Abstract
The presence of mixed-linkage (1,3;1,4)-β-d-glucan (MLG) in plant cell walls is a key feature of grass species such as cereals, the main source of calorie intake for humans and cattle. Accumulation of this polysaccharide involves the coordinated regulation of biosynthetic and metabolic machineries. While several components of the MLG biosynthesis machinery have been identified in diverse plant species, degradation of MLG is poorly understood. In this study, we performed a large-scale forward genetic screen for maize (Zea mays) mutants with altered cell wall polysaccharide structural properties. As a result, we identified a maize mutant with increased MLG content in several tissues, including adult leaves and senesced organs, where only trace amounts of MLG are usually detected. The causative mutation was found in the GRMZM2G137535 gene, encoding a GH17 licheninase as demonstrated by an in vitro activity assay of the heterologously expressed protein. In addition, maize plants overexpressing GRMZM2G137535 exhibit a 90% reduction in MLG content, indicating that the protein is not only required, but its expression is sufficient to degrade MLG. Accordingly, the mutant was named MLG hydrolase 1 (mlgh1). mlgh1 plants show increased saccharification yields upon enzymatic digestion. Stacking mlgh1 with lignin-deficient mutations results in synergistic increases in saccharification. Time profiling experiments indicate that wall MLG content is modulated during day/night cycles, inversely associated with MLGH1 transcript accumulation. This cycling is absent in the mlgh1 mutant, suggesting that the mechanism involved requires MLG degradation, which may in turn regulate MLGH1 gene expression.
Dark-induced degradation of the grass cell wall polysaccharide mixed-linkage glucan (MLG) is mediated by MLG HYDROLASE 1.
Introduction
Plant cells are surrounded by a cell wall made of complex networks of various polysaccharides, phenolic compounds, and proteins. A generic wall is based on cellulose microfibrils embedded in a matrix of various hemicelluloses, pectins, and/or lignin. However, the specific components, their relative abundance, and their biochemical interactions are specific to the plant species and the cell types (Pauly and Keegstra, 2010). Plant cell walls have been classified as type I wall (dicots and non-commelinid monocots) and a structurally different type II wall found in grasses (Poaceae; Carpita and Gibeaut, 1993; Somerville et al., 2004). In type II walls, the main hemicellulose found cross-linking cellulose microfibrils is glucuronoarabinoxylan (GAX), composed of a substituted β1,4-linked xylose backbone. Furthermore, compared with type I walls, grass walls contain very low amounts of xyloglucan, pectic polysaccharides, and structural proteins. Also, type II wall lignin contains ρ-hydroxyphenyl units that are only found in trace amounts in type I walls (Grabber et al., 2004). Unlike a type I wall, grass walls also contain high levels of hydroxycinnamates such as ferulic acid or p-coumaric acid, covalently linked to GAX and lignin (Hatfield et al., 1999; Saulnier et al., 1999). Another major difference of type II walls is the occurrence of mixed-linkage glucan (MLG), an unsubstituted homopolymer where β-1,4-linked glucose oligomers are connected by β-1,3 linkages (Carpita and Gibeaut, 1993; Carpita, 1996). The length and relative proportions of the oligomers vary among species but usually cellotrioses and cellotetraoses are dominant (Staudte et al., 1983). While MLG has not been observed in type I walls, it has been found in the walls of unrelated organisms such as ancient vascular plants, green algae, liverworts, bacteria, fungi, and lichens (Gorin et al., 1988; Stone and Clarke, 1992; Honegger and Haisch, 2001; Popper and Fry, 2003; Fry et al., 2008; Pettolino et al., 2009), suggesting independent evolution of this polysaccharide in multiple species (Fincher, 2009).
Advances have been made regarding the mechanism of MLG biosynthesis through the discovery and characterization of several cellulose synthase-like (CSL) proteins. Heterologous expression of various plant CSLF, CSLH, and CSLJ proteins results in the production and deposition of MLG in the walls of non-grass species that usually lack MLG (Burton et al., 2006; Doblin et al., 2009; Little et al., 2018). Multiple lines of evidence indicate that CSLF6 plays a major role in MLG biosynthesis in most grass species. CSLF6 transcripts are the most abundant among the MLG biosynthetic genes in barley (Hordeum vulgare), wheat (Triticum aestivum) and Brachypodium distachyon. Also, CSLF6 overexpression leads to increased MLG levels in transgenic plants, while CSLF6 loss-of-function results in a strong reduction in MLG abundance (Burton et al., 2008; Christensen et al., 2010).
MLG is considered a plant stage-specific polysaccharide and its accumulation appears to be regulated by synchronized mechanisms of biosynthesis and degradation. Several grass species accumulate large amounts of MLG (up to 20% dry mass) in actively growing tissues, and then degrade it once the growth/cell elongation stage ends (Carpita, 1996; Kim et al., 2000; Carpita and McCann, 2010). Due to its transitory nature, MLG has been considered a storage molecule for rapid carbon release in the growth phase of cells (Roulin et al., 2002). A handful of degrading enzymes with (1,3) (1,4)-β-d-glucan-4-glucanohydrolase activity or licheninases have been identified in barley, wheat, and rice and their respective biochemical activities characterized (Litts et al., 1990; Wolf et al., 1991; Lai et al., 1993). However, their specific biological role is largely speculative.
In the last 20 years, forward genetics has been used to identify mutants with altered plant cell walls. A myriad of large-scale screens has been performed, reaching near-saturation in model species such as Arabidopsis (Arabidopsis thaliana), a type I wall plant (e.g. Reiter et al., 1997; Turner and Somerville, 1997; Chen et al., 1998; Gille et al., 2009). These screens have been extremely useful in deciphering the mechanisms involved in the biosynthesis, deposition, turnover, and function of wall components. However, the number of high-throughput mutant screens performed in grass species is significantly lower (Carpita and McCann, 2002; Marriott et al., 2014). As a result, the catalog of wall-altered grass mutants is largely incomplete, and grass-specific aspects of the wall, such as MLG biology, remain underexplored.
Here, a forward genetic screen was performed on a chemically mutagenized maize (Zea mays) population, designed to identify mutants with altered cell wall structures and/or properties. We identified and characterized a loss-of-function mutant MLG hydrolase 1 (mlgh1), which is impaired in MLG degradation in a grass, here maize. We also show the potential advantage of mlgh1 or in combination with lignin-deficient mutations to improve the sugar yield obtained after enzymatic treatment of lignocellulosic biomass in a maize elite variety without impacting plant growth or grain yield (US Patent 13/152,219; US Patent 61/492,769; WO Patent 2,012,170,304; mlgh1 is termed candy-leaf 1 [cal1] in those patents). Our genetic and biochemical data demonstrate that the observed phenotypes are caused by a mutation in a specific (1,3;1,4)-β-d-glucanase that regulates MLG turnover following a circadian rhythm in corn.
Results
Identification and characterization of the maize mlgh1 mutant
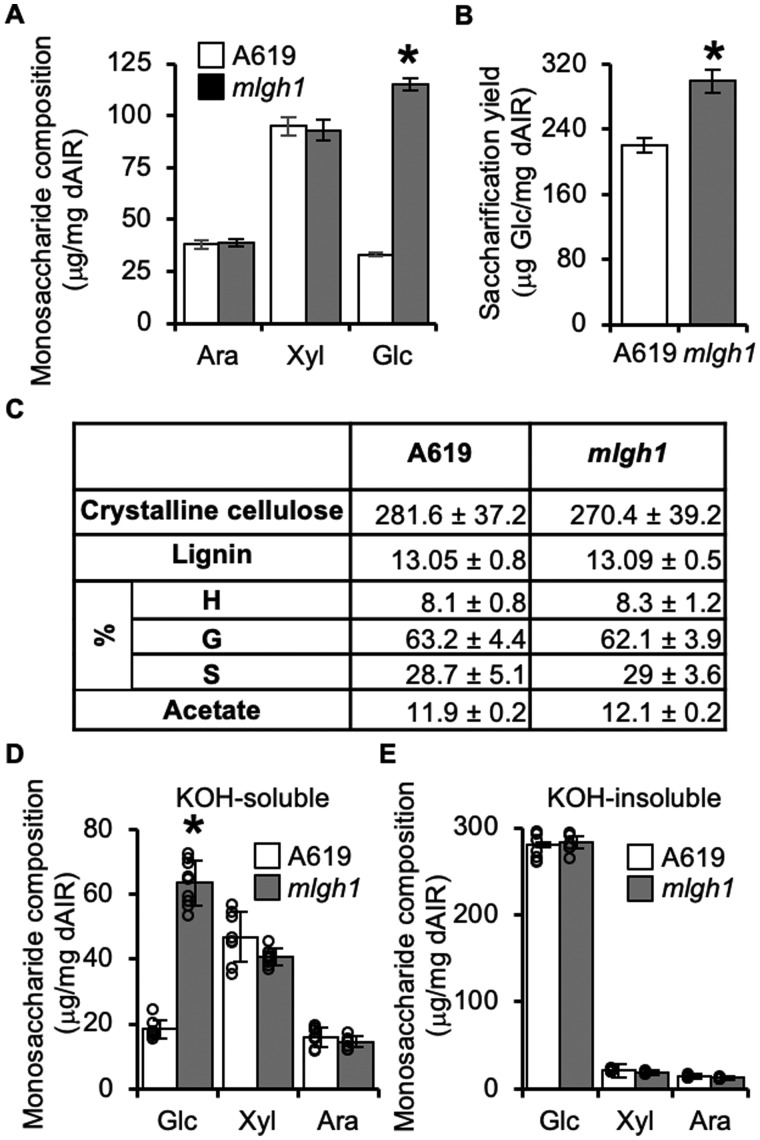
An EMS-mutagenized maize population was screened for individuals with altered cell wall attributes. For this purpose, destarched alcohol-insoluble residue (dAIR) was prepared from second leaves of individual 2-week-old mutagenized maize plants and its monosaccharide composition was determined. The mlgh1 mutant was identified because compared with the A619 non-mutagenized wild-type control, its wall material exhibited a 248.5% ± 8.5% increase in wall glucose (Glc) upon acid hydrolysis. In contrast, the content of two other abundant monosaccharides measured -xylose (Xyl) and arabinose (Ara)- remained unaltered (Figure 1, A). In addition, the high glucan content was accompanied by a 30.7% ± 6.2% increase in enzymatic saccharification yield compared with the wild type (Figure 1, B). A more detailed wall analysis of mlgh1 indicated no difference in cellulose abundance, wall-bound O-acetate, lignin content, or relative monolignol composition (Figure 1, C). The increased mlgh1-associated glucan content initially observed in young seedlings was also found in adult leaves (leaf above the ear), senesced flag leaves, and senesced stalks (internode above the ear), with increases of 51.3% ± 9.7%, 62.3% ± 11.2%, and 27.9% ± 7.6% in Glc content compared with A619 wild type, respectively. Similarly, saccharification yields were also significantly increased in adult and senesced leaves from mlgh1 individuals (Supplemental Table S1).
