Abstract
Anionic phospholipids include phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylinositol (PI), and its phosphorylated derivatives the phosphoinositides (e.g. phosphatidylinositol-4-phosphate [PI4P] and phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2]). Although anionic phospholipids are low-abundant lipids, they are particularly important for membrane functions. In particular, anionic lipids act as biochemical and biophysical landmarks that contribute to the establishment of membrane identity, signaling activities, and compartment morphodynamics. Each anionic lipid accumulates in different endomembranes according to a unique subcellular pattern, where they locally provide docking platforms for proteins. As such, they are mostly believed to act in the compartments in which they accumulate. However, mounting evidence throughout eukaryotes suggests that anionic lipids are not as compartment-specific as initially thought and that they are instead organized as concentration gradients across different organelles. In this update, we review the evidence for the existence of anionic lipid gradients in plants. We then discuss the possible implication of these gradients in lipid dynamics and homeostasis, and also in coordinating subcellular activities. Finally, we introduce the notion that anionic lipid gradients at the cellular scale may translate into gradients at the tissue level, which could have implications for plant development.
One-sentence summary
Mounting evidence reveals that anionic lipids exist in gradients within the plant endomembrane network that may have implications on lipid homeostasis and membrane dynamics.
Introduction
The endomembrane system is a network of membranes connected by vesicular trafficking and direct membrane contacts and is a characteristic of eukaryotic cells (Boutté and Jaillais, 2020). This network contains compartments derived from the endoplasmic reticulum (ER), including the nuclear membrane, autophagosomes, or the Golgi apparatus as well as post-Golgi compartments, such as the trans-Golgi network (TGN), the plasma membrane, endosomes, and lysosomes/vacuoles. Each of the organelles that makes the endomembrane system has a unique composition both in terms of proteins and lipids, which allows these compartments to achieve specific functions. At the same time, the endomembrane network is highly dynamic with constant exchanges of molecules between its different compartments. One of the key questions in cell biology is thus how individual compartments establish and maintain their unique identity (i.e. specific membrane properties) when faced with these constant exchanges.
Eukaryotic membranes are constituted by glycerophospholipids (hereafter referred to as phospholipids) which account for up to 75% of membrane lipids (Ejsing et al., 2009; Andreyev et al., 2010; Sampaio et al., 2011; Colin and Jaillais, 2019). Phosphatidic acid (PA) is the backbone of phospholipids. It is constituted by a glycerol group whose 1- and 2-hydroxy functions are esterified by two fatty acid chains and its 3-hydroxy function is esterified by a phosphate group. The PA backbone can be modified on the phosphate group by the addition of a choline, ethanolamine, serine, or inositol group to form phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI), respectively (Colin and Jaillais, 2019). The phosphorylation of the PI inositol group on the 3-, 4-, and/or 5-hydroxy functions can give up to seven phosphoinositide species distributed in three groups (monophosphates, biphosphates, and triphosphates), depending on the number and the position of the phosphorylations. Monophosphate phosphoinositides include PI3P, PI4P, and PI5P. Biphosphate phosphoinositides include PI(3,4)P2, PI(3,5)P2, and PI(4,5)P2, while there is only one triphosphate phosphoinositide species, PI(3,4,5)P3. Of note, plant genomes lack type-I and type-II PI3-kinases (PI3K), which phosphorylate PI3P and PI(4,5)P2 into PI(3,4)P2 and PI(3,4,5)P3, respectively (Mueller-Roeber and Pical, 2002; Noack and Jaillais, 2020). As a result, these phosphoinositide species have never been detected in plants and likely do not exist. The second major lipid class in eukaryotes is sphingolipids, which represent 8–12% mol of lipids (Drin, 2014). The last class is constituted by sterols, which constitute 12–14% mol of lipids (Drin, 2014). In mammals and yeast (Saccharomyces cerevisiae), cholesterol and ergosterol are the most common sterol species, respectively (Andreyev et al., 2010; Antonny et al., 2018). In plants, there is a large diversity of sterols (i.e. phytosterols), and the most abundant species are β-sitosterol, campesterol, and stigmasterol (Mamode Cassim et al., 2019; Boutté and Jaillais, 2020). Sterols are constituted by four fused cycloalkane rings. They are highly hydrophobic, rigid, and planar. As such, sterols impact the biophysical properties of the membrane they are embedded in by reducing the flexibility of the acyl chains of its surrounding lipid neighbors and thereby increasing the stability (and reducing the permeability) of the membrane (Boutte and Moreau, 2014; Mamode Cassim et al., 2019).
Advances
Plant cells, like yeast and animal cells, can be divided into two broad membrane territories: a territory of lipid packing defects and a territory of electrostatics.
Sharp gradients of anionic lipids exist between these two membrane territories.
Anionic lipid counter exchanges at membrane contact sites are powered by lipid gradients in animal and yeast systems and are often interdependent.
Anionic lipid gradients also exist within the plant electrostatic membrane territory.
The cellular gradients of anionic lipids are under developmental control, suggesting the existence of lipid gradients at the tissue level in plants.
Among lipids, anionic phospholipids are major determinants of membrane identity (Noack and Jaillais, 2017; Noack and Jaillais, 2020). Indeed, these lipids have several attributes that make them ideal landmarks at the surface of compartments. First, they are not abundant and so they do not constitute the bulk of membrane lipids (Colin and Jaillais, 2019). Second, they are mostly oriented toward the cytosolic membrane leaflet, allowing them to act as signals at the surface of compartments. Third, thanks to their anionic headgroups, they act as docking platforms for lipid-binding proteins, either through direct interaction with stereospecific lipid-binding domains or through electrostatic interactions with polybasic patches in proteins (Noack and Jaillais, 2020; de Jong and Munik, 2021). Fourth, they can be quite diverse, from PA and PS to several different species of phosphoinositides (i.e. PI3P, PI4P, PI5P, PI(3,5)P2 and PI(4,5)P2 in plants). This diversity in anionic lipid species potentially provides distinct lipid landmarks in each compartment. Fifth, most of these lipid species are interconvertible, in particular the phosphoinositides, which allows their rapid modification during membrane trafficking (Noack and Jaillais, 2017). Together, these attributes led to the formulation of the “lipid code hypothesis” (Kutateladze, 2010). This hypothesis proposes that each membrane compartment is enriched in a specific set of anionic lipids, which together represent an “identity signature” for this organelle. The idea of a “code,” as proposed in the histone code hypothesis, is supported by the existence of writer (i.e. kinases), eraser (i.e. phosphatases or phospholipases), and reader (i.e. stereospecific lipid-binding domain) modules. According to this model, proteins with stereospecific lipid-binding domains (reader module) localize to a specific compartment by registering the localization of their cognate lipid. In this model, certain anionic lipids are described as master regulators of specific compartments. For example, in animal cells, PI4P and PI3P are proposed to be major landmarks for Golgi and early endosome identity, respectively.
Although useful, this model is likely an oversimplification (Wang et al., 2019). First, lipid-binding domains are usually sensitive to several membrane features, which include their lipid ligand and also biophysical membrane properties, such as curvature, electrostatics, or lipid packing (Lemmon, 2008). Besides, these proteins often interact with additional membrane components, which can be other lipids and also proteins. Anionic lipids are therefore part of coincident signals for membrane localization. Furthermore, anionic lipids are not as organelle-specific as initially advertised (Wang et al., 2019). For example, in animal cells, PI4P was initially described as mainly being present in the Golgi. However, the use of immunolocalization and new lipid-binding sensors revealed that PI4P is also present at the plasma membrane and in endosomes (Hammond et al., 2009, 2014). Most importantly, a rigid identity code concept is unlikely because anionic lipids are highly dynamic. They can be produced or erased from compartment membranes in an acute manner, and their respective concentration varies according to the signaling activities of cells or their differentiation status.
In this update, we propose that on top of the classical lipid code model, we should overlay the concept of anionic lipid gradients as a key component of compartment identity and membrane dynamics. We review the evidence for the existence of such lipid gradients in plant cells and discuss their potential function in regulating lipid homeostasis and organelle biology.
Lipid gradients between membrane territories fuel lipid exchanges at membrane contact sites
The concept of membrane territories
Bigay and Antonny (2012) proposed that animal cells can be divided into two broad cellular territories (Figure 1). The first territory corresponds to ER-derived membranes and includes the ER, the nuclear envelope, and the cis-Golgi. Membranes from this territory are defined by their low electrostatic property (i.e. low abundance of anionic lipids on their cytosolic leaflets) and their loose lipid packing. This territory was coined the “territory of lipid packing defects” (Bigay and Antonny, 2012). The second territory corresponds to post-Golgi membranes and includes the trans-Golgi, the TGN, the plasma membrane, endosomes, and lysosomes. This territory is defined as the “territory of electrostatics” and by opposition to the territory of lipid packing defects, its membranes are electronegative (i.e. higher abundance of anionic lipids on their cytosolic leaflets) and have a more ordered lipid packing (Bigay and Antonny, 2012). The concept of membrane territories might have important functional implications. Indeed, loose lipid packing is more favorable to the translocation of protein across the membrane, which is relevant for the function of the ER (Holthuis and Menon, 2014). By contrast, a tighter lipid packing, together with higher electrostatic properties, will decrease membrane permeability, consistent with the barrier function of the plasma membrane (Bigay and Antonny, 2012). Besides, Bigay and Antonny (2012) proposed that certain combinations of membrane physicochemical properties could be “dangerous” because they would lead to the promiscuous recruitment of soluble peripheral proteins. This is notably the case, at least in vitro, when liposomes contain a high concentration of anionic lipids and large lipid packing defects. For example, peripheral proteins tend to bind promiscuously and strongly to liposomes made of pure C18:1-C18-1 PS (DOPS), in which every lipid carries a negative charge and two kinked acyl chains (Schulz et al., 2009; de Saint-Jean et al., 2011; Bigay and Antonny, 2012). By contrast, the recruitment of peripheral proteins tends to be inhibited when membranes are not electrostatic but tightly packed. Thus, the division of the cell in the two membrane territories discussed above seems to favor a certain degree of specificity and also leaves room for regulation (Bigay and Antonny, 2012). A consequence of this general organization is the creation of sharp lipid gradients between the two membrane territories. Using knowledge from animal and yeast systems, we will detail below how these gradients are established and also how they are used by the cells to fuel exchanges between the two membrane territories.
