TMAO functions as a protective osmolyte in plants, promoting abiotic stress tolerance.
Abstract
Trimethylamine N-oxide (TMAO) is a well-known naturally occurring osmolyte in animals that counteracts the effect of different denaturants related to environmental stress and has recently been associated with severe human chronic diseases. In plants, however, the presence of TMAO has not yet been reported. In this study, we demonstrate that plants contain endogenous levels of TMAO, that it is synthesized by flavin-containing monooxygenases, and that its levels increase in response to abiotic stress conditions. In addition, our results reveal that TMAO operates as a protective osmolyte in plants, promoting appropriate protein folding and as an activator of abiotic stress–induced gene expression. Consistent with these functions, we show that TMAO enhances plant adaptation to low temperatures, drought, and high salt. We have thus uncovered a previously unidentified plant molecule that positively regulates abiotic stress tolerance.
INTRODUCTION
Because of their sessile nature, plants have to be permanently exposed to changes in their environment. This constraint becomes particularly challenging when facing adverse environmental conditions such as extreme temperatures, water deficiency, or high salt in soils. To survive and reproduce under these conditions, plants activate adaptive responses that range from rapid protective mechanisms, such as osmolyte accumulation, to developmental modifications, such as reduction in the shoot/root ratio, that ultimately promote tolerance and survival (1). In the last years, experimental evidence has shown that most of these adaptive responses are controlled through extensive reprogramming of gene expression, and many genes involved in plant tolerance to abiotic stresses have been identified and characterized (1, 2).
Osmolytes are low–molecular weight, highly soluble compounds that are usually nontoxic at high cellular concentrations. In plants, the accumulation of osmolytes is the result of different signaling pathways and is required for survival under abiotic stress conditions. The best characterized plant osmolytes, i.e., sucrose, trehalose, proline, and methylammonium compounds such as glycine betaine, have been described to protect plants through several manners, including cellular osmotic adjustment and stabilization, detoxification of reactive oxygen species, and protection of membrane integrity (3–7). Osmolytes also act as chaperones preserving the folding status of proteins under stress conditions, thus maintaining their stability and function (3, 4, 6). Some osmolytes have been used to genetically engineering plants to improve their tolerance to abiotic stresses. Moreover, they have also been exogenously applied to plants growing under stress conditions. In both cases, the results obtained have been very diverse, depending on the osmolytes and plants used in the experiments (3, 4, 6, 8, 9).
In animals, trimethylamine N-oxide (TMAO) is the quintessential protein-stabilizing osmolyte (10, 11). It is generated by N-oxygenation of trimethylamine (TMA) by flavin-containing monooxygenases (FMOs) (12) and operates as a chaperone that retains the folding status of proteins and prevents the effect of denaturants such as pH, urea, high pressure, high salt, and low temperature (13–15). TMAO has recently gained a lot of attention due to its potential role as a promoter of atherosclerosis, which causes cardiovascular and kidney diseases, as well as diabetes in humans (10). Intriguingly, the presence of TMAO in plants has not yet been reported. In this study, we demonstrate that plants also contain endogenous levels of TMAO, that it is synthesized by plant FMOs, and that its levels increase under abiotic stress conditions. Furthermore, our results indicate that TMAO promotes adequate protein folding in plants and triggers an up-regulation of abiotic stress–induced gene expression. As expected from these results, we show that TMAO enhances plant tolerance to freezing temperatures, drought, and high salt.
RESULTS
FMOGS-OX5 positively regulates Arabidopsis tolerance to abiotic stress
We previously isolated a number of RARE COLD INDUCIBLE (RCI) genes by screening a complementary DNA (cDNA) library prepared from cold-acclimated (4°C, 7 days) Arabidopsis seedlings with a subtracted cDNA probe enriched in cold-induced transcripts (16). One of them, RCI5 (At1g12140), turned out to encode FMOGS-OX5, a FMO belonging to a subclade of the FMO phylogeny that includes seven proteins implicated in the S-oxygenation of glucosinolates (fig. S1A) (17). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analyses confirmed that the expression of RCI5/FMOGS-OX5 was induced at low levels in Col-0 [wild type (WT)] plants exposed to 4°C, this induction being transient (Fig. 1A) and mainly in leaves (Fig. 1B). Under control conditions, FMOGS-OX5 transcripts were detected at similar levels in all organs of Arabidopsis (Fig. 1B). FMOGS-OX5 expression was not affected in response to other cold-related abiotic stresses such as water deficiency and high salt (fig. S1B).
Fig. 1. FMOGS-OX5 positively regulates Arabidopsis tolerance to abiotic stress.
