Abstract
Knocking out SOBIR1 in Nicotiana benthamiana by CRISPR/Cas9, abolishes the functionality of the transgenic receptor-like protein Cf-4, recognizing the Avr4 effector of the fungus Cladosporium fulvum.
Dear Editor,
Plants are challenged by a plethora of agents causing biotic stress. To defend themselves, plants have developed a multi-layered immune system (Couto and Zipfel, 2016). The first layer is mediated by pattern recognition receptors (PRRs) that localize on the plasma membrane (PM) and perceive extracellular immunogenic patterns (ExIPs; van der Burgh and Joosten, 2019). So far, all known plant PRRs that carry an extracellular receptor domain consisting of leucine-rich repeats (LRRs) are either receptor-like kinases (RLKs) or receptor-like proteins (RLPs). They share the same overall structure; however, in contrast to RLKs, RLPs lack a cytoplasmic domain for downstream signaling (Couto and Zipfel, 2016). RLPs, such as the tomato (Solanum lycopersicum, Sl) PRR Cf-4 (Thomas et al., 1997) that mediates resistance against strains of the pathogenic extracellular fungus Cladosporium fulvum secreting the matching avirulence factor Avr4 (Joosten et al., 1994), constitutively interact with the RLK SUPPRESSOR OF BIR1-1 (SOBIR1; Gao et al., 2009; Liebrand et al., 2013). SOBIR1 is essential for Cf-4 accumulation and function (Liebrand et al., 2013), and upon recognition of Avr4 by Cf-4, the RLK BRI1-ASSOCIATED KINASE 1 (BAK1), which is a regulatory co-receptor involved in development and defense, is recruited by the activated Cf-4/SOBIR1 complex (Postma et al., 2016). BAK1 recruitment to the activated RLP/SOBIR1 complex appears to be a general process, as this was also shown for the RLP23/SOBIR1 complex (Albert et al., 2015). RLP23 from Arabidopsis (Arabidopsis thaliana, At) is involved in perception of the ExIP necrosis and ethylene-inducing peptide 1-like protein nlp20, which is produced by several bacterial, fungal, and oomycete species. It has been proposed that subsequent trans-phosphorylation events between the kinase domains of SOBIR1 and BAK1 eventually initiate downstream defense signaling (van der Burgh et al., 2019).
Although important advances have been made in deciphering RLK-mediated downstream immune signaling (Couto and Zipfel, 2016; van der Burgh and Joosten, 2019), little is known about how the RLP/SOBIR1/BAK1 complex functions at the level of complex formation and downstream signal initiation. What is known is that the tomato receptor-like cytoplasmic kinase (RLCK) AVR9/CF-9-INDUCED KINASE 1 (ACIK1) plays an essential role downstream of Cf-4 and the RLP Cf-9, which confers recognition of the secreted C. fulvum effector Avr9 (van Kan et al., 1991; Jones et al., 1994; Rowland et al., 2005). In Arabidopsis, the RLCK BOTRYTIS-INDUCED KINASE 1 (BIK1) is swiftly phosphorylated upon the perception of flg22, a peptide derived from bacterial flagellin, by the RLK FLAGELLIN-SENSITIVE 2 (FLS2; Gòmez-Gòmez and Boller, 2000). FLS2 also recruits BAK1 upon flg22 binding, and BIK1 phosphorylation is BAK1-dependent (Lu et al., 2010). It was concluded that, similar to ACIK1, BIK1 is a critical component, linking the PM-associated PRR complex to cytoplasmic immune signaling (Lu et al., 2010).
