Abstract
Parasitic plants that infect crops are devastating to agriculture throughout the world. These parasites develop a unique inducible organ called the haustorium that connects the vascular systems of the parasite and host to establish a flow of water and nutrients. Upon contact with the host, the haustorial epidermal cells at the interface with the host differentiate into specific cells called intrusive cells that grow endophytically toward the host vasculature. Following this, some of the intrusive cells re-differentiate to form a xylem bridge (XB) that connects the vasculatures of the parasite and host. Despite the prominent role of intrusive cells in host infection, the molecular mechanisms mediating parasitism in the intrusive cells remain poorly understood. In this study, we investigated differential gene expression in the intrusive cells of the facultative parasite Phtheirospermum japonicum in the family Orobanchaceae by RNA-sequencing of laser-microdissected haustoria. We then used promoter analyses to identify genes that are specifically induced in intrusive cells, and promoter fusions with genes encoding fluorescent proteins to develop intrusive cell-specific markers. Four of the identified intrusive cell-specific genes encode subtilisin-like serine proteases (SBTs), whose biological functions in parasitic plants are unknown. Expression of SBT inhibitors in intrusive cells inhibited both intrusive cell and XB development and reduced auxin response levels adjacent to the area of XB development. Therefore, we propose that subtilase activity plays an important role in haustorium development in P. japonicum.
Subtilases specifically expressed in intrusive cells regulate auxin-mediated host–parasite connections in the parasitic plant Phtheirospermum japonicum.
Introduction
There are about 4,500 species of parasitic plants; these parasites are widespread and those that infect crops are serious threats to agriculture (Yoshida et al., 2016; Clarke et al., 2019). In particular, members of the family Orobanchaceae, such as Striga spp. and Orobanche spp., are destructive root parasitic plants that invade major crops including rice, sorghum, and maize, often in resource-poor societies, and cause annual economic losses of over 1 billion US dollars (Parker, 2009; Runo and Kuria, 2018). Parasitic Orobanchaceae plants produce large numbers of tiny seeds that are spread widely by wind, water, and anthropogenic activity. To germinate, these seeds require host-derived stimulants such as strigolactones, which are a class of phytohormones (Yoneyama et al., 2010). These seeds can survive for decades in soil without germination, and thus it is difficult to eliminate parasitic plants from agricultural fields (Scholes and Press, 2008; Spallek et al., 2013; Gobena et al., 2017).
Parasitic plants develop a unique inducible organ called the haustorium that is used for invasion of the host plants. The haustorium connects the vasculature of the parasite with that of the host to establish a flow of nutrients and water from the host to the parasite (Yoshida et al., 2016; Clarke et al., 2019). Upon recognition of host-derived haustorium-inducing factors (Lynn and Chang, 1990), the parasite initiates organogenesis by activating cell division and cell expansion. In Orobanchaceae parasites, once the haustorium approaches the host, the epidermal cells in proximity to the host cells differentiate into intrusive cells, which have highly elongated shapes and function by intruding into the host (Musselman and Dickison, 1975). Once intrusive cells reach the host vasculature, some of the intrusive cells differentiate into xylem vessels, and subsequently formation of a xylem bridge (XB) between the parasite and host vasculature systems is initiated (Musselman and Dickison, 1975; Cui et al., 2016; Wakatake et al., 2018).
Despite many studies aimed at analyzing the transcriptional changes that occur during haustorium development and host infection in various species of parasitic plants (Ranjan et al., 2014; Ichihashi et al., 2015; Yang et al., 2015; Zhang et al., 2015; Sun et al., 2018; Yoshida et al., 2019), there have been few functional studies of these haustorium-specific genes. To explore the molecular mechanisms of parasitism, including haustorium organogenesis, we established a model parasitic plant system using Phtheirospermum japonicum, a facultative parasitic plant in the Orobanchaceae (Ishida et al., 2016; Spallek et al., 2017). Phtheirospermum japonicum is a self-fertilizing plant with a diploid genome, allowing forward genetics studies (Cui et al., 2016). In addition, an efficient root transformation system by Agrobacterium rhizogenes-mediated hairy root formation has been established, making functional studies of haustorial genes feasible (Ishida et al., 2011). To identify genes important for parasitism, we previously performed transcriptome analyses using rice-infecting P. japonicum and identified genes strongly expressed during the parasitic stage (Ishida et al., 2016). Among these was the auxin biosynthetic gene YUCCA3, which contributes to auxin biosynthesis in the haustorium. The resulting auxin undergoes intercellular transportation and leads to the differentiation of tracheary elements, resulting in the formation of the XB that connects the parasite with the host (Ishida et al., 2016; Wakatake et al., 2018; Wakatake et al., 2020).
In this study, we identified differentially expressed genes in P. japonicum intrusive cells by using a laser microdissection method (LMD) combined with transcriptome analysis. We then used temporal and spatial promoter analyses to establish intrusive cell-specific gene markers. Among the identified upregulated genes, we focused on four genes encoding subtilisin-like serine proteases (subtilases, SBTs) that were exclusively expressed in the intrusive cells. We found that expression of SBT inhibitor proteins in the intrusive cells inhibited the maturation of the haustorium. Thus, our findings provide molecular insight about how parasitic plants develop their haustoria via SBTs.
Results
Intrusive cell-specific gene expression
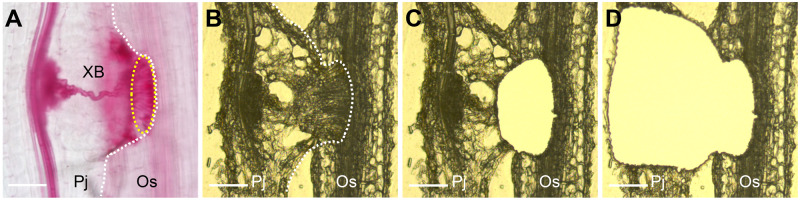
Intrusive cells only form at the interface between a parasite and a susceptible host and thus likely participate in the invasion into host tissues and the molecular dialogue between parasite and host (Goyet et al., 2019). Despite the distinctive nature of intrusive cells, they have not been studied functionally and in detail yet. To seek molecular markers of their function in P. japonicum, we performed LMD coupled with tissue-specific transcriptome analysis. We used rice (Oryza sativa cv Koshihikari) as the host plant because P. japonicum forms haustoria with relatively more intrusive cells on rice than on Arabidopsis roots (Figure 1, A and B). We separated the intrusive regions from other parts of the haustoria using LMD with cryosectioned haustoria (Figure 1, C and D), and obtained transcriptome profiles using the Illumina MiSeq system. After filtering out rice-derived sequences, the reads were mapped onto the draft genome of P. japoncium (Conn et al., 2015) and gene expression values were obtained. Whole-transcriptome data are listed in Supplemental Data Set S1. We detected a total of 3,079 differentially expressed genes between the intrusive cell region and the remainder of the haustorium (Supplemental Figure S1 and Supplemental Data Set S2). Subsequent Gene Ontology (GO) analysis revealed that nine GO terms, including “cell wall,” “response to biotic stimulus,” “transporter activity,” and “metabolic process” were enriched in both regions, whereas 15 GO terms, including “lipid metabolic process” and “carbohydrate metabolic process,” were enriched specifically in other parts of the haustorium (Supplemental Tables S1 and S2). Only one term, “response to stress” was enriched in intrusive cells but not in other parts of the haustorium (Supplemental Table S1). The other terms were not enriched in either region.
Figure 1.
Laser microdissection of the haustorium in P. japonicum. (A) A safranin-O-stained haustorium at 14 dpi. Within the haustorium, P. japonicum establishes a vascular connection with the rice root, called the XB. The dashed white line outlines the haustorium. Intrusive cells located at the interface with the host (dashed yellow circle). (B–D) Sample preparation for tissue-specific transcriptome analysis of a P. japonicum haustorium infecting a rice root. Example of a cryosectioned haustorium before laser microdissection (B), after dissecting the intrusive region (C), and after dissecting the other part of the haustorium (D). Pj, P. japonicum root; Os, Oryza satiνa root. Bars = 100 µm.