Figure 1.
Cell wall composition of the maize mlgh1 mutant. A, High performance anion exchange chromatography (HPAEC)-based quantification of monosaccharides released by acid hydrolysis. B, Glucose (Glc) released by enzymatic digestion of dAIR. C, Table summarizing the content in A619 and mlgh1 in terms of crystalline cellulose (mg/mg dAIR), lignin content (% of acetyl bromide soluble lignin) and lignin composition (%), and wall-bound O-acetate (mg/mg dAIR). (%) means lignin composition as the distribution of guaiacyl (G), p-hydroxyl phenol (H), and syringyl (S) monolignols. No significant differences were found between genotypes (Student’s t test, P < 0.05). D and E, HPAEC-based quantification of the monosaccharides present in KOH-soluble (D) and KOH-insoluble (E) fractions. All bar diagrams represent the mean ± standard deviation of four biological replicates. A619 (white bars) and mlgh1 (gray bars). Asterisks indicate statistical difference between the two genotypes according to Student’s t test (P < 0.05). Ara, Arabinose; Xyl, Xylose; and Glc, Glucose.
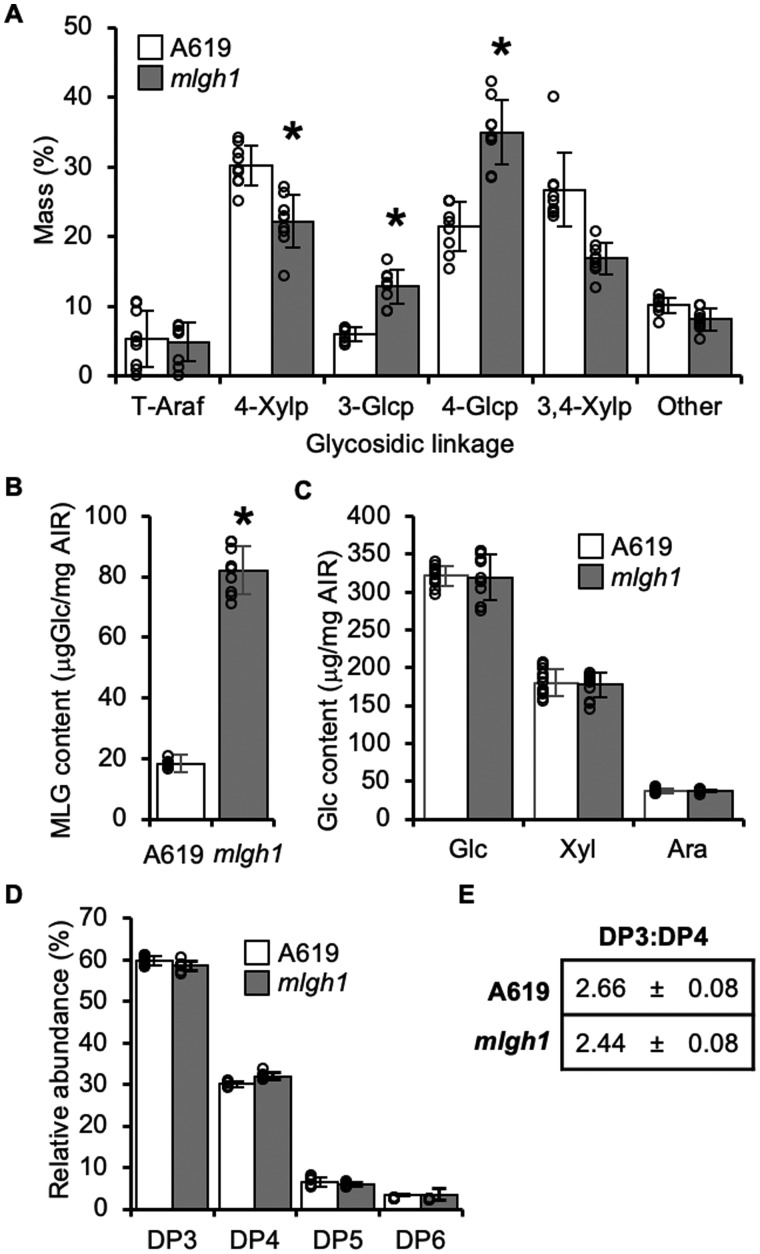
To further investigate the details of mlgh1-associated wall alterations, we focused on the second leaf of 2-week-old plants. An alkali extracted hemicellulosic fraction of mlgh1 and wild-type A619 walls was subjected to quantitative monosaccharide composition analysis. This KOH-soluble, hemicellulose-enriched mlgh1 fraction showed a 293.8% ± 17.1% increase in Glc content compared with A619 (Figure 1, D). In contrast, no differences were found in the abundance of Glc, Xyl, or Ara in the remaining KOH-insoluble residue (Figure 1, E). These results suggest that a Glc-containing hemicellulose polymer is likely affected in mlgh1 rather than cellulose. In order to determine its identity, glycosyl-linkage analysis was performed on the KOH-soluble fraction. An increase in the relative abundance of 3- and 4-linked glucopyranosyl (Glcp) residues in the mlgh1 hemicellulose fraction was observed when compared with A619 indicative of a MLG, while the remaining linkages were slightly decreased or unchanged (Figure 2, A). Hence, mlgh1 walls seem to have a higher abundance of MLG. To explore this hypothesis, the amount of MLG in the plant tissue was determined using a specific (1,3) (1,4)-β-d-glucan endoglucanase (licheninase). Digestion of AIR with this enzyme results in the release of β-glucooligosaccharides specifically derived from MLG that can then be hydrolyzed into Glc following glucosidase treatment (McCleary and Glennie‐Holmes, 1984). Using this method, mlgh1 walls showed a 229.2% ± 41.9% increase in MLG content compared with A619 (Figure 2, B). Further analysis of the licheninase-resistant residue showed no differences in monosaccharide composition among the genotypes (Figure 2, C). Combined, these results confirm that the high glucan content associated with mlgh1 results from a hyperaccumulation of MLG.
Figure 2.
MLG content and structure in mlgh1. A, Glycosidic linkage analysis of released oligosaccharides after licheninase treatment of AIR. Values represent the percentage of the total ion chromatogram peak area of the indicated partially methylated, acetylated alditols. t-Araf = terminal arabinofuranosyl; 4-Xylp = 4-linked xylopyranosyl; 3-Glcp = 3-linked glucopyranosyl; 4-Glcp = 4-linked glucopyranosyl; 3,4-Xylp = 3,4-linked xylopyranosyl. B, MLG content measured as glucose (Glc) released after licheninase digestion of AIR, followed by glucosidase treatment of the resulting (1,3;1,4) β‐glucan oligosaccharides. C, Glc content released by Saeman hydrolysis of the insoluble residue left after licheninase treatment in (B). Ara: Arabinose; Xyl: Xylose; Glc: Glucose. D, High performance anion exchange chromatography (HPAEC)-based quantification of (1,3;1,4) β‐glucan oligosaccharide products released by licheninase treatment of AIR. DP = DP of detected hexose oligomers. E, Molar ratio of cellotriosyl:cellotetraosyl units of MLG (DP3:DP4) in A619 and mlgh1. Data represents mean ± sd. All bar diagrams represent the mean ± standard deviation of n > 5 biological replicates. A619 (white bars) and mlgh1 (gray bars). Asterisks indicate statistical difference between the two genotypes according to Student’s t test (P < 0.05).
The β-glucooligosaccharides generated by licheninase treatment of AIR from mlgh1 and A619 were also quantified. Mainly hexose oligomers with degrees of polymerization (DP) of 3 and 4 were present, although small amounts of oligomers with a DP5 and DP6 were also detected (Figure 2, D). No differences between mlgh1 and A619 were found in the relative distribution of individual β-glucooligosaccharides. However, the DP3:DP4 molar ratio in the mlgh1 mutant is 2.44 ± 0.08, slightly reduced compared with A619 (2.66 ± 0.08). That difference is significant (P = 0.0001, two-tailed T test) indicating that the fine structure of the over-accumulated MLG in mlgh1 walls might be altered.
