Abstract
Seeds of the root parasitic plant Striga hermonthica can sense very low concentrations of strigolactones (SLs) exuded from host roots. The S. hermonthica hyposensitive to light (ShHTL) proteins are putative SL receptors, among which ShHTL7 reportedly confers sensitivity to picomolar levels of SL when expressed in Arabidopsis thaliana. However, the molecular mechanism underlying ShHTL7 sensitivity is unknown. Here we determined the ShHTL7 crystal structure and quantified its interactions with various SLs and key interacting proteins. We established that ShHTL7 has an active-site pocket with broad-spectrum response to different SLs and moderate affinity. However, in contrast to other ShHTLs, we observed particularly high affinity of ShHTL7 for F-box protein AtMAX2. Furthermore, ShHTL7 interacted with AtMAX2 and with transcriptional regulator AtSMAX1 in response to nanomolar SL concentration. ShHTL7 mutagenesis analyses identified surface residues that contribute to its high-affinity binding to AtMAX2 and residues in the ligand binding pocket that confer broad-spectrum response to SLs with various structures. Crucially, yeast-three hybrid experiments showed that AtMAX2 confers responsiveness of the ShHTL7–AtSMAX1 interaction to picomolar levels of SL in line with the previously reported physiological sensitivity. These findings highlight the key role of SL-induced MAX2–ShHTL7–SMAX1 complex formation in determining the sensitivity to SL. Moreover, these data suggest a strategy to screen for compounds that could promote suicidal seed germination at physiologically relevant levels.
The strigolactone receptor ShHTL7 of Striga hermonthica displays high-affinity interactions with two downstream signaling components, forming a highly sensitive tripartite complex.
Introduction
The parasitic plant Striga hermonthica produces seeds that can remain dormant in the soil for up to 15 years until they detect a suitable host to which they can attach, such as corn (Zea mays) and sorghum (Sorghum officianale; Nickrent, 2004). Striga withdraws water and nutrients from the host causing considerable damage, posing a serious threat to food security. Striga has been estimated to cost about $1 billion per year in crop losses and affects 100 million people in sub-Saharan Africa (Parker, 2009; Spallek et al., 2013). The germination of Striga seeds depends on strigolactones (SLs) released from the host, which also function as a signal to promote development of the interaction with symbiotic fungi to form arbuscular mycorrhiza (Cook et al., 1966; Akiyama et al., 2005). SLs were also identified as plant hormones that control shoot branching and other aspects of plant growth and development (Gomez-Roldan et al., 2008; Umehara et al., 2008; Brewer et al., 2013; Wang and Smith, 2016; Bouwmeester et al., 2019). The structure of canonical SLs consists of a butenolide ring (D-ring), which connects through an enol-ether bond to a tricyclic moiety (ABC rings). More recently, noncanonical SLs have been identified that lack an intact ABC ring (Waters et al., 2017; Wang and Bouwmeester, 2018; Aliche et al., 2020).
The receptor for SLs is DWARF14 (D14) in rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), which belongs to the α/β hydrolase family and also functions as an enzyme (Arite et al., 2009; Hamiaux et al., 2012; Waters et al., 2012; Nakamura et al., 2013; Zhao et al., 2013, 2015; Chevalier et al., 2014). Upon binding to the pocket of the receptor, SL hydrolysis is mediated by a conserved Ser–His–Asp catalytic triad, resulting in attachment of the D-ring to the active-site His as a covalently linked intermediate molecule (CLIM), which is considered crucial for receptor activation (de Saint Germain et al., 2016; Yao et al., 2016, 2018; Burger and Chory, 2020). Another highly conserved component in SL signaling is an F-box protein from the SKP1-CULLIN-F-BOX complex, known as MORE AXILLARY GROWTH2 (MAX2) in Arabidopsis (Stirnberg et al., 2002, 2007; Shen et al., 2007) or D3 in rice (Ishikawa et al., 2005). D14 undergoes SL-triggered conformational changes and subsequently interacts with MAX2/D3, causing the ubiquitination and degradation of four members of a family of transcriptional repressors known as SUPPRESSOR OF MAX2 1 (SMAX1)-LIKE (SMXL) in Arabidopsis and DWARF53 (D53) in rice (Jiang et al., 2013; Stanga et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015, 2020a, 2020b; Liang et al., 2016; Fang et al., 2020; Hu et al., 2020; Xie et al., 2020,). Among the eight-member SMXL family, SMAX1 is another important repressor that controls seed germination and seedling development in the karrikin (KAR) signaling pathway (,Stanga et al., 2013; Morffy et al., 2016; Khosla et al., 2020,), which occurs via the D14 paralog known as KARRIKIN-INSENSITIVE2 (KAI2) or HYPOSENSITIVE TO LIGHT (HTL) in a MAX2-dependent way (Nelson et al., 2011; Waters et al., 2012; Lumba et al., 2017a,).
In S. hermonthica, 11 diverse KAI2/HTL paralogs have been reported to encode putative SL receptors (ShHTLs), and are distributed into three phylogenetic clades: the conserved KAI2-like clade, including ShHTL1; the intermediate clade comprising ShHTL2 and ShHTL3; and the divergent clade containing ShHTL4 to ShHTL11, which have ligand binding pockets resembling D14 according to homology models and crystal structures (Conn et al., 2015; Toh et al., 2015; Xu et al., 2018; Zhang et al., 2020). Due to the technical limitations of genetic manipulation in Striga, investigations of ShHTL function have been carried out in Arabidopsis by transformation of Athtl or Atkai2 mutants with ShHTL genes. Germination assays of ShHTL activity in such transgenic Arabidopsis plants demonstrated that ShHTL2 and ShHTL3 respond to KAR but not SL, whereas ShHTL10 and ShHTL11 show no germination with either SL or KAR (Conn et al., 2015; Toh et al., 2015), which suggests that these ShHTLs do not participate in germination but might have other functions in Striga. The divergent clade family members, ShHTL4–ShHTL9, do not result in KAR-induced germination but respond to various SLs when expressed in Arabidopsis (Toh et al., 2015). ShHTL7 is particularly notable because Arabidopsis seeds expressing this protein respond to extremely low concentrations of SLs at picomolar levels, which has only been seen for the germination of weeds such as Striga (Toh et al., 2015; Bunsick et al., 2020).
ShHTL7 was further confirmed as an SL receptor that forms the ShHTL7–CLIM complex after hydrolyzing SL and that interacts with both ShMAX2 and AtMAX2 and with SMAX1 in a SL-dependent manner (Yao et al., 2017; Uraguchi et al., 2018). Importantly, SL hydrolysis and CLIM modification on ShHTL7 were demonstrated to be essential for full activation of the receptor (Uraguchi et al., 2018), although another model proposed that hydrolysis may not be a requirement for SL signaling (Seto et al., 2019). These observations imply that SL perception and signal transduction mechanisms are conserved between Striga and Arabidopsis (Lumba et al., 2017b). Due to the high sensitivity of ShHTL7 in the SL-induced germination assay, searching for ligands that could be perceived by ShHTL7 to trigger germination is a promising approach to combat Striga. Recently, several compounds have been identified that bind with ShHTL7, such as Triton X-100 and Triton X-100-like molecules, which could block ShHTL7 activity but have no effect on the growth of rice (Shahul Hameed et al., 2018). Furthermore, a synthetic compound named SPL7 has been developed to specifically target ShHTL7 for the germination of Striga, whose potency is comparable with that of (+)-5-deoxystrigol (5DS), a natural SL that was considered to be the most potent commercially available Striga germination stimulant (Uraguchi et al., 2018). These results suggest that ShHTL7 has high sensitivity in SL-induced Striga seed germination as well as broad-spectrum recognition capability for various SL molecules and related compounds.
Investigating the molecular basis for the sensitivity and selectivity of SL receptors toward different SLs would provide a means to understand the mechanism and evolution of signal perception by SL receptors as well as novel strategies for the control of root parasitic plants including Striga. Studies of amino acid residues that determine active site pocket size and topography of the receptor alone have so far failed to explain the very high sensitivity of ShHTL7 to SL in planta (Conn et al., 2015; Shahul Hameed et al., 2018; Xu et al., 2018). Thus, the aim of the present study was to investigate the SL sensitivity and selectivity of ShHTL7 during the formation of SL signaling protein complexes. Here, we report the crucial role of SL-induced MAX2–ShHTL7–SMAX1 complex formation in determining the sensitivity to SL. These results provide insight into the mechanism of SL action and rational design of a potent suicidal germination stimulant for combating Striga.
Results
ShHTL7 ligand binding affinity and hydrolysis activity
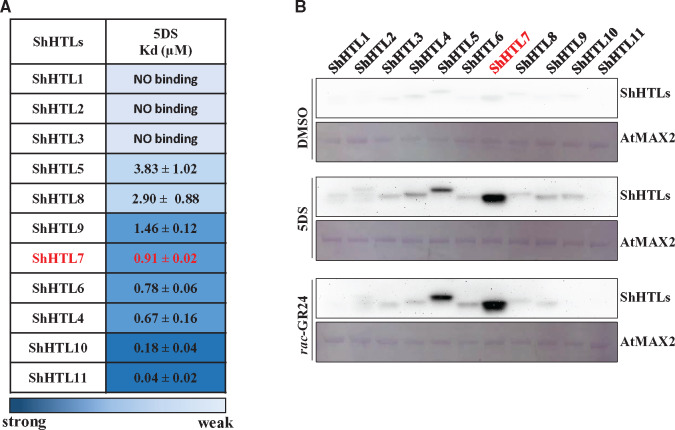
Eleven ShHTL proteins (ShHTL1–11; Toh et al., 2015) were expressed in Escherichia coli, purified to homogeneity, and characterized (Supplemental Figure S1A). Binding affinities of these proteins for 5DS were measured using isothermal titration calorimetry (ITC; Figure 1A;Supplemental Figure S2 and Supplemental Table S2). Unsurprisingly, no significant binding was detected for the highly conserved and intermediate clade members ShHTL1, ShHTL2, and ShHTL3, which are reported to respond to KAR but not to 5DS (Toh et al., 2015; Tsuchiya et al., 2015). Unexpectedly, the strongest binding affinity with 5DS was observed for ShHTL11 (Kd = 0.04 µM) and followed by ShHTL10 (Kd = 0.18 µM), and the weakest binding was observed for ShHTL5 and ShHTL8 (Kd = 3.83 and 2.90 µM, respectively). ShHTL4, ShHTL6, ShHTL7, and ShHTL9 all showed an intermediate level of binding with 5DS, from 0.67 µM (ShHTL4) to 1.46 µM (ShHTL9). The 5DS-binding affinities of ShHTLs were not consistent with the germination assays in Arabidopsis, in which ShHTL4, ShHTL5, and ShHTL7 showed the most sensitive germination responses but ShHTL10 and ShHTL11 exhibited very weak or no response to 5DS (Toh et al., 2015). These observations imply that the ligand-binding affinity is not the determinant of the germination activity of these receptors.