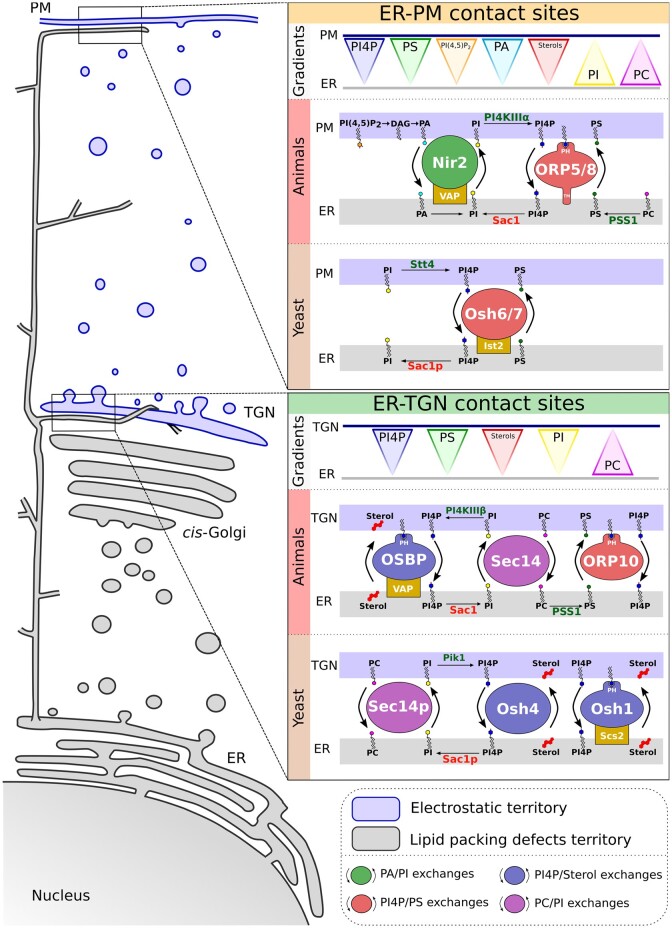
Figure 1.
Schematic representation of the two membrane territories and gradient-powered lipid exchanges at ER–plasma membrane and ER–TGN contact sites in yeast and animal cells. On the left, the electrostatic and the lipid packing defects territories are represented in blue and gray, respectively. On the right, ER–plasma membrane and ER–Golgi lipid gradients and lipid exchanges at contact sites in yeast and animal cells. Lipid transfer proteins that counter transport PA/PI are in green, PI4P/sterol are in lavender, PI4P/PS are in coral, and PC/PI are in mauve. Cellular compartment abbreviations: PM, plasma membrane. Protein abbreviations: Nir2, N-terminal domain-interacting receptor 2; PI4KIIIα/β, class III phosphatidylinositol 4-kinase alpha and beta; VAMP, vesicle-associated membrane protein; VAP, VAMP-associated protein; Ist2, increased sodium tolerance protein 2; Scs2, choline sensitivity suppressor 2; Sac1, suppressor of actin 1; sec14, secretion 14 protein; ORP, oxysterol-related protein; Osh, OSBP homology; PSS1, PS synthase 1; Stt4, staurosporine and temperature-sensitive 4; Pik1, phosphatidylinositol 4-kinase 1. Protein domain abbreviations: PH, pleckstrin homology; TM, transmembrane region.
PI4P gradients fuel cholesterol export from the ER to the Golgi in animal and yeast cells
Anionic lipids segregate within the endomembrane system by the localized action of enzymatic activities (e.g. the combinatorial subcellular localization of the phosphoinositide phosphatases and kinases is critical for the segregated membrane localization of their products) and membrane exchanges between compartments. For a long time, these exchanges were thought to be mainly mediated through vesicular transport. However, lipid research took a turning point in recent years by appreciating the central role of lipid transfer at membrane contact sites (Mesmin et al., 2019). Membrane contact sites refer to the tethered proximity (10–80 nm) between two membrane-bound organelles in the absence of fusion, with a specific function and a defined lipidome/proteome (Scorrano et al., 2019).
For this review, we will mainly concentrate on non-vesicular counter-transport exchanges of lipids at ER–Golgi (Mesmin et al., 2019) and ER–PM contacts (Saheki and De Camilli, 2017). Before its role in lipid exchanges at contact sites was revealed, it was thought that PI4P was primarily used as a precursor for PI(4,5)P2 synthesis, which is the target of the G-protein activated phospholipase C (PLC) at the plasma membrane, an important signaling process in metazoans (Balla, 2013). Observations in yeast showed that suppressor of actin1 (Sac1), an ER-localized PI4P phosphatase that dephosphorylates PI4P into PI, influences the pool of PI4P at the plasma membrane (Foti et al., 2001). Furthermore, sac1 yeast mutants show abnormal augmentation of cellular PI4P, PI3P, and PI(3,5)P2, but no significant augmentation of PI(4,5)P2 (Guo et al., 1999). These observations indicated that PI4P seemed to have other functions than being only the precursor of PI(4,5)P2, which was also confirmed using acute depletion of PI4P, PI(4,5)P2, or both at the plasma membrane (Hammond et al., 2012).
The elegant demonstration from Bruno Antonny’s group that human oxysterol-binding protein (OSBP) can drive the transport of cholesterol from the ER to the Golgi at ER–Golgi contact sites through coupled counter-transport of PI4P opened new outlooks on PI4P distribution and function in the cell (Mesmin et al., 2013; Figure 1). In this model, OSBP binds to the ER proteins vesicle-associated membrane proteins (VAPs) via its FFAT (diphenylalanine in an acidic tract) motif and the PI4P-enriched TGN membrane via its PH domain (Figure 2, A). As such, OSBP can be seen as a tethering molecule that drives the close apposition between the ER and the Golgi. In addition, the density and orientation of the intrinsically disordered N-terminus of OSBP controls the mobility of molecules within ER–Golgi contacts and the geometry of these contacts (Jamecna et al., 2019; Rosado and Bayer, 2021). The lipid transport domain of OSBP can bind as a cargo either PI4P or cholesterol (Mesmin et al., 2013). In this model, the lipid transport domain is more likely to bind to and extract cholesterol from the ER membrane, because PI4P is maintained at a low level through the action of Sac1 in this membrane. Once it arrives at the Golgi membrane, the cholesterol-loaded lipid transport domain releases its cholesterol ligand in exchange for PI4P. Indeed, on the Golgi membrane, PI4P is maintained at a high level by the action of two PI 4-kinases (PI4K), the type-III PI4Kβ (hereafter referred to as PI4KIIIβ) and, to a lesser extent, the type-II PI4Kα (PI4KIIα; Mesmin et al., 2017). The PI4P-loaded lipid transport domain will then transport PI4P back from the Golgi to the ER membrane, where it is dephosphorylated by Sac1 (Figures 1, 2, A). This cycle of exchanges between the ER and the TGN is therefore dependent on the PI4P gradient established by the coordinated action of Sac1 and PI4Ks (Mesmin et al., 2013; Figures 1, 2, A). Eventually, PI4P will be locally consumed from the Golgi membrane, which will induce the release of the PI4P-binding PH domain of OSBP and a relaxed tethering between the ER and Golgi membranes.
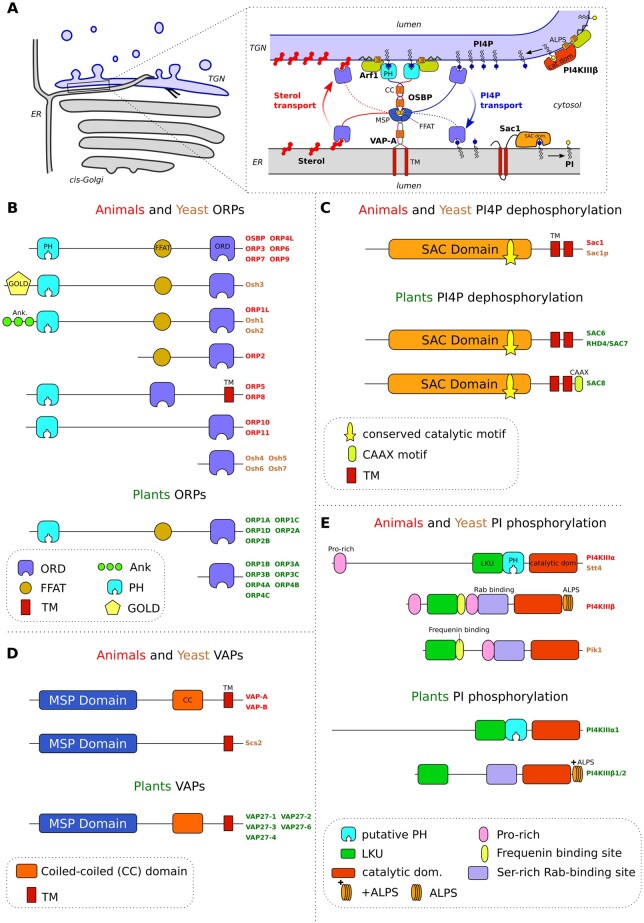
Figure 2.
