Abstract
The Raf-like protein kinase abscisic acid (ABA) and abiotic stress-responsive Raf-like kinase (ARK) previously identified in the moss Physcomitrium (Physcomitrella) patens acts as an upstream regulator of subgroup III SNF1-related protein kinase2 (SnRK2), the key regulator of ABA and abiotic stress responses. However, the mechanisms underlying activation of ARK by ABA and abiotic stress for the regulation of SnRK2, including the role of ABA receptor-associated group A PP2C (PP2C-A), are not understood. We identified Ser1029 as the phosphorylation site in the activation loop of ARK, which provided a possible mechanism for regulation of its activity. Analysis of transgenic P. patens ark lines expressing ARK-GFP with Ser1029-to-Ala mutation indicated that this replacement causes reductions in ABA-induced gene expression, stress tolerance, and SnRK2 activity. Immunoblot analysis using an anti-phosphopeptide antibody indicated that ABA treatments rapidly stimulate Ser1029 phosphorylation in the wild type (WT). The phosphorylation profile of Ser1029 in ABA-hypersensitive ppabi1 lacking protein phosphatase 2C-A (PP2C-A) was similar to that in the WT, whereas little Ser1029 phosphorylation was observed in ABA-insensitive ark missense mutant lines. Furthermore, newly isolated ppabi1 ark lines showed ABA-insensitive phenotypes similar to those of ark lines. Therefore, ARK is a primary activator of SnRK2, preceding negative regulation by PP2C-A in bryophytes, which provides a prototype mechanism for ABA and abiotic stress responses in plants.
Phosphorylation in the activation loop of the Raf-like kinase ARK is critical for SNF1-related protein kinase2 regulation during abscisic acid responses in the moss Physcomitrium (Physcomitrella) patens.
Introduction
The phytohormone abscisic acid (ABA) regulates a variety of developmental as well as physiological processes in plants. In vegetative tissues, elevated endogenous ABA in response to water deficit in shoots contributes to stomatal closure and tolerance to environmental abiotic stresses such as drought and cold (Rock et al., 2010). Studies on Arabidopsis (Arabidopsis thaliana) have revealed that ABA elicits these cellular responses by binding to the intracellular ABA receptor PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PYL/RCAR; Ma et al., 2009; Park et al., 2009). The ABA-PYR/PYL/RCAR complex binds to group A protein phosphatase 2C (PP2C-A) to inhibit its phosphatase activity, which leads to activation of subclass III SNF1-related protein kinase2 (SnRK2; Umezawa et al., 2009; Vlad et al., 2009; Nishimura et al., 2010; Rodrigues et al., 2013).
Of the three SnRK2 subgroups, subclass III is recognized as the key regulator of ABA signaling since it phosphorylates various cellular substrates, including bZIP transcription factors that activate a number of ABA-induced genes (Marcotte et al., 1989; Uno et al., 2000; Fujita et al., 2005). Activity of subclass III SnRK2 is stimulated by ABA-induced phosphorylation at specific Ser residues in the “activation loop” located between subdomains VII and VIII of the kinase domain (Boudsocq et al., 2004; Kornev et al., 2006; Boudsocq et al., 2007). This phosphorylation event is operated by either autophosphorylation by SnRK2 itself or by other protein kinases, though detailed mechanisms have not been clarified.
The roles of ABA in stress responses have been demonstrated not only in angiosperms but also in basal embryophytes, mosses, and liverworts. Protonemata of the moss Physcomitrium (Physcomitrella) patens acquire tolerance to desiccation, hyperosmosis, and freezing upon exogenous ABA treatment (Minami et al., 2003; Khandelwal et al., 2010; Koster et al., 2010). ABA induces expression of genes for late embryogenesis abundant (LEA)-like proteins (Cuming and Lane, 1979) with boiling-soluble characteristics mediated by conserved ABA-responsive promoter elements in the moss protonemata (Knight et al., 1995). The P. patens genome has four putative genes for PYR/PYL/RCAR, two genes for PP2C-A, and four genes for subclass III SnRK2 but no SnRK2 of other subclasses (Rensing et al., 2008; Sakata et al., 2014). Disruption of genes for PP2C-A (PpABI1A and PpABI1B) in the ppabi1 line results in an ABA-hypersensitive response and constitutive desiccation tolerance (Komatsu et al., 2013). Furthermore, disruption of all four subgroup III SnRK2 genes (PpSnRK2A to PpSnRK2D) results in a loss of ABA sensitivity (Shinozawa et al., 2019). These findings suggest that core signaling mechanisms for ABA signaling are common in embryophytes including bryophytes and angiosperms.
While the role of PP2C-A in negative regulation of subgroup III SnRK2 has been well documented, the molecules responsible for positive regulation of SnRK2 have not been clarified. By analysis of a P. patens mutant designated AR7 that shows reduced SnRK2 activity, we previously reported that the ABA- and abiotic stress-responsive Raf-like kinase (ARK) plays a crucial role in integration of ABA and abiotic stress response (Minami et al., 2006; Saruhashi et al., 2015). ARK with 1,148 amino acids consists of the C-terminal protein kinase domain with similarity to the group B3 Raf-like protein kinase (B3-Raf) and a large non-kinase region with unknown function toward the N-terminus. AR7 was not only insensitive to ABA but also less responsive to hyperosmotic conditions and cold, indicating that ARK might play a role in the integration of ABA and these abiotic signals. AR7 has a missense mutation in Ser532 changed to Phe in the non-kinase region of ARK, suggesting that the region toward the N-terminus to the kinase domain might play a role in the regulation of ARK activity. Null mutations of ARK causing loss of ABA-induced desiccation tolerance have also been reported (Yasumura et al., 2015; Stevenson et al., 2016). The kinase domain of ARK fused to glutathione-S-transferase phosphorylates and activates subclass III SnRK2 (PpSnRK2B and PpSnRK2D) of P. patens in vitro, indicating that ARK acts as a positive regulator of SnRK2 in the ABA signaling process in bryophytes (Saruhashi et al., 2015; Shinozawa et al., 2019).
Although our studies have indicated that ARK is one of the key regulators of abiotic stress signaling, little is known about how ARK activity is regulated by ABA. Studies on various eukaryotic Raf-related protein kinases highlight phosphorylation-mediated activation of the kinases and the role of their N-terminal domains in the regulation of phosphorylation (Köhlera and Brummer, 2016). By phosphopeptide mapping of P. patens ARK, we previously showed that ARK is phosphorylated in the activation loop of the kinase domain, providing a possible mechanism for regulation of ARK (Saruhashi et al., 2015). In this study, we therefore focused on phosphorylation and activation of ARK during ABA and stress response using P. patens lines for which site-specific mutations have been introduced. The results of our analysis indicated that ARK phosphorylation in the activation loop is critical for SnRK2 regulation during ABA response. We also analyzed changes in ARK phosphorylation during ABA and abiotic stress treatments using an anti-phosphopeptide antibody in various mutants as well as in wild-type (WT) P. patens.