Effect of altered lignin on the increased saccharification yield associated with mlgh1
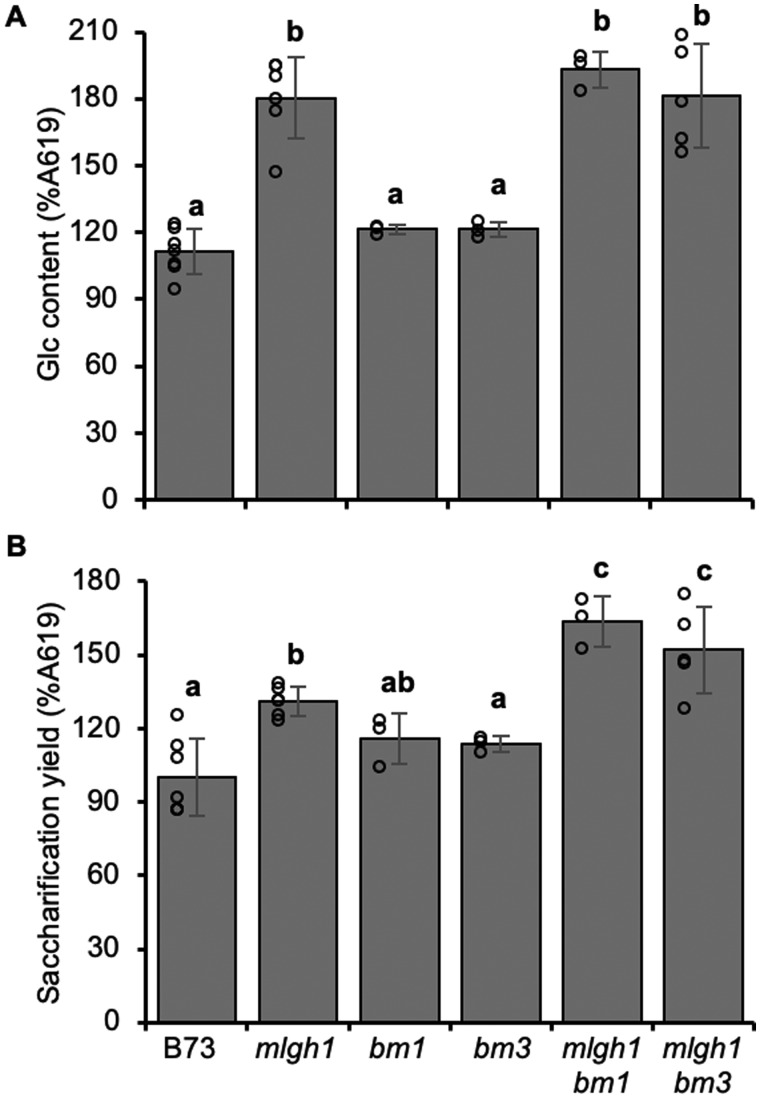
One hypothesis is that the enhanced saccharification yield observed in mlgh1 is caused by a high MLG content. No additional wall compositional differences were detected in mlgh1 walls, including crystalline cellulose content or the amount and composition of lignin, both factors reported to strongly influence saccharification yield (Grabber, 2005; Himmel et al., 2007). Lignin negatively impacts saccharification efficiency by hindering the action of hydrolytic glycanases and glycosidases. Accordingly, reduced lignin content and changes in the lignin structure, as those reported in some brown-midrib (bm) mutants, have a positive influence on wall digestibility (Saballos et al., 2008; Dien et al., 2009; Xiong et al., 2019). To study the effect of altered lignin content and composition in the enhanced saccharification yield exhibited by mlgh1, the mlgh1 mutant was crossed with individual bm lines affected in the lignin biosynthetic pathway, and the resulting double mutants were analyzed. Maize bm1 and bm3 encode a cinnamyl alcohol dehydrogenase (CAD) and caffeoyl-O-methyltransferase (COMT), respectively (Vignols et al., 1995, TPJ; Halpin et al., 1998). Comparative analyses of the saccharification yield and related wall composition traits (i.e. glucan content, total lignin content, and lignin composition) were performed (Figure 3 and Supplemental Table S2). As previously observed, mlgh1 exhibited increased saccharification values (≈25%). Interestingly, under the experimental conditions used here, no differences were observed for the saccharification yield of individual bm1 and bm3 single mutant lines compared with the B73 control. However, in the case of the mlgh1 bm1 and mlgh1 bm3 double mutants, 51% and 59% saccharification yield increases were noted compared with the wild type, respectively, suggesting a synergistic effect on saccharification efficiency between mlgh1 and each of the bm mutations used (Figure 3, B). The two mlgh1 bm double mutants and mlgh1 single mutant exhibited similar increases in wall Glc content compared with the wild type confirming the high MLG abundance (Figure 3, A). In terms of lignin content only a slight reduction in lignin content was observed in the mlgh1 bm1 line but not in the single mutant lines likely due to the developmental stage of the plants analyzed, i.e. second leaf of only 2-week-old seedlings. However, both bm3 and mlgh1 bm3 plants showed increased relative amounts of guaiacyl (G) and decreased syringyl (S) monolignol units (Supplemental Table S2). These results suggest that the increased saccharification yield observed in the mlgh1 bm1 and mlgh1 bm3 lines compared with mlgh1 are not correlated with differences in lignin or MLG contents. Instead, in the mlgh1 bm double mutant lines the combined increase of MLG content and altered lignin may lead to a different wall architecture resulting in the observed synergistic enhancement of wall digestibility.
Figure 3.
Effect of altered lignin on mlgh1 phenotype. A, Glucose (Glc) content of the hemicellulosic wall fraction (acid hydrolysis). B, Glc released by enzymatic digestion of dAIR. All bar diagrams represent the mean ± sd of n ≥ 3 biological replicates. Different letters indicate statistical differences based on ANOVA with post-hoc Tukey Honest Significance Difference test (Significant P < 0.05).
Effect of mlgh1 on plant growth and interaction with a pathogenic fungus
A detailed analysis of the seedling growth habit under controlled conditions did not reveal any difference between mlgh1 and the non-mutagenized control (Supplemental Figure S1). When grown in the field, no significant differences were found in overall plant architecture between wildtype and mlgh1 plants (Supplemental Figure S2). Diverse plant fitness-related traits were also analyzed (Supplemental Table S3). No differences were found in the moisture content in whole above-ground plant biomass or grain between the mlgh1/mlgh1 mutant in an inbred A619 background and wild-type (Mlgh1/mlgh1) Mo17/A619 or B73/A619 hybrids. Only marginal differences in grain dry weight were found between the wild-type Mo17/A619 hybrid and the mlgh1/mlgh1 mutant (P = 0.04) and differences in biomass dry weight between these two were not significant (P = 0.46). As expected, the B73/A619 hybrid, which is a much more heterotic combination than Mo17/A619 (Melchinger et al., 1991), yielded significantly more grain and biomass than either of the other two entries. Overall, these results indicate no major impact of the mlgh1 mutation on plant development and reproduction.
A higher abundance of a more easily degradable glucan in the walls of mlgh1 might lead to a higher susceptibility of plant pathogens. This does not seem to be the case. When mlgh1 and wild-type seedlings were challenged with the pathogenic fungus Ustilago maydis, no differences were found on symptom development in terms of tumor formation, anthocyanin accumulation, or appearance of chlorotic lesions (Supplemental Figure S3).
Mapping of mlgh1 mutation
To identify the causative mutation responsible for the mlgh1 chemotype, a mapping population was obtained by crossing mlgh1 in the A619 inbred by the Mo17 inbred and self-pollinating that hybrid. The glucan content of 101 individuals of the F2 mapping population was determined. From those, 25.25% of the individuals displayed a ≥200% increase in glucan content compared with A619, consistent with a recessive mutation. A Chi-square statistical analysis supported mlgh1 being a recessive mutation (Figure 4, A). The causative mutation was mapped to the region between 152.6 and 154.0 Mb on Chromosome 6 (Supplemental Figure S4). After sequencing of selected candidates among the gene models in this region (Supplemental Table S4), a G to A change was found in the second exon of gene GRMZM2G137535 causing a putative glutamic acid (E) to lysine (K) conversion in the encoded protein (Figure 4, B). A BLAST search resulted in the identification of multiple proteins with high sequence similarity and identity to GRMZM2G137535 within grass species. Phylogeny analysis showed that GRMZM2G137535 clusters together with proteins with predicted licheninase activity from the Glycoside Hydrolase Family 17 (GH17) according to CAZy classification (www.cazy.org; Supplemental Figure S5). Among them, GRMZM2G137535 shares 86.6%, 84.9%, and 86.3% identity with biochemically characterized licheninases isolated from germinated barley (EI and λHv29) and wheat (λLW2) grains, respectively (Litts et al., 1990; Wolf et al., 1991; Lai et al., 1993). The E262K substitution in mlgh1 is located in a residue highly conserved among all four proteins (Figure 4, C). Moreover, the glutamic acid E262 is predicted to be a part of the catalytic triad required for catalytic activity of the enzyme. Besides the barley and wheat putative homologs, other GH17 proteins are found in the same cluster with high identity (>80%) to GRMZM2G137535 encoded by the sorghum, oat, rice, B. distachyon, Panicum hallii, and Setaria italica genomes (Supplemental Figure S5). These results suggest that GRMZM2G137535 may encode a licheninase highly conserved among grasses.
Figure 4.
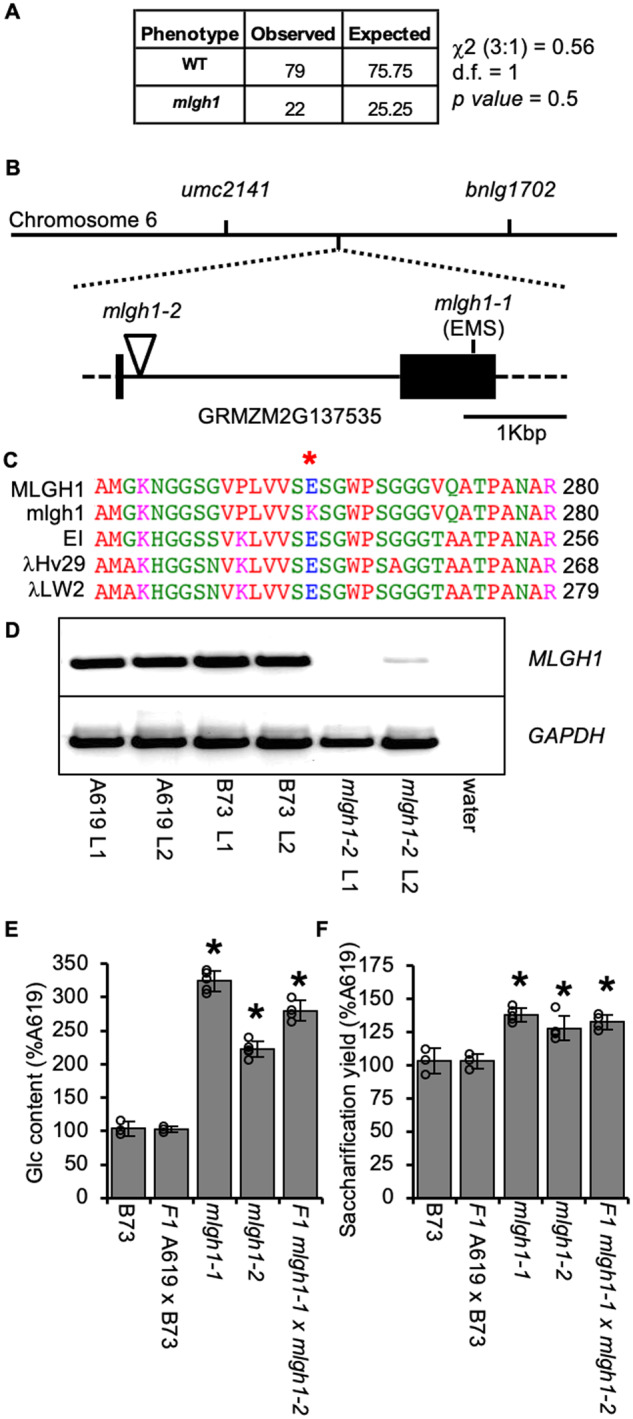
Two allelic mutants affected in the maize MLGH1 grass-specific gene encoding a putative licheninase. A, Chi-square (c2) test for a recessive segregation (3:1) of mlgh1-1 in a F2 cross to B73. d.f. = degree of freedom. P values > 0.05 means hypothesis must be accepted. B, Schematic representation of mlgh1-1 mapping on chromosome 6 between the umc2141 and bnlg1702 markers, and structure of the MLGH1 gene. Black boxes = exons; lines = introns; dashed lines = untranslated regions. Position of the mlgh1-1 and mlgh1-2 mutations is indicated. C, Detail of the protein alignment of maize MLGH1, mlgh1-1, and three highly similar licheninases from barley (EI and λHv29) and wheat (λLW2). Red asterisk indicates the mutated amino acid residue in mlgh1-1, an E262K substitution. Numbers to the right indicate the position of the last amino acid residue shown (R) related to the starting methionine. D, Agarose electrophoresis gel showing transcript level of MLGH1. RT-PCR with MLGH1-specific primers using cDNA synthetized from RNA extracted from the first (L1) and second (L2) leaf of the indicated genotypes. A negative control was included using water as template. Amplification with glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-specific primers was used as housekeeping control. E, Quantification of the hemicellulosic glucose (Glc) released after acid hydrolysis of dAIR material from the indicated genotypes. F, Glucose released by enzymatic digestion of dAIR from the indicated genotypes. Bars in E and F show the percentage of the A619 value. Values represent mean ± sd. Black asterisks indicate statistical differences to B73 according to Student’s t test (P < 0.05).
