Abstract
The lipid bilayer of biological membranes has a complex composition, including high chemical heterogeneity, the presence of nanodomains of specific lipids, and asymmetry with respect to lipid composition between the two membrane leaflets. In membrane trafficking, membrane vesicles constantly bud off from one membrane compartment and fuse with another, and both budding and fusion events have been proposed to require membrane lipid asymmetry. One mechanism for generating asymmetry in lipid bilayers involves the action of the P4 ATPase family of lipid flippases; these are biological pumps that use ATP as an energy source to flip lipids from one leaflet to the other. The model plant Arabidopsis (Arabidopsis thaliana) contains 12 P4 ATPases (AMINOPHOSPHOLIPID ATPASE1–12; ALA1–12), many of which are functionally redundant. Studies of P4 ATPase mutants have confirmed the essential physiological functions of these pumps and pleiotropic mutant phenotypes have been observed, as expected when genes required for basal cellular functions are disrupted. For instance, phenotypes associated with ala3 (dwarfism, pollen defects, sensitivity to pathogens and cold, and reduced polar cell growth) can be related to membrane trafficking problems. P5 ATPases are evolutionarily related to P4 ATPases, and may be the counterpart of P4 ATPases in the endoplasmic reticulum. The absence of P4 and P5 ATPases from prokaryotes and their ubiquitous presence in eukaryotes make these biological pumps a defining feature of eukaryotic cells. Here, we review recent advances in the field of plant P4 and P5 ATPases.
Membrane properties depend on the active translocation of lipids between leaflets by P4 ATPases, and perhaps also P5 ATPases, affecting a plethora of cellular functions and plant phenotypes.
Biological membrane properties
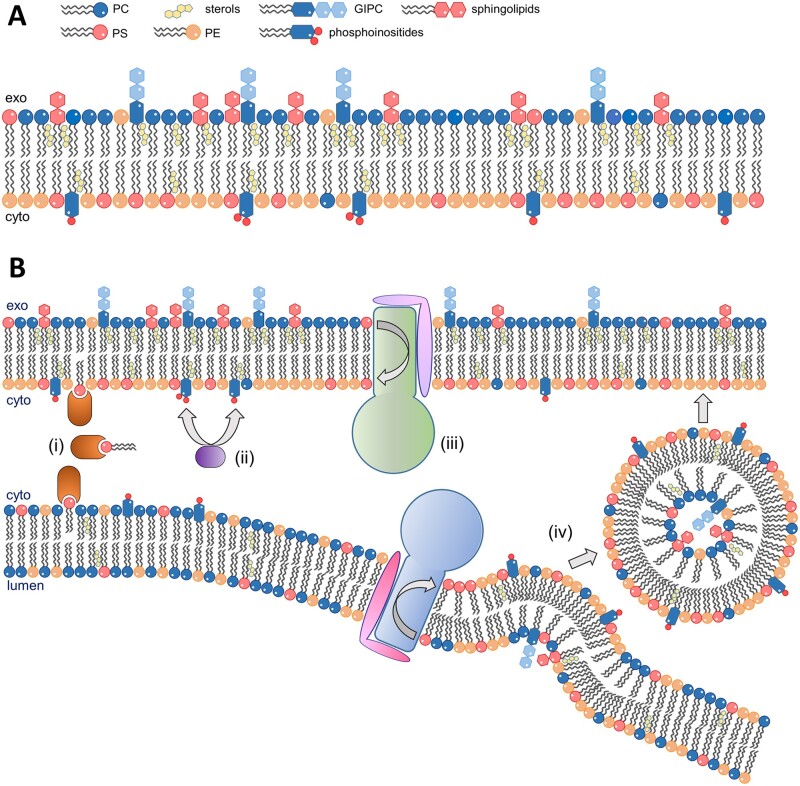
Like all eukaryotic cells, plant cells are enclosed by a plasma membrane, and the cytoplasm includes multiple membrane-delimited compartments (i.e. organelles). Biological membranes are characterized by a bilayer of lipids of different nature (for lipid structures, see Figure 1) that have their hydrophobic parts exposed to the membrane interior and their hydrophilic parts exposed to the aqueous environment on either side of the membrane (Figure 2, A). Eukaryotic plasma membranes present an asymmetric lipid distribution, a phenomenon that is collectively referred to as “lipid asymmetry.” For example, phosphatidylserine (PS) and phosphatidylinositol phosphates (PIPs), two negatively charged phospholipids, as well as the zwitterionic phospholipid phosphatidylethanolamine (PE) are preferentially located on the cytosolic side of the plasma membrane (Figure 2, A). By contrast, the extracytosolic side is enriched in phosphatidylcholine (PC), also a zwitterion, and ceramide-derived glycosphingolipids such as glucosylceramide (GlcCer), a polar neutral sphingolipid (Figure 2, A; van Meer et al., 2008; Zachowski, 2012; Nicolson, 2014). Whereas plasma membranes are rich in sterols that impose tight lipid packing, making the membrane relatively rigid and impermeable to its surroundings (Dufourc, 2008), the endoplasmic reticulum (ER) has a more flexible membrane with very low sterol content (Casares et al., 2019).
Figure 1.
Representative lipids of plant plasma membranes. A, Sterols. B, Glycerophospholipids. The glycerol backbone is marked in bold. C, Ceramide-derived lipids (glycosphingolipids). The ceramide backbone is marked in bold. A representative glycosylinositolphosphoceramide containing glucuronic acid and mannose is depicted.
Figure 2.
Membrane lipid asymmetry. A, The plasma membrane of eukaryotic cells presents an asymmetric lipid distribution where phosphatidylserine (PS), phosphatidylethanolamine (PE), and phosphatidylinositol phosphates (PIPs) are concentrated in the inner leaflet, and phosphatidylcholine (PC), glycosylinositolphosphorylceramides (GIPCs), and other sphingolipids are concentrated in the outer leaflet. Sterols give the membrane rigidity. B, Several cellular mechanisms coordinate to establish membrane lipid asymmetry. (i) Lipid transfer proteins move lipids between the cytosolic leaflets of two membranes at membrane contact sites. (ii) Phosphatidylinositol kinases and phosphatases phosphorylate/dephosphorylate their targets to generate specific PIPs at the cytosolic leaflet of cellular membranes. (iii) ATP-dependent transporters, such as P4 ATPases (depicted), translocate specific lipids between the two leaflets of the membranes in the secretory pathway. (iv) The activity of P4 ATPases contributes to vesicle budding in the secretory pathway and the subsequent flow of vesicles transports lipids among membranes. Lipids that are synthesized in the lumen of organelles, such as sphingolipids, are confined to the inner leaflet of the lipid bilayer in vesicles and are exposed to the extracellular/lumenal side of the membrane after vesicle fusion to its final destination.
Advances
P4 ATPases are involved in multiple cellular processes. Mutating the genes encoding these pumps gives rise to pleiotropic phenotypes, all of which might be the result of membrane trafficking defects.
P4A-e flippases appear to be essential to Arabidopsis (Arabidopsis thaliana), and likely other plant life.
Arabidopsis AMINOPHOSPHOLIPID ATPASE4 (AtALA4) and AtALA5 are P4A-e flippases that are critical for plant growth, potentially by facilitating the catabolism of glucosylceramide (GlcCer) sphingolipids.
AtALA3 helps regulate polar growth by controlling phosphatidylserine (PS) exposure at the cytoplasmic face of the plasma membrane.
