Abstract
Plants perceive dynamic light conditions and optimize their growth and development accordingly by regulating gene expression at multiple levels. Alternative splicing (AS), a widespread mechanism in eukaryotes that post-transcriptionally generates two or more messenger RNAs (mRNAs) from the same pre-mRNA, is rapidly controlled by light. However, a detailed mechanism of light-regulated AS is still not clear. In this study, we demonstrate that histone 3 lysine 36 trimethylation (H3K36me3) rapidly and differentially responds to light at specific gene loci with light-regulated intron retention (IR) of their transcripts in the moss Physcomitrella patens. However, the level of H3K36me3 following exposure to light is inversely related to that of IR events. Physcomitrella patens MORF-related gene 1 (PpMRG1), a chromatin adaptor, bound with higher affinity to H3K36me3 in light conditions than in darkness and was differentially targeted to gene loci showing light-responsive IR. Transcriptome analysis indicated that PpMRG1 functions in the regulation of light-mediated AS. Furthermore, PpMRG1 was also involved in red light-mediated phototropic responses. Our results suggest that light regulates histone methylation, which leads to alterations of AS patterns. The chromatin adaptor PpMRG1 potentially participates in light-mediated AS, revealing that chromatin-coupled regulation of pre-mRNA splicing is an important aspect of the plant’s response to environmental changes.
Light modulates the status of histone 3 lysine 36 trimethylation and the association of a chromatin adaptor to regulate alternative splicing of specific genes in Physcomitrella patens.
Introduction
Light is a key environmental factor for plant growth and development. Plants have developed sophisticated light sensing and signaling systems to control photomorphological changes and optimize light absorption. To initiate morphological changes and biological processes in response to light, gene expression in plants is precisely regulated. Accumulating data suggest that light regulation can occur at almost every stage of gene expression from alteration of chromatin status to post-translational modification to control the abundance of functional gene products (see review in Wu, 2014; Cheng and Tu, 2018). Whether light coordinates between different layers of gene expression to control plant growth and development is still unclear.
Positively charged lysine and arginine residues in the N-terminal part of histone are highly accessible for modifications such as methylation, acetylation, phosphorylation, ubiquitination, and sumoylation. Differential modifications of histones form a complex regulatory network to alter chromatin status and control gene expression (see review in Hansen et al., 1998; Jenuwein and Allis, 2001; Liu et al., 2010). In the case of histone methylation, methylated 9th and 27th lysine residues of histone H3 (H3K9 and H3K27) are considered repressive marks to suppress transcription by packaging chromatin, while methylated 4th and 36th lysine residues (H3K4 and H3K36) are suggested to be activating marks to unfold chromatin in both animals and vascular plants (see review in Martin and Zhang, 2005; Liu et al., 2010). In plants, histone methylation also plays critical roles in controlling genome arrangement, transcription, and development. In terms of light responses, histone methylation was shown to be involved in regulation of shade avoidance and gibberellin metabolism (Charron et al., 2009; Peng et al., 2018). Whether it participates in other steps of gene expression in response to light is an open question.
Alternative splicing (AS) is a widespread mechanism among eukaryotes that produces different mature messenger RNAs (mRNAs) from a single pre-mRNA. This rapid and regulated process increases plasticity in plant development and allows an immediate response to environmental changes. Several groups have observed global changes in AS patterns in response to light in plants (Shikata et al., 2012; Wu et al., 2014; Hartmann et al., 2016; Mancini et al., 2016). Molecular studies also showed that photoreceptors, splicing regulators, and spliceosomal components participate in light-dependent regulatory processes (Shikata et al., 2012; Xin et al., 2017; Lin et al., 2019; Shih et al., 2019). Our previous study suggested that light-dependent AS affects specific transcripts and thereby controls particular biological activities (Wu et al., 2014). We found many of the transcripts encoding ribosomal proteins undergo light-dependent intron retention (IR) within 1 h of red light (RL) illumination. The mechanism underlying this regulatory process requires further investigation.
Increasing evidence has shown that AS can occur cotranscriptionally (Luco et al., 2011). The cotranscriptional splicing is regulated by chromatin landscapes. Two models, the kinetic-coupling and chromatin-adaptor models, have been proposed to explain the connection between chromatin landscapes and AS (Luco et al., 2011). In the kinetic-coupling model, chromatin status affects the transcription elongation rate, which regulates AS. For example, splicing factors interact with the C-terminal domain of RNA polymerase II to couple RNA processing with transcription in yeast and human (Hsin and Manley, 2012; Gomez Acuna et al., 2013). In Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), IR occurs more frequently in DNase I hypersensitive sites, which indicates that increasing chromatin accessibility promotes splice site skipping (Ullah et al., 2018). In the chromatin-adaptor model, histones serve as anchors for adaptors to recruit splicing regulators (Luco et al., 2010). Several factors, such as MORF-related gene 15 (MRG15), General Control Nonderepressible-5 (GCN5), Chromodomain Helicase DNA binding protein 1 (CHD1), and heterochromatin protein 1α (HP1α), are chromatin adaptors associated both with histone and splicing machinery in higher eukaryotes (Luco et al., 2011).
MRG proteins were first identified to bind DNA and function in cell proliferation and senescence in human (Bertram et al., 1999). One of the members, MRG15, was shown to recognize H3K36me3 marks and recruit the splicing regulator polypyrimidine tract-binding protein (PTB) to regulate pre-mRNA splicing (Luco et al., 2010). In plants, MRG15 has been shown to associate with pre-mRNA splicing. MRG proteins are localized to the nucleus and function as chromatin remodeling complex components in Arabidopsis to recognize H3K36 trimethylation (H3K36me3) and control the expression of flowering time genes (Xu et al., 2014). A recent study also established that MRG binds with the H3K36me3 mark to mediate differential AS in response to temperature changes in Arabidopsis (Pajoro et al., 2017). These results raise the possibility that the H3K36me3-MRG interaction participates in the cotranscriptional regulation of gene expression in plants. Although evidence supporting a relationship between AS and histone modification in light responses is lacking, several studies focusing on the relationship between histone modification and AS have suggested this possibility (Guo et al., 2008; de Almeida et al., 2011; Kim et al., 2011; Pradeepa et al., 2012).
In this study, we investigated whether histone modification participates in the regulation of light-responsive AS in Physcomitrella patens. We first found that the level of H3K36me3 is responsive to RL. From determining the H3K36me3 level in several light-responsive IR genes, we observed IR and H3K36me3 had inverse relationships in response to RL. Intriguingly, the chromatin adaptor Physcomitrella patens MORF-related gene 1 (PpMRG1) quickly associated with the H3K36me3 mark upon RL exposure. The light-dependent interaction between PpMRG1 and the H3K36me3 mark differentially occurred at different loci. Moreover, the ppmrg1 mutant showed defects in AS and phototropic responses in P. patens, suggesting that PpMRG1 is involved in both splicing regulation and photomorphological control. These results indicate that PpMRG1 is important for H3K36me3-mediated IR regulation under light and provide detailed evidence to suggest a chromatin-coupled mechanism for regulation of pre-mRNA splicing in response to light.
Results
H3K36 is differentially trimethylated in response to RL
To determine whether histone modification can be modulated by RL in P. patens, we analyzed the effect of RL on the levels of several histone marks involved in splicing regulation in animals and yeasts, including H3K4me3, H3K36me3, and H3KAc. After P. patens protonemal cells were irradiated with RL for 1 and 4 h, total proteins were extracted for immunoblotting and hybridized with anti-histone H3, H3KAc, H3K4me3, and H3K36me3 antibodies. We found histone acetylation did not respond to RL (Supplemental Figure S1). However, the levels of H3K4me3 and H3K36me3 were slightly changed from the dark to the RL condition, suggesting that histone methylation can be rapidly modulated by RL.
