Abstract
Recent studies on membrane imaging in the plant endomembrane system by 2-D/3-D CLSM and TEM provide future perspectives of whole-cell ET and cryo-FIB-aided cryo-ET analysis.
Introduction
All eukaryotic cells contain an endomembrane system consisting of functionally distinct membranes and organelles that work together to mediate accurate and efficient transport of proteins and lipids, which is critical for cellular homeostasis (Stefan et al., 2017). A fundamental goal for cell biologists is to investigate the structure and dynamics of the endomembrane system in order to better understand its biological function (Carlton et al., 2020). Visualization of membrane structure and dynamics requires high spatial and temporal resolution, which has been recently achieved through various imaging approaches (Sezgin, 2017). Previously, protein localization and membrane dynamics have mainly been studied via two-dimensional (2-D) and 3-D confocal laser scanning microscopy (CLSM) live-cell imaging analysis, together with 2-D transmission electron microscope (TEM). However, these traditional techniques cannot overcome the 200-nm resolution limitation for CLSM and a lack of 3-D information for 2-D TEM. To get around these drawbacks, multiple super-resolution confocal systems with resolutions of 20–100 nm have recently been developed, including super-resolution confocal live imaging microscopy (SCLIM; Nakano, 2013), Airyscan confocal microscopy, structured illumination microscopy (SIM), stimulated emission depletion (STED) microscopy, photoactivated localization microscopy (PALM), and stochastic optical reconstruction microscopy (STORM; Schermelleh et al., 2019; Jacquemet et al., 2020). On the other hand, electron tomography (ET) can provide 3-D information of cellular structures with nanometer resolution, albeit with its own drawbacks, such as the limited sample section thickness (around 200–300 nm) for electrons to pass through (Otegui and Pennington, 2019). To obtain and construct 3-D models of large structures, given the limited section thickness, serial section ET can be used for 3-D reconstruction of large structures inside a cell. Recent studies on plant organelles have demonstrated that whole-cell ET is a powerful tool for large organelle modeling and reconstruction (Zhuang et al., 2017; Cui et al., 2019, 2020a). In addition, serial block-face scanning electron microscopy (SBF-SEM) and focused ion beam SEM (FIB-SEM) with image collection in a fully automated manner are also applicable to efficiently resolve 3-D cellular structures, using either chemically fixed, or high-pressure freezing (HPF)-prepared samples (Cocks et al., 2018; Cui et al., 2019; Spehner et al., 2020). Up till now, the best way to keep samples in their native condition is via cryo-fixation followed by subsequent cryo-EM/ET analysis, techniques that have been widely used to study the 3-D structure of biomacromolecules (Lyumkis, 2019). Combined with the newly developed cryo-FIB techniques, cryo-EM/ET with a direct electron detector (DED) and phase plate (Dai et al., 2013; Rast et al., 2019), a device that can enhance image contrast by increasing phase shift between diffracted electron waves and transmitted electron waves (Danev and Nagayama, 2010; Koning et al., 2018), are now the “Method of Choice” for visualization and analysis of cellular structures in vivo (Schaffer et al., 2019). Furthermore, recent advances in correlative light and electron microscopy (CLEM) have created opportunities to combine the unique capabilities of CLSM and EM/ET, by investigating the same section sequentially with cryo-CLSM and cryo-EM/ET (de Boer et al., 2015). With the maturation and timely application of these new techniques, research on cellular structures and their dynamics has reached a golden era with great promise for the future.
Advances
Room-temperature ET resolves plant membrane and organelle structures at nanoscale resolution such as ER-autophagosome contacts, plasmodesmata maturation, and symbiont membrane architecture.
Whole-cell ET analysis reveals that central vacuoles are derived from SVs after their fusion with MVBs. SVs then fuse to form large vacuoles.
Cryo-FIB-aided cryo-ET has become the method-of-choice in visualizing 3-D membrane structures and macromolecular assemblies within the native cellular environment, albeit constituting technical challenges when applied to plant materials.
Cryo-lift-out is a feasible method to prepare cellular samples for cryo-ET analysis from large tissues after HPF.
Cryo-CLEM can resolve protein–ultrastructure relationships at the whole-cell level with 4-nm isotropic resolution in mammalian cells with great promise in plant cells.
Most of these advanced techniques have been frequently used in research on yeast and mammalian cells, but have seen only limited application in plant cells. This is largely due to the thickness of plant cell walls and the autofluorescence of plant organelles such as chloroplasts (Tuijtel et al., 2019). In this update, we first summarize recent studies on membrane dynamics and structures in the plant endomembrane system using conventional 2-D CLSM, super-resolution microscopy (SRM), and EM approaches. We will then further discuss the latest findings using ET. Finally, we will use investigations on tip-vesicles (TVs) in pollen tubes as a proof-of-concept example to elaborate on the objectives, feasible approaches, and the challenges in sample preparation for ET, Cryo-FIB, and Cryo-ET analysis.
SRM for studying membrane dynamics in plants
Live-cell imaging plays a most important role in the investigation of membrane dynamics. Recent developments in SRM techniques have enabled unprecedented subcellular imaging at a nanoscale level, leading to an improved understanding of membrane structures and functions (Sezgin, 2017; Schermelleh et al., 2019). There are two main categories of SRM, including techniques that illuminate the sample by using patterned light (e.g. SIM and STED, Table 1; Chen et al., 2014) and single-molecule localization strategies that determine the position of individual fluorophores with sub-diffraction accuracy (e.g. PALM and STORM, Table 1; Komis et al., 2018). SRM approaches have been widely used in mammalian cells, including a recent study of the dynamic organization of chromatin domains (Nozaki et al., 2017; Xu et al., 2020) and the visualization of interactions between organelles and the cytoskeleton (Guo et al., 2018). However, the application of SRM in plant cells has been challenging because of a high-fluorescence background and the presence of a cell wall (Komis et al., 2015). Until now, most SRM studies in plants have been derived from SIM with a few examples of STED, PALM, and STORM (Kleine-Vehn et al., 2011; Demir et al., 2013; Dong et al., 2015; Wang et al., 2016; Martinière e,t al., 2019; Platre et al., 2019; Wang LH et al., 2019).
Table 1.
Examples of SRM and EM analysis on membrane dynamics and structures in plant cells
| Approaches | Methods | Membrane systems | Research highlights | References |
|---|---|---|---|---|
| SRM | SIM | Endosomes | Super-resolution study of dynamic behaviors of different stage endosomes in Arabidopsis root hairs | von Wangenheim (2016) |
| 3-D SIM | Microtubules | Super-resolution imaging of plant cortical microtubule and microtubule associated proteins using 3-D SIM | Vavrdová (2019) | |
| STED | PM | Super-resolution imaging of fluorescence-tagged PIN2 clusters in PM | Kleine-Vehn (2011) | |
| PALM | PM | Live super-resolution imaging with sptPALM to monitor ROP6, which is stabilized by phosphatidylserine on PM nanodomains and required for downstream auxin signaling | Platre (2019) | |
| PALM | PM | Dynamic study of PIP2;1, AHA2, LTI6b, and CLC2 particles within PM by sptPALM | Martinière (2019) | |
| STORM | / | Identifying NtPPME1-GFP-positive intracellular compartments that are distinct from various endosomal markers with STORM | Wang (2016) | |
| Airyscan | ER | Semi-high-resolution imaging of dynamic ER structures for quantitative analysis | Pain (2019) | |
| Airyscan | PM | Subdiffractional analysis of PM nanodomains for proteins involved in morphogenesis (PIN3) and pathogen perception (FLS2) | McKenna (2019) | |
| SCLIM | Golgi and ER | 4-D live-cell imaging of Golgi- and ER-localized proteins dynamics within Golgi entry core compartment | Ito (2018) | |
|
| ||||
| EM | SEM | / | Hierarchical imaging concept for targeted imaging of large volumes from cells to tissues | Wacker (2016) |
| SBF-SEM | Vacuole | SBF-SEM analysis of vacuole constriction upon auxin induction | Scheuring (2016) | |
| SBF-SEM | PSV | SBF-SEM analysis of the 3-D architecture of forming PSVs | Feeney (2018) | |
| FIB-SEM | Chloroplast | FIB-SEM analysis of cytoplasmic invaginations formation in chloroplast upon virus infection | Jin (2018) | |
| ET | TGN vesicles | ET analysis of TGN vesicles that transport specific materials in root border cells | Wang (2017) | |
| ET | Plasmodesmata | ET analysis of architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves | Nicolas (2017) | |
| ET | EV | ET analysis of EVs and membrane tubules during arbuscular invasion | Roth (2019) | |
| STEM-ET | Extensive membranes | Extensive membrane systems at the host–arbuscular mycorrhizal fungus interface revealed by STEM-ET | Ivanov (2019) | |
| ET | Vacuole | Vacuole biogenesis study using whole-cell ET in Arabidopsis root cells | Cui (2019) | |
| Cryo-ET | Thylakoid membranes | Native architecture of the Chlamydomonas thylakoid membranes revealed by in situ cryo-ET | Engel (2015a) | |
EM, electron microscopy; EV, extracellular vesicle; PSV, protein storage vacuole; sptPALM, single-particle tracking PALM.