Plant flippases appear to interact with guanine-nucleotide exchange factors for ADP-ribosylation factor GTPases (ARF-GEFs), like the yeast flippase DEFICIENT FOR RIBOSOMAL SUBUNITS2 (Drs2p). AtALA3 interacts with at least two ARF-GEFs, BREFELDIN A-INHIBITED GUANINE NUCLEOTIDE-EXCHANGE PROTEIN3 (BIG3) and GNOM.
Localization of AtALA10, and potentially other ALAs, might be regulated via ubiquitination events.
Compositional imbalances between each membrane leaflet are thought to be critical for proper cellular function. Negative lipids on the cytosolic side of the membrane (e.g. PS/PIPs) can bind membrane-associated proteins and allow for the fusion of membrane vesicles (Platre et al., 2018; Colin and Jaillais, 2020; Noack and Jaillais, 2020). On the cell exterior, sugar-modified lipids (e.g. gangliosides, GlcCers) are often important for cell-to-cell recognition (Lopez and Schnaar, 2009; Msanne et al., 2015; Rodriguez-Emmenegger et al., 2019), and PC is a precursor of signaling lipids generated by external threats, such as pathogens (Zhang and Xiao, 2015; Lim et al., 2017).
Establishment and maintenance of lipid asymmetry
In eukaryotic cells, many membrane constituents (e.g. phospholipids, sterols, and membrane proteins) are synthesized at the ER. From there, both lipids and proteins are trafficked by membrane vesicles moving among cellular compartments. In this transport system, the Golgi apparatus can be viewed as a station on the road that ultimately leads to the plasma membrane, to which vesicles fuse by exocytosis. Membrane vesicles can also originate via endocytosis from the plasma membrane, from where their cargo passes to endosomes that mature and fuse to the vacuole or plasma membrane, allowing protein degradation or recycling, as necessary. This flow of membrane vesicles in opposite directions is a hallmark of eukaryotic cells and is collectively known as the secretory pathway (Paez Valencia et al., 2016; Boncompain and Weigel, 2018; Brandizzi, 2018).
The asymmetric distribution of lipids across the bilayers of cellular membranes is partially established by the secretory pathway (Figure 2, B). For instance, many glycosphingolipids are attached to their sugar moieties on the lumenal side of the Golgi and are then transported to the plasma membrane via vesicles (Lopez and Schnaar, 2009; Mortimer et al., 2013; Cacas et al., 2016; Fang et al., 2016). After fusion, the vesicle inner side is exposed to the extracytosolic environment, placing the sugar-containing lipids on the plasma membrane outer leaflet. Other lipids, such as PIPs, are synthesized by cytosolic enzymes at the membrane where they reside, which confines them to the cytosolic membrane leaflet in the absence of a transporter (Adhikari and Cullen, 2015). Lipids can also be rapidly shuffled between the cytosolic leaflets of two biological membranes by the action of lipid transfer proteins (Wong et al., 2019). However, moving lipids across membranes against a concentration gradient, to establish and maintain an asymmetric lipid distribution between the two leaflets, is an energy-consuming process that involves specific ATP-hydrolyzing enzymes (Lopez-Marques et al., 2014).
P4 and P5 ATPases
P-type ATPases constitute a superfamily of membrane-spanning pumps that move ions and macromolecules from one side of a biological membrane to the other at the expense of ATP hydrolysis (Palmgren and Nissen, 2011). A characteristic of these pumps is the formation of a phosphorylated reaction cycle intermediate, hence the name P-type. Phylogenetically, they can be divided into five major families, P1–P5 (Axelsen and Palmgren, 1998). P1–P3 ATPases evolved in prokaryotes, and combinations of these pumps are present in all forms of life, where they transport a variety of cations such as calcium, zinc, copper, sodium, potassium, and protons. By contrast, P4 and P5 ATPases are absent from prokaryotes but ubiquitous in eukaryotes across all phyla. This would suggest that these pumps were present in a common eukaryotic ancestor and represent a fundamental feature that defines eukaryotic cells. Unlike cation-transporting P-type ATPases, P4 ATPases (named flippases) transport phospholipids across biological membranes, transferring their substrate from the extracytosolic to cytosolic-facing leaflets.
P4 ATPases are present in all membranes of the secretory pathway. However, they do not appear to accumulate as ER resident proteins, and instead—following their association with a β-subunit—traffic from the ER to other locations in the plant cell (Poulsen et al., 2008a; López-Marqués et al., 2010; Botella et al., 2016). On the contrary, the ER is the only membrane system in plants where the poorly characterized P5 ATPases are present. P4 and P5 ATPases form a monophyletic group and therefore have evolved together and possibly share common features (Palmgren et al., 2019).
Other enzyme families known to transport membrane lipids in animals and fungi include ATP-binding cassette (ABC) transporters (that actively transport lipids but mainly towards the extracytosolic side of membranes) and scramblases (that facilitate passive diffusion of lipids down a concentration gradient). However, no lipid transport activity has been characterized for members of these families in plants, and they are not discussed in this review.
Evolution of P4 ATPases and their substrate specificities
An analysis of P4 ATPases in protozoan, fungal, plant, and animal species points to the presence of three major clades of P4 ATPases (P4A, P4B, and P4C), with the P4A clade being further divided into six subclades (P4A-a, P4A-b, P4A-c, P4A-d, P4A-e, and P4A-f; Palmgren et al., 2019). While the transport activities of many flippases have yet to be characterized, the phylogenetic clades do not appear to correspond with known lipid transport profiles. In plants, P4 ATPases (aminophospholipid ATPases; ALAs) belong to either the P4A-b, P4A-e, or P4C clades. There is some debate as to whether the P4A-e clade is further divided into three smaller clusters (clusters 2–4, out of 5 total; Baxter et al., 2003). Regardless, the P4A-e clade is exclusive to plants, from green algae to angiosperms, suggesting the potential for unique roles in plant-specific cellular functions. Arabidopsis (Arabidopsis thaliana) mutants lacking multiple P4A-e members appear to be non-viable and sterile (Zhang et al., 2020), providing evidence that this plant-specific subclade is essential to plant life.
Uptake assays using fluorescently labeled lipids in the yeast Saccharomyces cerevisiae have been used to determine the substrate specificities of heterologously expressed AtALAs (Table 1). Again, the evolutionary organization of ALAs was not a strong predictor for conservation in transport profiles. AtALA1 (P4A-b) and AtALA2 (P4C) have highly similar substrate specificities that appear to be primarily restricted to PS transport, despite belonging to divergent subclades. Within the plant-specific P4A-e clade, differences have been observed between AtALA3, AtALA5, and AtALA10, most notably in the ability to transport lipids containing choline headgroups. While all three flippases transport PC, only AtALA5 and AtALA10 are capable of transporting sphingomyelin (SM), a sphingolipid containing a choline headgroup, and only AtALA10 can transport lysophosphatidylcholine (lyso-PC), a mono-acyl derivative of PC (Poulsen et al., 2015; Davis et al., 2020a). The discrepancy between phylogenetic organization and substrate specificity is not fully understood, but might be connected to the mechanism underlying P4 ATPase selectivity filters, in which different lipid flippases use different amino acid combinations to coordinate and transport the same lipid substrate (Palmgren et al., 2019). Likewise, seemingly minor residue changes can prevent similar P4 ATPases from being able to transport the same lipid.
Table 1.
