Abstract
Ligand-induced endocytosis of the immune receptor FLAGELLIN SENSING2 (FLS2) is critical for maintaining its proper abundance in the plasma membrane (PM) to initiate and subsequently down regulate cellular immune responses to bacterial flagellin or flg22-peptide. The molecular components governing PM abundance of FLS2, however, remain mostly unknown. Here, we identified Arabidopsis (Arabidopsis thaliana) DYNAMIN-RELATED PROTEIN1A (DRP1A), a member of a plant-specific family of large dynamin GTPases, as a critical contributor to ligand-induced endocytosis of FLS2 and its physiological roles in flg22-signaling and immunity against Pseudomonas syringae pv. tomato DC3000 bacteria in leaves. Notably, drp1a single mutants displayed similar flg22-defects as those previously reported for mutants in another dynamin-related protein, DRP2B, that was previously shown to colocalize with DRP1A. Our study also uncovered synergistic roles of DRP1A and DRP2B in plant growth and development as drp1a drp2b double mutants exhibited severely stunted roots and cotyledons, as well as defective cell shape, cytokinesis, and seedling lethality. Furthermore, drp1a drp2b double mutants hyperaccumulated FLS2 in the PM prior to flg22-treatment and exhibited a block in ligand-induced endocytosis of FLS2, indicating combinatorial roles for DRP1A and DRP1B in governing PM abundance of FLS2. However, the increased steady-state PM accumulation of FLS2 in drp1a drp2b double mutants did not result in increased flg22 responses. We propose that DRP1A and DRP2B are important for the regulation of PM-associated levels of FLS2 necessary to attain signaling competency to initiate distinct flg22 responses, potentially through modulating the lipid environment in defined PM domains.
A plant-specific large dynamin GTPase is required for plant responses against bacterial pathogens and, with another dynamin, regulates the cell surface composition for plant growth and defense.
Introduction
Plant proteins in the plasma membrane (PM) contribute to many cellular functions, including the ability to perceive environmental changes and evoke appropriate cellular responses. During infection by Pseudomonas syringae pv. tomato (Pto) DC3000, a pathogenic flagellated bacteria that colonizes and propagates within the extracellular space of leaves (Yu et al., 2017; Xin et al., 2018), plant PM proteins have many roles in host defense, from bacterial perception to initiation, amplification, and attenuation of immune responses (Boutrot and Zipfel, 2017; Gu et al., 2017). In addition, to help halt bacterial infection, plant cells rapidly remodel their cell surface composition through various cellular mechanisms, including through endocytosis (Ben Khaled et al., 2015; Ekanayake et al., 2019; Gu et al., 2017).
Endocytosis is a process by which PM proteins are internalized into small membrane-bound vesicles that bud from the PM into the cytoplasm (Gadeyne et al., 2014; Paez Valencia et al., 2016; Reynolds et al., 2018). The contents of endocytic vesicles are either recycled to the PM or delivered to the vacuole for degradation. Perturbation in endocytosis results in altered PM protein levels, which in turn can affect cellular responses (Claus et al., 2018; Reynolds et al., 2018; Ekanayake et al., 2019). Most plant studies have focused on constitutive endocytosis that occurs in the absence of any stimulus and serves as a general quality control of the PM composition (Gadeyne et al., 2014; Paez Valencia et al., 2016; Reynolds et al., 2018). Much less is known about the underlying molecular mechanisms and components that govern ligand-induced endocytosis, likely because relatively few plant PM cargo proteins are known to undergo stimulus-dependent endocytosis (Claus et al., 2018; Ekanayake et al., 2019). During ligand-induced endocytosis, binding of a ligand to its cognate PM receptor results in receptor internalization as a means to desensitize cells to the stimulus and attenuate stimulus-dependent signaling (Claus et al., 2018; Ekanayake et al., 2019).
Clathrin-mediated endocytosis (CME) has emerged as the predominant pathway of constitutive and ligand-induced endocytosis in plants (Reynolds et al., 2018; Ekanayake et al., 2019). After clathrin-coated vesicle (CCV) formation and maturation using clathrin and endocytic accessory proteins, CCVs are released from the PM through the action of large dynamin GTPases (Bednarek and Backues, 2010; Fujimoto and Tsutsumi, 2014). Arabidopsis (Arabidopsis thaliana) encodes 16 DYNAMIN RELATED PROTEIN (DRP)s that fall into six subfamilies (DRP1-6) based on their domain structure and function in fission of diverse membranes and/or organelles (Hong et al., 2003), with members of DRP1 and DRP2 subfamilies being implicated in CME (Bednarek and Backues, 2010; Fujimoto and Tsutsumi, 2014). DRP1s represent plant-specific dynamin GTPases that lack the characteristic pleckstrin-homology (PH)-domain (for membrane association) and the proline/arginine-rich domain (PRD; for protein interaction) present in mammalian dynamins and members of the plant DRP2 subfamily (for review, see Bednarek and Backues, 2010; Fujimoto and Tsutsumi, 2014; Ramachandran and Schmid, 2018). Specifically, DRP1A functions in cytokinesis and cell expansion, likely in part through its role in constitutive endocytosis of bulk membrane, cellulose accumulation, and polar localization of the auxin efflux carrier PIN-FORMED (PIN) proteins and the BORON TRANSPORTER1 (BOR1; Kang et al., 2001; Collings et al., 2008; Mravec et al., 2011; Yoshinari et al., 2016; Marhava et al., 2020). DRP1A interacts in yeast two-hybrid assays and colocalizes with DRP2B in plant cells (Fujimoto et al., 2008, 2010). Both DRP1A and DRP2B accumulate in a sterol-enriched, polar membrane domain during root hair initiation, with DRP1A contributing to high-lipid order at the cell plate (Frescatada-Rosa et al., 2014; Stanislas et al., 2015). DRP1A’s roles beyond plant growth and development, however, remain largely unknown. It is also unclear whether DRP1A interacts genetically with DRP2B for effective CME and/or physiological responses.
In Arabidopsis, the immune receptor FLAGELLIN SENSING2 (FLS2) has emerged as a model PM protein to study ligand-induced endocytosis (for review, see Ben Khaled et al., 2015; Ekanayake et al., 2019). Predominantly functioning in aerial tissues (Robatzek and Wirthmueller, 2013), FLS2 must reside in the PM to perceive bacterial flagellin (or its active peptide derivative flg22) in the apoplast and initiate cellular responses that help provide immunity against Pto DC3000 (for review see Robatzek and Wirthmueller, 2013; Yu et al., 2017). Notably, flg22-elicited responses do not form a single linear signaling pathway but rather a flg22-signaling network that consists of multiple signaling branches (Korasick et al., 2010; Tena et al., 2011; Smith et al., 2014a, 2014b). How these different branches are regulated and integrated into effective immunity remains largely unknown. Ligand-induced endocytosis of FLS2 removes the activated receptor from the site of stimulus-perception (the PM) to desensitize cells to flg22, contributing to an attenuation of immune signaling (Smith et al., 2014b). FLS2 also undergoes constitutive endocytosis to fine-tune FLS2 abundance in the PM in the absence of flg22 (Beck et al., 2012). Recent studies show that clathrin, CCV adaptors, and accessory proteins are necessary for ensuring the correct PM abundance of FLS2 for effective immune responses (Smith et al., 2014a; Mbengue et al., 2016; Collins et al., 2020). Specifically, loss of DRP2B, but not its close paralog DRP2A, leads to reduced ligand-induced endocytosis of FLS2 (Smith et al., 2014a). For drp2b but not drp2a, the resulting delay in FLS2 removal from the PM links to increased flg22-induced production of apoplastic reactive oxygen species (ROS; Smith et al., 2014a). Roles of DRP2 protein orthologs in flg22-induced endocytosis of FLS2 extend to other plant species (Chaparro-Garcia et al., 2015).
Given that drp2b mutants exhibit only a modest decrease in ligand-induced endocytosis of FLS2 (Smith et al., 2014a), we reasoned that DRP2B may function with DRP1A in flg22-induced internalization of FLS2. In this study, we expanded our limited understanding of DRP1A’s contribution to physiological responses in aerial tissue. We demonstrated that DRP1A is required for effective immunity against Pto DC3000 bacteria and flg22-signaling, and that DRP1A functions as an important regulator of flg22-induced endocytosis of FLS2. The isolation of drp1a drp2b double mutants also enabled us to address the genetic interaction of DRP1A and DRP2B and their physiological relevance in plant growth and development, immune signaling, and endocytosis.
Results
Loss of DRP1A in Arabidopsis Col-0 ecotype leads to growth and developmental defects
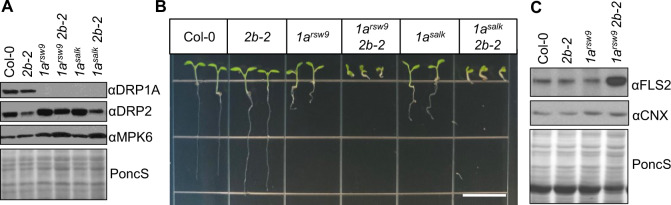
To assess whether DRP1A contributes to plant immune responses and FLS2 endocytosis after flg22 elicitation, we utilized two independent drp1a mutant alleles in the Arabidopsis ecotype Columbia (Col-0, wild-type (WT)) background (Supplemental Figure S1, A). First, we confirmed that drp1arsw9 (Collings et al., 2008) and drp1a069077 (Boutté et al., 2010; Mravec et al., 2011; referred to as drp1asalk from here on) are, as previously reported, null mutants for DRP1A. To streamline genotyping for drp1arsw9, we established Cleaved Amplified Polymorphic Sequence (CAPS) analysis (Supplemental Figure S1, B) to confirm the presence of the point mutation in drp1arsw9 that results in a premature stop codon (Collings et al., 2008). As the drp1asalk mutant is a SALK T-DNA insertion allele, standard PCR amplification with gene- and T-DNA-specific primers was utilized to confirm its genotype (Supplemental Figure S1, C and Supplemental Table S1). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and immunoblot analysis with a previously published αDRP1A antibody (Kang et al., 2003) confirmed that both drp1a mutant alleles are null mutants based on highly reduced accumulation of DRP1A mRNA (Figure 1, A) and no detectable DRP1A protein accumulation (Figure 1, B), respectively, compared with Col-0. Both drp1arsw9 and drp1asalk mutants exhibited similar growth and development defects, including reduced seedling root length (Figure 1, C and D), seedling weight (Figure 1, E), rosette size of 5-week-old plants grown in soil (Figure 1, F, see also Supplemental Figure S2 for comparison to drp2b-2), and silique length (Supplemental Figure S1, D). Albeit not as severe, phenotypic defects for Col-0 drp1a null mutants were reminiscent of those reported for Arabidopsis ecotype Wassilewskija drp1a mutants (Kang et al., 2001; Konopka and Bednarek, 2008). Taken together, results from two independent mutant alleles are consistent with the mutation in DRP1A being responsible for these growth and developmental defects in the Col-0 background. Having two independent drp1a mutant alleles enabled us to assess the role(s) of DRP1A in immune responses.
Figure 1.
