Abstract
One-sentence summary
Recent advances provide insight into the molecular mechanisms underlying host-dependent seed germination and haustorium formation in parasitic plants.
Advances
The conditioning period revealed extensive DNA demethylation required for parasitic weeds to germinate in response to host-exuded chemicals.
The diversification and subsequent neofunctionalization of HTL/KAI2 proteins have provided parasitic weeds with hypersensitivity to host-exuded SLs.
Both SLs and non-SLs induce similar molecular responses in parasitic weeds, analogous to nonparasitic plants.
Transcriptomes underlying haustoriogenesis in different lineages of parasitic plants converged to a similar list of genes that regulate root development, carbohydrate-active enzymes (CAZymes) activity, and hormone homeostasis in nonparasitic plants.
Vascularization of the haustorium depends on genes that regulate vascularization in nonparasitic plants
Parasitic plants sensu stricto are defined by their ability to tap nutrients requiredfor their growth directly from other plants, by invading the stems or the roots of their hosts through specific structures called haustoria (Wicke and Naumann, 2018). First records mentioning parasitic plants date back to the third century BC. Nevertheless, it is only in the 1950s that intensive research in their physiology began, when Striga asiatica was accidentally introduced into the United States where it rapidly and intensely altered maize (Zea mays) crops, then in the 1970s and 1980s when many African countries suffered from severe famines (Heide-Jørgensen, 2008). Research on parasitic plant biology is today experiencing a new boom, as a handful of species are increasingly damaging crops worldwide.
The world apart of parasitic plants
There are ∼4,750 parasitic plants species, all grouped within dicotyledonous Angiosperms (Nickrent, 2020) at the exception of Parasitaxus usta that belongs to the Gymnosperms group (Feild and Brodribb, 2005). Parasitic plants represent 292 genera, grouped in c. 20 families, distributed on all continents except Antarctica (Westwood et al., 2010; Nickrent, 2020). However, parasitic plants have been and still are subjected to extensive taxonomic rearrangements. Most recent phylogenetic analyses indicate 12 independent acquisitions of a parasitic lifestyle (Bromham et al., 2013). Consequently, parasitic plants display very high morphological variability and extremely diverse parasite strategies.
Parasitic plants are distinguished according to their site of attachment to the host. Thus, 40% are stem parasites that invade the stem of their host(s) (e.g., mistletoes and dodders), while 60% are root parasites that penetrate the host root system (e.g., broomrapes and witchweeds; Westwood et al., 2010). A second dichotomy separates facultative from obligate parasitic plants according to their degree of dependency to a host. Facultative parasites (e.g., Triphysaria, Phtheirospermum, and Rhinanthus spp.) are chlorophyllous organisms able to complete their life cycle in the absence of a host. However, in natural environments, most facultative parasites are likely to engage in parasitism as solitarily growing parasites suffer from impaired growth and reduced reproductive fitness as described in laboratory experiments (Kabiri et al., 2017; Honaas et al., 2019). Inversely, obligate parasites cannot survive without a host during at least part of their lifecycle (Irving and Cameron, 2009). Finally, most researchers differentiate flowering parasitic plants on the basis of their photosynthetic capacity. Approximately 10% of all flowering parasitic plants are achlorophyllous organisms. These so-called holoparasites are thus necessarily obligate parasites, and must take up water and nutrients (mainly reduced carbon and nitrogen) required for their growth by connecting to the xylem and phloem of their host through haustorial structures. By contrast, hemiparasites, whether facultative or obligate, are chlorophyllous and thus retain photosynthetic ability. Obligate hemiparasites can limit their spoliation to mainly water and mineral compounds from the xylem of the host once they reach photosynthetic maturity.
The Orobanchaceae family is the largest family of parasitic plants (Heide-Jørgensen, 2008). Orobanchaceae species are root parasites, presumably arisen from a single gain of the ability to form haustoria (Schneeweiss, 2013). It comprises the full spectrum of trophic specialization, with a broad range of host preferences and specificities. Most importantly, it contains species that have turned into weeds causing 20%–100% of crop yield losses at the expense of several billion dollars a year (Parker, 2012; Rodenburg et al., 2016). Among these pests are holoparasites of the Orobanche and Phelipanche genera (broomrapes), which together affect a wide spectrum of hosts (e.g., legumes, sunflower, tomato, oilseed rape, etc.) in Europe, North Africa, and Asia (Parker, 2012). In addition, Striga species (witchweeds) are considered the major biotic constraint to cereal production in sub-Saharan agriculture, with over 50 million hectares infested annually (Ejeta, 2007).
Broomrapes and witchweeds have acquired numerous functional adaptations allowing them to compete with crops for resources. As compared to nonparasitic plants and nonweedy parasites, they have adapted to sense the presence of host roots in their close vicinity to trigger seed germination and, subsequently, differentiation of early haustorial structures (Delavault et al., 2017). After successful establishment of a vascular continuum with the host, the parasite acts as a supernumerary and dominant sink whereby it withdraws water, nutrients, and hormones necessary for its own, at the expense of the host’s development (Péron et al., 2017). As the reallocated resources accumulate, the parasite develops an external bulbous structure, the tubercle. This structure then differentiates into an underground stem, from which an inflorescence emerges aboveground within a few weeks.
Broomrapes and witchweeds sustainability is ensured by a very high reproduction rate. Indeed, each mature flowering plant produces up to hundreds of thousands of microscopic seeds easily spread from one location to another. Most of the current dreadful parasitic weeds thrive well in temperate areas and they have remarkably expanded in response to global warming (Phoenix and Press, 2005). Consequently, the rhizosphere of host plants is easily replenished in the span of only one generation, and parasitic weeds are expected to dramatically expand to new territories in the near future (Fernández-Aparicio et al., 2016b).
This review intends to describe recent progress in understanding how parasitic weeds sense surrounding hosts and highlights advancements in their genetics.
Seed dormancy and germination in root parasitic weeds
The successful establishment of obligate parasitic weeds of the Orobanchaceae family in agro-ecosystems partly relies on a particular seed physiology. As compared to most facultative hemiparasites and nonparasites (except Orchidaceae species), broomrapes and witchweeds produce small seeds with an average size of 200 µm (Joel, 2013). A single mature plant annually releases tens to hundreds of thousands of these dust-like seeds, which can remain dormant and viable in the soil for more than a decade (Yoder and Scholes, 2010; Joel, 2013). The progressive release from the dormant state occurs, as for the many nonparasitic species displaying a physiological dormancy, upon perception of stable and adequate environmental conditions (Bewley et al., 2013; Baskin and Baskin, 2014).
Decades of research on nonparasitic models have identified multiple dormancy-breaking signals, among which appropriate temperatures and water and oxygen availability are arguably universal prerequisites in the decision to maintain or release physiological dormancy. The same rules perfectly apply to obligate parasitic weeds, whose dormancy release takes the form of a “warm stratification.” Also referred to as the “conditioning period,” it indeed requires a moist environment that initiates seed imbibition, and lasts from few days to several weeks at 18°C–30°C in in vitro conditions (Matusova et al., 2004; Lechat et al., 2012; Brun et al., 2019). A recent study identified ∼15,000 differentially expressed genes that correlate with the acquisition of a conditioned state in Phelipanche aegyptiaca (Bao et al., 2017). The authors found that key genes encoding phosphofructokinase (PFK) and pyruvate kinase (PK) involved in the glycolysis pathway are downregulated in conditioned seeds compared with unconditioned seeds (Bao et al., 2017). In the closely related species P. ramosa, the adenylate energy charge reaches a maximum of 0.9 within a day of conditioning (Lechat et al., 2012). These results together suggest that most of the energy required for subsequent germination is produced through the glycolysis pathway early during the conditioning phase, which constitutes a major response to water uptake. In addition, Lechat et al. (2015) demonstrated through a pharmacological approach in P. ramosa that global DNA demethylation must occur for the conditioning to be achieved. Similar patterns are observed upon early imbibition and in germinating seeds of multiple nonparasitic species (Portis et al., 2004; Lu et al., 2006; Meng et al., 2012; Bouyer et al., 2017; Kawakatsu et al., 2017; Narsai et al., 2017).
