Abstract
A vast majority of cellular processes take root at the surface of biological membranes. By providing a two-dimensional platform with limited diffusion, membranes are, by nature, perfect devices to concentrate signaling and metabolic components. As such, membranes often act as “key processors” of cellular information. Biological membranes are highly dynamic and deformable and can be shaped into curved, tubular, or flat conformations, resulting in differentiated biophysical properties. At membrane contact sites, membranes from adjacent organelles come together into a unique 3D configuration, forming functionally distinct microdomains, which facilitate spatially regulated functions, such as organelle communication. Here, we describe the diversity of geometries of contact site-forming membranes in different eukaryotic organisms and explore the emerging notion that their shape, 3D architecture, and remodeling jointly define their cellular activity. The review also provides selected examples highlighting changes in membrane contact site architecture acting as rapid and local responses to cellular perturbations, and summarizes our current understanding of how those structural changes confer functional specificity to those cellular territories.
Membrane shape and dynamics facilitate specialized responses to cellular perturbations within the structural framework of the endoplasmic reticulum-membrane contact sites.
Introduction
Cellular membranes occupy a large fraction of the eukaryotic cellular space and enclose between 30 and 90% of the cell volume in separate intracellular compartments (Diekmann and Pereira-Leal, 2013). These membranes consist of lipids and proteins of different shapes, sizes, forms, and functions, assembled together in a meticulous arrangement leading to the formation of semi-permeable bilayers (Sonnino and Prinetti, 2010; Cheng and Smith, 2019). The molecular organization of membranes and their physical properties confer biological membranes the ability to set apart different subcellular compartments, and as such to define functional territories within the cell (Bigay and Antonny, 2012; Ernst et al., 2018). Because of the space they occupy, as well as their intrinsic physical and chemical properties, membranes tend to congregate a large repertoire of activities, ranging from signal perception, integration and transduction to lipid metabolism, cytoskeleton remodeling, and the formation of transport vesicles (Schmick and Bastiaens, 2014; Bozelli and Epand, 2020). Membranes operate by concentrating functional elements at strategic points within the cell, acting as information centralizers and multimodal processing cores. The activities at those cores often require bidirectional information processing across lipid bilayers (Hurtley, 2005), but also between juxtaposed organelle membranes at specialized subdomains known as membrane contact sites (MCSs; Scorrano et al., 2019).
Advances
MCS molecular and membrane architecture is diverse.
Membrane geometry is emerging as a critical parameter for MCS function and specificity.
Changes in MCS architecture, and underlying modification in molecular and cellular activities, are essential for cellular adaptation including sensing and responding to environmental stresses.
The functional diversity of cellular membranes comes along with the broad multiscale structuring and forms they take, which range from local nanometer-scale lipid–protein domain organizations to broad micrometer-scale organelles with configurations stretching from flat-, curved-, and tubular-shape structures (Jarsch et al., 2016). Membranes also present differences in terms of charge densities, lipid packing, and membrane thickness (Bigay and Antonny, 2012), and these simple physical and geometrical parameters are used by the cell to generate sharp spatial–temporal responses (Bozelli and Epand, 2020; Noack and Jaillais, 2020; For a detailed description of anionic lipid gradient, subcellular activities, and plant development, see Dubois and Jaillais in this focus issue). In this review, we bring into focus how membrane shape and dynamics facilitate specialized responses to cellular perturbations within the structural framework of the endoplasmic reticulum (ER)-MCSs. These are ubiquitous membranous structures in eukaryotic cells that represent seemly examples of how membrane geometrical attributes are intimately intertwined with specialized cellular functions.
Before leaping further into this review, we direct the reader to the unifying features and essential functions assigned to MCSs. MCSs can be defined as sites of close membrane apposition between different organelles (although examples of homotypic MCSs between membranes within the same organelle also exist; Scorrano et al., 2019). The term “contact” conveys the notion of physical interaction between membrane-bound organelles, whether bi- or monolayered [as for lipid droplets (LDs)], which is usually achieved by supramolecular protein assemblies that populate the intermembrane gap and generate tethering forces through protein–protein or protein–lipid interactions (For a detailed description of protein–lipid interactions in plants, see de Jong and Munnik in this focus issue). These protein assemblies confer MCSs with functional specificity and prevent membrane fusion by maintaining an intermembrane distance in the 10–80 nm range (although larger or even smaller MCS intermembrane distances have also been described; Ping et al., 2016; Nicolas et al., 2017). The vast majority of MCSs that were initially described characterized interactions between the ER and other membrane compartments [ER–Plasma membrane (PM); ER–Golgi; ER-LDs; ER-mitochondria, etc.], but over the years, it has become clear that most organelles establish MCSs, and these structures are dynamic in time and space. The dominant view in the field has long been that MCSs were solely dedicated to direct nonvesicular exchange of molecules, such as lipids or Ca2+ (Phillips and Voeltz, 2016; Wu et al., 2018). The latest research in the field, however, has proven this concept too restrictive, and MCS functions expand much wider than originally thought. These functions now include, in addition to the bidirectional transport of molecules, membrane remodeling activities, such as organelle biogenesis and dynamics, autophagy, organelle fission, and regulation of enzymatic activities (Scorrano et al., 2019). This multiplicity of functions comes along with a diversity in MCS membrane geometry, shape, and dynamics, which is the focus of this review.
Cellular function depends on membrane geometrical parameters
As stated earlier, membrane geometry and cellular processes are intimately linked. The former helps to define local membrane environments holding unique properties through curved configurations (whether positive or negative, sharp or blunt), overall shape, and 3D architecture (McMahon and Boucrot, 2015). By building up microenvironments, which concentrate macromolecular assemblies dedicated to specific functions and assist protein activity, membrane geometry directly impacts cellular activities (Aimon et al., 2014; Schmick and Bastiaens, 2014; Iversen et al., 2015; Bozelli et al., 2018; Haucke and Kozlov, 2018; Lou et al., 2019; Bozelli and Epand, 2020; Larsen et al., 2020). MCSs are excellent examples of how membrane geometry influences cellular function. Their 3D organization enables the juxtaposition of two membranes over a short distance, and this tethered apposition is central to their function, for instance (but not only) for lipid transfer (Lahiri et al., 2015). Local membrane shaping and curvature also modifies MCS interfacial properties (Vanni et al., 2014), which in turn strongly impact protein recruitment, membrane deformation and budding, and lipid distribution within the same membrane and even across MCSs (as illustrated below). In other words, MCS membrane geometry and 3D organization are fundamental factors clenching on both specificity and versatility of function. Below are some examples, mainly extracted from yeast and mammals, which illustrate our point.
Autophagy is an evolutionarily conserved catabolic process characterized by the formation of a double-membrane vesicle, the autophagosome, which engulfs cytoplasmic components and delivers them to the lysosome or vacuole for recycling (Enrique Gomez et al., 2018; Nakatogawa, 2020). This essential cellular process requires the establishment of MCSs and heavily relies on membrane geometrical parameters such as curvature induction. For instance in mammals, and within the frame of selective ER-phagy (also called reticulophagy), several receptors acting on selective ER-phagy display membrane-shaping functions (Grumati et al., 2017; Bhaskara et al., 2019; Chen et al., 2019). Among the best studied examples, the mammalian receptor for ER turnover, Family with sequence similarity 134 member B (FAM134B), oligomerizes at autophagy puncta and induces membrane curvature in the ER while physically bridging the ER and the phagophore membranes through its interaction with MAP1LC3B (Bhaskara et al., 2019). By combining membrane tethering and curvature, FAM134B oligomerization promotes the recruitment of the autophagy machinery at these transitory MCSs and helps pinching-off targeted ER membranes for subsequent degradation (Jiang et al., 2020). In other words, through their membrane remodeling function, membrane-shaping proteins set the stage for creating localized and temporally regulated organelle responses.
