figure S3). Tracking the development of the fifth and sixth leaves between weeks 4 and 8 demonstrated an earlier initiation of senescence in axp6 plants compared with the wild type (Supplemental Figure S4). Correspondingly, the expression of WRKY53, SAG12, and SAG13, evaluated in semi-quantitative RT-PCR, accumulated earlier in the leaves of apx6-1 mutant (Supplemental Figure S5). In 5-week-old plants, Chl concentration was comparable between the wild type and mutants at leaf number 6 but showed a more significant decrease in the mutants in the older fifth leaf (Figure 3, E). This more pronounced loss of Chl in the mutants was also visualized and measured using delayed fluorescence imaging of whole rosettes (Figure 3, F and G). In contrast with the apx6 mutants, apx1-mutant plants do not show early senescence-related phenotype (Supplemental Figure S6). Overall, these results suggest that the activity of APX6 is required for preventing accelerated loss of Chl and premature onset of senescence.
Figure 3.
Early senescence phenotypes in the apx6 mutants. (A) 4-week-old wild-type and apx6 plants show no phenotypic differences. (B) The relative diameter of the rosettes of 4-week-old plants. The wild-type diameter was set to 100%. SDs represent averages of n = 9 replicates. (C) 7-week-old flowering wild-type and apx6 plants grown under normal condition. The inflorescence stems were removed to capture only the rosettes. (D) The relative diameter of 7-week-old plants. SDs represent averages of n = 9 replicates. (E) Relative chlorophyll content, measured by optical density, in the fifth and sixth leaves of 5-weeks-old wild-type and apx6 plants. SEs represent averages of n = 7 and n = 5 replicates for leaves 5 and 6, respectively. (F) Chl imaging in 7-weeks-old rosettes using delayed fluorescence imaging with the NightShade imager. (G) The relative Chl level measured by the delayed fluorescence imaging assay, shown in F. CPS = photons counts per second. SDs represent averages of n = 4 replicates. * and ** indicate Student’s t test significance at P < 0.05 at P < 0.01, respectively.
Accelerated senescence in apx6 mutants triggered by external stimuli
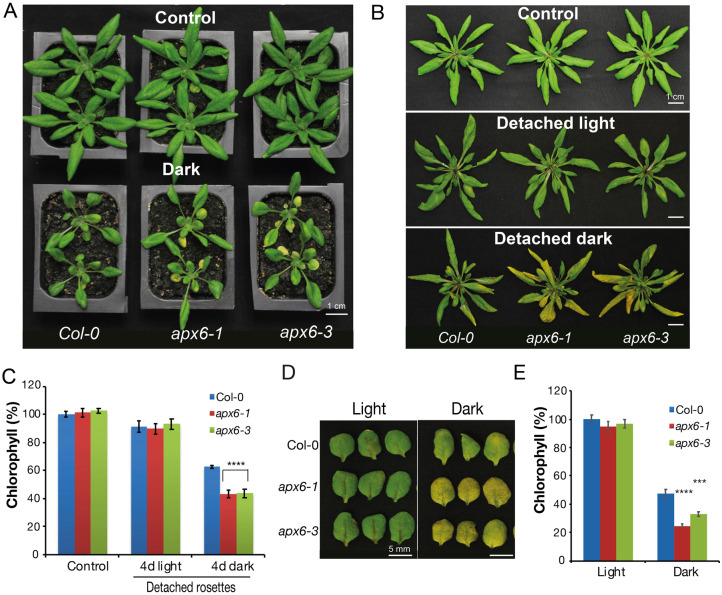
One of the best-known external cues for triggering senescence is extended darkness (Liebsch and Keech, 2016). Therefore, the senescence phenotype of apx6 plants was tested in response to dark stress. To this end, 3.5-week-old soil-grown plants were incubated for 7 d in complete darkness. In response to dark stress, the cotyledons and older leaves in the apx6 mutants were yellowing faster than in the wild type, which was still mostly green (Figure 4, A). Accelerated loss of Chl was further shown in detached rosettes and detached leaves of 4.5-week-old apx6 plants following 4 d of darkness (Figure 4, B–E). The increased senescence rate in the mutants was independent of injury stress, as light-incubated severed rosettes and leaves did not show notable differences from the wild-type (Figure 4, B–E).
Figure 4.
Dark stress-induced senescence phenotype of apx6 mutants. (A) Phenotype of 4.5-week-old plants grown under control (16/8-h light/dark) or after 7 d in dark stress conditions. (B) A photograph of detached rosettes of 4.5-week-old grown under control (detached just before photographing) and following 4 d incubation under light/dark cycle (detached light) or under dark conditions (detached dark). (C) The relative chlorophyll content of 4.5-week-old detached rosettes shown in B. The chlorophyll values, measured by absorbance, are presented relative to the control wild-type samples. SEs represent an average of n = 6 replicates. (D) A representative photograph of detached third or fourth rosette leaves of 4.5-week-old plants that were incubated for 4 d under light conditions or in the dark. (E) The relative chlorophyll content in the third or fourth detached leaves in control and after 4 d in dark stress. The chlorophyll values, measured by absorbance, are presented relative to the control wild-type samples. SEs in C and E represent an average of n = 8 replicates, *** and **** indicate Student’s t test significance at P < 0.005 and P < 10−4.
We further tested whether the acceleration of senescence in response to ethylene is higher in the apx6 mutants compared with the wild type. To this end, detached leaves of 4-week-old plants were incubated with 100 µM of the ethylene precursor aminocyclopropane-1-carboxylic acid (ACC) under continuous light for 4 d. Similar to the effect of dark stress, ACC treatment accelerated the loss of Chl in the mutants compared with the wild-type and the mock treatment (Supplemental Figure S7).
These results indicated that APX6 mitigates the activation of leaf senescence pathways induced by external stimuli such as darkness or ethylene.
Altered ROS level and ascorbic acid redox state in mature leaves of apx6 under light and dark conditions
To evaluate the impact of APX6 activity on ROS and redox metabolism associated with the initiation of leaf senescence, we compared the level of H2O2 and the ascorbic acid (AsA) pool in the wild-type and apx6 mutant under control light and dark stress conditions. To this end, we subjected 6-week-old plants to dark stress for 2 d, a period sufficient to induce pro-senescence signals, yet too short to result in senescence symptoms, and sampled mature fully developed green leaves.
