Abstract
Rhizobial infection of legume roots during the development of nitrogen-fixing root nodules can occur intracellularly, through plant-derived infection threads traversing cells, or intercellularly, via bacterial entry between epidermal plant cells. Although it is estimated that around 25% of all legume genera are intercellularly infected, the pathways and mechanisms supporting this process have remained virtually unexplored due to a lack of genetically amenable legumes that exhibit this form of infection. In this study, we report that the model legume Lotus japonicus is infected intercellularly by the IRBG74 strain, recently proposed to belong to the Agrobacterium clade of the Rhizobiaceae. We demonstrate that the resources available for L. japonicus enable insight into the genetic requirements and fine-tuning of the pathway governing intercellular infection in this species. Inoculation of L. japonicus mutants shows that Ethylene-responsive factor required for nodulation 1 (Ern1) and Leu-rich Repeat Receptor-Like Kinase (RinRK1) are dispensable for intercellular infection in contrast to intracellular infection. Other symbiotic genes, including nod factor receptor 5 (NFR5), symbiosis receptor-like kinase (SymRK), Ca2+/calmodulin dependent kinase (CCaMK), exopolysaccharide receptor 3 (Epr3), Cyclops, nodule inception (Nin), nodulation signaling pathway 1 (Nsp1), nodulation signaling pathway 2 (Nsp2), cystathionine-β-synthase (Cbs), and Vapyrin are equally important for both entry modes. Comparative RNAseq analysis of roots inoculated with IRBG74 revealed a distinctive transcriptome response compared with intracellular colonization. In particular, several cytokinin-related genes were differentially regulated. Corroborating this observation, cyp735A and ipt4 cytokinin biosynthesis mutants were significantly affected in their nodulation with IRBG74, whereas lhk1 cytokinin receptor mutants formed no nodules. These results indicate a differential requirement for cytokinin signaling during intercellular rhizobial entry and highlight distinct modalities of inter- and intracellular infection mechanisms in L. japonicus.
Genetics and transcriptomics uncovers modalities of intercellular infection in legume-rhizobium symbiosis.
Introduction
Legumes constitute a large and diverse plant family and most legumes are able to develop nitrogen-fixing root nodules in symbiosis with soil bacteria commonly referred to as rhizobia. Bacterial infection of roots and root nodules through intracellular infection threads (ITs) has been extensively researched in the model legumes Lotus japonicus (a relative of birdsfoot trefoil) and barrel medic (Medicago truncatula), as well as crop legumes like soybean (Glycine max L.), pea (Pisum sativum L.), and common bean (Phaseolus vulgaris L.). However, an alternative mechanism of intercellular infection is widespread in different genera of the Fabaceae family (Sprent et al., 2017) and proposed to represent an ancient and less sophisticated mechanism of rhizobial colonization (Sprent, 2007). In this process, rhizobia invade the legume roots between epidermal/root hair cells or by crack entry, during the protrusion of lateral roots (Coba de la Peña et al., 2017). Intercellular infection processes have been described in detail by microscopy in different legumes, such as Mimosa, Neptunia, Stylosanthes, Cytisus, and Lupinus (de Faria et al., 1988; James et al., 1992; Subba-Rao et al., 1995; Vega-Hernandez et al., 2001; Gonzalez-Sama et al., 2004; Goormachtig et al., 2004). Special attention has been dedicated to characterize the histology of intercellular infection and nodulation processes in Arachis hypogaea, Aeschynomene spp. and the semiaquatic legume Sesbania rostrata (Chandler, 1978; Boogerd and Van Rossum, 1997; Bonaldi et al., 2011; Ibañez et al., 2017). Under flooded conditions, rhizobial colonization takes places via infection pockets in S. rostrata, formed by a cell death process that depends on nodulation factors (NFs), perception, and localized formation of reactive oxygen species (D'Haeze et al., 2003). From such infection pockets, cortical ITs are formed and migrate to the nodule primordium, where the bacteria are released from the ITs and colonize the nodule cells in symbiosomes (Capoen et al., 2010). Unlike intracellular colonization, the symbiosis receptor-like kinase (SymRK) gene and the Ca2+/calmodulin-dependent protein kinase (CCaMK) gene are dispensable for intercellular infection in S. rostrata by Azorhizobium caulinodans. However, these genes are required for the subsequent intracellular cortical ITs (Capoen et al., 2005, 2009). In peanut (Arachis hypogaea), bradyrhizobia enter through the middle lamellae of two adjacent root hairs and spread intercellularly between epidermal and cortical cells. In parallel, adjacent axillary root hair basal cells become enlarged and infected by the microsymbiont (Chandler, 1978; Boogerd and Van Rossum, 1997; Guha et al., 2016). Both nodule formation and nodule cell colonization require proper exopolysaccharide production by rhizobia (Morgante et al., 2007). Similarly, an NF-independent nodulation program has been described in certain Aeschynomene spp. (Giraud et al., 2007). Currently, the Aeschynomene evenia–Bradyrhizobium symbiosis has been employed to study the molecular genetics of this unusual NF-independent symbiosis (Arrighi et al., 2012). Recent findings show that during this peculiar mechanism, several components of the NF-dependent process are also recruited, such as SYMRK, CCaMK, and the Histidine Kinase (HK1) cytokinin receptor (Fabre et al., 2015). Additionally, the structural requirements to perceive NF in S. rostrata are more permissive in intercellular infection compared with the intracellular infection (Goormachtig et al., 2004). Intercellular infection occurs in several Sesbania spp. by IRBG74 (Cummings et al., 2009), a nodulating strain that belongs to the Agrobacterium clade of the Rhizobiaceae (Aguilar et al., 2016), which is also able to colonize rice and Arabidopsis roots as an endophyte (Biswas et al., 2000; Mitra et al., 2016; Zhao et al., 2017). Therefore, a better understanding of intercellular colonization could facilitate the engineering of nonlegume crops for colonization by nitrogen-fixing bacteria.
In Lotus, NFs are recognized in the plasma membrane of the root hairs by nod factor receptors (NFR1, NFR5, and NFRe; Madsen et al., 2003; Radutoiu et al., 2003; Murakami et al., 2018). A compatible recognition leads to rhizobial attachment to the root hair tip, promoting its curling to trap the bacteria within an infection pocket. This gives rise to the formation of an IT, a tubular structure with an inward growth that originates from invagination of the plasma membrane of the root hair. The IT follows a polar growth toward inner root cell layers, reaching the nodule primordia, formed by the activation of cell division in the cortical cells. The nodule primordia give rise to mature nodules, wherein the bacteria are released from the IT into symbiosomes and differentiate to become nitrogen-fixing bacteroids (Downie, 2014).
The intracellular infection of rhizobia via ITs in the root hairs has been extensively investigated in Lotus and M. truncatula (Lace and Ott, 2018). In these legumes, the infection is orchestrated by several transcription factors, including nodule inception (Nin; Schauser et al., 1999; Marsh et al., 2007), Nsp1/Nsp2 (Kalo et al., 2005; Heckmann et al., 2006), Cyclops (Yano et al., 2008; Singh et al., 2014), and ERF required for nodulation (Ern1; (Cerri et al., 2012; Kawaharada et al., 2017). The latter is required for activation of the expression of the cytokinin-biosynthesis genes Ipt2 and Log4 that are major contributors to the initial symbiotic cytokinin response in Lotus (Reid et al., 2017). Cytokinin is necessary for nodule organogenesis but plays a negative role during rhizobial invasion. In the cytokinin oxidase/dehydrogenase 3 mutant (ckx3), where cytokinin levels in the roots are increased, rhizobial infection is significantly reduced (Reid et al., 2016). In contrast, the roots of cytokinin receptor Lhk1 mutants are hyperinfected by rhizobia (Murray et al., 2007). Recent reports show that additional signaling pathways play a positive role in infection, like the exopolysaccharide receptor EPR3 (Kawaharada et al., 2015) and the Leu-rich repeat receptor-like kinase (RINRK1; Li et al., 2019). In addition, several molecular components are required. Mutants disrupted in the E3 ligase Cerberus (Yano et al., 2009), the nodule pectate lyase Npl1 (Xie et al., 2012) or Arpc1, ScarN, Nap1, Pir1 involved in actin rearrangements (Yokota et al., 2009; Hossain et al., 2012; Qiu et al., 2015), show defects in IT development and abortion of the infection process. In Medicago, the IT localized cystathionine-β-synthase-like 1 (CBS1; Sinharoy et al., 2013), coiled-coil RPG protein (Arrighi et al., 2008) and Vapyrin (Murray et al., 2011) are crucial components of the root hair infectome (Liu et al., 2019).
Rhizobia cross the epidermal cell layer of legume roots intracellularly through root hair ITs, or by intercellular infection. The plant genetic basis for the latter is largely unexplored despite its estimated prevalence in 25% of all legumes and its engineering potential to achieve rhizobial colonization in nonlegume crops (Sprent, 2007; Oldroyd and Dixon, 2014). To understand and compare the genetic programs controlling intracellular and intercellular infection in Lotus roots, we have analyzed the infective capacity of Rhizobium sp. strain IRBG74, where a wide range of genetic, genomic, and transcriptomic resources are available. Crucial genes for both modalities of rhizobial infection were identified along with distinctive cellular, transcriptome, and genetic requirements for intercellular colonization.
