Abstract
Root system architecture (RSA) is a key factor in the efficiency of nutrient capture and water uptake in plants. Understanding the genetic control of RSA will be useful in minimizing fertilizer and water usage in agricultural cropping systems. Using a hydroponic screen and a gel-based imaging system, we identified a rice (Oryza sativa) gene, VAP-RELATED SUPPRESSOR OF TOO MANY MOUTHS1 (OsVST1), which plays a key role in controlling RSA. This gene encodes a homolog of the VAP-RELATED SUPPRESSORS OF TOO MANY MOUTHS (VST) proteins in Arabidopsis (Arabidopsis thaliana), which promote signaling in stomata by mediating plasma membrane–endoplasmic reticulum contacts. OsVST1 mutants have shorter primary roots, decreased root meristem size, and a more compact RSA. We show that the Arabidopsis VST triple mutants have similar phenotypes, with reduced primary root growth and smaller root meristems. Expression of OsVST1 largely complements the short root length and reduced plant height in the Arabidopsis triple mutant, supporting conservation of function between rice and Arabidopsis VST proteins. In a field trial, mutations in OsVST1 did not adversely affect grain yield, suggesting that modulation of this gene could be used as a way to optimize RSA without an inherent yield penalty.
Root meristem size and root system architecture in both rice and Arabidopsis are regulated by proteins related to mediators plasma membrane–endoplasmic reticulum contact.
Introduction
As living organs, plant roots play crucial roles in water and nutrient acquisition as well as in providing support and anchorage in soil. Root system architecture (RSA), the spatial arrangement of roots for a given plant, reflects these critical functions (Lynch, 1995). In the model dicot Arabidopsis (Arabidopsis thaliana), RSA is primarily determined by growth rates and the branching patterns of lateral roots (Smith and De Smet, 2012). Asian rice (Oryza sativa) and other agronomically important monocots, possess more complex RSA profiles, dominated by the contribution from shoot-derived roots (Hochholdinger and Zimmermann, 2009).
There is strong interest in selecting for specific RSA phenotypes in crops to increase yield and promote stability under stress. Yet, due mainly to the inherent difficulties in measuring largely underground root phenotypes, the genetics underlying RSA have remained elusive. While many different root imaging modalities are being developed, a single one with an ideal combination of throughput, accuracy, and realistic growth conditions has yet to be produced (Atkinson et al., 2019). In this study, we combined two approaches to identify a regulator of RSA in rice. We found a line having decreased root length in a high-throughput mutant screen in hydroponics. We then used a non-invasive, gel-based imaging platform to map and characterize the underlying mutation.
Optimizing RSA for agriculture will require careful consideration of target environments and crop-specific management practices (Bishopp and Lynch, 2015). Depending on soil conditions, fertilizer inputs, and water use, an ideal arrangement of roots may lie anywhere on a continuum from deep and expansive to shallow and dense (Morris et al., 2017). With this in mind, it will be useful to identify the genetics underlying a range of RSA phenotypes.
Despite the challenges of identifying RSA genes in monocots, there have been a few successes. Particularly notable in rice is the cloning of two genes, DEEPER ROOTING 1 and PHOSPHORUS STARVATION TOLERANCE 1, that underlie quantitative trait loci for root phenotypes (Gamuyao et al., 2012; Uga et al., 2013). The identification of favorable alleles of these genes has been followed by successful deployment into field trials for increased stress resilience. This suggests the potential utility of harnessing beneficial root architecture traits for yield improvement and stability.
In addition, a number of genes affecting RSA in rice have been identified in genetic screens using mutagenized populations (Mai et al., 2014). Most of these have strong effects on root traits due to the lack of certain root types or other detrimental defects. Many of these mutants also have highly pleiotropic phenotypes. While contributing to our understanding of the genetic requirements of root development, highly reduced grain yield serves as a barrier to the use of these alleles in breeding (Wissuwa et al., 2016).
A common theme of growth regulation in plants is signaling through extracellular receptors, such as receptor-like kinases (RLKs; De Smet et al., 2009). These are often plasma membrane proteins that bind extracellular ligands and affect downstream transcriptional responses. RLKs in particular are predicted to comprise ∼2% of protein coding genes in rice and Arabidopsis (Shiu et al., 2004). With such a large diversity of RLKs and downstream proteins, a developing theme is the importance of spatial proximity of protein complexes to properly initiate and propagate a signal transduction cascade (Qi and Torii, 2018).
Recent work uncovered the key roles of a family of Arabidopsis genes encoding proteins containing major sperm protein (MSP) domains in stomatal patterning and above-ground plant architecture (Ho et al., 2016). These proteins were found to interact with ERECTA family RLKs in stomatal development. They were named VAP-RELATED SUPPRESSORS OF TOO MANY MOUTHS (TMM), or VSTs. The VSTs were proposed to regulate signaling through establishment or maintenance of plasma membrane–endoplasmic reticulum (PM–ER) contact sites. Here, we identify genes encoding rice and Arabidopsis VSTs as key regulators of RSA.
Results
An RSA mutant with reduced root length and altered growth dynamics
We used a high-throughput hydroponic screening system to identify lines with altered root phenotypes within an ethyl methanesulfonate (EMS)-mutagenized rice population. We discovered a mutant (designated E104-SR) that had reduced root length compared with the unmutagenized, wild-type parent at 7 d after germination (Supplemental Figure S1, A and C). The mutant also showed reduced primary and crown root length at 28 d after germination (Supplemental Figure S1, B and C). Additionally, E104-SR had increased crown root number and slight reductions in shoot height at later developmental stages (Supplemental Figure S1, D and E).