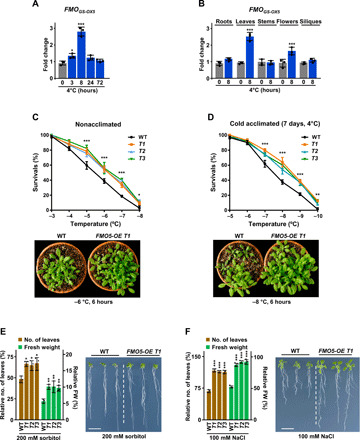
(A and B) Accumulation of FMOGS-OX5 transcripts in leaves of WT plants exposed different times to 4°C (A) and in different organs of WT plants exposed 8 hours to 4°C (B). (C and D) Freezing tolerance of nonacclimated (C) and cold-acclimated (D) WT and FMO5-OE plants (top panels). Tolerances are represented as the percentage of plants surviving each temperature. Bottom panels show representative plants after freezing. (E and F) Tolerance to water (E) and salt (F) stress of WT and FMO5-OE plants (left panels). Tolerances are represented as the relative number of leaves and remaining fresh weigh (FW) in plants after treatments. Right panels show representative plants after treatments. Scale bars, 1 cm. Data represent the mean of at least three independent biological replicates (n > 30). Asterisks indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001) between control (0 hours) and treated plants (A and B) or between WT and FMO5-OE lines (C to F), as determined by one-way (A, B, E, and F) or two-way (C and D) ANOVA (Dunnett’s post hoc test). Error bars indicate the SD. Photo credit: Rosa López-Cobollo, Centro de Investigaciones Biológicas Margarita Salas-CSIC.
To investigate a possible role of FMOGS-OX5 in plant response to low temperature, an Arabidopsis transgenic line containing a single T-DNA insertion in the fourth exon of FMOGS-OX5 was identified (fig. S1C). We used primers designed 3′ and 5′ from the T-DNA (fig. S1C and table S1) to assess the levels of FMOGS-OX5 transcripts in homozygous plants for the insertion. In both cases, the transcript levels were severely reduced compared to those in WT plants under control and low temperature conditions (fig. S1D), indicating that this FMOGS-OX5 allele (fmo5-1) was null or highly hypomorphic. No obvious morphological differences were found between fmo5-1 and WT plants (fig. S1E). The involvement of FMOGS-OX5 in cold response was assessed by analyzing the capacity of nonacclimated and cold-acclimated (7 days, 4°C) fmo5-1 mutants to tolerate freezing temperatures. As shown in fig. S1F, no significant differences between fmo5-1 and WT plants were found in any case.
Intriguingly, the expressions of genes corresponding to FMOGS-OX1, FMOGS-OX3, FMOGS-OX4, and FMOGS-OX6 proteins, which cluster to the same subclade as FMOGS-OX5 (fig. S1A), were also induced by cold stress (fig. S2), suggesting that the WT freezing tolerance phenotypes of fmo5-1 mutants could be due to functional redundancy between these FMOs. To overcome this potential issue, we generated Arabidopsis transgenic plants constitutively overexpressing FMOGS-OX5. Three independent lines (FMO5-OE T1, T2, and T3), homozygous for a single copy of the 35S::FMOGS-OX5 transgene and displaying high levels of FMOGS-OX5 expression when growing under control conditions (fig. S3A), were selected and evaluated for their freezing tolerance. All lines grew and developed indistinguishable from WTs under regular growing conditions (fig. S3B). However, the overexpression of FMOGS-OX5 significantly increased the freezing tolerance of both nonacclimated and cold-acclimated (7 days, 4°C) Arabidopsis (Fig. 1, C and D). Furthermore, all FMO5-OE lines also displayed increased tolerance to other cold-related abiotic stresses, such as water deficiency and high salt (Fig. 1, E and F). These results indicated that FMOGS-OX5 acts as a positive regulator of abiotic stress tolerance in Arabidopsis.