Organization and substrate preferences of the ALA family of P4 ATPases in Arabidopsis
| Eukaryotic clade | Plant cluster | ALA # | Substrates transported | Substrates failing to show transport | References |
|---|---|---|---|---|---|
| P4A-b | 1 | ALA1 | PSa, PEa | PC, lyso-PCb | Gomès et al. (2000); López-Marqués et al. (2012) |
| P4A-e | 2 | ALA8 | Untested | Untested | n.a. |
| ALA9 | Untested | Untested | n.a. | ||
| ALA10 | PC, PE, PS, PG, lyso-PC, SM, GlcCer | PA, DAG, Cer, LacCer, Sph-1-P, Sph, lyso-SM | Poulsen et al. (2015); Jensen et al. (2017) | ||
| ALA11 | Untested | Untested | n.a. | ||
| ALA12 | Untested | Untested | n.a. | ||
| 3 | ALA4 | Untested | Untested | n.a. | |
| ALA5 | PC, PE, PSc, SM | PG, lyso-PC, Cer, GlcCer, GalCer, LacCer | Davis et al. (2020a) | ||
| ALA6 | Failed to detect transport | PC, PE, PS, PA | McDowell et al. (2015) | ||
| ALA7 | Untested | Untested | n.a. | ||
| 4 | ALA3 | PC, PE, PS, PG | lyso-PC, SM | Poulsen et al. (2008a, 2015); López-Marqués et al. (2010, 2012); Jensen et al. (2017) | |
| P4C | 5 | ALA2 | PS | PC, PE, lyso-PCb | López-Marqués et al. (2010); Poulsen et al. (2015) |
Arabidopsis P4 ATPases (ALAs) are organized into their respective eukaryotic clades and plant-centric clusters.
PS and weak PE transport were detected for ALA1 in yeast microsomes (Gomès et al., 2000), but this result could not be confirmed through assays in living yeast cells (López-Marqués et al., 2012).
Indirect growth assays on plates containing the toxic lyso-PC analog miltefosine.
ALA5 showed no transport of fluorescently labeled PS but did convey resistance to Papuamide A, suggesting transport of endogenous PS.
PG, phosphatidylglycerol; PA, phosphatidic acid; Sph, sphingosine; GalCer, galactosylceramide; LacCer, lactosylceramide; DAG, diacylglycerol; n.a., not available.
P4 ATPases and the secretory pathway
P4 ATPases are present in membrane systems throughout the secretory pathway (Nintemann et al., 2019), where they appear to contribute to secretory vesicle formation. Such a function has been described in plants for AtALA3, which localizes to the Golgi, where it assists in the formation of slime vesicles in root tip cells (Poulsen et al., 2008a). Local and unidirectional flipping of membrane phospholipids by P4 ATPases generates the tension and curvature required for membrane vesicle budding from the flat sheets of biological membranes (Takada et al., 2018). In addition, some P4 ATPases interact with proteins involved in vesicle formation, such as ADP-ribosylation factor-guanine nucleotide exchange factors (ARF-GEFs) that activate small GTPases involved in recruiting coat proteins and cargo receptors to vesicles (Wicky et al., 2004; Natarajan et al., 2009; Tsai et al., 2013). Hence, P4 ATPases have been suggested to act as scaffolds that attract the correct machinery to the places of vesicle formation (Natarajan et al., 2009).
A potential scaffold interaction has been described in plants for AtALA3 (Zhang et al., 2020). Recently, the subcellular localization of this protein was extended to include the Golgi, trans-Golgi network, plasma membrane, and the vesicles that circulate between them (Zhang et al., 2020). In endomembranes, AtALA3 physically interacts with two ARF-GEFs, GNOM and BIG3, which regulate trafficking events in the late secretory pathway (Zhang et al., 2020). Deletion of AtALA3 generates several membrane trafficking defects at the root tip, increasing endocytosis rates from the plasma membrane and decreasing vesicle formation from the Golgi and trans-Golgi networks, resulting in the formation of intracellular membrane aggregates. Similar phenotypes are observed in yeast mutants devoid of the trans-Golgi network P4 ATPase Drs2p, which also transports PS (Natarajan et al., 2009). Such mutants fail to recruit small GTPases to endomembrane surfaces, resulting in cells devoid of trans-Golgi-derived secretory vesicles. Together, these results suggest that the scaffolding function for P4 ATPases in membrane trafficking is conserved between organisms.
Trafficking defects are also evident in ala3 mutant pollen tubes (McDowell et al., 2013). Pollen tube growth requires the mobilization of enormous numbers of vesicles that travel along the tube and fuse to the tip, allowing fast expansion in the apical direction. ala3 mutants have much shorter pollen tubes than the wild type, resulting in fertilization defects. While the underlying cellular defect is not clear, vesicle movement in growing ala3 pollen tubes is slower and more chaotic. A possible mechanism might be that ARF-GEF interacts with ALA3, causing flippase activation, which induces membrane curvature and allows vesicle formation.
AtALA2 was also suggested to be required for proper secretory pathway functioning. AtALA2 is a PS transporter located at the multivesicular bodies/prevacuolar compartment (MVBs/PVC; López-Marqués et al., 2010). The MVBs, which correspond to the late endosome in plants, play a central role in a plethora of essential cellular processes that require fine-tuned protein trafficking (Piper and Katzmann, 2007; Richter et al., 2009). Cellular responses to many external stimuli require the action of membrane receptors that bind to a ligand in the extracellular medium and subsequently trigger a signaling cascade, resulting in adaptation/defense responses appropriate to the stimulus (Gu et al., 2017). After signaling, these receptors are endocytosed and sent through endosomes to the MVBs. In some cases, the drop in luminal pH inside the MVBs is sufficient to trigger ligand release, readying the receptor to be recycled for another round of signaling at the plasma membrane. In others, the receptor is further internalized into smaller vesicles inside the MVBs (hence, the name MVB) and targeted for degradation in the lytic vacuole. Thus, MVBs are a central point in the secretory pathway where the fate of many membrane proteins is decided after endocytosis (Gao et al., 2017; Cui et al., 2020b). While no evidence of a secretory role for AtALA2 exists in its original Arabidopsis background, overexpression of AtALA2 in Nicotiana benthamiana cells results in enlarged MVBs structures (López-Marqués et al., 2010). This phenotype resembles that of lines overexpressing hyper-activated small GTPases involved in vesicle formation at the MVBs (e.g. ARA7), suggesting a possible coordinated action of AtALA2 with small GTPases (Jia et al., 2013).
P4 ATPases and polar growth
ALA deficiencies are often associated with defects related to polar cell growth. Mutants lacking AtALA3 were identified in a screen for altered trichome shapes (Zhang and Oppenheimer, 2009), and ala3 plants also display reduced pollen tube growth (McDowell et al., 2013). Mutants simultaneously lacking AtALA6 and AtALA7 also display pollen tube growth defects (McDowell et al., 2015). Double mutants lacking AtALA4 and AtALA5 are dwarfed and display multiple cell growth defects, including reduced root hair growth, impaired trichome formation and growth, and improperly expanded hypocotyl and leaf epidermal cells (Davis et al., 2020a).
The mechanism by which ALA proteins contribute to polar growth remains unclear. An important characteristic of some P4 ATPases is that they appear to be “electrogenic,” i.e. the charged headgroup of the lipid is flipped without a counter ion moving in the opposite direction (Tadini-Buoninsegni et al., 2019). In the case of PS transport, this flux would bring more negative charge to the cytosolic surfaces. While this transport may not be sufficient to influence membrane potential, anionic lipids can cluster in nanodomains at the inner leaflet of the plasma membrane, where the local negative charge is used to recruit soluble or lipid-anchored proteins (Platre et al., 2019; Colin and Jaillais, 2020; Noack and Jaillais, 2020). This has been demonstrated recently in pollen tubes, where AtALA3 plays roles in maintaining PS tip localization and potentially its asymmetry, which is critical for recruiting Rab GTPases and triggering polarized growth (Zhou et al., 2020).