A second Mutator allele, mlgh1-2, was found in the Trait Utility System for Corn (TUSC) collection developed by Pioneer Hi-Bred International (Meeley and Briggs, 1995). The mlgh1-2 Mutator line contained an insertion in the +178-bp position into the first intron, causing a reduction in GRMZM2G137535 transcript levels in the first and second leaves (Figure 4, D). Both glucan content and saccharification yield were increased in mlgh1-2 compared with the wild-type control, although to a lesser extent than in the EMS mlgh1 allele (now termed mlgh1-1). F1 plants generated by crossing mlgh1-1 to mlgh1-2 exhibited a mutant phenotype (i.e. increased glucan content and saccharification yield), indicating that the mutants are allelic (Figure 4, E and F). Together these results indicate that MLGH1 is GRMZM2G137535.
MLGH1 overexpression in maize plants
MLGH1 overexpression (MLGH1ox) lines were generated with increased MLGH1 transcript levels (Supplemental Figure S6, A). Three independent MLGH1ox lines were selected and crossed to B73 and the resulting F1 self-pollinated. In the segregating F2 populations, glucan content and saccharification were determined. Homozygous mlgh1-1 plants were used as a control, exhibiting a 256% increase in glucan amounts compared with A619. After individual genotyping, plants negative for the presence of the MLGH1ox transgene displayed a glucan content ranging from 98% to 123.8% of the A619 value, while MLGH1ox-containing plants showed significant reductions, showing values from 49.0% to 59.4% compared with A619 (Supplemental Figure S6, B). Similarly, saccharification yields were also reduced by 27.6%–42.7% correlating with the presence of the MLGH1ox transgene, in contrast to the MLGH1ox-negative plants exhibiting values similar to A619 (106%–116.4%). As expected, the mlgh1-1 control showed an increased saccharification yield (130.5%; Supplemental Figure S6, C). These results indicate that overexpression of MLGH1 results in low MLG content and decreased enzymatic saccharification yield, the opposite chemotype in the MLGH1 loss of function alleles mlgh1-1 and mlgh1-2. We could not identify any developmental phenotype associated to MLGH1 overexpression neither in terms of node/leaf growth (Supplemental Figure S6, D–G) nor overall seedling architecture (Supplemental Figure S6, H).
Characterization of MLGH1 enzymatic activity
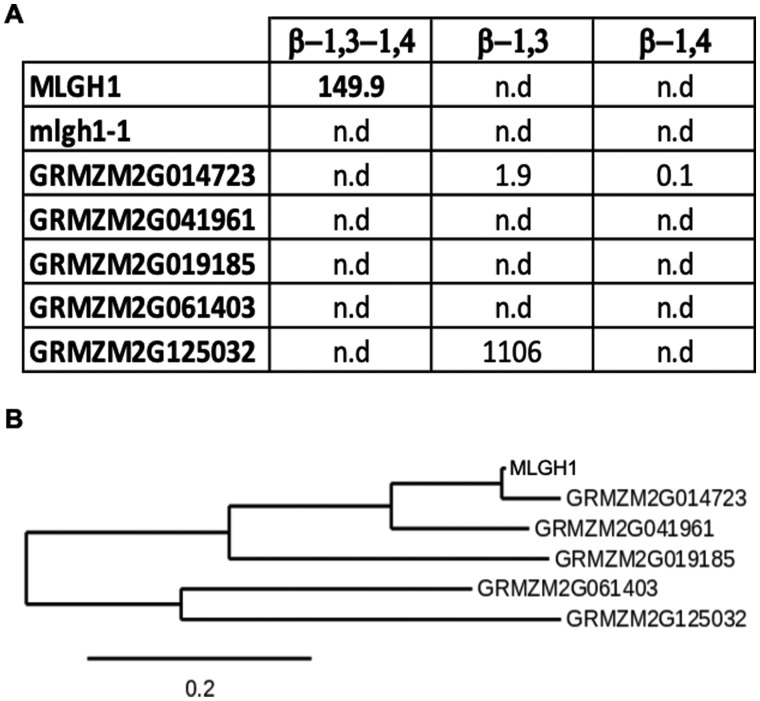
Wild-type (MLGH1) and mutant (MLGH1E262K) C-terminal His-tagged recombinant protein versions were produced in Nicotiana benthamiana by an Agrobacterium tumefaciens-mediated transient expression system. After affinity purification, the licheninase activity of both recombinant proteins was assayed using commercial MLG from barley flour as substrate (Figure 5, A). Laminarin and cello-oligosaccharides were also used as potential substrates to test the β-1,3- and β-1,4-glucanase activities, respectively. Under the conditions used, MLGH1 was only able to degrade MLG indicating its specific licheninase activity. The activity was abolished in the MLGH1E262K mutant version, indicating that the E262 residue is indeed required for protein activity and suggesting that mlgh1-1 is a loss-of-function allele (Figure 5, A).
Figure 5.
In vitro licheninase activity test of maize MLGH1 paralogs. A, Table showing the activity of MLGH1, MLGH1E262K mutant version (mlgh1-1), and five selected maize paralogs. b-1,3-1,4: licheninase activity using MLG as substrate. b-1,3: 1,3-b-glucanase activity using laminarin as substrate. b-1,4: 1,4-b-glucanase activity using cellooligosaccharides as substrate. Data shown in nKat/mg. n.d. = no activity was detected. B, Detail of the maximum-likelihood phylogenetic tree of MLGH1 and its closest paralogs in maize. See Supplemental Figure S7 for the complete phylogenetic tree.
A survey of proteins encoded in the maize genome with sequence similarity to MLGH1 resulted in 66 matches with identities ≥30%. From those, 23 proteins belong to GH17 and 43 to GH1 (Supplemental Figure S7). Only MLGH1 and the three closest putative paralogs—two GH17 proteins (GRMZM2G014723 and GRMZM2G041961) and one GH1 protein (GRMZM2G019185)—have predicted licheninase activity according to the CAZy database, while the others were classified as glucan endo-1,3-β-glucosidases (34 total, 11 GH17 and 23 GH1), O-glycosyl hydrolases (18 total, 9 GH17 and 9 GH1), or with unknown function (10 total, 1 GH17 and 9 GH1). The five closest proteins to MLGH1 were selected to test for potential licheninase activity, including the three mentioned putative licheninases (GRMZM2G014723, GRMZM2G041961, and GRMZM2G019185) and two other GH17 members with predicted glucan endo-1,3-β-glucosidase activity (GRMZM2G061403 and GRMZM2G125032; Figure 5, B). In contrast to MLGH1, none of the recombinant proteins assayed were able to degrade MLG under the assay conditions tested and only GRMZM2g125032 showed 1,3-β-glucosidase activity (Figure 5, A). This in vitro data suggest no apparent genetic redundancy of the maize MLGH1 licheninase explaining the observed drastic MLG chemotype in the mlgh1-1 mutant.
Dark-induced degradation of MLG
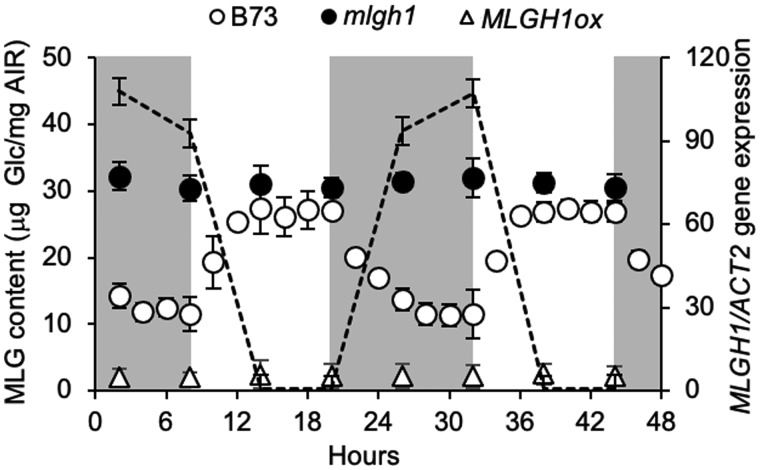
The accumulation of MLG in MLGH1-deficient and MLGH1-overexpressing plants compared with wild type was further investigated over a 48-h period. Plants were grown in a 12-h d, 24°C/12 h, 20°C night photoperiod, and the second leaf blade of 2-week-old plants was collected at different timepoints. As shown in Figure 6, MLG content in wild-type plants progressively increases during the day hours, peaking at approximately 6 h after the light cycle begins and remaining constant until dark. During the dark period, MLG content decreased, reaching minimum values after approximately 8 h. In contrast, MLG content in mlgh1-1 samples did not fluctuate but remained constant over the 2 day/night cycles studied here, with values similar to the maximum observed in wild-type samples. On the other hand, MLG content in MLGH1ox plants remained low at all timepoints, with only 10% of the highest content found in wild-type plants at the end of the day.
Figure 6.
Time course of MLGH1 gene expression and MLG content. Left axis shows MLG content measured as glucose (Glc) released after licheninase digestion of AIR followed by glucosidase treatment of the resulting (1,3;1,4) β‐glucan oligosaccharides. Wild-type (B73, empty circles), mlgh1-1 mutant (black circles) and MLGH1ox (empty triangles) values shown as mean ± sd of five biological replicates. Dashed line shows the MLGH1 gene expression in B73 measured by RT-qPCR (right axis) as mean ± sd of three biological replicates. Actin2 (ACT2) gene expression was used for normalization. Samples for both analyses were taken at the indicated time points. Gray/white areas indicate subjective night and day, respectively.
The fluctuation in MLG content in wild-type plants was inversely correlated with MLGH1 gene expression (Figure 6). MLGH1 expression was at a maximum around the end of the dark period, and its transcription was rapidly downregulated once the light cycle started. A 20,000-fold night-triggered MLGH1 transcript upregulation was observed comparing the values at the end of the night period (8 and 32 h) with the end of the day (20 and 44 h). Thus, MLGH1 transcription was maximal when MLG levels are lowest in a circadian rhythm.