FMOGS-OX5 is involved in TMAO biosynthesis in Arabidopsis
The question arising from the results described above was how FMOGS-OX5 could regulate Arabidopsis tolerance to abiotic stress. As mentioned before, FMOGS-OX5 has been involved in the S-oxygenation of glucosinolates (17), which in plants are mainly related to pest and pathogen responses (18, 19). However, in animals, in addition to S-oxygenate many biological compounds, FMOs also have the ability to N-oxygenate a vast spectrum of substrates, including TMA to generate the osmolyte TMAO (12). Given the role of TMAO in preserving the deleterious effects of adverse environments in animals (see above) and the implication of FMOGS-OX5 in Arabidopsis tolerance to abiotic stresses, we decided to investigate the possibility that FMOGS-OX5 could also be involved in generating TMAO. Because, to our knowledge, the presence of TMAO in plants has never been reported, we first tried to detect TMAO in WT plants grown under control conditions. Nuclear magnetic resonance (NMR) analyses revealed that unstressed Arabidopsis have noticeable [≈100 μmol kg−1 fresh weight (FW)] TMAO levels (Fig. 2A). The presence of TMAO in Arabidopsis was corroborated by means of liquid chromatography–tandem mass spectrometry (LC-MS/MS) (fig. S4A). As liquid chromatography–based methods may present some difficulties in determining accurate levels of metabolites, including TMAO (20), we decided to use NMR in further assays.
Fig. 2. FMOGS-OX5 catalyzes TMAO biosynthesis in Arabidopsis.
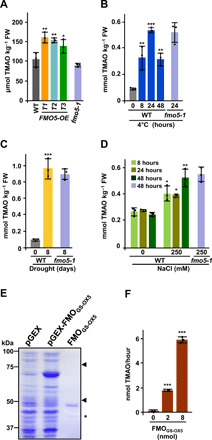
(A) TMAO content in WT, FMO5-OE, and fmo5-1 plants grown under control conditions. (B to D) Accumulation of TMAO in WT plants exposed during the indicated times to 4°C (B), to water privation for 0 or 8 days (C), or to 250 mM NaCl during the indicated times (D). The accumulation of TMAO in fmo5-1 mutants exposed to 4°C (B), water privation (C), and 250 mM NaCl (D) is also shown. (E) SDS–polyacrylamide gel electrophoresis gel containing extracts from E. coli expressing the empty vector (pGEX) and the vector with the FMOGS-OX5 cDNA (pGEX-FMOGS-OX5), and the purified FMOGS-OX5 protein. Arrowheads indicate the predicted molecular weight of FMOGS-OX5-GST (top) and FMOGS-OX5 (bottom) proteins. Asterisk indicates a protein contaminant. (F) TMAO synthesis from TMA by the indicated amounts of FMOGS-OX5. Data represent the mean of three independent biological replicates [in (A) to (D), n > 40]. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001) between WT and FMO5-OE lines (A), between untreated and treated plants (B to D), or between the assays with and without FMOGS-OX5 (F), as determined by one-way ANOVA (Dunnett’s post hoc test). No significant differences between the fmo5-1 mutants and their corresponding WT plants were found in any case. Error bars indicate the SD.
The three FMO5-OE transgenic lines showed significantly higher content of TMAO (≈150 μmol kg−1 FW) than WT plants (Fig. 2A). A transgenic line overexpressing FMOGS-OX6 (FMO6-OE) (21) also displayed higher levels of TMAO and enhanced tolerance to freezing, water deficiency, and salt stress than WT plants (fig. S4, B to E), supporting the proposed functional redundancy between the FMOs clustering to the same subclade that FMOGS-OX5. Consistent with this redundancy and with the expression patterns of FMOGS-OX1-FMOGS-OX6 genes in response to abiotic stresses (figs. S1A and S2), the content of TMAO increased to very high levels (0.5 to 1.0 mmol kg−1 FW) when WT plants were subjected to low-temperature, water stress, or high-salt conditions (Fig. 2, B to D). TMAO content, however, did not increase significantly in FMO-OE lines exposed to these adverse environments (fig. S5), suggesting that levels of this molecule are subjected to strong regulation. As expected, considering the suggested functional redundancy, TMAO levels in the fmo5-1 mutant were similar to those of WT plants under control and stress conditions (Fig. 2, A to D). All these data demonstrated that Arabidopsis contains constitutive levels of TMAO, that these levels significantly increase in response to abiotic stress, and that FMOGS-OX5 and FMOGS-OX6 promote TMAO accumulation. Definitive evidence on the implication of FMOGS-OX5 in TMAO biosynthesis was obtained from biochemical assays in which its capacity to oxygenate TMA to TMAO was studied. A recombinant FMOGS-OX5 protein expressed and purified from Escherichia coli was able to oxygenate TMA in a dose-dependent manner, generating substantial amounts of TMAO (Fig. 2, E and F). These results confirmed that plant FMOs catalyze TMAO synthesis.