P4 ATPases and plant development
While most mutant plant lines lacking individual P4 ATPases do not show any aberrant phenotypes in the absence of stressors, lines deficient in multiple P4 ATPases show severe fertility defects and dwarfism (McDowell et al., 2015; Davis et al., 2020a; Zhang et al, 2020). The only known exceptions to this rule are ala3 mutants, which display a dwarfism dependent on growth conditions, e.g. low temperature (Poulsen et al., 2008a; McDowell et al., 2013). Two models were proposed to explain this observation. First, disruption of membrane asymmetry was suggested to affect the activity of membrane transporters, resulting in defective nutrient uptake by roots. Second, lack of AtALA3-dependent vesicle production at the Golgi was suggested to result in the mistargeting of root membrane transporters required for nutrient uptake.
Recently, a link between AtALA3 and hormone-dependent plant growth was demonstrated (Zhang et al., 2020). Auxin is a plant growth hormone that controls a myriad of processes in plant cells. PIN-formed (PIN) auxin efflux carriers show a polar distribution in cells, which allows for directional cell-to-cell transport of auxin and results in gradients of growth (Steinmann, 1999; Luschnig and Vert, 2014). Loss of AtALA3 results in altered PIN1 trafficking in root epidermal cells, shifting PIN1 localization from the basal to the apical side of cells (Zhang et al., 2020). Additionally, PIN2 shows increased aggregation in intracellular locations in ala3 plants, but is still predominantly present on the basal side of cells at the root cortex. Plant lines containing multiple deletions of P4A-e ATPases, such as ala3/5/9 and ala3/4/5/9/11, show larger intracellular PIN2 aggregations, and in the sextuple mutant ala3/4/5/9/10/11, PIN2 localization is completely shifted to the apical side of cells. In agreement with cellular phenotypes, multiple deletions of P4-e ATPases result in severe plant developmental defects and the lack of gravitropic responses that can be linked to a lack of amyloplasts (Zhang et al., 2020). Thus, plant P4 ATPases have overlapping functions in promoting plant cell polarity. Disruption of the direct physical interaction of AtALA3 with ARF-GEFs seems to be the cause underlying PIN trafficking defects in ala3 mutants (Zhang et al., 2020), supporting a role for AtALA3 (and other P4 ATPases) as scaffold proteins that recruit vesicular machinery to specific cellular locations. This scaffolding role has also been proposed for P4 ATPases from yeast, such as endosomal Neo1p and trans-Golgi network Drs2p (Wicky et al., 2004; Natarajan et al., 2009). Thus, scaffolding of the vesiculation machinery might be a conserved function of flippases across organisms.
P4 ATPases and the regulation of cellular lipid homeostasis
Unidirectional flipping of phospholipids across cellular membranes affects lipid homeostasis in plant cells and alters the distribution of lipids between organelles. For example, disruption of AtALA10 affects the lipid composition of the chloroplast outer envelope (Botella et al., 2016; Salvaing et al., 2020). An altered membrane lipid composition might explain the increased low temperature sensitivity of plants with reduced expression of P4 ATPases such as AtALA1 (Gomès et al., 2000) and AtALA3 (McDowell et al., 2013), or the high-temperature sensitivity of lines deficient in AtALA6 (Niu et al., 2017) or both AtALA6 and -7 (McDowell et al., 2015). Whether this reflects a direct role of P4 ATPases in membrane adaptation to temperature changes is not known.
The developmental defects observed in mutant lines lacking both AtALA4 and -5 have been linked to a potential perturbation in sphingolipid homeostasis. The involvement of sphingolipid homeostasis in plant development and biotic and abiotic stress responses is well known (for reviews, see Grison et al., 2015; Michaelson et al., 2016; Ali et al., 2018). Of all sphingolipids, GlcCer is the only glycosphingolipid present in yeast, animals, and plants (Warnecke and Heinz, 2003). However, desaturation levels and fatty acid tail length vary between organisms. For plants, GlcCers typically contain at least one hydroxyl-group modification and a specific C8 cis double-bond on the sphingoid base (Sph), as well as a very-long-chain fatty acid hydroxylated at the C2 position. Transport of GlcCers has been demonstrated for yeast and mammalian P4 ATPases (Roland et al., 2019; Martin et al., 2020). While there is evidence for a weak GlcCer transport activity by plant AtALA10 (Jensen et al., 2017), the same fluorescently labeled GlcCer substrates were not transported by AtALA5 (Davis et al., 2020a). Nonetheless, ala4/5 double mutants have a defect in glycosphingolipid homeostasis that results in an increased accumulation of GlcCers and a reduction of glycosylinositolphosphorylceramides (GIPCs; Davis et al., 2020a), both of which are major components of plant plasma membranes (Cacas et al., 2016).
A working model for the role of flippases in GlcCer-related development was proposed (Davis et al., 2020b). In this model, flippases are required to transport GlcCer from extracellular/lumenal leaflets to cytosolic leaflets of membranes, where cytosolic glucosylceramidases can remove the glucose headgroup. After glucose removal, the ceramide portion has the potential to be further degraded to a free fatty acid and a Sph (e.g. phytosphingosine), which can then be phosphorylated to produce sphingosine-1-phosphate (Sph-1-P, e.g. phytosphingosine-1-P). Free fatty acids, Sph, and Sph-1-P are all secondary messengers (Okazaki and Saito, 2014; Lim et al., 2017; Huby et al., 2020). Thus, the lack of GlcCer translocation across membranes in the absence of AtALA4 and -5 could result in GlcCer accumulation in the extracytosolic leaflet or a reduction in essential signaling lipids, either of which might lead to the severe dwarfism observed in ala4/5 plants (Davis et al., 2020a, 2020b).
P4 ATPases and plant defense
Expression data in public databases suggest that a number of P4 ATPases in Arabidopsis are upregulated under pathogen attack. The Arabidopsis P4 ATPases AtALA1 and -2 are both required for antiviral RNA interference (RNAi) by a mechanism that remains unclear (Guo et al., 2017; Zhu et al., 2017). Plant viruses alter the organization of the ER to form membrane invaginations, so-called spherules, where viral replication occurs. AtALA1 and -2 have been proposed to help generate similar subcellular membrane microdomains in which RNAi takes place (Guo et al., 2017). Alternatively, AtALA1 and -2 might disrupt spherule formation leaving the virus exposed to the cytosol, so that RNAi becomes possible.
In response to infection by several different pathogens, the Arabidopsis PENETRATION 3 (PEN3) ABC transporter accumulates at the plasma membrane in papillae, where it mediates the accumulation of secondary metabolites (Underwood and Somerville, 2013; Underwood et al., 2017). Loss of AtALA3 results in mislocalization of PEN3, which accumulates in internal membranes instead of traveling in secretory vesicles to the plasma membrane (Underwood et al., 2017). Thus, AtALA3 is required for proper exit from the Golgi of a membrane protein important for the plant immune response. Since AtALA3 interacts with ARF-GEFs (Zhang et al., 2020), mislocalization of PEN3 might be the result of an ala3-scaffold deficiency that disrupts the recruitment of the vesiculation machinery. Interestingly, PEN3 trafficking seems to be unaffected by treatment with brefeldin A (BFA), a chemical compound that inhibits the activity of some ARF-GEFs and subsequently small GTPases. Of the two ARF-GEFs known to interact with AtALA3, GNOM is BFA-sensitive, while BIG3 is not (Nebenfuhr et al., 2002; Nielsen et al., 2012), suggesting that failed recruitment of BIG3 might be the cause for PEN3 mislocalization in ala3 plants.