Results
Analysis of ARK-GFP lines with mutations in putative phosphorylation sites
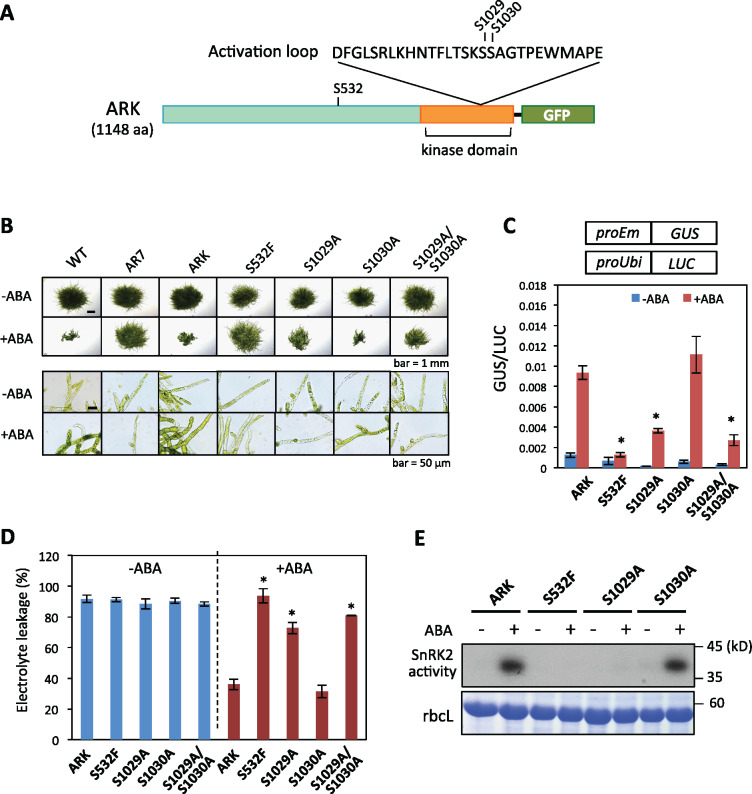
Our previous study showed by phosphopeptide mapping that Ser1029 and Ser1030 in the activation loop of the ARK kinase domain are putative phosphorylation sites (Saruhashi et al., 2015). To determine which serine residues are critical for activation of ARK, ARK-GFP constructs with or without mutations in these phosphorylation sites (ARKS1029A-GFP ARKS1030A-GFP ARKS1029A/S1030A-GFP; Figure 1, A) were generated and introduced into AR7 with the S532F mutation in ARK. Growth tests on a medium containing 10-µM ABA indicated that protonemal growth of AR7 and AR7 expressing non-mutated ARK-GFP and ARKS1030A-GFP was severely inhibited by ABA. In contrast, growth of AR7 expressing ARKS1029A-GFP and ARKS1029A/S1030A-GFP was less sensitive to ABA. We also generated AR7 expressing ARKS532F-GFP for comparison. As expected, there was little inhibition of growth of AR7 expressing ARKS532F-GFP by ABA (Figure 1, B and Supplemental Figure S1).
Figure 1.
Analysis of P. patens lines expressing ARK-GFP constructs. (A) Schematic representation of ARK-GFP showing positions of Ser532 (S532) in the non-kinase domain and putative phosphorylation sites Ser1029 (S1029) and Ser1030 (S1030) in the activation loop of the kinase domain. (B) Growth responses to ABA of WT, AR7, and AR7 lines expressing ARK-GFP without mutation (ARK) and with S532F, S1029A, S1030A, and S1029A/S1030A mutations. Protonemata of these lines were cultured with or without 10-µM ABA for 1 week. (C) ABA-induced gene expression in transgenic lines. Protonemata were bombarded with plasmid constructs of the Em promoter fused with the beta-glucuronidase (proEm-GUS) gene and the rice ubiquitin promoter fused with the luciferase gene (proUbi-LUC) and were cultured with or without 10-µM ABA for 1 d before GUS and LUC assays. Error bars indicate standard error of the mean. *P < 0.05 in the t test (n = 3) compared with the ABA-treated ARK line. (D) Freezing tolerance of ARK-GFP lines. Protonemata were cultured with or without 10-µM ABA for 1 d and subjected to freezing at 10°C. After thawing, electrolyte leakage was determined to estimate the damage caused by freezing. Error bars indicate standard error of the mean. *P < 0.05 in the t test (n = 3) compared with the ABA-treated ARK line. (E) In-gel kinase assays for detection of SnRK2 activity. Proteins extracted from protonemata were electrophoresed using SDS-polyacrylamide gel polymerized with histone IIIS as a substrate. After denaturation and renaturation processes, the proteins were reacted with 32P-ATP and radioactive signals were detected by exposure to X-ray film. Staining of the large subunit of ribulose bisphosphate carboxylase (rbcL) is shown as a control.
Using these lines, we analyzed ABA-induced gene expression by transient assays using the beta-glucuronidase (GUS) reporter gene fused to the ABA-inducible Em promoter (proEm; Knight et al., 1995; Sakata et al., 2010). AR7 lines expressing ARKS1029A-GFP and ARKS1029A/S1030A-GFP showed only a partial ABA response compared with the lines with ARK-GFP and ARKS1030A-GFP (Figure 1, C). We also analyzed ABA-induced freezing tolerance in these lines. We previously showed that ABA treatment of protonemata increases freezing tolerance of the cells in WT but not in AR7 (Minami et al., 2006; Bhyan et al., 2011). Tests for freezing tolerance indicated that AR7 lines with ARK-GFP and ARKS1030A-GFP showed similar levels of freezing tolerance induced by ABA treatment, whereas the lines with ARKS1029A-GFP and ARKS1029A/S1030A-GFP showed only a slight enhancement in the tolerance induced by ABA treatment (Figure 1, D). These results indicated that the mutation in Ser1029 to Ala causes reductions in ABA responses for gene expression and stress tolerance.
Since ARK is postulated to be an activator of SnRK2, we carried out in-gel kinase assays of ARK-GFP mutant lines using a gel containing histone IIIS, by which we previously showed that AR7 has little SnRK2 kinase activity (Saruhashi et al., 2015). Lines with ARKS532F-GFP and ARKS1029A-GFP showed little SnRK2 activity, while the ARKS1030A-GFP line showed ABA-induced SnRK2 activity similar to that of the ARK-GFP line (Figure 1, E). These results indicated that Ser1029 of ARK might be a critical phosphorylation site for activation of SnRK2 in P. patens.
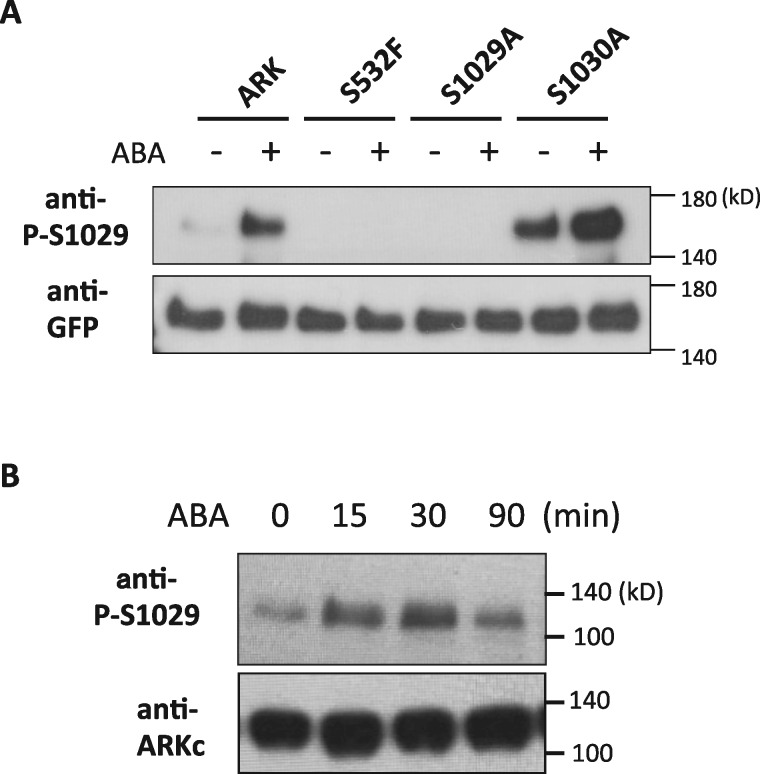
To determine the importance of Ser1029 phosphorylation, anti-phosphopeptide antibody specific to Ser1029 phosphorylation (anti-P-S1029) was raised and used for immunoblot analysis (Figure 2, A). We detected immuno-reacting signals around the expected molecular mass of ARK-GFP (157 kDa) that were enhanced by 15-min treatment with 10-µM ABA in the non-mutated ARK-GFP line. Similar enhancement of signals by ABA was also detected in the ARKS1030A-GFP lines. In contrast, the signals were very faint in ARKS532F-GFP and ARKS1029A-GFP lines (Figure 2, A). Phosphopeptide analysis of mutated and non-mutated ARK-GFP proteins immunoprecipitated from ABA-treated protonemata using anti-GFP antibody microbeads indicated that Ser phosphorylation in the peptide of amino acids 1029–1043 occurred in the ARK-GFP and ARKS1030A-GFP lines but not in the ARKS1029A-GFP and ARKS532F-GFP lines (Supplemental Table S1). The anti-P-S1029 antibody was also used for detection of phosphorylation of native ARK (Figure 2, B). The signal detected at around 130 kDa by the anti-P-S1029 antibody was increased after 15 min of ABA treatment, while the levels of ARK polypeptide detected by an antibody against ARK C-terminal peptide (anti-ARKc) were unchanged. The level of Ser1029 phosphorylation was highest at 30 min and gradually decreased with time, but the enhanced phosphorylation was still observed even after 3 h (Supplemental Figure S2). We also found that osmotic treatment with 0.5-M mannitol stimulates the Ser1029 phosphorylation, though to a lesser extent than that observed with ABA treatment (Supplemental Figure S3).
