Abstract
The enzymes involved in l-ascorbate biosynthesis in photosynthetic organisms (the Smirnoff–Wheeler [SW] pathway) are well established. Here, we analyzed their subcellular localizations and potential physical interactions and assessed their role in the control of ascorbate synthesis. Transient expression of C terminal-tagged fusions of SW genes in Nicotiana benthamiana and Arabidopsis thaliana mutants complemented with genomic constructs showed that while GDP-d-mannose epimerase is cytosolic, all the enzymes from GDP-d-mannose pyrophosphorylase (GMP) to l-galactose dehydrogenase (l-GalDH) show a dual cytosolic/nuclear localization. All transgenic lines expressing functional SW protein green fluorescent protein fusions driven by their endogenous promoters showed a high accumulation of the fusion proteins, with the exception of those lines expressing GDP-l-galactose phosphorylase (GGP) protein, which had very low abundance. Transient expression of individual or combinations of SW pathway enzymes in N. benthamiana only increased ascorbate concentration if GGP was included. Although we did not detect direct interaction between the different enzymes of the pathway using yeast-two hybrid analysis, consecutive SW enzymes, as well as the first and last enzymes (GMP and l-GalDH) associated in coimmunoprecipitation studies. This association was supported by gel filtration chromatography, showing the presence of SW proteins in high-molecular weight fractions. Finally, metabolic control analysis incorporating known kinetic characteristics showed that previously reported feedback repression at the GGP step, combined with its relatively low abundance, confers a high-flux control coefficient and rationalizes why manipulation of other enzymes has little effect on ascorbate concentration.
Metabolic engineering, genetic analysis, and functional mutant complementation identify GDP-l-galactose phosphorylase as the main control point in ascorbate biosynthesis in green tissues.
Introduction
L-Ascorbate (ascorbate, vitamin C) is the most abundant soluble antioxidant in plants and functions as a cofactor in many enzymatic reactions (Smirnoff, 2018; Fenech et al., 2019). Some groups of animals, among which humans are included, lost the ability to synthesize ascorbate (Smirnoff, 2018). Therefore, we need to incorporate this essential nutrient through the diet, with fruits and vegetables being the major source. In plants, ascorbate is synthesized from the hexose phosphate pool via D-mannose/L-galactose through the Smirnoff–Wheeler (SW) pathway (Wheeler et al., 1998; Figure 1). The pathway is supported by abundant biochemical and genetic studies (Wheeler et al., 2015; Bulley and Laing, 2016). D-Glucose 6-P is sequentially transformed into D-fructose 6-P, D-mannose 6-P, and D-mannose 1-P by phosphoglucose isomerase, phosphomannose isomerase (PMI), and phosphomannomutase. GDP-D-mannose pyrophosphorylase (GMP) then converts D-mannose 1-P into GDP-D-mannose, which is subsequently epimerized by GDP-D-mannose epimerase (GME) into GDP-L-galactose. GDP-D-mannose and GDP-L-galactose are additionally used in the synthesis of cell wall polysaccharides and protein glycosylation, thereby interconnecting the SW pathway with cell wall biosynthesis (Conklin et al., 1999; Reiter and Vanzin, 2001; Fenech et al., 2019). The next step is catalyzed by GDP-L-galactose phosphorylase (GGP), producing L-galactose 1-P. This step is proposed to be the main control point of the pathway (Dowdle et al., 2007; Linster et al., 2007; Yoshimura et al., 2014; Laing et al., 2015, 2007). In Arabidopsis (Arabidopsis thaliana), GGP activity is encoded by two paralogs, Vitamin C 2 and 5 (VTC2 and VTC5). Next, L-galactose 1-phosphate phosphatase (GPP) and other unidentified phosphatases (Laing et al., 2004; Conklin et al., 2006; Torabinejad et al., 2009), transform L-galactose 1-phosphate into L-galactose. L-Galactose is then oxidized to L-galactono-1,4-lactone by L-galactose dehydrogenase (L-GalDH). All these steps are believed to occur in the cytosol while L-galactono-1,4-lactone crosses the outer membrane of the mitochondria to be oxidized by L-galactono-1,4-lactone dehydrogenase (GLDH), which is an integral component of mitochondrial electron transport Complex 1 on the inner membrane (Pineau et al., 2008; Schimmeyer et al., 2016).
Figure 1.
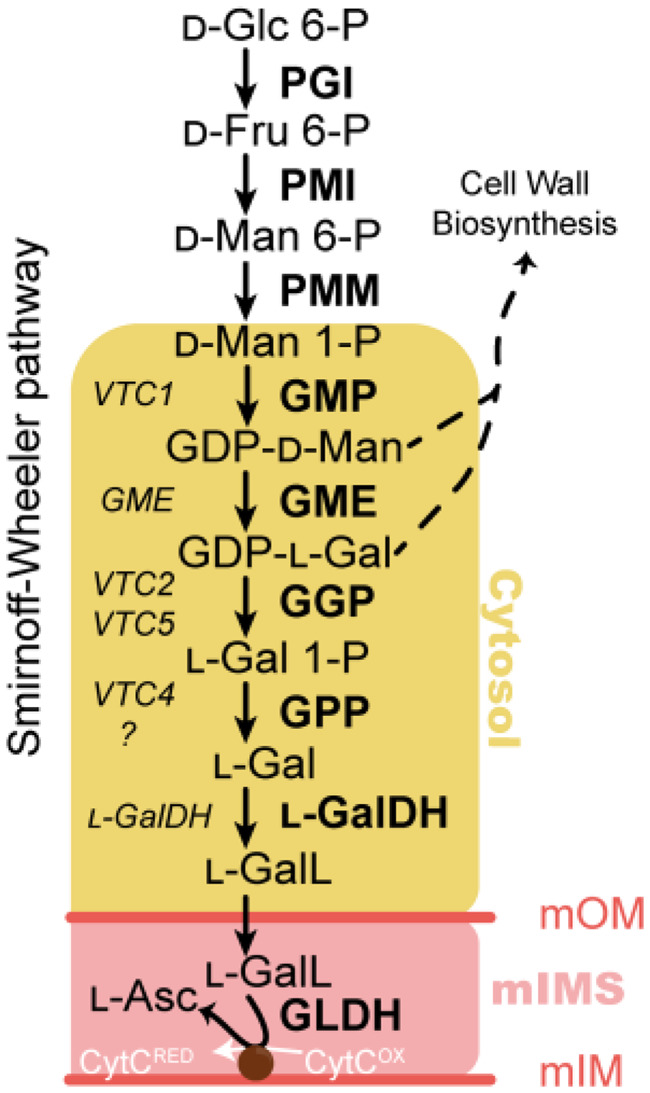
The ascorbate biosynthesis via GDP-D-mannose and L-galactose (the SW pathway) in A. thaliana. Yellow area refers to the cytosolic enzymes used in this work. mOM, mitochondrion outer membrane; mIMS, mitochondrion intermembrane space; mIM, mitochondrion inner membrane. In the left-hand side of the arrows, the genes encoding each enzyme are displayed, while in the right-hand side the encoded proteins/enzymatic activities are shown. Glc=glucose; Fru=fructose; Man=mannose; Gal=galactose; GalL=galactono-1,4-lactone; Asc=ascorbic acid; PGI=phosphoglucose isomerase; PMI=phosphomannose isomerase; PMM=phosphomannomutase; GMP=GDP-D-mannose pyrophosphorylase; GME=GDP-D-mannose 3′,5′ epimerase; GGP=GDP-L-galactose phosphorylase; GPP=L-galactose 1-phosphate phosphatase; L-GalDH=L-galactose dehydrogenase; L-GalLDH=L-galactono-1,4-lactone dehydrogenase; CytRED =cytochrome c reduced; CytOX =cytochrome c oxidized.
The ascorbate concentration is dependent on the tissue type (Franceschi and Tarlyn, 2002; Lorence et al., 2004; Müller-Moulé, 2008; Zhang et al., 2011) and it adjusts to changes in environmental conditions, particularly light (Bartoli, 2006; Page et al., 2012; Plumb et al., 2018). Surprisingly, despite being the most abundant soluble antioxidant, a good understanding of the mechanisms that control ascorbate concentration remains limited. While mutant analyses of genes involved in the biosynthetic pathway have corroborated the involvement of the various enzymes (Table 1), they provide little information on how the pathway is controlled. This is further complicated because the ascorbate pool is determined by the balance between synthesis, oxidation, recycling, and breakdown. The oxidation product of ascorbate is the monodehydroascorbate radical. This is either directly reduced back to ascorbate or can give rise to dehydroascorbate (DHA) in its hydrated bicyclic hemiketal form, which is reduced back to ascorbate through the Halliwell–Foyer–Asada cycle (Asada, 1999). In addition, a proportion of ascorbate and DHA can be degraded to various products such as oxalate, threonate, and tartrate (Pallanca and Smirnoff, 2000; Green and Fry, 2005; DeBolt et al., 2006, 2007; Dewhirst et al., 2017; Truffault et al., 2017; Terai et al., 2020). These recycling and breakdown reactions are evident under severe oxidative stress, which results in decreased ascorbate concentration (Waszczak et al., 2018; Terai et al., 2020).
Table 1.
Phenotypes associated with Arabidopsis mutants of the genes involved in ascorbate biosynthesis available in the literature
| Enzyme | Mutant | Allele | Phenotype | Reference |
|---|---|---|---|---|
| GMP | cyt1-1 | Knock-out | Embryo lethal | Lukowitz et al. (2001) |
| vtc1-1 | Knock-down | 30% of WT ascorbate | Conklin et al. (1996, 2000) | |
| GME | gme-1 | Knock-out | Male gametophyte lethal. Growth defect, rescued by boron but not by ascorbate supplementation. | Qi et al. (2017) |
| gme-2 | Knock-down | 30% of WT ascorbate | ||
| GGP | vtc2-4 | Single knock-out | 20% of WT ascorbate | Lim et al. (2016) |
| vtc5-2 | Single knock-out | 80% of WT ascorbate | Dowdle et al. (2007); Lim et al. (2016) | |
| vtc2/vtc5 | Double knock-out | Growth arrest, rescued by ascorbate supplementation | ||
| GPP | vtc4-4 | Knock-out | 65% of WT ascorbate | Torabinejad et al. (2009) |
| L-GalDH | lgaldh | Not reported | 30% of WT ascorbate in antisense suppression lines | Gatzek et al. (2002) |
| GLDH | gldh | Knock-out | Growth arrest, rescued by ascorbate supplementation | Pineau et al. (2008) |
Phenotype analysis shows that GGP is the first enzyme fully dedicated to ascorbate synthesis since growth arrest occurring in the vtc2/vtc5 double mutant and downstream mutants is rescued by ascorbate supplementation. Although a reduced function (knockdown mutation) of genes upstream of GGP also results in lower ascorbate concentration, the lethality occurring in knockout mutants cannot be prevented by exogenous ascorbate supplementation.
Multiple lines of evidence point to GGP as the critical controlling step of ascorbate biosynthesis. The transcription of the GGP gene (Dowdle et al., 2007; Müller-Moulé, 2008; Urzica et al., 2012) and the activity of the encoded enzyme (Dowdle et al., 2007) is highly responsive to environmental factors. Furthermore, GGP translation is subject to feedback repression via a conserved upstream open reading frame (uORF) in the 5′-Untranslated Region (5′-UTR; Laing et al., 2015). As a consequence, removing the uORF results in increased ascorbate in Arabidopsis (Laing et al., 2015), tomato (Solanum lycopersicum; Li et al., 2018), and lettuce (Lactuca sativa; Zhang et al., 2018). In addition, GGP is the only gene of the pathway whose overexpression consistently increases the ascorbate content (Bulley et al., 2012; Yoshimura et al., 2014) and quantitative trait loci analysis shows that GGP paralogs are located within regions of the genome associated with high ascorbate content in fruits (Mellidou et al., 2012). There have been reports that overexpression of other genes of the pathway, such as GMP or GME, also modestly increase ascorbate concentration in Arabidopsis [Zhou et al., 2012 {1.3-fold}; Li et al., 2016 {1.5-fold}], rice (Oryza sativa) [Zhang et al., 2015 {1.4-fold}], tobacco (Nicotiana tabacum) [Bulley et al., 2009 {1.2-fold}; Wang et al., 2011 {2-4-fold}), and tomato [Cronje et al., 2012 {1.7-fold}; Zhang et al., 2011 {1.4-fold}]. However, overexpression of these genes does not always have an effect on ascorbate concentration (Sawake et al., 2015; Yoshimura et al., 2014). In addition, various studies have reported ascorbate feedback inhibition of the biosynthetic enzymes PMI (Maruta et al., 2008), GME (Wolucka and Van Montagu, 2003), and L-GalDH (Mieda et al., 2004), although the latter remains controversial (Laing et al., 2004). This feedback control is further supported by the decreased incorporation of 14C-labelled sugars into ascorbate in vivo after feeding ascorbate (Pallanca and Smirnoff, 2000; Wolucka and Van Montagu, 2003).