H3K36me3 is involved in AS regulation in Arabidopsis (Pajoro et al., 2017). Since H3K36me3 levels also changed rapidly in response to RL, we hypothesized that H3K36 trimethylation was involved in regulating light-responsive AS. To test this possibility, we examined the H3K36me3 status of specific gene loci for light responsiveness using chromatin coimmunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR) with an antibody specific for H3K36me3.
We selected two marker genes previously shown to undergo light-responsive AS (Wu et al., 2014). Small Subunit Ribosomal Protein S8e (PpRS8B, p3c13_20020v3) is a representative gene that shows light-induced IR. HY5 Homolog (PpHYH, Pp3c11_17710v3) produces a bZIP TF that acts downstream of light signaling pathways to regulate light-responsive gene expression. PpHYH transcripts show light-repressed IR in the 5′-UTR and potentially produce more functional proteins under light (Wu et al., 2014). Both genes are well-established examples of light-regulated AS. We designed primer sets to examine different regions of PpRS8B and PpHYH genes across coding sequences (Figure 1). Using an anti-H3K36me3 antibody for ChIP, we found that H3K36me3 rapidly decreased on the PpRS8B gene locus in response to 1 h of RL and the effect became more pronounced at 4 h. However, the PpHYH gene showed an opposite pattern with a rapid increase of the H3K36me3 level upon light irradiation. Two other loci (Pp3c22_2920V3 and Pp3c17_21890V3) we tested also showed light-dependent changes (Supplemental Figure S2). These results indicate that the H3K36me3 status is differentially responsive to RL in loci showing distinct splicing patterns.
Figure 1.
H3K36 trimethylation responds to light. Light responsiveness of H3K36me3 enrichment on light-responsive IR gene loci. H3K36me3 enrichment on PpRS8B (upper) and PpHYH (bottom) gene loci was detected using ChIP-qPCR for 7-d-old protonema irradiated with RL (5 μmol m−2 s−1) for 0 (D), 1(R1), and 4 (R4) h. The gene model and PCR fragments designed for ChIP-qPCR are shown at the top of each figure. Gray bars, UTR; blue bars, exon; solid line, introns. The IR site is labeled on the top of the gene structure. H3K36me3 enrichment was normalized with data from input and H3 to obtain relative H3K36me3 levels. Values presented are mean with standard error of the mean (sem) (n = 3, biological replicates). *P < 0.05 (Student’s t test).
To find out whether the light-dependent changes of H3K36me3 level is RL specific, moss protonema were irradiated with blue light (BL) for 1 and 4 h and tested for light responsiveness of H3K36me3 on PpRS8B and PpHYH gene loci by ChIP-qPCR. Compared to the RL data, data from BL experiments showed different light-dependent changes of H3K36me3 with less consistency (Supplemental Figure S3). These results confirmed that light-dependent changes of H3K36me3 occur under RL in the PpRS8B and PpHYH loci.
IR events and H3K36me3 levels have opposite responses to RL
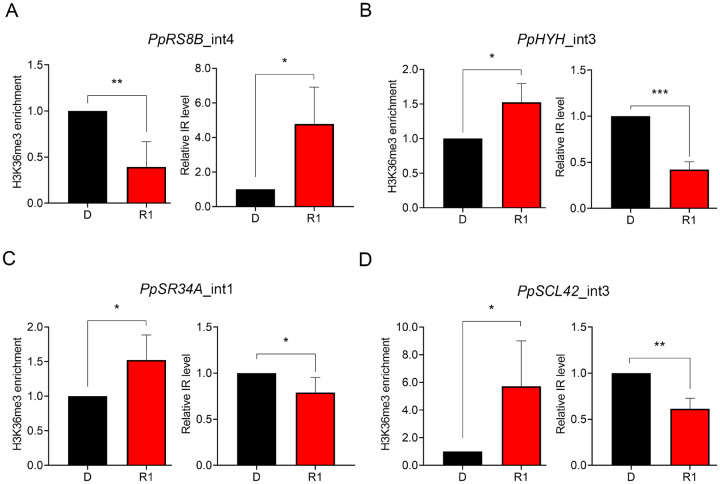
Based on ChIP-qPCR results of the two marker genes and previous analyses, we found that the light responsiveness of H3K36me3 and IR showed inverse patterns. For example, IR of PpHYH gene transcripts is rapidly repressed by RL within 1 h (Wu et al., 2014). However, the H3K36me3 mark was enriched by RL on this gene locus. To determine whether this pattern is common to other genes, we assessed the H3K36me3 status and IR level for additional gene loci by ChIP-qPCR and RT-qPCR, respectively (Figure 2). We tested several IR genes identified in our previous study, including the two genes showing RL-responsive H3K36me3 modification, PpRS8B and PpHYH (Figure 1), as well as other two genes PpSR34A (Pp3c1_35700V3) and PpSCL42 (Pp3s152_10v3; Wu et al., 2014). Using the same sets of primers to detect the IR level by RT-qPCR and H3K36me3 status by ChIP-qPCR, we observed that RL-dependent H3K36me3 changes occurs rapidly within 1 h and indeed showed inverse relationships with IR among the tested genes (Figure 2). These results revealed that the H3K36me3 status at specific gene loci is related to splicing pattern.
Figure 2.
Light-dependent changes in H3K36me3 enrichment and IR level have opposite patterns. Nuclear extracts from 7-d-old protonema irradiated with RL (5 μmol m−2 s−1) for 0 (D) or 1 (R1) h were used for determining H3K36me3 enrichment and IR level at the intron 4 of PpRS8B (PpRS8B_int4) (A), intron 3 of PpHYH (PpHYH_int3) (B), intron 1 of PpSR34A (PpSR34A_int1) (C), and intron 3 of PpSCL42 (PpSCL42_int3) (D), where light-responsive IR were found (Wu et al., 2014). For detection of H3K36me3, the H3K36me3-specific antibody was used to immunoprecipitate H3K36me3-enriched chromatin for CHIP-qPCR (left). Primers used in ChIP-qPCR experiments were the same as those used in RT-qPCR (right) for IR detection. For RT-qPCR, another set of primers was designed as the control for the detection of the overall expression near the IR region. Intensity of IR fragments was compared to the intensity of control fragments to calculate the relative IR level. Data from R1 samples were normalized with that of the D condition. Values presented are mean with sem (n = 3, biological replicates). *, **, ***, and **** indicate P <0.05, P <0.01, P <0.001, and P <0.0001, respectively (Student’s t test).
PpMRG1 differentially associates with H3K36me3 in response to RL
The correlation between histone methylation and AS led us to hypothesize that an unknown factor could function in linking the two processes. We focused on MRG proteins, which act as chromatin adaptors linking the H3K36me3 mark and splicing regulators in mammalian systems and associate with H3K36me3 to control flowering gene expression in Arabidopsis (Kornblihtt et al., 2013; Xu et al., 2014). In P. patens, two putative MRG paralogs (named PpMRG1 and 2) exist with nearly 85% shared amino acid identity and are presumedly derived from gene duplication. To determine whether P. patens MRGs also function as the chromatin adaptor, we generated transgenic plants expressing a c-Myc-tagged version of PpMRG1 (cMyc-MRG1; Supplemental Figure S4). Anti-cMyc antibody was used to immunoprecipitate PpMRG1, and potentially associated histone marks were detected by immunoblotting. This analysis revealed that PpMRG1 interacted with H3K36me3 and H3K4me3 marks in P. patens (Supplemental Figure S5).