Figure 2.
ABA-stimulated phosphorylation of Ser1029 detected by anti-phosphopeptide antibody. (A) Proteins from protonemata of transgenic P. patens AR7 lines expressing ARK-GFP (ARK) and ARK-GFP with S532F, S1029A, and S1030A mutations with or without treatment with 10-µM ABA for 15 min were subjected to immunoblot analysis using an anti-phospho-Ser1029 (P-S1029) antibody and an anti-GFP antibody. Since levels of ARK-GFP accumulation vary among these lines, the amount of proteins loaded per lane was adjusted using the anti-GFP antibody. Note that ARKS1030A-GFP tends to give enhanced signals for an unknown reason. (B) Detection of native ARK in WT P. patens. Protonemata were treated with 10-µM ABA for the indicated time periods, and extracted proteins were reacted with the anti-P-S1029 antibody and an antibody that recognizes the C-terminal 15-amino-acid peptide of ARK (anti-ARKc).
Analysis of AR144 (arkD995N) with mutation in the kinase catalytic core
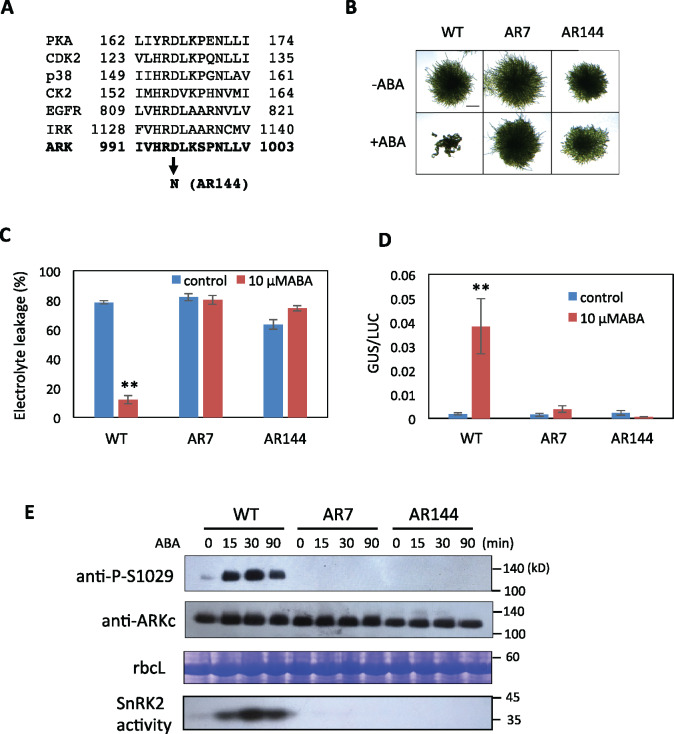
The fact that AR7 with the S523F mutation in the non-kinase region but with an intact kinase domain showed little Ser1029 phosphorylation and ABA response raised the question about whether Ser1029 phosphorylation requires catalytic activity of ARK itself. To determine the role of the kinase domain of ARK, we searched for new ark alleles by screening more than 6 × 106 mutagenized protonema cells, and we isolated the ABA-insensitive AR144 line, in which Asp995 in the kinase domain of ARK has been changed to Asn (Figure 3, A). This Asp residue corresponds to a highly conserved Asp in the “HRD motif” in the catalytic loop of various eukaryotic protein kinases (Figure 3, A). This Asp residue is essential for the kinase activity since it is assigned to be responsible for correct orientation of the target hydroxyl group of Ser in the substrate peptide (Kannan and Neuwald, 2005; Kornev et al., 2006).
Figure 3.
ABA response of AR144 of P. patens with a mutation in the catalytic core of the kinase domain of ARK. (A) Comparison of amino acid sequences in the catalytic core of protein kinase A (PKA), cyclin-dependent protein kinase2 (CDK2), p38, casein kinase2 (CK2), and epidermal growth factor receptor (EGFR) (adapted from Nolen et al., 2004) with ARK. The conserved Asp (D) has been changed to Asn (N) in AR144. (B) Growth responses to ABA. Protonemata of WT, AR7, and AR144 were grown on a medium with 10-µM ABA for 1 week. (C) Comparison of freezing tolerance. Protonemata incubated with or without 10-µM ABA for 1 d were frozen to −4°C. After thawing, electrolyte leakage was measured to estimate the extent of freezing injury. Error bars indicate standard error of the mean. (D) Transient gene expression assays of WT, AR7, and AR144. Protonemata were bombarded with plasmid constructs of proEm-GUS and proUbi-LUC and were cultured with or without 10-µM ABA for 1 d for GUS and LUC assays. Error bars indicate standard error of the mean. **P < 0.01 in the t test (n = 3) compared with control WT without ABA treatment for (C) and (D). (E) Effect of ABA on Ser1029 phosphorylation in WT, AR7, and AR144. Protonemata were treated with 10-µM ABA for different times and used for immunoblot analysis with anti-phospho-Ser1029 (P-S1029) and anti-ARKc antibodies. Staining of the large subunit of ribulose bisphosphate carboxylase (rbcL) is shown as a control. Results of an in-gel kinase assay for detection of SnRK2 are also shown.
When grown on medium containing 10-µM ABA, protonemata of AR144 showed ABA-insensitive growth similar to that of AR7 (Figure 3, B). In freezing tolerance tests, freezing-induced leakage of electrolytes in WT protonemata was remarkably reduced by pretreatment with ABA, similar to AR7, whereas the leakage was not mitigated by ABA in AR144 protonemata (Figure 3, C). Transient reporter assays indicated that there was no ABA-increased gene expression in AR144 (Figure 3, D), in contrast to AR7 with an intact kinase domain showing a slight increase. We analyzed phosphorylation of ARK in AR144 using the anti-P-Ser1029 antibody to examine whether the ARK kinase activity is necessary for its phosphorylation and is associated with activation of SnRK2. The results of analyses indicated that while both Ser1029 phosphorylation and SnRK2 activity were enhanced by ABA in WT, neither Ser1029 phosphorylation nor SnRK2 activity was detected in AR144 as well as in AR7 (Supplemental Figure 3, E). These results indicated that the catalytic activity of ARK is necessary for Ser1029 phosphorylation and cellular ABA response.