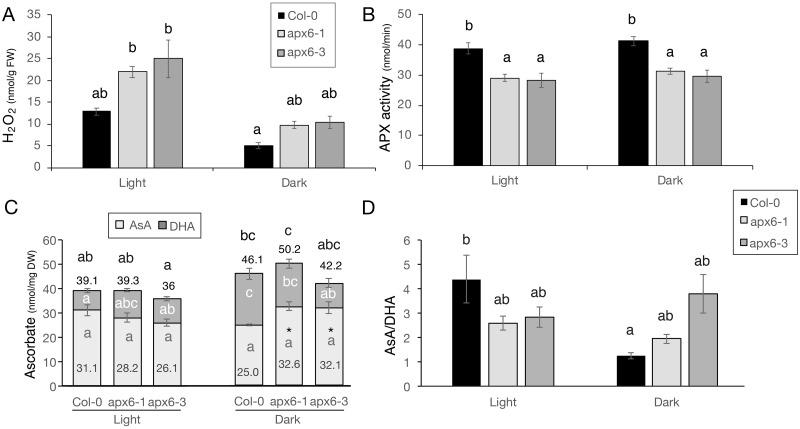
The level of H2O2, measured using the Amplex Red reagent, was 70% and 90% higher in apx6-1 and apx6-3 in the light, respectively, compared with the wild type (Figure 5, A). In response to dark stress, the overall H2O2 level was lower compared with the light conditions, as expected, yet the mutants maintained a two-fold higher level of H2O2 compared with the wild-type (Figure 5, A). Correspondingly, APX activity was 25% lower in both mutants compared with the wild type (Figure 5, B). In contrast, in 2-week-old plants, the level of H2O2 of the whole rosette was similar in both the wild-type and apx6 plants under control light conditions and was higher in the mutants during dark stress (Supplemental Figure S8).
Figure 5.
ROS and ascorbate levels in plants under light and dark conditions. All the assays were conducted in mature fully developed green leaves of 6-week-old plants, grown under light conditions, or following 2 d of dark stress. (A) H2O2 level measured by the Amplex red reaction. SEs represent an average of n = 5 replicates and three technical repeats. (B) APX activity. APX activity is expressed in nanomoles of AsA oxidized per minute per milligram protein. SEs represent an average of n = 4 biological replicates in three technical repeats. (C) AsA level. The values above the bars in C indicate the total ascorbate level, which is comprised of the reduced (AsA) and oxidized form (DHA). (D) The ratio between AsA and DHA that are presented in C. SEs represents an average of n = 5 replicates. Letters above each bar indicate statistical significance at the level of P < 0.05 as indicated by two-way ANOVA Tukey’s tests.
The level of total AsA was comparable between the wild type and the mutants in the light as well as under dark stress, which increased the level of the total AsA pool (Figure 5, C), as recently reported (Zhang et al., 2016). Yet, in the wild type, the level of the reduced AsA decreased by 20%, and the level of its oxidized form, dehydro-ascorbate (DHA), increased 2.6-fold. In contrast, the level of reduced AsA in apx6-1 and apx6-3 increased by 16% and 23%, respectively, during dark stress (Figure 5, C).
Consequently, the ratio of reduced AsA to DHA between the wild-type and the apx6 mutants was significantly different (Figure 5, D); the AsA/DHA ratio in the light was lower in apx6 leaves and remained similar under dark conditions, whereas in the wild type it decreased 3.5-fold compared with the control. These findings suggested that aging leaves of the apx6 mutants have altered redox state.
Collectively, the above-presented results suggest that the APX6 activity in Arabidopsis leaves facing senescence-promoting cues, such as aging and extended darkness, is needed for curbing the level of H2O2 and redox state.
Stress-induced expression of APX6
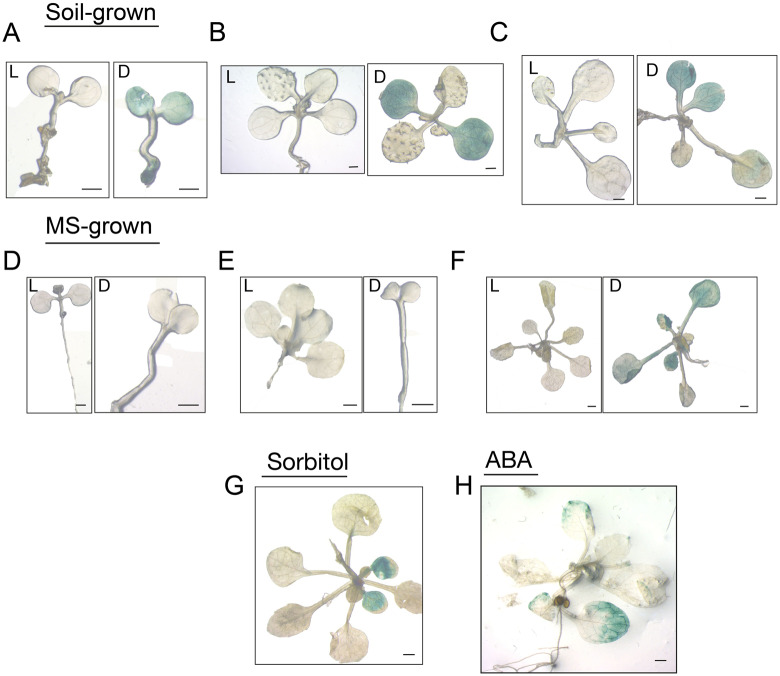
Using APX6pro::GUS reporter line, we investigated whether APX6 is dark stress-responsive. GUS expression was tested in 5 d- and 2-weeks-old seedlings, grown on solid Murashige and Skoog (MS) medium or soil, were dark-stressed for 4 or 7 d, and compared with light-grown plants of the same age. In the younger seedlings, GUS activity was detected only in the soil-grown (Figure 6, A and B) but not in MS grown (Figure 6, D and E) dark-stressed plants, regardless of the duration in the dark. Furthermore, GUS expression in the soil-grown seedlings was restricted to cotyledons. In contrast, in 2-week-old plants exposed to dark stress for 4 d, GUS activity was detected in both cotyledons and leaves in both soil-, and MS agar-grown plants (Figure 6, C and F). These results suggested that dark stress activation of APX6 expression is dependent on the age of the tissue and the plant.
Figure 6.
Age-dependent activity of the APX6 promoter::GUS reporter under control and stress conditions. (A–F) GUS stained plants kept under light (L) or dark (D) stress conditions. (A–C) Plants grown in soil. (D–H) Plants were grown on MS agar plates. A and D, 9-d-old plants after 4 d in dark stress. B and E, 12-d-old after 7 d in dark stress. C and F, 16-d-old plants after 4 in dark stress or under light conditions. Control (L) seedings of the same age are shown to the right of the dark-stressed plants. (G) 16-old-day plant after 4 d osmotic stress in plates containing 200-mM sorbitol. (H) 15-d-old plant after 24 h treatment with 1-µM ABA. Scale bar = 1 mm.
The differences in APX6 promoter activity between soil- and MS-grown seedlings prompted us to test whether low water potential triggers its expression. To this end, 2-weeks-old plants were transferred to MS-agar supplemented with 200-mM sorbitol under light conditions for 4 d. A strong GUS activity was observed in cotyledons but not in the true leaves, suggesting that osmotic-stress induction of APX6 expression is also age-dependent (Figure 6, G). The APX6 promoter contains two putative ABA responsive elements (Austin et al., 2016; Supplemental Figure S9). Accordingly, treatment of 2-week-old plants with exogenous ABA (1 µM, 24 h) resulted in the appearance of GUS activity in the edge of the cotyledons and of the leaves (Figure 6, H). This result suggests the involvement of ABA in the age-dependent and osmotic/stress activation of the APX6 promoter.