Since both H3K36me3 and IR respond to RL rapidly, we further tested whether PpMRG1 associates with the histone mark in a RL-dependent manner. Protonemal cells from wild-type (WT) and cMyc-MRG1-expressing lines were dark-adapted for 3 d, irradiated with RL for 1 h, and collected for coimmunoprecipitation experiments. Surprisingly, the association between PpMRG1 and the H3K36me3 mark was absent in the dark but was quickly induced by RL (Figure 3A; Supplemental Figure S6), suggesting that the interaction between PpMRG1 and the histone mark is dynamic and can be regulated by light.
Figure 3.
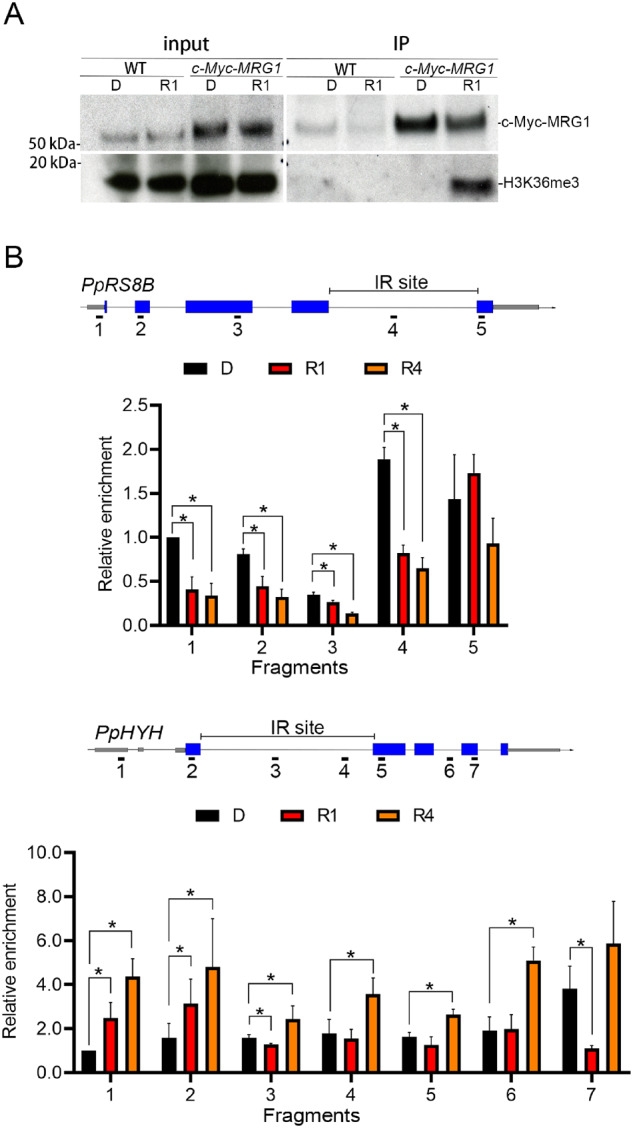
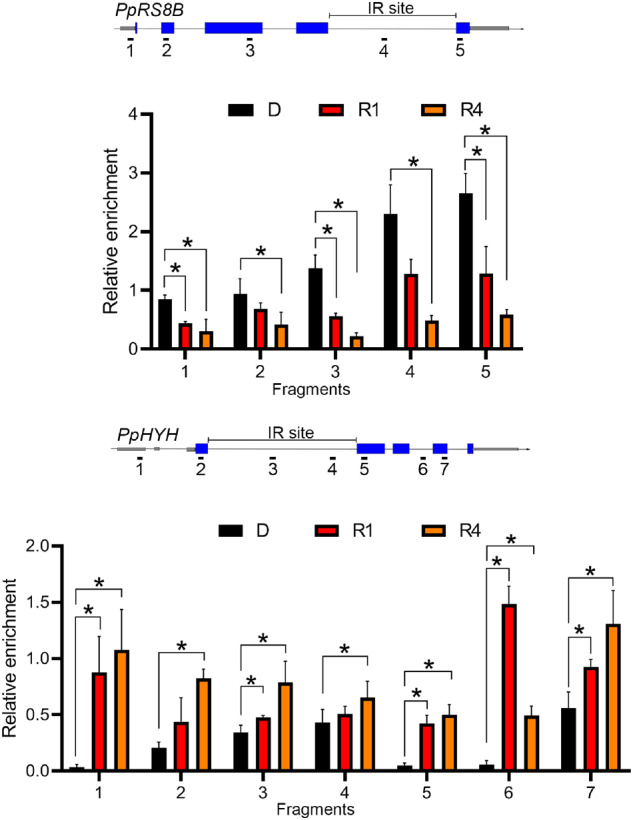
MRG1 associates with H3K36me3 under light. A, Co-immunoprecipitation (Co-IP) of MRG1 and K36 trimethylated histone. Co-IP was performed using anti-c-Myc antibody to pull down associated protein complexes from lysates of 7-d-old protonema ectopically expressing cMyc-PpMRG1 (c-Myc-MRG1). Lysates from the WT were used as the control. Lysates were prepared from plants irradiated with RL (5 μmol m−2 s−1) for 0 (D), 1 (R1), and 4 (R4) h. Fifteen micrograms of total lysates were used as the input. Immunoprecipitated proteins were visualized by immunoblotting with anti-c-Myc and anti-H3K36me3 antibodies. Three independent experiments were performed (Supplemental Figure S5). B, Validation of PpMRG1 enrichment at specific gene loci. PpRS8B and PpHYH genes were tested under 0 (D), 1 (R1), and 4 (R4) h of RL. MRG1 enriched across the gene regions were measured by ChIP-qPCR. Anti-c-Myc antibody was used to pull down the c-Myc-PpMRG1 protein complex. Gray bars, UTR; blue bars, exon; solid line, introns. Values presented are mean with SEM (n = 3, biological replicates). * and ** indicate P <0.05 and P <0.01, respectively (Student’s t test).
Although the interaction between PpMRG1 and the H3K36me3 mark is induced by light, the status of H3K36me3 on specific gene loci could be different. To determine whether PpMRG1 is differentially recruited to chromatin regions by H3K36me3, we performed ChIP-qPCR to validate PpMRG1 status on PpRS8B and PpHYH loci. As shown in Figure 3B, PpMRG1 are enriched on chromatin of the two IR genes and are responsive to RL. In comparison to results from Figure 1, the level of PpMRG1 enrichment on each locus near the IR region was associated with the abundance of H3K36me3, where RL simultaneously decreased the level of H3K36me3 and PpMRG1 on the PpRS8B locus or increased those of the PpHYH locus. We also assessed the PpMRG1 level under BL condition on these loci (Supplemental Figure S7) and found a distinct pattern of the PpMRG1 level in response to BL compared to that of RL, indicating potential wavelength-specific regulation. In summary, these results suggest that MRG1 availability may be coordinated with H3K36 methylation at specific gene loci to regulate RL-responsive IR.
Loss of MRG1 affects RL-responsive IR
To further determine whether PpMRG1 functions in light-responsive IR, we generated a ppmrg1 knockout mutant by gene targeting (Supplemental Figure S8). We then performed RNA sequencing (RNA-seq) on the WT and ppmrg1 mutant. Protonemata of WT and ppmrg1 plants were grown in the dark for 3 d and exposed to constant RL for 1 h. For RNA-seq, total RNA was subjected (three biological replicates per sample) to library preparation and sequencing. After data trimming and filtering, more than 1.02 billion reads were generated (Supplemental Table S1). The sequence reads were mapped to the P. patens genome annotation V3.3 using BLAT (Kent, 2002) and Bowtie 2 (Langmead and Salzberg, 2012). Approximately 95% of the reads were perfectly aligned to the reference genome. We then analyzed three major types of AS events: IR (Supplemental Data Set S1), exon skipping (ES; Supplemental Data Set S2), and alternative donor and/or acceptor site (AltD/A; Supplemental Data Set S3).
