Abstract
Mammalian phase II metabolism of dietary plant flavonoid compounds generally involves substitution with glucuronic acid. In contrast, flavonoids mainly exist as glucose conjugates in plants, and few plant UDP-glucuronosyltransferase enzymes have been identified to date. In the model legume Medicago truncatula, the major flavonoid compounds in the aerial parts of the plant are glucuronides of the flavones apigenin and luteolin. Here we show that the M. truncatula glycosyltransferase UGT84F9 is a bi-functional glucosyl/glucuronosyl transferase in vitro, with activity against a wide range of flavonoid acceptor molecules including flavones. However, analysis of metabolite profiles in leaves and roots of M. truncatula ugt84f9 loss of function mutants revealed that the enzyme is essential for formation of flavonoid glucuronides, but not most flavonoid glucosides, in planta. We discuss the use of plant UGATs for the semi-synthesis of flavonoid phase II metabolites for clinical studies.
UGT84F9 is a bifunctional glucuronosyltransferase/glucosyltransferase that is necessary for the glucuronidation of a wide range of flavonoid natural products in Medicago truncatula.
Introduction
UDP-dependent glycosyltransferases (UGTs) are members of an enzyme family that plays a central role in glycosylating small lipophilic compounds in both plants and animals (Bowles et al., 2006). In mammals including humans, UGTs essentially exist as glucuronosyltransferases (UGATs, EC 2.4.1.17) that use uridine diphosphate glucuronic acid (UDP-GlcA) as the sugar donor. These UGATs are the major phase II metabolizing enzymes responsible for conjugating toxic endo- and xeno-biotics to make them more soluble and improve their transport for renal elimination.
Epidemiological studies have shown that dietary intake of flavonoid-rich food sources has an inverse relationship to the risk of dementia in humans (Commenges et al., 2000), and an increasing body of experimental evidence suggests that flavonoids and their derivatives may have beneficial effects for improving cognitive function and neurological resilience in animal models (Wang et al., 2008, 2012; Ho et al., 2013). Following oral administration of grape (Vitis vinifera) seed extract to mice, constituent flavonoids are metabolized into glucuronidated and methylated derivatives that can accumulate in brain tissue (Wang et al., 2012, 2014, 2015). Such compounds are targets for mode-of-action studies to address the mechanisms involved in neuroprotection and psychological resiliency, but, unfortunately, most glucuronidated flavonoids are not commercially available. As their chemical synthesis is both complex and costly (de Fátima et al., 2019; Docampo-Palacios et al., 2020a, 2020b), enzymatic approaches require investigation.
Mammalian UGATs are membrane-bound enzymes that are largely localized in the lumen of the endoplasmic reticulum (Tukey and Strassburg, 2000) and as a result, heterologous expression using traditional microbial systems for further application is difficult. This makes it attractive to explore other biological sources of UGATs, such as plants, where glycosyltransferases are generally soluble enzymes, and, like the mammalian UGATs (Radominska-Pandya et al., 2010), are inverting glycosyltransferases and will therefore provide the correct stereochemistry at the anomeric carbon of the attached sugar for clinical studies. However, only a few UGATs have been identified and functionally characterized from a few plant species. These include flavonoid 7-O-glucuronosyltransferase (F7GAT, comprising UGT88D4, UGT88D6, and UGT88D7) from members of the order Lamiales (Noguchi et al., 2009), BpUGT94B1 from red daisy which conjugates sugar/sugar branching of cyanidin glycosides (Sawada et al., 2005), UBGAT (UGT88D1) from cultured cells of Scutellaria baicalensis Georgi which conjugates baicalein at the 7-OH position (Nagashima et al., 2000), VvGT5, a flavonol-3-O-glucuronosyltransferase from grapevine (V. vinifera; Ono et al., 2010), and UGT72P12-canonical from Glycyrrhiza uralensis which catalyzes sugar/sugar branching at the 3-O-glucuronosyl unit of the pentacyclic triterpene glycyrrhetinic acid (Nomura et al., 2019).
Medicago truncatula is an extensively studied model legume with a sequenced genome and available mutant collection resources (Young and Udvardi, 2009; Young et al., 2011). Given the diverse range of glycosylated natural products in M. truncatula, which includes not only flavonoids but also many triterpene saponins (Gholami et al., 2014), the UGT multigene family is expanded in this species. Thus, whereas about 107 UGT genes exist in the Arabidopsis (Arabidopsis thaliana) genome (Li et al., 2001), over 300 UGT genes have been annotated in M. truncatula (Young et al., 2011). The major flavonoid glycoconjugates detected in the aerial tissues of M. truncatula include glucuronides of the flavones tricin, apigenin, chrysoeriol, and luteolin (Kowalska et al., 2007; Marczak et al., 2010; Staszkow et al., 2011), indicating the presence of UGATs in M. truncatula. However, although a number of glucosyltransferase enzymes capable of glycosylating flavonoid aglycones with uridine diphosphate glucose (UDP-Glc) as sugar donor have been identified from M. truncatula (Shao et al., 2005; Li et al., 2007; Modolo et al., 2007), as of yet no UDP-GlcA dependent glycosyltransferase has been reported from this species.
Crystal structures of several plant UGTs have been obtained, including M. truncatula UGT71G1, UGT85H2, and UGT78G1 (Shao et al., 2005; Li et al., 2007; Modolo et al., 2009), grape (V. vinifera) VvGT1, (Offen et al., 2006), A. thaliana UGT72B1 (Brazier-Hicks et al., 2007), and Clitoria ternatea UGT78K6 (Hiromoto et al., 2015), all of which recognize only UDP-Glc as sugar donor. Recently, the structure of a plant flavonoid rhamnosyltransferase has been obtained, illuminating the molecular basis for specificity for UDP-Rha as sugar donor (Zong et al., 2019). As of yet, however, no complete UGAT crystal structure has been solved except for the C-terminal domain of human UGT2B7; only the partial structure could be obtained due to solubility problems (Miley et al., 2007; Radominska-Pandya et al., 2010). Identification and characterization of new UGATs from plants is therefore important for understanding the mode of action of these enzymes, as well as for increasing the UGAT repertoire for developing and modifying biologically relevant bioactive compounds useful for human health research and other biotechnological applications. Here, we identify the major flavonoid UGAT from M. truncatula, address its catalytic activities both in vitro and in vivo, and discuss its usefulness as a basis for catalyst design for generation of bioactive flavonoid glucuronides.
Results
Screening of mammalian UGATs for flavonoid acceptor specificity
To first obtain a picture of the range of glucuronidation activities of mammalian phase II enzymes for flavonoids, we evaluated commercially available UGT1A and UGT2B subfamily members for glucuronidation of apigenin, luteolin, quercetin, and epicatechin. Structures of these and other flavonoids discussed in this paper are presented in Supplemental Figure S1. Analysis of the conversion of aglycone to glucuronidated products by reverse phase HPLC showed that these enzymes exhibited wide variation of activity, with UGT1A9 having the highest activity for all acceptor substrates tested (Supplemental Figure S2). The relative preference for flavone or flavonol (quercetin) varied among the enzymes, and the UGT2B enzymes exhibited poor activity against all the flavonoids tested. These findings reflect previous studies on individual enzymes or groups of substrates (Wu et al., 2011). Further analysis of reaction products by HPLC and LC/MS indicated that multiple positional monoglucuronide isomers, including the 7-O-glucuronide of luteolin, were generated by the mammalian enzymes (Supplemental Figure S3 and Supplemental Table S1). Additional di-glucuronides were seen as minor products only in the case of UGT1A7 and 1A10 with 4′-O-methyl-quercetin as substrate.
Selection of candidate UGATs from M. truncatula
To explore plants as a source of soluble and therefore more easily deployable UGATs, we selected the model legume M. truncatula in view of its known accumulation of a range of flavonoid glucuronides (Kowalska et al., 2007; Marczak et al., 2010b; Staszkow et al., 2011). Members of the UGT enzyme family consist of two similar (N- and C-terminal) domains, each possessing several alpha helices and beta strands. Catalytically important motifs and residues include a conserved motif of about 44 amino acids referred to as the plant secondary product glycosyltransferase (PSPG) domain at the C-terminus which binds with the UDP portion of the sugar donor during catalysis, and residues at the N-terminus that are either crucial for interacting with the acceptor substrate during catalysis or can partly define donor substrate acceptability as described below (Shao et al., 2005; Osmani et al., 2008; Nomura et al., 2019). Using these characteristic features as a starting point, we utilized bioinformatics tools to mine the M. truncatula genome (Young et al., 2011) for in silico identification of candidate UGT genes which have not been hitherto characterized from this species. The strategy is summarized in Supplemental Figure S4.
We first used Hidden Markov Model-based software (HMMER) to search UGT pfam protein domains in the M. truncatula genome, using the query UDPGT and accession PF00201.17 for broad coverage of UGT candidates belonging to the UDP-glucuronosyl and UDP-glucosyl transferase gene families. The resulting top 280 UGT sequences from this search were selected. We next used the protein sequences of the previously identified plant UGATs BpUGT94B1, F7GAT, and UGT73P12 (Nagashima et al., 2000; Sawada et al., 2005; Noguchi et al., 2009; Nomura et al., 2019) as templates to search for orthologs among the HMMER filtered UGT candidates. Members of the plant UGT protein family usually possess a highly conserved catalytic histidine residue (e.g. His22 in UGT71G1) which is crucial for interaction with the acceptor substrate during catalysis. This residue is known to act by deprotonating the acceptor substrate, while being stabilized by a generally conserved aspartic acid (e.g. Asp121 in UGT71G1; Supplemental Figure S5; Shao et al., 2005). These two residues are located in the N-terminal domain. Similarly, amino acid residues that are considered to contribute, in part, to UDP-GlcA recognition are usually positively charged or polar residues such as Arg25 in BpUGT94B1 (Osmani et al., 2008) or Arg32 in licorice (G. uralensis) UGT73P12_cannonical (Nomura et al., 2019), and these Arg residues are proximal to the catalytic histidine in both the primary and predicted tertiary structures of BpUGT94B1 and UGT73P12_cannonical. In the predicted tertiary structure of Lamiales F7GAT (UGT88D7), Arg350 is near to His13 in the active site pocket (Noguchi et al., 2009; Supplemental Figure S6).
On the basis of these criteria, we identified a number of Medicago UGTs (25 candidates) that possess structural motifs or key residues that are typical of UG(A)Ts (Figure 1 and Supplemental Table S2). One candidate UGAT (initially annotated as C7, now officially named UGT84F9) possessed a positively charged residue, His22, which corresponded to Arg25 in BpUGT94B1 or Arg32 in UGT73P12_cannonical (Figure 1). The above search results were also aligned with the structurally characterized UGT71G1 and UGT78G1 (both UDP-glucosyltransferases) and BpUGT94B1, F7GAT, and UGT73P12 using MUSCLE, and the PhyML 3.0 algorithm was then used to construct the maximum-likelihood phylogenetic tree (Supplemental Figure S7). Clearly, in spite of the common residue features, UGT84F9 does not fall into any clade that contains functionally characterized UGATs. Together, these filters led to the identification of 25 candidate UGATs.
Figure 1.
Amino acid sequence alignment of the N-terminal domains of selected plant UGTs, indicating the presence of a critical catalytic His (indicated by red arrow) that is universally conserved across all plant UGTs. A neighboring residue believed to be important in contributing to sugar donor recognition is indicated with the green arrow. Blue boxes in the left hand column indicated previously identified UDP-glucuronosyltransferases. Orange boxes and binary names (e.g. C3, J5) indicate M. truncatula UG(A)T candidates in this study. Sugar–sugar branching glucuronidating enzymes such as BpUGT94B1 and UGT73P12_canonical have an Arg substitution at this position instead of Ser, Thr, or Pro found in UDP-Glc dependent UGTs. Other amino acid substitutions (His, Gln, or Phe) were found at this location for some of the M. truncatula UGAT candidates we identified. Sequences are: BpUGT94B1, UDP-GlcA dependent UGT from red daisy (Bellis perennis); C7 (UGT84F9), UDP-GlcA and UDP-Glc specific UGT from M. truncatula (this study); VvGT5, UDP-GlcA dependent UGT from V. vinifera, UGT73P12_canonical and UGT73P12_variant are, respectively, UDP-GlcA and UDP-Glc dependent UGTs from licorice (G. uralensis); UGT78G1, UGT71G1, and UGT72L1 are UDP-Glc dependent UGTs from M. truncatula; GuUGAT, UDP-GlcA dependent UGT from licorice; UGT88D-1, 4, 5, 6, 7, and UGT88A7 are UDP-GlcA dependent UGTs from Lamiales species. Blue boxes show previously identified UDP-GlcA-specific enzymes, brown boxes UGAT candidates that utilize UDP-Glc. GenBank accession numbers for these sequences are given in Supplemental Table S2.