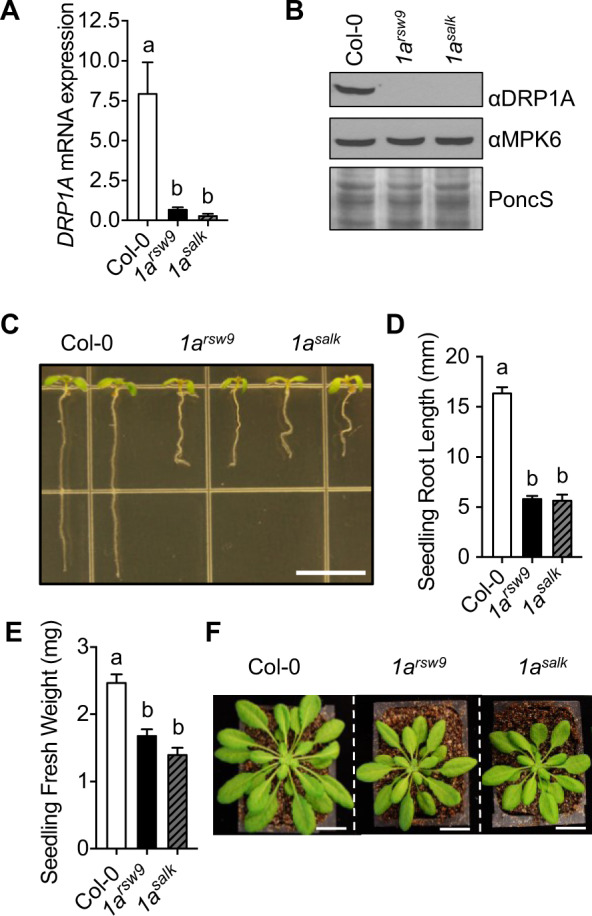
Morphological defects in drp1a null mutant alleles (ecotype Col-0). A, Using RT-qPCR, relative mRNA levels of DRP1A were measured in 7-d-old Col-0 (WT), drp1arsw9 (1arsw9) and drp1a069077 (1asalk) seedlings, and normalized to the reference SAND gene At2g28390. n = 3–4 samples/genotype, with each n representing a biological sample that contained four seedlings. B, Using immunoblot analysis, total protein extracts from 7-d-old seedlings of drp1arsw9 (1arsw9), drp1a069077 (1asalk), and Col-0 were probed with αDRP1A antibodies. αMPK6 and PonceauS served as loading controls. C, 7-d-old drp1arsw9 (1arsw9) and drp1a069077 (1asalk) seedling roots compared with Col-0 when grown vertically. Bar = 5 mm. D, Root length measurements (mm) of 8-d-old seedlings (n ≥ 10 seedlings/genotype) of drp1a null mutant alleles compared with Col-0. E, Fresh weight measurements of 8-d-old seedlings (n ≥ 20 seedlings/genotype) of drp1a null mutant alleles compared with Col-0. F, Rosettes of 5-week-old, soil-grown drp1a null mutant plants relative to Col-0 were digitally extracted from the same image and aligned for comparison. Scale bar = 2 cm. All experiments were repeated at least three times with similar results using biologically distinct samples for each biological replicate. Values are means ± SE with different letters indicating statistically significant differences and with same letters indicating no statistically significant differences based on ordinary one-way ANOVA (P < 0.0001). Col-0, WT (white bar); 1arsw9, drp1arsw9 (black bar); 1asalk, drp1a069077 (gray bar with black stripes).
Loss of DRP1A results in increased susceptibility against Pto DC3000 bacterial strains
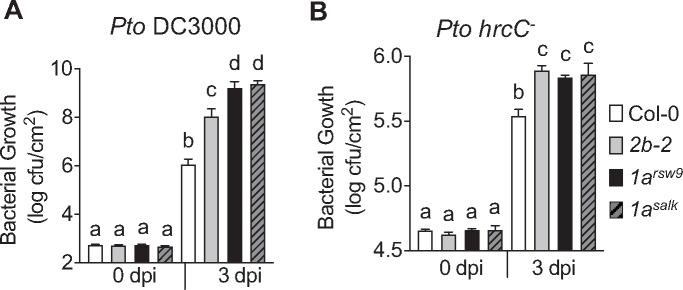
To assess whether DRP1A is required for effective immunity against pathogenic bacteria, we compared the growth of the pathogenic flagellated bacteria Pto DC3000 in drp1a mutant alleles to Col-0 plants. After syringe infiltration into leaves of 5- to 6-week-old plants, bacterial growth was assessed by bacterial dilution plating at 0 and 3 d post-infiltration (dpi) using established protocols (Korasick et al., 2010; Smith et al., 2014a). In control experiments, no difference in Pto DC3000 colony forming units (cfu) was observed at 0 dpi, indicating that similar amounts of bacteria were initially delivered into leaves of drp1a mutant alleles and Col-0 (Figure 2, A). At 3 dpi, however, both drp1arsw9 and drp1asalk mutant plants showed significant increases in Pto DC3000 growth compared with Col-0 (Figure 2, A), indicating that drp1a mutants were more susceptible to Pto DC3000. Notably, the growth of Pto DC3000 was significantly higher in both drp1a mutant alleles relative to the drp2b-2 mutant (Figure 2, A), a loss-of-function allele in the gene encoding DRP2B, previously shown by us to have a positive function in resistance against Pto DC3000 (Smith et al., 2014a). We also tested whether loss of DRP1A resulted in altered susceptibility to Pto DC3000 hrcC−, a hypovirulent bacterial strain that has a defective bacterial type 3 secretion system (T3SS), thus cannot inject bacterial effectors into host cells to suppress host immune responses (Xin et al., 2018). Indeed, both drp1arsw9 and drp1asalk mutant alleles showed increased susceptibility compared with Col-0 (Figure 2, B). In contrast to infection with the pathogenic Pto DC3000 (Figure 2, A), both drp1a alleles supported growth of non-pathogenic Pto DC3000 hrcC− to levels similar to that observed for drp2b-2 (Figure 2, B), indicating that DRP1A functions similarly to DRP2B in contributing positively to pattern-triggered immunity (PTI).
Figure 2.
DRP1A contributes positively to plant immunity against Pto DC3000 bacteria. A and B, Leaves of 5-week-old Col-0, drp1asalk, drp1arsw9, and drp2b-2 plants were syringe-infiltrated with (A) virulent Pto DC3000 (OD600 = 0.0005); or (B) hypovirulent Pto DC3000 hrcC- (OD600 = 0.02). Bacterial growth was assessed by serial dilution plating as cfu at 0 and 3 d post-infection (dpi). For 3 dpi, n = 8 different plants/genotype, and for 0 dpi, n = 4 different plants/genotype; with each n consisting of three leaf discs taken from three different leaves/plant for each genotype and time-point. Values are means ± se with different letters indicating statistically significant differences and with same letters indicating no statistically significant differences based on ordinary one-way ANOVA (P < 0.0001). All experiments were performed at least three times with similar results using biologically distinct samples for each biological replicate. dpi, days post-infection; OD, optical density. Col-0, WT (white bar); 2b-2, drp2b-2 (gray bar); 1arsw9, drp1arsw9 (black bar); 1asalk, drp1a069077 (gray bar with black stripes).
Taken together, we identified additional roles for DRP1A, in that this endocytic accessory protein has positive functions in plant immunity against Pto DC3000 strains. However, given that two independent drp1a mutant alleles showed increased susceptibility to the pathogenic Pto DC3000 strain relative to drp2b-2, it is likely that DRP1A may have additional roles in immune responses against Pto DC3000 that may depend on bacterial effector delivery compared with DRP2B.
Loss of DRP1A affects the three branches of the flg22-signaling network differently
As Pto DC3000 strains are flagellated bacteria (Xin et al., 2018), we next assessed whether drp1a mutant plants showed altered immune responses to bacterial flg22, which is the pathogen-associated molecular pattern derived from Pseudomonas flagellin. Notably, flg22 perception of FLS2 initiates a signaling network that consists of multiple, parallel signaling branches, including the ROS/CALCIUM-DEPENDENT PROTEIN KINASE (CDPK) branch, the MITOGEN-ACTIVATED PROTEIN KINASE (MAPK)-dependent branch, and the defense hormone salicylic acid (SA)-dependent branch (Korasick et al., 2010; Tena et al., 2011; Smith et al., 2014a, 2014b). As we have previously shown, loss of DRP2B has differential effects on these three distinct branches of the flg22-signaling network (Smith et al., 2014a). Thus, we used established flg22-induced marker assays (Boudsocq et al., 2010; Korasick et al., 2010; Ranf et al., 2011; Smith et al., 2014a) to interrogate the activity of the CDPK, MAPK, and SA signaling pathways in the drp1a mutants. Similar to Pto DC3000 infection, these flg22-induced signaling assays were performed in mature leaves.
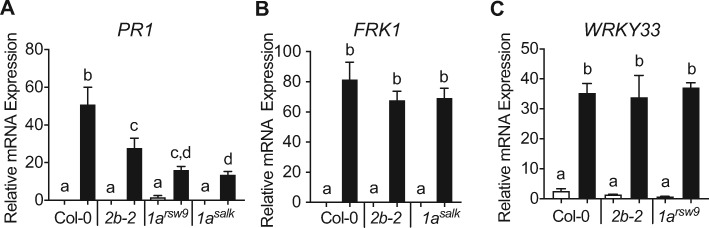
Using RT-qPCR, both drp1a mutant alleles showed reduced mRNA accumulation of PATHOGENESIS RELATED1 (PR1), the late marker downstream of the defense hormone SA, after flg22 elicitation (Figure 3, A). In contrast, no difference in flg22-induced mRNA accumulation was observed between for drp1a mutants compared with Col-0 and drp2b for any of the MAKP-dependent marker genes FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1; Figure 3, B), WRKY33 (Figure 3, C andSupplemental Figure S3, A), and WRKY40 (Supplemental Figure S3, B). These results indicated that, similar to DRP2B (Smith et al., 2014a; Figure 3 and Supplemental Figure S3, A and B), DRP1A has a positive function in flg22-induced PR1 mRNA accumulation and no discernable role in MAPK-signaling dependent expression of flg22-induced marker genes.
Figure 3.
DRP1A plays a positive role in flg22-induced mRNA accumulation of SA-dependent but has no apparent role in MAPK-dependent marker genes. A–C, Leaves of 5- to 6-week-old plants were syringe-infiltrated with 1 µM flg22 for 0 (□) or 24 h (■) for PR1 (A); 0.1 µM flg22 for 0 (□) or 3 h (■) for FRK1 (B); 0.1 µM flg22 for 0 h (□) or 30 min (■) for WRKY33 (C). Relative mRNA levels for each marker gene were measured using RT-qPCR and normalized to the reference SAND gene At2g28390. n = 4–6 plants/genotype/treatment for PR1 and FRK1; and n = 3–6 plants/genotype/treatment for WRKY33. Each n is a biological sample that consisted of three 0.2 mm2 leaf discs collected from the same plant for Col-0 and drp2b-2 or collected from one to two plants for the drp1a alleles due to their smaller leaf size. Values are means ± se with different letters denoting statistically significant difference and with same letters indicating no statistically significant differences based on ordinary one-way ANOVA. All experiments were performed at least three times with similar results using biologically distinct samples for each biological replicate. Col-0, WT; 2b-2, drp2b-2; 1arsw9, drp1arsw9; 1asalk, drp1a069077.