Next, we aimed to identify marker genes for intrusive cells as tools to investigate this cell type further. We selected three candidates among the differentially expressed genes that showed strong and specific expression in the intrusive cells: a homolog of Haesa-like1 (HSL1) that we named Intrusive Cell-Specific Leucine-rich repeat receptor-like kinase1 (ICSL1), Germin-Like Protein1 (GLP1), and Constitutive Disease Resistance1 (CDR1). These genes encode a leucine-rich repeat receptor-like kinase, a germin-like protein, and an aspartic protease, respectively (Xia et al., 2004; Ham et al., 2012; Qian et al., 2018). To test whether these genes show specific expression in intrusive cells, we made constructs containing each gene promoter linked to the sequence encoding a nuclear-localized fluorescent protein [3xVenus-nuclear localization signal {NLS}]. We used the constructs to transform P. japoncium and analyzed the Venus fluorescence in P. japoncium haustoria formed after infection of Arabidopsis thaliana roots. For all constructs, fluorescence was detected specifically in the intrusive cells at 2 d post-infection (dpi; Figure 2, A, C and E), and was stronger at 3 dpi (Figure 2, B, D and F). Intrusive cells are derived from epidermal cells, but an epidermis marker construct (pAtPGP4::3xVenus-NLS) is not expressed in the intrusive region (Wakatake et al., 2018). To further verify that ICSL1 expression is specific to intrusive cells, we used the ICSL1 promoter to drive a fluorescent marker module that localizes to the plasma membrane (3xmCherry-SYP; Wakatake et al., 2018). In P. japoncium haustoria that were transformed with both pAtPGP4::3xVenus-NLS and pICSL1::3xmCherry-SYP, we found mutually exclusive expression patterns for the two constructs at 4 dpi (Supplemental Figure S2), with only pICSL1::3xmCherry-SYP expression in the intrusive cells. Based on these analyses, we defined ICSL1 as a reliable intrusive cell marker for further analyses.
Figure 2.
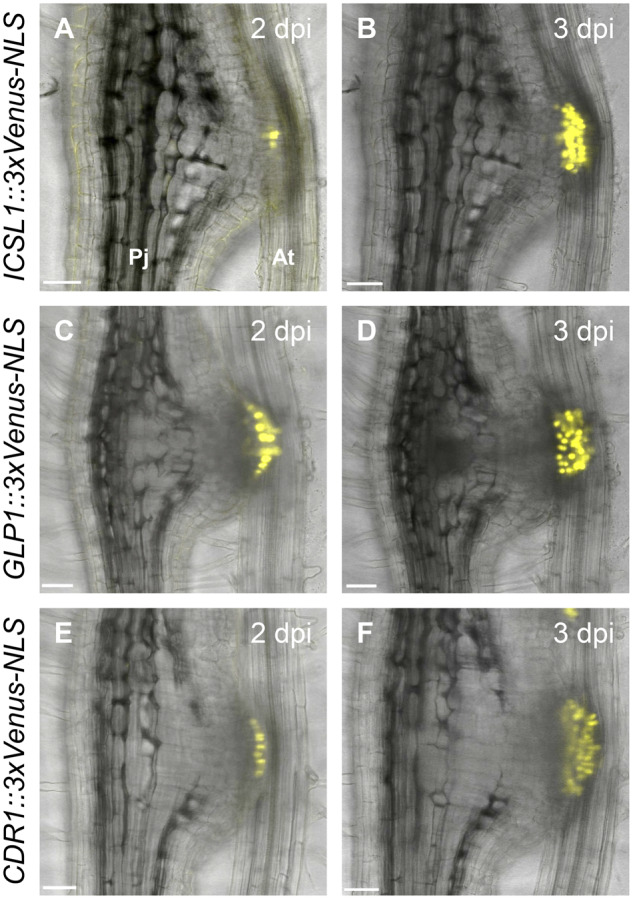
Expression dynamics of intrusive cell markers during haustorium development. Expression patterns of the ICSL1 (A, B), GLP1 (C, D), and CDR1 (E, F) promoters driving a fluorescent marker gene during haustorium development at the indicated dpi time points in P. japonicum. Bright-field and Venus fluorescent images were merged. Pj, P. japonicum root; At, A. thaliana root. Bars = 50 µm.
Phylogeny and expression patterns of SBTs in P. japonicum
Among the genes that were expressed at higher levels in intrusive cells than in the remainder of the haustorium, we found five genes encoding SBTs. This was consistent with our previous report that SBTs are highly expressed during the parasitic stage in P. japonicum (Ishida et al., 2016). We therefore hypothesized that SBTs in intrusive cells may contribute to the host invasion process. To classify the SBT genes expressed in intrusive cells, we first identified all SBTs in the P. japonicum genome (Conn et al., 2015) on the basis of their Asp–His–Ser catalytic triad and their peptidase S8 family domain (Smith et al., 1966; Wright et al., 1969). As a result, 97 putative SBTs met these criteria (Figure 3). A phylogenetic analysis revealed that the five SBTs upregulated in intrusive cells all belong to Group 1 (Taylor and Qiu, 2017; Reichardt et al., 2018; Figure 3), which contains many SBTs involved in biotic interactions. These genes were thus designated as SBT1.1.1, SBT1.2.3, SBT1.5.2, SBT1.7.2, and SBT1.7.3. We also found that many of the 97 SBT genes in P. japonicum were induced in the haustorium at 3 dpi or later (Figure 3; transcriptome data from Kurotani et al., 2020, available from the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/) under the accession number, DRA010010), indicating that these SBTs were activated after attachment to the host.
Figure 3.
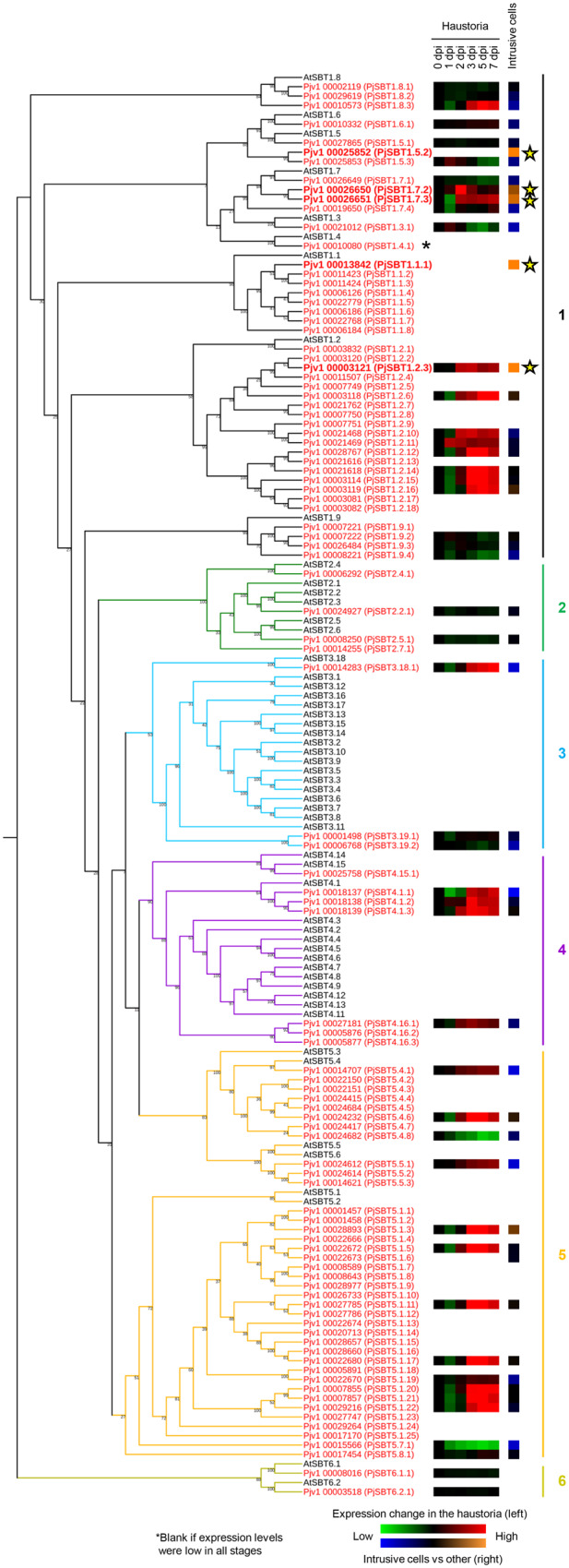
Phylogeny of the SBTs in P. japonicum and Arabidopsis. The 97 SBTs in P. japonicum are shown in red and the 55 SBTs in Arabidopsis are shown in black. According to Rautengarten et al. (2005), the SBTs are categorized into six groups. The Group-6 SBTs represent the outgroup. The green/red squares indicate the SBT gene expression levels at different time points in the haustorium of P. japonicum (Kurotani et al., 2020). The blue/orange squares indicate the SBT gene expression levels in the intrusive cells relative to their expression in other haustorial parts. Stars indicate the SBTs with higher expression in the intrusive cells than in the other haustorial parts.