TMAO enhances appropriate protein folding and abiotic stress tolerance in Arabidopsis
To assess whether TMAO functions as a chaperone in plants ensuring the folding status of proteins, we analyzed its capacity to alleviate the effects of tunicamycin (Tm), an inducer of unfolded protein stress (22), on main root growth. Figure 3A shows that the inhibition of root growth after 2 weeks of exposition to Tm was significantly mitigated in all FMO-OE lines compared to WT plants. Furthermore, the unfavorable effect of Tm on root growth in WT plants was significantly relieved in the presence of 500 μM TMAO (Fig. 3B), the concentration at which the endogenous levels were within the range detected in response to abiotic stresses (fig. S6A and Fig. 2, B to D). These results indicated that TMAO acts as a protein-stabilizing osmolyte in Arabidopsis promoting adequate protein folding.
Fig. 3. TMAO alleviates unfolded protein stress and promotes Arabidopsis abiotic stress tolerance.
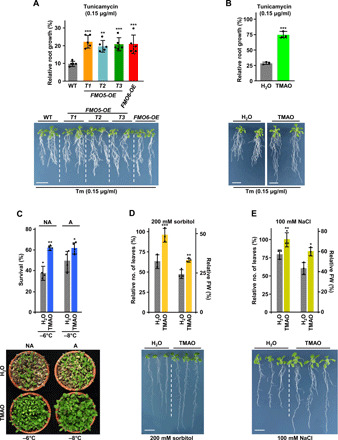
(A and B) Tolerance to unfolded protein stress of WT, FMO5-OE, and FMO6-OE plants (A), and untreated (H2O) and TMAO-treated WT plants (B) (top panels). Tolerances are represented as the relative main root growth in plants after Tm exposure. Bottom panels show representative plants after Tm exposure. (C) Freezing tolerance of nonacclimated (NA) and cold-acclimated (A) untreated (H2O) and TMAO-treated WT plants (top panel). Tolerances are represented as the percentage of surviving plants. The bottom panel shows representative plants after freezing. (D and E) Tolerance to water (D) and salt (E) stress of untreated (H2O) and TMAO-treated WT plants (top panel). Tolerances are represented as the relative number of leaves and remaining FW in plants after treatments. Bottom panels show representative plants after treatments. Data represent the mean of at least three independent biological replicates (n > 20). Asterisks indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001) between WT and transgenic plants (A), or untreated and TMAO-treated plants (B to E), as determined by one-way ANOVA (Dunnett’s post hoc test) (A) or one-sided t test (B to E). Error bars indicate the SD. Scale bars, 1 cm. Photo credit: Rafael Catalá, Centro de Investigaciones Biológicas Margarita Salas-CSIC.
Collectively, the findings described so far strongly suggested that TMAO could be involved in enhancing abiotic stresses tolerance in Arabidopsis. To test this hypothesis, WT plants were treated with TMAO and their tolerance to freezing temperatures, before and after cold acclimation, water deficiency, and high salt was determined. As shown in Fig. 3 (C to E), the application of 500 μM TMAO significantly enhanced the tolerance of Arabidopsis to all adverse conditions analyzed. We concluded that, as anticipated, TMAO functions as a positive regulator of Arabidopsis tolerance to abiotic stresses.
TMAO is present and improves abiotic stress tolerance in other plants than Arabidopsis
Next, we investigated whether the presence of TMAO was specific to Arabidopsis. To this, we analyzed the endogenous levels of TMAO in different plant species belonging to both dicots (i.e., tomato and Nicotiana benthamiana) and monocots (i.e., barley and maize). NMR analyses revealed that all species studied contained TMAO at similar levels as those detected in Arabidopsis, except in barley that were much higher (Fig. 4A). Upon exposure to low temperature, water deficiency, or salt stress, the content of TMAO raised significantly in tomato, N. benthamiana, and maize (Fig. 4A). In barley, we detect an increase in TMAO in response to cold and water stress but not in response to high salt (Fig. 4A). These data demonstrated that TMAO is widely distributed among plants, including important crops, and accumulates when they are exposed to abiotic stress conditions.
Fig. 4. TMAO is present and promotes abiotic stress tolerance in other plant species than Arabidopsis.