As an alternative energy store in plants, starch turnover was also investigated in MLGH1-deficient or MLGH1-overexpressing plants. Starch is the most abundant carbohydrate reserve in plants and is accumulated during the day and broken down at night (reviewed in Zeeman et al., 2010). Both starch and MLG contents were quantified in 2-week-old plants after 8 and 16 h of being transferred to dark conditions and compared with plants kept in constant light. As shown in Supplemental Figure S8, degradation of MLG in the dark is completely blocked in mlgh1-1 mutant plants, while neither the total starch accumulation nor the dark-induced starch degradation seems to be affected. In MLGH1ox plants, MLG is constitutively degraded preventing its accumulation during the day and the content remains very low in all conditions tested. However, no differences were found compared with the control (MLGH1ox-negative plants) in the amount of total starch in any of the conditions analyzed here. These observations suggest that MLGH1-dependent MLG turnover does not impact starch metabolism under normal conditions.
Taken together, we propose a model where maize plants regulate the accumulation of MLG during the day time by blocking the expression of the MLGH1 (1,3;1,4)-β-glucanase. During the night, MLGH1 expression is induced/de-repressed to degrade MLG. The lack of obvious developmental or reproductive effects in plants over- or under-accumulating MLG (i.e. mlgh1 and MLGH1ox plants, respectively) grown under the conditions used here does not give further insights into the physiological role of MLG turnover. However, the extended conservation of MLGH1-like proteins among diverse grass species suggests an evolutionary advantage of the mechanism of MLG degradation. Indeed, the existence of such precise regulation of MLG content suggests these enzymes have an important biological role.
Discussion
MLG degradation
Grass cell walls accumulate large amounts of MLG in different tissues. Synthesis of this polysaccharide is performed by members of the CSLH and CSLF protein families. However, accumulation of MLG is not only affected by the efficiency of its biosynthesis. For example, no direct relationship was found between either the expression of MLG synthase genes or the synthase activity with the level of MLG in barley varieties (Tsuchiya et al., 2005; Burton et al., 2008). Instead, degradation of MLG through specific licheninases is also genetically determined, and has a strong impact on the accumulation of MLG in a particular tissue, developmental stage, and growing conditions (Roulin et al., 2002; García-Gimenez et al., 2019). QTL and GWAS studies have identified licheninases as important factors to explain the variation in the grain MLG content among diverse barley varieties (Han et al., 1995; Houston et al., 2014). Licheninase gene expression, protein accumulation, and activity have been found in germinating barley and wheat seeds (Fincher et al., 1986; Litts et al., 1990; Wolf et al., 1991; Lai et al., 1993), suggesting a common mechanism among grasses. Here, we have identified mlgh1, a maize mutant impaired in MLG degradation. In vitro, MLGH1 exhibits specific licheninase activity, and its overexpression in maize results in a virtual lack of MLG accumulation. Interestingly, the maize genome encodes multiple proteins with predicted licheninase activity belonging to the GH17 and GH1 families, showing high sequence similarity to MLGH1. However, our data strongly suggest limited functional redundancy for MLG degradation in maize seedlings, as the MLG content found in mlgh1 mutant seedlings is similar to the maximum accumulation measured in the wild-type. Additionally, the four recombinant proteins showing the highest protein sequence similarity to MLGH1 failed to degrade MLG in our in vitro activity assays despite the predicted licheninase activity. Although we cannot rule out the possibility that these or other proteins are involved in MLG degradation under specific conditions, developmental stages, and/or specific cell-types, our data suggest that MLGH1 is not only necessary but sufficient for MLG degradation.
Regulation of MLG accumulation
MLG accumulation has long been known to be stage-specific, its maximum abundance occurring during phases of rapid elongation and diminished in adult tissues (Carpita, 1996; Kim et al., 2000). In addition, our results show that MLG content also changes during day/night cycles in maize seedlings. Under the conditions tested, maximum MLG abundance is observed at the end of the day and rapidly decreases during the night. This MLG dark-induced degradation of MLG is dependent on MLGH1 expression as it is absent in the mlgh1 mutant. Proteins with high sequence similarity to MLGH1 can be found encoded in the genomes of many grass species, both cereal and non-cereal, suggesting a conserved mechanism. In fact, many previous observations are in line with the existence of a dark-induced MLG degradation in barley, rice, and wheat. For example, MLGH1-like protein levels and licheninase activity significantly increase in barley and wheat coleoptiles after prolonged dark incubation periods (Roulin et al., 2001; Roulin et al., 2002). Similarly, MLG content is lower in rice dark-grown coleoptiles compared with those grown in light, coincident with increased licheninase activity (Chen et al., 1999). Our results expand on the idea that MLG may serve as an energy storage polymer, where in young growing tissue MLG accumulates during the day, potentially as a way to store excess carbon derived from photosynthesis, and degraded during the night, providing glucose molecules under conditions of sugar depletion. However, blocking MLG accumulation and turnover as in the mlgh1 mutant does not seem to affect the developmental phenotype of the maize seedlings at least under the growth conditions used here. It is likely that based on the amount of MLG that is turned-over per day (around 25 µg/mg AIR) starch seems to be the dominant carbon storage polymer with approximately 100 µg/mg dry weight turnover per day (Supplemental Figure S8), thus allowing the plant to maintain its growth rate. However, MLG turnover may make a difference in specific circumstances such as plants grown under biotic/abiotic stresses.
Engineering of MLG content in crops
Composed of (1,3)-β-linked cellotriosyl and tetraosyl units, MLG is efficiently hydrolyzed by glucanases present in commercial enzymatic cocktails (e.g. Accelerase 1500). Thus, the enhanced saccharification yield observed when processing mlgh1 plant material is not surprising. However, the increase in MLG content alone (around 20 µg/mg AIR; Figure 2) does not account for the enhanced saccharification yield (around 80 µg/mg AIR; Figure 1). As no other polymer seems to be affected, one possible explanation could be that the high MLG content in mlgh1 affects the accessibility of the saccharification enzymes to cellulose. The amount of Glc released in mlgh1 walls after mild acid treatment (TFA) is also higher than one would expect based only on the difference in MLG content (33 µg/mg AIR in wild-type plants and 115 µg/mg AIR in mlgh1; Figure 1). It has been hypothesized that MLG might form a gel-like matrix interacting with cellulose and assisting in the proper bundle organization of microfibrils (Fincher, 2009; Smith-Moritz et al., 2015; Kim et al., 2018). As TFA can hydrolyze amorphous cellulose but not crystalline cellulose, one possible explanation could be that the high MLG content in mlgh1 somehow alters cellulose–MLG interaction improving the accessibility of the saccharification enzymes and/or altering the crystallinity of cellulose. The slight decrease in the ratio of cellotriosyl:cellotetraosyl units of MLG (DP3:DP4) observed in mlgh1 walls might result in sufficient changes in the physiochemical properties of the polymer, e.g. increasing solubility, to lead to more easily degradable walls (Fincher, 2009; Pettolino et al., 2009). Reductions of the DP3:DP4 ratio have also been reported for transgenic barley lines overexpressing the CSLF6 MLG synthase (Burton et al., 2011). As in the mlgh1 mutant, CSLF6 overexpressing lines accumulate higher amounts of MLG with a smaller DP3:DP4 ratio. The differences in the DP3:DP4 ratio reported for CSLF6 overexpressing plants (2.1 versus 2.6) are much higher compared with mlgh1 (2.44 versus 2.66). Unfortunately, saccharification data for these lines have not been reported, so it is not possible to establish any correlation.
Also unexpected was the synergistic effect observed when stacking mlgh1 with individual mutations affecting lignin (i.e. bm1 or bm3). Lignin negatively impacts lignocellulosic enzymatic hydrolysis in two ways. First, aromatic compounds bind enzymes, removing them from their substrate. Second, self-aggregation of lignin polymers results in a highly hydrophobic composite, reducing the accessibility of the enzymes and the required water to the polysaccharide substrate. Changes in the lignin content or the modification of the monolignol composition alter also the interaction between lignin and other wall polymers (e.g. cellulose). The combination of high MLG content with altered lignin in the mlgh1 bm double mutants results in higher digestibility than the addition of both individual effects. This synergy may be explained by the generation of an overall cell wall architecture that results in better enzyme substrate accessibility, as no additional compositional differences were observed. This combination thus has the potential to improve plant feedstocks for the conversion of lignocellulosic biomass into renewable biofuels and other commodity chemicals.
High digestibility in maize is a desirable trait for the feed industry. In fact, commercial maize bm3 hybrid lines showing reduced lignin content and increased digestibility are currently being marketed for silage used in dairy production, as these traits have been associated with increased nutrient intake and higher milk production (Oba and Allen, 1999; Coons et al., 2019). High MLG content also enhances nutrient intake in cattle as rumen microbes can hydrolyze MLG (Grove et al., 2006). The mlgh1 bm lines thus have the potential of combining both advantages. Furthermore, MLG content is also a potential target to improve the characteristics of grass crops as feedstocks for food, as MLG is an important source of fiber intake in the human diet. Its uptake has been associated with benefits to gut microbiota and human health, including hypercholesterolemia, diabetes, obesity, and cancer (reviewed in Jayachandran et al., 2018).
Multiple approaches aiming at increasing MLG levels in several grass species by over-expressing synthase genes turned out to show only limited success due to associated developmental defects. For example, although CslF6 overexpression results in an increased accumulation of MLG in barley or Brachypodium, severe growth problems leading to lethality become apparent. Detailed analyses of these plants showed abnormal vascular development, affecting transport of water and nutrients (Burton et al., 2011; Kim et al., 2015, 2019; Fan et al, 2018).
Our results indicate that maize plants with high MLG content can be obtained by blocking MLG degradation mediated by MLGH1. MLGH1-deficient plants do not show any obvious developmental or reproductive phenotypes in either greenhouse or field conditions. This approach likely avoids the problems of ectopic deposition of MLG or absence of auxiliary enzymes required for the correct deposition of MLG in the wall as a suggested cause of the “vascular suffocation” in overexpressing MLG-synthase strategies (Burton et al., 2011; Kim et al., 2015). Accordingly, the high MLG content associated with mlgh1 does not alter biomass or grain yields. In addition, the disease susceptibility against U. maydis in mlgh1 seems to be unaffected suggesting that the increase in MLG content may not change the overall cell wall architecture in a way that favors the penetration of specialized wall-degrading fungi.