A newly reported line of plant defense involves the exudation of extracellular vesicles (named exosomes) into the apoplast (Cui et al., 2020a), that can, for instance, deliver small RNAs to pathogens for silencing of virulence genes (Cai et al., 2018). It remains to be determined whether P4 ATPases play a role in forming these membrane structures.
P4 ATPases and signal transduction
Flipping of phospholipids from the extracytosolic leaflet of the plasma membrane has been implicated as a means of communication between plants and their environment. For example, arbuscular mycorrhizae secrete the water-soluble lipid lyso-PC, which is perceived by roots as a signal that induces expression of phosphate transporter genes (Drissner et al., 2007). In Arabidopsis, lyso-PC is taken up from the environment (e.g. rhizosphere) and flipped across the plasma membrane by AtALA10 (Poulsen et al., 2015). Expression of AtALA10 is upregulated under phosphate starvation and ala10 mutants show longer primary roots than wild type, indicating that the plants fail to sense phosphate starvation and trigger the stress adaptation program that results in primary root growth arrest combined with the development of a higher number of secondary roots (Poulsen et al., 2015). While wild-type plants respond to the exogenous addition of lyso-PC by abandoning the root growth response to phosphate starvation, ala10 mutant plants fail to do so, indicating a lack of lyso-PC signaling. Whether this lack of signaling affects colonization by mycorrhizae remains to be tested.
Regulation of P4 ATPases
Due to their function in processes essential for cell survival, P4 ATPases are expected to be heavily regulated. Plant P4 ATPases associate with a β-subunit (proteins known as ALA interacting subunits, ALISs) to leave the ER (Poulsen et al., 2008a, 2015; López-Marqués et al., 2010, 2012; Botella et al., 2016). Independent of the 26S proteasome pathway, ubiquitination of AtALA10 by the plant U-box type 11 (PUB11) E3 ubiquitin ligase appears to determine its association with specific ALISs and the final cellular localization of the complex (Salvaing et al., 2020). Virtually nothing else is known about the regulation of plant P4 ATPases, but a picture is beginning to emerge for fungal and animal P4 ATPases. Two main types of regulatory mechanisms have been identified for yeast and mammalian P4 ATPases: autoinhibition by their C-terminal end and activation by protein kinases (Nakano et al., 2008; Zhou et al., 2013; Chalat et al., 2016; Bai et al., 2019; Timcenko et al., 2019).
Yeast Drs2p and mammalian ATP8A2 both have autoinhibitory C-terminal domains (Zhou et al., 2013; Chalat et al., 2016). Recent structural information obtained for several P4 ATPases in fungi and humans suggests that the C-terminal domain interacts with the cytosolic soluble domains of the protein, physically preventing the structural movements that allow ATP binding and hydrolysis (Bai et al., 2019; Hiraizumi et al., 2019; Timcenko et al., 2019; He et al., 2020; Nakanishi et al., 2020). The C-terminal domain of Drs2p interacts with an ARF-GEF (Gea2p) and the anionic phospholipid phosphatidylinositol-4-phosphate (PI4P; Natarajan et al., 2009). The cooperative interaction of both factors is necessary to release autoinhibition, and full Drs2p activation requires further interaction of the N-terminal domain with a small GTPase (Arl1p; Tsai et al., 2013; Timcenko et al., 2019). Structural information reveals that the binding site for PI4P is between several transmembrane helixes and residues at the cytosolic C terminus that are necessary for specific recognition of the PI4P headgroup (Timcenko et al., 2019). In general, the N- and C-termini of plant P4 ATPases are shorter than those of their fungal and mammalian counterparts, and evidence that plant P4 ATPases are autoinhibited is lacking. However, interaction with ARF-GEFs has been shown for AtALA3 (see above) and various autoinhibitory mechanisms have been identified for many members of the P-type ATPase superfamily (Palmgren and Nissen, 2011; Zhang et al., 2020).
Yeast P4 ATPases are also regulated by the so-called flippase kinases (Fpks). Fpks phosphorylate four of the five yeast P4 ATPases (Drs2p and Dnf1–3p; the exception being Neo1p; Nakano et al., 2008). Dnf1–3p are activated by Fpk-dependent phosphorylation (Nakano et al., 2008; Frøsig et al., 2020). Coordinated regulation of the Dnf proteins by the same kinases was suggested to be necessary for the control of polarized growth events, based on the observation that triple mutants lacking DNF1–3 show exacerbated polarized growth defects relative to mutants lacking only two DNFs (Frøsig et al., 2020). So far, evidence of kinase regulation of plant P4 ATPases is lacking, but numerous homologs of yeast Fpks exist in plants.
P5 ATPases
P5 ATPases are divided into two subfamilies, P5A and P5B, among which only P5A is present in plants. As in other eukaryotic cells, plant P5A ATPases localize to the ER (Jakobsen et al., 2005; Dunkley et al., 2006; Ticconi et al., 2009; Sørensen et al., 2012) and the C-terminus is equipped with a KKXX-type ER retrieval signal, a characteristic of proteins that undergo retrograde Golgi-to-ER transport (Stornaiuolo et al., 2003; Ticconi et al., 2009). As P5 ATPases are phylogenetically related to P4 ATPases, which normally do not accumulate in the ER, it has been suggested that P5 ATPases serve as the ER counterpart of P4 ATPases (Poulsen et al., 2008b). However, the transported substrate of a P5A ATPase has not been identified and biochemically verified in any organism. A characteristic feature of P-type ATPases is that ATP turnover and transport are tightly coupled, i.e. the pumps are not active in the absence of their transported ligand(s) and vice versa. Strikingly, even purified P5A ATPase preparations are catalytically active, suggesting that the substrate follows the protein during purification (Sørensen et al., 2019). Dephosphorylation of the catalytic phosphoenzyme intermediate has been reported to be stimulated by elevated Ca2+ concentrations (Sørensen et al., 2012), but this is an indirect effect not linked to transport (Corradi et al., 2020). A human P5B ATPase has been implicated in the transport of polyamines (van Veen et al., 2020); however, P5A and P5B ATPases have a different number of transmembrane segments and vary with respect to conserved residues in the membrane domain, which suggests they have different transport specificities (Sørensen et al., 2010).
The best characterized P5A ATPase is Spf1p from S. cerevisiae. spf1 cells have a complex pleiotropic phenotype, including disrupted ER functions such as protein folding and processing (Cronin et al., 2002). Evidence suggests that Spf1p is involved in charge separation at the ER, insertion of tail-anchored (TA) proteins in the ER membrane, and maintenance of cellular sterol homeostasis (Sørensen et al., 2015, 2019), and it was proposed that Spf1p assists in sterol export from the ER to other cellular membranes (Sørensen et al., 2019). Insertion of TA proteins into membranes is inhibited by ergosterol (Kemper et al., 2008), and the ER-accumulation of sterols in spf1 cells may provide a mechanism for the mishandling of TA proteins in this background. Recently, it was shown that Spf1p interacts physically with a TA protein (McKenna et al., 2020). Of the two low-resolution Spf1p cryo-EM structures resolved in this work, one shows the presence of a mass resembling an alpha-helix half-way out of the membrane that was suggested to represent a bound TA protein, and it was proposed that Spf1p is a transmembrane helix dislocase involved in removing mistargeted transmembrane proteins from the ER (McKenna et al., 2020). Evidence for a similar P5 ATPase-dependent protein removal system has been recently found in the worm Caenorhabditis elegans (Qin et al., 2020). Future work is required to determine the specificity of TA protein binding to Spf1p and its impact on catalytic turnover.