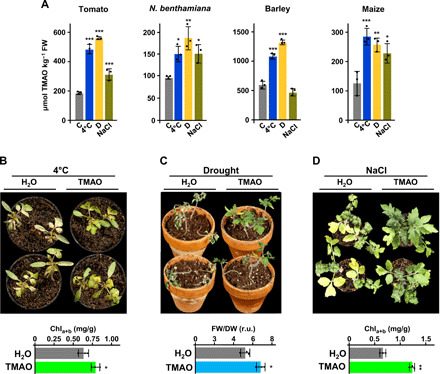
(A) TMAO content in tomato, N. benthamiana, barley, and maize plants grown under control conditions (C), exposed 1 day to 4°C (4°C), deprived of water for 8 days (D), or watered 1 day with 250 mM NaCl (NaCl). (B to D) Tolerance of tomato plants to low temperature (B), drought (C), or high salt (D) after TMAO treatment. Top panels show representative tomato plants untreated (H2O) or treated with TMAO after being exposed 2 weeks to 4°C (B), deprived of water for 2 weeks (C), or watered with increasing concentrations of NaCl (50 to 400 mM) during 2 weeks (D). Tolerances are represented in the bottom panels as the chlorophyll (B and D) or water [FW/dry weight (DW) ratio] content (C) in plants after stress treatments. Data represent the mean of three independent biological replicates (n > 20). Asterisks indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001) between control and stressed plants (A) or between untreated and treated plants (B to D), as determined by one-way ANOVA (Dunnett’s post hoc test) (A) or one-sided t test (B to D). Error bars indicate the SD. Photo credit: Rafael Catalá, Centro de Investigaciones Biológicas Margarita Salas-CSIC. r.u., relative units.
Then, we assessed whether TMAO also had the capacity to improve abiotic stress tolerance in other species than Arabidopsis. Tomato, one of the most popular and widely grown vegetable worldwide that, furthermore, is considered as a crop model system, was chosen for this study. Exogenous application of 10 mM TMAO, the concentration that originates a rise in the endogenous levels of TMAO in tomato plants similar to that caused by abiotic stress exposure (fig. S6B and Fig. 4A), significantly increased the tolerance of tomato plants to low temperature, water deficiency, and salt stress (Fig. 4, B to D). These results evidenced that TMAO promotes abiotic stress tolerance in other plant species than Arabidopsis and can be used to improve crop tolerance to environmental challenging conditions.
TMAO promotes abiotic stress–induced gene expression
As already mentioned, in plants, most adaptive responses to survive and reproduce under adverse environments involve transcriptome reprogramming (1). To determine whether the adaptive response generated by TMAO also involves extensive changes in gene expression, we first examined the transcriptomic profile of the FMO5-OE T1 line grown under control conditions by means of high-throughput RNA-sequencing (RNA-seq) experiments. Results revealed that the overexpression of FMOGS-OX5 altered the expression levels of 230 genes, 184 being up-regulated [≥2-fold; false discovery rate (FDR) < 0.001] (Fig. 5A and table S2). A high number of these genes (140; 76.1%) had been described to be induced in response to abiotic stresses (Fig. 5A and table S3) (23), and some of them—such as MPK3, ERF6, WRKY33, and ACS6, implicated in ethylene biosynthesis or signaling (24–27); STZ, MYB51, CZF1, and ANAC3, which encode transcription factors (28–31); or GOLS2, a gene encoding the galactinol synthase 2 (32)—to play a positive role in Arabidopsis tolerance to freezing temperature, drought, and/or salt stress. The up-regulation of these genes was verified in independent RNA samples from the FMO5-OE T1 line by RT-qPCR, validating the RNA-seq data (Fig. 5B). Furthermore, similar results were obtained with RNA samples from the FMO5-OE T2 and T3 lines (Fig. 5B). These data indicated that FMOGS-OX5 positively regulates abiotic stress–induced gene expression, in all likelihood, via TMAO. Exogenous application of 500 μM TMAO to WT plants grown under control conditions significantly increased the expression levels of the genes that were up-regulated in FMO5-OE lines (Fig. 5C), confirming that TMAO promotes abiotic stress–induced gene expression. The accumulation of FMOGS-OX5 transcripts did not show any significant change in response to TMAO (fig. S7), suggesting that the cold induction of FMOGS-OX5 is not mediated by this molecule.
Fig. 5. TMAO promotes abiotic stress–induced gene expression in Arabidopsis.
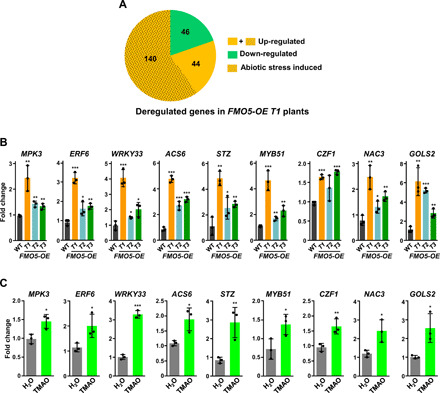
(A) Pie chart representing the number of deregulated genes in FMO5-OE T1 plants, including those up-regulated that have been described to be induced in response to low temperature, drought, and/or high salt. (B and C) Expression levels of different abiotic stress–induced genes in WT and FMO5-OE plants grown under control conditions (B), or in untreated (H2O) and TMAO-treated WT plants (C). Data represent the mean of three independent biological replicates (n > 40). Asterisks indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001) between WT and FMO5-OE plants (B) or between untreated and TMAO-treated plants (C), as determined by one-way ANOVA (Dunnett’s post hoc test) (B) or one-sided t test (C). Error bars indicate the SD.