SIM, with a lateral resolution of around 100 nm, allows visualization of various subcellular structures, including the endoplasmic reticulum (ER), ER exit sites (ERESs), endosomes, plasmodesmata, as well as microdomians or more precisely nanodomains (Ott, 2017), on the plasma membrane (PM; Knox et al., 2015; Zeng et al., 2015; von Wangenheim et al., 2016; Komis et al., 2018). 3-D SIM was recently used to visualize cortical microtubules and illustrate their interactions with microtubule-associated proteins in plants (Vavrdová et al., 2019).
In STED imaging, samples are scanned with a central activation beam overlapped by an annular-shaped, high-power depletion beam to achieve high-resolution imaging. Application of STED in plants has been limited to tissues or cells without light-absorbing chloroplasts because such absorbance would lead to cell damage (Schubert, 2017; McKenna et al., 2019). For example, STED was successfully used to visualize fluorescence-tagged PIN2 clustering in the PM of transgenic Arabidopsis root cells (Kleine-Vehn et al., 2011). In addition, the recently improved STED system allowed visualization of YFP-RABG3f on the membrane of multivesicular bodies (MVBs; ∼100–400 nm in diameter) in living root cells of transgenic Arabidopsis plants (Figure 1, unpublished results).
Figure 1.
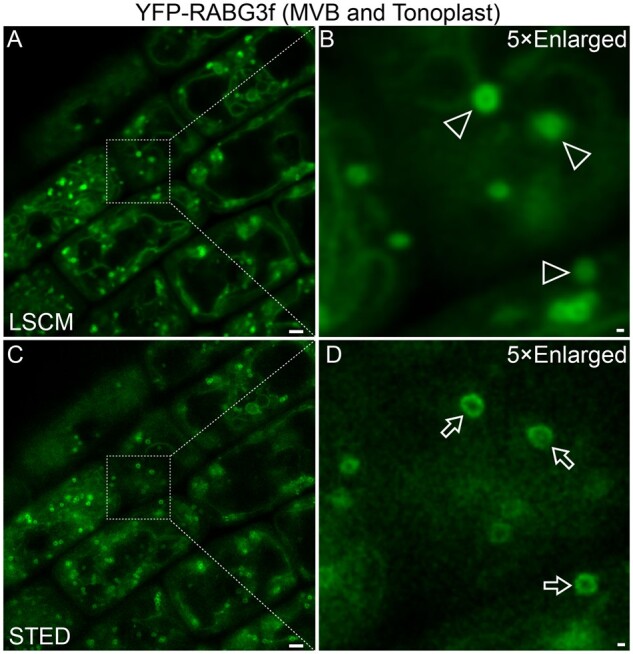
Visualization of MVBs in plant root cells: CLSM versus STED. Root cells of transgenic Arabidopsis plants expressing YFP-RABG3f were subjected to live-cell imaging at identical position via conventional CLSM (A and B) and improved STED (C and D). YFP-RABG3f is known to locate on both MVBs and the tonoplast. Note the sharp ring-like structures (arrows) in STED-derived C/D versus the corresponding dots (arrowheads) in CLSM-derived A/B. Scale bars: 1 μm in A/C; 100 nm in B/D.
Both PALM and STORM can achieve very high (∼20 nm) spatial resolution through collecting information on the precise localization of many individual molecules. The acquisition step in PALM and STORM is a rather static process with respect to the time window required to record all the events to build the super-resolution image. Besides, a relatively low-fluorescence background is crucial for these techniques. Thus, both PALM and STORM are commonly combined with TIRF microscopy to restrict the thickness of the excitation field so that only fluorophores within 100–200 nm of the sample near coverslip are efficiently excited. Owing to the thick cell wall (100–1,000 nm), it is therefore very challenging to apply PALM and STORM in studying plant cells albeit with examples of success. A recent live PALM analysis revealed that ROP6 is stabilized by phosphatidylserine into PM nanodomains, which is required for auxin signaling (Platre et al., 2019; Jaillais and Ott, 2020). In another study, the diffusion of PIP2;1, AHA2, LTI6b, and CLC2 within the PM has been studied through single-particle-tracking PALM (Martinière et a,l., 2019). STORM has also been used to visualize microtubules close to the PM (Dong et al., 2015). Finally, a modified STORM system has been used to distinguish NtPPME1-GFP positive secretory vesicles from the Golgi apparatus and trans-Golgi network (TGN) in plant cells (Wang et al., 2016).
The Airyscanning technique, which shares the same principle as CLSM but with an “Airyscan” detector concept, also drastically improves imaging resolution (about 140 nm in lateral resolution; Weisshart, 2014). This method complements some limitations in SIM, STED, and PALM (Weisshart, 2014; McKenna et al., 2019). For example, an Airyscan confocal system was used to obtain high-resolution information on dynamic ER structures with quantitative analysis in tobacco (Nicotiana tabacum) leaf epidermal cells (Pain et al., 2019). In another study, Airyscan imaging of the membrane protein nanodomains of PIN3 and FLS2 demonstrated that their size and dynamics are significantly affected upon perturbation of the plant cell wall (McKenna et al., 2019).
SCLIM is another super-resolution imaging system sharing the same principle as CLSM. Upon deconvolution, small vesicles of 50–60-nm diameter and a tubular structure of 50-nm diameter can be clearly captured in a 3-D movie by SCLIM (Nakano, 2013). In addition, such a system enables 4-D imaging with extremely high speed (e.g. 1,000 frames per second in 2D, 50–100 Z-stacks in a few seconds, at the completely simultaneous two-color separation) and high resolution in multiple colors at the same time (Nakano, 2013; Ito et al., 2018).
ET provides new insights into membrane dynamics
Although SRM imaging techniques have greatly promoted studies on membrane dynamics in living cells, they still have a relatively limited resolution of 20–100 nm when compared with conventional TEM, which can resolve cellular structures at nanometer resolution (Winey et al., 2014; Miranda et al., 2015), albeit with the drawback of 2-D images. Two approaches are commonly used to obtain 3-D volumes. An intuitive operation is to stack serial 2-D (XY) images together, which essentially represents SEM in 2-D image acquisition. The serial sections can be either preserved for imaging multiple times (array tomography; Wacker et al., 2016), or subjected to only one image cycle before being destroyed by SBF imaging methods (Denk and Horstmann, 2004; Jin et al., 2018; Hoffman et al., 2020). This method is particularly competitive in terms of imaging large volumes (Scheuring et al., 2016; Feeney et al., 2018), but the resolution in the Z dimension is relatively limited by the section thickness. The other promising technique, ET, first takes multiple 2-D projection images under TEM over a wide range of viewing directions (a tilt series), followed by back-projecting these 2-D images with appropriate weighting to generate a 3-D volume (McIntosh et al., 2005). Compared with sequential stacked 2-D images, ET is more powerful in getting 3-D representations of macromolecules and organelles in their cellular environment at significantly higher resolution. Thus, it has become a tempting option in 3-D membrane morphology and structure studies. When the cellular structure is well preserved, fine ultrastructure at a nanometer scale can be morphometrically analyzed by ET. Compared with 2-D TEM analysis, a complete 3-D ultrastructure allows an unbiased full view of the compartment of interest. Moreover, specific slice views of certain slicing orientations can be extracted from any part of the full tomogram for model building and further analysis.
In combination with HPF and freeze substitution (Karahara and Kang, 2014), ET was adapted and used to study cytokinesis in plant cells (Otegui et al., 2001). It has since become the method of choice to study various organelles and transport vesicles in the plant endomembrane system, including the ER, the Golgi apparatus, and the TGN (Otegui et al., 2006; Leitz et al., 2009; Kang et al., 2011). Several recent studies have extended ET analysis nicely to other complex membrane structures in plant cells, including the identification of a distinct class of vesicles derived from the TGN that mediate secretion of xylogalacturonan in alfalfa (Medicago sativa) root border cells (Wang et al., 2017), and the characterization of a linear developmental sequence of thylakoid formation and the involvement of thylakoid-bound polysomes and the dynamin-related protein FZL in Arabidopsis (Liang et al., 2018). In recent years, the power of ET analysis has been demondstrated for other organelles in plant responses and development, including autophagosomes during autophagy, plasmodesmata in cell–cell communication, membrane morphologies during plant–microbe interactions, and vacuole formation in root growth (Zhuang et al., 2017; Cui et al., 2019; Huang et al., 2019; Ivanov et al., 2019; Roth et al., 2019).