ORPs, Sac1, VAPs, and PI4KIIIs in animals and yeast and their structural homologs in plants. (A) Model for OSBP-mediated cholesterol/PI4P transfer at ER/TGN contact sites in humans with some functional domains found in ORP, SAC1, VAP, and PI4K indicated. The schematic of lipid transfer is inspired by Antonny et al. (2018). (B) Animal (Homo sapiens) and yeast (S. cerevisiae) ORPs and structural homologs in plants. (C) Animal (H. sapiens) and yeast (S. cerevisiae) Sac1 proteins and structural homologs in plants. (D) Animal (H. sapiens) and yeast (S. cerevisiae) VAPs and structural homologs in plants. (E) Animal (H. sapiens) and yeast (S. cerevisiae) PI4KIIIs and structural homologs in plants. Abbreviations: OR dom., oxysterol-related domain (function in lipid transfer); FFAT, two phenylalanine in an acidic tract domain (interacts with VAP); TM, transmembrane domain; PH, pleckstrin homology domain (interaction with lipids, and with ARF1 in some cases); CAAX, putative prenylation site (lipid anchoring); MSP, major sperm protein domain (interaction with FFAT motif); ALPS, amphipathic lipid packing sensors domain (curvature sensor); +ALPS, amphipathic lipid packing sensors with adjacent cationic residues. The following domains are shown for comparison purposes between different family members but their functions are not discussed in this review: Pro-rich, proline-rich domain; Ser rich, serine-rich domain. Ank, Ankyrin domain; GOLD, Golgi-localization domain; LKU, lipid kinase unique domain.
In vitro, Sac1 works in cis not trans (i.e. it dephosphorylates PI4P in the membrane in which it localizes, but cannot dephosphorylate PI4P in the second membrane within the contact site; Mesmin et al., 2013). In vivo, Sac1 has no activity in trans unless a linker is added between its transmembrane and catalytic domains (Zewe et al., 2018). However, recent work shows that the PI4P-binding protein four-phosphate-adaptor protein 1 (FAPP1), simultaneously binds VAP and Sac1 and stimulates the in trans phosphatase activity of the latter toward its substrate at the TGN (Venditti et al., 2019). Furthermore, Venditti et al. (2019) propose that FAPP1-mediated stimulation of Sac1 in trans only occurs at the level of tighter contact sites and only in domains with high PI4P concentration. Thus overall, Sac1 degrades PI4P in the ER to maintain a steep PI4P gradient with the donor membranes (Zewe et al., 2018) but may locally act in trans thanks to “helper” proteins such as FAPP1 (Venditti et al., 2019).
Note that the sterol/PI4P transport system is conserved in yeast, with the protein Osh1p acting similarly as OSBP (Loewen et al., 2003; Manik et al., 2017; Antonny et al., 2018; Figures 1, 2). Osh4p (also known as Kes1p), also countertransports ergosterol/PI4P between the ER and the Golgi (de Saint-Jean et al., 2011; Stefan et al., 2011; Moser von Filseck et al., 2015; Antonny et al., 2018; Figure 1). Like all Osh proteins, Osp4p has an OSBP-related (OR) domain, but unlike OSBP and Osh1p, it lacks a FFAT motif and a PH domain and is instead proposed to shuttle between the ER and Golgi membrane (Moser von Filseck et al., 2015; Figure 2, B).
PI4P gradients are required for PS localization in animal and yeast cells
This PI4P-sterol counter-exchange model highlights the importance of lipid gradients in the endomembrane system (Antonny et al., 2018). Increasing evidence suggests that gradient-fueled lipid exchanges at membrane contact sites are an evolutionarily conserved mechanism in eukaryotic cells. Indeed, these processes are found at membrane contact sites between different organelles and can involve different lipid substrates implicated in the maintenance of the physicochemical properties of membranes (Balla et al., 2019; Prinz et al., 2020). A striking example is the PS-PI4P counterflow at ER–plasma membrane contacts (Prinz et al., 2020; Figure 1). PS is synthesized at the ER in eukaryotic cells, like sterols, but is found in much greater abundance at the plasma membrane (Yeung et al., 2008; Fairn et al., 2011). PS is a hallmark lipid of the plasma membrane and is a major actor of its electrostatic signature allowing the recruitment of specific effectors (Yeung et al., 2008; Platre and Jaillais, 2017). The Gavin group showed that yeast Osh6p and Osh7p bind PS (not sterols) through their OR domains and are responsible for PS transport from the ER to the plasma membrane (Maeda et al., 2013). Further research then showed that Osh6p/Osh7p exchange PS and PI4P between membranes in vitro and in vivo (Moser von Filseck et al., 2015). Thus, the coordinated action of Sac1p at the ER and the PI4KIII Stt4p at the plasma membrane sustains a PI4P gradient at the ER/plasma membrane interface which maintains a directional transport of PS to the cell surface (Figure 1). Similar to Osh4p, Osh6p/Osh7p lacks a PH domain and FFAT motif (Figure 2, B). Nonetheless, they both localize to ER/plasma membrane contact sites through interaction with Ist2p (D'Ambrosio et al., 2020), one of the key tethers in yeast that drives the close apposition of these two membranes (Manford et al., 2012; Figure 1). Parallel studies showed that this process is conserved in human cells at both the ER–plasma membrane and the ER–Golgi contacts (Figure 1). Similar to OSBP at ER/Golgi contacts, ORP5 and ORP8 act as tethers between the ER and the plasma membrane (Chung et al., 2015). They are integral membrane proteins with a transmembrane domain integrated into the ER membrane (Figures 1, 2, B). Furthermore, they dock the ER with the plasma membrane via their PH domain, which binds plasma membrane PI4P and PI(4,5)P2 (Chung et al., 2015; Sohn et al., 2018). ORP5 and ORP8 transport PS from the ER to the plasma membrane through PS/PI4P counter-exchanges allowed by the PI4P gradient maintained by ER Sac1 and plasma membrane PI4KIIIα (Chung et al., 2015; Sohn et al., 2016, 2018; Balla et al., 2019; Figure 1). Besides, human ORP10 also transports PS/PI4P between the ER and the TGN (Venditti et al., 2019; Figure 1).
Gradient-dependent lipid transfer is required to regulate PI homeostasis
All of these processes indicate that PI4P has a central role in lipid exchange at membrane contact sites. Considering that PI4P is created from PI by PI4K-mediated ATP consumption, the analogy with other ATP-dependent transport mechanisms, like molecule transport by ion pumps against concentration gradients, is tempting (Antonny et al., 2018). It is estimated that in human cells, OSBP, at a stationary state, consumes up to 50% of cellular PI4P for sterol transport at the TGN (Mesmin et al., 2017; Antonny et al., 2018). Importantly, the presence of PI4KIIIα at the plasma membrane is not limiting for the production of PI4P. It appears that instead, it is the presence of PI in this membrane that is highly regulated and rate limiting for the production of PI4P (Pemberton et al., 2020; Zewe et al., 2020). Indeed, PI does not accumulate at the plasma membrane (Figure 1), likely because it is immediately phosphorylated into PI4P. Interestingly, PI is produced in the ER, where it accumulates to some extent, but it is mainly found in the cytosolic face of the Golgi, peroxisomes, and mitochondria (Pemberton et al., 2020; Zewe et al., 2020; Figure 1). SEC14 domain-containing proteins, which are lipid transfer proteins, have been involved in PI transfer from the ER to the Golgi (Wang et al., 2019). The SEC14 domain binds both PI and PC and thus might countertransport both lipids (Schaaf et al., 2008; Figure 1). The SEC14 domain is also thought to extract PI from the membrane to present it to PI4K, which might facilitate its phosphorylation into PI4P (de Campos and Schaaf, 2017; Noack and Jaillais, 2020). Besides, PI reaches the plasma membrane via the Nir2 lipid transfer protein, which countertransports PA from the plasma membrane to the ER membrane (Kim et al., 2015; Figure 1). As mentioned above, PI accumulates, at least to some extent, on the cytosolic face of the ER. Because it is negatively charged and relatively abundant, this appears as paradoxical with the idea that the cytosolic leaflet of the ER is electroneutral (Bigay and Antonny, 2012). However, PI is not present throughout the ER, but only in very specialized ER-subdomains (Kim et al., 2011). Besides, the charges generated by PI may be locally shielded by interaction with proteins.
Altogether, the examples described in this section clearly show that lipid gradients between the territory of lipid packing defect and the territory of electrostatics are key to power lipid exchanges and to regulate membrane lipid homeostasis. These gradients appear to often depend upon each other, further illustrating the elegant intricacy and spatial regulation of lipid metabolism (Figure 1).