Here, we took advantage of the CRISPR/Cas9 system to knock out SOBIR1 and its close homolog SOBIR1-like in the model plant Nicotiana benthamiana (Nb), as well as in N. benthamiana stably expressing the Cf-4 transgene. Cf-4 is functional in N. benthamiana, and we demonstrate that Cf-4 function is completely abolished in N. benthamiana:Cf-4 sobir1(/sobir1-like) knock-out mutants. We anticipate that these mutant lines will be important materials for studying the fundamentals of plant immunity mediated by RLPs. SOBIR1 is a positive regulator of plant immunity, as overexpression of AtSOBIR1 in Arabidopsis, as well as in N. benthamiana, leads to constitutive activation of cell death and defense responses (Gao et al., 2009; Wu et al., 2017; van der Burgh et al., 2019). Surprisingly, no symptoms of constitutive immunity were observed when tomato SlSOBIR1 or NbSOBIR1 was overexpressed in N. benthamiana (Wu et al., 2017). Therefore, over the past years, the AtSOBIR1-induced constitutive immunity in N. benthamiana, visible as a hypersensitive response (HR) at the site of agro-infiltration (transient expression) of AtSOBIR1, is commonly employed to decipher the mechanism behind RLP/SOBIR1-mediated plant immunity. For this, endogenous NbSOBIR1(-like) genes are silenced in N. benthamiana:Cf-4 by virus-induced gene silencing (VIGS), and subsequent complementation studies are performed by transiently expressing AtSOBIR1 and various mutants of this RLK (Liebrand et al., 2013; van der Burgh, 2018). However, as VIGS only generates a gene knock-down, such complementation experiments require high amounts of repetition due to variation caused by the presence of varying background levels that remain of the endogenous NbSOBIR1(-like) protein. With the advent of the CRISPR/Cas9 gene-editing system, it is now possible to generate stable gene knock-outs in various plant species (Belhaj et al., 2015), and this development prompted us to knock out functional SOBIR1 in N. benthamiana and N. benthamiana:Cf-4, thereby generating a perfect system for complementation studies with mutants of SOBIR1.
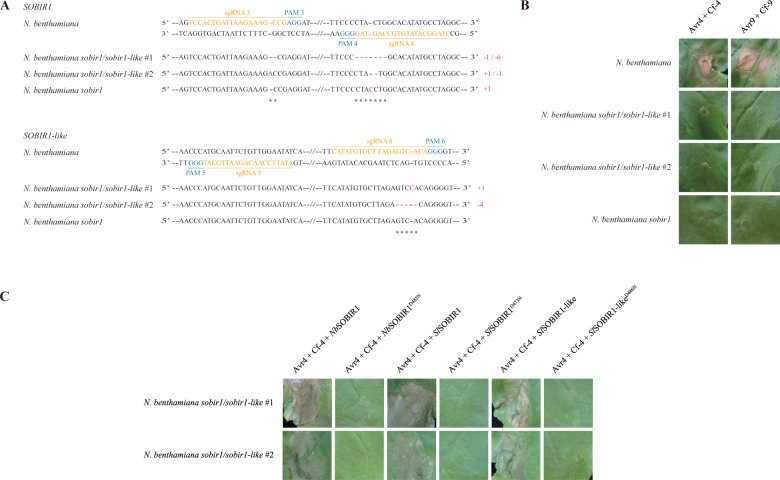
To knock out both SOBIR1 and SOBIR1-like in N. benthamiana, six single-guide RNAs (sgRNAs) targeting the open reading frames (ORFs) of both genes (Supplemental Figure S1 and Table S1) were designed using CRISPR-P 2.0 (Liu et al., 2017). Together with Cas9 and the selection marker BIALAPHOS RESISTANCE (BAR), sgRNA1, 2, 5, and 6 were assembled into the acceptor backbone (pAGM4723), referred to as Construct 1. In addition, sgRNA3, 4, 5, and 6 were cloned into the same acceptor backbone, referred to as Construct 2. The effectiveness and efficiency of the generated constructs were confirmed by studies based on their transient expression (Supplemental Figure S2), before stable transformation to explants of N. benthamiana. CRISPR/Cas9-induced mutations were detected by amplifying and sequencing the targeted gene regions, using isolated genomic DNA of the generated transformants as a template (Supplemental Table S3). Two homozygous sobir1/sobir1-like double knock-out lines (Supplemental Figure S3), and also one single sobir1 knock-out line, generated by Construct 2, were obtained. N. benthamiana sobir1/sobir1-like line #1 contains a 1 bp deletion in the sgRNA3-target region and a 6 bp deletion in the sgRNA4-target region in the ORF of SOBIR1, whereas a 1 bp insertion is present in the ORF of SOBIR1-like (Figure 1A). Similarly, there is a 1 bp insertion and a 1 bp deletion in SOBIR1, and a 4 bp deletion in SOBIR1-like in the N. benthamiana sobir1/sobir1-like line #2 (Figure 1A). The N. benthamiana single sobir1 mutant only contains a 1 bp insertion in SOBIR1 (Figure 1A).
Figure 1.
