We demonstrated nanoscale MRI of metabolism in single isolated mitochondria and within living cells using NV centers in diamond.
Abstract
Free radicals play a vital role in all kinds of biological processes including immune responses. However, free radicals have short lifetimes and are highly reactive, making them difficult to measure using current methods. Here, we demonstrate that relaxometry measurement, or T1, inherited from the field of diamond magnetometry can be used to detect free radicals in living cells with subcellular resolution. This quantum sensing technique is based on defects in diamond, which convert a magnetic signal into an optical signal, allowing nanoscale magnetic resonance measurements. We functionalized fluorescent nanodiamonds (FNDs) to target single mitochondria within macrophage cells to detect the metabolic activity. In addition, we performed measurements on single isolated mitochondria. We were able to detect free radicals generated by individual mitochondria in either living cells or isolated mitochondria after stimulation or inhibition.
INTRODUCTION
Mitochondria are the cell’s powerhouses and are considered the dominant organelles for producing free radicals within most mammalian cells (1–3). Mitochondrial free radicals are generated from the electron transport chain by electron escape during adenosine triphosphate synthesis. Free radicals are highly reactive and short lived. Several methods that allow detection of free radicals exist (4). Roughly, they can be divided into direct and indirect methods. The former is the indirect quantitative polymerase chain reaction. It is able to detect specific enzymes, which are generated in response to free radical generation (5). This method has the striking advantage that it is specific. Unfortunately, it is not possible to obtain any spatial information or single-cell resolution and requires a lot of background information. Lipid oxidation and DNA damage belong to the latter group and give an indication of the damage done by radicals (or other reactive molecules) (6, 7). So far, the best method for radical detection, which provides at least some spatial resolution, is fluorescence detection using organic dyes. Typically, these dyes react with reactive oxygen species (ROS) and form a fluorescent molecule. Some examples of popular dyes are 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA; used as comparison in this article), MitoSOX, or MitoTracker red CM-H2XRos (8, 9). In addition, these dyes usually react with all kinds of ROS and thus are not specific for radicals. Nonparamagnetic ROS usually dominate the signal, given that they are more abundant than free radicals. However, a few dyes are specific for certain radicals (rather than all radicals) as, for instance, hydroxyterephthalic acid (HTA) (10) or 4,5-diaminofluorescein (DAF-2) (11). Unfortunately, all these dyes suffer from photobleaching and react reversibly with ROS. As a result, they reveal the history of the sample rather than its current state. So far, no method is available to simultaneously localize, quantify, and identify them (12–14). Thus, new methods assessing ROS can contribute to our understanding of radical production at the nanoscale level (15, 16). Fluorescent nanodiamonds (FNDs) are a very promising tool for use in the biomedical fields because of their outstanding photostability (17, 18) and biocompatibility (19–26). FNDs enter cells in vesicles via clathrin-mediated endocytosis or macropinocytosis (27, 28). Another benefit of FNDs (at least in some cell types) is that they can escape endosomes. This has been attributed to their sharp edges, which allow them to penetrate the endosomal membrane (29, 30). Arguably most interesting is that the defects convert magnetic signals from the surroundings into optical signals. This makes it possible to perform highly sensitive nanoscale magnetic resonance measurements (16, 31, 32). So far, this technique has predominantly been used in physics or to measure spin labels (33, 34). In biomedical fields, tracking the particle orientation or measuring temperature in cells has been previously achieved (35–37). Last, more complex hybrid particles between FNDs and pH-responsive polymers have been used to measure pH changes (38).
Here, we make use of a specific way to conduct magnetometry experiments called relaxometry. This sequence is especially convenient because it does not require microwaves. The technique has already been used for several different applications including detection of gadolinium (39), magnetic structures (40), nanoparticles (41–43), and, most recently, free radicals (44, 45). We demonstrate their use for monitoring metabolic activity in single cells or organelles. More specifically, we use FNDs containing nitrogen-vacancy (NV) centers to detect spin noise caused by free radicals.
RESULTS
Figure 1 shows an outline of the magnetometry experiments that were performed in this study on living cells and functional isolated mitochondria.
Fig. 1. Schematic summary of the experimental conditions we investigated using diamond magnetometry (and compared to a conventional dye).
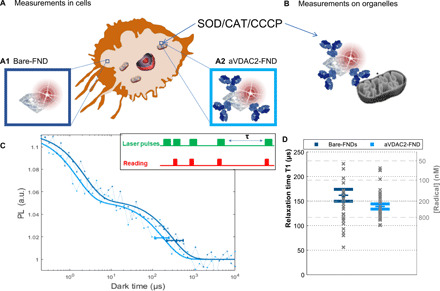
(A) First, we investigated free radical formation in single cells. Here, we differentiated between two subcellular locations: the cytosol reached by uncoated FNDs (A1) and the mitochondria reached by aVDAC2 antibody–coated FNDs (A2). (B) Measurements on single isolated mitochondria. In all conditions, we compared measurements before and after stimulation with CCCP in different concentrations: CAT (final concentration of 1000 U/ml) and SOD (final concentration of 600 U/ml). (C) The T1 relaxation curve is generated from different dark times plotted against the fluorescence intensity. The inset shows the pulsing sequence used in relaxometry experiments. The green blocks indicate when the laser was on, while the red blocks indicate when we read out the photoluminescence (PL) from the FND. The horizontal error bar indicates the position of the T1 time and the 95% confidence interval obtained with the fitting procedure. This T1 time gives a measure of radical concentration in the surroundings. The representative curves shown are from bare FNDs, or aVDAC2-coated FNDs, before applying any treatment. a.u., arbitrary unit. (D) Statistical distribution (mean and SE) of T1 from 26 and 36 FNDs (all the “initial conditions” are represented hereafter).