Figure 1.
ShHTL7 has moderate affinity for SL but high affinity for AtMAX2. (A) Dissociation constants (Kd) for 5DS binding to each ShHTL protein determined by ITC. Data are means sd (n = 3). (B) In vitro pull-down assays of the interactions between each ShHTL protein and AtMAX2 in the absence or presence of 1 μM 5DS or rac-GR24. The GST-AtMAX2 bait protein was visualized by staining the PVDF membrane with Memstain to show equal loading. The ShHTL proteins with His6-Flag tag were detected with anti-Flag antibody.
We further investigated ShHTL6 and ShHTL7 since they showed similar affinity for 5DS (Kd = 0.78 and 0.91 µM, respectively), yet ShHTL7 provided Arabidopsis seeds with much greater (about 1,700-fold higher) sensitivity than ShHTL6 (Toh et al., 2015). We assayed the hydrolytic activities of these two proteins using the bioactive pro-fluorescent substrate Yoshimulactone Green (YLG) and obtained similar Km values for ShHTL6 and ShHTL7 of 0.85 µM and 1.17 µM, respectively, whereas ShHTL6 had a five-fold higher kcat value than ShHTL7 (Supplemental Figure S3). This result is consistent with data showing that GR24 is hydrolyzed more rapidly by ShHTL6 than by ShHTL7 (Tsuchiya et al., 2015). Potentially, this could be explained by the formation of a relatively more stable CLIM intermediate by ShHTL7. However, overall the ligand binding affinity and hydrolytic activity of ShHTL7 are not sufficiently different to those of other ShHTL proteins to explain the extremely high germination sensitivity to SL when ShHTL7 is expressed in Arabidopsis.
Binding of ShHTLs to interacting signaling proteins
We investigated the interactions between ShHTLs and F-box protein AtMAX2 using in vitro pull-down assays. Purified GST-AtMAX2 was incubated with each purified ShHTL-His6-Flag protein in the presence of 1 µM rac-GR24 or 5DS followed by glutathione–sepharose affinity chromatography and immunoblotting using anti-Flag antibody. ShHTL7 had the strongest interaction with AtMAX2, whereas ShHTL5 also showed obvious interaction, and both were dependent on rac-GR24 or 5DS (Figure 1B).
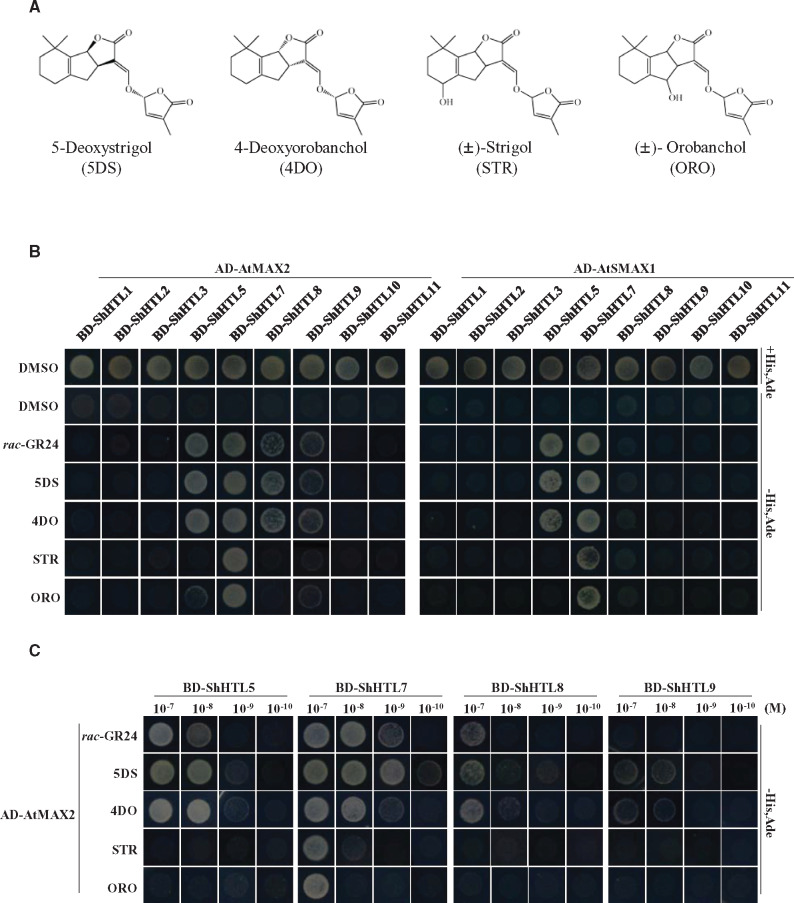
To further investigate such interactions, we turned to the relatively more efficient yeast two-hybrid (Y2H) assays to examine the interaction of ShHTL proteins individually with AtMAX2 and with AtSMAX1. Each of the ShHTL proteins was expressed as a fusion with the GAL4 DNA-binding domain (BD), whereas AtMAX2 and AtSMAX1 were each fused to the activation domain (AD). We were unable to evaluate interactions with ShHTL4 and ShHTL6 by Y2H because these proteins exhibited autoactivation. The AtMAX2 construct included a C-terminal fusion with AtASK1 to stabilize AtMAX2 (Yao et al., 2016; Xu et al., 2018). First, we tested the effects of 1 µM rac-GR24 and each of four natural SLs, specifically 5DS, 4-deoxyorobanchol (4DO), (±)-strigol (STR), and (±)-orobanchol (ORO; Figure 2A), on the protein–protein interactions. ShHTL7 interacted strongly with AtMAX2 and with AtSMAX1 in response to all SLs (Figure 2B). Interaction with AtMAX2 was also observed with ShHTL5, ShHTL8, and ShHTL9 in response to rac-GR24, 5DS, and 4DO. These results are broadly consistent with results of the pull-down experiments showing very strong interaction of AtMAX2 with ShHTL7 and moderate interaction with ShHTL5. Interaction of ShHTL5 with AtSMAX1 was also seen in response to rac-GR24, 5DS, and 4DO (Figure 2B). The results further indicate that ShHTL7 has broad-spectrum response to various SLs in relation to the binding to AtMAX2 and to AtSMAX1.
Figure 2.
ShHTL7 interacts with AtMAX2 and AtSMAX1 with high SL sensitivity and low SL specificity. (A) The structures of the four natural SLs used in this and subsequent assays. STR and ORO were used as racemic mixtures. (B) The interactions between ShHTLs and AtMAX2 or AtSMAX1 separately in the presence of 1 µM of rac-GR24, 5DS, 4DO, STR, and ORO were assessed in Y2H assays. AtMAX2 and AtSMAX1 were used as prey fused with Gal4 AD AD-AtMAX2 and AD-SMAX1 and ShHTL proteins were fused with the BD (BD-ShHTL). DMSO was used as solvent for SLs and as negative control in assays. (C) The Y2H assay was used to detect the binding of selected BD-ShHTLs with AD-AtMAX2 in the presence of 10−7 to 10−10 M rac-GR24, 5DS, 4DO, STR, and ORO. Interactions were detected in the control medium and the selective medium SD/-Leu/-Trp/-His/-Ade (-His, Ade).
To investigate the sensitivity of ShHTLs to these five SLs, we carried out Y2H assays of interactions between ShHTLs and AtMAX2 with SLs at concentrations from 0.1 nM to 0.1 µM. We focused on ShHTL5, ShHTL7, ShHTL8, and ShHTL9, each of which had shown interaction with AtMAX2 in the presence of SLs at 1 µM. Importantly, ShHTL7 interacted with AtMAX2 in response to all five SLs and at lower concentrations than that observed for the other ShHTL proteins (Figure 2C). Specifically, ShHTL7 interacted with AtMAX2 at 1 nM rac-GR24 and 5DS, 10 nM 4DO, and 100 nM STR and ORO (Figure 2C). ShHTL5 showed clear interaction in response to 10 nM rac-GR24, 5DS, or 4DO, whereas ShHTL8 and ShHTL9 required 10- or 100-fold higher concentration. The interaction of these ShHTLs with AtMAX2 correlated very well with the bioactivity of these receptors in the Arabidopsis germination assay, in which ShHTL7 showed greatest sensitivity to GR24 followed by ShHTL5, with ShHTL8 and ShHTL9 showing least sensitivity (Toh et al., 2015). These results further indicate that ShHTL7 has a broad-spectrum response to SLs for binding with AtMAX2. The higher SL sensitivity in AtMAX2 interaction suggested that the very high bioactivity of ShHTL7 might be attributed by its binding affinity with AtMAX2.
Since we were unable to obtain results for ShHTL4 and ShHTL6 in yeast, a quantitative ‘proximity fluorescence’ (AlphaScreen) assay was used to measure the rac-GR24-dependent binding of AtMAX2 to ShHTL4, ShHTL6, and ShHTL7. These experiments showed that ShHTL7 binding to AtMAX2 was significantly stronger than that of ShHTL4 or ShHTL6 and it responded more sensitively to rac-GR24 (Supplemental Figure S4, A and B), consistent with the pull-down experiments (Figure 1B).