In order to increase our understanding of the organization of the ascorbate biosynthesis pathway, we performed systematic over-expression of single and multiple combinations of all the enzymes of the pathway from GMP downstream in Nicotiana benthamiana. This allowed the systematic investigation of their effects on the ascorbate pool and supported the formation of a multiprotein complex. In addition, complementation of Arabidopsis mutants with the respective green fluorescent protein (GFP)-tagged enzymes confirmed the functionality of the constructs and provided information on their subcellular localizations. Finally, we constructed a kinetic model based on the known properties of the pathway whose predictions support that activity of GGP is the only significant controlling step and which also explains other observed features of the pathway. In addition to this information, this work provides a set of tools, which will allow a better understanding of ascorbate biosynthesis at the molecular and cellular level.
Results
Generation and analysis of GFP-tagged ascorbate biosynthesis Arabidopsis complementation lines reveals major differences in protein expression along the pathway
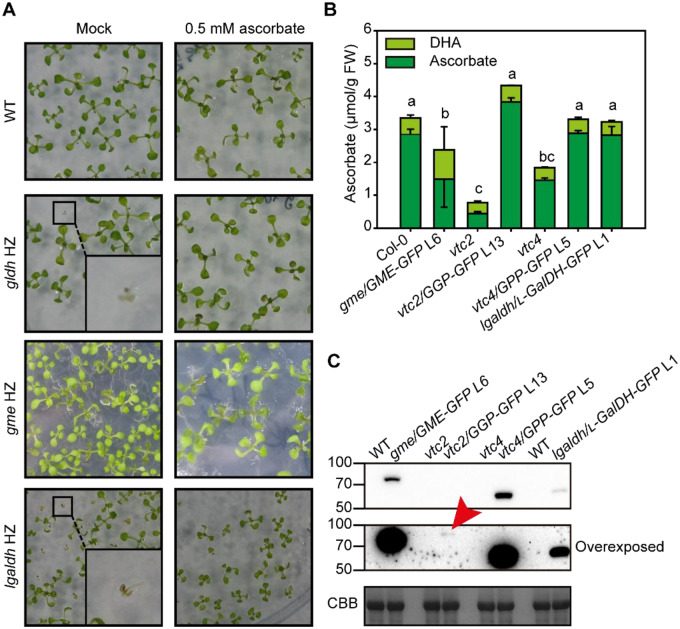
To study the function of SW proteins, we generated Arabidopsis lines transformed with genomic regions (including their promoters) of ascorbate biosynthesis genes that resulted in proteins fused to GFP at their C-terminus. To investigate the functionality of these constructs, they were introduced into Arabidopsis mutants and the complementation of their phenotypes was analyzed. Arabidopsis mutants in SW genes typically show lower ascorbate concentration and/or lethality in early stages of development (Table 1). Selfing of Arabidopsis heterozygous mutants for GME (SALK_150208, gme-3, hereafter gme; Supplemental Figure S1A) and L-GalDH (SALK_056664, Supplemental Figure S1B) did not produce homozygous mutants (Supplemental Figure S2). As exogenous supplementation of ascorbate prevented growth arrest of several SW mutants (Dowdle et al., 2007; Pineau et al., 2008), we germinated seeds from heterozygous gme-3 and gldh plants on solid media without and supplemented with 0.5-mM ascorbate (Figure 2A). As a control, we included seeds from a heterozygous gldh mutant plant, whose growth arrest is prevented by exogenous ascorbate (Pineau et al., 2008). The analysis of heterozygous gme progeny identified 40 wild-type (WT) seedlings, 46 heterozygous seedlings, and no homozygous for the T-DNA, confirming the lethality of a loss-of-function allele of GME (Qi et al., 2017; Voxeur et al., 2011; Figure 2A;Supplemental Figure S2A). Similarly, no homozygous gme seedlings were identified when the growth medium was supplemented with 0.5-mM ascorbate in contrast to gldh seedlings (Pineau et al., 2008; SupplementalFigure S2B), confirming that GME, similar to GMP (Lukowitz et al., 2001), has additional roles to that of ascorbate biosynthesis (SupplementalFigure S2A; Gilbert et al., 2009; Qi et al., 2017; Voxeur et al., 2011). On the other hand, diagnostic PCR of heterozygous lgaldh progeny identified 23 WT seedlings and 69 heterozygous T-DNA seedlings, while no homozygous mutants were found in the medium not supplemented with ascorbate. A number of seedlings showed growth arrest and yellowing similar to gldh progeny (Figure 2A, magnified square). In contrast, we identified 25 WT, 41 heterozygous T-DNA and 25 homozygous T-DNA seedlings by diagnostic PCR in ascorbate containing medium (Figure 2A, SupplementalFigure S2B). Therefore, in contrast to gme mutants, Igaldh mutants could be rescued by supplementation with exogenous ascorbate (Figure 2, A; SupplementalFigure S2, B). This supports the expression product of L-GalDH as being responsible for the L-galactose dehydrogenase activity present in Arabidopsis. In addition, the finding that, like gldh, Igaldh is an ascorbate auxotroph indicates that the activity of this enzyme is essential for ascorbate biosynthesis.
Figure 2.
Characterization of C-terminal GFP fusions of ascorbate biosynthesis enzymes. A, The ascorbate complementation assay rescues growth arrest from gldh and lgaldh mutants but not gme lethality. Insets show homozygous mutants with arrested growth in media lacking ascorbate, but not in media supplemented with ascorbate. HZ, heterozygous. B, Complementation of ascorbate concentration by expression of GFP fusion proteins in ascorbate deficient Arabidopsis mutants. Data displayed correspond to mean±SD from three independent samples composed of three 6-week-old fully expanded rosettes grown in a short-day regime at 150-µmol photons m−2 s−1, one and a half hours after lights turned on. Different letters denote statistically significant differences for ascorbate concentration (one-way ANOVA, α = 0.05, post-hoc Tukey test). C, Immunoblot (α-GFP) of complemented lines shows that a small amount of GGP compared with other components restores ascorbate content. Red arrowhead indicates GGP-GFP. CBB, Coomassie Brilliant Blue.
To investigate the functionality of a GME protein fused with GFP at the C-terminus, we transformed WT plants with a GMEp:GME-GFP construct. Of the several transformants showing high expression of GME-GFP and containing a single insertion (SupplementalFigure S3A), line 6 (L6) was selected to introduce the transgene into the gme mutant background using reciprocal crosses. Using heterozygous gme plants as the male parent, we did not obtain F1 gme/GMEp:GME-GFP (hereafter named as gme/GME-GFP L6) descendants, supporting the role of GME in pollen development and pollen tube elongation (Qi et al., 2017). However, when line L6 was used as a male parent, we obtained viable gme/GME-GFP F1 seedlings that allowed the identification in the F2 of homozygous gme/GME-GFP L6 seedlings (SupplementalFigure S3B). Surprisingly, while the GME–GFP fusion protein complemented the sterility defects, the gme/GME-GFP lines contained ∼55% of the ascorbate content of WT plants (Figure 2B) which, considering the lower ascorbate content of knock-down gme Arabidopsis mutants (Qi et al., 2017), indicates that it only partially complements the ascorbate content. Similarly, a L-GalDHp:L-GalDH-GFP construct was transformed into WT plants and we selected line 1 (L1) that showed high expression and a single insertion (SupplementalFigure S3C). Then, reciprocal crosses between heterozygous lgaldh plants and L-GalDHp:L-GalDH-GFP L1 were performed. On this occasion, we did obtain F1 plants when lgaldh plants were used as the male or female parent. Homozygous lgaldh/GalDH-GFP L1 seedlings were identified in F2 progeny using diagnostic PCR in medium lacking ascorbate (SupplementalFigure S3D), indicating that the L-GalDH-GFP protein driven by the L-GalDH promoter restores the enzyme activity lost in the lgaldh mutant. In addition, lgaldh/L-GalDH-GFP L1 plants contained WT levels of ascorbate (Figure 2B).
In contrast to gme and lgaldh, loss-of-function vtc2 and vtc4 homozygous mutants are viable and show WT growth under standard conditions despite containing 20 and 55% of the WT ascorbate content, respectively (Figure 2B; Conklin et al., 2000; Dowdle et al., 2007; Torabinejad et al., 2009). In the case of the vtc2 mutant, GGP activity is also provided by the paralogous VTC5 gene. Consistently, vtc2/vtc5 double mutants, like lgaldh and gldh mutants (Figure 2A;SupplementalFigure S2, B and C), show growth arrest that can be complemented with exogenous ascorbate (Dowdle et al., 2007; Lim et al., 2016). However, considering the phenotypes described (Table 1), the viability of the vtc4 mutant must rely on other enzymes with GPP activity (Nourbakhsh et al., 2015; Torabinejad et al., 2009). In order to test the functionality of GFP-tagged GGP and GPP, we restored WT ascorbate content by introducing GGPp:GGP-GFP and GPPp:GPP-GFP constructs into their respective mutants. The T-DNA knockout mutant vtc2-4 (SAIL_769_H05; hereafter named as vtc2; SupplementalFigure S1C; Lim et al., 2016) was directly transformed and lines with a single insertion were selected (SupplementalFigure S3E). Line 13 (L13) was selected because it showed the highest accumulation of GGP–GFP (SupplementalFigure S3F). Also, GPPp:GPP–GFP was introduced into the vtc4-4 mutant (SupplementalFigure S2D) following the same strategy described for GME and L-GalDH (SupplementalFigure S3G). A single-insertion GPPp:GPP-GFP line 5 was selected to cross with the vtc4 homozygous mutant and the homozygous GPP/GPP-GFP line was isolated by diagnostic PCR (SupplementalFigure S3H). The ascorbate concentration of vtc2/GGP-GFP and vtc4GPP/GPP-GFP lines was restored to WT levels, validating the functionally of the GFP-tagged enzyme (Figure 2B).
Immunoblot analysis indicated that GME–GFP and GPP–GFP were easily detectable, while L-GalDH-GFP showed a lower concentration (Figure 3C). Interestingly, despite the complementation of ascorbate levels in L13, GGP–GFP protein could be detected only after overexposure of the blot using a high sensitivity ECL substrate (see Materials and Methods section) for the detection of femtogram amounts of protein (Figure 2C, middle panel). This low amount of GGP protein cannot be explained by a low transcript level of GGP because, based on publicly available transcriptomic databases, GGP and GME showed the highest expression of all SW genes in vegetative tissue (SupplementalFigure S4).
Figure 3.
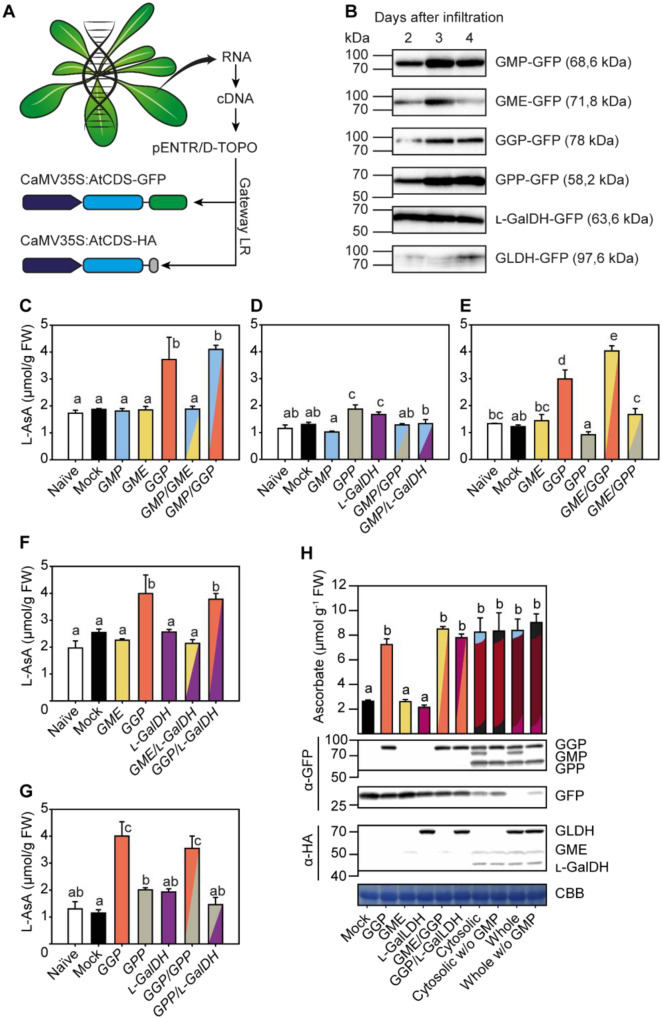
The effect of transient expression of C-terminal GFP and HA fusions of ascorbate biosynthesis enzymes on ascorbate concentration in N. benthamiana leaves. A, Strategy followed to clone and to overexpress ascorbate biosynthesis genes from Arabidopsis translationally fused to GFP and HA at the protein C-terminus in N. benthamiana leaves. B, Immunoblots (α-GFP) of fusion protein accumulation 2, 3, and 4 d after agroinfiltration. Expression was driven by the 35S promoter. C–H, Leaf ascorbate concentration 3 d after agroinfiltration with different combinations of C-terminal GFP ad HA fusions of ascorbate biosynthesis enzymes. In H, “Cytosolic” refers to the coexpression of GMP, GME, GGP, GPP, and L-GalDH genes; “Whole” refers to the coexpression of those mentioned cytosolic genes plus mitochondrial L-GalLDH. For further details, constructs used in each infiltration are shown in Supplementary Table S1. Data displayed correspond to mean±SD from two leaves of at least three N. benthamiana plants infiltrated, collected at 3 d after infiltration. Different letters denote statistically significant differences for ascorbate concentration (one-way ANOVA, α = 0.05, post hoc Tukey test).