SBTs specifically expressed in intrusive cells
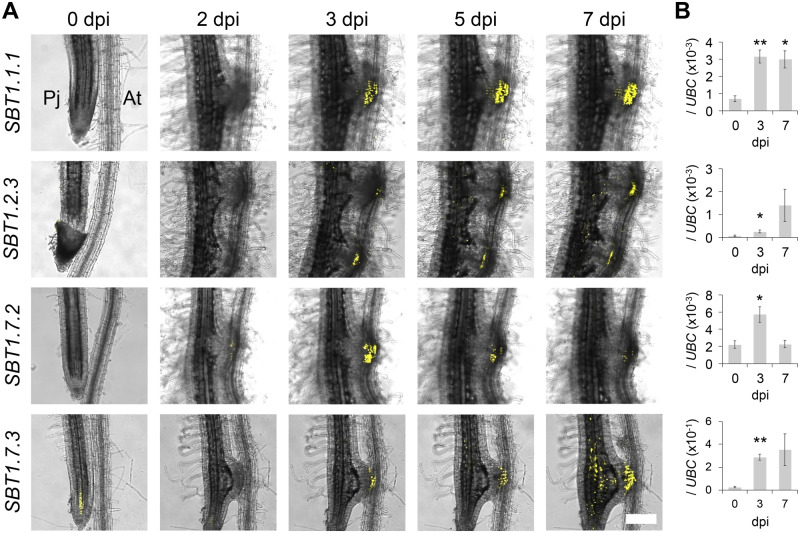
To confirm the expression patterns of the five SBTs upregulated in intrusive cells, we made constructs containing each gene promoter linked to the 3xVenus-NLS module, transformed P. japoncium with the constructs, and analyzed Venus fluorescence in P. japoncium haustoria after infection of A. thaliana roots with a confocal microscope. The Venus signal driven by the putative SBT1.5.2 promoter was not detected at selected time points. We thus focused on the remaining four SBTs in further analyses. An alignment of their protein products is shown in Supplemental Figure S3. Since these SBT proteins contain signal peptides, it is likely that they are localized extracellularly. The promoters of SBT1.1.1, SBT1.2.3, and SBT1.7.3 were sufficient to drive detectable Venus expression in the intrusive cells at 3–7 dpi (Figure 4A). Expression of SBT1.7.2 was more transient, with weaker signal at 7 dpi as compared with at 3 and 5 dpi. For the SBT1.7.3 promoter, signals were also detected in vascular cells in the meristematic region (Figure 4A). We used reverse transcription quantitative polymerase chain reaction (RT-qPCR) to analyze induction of the four SBTs in whole haustoria, and found that the levels of induction at 3 and 7 dpi were consistent with the results from the Venus fluorescence analysis (Figure 4B). We also analyzed expression of a 3xVenus-NLS construct driven by the SBT1.7.1 gene, which is phylogenetically close to SBT1.7.2 and SBT1.7.3 (Figure 3). Florescence from this construct was observed in the epidermal cells but not the intrusive cells (Supplemental Figure S4). Since the intrusive cells are uniquely found in parasitic plants, SBT1.1.1, SBT1.2.3, SBT1.7.2, and SBT1.7.3 expression in this cell type suggests that these SBTs function in parasitism.
Figure 4.
Expression dynamics of the SBTs during haustorium development. (A) Expression patterns of SBT promoters in P. japonicum during haustorium development at the indicated time points. Bright-field and Venus fluorescent images were merged. Pj, P. japonicum root; At, A.thaliana root. The scale bar in the right panel refers to all other panels too. Bar = 200 µm. (B) The relative expression level of each SBT at 0, 3, and 7 dpi in isolated haustoria. “0 dpi” values represent the expression of the marker genes in the root elongation zones of noninfecting P. japonicum roots. Representative data are shown (mean ± se of four technical replicates). We used PjUBC as a reference gene. The experiments were performed three times with similar results. Asterisks indicate statistical significance (Welch’s t test, *P < 0.05, **P < 0.01).
SBTs play important roles in development of the host–parasite connection via auxin response
The four SBT genes discussed above may be functionally redundant, and silencing multiple genes in P. japonicum is challenging due to the lack of a transgenerational transformation method. Therefore, we used an SBT inhibitor protein to analyze the function of the intrusive cell-specific SBTs. For this purpose, we chose extracellular proteinase inhibitor 10 (Epi10) from Phytophthora infestans. Epi10 inhibits bacterial and plant SBTs but does not inhibit the other major serine proteases tested, namely trypsin and chymotrypsin (Tian et al., 2005; Schardon et al., 2016). In Arabidopsis, the tissue-specific expression of Epi10 results in the efficient inhibition of partly redundant SBTs (Schardon et al., 2016; Stührwohldt et al., 2020). Therefore, we considered that Epi10 is a suitable tool to inhibit several PjSBTs (Pj, P. japonicum) simultaneously in a tissue-specific manner. To specifically inhibit the SBTs expressed in developing haustoria, we used the promoter sequences of SBT1.1.1 and SBT1.2.3 to drive expression of the Epi10 coding region. We compared the development of haustoria in P. japonicum roots transformed with these constructs with the development of haustoria in control roots transformed with an empty vector. We found that hairy roots transformed with the Epi10 constructs showed reduced XB formation in the haustoria at 5 dpi after infection of Arabidopsis roots when compared with control hairy roots (Figure 5, A–D and Supplemental Figure S5–7). The same effect on XB formation was also observed in haustoria expressing the SBT inhibitor AtSPI-1 (Supplemental Figure S7; Hohl et al., 2017). EPI10 and AtSPI-1 belong to different families of proteinase inhibitors, the I1 and I9 families, respectively. Supported by the use of two structurally unrelated yet specific proteinase inhibitors, we conclude that SBT activity is required in haustoria for normal XB development. This conclusion is also supported by the previous report of the potential involvement of SBT in xylem formation (Zhao et al., 2000).
Figure 5.
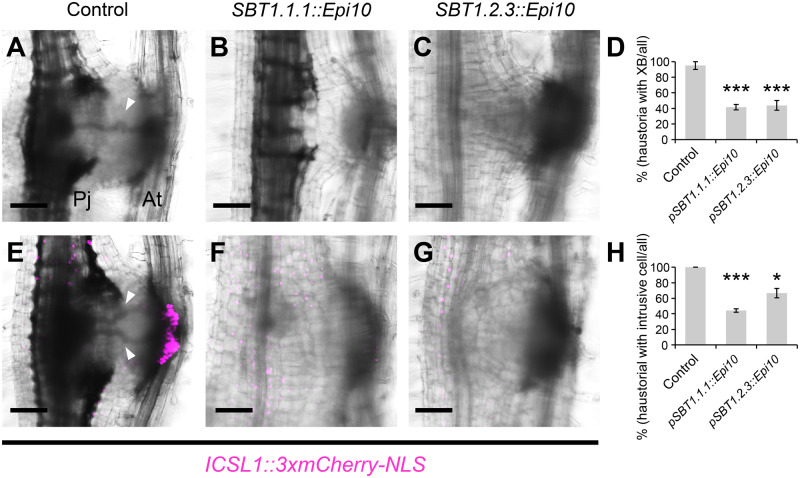
Effect of the SBT inhibitor on haustorial formation. (A–C) Representative images of the haustoria that did (A) and did not (B, C) form a XB at 5 dpi. (A) control; (B) construct containing SBT1.1.1::Epi10; (C) construct containing SBT1.2.3::Epi10. (D) Ratio of the number of haustoria that formed an XB to the number of all haustoria for each construct (means ± SE of 4 replicates, n = 4–9). (E–G) Representative images of the haustoria that did (E) and did not (F, G) form the intrusive cells at 5 dpi. (B) Control, (F) construct containing SBT1.1.1::Epi10, (G) construct containing SBT1.2.3::Epi10. ICSL1 promoter was used as an intrusive cell marker. Bright-field and mCherry fluorescent images are merged. (H) Ratio of the number of haustoria that formed intrusive cells to the number of all haustoria for each construct (means ± se of four replicates, n = 4–9). Arrowheads point to XB. Asterisks indicate statistical significance (Welch’s t test, *P < 0.05, **P < 0.01, ***P < 0.001). Pj, P. japonicum; At, A. thaliana;. Bars = 100 µm.