Measurements in cells
For measurements in cells, we used two kinds of particles: bare (uncoated) FNDs and FNDs coated with physically adsorbed anti-VDAC2 antibodies (aVDAC2-FNDs). While bare FNDs are expected to be in the cytosol, the coated ones are targeted to mitochondria in macrophages. [Results for characterization (fig. S1) and internalization (determined by particle counting; fig. S2) can be found in the Supplementary Materials.] To confirm that aVDAC2-FNDs did bind to the mitochondria, we performed the uptake protocol with aVDAC2-FND (using FNDs as a control) and isolated the mitochondria. In the aVDAC2-FND group, we were still able to find the FNDs in the fraction with the isolated mitochondria, while for the FND fraction we did not (for details, see the Supplementary Materials: Counting the number diamond attached to mitochondrial surface inside macrophage cells). From fig. S5C, we can conclude that diamond particles were attached to mitochondrial surface or close to the mitochondria. Figures S15 and S16 show that FNDs can escape from the endocytosis or phagocytosis pathway.
Before relaxometry experiments, we measured confocal z-scans of cells with ingested FNDs or aVDAC2-FNDs. Typical images for mitochondria-targeted aVDAC2-FNDs (in red) are shown in Fig. 2 (A to C) (images for the controls can be found in the Supplementary Materials). After identifying a particle, we performed T1 (relaxometry) measurements. These produced a signal that was equivalent to T1 in conventional magnetic resonance imaging (MRI) but from nanoscale voxels. To perform such a measurement, we used a laser pulsing sequence, which is shown and described in Fig. 1C. During this sequence, the NV centers were brought into their (bright) ground state. We then probed how long they could remain in this state. This time, called relaxation time or T1, is shortened by the presence of spin noise (46) from free radicals. In a previous work (45), we found that, under controlled conditions, the obtained plots could be fitted with the following equation
| (1) |
Fig. 2. Free radical detection.
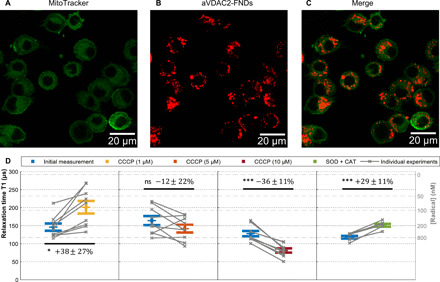
(A to C) Confocal images of FNDs (1 μg/ml) with adsorbed aVDAC2 antibodies incubated with cells (104 cells/ml) for 14 hours. Green, MitoTracker Green–stained mitochondria; red, aVDAC2-FND. (D) Single-cell free radical measurement by T1. After recording the initial T1 (blue), 1, 5, and 10 μM CCCP as well as SOD (1000 U/ml) and CAT (600 U/ml) were tested on the same samples. The error bars represent SEs. Gray lines show individual experiments on single cells. The left gray axis represents the approximated concentration determined from a calibration with *OH radicals in solution from the previous work (45). The experiments were repeated nine times. Data were analyzed using a paired t test to compare between groups with a *P ≤ 0.05 and ***P ≤ 0.001 significance.
This suggests that the NV centers could approximately be grouped into two categories of different relaxation time, Ta < Tb. Because surface paramagnetic impurities seem to play a dominant role in the FND relaxation process (47), we hypothesize that such a distinction could be linked to the direction of the NV center compared to most proximal crystallographic surface. This could make them more or less sensitive to magnetic dipole located on it. While both constants were found dependent on the surrounding free radical concentration, the longer one Tb was more susceptible to changes in magnetic noise and thus is more sensitive to changes in the surrounding radical concentration (45). For this reason, Tb was selected from these two relaxation constants for analysis and quantification, as it has proven to be more sensitive to changes from the surrounding
| (2) |
Furthermore, it is worth noting that NV centers only “feel” spin noise from their immediate surrounding, up to some tens of nanometers at the most (31, 48).
In Fig. 1D, we can compare FNDs and VDAC2-FNDs, which were targeted to the mitochondrial surface when internalized in macrophages. As expected, T1 values were significantly lower (uncorrelated t test, P < 0.001) when the nanodiamonds were attached to mitochondria (aVDAC2-FNDs) than when they were not (FNDs). This can be attributed to the mitochondrial radical production. In addition, T1 variability is higher in the cytosol. This could be attributed to the fact that the particles can be in vicinity of different organelles that may or may not generate free radicals. To estimate the radical concentrations from T1 values, we used a calibration in a controlled environment. This calibration was obtained from measurements of known concentrations of *OH radicals [for more information, see (45)].
Figure 2D shows the evolution of T1, when adding different doses of carbonyl cyanide 3-chlorophenylhydrazone (CCCP). While the colored bars show averages, single experiments are represented in gray. We monitored the T1 of individual FNDs while changing the biological environment. The paired t test was used for assessing the significance of the data. The uncertainty when determining the T1 by fitting always remains much smaller than the variability of T1 taken from different particles. The error bars represent the SE caused by this heterogeneous variability. In addition to showing T1 data as in Fig. 2, it was also possible to generate time-resolved T1 evolution curves or spatially resolved T1 maps. This is demonstrated in figs. S7 and S14. In principle, it is possible to generate a data point every hundred microseconds for time-resolved data. However, to measure radicals in the nanomolar range, it is necessary to integrate over approximately 2 min. For spatially resolved T1, there is a trade-off between measuring the position and measuring T1. For the current tracks, we received a coordinate every 5 s. According to the literature, the metabolic processes we are interested in here occur within minutes and persist for around 30 min up to an hour (49).