Arabidopsis has a sole P5A ATPase, MALE IMAPIRED ANTHERS (MIA)/PHOSPHATE DEFICIENCY RESPONSE 2 (PDR2; Jakobsen et al., 2005; Ticconi et al., 2009), and in most other plants only a single P5A is present (Palmgren et al., 2020). Disruption of MIA/PDR2 disturbs ER homoeostasis and upregulates proteins involved in the unfolded protein response (Jakobsen et al., 2005; Ticconi et al., 2009). This observation suggests that MIA/PDR2, like its yeast counterpart Spf1p (Sørensen et al., 2015), is involved in a basal ER function related to general secretion. The mia/pdr2 mutants have collapsed pollen grains, which may be the result of inefficient export of cell wall materials (Jakobsen et al., 2005), and a short-root phenotype similar to that observed in wild-type plants subjected to phosphate deficiency (Ticconi et al., 2004, 2009), which may result from ER stress-activated autophagy in root meristematic cells (Naumann et al., 2019).
Concluding remarks
Plant P4 ATPases are emerging as master players in a number of essential cellular processes, including membrane trafficking, membrane lipid homeostasis, and lipid signaling (Figure 3). In agreement, plants lacking P4 ATPases show defects in polarized cell growth, temperature tolerance, pathogen defenses, and development that can be related to defects in membrane trafficking or lipid biogenesis and catabolism. While multiple recent studies have contributed to our understanding of the physiological roles of P4 ATPases in plants, several aspects remain to be studied (Outstanding Questions Box). Only limited information about how plant P4 ATPases are regulated is available, in contrast to the situation for their yeast and mammalian counterparts. In addition, most plant P4 ATPases do not have an assigned substrate, and it is possible that at least some of these proteins transport lipids specific for plant membranes. Finally, the function of P5 ATPases, which are closely related to P4 ATPases, is unknown and their possible role as lipid translocators remains to be determined. Other potential lipid translocators in plants include ABC transporters and scramblases, but their lipid transport capabilities remain to be tested experimentally in plant cells.
Figure 3.

Functions of plant P4 ATPases. Lipid translocation by P4 ATPases is essential for vesicle formation in the secretory pathway, which might be required for plasma membrane protein recycling (i) and is essential for delivery of proteins and material to the cellular surface (ii). Transport of specific lipids can be used to trigger signaling cascades (iii) or to generate a local lipid accumulation that can allow recruitment of lipid-binding proteins and their effectors to the membrane (iv). In addition, this lipid accumulation on the cytosolic leaflet of a biological membrane could be sensed by other proteins, affecting lipid homeostasis. As a result of their involvement in these crucial cellular functions, P4 ATPases affect numerous plant responses to their environment and development processes.
Outstanding questions box
Are plant P4 ATPases constitutively active as part of the basal cellular machinery, or are they first activated when required for specific cellular processes?
How are plant P4 ATPases regulated? Are they autoinhibited? Do regulatory events release this autoinhibition?
Do some plant P4 ATPases have highly specialized functions while others have general functions?
Are P4 ATPases directly involved in the formation of membrane structures that function in plant defense, such as exosomes or spherules?
What is the role of P5 ATPases? Are they the flippases of the endoplasmic reticulum (ER)? Do they assist in the export of sterols or in the removal of mistargeted transmembrane proteins from the ER?
Do plant ATP-binding cassette (ABC) transporters and scramblases also regulate membrane lipid asymmetries?
Funding
This work was supported by the Innovation Fund Denmark (LESSISMORE; to M.P.), the Carlsberg Foundation (RaisingQuinoa; project number CF18-1113; to M.P.), the Novo Nordisk Foundation (NovoCrops; project number 2019OC53580; to M.P. and R.L.L.-M.), the United States Department of Agriculture (HATCH grant no. NEV00384; to J.F.H.), and the National Science Foundation (IOS grant no. 1656774; to J.F.H.).
Conflict of interest statement. None declared.
All authors played equal roles in authoring the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Rosa L. López-Marqués (rlo@plen.ku.dk).
References
- Adhikari H, Cullen PJ (2015) Role of phosphatidylinositol phosphate signaling in the regulation of the filamentous-growth mitogen-activated protein kinase pathway. Eukaryot Cell 14:427–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali U, Li H, Wang X, Guo L (2018) Emerging roles of sphingolipid signaling in plant response to biotic and abiotic stresses. Mol Plant 11:1328–1343 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46:84–101 [DOI] [PubMed] [Google Scholar]
- Bai L, Kovach A, You Q, Hsu H, Zhao G, Li H (2019) Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p. Nat Commun 10:4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132:618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G, Weigel A V. (2018) Transport and sorting in the Golgi complex: multiple mechanisms sort diverse cargo. Curr Opin Cell Biol 50:94–101 [DOI] [PubMed] [Google Scholar]
- Botella C, Sautron E, Boudiere L, Michaud M, Dubots E, Yamaryo-Botté Y, Albrieux C, Marechal E, Block MA, Jouhet J. et al. (2016) ALA10, a phospholipid flippase, controls FAD2/FAD3 desaturation of phosphatidylcholine in the ER, and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiol 170:1300–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F (2018) Transport from the endoplasmic reticulum to the Golgi in plants: where are we now? Semin Cell Dev Biol 80:94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas JL, Buré C, Grosjean K, Gerbeau-Pissot P, Lherminier J, Rombouts Y, Maes E, Bossard C, Gronnier J, Furt F, et al. (2016) Revisiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol 170:367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Da Huang S, Jin H (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360:1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares D, Escribá PV, Rosselló CA (2019) Membrane lipid composition: effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int J Mol Sci 20:2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalat M, Moleschi K, Molday RS (2016) C-terminus of the P4-ATPase ATP8A2 functions in protein folding and regulation of phospholipid flippase activity. Mol Biol Cell 28:452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin LA, Jaillais Y (2020) Phospholipids across scales: lipid patterns and plant development. Curr Opin Plant Biol 53:1–9 [DOI] [PubMed] [Google Scholar]
- Corradi GR, Mazzitelli LR, Petrovich GD, Grenon P, Sørensen DM, Palmgren M, de Tezanos Pinto F, Adamo HP (2020) Reduction of the P5A-ATPase Spf1p phosphoenzyme by a Ca2+-dependent phosphatase. PLoS ONE 15:e0232476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SR, Rao R, Hampton RY (2002) Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J Cell Biol 157:1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Gao J, He Y, Jiang L (2020a) Plant extracellular vesicles. Protoplasma 257:3–12 [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Hu S, Jiang L (2020b) Vacuole biogenesis in plants: how many vacuoles, how many models? Trends Plant Sci 25:538–548 [DOI] [PubMed] [Google Scholar]
- Davis JA, Pares RB, Bernstein T, McDowell SC, Brown E, Stubrich J, Rosenberg A, Cahoon EB, Cahoon RE, Poulsen LR, et al. (2020a) The lipid flippases ALA4 and ALA5 play critical roles in cell expansion and plant growth. Plant Physiol 182:2111–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JA, Pares RB, Palmgren M, López-Marqués RL, Harper JF (2020b) A potential pathway for flippase-facilitated glucosylceramide catabolism in plants. Plant Signal Behav 15: 1783486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissner D, Kunze G, Callewaert N, Gehrig P, Tamasloukht M, Boller T, Felix G, Amrhein N, Bucher M (2007) Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318:265–268 [DOI] [PubMed] [Google Scholar]