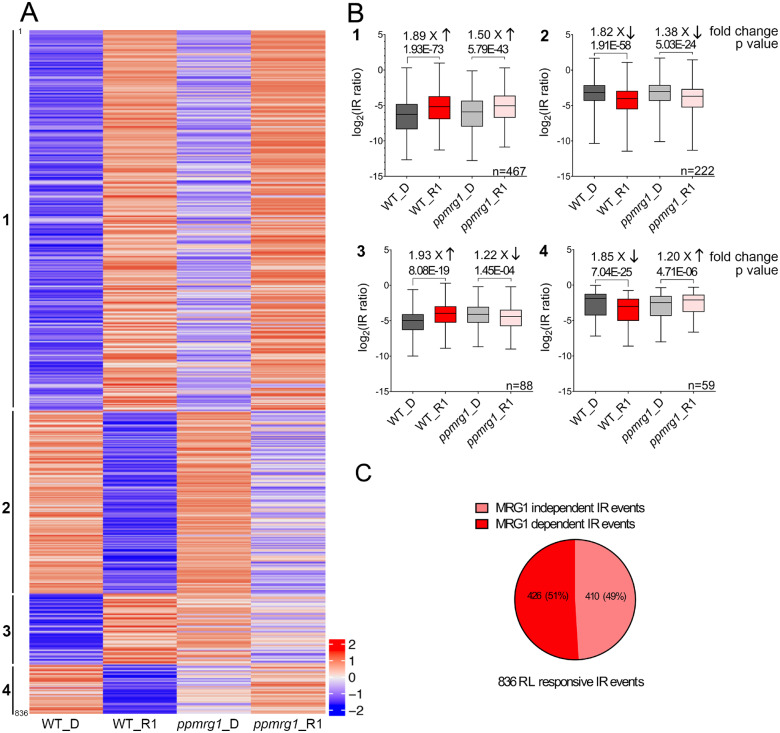
We first examined the RL-responsive IR events in the WT and ppmrg1. Of the 44,923 IR events found in the WT, we identified 836 RL-responsive IR events. RL responsiveness of these IR events in ppmrg1 was further determined by comparing the IR pattern between the WT and the mutant. We performed Pearson correlation analysis to categorize the events and construct a heatmap (Figure 4A; Supplemental Data Set S4). Four type of regulations were defined: type 1, IR are upregulated by light in both WT and ppmrg1; type 2, IR are downregulated by light in both WT and ppmrg1; type 3, IR are upregulated by light in the WT but downregulated in the ppmrg1; type 4, IR are downregulated by light in the WT but upregulated in the ppmrg1. Among the four types of event classes identified, types 1 and 2 showed similar IR patterns in both lines, but RL-induced changes were more pronounced in the WT than in the mutant. As indicated in the box plot (Figure 4B), the median fold-change values between darkness and RL conditions was higher in the WT than in ppmrg1 for both types 1 and 2 events. The P-value from a statistical comparison was also lower in the WT than in ppmrg1. These findings suggest that PpMRG1 plays a role in modulating RL-responsive IR.
Figure 4.
PpMRG1 is involved in RL-regulated IR. A, Patterns of IR levels among the 836 RL-responsive IR events in the WT. IR levels after dark (D) and 1 h of RL irradiation (R1) in the WT and ppmrg1 were log2 transformed. Events with an IR level of 0 were removed from the list; 836 events are shown in the heatmap. The events were grouped by Pearson correlation analysis of the D to R1 pattern in ppmrg1 and sorted by the R1/D fold changes in the WT. Four categories based on the correlation between IR pattern of the WT and ppmrg1 were identified. The scale of fold change is shown on the lower right side of the heatmap. B, Box plots of IR events in four categories. The log2 IR levels of the events were plotted. The IR levels of each line were used to determine fold changes in response to RL and statistical significance by two-tailed Student’s t test. C, MRG-dependent (426) and -independent (410) IR events are shown in the pie chart. Error bars show the SEM values.
To investigate the involvement of PpMRG1 in this process, we examined whether these events were still significant in the ppmrg1 mutant. Of the 836 IR events examined, 426 (51%) events were no longer RL responsive in the mutant and can be defined as MRG1-dependent IR events regulated by RL, while 410 (49%) events were RL responsive but MRG1-independent (Figure 4C). To further determine whether PpMRG1 functions independent of light, we compared RNA-seq data of WT and the ppmrg1 mutant in the dark-grown samples and identified 1,579 IR events that were regulated by PpMRG1 in the dark. From the WT data, we also found 23,112 IR events that were not light responsive. As shown in Supplemental Figure S9, we found a total of 546 IR events that are potentially controlled by PpMRG1 but not light-responsive, suggesting multiple functions of MRG1 in regulating AS.
The functions enriched from PpMRG1-dependent IR genes were further determined. Ribosomal protein genes again were highly enriched, which is consistent with our previous results that AS of many ribosomal protein gene transcripts is rapidly responsive to light (Supplemental Table S2; Shikata et al., 2012; Wu et al., 2014; Hartmann et al., 2016; Mancini et al., 2016). This finding further supports that PpMRG1 is involved in regulating RL-responsive IR.
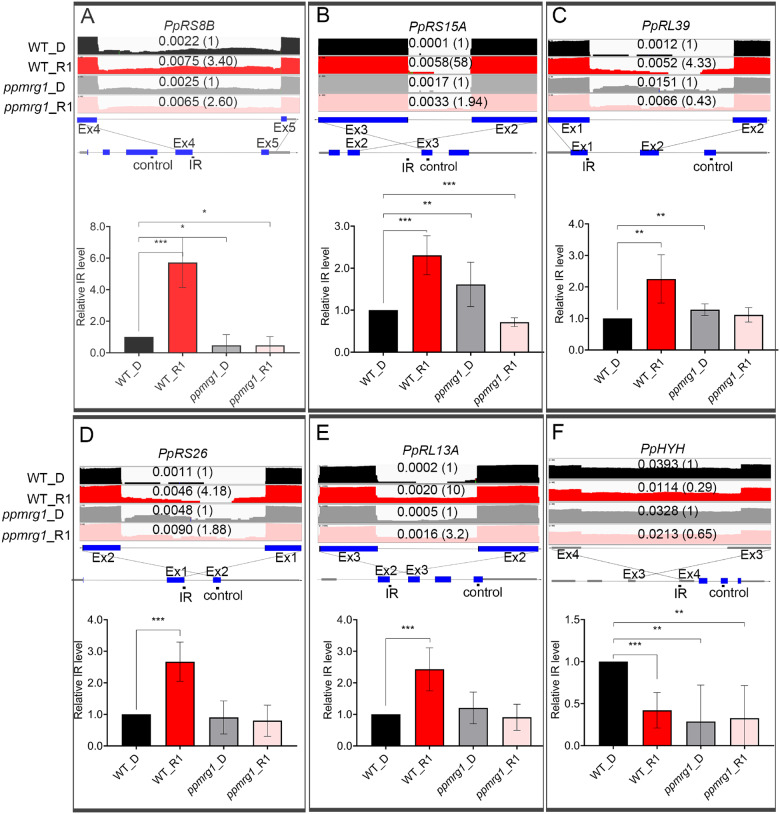
We validated IR events with RT-qPCR in the WT and ppmrg1. While comparing the IR level of several ribosomal protein genes (Figure 5, A–E; Supplemental Figure S10) from RNA-seq (three biological replicates), we confirmed that the IR level more frequently increased after 1 h of RL treatment in the WT than in ppmrg1. In the case of PpHYH intron 3, IR showed downregulation in response to RL (as we observed above) in the WT, but lost light responsiveness in ppmrg1 (Figure 5F). We also identified RL-regulated AltD/A and ES events in the WT and checked the splicing patterns of these events in ppmrg1 (Supplemental Figure S11). AltD/A and ES were also defective in the mutant, as we found that the majority of RL-responsive AltD/A and ES events are misregulated in the mutant and can be defined as MRG1-dependent. These results further confirm the importance of PpMRG1 in regulating light-mediated AS.