After recording the initial T1, we exposed the cells to different concentrations of CCCP (1, 5, and 10 μM) or catalase (CAT) and superoxide dismutase (SOD). CCCP interferes with the membrane potential. All three chemicals affect radical formation in mitochondria (49–52). Because these chemicals only influence the mitochondria locally, the aVDAC2-FNDs respond to the stimuli, while FNDs that reside in the cytosol do not. In the presence of low concentrations (1 μM) of CCCP, aVDAC2-FND–incubated cells decreased their radical production (paired t test, P < 0.05), whereas 5 μM CCCP did not show any differences in radical production. At 10 μM, CCCP enhanced mitochondrial radical generation (paired t test, P < 0.001). These results confirm former findings where CCCP altered mitochondrial membrane potential (MMP) and ROS production in a dose-dependent manner (49). Xiao et al. explain this concentration dependency, which we also observed, by changes in the membrane potential. While treatment of cells with low concentrations (up to 2.5 μM) of CCCP enhanced mitochondrial potential, CCCP treatment at concentrations higher than 5 μM depolarized mitochondria. MMP is directly related to electron transfer; hence, it is expected to alter radical generation (50). SOD/CAT convert radicals to H2O2 and H2O. Eventually, this resulted in increased T1 values (paired t test, P < 0.001).
The abovementioned results were also confirmed by data from an Amplex/horseradish peroxidase (HRP) assay. This assay, which responds similarly to CCCP and SOD/CAT, measures cellular ROS production. The main contributors to this measurement in cells are O2*− and H2O2. Here, we observed fluorescence signal changes indicating the amount of ROS production, in response to different concentrations of CCCP and SOD/CAT. Notably, Amplex/HRP measured ROS released from all cells in the well. In addition, the assay revealed the concentration of ROS that was generated until the measurement, while diamond magnetometry reflects the current situation. Diamond magnetometry detects free radicals from single cells with subcellular resolution. In this case, we can differentiate between free radical levels on the mitochondria and in the cytosol. Despite these differences in what exactly is measured, and at which time and length scale, the results from conventional assays present similar trends to our T1 measurements.
As a control, we investigated the response of uncoated FNDs and aVDAC2-FNDs to CCCP and SOD/CAT in the absence of cells. FND or aVDAC2-FND in cell culture medium without cells was tested before and after adding CCCP and SOD/CAT. The results were that no radicals were created and neither FND nor aVDAC2-FNDs responded (see fig. S8A). A concern was that the time in the measurement setup itself would cause the cells to be stressed and produce radicals. To exclude this, we did an additional control measurement. Here, we performed two consecutive measurements without adding any stimulus (see fig. S8B). Similarly, we did not see a significant response.
Isolated mitochondria
First, mitochondria were isolated from living cells. Then, we characterized the mitochondria by determining size and zeta potential (see fig. S4). Our findings were in good agreement with the literature (53). After having confirmed that we isolated mitochondria, we tested their metabolic activity to prove that they had retained their function. As fig. S6 indicates, the mitochondria responded to CCCP after staining with the cationic dye rhodamine 123. This assay determines whether there is a membrane potential. Because we observed changes in MMP, we can conclude mitochondrial functionality (49, 54).
Next, we detected free radical activity in single mitochondria. First, confocal imaging was performed to find an aVDAC2-FND on a mitochondrion. A typical result of this imaging process is shown in Fig. 3 (A to C). Once we had identified an aVDAC2-FND on the mitochondrial surface, we performed a T1 measurement.
Fig. 3. Free radical measurements in single mitochondria.
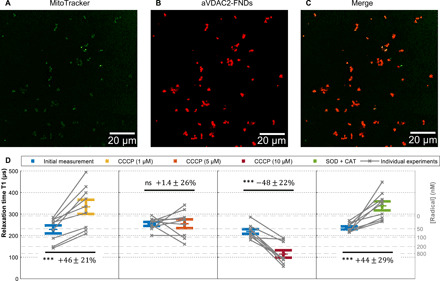
(A to C) Confocal images of aVDAC2-FND targeted to isolated mitochondria. Green, MitoTracker Green–stained mitochondria; red, FNDs. Scale bar, 20 μm. (D) Free radicals detected on single isolated mitochondria by T1 measurements. After recording an initial T1 measurement, 1, 5, and 10 CCCP as well as SOD (1000 U/ml) and CAT (600 U/ml) were tested on the same samples. The error bars represent SEs. The left gray axis represents the approximated concentration determined from a calibration with *OH radicals in solution from the previous work (45). The experiments were repeated nine times. The data were analyzed by using a paired t test against the previous group. ***P ≤ 0.001.
Figure 3D first compares the initial T1 values from mitochondria with T1 after adding different concentrations of CCCP. At low concentrations (1 μM) of CCCP, radical generation decreased slightly (paired t test, P < 0.001). Similar to mitochondria in cells, isolated mitochondria treated with 5 μM CCCP did not show any effect on radical generation. However, at 10 μM CCCP, radical production increased strongly (paired t test, P < 0.001). SOD/CAT decreased radical production. Data from isolated mitochondria are in good agreement with T1 measurements in cells (aVDAC2-FNDs).