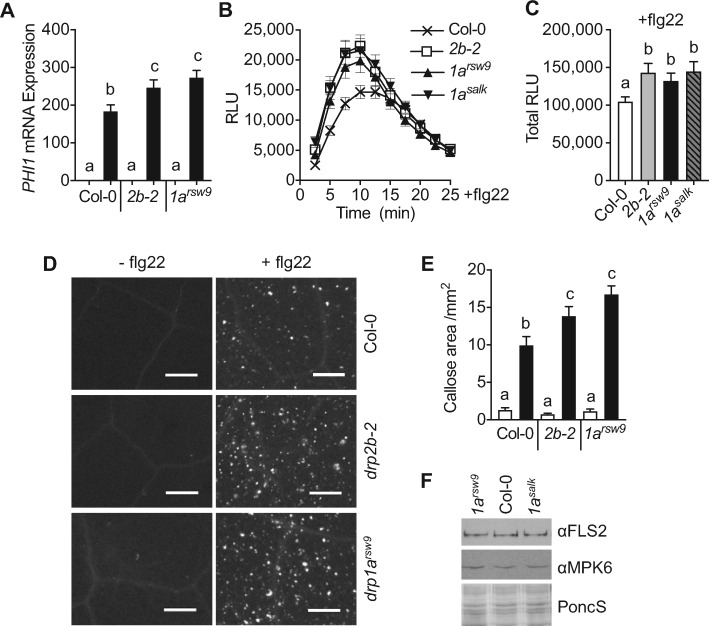
Analysis of flg22-induced mRNA accumulation of PHOSPHATE INDUCED1 (PHI1), a marker gene downstream of CDPKs, demonstrated that PHI1 mRNA expression increased in drp1a mutant plants relative to Col-0 (Figure 4, A). To investigate DRP1A’s roles in flg22 responses beyond mRNA accumulation, we tested drp1a mutant leaves for ROS production and callose deposition using luminol-based and aniline-blue staining assays, respectively. drp1a mutant plants exhibited increased flg22-induced ROS production when measured over time (Figure 4, B) and as total ROS (Figure 4, C) compared with Col-0. Loss of DRP1A also resulted in elevated callose deposits in response to flg22 that were statistically similar to drp2b-2 but increased compared with Col-0 (Figure 4, D and E). In control experiments, no differences were observed between drp mutants and Col-0 after mock treatment (Figure 4, D and E). We conclude that, similar to DRP2B, DRP1A contributes negatively to ROS/CDPK-dependent responses upon flg22 elicitation. As no apparent differences in steady-state FLS2 mRNA (Supplemental Figure S3, C) or steady-state FLS2 protein (Figure 4, F andSupplemental Figure S3, D) accumulation were detected in total leaf extracts of drp1a mutant alleles compared with drp2b-2 and Col-0, the flg22-response defects in drp1a mutants were unlikely due to altered steady-state FLS2 accumulation.
Figure 4.
DRP1A contributes negatively to flg22-induced ROS/CDPK-dependent responses. A, Using RT-qPCR, PHI1 mRNA accumulation was compared between 5- and 6-week-old Col-0, drp1a and drp2b single mutant plants at 30 min after infiltration with 0.1 µM flg22 (■, + flg22) or mock (□, − flg22). Relative PHI1 mRNA levels were measured and normalized to the reference SAND gene At2g28390. n = 3 different plants/genotype/treatment with each n representing a biological sample that contains three leaf punches collected from the same plant. B, Time-course of 0.1 µM flg22-induced ROS production in leaf tissue of 5- to 6-week-old drp mutant and WT plants. n = 24 leaf samples/genotype collected from three to four different plants with each n representing a biological sample consisting of a leaf disc half. C, Total 0.1 µM flg22-induced ROS production from time-course shown in B. D and E, For callose deposition, leaves of 5- to 6-week-old plants were infiltrated with 1 µM flg22 or DMSO. After 24 h, leaf punches were collected and processed for aniline blue staining and imaging. D, Representative leaf image for callose depositions of each genotype and treatment from the same experiment shown in E. Scale bar = 0.5 mm. E, Percentage of total leaf surface area covered by aniline blue-stained fluorescent callose at 24 h after infiltration of 1 µM flg22 (■) or mock (DMSO, □) for n > 20 leaf punches collected from three to four plants/genotype/treatment. F, Using immunoblot analysis, total protein extracts from 7-d-old seedlings of drp1arsw9, drp1asalk, and Col-0 were probed with αFLS2 antibodies. αMPK6 and PonceauS served as loading controls. Values are means ± se with different letters denoting statistically significant difference and with same letters indicating no significant differences based on ordinary one-way ANOVA. All experiments were performed at least three times with similar results using biologically distinct samples for each biological replicate. Col-0, WT; 2b-2, drp2b-2; 1arsw9, drp1arsw9; 1asalk, drp1a069077; min, minutes; RLU, relative light units.
Taken together, our findings that DRP1A has a differential impact on the three different branches of the flg22-signaling network indicate that DRP1A has negative function(s) in the ROS/CDPK-branch, positive function(s) in the SA-branch but no apparent role in the MAPK-branch. Notably, the combination of immune defects in drp1a was similar to those observed for drp2b mutants, indicating that these two DRP proteins may modulate component(s) of the flg22-signaling network in a similar manner.
DRP1A plays a significant role in ligand-induced endocytosis of FLS2
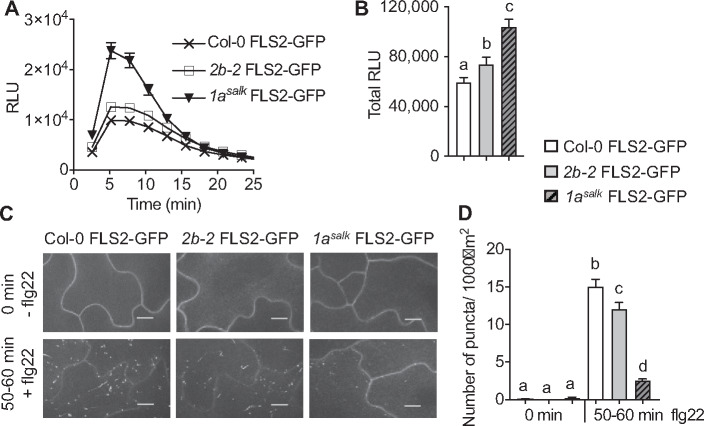
Next, we addressed whether DRP1A may contribute to ligand-induced endocytosis of FLS2 using live-cell imaging. To this end, Col-0 expressing FLS2-green fluorescent protein (GFP) under the control of its own promoter (pFLS2::FLS2-3xMyc-EGFP; subsequently referred to as FLS2-GFP; Beck et al., 2012; Smith et al., 2014a) was crossed with drp1asalk mutant plants to generate homozygous drp1asalk FLS2-GFP. Similar to drp1a leaf tissue (Figure 3, B and C), cotyledons of drp1asalk FLS2-GFP displayed increased flg22-elicited ROS production compared with Col-0 FLS2-GFP (Figure 5, A, time-course; Figure 5, B, total ROS production). Notably, the flg22-dependent increase in ROS was significantly higher in drp1asalk FLS2-GFP cotyledons compared with that in drp2b-2 FLS2-GFP cotyledons. We concluded that cotyledons of drp1asalk FLS2-GFP are a biologically relevant tissue for live-cell imaging studies. Elevated ROS production was unlikely due to increased steady-state levels of FLS2 because compared with Col-0 FLS2-GFP, no apparent increase in endogenous FLS2 or ectopically expressed FLS2-GFP protein accumulation was observed when total protein seedling extracts were probed with antibodies against FLS2 and GFP (Supplemental Figure S4; endogenous FLS2, white arrowhead; FLS2-GFP, black arrowhead).
Figure 5.
DRP1A has a prominent role in flg22-induced endocytosis of FLS2-GFP. A, Time-course of 1 µM flg22-induced ROS production in cotyledons of 7-d-old Col-0 FLS2-GFP, drp1asalk FLS2-GFP and drp2b-2 FLS2-GFP mutant seedlings. n = 24 biological samples/genotype, with each n consisting of two halves of a cotyledon. B, Total 1 µM flg22-induced ROS production from time-course shown in A. For C and D, Col-0 FLS2-GFP, drp2b-2 FLS2-GFP, and drp1asalk FLS2-GFP homozygous seedlings were treated with 1 µM flg22 for 0 and 50–60 min to examine ligand-induced endocytosis of FLS2-GFP on the adaxial surface of the cotyledon epidermis using SDCM. C, Representative maximum-intensity projection images of FLS2-GFP fluorescence. Scale bars = 10 µm. D, Quantification of FLS2-GFP puncta at indicated times post-elicitation with n 18 images/genotype/treatment with at least six images each taken from three different seedlings per genotype and treatment. Values are means ± se with different letters denoting statistically significant difference and with same letters indicating no statistically significant differences based on ordinary one-way ANOVA (P < 0.05). All experiments were performed at least five times, with similar results using biologically distinct samples for each biological replicate. Col-0 FLS2-GFP, WT FLS2-GFP; 2b-2 FLS2-GFP, drp2b-2 FLS2-GFP; 1asalk FLS2-GFP, drp1a069077 FLS2-GFP; min, minutes; and RLU, relative light units.
We performed quantitative live-cell imaging in pavement cells on the adaxial surface of cotyledons using Spinning Disc Confocal Microscopy (SDCM), a well-established technique to quantify ligand-induced endocytosis of FLS2-GFP Arabidopsis cotyledons (Beck et al., 2012; Smith et al., 2014a; Leslie and Heese, 2017). Consistent with FLS2 being a PM-localized receptor in the absence of any stimulus, FLS2-GFP localized predominantly at the PM of Col-0, drp2b-2, and drp1asalk mutants with some FLS2-GFP in small intracellular puncta prior to flg22-elicitation (Figure 5, C; −flg22, 0 min). When quantifying the number of intracellular FLS2-GFP puncta using Fiji software (Leslie and Heese, 2017), we observed no statistically significant difference between the drp2b-2 and drp1asalk single mutants and Col-0 at 0 min (Figure 5, D). In response to flg22, FLS2-GFP in Col-0 undergoes ligand-induced internalization from the PM, resulting in endocytic movement to endosomal compartments that appear as intracellular FLS2-GFP puncta around 50–60 min post-elicitation (Figure 5, C and D, +flg22; Beck et al., 2012; Smith et al., 2014a; Leslie and Heese, 2017). In drp1a, ligand-induced endocytosis of FLS2-GFP was strongly impaired (Figure 5, C and D, +flg22). Notably, FLS2 endocytosis was more compromised in drp1asalk relative to drp2b (Figure 5, C and D, +flg22). Similar to drp1asalk, flg22-elicited endocytosis of FLS2-GFP was inhibited in the independent drp1arsw9 mutant allele (Supplemental Figure S5). Following flg22 elicitation for 50–60 min, we observed a 60%–70% reduction in the levels of FLS2-GFP intracellular puncta in drp1asalk and drp1arsw9 cotyledon cells (Figure 5, D and Supplemental Figure S5, 50–60 min flg22), whereas drp2b-2 single mutant showed only a ∼20% reduction (Figure 5, D, 50–60 min flg22). These results indicate that DRP1A has a more prominent role than DRP2B in flg22-induced endocytosis of FLS2.