Combinatorial expression of ascorbate biosynthesis genes in N. benthamiana leaves identifies GGP as the main control point of ascorbate biosynthesis
We have shown that SW proteins are functional when a tag is added to their C-terminus and that a small amount of GGP–GFP protein is sufficient to increase the content of ascorbate in the vtc2 mutant to WT levels. In order to investigate the effect of the activity of ascorbate biosynthesis genes on ascorbate concentration we cloned the coding DNA sequences (CDS) of Arabidopsis GMP, GME, GGP, GPP, L-GalDH, and L-GalLDH with C-terminal GFP and HA, driven by the CaMV35S promoter (Figure 3A) and transiently expressed them in N. benthamiana leaves. Immunoblot analysis of the GFP-tagged SW proteins showed that the molecular masses of the different fusion proteins were consistent with their predicted sizes (Figure 3B). Moreover, all the fusion proteins showed little degradation at all time points tested (SupplementalFigure S5). Three days after agroinfiltration, GMP–GFP, GME–GFP, GGP–GFP, and GPP–GFP proteins showed the highest expression, while L-GalDH-GFP and L-GalLDH-GFP were also well expressed (Figure 3B). Therefore, we performed further analyses after 3 d of infiltration
We performed a systematic analysis of the effect of transient overexpression of individual or multiple combinations of these SW Arabidopsis genes on the ascorbate concentration in N. benthamiana leaves. On the basis of our previous data (Figure 3B), we analyzed the ascorbate content 3 d after agroinfiltration. All genes were expressed either individually or in pairs (Figure 3, C–G; SupplementalTable S1), and due to the large number of possible combinations, three genes were expressed individually and two genes out of the three were co-expressed to analyze the ascorbate content in the same experiment (Figure 3, C–G). This experimental design validated the reproducibility of our experimental system since individual genes have been analyzed three times with their proper controls (except for GMP, with only two). As shown in Figure 3C, the transient expression of individual GMP–GFP and GME–GFP genes did not increase ascorbate content relative to the naïve (not agroinfiltrated) or control (infiltrated with GFP) N. benthamiana leaves. However, expression of GGP–GFP increased ascorbate concentration between 2- and 3-fold. The expression of individual GMP, GME, and L-GalDH genes or any combination of these three genes without GGP-GFP did not significantly increase ascorbate concentration in any of the multiple assays (Figure 3, D–G). It also demonstrates that higher levels of GMP (Figure 3C), GPP (Figure 3G), and L-GalDH (Figure 3F) do not increase the ascorbate content, either individually or when combined. The combination of GGP with GME caused a significant increase in ascorbate relative to GGP (Figure 3E), although the effect was not consistent in all experiments (Figure 3, E and H). An increased ascorbate content by combining GME and GGP has been previously reported using transient overexpression of kiwifruit (Actinidia deliciosa) GME and GGP homologs in tobacco (N. tabacum) leaves (Bulley et al., 2009).
Finally, we tested the effect of the simultaneous transient overexpression of all SW cytosolic Arabidopsis genes on the ascorbate concentration in N. benthamiana leaves. The expression of all cytosolic components was confirmed by immunoblot analysis (Figure 3H) and increased the ascorbate content to similar levels as produced by expressing GGP alone (Figure 3H). Expressing all the cytosolic enzymes together with the final mitochondrial enzyme L-GalLDH also did not increase the ascorbate concentration (Figure 3H). Overall, these results point to GGP as the main control point in the biosynthesis of ascorbate.
Subcellular localization of ascorbate biosynthesis enzymes using GFP-tagged proteins
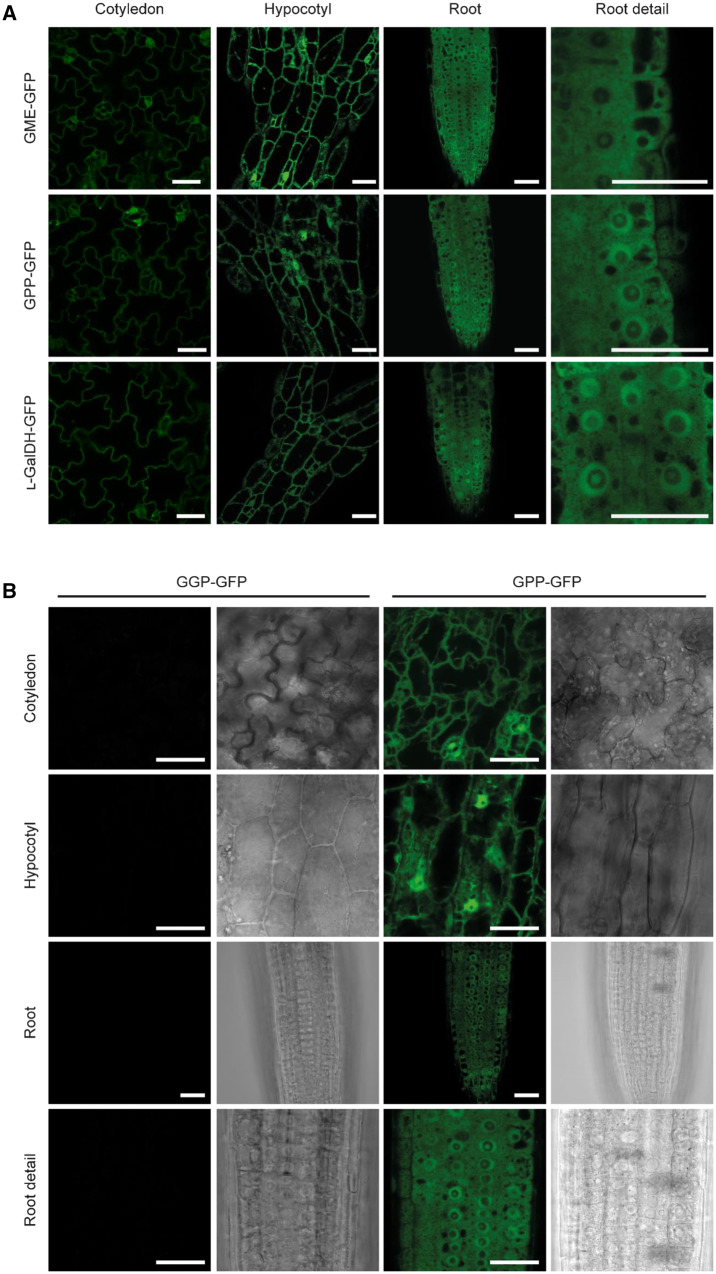
The biosynthesis of ascorbate up to the L-galactose dehydrogenase step is proposed to take place in the cytosol with the exception of the last step that occurs at the intermembrane space of the mitochondria (Østergaard et al., 1997; Heazlewood et al., 2004; Dunkley et al., 2006; Pineau et al., 2008; Schimmeyer et al., 2016). In silico analysis using the software Compartments (https://compartments.jensenlab.org/Search) predicted a cytosolic subcellular localization for GME, GPP, and L-GalDH, a dual cytosolic–nuclear localization of GMP and GGP and a mitochondrial localization of L-GalLDH (SupplementalFigure S6). Confocal microscopy imaging of C-terminal GFP-tagged proteins transiently expressed in N. benthamiana revealed a cytosolic subcellular localization for GMP, GME, GGP, GPP, and L-GalDH, while L-GalLDH-GFP appeared in structures that resembled mitochondria (Figure 4). Interestingly, GMP–GFP, GGP–GFP, GPP–GFP, and L-GalDH–GFP also showed a nuclear localization in addition to their cytoplasmic localization, whereas we observed a GME–GFP signal surrounding the nuclei (Figure 4). Evidence for a dual cytosolic–nuclear localization has been provided for GMP and GGP using stable transgenic Arabidopsis lines (Müller-Moulé, 2008; Wang et al., 2013), suggesting a potential nuclear function of these proteins. Remarkably, similarly to GMP, nuclear localization signals are not predicted for GPP or L-GalDH and, therefore, the localization reported here was unexpected (SupplementalTable S2).
Figure 4.
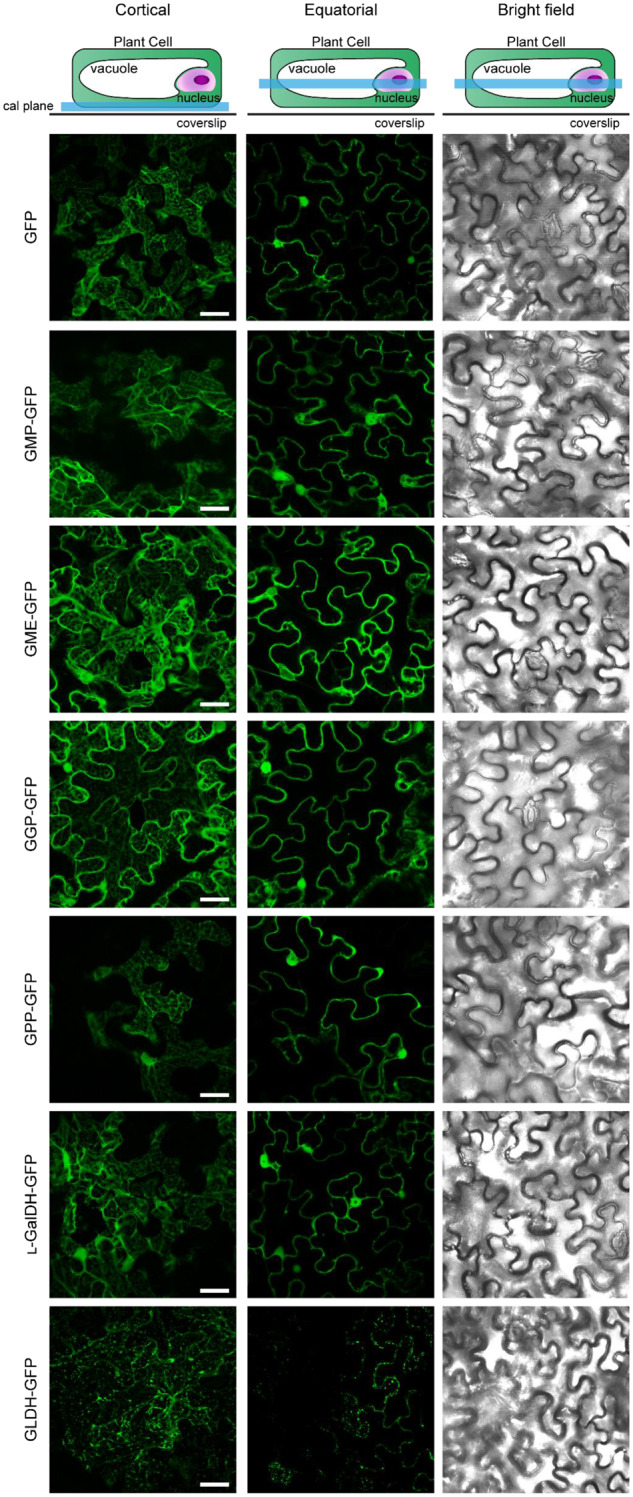
Subcellular localization of free GFP and C-terminal GFP fusions of ascorbate biosynthesis enzymes transiently expressed in N. benthamiana leaves under the control of the 35S promoter 3 d after agroinfiltration. GFP was visualized by laser scanning confocal microscopy. Scale bar = 30 µm.
Because overexpression can result in ectopic localization of proteins, we used the Arabidopsis transgenic lines that carry functional biosynthetic ascorbate proteins C-terminally fused to GFP and driven by their own promoters to analyze their in vivo subcellular localization as well as their tissue distribution. Cotyledons, cotyledon–hypocotyl junctions, hypocotyls and root tips of 4-d-old Arabidopsis seedlings were analyzed. GME–GFP, GPP–GFP, and L-GalDH–GFP lines showed a strong fluorescence signal in the cytoplasm (Figure 5A). In addition, GPP–GFP and L-GalDH–GFP were present within nuclei-like structures. In roots, these two proteins were present within the nucleus as a dot that suggests location in the nucleolus, but they also displayed a ring-like pattern in the nucleus, whereas GME–GFP showed an even distribution across the cytosol (Figure 5A). We did not detect fluorescence signals for GGP–GFP above the background level (Figure 5B) consistent with the weak signal previously observed in immunoblot analysis (Figure 2H). Except for GGP, these data are consistent with the over-expression data of SW-GFP constructs in N. benthamiana.
Figure 5.
Subcellular localization of ascorbate biosynthesis enzymes fused to GFP at the C-terminus and expressed under the control of their native promoters in Arabidopsis. A, The expression of GFP-tagged enzymes is ubiquitous in 4-d-old seedlings and their subcellular localization is compatible with the cytosol and nucleus except for GME, which only locates in cytosol. B, GFP signal of a functional GGP-GFP (line 13) protein was not detectable. GFP was visualized by laser scanning confocal microscopy. Scale bar = 30 µm.