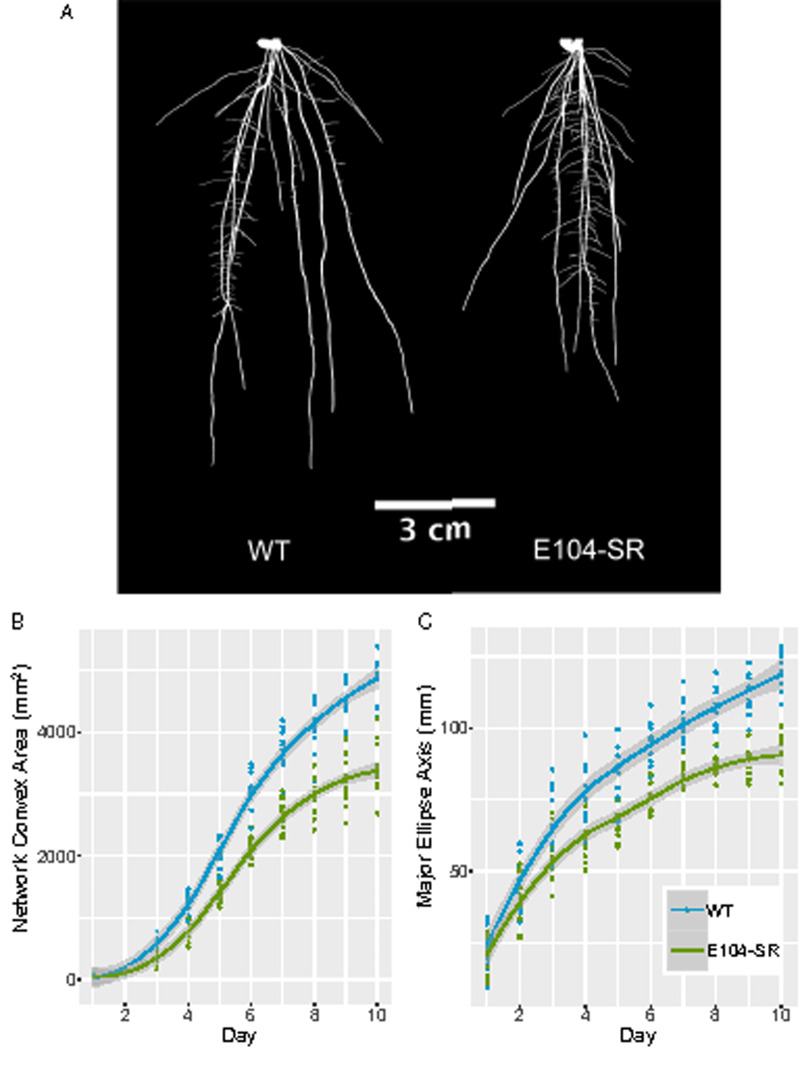
Using a non-destructive, gel-based root imaging system (Iyer-Pascuzzi et al., 2010; Galkovskyi et al., 2012; Topp et al., 2013), we examined differences in RSA between E104-SR and wild type. At 10 d after transplanting from plates into gel media, the mutant had a different RSA profile, characterized by a shorter and more compact root system (Figure 1, A). Imaging at 24-h intervals across 10 d of growth allowed us to monitor the rates of change of RSA traits throughout early development. Network convex area is defined as the area of the convex hull encompassing the root system and is a measure of RSA extent (Iyer-Pascuzzi et al., 2010). Major ellipse axis is the length of the major axis of the best-fitting ellipse to the root system and in these plants are highly correlated with RSA depth (Galkovskyi et al., 2012; Topp et al., 2013). Both network convex area and major ellipse axis increase throughout the 10-d time-course in wild-type plants, while the E104-SR mutant shows a gradual flattening in the rates of change of these traits (Figure 1, B and C).
Figure 1.
E104-SR mutant plants have shorter, more compact root systems and altered growth dynamics. (A) Representative thresholded images of gel-grown roots at 10 d after transplanting. Images were digitally extracted and pasted on a black background. (B) Network convex area of wild-type (n = 10) and E104-SR (n = 11) roots throughout 10 d of imaging after transplanting. E104-SR is significantly different from wild-type by two-sample t test (P < 0.05) starting at day 2. (C) Major ellipse axis of wild-type (n = 10) and E104-SR (n = 11) roots throughout 10 d of imaging. E104-SR is significantly different from wild-type by two-sample t test (P < 0.05) starting at day 3. Lines are fitted with the “geom_smooth” function in ggplot2 using method “loess.”
E104-SR roots have smaller meristems
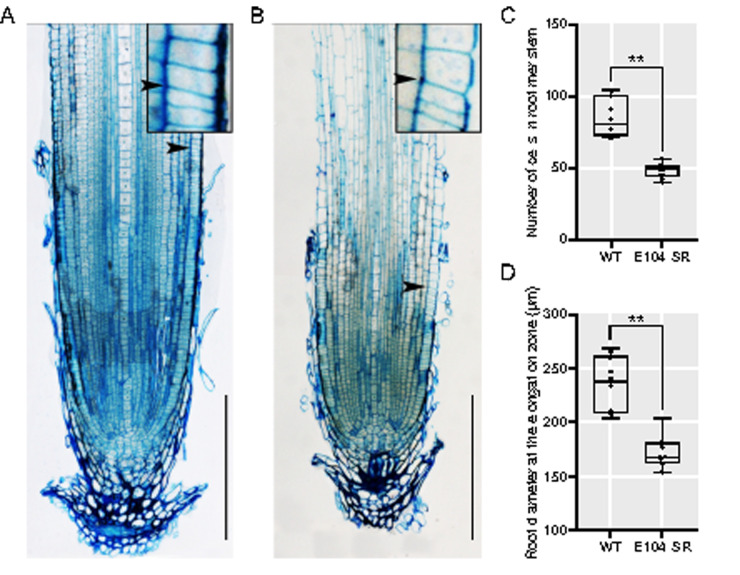
To better understand the nature of the E104-SR mutant phenotype, we examined longitudinal sections of E104-SR and wild-type primary roots. The mutant roots have significantly smaller meristematic regions during early growth (Figure 2). Additionally, primary root width is reduced in mutants (Figure 2, D).
Figure 2.
E104-SR has reduced root meristem size and decreased root width. (A) and (B) Longitudinal sections of the primary root of 5-d-old seedlings of (A) wild-type and (B) E104-SR. Arrow indicates location of junction of meristem and elongation zone. Bars = 200 μm. Inset images are enlarged 450×. (C) and (D) Quantification of (C) root meristem cell number and (D) root diameter at the elongation zone. Data are shown as box plots (n = 10). Median is represented by horizontal line in the box plot, box ranges represent quartiles 1 and 3, and minimum and maximum values are represented by error bars. Data significantly different from the corresponding wild type are indicated (**P < 0.01; Student’s t test).
The causative mutation in E104-SR maps to an 11-kb deletion
To identify the mutation underlying the mutant RSA phenotype, we used a mapping-by-sequencing approach based on bulked segregant analysis (Michelmore et al., 1991). We backcrossed the E104-SR mutant to the unmutagenized parental cultivar and selfed the F1 to generate a BC1F2 mapping population. Using our gel imaging platform, we phenotyped ∼120 BC1F2 plants, isolating DNA from 20 plants with the deepest and 20 plants with the most shallow root systems. Each of these groups of DNA samples was pooled separately, making up the mutant and wild-type bulks, respectively.