The current lack of genetically tractable systems along with the relatively low amount of “omics” datasets for Orobanchaceae species still hinder our understanding of the molecular mechanisms at play during the conditioning period. For instance, which molecular actors contribute to such extensive DNA demethylation, and what are the targeted genomic regions? The current lines of evidence indicate that the conditioning period in parasitic weeds resembles the process of dormancy release of nonparasites to a reasonable extent, which suggests that similar genetic and epigenetic programs might regulate both developmental frameworks. Accordingly, the purpose of dormancy release in both cases is to allow seeds to germinate at the proper time and place, hence upon appropriate germination factors. Broomrapes and witchweeds are unique in their ability to sense the presence of potential host roots in their close vicinity, which ensures that conditioned seeds do not germinate unless they perceive chemicals exuded by surrounding potential host roots into the rhizosphere. The functional gain of such a recognition system is critical to their survival since it prevents microscopic seeds from wasting their limited resources in the attempt to reach a suitable host. The most characterized germination stimulants are strigolactones (SLs), a family of plant hormones that also act in planta throughout the development of the host (Waters et al., 2017). More than 25 SLs have been identified since the first discovery of strigol as a germination stimulant for S. lutea (Cook et al., 1966), and recent reviews exhaustively describe SL chemical diversity and activities on the germination of parasitic weeds (Al-Babili and Bouwmeester, 2015; Ćavar et al., 2015; Brun et al., 2018). There is a clear correlation between the ability of the parasitic weed to germinate upon a certain SL structure and its occurrence in the parasite’s host(s) exudates (Fernández-Aparicio et al., 2009; Fernández-Aparicio et al., 2011; Ueno et al., 2014), suggesting that host specificity and range are at least partly determined by this chemical interaction. In some instances, the use of low SL producers has proven successful in inhibiting the germination of parasitic weeds (Gobena et al., 2017). However, it is not universally practicable as many of them also exhibit submicromolar sensitivities to chemicals whose structures deviate from canonical SLs (Brun et al., 2018). For example, dehydrocostus lactone (DCL) exuded by sunflower stimulates germination of the sunflower-specific parasite Orobanche cumana, but not of other incompatible Phelipanche or Striga species (Joel et al., 2011; Brun et al., 2019). Similarly, a specific strain of P. ramosa displays a high sensitivity to 2-phenylethyl isothiocyanate (2-PEITC) present in the rhizosphere of oilseed rape on which it has adapted exceptionally well in the span of a decade (Auger et al., 2012; Brun et al., 2019; Stojanova et al., 2019).
Recent studies conducted in P. aegyptiaca and S. hermonthica have revealed that perception of such diversity of SLs occurs through multiple α/β hydrolases of the HTL/KAI2 family (HYPOSENSITIVE TO LIGHT/KARRIKIN INSENSITIVE2), with each protein displaying its own affinity toward specific SLs (Conn et al., 2015; Toh et al., 2015; Zhang et al., 2020). Interestingly, weedy Orobanchaceae species contain a higher number of HTL/KAI2 genes that display a faster rate of molecular evolution compared with nonweeds and nonparasitic species (Conn et al., 2015). This has probably led to a neofunctionalization of HTL/KAI2 proteins, since HTL/KAI2 in nonparasitic species perceive karrikins (KARs) but not SLs (Flematti et al., 2004; Nelson et al., 2009). KARs share partial structural similarity with SLs, although they are produced through combustion of plant material. This family of compounds acts as smoke-derived signals to promote seed germination of numerous nonparasitic species after forest burning, thus favoring recolonization of devastated ecosystems (Nelson et al., 2012; Flematti et al., 2015). While SLs and KARs are different signals leading to seed germination of either parasitic weeds or nonparasitic plants, both share signaling analogies. Upon KAR perception in nonparasites, the HTL/KAI2 receptor binds the F-box protein MORE AXILLARY BRANCHES2 (MAX2), which supposedly ubiquitinates the transcriptional repressor SUPPRESSOR OF MAX2-1 (SMAX1) for proteasomal degradation (Nelson et al., 2011; Stanga et al., 2013). Interestingly, parasitic weeds also contain functional MAX2 proteins that can rescue the Arabidopsis max2 mutant (Liu et al., 2014; Li et al., 2016). In addition, several studies indicate that KAR and SL signaling pathways similarly lead to altered abscisic acid (ABA) and gibberellins (GA) homeostasis. Notably, smoke-treated seeds of Nicotiana attenuata display lower ABA levels and higher GA levels (Schwachtje and Baldwin, 2004), which corroborates results obtained with A. thaliana and Lactuca sativa seeds treated with purified KARs (Nelson et al., 2009; Gupta et al., 2019). Similarly, exogenous application of the synthetic SL analog rac-GR24, which induces both SL and KAR signaling in nonparasitic plants (Scaffidi et al., 2014), promotes ABA decrease and GA increase in the obligate root parasitic plants S. hermonthica, P. ramosa, and P. aegyptiaca (Lechat et al., 2012; Toh et al., 2012; Bao et al., 2017). Such hormonal changes are consistently correlated with differential regulation of genes encoding ABA and GA biosynthetic and catabolic enzymes (Nelson et al., 2009; Lechat et al., 2012; Toh et al., 2012; Bao et al., 2017; Brun et al., 2019). These findings together illustrate that HTL/KAI2 signaling, although activated by distinct chemical cues in parasitic weeds and nonparasitic species, similarly stimulates a decrease in the ABA/GA balance, as happens in response to other major environmental cues favoring seed germination over dormancy (Finch-Savage and Leubner-Metzger, 2006; Holdsworth et al., 2008; Finkelstein, 2013).