Protein–protein interaction assays support a physical association of proteins involved in ascorbate biosynthesis
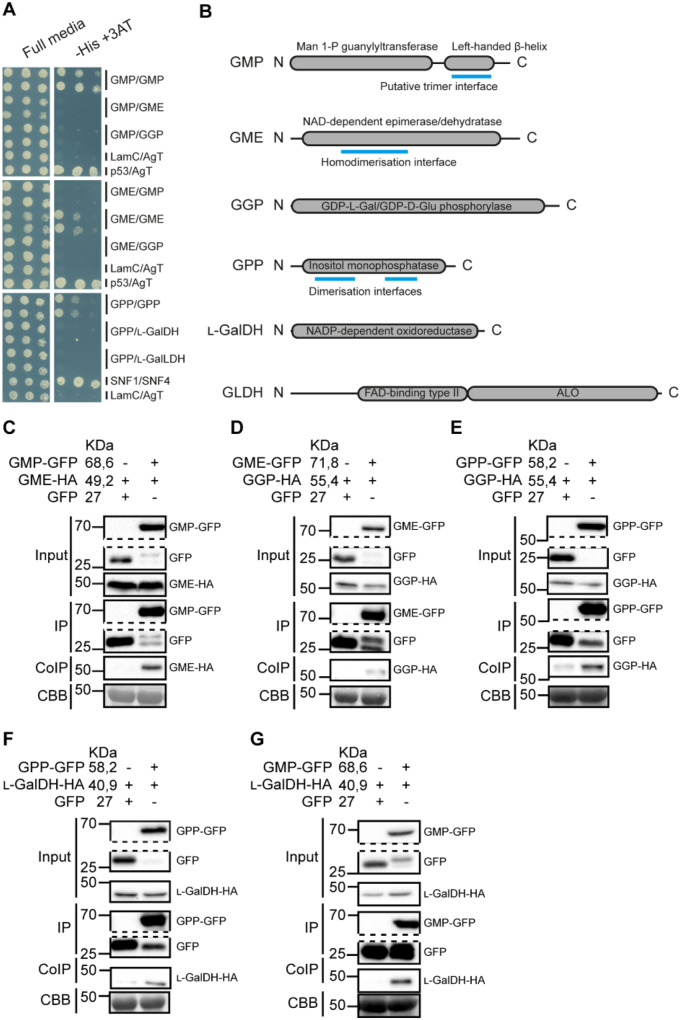
Enzymes involved in metabolic pathways are often physically associated (Smirnoff, 2019; Sweetlove and Fernie, 2018). Since GMP, GME, GGP, GPP, and L-GalDH proteins showed cytoplasmic localization (Figures 4 and 5), we investigated their interaction in vivo using a yeast two-hybrid (Y2H) assay (Figure 6A;SupplementalFigure S7). To achieve this, all CDSs of ascorbate biosynthesis genes (including the mitochondria-localized L-GalLDH as a negative control) were cloned into both the binding domain (BD, pGBKT7) and activation domain (AD, pGADT7) yeast vectors and all 36 possible interactions resulting from the combination of all the enzymes of the pathway in both directions were tested (fused to AD or to BD; SupplementalFigure S7). As positive controls, we included SNF1/SNF2 (Fields and Song, 1989) and p53/SV40-Large AgT (Iwabuchi et al., 1993; Li and Fields, 1993) interactions, whereas Laminin C/SV40-Large AgT was used as a negative control (Bartel et al., 1993; Ye and Worman, 1995). This analysis did not provide evidence of direct interactions between the enzymes of the pathway although GMP, GME, and GPP showed self-interaction, showing that these proteins were expressed in yeast (Figure 6A). A dimerization interface is predicted for GME and GPP, while a trimerization domain is predicted for GMP (Figure 6B), consistent with our Y2HY2H interaction results.
Figure 6.
Interaction of ascorbate biosynthesis enzymes assessed by Y2H analysis and Co-IP. A, The Y2HY2H assay detected no direct interaction between the enzymes, but GMP, GME, and GPP, which contain predicted dimerization/trimerization domains (B), form dimers/trimers. B, Protein scheme for each of the enzymes assayed in the Y2H assay showing length (in amino acids), annotated domains and predicted dimer/trimerization interfaces. C–G, The Co-IP assay reported association in vivo in N. benthamiana between consecutive steps as well as between the first (GMP) and the last (L-GalDH) steps occurring in the cytosol. GFP-tagged enzyme from N. benthamiana crude protein extracts containing two overexpressed consecutive enzymes, GFP and HA-tagged, was pulled-down using α-GFP agarose beads and coIP protein was detected using α-HA antibody. CBB: Coomassie Brilliant Blue.
Despite the apparent lack of direct interactions of ascorbate biosynthesis enzymes in yeast, we investigated whether these proteins could associate in vivo. For this, we co-expressed biosynthesis enzymes of the pathway tagged with C-terminal GFP and HA in N. benthamiana and performed coimmunoprecipitation (Co-IP) assays. We used GFP-Trap beads to immunoprecipitate GFP-tagged proteins and α-HA antibodies to detect co-immunoprecipitated (IP) HA-fused proteins using immunoblot. We first tested biosynthesis proteins that are consecutive in the pathway (Figure 6, C–F), that is, GMP/GME, GME/GGP, GGP/GPP, and GPP/L-GalDH. We co-expressed GMP–GFP and GME–HA to test the interaction, and GFP together with GME–HA as a negative control (Figure 6C) and checked by immunoblot if protein expression was suitable for immunoprecipitation prior to the assay (SupplementalFigure S8). After immunoprecipitation of GMP–GFP and free GFP, we detected a specific association between GME–HA with GMP–GFP but not with free GFP (Figure 6C). Following the same strategy, we found associations between all consecutive enzymes of the pathway (Figure 6, D–F). In addition, we also performed Co-IP between the first and last cytosolic enzymes of the pathway, that is, GMP and L-GalDH found a specific association between GMP–GFP and L-GalDH–HA (Figure 6G).
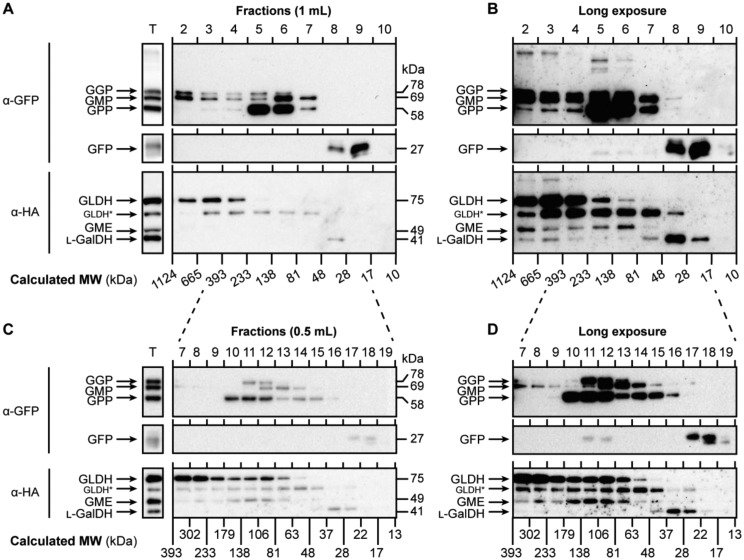
The finding that all SW proteins associate using Co-IP experiments suggests that they are part of a large enzymatic complex. In order to investigate this possibility, we expressed all SW enzymes in N. benthamiana leaves (see SupplementalTable S1) and extracted proteins in their native state. This extraction produced all proteins of the expected size (Figure 7A, left panel) with the exception of a protein that is likely a degradation product of GLDH, since this band was not detected after a denaturing extraction using Laemmli buffer (SupplementalFigure S9). This crude extract was then fractionated using gel filtration chromatography under nondenaturing conditions. We discarded the first fraction since it typically contains protein aggregates and detected by immunoblots the SW proteins in the collected fractions (0.5 or 1 mL) ranging from 10 to 1,124 kDa. Interestingly, all SW proteins eluted in high-molecular-weight fractions (Figure 7, A–D). In addition to their presence in high-molecular weight fractions, most proteins eluted in a size between 63 and 233 kDa except for GLDH, which accumulated in fractions over 233 kDa in size (Figure 7A) and L-GalDH, which eluted in fractions consistent with its predicted molecular weight. The elution of GLDH in high-molecular weight fractions is consistent with it being in complex I of the mitochondrial electron transport chain (Schimmeyer et al., 2016).
Figure 7.
The enzymes of the Smirnoff-Wheeler pathway elute together in high-molecular weight fractions of N. benthamiana protein extract. A, All the proteins transiently expressed were separated in 1 mL fractions and mainly accumulated between 393 and 48 kDa. B, Longer exposure of the blot presented in (A) shows that all the proteins eluted in high-molecular weight fractions. C, Fractions collected every 0.5 mL more accurately determined that the accumulation of proteins mainly occurs between 81 and 138 kDa but a longer exposure (D) enabled the detection of proteins in higher-molecular weight fractions than that. Proteins from N. benthamiana leaves were extracted as described in “Co-IP assay” section and separated as described in “Gel filtration chromatography” section. T=Total protein extract; MW=molecular weight. Calculated MWs were estimated using Dextran blue (2,000 kDa), Alcohol dehydrogenase (150 kDa), Albumin (66 kDa), and Carbonic Anhydrase (29 kDa); GLDH* points to a degradation product of GLDH (see Supplemental Figure S9).
A kinetic model of ascorbate biosynthesis indicates GGP as the main control point
To further understand the results of over-expression, a kinetic model of ascorbate biosynthesis was constructed using Complex Pathway Simulator (COPASI; Hoops et al., 2006). Figure 8A shows the reactions included in the pathway. The model carried out metabolic control analysis (MCA) using parameters described in Materials and Methods section and Supplementary Table S3. It incorporated the observed low expression of GGP with the reported characteristics of feedback inhibition. Evidence from radiolabeling suggests that ascorbate synthesis is subject to feedback inhibition by ascorbate (Conklin et al., 1997; Pallanca and Smirnoff, 2000; Wolucka and Van Montagu, 2003; Laing et al., 2015). The proposed uORF-mediated feedback repression of GGP translation (Laing et al., 2015) was modeled by including noncompetitive inhibition by ascorbate. Competitive inhibition of PMI and L-GalDH by ascorbate (Mieda et al., 2004; Maruta et al., 2008) were included, although in the case of L-GalDH, the inhibition could be an assay artifact (Laing et al., 2004).
Figure 8.
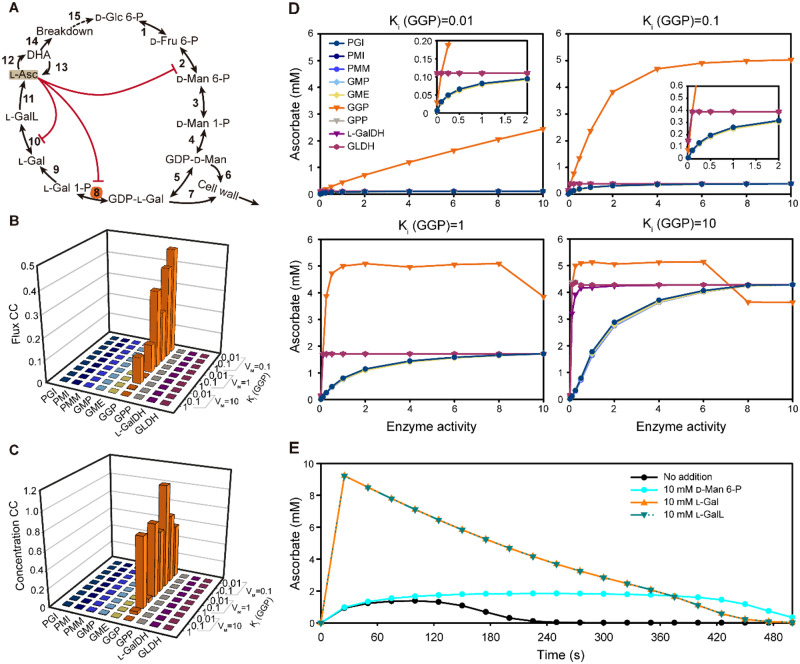
A kinetic model of ascorbate biosynthesis and turnover. A, Scheme of the pathway modeled in COPASI showing intermediates catalytic steps (arrows) and feedback (red lines). The initial model parameters are shown in Supplementary Table S3. B, Metabolic control analysis at steady state was used to calculate flux control coefficients for each enzyme with various strengths of noncompetitive inhibition exerted by ascorbate (indicated by Ki values) on GGP (Step 8). C, Metabolic control analysis at steady state was used to calculate concentration control coefficients for ascorbate with various strengths of noncompetitive inhibition exerted by ascorbate (indicated by Ki values) on GGP (Step 8). D, The effect of varying enzyme activity on ascorbate concentration with various strengths of noncompetitive inhibition exerted by ascorbate (indicated by Ki values) on GGP (Step 8). E, Time course of the change in ascorbate concentration in response to the addition of D-mannose 6-phosphate (D-Man 6-P), L-galactose (L-Gal), and L-galactono-1,4-lactone (L-GalL). 1: PGI, 2: PMI, 3: PMM, 4: GMP, 5: GME, 6:GDP-D-mannose to cell wall polymers, 7: GDP-L-galactose to cell wall polymers, 8: GGP, 9: GPP, 10: L-GalDH, 11: GLDH, 12: Oxidation, 13: Reduction, 14: Turnover, and 15: Recycle.