Transient expression of APX1 and APX6 proteins in Nicotiana benthamiana tobacco leaves
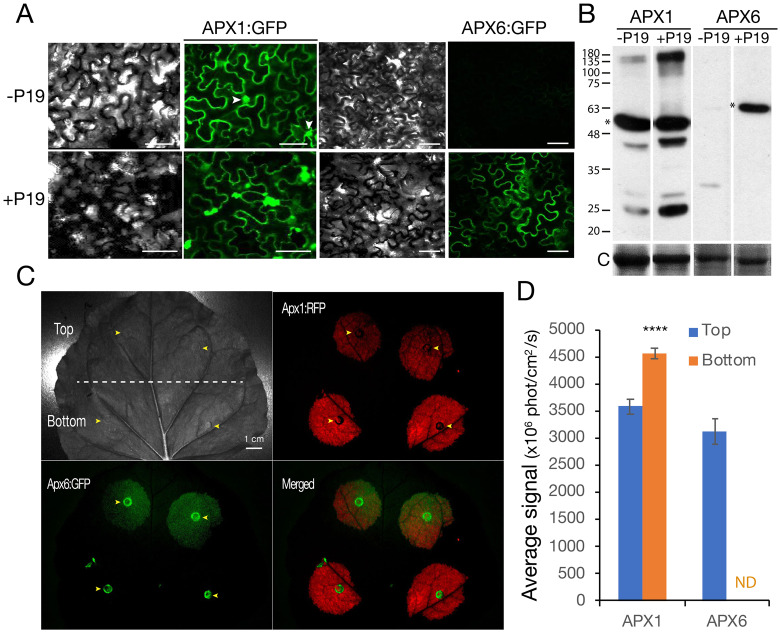
As APX1 and APX6 are both cytosolic, and because partial redundancy in their activity is assumed, we set to verify their cellular distribution. To this end, we conducted transient expression of CaMV35S promoter-controlled APX1 or APX6 fused to GFP in N. benthamiana leaves and detected their level under a confocal microscope or with in vivo live-imaging of whole leaves. Surprisingly, APX6::GFP was undetectable in leaves of 4–6-week-old plants, compared with the high fluorescence of APX1::GFP. Therefore, we co-infiltrated APX6::GFP with the RNA silencing suppressor p19 protein. Co-expression with P19 facilitated the accumulation of the APX6::GFP chimeric protein, detected by microscopy and immunoblot with GFP antibodies (Figures 7, A and B and Supplemental Figure S10), but still it was five-fold lower than that of APX1::GFP (Figure 7, B). These results suggested that APX6 is subjected to post-transcriptional regulation at the level of the RNA, protein, or both. Interestingly, the immunoblot with anti-GFP antibodies identified additional recombinant APX1::GFP protein forms of higher or lower masses than the expected full-size form, suggesting the existence of a multimeric complex and degradation forms, respectively (Figure 7, B). In addition, the APX1::GFP was localized to the nucleus as well as the expected cytosolic accumulation, whereas APX6::GFP was restricted to the cytosol (Figure 7, A). We argue that this confinement of APX6::GFP intracellular localization is due to the large size of this chimeric protein (63.2 kDa), 8.68 kDa larger than APX1::GFP and 3.2 kDa larger than the size exclusion limit of the nuclear pore (Haasen et al., 1999). These results suggest that cytosolic APXs may function in the nucleus.
Figure 7.
Protein abundance of APX1 and APX6 fluorescent-tagged-proteins in N. benthamiana leaves. (A) Confocal microscopy pictures of transiently expressed APX1::GFP and APX6::GFP controlled by the CaMV 35S promoter in leaves of 4-week-old plant. The lower panel pictures show results after co-infiltration with the P19 protein, which suppresses the RNA silencing apparatus. White arrowheads in the APX1:GFP upper (-P19) image points to GFP in nuclei. Bar = 50 µM. (B) Protein blot analysis of the chimeric APX::GFP proteins expressed in leaves of 4-weeks-old N. Benthamiana plants. Anti-GFP antibodies were used for the immunoblot, and coomassie staining was used as a loading control. Asterisks mark the full-size APX1:GFP and APX6:GFP chimeric proteins. (C) Imaging of co-expression experiment of APX1::RFP and APX6::GFP in mature fully developed leaves of 2-month-old flowering plants. Yellow arrowheads point to the marks left by the syringe at the sites of agrobacterium infiltration. Bar = 1 cm. (D) The average intensity of the fluorescence signal in the co-infiltrated leaves, represented in C. The strong APX6::GFP signal at the sites of infiltration was not included in the measurements. SEs represent n = 12 injected leaf areas each of the leaf sections (Top and bottom), in six leaves of a similar developmental stage, as shown in C. The expression level of APX1::RFP differed significantly between the top and bottom parts of the leaf: **** indicate Student’s t test significance at P < 10−4. ND, not detected.
To further compare the steady-state level of the two APXs, we co-expressed APX1::RFP and APX6::GFP under CaMV 35S promoter in fully expanded leaves of mature flowering 8-weeks-old N. benthamiana. Surprisingly, whole-leaf fluorescence imaging revealed that the level of APX1::RFP is higher at the bottom part of the leaf blade compared with the upper portion that is closer to the tip. In contrast, the level of APX6::GFP was almost the complete opposite; high at the top but undetectable at the bottom (Figure 7, C and D). Transient expression with APX1::GFP resulted in similar patterns (Supplemental Figure S11), indicating the results were independent of the identity of the fluorescent protein. Interestingly, at the center of each agrobacterium infiltration site, there was a strong expression of APX6::GFP compared with the undetectable APX1::RFP (Figure 7, C). Because these imprints left by the tip of the syringe during injection become yellow and necrotic within 3–4 d, these results suggested that program cell death is also involved in the regulation of APX6 abundance. Taken together, the transient expression results indicated that APX6 and APX1 are regulated post-transcriptionally in an age- and cell-fate status manner, but in opposite directions.
Identification of potential small RNAs involved in regulating APX6 level
To identify small RNAs that may be targeting the APX6 CDS, we used the plant small RNA target analysis (psRNATarget) server, which includes published microRNAs (miRs) from 72 plant species (Dai and Zhao, 2011; Dai et al., 2018). The analysis identified miR398 as a potential regulator of APX6 in A. thaliana and 25 other plants species from various families of eudicots and monocots, including Solanaceae, Cucurbitaceae, Fabaceae, and Poaceae to name a few (Supplemental Figure S12 and Supplemental Table S2). miR398 is a conserved plant miRNA that regulates copper redistribution by targeting mRNAs of copper-binding proteins. Two of the canonical targets of miR398 include copper/zinc superoxide dismutase 1 (CSD1) and plastid CSD2 (Zhu et al., 2011; Sunkar et al., 2012). The expression of miR398 in Arabidopsis is dependent on the transcription factor SQUAMOSA promoter binding protein-like 7 (SPL7). Consequently, spl7 mutants do not express miR398a-c (Yamasaki et al., 2009; Bernal et al., 2012; Brousse et al., 2014).