CRISPR/Cas9-induced targeted knockout of SOBIR1 in N. benthamiana abolishes the responsiveness to matching Avr/Cf combinations. A, Nucleotide sequence alignment of the regions in SOBIR1 (upper panel) and SOBIR1-like (lower panel), targeted by single-guide RNAs (sgRNAs) in the two N. benthamiana double sobir1/sobir1-like knock-out lines and the N. benthamiana single sobir1 knock-out line, with wild-type SOBIR1 and SOBIR1-like sequences, respectively. The sgRNA sequences are indicated in orange, and the protospacer-adjacent motifs (PAMs) are indicated in blue. The deleted nucleotides in the generated transformants are indicated with carmine dashes, and the inserted nucleotides are denoted with carmine letters. The type of mutations and the numbers of deleted/inserted nucleotides are shown on the right. B, Transient co-expression of Cf-4 with the matching C. fulvum effector Avr4, or of Cf-9 with its matching C. fulvum effector Avr9, by Agrobacterium-mediated transient expression, triggers a rapid HR in the leaves of wild-type N. benthamiana plants (upper two panels), whereas neither Avr4/Cf-4-, nor Avr9/Cf-9-induced cell death was observed in the two double sobir1/sobir1-like knock-out lines (middle four panels) and in the single sobir1 knock-out line (lower two panels). C, Complementation by transient expression of NbSOBIR1, SlSOBIR1, or SlSOBIR1-like, restores the Avr4/Cf-4-specific HR in the N. benthamiana sobir1/sobir1-like mutants, whereas this complementation does not take place upon transient expression of the corresponding kinase-dead mutants. NbSOBIR1, SlSOBIR1, or SlSOBIR1-like, as well as their corresponding kinase-dead mutants (negative controls), was transiently co-expressed with Avr4/Cf-4, in the leaves of the N. benthamiana sobir1/sobir1-like knock-out lines. Each construct was agro-infiltrated at an optical density at 600 nm (OD600) of 0.5, and all leaves were photographed at 5 d post infiltration (dpi). Experiments were repeated at least three times, and similar results were obtained. Representative pictures are shown.
Transient co-expression of Cf proteins with their matching Avr ligands in N. benthamiana triggers a typical HR (Figure 1B; van der Hoorn et al., 2000). Compared to the wild-type plant, none of the double knock-out lines was responsive to the Avr4/Cf-4 or Avr9/Cf-9 combination (Figure 1B), indicating that SOBIR1 and SOBIR1-like are indeed non-functional in the two mutant lines, due to disruption of their ORFs. Additionally, the N. benthamiana sobir1 single knock-out mutant was non-responsive to the Avr4/Cf-4 or Avr9/Cf-9 combination (Figure 1B), similar to the two double knock-out mutants. This observation is in accordance with our earlier finding that NbSOBIR1-like is not expressed or expressed only at a very low level, indicating that this gene is not functional (Liebrand et al., 2013). Interestingly, complementation of the SOBIR1(-like) knock-outs through transient expression of NbSOBIR1, SlSOBIR1, or SlSOBIR1-like, together with the Avr4/Cf-4 combination, restored the HR (Figure 1C). Complementation did not take place upon co-expression of the corresponding kinase-dead mutants of SOBIR1, as in this case, the leaf tissue remained non-responsive to the Avr4/Cf-4 combination (Figure 1C). These results reinforce the conclusion that SOBIR1/SOBIR1-like plays a pivotal role in RLP-mediated immunity and that the N. benthamiana sobir1/sobir1-like mutant plants form a robust basis for complementation studies.
We use N. benthamiana:Cf-4 for the elucidation of the molecular mechanisms of signal transduction events triggered by Cf-4 upon Avr4 recognition (Liebrand et al., 2013; Wu et al., 2017; van der Burgh et al., 2019). Therefore, SOBIR1 and SOBIR1-like were also knocked out in N. benthamiana:Cf-4 (Supplemental Table S3). Two homozygous double knock-out mutant lines were obtained with disruptions in both the ORF of SOBIR1 and SOBIR1-like, which were introduced by Construct 1 (Figures 2A and Supplemental Figure S3). In addition, one single sobir1 knock-out line, generated by Construct 2, was also obtained (Figure 2B). In agreement with our previous finding, transient expression of Avr4 triggered an HR in N. benthamiana:Cf-4 plants; however, the knock-out lines all were non-responsive to Avr4 (Figure 2C), again confirming that Cf-4 functionality requires functional SOBIR1(-like). Complementation with NbSOBIR1, SlSOBIR1, or SlSOBIR1-like in the double knock-out N. benthamiana:Cf-4 plants, in combination with transient expression of Avr4, again resulted in a Cf-4-mediated HR, whereas complementation did not take place upon co-expression of the corresponding kinase-dead mutants of SOBIR1 (Figure 2D).
Figure 2.