Automated protein modeling, using SWISS-MODEL, of the candidate UGATs was then carried out. WinCoot software was used to determine if the presumed active site would be exposed in the three-dimensional model structures for substrate binding and catalysis. A total of 19 UGATs passed this filter. More detailed structural analysis of the confirmed UGAT is shown below.
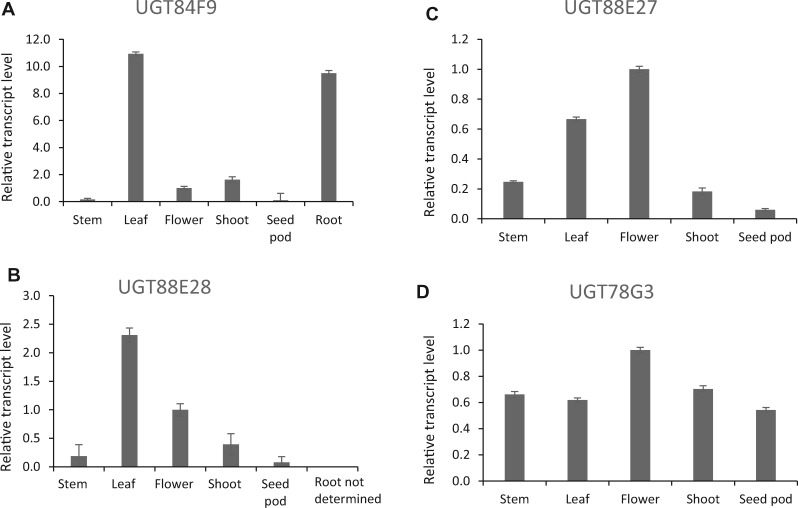
We finally analyzed tissue-specific expression of candidate UGATs in silico using the available M. truncatula Gene Expression Atlas (https://mtgea.noble.org/v3/; Supplemental Figure S8). UGT84F9 was the most highly expressed UGAT candidate overall, specifically in both leaf and root tissues where flavone glucuronides accumulate (Supplemental Figure S8, A and C; Marczak et al., 2010; Staszkow et al., 2011). It was not expressed in root nodules, or cell suspension cultures (Supplemental Figure S8, A), and its expression pattern was distinct from that of UGT71G1, a relatively promiscuous UDP-glucosyltransferase active with several classes of flavonoid acceptors (Modolo et al., 2007) that is expressed at somewhat lower levels in a similarly broad range of tissues and treatments (Supplemental Figure S8, B). RT-qPCR analysis confirmed the high expression of UGT84F9 in leaf tissue compared with three other UGAT candidates that showed low expression in the M. truncatula Gene Expression Atlas (Figure 2). Based on all the criteria above, we selected UGT84F9 as the candidate for further functional characterization. The other 18 UG(A)T candidates were, however, tested with apigenin and quercetin as acceptor substrates and UDP-GlucA and UDP-Glc as sugar donors. G1, G2, and G3 exhibited very weak activity with UDP-GlcA as sugar donor that required sensitive LC–MS analysis for detection; however, these enzymes appeared to have very low, if any, expression in leaf tissue (Supplemental Figure S8, C). The remainder, including UGTs 88E27, 88E28, 72Y5, and 78G3, all appeared to be mono-functional UDP-glucosyl transferases. The position of glycosylation was not determined for candidates other than UGT84F9 (see below).
Figure 2.
Expression analysis of selected candidate UGAT transcripts in different tissues of M. truncatula ecotype R108. A–D, Quantitative real-time PCR analysis of relative transcript levels of UGT84F9 (A), UGT88E28 (B), UGT88E27 (C), and UGT78G3 (D). The latter three were expressed at lower levels in roots than in leaves based on transcriptome analysis in the M. truncatula Gene Expression Atlas (https://mtgea.noble.org/v3/). Transcript levels are expressed relative to those of the housekeeping gene tubulin. Error bars represent standard deviation, n = 3 analytical replicates.
Biochemical characterization of UGT84F9
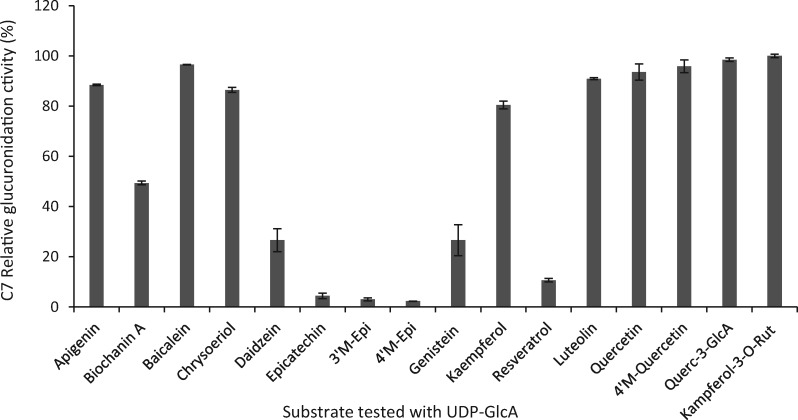
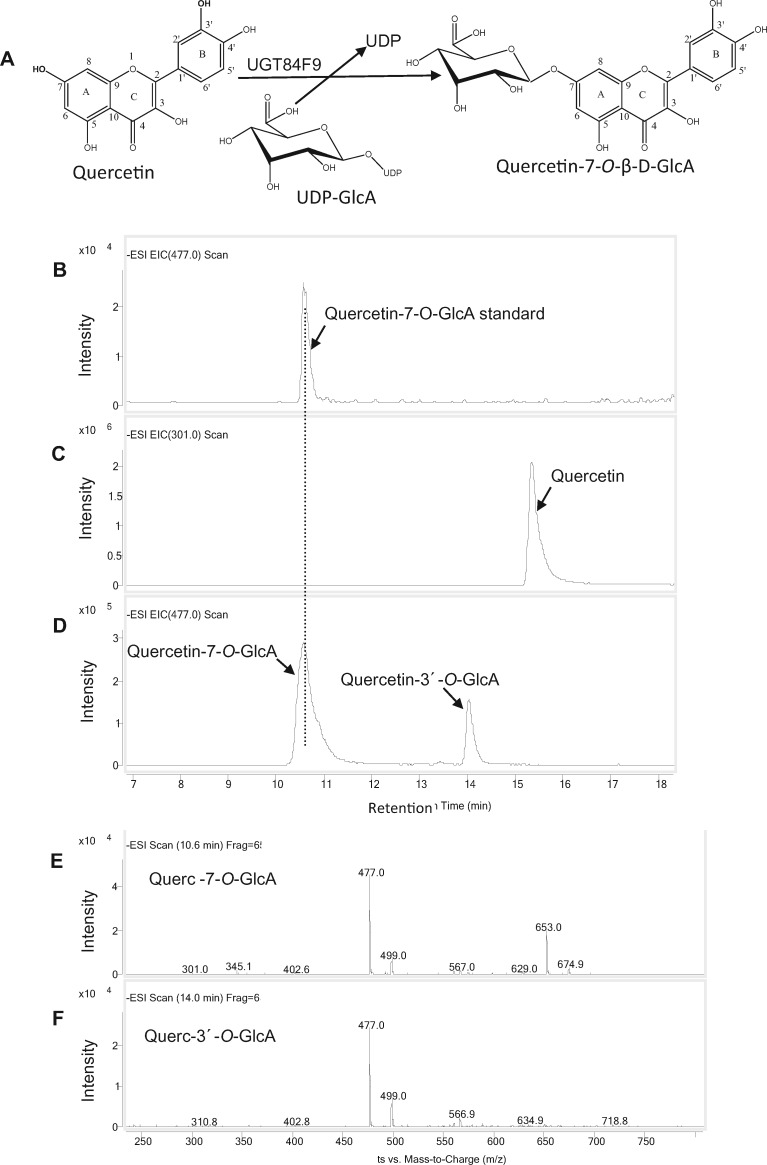
To test the substrate specificities of UGT84F9, we amplified its open-reading frame from mRNA from leaf tissue, and expressed the recombinant N-terminal maltose binding protein (MBP)-tagged enzyme in Escherichia coli. The protein was purified using amylose resin and protein purity assessed by SDS–PAGE (Laemmli, 1970; Supplemental Figure S9). Enzyme assays were first carried out with a range of flavonoid compounds as sugar acceptors and UDP-GlcA as sugar donor, and reactions monitored by reverse-phase HPLC analysis. UGT84F9 exhibited broad acceptor specificity as a UDP-glucuronosyltransferase (Figure 3). Flavones and flavonols or their derivatives were the preferred substrates, with epicatechin and its methylated derivatives the poorest substrates (Figure 3). Incubation of UGT84F9 with quercetin, which has five free -OH positions that can potentially serve as glucuronidation sites, yielded a major product that co-chromatographed with quercetin 7-O-glucuronide, along with an additional peak with the molecular mass of a second positional isomer of quercetin mono-glucuronide (Figure 4). LC–MS analysis indicated that a minor product with the molecular mass of a diglucuronide co-chromatographed with the peak of quercetin 7-O-glucuronide (Figure 4). Among the flavones tested, apigenin and chrysoeriol yielded one major glucuronidated product whereas two monoglucuronides, as well as a minor diglucuronide product, were generated with luteolin as the acceptor substrate (Supplemental Figures S10, S11). Epicatechin and its 3′- and 4′-O-methyl derivatives were each converted to one major product, suggesting that glucuronidation is at the 3, 5, or 7 position.
Figure 3.
Relative glucuronidation activity of UGT84F9 toward flavonoids and other compounds. Recombinant UGT84F9 was incubated with the individual acceptor substrates (750 µM) and UDP-GlcA (2 mM) for 2 h at 30°C. The activity is expressed as percentage relative to that of the most favored substrate, kaempferol-3-O-rutinoside. 100% activity = 40.055 nmol/min/mg protein. Data are means for three technical replicates, with error bars representing the standard deviation.
Figure 4.
UGT84F9 catalyzes the glucuronidation of quercetin. A, The enzymatic reaction catalyzed by UGT84F9. B–D, LC–MS analysis of the enzymatic reaction of UGT84F9 to generate Q-7-O-GlcA. B, Q-7-O-GlcA standard. C, Remaining Q substrate. D, Reaction products of UGT84F9 incubated with quercetin. E, Mass spectrum showing the molecular ions of the glucuronidated product of quercetin at retention time 10. 6 min (m/z = 477). A molecular ion (m/z = 653) matching that of di-glucuronides of Q is also seen. F, Mass spectrum of molecular ions of quercetin glucuronide at retention time 14 min (m/z = 477). The LC–MS was performed in negative ion mode. All reactions were incubated for 2 h.
To determine whether UGT84F9 can also glucuronidate diglycosides, kaempferol-3-rutinoside and quercetin-3-O-GlcA were tested as acceptor substrates. The enzyme was highly active with these substrates, particularly the former. The detection of single triglycoside products from these substrates (Supplemental Figure S12) indicates that UGT84F9 introduces a single additional sugar moiety at a single position. The position of attachment of the additional sugar was not determined.
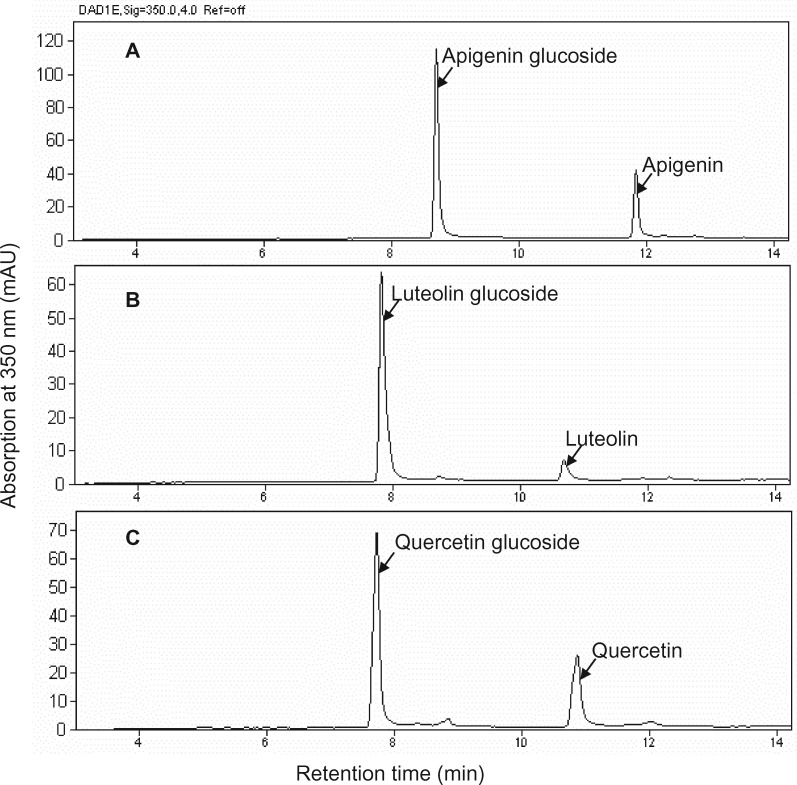
To determine whether UGT84F9 also exhibits promiscuity with regards to the sugar donor, the enzyme was assayed with apigenin, luteolin, and quercetin with UDG-Glc as sugar donor (Figure 5). The in vitro enzyme reactions were efficient and yielded one positional isomer of the corresponding flavonoid glucoside from all three substrates. No diglucosides were observed. No products were obtained with UDP-galactose as sugar donor and quercetin as acceptor substrate.
