Abstract
When germinating in the light, Arabidopsis (Arabidopsis thaliana) seedlings undergo photomorphogenic development, characterized by short hypocotyls, greening, and expanded cotyledons. Stressed chloroplasts emit retrograde signals to the nucleus that induce developmental responses and repress photomorphogenesis. The nuclear targets of these retrograde signals are not yet fully known. Here, we show that lincomycin-treated seedlings (which lack developed chloroplasts) show strong phenotypic similarities to seedlings treated with ethylene (ET) precursor 1-aminocyclopropane-1-carboxylic acid, as both signals inhibit cotyledon separation in the light. We show that the lincomycin-induced phenotype partly requires a functioning ET signaling pathway, but could not detect increased ET emissions in response to the lincomycin treatment. The two treatments show overlap in upregulated gene transcripts, downstream of transcription factors ETHYLENE INSENSITIVE3 and EIN3-LIKE1. The induction of the ET signaling pathway is triggered by an unknown retrograde signal acting independently of GENOMES UNCOUPLED1. Our data show how two apparently different stress responses converge to optimize photomorphogenesis.
Chloroplast retrograde signaling targets the ethylene-regulated gene network to repress photomorphogenesis in Arabidopsis (Arabidopsis thaliana).
Introduction
As photoautotrophs, plants depend on light for growth and survival. Chloroplasts absorb photons to fuel the photosynthesis reaction, which is the basis of all carbon sources on earth. Most chloroplast proteins are encoded in the nuclear genome, so tight communication between chloroplasts and nucleus is essential to build and maintain the photosynthesis apparatus and to accurately adjust to environmental stresses with potential damage to the photosystems. Retrograde signals from the plastid inform the nucleus about disruptions in its function or development (reviewed in Chan et al., 2016; Crawford et al., 2018).
Quickly after germination, light triggers a developmental program, which includes the opening and expansion of cotyledons, chloroplast maturation, reduced hypocotyl elongation, and promoted root growth called photomorphogenesis (Gommers and Monte, 2018). The initiation of photomorphogenesis depends on the activation of a set of photoreceptors, sensitive to red and far-red (phytochromes, phys), or blue light (cryptochromes, crys). These repress a set of transcription factors, including PHYTOCHROME INTERACTING FACTORS (PIFs) and ETHYLENE INSENSITIVE3 (EIN3), which are repressors of photomorphogenesis in darkness (Leivar et al., 2008; Shi et al., 2018).
When chloroplast development is interrupted, by chemicals such as lincomycin (inhibitor of chloroplast translation) or norflurazon (inhibitor of carotenoid synthesis), or by high light, this induces a retrograde signaling (RS) cascade and inhibits cotyledon separation, possibly to minimize the stress-exposed area and protect the apical meristem (Ruckle and Larkin, 2009; Martín et al., 2016). This RS pathway acts via the chloroplast localized, nuclear encoded, protein GENOMES UNCOUPLED1 (GUN1), a master integrator of RS. GUN1 is a plastid-localized member of the pentatricopeptide repeat family involved in chloroplast synthesis as well as stress signaling, especially early during the leaf development (Koussevitzky et al., 2007; Jia et al., 2019; Pesaresi and Kim, 2019). The lincomycin-induced, GUN1-mediated, RS pathway antagonizes phytochrome signaling and targets genes, which encode proteins that promote photomorphogenesis and are repressed by PIFs in darkness. One of such genes encodes the transcription factor GOLDEN2 LIKE1 (GLK1). GLK1 transcriptional repression by GUN1-mediated RS contributes to the closed cotyledon-phenotype in lincomycin-treated seedlings (Martín et al., 2016). An additional RS pathway with direct effect on light signaling is mediated by the plastid stress molecule methylerythritol cyclodiphosphate (MEcPP), which promotes phyB accumulation and suppresses auxin and ethylene (ET)-mediated hypocotyl elongation in red light (Jiang et al., 2020).
Another stress signal that affects photomorphogenesis is the accumulation of the gaseous phytohormone ET. ET biosynthesis is induced during environmental stresses such as pathogen attack or vegetation shade, it accumulates in the plant in situations where the gas-flow is limited (flooding, under a pressing soil layer, or in close canopies), and acts as a neighbor detection molecule (Dubois et al., 2018). Depending on the concentration, light availability, and species, ET can either suppress or induce plant growth (Pierik et al., 2006; Dubois et al., 2018). In young Arabidopsis seedlings, it represses several aspects of photomorphogenesis, such as hypocotyl growth arrest and cotyledon separation and expansion (Das et al., 2016; Shi et al., 2016, 2018).
ET is perceived by multiple ET receptors located at the golgi and ER membranes: ETHYLENE RESPONSE SENSOR 1 (ERS1), ERS2, ETHYLENE RESISTANCE 1 (ETR1), ETR2, and ETHYLENE INSENSITIVE 4 (EIN4; Lacey and Binder, 2014). In the absence of ET, these receptors are active and activate the kinase CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), an inhibitor of the ER-localized, NRAMP-like protein ETHYLENE INSENSITIVE2 (EIN2). In the nucleus, EIN3-BINDING F-BOX1 (EBF1) and EBF2 bind and degrade the transcription factors EIN3 and EIN3-LIKE1 (EIL1). In the presence of ET, the receptors and therefore CTR1 are inactive. This renders active EIN2 that inhibits the translation of EBF1 and EBF2 mRNA. As a consequence, EIN3 and EIL1 accumulate and target ET-regulated genes to optimize development to the stress situation that caused ET to accumulate (Binder, 2020).
