Abstract
During drought stress, cellular proteostasis on the one hand and amino acid homeostasis on the other hand are severely challenged, because the decrease in photosynthesis induces massive proteolysis, leading to drastic changes in both the proteome and the free amino acid pool. Thus, we selected progressive drought stress in Arabidopsis (Arabidopsis thaliana) as a model to investigate on a quantitative level the balance between protein and free amino acid homeostasis. We analyzed the mass composition of the leaf proteome based on proteomics datasets, and estimated how many protein molecules are present in a plant cell and its subcellular compartments. In addition, we calculated stress-induced changes in the distribution of individual amino acids between the free and protein-bound pools. Under control conditions, an average Arabidopsis mesophyll cell contains about 25 billion protein molecules, of which 80% are localized in chloroplasts. Severe water deficiency leads to degradation of more than 40% of the leaf protein mass, and thus causes a drastic shift in distribution toward the free amino acid pool. Stress-induced proteolysis of just half of the 340 million RubisCO hexadecamers present in the chloroplasts of a single mesophyll cell doubles the cellular content of free amino acids. A major fraction of the amino acids released from proteins is channeled into synthesis of proline, which is a compatible osmolyte. Complete oxidation of the remaining fraction as an alternative respiratory substrate can fully compensate for the lack of photosynthesis-derived carbohydrates for several hours.
A quantitative perspective on the protein and amino acid composition of a leaf cell illustrates the dimension and specific consequences of massive proteolysis induced by severe water deficit.
Introduction
Proteostasis (protein homeostasis) is essential for maintaining normal cellular functions, which rely on an appropriate composition as well as correct folding of the proteome. Plant cells contain several thousand different proteins that are highly diverse—not only in terms of their function but also in size and abundance. RubisCO has to be present in large quantities in leaf cells due to its low enzymatic activity and carbon fixation efficiency, whereas hardly detectable amounts of, e.g. signaling molecules or transcription factors efficiently fulfil their functions. The protein composition of other tissues, such as roots or seeds, again, is completely different (Baerenfaller et al., 2008; Mergner et al., 2020). In addition, 1 mg of a large protein such as glutamate synthase contains only 4 nmol active sites compared with 83 nmol for the small protein glutaredoxin. Thus, the investment of resources (energy and nutrients) required for the synthesis of large and/or high abundance proteins is by several magnitudes higher than for small proteins of low abundance.
Not surprisingly, cells contain several sophisticated systems to control proteostasis and are able to recycle the resources needed for new growth. Protein synthesis is catalyzed by the ribosomes in the cytosol, plastids, and mitochondria. The synthesis rate is regulated on different levels in response to the energy status of the cell, e.g. via mRNA availability, the GDP and GTP pools, and posttranslational modifications of the ribosome (Merchante et al., 2017). The two major protein recycling systems in eukaryotes are autophagy and the ubiquitin-proteasome system (reviewed by Vierstra, 2009; Dikic, 2017; Marshall and Vierstra, 2018). During autophagy, cytoplasmic constituents, including large protein and nucleic acid aggregates, lipid bodies, and even entire organelles, are sequestered into a double membrane vesicle, the autophagosome, and delivered to the vacuole for breakdown. Thus, autophagy, in addition to proteins, digests nucleic acids, lipids, and carbohydrates. Autophagosome formation is controlled by a highly conserved set of 40 autophagy-related (ATG) proteins. These include receptors that recognize specific cellular components and tether them to the enveloping autophagic membrane to target them for destruction. In contrast, the ubiquitin-proteasome system localized in the cytosol catabolizes proteins individually. Substrates are marked for degradation by a poly ubiquitin tag that enables their recognition and hydrolysis by the proteasome, a large protein complex composed of a 20S catalytic core and two regulatory 19S lids. Several molecules of the 8.5-kDa protein ubiquitin are covalently conjugated to a lysine residue of the target protein by an enzymatic cascade consisting of ubiquitin activating (E1), conjugating (E2), and ligating (E3) enzymes. Substrate specificity is provided by a high number of different E3 ubiquitin ligases (>1,400 in the Arabidopsis genome). In addition to the bulk degradation systems, plants contain hundreds of individual proteases from several unrelated families. They can be grouped into four major classes according to the nature of the nucleophile used for proteolytic cleavage of the peptide bond. Cysteine and serine proteases use a Cys or Ser activated by His as a nucleophile whereas metalloproteases and aspartic proteases activate water using a metal ion or Asp, respectively (van der Hoorn, 2008). Proteases are present in all the different subcellular compartments. Plastids and mitochondria contain distinctive proteolytic systems from prokaryotic origin such as AAA-class (ATPases associated with diverse cellular activities), Lon, FtsH (filamentation temperature sensitive H), and Clp (caseinolytic protease) proteases (Kwasniak et al., 2012; Nishimura et al., 2016).
The accumulation of non-functional and misfolded proteins would lead to the formation of large protein aggregates that are detrimental to cellular function (McClellan et al., 2005). Thus, damaged proteins are efficiently detected and eliminated by the two main protein quality control systems, the ubiquitin-proteasome system and autophagy, to avoid proteotoxic stress (Dikic, 2017). Even under steady-state conditions, the turnover rates of individual proteins are highly diverse: a more than 150-fold variation in protein degradation has been reported (Li et al., 2017). The D1 protein localized in the reaction center of photosystem II is replaced on a daily basis since it is frequently damaged by reactive oxygen species as a result of photosynthetic activity. Also, regulatory proteins such as hormone response factors usually have a short half-life to allow rapid responses to a changing environment (Nelson and Millar, 2015). In contrast, ribosomal subunits are among the most stable proteins in Arabidopsis and remain functional for several months (Li et al., 2017). Protein stability is defined by different factors such as the physical location of the protein, interactions with cofactors or other proteins, and post-translational modifications (Nelson and Millar, 2015).
Proteostasis is closely connected to amino acid homeostasis since protein synthesis requires a sufficient supply of loaded t-RNAs whereas proteolysis releases free amino acids. The effect of protein metabolism on the relative contents of free amino acids can be substantial, in particular, for low abundance amino acids such as the sulfur containing, aromatic, and branched chain amino acids (Hildebrandt, 2018). In yeast and animal cells, proteasome inhibition leads to cell death, which is primarily caused not by the accumulation of misfolded proteins, but by a detrimental deficiency in free amino acids (Suraweera et al., 2012). Apart from serving as building blocks for proteins, free amino acids have several additional functions in plant metabolism. They are precursors for the synthesis of secondary metabolites, hormones, and signaling molecules, and also act as transport and storage forms for organic nitrogen (Lam et al., 2003; Alcázar et al., 2006; Tzin and Galili, 2010). During drought and salt stress, Pro and the non-proteinogenic amino acid γ-aminobutyric acid (GABA) function as compatible osmolytes (Krasensky and Jonak, 2012). Proteolysis is increased in response to adverse environmental conditions to provide amino acids as precursors for these defense-related metabolites and also as alternative substrates for ATP production when photosynthetic activity is low (Araújo et al., 2011; Hildebrandt et al., 2015). In this study, we use progressive drought stress in Arabidopsis as a model to investigate the balance between protein and free amino acid homeostasis on a quantitative level. We estimate the molecular as well as the mass protein composition of an average rosette leaf and an individual mesophyll cell. How many protein molecules are present in a plant cell and its subcellular compartments? Which fraction of their leaf proteome do plants degrade maximally under severe drought stress? How is proteostasis controlled under these conditions? Do cells just eat anything when they are really starved or are they still picky? Are the proteins that are essential for stress tolerance synthesized or, rather, spared from degradation? Which proteins contribute to the free amino acid pool and what happens to the amino acids released during proteolysis?
Results
Quantitative composition of the leaf proteome
As a starting point for investigating protein homeostasis during drought stress, we focused on the proteome of control plants grown under optimal conditions to gain an impression of their status in the absence of stress (Figure 1, A). Intensity-based absolute quantification (iBAQ; Schwanhäusser et al., 2011) was used for calculating the absolute content [µg protein · g−1 dry weight (DW)] of each of the 1,399 different proteins detected by our shotgun-mass spectrometry (MS) approach. The complete MS dataset, as well as detailed information on the calculation methods, can be found in the Supplemental Material (Supplemental Dataset S1, A and Supplemental Figure S1). In order to estimate which mass fraction of the total leaf proteome is covered by our MS dataset, we used labeled peptides for absolute quantification of RubisCO large subunit (Supplemental Dataset S2 and Supplemental Figure S2). This approach revealed that the 1,399 proteins in our dataset represent 82.4% of the leaf protein mass. We included this factor in all calculations to provide the best possible estimation of individual protein contents (Supplemental Dataset S1, B). However, some questions, such as calculating the total number of protein molecules in a cell or the amount of amino acids produced by protein degradation during stress, require making the best possible assumptions about the composition of the 17.6% of the leaf proteome not visible in our proteomics dataset. For these aspects we assumed that the composition of this invisible portion corresponds to the rest of the proteome, and thus used the original dataset as a basis for calculations (Supplemental Dataset S1, A).
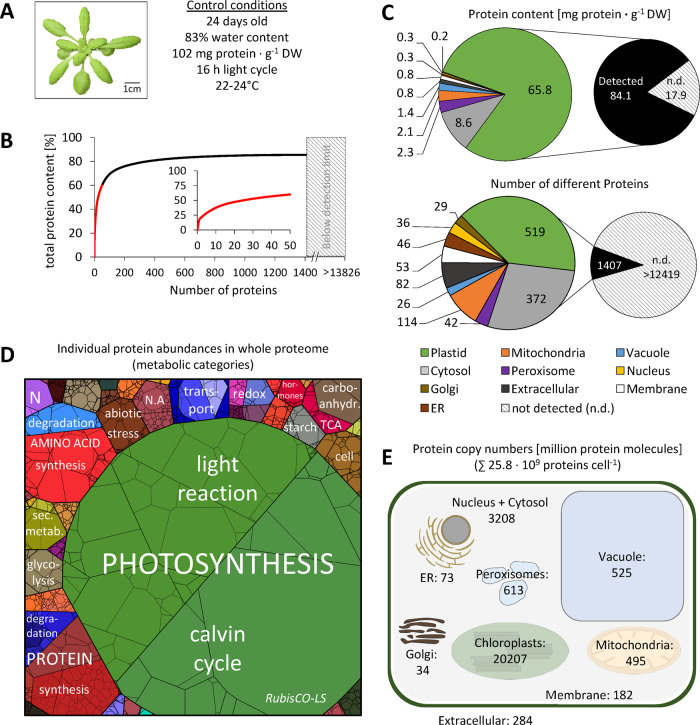
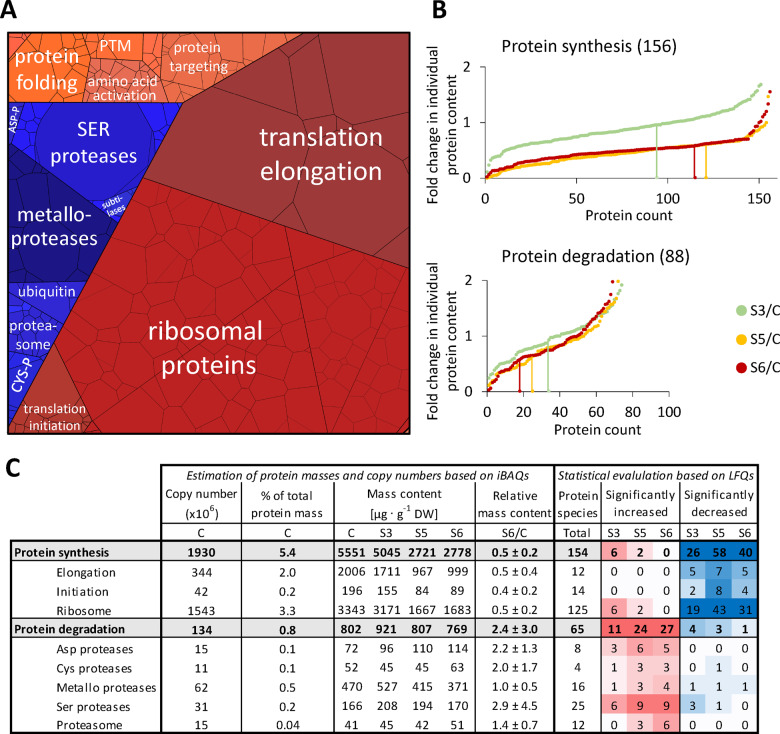
Figure 1.