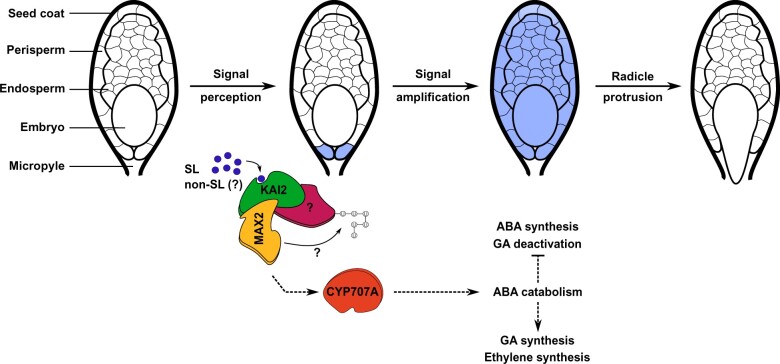
ABA catabolism seems especially key to germination of parasitic weeds. Indeed, CYP707A genes encoding ABA catabolic enzymes are consistently upregulated early upon stimulation in multiple species of broomrapes and witchweeds (Lechat et al., 2012; Brun et al., 2019). In P. ramosa, PrCYP707A1 is upregulated as soon as 30 min following exogenous treatment with rac-GR24, and up to 100-fold after 24 h (Lechat et al., 2012). Moreover, exogenous application of Abz-E2B, a specific inhibitor of CYP707A enzymes (Okazaki et al., 2012) completely abolishes germination of broomrapes and witchweeds (Brun et al., 2019). This suggests that early induction of ABA catabolism by CYP707A enzymes is a prerequisite for seed germination of parasitic weeds. Opposite to this is the recent demonstration that seeds of S. hermonthica are insensitive to 100 µM ABA during germination (Fujioka et al., 2019). Due to the high genetic variability within and between Striga populations (Bozkurt et al., 2015; Unachukwu et al., 2017), these controversial results might simply be explained by the use of different genetic variants between studies. As compared to the early induction of ABA-related genes in obligate root parasites, the actual decrease in ABA content rather constitutes one of the late germination events when the radicle is about to protrude out of the seed coat (Lechat et al., 2012; Toh et al., 2012). Lechat et al. (2012) observed that early induction of PrCYP707A1 expression occurs in two cells beneath the micropyle, which colocalizes with the putative site of SL perception (Plakhine et al., 2012). Moreover, only 58 genes are differentially expressed after 6 h of rac-GR24 treatment in P. ramosa (Lechat et al., 2012), which corroborates studies conducted in Arabidopsis showing that SL application does not induce massive changes at the transcript level (Mashiguchi et al., 2009). By contrast, exogenous application of rac-GR24 on P. aegyptiaca seeds for 48 h results in thousands of differentially expressed genes, among which ABA biosynthetic and GA degradation genes are downregulated, while GA biosynthetic genes are upregulated (Bao et al., 2017). These results suggest that early events of SL signaling consist of rapid and localized—yet undetected so far—ABA degradation. This would act as a signal that propagates throughout the seed and activates transcriptional responses in favor of a decrease in ABA/GA balance at a larger scale to induce germination (Figure 1).
Figure 1.
Hypothetical model for stimulant-induced seed germination of root parasitic weeds of the Orobanchaceae family. The perception of SLs or structurally divergent (non-SL) germination stimulants occurs in two cells beneath the micropyle. Perception of SL signals (and perhaps non-SL) by KAI2 proteins triggers the recruitment of the F-box protein MAX2. In turn, MAX2 supposedly ubiquitinates other proteins still unknown in parasitic plants. This signaling pathway leads to limited transcriptional responses (indicated in blue) in these two cells, among which CYP707A transcripts accumulate. The localized degradation of ABA through CYP707A enzymes at early time points is crucial for the signal to amplify throughout the seed at later time points, whereby strong transcriptional responses and extensive modulation of ABA, GA, and probably ethylene homeostasis occur. The combined decrease in ABA contents and increase in GA and probably ethylene contents enables the radicle to protrude out of the seed coat. Dashed arrows indicate unknown intermediate steps in between molecular events.
At present, information on parasitic weed seed germination is limited and most often consists of finding similarities to major regulatory pathways found in nonparasitic species. The fact that parasitic weeds adapt to their hosts’ environment by responding to non-SL molecules raises the question of how their perception systems have evolved. Brun et al. (2019) demonstrated that DCL and 2-PEITC also trigger upregulation of CYP707A genes in broomrapes, as observed for rac-GR24 (Lechat et al., 2012). While this supposes that HTL/KAI2 proteins can also perceive DCL and 2-PEITC, it remains plausible that these are perceived by completely different receptors. In addition, exogenous application of Abz-E2B fully inhibits seed germination in broomrapes and witchweeds (Brun et al., 2019) but not in nonparasitic species like A. thaliana (Okazaki et al., 2012). Similarly, Arabidopsis cyp707a mutants, although hyperdormant, ultimately germinate under appropriate light and temperature conditions (Kushiro et al., 2004; Okamoto et al., 2006). This suggests that alternate pathways must be able to compensate the loss of function of CYP707A enzymes in nonparasitic species only.
Such differences between parasitic and nonparasitic species correlate with the relative insensitivity of most parasitic weeds toward cues known to influence physiological dormancy and germination of nonparasitic plants, e.g., light, nitrates, and gibberellins (Zehhar et al., 2002; Chae et al., 2004; Takagi et al., 2009; Yoneyama et al., 2013). In line with this, Bunsick et al. (2020) recently reported that the inability of Arabidopsis GA-depleted or GA-insensitive mutants to germinate is rescued when complemented with certain S. hermonthica HTL/KAI2 receptors. The authors also found that loss of SMAX1 was sufficient to observe germination in Arabidopsis GA-depleted mutants, which suggests that recruitment of the HTL/KAI2 pathway and subsequent SMAX1 degradation bypasses GA signaling, and hence GA requirement for germination, in nonparasitic species. By contrast, S. hermonthica seeds do contain much higher levels of HTL/KAI2 expression, and GR24-treated seeds germinate even on high concentrations of paclobutrazol, a chemical inhibitor of GA synthesis. These results are in accordance with the idea that parasitic weeds have become dependent on solely host-derived signals to germinate, while “differential wiring” (Machin and Bennett, 2020) of independent pathways allows nonparasitic species to respond to a wider range of exogenous cues. Although pioneering, this study does not explain why parasitic weeds do not germinate upon exogenous GA alone. Therefore, collection of genomic and transcriptomic data would be a major step forward in identifying the molecular mechanisms gained and lost during the evolution toward such a restricted germination process.
Haustoriogenesis—the A-P-C of haustorium development
Most molecular studies on haustoriogenesis focus on a relatively small number of species that cover less than 0.5% of the full parasitic plant species range and diversity (Clarke et al., 2019). Newly released genomes and whole transcriptome profiles of these species portray a familiar picture across different evolutionary lineages: many genes and molecular pathways of haustoriogenesis are rooted in the developmental processes of their nonparasitic past (Figure 2).
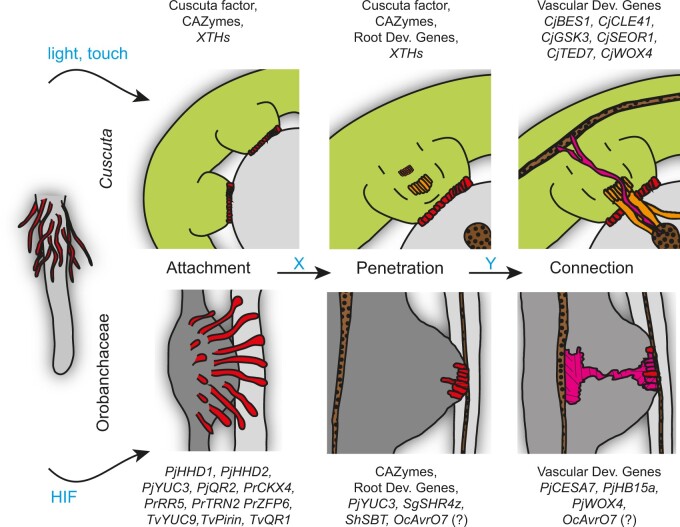
Figure 2.
Checkpoints during haustoriogenesis in Cuscuta spp. and Orobanchaceae. Attachment structures (red) such as holdfasts and haustorial hairs share similarities to root hairs of nonparasitic plants (left). External factors (blue) trigger the formation of these attachment structures. Other, yet unknown, factors (X, Y) initiate the progression through the different phases of haustoriogenesis. Genes and proteins from C. japonica (Cj), O. cumana (Oc), Phtheirospermum japonicum (Pj), P. ramosa (Pr), T. versicolor (Tv), and S. hermonthica (Sh) that accompany the different phases of haustoriogenesis and that have been studied in more detail are highlighted above and below the schematic images. Vascularization of the haustoria is shown in pink, the searching hyphae of Cuscuta in orange. The representations of haustoria are loosely based on micrographs published in Heide-Jørgensen and Kuijt (1993); Svubova et al. (2017), and Wakatake et al. (2018). CAZymes, carbohydrate-active enzymes; Dev., developmental; XTH, xyloglucan endotransglucosylases/hydrolases.