LD biogenesis provides another example of how membrane shaping and MCS activity are intrinsically linked. In mammals, the LD biogenesis process initiates at localized ER subdomains where neutral lipids accumulate between the ER leaflets and coalesce to form lens-like structures (nascent LDs; Walther et al., 2017). These first steps of LD initiation have been imputed to the high membrane curvature of tubular ER, which favors the condensation of triglycerides by lowering the energetic barrier for lipid de-mixing, thereby leading to neutral lipid accumulation in nascent LDs (Roberts et al., 2020; Santinho et al., 2020). Shortly after initiation, LDs grow and extend toward the cytosol through a continuous neutral lipid flux from the ER to the LD, while remaining connected to the ER. This process involves the oligomeric seipin complex, which accumulates at LD lens initiation sites, possibly owing to its ability to recognize phospholipid packing defects (i.e. when lipids are loosely packed) generated by local membrane deformation (Sui et al., 2018). Seipin acts in anchoring and stabilizing nascent LDs to the ER and generates a neck region of high membrane curvature between the two structures (Salo et al., 2016, 2019; Greer et al., 2020). This process might be linked to seipin’s propensity to bind and locally stabilize the cone-shaped lipid phosphatidic acid (Yan et al., 2018). Although this remains to be experimentally confirmed, these ER tubes have been proposed to facilitate flow of neutral lipids from the ER to the LD (Salo et al., 2019; Santinho et al., 2020). Despite being structurally unrelated, the biogenesis of peroxisomes from the ER in mammals similarly requires local ER remodeling by membrane-shaping proteins (Joshi et al., 2018; Wang et al., 2018), suggesting that organelle buddings from the ER share common structural and molecular bases. Remarkably, the spatial organization of mammalian LD tethers goes beyond the formation of bi-organellar contacts during LD biogenesis. Indeed, mature LDs display the structural capacity to form tri-organellar MCSs that facilitate bulk lipid flux by positioning lipid metabolism enzymes next to LD contacts (Bohnert, 2020). This is exemplified by the mitoguardin 2 tether in mammalian adipocytes that interacts with ER-localized vesicle-associated membrane protein-associated proteins (VAPs) and coordinates the lipid transfer activities of mitochondria, LDs, and the ER within a single MCS (Freyre et al., 2019)
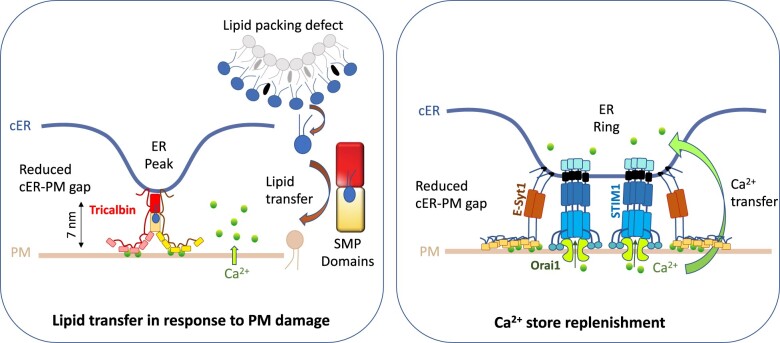
The structural mechanism by which membrane curvature could facilitate lipid exchanges in trans between two organelles at MCSs is illustrated by recent analyses of ER shaping functions mediated by the yeast tricalbins, which are homologous to the animal extended synaptotagmins (E-Syts) and plant synaptotagmin ER–PM MCS tethers. These studies showed that, in addition to tethering the cortical ER (cER) to the PM, tricalbins create peaks of high membrane curvature which extrude from the cER facing the PM, with the consequence of reducing the cER–PM gap from 22 to 7 nm (Collado et al., 2019; Hoffmann et al., 2019). These peaks would promote lipid export between membranes by first facilitating lipid extraction from the positively curved cER, and second, reducing the intermembrane gap, hence the transport distance (Figure 1). The importance of membrane curvature in nonvesicular lipid transfer was previously discussed and experimentally tested within the frame of the sterol/PI4P lipid exchange at mammalian ER–trans–Golgi MCSs (Von Filseck et al., 2015). In this example, the mammalian oxysterol-binding homology protein Osh4p, which mediates sterol/PI4P exchange between the ER and trans-Golgi network, shows faster transport efficiency in vitro when lipid-packing defects (which depend on membrane curvature and lipid composition) increase. The proposed mechanism underlying this observation is that sterol desorption from the ER is facilitated by lipid-packing defects that make the membrane more permissive for lipid extraction. By generating an asymmetrical geometry of the membranes positioned face to face at MCSs (for instance, curved membrane at the ER and flat at the PM or Golgi), membrane shape could theoretically regulate directional lipid transfer (For details of how this process is powered by lipid gradients between the closely apposed membranes, see Dubois and Jaillais in this focus issue). It is currently unknown whether ER shaping is a conserved function of plant tethers, but the evolutionary conservation of the proteins involved in the process, and the multiple phenotypes and activities associated with their loss of function strongly suggest that the actions of MCS tethers in plants go beyond mechanical anchoring.
Figure 1.
ER shaping accompanies lipid and Ca2+ transfer at ER–PM MCSs. Left: The yeast tethers tricalbins generate high curvature at the cER (cER peaks) through their transmembrane domains and reduce inter-organelle distance possibly by binding to the PM in a Ca2+-dependent manner (most likely not all C2s interact with the PM). High membrane curvature at cER peaks induces lipid packing defects and may facilitate lipid extraction by the tricalbin SMP lipid transfer domains, which would then transfer phospholipids to the PM from the ER. This response may occur during heat stress, when PM integrity has lost generating Ca2+ entry in the cell and subsequent tricalbin-mediated cER peak at ER–PM MCS to facilitate lipid transport and PM repair. Adapted from Hoffmann et al. (2019) and Collado et al. (2019). Right: In nonexcitable animal cells, depletion of the ER Ca2+ stores induces the formation of cER–PM MCS to activate SOCE and refill the ER. The drop in luminal ER Ca2+ causes STIM1 to oligomerize and to transfer to ER–PM MCS where it interacts with and traps the Ca2+ channel Orai1. Subsequently, Ca2+ influx via SOCE induces E-Syt1 to reshape the originally formed MCSs into ring structures. These rings have been proposed to facilitate Ca2+ store replenishment by reducing the ER–PM gap and stabilizing the structure. Adapted from Kang et al. (2019).
Highly specialized protein arrangements underlie ER–MCS architecture and function
The wide variety of cellular and physiological processes that take place at ER-MCSs demonstrates their importance in the normal development and physiology of eukaryotic organisms. The emerging picture in the field, whether in yeast, mammal, or plant, is that MCS functions are essentially determined by their molecular composition (Perez-Sancho et al., 2016; Scorrano et al., 2019). Hence, the importance to characterize the unique protein (and lipid) make up that shapes those cellular microdomains and confers them with functional specificity. In an attempt to develop a unified terminology for the MCS field, Scorrano et al. (2019) classified the putative MCS protein residents into four possible categories based on molecular functions either assigned or predicted for those MCS components. In this classification, MCS proteins could have: (1) structural roles carried out by tethers that hold the two organelles together or spacers that keep the two membranes at a defined distance; (2) sorting roles carried out by proteins that define the contact site proteome and lipidome through active recruitment or exclusion of proteins into the contact site microdomain; (3) roles in the bidirectional transfer of materials mediated by ion channels, lipid transfer proteins (LTPs), or metabolite transporters; and (4) signaling roles mediated by sensors that integrate environmental and developmental cues and modulate the extent of the MCS itself, as well as the relative positioning and activity of the proteins within (Scorrano et al., 2019).
Remarkably, the repertoire of structural functions and biochemical activities assigned to MCS components keeps growing in the literature, making their classification into a single category increasingly difficult. This difficulty is clearly exemplified by the multiplicity of functions associated with the activity of the E-Syts at mammalian ER–PM contact sites. Early genetic and cell biology analyses showed that E-Syts form heteromeric complexes that act as tethers regulating the establishment of ER–PM contacts, a process which depends on both cytosolic Ca2+ and the anionic lipid PI(4,5)P2 (Giordano et al., 2013; Idevall-Hagren et al., 2015). Subsequent cryoelectron tomography studies outlined E-Syts as spacers capable of maintaining the intermembrane cER–PM distance of about 20 nm (Fernández-Busnadiego et al., 2015; Collado et al., 2019). Besides their function as tethers/spacers, E-Syt1 and E-Syt2 also facilitate the nonvesicular transfer of phospholipids and diacylglycerol between the cER and the PM through their synaptotagmin-like mitochondrial lipid-binding protein (SMP) domains (Schauder et al., 2014; Saheki et al., 2016; Yu et al., 2016; Jeyasimman and Saheki, 2020). During this process, E-Syt1 complexes release the auto inhibitory conformation of their SMP domains, reducing the cER-PM intermembrane gap (Saheki et al., 2016; Bian et al., 2018), and the E-Syt2 SMP domain dimerizes to form a 9-nm-long hydrophobic cylinder that enables the nonvesicular transfer of glycerophospholipids (Schauder et al., 2014). These findings illustrate how the E-Syt activity in mammals results from a combination of molecular actions that secure lipid transfers at cER–PM MCSs, presumably as a specific inter-organelle response to elevated Ca2+ (Chang et al., 2013; Idevall‐Hagren et al., 2015; Bian et al., 2018; Kang et al., 2019). Adding to this complexity, the internal MCS architecture often creates asymmetric distribution of proteins along the membrane interface that generates territories with varied composition and specific function (Pérez-SancHo et al., 2016; Perico and Sparkes, 2018; Baillie et al., 2020; Prinz et al., 2020; Zaman et al., 2020). This is exemplified by the yeast ER–PM contact sites where membrane curvature restricts the localization of VAP tethers to relatively flat cER sheets and the localization of tricalbin tethers to the tubular cER and curved edges of the cER sheets (Collado et al., 2019; Hoffmann et al., 2019). The information gathered from yeast and mammals suggests that to fully understand how MCS components trigger specific and conditionally regulated ER activities in plants, we will need to first identify their repertoire of activities, but also define the structural parameters conferring functional specificity within the plant MCS territories.