We performed paired-end, short read sequencing on the two contrasting bulks and used the R package QTLseqr (Mansfeld and Grumet, 2018), with an approach similar in concept to MUT-map (Abe et al., 2012). This analysis identified a candidate region on chromosome 8 that contained the likely causative mutation underlying the E104-SR root phenotype (Supplemental Figure S2). Further genotyping of the individuals that comprised the mutant bulk allowed for fine-mapping of the causative region to an interval between approximately 2.2–3.5 Mb on chromosome 8.
Deletion of a single gene is responsible for the E104-SR phenotype
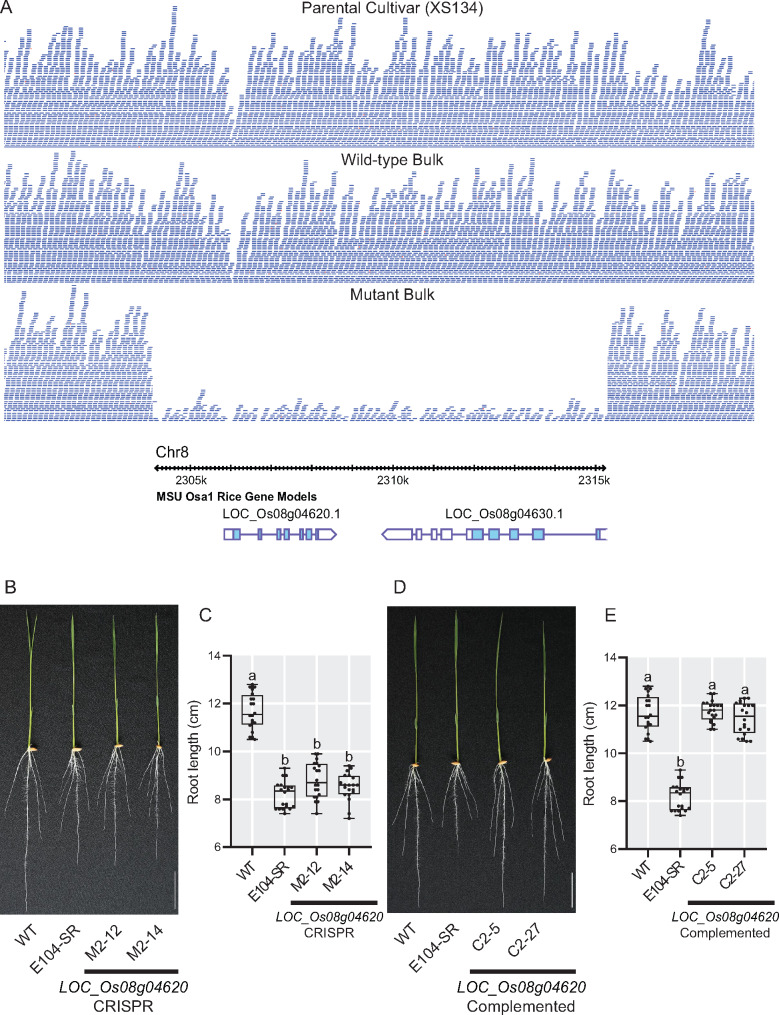
Although no EMS-induced SNPs were found within the interval on chromosome 8 between 2.2 and 3.5 Mb, there was an 11-kb deletion located at 2.3 Mb that was highly enriched in the mutant bulk (Figure 3, A). The deletion results in the complete loss of two genes. The first, LOC_Os08g04620, is predicted to encode a MSP domain containing protein. The second, LOC_Os08g04630, is annotated as encoding a mitochondrial-localized protein, an external NADH-ubiquinone oxidoreductase. Both of these genes are expressed in roots, and at higher levels than in shoots (Raines et al., 2016).
Figure 3.
LOC_Os08g04620 is located within a deletion in E104-SR and is the causative gene for the reduced root length phenotype. (A) Sequence read depth from the parental cultivar, wild-type, and mutant bulks with the MSU v7 gene models within the deleted region shown below. (B) and (C) Phenotypes (B) and root lengths (C) of 7-d-old knockout transgenic lines grown in hydroponic culture. M2–12 and M2–14 are two independent CRISPR-Cas9-generated LOC_Os08g04620 knockout T1 lines. (D) and (E) Phenotypes (D) and root lengths (E) of 7-d-old complementation lines grown in hydroponic culture. C2–5 and C2–27 are two independent T2 complementation lines of E104-SR containing a genomic fragment with only LOC_Os08g04620. Bars = 3 cm. Data are shown as box plots (n = 20). Median is represented by horizontal line in the box plot, box ranges represent quartiles 1 and 3, and minimum and maximum values are represented by error bars. Significantly different values are indicated by different letters (P < 0.01; one-way ANOVA with Tukey’s test).
To determine which of these candidates is responsible for the E104-SR root phenotype, we took two complementary approaches. First, we generated additional loss-of-function alleles of LOC_Os08g04620 and LOC_Os08g04630 using CRISPR-Cas9 (Supplemental Figures S3, S4). In hydroponics, two independent CRISPR-generated mutant alleles of LOC_Os08g04620 (designated M2-12 and M2-14) had root lengths that were shorter than the wild-type parent and indistinguishable from that of E104-SR (Figure 3, B and C). In contrast, plants with two independent CRISPR-generated mutant alleles of LOC_Os08g04630 (designated M3–6 and M3–7) had root lengths similar to wild type (Supplemental Figure S5). Next, we constructed transgenic lines with genomic fragments containing either LOC_Os08g04620 or LOC_Os08g04630 in the E104-SR mutant background to test for complementation of the reduced root length phenotype. Two complementation lines with LOC_Os08g04620 (designated C2–5 and C2–27) in the mutant background had root lengths comparable to wild type (Figures 3, D and E). Lines containing LOC_Os08g04630 (designated C3–3 and C3–5), however, failed to complement the mutant phenotype (Supplemental Figure S6). These data support the conclusion that deletion of LOC_Os08g04620 is responsible for the reduced root length phenotype in E104-SR.