Involvement of SPL7 and miR398 in APX6 post-transcriptional regulation
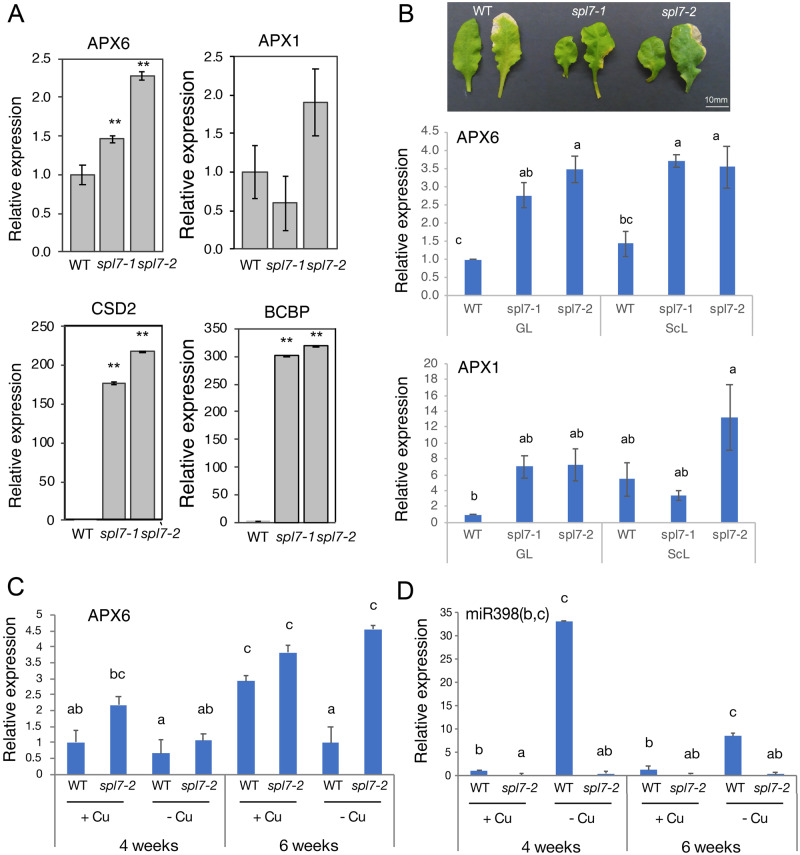
To test whether APX6 is a target of miR398, we used the two spl7 T-DNA insertion mutant alleles, spl7-1 and spl7-2 (Yamasaki et al., 2009; Bernal et al., 2012; Brousse et al., 2014). RT-qPCR in mature green leaves of 6-week-old plants grown in soil under normal conditions (i.e. without the addition of copper) showed that APX6 was 1.5- and 2.3-fold higher in spl7-1 and spl7-2, respectively, compared with the wild type (Figure 8, A). The higher APX6 abundance in the leaves of spl7 mutants was in correlation with the increased levels of CSD2 and blue copper-binding protein (BCBP), two canonical miR398 targets (Figure 8, A). In contrast, the level of APX1 did not change significantly in the spl7 lines.
Figure 8.
SPL7 negatively regulates APX6 transcript accumulation. (A) Real-time PCR results of APX1 and APX6 level in 6-week-old spl7 mutants and wild-type plants grown on soil under control conditions, without copper supplementation. CSD2 and BCBP served as typical miR398 gene targets reference. Real-time PCR in 15-week-old spl7 mutants and wild-type grown under copper sufficient conditions (irrigated with 25-µM CuSO4). SEs represent an average of n = 3 replicates, ** indicate Student’s t test significance at P < 0.01. (B) Real-time PCR of APX6 and APX1 in fully mature green (left side) and senescing (right side) leaves of 15-week-old spl7 mutants and wild type, as shown in B. Plants were grown in soil irrigated with 25-µM Cu. GL and ScL refer to green and senescing leaves, respectively. (C) and (D) Real-time PCR of APX6 and miR398, respectively, in fully mature green leaves of 4-week- and 6-week-old spl7 mutant and wild-type plants, grown hydroponically with or without 250-nmol CuSO4. The miR398 primers are specific to miR398a and miR398b, the two miR398 showing expression. SEs indicate an average of n = 3 independent replicates. Letters above each bar in B–D indicate statistical significance at the level of P < 0.05 as indicated by two-way ANOVA Tukey’s tests.
Because spl7 mutants are deficient in copper distribution, they require copper supplementation in the growth medium to grow in size and develop into fertile adults (Yan et al., 2017). Therefore, to further test the expression of APX1 and APX6 in old senescing spl7 mutants, plants were grown in soil irrigated with 25-µM CuSO4, as previously described (Yan et al., 2017). To this end, we sampled still green and early senescing inner whorl leaves of 15-week-old copper-supplemented plants. In green leaves, the level of APX6 was 2.75- and 3.5-fold higher in spl7-1 and spl7-2, respectively, compared with the wild type. A similar increased level of APX6 expression was observed in senescing leaves of the spl7 lines (Figure 8, B).
These results, which are consistent with the results presented in Figure 8, A, suggested that APX6 transcripts accumulation depends on the presence of SPL7. In contrast, the level APX1 increased in the green leaves of both spl7 lines but decreased in the senescing leaves of the spl7-1 line compared with the wild type level (Figure 8, B), indicating that APX1 expression does not depend on SPL7.
miRNA398 expression increase during copper deficiency (Zhu et al., 2011; Bernal et al., 2012; Waters et al., 2012; Pilon, 2017). Therefore, we tested whether the age-dependent transcript accumulation of APX6 is affected by the copper availability status of the plant. To this end, the level of APX6 was examined in 4–6-week-old plants grown hydroponically with or without 250-nmol CuSO4. As expected, spl7-2 consistently showed higher levels of APX6 compared with the corresponding wild-type sample. Also, as expected, an age-dependent increase in APX6 expression was obtained under copper sufficient conditions in both wild type and spl7-2 (Figure 8, C). The abundance of APX6 was inversely correlated with the expression level of miR398 under both conditions and plant age (Figure 8, D). As expected, the expression of miR398 was negligible in the spl7-2 under all conditions and highly induced in the wild type under copper deficiency, However, in the 6-week-old plant, the induction level of miR398 under copper deficiency was substantially reduced compared with the 4-week-old, suggesting this decrease is age-dependent. This result is consistent with the age-dependent reduction in the expression of SPL7 in 6-week compared with 4-week-old plant (Supplemental Figure S13), the Genevestigator developmental expression profile (Hruz et al., 2008; Supplemental Figure S14), and along the blade of senescing leaves (Supplemental Figure S15).