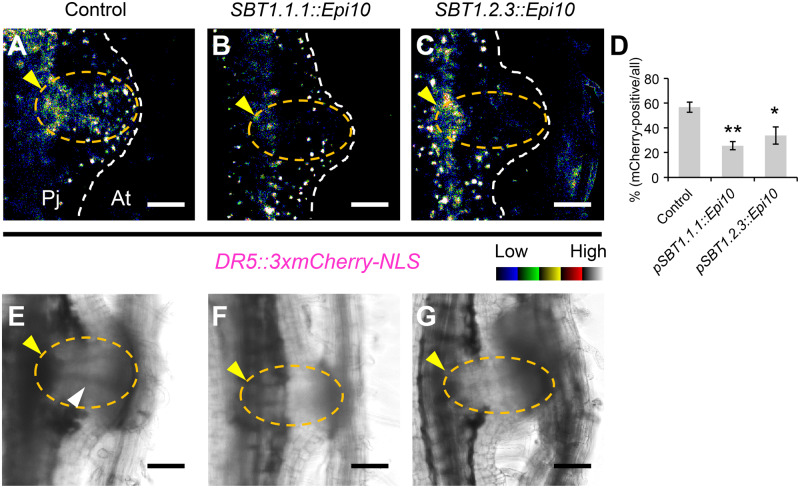
Next, we investigated whether the Epi10-transformed hairy roots would show other developmental abnormalities. We were particularly interested in the effects on intrusive cells, given the specific expression of SBT1.1.1 and SBT1.2.3 in intrusive cells. Therefore, we monitored expression of the intrusive cell marker ICSL1 (Figure 2, A and B) in the Epi10-expressing haustoria. To accomplish this, we transformed P. japonicum roots with each of the Epi10 constructs and with a construct encoding the mCherry fluorescent protein with a 3xmCherry-NLS driven by the ICSL1 promoter. In control roots transformed with pICSL1::3xmCherry-NLS but not with Epi10, all haustoria showed specific mCherry fluorescence in the intrusive cells. In contrast, <45% of the pSBT1.1.1::Epi10 haustoria, and ∼67% of the pSBT1.2.3::Epi10 haustoria showed mCherry fluorescence at 5 dpi (Figure 5, E–H). We noticed that some, but not all, of the pSBT1.1.1::Epi10 and pSBT1.2.3::Epi10 haustoria recovered intrusive cells and XB at 9 dpi, indicating that either Epi10 causes delay of haustorium maturation, or Epi10 effects are reduced and variable at 9 dpi due to the variation of promoter activity at this time point (Supplemental Figure S6). These results suggest that the intrusive cell-specific SBT activities promote the maturation of haustoria by regulating the development of intrusive cells and the subsequent XB formation. Lack of intrusive cell identity may affect auxin distribution (Ishida et al., 2016; Wakatake et al., 2020). Therefore, we investigated whether Epi10 expression alters auxin response within the haustorium by using the 3xmCherry-NLS module controlled by the synthetic, auxin-responsive DR5 promoter (Ulmasov et al., 1997). Most of the auxin response in the central region of the haustoria, but not around the xylem plate, was diminished by Epi10 (Figure 6). As well as the expression analysis of the ICSL1, some, but not all, of the pSBT1.1.1::Epi10 and pSBT1.2.3::Epi10 haustoria recovered auxin response at 9 dpi (Supplemental Figure S8) Taken together, our results suggest that the SBT activities regulate auxin-dependent maturation of P. japonicum haustoria.
Figure 6.
Effect of the SBT inhibitor on auxin response. (A–C) Representative images of the haustoria in which auxin response was observed (A) and not observed (B, C) at the center of haustorium at 5 d dpi. (A) Control, (B) construct containing SBT1.1.1::Epi10, (C) construct containing SBT1.2.3::Epi10. DR5 promoter was used as an auxin response marker. mCherry fluorescence intensity is depicted in a 5 ramps spectrum. (D) Ratio of the number of haustoria in which auxin signaling was observed around the XB to the number of all haustoria for each construct (means ± SE of four replicates, n = 2–8). Asterisks indicate statistical significance (Welch’s t test, *P < 0.05, **P < 0.01). (E–G) Bright field images corresponding with (A–C). Dashed white lines, dashed orange lines, yellow arrowheads, and a white arrowhead indicate the edge of the haustorium, the center of haustorium, xylem plates, and an XB, respectively. Pj, P. japonicum; At, A. thaliana. Bars = 100 µm.
Discussion
We used P. japonicum as a model parasitic plant to elucidate the molecular mechanisms that regulate parasitic functions in the intrusive cells of the haustoria. By using tissue-specific RNA-seq analysis coupled with LMD, we identified a number of genes that are upregulated in intrusive cells (Supplemental Data Set S2). A previous study used the LMD method to reveal genes that are specifically expressed at the host–parasite interface, which includes the intrusive cells, in the facultative hemiparasite Tryphisaria versicolor infecting Zea mays or Medicago truncatula (Honaas et al., 2013). Here, the GO term “transcription factor activity” was overrepresented, whereas the term “transporter activity” was under-represented at the host–parasite interface. In contrast, we found that the term “transporter activity” was enriched in the intrusive cells in P. japonicum (Supplemental Table S1). Although a direct comparison of the two experiments is difficult, the results may indicate that T. versicolor and P. japonicum employ different strategies to invade their particular hosts. Enrichment of “transporter activity” in intrusive cells also suggests that these cells may have a function in nutrient transfer, which would be consistent with their position at the interface between host vasculature and parasite haustorium. We found that several GO terms, such as “lipid metabolic process” and “carbohydrate metabolic process,” are strongly enriched in the rest of the haustorium but not in the intrusive cells (Supplemental Tables S1 and S2). Yoshida et al. (2019) showed that genes categorized under the GO terms “protein metabolic process,” “carbohydrate metabolic process,” and “catabolic process” are upregulated in rice-infecting Striga hermonthica at 7 dpi, which is when the host–parasite connections in the haustorium are established. This result indicates that metabolically demanding processes such as morphological changes are activated in the haustoria in the family Orobanchaceae.
Based on our intrusive cell-specific transcriptome, we established that the three P. japonicum genes ICSL1, GLP1, and CDR1 showed strong and specific expression in intrusive cells and could be used as molecular markers for these cells (Figure 2). ICSL1 is homologous to the Arabidopsis HSL1 receptor, which localizes to the plasma membrane and recognizes peptide hormones (Jinn et al., 2000; Stenvik et al., 2008). The phylogenetically closest Arabidopsis ICSL1 homolog, however, is AtRLP52, a receptor-like kinase associated with disease resistance and an unknown ligand (Ramonell et al., 2005; Ellendorff et al., 2008; Supplemental Figure S9). Thus, it is possible that ICSL1 also recognizes peptide hormones. Further experiments are required to identify the unknown ICSL1 ligand and to determine if it originates from the parasite or the host. The second marker gene encodes GLP1, which belongs to a GLP superfamily that consists of structurally diverse plant glycoproteins including enzymes such as oxalate oxidases and superoxide dismutases (Rietz et al., 2012; Sakamoto et al., 2015). Phylogenetic analysis revealed that GLP1 in P. japonicum is closely related to Arabidopsis GLP1 and GLP3, which lack oxalate oxidase activity, and to GhABP19 in Gossypium hirsutum, a superoxide dismutase potentially regulating redox status (Pei et al., 2019; Supplemental Figure S10). Interestingly, the only other gene with experimentally confirmed expression in intrusive cells encodes a peroxidase in S. hermonthica (Yoshida et al., 2019). Also, chemically inhibiting peroxidase activity, and thus altering the redox homeostasis, reduces haustorium formation in Striga spp. and Triphysaria (Wada et al., 2019; Wang et al., 2019). The expression of a superoxide dismutase in P. japonicum intrusive cells further supports a role for redox regulating enzymes in haustorium development. The third intrusive cell-specific gene that we identified encodes CDR1, which belongs to a family of aspartic proteases. The P. japonicum CDR1 is a homolog of aspartic proteases that regulate disease resistance signaling in Arabidopsis (Xia et al., 2004; Supplemental Figure S11). It is currently not known if CDR1 regulates defense responses in P. japonicum; however, the expression of defense-related genes in P. japonicum haustoria was seen in a previous microarray study (Ishida et al., 2016).