LOC_Os08g04620 encodes a homolog of the Arabidopsis VST proteins
LOC_Os08g04620 encodes a protein that has a domain architecture characterized by a central MSP domain and a C-terminal coiled-coil domain (Supplemental Figure S7, B). The protein has a predicted nuclear localization sequence, but lacks a transmembrane domain. This structure is similar to a recently characterized family of proteins in Arabidopsis, the VAP-RELATED SUPPRESSORS OF TMM (VSTs; Ho et al., 2016). The Arabidopsis VST proteins were identified as key regulators of stomatal patterning. They were shown to function as peripheral plasma membrane proteins, promoting PM-ER contacts to facilitate signaling. Consistent with this, despite not having any predicted transmembrane domains, the rice protein encoded by LOC_Os08g04620 was detected as PM-localized in a proteomics study (Natera et al., 2008). In a multiple sequence alignment of MSP domain-containing proteins in Arabidopsis and rice, LOC_Os08g04620 clusters with the three Arabidopsis VST proteins and a rice putative paralog (LOC_Os07g37270; Supplemental Figure S7, A). The rice genes LOC_Os08g04620 and LOC_Os07g37270 have distinct organ-level expression profiles, with LOC_Os08g04620 notably being more highly expressed in roots than LOC_Os07g37270 (Raines et al., 2016). As LOC_Os08g04620 and LOC_Os07g37270 encode proteins that have high degrees of sequence similarity with the Arabidopsis VSTs, we will refer to them as OsVST1 and OsVST2, respectively.
OsVST1 is expressed broadly in roots
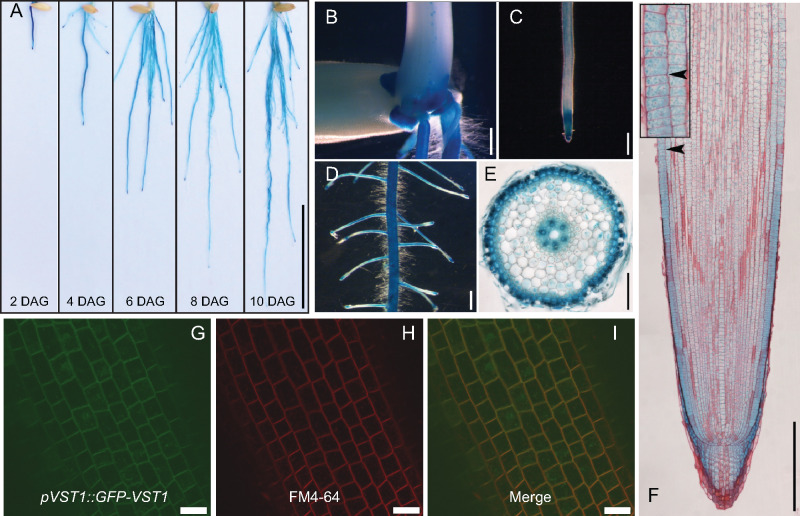
We assessed expression of OsVST1 using a line containing a transcriptional reporter with the OsVST1 promoter fused to the beta-glucuronidase gene (GUS). OsVST1 is expressed throughout the root system at 2, 4, 6, 8, and 10 d after germination (Figure 4, A). OsVST1 showed high levels of staining at the stem base (the region of crown root initiation; Figure 4, B); is expressed highly in the tips of primary, crown, and lateral roots (Figure 4, C, D, and F); and is present in outer cell layers as well as in the vasculature of the root maturation zone (Figure 4, E).
Figure 4.
Expression and protein accumulation pattern of OsVST1 are consistent with mutant RSA phenotype of E104-SR. (A) GUS staining of root systems of ProVST1:GUS transgenic seedlings at 2, 4, 6, 8, and 10 ds after germination. (B)–(F) GUS staining of 5-d-old ProVST1:GUS transgenic seedlings. Stem base (B), root tip (C), primary root with lateral roots (D), cross section of the primary root maturation zone (E), and root tip costained with safranin (F). (G)–(I) Colocalization of GFP-VST1 protein with the plasma membrane marker FM4-64 in primary root cells. GFP-VST1 (G), FM4-64 (H), and merged image (I). Bars, 3 cm in (A); 1 mm in (B)–(D); 100 μm in (E); 200 μm in (F); and 20 μm in (G)–(I). Inset image in (F) is enlarged 300x.
To analyze patterns of OsVST1 protein accumulation, we produced transgenic plants expressing OsVST1 fused to either a C-terminal or N-terminal GFP (VST1-GFP or GFP-VST1, respectively). Each of these constructs is driven by the OsVST1 promoter and was transformed into E104-SR mutant plants. Although each of the two transgenic constructs can only partially complement the short root phenotype of E104-SR mutant (Supplemental Figure S8), the pVST1::GFP-VST1 transgenic plants had longer primary roots than the pVST1::VST1-GFP transgenic plants. Therefore, we selected a pVST1::GFP-VST1 transgenic plant for further analysis. Using this line, we examined protein localization within root cells. We found that GFP-VST1 colocalizes with the plasma membrane marker FM4-64 (Figure 4, I and Supplemental Figure S9). This suggests that OsVST1 protein is present at or near the plasma membrane in rice root cells, a pattern similar to that of Arabidopsis VST1 in leaf epidermal cells (Ho et al., 2016).
The Arabidopsis vst1/2/3 triple mutant has reduced root length and decreased meristem size
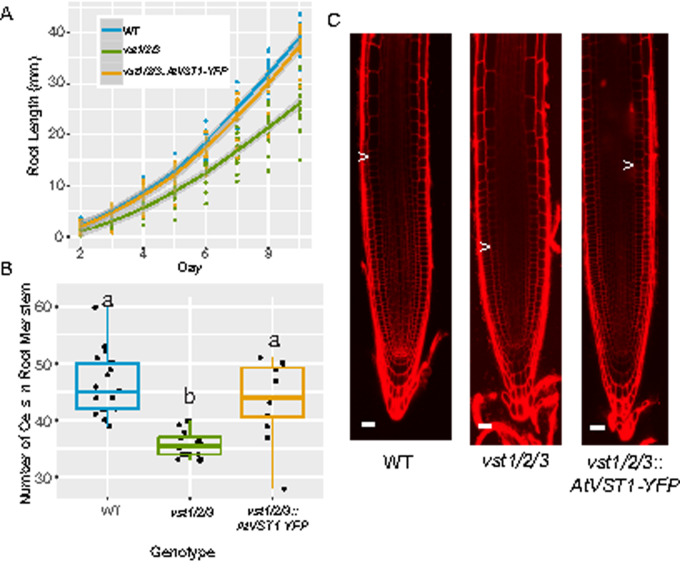
In the previous characterization of the Arabidopsis VST genes, the authors described a compact aboveground growth pattern in the vst1/2/3 triple mutant (Ho et al., 2016). They showed that expression of AtVST1 was able to rescue the plant height phenotype. As their analysis did not include root phenotyping of the vst1/2/3 mutant, we characterized it and found that primary root length is reduced (Figure 5, A). Root length was restored to wild-type levels when AtVST1 was expressed. Similar to that of the rice line E104-SR, the vst1/2/3 triple mutant has a shorter root meristem (Figure 5, B and C). The reduced meristem size was also rescued by expression of AtVST1.