Even though lincomycin-induced RS and ET cause similar phenotypes in light-grown seedlings, with MEcPP affecting ET-mediated hypocotyl elongation (Vogel et al., 2014; Jiang et al., 2020), and high light has been shown to induce a stress-pathway via ETHYLENE RESPONSE FACTORS (ERFs; Vogel et al., 2014), very little is known about a possible overlap between these stress signaling pathways. Here, we show that chloroplast RS promotes the expression of ET-responsive genes, which causes the delayed photomorphogenesis in light-grown plants. We present evidence that this RS-pathway acts independent of the well-studied GUN1-mediated signaling cascade. Our findings additionally suggest that various environmental stresses converge at a similar core-set of nuclear genes to change plant development and prevent further damage.
Results
ET affects photomorphogenesis in continuous light
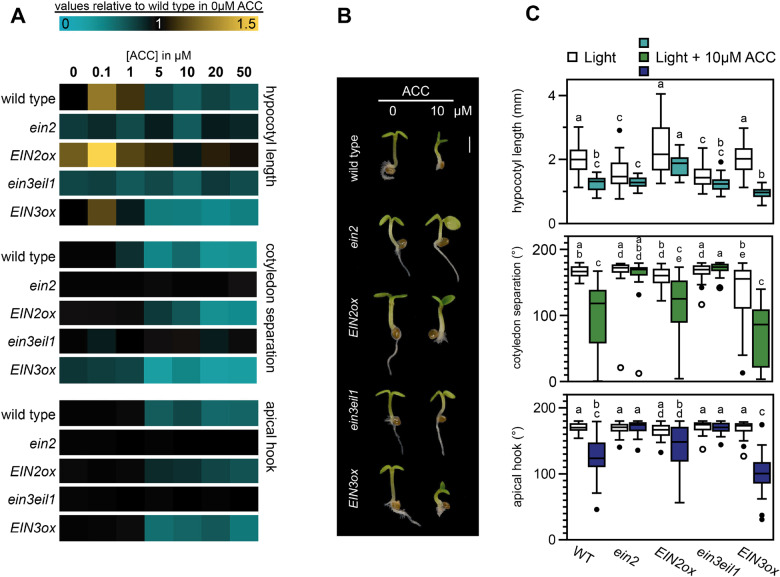
Previous work has shown that ET can affect seedling photomorphogenesis. Dark-grown EIN3/pifqein3eil1 seedlings treated with ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC) had small, partly unseparated cotyledons but short hypocotyls compared to seedlings grown on regular MS medium (Shi et al., 2018). Dark-grown ein3eil1 seedlings transferred to light had larger and more separated cotyledons, while EIN3-ox displayed unopened and smaller cotyledons. In continuous red light, ACC triggered hypocotyl elongation, whilst inhibiting cotyledon expansion in wild type (WT) and EIN3-ox, but not in ein3eil1 (Shi et al., 2016). Similar phenotypes were found when short day-grown WT seedlings were treated with ET gas (Das et al., 2016). To confirm the effect of ET on several aspects of photomorphogenic growth in continuous white light-grown Arabidopsis thaliana seedlings, we germinated seeds on media with different concentrations of ACC and analyzed hypocotyl length, cotyledon unfolding, and apical hook opening. Compared to the control, low levels of ACC caused a slight increase of hypocotyl length in WT seedlings. This effect was reversed by higher concentrations (> 5 µM), as visualized by the heatmap in Figure 1A, which represents relative values compared to WT without ACC (Supplemental Figure S1). At these higher concentrations, other aspects of photomorphogenesis were repressed, resulting in partly unseparated cotyledons and apical hook formation (Figure 1, A–C, Supplemental Figure S1). ACC additionally strongly inhibited primary root growth (Figure 1B). All these phenotypes depended on the activation of the ET pathway via EIN2 and EIN3/EIL1, shown by the ACC-insensitivity of ein2-5 and ein3eil1 mutants. For further experiments we focused on the cotyledon phenotype using 10 µM ACC, a concentration that strongly prevented cotyledon separation in continuous light without compromising on the overall growth.
Figure 1.
ACC inhibits photomorphogenesis in the light via ETHYLENE INSENSITIVE2 (EIN2) and ETHYLENE INSENSITIVE3 (EIN3). A, Relative hypocotyl length, cotyledon separation (presented as the angle between cotyledons), and apical hook angle of 3-d-old WT Columbia-0 (Col-0), ein2, 35S::EIN2-GFP/ein2 (EIN2ox), ein3eil1, and 35S::EIN3-GFP/ein3eil1 (EIN3ox) seedlings grown in continuous low light (approx. 1.5 µmol m−2 s−1) on medium supplemented with different concentrations of ACC as compared to normal growth medium (complete dataset in Supplemental Figure S1). B, Representative seedlings grown in control medium or supplemented with 10 µM ACC. Scale bar = 1 mm. Images of plants were digitally extracted for comparison. C, Boxplots representing the absolute data for control and 10 µM ACC-treated seedlings. Same data as in (A). Different letters indicate significant differences, p < 0.05, Kruskal–Wallis with post hoc Dunn test. In (A) and (C), biological replicates n = approx. 40 seedlings.