Organelle biogenesis in plants revealed by ET
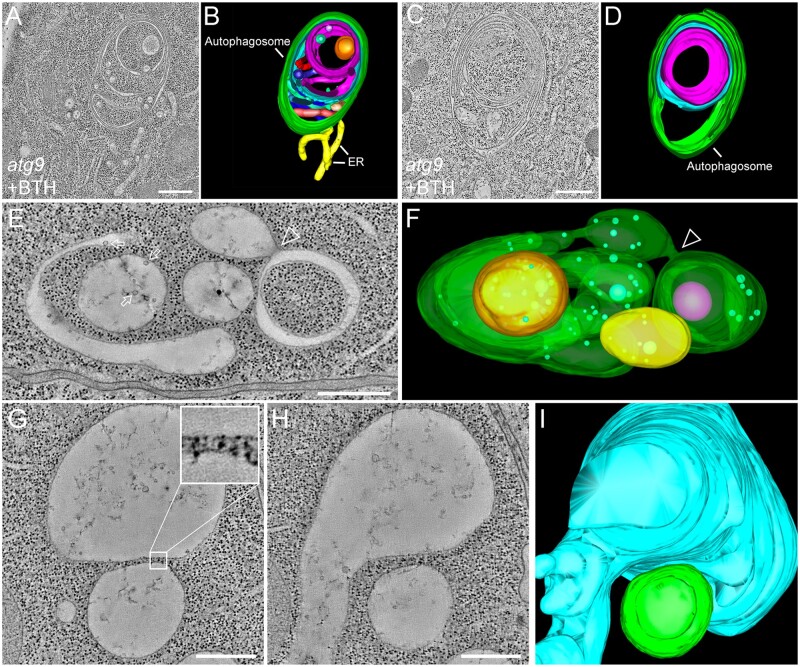
Autophagy is a highly conserved metabolic process in eukaryotic cells that is essential for plant development and growth as well as in response to various stresses. The spatiotemporal formation of autophagosomes is an example for exquisite membrane dynamics in organelle biogenesis. As the sole transmembrane autophagy-related (ATG) protein, ATG9 has long been speculated to play an essential role in ER-derived autophagosome formation. This is supported by extensive time-lapse live-cell CLSM imaging showing the dynamic association of the ER with autophagosomal markers in yeast and mammalian cells in 4D (Kakuta et al., 2012; Orsi et al., 2012; Karanasios et al., 2016). A direct connection between an autophagosome and ER has also been visualized in a recent study on the role of Arabidopsis ATG9 in regulating autophagosome formation in plants (Zhuang et al., 2017). Here, an ET analysis revealed a direct membrane connection between the ER and the abnormal autophagosome with a complicated multi-layer structure in atg9 mutants (Figure 2, A–D; Zhuang et al., 2017).
Figure 2.
The ET analysis of cellular organelles in plant cells. (A–D) Tomographic slices (A and C) and their corresponding 3-D models (B and D), respectively, showing the multi-layer autophagosomes (green) and its direct connection to the ER (yellow) in the Arabidopsis atg9 mutants after BTH treatment to induce autophagy. (E) and (F) The ET image and the corresponding model show complicated vacuole formation at the early stage of Arabidopsis root cortex. The arrows indicate examples of ILVs inside vacuoles. The arrowhead indicates the fusion between SV and cup-shaped vacuoles. (G–I) Tomographic slice (G and H) and 3-D model (I) show the closely associated relationship between two vacuoles with a minimum distance of 20 nm between the tonoplasts of the two vacuoles (e.g. enlarged insert in G). Bars = 500 nm. (A) and (B) are adapted from (Zhuang et al. 2017; Copyright 2017 National Academy of Sciences); (D) is adapted from (Jiang 2017; Copyright 2017 Springer Science + Business Media LLC).
The fine structure of plasmodesma—a model for ET
Plasmodesmata are plant-unique structures consisting of small channels that traverse cell walls for communication and transport between cells. For a long time, it has been challenging to obtain satisfying details of plasmodesmata due to their special position and nanoscopic size. Extensive conventional 2-D TEM analyses defined the plasmodesma as a micro channel with a cytoplasmic sleeve, a space between the tubular ER and the PM, spanned by spoke-elements (Overall et al., 1982; Ding et al., 1992; Badelt et al., 1994). However, a recent ET analysis on plasmodesmata in root tip columella tissue showed clear traceability of cell lineage, thus defining a new type of plasmodesma without a visible intermembrane space between the ER and the PM. This has led to a substantial remodeling of the ER–PM connections ranging from very tight contacts to intermembrane gaps with spokes during plasmodesmal maturation (Nicolas et al., 2017). In addition, ET was also recently used to calculate the plasmodesmal aperture size in Arabidopsis mutants and overexpression lines of the lipid raft regulatory protein Remorin (REM1.2 and REM1.3), demonstrating their important role in regulating plasmodesmal aperture (Huang et al., 2019).
Symbiosis-membrane morphologies between species by ET
ET has also been extended to study the ephemeral membrane profile of plant–microbe symbionts (Roth et al., 2019). Taking the inherent advantages of 2-D TEM, Roth et al. (2019) first obtained confirmative evidence of extensive membrane tubules (memtubs) in the extracellular space between the fungal PM and cell wall as previously described (Marchant and Moore, 1973). With additional support from ET data, they demonstrated continuity between the memtubs and the fungal PM. Furthermore, a connection between vesicles within the peri-arbuscular space and the plant PM was also revealed by ET (Roth et al., 2019). Such detailed and covert membrane connections caused by the dynamic and convoluted nature of the membranes involved, can now be neatly illustrated by ET, and are hardly observed in the projected and compressed 2-D micrographs.
Another ET study on arbuscular mycorrhiza focused more on the classification of the membrane compartments that lie within the peri-arbuscular space using another plant host species (Ivanov et al., 2019). This study adopted scanning TEM-ET (STEM-ET) to image larger intact volumes (section thickness up to 1 μm) when compared with ET (250–300-nm section thickness). In STEM, the electron beam is focused into a fine spot for scanning over the sample, which makes it more resistant to the energy loss of the electrons. Therefore, STEM is less limited with respect to the imaging area and hence capable of imaging larger volumes (Aoyama et al., 2008; Hohmann-Marriott et al., 2009; Wolf et al., 2014), albeit with the potential risk of losing information caused by long periods of exposure to the intense electron beam.
Whole-cell ET
ET analysis for small cellular structures has considerably advanced our understanding of intra- and intercellular membrane dynamics at nanometer resolution. However, it is challenging to use ET analysis on single sections to investigate large-size organelles such as vacuoles or membrane morphologies of multiple organelles with significant size differences in plant cells, which can be addressed using whole-cell ET analysis. Whole-cell morphological analysis on vacuoles has been performed by SBF-SEM on Arabidopsis embryo cells, and showed that the Arabidopsis protein storage vacuoles arise by the remodeling of preexisting vacuoles rather than by de novo biogenesis (Feeney et al., 2018). A similar approach was applied to analyze auxin-induced vacuolar constrictions in Arabidopsis root cells (Scheuring et al., 2016). Although the SBF-SEM data were very informative at the whole-cell level, the 100-nm serial section thickness restricts the Z-axis resolution of the rendered model, whereas the consumption of samples also prevents re-imaging of the targeted areas at higher magnification. Therefore, ET analysis at a whole-cell level is needed to completely reflect the large-size organelles or multiple organelles of different size within cells. Indeed, despite the technical challenge involved, recent whole-cell ET analysis of vacuole formation in Arabidopsis root cells has allowed a new model of vacuole biogenesis to be formulated (Cui et al., 2019). In this study, a whole-cell vacuole model was reconstructed by ET, which demonstrated that the nascent small vacuoles (SVs) contain intraluminal vesicles (ILVs) that are derived from MVBs. The fusion among SVs and other tubular vacuoles leads to the formation of larger vacuoles (Figure 2, E and F; Cui et al., 2019). Of note, the vacuoles in root cells of wild-type and the fragmented vacuoles in free1-mutant cells were shown to be distinct separated structures (Cui et al., 2019). Indeed, as revealed by ET, the distance between two vacuoles can be as close as 20 nm (Figure 2, G–I), which can hardly be resolved by CLSM (Cui et al., 2019).
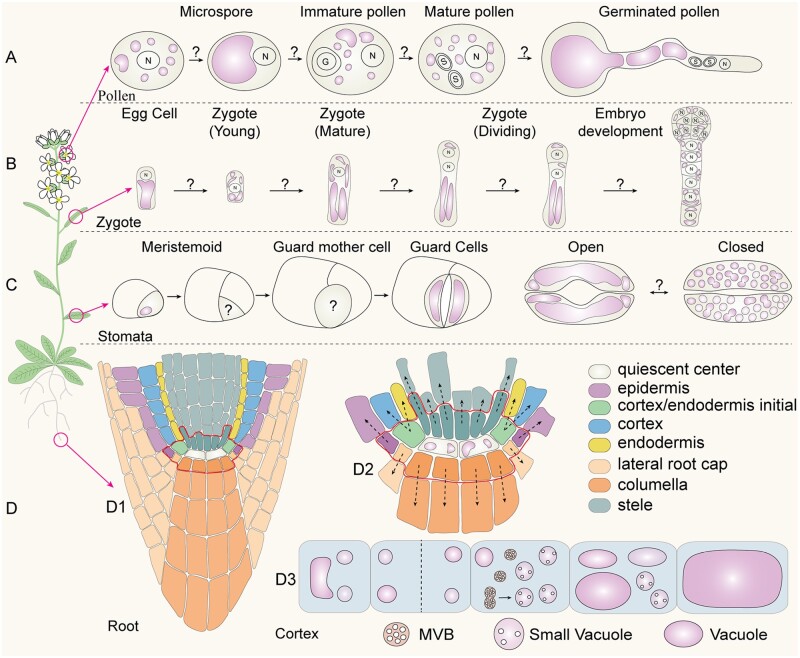
The model was based on data from a whole-cell ET analysis of three cortex cells with different vacuole sizes (Cui et al., 2019). Interestingly, in addition to the cortex layer cells showing developmental profiles of vacuole formation, other cell layers in the Arabidopsis root tip may represent excellent alternative systems for investigating models of vacuole biogenesis and function in plants via whole-cell ET, including epidermis, endodermis, root cap, lateral root cap, columella, and stele. Although vacuoles already exist in the quiescent center (QC) and initial cells (Figure 3, D; Viotti et al., 2013; Cui et al., 2020a), plant cells can also synthesize vacuoles de novo from MVBs in the root cortex (Cui et al., 2019). It remains unknown whether vacuoles can be generated de novo in different root cell layers and how these cells coordinate the inherited vacuoles (e.g. in the QC) with the newly formed ones (e.g. in cortex cells). Besides root cells, vacuoles also exist and rapidly change their morphology during various cellular processes in plants, including pollen development and germination (Figure 3, A), embryogenesis (Figure 3, B), stomatal development, and stomatal opening and closing (Figure 3, C). Most previous studies on vacuole dynamics and formation were mainly performed using CLSM and 2-D TEM. It will be of great interest in future studies to use the whole-cell ET analysis to further illustrate vacuole dynamics and formation as well as the well-coordinated cycles of vacuole fusion and fission processes and vacuolar convolution during these important biological processes (Figure 3, A–D).