Architectural changes of cellular membranes act as a dynamic response to the environment
Cellular membranes are deformable material that can be quickly and easily remodeled. This property represents a significant advantage when it comes to physiological adaptations at the cellular level. In fact, cells commonly modify the geometry of their membranes in response to physiological perturbations with the consequence of directly affecting membrane-based signaling and metabolic reactions. A prime example of membrane re-shaping is the profound remodeling of the cER polygonal network in response to the developmental cues or external perturbations. In plant cells, the remodeling of cER membranes is largely driven by cER–cytoskeleton interactions (Wang et al., 2014); ER-shaping and curvature stabilizing proteins, such as reticulons (Sparkes et al., 2010; Breeze et al., 2016; Brooks and Dixon, 2020) and lunaparks (Kriechbaumer et al., 2018; Sun et al., 2020); ER fusogens, such as the membrane-anchored root hair defective 3 family of GTPases (Chen et al., 2011; Zhang et al., 2013; Ueda et al., 2016); and molecular anchors, such as Synaptotagmin1 (SYT1) and Vesicle-Associated membrane Protein-associated 27 (VAP27) tethering complexes. The coordinated action of some of these components has been deemed essential to maintain the structural stability of the polygonal ER network (Siao et al., 2016), and to create remodeling nodes (ER–PM MCSs) where ER tubules connect and “wrap” around (Bayer et al., 2017; Lin et al., 2017).
Like any cellular membranes, the ER–PM MCS diversity in shapes, hence functions, directly relates to their plastic nature. As previously described, the emerging consensus is that the specialized functions assigned to a particular MCS are determined by a cooperative phenomenon involving changes in its 3D membrane architecture and molecular organization (Fernández-Busnadiego, 2016; Petit et al., 2019). This dynamic interplay involves, among others, cytoskeletal components, specific membrane lipids, and MCS protein residents, which altogether modulate the MCS biochemical activities at the membrane interface and ultimately determine its functional specificity. For instance, multiple studies in eukaryotes, including yeast, mammals and plants, have shown that ER–PM MCSs have the ability to regulate the MCS intermembrane gap and to change their overall architecture in response to cellular perturbations. Some of these studies relied on super-resolution microscopy and single-particle tracking techniques to analyze the nanoscale dynamics of MCS-resident proteins, as well as the subsequent changes in MCS membrane architecture driven by their activity. For example, in mammalian cells, single particle tracking was used to study changes in the patterns of diffusion and trapping of stromal interaction molecule 1 (STIM1) and Orai1 Ca2+ channels (named “Orai” after the Greek keepers of the gates of heaven) during stress-induced store-operated calcium entry (SOCE) at ER–PM MCSs (Wu et al., 2014). In yeast, cryoelectron tomography coupled with correlative light and electron microscopy was used to show that tricalbins generate peaks of extreme curvature on the cortical ER, presumably to sustain lipid transfer and maintain PM integrity during heat stress (Collado et al., 2019). Finally, in plants, time lapse imaging techniques and persistence mapping were used to analyze the changes in architecture and dynamics of ER–PM MCSs in response to ionic stress (Lee et al., 2019; For detailed information on membrane imaging techniques in plant systems see Liu et al., in this focus issue). The plethora of structural data obtained in these studies, combined with genetic and functional analyses of MCS components, clearly establish that MCSs serve as dynamic platforms hosting a variety of regulatory activities (Lewis and Lazarowitz, 2010; Mehrshahi et al., 2013; Levy et al., 2015; Pérez-Sancho et al., 2015; Ishikawa et al., 2018, 2020; Lee et al., 2019; Wang et al., 2019). In the next section, we will provide mechanistic information of the multiple factors that contribute to the establishment, stabilization, tightening, and remodeling of different ER–MCS structures in response to cellular and environmental perturbations.
Changes in MCS membrane shape and intermembrane distance allow rapid and local responses to cellular perturbations
By using examples from selected eukaryotic organisms, we now illustrate our current understanding of how the cortical cytoskeleton, lipids, and cellular and environmental signals influence MCS architecture and re-shaping activity and how this relates to MCS-specialized functions.
Regulation of MCS architecture by the cytoskeleton
The narrow width of the cytosolic sleeve, combined with the molecular crowding at ER–PM MCSs, can impose steric restrictions on the cortical cytoskeleton network. This phenomenon has been described structurally for the contractile ring assembly in the fission yeast Schizosaccharomyces pombe, where prominent ER–PM contacts restrict actomyosin kinetics and limit the PM accessibility for the initial allocation of ring precursors along the cell cortex (Zhang et al., 2016; Zhang, 2020). Conversely, cortical cytoskeleton components can create trapping mechanisms regulating MCS architecture and expansion. For example, in HeLa cells, the microtubule (MT) plus-end binding protein EB1 traps and delays the translocation of the ER-localized Ca2+ sensor STIM1 to ER–PM MCSs, effectively preventing Ca2+ overaccumulation at the ER lumen (Chang et al., 2018). The extension of ER–PM membrane contacts in mammals also depends on F-actin dynamics. Thus, following Ca2+depletion in ER and subsequent cytosolic Ca2+ elevation, the ER stress sensor PKR-related endoplasmic reticulum kinase (PERK) dimerizes and forms a complex with the actin-binding protein Filamin A. This complex modifies F-actin remodeling and polymerization dynamics with the consequence of favoring the expansion of ER–PM MCSs. Cells that do not express PERK accumulate F-actin at the cell edges, which obstructs ER–PM contact expansion (Van Vliet and Agostinis, 2016).
In plants, the ER–PM MCSs and the cortical MT network create mutual exclusion zones (Pérez-Sancho et al., 2015; Wang et al., 2016; McFarlane et al., 2017). This exclusion is likely determined by the narrow ER–PM MCS intermembrane gap, as well as by the molecular crowding at the ER–PM MCS interface. Remarkably, MT depolymerization (stress or pharmacologically induced) influences the lateral diffusion of SYT1 tethers within the ER–PM MCS but does not affect the morphology and/or expansion of pre-existing ER–PM MCSs (Pérez-SancHo et al., 2016; Lee et al., 2019). Furthermore, ER–PM MCSs expand in response to the PM lipid composition in a process that does not disrupt the cortical MTs, suggesting that the cortical MT network does not create physical constraints to ER–PM MCS expansion (Lee et al., 2019). Although these studies propose additional processes (e.g. local Ca2+ or lipid signaling) as the drivers of ER–PM MCS remodeling in response to stress, they cannot rule out that a functional MT network could still be required for the establishment and/or formation of de novo ER–PM MCSs. For example, these studies did not address whether MT disruption during cell division, a process that heavily relies on membrane trafficking and remodeling through MT-associated proteins, influences the generation of ER–PM MCS at the nascent cell plate.
In contrast, the spatial proximity and physical associations between ER–PM MCS components and the actin cytoskeleton in plants are essential for the establishment and function of ER–PM MCSs (Lewis and Lazarowitz, 2010; Stefano et al., 2018; Wang et al., 2019). These interactions are mediated by members of the plant-specific NETWORKED (NET) family that act as membrane cytoskeleton adaptors (Deeks et al., 2012) and localize actin filaments in close proximity to ER–PM MCSs (Wang et al., 2014). For instance, the Arabidopsis (Arabidopsis thaliana) actin-binding protein NET3C interacts with the lipid-binding protein VAP27-1 at punctate ER–PM junctions creating actin patch assemblies where autophagy intersects with endocytic processes (Stefano et al., 2018; Wang et al., 2019). In the next section, we describe how these and additional factors, such as local lipid composition and the activity of LTPs, establish the MCS architecture that facilitates the assembly of autophagy and endocytosis machineries in plants.