DISCUSSION
In this study, we report the identification and functional characterization of a previously unknown plant molecule, TMAO. We have found that FMOGS-OX5, FMOGS-OX6, and, in all likelihood, other FMO proteins clustering in the same subclade mediate TMAO accumulation in Arabidopsis. In contrast to other plant osmolytes (e.g., glycine betaine), TMAO is widely distributed among plants, suggesting that it should have a substantial role in plant physiology. In this sense, the levels of TMAO significantly increase in all species analyzed when challenged by low temperature, water deficiency, or high salt, and increased TMAO levels in genetically engineered plants or in plants exogenously treated with the osmolyte display enhanced tolerance to those adverse situations. These findings indicate that TMAO plays an essential role in plant physiology, particularly in plant adaptation to abiotic stress. In animals, TMAO counteracts the effect of different denaturants by retaining the folding status of proteins (13–15). Our data indicate that TMAO also acts as a chaperone in plant cells, promoting adequate protein folding, and, therefore, that the ability of TMAO to function as a protein-stabilizing osmolyte has been evolutionary conserved between plants and animals. This function, furthermore, must account for the capacity of this molecule to enhance plant tolerance to abiotic stress.
Osmolyte accumulation is an important response in plants to adapt and survive under abiotic stress conditions and, consequently, osmolytes have long been considered as potential biotechnological tools to improve the tolerance of crops to such adverse environmental situations. Increased levels of some osmolytes in plants, either by genetic engineering or exogenous application, have been shown to improve their tolerance to abiotic stresses (3, 4, 6, 8, 9). In this context, it is noteworthy that the overexpression of just one gene is sufficient for genetically engineering plants containing high levels of TMAO and displaying increased tolerance to freezing temperatures, drought, and high salt. Moreover, TMAO can be applied exogenously by spraying or irrigation, both types of administration triggering similar adaptive responses. Although at this point we cannot conclude that exogenously applied TMAO is imported into plant cells, the fact that (i) the application of TMAO to Arabidopsis plants reproduces all the physiological and molecular effects caused by high intracellular TMAO levels (see above), (ii) exogenously applied glycine betaine, a plant quaternary amine like the TMAO that also functions as an osmolyte, is imported into tomato cells (33), and (iii) exogenously applied TMAO can enter into HeLa cells (34) strongly suggests that it can be the case. On the other hand, it should be noted that high levels of TMAO do not have any noticeable effect on plant growth and development. All these characteristics make TMAO a very suitable molecule to improve crop tolerance to abiotic stress.
Our transcriptome analyses indicate that TMAO promotes abiotic stress–induced gene expression, which would also contribute to its ability to enhance plant tolerance to abiotic stresses. Several abiotic stress–induced genes whose expression is up-regulated in Arabidopsis plants having elevated TMAO levels (i.e., the FMO5-OE lines and plants exogenously treated with the osmolyte), such as ERF6, WRKY33, ACS6, STZ, CZF1, or NAC3, have been reported to encode positive regulators of plant tolerance to low temperature, drought, and/or high salt (24, 26–29, 31). Other plant osmolytes, besides their protective function, also act as signaling molecules to modulate specific abiotic stress–related gene expression that can be essential for plant adaptation, which suggests that this may be a rather generalized role among these kinds of molecules. The levels of glycine betaine augment under situations of abiotic stress in a variety of plant species (35), affecting the expression patterns of numerous stress-regulated genes (33, 36, 37). Similarly, trehalose-6-phosphate and proline have also arisen as important plant osmolytes whose levels accumulate in different species in response to abiotic stresses and control the expression of many genes involved in abiotic stress tolerance (9, 38). The molecular mechanisms and signaling pathways through which osmolytes promote gene expression, however, remain practically unknown.
TMAO has emerged as a potential promoter of chronic diseases in humans, including atherosclerosis and diabetes (10). Nonetheless, a clear mechanistic link between TMAO and such diseases has not yet been established and whether increased TMAO concentrations are the cause or the result of them is still under discussion. Elevated levels of TMAO in plasma have been associated with diets entailing a high intake of animal products rich in carnitine and choline, while vegan and vegetarian diets have been suggested to reduce plasma TMAO levels (10). Our results should contribute to better understand the influence of diets on the levels of TMAO in human plasma.