Figure 5.
Validation of IR pattern in the WT and ppmrg1. Five genes previously identified with RL responsive IR (PpRS8B, PpRL39, PpRS26, PpRL13A, and PpHYH) (Wu et al., 2014) and one found in this study (PpRS15A) were selected for experimental validation. RNA-seq tracks showing read coverage (log2 transformed) for six IR events (A, PpRS8B intron 4. B, PpRS15A intron 2. C, PpRL39 intron 1. D, PpRS26 intron 1. E, PpRL13A intron 2. F, PpHYH intron 3) in WT and ppmrg1 samples from plants irradiated with RL (5 μmol m−2 s−1) for 0 (D) and 1 (R1) h. Relative IR ratio compared to the dark is shown in parentheses. Corresponding gene models with PCR-amplified regions for IR and control are below the RNA-seq tracks. Validation was performed in WT and ppmrg1 samples under D and R1. For RT-qPCR, CT values of IR fragments were compared with that of control fragments to calculate relative IR levels. Data from RL samples were normalized with that of the D condition. Values presented are mean with sem (n = 3, biological replicates). *, **, and *** indicate P <0.05, P< 0.01, and P< 0.001, respectively (Student’s t test).
We selected one of light-responsive IR genes Pp3c15_14860 (PpRL39; Figure 5C) to test for H3K36me3 and PpMRG1 enrichment. The results are consistent with our prediction that indeed H3K36me3 and MRG1 levels are responsive to RL and have opposite patterns to IR (Supplemental Figure S12A). In comparison, another gene (PpRPS7) showing IR but without RL responsiveness had no significant RL-induced changes in H3K36me3 and MRG1 levels (Supplemental Figure S12B). In summary, our findings indicate that PpMRG1 is involved in the IR regulation of specific genes under RL.
PpMRG1 positively regulates phototropic responses in P. patens
RL triggers directional protonemal cell growth in mosses and ferns (Burgess and Linstead, 1981; Jenkins and Cove, 1983). Phytochromes are the main photoreceptors that sense the direction of light in P. patens (Mittmann et al., 2004). However, downstream signaling for controlling directional protonemal cell growth is unknown. We recently identified one splicing regulator, PphnRNP-H1, potentially involved in downstream signaling (Shih et al., 2019). The finding supports that regulation at the pre-mRNA splicing step may modulate RL-promoted phototropic responses. To investigate whether MRG proteins also function in photomorphogenic responses in P. patens, we examined RL-mediated phototropic responses in the WT and ppmrg1. We initially grew the protonemata under negative gravitropic conditions in the dark and measured bending angles of the protonemata after irradiating with unilateral RL (1 µmol m−2 s−1) for 1–8 h to induce positive phototropism. The average bending angle increased during RL irradiation but to different degrees among the lines (Figure 6A; Supplemental Figure S13 and Supplemental Table S3). After the 1-h RL treatment, WT protonemata started to bend 10°–20° toward the light, whereas the majority of ppmrg1 protonemata remained at a similar angle as the dark sample. With longer RL exposure, the majority of WT protonemata clearly bent toward the light. However, ppmrg1 protonemata grew without directional preference. The average bending angle of ppmrg1 protonemata was significantly smaller than that of the WT after lateral RL irradiation (Figure 6B). These observations indicate that PpMRG1 is involved in regulating RL-induced phototropic responses, especially during the early step of direction sensing. It thus appears that PpMRG1 promotes the phototropic response in P. patens.
Figure 6.
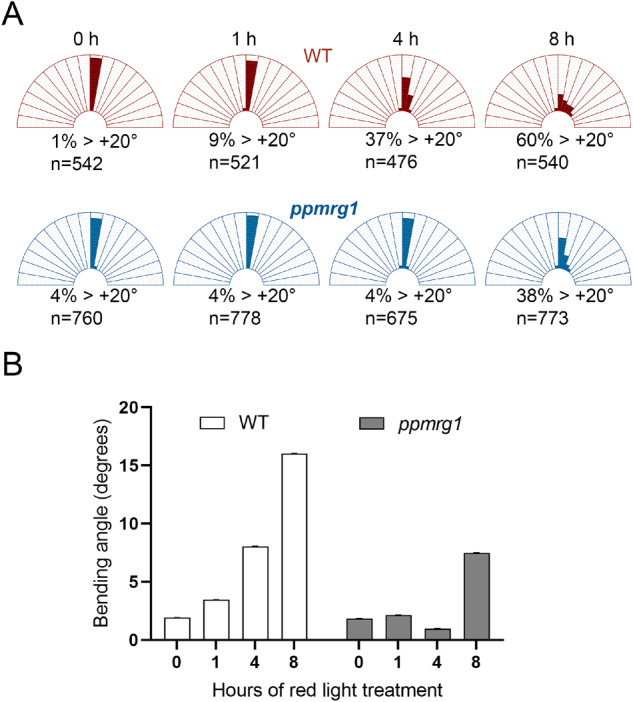
PpMRG1 functions in RL-dependent phototropic response. Seven-day-old protonema were grown vertically in the dark for 3–5 d to induce negatively gravitropic growth of caulonema and then illuminated with lateral RL for 0, 1, 4, and 8 h. Bending angles of WT and ppmrg1 caulonema at each time point were observed under a microscope. A, The distribution (%) of bending plants. Numbers under the figure show the percentage of caulonema at each time point with bending angle > 20°. B, Average bending angles at 0 (n =542), 1 (n =521), 4 (n = 476), and 8 (n = 540) h in the WT and 0 (n = 760), 1 (n = 778), 4 (n = 675), and 8 (n = 773) h in the ppmrg1 are shown. Error bars show the sem values.
Discussion
In this study, we investigated the relationship between histone methylation and AS in response to light. We found that H3K36 is differentially trimethylated at specific gene loci encoding transcripts with light-responsive IR. However, H3K36me3 and IR levels showed opposite patterns of light responsiveness. We further identified a chromatin adaptor, PpMRG1, that associates with the H3K36me3 histone mark more strongly under RL. Loss of PpMRG1 in P. patens not only reduced light responsiveness in AS but also led to a defective RL-mediated phototropic response. Building on the current understanding of MRG function, we therefore propose that MRG1 associates with H3K36me3 and is involved in light regulation of IR in plants.
Based on this hypothesis, MRG1 may act at different loci with different activities in terms of splicing regulation. As illustrated in Figure 7, in the case of genes with transcripts showing light-induced IR, such as PpRS8B, intron splicing is relatively active in darkness. Under light conditions, the MRG potentially associates with H3K36me3 even when H3K36me3 decreases on these loci. The chromatin-anchored MRG could dissociate the unknown splicing regulator from the spliceosome and eventually silence splicing activity (produce IR). For genes with transcripts showing light-repressed IR, such as PpHYH, H3K36me3-associated MRG may help recruit unknown regulators that promote splicing under light conditions to promote splicing.
Figure 7.