Regulation of MCS architecture by lipids and LTPs
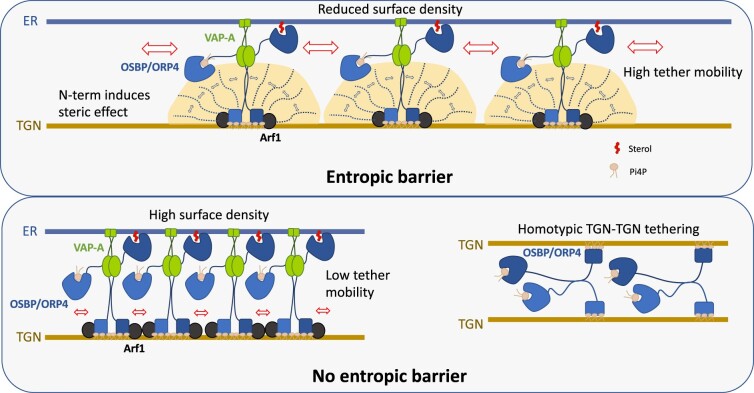
In mammals, lipid transfer activities are required at ER-autophagosome MCSs to sustain the omegasome growth into its typical highly curved structure (Axe et al., 2008; Biazik et al., 2015). This process necessitates the regulated partition of incoming lipids in time and space (Von Filseck et al., 2015; Nascimbeni et al., 2017; Hsieh and Yang, 2019). The regulatory role of MCSs in the process has been inferred by the suppression of autophagosome formation upon knockdown of tethering factors at ER–mitochondria MCSs (e.g. mitofusin 2; Hamasaki et al., 2013) or ER–PM MCSs (e.g. E-Syts1/2/3; Nascimbeni et al., 2017) which presumably prevents, among other activities, lipid intake (Schütter et al., 2020). Multiple studies in mammals also point toward MCS-localized LTPs, which contain two phenylalanines in an acidic tract motif and oxysterol-binding protein (OSBP)-related proteins (ORPs), as essential contributors for the correct positioning and transfer of different lipid species onto the autophagosome membrane (Hammond and Pacheco, 2019; Ye et al., 2020). In this context, the activity of intrinsically disordered regions within the LTPs’ 3D structure and their interactions with specific phosphoinositides underlie the MCS’ ability to change its geometry and regulate lipid transfer. For example, in HeLa cells, the unfolded N-terminus of the two related oxysterol-binding proteins, OSBP and ORP4, acts as entropic barrier whose thermal motion limits OSBP surface density at ER–trans-Golgi network MCSs, preventing their accumulation and immobilization by protein crowding. A second effect of the N-terminus is to prevent the two PH domains from simultaneously bridging over the two membranes through PI4P interaction and to promote heterotypic membrane tethering between VAP-containing and PI4P-containing membranes at the ER–trans-Golgi network interface (Jamecna et al., 2019; Figure 2).
Figure 2.
An entropic barrier controls membrane tethering geometry. Top: OSBP/ORP4 (blue dimers) transports cholesterol from the ER to the trans-Golgi network (TGN) and back transfer PI4P in exchange. They interact via their PH domain, with PI4P and the small G protein Arf1 (black) on the TGN membrane and with the ER-resident protein VAP-A (Green) via their FFAT motif. Both OSBP and ORP4 present a long N-terminal unstructured region (N-term), which through steric effect controls OSBP/ORP4 density at MCS by limiting the surface density of the downstream PH domain. This in turn facilitates protein diffusion within MCS and protein recycling between MCS. The N-term region also controls the geometry of ER–TGN MCSs and protein orientation by favoring heterotypic membrane tethering (TGN–ER) between VAP-containing and PI4P-containing membranes. Bottom: In the absence of the N-term region, protein density is higher and mobility reduced (left). Homotypic configuration (TGN–TGN), where two PI4P-rich TGN membranes are tethered by the two PH domains of OSBP/ORP4, is now favored. Adapted from Jamecna et al. (2019). Arf1: adenosine diphosphate-ribosylation factor; FFAT: two phenylalanines in an acidic tract; PH: pleckstrin homology; TGN: trans-Golgi network; VAP-A: vesicle-associated membrane protein-associated protein A.
In yeast and mammals, MCS-localized ORPs and additional components, such as the StARkin family of sterol-binding proteins, regulate the local accumulation of phospholipids and sterols in lipid membranes (Schulz et al., 2009; Gatta et al., 2015; Quon et al., 2018; Tong et al., 2018). These accumulations generate liquid-ordered membrane micro-domains exhibiting positive and negative membrane curvature (Bacia et al., 2005; Bigay and Antonny, 2012) that could serve as positional cues for the assembly of autophagy and/or endocytosis machineries (Hirama et al., 2017; Nguyen et al., 2017). Studies in yeast have shown that the interaction between ORPs and VAP27 homologs is a requirement for the generation of ER-derived autophagic or endocytic structures (Encinar del Dedo et al., 2017). Through these interactions, ORPs could provide the lipid environment that generates the shape and curvature of the nascent autophagosome/endocytic membrane, and VAPs could act as membrane anchors that attach those nascent structures to the contact site (Murphy and Levine, 2016). This model has a mechanistic precedent in plants where tomboviruses co-opt ORP and VAP proteins from ER–PM MCSs to generate sterol-rich membrane surfaces with high curvature, presumably to facilitate virus replication through local lipid channeling (Barajas et al., 2014). The ER–PM MCSs in plants also host an alternative autophagy-dependent pathway that is activated during nutrient starvation (Stefano et al., 2018; Wang et al., 2019) and salt stress (Fox et al., 2020). In these pathways, VAP27s associate with essential endocytic machineries, such as the AtEH1/Pan1 and AtEH2/Pan1 components of the TPLATE complex (Stefano et al., 2018; Wang et al., 2019), as well as water transporters, such as aquaporins (Fox et al., 2020), to target stress-induced autophagy and endocytosis to ER–PM MCSs (Stefano et al., 2018; Wang et al., 2019). The examples above clearly illustrate how MCS machineries act as multi-functional scaffolds generating compatible membrane geometries that can be used by conventional endocytic trafficking pathways (Stanhope and Derré, 2018; Wang and Hussey, 2019) and enable localized water transport in response to stress (Fox et al., 2020).
Regulation of MCS architecture by calcium and mechanical signals
At the core of many stress-induced changes in ER–MCS architecture, dynamics, and function is the sensing and transduction of stress-specific Ca2+ signatures achieved by an extensive array of MCS-localized proteins. In mammalian cells, the Ca2+-dependent changes in ER–PM MCS architecture are mediated by families of Ca2+ sensing proteins that are localized either in the ER lumen (e.g. STIMs, (Liou et al., 2005) or the cytosol (e.g. E-Syts, (Giordano et al., 2013) and TMEM24 (Lees et al., 2017)). A classical example of Ca2+/MCS interplay is the previously mentioned SOCE in mammalian cells. During stress episodes, Ca2+ depletion in the ER lumen induces changes in the ER–PM MCS architecture, and these changes subsequently modulate the activity of ER–PM MCS-localized Ca2+ transporters. At the molecular level, Ca2+ depletion induces a conformational change in STIM1 that exposes its polybasic C-terminus. This change in architecture promotes its oligomerization and translocation to ER–PM MCSs (Liou et al., 2007; Gudlur et al., 2019) where it recruits the PM Ca2+ channel Orai1 (Park et al., 2009; Gudlur et al., 2019). In parallel, the E-Syt1 tethers reshape the originally formed ER–PM MCSs into ring-shaped structures. This ring-shaped configuration is thought to facilitate STIM1/Orai1 Ca2+ gating and Ca2+ store replenishment through the stabilization of the ER–PM MCS structure and the reduction of the cER–PM intermembrane distance (Kang et al., 2019; Figure 1).