ICSL1, GLP1, and CDR1 show the same spatio-temporal expression pattern, with detectable expression beginning at 2 dpi at the interface with the host (Figure 2, A, C and E). This is the time point when expression of an epidermis marker ceases in the same region (Wakatake et al., 2018). Thus, the developmental switch from epidermis to intrusive cell is likely to be activated around this time point. Considering the mutually exclusive expression patterns of the epidermis marker gene and the intrusive cell marker gene (Supplemental Figure S2), we would expect that the transcriptional landscapes of these two cell types are substantially different. Specific expression of SBT1.7.1 in the epidermal cells, but not in the intrusive cells, further supports this idea (Supplemental Figure S4). The intrusive cell-specific markers identified in this study were expressed uniformly in the entire intrusive region (Figure 2). However, only a fraction of those cells differentiate into tracheary elements to be part of the XB (Wakatake et al., 2020). Thus, it seems that there are different types of cells in the intrusive cell population. This is likely due to nonuniform auxin response in the intrusive region. Thus, further detailed analyses are required to reveal the mechanisms by which auxin responses are controlled in intrusive cells.
A transcriptome analysis of P. japonicum haustoria in our previous study revealed that 7 of the 10 genes with the highest, exclusive expression in the parasitic stage were SBTs (Ishida et al., 2016; Supplemental Table S3). Many SBT genes are also upregulated in Striga spp. upon infection (Yoshida et al., 2019). SBTs are a widespread protein family existing in eubacteria, archaebacteria, eukaryotes, and viruses (Rawlings and Barrett, 1994; Schaller et al., 2018). In plants, SBTs are required for the maturation of plant peptide hormones, leading to phenotypic changes such as root elongation, abscission of floral organs, and embryonic cuticle integrity (Matsubayashi, 2014; Ghorbani et al., 2016; Schardon et al., 2016; Doll et al., 2020; Reichardt et al., 2020). Here, we identified four SBTs that are exclusively expressed in intrusive cells (Figures 3 and 4), and they all belong to Group 1. Groups 1 and 5 SBTs are highly expanded in parasitic plants compared with those in Arabidopsis, whereas Groups 3 and 4 are much smaller in P. japonicum than in Arabidopsis (Figure 3 and Supplemental Figure S12). Such apparent species-specific expansion of SBTs might indicate distinct biological function for each species. In particular, Clade 6 is highly expanded in S. hermonthica but not in other species tested. Further analyses are required for elucidating such expanded clades in particular species. Importantly, more than 40% of SBTs in the parasites P. japonicum, Striga asiatica, and S. hermonthica belong to Group-1 SBTs. In addition, Group-1 SBTs also expanded in plants that undergo symbiosis with nitrogen-fixing bacteria. Group-1 SBTs also include many that are involved in plant defense (Taylor and Qiu, 2017; Reichardt et al., 2018). These findings indicate that Group-1 SBTs may have evolved for biotic interactions, including parasitism. The molecular functions and substrates of several Group-1 SBTs in nonparasitic plants have been investigated. For example, Phytaspase 2, a Group-I SBT in tomato (Solanum lycopersicum), cleaves and activates the peptide hormone PHYTOSULFOKINE (PSK), which induces stress-induced flower drop in tomato, in addition to its well-established growth regulatory and immune-modulating activities (Reichardt et al., 2020). Genes for PSK and its candidate receptor are present in the P. japonicum genome. Interestingly, expression of these two genes is upregulated in the haustoria but not in the intrusive cells. If SBT1.1.1 or SBT1.2.3 is involved in PSK precursor processing, the expression of these two proteins in two different cell types would suggest that they may facilitate the communication between haustorial tissues. A similar tissue–tissue dialogue mediated by SBTs was recently shown to operate during Arabidopsis seed development (Doll et al., 2020). In contrast, Arabidopsis SBT1.2 (alias SDD1), a homolog of P. japonicum SBT1.2.3, contributes to stomatal development (Von Groll et al., 2002). The substrates of SDD1 have not been identified, but were suggested to also include plant peptide hormones.
We showed that SBT activity in intrusive cells contributes to haustorium development (Figures 4–6; Supplemental Figure S6 and S7). Intrusive cells per se were still formed in Epi10-transgenic hairy roots (Figure 5, B and E; Supplemental Figure S5). Epi10 was previously used to overcome genetic redundancy in SBT loss-of-function studies (Schardon et al., 2016; Stührwohldt et al., 2017, 2020). The delay in haustorium development observed in Epi10-transgenic hairy roots (Supplemental Figure S6), together with the specific expression of several SBTs in intrusive cells, supports our hypothesis that SBTs contribute to the differentiation of intrusive cells into xylem vessels, leading to XB formation. However, Epi10 comprises three inhibitor domains, only one of which (EPI10b, the so-called atypical Kazal domain) is SBT-specific (Tian and Kamoun, 2005; Tian et al., 2005). Therefore, to exclude the possibility that Epi10-mediated inhibition of other, as yet unknown, serine proteases is responsible for the delay in haustorium development, we repeated the experiment using AtSPI-1 (At, A. thaliana) as a structurally unrelated yet specific SBT inhibitor (Hohl et al., 2017). The same effect on haustorium development was observed in AtSPI-1-transgenic hairy roots as for Epi10, confirming a role for SBTs in XB formation (Supplemental Figure S7). The specific expression of SBTs during the parasitic stage is shared between P. japonicum and S. hermonthica, suggesting that SBTs are important for parasitism in the family Orobanchaceae. Intrusive cell-specific SBTs have not yet been identified in S. hermonthica. However, an S. hermonthica SBT is expressed specifically in the haustorial hyaline body (Yoshida et al., 2019). The hyaline body consists of parenchymatic tissue in the central region of the haustorium and is characterized by dense, organelle-rich cytoplasm, abundant paramural deposits, and high metabolic activity (Visser et al., 1984). The hyaline body has not yet been identified in P. japonicum and it may be morphologically distinct from that in S. hermonthica. The further identification of cell type-specific SBTs in haustoria may facilitate the identification and functional studies of the hyaline body in P. japonicum.
Our data suggest that expression of the SBTs may be initiated in cells that eventually become intrusive cells, then the SBT activities contribute to the maturation of the intrusive cells, where the marker gene ICSL1 is expressed. After expression of intrusive cell-specific markers, intrusive cells may invade host tissue, intrusive cells reach the host vasculature, auxin is transported inward toward the root vasculature, and then the XB is formed (Wakatake et al., 2020). Importantly, treatment with haustorium-inducing factors induces haustorium organogenesis in P. japonicum without hosts, but the intrusive cells and XB are not formed in these haustoria (Ishida et al., 2016; Goyet et al., 2019). Identification of the unknown, host-derived signals required for intrusive cell-specific SBT induction will provide insight into the mechanisms by which Orobanchaceae parasites invade the host plants.
We also found that several SBTs were induced at the later stages of the infection both in P. japonicum and in S. hermonthica (Supplemental Figure S12). The late expression of SBTs during the infection indicates that parasitic plants utilize SBTs also after attachment to the host, possibly in regulating parasitism. We focused our study on SBTs with a role in haustorium development that can be studied with transgenic P. japonicum hairy roots (Ishida et al., 2016; Wakatake et al., 2020). To address the role of SBTs in later stages of the infection would require the generation of stable transgenic plants. In addition, many SBT clades were found to be species-specific, suggesting that each parasite has recruited SBTs independently to promote parasitism. Since parasitic plants are able to transfer molecules such as phytohormones and microRNAs (Spallek et al., 2017; Shahid et al., 2018), it is possible that peptides processed by SBTs in the haustorium can be transported from the parasite into the host. Further analyses of peptides in infected hosts will be required to assess this hypothesis. In summary, our study showed that SBTs are required for haustorium development. Functional studies of other parasite SBTs and their targets will provide important insights into plant parasitism in future studies.
Materials and methods
Plant materials and growth conditions
Phtheirospermum japonicum (Thunb.) Kanitz and rice (Oryza sativa L. subspecies japonica, cv Koshihikari) seeds were handled as described previously (Yoshida and Shirasu, 2009; Ishida et al., 2011). For in vitro germination, P. japonicum seeds were surface sterilized with 10% (v/v) commercial bleach solution (Kao, Tokyo, Japan) for 5 min, followed by five rinses with sterilized water. Seeds were then sown on solid half-strength MS medium (0.8% (w/v) Bacto agar, 1% (w/v) sucrose, pH 5.8). After stratification at 4°C in the dark overnight, plants were grown either vertically for infection assays or horizontally for transformation, at 25°C under long-day conditions (16-h light, 8-h dark). Arabidopsis (A. thaliana, ecotype Col-0) seeds were surface sterilized with 5% (v/v) commercial bleach solution for 5 min, followed by five rinses with sterilized water. Seeds were then sown on solid half-strength MS medium. After stratification at 4°C in the dark overnight, plants were grown vertically at 22°C under long-day conditions. Rice seeds were sterilized with 70% (v/v) ethanol for 3 min, followed by incubation in a 50% (v/v) commercial bleach solution for 20 min. After five rinses with sterilized water, seeds were sown on quarter-strength Gamborg’s B5 medium (Sigma) with 0.8% (w/v) agar (INA). Plates were kept vertically at 26°C under long-day conditions.