Figure 5.
The reduced root length and meristem size in Arabidopsis vst1/2/3 triple mutants is rescued by expression of AtVST1. (A) Timecourse of growth of Arabidopsis roots on agar plates (n = 16–19 plants). Line is fitted with the “geom_smooth” function in ggplot2 using the method “loess.” (B) Quantification of root meristem cell number (n = 12–21 plants) and (C) representative confocal images of 7-d-old Arabidopsis roots. Arrow indicates location of junction of meristem and elongation zone. In boxplots, median is represented by horizontal line in the boxplot, box ranges represent quartiles 1 and 3, lines indicate points within 1.5 times the interquartile range. Points beyond lines are considered outliers. Significantly different values are indicated by different letters (P < 0.05; one-way ANOVA with Tukey’s test). Bars = 25 μm.
OsVST1 can complement the Arabidopsis VST mutant root length and aerial phenotypes
We hypothesized that OsVST1 might function in a similar manner as that of the Arabidopsis VSTs. To determine if OsVST1 can complement the Arabidopsis vst1/2/3 triple mutant we expressed OsVST1 in the vst1/2/3 background and found an increase in root length and plant height (Supplementary Figure S10). These results indicate that OsVST1 can functionally complement these Arabidopsis triple mutant phenotypes.
OsVST1 mutant tested in field conditions shows stable grain yield properties
Using hydroponic and gel-based imaging systems, we found that OsVST1 is required for wild-type root system depth and area. Many RSA genes that have been cloned in monocot crops have highly pleiotropic phenotypes that would result in considerable yield drag (Wissuwa et al., 2016). Our initial characterization of the OsVST1 mutant indicated that its alleles have modest effects on shoot architecture (Supplemental Figure S1, D). This suggests that varying expression of OsVST1 may be a means of modulating root architecture in the field without encountering a strong yield penalty.
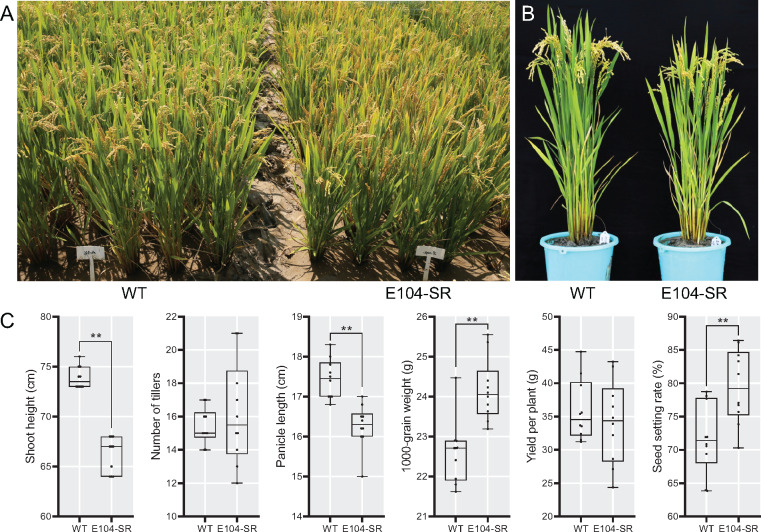
We tested the E104-SR mutant in field trials in southern China. We found that plant height was shorter, yield per plant and tiller number were unaffected, but 1,000-grain weight and seed setting rate were significantly higher in the mutant (Figure 6).
Figure 6.
E104-SR has decreased plant height and slightly increased 1,000-grain weight when grown in field conditions. (A) and (B) Morphological phenotypes of wild type and E104-SR growing in Hainan province. (C) Statistical analysis of agronomic traits of wild-type and E104-SR growing field. Data are shown as box plots (n = 10). Median is represented by horizontal line in the box plot, box ranges represent quartiles 1 and 3, and minimum and maximum values are represented by error bars. **Significant difference between wild type and E104-SR (P < 0.01; Student’s t test); *Significant difference between wild type and E104-SR (P < 0.05; Student’s t test).
Discussion
Using two different non-destructive imaging platforms, we identified a regulator of RSA. We mapped the causal lesion underlying the E104-SR mutant root phenotype in an EMS-treated rice population to an 11-kb deletion (Supplementary Figure S2). This is surprising considering the strong bias of EMS for transition mutations (Greene et al., 2003; Henry et al., 2014). However, larger deletions have been identified as likely causal mutations in EMS-mutagenized populations in maize and wheat (Okagaki et al., 1991; Mo et al., 2018).
The OsVST1 gene encodes a protein with high sequence similarity to the Arabidopsis VSTs. Coupled with the complementation of the root length and aerial phenotypes of vst1/2/3 by OsVST1, this points to a potential role of this protein in regulating ER–PM contact sites. ER–PM interactions have been implicated in processes other than developmental signaling, including response to stress and establishment of cell-to-cell communication through plasmodesmata (Bayer et al., 2017; Lee et al., 2019).
Important open questions remain concerning the specific molecular pathways through which OsVST1 regulates RSA. OsVST1 could influence meristem size through broad modulation of cell–cell communication or through specific signaling complexes. In the context of Arabidopsis stomatal patterning, signaling through complexes specifically involving the ERECTA family of RLKs is affected in VST mutants (Ho et al., 2016). Considering the large number of RLKs that could control root meristem growth, it will be challenging to determine the specific signaling complexes affected in OsVST1 mutants. The similarity of root meristem phenotypes between Arabidopsis and rice VST mutants may allow for use of this more genetically tractable system to understand the molecular pathways affected by VST proteins in roots.
VST homologs in Poplar were identified as differentially expressed during cell proliferation and radial growth in wood formation (Hertzberg et al., 2001; Schrader et al., 2004). In a patent application (US20170349910A1), overexpression of these genes resulted in trees with increased biomass. Our field results suggest that modulation of OsVST1 does not have an overall detrimental effect on yield and in fact may positively impact some yield phenotypes (Figure 6). Thus, there may be field conditions in which the shallow root architecture of the OsVST1 mutant may be agronomically beneficial.