Figure 3.
Vacuole biogenesis and dynamics in plant growth and development. (A) Model of vacuole morphology and distribution during different stages (microspore, immature, and mature) of pollen development and germination in Arabidopsis. This model is based on data derived from 2-D TEM analysis (Backues et al., 2010). The mechanism underlying vacuole dynamics remains unknown. (B) Vacuole dynamics in egg cell, zygote, and embryo development, which is based on CLSM data (Kimata et al., 2019). (C) Vacuole morphology in stomata development as well as stomata opening and closing, which is based on both CLSM and TEM data (Serna et al., 2002; Gao et al., 2005). (D) Model of vacuole formation in Arabidopsis root cells. D1, Different cell layers of the root. D2, The QC cells contain visible large vacuoles. D3, In cortex, SVs are derived from MVB fusion (Cui et al., 2019).
Future perspectives: state-of-the-art and proof-of-concept
Room-temperature ET combined with conventional CLSM have greatly advanced plant membrane dynamics studies especially in regard to fine and transient structures (Nicolas et al., 2017; Yan et al., 2019), and have enabled new whole-cell models to be presented (Cui et al., 2019). More recently, new exciting technologies for sample preparation and visualization under cryogenic conditions have been developed for studying cellular structures in both yeast and animal systems. These include cryo-FIB milling, cryo-lift-out, cryo-CLEM, and cryo-ET. These new techniques have provided an unprecedented opportunity to analyze cellular structures and molecules in their native state in various organisms.
In situ cryo-ET
In situ cryo-ET is a powerful tool for structural biology because it can directly visualize frozen-hydrated cellular samples, offering the best possible means to analyze 3-D morphological structures of membranous organelles and macromolecular assemblies in their native state (Beck and Baumeister, 2016; Erdmann et al., 2018). In general, cryo-fixed samples, either by plunge freezing or HPF, are imaged under EM by tomography after proper trimming on a FIB, followed by a series of image processing steps (Asano et al., 2016; Koning et al., 2018). With the recent resolution, revolution of DEDs (McMullan et al., 2014; Kuijper et al., 2015) and contrast-enhancing devices such as the Volta phase plates (VPP; Danev et al., 2014), the resolution of cryo-EM has greatly improved. These developments have not only allowed single-particle analysis of protein structure at atomic resolution (Danev et al., 2017), but also facilitated the superior performance of in situ cryo-ET (Fukuda et al., 2015). In this workflow, computational approaches like subtomogram averaging (STA) play important roles in improving the resolution (Briggs, 2013; Asano et al., 2016; Schur, 2019). Using purified virus samples, cryo-ET has achieved sub-nanometer resolution allowing the construction of atomic models (Schur et al., 2016). When using unpurified cellular samples, structures of macromolecular assemblies (e.g. ribosomes, proteasomes, and coat assemblies) can now be resolved within complex, heterogeneous cell environments at a resolution of 10–20 Å (Albert et al., 2017, 2020; Bykov et al., 2017; Guo et al., 2018). Most recently, taking advantage of DED, VPP, and STA, a new visualization approach called membranogram has been developed and used to map the densities of protein complexes decorating thylakoid membranes into the native thylakoid architecture of Chlamydomonas (Wietrzynski et al., 2020).
Cryo-FIB milling and cryo-lift-out
Cryo-FIB milling is a technology used to produce lamellae thin enough for EM analysis (Marko et al., 2007). The major challenge of cryo-ET is the sample thickness, with an optimal sample thickness of less than 300 nm (Schaffer et al., 2017). Therefore, with a few exceptions of small-sized prokaryotic cells that can be imaged as a whole, all other cells and tissues must be trimmed down. This was traditionally achieved by cryo-sectioning (CEMOVIS; Al-Amoudi et al., 2004), but was recently replaced by cryo-FIB milling with less artifacts (Hayles et al., 2010). With the aid of cryo-FIB milling, Cryo-ET has been extensively used to solve various native ultrastructure in yeast and mammalian cells, from membrane contact sites (Collado and Fernández-Busnadiego, 2017) and mitochondrial membrane remodeling (Ader et al., 2019) to nuclear pore complex (Mahamid et al., 2016) and protein aggregates (Guo et al., 2018). Most recently, cryo-lift-out has been identified as a viable option to process much larger HPF-prepared samples (up to ∼200 µm) such as C. elegans for cryo-ET (Schaffer et al., 2019).
Cryo-CLEM
The CLEM technology provides an effective solution to link fluorescent protein-tagged molecular identities (e.g. regular, rare, dispersed, or transient events) with contextual high-resolution ultrastructural information in situ (de Boer et al., 2015). Traditionally, fiducial beads are required to achieve faithful target recognition and the correlation accuracy in CLEM (Arnold et al., 2016), with successful examples in plant cells (Wang PF et al., 2019). Recently, a post-correlation on-lamella cryo-CLEM approach has been established to reduce the dependency on randomly distributed fiducial beads in correlation accuracy (Klein et al., 2020). Indeed, the combination of cryo-CLEM with cryo-FIB milling and cryo-ET inherits the outstanding performance of cryo-ET in extracting high-resolution structural information in situ where individual ribosomal translocation states can be distinguished (Schaffer et al., 2019). In addition, FIB-SEM-based cryo-CLEM has been capable of resolving protein–ultrastructure relationships in mammalian cells at a whole-cell level with 4–8-nm isotropic resolution (Hoffman et al., 2020).
Challenges and application in plant cells
Despite the recent development and demonstration of using these state-of-the-art in situ cryo-ET and its auxiliary technologies in yeast and animal systems, as well as in green algae (Engel et al., 2015a; Wietrzynski et al., 2020), their application to vascular plant cells remains a challenge because of their large size, the presence of the cell wall, as well as the complex cell types and large organelles such as the vacuoles. Thus, optional workflows and specific protocols need to be developed and tested for different plant cell types in order to address various biological questions in plants. Below we will use a preliminary study on pollen tube as a proof-of-concept to demonstrate the power of the discussed technologies.
Proof-of-concept: vesicles at the pollen tube tip
The rapid polar growth of pollen tube is maintained by highly dynamic transport vesicles participating in both endocytosis and exocytosis in the tip region (Grebnev et al., 2017), also known as the clear zone (Hepler and Winship, 2015). The dynamics and distributions of TVs have been extensively studied by CLSM and 2-D TEM (Figure 4, A and B), pointing to the multiple roles of TVs in both endocytosis and exocytosis with different models (Wang et al., 2010; Li et al., 2017; Luo et al., 2017; Li et al., 2018; Do et al., 2019; Meng et al., 2020). For example, some CLSM studies on the dynamics of GFP-tagged proteins in growing pollen tube suggested that the active site for exocytosis is in the apex region (Lee et al., 2008; Wang et al., 2013), whereas other studies using pollen tubes stained with styryl FM dyes place it at a shoulder sup-apical to the tip (Bove et al., 2008; Zonia and Munnik, 2008). In addition, using 2-D TEM analysis, morphologically distinct vesicles in the tip regions have been identified (Lancelle and Hepler, 1992; Derksen et al., 1995; Derksen et al., 2002; Figure 4, B). However, the nature, identity, and function of these TVs remain elusive, suggesting that a high-resolution 3-D map of the pollen-tube tip would provide the basis for understanding the nature and function of TVs in a growing pollen tube.
Figure 4.