To compare quantum sensing measurements to a conventional technique, we measured the ROS production of isolated mitochondria using a well-established Amplex/HRP assay (55). As shown in fig. S9, the Amplex/HRP assay on isolated mitochondria agrees very well with our findings.
DISCUSSION
In principle, there are several factors that might potentially disturb T1 measurements. To determine the contribution of other factors besides radical concentrations, we measured the dependence of T1 on temperature (fig. S12), viscosity (fig. S11), and pH (fig. S13). First, for temperature, we know from the literature that mitochondria can function up to 50°C (56). With temperatures ranging from 37° to 55°C, we only observed a slight decrease in T1 with rising temperatures. Thus, we can safely neglect this effect. Second, viscosity is relevant in the mammalian cells in the range of 2 to 5 mPa·s (57). Within this range and from viscosity changes alone, we did not observe any changes in T1 [we compared water (1 mPa·s) and glucose solutions at 27.73 and 945 mPa·s]. Last, concerning pH, we did observe significant changes in T1 when varying the pH. These changes can be attributed to changes in conductivity in the surrounding medium. However, the most extreme pH changes in a cellular environment would occur in the lysosomes. Estimates of pH variations vary greatly in the literature depending on the exact conditions and cell types; however, they always remain below one pH unit. For this reason, we did not expect pH changes to occur, which were drastic enough to cause a change in T1 (58, 59).
While the conventional assay- and diamond-based quantum sensing is in good agreement here, it is necessary to mention that important differences prevail. First, the signals from the conventional dyes are taken from an ensemble of cells and do not reveal subcellular resolution or even single-cell resolution. Second, the conventional method requires a individual measurement with a different population for every time point. In comparison, the FND measurements before and after were from the exact cells at the exact location and data were collected in real time. This is important in case there was a biological variability between cells. While the conventional dyes measure the concentration of ROS that has been generated until the point of the measurement, FNDs provide the current status. Last, FNDs use a different mechanism to be specific. Namely, they are specific for radicals, while most dyes are specific for ROS or for certain molecules. Because nonradical ROS are more abundant, these usually dominate the conventional signal. For this reason, relaxometry complements the existing techniques very well.
In sum, relaxometry measurements with nanodiamond are allowed free radical detection in both cells and isolated organelles (mitochondria). The measurements were nondestructive and could be repeated over time on the same individual cell or organelle, while a change in biological state was triggered. This unique capability allowed us to differentiate between biological variability and changes caused by the intervention.
MATERIALS AND METHODS
Materials
FNDs with a hydrodynamic diameter of 70 nm (FND70) containing >300 NV centers were purchased from Adams Nanotechnologies (NC, USA). We found these particles to have an ideal size for this purpose. Smaller particles are less bright, so it takes longer to obtain a good signal-to-noise ratio. In addition, they move faster and are therefore more difficult to track. However, it is probably most important that the larger particles contain more NV centers, and thus, that each measurement is already an average of all these NV centers. Hence, signals from these particles are more reproducible. The drawback of even larger particles is that NV centers in the core of these particles are too distant from the surface to sense the spin noise from radicals. They are oxygen-terminated and have already been characterized extensively before (39, 60). MitoTracker Green was purchased from Gibco, Thermo Fisher Scientific (The Netherlands). Anti-VDAC2 antibody ([C2C3], C-term, catalog no. GTX104745) was obtained from GeneTex (The Netherlands). CCCP, rhodamine 123, glycerol, SOD from bovine erythrocytes, and CAT from bovine liver were purchased from Sigma-Aldrich (The Netherlands). Chemicals were stored and used according to the instructions from the manufacturers. Amplex UltraRed reagent, pHrodo Green E. coli BioParticles Conjugate for Phagocytosis, and pHrodo Green Dextran, 10,000 MW, for Endocytosis and Live Cell Imaging Solution were obtained from Thermo Fisher Scientific. The gelatin solution (2% in H2O) was purchased from Merck and stored at 4°C in the fridge. LysoView 405 was purchased from Biotium (https://biotium.com/).
Diamond preparation
Antibody attachment was already established earlier for various antibodies (21). Here, anti-VDAC2 antibodies (Gibco, Thermo Fisher Scientific, The Netherlands) were diluted to a concentration of 0.089 mg/ml (1:100 dilution, as suggested by the manufacturer). The antibodies were mixed with FNDs (1 μg/ml) in a 1:4 ratio for 1 to 2 min while vortexing. This was followed by incubation at room temperature (RT) for 15 min to allow antibodies to adsorb on the FNDs. The Malvern ZetaSizer Nanosystem (Dynamic Light Scattering; Malvern Instruments Ltd., Malvern, UK; www.malvern.com) was used to evaluate size changes after the modification of nanodiamonds.
Diamond uptake by macrophage cells
J774A.1 Macrophages (LGC Standards, Germany), immune cells from a mouse cell line, were cultured in complete Dulbecco’s modified Eagle’s medium with high glucose, supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% GlutaMAX (Gibco, Thermo Fisher Scientific, The Netherlands). Cells were incubated with bare FND70 (1 μg/ml) in cell culture medium for 30 min. Separate samples were prepared with incubation times between 2 and 24 hours. Cells were incubated at 37°C and 5% CO2. At each time point, the cell culture medium with FNDs was removed, followed by washing with 1× phosphate-buffered saline (PBS). Subsequently, cells were fixed with 3.7% paraformaldehyde for 10 min at RT. After fixation, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and phalloidin–fluorescein isothiocyanate (FITC) (Sigma-Aldrich, The Netherlands) for nuclei and F-actin. Images were taken with a Zeiss LSM780 confocal microscope (Zeiss, Jena, Germany; www.zeiss.com) using a 405-nm laser to detect DAPI, a 488-nm laser to measure phalloidin-FITC, and a 561-nm laser to detect FNDs. Images were analyzed with a home-written FIJI plugin [the method used in this plugin is explained by Hemelaar et al. (21)]. Three independent experiments were performed, and 100 cells were quantified for each time point.