Quantitative composition of the Arabidopsis leaf proteome. (A) Phenotype of a representative control plant used for MS analysis (the rosette was digitally extracted from the soil background). (B) Fraction of total protein content contributed by each of the 1,399 proteins detected by shotgun proteomics. Proteins were sorted according to their absolute content in descending order and added up. The 50 most abundant proteins (red graph) are shown in the inset (the axis label is identical to the main graph). (C) Distribution of the proteins detected in control samples in the different subcellular compartments according to SUBA4 prediction (Hooper et al., 2017). Protein content (sum of all individual protein contents calculated from iBAQs) versus number of different protein species per subcellular compartment. Hatched areas in B and C indicate the protein mass and estimated number of protein groups not detectable by our MS approach. The invisible mass fraction has been calculated on the basis of labeled peptides (Supplemental Figure S2 and Supplemental Dataset S2); the estimated number of leaf protein groups is taken from Mergner et al. (2020). (D) Proteomap illustrating the quantitative composition of the leaf proteome under control conditions. Proteins are shown as polygons whose sizes represent the mass fractions (protein abundances obtained by MS [iBAQ], multiplied with protein molecular weight). Proteins involved in similar cellular functions according to the MapMan annotation file (version Ath_AGI_LOCUS_TAIR10_Aug2012, Thimm et al., 2004) are arranged in adjacent locations and visualized by colors. Mass fractions of the functional categories [%] are provided in Supplemental Dataset S3. (E) Number of protein molecules [million proteins] present in the subcellular compartments of an average Arabidopsis mesophyll cell. Copy numbers represent the sum of protein molecules present in all chloroplasts (ca. 100 per cell; Königer et al., 2008), mitochondria (300–450 per cell; Preuten et al., 2010), or peroxisomes in the cell. Copy numbers for all individual proteins detected in our MS approach are given in Supplemental Dataset S1. Only proteins with unambiguous assignments are shown. N.A., not annotated; N, nitrogen metabolism; LS, large subunit.
The leaf proteome is dominated by a limited number of very high abundance proteins (Figure 1, B). RubisCO alone, which is well known for being one of the most abundant proteins on earth (Bar-On and Milo, 2019), constitutes about one-fifth of the leaf protein mass, corresponding to 21 mg · g−1 DW under control conditions (Supplemental Dataset S1, B). Another fifth consists of 11 other photosynthesis-related proteins, and in total, about 80% of the leaf protein mass is found in the chloroplasts (Figure 1, C, top; subcellular protein localization predicted by SUBA4, Hooper et al., 2017). Without taking absolute quantities into account, the distribution of the proteins detected by MS on subcellular compartments looks markedly different, with only 37% chloroplast protein species (Figure 1, C, bottom). It also has to be considered that the 18% of protein mass not covered by our MS approach contains a high number of very low abundance proteins. The protein investment of a leaf cell into different functions can be visualized on a PROTEOmap (Figure 1, D; Liebermeister et al., 2014). Under control conditions, the major part of the leaf protein mass (66%) is dedicated to photosynthesis, followed by protein metabolism (7.5%) and amino acid metabolism (6%; the relative values [%] for all pathways shown are listed in Supplemental Dataset S3).
Estimating protein copy numbers in a plant cell
We used two different approaches to estimate how many protein molecules are actually present in a plant cell, based on cell number and cell size, respectively (Supplemental Figure S1). We selected mesophyll cells as the representative leaf cell for this estimation, since they are photosynthetically active and constitute the major part of the leaf volume. Total protein copy numbers have already been reported for yeast cells and different animal cell lines. A haploid cell of budding yeast (Saccharomyces cerevisiae) has a volume of 42 µm3 (Jorgensen et al., 2002) and contains about 42 million proteins (Ho et al., 2018), whereas for human cells with a volume of about 4,200 µm3, three billion protein molecules have been calculated (Kulak et al., 2014). Thus, yeast and human cells contain 1.0 and 0.7 million proteins per µm3, respectively. An average mature leaf cell has a volume of approximately 150,000 µm3 (Supplemental Figure S1). Assuming an average protein abundance of 0.85 × 106 molecules per µm3 and subtracting the volume of the central vacuole that typically covers about 80% of a plant cell we postulate that a leaf mesophyll cell contains close to 25 billion proteins (Table 1 and Supplemental Figure S1).
Table 1.
Total number of protein molecules in an average Arabidopsis leaf mesophyll cell and its subcellular compartments under control conditions (C) and during progressive drought stress (S3, moderate stress; S5, severe stress; and S6, maximum tolerable stress)
| Protein numbers (× 106) |
||||
|---|---|---|---|---|
| Subcellular compartment | C | S3 | S5 | S6 |
| Number of proteins in a mesophyll cell | 25,766 | 24,309 | 15,262 | 15,337 |
| Number of proteins in all chloroplasts (∼100) in a cell | 20,208 | 18,984 | 11,603 | 11,468 |
| Number of RubisCO LS (AtCg00490) per cell | 3,191 | 3,393 | 2,024 | 1,724 |
| Number of proteins in an individual chloroplast | 202 | 190 | 116 | 115 |
| Number of RubisCO LS (AtCg00490) per chloroplast | 32 | 34 | 20 | 17 |
| Number of proteins in all mitochondria (∼400) in a cell | 495 | 480 | 385 | 407 |
| Number of serine hydroxymethyltransferase 1 (At4g37930) per cell | 54 | 42 | 26 | 24 |
| Number of proteins in an individual mitochondrion | 1.2 | 1.2 | 1.0 | 1.0 |
| Number of serine hydroxymethyltransferase 1 (At4g37930) per mito. | 0.14 | 0.11 | 0.06 | 0.06 |
| Number of proteins in the cytosol per cell | 2,782 | 2,575 | 1,602 | 1,696 |
| Number of GTP binding EF Tu (At5g60390) per cell | 116 | 108 | 62 | 69 |
| Number of proteins in the vacuole | 526 | 443 | 326 | 310 |
| Number of tonoplast intrinsic protein 2 (At3g26520) per cell | 161 | 122 | 92 | 75 |
| Number of proteins in the extracellular space per cell | 285 | 313 | 287 | 338 |
| Number of germin-like protein 1 (At1g72610) | 89 | 50 | 38 | 37 |
| Number of proteins per nucleus | 160 | 206 | 93 | 131 |
| Number of ubiquitin 5 (At3g62250) per nucleus | 31 | 41 | 18 | 18 |
| Number of proteins in all peroxisomes of a cell | 614 | 650 | 498 | 533 |
| Number of Aldolase-type TIM barrel protein (At3g14415) per cell | 68 | 57 | 27 | 26 |
| Number of proteins in the endoplasmic reticulum per cell | 74 | 70 | 58 | 57 |
| Number of ADP-ribosylation factor 1 (At1g70490) per cell | 17 | 12 | 8 | 6 |
| Number of proteins in the Golgi apparatus per cell | 34 | 35 | 23 | 24 |
| Number of RGP2; UDP-arabinose mutase (At5g15650) per cell | 4 | 5 | 5 | 5 |
| Number of proteins in the plasma membrane per cell | 183 | 179 | 130 | 104 |
| Number of plasma membrane intrinsic protein 2A (At3g53420) | 32 | 29 | 17 | 12 |
For each compartment, the copy number of the most abundant protein is listed individually. All numbers are based on estimations as discussed in the text (see also Supplemental Figure S1). The total number of proteins in a cell/subcellular compartment is highlighted in bold.
An alternative, completely independent way to calculate protein copy numbers is based on an average number of 300,000 mesophyll cells (Wuyts et al., 2010) in a mature rosette leaf of 5 mg DW with a protein content of 102 mg · g−1 DW. A total of 1.7-ng protein per cell would add up to 20.5 billion protein molecules with an average molecular weight of 50 kDa. Quantitative proteomics, irrespective of its intrinsic limitations, makes it possible to deduce a more precise estimate of 25.8 billion proteins per cell. In addition, this approach provides information about the copy numbers of individual proteins (Figure 1, E, Table 1, and Supplemental Dataset S1, B). It has to be kept in mind that these results are estimations based on the average characteristics of an Arabidopsis mesophyll cell combined with the proteome composition of total rosette leaf material, and thus cannot be regarded as exact, statistically firm numbers. Clearly, the largest share of the protein molecules in a mesophyll cell (∼20 billion) are located in the chloroplasts, 3.2 billion in the cytosol, and 0.5 billion in the mitochondria. Copy numbers range from 3.2 billion molecules of RubisCO large subunit to 2,039 acetyl-CoA carboxylase 1 molecules, which is the detection limit of our MS approach. Thus, an average Arabidopsis leaf mesophyll cell contains about 340 million RubisCO hexadecamers under optimal growth conditions.
