Abstract
Deglycosylation is a key step in the activation of specialized metabolites involved in plant defense mechanisms. This reaction is notably catalyzed by β-glucosidases of the glycosyl hydrolase 1 (GH1) family such as strictosidine β-d-glucosidase (SGD) from Catharanthus roseus. SGD catalyzes the deglycosylation of strictosidine, forming a highly reactive aglycone involved in the synthesis of cytotoxic monoterpene indole alkaloids (MIAs) and in the crosslinking of aggressor proteins. By exploring C. roseus transcriptomic resources, we identified an alternative splicing event of the SGD gene leading to the formation of a shorter isoform of this enzyme (shSGD) that lacks the last 71-residues and whose transcript ratio with SGD ranges from 1.7% up to 42.8%, depending on organs and conditions. Whereas it completely lacks β-glucosidase activity, shSGD interacts with SGD and causes the disruption of SGD multimers. Such disorganization drastically inhibits SGD activity and impacts downstream MIA synthesis. In addition, shSGD disrupts the metabolic channeling of downstream biosynthetic steps by hampering the recruitment of tetrahydroalstonine synthase in cell nuclei. shSGD thus corresponds to a pseudo-enzyme acting as a regulator of MIA biosynthesis. These data shed light on a peculiar control mechanism of β-glucosidase multimerization, an organization common to many defensive GH1 members.
A C. roseus pseudo-enzyme created by alternative splicing modulates alkaloid production through inhibition of the central step of the biosynthetic pathway catalyzed by strictosidine β-glucosidase.
Introduction
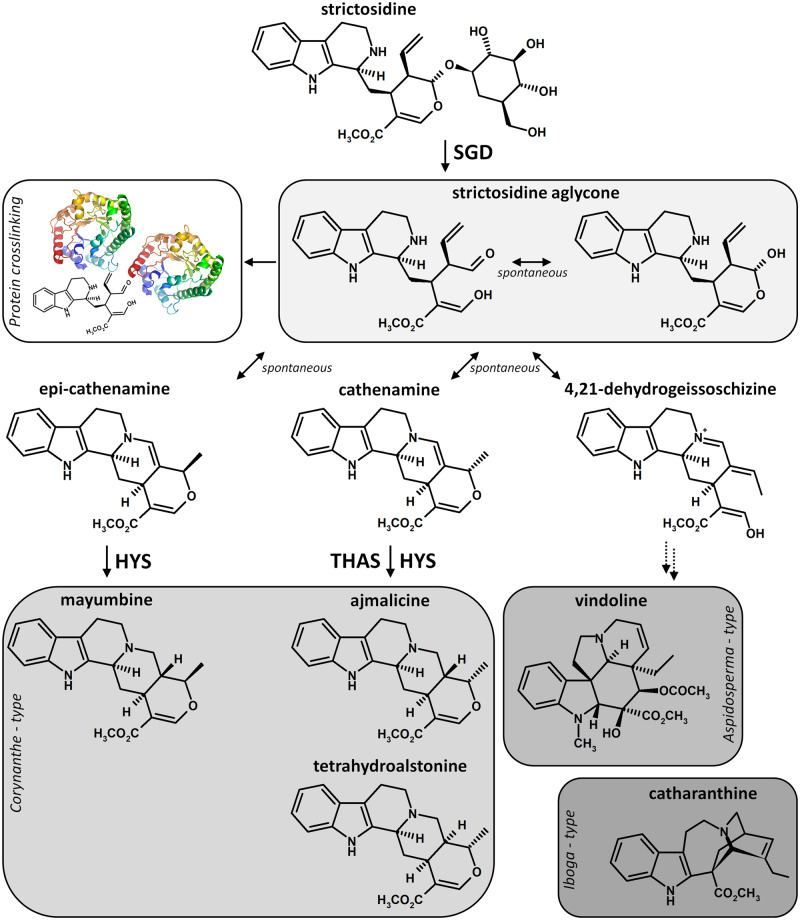
Monoterpene indole alkaloids (MIAs) are nitrogen-containing molecules mainly found in the Apocynaceae, Loganiaceae, and Rubiaceae families and belong to the chemical arsenal that plants can deploy to face biotic challenges, notably pest or herbivore attacks (O’Connor and Maresh, 2006; St-Pierre et al., 2013). Due to their intrinsic toxicity, these specialized metabolites also display valuable pharmaceutical properties such as the antineoplastic vinblastine and vincristine produced in the Madagascar periwinkle (Catharanthus roseus) or the antiarrhythmic ajmaline synthesized in Rauwolfia serpentina (Muto et al., 2005). Almost all MIAs derive from strictosidine that arises from the condensation of indole and monoterpene precursors, namely tryptamine and secologanin. The subsequent deglycosylation of strictosidine is catalyzed by strictosidine-β-d-glucosidase (SGD), a prominent member of the glycosyl hydrolase 1 (GH1) family and leads to the formation of a highly reactive aglycone that spontaneously rearranges into the more stable compounds cathenamine, epi-cathenamine, and 4,21-dehydrogeissoschizine (Hemscheidt and Zenk, 1980; Geerlings et al., 2000; Gerasimenko et al., 2002; Figure 1). This mechanism constitutes the entry-point to the syntheses of the different MIA sub-families that are further processed according to the downstream enzyme. This is exemplified by the characterization of tetrahydroalstonine synthase isoforms (THAS1–4) and heteroyohimbine synthase (HYS) responsible for the formation of the corynanthe MIAs including tetrahydroalstonine, ajmalicine, or mayumbine from cathenamine, or illustrated by the recent elucidation of the complete set of enzymes ensuring the synthesis of both Aspidosperma (tabersonine) and Iboga (catharanthine) MIAs from 4,21-dehydrogeissoschizine (Rueffer et al., 1978; Stoeckigt et al., 1983; Stavrinides et al., 2015, 2016; Tatsis et al., 2017; Caputi et al., 2018; Qu et al., 2018).
Figure 1.
Deglycosylation of strictosidine and associated arising reactions. In C. roseus, the deglycosylation of strictosidine by strictosidine β-d-glucosidase (SGD) leads to the formation of a reactive aglycone that causes protein crosslinking during the “nuclear-time bomb” defense process or undergoes a spontaneous rearrangement initiating the synthesis of downstream MIAs. Enzymatic and spontaneous reactions are indicated by enzyme name or blank in front of arrows. Double dashed arrows represent multiple biosynthetic reactions. HYS, heteroyohimbine synthase; THAS, tetrahydroalstonine synthase.
Up to now, SGD has only been characterized in a small number of plant species, including R. serpentina and C. roseus, of which for the latter a single-copy gene has been identified in the recently published genomic sequence (Geerlings et al., 2000; Gerasimenko et al., 2002; Kellner et al., 2015). In both species, SGD catalyzes a similar reaction of deglycosylation with high specificity toward strictosidine and self-organizes as supramolecular complexes located in cell nuclei (Luijendijk et al., 1998; Guirimand et al., 2010). In C. roseus leaves, this peculiar organization specifically takes place in the epidermis and ensures a total separation of SGD from strictosidine that is synthesized and accumulated in the vacuole (McKnight et al., 1991). Based on this enzyme/substrate segregation, C. roseus evolved a defense mechanism similar to the glucosinolate/myrosinase system. This process relies on the capacity of the strictosidine aglycone to crosslink proteins when high concentrations of strictosidine come into contact with SGD following intracellular membrane rupture upon attacks, which is the so-called “nuclear-time bomb” mechanism (Luijendijk et al., 1996; Guirimand et al., 2010; Figure 1). In this system, the formation of SGD supramolecular complexes prevents SGD from diffusing out of the nucleus and protects this enzyme from a potential degradation by exogenous proteases (Guirimand et al., 2010). The efficiency of this “nuclear-time bomb” is likely reinforced by the fast and massive local increases in SGD gene expression and strictosidine content in C. roseus leaves consumed by the non-host interaction-inducing hornworm Manduca sexta (Dugé de Bernonville et al., 2017). In addition, the recent identification of THAS isoforms and HYS also shed light on a new role of SGD multimers that can recruit downstream enzymes to favor enzyme channeling and to prevent the release of the reactive aglycone before its conversion in tetrahydroalstonine or ajmalicine (Stavrinides et al., 2015, 2016). The recruitments of THAS isoforms and HYS close to SGD complexes were observed for both nuclear and cytosolic enzymes, suggesting that synthesis of the corresponding MIAs only occurs in the nucleus. Subcellular localization and protein (self)-interactions thus appear as two processes directly involved in the regulation of SGD activity up to MIA synthesis, which can be extended to other β-glucosidases of the GH1 family involved in defense mechanisms such as in olive tree (Olea europaea), wheat (Triticum aestivum), oat (Avena sativa), and sorghum (Sorghum bicolor; Verdoucq et al., 2004; Sue et al., 2006; Kwak et al., 2009; Koudounas et al., 2017). However, the molecular events presently known to occur within both processes have not entirely explained the fine-tuned control of deglycosylation as the central step in MIA production that leads to both the synthesis of a myriad of downstream MIAs and to the “nuclear-time bomb” mechanism. A trade-off between the release of reactive aglycones and the management of their toxic activity is likely to occur, thus suggesting the existence of complementary molecular events such as the β-glucosidase aggregating factor (BGAF) involved in maize (Zea mays) β-glucosidases multimerization (Blanchard et al., 2001).
In this study, we took advantage of the recently assembled C. roseus consensus transcriptome to identify an alternative splicing event of the SGD-encoding gene that led to the formation of a shorter isoform of SGD (shSGD; Dugé de Bernonville et al., 2015). Although this isoform did not show any deglycosylation activity toward strictosidine, we demonstrated that the corresponding transcript is always co-expressed along with the SGD transcript and the presence of this protein has substantial effects on SGD multimerization and strictosidine deglycosylation activity as well as THAS subcellular localization and MIA synthesis. As such, the existence of shSGD that meets the definition of the concept of pseudo-enzyme, also known as zombie enzyme, highlights the involvement of a pseudo-enzyme generated by an alternative splicing event in the regulation of β-glucosidase activity and downstream processes (Eyers and Murphy, 2016; Murphy et al., 2017a).
Results
Identification of alternative SGD transcripts in the C. roseus consensus transcriptome
Whereas SGD is encoded by a single-copy gene in C. roseus, a BLAST analysis performed on the CDF97 consensus assembly (C. roseus consensus transcriptome) revealed the presence of several sequences closely related to SGD, and displaying a local identity superior to 95% and a hitscore of at least 1100 (Supplemental Figure S1, A; Dugé de Bernonville et al., 2015; Kellner et al., 2015). Besides a transcript (SRR648705_TR21994_c0_g4_i4_len=2338) of very high identity (99.94%) and high expression level that is likely to correspond to the original SGD sequence (Supplemental Figure S1, B–F), we detected two fusion transcripts SRR924147_TR30389_c0_g2_i2_len=3912 containing two inverted tandem copies of SGD and SRR648707_TR25285_c3_g4_i4_len=4173 resulting from a combination of a major facilitator superfamily (MFS) transporter at its 5′ extremity and a SGD copy at its 3′-end, which may have both resulted from artifactual transcript reconstruction. In addition, some transcripts were assembled as antisense by Trinity although no strand-specific libraries were used. These transcripts were then considered in the forward orientation and three other sequences with homology with SGD (hitscore > 2,800), namely SRR342023_TR15323_c0_g2_i2_len=2096, SRR646572_TR28058_c0_g1_i3_len=2050, and SRR646572_TR28058_c0_g1_i2_len=2029, were thus identified in CDF97 and mainly differed by their 5′-untranslated region (5′-UTR; Supplemental Figure S1, A–C and F). Although both transcript types were detected in almost all experimental conditions (Supplemental Figure S1, G), a marked study-dependent effect was also observed (e.g. SRR646572_TR28058_c0_g1_i3_len=2050 was only expressed in the CathaCyc samples, Van Moerkercke et al., 2013), leading us to speculate that these transcripts may also correspond to partially inaccurate reconstitution. Finally, a last transcript was found (SRR924148_TR34256_c6_g1_i5_len=2115) that strongly differed from the original SGD sequence by its 3′-end (Supplemental Figure S1, D). Although expressed at a lower level than SGD, this potential variant caught our attention for further analysis.