Root parasites form haustoria laterally or terminally. In either case, their development is generally induced by host-related chemical signals, called haustorium-inducing factors (HIFs). The efficiency of phenolic compounds or cytokinins (CKs) as HIFs, in addition to the subsequent redox or cytokinin signaling pathways, is well established in many Orobanchaceae (Goyet et al., 2017; Wada et al., 2019; Billard et al., 2020). CKs are efficient in haustoriogenesis induction in many root holoparasites such as O. cumana, O. aegyptiaca, and P. ramosa, suggesting the loss of sensitivity to phenolics in these species during evolution toward holoparasitism. Unlike in Orobanchaceae, haustoria in the stem parasite Cuscuta (dodders) form in response to light and mechanical stimuli, indicating that the most upstream molecular events in haustorium initiation differ between Cuscuta and Orobanchaceae species. Exogenously applied CKs, however, bypasses the need for light and a mechanical stimulus to induce haustoriogenesis in Cuscuta, suggesting a broader role of cytokinin signaling in haustoriogenesis (Haidar et al., 1998; Ramasubramanian et al., 1988). Notably, not only plant-derived chemicals serve as HIFs. Sphaeropsidones, phytotoxins of the fungal plant pathogen Diplodia cupressi, are potent HIFs in S. hermonthica, O. cumana, and O. crenata (Fernández-Aparicio et al., 2016a).
Haustoriogenesis can be divided into at least three phases (Figure 2): (1) the Attachment to host tissue is followed by (2) the Penetration of the host, and (3) the Connection to the host vasculature, altogether the “A-P-C” of haustorium development. Haustorial structures such as haustorial hairs, haustorial papillae, and holdfasts facilitate attachment. Interestingly, these adhesive structures share developmental mechanisms with root hairs or leaf trichomes. In Cuscuta, localized swellings termed holdfasts (prehaustoria) form by elongation of epidermal and cortical cells toward the host surface and adhere tightly to it (Shimizu and Aoki, 2019). Such adhesive organs also occur in root parasites, for example, in Nuytsia (Heide-Jørgensen, 2008). In Orobanchaceae, HIFs induce prehaustoria in successive steps that include a temporary (lateral haustoria) or a complete (terminal haustoria) arrest of root growth, which is then followed by localized root swellings and the formation of haustorial hairs or papillae. Like regular root hairs, haustorial hairs are filamentous single-cell protuberances of the root epidermis, while haustorial papillae are short extensions of epidermal cells. In the facultative parasites Triphysaria and Phtheirospermum (Orobanchaceae), haustorial hairs proliferate from the surface of the developing haustoria within 12 h after HIF application (Ishida et al., 2016; Wang et al., 2019). Blocking haustorium initiation by the coapplication of HIFs and redox inhibitors prevents the emergence of haustorial hairs, indicating that redox-regulated processes control both: the haustorial hair formation and the initiation of haustorium development (Wang et al., 2019). Similar effects were reported in S. hermonthica and other parasitic plants (Goyet et al., 2019; Wada et al., 2019). Downstream of HIF perception, haustorial hair formation and haustorium development diverge, as Phtheirospermum japonicum haustorial hair defective (hhd) mutants form wild type like haustoria (Cui et al., 2016). The absence of root hairs in Phtheirospermum hhd mutant shows that once initiated, haustorial hair formation depends on genes that also regulate root hair formation outside the haustorium surface. Knowledge is much lower in the obligate root parasites, mainly due to the absence to date of mutants affected in haustoriogenesis. In P. ramosa, haustorial papillae formation, which becomes evident by the proliferation of cells at the tip of germinating seeds, is similar in its kinetics to facultative parasites (Goyet et al., 2017). In addition, blocking haustoriogenesis by the coapplication of HIFs and an inhibitor of cytokinin perception, PI-55, prevents the HIF-induced root growth arrest and the formation of haustorial papillae, indicating that CKs control both. As a result, PI-55-treated P. ramosa seeds are less aggressive toward host roots.
Host attachment mediated by haustorial hairs or papillae in the Orobanchaceae and by epidermal cells of holdfasts in Cuscuta goes beyond a mechanical clasping of the host plant. These specialized cells also produce adhesive substances that further facilitate close adherence to the infected host organ (Yoshida et al., 2016). In Cuscuta, mechanical stimuli induce the secretion of polysaccharides and cell wall degrading enzymes, even in the absence of a living host (Olsen and Krause, 2017). The activity of cell wall degrading enzymes such as xyloglucan endotransglucosylases/hydrolases (XTHs) outlines the invading Cuscuta reflexa haustorium (Olsen and Krause, 2017). Chemically inhibiting XTH activity blocks penetration, thus arguing for the importance of cell wall degrading enzymes in aiding endophytic growth of the parasite (Olsen and Krause, 2017). Concurrently, transcriptomes of C. australis prehaustoria are overrepresented by genes linked to lignin and xyloglucan metabolism (Sun et al., 2018; Vogel et al., 2018). Likewise, ∼20% of identified carbohydrate-active enzyme-categorized genes (CAZyme) in S. hermonthica show differential expression during host invasion (Yoshida et al., 2019). In particular, genes associated with pectin metabolism, typically found in primary cell walls, are expressed in an early stage of haustorium development. The prevalence of genes linked to cell wall remodeling in early haustorium development extends to transcriptomes of other Orobanchaceae, Cuscuta, and parasitic Santalaceae species (Yang et al., 2015; Zhang et al., 2015; Ichihashi et al., 2018). Intriguingly, two CAZymes that are expressed in C. campestris haustoria show strong indications of being acquired by horizontal gene transfers early in the evolution of the Cuscuta genus (Yang et al., 2016).
Behind the epidermal cell layer, the Cuscuta prehaustoria contain disk-like meristems that locate near the vascular cylinder. Cells divide first anticlinally and then arrange into an array of digitate cells prior to tissue invasion (Svubova et al., 2017). These digitate cells develop into intrusive cells within the searching hyphae. Searching hyphae then grow endophytically through the host tissue until they reach the host vasculature. Additionally, searching hyphae may also incorporate epidermal cells, as seen for C. europaea (Svubova et al., 2017). In Phtheirospermum, intrusive cells transdifferentiate from epidermal root cells (Wakatake et al., 2018). Epidermal root cells generally play a crucial role in the development of Phtheirospermum haustoria. With the initiation of haustorium development, epidermal cells at the haustorium initiation site express an enzyme that catalyzes the rate-limiting step in de novo auxin synthesis, PjYUC3 (Ishida et al., 2016). Silencing PjYUC3 expression lowers infection rates and ectopic expression of PjYUC3 produces swellings of the root that resemble prehaustoria. Parallel to PjYUC3 expression, the auxin response reporter DR5 is highly active in developing Phtheirospermum haustoria (Ishida et al., 2016). Interestingly, auxin alone triggers the expression of CAZymes in nonparasitic plants, many of which have homology to the CAZymes upregulated in prehaustorial tissue (Majda and Robert, 2018).
Given the central role of auxin in root development, it may be of little surprise that the transcriptomes of developing haustoria in Orobanchaceae and Santalaceae root parasites are similar to root expression profiles in nonparasitic plants. The root developmental program is thereby not simply blueprinted onto haustorial development. For example, S. hermonthica radicles infecting rice do not express the key components of the lateral root development program ALF4 and ACR4 (Yoshida et al., 2019). By contrast, host plants expressed ALF4 and ACR4 coincidentally. The relevance of parasite-induced ALF4 and ACR4 expression in host plants is currently not known, but it suggests intertwined signaling between parasite and host.
