Abstract
Chemical signals known as strigolactones (SLs) were discovered more than 50 years ago as host-derived germination stimulants of parasitic plants in the Orobanchaceae. Strigolactone-responsive germination is an essential adaptation of obligate parasites in this family, which depend upon a host for survival. Several species of obligate parasites, including witchweeds (Striga, Alectra spp.) and broomrapes (Orobanche, Phelipanche spp.), are highly destructive agricultural weeds that pose a significant threat to global food security. Understanding how parasites sense SLs and other host-derived stimulants will catalyze the development of innovative chemical and biological control methods. This review synthesizes the recent discoveries of strigolactone receptors in parasitic Orobanchaceae, their signaling mechanism, and key steps in their evolution.
A family of receptors that evolved in the Orobanchaceae family enable seeds of parasitic plants to sense strigolactones from a nearby host root and germinate.
Advances
Strigolactone perception by parasite seed is mediated by a clade of neofunctionalized KAI2d proteins that evolved from a receptor that mediates karrikin responses in other plants.
KAI2d proteins use a similar mechanism to perceive SLs as D14, which mediates growth responses to SLs in nonparasites, but activate different signaling pathways.
Crystal structure analyses and chemical probes reveal features of KAI2d ligand-binding pockets that contribute to their specificity.
Strigolactones, multifaceted signals in plants and soil
The seed of many parasitic species in the Orobanchaceae can lie dormant for years or decades in soil until chemical signals from a nearby host root activate their germination. This remarkable adaptation is critical for obligate parasites such as witchweeds (Striga, Alectra spp.) and broomrapes (Orobanche, Phelipanche spp.), whose survival depends upon attaching to a host soon after germination. Witchweeds and broomrapes are major constraints to crop production and food security in sub-Saharan Africa, Asia, and the Mediterranean region (Xie et al., 2010). The need to develop chemical or biological solutions for this multibillion-dollar agricultural problem has driven a quest to understand how host-triggered germination occurs. Several questions are prominent: (1) What host-derived cues are detected by parasite seeds? (2) How are those cues perceived and translated into a germination response? and (3) How did this adaptation evolve and continue to evolve?
Answers to the first question began in 1966 with the discovery of strigol, a potent germination stimulant of Striga lutea that was isolated from cotton (Gossypium) root exudates (Cook et al., 1966). In the following decades, more than 20 other parasite germination stimulants with similar chemical structures to strigol have been identified from root exudates (Yoneyama et al., 2018b; Bouwmeester et al., 2020). These molecules compose the family of strigolactones (SLs). Key structural features of SLs and their major classifications are discussed in Box 1.
BOX 1.
The structure of strigolactones affects their activity in plants
Strigolactones are synthesized from β-carotene via an intermediate molecule, carlactonoic acid (Alder et al., 2012; Seto etal., 2014; Bouwmeester et al., 2020). Carlactonoic acid is converted into either canonical or noncanonical SLs, or both types, depending on the species (Yoneyama et al., 2018b). Canonical SLs are composed of a tricyclic ABC-ring system connected by an enol-ether bridge to a methylbutenolide D-ring. Based upon their stereochemical configuration at the B-C ring juncture, canonical SLs can be subdivided further into orobanchol-type and strigol-type molecules (Figure 1). By contrast, noncanonical SLs have diverse alternatives to the ABC-ring structure, while retaining an enol-ether-linked methylbutenolide D-ring. It is important to note that all known naturally occurring SLs have a 2′R-configured D-ring.
The structure and stereochemistry of SLs often influences their activity as parasite germination stimulants. For example, Striga gesnerioides selectively germinates in response to orobanchol-type SLs but its germination is inhibited by strigol-type SLs (Nomura et al., 2013). Many synthetic SL analogs have been developed during the search for inexpensive, stable compounds that can trigger suicidal germination of parasites in the absence of host. One such analog, GR24, is commonly used to study SL signaling and SL roles in plant development. However, GR24 is frequently used as a racemic mixture (rac-GR24) of a 2′R configured molecule known as GR245DS or (+)-GR24, and its unnaturally configured 2′S enantiomer, GR24ent-5DS or (-)-GR24 (Figure 1). These molecules activate two different pathways (Scaffidi et al., 2014; Flematti et al., 2016). In Arabidopsis thaliana, GR245DS signals through D14, while GR24ent-5DS signals predominantly through KAI2.
Nearly 40 years after the isolation of strigol, an explanation emerged for why plants exude SLs. SLs promote hyphal branching, metabolic activity, and hyphopodium formation of arbuscular mycorrhizal (AM) fungi, enhancing the ability of the fungi to colonize roots (Akiyama et al., 2005; Besserer et al., 2008; Gomez-Roldan et al., 2008; Kobae et al., 2018). Plants supply AM fungi with carbon in exchange for mineral nutrients. When inorganic phosphate or nitrogen availability is low, symbiosis with AM fungi is particularly beneficial and SL production increases (Yoneyama et al., 2007; López-Ráez et al., 2008; Umehara et al., 2008). A few years after the discovery that SLs affect growth of AM fungi, it was found that SLs are not just signals to the rhizosphere but are also plant hormones that regulate the outgrowth potential of axillary buds, or tillers (Gomez-Roldan et al., 2008; Umehara et al., 2008). Diverse roles for SLs in plant development have since been identified through studies of SL biosynthesis and signaling mutants and application of SL analogs. In addition to shoot branching, SLs regulate stem elongation, auxin transport, root elongation, leaf shape and angle, leaf senescence, secondary growth of the cambium, defense against pathogens and nematodes, stomatal closure, and drought tolerance (Gomez-Roldan et al., 2008; Umehara et al., 2008; Agusti et al., 2011; Kapulnik et al., 2011; Ruyter-Spira et al., 2011; Scaffidi et al., 2013; Shinohara et al., 2013; Bu et al., 2014; Van Ha et al., 2014; Yamada et al., 2014; Lauressergues et al., 2015; Soundappan et al., 2015; Ueda and Kusaba, 2015; Bennett et al., 2016; Lahari et al., 2019; Nasir et al., 2019; Kalliola et al., 2020; Li et al., 2020; Shindo et al., 2020). It may be that noncanonical SLs (see Box 1) function as plant hormones, whereas canonical SLs have external roles primarily (Yoneyama et al., 2018b). In tomato (Solanum lycopersicum) at least, loss of both SL types causes obvious developmental phenotypes, but loss of canonical SL production alone does not (Wakabayashi et al., 2019).
Strigolactone perception in nonparasitic angiosperms
To understand how SLs are recognized by the seed of root parasites, it is useful to first discuss how SLs are perceived as hormones by nonparasitic plants. SLs are recognized by DWARF14 (D14)/DECREASED APICAL DOMINANCE2 (DAD2)/RAMOSUS3 (RMS3), which is both an α/β-hydrolase protein and a receptor (Arite et al., 2009; Hamiaux et al., 2012; Waters et al., 2012; de Saint Germain et al., 2016; Yao et al., 2016). SL causes D14 to interact with MORE AXILLARY GROWTH (MAX2)/DWARF3 (D3; Hamiaux et al., 2012; Zhao et al., 2015; Yao et al., 2016). As an F-box protein, MAX2/D3 confers substrate specificity to an SCF (Skp1, Cullin, F-box) E3 ubiquitin ligase complex. SCF complexes attach polyubiquitin chains to target proteins, which are then rapidly degraded by the 26S proteasome. The targets of D14-SCFMAX2 are a subset of proteins in the SUPPRESSOR OF MAX2 1 (SMAX1)-LIKE (SMXL) family that are known as DWARF53 (D53) in rice (Oryza sativa) and petunia (Petunia hybrida), or SMXL6, SMXL7, and SMXL8 in Arabidopsis thaliana. SL activates association of D14 with these targets. Without D14, MAX2 is likely to have little or no interaction with SMXLs (Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015; Liang et al., 2016; Shabek et al., 2018; Lee et al., 2020).
The SL signaling mechanism is analogous to gibberellin, auxin, and jasmonate signaling mechanisms. In each of these pathways, hormone perception triggers SCF-mediated degradation of protein targets that indirectly regulate gene expression through association with transcription factors. The targets of auxin and jasmonate signaling also interact directly or indirectly with transcriptional corepressors in the TOPLESS/TOPLESS-RELATED (TPL/TPR) family. Thus, target degradation triggers downstream transcriptional responses (Blázquez et al., 2020). The prevailing hypothesis for SL signaling is that SMXL proteins function similarly as transcriptional co-repressors. SMXL proteins have a conserved C-terminal EAR motif that mediates interactions with TPL/TPR proteins (Jiang et al., 2013; Soundappan et al., 2015; Wang et al., 2015; Ma et al., 2017). In rice, D53 also interacts with the transcription factor IDEAL PLANT ARCHITECTURE1 (IPA1) to regulate expression of TEOSINTE BRANCHED1 (OsTB1), D53 itself, and other downstream genes (Lu et al., 2013; Song et al., 2017). A similar mechanism is found in wheat. TaD53 interacts with two homologs of IPA1, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE17 (SPL17) and SPL3, to repress TB1 expression (Liu et al., 2017). However, partnering with SPL transcription factors may be specific to monocots (Bennett et al., 2016; Wang et al., 2020a). Other transcription factors that presumably associate with SMXL proteins await discovery. Unexpectedly, the model of SMXL function has recently been extended to include SMXL proteins themselves as transcription factors (Wang et al., 2020a). Arabidopsis SMXL6 can bind DNA directly to regulate its own expression as well as that of SMXL7 and SMX8. In addition, SMXL6 works with unknown partners to regulate expression of BRANCHED1 (BRC1), TCP DOMAIN PROTEIN 1 (TCP1), and PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1), which encode key regulators of downstream growth responses to SL. Of the 401 SL-responsive genes that have been identified in Arabidopsis seedlings, 28 genes are directly bound by SMXL6 (Wang et al., 2020a). Putatively, other SMXL family proteins function as transcription factors too, adding an unusual twist to the typical model of F-box-mediated phytohormone signaling mechanisms.
Activation of the strigolactone receptor D14
D14 has a strictly conserved Ser-His-Asp catalytic triad that is a common feature of α/β-hydrolase proteins. D14 hydrolyzes SL slowly, and this activity requires an intact catalytic triad (Hamiaux et al., 2012; Seto et al., 2019). Although the byproducts of SL hydrolysis are not thought to be active, catalytic triad mutants demonstrate that hydrolysis is important for D14 signaling activity, with the notable exception of a D218A substitution (Hamiaux et al., 2012; Seto et al., 2019). During SL hydrolysis, the methylbutenolide D-ring is opened and cleaved through nucleophilic attack by the catalytic Ser, and ultimately transferred to the catalytic His residue (de Saint Germain et al., 2016; Yao et al., 2016). A crystal structure of Arabidopsis D14 (AtD14) in complex with rice D3 (OsD3) and Arabidopsis Skp1 (ASK1) led to the proposal that the opened D-ring bridges the Ser and His residues in the activated form of D14, forming a covalently linked intermediate molecule (CLIM; Yao et al., 2016). However, further analyses of the electron density at this site have challenged this interpretation. An iodide ion has been proposed to explain the electron density present in the pocket of activated D14, but a methylbutenolide-His complex seems to provide an even better fit (Carlsson et al., 2018; Bürger and Chory, 2020a). Covalent modification of the catalytic His residue by the D-ring is supported by tandem mass spectrometry analysis of D14 orthologs from Arabidopsis, pea (Pisum sativum), and rice (de Saint Germain et al., 2016; Yao et al., 2016; 2018b). Eventually, the D-ring can be released, enabling a new round of SL hydrolysis at least in vitro. However, D14 is degraded within a few hours after SL treatment in Arabidopsis and rice (Chevalier et al., 2014; Hu et al., 2017). If this is a common feature of angiosperms, D14 proteins that have a slow rate of D-ring release, such as RMS3 in pea, are likely to function as “one-shot” enzymes (de Saint Germain et al., 2016).
Pea and rice D14 proteins show biphasic SL hydrolysis activity in vitro consisting of a brief “burst” phase of rapid hydrolysis followed by a “plateau” phase of slow hydrolysis (de Saint Germain et al., 2016; Shabek et al., 2018). Putatively, the plateau phase is due to the rate at which the enzyme can discharge the SL byproducts (especially the D-ring) and reset. However, Arabidopsis D14 has not shown a biphasic hydrolysis response to the synthetic SL analog GR24 (Box 1; Seto et al., 2019). It is not clear whether this difference reflects the species origin of the D14 or the ligands used. These observations of D14 enzymatic activity are potentially complicated by the use of fluorogenic reporter molecules and SL analogs whose byproducts after hydrolysis may dissociate at different rates than those of natural SLs. The ratio of enzyme to ligand also affects the ability to observe the biphasic response (Shabek et al., 2018). Finally, the presence of D3 and D53 can influence the in vitro rates of SL hydrolysis by D14, potentially by stabilizing D14 in different conformations (Shabek et al., 2018). Notably, a major conformational change to D14 and a covalently attached SL byproduct have only been observed when D3 was co-crystallized (Yao et al., 2016). It is unknown how association of D14 with SCFMAX2 and SMXL proteins influences its reaction kinetics in vivo. It is also currently under debate which stage of the process of SL binding and hydrolysis by D14 activates signal transduction (de Saint Germain et al., 2016; Yao et al., 2016; Shabek et al., 2018; Seto et al., 2019). For an excellent critical discussion of the several models for D14 activation that have been proposed, see Bürger and Chory (2020a).
The mechanism of karrikin perception
Remarkably, the SL signaling pathway is an evolutionary innovation of a pathway that mediates responses to karrikins (KARs; see excellent review by Machin et al., 2020). KARs are a class of butenolide molecules found in smoke, biochar, and soil after a fire that act as plant growth regulators (Figure 1; Flematti et al., 2004; Kochanek et al., 2016; Hrdlička et al., 2019). The capacity to respond to KARs is likely to be widespread among angiosperms, and is not restricted to species from fire-prone ecosystems (Nelson et al., 2012).