Identification of an alternative splicing event leading to the translation of a truncated SGD
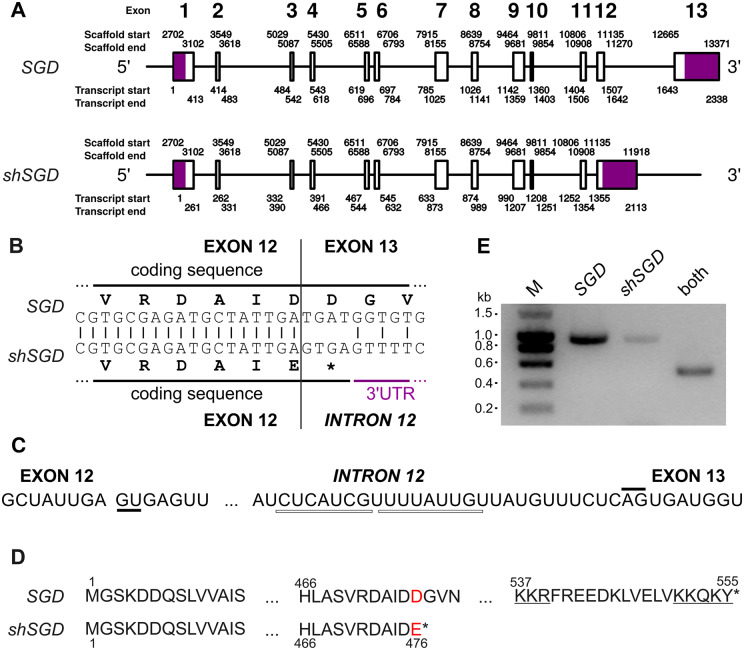
In order to identify the origin of the selected alternative transcript of SGD, we compared this transcript sequence to the SGD genomic region. However, the current version of the C. roseus genome displays a scaffolding problem in the region containing the SGD locus rendering its study difficult to perform. To circumvent this problem, we assembled de novo raw sequences obtained from a BAC construction deposited in NCBI SRA and which contains the SGD locus (run accession number ERR599180; Kellner et al., 2015). We generated a 22-kb scaffold containing the SGD reference sequence (Supplemental Figure S2). This SGD sequence was composed of 13 exons of different sizes ranging from 43 to 243 nt (Figure 2, A). Interestingly, BLAST analyses revealed that the 3′-end of SRR924148_TR34256_c6_g1_i5_len=2115 corresponds to the intronic sequence localized between exons 12 and 13 of the reconstituted gene and of SRR648705_TR21994_c0_g4_i4_len=2338 (Figure 2, A–C). More precisely, such intronic sequence is located at the end of exon 12 and is surrounded by donor and acceptor splicing sites (Figure 2, C). This suggests that the formation of SRR924148_TR34256_c6_g1_i5_len=2115 resulted from an alternative splicing event of the SGD-encoding gene leading to intron retention, causing the appearance of a stop codon in frame within the coding sequence of exon 12 and resulting in the expression of a truncated protein lacking the last 71 residues (Figure 2, B and D). Multiple reads of this intronic sequence were found in the consensus transcriptome potentially confirming the occurrence of this phenomenon in vivo. However, the read coverage in this region was low in comparison to the other exons explaining the low expression level observed for SRR924148_TR34256_c6_g1_i5_len=2115 (Supplemental Figures S1, G and S2). A definitive evidence of this alternative splicing was obtained by amplification of the truncated form by a PCR performed on cDNA from the Apricot Sunstorm cultivar, with primers designed on its specific 3′-UTR sequence, even if this amplification was always lower than those of the original SGD transcript and a sequence common to both transcripts (Figure 2, E and Supplemental Figure S1). Evidence for the presence of shSGD transcript was also obtained in three other cultivars (Supplemental Figure S3). Based on these results, we concluded that an alternative splicing event occurs at the end of the SGD gene that leads to the formation of a shorter sequence that is called hereafter short SGD (shSGD; GenBank accession number MG520182).
Figure 2.
Alternative splicing generates shSGD by intron retention. A, Genomic organization of SGD and shSGD depicting the general exon/intron junctions. The genomic coordinates were obtained with a reconstructed scaffold from the sequencing of a genomic BAC containing the SGD locus. B, Focus on the 3′-end of exon 12 and on the consequence of intron retention. C, Donor and acceptor sites of splicing of intron 12 are highlighted by solid lines and putative branch points are shown by white rectangles. D, Amino acid sequences of SGD and the truncated protein encoded by SRR924148_TR34256_c6_g1_i5_len=2115. Red letters and stars indicate mutated amino acids and protein ends, respectively. The bipartite NLS of SGD is underlined. E, Amplification of specific fragments of SGD and shSGD transcripts by RT-PCR performed on RNA from leaves. A combination of primers common to both transcripts was used as a control.
shSGD encodes a nonfunctional glucosidase displaying a nucleocytosolic localization
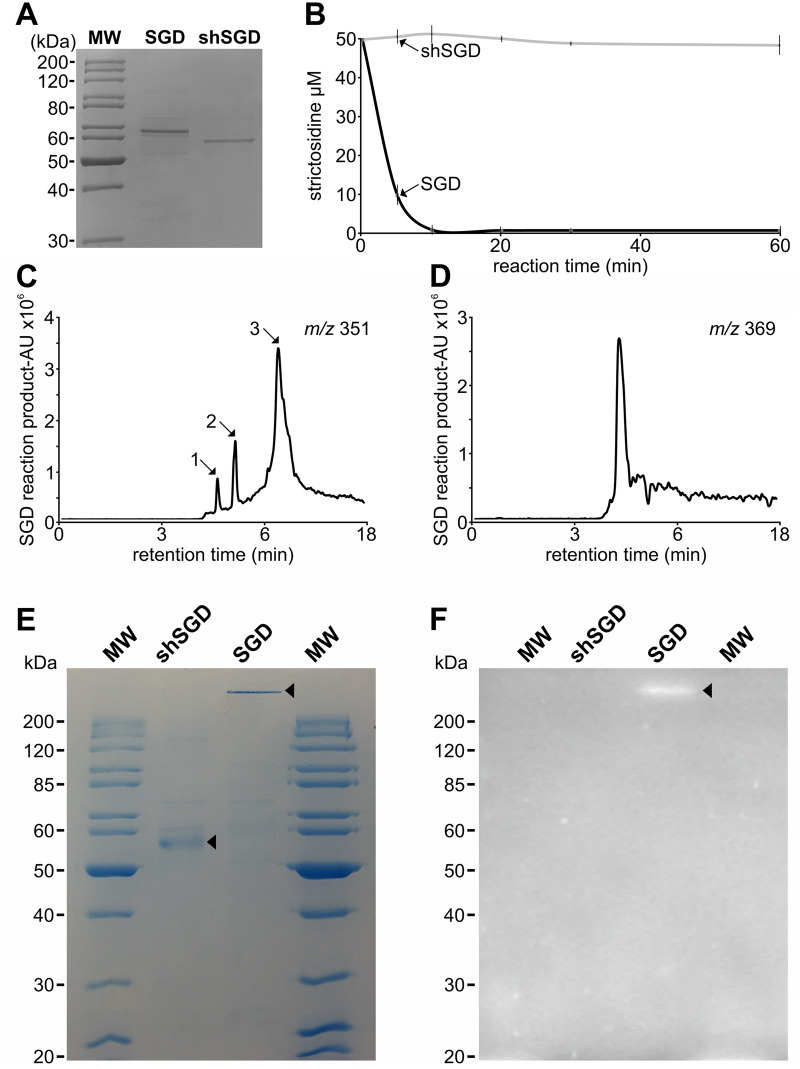
Amino acid sequence alignment of shSGD with SGDs from C. roseus and Rauwolfia species showed that the deletion of the last 71 residues of shSGD caused the disappearance of at least four amino acids involved in strictosidine binding as defined by (Barleben et al. 2007; Supplemental Figure S4). In addition, this also resulted in the elimination of the bipartite nuclear localization signal (NLS) located at the extreme C-terminal part of the protein (Guirimand et al., 2010). To gain insight into the consequences of this sequence modification, recombinant shSGD was expressed in Escherichia coli and its activity was compared with that of recombinant SGD (Figure 3, A). Incubation of purified SGD with strictosidine quickly resulted in substrate consumption (Figure 3, B) accompanied by the formation of cathenamine, epi-cathenamine, and dehydrogeissoschizine (m/z 351, Figure 3, C), as well as synthesis of the reactive aglycone (m/z 369, Figure 3, D). By contrast, no substantial strictosidine deglycosylation was observed with the purified recombinant shSGD in these conditions, indicating this protein was not functional against this substrate (Figure 3, B). An additional assay conducted with 4-methylumbelliferyl-β-d-glucoside (MUGlc), a general artificial substrate of β-glucosidases, confirmed that only SGD was active as revealed on zymogram (Figure 3, E and F). Because of its lack of catalytic activity is likely caused by deleted key amino acids, shSGD may be considered as a pseudo-enzyme (pseudo-glucosidase) or otherwise the “zombie” version of SGD (Eyers and Murphy, 2016; Murphy et al., 2017a).
Figure 3.
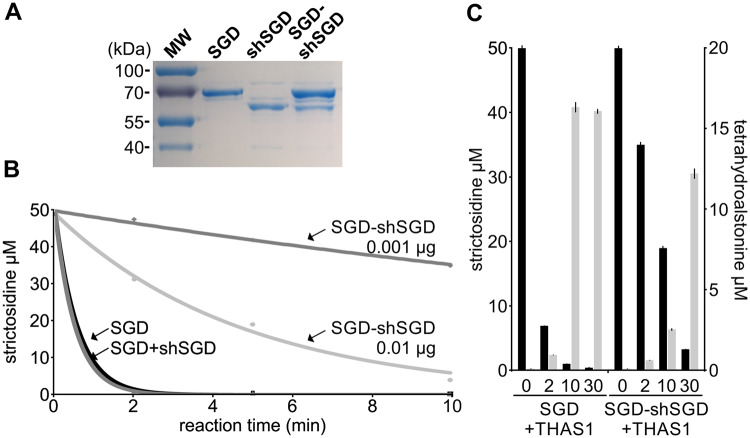
shSGD does not deglycosylate strictosidine. A, SDS-PAGE analysis of the purified SGD and shSGD. B, Time-course analysis of a representative strictosidine deglycosylation conducted by incubating 50 µM of strictosidine with SGD or shSGD. C, MIAs formed through deglycosylation of strictosidine by SGD. 1, epi-cathenamine; 2, dehydrogeissoschizine; 3, cathenamine. D, The aglycone crosslinking product resulting from SGD activity. E, Coomassie blue staining and F, zymogram of SGD and shSGD activities. Non-heat-treated recombinant proteins were separated on a semi-native PAGE before incubation in a MUGlc containing solution and UV-visualization.
The possible outcome of the bipartite NLS deletion on protein targeting was also evaluated by studying the subcellular localization of shSGD fused at the N- or C-terminus to the yellow fluorescent protein (YFP). In transiently transformed C. roseus cells, shSGD-YFP and YFP-shSGD displayed a similar distribution in both the cytosol and the nucleus as revealed by the perfect superimposition of the fluorescent signal with that of the CFP-nucleocytosolic marker (Figure 4, A–H). Similar localization of shSGD-YFP was also observed in onion cells (Figure 4, M–P).
Figure 4.
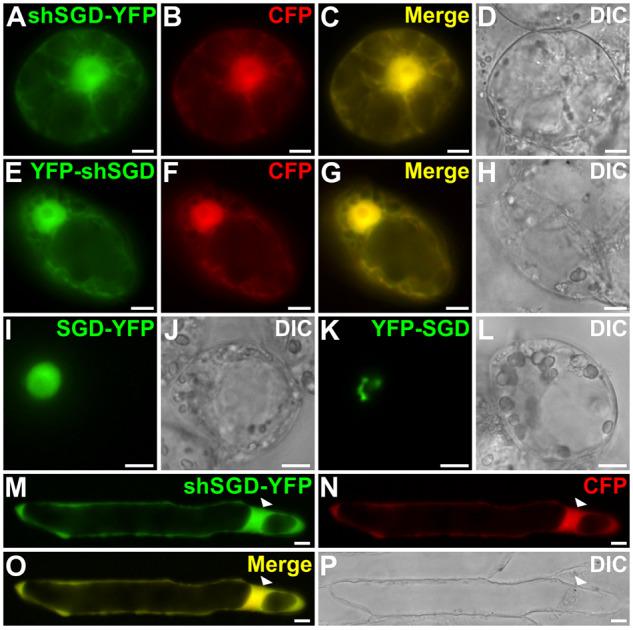
shSGD displays a nucleocytosolic localization. Catharanthus roseus (A–L) and onion (M–P) cells were transiently transformed with plasmids expressing either shSGD–YFP (A, M), YFP-shSGD (E), SGD-YFP (I), or YFP-SGD (K) and a plasmid encoding the nucleocytosolic CFP marker (B, F, and N). Co-localization of the fluorescence signals appears in yellow when merging the two individual (green/red) false color images (C, G, and O). Cell morphology is observed with differential interference contrast (DIC) (D, H, J, L, and P). Bars, 10 µm.