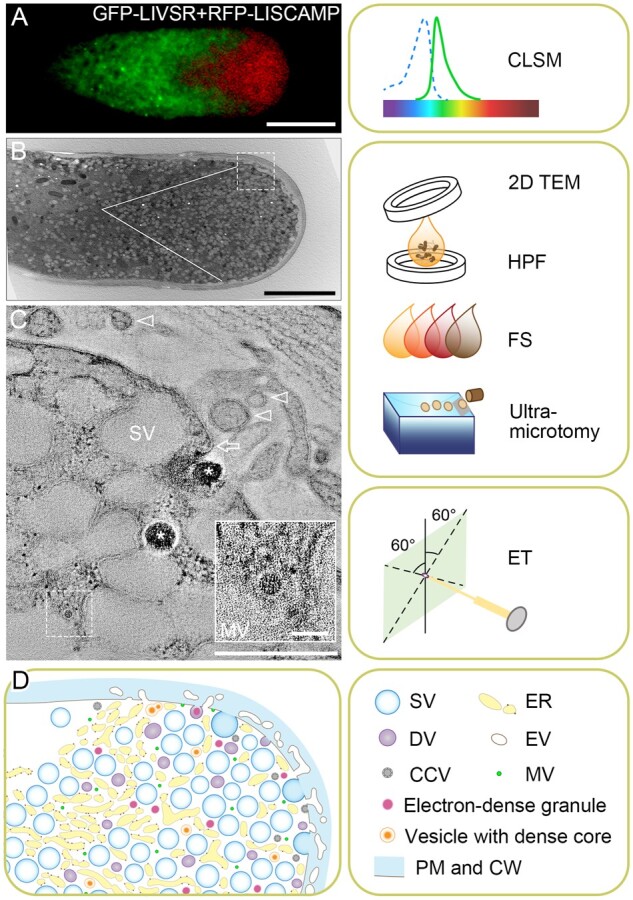
Membrane dynamics and TVs in germinating pollen tube. (A) Membrane dynamics as revealed by 2-D CLSM imaging of a lily (LI) pollen tube co-expressing the GFP-tagged lily vacuolar sorting receptor (GFP-LIVSR); and RFP-tagged lily secretory carrier membrane protein (RFP-LISCAMP). Bar, 10 μm. (B) Traditional 2-D transmission electron microscopy (TEM) image showing a growing lily pollen tube tip region. The outlined V-shaped area is rich in vesicles. Bar, 5 μm. Right panel shows the sample preparation workflow for 2-D/3-D TEM analysis, in which pollen tubes suspended in in vitro germination media are first loaded in a planchette and capped with another one prior to HPF. Frozen samples are then freeze-substituted (FS) in FS cocktail in low temperature, followed by resin embedding and curing. Cured resin blocks containing samples are then sliced to ultrathin sections (<300 nm) by an ultra-microtome for subsequent TEM analysis. (C) A xy slice of the tomogram reconstructed from the box area in B, showing the complexity of a tip region with electron-translucent large secretory vesicles (SV), electron dense granules (asterisks), mini vesicle (MV; dashed box, the lower right inlet showing enlarged view; bar of inlet, 50 nm), possible vesicle fusion with the PM (arrow) and possible extracellular vesicles (EVs, arrowheads). The nature of PM fusion and EVs can only be defined via further ET analysis. Bar, 500 nm. Right panel shows the ET method of capturing images at angles typically varied from −60° to 60°. (D) A proposed model of TVs in growing pollen tube according to multiple 3-D volumes generated by ET method. The right panel shows the legend. DV, dense vesicle; CCV, clathrin-coated vesicle; CW, cell wall. (A) is adapted from (Wang et al. 2010; Copyright 2010 Blackwell Publishing Ltd.).
As a proof-of-concept, a “whole-cell” ET analysis was performed on HPF-prepared, freeze-substituted lily pollen tubes with a clear zone (Figure 4, B), resulting in an observation of distinct transport vesicles (Figure 4, C) and a hypothetical model of the pollen tube tip (Figure 4, D). Interestingly, in addition to the presence of various vesicles, including electron-translucent large secretory vesicles, dense vesicles, and mini vesicles (Figure 4, C), a possible fusion event between a dense vesicle and the PM can also be observed in this tomographic slice view (Figure 4, C, arrow). Whereas extensive apoplastic tubular structures, probably contributing to the formation of extracellular vesicles (Cui et al., 2020b), were also observed along the apical dome of the PM (Figure 4, B and C). The outcome of these “whole-cell” ET analyses will hopefully provide answers to these questions.
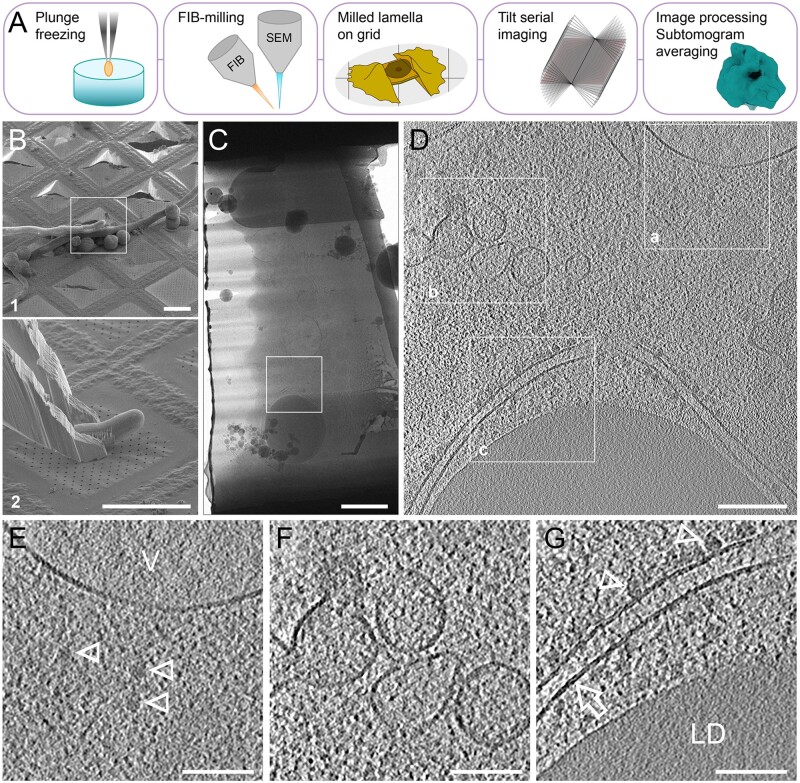
To obtain the most native ultrastructure of TVs from a truly cryogenic image, we recently tested and used the cryo-FIB milling procedure to prepare lamellae from germinating tobacco pollen tubes after plunge freezing and cryo-lift-out (Schaffer et al., 2019) for cryo-ET analysis (Figure 5, A–C). This technique shows the in situ structures of ribosomes, ER membranes, vacuolar membranes, and lipid droplets (Figure 5, D–G) at an unprecedented resolution and clarity. Therefore, through a combination of whole-cell ET analysis of HPF-prepared pollen tubes, cryo-ET analysis of cryo-FIB milled lamellae from the target pollen tube tip region, and further developments in cryo-CLEM, the identity and function of TVs should be resolvable in the not too distant future.
Figure 5.
Cryo-ET analysis of germinating tobacco pollen tube prepared by cryo-lift-out and cryo-FIB-milling. (A) General workflow of cell-sample preparation for cryo-ET imaging analysis via Cryo-FIB. In plunge freezing using Vitrobot, a silica-coated grid loaded with pollen tube suspension is picked up by a tweezer and rapidly plunged into liquid ethane or a mixture of liquid ethane and propane. In FIB-milling, the electron beam for SEM imaging/monitoring and the focused gallium ion beam for milling are placed at certain geometry. Milling is achieved by multiple steps and finally generates a thin lamella (<300 nm) bridged between the remnants of the milled cell on the grid. Tilt serial imaging can be implemented after the sample is transferred from the cryo-FIB to cryo-TEM. Raw tilt images are next pre-processed and reconstructed to generate a tomogram. Further analysis such as STA is optional for structural analysis on macromolecular assemblies (e.g. ribosomes). (B) SEM image of a targeted pollen tube on the grid to be translocated (B1), with its tip region sticking out (white box), and the landing of the excised tip region held by a cryo-gripper (B2). Bar, 50 μm. (C) TEM image of the milled lamella from the excised part of the pollen tube in B. Bar, 1 μm. (D) A xy slice of the tomogram reconstructed from box area in C. Bar, 200 nm. (E–G) Detailed features of box areas 1–3 in D, showing free ribosomes (arrowheads) near vacuole (V) membrane (E, note the different textures between the vacuole lumen and cytoplasm), vesicles profile (F), rough ER (arrow) with ribosomes (arrowheads), and surrounded lipid droplet (LD; G, note the differences in lipid bilayer of ER and the phospholipid monolayer boundary of the lipid droplet). Bar, 100 nm. V, vacuole; LD, lipid droplet.
Outstanding questions
How can SRM, together with (cryo-)CLEM and (cryo-)ET, be used to unveil the nature of the long-debated plant extracellular vesicles in situ?
Can single-particle tracking methods be used to assign molecular identity to different populations of vesicles at the pollen tube tip and monitor their dynamic behaviors?
Can correlative SRM and EM be applied to study vacuole biogenesis and dynamics in various cell types during plant growth and development?
Can whole-cell ET analysis be extended to map vacuoles in different plant cell types (e.g. pollen and stomata development) to understand the dynamic roles of vacuoles in plant growth and development?
Can cryo-FIB lift-out technique enable molecular-resolution cryo-ET within native plant tissue?
Will the combination of whole-cell ET and cryo-ET be sufficient to answer questions on the identity and function of pollen tube TVs?
The above preliminary trials demonstrated that most of the newly developed cryogenic technologies can be applied to plant cells, which opens up new avenues for answering the questions mentioned above. Most important in this regard will be the discovery of structures, distributions, conformations, and interplays of membrane-related macromolecules in their native cellular environment, which cannot be attained by in vitro methods and room-temperature ET (Engel et al., 2015b; Albert et al., 2020). In addition, this dense, stereoscopic, and comprehensive information can further lead us to their functions and mechanisms. For example, cryo-ET can answer questions as to how TVs interact with actin filaments (Stephan et al., 2014; Li et al., 2017) and whether the rough ER and ribosomes in the pollen tube tip (Lovy-Wheeler et al., 2007; Hepler and Winship, 2015; e.g. Figure 5, D and G) are translationally active. Cryo-ET can be a new approach to investigate the “kiss-and-run” model of exocytosis in the pollen tube tip (Guo and Yang, 2020). However, of course both biochemical and genetic approaches will be needed for further functional characterization of molecules at these membranes.