Formation of a ShHTL–MAX2–SMAX1 complex in response to SL
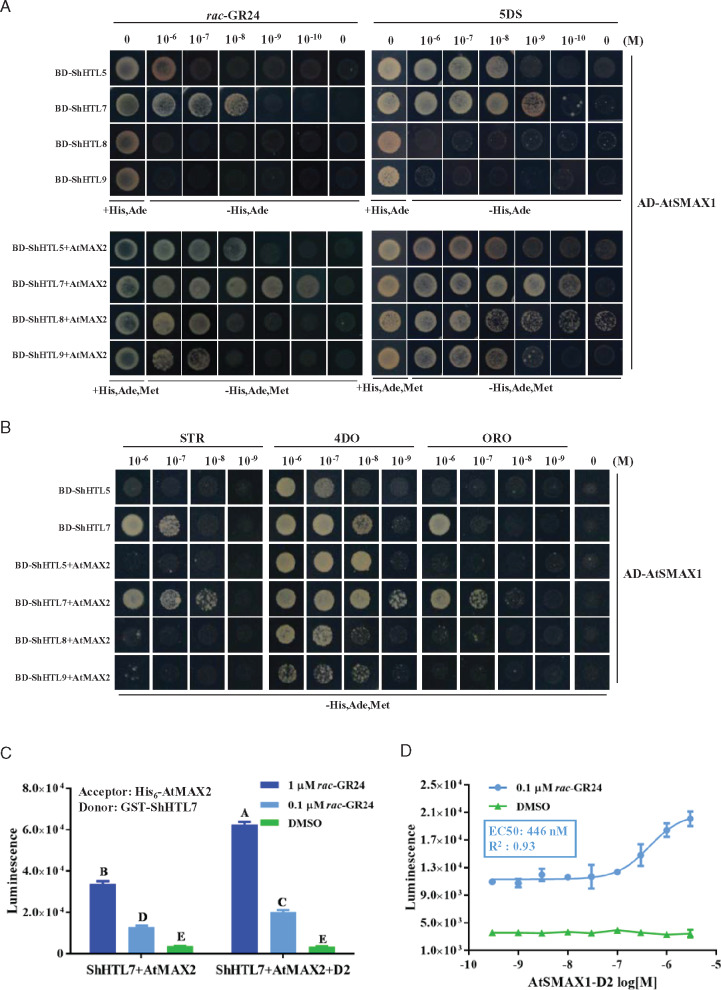
We observed that there was no significant interaction between AtSMAX1 and ShHTL8 or ShHTL9 in the presence of 1 µM rac-GR24, at which concentration seed germination in the ShHTL8 and ShHTL9 transgenic Arabidopsis plants was stimulated (Toh et al., 2015). We therefore considered whether AtMAX2 and AtSMAX1 could act in a concerted or synergistic manner in the binding to ShHTL. To measure the tripartite interaction, a yeast three-hybrid assay (Y3H) was employed. We focussed on ShHTL5, ShHTL7, ShHTL8, and ShHTL9 and analyzed their interaction with AtSMAX1 separately and in combination with AtMAX2. The interactions between BD-fused ShHTLs and AD-fused AtSMAX1 were analyzed with rac-GR24 and 5DS at concentrations over five orders of magnitude from 0.1 nM to 1 µM. ShHTL7 provided the greatest sensitivity to rac-GR24 and 5DS for the interactions, and ShHTL7 could bind to AtSMAX1 with 10 nM rac-GR24 or 1 nM 5DS, which is 100- and 10-fold lower than ShHTL5, respectively (Figure 3A). Furthermore, when BD-fused ShHTL proteins were co-expressed with AtMAX2 from a single plasmid, interaction with AD-fused AtSMAX1 was much more sensitive to rac-GR24 and 5DS for each ShHTL protein (Figure 3A). ShHTL7 interactions were observed at as low as 100 pM rac-GR24 and 5DS, which was two orders of magnitude lower than that for ShHTL5 and two or three orders of magnitude lower than that for ShHTL8 and ShHTL9 (Figure 3A). The sensitivity of these interactions is in accordance with the seed germination assays of Arabidopsis transgenic plants expressing these ShHTL genes, and strongly suggests that the tripartite interaction of ShHTL, AtSMAX1, and AtMAX2 might be the key factor contributing to SL sensitivity in germination. Besides, we observed that the interaction of AtSMAX1 with ShHTL7 was more sensitive to STR, 4DO, and ORO in the presence of AtMAX2 (Figure 3B). Similar results were obtained for ShHTL5 with 4DO, but STR and ORO did not stimulate its interaction with AtSMAX1 even in the presence of AtMAX2 (Figure 3B).
Figure 3.
AtMAX2 and AtSMAX1 enhance the SL-induced binding of each other to ShHTL. (A) Y2H and Y3H assay of interactions between AtSMAX1 and ShHTL5, ShHTL7, ShHTL8, and ShHTL9 in the absence or presence of AtMAX2 were detected in the presence of 10−6 to 10−10 M rac-GR24 (left panel) and 5DS (right panel). AtSMAX1 was fused with the Gal4 AD-SMAX1 and ShHTLs were fused with Gal4 DNA BD as bait (BD-ShHTLs). AtMAX2 fused with AtASK1 at its N-terminus was co-expressed with ShHTL5, ShHTL7, ShHTL8, and ShHTL9. (B) Y2H assay of the interaction of AD-AtSMAX1 with ShHTL5 and ShHTL7 and interaction of AD-AtSMAX1 with ShHTL5, ShHTL7, ShHTL8, and ShHTL9 in the presence of AtMAX2 were assessed in response to 4DO, STR, and ORO in the concentration range 10−6 to 10−9 M. Interactions were detected in the control medium and the selective medium SD/-Leu/-Trp/-His/-Ade (-His, Ade) for Y2H and SD/-Leu/-Trp/-His/-Ade/-Met (-His, Ade, Met) for Y3H. (C) The interaction of GST-ShHTL7 and HIS6-AtMAX2 were tested in the presence or absence of AtSMAX1-D2 in the AlphaScreen assay in response to 0.1 and 1 μM rac-GR24, with DMSO as the negative control. Error bars indicate se (n = 3). Two-way ANOVA analysis revealed significant differences between rac-GR24 concentrations and between presence and absence of AtSMAX1-D2 (P < 0.001). (D) AlphaScreen assay of the effect of AtSMAX1-D2 on the binding of GST-ShHTL7 to His6-AtMAX2 in response to GR24. A gradient concentration ranging from 3 to 3 × 10−5μM AtSMAX1-D2 was applied in the presence of 0.1 µM rac-GR24. The assay of GST-ShHTL7 and His6-AtMAX2 incubated with AtSMAX1-D2 in the presence of DMSO as the negative control. Error bars indicate se (n = 3). EC50 value was determined using nonlinear curve-fitting of graphs generated with Prism6 (GraphPad).
We further investigated whether ShHTL7 has high affinity for the Striga protein ShMAX2, which is the ortholog of AtMAX2. The function of ShMAX2 is conserved as it can replace AtMAX2 in the regulation of germination of Arabidopsis seeds (Liu et al., 2014). We first used the Y2H assay to test the interaction of ShHTLs with ShMAX2 in response to 1 μM 5DS, 4DO, STR, and ORO. The results are strikingly similar to those using AtMAX2, in that ShHTL7 interacted with ShMAX2 with all four SLs, and ShHTL5, ShHTL8, and ShHTL9 interacted mainly in response to 5DS and 4DO (Supplemental Figure S5A). Next, we investigated the interactions of ShHTL5, ShHTL7, ShHTL8, and ShHTL9 with ShMAX2 in response to 5DS, 4DO, STR, and ORO at concentrations over four orders of magnitude. We observed that ShHTL7 showed greatest sensitivity and broadest selectivity to these four SLs, which is similar to results with AtMAX2 except that ShHTL8 and ShHTL9 interacted more effectively with ShMAX2 than with AtMAX2 (Supplemental Figure S5B). We were unable to address ShHTL interaction with ShSMAX1 because the appropriate protein has not been identified. However, we did investigate whether ShMAX2 behaved in the same manner as AtMAX2 in a tripartite interaction with AtSMAX1 and ShHTLs. We examined the tripartite interaction of the same four ShHTLs with AtSMAX1, alongside ShMAX2, in response to 5DS at concentrations over five orders of magnitude. The results confirm that ShHTL7 responds most sensitively to 5DS in its interaction with AtSMAX1, whereas addition of ShMAX2 further enhances the sensitivity, which can be observed at 100 pM (Supplemental Figure S5C). It was also seen that ShHTL5, ShHTL8, and ShHTL9 interact with AtSMAX1 in combination with ShMAX2, with interaction seen at 1 nM (Supplemental Figure S5C). In summary, the studies in yeast establish that tripartite complexes are formed at lower SL concentrations and this could explain the very high sensitivity to SL provided by ShHTL7 in planta.
We found no evidence for direct interaction of AtMAX2 or ShMAX2 with AtSMAX1 in the Y2H system either in the presence or in the absence of rac-GR24 (Supplemental Figure S5D). Therefore, the strength of interactions between the proteins in the tripartite SL signaling complex is presumably a result of the affinity of the ShHTL protein for AtMAX2 and for AtSMAX1.
Previous studies with proteins from rice had suggested that the binding of MAX2 ortholog D3 to SL receptor D14 could stabilize D14 interaction with SMAX1-like protein D53 via its D2 domain (Shabek et al., 2018). It is potentially a general mechanism in SL signalling that MAX2 (or D3) and SMAX1-type proteins stabilize or enhance the binding of each other to the receptor. To further investigate the role of SMAX1 in the MAX2–ShHTL7 interaction, we identified the D2 domain of AtSMAX1 based on previous studies (Supplemental Figure S6A; Zhou et al., 2013; Shabek and Zheng, 2014; Shabek et al., 2018). We expressed and purified this 420-aa AtSMAX1-D2 protein for AlphaScreen assays to determine its effect on interaction between AtMAX2 and ShHTL7. In a SL-dependent manner, AtSMAX1-D2 was able to significantly enhance the binding of AtMAX2 and ShHTL7 (Figure 3, C and D; Supplemental Figure S6B). These results support the view that ShHTL7, AtMAX2, and AtSMAX1 bind cooperatively to form a stable ternary complex. However, we discovered surprisingly that the AtSMAX1-D2 domain alone could not interact with either AtMAX2 or ShHTL7 (Supplemental Figure S6C). This suggests that the interaction of AtMAX2 with ShHTL7 might create a new interface to which AtSMAX1-D2 binds, making the interaction of AtMAX2 with ShHTL7 stronger, which in turn contributes to the binding of AtSMAX1–ShHTL7. This model could explain why the binding of both AtMAX2 and AtSMAX1 makes the complex more stable.
Overall structure of ShHTL7
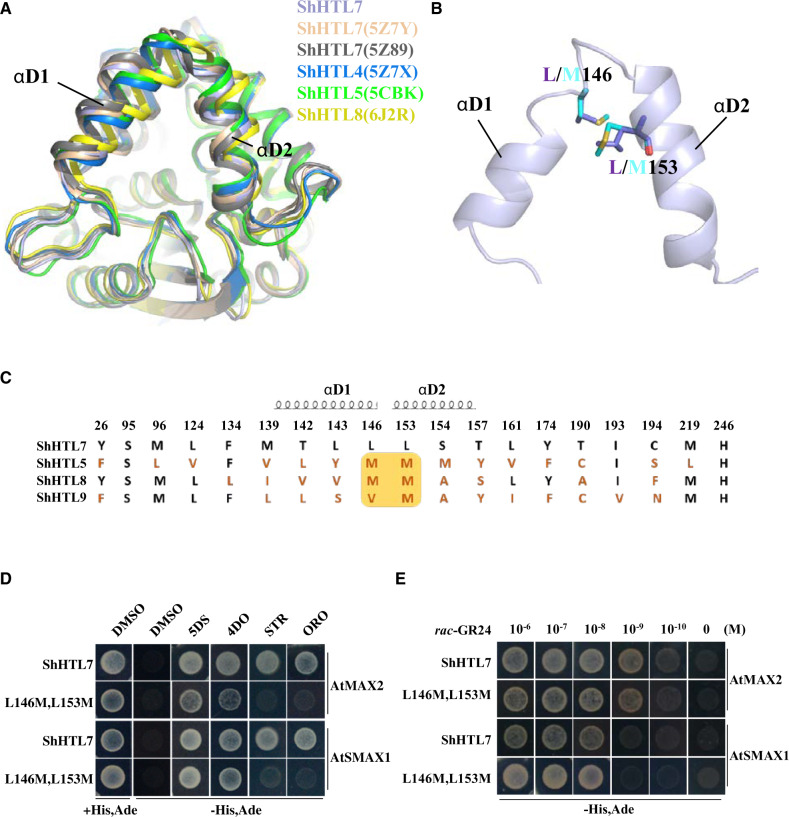
We determined the crystal structure of ShHTL7 to help understand its high sensitivity and low selectivity. The purified ShHTL7 protein was crystallized using the hanging-drop vapor diffusion method and was subjected to X-ray diffraction analysis. Data analysis and refinement led to determination of the structure at 2.33Å resolution (Supplemental Table S2). This structure shows almost the same topology as two others determined recently for ShHTL7 (Shahul Hameed et al., 2018; Xu et al., 2018; Figure 4A;Supplemental Figure S7A). Comparison of ShHTL7 structures with ShHTL4 (Xu et al., 2018), ShHTL5 (Toh et al., 2015), and ShHTL8 (Zhang et al., 2020) showed significant differences in αD1 and αD2 helices (Figure 4A), which were proposed to undergo a conformational change for D14 interaction with AtMAX2/D3 (Yao et al., 2016). The looser organization of αD1 and αD2 helices in ShHTL7 makes the entrance of the pocket bigger than that of other ShHTLs, which might provide ShHTL7 with broad-spectrum response to various SLs with different structures.