Effect of cold on ARK phosphorylation
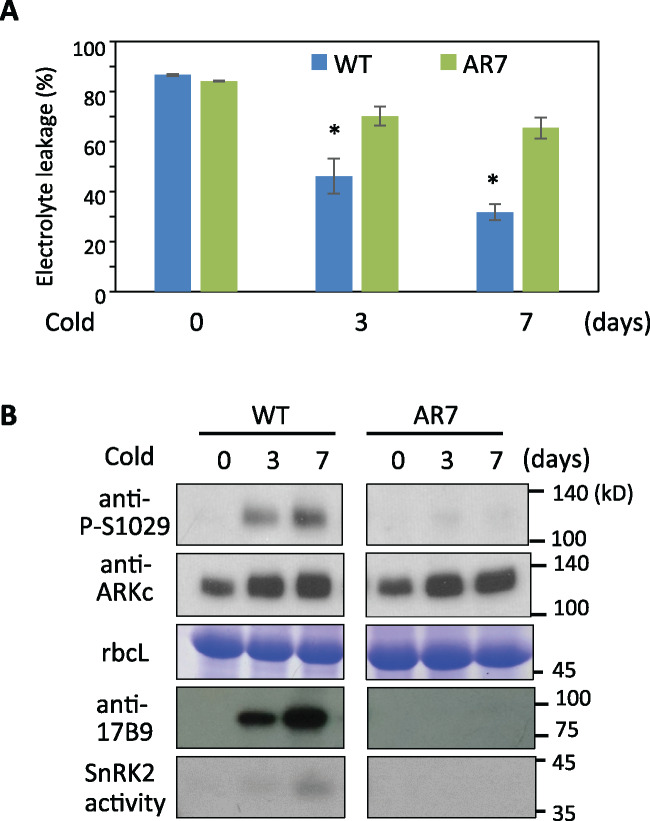
We previously reported that the freezing tolerance of P. patens protonema cells is enhanced by exposure to low temperatures (Minami et al., 2005). This cold acclimation process accompanies accumulation of LEA-like proteins that mitigate freezing damage (Minami et al., 2005; Sasaki et al., 2013). Comparison of freezing-induced electrolyte leakage in WT and AR7 indicated that cold treatment (0°C) enhanced freezing tolerance in WT but only slightly in AR7 (Figure 4, A), which was associated with accumulation of the LEA-like 17B9 protein in boiling-soluble fractions of the WT (Figure 4, B). Immunoblot analysis of WT and AR7 protonemata with anti-P-S1029 and anti-ARKc antibodies revealed that phosphorylation of Ser1029 was enhanced in WT with increased abundance of ARK polypeptide during cold acclimation (Figure 4, B). The Ser1029 phosphorylation was associated with enhancement of SnRK2 activity. In AR7, however, little enhancement of Ser1029 phosphorylation and SnRK2 activity was observed during cold acclimation, although cold-enhanced accumulation of ARK polypeptide was observed. These results suggest that Ser1029 phosphorylation of ARK plays a role in the cold acclimation process in the moss cells.
Figure 4.
Cold responses mediated by ARK in P. patens. (A) Changes in freezing tolerance during cold acclimation in WT and AR7 (arkS532F). Protonemata that had been cold-acclimated for the indicated days were frozen to −3°C and injury rate was estimated by measurement of electrolyte leakage after thawing. Error bars indicate standard error of the mean. *P < 0.05 in the t test (n = 3) compared with non-acclimated protonemata of each line. (B) Immunoblot analysis with anti-phospho-Ser1029 (P-S1029) and anti-ARKc antibodies. Coomassie Brilliant Blue staining of the large subunit of ribulose bisphosphate carboxylase (rbcL), results of immunoblot analysis for cold-induced 17B9 protein, and SnRK2 activity analyzed by an in-gel kinase assay are also shown.
Roles of PP2C-A in regulation of ARK phosphorylation and SnRK2 activity
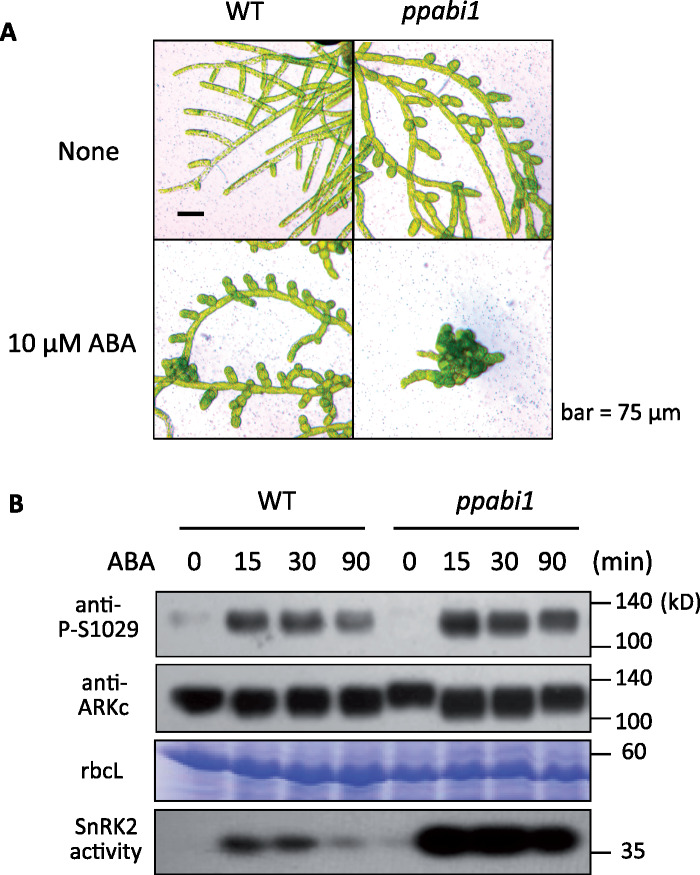
PP2C-A association with the PYR/PYL/RCAR receptor is thought to be a critical regulator of SnRK2 in Arabidopsis (Park et al., 2009; Umezawa et al., 2009). Komatsu et al. (2013) reported that the P. patens ppabi1 line lacking both of the two genes for PP2C-A showed desiccation tolerance without ABA treatment. Protonemata of ppabi1 frequently formed spherical “brood cells” (Arif et al., 2019) under normal growth conditions, while WT protonemata formed these cells in response to ABA treatment for several days (Figure 5, A). Interestingly, the ppabi1 protonemata were still responsive to ABA and formed more brood cells upon ABA treatment (Figure 5, A). In-gel kinase assays of ppabi1 indicated that ABA-dependent SnRK2 activity in ppabi1 was greater than that in WT, though the activity remained low without exogenous ABA (Komatsu et al., 2013), indicating that PP2C-A is not the only regulator of SnRK2. Since ARK has been identified as a positive regulator of SnRK2, we suspected that PP2C-A might affect SnRK2 through regulation of ARK. Comparison of ABA-induced changes in Ser1029 phosphorylation of ARK and SnRK2 activity in the same plant extract indicated that phosphorylation of ARK was stimulated by ABA in ppabi1 in a manner similar to that of WT, in contrast to a remarkable enhancement of SnRK2 activity in ppabi1 (Figure 5, B). These results indicated that PP2C-A is not required for ABA-induced activation of ARK phosphorylation.
Figure 5.
ABA response of the ppabi1 line of P. patens. (A) Growth responses to ABA in protonemata of WT and ppabi1. The protonemata were grown on medium with or without 10-µM ABA for 1 week. (B) Effect of ABA treatment on Ser-1029 phosphorylation in WT and ppabi1. Protonemata were treated with 10-µM ABA for different times and used for immunoblot analysis with anti-ARKc and anti-phospho-Ser1029 (P-S1029) antibodies. Coomassie Brilliant Blue staining of the large subunit of ribulose bisphosphate carboxylase (rbcL) and SnRK2 activity analyzed by an in-gel kinase are also shown.