Results
IRBG74 induces nitrogen-fixing nodules in Lotus
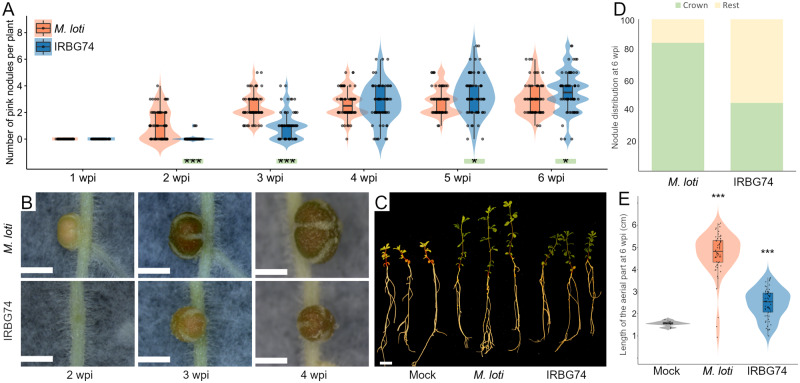
The rhizobial strain IRBG74, infects Sesbania cannabina intercellularly (Cummings et al., 2009; Mitra et al., 2016) and interestingly it is also capable of colonizing Oryza sativa and Arabidopsis thaliana roots as an endophyte (Mitra et al., 2016; Zhao et al., 2017). In order to evaluate the infective capacity of IRBG74 in Lotus, nodulation kinetics was recorded from 1- to 6-week postinoculation (wpi) of the Gifu accession. As a control, L. japonicus seedlings were inoculated with its customary symbiont Mesorhizobium loti R7A that infects intracellularly. Nodule primordia were observed at 1 wpi on Lotus roots inoculated with M. loti, with mature pink nodules evident by 2 wpi (Figure 1A). IRBG74 also induced nodule organogenesis in Lotus, but the first nodule primordia were not observed until 2 wpi (Figure 1, A and B). The first mature pink nodules on plants inoculated with IRBG74 usually appeared at 3 wpi, but these were evidently smaller compared with the pink nodules induced by M. loti at the same time point (Figure 1B). In addition, the number of pink and total nodules was significantly lower at 2 and 3 wpi in plants inoculated with IRBG74 compared with plants inoculated with M. loti (Figure 1A;Supplemental Figure 1B), but after 4–6 wpi, the number of nodules on plants inoculated with M. loti and IRBG74 was comparable (Figure 1A;Supplemental Figure 1B). The delay in the IRBG74 nodulation was reflected by the distribution pattern of the nodules in the root system, since 45% of the nodules were found in the root crown (upper 2 cm of the root system), whereas plants inoculated with M. loti had 84% of the nodules in this segment of the root (Figure 1, C and D). Nodules induced by IRBG74 were pink and the plant shoots were green (Figure 1, B and C), indicative of nitrogen fixation. At 6 wpi, shoot lengths were significantly lower compared with plants inoculated with M. loti, reflecting the delayed nodule formation, but higher compared with mock-treated plants (Figure 1, C and E). These results show that IRBG74 is able to induce nitrogen-fixing nodules in Lotus, albeit with a delay.
Figure 1.
Nodulation phenotype of Lotus plants inoculated with M. loti or IRBG74. (A) Nodule numbers at 1–6 wpi. Mann–Whitney U-test of pink and total number of nodules (asterisks below the violin graphs indicates significant difference: *P < 0.05; ***P < 0.001) between Gifu plants inoculated with M. loti and IRBG74. (B) Images of nodules at 2–4 wpi with M. loti or IRBG74 (upper and lower panels, respectively). Scale bars = 1 mm. (C) Representative plants at 6 wpi with M. loti, IRBG74 or uninoculated (mock). Images were digitally extracted for comparison. Scale bar = 1 cm. (D) Nodule distribution in Lotus roots at 6 wpi with M. loti or IRBG74. (E) Shoot length of Lotus plants harvested at 6 wpi with M. loti, IRBG74 or uninoculated (mock). Student’s t test of length of the aerial part between mock-treated plants and inoculated with M. loti or IRBG74. *P < 0.05; **P < 0.01; ***P < 0.001; n ≥ 66. Violin boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, individual data points.
To determine if the delay in the nodulation program by IRBG74 was restricted to the accession Gifu, the nodulation kinetics was recorded at 1–3 wpi with IRBG74 in three different Lotus ecotypes (MG134, MG144, and MG145; Hashiguchi et al., 2012), testing in parallel plants inoculated with M. loti. The first nodule primordia were observed at 1 wpi with M. loti in the different Lotus accessions, whereas in response to IRBG74 primordia emerged from the second week. This analysis shows that the delay in the nodulation process by IRBG74 is not restricted to accession Gifu (Supplemental Figure 1A).
Lotus is intercellularly infected by IRBG74
The delay in organogenesis following inoculation with IRBG74 compared with M. loti prompted us to explore the infection process. For this purpose, the constitutive DsRED expressing plasmid pSKDSRED was transformed into IRBG74 in order to monitor the early infection process by confocal microscopy. Mesorhizobium loti-DsRed was used as a control. Infection and nodule organogenesis were unaltered with these engineered strains. Typical intracellular ITs in long root hairs were abundant at 7-d postinoculation (dpi) with M. loti-DsRed (Figure 2A). In contrast, no IRBG74-DsRed ITs were observed. IRBG74 was attached to the surface of the roots, mainly associated with the boundaries of the epidermal and root hair cells (Figure 2, B–D; Supplemental Video 1). A detailed and quantitative inspection revealed an average of 35 root hair ITs in response to M. loti-DsRed, whereas none were found in the entire root system of plants inoculated with IRBG74-DsRed at 10 and 21 dpi (Figure 2H). IRBG74-DsRed was observed on the epidermis of emergent nodule primordium, associated to highly deformed and twisted root hairs (Figure 2, C and D; Supplemental Movie S1). IRBG74, therefore, elicits a root hair response but no root hair ITs were observed in these deformed root hairs. In nodule primordia at a more advanced developmental stage, IRBG74-DsRed infected the epidermal cells intercellularly (Figure 2, E). The intercellular infection was followed by the formation of infection pockets accumulating IRBG74 in the subepidermal root cell layers (Figure 2, E–G). From these infection pockets, rhizobial colonization progressed either transcellularly or intercellularly into inner root cell layers (Figure 2, E and F; Supplemental Movie 1).
Figure 2.

Intracellular and intercellular rhizobial infection of L. japonicus roots. (A) L. japonicus root with root hair ITs at 1 wpi with M. loti-DsRed. (B) IRBG74-DsRed at 1 wpi associated with the surface of the root epidermal cells (boundaries highlighted with dashed lines). (C–G) Nodule primordia of 2–3 wpi with IRBG74-DsRed at progressive colonization and developmental stages. Highly deformed and twisted root hairs in emerging nodule primordia (C and D). Intercellular infection of IRBG74-DsRed at the root surface (E) is followed by infection pocket formation in subepidermal cell layers and intercellular progression (E–G). Scale bar, 50 µm (A–D) and 20 µm (E–G). (H) Number of root hair ITs per plant at different time points (Error bars mean SE; n = 14). rh-IT, root hair IT; T-rh, twisted root hairs; e, epidermis; c, cortex; arrows, intercellular infection; arrowheads, infection pockets. Dashed lines indicate the surface and some of the boundaries of root epidermal cells in B and E (see Supplemental Movie 1).
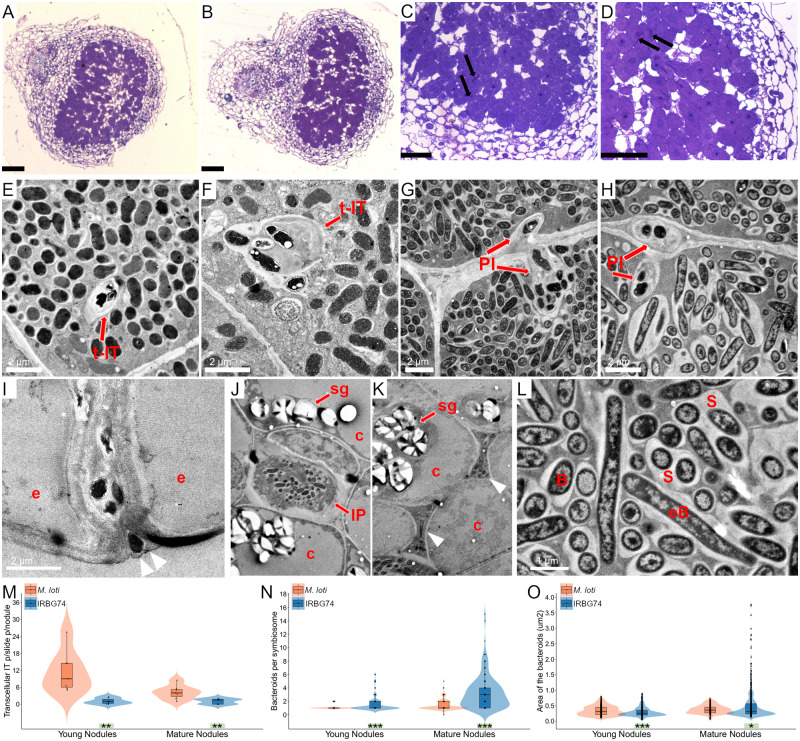
The confocal microscopy analysis revealed an intercellular infection of IRBG74 in Lotus roots. To characterize the progression of the IRBG74 infection process in more detail, the histology of young and mature nodules was analyzed by light microscopy and compared with nodules of similar developmental stage induced by M. loti at 3 wpi. Symbiosome-containing nodule cells and transcellular ITs were observed in both young and mature nodules of Lotus with both rhizobial strains (Figure3, A–D; Supplemental Figure 2, A–C). These structures were remarkably more numerous in nodules colonized by M. loti, compared with IRBG74, particularly in young nodules (Figure 3M). To discern differences in infection mechanisms and structures with more resolution, we decided to evaluate the bacteroid occupancy and infection within the nodule cells by transmission electron microscopy (TEM). Nodule cells were successfully occupied by M. loti and IRBG74 in young and mature nodules (Figure 3, E–H). The symbiosomes commonly contained one or two bacteroids in young nodules colonized by M. loti and IRBG74 (Figure 3, E–G and N). However, mature nodules infected by IRBG74 contained large symbiosomes with multiple bacteroids (Figure 3, H, L, N, and O). Similar to what was observed by confocal and light microscopy, transcellular ITs were found in nodules inoculated with this symbiont (Supplemental Figure 2E). Additionally, the TEM micrographs clearly showed a contrasting infection mechanism between nodules colonized by M. loti and IRBG74. Only the typical transcellular ITs were abundant in nodules infected by M. loti (Figure 3, E and F). In contrast, butterfly-shaped infection pegs, originated from the intercellular space of nodule cells was observed with IRBG74 (Figure 3, G and H). TEM images also documented progression of IRBG74 infection, from the intercellular infection site in the epidermis of young nodules (Figure 3I;Supplemental Figure 2D), followed by the formation of infection pockets, where IRBG74 was accumulated both in these structures and the intercellular space between uninfected cells in the cortex (Figure 3, J and K; Supplemental Figure 2D), and ultimately colonization of infected cells via infection pegs or transcellular ITs (Figure 3G;Supplemental Figure 2, E and F). In summary, using complementary approaches, we found that IRBG74 transgresses the epidermis intercellularly and forms infection pockets; subsequent progression is either via transcellular ITs or intercellularly, resulting in the formation of symbiosomes by peg infection. Thus, representing a substantial difference in infection mechanism compared with the “standard” intracellular M. loti infection process in Lotus.
Figure 3.