Evidence for gradient-powered lipid exchanges in plants
A first preprint recently established the importance of ER/plasma membrane contacts for diacylglycerol (DAG) homeostasis in Arabidopsis (Arabidopsis thaliana; Ruiz-Lopez et al., 2020). This transfer of DAG is mediated by Synaptotagmin1 (SYT1) and SYT3 notably during cold stress episodes (Ruiz-Lopez et al., 2020). However, there is very little direct experimental evidence for the exchanges of anionic phospholipids at membrane contact sites in plants. Nonetheless, it is reasonable to assume that the exchanges of anionic lipids at membrane contacts are likely happening. First, the two-membrane territory model is conserved in Arabidopsis. While probes to measure packing defects are still lacking, the notion that the plant ER has a loose lipid packing compared with the plasma membrane is substantiated by their respective lipid composition. Indeed, like in animal and yeast systems, the plant ER is rich in unsaturated lipids and poor in sterols, which favor lipid packing defects. By contrast, the plant plasma membrane is rich in phytosterols and sphingolipids (Mamode Cassim et al., 2019). Sterols interact with sphingolipids containing saturated chains, which increases lipid packing (Boutté and Jaillais, 2020). Furthermore, a set of genetically encoded biosensors with a defined number of positive charges allowed mapping of the electrostatic landscape of endomembrane compartments in Arabidopsis (Simon et al., 2016; Platre et al., 2018). This showed that ER-derived membranes are not electrostatic, while post-Golgi membranes are (Platre et al., 2018). These results are coherent with the presence of anionic lipids in post-Golgi but not ER-derived membranes in plants (Vermeer et al., 2006, 2009; van Leeuwen et al., 2007; Simon et al., 2014). Thus, sharp gradients of anionic lipids exist between the territory of lipid packing defects and the territory of electrostatics in plant cells.
Furthermore, the lipid counter flow mechanisms described above are conserved from yeast to humans (Balla et al., 2019; Prinz et al., 2020), making it a real possibility that they also exist in plants. Most of the enzymes involved in the generation of anionic lipid gradients are conserved in plants (Heilmann, 2016; Noack and Jaillais, 2020; Figure 2). This is notably true for the PI4Ks, with the Arabidopsis genome encoding two PI4KIIIIβs and one PI4KIIIIα (Mueller-Roeber and Pical, 2002; Heilmann, 2016; Figure 2, E). The Arabidopsis genome contains three putative orthologs of Sac1 called SAC6 (also called Sac1b), SAC7 (also called Sac1c and ROOT HAIR DEFECTIVE4), and SAC8 (also called Sac1a; Despres et al., 2003; Thole et al., 2008; Heilmann, 2016; Figure 2, C). All three SACs complement the yeast sac1 mutant suggesting that they are likely PI4P phosphatases (Despres et al., 2003). Furthermore, SAC7 specifically hydrolyzes PI4P in vitro (Thole et al., 2008). Arabidopsis SAC6, SAC7, and SAC8 all localize to the ER when expressed in tobacco (Nicotiana tabacum cv BY-2) BY-2 cells and fused to GFP at their C-terminus, suggesting that they could indeed be functional homologs of yeast and animal Sac1 (Despres et al., 2003). However, SAC7 localizes at the TGN, not the ER, in Arabidopsis root hairs when YFP is fused at its N-terminus (Thole et al., 2008). YFP-SAC7 complements the sac7 loss-of-function phenotype (Thole et al., 2008), showing that this fusion is functional and questioning whether SAC7 is indeed acting at the ER in plants. Future experimental validations are thus clearly required to address whether Arabidopsis SAC6, SAC7, and/or SAC8 could act similarly as Sac1 in other eukaryotes. Plant genomes also encode for 12 proteins with an OR domain that could transport lipids, including PI4P (Umate, 2011; Ye et al., 2020; Figure 2, B). The Arabidopsis ORP3a protein localizes in the ER when transiently expressed in Nicotiana benthamiana and binds to sterol (Saravanan et al., 2009). However, it is unknown whether it can counter transport PI4P, like OSBP in human cells. Plants also have functional homologs of VAP, called the VAP27 family, that localize at membrane contact sites (Wang et al., 2014, 2016, 2017; Stefano et al., 2018; Figure 2, D). Finally, AtSac1, AtORP3a, and AtVAP27s are involved in rearranging ER-peroxisome membranes to accommodate the replication of positive-strand RNA viruses when expressed heterologously in N. benthamiana or yeast cells (Barajas et al., 2014; Sasvari et al., 2020). Altogether, the presence of these proteins and their preliminary functional characterization suggest that gradient-powered anionic lipid exchanges at membrane contacts likely occur in plants. Of note, we could not find clear orthologs of the PI/PA transporter Nir2 in plants. The absence of Nir2 might be functionally compensated by the presence of many Sec 14 homolog (SFH) proteins, some of which localize at the plasma membrane and cotransport both PI and PC, like their yeast counterpart Sec14p (Vincent et al., 2005; Ghosh et al., 2015; Huang et al., 2016; de Campos and Schaaf, 2017). In any case, the functional importance of gradient-powered lipid exchanges at membrane contact sites remains to be determined, notably for lipid cellular patterning and also plant development and environmental interactions.
Lipid gradients within the electrostatic territory
Evidence for the existence of anionic lipid gradients within the electrostatic territory
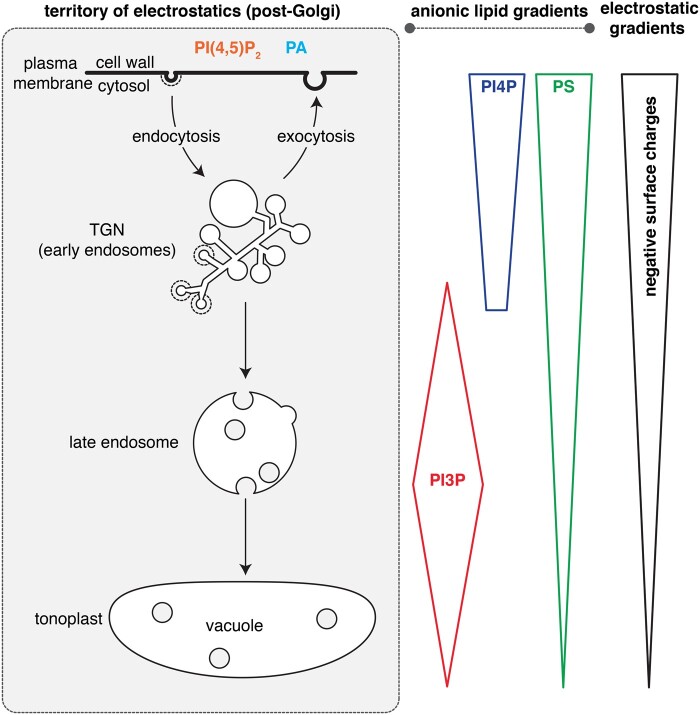
Mapping of the subcellular accumulation of anionic lipids within the plant endomembrane system revealed that they mostly localized in post-Golgi compartments (i.e. corresponding to the electrostatic territory; Vermeer et al., 2006, 2009; van Leeuwen et al., 2007; Simon et al., 2014, 2016; Noack and Jaillais, 2020). However, within the electrostatic territory, several anionic lipids do not have compartment-specific localization. Rather, they accumulate according to concentration gradients (Simon et al., 2014; Platre et al., 2018; Figure 3). For example, this is the case for PI4P, which massively accumulates at the plasma membrane and to a lesser extent at the TGN (Vermeer et al., 2009; Simon et al., 2014, 2016). Similarly, PS accumulates at the plasma membrane but is also present all along the endocytic pathway, including the TGN (which is also an early endosome in plants [Dettmer et al., 2006]), and to a lesser extent the late endosomes and the vacuolar membranes (Platre et al., 2018). PI3P is also present as a gradient within the late endocytic pathway with maximum PI3P concentration in late endosomes and also a presence in the vacuolar membrane (i.e. the tonoplast) and the TGN (Vermeer et al., 2006; Simon et al., 2014; Singh et al., 2014; Figure 3).
Figure 3.
Schematic representation of anionic lipid gradients within the territory of electrostatics.
Analyzing and concluding on lipid localization and accumulation at the subcellular level is difficult because there is no straightforward technique to do it (Platre and Jaillais, 2016). The existence of lipid gradients at the subcellular levels was revealed by the use of fluorescent genetically encoded lipid biosensors in Arabidopsis roots (Simon et al., 2014, 2016; Platre et al., 2018). These sensors correspond to the transgenic expression of stereospecific lipid-binding domains fused to fluorescent proteins and are an indirect way of analyzing lipid localization (Platre and Jaillais, 2016). However, these sensors have limitations, which complicates their analysis (Heilmann, 2016; Platre and Jaillais, 2016). First, they may be outcompeted by endogenous proteins for lipid binding (i.e. the presence of their cognate lipid is locally masked by endogenous proteins). Second, their localization will depend on their affinity for the lipid (i.e. domains with different lipid affinity may have different localization). Third, their localization may rely on additional membrane features, which are not always known beforehand (e.g. coincident detection of other lipids or proteins, the requirement for specific membrane shape or charges). A famous example is the PH domain of FAPP1 and OSBP, which bind to PI4P and were the first domains used as PI4P sensors (Levine and Munro, 1998, 2001). In both yeast and animal cells, PH(FAPP1) and PH(OSBP) localize to the Golgi leading to the conclusion that PI4P mainly accumulates in this compartment. However, these PH domains bind not only PI4P but also the small GTPase ARF1, and the strong Golgi association of this domain is conferred by the coincident detection of both PI4P and ARF1 (Levine and Munro, 2001; Godi et al., 2004). The use of additional biosensors later revealed that PI4P indeed localizes to the Golgi but is not specific to this compartment and is also present at the plasma membrane and in late endosomes (Hammond et al., 2014). This example nicely illustrates the inherent difficulties in using lipid sensors to study lipid localization (Platre and Jaillais, 2016). Thus, the evidence that anionic lipids are present as concentration gradients in post-Golgi compartments in plant cells should be examined with care. Overall, such conclusions should be supported by several independent evidences, which could include additional techniques (e.g. lipid immunolocalization and organelle fractionation followed by lipid measurement; Platre and Jaillais, 2016). Anionic lipid immunolocalization has been used in plant tissues (Tejos et al., 2014; Gerth et al., 2017) but not extensively, and the subcellular localization of PI4P, PI3P, or PS in intracellular compartments has not been analyzed with this technique. Biochemical measurements of anionic lipids have been performed on whole plants or cell extracts but not on purified compartments (Konig et al., 2008; Munnik and Zarza, 2013).