Determination of the flux control coefficients for each step in the biosynthesis pathway showed that only GGP contributes significantly, having values of up to 0.5. Simulation using various combinations of GGP activity (V) and Ki (smaller values indicate stronger feedback inhibition) showed that low activity and small Ki increases the flux control coefficient of GGP (Figure 8B). The remainder of flux control resided in the turnover reactions (SupplementalTable S4). Therefore, for the biosynthesis steps, the model predicts that only GGP controls the flux through the pathway if feedback repression is sufficiently strong. Correspondingly, concentration control coefficients (i.e. the change in the concentration of each pathway intermediate caused by a change in enzyme activity) for each step were calculated. The results for ascorbate are shown in Figure 8C and for all intermediates in SupplementalTable S5. As with the flux control coefficient, only GGP influenced ascorbate concentration; the control increased with decreased Ki for ascorbate. The outcome could be affected by the starting conditions (relative enzyme activities and kinetic constants) but testing the model with varying Km values or with and without competitive feedback inhibition of PMI and L-GalDH had little effect on ascorbate concentration (see Supplementary Material 1 for details).
To simulate the metabolic engineering experiments, the enzyme activity of each step was increased over a large range and the predicted change in ascorbate concentration at various feedback repression strengths was determined (Figure 8D and SupplementalTable S6). As expected from the MCA, only varying GGP activity had a significant effect on ascorbate over a very wide range of activities and this effect was strongly dependent on the extent of feedback repression. In comparison, it was necessary to decrease the activity of the other enzymes to very low values before ascorbate concentration was affected and this behavior was particularly marked for the post-GGP steps. The kinetic model therefore reflects the effect of transient expression of the pathway enzymes in N. benthamiana in which only increasing GGP activity increases ascorbate concentration. Simulation of mannose 6-P addition caused a small transient increase in ascorbate accumulation, while L-galactose addition had a large effect (Figure 8E). This differential response reflects the operation of feedback repression at the GGP step. GDP-D-Man and GDP-L-Gal are also substrates for polysaccharide synthesis. To test the influence of draining these intermediates from the pathway, irreversible incorporation into cell wall components was modeled. This had no effect on the relative control of other pathway enzymes, including GGP (SupplementalTable S7). Finally, the model correctly simulates the effect of knocking out the two GGP isoforms (VTC2 and VTC5) which were introduced into the model as parallel reactions. Ascorbate concentration in vtc2 and vtc5 mutants is not compensated (Dowdle et al., 2007) and the model recapitulated this result (SupplementalTable S8).
Discussion
We prepared GFP-tagged versions of all the enzymes in the SW ascorbate biosynthesis pathway and showed that they were functional by complementation of ascorbate concentration in their respective Arabidopsis mutants. The GFP-tagged enzymes were transiently expressed, individually and in combination, in N. benthamiana leaves. Only GGP over-expression was able to increase the ascorbate concentration, which confirms previous studies (Bulley et al., 2009, 2012; Yoshimura et al., 2014; Laing et al., 2015; Li et al., 2018; Zhang et al., 2018; ALi et al., 2019). Interestingly, a vtc2 mutant containing a GGP–GFP construct driven by its own promoter and including the 5′-UTR showed an extremely low GFP fluorescence compared with other GFP-tagged SW proteins. This low expression of GGP–GFP was also reflected in the immunoblot comparing the expression of all the pathway enzymes (Figure 2E), likely reflecting the translational repression of GGP via the conserved uORF in the 5′-UTR (Laing et al., 2015). This is further confirmed by the finding that the overexpression of GGP in N. benthamiana without the uORF resulted in a similar amount of GGP protein to other proteins of the pathway.
On the basis of the data presented here and other reports, we constructed a kinetic model based on the known properties of the pathway enzymes that includes the noncompetitive inhibition of GGP activity by ascorbate. This approach simulates the proposed translational repression via the 5′-uORF since it results in an ascorbate-dependent decrease in activity without changing its kinetic properties. The model was used for MCA, an approach that determines the distribution of the control of flux and concentration of pathway intermediates in response to small changes in activity of each enzyme in the pathway by calculating flux control and concentration control coefficients for each step (Fell, 1992). This analysis showed that the only step in the biosynthesis pathway that controls pathway flux and ascorbate pool size is GGP, if feedback repression is sufficiently strong. Decreasing the strength of feedback repression by ascorbate on GGP eventually decreased its flux and concentration control coefficients. The model predicts that the steps after GGP exert little control and that in the presence of feedback repression, GGP is the only critical step. Consequently, in virtual metabolic engineering experiments, in which each enzyme was changed over a large range, only GGP affected the ascorbate concentration. The model prediction is consistent with the relatively small effect of over-expressing enzymes other than GGP. The model outcomes were robust over a range of starting conditions and are minimally affected by the Km values or by the presence/absence of competitive inhibition of PMI, GME, and L-GalDH. The model also explains another feature of the pathway, which is that VTC2 and VTC5, the paralogs of GGP in Arabidopsis, do not compensate each other when knocked out (Dowdle et al., 2007). The model also shows that adding exogenous mannose 6-P has no effect on ascorbate synthesis, while the downstream intermediates L-Gal and L-GalL do, corresponding with other experimental results (Gatzek et al., 2002). The control features of the pathway appear to emerge from its architecture in which a strongly repressible GGP step is followed by the irreversible L-Gal 1-P phosphatase and L-galactonolactone dehydrogenase steps. In a previous study of transient expression in tobacco GME alone did not affect ascorbate, in accordance with the model, but also enhanced the effect of expressing GGP (Bulley et al., 2009), suggesting that the outcome of metabolic engineering may be dependent on other factors. Stable over-expression of GMP and GME has been reported to increase ascorbate in tomato and tobacco (Badejo et al., 2008, 2007; Li et al., 2019) and the model predicts that these enzymes could be effective if endogenous activity is low. Similarly, increasing the expression of L-GalDH in tobacco had no effect on ascorbate, while antisense suppression in Arabidopsis only had a small effect in high light (Gatzek et al., 2002). Comparison of the activity of SW enzymes where the ascorbate pool size differs under low- and high-light intensity showed that only GGP had increased activity in high light in Arabidopsis (Dowdle et al., 2007). Transcript levels suggest that only VTC2 and VTC5 (and sometimes GME) are responsive to environmental conditions (Li et al., 2013), and that there is not a coordinated induction of SW genes under high light and low temperature, conditions that increase ascorbate concentration (Laing et al., 2017). The control of ascorbate biosynthesis therefore differs from pathways in which there is coordinated induction of multiple pathway enzymes by transcription factors, for example, anthocyanin synthesis (Zhang et al., 2014). Importantly, the model shows that the ascorbate pool size responds to GGP over a wide range and thereby explains how a combination of transcription and feedback repression controls the amount/activity of GGP and provides a mechanism to adjust the ascorbate pool size to prevailing conditions. Light is the best studied factor, and ascorbate levels adjust to the prevailing light intensity over several days (Page et al., 2012). The mechanism likely involves a changed balance between increased transcription of GGP via a possible photosynthesis-derived signal and light response promoter elements (Gao et al., 2011; Yabuta et al., 2007) and ascorbate repression of translation via the uORF. In addition to the transient expression assays showing the role of the uORF in translational repression of GGP (Laing et al., 2015), clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 editing of the uORF in Arabidopsis, lettuce and tomato increases ascorbate concentration, confirming its role in repression of ascorbate synthesis in diverse species (Li et al., 2018; Zhang et al., 2018). Faster control could also occur if ascorbate status more directly affects GGP activity via direct inhibition or post-translational modification. These latter possibilities have not been investigated but could operate since ascorbate addition relatively rapidly decreases incorporation of 14C-glucose or mannose into ascorbate (Pallanca and Smirnoff, 2000; Wolucka and Van Montagu, 2003). It is not known if ascorbate or a proxy of ascorbate concentration exerts repression and, also, the mechanism by which the uORF controls GGP translation is not known.
In addition to investigating the effect of enzyme overexpression on ascorbate accumulation, we used tagged enzymes to investigate subcellular localization and the formation of enzymatic complexes. Except for L-GalLDH, the cytosolic localization of the SW enzymes is consistent with the in silico prediction. In this study, we have shown that all SW proteins are present in the cytosol. Interestingly, in addition to GMP and GGP that were previously reported to localize in the nucleus (Müller-Moulé, 2008; Wang et al., 2013), GPP and L-GalDH also showed nuclear localization despite the lack of nuclear localization signals. Whether or not this nuclear localization is functionally important remains to be determined, but several metabolic enzymes moonlighting in the nucleus have been previously reported (Boukouris et al., 2016; Zhang et al., 2020).
Physical association between enzymes in metabolic pathways and proteins in signaling pathways has been frequently reported (Zhang et al., 2017, 2020; Sweetlove and Fernie, 2018; Amorim-Silva et al., 2019; Smirnoff, 2019). We investigated this possibility for ascorbate biosynthesis enzymes using Y2HY2H analysis, immunoprecipitation and gel filtration chromatography. Co-IP analysis using GFP-tagged enzymes expressed in N. benthamiana indicated that consecutive SW enzymes, as well as the first and last enzymes (GMP and L-GalDH) are associated. Furthermore, gel filtration revealed that a population of all the enzymes elute as a higher molecular weight complex. These assays therefore reveal relatively stable interactions between the ascorbate biosynthesis enzymes from GMP through to L-GalDH in the cytosol (although some also have dual cytosolic/nuclear localization). However, in contrast, Y2HY2H analysis failed to detect pairwise interaction of the enzymes. Although the N-terminal position of both activation and binding domains could be impairing the interactions involving GGP or L-GalDH, the self-interactions found in GMP, GME, and GPP suggest that this possibility is unlikely. Furthermore, previous studies reported ascorbate-producing yeasts by heterologous expression of the plant pathway in Saccharomyces cerevisiae, proving that these proteins are stable in yeast (Branduardi et al., 2007; Fossati et al., 2011). The lack of direct interaction in yeast but the associations found in N. benthamiana may indicate that the enzymes might require the presence of each other so that the interaction would not be detectable in the Y2HY2H assay. Based on these results, it is tempting to speculate that some of the ascorbate biosynthesis enzymes might form a functional enzymatic complex (metabolon) channeling the ascorbate precursors, hence increasing the metabolic flux along the pathway. This complex could also assist partitioning of GDP-D-mannose and GDP-L-galactose between ascorbate and polysaccharide synthesis. However, assessing the functional significance of enzyme complexes is challenging (Smirnoff, 2019) and further work is needed.
In conclusion, metabolic engineering experiments using the last six enzymes of the SW pathway show that only GGP has significant control over the pathway, and we have developed a kinetic model that provides a rationale for this result. It is clear that the balance of GGP amount is controlled by a combination of transcription and translation repression, largely, but not entirely mediated by the uORF mechanism. In addition, we have produced a set of complemented mutants from GME to L-GalDH tagged with GFP that provides a resource for further investigation of the SW pathway.
Materials and methods
Plant material
Arabidopsis thaliana (L.) Heynh WT and transgenic lines generated were in the Col-0 ecotype. Arabidopsis mutants vtc2-4 (SAIL_769_H05; Lim et al., 2016), vtc4-4 (SALK_077222; Torabinejad et al., 2009), and gldh (SALK_060087; Pineau et al., 2008) used in this study have been previously described. However, to the best of our knowledge, this is the first time that SALK_150208 (here as named gme-3) and SALK_056664 (here named as lgaldh-1), obtained from The European Arabidopsis Stock Center (NASC: http://arabidopsis.info/), were described (SupplementalTable S9). Diagnostic PCR was performed to identify the presence of the T-DNA insertion in individual plants using the allele-specific primers listed in SupplementalTable S10A. The generation of stable transgenic lines either in the WT or in the respective mutant backgrounds is described in “Generation of transgenic lines” section and lines are listed in M&M, SupplementalTable S1.
Standard growth conditions
Arabidopsis seeds were surface sterilized using chlorine gas by pouring 3 mL of 37% [w/w] HCl into 100 mL of commercial bleach and airtight sealed for 4 h. Then, seeds were air-cleared for at least 4 h in a laminar flow cabinet and stored at 4°C for a 3-d stratification. Seeds were sown on half-strength Murashige–Skoog agar-solidified (0.6% [w/v] agar) medium supplemented with 1.5% [w/v] sucrose under sterile conditions and grown with a long-day photoperiod (16-h light/8-h darkness cycle, 22 ± 1°C, 150 ± 50-µmol photons m−2 s−1) for 7–10 d. Seedlings were then collected for analysis and transferred to liquid media or to soil (4:1 [v/v] soil:vermiculite).