Histology and intercellular path of L. japonicus nodules colonized by IRBG74. Nodule sections stained with toluidine blue and visualized by light microscopy illustrate the structure and presence of transcellular ITs in young (A and C) and mature nodules (B and D) colonized by M. loti (A and B) and IRBG74 (C and D). TEM micrographs of young (E and G) and mature nodules (F and H) reveal transcellular ITs (E and F) and peg-infection (G and H) by M. loti and IRBG74, respectively. IRBG74 infection between the epidermal cells of a young nodule (I) is followed by infection pocket formation (J) and intercellular progression in the cortex (K). (L) Magnification of a nodule cell with symbiosomes and elongated IRBG74 bacteroids. Number of transcellular ITs per nodule (M), number of bacteroids per symbiosome (N) and area of the bacteroids in young and mature nodules (O) colonized by M. loti and IRBG74. Student’s t test (M) and Mann–Whitney U-test (N and O). *P<0.05; ** P<0.01; and ***P<0.001. Violin boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, individual data points. Black arrows, transcellular ITs (t-IT); Red arrows, highlight different structures. Arrowheads, intercellular colonization; IP, infection peg; PI, peg-infection; e, epidermis; c, cortex; sg, starch granules; B, bacteroids; eB, elongated bacterioids. Scale bar, 100 µm (A and B) and 50 µm (C and D).
rinrk1 and ern1 mutants show contrasting symbiotic phenotypes with IRBG74 and M. loti
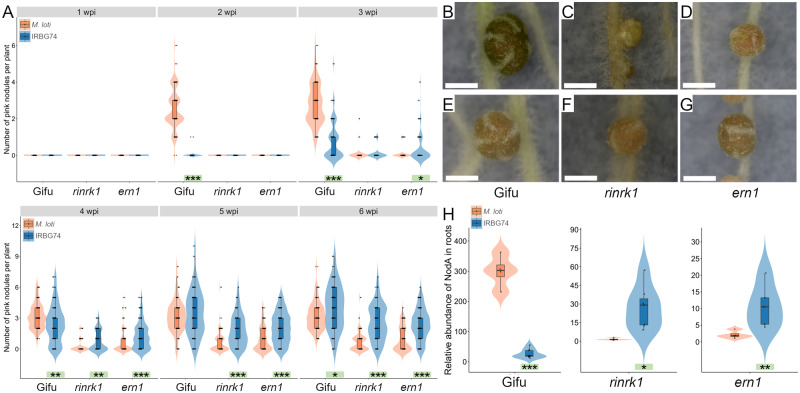
Root infection and nodule organogenesis are highly coordinated multistep processes and nodule organogenesis is affected in mutants interrupted in rhizobial infection (Oldroyd and Downie, 2008; Madsen et al., 2010). Nodulation kinetics was, therefore, a suitable readout to determine the genetic dependency of the Lotus-IRBG74 intercellular process. IRBG74- and M. loti-induced nodulations of a set of previously identified Lotus symbiotic mutants were scored at 1–6 wpi. First, mutants affected in the symbiotic receptor genes Nfr5, SymRK, RinRk1, and Epr3 were tested. Both M. loti and IRBG74 were unable to form nodules in the nfr5 and symrk plants (Supplemental Table 2), indicating that IRBG74 nodulation is NF-dependent and requires functional NF receptors for recognition of the NF produced by IRBG74 (Crook et al., 2013; Poinsot et al., 2016) to trigger downstream signal transduction. This latter was further confirmed by the Nod− phenotype observed in Lotus plants inoculated with an IRBG74 nodA mutant strain (Supplemental Table 2). However, the nodulation performance of the rinrk1 mutant was different between plants inoculated with IRBG74 and M. loti. In rinrk1 plants inoculated with M. loti, many white uninfected nodules were formed, with very few pink nodules at the different time points analyzed (Figure 4, A and C; Supplemental Figure 1C). In contrast, this hypernodulated-uninfected phenotype was not observed with IRBG74. The rinrk1 plants infected by IRBG74 developed similar numbers of nodule structures to w.t. plants inoculated with M. loti or IRBG74 at 4–6 wpi, the majority of them pink, indicating effective rhizobial colonization and symbiosis (Figure 4, A and F; Supplemental Figure 1C). The number of pink nodules was significantly higher in plants infected by IRBG74 at 3–6 wpi compared with plants colonized by M. loti, indicating that intercellular infection was not impaired in the rinrk1 mutant. The role of the expolysaccharide receptor EPR3 is apparently important for both types of infection in L. japonicus, since a comparable delayed nodulation phenotype of the epr3 mutant was observed following inoculation with M. loti or IRBG74 (Supplemental Figure 3A).
Figure 4.
Nodulation phenotype of rinrk1 and ern1 mutants. (A) Nodulation kinetics from 1 to 6 wpi with M. loti or IRBG74 in rinrk1 and ern1. Mann–Whitney U-test of pink nodules between plants inoculated with M. loti or IRBG74 in the same genetic background. n = Gifu ≥ 59; rinrk1 ≥ 49; ern1 ≥ 77. Representative images of nodules at 4 wpi with IRBG74 (E–G) or M. loti (B, C, and D) from Gifu (B and E), rinrk1 (C and F) and ern1 (D and G). Scale bar 1 mm. H, rhizobial occupancy of L. japonicus roots by qPCR. The abundance of M. loti and IRBG74 nodA gene was determined by qPCR using total DNA isolated from Gifu, rinrk1 and ern1 roots at 4 wpi with M. loti or IRBG74. The nodA accumulation was normalized to the LotjaGi1g1v0152000 gene abundance. Student’s t test of nodA abundance between roots inoculated with M. loti and IRBG74 in the same genetic background. * P<0.05; **P<0.01; and *** P<0.001. Violin boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, individual data points.
Since the infection process is also controlled by several transcriptional regulators, the role of Nin, Cyclops, Ern1, Nsp1, and Nsp2 were tested with IRBG74. The transcription factors Nin, Nsp1, and Nsp2 are indispensable for the symbiotic process established by IRBG74 and M. loti, since the mutants affected in these genes were unable to form nodules (Supplemental Table 2). The nodulation phenotype of cyclops plants inoculated with IRBG74 and M. loti was similar, and only uninfected white nodules developed (Supplemental Figure 3, B). However, a clear difference was found in the symbiotic performance of the ern1 mutant. The first pink nodules appeared at 4 wpi with M. loti (Figure 4, A and D), whereas in the presence of IRBG74, these were detected at 3 wpi with fully developed pink nodules at 4 wpi (Figure 4, A and G). This is the reverse of the kinetics shown by w.t. plants wherein pink nodules emerged at 2 and 3 wpi with M. loti and IRBG74, respectively (Figure 1, A and B). Additionally, both the total number of nodules and pink nodules were higher in ern1 mutants with IRBG74 at 3–6 wpi, compared with M. loti (Figure 4A;Supplemental Figure 1D). This difference in the symbiotic performance suggests a distinct genetic program promoting the intercellular colonization by IRBG74 in Lotus.
Both ern1 and rinrk1 mutants showed a greater ability to develop pink nodules in response to IRBG74 inoculation compared with M. loti. To assess if this response was accompanied by rhizobial infection, the root endosphere colonization was measured. The abundance of the rhizobial nodA gene was determined by quantitative real-time PCR, using total DNA extracted from roots at 4 wpi with M. loti and IRBG74 (Figure 4H). This analysis revealed that Gifu roots colonized by M. loti contained greater nodA abundance than plants inoculated with IRBG74: 299 versus 26 relative abundance units (rau), respectively. However, both the rinrk1 and ern1 mutants showed significantly lower nodA abundance in roots colonized by M. loti compared with roots inoculated with IRBG74. Interestingly, Gifu and rinrk1 roots infected by IRBG74 showed comparable levels of nodA abundance in the roots, with 26 and 27 rau, respectively (Figure 4H). These results confirm a better colonization of rinrk1 and ern1 mutants through intercellular infection by IRBG74 compared with the root hair infection by M. loti.
Nodulation by IRBG74 is negatively impacted in root hair IT mutants
In Lotus, several genes participating in a wide variety of molecular processes have been described as important for IT progression. For instance, the Lotus mutants affected in the U-box protein Cerberus (Yano et al., 2009), the nodule pectate lyase Npl1 (Xie et al., 2012) or the cytoskeleton component ScarN (Qiu et al., 2015), show defects in IT growth and progression. The contribution of these genes to the intercellular infection of IRBG74 was therefore assessed. These mutants were characterized by the formation of a high proportion of white nodules both with M. loti and IRBG74 (Supplemental Figure 3, C–E), and few pink nodules were developed in the npl1 and scarN mutants (Supplemental Figure 2, D and E). In all these mutants, the number of nodule structures was reduced when IRBG74 was used as inoculum (Supplemental Figure 3, C–E).
In Medicago, IT development has been shown to require the function of the Vapyrin, RPG, and Cbs genes. Since the participation of these genes has not been reported in Lotus–rhizobia symbiosis, their homolog counterparts were identified in the Lotus genome (https://lotus.au.dk/). For Vapyrin, two homologous genes were named LjVpy1 (LotjaGi2g1v0091200) and LjVpy2 (LotjaGi1g1v0646300; Supplemental Table 1), encoding proteins with 77% and 69% identity to the MtVPY protein, respectively. Similarly, two genes encoding proteins with 61% and 31% amino acid identities to MtRPG were named LjRPG (LotjaGi5g1v0253300) and LjRPG-like (LotjaGi5g1v0086600), respectively (Supplemental Table 1). The LjCBS found (LotjaGi2g1v0126500) showed 78% of protein similarity to MtCBS. Using the LORE1 database, mutant lines with LORE1 insertions were identified and genotyped to obtain homozygous mutant plants. A delay in nodule organogenesis was observed for all the mutants inoculated with M. loti, reflected by a reduced number of nodules within the first week after rhizobial inoculation (Supplemental Figure 3, F–J). In response to IRBG74 inoculation, nodulation was also delayed in vpy1 and vpy2 (Supplemental Fig. 3, G and H), but not in the rpg and rpg-l mutants, where the number of nodules were similar to the w. t. plants at different time points (Supplemental Fig. 3, I and J). Based on the nodule numbers, vpy1 plants were more severely impacted with IRBG74 (Supplemental Fig. 3G), whereas the vpy2 mutant was more affected with M. loti (Supplemental Fig. 3H). However, at 5 and 6 wpi, both the total number of nodules and pink nodules, tended to be similar between the w.t. and mutants. Since MtVpy and MtRPG have been described as important for IT development in Medicago, the number of root hair ITs was recorded in mutants affected in these genes in Lotus at 1 wpi with M. loti-DsRed. A significant reduction of IT numbers of around 50% in the vpy1, vpy2, and rpg mutants compared with w.t. was observed (Supplemental Table 3).