Low MLG content is also a desirable trait for the brewing and distilling industries, as it has been associated with many quality parameters (Wang et al., 2004). Different reports describe successful strategies to reduce MLG content in diverse grass species by mutating MLG synthases. However, that reduction in MLG content has been associated with developmental- and stress-related phenotypes (Taketa et al., 2012; Vega-Sanchez et al., 2012; Garcia-Gimenez et al., 2020). For example, loss-of-function mutations in the CSLF6 MLG synthase in barley and rice results in extremely low amounts of MLG accompanied by increased sensitivity to chilling temperatures, reduction of plant height, stem diameter, spike development, or grain morphology. Interestingly, overexpression of MLGH1 seems a promising alternative strategy, as it results in a similar reduction of MLG content, but no obvious developmental phenotypes under the conditions tested (Supplemental Figure S6). Future work is required to determine if MLGH1 overexpression impacts adult plants, which might give insights into a possible structural role for MLG. However, the remaining trace amounts of MLG detected in MLGH1ox plants could still play a functional role in certain cell types where they are protected from licheninase degradation, as proposed for some specific cell types such as the vascular tissue (Trethewey et al., 2005; Fincher, 2009; Vega-Sanchez et al., 2015).
Transference of the mlgh1 traits to other grasses is possible, as MLGH1 is highly conserved in cereals including rice, wheat, barley, and millet, and also energy crops as such as miscanthus, switchgrass, and sugarcane.
Materials and methods
Mapping, plant materials, and growth
The mlgh1-1 mutant of maize (Z. mays) was identified from a F2 mutagenized population generated by ethylmethane sulfonate (EMS) mutagenesis in the A619 inbred (Neuffer, 1982; Lewis et al., 2014). The F1 plants were self-pollinated and 12 seeds per F2 family were planted in the greenhouse. F2 plants were grown under 16-h d/8-h night photoperiod, temperatures of low 20°C/high 25.6°C and watered two times per day with 100 ppm M, W, F w 20/20/20 fertilizer (Peters Professional). After 2 weeks, plants were transferred to a dark room for 20 h and the second leaf blade collected for cell wall analyses. Only normal-appearing seedlings were analyzed. Putative mutants were transplanted and grown to maturity to cross with a different inbred for mapping purposes. mlgh1 was crossed to B73 and then self-pollinated to generate an F2 mapping population. The map position was determined by pooling 30 mlgh1-1 individuals and a similar number of normal siblings and performing bulked segregant DNA mapping (Michelmore et al., 1991) which identified a 12.3-Mb interval on chromosome 6 between markers umc2141 and bnlg1702. The mlgh1 causal locus was narrowed to a region between 152.6 and 154 Mb of Chromosome 6 (Supplemental Figure S4). Sequencing of the endoglucanase candidate gene (GRMZM2G137535) found within this interval showed a base pair change consistent with EMS mutagenesis (Primer sequences can be found in Supplemental Table S5). A list of all predicted genes contained in the mapping interval according to the Zm-B73-REFERENCE-NAM-5.0 genome assembly release is provided in Supplemental Table S4. mlgh1-2 (identification number PV03 43 H-03) was obtained from the TUSC collection developed by Pioneer Hi-Bred International (Meeley and Briggs, 1995). Primers listed in Supplemental Table S5 were used to genotype these plants. bm1 and bm3 mutants were kindly provided by the Maize Genetics Coop (Catalog # 515D and 408E, respectively). A mlgh1-1 dCAP marker was used to identify the mutation in segregating populations. mlgh1MmeIF and R primers were used to amplify a 862-bp amplicon. PCR fragments were treated with MmeI restriction enzyme (Thermo) according to manufacturer’s instructions. Wild-type-derived PCR amplicons resulted in three bands of 330, 316, and 127 bp, while mlgh1-1 mutant samples showed only two bands (647 and 127 bp) due to a G–A nucleotide change in one of the MmeI restriction sites.
Generation of MLGH1 overexpression lines
For the generation of MLGH1 overexpression lines, the full-length coding sequence from MLGH1 was obtained from GeneArt (www.thermofisher.com) and then subcloned into the pUbi-BASK vector using BamHI and SpeI restriction enzymes. The expression cassette including UBI5 promoter, UBI5' intron, MLGH1 coding region, and a NOS terminator was excised using EcoRI and HindIII restriction enzymes and cloned into binary pTF101 vector (Paz et al., 2004; Addgene plasmid #134770; http://n2t.net/addgene:134770; RRID: Addgene_134770).
Hi Type II hybrid immature zygotic embryos were transformed with the pTF101:MLGH1 construct via Agrobacterium-mediated transformation at the Iowa State University Plant Transformation Facility (Frame et al., 2002; http://agron-www.agron.iastate.edu/ptf/service/agromaize.aspx). T0 transformant plantlets were assayed for transgene presence by screening for BAR marker gene-conferred resistance. A 5-cm diameter circle was drawn on an adult leaf (usually leaf 10) with a marker and applying 20 μL/mL Finale (11% w/w ammonium glufosinate) with 1 µL/mL Tween20 detergent with a cotton swab. Expression of resistance or susceptibility was studied by visual inspection after 5–7 d. Plants were genotyped for the presence of the transgene using primers newPTF101R and CL 391. To verify transgene expression, RNA was extracted from second leaves of three plants showing a positive genotyping result for each independent line and reverse-transcribed. Two pools of three plants negative for the transgene were used as controls. RT-qPCR was used to test relative expression in the different lines by amplifying the resulting cDNA using primers CL 407/CL 408.
Cell wall analyses
Plant tissues were lyophilized in a ScanVac CoolSafe Freeze‐dryer (Labogene) for 48 h and homogenized in 2 mL screw cap tubes containing two 5-mm steel beads for 2 min at 30 Hz in a MM400 mixer mill (Retsch Technology). Preparation of destarched AIR and determinations of crystalline cellulose content, total lignin content, and monolignol composition were performed as indicated in Foster et al. (2010a, 2010b). For the biomass monosaccharide composition to screen for maize mutants, a Shimadzu Prominence HPLC system equipped with a refractive index detector was used to measure glucose, xylose, and arabinose in the TFA hydrolysates. Samples were separated with a Phenomenex Rezex RFQ-Fast Acid H+ (8%) ion exchange column (100 × 7.8 mm) and a Bio-Rad Cation H guard column (30 × 4.6 mm) with 5 mM sulfuric acid as the mobile phase and a flow rate of 1.0 mL min−1 at 55°C for 5 min. The determination of total wall acetate content was performed as described in Ramirez et al. (2018).
For saccharification assays, 1 mg dAIR was incubated in the presence of 0.5 µL of Accelerase 1500 (Genencor) in 0.1 M citrate buffer pH 4.5 and 0.15 mM NaN3 in a 1.2-mL final volume reaction (Santoro et al., 2010). One 5-mm steel bead was added and reactions were incubated at 50°C for 20 h shaking at 250 rpm. Released Glc was measured in a YSI 2900 biochemistry analyzer (Xylem Inc.) following manufacturer’s instructions.
Determination of the MLG content was performed using a mixed-linkage beta-glucan assay kit (K-GLU kit, Megazyme) with modifications. In brief, 2 mg of AIR was resuspended in 200 µL 20 mM NaPO4 buffer pH 6.5 and incubated 5 min at 100°C. After cooling down to room temperature, 20 µL licheninase [specific, endo-(1-3); (1-4)-β-d-glucan 4-glucanohydrolase; Megazyme] was added and the mix incubated at 40°C for 1 h. After centrifugation 10 min at 12,000 rpm, 10 µL of the supernatant containing MLG-derived oligosaccharides was mixed with 10 µL β-glucosidase (Megazyme) and incubated 15 min at 50°C. After adding 300 µL GOPOD reagent, the mix was incubated 20 min at 50°C. OD510 was measured and the Glc content in the samples were calculated using a Glc standard curve. Glc content obtained from samples not treated with licheninase but treated with β-glucosidase was subtracted.
For the determination of MLG fine structure, 2 mg of AIR was treated with 4 M KOH for 4 h under constant shaking. Samples were centrifuged for 10 min at 12,000 rpm and supernatants transferred to a new tube and pellets washed three times with 1.5 mL water. Supernatants from all steps were combined resulting in the 4-M KOH-soluble fraction and neutralized with concentrated HCl. KOH-soluble fraction was then digested with licheninase (Megazyme) as described before. The supernatant obtained was then filtered through 0.45 µm PTFE filter units (Millipore), diluted 1:10, and injected in an ICS-3000 Dionex chromatography system (Dionex, Sunnyvale, CA, USA) equipped with a CarboPac PA200 anion-exchange column. Identification and quantification of the present cello-oligosaccharides (DP3-DP6) was performed using cellotriose, cellotetraose, cellopentaose, and cellohexaose commercial standards (Megazyme).
For the Saeman hydrolysis (Saeman, 1945) of licheninase-resistant residues, samples were dried in a Speed Vac before adding 175 µL of 72% (w/w) H2SO4. Samples were incubated at room temperature for 45 min. Glc content of the supernatant was determined by anthrone assay (Dische, 1962). Briefly, 10 µL sample was mixed with 90 µL water and 250 µL anthrone reagent (0.2% [w/w] anthrone [Sigma] in concentrated H2SO4). Glc concentration was quantified by reading the absorbance at 620 nm.
Starch analysis
Starch content was determined using the Total Starch Assay Kit (Megazyme) with the following modifications. A total of 1 mg AIR was resuspended in 100 µL thermostable amylase and incubated 15 min at 100°C. Samples were allowed to cool down and 10 µL supernatant was mixed with 10 µL α-glucosidase and incubated 30 min at 50°C. Glc content was determined using the GOPOD method as described above.
Production and purification of His-tagged proteins
Full-length coding sequences from MLGH1, mlgh1, and maize paralogs were obtained from GeneArt (www.thermofisher.com). A 3′-Histidine tag coding sequence was added using primers listed in Supplemental Table S5 and the resulting PCR products cloned in TOPO/TA vector according to manufacturer’s instructions (www.thermofischer.com). His tag-containing fragments were excised from pTOPO/TA and cloned into pBART27 (Stintzi and Browse, 2000) vector using XhoI and BamHI restriction sites.
pBART27 constructs were transformed into A. tumefaciens strain GV3101 following the protocol described in Höfgen and Willmitzer (1988). Transient transformation of N. benthamiana was performed as described in Ramirez et al. (2013). Infiltrated leaves were collected after 3 d and immediately frozen in liquid nitrogen. Frozen leaves were milled to a fine powder using mortar and pestle. Extraction buffer (50 mM sodium phosphate buffer, 1 M NaCl, pH 8, 10 μL/mL of Protease Inhibitor Cocktail [Sigma], and 0.14 μL/mL of β-mercaptoethanol) was added in a 1:3 ratio (powder/buffer) and incubated 1 h at 4°C in a rotatory wheel. After filtering through 0.45 μm filter units (Merck), His-tagged proteins were washed and purified using the Amicon Pro System (Merck) according to manufacturer’s instructions. Purified proteins were eluted twice using 100 μL elution buffer (50 mM, 300 mM NaCl, 350 mM imidazole pH 8.0).