Determining targeting efficiency
To count the number of diamonds attached to the mitochondria surface, mitochondria were isolated from cells (~1 × 105 cells/ml) containing aVDAC2-FND or FND (1 μg/ml) as a separate control. We investigated incubation times between 30 min and 24 hours. To take confocal images, the mitochondria solution was attached to gelatin-coated glass-bottom petri dishes. Confocal images were taken with a Zeiss 780 microscope and analyzed using FIJI software to determine the number of FNDs attached to the mitochondrial surface.
Measurement of free radical production in cells using relaxometry
For T1 measurements, we used a homemade magnetometer setup. In principle, this setup is a confocal microscope with a few changes as described in the following. A ×100 magnification oil objective (Olympus, UPLSAPO 100XO) was used for light collection. An acousto-optical modulator (Gooch & Housego, model 3350-199) was implemented to conduct the pulsing sequence that is shown in Fig. 1D. This optically detected T1 time is equivalent to a T1 signal in conventional MRI.
To start a T1 measurement of cellular-free radical production, the location of the FND inside a cell was confirmed using Z-stack imaging. After a particle was found, we started the actual T1 measurement. The pulsing sequence was repeated 10,000 times for each measurement to obtain a sufficient signal-to-noise ratio. Each T1 measurement took around 10 min. A tracking algorithm was used to track the movement of a diamond particle inside the cell. The laser was attenuated to 50 μW at the location of the sample. This laser power was chosen with the aim of minimizing damage to cells while simultaneously providing enough power to polarize the NV centers. This polarization was achieved by laser pulses separated by variable dark times τ from 0.2 to 10 μs, as shown in fig. S1. The 532-nm green laser pulse length was set to 5 μs to ensure the polarization of the NV centers. During this time, the NV centers are pumped into the (bright) ground state from the (dark) excited state. The T1 time is a measure for how long it takes until they reach equilibrium. Moreover, it gives a quantitative measure of radicals by detecting the brightness.
For T1 measurements of free radical production in cells, macrophage cells in glass-bottom petri dishes were incubated with aVDAC2-FND or FND for 14 hours (40,000 cells were seeded to the dish). After incubation, cells were washed once with PBS and the medium was changed to fresh diamond free cell culture medium at 37°C. After an initial T1 measurement was recorded, different concentrations of CCCP (1, 5, and 10 μM) were added to test their effect on mitochondrial radical production. In a separate experiment, SOD (final concentration of 1000 U/ml) and CAT (final concentration of 600 U/ml) were used, which lower the radical load. Each condition was repeated in triplicate.
Estimating radical concentrations from T1
To estimate radical concentrations from T1 values, we used a calibration with known concentrations of radicals that we had established earlier (45). The known concentrations were produced by ultraviolet (UV) irradiation of H2O2 at a fixed UV light intensity. The concentrations were confirmed using the conventional HTA assay.
For the relationship between T1 and C*OH (concentration of *OH), we obtained an exponential relationship in the form
| (3) |
where T1∞ is expressed in μs, C0 is expressed in μM, and A is dimensionless.
The parameters of the model in Eq. 2 are calculated by a nonlinear fitting procedure
The confidence intervals of our T1 measurement (dispersion of the measurements) were concentration dependent. More specifically, we found a 95% confidence interval of ±62 μs for water, ±17 μs for 0.3 μM, ±21 μs for 0.6 μM, and ±18 μs for 0.9 μM.
Isolating mitochondria
The protocol of mitochondria isolation used in this paper is based on the protocol of Clayton and Shadel (61) and optimized by our group. Briefly, mitochondria were isolated from J744 macrophage cells by different centrifugation steps. The following steps were all performed at 4°C or on ice. Cells were scraped into 3 ml of cell culture medium and spun down at 500g for 5 min. Cell pellets were then resuspended in cold hypotonic reticulocyte standard buffer (RSB) hypo buffer (10 mM NaCl, 1.5 mM MgCl2, and 10 mM tris-HCl) and milli-Q water for 10 min before transferring to a Dounce homogenizer to allow the cells to swell. In a Dounce homogenizer, cells were broken mechanically by stroking 10 times. Next, we added the homogenization buffer (525 mM mannitol, 175 mM sucrose, 12.5 mM tris-HCl, and 2.5 mM EDTA). The liquid was transferred to a new tube and centrifuged for 5 min at 4°C at 1000g. The supernatant was collected and followed by two more centrifugation steps at 1000g. Again, the supernatant was collected and spun down at 12,000g and 4°C for 15 min. This step was repeated once. In the end, the isolated mitochondria solution was added to gelatin (2% in H2O)–coated glass-bottom petri dishes for image acquisition. To evaluate the purity of isolated mitochondria, size and zeta potential of mitochondria were measured by a Malvern Zetasizer Nano system (53). The concentration of isolated mitochondria was measured directly after isolation from cells by using a Bio-Rad protein assay kit (Bio-Rad, Richmond, CA, USA) (53).