Dispensable role of ROS and ethylene in the Lotus-IRBG74 symbiosis
It has been described that ethylene plays a positive role in the intercellular infection and nodulation program in the S. rostrata–Azorhizobium symbiosis (D'Haeze et al., 2003), which occurs under flooded conditions while infection in aerated soil occurs through root hair ITs (Ndoye et al., 1994; Goormachtig et al., 1998). To determine the role of this phytohormone in the Lotus-IRBG74 symbiotic process, the nodulation kinetics of the double mutant insensitive to ethylene ein2a ein2b (Reid et al., 2018) was recorded. The first nodule structures appeared at 2 wpi in the ein2a ein2b mutant inoculated with IRBG74, a similar kinetics to that observed in w.t. plants. The mutant showed a hypernodulation phenotype in the subsequent weeks, with 6, 12, and 16 nodules in average per plant at 4–6 wpi, which contrasts with the average of 2–4 nodules per plant developed in the w.t. background. However, the total number of nodules induced by IRBG74 was considerably lower than M. loti (Supplemental Fig. 3K). In addition, ethylene production was lower in w.t. Lotus roots inoculated with IRBG74 in comparison to plants treated with M. loti (Supplemental Fig. 4). Taken together, these results suggest that ethylene is not playing a positive role in the infection and organogenesis program triggered by IRBG74 in Lotus, and more likely, acts as a negative regulator of these processes in a manner similar to intracellular infection.
In several legumes, it has been shown that reactive oxygen species (ROS) produced by respiratory burst oxidase homolog (RBOH) enzymes are required for intracellular and intercellular infection (Peleg-Grossman et al., 2007; Montiel et al., 2012; Montiel et al., 2016; Arthikala et al., 2017). To address the involvement of these compounds in the symbiotic process induced by IRBG74 in Lotus, homozygous mutant lines affected in two Rboh isoforms, LjRbohE (LotjaGi5g1v0224200) and LjRbohG (LotjaGi1g1v0771200), were obtained from the LORE1 database. LjRbohE and LjRbohG are putative orthologs of MtRbohA and PvRbohB, previously characterized genes, required for nodule functioning and rhizobial infection in Medicago and common bean, respectively (Marino et al., 2011; Montiel et al., 2012). In response to M. loti inoculation, both rbohE and rbohG showed a reduced number of nodule primordia and pink nodules at 1 wpi (Supplemental Fig. 3, L and M). However, in the ensuing weeks, these nodule structures attained similar numbers to w. t. plants at all timepoints tested. The nodulation kinetics of these rboh mutants in response to IRBG74 infection was comparable to the w.t. plants (Supplemental Fig. 3, L and M), which indicates that these genes are not playing an important role in the intercellular colonization by IRBG74 in Lotus.
Intercellular infection by IRBG74 promotes a distinct transcriptional reprogramming
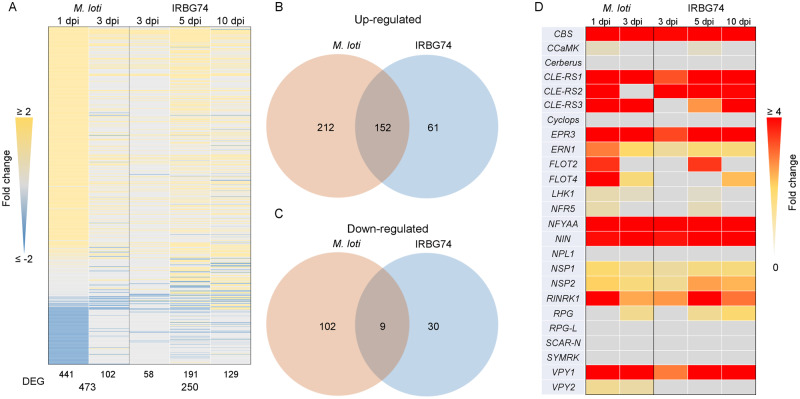
The nodulation assays of the Lotus mutants inoculated with IRBG74 showed that the genetic dependencies for intercellular and intracellular infection modes differ. To further evaluate the signaling pathways involved in intercellular infection, RNAseq transcriptome data were collected and analyzed from the susceptible infection zone of IRBG74-inoculated Lotus roots at 3, 5, and 10 dpi, to cover the infection phase preceding nodule organogenesis. We decided to avoid the massive transcriptome program triggered during nodule development after 10 dpi, since this would hinder the identification of genes regulated during intercellular infection. These data were compared with available RNAseq information on the Lotus root susceptible zone and whole roots at 1 and 3 dpi with M. loti, respectively (Mun et al., 2016; Kamal et al., 2020), since these time points encompass different rhizobial infection stages in the root hairs, prior to the nodule organogenesis program. Due to the lack of information on the transcriptomic response in Lotus roots during intercellular infection and its notorious nodulation delay, first a low stringency analysis of differentially expressed genes (DEGs) was performed (P-adjust <0.5). In response to M. loti and IRBG74 inoculation, a total of 12,637 and 10,947 DEG were identified with these criteria, respectively (Supplemental Figure 5A; Supplemental Table 4). The largest transcriptome responses were observed at 1 dpi with M. loti (12,534) and at 10 dpi with IRBG74 (9,438) DEG (Supplemental Figure 5B). A more stringent analysis, with the DEG showing a LOG2FC ≥2, revealed that the most important transcriptome response was triggered at 5 dpi IRBG74 (Figure 5A). Interestingly, only 33% of the DEG by M. loti were similarly affected by IRBG74 and a large proportion of the up/downregulated genes during M. loti infection (314: sum of the up- and downregulated) were not similarly affected by IRBG74 (Figure 5, B and C). Additionally, the majority of the genes downregulated in response to IRBG74 inoculation were not repressed in the M. loti transcriptome (Figure 5, B and C). Through different stringency criteria, we found that IRBG74 induces a progressive increase in the number of DEG from 3 to 10 dpi, where only 1.3% (129 genes) showed >two-fold change at 10 dpi. In contrast, at 3 dpi, 17% (58) of the DEG had a more than two-fold change, showing a similar response to roots inoculated with M. loti at the same time point (Supplemental Figure 5C).
Figure 5.
Comparison of the transcriptomic profile of L. japonicus roots during intracellular and intercellular rhizobial infection. (A) Heat-map expression of DEG [Log2 fold change {FC} ≥2 and a P-adjust <0.5] in Lotus roots after M. loti or IRBG74 inoculation. (B and C) Venn diagrams with total number (M. loti: 1 and 3 dpi; IRBG74: 3, 5, and 10 dpi) of the up/downregulated DEG during intracellular (M. loti) and intercellular (IRBG74) colonization. (D) Heat-map expression of known symbiotic genes significantly (P-adjust <0.5) induced after rhizobial perception.
The nodulation kinetics of Lotus inoculated with IRBG74 revealed that nodule organogenesis was delayed with respect to plants inoculated with M. loti. To determine if this delay was linked to a deficient induction of the early symbiotic signaling pathway, the expression profile of several genes known to be involved in infection and/or nodule organogenesis were analyzed using the transcriptome data set. Most of the known symbiotic genes tested were induced by M. loti or IRBG74 (Figure 5D), but when the same time point was compared (3 dpi), several genes were less upregulated in response to IRBG74 inoculation (Figure 5D).
Cytokinin signaling is differentially regulated in response to IRBG74
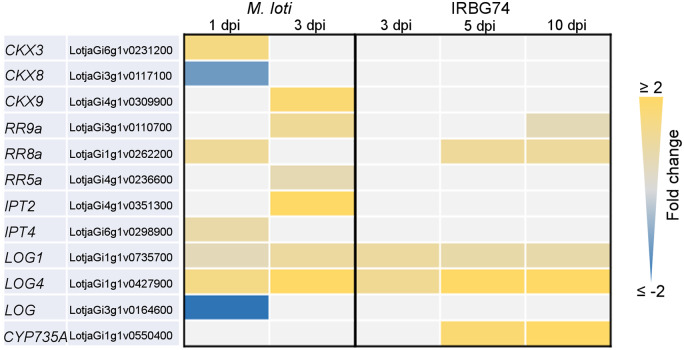
This work showed that Ern1, a transcription factor implicated in cytokinin signaling (Cerri et al., 2017; Kawaharada et al., 2017), has a less relevant role during intercellular infection by IRBG74. These results suggest that this mode of infection triggers a different transcriptional response of cytokinin-related genes compared with intracellular infection. To validate this hypothesis, a transcriptome heat-map was created for genes involved in cytokinin synthesis and regulation and significantly affected by M. loti or IRBG74 inoculation (Figure 6). This approach confirmed the different gene-expression response of several components of cytokinin regulation during IRBG74 infection. Particularly, genes encoding the cytokinin degrading enzymes Ckx3, Ckx9, the response regulator involved in cytokinin signaling RR11a, and the cytokinin biosynthesis gene Ipt2 were poorly or insignificantly induced during intercellular infection, relative to their evident upregulation after M. loti inoculation (Figure 6). Mesorhizobium loti triggered a significant reduction in the expression levels of Ckx2, Ckx8, Lhk2, RR3a, RR19, and Log. However, most of these genes were not significantly downregulated by IRBG74 colonization. In contrast, the cytokinin biosynthesis gene Cyp735a, which converts iP to tZ type cytokinins, showed a strong upregulation after IRBG74 colonization at 5 and 10 dpi, whereas its gene expression was only slightly altered during intracellular infection at early timepoints (Figure 6).
Figure 6.
Heatmap of DEGs encoding cytokinin-related proteins in L. japonicus roots after rhizobial inoculation. The heatmap highlights the differences in the expression profile of DEG [log2-fold change {FC} ≥0.5, P-adjust <0.5) involved in the metabolism, perception, signaling and synthesis of cytokinins in response to M. loti or IRBG74 infection.