This localization differs from the exclusive nuclear targeting of the SGD-YFP fusion protein that appears diffuse in the nucleus due to YFP fusion constraints (Figure 4, I and J). This suggests, together with the lack of bipartite NLS, that the presence of shSGD in the nucleus is likely to be due to passive diffusion (Figure 4, I and J). The nucleocytosolic localization of shSGD in cells of onion, a non-MIA-producing plant that does not possess endogenous SGD, further suggests that the nuclear localization of shSGD is the result of passive diffusion rather than a potential piggyback transport involving interactions with SGD (Figure 4, M–P). Furthermore, we noted that neither the shSGD-YFP nor the YFP-shSGD fusion proteins were able to adopt the fusiform conformation observed for the YFP-SGD protein (Figure 4, A–H, K, and L). This kind of structure formation relies on the accessibility of the C-terminal end of SGD and is a consequence of protein multimerization, thus suggesting that shSGD does not undergo protein self-interactions (Guirimand et al., 2010).
shSGD is strictly co-expressed with SGD but in variable proportions
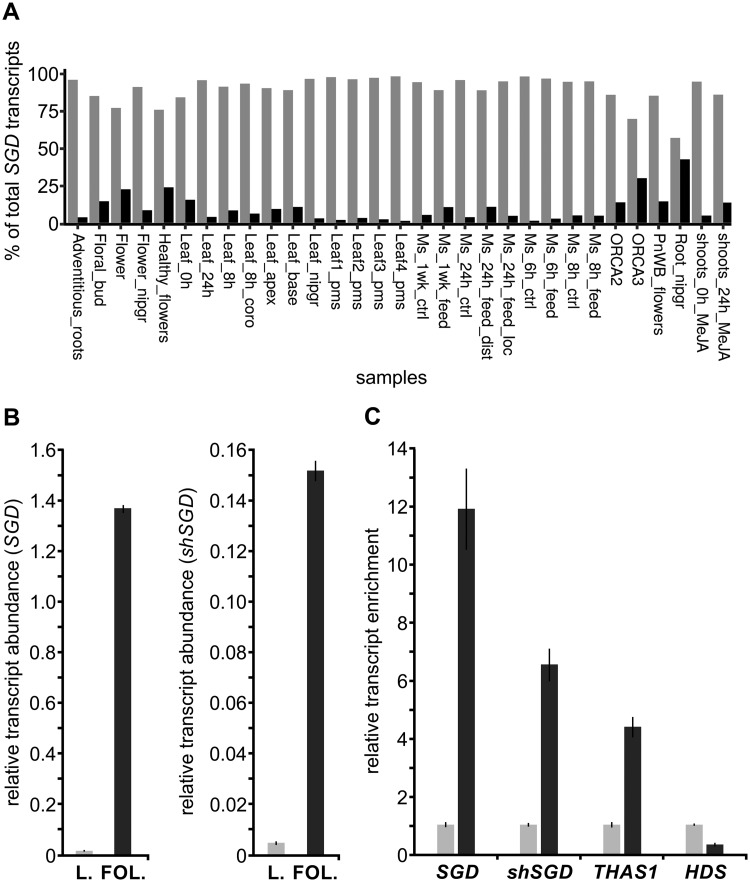
Besides a high compartmentalization at the subcellular level, the MIA biosynthetic pathway also displays a complex organization at the cellular level relying on the dedicated expression of genes in specific organs and tissues with SGD expressed in all organs and notably in the leaf epidermis (Courdavault et al., 2014). A careful re-examination of transcript abundance in the C. roseus consensus transcriptome revealed that shSGD is expressed at basal levels in all conditions/organs including leaves, one of the main sites of MIA biosynthesis (Supplemental Figure S1, G). Interestingly, this expression profile was similar to that of SGD (Spearman’s rho = 0.71; P-value = 10−6) and the ratio between both transcript types reached a mean of around 1/10 but displayed huge variations, ranging from 1.7% to 42.8% depending on organs and conditions (Figure 5, A). Ratios between both transcripts were confirmed by reverse transcription quantitative PCR (RT-qPCR) in C. roseus leaves subjected to M. sexta hornworm attacks known to cause a huge enhancement of SGD expression (Dugé de Bernonville et al., 2017). After confirming this positive effect on SGD expression with a 36-fold enhancement of expression in leaves consumed by larvae, we indeed established that shSGD followed a similar pattern and remained in a 1/10 ratio with SGD (Figure 5, B andSupplemental Figure S1, G). shSGD response to environmental cues was also supported by its jasmonate responsiveness as revealed by RNA-seq results (Supplemental Figure S1, G). Finally, an epidermis-enriched fraction of leaves was prepared and expression levels of both SGD and shSGD were analyzed (Figure 5, C). As observed for SGD transcripts showing a roughly 12-fold enrichment in the epidermal fraction compared with the whole leaf, shSGD transcripts were also enriched in a similar range (almost seven-fold) in the epidermis, thus demonstrating that shSGD and SGD strictly co-exist in this tissue. As a positive control, we also demonstrated that these transcripts were accompanied by transcripts of THAS1 encoding a direct subsequent enzyme in the biosynthetic pathway. By contrast, we showed that transcripts of HYDROXYMETHYLBUTENYL 4-DIPHOSPHATE SYNTHASE (HDS), known to accumulate in internal phloem-associated parenchyma, were not enriched in the epidermal fraction (Oudin et al., 2007). All these results thus establish that shSGD expression is strictly concomitant to that of SGD but displays considerable variation depending on organ type and conditions. This is in agreement with an action mode of a pseudo-enzyme but leaves open the question of shSGD function in planta.
Figure 5.
shSGD and SGD are co-expressed in variable proportions. A, Relative abundance of SGD (light gray; SRR342023_TR15323_c0_g2_i2_len=2096) and shSGD (dark gray; SRR924148_TR34256_c6_g1_i5_len=2115) transcripts in paired-end samples available in public databases, including multiple organs and experimental conditions. Each entity is expressed as a percentage of the total SGD transcript (shSGD + SGD) amount. B, Relative quantification of SGD and shSGD transcripts in intact C. roseus leaves (light gray) and in C. roseus leaves (L.) subjected to folivory (FOL.) by M. sexta (dark gray). RNAs were extracted from both leaf treatments and reverse-transcribed before determination of relative gene expression by qPCR. Transcript copy numbers were normalized using CrRPS9. For both experiments, assays were performed in triplicate, and expression measurements were performed at least twice with independent experimental replicates (data represents means ± SE). C, Relative expression of SGD, shSGD, THAS1, and HDS in epidermis-enriched fractions of C. roseus leaves (dark gray) compared with the whole-leaf fraction (light gray). Epidermis-enriched transcript fractions were generated by a carborundum abrasion and both fraction types were reverse-transcribed before determination of relative gene expression by qPCR. Transcript copy numbers were normalized using CrRPS9 and expressed relative to the amount of transcript measured in the whole-leaf fraction. Data represent means ± SE of three technical replicates of two biological replicates.
shSGD is not capable of self-interactions but disrupts SGD high-molecular-weight complexes
To determine a possible function of shSGD, we investigated the existence of potential (self)-interactions between shSGD and SGD since protein multimerization is a well-known mechanism controlling SGD activity (Guirimand et al., 2010).
In C. roseus cells, SGD is organized in high-molecular-weight complexes in the nucleus (Figure 4, K), resulting from protein self-interactions as confirmed by bimolecular fluorescence complementation (BiFC) assays performed through the expression of SGD in fusion with N- or C-terminal split YFP fragments (YFPN, YFPC; Guirimand et al., 2010; Figure 6, A–F). Careful examination by negative staining electron microscopy of the purified recombinant protein revealed that SGD self-organized as long fibrillary multimers of various lengths up to 300 nm long and ca. 10 nm in diameter (Figure 6, G–I). In addition, proteins seemed to stack in a tubular conformation to form a tunnel that may control aglycone reactivity during strictosidine deglycosylation (Figure 6, H). A similar mechanistic scenario has been proposed for the fibrillar tunnel-like structure observed in oat avenacosidase (Kim et al., 2000, 2005; Kwak et al., 2009).
Figure 6.
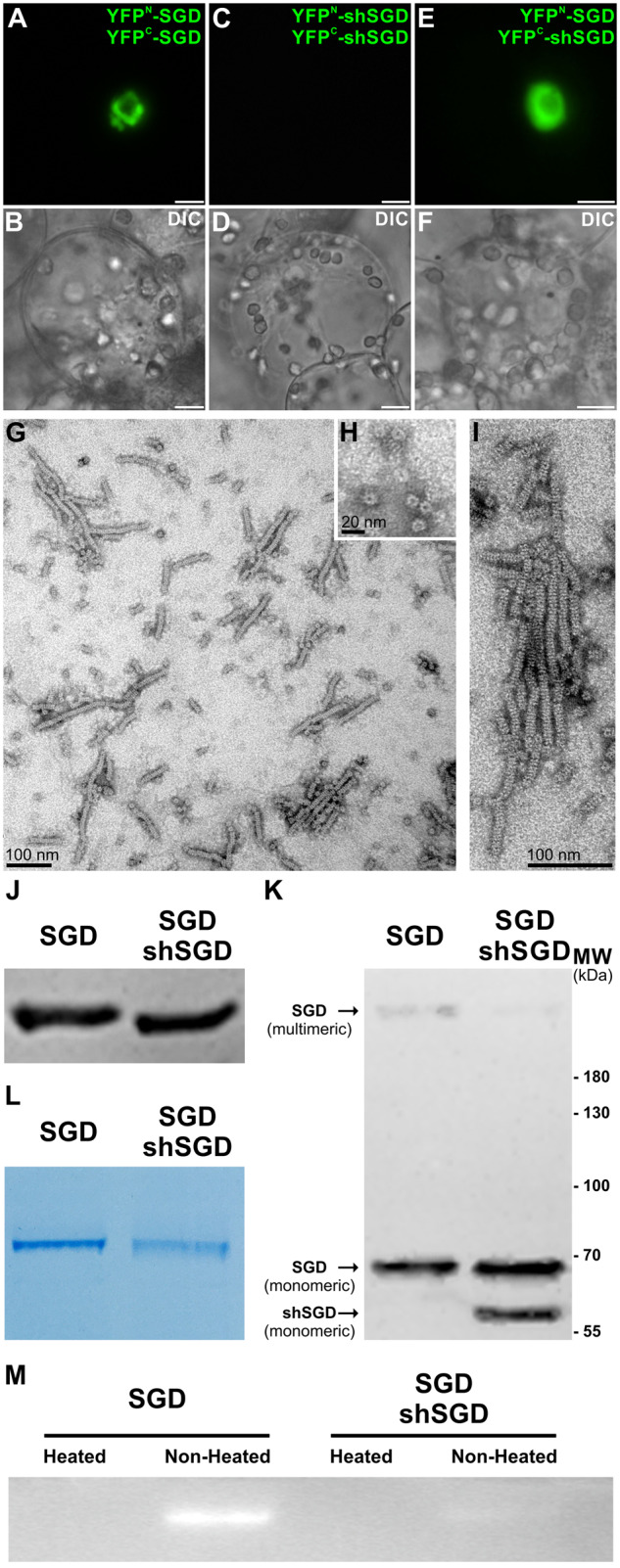
shSGD is not capable of self-interactions but affects the enzymatically active multimeric conformation of SGD by disrupting SGD high-molecular-weight complexes. A, SGD; C, shSGD self-interactions; and E, shSGD/SGD interactions were analyzed by BiFC in C. roseus cells transiently transformed by appropriate combinations of plasmids encoding fusions with the two split YFP fragments, as indicated on the right of each fluorescence picture. Cell morphology is observed with differential interference contrast (DIC) (B, D, and F). Bars, 10 µm. G, Observation of the recombinant purified SGD multimers purified from E. coli by negative staining electron microscopy, (H) transversal observation of the multimers, and (I) multimers at a higher magnification. J, Immunoblot analysis of denatured protein extracts of E. coli expressing either SGD or SGD-shSGD constructs normalized based on the monomeric conformation of SGD. K, The same quantity of proteins was then resolved in acrylamide gels without denaturing the extracts (i.e. non-heat treatment) followed by immunoblotting. L, Since the supramolecular structure of the multimeric conformation of SGD barely migrated to the PVDF membrane, Coomassie brilliant blue-staining was used to highlight the difference in the assembly of the multimeric conformation of SGD. M, The enzymatic activity of extracts co-expressing SGD-shSGD was barely detected compared with extracts expressing SGD as revealed by zymography.
In this context, the self-interacting capacities of shSGD were tested through BiFC assays. These were performed by generating fusions with the C-terminal extremity of the split YFP fragments to ensure the accessibility of the C-terminal end of shSGD since this domain is required for the formation of high-molecular-weight complexes of SGD (Guirimand et al., 2010). Interestingly, co-expression of YFPN-shSGD and YFPC-shSGD did not result in the formation of a BiFC complex (Figure 6, C and D) in contrast to that of YFPN-SGD and YFPC-SGD (Figure 6, A and B). This suggests that shSGD is not capable of self-interaction as hypothesized on the basis of its subcellular localization (Figure 4, A–H). Such a result is also in agreement with the migration profile of both shSGD and SGD on a semi-native polyacrylamide gel (i.e. non-heat-treated samples), which clearly displayed a monomeric and high multimeric conformation, respectively (Figure 3, E and F). However, when both SGD isoforms were co-expressed (YFPN-SGD and YFPC-shSGD), a fluorescent signal was observed, indicating cross-interactions between shSGD and SGD (Figure 6, E and F). Interestingly, this signal did not exhibit the fusiform conformation as observed for SGD self-interactions (Figure 6, A and B), but rather remained diffuse in the nucleus suggesting that shSGD disrupted the formation of the SGD high-molecular-weight complexes. Such a disruptive effect was also studied by comparing the homo/hetero-multimeric status of SGD expressed alone in E. coli or co-expressed with shSGD (Figure 6, J–L and Supplemental Figure S5, A and B). Immunoblot analysis and CBB (Coomassie Brilliant Blue R-250) staining of crude protein extracts from E. coli on a semi-native SDS-PAGE revealed the presence of high-molecular-weight multimers for SGD that partially disappeared with shSGD co-expression, thus confirming that shSGD affects the level of SGD multimerization. This partial disruption of the multimeric conformation of SGD when co-expressed with shSGD also had an impact on the activity of SGD against MUGlc, as revealed by zymography (Figure 6, M). A similar disappearance of SGD multimer was also observed by negative staining electron microscopy when shSGD was co-expressed with SGD, albeit potential sample treatment effects cannot be excluded in these conditions (Supplemental Figure S5, C and D). In order to identify amino acids potentially involved in the assembly of the quaternary structure of SGD, we performed homology-based modeling. As expected, the predicted tertiary structure of C. roseus SGD resembled the characteristic (β/α)8 TIM barrel fold of the GH1 family and perfectly superimposed with the resolved crystal structure of R. serpentina (RsSGD; Barleben et al., 2007), thus demonstrating the accuracy of the in silico prediction (Supplemental Figure S6). The 3-D model was further submitted for prediction of the quaternary structure and the pose with the highest similarity was the one based on the homo-octameric conformation of RsSGD (Supplemental Figure S7). Further analysis of the interacting residues among the monomers of the putative homo-octameric architecture revealed that the majority of interactions actively engage amino acids located at the C-terminus of SGD (Supplemental Figure S8), in agreement with our previous experimentally validated results (Guirimand et al., 2010). Thus, the C-terminus of SGD plays a critical role in the assembly of the multimeric conformation. Interestingly, in 4 of the 16 interfaces of the predicted pose (i.e. chains A:D, B:E, C:H, and F:G), no residues located at the C-terminus of SGD are engaged in interactions among the chains but rather residues from the N-terminus and core region of SGD. This result suggests that although shSGD, which lacks residues 475–555, is not able to homo-multimerize, it could potentially be incorporated in a hetero-multimeric assembly with SGD through its N-terminal extremity.
shSGD inhibits SGD activity
In order to study in detail the effect of shSGD on SGD activity, a comparative deglycosylation assay was performed by mixing strictosidine with purified SGD, individually expressed and purified SGD and shSGD (SGD + shSGD, combined post-purification), or purified SGD and shSGD (SGD−shSGD) co-expressed by the same plasmid (Figure 7, A).