Severe drought stress leads to a massive decrease in leaf protein content
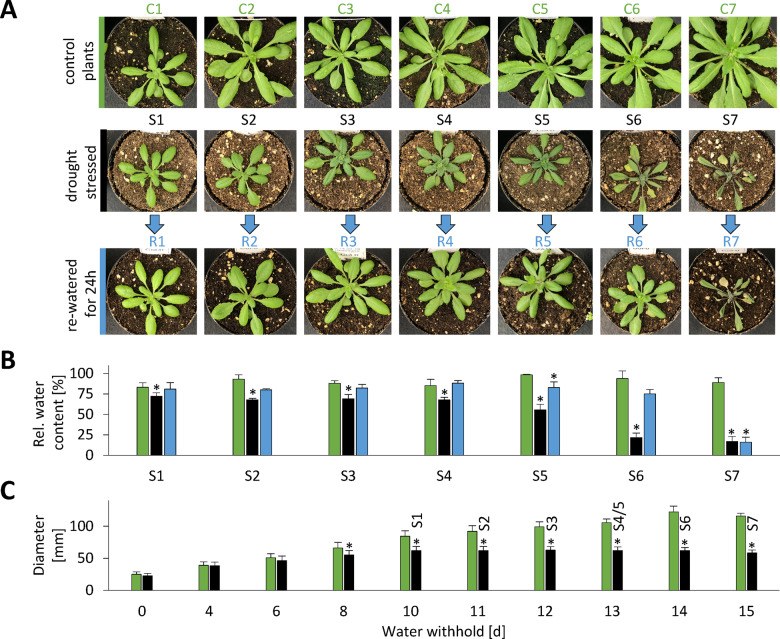
We carefully established an experimental setup that mimicked physiological drought stress conditions as closely as possible and at the same time led to a highly reproducible stress phenotype (Figure 2, a detailed description of the drought treatment is given in the “Materials and methods” section). In brief, plants were grown under long-day control conditions for 2 weeks and watered to the same level. The dehydration process was then monitored on a daily basis and leaf samples were taken at different time points during the desiccation process from beginning to moderate and severe drought stress until recovery was no longer possible. Rosette growth gradually declined and stopped after 10 d without water (Figure 2, C). We defined this time point as stress level S1 and numbered the following days of progressive drought stress consecutively. First indications of a loss in leaf turgor became visible in some of the plants after 12 d without water (S3) and complete wilting until death occurred within the following 72 h. These late stages of severe drought stress (S4–S7) were classified according to their leaf phenotype: number of rolled leaves, relative water content (RWC), and potential to recover after re-watering. The leaf protein content remained stable (109 ± 13 mg · g−1 DW) during the first 12 d without watering (S1–S3), but then rapidly decreased by 39% within 24 h (S5).
Figure 2.
Complete setup of the progressive drought stress experiment. Arabidopsis plants were grown in soil under long-day conditions for 2 weeks. All pots were then brought to the same weight and the stress group was not watered for up to 15 d while the control group was kept at a constant water level. Leaf samples were first collected after 10 d without water (stress level S1) and continued each following day until recovery of the plants was no longer possible (stress level S7). (A) Phenotype of representative plants (pot diameter = 8 cm). (B) RWC (%) in rosette leaves of control plants (green bars), stressed plants (black bars), and stressed plants 24 h after re-watering (blue bars) at the different stress levels. (C) Rosette diameter [mm] of control plants (green bars) and stressed plants (black bars) at 0–15 d after the beginning of the stress treatment. The corresponding stress levels of the plants are indicated on top of the black bars. A detailed description of the drought treatment is given in the “Materials and methods” section. Values are means ± SD of the biological replicates. S1–S7, n = 7; C1–C7, n = 3; R1–R7, n = 3. Significant differences were determined by Student’s t test P < 0.01. Starting material (stress levels) for experimental analyses: S1, 10 d after end of watering; S2, 11 d after end of watering; S3, 12 d after end of watering, first signs of stress (rolled/wrinkled leafs); S4, 13 d after end of watering, 4–7 rolled leaves; S5, ∼13 d after end of watering, 8–10 rolled leaves; S6, ∼14 d after end of watering, >10 rolled leaves; recovery of plants still possible; S7, ∼15 d after end of watering, >10 rolled leaves; recovery of plants not possible; C1–C7, control plants (watering continued); R1–R7: same as S1–S7, but re-watered for 24 h.
Patterns of stress-induced proteome changes in subcellular compartments
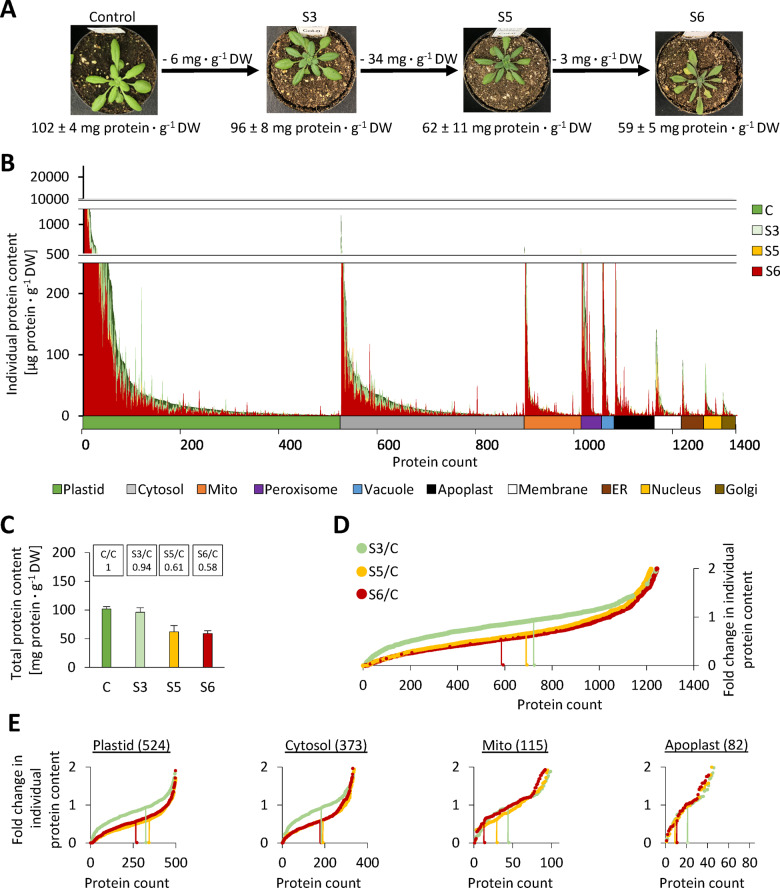
Four stress levels were selected for leaf proteome analysis by shotgun MS (Figure 3, A, Supplemental Dataset S1, and Supplemental Figure S3): control (RWC = 88 ± 5%), S3 (moderate stress, no wilting, RWC = 69 ± 5%), S5 (severe stress, RWC = 55 ± 7%), and S6 (maximum tolerable stress, RWC = 22 ± 5%). Statistical evaluation of the proteomics dataset based on label-free quantification (LFQ)-values indicates significant changes in the relative abundance of 291, 523, and 517 protein species in relation to the control at stress levels S3, S5, and S6, respectively (Supplemental Dataset S1, columns X-AC). These results are provided in the Supplemental Material and can be used for data mining. However, since the major focus of this study is a quantitative perspective, we will not evaluate significant changes in individual protein levels in detail. To provide a first impression of quantitative changes in the leaf proteome during progressive drought stress, we sorted all detected proteins according to their absolute content under control conditions for each compartment individually. The contents of each individual protein during progressive drought stress were then plotted in superimposing graphs (Figure 3, B). The fraction of proteins degraded in the course of the stress treatment becomes visible as green or orange area. Interestingly, there are clear differences between the compartments. A large fraction of proteins localized in chloroplasts, the cytosol, the plasma membrane, or the Golgi apparatus shows roughly homogenous decrease rates. In contrast, hardly any green areas are visible for mitochondrial and extracellular proteins, indicating a lower degradation rate (Supplemental Figure S4). In order to quantify this observation, we calculated fold change ratios of individual protein contents in stressed versus control plants using the iBAQ-based protein contents shown in Figure 3, B, and sorted them in ascending order for each stress level individually (Figure 3, D and E and Supplemental Figure S5). The total leaf protein content decreased to 94% of control values at stress level S3, to 61% at S5, and to 58% at S6 (Figure 3, C). Therefore, individual proteins with an average decrease rate during stress are localized at approximately 0.94, 0.61, and 0.58 (marked by vertical green, yellow, and red lines in Figure 3, D and E). In the complete dataset (Figure 3, D), and in the subsets of plastid and cytosolic proteins (Figure 3, E), there is a large area with almost horizontal graphs representing proteins with roughly average degradation rates. In contrast, the slopes of the mitochondrial and extracellular graphs are much steeper and only 11–12% of the proteins show average or increased degradation rates in severely stressed plants (Figure 3, E, vertical red lines in the graphs “Mito” and “Apoplast”).
Figure 3.
Compartment-specific patterns of stress-induced changes in individual protein abundances. (A) Phenotype and protein content of the plants used for proteome analysis. The representative images are the same as in Figure 2. Complete rosettes of four plants were harvested at the beginning of the stress treatment (parallel to S1–S3) (control), and at three defined stress stages (S3, S5, and S6), respectively. (B) Absolute contents [μg protein · g−1 DW] of all individual proteins detected by shotgun proteomics in descending order (under control conditions), sorted by subcellular compartments according to SUBA4 prediction (Hooper et al., 2017). Protein contents under control and stress conditions are shown in superimposed graphs. Individual plots for each subcellular compartment are shown in Supplemental Figure S4. (C) Total leaf protein contents [mg protein ⋅ g−1 DW] at the different stress levels. Values are means ± sd (n = 4). The relative protein contents compared with the control group are indicated at the top. (D) and (E) Fold change ratios of individual protein contents in stressed versus control plants for all proteins (D) and individual subcellular compartments (E). In order to visualize the fraction of proteins with average, high, or low degradation rates, changes in individual protein contents were sorted in ascending order for each stress level. Vertical lines indicate proteins that correspond exactly to the decrease in total protein content, i.e. 0.94 for stress level S3 (light green), 0.61 for S5 (orange), and 0.58 at S6 (red). Changes in individual protein contents were calculated using the absolute protein contents shown in B (Supplemental Dataset S1), and are thus based on iBAQ data. Additional individual plots for subcellular compartments are provided in Supplemental Figure S5.
Regulation of protein abundance via synthesis and degradation
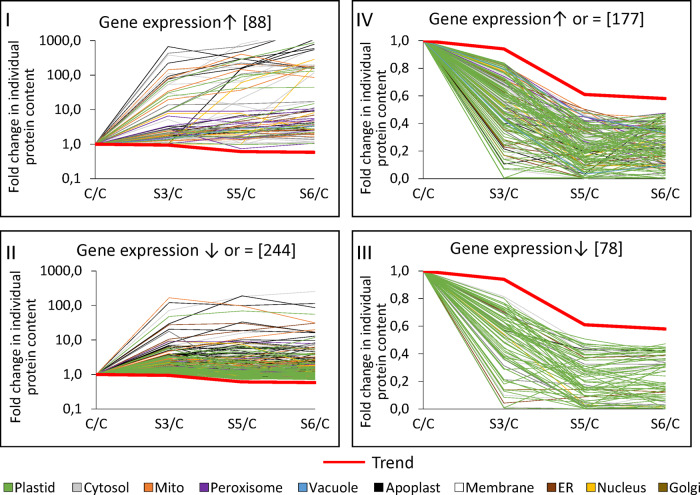
Protein abundance can be regulated at the level of synthesis and/or degradation. We used genevestigator (Hruz et al., 2008) to estimate gene expression levels during drought stress and combined this information with the relative protein abundances detected by our proteomics approach (Supplemental Dataset S4). We filtered the proteomics dataset for proteins of consistently increased abundance and divided the resulting list of 332 proteins in two subgroups: Group I contained the proteins with significantly increased expression levels (88 proteins) and group II contained proteins with decreased or unaffected expression levels (244 proteins), indicating that regulation might rather be achieved at a posttranscriptional level, e.g. via decreased proteolysis (Figure 4). In order to determine the metabolic pathways preferentially regulated by these strategies, ratios between the observed frequencies in each group and the observed frequency in the whole MS dataset were calculated (Table 2). The proteins up-regulated via gene expression (group I) were mainly involved in protein, lipid, or amino acid degradation; stress response; and secondary metabolism. Energy metabolism (glycolysis and respiratory chain) and extracellular proteins required for cell wall metabolism and proteolysis were prevalent in group II, and thus might be regulated by decreased degradation rates. The proteins of consistently decreased relative abundance (255 proteins) were also subdivided into those with decreased expression rates (group III, 78 proteins) and those with increased or unaffected expression rates (group IV, 177 proteins; Figure 4, right part). Group III (down-regulation on expression level) contains specific vacuolar proteins and enzymes catalyzing lipid or tetrapyrrole synthesis (Table 2). No particular enrichment in subcellular compartments or functional categories was detected for proteins potentially down-regulated by increased proteolysis (group IV).