Suppression of photomorphogenesis by retrograde signals requires the ET signaling pathway
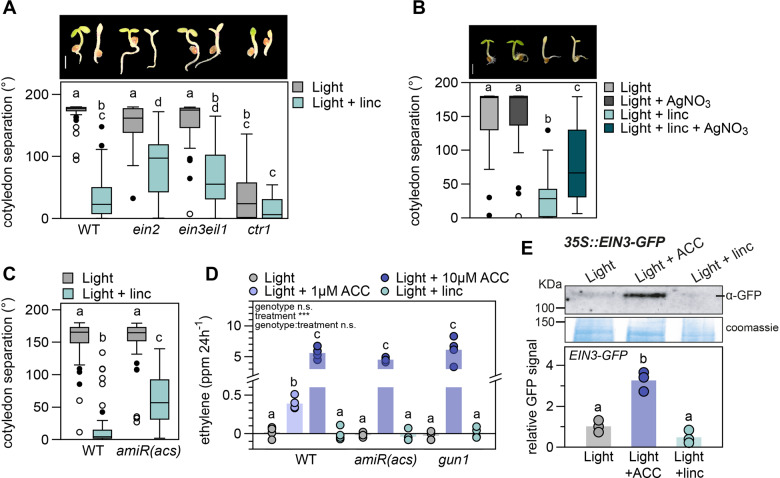
The closed cotyledon phenotype caused by ACC resembles the phenotype seen in light-grown Arabidopsis seedlings treated with chloroplast inhibitor lincomycin (Figure 2A; Martín et al., 2016), suggesting that both might be regulated by a similar pathway. To test this possibility, we treated ein2 and ein3eil1 mutant seedlings with lincomycin. As shown in Figure 2A, these ET mutants were partially insensitive to the lincomycin treatment and cotyledon separation was only partially inhibited compared to WT. The WT-like phenotype of the single ein3 and eil1 mutants as compared to the significantly distinct cotyledon phenotype of ein3eil1 in lincomycin, shows that EIN3 and EIL1 act redundant in the repression of cotyledon opening (Supplemental Figure S2A). Overexpression of EIN3 slightly, but significantly, enhanced the lincomycin-induced phenotype compared to the WT control. Contrastingly, the ctr1 mutant, with a constitutive ET response, displayed constitutively closed cotyledons and was insensitive to lincomycin (Figure 2A). Together, these results suggest that lincomycin-induced inhibition of cotyledon separation in light-grown seedlings requires ET signaling. These findings were further confirmed by chemical inhibition of ET perception by silver (AgNO3), which did not affect WT seedlings under normal light conditions but partially blocked the lincomycin-induced inhibition of cotyledon separation (Figure 2B). Although less robustly than lincomycin, norflurazon treatment also inhibited cotyledon separation significantly, an effect that was blocked by inhibition of ET perception using AgNO3 as well (Supplemental Figure S2B).
Figure 2.
Lincomycin-mediated repression of cotyledon separation acts via the ET signaling pathway without altering ET emission and ETHYLENE INSENSITIVE3 (EIN3) levels. A, Cotyledon separation (degrees) of 3-d-old, light-grown WT, ein2, ein3eil1, and ctr1 seedlings on regular growth medium or medium supplemented with 0.5 mM lincomycin (linc). Pictures are representative seedlings, scale bar = 1 mm. B, Cotyledon separation of 3-d-old, light-grown WT seedlings on medium supplemented with 5 µM AgNO3, 0.5 mM linc, or a combination of both. Pictures are representative seedlings, scale bar = 1 mm. C, Cotyledon separation of WT and ET-deficient amiR(acs) mutants, grown as in (A). For (A–C), biological replicates n = approx. 40 seedlings. D, ET emission (in ppm/24 h) of 120 WT, amiR(acs) and gun1 seedlings grown in light, on regular growth medium, or supplemented with 1 µM (WT only) or 10 µM ACC, or 0.5 mM linc. Dots represent individual measurements (n = 4), bars are averages. E, EIN3 protein accumulation in 3-d-old 35S::EIN3-GFP/ein3eil1 seedlings grown in low light on regular growth medium, or medium supplemented with 10 µM ACC or 0.5 mM linc, detected by anti-GFP antibodies. Data are relative to light control and Coomassie staining, dots represent biological replicates (n = 3), bars are averages. Complete immunoblot image presented in Supplemental Figure S3A. In (A–E), different letters mark significant differences, p < 0.05 (A and D: two-way ANOVA and post hoc Tukey, B and C: Kruskal–Wallis with post hoc Dunn, E: one-way ANOVA with post hoc Tukey). Images of plants in (A) and (B) were digitally extracted for comparison.
Our findings above suggested that the suppression of cotyledon separation in light induced by RS might be a consequence of ET accumulation. To test this end, we first examined the response of the ET-deficient mutant amiR(acs;Tsuchisaka et al., 2009). The conversion of S-adenosylmethionine to ACC is the rate-limiting step during ET production and is catalyzed by ACC-synthase (ACS), which acts as a dimer of various combinations of ACS isoforms (Tsuchisaka et al., 2009). ACC is further oxidized to C2H4 (ET) by ACC oxidase (ACO). Tsuchisaka et al. described that the amiR(acs) octuple mutant, deficient in nine ACS isoforms, is unable to form functional dimers and as a consequence produces extremely low levels of ET. We exposed this mutant to lincomycin and found that the synthesis of ET is essential for the lincomycin-induced repression of cotyledon separation in the light (Figure 2C). Next, we used gas chromatography followed by mass spectrometry (GC–MS) to measure ET emission by seedlings grown on low (1 µM) and higher (10 µM) levels of ACC and compared this to seedlings treated with lincomycin (Figure 2D). We included amiR(acs) and gun1 mutants as controls for seedlings that lack ACC-synthesis or retrograde signals, respectively. As expected, 10 µM ACC strongly induced ET emission in all three genotypes. WT seedlings treated with 1 µM ACC produced less, but still significant levels of ET compared to control without ACC. Nevertheless, lincomycin did not affect ET emissions in any of the genotypes.
Coherent with the lack of increased ET synthesis, lincomycin did not stabilize EIN3. We used the GFP-tagged, overexpressed EIN3 protein (p35S::EIN3-GFP/ein3eil1) to measure protein stability in our experimental set-ups by immuno-blotting. Figure 2E shows that EIN3-GFP is stabilized by 10 µM ACC but not by lincomycin. A smaller protein detected in lincomycin-treated seedlings corresponds to a nonspecific product as it was also present in the ein3eil1 mutant background samples (Supplemental Figure S3).