DRP1A and DRP2B function synergistically in plant growth and development
As described above, drp1a and drp2b single mutants showed similar phenotypic flg22−defects and were impaired in ligand-induced endocytosis of FLS2-GFP. To test whether DRP1A and DRP2B genetically interact, we crossed drp2b-2 with drp1asalk or with drp1arsw9 single mutant alleles to generate homozygous drp1asalkdrp2b-2 or drp1arsw9drp2b-2 double mutants, respectively. Using gene-specific primers (Supplemental Table S1), we confirmed by RT-qPCR that expression levels of DRP1A and DRP2B mRNA in the double homozygous drp1a drp2b lines were significantly reduced relative to Col-0 (Supplemental Figure S6, A). Furthermore, DRP1A protein was not detected in total protein extracts of drp1a single and drp1a drp2b double mutants analyzed by immunoblotting using αDRP1A antibodies (Figure 6, A). Similarly, immunoblot analysis using affinity purified polyclonal αDRP2 peptide antibody, which detects both DRP2A and DRP2B proteins due to their high amino acid sequence identity (Backues et al., 2010; Smith et al., 2014a), demonstrated that the levels of DRP2 proteins were substantially reduced in drp2b single and drp1a drp2b double mutant total protein extracts (Figure 6, A). The residual DRP2 protein detected by the αDRP2 antibody in the drp2b-2 mutant likely corresponds to the protein encoded by DRP2A (Backues et al., 2010; Smith et al., 2014a). In contrast to drp1asalk and drp1arsw9 single mutants (Figures 1, C–E and 6, B), drp2b-2 single mutants did not display obvious differences in seedling growth relative to Col-0 (Figure 6, B) as previously described (Backues et al., 2010; Smith et al., 2014a). However, growth of both drp1asalkdrp2b-2 and drp1arsw9drp2b-2 double mutant seedlings was more severely stunted than either of the drp1a or drp2b single mutants (Figure 6, B). The drp1a drp2b double mutants survived on MS plates for up to 21 d but were unable to grow on soil and reproduce, indicating that loss of both DRP1A and DRP2B resulted in a seedling lethal phenotype.
Figure 6.
DRP1A and DRP2B act synergistically in plant growth and in hyperaccumulation of FLS2. A, Using immunoblot analysis, total protein extracts from 7-d-old seedlings of drp2b-2, drp1arsw9, drp1arsw9 drp2b-2, drp1asalk, drp1asalk drp2b-2, and Col-0 were probed with αDRP1A and αDRP2 antibodies. αMPK6 and PonceauS served as loading controls. B, Representative image of 7-d-old seedlings. Scale bar = 1 cm. C, Using immunoblot analysis, total protein extracts from 7-d-old seedlings of drp2b-2, drp1arsw9, drp1arsw9 drp2b-2, drp1asalk, drp1asalk drp2b-2, and Col-0 were probed with αFLS2 antibodies. αCNX and PonceauS served as loading controls. For immunoblot analyses (A and C), each sample of Col-0, drp2b-2, and drp1a single mutants consisted of 10 seedlings; and for drp1a drp2b double mutants, each sample consisted of 30 seedlings. All experiments were performed at least three times with similar results using biologically distinct samples for each biological replicate. CNX, CALNEXIN1/2; PoncS, PonceauS; Col-0, WT; 2b-2, drp2b-2; 1arsw9, drp1arsw9; 1asalk, drp1a069077; 1arsw9 2b-2, drp1arsw9 drp2b-2; 1asalk 2b-2, drp1a069077 drp2b-2.
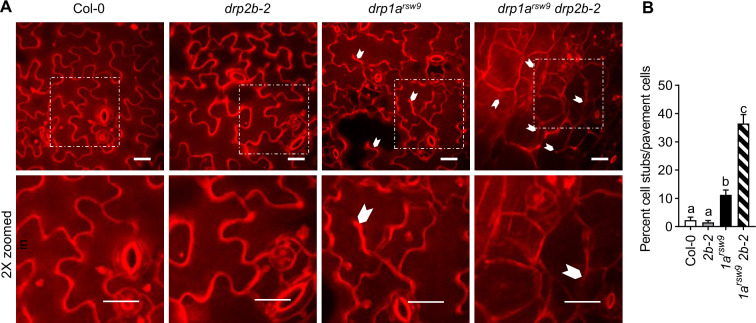
Previous studies have shown that drp1a mutant seedlings exhibit cytokinesis and cell enlargement defects in multiple tissues (Kang et al., 2001, 2003; Collings et al., 2008; Mravec et al., 2011). As FLS2 predominately functions in cotyledons at the seedling stage, we focused on examining cotyledon epidermal cells in Col-0, drp single and double mutants. After staining with propidium iodide (PI), drp1a cotyledons revealed altered cell size (Figure 7, A) reminiscent of those reported in other tissues for drp1a single mutants (Kang et al., 2001, 2003; Collings et al., 2008; Mravec et al., 2011). No apparent cell size reduction or misshapen cells were observed for cotyledon epidermal cells in the drp2b-2 single mutant compared with Col-0 (Figure 7, A). The combined loss of DRP1A and DRP2B caused small, round, and mis-shaped epidermal cells with significantly increased number of cell wall stubs that were more severe than those for the drp1a single mutant (Figure 7, B). We concluded that the severely stunted areal tissue observed in the drp1a drp2b double mutant was consistent with potential synergistic role(s) of DRP1A and DRP2B in cell expansion and cytokinesis.
Figure 7.
Loss of both DRP1A and DRP2B results in cell morphology defects and cytokinesis defects of cotyledon pavement cells. A, After staining of 7-d-old seedlings with 200 μM PI for 20 min, epidermal cells of cotyledons were imaged using SDCM. Representative maximum-intensity projection images of PI fluorescence are shown. Partial cell divisions are indicated with white arrowheads. 2× zoomed in regions are marked with dotted squares in the top images. Scale bars = 10 µm. B, The severity of cytokinesis defects in pavement cells was quantified as percentage of cells with cell wall stubs per total number of pavement cells. n = 10–17 images containing multiple pavement cells/genotype with 2 to 3 images taken from 8 to 10 different seedlings/genotype. Values are means ± se with different letters denoting statistically significant difference and with same letters indicating no statistically significant differences based on ordinary one-way ANOVA (P < 0.05). The experiment was repeated three times with similar results using biologically distinct samples for each biological replicate. Col-0, WT; 1arsw9, drp1arsw9; 2b-2, drp2b-2; and 1arsw9 2b-2, drp1arsw9 drp2b-2.
PM hyperaccumulation of FLS2 in drp1a drp2b double mutant seedlings does not induce increased flg22 responses
As shown previously (Smith et al., 2014a) and in this study (Figure 4, F and Supplemental Figures S3, C and D), FLS2 mRNA and FLS2 protein accumulation were similar in total seedling extracts of drp1a and drp2b single mutants to Col-0. In contrast, immunoblot analysis demonstrated that the FLS2 protein hyperaccumulated in the drp1arsw9 drp2b double mutant seedlings (Figure 6, C and Supplemental Figure S7, A), potentially due to increased expression of FLS2 mRNA as determined by RT-qPCR (Supplemental Figure S7, B). As a negative control, we created a drp1arsw9 drp2b fls2Δ triple mutant that did not show any detectable FLS2 protein or FLS2 mRNA accumulation (Supplemental Figures S7, A or B, respectively). Additionally, loss of FLS2 did not alleviate the stunted growth of the drp1arsw9 drp2b-2 double mutant seedlings (Supplemental Figure S7, C), indicating that synergistic role(s) of DRP1A and DRP2B in plant growth and development were independent of FLS2.
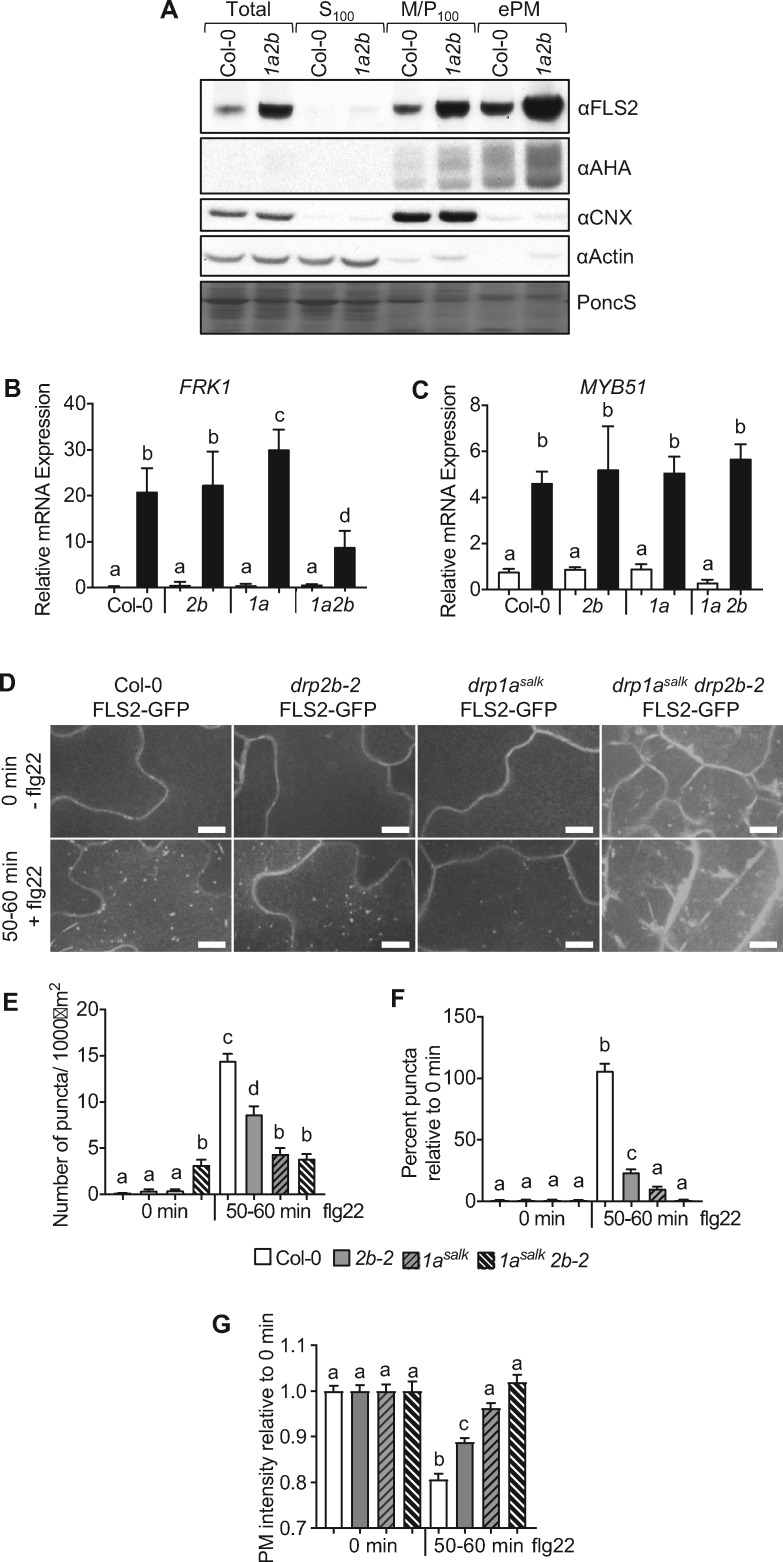
To assess whether the hyperaccumulation of FLS2 protein in the drp1a drp2b double mutant is associated with increased levels of the receptor at the PM, we enriched for PM proteins by depleting contaminating organelles through a combination of differential centrifugation and Brij58-treatment (Zhang and Peck, 2011; Collins et al., 2017, 2020). Fractionation efficacy for seedling extracts was confirmed by probing protein fractions from Col-0, drp single and double mutants with organelle-specific antibodies (Figure 8, A and Supplemental Figure S8). The soluble marker proteins actin or MPK6 were present in total and soluble (S100) but depleted from microsomal (M/P100) and enriched PM (ePM) fractions; and the endoplasmic reticulum membrane marker CALNEXIN1/2 (CNX) was associated with total and M/P100 but depleted from ePM fractions. The AHA H+-ATPases or BRASSINOSTEROID INSENSITIVE 1 (BRI1) represented PM marker proteins that were present in microsomes (M/P100) and ePM but not in soluble (S100) fractions. In addition to hyperaccumulation of FLS2 in total and microsomal (M/P100) fractions, drp1a drp2b double mutants accumulated FLS2 protein in the PM to higher levels relative to Col-0 (Figure 8 and Supplemental Figure S8). No apparent difference in FLS2 protein abundance in total, M/P100, and ePM fractions was observed in drp1a and drp2b single mutants compared with Col-0 seedling extract (Supplemental Figure S8, A). drp1a drp2b double mutant seedlings also showed an apparent increased abundance for AHA H+-ATPases in M/P100 and ePM fractions (Figure 8, A). However, the combinatorial loss of DRP1A and DRP2B did not result in hyperaccumulation of all PM proteins as BRI1, the PM-localized receptor kinase for brassinosteroid signaling and implicated in plant immunity (Albrecht et al., 2012; Belkhadir et al., 2012) showed similar protein accumulation in microsomes (M/P100) and ePM in the drp1a drp2b double mutant compared with Col-0 (Supplemental Figure S8, B).