Figure 7.
shSGD decreases SGD deglycosylation activity and production of tetrahydroalstonine in vitro. A, Integrity of the recombinant proteins used for the assays was checked by PAGE analysis after loading 0.5 µg per lane. B, Time-course analysis of strictosidine deglycosylation assays conducted by incubating 50 µM of strictosidine with SGD (0.001 µg), individually expressed SGD and shSGD (SGD+shSGD; 0.001 µg each), and co-expressed SGD and shSGD (SGD-shSGD; 0.001 and 0.01 µg) for 0, 2, 5, and 10 min. Assays were conducted in triplicate. Error bars are sd, but are too small to being visible. C, Effect of shSGD on downstream MIA production measured in vitro by tetrahydroalstonine production (light gray) by incubating 50 µM of strictosidine (dark gray) with SGD (0.001 µg) plus THAS (0.1 µg) or with co-expressed SGD and shSGD (SGD-shSGD; 0.01 µg) plus THAS (0.1 µg). Assays were conducted in technical triplicates and were repeated twice.
Interestingly, whereas SGD and SGD + shSGD (combined post-purification) catalyze a similar strictosidine deglycosylation, a marked decrease of enzymatic activity against strictosidine was observed for co-expressed SGD−shSGD (Figure 7, B). Two minutes after initiating the reaction, SGD and SGD + shSGD (combined post-purification) consumed around 98% of strictosidine whereas SGD−shSGD (co-expressed by the same plasmid) only deglycosylated 36%. This reduction of activity was observed throughout the reaction and is in agreement with the zymogram (Figure 6, M). This further confirms the inhibitory effect of shSGD on SGD when the two isoforms are co-expressed, due to disruption of SGD multimers. In addition, this decrease of SGD activity was accompanied by a reduction of downstream MIA synthesis in vitro (Figure 7, C). When SGD was mixed with THAS1, around 85% of the starting amount of strictosidine was consumed after 2 min of reaction and tetrahydroalstonine formation reached a maximum of 16.03 ± 0.32 µM after 10 min. By contrast, consumption of strictosidine was markedly lower when SGD−shSGD was co-incubated with THAS1, with only 2.32 ± 0.22 and 12.29 ± 1.97 µM of tetrahydroalstonine produced after 10 and 30 min, respectively.
To potentially extend these biochemical results to the whole plant, we next conducted in planta functional assays. We first initiated a virus-induced gene silencing (VIGS) approach selectively targeting each isoform. Since shSGD and SGD only differ in the 3′-end of their coding sequence and their 3′-UTR, the selection of the sequence used for shSGD silencing was specifically restricted to this area (Supplemental Figure S9). After amplifying and cloning these sequences in pTRV2, C. roseus plantlets were inoculated with viral vectors via a biolistic-mediated VIGS procedure (Carqueijeiro et al., 2015; Foureau et al., 2016). Interestingly, around 1 week post-transformation, the SGD-silenced plants began to exhibit strong necrotic symptoms that quickly led to whole-plant death in contrast to control plants transformed with empty vector (Supplemental Figure S10). Whereas more pronounced in the present assays, this phenomenon was quite similar to the local necrosis caused by the silencing of the strictosidine transporter, suggesting high toxicity of strictosidine when accumulated in high proportions (Payne et al., 2017). By contrast, no visible phenotype was observed in shSGD-silenced plants (Supplemental Figure S10). Whereas gene silencing remained limited (around 60% of transcript amount decrease compared with plants transformed with empty vectors), no cross-silencing of SGD was observed (Figure 8, A). Nevertheless, this shSGD silencing resulted in a significant increase of the vindoline amount in agreement with the inhibitory effect of shSGD measured in vitro (Figure 8, B).
Figure 8.
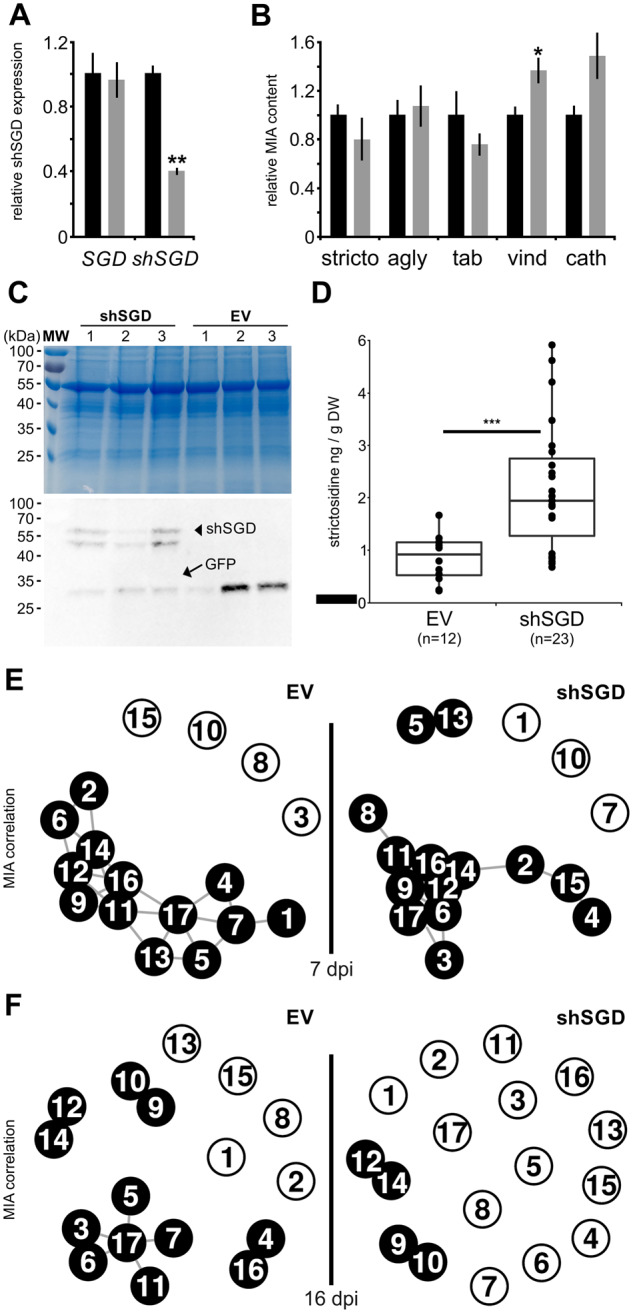
shSGD negatively impacts SGD deglycosylation activity. A, Relative expression of SGD and shSGD in leaves of plants transformed by empty vector (EV, black bar) or by the pTRV2-shSGD silencing construct (shSGD, gray bar) measured by RT-qPCR. B, Relative means of strictosidine (stricto), strictosidine aglycone (agly), tabersonine (tab), vindoline (vind), and catharanthine (cath) contents in leaves of EV (black bar) and shSGD (gray bar) transformed plants. Data represent means ± se of three technical replicates performed on four plants transformed with EV or shSGD (see Supplemental Material and Methods). Asterisks denote statistical significance (*P < 0.005; **P < 0.001; by Student’s t test). C, Coomassie blue staining and immunoblot analysis of crude protein extracts (15 µg per lane) from C. roseus leaves agroinfiltrated with pEAQ-HT:GFP-6His and either pEAQ-HT:shSGD-6His or empty vector (EV). D, Strictosidine concentration in infiltrated leaves (three independent biological replicates consisting of three plants transformed with EV, six plants transformed with shSGD construct, and for each plant four infiltrated leaves except for one sample of shSGD were analyzed after 7 d, thus totaling 12 EV samples and 23 shSGD samples). Points correspond to individual samples. Asterisks denote statistical significance in difference between the means (Wilcoxon rank sum test, P < 0.001). E and F, Metabolite correlation networks in control and shSGD overexpressing plants. Correlations among metabolite concentrations were calculated over 87 and 96 samples, respectively, from 10 plants and at least 8 infiltrated leaves for each construct at 7 d (E) and 16 d (F) post-infiltration (dpi). Nodes correspond to metabolites: 1, secologanin; 2, strictosidine; 3, tabersonine; 4, tabersonine imine alcohol; 5, 16-hydroxytabersonine; 6, 16-methoxytabersonine; 7, 16-methoxytabersonine epoxide; 8, 16-methoxy-2,3-dihydrotabersonine; 9, desacetoxyvindoline; 10, desacetoxyvindorosine; 11, desacetylvindoline; 12, vindoline; 13, demethylvindorosine; 14, catharanthine; 15, vinblastine; 16, ajmalicine; 17, serpentine. Node pairs connected by black lines indicate a Spearman rho >0.6 (E) and rho >0.7 (F) for these pairs, the higher the correlation, the shorter the line.
To confirm this potential effect of shSGD on MIA metabolism, we further evaluated shSGD function through a transient overexpression assay. Consequently, agro-infiltration of a GFP-expressing vector, used as a visual marker of transformation, and shSGD-expressing constructs or empty vector were achieved in C. roseus plants leading to the overexpression of both proteins in leaves as observed in the protein extracts of transformed plants. Immunoblot analyses performed on three representative plants of each combination of transformation revealed the presence of GFP in empty-vector samples and the concomitance of GFP and shSGD in plants transformed by the two corresponding constructs (Figure 8, C). Although immunodetection of shSGD resulted in unexpected double bands at ∼55 kDa, which could be indicative of possible post-translational modifications or proteolysis, the presence of the respective bands in GFP/shSGD infiltrated plants confirmed the successful overexpression of the protein in leaves. Furthermore, we observed that shSGD overexpression in these plants had no impact on the expression levels of SGD and other MIA biosynthetic genes engaged in either upstream (i.e. SLS1 and STR) or downstream (i.e. HYS, T16H2, OMT, and DAT) enzymatic steps (Supplemental Figure S11).
In agreement with our in vitro assays, we observed that increasing amounts of shSGD in planta led to a concomitant and significant accumulation of strictosidine 7 d post-infiltration compared with plants transformed with an empty vector (Figure 8, D). This reflects again the inhibitory effect of shSGD on SGD activity. In addition, this enhanced accumulation of strictosidine was accompanied by a noticeable change in MIA correlations in aerial parts of plants overexpressing shSGD. This was notably characterized by a change in strictosidine relationship with downstream MIAs (Figure 8, E andSupplemental Figure S12). For longer transient overexpression (16 dpi; Supplemental Figure S13), no substantial alterations in the final MIA amounts could be observed, as a probable consequence of the slower MIA metabolism taking place in fully expanded leaves used in our assays (Supplemental Figure S14). We rather observed a more pronounced shift in correlation with MIAs (Figure 8, F). In control plants transformed with the empty vector, strong dependencies were observed among MIAs especially around strictosidine and tabersonine. By contrast, MIA concentrations in plants overexpressing shSGD were less correlated as shown by a loss in correlations >0.7, suggesting an impact of altered control of SGD activity over the whole MIA metabolism.
