The shapes of leaves are crucial for effectively capturing light and minimizing transpiration, and, therefore, for optimal functioning of the plant (Lang et al. 2004; Moon and Hake 2011). Many different leaf structures and shapes exist. Due to this variety, as well as it being a visible plant feature, leaf shape is often used to identify and describe plant species. Leaves are typically composed of a relatively flat surface called the blade that is specialized for photosynthesis. However, leaf flatness varies greatly depending on leaf developmental stage, species, environmental conditions, and even disease, and is produced by specific cell types on the leaf surface.
Bulliform cells are large, specialized epidermal cells that occur on the adaxial (top) side of leaves in monocotyledonous plants (Itoh et al. 2005; Xiang et al. 2012). In drier conditions, dehydration of these cells causes the leaf to fold adaxially (curling upwards), resulting in less exposure to sunlight and, therefore, less water loss. In rice, several genes have been identified to control adaxial or abaxial leaf rolling by increasing or decreasing the size or number of bulliform cells (Hibara et al. 2009; Li et al. 2010). One of these is ABAXIALLY CURLED LEAF 1 (ACL1), which encodes a small protein of unknown function. As the name indicates, loss-of-function of ACL1 results in abaxially curled leaves.
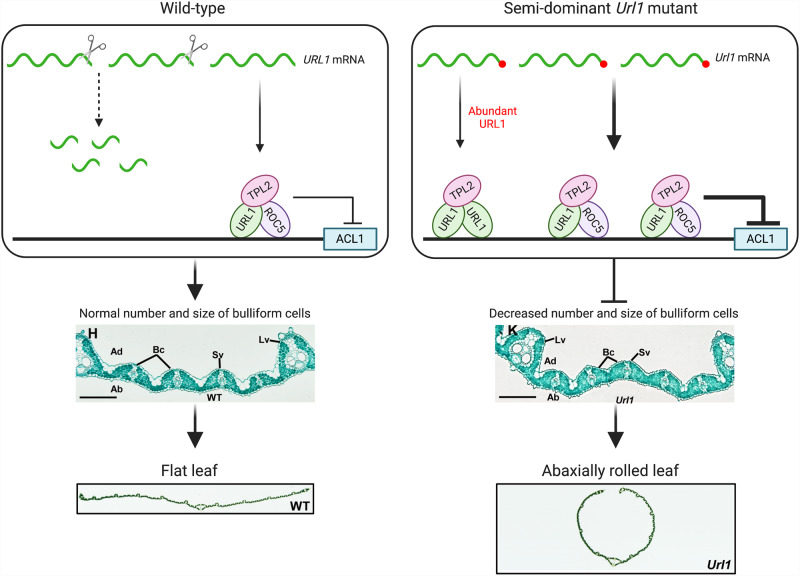
In this issue, Fang et al. (2021) showed that a complex consisting of UPWARD ROLLED LEAF (URL) 1, RICE OUTERMOST CELL-SPECIFIC (ROC) 5, and the Topless corepressor TPL2 represses the expression of ACL1, resulting in a reduced number and size of bulliform cells, and rice leaves that are rolled as opposed to flat. URL1 (also known as ROC8) and ROC5 are both homeodomain-leucine Zipper (HD-Zip) IV family members. Previous studies have shown that ROC5 negatively regulates bulliform cell development and influences leaf rolling (Zou et al. 2011). Moreover, Topless and topless-related proteins have been shown to interact with transcription factors to control transcriptional repression in many developmental processes (Causier et al. 2012). The authors demonstrated the role of URL1 in mediating leaf rolling by interacting with ROC5 and TPL2 through multiple lines of evidence.