Histological staining
Safranin-O staining of whole haustorium was performed as described previously (Wakatake et al., 2018).
Sample preparation for RNA-seq
Ten-day-old P. japonicum seedlings were transferred to quarter-strength Gamborg’s B5 medium (0.7% (w/v) agar; INA) and grown vertically at 25°C under long-day conditions for 2 d. These seedlings and 7-d-old rice seedlings were transferred together to new quarter-strength Gamborg’s B5 plates for infection at 25°C under long-day conditions. At 5 dpi, haustoria were excised and immediately soaked in chilled RNAlater (Sigma) and stored at 4°C. Samples were embedded in FSC 22 frozen section media (Leica biosystems) in self-made aluminum molds in an acetone bath at −75°C. Frozen blocks were sectioned to 20 µm thickness using a cryostat (Leica CM3050S) with adhesive seals at −30°C. Sections were transferred to room temperature and immediately air-dried. The intrusive regions and the other parts of the haustorium were dissected using a Leica LMD7000. Dissected tissues were collected in the lids of 0.5-mL microtubes filled with RNA extraction buffer. Approximately 20 haustoria were used for one biological replicate. Total RNAs were extracted using the Picopure RNA isolation kit (Arcturus) according to the manufacturer’s instructions. DNase I (Qiagen) was applied to the column during the procedure to digest genomic DNA. Elution buffer (11 µL) was used to elute the total RNA. The quality and quantity of the total RNA were assessed using a Bioanalyzer (Agilent Technologies) and the RNA 6000 pico kit.
Whole-Transcripts Amplification and Library Preparation
The procedure for whole-transcript amplification was based on the Quartz-seq method (Sasagawa et al., 2013). Approximately 1 ng of total RNA was used as a starting material. Total RNA was denatured (70°C for 90 s) and primed (35°C for 15 s) followed by first-strand synthesis (35°C for 5 min; 45°C for 20 min; 70°C for 10 min) with reverse transcriptase and oligo-dT-containing RT primers, using the SuperScript III system (Life Technologies). Single-stranded cDNA was purified using AMPure XP magnetic beads (Beckman Coulter). The remaining RT primers were digested on the beads with Exonuclease I (TAKARA; 37°C for 30 min; 80°C for 20 min). Subsequently, the poly-A-tailing reaction was performed with terminal transferase (Roche; 37°C for 50 s; 65°C for 10 min) followed by second-strand synthesis using a tagging primer with MightyAmp DNA polymerase (TAKARA; 98°C for 130 s; 40°C for 1 min; 68°C for 5 min). PCR enrichment was performed using the enrichment primers and MightyAmp DNA polymerase (98°C for 10 s; 65°C for 15 s; 68°C for 5 min). The total number of PCR cycles was either 14 or 15, depending on the amount of total RNA input. The amplified cDNA was purified using DNA concentrator-5 (Zymo Research) according to the manufacturer’s instructions. The size distribution of the amplified cDNA was assessed using the Bioanalyzer with a High sensitivity DNA kit. The amount of cDNA in each sample was measured using the Qubit dsDNA HS assay kit (Thermo Fisher). Library preparation was performed using the Nextera XT kit (Illumina) according to the manufacturer’s instructions. The KAPA LA kit (Nippon Genetics) was used for library amplification after fragmentation. PCR cycles were adjusted to eight or nine depending on the amount of input cDNA. Libraries were pooled and sequenced with three runs on the MiSeq using the reagent kit V2 (Illumina).
Bioinformatics Analysis
The adapter sequences in the primers for library preparation and whole transcript amplification were trimmed and low-quality sequences were removed. Quality-filtered reads were mapped to Nipponbare-reference-IRGSP-1.0 pseudomolecules (Kawahara et al., 2013) using the CLC genomics workbench (ver. 8.0, Qiagen) with a threshold setting of 95% match. The remaining unmapped reads were considered as P. japonicum-derived sequences and mapped to the P. japonicum draft genome with a threshold setting of 90% match. Unique read counts obtained for each gene model were used for further analysis. Differential gene expression analysis was performed in R with the TCC package (Sun et al., 2013; https://www.R-project.org/.). GO analysis was performed with GO seq using the results of the differential gene expression analysis (Young et al., 2010). We used the CLC Main Workbench (ver. 8.0.1, Qiagen) for identification of the putative SBTs in P. japonicum, vector design, and sequence analyses.
Phylogenetic Analysis
Phylogenetic analyses were performed using the CLC Genomics Workbench (ver. 8.0, Qiagen). Predicted amino acid sequences were trimmed using trimAL (Capella-Gutiérrez et al., 2009), followed by alignment. Based on the alignment, the phylogenetic tree was drawn using the maximum-likelihood method. For comparing the SBTs in P. japonicum and Arabidopsis, the reliability of the trees was tested by bootstrap analysis with 1,000 resamplings. For comparing the SBTs in P. japonicum, Striga asiatica, and Striga hermonthica, the reliability of the trees was tested by bootstrap analysis with 100 resamplings. The figure was generated by iTOL (ver. 5; https://itol.embl.de/; Letunic and Bork, 2007).
Cloning
Golden Gate cloning technology was used for cloning (Engler et al., 2014). All the BpiI and BsaI restriction sites within the cloned DNA sequences were mutated. The golden gate modules 3xVenus-NLS, 3xmCherry-SYP, pACT::3xmCherry-NLS, and pAtPGP4::3xVenus-NLS were described previously (Ishida et al., 2016; Wakatake et al., 2018).
Vectors Containing Intrusive Cell Markers. The PjICSL1 (2,652 bp), PjGLP1 (2,634 bp), and PjCDR1 (2,496 bp) promoter regions were each PCR-amplified as two fragments from P. japonicum genomic DNA and cloned separately into pAGM1311. The fragments were then combined into the pICH41295 level-0 vector. The promoter sequences were next assembled into level-1 vectors together with the fluorescent protein module and the 3ʹ-UTR and HSP18.2 terminator module (UTR, untranslated region; HSP, Heat Shock Protein). pICSL1::3xmCherry-SYP was further combined with pAtPGP4::3xVenus-NLS in the binary vector pAGM4723 (Engler et al., 2014).
Vectors Containing the SBT Promoters. The SBT1.5.2 (2,000 bp) and SBT1.7.3 (1,799 bp) promoter regions were each PCR-amplified as three fragments from P. japonicum genomic DNA and cloned separately into pAGM1311. The fragments were then combined into the pICH41295 level-0 vector. The SBT1.1.1 (1,691 bp), SBT1.2.3 (1,955 bp), SBT1.7.2 (1,802 bp), and SBT1.7.1 (1,864 bp) promoter regions were each PCR-amplified as one fragment from P. japonicum genomic DNA and cloned into the pICH41295 level-0 vector. The promoter sequences were then assembled into level-1 vectors together with the Venus protein module fused with the NLS, and the 3ʹ-UTR and terminator module. We also assembled the actin promoter (Wakatake et al., 2018) into a level-1 vector together with the mCherry protein module fused with the membrane localization signal (Syntaxin of Plant 122, SYP122), and the 3ʹ-UTR and HSP terminator module, to generate the pACT::3xmCherry-SYP122 transcription unit. Each pSBT::3xVenus-NLS unit was further combined with pACT::3xmCherry-SYP122 in the binary vector pAGM4723.
Vectors Containing Epi10 Sequence. A codon-optimized Epi10 construct containing the signal peptide-encoding region of AtSBT1.7 (At5g67360; Schardon et al., 2016) was amplified with Golden Gate-compatible primers and cloned into pAGM9121 to generate a level-0 CDS1 module (Engler et al., 2014). The Epi10 level-0 CDS1 module was then assembled between the PjSBT promoter modules and an HSP terminator sequence (Wakatake et al., 2018). The final level-1 constructs were combined with the fluorescent transformation marker p35S:3xVenus-NLS in the binary vector pAGM4723. Using previously described AtSPI-1 constructs (Hohl et al., 2017), Golden Gate-compatible, AtSPI-1-containing plasmids were generated similarly to the Epi10 constructs, except that we introduced a synonymous mutation to the AtSPI-1 sequence to remove the endogenous BpiI restriction site.