However, results obtained with one lipid biosensor can also be validated by using additional, independent lipid-binding domains, and such experiments were carried out in plants. Strikingly, when different biosensors for the same cognate lipid were expressed in transgenic Arabidopsis, they showed somewhat different subcellular localizations (Simon et al., 2014, 2016; Platre et al., 2018). Given the fact that these domains likely have different affinities for their cognate lipid and may also recognize additional membrane features, this result is not surprising but further exemplifies the difficulty in using sensors for localizing lipids. However, despite these differences, this also allowed building a more robust picture of anionic lipid localization within plant cells (Noack and Jaillais, 2017, 2020). In the case of PI3P sensors, quantitative colocalization with compartment markers showed that two independent PI3P-binding domains (the FYVE domain of the mouse hepatocyte growth factor-regulated tyrosine kinase substrate [HRS] protein and the PHOX domain of the human p40 protein) both localized to late endosomes, with a weaker association to the TGN and tonoplast also recorded (Simon et al., 2014). Similarly, the gradient of PS from the plasma membrane and along the endocytic pathway was confirmed with two independent PS-binding domains, the C2 domain of the bovine Lactadherin (Lact) protein and the PH domain of the human EVECTIN2 (EVCT2) protein (Platre et al., 2018). However, the C2(Lact) association with the plasma membrane was more pronounced than that of the PH(EVCT2) domain (Platre et al., 2018), a difference in localization that is conserved in animal cells (Yeung et al., 2008; Uchida et al., 2011; Chung et al., 2015). This difference likely reflects different PS affinity between these two domains and/or their reliance on additional membrane features. In any case, the fact that different lipid-binding domains, with apparently different properties, consistently show the existence of a PS gradient at the subcellular level in plants suggests that such a gradient might indeed exist (Figure 3).
The localization of PI4P was also the subject of intense experimental validation. As mentioned above, early work in yeast and animals using the PH(FAPP1) and PH(OSBP) sensors suggested that the main localization (and assumed function) of PI4P was at the Golgi (Levine and Munro, 1998, 2001). By contrast to yeast and animal cells, PH(FAPP1) not only localizes to intracellular compartments but also the plasma membrane when expressed in plant cells (Vermeer et al., 2009). Using colocalization in cowpea (Vigna unguiculata L.) protoplasts, the intracellular compartments labeled by PH(FAPP1) were initially thought to be Golgi (Vermeer et al., 2009). However, quantitative colocalization in Arabidopsis roots later revealed that they mostly correspond to TGN rather than Golgi (Simon et al., 2014), which in plants are two distinct compartments (Dettmer et al., 2006; Kang et al., 2011). The prominent plasma membrane labeling of PH(FAPP1), when expressed in plants, could suggest that (i) this domain does not naturally interact with the plant ARF1 GTPase or (ii) that PI4P massively accumulates at the plasma membrane compared with other eukaryotic organisms. Mutant versions of the FAPP1 PH domain, which do not interact with ARF1 anymore but retain their ability to interact with PI4P, are strongly targeted to the plasma membrane and only marginally to the TGN in both N. benthamiana leaves and Arabidopsis roots (Simon et al., 2016; Ito et al., 2020). This suggests that PH(FAPP1) indeed interacts with the plant ARF1 and that this interaction contributes to its TGN localization (likely as coincident detection between PI4P and ARF1). By extension, this result suggests that there is quantitatively a minor pool of PI4P in the TGN compared with the plasma membrane in plants, which opposes what has been found in animal cells. This massive accumulation of PI4P at the plant plasma membrane is further validated by additional sensors, which do not interact with ARF1, including the P4M domain from the Legionella pneumophila SidM protein (Simon et al., 2016). This evidence is also substantiated by genetic experiments. First, P4M relocalizes to intracellular compartments upon depletion of PI4P specifically at the plasma membrane (Simon et al., 2016; Platre et al., 2018). This demonstrates that the specific localization of P4M at the plasma membrane is not due to its inability to interact with the TGN membrane—for example, because it is more curved or because PI4P is shielded by endogenous proteins—but rather because the pool of PI4P at the TGN is much smaller than at the plasma membrane (Doumane and Caillaud, 2020). Similar results were obtained with the PH(FAPP1) sensor with an increased PI4P localization in intracellular compartments upon depletion of PI4P specifically at the plasma membrane, again suggesting that interfering with the balance of PI4P between the TGN and the plasma membrane directly impact the localization of the sensor (Simon et al., 2016). Finally, there is also evidence—which will be detailed in the next paragraph—that the PI4P gradient between the plasma membrane and the TGN is actively maintained and which further substantiates the existence of this gradient.
Maintenance of anionic lipid gradients
At the mechanistic level, how the anionic lipid gradients within the electrostatic territory of plant cells are established and maintained is only partially understood. Indeed, it is unclear to what extent the mechanisms described in other eukaryotes can be applied to plant cells. For example and as mentioned above, we currently do not fully appreciate the importance of membrane contact sites in patterning anionic phospholipids accumulation in plant cells.
In yeast, the pools of PI4P at the plasma membrane and in the Golgi are believed to be largely independent (Roy and Levine, 2004). However, it is unclear whether this is also valid for plant cells. In yeast, there are two PI4Ks, Pik1p, and Stt4p, which localize at the Golgi and plasma membrane, respectively (Figure 1). A PI4P biosensor consisting of a tandem dimer of PH(Osh2) localizes at the plasma membrane and in the Golgi (Roy and Levine, 2004). In the temperature-sensitive pik1 mutant and at restricting temperature, 2×PH(Osh2) localizes only at the plasma membrane, suggesting that Pik1p is responsible for the production and maintenance of the Golgi pool of PI4P (Roy and Levine, 2004). Conversely, in the temperature-sensitive stt4 mutant and at restricting temperature, 2×PH(Osh2) localizes in the Golgi but not the plasma membrane. This result suggests that Stt4p is involved in the production of PI4P at the plasma membrane but not in the Golgi (Roy and Levine, 2004). Together, these results suggest that the PI4P pools are largely independent in yeast. In plants, two PI4Ks (PI4KIIIβ1 and PI4KIIIβ2) that localize in the TGN have been identified (Preuss et al., 2006; Kang et al., 2011). They act redundantly to regulate TGN morphology and function (Preuss et al., 2006; Kang et al., 2011). The corresponding double mutant do not show a significant reduction in overall PI4P levels. Nonetheless, quantitative analyses revealed a slight decreased of the PH(FAPP1) sensor targeting to the TGN (Lin et al., 2019). Taken together these results suggest that PI4Kβs are involved in PI4P production at the TGN in plants and also confirm that this compartment accumulates only a minor pool of this lipid compared to the plasma membrane. There is a plant ortholog of Stt4p, PI4KIIIα1, which is an essential gene (Delage et al., 2012) and localizes at the plasma membrane (Noack et al., 2020). However, it is involved in PI4P production in chloroplast and its importance for PI4P homeostasis at the plasma membrane and the TGN are currently unknown (Okazaki et al., 2015).
In addition to the localized production of PI4P by kinases, localized PI4P breakdown appears critical to maintaining the PI4P gradients (Noack and Jaillais, 2020). Indeed, as previously mentioned, the PI4P phosphatase SAC7/RHD4—an ortholog of mammalian and yeast Sac1 (Figure 2)—localizes at the TGN at the tip of root hairs (Thole et al., 2008). In the rhd4 mutant, the PH(FAPP1) sensor accumulates more strongly in this compartment than in the wild type (Thole et al., 2008). This suggests that the sharp PI4P gradient between the plasma membrane and the TGN is actively maintained by local phosphatase activities directly in the TGN, at least in this cell type. Furthermore, phosphoinositide-specific PLCs, which use PI(4,5)P2 and also PI4P as substrates, are important to maintain the proper PI4P balance between the TGN and the plasma membrane. Indeed, a recent preprint suggests that the pharmacological inhibition of PLC activity increases the pool of PI4P at the TGN (Ito et al., 2020). A subcellular proteomic analysis found that two PI-PLCs, PLC2 and PLC7, localize at the TGN and that this localization is dependent upon the chain length of sphingolipids. Thus, this preprint suggests that sphingolipids at the TGN play a critical role in the establishment of the PI4P gradient via the recruitment of PI-PLCs and thus the local consumption of PI4P at the TGN (Ito et al., 2020).