In the study, a semidominant mutant, Url1, with incurved leaves was identified from a population of Oryza sativa japonica. The number and size of bulliform cells of Url1 were significantly reduced compared to wild-type plants and were responsible for the adaxially rolled leaves of the mutant plant. The authors showed that, like the Url1 mutant, increased expression of URL1 led to rolled leaves, and that the mutant had a single nucleotide substitution in the 3′-untranslated region (UTR) of the URL1 gene, altering the stem-loop structure formed in the mRNA post-transcriptionally. In the wild-type URL1 plant, this stem-loop triggers RNA degradation, while the point mutation in the Url1 mutant resulted in enhanced mRNA stability and accumulation of mRNA, and subsequently increased amounts of URL1 protein. Inversely, knockout of URL1 led to outcurved, abaxially rolled leaves due to an increase in bulliform cells. The authors also showed that URL1 contains an EAR(Ethylene-responsive element binding factor-associated Amphiphilic Repression) motif, a transcriptional repression domain (Kagale et al. 2010), which it uses to recruit TPL2, and that URL1 physically interacts with ROC5, forming a heterodimer protein. Finally, the authors verified that ACL1 was the downstream target of URL1, and that URL1 transcriptionally represses the expression of ACL1. Furthermore, overexpression of ACL1 resulted in an increased number and size of bulliform cells, partially alleviating the leaf rolling phenotype in Url1 mutants.
In this study, Fang et al. (2021) elegantly elucidated the roles of genes involved in leaf rolling in rice, clarifying precisely how the products of three genes interact to repress the expression of a fourth in a regulatory network that directly affects the development of bulliform cells in the leaves of rice (Figure 1). Since knowledge of the molecular mechanisms of bulliform cell formation and development is limited, these findings shed light on how these cells mediate leaf flatness and how these mechanisms affect transpiration and light absorption in the plant. Taken together, insights from this research contribute to fine-tuning plant productivity, especially in changing climatic environments.
Figure 1.
Working model for the URL1-TPL2-ROC5 transcriptional repression complex. In the wild-type plant, some URL1 transcripts are cleaved due to the stem-loop structure rendering the mRNA prone to degradation. Protein from the remaining transcripts form a heterodimer with ROC5 and interact with TPL2 to inhibit ACL1 expression, mediating bulliform cell development and maintaining flat leaves. In the Url1 semi-dominant mutant, a single nucleotide substitution in the 3′-UTR of URL1 increases the stability of its mRNA, limiting degradation. The more abundant URL1 forms a heterodimer with ROC5 as well as a homodimer, further repressing the expression of the target genes, resulting in a reduction in number and size of bulliform cells, and abaxially rolled leaves. Ad, adaxial; Ab, abaxial; Lv, large vein; Sv, small vein; Bc, bulliform cell. Adapted from Fang et al (2021), Figures 1 and 11. Created with Biorender.com.
References
- Causier BAshworth MGuo W, Davies B (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Guo T, Xie Z, Chun Y, Zhao J, Peng L, Zafar SA, Yuan S, Xiao L, Li X (2021) The URL1-ROC5-TPL2 transcriptional repressor complex represses the ACL1 gene to modulate leaf rolling in rice. Plant Physiol 185: 1722–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara KObara MHayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y (2009) The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev Biol 334: 345–354 [DOI] [PubMed] [Google Scholar]
- Itoh JNonomura KIkeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, NagatoY ( . 2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K ( 2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Y-Z, Zhang Z-J, Gu X-Y, Yang J-C, Zhu Q-S (2004) Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) II. Photosynthetic character, dry mass production and yield forming. Acta Agronomica Sinica 30: 739–744 [Google Scholar]
- Li LShi ZYLi L, Shen GZ, Wang XQ, An LS, ZhangJL ( . 2010) Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol Plant 3: 807–817 [DOI] [PubMed] [Google Scholar]
- Moon J, Hake S (2011) How a leaf gets its shape. Curr Opin Plant Biol 14: 24–30 [DOI] [PubMed] [Google Scholar]
- Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) Semi-rolled leaf1 encodes putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159:1488–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou LPSun XHZhang ZG, Liu P, Wu JX, Tian CJ, Qiu JL, LuTG ( . 2011) Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol 156: 1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]