Figure 4.
ShHTL7 residues within helices αD1 and αD2 at the pocket entrance influence SL selectivity but not SL sensitivity in binding to AtMAX2 and AtSMAX1. (A) Alignment of ShHTL7 structures from this work (blue), previously determined ShHTL7 structures 5Z89 (gray) (Shahul Hameed et al., 2018) and 5Z7Y (pink) (Xu et al., 2018), ShHTL4 (5Z7X in dark blue), ShHTL5 (5CBK in green) (Xu et al., 2018), and ShHTL8 (6J2R in yellow) (Zhang et al., 2020). There are different shifts in the αD1 and αD2 helixes among the structures. (B) Sequence alignment of pocket amino acid residues for ShHTL7, ShHTL5, ShHTL8, and ShHTL9. Residues in red differ from those in ShHTL7. The yellow box highlights positions 146 and 153 that are most commonly M in other ShHTL proteins, in contrast to ShHTL7. Residues contained within helices αD1 and αD2 are indicated. (C) Location of residues L146 and L153 on αD1 and αD2 helices near the pocket entrance, and the structural changes associated with the L146M L153M double mutation. The ShHTL7 residues (L146 and L153) are shown in purple and mutant forms (M residues) in cyan. (D) Binding of BD-ShHTL7 and its double mutant (BD-L146M, L153M) to AD-AtMAX2 and AD-AtSMAX1 in response to 1 μM 5DS, 4DO, STR, and ORO in Y2H assays. (E) Interactions between BD-ShHTL7 or its double mutant BD-L146M, L153M with AD-AtSMAX1 or AD-AtMAX2 in response to rac-GR24 in the concentration range 10−6 to 10−10 M in Y2H assays.
Ligand selectivity of ShHTLs
It is striking that ShHTL7 exhibits broad-spectrum response to SLs and has strong affinity for interacting proteins, so we first wanted to examine whether these two features are linked. We located residues in the ligand-binding pocket and established the topography of the pocket of the receptor protein (Supplemental Figure S7B). ShHTL7 is much more responsive than any other ShHTL to STR and ORO (Figure 3B). As ShHTL5, ShHTL8, and ShHTL9 interacted with 5DS and 4DO but had low affinities to ORO and STR (Figures 2, B and C and 3B), we aligned amino acid residues comprising the ligand binding pocket of these ShHTLs so that conserved and variant residues could be identified (Figure 4C). We focused attention on L146 and L153 of ShHTL7, where other ShHTLs commonly have M. These two sites are close to the pocket entrance and at the junction of αD1 and αD2 helices (Figure 4B) and so might have an impact on the conformation of the helixes and ligand selectivity.
We therefore created a L146M L153M double mutation in ShHTL7 to test the resultant binding to AtMAX2 and AtSMAX1 and the responsiveness to different SLs using the Y2H system. We observed that ShHTL7 with the L146M L153M double mutation still responded to 5DS and 4DO but lost the ability to bind to AtMAX2 and SMAX1 in response to ORO and STR (Figure 4D). We also measured the interaction between the L146M L153M double mutant and AtMAX2 or AtSMAX1 in response to rac-GR24 over a concentration range of five orders of magnitude. The double mutant appeared to be as responsive to rac-GR24 as wild-type ShHTL7 protein, with clear interaction seen at 10−9 M for AtMAX2 and 10−8 M for AtSMAX1 (Figure 4E). Therefore, L146 and L153 confer upon ShHTL7 lower ligand selectivity but have no apparent effect on the binding of ShHTL7 to AtMAX2 or to AtSMAX1. It has previously been noted that the pocket of ShHTL7 has a larger volume than that of most other ShHTL proteins and L146 and L153 contribute to the larger pocket (Xu et al., 2018). The change of L to M at both positions reduces the volume of the pocket (Supplemental Figure S8, A and B) and M might provide greater steric hindrance and so restrict ligand binding.
Structure-based mutagenesis of the pocket of ShHTL7
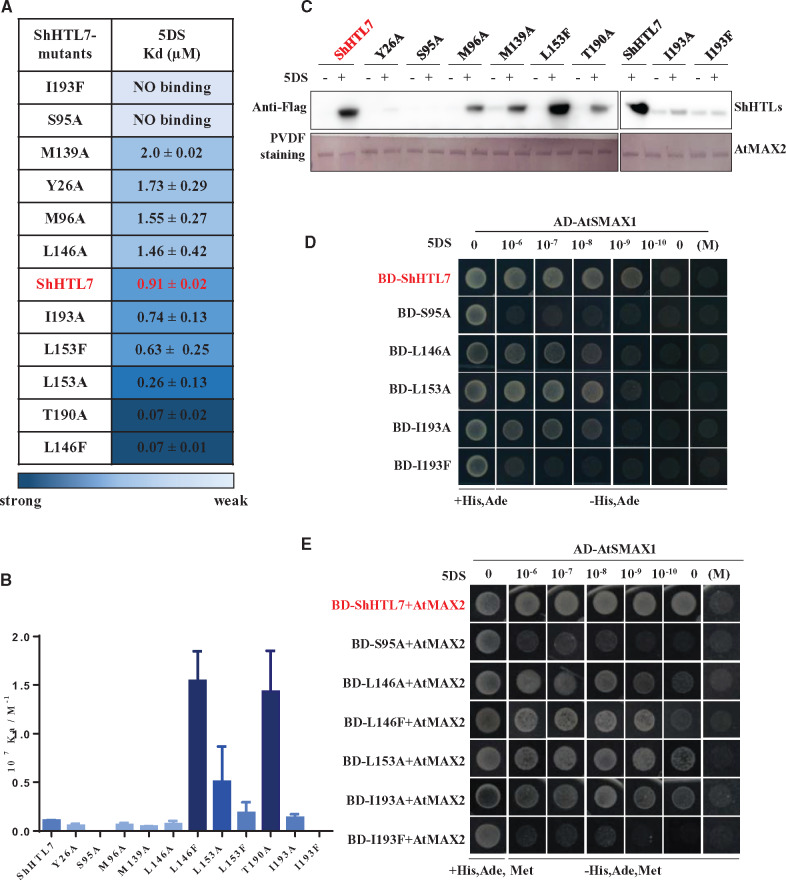
To further understand the relationship between the ligand binding pocket and the bioactivity of ShHTL7, we analyzed the sequence alignments of these residues in the pocket among the ShHTLs (Supplemental Figure S7C). Residues at some positions are well conserved whereas others are more variable. The more variable sites in ShHTL7 were identified as L124, M139, T142, L146, L153, T157, T190, and C194, suggesting that they likely endow ShHTL7 with its particular features including the bigger pocket and lower ligand selectivity. Alongside the Ser–His–Asp catalytic triad, F134 and I193 are highly conserved, whereas Y26 and Y174 are relatively well conserved (Supplemental Figure S7C). Therefore, we conducted mutagenesis to alter some of these pocket residues then prepared mutated proteins for in vitro assays and expressed them in yeast for Y2H assays. We created L146F and L153F because both could be expected to introduce steric hindrance and reduce pocket volume. We also changed several amino acids to alanine (A) to change the pocket size since alanine is small and neutral and would not be expected to result in a gain-of-function. It is known that replacement of the active site serine S95 with alanine renders the protein inactive, so this change was made as a control.
Mutated proteins were analyzed for 5DS binding affinity using ITC (Figure 5A, Supplemental Figure S9, A–C). Replacement of the catalytic serine residue in the S95A mutant abolished 5DS binding in this assay. This mutation is well known to abolish hydrolytic activity and ligand binding (Waters et al., 2015; de Saint Germain et al., 2016). The results also show that the I193F mutation, which introduces steric hindrance deep in the pocket, abolished 5DS binding whereas I193A had no significant effect (Figure 5A). Of the mutations expected to increase the size of the pocket, several (Y26A, M96A, F134A, M139A, T142A, L146A) slightly decreased 5DS affinity whereas others (I193A, C194A) had no obvious effect. In contrast, L153A and T190A increased ligand affinity.
Figure 5.
ShHTL7 ligand binding affinity is not consistent with binding to AtMAX2 and AtSMAX1 or sensitivity to SL. A, Dissociation constants (Kd) for the binding of 5DS to ShHTL7 with mutations of pocket residues were measured by ITC assays. Kd values are means sd (n = >2). (B) Association constants (Ka) derived from (A). Values are means ± sd (n = >2). (C) In vitro pull-down assays of the interactions of His6-Flag-tagged ShHTL7 and its mutant forms with GST-AtMAX2 in response to 10 μM 5DS. The GST-AtMAX2 as bait was visualized by staining the PVDF membrane with Memstain to show loadings and the ShHTL7 proteins were detected with anti-Flag antibody. (D) Y2H assays of interactions between AD-AtSMAX1 and BD-ShHTL7 and its mutant forms in response to 5DS in the concentration range 10−6 to 10−10 M. (E) Y3H assays to determine the effect of AtMAX2 on the interactions shown in (D).
When the bulky Phe residue was introduced near the pocket entrance in L153F, it had no effect in the ITC assay but L146F had stronger binding affinity for 5DS than wild-type ShHTL7. In contrast, introducing the bulky residue Phe residue into the pocket can inhibit the 5DS binding as seen with I193F (Supplemental Figure S9D). However, increasing the pocket size tended to weaken binding affinity as seen with Y26A and L146A, potentially because 5DS was less well able to interact with the catalytic triad residues (Supplemental Figure S9D). These results demonstrate that residues inside and at the pocket entrance are important for ligand binding. However, substitutions aimed simply at increasing or decreasing the size of the pocket did not suggest that pocket volume per se is important for ligand binding affinity, implying that pocket conformation and/or chemical interactions with the ligand are important.