Figure 1.
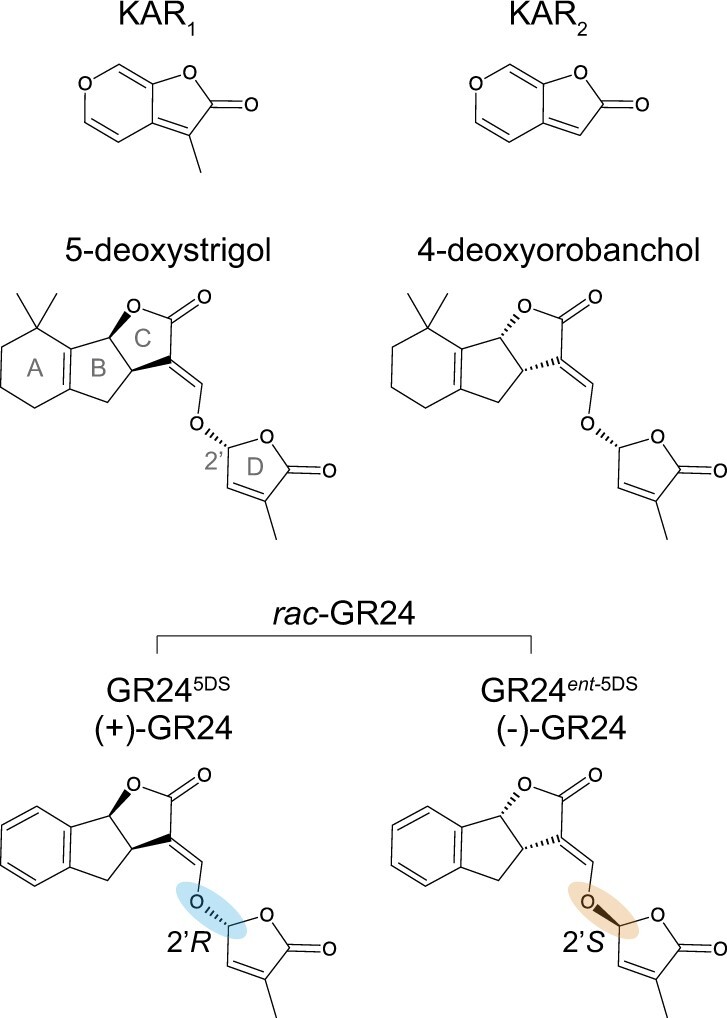
Chemical structures of representative karrikins, strigolactones, and GR24. KAR1 and KAR2 are naturally occurring karrikins in smoke. 5-deoxystrigol (5DS) and 4-deoxyorobanchol are representatives of the strigol-type and orobanchol-type strigolactone classes, respectively, which differ in the stereochemical configuration of the B–C ring junction. rac-GR24 is a racemic mixture of a synthetic analog of 5DS and its enantiomer. The D-ring of GR24ent-5DS has a 2′S configuration that has not been found in naturally occurring strigolactones.
KAR signaling is similar to SL signaling in several ways. KAR responses are mediated by SCFMAX2 and a D14 paralog known as KARRIKIN INSENSITIVE 2 (KAI2), HYPOSENSITIVE TO LIGHT (HTL), or D14-LIKE (D14L; Nelson et al., 2011; Sun and Ni, 2011; Waters et al., 2012). SMAX1 and SMXL2, which are paralogs of D53-type SMXL proteins, act downstream of MAX2 and KAI2 (Stanga et al., 2013; 2016). Upon activation, KAI2 interacts with SCFMAX2 and SMAX1 or SMXL2, triggering polyubiquitination and degradation of the SMXL proteins (Figure 2; Zheng et al., 2020; Khosla et al., 2020a; Wang et al., 2020b). This pathway regulates many processes in plants, including seed germination, hypocotyl or mesocotyl elongation, cotyledon expansion, seedling responses to light, leaf shape, cuticle development, drought tolerance, root skewing, root hair density and elongation, and the capacity for AM fungal symbiosis (Shen et al., 2007; Nelson et al., 2009; 2010; Sun and Ni, 2011; Stanga et al., 2013; Gutjahr et al., 2015; Soundappan et al., 2015; Stanga et al., 2016; Li et al., 2017; Swarbreck et al., 2019; Villaécija-Aguilar et al., 2019; Carbonnel et al., 2020a; Choi et al., 2020; Zheng et al., 2020).
Figure 2.
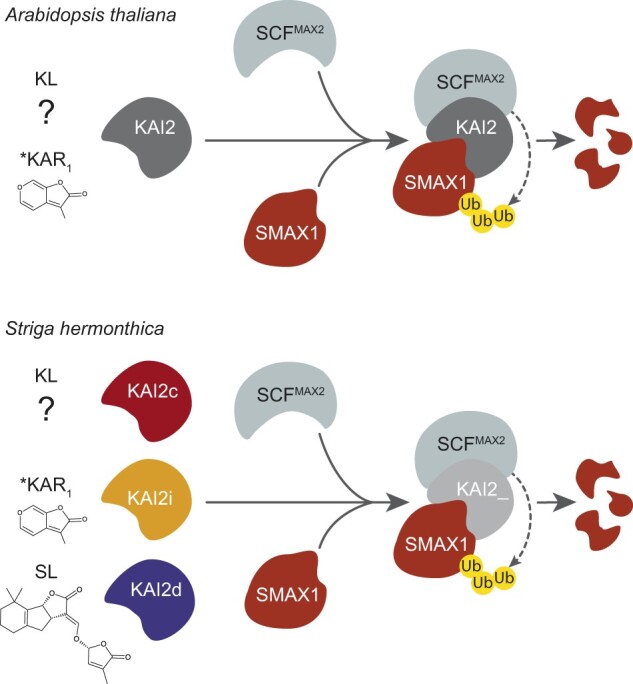
Representative models of KAI2-SCFMAX2 signaling in nonparasitic and parasitic plants. In Arabidopsis thaliana, KAI2 mediates responses to a putative KAI2 ligand (KL) and an unknown modified form of KAR1. In many parasitic Orobanchaceae such as Striga hermonthica, an increase in gene copies of KAI2 have led to diversified ligand preferences. Cross-species complementation assays suggest that KAI2c proteins preferentially mediate responses to KL, and KAI2i proteins can mediate responses to KARs. However, for unknown reasons obligate parasites that have a KAI2i gene, such as Striga hermonthica, do not germinate in response to KAR treatments. A diverse collection of KAI2d proteins function as strigolactone (SL) receptors that can regulate seed germination. Activation of a KAI2 protein induces its association with SCFMAX2 and a SMAX1-type SMXL protein (e.g. SMAX1 or SMXL2 in Arabidopsis thaliana). The SMXL protein is then polyubiquitinated by SCFMAX2 and degraded by the 26S proteasome, triggering downstream responses.
The details of KAI2 activation are less understood than for D14, partly because it remains unclear what ligand(s) KAI2 perceives. Genetic studies clearly show that KAI2 is necessary for KAR responses (Waters et al., 2012). There is also ample biochemical evidence that KAI2 from several species can bind KAR1 in vitro (Guo et al., 2013; Kagiyama et al., 2013; Toh et al., 2014; Xu et al., 2016; Lee et al., 2018; Bürger et al., 2019). These observations have supported the idea that KAI2 is a KAR receptor. However, other data strongly suggest that KAR1 requires metabolism by plants to become a ligand for KAI2. First, the affinity of KAI2 for KAR1 in vitro is typically one to two orders of magnitude lower than the biologically effective concentrations. Second, KAI2-KAR1 crystal structures from two species have not shown a consistent orientation of KAR1 in the ligand-binding pocket, so it is unclear which, if either, captures a true binding pose (Guo et al., 2013; Xu et al., 2016). Third, in multiple assays in vitro and in vivo, KAI2 is unresponsive to KARs but is activated by GR24ent-5DS, which has a stereochemical configuration not found in natural SLs (see Box 1; Figure 1; Flematti et al., 2016). Differential scanning fluorimetry (DSF) can detect shifts in the melting temperature (Tm) of a protein in response to a candidate ligand, and has emerged as a very useful tool for studying activation of D14 in vitro (Hamiaux et al., 2012; Abe et al., 2014; Hamiaux et al., 2018; Seto et al., 2019; Yasui et al., 2019). In DSF assays of AtKAI2, GR24ent-5DS but not KAR1, KAR2, or GR245DS triggers a Tm decrease (Waters et al., 2015; Yao et al., 2018a). Corresponding with this putative readout of KAI2 activation, yeast two-hybrid interactions between KAI2 and SMAX1 are stimulated by GR24ent-5DS but not by KAR1, KAR2, or GR245DS (Khosla et al., 2020a). KAI2 can pull down SMAX1 and SMXL2 expressed in protoplasts in the presence of GR24ent-5DS. However, KAI2 does not pull down SMXL2 in the presence of KAR1 (Wang et al., 2020b). Similar results are obtained for KAI2-MAX2 interactions. Yeast two-hybrid interactions between KAI2 and MAX2 are weakly enhanced in the presence of rac-GR24 (a racemic mixture of GR245DS and GR24ent-5DS), but not KAR1. In vitro pulldown interactions between KAI2 and MAX2 are stimulated by rac-GR24 but not KAR1 (Xu et al., 2018). Finally, KAR1 treatments require several-fold longer incubations than GR24ent-5DS to stimulate polyubiquitination and degradation of SMXL2 protein in Arabidopsis seedlings (Wang et al., 2020b). Therefore, KAR1 and GR24ent-5DS are not equivalent agonists of KAI2; while GR24ent-5DS is “ready-to-go,” KAR1 clearly is not. It is currently hypothesized that the normal function of KAI2 in plants is not transducing KAR signals, but sensing an unknown endogenous signal known as KAI2 ligand (KL; Nelson et al., 2011; Conn and Nelson, 2015; Waters et al., 2015; Sun et al., 2016). It may be that the hydrolyzable D-ring of GR24ent-5DS makes it a better substitute for KL than unmetabolized KARs.
Evolution of strigolactone perception in angiosperms
SL biosynthesis has an ancient origin in land plants. The complete set of SL biosynthetic pathway enzymes is found in all major land plant lineages, with the possible exception of hornworts. Physcomitrium (formerly Physcomitrella) patens and Marchantia polymorpha, which are used as models of basally diverged land plants, have lost one or more SL biosynthesis genes, but this is not representative of bryophytes (Walker et al., 2019). Sampling for SLs outside of angiosperms is still somewhat limited, but orobanchol-type SLs have been reported in the lycophyte Selaginella moellendorffii and gymnosperms (Yoneyama et al., 2018a; 2018b). The moss P. patens is reported to only produce carlactone (the biosynthetic precursor of carlactonoic acid and SLs), and not canonical SLs (Alder et al., 2012; Seto et al., 2014; Yoneyama et al., 2018b). However, an unknown SL-like signal(s) derived from carlactone seems likely. This signal inhibits protonemal growth of moss and can also stimulate germination of the parasitic plant Phelipanche ramosa (Proust et al., 2011; Lopez-Obando et al., 2020).
The emergence of SL signaling in land plants is less clear (Walker et al., 2019; Blázquez et al., 2020; Machin et al., 2020). In terms of MAX2-associated receptors, KAI2 orthologs are found in early diverging land plant lineages, such as Physcomitrium, Selaginella, and Marchantia. Phylogenetic analysis indicates that D14 arose from an early duplication of KAI2, but clear D14 orthologs are apparent only in seed-bearing plants (spermatophytes, i.e. gymnosperms and angiosperms; Bythell-Douglas et al., 2017). Similar to D14, the proteins targeted by SL signaling emerged during later phases of land plant evolution. SMXL genes are found throughout land plants. However, SMAX1 orthologs emerged in spermatophytes, and D53 orthologs (e.g. SMXL6, SMXL7, and SMXL8) evolved later, after the angiosperm lineage diverged (Walker et al., 2019). Divergence within the SMXL family may be associated with specialized functions in plant development and the co-evolution of receptor-target pairs (Waters et al., 2017; Blázquez et al., 2020). Notably, SMAX1-type and D53-type SMXL proteins typically have different roles. For example, in Arabidopsis SMAX1 and its paralog SMXL2 regulate germination and hypocotyl elongation, whereas SMXL6, SMXL7, and SMXL8 regulate axillary bud outgrowth (Soundappan et al., 2015; Stanga et al., 2016). However, some developmental processes, such as mesocotyl elongation in rice, are regulated by both types of SMXL proteins (Zheng et al., 2020).
Although the genetic components of canonical SL signaling are angiosperm-specific, it has seemed plausible that an analogous SCFMAX2-dependent signaling mechanism mediates SL responses in other land plant clades. However, recent studies provide support for a KAI2-SCFMAX2 signaling system that mediates responses to KL but not necessarily SL in Marchantia polymorpha and Physcomitrium patens. Marchantia polymorpha has a relatively simple set of genes for this pathway, with only two KAI2, one MAX2, and one SMXL. Mpkai2a and Mpmax2 mutants have highly similar phenotypes that are suppressed by loss of MpSMXL, as expected from the pathway found in angiosperms, whereas the Mpkai2b mutant has no obvious phenotype (Mizuno et al., 2020). High concentrations of rac-GR24 affect thalli growth, but in a MpMAX2-, MpKAI2a-, and MpKAI2b-independent manner. Only a 2′S-configured GR24 enantiomer, GR24ent-4DO, induces thermal destabilization of the MpKAI2 proteins in vitro, supporting that they do not respond to naturally configured SLs (Mizuno et al., 2020). The signaling system is more complex in Physcomitrium patens, which has 13 KAI2-LIKE (PpKAI2L) genes. It was initially proposed that some of the less-conserved PpKAI2L proteins might function as SL receptors in Physcomitrium patens, which makes a carotenoid-derived transmissible signal and responds to rac-GR24 and GR245DS (Proust et al., 2011; Hoffmann et al., 2014; Lopez-Obando et al., 2016; 2020). However, a biochemical analysis of several PpKAI2L proteins only showed evidence of binding KAR1 or unnaturally configured 2′S SLs (Bürger et al., 2019). Moreover, the Ppmax2 mutant is phenotypically different from a carlactone-deficient mutant (Lopez-Obando et al., 2018). An extensive genetic analysis of the PpKAI2L family now shows that the five members of the PpKAI2L(A-E) clade, which are grouped among the eu-KAI2 proteins that are conserved in all land plants, are likely to control growth in coordination with PpMAX2. On the other hand, proteins in the PpKAI2L(JGM) clade are proposed to mediate responses to carlactone-derived signals in a PpMAX2-independent manner (Lopez-Obando et al., 2020). Studies of additional species outside the angiosperms will be required to determine whether these results are representative.