Transformation of P. japonicum
Transformation of P. japonicum was performed as previously described by Ishida et al. (2011) with several modifications. Silwet L-77 (Bio Medical Science) was added to an A. rhizogenes bacterial solution (OD600 = 0.1) to a final concentration of 0.02% (v/v) just prior to transformation. Six-day-old P. japonicum seedlings were immersed in the bacterial/Silwet L-77 solution and submitted to ultrasonication using a bath sonicator (Ultrasonic Automatic Washer; AS ONE) for 10–15 s. The sonicated seedlings were vacuum infiltrated for 5 min. The seedlings were transferred to freshly made co-cultivation medium [Gamborg B5 agar medium with 1% {w/v} sucrose and 450 µM acetosyringone] and kept in the dark at 22°C for 2 d. After co-cultivation, the seedlings were transferred to B5 agar medium containing cefotaxime (300 µg mL−1). After 3–4 weeks, the transformed roots were used for infection. Identification of the transgenic roots was performed as previously described by Ishida et al. (2016).
Microscopy
Microscopy with transformed P. japonicum was performed as previously described by Wakatake et al. (2018) with several modifications. In brief, we selected P. japonicum plants with hairy roots generated from aerial parts as candidate transformants. We transferred them from B5 media to 0.7% agar (Ina food industry) without any nutrient to induce root elongation during 1-d incubation at 25°C under long-day conditions. We then selected hairy roots showing fluorescence as transformants using a dissection microscope (E165). We transferred the transgenic hairy roots onto glass-bottomed petri dishes (IWAKI, Japan) and covered them with thin 0.7% (w/v) agar containing cefotaxime (300 µg mL−1), followed by 1-d incubation at 25°C under long-day conditions for acclimation. To initiate parasitism, we placed 7-d-old Arabidopsis seedlings next to the transgenic roots between glass bottom and thin agar, followed by incubation at 25°C under long-day conditions. We observed fluorescence in the induced haustoria with an inverted confocal microscope (Leica, TCS SP5 II). For Venus fluorescence, we used a 514-nm laser for excitation and detected emission at 525–560 nm. For mCherry fluorescence, we used a 543-nm laser for excitation and detected emission at 570–640 nm. We selected the expression pattern observed in the majority of the transgenic hairy roots transformed with the same construct as the representative phenotype, after exclusion of the transgenic roots that did not establish haustorium development. For the figures in this study, we selected the haustoria exhibiting strong fluorescence to clearly show the expression patterns in the cells of the deeper layers of the haustoria.
RT-qPCR
For extraction of total RNA from the haustoria, P. japonicum seedlings were grown vertically for 9 d followed by incubation for 2 d on water agar plates before infection of 7-d-old A. thaliana seedlings. At 3 and 7 dpi, haustoria were excised and immediately frozen in liquid nitrogen. We removed the A. thaliana roots as much as possible. Ten to twenty haustoria were used for each sample. For the 0-dpi samples, we used the root elongation zones from P. japonicum seedlings without infection. Total RNA was extracted using the RNeasy plant mini kit (QIAGEN), followed by cDNA synthesis using the ReverTra Ace qPCR RT Kit (TOYOBO). During RNA extraction, we treated with DNase to remove residual genomic DNA. RT-qPCR was performed as previously described by Spallek et al. (2017). PjUBC2 was used as a reference gene. The expression level of each gene was quantified using the ddCt method (dd, delta-delta).
Primers
All primers used for library preparation, cloning, and RT-qPCR are listed in Supplemental Table S4.
Statistics
Welch’s t test was performed in Microsoft Excel 2016.
Accession numbers
Assembled P. japonicum draft genome and annotation are available in the DNA Data Bank of Japan (https://www.ddbj.nig.ac.jp/index-e.html) with accession numbers BMAC01000001-BMAC01010559 and also available at Dryad database (https://doi.org/10.5061/dryad.vt4b8gtpt).
Sequence data from this article can be found in the GenBank/EMBL libraries under accession number BankIt2316603: MT149970-MT150066 (SBTs); BankIt2324477: MT226912 (ICSL1), MT226913 (GLP1), MT226914 (CDR1).
MIAME-compliant (minimum information about a microarray experiment) raw RNA-seq data were deposited at the DNA Data Bank of Japan under accession number DRA010238.
Gene ID: ICSL1, Pjv1_20230; GLP1: Pjv1_12278; CDR1, Pjv1_10273. Gene ID of the 97 SBTs in P. japonicum are provided in Supplemental Data Set S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Differentially expressed genes between the intrusive cell region and the remainder of the haustorium.
Supplemental Figure S2. Expression of epidermis and intrusive cell markers during haustorium development.
Supplemental Figure S3. Alignment of amino acid sequences of four SBTs whose gene expression was high in intrusive cells.
Supplemental Figure S4. Expression dynamics of the SBT1.7.1 promoter during haustorium development.
Supplemental Figure S5. Xylem bridge in the haustoria.
Supplemental Figure S6. Effect of Epi10 on haustorial formation at a later stage.
Supplemental Figure S7. Effect of the two structurally unrelated SBT inhibitors from different species on xylem bridge formation.
Supplemental Figure S8. Effect of Epi10 on haustorial formation and auxin response at a later stage.
Supplemental Figure S9. Phylogeny of ICSL1 in P. japonicum and the related LRR-RLKs in Arabidopsis.
Supplemental Figure S10. Phylogeny of GLP1 in P. japonicum, GhABP19 in Gossypium hirsutum, and the GLPs in Arabidopsis.
Supplemental Figure S11. Phylogeny of CDR1 in P. japonicum and aspartic proteases in Arabidopsis.
Supplemental Figure S12. Phylogeny of the SBTs in P. japonicum, S. asiatica and S. hermontica.
Supplemental Table S1. List of GO terms enriched in intrusive cells.
Supplemental Table S2. List of GO terms enriched in the haustorial parts, excluding the intrusive cells.
Supplemental Table S3. Genes exclusively expressed during the parasitic stage.
Supplemental Table S4. Primers used in this study.
Supplemental Data set S1. List of the whole transcriptome data.
Supplemental Data set S2. List of the 3,079 differentially expressed genes between the intrusive cell region and the remainder of the haustorium.
Supplemental Data set S3. Gene IDs of the 97 SBTs in P. japonicum.
Supplementary Material
Funding
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology KAKENHI grants (18H02464 and 18H04838 to S.Y., 15H05959 and 17H06172 to K.S.); Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship (to T.S.); JSPS Research Fellowship for Young Scientist (to T.W.); JST PRESTO (JPMJPR194D to S.Y.); and the RIKEN Special Postdoctoral Researchers Program and the German Research Foundation (DFG, 424122841 to T.S.).
Conflict of interest statement: The authors declare no conflict of interest related to this manuscript.
S.O., T.W., T.S., T.D., S.Y., A.S., and K.S. conceived and designed the study; K.S. supervised the experiments; S.O., T.W., T.S., J.K.I., R.S., T.K., S.Y., and Y.I. performed the experiments; S.O., T.W., T.S., T.D., S.Y., Y.I., A.S., and K.S. analyzed the data; S.O., T.W., T.S., and K.S. drafted the article; all authors critically revised the article and approved the final version.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ken Shirasu (ken.shirasu@riken.jp).