To analyze the relationship between ligand binding affinity and the interaction with downstream proteins, we investigated the interaction of variant forms of ShHTL7 with AtMAX2 and AtSMAX1 by in vitro pull-down and Y2H or Y3H assays (Figure 5, B–D). Several mutations resulted in decreased binding of ShHTL7 to AtMAX2 in pull-down experiments using 10 µM 5DS (Figure 5C). These included Y26A, S95A, M96A, and M139A, each of which had lower affinity for 5DS in ITC, which could potentially explain the lower affinity for AtMAX2. Therefore, ligand binding is important for downstream protein interaction. However, I193A and I193F proteins both had very weak binding to AtMAX2, yet I193A had no effect on 5DS affinity. Similarly, T190A had stronger binding affinity to 5DS whereas the interaction with AtMAX2 was apparently weaker (Figure 5, B and C). Some of these mutations were further investigated in Y2H and Y3H experiment using 5DS at concentrations over five orders of magnitude to detect the interaction of ShHTL7 with AtSMAX1 separately and in the presence of AtMAX2 (Figure 5, D and E). The results established that I193F protein does not interact with AtSMAX1 at any concentration of 5DS (similar to the S95A mutant) in both Y2H and Y3H. L146A, L153A, and I193A require approximately 10-fold higher concentration of 5DS than the ShHTL7 control for equivalent interaction in Y2H, whereas they are similar to ShHTL7 in Y3H. We also observed that the sensitivity of L146F to 5DS concentration in the Y3H assay is similar to that of L146A, although its affinity for 5DS in ITC is 20-fold stronger. Thus, high sensitivity of 5DS-induced interaction of ShHTL7 with AtMAX2 and AtSMAX1 is not determined simply by the 5DS binding affinity of ShHTL7. However, the L153F mutant appeared to interact very strongly with AtMAX2 in pull-down experiments (Figure 5C), which led us to investigate this further (see below).
Next, we analyzed the hydrolytic activity of three of the ShHTL7 mutants using YLG as a convenient substrate to compare these proteins. We found that the T190A and I193A mutants had lower affinity for YLG (higher Km) but higher maximum rate of hydrolysis (kcat) than ShHTL7 (Supplemental Figure S3). These data indicate that residues within the pocket that can influence ligand binding affinity or hydrolysis activity can also influence binding of ShHTL7 to AtMAX2 or SMAX1. Further experiments are needed to determine how such pocket residues influence this binding. Comparing the affinity of ShHTLs for 5DS in ITC assays with results from pull-down and Y2H assays of 5DS-dependent interaction of ShHTL with AtMAX2 or AtSMAX1, the higher binding affinity for 5DS, as observed for ShHTL8, ShHTL9, ShHTL10, and ShHTL11, was not matched by higher sensitivity to 5DS in the interaction with AtMAX2 or AtSMAX1. Furthermore, ShHTL6 had a stronger 5DS-binding affinity than ShHTL5 or ShHTL7 in ITC assays (Figure 1A), but the germination of transgenic Arabidopsis seeds expressing ShHTL6 in the htl-3 background was less sensitive to 5DS (Toh et al., 2015). Thus, whereas we should be cautious about direct comparisons between different experimental systems, higher ligand-binding affinity does not appear to confer the SL receptor with high sensitivity to SL for signal transduction in planta.
Structural basis for interaction of ShHTL7 with AtMAX2
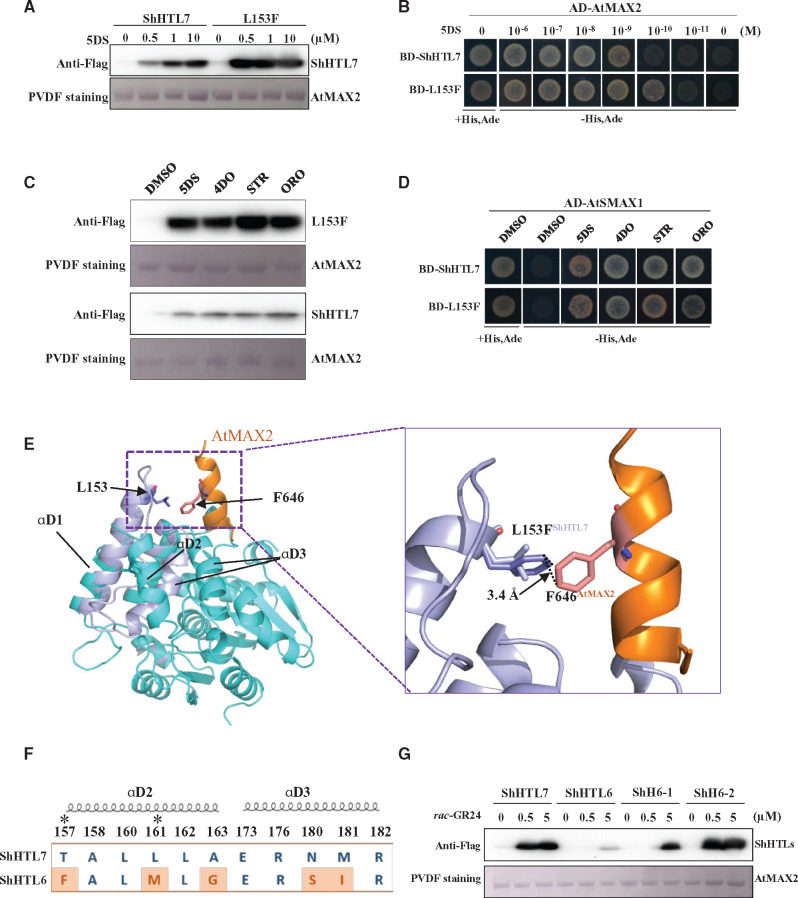
We observed that the binding of L153F protein to AtMAX2 in response to 5DS appears to be stronger than wild-type ShHTL7 in pull-down experiments (Figure 5C), yet its affinity for 5DS is not significantly different in ITC assays (Figure 5A), and its Km for YLG is significantly higher (Supplemental Figure S3). To further investigate the affinity of ShHTL7 and the L153F mutant for AtMAX2, we tested their binding in response to 0, 0.5, 1, and 10 µM 5DS by pull-down assay and the response to 5DS over six orders of magnitude (1 µM to 10 pM) in the Y2H assay (Figure 6, A and B). Clearly, the L153F mutant bound to AtMAX2 at a lower concentration of 5DS in both assays. We subsequently investigated whether the L153F mutation had a similar effect on the binding to ShMAX2, for which the microscale thermophoresis (MST) assay was employed. The interaction of ShMAX2 with L153F occurred at lower concentration of 5DS and was also stronger than with wild-type ShHTL7 (Supplemental Figure S10). We also observed in pull-down experiments that 5DS, 4DO, STR, and ORO were all effective at stimulating interaction of L153F with AtMAX2 (Figure 6C). Finally, we showed in Y2H assays that L153F still has the ability to interact with AtSMAX1 in response to 5DS, 4DO, STR, and ORO (Figure 6D). Overall, the L153F mutant exhibited greater SL sensitivity and stronger binding to AtMAX2 than wild-type ShHTL7 in several different assays with no apparent loss of its broad-spectrum response to SLs.
Figure 6.
Residues at the interface surface of ShHTLs determine the affinity of binding to AtMAX2. (A) In vitro pull-down assay to detect the interaction between AtMAX2 and ShHTL7 or ShHTL7-L153F (labeled L153F) in response to 0, 0.5, 1, and 10 μM 5DS. ShHTL7 and ShHTL7-L153F mutant with His6-Flag tag were detected by immunoblotting with anti-Flag antibody. GST-AtMAX2 was detected by staining the PVDF membrane with Memstain to show loading. (B) Y2H assays of the interaction between AD-AtMAX2 and BD-ShHTL7 or BD-ShHTL7-L153F (L153F) with 10−6 to 10−11 M 5DS. (C) In vitro pull-down assay to detect the interaction between GST-AtMAX2 and ShHTL7 or ShHTL7-L153F (labeled L153F) in response to 1 µM 5DS, 4DO, STR, and ORO. Protein detection as in (A). (D) The interaction between AD-AtSMAX1 and BD-ShHTL7 or BD-ShHTL7-L153F in response to 1 µM 5DS, 4DO, STR, and ORO in Y2H assays. (E) Structural alignment of open-ShHTL7 (cyan) and ShHTL7 (lilac) after binding to AtMAX2 (orange). The box to the right shows the positions of L153 (lilac) and F153 (purple) and interaction between F153 and F618 of AtMAX2 (pink). (F) Sequence alignment of residues from ShHTL7 and ShHTL6 in the interface domain. Residues that differ are highlighted in red. Asterisks indicate residues with no direct interaction with AtMAX2 in the ShHTL7–AtMAX2 complex model. Helices αD2 and αD3 are indicated. (G) In vitro pull-down assay to show interaction of ShHTL7 and ShHTL6 with GST-AtMAX2 in the presence of 0, 0.5, and 5 μM rac-GR24. ShH6-1 is ShHTL6 with S180N, I181M double mutation. ShH6-2 has F157T, M161L, G163A, S180N, I181M quintuple mutation. Protein detection as in (A).
To understand the interaction between ShHTL7 and AtMAX2, we modeled the ShHTL7–AtMAX2–ASK1 complex based on our structures of ShHTL7 and the CLIM–AtD14–D3–ASK1 complex (PDB code: 5HZG; B chain). This enabled us to model the conformational change occurring in ShHTL7 upon SL-induced AtMAX2 binding and to identify residues at the interface between ShHTL7 and AtMAX2 (Supplemental Figure S11A). After aligning the structures of ShHTL7 before and after binding with AtMAX2, we noticed that AtMAX2 caused a conformational change in which αD2 is released and shifted inward toward the entrance of the pocket (Supplemental Figure S11C). As a result, L153 is located at the top of the new helix (Figure 6E). We modeled the putative interface between the L153F protein and AtMAX2 and discovered that F153 can interact with F646 of AtMAX2 by a π–π interaction after conformational change (Figure 6E;Supplemental Figure S11A). This could potentially explain the stronger protein–protein interaction of L153F mutant with AtMAX2.