Finally, we tested if the inhibition of cotyledon separation caused by ET is regulated via GUN1-mediated chloroplast signals. We used gun1 knock-out and GLK1-overexpressing seedlings, both with severely reduce sensitivity to lincomycin (Martín et al., 2016). In contrast to lincomycin, ACC could inhibit cotyledon separation in gun1 and GLK1-ox, similar to WT (Supplemental Figure S4).
Together, these results indicate that: (1) lincomycin-induced RS and ET inhibit cotyledon separation in light-grown seedlings; (2) chloroplast disruption by lincomycin does not cause changes in ET emissions; (3) GUN1 and GLK1 are not required for the ACC-induced inhibition of cotyledon separation; and (4) ET signaling downstream of EIN3 is required for the lincomycin-mediated inhibition of cotyledon separation in light.
Lincomycin and ET co-target photomorphogenesis-repressing genes
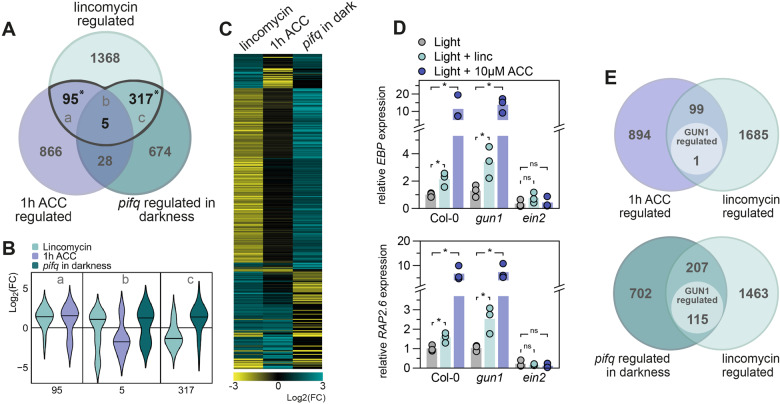
Because lincomycin and ET regulate similar phenotypes, but RS did not affect ET emissions, we hypothesized that both pathways might co-target a group of photomorphogenesis repressing genes downstream of EIN3. We compared previously published microarray data of low light-grown lincomycin-treated seedlings and ACC-treated seedlings and additionally included a dataset of pif quadruple (pifq; pif1pif3pif4pif5) seedlings grown in darkness (Goda et al., 2008; Leivar et al., 2009; Ruckle et al., 2012). Lincomycin-regulated genes showed strong overlap with either ACC-regulated, or PIF-regulated genes, but only five genes were differentially regulated in all three datasets (Figure 3A). The violin plot in Figure 3B represents the fold change of the genes in the three Venn-intersections in Figure 3A. It shows that lincomycin-repressed genes are PIF-repressed in darkness (positive Log2(FC) in pifq; 317-gene subset), like we previously concluded (Martín et al., 2016). Interestingly, and in contrast to the PIF overlap, it is predominantly the lincomycin-induced gene set that strongly overlaps with ACC-induced genes (95-gene subset). Within this 95-gene subset, 66 genes are co-upregulated by lincomycin and ACC, which is a substantial fraction of the complete lincomycin upregulated gene set. These results are visualized together in the heatmap in Figure 3C, which presents the same pattern showing that the lincomycin-repressed (negative Log2(FC) depicted in yellow)/PIF-repressed (depicted in turquoise) gene set is indeed not regulated by ACC, and that many of the lincomycin-induced genes (positive Log2(FC) depicted in turquoise) overlap with the ACC-upregulated set. To explore the role of the EIN3-signaling pathway in the lincomycin-induced RS, we performed additional data comparisons that show no significant enrichment of EIN3 targets among the lincomycin-regulated genes, but does show a significant co-regulation with ein3eil1 differentially expressed genes (Supplemental Figure S5A; Chang et al., 2013). The lincomycin/ACC co-induced genes do not significantly overlap with PIF-regulated genes (Figure 3C), as also shown in Figure 3A. Further comparison to light regulated genes (continuous red light versus continuous darkness (Leivar et al., 2009)) shows that, as expected from the lack of PIF-overlap, 94 of the 100 lincomycin/ACC co-regulated genes are not regulated by red light (Supplemental Figure S5B, Supplemental Table S1). We confirmed that the pifq mutant has a WT-like phenotype when grown in lincomycin that can be partly rescued by AgNO3 (Supplemental Figure S6), which supports that RS via the ET signaling cascade acts independently of PIF-mediated signaling.
Figure 3.
Lincomycin and ACC signaling pathways converge downstream of ETHYLENE INSENSITIVE3 (EIN3). A, Overlap of differentially expressed genes (p < 0.05, FC > |1|) in 6-d-old low-light grown 0.5 mM linc-treated seedlings, 7-d-old long-day grown 10 µM ACC-treated seedlings, and 3-d-old dark-grown pif quadruple (pifq) mutants (Goda et al., 2008; Leivar et al., 2009; Ruckle and Larkin, 2009). Letters (a, b, c) refer to B. Asterisks represent significant overlap (hypergeometric distribution, p < 0.05). B, Violin plots representing the Log2(fold change) of the genes differentially expressed by linc and at least one other condition (a, b, c denote the gene sets defined in A). C, Heatmap representing the Log2(fold change) of the genes in (B). D, Relative expression of EBP and RAP2.6 in WT Columbia-0 (Col-0), gun1 and ein2, grown in low light on regular growth medium, or medium supplemented with 10 µM ACC (left) or 0.5 mM linc (right), measured by RT-qPCR. Light control data are similar for left and right graphs. Dots represent biological replicates (n = 3), bars are averages. Significant differences are indicated by asterisks (Student’s t test, p < 0.05). E, Overlap of differentially expressed genes (p < 0.05, FC > |1|). Lincomycin, ACC and pifq datasets (as in A), are compared to 5-d-old light-grown and lincomycin-treated (200 μg/mL) gun1 mutants as compared to WT (A and B; GSE5770).