Concluding remarks
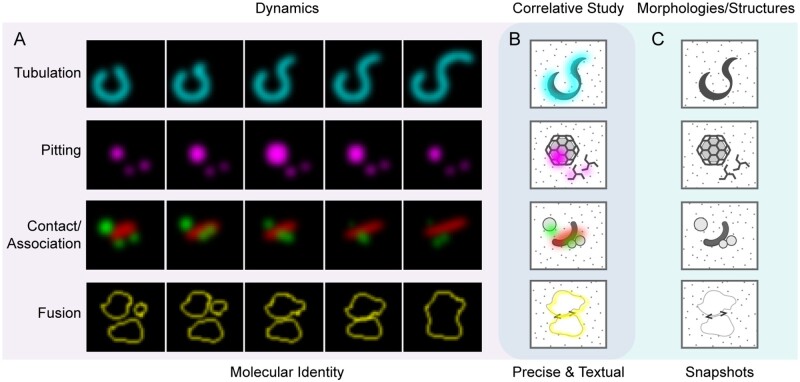
Protein trafficking and organelle biogenesis are highly dynamic events in plant cells and play important roles in regulating plant growth and development, as well as responses to environments. Recent advances in plant membrane dynamics by SRM have led to substantial improvements in spatial resolution (Komis et al., 2018). The rich structural information generated by room-temperature ET combined with CLSM keeps surprising us with new answers to old questions. However, with the maturation of cryo-FIB lift-out and cryo-ET techniques, new findings in plant organelle biogenesis, membrane dynamics, and structures can be expected: a new era in plant membrane imaging has begun (see the “Outstanding Questions”). In summary, Figure 6 illustrates the relationship and potential applications with hypothetical examples of various live-cell imaging techniques for membrane dynamics and correlative (cryo-)EM/ET analysis on corresponding membrane structures in plant cells.
Figure 6.
Application of live-cell imaging and correlative EM/ET analysis in studying membrane dynamics and structures in plant cells. (A) Typical membrane dynamics reflected by live-cell imaging, with black background and fluorescent signals. Shown patterns are hypothetical and supported by examples. The first panel illustrates membrane tubulation, e.g. peroxisome extension (Thazar-Poulot et al., 2015). The second panel shows vesicle pitting, e.g. clathrin-coated vesicle (Sochacki et al., 2017; Narasimhan et al., 2020). The third panel indicates transient membrane association, e.g. autophagosome progression (Zhuang et al., 2017). The last panel shows membrane fusion, e.g. vacuole fusion (Takemoto et al., 2018; Cui et al., 2019). (B) Correlative study involves both molecular identities and in situ ultrastructures, which precisely navigates the events of interest and provides an informative contextual cellular environment. (C) Morphologies and structures obtained by EM-based imaging showing snapshots of cellular events in grayscale.
Acknowledgments
The authors apologize to colleagues whose work could not be included in this review because of space limitations. No conflict of interest declared.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31670179 and 91854201), the Research Grants Council of Hong Kong (AoE/M-05/12, G-CUHK410/19, CUHK14130716, 14104716, 14102417, 14100818, 14101219, C4012-16E, C4002-17G, R4005-18, and C4033-19E), and the CUHK Research Committee to L.J.
Conflict of interest statement. None declared.
Z.L, J.G, Y.C., P.S.E., and L.J. wrote and edited the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Liwen Jiang (ljiang@cuhk.edu.hk).
References
- Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang CX, Youle RJ, Kukulski W (2019) Molecular and topological reorganizations in mitochondrial architecture interplay during Bax-mediated steps of apoptosis. eLife 8: e40712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A, Norlen LPO, Dubochet J (2004) Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J Struct Biol 148: 131–135 [DOI] [PubMed] [Google Scholar]
- Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W, Engel BD (2017) Proteasomes tether to two distinct sites at the nuclear pore complex. Proc Natl Acad Sci U S A 114: 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Wietrzynski W, Lee CW, Schaffer M, Beck F, Schuller JM, Salomé PA, Plitzko JM, Baumeister W, Engel BD (2020) Direct visualization of degradation microcompartments at the ER membrane. Proc Natl Acad Sci U S A 117: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama K, Takagi T, Hirase A, Miyazawa A (2008) STEM tomography for thick biological specimens. Ultramicroscopy 109: 70–80 [DOI] [PubMed] [Google Scholar]
- Arnold J,, Mahamid J, Lucic V, de Marco A, Fernandez JJ, Laugks T, Mayer T, Hyman AA, Baumeister W, Plitzko JM (2016) Site-specific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys J 110: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S, Engel BD, Baumeister W (2016) In situ cryo-electron tomography: a post-reductionist approach to structural biology. J Mol Biol 428: 332–343 [DOI] [PubMed] [Google Scholar]
- Backues SK, Korasick DA, Heese A, Bednarek SY (2010) The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22: 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badelt K, White RG, Overall RL, Vesk M (1994) Ultrastructural specializations of the cell-wall sleeve around plasmodesmata. Am J Bot 81: 1422–1427 [Google Scholar]
- Beck M, Baumeister W (2016) Cryo-electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol 26: 825–837 [DOI] [PubMed] [Google Scholar]
- Bove J, Vaillancourt B, Kroeger J, Hepler PK, Wiseman PW, Geitmann A (2008) Magnitude and direction of vesicle dynamics in growing pollen tubes using spatiotemporal image correlation spectroscopy and fluorescence recovery after photobleaching. Plant Physiol 147: 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JAG (2013) Structural biology in situ—the potential of subtomogram averaging. Curr Opin Struc Biol 23: 261–267 [DOI] [PubMed] [Google Scholar]
- Bykov YS, Schaffer M, Dodonova SO, Albert S, Plitzko JM, Baumeister W, Engel BD, Briggs JA (2017) The structure of the COPI coat determined within the cell. eLife 6: e32493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Jones H, Eggert US (2020) Membrane and organelle dynamics during cell division. Nat Rev Mol Cell Biol 21: 151–166 [DOI] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XFS, Hammer JA, Liu Z, et al. (2014) Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346: 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocks E, Taggart M, Rind FC, White K (2018) A guide to analysis and reconstruction of serial block face scanning electron microscopy data. J Microsc 270: 217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado J, Fernández-Busnadiego R (2017) Deciphering the molecular architecture of membrane contact sites by cryo-electron tomography. Biochim Biophys Acta Mol Cell Res 1864: 1507–1512 [DOI] [PubMed] [Google Scholar]
- Cui Y, Cao WH, He YL, Zhao Q, Wakazaki M, Zhuang XH, Gao JY, Zeng YL, Gao CJ, Ding Y, et al. (2019) A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat Plants 5: 95–105 [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Hu S, Jiang L (2020a) Vacuole biogenesis in plants: how many vacuoles, how many models? Trends Plant Sci 25: 538–548 [DOI] [PubMed] [Google Scholar]
- Cui Y, Gao J, He Y, Jiang L (2020b) Plant extracellular vesicles. Protoplasma 257: 3–12 [DOI] [PubMed] [Google Scholar]
- Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu XG, Rochat RH, Haase-Pettingell C, Piret J, Ludtke SJ, et al. (2013) Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 502: 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W (2014) Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci U S A 111: 15635–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Nagayama K (2010) Phase plates for transmission electron microscopy. Methods Enzymol 481: 343–369 [DOI] [PubMed] [Google Scholar]
- Danev R, Tegunov D, Baumeister W (2017) Using the Volta phase plate with defocus for cryo-EM single particle analysis. eLife 6: e23006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Hoogenboom JP, Giepmans BNG (2015) Correlated light and electron microscopy: ultrastructure lights up! Nat Methods 12: 503–513 [DOI] [PubMed] [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D, et al. (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci U S A 110: 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Horstmann H (2004) Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol 2: 1900–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen J, Knuiman B, Hoedemaekers K, Guyon A, Bonhomme S, Pierson ES (2002) Growth and cellular organization of Arabidopsis pollen tubes in vitro. Sex Plant Reprod 15: 133–139 [Google Scholar]
- Derksen J, Rutten T, Lichtscheidl IK, Dewin AHN, Pierson ES, Rongen G (1995) Quantitative-analysis of the distribution of organelles in tobacco pollen tubes—implications for exocytosis and endocytosis. Protoplasma 188: 267–276 [Google Scholar]
- Ding B, Turgeon R, Parthasarathy MV (1992) Substructure of freeze-substituted plasmodesmata. Protoplasma 169: 28–41 [Google Scholar]
- Do THT, Choi H, Palmgren M, Martinoia E, Hwang JU, Lee Y (2019) Arabidopsis ABCG28 is required for the apical accumulation of reactive oxygen species in growing pollen tubes. Proc Natl Acad Sci U S A 116: 12540–12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Yang XC, Zhu SB, Bassham DC, Fang N (2015) Stochastic optical reconstruction microscopy imaging of microtubule arrays in intact Arabidopsis thaliana seedling roots. Sci Rep 5:15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Cuellar LK, Villa E, Plitzko JM, Baumeister W (2015a) Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: e04889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Albert S, Asano S, Plitzko JM, Baumeister W (2015b) In situ structural analysis of Golgi intracisternal protein arrays. Proc Natl Acad Sci U S A 112: 11264–11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann PS, Plitzko JM, Baumeister W (2018) Addressing cellular compartmentalization by in situ cryo-electron tomography. Curr Opin Colloid Interface Sci 34: 89–99 [Google Scholar]