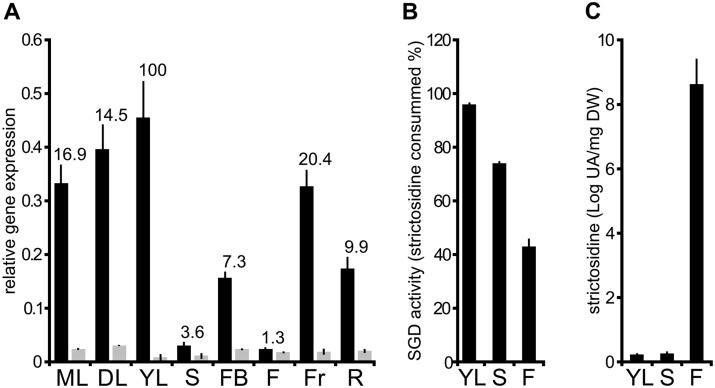
To determine the impact of shSGD on SGD activity in planta, we compared the global transcript level of SGD and shSGD in different C. roseus organs (Figure 9, A) by RT-qPCR. Whereas we observed that shSGD transcripts were always detected at a low level, the amount of SGD transcripts was highly variable among organs, with the highest and lowest accumulation in young leaves and flowers, respectively (Figure 9, A). Consequently, the variations of SGD transcript accumulation probably constitute the main cause of the huge differences in SGD/shSGD transcript ratio, ranging from 1.3 in flowers up to 100 in young leaves from the batch of analyzed plants. Since the dramatic changes in SGD expression render comparison of SGD activity among organs not easily interpretable, we quantified this enzymatic activity by monitoring in vitro the strictosidine consumption of protein extracts from either stems or flowers. These tissues display an almost identical amount of SGD transcript but a distinct amount of shSGD transcripts (two-fold higher in flowers). Additionally, protein extracts from young leaves were also analyzed as a positive control (Figure 9, B). Whereas high SGD activity was observed in young leaves after 30 min of incubation with strictosidine (around 95% of strictosidine consumption), lower activities were monitored in stem and flowers. The relative enzymatic activity was reduced to ca. 73% and 44%, respectively. Furthermore, we also noted that SGD activity was two-fold higher in flowers than in stem tissue, in agreement with SGD/shSGD ratio. Such decreased SGD activity was specifically accompanied by an increase of the in planta strictosidine amount in flowers, which may reinforce the proposed regulatory role of shSGD (Figure 9, C). Taken together, in vitro and in vivo results both confirm that shSGD acts de facto as a pseudo-enzyme through an inhibitory effect on SGD. Such an effect thus suggests a physiological role of shSGD in the whole plant for proper MIA production that could involve the negative regulation of SGD activity.
Figure 9.
In planta correlation of shSGD and SGD transcript amounts, SGD activity, and strictosidine accumulation. A, Relative quantification of SGD (dark) and shSGD (gray) transcripts in different C. roseus organs including mature leaves (ML), developing leaves (DL), young leaves (YL), stems (S), flower buds (FB), flowers (F), fruits (Fr), and roots (R). RNAs were extracted from organs and reverse-transcribed before determination of gene expression by qPCR. Transcript copy numbers were normalized using CrRPS9. Values correspond to SGD/shSGD rations in the different organ types. B, SGD activity was measured in YL, S, and F by incubating 15 µg of each crude protein extract with 50 µM strictosidine for 30 min. SGD activity is expressed as the percent of consumed strictosidine. C, Quantification of in planta strictosidine in YL, S, and F. For all experiments in G, H, and I, assays were performed in triplicate, and expression measurements were performed at least twice with independent experimental replicates.
shSGD causes the delocalization of THAS1 in the cytosol
Finally, since SGD multimers are known to recruit downstream MIA biosynthetic enzymes including THAS1 (Stavrinides et al., 2015, 2016), the existence of possible interactions between shSGD and THAS1 was investigated through BiFC assays.
The occurrence of such interactions was plausible since THAS1 transcripts were enriched in leaf epidermis in an order of magnitude comparable to those of SGD and shSGD, strongly suggesting that the corresponding proteins were all present in the same cell type (Figure 5, A and C). After cloning the THAS1 coding sequence into BiFC vectors to express YFPN-THAS1 and YFPC-THAS1, we restored the dimerization of THAS1 in the nucleus (Figure 10, A and B). As previously demonstrated, we also re-established, as a control, the recruitment of THAS1 in SGD high-molecular-weight complexes, which is supposed to channel the MIA biosynthetic flux (Figure 10, C and D). Surprisingly, whereas shSGD was not capable of self-interaction (Figures 3, E and 6, C and D), evident interactions with THAS1 were observed (Figure 10, E and F). Moreover, these interactions led to the delocalization of high amounts of THAS1 from the nucleus to the cytosol since BiFC complexes displayed a nucleocytosolic localization resembling that of shSGD localization (Figures 4, A–H and 10, E and F). To analyze the consequences of a possible co-occurrence of interactions between THAS1 or SGD and shSGD, we next performed multicolor BiFC (mcBiFC) assays that allow analysis of an assembly of proteins by pairs within a 3-protein set through the reformation of two distinct fluorophores (Hu and Kerppola, 2003). After cloning the SGD coding sequence in fusion with that of a split-Cyan Fluorescent Protein (CFP) fragment, YFPN-THAS1, YFPC-shSGD, and CFPN-SGD were co-expressed in C. roseus cells (Figure 10, G–I). The observation of CFP appearing as a diffuse nuclear signal first confirmed the previously observed interactions of shSGD and SGD (Figures 6, E and F and 10, H). More interestingly, the observation of the YFP BiFC complexes highlighted the interactions between THAS1 and shSGD (Figure 10, G–I) that displayed the nucleocytosolic localization observed during BiFC assays (Figure 10, E and F). In addition, the presence of shSGD also affected the distribution of SGD and THAS1 interactions that appeared diffuse in the nucleus similarly to SGD and shSGD interactions (Figure 10, J–L), as observed previously (Figure 6, E and F). Finally, as controls, the recruitment of THAS1 by SGD high-molecular-weight complexes (Figure 10, M–O) as well as the lack of interaction of shSGD and SGD with a distinct MIA biosynthetic enzyme, namely 16-hydroxytabersonine 16-O-methyltransferase (16OMT), confirmed the specificity of the observed interactions (Figure 10, P–U). All together, these results demonstrated that shSGD is capable of interactions with THAS1 even in the presence of SGD, and that this causes the partial delocalization of THAS1 to the cytosol.
Figure 10.
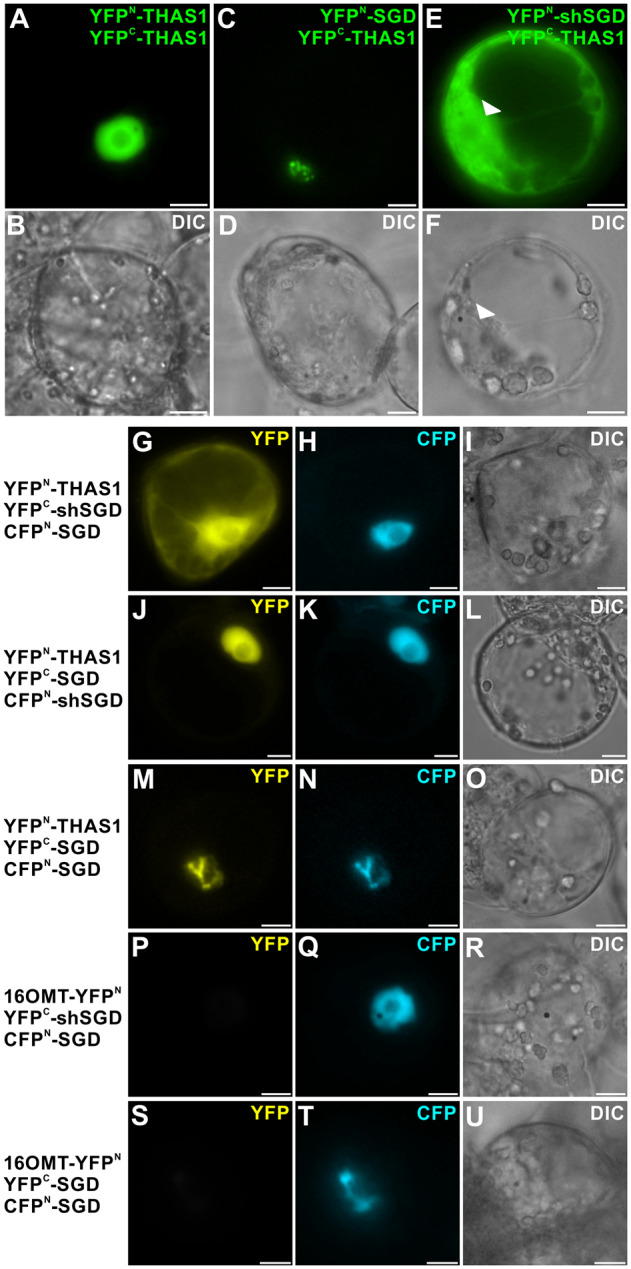
shSGD/THAS1 interaction causes partial delocalization of THAS1 to the cytosol and impacts the distribution of SGD/THAS1 complexes. A and B, THAS1 self-interactions; C and D, SGD/THAS1; and E and F, shSGD/THAS1 interactions were analyzed by BiFC in C. roseus cells transiently transformed by appropriated combinations of plasmids encoding fusions with the two split YFP fragments, as indicated on the right of each fluorescence picture. Arrowheads label cell nucleus. G–O, C. roseus cells were transiently co-transformed by the combinations of plasmids described on the left margin, expressing THAS1, shSGD, or SGD fused to split YFP fragment (YFPN and YFPC) and SGD or shSGD fused to a split CFP fragment (CFPN-SGD). P–R, 16-Hydroxytabersonine 16OMT/SGD/shSGD self-interactions and S–U, 16OMT/SGD/SGD interactions were studied to evaluate the specificity of THAS1/SGD and THAS1/shSGD interactions. Association of the two split YFP fragments results in the emission of a yellow fluorescent signal (G, J, and M), whereas association of split CFPN and split YFPC (H, K, N, Q, and T) allows the emission of a blue fluorescent signal. Cell morphology is observed with differential interference contrast (DIC) (B, D, F, I, L, O, R, and U). Bars, 10 µm.
Discussion
Alternative splicing produces a regulator of MIA biosynthesis
Whereas genome sequencing and high-throughput transcriptomics have hugely contributed to the elucidation of plant specialized metabolisms and their regulation, the potential involvement of alternative splicing events has been under-explored to date. However, recent studies demonstrated the unexpected importance of splicing with the existence of more than 150,000 splicing junctions in the Arabidopsis genome, which may lead to as many splicing events and distinct coding sequences (Klepikova et al., 2016). Furthermore, a growing body of evidence indicates the crucial importance of alternative splicing in plant response to abiotic and biotic stresses as well as in plant defenses (Staiger and Brown, 2013). For instance, the Arabidopsis RESISTANCE TO PSEUDOMONAS SYRINGAE4 (RPS4) that activates defense response exists in several forms, which are the result of alternative splicing including an intron-retaining form highly induced upon Pseudomonas inoculation and strictly required for resistance (Zhang and Gassmann, 2003, 2007). A similar phenomenon of intron retention also affects jasmonate signaling through the consequent generation of truncated variants of Jasmonate ZIM-domain (JAZ) that display reduced interaction with Coronatine Insensitive 1 (COI1) and increased resistance to proteosomal degradation (Moreno et al., 2013). In this work, we identified an alternative splicing event affecting SGD, a gene that encodes a central enzyme of the MIA biosynthetic pathway acting at the cross-road of defense processes and biosynthesis of distinct MIA sub-families (Figure 1). Such alternative splicing relies on an intron retention (Figure 2, A–C), which corresponds to the most frequent alternative splicing type found in Arabidopsis (Reddy et al., 2013). Due to the appearance of a premature termination codon in the protein primary sequence, it results in the formation of a shorter SGD version truncated at the C-terminal end, named shSGD. The corresponding shSGD transcript variant is barely detectable in all plant organs (Supplemental Figure S1, G) and notably co-accumulated in leaf epidermis with that of SGD strongly suggesting that both corresponding proteins are co-localized in this plant tissue (Figure 5, C). Whereas the relative abundance of shSGD and SGD considerably varies according to organ type and stimuli (Figures 5, A and B and 9, A), we noted that the mean amount of shSGD transcript reached around 10% of the original SGD variant type, a ratio corresponding to the observed frequencies of alternative variants of isochorismate synthase in Populus (Yuan et al., 2009). Furthermore, shSGD transcript production is strictly concomitant to SGD expression in agreement with the effect of shSGD on SGD activity. Due to the inhibitory effect of shSGD, this alternative splicing event engenders a possible negative control around strictosidine deglycosylation and SGD activity.
Inhibition of strictosidine deglycosylation by a pseudo-enzyme
In our experimental conditions, shSGD never exhibited strictosidine deglycosylation activity, as an expected consequence of the loss of key residues involved in substrate binding due to the alternative splicing (Figure 3 and Supplemental Figure S4; Barleben et al., 2007). However, interaction studies performed through BiFC analyses or protein co-expression in E. coli demonstrated that interaction of shSGD with SGD partially disrupts SGD multimers and results in a marked decrease of SGD deglycosylation activity both in vitro and in planta (Figures 6, A–F, J–M, 7, B, 8, D, and 9, A–C). Since the relative abundance of shSGD and SGD transcripts oscillates between 1.7% and 43%, a similar ratio between shSGD and SGD can be expected at the protein level; however, complementary assays with antibodies raised specifically against SGD are needed to confirm this observation in planta. Therefore, the pronounced inhibitory effect observed in our assays in which shSGD and SGD are produced in almost equivalent proportion in E. coli thus represents the ultimate stage of the inhibition that may exist in planta. shSGD meets the concept of pseudo-enzymes corresponding to dead or zombie versions of proteins (Eyers and Murphy, 2016; Murphy et al., 2017a). These pseudo-enzymes are catalytically deficient variants of enzymes lacking essential catalytic amino acids and their prevalence may represent up to 10%–15% of proteomes (Pils and Schultz, 2004). They are conserved in more than 20 different protein families and exert their effect on signal transduction or metabolism through specific protein/protein interactions, ranging from allosteric regulation of active enzymes, regulation of protein localization, and substrate sequestration to competition for complex assembly, as observed for shSGD (Figures 6, 10; Murphy et al., 2017a, 2017b). In plants, a growing body of evidence indicates the role of pseudo-enzymes in key physiological processes ranging from starch hydrolysis to vitamin B6 synthesis (Li et al., 2009; Moccand et al., 2014; Dell’Aglio et al., 2017; Monroe and Storm, 2018). The existence of shSGD now broadens the involvement of pseudo-enzymes to specialized metabolisms as illustrated for MIA synthesis. In addition, whereas pseudo-enzymes are usually transcribed from genes duplicated from their catalytically active counterparts, the production of pseudo-enzymes from alternatively spliced genes has not been explored to the best of our knowledge (Murphy et al., 2017b). As such, shSGD could represent a previously uncharacterized type of pseudo-enzyme/pseudo-glucosidase resulting from alternative splicing, thus broadening the physiological role of this process.