Materials and methods
Plant materials and growth conditions
For hydroponic growth experiments, germinated rice (O. sativa) seeds were sown on floating nets and grown in full-strength Kimura nutrient solution (pH, 5.6) as described previously (Chen et al., 2013). The seedlings were photographed at 7 d after germination and transferred to a tank (61 cm in length, 41 cm in width and 21 cm in height) with full-strength nutrient solution to monitor plant growth. Nutrient solution was changed every 7 d. Root length, crown root number, and shoot length were measured at 7, 14, and 28 d after transferred to tank. The phenotypic characterization of plants was performed in a growth room under 14-h d length at 60%–70% humidity and 30°C/22°C (day/night). The growth room uses bulb-type light with a photon density of ∼300 μmol m−2 s−1.
For gel imaging experiments, plants were grown in solidified Yoshida’s Nutrient Solution (pH 5.8). After 2 d of pregrowth on petri plates in the dark at 30°C, seedlings were transplanted to sterile glass jars containing media. Plants were largely shielded from light and grown in a growth chamber under 12-h d length at 30°C/27°C.
For field experiments, plants were grown at the Zhejiang University Experimental station in Hainan Province, China. Seeds were sown in early December, seedlings were transplanted after a month, and harvesting and agronomic trait analysis was performed after maturation in mid-April of the next year.
Mutagenesis and screening
Seeds of rice cultivar Xiushui 134 (XS134) were soaked in water for 2 d at 37°C and then treated with 0.8% (w/v) EMS for 10 h at 25°C. Treated seeds were sown and planted in field to harvest seeds of 12,000 M1 plants lines. To identify lines with aberrant root length phenotypes, germinated seeds of M2 populations were sown on floating nets and grown in a tank (61 cm in length, 41 cm in width, and 21 cm in height) in hydroponics with a 1/2 strength nutrient solution. After growing 7 d, root length of seedlings was measured and photographed. E104-SR was identified as a mutant segregating for a shorter root length compared with wild-type.
Gel-based imaging and analysis
Gel-grown plants were imaged daily using a previously described imaging system (Topp et al., 2013). Twenty to forty images were taken for each plant at each timepoint. Images were analyzed with the GiaRoots software (Galkovskyi et al., 2012).
Mutation mapping
The E104-SR mutant was backcrossed to the unmutagenized XS134 parent and the resulting F1 plant was selfed to produce an F2 mapping population segregating for the shorter root phenotype. Over 120 F2 plants were screened using the gel imaging system. Twenty plants with the shortest and most compact root systems were selected as the mutant class, while the 20 plants with the largest root systems made up the wild-type class.
DNA was isolated from leaf tissue from these individuals and the parental line XS134 using the Qiagen DNAeasy Plant Kit. These were collected in equal concentrations to make up the mutant bulk, wild-type bulk, and the XS134 sample. Library preparation and paired-end sequencing with the Illumina HiSeq 2500 were done at the Duke University Genomics Core Facility.
Reads were mapped to the Nipponbare (Oryza_sativa.IRGSP-1.0.31) reference genome using the “aln” function in BWA (Li and Durbin, 2009). SNPs and small indels were called using the “mpileup” function in SAMTools (Li, 2011). Allele counts we extracted from the resulting VCF file using an awk script from Mascher et al. (2014). Background SNPs (also present in the XS134 sequence) were removed. The final VCF file was imported into the QTLseqr R package (Mansfeld and Grumet, 2018) and the “deltaSNP” index was calculated and plotted.
Mapped sequence reads were visualized with the Integrated Genome Browser (Freese et al., 2016).
Vectors construction and transformation
CRISPR-Cas9-mediated gene editing was used to generate additional mutant alleles of LOC_Os08g04620 and LOC_Os08g04630. Callus from XS134 was transformed with agrobacterium strain EHA105 containing constructs based on the vector pYLCRISPR/Cas9-MH containing gRNAs targeting exons of LOC_Os08g04620 or LOC_Os08g04630. Diagrams are in Supplemental Figures S3, S4. Deletions were confirmed by Sanger sequencing. Two independent loss-of-function mutants were identified and phenotyped for each candidate gene.
Callus from E104-SR was transformed with agrobacterium strain EHA105 containing pCAMBIA1300 constructs with the LOC_Os08g04620 (an 6896-bp DNA fragment containing a 3,144-bp sequence upstream of the start codon, the entire gene sequence, and a 1,651-bp sequence downstream of the stop codon) or LOC_Os08g04630 (a 9,466-bp DNA fragment containing a 3,113-bp sequence upstream of the start codon, the entire gene sequence, and a 3,199-bp sequence downstream of the stop codon) open-reading frames. Two independent potential complementation lines were identified and phenotyped for each candidate gene.
To construct the promoter: GUS reporter lines, callus from XS134 was transformed with agrobacterium strain EHA105 containing pCAMBIA1300 construct with GUS reporter gene driven by the LOC_Os08g04620 promoter (the 3,139-bp sequence upstream of the start codon).
To construct the OsVST1 and GFP fusion protein lines. Two constructs of pVST1::VST1-GFP (VST1-GFP) and pVST1::GFP-VST1 (GFP-VST1) were generated. A 5,237-bp DNA fragment containing genomic OsVST1 and the 3,139-bp region upstream of the OsVST1 start codon fused in-frame to the 5′-end of GFP in the modified pCAMBIA1300-GFP vector to generate pVST1::VST1-GFP. A 3,747-bp DNA fragment containing genomic OsVST1 and the 1,646-bp sequence downstream of the stop codon fused in-frame to the 3′-end of GFP, and driven by the OsVST1 promoter (the 3,139-bp sequence upstream of the start codon) to generate pVST1::GFP-VST1. Callus from E104-SR was transformed with agrobacterium strain EHA105 containing pVST1::VST1-GFP or pVST1::GFP-VST1 constructs. The primers used (including the restriction enzyme sites used in the cloning) are listed in Supplemental Table S1.
Protein sequence alignment
Proteins containing a MSP domain were downloaded from the MSU Rice Genome Database and The Arabidopsis Information Resource. Proteins encoding the longest isoforms of each gene were used. Sequences were aligned using MAFFT (Madeira et al., 2019).
Histological observation, GUS staining, and fluorescence imaging
To observe meristem and elongation zone of root tip in mutant, rice root tips were fixed in 2.5% glutaric dialdehyde fixation solution overnight, dehydrated using acetone, infiltrated and embedded in Spurr’s resin, and then sectioned into 2-μm sections. Sections were mounted on slides and stained with methylene blue solution. Images were taken using a microscope (Nikon).