Figure 8.
Loss of both DRP1A and DRP2B impairs some but not all flg22 responses and blocks ligand-induced endocytosis of FLS2-GFP. A, Using differential centrifugation and immunoblot analysis, total homogenate (Total), soluble (S100), microsomal (M/P100), and ePM protein fractions from drp1arsw9 drp2b-2 (1a2b) and Col-0 were probed with αFLS2 antibodies. AHA, CALNEXIN1/2 (CNX) and ACTIN served as markers for PM, endoplasmic reticulum membrane, and soluble fractions, respectively, to confirm cellular fractionation. B and C, Seven-day-old seedlings of Col-0, drp2b-2 (2b), drp1asalk (1a), drp1asalk drp2b-2 (1a 2b) were treated with 1 µM flg22 for 0 (□) or 2 h (■) to test for FRK1 (B) or MYB51 (C) mRNA accumulation. Using RT-qPCR, relative mRNA levels for marker genes were measured and normalized to the reference SAND gene At2g28390. Analysis included data pooled from two independent experiments with n = 8 samples/genotype for +flg22 and n = 7 samples/genotype for –flg22. Each sample (n) consisted of four seedlings/biological sample for Col-0, drp2b, and drp1a single mutants and 15 seedlings/biological sample for drp1a drp2b double mutants. For D–F, Col-0 FLS2-GFP, drp2b-2 FLS2-GFP, drp1asalk FLS2-GFP, and drp1asalk drp2b-2 FLS2-GFP homozygous seedlings were treated with 1 µM flg22 for 0 min (−) and 50–60 min (+) to examine ligand-induced endocytosis on the adaxial surface of the cotyledon epidermis using SDCM. D, Representative maximum-intensity projection images of FLS2-GFP fluorescence. Scale bars = 10 µm. E, Quantification of FLS2-GFP puncta as number of puncta per 1,000 µm2. F, Quantification of FLS2-GFP puncta as percent FLS2-GFP puncta relative to 0 min puncta for respective genotype. G, Quantification of PM intensity of FLS2-GFP at 50–60 min relative to the 0 min PM intensity for respective genotypes. For E–G, n 18 images/genotype/treatment with at least six images each taken from three different seedlings/genotype/treatment; and for the drp1a drp2b double mutant, 6–12 images each were taken from four to five different seedlings/treatment. Values are means ± se with different letters denoting statistically significant difference and with same letters indicate no significant differences based on ordinary one-way ANOVA (P < 0.05). All experiments were performed at least three times with similar results using biologically distinct samples for each biological replicate. PoncS, PonceauS; Col-0, WT; 2b-2, drp2b-2; 1asalk, drp1a069077; 1asalk 2b-2, drp1a069077drp2b-2; SDCM, spinning disc confocal microscope; min, minutes.
Notably, the increased accumulation of FLS2 in the PM in drp1a drp2b double mutant seedlings did not result in increased flg22 responses. Indeed, flg22-induced FRK1 mRNA levels were significantly decreased in drp1a drp2b double mutant seedlings compared with the respective single drp mutants and Col-0 (Figure 8, B, drp1asalk drp2b-2;Supplemental Figure S9, A, drp1arsw9 drp2b-2). This analysis also uncovered tissue-specific differences in DRP1A function for FRK1 mRNA accumulation. Flg22-induced FRK1 mRNA levels in drp1a seedlings were increased relative to Col-0 and drp2b (Figure 8, B and Supplemental Figure S9, A) while FRK mRNA accumulation in mature leaves was similar between drp1a, drp2b, and Col-0 (Figure 3, B). Notably, FLS2 appeared to be signaling competent in drp1a drp2b seedlings for at least some responses because flg22-induced mRNA accumulation for MYB51, an established flg22-response marker gene in seedlings (Orosa et al., 2018), did not show any apparent differences between Col-0, drp1a drp2b double and drp single mutant seedlings (Figure 8, C, drp1asalk drp2b-2 and Supplemental Figure S9, B, drp1arsw9 drp2b-2).
Given the increased level of FLS2 observed in the ePM fraction of the drp1a drp2b double mutant (Figure 8 and Supplemental Figure S8), we examined the combined contribution of DRP1A and DRP2B in ligand-induced FLS2 endocytosis. Homozygous drp1a drp2b double mutant seedlings expressing FLS2-GFP were created by crossing drp1asalk/DRP1A drp2b-2/drp2b-2 with Col-0 FLS2-GFP. Based on immunoblot analysis, drp1asalk drp2b-2 double mutant seedlings accumulated higher levels of FLS2-GFP compared with Col-0, drp1asalk, and drp2b-2 single mutants (Supplemental Figure S10). In agreement, drp1asalk drp2b-2 double mutants showed an overall higher GFP fluorescence in cotyledon pavement cells when utilizing the same SDCM imaging conditions to compare Col-0, single and double drp mutants expressing FLS2-GFP (Figure 8, D).
In contrast to WT cells that showed a robust increase in intracellular FLS2-GFP puncta upon flg22 stimulation, we did not observe an increase in the levels of FLS2-GFP puncta in the drp1asalk drp2b-2 double mutant cotyledons in response to flg22 as assessed by quantitative analysis of FLS2-GFP puncta before (0 min) and after flg22 treatment (50–60 min; Figure 8, E). This result was different from that in drp1asalk and drp2b-2 single mutants, which displayed a flg22-induced increase in intracellular FLS2-GFP puncta, albeit to statistically reduced levels compared with Col-0 (Figure 8, D and E; see also Figure 5, C and D and Supplemental Figure S5). Interpreting the ligand-induced endocytosis defect for the drp1a drp2b double mutant, however, was complicated by the presence of preexisting FLS2-GFP-labeled structures of unknown origin in the drp1a drp2b double mutant cells in the absence of any stimulus (Figure 8, E; 0 min, −flg22). To address this issue, we calculated the number of FLS2-GFP puncta at 50–60 min as a percentage of those at 0 min of the respective genotype. Results from this analysis showed that flg22 elicitation did not lead to any detectable increase in flg22-induced FLS2-GFP puncta in drp1asalk drp2b-2 double mutant seedlings (Figure 8, F). Consistent with a block in ligand-induced endocytosis and lack of FLS2-GFP removal from the PM, we did not observe any apparent change in the PM intensity at 0 and 50–60 min after flg22 elicitation in the drp1asalk drp2b-2 double mutant seedlings (Figure 8, G). In contrast, the FLS2-GFP PM intensity for Col-0 was greatly reduced at 50–60 min after flg22 elicitation, which corresponded with the increased number of intracellular FLS2-GFP puncta and was in agreement with removal of activated FLS2-GFP from the PM by ligand-induced endocytosis. Notably, both drp1a and drp2b single mutants also showed a reduction in the loss of PM fluorescence intensity relative to the WT Col-0 control; but consistent with the less severe endocytic defects observed in the drp2b single mutant, there was a larger decrease in PM fluorescence signal intensity in the cells of the drp2b mutant after flg22 treatment relative to the drp1a single and drp1a drp2b double mutants (Figures 5, C and D, and 8, D–G). Therefore, we conclude that loss of DRP1A alone as well as the combinatorial loss of DRP1A and DRP2B resulted in a severe defect in ligand-induced endocytosis for FLS2-GFP.
Discussion
The ability of host cells to modulate their PM composition is pivotal in regulating the strength, duration, and integration of host immune responses to microbial pathogens that colonize the plant apoplast. In the case of the FLS2 immune receptors, ligand-induced endocytosis has emerged as an effective means to attenuate a subset of flg22-signaling responses (Smith et al., 2014a, 2014b). Similar to other plant PM proteins that undergo endocytosis (Claus et al., 2018; Reynolds et al., 2018; Ekanayake et al., 2019), the processes of constitutive and ligand-induced internalization of FLS2 from the PM are likely complex and require the careful spatiotemporal coordination of many vesicle components for budding, maturation, and scission of the endocytic vesicles. However, with the exception of the clathrin heavy chain isoform CHC2 (Mbengue et al., 2016) and DRP2B (Smith et al., 2014a), little is known about the molecular machinery and mechanisms involved in FLS2 endocytosis and signaling.
In this study, we demonstrated that a member of the plant-specific DRP1 subfamily of GTPases, namely DRP1A, previously implicated in CME in root cells (Bednarek and Backues, 2010; Fujimoto and Tsutsumi, 2014), functions in flg22-induced immune signaling, immunity against flagellated Pto DC3000 bacteria, and ligand-induced endocytosis of FLS2 (Figure 9). This work was primarily performed in leaves and cotyledons because these are the biologically relevant tissues that are primarily infected by Pto DC3000 (Xin et al., 2018) and in which FLS2 is expressed and functions in plant immunity (Robatzek and Wirthmueller, 2013; Yu et al., 2017). Thus, the presented results advanced the limited understanding of DRP1A’s physiological relevance in areal tissues. Our genetic interaction studies indicated that DRP1A functions synergistically with DRP2B in regulating the PM abundance of FLS2 as well as in plant growth and development, likely due to their roles in membrane dynamics required for cytokinesis, cell expansion, trafficking of PM proteins, and/or cellulose accumulation (Kang et al., 2001; Collings et al., 2008; Mravec et al., 2011; Yoshinari et al., 2016; Marhava et al., 2020).