Activity assays
Glycanase activity assays were performed in 20 mM sodium citrate buffer pH 4.5 at 37°C for 1 h using 2 μL elution solution diluted 1:20 (1.2 μg total protein) and 25 μg substrate. 150 μL water were added and reactions heat-inactivated at 100°C for 10 min. The amount of reducing ends generated was determined using the PAHBAH assay (Lever et al., 1973). Briefly, PAHBAH reagent was prepared by dissolving 5% (w/v) 4-hydroxybenzoic acid hydrazide in 0.5 N HCl and mixed 1:5 (v/v) with 5 M NaOH. 750 μL PAHBAH reagent was added to the reaction mix and incubated 5 min at 100°C. 200 μL aliquots were then transferred to 96-well UV-plates and Abs410 determined.
All substrates, (1,3-1,4)-β-d-Glucan from barley flour, and laminarin were obtained from Megazyme.
Gene expression analyses
Total RNA was extracted from pools of 3–5 s leaves using the Plant RNAeasy kit (Qiagen) using manufacturer’s instructions. DNAse-treated RNA (Turbo DNAse, Ambion) was used for cDNA synthesis (iScript, Biorad). Semi-quantitative RT-PCR was performed on 0.8 μL of cDNA resulting from reverse transcription of 500 ng of RNA. Specific primers were used to amplify MLGH1 and GAPDH as housekeeping control using Phusion Taq polymerase (Thermo Scientific). Reaction conditions and annealing temperatures followed the manufacturer’s suggestions. Products were resolved using 1% agarose gels and visualized using 0.5 μg/mL ethidium bromide. RT-qPCRs were performed in a Biorad CFX96 system using recommended parameters and data analyzed by the CFX Master Software. Actin2 or GAPDH were used as housekeeping genes. Primer sequences can be found in Supplemental Table S5.
Sequence alignments and phylogeny
Protein sequences were obtained from the Maize Genetics and Genomics Database (www.maizedgb.org). Protein alignments were performed using Clustal Omega software with default settings (Sievers and Higgins, 2018). Phylogenetic trees were constructed by using the Phylogeny.fr web service (Dereeper et al., 2008).
U. maydis infection assay
The sensitivity of the mlgh1-1 line to smut fungal infection was tested in established seedling infection assays as described before (Bösch, 2016). The solopathogenic U. maydis strain SG200 (Kämper, 2006) was grown to an OD600 of 0.8 in complete medium, washed three times with H2O, and resuspended to an OD600 of 3 in H2O. The cell suspension was injected into 7-d-old maize seedlings. Plants at 7 and 14 dpi were scored for symptom formation. Categories for disease rating were the following: 1, no symptoms; 2, chlorosis; 3, anthocyanin accumulation; 4, small tumors (<1 mm); 5, medium tumors (1–5 mm); and 6, heavy tumors (>5 mm) associated with bending of the stem.
Yield determinations
Seeds of mlgh1/mlgh1 mutants in an inbred A619 background were crossed to B73 and Mo17 to obtain wild-type (Mlgh1/mlgh1) B73/A619 and Mo17/A619 hybrids, respectively. All three entries were planted in Urbana in summer 2011 in 4 m rows spaced 0.76 m apart with 112 kg N/ha applied prior to planting. Each of the three entries was planted in eight replicates, for a total of 24 rows. Seeds were hand-planted every 15 cm. At grain maturity, a single representative plant from each row was harvested, wet grain and wet biomass were weighed separately, and a weighed subsample of grain and biomass was oven-dried at 80°C for 3 d and re-weighed to determine moisture content.
Accession numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers: MLGH1 (GRMZM2G137535): AFW77805; GRMZM2G014723: AFW81474; GRMZM2G041961: AFW77809; GRMZM2G019185: DAA55871; GRMZM2G061403: AFW83992; GRMZM2G125032: DAA55870.
Supplemental data
Supplemental Figure S1. mlgh1 plants exhibit normal growth habit.
Supplemental Figure S2. mlgh1 plants exhibit normal growth habit in the field.
Supplemental Figure S3. Ustilago maydis infection of mlgh1.
Supplemental Figure S4. Fine-mapping of mlgh1 mutation on Chromosome 6.
Supplemental Figure S5. A maximum likelihood protein sequence phylogenetic tree of MLGH1.
Supplemental Figure S6. Overexpression of MLGH1 in maize.
Supplemental Figure S7. Phylogenetic tree of maize proteins with similarity to MLGH1.
Supplemental Figure S8. Dark-induced MLG and starch degradation.
Supplemental Table S1. Cell wall composition of various tissues of the maize mlgh1 mutant.
Supplemental Table S2. Lignin content and composition in mlgh1 bm double mutants.
Supplemental Table S3. Total weight and moisture content in mlgh1 maize mutant plants.
Supplemental Table S4. List of genes included in the mlgh1 mapping interval.
Supplemental Table S5. Primers used in this study.
Supplementary Material
Acknowledgments
The authors would like to thank Grace Kayser, Jiayin Shum, Yadanar Htike, and Cesar Morphin for assistance in mapping mlgh1; Nathalie Bolduc, George Chuck, and Devi Santosh for primer design; Vincent Wu for the analysis of MLGH1ox saccharification; Anand Narayanan for help characterizing the mlgh1-2 allele; and Katharina Lufen for technical support on the cell wall analyses.
Funding
This work has been funded by the Germany’s Federal Ministry of Education and Research (BMBF) grant “Cornwall,” 031B0193A to M.P. Additional funding was provided by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2048/1—Project ID: 390686111 to M.P., Marie Curie PIOF‐GA‐2013‐623553 to V.R., and USDA-ARS CRIS 2030-21000-051-00D to S.H.
Conflict of interest statement. The authors declare that there is no conflict of interest.
S.H., M.P., and V.R. conceived and planned the experiments. F.J.K. and C.L. identified, characterized, and mapped mlgh1. M.K. and B.M.K. characterized the protein activities, MLGH1ox lines, and mlgh1 bm double mutants. C.R. carried out the seedling growth measurements. P.J.B. analyzed plant yield in a field trial. P.H. and V.G. performed the U. maydis infection experiments. V.R. and M.P. wrote the manuscript with input from all authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Vicente Ramírez (ramirezg@hhu.de).
References
- Bösch K, Frantzeskakis L, Vraneš M, Kämper J, Schipper K, Göhre V (2016) Genetic manipulation of the plant pathogen Ustilago maydis to study fungal biology and plant microbe interactions. J Vis Exp (115): 54522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Collins HM, Kibble NAJ, Smith JA, Shirley NJ, Jobling SA, Henderson M, Singh RR, Pettolino F, Wilson SM, et al. (2011) Over‐expression of specific HvCslF cellulose synthase‐like genes in transgenic barley increases the levels of cell wall (1,3;1,4)‐β‐d‐glucans and alters their fine structure. Plant Biotechnol J 9: 117–135 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol 146: 1821–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase‐like CslF genes mediate the synthesis of cell wall (1,3;1,4)‐β‐d‐Glucans. Science 311: 1940–1942 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC (2010) The maize mixed-linkage (1→3),(1→4)-β-d-glucan polysaccharide is synthesized at the Golgi membrane. Plant Physiol 153: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, McCann MC (2015) Characterizing visible and invisible cell wall mutant phenotypes. J Exp Bot 66: 4145–4163 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Chen L-M, Carpita NC, Reiter W-D, Wilson RW, Jeffries C, McCann MC (1998) A rapid method to screen for cell wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J 8: 375–382 [DOI] [PubMed] [Google Scholar]
- Chen L, Kamisaka S, Hoson T (1999) Suppression of (1->3),(1->4)-beta-D-glucan during light-induced inhibition of rice coleoptile growth. J Plant Res 112: 7–13 [Google Scholar]
- Christensen U, Alonso-Simon A, Scheller HV, Willats WGT, Harholt J (2010) Characterization of the primary cell walls of seedlings of Brachypodium distachyon—a potential model plant for temperate grasses. Phytochemistry 71: 62–69 [DOI] [PubMed] [Google Scholar]
- Coons EM, Fredin SM, Cotanch KW, Dann HM, Ballard CS, Brouillette J, Grant RJ (2019) Influence of a novel bm3 corn silage hybrid with floury kernel genetics on lactational performance and feed efficiency of Holstein cows. J Dairy Sci 102: 9814–9826 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 1: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dien BS, Sarath G, Pedersen JF, Sattler SE, Chen H, Funnell-Harris DL, Nichols NN, Cotta MA (2009) Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. Bioenergy Res 2: 153–164 [Google Scholar]
- Dische Z (1962) Color reactions of carbohydrates. In Whistler RL, Wolfram ML, eds, Methods in Carbohydrate Chemistry, Vol. 1, Academic Press, New York, pp. 478–481 [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci U S A 106: 5996–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Herburger K, Jensen JK, Zemelis-Durfee S, Brandizzi F, Fry SC, Wilkerson CG (2018) A trihelix family transcription factor is associated with key genes in mixed-linkage glucan accumulation. Plant Physiol 178: 1207–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB, Lock PA, Morgan MM, Lingelbach K, Wettenhall REH, Mercer JFB, Brandt A, Thomsen KK (1986) Primary structure of the (1–>3,1–>4)-beta-D-glucan 4-glucohydrolase from barley aleurone. Proc Natl Acad Sci U S A 83: 2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB (2009) Exploring the evolution of (1,3;1,4)-β-D-glucans in plant cell walls: comparative genomics can help! Curr Opin Plant Biol 12: 140–147 [DOI] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) part I: lignin. J Vis Exp (37): 1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp (37): 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SEK, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR (2008) Mixed-linkage (1,3;1,4)-β-D-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol 179: 104–115 [DOI] [PubMed] [Google Scholar]
- Garcia-Gimenez G, Russell J, Aubert MK, Fincher GB, Burton RA, Waugh R, Tucker MR, Houston K (2019) Barley grain (1,3;1,4)-β-glucan content: effects of transcript and sequence variation in genes encoding the corresponding synthase and endohydrolase enzymes. Sci Rep 9: 17250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gimenez G,, Barakate A, Smith P, Stephens J, Khor SF, Doblin MS, Hao P, Bacic A, Fincher GB, Burton RA, et al. (2020) Targeted mutation of barley (1,3;1,4)-β-glucan synthases reveals complex relationships between the storage and cell wall polysaccharide content. Plant J 104: 1009–1022 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC (1993) Synthesis of (1→3), (1→4)-β-d-glucan in the Golgi apparatus of maize coleoptiles. Proc Natl Acad Sci U S A 90: 3850–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci U S A 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin P, Baron M, Iacomini M (1998) Storage products of lichens. In Galun M, ed., Handbook of Lichenology, CRC Press, Boca Raton, FL, USA. pp. 9–23 [Google Scholar]