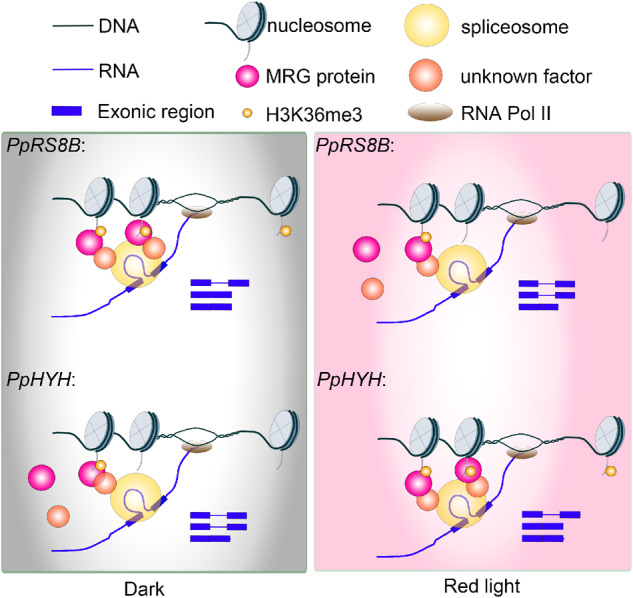
Proposed model for chromatin-coupled regulation of AS. Light may alter H3K36 trimethylation at different gene loci such as PpRS8B and PpHYH or the affinity of the chromatin adaptor MRG1 to have higher or lower occupancy of MRG on chromatin. Unknown factors, which could be splicing regulators, may be differentially recruited by MRG to specific gene loci for AS regulation.
Previous studies have suggested that pre-mRNA splicing is coupled with transcription in mammalian systems (see review in Naftelberg et al., 2015). Accumulating data have shown that histone modification controls the chromatin structure and contributes to recruitment of chromatin adaptors that interact with splicing factors. Chromatin adaptors including MRG15, GCN5, CHD1, and HP1α have been reported to interact with different histone marks and splicing factors in metazoans (Martinez et al., 2001; Bowman et al., 2006; Sims et al., 2007; Gunderson and Johnson, 2009; Piacentini et al., 2009).
MRG proteins were first identified as having a TF function in the chromatin remodeling complex for cell senescence in human (Bertram et al., 1999). The MRG protein MRG15 is of interest because it is conserved across different species from animals to plants (Bertram and Pereira-Smith, 2001; Bu et al., 2014; Xu et al., 2014). It was later found to recognize the H3K36me3 mark and to recruit the splicing regulator PTB to alternatively spliced exons, thereby potentially regulating pre-mRNA splicing (Luco et al., 2010). In plants, increasing evidence has shown that MRG15 also associates with the H3K36me3 mark (Xu et al., 2014; Pajoro et al., 2017). Arabidopsis MRGs regulating AS in response to temperature changes further suggest that an adaptor recruitment mechanism for splicing regulation can control environmental responses in plants (Pajoro et al., 2017). Another mechanism that recruits protein factors for splicing regulation involves the C-terminal domain (CTD) of RNA polymerase II (RNAPII). Studies from animal systems indicate phosphorylation of different sites in the CTD of RNAPII is important to specifically recruit TFs and splicing factors (see review in Hsin and Manley, 2012). A similar mechanism also exists in plants (Hajheidari et al., 2013; Grasser and Grasser, 2018).
Besides the recruitment model, the kinetic model that considers the elongation rate of RNAPII may also influence the activity of splicing machinery on target transcripts (Luco et al., 2011). The model suggests that the lower speed of RNAPII recruits spliceosomes more efficiently to the splice sites. Notably, a recent study indicates that light regulates AS by controlling the elongation rate of RNAPII (Godoy Herz et al., 2019). RNAPII elongation is faster under light conditions than in darkness. This could potentially decrease the chance of spliceosome association to exons and increase the possibility of IR. The detailed mechanism still requires further investigation.
Although both recruitment and kinetic models explain the basic mechanism by which cotranscriptional regulation occurs during pre-mRNA splicing, one open question is how transcript selectivity for AS is determined, especially when differential splicing occurs under changing conditions. This could depend on the availability and activities of splicing-related factors in response to changing conditions. In the case of light, several recent studies indicate that splicing regulators and spliceosome components are modulated to regulate light-responsive AS in Arabidopsis and Physcomitrella (Shikata et al., 2012; Xin et al., 2017; Lin et al., 2019; Shih et al., 2019). Activities of these factors, such as RNA binding and protein interactions, may be modulated by light, thereby generating different transcript specificities. Results from this study provide an alternative explanation on how splicing regulation associates with chromatin status to determine transcript specificity. Since the association of adaptor proteins depends on the status of histone modification, transcript selectivity can be adjusted by altering histone modification or the affinity of adaptor proteins to histone marks in response to light. Additional experiments are needed to confirm this hypothesis.
Recently, the relationship between IR and chromatin accessibility was explored in Arabidopsis and rice. IR is enriched in DNase I hypersensitive sites, which indicates that chromatin is more open in retained introns (Ullah et al., 2018). Increasing chromatin accessibility may allow for fast elongation of transcription and promotes splice site skipping. As light increases the elongation rate of RNAPII (Godoy Herz et al., 2019), which potentially occurs on open chromatin, light could then promote IR at these regions. Because H3K36me3 is mainly distributed on actively transcribed regions, which could include the gene body where chromatin is relatively open, H3K36me3 may participate in the regulation of light-responsive IR through recruiting protein factors such as MRGs. Future work such as investigation of light-responsive chromatin accessibility as well as H3K36me3 status and MRG occupancy, especially in gene body regions at the genome-wide scale, may help to strengthen the hypothesis.
AS largely contributes to transcriptome and proteome complexity in eukaryotes and therefore provides an additional layer of regulation to reprogram biological activities in the cell. AS plays an important role in the plant response to changing environmental conditions, as posttranscriptional regulation allows rapid actions (Staiger and Brown, 2013). Numerous studies have demonstrated that AS of specific gene transcripts alters plant light responses (see review in Cheng and Tu, 2018). How transcript specificity is determined in response to changing light conditions is largely unknown. In this study, we provided evidence that a chromatin adaptor associates with histones and participates in light-mediated AS. Further experiments are needed to provide additional details about this mechanism, such as the involvement of splicing factors in associating with the chromatin adaptor to determine transcript specificity of AS. By further deciphering the relationship between the adaptor protein and histone modification in response to light and their roles in splicing regulation, we hope to elucidate the intricate mechanism underlying pre-mRNA splicing in plants.
Materials and methods
Plant materials and growth conditions
Protonemata of P. patens were grown on solid BCDAT medium (BCD medium supplemented with 5 mM ammonium tartrate) overlaid with cellophane for approximately 7–14 d (Cove et al., 2009). Plants were cultured at 25°C under continuous white light (80–100 μmol m−2 s−1). Seven-day-old protonemata were collected, blended in sterile water with a tissue grinder (Kurabo, Japan) at 1,200 rpm for 5 min, and spread onto solid BCDAT medium overlaid with cellophane. For light treatment, 7-d-old protonemata of the corresponding plants were grown in the dark for 3 d, followed by exposure to liquid nitrogen, and then stored at –80°C for further use.
Plasmid construction
Plasmids used for P. patens transformation were constructed using an In-Fusion HD Cloning kit (Clontech, USA). In brief, complementary DNA (cDNA) sequences of the desired genes were amplified using primers containing 12–15 nucleotides identical to the cDNA ends and 12–15 nucleotide extensions homologous to the vector ends according to the manufacturer’s instructions. PCR-generated sequences were ligated with linearized vectors using the In-Fusion enzyme and transformed into Escherichia coli for plasmid amplification. To generate constructs for the PpMRG1 overexpression line, the PpActin promoter-driven c-Myc-PpMRG1 was constructed into pTN80-APH4 vector containing an APH4 (for hygromycin resistance) cassette and two fragments from an intergenic region of P. patens chromosome 2 as the downstream and upstream sequences for homologous recombination. For generating the mutant line, about 1 kb upstream and downstream of target gene regions was amplified and constructed into the pTN80-nptII vector (including a G418-resistance nptII gene) for gene targeting.