Taken together, the results presented above implicate SPL7 as a negative regulator of APX6 transcript accumulation in an age-dependent manner, most likely via the activation of miR398.
Discussion
A recent bioinformatic evaluation of APX family gene expression indicated that AtAPX6 is relatively highly abundant during senescence (Ozyigit et al., 2016). Curiously, a previous analysis of age-related expression changes in all eight Arabidopsis APX family gene members reported age-dependent reduction for APX4 and thylakoid (tyl)APX, but no remarkable changes in the expression of all others (Panchuk et al., 2005).
In this study, we present a body of evidence showing that APX6 is a bona fide senescence-associated gene which function delays the onset of senescence. The fact that apx6 mutants display early developmental senescence phenotype, whereas apx1 mutant does not (Supplemental Figure S6; Miller et al., 2007), underline the distinct function of APX6. In line with the paradigm that ROS activates specific-signaling pathways (Foyer and Noctor, 2009), we postulate that the accumulation of H2O2 in the absence of APX6 activity in mature leaves of the mutants (Figure 5) acts as specific senescence promoting signal. Accordingly, the increased H2O2 level and the altered redox state in mature leaves of the apx6 mutants rendered them more sensitive to senescence-promoting stimuli such as aging cues, extended darkness, and ethylene (ACC treatment). Thus, it may be that the activity of APX6 mitigates age-related changes and delays the onset of senescence.
At the systemic level, plants induce leaf senescence to provide carbon, nitrogen, and mineral resources to the developing fruits and seeds. Remobilization of the degradation products of DNA, ribosomal RNA, and other molecules, as well as micronutrients during reproductive senescence, supports the viability of newly developing seeds (Schippers, 2015). These processes must be tightly coordinated under optimal conditions, and more so during stress, to achieve maximal productivity. Previously, we showed that APX6 also functions in mature drying seeds, protecting them from excessive oxidative stress and acting in the regulation of ROS signals (Chen et al., 2014a, 2014b). The distinct function of APX6 during leaf senescence and seed maturation underlines its importance during the final reproductive-development stages of the plant. Thus, functioning at both ends of the sink and source conduit, APX6 may provide extra protection, preventing excessive oxidation of the bulk of nutrients transported to the reproductive tissues, and protecting seeds during their desiccation stage.
We showed that APX6 promoter activity is triggered by ABA, extended darkness, and osmotic stress (sorbitol) in an age-dependent manner (Figure 6), suggesting that ABA governs the transcriptional activation of APX6. During leaf senescence or prolonged dark incubation, ABA biosynthesis rate and concentration increase, further enhancing the senescence process (Zhang and Zhou, 2013). Furthermore, ABA was shown to promote drought tolerance and leaf senescence, allowing proper reallocation of nutrients from source-to-sink tissues (Zhao et al., 2016). It has been suggested that the impact of ABA on leaf senescence is mediated through ABA insensitive 5 (ABI5) transcription factor. ABI5 directly activates the expression of NON-YELLOW COLORING1 (NYC1) and STAYGREEN1 (SGR1) that function in Chl catabolism, whereas its deficiency delayed leaf senescence (Sakuraba et al., 2014; Su et al., 2016). Other studies have reported the involvement of ABI5 in dark-induced senescence (Lee et al., 2015; Su et al., 2016). ABI5 directly suppressed the activation of the LEA protein ABR, which activity inhibits dark-induced senescence in Arabidopsis (Su et al., 2016). Similar to APX6, ABR expression is induced by ABA, low water potentials, and darkness. Previously we showed that apx6 seeds are hypersensitive to ABA, which may be related to their altered ABI5 expression (Chen et al., 2014a, 2014b). Thus, ABI5 may link between ABA- and stress-activated senescence programs and the induction of APX6 (Figure 9). The involvement of ABI5 in regulating APX6 transcriptional activation is currently under investigation.
Figure 9.
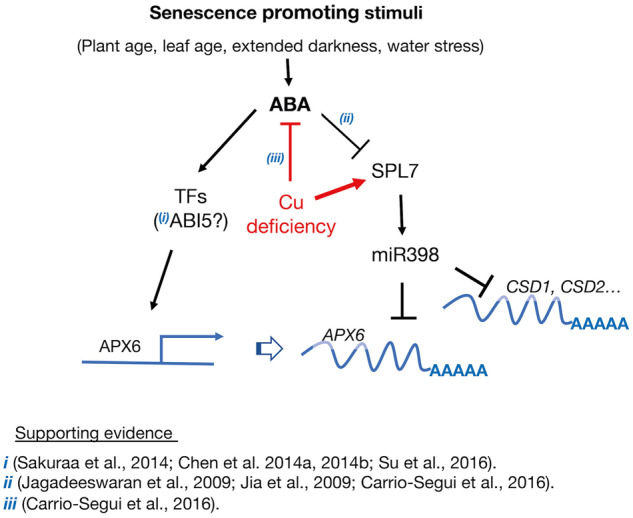
Proposed model for the induction of APX6 transcript accumulation responding to senescence promoting stimuli via the ABA-dependent pathway. ABA induces the transcriptional activation of the APX6 promoter, directly or indirectly, by yet unknown transcription factors (TFs). Previous studies (i) provided evidence indirectly linking ABA INSENSITIVE5 (ABI5) as a plausible upstream regulator of APX6 expression—thus, the question mark. APX6 is also negatively regulated transcriptionally by miR398 via the activity of its master regulator SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7). We suggest that while miR398 suppresses its canonical targets COPPER/ZINC SUPEROXIDE DISMUTASE1 (CSD1), CSD2, and others, it is also moonlighting on APX6, and perhaps other targets not directly linked to copper distribution control. Under copper sufficient conditions, the expression of SPL7 decreases with the age of the plant and leaves promoting the accumulation of APX6 transcripts. ABA suppresses the expression of SPL7, further enhancing the age- and stress-dependent expression of APX6 (ii). In contrast, Cu deficiency, inhibiting ABA biosynthesis and signaling (iii), decreasing APX6 expression in a two-prong manner, suppressing the ABA signaling pathway while increasing the SPL7-mediated Cu-redistribution response.