Figure 5.
HPLC-UV analysis of the products of UGT84F9 activity with three flavonoid acceptors using UDP-Glc as sugar donor. A, apigenin. B, luteolin. C, quercetin.
Kinetics of UGT84F9 for acceptor and donor substrates
To determine the catalytic efficiencies of UGT84F9, we performed kinetic analyses of the reactions with luteolin, apigenin, kaempferol-3-O-rutinoside, and quercetin as acceptor substrates with UDP-GlcA as sugar donor, and quercetin as acceptor with UDP-Glc as sugar donor. In the case of luteolin and quercetin, we included both monoglucuronide products determined by HPLC for calculation of reaction rates. The amount of diglucuronide product from luteolin and quercetin was less than 1% of the total products in the short time incubations used for initial rate measurements. All combinations exhibited classical Michaelis–Menten kinetics (Supplemental Figure S13). As suggested by the initial activity screening (Figure 3), the most efficient acceptor substrate with UDP-GlcA as sugar donor was kaempferol-3-O-rutinoside (Kcat/Km of 2.314 s−1 mM−1) followed by luteolin (Kcat/Km = 1.04 s−1 mM−1), quercetin (Kcat/Km = 0.672 s−1 mM−1), and apigenin (Kcat/Km = 0.635 s−1 mM−1; Supplemental Table S3). With UDP-Glc as sugar donor, the Kcat/Km with quercetin as acceptor substrate was 0.782 s−1 mM−1, slightly higher than with UDP-GlcA. This is largely because of the increased affinity for UDP-Glc, as the turnover rate was significantly lower (Supplemental Table S3).
We then compared the kinetics of UGT84F9 for UDP-GlcA or UDP-Glc at a constant saturating concentration of quercetin or apigenin. The reactions again followed classical Michaelis–Menten kinetics (Supplemental Figure S14). UGT84F9 had higher affinity for UDP-Glc than for UDP-GlcA based on the Km values (Table 1). However, the Kcat values with respect to UDP-GlcA as sugar donor were higher than with UDP-Glc, resulting in an overall Kcat/Km for UDP-GlcA that was higher than that for UDP-Glc (Table 1).
Table 1.
Kinetic parameters of wild-type and mutant recombinant UGT84F9 for UDP-Glc or UDP-GlcA as sugar donor in the presence of a saturating concentration of quercetin or apigenin (2 mM) as acceptor substrate
| Enzyme (UGT84F9) | Substrate | Sugar donor | K m (mM) | K cat (s−1) | K cat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| Wild type | Quercetin | UDP-GlcA | 0.90 ± 0.12 | 0.84 ± 0.00 | 0.94 |
|
Wild type |
Quercetin |
UDP-Glc |
0.38 ± 0.05 |
0.23 ± 0.01 |
0.61 |
| Wild type | Apigenin | UDP-GlcA | 0.51 ± 0.10 | 1.70 ± 0.09 | 3.30 |
| Wild type | Apigenin | UDP-Glc | 0.41 ± 0.07 | 0.13 ± 0.00 | 0.37 |
| H22S | Quercetin | UDP-Glc | 0.29 ± 0.04 | 0.20 ± 0.01 | 0.69 |
| H22S | Quercetin | UDP-GlcA | 0.12 ± 0.01 | 0.02 ± 0.00 | 0.14 |
| H22R | Quercetin | UDP-GlcA | 0.25 ± 0.05 | 0.02 ± 0.00 | 0.06 |
| H22R-N121D | Quercetin | UDP-GlcA | 0.69 ± 0.12 | 0.01 ± 0.00 | 0.01 |
Representative kinetic plots are shown in Supplemental Figure S14.
Identification of residues contributing to sugar specificity of UGT84F9
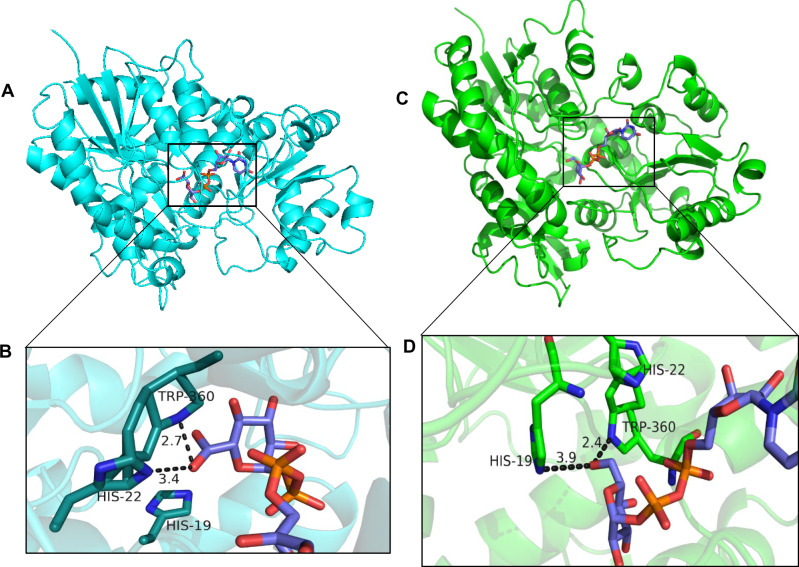
To begin to address the basis for the sugar donor specificity of UGT84F9, we first performed automated protein modeling, using SWISS-MODEL, to predict the 3-D structure of the enzyme, using the crystal structure of UGT71G1 as template; WinCoot software was then used to analyze the architecture of the active site of UGT84F9 and enzyme–substrate interactions (Figure 6). The analysis showed that His22 and Trp360 can potentially interact with the sugar donor (UDP-GlcA or UDP-Glc) during catalysis, being, respectively, within 3.4 and 2.7 Å distance from the bound ligand (Figure 6, A and B). His19 in the pocket is conserved among UGT family members as it acts as a catalytic residue and interacts with the acceptor substrate.
Figure 6.
The modeled three-dimensional structure of UGT84F9 with UDP-GlcA and UDP-Glc, showing the possible interactions between the enzyme residues and sugar donor. A, The predicted protein structure of UGT84F9 represented by cartoon (cyan) with UDP-GlcA shown as a stick model (carbon atom in slate blue, oxygen in red, phosphorus in orange). B, The putative active site and substrate binding pocket. Key residues His22 and Trp360, shown as sticks (carbon atom in cyan, nitrogen atom in blue), can potentially interact with the sugar donor, UDP-GlcA, that is also represented in stick model (carbon atom in slate blue, oxygen in red, phosphorus in orange). Catalytic residue His19, shown as a stick model, is in the active pocket and interacts with the acceptor substrate. C and D, The potential three-dimensional structure of UGT84F9 with superimposition of bound UDP-Glc, showing the possible residues interacting with the sugar donor. C, The predicted protein structure of UGT84F9 represented by cartoon (green) with bound UDP-Glc represented in stick model (carbon atoms in slate, oxygen in red, phosphorus in orange, and nitrogen in blue). D, The presumed active site pocket with the bound UDP-Glc showing the possible interaction with residues including His19, Trp360, and His22. SWISS-MODEL software was used to build the 3-dimensional structure of UGT84F9 in comparison to UGT71G1, and the ligands docked into UGT71G1 were superimposed onto UGT84F9 using Wincoot software. PyMOL software was used to visualize the possible catalytic residues interacting with the ligand.
As described above, key residues that may be important for catalysis were previously identified based on comparisons of the amino acid sequence of UGT84F9 with the two previously identified plant UGATs (BpUGT94B1 and UGT72P12-canonical) and the glucosyltransferase UGT71G1 for which a crystal structure is available (Figure 1 and Supplemental Figures S5, S6). His22 in UGT84F9 was identified as corresponding to Arg25 in BpUGT94B1 and UGT72P12-canonical, whereas serine, threonine, proline, or similar residues in a range of UDP-Glc dependent UGTs (e.g. UGT72P12-variant or UGT71G1) were found to correspond to Arg-25 in BpUGT94B1 and UGT72P12-canonical and to His22 in UGT84F9 (Figure 1). Additionally, a conserved Asp among UGTs (e.g. Asp121 in UGT71G1) that was described as contributing to UGT catalysis by stabilizing the catalytic His (e.g. His22 in UGT71G1; Shao et al., 2005) was found to correspond to Asn121 in UGT84F9 (Supplemental Figure S5).
To test the significance of residues in contributing to the capability of UGT84F9 to use both UDP-GlcA and UDP-Glc, site-directed mutagenesis was performed to generate single (H22S and H22R) and double (H22S-N121D and H22R-N121D) mutants that were expressed in E. coli (Supplemental Figure S9) and evaluated with quercetin as acceptor substrate (Supplemental Figures S14, S15). The H22S and H22R substitution in UGT84F9 greatly compromised the turnover of UDP-GlcA by a factor of up to 40-fold, while increasing the affinity of the enzyme for this sugar donor by two- to four-fold. Likewise, H22S exhibited a slightly higher affinity for UDP-Glc, but the decrease in Kcat was much less with UDP-Glc than with UDP-GlcA as sugar donor (Supplemental Figure S14, C and Table 1). The double mutation H22R-N121D further diminished the sugar donor recognition by a factor of 87-fold. Importantly, nearly all the mutations tested led to a greater reduction in activity with UDP-GlcA than with UDP-Glc (Supplemental Figure S15). These data confirm that His-22 in UGT84F9 may indeed function similarly to Arg-25 in BpUGT94B1 and UGT72P12-canonical to contribute to recognition of UDP-GlcA as sugar donor, and a similar function can be inferred for Asn121.
Functional identification of UGT84F9 in planta
To address the in planta function of UGT84F9, we searched for potential knock-out mutations in the M. truncatula Tnt1 retrotransposon insertion mutant collection available at the Noble Research Institute (medicago-mutant.noble.org; Tadege et al., 2008). Four independent lines, NF8294A-R1, NF5230-R1, NF1799-R0, and NF11932, were identified in which the UGT84F9 gene was disrupted by a Tnt1 insertion in the N-terminal region. Seeds of each line were germinated, and genotyped by PCR to find ugt84f9 homozygous mutant lines (Figure 7, A–C). Two of the NF8294A-R2 lines were found to be homozygotes (Figure 7, B and C), and showed clearly observable phenotypic abnormalities in the form of stunted growth compared with the heterozygote or the wild type (Figure 7, A). The remaining heterozygotes and lines that were wild type at the UGT84F9 locus were maintained as controls. We were only able to find one additional homozygous R3 generation plant arising from NF5230-R2 seed, which again showed a stunted growth phenotype, along with heterozygous and null control lines (Supplemental Figure S16). The NF8294A-R2 and NF5230-R3 plants were therefore analyzed as two independent homozygous mutant alleles for the ugt84f9 knock-out mutation.
Figure 7.
Characterization of Tnt1 transposon insertion mutations in UGT84F9. A, Seven-weeks-old plants with ugt84f9 Tnt1 insertions in NF8294A mutant lines showing smaller size of the homozygote compared with the corresponding heterozygote at the UGT84F9 locus. B, Molecular characterization of the mutation at the UGT84F9 locus in NF8294A and NF5230. Agarose gel electrophoresis analysis of PCR products resulting from genotyping the Tnt1 insertion mutant lines for UGT84F9 gene disruption, compared with the wild-type plant. The first and second columns show the presence of intact UGT84F9 gene, with absence of the Tnt1 insertion (UGT84F9/tnt) in the R108 wild type (the genetic background used for the transposon insertion mutagenesis) and the null-segregant at the UGT84F9 locus. The third column shows the presence of the wild-type and mutant alleles at the UGT84F9 locus indicative of the heterozygote. The fourth and fifth columns show loss of the wild-type allele and presence of the Tnt1 insertion in two independent homozygous lines of NF8294A. In the last two columns, NF5230-WT shows the intact UGT84F9 gene without Tnt1 insertion and NF5230-Homo shows the Tnt1 insertion in the homozygous line of the second allele. UGT88E28 and tubulin served as controls. C, Model showing the Tnt1 insertion in the N-terminal region of the UGT84F9 gene in the NF8294A and NF5230 lines.