Transformation and generation of knockout and overexpression lines
Transformation of P. patens was performed with the polyethylene glycol (PEG)-mediated DNA method similar to that described in PHYSCOBase (http://www.nibb.ac.jp/evodevo/PHYSCOmanual/9.1.htm). Seven-day-old protonemata generated from sporophytes or subcultures were collected for protoplast isolation and PEG-mediated plasmid transformation. The transformants were selected on BCDAT medium with respective antibiotics (for mutant line, 20 mg/L of G418; for overexpression line, 20 mg/L of hygromycin). Gene-specific insertion in stable transformants was verified by PCR using specific primers (Supplemental Data Set S5).
RT-qPCR analysis
Plant tissues collected before and after light treatment were frozen in liquid nitrogen for total RNA extraction with a Plant Total RNA Miniprep Purification kit (GeneMark, Taiwan) following the manufacturer’s instructions. On-column DNase digestion was performed to remove any genomic DNA contamination prior to cDNA synthesis. First-strand cDNA was synthesized using 4.5 μg of RNA with a SuperScript III RT kit (Invitrogen, USA) according to the standard protocol. Diluted cDNA was subjected to RT-qPCR on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific, USA) using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA). Primers for RT-qPCR analysis were designed based on the sequence of the corresponding region (Supplemental Data Set S5). PpACT7 (Pp3c3_33440V3) was used as the control for normalization in RT-qPCR. To calculate the relative IR level, signal intensities from IR fragments were normalized with the control. IR level under RL was then normalized with that in the dark condition. RT-qPCR was performed with three biological replicates.
ChIP
ChIP was performed according to Widiez et al. (2014) with some modification. For each chromatin extraction, 1 g of tissue was pooled and ground under liquid nitrogen for nuclei isolation. Chromatin extractions from the same tissue/condition were pooled into a 1.5-mL tube and sonicated 45 min at 4°C on the bioruptor sonicator (Diagenode, USA). Fragmented chromatin samples were clarified by 13,000g centrifugation for 5 min at 4°C. The pelleted nuclei were suspended in 100 µL of nuclear lysis buffer (50-mM Tris–HCl, pH 7.5, 1% w/v sodium dodecyl sulfate ( SDS), 10-mM EDTA, pH 8 supplemented with protease inhibitor cocktail from Roche) at 65°C for 5 min and then centrifuged at 13,000g for 5 min to collect supernatants. Chromatin samples were diluted 10 times with protein IP buffer (1% v/v Triton X-100, 8-mM Tris–HCl pH 7.5, 0.5-mM EDTA pH 8, 120-mM NaCl, 1-mM phenylmethylsulfonyl fluoride (PMSF), supplemented with protease inhibitor cocktail from Roche). Two milliliters of chromatin samples was incubated with 50 µL of protein A magnetic beads (Millipore) for 2 h, followed by incubation with 10 µL of antibodies overnight at 4°C. Protein A beads were washed three times at 4°C. Chromatin was reverse-cross-linked by adding 100 µL of elution buffer (1% w/v SDS, 0.1-M NaHCO3) and incubated overnight at 65°C under agitation. “Input DNA” was prepared in the same way as described above without the immunoprecipitation step.
Immunoprecipitation
One gram of tissue was pooled, ground under liquid nitrogen, and suspended in 25 mL of nuclei isolation buffer I (10-mM, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.6, 0.4-M sucrose, 5-mM KCl, 5-mM MgCl2, 5-mM EDTA, pH 8, 0.6% v/v Triton X-100, 14-mM β-mercaptoethanol, and 1-mM PMSF) for 5 min at room temperature. Samples were centrifuged at 3,000g for 10 min at 4°C. The pelleted nuclei were lysed with 100 μL of nuclear lysis buffer (50-mM Tris–HCl, pH 7.5, 1% w/v SDS, 10-mM EDTA, pH 8 supplemented with protease inhibitor cocktail from Roche). Supernatants were collected after centrifugation and then diluted with 1 mL of protein IP buffer (1% v/v Triton X-100, 8-mM Tris–Cl, pH 7.5, 0.5-mM EDTA, pH 8, 120-mM NaCl, 1-mM PMSF, supplemented with protease inhibitor cocktail from Roche). The c-Myc-specific antibody conjugated with agarose beads was incubated with nucleus protein extract, washed three times with protein IP buffer, boiled in sample buffer for 5 min, and then detected as pulled-down proteins by immunoblotting.
RNA-seq and data analysis
RNA-seq was performed as described with minor modifications (Shih et al., 2019). Seven-day-old WT and mutant protonemata were grown in the dark for 3 d, followed by 1 h (R1) of RL treatment (660-nm LED, 5 μmol m−2 s−1) at 25°C. Dark-grown protonemal cells were collected as the dark control. RNA isolation and library construction of three biological replicates sampled at different times were performed independently. Sequencing was performed on the HiSeq 4000 platform at Yourgene Bioscience. On average, 95 million 100-nucleotide paired-end reads were obtained per library (Supplemental Table S1). Sequence reads were mapped to the Physcomitrella genome annotation V3.3 (JGI, https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppatens) using BLAT (Kent, 2002) and Bowtie 2 (Langmead and Salzberg, 2012). More than 95% of the reads were perfectly aligned to the reference genome. The three major types of AS events, IR, AltD/A, and ES, were analyzed as described previously with minor modifications (Kanno et al., 2017). In brief, IR was measured by comparing IR levels of each individual event in the dark controls (biological repeats from three independent experiments) with those in the R1 replicates using a Student’s t test, where the IR level was defined as the average read coverage depth of the intron divided by the average read coverage depth of the adjacent exons. IR events were filtered out using the following criteria: no replicates in two samples with 100% read coverage for the retained intron and no replicates in two samples with average read coverage depth >1 for the retained intron. RL-regulated IR events were considered when an event had a P ≤ 0.05. To identify MRG1-dependent IR events, fold changes (R1/D) of IR levels for each RL-regulated IR events were calculated. Those events with the fold change in WT larger than that of ppmrg1 were defined as MRG1-dependent IR events. RL-responsive ES and AltD/A events were determined using a previously described method (Shih et al., 2019). ES and AltD/A events with a P ≤0.05 in WT but ≥0.05 in ppmrg1 were defined as MRG1-dependent events.
Phototropism assay
Phototropism assay was performed as described with minor modifications (Shih et al., 2019). Physcomitrella patens protonemata were grown on BCDAT with 0.8% agar (w/v) under continuous white light. The protonemata were collected and transferred to vertically placed Petri dishes, sandwiched between two layers of cellophane, and incubated in the dark for 3 d to induce caulonema formation. In the dark, caulonema filaments grow vertically in response to negative gravitropism. To analyze phototropism, dark-incubated protonemata were treated with lateral monochromatic RL (660-nm LED, 1 μmol m−2 s−1). The protonemata were photographed under a Lumar V12 stereomicroscope (Zeiss), and the bending angle was calculated with ImageJ. The average bending angle plus SEM of dark-incubated plants (either WT or mutant) was defined as straight, and protonemata with larger bending angles than those of dark-incubated plants were categorized as exhibiting positive phototropism.
Accession numbers
RNA-seq data from this publication have been submitted to the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the accession number PRJNA398698. Gene information described in this article can be found in the Phytozome JGI (http://www.phytozome.net/physcomitrella.php) under Supplemental Table S4.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. H3K36 trimethylation responds to light.
Supplemental Figure S2. Validation of H3K36me3 enrichment on additional genes using ChIP-qPCR.
Supplemental Figure S3. Validation of H3K36me3 enrichment under blue light condition.
Supplemental Figure S4. Generation of c-Myc-PpMRG1 transgenic plants by homozygous recombination.
Supplemental Figure S5. Association of PpMRG1 with histone lysine trimethylations.