In this work, we also uncovered the age- and cell death-dependent post-transcriptional regulation of APX6 (Figure 7). We then identified the conserved miR398 as the plausible small RNA targeting APX6 transcripts (Supplemental Figure S12). By interrogating the spl7 mutants, which do not induce the expression of miR398, we provided evidence that SPL7, most likely through miR398, is involved in suppressing APX6 transcript accumulation in Arabidopsis leaves in an age-dependent manner (Figure 8 and Supplemental Figure S13–S15). In support of this conclusion, under copper deficiency conditions that strongly induce miR398 expression (Yamasaki et al., 2009), the age-dependent increase in APX6 was suppressed in the wild type but not in the spl7-2 mutant (Figure 8, C). Thus, we propose that while miR398 is primarily engaged in fine-tuning the expression of CSD1, CSD2, and other copper-binding proteins (Sunkar et al., 2006), it is also suppressing the accumulation of APX6 transcripts and perhaps other non-Cu-binding enzymes (Figure 9).
The age-related decrease in SPL7, which facilitated the accumulation of APX6 transcripts, may be associated with the senescence-mediated increase in ABA, as ABA and salinity treatments suppressed miR398 expression in Arabidopsis seedlings (Jagadeeswaran et al., 2009; Jia et al., 2009). It is noteworthy that copper deficiency negatively impacts ABA accumulation, while ABA significantly decreases SPL7 expression (Carrio-Segui et al., 2016). Thus, it is possible that ABA signaling also facilitates the accumulation of APX6 transcripts by inhibiting the expression of SPL7/miR398 (Figure 9).
Overall, in this work, we discovered the role of APX6 in delaying the early onset of age-dependent and stress-induced senescence programs in A. thaliana and showed that its activity is modulated transcriptionally and post-transcriptionally by ABA and SPL7/miR398, respectively (Figure 9).
Materials and methods
Plant materials, growth conditions, and treatments
All Arabidopsis (A. thaliana) mutant lines used in this study are in the background of Columbia (Col-0) ecotype. APX6 T-DNA insertion lines (apx6-1, WiscDsLox321C09; and apx6-3, WiscDsLox337H07) were previously described by Chen et al. (2014a), spl7-1 (SALK_093849) and spl7-2 (SALK_125385) previously described by Brousse et al. (2014).
Plants were grown in growth chambers (Percival AR-66; Percival Scientific) at 23°C under long day 16-/8-h light/dark cycle, 80 µmol m−2 s−1 photosynthetic flux density, and with 70% relative humidity, or under continuous light (50 µmol m−2 s−1) in a growth room with 65% relative humidity. For experiments with spl7 mutants grown in soil (Figure 8, B), plants were irrigated with 25-µM CuSO4. For hydroponically grown plants (Figure 8, C and Supplemental Figure S13), plants were grown with or without 250-nmol CuSO4 as previously described (Yan et al., 2017).
Dark stress treatment
For phenotypic comparison of apx6 mutants with the wild type, 3.5-week-old soil-grown plants were transferred to dark growth chamber for 7 d or kept under 16-/8-h regime for the same duration, as control. Additionally, detached third and fourth leaves of 4.5-week-old plants were incubated in double distilled water under constant darkness or light for 4 d before analyses.
ACC treatment
The detached third and fourth leaves of 4-week-old plants grown under control condition as above were incubated in 10-µM ACC for 4 d before analyses, as previously described (Li et al., 2013).
Chl measurements
Preparative Chl determination
Chl was extracted from the third and or fourth rosette leaves using 99.8% (v/v) ethanol for 20 min at 80°C as previously described (Cui et al., 2013). Chl content per fresh weight (FW) of a leaf was calculated as described by Lichtenthaler et al. (1986). Absorbance measured at 664.5 and 647 nm and the Chl content in micrograms was calculated per gram FW as followed: Chl a = 12.70A (664.5)–2.79A (647); Chl b = 20.70 A (647)–4.62A (664.5); total Chl = 17.90A (647) + 8.08A (664.5).
Determination of Chl in live plants
Chl was evaluated in the entire rosette of 7-week-old plants or detached leaves by measuring delayed fluorescence using the NightSHADE LB985 device (Berthold Technologies, Bad Wildbad, Germany) according to Gould et al. (2009).
H2O2 measurements
Quantification of H2O2 in leaves extracts was performed according to the Amplex Red (Molecular Probes, Invitrogen) Hydrogen/Peroxidase assay kit protocol. The reaction buffer: 25-mM sodium phosphate, pH 7 10-mM Amplex Red reagent and 1 U/mL of horseradish peroxidase (HRP). Three technical repeats in four biological replicates were used for the assay. To avoid the possibility of residual H2O2-reducing enzymatic activity before the Amplex Red reaction, all the samples were kept on ice up until the addition of the HRP enzyme. The Amplex Red reagent reacts with H2O2 in a 1:1 stoichiometry to produce the red-fluorescent oxidation product, resorufin. The resorufin fluorescent signal was measured in the Synergy 4 fluorescence plate reader (Bio-Tek) using 530-/590-nm excitation/emission.
APX activity measurements
APX activity assay was done as previously described (Panchuk et al., 2002); 10-µl extract from 6-week-old plants in a reaction buffer containing 25-mM Na-phosphate (Ph 7.0), 0.1-mM EDTA, 1-mM H2O2, and 0.25-mM ASA in a total volume of 1 mL. Changes in AsA absorbance at OD 265 nm as previously described (Yamasaki et al., 1997) with the absorption coefficient 14,500 M−1 cm−1 (Witmer et al., 2016) were recorded every 10 s over 5 min and normalized to the protein level that was quantified by Bradford assay. We used OD265 instead of the previously used OD290 (Nakano and Asada, 1981) because 265 nm is the absorption maximum of AsA, and DHA had significant absorption at 290 nm in our assay mixture. The experiment was performed with three technical repeats and three biological replicates. In Figure 5, the activity values are presented in µmol normalized to the total protein per time (t = 60 s).
AsA measurements
The concentrations of total ascorbate and the reduced AsA were determined using the ferrous ion reaction with α-α′-bipyridyl according to Gillespie and Ainsworth (2007). The level of DHA in each of the samples was deduced by subtracting the reduced AsA from the total ascorbate content. Measurements in green leaves of 6-week-old plants grown under control light conditions and following dark stress were performed with five biological replicates with three technical repeats.
Molecular analyses
Gene expression analysis
RNA extraction from fresh leaves was carried with the TRIZOL reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s recommendations. PCR, complementary DNA synthesis, quantitative real-time PCR, and semi-quantitative PCR were done as described previously (Chen et al., 2014a). The primers’ names and sequences are listed in Supplemental Table S1. Quantification of the bands’ intensity in the semi-quantitative PCR in Supplemental Figure S5 was conducted using ImageJ software.
The extraction of total protein, immunoblotting, and protein oxidation analyses was performed as previously described (Miller et al., 2007). Mouse anti-GFP antibodies and goat-anti mouse IgG HRP (Santa Cruz Biotechnology, USA) were used for the detection of APX::GFP recombinant proteins.
Cloning
The genomic fragment of ∼1000-bp upstream of start codons of APX6, covering the entire intergenic up-stream region, was cloned to generate APX6pro-GUS fusions (Supplemental Figure S16).