Distinct germination responses to KARs and SLs in autotrophs and parasites
There are clear differences in the germination responses of Arabidopsis thaliana and parasitic Orobanchaceae seed to KAR and SL. Arabidopsis seed germinates in response to both treatments, but rac-GR24 is clearly less potent than KARs (Nelson et al., 2009). By contrast, the obligate parasites Orobanche cernua, O. crenata, O. cumana, O. minor, Phelipanche aegyptiaca, P. ramosa, or Striga hermonthica respond well to rac-GR24 but are altogether insensitive to KARs (Fernández-Aparicio et al., 2009; Nelson et al., 2009; Scaffidi et al., 2014; Conn et al., 2015; Brun et al., 2019). If we consider KARs as indicators of low competition (due to fire) and SLs as indicators of nearby plants, these selective germination responses seem well-suited for autotrophic and auxotrophic plants, respectively. Germination of obligate parasites in response to KARs that appear after fire would likely be suicidal, for example.
Several lines of evidence indicate that SLs do not regulate Arabidopsis germination. Primary dormant seed of the SL-deficient mutants max1, max3, and max4 and the SL-insensitive mutant d14 have normal germination. In contrast, kai2 and max2 mutants have clearly enhanced dormancy (Nelson et al., 2011; Waters et al., 2012). Similar germination trends are observed among max mutants under light-restricted conditions (Shen et al., 2012). Therefore, Arabidopsis germination is controlled by KAI2-SCFMAX2-dependent signaling, but not by SL signaling through D14. Indeed, the 2′S configured molecules GR24ent-5DS and ent-5-deoxystrigol (ent-5DS, an enantiomer of the canonical SL 5-deoxystrigol) promote Arabidopsis germination, but other stereoisomers of GR24 and natural SLs do not (Scaffidi et al., 2014). These responses correspond to the stereoselective activation of KAI2 in vitro and in vivo (Scaffidi et al., 2014; Waters et al., 2015; Khosla et al., 2020a; Wang et al., 2020b).
It might be argued, however, that the exclusive control of germination by KAI2 is only a feature of primary dormant seed. For the commonly used Arabidopsis ecotype Col-0, primary dormancy typically is lost within several days of afterripening and varies between seed batches, making this a difficult trait to study. Imbibition of after-ripened Arabidopsis seed at supraoptimal temperatures imposes thermoinhibition, a form of secondary dormancy (Toh et al., 2012). This approach has become a powerful tool for evaluating MAX2-dependent germination responses in Arabidopsis (Toh et al., 2015; Uraguchi et al., 2018; de Saint Germain et al., 2020). SL biosynthesis mutants have been reported to be hypersensitive to seed thermoinhibition, suggesting that SLs contribute to germination under these conditions (Tsuchiya et al., 2010; Toh et al., 2012). However, an Arabidopsis kai2/htl line that has acquired extraordinary, picomolar sensitivity to applied SLs through introduction of ShHTL7, a SL receptor transgene from Striga hermonthica (see below), still does not germinate under thermoinhibited conditions without a SL treatment (Toh et al., 2015). If ShHTL7 can sense the noncanonical SLs produced by Arabidopsis (Abe et al., 2014; Seto et al., 2014; Brewer et al., 2016; Yoneyama et al., 2018a; 2018b), this suggests that there is little or no SL in Arabidopsis seed, even under thermoinhibition conditions. It is not yet clear whether a lack of germination responses to SLs is peculiar to Arabidopsis or a common feature of nonparasitic angiosperms. However, it seems that autotrophic plants would be at a competitive disadvantage when germinating in response to SLs, which are exuded from established plants already starved for nutrients.
Identification of novel SL receptors in parasitic Orobanchaceae
The discoveries that D14 was likely a SL receptor and its homolog KAI2 regulates seed germination set the stage for understanding SL perception in parasitic Orobanchaceae. It was hypothesized that in parasites a MAX2-dependent mechanism had been co-opted for SL-responsive germination (Nelson, 2013). This could occur if D14 became a germination regulator, or if KAI2 evolved the ability to recognize SLs. Supporting the first idea, Arabidopsis D14 can crosstalk with SMXL2 when an analog of 4-deoxyorobanchol, GR244DO, is supplied (Wang et al., 2020b). Putatively D14 can also crosstalk with SMAX1, which has a bigger role than SMXL2 in hypocotyl elongation, as kai2 seedlings respond to rac-GR24 treatment but kai2 d14 seedlings do not (Waters et al., 2012; Scaffidi et al., 2014; Stanga et al., 2016). Because the d14 mutant does not show phenotypes consistent with SMAX1 or SMXL2 overaccumulation, however, it is unlikely that D14 normally regulates these proteins. Furthermore, expression of D14 under the control of a KAI2 promoter does not rescue germination of kai2, even with rac-GR24 treatment (Conn et al., 2015; Waters et al., 2015). Therefore, to regulate germination, D14 might require changes that enhance its expression in seed as well as its affinity for SMAX1-type SMXL proteins. By contrast, evolution of SL perception in a KAI2 protein would require a switch in stereochemical selectivity to accommodate 2′R configured SL molecules as ligands.
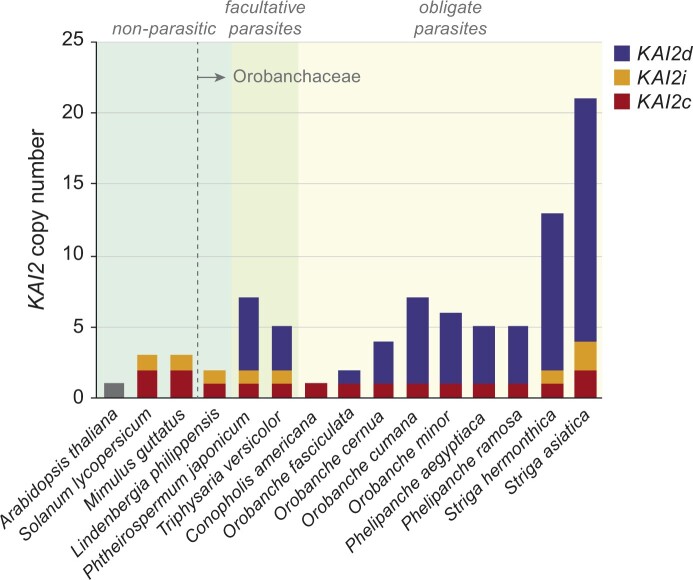
An investigation of D14 and KAI2 genes in parasitic plant genomes and transcriptomes was carried out, thanks largely to resources developed by the Parasitic Plant Genome Project and 1000 Plants Initiative (Westwood et al., 2012; Conn et al., 2015; One Thousand Plant Transcriptomes Initiative, 2019). This revealed an unexpected, and sometimes dramatic, expansion of KAI2 copy number in several parasites (Conn et al., 2015). Although D14 appears to be a single copy gene in nonparasitic and parasitic species in the Lamiids, KAI2 has undergone extensive duplication in many facultative and obligate parasite genomes (Figure 3). The Striga asiatica genome has 21 KAI2 paralogs, and Striga hermonthica is likely to have at least 13 (Conn et al., 2015; Toh et al., 2015; Tsuchiya et al., 2015; Yoshida et al., 2019). Lamiid KAI2 genes are distributed into three phylogenetic groups undergoing different rates of evolution. Both parasitic and nonparasitic Lamiids have one or two KAI2 copies that are grouped within a “conserved” clade (KAI2c). Many nonparasitic Lamiids and Striga spp. have one or two copies of KAI2 that are grouped within a “intermediate” grade (KAI2i) under weaker purifying selection than KAI2c. Parasitic Orobanchaceae uniquely carry a third type of rapidly evolving, “divergent” KAI2 (KAI2d) that often comprise the majority of KAI2 paralogs in a species (Figure 3; Conn et al., 2015). Because gene duplication can enable the evolution of different gene functions, KAI2d proteins were attractive candidates for SL receptors.
Figure 3.
Diversification of KAI2 in parasitic Orobanchaceae. Bar plot of the number of KAI2 genes and their types detected in Orobanchaceae species and nonparasitic dicots. Obligate parasites require a host to complete their life cycle. Facultative hemiparasites retain photosynthetic capacity and do not require a host for survival. Lindenberghia philippensis is a basally diverged nonparasite in the Orobanchaceae. Gene copy estimates are from Conn et al.(2015),Yoshida et al.(2019), and de Saint Germain et al., 2020.
Indeed, biochemical studies have provided compelling evidence that KAI2d genes encode SL receptors. A fluorogenic agonist, Yoshimulactone Green (YLG), was developed to monitor the hydrolytic activity of SL receptors (Tsuchiya et al., 2015). This enabled in vitro YLG competition assays, which test the inhibitory effects of a candidate ligand on the rate of YLG hydrolysis by a SL receptor. The half-maximal inhibitory concentration (IC50) value provides an indirect assessment of the affinity a receptor has for a given SL. Of the two KAI2i- and eight KAI2d-class HTL proteins tested from Striga hermonthica, ShHTL6 (KAI2d-class) and ShHTL7 (KAI2d-class) have low IC50 (< 500 nM) for several SLs. This likely indicates high affinity for SLs, as ShHTL7 has similar Km and IC50 values for strigol of 57 nM and 120 nM, respectively. Other KAI2d-class ShHTL proteins show more selective ligand preferences among five SLs tested in YLG competition assays (Tsuchiya et al., 2015). Isothermal calorimetry assays provide further support that KAI2d proteins have high affinities for SL. Several KAI2d-class ShHTL proteins bind 5-deoxystrigol with Kd values in the ∼40 nM to 4 µM range (Wang et al., 2021).
Further evidence that KAI2d proteins are SL receptors comes from cross-species complementation experiments, which provide a convenient way to evaluate the function of an individual KAI2 gene in vivo. Arabidopsis thaliana is well-suited for this purpose due to the availability of KAR and SL pathway mutants, easy transformation, and a 2-month generation time. In these assays, Arabidopsis kai2 mutants are transformed with KAI2 transgenes from parasites and tested for responses to SLs and other germination stimulants. Although some transgenes are not functional—which could reflect a loss of function in their native context, or incompatible interactions with signaling partners in Arabidopsis—many KAI2d transgenes from Striga hermonthica, Phelipanche aegyptiaca, and Phelipanche ramosa confer clear germination responses to SLs, rac-GR24, and/or 2′R configured GR24 stereoisomers (Conn et al., 2015; Toh et al., 2015; Khosla and Nelson, 2016; de Saint Germain et al., 2020). Transgenic Arabidopsis lines carrying ShHTL7 are striking examples, with germination responses to picomolar concentrations of several SLs and rac-GR24 (Toh et al., 2015). Similarly, a fusion protein of GFP and Phelipanche ramosa KAI2d3 confers germination responses to ∼10 picomolar (+)-GR24 to the Arabidopsis kai2 mutant (de Saint Germain et al., 2020). PrKAI2d3 shows a clear preference for 2′R configured SL analogs, and is several orders of magnitude less responsive to 2′S configured analogs.
The ability of a parasite D14 gene to regulate Arabidopsis seed germination has not been reported. Therefore, while there are no obvious evolutionary signatures, such as gene duplication or evidence of positive selection, to suggest that D14 may have gained new functions in parasites, it is still formally possible that it could contribute to SL-responsive germination. Although loss-of-function mutations are not available to test the roles of KAI2 and D14 genes in parasites directly, chemical tools have provided a way forward. Sphynolactone-7 (SPL7, not to be confused with SPL transcription factors) was developed as a synthetic agonist of Striga hermonthica germination (Uraguchi et al., 2018). The potency of SPL7 is very similar to that of the natural SL, 5-deoxystrigol (5DS); both can induce germination effectively at concentrations of ∼100 fM. SPL7 is different from 5DS, however, because it shows a high degree of selectivity for ShHTL7 and, to a lesser extent, ShHTL8. In vitro YLG competition assays show that SPL7 has an IC50 of 0.31 µM for ShHTL7, 1.2 µM for ShHTL8, and 7.8 µM for ShHTL11. By contrast, SPL7 has an IC50>10 µM for D14, the KAI2i-class proteins ShHTL2 and 3, and the KAI2d-class proteins ShHTL4, ShHTL5, ShHTL6, ShHTL9, and ShHTL10 in vitro (Uraguchi et al., 2018). Although not all ShHTL proteins have been tested and their relative abundance in Striga seed is unknown, this suggests that ShHTL7 is primarily responsible for detecting SPL7. If so, then activation of ShHTL7 is apparently sufficient to trigger Striga hermonthica germination. By contrast, ShD14 is not required, at least for responses to SPL7.
Further support for the importance of ShHTL7 in SL-responsive germination is potentially provided by the nonionic surfactant Triton X-100, which reduces germination responses of Striga hermonthica to rac-GR24 (Shahul Hameed et al., 2018). Triton X-100 binds to ShHTL7 in vitro, blocking rac-GR24-induced structural rearrangements of ShHTL7 and its interaction with ShMAX2. Homology modeling suggests that ShHTL7, but not other ShHTL proteins, can accommodate Triton X-100 in their active sites (Shahul Hameed et al., 2018). Therefore, if the inhibition of Striga hermonthica germination by Triton X-100 is due to interfering with SL perception and not a nonspecific effect, these data indicate that ShHTL7 is critical for GR24 responses. This does not exclude the possibility that other ShHTL proteins contribute to germination responses to 5DS or other SLs. If selective inhibitors can be developed for other ShHTL proteins, their individual contributions to host-induced germination could be evaluated.