Supplemental Figure S6. RL-dependent association of PpMRG1 with H3K36me3.
Supplemental Figure S7. PpMRG1 enrichment under blue light condition.
Supplemental Figure S8. Generation of ppmrg1 knockout mutant by homozygous recombination.
Supplemental Figure S9. PpMRG1 also regulates IR independent of RL.
Supplemental Figure S10. Validation of IR pattern in the WT and ppmrg1.
Supplemental Figure S11. PpMRG1 is involved in regulating RL-mediated AS.
Supplemental Figure S12. Validation of H3K36me3 and PpMRG1 enrichment on additional genes by using ChIP-qPCR.
Supplemental Figure S13. Phototropism in the WT and ppmrg1.
Supplemental Table S1. Read mapping statistics of RNA-seq data.
Supplemental Table S2. Functional enrichment of IR genes regulated by PpMRG1.
Supplemental Table S3. Distribution of bending angles in the WT and ppmrg1.
Supplemental Table S4. IDs and annotations of genes used in this study.
Supplemental Data Set S1. List of light-regulated IR events in the WT and ppmrg1.
Supplemental Data Set S2. List of light-regulated ES events in the WT and ppmrg1.
Supplemental Data Set S3. List of light-regulated AltD/A events in the WT and ppmrg1.
Supplemental Data Set S4. IR, ES, and AltD/A levels in the WT and ppmrg1 for plotting heatmaps.
Supplemental Data Set S5. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Kathleen Farquharson for critically reading the manuscript, Wen-Dar Lin in the Bioinformatics Core Laboratory, Shu-Jen Chou and Mei-Jane Fang in the Genomic Technology Core Laboratory of the Institute of Plant and Microbial Biology, Academia Sinica, Taiwan for technical assistance.
Funding
This work was supported by the grant to S.-L.T. from the Ministry of Science and Technology (Grant No: MOST 106-2311-B-001 -033 -MY3) and Academia Sinica, Taiwan.
Conflict of interest statement. The authors declare that they have no competing interests.
C.-C.W., H.-L.H., and S.-L.T. designed the research. C.-C.W. and H.-Y.H. performed the experiments. C.-C.W., H.-Y.H., and S.-L.T. analyzed the data. C.-C.W. and S.-L.T. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Shih-Long Tu (tsl@gate.sinica.edu.tw).
References
- Bertram MJ, Pereira-Smith OM (2001) Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene 266: 111–121 [DOI] [PubMed] [Google Scholar]
- Bertram MJ, Bérubé NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, et al. (1999) Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol 19: 1479–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BR, Moure CM, Kirtane BM, Welschhans RL, Tominaga K, Pereira-Smith OM, Quiocho FA (2006) Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure 14: 151–158 [DOI] [PubMed] [Google Scholar]
- Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong AW (2014) Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess J, Linstead PJ (1981) Studies on the growth and development of protoplasts of the moss, Physcomitrella patens, and its control by light. Planta 151: 331–338 [DOI] [PubMed] [Google Scholar]
- Charron JB, He H, Elling AA, Deng XW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YL, Tu SL (2018) Alternative splicing and cross-talk with light signaling. Plant Cell Physiol 59: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Cove DJ, Perroud P-F, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS (2009) Culturing the moss Physcomitrella patens. Cold Spring Harbor Protocols 2009, pdb.prot5136. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. (2011). Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–983 [DOI] [PubMed] [Google Scholar]
- Godoy Herz MA, Kubaczka MG, Brzyzek G, Servi L, Krzyszton M, Simpson C, Brown J, Swiezewski S, Petrillo E, Kornblihtt AR (2019) Light regulates plant alternative splicing through the control of transcriptional elongation. Mol Cell 73: 1066–1074 [DOI] [PubMed] [Google Scholar]
- Gómez Acuña LI, Fiszbein A,, Allo M, Schor I, Kornblihtt A. (2013) Connections between chromatin signatures and splicing. WIREs RNA 4: 77–91 [DOI] [PubMed] [Google Scholar]
- Grasser M, Grasser KD (2018) The plant RNA polymerase II elongation complex: a hub coordinating transcript elongation and mRNA processing. Transcription 9: 117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson FQ, Johnson TL (2009) Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet 5: e1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Zhou J, Elling AA, Charron J-BF, Deng X (2008) Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol 147: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M, Koncz C, Eick D (2013) Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci 18: 633–643 [DOI] [PubMed] [Google Scholar]
- Hansen JC, Tse C, Wolffe AP (1998) Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37: 17637–17641 [DOI] [PubMed] [Google Scholar]
- Hartmann L, Drewe-Boss P, Wiessner T, Wagner G, Geue S, Lee HC, Obermuller DM, Kahles A, Behr J, Sinz FH, et al. (2016). Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 28: 2715–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL (2012) The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26: 2119–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI, Cove DJ (1983) Phototropism and polarotropism of primary chloronemata of the moss Physcomitrella patens: responses of mutant strains. Planta 159: 432–438 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kanno T, Lin WD, Fu JL, Chang CL, Matzke AJM, Matzke M (2017) A genetic screen for pre-mRNA splicing mutants of Arabidopsis thaliana identifies putative U1 snRNP components RBM25 and PRP39a. Genetics 207: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—The BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL (2011) Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA 108: 13564–13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165 [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Negri R, Del Vescovo V, Fatica A, Altieri F, Pimpinelli S (2009) Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet 5: e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BY, Shih CJ, Hsieh HY, Chen HC, Tu SL (2019) Phytochrome coordinates with a hnRNP to regulate alternative splicing via an exonic splicing silencer. Plant Physiol 182: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Lu FL, Cui X, Cao XF (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T (2011) Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T (2010) Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E, Sanchez SE, Romanowski A, Schlaen RG, Sanchez-Lamas M, Cerdan PD, Yanovsky MJ (2016) Acute effects of light on alternative splicing in light-grown plants. Photochem Photobiol 92: 126–133 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol 21: 6782–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann F, Brucker G, Zeidler M, Repp A, Abts T, Hartmann E, Hughes J (2004). Targeted knockout in Physcomitrella reveals direct actions of phytochrome in the cytoplasm. Proc Natl Acad Sci USA 101: 13939–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, Kornblihtt AR (2015) Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 84: 165–198 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Severing E, Angenent GC, Immink RGH (2017) Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol 18: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Li Z, Zhou N, Ma M, Jiang Y, Dong A, Shen WH, Li L (2018) Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol 176: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA (2012) Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet 8: e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CJ, Chen HW, Hsieh HY, Lai YH, Chiu FY, Chen YR, Tu SL (2019) Heterogeneous nuclear ribonucleoprotein H1 coordinates with phytochrome and the U1 snRNP complex to regulate alternative splicing in Physcomitrella patens. Plant Cell 31: 2510–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Nakashima M, Matsuoka K, Matsushita T (2012) Deletion of the RS domain of RRC1 impairs phytochrome B signaling in Arabidopsis. Plant Signal Behav 7: 933–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Brown JW (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah F, Hamilton M, Reddy ASN, Ben-Hur A (2018) Exploring the relationship between intron retention and chromatin accessibility in plants. BMC Genomics 19: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widiez T, Symeonidi A, Luo C, Lam E, Lawton M, Rensing SA (2014) The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. Plant J 79: 67–81 [DOI] [PubMed] [Google Scholar]
- Wu SH (2014) Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu Rev Plant Biol 65: 311–333 [DOI] [PubMed] [Google Scholar]
- Wu HP, Su YS, Chen HC, Chen YR, Wu CC, Lin WD, Tu SL (2014) Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol 15: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E (2017) SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc Natl Acad Sci USA 114: E7018–E7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gan ES, Zhou J, Wee WY, Zhang X, Ito T (2014) Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res 42: 10960–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.