We next considered how the conformational change at the interface of ShHTL proteins and AtMAX2 could influence the binding between them. We first modeled the surface residues of ShHTL7 that potentially interact with AtMAX2 by finding contacts between chains within 3.5 Å (Supplemental Figure S11A). Then we aligned and compared the interface residues in ShHTL proteins (Supplemental Figure S11B). Through this sequence alignment, we found that most of the interface amino acids are well conserved but ShHTL10 and ShHTL11 contained significant differences, specifically in residues 58, 161, 165, 180, and 181 that are on the αD2 and αD3 helices (Supplemental Figure S11B). Considering that ShHTL10 and ShHTL11 could not respond to SL in the germination assay in Arabidopsis (Toh et al., 2015) and the Y2H and pull-down assay showed ShHTL10 and 11 had no interaction with AtMAX2 (Figure 2B), we speculate that these residues are particularly important for the interaction with AtMAX2.
To verify the influence of the interface structure, we adopted a “gain-of-function” approach on ShHTL6, which is phylogenetically most closely related to ShHTL7 and has a very similar Km for YLG hydrolysis and a similar affinity for 5DS in ITC but binds weakly to AtMAX2 in pull-down experiments (Figure 1, A and B, Supplemental Figure S3). We found five interface residues that distinguish ShHTL6 from ShHTL7 by sequence alignment and note that ShHTL10 and 11 also differ from ShHTL7 in five and four of these residues, respectively (Figure 6F;Supplemental Figure S11B). We made two ShHTL6 mutants, one with two substitutions (S180N and I181M, named ShH6-1), and one with all five of the interface residues changed (F157T, M161L, G163A, S180N, and I181M) named ShH6-2. We expressed and purified these proteins and tested their binding to AtMAX2 in response to rac-GR24 at three concentrations in a pull-down assay, and found that binding of ShH6-2 was now equivalent to that of ShHTL7, whereas ShH6-1 showed intermediate binding (Figure 6G). These results establish that specific residues at the surface of ShHTL proteins determine the strength of interaction with AtMAX2, which raises the possibility that this influences the sensitivity of the ShHTL–AtMAX2 complex to SLs. Therefore, the interface interactions of ShHTL6 with AtMAX2 could potentially explain why ShHTL6 had lower sensitivity to 5DS and GR24 at 1 µM and 10 µM in the Arabidopsis germination assay (Toh et al., 2015), even though ShHTL6 has similar or stronger binding affinity for 5DS in ITC (Figure 1A) and for GR24 (Tsuchiya et al., 2015), and higher hydrolytic activity than ShHTL7 for YLG (Supplemental Figure S3) and for GR24 (Tsuchiya et al., 2015).
Although some phylogenetic analyses place ShHTL6 and ShHTL7 in the same sub-clade (Toh et al., 2015), we also see that ShHTL6 has a close relationship with ShHTL10 and ShHTL11, which could represent an alternative branch. We noticed that ShHTL10 and ShHTL11 had strong binding affinity to GR24 or 5DS, as well as relatively high hydrolytic activity (Tsuchiya et al., 2015), but they do not respond to GR24 or 5DS in Arabidopsis seed germination assays (Toh et al., 2015). Furthermore, they did not interact with MAX2 or AtSMAX1 in Y2H and pull-down assays, which is consistent with their responses in seed germination (Figure 2B). Compared with other ShHTLs in the MAX2 interface sequence, ShHTL10 and ShHTL11 are appreciably different, which implies that they might not function through MAX2 and potentially do not contribute to seed germination, although this needs further investigation. In contrast, ShHTL1, ShHTL2, and ShHTL3 have similar proposed interface sequences to that of ShHTL7 (Supplemental Figure S11B). This observed similarity is consistent with reports that AtHTL (AtKAI2) and members of the ShHTL1 clade of proteins exhibit MAX2-dependence in their response to KARs (Nelson et al., 2011; Conn et al., 2015; Toh et al., 2015). Previous work reported that ShHTL3 could respond to KAR1 and KAR2 in the Arabidopsis germination assay implying that it interacts functionally with AtMAX2 in planta (Conn et al., 2015). In summary, the sensitivity to SLs is determined not simply by the SL receptor protein in isolation, but by the SL receptor in a complex with its interacting proteins in signal transduction.
Discussion and conclusion
Importance of protein complexes
Consistent with other studies, we observed that the affinity of ShHTL7 for SLs and its SL hydrolytic activity are not significantly different from those of other ShHTL proteins (Tsuchiya et al., 2015). Therefore, the very high SL sensitivity of seeds expressing ShHTL7 is not apparently due to the affinity of ShHTL7 for SLs. Instead, we discovered that ShHTL7 has a high affinity for AtMAX2 in pull-down experiments in response to SL in vitro. We subsequently used Y2H assays to show that SL induces binding of ShHTL7 to AtMAX2 and ShMAX2, and also to AtSMAX1, and that these interactions occurred at SL concentrations lower than for other ShHTL proteins. Furthermore, we used Y3H assays to show that binding of ShHTL7 to AtSMAX1 becomes even more sensitive to SL when AtMAX2 or ShMAX2 is co-expressed with ShHTL7. We conclude that formation of the ternary protein complex of ShHTL7 with MAX2 and AtSMAX1 is triggered by a lower concentration of SL than occurs with other ShHTL proteins. We propose that this determines the SL sensitivity of seeds expressing ShHTL7, and that this could enable seeds of Striga to respond to very low concentrations of SL in the rhizosphere.
We observed that the D2 domain of SMAX1 (SMAX1-D2) had the ability to enhance interaction of MAX2 with ShHTL7 even though no significant SMAX1-D2 binding was detected with either ShHTL7 or MAX2. This suggests that each component could contribute to the stabilization of the ternary complex and make SL perception more sensitive. Potentially such interactions might also lead to higher affinity between interacting proteins endowing the receptor complex with greater activity. Previous work has shown that rice F-box protein D3 could stabilize the interaction of D14 with the D2 domain of D53 (D53-D2), whereas no significant binding was observed between D14 and D53-D2 in the absence of D3 (Shabek et al., 2018). It is possible that the interaction of ShHTL/D14 with MAX2/D3 could create a new interface for the D2 domain of AtSMAX1/D53, which could make it more stable, thus enhancing the interaction of AtSMAX1 with ShHTLs. Very recent work indicates that the D1-M domain rather than the D2 domain of SMAX1 or SMXL7 interacts with KAI2 or D14 proteins (Zhou et al., 2013; Khosla et al., 2020). However, Khosla et al. (2020) acknowledge that their results contradict those of previous studies and suggest that the discrepancy could be due to the fact that their experiments did not include D3 or MAX2 in the protein interaction assays. Our results support the conclusion that the C-terminal end (D2 domain) interacts with the interface surface formed by D14/HTL binding to D3/MAX2, but the role of the D1-M domains in this interaction remains to be determined.
Ligand specificity
The factors that determine the activity of different ligands with ShHTL proteins are unknown, but are very important for understanding Striga evolution and ecology and for the discovery of potentially useful SL analogs. We observed that the αD1 and αD2 helices that reside at the pocket entrance show different positions when the different crystal structures of ShHTLs are compared. This implies that the α-helices of the receptor might provide flexibility to the pocket and accommodate various SLs. Comparison of different ShHTLs shows that these two α-helices have an impact on the size and structure of pocket and so could play a vital role in distinguishing diverse SLs. Creating the L146M and L153M mutations, which are located in αD1 and αD2 helices, resulted in loss of response to ORO and STR, thus indicating that these two sites are very important in determining ligand selectivity of ShHTL7. However, we do not yet know if these mutations cause steric hindrance, which influences the shift of αD1 and αD2 helices after binding ligands, or if they simply result in a smaller pocket size. Previous studies suggested that the ligand selectivity of ShHTLs is due to the shape and size of the pocket, since the highly variable pocket of the KAI2d clade in parasitic plants is consistent with altered ligand selectivity (Conn et al., 2015). The pocket size of ShHTL7 is larger than that of other SL receptors (Xu et al., 2018), which might endow it with low selectivity to SLs. This feature potentially also makes ShHTL7 more amenable to the discovery of further ligands that have no interaction with other ShHTLs or D14 of the host plant (Holbrook-Smith et al., 2016; Shahul Hameed et al., 2018; Uraguchi et al., 2018).
The functions of different ShHTLs in Striga
It might be anticipated that there would be a trade-off between SL selectivity and sensitivity. However, ShHTL7 shows a broad-spectrum response (low selectivity) to natural SLs while conferring very high sensitivity in triggering interaction with MAX2 and SMAX1, as well as in seed germination. One hypothesis is that the ShHTL7 has a selective advantage as it could detect most SLs secreted by potential host plants and respond to the very low SL concentrations in the soil to trigger germination. Consistency of results for ShMAX2 and AtMAX2 leads us to suggest that ShHTL7 is highly sensitive to SLs in both Arabidopsis and Striga. Therefore, ShHTL7 might be a pivotal SL receptor in Striga, which suggests that its structure–function properties and its evolution deserve detailed scrutiny. However, the presence of several other functional ShHTLs implies that they also play important roles in Striga. Arguably, ShHTL7 alone could make seeds susceptible to trace amounts of SL-like compounds in the soil, which are produced by diverse soil organisms rather than by potential host plants. Conceivably, other ShHTLs could provide a type of validation of SL identity. Another possibility is that divergent ShHTLs enable Striga to recognize different host plants through the recognition of very specific SLs or combinations of SLs from those different hosts. We have only investigated a small number of known canonical SLs; however, there are many others that could potentially be recognized by ShHTL4–9 and so enable seeds to respond to different germination stimulants from different host plants. We are also conscious that much of this research has been conducted with heterologous but powerful systems and in vitro methods, and that in future it must be extended into studies in Striga (Bunsick et al., 2020).
Practical applications
Our results showing that the sensitivity to SLs is determined by the formation of the ShHTL–MAX2–SMAX1 complex rather than by ligand binding affinity of ShHTL proteins suggests that future screening systems for further SLs or SL analogues should employ such tripartite complexes. For example, the sensitivity of such a system might provide the basis for an assay to detect and purify noncanonical SLs that function at very low concentrations in the rhizosphere. Such assays would also provide a way to determine the ligand selectivity of different members of the ShHTL family. Considering that ShHTL7 is the primary contributor to Striga seed germination (Uraguchi et al., 2018), a system based specifically on the formation of a SMAX1–ShHTL7–MAX2 complex offers the potential to discover compounds that could either act as agonists to stimulate suicidal Striga germination before crop planting, or to act as antagonists to block Striga seed germination within a crop (Okazawa and Wakabayashi, 2015; Holbrook-Smith et al., 2016; Hamiaux et al., 2019; Nakamura et al., 2019). We therefore believe that our findings now open up research pathways to discover the functions of each ShHTL and identify noncanonical ligands that can be used in the control of Striga. A further outcome from this research is the opportunity to explore potentially increasing the SL sensitivity in crop plants by modifying amino acids to enhance the interaction between D14 and its interacting proteins. Such an outcome could have benefits in the control of shoot development or in responses to environmental stresses.