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CR, Timko MP, Yoder JI, Axtell MJ, Westwood JH (2019) Molecular dialog between parasitic plants and their hosts. Annu Rev Phytopathol 57:279–299 [DOI] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349:540–543 [DOI] [PubMed] [Google Scholar]
- Cui S, Wakatake T, Hashimoto K, Saucet SB, Toyooka K, Yoshida S, Shirasu K (2016) Haustorial hairs are specialized root hairs that support parasitism in the facultative parasitic plant Phtheirospermum japonicum. Plant Physiol 170:1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll NM, Royek S, Fujita S, Okuda S, Chamot S, Stintzi A, Widiez T, Hothorn M, Schaller A, Geldner N, et al. (2020) A two-way molecular dialogue between embryo and endosperm is required for seed development. Science 367:431–435 [DOI] [PubMed] [Google Scholar]
- Ellendorff U, Zhang Z, Thomma BP (2008) Gene silencing to investigate the roles of receptor-like proteins in Arabidopsis. Plant Signal Behav 3:893–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3:839–843 [DOI] [PubMed] [Google Scholar]
- Ghorbani S, Hoogewijs K, Pečenková T, Fernandez A, Inzé A, Eeckhout D, Kawa D, De Jaeger G, Beeckman T, Madder A, et al. (2016) The SBT6.1 subtilase processes the GOLVEN1 peptide controlling cell elongation. J Exp Bot 67:4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobena D, Shimels M, Rich PJ, Ruyter-Spira C, Bouwmeester H, Kanuganti S, Mengiste T, Ejeta G (2017) Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc Natl Acad Sci U S A 114:4471–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyet V, Wada S, Cui S, Wakatake T, Shirasu K, Montiel G, Simier P, Yoshida S (2019) Haustorium inducing factors for parasitic Orobanchaceae. Front Plant Sci 10:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham BK, Li G, Kang BH, Zeng F, Lucas WJ (2012) Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development. Plant Cell 24:3630–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Stintzi A, Schaller A. (2017) A novel subtilase inhibitor in plants shows structural and functional similarities to protease propeptides. J Biol Chem 292:6389–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaas LA, Wafula EK, Yang Z, Der JP, Wickett NJ, Altman NS, Taylor CG, Yoder JI, Timko MP, Westwood JH, dePamphilis CW (2013) Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol 13:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Mutuku JM, Yoshida S, Shirasu K (2015) Transcriptomics exposes the uniqueness of parasitic plants. Brief Funct Genomics 14:275–282 [DOI] [PubMed] [Google Scholar]
- Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Kasahara H, Wafula E, dePamphilis CW, Namba S, Shirasu K (2016) Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell 28:1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Yoshida S, Ito M, Namba S, Shirasu K (2011) Agrobacterium rhizogenes-mediated transformation of the parasitic plant Phtheirospermum japonicum. PLoS One 6:e25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14:108–117 [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S,, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotani KI, Wakatake T, Ichihashi Y, Okayasu K, Sawai Y, Ogawa S, Suzuki T, Shirasu K, Notaguchi M (2020) Host-parasite tissue adhesion by a secreted type of β-1,4-glucanase in the parasitic plant Phtheirospermum japonicum. Comm Biol 3:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- Lynn DG, Chang M (1990) Phenolic signals in cohabitation: implications for plant development. Annu Rev Plant Physiol Plant Mol Biol 41:497–526 [Google Scholar]
- Matsubayashi Y (2014) Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol 65:385–413 [DOI] [PubMed] [Google Scholar]
- Musselman L, Dickison W (1975) The structure and development of the haustorium in parasitic Scrophulariaceae. Bot J Linn Soc 70:183–212 [Google Scholar]
- Parker C (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci 65:453–459 [DOI] [PubMed] [Google Scholar]
- Pei Y, Li X, Zhu Y, Ge X, Sun Y, Liu N, Jia Y, Li F, Hou Y (2019) GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium wilt pathogens. Front Plant Sci 10:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, Song W, Yokoo T, Minobe A, Wang G, Ishida T, Sawa S, Chai J, Kakimoto T (2018) The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat Plants 4:1071–1081 [DOI] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138:1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Ichihashi Y, Farhi M, Zumstein K, Townsley B, David-Schwartz R, Sinha NR (2014) De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol 166:1186–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Steinhauser D, Büssis D, Stintzi A, Schaller A, Kopka J, Altmann T (2005) Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol 1:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ (1994) Families of serine peptidases. Methods Enzymol 244:19–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt S, Piepho HP, Stintzi A, Schaller A (2020) Peptide signaling for drought-induced tomato flower drop. Science 367:1482–1485 [DOI] [PubMed] [Google Scholar]
- Reichardt S, Repper D, Tuzhikov AI, Galiullina RA, Planas-Marquès M, Chichkova NV, Vartapetian AB, Stintzi A, Schaller A (2018) The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Sci Rep 8:10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S, Bernsdorff FE, Cai D (2012) Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J Exp Bot 63:5507–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runo S, Kuria EK (2018) Habits of a highly successful cereal killer, Striga. PLoS Pathog 14:e1006731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Nishimura T, Miyaki YI, Watanabe S, Takagi H, Izumi S, Shimada H (2015) In vitro and in vivo evidence for oxalate oxidase activity of a germin-like protein from azalea. Biochem Biophys Res Commun 458:536–542 [DOI] [PubMed] [Google Scholar]
- Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR (2013) Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol 14:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Stintzi A, Rivas S, Serrano I, Chichkova NV, Vartapetian AB, Martínez D, Guiamét JJ, Sueldo DJ, van der Hoorn RAL, et al. (2018) From structure to function—a family portrait of plant subtilases. New Phytol 218:901–915 [DOI] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A (2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354:1594–1597 [DOI] [PubMed] [Google Scholar]
- Scholes JD, Press MC (2008) Striga infestation of cereal crops—an unsolved problem in resource limited agriculture. Curr Opin Plant Biol 11:180–186 [DOI] [PubMed] [Google Scholar]
- Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, dePamphilis CW, Westwood JH, Axtell MJ (2018) MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553:82–85 [DOI] [PubMed] [Google Scholar]
- Smith EL, Markland FS, Kasper CB, DeLange RJ, Landon M, Evans WH (1966) The complete amino acid sequence of two types of subtilisin, BPN' and Carlsberg. J Biol Chem 241:5974–5976 [PubMed] [Google Scholar]
- Spallek T, Melnyk CW, Wakatake T, Zhang J, Sakamoto Y, Kiba T, Yoshida S, Matsunaga S, Sakakibara H, Shirasu K (2017) Interspecies hormonal control of host root morphology by parasitic plants. Proc Natl Acad Sci USA 114:5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T, Mutuku M, Shirasu K (2013) The genus Striga: a witch profile. Mol Plant Pathol 14:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of inflorescence deficient in abscission is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20:1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N, Scholl S, Lang L, Katzenberger J, Schumacher K, Schaller A (2020) The biogenesis of CLEL peptides involves several processing events in consecutive compartments of the secretory pathway. Elife 9:e55580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N, Schardon K, Stintzi A, Schaller A. (2017) A toolbox for the analysis of peptide signal biogenesis. Mol Plant 10:1023–1025 [DOI] [PubMed] [Google Scholar]
- Sun G, Xu Y, Liu H, Sun T, Zhang J, Hettenhausen C, Shen G, Qi J, Qin Y, Li J, Wang L, Chang W, Guo Z, Baldwin IT, Wu J (2018) Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat Commun 9:2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Nishiyama T, Shimizu K, Kadota K (2013) TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A, Qiu YL (2017) Evolutionary history of subtilases in land plants and their involvement in symbiotic interactions. Mol Plant Microbe Interact 30:489–501 [DOI] [PubMed] [Google Scholar]
- Tian M, Benedetti B, Kamoun S (2005) A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol 138:1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Kamoun S (2005) A two disulfide bridge Kazal domain from Phytophthora exhibits stable inhibitory activity against serine proteases of the subtilisin family. BMC Biochem 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser JH, Inge D, Kollmann R (1984) The "hyaline body" of the root parasiteAlectra orobanchoides benth. (Scrophulariaceae)—Its anatomy, ultrastructure and histochemistry. Protoplasma 121:146–156 [Google Scholar]
- Von Groll U, Berger D, Altmann T (2002) The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14:1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Cui S, Yoshida S (2019) Reactive oxygen species (ROS) generation is indispensable for haustorium formation of the root parasitic plant Striga hermonthica. Front Plant Sci 10:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatake T, Yoshida S, Shirasu K (2018) Induced cell fate transitions at multiple cell layers configure haustorium development in parasitic plants. Development 145: dev164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatake T, Ogawa S, Yoshida S, Shirasu K (2020) Auxin transport network underlies xylem bridge formation between the hemi-parasitic plant Phtheirospermum japonicum and host Arabidopsis. Development 147: dev.187781 [DOI] [PubMed] [Google Scholar]
- Wang Y, Steele D, Murdock M, Lai S, Yoder J (2019) Small-molecule screens reveal novel haustorium inhibitors in the root parasitic plant. Phytopathology 109:1878–1887 [DOI] [PubMed] [Google Scholar]
- Wright CS, Alden RA, Kraut J (1969) Structure of subtilisin BPN' at 2.5 angström resolution. Nature 221:235–242 [DOI] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23:980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PC,, Wu B, Der JP,. et al. (2015) Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol 32:767–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Awad AA, Xie X, Takeuchi Y (2010) Strigolactones as germination stimulants for root parasitic plants. Plant Cell Physiol 51:1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Cui S, Ichihashi Y, Shirasu K (2016) The haustorium, a specialized invasive organ in parasitic plants. Annu Rev Plant Biol 67:643–667 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kim S, Wafula EK, Tanskanen J,, Kim YM, Honaas L, Yang Z, Spallek T, Conn CE, Ichihashi Y, et al. (2019) Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr Biol 29:3041–3052.e3044 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K (2009) Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol 183:180–189 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Berkowitz O, Teixeira da Silva JA, Zhang M, Ma G, Whelan J, Duan J (2015) RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album. Front Plant Sci 6:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP (2000) Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol 123:1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.