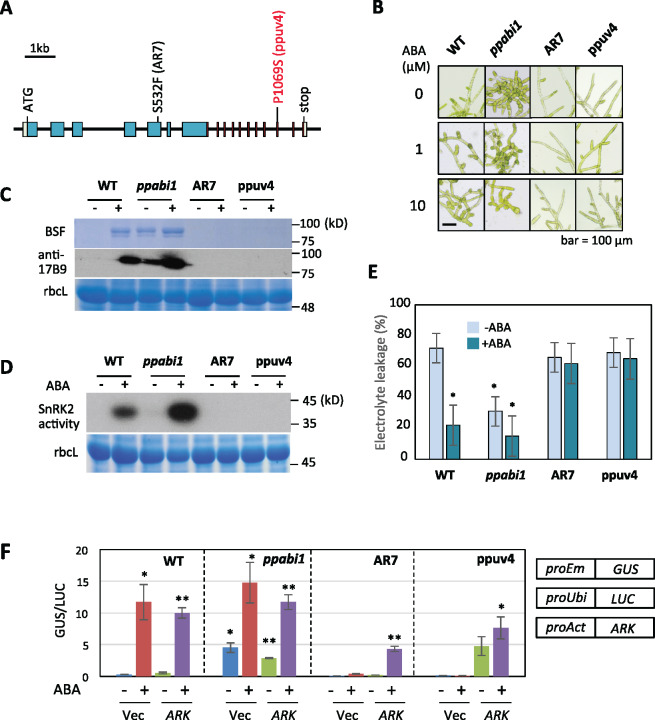
To determine the roles of ARK in the ppabi1 background, we mutagenized protonemata of ppabi1 by ultraviolet light and isolated lines that grow well on medium containing 1-µM ABA, which severely inhibits growth of ppabi1. Of several isolated mutant lines, we identified ppuv4 (arkP1069S), ppuv6 (arkP1069L), and ppuv9 (arkS998L) lines (Figure 6, A and Supplemental Figure S4, A) that all had missense mutations in the ARK kinase domain. These mutant lines showed ABA-insensitive growth similar to that of AR7 (arkS532F), forming no brood cells upon ABA treatment (Figure 6, B and Supplemental Figure S4, B). We found that ppuv4 accumulates reduced amounts of ARK with little phosphorylation at Ser1029 (Supplemental Figure S5). A comparison of ABA-induced accumulation of boiling-soluble proteins and the LEA-like 17B9 protein therein in WT, ppabi1, AR7, and ppuv4 indicated that AR7 and ppuv4 accumulated little of these proteins with or without ABA treatment (Figure 6, C). Furthermore, in-gel kinase assays for SnRK2 activity (Figure 6, D), freezing tolerance tests (Figure 6, E), and gene expression studies (Supplemental Figure S6) indicated that ppuv4 is insensitive to ABA. Transient reporter assays using proEm-GUS for determination of exogenous ABA- and ARK-dependent gene expression indicated that, while ppabi1 shows GUS expression even without exogenous ABA due to its hypersensitivity, ppuv4 shows little expression with or without exogenous ABA, which can be only restored by introduction of ARK cDNA (Figure 6, F). These results indicated that mutations in ARK cause ABA insensitivity even in the ppabi1 background.
Figure 6.
ABA response of ppabi1 ark lines of P. patens. (A) Positions of mutations found in the ABA-insensitive ppuv4 line isolated by ultraviolet mutagenesis of ppabi1. (B) Effects of ABA on growth of WT, ppabi1, AR7, and ppuv4. (C) ABA-induced accumulation of boiling-soluble LEA-like proteins in WT, ppabi1, AR7, and ppuv4. Total soluble proteins of these lines were boiled for 1 min and centrifuged, and the boiling-soluble fraction (BSF) in the supernatant was analyzed by SDS-PAGE. The proteins were either stained with Coomassie Brilliant Blue or used for immunoblot analysis using an antibody against the LEA-like 17B9 protein. Staining of the large subunit of ribulose bisphosphate carboxylase (rbcL) of the total soluble proteins is shown as a control. (D) SnRK2 activity in WT and the mutant lines analyzed by in-gel kinase assays using histone IIIS as a substrate. (E) Freezing tolerance tests of WT and mutant lines. Protonemata were treated with or without 10-µM ABA for 1 d and frozen to −4°C. Electrolyte leakage (%) was measured after thawing to determine freezing injury. Error bars indicate standard error of the mean. *P < 0.05 in the t test (n = 3) compared with ABA-non-treated WT. (F) Gene expression with or without 10-µM ABA treatment in WT and the mutant lines. Transient gene expression analysis was carried out using the construct of ARK fused to the rice actin promoter (proAct) or that without ARK (Vec). Error bars indicate standard error of the mean. *P < 0.05, **P < 0.01 in the t test (n = 4) compared with the values of WT (Vec, -ABA).
Discussion
Activation of the Raf-like kinase ARK by Ser1029 phosphorylation is a key event in ABA response
We demonstrated in this study that ABA and abiotic stress responses in P. patens require activation of ARK, which belongs to the family of B3-Raf kinases and has been suggested to be an upstream activator of subclass III SnRK2. Previous studies indicate that nearly 90% and 80% of ABA-regulated gene expression is under the control of ARK and subgroup III SnRK2, respectively (Saruhashi et al., 2015; Shinozawa et al., 2019). Consistent with this, approximately 88% of ABA-increased phosphorylation is ARK-dependent (Amagai et al., 2018), where drastic reduction in the levels of ABA-activated phosphorylation of both SnRK2 and ABA-responsive element binding (AREB) transcription factors in AR7 was observed. Our results showing association of Ser1029 phosphorylation with ABA-induced gene expression, stress tolerance, and SnRK2 activity indicate that activation of ARK by phosphorylation is a crucial event during the ABA response in P. patens. The role of ARK in SnRK2 activation may not be restricted to bryophytes; we previously showed that ectopic expression of three Arabidopsis B3-Raf kinases (At1G18160, At1G73660, and At4G24480) can restore ABA sensitivity of AR7 (Saruhashi et al., 2015). It has recently been reported that these B3-Raf kinases are necessary for activation of SnRK2 kinases during ABA and osmotic responses in Arabidopsis (Takahashi et al., 2020; Lin et al., 2020; Katsuta et al., 2020). The serine residue corresponding to Ser1029 of ARK is conserved in Arabidopsis B3-Raf kinases (Supplemental Figure S7). Considering the functional conservation of subclass III SnRK2 in P. patens and Arabidopsis (Chater and Gray, 2011; Shinozawa et al., 2019), the results of this study suggest that sequential activation of B3-Raf kinase and subclass III SnRK2 might be an evolutionarily conserved mechanism for ABA and stress responses in embryophytes, which requires experimental confirmation in Arabidopsis. The presence of Arabidopsis B3-Raf kinases with specific functions, such as CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) for ethylene signaling (Kieber et al., 1993), ENHANCED DISEASE RESISTANCE1 (EDR1) for non-host resistance (Frye and Innes, 1998; Hiruma et al., 2011), and SUGAR INSENSITIVE8 (SIS8) for sugar sensing (Huang et al., 2014), suggests functional diversification of B3-Raf kinases in angiosperms.
Ser1029 phosphorylation of ARK requires a functional kinase domain
This study revealed the importance of both the kinase domain and the N-terminal region of ARK for its activation (Figure 3, E). AR144 lacking the critical Asp residue in the catalytic loop of the kinase domain showed no ABA response or Ser1029 phosphorylation, suggesting that the phosphorylation requires a functional kinase domain. This indicates that Ser1029 is not phosphorylated by other protein kinases but is phosphorylated by ARK itself (i.e. autophosphorylation). Consistent with this, bacterially expressed recombinant GST-ARK proteins readily undergo autophosphorylation in the activation loop (Supplemental Table S2). What controls this autophosphorylation upon ABA and cold treatment in vivo is yet to be clarified. In animal Raf kinases, the N-terminal domains play a crucial role in regulation of the catalytic activity (Matallanas et al., 2011). For human c-Raf, the “autoinhibitory block” consisting of the Ras-binding domain and the C1 domain directly interacts with the C-terminal kinase domain and inhibits its catalytic activity, and upon stimulation, binding of Ras-GTP releases the kinase domain from this autoinhibition. The partially activated c-Raf undergoes autophosphorylation in the activation loop, which fully activates the kinase activity. For P. patens ARK, the N-terminal non-kinase region might also play a role in regulation of the kinase domain, since AR7 with the S532F mutation showed little Ser1029 phosphorylation and catalytic activity toward SnRK2 (Figure 2, A and Figure 3, E). The negative effect of the S532F mutation on ABA-induced gene expression was compromised by introduction of phosphomimetic Asp residues in the activation loop (Supplemental Figure S8). This indicates that the N-terminal region of ARK might play a role in negative regulation of the kinase domain. Overexpression of the N-terminal region fused to GFP resulted in reductions in ABA responses, further supporting this possibility (Supplemental Figure S9). Given that Ser1029 phosphorylation occurs by autophosphorylation, activity of ARK must be controlled by the action of an unidentified negative regulatory factor that inhibits the catalytic activity of the kinase domain, possibly through interaction with a specific domain in the N-terminal region, and by ABA-dependent removal of the negative regulatory factors. It is also possible that phosphorylation in the N-terminal region affects ARK activity, since we identified over 10 residues in the N-terminal region as phosphorylation sites, and that the levels of phosphorylation in some residues are changed by ABA treatment (Supplemental Table S3 and Supplemental Figure S10). Most of these residues, however, were also phosphorylated in the ARKS532F-GFP line (Supplemental Table S3), and the roles of the phosphorylation in the regulation of ARK activity are not clear.