Free radical production in isolated mitochondria by relaxometry
To start T1 measurements on single mitochondria, the FNDs were attached to their surface. Again, we used aVDAC2-FND (60 μg/ml) (or FND as control), which was added to isolated mitochondria in the respiratory buffer for 20 min at 37°C. Then, the solution was transferred to a glass-bottom petri dish. Beforehand, these dishes had been coated with 200 μl of gelatin solution to immobilize the mitochondria.
T1 measurements were performed on isolated mitochondria in the same way as mentioned before for cells. T1 measurements started with mitochondria to record a baseline, and then we added 1, 5, and 10 μM CCCP or SOD (final concentration of 1000 U/ml) and CAT (final concentration of 600 U/ml). All experiments were performed in triplicate.
Acknowledgments
We would like to thank K. Sjollema for help with imaging and K. Holczer for fruitful discussions and for helping us to interpret T1 results. Funding: This work was financially supported by an ERC starting grant (ERC-2016-STG Stress Imaging 714289) and a VIDI grant (016.Vidi.189.002). We would also like to thank the China Scholarships Council for supporting us through a scholarship (No.201706170089) for L.N. R.S. acknowledges financial support from FOM via the projectruimte grant 15PR3229. F.P.P.M. acknowledges support from the Chilean government via a CONICYT scholarship (grant number 72160222). A.C.N. acknowledges the Kolff Institute for her PhD scholarship. Confocal images shown in this paper were acquired from UMCG Imaging and Microscopy Center (UMIC) under NWO grant 175-010-2009-023. Author contributions: L.N. designed the study under the supervision of R.Sc. L.N. and A.C.N. conceived the study and acquired and analyzed the data. L.N., A.C.N., K.J.v.d.L., M.C., and R.Sc. wrote the manuscript. E.P.P.E. edited the manuscript. S.R.H. optimized mitochondrial isolation. F.P.P.M. designed the measurement method and built the equipment under the supervision of M.C. and R.Sc. M.C. analyzed T1 data. V.G.D. and T.V. performed some T1 measurements in this study, R.L. and R.Sh. helped with some sample preparation. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/21/eabf0573/DC1
REFERENCES AND NOTES
- 1.Quinlan C. L., Perevoschikova I. V., Goncalves R. L. S., Hey-Mogensen M., Brand M. D., The determination and analysis of site-specific rates of mitochondrial reactive oxygen species production. Methods Enzymol. 526, 189–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W., Karamanlidis G., Tian R., Novel targets for mitochondrial medicine. Sci. Transl. Med. 8, 326rv3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith R. A. J., Hartley R. C., Murphy M. P., Mitochondria-targeted small molecule therapeutics and probes. Antioxid. Redox Signal. 15, 3021–3038 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Sigaeva A., Ong Y., Damle V. G., Morita A., van der Laan K. J., Schirhagl R., Optical detection of intracellular quantities using nanoscale technologies. Acc. Chem. Res. 52, 1739–1749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi J., Liu M., Shi J., Zheng G., Wang Y., Wang J., Chen Y., Lu C., Yin W., Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep. 31, 1245–1254 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Jambunathan N., Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol. Biol. 639, 292–298 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Barzilai A., Yamamoto K.-I., DNA damage responses to oxidative stress. DNA Repair 3, 1109–1115 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Eruslanov E., Kusmartsev S., Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594, 57–72 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Kuznetsov A. V., Kehrer I., Kozlov A. V., Haller M., Redl H., Hermann M., Grimm M., Troppmair J., Mitochondrial ROS production under cellular stress: Comparison of different detection methods. Anal. Bioanal. Chem. 400, 2383–2390 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Bubacz K., Kusiak-Nejman E., Tryba B., Morawski A. W., Investigation of OH radicals formation on the surface of TiO2/N photocatalyst at the presence of terephthalic acid solution. Estimation of optimal conditions. J. Photochem. Photobiol. A Chem. 261, 7–11 (2013). [Google Scholar]
- 11.Nagata N., Momose K., Ishida Y., Inhibitory effects of catecholamines and anti-oxidants on the fluorescence reaction of 4,5-diaminofluorescein, DAF-2, a novel indicator of nitric oxide. J. Biochem. 125, 658–661 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Holley A. E., Cheeseman K. H., Measuring free radical reactions in vivo. Br. Med. Bull. 49, 494–505 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Saran M., Bors W., Direct and indirect measurements of oxygen radicals. Klin. Wochenschr. 69, 957–964 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Dikalov S. I., Harrison D. G., Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid. Redox Signal. 20, 372–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zurbuchen M. A., Lake M. P., Kohan S. A., Leung B., Bouchard L.-S., Nanodiamond landmarks for subcellular multimodal optical and electron imaging. Sci. Rep. 3, 2668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann P., Jakobi I., Dolde F., Burk C., Reuter R., Waldherr G., Honert J., Wolf T., Brunner A., Shim J. H., Suter D., Sumiya H., Isoya J., Wrachtrup J., High-precision nanoscale temperature sensing using single defects in diamond. Nano Lett. 13, 2738–2742 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Bolshedvorskii S. V., Vorobyov V. V., Soshenko V. V., Shershulin V. A., Javadzade J., Zeleneev A. I., Komrakova S. A., Sorokin V. N., Belobrov P. I., Smolyaninov A. N., Akimov A. V., Single bright NV centers in aggregates of detonation nanodiamonds. Opt. Mater. Express 7, 4038–4049 (2017). [Google Scholar]