Where strigolactone perception occurs
Based on a detailed anatomical and physiological analysis, perisperm cells adjacent to the micropyle have been proposed to be the site of host-chemical detection in Phelipanche aegyptiaca (Joel et al., 2012). In support of this, transcripts of CYP707A1, an ABA 8′-hydroxylase that acts downstream of SL signaling, accumulate in these perisperm cells specifically after several hours of GR24 treatment (Lechat et al., 2012).
A fluorogenic chemical probe has also been used to examine the sites and timing of SL perception in Striga hermonthica (Tsuchiya et al., 2015). YLGW is a brighter, but less specific, form of YLG. It is hydrolyzed in vitro by several ShHTL proteins but not by ShD14, ShHTL7, and a few other ShHTL proteins. After seed coat removal, fluorescence microscopy of Striga hermonthica embryos reveals two waves of YLGW hydrolysis that putatively report the activity of SL receptors. Fluorescence is observed in the embryonic root tip within minutes, then diffuses apically during a “wake-up” phase that occurs over several hours. After a pause during which the fluorescence signal dissipates, germination begins. As the root grows, an “elongation tide” of fluorescence emerges in what appears to be the elongation and differentiation zones. These fluorescence patterns were not observed in nonconditioned seeds, which do not respond to SL (Tsuchiya et al., 2015). Because ShHTL7 and ShD14 do not hydrolyze YLGW, this implies that neither is required for the wake-up phase of SL perception. If YLGW hydrolysis is ShHTL-dependent, these results imply the enzymatic activity of other ShHTL proteins.
The mechanism of strigolactone perception in parasitic seed
KAI2d proteins in parasites perceive SL in a highly similar manner to D14 (Figure 2). ShHTL7 and ShHTL4 hydrolyze rac-GR24 and 5DS, whereas ShHTL1 (KAI2c-class) and ShHTL3 (KAI2i-class) have little or no hydrolytic activity on these substrates (Xu et al., 2018). As a consequence of SL hydrolysis, the methylbutenolide D-ring becomes attached to the catalytic His residue of KAI2d proteins, as was observed for D14. This was shown through mass spectrometry of ShHTL7 after 5DS and rac-GR24 hydrolysis, and PrKAI2d3 in the presence of rac-GR24 (Yao et al., 2017; Uraguchi et al., 2018; de Saint Germain et al., 2020). Similar covalent modification of the catalytic His residue in ShHTL7 occurs after treatment with a broad range of SPL7 analogs (Uraguchi et al., 2018). Interestingly, PrKAI2d3 also confers weak responses to the hydroxymethylbutenolide product of SL hydrolysis (D-OH), which is ineffective at activating SL signaling (Hamiaux et al., 2012; de Saint Germain et al., 2020). However, because D-OH is ∼10,000 times less potent than rac-GR24 at activating Striga hermonthica germination, this may not be a biologically significant reaction.
KAI2d proteins very likely function in cooperation with MAX2, as is the case for KAI2 in nonparasitic species. ShMAX2 is able to rescue many, but not all, mutant phenotypes of Arabidopsis max2, demonstrating at least partially conserved functions (Liu et al., 2014). Likewise, the ability of ShHTL transgenes to stimulate seed germination or regulate hypocotyl elongation of Arabidopsis thaliana is MAX2-dependent (Bunsick et al., 2020). Homology modeling of the ShHTL7-ShMAX2 complex shows an interface that is well-conserved with that of AtD14-MAX2 (Shahul Hameed et al., 2018). Many of the amino acids at this interface are also highly conserved in the broader eu-KAI2 protein family in land plants (Bythell-Douglas et al., 2017). Finally, ShHTL4, ShHTL5, ShHTL7, ShHTL8, and ShHTL9 physically interact with ShMAX2 in the presence of rac-GR24 and natural SLs, similar to ShD14 (Yao et al., 2017; Shahul Hameed et al., 2018; Xu et al., 2018; Wang et al., 2021).
Interactions between KAI2 and SMAX1 proteins in parasites have not been tested, however, there is indirect evidence that this is likely to occur. First, KAI2d transgenes from parasites are able to regulate Arabidopsis germination and seedling growth, which are exclusively or predominantly controlled by SMAX1 among the SMXL family members in Arabidopsis (Stanga et al., 2016). Second, GR24 enhances pull-down interactions between ShHTL7 and Arabidopsis SMAX1, but not Arabidopsis SMXL6 (Yao et al., 2017). This is consistent with the preference that AtKAI2 shows for SMXL partners, and suggests that only the ligand-binding capacity of KAI2d proteins has changed in parasites (Soundappan et al., 2015; Khosla et al., 2020a). Third, ShHTL5, ShHTL7, ShHTL8, and ShHTL9 proteins interact with Arabidopsis SMAX1 in yeast two-hybrid assays in the presence of rac-GR24 or natural SLs. When AtMAX2 is coexpressed, ShHTL interactions with SMAX1 are strengthened. Likewise, the C-terminal “D2” domain of Arabidopsis SMAX1 enhances in vitro interactions between ShHTL7 and AtMAX2 (Wang et al., 2021).
These observations collectively indicate that KAI2d proteins in parasites have retained the function of KAI2 proteins in stimulating degradation of SMAX1 via SCFMAX2, but are activated by SLs instead of KL or KAR. Thus, D14 and KAI2d have convergently evolved into SL receptors from duplicated KAI2.
Downstream effects of KAI2 activation
Although activation of KAI2d proteins can reasonably be expected to cause SMAX1 degradation in parasites, how this leads to germination has been less clear. Recent studies have made progress toward understanding crosstalk between SMAX1 degradation and other hormone pathways that regulate seed germination. Two major players in control of physiological seed dormancy are abscisic acid (ABA) and gibberellic acid (GA), which have antagonistic effects as inhibitors and promoters of germination, respectively. Ethylene also promotes germination of many species, including some parasites, at least in part through inhibition of ABA levels and signaling (Arc et al., 2013). Crosstalk between these hormones is complex.
ABA degradation is an important component of SL-induced germination of Phelipanche ramosa seed. Application of rac-GR24 quickly and transiently induces expression of CYP707A1, which encodes an ABA-catabolizing enzyme. Corresponding with this, ABA levels decline several-fold in conditioned seeds after GR24 treatment. Inhibition of ABA catabolism with the CYP707A inhibitor abscinazole-E2B reversibly blocks GR24-induced germination (Lechat et al., 2012). A similar induction of CYP707A1 occurs in the obligate parasites Orobanche cumana, O. minor, and Striga hermonthica upon treatment with rac-GR24. In contrast, the facultative hemiparasite Triphysaria versicolor does not require host-derived stimulants and has only a modest germination response to rac-GR24. Induction of CYP707A1 in T. versicolor seed by rac-GR24 is similarly low. Thus, SL-induced catabolism of ABA may be more important for germination of obligate parasites than facultative hemiparasites. Indeed, the enhanced seed dormancy of obligate parasites relative to T. versicolor may be due to their increased sensitivity to ABA (Brun et al., 2019).
Initial physiological studies of KAR responses in Arabidopsis showed that KAR1 could not recover germination of GA-deficient mutants. KAR1 also induces expression of GA biosynthesis genes GA3ox1 and GA3ox2 in seed, while not affecting GA sensitivity, suggesting that GA is required for KARs to stimulate Arabidopsis germination (Nelson et al., 2009). However, when activated with rac-GR24, several KAI2d-class ShHTL proteins expressed in Arabidopsis are able to override the GA requirement (Bunsick et al., 2020). A similar effect is achieved through a smax1 loss-of-function mutation, which can overcome germination inhibition by the GA biosynthesis inhibitor paclobutrazol. The differences between these studies might be due to the degree to which SMAX1 is removed from Arabidopsis seeds; i.e. KAR1 activation of AtKAI2 may not be as effective at reducing SMAX1 protein levels as rac-GR24 activation of ShHTL or a smax1 mutation. Regardless, experiments with Striga hermonthica support the idea that SL signaling can activate germination independently of GA in parasitic plants. GA had little effect on promoting Striga hermonthica germination, while GR24 stimulated Striga germination even in the presence of paclobutrazol (Bunsick et al., 2020).
It is possible that KAI2d proteins stimulate germination in parasites by inducing production of ethylene. In contrast to paclobutrazol, the ethylene biosynthesis inhibitor aminoethoxyvinylglycine is effective at blocking GR24-stimulated germination of Striga hermonthica (Tsuchiya et al., 2015). In Arabidopsis, SL signaling through D14 triggers dark-induced leaf senescence in a feed-forward loop with ethylene signaling (Ueda and Kusaba, 2015). This invites speculation that KAI2-regulated SMXL proteins might also control growth, at least partially, through ethylene. Notably, under light-grown conditions ethylene can overcome a GA deficiency to stimulate Arabidopsis germination (Karssen et al., 1989). Furthermore, regulation of root and root hair development in Lotus japonicus and Arabidopsis by KAR/KL is due to upregulation of ethylene biosynthesis after degradation of SMAX1 (Carbonnel et al., 2020a).
How KAI2 diversification in parasites may affect host range
During the antagonistic coevolution of parasitic Orobanchaceae and their host plants, an increased capacity for SL perception is likely to have been valuable for a parasite. This could enable the parasite to expand its host range, counterbalance evolution of alternative SLs in hosts, or prevent an evolutionary dead-end when a host species becomes extinct locally. For obligate parasites of crops (i.e. weeds), whose potential hosts may change dramatically from year to year, a broadened SL response could be a key adaptation. For example, this likely enabled the recent expansion in host range for a new race of Orobanche cumana (Dor et al., 2020). Expansion of the KAI2d gene family in parasitic Orobanchaceae potentially enables detection of a greater variety of SLs. Indeed, although the dataset is limited, there seems to be a trend of more KAI2d paralogs in the genomes of weedy parasites, which can attack many crop species, compared to species with specialized parasitic relationships (Figure 3; Conn et al., 2015). At the other end of the spectrum, one KAI2c gene but no KAI2d gene was able to be identified in a de novo transcriptome assembly for Conopholis americana, which parasitizes oak and beech trees (Figure 3; Conn et al., 2015). Perhaps this parasite does not require a mechanism for its seed to detect the presence of a host because its hosts are so long-lived.
Because so much emphasis has been placed on studying parasitic weeds, however, it is important to remember that many parasitic Orobanchaceae are not generalists, but instead have a narrow host range. From the point of view of an obligate parasite, an inherent danger of SL-responsive germination is that not all SL-exuding plants are compatible hosts. Plants mount an array of barriers against parasitism, ranging from physical fortifications to immune responses (Clarke et al., 2019). Specialization for different hosts can be observed even among races of a parasitic species, and likely drives speciation (Thorogood et al., 2009). As germination responses are one component of host-specialization, it is not surprising that obligate parasites can show strong germination response preferences for root exudates from specific species (Fernández-Aparicio et al., 2009; 2011b). Presumably, this is due to the presence of specific SLs in some exudates but not others, as obligate parasites show specific germination responses to different SLs (Fernández-Aparicio et al., 2011b; Nomura et al., 2013).
Putatively, some KAI2d proteins have evolved the ability to detect or prefer specific SL(s). Alternatively, some KAI2d proteins may have become highly sensitive to a broad range of ligands. Both cases have been observed among ShHTL proteins through YLG competition assays and cross-species complementation experiments (Toh et al., 2015; Tsuchiya et al., 2015). It should be noted, however, that the SL preferences of KAI2d proteins in vitro do not always reflect which SLs are effective stimulants of seed germination. For example, ShHTL6 and ShHTL10 have a very low IC50 for orobanchol (58 nM and 390 nM, respectively), yet Striga hermonthica seed are relatively unresponsive to orobanchol (Tsuchiya et al., 2015). Thus, the emergent germination response to host-derived stimuli may depend upon the relative abundance and activities of the suite of available KAI2d proteins. Although parasite KAI2d proteins have so far been considered activators of germination, I hypothesize that the function of some KAI2d proteins could be to inhibit germination after perception of a specific non-host SL (see Box 2). A combination of positive and negative responses to different SLs could produce finely tuned host-specific germination responses.
BOX 2.
How do host-specific germination responses evolve?
Even parasitic weeds with broad host ranges can show selective germination responses to SLs. For example, Striga hermonthica seed are much more sensitive to strigol-type SLs than to orobanchol-type SLs (Tsuchiya et al., 2015). By contrast, germination of Striga gesnerioides is inhibited by strigol-type SLs and promoted by orobanchol-type SLs (Nomura et al., 2013). This raises the question of how stereoselective germination responses evolved in these species, which have many KAI2d paralogs. Gaining a response to a specific SL could be a straightforward process in which a KAI2d paralog acquires a mutation(s) that alters its ligand-specificity. Gaining inhibited germination responses or insensitivity to a SL may be more difficult to achieve, particularly if multiple KAI2d proteins can perceive it. Nonetheless, there is a selective advantage for this to occur when a SL from a nonhost plant triggers suicidal germination. Insensitivity to the disadvantageous SL could emerge through the gradual acquisition of mutations that deactivate the expression or function of all KAI2d that perceive that SL in seed. Alternatively, the same effect could be rapidly achieved through an antimorphic (dominant-negative) mutation of a KAI2d protein that recognizes the disadvantageous SL. Such a mutation would prevent the affected KAI2d protein from interacting with either MAX2 or SMAX1, but not both proteins. This could enable competitive sequestration of SMAX1 or MAX2 from other KAI2d proteins, putatively protecting SMAX1 from degradation and inhibiting germination (Figure 4). Such mutations are plausible, as single amino acid substitutions that affect D14 interactions with MAX2 or D53-type SMXL proteins have been identified (Zhao et al., 2015; Yao et al., 2016; Seto et al., 2019; Lee et al., 2020).