Roles of ARK in cold responses
Many plant species can acquire tolerance to freezing upon exposure to low, non-freezing temperatures. In Arabidopsis, cold provokes expression of a number of transcripts such as those encoding cold-related/dehydrin proteins, which have characteristics similar to LEA proteins. Expression of these transcripts is driven by C-repeat binding factor (CBF) transcription factors, genes for which are also up-regulated by cold (Thomashow, 2010; Shi et al., 2018). Several distinct mechanisms for regulation of CBF genes have been proposed, although the molecules for cold sensing and early signal transduction have not been identified. It has been proposed in Arabidopsis that subclass III SnRK2 OST1 activated by cold positively regulates CBF gene expression by phosphorylation of the transcriptional regulators INDUCER OF CBF EXPRESSION 1 and BASIC TRANSCRIPTION FACTOR 3 (Ding et al., 2015, 2018). Bryophytes also undergo cold acclimation accompanied by enhancement of SnRK2 activity (Saruhashi et al., 2015). The fact that cold-enhanced Ser1029 phosphorylation of ARK and activation of SnRK2 were not observed in AR7 (Figure 4, B) therefore indicates the role of ARK in activation of SnRK2 for the cold response in P. patens. We also showed an increased accumulation of ARK polypeptide during the cold treatment (Figure 4, B), which might provide a mechanism for the control of ARK by cold. Interestingly, ARK transcript levels are apparently not affected by cold (Supplemental Figure S11). It is also likely that this process does not require functional ARK, since accumulation of ARK polypeptide in response to cold was also observed in AR7, which accumulated reduced amounts of the 17B9 protein and transcripts (Figure 4, B andSupplemental Figure S11).
PP2C-A functions downstream to ARK
The PP2C-A family, which has nine members in Arabidopsis, is recognized as a primary regulator of SnRK2 (Umezawa et al., 2009; Vlad et al., 2009). The role of PP2C-A in negative regulation of ABA signaling is likely conserved in both mosses and liverworts (the basal lineage of embryophytes), because overexpression of PP2C-A genes of these plants causes abolishment of ABA-responsive gene expression and loss of abiotic stress tolerance (Komatsu et al., 2009; Tougane et al., 2010). Furthermore, the fact that the ppabi1 plants still respond to ABA and activate SnRK2 (Komatsu et al., 2013) indicates the presence of PP2C-A-independent ABA-response mechanisms in bryophytes. Consistent with this, phosphoproteome analysis of P. patens indicated that only a small portion (23 out of 143) of ABA-stimulated phosphoproteins detected in WT were hyperphosphorylated in ppabi1, and 126 of those were dependent on ARK (Amagai et al., 2018).
Whether SnRK2 is activated by autophosphorylation or phosphorylation by upstream kinases has been controversial (Boudsocq et al., 2007). This study on ppuv4 (ppabi1 ark), however, suggests that SnRK2 must be activated by ARK-dependent phosphorylation before SnRK2 can be dephosphorylated and inactivated by PP2Cs (Figure 6, A–F). Furthermore, our results suggested that PP2C-A may not be a primary regulator of ARK phosphorylation (Figure 5, B). Figure 7 and Supplemental Figure S12 illustrate working models for the control of SnRK2 in bryophytes, focusing on the role of ARK for positive regulation and PP2C-A for subsequent negative regulation. These models are supported not only by abolishment of ABA responses in ppuv4 but also by the results of our experiment with MpPYL1 (Supplemental Figure S13). MpPYL1 is a major ABA receptor in vegetative tissues of Marchantia polymorpha and has recently been shown to have ABA-independent inhibitory activity toward PP2C-A (Jahan et al., 2019; Sun et al., 2019). Our experiment indicated that transient overexpression of MpPYL1 enhances the activity of the GUS reporter gene fused to the ABA-inducible Em promoter without ABA treatment, but such enhancement was not observed in AR7 with or without ABA (Supplemental Figure S13), suggesting that inhibition of PP2C-A by PYR/PYL/RCAR causes activation of the promoter only in the presence of functional ARK. Our previous analysis indicated spatial separation of ARK and PP2C-A in bryophytes; the ARK-GFP fusion protein was mainly in the cytoplasm and partly in the endoplasmic reticulum, while PP2C-A-GFP was only in the nucleus (Komatsu et al., 2009; Tougane et al., 2010; Saruhashi et al., 2015).
Figure 7.
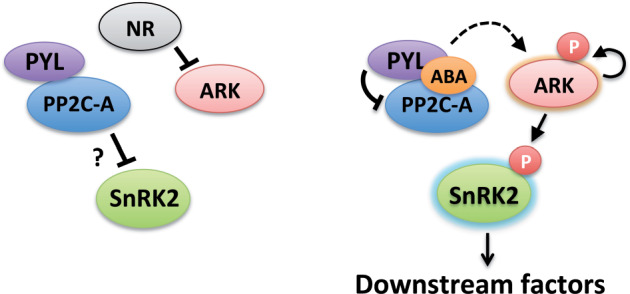
Working models showing ARK-mediated ABA responses in P. patens. ABA stimulates ARK autophosphorylation and inhibits PP2C-A, causing activation of SnRK2 regulating downstream factors, in which phosphorylation of SnRK2 by ARK precedes negative regulation by PP2C-A. ABA-stimulated activation of both ARK and SnRK2 occurs without PP2C-A, suggesting the presence of an unidentified negative regulator (NR) and PP2C-A-independent mechanisms for activation of ARK (dashed arrow). Details of these models are shown in Supplemental Figure S12.
The working models we propose for bryophytes in this study represent the basic ABA response machinery, a prototype to that in angiosperms possessing diverse PP2C-A molecules. Evolutionary diversification of PP2C-A molecules in angiosperms resulted in localization of some PP2C-A molecules in the nucleus for regulation of transcription factors, while others are in the cytoplasm for regulation of angiosperm-specific substrates, such as plasma membrane-localized ion channels. Takahashi et al. (2020) hypothesized from the results of in vitro and Xenopus laevis oocyte reconstitution assays that Arabidopsis B3-Raf kinases play a role in reactivation of SnRK2 that had been dephosphorylated by PP2C-A for regulation of the AKS1 transcription factor and the S-type ion channel SLAC1. The results of our experiments on ppuv4 strongly suggest that P. patens SnRK2 does not activate by autophosphorylation but requires ARK for activation. We do not, however, rule out the possibility of a role of P. patens ARK in reactivation of SnRK2 because P. patens SnRK2 is localized in both the cytoplasm and nucleus (Saruhashi et al., 2015).
Materials and methods
Chemicals and plant materials
Chemicals were purchased from Wako Jun-yaku (Osaka, Japan) unless otherwise stated. ABA was from Sigma (A4906, St. Louis, MO, USA). Culture and ABA, osmotic and cold treatments of protonemata of P. (Physcomitrella) patens WT and mutant lines were carried out as described previously (Minami et al., 2003, 2005).
Transient reporter assays
Physcomitrium patens protonemata cultured on cellophane-overlaid BCDAT agar medium (BCD medium containing 1-mM CaCl2, 5-mM ammonium tartrate, and 0.8% agar; Nishiyama et al., 2000) for 5 d were used for particle bombardment (Marella et al., 2006). The protonema cells were bombarded with DNAs of the proEm-GUS reporter construct (Marcotte et al., 1989) and the proUbi-LUC reference construct (Bruce et al., 1989) with or without ARK effector constructs using the PDS-1000He particle delivery system (Bio-Rad, Hercules, CA, USA). The bombarded cells were incubated in a medium with or without ABA and were used for GUS and LUC assays (Bruce et al., 1989; Jefferson et al., 1987; Marella et al., 2006; Komatsu et al., 2009).
Generation of transgenic P. patens
Physcomitrium patens protonemata were subjected to PEG-mediated transformation according to the protocol described by Nishiyama et al. (2000). Constructs of ARK-green fluorescent protein (ARK-GFP) without mutation and that with mutations in Ser532 to Phe (ARKS532F-GFP), Ser1029 to Ala (ARKS1029A-GFP), Ser1030 to Ala (ARKS1030A-GFP), and Ser1029/Ser1030 to Ala/Ala (ARKS1029A/S1030A-GFP) driven by the rice (Oryza sativa) ACTIN promoter were made using the pFL-Not vector (Saruhashi et al., 2015).