Extracted flavonoid metabolites from leaf and root tissues of M. truncatula wild-type, null segregant, heterozygote, and homozygote for the Tnt1 insertion in UGT84F9 were then analyzed by LC–MS using an Agilent 6460 QQQ triple quadrupole mass spectrometer (Figures 8, A–J, 9 and Supplemental Figures S17–S29). The samples analyzed were four from NF1799-null segregant; three from NF5230 heterozygote; one from R108 WT; two from NF5230 null segregant and NF8294A homozygote; and one from each of NF8294A null segregant, NF8294A heterozygote, and NF5230 homozygote. The metabolites targeted included mono-, di-, and triglycosides (glucuronides) of the flavones apigenin, luteolin, tricin, and chrysoeriol, the flavonols laricitrin, quercetin, and kaempferol and the isoflavone formononetin, with analysis based on previously published MS characteristics of these compounds (Kowalska et al., 2007; Marczak et al., 2010; Staszkow et al., 2011). Mass spectra were scanned for the [M–H]+ molecular ions. The conclusions from these analyses are summarized in Table 2.
Figure 8.
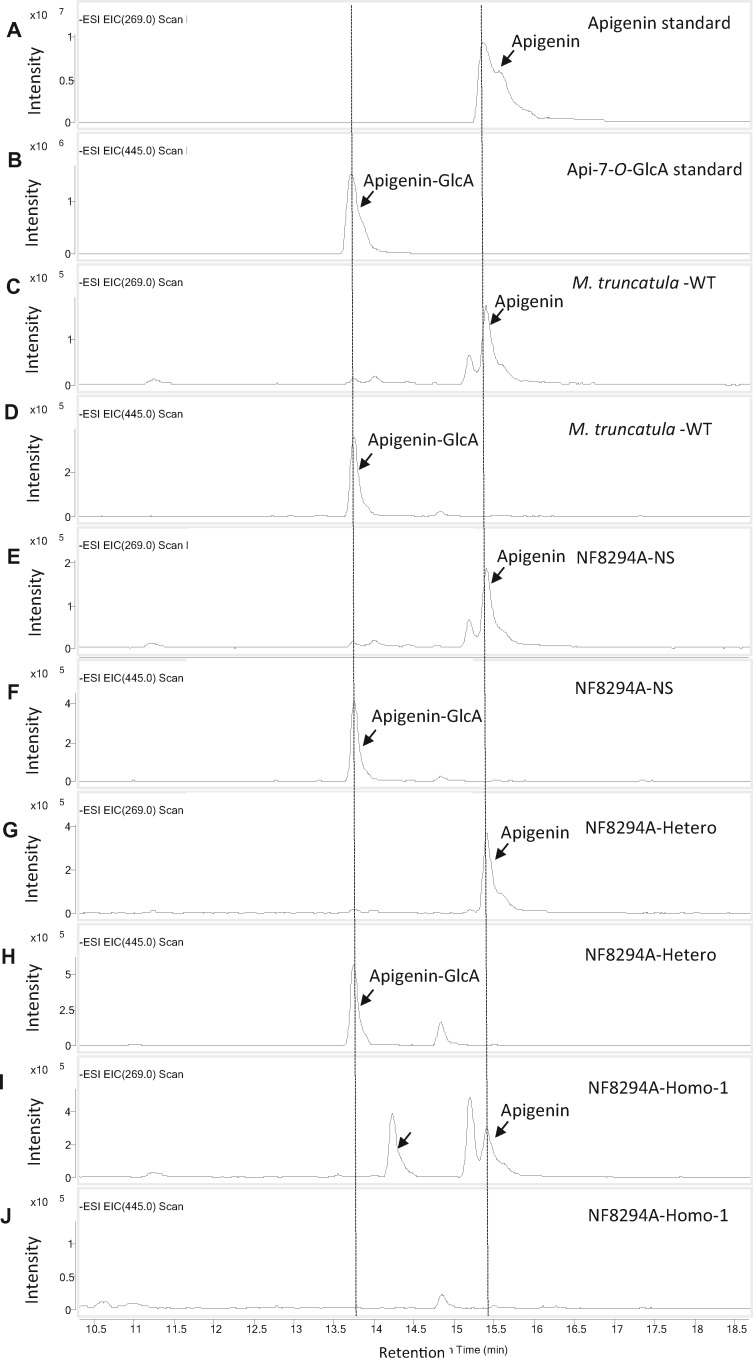
Loss of function of UGT84F9 leads to disappearance of apigenin glucuronide in M. truncatula. A–F, LC–MS analysis of flavonoid metabolites in extracts from wild-type and mutant M. truncatula leaf tissue for detection of the flavone apigenin and its mono-glucuronide apigenin-7-O-GlcA. A, Apigenin standard (m/z = 269). D, Apigenin-7-O-GlcA standard (m/z 445). C, E, G, and I, The extracted ion chromatograms from scanning the M. truncatula wild-type R108, the NF8294A-null segregant, the heterozygote and the homozygote extracts, respectively, for apigenin (m/z = 269). D, F, H, and J, The extracted ion chromatograms from scanning the M. truncatula wild-type R108, the NF8294A-null segregant, the heterozygote and the homozygote extracts, respectively, for apigenin-7-O-GlcA (m/z 445). The LC–MS was performed in negative mode.
Figure 9.
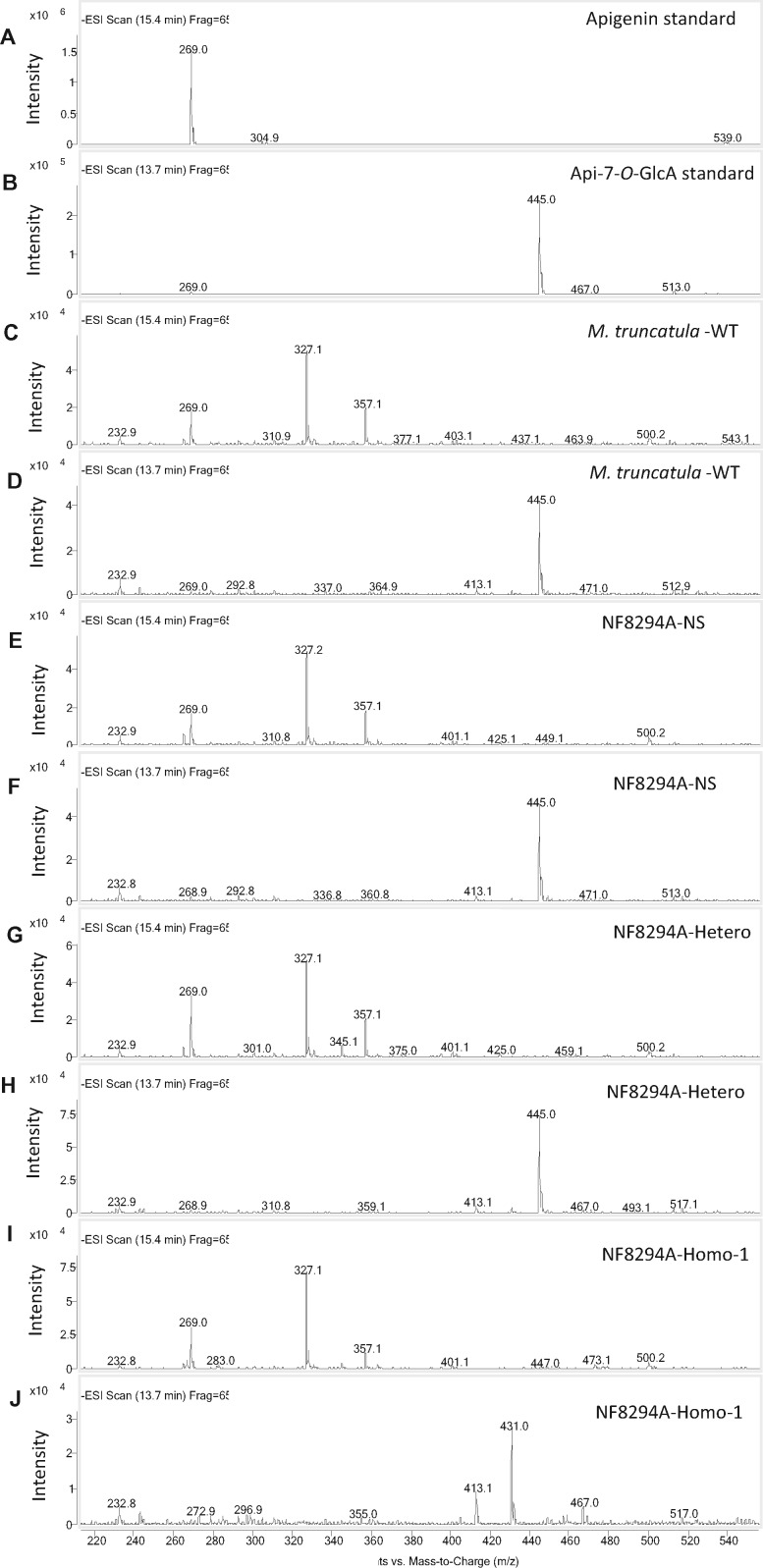
MS spectra for the metabolites in Figure 8. The peak at retention time 15.4 min is apigenin and the peak at 13.7 min Api-7-O-GlcA. A and B, Apigenin and Api-7-O-GlcA standards. Apigenin, with molecular mass of the parent ion at [M–H]+ = 269, was seen in all the samples (C, E, G, and I); apigenin-7-O-GlcA, [M–H]+ = 455, was present in the extract from M. truncatula wild-type R108, the NF8294A-null segregant, and the heterozygote (D, F, and H) but was not detected in the extract from NF8294A-homozygote (J). The LC–MS was performed in negative mode.
Table 2.
Flavonoid metabolites detected by LC–MS from leaf tissue extracts of M. truncatula R108 wild type (WT), NF1799-, NF5230-, and NF8294A-null segregants (NS), NF5230- and NF8294A-heterozygotes, and NF5230- and NF8294A-homozygotes
| Compound | Elemental composition | MW (g/mol) | Observed [M – H]+ ions | R108-, NF1799-, NF5230-, NF8294A-NS and Hetero | NF8294A- and NF5230-Homo |
|---|---|---|---|---|---|
| Apigenin | C15 H10 O5 | 270 | 269 | + | + |
| Chrysoeriol | C16 H12 O6 | 300 | 299 | + | + |
| Quercetin | C15 H10 O7 | 302 | 301 | + | + |
| Luteolin | C15 H10 O6 | 286 | 285 | + | + |
| Tricin | C17 H14 O7 | 330 | 329 | + | + |
| Apigenin-7-O-GlcA | C21 H18 O11 | 446 | 445 | + | − |
| Chrysoeriol-GlcA | C22 H20 O12 | 476 | 475 | + | − |
| Quercetin-GlcA | C21 H18 O13 | 478 | 477 | + | − |
| Luteolin-7-O-GlcAa | C21H18 O12 | 462 | 461 | + | − |
| Tricin-GlcA | C23 H22 O13 | 506 | 505 | + | − |
| Apigenin-glucoside | C21 H20 O10 | 432 | 431 | + | + |
| Chrysoeriol-7-O-GlcAa | C22 H22 O11 | 462 | 461 | + | + |
| Quercetin-glucoside | C21 H20 O12 | 464 | 463 | + | + |
| Luteolin-glucoside | C21 H20 O11 | 448 | 447 | + | + |
| Tricin-glucoside | C23 H24 O12 | 492 | 491 | + | + |
| Kaempferol-3-O-rutinoside | C27 H30 O15 | 594 | 593 | + | + |
| Kaempferol-3-O-rutinoside-glucoside | C33 H40 O20 | 756 | 755 | + | + |
| Laricitrin-3-O-glucopyranoside-5′-O-glucopyranosyl-7-O-glucosidea | C34 H42 O23 | 818 | 817 | + | − |
| Apigenin-7-O-glucuronopyranosyl-(1-3)-O-glucuronopyranosyl-(1-2)- O-glucuronopyranosidea | C33 H34 O23 | 798 | 797 | + | − |
| Apigenin-7-O-glucuronopyranosyl-(1-2)-O-glucuronopyranosyl-(1-2)- O-glucosidea | C33 H36 O22 | 784 | 783 | + | − |
| Apigenin-7-O-glucuronopyranosyl-(1-2)-O-glucuronopyranosidea | C27 H26 O17 | 622 | 621 | + | − |
| Apigenin-7-O-glucuronopyranosyl-(1-2)-O-glucopyranosidea | C27 H28 O16 | 608 | 607 | + | − |
| Chrysoeriol-7-O-glucuronopyranosyl-(1-2)-O-glucuronopyranosidea | C28 H28 O18 | 652 | 651 | + | − |
| Tricin-7-O-glucuronopyranosyl-(1-2)-O-glucuronopyranosidea | C29 H30 O19 | 682 | 681 | + | − |
| Chrysoeriol-7-O-glucuronopyranosyl-(1-2)-O-glucopyranosidea | C28 H30 O17 | 638 | 637 | + | − |
| Tricin-7-O-glucuronopyranosyl-(1-2)-O-glucopyranosidea | C29 H32 O18 | 668 | 667 | + | − |
| Apigenin-7-O-[2′-O-feruloyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C37 H34 O20 | 798 | 797 | + | − |
| Apigenin-7-O-[2′-O-coumaroyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C36 H32 O19 | 768 | 767 | + | − |
| Apigenin-7-O-[2′-O-feruloyl-glucuronopyranosyl-(1-2)- O-glucopyranoside]a | C37 H36 O19 | 784 | 783 | + | − |
| Chrysoeriol-7-O-[2′-O-feruloyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C38 H36 O21 | 828 | 827 | + | − |
| Tricin-7-O-[2′-O-feruloyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C39 H38 O22 | 858 | 857 | + | − |
| Apigenin-7-O-[2′-O-coumaroyl-glucuronopyranosyl-(1-2)- O-glucopyranosidea | C36 H34 O18 | 754 | 753 | + | − |
| Apigenin-7-O-[2′-O-sinapoyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C38 H36 O21 | 828 | 827 | + | − |
| Chrysoeriol-7-O-[2′-O-coumaroyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C37 H34 O20 | 798 | 797 | + | − |
| Chrysoeriol-7-O-[2′-O-feruloyl-glucuronopyranosyl-(1-2)- O-glucopyranoside]a | C38 H38 O20 | 814 | 813 | + | – |
| Tricin-7-O-[2′-O-feruloyl-glucuronopyranosyl-(1-2)-O-glucopyranoside]a | C39 H40 O21 | 844 | 843 | + | − |
| Chrysoeriol-7-O-[2′-O-coumaroyl-glucuronopyranosyl-(1-2)- O-glucopyranoside]a | C37 H36 O19 | 784 | 783 | + | − |
| Apigenin-7-O-[2′-O-feruloyl-[glucuronopyranosyl-(1-3)]- O-glucuronopyranosyl-(1-2)-O-glucuronopyranoside]a | C43 H42 O26 | 974 | 973 | + | – |
| Tricin-7-O-[2′-O-coumaroyl-glucuronopyranosyl-(1-2)- O-glucuronopyranoside]a | C38 H36 O21 | 828 | 827 | + | – |
| Formononetin-7-O-glucosidea | C22 H22 O9 | 430 | 429 | + | + |
| Formononetin-7-O-glucoside malonatea | C25 H24 O12 | 516 | 515 | + | + |
Compounds identified previously (Kowalska et al., 2007; Marczak et al., 2010). (+), detected; (−), not detected. Numbers of samples analyzed R108 WT (1), NF8294A NS (1), NF8294A heterozygote (1), NF8294A homozygote (2), NF1799-NS (4), NF5230 NS (2), and NF5230 heterozygote (3) NF5230 homozygote (1). Supporting information (extracted ion chromatograms and mass spectra) are available in Supplemental Figures S16–S25.