This hypothesis awaits investigation. Meanwhile, it is notable that ShHTL10 and ShHTL11 have high affinities for SLs in vitro but are inactive when expressed in Arabidopsis (Toh et al., 2015; Tsuchiya et al., 2015; Wang et al., 2021). ShHTL10 and ShHTL11 are grouped in a Striga-specific KAI2d subclade with ShKAI2d2, which is also inactive in Arabidopsis (Conn et al., 2015). These three proteins have substitutions at several well-conserved surface residues that may affect interactions with MAX2 or SMAX1 (Khosla and Nelson, 2016; Wang et al., 2021). Indeed, ShHTL10 and ShHTL11 do not interact with AtMAX2 or ShMAX2 in yeast two-hybrid assays in the presence of SLs (Wang et al., 2021). The activity of these proteins might explain the reduced germination responses of Striga hermonthica to orobanchol-type SLs.
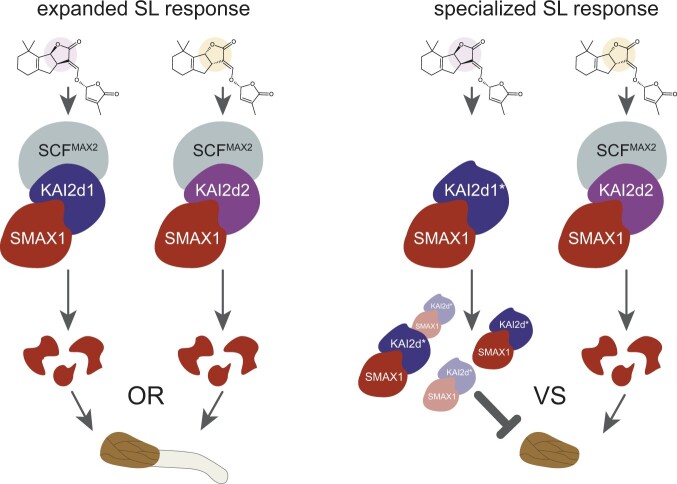
Figure 4 A.
hypothesis for selective germination responses to strigolactones. In this model, KAI2d1 and KAI2d2 represent two paralogous receptors that respectively prefer either strigol-type strigolactone (SL; B–C ring configuration highlighted in purple) or orobanchol-type SL (B–C ring configuration highlighted in orange) as ligands. For some parasites, expansion of the KAI2d family may enable responses to a broader range of SLs (left). Activation of either the KAI2d1 or KAI2d2 receptor by the presence of strigol-type SL or orobanchol-type SL may cause sufficient SMAX1 degradation to activate germination. This is equivalent to an OR logic gate. However, the seed of some parasites, such as Striga gesnerioides, respond positively to some SLs but are inhibited by other SLs. This specificity could fine-tune germination responses to exudates from compatible hosts. In the specialized SL response hypothesis (right), a dominant-negative mutation (*) causes a KAI2d protein to lose interactions with either MAX2 or SMAX1, but not both, proteins. In this example, the mutant KAI2d1* protein can interact with SMAX1 but not MAX2 upon activation. This sequesters SMAX1 and prevents it from being targeted for degradation by other KAI2d-SCFMAX2 complexes. Competition between KAI2d1* and fully functional KAI2d proteins that can trigger SMAX1 degradation affects the overall abundance of SMAX1 protein. If SMAX1 levels remain high, germination is blocked. This competition is denoted by VERSUS (VS) and is affected by the presence of SLs that activate KAI2d1* or KAI2d2.
Several noncanonical SLs and lactone molecules that can stimulate germination of parasitic Orobanchaceae have been identified (reviewed in Yoneyama et al., 2018b; Bouwmeester et al., 2020). These include zealactone, avenaol, heliolactone, dehydrocostus lactone, and peagoldione (Evidente et al., 2009; Joel et al., 2011; Kim et al., 2014; Ueno et al., 2014; Charnikhova et al., 2017). It is plausible that these molecules are also perceived by KAI2d proteins, but their specific receptors await discovery. Notably, dehydrocostus lactone induces CYP707A1 expression in seed of the sunflower (Helianthus annuus) parasite Orobanche cumana, similar to rac-GR24, implying it is perceived through a similar system (Brun et al., 2019). Like KARs, dehydrocostus lactone lacks a cleavable D-ring; it may be that it also requires metabolism before perception by Orobanche cumana.
KAI2d agonists are not necessarily restricted to lactones. Phelipanche ramosa is a widespread parasitic weed that has recently expanded its host range to include oilseed rape (Brassica napus). Phelipanche ramosa seed germinate in response to isothiocyanates derived from glucosinolate breakdown in Brassica napus root exudates, albeit with about four orders of magnitude less sensitivity than to SLs (Auger et al., 2012). Triton X-100 and KK094 inhibit the ability of GR24 to induce Striga hermonthica germination, and at least partially interfere with SL perception by ShHTL7 (Shahul Hameed et al., 2018; Nakamura et al., 2019). Addition of these antagonists to Phelipanche ramosa seed blocks the germination-stimulating effect of 2-phenethyl isothiocyanate (2-PEITC), suggesting that 2-PEITC may signal through KAI2d protein(s) (de Saint Germain et al., 2020). PrKAI2d3 has recently been reported to undergo a Tm shift in the presence of isothiocyanates, although it is less substantial than the shift induced by GR24. Remarkably, 2-PEITC becomes covalently bound to the catalytic Ser of PrKAI2d3, suggesting a mechanism for activating the receptor (de Saint Germain et al., 2020). The sensitivity of Phelipanche ramosa populations to different germination stimulants varies according to the host they were isolated from. Phelipanche ramosa seed sourced from Brassica napus fields are uniformly responsive to 2-PEITC, whereas Phelipanche ramosa seed sourced from tobacco (Nicotiana tabacum) or hemp fields show reduced or heterogeneous responses to 2-PEITC (Huet et al., 2020). Comparisons of KAI2d sequences and expression levels among these populations should provide fascinating insights into how rapid shifts in host-stimulant perception can evolve. Assuming dehydrocostus lactone is perceived by a KAI2d protein(s), a similar comparison of KAI2 evolution in the recently diverged species Orobanche cumana and Orobanche cernua will also be informative.
Structural basis of ligand-specificity and SL sensitivity in parasitic KAI2d
While there is substantial evidence that KAI2d proteins are responsible for SL perception, very limited surveys suggest that KAI2c and KAI2i proteins in parasitic plants mediate KL and/or KAR perception instead (Conn et al., 2015; Conn and Nelson, 2015; Toh et al., 2015). Therefore, comparisons of the amino acid sequences and three-dimensional structures of parasitic KAI2 are expected to reveal how different ligand-specificities and affinities are achieved within this family. Crystal structures have been solved for ShD14 and several KAI2/HTL proteins from Striga hermonthica, including ShHTL1/ShKAI2c, ShHTL3/ShKAI2i, ShHTL4, ShHTL5, ShHTL7, and ShHTL8 (Toh et al., 2015; Xu et al., 2016; 2018; Zhang et al., 2020). This wealth of information, as well as homology modeling and mutational analysis, have revealed several factors that influence ligand recognition: pocket size, ligand-positioning residues, and residues that guard the pocket entrance. In the following discussion of these insights, it is important to remember that some mechanistic details derived from in vitro or in silico single-protein experiments with KAI2/HTL may differ in vivo, where signaling partners or other cellular factors may affect a receptor’s structure and activity.
Homology modeling predicts that the ligand-binding pockets of most KAI2d proteins are unusually large compared to KAI2c or KAI2i proteins (Conn et al., 2015; de Saint Germain et al., 2020). Indeed, crystal structures of ShHTL4, ShHTL5, ShHTL7, and ShHTL8 reveal pockets with more than twice the volume of AtKAI2, ShHTL1/ShKAI2c, or ShHTL3/ShKAI2i. These pockets are also larger than those found in D14 proteins from rice and Striga hermonthica (Xu et al., 2018; Zhang et al., 2020). Residues at positions Y124, W153, F157, and F194 are among those that influence pocket volume (n.b., residue identifiers are based on AtKAI2). These residues are usually highly conserved in angiosperm KAI2 proteins, but have undergone extensive substitutions to less bulky amino acids in KAI2d proteins (Conn et al., 2015). The substitutions that have occurred at Y124 and S196 in KAI2d proteins are predicted to better accommodate the D-ring of GR24, while nonconservative changes at W153, F194, and A219 are proposed to affect ligand positioning (Toh et al., 2015). The pocket volume is also influenced by shifts in the positioning of the lid helix αD1, which forms part of the pocket entrance (Xu et al., 2018). In ShHTL7, helix αD1 tilts away from the pocket entrance more than in ShHTL/KAI2 proteins that have smaller pockets. The shift of helices αD1 and αD2 away from αD3, which enlarges the pocket, is proposed to be due to an Y150F substitution in the αD1–αD2 loop (Xu et al., 2018). Variable substitutions at this position occur in some, but not all, KAI2d proteins in parasites.
By contrast, the pocket of ShHTL1/ShKAI2c has a smaller volume than AtKAI2, and ShHTL3/ShKAI2iB is smaller yet (Xu et al., 2016; 2018). While these proteins can bind KAR1 in vitro, their pockets are likely to be too small to accommodate SLs without substantial conformational changes upon binding. Indeed, ShHTL1 and ShHTL3 do not hydrolyze rac-GR24, and ShHTL1 only has very weak hydrolytic activity against 5DS (Xu et al., 2016; 2018). Likewise, Arabidopsis lines carrying ShHTL1/ShKAI2c and ShHTL3/ShKAI2i transgenes are not responsive to rac-GR24 or SLs, but ShKAI2i lines are responsive to KARs (Conn et al., 2015; Conn and Nelson, 2015; Toh et al., 2015). The small pocket of ShHTL1 is a product of bulky residues, for example at position 190 (Xu et al., 2018). The ShHTL3 pocket is influenced more by an inward shift of helix αD1 that reduces the pocket volume and closes the entrance. Conformational shifts have been observed for αD1 in different ShKAI2iB structures, suggesting this helix could act as a gatekeeper for ligand entry and exit (Xu et al., 2016).
The significance of many binding pocket residues for the ligand affinity of KAI2 proteins has been examined through site-directed mutations. Substitution of residues 124, 190, and 194 in ShHTL7 with bulky amino acids that reduce pocket volume causes a >100-fold decrease in rac-GR24 binding affinity in vitro (Xu et al., 2018). Position 124 may be particularly important. This residue is highly conserved as Tyr in KAI2c and Phe in KAI2i, but is substituted with smaller hydrophobic amino acids in KAI2d (Conn et al., 2015). Substitutions of the Leu at this position with Phe or Tyr in ShHTL8 caused 10- to 100-fold increases in IC50 for rac-GR24 in YLG competition assays and disrupted rac-GR24 hydrolysis (Zhang et al., 2020). Hydrolysis of rac-GR24 was also decreased by I124F substitutions in ShHTL6, but not ShHTL4. Conversely, substitutions of Tyr or Phe with Leu at this position enabled rac-GR24 hydrolysis by ShHTL1/ShKAI2c and ShHTL2/ShKAI2i (Zhang et al., 2020). A recent in-depth biochemical analysis of the ShHTL7 binding pocket has revealed substitutions at 18 residues that reduce the binding affinity for rac-GR24 more than 10-fold. G25A, M139K, and mutation of the catalytic Ser (S95A) have particularly strong effects, with at least a 200-fold reduction in rac-GR24 affinity. Interestingly, T157Y appears to cause a ∼10-fold increase in affinity for rac-GR24 in vitro (Pang et al., 2020).
A more refined understanding of how ligand specificity is determined in KAI2 proteins is beginning to emerge. Due to potential steric clashes, residues 142, 157, 218, and 219 are thought to contribute to specificity in binding 2′R versus 2′S stereoisomers of GR24 and KAR1 (Xu et al., 2018). Residues at positions 96 and 189 are important contributors to KAR1 versus KAR2 selectivity in Brassicaceae KAI2 proteins (Sun et al., 2020). Likewise, amino acid identity at positions 157, 160, and 190 influences the ability of KAI2 paralogs in Lotus japonicus to recognize GR24ent-5DS (Carbonnel et al., 2020b). In Physcomitrium patens KAI2L proteins, the loop between αD2 and αD3 contributes to ligand specificity, putatively by influencing the formation and rigidity of the ligand-binding pocket (Bürger et al., 2019). A combination of molecular docking and molecular dynamics simulations was used recently to examine interactions between eight SL receptors (including four D14 and four ShHTL proteins) and 20 canonical and noncanonical SLs (Bürger and Chory, 2020b). This in silico approach builds upon the static snapshots of protein structure provided by X-ray crystallography to provide detailed predictions of a protein’s conformational dynamics and substrate-binding behaviors “in solution.” The analysis suggested that an inflexible internal bottleneck can limit access of some SL molecules to the binding pocket of SL receptors and therefore may be an important component of ligand specificity. By contrast, the outer entrance to the pocket does not appear to be a limiting factor for SL compatibility. The pocket itself has enough volume and flexibility to accommodate internal rotation of SLs to optimal binding poses for SL hydrolysis. Interestingly, molecular dynamics simulations predict that the catalytic Ser and His residues are important for positioning SL in the pocket correctly. Pocket residues at KAI2-equivalent positions 157, 134, 142, 193, and, to a lesser degree, positions 26 and 124 were implicated in frequently forming contacts with docked SLs (Bürger and Chory, 2020b).