The pseudo-glucosidase shSGD as an adjustment variable of MIA metabolism
Strictosidine is a metabolic hub molecule involved both in the synthesis of MIA under physiological conditions and in a defense mechanism based on protein crosslinking when huge amounts of strictosidine are massively deglycosylated (Guirimand et al., 2010). Whereas a toxic effect is beneficial in the latter case, the biosynthesis of MIAs requires a more restricted flow of deglycosylation linked to complementary processes to manage the reactivity of the aglycone and avoid harmful consequences due to self-protein crosslinking. For instance, multimerization of SGD and the formation of a fibrillar tunnel-like structure, similar to the one observed in oat avenacosidase (Kim et al., 2000, 2005; Kwak et al., 2009), probably allows aglycone to spontaneously rearrange in non-toxic MIAs such as cathenamine or 4,21-dehydrogeissoschizine (Figure 6, G–I). Depending on conditions, the differential translation of shSGD resulting in a proportional inhibition of SGD activity may constitute an adjustment variable for managing further the aglycone toxicity and preventing plant cell damage. As such, one of the main roles of the pseudo-enzyme shSGD could be to maintain strictosidine deglycosylation under a critical threshold to avoid undesirable effects of aglycone production. Production of both shSGD and SGD through alternative splicing of the same gene may establish a basal and constant negative feedback, whereas mechanisms regulating splicing efficiency may increase shSGD/SGD ratio when less strictosidine deglycosylation is required.
Furthermore, deglycosylation of strictosidine by SGD is the central entry point to downstream MIA synthesis. Whereas the control of this reaction is crucial, only few factors impacting SGD expression have been identified including jasmonate, folivory (Geerlings et al., 2000; Dugé de Bernonville et al., 2017), as well as transcription factors that, except for the artificially de-repressed version of MYC2, only modestly trigger or inhibit its expression (Liu et al., 2017; Schweizer et al., 2018; Colinas et al., 2020). The production of a pseudo-enzyme negatively affecting SGD activity could thus act as an additional adjustment variable of MIA metabolism in organs, as measured in planta (Figure 9, A–C). The increased strictosidine accumulation combined with apparent MIA synthesis de-synchronization caused by shSGD overexpression in C. roseus leaves, as well as the increased vindoline accumulation in shSGD-silenced plants, strongly support this statement (Figure 8). Finally, since SGD is a highly efficient and stable enzyme that is difficult to inhibit (Guirimand et al., 2010), decreasing SGD transcription may not be sufficient to dramatically reduce strictosidine deglycosylation when required, such as in flowers. Maintaining low-level production of shSGD through basal expression of shSGD transcripts (Figure 9) may contribute to this fine-tuning due to the strong inhibitory effect of shSGD. The molecular mechanism governing the alternative splicing of the single-copy gene encoding SGD in apparent organ-specific differential ratios remains to be addressed.
The pseudo-glucosidase shSGD reorganizes the subcellular localization of MIA biosynthetic enzymes
The deletion of the C-terminal end of shSGD results in the loss of the bipartite NLS normally targeting SGD to the nucleus and thus explains the nucleocytosolic localization of shSGD (Figure 4, A–H). A similar splicing event in rice (Oryza sativa) impacts the localization of proteins encoded by the three transcript variants of the mitogen-activated protein kinase (MAPK) 1, which are preferentially located to the cytosol or the nucleus (Koo et al., 2007). However, shSGD localization did not result in an extension of the strictosidine glucosidase activity area to other subcellular compartments, as proposed previously for the cytosol by indirect methodologies (i.e. activity assays after subcellular fractionation or microscopic observation after strictosidine activity staining; Stevens et al., 1993; Geerlings et al., 2000). By contrast, the deletion of the C-terminal end of shSGD directly impacts the quaternary structure of SGD and results in a partial disruption of SGD multimers causing, in turn, a diffused nuclear localization of SGD (Figure 6, E–L). Whereas the reorganization of the subcellular distribution of MIA biosynthetic enzymes remains a tricky issue to deal with, one can consider that it could be associated with the defense mechanisms involving SGD. The massive strictosidine deglycosylation by SGD is likely to cause protein crosslinking to protect plant leaves during attacks. Whereas SGD multimers are protease-resistant structures that can ensure deglycosylation over time, more soluble forms, as released by shSGD, can diffuse rapidly throughout the cell and broadens aglycone outflow to ensure a first quick protein crosslinking, even with a reduced intrinsic glucosidase activity. Furthermore, shSGD also interacted with THAS1 causing its cytosolic retention (Figure 10, A–F) and restricting its sequestration by SGD into the nucleus (Figure 10, G–L). Besides contributing to funnel aglycone reactivity (Stavrinides et al., 2015, 2016), this recruitment also favors a main synthesis of tetrahydroalstonine. Given the diversity of enzymes potentially interacting with SGD, a fine adjustment of the proteins recruited by SGD multimers is required to guide the MIA biosynthetic flux. The pseudo-glucosidase shSGD could be part of this process by selectively re-localizing enzymes in the cytosol to limit the synthesis of the corresponding MIA as suggested again by the de-synchronization of MIA synthesis after shSGD overexpression or silencing (Figure 8, A, B, E, and F).
Managing protein interactions in the GH1 family
Overall, the management of protein interactions constitutes a crucial element of the defense mechanism involving β-glucosidases from the GH1 family. In oat, β-glucosidases are engaged in preformed defense mechanism against fungi by being targeted to plastids while their glycosidic substrate, avenacoside A, is sequestrated in the vacuole (Nisius, 1988). Interestingly, two β-glucosidases isoforms exist. Whereas only one (As-Glu1) forms high-molecular-weight multimers adopting a typical fibrillar structure, the second isoform (As-Glu2) rather dimerizes and also form heterodimers with As-Glu1 displaying heterohexameric ring structures, thus resembling the effect of the pseudo-glucosidase shSGD (Kim et al., 2000). In the myrosinase system, the PYK10 glucosidase localized in endoplasmic reticulum (ER) bodies oscillates between soluble inactive forms and active aggregated forms through the recruitment of PYK10-binding protein-1 (PBP1; Nagano et al., 2005). Interestingly, PBP1 is not localized in ER bodies and only meets PYK10 following the destruction of subcellular structures by pests as proposed in the “nuclear time bomb” system. Another example is the maize BGAF, a 35-kDa protein that specifically binds to the defensive ZmGlu1 and ZmGlu2 forming supramolecular aggregates of BGAF/β-glucosidase and both the N- and C-termini of ZmGLU1 play a definitive role in the degree of multimerization (Yu et al., 2009). It is worth noting that mutations on either the N- or C-termini of defensive β-glucosidases are known to affect both the proper assembly of quaternary structures and the enzymatic activity of the complexes (Sue et al., 2006; Kwak et al., 2009; Koudounas et al., 2017). Although SGD does not require a protein partner for self-interactions, the adjustment of SGD multimerization as well as interactions with THAS1 ensured by shSGD could represent a similar type of regulatory processes between Apocynaceae and Brassicaceae. Furthermore, the parallel between myrosinase and “nuclear-time bomb” systems also emerges through the control of the toxicity of the deglycosylation products. Hydrolysis of glucosinolates by myrosinases leads to the formation of unstable aglycones that rearrange to form the defensive isothiocyanates (Kliebenstein et al., 2005). However, epithiospecifier proteins (ESPs) can redirect this reaction to form less toxic compounds including organic thiocyanates, epithionitriles, or simple nitriles as similarly described for the THAS1/SGD/shSGD interaction and the formation of tetrahydroalstonine (Mumm et al., 2008; Stavrinides et al., 2015, 2016). Interestingly, alternative splicing of EPS has also been shown to act as a regulator of EPS protein amount and activity, thus providing further strong evidence for the role of gene splicing in plant defense regulation (Kissen et al., 2012). Overall, the recruitment of a pseudo-enzyme to manage protein interactions is a further regulatory process that deserves to be studied in the GH1 family.
Conclusion
In conclusion, we report the alternative splicing of a MIA biosynthetic gene in C. roseus which belongs to the GH1 family. Beyond the recently discovered multiple isoforms of MIA biosynthetic enzymes (Besseau et al., 2013, Dugé de Bernonville et al., 2015, Kellner et al., 2015), this study sheds light on the possible implication of unexpected molecular processes in the global regulation of MIA biosynthesis and plant defense mechanisms. The splicing-mediated synthesis of the pseudo-glucosidase shSGD impacting strictosidine glucosidase activity and protein subcellular localizations could thus be one such process. Identification of such a fine-tuned control of SGD activity will undoubtedly guide efficient MIA production through metabolic engineering and pathway reconstitution in heterologous organisms (Cravens et al., 2019).
Materials and methods
Data mining for alternative transcript identification
In a search for alternative splicing events in stressed leaves of C. roseus (cv Apricot Sunstorm), transcript abundance was monitored in treated and control leaves in diverse conditions. In our previous work, we set up an experimental device allowing M. sexta larvae to feed on C. roseus leaves (Dugé de Bernonville et al., 2017). The subsequent folivory has been shown to trigger a strong defense reaction involving jasmonate signaling. RNA-seq runs of control and consumed leaves of C. roseus were included in a data mining approach to identify alternatively spliced isoforms. In addition, we included RNA-seq runs of shoots (SRR646572 and SRR646596) and suspension cells (SRR648705 and SRR648707) elicited with methyl-jasmonate that have been previously deposited by the SmartCell European consortium (Van Moerkercke et al., 2013). All sequencing data were downloaded from the public database EBI ENA. Transcript abundance was obtained in the different samples after quasi-mapping trimmed reads (Bolger et al., 2014) on the CDF97 transcriptome assembly (Dugé de Bernonville et al., 2015) with Salmon v0.8.2 (Patro et al., 2017). Differentially expressed genes between control (SRR648705, SRR646572, ERR1512372, and ERR1512372) and treated samples (SRR646596, SRR648707, ERR1512372, and ERR1512371) were identified with EdgeR (Robinson et al., 2010) R package. Candidate splicing events were visualized with Splign (Kapustin et al., 2008).
SGD scaffold reconstitution
A BAC clone of a C. roseus (cv Apricot Sunstorm) genomic DNA BAC library containing the SGD-encoding gene has been previously sequenced and the corresponding Illumina MiSeq sequencing run has been deposited in the SRA database under accession number ERR599180. We thus used these raw data to generate a large genomic scaffold containing the SGD gene. Raw MiSeq reads were obtained from SRA, converted into fastq files with SRA Toolkit v2.3.4, and trimmed with Trimmomatic v0.32 (Bolger et al., 2014) with default parameters. Contigs were de novo assembled with Trinity v2.1.1 (Grabherr et al., 2011) without prior in silico normalization and with other default parameters.
Preparation of leaf epidermis-enriched fraction of RNA
Epidermis-enriched fraction was obtained from a modified version of a procedure previously described by Murata et al. (2008). Further information is provided in the Supplemental Materials and Methods.
C. roseus/M. sexta interaction and RNA extraction
The C. roseus/M. sexta interactions were conducted as described in Dugé de Bernonville et al. (2017). RNAs from intact and attacked leaves were extracted and reverse-transcribed as described in Parage et al. (2016).
Validation of the presence of SGD and shSGD transcripts in planta
RNA was extracted with NucleoSpin RNA Plant and Fungi kit (Macherey Nagel, Germany) followed by rDNase Set (Macherey Nagel, Germany) treatment following the manufacturer’s instruction. Presence of SGD and shSGD transcripts (Figure 2, D) was confirmed by PCR performed on reverse-transcribed RNAs from C. roseus (cv Apricot Sunstorm) intact leaves using the SGD-F (5′-AACCCTGGCAAAGAACCCTA-3′; hybridizing at the transcribed region of Exon 7) forward primer and SGD-R_long (5′-TGTCATCAGATGCAAATTCGCT-3′; hybridizing at the transcribed 3′-UTR of Exon 13) for SGD, SGD-R_short (5′-TGATCGCGAGGGAAGGTATT-3′; hybridizing at the transcribed 3′-UTR of the non-spliced Intron 12) for shSGD, or SGD-R common (5′-ACCAAGAGATTGTAGAGTCCAGA-3′; hybridizing at the transcribed region of Exon 11) for both SGD and shSGD. Presence of SGD and shSGD transcripts (Supplemental Figure S3) was further confirmed in seedlings of other C. roseus cultivars using either the SGD-F and SGD-R_long primers or the SGD-F2 (5′-GTGGATGGAGCCTCTCAATGAAACC-3′; hybridizing at the transcribed region of Exon 8) and SGD-R_short2 (5′-CCTATTGAATATCACTTTGTCACGACG-3′; hybridizing at the transcribed 3′-UTR of the non-spliced Intron 12) primers, respectively. Amplifications were analyzed by electrophoresis on a 1% agarose gel and the resulting PCR products from cv Apricot Sunstorm were cloned in the pGEM-T plasmid (Promega) and sequenced to confirm sequence identity.