The mammalian E-Syts and the plant orthologs SYTs share a common structural organization and regulation. Both use homotypic and heterotypic tethering complexes with their N-terminal domains anchoring the ER and their C-terminal C2 domains establishing Ca2+-dependent interactions with PM phospholipids (Schapire et al., 2008; Yamazaki et al., 2010; Giordano et al., 2013; Xu et al., 2014; Pérez-Sancho et al., 2015). Despite the mechanistic similarities, clear differences in the temporal regulation of ER–PM MCS remodeling between these organisms exist. For example, following an stress-induced increase in [Ca2+]cyt, the mammalian E-Syts aggregate and concentrate at membrane junctions within minutes (Giordano et al., 2013; Idevall‐Hagren et al., 2015; Saheki et al., 2016), but similar changes in MCS architecture require hours in plants (Ishikawa et al., 2018; Lee et al., 2019; Lee et al., 2020). This difference suggests that additional processes, such as stress-induced changes in the lipid composition in the inner leaflet of the PM (which control E-Syts and SYTs localization), could be a limiting factor controlling the dynamics of the ER–PM remodeling in plants. In agreement with this model, recent studies in Arabidopsis have shown that the slow changes in ER–PM connectivity in response to different stresses are reversible and correspond with the kinetics of accumulation of specific PM phosphoinositides, such as PI(4,5)P2 in the cytoskeleton-independent responses to NaCl stress (Lee et al., 2019) or PI4P in the cytoskeleton-dependent responses to Ca2+ signaling surrogates in the cytosol (Lee et al., 2020; Figure 3). In addition, biochemical studies and structural analyses in vitro suggest that this process is mediated by Ca2+-dependent electrostatic interactions between negatively charged phosphoinositides at the PM and Lysine/Arginine-rich polybasic clusters conferring electropositive potential to C2-containing SYT tethers (Schapire et al., 2008; Pérez-Sancho et al., 2015; Lee et al., 2019, 2020). Last, genetic and physiological studies using syt loss-of-function mutants and SYT fluorescent markers suggest that these connectivity changes could function as an adaptive response providing mechanical stability to the PM during sustained periods of stress (Pérez-Sancho et al., 2015; Lee et al., 2019). Changes in connectivity could also create a molecular platform for unconventional secretion or targeted exocytosis (Harrison and Ivanov, 2017; Bellucci et al., 2018) or regulate cell-to-cell communication in plants through plasmodesmata (Uchiyama et al., 2014; Levy et al., 2015; Ishikawa et al., 2020; Brault et al., 2019). Although the common outcome of different plant stress responses is clear (changes in ER–PM connectivity), the specific sequence of events underlying this remodeling process (changes in PM phospholipid composition, ER–PM MCS tether re-localization, changes in ER morphology, and cytoskeleton re-organization) has not been established and remains an open question in the plant MCS field. In structural terms, NaCl stress expands the cER localization of SYT1 tethering complexes, which are normally restricted to immobile ER tubules (Ishikawa et al., 2018), to the whole cER tubular network (Lee et al., 2019). A consequence of this process is the overall reduction of the cER–PM intermembrane distance along the cortical ER tubules (Lee et al., 2019). This process potentially facilitates the nonvesicular transfer of lipids via their SMP domains as described in mammalian E-Syts (Schauder et al., 2014; Reinisch and De Camilli, 2016; Saheki et al., 2016), the cis/trans activity of MCS-localized enzymes as discussed for the PI4P phosphatase Sac1p in yeast (Manford et al., 2010; Stefan et al., 2011; Zewe et al., 2018; Venditti et al., 2019), or the activation receptor-mediated signaling pathways through physical interactions with receptor-like kinases as described in the plant stomatal lineage (Ho et al., 2016). It is still unknown whether NaCl stress also creates physical forces (e.g. tensile stress) that could contribute to the observed changes in MCS architecture. Indeed, the influence of physical forces in MCS remodeling has been illustrated by changes in ER–PM MCS architecture in response to external pressure (Pérez-Sancho et al., 2015). Another example is the accumulation of ER–PM MCS components around rapidly expanding symbiosomes where membrane tension directs targeted lipid transfer and exocytosis toward the symbiotic interface (Gavrin et al., 2017; Harrison and Ivanov, 2017; Bellucci et al., 2018). Those studies clearly convey the importance of creating mechanically sensitive MCS assemblies to properly sense and integrate a wide variety of physical/mechanical cues; however, the specific architectural changes induced by physical forces and the putative role of MCS-localized and/or MCS-associated Ca2+ mechanosensors in the process are currently unknown and represent an exciting field of study in plant MCS research.
Figure 3.
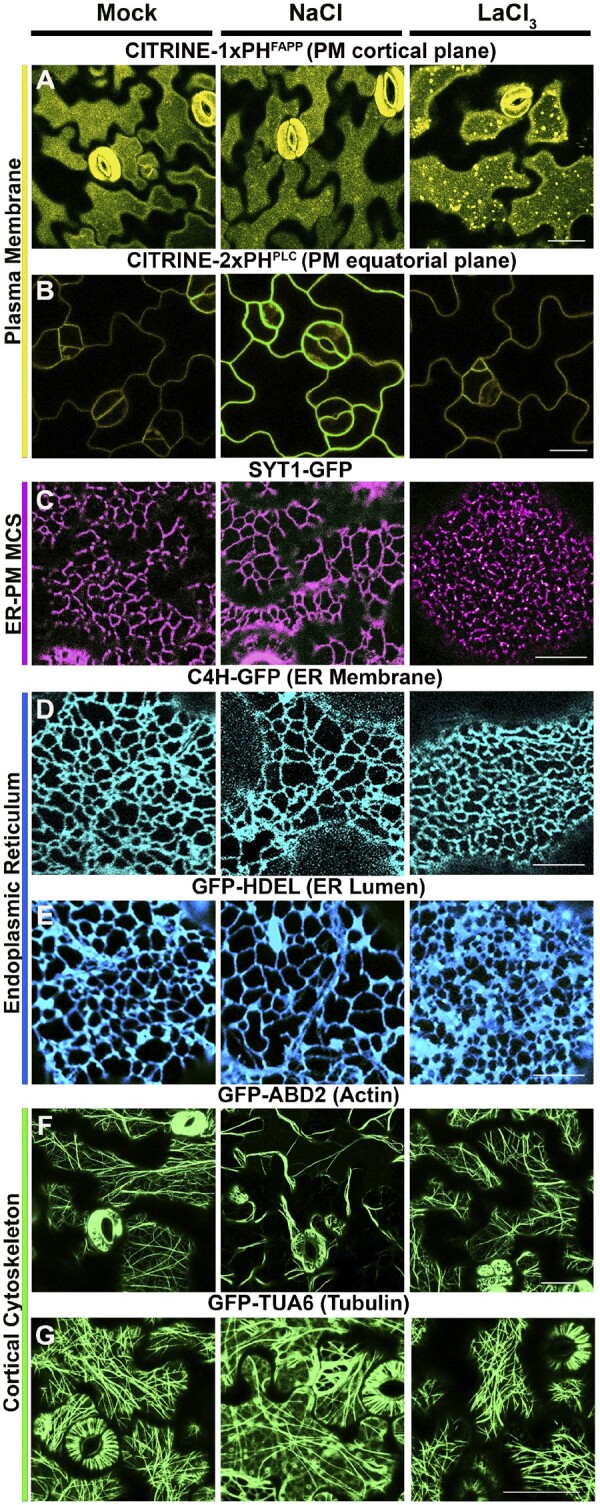
Architectural and compositional changes at the ER-PM MCS interface. Confocal microscopy images of Arabidopsis cotyledons expressing the indicated fluorescent markers in Mock (water or 5mM EGTA) treatments (left), 100 mM NaCl (center) and 500 µM LaCl3 (right). For all treatments, the CITRINE-1xPHFAPP marker indicates the accumulation of PI4P at the PM (Simon et al., 2016) (A). The CITRINE-2xPHPLC marker indicates the accumulation of PI(4,5)P2 at the PM (Simon et al., 2016) (B). The SYT1-GFP marker indicates the distribution of SYT1 tethers at ER–PM MCS (Perez-Sancho et al., 2015) (C). The GFP-HDEL marker indicates the morphology of the ER lumen (Batoko et al., 2000) (D). The GFP-ABD2 marker labels actin filaments (Sheahan et al., 2004) (E). The TUA6-GFP maker labels the microtubules network (Ueda et al., 1999) (F). Compared to Mock (left), the NaCl treatment (center) induces the homogeneous accumulation of PI(4,5)P2 at the PM, promotes the formation of ER–PM MCS along cortical ER tubules, increases the average area of the cortical ER polygons, and induces actin filament bundling and cortical microtubules depolymerization. Compared to Mock (left), the LaCl3 treatment (right) induces the heterogeneous accumulation of PI4P labeled vesicles at the PM, increases the number of ER–PM MCS puncta, reduces the number of tubular-shape ER–PM MCSs, reduces the average area of the cortical ER polygons, and does not cause visible morphological defects in the actin filaments or microtubule networks. Scale bars = 50 µm. Adapted from Lee et al., (2019, 2020). ABD2: actin binding domain 2; FAPP: four-phosphate-adaptor protein; GFP: green fluorescent protein; HDEL: ER retention motif; LaCl3: lanthanum chloride; NaCl: sodium chloride; PH: pleckstrin homology; PM: plasma membrane; PLC: phospholipase C; TUA6: tubulin alpha-6 chain.