A major question to be addressed is how some SL receptors in parasites, such as ShHTL7 and PrKAI2d3, are able to achieve such extraordinary sensitivity to SLs. It is important to note that ShHTL7 is not particularly remarkable compared to other KAI2d proteins in vitro. In YLG competition assays, ShHTL7 has IC50 values for various SLs in the ∼0.1 to 1 µM range, comparable to ShHTL6 and ShHTL8 (Tsuchiya et al., 2015). Neither does ShHTL7 show an unusually high or low rate of GR24 hydrolysis in vitro compared to other KAI2d proteins in Striga hermonthica (Tsuchiya et al., 2015). However, in germination assays of transgenic Arabidopsis, the EC50 of rac-GR24 for ShHTL7 lines is ∼20 pM, about 1,000 times lower than the ∼30–100 nM EC50 observed in lines carrying ShHTL6 or ShHTL8 (Toh et al., 2015). ShHTL4 and ShHTL5 also confer higher sensitivity to SL in vivo than would be expected from YLG competition assays (Toh et al., 2015; Tsuchiya et al., 2015). Clearly, the affinities that KAI2d proteins show for SLs in vitro are not sufficient to account for their germination-promoting activities in vivo (Shahul Hameed et al., 2018). Instead, KAI2d proteins that confer particularly sensitive germination responses to SL must be more effective at activating downstream signal transduction. This could occur if a receptor is more readily activated or its activated state more persistent. This is a difficult mystery to resolve, not least because of the current disagreements about what constitutes activation of a SL receptor. While some models for D14 have focused on formation of CLIM (or rather, a covalently modified catalytic His residue) during SL hydrolysis, others propose that SL binding is sufficient for signaling and hydrolysis is a subsequent deactivation step (Yao et al., 2016; Seto et al., 2019). Alternatively, enhanced signal transduction could occur if a receptor has higher affinity for MAX2 and/or SMAX1 upon activation than other KAI2d proteins. New evidence supports this hypothesis (Wang et al., 2021). Among 11 ShHTL proteins tested with in vitro pull-down assays, ShHTL7 shows a clearly enhanced ability to interact with AtMAX2 in the presence of SL. Substituting five residues at the MAX2 interface of ShHTL6 with ShHTL7 amino acid identities dramatically increases ShHTL6 affinity for AtMAX2 in pull-downs to ShHTL7 levels (Wang et al., 2021). In vivo assays of this ShHTL6 mutant are needed to establish whether it can confer germination responses to picomolar SL concentrations, similar to ShHTL7.
The ShHTL7-specific agonist SPL7 and its analogs show that hydrolysis of the D-ring is dispensable for SL signaling, but comes at a very high cost in potency (Uraguchi et al., 2018). SPL7 and its demethylated analog H-SPL7 have minimum effective concentrations on Striga hermonthica germination of 10 fM and 10 pM, respectively. By contrast, their hydrolysis-resistant analogs require six to eight orders of magnitude higher concentrations to achieve a similar effect. Thus CLIM formation is important for effective signaling. However, the correlation between the rate of CLIM formation in vitro and potency in vivo is weak. This is illustrated by a series of SPL7 analogs that separately modify the scaffold and D-ring, or by a direct comparison of SPL7 and GR24, which have identical D-rings. Although CLIM formation in ShHTL7 is about 10 times slower with SPL7 than GR24, SPL7 is ∼1,000 times more potent for Striga germination. Therefore, interaction of the ABC-ring of SLs or the scaffold of SL analogs with the pocket of KAI2d, presumably after hydrolysis, also appears to be important for highly effective signal transduction (Uraguchi et al., 2018).
Long-timescale molecular dynamics simulations have been used to compare SL perception in ShHTL7 and AtD14, and identify potential explanations for their different levels of SL sensitivity (Chen et al., 2020). These in silico simulations predict that ShHTL7 is more efficient than AtD14 at binding GR24 in a productive pose that favors hydrolysis. In part this is due to more stable associations of GR24 with hydrophobic residues at the entrance to the D14 pocket that slow its binding. Also, D14 is more prone to dwelling in nonproductive conformations in which the D-loop of D14, which contains the catalytic Asp residue, extends outside the core of the protein. Finally, the volume of the ShHTL7 pocket fluctuates within a narrower range than D14, reducing small-volume conformations that prevent GR24 binding or large-volume conformations that allow nonproductive orientations of the ligand (Chen et al., 2020). These exciting hypotheses may be able to explain at least some of the SL hypersensitivity of ShHTL7. Mutations of ShHTL7 have demonstrated which residues are important for maintaining high sensitivity to agonists (Uraguchi et al., 2018; Xu et al., 2018; Pang et al., 2020). However, specific mutations that cause a KAI2 protein to mimic ShHTL7 by increasing its affinity for SL in vitro or SL-signaling activity in vivo have not been reported. This will be an important goal to demonstrate a true understanding of how highly sensitive SL perception occurs in parasite seed.
Origins and implications of the KAI2 expansion in Orobanchaceae
The dramatic KAI2 expansion observed in several parasite genomes is likely due to unequal crossover events, which cause localized cis-duplications. These duplications originated before the parasitic lineage diverged, as tandem or near-tandem KAI2 duplications can be observed in nonparasitic Asterid relatives of the Orobanchaceae. In tomato (Solanum lycopersicum), two pairs of tandem copies of KAI2 are located on chromosome 2. One pair has a KAI2c adjacent to a KAI2i, and the other pair has a likely pseudogene KAI2 adjacent to a KAI2c. In Mimulus guttatus, two KAI2c are separated by two genes on the same scaffold. The third KAI2 paralog, a KAI2i is found on another scaffold. The first draft genome sequence for a parasitic plant, Striga asiatica, has provided further insights into the genomic distribution of the amplified KAI2 family. The Striga asiatica genome has several examples of functional KAI2 paralogs and KAI2 pseudogenes that are linked on the same scaffold (Yoshida et al., 2019). Curiously, duplication of D14 is relatively uncommon in angiosperms (Conn et al., 2015). It is possible that there is purifying selection to maintain a single copy of D14. Alternatively, KAI2 may be surrounded by sequences that make it more prone to unequal crossover events. Genome sequences for Striga hermonthica and Orobanche cumana are expected to be released soon, and will provide useful points of comparison.
Genetic linkage between KAI2d paralogs has several implications. First, the capacity to perceive a set of SLs may be heritable as a haplotype block, potentially enabling rapid spread of a beneficial, multigenic trait through a parasitic population. Second, the KAI2d family may be able to expand or contract relatively rapidly through additional unequal crossovers. Gene conversion may also influence the diversity of the KAI2d repertoire. Third, it will likely be challenging to connect perception of a specific SL to a single KAI2d through recombination-based trait mapping.
Overcoming limitations to genetic studies of KAI2 function in parasites
KAI2d proteins in parasites have been convincingly implicated in the perception of host-derived germination stimulants through in vitro SL-binding and SL-hydrolysis assays, cross-species complementation, and application of KAI2-specific inhibitors. However, genetic analysis in parasitic plants would provide a more conclusive and direct evaluation of the functional contributions of individual KAI2d genes. This would be a formidable pursuit through classical genetic methods due to functional redundancy and genetic linkage among parasite KAI2 paralogs. Reverse genetic approaches, particularly the use of CRISPR-Cas9 gene editing technology, offer a way forward if efficient methods for parasite transformation are available.
Transient and stable transformation of the facultative hemiparasites Triphysaria versicolor and Phtheirospermum japonicum has been achieved with Agrobacterium tumefaciens or A. rhizogenes (Tomilov et al., 2007; Ishida et al., 2011; Bandaranayake and Yoder, 2018). A method for stable transformation of the obligate holoparasite Phelipanche aegyptiaca has also been developed, producing transgenic roots and shoot buds (Fernández-Aparicio et al., 2011a). These approaches enable evaluation of gene function in parasite roots. However, fertile plants have not been regenerated, so the ability to induce the heritable genetic changes needed for evaluation of germination phenotypes is still lacking.
A breakthrough in transformation of recalcitrant dicots was achieved recently (Maher et al., 2020). Expression of developmental regulators such as WUSCHEL, BABYBOOM, and SHOOT MERISTEMLESS during Agrobacterium-mediated transformation can induce de novo formation of meristems from leaf and stem tissues. This method successfully generates heritable transgenic or gene-edited events from plants grown in sterile culture or in soil. Because this approach bypasses the need to regenerate shoots and roots from callus tissue, which is a common roadblock to tissue culture-based transformations, it may be a promising tool for genetic manipulation of parasitic plants.
In terms of prioritizing targets, gene expression patterns may provide clues about which KAI2 paralogs have major roles in germination control. So far, comparisons of the expression of KAI2d paralogs in different parasite tissues are limited to Striga hermonthica and Striga asiatica. Interestingly, while the transcripts of some KAI2d paralogs are enriched in seeds or induced during seed conditioning, others show seedling-specific expression patterns (Tsuchiya et al., 2015; Yoshida et al., 2019). This raises the possibility that there may be a role for host-derived SL perception after germination.
Translational outcomes of understanding the host-detection mechanism
An ongoing goal of the SL field is the discovery of stable and inexpensive chemical substitutes for SLs that can either stimulate suicidal germination of parasitic weeds or block parasite germination in the field. Many approaches to finding parasite germination regulators have been guided initially by assays of D14-mediated SL activities, such as tillering/shoot branching repression in nonparasitic angiosperms or yeast two-hybrid interactions between D14 and D53 (e.g. Nakamura et al., 2019). Others have used assays for activation of Arabidopsis KAI2 (AtKAI2). For example, a yeast two-hybrid assay for AtKAI2 interaction with MAX2 was used to screen a chemical library of 4,182 compounds. Forty-two compounds that promoted AtKAI2-MAX2 interactions in yeast were further screened with seedling growth and CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)-localization assays in Arabidopsis, leading to the identification of three lead compounds. All three compounds promoted germination of Striga hermonthica, albeit with much lower potency than GR24 (Toh et al., 2014). A screen for antagonists that interfere with GR24 perception in Arabidopsis seedlings, which occurs through AtKAI2 and D14, was performed with the same chemical library (Holbrook-Smith et al., 2016). Thirty-seven compounds blocked the inhibitory effect of rac-GR24 on Arabidopsis hypocotyl elongation, of which seven inhibited germination of Arabidopsis seed. The best performing lead compound, termed soporidine, was shown to bind AtKAI2 and ShHTL7. Micromolar concentrations of soporidine can block the stimulation of Striga hermonthica germination by rac-GR24 (Holbrook-Smith et al., 2016).
These strategies may have benefited from greater specificity for parasite KAI2d. Although parasite KAI2d proteins have converged on SL perception with D14, there are substantial differences in their ligand-binding pockets and downstream signaling partners. KAI2d proteins are more similar to AtKAI2 than D14 proteins, of course with obvious differences in their ligand specificities and pockets. Therefore, chemicals that work on D14 or AtKAI2 may not be effective on KAI2d proteins, and vice versa. The most effective screens for KAI2d agonists and antagonists will likely be based upon direct tests of parasite germination, high-throughput assays for KAI2d activation, or structures of KAI2d proteins. For example, a chemical library screen of 12,000 synthetic molecules for Striga hermonthica germination stimulants led to the identification of N-arylsulfonylpiperazine as a molecular scaffold that could replace the ABC-ring of SL (Uraguchi et al., 2018). Joining this scaffold to a methylbutenolide D-ring formed SPL7, a specific agonist of ShHTL7 that can trigger S. hermonthica germination at femtomolar concentrations. SPL7 has similar potency to the natural SL 5-deoxystrigol, and is at least 100 times more potent than (+)-GR24 on S. hermonthica (Uraguchi et al., 2018).
Variability among parasite seed populations, access to parasite seed, and biosafety considerations pose challenges to chemical screens based on parasite germination responses, however. Therefore, there is a need for simple, consistent assays to measure the effects of different compounds on KAI2d activity. Yeast two-hybrid assays between parasite KAI2d proteins and MAX2 or SMAX1 might be an effective primary screen for chemical regulators of KAI2d. Arabidopsis KAI2 is prone to nonspecific interactions with MAX2 in yeast two-hybrid, however (Yao et al., 2018a). The middle domains of SMAX1 mediate KAI2 interactions in Arabidopsis, and might provide a clearer interaction assay (Khosla et al., 2020a). Putatively, FRET-based biosensors for SL could also be developed based upon protein–protein interactions with KAI2d, as have already been accomplished for ABA and GA (Jones et al., 2014; Waadt et al., 2014; Rizza et al., 2017). A functional assay that measures the ability of a parasite KAI2d to target SMAX1 for degradation would be ideal, as the binding or hydrolysis activity of KAI2d proteins for a compound in vitro does not always reflect its potency in vivo. A highly sensitive and specific assay for SL activity, StrigoQuant, measures degradation of a ratiometric bioluminescent SMXL6 reporter in Arabidopsis protoplasts (Samodelov et al., 2016). Potentially, a similar system that measures SMAX1 degradation could enable characterization of the ligand preferences of a coexpressed parasite KAI2d protein. A ratiometric system that has a similar design principle to StrigoQuant has been used to assay SMAX1 degradation in Nicotiana benthamiana leaves (Khosla et al., 2020a; 2020b). This may be useful as a medium-throughput in vivo assay for agonists/antagonists of a KAI2d protein. Another exciting possibility has come from the development of SL biosensors that integrate circular-permutated GFP into DAD2 and ShHTL7 proteins (Chesterfield et al., 2020). A C-terminal fusion of the biosensor to a second fluorescent protein, LSSmOrange, provides an internal control that normalizes for changes in biosensor abundance. These single-protein sensors show a two-fold decrease in GFP fluorescence relative to LSSmOrange in the presence of SLs due to conformational changes that occur in the receptor during SL binding and/or hydrolysis. The cpGFP-ShHTL7 biosensor shows high sensitivity to SLs, with EC50 values ranging from 9 nM for rac-GR24 to 116 nM for rac-5DS (Chesterfield et al., 2020). However, the relative sensitivity of ShHTL7 to rac-GR24, rac-orobanchol, and rac-5DS differs in biosensor assays versus YLG competition assays (Tsuchiya et al., 2015; Chesterfield et al., 2020).