- Feeney M, Kittelmann M, Menassa R, Hawes C, Frigerio L (2018) Protein storage vacuoles originate from remodeled preexisting vacuoles in Arabidopsis thaliana. Plant Physiol 177: 241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Laugks U, Lučić V, Baumeister W, Danev R (2015) Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol 190: 143–154 [DOI] [PubMed] [Google Scholar]
- Gao XQ, Li CG, Wei PC, Zhang XY, Chen J, Wang XC (2005) The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol 139: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebnev G, Ntefidou M, Kost B (2017) Secretion and endocytosis in pollen tubes: models of tip growth in the spot light. Front Plant Sci 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Yang Z (2020) Exocytosis and endocytosis: coordinating and fine-tuning the polar tip growth domain in pollen tubes. J Exp Bot 71: 2428–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martínez-Sánchez A, Rudack T, Beck F, Hartmann H, Pérez-Berlanga M, Frottin F, Hipp MS, Hartl FU, et al. (2018) In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172: 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YT, Li D, Zhang SW, Yang YR, Liu JJ, Wang XY, Liu C, Milkie DE, Moore RP, Tulu US, et al. (2018) Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell 175: 1430–1442 [DOI] [PubMed] [Google Scholar]
- Hayles MF, de Winter DAM, Schneijdenberg CTWM, Meeldijk JD, Luecken U, Persoon H, de Water J, de Jong F, Humbel BM, Verkleij AJ (2010) The making of frozen-hydrated, vitreous lamellas from cells for cryo-electron microscopy. J Struct Biol 172: 180–190 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Winship LJ (2015) The pollen tube clear zone: clues to the mechanism of polarized growth. J Integr Plant Biol 57: 79–92 [DOI] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. (2020) Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367: eaaz5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Sousa AA,, Azari AA, Glushakova S, Zhang GF, Zimmerberg J, Leapman RD (2009) Nanoscale 3D cellular imaging by axial scanning transmission electron tomography. Nat Methods 6: 729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DQ, Sun YB, Ma ZM, Ke MY, Cui Y, Chen ZC, Chen CF, Ji CY, Tran TM,, Yang L, et al. (2019) Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc Natl Acad Sci U S A 116: 21274–21284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Uemura T, Nakano A (2018) The Golgi entry core compartment functions as a COPII-independent scaffold for ER-to-Golgi transport in plant cells. J Cell Sci 131: jcs203893 [DOI] [PubMed] [Google Scholar]
- Ivanov S, Austin J, Berg RH, Harrison MJ (2019) Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat Plants 5: 194–203 [DOI] [PubMed] [Google Scholar]
- Jacquemet G, Carisey AF, Hamidi H, Henriques R, Leterrier C (2020) The cell biologist’s guide to super-resolution microscopy. J Cell Sci 133: jcs240713. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Ott T (2020) The nanoscale organization of the plasma membrane and its importance in signaling: a proteolipid perspective. Plant Physiol 182: 1682–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L (2017) Plant Protein Secretion: Methods and Protocols. New York: Humana Press [Google Scholar]
- Jin X, Jiang Z, Zhang K, Wang P, Cao X, Yue N, Wang X, Zhang X, Li Y, Li D, et al. (2018) Three-dimensional analysis of chloroplast structures associated with virus infection. Plant Physiol 176: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y (2012) Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem 287: 44261–44269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4K beta 1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Karahara I, Kang BH (2014) High-pressure freezing and low-temperature processing of plant tissue samples for electron microscopy. Methods Mol Biol 1080: 147–157 [DOI] [PubMed] [Google Scholar]
- Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT (2016) Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun 7:12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Kato T, Higaki T, Kurihara D, Yamada T, Segami S, Morita MT, Maeshima M, Hasezawa S, Higashiyama T, et al. (2019) Polar vacuolar distribution is essential for accurate asymmetric division of Arabidopsis zygotes. Proc Natl Acad Sci U S A 116: 2338–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Wimmer BH, Winter SL, Kolovou A, Laketa V, Chlanda P (2020) Membrane architecture of pulmonary lamellar bodies revealed by post-correlation on-lamella cryo-CLEM. bioRxiv: 2020.2002.2027.966739
- Kleine-Vehn J, Wabnik K, Martinière A, Langowski L, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. (2011) Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Wang PW, Kriechbaumer V, Tilsner J, Frigerio L, Sparkes I, Hawes C, Oparka K (2015) Putting the squeeze on plasmodesmata: a role for reticulons in primary plasmodesmata formation. Plant Physiol 168: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Novák D, Ovečka M, Šamajová O, Šamaj J (2018) Advances in imaging plant cell dynamics. Plant Physiol 176: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Šamajová O, Ovečka M, Šamaj J (2015) Super-resolution microscopy in plant cell imaging. Trends Plant Sci 20: 834–843 [DOI] [PubMed] [Google Scholar]
- Koning RI, Koster AJ, Sharp TH (2018) Advances in cryo-electron tomography for biology and medicine. Ann Anat 217: 82–96 [DOI] [PubMed] [Google Scholar]
- Kuijper M, van Hoften G, Janssen B, Geurink R, De Carlo S, Vos M, van Duinen G, van Haeringen B, Storms M (2015) FEI’s direct electron detector developments: embarking on a revolution in cryo-TEM. J Struct Biol 192: 179–187 [DOI] [PubMed] [Google Scholar]
- Lancelle SA, Hepler PK (1992) Ultrastructure of freeze-substituted pollen tubes of Lilium longiflorum. Protoplasma 167: 215–230 [Google Scholar]
- Lee YJ, Szumlanski A, Nielsen E, Yang ZB (2008) Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J Cell Biol 181: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitz G, Kang BH, Schoenwaelder MEA, Staehelin LA (2009) Statolith sedimentation kinetics and force transduction to the cortical endoplasmic reticulum in gravity-sensing Arabidopsis columella cells. Plant Cell 21: 843–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Luo N, Wang WD, Liu ZY, Chen JS, Zhao LT, Tan L, Wang CY, Qin Y, Li C, et al. (2018) The REN4 rheostat dynamically coordinates the apical and lateral domains of Arabidopsis pollen tubes. Nat Commun 9:2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SW, Dong HJ, Pei WK, Liu CN, Zhang S, Sun TT, Xue XH, Ren HY (2017) LlFH1-mediated interaction between actin fringe and exocytic vesicles is involved in pollen tube tip growth. New Phytol 214: 745–761 [DOI] [PubMed] [Google Scholar]
- Liang ZZ,, Zhu N, Mai KK, Liu ZY, Tzeng D, Osteryoung KW, Zhong SL, Staehelin LA, Kang BH (2018) Thylakoid-bound polysomes and a dynamin-related protein, FZL, mediate critical stages of the linear chloroplast biogenesis program in greening Arabidopsis cotyledons. Plant Cell 30: 1476–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A, Cárdenas L, Kunkel JG, Hepler PK (2007) Differential organelle movement on the actin cytoskeleton in lily pollen tubes. Cell Motil Cytoskeleton 64: 217–232 [DOI] [PubMed] [Google Scholar]
- Luo N, Yan A, Liu G, Guo JZ, Rong DY, Kanaoka MM, Xiao Z, Xu GS, Higashiyama T, Cui XP, et al. (2017) Exocytosis-coordinated mechanisms for tip growth underlie pollen tube growth guidance. Nat Commun 8:1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D (2019) Challenges and opportunities in cryo-EM single-particle analysis. J Biol Chem 294: 5181–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, Baumeister W (2016) Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351: 969–972 [DOI] [PubMed] [Google Scholar]
- Marchant R, Moore RT (1973) Lomasomes and plasmalemmasomes in fungi. Protoplasma 76: 235–247 [Google Scholar]
- Marko M, Hsieh C, Schalek R, Frank J, Mannella C (2007) Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods 4: 215–217 [DOI] [PubMed] [Google Scholar]
- Martinière A, Fiche JB, Smokvarska M, Mari S, Alcon C, Dumont X, Hematy K, Jaillais Y, Nollmann M, Maurel C (2019) Osmotic stress activates two reactive oxygen species pathways with distinct effects on protein nanodomains and diffusion. Plant Physiol 179: 1581–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh R, Nicastro D, Mastronarde D (2005) New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol 15: 43–51 [DOI] [PubMed] [Google Scholar]
- McKenna JF, Rolfe DJ, Webb SED, Tolmie AF, Botchway SW, Martin-Fernandez ML, Hawes C, Runions J (2019) The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc Natl Acad Sci U S A 116: 12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Faruqi AR, Clare D, Henderson R (2014) Comparison of optimal performance at 300 keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy 147: 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng JG, Liang L, Jia PF, Wang YC, Li HJ, Yang WC (2020) Integration of ovular signals and exocytosis of a Ca2+ channel by MLOs in pollen tube guidance. Nat Plants 6: 143–153 [DOI] [PubMed] [Google Scholar]
- Miranda K, Girard-Dias W, Attias M, de Souza W, Ramos I (2015) Three dimensional reconstruction by electron microscopy in the life sciences: an introduction for cell and tissue biologists. Mol Reprod Dev 82: 530–547 [DOI] [PubMed] [Google Scholar]