CRISPR/Cas9-induced targeted knockout of SOBIR1 in transgenic N. benthamiana:Cf-4 abolishes the functionality of the Cf-4 transgene. A, Nucleotide sequence alignment of the regions in SOBIR1 (upper panel) and SOBIR1-like (lower panel) targeted by sgRNAs in the two N. benthamiana:Cf-4 double sobir1/sobir1-like knock-out lines, with wild-type SOBIR1 and SOBIR1-like sequences, respectively. The sgRNA sequences are shown in orange, and the PAM sites are indicated in blue. The deleted nucleotides in the generated transformant are indicated with carmine dashes, and the inserted nucleotides are denoted with carmine letters. The type of mutations and the numbers of deleted/inserted nucleotides are shown on the right. B, Nucleotide sequence alignment of the regions in SOBIR1 (upper panel) and SOBIR1-like (lower panel) targeted by sgRNAs in the N. benthamiana:Cf-4 single sobir1 knock-out line, with wild-type SOBIR1 and SOBIR1-like sequences, respectively. The sgRNA sequences are shown in orange, and the PAM sites are indicated in blue. The deleted nucleotides in the generated transformant are indicated with carmine dashes, and the inserted nucleotides are denoted with carmine letters. The type of mutations and the numbers of deleted/inserted nucleotides are shown on the right. C, Agrobacterium-mediated expression of Avr4 in N. benthamiana:Cf-4 plants results in a rapid HR at the site of infiltration (upper panel), whereas agro-infiltration of Avr4 failed to induce cell death in the two N. benthamiana:Cf-4 double sobir1/sobir1-like knock-out lines (middle two panels) and in the N. benthamiana:Cf-4 single sobir1 knock-out line (lower panel). D, Complementation by transient expression of NbSOBIR1, SlSOBIR1, or SlSOBIR1-like, restores the Avr4/Cf-4-specific HR in the two N. benthamiana:Cf-4 sobir1/sobir1-like mutants, whereas this complementation does not take place upon transient expression of the corresponding kinase-dead mutants. NbSOBIR1, SlSOBIR1, or SlSOBIR1-like, as well as their corresponding kinase-dead mutants (negative controls), was transiently co-expressed with Avr4, in the leaves of the two N. benthamiana:Cf-4 sobir1/sobir1-like knock-out lines. Each construct was agroinfiltrated at an OD600 of 0.5, and the leaves were photographed at 5 dpi. E, Avr4 fails to induce a ROS burst in the two N. benthamiana:Cf-4 sobir1/sobir1-like knock-out lines and in the N. benthamiana:Cf-4 single sobir1 knock-out line. Leaf discs of N. benthamiana:Cf-4 (upper panel), the two N. benthamiana:Cf-4 double sobir1/sobir1-like knock-out lines (middle two panels), and the N. benthamiana:Cf-4 single sobir1 knock-out line (lower panel), were treated with 0.1 μM Avr4 or 0.1 μM flg22 (positive control), or with water (mock) (negative control). ROS production is expressed as relative light units (RLUs), and the data are represented as mean + SD. Experiments were repeated at least three times, and similar results were obtained. Representative pictures are shown. Note that in the N. benthamiana:Cf-4 sobir1/sobir1-like knock-out lines, as well as in the N. benthamiana:Cf-4 single sobir1 knock-out line, the response to flg22 manifests itself as a biphasic ROS burst, whereas in N. benthamiana:Cf-4 the flg22-triggered ROS burst is monophasic.
These mutant lines were further validated by monitoring the production of reactive oxygen species (ROS), which is a very early downstream response upon immune activation, upon treatment with either Avr4 or flg22. Note that FLS2 is also present in N. benthamiana and does not interact with SOBIR1 and does not require SOBIR1 for its functionality (Liebrand et al., 2013; Albert et al., 2015). Unlike the rapid and monophasic ROS burst induced by the flg22 peptide, a biphasic ROS accumulation was observed when leaf discs of N. benthamiana:Cf-4 were treated with Avr4 protein. In the latter case, the first transitory response was followed by a second, sustained ROS burst, which was of higher amplitude when compared to the initial ROS burst (Figure 2E). As expected, the biphasic Avr4-triggered ROS burst was completely abolished in all mutant lines (Figure 2E), which further verifies that these mutant lines have become non-responsive to Avr4. Intriguingly, an unexpected second sustained ROS burst, triggered by flg22, was observed in the two N. benthamiana:Cf-4 double sobir1/sobir1-like mutants, as well as in the single N. benthamiana:Cf-4 sobir1 mutant (Figure 2E). Our observation suggests that there is potential crosstalk taking place between RLP/SOBIR1- and FLS2-triggered pathways.