Crystal structures of receptors unlock additional approaches to the discovery and design of novel chemical regulators. For example, molecular docking can be used to perform in silico screens for potential agonists or antagonists that are refined further through in vitro assays and structure-guided optimization. This approach was employed with great success to design a headgroup that dramatically improves the affinity of an ABA receptor agonist, opabactin (Vaidya et al., 2019). In another example, pharmacophore models developed from crystal structures of rice D14 were used to perform in silico screening of 4.7 million compounds for potential SL signaling inhibitors (Mashita et al., 2016). Further screening of 61 commercially available compounds for the ability to block GR24-induced yeast two-hybrid interactions between D14 and D53 or SLR1 led to the identification of 2-methoxy-1-naphthaldehyde (2-MN). Interestingly, 2-MN was able to inhibit or partially inhibit germination of Arabidopsis thaliana and Striga hermonthica in the presence of GR24, suggesting it is a KAI2 antagonist as well (Mashita et al., 2016). If applied against KAI2d structures from parasites, this strategy may prove even more effective for identifying regulators of parasite germination. This is illustrated by molecular docking experiments that examined the placement of 4-bromodebranone (4BD) in the ShHTL5 pocket (Fukui et al., 2017). 4BD was originally identified as a D14 agonist with a simple structure that suppresses tillering, but it only has weak activity on Striga hermonthica germination (Fukui et al., 2013). The docking analysis enabled rational design of 4BD analogs that had improved activity and specificity for Striga germination (Fukui et al., 2017). A recent biochemical characterization of 60 ShHTL7 proteins with substitutions in ligand-binding pocket residues enabled the development of a mutation-dependent biomacromolecular quantitative structure−activity relationship (MB-QSAR) model. This model has a strong ability to predict the binding affinities of ShHTL7 mutants and other ShHTL proteins for GR24, and may prove useful for the rational design of new ShHTL regulators (Pang et al., 2020).
Concluding Remarks
The discovery of novel receptors for SLs in parasites was an important step toward understanding the dynamic relationship between parasitic Orobanchaceae and their hosts. Further questions about the evolution of this key parasitic adaptation are now raised (see Outstanding Questions). Over the past several years, a remarkable combination of approaches from the disciplines of chemistry, biochemistry, structural biology, evolution, and genetics have rapidly led to detailed mechanistic insights into strigolactone signaling in parasites. With the development of new tools to probe activation of KAI2d proteins in vitro, in vivo, and in silico, there is an unprecedented opportunity to translate these discoveries into better ways to combat parasitic weed infestations and improve crop yields.
Funding
This work was supported by the National Science Foundation (IOS-1856741) and the USDA National Institute of Food and Agriculture (Hatch project 1023345).
Conflict of interest statement. None declared.
Outstanding Questions
How are noncanonical SLs and other lactone molecules that stimulate parasite germination perceived?
Do KAI2d proteins have adaptive value as SL receptors in facultative hemiparasites that do not show host-activated germination?
Do some KAI2d proteins negatively regulate germination after detecting specific SLs?
Is expansion of the KAI2d clade adaptive in the transition to a weedy lifestyle?
Is host-triggered germination used by obligate parasites of long-lived perennial hosts?
What are the roles of KAI2c and KAI2i proteins in parasitic plants? Why do KARs not activate germination of S. hermonthica, which expresses KAI2i?
How does SMAX1 impose seed dormancy, and how has this system become so important to parasite dormancy?
Which cells in parasite seed control germination? How do parasite seed maintain long-term viability while supporting a system for detecting SLs?
References
- Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T, et al. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA 111: 18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, et al. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108: 20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K-I, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Arc E, Sechet J, Corbineau F, Rajjou L, Marion-Poll A (2013) ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front Plant Sci 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Auger B, Pouvreau J-B, Pouponneau K, Yoneyama K, Montiel G, Le Bizec B, Yoneyama K, Delavault P, Delourme R, Simier P (2012) Germination stimulants of Phelipanche ramosa in the rhizosphere of Brassica napus are derived from the glucosinolate pathway. Mol Plant Microbe Interact 25: 993–1004 [DOI] [PubMed] [Google Scholar]
- Bandaranayake PCG, Yoder JI (2018) Factors affecting the efficiency of Rhizobium rhizogenes root transformation of the root parasitic plant Triphysaria versicolor and its host Arabidopsis thaliana. Plant Methods 14: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Liang Y, Seale M, Ward S, Müller D, Leyser O (2016) Strigolactone regulates shoot development through a core signalling pathway. Biol Open 5: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Bécard G, Jauneau A, Roux C, Séjalon-Delmas N (2008) GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol 148: 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Nelson DC, Weijers D (2020) Evolution of plant hormone response pathways. Annu Rev Plant Biol 10.1146/annurev-arplant-050718-100309 [DOI] [PubMed] [Google Scholar]
- Bouwmeester H, Li C, Thiombiano B, Rahimi M, Dong L (2020) Adaptation of the parasitic plant lifecycle: germination is controlled by essential host signaling molecules. Plant Physiol 10.1093/plphys/kiaa066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Yoneyama K, Filardo F, Meyers E, Scaffidi A, Frickey T, Akiyama K, Seto Y, Dun EA, Cremer JE, et al. (2016) LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 113: 6301–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun G, Thoiron S, Braem L, Pouvreau J-B, Montiel G, Lechat M-M, Simier P, Gevaert K, Goormachtig S, Delavault P (2019) CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants. Plant Cell Environ 42: 2612–2626 [DOI] [PubMed] [Google Scholar]
- Bunsick M, Toh S, Wong C, Xu Z, Ly G, McErlean CSP, Pescetto G, Nemrish KE, Sung P, Li JD, et al. (2020) SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat Plants 6: 646–652 [DOI] [PubMed] [Google Scholar]
- Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH, et al. (2014) Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol 164: 424–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Chory J (2020a) The many models of strigolactone signaling. Trends Plant Sci 25: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Chory J (2020b) In-silico analysis of the strigolactone ligand-receptor system. Plant Direct 4: e00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Mashiguchi K, Lee HJ, Nakano M, Takemoto K, Seto Y, Yamaguchi S, Chory J (2019) Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep 26: 855–865.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell-Douglas R, Rothfels CJ, Stevenson DWD, Graham SW, Wong GK-S, Nelson DC, Bennett T (2017) Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol 15: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel S, Das D, Varshney K, Kolodziej MC, Villaécija-Aguilar JA, Gutjahr C (2020a) The karrikin signaling regulator SMAX1 controls Lotus japonicus root and root hair development by suppressing ethylene biosynthesis. Proc Natl Acad Sci USA 10.1073/pnas.2006111117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel S, Torabi S, Griesmann M, Bleek E, Tang Y, Buchka S, et al (2020b) Lotus japonicus karrikin receptors display divergent ligand-binding specificities and organ-dependent redundancy. PLoS Genet 16(12): e1009249. 10.1371/journal.pgen.1009249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson GH, Hasse D, Cardinale F, Prandi C, Andersson I (2018) The elusive ligand complexes of the DWARF14 strigolactone receptor. J Exp Bot 69: 2345–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnikhova TV, Gaus K, Lumbroso A, Sanders M, Vincken J-P, De Mesmaeker A, Ruyter-Spira CP, Screpanti C, Bouwmeester HJ (2017) Zealactones. Novel natural strigolactones from maize. Phytochemistry 137: 123–131 [DOI] [PubMed] [Google Scholar]
- Chen J, White A, Nelson DC, Shukla D (2020) Role of substrate recognition in modulating strigolactone receptor selectivity in witchweed. bioRxiv. 10.1101/2020.07.28.225722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesterfield RJ, Whitfield JH, Pouvreau B, Cao D, Alexandrov K, Beveridge CA, Vickers CE (2020) Rational design of novel fluorescent enzyme biosensors for direct detection of strigolactones. ACS Synth Biol 9: 2107–2118 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lee T, Cho J, Servante EK, Pucker B, Summers W, Bowden S, Rahimi M, An K, An G, et al. (2020) The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat Commun 11: 2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CR, Timko MP, Yoder JI, Axtell MJ, Westwood JH (2019) Molecular dialog between parasitic plants and their hosts. Annu Rev Phytopathol 57: 279–299 [DOI] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349: 540–543 [DOI] [PubMed] [Google Scholar]
- Conn CE, Nelson DC (2015) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6: 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of Witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Dor E, Plakhine D, Joel DM, Larose H, Westwood JH, Smirnov E, Ziadna H, Hershenhorn J (2020) A new race of sunflower broomrape (Orobanche cumana) with a wider host range due to changes in seed response to strigolactones. Weed Sci 68: 134–142 [Google Scholar]
- Evidente A, Fernández-Aparicio M, Cimmino A, Rubiales D, Andolfi A, Motta A (2009) Peagol and peagoldione, two new strigolactone-like metabolites isolated from pea root exudates. Tetrahedron Lett 50: 6955–6958 [Google Scholar]
- Fernández-Aparicio M, Flores F, Rubiales D (2009) Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann Bot 103: 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Rubiales D, Bandaranayake PC, Yoder JI, Westwood JH (2011a) Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Yoneyama K, Rubiales D (2011b) The role of strigolactones in host specificity of Orobanche and Phelipanche seed germination. Seed Sci Res 21: 55–61 [Google Scholar]
- Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD (2004) A compound from smoke that promotes seed germination. Science 305: 977. [DOI] [PubMed] [Google Scholar]
- Flematti GR, Scaffidi A, Waters MT, Smith SM (2016) Stereospecificity in strigolactone biosynthesis and perception. Planta 243: 1361–1373 [DOI] [PubMed] [Google Scholar]
- Fukui K, Ito S, Asami T (2013) Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol Plant 6: 88–99 [DOI] [PubMed] [Google Scholar]
- Fukui K, Yamagami D, Ito S, Asami T (2017) A Taylor-made design of phenoxyfuranone-type strigolactone mimic. Front Plant Sci 8: 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP (2013) Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc Natl Acad Sci USA 110: 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang S-Y, Nadal M, et al. (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RSM, Luo Z, Lee HW, Sharma P, Janssen BJ, Perry NB, Denny WA, Snowden KC (2018) Inhibition of strigolactone receptors by N-phenylanthranilic acid derivatives: structural and functional insights. J Biol Chem 293: 6530–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Proust H, Belcram K, Labrune C, Boyer F-D, Rameau C, Bonhomme S (2014) Strigolactones inhibit caulonema elongation and cell division in the moss Physcomitrella patens. PLoS One 9: e99206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook-Smith D, Toh S, Tsuchiya Y, McCourt P (2016) Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat Chem Biol 12: 724–729 [DOI] [PubMed] [Google Scholar]
- Hrdlička J, Gucký T, Novák O, Kulkarni M, Gupta S, van Staden J, Doležal K (2019) Quantification of karrikins in smoke water using ultra-high performance liquid chromatography–tandem mass spectrometry. Plant Methods 15: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, He Y, Wang L, Liu S, Meng X, Liu G, Jing Y, Chen M, Song X, Jiang L, et al. (2017) DWARF14, areceptor covalently linked with the active form of strigolactones, undergoes strigolactone-dependent degradation in rice. Front Plant Sci 8: 1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet S, Pouvreau J-B, Delage E, Delgrange S, Marais C, Bahut M, Delavault P, Simier P, Poulin L (2020) Populations of the parasitic plant Phelipanche ramosainfluence their seed microbiota. Front Plant Sci 11: 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Yoshida S, Ito M, Namba S, Shirasu K (2011) Agrobacterium rhizogenes-mediated transformation of the parasitic plant Phtheirospermum japonicum. PLoS One 6: e25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel DM, Bar H, Mayer AM, Plakhine D, Ziadne H, Westwood JH, Welbaum GE (2012) Seed ultrastructure and water absorption pathway of the root-parasitic plant Phelipanche aegyptiaca (Orobanchaceae). Ann Bot 109: 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel DM, Chaudhuri SK, Plakhine D, Ziadna H, Steffens JC (2011) Dehydrocostus lactone is exuded from sunflower roots and stimulates germination of the root parasite Orobanche cumana. Phytochemistry 72: 624–634 [DOI] [PubMed] [Google Scholar]
- Jones AM, Danielson JA,, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim S-Y, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18: 147–160 [DOI] [PubMed] [Google Scholar]
- Kalliola M, Jakobson L, Davidsson P, Pennanen V, Waszczak C, Yarmolinsky D, Zamora O, Palva ET, Kariola T, Kollist H, et al. (2020) Differential role of MAX2 and strigolactones in pathogen, ozone, and stomatal responses. Plant Direct 4: e00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux P-M, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Séjalon-Delmas N, Combier J-P, Bécard G, Belausov E, et al. (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233: 209–216 [DOI] [PubMed] [Google Scholar]
- Karssen CM, Zagorski S, Kepczynski J, Groot SPC (1989) Key role for endogenous gibberellins in the control of seed germination. Ann Bot 63: 71–80 [Google Scholar]
- Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, Cai ML, Stanga JP, Flematti GR, et al. (2020a) Structure-function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell 10.1105/tpc.19.00752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Nelson DC (2016) Strigolactones, super hormones in the fight against Striga. Curr Opin Plant Biol 33: 57–63 [DOI] [PubMed] [Google Scholar]
- Khosla A, Rodriguez‐Furlan C, Kapoor S, Van Norman JM, Nelson DC (2020b) A series of dual‐reporter vectors for ratiometric analysis of protein abundance in plants. Plant Direct 10.1002/pld3.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HI, Kisugi T, Khetkam P, Xie X, Yoneyama K, Uchida K, Yokota T, Nomura T, McErlean CSP, Yoneyama K (2014) Avenaol, a germination stimulant for root parasitic plants from Avena strigosa. Phytochemistry 103: 85–88 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Kameoka H, Sugimura Y, Saito K, Ohtomo R, Fujiwara T, Kyozuka J (2018) Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol 59: 544–553 [DOI] [PubMed] [Google Scholar]