Mutant screening
Partially homogenized protonemata spread on a cellophane-overlaid agar medium were cultured for 2 d prior to irradiation of ultraviolet light under conditions described by Minami et al. (2006). After incubation in the dark at 25°C for 1 d, the protonemata were cultured under continuous light for 1 or 2 d at 25°C. They were then transferred with cellophane onto a medium containing 10-µM ABA for WT and onto a medium containing 1-µM ABA for ppabi1.
Protein gel electrophoresis and immunoblot analysis
For protein extraction, protonemata were homogenized in a buffer containing 50-mM Tris-Cl (pH 8.0), 150-mM NaCl, 1% (v/v) Triton X-100, 25-mM NaF, 2-mM dithiothreitol, 1-mM o-vanadate, and 50-mM β-glycerophosphate and a 1/100 volume of proteinase inhibitor mixture (P9599; Sigma, MO, USA) on ice. After centrifugation at 14,000 × g for 10 min at 4°C, the supernatants were used as protein samples for electrophoresis. The proteins (30 μg) were electrophoresed on 8% SDS-polyacrylamide gel and blotted onto a polyvinylidene fluoride membrane for immunoblot analysis. After blocking with 1% bovine serum albumin (for the anti-phosphopeptide antibody) or 3% skim milk (for other antibodies) in TBS-T (25-mM Tris-Cl, pH 7.5, 150-mM NaCl, and 0.05% [v/v] Tween-20), the proteins were reacted with the following primary antibodies: anti-GFP (MBL, Nagoya, Japan), anti-ARK C-terminal 15 amino acids (LGGTPKSGLSDRDL; anti-ARKc; Saruhashi et al., 2015), and anti-Ser1029-phosphorylated peptide (FLTSKpSSAGTPEWMAPE; anti-P-Ser1029). The anti-P-Ser1029 antibody raised in rabbits was affinity-purified using the same phosphopeptide and then cross-absorbed against the dephosphorylated GST-ARK protein to remove antibody that might recognize non-phosphorylated ARK. The membrane that had been reacted with a primary antibody was reacted with a 1:10,000 dilution of horseradish peroxidase-conjugated secondary antibody (MBL, Nagoya, Japan). The membrane was then immersed in chemiluminescence reagent (Chemi-Lumi One, Nacalai Tesque, Kyoto, Japan) and exposed to X-ray film for signal detection.
Analysis of phosphopeptides
Protein extracts were prepared from protonemata of AR7 expressing ARK-GFP constructs using the buffer used for immunoblot analysis. Immunoprecipitation was carried out using the µMACS GFP Tagged Protein Isolation Kit (Miltenyi Biotech). Protein samples were electrophoresed using 8% SDS polyacrylamide gel and stained with Coomassie Brilliant Blue, and stained bands corresponding to ARK-GFP near 150 kilodaltons were excised. After proteolytic enzyme digestion, the phosphopeptides were analyzed by nano-flow reverse-phase liquid chromatography followed by tandem MS using a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific).
Extraction of total RNA and reverse transcription quantitative PCR (RT-qPCR) analysis
RNA extraction, first-strand cDNA synthesis, and PCR reactions were carried out as described previously (Saruhashi et al., 2015). The RNA was reverse-transcribed with oligo-dT and used for semi-quantitative PCR and SYBR Green-based quantitative PCR. Sequences of oligonucleotide primers are listed in Supplemental Table S4.
In-gel kinase assay for SnRK2 activity
An in-gel kinase assay was carried out according to the protocol described by Suzuki and Shinshi (1995). Protein extracts used for immunoblot analysis were subjected to electrophoresis using a 10% SDS-polyacrylamide gel containing 0.5 mg/mL histone IIIS (Sigma–Aldrich) as a substrate. The gel was successively washed for 30 min twice in buffer I (50-mM Tris–Cl, pH 8.0, and 20% isopropanol) and buffer II (50-mM Tris–Cl, pH 8.0, and 5-mM 2-mercaptoethanol), denaturation buffer (buffer II containing 6-M guanidine hydrochloride), and renaturation buffer (buffer II containing 0.04% Tween 40) at room temperature. The gel was then incubated in renaturation buffer at 4°C for 16 h and again washed once in renaturation buffer at room temperature for 30 min. After incubation in buffer III (40-mM HEPES-KOH, pH 7.5, 15-mM MgCl2, 0.1-mM ethylene glycol tetraacetic acid, and 2-mM dithiothreitol) for 30 min at room temperature, the gel was reacted with 50-µM [γ-32P] ATP (1.5 TBq/mmol) in buffer III for 1 h at 25°C. After removal of the reaction solution, the gel was washed for 30 min for at least six times with washing solution (5% trichloroacetic acid and 1% sodium pyrophosphate) before drying and exposure to X-ray film.
Tests for freezing tolerance
Protonema tissues placed in a glass test tube containing 0.5 mL of distilled water were set in a programmable cooling bath. The tissues were seeded with ice at −1°C, kept at −1°C for 50 min, and then cooled at a rate of −2.4°C h−1 to desired temperatures. Electrolyte leakage from the damaged tissues was determined after thawing by measurement of conductivity in the water and represented as the percentage against conductivity of total ions released by subsequent boiling.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number ARK, Pp3c12_3550V3.1.
Supplemental data
Supplemental Figure S1. Growth responses to ABA of transgenic AR7 lines expressing various ARK-GFP constructs.
Supplemental Figure S2. Detection of Ser1029 phosphorylation of native ARK.
Supplemental Figure S3. Effects of hyperosmotic mannitol on ARK Ser1029 phosphorylation.
Supplemental Figure S4. Growth responses to ABA of three ppuv mutants isolated by ultraviolet light mutagenesis of ppabi1.
Supplemental Figure S5. Accumulation and Ser1029 phosphorylation of ARK in ppuv4.
Supplemental Figure S6. ABA-induced gene expression of LEA-like transcripts in WT, ppabi1, AR7, and ppuv4.
Supplemental Figure S7. Amino acid sequence alignment in the activation loops of P. patens ARK and Arabidopsis B3-Raf kinases.
Supplemental Figure S8. Effect of introduction of phosphomimetic Asp residues in the activation loop on ABA-induced gene expression.
Supplemental Figure S9. Effect of overexpression of the ARK N-terminal region on sensitivity to ABA.
Supplemental Figure S10. ABA-induced changes in the levels of phosphorylation at various amino acid residues in ARK.
Supplemental Figure S11. Expression of ARK and LEA-like 17B9 transcripts during cold acclimation in WT and AR7.
Supplemental Figure S12. Details of the working model shown in Figure 7.
Supplemental Figure S13. Effect of MpPYL1 overexpression on ABA-inducible promoter in WT and AR7.
Supplemental Table S1. Analysis of phosphopeptides immunoprecipitated from AR7 expressing ARK-GFP with or without mutations in Ser1029 and Ser1030
Supplemental Table S2. Autophosphorylation in the peptide SSAGTPEWMAPEVLR in the activation loop of recombinant GST-ARK
Supplemental Table S3. Phosphopeptide analysis of immunoprecipitates of transgenic lines expressing ARK-GFP, focusing on phosphorylation of the N-terminal region.
Supplemental Table S4. Oligonucleotide primers used for gene expression studies
Supplementary Material
Acknowledgements
The authors thank Yasuo Niwa for GFP(S65T; Niwa, 2003), Ralph Quatrano for proEm-GUS, and Tuan-hua David Ho for Ubi-LUC constructs. They also thank Masashi Saruhashi and Marina Fujita for technical assistance.
Funding
This work was supported by the Program for the Strategic Research Foundation at Private Universities [S1311017] and Grant-in-Aid for Scientific Research [Nos. 26291054, 18H04774, and JP15H05955] of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. ITbM was supported by the World Premier International Research Center Initiative (WPI) of MEXT.
Conflict of interest statement. The authors declare no conflicts of interest.
M.I. and T.I. conducted most of the experiments and organized the entire manuscript with M.H. and D.T. N.K. contributed to the cold acclimation experiments. K.K. and S.K. analyzed the phosphopeptides. A.J. and I.Y. contributed to the transient gene expression assays. T.U. and Y.S. contributed to experimental design in the transgenic study.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Daisuke Takezawa (takezawa@mail.saitama-u.ac.jp).