Gene expression measurements
RNAs from C. roseus (cv Little Bright Eye) plants grown at 28°C were extracted and reverse transcribed as described on the previous section. SGD, shSGD, THAS1, and HDS gene expression was measured by RT-qPCR using primers qSGD-For (5′-CTTCGACAACTTCGAATGGAA-3′)/qSGD-Rev (5′-CTTCTTGACTAACTCAACTAGT-3′), qSGDsh2-For (5′-CAACATATTACTTACAGAAGCTCGT-3′)/qSGDsh2-Rev (5′-ATGGTCTACGGTCTACCTTCTTCT-3′), qCrTHAS-For (5′-AGTATGAAGGAAATACAAGAGATGATT-3′)/qCrTHAS-Rev (5′-AGTATGAAGGAAATACAAGAGATGATT-3′), and qHDS-For (5′-GTCCCTTACTGAACCTCCAGAG-3′)/qHDS-Rev (5′-AATCACCTGTCCTGCGTGG-3′). The 40S ribosomal RPS9 (CrRPS9; AJ749993.1) gene was chosen as a housekeeping gene and measured by using primers qRPS9-For (5′-TTACAAGTCCCTTCGGTGGT-3′)/qRPS9-Rev (5′-TGCTTATTCTTCATCCTCTTCATC-3′). Further information concerning the RT-qPCR analyses is provided in the Supplemental Materials and Methods.
Production of the SGD and shSGD recombinant proteins and test of activity
SGD, shSGD, and THAS coding sequences were amplified using the following primers: SGD-BamHI-For (5′-CTGAGAGGATCCATGGGATCTAAAGATGATCAGTCCCT-3′)/SGD-HindIII-Rev (5′-CTGAGAAAGCTTTTAGTATTTTTGCTTCTTGACTAACTCAACTC-3′), shSGD-BamHI-For (5′-CTGAGAGGATCCATGGGATCTAAAGATGATCAGTCCCT-3′)/shSGD-BamHI-Rev (5′-CTGAGAGGATCCTCACTCAATAGCATCTCGCACG-3′), and THAS1-BamHI-For (5′-CTGAGAGGATCCATGGCAATGGCTTCAAAG-3′)/THAS1-BamHI-Rev (5′-CTGAGAGGATCCATTTGATTTCAGAGTGTT-3′), respectively. The resulting PCR product of SGD was cloned into the pET28a (Novagen) plasmid using BamHI and HindIII, shSGD PCR product was cloned in the BamHI restriction site of pET28a, whereas THAS1 was cloned into the pQE-30 (Qiagen). In order to co-express both shSGD and SGD from the same E. coli cells, the SGD expression cassette was excised from the pET28a-SGD vector after a SmaI and HincII digestion and then ligated into a SmaI-digested pET28a-shSGD vector to generate the pET28a-SGD-shSGD construct. Further information concerning heterologous expression, purification, activity assays, and UPLC-MS analyses is provided in the Supplemental Materials and Methods.
In silico prediction of the quaternary structure of CrSGD
The tertiary protein structure prediction of CrSGD (NCBI accession no. AAF28800) was generated with the Phyre2 application (Kelley et al., 2015) using the “intensive” modeling mode. The quaternary protein structure of CrSGD was predicted by submitting the tertiary model to the GalaxyHomomer server (Baek et al., 2017). The resulting poses were analyzed with PyMOL (DeLano, 2002) and protein–protein interactions were identified with PDBsum (Laskowski, 2001).
Analysis of SGD multimers by negative staining in transmission electron microscopy
Formvar/carbon-coated nickel grids were deposited on a drop of SGD multimers, purified as described previously, for 5 min and rinsed three times with phosphate-buffered saline (PBS). The grids were washed in distilled water and then negative straining was performed with three consecutive contrasting steps using 2% (w/v) uranyl acetate (Agar Scientific, Stansted, UK) before observation under the transmission electron microscope (JEOL JEM-1400 Plus, Tokyo, Japan).
Analysis of SGD and shSGD interactions in native PAGE
To analyze the consequences of shSGD and SGD interactions, soluble crude protein extracts from E. coli expressing either SGD or both SGD and shSGD were resolved in 8% SDS-PAGE after heating at 90°C for 5 min. Immunoblot analysis was performed with an anti-HIS tag primary antibody and a horseradish peroxidase-conjugated secondary antibody following standard protocols. The protein extracts were normalized based on the intensity of the immunodetected monomeric conformation of SGD. The same amount of protein extracts was then resolved without heat-treatment under semi-native conditions followed by zymogram and immunoblotting as described in Guirimand et al. (2010) and Koudounas et al. (2017).
Virus-induced gene silencing
VIGS studies were performed according to Carqueijeiro et al. (2015, 2018) using C. roseus (cv Little Bright Eye) plants with slight modifications. Further information is provided in the Supplemental Materials and Methods.
Subcellular localization studies and analysis of protein–protein interactions by BiFC
The subcellular localization of shSGD was studied by creating a fluorescent fusion protein using the pSCA-cassette YFPi plasmid (Guirimand et al., 2009). For this purpose, the full-length open-reading frame of shSGD was amplified using the specific couple of primers shSGD-For (5′-CTGAGAACTAGTATGGGATCTAAAGATGATCAGTCCCT-3′) and shSGD-rev3 (5′-CTGAGAACTAGTCTCAATAGCATCTCGCACGCTAG-3′) without stop codon or shSGD-For and shSGD-Rev2 (5′-CTGAGAACTAGTGAAAACTCACTCAATAGCATCTCGCACG-3′), which were designed to introduce the SpeI restriction site at cDNA extremities. PCR products were cloned at the 5′-end of the YFP coding sequence to generate the shSGD-YFP fusion protein, or at the 3′-end to express the YFP-shSGD fusion. All constructs were verified by sequencing. SGD localization was studied using the plasmids described in Guirimand et al. (2010).
Interactions with THAS1 were tested using the previously described plasmids (YFPN-THAS1 and YFPC-THAS1) and plasmids expressing 16OMT-YFPN were used as controls (Guirimand et al., 2011; Stavrinides et al., 2015). Further information concerning BiFC analysis is provided in the Supplemental Materials and Methods.
shSGD, SGD, and THAS1 multiple interactions were analyzed by multicolor mBIFC using the plasmid expressing SGD fused to the C-terminal extremity of the split CFPN fragment (CFPN-SGD) as described in Stavrinides et al. (2016).
Transient transformations of C. roseus cells by particle bombardment and fluorescence imaging were performed following the procedures previously described (Foureau et al., 2016).
Transient overexpression of shSGD in C. roseus
The coding sequence of shSGD without a stop codon was amplified with primers ShSGD-AgeI-For (5′-CTGAGAACCGGTATGGGATCTAAAGATGATCAGTCC-3′) and ShSGD-AgeI-Rev (5′-CTGAGAACCGGTCTCAATAGCATCTCGCACGCTA-3′), which introduce an AgeI restriction site in both extremities and the product was ligated in a AgeI/XmaI digested pEAQ-HT plasmid in order to produce the pEAQ-HT:shSGD-6His construct. Agroinfiltration was performed as described in Koudounas et al. (2015) with some modifications. Further information is provided in the Supplemental Materials and Methods.
Statistical procedures
All statistical comparisons were performed in R v3.6. Means were compared using non-parametric tests because of heteroscedasticity and non-normality of the residues. Metabolite correlation networks were constructed by calculating a distance matrix with Spearman’s rho. Gene pairs displaying a rho > 0.7 were further retained and all resulting significant associations were visualized in a graph containing nodes (metabolites) connected by edges with the “igraph” package.
Accession number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number: GenBank accession number of shSGD is MG520182.
Supplemental data
Supplemental Figure S1. Characterization of predicted SGD transcripts in CDF97.
Supplemental Figure S2. Genomic organization and read coverage of SGD and shSGD.
Supplemental Figure S3. SGD and shSGD are co-expressed in four distinct C. roseus cultivars.
Supplemental Figure S4. Amino acid sequence alignment of strictosidine glucosidases.
Supplemental Figure S5. Analysis of recombinant SGD and co-expressed SGD/shSGD.
Supplemental Figure S6. In silico prediction of CrSGD tertiary structure.
Supplemental Figure S7. Overall architecture of the putative quaternary structure of CrSGD.
Supplemental Figure S8. Residue interactions across all the interfaces.
Supplemental Figure S9. Nucleotide sequence alignment of the 3′-UTRs of SGD and shSGD.
Supplemental Figure S10. Silencing of SGD leads to plant death.
Supplemental Figure S11. Relative expression of MIA biosynthetic genes in C. roseus plants agroinfiltrated with an empty vector (black) or overexpressing shSGD (gray).
Supplemental Figure S12. MIA accumulation in plants transformed with empty vector (EV) or overexpressing shSGD (shSGD) after 7 d.
Supplemental Figure S13. Detection of transiently overexpressed shSGD in C. roseus leaves.
Supplemental Figure S14. MIA accumulation in plants transformed with empty vector (EV) or overexpressing shSGD (shSGD) after 16 d.
Supplementary Material
Acknowledgments
The authors thank Marie-Antoinette Marquet, Evelyne Danos, Emeline Marais, and Cédric Labarre (EA2106 Biomolécules et Biotechnologies Végétales) for help in maintaining cell cultures/plants and for valuable technical assistance. They acknowledge the Fédération CaSciModOT for access to the computing grid. They also acknowledge funding from ARD2020 Biopharmaceutical program of the Région Centre Val de Loire (BioPROPHARM, CatharSIS, ETOPOCentre projects) and from le Studium (Consortium fellowship).
Funding
This research was supported by “Région Centre-Val de Loire” (France) by funding the CatharSIS program, BioPROPHARM and ETPOCentre projects—ARD2020 Biomédicaments. V.C. acknowledges “le STUDIUM” for a STUDIUM Research Consortium grant.
Conflict of interest statement: The authors declare no conflict of interest.
I.C. performed the recombinant protein purification, activity tests, and contributed to SGD short transcript identification. K.K. performed the zymograms, biochemical assays, and protein overexpression assays. T.D.d.B. reconstituted the SGD genomic scaffold, identified the short SGD transcript, and performed statistical analysis. A.M., L.J.S., and P.L.C studied the protein subcellular localization and interactions. D.K.P. and D.A.N. performed the VIGS analysis. A.O. performed the qPCR analysis. A.L. carried out the LC–MS analyses. S.B. assisted in activity testing. N.K. and E.A.S. assisted in overexpression experiments. S.E., J.B.-G., and E.B. performed the electron microscopy. N.P. and M.C. assisted in the biochemical characterization of enzymes and data interpretation. L.A., N.G.-G., and B.S.-P. analyzed the data. S.E.O. and V.C. designed the research and wrote the manuscript. V.C. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Vincent Courdavault (vincent.courdavault@univ-tours.fr).
References
- Baek M, Park T, Heo L, Park C, Seok C (2017) GalaxyHomomer: a web server for protein homo-oligomer structure prediction from a monomer sequence or structure. Nucleic Acids Res 45:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barleben L, Panjikar S, Ruppert M, Koepke J, Stöckigt J (2007) Molecular architecture of strictosidine glucosidase: the gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell 19:2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Kellner F, Lanoue A, Thamm AM, Salim V, Schneider B, Geu-Flores F, Höfer R, Guirimand G, Guihur A, et al. (2013) A pair of tabersonine 16-hydroxylases initiates the synthesis of vindoline in an organ-dependent manner in Catharanthus roseus. Plant Physiol 163:1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard DJ, Cicek M, Chen J, Esen A (2001) Identification of β-glucosidase aggregating factor (BGAF) and mapping of BGAF binding regions on maize β-glucosidase. J Biol Chem 276:11895–11901. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L, Franke J, Farrow SC, Chung K, Payne RME, Nguyen TD, Dang TT, Soares Teto Carqueijeiro I, Koudounas K, et al. (2018) Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 360:1235–1239. [DOI] [PubMed] [Google Scholar]
- Carqueijeiro I, Dugé de Bernonville T, Lanoue A, Dang TT, Teijaro CN, Paetz C, Billet K, Mosquera A, Oudin A, Besseau S, et al. (2018) A BAHD acyltransferase catalyzing 19-O-acetylation of tabersonine derivatives in roots of Catharanthus roseus enables combinatorial synthesis of monoterpene indole alkaloids. Plant J 94:469–484. [DOI] [PubMed] [Google Scholar]
- Carqueijeiro I, Masini E, Foureau E, Sepúlveda LJ, Marais E, Lanoue A, Besseau S, Papon N, Clastre M, Dugé de Bernonville T, et al. (2015) Virus-induced gene silencing in Catharanthus roseus by biolistic inoculation of tobacco rattle virus vectors. Plant Biol (Stuttg) 17:1242–1246. [DOI] [PubMed] [Google Scholar]
- Colinas M, Pollier J, Vaneechoutte D, Malat DG, Schweizer F, De Clercq R, Guedes JG, Martínez-Cortés T, Molina Hidalgo FJ, Sottomayor M, et al. (2020) A modular system regulates specialized metabolite pathway branch choice in the medicinal plant Catharanthus roseus. BioRxiv 20200504075671.