Visual analysis of the lincomycin/ACC upregulated gene set revealed several transcription factors members of the MYB, bZIP and NAC protein families (Table S1).
A GUN1-independent retrograde signal targets ET signaling components
To investigate whether these lincomycin and ET co-induced genes are possibly regulated by a GUN1-mediated retrograde signal, we analyzed the expression of two genes encoding ET response factors: ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN (EBP) and RELATED TO AP2 6 (RAP2.6). Both genes were significantly induced by 10 µM ACC and by 0.5 mM lincomycin (Figure 3D). The induction of EBP and RAP2.6 expression by both treatments was lost in the ein2 mutant, which confirms that the induction of ET response factors by lincomycin-induced retrograde signals requires the functional ET-response pathway. Interestingly, induced expression by lincomycin was maintained in the gun1 mutant, which hints towards a GUN1-independent retrograde pathway affecting ET-regulated seedling development.
To further explore this possible GUN1-independence, we extended our analysis and compared the genes co-targeted by lincomycin and ACC (Figure 3A) to a previously defined list of GUN1-dependent genes. This gene list was derived from light grown, lincomycin-treated seedlings of WT vs. gun1 mutants (Koussevitzky et al., 2007). Only 1 out of the 100 lincomycin and ACC co-regulated genes is regulated by GUN1-dependent retrograde signals according to the data of Koussevitzky et al. (Figure 3E). Contrastingly, out of the 322 pifq and lincomycin co-targeted genes, 115 (35.7%) are regulated via GUN1 (Figure 3E). These data strongly suggest that lincomycin induces ET-responsive genes independently of GUN1 (Figure 4). To explore if this GUN1-independent pathway is functionally involved in the closed-cotyledon phenotype, we treated gun1 mutants with AgNO3 to simultaneously repress the ET signaling pathway. Even though gun1 seedlings show almost completely separated cotyledons in the presence of lincomycin, the inhibition of ET perception significantly increased the angle between the cotyledons towards control-level (Supplemental Figure S6). This supports the presence of a GUN1-independent, ET-dependent, RS cascade, which inhibits photomorphogenesis in light-grown seedlings.
Figure 4.
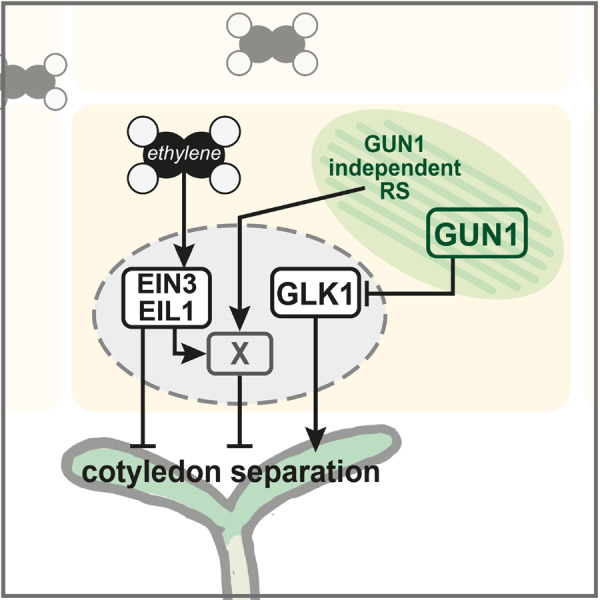
Graphic summary. ET represses cotyledon opening via ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (EIL1). Retrograde signals, induced by lincomycin, repress cotyledon separation via two pathways: a GENOMES UNCOUPLED1 (GUN1)-dependent repression of photomorphogenesis-inducing factors (e.g. GOLDEN2-LIKE1, GLK1), and a GUN1-independent induction of EIN3/EIL1 co-targets. “X” represents a repressor of cotyledon separation downstream of EIN3/EIL1 that is induced by ET and by chloroplast signals independently of GUN1. X is likely a gene (or set of genes) regulated redundantly by EIN3 and EIL1.
Discussion
Light strongly drives plant development and promotes photomorphogenesis during seedling establishment. Activated light receptors promote greening and expansion of cotyledons, opening of the apical hook, and inhibition of stem growth (Gommers and Monte, 2018). Nevertheless, this process is inhibited when the development of chloroplasts is blocked by drugs such as lincomycin (Ruckle and Larkin, 2009). The retrograde signals released by disrupted chloroplasts target the PIF-repressed phy-induced transcriptome including genes like GLK1, to inhibit photomorphogenesis in the light (Martín et al., 2016). Our findings here support a model whereby ET prevents excessive cotyledon separation under continuous light by regulating a set of ET-induced genes downstream of the transcription factor ETHYLENE INSENSITIVE3 (EIN3). In turn, when retrograde signaling (RS) is induced during early photomorphogenesis, inhibition of cotyledon separation by chloroplast signals takes place at least in part by further inducing this same ET signaling pathway. Both signals converge to co-regulate a set of RS/ET-induced genes. The chloroplast signal is yet undefined, but it does not involve GUN1 (Figure 4).