We propose that the appearance of a biphasic ROS burst triggered by flg22 in the SOBIR1(-like) knock-out mutants uncovers the presence of inhibitory activity of the RLP/SOBIR1 signal transduction pathway on the signaling route employed by FLS2. In addition to the work presented here, recently, a large number of immune-related genes, including SOBIR1 and SOBIR1-like, were targeted by CRISPR/Cas9 technology in tomato (Zhang et al., 2020). These resources will allow the study of the role of these genes in resistance of tomato to C. fulvum, by crossing these mutants to tomato carrying the Cf-4 resistance gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Methods
Supplemental Figure S1 Nucleotide sequence of NbSOBIR1 and NbSOBIR1-like.
Supplemental Figure S2 Determination of the effectiveness and efficiency of the generated CRISPR/Cas9 constructs.
Supplemental Figure S3 Phenotypes of wild-type N. benthamiana and the various mutant lines. The plants were extracted from different photos and placed on a black background.
Supplemental Table S1 Nucleotide sequences of the six single-guide RNAs.
Supplemental Table S2 Nucleotide sequences of the primers used in this study.
Supplemental Table S3 Number of transgenic N. benthamiana lines screened and the types of mutations that were obtained.
Supplementary Material
Acknowledgments
We thank Bert Essenstam and Henk Smid from Unifarm for excellent plant care.
Funding
This research was supported by the China Scholarship Council (W.R.H.H.) and the Peruvian Council for Science, Technology and Technological Innovation (CONCYTEC) and its executive unit FONDECYT (S.L.V.).
Conflict of interest statement. The authors declare that there is no conflict of interest.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Matthieu H. A. J. Joosten (matthieu.joosten@wur.nl)
W.R.H.H., C.S., and S.L.V. performed experiments. R.H. provided the CRISPR/Cas9 constructs. All authors designed the research and analyzed data. W.R.H.H., C.S., and M.H.A.J.J. wrote the letter, with contributions from all the authors.
References
- Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V (2015) Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 32: 76–84 [DOI] [PubMed] [Google Scholar]
- Couto D,, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gòmez-Gòmez L,, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Joosten MHAJ, Cozijnsen TJ, De Wit PJGM (1994) Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386 [DOI] [PubMed] [Google Scholar]
- Liebrand TWH, van den Berg GCM, Zhang Z, Smit P, Cordewener JHG, America AHP, Sklenar J, Jones AME, Tameling WIL, Robatzek S, et al. (2013) Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc Natl Acad Sci USA 110: 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10: 530–532 [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Liebrand TWH, Bi G, Evrard A, Bye RR, Mbengue M, Kuhn H, Joosten MHAJ, Robatzek S (2016) Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol 210: 627–642 [DOI] [PubMed] [Google Scholar]
- Rowland O, Ludwig AA, Merrick CJ, Baillieul F, Tracy FE, Durrant WE, Fritz-Laylin L, Nekrasov V, Sjölander K, Yoshioka H, et al. (2005) Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9–dependent disease resistance in tomato. Plant Cell 17: 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JDG (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9:2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgh AM (2018) SOBIR1-containing immune complexes at the plant cell surface: partners and signalling. Doctoral dissertation. Wageningen University and Research, The Netherlands
- van der Burgh AM,, Joosten MHAJ (2019) Plant immunity: thinking outside and inside the box. Trends Plant Sci 24: 587–601 [DOI] [PubMed] [Google Scholar]
- van der Burgh AM, Postma J, Robatzek S, Joosten MHAJ (2019) Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol Plant Pathol 20: 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RAL, Laurent F, Roth R, De Wit PJGM (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol Plant Microbe Interact 13: 439–446 [DOI] [PubMed] [Google Scholar]
- van Kan JAL, van den Ackerveken GFJM, de Wit PJGM (1991) Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol Plant Microbe Interact 4: 52–59 [DOI] [PubMed] [Google Scholar]
- Wu J, van der Burgh AM, Bi G, Zhang L, Alfano JR, Martin GB, Joosten MHAJ (2017) The bacterial effector AvrPto targets the regulatory coreceptor SOBIR1 and suppresses defense signaling mediated by the receptor-like protein Cf-4. Mol Plant Microbe Interact 31: 75–85 [DOI] [PubMed] [Google Scholar]
- Zhang N, Roberts HM, Van Eck J, Martin GB (2020) Generation and molecular characterization of CRISPR/Cas9-induced mutations in 63 immunity-associated genes in tomato reveals specificity and a range of gene modifications. Front Plant Sci 10.3389/fpls.2020.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.