On the basis of the data described in this study, a hypothetical model for TMAO function in Arabidopsis tolerance to abiotic stresses is proposed in Fig. 6. Unfavorable environmental conditions, such as low temperature, water deficiency, or high salt, would originate unfolding and, therefore, denaturation of protoplasmic proteins. Concomitantly, the expression of several FMOGS-OX genes would be induced, prompting the accumulation of the corresponding FMO proteins and the subsequent increase in TMAO levels that, in turn, would contribute to minimize the impact of the stress conditions by recovering the folding status of proteins. In addition, TMAO would mediate abiotic stress–induced gene expression through pathways not yet identified, which would also contribute to guarantee the precise development of Arabidopsis tolerance to freezing temperatures, drought, and high salt. Identifying the signaling pathways through which TMAO mediates stress-induced gene expression constitutes a remarkable challenge for future studies that will provide new insights on how plants respond and adapt to adverse environments.
Fig. 6. Proposed model for the function of TMAO in response to abiotic stress.
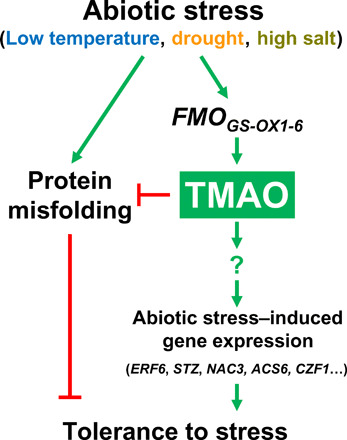
Arrows and end lines indicate positive and negative regulation, respectively.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana (L.) ecotype Columbia (Col-0) was used in all experiments as WT plant. The fmo5-1 mutant (SAIL_666_E10) was obtained from the Nottingham Arabidopsis Stock Centre. A fmo5-1 homozygous line was identified by PCR amplification with suitable primers (table S1), and DNA sequencing confirmed that the T-DNA insertion was 1358 base pairs (bp) downstream of the starting codon of FMOGS-OX5. The FMO5-OE lines were obtained by amplifying the full coding sequence of FMOGS-OX5 by PCR with appropriate primers (table S1) and transferring the resulting PCR product to the Sma I site downstream of the CaMv35S promoter in the pROK2 vector (39). The recombinant plasmid, once the construct by DNA sequencing verified, was introduced into the Agrobacterium tumefaciens (GV3101 strain). Transformation of Arabidopsis Col-0 was performed following the floral dip method (40). Three independent transgenic lines (FMO5-OE T1, T2, and T3) were used for analyses, all of them being genetically determined to have the 35S::FMOGS-OX5 fusion integrated at a single locus in homozygosis. The Arabidopsis FMO6-OE transgenic line was provided by W. Ji. Plants of N. benthamiana, tomato (Solanum lycopersicum, variety Moneymaker), barley (Hordeum vulgare, cultivar Sonora), and maize (Zea mays, hybrid DKC4795) were also used in this study.
Arabidopsis seeds were germinated and grown under standard conditions [20°C under long-day photoperiod (16 hours of cool-white fluorescent light, photon flux of 90 μmol m−2 s−1)] in pots containing a mixture of organic substrate and vermiculite (3/1, v/v) or in petri dishes containing Murashige and Skoog medium supplemented with 1% sucrose [germination media (GM)] and solidified with 0.9% (w/v) plant agar. Tomato, N. benthamiana, barley, and corn seeds were germinated on moistened filter paper in petri dishes at 25°C for 4 days in the dark, before being transferred to pots containing a COMPO SANA Universal substrate (COMPO GmbH). Seedlings were grown in a chamber set at 25°C under long-day photoperiod with cool-white fluorescent light, photon flux of 90 μmol m−2 s−1.
Plant treatments
Low-temperature treatments for gene expression analyses were performed by transferring 12-day-old or 8-week-old Arabidopsis plants growing in petri dishes or in soil, respectively, under control conditions to a growth chamber set at 4°C for different times under a long-day photoperiod with 16 hours of cool-white fluorescent light, photon flux of 40 μmol m−2 s−1. Gene expression analyses in response to water and salt stresses were accomplished on 2-week-old Arabidopsis plants growing in soil under standard conditions that were deprived of water for 8 days or watered 1 day with 250 mM NaCl, respectively. Low-temperature, water stress, and high-salt treatments to determine their effects on TMAO content were carried out in 2-week-old Arabidopsis, N. benthamiana, tomato, barley, and maize plants growing in soil under control conditions. For cold stress, plants were exposed different times to 4°C. Water stress was imposed by stopping watering for 8 days. For salt stress, plants were watered with 250 mM NaCl during different times. TMAO (Sigma-Aldrich, catalog no. T0514) treatments for gene expression were accomplished by transferring 12-day-old Arabidopsis plants growing in petri dishes under control conditions to filter papers soaked with 500 μM TMAO for 8 hours. Treatments with TMAO to increase the endogenous levels were carried out by spraying 2-week-old Arabidopsis and tomato plants growing in soil under standard conditions with different concentrations (100 and 500 μM for Arabidopsis; 0.1, 1.0, and 10 mM for tomato) of TMAO. Arabidopsis plants were harvested 8 and 24 hours after treatments, while tomato plants after 24 hours. TMAO was replaced by water in control treatments of all experiments involving treatment with TMAO. For gene expression analyses and TMAO measurements, tissue samples were frozen in liquid nitrogen immediately after treatments and stored at −80°C until use.