- 18.Purdey M. S., Capon P. K., Pullen B. J., Reineck P., Schwarz N., Psaltis P. J., Nicholls S. J., Gibson B. C., Abell A. D., An organic fluorophore-nanodiamond hybrid sensor for photostable imaging and orthogonal, on-demand biosensing. Sci. Rep. 7, 15967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claveau S., Bertrand J.-R., Treussart F., Fluorescent nanodiamond applications for cellular process sensing and cell tracking. Micromachines 9, 247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemelaar S. R., Saspaanithy B., L’Hommelet S. R. M., Perona Martinez F. P., Van der Laan K. J., Schirhagl R., The response of HeLa cells to fluorescent nanodiamond uptake. Sensors 18, 355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemelaar S. R., van der Laan K. J., Hinterding S. R., Koot M. V., Ellermann E., Perona-Martinez F. P., Roig D., Hommelet S., Novarina D., Takahashi H., Chang M., Schirhagl R., Generally applicable transformation protocols for fluorescent nanodiamond internalization into cells. Sci. Rep. 7, 5862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chipaux M., van der Laan K., Hemelaar S. R., Hasani M., Zheng T., Schirhagl R., Nanodiamonds and their applications in cells. Small 14, e1704263 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Prabhakar N., Khan M. H., Peurla M., Chang H.-C., Hänninen P. E., Rosenholm J. M., Intracellular trafficking of fluorescent nanodiamonds and regulation of their cellular toxicity. ACS Omega 2, 2689–2693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K.-K., Cheng C.-L., Chang C.-C., Chao J.-I., Biocompatible and detectable carboxylated nanodiamond on human cell. Nanotechnology 18, 325102 (2007). [Google Scholar]
- 25.Vaijayanthimala V., Cheng P.-Y., Yeh S.-H., Liu K.-K., Hsiao C.-H., Chao J.-I., Chang H.-C., The long-term stability and biocompatibility of fluorescent nanodiamond as an in vivo contrast agent. Biomaterials 33, 7794–7802 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Kuo Y., Hsu T.-Y., Wu Y.-C., Chang H.-C., Fluorescent nanodiamond as a probe for the intercellular transport of proteins in vivo. Biomaterials 34, 8352–8360 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Sigaeva A., Morita A., Hemelaar S. R., Schirhagl R., Nanodiamond uptake in colon cancer cells: The influence of direction and trypsin-EDTA treatment. Nanoscale 11, 17357–17367 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Perevedentseva E., Hong S.-F., Huang K.-J., Chiang I.-T., Lee C.-Y., Tseng Y.-T., Cheng C.-L., Nanodiamond internalization in cells and the cell uptake mechanism. J. Nanopart. Res. 15, 1834 (2013). [Google Scholar]
- 29.Chu Z., Zhang S., Zhang B., Zhang C., Fang C.-Y., Rehor I., Cigler P., Chang H.-C., Lin G., Liu R., Li Q., Unambiguous observation of shape effects on cellular fate of nanoparticles. Sci. Rep. 4, 4495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu Z., Miu K., Lung P., Zhang S., Zhao S., Chang H.-C., Lin G., Li Q., Rapid endosomal escape of prickly nanodiamonds: Implications for gene delivery. Sci. Rep. 5, 11661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mamin H. J., Kim M., Sherwood M. H., Rettner C. T., Ohno K., Awschalom D. D., Rugar D., Nanoscale nuclear magnetic resonance with a nitrogen-vacancy spin sensor. Science 339, 557–560 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Staudacher T., Shi F., Pezzagna S., Meijer J., Du J., Meriles C. A., Reinhard F., Wrachtrup J., Nuclear magnetic resonance spectroscopy on a (5-Nanometer)3 sample volume science. Science 339, 561–563 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Glenn D. R., Lee K., Park H., Weissleder R., Yacoby A., Lukin M. D., Lee H., Walsworth R. L., Connolly C. B., Single-cell magnetic imaging using a quantum diamond microscope. Nat. Methods 12, 736–738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinert S., Ziem F., Hall L. T., Zappe A., Schweikert M., Götz N., Aird A., Balasubramanian G., Hollenberg L., Wrachtrup J., Magnetic spin imaging under ambient conditions with sub-cellular resolution. Nat. Commun. 4, 1607 (2013). [DOI] [PubMed] [Google Scholar]
- 35.McGuinness L. P., Yan Y., Stacey A., Simpson D. A., Hall L. T., Maclaurin D., Prawer S., Mulvaney P., Wrachtrup J., Caruso F., Scholten R. E., Hollenberg L. C. L., Quantum measurement and orientation tracking of fluorescent nanodiamonds inside living cells. Nat. Nanotechnol. 6, 358–363 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Kucsko G., Maurer P. C., Yao N. Y., Kubo M., Noh H. J., Lo P. K., Park H., Lukin M. D., Nanometre-scale thermometry in a living cell. Nature 500, 54–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiguchi T., Sotoma S., Harada Y., Fluorescent nanodiamonds as a robust temperature sensor inside a single cell. Biophys. Physicobiol. 15, 229–234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rendler T., Neburkova J., Zemek O., Kotek J., Zappe A., Chu Z., Cigler P., Wrachtrup J., Optical imaging of localized chemical events using programmable diamond quantum nanosensors. Nat. Commun. 8, 14701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tetienne J.-P., Hingant T., Rondin L., Cavaillès A., Mayer L., Dantelle G., Gacoin T., Wrachtrup J., Roch J.-F., Jacques V., Spin relaxometry of single nitrogen-vacancy defects in diamond nanocrystals for magnetic noise sensing. Phys. Rev. B 87, 235436 (2013). [Google Scholar]