- Kochanek J, Long RL, Lisle AT, Flematti GR (2016) karrikins identified in biochars indicate post-fire chemical cues can influence community diversity and plant development. PLoS One 11: e0161234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahari Z, Ullah C, Kyndt T, Gershenzon J, Gheysen G (2019) Strigolactones enhance root-knot nematode (Meloidogyne graminicola) infection in rice by antagonizing the jasmonate pathway. New Phytol 224: 454–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, André O, Peng J, Wen J, Chen R, Ratet P, Tadege M, Mysore KS, Rochange SF (2015) Strigolactones contribute to shoot elongation and to the formation of leaf margin serrations in Medicago truncatula R108. J Exp Bot 66: 1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat M-M, Pouvreau J-B, Péron T, Gauthier M, Montiel G, Véronési C, Todoroki Y, Le Bizec B, Monteau F, Macherel D, et al. (2012) PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J Exp Bot 63: 5311–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Sharma P, Janssen BJ, Drummond RSM, Luo Z, Hamiaux C, Collier T, Allison JR, Newcomb RD, Snowden KC (2020) Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. J Biol Chem 295: 4181–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Kim K, Lee S, Lee S, Hwang E, Shin K, Kim D, Choi J, Choi H, Seok Cha J, et al. (2018) Functional analysis of a missense allele of KARRIKIN-INSENSITIVE2 that impairs ligand-binding and downstream signaling in Arabidopsis thaliana. J Exp Bot 69:3609–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28: 1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Sun J (2017) miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol 174: 1931–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang Y, Matusova R, Charnikhova T, Amini M, Jamil M, Fernandez-Aparicio M, Huang K, Timko MP, Westwood JH, et al. (2014) Striga hermonthica MAX2 restores branching but not the very low fluence response in the Arabidopsis thaliana max2 mutant. New Phytol 202: 531–541 [DOI] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Chu HD, Van Ha C, Watanabe Y, Osakabe Y, Leyva-González MA, Sato M, Toyooka K, Voges L, et al. (2017) The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet 13: e1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Chu HD, Watanabe Y, Osakabe Y, Sato M, Toyooka K, Seo M, Tian L, Tian C, et al. (2020) Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE2 in drought adaptation of Arabidopsis thaliana. Plant J 103: 111–127 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S (2016) Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta 243: 1441–1453 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, de Villiers R, Hoffmann B, Ma L, de Saint Germain A, Kossmann J, Coudert Y, Harrison CJ, Rameau C, Hills P, et al. (2018) Physcomitrella patens MAX2 characterization suggests an ancient role for this F-box protein in photomorphogenesis rather than strigolactone signalling. New Phytol 219: 743–756 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, Guillory A, Boyer F-D, Cornu D, Hoffmann B, Le Bris P, Pouvreau J-B, Delavault P, Rameau C, de Saint Germain A, et al. (2020) The Physcomitrium (Physcomitrella) patens PpKAI2L receptors for strigolactones and related compounds highlight MAX2 dependent and independent pathways. bioRxiv 10.1101/2020.11.24.395954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q, et al. (2013) Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin DC, Hamon-Josse M, Bennett T (2020) Fellowship of the rings: a saga of strigolactones and other small signals. New Phytol 225: 621–636 [DOI] [PubMed] [Google Scholar]
- Ma H, Duan J, Ke J, He Y, Gu X, Xu T-H, Yu H, Wang Y, Brunzelle JS, Jiang Y, et al. (2017) A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv 3: e1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF (2020) Plant gene editing through de novo induction of meristems. Nat Biotechnol 38: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashita O, Koishihara H, Fukui K, Nakamura H, Asami T (2016) Discovery and identification of 2-methoxy-1-naphthaldehyde as a novel strigolactone-signaling inhibitor. J Pestic Sci 41: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Komatsu A, Simazaki S, Xie X, Ishizaki K (2020) Major components in the KARRIKIN INSENSITIVE2-ligand signaling pathway are conserved in the liverwort, Marchantia polymorpha. bioRxiv 10.1101/2020.11.17.387852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Hirabayashi K, Miyakawa T, Kikuzato K, Hu W, Xu Y, Jiang K, Takahashi I, Niiyama R, Dohmae N, et al. (2019) Triazole ureas covalently bind to strigolactone receptor and antagonize strigolactone responses. Mol Plant 12: 44–58 [DOI] [PubMed] [Google Scholar]
- Nasir F, Tian L, Shi S, Chang C, Ma L, Gao Y, Tian C (2019) Strigolactones positively regulate defense against Magnaporthe oryzae in rice (Oryza sativa). Plant Physiol Biochem 142: 106–116 [DOI] [PubMed] [Google Scholar]
- Nelson DC (2013) Are karrikin signaling mechanisms relevant to strigolactone perception? InJoel DM, Gressel J, Musselman LJ, eds, Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Springer, Berlin, Heidelberg, pp 221–230 [Google Scholar]
- Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM (2012) Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu Rev Plant Biol 63: 107–130 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough J-A, Ghisalberti EL, Dixon KW, Smith SM (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Riseborough J-A, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, Smith SM (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol 149: 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S, Nakashima H, Mizutani M, Takikawa H, Sugimoto Y (2013) Structural requirements of strigolactones for germination induction and inhibition of Striga gesnerioides seeds. Plant Cell Rep 32: 829–838 [DOI] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative(2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Zhang X, Ma F, Liu J, Zhang H, Wang J, Wen X, Xi Z (2020) Comparative studies of potential binding pocket residues reveal the molecular basis of ShHTL receptors in the perception of GR24 in Striga. J Agric Food Chem 68: 12729–12737 [DOI] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, Nogué F, Rameau C (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138: 1531–1539 [DOI] [PubMed] [Google Scholar]
- Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants 3: 803–813 [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot M-A, Pillot J-P, Cornu D, Le Caer J-P, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Jacobs A, Brun G, Pouvreau J-B, Braem L, Cornu D, Clavé G, Baudu E, Steinmetz V, Servajean V, et al. (2020) A Phelipanche ramosa KAI2 protein perceives enzymatically strigolactones and isothiocyanates. bioRxiv 10.1101/2020.06.09.136473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samodelov SL, Beyer HM, Guo X, Augustin M, Jia K-P, Baz L, Ebenhöh O, Beyer P, Weber W, Al-Babili S, et al. (2016) StrigoQuant: a genetically encoded biosensor for quantifying strigolactone activity and specificity. Sci Adv 2: e1601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Ghisalberti EL, Dixon KW, Flematti GR, Smith SM (2013) Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J 76: 1–9 [DOI] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165: 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Sado A, Asami K, Hanada A, Umehara M, Akiyama K, Yamaguchi S (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc Natl Acad Sci USA 111: 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Ticchiarelli F, Mao H, Hinds TR, Leyser O, Zheng N (2018) Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 563: 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahul Hameed U, Haider I, Jamil M, Kountche BA, Guo X, Zarban RA, Kim D, Al-Babili S, Arold ST (2018) Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor. EMBO Rep 10.15252/embr.201745619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E (2007) The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Bu Q-Y, Huq E (2012) MAX2 affects multiple hormones to promote photomorphogenesis. Mol Plant 5: 750–762 [DOI] [PubMed] [Google Scholar]
- Shindo M, Yamamoto S, Shimomura K, Umehara M (2020) Strigolactones decrease leaf angle in response to nutrient deficiencies in rice. Front Plant Sci 11: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lu Z, Yu H, Shao G, Xiong J, Meng X, Jing Y, Liu G, Xiong G, Duan J, et al. (2017) IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res 27: 1128–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Morffy N, Nelson DC (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X-D, Ni M (2011) HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol Plant 4: 116–126 [DOI] [PubMed] [Google Scholar]
- Sun YK, Flematti GR, Smith SM, Waters MT (2016) Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front Plant Sci 7: 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YK, Yao J, Scaffidi A, Melville KT, Davies SF, Bond CS, Smith SM, Flematti GR, Waters MT (2020) Divergent receptor proteins confer responses to different karrikins in two ephemeral weeds. Nat Commun 11: 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck SM, Guerringue Y, Matthus E, Jamieson FJC, Davies JM (2019) Impairment in karrikin but not strigolactone sensing enhances root skewing in Arabidopsis thaliana. Plant J 98: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood CJ, Rumsey FJ, Hiscock SJ (2009) Host-specific races in the holoparasitic angiosperm Orobanche minor: implications for speciation in parasitic plants. Ann Bot 103: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350: 203–207 [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stokes ME, Tsuchiya Y, McCourt P (2014) Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem Biol 21: 988–998 [DOI] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y (2012) Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol 53: 107–117 [DOI] [PubMed] [Google Scholar]
- Tomilov A, Tomilova N, Yoder JI (2007) Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 225: 1059–1071 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P (2010) A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol 6: 741–749 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook-Smith D, Zhang H, McCourt P, Itami K, Kinoshita T, et al. (2015) PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349: 864–868 [DOI] [PubMed] [Google Scholar]
- Ueda H, Kusaba M (2015) Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol 169: 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Furumoto T, Umeda S, Mizutani M, Takikawa H, Batchvarova R, Sugimoto Y (2014) Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry 108: 122–128 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Uraguchi D, Kuwata K, Hijikata Y, Yamaguchi R, Imaizumi H, Am S, Rakers C, Mori N, Akiyama K, Irle S, et al. (2018) A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science 362: 1301–1305 [DOI] [PubMed] [Google Scholar]
- Vaidya AS, Helander JDM, Peterson FC, Elzinga D, Dejonghe W, Kaundal A, Park S-Y, Xing Z, Mega R, Takeuchi J, et al. (2019) Dynamic control of plant water use using designed ABA receptor agonists. Science 10.1126/science.aaw8848 [DOI] [PubMed] [Google Scholar]
- Van Ha C, Leyva-González MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Yamaguchi S, Van Dong N, et al. (2014) Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci USA 111: 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaécija-Aguilar JA, Hamon-Josse M, Carbonnel S, Kretschmar A, Schmid C, Dawid C, Bennett T, Gutjahr C (2019) SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet 15: e1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi T, Hamana M, Mori A, Akiyama R, Ueno K, Osakabe K, Osakabe Y, Suzuki H, Takikawa H, Mizutani M, et al. (2019) Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Sci Adv 5: eaax9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CH, Siu-Ting K, Taylor A, O’Connell MJ, Bennett T (2019) Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol 17: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang A, Shao N, Ma H, Xiong G, et al. (2020a) Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583: 277–281 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu Q, Yu H, Ma H, Li X, Yang J, Chu J, Xie Q, Wang Y, Smith SM, et al. (2020b) Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation inArabidopsis thaliana. Plant Cell 10.1105/tpc.20.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yao R, Du X, Guo L, Chen L, Xie D, Smith SM, et al. (2021) Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga. Plant Physiol 10.1093/plphys/kiaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Annu Rev Plant Biol 68: 291–322 [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Moulin SLY, Sun YK, Flematti GR, Smith SM (2015) A Selaginella moellendorffii Ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27: 1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, dePamphilis CW, Das M, Fernández-Aparicio M, Honaas LA, Timko MP, Wafula EK, Wickett NJ, Yoder JI (2012) The parasitic plant genome project: new tools for understanding the biology of Orobanche and Striga. Weed Science 60: 295–306 [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K (2010) The strigolactone story. Annu Rev Phytopathol 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Xu Y, Miyakawa T, Nakamura H, Nakamura A, Imamura Y, Asami T, Tanokura M (2016) Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Sci Rep 6: 31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Miyakawa T, Nosaki S, Nakamura A, Lyu Y, Nakamura H, Ohto U, Ishida H, Shimizu T, Asami T, et al. (2018) Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat Commun 9: 3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240: 399–408 [DOI] [PubMed] [Google Scholar]
- Yao J, Mashiguchi K, Scaffidi A, Akatsu T, Melville KT, Morita R, Morimoto Y, Smith SM, Seto Y, Flematti GR, et al. (2018a) An allelic series at the KARRIKIN INSENSITIVE 2 locus of Arabidopsis thaliana decouples ligand hydrolysis and receptor degradation from downstream signalling. Plant J 96: 75–89 [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473 [DOI] [PubMed] [Google Scholar]
- Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Zhang W, Deng H, Xie D (2017) ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res 27: 838–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Wang L, Li Y, Chen L, Li S, Du X, Wang B, Yan J, Li J, Xie D (2018b) Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J Exp Bot 69: 2355–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui R, Seto Y, Ito S, Kawada K, Itto-Nakama K, Mashiguchi K, Yamaguchi S (2019) Chemical screening of novel strigolactone agonists that specifically interact with DWARF14 protein. Bioorg Med Chem Lett 29: 938–942 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Mori N, Sato T, Yoda A, Xie X, Okamoto M, Iwanaga M, Ohnishi T, Nishiwaki H, Asami T, et al. (2018a) Conversion of carlactone to carlactonoic acid is a conserved function of MAX 1 homologs in strigolactone biosynthesis. New Phytol 218: 1522–1533 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K (2007) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227: 125–132 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Yoneyama K, Kisugi T, Nomura T, Nakatani Y, Akiyama K, McErlean CSP (2018b) Which are the major players, canonical or non-canonical strigolactones? J Exp Bot 69: 2231–2239 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim Y-M, Honaas L, Yang Z, Spallek T, Conn CE, Ichihashi Y, et al. (2019) Genome Sequence of Striga asiaticaprovides insight into the evolution of plant parasitism. Curr Biol 29: 3041–3052.e4 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Shen Y, Xi Z (2020) Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway. Biochem Biophys Res Commun 523: 1040–1045 [DOI] [PubMed] [Google Scholar]
- Zhao L-H, Zhou XE, Yi W, Wu Z, Liu Y, Kang Y, Hou L, de Waal PW, Li S, Jiang Y, et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res 25: 1219–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Hong K, Zeng L, Wang L, Kang S, Qu M, Dai J, Zou L, Zhu L, Tang Z, et al. (2020) Karrikin Signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell 10.1105/tpc.20.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]