Figure 4.
Transcriptional and post-translational regulation of protein abundances during progressive drought stress. Fold change ratios (stress/control) of individual protein contents during progressive drought stress. Red graphs (trend lines) indicate the fold change in total leaf protein content at each stress level (S3/C: 0.94; S5/C: 0.61; S6/C: 0.58). Three microarray datasets available via Genevestigator were used to estimate gene expression levels during drought stress (see the “Materials and methods” section). The proteomics dataset was filtered for proteins that were of increased relative abundance according to both iBAQ-based and LFQ-based data interpretation at each stress level (more details on the filter criteria are provided in Supplemental Dataset S4). These proteins of increased abundance (left part of the figure) were divided into two groups: proteins with increased gene expression levels according to the microarray datasets (group I) and proteins with unaffected or decreased gene expression levels during drought stress (group II). Proteins of consistently decreased abundance (right part of the figure) were also filtered for decreased (group III) and increased or unaffected expression levels (group IV). Colors indicate the subcellular localization of the individual proteins according to SUBA4 prediction (Hooper et al., 2017). Enrichment of compartments and functional categories in the different regulation groups is listed in Table 2.
Table 2.
Estimating the enrichment of specific compartments (left) or metabolic pathways (right) in groups of proteins regulated on a transcriptional (I, III) or post-translational (II, IV) level
| Compartment | I | II | III | IV | Pathway | I | II | III | IV |
|---|---|---|---|---|---|---|---|---|---|
| Cytosol | 1.0 | 0.8 | 0.5 | 1.1 | AA degradation | 2.6 | 0.6 | 0.6 | 1.0 |
| ER | 1.5 | 1.9 | 0.8 | 0.7 | Cell wall | 0.7 | 2.6 | 0.8 | 1.3 |
| Extracellular | 2.3 | 3.1 | 0.2 | 0.4 | Glycolysis | 1.0 | 2.2 | 0.0 | 0.0 |
| Golgi | 0.0 | 0.8 | 0.6 | 1.6 | Lipid degradation | 6.4 | 1.9 | 0.0 | 0.0 |
| Mitochondria | 1.3 | 1.8 | 0.0 | 0.3 | Lipid synthesis | 0.6 | 1.2 | 2.2 | 1.0 |
| Nucleus | 2.3 | 0.8 | 0.5 | 0.7 | mETC | 0.0 | 2.2 | 0.0 | 0.3 |
| Peroxisome | 3.2 | 0.8 | 0.0 | 1.1 | Protein degradation | 2.0 | 1.5 | 0.0 | 0.8 |
| Plasma membrane | 2.5 | 1.0 | 1.6 | 1.6 | Protein handling | 0.5 | 2.3 | 0.0 | 1.2 |
| Chloroplast | 0.3 | 0.6 | 1.7 | 1.1 | Secondary metabolism | 2.3 | 1.1 | 1.3 | 0.6 |
| Vacuole | 0.6 | 1.1 | 2.7 | 0.6 | Stress | 3.0 | 1.5 | 0.3 | 0.4 |
| Tetrapyrrole synthesis | 0.0 | 0.0 | 9.0 | 1.2 |
Numbers indicate the quotient of the fraction of proteins localized in specific compartments (left) or attributed to metabolic pathways (right) in the regulation groups (I–IV) divided by the fraction of the respective proteins in the total proteomics dataset. Group I: increased protein abundance, increased expression; Group II: increased protein abundance, unaffected or decreased expression; Group III: decreased protein abundance, decreased expression; Group IV: decreased protein abundance, unaffected or increased expression. Only metabolic pathways with quotients ≥2 (bold) in at least one regulation group are shown. The complete dataset used for enrichment analysis is provided in Supplemental Dataset S4.
Adaptations of the protein synthesis and degradation machineries during progressive drought stress
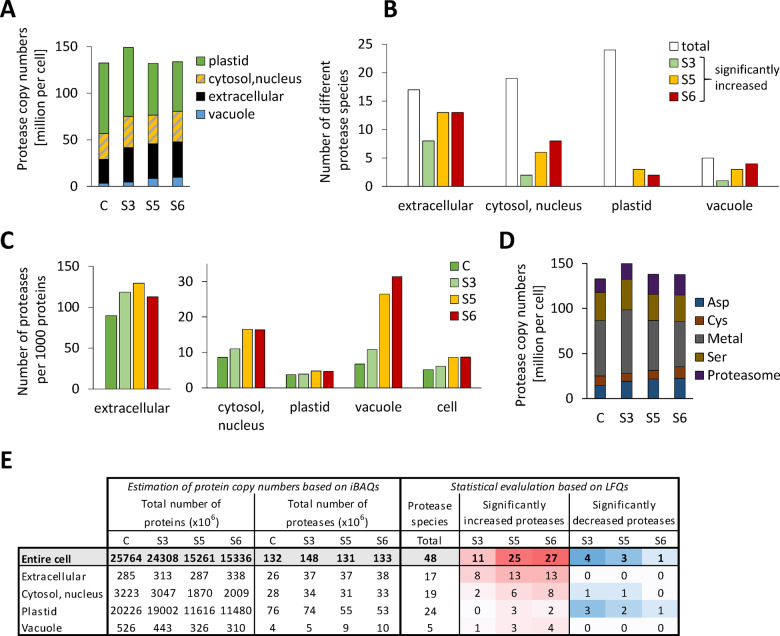
Under control conditions ∼5.4% of the leaf proteome detected by our MS approach is dedicated to protein synthesis (ribosomal proteins, translation initiation, and elongation factors) compared with 1.4% involved in proteolysis (proteasomes, autophagy proteins, proteases, and regulatory proteins; Figure 5, A and C). During progressive drought stress a majority of the proteins involved in protein synthesis (∼75%) decreased more than average (Figure 5, B, proteins on the left side of the vertical lines that mark the average fold changes in leaf protein content). In particular, the large group of ribosomal proteins (125 proteins, contributing 3.3 mg protein · g−1 DW under control conditions) had strikingly homogenous degradation rates (mass ratio S6/C = 0.5 ± 0.2; Figure 5, C). In contrast, the total leaf content of proteolytic enzymes remained stable (0.8–0.9 mg protein · g−1 DW; Figure 5, C) but changed drastically in its composition, reflected by variations in the total mass content of the different classes of proteolytic enzymes and also by the high number of significant changes in the abundance of individual protease species (Figure 5, C; the proteomics dataset for all individual proteins involved in protein synthesis and degradation is provided in Supplemental Dataset S5). Protease copy numbers in the cytosol, the vacuole, and in the apoplast increased progressively (Figure 6, A and E), and after severe stress, most of the vacuolar and extracellular proteases were of significantly increased abundance compared with control conditions indicating their specific relevance for drought response (Figure 6, B and E and Supplemental Dataset S5). In order to estimate the mean workload of the proteolytic system in the individual subcellular compartments, we calculated the number of proteases per 1,000 protein molecules (Figure 6, C) on the basis of the estimated total numbers of protein molecules and protease molecules in each subcellular compartment (Figure 6, E). The relative abundance of proteases per potential substrate was at least 10-fold higher in the apoplast than in any other compartment, even under control conditions, and further increased during moderate stress (S3; Figure 6, C). Vacuolar proteases strongly accumulated during severe stress, and also in the cytosol plus nucleus, the relative capacity of proteases approximately doubled (from 8.6 to 16.4 proteases per 1,000 protein molecules), although only a specific subset of proteolytic enzymes (mainly subunits of the proteasome) was significantly increased (Figure 6, B and C and Supplemental Dataset S5). Due to their high abundance, chloroplasts contained the major fraction of cellular proteases (58%) in the leaves of non-stressed plants (Figure 6, A and E). However, proteases constituted less than 0.5% of all plastid proteins (compared with 9–13% in the apoplast, Figure 6, E) and decreased during stress to a similar extent as the majority of chloroplast proteins, resulting in a low number of significant changes in the relative abundance of plastid proteases during severe drought stress (Figure 6, B and E). The largest increase in protease copy numbers during stress (from 14.9 to 22.8 million molecules per cell) can be attributed to the aspartate class of proteases, which includes mainly extracellular subtilases (Figure 6, D andSupplemental Dataset S5).
Figure 5.
Abundance of the proteostasis apparatus during drought stress. (A) Proteomap illustrating the quantitative composition of the proteostasis apparatus under control conditions. Proteins are shown as polygons whose sizes represent the mass fractions (protein abundances obtained by MS [iBAQ], multiplied by protein molecular weight). Proteins involved in similar cellular functions according to the MapMan annotation file (version Ath_AGI_LOCUS_TAIR10_Aug2012) are arranged in adjacent locations and visualized by colors. The total protein fraction represented in the Proteomap is 6.9 mg · g−1 DW, corresponding to 6.7% of the leaf proteome. (B) Fold change ratios of the individual contents of proteins involved in protein synthesis (top) or proteolysis (bottom) in stressed versus control plants. In order to visualize the fraction of proteins with average, high, or low degradation rates, changes in individual protein contents were sorted in ascending order for each stress level. Vertical lines indicate proteins that correspond exactly to the decrease in total protein content, i.e. 0.94 for stress level S3 (light green), 0.61 for S5 (orange), and 0.58 at S6 (red). (C) Accumulated copy numbers and mass contents of proteins assigned to the functional sub-categories of protein metabolism. Significant changes in the relative abundance of individual protein species during stress were identified based on LFQ values (Student’s t test, P < 0.05). Color intensities correspond to the number of significant changes (red shading: increase, blue shading: decrease). PTM, post-translational modification; ASP-P, aspartate protease; and CYS-P, cysteine protease.
Figure 6.
Adaptation of the proteolytic apparatus during progressive drought stress. (A) Estimated total number and subcellular distribution of protease molecules in an average leaf mesophyll cell under control conditions and during stress. The proteomics dataset (Supplemental Dataset S1) was filtered for the MapMan category “protein.degradation” (Thimm et al., 2004, the filtered list is provided in Supplemental Dataset S5). Protein copy numbers of all enzymes with proteolytic activity (without regulatory proteins and inhibitors) were added up for each subcellular compartment individually (see also E). (B) Significant increase in the abundance of individual protease species during drought stress in the different subcellular compartments. White bars indicate the total number of different proteases detected and colored bars illustrate how many of them were significantly increased based on LFQ values at the respective stress level (Student’s t test, P < 0.05, see also E). (C) Copy numbers of protease molecules per 1,000 proteins in the subcellular compartments of an average mesophyll cell under control conditions and during stress. The protease copy numbers (A) were divided by the total number of protein molecules in the respective subcellular compartment (see E) and multiplied by 1,000. (D) Number of protease molecules sorted by their functional classes. (E) Accumulated copy numbers of all detected proteins and of proteases in the different subcellular compartments. Significant changes in the relative abundance of individual protease species during stress were identified based on LFQ values (Student’s t test, P < 0.05). Color intensities correspond to the number of significant changes (red shading: increase, blue shading: decrease).