Similarities between haustoriogenesis and the gene expression in nonparasitic roots were also observed in C. australis (Sun et al., 2018). This observation is surprising as Cuscuta species are rootless plants that lost and pseudogenized many root-associated genes (Sun et al., 2018; Vogel et al., 2018). To which extent Cuscuta haustoriogenesis interconnects to host responses has also been a continuous subject of interest.
An emerging aspect of these studies is the question of how parasitic plants avoid detection by the host immune system. When parasitic plants trigger an elevated host immune response, haustoriogenesis aborts, and haustoria degenerate. For example, in tomato (Solanum lycopersicum), the plasma membrane receptor CuRe1 recognizes a Cuscuta conserved peptide factor (Hegenauer et al., 2016). Recognition of the Cuscuta factor triggers a robust immune response, visible as brownish dead tissue around the penetration. Similarly, the recently cloned HAOR7 receptor-like kinase in sunflower detects AvrO7-containing O. cumuna races (Duriez et al., 2019). HaOR7-induced resistance leads to termination of the infection process when O. cumuna penetrates the outer sunflower root layers and prevents its connection to the host vasculature. S. hermonthica haustoria also fail to connect to the vasculature of resistant rice varieties (Cissoko et al., 2011). Overcoming resistance to establish parasitism successfully is a matter of life and death for obligate parasites and likely involves effector proteins as recently shown for S. gesnerioides (Su et al., 2019), also highlighted in Delavault’s commentary (2020).
For parasitic plants that slip through host detection, haustoria mature and enclose “the most general anatomic feature of the haustorium”—the xylem bridge (Heide-Jørgensen, 2008). Xylem bridges comprise lignified xylem vessels that span the entire haustorium and connect parasite to host vasculature. The (pro-)cambium orchestrates vascular development in plants (Jouannet et al., 2015). (Pro-)cambial-like domains within the haustorium cortex express cambial marker genes such as CLE41, GSK3, BES1, and WOX4 in C. japonica (Shimizu et al., 2017). The expression of these marker genes suggests that the default vascular development program is also active in maturing Cuscuta haustoria. Unlike in nonparasitic plants, where phloem cells express and secrete CLE41 and cambial cells respond to the perceived CLE41 peptide by expressing CjWOX4, Cuscuta haustoria show overlapping CjCLE41 and CjWOX4 expression domains. The authors speculate that the lack of distinct CLE41 and WOX4 expression domains points at additional host cues that could participate in the organization of the vasculature in Cuscuta (Shimizu et al., 2017).
In Phtheirospermum, (pro)cambial-like cells within the haustorium core express PjWOX4 in a similar temporal and spatial manner to Cuscuta haustoria (Wakatake et al., 2018). Phtheirospermum (pro)cambial-like cells transdifferentiate from endodermal, cortical, and epidermal cells, but not from procambial stele tissue of the main root. The de novo formation of the haustorial (pro)cambium further supports a more than hundred-year-old conclusion by Stephens that haustoria are not modified lateral roots, but “organs sui generis” (Stephens, 1912).
CKs play another critical role in vascular development. CKs promote radial growth through periclinal divisions (De Rybel et al., 2015). CKs increase during the transition from pre- to mature haustoria in different lineages of parasitic plants (Zhang et al., 2012; Furuhashi et al., 2013; Spallek et al., 2017). In Phtheirospermum, CK responses occur not only in (pro-)cambial tissue of the haustorium cortex but also in intrusive cells at the haustorial apex (Spallek et al., 2017). The proximity of intrusive cells to host vessels allows the transfer of parasite-derived CKs to the host to trigger hypertrophy in host plants.
Hemiparasitic plants only connect through xylem bridges to the host vasculature. Holoparasites such as Cuscuta and broomrapes, by contrast, also withdraw phloem-mobile molecules from their host. Phloem structures within haustoria are far less distinctive compared to xylem vessels of xylem bridges. Cuscuta haustoria express phloem companion marker genes such as CjAPL and CjSEOR1 (Shimizu et al., 2017). Still, no phloem companion cells were detected adjacent to cells that symplastically transported a fluorescent dye that was fed to the host. Also, P. aegyptiaca phloem conducting cells differ from regular phloem conducting cells in that they contain nuclei (Ekawa and Aoki, 2017). Of particular interest is the interface between host and parasite phloem. Electro-microscopic studies of O. cumana infecting sunflower roots show direct sieve-element connections and share interspecies sieve plates. Interspecies sieve plates show an uneven callose deposition, with more callose on the host side (Krupp et al., 2019). O. cumuna sieve-tube elements connected to sunflower sieve-tubes lack nuclei and vacuoles and are accompanied by companion cells, thus resembling sieve elements-companion cell complexes of nonparasitic plants. The differences in phloem morphology, even within the Orobanchaceae, further support the need to study a broader range of diverse parasitic plant species at cellular resolution, using, among others, methods that have been previously successfully employed to analyze parasitic plants–host interactions (Honaas et al., 2013).
Concluding remarks and perspectives
Overall, in comparison with many other organisms, the progress of knowledge in the biology of parasitic plants, especially obligate root parasites, has been limited until recently mainly due to the lack of data on their genetics. In 2018, Vogel et al. and Sun et al. sequenced for the first time the genomes of field dodder (C. campestris) and Australian dodder (C. australis), respectively. In 2019, Yoshida et al. published the complete genome sequence of the witchweed S. asiatica. The genomes of two broomrape species, P. ramosa and O. cumana (http://www.heliagene.org), are in the process of finalization. In addition, several transcriptomic datasets are already available for P. japonicum, P. ramosa, P. aegyptiaca, S. gesnerioides, S. hermonthica, and T. versicolor (Yang et al., 2015; Ishida et al., 2016; Goyet et al., 2017; Yoshida et al., 2019). Combined, these datasets provide the raw material for future studies that must now employ approaches to validate the function of key candidate genes involved in critical processes of parasitic plant development. At present, several transformation protocols have been implemented in Triphysaria versicolor (Tomilov et al., 2007), P. aegyptiaca (Fernández-Aparicio et al., 2011), P. japonicum (Ishida et al., 2011), S. hermonthica (Kirigia et al., 2014), and P. ramosa (Libiaková et al., 2018). However, there is still a long way to go before being able to generate stable transgenic obligate parasitic plants, and thus before applying high-throughput reverse genetics. Yet, this remains a crucial step in the objective of developing methods to control parasitic weeds. Although several physical, cultural, chemical, and biological approaches have been explored (Fernández-Aparicio et al., 2016b), none of them has proven unequivocal success. The fact that the root parasitic weed withers its host long before it emerges aboveground suggests that the early stages of its development are interesting targets to design control strategies. This has become even more relevant in recent years, with the identification of key molecular components involved in parasitic weeds recognition of host-derived chemicals required for seed germination and haustorium formation.
In the Outstanding Questions Box, we present some examples of challenges faced in deciphering the biology of parasitic plants. Answers to these questions will fill important knowledge gaps and can be used to develop new strategies for controlling parasitic plants.
Funding
This work was supported by the German Research Foundation (DFG, 424122841) to T.S. and the French National Research Agency (ANR-16-CE20-0004) to P.D.
Outstanding questions
What are the molecular actors responsible for and subjected to extensive epigenetic remodeling during the conditioning period?
What genomic reconfigurations underlie the acquisition of a seed germination process that is strictly dependent on perception of host-exuded signals in parasitic weeds?
Is the ability to germinate upon non-SLs signals driven by the neofunctionalization of HTL/KAI2 proteins or the acquisition of unique perception pathways?
Which host-derived signals control haustoriogenesis and how are they perceived?