Using apigenin and apigenin-7-O-β-d-glucuronide as standards, LC–MS analysis showed the presence of apigenin aglycone in all of the samples analyzed, whereas apigenin-GlcA was detected in the WT, null segregants, and all UGT84F9 heterozygous transposon insertion plants, but not in the homozygous UGT84F9 knockout mutants (Table 2 and Figures 8, A–J, 9). However, glucosides of apigenin were detected in all the analyzed samples including the homozygous ugt84f9 knockout mutants (Supplemental Figure S17). Loss of function of UGT84F9 in the homozygous Tnt1 insertion line also led to loss of apigenin-7-O-GlcA-(1-2)-O-GlcA and apigenin 7-O-[2′-O-coumaroyl-GlcA-(1-2)-O-GlcA (Supplemental Figures S18, S19). The same conclusion was reached from analysis of tricin, luteolin, chrysoeriol, laricitrin, quercetin, and formononetin aglycone and glucuronide molecular ions (Table 2); the aglycones and corresponding flavonoid glucosides (with one exception, see below) were present in all samples analyzed (Supplemental Figures S20, S21), whereas the glucuronides were not detected in the homozygous ugt84f9 knockout mutants (Table 2). Together, Supplemental Figures S17–S26 document that loss of function of UGT84F9 leads to loss of glucuronidated flavonoids in both leaves and roots of the two independent ugt84f9 mutant alleles, whereas most glucosylated flavonoids were still present. The only exception was the loss of laricitrin-3-O-glucopyranoside-5′-O-glucopyranosyl-7-O-glucoside in homozygous ugt84f9 mutant lines (Supplemental Figures S27, S28).
To further examine the effect of loss of function of UGT84F9 on flavonoid glucoside levels, we analyzed leaves of 4-weeks-old plants of homozygous, heterozygous, and null segregant ugat transposon insertion lines for the levels of apigenin, luteolin, and formononetin monoglucosides. There was no significant change in the levels of apigenin and formononetin glucosides at the P = 0.05 level, but the levels of luteolin glucoside were reduced by more than 90% (Supplemental Figure S29).
Discussion
Selection of UGATs from M. truncatula
The PSPG motif is conserved across the plant UGT family and has therefore been used as an identifying signature of plant UGTs (Bowles et al., 2005, 2006). However, it is very difficult to predict substrate specificity, particularly the sugar donor preference, based on the UGT candidate sequence alone. Indeed, it has been proposed that plant UGT clades may organize in terms of regio-specificity on acceptor substrates without necessarily defining the nature of that acceptor, and this regio-specificity is considered to have been established among plant lineages prior to speciation whereas, in contrast, sugar recognition is believed to be a local adaptation among plant species (Noguchi et al., 2009).
The overall sequence identities or similarities of annotated UGT genes from M. truncatula to the previously identified plant UGATs (e.g. F7GAT, BpUGT94B1, and UGT73P12) clearly could not be used as a basis for candidate selection due to the low sequence identity between UGTs from M. truncatula compared with the previously characterized UGATs from other plant species. For example, UGT88E28 is the closest candidate from M. truncatula to the Lamiales F7GAT UGT88D7, with 42% protein sequence identity, whereas the sequence identity of UGT84F9 to UGT88D7 is only 23%.
In virtually all of the previously studied plant UGATs, a positively charged residue such as Arg, e.g. Arg25 in BpUGT94B1 and UGT73P12, is found proximal to a highly conserved catalytic His in the N-terminus, or the Arg residue is found within the PSPG box, e.g. Arg350 in UGT88D7. This Arg residue was considered to be involved in recognition of UDP-GlcA as sugar donor (Sawada et al., 2005; Noguchi et al., 2009; Nomura et al., 2019). However, we did not find any UGAT candidate from M. truncatula having Arg in a corresponding position; instead, similarly charged residues such as His or Gln were found in such positions, and we used these features to arrive at our final list of candidate UGATs. However, from 19 candidate UGATs, only one, UGT84F9, exhibited significant UDP-glucuronosyltransferase activity against flavonoid acceptors.
UGT84F9 is a bifunctional UDP-glucosyl/glucuronosyl transferase that is necessary and sufficient for synthesis of multiple flavonoid glucuronides in M. truncatula
Unlike most of the plant UGATs characterized to date, UGT84F9 glucuronidates a wide range of flavonoid acceptor substrates in vitro, including flavones, isoflavones, flavonols, flavan-3-ols, and stilbenes, as well as monoglycosides of some of these compounds. The biochemical activity of UGT84F9 in vitro was most favorable toward the flavones that are known to be glucuronidated in M. truncatula (Kowalska et al., 2007; Marczak et al., 2010; Staszkow et al., 2011), suggesting an in planta function. The enzyme also efficiently glucosylates flavones and the flavonol quercetin, and likely other flavonoid classes, in vitro. It is interesting that the enzyme produced two monoglucuronide products in equal amounts from luteolin, but only a single major glucoside product. Further structural studies are needed to explain this phenomenon.
Utilization of UDP-Glc as sugar donor is considered the default mechanism of glycosylation in many plant species for detoxification of phytochemicals (Kitamura, 2006). In this regard, the overall catalytic efficiency of UGT84F9 in terms of Kcat/Km is largely in favor of glucosylation for the flavone substrates tested here. Although the Kcat values using UDP-GlcA as sugar donor for UGT84F9 are generally higher compared with UDP-Glc, the higher Km values for UDP-GlcA reverse the overall Kcat/Km ratio toward favoring UDP-Glc as sugar donor, at least in vitro. The generally high Km values for UGT84F9 are consistent with the kinetic parameters reported for Lamiales F7GATs such as UGT88D4 (Noguchi et al., 2009). Site-directed mutagenesis confirmed that His22 is functionally equivalent to Arg25 in UGATs from the Lamiales family, and conversion of His 22 to Ser strongly inhibited UGAT activity while maintaining glucosyltransferase activity.
It is perhaps surprising that the plant UGATs have relatively high Km values for UDP-GlcA, as the levels of this nucleotide sugar in plant tissues appear to be much lower than the levels of UDP-Glc (Alonso et al., 2010). It was also interesting that the Km of UGT84F9 for the acceptor quercetin was lower with the sugar donor UDP-Glc compared with UDP-GlcA. Grapevine VvGT5 is a monofunctional flavonoid glucuronosyltransferase with high activity with quercetin and kaempferol; the Arg140Trp mutation almost completely abolishes activity with UDP-GlcA as sugar donor, but this mutant enzyme is then active with UDP-Glc and UDP-galactose (UDP-Gal). In this case, the Km for kaempferol is also affected by the sugar donor, being approximately 2.5-fold lower with UDP-Glc than UDP-Gal (Ono et al., 2010). Similar differences in Km for the flavonoid acceptor were seen with the grapevine bifunctional glucosyl/galactosyltransferase VvGT6 with UDP-Glc and UDP-Gal (Ono et al., 2010). The structural basis for such effects of sugar donor on the affinity for the sugar acceptor requires further investigation.
Loss of function of UGT84F9 through transposon insertion mutagenesis resulted in the disappearance of all the documented flavonoid glucuronides, including di- and triglucuronides, of M. truncatula, but did not affect the presence of most flavone, isoflavone, and flavonol glucosides. All the di- and triglucuronides analyzed were sugar substituted at a single position of the flavonoid ring, so it was not possible to tell without additional analysis whether the enzyme is catalyzing sugar–sugar conjugation in planta. Although the function of UGT84F9 as a flavonoid glucuronosyltransferase in planta was confirmed, the glucosyltransferase activity of the enzyme in planta, at least for the majority of the compounds tested, remains unclear. Medicago truncatula contains a number of enzymes that can collectively glucosylate the compounds tested here, perhaps redundantly (Modolo et al., 2007). Nevertheless, it does appear that UGAT84F9 is important, but not essential, for the glucosylation of luteolin, whereas loss of function of UGT84F9 does not reduce apigenin glucoside levels. Apigenin and luteolin differ in the presence of the additional OH group at the 3′-position in luteolin, which is therefore the likely site of attachment of the glucose moiety affected in the knock-out line. The one exception where UGT84F9 appeared essential for flavonoid glucosylation was the case of laricitrin-3-O-glucopyranoside-5′-O-glucopyranosyl-7-O-glucoside. This unusual triglycosidic conjugate of a 3′-O-methylated flavonol contains glucose residues attached to the A ring and the 3-position on the central heterocyclic C ring. Further work will be necessary to determine which of these three positions is/are conjugated by the enzyme.
The above conclusions should be qualified by the fact that we did not analyze the total flavonoid glucoside pools by LC–MS; several of these compounds are further substituted with malonate on the sugar moiety to provide acidity to aid in retention in the vacuole (Matern et al., 1986); it is assumed that the glucuronides require no further substitution for efficient vacuolar sequestration.
The phenomenon whereby the in vitro substrate preference of an enzyme does not reflect its in vivo function is not uncommon among plant biosynthetic enzymes, and certainly among the plant UGT enzyme family. For example, UGT71G1 from M. truncatula prefers quercetin and 5-hydroxyisoflavone in vitro more than its in vivo preferred triterpene substrate (Achnine et al., 2005), and UGT78G1 shows a strong preference for isoflavones in vitro (Modolo et al., 2007) but functions as an anthocyanidin glucosyltransferase in planta (Peel et al., 2009). Such apparent inconsistencies may reflect substrate accessibility (Jørgensen et al., 2005), substrate availability/concentration in planta, or, as is most likely in the present case of sugar donor specificity, the presence of redundant enzymes.
Irrespective of the mechanistic aspects of UGT84F9 specificity, the present loss of function lines provides a useful tool for future studies to address the roles of flavonoid glucuronidation in M. truncatula under biotic or abiotic stress, and the reason why loss of function of this one enzyme leads to negative growth impacts. Growth may be affected by loss of flavonoid glycosides (Taylor and Grotewold, 2005), or could perhaps reflect loss or changes of other potential glucuronidated products such as triterpene saponins (Naoumkina et al., 2010). Further study of additional candidate UGATs such as J2, 3, 7, and 8 with expression in leaf and root tissue is also warranted. Although they clearly do not possess redundant functions with UGT84F9 for flavonoid glucuronidation, they could potentially glucuronidate alternative acceptors. It will also be important to determine the structures of the di-glucuronides produced by UGT84F9 with substrates such as quercetin and luteolin.