Dynamics in free and protein-bound amino acid pools
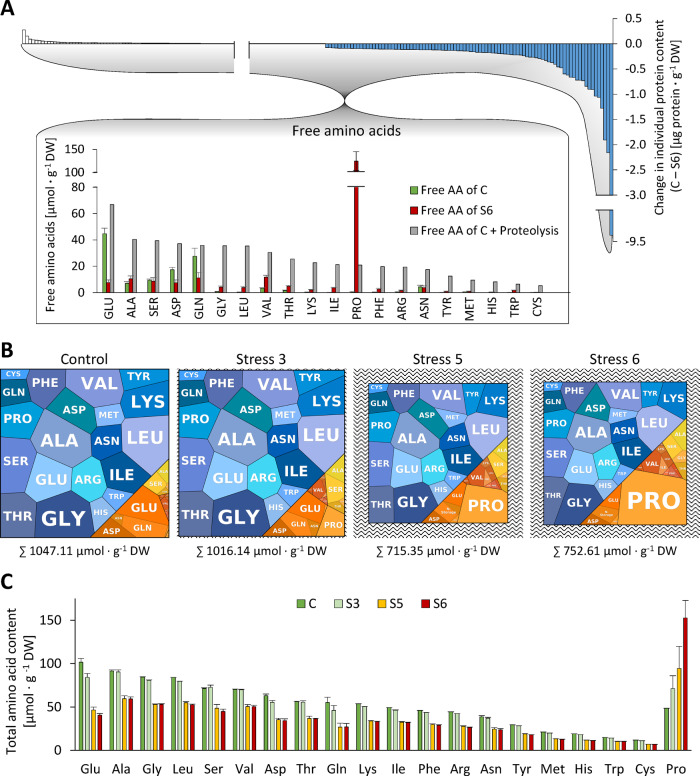
Massive proteolysis during severe drought stress inevitably leads to liberation of large amounts of amino acids. We thus changed perspective and focused on the fate of the degraded part of the proteome and its effect on free amino acid homeostasis. For each individual protein, we calculated the difference in absolute content in control versus stressed plants (Figure 7, A, top and Supplemental Dataset S1). It immediately becomes obvious that the amino acids added to the free pool are quantitatively derived from a limited number of very high abundance proteins (blue bars). Degradation of about 170 million RubisCO hexadecamers per cell alone accounts for 23% of the total amino acid release during stress. The profiles of free amino acids in the leaves of control and stressed plants were quantified by HPLC (Supplemental Dataset S6). In addition, we estimated the total amount of each individual amino acid bound in proteins on the basis of the leaf protein content and the quantitative composition of the proteome (Supplemental Dataset S7). The pool sizes and compositions of the free and protein-bound amino acid pools can be visualized using a modified version of PROTEOmaps (Figure 7, B, orange: free pool, blue: protein-bound pool). Under control conditions, the Arabidopsis leaves contained 1.05 mmol · g−1 DW amino acids of which 0.93 mmol · g−1 DW were bound in proteins (Supplemental Dataset S7). Drought stress led to a decrease of the total amino content by 28% (to 0.75 mmol · g−1 DW at stress level S6; Supplemental Dataset S7). Also, the ratio between free and protein-bound amino acids (orange/blue area in Figure 7, B; data provided in Supplemental Dataset S7) shifted from 0.13 to 0.39 due to massive proteolysis. The amino acid composition of the proteome did not change considerably during stress. The molar share of the 20 proteinogenic amino acid was in the range of 1.3% (Cys) to 9.0% (Ala; Supplemental Figure S6, A). In contrast, the free amino acid pool strongly reacted to drought stress (orange areas in Figure 7, B, Supplemental Figure S6, B, and Supplemental Dataset S7), and also the concentrations of high and low abundance amino acids differed up to 460-fold (0.27 ± 0.11 µmol ⋅ g−1 DW Cys versus 125 ± 20 µmol ⋅ g−1 DW Pro at stress level S6; Supplemental Figure S6, B and Supplemental Dataset S6). Under control conditions, the free amino acid pool was dominated by Glu, Gln, and Asp (Figure 7, B and Supplemental Dataset S6). Water deficiency led to progressive accumulation of Pro (Figure 7, B), which in the leaves of severely stressed plants represented 59% of the free and 20% of the total amino acid pool (Supplemental Dataset S7).
Figure 7.
Interconnection of amino acid pools during progressive drought stress. (A) Effect of proteolysis on free amino acid homeostasis. The quantitative composition of the degraded fraction of the proteome (blue bars) was used to calculate the theoretical composition of the free amino acid pool (gray bars) that would result from massive proteolysis during drought stress (control versus maximum tolerable stress) without any metabolic conversion of the amino acids produced. The actual free amino acid profiles in the leaves of control plants (green bars) and of severely stressed plants (red bars) were analyzed by HPLC. (B) “AMINOmaps” illustrating pool sizes and compositions of the free (orange colors) and protein-bound (blue colors) amino acid pools during progressive drought stress. Amino acids are shown as polygons whose sizes represent the molar fractions. Free amino acid contents were quantified by HPLC, and quantitative amino acid composition of the proteome was calculated on the basis of molar composition of the proteome (see Supplemental Dataset S1) as detailed in the “Materials and methods” section. (C) Total amino acid contents (protein bound plus free) during progressive drought stress. The amino acid contents in proteins were calculated based on iBAQ values. Free amino acid profiles were analyzed by HPLC. Error bars indicate the variability (sd, n = 7) in the free amino acid pool (see Supplemental Datasets S6, S7). AA, amino acid.
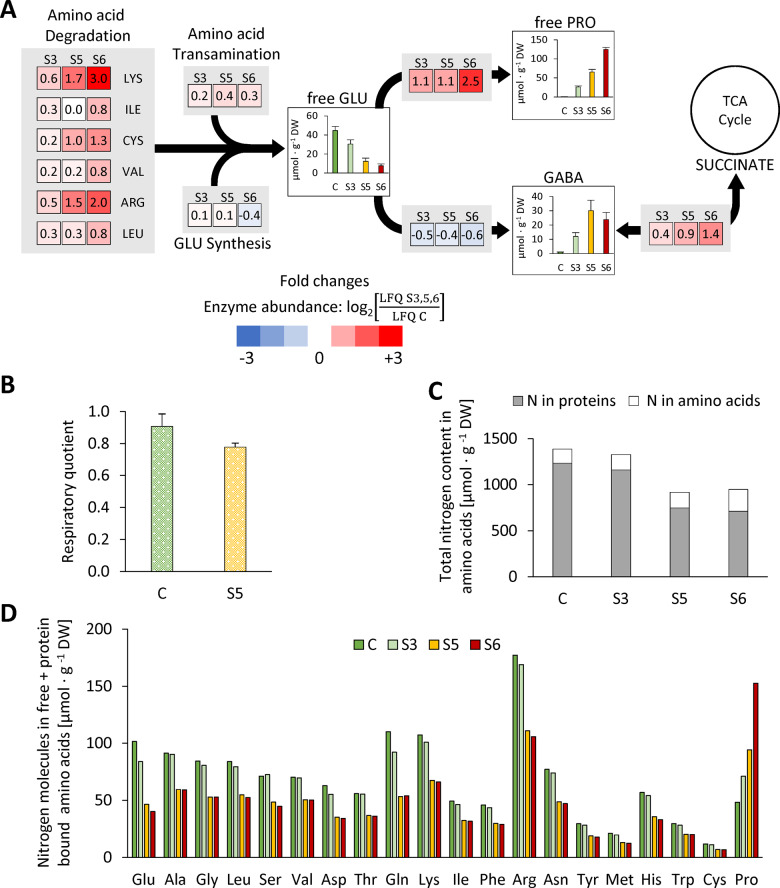
In order to estimate the role of proteolysis in amino acid homeostasis, we calculated the theoretical composition of the free amino acid pool that would result from partial degradation of the proteome (as detected by our proteomics approach) without any metabolic conversion of the amino acids produced (Figure 7, A, gray bars). With the clear exception of Pro, the free amino acid contents actually detected in severely stressed leaves (Figure 7, A, red bars) were several fold lower than the calculated ones, indicating their degradation or conversion to other metabolites. Enzymes involved in the degradation of branched-chain amino acids, Cys, Lys, and Arg were indeed increased by drought stress, as were Pro and GABA metabolism (Figure 8, A and Supplemental Dataset S8). Statistical evaluation based on LFQ values showed that 8 of the 31 amino acid catabolic enzymes included in our dataset increased significantly in their relative abundance during stress and none significantly decreased (Supplemental Dataset S8, column W). In contrast, enzymes involved in amino acid synthesis and primary nitrogen assimilation (e.g. nitrate reductase, nitrite reductase, and Gln synthase) were significantly decreased, indicating that de novo synthesis of amino acids from inorganic nitrogen is negligible. The respiratory quotient (RQ; RC = carbon dioxide production/oxygen consumption) can be used as an indicator of which substrates are mainly being metabolized, since complete oxidation of carbohydrates consumes less oxygen per carbon dioxide released (RC = 1.0) than the oxidation of proteins (RC = 0.8) or lipids (RC = 0.7). We measured a RQ of 0.91 ± 0.08 in the leaves of control plants, which decreased to 0.78 ± 0.03 in severely stressed plants (stress level S5; Figure 8, B). This result thus supports a substrate shift from mainly carbohydrate-based respiration to a larger contribution of proteins and lipids during drought stress. In order to develop an idea about how long plants would be able to keep up their regular mitochondrial respiration rate when using exclusively the amino acids released by protein degradation as substrates, we calculated the total number of electrons that would be transferred to oxygen via the mitochondrial respiratory chain during complete oxidation of the specific set of amino acids released during drought stress (Supplemental Dataset S7; Hildebrandt et al., 2015). This oxidation process would lead to a total oxygen consumption of 1,062 µmol O2 · g−1 DW and thus, on the basis of a mean leaf respiration rate of 3.4 nmol O2 · g−1 fresh weight (FW) · s−1 (O’Leary et al., 2017), could fully sustain leaf energy metabolism for about 7 h.
Figure 8.
Drought stress-induced amino acid degradation delivers nitrogen and glutamate for the production of proline and GABA as osmolytes as well as alternative substrates for mitochondrial respiration. (A) Heatmap squares show LFQ-based log2‐fold changes in abundances of enzymes involved in amino acid metabolism at the selected stress levels. Red colors: increase with respect to the control fraction. Blue colors: decreased with respect to the control fraction. Abundance of currently known enzymes involved in amino acid synthesis and degradation pathways were extracted from the proteomics dataset (Supplemental Dataset S1, column “Amino acid pathway”). The colored squares represent the means of all changes in the abundance of enzymes involved in the respective branch of the pathway (see Batista-Silva et al. [2019] for a complete map of plant amino acid metabolism). Bar charts show free amino acid contents (means ± sd, n = 7) in the leaves [µmol · g−1 DW]. (B) RQ (CO2 production/O2 consumption) of rosette leaves under control conditions (green bar) and at stress level S5 (yellow bar). Values are means ± sd, n = 3. (C) Total leaf nitrogen content [μmol ⋅ g−1 DW] included in the free and protein-bound amino acid pools of control and stressed plants. (D) Estimation of the nitrogen content of individual (bound and free) amino acids [μmol ⋅ g−1 DW] (Supplemental Dataset S7). GABA, γ‐aminobutyric acid; GLU, l-glutamate; and PRO: l-proline.