- Nakano A (2013) Super-resolution confocal live imaging microscopy (SCLIM)—cutting-edge technology in cell biology. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). New York: IEEE, pp. 133–135 [DOI] [PubMed]
- Narasimhan M, Johnson A, Prizak R, Kaufmann WA, Tan ST, Casillas-Pérez B, Friml J (2020) Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife 9: e52067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, Trépout S, Gaston A, Fouché M, Cordelières FP, Oparka K, Tilsner J, Brocard L, Bayer EM (2017) Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat Plants 3: 17082. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, et al. (2017) Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol Cell 67: 282–293 [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley H, Robinson D, Weston A, Collinson L, Tooze S (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23: 1860–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA (2001) Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13: 2033–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Pennington JG (2019) Electron tomography in plant cell biology. Microscopy 68: 69–79 [DOI] [PubMed] [Google Scholar]
- Ott T (2017) Membrane nanodomains and microdomains in plant–microbe interactions. Curr Opin Plant Biol 40: 82–88 [DOI] [PubMed] [Google Scholar]
- Overall RL, Wolfe J, Gunning BES (1982) Intercellular communication in Azolla roots: I. Ultrastructure of plasmodesmata. Protoplasma 111: 134–150 [Google Scholar]
- Pain C, Kriechbaumer V, Kittelmann M, Hawes C, Fricker M (2019) Quantitative analysis of plant ER architecture and dynamics. Nat Commun 10: 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, Marquès-Bueno MD, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, et al. (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62 [DOI] [PubMed] [Google Scholar]
- Rast A, Schaffer M, Albert S, Wan W, Pfeffer S, Beck F, Plitzko JM, Nickelsen J, Engel BD (2019) Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat Plants 5: 436–446 [DOI] [PubMed] [Google Scholar]
- Roth R, Hillmer S, Funaya C, Chiapello M, Schumacher K, Lo Presti L, Kahmann R, Paszkowski U (2019) Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat Plants 5: 204–211 [DOI] [PubMed] [Google Scholar]
- Schaffer M, Mahamid J, Engel BD, Laugks T, Baumeister W, Plitzko JM (2017) Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J Struct Biol 197: 73–82 [DOI] [PubMed] [Google Scholar]
- Schaffer M, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, Albert S, Engel BD, Rummel A, Smith AJ, Baumeister W, et al. (2019) A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat Methods 16: 757–762 [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Ferrand A, Huser T, Eggeling C, Sauer M, Biehlmaier O, Drummen GPC (2019) Super-resolution microscopy demystified. Nat Cell Biol 21: 72–84 [DOI] [PubMed] [Google Scholar]
- Scheuring D, Löfke C, Krüger F, Kittelmann M, Eisa A, Hughes L, Smith RS, Hawes C, Schumacher K, Kleine-Vehn J (2016) Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc Natl Acad Sci U S A 113: 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V (2017) Super-resolution microscopy—applications in plant cell research. Front Plant Sci 8:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schur FKM (2019) Toward high-resolution in situ structural biology with cryo-electron tomography and subtomogram averaging. Curr Opin Struc Biol 58: 1–9 [DOI] [PubMed] [Google Scholar]
- Schur FKM, Obr M, Hagen WJH, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Kräusslich HG, Briggs JAG (2016) An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353: 506–508 [DOI] [PubMed] [Google Scholar]
- Serna L, Torres-Contreras J, Fenoll C (2002) Clonal analysis of stomatal development and patterning in Arabidopsis leaves. Dev Biol 241: 24–33 [DOI] [PubMed] [Google Scholar]
- Sezgin E (2017) Super-resolution optical microscopy for studying membrane structure and dynamics. J Phys Condens Matter 29:273001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacki KA, Dickey AM, Strubl MP, Taraska JW (2017) Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat Cell Biol 19: 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehner D, Steyer AM, Bertinetti L, Orlov I, Benoit L, Pernet-Gallay K, Schertel A, Schultz P (2020) Cryo-FIB-SEM as a promising tool for localizing proteins in 3D. J Struct Biol 211:107528. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Trimble WS, Grinstein S, Drin G, Reinisch K, De Camilli P, Cohen S, Valm AM, Lippincott-Schwartz J, Levine TP, et al. (2017) Membrane dynamics and organelle biogenesis-lipid pipelines and vesicular carriers. BMC Biol 15:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan O, Cottier S, Fahlén S, Montes-Rodriguez A, Sun J, Eklund DM, Klahre U, Kost B (2014) RISAP is a TGN-associated RAC5 effector regulating membrane traffic during polar cell growth in Tobacco. Plant Cell 26: 4426–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Ebine K, Askani JC, Krüger F, Gonzalez ZA, Ito E, Goh T, Schumacher K, Nakano A, Ueda T (2018) Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc Natl Acad Sci U S A 115: E2457–E2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazar-Poulot N, Miquel M, Fobis-Loisy I, Gaude T (2015) Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc Natl Acad Sci U S A 112: 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuijtel MW, Koster AJ, Faas FGA, Sharp TH (2019) Correlated cryo super-resolution light and cryo-electron microscopy on mammalian cells expressing the fluorescent protein rsEGFP2. Small Methods 3: 1900425 [Google Scholar]
- Vavrdová T, Šamajová O, Křenek P, Ovečka M, Floková P, Šnaurová R, Šamaj J, Komis G (2019) Multicolour three dimensional structured illumination microscopy of immunolabeled plant microtubules and associated proteins. Plant Methods 15:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25: 3434–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D, Rosero A, Komis G, Šamajová O, Ovečka M, Voigt B, Šamaj J (2016) Endosomal interactions during root hair growth. Front Plant Sci 6:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker I, Spomer W, Hofmann A, Thaler M, Hillmer S, Gengenbach U, Schröder RR (2016) Hierarchical imaging: a new concept for targeted imaging of large volumes from cells to tissues. BMC Cell Biol 17:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tse YC, Law AHY, Sun SSM, Sun YB, Xu ZF, Hillmer S, Robinson DG, Jiang LW (2010) Vacuolar sorting receptors (VSRs) and secretory carrier membrane proteins (SCAMPs) are essential for pollen tube growth. Plant J 61: 826–838 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhuang X, Wang X, Law AH, Zhao T, Du S, Loy MM, Jiang L (2016) A distinct pathway for polar exocytosis in plant cell wall formation. Plant Physiol 172: 1003–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhuang XH, Cai Y, Cheung AY, Jiang LW (2013) Apical F-actin-regulated exocytic targeting of NtPPME1 is essential for construction and rigidity of the pollen tube cell wall. Plant J 76: 367–379 [DOI] [PubMed] [Google Scholar]
- Wang LH, Cheng MZ, Yang Q, Li JG, Wang X, Zhou Q, Nagawa S, Xia BX, Xu TD, Huang RF, et al. (2019) Arabinogalactan protein-rare earth element complexes activate plant endocytosis. Proc Natl Acad Sci U S A 116: 14349–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PF, Chen XS, Goldbeck C, Chung E, Kang BH (2017) A distinct class of vesicles derived from the trans-Golgi mediates secretion of xylogalacturonan in the root border cell. Plant J 92: 596–610 [DOI] [PubMed] [Google Scholar]
- Wang PF, Liang ZZ, Kang BH (2019) Electron tomography of plant organelles and the outlook for correlative microscopic approaches. New Phytol 223: 1756–1761 [DOI] [PubMed] [Google Scholar]
- Weisshart K (2014) The basic principle of airyscanning. Zeiss Technol Note 22 [Google Scholar]
- Wietrzynski W, Schaffer M, Tegunov D, Albert S, Kanazawa A, Plitzko JM, Baumeister W, Engel BD (2020) Charting the native architecture of Chlamydomonas thylakoid membranes with single-molecule precision. eLife 9: e53740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Meehl JB, O’Toole ET, Giddings TH (2014) Conventional transmission electron microscopy. Mol Biol Cell 25: 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SG, Houben L, Elbaum M (2014) Cryo-scanning transmission electron tomography of vitrified cells. Nat Methods 11: 423–428 [DOI] [PubMed] [Google Scholar]
- Xu J, Ma H, Ma H, Jiang W, Mela CA, Duan M, Zhao S, Gao C, Hahm ER, Lardo SM. (2020) Super-resolution imaging reveals the evolution of higher-order chromatin folding in early carcinogenesis. Nat Commun 11: 1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Yadav SR, Paterlini A, Nicolas WJ, Petit JD, Brocard L, Belevich I, Grison MS, Vaten A, Karami L, et al. (2019) Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat Plants 5: 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Chung KP, Li B, Lai CM, Lam SK, Wang X, Cui Y, Gao C, Luo M, Wong KB, et al. (2015) Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc Natl Acad Sci U S A 112: 14360–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang XH, Chung KP, Cui Y, Lin WL, Gao CJ, Kang BH, Jiang LW (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A 114: E426–E435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T (2008) Vesicle trafficking dynamics and visualization of zones of exocytosis and endocytosis in tobacco pollen tubes. J Exp Bot 59: 861–873 [DOI] [PubMed] [Google Scholar]