For the cloning, we used PCR-amplification with Herculase II Fusion DNA Polymerase enzyme (Agilent Genomics, USA) using specific primers (Supplemental Table S1), and introduction into pDONR221 entry vector by recombination using the Gateway BP clonase reaction (Invitrogen, USA). The APX6 promoter was fused to the GUS reporter gene by recombination into the pHGWFS7,0 binary destination vector that contains the β-glucuronidase (GUS) reporter gene (Karimi et al., 2002), using the Gateway LR clonase reaction.
To generate APX1 or APX6 fluorescent protein fusions, the CDS of both genes was PCR amplified and cloned into pDONR221, as described above. The recombinant CDS were cloned into pH7FWG2,0 or pH7RWG2,0 binary vectors containing GFP and RFP, respectively (Karimi et al., 2002). The CDS of the tombusviral P19 protein was cloned into the pGreen vector downstream to the CaMV35S promoter. Agrobacterium tumefaciens strain GV3101 was used for the introduction of all gene cassettes into Arabidopsis in stable, and Nicotiana benthamiana, transiently.
Transient expression in N. benthamiana
Agrobacterium infiltration was performed according to Sparkes et al. (2006). The final concentration of the infiltrated agrobacterium was 0.2 OD600. In the co-expression experiments, the APX1 and APX6 agrobacteria were mixed in a 1:1 ratio, and when P19 use with APX1/6, the ratio was 1:3.
GUS staining
GUS staining was performed in young seedlings, plants, and leaves as previously described (Miller et al., 2005), following treatments. For dark stress treatment, 5-d, 10-d, or 2-week-old plants were incubated in the dark for 4 or 7 d. Control plants of the same age were kept for the same duration under constant light. For osmotic stress and ABA treatments, 1–2-week-old plants germinated on MS agar were transferred to new plates containing 200-mM sorbitol or 1-µM ABA for 24–48 h.
Microscopy and imaging
GFP fluorescence was imaged in Agrobacterium-infiltrated leaves sections using the Olympus-FV1000 confocal microscope using multi-Argon laser with excitation at 488 nm and emission at 520 nm with a long-pass filter. For GFP and RFP imaging in the entire leaf blades, either the Cri Maestro II (PerkinElmer, USA) or NightShade LB 985 (Berthold Technologies, Germany) live imaging systems were used.
Statistical analysis
For the results of APX activity, H2O2 content, AsA, and DHA content, the differences between mean values were analyzed for significance using two-way ANOVA and Tukey’s test in RStudio (R. RStudio, Inc., Boston, MA, USA; http://www.rstudio.com/). In real-time PCR experiments, the relative concentrations of PCR products were statistically significant according to the t test or ANOVA test (one-way or two-way) by RStudio.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: APX1 (At1g07890), APX2 (At3g09640), APX6 (At4g32320), SPL7 (AT5G18830), miRNA398b (AT5G14545), miRNA398c (AT5G14565), SAG12 (AT5G45890), SAG13 (AT2G29350), WEKY53 (AT4G23810), CSD2 (AT2G28190), BCBP (AT5G20230), ACTIN2 (AT3G18780).
Supplemental data
Supplemental Figure S1. Age-dependent expression of APX1pro:GUS.
Supplemental Figure S2. GUS staining in 9-week old APX6pro::GUS plants.
Supplemental Figure S3. Detached leaves of 6-week-old wild-type and apx6 plants.
Supplemental Figure S4. Week by week aging progress of leaves 5 and 6.
Supplemental Figure S5. Semi-quantitative PCR of SAG12, SAG13, and WRKY53.
Supplemental Figure S6. APX1 knockout does not show increased senescence phenotype.
Supplemental Figure S7. Ethylene-induced senescence phenotype of apx6 mutants.
Supplemental Figure S8. H2O2 level in 2-week-old light-grown and dark stressed plants.
Supplemental Figure S9. APX6 promoter contains two putative novel ABA-responsive elements.
Supplemental Figure S10. Co-expression of APX1::GFP or APX6::GFP with or without P19 protein in the whole leaf.
Supplemental Figure S11. APX1::GFP transient expression in fully developed Nicotiana benthamiana leaf.
Supplemental Figure S12. Top hit results of microRNA targeting APX6 coding sequences in the psRNATarget web tool.
Supplemental Figure S13. Age-dependent expression of SPL7.
Supplemental Figure S14. Genevestigator developmental expression of SPL7, APX1, and APX6.
Supplemental Figure S15. Relative expression of SPL7 in the different sections of a senescing leaf.
Supplemental Figure S16. Genome browser snapshot of APX1 and APX6 genes.
Supplemental Table S1. List of primers
Supplemental Table S2. Full list results of microRNA targeting APX6 coding sequences in the psRNATarget web tool
Supplementary Material
Acknowledgments
The authors thank the Bar Ilan University Core Facility unit for providing RT-qPCR, microscopy, and cytometry services.
Funding
This work was supported by the Israel Science Foundation [ISF Grant No. 938/11], the Marie Curie Actions—International Career Integration Grant [No. 293999], and the Bi-national Agricultural Research & Development Fund [BARD, Project No. IS-4652-13 R]. The work in the Vatamaniuk Laboratory was supported by the US National Science Foundation [Grant IOS 1656321].
Conflict of interest statement. The authors declare no conflict of interest.
C.C., Y.G., O.K.V., and G.M. designed the experiments. C.C. conducted phenotypic and physiological evaluations of the apx6 mutants, Y.G., M.R.I., and C.C. performed the supervised qPCRs. Y.G. evaluated the APX activity assays. A.R. measured the H2O2 levels. S.M. and V.S. were responsible for the GUS histochemical assays and microscopy. S.T. measured the AsA. S.M. performed the transient expression assays. Y.G. and M.R.I. conducted the spl7 experiments, and prepared the figures. G.M. and O.K.V. provided the laboratory infrastructure and funding, and designed the experiments. G.M. conceived the study, wrote the manuscript, and prepared the figures.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Gad Miller (gad.miller@biu.ac.il).