Cyp735a, Ipt4, and Lhk1 are relevant players in the Lotus-IRBG74 symbiosis
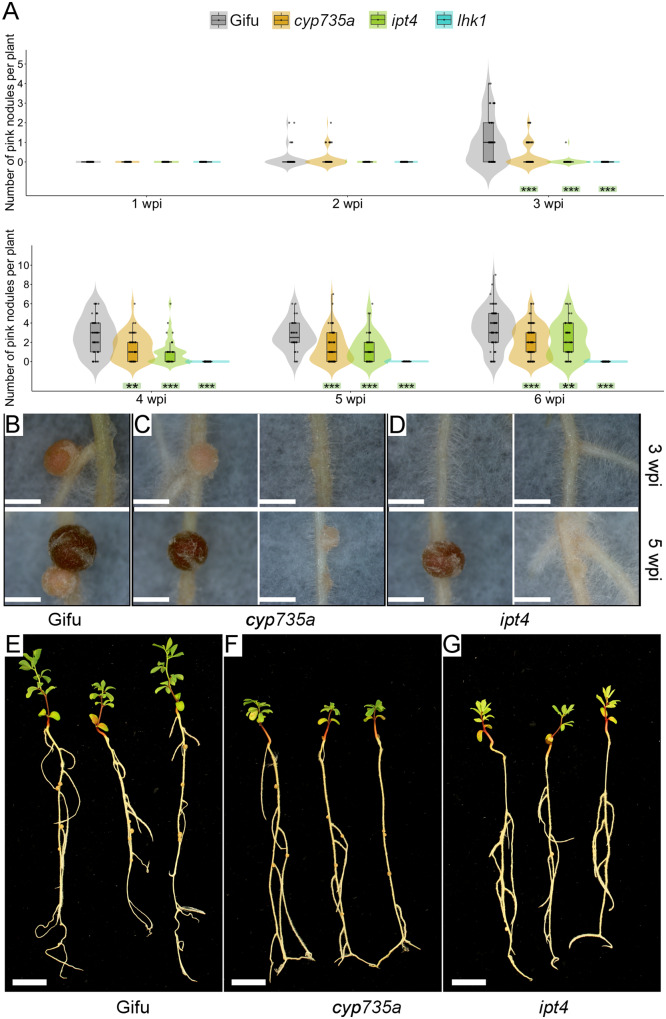
In the Lotus–M. loti symbiosis, the cytokinin receptor mutant lhk1 shows a delayed and reduced nodulation (Murray et al., 2007), whereas the ipt4 and cyp735a mutants show minor or insignificant phenotypes, respectively (Reid et al., 2017; Supplemental Fig. 6). The RNAseq data presented in this study indicates that these genes are differentially regulated by M. loti or IRBG74. In order to determine the relevance of these cytokinin-related genes in the Lotus-IRBG74 symbiosis, nodulation kinetics at 1–6 wpi were scored for the cyp735a, ipt4, and lhk1 mutants after IRBG74 inoculation. The nodulation capacity of both cyp735a and ipt4 mutants was substantially reduced, although with different symbiotic phenotypes. The cyp735a mutant developed similar numbers of nodules to w. t. plants, at 2 and 3 wpi (Figure 7A;Supplemental Figure 1E), but at 4–6 wpi cyp735a formed more nodule-like structures, most of them uninfected white nodules (Figure 7C, right panel). In contrast, at 2–4 wpi, the total number of nodules in ipt4 was lower compared with w.t. plants (Supplemental Figure 1E). The number of pink nodules was reduced at all timepoints tested and these comprised a mixture of pink and pale pink nodules (Figure 7, A and D, right panel). Interestingly the lhk1 mutant was unable to develop any nodule structure in the presence of IRBG74 (Figure 7A;Supplemental Figure 1E). This drastic symbiotic phenotype contrasts with the lhk1 plants inoculated with M. loti, where several pink nodules were observed at 6 wpi (Supplemental Figure 6). These results further demonstrate the different cytokinin regulation during intercellular infection in Lotus, whereby Cyp735a, Ipt4, and Lhk1 are important players for this type of rhizobial infection (Figure 8).
Figure 7.
Symbiotic phenotype of L. japonicus mutants affected in cytokinin-related genes. (A) Nodulation kinetics of cyp735a, ipt4, and lhk1 plants from 1 to 6 wpi with IRBG74. Mann–Whitney U-test of pink nodules. **P<0.01; ***P<0.001. n = 49 (Gifu), 86 (cyp735a), 53 (ipt4), and 55 (lhk1). Phenotype of nodules developed in wild-type Gifu (B), cyp735a (C) and ipt4 (D) plants at 3 (upper panel) and 5 wpi (lower panel) with IRBG74. Representative images of wild type Gifu (E), cyp735a (F), and ipt4 (G) plants at 6 wpi with IRBG74. Scale bar, 1 mm (B–D) and 1 cm (E–G). Violin boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, individual data points.
Figure 8.
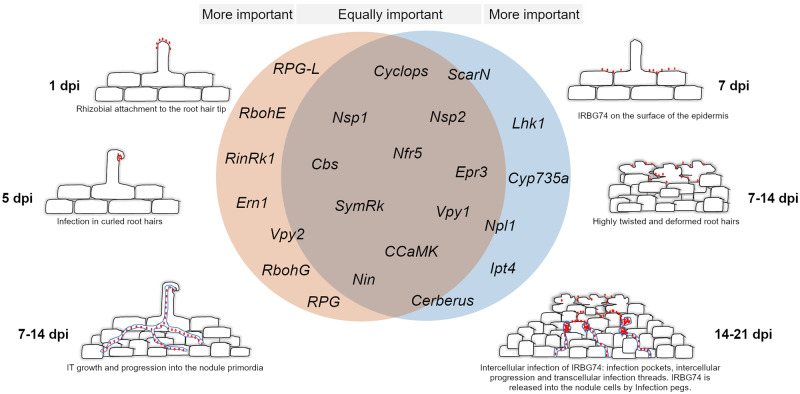
Comparative model of gene requirements in the intracellular and intercellular symbiotic program in L. japonicus. A core of essential genes is recruited by L. japonicus for root nodule symbiosis, regardless the type of infection mechanism employed by rhizobia. Nonetheless, certain players are particularly more relevant depending on the mode of colonization. Intercellular infection in L. japonicus appears to be more sensitive to the absence of certain cytokinin-related genes. IRBG74 colonizes L. japonicus roots intercellularly, provoking massive root hair deformation. The intercellular progression of IRBG74 is followed by transcellular and infection peg in the nodule cells. Red dots: M. loti (left panel) and IRBG74 (right panel).
Discussion
Lotus-IRBG74 symbiosis: a working model to study intercellular infection
Intercellular colonization of legumes by rhizobia occurs via a variety of entry modes. Bacteria can penetrate through middle lamellae of root hairs, cracks at emergent lateral roots or between epidermal cells (Subba-Rao et al., 1995; Gonzalez-Sama et al., 2004; Goormachtig et al., 2004; Bonaldi et al., 2011). Diverse Lotus spp. exhibit this infection mechanism, which leads to the formation of either ineffective or nitrogen-fixing nodules, depending on the growth conditions and rhizobial partner (Ranga Rao, 1977; James and Sprent, 1999; Liang et al., 2019). Previously, it was described that under certain mutant background conditions and with a low frequency, Lotus roots can be intercellularly colonized by M. loti (Madsen et al., 2010) by an NF-independent mechanism. In our study, we have formulated a working model in which intercellular infection can be effectively studied in a model legume, and therefore, our approaches were focused on this process. The compatibility and nodulation rate of the L. japonicus–IRBG74 symbiosis represents an optimal system to analyze the intercellular infection, taking advantage of a comprehensive set of methodologies, techniques, and resources (Sandal et al., 2002; Malolepszy et al., 2016; Mun et al., 2016; Kelly et al., 2018 ; Kamal et al., 2020; Shah et al., 2020). These resources have not been developed for the Lotus burttii– Rhizobium leguminosarum Norway interaction (Liang et al., 2019), whereas the L. japonicus–R. leguminosarum Norway approach is excluded by the inability of R. leguminosarum Norway to induce nodules on L. japonicus (Liang et al., 2018). We show that IRBG74, a nitrogen-fixing strain originally isolated from Sesbania spp., massively accumulates on the root surface of Lotus roots and invades the roots between the epidermal and root hair cells. This intercellular invasion transforms into transcellular ITs or peg infection structures, which allows the gene-dependencies of these processes to be distinguished from the genes required for root hair ITs. Intercellularly infection by IRBG74 at lateral root junctions of Sesbania spp. has also been observed under flooded conditions (Cummings et al., 2009) and in S. rostrata, where the infection pocket, formed by an intercellular invasion of certain Azorhizobium spp., is followed by transcellular ITs (Goormachtig et al., 2004). Although IRBG74 promoted nodule formation in Lotus, the organogenesis and infection programs were delayed, compared with the symbiotic proficient M. loti. This was evidenced by a reduced plant growth promotion, compared with that obtained by M. loti, which indicates that IRBG74 might not be fully compatible with Lotus. The delay could be explained by differences in NF composition and/or abundance, produced by M. loti R7A and IRBG74. More than 40 different NF structures are synthesized by IRBG74, mostly pentasaccharides, carbamoylated, and fucosylated, with N-methylation at the nonreducing end (Poinsot et al., 2016). These chemical modifications can be found in the NF synthesized by M. loti (Lopez-Lara et al. 1995; Bek et al., 2010); however, the terminal decorations in the reducing end executed by nodZ and nolL are particularly relevant, since Lotus spp. inoculated with R7AΔnodZ and R7AΔnolL mutants, exhibit delayed nodulation development (Rodpothong et al., 2009). Likewise, the timing could be impacted by differences in the cell surface composition of IRBG74 since production and composition of EPS, LPS, and KPS affect the nodulation kinetics of rhizobial strains in Lotus (D'antuono et al., 2005; Townsend et al., 2006; Kelly et al., 2013).