- Grabber JH, Ralph J, Lapierre C, Barrière Y (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. C R Biol 327: 455–465 [DOI] [PubMed] [Google Scholar]
- Grabber JH (2005) How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci 45: 820–831 [Google Scholar]
- Grove AV, Kaiser CR, Iversen N, Hafla A., Robinson BL, Bowman JGP (2006) Digestibility of barley beta‐glucan in beef cattle. Proc West Sect Am Soc Anim Sci 57: 367–369 [Google Scholar]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1)—a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14: 545–553 [DOI] [PubMed] [Google Scholar]
- Han F, Ullrich SE, Chirat S, Menteur S, Jestin L, Sarrafi A, Hayes PM, Jones BL, Blake TK, Wesenberg DM, et al. (1995) Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor Appl Genet 91: 921–927 [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Ralph J, Grabber JH (. 1999) Cell wall cross-linking by ferulates and diferulates in grasses. J Sci Food Agric 79: 403–407 [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807 [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (. 1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger R, Haisch A (2001) Immunocytochemical location of the (1,3;1,4)-β-glucan lichenin in the lichen-forming ascomycete Cetraria islandica (Icelandic moss). New Phytol 150: 739–746 [Google Scholar]
- Houston K, Russell J, Schreiber M, Halpin C, Oakey H, Washington JM, Booth A, Shirley N, Burton RA, Fincher GB, et al. (2014) A genome wide association scan for (1,3;1,4)-β-glucan content in the grain of contemporary 2-row Spring and Winter barleys. BMC Genomics 15: 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran M, Chen J, Chung SSM, Xu B (2018) A critical review on the impacts of β-glucans on gut microbiota and human health. J Nutr Biochem 61: 101–110 [DOI] [PubMed] [Google Scholar]
- Kämper J, Kahmann R, Bölker M, Ma L-J, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Müller O, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kim JB, Olek AT, Carpita NC (2000) Cell wall and membrane-associated exo-β-D-glucanases from developing maize seedlings. Plant Physiol 123: 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Zemelis S, Keegstra K, Brandizzi F (2015) The cytoplasmic localization of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan. Plant J 81: 537–547 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Zemelis-Durfee S,, Jensen JK, Wilkerson CG, Keegstra K, Brandizzi F (2018) In the grass species Brachypodium distachyon, the production of mixed-linkage (1,3;1,4)-β-glucan (MLG) occurs in the Golgi apparatus. Plant J 93: 1062–1075 [DOI] [PubMed] [Google Scholar]
- Lai DM, Høj PB, Fincher GB (1993) Purification and characterization of (1–>3, 1–>4)-beta-glucan endohydrolases from germinated wheat (Triticum aestivum). Plant Mol Biol 22: 847–859 [DOI] [PubMed] [Google Scholar]
- Lever M, Powell JC, Killip M, Small CW (1973) A comparison of 4-hydroxybenzoic acid hydrazide (PAHBAH) with other reagents for the determination of glucose. J Lab Clin Med 82: 649–655 [PubMed] [Google Scholar]
- Lewis MW, Bolduc N, Hake K, Htike Y, Hay A, Candela H, Hake S (2014) Gene regulatory interactions at lateral organ boundaries in maize. Development 141: 4590–4597 [DOI] [PubMed] [Google Scholar]
- Little A, Schwerdt JG, Shirley NJ, Khor SF, Neumann K, O’Donovan LA, Lahnstein J, Collins HM, Henderson M, Fincher GB, et al. (2018) Revised phylogeny of the cellulose synthase gene superfamily: insights into cell wall evolution. Plant Physiol 177: 1124–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litts JC, Simmons CR, Karrer EE, Huang N, Rodriguez RL (1990) The isolation and characterization of a barley 1,3-1,4-beta-glucanase gene. Eur J Biochem 194: 831–838 [DOI] [PubMed] [Google Scholar]
- Marriott PE, Sibout R, Lapierre C, Fangel JU, Willats WGT, Hofte H, Gomez LD, McQueen‐Mason SJ (2014) Range of cell‐wall alterations enhance saccharification in Brachypodium distachyon mutants. Proc Natl Acad Sci; U S A: 111: 14601–14606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary BV, Glennie‐Holmes M (1985) Enzymic quantification of (1→3)(1→4)‐β‐d‐glucan in barley and malt. J Inst Brew 91: 285–295 [Google Scholar]
- Meeley RB, Briggs SP (1995) Reverse genetics for maize. Maize Genet Coop Newslett 69: 67–82 [Google Scholar]
- Melchinger AE, Messmer MM, Lee M, Woodman WL, Lamkey KR (1991) Diversity and relationships among U.S. maize inbreds revealed by restriction fragment length polymorphisms. Crop Sci 31: 669–678 [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (. 1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer MG ( 1982) Mutant induction in maize. In: Sheridan WF, ed, Maize for Biological Research, Plant Mol. Biol. Assoc., Charlottesville, VA, pp. 61–64 [Google Scholar]
- Oba M, Allen MS (1999) Effects of brown midrib 3 mutation in corn silage on dry matter intake and productivity of high yielding dairy cows. J Dairy Sci 82: 135–142 [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2010) Plant cell wall polymers as precursors for biofuels. Curr Opin Plant Biol 13: 305–312 [DOI] [PubMed] [Google Scholar]
- Paz MM, Shou H, Guo Z, Zhang Z, Banerjee AK, Wang K (2004) Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136: 167–179 [Google Scholar]
- Pettolino F, Sasaki I, Turbic A, Wilson SM, Bacic A, Hrmova M, Fincher GB (2009) Hyphal cell walls from the plant pathogen Rhynchosporium secalis contain (1,3;1,6)-β-D-glucans, galacto- and rhamnomannans, (1,3;1,4)-β-D-glucans and chitin. FEBS J 276: 4122–4133 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2003) Primary cell wall composition of bryophytes and charophytes. Ann Bot Lond 91: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez V, López A, Mauch-Mani B, Gil MJ, Vera P (2013) An extracellular subtilase switch for immune priming in Arabidopsis [published correction appears in PLoS Pathog. 2016 Nov 2;12 (11):e1006003]. PLoS Pathog 9: e1003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez V, Xiong G, Mashiguchi K, Yamaguchi S, Pauly M (2018) Growth- and stress-related defects associated with wall hypoacetylation are strigolactone-dependent. Plant Direct 2: e00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W-D, Chapple C, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Roulin S, Buchala AJ, Fincher GB (2002) Induction of (1->3,1->4)-β-D-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta 215: 51–59 [DOI] [PubMed] [Google Scholar]
- Roulin S, Feller U (2001) Reversible accumulation of (1–>3,1–>4)-beta-glucan endohydrolase in wheat leaves under sugar depletion. J Exp Bot 52: 2323–2332 [DOI] [PubMed] [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeman JF (1945) Kinetics of wood saccharification—hydrolysis of cellulose and decomposition of sugars in dilute acid at high temperature. Ind Eng Chem 37: 43–52 [Google Scholar]
- Santoro N, Cantu SL, Tornqvist C, Falbel TG, Bolivar JL, Patterson SE, Pauly M, Walton JD (2010) A high-throughput platform for screening milligram quantities of plant biomass for lignocellulose digestibility. Bioenerg Res 3: 93–102 [Google Scholar]
- Saulnier L, Crépeau MJ, Lahaye M,, Thibault JF, Garcia-Conesa MT, Kroon PA, Williamson G (1999) Isolation and structural determination of two 5–5′-diferuloyl oligosaccharides indicate that maize heteroxylans are covalently cross-linked by oxidatively coupled ferulates. Carbohydr Res 320: 82–92 [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, Li L, O’Brien BA, Fajardo DS, Wroclawska E, et al. (2007) Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, Gardiner J, Bergstrom D, Houchins K, Melia-Hancock S, Musket T, et al. (2002) Development and mapping of SSR markers for maize. Plant Mol Biol 48: 463–481 [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG (2018) Clustal Omega for making accurate alignments of many protein sciences. Protein Sci 27: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Moritz AM, Hao Z, Fernández-Niño SG, Fangel JU, Verhertbruggen Y, Holman H-YN, Willats WGT, Ronald PC, Scheller HV, Heazlewood JL, et al. (2015) Structural characterization of a mixed-linkage glucan deficient mutant reveals alteration in cellulose microfibril orientation in rice coleoptile mesophyll cell walls. Front Plant Sci 6: 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Staudte RG, Woodward JR, Fincher GB, Stone BA (1983) Water-soluble (1→3), (1→4)-β-d-glucans from barley (Hordeum vulgare) endosperm. III. Distribution of cellotriosyl and cellotetraosyl residues. Carbohydr Polym 3: 299–312 [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci U S A 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BA, Clarke AE (1992) Chemistry and biology of (1,3)-β-D-glucan. La Trobe University Press, Victoria, Australia [Google Scholar]
- Taketa S, Yuo T, Tonooka T, Tsumuraya Y, Inagaki Y, Haruyama N, Larroque O, Jobling SA (2012) Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-D-glucan biosynthesis. J Exp Bot 63: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey JA, Campbell LM, Harris PJ (2005) (1->3),(1->4)-beta-d-Glucans in the cell walls of the Poales (sensu lato): an immunogold labeling study using a monoclonal antibody. Am J Bot 92: 1660–1674 [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Urahara T, Konishi T, Kotake T, Tohno-oka T, Komae K, Kawada N, Tsumuraya Y (2005) Biosynthesis of (1→3),(1→4)-β-glucan in developing endosperms of barley (Hordeum vulgare L. Physiol Plant 125: 181–191 [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Verhertbruggen Y, Christensen U, Chen X, Sharma V, Varanasi P, Jobling SA, Talbot M, White RG, Joo M, et al. (2012) Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol 159: 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomènech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang G, Chen J, Wu F (2004) The changes of β‐glucan content and β‐glucanase activity in barley before and after malting and their relationships to malt qualities. Food Chem 86: 223–228 [Google Scholar]
- Wolf N (1991) Complete nucleotide sequence of a Hordeum vulgare gene encoding (1–>3, 1–>4)-beta-glucanase isoenzyme II. Plant Physiol 96: 1382–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Wu Z, Liu Y, Li Y, Su K, Bai Z, Guo S, Hu Z, Zhang Z, Bao Y, et al. (2019) Mutation of 4-coumarate: coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol Biofuels 12: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.