Concluding remarks
Despite the evolutionary advantages of subcellular compartmentation, the successful metabolic activity of eukaryotic cells requires spatially controlled and integrated interactions between organelles in response to environmental and developmental cues. The eukaryotic ER–MCSs provide such highly specialized microenvironments for Ca2+ homeostasis regulation (ER–PM MCSs), inter-organelle lipid transfer (ER–PM, ER–mitochondria, ER–Golgi, ER-LDs), organelle biogenesis (ER–peroxisome, ER–LD MCSs), and endocytic and autophagy processes (ER–PM, ER–mitochondria MCSs). Discoveries spanning several decades have identified an assortment of factors ranging from cytoskeletal components to Ca2+ sensors, LTPs, and tethering factors as modulators that enable the fast and dynamic rearrangement of the organelle membrane interface in response to cellular perturbations. What transpires from these studies is that MCS structures are highly sophisticated, dynamic, and responsive to environmental and developmental cues, and that the MCS individual molecular and 3D architecture preludes functional specialization. We envision that a deeper understanding of the regulatory and functional activities coordinating the remodeling and 3D architecture of membrane interfaces at MCSs (See outstanding questions box) will create new paradigms about fundamental communication mechanisms that enabled organelle specialization and functional diversification in eukaryotic lineages.
Outstanding questions
What are the molecular mechanisms controlling MCS shape, intermembrane distance, and dynamics in plants?
Is lipid transfer activity at MCS directly related to membrane curvature?
What is the relationship between MCS dynamics and physical forces or membrane tension?
Do changes in MCS geometry and dynamics regulate biochemical activities in cis/trans between apposed organelles?
Can geometrical parameters and functions derived from yeast/mammalian MCS be extrapolated to characterize plant MCS?
Acknowledgments
We would like to thank Dr Eunkyoung Lee (Botany Department, University of British Columbia) for the experimental data provided in Figure 3.
Funding
Funding was provided by Agence Nationale de la Recherche (L' Agence Nationale de la Recherche) to Abel Rosado (ANR-18-CE13-0016 STAYING-TIGHT), European Research Council (772103-BRIDGING), and the Government of Canada Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada Discovery Grant RGPIN-2019-05568).
Conflict of interest statement: The authors declare no competing interests.
A.R. and E.M.B. conceived and wrote the manuscript, and generated the figures.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Emmanuelle M. Bayer (emmanuelle.bayer@u-bordeaux.fr).
References
- Aimon S, Callan-Jones A, Berthaud A, Pinot M, Toombes GES, Bassereau P (2014) Membrane shape modulates transmembrane protein distribution. Dev Cell 28: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K, Schwille P, Kurzchalia T (2005) Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc Natl Acad Sci USA 102: 3272–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie AL, Falz AL, Müller-Schüssele SJ, Sparkes I (2020) It started with a kiss: Monitoring organelle interactions and identifying membrane contact site components in plants. Front Plant Sci 11: 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D, Xu K, de Castro Martín IF, Sasvari Z, Brandizzi F, Risco C, Nagy PD (2014) Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog 10: e1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, , Zheng HQ, , Hawes C, , Moore I ( 2000) Arab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Sparkes I, Vanneste S, Rosado A (2017) From shaping organelles to signalling platforms: the emerging functions of plant ER–PM contact sites. Curr Opin Plant Biol 40: 89–96 [DOI] [PubMed] [Google Scholar]
- Bellucci M, De Marchis F, Pompa A (2018) The endoplasmic reticulum is a hub to sort proteins toward unconventional traffic pathways and endosymbiotic organelles. J Exp Bot 69: 7–20 [DOI] [PubMed] [Google Scholar]
- Bhaskara RM, Grumati P, Garcia-Pardo J, Kalayil S, Covarrubias-Pinto A, Chen W, Kudryashev M, Dikic I, Hummer G (2019) Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun 10: 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Saheki Y, De Camilli P (2018) Ca2+ releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J 37: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazik J, Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2015) Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 11: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Antonny B (2012) Curvature, lipid packing, and electrostatics of membrane organelles: Defining cellular territories in determining specificity. Dev Cell 23: 886–895 [DOI] [PubMed] [Google Scholar]
- Bohnert M (2020) Tethering fat: Tethers in lipid droplet contact sites. Contact 3: 1–1734113777 [Google Scholar]
- Bozelli JC, Epand RM (2020) Membrane shape and the regulation of biological processes. J Mol Biol 432: 5124–5136 [DOI] [PubMed] [Google Scholar]
- Bozelli JC, Jennings W, Black S, Hou YH, Lameire D, Chatha P, Kimura T, Berno B, Khondker A, Rheinstädter MC, et al. (2018) Membrane curvature allosterically regulates the phosphatidylinositol cycle, controlling its rate and acyl-chain composition of its lipid intermediates. J Biol Chem 293: 17780–17791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault ML, Petit JD, Immel F, Nicolas WJ, Glavier M, Brocard L, Gaston A, Fouché M, Hawkins TJ, Crowet J. et al. (2019) Multiple C2 domains and transmembrane region proteins (MCTP) tether membranes at plasmodesmata. EMBO Rep e47182: 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Dzimitrowicz N, Kriechbaumer V, Brooks R, Botchway SW, Brady JP, Hawes C, Dixon AM, Schnell JR, Fricker MD, et al. (2016) A C-terminal amphipathic helix is necessary for the in vivo tubule-shaping function of a plant reticulon. Proc Natl Acad Sci USA 113: 10902–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks RL, Dixon AM (2020) Revealing the mechanism of protein-lipid interactions for a putative membrane curvature sensor in plant endoplasmic reticulum. Biochim Biophys Acta Biomembr 1862: 183160. [DOI] [PubMed] [Google Scholar]
- Chang CL, Chen YJ, Quintanilla CG, Hsieh TS, Liou J (2018) EB1 binding restricts STIM1 translocation to ER-PM junctions and regulates store-operated Ca2+ entry. J Cell Biol 217: 2047–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, Liou J (2013) Feedback regulation of receptor-induced Ca2+ signaling mediated by e-syt1 and nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5: 813–825 [DOI] [PubMed] [Google Scholar]
- Chen J, Stefano G, Brandizzi F, Zheng H (2011) Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J Cell Sci 124: 2241–2252 [DOI] [PubMed] [Google Scholar]
- Chen Q, Xiao Y, Chai P, Zheng P, Teng J, Chen J (2019) ATL3 Is a tubular ER-Phagy receptor for GABARAP-mediated selective autophagy. Curr Biol 29: 846–855 [DOI] [PubMed] [Google Scholar]
- Cheng X, Smith JC (2019) Biological membrane organization and cellular signaling. Chem Rev 119: 5849–5880 [DOI] [PubMed] [Google Scholar]
- Collado J, Kalemanov M, Campelo F, Bourgoint C, Thomas F, Loewith R, Martínez-Sánchez A, Baumeister W, Stefan CJ, Fernández-Busnadiego R (2019) Tricalbin-mediated contact sites control ER curvature to maintain plasma membrane integrity. Dev Cell 51: 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Calcutt JR, Ingle EKS, Hawkins TJ, Chapman S, Richardson AC, Mentlak DA, Dixon MR, Cartwright F, Smertenko AP, et al. (2012) A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr Biol 22: 1595–1600 [DOI] [PubMed] [Google Scholar]
- Diekmann Y, Pereira-Leal JB (2013) Evolution of intracellular compartmentalization. Biochem J 449: 319–331 [DOI] [PubMed] [Google Scholar]
- Encinar del Dedo J, Idrissi FZ, Fernandez-Golbano IM, Garcia P, Rebollo E, Krzyzanowski MK, Grötsch H, Geli MI (2017) ORP-mediated ER contact with endocytic sites facilitates actin polymerization. Dev Cell 43: 588–602 [DOI] [PubMed] [Google Scholar]
- Enrique Gomez R, Joubès J, Valentin N, Batoko H, Satiat-Jeunemaître B, Bernard A (2018) Lipids in membrane dynamics during autophagy in plants. J Exp Bot 69: 1287–1299 [DOI] [PubMed] [Google Scholar]
- Ernst R, Ballweg S, Levental I (2018) Cellular mechanisms of physicochemical membrane homeostasis. Curr Opin Cell Biol 53: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Busnadiego R (2016) Supramolecular architecture of endoplasmic reticulum – Plasma membrane contact sites. Biochem Soc Trans 44: 534–540 [DOI] [PubMed] [Google Scholar]
- Fernández-Busnadiego R, Saheki Y, De Camilli P (2015) Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc Natl Acad Sci USA 112: E2004–E2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AR, Scochera F, Laloux T, Filik K, Degand H, Morsomme P, Alleva K, Chaumont F (2020) Plasma membrane aquaporins interact with the endoplasmic reticulum resident VAP27 proteins at ER–PM contact sites and endocytic structures. New Phytol 228: 973–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre CAC, Rauher PC, Ejsing CS, Klemm RW (2019) MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol Cell 76: 811–825 [DOI] [PubMed] [Google Scholar]