- Dufourc EJ (2008) Sterols and membrane dynamics. J Chem Biol 1:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci U S A 103:6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Ishikawa T, Rennie EA, Murawska GM, Lao J, Yan J, Tsai AYL, Baidoo EEK, Xu J, Keasling JD, et al. (2016) Loss of inositol phosphorylceramide sphingolipid mannosylation induces plant immune responses and reduces cellulose content in Arabidopsis. Plant Cell 28:2991–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøsig MM, Costa SR, Liesche J, Østerberg JT, Hanisch S, Nintemann S, Sørensen H, Palmgren M, Pomorski TG, López-Marqués RL (2020) Pseudohyphal growth in Saccharomyces cerevisiae involves protein kinase-regulated lipid flippases. J Cell Sci 133:jcs235994. [DOI] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Shen J, Jiang L (2017) Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci 22:986–998 [DOI] [PubMed] [Google Scholar]
- Gomès E, Jakobsen MK, Axelsen KB, Geisler M, Palmgren MG (2000) Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell 12:2441–2453 [PMC free article] [PubMed] [Google Scholar]
- Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dörmann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V, et al. (2015) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27:1228–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Zavaliev R, Dong X (2017) Membrane trafficking in plant immunity. Mol Plant 10:1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Lu J, Wang X, Zhan B, Li W, Ding S-W (2017) Lipid flippases promote antiviral silencing and the biogenesis of viral and host siRNAs in Arabidopsis. Proc Natl Acad Sci U S A 114:1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xu J, Wu X, Li L (2020) Structures of a P4-ATPase lipid flippase in lipid bilayers. Protein Cell 11:458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraizumi M, Yamashita K, Nishizawa T, Nureki O (2019) Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365:1149–1155 [DOI] [PubMed] [Google Scholar]
- Huby E, Napier JA, Baillieul F, Michaelson LV, Dhondt‐Cordelier S (2020) Sphingolipids: towards an integrated view of metabolism during the plant stress response. New Phytol 225:659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen MK, Poulsen LR, Schulz A, Fleurat-Lessard P, Møller A, Husted S, Schiøtt M, Amtmann A, Palmgren MG (2005) Pollen development and fertilization in Arabidopsis is dependent on the MALE GAMETOGENESIS IMPAIRED ANTHERS gene encoding a type V P-type ATPase. Genes Dev 19:2757–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MS, Costa SR, Duelli AS, Andersen PA, Poulsen LR, Stanchev LD, Gourdon P, Palmgren M, Pomorski TG, López-Marqués RL (2017) Phospholipid flipping involves a central cavity in P4 ATPases. Sci Rep 7:17621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T, Gao C, Cui Y, Wang J, Ding Y, Cai Y, Ueda T, Nakano A, Jiang L (2013) ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. J Exp Bot 64:2817–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D (2008) Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci 121:1990–1998 [DOI] [PubMed] [Google Scholar]
- Lim G-H, Singhal R, Kachroo A, Kachroo P (2017) Fatty acid- and lipid-mediated signaling in plant defense. Annu Rev Phytopathol 55:505–536 [DOI] [PubMed] [Google Scholar]
- López-Marqués RL, Poulsen LR, Hanisch S, Meffert K, Buch-Pedersen MJ, Jakobsen MK, Pomorski TG, Palmgren MG (2010) Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA alpha-subunit. Mol Biol Cell 21:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués RL, Poulsen LR, Palmgren MG (2012) A putative plant aminophospholipid flippase, the Arabidopsis P4 ATPase ALA1, localizes to the plasma membrane following association with a β-subunit. PLoS ONE 7:e33042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG (2014) P4-ATPases: lipid flippases in cell membranes. Pflügers Arch 466:1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez PH, Schnaar RL (2009) Gangliosides in cell recognition and membrane protein regulation. Curr Opin Struct Biol 19:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Vert G (2014) The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141:2924–2938 [DOI] [PubMed] [Google Scholar]
- Martin S, Smolders S, Van den Haute C, Heeman B, van Veen S, Crosiers D, Beletchi I, Verstraeten A, Gossye H, Gelders G, et al. (2020) Mutated ATP10B increases Parkinson’s disease risk by compromising lysosomal glucosylceramide export. Acta Neuropathol 139:1001–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell SC, López-Marqués RL, Cohen T, Brown E, Rosenberg A, Palmgren MG, Harper JF (2015) Loss of the Arabidopsis thaliana P4-ATPases ALA6 and ALA7 impairs pollen fitness and alters the pollen tube plasma membrane. Front Plant Sci 6:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell SC, López-Marqués RL, Poulsen LR, Palmgren MG, Harper JF (2013) Loss of the Arabidopsis thaliana P4-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development. PLoS ONE 8:e62577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MJ, Sim SI, Ordureau A, Wei L, Harper JW, Shao S, Park E (2020) The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 369: eabc5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson LV, Napier JA, Molino D, Faure JD (2016) Plant sphingolipids: their importance in cellular organization and adaption. Biochim Biophys Acta 1861:1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer JC, Yu X, Albrecht S, Sicilia F, Huichalaf M, Ampuero D, Michaelson LV, Murphy AM, Matsunaga T, Kurz S, et al. (2013) Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25:1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msanne J, Chen M, Luttgeharm KD, Bradley AM, Mays ES, Paper JM, Boyle DL, Cahoon RE,, Schrick K, Cahoon EB (2015) Glucosylceramides are critical for cell-type differentiation and organogenesis, but not for cell viability in Arabidopsis. Plant J 84:188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Irie K, Segawa K, Hasegawa K, Fujiyoshi Y, Nagata S, Abe K (2020) Crystal structure of a human plasma membrane phospholipid flippase. J Biol Chem 295:10180–10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K (2008) Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol Biol Cell 19:1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Liu K, Patil D V, Sciorra VA, Jackson CL, Graham TR (2009) Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol 11:1421–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann C, Müller J, Sakhonwasee S, Wieghaus A, Hause G, Heisters M, Bürstenbinder K, Abel S (2019) The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol 179:460–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr A, Ritzenthaler C, Robinson DG (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130:1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson GL (2014) The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim Biophys Acta 1838:1451–1466 [DOI] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. et al. (2012) Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci U S A 109:11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nintemann SJ, Palmgren M, López-Marqués RL (2019) Catch you on the flip side: a critical review of flippase mutant phenotypes. Trends Plant Sci 24:468–478 [DOI] [PubMed] [Google Scholar]
- Niu Y, Qian D, Liu B, Ma J, Wan D, Wang X, He W, Xiang Y (2017) ALA6, a P4-type ATPase, is involved in heat stress responses in Arabidopsis thaliana. Front Plant Sci 8:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2020) Functions of anionic lipids in plants. Annu Rev Plant Biol 71:71–102 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Saito K (2014) Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J 79:584–596 [DOI] [PubMed] [Google Scholar]
- Paez Valencia J, Goodman K, Otegui MS (2016) Endocytosis and endosomal trafficking in plants. Annu Rev Plant Biol 67:309–335 [DOI] [PubMed] [Google Scholar]