- Courdavault V, Papon N, Clastre M, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2014) A look inside an alkaloid multisite plant: the Catharanthus logistics. Curr Opin Plant Biol 19:43–50. [DOI] [PubMed] [Google Scholar]
- Cravens A, Payne J, Smolke CD (2019) Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat Commun 10:2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL molecular graphics system. San Carlos (CA: ): Delano Scientific. [Google Scholar]
- Dell’Aglio E, Boycheva S, Fitzpatrick TB (2017) The Pseudoenzyme PDX12 sustains vitamin B6 biosynthesis as a function of heat stress. Plant Physiol 174:2098–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugé de Bernonville T, Carqueijeiro I, Lanoue A, Lafontaine F, Sánchez Bel P, Liesecke F, Musset K, Oudin A, Glévarec G, Pichon O, et al. (2017) Folivory elicits a strong defense reaction in Catharanthus roseus: metabolomic and transcriptomic analyses reveal distinct local and systemic responses Sci Rep 7:40453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugé de Bernonville T, Foureau E, Parage C, Lanoue A, Clastre M, Londono MA, Oudin A, Houillé B, Papon N, Besseau S, et al. (2015) Characterization of a second secologanin synthase isoform producing both secologanin and secoxyloganin allows enhanced de novo assembly of a Catharanthus roseus transcriptome. BMC Genomics 19:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers PA, Murphy JM (2016) The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol 14:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foureau E, Carqueijeiro I, Dugé de Bernonville T, Melin C, Lafontaine F, Besseau S, Lanoue A, Papon N, Oudin A, Glévarec G, et al. (2016) Prequels to synthetic biology: from candidate gene identification and validation to enzyme subcellular localization in plant and yeast cells. Methods Enzymol 576:167–206. [DOI] [PubMed] [Google Scholar]
- Geerlings A, Ibañez MM, Memelink J, van Der Heijden R, Verpoorte R (2000) Molecular cloning and analysis of strictosidine beta-D-glucosidase, an enzyme in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J Biol Chem 275:3051–3056. [DOI] [PubMed] [Google Scholar]
- Gerasimenko I, Sheludko Y, Ma X, Stöckigt J (2002) Heterologous expression of a Rauvolfia cDNA encoding strictosidine glucosidase, a biosynthetic key to over 2000 monoterpenoid indole alkaloids. Eur J Biochem 269:2204–2213. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G, Burlat V, Oudin A, Lanoue A, St-Pierre B, Courdavault V (2009) Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep 28:1215–1234. [DOI] [PubMed] [Google Scholar]
- Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2010) Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol 10:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G, Guihur A, Poutrain P, Héricourt F, Mahroug S, St-Pierre B, Burlat V, Courdavault V (2011) Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J Plant Physiol 168:549–557. [DOI] [PubMed] [Google Scholar]
- Hemscheidt T, Zenk MH (1980) Glucosidases involved in indole alkaloid biosynthesis of Catharanthus cell cultures. FEBS Lett 110:187–191. [DOI] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21:539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustin Y, Souvorov A, Tatusova T, Lipman D (2008) Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner F, Kim J, Clavijo BJ, Hamilton JP, Childs KL, Vaillancourt B, Cepela J, Habermann M, Steuernagel B, Clissold L, et al. (2015) Genome-guided investigation of plant natural product biosynthesis. Plant J 82:680–692. [DOI] [PubMed] [Google Scholar]
- Kim YW, Kang KS, Kim SY, Kim IS (2000) Formation of fibrillar multimers of oat beta-glucosidase isoenzymes is mediated by the As-Glu1 monomer. J Mol Biol 303:831–842. [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim YW, Hegerl R, Cyrklaff M, Kim IS (2005) Novel type of enzyme multimerization enhances substrate affinity of oat β-glucosidase J Struct Biol 150:1–10. [DOI] [PubMed] [Google Scholar]
- Kissen R, Hyldbakk E, Wang CW, Sørmo CG, Rossiter JT, Bones AM (2012) Ecotype dependent expression and alternative splicing of epithiospecifier protein (ESP) in Arabidopsis thaliana. Plant Mol Biol 78:361–375. [DOI] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88:1058–1070. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Mitchell-Olds T (2005) The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr Opin Plant Biol 8:264–271. [DOI] [PubMed] [Google Scholar]
- Koo SC, Yoon HW, Kim CY, Moon BC, Cheong YH, Han HJ, Lee SM, Kang KY, Kim MC, Lee SY, et al. (2007) Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochem Biophys Res Commun 360:188–193. [DOI] [PubMed] [Google Scholar]
- Koudounas K, Banilas G, Michaelidis C, Demoliou C, Rigas S, Hatzopoulos P (2015) A defence-related Olea europaea β-glucosidase hydrolyses and activates oleuropein into a potent protein cross-linking agent. J Exp Bot 66:2093–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudounas K, Thomopoulou M, Michaelidis C, Zevgiti E, Papakostas G, Tserou P, Daras G, Hatzopoulos P (2017) The C-domain of oleuropein β-glucosidase assists in protein folding and sequesters the enzyme in nucleus. Plant Physiol 174:1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SN, Kim SY, Choi SR, Kim IS (2009) Assembly and function of AsGlu2 fibrillar multimer of oat β-glucosidase. BBA Proteins Proteom 1794:526–531. [DOI] [PubMed] [Google Scholar]
- Laskowski RA (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res 29:221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Francisco P, Zhou W, Edner C,, Steup M, Ritte G, Bond CS, Smith SM (2009) Catalytically-inactive beta-amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch-binding-protein. Arch Biochem Biophys 489:92–98. [DOI] [PubMed] [Google Scholar]
- Liu J, Gao F, Ren J, Lu X, Ren G, Wang RA (2017) Novel AP2/ERF transcription factor CR1 regulates the accumulation of vindoline and serpentine in Catharanthus roseus. Front Plant Sci 8:2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijendijk TJ, van der Meijden E, Verpoorte R (1996) Involvement of strictosidine as a defensive chemical in Catharanthus roseus. J Chem Ecol 22:1355–1366. [DOI] [PubMed] [Google Scholar]
- Luijendijk TJ, Stevens LH, Verpoorte R (1998) Purification and characterisation of strictosidine b-D-glucosidase from Catharanthus roseus cell suspension cultures. Plant Physiol Biochem 36:419–425. [Google Scholar]
- McKnight TD, Bergey DR, Burnett RJ, Nessler CL (1991) Expression of enzymatically active and correctly targeted strictosidine synthase in transgenic tobacco plants. Planta 185:148–152. [DOI] [PubMed] [Google Scholar]
- Moccand C, Boycheva S, Surriabre P, Tambasco-Studart M, Raschke M, Kaufmann M, Fitzpatrick TB (2014) The pseudoenzyme PDX12 boosts vitamin B6 biosynthesis under heat and oxidative stress in Arabidopsis. J Biol Chem 289:8203–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Storm AR (2018) The Arabidopsis β-amylase (BAM) gene family: diversity of form and function. Plant Sci 276:163–170. [DOI] [PubMed] [Google Scholar]
- Moreno JE, Shyu C, Campos ML, Patel LC, Chung HS,, Yao J, He SY, Howe GA (2013) Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol 162:1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm R, Burow M, Bukovinszkine’kiss G, Kazantzidou E, Wittstock U, Dicke M, Gershenzon J (2008) Formation of simple nitriles upon glucosinolate hydrolysis affects direct and indirect defense against the specialist herbivore, Pieris rapae. J Chem Ecol 34:1311–1321. [DOI] [PubMed] [Google Scholar]
- Murata J, Roepke J, Gordon H, De Luca V (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20:524–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Farhan H, Eyers PA (2017a) Bio-Zombie: the rise of pseudoenzymes in biology. Biochem Soc Trans 45:537–544. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Mace PD, Eyers PA (2017b) Live and let die: insights into pseudoenzyme mechanisms from structure. Curr Opin Struct Biol 47:95–104. [DOI] [PubMed] [Google Scholar]
- Muto A, Nakagawa A, Shimomura Y, Kitagawa Y, Tsurusawa M (2005) Antineoplastic agents for pediatric anaplastic large cell lymphoma: vinblastine is the most effective in vitro. Leuk Lymphoma 46:1489–1496. [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Matsushima R, Hara-Nishimura I (2005) Activation of an ER-body-localized beta-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol 46:1140–1148. [DOI] [PubMed] [Google Scholar]
- Nisius A (1988) The stromacentre in Avena plastids: an aggregation of β-glucosidase responsible for the activation of oat-leaf saponins. Planta 173:474–481. [DOI] [PubMed] [Google Scholar]
- O’Connor SE, Maresh JJ (2006) Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep 23:532–547. [DOI] [PubMed] [Google Scholar]
- Oudin A, Mahroug S, Courdavault V, Hervouet N, Zelwer C, Rodríguez-Concepción M, St-Pierre B, Burlat V (2007) Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol Biol 65:13–30. [DOI] [PubMed] [Google Scholar]
- Parage C, Foureau E, Kellner F, Burlat V, Mahroug S, Lanoue A, Dugé de Bernonville T, Londono MA, Carqueijeiro I, Oudin A, et al. (2016) Class II Cytochrome P450 reductase governs the biosynthesis of alkaloids. Plant Physiol 172:1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RM, Xu D, Foureau E, Teto Carqueijeiro MI, Oudin A, Bernonville TD, Novak V, Burow M, Olsen CE, Jones DM, et al. (2017) An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat Plants 13:16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils B, Schultz J (2004) Inactive enzyme-homologues find new function in regulatory processes. J Mol Biol 340:399–404. [DOI] [PubMed] [Google Scholar]
- Qu Y, Easson MEAM, Simionescu R, Hajicek J, Thamm AMK, Salim V, De Luca V (2018) Solution of the multistep pathway for assembly of corynanthean, strychnos, iboga, and aspidosperma monoterpenoid indole alkaloids from 19E-geissoschizine. Proc Natl Acad Sci U S A 115:3180–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA (2013) Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol 163:1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) AdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueffer M, Nagakura N, Zenk MH (1978) Strictosidine, the common precursor for monoterpenoid indole alkaloids with 3α and 3β configuration. Tetrahedron Lett 18:1593–1596. [Google Scholar]
- Schweizer F, Colinas M, Pollier J, Van Moerkercke A, Vanden Bossche R, de Clercq R, Goossens A (2018) An engineered combinatorial module of transcription factors boosts production of monoterpenoid indole alkaloids in Catharanthus roseus. Metab Eng 48:150–162. [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Besseau S, Clastre M, Courdavault V, Courtois M, Crèche J, Ducos E, Dugé de Bernonville T, Dutilleul C, Glévarec G, et al. (2013) Deciphering the evolution, cell biology and regulation of monoterpene indole alkaloids. Adv Bot Res 68:73–109. [Google Scholar]
- Staiger D, Brown JW (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25:3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides A, Tatsis EC, Caputi L, Foureau E, Stevenson CE, Lawson DM, Courdavault V, O’Connor SE (2016) Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity. Nat Commun 7:12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides A, Tatsis EC, Foureau E,, Caputi L, Kellner F, Courdavault V, O’Connor SE (2015) Unlocking the diversity of alkaloids in Catharanthus roseus: nuclear localization suggests metabolic channeling in secondary metabolism. Chem Biol 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LH, Blom TJ, Verpoorte R (1993) Subcellular localization of tryptophan decarboxylase, strictosidine synthase and strictosidine glucosidase in suspension cultured cells of Catharanthus roseus and Tabernaemontana divaricata. Plant Cell Rep 12:573–576. [DOI] [PubMed] [Google Scholar]
- Stoeckigt J, Hemscheidt T, Hoefle G, Heinstein P, Formacek V (1983) Steric course of hydrogen transfer during enzymatic formation of 3α-heteroyohimbine alkaloids. Biochemistry 22:3448–3452. [Google Scholar]
- Sue M, Yamazaki K, Yajima S, Nomura T, Matsukawa T, Iwamura H, Miyamoto T (2006) Molecular and structural characterization of hexameric β-D-glucosidases in wheat and rye. Plant Physiol 141:1237–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsis EC, Carqueijeiro I, Dugé de Bernonville T, Franke J, Dang TT, Oudin A, Lanoue A, Lafontaine F, Stavrinides AK, Clastre M, et al. (2017) A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat Commun 8:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Moerkercke A, Fabris M, Pollier J, Baart GJ, Rombauts S, Hasnain G, Rischer H, Memelink J, Oksman-Caldentey KM, Goossens A (2013) CathaCyc, a metabolic pathway database built from Catharanthus roseus RNA-Seq data. Plant Cell Physiol 54:673–685. [DOI] [PubMed] [Google Scholar]
- Verdoucq L, Moriniere J, Bevan DR, Esen A, Vasella A, Henrissat B, Czjze M (2004) Structural determinants of substrate specificity in family 1 β‐glucosidases: novel insights from the crystal structure of sorghum dhurrinase‐1, a plant β‐glucosidase with strict specificity, in complex with its natural substrate. J Biol Chem 279:31796–31803. [DOI] [PubMed] [Google Scholar]
- Yu HY, Kittur FS, Bevan DR, Esen A (2009) Determination of β-glucosidase aggregating factor (BGAF) binding and polymerization regions on the maize β-glucosidase isozyme Glu1. Phytochemistry 70:1355–1365. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Chung JD, Fu X, Johnson VE, Ranjan P, Booth SL, Harding SA, Tsai CJ (2009) Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. Proc Natl Acad Sci U S A 106:22020–22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Gassmann W (2003) RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 15:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Gassmann W (2007) Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 145:1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.