Common gene dependencies of intercellular and intracellular colonization
The characterization of several legume mutants has identified the molecular players of the symbiotic pathway, from the early signaling to nodule organogenesis. NF receptors are indispensable for initiating symbiotic signaling, since Lotus mutants disrupted in these genes do not show any symbiotic response (Madsen et al., 2003; Radutoiu et al., 2003). The nodulation tests of nfr5 with IRBG74 and w.t. plants with an IRBG74-nodA mutant revealed that an NF signaling pathway is required for the Lotus–IRBG74 symbiosis. Likewise, both intercellular rhizobial infection and nodule organogenesis is an NF-dependent process in the S. rostrata–A. caulinodans relationship (Capoen et al., 2010), but there are also intercellular processes whereby an NF-independent mechanism can lead to nitrogen-fixing nodules in certain legumes (Giraud et al., 2007; Madsen et al., 2010; Ibañez and Fabra, 2011). This study revealed a genetic machinery that is equally important for both types of infection modes, since a similar detrimental impact on the nodulation process was observed in the nfr5, symrk, ccamk, cyclops, nin, nsp1, nsp2 epr3, cbs, and vpy1 mutants whether M. loti or IRBG74 were used as inoculum in Lotus (Figure 8). This set of results shows that these genes participate in the formation of root hair ITs, transcellular ITs, and peg structures. However, the intracellular symbiotic program was more affected in the rinrk1, ern1, rbohE, rbohG, rpg, rpg-like, and vpy2 mutants. One interpretation is that these genes are more important for formation of root hair ITs than transcellular ITs and peg structures. Previously, it was described in A. evenia that SYMRK, CCaMK, and the HK1 were required both in the intracellular and intercellular infection by Bradyrhizobium (Fabre et al., 2015), reinforcing the notion of a common genetic repertoire for these types of rhizobial infection. Likewise, SYMRK and CCaMK are required for intercellular infection and nodulation in the IRBG74–Lotus symbiosis. However, intercellular colonization is apparently more sensitive to the absence of certain genes. cerberus, npl1, and scarN mutants developed numerous white nodules, frequently uninfected after M. loti inoculation (Yano et al., 2009; Xie et al., 2012; Qiu et al., 2015), but when IRBG74 was used as inoculum, nodule development and the number of pink nodules were severely reduced in these mutants. This suggests that actin rearrangement plays an important role in formation of cortical and transcellular ITs and that initiation of ITs from intercellular infection pockets is more dependent on actin rearrangement.
As mentioned above, there are few reports describing the molecular components required for intercellular infection. One of them, revealed the positive role of ROS to induce cell death during the crack entry infection of Azorhizobium in S. rostrata. Deprivation of ROS production by applying diphenyleneiodonium chloride, an inhibitor of the ROS-producing enzymes RBOHs, prevents rhizobial colonization in this legume (D'Haeze et al., 2003). Similarly, these genes have been implicated in the IT development during intracellular rhizobial infection in Medicago and P. vulgaris (Peleg-Grossman et al., 2007; Montiel et al., 2012; Arthikala et al., 2017). Likewise, the nodulation program was delayed in Lotus mutants disrupted in RbohE or RbohG genes after inoculation with M. loti. The intercellular symbiotic performance by IRBG74 was not negatively impacted in the rbohE and rbohG mutants, which indicates that ROS produced by these isoforms is not relevant in this process, however since the LjRboh gene family is composed by nine members (Montiel et al., 2018), other isoforms could contribute to the intercellular infection by IRBG74. Inhibition of ethylene synthesis or perception has a negative effect on the nodulation and intercellular infection induced by Azorhizobium in S. rostrata (D'Haeze et al., 2003). In contrast, ethylene plays a negative role in the Lotus–M. loti symbiosis. The ein2a ein2b double mutant that exhibits complete ethylene insensitivity is hyperinfected and hypernodulated by M. loti (Reid et al., 2018). Unlike, the intercellular symbiotic process in S. rostrata, where ethylene is required for nodulation, the ein2a ein2b mutant was hypernodulated by IRBG74, indicating that this phytohormone is not essential for the IRBG74 intercellular infection. Consequently, the positive role of ethylene in the intercellular infection is not conserved in Lotus, which highlights the relevance of a better understanding of the intercellular colonization in legumes.
Distinctive cytokinin signaling program during intercellular infection
The dual role of cytokinins, as positive and negative regulators of nodule development and rhizobial infection, respectively, makes them key phytohormones in the legume–rhizobia symbiosis (Miri et al., 2016). The lhk1 mutant belatedly develops a reduced number of pink nodules in response to M. loti infection, but when IRBG74 is used as inoculum the mutant is unable to form nodules up to 6 wpi. In the intracellular infection mediated by M. loti, it has been suggested that other cytokinin receptors are sufficient to induce nodule organogenesis in the absence of Lhk1 (Murray et al., 2007). However, the signaling pathway triggered by LHK1 is indispensable in the Lotus–IRBG74 symbiosis. Conversely, the ern1 mutant displayed improved symbiotic performance with IRBG74, which further confirms that depending on the type of rhizobial infection program, distinct signaling pathways are triggered. This was reflected in the different transcriptomic responses of genes involved in the synthesis, perception signaling and metabolism of cytokinin induced by M. loti or IRBG74. It has been shown that different cytokinin biosynthesis genes are induced during intracellular colonization of M. loti in Lotus roots (Reid et al., 2016, 2017). Although the cytokinin trans-hydroxylase Cyp735a is highly induced by M. loti, the nodulation performance is not significantly affected in Lotus plants disrupted in this gene (Reid et al., 2017). In contrast, nodule organogenesis is delayed and reduced in cyp735a mutants inoculated with IRBG74. Cyp735a encodes cytochrome P450 monooxygenases (P450s) that catalyze the biosynthesis of trans-Zeatin, an isoprenoid cytokinin compound (Takei et al., 2004), indicating that tZ cytokinins play a more relevant role during intercellular colonization. The mutant affected in the isopentenyl transferase 4 (Ipt4) gene showed a mild impact on the nodulation capacity with M. loti (Reid et al., 2017). However, in response to IRBG74 inoculation, the development of nodules was evidently delayed, and the number of pink nodules reduced. IPT is placed in the first step during isoprenoid cytokinin biosynthesis, giving rise to iP riboside 50-diphosphate or iP riboside 50-triphosphate intermediates, which can be converted by CYP735a to tZ cytokinins (Hwang and Sakakibara, 2006). The delayed nodulation and enhanced phenotypes of the cytokinin-related mutants may indicate that the peak of cytokinin triggered by the intercellular program is lower or more dispersed in the root relative to the highly localized cytokinin signaling achieved in the intracellular infection modes.
Conclusions
The intracellular colonization has been extensively described at the cellular and molecular levels in recent decades, but there is little knowledge about the molecular players controlling the intercellular invasion. In this study, we find that Lotus has a genetic repertoire that confers on it the ability to establish effective symbiotic processes with rhizobia, both intracellularly and intercellularly, using a core genetic machinery. However, there is a set of molecular players that are dispensable or recruited, depending on the mode of infection. Different approaches revealed that the cytokinin signaling pathway is apparently a key difference to be further analyzed. Similarly, other components seem to be differentially relevant for this type of infection. For instance, the rinrk1 receptor mutant showed a better nodulation performance with IRBG74 compared with M. loti. The latter indicates that during intercellular infection, certain uncharacterized ligands are not entirely necessary for rhizobial colonization. This study opens new research questions, for instance, the identification of novel players exclusively required for intercellular infection using the Lotus mutant collections. A comparative analysis of the molecular components required for the different intercellular colonization modes in Robinoid, Genistoid, and Dalbergoid legumes would also be interesting and might contribute to our understanding of symbiosis from an evolutionary perspective. Although the plant genes are crucial for this process, the identification of the bacterial genes promoting intercellular infection would have an impact on microbiome studies.
Materials and methods
Germination and nodulation assays
Lotus seeds of accession Gifu (Handberg and Stougaard, 1992) were scarified with sandpaper, surface disinfected with 0.3% (v/v) of sodium hypochlorite for 10 min and then washed five times with autoclaved distilled water, to remove traces of chlorine. The washed seeds were incubated overnight at 4°C and then transferred to square Petri dishes for germination at 21°C. For monitoring nodulation kinetics, 3-d postgermination seedlings (n ≥ 20 plants per condition) were placed into square Petri dishes with 1.4% B&D agar slant covered with filter paper and inoculated with the respective rhizobial strains (1 mL of bacterial culture per plate; OD600 = 0.05). Through this method, Lotus roots were kept in contact with the inoculum throughout the experiment. After rhizobial inoculation the number of white (bumps and nodule primordia) and pink nodules were recorded weekly until 6 wpi with a stereomicroscope. The plants were harvested at 6 wpi to measure the fresh weight and length of the aerial part. LORE1 lines (Urbanski et al., 2012; Malolepszy et al., 2016) disrupted in the genes of interest were ordered from the LORE1 database (https://lotus.au.dk/) and genotyped with allele-specific primers (Supplemental Table 5) to obtain homozygous mutants following the database instructions (Mun et al., 2016). The gene IDs and the corresponding LORE1 IDs are shown in Supplemental Table 1. Both the w.t. and mutant plants used in this study are in the Gifu accession, except for the lines MG-134, MG-144, and MG-145 used in Supplemental Figure 1.
Infection phenotyping using confocal microscopy
Three-day-old seedlings of Lotus (accession Gifu; Handberg and Stougaard, 1992) were placed on 1/4 B&D plates and inoculated with fluorescently labelled M. loti R7A (Kelly et al., 2013) or IRBG74 strains (Cummings et al., 2009), obtained through transformation with the constitutive DsRED expressing plasmid pSKDSRED (Kelly et al., 2013). The roots were harvested from the plates at different time points. To enable observations in deeper parts of the tissue, a fluorescent compatible clearing protocol was used as described before (Nadzieja et al., 2019). Cleared roots were visualized by confocal microscopy with the following excitation lasers/emission cut-offs: 405/408–498 nm (autofluorescence: intensity, 3%–11%; Gain, 570–620), 561/517–635 nm (DsRed: intensity, 5%–12%; Gain, 620–640). For 3D projections, Fiji ImageJ (Schindelin et al., 2012) was used to create animation frames, which then were rearranged using Adobe Photoshop CC into final projections.
Nodule histology analysis
Young and mature nodules at 3 wpi with IRBG74 or M. loti were detached from Lotus roots, sliced in half, and incubated overnight in fixative solution (2.5% glutaraldehyde, 0.1 M sodium cacodylate pH 7). The fixed nodules slices were embedded in acrylic resin and sectioned for light and TEM. The nodule slices were stained with Toluidine blue for light microscopy analysis (James and Sprent, 1999; Madsen et al., 2010). The number of transcellular IT, bacteroids per symbiosome and area of the bacteroids were recorded from images taken of 10 slides from 7 different young and mature nodules colonized by IRBG74 or M. loti.
Rhizobial occupancy by qPCR
Four wpi roots (including nodules) were surface disinfected with 0.3% of sodium hypochlorite and 70% EtOH solution for 1 min to remove rhizobia attached to the root surface, and then washed five times with distilled water. Later, DNA was isolated from individual roots (Chomczynski and Rymaszewski 2006) and the concentration adjusted to be used as a template (10 ng/µL). NodA abundance from M. loti R7A (forward: TATGAGCCGACCGGAGCCTTCAAT and reverse: CCGTATAGACCGAGTTCAGCGACCA) and IRBG74 (forward: GAACTGCAAGTTGACGATCACGC and reverse: AAACGTCGTAACAAGCCCATGTGG) was measured by qPCR. The expression values were normalized to the abundance of the Lotus gene (LotjaGi1g1v0152000.1; forward: GAAGGACCCAGAGGATCACA and reverse: CGGTCTTCGTACTTCTTCGC) using the delta Ct method (Pfaffl, 2001).