- Palmgren M, Østerberg JT, Nintemann SJ, Poulsen LR, López-Marqués RL (2019) Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim Biophys Acta 1861:1135–1151 [DOI] [PubMed] [Google Scholar]
- Palmgren M, Sørensen DM, Hallström BM, Säll T, Broberg K (2020) Evolution of P2A and P5A ATPases: ancient gene duplications and the red algal connection to green plants revisited. Physiol Plant 168:630–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P (2011) P-type ATPases. Annu Rev Biophys 40:243–266 [DOI] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ (2007) Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 23:519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, del Mar Marquès-Bueno M, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, et al. (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364:57–62 [DOI] [PubMed] [Google Scholar]
- Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P, et al. (2018) A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev Cell 45:465–480.e11 [DOI] [PubMed] [Google Scholar]
- Poulsen LR,, López-Marqués RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG (2008a) The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20:658–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LR,, López-Marqués RL, Palmgren MG (2008b) Flippases: still more questions than answers. Cell Mol Life Sci 65:3119–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LR,, López-Marqués RL, Pedas PR, McDowell SC, Brown E, Kunze R, Harper JF, Pomorski TG, Palmgren M (2015) A phospholipid uptake system in the model plant Arabidopsis thaliana. Nat Commun 6:7649. [DOI] [PubMed] [Google Scholar]
- Qin Q, Zhao T, Zou W, Shen K, Wang X (2020) An endoplasmic reticulum ATPase safeguards endoplasmic reticulum identity by removing ectopically localized mitochondrial proteins. Cell Reports 33: 108363 [DOI] [PubMed] [Google Scholar]
- Richter S, Voss U, Jurgens G (2009) Post-Golgi traffic in plants. Traffic 10:819–828 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Emmenegger C, Xiao Q, Kostina NY, Sherman SE, Rahimi K, Partridge BE, Li S, Sahoo D, Reveron Perez AM, Buzzacchera I, et al. (2019) Encoding biological recognition in a bicomponent cell-membrane mimic. Proc Natl Acad Sci U S A 116:5376–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland BP, Naito T, Best JT, Arnaiz-Yépez C, Takatsu H, Yu RJ, Shin H-WW, Graham TR (2019) Yeast and human P4-ATPases transport glycosphingolipids using conserved structural motifs. J Biol Chem 294:1794–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaing J, Botella C, Albrieux C, Gros V, Block MA, Jouhet J (2020) PUB11-dependent ubiquitination of the phospholipid flippase ALA10 modifies ALA10 localization and affects the pool of linolenic phosphatidylcholine. Front Plant Sci 11:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen DM, Buch-Pedersen MJ, Palmgren MG (2010) Structural divergence between the two subgroups of P5 ATPases. Biochim Biophys Acta 1797:846–855 [DOI] [PubMed] [Google Scholar]
- Sørensen DM, Holen HW, Holemans T, Vangheluwe P, Palmgren MG (2015) Towards defining the substrate of orphan P5A-ATPases. Biochim Biophys Acta 1850:524–535 [DOI] [PubMed] [Google Scholar]
- Sørensen DM, Holen HW, Pedersen JT, Martens HJ, Silvestro D, Stanchev LD, Costa SR, Pomorski TG, López-Marqués RL, Palmgren M (2019) The P5A ATPase Spf1p is stimulated by phosphatidylinositol 4-phosphate and influences cellular sterol homeostasis. Mol Biol Cell 30:1069–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen DM, Møller AB, Jakobsen MK, Jensen MK, Vangheluwe P, Buch-Pedersen MJ, Palmgren MG (2012) Ca2+ induces spontaneous dephosphorylation of a novel P5A-type ATPase. J Biol Chem 287:28336–28348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann T (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286:316–318 [DOI] [PubMed] [Google Scholar]
- Stornaiuolo M, Lotti LV, Borgese N, Torrisi M-R, Mottola G, Martire G, Bonatti S (2003) KDEL and KKXX retrieval signals appended to the same reporter protein determine different trafficking between endoplasmic reticulum, intermediate compartment, and Golgi complex. Mol Biol Cell 14:889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadini-Buoninsegni F, Mikkelsen SA, Mogensen LS, Molday RS, Andersen JP (2019) Phosphatidylserine flipping by the P4-ATPase ATP8A2 is electrogenic. Proc Natl Acad Sci U S A 116:16332–16337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Naito T, Inoue T, Nakayama K, Takatsu H, Shin H-WH (2018) Phospholipid‐flipping activity of P4‐ATPase drives membrane curvature. EMBO J 37:e97705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37:801–814 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S (2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci U S A 106:14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timcenko M, Lyons JA, Januliene D, Ulstrup JJ, Dieudonné T, Montigny C, Ash M-RR, Karlsen JL, Boesen T, Kühlbrandt W, et al. (2019) Structure and autoregulation of a P4-ATPase lipid flippase. Nature 571:366–370 [DOI] [PubMed] [Google Scholar]
- Tsai P-C, Hsu J-W, Liu Y-W, Chen K-Y, Lee F-JSF (2013) Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc Natl Acad Sci U S A 110:E668–E677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W, Ryan A, Somerville SC (2017) An Arabidopsis lipid flippase is required for timely recruitment of defenses to the host–pathogen interface at the plant cell surface. Mol Plant 10:805–820 [DOI] [PubMed] [Google Scholar]
- Underwood W, Somerville SC (2013) Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc Natl Acad Sci U S A 110:12492–12497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen S, Martin S, Van den Haute C, Benoy V, Lyons J, Vanhoutte R, Kahler JP, Decuypere JP, Gelders G, Lambie E, et al. (2020) ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 578:419–424 [DOI] [PubMed] [Google Scholar]
- Warnecke D, Heinz E (2003) Recently discovered functions of glucosylceramides in plants and fungi. Cell Mol Life Sci 60:919–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S, Schwarz H, Singer-Krüger B (2004) Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol Cell Biol 24:7402–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Gatta AT, Levine TP (2019) Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol 20:85–101 [DOI] [PubMed] [Google Scholar]
- Zachowski A (2012) New insights in membrane lipid asymmetry in animal and plant cells. In Devaux PF, Herrmann A, eds, Transmembrane Dynamics of Lipids, John Wiley & Sons, Inc., Hoboken, NJ, USA, pp. 45–64 [Google Scholar]
- Zhang Q, Xiao S (2015) Lipids in salicylic acid-mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidylinositol 4-phosphate. Front Plant Sci 6:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Adamowski M, Marhava P, Tan S, Zhang Y, Rodriguez L, Zwiewka M, Pukyšová V, Sánchez AS, Raxwal VK, et al. (2020) Arabidopsis flippases cooperate with Arf GTPase exchange factors to regulate the trafficking and polarity of PIN auxin transporters. Plant Cell 32:1644–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XG, Oppenheimer DG (2009) Irregular Trichome Branch 2 (ITB2) encodes a putative aminophospholipid translocase that regulates trichome branch elongation in Arabidopsis. Plant J 60:195–206 [DOI] [PubMed] [Google Scholar]
- Zhou X, Sebastian TT, Graham TR (2013) Auto-inhibition of Drs2p, a yeast phospholipid flippase, by its carboxyl-terminal tail. J Biol Chem 288:31807–31815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang Y, Niu Y, Fan T, Qian D, Luo C, Shi Y, Li S, An L, Xiang Y (2020) The tip-localized phosphatidylserine established by Arabidopsis ALA3 is crucial for Rab GTPase-mediated vesicle trafficking and pollen tube growth. Plant Cell 32: 3170–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gao H, Xu G, Wu D, Song S, Jiang H, Zhu S, Qi T, Xie D (2017) Arabidopsis ALA1 and ALA2 mediate RNAi-based antiviral immunity. Front Plant Sci 8: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]