- Gatta AT, Wong LH, Sere YY, Calderón-Noreña DM, Cockcroft S, Menon AK, Levine TP (2015) A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 4: e07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrin A, Kulikova O, Bisseling T, Fedorova EE (2017) Interface symbiotic membrane formation in root nodules of Medicago truncatula: The role of Synaptotagmins MtSyt1, MtSyt2 and MtSyt3. Front Plant Sci 8: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P (2013) PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153: 1494–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer MS, Cai Y, Gidda SK, Esnay N, Kretzschmar FK, Seay D, Mcclinchie E, Mullen RT, Dyer JM, Chapman KD, et al. (2020) SEIPIN isoforms interact with the membrane-tethering protein VAP27-1 for lipid droplet formation. Plant Cell 32: 2932–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Hölper S, Mari M, Harwardt MLIE, Yan R, Müller S, Reggiori F, Heilemann M, Dikic I (2017) Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 6: e25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudlur A, Zeraik AE, Hirve N, Hogan PG (2019) STIM calcium sensing and conformational change. J Physiol 598: 1695–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495: 389–393 [DOI] [PubMed] [Google Scholar]
- Hammond GRV, Pacheco J (2019) Oxysterol binding protein: Tether, transporter… and flux capacitor? Trends Cell Biol 29: 531–533 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Ivanov S (2017) Exocytosis for endosymbiosis: membrane trafficking pathways for development of symbiotic membrane compartments. Curr Opin Plant Biol 38: 101–108 [DOI] [PubMed] [Google Scholar]
- Haucke V, Kozlov MM (2018) Membrane remodeling in clathrin-mediated endocytosis. J Cell Sci 131: jcs216812. [DOI] [PubMed] [Google Scholar]
- Hirama T, Lu SM, Kay JG, Maekawa M, Kozlov MM, Grinstein S, Fairn GD (2017) Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat Commun 8: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CMK, Paciorek T, Abrash E, Bergmann DC (2016) Modulators of stomatal lineage signal transduction alter membrane contact sites and reveal specialization among ERECTA kinases. Dev Cell 38: 345–357 [DOI] [PubMed] [Google Scholar]
- Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W (2019) Tricalbins contribute to cellular lipid flux and form curved ER-PM contacts that are bridged by rod-shaped structures. Dev Cell 51: 488–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CW, Yang WY (2019) Omegasome-proximal PtdIns(4,5)P 2 couples F-actin mediated mitoaggregate disassembly with autophagosome formation during mitophagy. Nat Commun 10: 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley SM (2005) Crossing the bilayer. Science 310: 1451 [Google Scholar]
- Idevall‐Hagren O, Lü A, Xie B, De Camilli P (2015) Triggered Ca2+ influx is required for extended synaptotagmin 1‐induced ER ‐plasma membrane tethering. EMBO J 34: 2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Tamura K, Fukao Y, Shimada T (2020) Structural and functional relationships between plasmodesmata and plant endoplasmic reticulum–plasma membrane contact sites consisting of three synaptotagmins. New Phytol 226: 798–808 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Tamura K, Ueda H, Ito Y, Nakano A, Hara-Nishimura I, Shimada T (2018) Synaptotagmin-associated endoplasmic reticulum-plasma membrane contact sites are localized to immobile ER tubules. Plant Physiol 178: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen L, Mathiasen S, Larsen JB, Stamou D (2015) Membrane curvature bends the laws of physics and chemistry. Nat Chem Biol 11: 822–825 [DOI] [PubMed] [Google Scholar]
- Jamecna D, Polidori J, Mesmin B, Dezi M, Levy D, Bigay J, Antonny B (2019) An intrinsically disordered region in OSBP acts as an entropic barrier to control protein dynamics and orientation at membrane contact sites. Dev Cell 49: 220–234 [DOI] [PubMed] [Google Scholar]
- Jarsch IK, Daste F, Gallop JL (2016) Membrane curvature in cell biology: An integration of molecular mechanisms. J Cell Biol 214: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyasimman D, Saheki Y (2020) SMP domain proteins in membrane lipid dynamics. Biochim Biophys Acta Mol Cell Biol Lipids 1865: 158447. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X, Ding X, Du M, Li B, Weng X, Zhang J, Li L, Tian R, Zhu Q, et al. (2020) FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER‐phagy. EMBO J 39: e102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Nebenfuehr B, Choudhary V, Satpute-Krishnan P, Levine TP, Golden A, Prinz WA (2018) Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat Commun 9: 2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F, Zhou M, Huang X, Fan J, Wei L, Boulanger J, Liu Z, Salamero J, Liu Y, Chen L (2019) E-syt1 re-arranges STIM1 clusters to stabilize ring-shaped ER-PM contact sites and accelerate Ca2+ store replenishment. Sci Rep 9: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Breeze E, Pain C, Tolmie F, Frigerio L, Hawes C (2018) Arabidopsis lunapark proteins are involved in ER cisternae formation. New Phytol 219: 990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Toulmay A, Prinz WA (2015) Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol 33: 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JB, Rosholm KR, Kennard C, Pedersen SL, Munch HK, Tkach V, Sakon JJ,, Bjørnholm T, Weninger KR, Bendix PM, et al. (2020) How membrane geometry regulates protein sorting independently of mean curvature. ACS Cent Sci 2020: 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Santana BVN, Samuels E, Benitez-Fuente F, Corsi E, Botella MA, Perez-Sancho J, Vanneste S, Friml J, Macho A, et al. (2020) Rare earth elements induce cytoskeleton-dependent and PI4P-associated rearrangement of SYT1/SYT5 endoplasmic reticulum–plasma membrane contact site complexes in Arabidopsis. J Exp Bot 71: 3986–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Vanneste S, Pérez-Sancho J, Benitez-Fuente F, Strelau M, Macho AP, Botella MA, Friml J, Rosado A (2019) Ionic stress enhances ER–PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc Natl Acad Sci USA 116: 1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Messa M, Sun EW, Wheeler H, Torta F, Wenk MR, De Camilli P, Reinisch KM (2017) Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science 355: eaah6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Zheng JY, Lazarowitz SG (2015) Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 25: 2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci USA 107: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, White RR, Sparkes I, Ashwin P (2017) Modeling endoplasmic reticulum network maintenance in a plant cell. Biophys J 113: 214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Fivaz M, Inoue T, Meyer T (2007) Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA 104: 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Won DH, Jones JT, Myers JW, Ferrell JE, Meyer T (2005) STIM is a Ca2+ sensor essential for Ca2+-store- depletion-triggered Ca2+ influx. Curr Biol 15: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou HY, Zhao W, Li X, Duan L, Powers A, Akamatsu M, Santoro F, McGuire AF, Cui Y, Drubin DG, et al. (2019) Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proc Natl Acad Sci USA 116: 23143–23151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD, Mao Y (2010) Crystal structure of the yeast Sac1: Implications for its phosphoinositide phosphatase function. EMBO J 29: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HE, Lee EK, Van Bezouwen LS, Ross B, Rosado A, Samuels AL (2017) Multiscale structural analysis of plant ER-PM contact sites. Plant Cell Physiol 58: 478–484 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E (2015) Membrane curvature at a glance. J Cell Sci 128: 1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrshahi P, Stefano G, Andaloro JM, Brandizzi F, Froehlich JE, DellaPenna D (2013) Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc Natl Acad Sci USA 110: 12126–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Levine TP (2016) VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta Mol Cell Biol Lipids 1861: 952–961 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H (2020) Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21: 439–458 [DOI] [PubMed] [Google Scholar]
- Nascimbeni AC, Giordano F, Dupont N, Grasso D, Vaccaro MI, Codogno P, Morel E (2017) ER – plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J 36: 2018–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N, Shteyn V, Melia TJ (2017) Sensing membrane curvature in macroautophagy. J Mol Biol 429: 457–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, Trépout S, Gaston A, Fouché M, Cordelières FP, Oparka K, Tilsner J, Brocard L, Bayer EM (2017) Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat Plants 3: 17082. [DOI] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2020) Functions of anionic lipids in plants. Annu Rev Plant Biol 71: 71–102 [DOI] [PubMed] [Google Scholar]
- Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS (2009) STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136: 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sancho J, Tilsner J, Samuels AL, Botella MA, Bayer EM, Rosado A (2016) Stitching organelles: Organization and function of specialized membrane contact sites in plants. Trends Cell Biol 26: 705–717 [DOI] [PubMed] [Google Scholar]
- Pérez-Sancho J, Vanneste S, Lee E, McFarlane HE, del Valle AE, Valpuesta V, Friml J, Botella MA, Rosado A (2015) The arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol 168: 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico C, Sparkes I (2018) Plant organelle dynamics: cytoskeletal control and membrane contact sites. New Phytol 220: 381–394 [DOI] [PubMed] [Google Scholar]