To observe expression pattern of OsVST1, seedlings were stained with beta-galactosidase solution for 6 h at 37°C. Images of root system or stem base, primary root with lateral roots, and root tips were taken using a camera (Nikon) or stereomicroscope (Leica), respectively.
To observe OsVST1 expression in detail in the root tip regions, root tips of GUS staining were fixed in FAA (5 mL of 38% formaldehyde mixed with 5 mL of acetic acid and 90 mL of 70% ethanol) fixation solution overnight, dehydrated using acetone, infiltrated, and embedded in Spurr’s resin, and then sectioned into 2-μm sections. Sections were mounted on slides and taken images using a microscope to observe GUS staining. To observe cell wall structure, the same slides were stained using safranin solution and then images were taken using microscope.
For the OsVST1 subcellular localization analyses, 3-d-old rice seedlings containing the pVST1::GFP-VST1 transgene were stained with 2 μg/mL FM4-64 staining solution for 3 min at room temperature. The stained roots were then imaged using a Zeiss 510 confocal microscope. Confocal experimental set-up: lasers: HFT488 nm. Intensity: for FM4-64, 40%; for GFP 90%. Collection bandwidth: for FM4-64, 590–750 nm; for GFP, 505–530 nm. Master gain: for FM4-64, 600; for GFP, 650. Digital gain for both: 1. Pinhole: for FM4-64, 34.0; for GFP, 90.
Arabidopsis (A. thaliana) root growth assays
For Arabidopsis experiments, seeds were surface sterilized with 50% bleach for 10 min, rinsed 5× with sterile water, then sown on half MS 1% sucrose plates solidified with 1% agar in square petri dishes, sealed with micropore tape, stratified at 4°C for 2 d in the dark before moving to a 22°C, 16-h light cycle growth chamber vertically. Under these conditions, germination occurred approximately 1 d after moving to the chamber. For analysis of root growth dynamics of WT Arabidopsis, the vst1/2/3 triple mutant, and the vst1/2/3::AtVST1-YFP complemented line, plates were imaged every d between 2 and 9 post-germination, and root lengths for plants were measured in imageJ. In a separate experiment, cortical meristem size was measured for WT, vst1/2/3, and vst1/2/3::AtVST1-YFP 7 d after germination by staining with 10 mg/mL propidium iodide for approximately 2 min before imaging with a Zeiss 510 confocal microscope. Meristem cells were counted from the first cortical cell after the cortex/endodermal initial or quiescent center (in the case where the initial had asymmetrically divided) until the abrupt increase in cell length observed in the transition zone. Counting was performed in imageJ, and two independent double-blinded counts were performed with nearly identical results in order to validate the apparent difference in meristem size.
For the OsVST1 complementation experiment, we synthesized the OsVST1 cDNA sequence (GenScript) to include attL1/L2 sites for direct gateway recombination into pGWB502-omega (Nakagawa et al., 2007), which contains a 35S promoter and translational enhancer for overexpression. The vst1/2/3 mutant was transformed by floral dip and transformants were selected on hygromycin. We selected two independent lines in the T2 that appeared to be segregating for an increase in plant height. We isolated homozygous lines for each in the T4 based on uniform hygromycin resistance. To test complementation of root length, we grew WT, vst1/2/3, and vst1/2/3::35s:OsVST1 in the same manner as above, and measured root length was measured at 8 d post-germination. Seedlings of these plants were transferred to soil and plant height measured approximately 5 weeks later.
Statistical analysis
Student’s t test or one-way ANOVA with post hoc Tukey’s honest significance difference tests were used to assess statistical significance. Statistical significance levels are indicated in the figure legends.
Accession numbers
Sequence data from this article can be found in the NCBI SRA under Bioproject PRJNA631002.
Supplemental data
Supplemental Figure S1. E104-SR is a mutant with decreased root length and slightly decreased shoot height.
Supplemental Figure S2. Bulked-segregant analysis identifies a peak on chromosome 8 associated with the E104-SR root length phenotype.
Supplemental Figure S3. CRISPR-Cas9-mediated target mutagenesis of LOC_Os08g04620.
Supplemental Figure S4. CRISPR-Cas9-mediated target mutagenesis of LOC_Os08g04630.
Supplemental Figure S5. CRISPR-mediated deletion of LOC_Os08g04630 does not result in decreased root length.
Supplemental Figure S6. Insertion of genomic fragment containing LOC_Os08g04630 into E104-SR does not complement the root length phenotype.
Supplemental Figure S7. Sequence alignment of MSP domain-containing proteins in Rice and Arabidopsis shows clustering of OsVST1 with AtVST proteins.
Supplemental Figure S8. GFP-VST1 complements the short root phenotype of E104-SR better than VST1-GFP.
Supplemental Figure S9. OsVST1 is localized at or near the plasma membrane in rice root cells.
Supplemental Figure S10. Expression of OsVST1 can partially rescue the decreased root length and plant height phenotypes in the Arabidopsis vst1/2/3 triple mutant.
Supplemental Table S1. Primer sequences used for genotyping and plasmid construction.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Dominique Bergmann for the vst1/2/3 and complementation lines, Dr. Colleen Drapek for critical reading of this manuscript, members of the Benfey and Mao Laboratories for helpful discussions, and Alan Tang, Medhavinee Mijar, and Trevor Gannalo for their help with plant care and image processing. Listing order for co-lead authors was determined by the last digit of the closing price (odd or even) of the Shanghai Stock Exchange Composite Index on May 12, 2020.
Funding
This work was supported by a National Science Foundation Graduate Research Fellowship to K.R.L. and National Science Foundation grant PHY-1915445 to P.N.B.; the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation [through Grant GBMF3405] to P.N.B.; and the National Key Research and Development Program of China [2016YFD0100700] and the Ministry of Education and Bureau of Foreign Experts of China [B14027] to C.M.
Conflict of interest statement. None declared.
Y.S., K.R.L., C.M., and P.N.B.: conceptualization. Y.S., K.R.L., H.Z., I.T., M.Z., C.M., and P.N.B.: methodology. Y.S., K.R.L., H.Z., I.T., and M.Z.: investigation. Y.S. and K.R.L.: writing—original draft preparation. All authors were involved in writing—review and editing. C.M. and P.N.B.: supervision.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Philip N. Benfey (philip.benfey@duke.edu).