While it is likely that a combination of localized production and breakdown is involved in the establishment of the phosphoinositide cellular gradients, this is probably not the case for PS. Indeed, in Arabidopsis, PS is produced in the lumen of the ER by a single enzyme called PS SYNTHASE1 (PSS1; Yamaoka et al., 2011; Platre et al., 2018). Thus, PS has to flip to the cytosolic membrane leaflets via flippases and needs to be transported to the plasma membrane and endosomal compartments (Noack and Jaillais, 2020). Several PS flippases have been identified in Arabidopsis and are involved in the plasma membrane and endosomal accumulation of PS (Nintemann et al., 2019; Zhang et al., 2020; Zhou et al., 2020). However, in plants, it is still unclear whether PS are mostly transported by vesicular trafficking and/or by lipid transfer at membrane contact sites (see above) and whether these pathways are important to control the cellular PS gradient. There is one pathway for PS breakdown via PS DECARBOXYLASEs (PSDs), which decarboxylate PS into PE (Noack and Jaillais, 2020). However, the importance of these enzymes in PS homeostasis and subcellular patterning within the plant endomembrane system is unclear. Indeed, the Arabidopsis genome encodes three PSD genes and the corresponding triple psd mutant completely lacks PSD activity (Nerlich et al., 2007). Unlike the Arabidopsis pss1 mutant (Yamaoka et al., 2011; Liu et al., 2013; Platre et al., 2018, 2019), the psd triple mutant does not show any visible developmental phenotypes (Nerlich et al., 2007). Furthermore, it seems to mostly affect the relative accumulation of PE and PS in mitochondria. This is surprising since only PSD1 is found in mitochondria in transiently transfected N. benthamiana cells, while PSD2 and PSD3 localize in the tonoplast and ER, respectively (Nerlich et al., 2007). Overall, the mechanisms behind the establishment of the cellular PS gradient in plants remain to be investigated.
Functional implications of lipid gradients within the electrostatic territory
The fact that PI4P and PS, which are among the most abundant anionic lipids, are organized according to concentration gradients that are highest at the plasma membrane, intermediate in the TGN, and low in late endosomes and the tonoplast has a direct consequence on the landscape of the electrostatic territory. Indeed, quantitative colocalization between biosensors with varying net positive charges and compartment markers reveals that within the electrostatic territory, not all membranes have a similar electrostatic field (Simon et al., 2016; Platre et al., 2018; Figure 3). These analyses show that the plasma membrane is the most electrostatic cytosolic-facing membrane in plant cells (Simon et al., 2016). The TGN has an intermediate electrostatic property, while late endosomes and the tonoplast are weakly electrostatic (Platre et al., 2018). Thus, the amount of negative membrane surface charges corresponds to the gradients of PI4P and PS. In other words, the PI4P and PS gradients are directly translated into an electrostatic gradient along the plant endocytic pathway (Figure 3). As a result, peripheral proteins with very strongly cationic regions (e.g. >8 net positive charges) tend to localize specifically at the plasma membrane, owing to its very high electronegativity (Simon et al., 2016). By contrast, proteins with lower cationic regions (e.g. three–six net positive charges) will have a broader localization within the electrostatic territory by localizing both to the plasma membrane and the TGN (Simon et al., 2016). The coincident detection of additional membrane features might provide further localization specificity. For example, the Arabidopsis PI4Kβs have a +ALPS domain at their C-terminus (Platre et al., 2018; Figure 2). This motif recognizes a combination of curved and electrostatic membranes, which corresponds to the TGN, the compartment where PI4Kβs are located (Platre et al., 2018).
As mentioned in the “Introduction” section, anionic lipids act as landmarks in the membrane in which they reside (Noack and Jaillais, 2017). As such, anionic lipids are mostly believed to act locally in these membranes. However, the fact that anionic lipids are organized in gradients within the electrostatic territory suggests that they may also impact membrane properties at a distance. Indeed, local variations in a given lipid are expected to impact the localization of proteins that binds this lipid within that membrane. However, such variations will also affect the relative balance of this lipid between compartments. In other words, a local variation in the concentration of one lipid will also impact the sharpness of the lipid gradient between compartments. As such, this could impact the localization of proteins at a distance. As a theoretical example, we will consider a PI4P binding protein that localizes at the plant plasma membrane due to the high accumulation of PI4P in this membrane. A diminution of PI4P at the plasma membrane will change the balance of PI4P between the plasma membrane and the TGN, as the corresponding gradient will become less sharp. Such PI4P modification might thus induce a concomitant reduction of that protein at the plasma membrane with an increased targeting to the TGN. Therefore, such modification of PI4P at the plasma membrane might impact the membrane composition locally (i.e. at the plasma membrane) and also at a distance (i.e. in the TGN). Conceptually, this is what is observed with lipid sensors that switch localization from the plasma membrane to the TGN upon manipulation of the PI4P plasma membrane pool (Doumane and Caillaud, 2020). However, such “long-range” effects are purely conceptual at the moment, and it is currently not known whether the localization of plant peripheral proteins could be sensitive to the lipid balance between two compartments. Future works are required to determine whether endogenous proteins also behave similarly to lipid sensors and whether this is functionally relevant during signaling and plant development. A prerequisite for such a hypothesis is that anionic lipid gradients are dynamic and not fixed within cells. This appears to be the case, at least for PS. Indeed, the localization of PS biosensors at the plasma membrane compared with the TGN is enhanced in response to the plant hormone auxin, one of the most potent regulators of plant developmental responses (Platre et al., 2019). Besides, the analysis of lipid gradients at the tissue and organ scales supports the notion that anionic lipid gradients within the electrostatic territory are under developmental control (Colin and Jaillais, 2019).
Anionic lipid gradients at the organismal scale
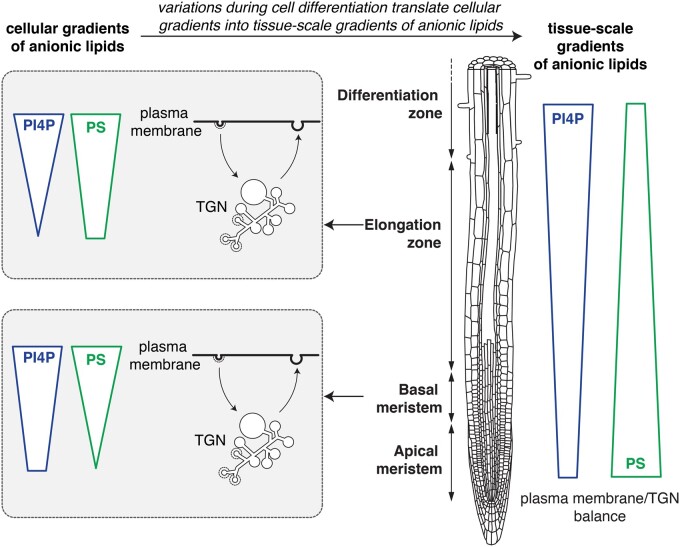
Transgenic lines that stably express anionic lipid biosensors allow analyses of the localization and dynamics of their cognate lipids in many different cell types and during cellular differentiation (Colin and Jaillais, 2019; Figure 4). Such analyses at the organismal level are not often carried out in animals, because most studies express genetically encoded biosensors in cell lines in vitro. Interestingly, the relative accumulation of PS between the plasma membrane and the TGN varies during root epidermis differentiation (Platre et al., 2019). Indeed, two independent PS sensors localize extensively at the plasma membrane but marginally in the TGN in the root meristem (Figure 4). However, in the elongation zone, both sensors partition more evenly between the plasma membrane and the TGN (Figure 4). This analysis suggests that the relative balance of PS between these two compartments is regulated during cell differentiation and that there is not only a cellular gradient of PS but that this gradient can be translated also at the developmental level (Boutté and Jaillais, 2020; Figure 4). While PI4P and PS gradients at the cellular level largely overlap (Figure 3), the concentration of both lipids being high at the plasma membrane and lower in the TGN, PI4P, and PS have opposite gradients at the organ level in the root (Platre et al., 2019). Indeed, the localization of PI4P sensors at the plasma membrane is more strongly pronounced in the elongation zone than in the root meristem (Platre et al., 2019; Figure 4).
Figure 4.
Schematic representation of anionic lipid gradients at the tissue scale. Root schematic was modified from B. Peret: https://figshare.com/articles/Primary_and_lateral_root_ai/5143987.
The observed variation of accumulation of PS at the plasma membrane during epidermis differentiation are likely physiologically relevant. Indeed, PS is rate-limiting for the signaling activity of the Rho GTPase RHO OF PLANTS6 (ROP6) during cell surface auxin signaling (Platre et al., 2019; Smokvarska et al., 2021). A high accumulation of PS at the plasma membrane, like in the meristematic zone, promotes ROP6 nanoclustering, which is required for signaling. By contrast, a lower concentration of PS at the plasma membrane dampens ROP6 signaling by limiting the amount of ROP6 recruited into nanoclusters, even when the concentrations of auxin are high (Platre et al., 2019; Boutté and Jaillais, 2020). It is known that fast auxin responses at the plasma membrane regulate many cellular functions, including cytoskeleton dynamics and intracellular trafficking (Armengot et al., 2016). While some of these responses can be attributed to ROP6 signaling, others may not. In particular, how auxin signaling at the plasma membrane can rapidly regulate TGN functions is unknown. Importantly, the PS variations observed during epidermis differentiation impact not only the PS plasma membrane pool but the very gradient of PS between the plasma membrane and the TGN (Platre et al., 2019). It is thus conceivable that the concept of the “long-range” effects of anionic lipids discussed above could be relevant to understand some of the many coordinated responses triggered by auxin. Again, we would like to emphasize that no endogenous proteins have been shown to date to respond to variations of the PS (or PI4P) gradients between the plasma membrane and the TGN, and therefore such effects are discussed as a conceptual possibility that remains to be experimentally validated.