UGT84F9 as a chassis for design of UGATs for biochemical semi-synthesis
Understanding the mechanisms whereby polyphenol-rich botanical supplements ameliorate symptoms of neurological disorders will require the availability of the brain-targeted phase II metabolites derived from the constituent polyphenols. Because of the expense and complexity of current chemical synthetic approaches (Docampo-Palacios et al., 2020a, 2020b), enzyme-based approaches require consideration. Ideally, a single enzyme with wide acceptor substrate specificity would be advantageous. Overall, the acceptor specificity of UGT84F9 appears to be broader than that reported for other plant UGATs (Nagashima et al., 2000; Sawada et al., 2005; Noguchi et al., 2009; Ono et al., 2010). A comparison of the specific activity of recombinant UGT84F9 with the mammalian UGATs with the best activities with flavonoids (UGATs 1A7 and 1A9) reveals that all three have highest activity with quercetin and lowest activity with epicatechin, and that UGT84F9 has the highest activity with flavones, among its natural substrates in vivo. Because of its being an operationally soluble enzyme and therefore of easier expression/handling than the intrinsically membrane-bound mammalian UGATs, UGT84F9 would be a good candidate for semi-synthesis of potentially neuro-active quercetin and methyl-quercetin glucuronides, either in vitro or in a microbial cell-based system (Yang et al., 2016). However, such approaches would still require separation of the final products in view of the somewhat promiscuous regioselectivity of UGT84F9.
UGT84F9 was relatively inefficient for glucuronidation of flavan-3-ols and stilbenes, important compounds with pharmacological and neuroprotective activity (Wang et al., 2012; Gomes et al., 2018). Only one product was formed from epicatechin and its 3′- and 4′-O-methyl derivatives. Further studies are required to determine the position of glucuronidation. Obtaining a crystal structure for UGT84F9 will be important for protein engineering efforts to broaden its acceptor/regio-specificity and/or tailor its catalytic efficiency with different substrates. The alternative approach would be to attempt to modify existing plant UGTs with the desired acceptor specificity to work with UDP-GlcA as well as or in place of UDP-Glc. However, very little success has been achieved with such attempts, and it has been suggested that it is far easier to switch the donor specificity of UGTs from UDP-GlcA to UDP-Glc than vice versa (Noguchi et al., 2009). Clearly, the molecular basis for recognition of UDP-GlcA by glycosyltransferases requires further study.
Materials and methods
Plant materials and growth conditions
Seeds of M. truncatula (ecotype R108) wild type and transposon insertion mutant lines were obtained from Noble Research Institute, Ardmore, OK. The seeds were prepared for germination by suspending in 1 mL of 100% sulfuric acid in 2 mL Eppendorf tubes for 4 min, followed by three to four washes with sterile water. Bleach solution (40% (v/v)) was then used to sterilize for 10 min followed by three to four washes with sterile water. The seeds were then placed on B5 agar media plates, ranging from 7 to 10 seeds per plate, for germination. To induce rooting for germination, the plates were covered with aluminum foil and placed at 4°C for 5 days, after which they were transferred to constant light until germination. After germination, the seedlings were potted in soil (2:1 professional growing mix soil: vermiculite) and transferred to a Conviron growth chamber set at 22°C, with day/night settings of 16-h light and 8-h dark, with watering every 3–5 days.
Chemicals and enzymes
UDP-GlcA and UDP-Glc were purchased from Carbosynth Ltd, UK. Apigenin, luteolin, quercetin, epicatechin, kaempferol, and quercetin-3-O-GlcA (standard) were from Sigma–Aldrich. Kaempferol-3-O-rutinoside was from Extrasynthese (Z.I. Lyon Nord, France). Authentic standards of apigenin-7-O-GlcA, luteolin-7-O-GlcA, and quercetin-7-O-GlcA were generated using the previously characterized Lamiales F7GAT, UGT88D4 specific for glucuronidating only at the 7-OH (Noguchi et al., 2009).
All restriction enzymes, DNA ligase, and high-fidelity DNA polymerase for molecular cloning were purchased from New England Biolabs unless otherwise stated. Trizol reagent for RNA extraction, reverse transcriptase kit for first strand cDNA synthesis (superscript III reverse transcriptase kit), and the entry vector pENTR-D-TOPO kit were purchased from Thermo Fisher Scientific. For qPCR, SYBR Select master mix (Applied Biosystems) and the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific) were used.
Recombinant mammalian UGAT enzymes were purchased from Corning Life Sciences, NY, USA. These enzymes (protein stock concentration 5 mg/mL) were prepared from insect cell lines infected with baculovirus harboring cDNAs encoding human UGTs (UGT1A-1, 3, 4, 6, 7, 8, 9, 10 and UGT2B-4, 15, 17). The same supplier provided UDP-GlcA and the reaction buffer (250 mM Tris–HCl, pH 7.5).
Phylogenetic analysis and homology modeling
For phylogenetic analysis, sequences of candidate UG(A)Ts from M. truncatula were aligned with previously characterized UG(A)Ts using multiple sequence comparison by Log-Expectation (MUSCLE). The resulting alignment was used to construct the maximum-likelihood phylogeny tree by performing 1,000 bootstrap replicates using the PhyML 3.0 algorithm.
The available crystal structure of UGT71G1 (pdb code 2ACV) from M. truncatula was used as reference to generate three-dimensional structural models by automated protein modeling using SWISS-MODEL. The glucose component of the UDP-Glc bound in UGT71G1 was changed to UDP-GlcA which was then superimposed onto UGT84F9 using Wincoot software. WinCoot and PyMOL software were further used to analyze the structural model, the architecture of the active site, and enzyme–substrate interactions, facilitating the understanding of substrate binding and catalysis. The PyMOL software was also used for figure preparation.
DNA and RNA isolation and cDNA synthesis
Plant genomic DNA was isolated from leaf tissue using CTAB buffer (2% w/v cetyl trimethylammonium bromide [CTAB], 1% polyvinyl pyrrolidone, 100 mM Tris–HCl, 1.4 M NaCl, 20 mM EDTA). The extraction buffer (500 µL of CTAB) was used to homogenize 60–100 mg of pulverized plant tissue and the homogenate allowed to incubate at 60°C in a water bath for 30 min. The homogenate was then centrifuged at 14,000 × g for 5 min, after which the supernatant was transferred into a new tube, and 5 µL of RNase A solution was added and incubated at 32°C for 20 min. An equal volume of chloroform/isoamyl alcohol (24:1) was added to the mixture, and vortexed for 5 s before centrifuging at 14,000 × g for 1 min. The aqueous upper phase was then transferred to a new tube. To precipitate the DNA, 0.7 vol isopropanol was added and the mixture allowed to incubate at −20°C for 15 min. The samples were centrifuged at 14,000 × g for 10 min. The pellets were collected by gently decanting the supernatants, followed by washing with 500 µL of 70% ethanol. The DNA pellet was then air dried and the DNA dissolved in 20 µL TE buffer (10 mM Tris, pH 8, 1 mM EDTA).
To isolate total RNA, different tissues harvested from M. truncatula (ecotype R108) were ground in liquid nitrogen and stored at −80°C until use. Pulverized plant tissue (60–100 mg) was resuspended in 1 mL of Trizol by vortexing, followed by incubating at room temperature for 3 min, and then centrifuging at 12,000 × g for 15 min at 4°C. The separated upper aqueous phase was collected by pipetting into a clean 1.5 mL Eppendorf tube. Isopropanol (0.5 mL) was added, gently mixed, and incubated for 10 min prior to centrifugation at 12,000 × g for 10 min at 4°C. The resulting supernatant was discarded and the pellet was washed in 1 mL of DNAse/RNAse free 75% ethanol, air dried for 8 min, and then resuspended in DNAse/RNAse free water, after which a nanodrop was used to quantify the extracted RNA.
To remove genomic DNA, the extracted RNA was subjected to DNAse treatment. Three micrograms of extracted RNA was treated with 1 unit of DNAase (Ambion) and incubated at 37°C for 30 min after which the DNAse was inactivated by addition of DNAse-inactivation reagent (Ambion). The mixture was centrifuged at 10,000 × g for 5 min and the supernatant that was now free of contaminating genomic DNA was collected and used for reverse transcription for cDNA synthesis. Equal amounts of RNA extracted from different tissues (leaf, stem, root, flower, shoot, and seed pod) were used for first strand cDNA synthesis, using superscript III reverse transcriptase kit (Thermo Fisher Scientific) and oligo (dT)20 primer. The reaction mixture contained 1 µL of 50 µM oligo (dT)20, 2.5 µg of total RNA, and 1 µL of 10 mM dNTP in a reaction volume of 13 µL. This was incubated at 65°C for 5 min in a thermocycler followed by cooling on ice for 2 min, and briefly centrifuged to pull down the contents before adding 4 µL of 5× first strand buffer, 1 µL of 0.1 M DTT, 1 µL of (40 units/µL) RNaseOUT, and 1 µL of (200 units/µL) SuperScript III RT. The reaction was gently mixed by pipetting up and down and incubated in a thermocycler set at a temperature cycle of 25°C for 5 min and 50°C for 60 min with reaction inactivated at 70°C for 15 min. To remove RNA complementary to the cDNA, 1 µL (two units) of RNase H was added to the reaction mixture and incubated at 37°C for 20 min.
RT-qPCR analysis
The synthesized cDNAs from RNA extracted from each organ were used as templates for transcript level quantification by RT-qPCR using the RT-qPCR primers sets for the individual genes, as well as tubulin housekeeping gene (Supplemental Table S4). The specificity of the primers was validated on the basis of melting profiles that showed single products at specific melting temperatures. The qPCR reaction mix was set up in triplicate for each gene for a given tissue sample, following the SYBR Select master mix protocol (Applied Biosystems), and the reactions were run in a QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific). The results were normalized to tubulin as the reference gene and analyzed as expression ratios to a “control” using the ΔΔCt method (Thermo Fisher Scientific).
Molecular cloning
The candidate UGAT gene fragments were amplified from the generated cDNA template from the RNA isolated from M. truncatula using candidate UGAT gene-specific primer sets (Supplemental Table S4). Gateway Technology was then utilized to clone selected candidate UG(A)T genes into the entry vector pENTR-D-TOPO, according to the Gateway cloning kit manufacturer (Thermo Fisher Scientific). Inserts were verified by sequencing prior to subcloning of the entry clones into destination vectors by LR-based cloning into 6 × His tagged pDEST17 (Invitrogen) or traditional restriction enzyme-based cloning into MBP-tagged pMAL-c5X vector, the latter being used in cases of initial protein insolubility during expression in E. coli.
Heterologous expression and protein purification
Chemically competent E. coli BL21(DE3) cells (Novagen) were transformed with expression plasmid constructs harboring candidate UG(A)Ts (His-tagged pDEST17-UGT for UGT88E28, UGT88E27, UGT78G3, UGT72Y5, and UGT88E2) or MBP-tagged pMAL-UGT for UGT84F9 generated as described above. The transformation of chemically competent cells with the plasmid construct was carried out by the heat shock method (Huff et al., 1990), and the positive transformants were selected using ampicillin-/carbenicillin-supplemented LB agar media. A positive colony was then grown in ampicillin/carbenicillin supplemented Luria–Bertani (LB) broth medium at 37°C overnight. The overnight culture was subcultured into fresh LB medium supplemented with ampicillin/carbenicillin, and incubated at 37°C until OD600 reached 0.7–0.8, after which protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5–1 mM. At this point, the culture was further incubated at 16°C for 16–18 h. The bacterial cells were then harvested by centrifuging the culture at 4,500 × g for 15 min at 4°C, and the harvested cells stored at −80°C prior to protein extraction and purification.
For isolation and purification of induced protein, the harvested cells were first thawed on ice and resuspended in lysis buffer (50 mM Tris–HCl, pH 7.5, 10% glycerol, 0.5 M NaCl, 50 mM imidazole, and 5.88 µM β-mercaptoethanol), freeze-thawed in liquid nitrogen in a water bath at 42°C for three to four cycles, and then sonicated on ice (20 s pulse on, 10 s pulse off, frequency 25 Hz), for 2–10 min depending on the sample volume. The lysed cells were then centrifuged at 13,000 rpm for 2 min to sediment the cell debris and the soluble protein collected for purification. The protein was purified using either Ni2+-NTA resin for N-terminal His-tagged fusion proteins or amylose resin for MBP-tagged proteins. One milliliter of soluble protein was mixed with 120–250 μL Ni2+-NTA resin or amylose resin, previously equilibrated with the lysis buffer. The tagged protein was allowed to bind to immobilized metal (Ni2+) or amylose resin for 45 min under gentle rotation at 4°C in a cold room. The unbound protein was released by washing three times with lysis buffer, and the tagged protein was finally collected using elution buffer (lysis buffer supplemented with a final concentration of 250 mM imidazole for His-tagged protein or 10 mM amylose for MBP-tagged protein). The eluted proteins were collected, concentrated, and exchanged with 50 mM Tris–HCl buffer (pH 7.5) using Amicon ultracentrifuge filters (EMD Millipore) following the manufacturer’s recommended protocol, and finally quantified by Bradford assay (Bradford, 1976) and qualitatively checked by SDS–PAGE (Laemmli, 1970). Densitometric analysis by Image-j software was then used to estimate the amount of the protein band of interest relative to the contaminating protein bands to allow adjustment of the volume of recombinant UGT84F9 or subsequent mutants for enzyme assay.