Materials and methods
Protein preparation
For the pull-down assays, full-length S. hermonthica HTL1–HTL11 (ShHTL1–ShHTL11, GenBank accessions KR013121 to KR013131) and ShHTL7-mutants fused with a C-terminal His6-Flag tag were expressed in E. coli strain BL21 (DE3). After initial purification by Ni-NTA (Novagen) affinity chromatography, the ShHTL proteins were further purified by Source 15Q (GE Healthcare) anion exchange followed by Superdex 200 10/300 (GE Healthcare) gel filtration with TBS buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl). The full-length A. thaliana MAX2 (AtMAX2) and S. hermonthica MAX2 (ShMAX2) with N-terminal GST (Glutathione S-Transferase)-tags were co-expressed with AtASK1-Flag in Spodoptera frugiperda Sf9 insect cells. The AtASK1 protein was used to stabilize the F-box protein. Both AtMAX2 and ShMAX2 were co-purified with AtASK1 by Glutathione Sepharose 4B (GE Healthcare) affinity chromatography.
The ShHTL7 protein for crystallization, ShHTL1–ShHTL11 proteins, and mutated ShHTL7 protein for use in ITC or MST assays were fused with an N-terminal GST tag. AtSMAX1-D2 (571–990 aa) used for an AlphaScreen assay was also fused with an N-terminal GST tag. Proteins were first purified by Glutathione Sepharose 4B (GE Healthcare) affinity chromatography then GST tags were removed through on-column cleavage by PreScission Protease, which was also GST-tagged. The eluents were subsequently loaded to Source 15Q and further purified by Superdex 200 10/300 in buffer containing 20 mM Tris–HCl, pH 8.0, 150 mM NaCl (for crystallization and ITC) or buffer with 20 mM HEPES, pH 7.0, 150 mM NaCl (for AlphaScreen assays). GST-ShHTLs were used for AlphaScreen assays.
The full-length ShMAX2 protein was co-expressed with AtASK1 in insect cells as an N-terminal His6-Flag fusion protein for MST assays. The protein was purified by Ni-NTA and Glutathione Sepharose 4B affinity chromatography and Source 15Q. Finally, all proteins were transferred into a buffer containing 20 mM HEPES, pH 7.0, 150 mM NaCl using Superdex 200 10/300.
In vitro pull-down assays
To detect the interaction between ShHTL proteins (ShHTL1–ShHTL11, ShHTL7 mutants, and ShHTL6 mutants) and AtMAX2, we used GST-AtMAX2 co-expressed with AtASK1-His6 as bait and the ShHTL–His6–Flag proteins as prey. Purified GST–AtMAX2–AtASK1 (∼20 µg) and ShHTL–His6–Flag (12 µg) were incubated separately with 50 μL Glutathione Sepharose 4B at 4°C for 1 h in the presence of racemic (rac)-GR24, 5DS, or the dimethylsufoxide (DMSO) solvent control. The reaction buffer contained 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 0.5% (v/v) Tween-20, 20 mM 2-mercaptoethanol. After extensive washing with the reaction buffer to remove unbound protein, the protein complexes were released from the beads with SDS-gel loading buffer and subjected to immunoblot analysis. The ShHTL1–ShHTL11 His6-Flag proteins were detected by Anti-Flag antibody (Anti-DYKDDDDK-Tag Mouse mAb, Abmart, Shanghai) and GST-AtMAX2 was visualized by staining the polyvinylidene difluoride membrane with Memstain (Applygen Technologies Inc, Beijing).
ITC assays
In ITC assays, ShHTL (1–11) and ShHTL7 mutant proteins without tag were prepared in 20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% (v/v) DMSO. 5DS and rac-GR24 were diluted in 20 mM Tris–HCl, pH 8.0, 150 mM NaCl to a final concentration of 100 μM or 150 μM containing 1% (v/v) DMSO. About 200 μL 5DS or rac-GR24 was delivered into the sample cell and 1 or 2 μL of concentrated ShHTL protein (300–1,000 μM) was injected into the cell each time at 20°C after the first injection of 0.5 μL. The binding assays were carried out using a MicroCal ITC200 (GE Healthcare). Data fitting was performed in “one set of sites” mode and the results were calculated by Origin software of MicroCal. The concentrations of detected proteins were measured with a NanoDrop 2000 (Thermo Scientific, Shanghai).
Crystallization and structure determination
Crystallization trials were conducted at 18°C by the hanging-drop vapor diffusion method. Crystals of ShHTL7 were obtained in a reservoir solution containing 4 mM calcium chloride, 20 mM sodium acetate pH 4.6, 6% (v/v; +/-)-2-Methyl-2,4-pentanediol, 160 mM magnesium chloride, 80 mM Tris pH 8.5, and 20% (w/v) PEG 3350. Reservoir solution with 15% (v/v) glycerol was used to flash-cool the crystal for data collection.
The native datasets of ShHTL7 were collected on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF). The data were processed and scaled using the HKL2000 package (Navaza and Saludjian, 1997). The structure of ShHTL7 was determined by molecular replacement using a model of ShHTL7 (PDB 5Z7Y; Xu et al., 2018) as template. The modified experimental electron density was built by COOT (Emsley and Cowtan, 2004) and further refined in PHENIX (Adams et al., 2010). The final refinement data are presented in Supplemental Table S1.
Homology modeling and pocket size analysis
Homology modeling was performed using the SWISS-MODEL server (http://swissmodel.expasy.org), as previously described (Yao et al., 2016). The structure model of ShHTL7–AtMAX2 and ShHTL6–AtMAX2 are based on the structure of CLIM–AtD14–D3–ASK1 complex (PDB code: 5HZG; B chain) and ShHTL7 mutants are based on the structure of ShHTL7 from this work. The cavity volume was calculated using CASTp program server (http://sts.bioe.uic.edu/castp/index.html).
Yeast Y2H and yeast Y3H assays
In the Y2H assays, ShHTL (1–11) and ShHTL7 mutants were cloned into the pGBKT7 vector (Clontech) individually as bait. AtSMAX1, ShMAX2, and AtMAX2 were cloned into pGADT7 vector (Clontech) as prey. To stabilize ShMAX2 and AtMAX2, AtASK1 was fused at the N-terminal end of each with a 16-GGSG linker (Xu et al., 2018). The Y3H assay used AD-AtSMAX1 as a prey. The ShHTLs sequences were cloned into the pBridge vector (Clontech), fused at the N-terminus with the Gal4 DNA BD-ShHTLs. AtMAX2 or ShMAX2 fused with AtASK1 was cloned into the second multiple cloning site 2 of the pBridge vector and co-expressed with BD-ShHTLs. To detect the interaction between MAX2 proteins with AtSMAX1, AtSMAX1 was cloned into pGBKT7 fused with the Gal4 DNA BD and co-transformed with pGADT7-MAX2.
The bait and prey constructs, pGBKT7–ShHTL (1–11) and pGADT7–AtSMAX1, pGBKT7–ShHTL (1–11) and pGADT7–MAX2, or pBridge-ShHTL–MAX2 and pGADT7–AtSMAX1 were co-transformed in pairs into yeast strain Y2HGold (Clontech) cells by the lithium acetate-mediated method. The transformed yeast cells were plated on SD/-Leu/-Trp medium and incubated at 28°C for 3 d to confirm the presence of both plasmids. The interactions were detected in the presence or absence of rac-GR24, 5DS, 4DO, (±)-ORO, and (±)-STR in selective medium -Leu/-Trp/-His/-Ade (-His, Ade for short) for Y2H or -Leu/-Trp/-His/-Ade/-Met (-His, Ade, Met for short) for Y3H, with incubation for 3 d.
YLG hydrolysis assays
YLG hydrolysis assays were conducted with 1 μg purified recombinant GST-tagged ShHTL6, ShHTL7, or mutant proteins in reaction buffer 100 mM HEPES, pH 7.0, 150 mM NaCl. YLG was diluted in reaction buffer to give a concentration gradient of 5–0.15625 μM. The proteins and dilutions of YLG were mixed in a 96-well plate (PerkinElmer Optiplate-96F) in a 100 μL final volume. The measurements were performed on an EnSpire plate reader (PerkinElmer) at room temperature using a 480-nm excitation wavelength and a 520-nm emission wavelength. The background signal was provided by YLG in reaction buffer. A standard curve was established for fluorescin, the hydrolysis product of YLG. The fluorescence intensity was measured in 10-min intervals over 100 min. The Km and Vmax values of each protein were calculated from a nonlinear Michaelis–Menten curve-fitting based on relative fluorescence intensity of a concentration gradient of YLG by Prism 6 (GraphPad). The assays were repeated three times independently each with two replicates.
Amplified luminescent proximity homogeneous assay (AlphaScreen)
To determine and measure the interactions of AtMAX2 and ShHTL proteins, the purified GST-tagged ShHTL4, ShHTL6, ShHTL7, ShHTL7-S95A, and His6-Flag-AtMAX2 (which was co-expressed with AtASK1) were used in an AlphaScreen (Perkin Elmer) assay. The experiments were conducted with 200 nM GST-ShHTL4, GST-ShHTL6, GST-ShHTL7, GST-ShHTL7-S95A, and His6-Flag-AtMAX2 by mixing with 5 μg mL−1 GST donor beads and 5 μg mL−1 nickel chelate acceptor beads (PerkinElmer). Serial dilutions (10 μL) of rac-GR24 of 0.0001–10 μM were incubated with 30 μL mixture of proteins and beads at 4°C for 1 h. To detect the impact of AtSMAX1-D2 on the rac-GR24-induced interaction of MAX2 with ShHTL7, AtSMAX1-D2 with no tag protein in concentration gradient ranging from 3 µM to 0.3 nM was incubated with 200 nM ShHTL7 and His6-Flag-AtMAX2 in the presence of 0.1 µM and 1 µM rac-GR24 and mixing with 5 μg mL−1 GST donor beads and 5 μg mL−1 nickel-chelate acceptor beads (PerkinElmer). To test the interaction between AtSMAX1-D2 and ShHTL7, or AtSMAX1-D2 and AtMAX2, 200 nM GST-AtSMAX1-D2 was incubated with 200 nM ShHTL7-His6-Flag or His6-Flag-AtMAX2 individually for 1 h with 1 µM rac-GR24 and 5 μg mL−1 GST donor beads and 5 μg mL−1 nickel–chelate acceptor beads (PerkinElmer). After incubation, the binding signal was detected with an EnSpire plate reader (PerkinElmer) in the 1/2 AreaPlate-96 plate at room temperature. All dilution and reaction buffers contained 50 mM MES, pH 6.5, 150 mM NaCl, 1 mM dithiothreitol, and 0.1 mg·mL−1 bovine serum albumin. The results were based on an average of three experiments and the EC50 values were determined using nonlinear curve-fitting of graphs generated with Prism 6 (GraphPad).