Discussion
The number of protein molecules in a plant cell
Common sense indicates that cells require an adequate set of proteins to function properly. However, the literature did not allow us to deduce a comprehensive picture of what this protein infrastructure of a plant cell might look like. Thus, we calculated the average protein copy number in a plant cell based on published information about the size and number of cells in an average Arabidopsis leaf using two independent approaches. Both calculations consistently revealed that an average mesophyll cell in a mature Arabidopsis leaf contains ∼25 billion protein molecules.
A major function of leaf mesophyll cells is photosynthesis, and this is reflected by the large fraction of proteins (20 billion) localized in the ∼100 chloroplasts present in each cell, corresponding to ∼200 million proteins per chloroplast, and again, the largest fraction of these proteins are included in ∼3.4 million RubisCO hexadecamers (Königer et al., 2008). The high abundance of RubisCO in leaves is well established. Recent estimations suggest that its total mass on earth is ∼0.7 Gt, and RubisCO accounts for ∼3% of leaf DW (Bar-On and Milo, 2019). Adding up the mass content of all RubisCO subunits in our MS dataset (Supplemental Dataset S1, B) results in a very similar estimate of 21.4 mg · g DW−1. The dominant position of RubisCO within the leaf proteome is also illustrated by gel-based proteomic approaches (Supplemental Figure S2, B).
Interestingly, the protein copy number we calculated for the cytosol of a plant cell (∼3.2 billion) matches almost exactly the total number of proteins reported for animal cells (Kulak et al., 2014; Supplemental Figure S1). According to our estimation, a mesophyll cell contains about 495 million mitochondrial proteins (Table 1). Assuming that between 300 and 450 mitochondria are present in a plant cell, depending on the leaf age (Preuten et al., 2010), a single mitochondrion would harbor 1.1–1.7 million protein molecules, which is in perfect agreement with previous results (Fuchs et al., 2020).
Strengths and limitations of the proteomics approach and its different evaluation strategies
For statistical analysis to identify significant differences between the stress levels we used LFQ, an algorithm optimized for accurate horizontal comparisons between different samples including multiple levels of normalization (Cox et al., 2014). This approach helps to identify, e.g. a set of extracellular proteases that might be particularly relevant during drought stress response or to estimate the regulation of amino acid catabolic pathways (Figure 8). However, LFQ-based data interpretation is not suitable for vertical comparisons of the abundance of different protein species and therefore was not the major focus during our evaluation. Detailed information on significant changes in protein abundances at the different stress levels compared with the control are provided in the Supplementary Material (Supplemental Dataset S1, columns U-AC, S5, and S8). A quantitative perspective on the leaf proteome based on iBAQs enables calculation of mass fractions, molarities, and even copy numbers of individual proteins, but lacks statistical validation. Both quantitation methods are limited by the intrinsic shortcomings of shotgun proteomics, which cannot detect very low abundance proteins, and tends to underestimate membrane proteins since the biochemical properties of their peptides, such as high hydrophobicity, are unfavorable for ionization and detection (Schwanhäusser et al., 2011; Krey et al., 2014; Fabre et al., 2014). Furthermore, the extraction procedure may also introduce a bias against, e.g. membrane and membrane-associated proteins. Every proteomics dataset therefore has to be regarded as a representative fraction of the complete picture. We can estimate on the basis of labeled peptides that the fraction detectable by our approach covers more than 80% of the protein mass in a leaf. Thus, it is suitable to provide a realistic impression of the total number of protein molecules in a cell, as well as the general behavior of high to medium abundance proteins during drought stress. Recent large-scale MS surveys with a focus on covering the largest possible fraction of protein groups present in different Arabidopsis tissues were able to detect 8,700 or even 13,826 different proteins by fractionation of individual leaf samples (Zhang et al., 2019; Mergner et al., 2020). For studies focusing on individual pathways or low abundance proteins, it is thus reasonable to increase the sensitivity of the MS analysis by including sample fractionation or targeted approaches.
Proteostasis under challenging conditions—a focus on subcellular compartments and metabolic pathways
Plants use various physiological adaptation mechanisms to cope with insufficient water supply (Gupta et al., 2020). They restrict the number and size of leaves and close their stomata to reduce water consumption. In addition, they increase the root-to-shoot ratio to improve water uptake from the soil. Osmotic adjustment preserves the cell turgor, and antioxidants are produced to attenuate oxidative damage. Even at the lowest stress level analyzed in this study (S3), plant rosettes had already stopped growing, accumulated the osmoprotectant proline, and significantly changed the abundance of at least 291 proteins. The drastic effects on the total leaf protein content and amino acid profile analyzed here are characteristic for the late phase of severe dehydration (stress levels S5 and S6).
How to focus on the relevant pathways during severe drought stress
Combined information about protein abundance and expression levels illustrates the general strategies employed by leaf cells to adjust their protein setup to the challenges posed by insufficient water supply. Specific stress-related proteins, such as individual heat shock proteins and dehydrins and those involved in secondary metabolism, are induced at the expression level. Similarly, cells increase the abundance of pathways that are barely used under control conditions but important to make alternative energy sources accessible, such as protein, amino acid, and lipid catabolism by de novo synthesis of catabolic enzymes. In contrast, the basic mitochondrial functions fulfilled by TCA cycle and respiratory chain are not required to be more active during stress than under control conditions, they just change their initial substrate from carbohydrates to amino acids and lipids. Therefore, it makes perfect sense that these pathways are preserved from degradation rather than up-regulated at the transcriptional level. Protection from degradation might be achieved by selective autophagy of specific organelles. During developmental senescence, autophagic vesicles have been shown to preferentially contain RubisCO, entire chloroplasts, and also ribosomes, whereas mitochondrial integrity and function is preserved until very late stages (Chrobok et al., 2016; Marshall and Vierstra, 2018). Our results are in good agreement with these findings since we observed stronger than average decrease rates in plastid and ribosomal proteins during progressive drought stress but very little effect on mitochondrial proteins. Ribosomes are among the most stable proteins under control conditions (Li et al., 2017). However, they tie up a substantial fraction of cellular resources since they account for a majority of the cell’s RNA and also ∼3% of the protein mass. Thus, the turnover of ribosomes in eukaryotes is activated by nutritional stress such as carbon, nitrogen, or phosphate deficiency (Floyd et al., 2016). Conveniently, this measure also serves the purpose of down-regulating protein synthesis rates during stress. Apart from selective autophagy, the stability of individual proteins can be regulated via ubiquitinylation and is also affected by other post-translational modifications, substrate, or cofactor binding, so that less busy enzymes are degraded faster (Nelson and Millar, 2015).
Proteolytic systems and their contribution to stress-induced protein turnover
Autophagy and proteasomes are considered to be the two major proteolytic systems in a cell. However, due to the sheer abundance of chloroplasts, the plastid proteases, according to our evaluation, represent the major share of proteolytic enzymes in a leaf cell under control conditions. Thus, they would be suitable for contributing considerably to the regular turnover of chloroplast proteins. Since amino acid synthesis is also localized mainly in these organelles, they are perfectly equipped for exporting the amino acids resulting from proteolysis (Pottosin and Shabala, 2016). However, the frequency of proteases per total number of proteins is comparatively low in chloroplasts and in contrast to other subcellular compartments does not increase during stress. Bulk degradation of chloroplast proteins during severe dehydration therefore requires additional capacities outside the chloroplast, and these can be provided by the lytic vacuoles that strongly increase their protease content and are able to hydrolyze proteins delivered by autophagic vesicles (Michaeli and Galili, 2014; Marshall and Vierstra, 2018).
In contrast to plastids, the extracellular space is extremely rich in protease molecules per total proteins. Apart from maintaining the cell wall, major functions of the apoplast are signaling and defense against pathogens, which both involve proteolysis. Extracellular plant proteases hydrolyze proteins of invading pathogens to inactivate them and to release signal peptides, triggering immune reactions (Balakireva and Zamyatnin, 2018). Plant peptide hormones are usually produced as pre–pro-protein and need to be activated by proteolytic cleavage (Stührwohldt and Schaller, 2019). This function has been shown to be particularly relevant for drought tolerance. Extracellular subtilisin-like proteases are involved in the regulation of stomatal density and distribution in response to environmental stimuli (Berger and Altmann, 2000; Engineer et al., 2014). In addition, the subtilase Senescence-Associated Subtilisin Protease (SASP) degrades and thus inactivates OST1 (Open Stomata 1), a kinase activated by abscisic acid (ABA), and therefore acts as a negative regulator in ABA signaling (Wang et al., 2018). Our dataset shows a strong induction of SASP during drought stress and identifies 14 additional extracellular proteases that are significantly increased and thus might be relevant for stress tolerance. The apoplast proteome is remarkably stable even during severe dehydration. This finding might indicate a specific relevance of extracellular proteins during drought stress, which is clearly the case for proteases. An alternative explanation could be that apoplast proteins simply evade the intracellular bulk degradation systems autophagy and proteasome due to their remote localization. Since our proteomics dataset covers only a fraction of all cellular proteases, it has to be kept in mind that these insights do not show the complete picture. Additional stress relevant compounds of proteolytic systems, which were not detected here, may be involved.
Amino acid homeostasis under challenging conditions—massive adjustments to the free pool provide osmolytes and ATP
The free pool represents only about 11% of all cellular amino acids under control conditions but strongly gains impact in the course of the drought stress response. Also, despite massive proteolysis the relative composition of the proteome looks roughly similar before and after stress (Supplemental Figure S7), whereas changes on the metabolite level are rapid and drastic (Figure 7, B). Taken together these observations illustrate that homeostasis has a different meaning with regard to free amino acids and proteins. Proline is a well-known compatible osmolyte in plants and also in some euryhaline animals (Szabados and Savouré, 2010; Wiesenthal et al., 2019). Free proline accumulated 219-fold, and even its total amount (free plus bound in proteins) increased from 48 to 153 µmol ⋅ g−1 DW during progressive drought stress, indicating extensive de novo synthesis (Supplemental Dataset S7 and Figure 7, C). In contrast, the total contents of all other 19 proteinogenic amino acids (except Pro) clearly decreased during the stress phase indicating that they are most likely not synthesized during stress, but accumulate in the free pool as a consequence of proteolysis (Supplemental Figure S6, B). An exception might be those amino acids that serve as precursors for secondary metabolites such as the aromatic amino acids (Tzin and Galili, 2010). The total amount of nitrogen contained in amino acids decreases by about one-third during progressive drought stress (Figure 8, C). Taken together with the down-regulation of enzymes involved in primary nitrogen assimilation, it becomes clear that protein degradation can most likely cover the full nitrogen demand for the synthesis of proline (Figure 8, D), GABA, and additional metabolites involved in stress response.