Plasmid constructs
Two different types of DNA fragments were used as templates (genomic and coding DNA sequence) to generate the constructs used in this work. First, genomic fragments were amplified starting from at least 2-kb upstream of the +1 ATG site unless otherwise specified (SupplementalFigure S1) until the stop codon (not included) using genomic Col-0 DNA as template. Second, CDSs were amplified, starting at +1 ATG until the stop codon (not included), using as template Col-0 Arabidopsis cDNA that was generated using total RNA. In the case of GGP, due to a failure in amplifying the entire GGPp:GGP product, about 1.4 kb (according to Gao et al., 2011) of the GGP promoter including the 5′-UTR was amplified (SupplementalFigure S1 and Table S11). The primers used to generate these above-mentioned DNA fragments are detailed in SupplementalTable S10, B and C. A proofreading DNA polymerase was used for all these PCR amplifications (TAKARA PrimeSTAR Max DNA Polymerase, CAT. #R045A).
The GGP promoter was digested with HindIII and directly cloned into the pGWB504 vector. All other DNA fragments (excluding the GGP promoter) were cloned into pENTR/D-TOPO using the pENTR Directional TOPO cloning kit (Invitrogen). Next, DNA genomic fragments (which include the promoter and lack the stop codon) were recombined from pENTR/D-TOPO into pGWB4 by LR reaction (Invitrogen) to generate SWp:SW-GFP constructs for GME, GPP, and L-GalDH. Also, all CDS fragments (without the promoter, 5′-UTR and stop codon) were sub-cloned into pGWB5 and pGWB14 to generate CaMV35S:SWCDS-GFP and CaMV35S:SWCDS-HA, respectively. In the case of GGP, the CDS was also recombined into the GGPp-pGWB504 by LR reaction (Invitrogen), thus obtaining the GGPp:GGPCDS-GFP construct. Likewise, CDSs (pENTR/D-TOPO) were subcloned into both pGADT7-GW (activation domain, AD) and pGBKT7-GW (binding domain, BD) to generate SWCDS-AD and SWCDS-BD, respectively, for the Y2HY2H assay (see “Y2H assay” section). The expression vectors generated using the destination vectors pGWB5 and pGWB14 contain a kanamycin and a hygromycin resistance gene for bacteria and plant selection, respectively. The Y2HY2H vectors pGADT7 and pGBKT7 contain an ampicillin and kanamycin resistance gene, respectively, for selection in bacteria. A summary of the generated constructs is shown in SupplementalTable S11. All constructs were analyzed by colony PCR (using the primers indicated in SupplementalTable S10D), enzymatic DNA restriction and sequencing (using the primers indicated in SupplementalTable S10E).
The plasmids used from the pGWB vector series were provided by Tsuyoshi Nakagawa (Department of Molecular and Functional Genomics, Shimane University; Nakagawa et al., 2007). The pGADT7(GW) and pGBKT7(GW) destination vectors were provided by Salomé Prat (Nacional de Biotecnología-Consejo Superior de Investigaciones Científicas).
Generation of Arabidopsis transgenic lines
The constructs generated using the pGWB4 plasmid for native expression of the SW genes (pGWB504 for GGP) were transformed into Agrobacterium (Agrobacterium tumefaciens) GV3101::pMP90 by electroporation and confirmed by colony PCR using the primers indicated in SupplementalTable S10D. Arabidopsis WT (Col-0) plants were then transformed using these Agrobacterium strains by floral dipping (Clough and Bent, 1998). T2 plants were analyzed for hygromycin resistance ratio resistant:sensitive (R:S) and lines with single insertion (T2 HygR:HygS=3, χ2 (α = 0.05), n > 300) were selected. Selected T2 plants were grown in a long-day photoperiod and a sample of a rosette leaf was taken to perform immunoblot analysis (Figure 2; SupplementalFigure S3). Seeds from different T2 lines with a high expression were selected and homozygous T3 and T4 plants were used in this work.
For generation of the complementation lines, selected T2 plants (with a single insertion) were crossed with their respective mutants in both directions as male and female. In order to obtain homozygous plants for both the T-DNA insertion and the SWp:SW-GFP construct (HygR), we selected hygromycin resistant plants and performed diagnostic PCR to identify the T-DNA insertion using the primers in SupplementalTable S10A. Homozygous F3 plants for T-DNA and SWp:SW-GFP insertion were used in this work.
Ascorbate complementation assay
WT, gme (SALK_150208), lgaldh (SALK_056664), and gldh (SALK_060087) seeds were surface sterilized and stratified for 3 d at 4°C. Seeds were then sown in agar-solidified (0.5% [w/v] agar) modified Scholl medium with or without 0.5-mM ascorbate as described elsewhere (Lim et al., 2016) and grown with a long-day photoperiod. After 10 d, plants were individually collected and PCR-diagnosed to identify homozygous mutants.
Ascorbate assay for combinatorial expression in N. benthamiana
Approximately 250 mg fresh tissue was extracted with 2 mL of 2% (w/v) metaphosphoric acid, 2 mM EDTA ice-cold buffer (modified from Davey et al., 2006). Samples were centrifuged at 18,000g for 20 min at 4°C, and supernatants were filtered (0.45 µm) before injection into an HPLC (Agilent Technologies 1200 series; Rx-C18, 4.6 × 100 mm2 3.5 µm column (Agilent Technologies 861967-902) diode array detector (Agilent Technologies G1315D). Samples were measured at 254 nm using filtered (0.45 µm) 0.1 M NaH2PO4, 0.2 mM EDTA, pH 3.1 (orthophosphoric acid) as the mobile phase (Harapanhalli et al., 1993) at 4°C (column at 20°C; 5 µL sample injection, 0.7 mL/min flow, 200 bar). Standards were prepared using ascorbic acid dissolved in extraction buffer, with concentrations ranging from 0 to 2.5 mM.
Ascorbate assay for Arabidopsis transgenic lines
Approximately 80-mg fresh tissue was extracted with 1 mL ice cold 3% (w/v) metaphosphoric acid plus 1 mM EDTA. Samples were centrifuged at 16,000g for 10 min at 4°C. Absorbance measurements were carried out at 265 nm using 20 µL sample/standard aliquots in a 96 well UV-transparent plate (Greiner UV-Star 96-Well Microplate) mixed with 100 µL of either phosphate buffer (0.2 M KH2PO4, pH 7 to measure ascorbate) or Tris(2-carboxyethyl)phosphine (TCEP)-phosphate buffer (0.2 M KH2PO4, pH 7, 2 mM TCEP, to measure ascorbate and DHA). Then, 5 µL 40 U/mL AO was added to each well, mixed and incubated at room temperature for 20 min, when the absorbance was again measured. The difference in the absorbance was interpolated in the standard curve, using standards ranging from 0 to 1 mM in extraction buffer. TCEP and AO stocks were prepared in phosphate buffer.
Immunoblot
Proteins were separated by size through sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Millipore, 0.45 µm pore size; CAT. #IPVH00010) previously methanol-equilibrated. Next, the PVDF membrane was blocked with 5% (w/v) skimmed milk followed by incubation with a primary antibody synthesized in mouse against GFP (Santa Cruz Biotechnology, CAT. #sc-9996; 1:600) or HA (Sigma-Aldrich, CAT. #H3663; 1:3,000) epitopes. Then, the membrane was incubated with a horseradish peroxidase (HRP)-conjugated α-mouse whole IgG antibody produced in rabbit (Sigma-Aldrich, CAT. #A9044; 1:80,000). Protein-HRP chemiluminescence was detected using Chemidoc XRS1 System (Bio-Rad) after incubation with Clarity ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate according to the manufacturer’s instructions.
Transient expression in N. benthamiana leaves
Agrobacterium GV3101::pMP90 was transformed by electroporation with the constructs generated using pGWB5 and pGWB14 positive colonies were confirmed by colony PCR. Two leaves per plant (leaves 3 and 4 from the apex) of 4-week-old N. benthamiana were infiltrated in the abaxial side using a 1 mL syringe (without the needle). Agrobacterium cultures were grown overnight in liquid Luria-Bertani medium containing rifampicin (50 μg/mL), gentamycin (25 μg/mL), and the construct-specific antibiotic kanamycin (50 μg/mL). Cells were then harvested by centrifugation (for 15 min at 3,000g in 50-mL falcon tubes) at room temperature, pellets were resuspended in agroinfiltration solution (10 mM MES, pH 5.6, 10 mM MgCl2, and 1 mM acetosyringone), and kept for 2–3 h in dark conditions at room temperature. In order to avoid overexpression-associated gene silencing, an Agrobacterium strain containing a p19 expression vector was coinfiltrated. For single-gene expression, the gene of interest and p19 were mixed in such a way that the optical densities (600 nm, OD600) were 0.8 and 0.2, respectively. For multiple co-infiltration experiments, the genes of interest were diluted in the same proportion so the final OD600 was 1 (0.8 for the combination of genes of interest and 0.2 for p19). Agrobacterium strains expressing either free GFP or TTL3-HA [an ascorbate biosynthesis nonrelated protein (Amorim-Silva et al., 2019)] were used to compensate OD600 in co-infiltration experiments.
To find the optimal protein expression, immunoblot time course analysis was addressed 2, 3, and 4 d after infiltration (Figure 3B). Protein loads at 2 d after infiltration were adjusted by dilutions with Laemmli buffer so a similar concentration among constructs is detected at this timepoint. Then dilutions were calculated and applied so relative concentrations among timepoints of the same construct were conserved.
Confocal laser scanning microscopy
All confocal images of N. benthamiana (Figure 4) were obtained using a Leica TCS SP5 II confocal microscope equipped with a 488-nm argon laser for GFP excitation and a PMT for its detection using the HCX PL APO CS 40 × 1,25 OIL objective with additional 2× digital zoom. Arabidopsis imaging was carried out using a Leica TCS SP8 equipped with HC PL APO CS2 40×/1,30 OIL objective and a solid state 488-nm laser used to excite GFP and a high-sensitivity SP hybrid detector (Figure 5A) and a Zeiss LSM 880 (Figure 5B) with a 488-nm argon laser and PMT detectors for GFP and transmitted light using the C-Apochromat 40×/1.2 W Korr M27 objective. All image processing was performed using FIJI (Schindelin et al., 2012; Schneider et al., 2012).
Y2H assay
The Gal4-based Y2H system (Clontech Laboratories) was used for testing the interaction. Saccharomyces cerevisiae Meyen ex E. C. Hansen strain PJ69-4A was transformed with the constructs described in “Plasmid constructs” section (SW genes clones into pGADT7 and pGBKT7) as described elsewhere (Gietz and Schiestl, 1995). Constructs transformed are explained in “Plasmid constructs” section (pGADT7 and pGBKT7). Transformants were grown on plasmid-selective medium (synthetic defined [SD]/-Trp-Leu) and grown at 28°C for 4 d. Two independent colonies were taken per transformation event and resuspended in 200 µL of sterile water. Ten-fold serial dilutions were made and 5 µL of each dilution were spotted onto three different interaction-selective medium: SD-Trp (minus tryptophan) -Leu (minus leucin) -His (minus histidine) +2 mM 3-AT (3-amino-1,2,4-triazole); SD-Trp-Leu-Ade; SD-Trp-Leu-Ade +3-AT. Full SD medium was also used to check strain survival. Plates were grown at 28°C and pictures were taken after 3, 5, and 9 d (only 5 d is shown). Concentrations are available at Rodríguez-Negrete et al. (2014).
Bioinformatic analyses
Protein domain predictions were performed using Protein Basic Local Alignment Search from the National Center for Biotechnology Information (Altschul et al., 1997, BLASTp; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) followed by functional annotation of protein domains using the Conserved Domain Database (Marchler-Bauer and Bryant, 2004; Marchler-Bauer et al., 2011, 2015, 2017; https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Protein subcellular localization was predicted using COMPARTMENTS (Binder et al., 2014; https://compartments.jensenlab.org/Search). Predicted subcellular localization signals were performed using LOCALIZER (Sperschneider et al., 2017; http://localizer.csiro.au/). Metabolic ControlC Analysis was carried out using COPASI (Hoops et al., 2006; http://copasi.org/). All the details needed for the simulations are shown in SupplementalTables S3–S7). GGP mRNA expression datasets were extracted from Transcriptome Variation Analysis database (Klepikova et al., 2016, TRAVA; http://travadb.org/) and eFP-seq Browser (Sullivan et al., 2019; https://bar.utoronto.ca/eFP-Seq_Browser/).
Co-IP assay
Four-week-old N. benthamiana plants were used for transient expression assays as described above. Leaves were ground to fine powder in liquid nitrogen. Approximately 0.5 g of ground leaves per sample was used, and total proteins were extracted with 1 mL of nondenaturing extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% [v/v] Nonidet P-40, 10 mM EDTA, 1 mM Na2MoO4, 1 mM NaF, 10 mM DTT, 0.5 mM PMSF**, and 1% [v/v] protease inhibitor [Sigma P9599]) and incubated for 30 min at 4°C using an end-over-end rocker. Protein extracts were centrifuged at 20,000g for 20 min at 4°C and then filtered by gravity using Poly-Prep chromatography columns (731-1550, Bio-Rad). Supernatants were filtered by gravity through Poly-Prep chromatography columns (731-1550, Bio-Rad), and 100 mL was reserved for immunoblot analysis as input. The remaining supernatants were used for immunoprecipitation of GFP-fused proteins using GFP-Trap coupled to agarose beads (Chromotek) and following the manufacturer’s instructions. Total (input), IP, and Co-IP samples were resuspended with 2× concentrated Laemmli sample buffer and heated at 95°C for 5 min for protein denaturation. Finally, samples were separated in a 10% (w/v) SDS–PAGE gel and analyzed as described above.