References
- Amagai A, Honda Y, Ishikawa S, Hara Y, Takezawa D, Sakata Y, Shinozaki K, Umezawa T (2018) Phosphoproteomic profiling reveals ABA-responsive phosphosignaling pathways in Physcomitrella patens. Plant J 94:699–708 [DOI] [PubMed] [Google Scholar]
- Arif MA, Hiss M, Tomek M, Busch H, Meyberg R, Tintelnot S, Reski R, Rensing SA, Frank W (2019) ABA-induced vegetative diaspore formation in Physcomitrella patens. Front Plant Sci 10:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhyan SB, Minami A, Kaneko Y, Suzuki S, Arakawa K, Sakata Y, Takezawa D (2011) Cold acclimation in the moss Physcomitrella patens involves abscisic acid-dependent signaling. J Plant Physiol 169:137–145 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C (2004) Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 279:41758–41766 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63:491–503 [DOI] [PubMed] [Google Scholar]
- Bruce WB, Christensen AH, Klein T, Fromm M, Quail PH (1989) Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc Natl Acad Sci U S A 86:9692–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C, Gray JE. (2011) Stomatal closure: the old guard takes up the SLAC. Curr Biol 25:R271–R273 [DOI] [PubMed] [Google Scholar]
- Cuming AC, Lane BG (1979) Protein synthesis in imbibing wheat embryos. Eur J Biochem 99:217–224 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32:278–289 [DOI] [PubMed] [Google Scholar]
- Ding Y, Jia Y, Shi Y, Zhang X, Song C, Gong Z, Yang S (2018) OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J 37:e98228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10:947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y (2011) Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J 67:980–992 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li CY, Qi Y, Park S, Gibson SI (2014) SIS8, a putative mitogen-activated protein kinase kinase kinase, regulates sugar-resistant seedling development in Arabidopsis. Plant J 77:577–588 [DOI] [PubMed] [Google Scholar]
- Jahan A, Komatsu K, Wakida-Sekiya M, Hiraide M, Tanaka K, Ohtake R, Umezawa T, Toriyama T, Shinozawa A, Yotsui I, et al . (2019) Archetypal roles of the PYR/PYL/RCAR-like receptor in drought and sugar responses in liverworts. Plant Physiol 179:317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Neuwald AF (2005) Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol 351:956–972 [DOI] [PubMed] [Google Scholar]
- Katsuta S, Masuda G, Bak H, Shinozawa A, Kamiyama Y, Umezawa T, Takezawa D, Yotsui I, Taji T, Sakata Y (2020) Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J 103:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud PF, Pan A, Quatrano RS (2010) Role of ABA and ABI3 in desiccation tolerance. Science 327:546. [DOI] [PubMed] [Google Scholar]
- Kieber JJ,, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1,a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72:427–441 [DOI] [PubMed] [Google Scholar]
- Knight CD, Sehgal A, Atwal K, Wallace JC, Cove DJ,, Coates D, Quatrano RS, Bahadur S, Stockley PG, Cuming AC (1995) Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell 7:499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhlera M, Brummer T (2016) B-Raf activation loop phosphorylation revisited. Cell Cycle 15:1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, Sakata Y (2009) Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol 70:327–340 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Suzuki N, Kuwamura M, Nishikawa Y, Nakatani M, Ohtawa H, Takezawa D, Seki M, Tanaka M, Taji T,. et al. (2013) Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun 4:2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Ten Eyck LF (2006) Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A 103:17783–17788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster KL, Balsamo RA, Espinoza C, Oliver MJ (2010) Desiccation sensitivity and tolerance in the moss Physcomitrella patens: assessing limits and damage. Plant Growth Regul 62:293–302 [Google Scholar]
- Lin Z., Li Y., Zhang Z., Liu X, Hsu C-C, Du Y, Sang T, Zhu C, Wang Y, Satheesh V. et al. (2020) A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat Commun 11:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068 [DOI] [PubMed] [Google Scholar]
- Marcotte WR Jr, Russel SH, Quatrano RS (1989) Abscisic acid-responsive sequences from the Em gene of wheat. Plant Cell 1:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS (2006) Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J 46:1032–1044 [DOI] [PubMed] [Google Scholar]
- Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W (2011) Raf family kinases: old dogs have learned new tricks. Genes Cancer 2:232–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A, Nagao M, Arakawa K, Fujikawa S, Takezawa D (2003) Abscisic acid-induced freezing tolerance in the moss Physcomitrella patens is accompanied by increased expression of stress-related genes. J Plant Physiol 160:475–483 [DOI] [PubMed] [Google Scholar]
- Minami A, Nagao M, Ikegami K, Koshiba T, Arakawa K, Fujikawa S, Takezawa D (2005) Cold acclimation in bryophytes: low-temperature-induced freezing tolerance in Physcomitrella patens is associated with increases in expression levels of stress-related genes but not with increase in level of endogenous abscisic acid. Planta 220:414–423 [DOI] [PubMed] [Google Scholar]
- Minami A, Togawa S, Nagao M, Takezawa D (2006) Altered freezing tolerance in the Physcomitrella patens mutant with reduced sensitivity to abscisic acid. Cryobiol Cryotechnol 52:135–139 [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J,. et al. (2010) PYR/PYL/RCAR family members are major in‐vivo ABI1 protein phosphatase 2C‐interacting proteins in Arabidopsis. Plant J 61:290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Niwa Y (2003) A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol 20:1–11 [Google Scholar]
- Nolen B, Taylor S, Ghosh G (2004) Regulation of protein kinases: controlling activity through activation segment conformation. Mol Cell 15:661–675 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Tsz-fung FC, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y,. et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rock CD, Sakata Y, Quatrano RS (2010) Stress signaling I: the role of abscisic acid (ABA). In: A Pareek, SK Sopory, HJ Bohnert, eds, Abiotic Stress Adaptation in Plants. Springer, pp. 33–73 [Google Scholar]
- Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, Elias A, Rabissi A, Lumbreras V, González-Guzmán M, et al. (2013) ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25:3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata Y, Komatsu K, Takezawa D (2014) ABA as a universal plant hormone. Prog Bot 75:58–96 [Google Scholar]
- Sakata Y, Nakamura I, Taji T, Tanaka S, Quatrano RS (2010) Regulation of the ABA-responsive Em promoter by ABI3 in the moss Physcomitrella patens: role of the ABA response element and the RY element. Plant Signal Behav 5:1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saruhashi M, Ghosh TK, Arai K, Ishizaki Y, Hagiwara K, Komatsu K, Shiwa Y, Izumikawa K, Yoshikawa H, Umezawa T,. et al. (2015) Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc Natl Acad Sci U S A 112:E6388–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Christov NK, Tsuda S, Imai R. (2013) Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol 55:136–147 [DOI] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S (2018) Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci 23:623–637 [DOI] [PubMed] [Google Scholar]
- Shinozawa A, Otake R, Takezawa D, Umezawa T, Komatsu K, Tanaka T, Amagai A, Ishikawa S, Hara Y, Kamisugi Y,. et al. (2019) SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun Biol 2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson SR, Kamisugi Y, Trinh CH, Schmutz J, Jenkins JW, Grimwood J, Muchero W, Tuskan GA, Rensing SA, Lang D,. et al. (2016) Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE, a regulator of aba responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Harpazi B, Wijerathna-Yapa A, Merilo E, de Vries J, Michaeli D, Gal M, Cuming AC, Kollist H, Mosquna A (2019) A ligand-independent origin of abscisic acid perception. Proc Natl Acad Sci U S A 116:24892–24899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shinshi H. (1995) Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell 7:639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Zhang J, Hsu P, Ceciliato PHO, Zhang L, Dubeaux G, Munemasa S, Ge C, Zhao Y, Hauser F. et al. (2020) MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat Commun 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, Kohchi T, Takezawa D (2010) Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol 152:1529–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 106:17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K., Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A 97:11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Plant cell protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21:3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Pierik R, Kelly S, Sakuta M, Voesenek LA, Harberd NP (2015) An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol 169:283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.