Site-directed mutagenesis
Mutagenic primer sets (listed in Supplemental Table S4) were designed based on the codon of the amino acid substitution to be introduced and were ordered for synthesis (Sigma–Aldrich). The QuickChange Lightening Site-Directed Mutagenesis Kit (Agilent Technologies) was used to amplify the entire plasmid harboring the target UG(A)T using the corresponding mutagenic primer sets following the manufacturer’s protocol. The amplified product was checked on a 1% agarose gel and the non-mutated parental plasmid DNA was digested using DpnI restriction enzyme at 37°C for 1 h. Chemically competent E. coli cells were transformed with 2 µL of the reaction and plated on selection (ampicillin/carbenicillin) LB agar media from which a single colony was subcultured into 2 mL LB broth supplemented with ampicillin/carbenicillin. Plasmids were extracted from positive transformed E. coli cells and the mutation was verified by sequencing. The mutant protein was expressed in BL21 (DE3) E. coli (Novagen) and the UGT enzyme activity assay was carried out as described below.
Identification of glucuronide products
The position of glucuronidation of flavonoid acceptors was determined by comparison with authentic standards (either purchased, generated enzymatically, or chemically synthesized in our laboratory (Blount et al., 2012, 2015; Docampo-Palacios et al., 2020a, 2020b). For products of mammalian UGTs, glucuronidation positions in cases where standards were not available were assigned tentatively based on information from the literature (Docampo et al., 2017). For further details, see Supplemental Table S5.
Enzyme assays and kinetics
Enzyme assays were performed according to an established method (Blount et al., 2015). Briefly, different concentrations of acceptor substrates from 0.1 to 5 mM final concentration were mixed with 2 mM UDP-sugar in a final concentration of 50 mM Tris–HCl buffer (pH 7.5). The enzymatic reaction was started by the addition of enzyme (recombinant mammalian or candidate UG(A)T enzyme) with final protein concentration in the range of 0.06–0.153 mg/mL as determined to be optimal for individual reactions for plant UGT enzyme or 100 µg mammalian UGT enzyme. The final volume of the reaction mixture was 100 μL, and the enzyme assay was incubated at 30°C for a predetermined time period (10 min to 12 h depending on the reaction rate) and the reaction stopped by the addition of 100 μL MeOH. The reaction mixture was centrifuged at 10,000 × g for 10 min at 4°C, and the supernatant filtered through a 0.2-mm membrane filter and the filtrate analyzed by reverse phase HPLC and LC–MS, according to the protocol described below.
For kinetic analyses, the final reaction mixture of 50 μL consisted of various concentrations of the variable substrates (generally 5 μM to 1.5 mM for acceptor substrate and 0.25–8 mM for sugar donor) while the counter-substrate was maintained at saturating concentration (5 mM for UDP-GlcA or UDP-Glc and 2 mM for acceptor substrate). All reactions were performed according to the previously established reaction conditions with 50 mM Tris–HCl buffer pH 7.5 (Sawada et al., 2005; Noguchi et al., 2009; Ono et al., 2010; Liu et al., 2018; Nomura et al., 2019). The reaction was started by the addition of enzyme (1.5–2 μg), incubated at 30°C for 20 min, and stopped by addition of an equal volume of methanol. Products were detected and measured by HPLC. To determine the kinetic properties of the candidate UGTs, the apparent Vmax and Km values were determined by fitting the initial velocity to the Michaelis–Menten equation by non-linear regression using GraphPad Prism 7.04.
Screening M. truncatula Tnt1 retrotransposon mutants
Leaf tissue was harvested for DNA extraction using CTAB buffer as described above. Using gene-specific primers (UGT84F9-forward and UGT84F9-reverse) in combination with Tnt1 primers (Tnt1-forward and Tnt1-reverse), PCR screening was used to identify Tnt1 insertions in the gene of interest. Primer sets are listed in Supplemental Table S4.
PCR reaction mixtures containing 25 μL of 1× GoTaq Green Master Mix (Promega) and 1 μL of template DNA were set up to contain 1 μM of UGT84F9-forward/UGT84F9-reverse primer combination, UGT84F9-forward/Tnt1-reverse primer combination, or Tnt1-forward/UGT84F9-reverse primer combination primers. The thermocycler conditions were 1 cycle of 95°C for 3 min initial denaturation step, followed by 40 cycles of 95°C for 30 s denaturation step, 55°C for 30 s annealing step, and 72°C for 1.45 min extension step. A final extension was at 72°C for 5 min. The PCR products were resolved by electrophoresis in 1% agarose gels and visualized under the UV transilluminator.
Extraction of flavonoid metabolites
Leaf tissue of M. truncatula wild-type and Tnt1 insertion lines was harvested from 6-week-old plants and ground into powder under liquid nitrogen. The pulverized leaf tissue (100 mg fresh weight) was re-suspended in 80% (v/v) methanol for homogenization. Similarly, root tissues were harvested, washed in deionized water before pulverizing under liquid nitrogen, and homogenizing in 80% (v/v) methanol. The volume of MeOH was adapted based on the amount of tissue material, to give 100 mg sample in 1 mL of 80% methanol. The suspension was placed in an ultrasonic bath for 30 min followed by centrifuging at 10,000 × g for 10 min to remove cell debris, and the supernatant was then transferred into a new screw-capped tube. The solvent was evaporated to near dryness to remove methanol using a SpeedVac concentrator (Thermo Scientific) and the dried extract dissolved in 1 mL of milli-Q water according to a previously established method (Marczak et al., 2010).
A Water’s Sep pak C18 cartridge was used for further purification, sugar removal, and desalting. The C18 cartridge was conditioned by washing with 100% methanol followed by three to four washes with water before the sample was loaded onto the C18 cartridge, and washed two times with water prior to eluting using 1.3 mL of 100% methanol. The eluted extract was then used for LC–MS analysis.
HPLC analysis
Analytical HPLC was used to analyze products from enzyme assays or kinetic studies. A sample volume of 50 µL was applied to a reverse phase C18 Hypersil Gold column (250 × 4.6 mm dimensions, particle size 5 µm; Agilent Technology). The solvents used were 0.1% formic acid (solvent A) and 100% acetonitrile (solvent B). The solvent flow gradient was 95% A, 5% B for 5 min; 85% A, 15% B for 5 min; 77% A, 23% B for 15 min; 67% A, 33% B for 5 min; 60% A, 40% B for 5 min; 0% A, 100% B for 5 min; and 95% A, 5% B for 5 min. Chromatograms were monitored at 350 nm for flavones, flavonols and derivatives, and 280 nm for flavan-3-ol (epicatechin) and derivatives using an Agilent G1315C diode array detector. Products were verified by HPLC (for retention time and UV spectrum) and by LC–MS in comparison with chemically synthesized authentic standards whenever possible.
LC–MS analysis
An Agilent 1290 Infinity II system coupled with 6460 QQQ triple quadrupole mass spectrometer (Agilent) was used to analyze glucuronidated products from enzyme assays as well as the flavonoid extracts from M. truncatula tissues. This was set up with an electrospray ionization source (AJS ESI) operated in negative ionization mode. An XTerra MS C18 column, 2.1 × 250 mm dimensions, 5 µm particle size (Waters) was used for liquid chromatography separation. The solvents used as mobile phase were 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B), with the solvent flow rate set at 0.45 mL/min and the solvent flow gradient was 95% A, 5% B, 0–5 min; 90% A, 10% B, 5–10 min; 80% A, 20% B, 10–15 min; 40% A, 60% B, 15–20 min; 5% A, 95% B, 20–28 min; and 95% A, 5% B, 28–30 min. The mass spectrometer data were acquired, recorded, and processed using Agilent MassHunter Qualitative analysis B.06.00 and the mass spectrometer was set to scan m/z from 100 to 1,000. The conditions for ionization were 300°C for gas temperature (capillary temperature), 7 V for voltage used. The nitrogen (sheath gas) flow rate was at 3.0 L/min.
The targeted metabolites were profiled by scanning for their reported molecular ions (m/z) from the total ions acquisition, and confirmed by comparison of their extracted ion chromatograms to those previously established for Medicago flavonoid aglycones, glucuronides, and glucosides, including di- and triglucuronides of apigenin, luteolin, tricin, and chrysoeriol (Kowalska et al., 2007; Marczak et al., 2010; Staszkow et al., 2011).
Statistical analysis
Statistical analysis of flavonoid levels used three biological replicates, with comparison to null-segregant controls by one-way ANOVA with Tukey’s test (GraphPad Prism 7.04). P < 0.05 was taken as indicating significant differences.
Data deposition
All data needed to evaluate the conclusions in this article are present in the article and/or the Supplementary Materials.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S2.
Supplemental data
The following supplemental materials are available.
Supplemental Figure S1. Chemical structures of the major (iso)flavonoid compounds tested as substrates for UG(A)Ts in the present work.
Supplemental Figure S2 . Screening of mammalian UGATs for activity against flavonoid compounds.
Supplemental Figure S3 . LC–MS analysis of the reaction products from incubating UGT1A9 with luteolin as acceptor substrate and UDP-GlcA as sugar donor.
Supplemental Figure S4 . Outline of methods and criteria for selection of candidate UG(A)T genes from the genome of the model legume M. truncatula.
Supplemental Figure S5. Amino acid sequence alignment around the largely conserved aspartic acid residue in plant UGTs believed to be involved in stabilizing the catalytic histidine involved in deprotonation of the acceptor substrate during catalysis.
Supplemental Figure S6. Multiple sequence alignment of the conserved PSPG motif located in the C-terminal domain of plant UGTs.
Supplemental Figure S7. Phylogenetic analysis of selected UG(A)T candidates.
Supplemental Figure S8. Expression of UGAT candidates and previously characterized UGTs in M. truncatula.
Supplemental Figure S9. SDS–PAGE of purified recombinant UGT84F9 and its site-directed mutants.
Supplemental Figure S10. HPLC-UV analysis of conjugated flavones generated by recombinant UGT with UDP-GlcA as sugar donor.
Supplemental Figure S11. LC–MS analysis of glucuronidated products of luteolin generated by recombinant UGT84F9 with UDP-GlcA as sugar donor.
Supplemental Figure S12 . HPLC–UV and LC–MS analysis of glucuronidated products of UGT84F9 activity with the di-glycoside kaempferol-3-O-rutinoside and the mono-glucuronide Q-3-O-GlcA as sugar acceptors and UDP-GlcA as sugar donor.
Supplemental Figure S13. Michaelis–Menten curves for activity of UGT84F9 toward acceptor substrates.
Supplemental Figure S14. Michaelis–Menten curves for wild-type and mutant UGT84F9 toward sugar donor substrates with quercetin and apigenin as acceptors.
Supplemental Figure S15. Relative activity of UGT84F9 mutant and wild-type proteins toward UDP-GlcA and UDP-Glc.
Supplemental Figure S16 . Five-week-old plants of NF5230 mutant lines harboring a Tnt1 insertion in the UGT84F9 gene.
Supplemental Figure S17 . MS spectra of apigenin glucosides detected in extracts from M. truncatula leaf tissue of NF8294A null segregant, and heterozygous and homozygous knockout lines.
Supplemental Figure S18. LC–MS analysis of extracts from M. truncatula leaf tissue for detection of apigenin-7-O-GlcA-(1-2)-O-GlcA and apigenin-7-O-[2′-O-coumaroyl-GlcA-(1-2)-O-GlcA.
Supplemental Figure S19. Mass spectra of extracts from M. truncatula leaf tissue for detection of apigenin-7-O-GlcA-(1-2)-O-GlcA, apigenin-7-O-GlcA-(1-3)-O-GlcA-(1-2)-O-GlcA, chrysoeriol-7-O-[2′-O-coumaroyl-GlcA-(1-2)-O-GlcA], and apigenin-7-O-[2′-O-coumaroyl-GlcA-(1-2)-O-GlcA.
Supplemental Figure S20. LC–MS chromatograms of formononetin-7-O-glucoside and quercetin glucoside detected in extracts from M. truncatula leaf tissue of NF8294A UGT84F9 null segregant, heterozygote, and homozygous knockout lines.
Supplemental Figure S21. Mass spectra corresponding to the chromatograms showing formononetin-7-O-glucoside and quercetin glucoside in Supplemental Figure S20.
Supplemental Figure S22. Disappearance of apigenin glucuronide due to loss of function of UGT84F9 in the homozygote ugt84f9 Tnt1 insertion in NF5230 mutant lines of M. truncatula.