Gel filtration chromatography
Nicotiana benthamiana leaves transiently expressing all enzymes of the pathway from GMP to GLDH (see SupplementalTable S1) were collected 3 d after agroinfiltration and frozen in liquid nitrogen. Total proteins were extracted in their native state as described in “Co-IP assay” section and separated by gel filtration chromatography using an Akta Prime Plus, through a Superdex 200 Increase 10/300 GL column. Approximately 1.5 mg of protein extract was separated using a modified version of the extraction buffer as the mobile phase (50 mM Tris–HCl, 150 mM NaCl, 0.1% (v/v) Nonidet P-40, 10 mM DTT, 10 mM EDTA, 1 mM NaF, and 1 mM NaMoO4) at 0.2 mL/min and fractions were collected every 1 mL (Figure 7, A and C) or 0.5 mL (Figure 7, B and D). Next, 100 µL aliquots per fraction were denatured by adding 100 µL of Laemmli 2× buffer and boiled at 95°C for 10 min, followed by SDS–PAGE and immunoblotting of similar protein loads. Immunoblots were performed as described above.
Metabolic control analysis
A kinetic model of ascorbate biosynthesis was constructed using COPASI (http://copasi.org/; Hoops et al., 2006). Reactions were described by reversible Michaelis-Menten kinetics, except for L-Gal 1-P phosphatase/GPP and L-GalLDH which were described by irreversible Michaelis–Menten kinetics. Kinetic constants and competitive feedback inhibition (Ki) values were used where known (Østergaard et al., 1997; Gatzek et al., 2002; Wolucka and Van Montagu, 2003; Laing et al., 2004; Linster et al., 2007; Hoeberichts et al., 2008; Maruta et al., 2008). In other cases, Km was set to 0.1 mM. Forward and reverse rates (Vf/Vr) were set to 10 (or in cases where reverse rates are known, Vf was set to 10 and the reverse as the appropriate fraction). The activity of GGP was set to Vf = 0.1 and Vr = 0.02 in the initial model state to reflect the measured very low protein concentration. Variations in the Km and Ki (for competitive inhibition) values do not substantially affect model outcome. Competitive inhibition of PMI and L-GalDH by ascorbate (Mieda et al., 2004; Maruta et al., 2008) were included, although in the case of L-GalDH, the inhibition could be an assay artifact (Laing et al., 2004). To facilitate MCA, the pathway was made cyclic by incorporating a recycling element enabling the model to come to steady state. To incorporate recycling, ascorbate is oxidized to DHA (strictly the hydrated bicyclic form of DHA). DHA is either reduced back to ascorbate or broken down to products that are all recycled to the hexose phosphate pool (glucose 6-P). This approach very likely over-estimates the proportion recycled but will not affect conclusions on the control of the biosynthetic pathway. Turnover and recycle rates use a mass action law with K set to 0.1.
The feedback repression of GGP translation via the 5′-uORF was simulated by noncompetitive inhibition of GGP by ascorbate (Laing et al., 2015). This mechanism provides dynamic control over GGP activity as ascorbate accumulates. Noncompetitive inhibition decreases activity (V) without changing Km for the substrate (GDP-L-Gal). This exactly parallels the uORF feedback mechanism in that increasing ascorbate decreases enzyme amount (V), and therefore activity, but does not change its Km. The factor that might differ between the uORF-mediated response and noncompetitive inhibition is the shape of the ascorbate response curve. This is not known in detail in vivo so would be difficult to model explicitly and the Ki values would not be expected to fit in vivo ascorbate concentration. These considerations do not affect the principle of the response to ascorbate but may affect the exact dose response curve.
GDP-Man and GDP-L-Gal used for cell wall/glycoprotein synthesis can be simulated by irreversible incorporation into the cell wall. This was activated by changing the rates (V from 0 to 0.1). The pathway was then unable to come to steady state but was simulated in time course mode. Substrate addition was simulated by inactivating recycling (V = 0), altering the chosen substrate concentration and running a time course (over 500 s). GGP is encoded by VTC2 and VTC5 in Arabidopsis (Dowdle et al., 2007). To simulate knockouts of each gene (vtc2 and vtc5 mutants), the model was modified by running these in parallel (and assuming 80% contribution of activity from GGP (Dowdle et al., 2007). The models are supplied in Copasi format (*cps) and Systems Biology Markup Language formats (Supplementary Materials 1 and 2).
Statistical analysis
All analyses were performed using SigmaPlot v11 for Windows considering n ≥ 3 and α = 0.05.
Accession numbers
Sequence data were sourced from The Arabidopsis Information Resource (https://www.arabidopsis.org/) under the following accession numbers: AT2G39770 for GMP, AT5G28840 for GME, AT4G26850 for GGP, AT3G02870 for GPP, AT4G33670 for L-GalDH, and AT3G47930 for L-GalLDH.
Supplemental material
The following materials are available in the online version of this article.
Supplementary Figure S1. Gene structure, primers disposition and mutants’ T-DNA location.
Supplementary Figure S2. Diagnostic PCR of the gme, lgaldh, and gldh seedlings analyzed in the complementation assay described in Figure 2A.
Supplementary Figure S3. Generation of Arabidopsis transgenic lines containing ascorbate biosynthesis enzymes fused to GFP.
Supplementary Figure S4. GGP mRNA levels are similar to those of GMP and GME, although GGP protein is hardly detectable.
Supplementary Figure S5. Complete blots corresponding to Figure 2B (red rectangle).
Supplementary Figure S6. Predicted subcellular localization of ascorbate biosynthesis proteins in the plant cell according to COMPARTMENTS.
Supplementary Figure S7. Yeast two-hybrid whole screening.
Supplementary Figure S8. Immunoblot (α-GFP) of crude extracts of N. benthamiana leaves following agroinfiltration of the indicated combinations of proteins (also see Supplementary Table S1).
Supplementary Figure S9. Immunoblots (α-GFP upper panel; α-HA lower panel) of crude extracts of N. benthamiana leaves following agroinfiltration of all proteins in the pathway corresponding to Figure 7.
Supplementary Table S1. Infiltration in N. benthamiana corresponding to Figure 2, C–H.
Supplementary Table S2. Prediction of transit peptides and nuclear localization signals within VTC genes.
Supplementary Table S3. Initial conditions for the COPASI kinetic model of ascorbate biosynthesis and turnover.
Supplementary Table S4. Flux control coefficients for ascorbate biosynthesis and turnover with various strengths of noncompetitive feedback inhibition at the GDP-L-Gal phosphorylase step (Ki GGP) at different performing activities (Vm).
Supplementary Table S5. Concentration control coefficients at each step for pathway intermediates with various strengths of noncompetitive feedback inhibition at the GDP-L-Gal phosphorylase step (Ki GGP) under different GGP activity rates.
Supplementary Table S6. The effect of varying relative enzyme activity on the concentration of pathway intermediates with various strengths of noncompetitive feedback inhibition at the GDP-L-Gal phosphorylase step (Ki GGP).
Supplementary Table S7. Ascorbate concentration upon changes of flux in ascorbate precursors toward cell wall biosynthesis, under different GME activity rates.
Supplementary Table S8. Time course of ascorbate concentration modeled according to the reaction 8*) described in Supplementary Table S3.
Supplementary Table S9. Arabidopsis mutants and lines used in this work.
Supplementary Table S10. Arabidopsis stable lines used in this work.
Supplementary Table S11. (A) Sizes of constructs generated in this work using the vectors listed in (B) and the resultant construct.
Supplementary Material 1. Ascorbate biosynthesis and turnover kinetic model simulated in COPASI.
Supplementary Material 2. Ascorbate biosynthesis and turnover kinetic model simulated in COPASI considering the two isoforms of GGP activity, VTC2, and VTC5.
Supplementary Material
Acknowledgments
We acknowledge Glenda Gillaspy (Department of Biochemistry, Virginia Tech) for kindly providing us the vtc4-4 mutant used in this work.
Funding
This work was supported by a grant from the Spanish Ministerio de Educación, Cultura y Deporte para la formación del Profesorado Universitario (FPU014/01974), I Plan Propio de Investigación, Transferencia y Divulgación Científica de la Universidad de Málaga, The Ministerio de Economía, Industria y Competitividad (RYC-2013-12699), also co-financed by the European Regional Development Fund (BIO2016-81957-REDT, BIO2017-82609-R), the Spanish “Ministerio de Economía, Industria y Competitividad/FEDER” (AGL2016-75819-C2-1-R) and the Spanish Ministerio de Ciencia, Innovación y Universidades (PGC2018-098789-B-I00). N.S. was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) grants BB/G021678/1 and BB/N001311/1.
M.F., V.A.S., M.A.B., and N.S. conceived the project and designed the experiments; M.F. performed the experiments and analyzed the data; N.S. performed the MCA simulation; V.A.S., M.A.B., and N.S. supervised the experiments; D.A. generated vtc2/GGP-GFP transgenic lines and tested mutant complementation by AO assay; A.G.C. performed yeast two-hybrid assay with technical assistance by M.F.; A.E.V. provided technical assistance to M.F.; N.R.L. took part in the experimental design and performance of the gel filtration chromatography experiments; M.F., M.A.B., and N.S. wrote the article; V.A.S., M.A.B., and N.S. supervised and completed the writing; and M.A.B. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Miguel A. Botella (mabotella@uma.es).