Figure 9.
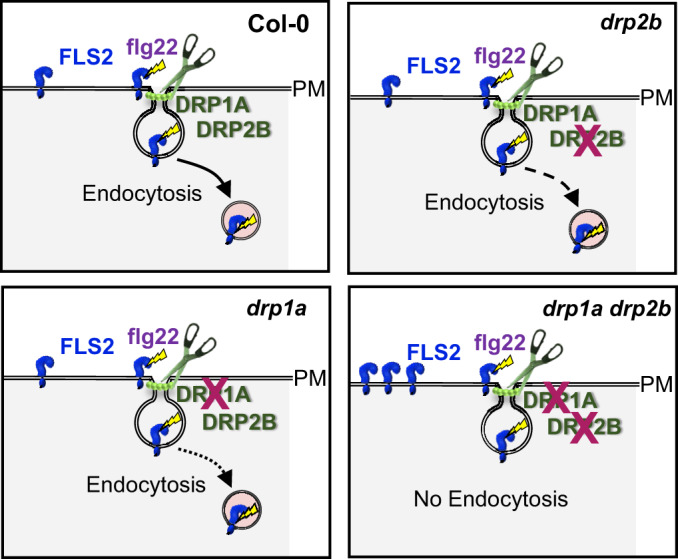
Model depicting DRP1A- and DRP2B-dependent ligand-induced endocytosis of FLS2. In Arabidopsis, loss of either DRP1A or DRP2B results in reduced flg22-induced endocytosis of the pattern recognition receptor FLS2 compared with WT Col-0. As indicated by the dotted and dashed arrows, respectively, the drp1a single mutant is more compromised than the drp2b single mutant in FLS2 endocytosis after elicitation with the bacterial PAMP flg22. The combinatorial loss of DRP1A and DRP2B leads to a block in ligand-induced endocytosis of FLS2. For the drp1a drp2b double mutant, impaired constitutive endocytosis likely contributes to hyperaccumulation of FLS2 at the PM in the absence of any stimulus. DRP, DYNAMIN RELATED PROTEIN; flg22, active 22-amino acid peptide derivative from bacterial flagellin; FLS2, FLAGELLIN SENSING2; PAMP, pathogen-associated molecular pattern.
We report here that loss of DRP1A function results in differential effects on flg22 signaling responses, providing additional support to the evolving concept that flg22 signaling is a signaling network that consists of at least three different branches rather than a simple linear pathway (Korasick et al., 2010; Tena et al., 2011; Smith et al., 2014a, 2014b). More specifically upon flg22 elicitation in leaf tissue (Figures 3 and 4 and Supplemental Figure S3), drp1a mutants exhibited increased accumulation of PHI1 mRNA, ROS, and callose, indicating that DRP1A contributes negatively to the ROS/CDPK-dependent branch. Conversely, loss of DRP1A caused a decrease in PR1 mRNA accumulation, indicating a positive role for DRP1A in the SA-dependent branch of flg22 responses. Lastly, we did not observe any apparent flg22-response defects in WRKY33, WRKY40, and FRK1 mRNA accumulation, indicating no apparent role of DRP1A in modulating the MAPK-dependent branch of the flg22-signaling network in leaf tissue. This combination of phenotypic defects was reminiscent to that previously described for drp2b mutant leaves (Smith et al., 2014a; Leslie et al., 2016), indicating that these two DRPs appear to modulate flg22 signaling in a similar manner, potentially by affecting a similar set of proteins with immune functions. Such a scenario is consistent with previous studies showing subcellular co-localization and intermolecular interaction between DRP1A and DRP2B (Fujimoto et al., 2008, 2010).
We propose several, not mutually exclusive, explanations for the observed differential effects of loss of DRP1A on distinct branches of the flg22-signaling network. First, DRP1A may participate in removing FLS2 from its site of function (the PM) to attenuate flg22-signaling. This hypothesis, in which ligand-induced endocytosis of FLS2 would serve to dampen immune signaling, is supported by our findings that impaired flg22-induced endocytosis of FLS2 was linked to increased early flg22-signaling responses in drp1a (Figures 4, 5, 8, B). Notably, the observed enhancement in flg22-signaling in drp1a mutants was apparent when assessing flg22 responses in the same tissue (cotyledons) and the same developmental stage (7-d-old seedlings) used for ligand-induced endocytosis of FLS2. drp1a mutant seedlings displayed a more severe defect in ligand-induced endocytosis of FLS2 than drp2b (Figures 5, C and D, 8, D–G and Supplemental Figure S5) that was accompanied by a more pronounced increase in flg22-induced ROS (Figure 5, A and B) and FRK1 mRNA accumulation (Figure 8, B) compared with those observed in drp2b seedlings. Another explanation for how loss of DRP1A may affect the distinct flg22-signaling branches differently is that DRP1A may modulate the localization and/or activity of other proteins that differ in their spatial localization and/or temporal contribution to flg22-signaling. Such a scenario may also explain why DRP1A had a positive role in mRNA accumulation of PR1 (Figure 3, A), a late flg22 response that is under the regulation of chloroplastic-derived SA (Yu et al., 2017; Xin et al., 2018); but it played a negative role in flg22-induced callose deposition (Figure 4, D and E), a late flg22 response previously shown to be dependent on PM-localized RESPIRATORY BURST OXIDASE PROTEIN D and CALLOSE SYNTHASE12 (Smith et al., 2014a; Leslie et al., 2016).
Our findings also raised the question as to why drp1a displayed a more severe defect in ligand-induced endocytosis of FLS2 relative to drp2b mutant seedlings (Figures 5, C and D, 7, D–F). A potential answer may be provided by previously described findings for DRP1A. As indicated in Fujimoto and Tsutsumi (2014), DRP1A’s GTPase domain (62%) shows higher amino acid identity to human dynamin than that of DRP2 (29%). Considering that GTPase activity of animal dynamins is critical for release of CCVs from the PM (Ramachandran and Schmid, 2018), DRP1A may contribute more substantially to the GTPase activity than DRP2B for scission of endocytic vesicles that contain FLS2. Another possibility may be that DRP1A’s function in maintaining high lipid-order sterol-rich domains (Frescatada-Rosa et al., 2014) adds to effective ligand-induced endocytosis of FLS2. In light of a recent report that implicates sterols in flg22-induced FLS2 endocytosis (Cui et al., 2018), it is tempting to speculate that loss of DRP1A causes a disruption in sterol-rich domains, which in turn may contribute to impaired FLS2 endocytosis after flg22 elicitation.
As shown by biochemical fractionation and live cell imaging (Figure 8, A, D–G and Supplemental Figure S10), the combinatorial loss of DRP1A and DRP2B resulted in hyperaccumulation of FLS2 in the PM in the absence of any stimulus, likely due to severely impaired constitutive endocytosis of FLS2. However, the elevated levels of PM-associated FLS2 were not linked to increased cellular responsiveness (Figure 8, B and C and Supplemental Figure S9). In fact, DRP1A and DRP2B showed a synergistic reduction in flg22-induced FRK1 mRNA accumulation in the drp1a drp2b double mutant (Figure 8, B and Supplemental Figure S9, A). Based on these findings, we speculate that the combined loss of DRP1A and DRP2B results in a change in some fraction of the total pool of FLS2 into a form that is unable to perceive and/or elicit flg22 responses; but the underlying cell biological mechanisms that allow FLS2 to be or become signaling competent remain largely unknown. Notably, PM abundance of another PM receptor, BRI1, was not apparently altered in drp1a drp2b double mutant seedlings despite the fact that both BRI1 and FLS2 have been shown to require clathrin for their internalization (Irani et al., 2012; Mbengue et al., 2016). In agreement with our findings that FLS2 but not BRI1 required DRP1/2 family members to modulate its PM accumulation, viral-induced silencing of multiple DRP2 paralogs in Nicotiana benthamiana impairs ligand-induced FLS2 internalization but does not affect internalization of BRI1 (Chaparro-Garcia et al., 2015). Previous studies show that in contrast to FLS2, BRI1 relies primarily on constitutive endocytosis for signal attenuation (Irani et al., 2012), and that FLS2 and BRI1 spatially separate into distinct PM nanodomains (Bücherl et al., 2017). Based on these differences between FLS2 and BRI1, it is conceivable that these two PM receptors may require distinct endocytosis accessory proteins to fine-tune and regulate their PM abundance for dampening responses through effective endocytosis.
Increasing evidence points at lipid microdomains as contributors to lateral mobility, distribution, and complex formation of FLS2 with other immune components within the plant PM, which in turn impact effective flg22-signaling, ligand-induced endocytosis, and/or protein accumulation of FLS2 in a spatiotemporal manner (Keinath et al., 2010; Bücherl et al., 2017; Cui et al., 2018; Yang et al., 2019). Considering that (a) DRP1A contributes to maintaining sterol-rich domains (Frescatada-Rosa et al., 2014) and (b) DRP1A, DRP2B, FLS2, and other PM proteins with immune functions are present in sterol-rich, detergent-resistant membranes (Keinath et al., 2010; Frescatada-Rosa et al., 2014), it will be interesting to assess in the future whether loss of DRP1A and/or DRP2B alters the spatiotemporal movement of FLS2 or other PM proteins functioning early with FLS2 in flg22-signaling (Tian et al., 2019; Lee et al., 2020; Ma et al., 2020) within the PM. Furthermore, the drp1a drp2b double and single mutants will serve as important genetic tools to understand how individual flg22-signaling branches are regulated and/or integrated into effective immune responses to restrict pathogen infection.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) mutants drp2b-2 (SALK_134887; Backues et al., 2010), drp1arsw9 (Collings et al., 2008), drp1asalk (SALK_069077; Boutté et al., 2010), and fls2Δ (SALK_93905; Smith et al., 2014b) have been previously described. All mutants were in the Col-0 ecotype background. Homozygous drp1asalk drp2b-2 double, drp1arsw9 drp2b-2 double, or drp1arsw9 drp2b-2 fls2Δ triple mutants were generated by pollinating drp2b-2 or drp2b-2 fls2Δ stigmas with pollen from respective drp1a mutant lines. drp1asalk drp2b-2 double, drp1arsw9 drp2b-2 double, or drp1arsw9 drp2b-2 fls2Δ triple mutant lines were propagated as DRP1A/drp1asalk drp2b-2/drp2b-2 double, DRP1A/drp1arsw9 drp2b-2/drp2b-2 double, or DRP1A/drp1arsw9 drp2b-2/drp2b-2 fls2Δ/fls2Δ mutant plants, respectively. drp1arsw9 FLS2-GFP, drp1asalk FLS2-GFP, and drp1asalk drp2b-2 FLS2-GFP lines were generated by pollinating stigma of FLS2Pro:FLS2-3xMyc-EGFP (Beck et al., 2012; Smith et al., 2014a) with pollen from DRP1A/drp1arsw9 drp2b-2/drp2b-2 or DRP1A/drp1asalk drp2b-2/drp2b-2 mutant plants.