Outstanding questions
Are lipid counter exchanges at membrane contact sites used to pattern anionic lipid accumulation in plant cells?
To what extent is the molecular machinery used for lipid exchanges in animals and yeasts functionally conserved in plants?
How are anionic lipid gradients within the electrostatic territory established and maintained? And how are these pathways differentially controlled during development and environmental interactions?
Can we find endogenous plant proteins that are sensitive to the balance of anionic lipids between the plasma membrane and the TGN, and which could substantiate the hypothesis of a “long-distance” impact of anionic lipids within the endomembrane system?
Conclusion
Throughout this review, we discussed the existence of gradients of anionic lipids in plants. These gradients exist at the junction of the two main membrane territories and also within the electrostatic territory itself. While evidence for the existence of these gradients exists, they still need to be consolidated using additional biochemical and genetic experiments. Furthermore, we understand very little about the function of these gradients (see Outstanding Questions). To tackle this question, we first need to understand how these gradients are established and maintained. We also need to address whether the gradients of anionic lipids that exist between the ER and electrostatic membranes are involved in the formation of the lipid gradients within the electrostatic territory. However, to date, the importance of lipid exchanges at membrane contact sites is not well understood in plants. Based on evidence from yeast and animal systems, lipid counter exchanges between the ER and the plasma membrane or the TGN are often coupled. For example, the Nir2 system delivers PI at the plasma membrane by counter-transporting PA (Kim et al., 2015; Balla et al., 2019). The presence of PI is then rate-limiting for the production of PI4P by PI4Ks at the plasma membrane (Pemberton et al., 2020; Zewe et al., 2020), but PI4P itself is a critical fuel to export PS out of the ER (Balla et al., 2019). It is thus not only important to consider the presence of anionic lipid gradients but also to envision that these gradients are likely interdependent (see Outstanding Questions; Wang et al., 2019; Prinz et al., 2020). We also discussed the possibility that anionic lipids may impact membrane properties both locally and also at a distance via the existence of gradients within the electrostatic territory. This is a non-intuitive outcome that is not often considered and is so far mainly theoretical. However, we believe that such outcomes should be taken into account in future studies when considering the function of anionic lipids both at the cellular and developmental levels.
Acknowledgments
The authors thank Benjamin Peret (Montpellier, France) for his root template used in Figure 4.
Funding
This work was funded by the French National Research Agency caLIPSO (ANR-18-CE13-0025-02 to Y.J.) and STAYING-TIGHT (ANR-18-CE13-0016-02 to Y.J.).
Conflict of interest statement. None declared.
G.A.D. and Y.J. wrote the manuscript and prepared the figures.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Yvon Jaillais (yvon.jaillais@ens-lyon.fr).
References
- Armengot L, Marques-Bueno MM, Jaillais Y (2016) Regulation of polar auxin transport by protein and lipid kinases. J Exp Bot 67: 4015–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93: 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Kim YJ, Alvarez-Prats A, Pemberton J (2019) Lipid dynamics at contact sites between the endoplasmic reticulum and other organelles. Annu Rev Cell Dev Biol 35: 85–109 [DOI] [PubMed] [Google Scholar]
- Barajas D, Xu K, de Castro Martin IF, Sasvari Z, Brandizzi F, Risco C, Nagy PD (2014) Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog 10: e1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Antonny B (2012) Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell 23: 886–895 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Jaillais Y (2020) Metabolic cellular communications: feedback mechanisms between membrane lipid homeostasis and plant development. Dev Cell 54: 171–182 [DOI] [PubMed] [Google Scholar]
- Boutte Y, Moreau P (2014) Modulation of endomembranes morphodynamics in the secretory/retrograde pathways depends on lipid diversity. Curr Opin Plant Biol 22: 22–29 [DOI] [PubMed] [Google Scholar]
- Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P (2015) PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin LA, Jaillais Y (2019) Phospholipids across scales: lipid patterns and plant development. Curr Opin Plant Biol 53: 1–9 [DOI] [PubMed] [Google Scholar]
- D’Ambrosio JM, Albanèse V, Lipp NF, Fleuriot L, Debayle D, Drin G, Čopič A (2020) Osh6 requires Ist2 for localization to ER–PM contacts and efficient phosphatidylserine transport in budding yeast. J Cell Sci 133: jcs243733 [DOI] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G (2011) Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol 195: 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage E, Ruelland E, Guillas I, Zachowski A, Puyaubert J (2012) Arabidopsis type-III phosphatidylinositol 4-kinases beta1 and beta2 are upstream of the phospholipase C pathway triggered by cold exposure. Plant Cell Physiol 53: 565–576 [DOI] [PubMed] [Google Scholar]
- Despres B, Bouissonnié F, Wu HJ, Gomord V, Guilleminot J, Grellet F, Berger F, Delseny M, Devic M (2003) Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J 34: 293–306 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumane M, Caillaud MC (2020) Assessing extrinsic membrane protein dependency to PI4P using a plasma membrane to endosome relocalization transient assay in Nicotiana benthamiana. Methods Mol Biol 2177: 95–108 [DOI] [PubMed] [Google Scholar]
- Drin G (2014) Topological regulation of lipid balance in cells. Annu Rev Biochem 83: 51–77 [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A 106: 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG (2011) High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol 194: 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD (2001) Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 12: 2396–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth K, Lin F, Daamen F, Menzel W, Heinrich F, Heilmann M (2017) Arabidopsis phosphatidylinositol 4-phosphate 5-kinase 2 contains a functional nuclear localization sequence and interacts with alpha-importins. Plant J 92: 862–878 [DOI] [PubMed] [Google Scholar]
- Ghosh R, de Campos MK, Huang J, Huh SK, Orlowski A, Yang Y, Tripathi A, Nile A, Lee HC, Dynowski M, et al. (2015) Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Mol Biol Cell 26: 1764–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA (2004) FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6: 393–404 [DOI] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD (1999) SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem 274: 12990–12995 [DOI] [PubMed] [Google Scholar]
- Hammond GR,, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337: 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T (2014) A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol 205: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Schiavo G, Irvine RF (2009) Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem J 422: 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I (2016) Phosphoinositide signaling in plant development. Development 143: 2044–2055 [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK (2014) Lipid landscapes and pipelines in membrane homeostasis. Nature 510: 48–57 [DOI] [PubMed] [Google Scholar]
- Huang J, Ghosh R, Tripathi A, Lönnfors M, Somerharju P, Bankaitis VA (2016) Two-ligand priming mechanism for potentiated phosphoinositide synthesis is an evolutionarily conserved feature of Sec14-like phosphatidylinositol and phosphatidylcholine exchange proteins. Mol Biol Cell 27: 2317–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Esnay N, Platre MP, Noack LC, Menzel W, Claverol S, Moreau P, Jaillais Y, Boutté Y (2020) Sphingolipids mediate polar sorting of PIN2 through phosphoinositide consumption at the trans-Golgi Network. bioRxiv: 2020.2005.2012.090399 [DOI] [PMC free article] [PubMed]
- Jamecna D, Polidori J, Mesmin B, Dezi M, Levy D, Bigay J, Antonny B (2019) An intrinsically disordered region in OSBP acts as an entropic barrier to control protein dynamics and orientation at membrane contact sites. Dev Cell 49: 220–234.e228 [DOI] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4Kbeta1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- de Campos Kf M, Schaaf G (2017) The regulation of cell polarity by lipid transfer proteins of the SEC14 family. Curr Opin Plant Biol 40: 158–168 [DOI] [PubMed] [Google Scholar]
- de Jong F,, Munnik T (2021) Attracted to Membranes: Lipid-Binding Domains in Plants. Plant Physiol 185: 707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Balla T (2011) A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell 21: 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T (2015) Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER–PM contact sites maintains phosphoinositide signaling competence. Dev Cell 33: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig S, Hoffmann M, Mosblech A, Heilmann I (2008) Determination of content and fatty acid composition of unlabeled phosphoinositide species by thin-layer chromatography and gas chromatography. Anal Biochem 378: 197–201 [DOI] [PubMed] [Google Scholar]
- Kutateladze TG (2010) Translation of the phosphoinositide code by PI effectors. Nat Chem Biol 6: 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA (2008) Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9: 99–111 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol 8: 729–739 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (2001) Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol Biol Cell 12: 1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F,Krishnamoorthy P,Schubert V,Hause G, Heilmann M, Heilmann I (2019) A dual role for cell plate-associated PI4Kβ in endocytosis and phragmoplast dynamics during plant somatic cytokinesis. The EMBO journal. 38: 30617084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yin H, Gao P, Hu X, Yang J, Liu Z, Fu X, Luo D (2013) Phosphatidylserine synthase 1 is required for inflorescence meristem and organ development in Arabidopsis. J Integr Plant Biol 55: 682–695 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 22: 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC (2013) Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501: 257–261 [DOI] [PubMed] [Google Scholar]
- Mamode Cassim A, Gouguet P, Gronnier J, Laurent N, Germain V, Grison M, Boutte Y, Gerbeau-Pissot P, Simon-Plas F, Mongrand S (2019) Plant lipids: key players of plasma membrane organization and function. Prog Lipid Res 73: 1–27 [DOI] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23: 1129–1140 [DOI] [PubMed] [Google Scholar]