The sum of all amino acids dropped by 29% during stress, most likely due to their use as alternative respiratory substrates and precursors for secondary metabolites (Araújo et al., 2011; Hildebrandt, 2018). The drop in the RQ in stressed compared with control leaves also suggests that respiratory substrates other than carbohydrates (i.e. proteins and lipids) are used during drought. Our estimation based on published respiration rates of Arabidopsis leaves (O’Leary et al., 2017) indicates that amino acid oxidation could fully sustain leaf energy metabolism for about 7 h. However, leaf respiration rates tend to decrease during dehydration (Pinheiro and Chaves, 2011), so that amino acid catabolism, in addition to some residual photosynthetic activity and the oxidation of lipids and chlorophyll, can be anticipated to make a substantial contribution to the ATP supply of drought stressed plants. A shift in the respiratory substrate can thus provide an efficient means to counterbalance carbohydrate limitation in Arabidopsis during drought stress.
Conclusion
Our estimation of the quantitative protein and amino acid composition of a plant leaf cell provides an initial idea of scales and dimensions. On this basis, the dynamic interconnection of protein and amino acid homeostasis during severe drought stress could be monitored and quantified on absolute scales. The current understanding of protein copy numbers in individual plant cells will be refined in the future based on single cell approaches and the analysis of different tissues, developmental stages, and stress conditions.
Materials and methods
Plant growth and drought stress treatment
Arabidopsis (Arabidopsis thaliana) Columbia-0 plants were grown for 2 weeks in pots (200 cm3) in a phytochamber (22–24°C, 16-h light, 8-h darkness, 110 μmol s−1 m−2 light). The stress treatment started with soaking the substrate (Steckmedium, Klasmann-Deilmann GmbH) with tap water to a distinct weight (150 g). A uniform desiccation process was achieved by monitoring pot weights and reorganizing the positions of the pots in the chamber every other day. After 10 d without watering, leaf material (complete rosettes) was harvested on a daily basis in the middle of the light period (Figure 2). During late stages of severe drought stress (S4–S7), plants were additionally sub-classified according to their leaf phenotype (S4: 4–7 rolled leaves S5: 8–10 rolled leaves, S6: >10 rolled leaves). For each stress level, seven stressed plants and three controls were harvested individually. The leaf material of each individual plant is considered as a biological replicate. In addition, three stressed plants were re-watered to test their viability and harvested after 24 h.
Determination of RWC
The method used is based on Smart and Bingham (1974). The weight of a leaf was measured immediately after harvest (FW), after overnight incubation in distilled water (turgor weight, TW), and after overnight drying at 37°C (DW). RWC was calculated according to the following formula: .
Determination of the RQ
Dark respiration of rosette leaf discs was measured at 25°C in a reaction mixture containing 1-mM NaHCO3 and 100-mM KCl using an O2K respirometer (Oroboros Instruments, Innsbruck, Austria). The oxygraph chamber was additionally equipped with a FiveEasy pH meter (Mettler Toledo) to detect O2 consumption and CO2 production simultaneously. The increase in CO2 was calculated on the basis of the pKs value 6.4 and divided by the concurrent decrease in the oxygen concentration of the reaction mixture to determine the RQ (RC).
Extraction and quantification of total protein
In total, 5 mg of lyophilized plant rosette powder was dissolved in 700 µL methanol and incubated for 20 min shaking at 80°C. After centrifugation (10 min, 4°C, 18,800 × g), the pellet was washed twice in 1-mL ethanol (70%, v/v) and resuspended in 400-µL NaOH (0.1 M). The solution was incubated for 1 h shaking at 95°C and centrifuged again. The protein content of the supernatant was quantified using Ready-to-use Coomassie Blue G-250 Protein Assay Reagent (ThermoFisher) and Albumin Standard 23209 (ThermoFisher).
Quantification of free amino acids by HPLC
Free amino acids were extracted as described in Batista-Silva et al. (2019). The pre-column derivatization with o-phthaldialdehyde (OPA) and fluorenylmethoxycarbonyl (FMOC) was based on the application note “Automated amino acids analysis using an Agilent Poroshell HPH-C18 Column” by Agilent. The samples were injected onto a 100 mm × 3 mm InfinityLab Poroshell HPH-C18 column (2.7 µm) using an Ultimate 3000 HPLC system (ThermoFisher). HPLC settings were set as described in Batista-Silva et al. (2019). Cysteine was quantified after derivatization with the fluorescent dye monobromobimane, using the same HPLC system (Fahey et al., 1980; Newton et al., 1981). Five milligrams of lyophilized plant powder was mixed with 10 µL bromobimane (46 mM in acetonitrile), 100 µL acetonitrile, and 200 µL buffer (160-mM HEPES, 16-mM EDTA, pH 8.0), and incubated on a shaker for 30 min in darkness before adding 100 µL methanesulfonic acid (65 mM). Samples were separated on a LiChrospher 60 RP-select Hibar RT 5 µm column (Merck) at 18°C using a gradient of two solvents (0.25% [v/v] acetic acid [pH 4] and methanol). Labeled thiols were detected using a fluorescence detector 3400 RS (ThermoFisher) at 380 nm for excitation and 480 nm for emission.
Protein extraction and label-free quantitative shotgun MS
For protein extraction, about 5 mg of the lyophilized rosette powder was used (C, S3, S5, S6; n = 4). Protein extraction, sample preparation, and LC–MS/MS were performed as previously described (Thal et al., 2018) using a Q‐Exactive mass spectrometer coupled to an Ultimate 3000 UPLC (ThermoFisher). Peptides were first bound to a 2-cm C18 reversed phase trap column (Acclaim PepMap100, diameter: 100 µm, granulometry: 5 µm, pore size: 100 Å; Thermo Fisher Scientific, Waltham, MA, USA). Separation took place on a 50-cm C18 reversed phase analytical column (Acclaim PepMap100, diameter: 75 µm, granulometry: 3 µm, pore size: 100 Å; Thermo Fisher Scientific) eluted using a non-linear 5–36% acetonitrile gradient containing 0.1% (v/v) formic acid. Peptides were transferred into a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Dreieich, Germany) by electrospray ionization (ESI) using a NSI source (Thermo Fisher Scientific, Dreieich, Germany) equipped with a stainless steel nano-bore emitter (Thermo Fisher Scientific, Dreieich, Germany). The data-dependent duty cycle involved a top-10 method, using resolutions set to 70,000 for MS1 (AGI set to 1,000,000) and 17,500 for MS2 (AGI set to 100,000). Profile mode was used during data acquisition.
Protein identification by MaxQuant and data processing via Perseus software
The LC–MS/MS spectra were analyzed using MaxQuant (Version 1.5.5.1, Cox and Mann, 2008) and protein identification was based on the TAIR10 database (35,387 proteins plus the common contaminants trypsin, bovine serum albumin (BSA), and keratin). The search parameters were set to: carbamidomethylation (C) as fixed modification, oxidation (M) and acetylation (protein N-term) as variable modifications. The specific digestion mode was set to trypsin (P) and a maximum of two missed cleavage sites was allowed. A positive peptide identification was required to contain a minimum of seven amino acids. The mass tolerances of the precursor ion were set to 20 and 4.5 ppm for the first and main searches, respectively. The mass tolerances of the fragment ions were set to 40 ppm. FDR at the protein and PSM level was set to 1%. The minimum number of unique peptides per protein group was 1. In total, 3,472 protein groups were identified (1,298–1,655 per sample). Unique and razor peptides were used for protein quantification. The iBAQ function of MaxQuant was enabled, “log fit” disabled. Further analyses and statistical evaluation based on LFQ and iBAQ values generated by MaxQuant were performed in Perseus (version 1.6.1.1), (Tyanova et al., 2016). Changes in the relative abundance of individual proteins were estimated via label-free quantification (LFQ; Cox et al., 2014). This approach is suitable for identifying proteins that are induced and thus might be particularly relevant during the conditions tested. iBAQs were used as a basis for calculating mass and molar contents of the individual proteins. MaxQuant output tables were filtered to remove non-plant contaminants, reversed sequences, and proteins that were only identified based on modified peptides. Proteins were excluded from further analysis if they were not detected in at least three of four replicates in at least one group (C, S3, S5, S6). Missing protein intensities were then considered as too low for proper quantification and replaced by very low values from a normal distribution. Finally, a list of 1,399 proteins (Supplemental Dataset S1) was used for all further calculations. Statistical analysis of the MS dataset was performed in Perseus using two‐sample t tests (P < 0.05).
Calculating absolute contents of individual proteins based on iBAQ values
Raw iBAQ values generated by MaxQuant were multiplied with the molecular weight of the respective protein [kDa]. These individual weighted iBAQs were then divided by the sum of weighted iBAQs of all detected proteins for normalization, and means of the four biological replicates in each sample group were calculated. The mean mass fractions were then multiplied with the total protein content of the sample [mg · g−1 DW] to determine the mass content of each individual protein [µg · g−1 DW]. The mass contents were divided by the molecular weight of the respective protein to calculate the molar protein contents [nmol · g−1 DW]. Protein copy numbers in an individual mesophyll cell were calculated by multiplying the molar protein contents with the mean leaf DW and the Avogadro constant, and dividing it by the mean number of mesophyll cells per leaf. A more detailed description of the calculation methods is provided in Supplemental Figure S1.
Calculating protein-bound amino acid contents based on individual protein contents
The amino acid composition of each protein was determined on the basis of its sequence. The molar content of the protein was then multiplied with the number of each of the 20 amino acids present in this protein to calculate the molar contents of the individual amino acids. The resulting molar amino acid contents were summed up for all identified proteins in a sample. The total numbers of amino acids released due to proteolysis were calculated by subtracting contents of protein-bound amino acids in stressed and control plants.
Quantification of RubisCO using parallel reaction monitoring targeted proteomics
In order to verify the iBAQ-based calculation of absolute amounts of individual proteins, the ribulose-bisphosphate carboxylase/oxygenase large subunit (RubisCO LS) was quantified in control samples via parallel reaction monitoring (PRM) in a targeted MS approach. Three unique peptides of RubisCO LS were selected, based on their intensity, peak symmetry, and the absence of miscleavages and modification sites (Rauniyar, 2015). The following isotopically heavy labeled peptides were provided by New England Peptides (Gardner, MA, USA): (1) DTDLILAAFR*, (2) LTYYTPEYETK*, (3) ESTLGFVDLLR* (R* = Arg, 13C6, 15N4; K* = Lys, 13C6, 15N2). The peptides were mixed equally and diluted with plant sample matrix for calibration (1,600–25 fmol/µL). An inclusion list containing the masses of the labeled and the natural peptides in addition to their retention times was implemented. For the absolute quantification of RubisCO LS, the digested plant samples were spiked with heavy peptides to a final concentration of 400 fmol/µL. Chromatograms were extracted and evaluated with Skyline (V20.1.0.155, MacLean et al., 2010). Peak areas of three transition fragments per peptides were quantified against the calibration of the heavy labeled peptides and subsequently divided by the total peptide content of the individual sample (Supplemental Dataset S2). Due to low and inconsistent signal intensity, the third peptide (ESTLGFVDLLR*) could not be used for quantitation. The total peptide content of the digested leaf samples was determined by using the Pierce Quantitative Colorimetric Peptide Assay (ThermoFisher).