Microscale thermophoresis
Purified ShHTL7 and ShHTL7 mutants were labeled by Monolith NT.115 Protein Labeling Kit Red NHS (MicroScale Thermophoresis grade). A Sephadex G-25 column was used to remove the free unbound dye; 2 μM labeled ShHTL7 or ShHTL7 mutants were mixed with 2 μM purified unlabeled His6-Flag-ShMAX2. The mixture was then incubated with a concentration gradient of 5DS (the final concentration ranging from 20 μM to 0.6 nM) for 1 h at 4°C. The reaction buffer contained 20 mM HEPES, pH 7.0, 150 mM NaCl, 0.005% (v/v) Tween-20. After loading the sample into premium monolith NT capillaries, the interactions were detected three independent times by a Monolith NT. 115 instrument (Nano Temper Monolith NT.115). The data were analyzed by NanoTemper analysis software.
Chemicals and reagents
The rac-GR24 (a racemic mixture), 5DS, 4DO, (±)-STR, and (±)-ORO were obtained from Strigolab (https://strigolab.eu/). YLG and fluorescein used in YLG hydrolysis assay were synthesized by HEOWNS (http://www.heowns.com/, Tianjing) according to the reported methods (Tsuchiya et al., 2015).
Accession numbers
Sequence information in this work can be found in GenBank or the Arabidopsis Genome Initiative databases under the following accession numbers: S. hermonthica HTL1–11 (KR013121–KR013131); Striga hermonthica MAX2 (JX565467.1), A. thaliana MAX2 (At2G42620), A. thaliana ASK1 (At1G75950), and A. thaliana SMAX1 (At5G57710).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. SDS–PAGE analysis of ShHTL proteins purified by size-exclusion chromatography.
Supplemental Figure S2. Isothermal titration calorimetry of 5DS binding to ShHTL proteins.
Supplemental Figure S3. Kinetics of YLG hydrolysis by ShHTL7, ShHTL7 mutations, and ShHTL6.
Supplemental Figure S4. Interaction of ShHTLs with AtMAX2 induced by rac-GR24.
Supplemental Figure S5. The interaction of ShHTLs with ShMAX2 and AtSMAX1 in response to different SLs.
Supplemental Figure S6. AtSMAX1 enhances the interactions between ShHTL7 and AtMAX2.
Supplemental Figure S7. Analysis of the pocket structure of ShHTL7.
Supplemental Figure S8. The influence of L146M and L153M on pocket volume.
Supplemental Figure S9. Analysis of 5DS binding affinity of ShHTL7 proteins with mutated pocket residues.
Supplemental Figure S10. The interaction of ShMAX2 with ShHTL7 and mutants in response to 5DS.
Supplemental Figure S11. Analysis of the interface structure in the complex of ShHTL7 with AtMAX2 and the conformational change in ShHTL7.
Supplemental Table S1. Binding affinities and thermodynamic parameters of interaction of ShHTL proteins with 5DS in ITC assays.
Supplemental Table S2. X-ray data collection and refinement statistics for ShHTL7.
Supplementary Material
Acknowledgments
We thank Dr Z. Ming (Guangxi University), Drs S. Fan and L. Yan (Tsinghua University), and the staff at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U for help with data collection and processing.
Funding
This work was supported by the Ministry of Science and Technology of China (2016YFA0500501), the National Natural Science Foundation of China (32070321) and China Hunan Provincial Science and Technology Department (2019RS2019 and 2020JJ3007). S.M.S. acknowledges the Foreign Experts Program of Beijing, the Chinese Academy of Sciences President's International Fellowship Initiative (2018VBA0025), and the Australian Research Council (CE200100015).
Conflict of interest statement. None declared.
R.Y., D.X., and S.M.S. conceived and supervised the research; Y.W. and R.Y. performed the experiments with help from X.D., L.G., and L.C.; Y.W., R.Y., D.X., and S.M.S. analyzed the data and wrote the manuscript with inputs from other authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ruifeng Yao (ryao@hnu.edu.cn).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW. , et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827 [DOI] [PubMed] [Google Scholar]
- Aliche EB, Screpanti C, De Mesmaeker A, Munnik T, Bouwmeester HJ (2020) Science and application of strigolactones. New Phytol 227:1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50:1416–1424 [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Fonne‐Pfister R, Screpanti C, De Mesmaeker A (2019) Strigolactones: plant hormones with promising features. Angew Chem Int Ed 58:12778–12786 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA (2013) Diverse roles of strigolactones in plant development. Mol Plant 6:18–28 [DOI] [PubMed] [Google Scholar]
- Bunsick M, Toh S, Wong C, Xu Z, Ly G, McErlean CSP, Pescetto G, Nemrish KE, Sung P, Li JD. , et al. (2020) SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat Plants 6:646–652 [DOI] [PubMed] [Google Scholar]
- Burger M, Chory J (2020) The many models of strigolactone signaling. Trends Plant Sci 25:395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sanchez-Ferrero JC, Rodriguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26:1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349:540–543 [DOI] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of Witchweed (Striga lutea Lour.): Isolation and Properties of a Potent Stimulant. Science 154:1189–1190 [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clave G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C. , et al. (2016). An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- Fang Z, Ji Y, Hu J, Guo R, Sun S, Wang X (2020) Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol Plant 13:586–597 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22:2032–2036 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Larsen L, Lee HW, Luo Z, Sharma P, Hawkins BC, Perry NB, Snowden KC (2019) Chemical synthesis and characterization of a new quinazolinedione competitive antagonist for strigolactone receptors with an unexpected binding mode. Biochem J 476:1843–1856 [DOI] [PubMed] [Google Scholar]
- Holbrook-Smith D, Toh S, Tsuchiya Y, McCourt P (2016) Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat Chem Biol 12:724–729 [DOI] [PubMed] [Google Scholar]
- Hu J, Ji Y, Hu X, Sun S, Wang X (2020) BES1 functions as the co-regulator of D53-like SMXLs to Inhibit BRC1 expression in strigolactone-regulated shoot branching in arabidopsis. Plant Commun 1:100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46:79–86 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y. , et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, Cai ML, Stanga JP, Flematti GR, et al. (2020) Structure-function analysis of SMAX1 reveals domains that mediate its Karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28:1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang Y, Matusova R, Charnikhova T, Amini M, Jamil M, Fernandez-Aparicio M, Huang K, Timko MP, Westwood JH, et al. (2014) Striga hermonthica MAX2 restores branching but not the very low fluence response in the Arabidopsis thaliana max2 mutant. New Phytol 202:531–541 [DOI] [PubMed] [Google Scholar]
- Lumba S, Holbrook-Smith D, McCourt P (2017a) The perception of strigolactones in vascular plants. Nat Chem Biol 13:599–606 [DOI] [PubMed] [Google Scholar]
- Lumba S, Subha A, McCourt P (2017b) Found in translation: applying lessons from model systems to strigolactone signaling in parasitic plants. Trends Biochem Sci 42:556–565 [DOI] [PubMed] [Google Scholar]
- Morffy N, Faure L, Nelson DC (2016) Smoke and hormone mirrors: action and evolution of karrikin and strigolactone signaling. Trends Genet 32:176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Hirabayashi K, Miyakawa T, Kikuzato K, Hu W, Xu Y, Jiang K, Takahashi I, Niiyama R, Dohmae N. , et al. (2019) Triazole ureas covalently bind to strigolactone receptor and antagonize strigolactone responses. Mol Plant 12:44–58 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH, et al. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4:2613. [DOI] [PubMed] [Google Scholar]
- Navaza J, Saludjian P (1997) [33] AMoRe: An automated molecular replacement program package. Methods Enzymol 276:581–594 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011a) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108:8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Musselman LJ (2004) Introduction to parasitic flowering plants. Plant Health Instructor 13: 300–315 [Google Scholar]
- Okazawa A, Wakabayashi T (2015) Chemical control of root parasitic weeds. ACS Symp Ser 1204: 317–330 [Google Scholar]
- Parker C (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manage Sci 65:453–459 [DOI] [PubMed] [Google Scholar]
- Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Ticchiarelli F, Mao H, Hinds TR, Leyser O, Zheng N (2018) Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 563:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Zheng N (2014) Plant ubiquitin ligases as signaling hubs. Nat Struct Mol Biol 21:293–296 [DOI] [PubMed] [Google Scholar]
- Shahul Hameed U, Haider I, Jamil M, Kountche BA, Guo X, Zarban RA, Kim D, Al-Babili S, Arold ST (2018) Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor. EMBO Rep 19: e45619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E (2007) The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145:1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27:3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T, Mutuku M, Shirasu K (2013) The genus Striga: a witch profile. Mol Plant Pathol 14:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163:318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50:80–94 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129:1131–1141 [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350:203–207 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook-Smith D, Zhang H, McCourt P, Itami K, Kinoshita T. , et al. (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349:864–868 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200 [DOI] [PubMed] [Google Scholar]
- Uraguchi D, Kuwata K, Hijikata Y, Yamaguchi R, Imaizumi H, Am S, Rakers C, Mori N, Akiyama K, Irle S. , et al. (2018) A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science 362:1301–1305 [DOI] [PubMed] [Google Scholar]
- Wang L, Smith SM (2016) Strigolactones redefine plant hormones. Sci China Life Sci 59:1083–1085 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27:3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang A, Shao N, Ma H, Xiong G. , et al. (2020a) Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583:277–281 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu Q, Yu H, Ma H, Li X, Yang J, Chu J, Xie Q, Wang Y, Smith SM, et al. (2020b). Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in arabidopsis thaliana. Plant Cell 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester HJ (2018) Structural diversity in the strigolactones. J Exp Bot 69:2219–2230 [DOI] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Annu Rev Plant Biol 68:291–322 [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139:1285–1295 [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Moulin SL, Sun YK, Flematti GR, Smith SM (2015) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27:1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Ma M, Zhou Q, Zhao Y, Zhao B, Wang B, Wei H, Wang H (2020) Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat Commun 11:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Miyakawa T, Nosaki S, Nakamura A, Lyu Y, Nakamura H, Ohto U, Ishida H, Shimizu T, Asami T. , et al. (2018) Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat Commun 9:3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Li J, Xie D (2018) Recent advances in molecular basis for strigolactone action. Sci China Life Sci 61:277–284 [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L. , et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536:469–473 [DOI] [PubMed] [Google Scholar]
- Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Zhang W, Deng H, Xie D (2017) ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res 27:838–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Shen Y, Xi Z (2020) Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway. Biochem Biophys Res Commun 523:1040–1045 [DOI] [PubMed] [Google Scholar]
- Zhao L-H, Zhou XE, Wu ZS, Yi W, Xu Y, Li S, Xu TH, Liu Y, Chen RZ, Kovach A, et al. (2013) Crystal structures of two phytohormone signal-transducing alpha/beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res 23:436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L-H, Zhou XE, Yi W, Wu Z, Liu Y, Kang Y, Hou L, de Waal PW, Li S, Jiang Y, et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res 25:1219–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504:406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.