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M. et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178 ( 10.1038/nbt.2095). [DOI] [PubMed] [Google Scholar]
- Atkinson JA, Pound MP, Bennett MJ, Wells DM (2019) Uncovering the hidden half of plants using new advances in root phenotyping. Curr Opin Biotechnol 55: 1–8 ( 10.1016/j.copbio.2018.06.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Sparkes I, Vanneste S, Rosado A (2017) From shaping organelles to signalling platforms: the emerging functions of plant ER–PM contact sites. Curr Opin Plant Biol 40:89–96 ( 10.1016/j.pbi.2017.08.006). [DOI] [PubMed] [Google Scholar]
- Bishopp A, Lynch JP (2015) The hidden half of crop yields. Nat Plants 1: Art. No. 15117 ( 10.1038/nplants.2015.117). [DOI] [PubMed] [Google Scholar]
- Chen X, Shi J, Hao X, Liu H, Shi J, Wu Y, Wu Z, Chen M, Wu P, Mao C (2013) OsORC3 is required for lateral root development in rice. Plant J 74:339–350 ( 10.1111/tpj.12126). [DOI] [PubMed] [Google Scholar]
- De Smet I, Voß U, Jürgens G, Beeckman T (2009) Receptor-like kinases shape the plant. Nat Cell Biol 11:1166–1173 ( 10.1038/ncb1009-1166). [DOI] [PubMed] [Google Scholar]
- Freese NH, Norris DC, Loraine AE. (2016) Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32:2089–2095 ( 10.1093/bioinformatics/btw069). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkovskyi T, Mileyko Y, Bucksch A, Moore B, Symonova O, Price CA, Topp CN, Iyer-Pascuzzi AS, Zurek PR, Fang S. et al. (2012) GiA roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol 12:116 ( 10.1186/1471-2229-12-116). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488:535–539 ( 10.1038/nature11346). [DOI] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Nagalakshmi U, Lieberman MC, Ngo KJ, Krasileva KV, Vasquez-Gross H, Akhunova A, Akhunov E, Dubcovsky J, Tai TH et al. (2014) Efficient genome-wide detection and cataloging of EMS-induced mutations using Exome capture and next-generation sequencing. Plant Cell 26:1382–1397 ( 10.1105/tpc.113.121590). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlén M, Teeri TT, Lundenberg J. et al. (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci U S A 98:14732–14737 ( 10.1073/pnas.261293398). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CMK, Paciorek T, Abrash E, Bergmann DC (2016) Modulators of stomatal lineage signal transduction alter membrane contact sites and reveal specialization among ERECTA kinases. Dev Cell 38:345–357 ( 10.1016/j.devcel.2016.07.016). [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Zimmermann R (2009) Molecular and genetic dissection of cereal root system development. Plant Mol Biol 37:175–191 (http://doi.org/10.1002/9781444310023.ch7). [Google Scholar]
- Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, Weitz JS,, Benfey PN (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 152:1148–1157 ( 10.1104/pp.109.150748). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Vanneste S, Pérez-Sancho J, Benitez-Fuente F, Strelau M, Macho AP, Botella MA, Frimi J, Rosado A. (2019) Ionic stress enhances ER–PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc Natl Acad Sci U S A 116:1420–1429 ( 10.1073/pnas.1818099116). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993 ( 10.1093/bioinformatics/btr509). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760 ( 10.1093/bioinformatics/btp324). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109:7–13 ( 10.1104/pp.109.1.7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD. et al. (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641 ( 10.1093/nar/gkz268). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai CD, Phung NTP, To HTM, Gonin M, Hoang GT, Nguyen KL, Do VN, Courtois B, Gantet P (2014) Genes controlling root development in rice. Rice 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld BN, Grumet R. (2018) QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. Plant Genome 11 ( 10.3835/plantgenome2018.01.0006). [DOI] [PubMed] [Google Scholar]
- Mascher M, Jost M, Kuon JE, Himmelbach A, Abfalg A, Beier S, Scholz U, Stein A, Graner N (2014). Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol 15 (h 10.1186/gb-2014-15-6-r78). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 88:9828–9832 ( 10.1073/pnas.88.21.9828). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Howell T, Vasquez-Gross H, de Haro LA, Dubcovsky J, Pearce S (2018) Mapping causal mutations by exome sequencing in a wheat TILLING population: a tall mutant case study. Mol Genet Genomics 293:463–477 ( 10.1007/s00438-017-1401-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr-Hersey J, Goh T, von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP. et al. (2017) Shaping 3D root system architecture. Curr Biol 27: PR919–R930 ( 10.1016/j.cub.2017.06.043). [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71:2095–2100. 10.1271/bbb.70216 [DOI] [PubMed] [Google Scholar]
- Natera SHA, Ford KL, Cassin AM, Patterson JH, Newbign EJ, Bacic A (2008) Analysis of the Oryza sativa plasma membrane proteome using combined protein and peptide fractionation approaches in conjunction with mass spectrometry. J Proteome Res 7:1159–1187 ( 10.1021/pr070255c). [DOI] [PubMed] [Google Scholar]
- Okagaki RJ, Neuffer MG, Wessler SR (1991) A deletion common to two independently derived waxy mutations of maize. Genetics 128:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Torii KU (2018) Hormonal and environmental signals guiding stomatal development. BMC Biol 16: Art. No. 21 ( 10.1186/s12915-018-0488-5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines T, Blakley IC, Tsai YC, Worthen JM, Franco-Zorrilla JM, Solano R, Eric Schaller G, Loraine AE, Kieber JJ (2016) Characterization of the cytokinin-responsive transcriptome in rice. BMC Plant Biol 16:1–16 ( 10.1186/s12870-016-0932-z). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. (2004) A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16:2278–2292 ( 10.1105/tpc.104.024190). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S, Karlowski W, Pan R. (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234 (http://doi.org/10.1105/tpc.020834.1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, De Smet I (2012) Root system architecture: insights from Arabidopsis and cereal crops. Phil Trans R Soc B Biol Sci 367:1441–1452 ( 10.1098/rstb.2011.0234). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee C-R, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci U S A 110:E1695–704 ( 10.1073/pnas.1304354110). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45:1097–102 ( 10.1038/ng.2725). [DOI] [PubMed] [Google Scholar]
- Wissuwa M, Kretzschmar T, Rose TJ (2016) From promise to application: root traits for enhanced nutrient capture in rice breeding. J Exp Bot 67:3605–3615 ( 10.1093/jxb/erw061). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.