Interestingly, this RS/ET-targeted gene set does not overlap with the RS/PIF-repressed network previously defined in our laboratory (Figure 3; Martín et al., 2016). This finding suggests that RS optimizes seedling development in response to chloroplast disruption (caused for example by high light) by targeting at least two distinct gene networks that independently inhibit cotyledon separation: (1) a light-regulated network downstream of the phytochrome/PIF system, and (2) a light-independent ET hormone-regulated pathway downstream of EIN3. Existence of these two separate regulatory pathways is supported by the strong photomorphogenic phenotype of pifqein3eil1 in darkness compared to pifq and ein3eil1 (Shi et al., 2018). Remarkably, whereas RS antagonizes the light-regulated network by repressing light-induced genes (Martín et al., 2016), our results here suggest that RS further induces an ET-induced network. This dichotomy resulting in the repression or promotion of nuclear gene expression might explain why the chloroplast utilizes two distinct signaling pathways: a GUN1-dependent to repress the light-induced gene network and a GUN1-independent pathway to induce ET-regulated genes. Although retrograde signals other than GUN1 have been described in response to different stresses (Chan et al., 2016), GUN1 is considered as a hub factor of RS during chloroplast biogenesis (Mochizuki et al., 1996; León et al., 2013; Hernandez-Verdeja and Strand, 2018; Wu et al., 2018). Our findings here suggest the existence of at least one additional GUN1-independent pathway to optimally adjust seedling development to the light environment.
Cotyledon closing by EIN3 in darkness requires activation of the ET signaling pathway by ACC (Shi et al., 2018). We showed genetically and chemically that the inhibition of cotyledon separation by lincomycin also requires functioning ET perception and EIN2 and EIN3/EIL1 signaling, but does not enhance ET emission. We did not see a stabilization of the EIN3 protein by lincomycin, which is coherent with the lack of induced ET synthesis.
Despite the undetectable stabilization of EIN3-GFP, transcriptional targets of the EIN2/EIN3 signaling module were upregulated by lincomycin treatment and ET signaling mutants showed reduced lincomycin sensitivity. Different scenarios could explain our results: (1) lincomycin might minimally increase ET synthesis, undetectable in our set-up but sufficient to allow the activation of the signaling cascade downstream of EIN3, (2) lincomycin might stabilize EIN3 transiently during seedling establishment and we might have missed it at the selected time point, or (3) lincomycin might promote an increase in EIN3 activity without affecting EIN3 levels. We consider the first two scenarios most likely, since overexpression of EIN3 could significantly enhance the unseparated cotyledon phenotype compared to the WT control (Supplemental Figure S2A).
The origin of the GUN1-independent retrograde signal that targets the ET signaling pathway remains speculative. However, it is interesting to note that the ET pathway has been associated with different stress-induced chloroplast signals before. The chloroplast stress-metabolite MEcPP was recently shown to repress ET synthesis and hypocotyl elongation in red light in an auxin-dependent fashion (Jiang et al., 2020). Additionally, reactive oxygen species (ROS) released by mature (green) chloroplasts upon high light stress induce expression of ET response factors in the nucleus (Vogel et al., 2014). Lastly, drought and high light stress induce 3′-phosphoadenisone 5′-phosphate (PAP) accumulation in mature chloroplasts that act as RS and inhibit exoribonucleases (XRNs) such as XRN4, involved in the ET signaling pathway as it targets EBF1 and EBF2 mRNA for degradation (Estavillo et al., 2011). Future work will be necessary to address whether these signals are also relevant in young seedlings grown in continuous white light, such as the ones used in our study. We speculate that young seedlings with developing chloroplasts might release biogenic signals via GUN1 as well as ROS and/or PAP to trigger ET responses.
To conclude, our results here together with previous work demonstrate that lincomycin represses photomorphogenesis via at least two separate pathways. One requires GUN1-mediated signals and represses PIF-repressed target genes. The other requires GUN1-independent retrograde signals and induces the ET response pathway. Future work will explore if this GUN1-independent pathway involves any of the other known retrograde signaling mechanisms, or if it represents a novel and yet undescribed pathway.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana seeds used here were all described before, including ein2-5, ein3-1/eil1-1, p35S::EIN2-GFP/ein2-5, p35S::EIN3-GFP/ein3-1/eil1-1, ctr1-1, amiR(acs), gun1-201, p35S::GLK1/glk1glk2, and pifq, all in the Columbia-0 WT background (Kieber et al., 1993; Leivar et al., 2008; Waters et al., 2008; Tsuchisaka et al., 2009; He et al., 2011; Ju et al., 2012; Wen et al., 2012; Xie et al., 2015; Martín et al., 2016). For all experiments, seeds were surface-sterilized and sowed on half-strength Murashige and Skoog (0.5 MS) medium (0.8% w/v plant agar), followed by a 4-d stratification treatment before moving to continuous low white light (approx. 1.5 µmol m−2 s−1, T8 LED tube 4000K, Systion Electronics) at 20°C. Phenotypes, as well as gene, protein, and ET quantification were analyzed after 3 d.
Pharmacological treatments
1-Aminocyclopropane-1-carboxylic acid (ACC; VWR P10007) was dissolved in sterile water (10 mM stock) and added to the growth medium to obtain different final concentrations (0.1, 1, 5, 10, 20, 50 µM). Lincomycin (Sigma-Aldrich L6004) was added directly to the growth medium to obtain 0.5 mM as described before (Martín et al., 2016). Silver nitrate (AgNO3; Sigma-Aldrich 209139) was dissolved in sterile water (1 mM stock) and added to the growth medium in a final concentration of 5 µM. Norflurazon (Sigma-Aldrich 34364) was added directly to the growth medium to obtain a 5 µM solution.
Phenotype analysis
After 3 d of growth, seedlings were photographed (Nikon D7000 camera) and pictures were analyzed with Fiji (Schindelin et al., 2012). For phenotyping experiments, n = 36–40 biological replicates (seedlings) per treatment and genotype, divided over two petri dishes (18–20 seedlings of each genotype per petri dish).