- 40.Casola F., van der Sar T., Walsworth R., Yacoby A., Single spin relaxometry of spin noise from a ferromagnet. APS 2015, J30-013 (2015). [Google Scholar]
- 41.Pelliccione M., Myers B. A., Pascal L. M. A., Das A., Jayich A. C. B., Two-dimensional nanoscale imaging of gadolinium spins via scanning probe relaxometry with a single spin in diamond. Phys. Rev. Applied 2, 054014 (2014). [Google Scholar]
- 42.Schmid-Lorch D., Häberle T., Reinhard F., Zappe A., Slota M., Bogani L., Finkler A., Wrachtrup J., Relaxometry and dephasing imaging of superparamagnetic magnetite nanoparticles using a single qubit. Nano Lett. 15, 4942–4947 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Sadzak N., Héritier M., Benson O., Coupling a single nitrogen-vacancy center in nanodiamond to superparamagnetic nanoparticles. Sci. Rep. 8, 8430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barton J., Gulka M., Tarabek J., Mindarava Y., Wang Z., Schimer J., Raabova H., Bednar J., Plenio M. B., Jelezko F., Nesladek M., Cigler P., Nanoscale dynamic readout of a chemical redox process using radicals coupled with nitrogen-vacancy centers in nanodiamonds. ACS Nano 14, 12938–12950 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Perona Martínez F., Nusantara A. C., Chipaux M., Padamati S. K., Schirhagl R., Nanodiamond relaxometry-based detection of free-radical species when produced in chemical reactions in biologically relevant conditions. ACS Sens. 5, 3862–3869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Rollo, A. Finco, R. Tanos, F. Fabre, T. Devolder, I. Robert-Philip, V. Jacques, Quantitative study of the response of a single NV defect in diamond to magnetic noise. arXiv:2101.00860 [cond-mat.mes-hall] (4 January 2021).
- 47.Peng Z., Dallas J., Takahashi S., Reduction of surface spin-induced electron spin relaxations in nanodiamonds. J. Appl. Phys. 128, 054301 (2020). [Google Scholar]
- 48.Staudacher T., Shi F., Pezzagna S., Meijer J., Du J., Meriles C. A., Reinhard F., Wrachtrup J., Nuclear magnetic resonance spectroscopy on a (5-nanometer)3 sample volume. Science 339, 561–563 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Xiao B., Goh J.-Y., Xiao L., Xian H., Lim K.-L., Liou Y.-C., Reactive oxygen species trigger Parkin/PINK1 pathway-dependent mitophagy by inducing mitochondrial recruitment of Parkin. J. Biol. Chem. 292, 16697–16708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Branicky R., Noë A., Hekimi S., Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 217, 1915–1928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roca J., Rodríguez M. J., Gil M. A., Carvajal G., Garcia E. M., Cuello C., Vazquez J. M., Martinez E. A., Survival and in vitro fertility of boar spermatozoa frozen in the presence of superoxide dismutase and/or catalase. J. Androl. 26, 15–24 (2005). [PubMed] [Google Scholar]
- 52.Bénit P., Kahn A., Chretien D., Bortoli S., Huc L., Schiff M., Gimenez-Roqueplo A.-P., Favier J., Gressens P., Rak M., Rustin P., Evolutionarily conserved susceptibility of the mitochondrial respiratory chain to SDHI pesticides and its consequence on the impact of SDHIs on human cultured cells. PLOS ONE 14, e0224132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitani T., Kami D., Matoba S., Gojo S., Internalization of isolated functional mitochondria: Involvement of macropinocytosis. J. Cell. Mol. Med. 18, 1694–1703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zorova L. D., Popkov V. A., Plotnikov E. Y., Silachev D. N., Pevzner I. B., Jankauskas S. S., Babenko V. A., Zorov S. D., Balakireva A. V., Juhaszova M., Sollott S. J., Zorov D. B., Mitochondrial membrane potential. Anal. Biochem. 552, 50–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starkov A. A., Measurement of mitochondrial ROS production. Methods Mol. Biol. 648, 245–255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chrétien D., Bénit P., Ha H.-H., Keipert S., El-Khoury R., Chang Y.-T., Jastroch M., Jacobs H. T., Rustin P., Rak M., Mitochondria are physiologically maintained at close to 50 °C. PLOS Biol. 16, e2003992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puchkov E. O., Intracellular viscosity: Methods of measurement and role in metabolism. Biochem. Moscow Suppl. Ser. A Membr. Cell Biol. 7, 270–279 (2013). [Google Scholar]
- 58.Santo-Domingo J., Demaurex N., The renaissance of mitochondrial pH. J. Gen. Physiol. 139, 415–423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei A.-C., Aon M. A., O’Rourke B., Winslow R. L., Cortassa S., Mitochondrial energetics, pH regulation, and ion dynamics: A computational-experimental approach. Biophys. J. 100, 2894–2903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ong S. Y., van Harmelen R. J. J., Norouzi N., Offens F., Venema I. M., Habibi Najafi M. B., Schirhagl R., Interaction of nanodiamonds with bacteria. Nanoscale 10, 17117–17124 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Clayton D. A., Shadel G. S., Isolation of mitochondria from tissue culture cells. Cold Spring Harb. Protoc. 2014, pdb.prot080002 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Abdul Kadir L., Stacey M., Barrett-Jolley R., Emerging roles of the membrane potential: Action beyond the action potential. Front. Physiol. 9, 1661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/21/eabf0573/DC1