- Manik MK, Yang H, Tong J, Im YJ (2017) Structure of yeast OSBP-related protein Osh1 reveals key determinants for lipid transport and protein targeting at the nucleus–vacuole junction. Structure 25: 617–629.e613 [DOI] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B (2013) A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER–Golgi tether OSBP. Cell 155: 830–843 [DOI] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B (2017) Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J 36: 3156–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Kovacs D, D’Angelo G (2019) Lipid exchange and signaling at ER–Golgi contact sites. Curr Opin Cell Biol 57: 8–15 [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G (2015) Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science 349: 432–436 [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, Vanni S, Mesmin B, Antonny B, Drin G (2015) A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun 6: 6671. [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Zarza X (2013) Analyzing plant signaling phospholipids through 32Pi-labeling and TLC. Methods Mol Biol 1009: 3–15 [DOI] [PubMed] [Google Scholar]
- Nerlich A, von Orlow M, Rontein D, Hanson AD, Dormann P (2007) Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol 144: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nintemann SJ, Palmgren M, Lopez-Marques RL (2019) Catch you on the flip side: a critical review of flippase mutant phenotypes. Trends Plant Sci 24: 468–478 [DOI] [PubMed] [Google Scholar]
- Noack LC, , Bayle V, , Armengot L, , Rozier F, , Mamode-Cassim A, , Stevens FD, , Caillaud MC, , Munnik T, , Mongrand S, , Jaillais Y (2020) A nanodomain anchored-scaffolding complex is required for PI4Kα function and localization in plants. bioRxiv : . 12.08.415711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2017) Precision targeting by phosphoinoistides: how PIs direct endomembrane trafficking in plants. Curr Opin Plant Biol 40: 22–33 [DOI] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2020) Functions of anionic lipids in plants. Annu Rev Plant Biol 71: 71–102 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Miyagishima SY, Wada H (2015) Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton JG, Kim YJ, Humpolickova J, Eisenreichova A, Sengupta N, Toth DJ, Boura E, Balla T (2020) Defining the subcellular distribution and metabolic channeling of phosphatidylinositol. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, Marques-Bueno MDM, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miege C, et al. (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62 [DOI] [PubMed] [Google Scholar]
- Platre MP, Jaillais Y (2016) Guidelines for the use of protein domains in acidic phospholipid imaging. Methods Mol Biol 1376: 175–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Jaillais Y (2017) Anionic lipids and the maintenance of membrane electrostatics in eukaryotes. Plant Signal Behav 12: e1282022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P, et al. (2018) A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev Cell 45: 465–480.e411 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E (2006) A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol 172: 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Toulmay A, Balla T (2020) The functional universe of membrane contact sites. Nat Rev Mol Cell Biol 21: 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, , Bayer E (2021) Geometry and cellular function of organelle membrane interfaces. Plant Physiol 185: 650–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Levine TP (2004) Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem 279: 44683–44689 [DOI] [PubMed] [Google Scholar]
- Ruiz-Lopez N, Pérez-Sancho J, del Valle AE, Haslam RP, Vanneste S, Catalá R, Perea-Resa C, Van Damme D, García-Hernández S, Albert A, et al. (2020) Synaptotagmins maintain diacylglycerol homeostasis at endoplasmic reticulum–plasma membrane contact sites during abiotic stress. bioRxiv: 2020.2007.2028.222919 [DOI] [PMC free article] [PubMed]
- Saheki Y, De Camilli P (2017) Endoplasmic reticulum–plasma membrane contact sites. Annu Rev Biochem 86: 659–684 [DOI] [PubMed] [Google Scholar]
- Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A (2011) Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A 108: 1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan RS, Slabaugh E, Singh VR, Lapidus LJ, Haas T, Brandizzi F (2009) The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J 58: 817–830 [DOI] [PubMed] [Google Scholar]
- Sasvari Z, Lin W, Inaba JI, Xu K, Kovalev N, Nagy PD (2020) Co-opted cellular Sac1 lipid phosphatase and PI(4)P phosphoinositide are key host factors during the biogenesis of the tombusvirus replication compartment. J Virol 94: e01979–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, et al. (2008) Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell 29: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA (2009) Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol 187: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnoczky G, Kornmann B, Lackner LL,, Levine TP, Pellegrini L, Reinisch K, et al. (2019) Coming together to define membrane contact sites. Nat Commun 10: 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y (2014) A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J 77: 322–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Marques-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud MC, Jaillais Y (2016) A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Kruger F, Beckmann H, Brumm S, Vermeer JE, Munnik T, Mayer U, Stierhof YD, Grefen C, Schumacher K, et al. (2014) Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol 24: 1383–1389 [DOI] [PubMed] [Google Scholar]
- Smokvarska M, , Jaillais Y, , Martinère A (2021) Function of membrane domains in Rho-Of-Plant signaling. Plant Physiology 185: 663–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn M, Ivanova P, Brown HA, Toth DJ, Varnai P, Kim YJ, Balla T (2016) Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER–PM and ER–Golgi junctions. Proc Natl Acad Sci U S A 113: 4314–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn M, Korzeniowski M, Zewe JP, Wills RC, Hammond GRV, Humpolickova J, Vrzal L, Chalupska D, Veverka V, Fairn GD, et al. (2018) PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER-PM contact sites. J Cell Biol 217: 1797–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Wormsbaecher C, Gamble J, Zienkiewicz K, Brandizzi F (2018) Plant endocytosis requires the ER membrane-anchored proteins VAP27-1 and VAP27-3. Cell Rep 23: 2299–2307 [DOI] [PubMed] [Google Scholar]
- Tejos R, Sauer M, Vanneste S, Palacios-Gomez M, Li H, Heilmann M, van Wijk R, Vermeer JE, Heilmann I, Munnik T, et al. (2014) Bipolar plasma membrane distribution of phosphoinositides and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26: 2114–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Vermeer JE, Zhang Y, Gadella TW Jr, Nielsen E (2008) Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, et al. (2011) Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc Natl Acad Sci U S A 108: 15846–15851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umate P (2011) Oxysterol binding proteins (OSBPs) and their encoding genes in Arabidopsis and rice. Steroids 76: 524–529 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JE, Gadella TW Jr, Munnik T (2007) Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J 52: 1014–1026 [DOI] [PubMed] [Google Scholar]
- Venditti R, Masone MC, Rega LR, Di Tullio G, Santoro M, Polishchuk E, Serrano IC, Olkkonen VM, Harada A, Medina DL, et al. (2019) The activity of Sac1 across ER-TGN contact sites requires the four-phosphate-adaptor-protein-1. J Cell Biol 218: 783–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti R, Rega LR, Masone MC, Santoro M, Polishchuk E, Sarnataro D, Paladino S, D’Auria S, Varriale A, Olkkonen VM, et al. (2019) Molecular determinants of ER–Golgi contacts identified through a new FRET-FLIM system. J Cell Biol 218: 1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella TW Jr (2009) Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J 57: 356–372 [DOI] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobena-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr, Munnik T (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA (2005) A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol 168: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hawes C, Hussey PJ (2017) Plant endoplasmic reticulum–plasma membrane contact sites. Trends Plant Sci 22: 289–297 [DOI] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C, Cummins I, Deeks MJ, Sparkes I, Hawes C, Hussey PJ (2014) The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr Biol 24: 1397–1405 [DOI] [PubMed] [Google Scholar]
- Wang P, Richardson C, Hawkins TJ, Sparkes I, Hawes C, Hussey PJ (2016) Plant VAP27 proteins: domain characterization, intracellular localization and role in plant development. New Phytol 210: 1311–1326 [DOI] [PubMed] [Google Scholar]
- Wang Y, Mousley CJ, Lete MG, Bankaitis VA (2019) An equal opportunity collaboration between lipid metabolism and proteins in the control of membrane trafficking in the trans-Golgi and endosomal systems. Curr Opin Cell Biol 59: 58–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Yu Y, Mizoi J, Fujiki Y, Saito K, Nishijima M, Lee Y, Nishida I (2011) PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J 67: 648–661 [DOI] [PubMed] [Google Scholar]
- Ye H, Ji C, Guo R, Jiang L (2020) Membrane contact sites and organelles interaction in plant autophagy. Front Plant Sci 11: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S (2008) Membrane phosphatidylserine regulates surface charge and protein localization. Science 319: 210–213 [DOI] [PubMed] [Google Scholar]
- Zewe JP, Miller AM, Sangappa S, Wills RC, Goulden BD, Hammond GRV (2020) Probing the subcellular distribution of phosphatidylinositol reveals a surprising lack at the plasma membrane. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewe JP, Wills RC, Sangappa S, Goulden BD, Hammond GR (2018) SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Adamowski M, Marhava P, Tan S, Zhang Y, Rodriguez L, Zwiewka M, Pukyšová V, Sánchez AS, Raxwal VK, et al. (2020) Arabidopsis flippases cooperate with ARF GTPase exchange factors to regulate the trafficking and polarity of PIN auxin transporters. Plant Cell 32: 1644–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang Y, Niu Y, Fan T, Qian D, Luo C, Shi Y, Li S, An L, Xiang Y (2020) The Tip-localized phosphatidylserine established by Arabidopsis ALA3 is crucial for Rab GTPase-mediated vesicle trafficking and pollen tube growth. Plant Cell [DOI] [PMC free article] [PubMed] [Google Scholar]