Genotyping was performed using standard PCR techniques with allele-specific primers (Supplemental Table S1). CAPS analysis was used to confirm the drp1arsw9 point mutation. A PCR fragment spanning the point mutation was PCR amplified using DRP1A primers (Supplemental Table S1; DRP1Arsw9 Short F and DRP1Arsw9 Short R) and directly subjected to restriction enzyme digest using HinfI with HinfI cleaving DRP1A WT (Col-0) but not drp1arsw9 mutant fragment due to a point mutation in drp1arsw9 at 2,314 bp (Collings et al., 2008; Supplemental Figure S1, B).
Seeds were surface-sterilized, germinated, and grown on 0.5× Murashige and Skoog (MS) medium +1% (w/v) sucrose solidified with 0.6% (w/v) agar (MS plates), in a 16-h light/8-h dark cycle photoperiod using 82 μmol m−2 s−1. Experiments utilizing double or triple mutants were performed on progeny derived from DRP1A/drp1a drp2b-2/drp2b-2 or DRP1A/drp1a drp2b-2/drp2b-2 fls2Δ/fls2Δ, respectively. Unless specified otherwise, 7-d-old seedlings were used for all seedling assays. For leaf assays, 7-d-old seedlings were transplanted into soil and grown at 22°C in a 8-h light/16-h dark cycle photoperiod at 82 μmol m−2 s−1 for indicated times. Except for tissue infiltration, seedlings or leaf tissues were floated on sterile dH2O overnight at 22°C to reduce wounding response prior to any flg22-assay.
Silique length, root length, and fresh weight measurements
For silique and root length measurements, self-pollinated siliques from 8- to 10-week-old plants and roots from 7-d-old seedlings grown at 24 h light, respectively, were traced using Fiji Free-hand tool. The same seedlings used for root measurements were utilized for fresh weight measurements.
Flg22 peptide
Active flg22 (QRLSTGSRINSAKDDAAGLQIA) peptide (Gómez-Gómez and Boller, 2000; Smith et al., 2014b) was made by GenScript (Scotch Plains, NJ) and used at indicated concentrations.
RNA isolation and RT-qPCR
For leaf assays in mature plants, three leaves of 5- to 6-week-old plants were syringe-infiltrated with flg22 at indicated concentrations and for indicated times for each genotype and treatment. For seedling assays, 4 seedlings of Col-0 or drp single mutants and 15 seedlings of drp1a drp2b double mutants were used. Total RNA was isolated from indicated tissue using Trizol Reagent (Sigma) according to the manufacturer’s protocol and processed for cDNA synthesis. Reverse transcription quantitative PCR (RT-qPCR) was performed as previously described (Anderson et al., 2011; Smith et al., 2014a) using gene-specific primers (Supplemental Table S1) with the SAND gene At2g28390 as a reference gene.
Apoplastic ROS production
Luminol-based ROS production in leaf tissue was performed as described (Heese et al., 2007; Smith et al., 2014a) using indicated flg22 concentrations. For FLS2-GFP tagged lines, ROS assays were performed on 7- to 8-d-old cotyledons that were cut in half and placed into the same well of a 96-well microplate for elicitation (Smith et al., 2014a). All ROS experiments shown in the same panel were performed in the same 96-well plate simultaneously to allow for direct comparison.
Bacterial pathogen assays
Syringe infiltration with P. syringae pv. tomato (Pto) DC3000 (OD600 = 0.0005) or Pto DC3000 hrcC− (OD600 = 0.02), collection of leaf discs, and quantification of bacterial growth using serial dilution plating were performed as previously described (Korasick et al., 2010), except that three leaf discs were ground in 300 μl dH2O before serial dilution plating.
Callose deposition and quantification
Leaves were syringe-infiltrated with indicated flg22 concentrations or DMSO (mock). At 24 h post-infiltration, leaf discs (0.2 cm2) were processed for aniline-blue staining as described previously (Leslie et al., 2016; Mason et al., 2020). Callose deposits were visualized by ultraviolet (UV) epifluorescence using a Leica M205 FA microscope (Leica Microsystems Inc.; Buffalo Grove, IL, USA). The percent area of the leaf disc covered by callose deposits was quantified using Fiji (Fiji is just ImageJ; NIH) software and the Trainable Weka Segmentation plug-in for Fiji/ImageJ as described in Leslie et al. (2016) and Mason et al. (2020).
SDCM for FLS2-GFP endocytosis and PI staining
Live-cell imaging for FLS2-GFP endocytosis and PI staining were carried out using a custom Olympus IX-71 inverted microscope (Center Valley, PA) equipped with a Yokogawa CSU-X1 5000 rpm spinning disc unit (Tokyo, Japan), Andor iXon Ultra 897 High-Speed EMCCD camera (Belfast, UK), PZ-2000 XYZ series automated stage with Piezo Z-axis top plate (Applied Scientific Instrumentation; Eugene, OR), and a 60×-silicon oil objective (Olympus UPlanSApo 60×/1.30 Sil). Images were captured using Andor iQ3 software (Belfast, UK).
For FLS2-GFP endocytosis experiments, 6-d-old Col-0 FLS2-GFP, drp2b-2 FLS2-GFP, drp1asalk FLS2-GFP, drp1arsw9 FLS2-GFP, or drp1asalk drp2b-2 FLS2-GFP seedlings were treated with 1 μM flg22 and assayed for indicated times as described previously in detail (Smith et al., 2014a; Leslie and Heese, 2017). The epidermal pavement cell layer of the adaxial cotyledon surface of at least four different seedlings was imaged per genotype/treatment with three to six fields of view for each cotelydon. GFP was excited with a Spectra Physics 488-nm diode laser (Santa Clara, CA), and fluorescence was collected through a series of Semrock Brightline 488-nm single-edge dichroic beam splitter and 500–550-nm bandpass filter (Rochester, NY). Camera exposure was set to 150 ms. For each image series, 68 consecutive images at a z-step interval of 0.31 μm (22 μm total depth) were captured. FLS2-GFP quantification was carried out using Fiji software and Advanced Weka Segmentation plug-ins for Fiji as previously described (Smith et al., 2014a; Leslie and Heese, 2017).
Measuring levels of FLS2-GFP at the PM (FLS2-GFP PM intensity) was performed as detailed previously (Smith et al., 2014a) with minor changes, in that the same maximum intensity projection images used for FLS2-GFP puncta quantification were utilized for FLS2-GFP PM intensity. PM regions were highlighted with the freehand tool using Fiji and subsequently analyzed for mean pixel intensity. For each image, the FLS2-GFP PM intensity was measured in five different PM regions, and the FLS2-GFP PM intensity was calculated as the average value of the pixel intensity. For each genotype and treatment, FLS2-GFP PM intensities were reported relative to the FLS2-GFP PM intensity of the un-elicited corresponding genotype at 0 min.
For PI staining, 7-d-old seedlings were rinsed in dH2O, stained with 200 μM PI (MP Biomedicals, Solon, OH) for 20 min at room temperature in the dark, rinsed again, and then imaged using SDCM. PI was excited with a Spectra Physics 561-nm diode laser (Santa Clara, CA), and fluorescence was collected through a series of Semrock Brightline 561-nm polychroic mirror and 561-nm bandpass filter (Rochester, NY). Camera exposure was set to 150 ms. A Z series of 40 images was taken with total Z depth of 12.4 μm and images of Z series were collapsed to obtain maximum intensity projections. Using the multipoint tool in Fiji, cell wall stubs were quantified by counting the number of cell wall stubs and the number of pavement cells in an image, which were then used to calculate the percent of cell wall stubs of the total number of pavement cells for each genotype.
Protein sample preparation, immunoblot analysis, and antibodies
Sample preparation and immunoblot analysis of total, soluble, and microsomal proteins were performed as previously described (Heese et al., 2007; Smith et al., 2014b; LaMontagne et al., 2016) with 25 or 20 µg loaded per well for leaf or seedling extracts, respectively. The following antibody dilutions were used: αDRP2, 1:4,000 (Backues et al., 2010); αDRP1A, 1:1,000 (Kang et al., 2003); 1:400 αFLS2 (Heese et al., 2007), 1:500 αBRI1 (kind gift of Marisa Otegui; Wu et al., 2011), 1:3,000 αMPK6 (Merkouropoulos et al., 2008), 1:500 αGFP (JL-8; Clontech Laboratories), 1:10,000 αAHA (AS07 260; Agrisera), 1:3,000 αCNX (AS12 2365; Agrisera; LaMontagne et al., 2016), and 1:300 actin (JLA20; Developmental Studies Hybridoma Bank (DSHB)),
PM enrichment from seedlings
Enrichment of PMs (ePM) from 7-d-old seedlings grown in 16-h light/8-h dark were performed as described (Collins et al., 2017, 2020) with the following changes. For Col-0, drp2b-2 and drp1a single mutants approximately 400 seedlings/genotype, and for the drp1a drp2b double mutants, 800 seedlings/genotype were flash-frozen in liquid nitrogen and processed for ePM using differential centrifugation and 0.02% (w/v) Brij-58 at a protein-to-detergent ratio of 1 μg microsomal protein to 2 μL 0.02% (w/v) Brij-58 solution. Final pellet fractions (ePM) were resuspended to 1.5 μg/μL for immunoblot analysis.
Statistical analysis
Each experiment represented a biological replicate that consisted of biologically distinct samples and was performed at least three independent times with similar results. For each experiment, mutant samples were compared with Col-0 WT samples, and statistical analyses were performed using n values with each n representing a biological sample as detailed in each figure legend. Statistical significances were based on ordinary one-way ANOVA as stated in the figure legends. Statistical significances were determined with GraphPad Prism 8.3.1 software (La Jolla, CA). Grubbs test with the Alpha = 0.05 (standard) was performed on the data sets to calculate outliers using GraphPad QuickCalcs outlier calculator (https://www.graphpad.com/quickcalcs/Grubbs1.cfm).
Accession numbers
DRP1A, AT5G42080; DRP2B, AT1G59610; FLS2, AT5G46330; WRKY33, AT2G38470; WRKY40, AT1G80840; PR1, AT2G14610; PHI1, AT1G35140; FRK1, AT2G19190; BRI1, AT4G394000.
Supplemental data
Supplemental Figure S1. Isolation and confirmation of drp1a mutant alleles.
Supplemental Figure S2. Loss of DRP1A results in reduced rosette leaf size compared with drp2b and Col-0 plants.
Supplemental Figure S3. drp1a mutant plants show similar accumulation of WRKY33 mRNA, WRKY40 mRNA, FLS2 mRNA, and FLS2 protein to Col-0.
Supplemental Figure S4 . FLS2-GFP and endogenous FLS2 protein accumulation is similar in drp1a and Col-0 seedlings.
Supplemental Figure S5. Similar to drp1asalk, cotyledons of drp1arsw9 mutant seedlings show impaired flg22-induced endocytosis of FLS2-GFP.
Supplemental Figure S6. Steady-state accumulation of DRP1A and DRP2B mRNA in drp single and double mutant seedlings.
Supplemental Figure S7. drp1a drp2b double mutants hyperaccumulate FLS2 protein and FLS2 mRNA.
Supplemental Figure S8. Combinatorial loss of DRP1A and DRP2B results in hyperaccumulation of FLS2 but not BRI1.
Supplemental Figure S9. Combinatorial loss of DRP1A and DRP2B results in reduced FRK1 but not MYB51 mRNA accumulation in response to flg22.
Supplemental Figure S10. FLS2-GFP and endogenous FLS2 proteins hyperaccumulate in drp1asalk drp2b-2 FLS2-GFP mutant lines.