ET measurements
To measure ET emissions, seeds were sown in sterilized glass vials (10 mL “Headspace Vials” with screw heads, Restek) which contained 7 mL of growth medium (with or without pharmacological treatment). Each vial contained 120 seeds. Seeds were cold-stratified and germinated as described above. Forty-eight hours after the transfer to light, the air headspace of the vials was flushed with clean air to set a starting point for ET accumulation. Twenty-four hours later, measurements were taken. Growth protocol was adapted from (Jeong et al., 2016). Three mL air was taken from the vial’s headspace and transferred to clean (flushed) Restek vials. ET concentrations were measured by GC–MS with an Agilent 7890A gas chromatograph coupled to a 5975C mass selective detector, as described before (Pereira et al., 2017).
Transcriptome re-analysis
Published and publicly available micro-array datasets (accession numbers: GSE24517, GSE17159, GSE39384, GSE5770, and GSE21762) were re-analyzed using the GEO2R online tool (NCBI). Differentially expressed genes were selected by 2-fold (Log2 fold > |1|) regulation and adjusted p< 0.05. For comparison with chromatin immunoprecipitation followed by sequencing (ChIP-seq) data (Supplemental Figure S5A), we have removed the EIN3 target genes that are not represented on the Affimetrix chip, which was used for the micro-array experiment.
Gene expression analysis
To analyze gene expression, 20 seedlings were harvested and pooled per sample, flash-frozen, homogenized and RNA was extracted using the Maxwell total RNA purification kit with DNAse treatment (Promega). cDNA was synthesized using the NZYtech first-strand cDNA synthesis kit with random primers and afterwards treated with RNAse. RT-qPCR was performed using a Roche Lightcycler 480, SYBR Green mix (Roche) and primers for EBP (AT3G16770; 5′-CCCACCAACCAAGTTAACGT-3′ and 5′-GTGGATCTCGAATCTCAGCC-3′), RAP2.6 (AT1G43160; 5′-TGATTACCGGTTCAGCTGTG-3′ and 5′-CTTGTGTGGGTCTCGAATCT-3′). Expression of PP2A was analyzed as a reference (AT1G13320; 5′-TATCGGATGACGATTCTTCGT-3′ and 5′-GCTTGGTCGACTATCGGAATG-3′). Relative expression was calculated as 2 ^(delta-delta CT).
Protein analysis
Protein extracts were prepared of 3-d-old seedlings, pooled per plate for each sample, flash-frozen, and homogenized by hand. Nuclear protein was extracted with an extraction buffer as described before (Soy et al., 2014; buffer: 100 mM MOPS (pH 7.6), 2% SDS, 10% glycerol, 4 mM EDTA, 50 mM Na2S2O5, 2 µgL−1 aprotinin, 3 µgL−1 leupeptine, 1 µgL−1 pepstatin, and 2 mM PMSF). Total protein of the samples was quantified using a Protein DC kit (Bio-Rad). β-Mercaptoethanol and loading dye was added to 125 µg of the samples, which were boiled at 95°C for 5 min before being loaded on a 7.5% SDS-PAGE gel. Proteins were then transferred to Immobilon-P membrane (Millipore) and EIN3-GFP was detected using an anti-GFP antibody (diluted 1:10,000; Invitrogen A11122). Anti-rabbit secondary antibody (Sigma NA934) and SuperSignal West Femto chemiluminescence kit (Pierce) were used for protein detection in a Bio-Rad imaging system. The ImageLab program (Bio-rad) was used to quantify band intensity and sizes from blot images, which was compared to a reference from the Coomassie-stained blot.
Statistical analysis
Multivariate comparisons were done in R and using the online MVApp (Julkowska et al., 2019). First, data were checked for equal variance by Levene’s test and when needed, data were LN-transformed to make variance equal. Multivariate analyses were done by one- or two-way ANOVA with a post hoc Tukey test for pairwise comparisons. For experiments with nonequal variances, the nonparametric Kruskal–Wallis test was applied, with a post hoc Dunn test or Mann–Whitney for pairwise comparison. Student’s t test pairwise comparisons were done in Microsoft Excel preceded by an F test to test for equal variances and LN-transformed when needed. Hypergeometric distribution tests for significant overlap in Venn-diagrams were done in R.
Accession numbers
Sequence data from this article can be found in the NCBI gene expression omnibus (GEO) data repository under accession numbers: GSE24517, GSE17159, GSE39384, GSE5770, and GSE21762. Arabidopsis gene identifiers for the major genes and proteins mentioned are: AT3G20770 (ETHYLENE INSENSITIVE3; EIN3), AT2G27050 (EIN3-LIKE1; EIL1), AT5G03280 (ETHYLENE INSENSITIVE2; EIN2), AT2G31400 (GENOMES UNCOUPLED1; GUN1), AT2G20180 (PHYTOCHROME INTERACTING FACTOR1; PIF1), AT1G09530 (PIF3), AT2G43010 (PIF4), AT3G59060 (PIF5), AT2G20570 (GOLDEN2-LIKE1; GLK1), AT5G03730 (CONSTITUTIVE TRIPLE RESPONSE1; CTR1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. ACC inhibits photomorphogenesis via ETHYLENE INSENSITIVE2 (EIN2) and ETHYLENE INSENSITIVE3 (EIN3).
Supplemental Figure S2. ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (EIL1) act redundantly to repress cotyledon separation during lincomycin-induced stress, and ET perception is required for norflurazon-mediated inhibition of cotyledon separation.
Supplemental Figure S3. Immunoblots for the EIN3-GFP protein.
Supplemental Figure S4. ACC inhibits photomorphogenesis independent of GENOMES UNCOUPLED1 (GUN1) and GOLDEN2-LIKE1 (GLK1).
Supplemental Figure S5. Transcriptional comparison of lincomycin-regulated genes with ETHYLENE INSENSITIVE3 (EIN3) targets, EIN3- and EIN3-LIKE1 (EIL1)-regulated genes, and red light regulated genes.