Calculating mitochondrial oxygen consumption with amino acids as alternative respiratory substrates
To estimate mitochondrial respiration in leaves that exclusively use the set of amino acids released by protein degradation during drought stress as substrates, total leaf amino acid contents of stressed plants were subtracted from those of control plants. For each amino acid this difference was multiplied with the number of electrons transferred to the respiratory chain during complete oxidation (Hildebrandt et al., 2015) and divided by four to calculate the total amount of oxygen consumed (Supplemental Dataset S7).
Genevestigator datasets
The following three microarray datasets were used for estimating gene expression levels during drought stress: (1) AT-00684_1 (Ludwików et al., 2009; long-day conditions, start: 3 weeks, samples after 5 d of dehydration in soil); (2) AT-00626_1 (Pandey et al., 2013: long-day conditions, start: 3 weeks, samples after 10 d of dehydration in soil); (3) AT-00292_1 (Perera et al., 2008: short-day conditions, start: 6 weeks, samples after 7 d of dehydration in soil).
Data availability statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Perez-Riverol et al., 2019) with the dataset identifier PXD021563.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Dataset S1.
Supplemental data
Supplemental Figure S1. Calculation of individual protein contents and copy numbers.
Supplemental Figure S2. Estimation of the absolute content of RubisCO large subunit.
Supplemental Figure S3. Principal component analysis of the MS dataset.
Supplemental Figure S4. Compartment-specific patterns of stress-induced changes in individual protein abundances:individual protein contents.
Supplemental Figure S5. Compartment-specific patterns of stress-induced changes in individual protein abundances:fold change ratios.
Supplemental Figure S6. Protein-bound and free amino acids during progressive drought stress in Arabidopsis rosette leaves.
Supplemental Figure S7. Changes in the quantitative composition of the leaf proteome during drought stress.
Supplemental Dataset S1. Complete MS dataset: LFQ and iBAQ values, relative protein abundances, mass contents [µg · g−1 DW], molar contents [nmol · g−1 DW], and copy numbers [million proteins per cell] of 1,399 protein species during progressive drought stress.
Supplemental Dataset S2. Quantification of RubisCO LS via PRM in a targeted MS approach.
Supplemental Dataset S3. Quantitative composition of the leaf proteome under control conditions. Mass fractions (%) of metabolic categories.
Supplemental Dataset S4. Combined analysis of protein abundances and expression levels to identify general strategies of leaf cells to adjust their protein setup to the challenges posed by insufficient water supply.
Supplemental Dataset S5. MS dataset of all proteins involved in protein metabolism (extracted from Supplemental Dataset S1).
Supplemental Dataset S6. Free amino acid contents in Arabidopsis rosette leaves during progressive drought stress.
Supplemental Dataset S7. Pools of bound and free proteinogenic amino acids (AA) in Arabidopsis rosette leaves during progressive drought stress.
Supplemental Dataset S8. MS dataset of all proteins involved in amino acid metabolism (extracted from Supplemental Dataset S1).
Supplementary Material
Acknowledgments
The authors thank Marianne Langer and Dagmar Lewejohann for skillful technical assistance during MS sample preparation and Michael Senkler for IT support.
Conflict of interest statement. The authors have no conflicts of interest to declare.
T.M.H. and H.-P.B. initiated the project. T.M.H. designed the research, wrote the manuscript with support from H.-P.B. and B.H., and agreed to serve as the author responsible for contact and to ensure communication. B.H. performed most experiments. B.H. and P.K. performed the shotgun proteomics experiments. B.H. and H.E. performed the absolute Rubisco quantification. T.M.H. and B.H. analyzed the data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Tatjana M. Hildebrandt (hildebrandt@genetik.uni-hannover.de).
References
- Alcázar R, Marco F, Cuevas JC, Patron M, Ferrando A, Carrasco P, Tiburcio AF, Altabella T (2006) Involvement of polyamines in plant response to abiotic stress. Biotechnol Lett 28:1867–1876. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation—an alternative respiratory substrate for stressed plants. Trends Plant Sci 16:489–498. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320:938–941. [DOI] [PubMed] [Google Scholar]
- Balakireva AV, Zamyatnin AA (2018) Indispensable role of proteases in plant innate immunity. Int J Mol Sci 19:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On YM, Milo R (2019) The global mass and average rate of rubisco. Proc Natl Acad Sci U S A 116:4738–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Silva W, Heinemann B, Rugen N, Nunes-Nesi A, Araújo WL, Braun H-P, Hildebrandt TM (2019) The role of amino acid metabolism during abiotic stress release. Plant Cell Environ 42:1630–1644. [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T (2000) A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, Lindén P, Ziolkowska A, Liebsch D, Narsai R, Szal B, Moritz T, Rouhier N, et al. (2016) Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant physiol 172:2132–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- Dikic I (2017) Proteasomal and autophagic degradation systems. Annu Rev Biochem 86:193–224 [DOI] [PubMed] [Google Scholar]
- Engineer CB, Ghassemian M, Anderson JC, Peck SC, Hu H, Schroeder JI (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre B, Lambour T, Bouyssié D, Menneteau T, Monsarrat B, Burlet-Schiltz O, Bousquet-Dubouch M-P (2014) Comparison of label-free quantification methods for the determination of protein complexes subunits stoichiometry. EuPA Open Proteomics 4:82–86. [Google Scholar]
- Fahey RC, Newton GL, Dorian R, Kosower EM (1980) Analysis of biological thiols: derivatization with monobromotrimethylammoniobimane and characterization by electrophoresis and chromatography. Anal Biochem 107:1–10 [DOI] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, MacIntosh GC, Bassham DC (2016) Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11:2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs P, Rugen N, Carrie C, Elsässer M, Finkemeier I, Giese J, Hildebrandt TM, Kühn K, Maurino VG, Ruberti C, et al. (2020) Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J 101:420–441. [DOI] [PubMed] [Google Scholar]
- Gupta A, Rico-Medina A, Caño-Delgado AI (2020) The physiology of plant responses to drought. Science 368:266–269 [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM (2018) Synthesis versus degradation: directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol Biol 98:121–135. [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun H-P (2015) Amino acid catabolism in plants. Mol Plant 8:1563–1579. [DOI] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW (2018) Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst 6:192–205.e3. [DOI] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45: D1064–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008:420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz B-J, Tyers M (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297:395–400 [DOI] [PubMed] [Google Scholar]
- Königer M, Delamaide JA, Marlow ED, Harris GC (2008) Arabidopsis thaliana leaves with altered chloroplast numbers and chloroplast movement exhibit impaired adjustments to both low and high light. J Exp Bot 59:2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Wilmarth PA, Shin J-B, Klimek J, Sherman NE, Jeffery ED, Choi D, David LL, Barr-Gillespie PG (2014) Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. J Proteome Res 13:1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11:319–324. [DOI] [PubMed] [Google Scholar]
- Kwasniak M, Pogorzelec L, Migdal I, Smakowska E, Janska H (2012) Proteolytic system of plant mitochondria. Physiol Plant 145:187–195. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Wong P, Chan H-K, Yam K-M, Chen L, Chow C-M, Coruzzi GM (2003) Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol 132:926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH (2017) Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29:207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R (2014) Visual account of protein investment in cellular functions. Proc Natl Acad Sci U S A 111:8488–8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwików A, Kierzek D, Gallois P, Zeef L, Sadowski J (2009) Gene expression profiling of ozone-treated Arabidopsis abi1td insertional mutant: protein phosphatase 2C ABI1 modulates biosynthesis ratio of ABA and ethylene. Planta 230:1003–1017. [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD (2018) Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 69:173–208. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Tam S, Kaganovich D, Frydman J (2005) Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol 7:736–741. [DOI] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM (2017) Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J 90:628–653. [DOI] [PubMed] [Google Scholar]
- Mergner J, Frejno M, List M, Papacek M, Chen X, Chaudhary A, Samaras P, Richter S, Shikata H, Messerer M, et al. (2020) Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579:409–414. [DOI] [PubMed] [Google Scholar]
- Michaeli S, Galili G (2014) Degradation of organelles or specific organelle components via selective autophagy in plant cells. Int J Mol Sci 15:7624–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CJ, Millar AH (2015) Protein turnover in plant biology. Nat Plants 1: 15017. [DOI] [PubMed] [Google Scholar]
- Newton GL, Dorian R, Fahey RC (1981) Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Anal Biochem 114:383–387. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W (2016) Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiol 171:2280–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary BM, Lee CP, Atkin OK, Cheng R, Brown TB, Millar AH (2017) Variation in leaf respiration rates at night correlates with carbohydrate and amino acid supply. Plant Physiol 174:2261–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N, Ranjan A, Pant P, Tripathi RK, Ateek F, Pandey HP, Patre UV, Sawant SV (2013) CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 8:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung C-Y, Moore CD, Stevenson-Paulik J, Boss WF (2008) Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20:2876–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro C, Chaves MM (2011) Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot 62:869–882. [DOI] [PubMed] [Google Scholar]
- Pottosin I, Shabala S (2016) Transport across chloroplast membranes: optimizing photosynthesis for adverse environmental conditions. Mol Plant 9:356–370. [DOI] [PubMed] [Google Scholar]
- Preuten T, Cincu E, Fuchs J, Zoschke R, Liere K, Börner T (2010) Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J 64:948–959. [DOI] [PubMed] [Google Scholar]
- Rauniyar N (2015) Parallel reaction monitoring: a targeted experiment performed using high resolution and high mass accuracy mass spectrometry. Int J Mol Sci 16:28566–28581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. [DOI] [PubMed] [Google Scholar]
- Smart RE, Bingham GE (1974) Rapid estimates of relative water content. Plant Physiol 53:258–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N,, Schaller A (2019) Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol 21(Suppl 1):, 49–63. [DOI] [PubMed] [Google Scholar]
- Suraweera A, Münch C, Hanssum A, Bertolotti A (2012) Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 48:242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97. [DOI] [PubMed] [Google Scholar]
- Thal B, Braun H-P, Eubel H (2018) Proteomic analysis dissects the impact of nodulation and biological nitrogen fixation on Vicia faba root nodule physiology. Plant Mol Biol 97:233–251. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3:956–972. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59:191–223. [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397. [DOI] [PubMed] [Google Scholar]
- Wang Q, Guo Q, Guo Y, Yang J, Wang M, Duan X, Niu J, Liu S, Zhang J, Lu Y. et al. (2018) Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OPEN STOMATA 1. J Exp Bot 69:4403–4417. [DOI] [PubMed] [Google Scholar]
- Wiesenthal AA, Müller C, Harder K, Hildebrandt J-P (2019) Alanine, proline and urea are major organic osmolytes in the snail Theodoxus fluviatilis under hyperosmotic stress. J Exp Biol 222: jeb193557. [DOI] [PubMed] [Google Scholar]
- Wuyts N, Palauqui J-C, Conejero G, Verdeil J-L, Granier C, Massonnet C (2010) High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods 6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu P, Guo T, Zhao H, Bensaddek D, Aebersold R, Xiong L (2019) Arabidopsis proteome and the mass spectral assay library. Sci Data 6:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Perez-Riverol et al., 2019) with the dataset identifier PXD021563.