References
- Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S (2014) Salt stress and senescence: identification of cross-talk regulatory components. J Exp Bot 65: 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RS, Hiu S, Waese J, Ierullo M, Pasha A, Wang TT, Fan J, Foong C, Breit R, Desveaux D, et al. (2016) New BAR tools for mining expression data and exploring Cis-elements in Arabidopsis thaliana. Plant J 88: 490–504. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H(2)O(2)-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson J, Bieker S, Riester L, Doll J, Zentgraf U (2017) A guideline for leaf senescence analyses: from quantification to physiological and molecular investigations. J Exp Bot 69: 769–786. [DOI] [PubMed] [Google Scholar]
- Brousse C, Liu Q, Beauclair L, Deremetz A, Axtell MJ, Bouche N (2014) A non-canonical plant microRNA target site. Nucleic Acids Res 42: 5270–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio-Segui A, Romero P, Sanz A, Penarrubia L (2016) Interaction between ABA signaling and copper homeostasis in Arabidopsis thaliana. Plant Cell Physiol 57: 1568–1582. [DOI] [PubMed] [Google Scholar]
- Chen C, Letnik I, Hacham Y, Dobrev P, Ben-Daniel BH, Vankova R, Amir R, Miller G (2014a) ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol 166: 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Twito S, Miller G (2014b) New cross talk between ROS, ABA and auxin controlling seed maturation and germination unraveled in APX6 deficient Arabidopsis seeds. Plant Signal Behav 9: e976489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, Liu CP, Chen SC, Wang LC (2012) Role of ARABIDOPSIS A-FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2. J Exp Bot 63: 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui MH, Ok SH, Yoo KS, Jung KW, Yoo SD, Shin JS (2013) An Arabidopsis cell growth defect factor-related protein, CRS, promotes plant senescence by increasing the production of hydrogen peroxide. Plant Cell Physiol 54: 155–167. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46: W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inze D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farage-Barhom S, Burd S, Sonego L, Perl-Treves R, Lers A (2008) Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J Exp Bot 59: 3247–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905. [DOI] [PubMed] [Google Scholar]
- Gillespie KM, Ainsworth EA (2007) Measurement of reduced, oxidized and total ascorbate content in plants. Nat Protoc 2: 871–874. [DOI] [PubMed] [Google Scholar]
- Gould PD, Diaz P, Hogben C, Kusakina J, Salem R, Hartwell J, Hall A (2009) Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J 58: 893–901. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612. [DOI] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229: 1009–1014. [DOI] [PubMed] [Google Scholar]
- Jajic I, Sarna T, Szewczyk G, Strzalka K (2015) Changes in production of reactive oxygen species in illuminated thylakoids isolated during development and senescence of barley. J Plant Physiol 184: 49–56. [DOI] [PubMed] [Google Scholar]
- Jia X, Wang WX, Ren L, Chen QJ, Mendu V, Willcut B, Dinkins R, Tang X, Tang G (2009) Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol Biol 71: 51–59. [DOI] [PubMed] [Google Scholar]
- Johansson E, Olsson O, Nystrom T (2004) Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 279: 22204–22208. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Khanna-Chopra R (2012) Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249: 469–481. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo HR, Nam HG (2016) Toward systems understanding of leaf senescence: an integrated multi-omics perspective on leaf senescence research. Mol Plant 9: 813–825. [DOI] [PubMed] [Google Scholar]
- Lee HN, Lee KH, Kim CS (2015) Abscisic acid receptor PYRABACTIN RESISTANCE-LIKE 8, PYL8, is involved in glucose response and dark-induced leaf senescence in Arabidopsis. Biochem Biophys Res Commun 463: 24–28. [DOI] [PubMed] [Google Scholar]
- Leshem YY (1988) Plant senescence processes and free radicals. Free Radic Biol Med 5: 39–49. [DOI] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H (2013) Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Buschmann C, Rinderle U, Schmuck G (1986) Application of chlorophyll fluorescence in ecophysiology. Radiat Environ Biophys 25: 297–308. [DOI] [PubMed] [Google Scholar]
- Liebsch D, Keech O (2016) Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol 212: 563–570. [DOI] [PubMed] [Google Scholar]
- Miller G, Stein H, Honig A, Kapulnik Y, Zilberstein A (2005) Responsive modes of Medicago sativa proline dehydrogenase genes during salt stress and recovery dictate free proline accumulation. Planta 222: 70–79. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R (2007) Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol 144: 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22: 867–880. [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, AH-Mackerness S, Buchanan-Wollaston V (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54: 2285–2292. [DOI] [PubMed] [Google Scholar]
- Orendi G, Zimmermann P, Baar C, Zentgraf U (2001) Loss of stress-induced expression of catalase3 during leaf senescence in Arabidopsis thaliana is restricted to oxidative stress. Plant Sci 161: 301–314. [DOI] [PubMed] [Google Scholar]
- Ozyigit II, Filiz E, Vatansever R, Kurtoglu KY, Koc I, Ozturk MX, Anjum NA (2016) Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front Plant Sci 7: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schoffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Zentgraf U, Volkov RA (2005) Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 222: 926–932. [DOI] [PubMed] [Google Scholar]
- Pilon M (2017) The copper microRNAs. New Phytol 213: 1030–1035. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Schippers JH (2015) Transcriptional networks in leaf senescence. Curr Opin Plant Biol 27: 77–83. [DOI] [PubMed] [Google Scholar]
- Smykowski A, Zimmermann P, Zentgraf U (2010) G-box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol 153: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Su M, Huang G, Zhang Q, Wang X, Li C, Tao Y, Zhang S, Lai J, Yang C, Wang Y (2016) The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci 247: 93–103. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Sejima H, Harper J, Mittler R (2013) Enhanced seed production under prolonged heat stress conditions in Arabidopsis thaliana plants deficient in cytosolic ascorbate peroxidase 2. J Exp Bot 64: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H (2013) Senescence, ageing and death of the whole plant. New Phytol 197: 696–711. [DOI] [PubMed] [Google Scholar]
- Waters BM, McInturf SA, Stein RJ (2012) Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J Exp Bot 63: 5903–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer JR, Wetherell BJ, Wagner BA, Du J, Cullen JJ, Buettner GR (2016) Direct spectrophotometric measurement of supra-physiological levels of ascorbate in plasma. Redox Biol 8: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Huhn K, Brandt R, Potschin M, Bieker S, Straub D, Doll J, Drechsler T, Zentgraf U, Wenkel S (2014) REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 141: 4772–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Ikehara N (1997) Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol 115: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J,, Chia JC, Sheng H, Jung HI, Zavodna TO, Zhang L, Huang R, Jiao C, Craft EJ, Fei Z. et al. (2017) Arabidopsis pollen fertility requires the transcription factors CITF1 and SPL7 that regulate copper delivery to anthers and jasmonic acid synthesis. Plant Cell 29: 3012–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Rodriguez R, Tran A, Hoang H, de los Santos D, Brown S, Vellanoweth RL (2000) The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci 158: 115–127. [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Laun T, Miao Y (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89: 133–137. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou C (2013) Signal transduction in leaf senescence. Plant Mol Biol 82: 539–545. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li H, Xu B, Li J, Huang B (2016) Exogenous melatonin suppresses dark-induced leaf senescence by activating the superoxide dismutase-catalase antioxidant pathway and down-regulating chlorophyll degradation in excised leaves of perennial ryegrass (Lolium perenne L.). Front Plant Sci 7: 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci U S A 113: 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ding Y, Liu H (2011) MiR398 and plant stress responses. Physiol Plant 143: 1–9. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Heinlein C, Orendi G, Zentgraf U (2006) Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ 29: 1049–1060. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Zentgraf U (2005) The correlation between oxidative stress and leaf senescence during plant development. Cell Mol Biol Lett 10: 515–534. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.