RNAseq of Lotus roots and bioinformatics
The susceptible infection zone of L. japonicus roots by IRBG74 (elongation and maturation zone of the root) was cut and then freezed in liquid nitrogen from seedlings at 3, 5, and 10 dpi with IRBG74 (OD600 = 0.05) or mock-treated (water) at the same time points. Total RNA was isolated and DNA contamination was removed by DNAse treatment. Library preparations using randomly fragmented mRNA were performed by IMGM laboratories (Martinsried, Germany) and sequenced in paired-end 150-bp mode on a Illumina NovaSeq 6000 instrument.
A decoy-aware index was built for Gifu transcripts using default Salmon parameters and reads were quantified using the validateMappings flag (Salmon version 0.14.1; (Patro et al., 2017). Normalized expression levels and differential expression testing were performed using the R-package DESeq2 version 1.20 (Love et al., 2014) after summarizing gene level abundance using the R-package tximport (version 1.8.0).
Statistical analysis
Significant differences of pink and total number of nodules among different inoculums and genetic backgrounds were calculated by Mann–Whitney U-test. Other parameters evaluated in this study were determined by Student’s t test. The calculated expression values and statistics of the RNAseq data are included as Supplemental Table 4.
Accession numbers
The RNAseq reads associated with this study are available in the SRA under bioproject accession number PRJNA632725. The L. japonicus (accession Gifu and MG20) gene identifiers are shown in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nodulation kinetics in different L. japonicus accessions and mutants.
Supplemental Figure S2. Nodule cell occupancy and infection in L. japonicus nodules colonized by Mesorhizobium loti or IRBG74.
Supplemental Figure S3. Nodulation performance of L. japonicus mutants.
Supplemental Figure S4. Ethylene production in L. japonicus roots inoculated with rhizobia.
Supplemental Figure S5. Differentially expressed genes (DEG) in L. japonicus roots after rhizobial inoculation.
Supplemental Figure S6. Phenotype of L. japonicus mutants at 6 wpi with Mesorhizobium loti or IRBG74.
Supplemental Table S1. List of L. japonicus mutants used in this study.
Supplemental Table S2. Mutants with a Nod-phenotype in response to Mesorhizobium loti or IRBG74 inoculation.
Supplemental Table S3. Number of ITs per plant at 1 wpi with Mesorhizobium loti.
Supplemental Table S4. RNAseq expression data of L. japonicus roots inoculated with Mesorhizobum loti (1 and 3 dpi) or IRBG74 (3, 5, and 10 dpi).
Supplemental Table S5. List of primers used for genotyping.
Supplemental Movie S1. 3D-projection of L. japonicus roots infected by IRBG74 at 1–3 wpi.
Supplementary Material
Acknowledgments
We thank the assistance in the greenhouse of Finn Pedersen and Nanna Walther. We thank Dr. Fang Xie for providing rinrk1 seeds.
Funding
This work was supported by the grant Engineering the Nitrogen Symbiosis for Africa made to the University of Cambridge by the Bill & Melinda Gates Foundation (ENSA; OPP11772165). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 834221).
Conflict of interest statement. The authors declare no competing financial interest or other conflict of interest.
J.M., D.R., C.M.B., and T.H.G. characterized and isolated the mutants. D.R. and J.M. processed and analyzed the RNAseq data, respectively. M.N., F.A.D., and E.K.J. performed the microscopy analyses. J.M. wrote the manuscript and prepared the figures with contributions of the co-authors. J.M., S.K., T.O., and J.S. conceived the research plan. J.S. and S. K. coordinated and guided the research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Jens Stougaard (stougaard@mbg.au.dk).
References
- Aguilar A, Peralta H, Mora Y, Díaz R, Vargas-Lagunas C, Girard L, Mora J (2016) Genomic comparison of Agrobacterium pusense strains isolated from bean nodules. Front Microbiol 7:1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Cartieaux F, Brown SC, Rodier-Goud M, Boursot M, Fardoux J, Patrel D, Gully D, Fabre S,, Chaintreuil C, et al. (2012) Aeschynomene evenia, a model plant for studying the molecular genetics of the nod-independent rhizobium-legume symbiosis. Mol Plant Microbe Interact 25:851–861 [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Godfroy O, de Billy F, Saurat O, Jauneau A, Gough C (2008) The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc Natl Acad Sci U S A 105:9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthikala MK, Montiel J, Sanchez-Lopez R, Nava N, Cardenas L, Quinto C (2017) Respiratory burst oxidase homolog gene A is crucial for rhizobium infection and nodule maturation and function in common bean. Front Plant Sci 8:2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek AS, Saure J, Thygesen MB, Duus JO, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S (2010) Genetically based variation in LysM receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol Plant Microbe Interact 23:58–66 [DOI] [PubMed] [Google Scholar]
- Biswas JB, Ladha JK, Dazzo FB, Yanni YG, Rolfe BG (2000) Rhizobial inoculation influences seedling vigor and yield of rice. Agron J 92:880–886 [Google Scholar]
- Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N, Goormachtig S, Giraud E (2011) Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol Plant Microbe Interact 24:1359–1371 [DOI] [PubMed] [Google Scholar]
- Boogerd FC, Van Rossum D (1997) Nodulation of groundnut by Bradyrhizobium: a simple infection process by crack entry. FEMS Microbiol Rev 21:5–7 [Google Scholar]
- Capoen W, Den Herder J, Sun J, Verplancke C, De Keyser A, De Rycke R, Goormachtig S, Oldroyd G, Holsters M (2009) Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell 21:1526–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M (2005) SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci U S A 102:10369–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Oldroyd G, Goormachtig S, Holsters M (2010) Sesbania rostrata: a case study of natural variation in legume nodulation. New Phytol 186:340–345 [DOI] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160:2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Wang Q, Stolz P, Folgmann J, Frances L, Katzer K, Li X, Heckmann AB, Wang TL, Downie JA, et al. (2017) The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol 215:323–337 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Rymaszewski M (2006) Alkaline polyethylene glycol‐based method for direct PCR from bacteria, eukaryotic tissue samples, and whole blood. BioTechniques 40:454–458 [DOI] [PubMed] [Google Scholar]
- Coba de la Peña T, Fedorova E, Pueyo JJ, Lucas MM (2017) The symbiosome: legume and rhizobia co-evolution toward a nitrogen-fixing organelle? Front Plant Sci 8:2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook MB, Mitra S, Ane JM, Sadowsky MJ, Gyaneshwar P (2013) Complete genome sequence of the sesbania symbiont and rice growth-promoting endophyte Rhizobium sp. Strain IRBG74. Genome Announc 1: e00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings SP, Gyaneshwar P, Vinuesa P, Farruggia FT, Andrews M, Humphry D, Elliott GN, Nelson A, Orr C, Pettitt D, et al. (2009) Nodulation of sesbania species by Rhizobium (Agrobacterium) strain IRBG74 and other rhizobia. Environ Microbiol 11:2510–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler MR (1978) Some observations on infection of Arachis hypogaea L. by rhizobium. J Exp Bot 29:749–755 [Google Scholar]
- D'antuono AL, Casabuono A, Couto A, Ugalde RA, Lepek VC (2005) Nodule development induced by Mesorhizobium loti mutant strains affected in polysaccharide synthesis. Mol Plant Microbe Interact 18:446–457 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, De Rycke R, Mathis R, Goormachtig S, Pagnotta S, Verplancke C, Capoen W, Holsters M (2003) Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc Natl Acad Sci U S A 100:11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Faria S, Hay G, Sprent JI (1988) Entry of rhizobia into roots of Mimosa scabrella Bentham occurs between epidermal cells. Journal of General Microbiology 134:2291–2296 [Google Scholar]
- Downie JA (2014) Legume nodulation. Curr Biol 24:R184–R190 [DOI] [PubMed] [Google Scholar]
- Fabre S, Gully D, Poitout A, Patrel D, Arrighi JF, Giraud E, Czernic P, Cartieaux F (2015) Nod factor-independent nodulation in Aeschynomene evenia required the common plant-microbe symbiotic toolkit. Plant Physiol 169:2654–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, et al. (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sama A, Mercedes-Lucas MM, De Felipe MR, Pueyo JJ (2004) An unusual infection mechanism and nodule morphogenesis in white lupin (Lupinus albus). New Phytol 163:371–380 [DOI] [PubMed] [Google Scholar]
- Goormachtig S, Mergaert P, Van Montagu M, Holsters M (1998) The symbiotic interaction between Azorhizobium caulinodans and Sesbania rostrata. Molecular cross‐talk in a beneficial plant–bacterium interaction. Plant–Microbe interactions 29:117–164 [DOI] [PubMed] [Google Scholar]
- Goormachtig S, Capoen W, James EK, Holsters M (2004) Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc Natl Acad Sci U S A 101:6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Sarkar M, Ganguly P, Uddin MR, Mandal S, DasGupta M (2016) Segregation of nod-containing and nod-deficient bradyrhizobia as endosymbionts of Arachis hypogaea and as endophytes of Oryza sativa in intercropped fields of Bengal Basin, India. Environ Microbiol 18:2575–2590 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2:487–496 [Google Scholar]
- Hashiguchi M, Abe J, Aoki T, Anai T, Suzuki A, Akashi R (2012) The National BioResource Project (NBRP) Lotus and Glycine in Japan. Breed Sci 61:453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA,, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142:1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Liao J, James EK, Sato S, Tabata S, Jurkiewicz A, Madsen LH, Stougaard J, Ross L, Szczyglowski K (2012) Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol 160:917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sakakibara H (2006) Cytokinin biosynthesis and perception. Physiologia Plantarum 126:528–538 [Google Scholar]
- Ibañez F, Fabra A (2011) Rhizobial Nod factors are required for cortical cell division in the nodule morphogenetic programme of the Aeschynomeneae legume Arachis. Plant Biol (Stuttg) 13:794–800 [DOI] [PubMed] [Google Scholar]
- Ibañez F, Wall L, Fabra A (2017) Starting points in plant-bacteria nitrogen-fixing symbioses: intercellular invasion of the roots. J Exp Bot 68:1905–1918 [DOI] [PubMed] [Google Scholar]
- James EK, Sprent JI (1999) Development of N2‐fixing nodules on the wetland legume Lotus uliginosus exposed to conditions of flooding. New Phytol 142:219–231 [Google Scholar]