Supplemental Figure S23. Mass spectra corresponding to the metabolites in Supplemental Figure S22.
Supplemental Figure S24. LC–MS analysis of flavonoid metabolites from root extracts of NF8294A-null segregant, -heterozygote, and -homozygote ugt84f9 Tnt1 insertion mutants of M. truncatula.
Supplemental Figure S25. Mass spectra corresponding to apigenin (at m/z 269) in Supplemental Figure S24.
Supplemental Figure S26. Mass spectra corresponding to apigenin-7-O-GlcA in Supplemental Figure S24.
Supplemental Figure S27. Disappearance of laricitrin-3-O-glucopyranoside-5′-O-glucopyranosyl-7-O-glucoside due to loss of function of UGT84F9 in the homozygous ugt84f9 Tnt1 insertion in NF8294A mutant lines M. truncatula.
Supplemental Figure S28. Mass spectra corresponding to laricitrin-3-O-glucopyranoside-5′-O-glucopyranosyl-7-O-glucoside in Supplemental Figure S27.
Supplemental Figure S29. Levels of apigenin, luteolin, and formononetin monoglucosides in ugt84f9 Tnt1 insertion lines.
Supplemental Table S1 . Screening of selected flavonoid compounds and their derivatives for glucuronidation by mammalian UGATs
Supplemental Table S2. GenBank accession numbers of the UG(A)T sequences analyzed in the present work
Supplemental Table S3 . Kinetic parameters of recombinant UGT84F9 with selected flavonoid substrates in the presence of saturating concentrations of sugar donor
Supplemental Table S4. Primer sets used in this study
Supplemental Table S5. List of standards or methods used for identification of glucuronide products
Supplementary Material
Acknowledgments
The authors thank Dr. Ji Hyung Jun for technical suggestions on tnt1 insertion mutant seed selection, growth and genotyping, and Dr. Xiaolan Rao for assistance with protein domain analysis. They acknowledge the BioAnalytical Facility at the University of North Texas for assistance with mass spectrometry analyses during this work. Finally, they acknowledge that the contents of this study do not represent the views of the NCCIH, the ODS, the NIH, the U.S. Department of Veterans Affairs, or the United States Government.
Funding
The M. truncatula plants utilized in this research project, which are jointly owned by the Centre National de la Recherche Scientifique and the Samuel Roberts Noble Foundation, Ardmore, Oklahoma, USA, were created through research funded, in part, by Grant No. 703285 from the National Science Foundation. This study was supported by the University of North Texas, and by Grant Number P50 AT008661-01 from the National Institutes of Health (NCCIH and ODS). Dr. G.M.P. holds a Senior VA Career Scientist Award.
Conflict of interest statement. None declared.
R.A.D., G.M.P., and X.W. conceived the research. R.A.D. supervised the experiments. O.A.A. performed all molecular, biological, and biochemical experiments. M.L.D.-P. and A.A.-H. performed the chemical syntheses. X.W. assisted with structural modeling. R.A.D. wrote the manuscript and agreed to serve as the author responsible for contact and ensured communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Richard A. Dixon (richard.dixon@unt.edu).
References
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41:875–887 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Piasecki RJ, Wang Y, LaClair RW, Shachar-Hill Y (2010) Quantifying the labeling and the levels of plant cell wall precursors using ion chromatography tandem mass spectrometry Plant Physiol 153:915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount JW, Ferruzzi M, Raftery D, Pasinetti GM, Dixon RA (2012) Enzymatic synthesis of substituted epicatechins for bioactivity studies in neurological disorders. Biochem Biophys Res Commun 417:457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount JW, Redan BW, Ferruzzi MG, Reuhs BL, Cooper BR, Harwood JS, Shulaev V, Pasinetti G, Dixon RA (2015) Synthesis and quantitative analysis of plasma-targeted metabolites of catechin and epicatechin. J Agric Food Chem 63:2233–2240 [DOI] [PubMed] [Google Scholar]
- Bowles D, Isayenkova J, Lim E-K, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8:254–263 [DOI] [PubMed] [Google Scholar]
- Bowles D, Lim E-K, Poppenberger B, Vaistij FE (2006) Glycosyltransferases of lipophilic small molecules. Annu Rev Plant Biol 57:567–597 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim E-K, Bowles DJ, Davies GJ, Edwards R (2007) Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci U S A 104:20238–20243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commenges D, Scotet V, Renaud S, Jacqmin-Gadda H, Barberger-Gateau P, Dartigues J-F (2000) Intake of flavonoids and risk of dementia. Eur J Epidemiol 16:357–363 [DOI] [PubMed] [Google Scholar]
- de Fátima Â, Docampo-Palacios M, Alvarez-Hernandez A, Pasinetti G, Dixon RA (2019) An efficient synthesis of deoxyrhapontigenin-3-O-β-D-glucuronide, a brain-targeted derivative of dietary resveratrol, and its precursor 4’-O-Me-resveratrol. ACS Omega 4:8222–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo M, Olubu A, Wang X, Pasinetti G, Dixon RA (2017). Glucuronidated flavonoids in neurological protection: structural analysis and approaches for chemical and biological synthesis. J Agric Food Chem 65:7607–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo-Palacios M, Alvarez A, Adiji O, de Fatima A, de Oliveira A, De Abreu H, Pasinetti G, Dixon RA (2020a) Glucuronidation of methylated quercetin derivatives: chemical and biochemical approaches. J Agric Food Chem 68: 14790–14807 ( 10.1021/acs.jafc.0c04500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo-Palacios ML, Alvarez-Hernández A, de Fátima A, Liao LM, Pasinetti GM, Dixon RA (2020b) Efficient chemical synthesis of (epi)catechin glucuronides, brain-targeted metabolites for treatment of Alzheimer’s disease and other neurological disorders. ACS Omega 5: 30095–30110 ( 10.1021/10.1021/acsomega.0c04512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholami A, De Geyter N, Pollier J, Goormachtig S, Goossens A (2014) Natural product biosynthesis in Medicago species. Nat Prod Rep 31:356–380 [DOI] [PubMed] [Google Scholar]
- Gomes BAQ, Silva JPB, Romeiro CFR, Dos Santos SM, Rodrigues CA, Gonçalves PR, Sakai JT, Mendes PFS, Varela ELP, Monteiro MC (2018) Neuroprotective mechanisms of resveratrol in Alzheimer’s disease: role of SIRT1. Oxid Med Cell Longev 2018:8152373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromoto T, Honjo E, Noda N, Tamada T, Kazuma K, Suzuki M, Blaber M, Kuroki R (2015) Structural basis for acceptor–substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci 24:395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ferruzzi MG, Janle EM, Wang J, Gong B, Chen T-Y, Lobo J, Cooper B, Wu QL, Talcott ST, et al. (2013) Identification of brain-targeted bioactive dietary quercetin-3-O-glucuronide as a novel intervention for Alzheimer’s disease. FASEB J 27:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JP, Grant BJ,, Penning CA, Sullivan KF (1990) Optimization of routine transformation of Escherichia coli with plasmid DNA. Biotechniques 9:570–572 [PubMed] [Google Scholar]
- Jørgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Møller BL (2005) Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 8:280–291 [DOI] [PubMed] [Google Scholar]
- Kitamura S (2006) Transport of flavonoids: from cytosolic synthesis to vacuolar accumulation. In Grotewold E, ed., The Science of Flavonoids. Springer, New York, NY, pp. 123–146 [Google Scholar]
- Kowalska I, Stochmal A, Kapusta I, Janda B, Pizza C, Piacente S, Oleszek W (2007) Flavonoids from barrel medic (Medicago truncatula) aerial parts. J Agric Food Chem 55:2645–2652 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) SDS-page Laemmli method. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- Li L, Modolo L V, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X (2007) Crystal structure of Medicago truncatula UGT85H2—insights into the structural basis of a multifunctional (iso) flavonoid glycosyltransferase. J Mol Biol 370:951–963 [DOI] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276:4338–4343 [DOI] [PubMed] [Google Scholar]
- Liu X, Cheng J, Zhang G, Ding W, Duan L, Yang J, Kui L, Cheng X, Ruan J, Fan W, et al. (2018) Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak L, Stobiecki M, Jasiński Michałand Oleszek W, Kachlicki P (2010) Fragmentation pathways of acylated flavonoid diglucuronides from leaves of Medicago truncatula. Phytochem Anal 21:224–233 [DOI] [PubMed] [Google Scholar]
- Matern U, Reichenbach C, Heller W (1986) Efficient uptake of flavonoids into parsley (Petroselinum hortense) vacuoles requires acylated glycosides. Planta 167:183–189 [DOI] [PubMed] [Google Scholar]
- Miley MJ, Zielinska AK, Keenan JE, Bratton SM,, Radominska-Pandya A, Redinbo MR (2007) Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J Mol Biol 369:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo LV, , Escamilla-Trevino LL, , Dixon RA,, Wang X ( 2009) Single amino acid mutations of Medicago glycosyltransferase UGT85H2 enhance activity and impart reversibility. FEBS Lett 583: 2131–2135 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang X, Dixon RA (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64:499–518 [DOI] [PubMed] [Google Scholar]
- Nagashima S, Hirotani M, Yoshikawa T (2000) Purification and characterization of UDP-glucuronate: baicalein 7-O-glucuronosyltransferase from Scutellaria baicalensis Georgi. cell suspension cultures. Phytochemistry 53:533–538 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, Modolo LV, Huhman DV, Urbanczyk-Wochniak E, Tang Y, Sumner LW, Dixon RA (2010) Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22:850–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A, Horikawa M, Fukui Y, Fukuchi-Mizutani M, Iuchi-Okada A, Ishiguro M, Kiso Y, Nakayama T, Ono E (2009) Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21:1556–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Seki H, Suzuki T, Ohyama K, Mizutani M, Kaku T, Tamura K, Ono E, Horikawa M, Sudo H, et al. (2019) Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J 99:1127–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25:1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Homma Y, Horikawa M, Kunikane-Doi S, Imai H, Takahashi S, Kawai Y, Ishiguro M, Fukui Y, Nakayama T (2010) Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22:2856–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Imberty A, Olsen CE, Moller BL (2008) Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol 148:1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel GJ, Pang Y, Modolo LV, Dixon RA (2009) The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J 59:136–149 [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Bratton SM,, Redinbo MR, Miley MJ (2010) The crystal structure of human UDP-glucuronosyltransferase 2B7 C-terminal end is the first mammalian UGT target to be revealed: the significance for human UGTs from both the 1A and 2B families. Drug Metab Rev 42:133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T (2005) UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers: enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem 280:899–906 [DOI] [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17:3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staszkow A, Swarcewicz B, Banasiak J, Muth D, Jasinski M, Stobiecki M (2011) LC/MS profiling of flavonoid glycoconjugates isolated from hairy roots, suspension root cell cultures and seedling roots of Medicago truncatula. Metabolomics 7:604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54:335–347 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8:317–323 [DOI] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP (2000) Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol 40:581–616 [DOI] [PubMed] [Google Scholar]
- Wang D, Ho L, Faith J, Ono K, Janle EM, Lachcik PJ, Cooper BR, Jannasch AH, D’arcy BR, Williams BA, et al. (2015) Role of intestinal microbiota in the generation of polyphenol-derived phenolic acid mediated attenuation of Alzheimer’s disease β-amyloid oligomerization. Mol Nutr Food Res 59:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bi W, Cheng A, Freire D, Vempati P, Zhao W, Gong B, Janle EM, Chen T-Y, Ferruzzi MG, et al. (2014) Targeting multiple pathogenic mechanisms with polyphenols for the treatment of Alzheimer’s disease-experimental approach and therapeutic implications. Front Aging Neurosci 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ferruzzi MG, Ho L, Blount J, Janle EM,, Gong B, Pan Y, Gowda GAN, Raftery D, Arrieta-Cruz I, et al. (2012) Brain-targeted proanthocyanidin metabolites for Alzheimer’s disease treatment. J Neurosci 32:5144–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ho L, Zhao W, Ono K, Rosensweig C, Chen L, Humala N, Teplow DB, Pasinetti GM (2008) Grape-derived polyphenolics prevent Aβ oligomerization and attenuate cognitive deterioration in a mouse model of Alzheimer’s disease. J Neurosci 28:6388–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Xu B, Hu M (2011) Regioselective glucuronidation of flavonols by six human UGT1A isoforms. Pharm Res 28:1905–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang H-M, Tong Y-F, Liu M-Z, Cheng KD, Wu S, Wang W (2016) Systems metabolic engineering of Escherichia coli to enhance the production of flavonoid glucuronides. RSC Adv 6:33622–33630 [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Udvardi M (2009) Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol 12:193–201 [DOI] [PubMed] [Google Scholar]
- Zong G, Fei S, Liu X, Li J, Gao Y, Yang X, Wang X, Shen Y (2019) Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-L-rhamnose and rhamnosylation mechanism. Plant J 99:257–269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.