References
- Ali B, Pantha S, Acharya R, Ueda Y, Wu LB, Ashrafuzzaman M, Ishizaki T, Wissuwa M, Bulley S, Frei M (2019) Enhanced ascorbate level improves multi-stress tolerance in a widely grown indica rice variety without compromising its agronomic characteristics. J Plant Physiol 240: 1–9 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim-Silva V, García-Moreno Á, Castillo AG, Lakhssassi N, Del Valle AE, Pérez-Sancho J, Li Y, Posé D, Pérez-Rodriguez J, Lin J, et al. (2019) TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in arabidopsis. Plant Cell 31: 1807–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Badejo AA, Jeong ST, Goto-Yamamoto N, Esaka M (2007) Cloning and expression of GDP-D-mannose pyrophosphorylase gene and ascorbic acid content of acerola (Malpighia glabra L.) fruit at ripening stages. Plant Physiol Biochem 45:665.– [DOI] [PubMed] [Google Scholar]
- Badejo AA, Tanaka N, Esaka M (2008) Analysis of GDP-D-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol 49: 126–132 [DOI] [PubMed] [Google Scholar]
- Bartel P, Chien CT, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques 14: 920–924 [PubMed] [Google Scholar]
- Bartoli CG (2006) Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J Exp Bot 57: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O’Donoghue SI, Schneider R, Jensen LJ, (2014) COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database 2014: bau012–bau012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouris AE, Zervopoulos SD, Michelakis ED (2016) Metabolic enzymes moonlighting in the nucleus: metabolic regulation of gene transcription. Trends Biochem Sci 41: 712–730 [DOI] [PubMed] [Google Scholar]
- Branduardi P, Fossati T, Sauer M, Pagani R, Mattanovich D, Porro D (2007) Biosynthesis of vitamin C by yeast leads to increased stress resistance. PLoS One 2: e1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley S, Laing W (2016) The regulation of ascorbate biosynthesis. Curr Opin Plant Biol 33: 15–22 [DOI] [PubMed] [Google Scholar]
- Bulley S, Wright M, Rommens C, Yan H, Rassam M, Lin-Wang K, Andre C, Brewster D, Karunairetnam S, Allan AC, et al. (2012) Enhancing ascorbate in fruits and tubers through over-expression of the l-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol J 10: 390–397 [DOI] [PubMed] [Google Scholar]
- Bulley SM, Rassam M, Hoser D, Otto W, Schünemann N, Wright M, MacRae E, Gleave A, Laing W (2009) Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-l-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J Exp Bot 60: 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method forAgrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N (2006) Arabidopsis thaliana VTC4 encodes l-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem 281: 15662–15670 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci 96: 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Pallanca JE, Last RL, Smirnoff N (1997) L-ascorbic acid metabolism in the ascorbate-deficient arabidopsis mutant vtc1. Plant Physiol 115: 1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Norris SR, Last RL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154: 847–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Willlams EH, Last RL (1996) Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc Natl Acad Sci 93: 9970–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronje C, George GM, Fernie AR, Bekker J, Kossmann J, Bauer R, (2012) Manipulation of l-ascorbic acid biosynthesis pathways in Solanum lycopersicum: elevated GDP-mannose pyrophosphorylase activity enhances L-ascorbate levels in red fruit. Planta 235: 553–564 [DOI] [PubMed] [Google Scholar]
- Davey MW, Kenis K, Keulemans J (2006) Genetic control of fruit vitamin C contents. Plant Physiol 142: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Cook DR, Ford CM (2006) L-Tartaric acid synthesis from vitamin C in higher plants. Proc Natl Acad Sci 103: 5608–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debolt S, Melino V, Ford CM (2007) Ascorbate as a biosynthetic precursor in plants. Ann Bot 99: 3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst RA, Clarkson GJJ, Rothwell SD, Fry SC (2017) Novel insights into ascorbate retention and degradation during the washing and post-harvest storage of spinach and other salad leaves. Food Chem 233: 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N (2007) Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 282: 18879–18885 [DOI] [PubMed] [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, Watson RB, Dupree P, Lilley KS (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA (1992) Metabolic control analysis: a survey of its theoretical and experimental development. Biochem J 286: 313–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M, Amaya I, Valpuesta V, Botella MA (2019) Vitamin C content in fruits: biosynthesis and regulation. Front Plant Sci 9: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O (1989) A novel genetic system to detect protein–protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Fossati T, Solinas N, Porro D, Branduardi P (2011) l-ascorbic acid producing yeasts learn from plants how to recycle it. Metab Eng 13: 177–185 [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Tarlyn NM (2002) l-ascorbic acid is accumulated in source leaf phloem and transported to sink tissues in plants. Plant Physiol 130: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Badejo AA, Shibata H, Sawa Y, Maruta T, Shigeoka S, Page M, Smirnoff N, Ishikawa T (2011) Expression analysis of the VTC2 and VTC5 genes encoding GDP-l-galactose phosphorylase, an enzyme involved in ascorbate biosynthesis, in Arabidopsis thaliana. Biosci Biotechnol Biochem 75: 1783–1788 [DOI] [PubMed] [Google Scholar]
- Gatzek S, Wheeler GL, Smirnoff N (2002) Antisense suppression of l-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. Plant J 30: 541–553 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH (1995) Transforming yeast with DNA. Methods Mol Cell Biol 5: 255–269 [Google Scholar]
- Gilbert L, Alhagdow M, Nunes-Nesi A, Quemener B, Guillon F, Bouchet B, Faurobert M, Gouble B, Page D, Garcia V, et al. (2009) GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J 60: 499–508 [DOI] [PubMed] [Google Scholar]
- Green MA, Fry SC (2005) Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-l-threonate. Nature 433: 83–87 [DOI] [PubMed] [Google Scholar]
- Harapanhalli RS, Howell RW, Rao DV (1993) Testicular and plasma ascorbic acid levels in mice following dietary intake: a high-performance liquid chromatographic analysis. J Chromatogr B Biomed Sci Appl 614: 233–243 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts FA, Vaeck E, Kiddle G, Coppens E, Van De Cotte B, Adamantidis A, Ormenese S, Foyer CH, Zabeau M, Inzé D, et al. (2008) A temperature-sensitive mutation in the Arabidopsis thaliana phosphomannomutase gene disrupts protein glycosylation and triggers cell death. J Biol Chem 283: 5708–5718 [DOI] [PubMed] [Google Scholar]
- Hoops S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U (2006) COPASI—a COmplex PAthway SImulator. Bioinformatics 22: 3067–3074 [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Li B, Bartel P, Fields S (1993) Use of the two-hybrid system to identify the domain of p53 involved in oligomerization. Oncogene 8: 1693–1696. [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Laing W, Norling C, Brewster D, Wright M, Bulley S (2017) Ascorbate concentration in Arabidopsis thaliana and expression of ascorbate related genes using RNAseq in response to light and the diurnal cycle. 2017 bioRxiv 138008. 10.1101/138008 [DOI] [Google Scholar]
- Laing WA, Bulley S, Wright M, Cooney J, Jensen D, Barraclough D, MacRae E (2004) A highly specific l-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc Natl Acad Sci 101: 16976–16981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing William A, Frearson N, Bulley S, MacRae E (2004) Kiwifruit l-galactose dehydrogenase: molecular, biochemical and physiological aspects of the enzyme. Funct Plant Biol 31: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. (2015) An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27: 772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Wright MA, Cooney J, Bulley SM (2007) The missing step of the l-galactose pathway of ascorbate biosynthesis in plants, an l-galactose guanyltransferase, increases leaf ascorbate content. Proc Natl Acad Sci 104: 9534–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Fields S (1993) Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two‐hybrid system. FASEB J 7:957.– [DOI] [PubMed] [Google Scholar]
- Li J, Liang D, Li M, Ma F (2013) Light and abiotic stresses regulate the expression of GDP-l-galactose phosphorylase and levels of ascorbic acid in two kiwifruit genotypes via light-responsive and stress-inducible cis-elements in their promoters. Planta 238: 535–547 [DOI] [PubMed] [Google Scholar]
- Li S, Wang J, Yu Y, Wang F, Dong J, Huang R (2016) D27E mutation of VTC1 impairs the interaction with CSN5B and enhances ascorbic acid biosynthesis and seedling growth in Arabidopsis. Plant Mol Biol 92: 473–482 [DOI] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C (2018) Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36: 1160–1163 [DOI] [PubMed] [Google Scholar]
- Li X, Ye J, Munir S, Yang T, Chen W, Liu G, Zheng W, Zhang Y (2019) Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato. Int J Mol Sci 20: 1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Smirnoff N, Cobbett CS, Golz JF (2016) Ascorbate-deficient vtc2 mutants in arabidopsis do not exhibit decreased growth. Front Plant Sci 7: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG (2007) Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the smirnoff-wheeler pathway to ascorbic acid in plants. J Biol Chem 282:18879–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134: 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR (2001) Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci 98: 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45: D200–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Yonemitsu M, Yabuta Y, Tamoi M, Ishikawa T, Shigeoka S (2008) Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis. J Biol Chem 283: 28842–28851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellidou I, Chagne D, Laing WA, Keulemans J, Davey MW (2012) Allelic variation in paralogs of GDP-L-galactose phosphorylase is a major determinant of vitamin C concentrations in apple fruit. Plant Physiol 160: 1613–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda T, Yabuta Y, Rapolu M, Motoki T, Takeda T, Yoshimura K, Ishikawa T, Shigeoka S (2004) Feedback inhibition of spinach l-galactose dehydrogenase by l-ascorbate. Plant Cell Physiol 45: 1271–1279 [DOI] [PubMed] [Google Scholar]
- Müller-Moulé P (2008) An expression analysis of the ascorbate biosynthesis enzyme VTC2. Plant Mol Biol 68: 31–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nourbakhsh A, Collakova E, Gillaspy GE (2015) Characterization of the inositol monophosphatase gene family in Arabidopsis. Front Plant Sci 5: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard J, Persiau G, Davey MW, Bauw G, Van Montagu M (1997) Isolation of a cDNA coding for l-galactono-gamma-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. Purification, characterization, cDNA cloning, and expression in yeast. J Biol Chem 272: 30009–30016 [DOI] [PubMed] [Google Scholar]
- Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N (2012) The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: further evidence for redox control of anthocyanin synthesis. Plant Cell Environ 35: 388–404 [DOI] [PubMed] [Google Scholar]
- Pallanca JE, Smirnoff N (2000) The control of ascorbic acid synthesis and turnover in pea seedlings. J Exp Bot 51: 669–674 [PubMed] [Google Scholar]
- Pineau B, Layoune O, Danon A, De Paepe R (2008) l-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J Biol Chem 283: 32500–32505 [DOI] [PubMed] [Google Scholar]
- Plumb W, Townsend AJ, Rasool B, Alomrani S, Razak N, Karpinska B, Ruban AV, Foyer CH (2018) Ascorbate-mediated regulation of growth, photoprotection, and photoinhibition in Arabidopsis thaliana. J Exp Bot 69: 2823–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Liu Z, Fan M, Chen Y, Tian H, Wu D, Gao H, Ren C, Song S, Xie D (2017) GDP-D-mannose epimerase regulates male gametophyte development, plant growth and leaf senescence in Arabidopsis. Sci Rep 7: 10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47: 95–113 [PubMed] [Google Scholar]
- Rodríguez-Negrete E, Bejarano ER, Castillo AG (2014) Using the yeast two-hybrid system to identify protein–protein interactions. In: Jorrin-Novo JV, Komatsu S, Weckwerth W, Wienkoop S (eds), Plant Proteomics: Methods and Protocols, Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 241–258 [DOI] [PubMed] [Google Scholar]
- Sawake S, Tajima N, Mortimer JC, Lao J, Ishikawa T, Yu X, Yamanashi Y, Yoshimi Y, Kawai-Yamada M, Dupree P. et al. (2015) KONJAC1 and 2 are key factors for GDP-mannose generation and affect l-ascorbic acid and glucomannan biosynthesis in Arabidopsis. Plant Cell 27: 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmeyer J, Bock R, Meyer EH (2016) l-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol Biol 90: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N (2019) Engineering of metabolic pathways using synthetic enzyme complexes. Plant Physiol 175: 6–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N (2018) Ascorbic acid metabolism and functions: a comparison of plants and mammals. Free Radic Biol Med 122: 116–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Catanzariti AM, Deboer K, Petre B, Gardiner DM, Singh KB, Dodds PN, Taylor JM (2017) LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep 7: 44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A, Purohit PK, Freese NH, Pasha A, Esteban E, Waese J, Wu A, Chen M, Chin CY, Song R, et al. (2019) An ‘eFP‐Seq Browser’ for visualizing and exploring ‘RNA’ sequencing data. Plant J 100: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Fernie AR (2018) The role of dynamic enzyme assemblies and substrate channeling in metabolic regulation. Nat Commun 9: 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y, Ueno H, Ogawa T, Sawa Y, Miyagi A, Kawai-Yamada M, Ishikawa T, Maruta T (2020) Dehydroascorbate reductases and glutathione set a threshold for high-light-induced ascorbate accumulation. Plant Physiol 183:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabinejad J, Donahue JL, Gunesekera BN, Allen-Daniels MJ, Gillaspy GE (2009) VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol 150: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truffault V, Fry SC, Stevens RG, Edinburgh T, Wall C, Building DR, Gautier H, Edinburgh T, Wall C, Building DR (2017) Ascorbate degradation in tomato leads to accumulation of oxalate, threonate and oxalyl threonate. Plant J 89: 996–1008 [DOI] [PubMed] [Google Scholar]
- Urzica EI, Adler LN, Page MD, Linster CL, Arbing MA, Casero D, Pellegrini M, Merchant SS, Clarke SG (2012) Impact of oxidative stress on ascorbate biosynthesis in Chlamydomonas via regulation of the VTC2 gene encoding a GDP-L-galactose phosphorylase. J Biol Chem 287: 14234–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Gilbert L, Rihouey C, Driouich A, Rothan C, Baldet P, Lerouge P (2011) Silencing of the GDP-D-mannose 3,5-epimerase affects the structure and cross-linking of the pectic polysaccharide rhamnogalacturonan II and plant growth in tomato. J Biol Chem 286: 8014–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, Huang R (2013) Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25:625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H Sen, Yu C, Zhu ZJ, Yu XC (2011) Overexpression in tobacco of a tomato GMPase gene improves tolerance to both low and high temperature stress by enhancing antioxidation capacity. Plant Cell Rep 30: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J (2018) Reactive oxygen species in plant signaling. Annu Rev Plant Biol 69: 209–236 [DOI] [PubMed] [Google Scholar]
- Wheeler G, Ishikawa T, Pornsaksit V, Smirnoff N (2015) Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes. Elife 2015: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393: 365–369 [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M (2003) GDP-mannose 3′,5′-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278: 47483–47490 [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Mieda T, Rapolu M, Nakamura A, Motoki T, Maruta T, Yoshimura K, Ishikawa T, Shigeoka S (2007) Light regulation of ascorbate biosynthesis is dependent on the photosynthetic electron transport chain but independent of sugars in Arabidopsis. J Exp Bot 58: 2661–2671 [DOI] [PubMed] [Google Scholar]
- Ye Q, Worman HJ (1995) Protein-protein interactions between human nuclear lamins expressed in yeast. Exp Cell Res 219:292–298 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Nakane T, Kume S, Shiomi Y, Maruta T, Ishikawa T, Shigeoka S (2014) Transient expression analysis revealed the importance of VTC2 expression level in light/dark regulation of ascorbate biosynthesis in Arabidopsis 78: 60–66 [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu J, Zhang Y, Cai X, Gong P, Zhang J, Wang T, Li H, Ye Z (2011) Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep 30: 389–398 [DOI] [PubMed] [Google Scholar]
- Zhang GY, Liu RR, Zhang CQ, Tang KX, Sun MF, Yan GH, Liu QQ (2015) Manipulation of the rice L-galactose pathway: evaluation of the effects of transgene overexpression on ascorbate accumulation and abiotic stress tolerance. PLoS One 10: e0125870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Si X, Ji X, Fan R, Liu J, Chen K, Wang D, Gao C (2018) Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol 36: 894–898 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Beard KFM, Swart C, Bergmann S, Krahnert I, Nikoloski Z, Graf A, George Ratcliffe R, Sweetlove LJ. et al. (2017) Protein-protein interactions and metabolite channeling in the plant tricarboxylic acid cycle. Nat Commun 8 10.1038/ncomms15212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 8:272–279 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sampathkumar A, Kerber SML, Swart C, Hille C, Seerangan K, Graf A, Sweetlove L, Fernie AR (2020) A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat Commun 11: 4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tao QC, Wang ZN, Fan R, Li Y, Sun XF, Tang KX (2012) Engineering ascorbic acid biosynthetic pathway in Arabidopsis leaves by single and double gene transformation. Biol Plant 56: 451–457 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.