- Petit JD, Immel F, Lins L, Bayer EM (2019) Lipids or proteins: Who is leading the dance at membrane contact sites? Front Plant Sci 10: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK (2016) Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping HA, Kraft LM, Chen WT, Nilles AE, Lackner LL (2016) Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J Cell Biol 213: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Toulmay A, Balla T (2020) The functional universe of membrane contact sites. Nat Rev Mol Cell Biol 21: 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon E, Sere YY, Chauhan N, Johansen J,, Sullivan DP, Dittman JS, Rice WJ, Chan RB, Di Paolo G, Beh CT, et al. (2018) Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol 16(5): e2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, De Camilli P (2016) SMP-domain proteins at membrane contact sites: Structure and function. Biochim Biophys Acta Mol Cell Biol Lipids 1861: 924–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MA, Segura-Roman A, Olzmann JA (2020) Organelle biogenesis: ER shape influences lipid droplet nucleation. Curr Biol 30: R770–R773 [DOI] [PubMed] [Google Scholar]
- Saheki Y, Bian X, Schauder CM, Sawaki Y, Surma MA, Klose C, Pincet F, Reinisch KM, De Camilli P (2016) Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat Cell Biol 18: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magré J, Thiele C, Hölttä‐Vuori M, Jokitalo E, et al. (2016) Seipin regulates ER –lipid droplet contacts and cargo delivery. EMBO J 35: 2699–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Li S, Vihinen H, Hölttä-Vuori M, Szkalisity A, Horvath P, Belevich I, Peränen J, Thiele C, Somerharju P, et al. (2019) Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev Cell 50: 478–493 [DOI] [PubMed] [Google Scholar]
- Santinho A, Salo VT, Chorlay A, Li S, Zhou X, Omrane M, Ikonen E, Thiam AR (2020) Membrane curvature catalyzes lipid droplet assembly. Curr Biol 30: 2481–2494 [DOI] [PubMed] [Google Scholar]
- Schapire AL, Voigt B, Jasik J, Rosado A,, Lopez-Cobollo R, Menzel D, Salinas J, Mancuso S, Valpuesta V, Baluska F, et al. (2008) Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM (2014) Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 510: 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmick M, Bastiaens PIH (2014) The interdependence of membrane shape and cellular signal processing. Cell 156: 1132–1138 [DOI] [PubMed] [Google Scholar]
- Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA (2009) Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol 187: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütter M, Giavalisco P, Brodesser S, Graef M (2020) Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell 180: 135–149 [DOI] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnóczky G, Kornmann B, Lackner LL,, Levine TP, Pellegrini L, Reinisch K, et al. (2019) Coming together to define membrane contact sites. Nat Commun 10: 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, , Staiger CJ, , Rose RJ, , McCurdy DW ( 2004) A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siao W, Wang P, Voigt B, Hussey PJ, Baluska F (2016) Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum-plasma membrane contact sites. J Exp Bot 67: 6161–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Laetitia A, Platre MP, Marqués-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud M-C, Jaillais Y (2016) A PI4P-driven electrostatic field controls cell membrane identity and signaling in plants. Nat Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnino S, Prinetti A (2010) Lipids and membrane lateral organization. Front Physiol 1: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Tolley N, Aller I, Svozil J, Osterrieder A, Botchway S, Mueller C, Frigerio L, Hawes C (2010) Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope R, Derré I (2018) Making contact: VAP targeting by intracellular pathogens. Contact 1: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Wormsbaecher C, Gamble J, Zienkiewicz K, Brandizzi F (2018) Plant endocytosis requires the ER membrane-anchored proteins VAP27-1 and VAP27-3. Cell Rep 23: 2299–2307 [DOI] [PubMed] [Google Scholar]
- Sui X, Arlt H, Brock KP, Lai ZW, DiMaio F, Marks DS, Liao M, Farese RV, Walther TC (2018) Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. J Cell Biol 217: 4080–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Movahed N, Zheng H (2020) Lunapark is an E3 ligase that mediates degradation of ROOT HAIR DEFECTIVE3 to maintain a tubular ER network in Arabidopsis. Plant Cell 32: 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Manik MK, Im YJ (2018) Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites. Proc Natl Acad Sci USA 115: E856–E865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama A, Shimada-Beltran H, Levy A, Zheng JY, Javia PA, Lazarowitz SG (2014) The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front Plant Sci 5: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, , Matsuyama T, , Hashimoto T ( 1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Ueda H, Yokota E, Kuwata K, Kutsuna N, Mano S, Shimada T, Tamura K, Stefano G, Fukao Y, Brandizzi F, et al. (2016) Phosphorylation of the C terminus of RHD3 has a critical role in homotypic ER membrane fusion in Arabidopsis. Plant Physiol 170: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet AR, Agostinis P (2016) When under pressure, get closer: PERKing up membrane contact sites during ER stress. Biochem Soc Trans 44: 499–504 [DOI] [PubMed] [Google Scholar]
- Vanni S, Hirose H, Barelli H, Antonny B, Gautier R (2014) A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun 5: 1–10 [DOI] [PubMed] [Google Scholar]
- Venditti R, Masone MC, Rega LR, Tullio G Di, Santoro M, Polishchuk E, Serrano IC, Olkkonen VM, Harada A, Medina DL, et al. (2019) The activity of Sac1 across ER–TGN contact sites requires the four-phosphate-adaptor-protein-1. J Cell Biol 218: 783–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Filseck JM, Vanni S, Mesmin B, Antonny B, Drin G (2015) A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun 6: 1–12 [DOI] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV (2017) Lipid droplet biogenesis. Annu Rev Cell Dev Biol 33: 491–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C, Cummins I, Deeks MJ, Sparkes I, Hawes C, Hussey PJ (2014) The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr Biol 24: 1397–1405 [DOI] [PubMed] [Google Scholar]
- Wang P, Hussey PJ (2019) Plant ER-PM contact sites in endocytosis and autophagy: Does the local composition of membrane phospholipid play a role? Front Plant Sci 10: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Idrissi FZ, Hermansson M, Grippa A, Ejsing CS, Carvalho P (2018) Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat Commun 9: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K, Gong J, Guan Y, et al. (2019) Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun 10: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Richardson C, Hawkins TJ, Sparkes I, Hawes C, Hussey PJ (2016) Plant VAP27 proteins: Domain characterization, intracellular localization and role in plant development. New Phytol 210: 1311–1326 [DOI] [PubMed] [Google Scholar]
- Wu H, Carvalho P, Voeltz GK (2018) Here, there, and everywhere: The importance of ER membrane contact sites. Science 36: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Covington ED, Lewis RS (2014) Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol Biol Cell 25: 3672–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bacaj T, Zhou A, Tomchick DR, Südhof TC, Rizo J (2014) Structure and Ca2+-binding properties of the tandem C 2 Domains of E-Syt2. Structure 22: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Takata N, Uemura M, Kawamura Y (2010) Arabidopsis synaptotagmin SYT1, a type I signal-anchor protein, requires tandem C2 domains for delivery to the plasma membrane. J Biol Chem 285: 23165–23176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Qian H, Lukmantara I, Gao M, Du X, Yan N, Yang H (2018) Human SEIPIN binds anionic phospholipids. Dev Cell 47: 248–256 [DOI] [PubMed] [Google Scholar]
- Ye H, Ji C, Guo R, Jiang L (2020) Membrane contact sites and organelles interaction in plant autophagy. Front Plant Sci 11: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Liu Y, Gulbranson DR, Paine A, Rathore SS, Shen J (2016) Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc Natl Acad Sci USA 113: 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MF, Nenadic A, Radojičić A, Rosado A, Beh CT (2020) Sticking with it: ER-PM membrane contact sites as a coordinating nexus for regulating lipids and proteins at the cell cortex. Front Cell Dev Biol 8: 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewe JP, Wills RC, Sangappa S, Goulden BD, Hammond GRV (2018) SAC1 degrades its lipid substrate Ptdins4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. Elife 7: e35588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D (2020) Interplay between endoplasmic reticulum membrane contacts and actomyosin cytoskeleton. Cytoskeleton 77: 241–248 [DOI] [PubMed] [Google Scholar]
- Zhang D, Bidone TC, Vavylonis D (2016) ER-PM contacts define actomyosin kinetics for proper contractile ring assembly. Curr Biol 26: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, Hu J (2013) ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol 163: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]