Which parasite gene products are recognized by host immunity and which ones evolved to subvert it?
How can we produce stable transgenic stem and root parasitic plants?
G.B., T.S., P.S., and P.D. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Philippe Delavault (philippe.delavault@univ-nantes.fr).
References
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 65:161–186 [DOI] [PubMed] [Google Scholar]
- Auger B, Pouvreau JB, Pouponneau K, Yoneyama K, Montiel G, Le Bizec B, Yoneyama K, Delavault P, Delourme R, Simier P (2012) Germination stimulants of Phelipanche ramosa in the rhizosphere of Brassica napus are derived from the glucosinolate pathway. Mol Plant Microbe Interact 25:993–1004 [DOI] [PubMed] [Google Scholar]
- Bao YZ, Yao ZQ, Cao XL, Peng JF, Xu Y, Chen MX, Zhao SF (2017) Transcriptome analysis of Phelipanche aegyptiaca seed germination mechanisms stimulated by fluridone, TIS108, and GR24. PLoS One 12:e0187539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin CC, Baskin JM (2014) Seeds, Ed 2. Academic Press, San Diego, CA [Google Scholar]
- Bewley JD, Bradford K, Hilhorst H, Nonogaki H (2013) Seeds – Physiology of Development, Germination and Dormancy, Ed 3. Springer, New York [Google Scholar]
- Billard E, Goyet V, Delavault P, Simier P, Montiel G (2020) Cytokinin treated microcalli of Phelipanche ramosa: an efficient model for studying haustorium formation in holoparasitic plants. Plant Cell Tiss Organ Cult 10.1007/s11240-020-01813-6 [Google Scholar]
- Bozkurt ML, Muth P, Parzies HK, Hausmann BIG (2015) Genetic diversity of East and West African Striga hermonthica populations and virulence effects on a contrasting set of sorghum cultivars. Weed Res 55: 71–81 [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V (2017) DNA methylation dynamics during early plant life. Genome Biol 18: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L, Cowman PF, Lanfear R (2013) Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evol Biol 13: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun G, Braem L, Thoiron S, Gevaert K, Goormachtig S, Delavault P (2018) Seed germination in parasitic plants: what insights can we expect from strigolactone research? J Exp Bot 69: 2265–2280 [DOI] [PubMed] [Google Scholar]
- Brun G, Thoiron S, Braem L, Pouvreau J-B, Montiel G, Lechat M-M, Simier P, Gevaert K, Goormachtig S, Delavault P (2019) CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants. Plant Cell Environ 42: 2612–2626 [DOI] [PubMed] [Google Scholar]
- Bunsick M, Toh S, Wong C, Xu Z, Ly G, McErlean CSP, Pescetto G, Nemrish KE, Sung P, Li JD, et al. (2020) SMAX1-dependent seed germination bypasses GA signaling in Arabidopsis and Striga. Nat Plants 6: 646–652 [DOI] [PubMed] [Google Scholar]
- Ćavar S, Zwanenburg B, Tarkowski P (2015) Strigolactones: occurrence, structure, and biological activity in the rhizosphere. Phytochem Rev 14: 691–711 [Google Scholar]
- Chae SH, Yoneyama K, Takeuchi Y, Joel DM (2004) Fluridone and norflurazon, carotenoid-biosynthesis inhibitors, promote seed conditioning and germination of the holoparasite Orobanche minor. Physiol Plant 120: 328–337. [DOI] [PubMed] [Google Scholar]
- Cissoko M, Boisnard A, Rodenburg J, Press MC, Scholes JD (2011) New rice for Africa (NERICA) cultivars exhibit different levels of post-attachment resistance against the parasitic weeds Striga hermonthica and Striga asiatica. New Phytol 192: 952–963. [DOI] [PubMed] [Google Scholar]
- Clarke CR, Timko MP, Yoder JI, Axtell MJ, Westwood JH (2019) Molecular dialog between parasitic plants and their hosts. Annu Rev Phytopathol 57: 279–299 [DOI] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349: 540–543 [DOI] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of Witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Cui S, Wakatake T, Hashimoto K, Saucet SB, Toyooka K, Yoshida S, Shirasu K (2016) Haustorial hairs are specialized root hairs that support parasitism in the facultative parasitic plant Phtheirospermum japonicum. Plant Physiol 170: 1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavault P, Montiel G, Brun G, Pouvreau JB, Thoiron S, Simier P (2017) Communication between host plants and parasitic plants.InBecard G, ed, Advances in Botanical Research. Academic Press, Oxford, pp 55–82 [Google Scholar]
- Delavault P (2020) Are root parasitic plants like any other plant pathogens?. New Phytol 226: 641–643 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Mähönen AP, Helariutta Y, Weijers D (2015) Plant vascular development: from early specification to differentiation. Nat Rev Mol Cell Biol 17: 30–40 [DOI] [PubMed] [Google Scholar]
- Duriez P, Vautrin S, Auriac M-C, Bazerque J, Boniface M-C, Callot C, Carrère S, Cauet S, Chabaud M, Gentou F, et al. (2019) A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nat Plants 5: 1211–1215 [DOI] [PubMed] [Google Scholar]
- Ejeta G (2007) Breeding for Striga resistance in sorghum: exploitation of an intricate host-parasite biology. Crop Sci 47: 216–227 [Google Scholar]
- Ekawa M, Aoki K (2017) Phloem-conducting cells in haustoria of the root-parasitic plant Phelipanche aegyptiaca retain nuclei and are not mature sieve elements. Plants 6: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feild TS, Brodribb TJ (2005) A unique mode of parasitism in the conifer coral tree Parasitaxus ustus (Podocarpaceae). Plant Cell Environ 28: 1316–1325 [Google Scholar]
- Fernández-Aparicio M, Flores F, Rubiales D (2009) Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann Bot 103: 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Rubiales D, Bandaranayake PC, Yoder JI, Westwood JH (2011) Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Yoneyama K, Rubiales D (2011) The role of strigolactones in host specificity of Orobanche and Phelipanche seed germination. Seed Sci Res 21: 55–61 [Google Scholar]
- Fernández-Aparicio M, Masi M, Maddau L, Cimmino A, Evidente M, Rubiales D, Evidente A (2016a) Induction of haustorium development by sphaeropsidones in radicles of the parasitic weeds Striga and Orobanche. A structure–activity relationship study. J Agric Food Chem 64: 5188–5196 [DOI] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Reboud X, Gibot-Leclerc S (2016b) Broomrape weeds. Underground mechanisms of parasitism and associated strategies for their control: a review. Front Plant Sci 7: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R (2013) Abscisic acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD (2004) A compound from smoke that promotes seed germination. Science 305: 977–977 [DOI] [PubMed] [Google Scholar]
- Flematti GR, Dixon KW, Smith SM (2015) What are karrikins and how were they “discovered” by plants?. BMC Biol 13: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka H, Samejima H, Suzuki H, Mizutani M, Okamoto M, Sugimoto Y (2019) Aberrant protein phosphate 2C leads to abscisic acid insensitivity and high transpiration in parasitic Striga. Nat Plants 5: 258–262 [DOI] [PubMed] [Google Scholar]
- Furuhashi T, Kojima M, Sakakibara H, Fukushima A, Hirai MY, Furuhashi K (2013) Morphological and plant hormonal changes during parasitization by Cuscuta japonica on Momordica charantia. J Plant Interact 9: 220–232 [Google Scholar]