Supplemental Figure S6. AgNO3 inhibits lincomycin-induced inhibition of cotyledon separation independent of PIFs and GENOMES UNCOUPLED1 (GUN1).
Supplemental Table S1. List of genes co-regulated by lincomycin and 1h ACC or pifq in darkness, which are marked in Figure 3A and plotted in Figure 3B, with fold-change and adjusted p-value (Goda et al., 2008; Leivar et al., 2009; Ruckle and Larkin, 2009).
Supplementary Material
Acknowledgments
We thank Jason Argyris (CRAG, IRTA) for his help to prepare the ET measurements, Zeguang Liu, Sjon Hartman, and Ronald Pierik (Utrecht University) for sharing seeds of the ET signaling mutants, and lab members Arnau Rovira, Nil Veciana, Liu Duan, and Ana Couso for their help to harvest materials and support.
Funding
This work was supported by grants from FEDER/Ministerio de Ciencia, Innovación y Universidades – Agencia Estatal de Investigación (Project References BIO2015-68460-P and PGC2018-099987-B-I00), from the Spanish Ministry of Economy and Competitiveness (FJCI-2016-30876 to C.G.) and from the CERCA Programme/Generalitat de Catalunya (Project Reference 2017SGR-718) to E.M. We acknowledge the financial support from the Spanish Ministry of Economy and Competitiveness, through the “Severo Ochoa Programme for Centres of Excellence in R&D” 2016-2019 (SEV‐2015‐0533).
Conflict of interest statement. We declare no conflicts of interest.
C.M.M.G. and E.M. conceived the project. C.M.M.G., M.A.R-S., A.A., L.P., and M.P. planned and performed the experiments. C.M.M.G., M.A.R-S., and A.A. analyzed the data. C.M.M.G. and E.M. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is (https://academic.oup.com/plphys): E.M. (elena.monte@cragenomica.es).
References
- Binder BM (2020) Ethylene signaling in plants. J Biol Chem 295: 7710–7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Carol Huang SS,, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford T, Lehotai N, Strand Å (2018) The role of retrograde signals during plant stress responses. J Exp Bot 69: 2783–2795 [DOI] [PubMed] [Google Scholar]
- Das D, St Onge KR, Voesenek LACJ, Pierik R, Sasidharan R (2016) Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol 172: 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Van den Broeck L, Inzé D (2018) The pivotal role of ethylene in plant growth. Trends Plant Sci 23: 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Marin E, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M,, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gommers CMM, Monte E (2018) Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol 176: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies L-Kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdeja T, Strand A (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Kim K, Kim ME, Kim HG, Heo GS, Park OK, Park Y-i, Choi G, Oh E (2016) Phytochrome and ethylene signaling integration in Arabidopsis occurs via the transcriptional regulation of genes co-targeted by PIFs and EIN3. Front Plant Sci 7: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Tian H, Zhang S, Ding Z, Ma C (2019) GUN1-interacting proteins open the door for retrograde signaling. Trends Plant Sci 24: 884–887 [DOI] [PubMed] [Google Scholar]
- Jiang J, Xiao Y, Chen H, Hu W, Zeng L, Ke H, Ditengou FA, Devisetty U, Palme K, Maloof J, et al. (2020) Retrograde induction of phyB orchestrates ethylene-auxin hierarchy to regulate growth. Plant Physiol 183: 1268–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM,, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska MM, Saade S, Agarwal G, Gao G, Pailles Y, Morton M, Awlia M, Tester M (2019) MVApp—multivariate analysis application for streamlined data analysis and curation. Plant Physiol 180: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ,, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Lacey RF, Binder BM (2014) How plants sense ethylene gas - the ethylene receptors. J Inorganic Biochem 133: 58–62 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León P, Gregorio J, Cordoba E (2013) ABI4 and its role in chloroplast retrograde communication. Front Plant Sci 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Susek R, Chory J (1996) An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol 112: 1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Pujol M, Garcia-Mas J, Phillips MA (2017) Non-invasive quantification of ethylene in attached fruit headspace at 1 p.p.b. by gas chromatography–mass spectrometry. Plant J 91: 172–183 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Kim C (2019) Current understanding of GUN1: A key mediator involved in biogenic retrograde signaling. Plant Cell Rep 38: 819–823 [DOI] [PubMed] [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJW, Voesenek LACJ (2006) The Janus face of ethylene: Growth inhibition and stimulation. Trends Plant Sci 11: 176–183 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, Burgoon LD, Lawrence LA, Sinkler CA, Larkin RM (2012) Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol 159: 366–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, Larkin RM (2009) Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol 182: 367–379 [DOI] [PubMed] [Google Scholar]
- Schindelin J, , Arganda-Carreras I, , Frise E, , Kaynig V, , Longair M, , Pietzsch T, , Preibisch S, , Rueden C, , Saalfeld S, , Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nature Methods 9: 676. 10.1038/nmeth.2019 22743772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lyu M, Luo Y, Liu S, Li Y, He H, Wei N, Wang X (2018) Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc Natl Acad Sci 115: 2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shen X, Liu R, Xue C, Wei N, Deng XW, Zhong S (2016) The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev Cell 39: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, Monte E (2014) PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J Exp Bot 65: 2925–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A (2009) A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183: 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MO, Moore M, Konig K, Pecher P, Alsharafa K, Lee J, Dietz K-J (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26: 1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22: 1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G-Z, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R (2018) Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol 176: 2472–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie LJ, Chen QF, Chen MX, Yu LJ,, Huang L, Chen L, Wang FZ, Xia FN, Zhu TR, Wu JX, et al. (2015) Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in Arabidopsis. PLoS Genet 11: 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.