- James EK, Sprent JI, Sutherland JM, Mcinroy SG, Minchin FR (1992) The structure of nitrogen fixing root nodules on the aquatic Mimosoid legume Neptunia plena. Ann Botany 69:173–180 [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308:1786–1789 [DOI] [PubMed] [Google Scholar]
- Kamal N, Mun T, Reid D, Lin J-S, Akyol TY, Sandal N, Asp T, Hirakawa H, Stougaard J, Mayer KFX, et al. (2020) A chromosome-scale Lotus japonicus Gifu genome assembly indicates that symbiotic islands are not general features of legume genomes. bioRxiv 2020.04.17.042473
- Kawaharada Y, James EK, Kelly S, Sandal N, Stougaard J (2017) The ethylene responsive factor required for nodulation 1 (ERN1) transcription factor is required for infection-thread formation in Lotus japonicus. Mol Plant Microbe Interact 30:194–204 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszynski A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523:308–312 [DOI] [PubMed] [Google Scholar]
- Kelly S, Mun T, Stougaard J, Ben C, Andersen SU (2018) Distinct Lotus japonicus Transcriptomic Responses to a Spectrum of Bacteria Ranging From Symbiotic to Pathogenic. Front in Plant Sci 9: 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Muszynski A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW (2013) Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol Plant Microbe Interact 26:319–329 [DOI] [PubMed] [Google Scholar]
- Lace B, Ott T (2018) Commonalities and differences in controlling multipartite intracellular infections of legume roots by symbiotic microbes. Plant Cell Physiol 59:661–672 [DOI] [PubMed] [Google Scholar]
- Li X, Zheng Z, Kong X, Xu J, Qiu L, Sun J, Reid D, Jin H, Andersen SU, Oldroyd GED, Stougaard J, Downie JA, Xie F (2019) Atypical receptor kinase RINRK1 required for rhizobial infection but not nodule development in Lotus japonicus. Plant Physiol 181:804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Hoffrichter A, Brachmann A, Marín M (2018) Complete genome of Rhizobium leguminosarum Norway, an ineffective Lotus micro-symbiont. Stand Genom Sci 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Klingl A, Lin YY, Boul E, Thomas-Oates J, Marin M (2019) A subcompatible rhizobium strain reveals infection duality in Lotus. J Exp Bot 70:1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Breakspear A, Stacey N, Findlay K, Nakashima J, Ramakrishnan K, Liu M, Xie F, Endre G, de Carvalho-Niebel F. et al. (2019) A protein complex required for polar growth of rhizobial infection threads. Nat Commun 10:2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lara IM, van den Berg JD, Thomas-Oates JE, Glushka J, Lugtenberg B J, Spaink HP (1995) Structural identification of the lipo-chitin oligosaccharide nodulation signals of Rhizobium loti. Mol Microbiol 15:627–638 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425:637–640 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A,, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malolepszy A, Mun T, Sandal N, Gupta V, Dubin M, Urbanski D, Shah N, Bachmann A, Fukai E, Hirakawa H, et al. (2016) The LORE1 insertion mutant resource. Plant J 88:306–317 [DOI] [PubMed] [Google Scholar]
- Marino D, Andrio E, Danchin EG, Oger E,, Gucciardo S, Lambert A, Puppo A, Pauly N (2011) A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol 189:580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144:324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri M, Janakirama P, Held M, Ross L, Szczyglowski K (2016) Into the root: how cytokinin controls rhizobial infection. Trends Plant Sci 21:178–186 [DOI] [PubMed] [Google Scholar]
- Mitra S, Mukherjee A, Wiley-Kalil A, Das S, Owen H, Reddy PM, Ane JM, James EK, Gyaneshwar P (2016) A rhamnose-deficient lipopolysaccharide mutant of Rhizobium sp. IRBG74 is defective in root colonization and beneficial interactions with its flooding-tolerant hosts Sesbania cannabina and wetland rice. J Exp Bot 67:5869–5884 [DOI] [PubMed] [Google Scholar]
- Montiel J, Nava N, Cardenas L, Sanchez-Lopez R, Arthikala MK, Santana O, Sanchez F, Quinto C (2012) A Phaseolus vulgaris NADPH oxidase gene is required for root infection by Rhizobia. Plant Cell Physiol 53:1751–1767 [DOI] [PubMed] [Google Scholar]
- Montiel J, Arthikala M-K, Cárdenas L, Quinto C (2016) Legume NADPH oxidases have crucial roles at different stages of nodulation. Int J Mol Sci 17:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J, Fonseca-Garcia C, Quinto C (2018) Phylogeny and expression of NADPH oxidases during symbiotic nodule formation. Agriculture 8:179 [Google Scholar]
- Morgante C, Castro S, Fabra A (2007) Role of rhizobial EPS in the evasion of peanut defense response during the crack-entry infection process. Soil Biol Biochem 39:1222–1225 [Google Scholar]
- Mun T, Bachmann A, Gupta V, Stougaard J, Andersen SU (2016) Lotus Base: An integrated information portal for the model legume Lotus japonicus. Sci Rep 6:39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E, Cheng J, Gysel K, Bozsoki Z, Kawaharada Y, Hjuler CT, Sorensen KK, Tao K,, Kelly S, Venice F, et al. (2018) Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315:101–104 [DOI] [PubMed] [Google Scholar]
- Murray JD, Muni RR, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, et al. (2011) Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65:244–252 [DOI] [PubMed] [Google Scholar]
- Nadzieja M, Stougaard J, Reid D (2019) A toolkit for high resolution imaging of cell division and phytohormone signaling in legume roots and root nodules. Front Plant Sci 10:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndoye I, de Billy F, Vasse J, Dreyfus B, Truchet G (1994) Root nodulation of Sesbania rostrata. J Bacteriol 176:1060–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59:519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Dixon R (2014) Biotechnological solutions to the nitrogen problem. Curr Opin Biotechnol 26:19–24 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg-Grossman S, Volpin H, Levine A (2007) Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J Exp Bot 58:1637–1649 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsot V, Crook MB, Erdn S, Maillet F, Bascaules A, Ane JM (2016) New insights into Nod factor biosynthesis: analyses of chitooligomers and lipo-chitooligomers of Rhizobium sp. IRBG74 mutants. Carbohydr Res 434:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Lin JS, Xu J,, Sato S, Parniske M, Wang TL, Downie JA, Xie F (2015) SCARN a novel class of SCAR protein that is required for root-hair infection during legume nodulation. PLoS Genet 11:e1005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585–592 [DOI] [PubMed] [Google Scholar]
- Ranga Rao V (1977) Effect of root temperature on the infection processes and nodulation in Lotus and Stylosanthes. J Exp Bot 28:241–259 [Google Scholar]
- Reid D, Liu H, Kelly S, Kawaharada Y, Mun T, Andersen SU, Desbrosses G, Stougaard J (2018) Dynamics of ethylene production in response to compatible Nod factor. Plant Physiol 176:1764–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D, Nadzieja M, Novak O, Heckmann AB, Sandal N, Stougaard J (2017) Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol 175:361–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Heckmann AB, Novak O, Kelly S, Stougaard J (2016) Cytokinin oxidase/Dehydrogenase3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol 170:1060–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodpothong P, Sullivan JT, Songsrirote K, Sumpton D, Cheung KWJT, Thomas-Oates J, Radutoiu S, Stougaard J, Ronson CW (2009) Nodulation gene mutants of Mesorhizobium loti R7A-nodZ and noIL mutants have host-specific phenotypes on Lotus spp. Mol Plant Microbe Interact 22:1546–1554 [DOI] [PubMed] [Google Scholar]
- Sandal N, Krusell L, Radutoiu S, Olbryt M, Pedrosa A, Stracke S, Sato S, Kato T, Tabata S, Parniske M, et al. (2002) A genetic linkage map of the model legume Lotus japonicus and strategies for fast mapping of new loci. Genetics 161:1673–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Wakabayashi T, Kawamura Y, Skovbjerg CK, Wang MZ, Mustamin Y, Isomura Y, Gupta V, Jin H, Mun T, et al. (2020) Extreme genetic signatures of local adaptation during Lotus japonicus colonization of Japan. Nat Commun 11:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402:191–195 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15:139–152 [DOI] [PubMed] [Google Scholar]
- Sinharoy S, Torres-Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK (2013) The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25:3584–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI (2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol 174:11–25 [DOI] [PubMed] [Google Scholar]
- Sprent JI, Ardley J, James EK (2017) Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol 215:40–56 [DOI] [PubMed] [Google Scholar]
- Subba-Rao NS, Mateos PF, Baker D, Pankratz HS, Palma J, Dazzo B, Sprent JI (1995) The unique root-nodule symbiosis between Rhizobium and the aquatic legume, Neptunia natans (L. f.) Druce. Planta 196:311–320 [Google Scholar]
- Takei K, Yamaya T, Sakakibara H (2004) Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J Biol Chem 279:41866–41872 [DOI] [PubMed] [Google Scholar]
- Townsend GE, Forsberg LS, Keating DH (2006) Mesorhizobium loti produces nodPQ-dependent sulfated cell surface polysaccharides. J Bacteriol 188:8560–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski DF, Malolepszy A, Stougaard J, Andersen SU (2012) Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J 69:731–741 [DOI] [PubMed] [Google Scholar]
- Vega-Hernandez MC, Perez-Galdona R, Dazzo FB, Jarabo-Lorenzo A, Alfayate MC, Leon-Barrios M (2001) Novel infection process in the indeterminate root nodule symbiosis between Chamaecytisus proliferus (tagasaste) and Bradyrhizobium sp. New Phytol 150:707–721 [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci U S A 109:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Shibata S, Chen WL, Sato S, Kaneko T, Jurkiewicz A, Sandal N, Banba M, Imaizumi-Anraku H, Kojima T, et al. (2009) CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume–Rhizobium symbiosis. Plant J 60:168–180 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci U S A 105:20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, Jurkiewicz A, Rueda P, Radutoiu S, Held M, Hossain MS, Szczyglowski K, Morieri G, et al. (2009) Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21:267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CZ, Huang J, Gyaneshwar P, Zhao D (2017) Rhizobium sp. IRBG74 alters arabidopsis root development by affecting auxin signaling. Front Microbiol 8:2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.