- Gobena D, Shimels M, Rich PJ, Ruyter-Spira C, Bouwmeester H, Kanuganti S, Mengiste T, Ejeta G (2017) Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc Natl Acad Sci USA 114: 4471–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyet V, , Billard E, , Pouvreau JB, , Lechat MM, , Pelletier S, , Bahut M, , Monteau F, , Spichal L, , Delavault P, , Montiel G, , Simier P ( 2017) Haustorium initiation in the obligate parasitic plant Phelipanche ramosa involves a host-exudated cytokinin signal. J Exp Bot 68: 5539–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyet V, Wada S, Cui S, Wakatake T, Shirasu K, Montiel G, Simier P, Yoshida S (2019) Haustorium inducing factors for parasitic Orobanchaceae . Front Plant Sci 10: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Plačková L, Kulkarni MG, Doležal K, Staden JV (2019) Role of smoke stimulatory and inhibitory biomolecules in phytochrome-regulated seed germination of Lactuca sativa. Plant Physiol 181: 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar MA, Orr GL, Westra P (1998). The response of dodder (Cuscuta spp.) seedlings to phytohormones under various light regimes. Ann Appl Biol 132: 331–338 [Google Scholar]
- Hegenauer V, Fürst UB, Kaiser B, Smoker M, Zipfel C, Felix G, Stahl M, Albert M, Spallek T, Mutuku M, et al. (2016) Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 353: 478–481 [DOI] [PubMed] [Google Scholar]
- Heide-Jørgensen H (2008) Parasitic Flowering Plants, Ed 1. Brill, Boston, MA [Google Scholar]
- Heide-Jørgensen HS,, Kuijt J (1993). Epidermal derivatives as xylem elements and transfer cells: a study of the host-parasite interface in two species of Triphysaria (Scrophulariaceae). Protoplasma 174: 173–183 [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Honaas LA, Wafula EK, Yang Z, Der JP, Wickett NJ, Altman NS, Taylor CG, Yoder JI, Timko MP, Westwood JH, DePamphilis CW (2013). Functional genomics of a generalist parasitic plant: Laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol 13: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaas LA, Jones S, Farrell N, Kamerow W, Zhang H, Vescio K, Altman NS, Yoder JI, dePamphilis CW (2019) Risk versus reward: host dependent parasite mortality rates and phenotypes in the facultative generalist Triphysaria versicolor. BMC Plant Biol 19: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Kusano M, Kobayashi M, Suetsugu K, Yoshida S, Wakatake T, Kumaishi K, Shibata A, Saito K, Shirasu K (2018) Transcriptomic and metabolomic reprogramming from roots to haustoria in the parasitic plant, Thesium chinense. Plant Cell Physiol 59: 724–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving LJ, Cameron DD (2009) You are what you eat: interactions between root parasitic plants and their hosts.InKader JC, Delseny M, eds, Advances in Botanical Research. Academic Press, Oxford, pp 87–138 [Google Scholar]
- Ishida JK, Yoshida S, Ito M, Namba S, Shirasu K (2011) Agrobacterium rhizogenes-mediated transformation of the parasitic plant Phtheirospermum japonicum. PLoS One 6: 25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Kasahara H, Wafula E, dePamphilis CW, Namba S, Shirasu K (2016) Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell 28: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel DM, Chaudhuri SK, Plakhine D, Ziadna H, Steffens JC (2011) Dehydrocostus lactone is exuded from sunflower roots and stimulates germination of the root parasite Orobanche cumana. Phytochemistry 72: 624–634 [DOI] [PubMed] [Google Scholar]
- Joel DM (2013) Seed production and dispersal in the Orobanchaceae. InJoel DM, Gressel J, Musselman LJ, eds, Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Springer, Berlin, pp 143–146 [Google Scholar]
- Jouannet V, Brackmann K, Greb T (2015) ( Pro)cambium formation and proliferation: two sides of the same coin?. Curr Opin Plant Biol 23: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabiri S, Rodenburg J, van Ast A, Bastiaans L (2017) Slavery in plants: how the facultative hemi‐parasitic plant Rhamphicarpa fistulosa can completely dominate its host. Ann Appl Biol 171: 353–363 [Google Scholar]
- Kawakatsu T, Nery JR, Castanon R, Ecker JR (2017) Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirigia D, Runo S, Alakonya A (2014) A virus-induced gene silencing (VIGS) system for functional genomics in the parasitic plant Striga hermonthica. Plant Methods 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp A, Heller A, Spring O (2019) Development of phloem connection between the parasitic plant Orobanche cumana and its host sunflower. Protoplasma 256: 1385–1397 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat M-M, Pouvreau J-B, Péron T, Gauthier M, Montiel G, Véronési C, Todoroki Y, Le Bizec B, Monteau F, Macherel D, et al. (2012) PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J Exp Bot 63: 5311–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat M-M, Brun G, Montiel G, Véronési C, Simier P, Thoiron S, Pouvreau J-B, Delavault P (2015) Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel. J Exp Bot 66: 3129–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Watanabe Y, Yamaguchi S, Tran L-SP (2016) OaMAX2 of Orobanche aegyptiaca and Arabidopsis AtMAX2 share conserved functions in both development and drought responses. Biochem Biophys Res Commun 478: 521–526 [DOI] [PubMed] [Google Scholar]
- Libiaková D, Ruyter-Spira C, Bouwmeester HJ, Matusova R (2018) Agrobacterium rhizogenes transformed calli of the holoparasitic plant Phelipanche ramosa maintain parasitic competence. Plant Cell Tissue Organ Cult 135: 321–329 [Google Scholar]
- Liu Q, Zhang Y, Matusova R, Charnikhova T, Amini M, Jamil M, Fernandez-Aparicio M, Huang K, Timko MP, Westwood JH, et al. (2014) Striga hermonthica MAX2 restores branching but not the Very Low Fluence Response in the Arabidopsis thaliana max2 mutant. New Phytol 202: 531–541 [DOI] [PubMed] [Google Scholar]
- Lu G, Wu X, Chen B, Gao G, Xu K, Li X (2006) Detection of DNA methylation changes during seed germination in rapeseed (Brassica napus). Chinese Sci Bull 51: 182–190 [Google Scholar]
- Machin DC, Bennett T (2020) Two routes to germinate a seed. Nat Plants 6: 602–603 [DOI] [PubMed] [Google Scholar]
- Majda M, Robert S (2018) The role of auxin in cell wall expansion. Int J Mol Sci 19: 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 73: 2460–2465 [DOI] [PubMed] [Google Scholar]
- Matusova R, van Mourik T, Bouwmeester HJ (2004) Changes in the sensitivity of parasitic weed seeds to germination stimulants. Seed Sci Res 14: 335–344 [Google Scholar]
- Meng F, Li Y, Yin J, Liu H, Chen X, Ni Z, Sun Q (2012) Analysis of DNA methylation during the germination of wheat seeds. Biol Plantarum 56: 269–275 [Google Scholar]
- Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J (2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol 18: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Riseborough J-A, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, Smith SM (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol 149: 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM (2012) Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu Rev Plant Biol 63: 107–130 [DOI] [PubMed] [Google Scholar]
- Nickrent D (2020) Parasitic angiosperms: how often and how many?. Taxon 69: 5–27 [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8’-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki M, Kittikorn M, Ueno K, Mizutani M, Hirai N, Kondo S, Ohnishi T, Todoroki Y (2012) Abscinazole-E2B, a practical and selective inhibitor of ABA 8’-hydroxylase CYP707A. Bioorg Med Chem 20: 3162–3172 [DOI] [PubMed] [Google Scholar]
- Olsen S, Krause K (2017) Activity of xyloglucan endotransglucosylases/hydrolases suggests a role during host invasion by the parasitic plant Cuscuta reflexa. PLoS One 12: e0176754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C (2012) Parasitic weeds: a world challenge. Weed Sci 60: 269–276 [Google Scholar]