Supplemental Table S1. Primer list.
Supplementary Material
Acknowledgments
The authors thank Dr. Scott Peck (University of Missouri–Columbia (MU)) for MPK6 antibody; Dr. Silke Robatzek (Ludwig Maximilian Universität München, Germany) for the Col-0 FLS2-GFP line; Dr. Marisa Otegui (University of Wisconsin–Madison) for BRI1 antibody; Drs. Walter Gassmann (MU), Scott Peck (MU), Jim Schoelz (MU), and present and former Heese laboratory members for discussions; and Dr. Michelle Leslie (MU) for help with SDCM. The callose images were acquired at the MU Molecular Cytology Core facility.
Funding
This work was supported by grants and fellowships from the National Science Foundation (NSF)-IOS 1147032 and NSF-IOS 1025837 (A.H.); NSF-GRF 1443129 (E.D.L.); NSF-MCB 1121998 and NSF-MCB 1614915 (S.Y.B.); National Institute of Health (NIH)-Initiative Maximizing Student Diversity (IMSD) Fellowship R25 GM056901 (G.E. and P.H.K.); University of Missouri (MU)-Diane P. and Robert E. Sharp Fund Fellowship (G.E.); Daniel F. Millikan Graduate Fellowship (MU-Division of Plant Sciences; J.M.S.); MU-College of Agriculture, Natural Food and Resources-Undergraduate Research Internship (H.M.S) and Dudley Alexander Undergraduate Research On-Campus Internships Gift Fund (S.J.R.); MU-Discovery Fellowship (K.B.J.); and MU-Life Sciences Undergraduate Research Opportunity Fellowship (K.B.J.).
Conflict of interest statement. None declared.
A.H. supervised the experiments. G.E. and J.M.S. performed the experiments with assistance from S.J.R., K.B.J., H.M.S., P.H.K., E.D.L., and P.H.K. P.V.C. provided the technical assistance for spinning disc confocal microscopy. S.Y.B. provided the material and technical assistance. A.H., G.E., and J.M.S. designed the experiments and analyzed the data. A.H. and G.E. wrote the article with the assistance of S.Y.B. and J.M.S. A.H. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Antje Heese (heesea@missouri.edu).
References
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S,, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci U S A 109:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Bartels S, Gonzalez Besteiro MA, Shahollari B, Ulm R, Peck SC (2011) Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J 67:258–268 [DOI] [PubMed] [Google Scholar]
- Backues SK, Korasick DA, Heese A, Bednarek SY (2010) The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22:3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S (2012) Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24:4205–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek SY, Backues SK (2010) Plant dynamin-related protein families DRP1 and DRP2 in plant development. Biochem Soc Trans 38:797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci U S A 109:297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khaled S, Postma J, Robatzek S (2015) A moving view: subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu Rev Phytopathol 53:379–402 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng S-H, Sheen J (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464:418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55:257–286 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Frescatada-Rosa M, Men S, Chow C-M, Ebine K, Gustavsson A, Johansson L, Ueda T, Moore I, Jürgens G. et al. (2010) Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J 29:546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6:e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Schwizer S, Sklenar J, Yoshida K, Petre B, Bos JI, Schornack S, Jones AM, Bozkurt TO, Kamoun S (2015) Phytophthora infestans RXLR-WY effector AVR3a associates with dynamin-related protein 2 required for endocytosis of the plant pattern recognition receptor FLS2. PLoS ONE 10:e0137071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus LAN, Savatin DV, Russinova E (2018) The crossroads of receptor-mediated signaling and endocytosis in plants. J Integr Plant Biol 60:827–840 [DOI] [PubMed] [Google Scholar]
- Collings DA, Gebbie LK, Howles PA,, Hurley UA, Birch RJ, Cork AH, Hocart CH, Arioli T, Williamson RE (2008) Arabidopsis dynamin-like protein DRP1A: a null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J Exp Bot 59:361–376 [DOI] [PubMed] [Google Scholar]
- Collins CA, LaMontagne ED, Anderson JC, Ekanayake G, Clarke AS, Bond LN, Salamango DJ,, Cornish PV, Peck SC, Heese A (2020) EPSIN1 modulates the plasma membrane abundance of FLAGELLIN SENSING2 for effective immune responses. Plant Physiol 182:1762–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Leslie ME, Peck SC, Heese A (2017) Simplified enrichment of plasma membrane proteins from Arabidopsis thaliana seedlings using differential centrifugation and Brij-58 treatment. Methods Mol Biol 1564:155–168 [DOI] [PubMed] [Google Scholar]
- Cui Y, Li X, Yu M, Li R, Fan L, Zhu Y, Lin J (2018) Sterols regulate endocytic pathways during flg22-induced defense responses in Arabidopsis. Development 145 [DOI] [PubMed] [Google Scholar]
- Ekanayake G, LaMontagne ED, Heese A (2019) Never walk alone: clathrin-coated vesicle (CCV) components in plant immunity. Annu Rev Phytopathol 57:387–409 [DOI] [PubMed] [Google Scholar]
- Frescatada-Rosa M, Stanislas T, Backues SK, Reichardt I, Men S, Boutté Y, Jürgens G, Moritz T, Bednarek SY, Grebe M (2014) High lipid order of Arabidopsis cell-plate membranes mediated by sterol and DYNAMIN-RELATED PROTEIN1A function. Plant J 80:745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Nakazono M, Tsutsumi N (2008) Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep 27:1581–1586 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N (2010) Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc Natl Acad Sci U S A 107:6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Tsutsumi N (2014) Dynamin-related proteins in plant post-Golgi traffic. Front Plant Sci 5:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G. et al. (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156:691–704 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011 [DOI] [PubMed] [Google Scholar]
- Gu Y, Zavaliev R, Dong X (2017) Membrane trafficking in plant immunity. Mol Plant 10:1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A 104:12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Bednarek SY, Blumwald E, Hwang I, Jurgens G, Menzel D, Osteryoung KW, Raikhel NV, Shinozaki K, Tsutsumi N.. et al. (2003) A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol Biol 53:261–265 [DOI] [PubMed] [Google Scholar]
- Irani NG, Di Rubbo S, Mylle E, Van den Begin J, Schneider-Pizoń J, Hniliková J, Šíša M, Buyst D, Vilarrasa-Blasi J, Szatmári A-M. et al. (2012) Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol 8:583–589 [DOI] [PubMed] [Google Scholar]
- Kang B-H, Busse J, Dickey C, Rancour D, Bednarek S (2001) The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol 126:47–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B-H, Busse JS, Bednarek SY (2003) Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15:899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285:39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY (2008) Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol 147:1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, McMichael C, Walker KA, Anderson JC, Bednarek SY, Heese A (2010) Novel functions of stomatal cytokinesis-defective 1 (SCD1) in innate immune responses against bacteria. J Biol Chem 285:23342–23350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMontagne ED, Collins CA, Peck SC, Heese A (2016) Isolation of microsomal membrane proteins from Arabidopsis thaliana. Curr Protoc Plant Biol 1:217–234 [DOI] [PubMed] [Google Scholar]
- Lee D, Lal NK, Lin ZD, Ma S, Liu J, Castro B, Toruño T, Dinesh-Kumar SP, Coaker G (2020) Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun 11:1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie ME, Heese A (2017) Quantitative analysis of ligand-induced endocytosis of FLAGELLIN-SENSING 2 using automated image segmentation. Methods Mol Biol 1578: 39–54 [DOI] [PubMed] [Google Scholar]
- Leslie ME, Rogers SW, Heese A (2016) Increased callose deposition in plants lacking DYNAMIN-RELATED PROTEIN 2B is dependent upon POWDERY MILDEW RESISTANT 4. Plant Signal Behav 11:e1244594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Claus LAN, Leslie ME, Tao K, Wu Z, Liu J, Yu X, Li B, Zhou J, Savatin DV. et al. (2020) Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581:199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Aliaga Fandino AC, Koh SWH, Jelínková A, Kolb M, Janacek DP, Breda AS, Cattaneo P, Hammes UZ, Petrášek J. et al. (2020) Plasma membrane domain patterning and self-reinforcing polarity in Arabidopsis. Dev Cell 52:223–235 [DOI] [PubMed] [Google Scholar]
- Mason KN, Ekanayake G, Heese A (2020) Staining and automated image quantification of callose in Arabidopsis cotyledons and leaves. Methods Cell Biol 160:181–199 [DOI] [PubMed] [Google Scholar]
- Mbengue M, Bourdais G, Gervasi F, Beck M, Zhou J, Spallek T, Bartels S, Boller T, Ueda T, Kuhn H. et al. (2016) Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci U S A 113:11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Andreasson E, Hess D, Boller T, Peck SC (2008) An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J Biol Chem 283:10493–10499 [DOI] [PubMed] [Google Scholar]
- Mravec J, Petrasek J, Li N, Boeren S, Karlova R, Kitakura S, Parezova M, Naramoto S, Nodzynski T, Dhonukshe P. et al. (2011) Cell plate restricted association of DRP1A and PIN proteins is required for cell polarity establishment in Arabidopsis. Curr Biol 21:1055–1060 [DOI] [PubMed] [Google Scholar]
- Orosa B, Yates G, Verma V, Srivastava AK, Srivastava M, Campanaro A, De Vega D, Fernandes A, Zhang C, Lee J. et al. (2018) SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat Commun 9:5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez Valencia J, Goodman K, Otegui MS (2016) Endocytosis and endosomal trafficking in plants. Annu Rev Plant Biol 67:309–335 [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL (2018) The dynamin superfamily. Curr Biol 28:R411–R416 [DOI] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68:100–113 [DOI] [PubMed] [Google Scholar]
- Reynolds GD, Wang C, Pan J, Bednarek SY (2018) Inroads into internalization: five years of endocytic exploration. Plant Physiol 176:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Wirthmueller L (2013) Mapping FLS2 function to structure: LRRs, kinase and its working bits. Protoplasma 250:671–681 [DOI] [PubMed] [Google Scholar]
- Smith JM, Leslie ME, Robinson SJ, Korasick DA, Zhang T, Backues SK, Cornish PV, Koo AJ, Bednarek SY, Heese A (2014a) Loss of Arabidopsis thaliana dynamin-related protein 2B reveals separation of innate immune signaling pathways. PLoS Pathog 10:e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A (2014b) Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol 164:440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T, Hüser A, Barbosa ICR, Kiefer CS, Brackmann K, Pietra S, Gustavsson A, Zourelidou M, Schwechheimer C, Grebe M (2015) Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat Plants 1:15162. [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14:519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L. et al. (2019) A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572:131–135 [DOI] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X-F, Kvitko B, He SY (2018) Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16:316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Kimberlin AN, Elowsky CG, Liu Y, Gonzalez-Solis A, Cahoon EB, Alfano JR (2019) A plant immune receptor degraded by selective autophagy. Mol Plant 12:113–123 [DOI] [PubMed] [Google Scholar]
- Yoshinari A, Fujimoto M, Ueda T, Inada N, Naito S, Takano J (2016) DRP1-dependent endocytosis is essential for polar localization and boron-induced degradation of the borate transporter BOR1 in Arabidopsis thaliana. Plant Cell Physiol 57:1985–2000 [DOI] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55:109–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Peck SC (2011) Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana. Proteomics 11:1780–1788 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.