- Péron T, Candat A, Montiel G, Veronesi C, Macherel D, Delavault P, Simier P (2017) New insights into phloem unloading and expression of sucrose transporters in vegetative sinks of the parasitic plant Phelipanche ramosa L. (Pomel). Front Plant Sci 7: 2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix GK, Press MC (2005) Effects of climate change on parasitic plants: the root hemiparasitic Orobanchaceae. Folia Geobot 40: 205–216 [Google Scholar]
- Plakhine D, Tadmor Y, Ziadne H, Joel DM (2012) Maternal tissue is involved in stimulant reception by seeds of the parasitic plant Orobanche. Ann Bot 109: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis E, Acquadro A, Comino C, Lanteri S (2004) Analysis of DNA methylation during germination of pepper (Capsicum annuum L.) seeds using methylation-sensitive amplification polymorphism (MSAP). Plant Sci 166: 169–178 [Google Scholar]
- Ramasubramanian TS, Paliyath G, Rajagopal I, Maheshwari R, Mahadevan S (1988) Hormones and Cuscuta development: In vitro induction of haustoria by cytokinin and its inhibition by other hormones. J Plant Growth Regul 7: 133–144 [Google Scholar]
- Rodenburg J, Demont M, Zwart SJ, Bastiaans L (2016) Parasitic weed incidence and related economic losses in rice in Africa. Agric Ecosyst Environ 235: 306–317 [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165: 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss GM (2013) Phylogenetic relationships and evolutionary trends in Orobanchaceae. InJoel DM, Gressel J, Musselman LJ, eds, Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Springer, Berlin, pp 243–265 [Google Scholar]
- Schwachtje J, Baldwin IT (2004) Smoke exposure alters endogenous gibberellin and abscisic acid pools and gibberellin sensitivity while eliciting germination in the post-fire annual, Nicotiana attenuata. Seed Sci Res 14: 51–60 [Google Scholar]
- Shimizu K, Hozumi A, Aoki K (2017) Organization of vascular cells in the haustorium of the parasitic flowering plant Cuscuta japonica. Plant Cell Physiol 59: 720–728 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Aoki K (2019) Development of parasitic organs of a stem holoparasitic plant in genus Cuscuta. Front Plant Sci 10:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T, Melnyk CW, Wakatake T, Zhang J, Sakamoto Y, Kiba T, Yoshida S, Matsunaga S, Sakakibara H, Shirasu K (2017) Interspecies hormonal control of host root morphology by parasitic plants. Proc Natl Acad Sci USA 114: 5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens EL (1912) The structure and development of the haustorium of Striga lutea. Ann Bot 26: 1067–1076 [Google Scholar]
- Stojanova B, Delourme R, Duffé P, Delavault P, Simier P (2019) Genetic differentiation and host preference reveal non-exclusive host races in the generalist parasitic weed, Phelipanche ramosa L. (Pomel). Weed Res 59: 107–118 [Google Scholar]
- Su C, Liu H, Wafula EK, Honaas L, de Pamphilis CW, Timko MP (2019) SHR4z, a novel decoy effector from the haustorium of the parasitic weed Striga gesnerioides, suppresses host plant immunity. New Phytol 226: 891–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Xu Y, Liu H, Sun T, Zhang J, Hettenhausen C, Shen G, Qi J, Qin Y, Li J, et al. (2018) Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat Commun 9: 2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svubova R, Lukacova Z, Kastier P, Blehova A (2017) New aspects of dodder–tobacco interactions during haustorium development. Acta Physiol Plant 39: 66 [Google Scholar]
- Takagi K, Okazawa A, Wada Y, Mongkolchaiyaphruek A, Fukusaki E, Yoneyama K, Takeuchi Y, Kobayashi A (2009) Unique phytochrome responses of the holoparasitic plant Orobanche minor. New Phytol 182: 965–974 [DOI] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y (2012) Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol 53: 107–117 [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350: 203–207 [DOI] [PubMed] [Google Scholar]
- Tomilov AA, Tomilova N, Yoder JI (2007) Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 225: 1059–1071 [DOI] [PubMed] [Google Scholar]
- Ueno K, Furumoto T, Umeda S, Mizutani M, Takikawa H, Batchvarova R, Sugimoto Y (2014) Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry 108: 122–128 [DOI] [PubMed] [Google Scholar]
- Unachukwu NN, Menkir A, Rabbi IY, Oluoch M, Muranaka S, Elzein A, Odhiambo G, Farombi EO, Gedil M (2017) Genetic diversity and population structure of Striga hermonthica populations from Kenya and Nigeria. Weed Res 57: 293–302 [Google Scholar]
- Vogel A, Schwacke R, Denton AK, Usadel B, Hollmann J, Fischer K, Bolger A, Schmidt MHW, Bolger ME, Gundlach H, et al. (2018) Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nat Commun 9: 2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Cui S, Yoshida S (2019) Reactive oxygen species (ROS) generation is indispensable for haustorium formation of the root parasitic plant Striga hermonthica. Front Plant Sci 10: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatake T, Yoshida S, Shirasu K (2018) Induced cell fate transitions at multiple cell layers configure haustorium development in parasitic plants. Development 145: dev164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Steele D, Murdock M, Lai S, Yoder J (2019) Small-molecule screens reveal novel haustorium inhibitors in the root parasitic plant Triphysaria versicolor. Phytopathology 109: 1878–1887 [DOI] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Annu Rev Plant Biol 68: 291–322 [DOI] [PubMed] [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW (2010) The evolution of parasitism in plants. Trends Plant Sci 15: 227–235 [DOI] [PubMed] [Google Scholar]
- Wicke S, Naumann J (2018) Molecular evolution of plastid genomes in parasitic flowering plants. InChaw SM, Jansen RK, eds, Advances in Botanical Research. Academic Press, Oxford, pp 315–347 [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PCG, Wu B, Der JP, et al. (2015) Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol 32: 767–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang Y, Wafula EK, Honaas LA, Ralph PE, Jones S, Clarke CR, Liu S, Su C, Zhang H, et al. (2016) Horizontal gene transfer is more frequent with increased heterotrophy and contributes to parasite adaptation. Proc Natl Acad Sci USA 113: 7010–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JI, Scholes JD (2010) Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr Opin Plant Biol 13: 478–484 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Ruyter-Spira C, Bouwmeester H (2013) Induction of Germination. InJoel DM, Gressel J, Musselman LJ, eds, Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Springer, Berlin, pp 167–194 [Google Scholar]
- Yoshida S, Cui S, Ichihashi Y, Shirasu K (2016) The haustorium, a specialized invasive organ in parasitic plants. Annu Rev Plant Biol 67: 643–667 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim YM, Honaas L, Yang Z, Spallek T, Conn CE, Ichihashi Y. et al. (2019) Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr Biol 29: 3041–3052 [DOI] [PubMed] [Google Scholar]
- Zehhar N, Ingouff M, Bouya D, Fer A (2002) Possible involvement of gibberellins and ethylene in Orobanche ramosa germination. Weed Res 42: 464–469 [Google Scholar]
- Zhang X, Teixeira da Silva JA, Duan J, Deng R, Xu X, Ma G (2012) Endogenous hormone levels and anatomical characters of haustoria in Santalum album L. seedlings before and after attachment to the host. J Plant Physiol 169: 859–866 [DOI] [PubMed] [Google Scholar]
- Zhang X, Berkowitz O, Teixeira da Silva JA, Zhang M, Ma G, Whelan J, Duan J (2015) RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album. Front Plant Sci 6: 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Shen Y, Xi Z (2020). Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathways. Biochem Biophys Res Commun 523: 1040–1045 [DOI] [PubMed] [Google Scholar]