For freezing tolerance assays, 2-week-old Arabidopsis plants growing in soil under standard conditions (nonacclimated) or exposed seven additional days to 4°C (cold acclimated) were transferred to a freezing chamber set to 4°C for 30 min in darkness. Subsequently, temperature was allowed to decrease at a rate of −1°C per 30 min until reaching the final desired freezing temperature, which was maintained for 6 hours. Then, temperature was increased to 4°C at the same rate, and thawing was allowed for 12 hours before returning plants to control conditions for recovering. Tolerance to freezing was determined as the percentage of plants surviving after 2 weeks of recovery under control conditions. The effect of TMAO on the constitutive freezing tolerance of Arabidopsis and on its capacity to cold acclimate was examined by spraying plants with 500 μM TMAO 1 day before being exposed to freezing temperatures. Arabidopsis tolerance to water and salt stresses was analyzed on 5-day-old seedlings grown in petri dishes under standard conditions and then transferred to new plates supplemented with 200 mM sorbitol or 100 mM NaCl, respectively, for 2 weeks. In both cases, tolerance was estimated as the relative number of leaves and remaining FW of the plants after treatments. To study the effect of TMAO on Arabidopsis tolerance to water and salt stresses, 500 μM TMAO was added to plates containing sorbitol or NaCl. The tolerance of tomato plants to low temperature after TMAO treatment was assessed by spraying 2-week-old plants growing under control conditions with 10 mM TMAO and then exposing them to 4°C during 2 weeks under long-day photoperiods with cool-white fluorescent light, photon flux of 40 μmol m−2 s−1. The effect of TMAO on the tolerance of tomato plants to water stress was tested by watering 2-week-old plants growing under control conditions with 50 ml of 10 mM TMAO before stopping watering for 2 weeks. The tolerance of tomato plants to salt stress was evaluated by watering 2-week-old tomato plants growing under control conditions with increased concentrations (50, 100, 200, 300, and 400 mM) of NaCl supplemented with 10 mM TMAO every 3 days. Tomato tolerance to cold and salt stresses was estimated as the chlorophyll content in plants after treatments, which was quantified as described by Purdy et al. (41). The tolerance of tomato plants to water stress was determined by calculating their FW/dry weight (DW) ratios at the end of treatments. Tm (Sigma-Aldrich, catalog no. T7765) treatments to provoke unfolded protein stress were achieved by transferring 5-day-old Arabidopsis seedlings growing in petri dishes under control conditions to new plates supplemented with Tm (0.15 μg/ml) for 2 weeks. The negative impact of unfolded protein stress was assessed as the percentage of main root growth in plants exposed to Tm with respect to not exposed plants. Treatments with TMAO to examine its effect on the unfolded protein stress were performed by transferring 5-day-old Arabidopsis seedlings growing in petri dishes under control conditions to new plates supplemented with Tm (0.15 μg/ml) and 500 μM TMAO for 2 weeks. In all experiments involving treatments with TMAO, control treatments were performed with water instead of TMAO.
Quantification of TMAO content
TMAO content was quantified by 1H-NMR spectrometry, essentially as described by Shumilina et al. (42). In brief, 100 mg of frozen plant material was incubated overnight with 1 ml of 7% trichloroacetic acid (Sigma-Aldrich, catalog no. T6399) prepared with deuterated water. After incubation, extracts were centrifuged and the resulting supernatants filtered through miracloth filters. An aliquot of the extracts (900 μl) was mixed with 100 μl of 1 mM 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP-d4) (Sigma-Aldrich, catalog no. 269913), which was used as internal reference. 1H-NMR was performed using a Bruker Advance DRX 500 MHz spectrometer equipped with a 5-mm inverse triple resonance probe head (Bruker BioSpin). All experiments were conducted at 298 K, and data were acquired and processed using the same parameters. Spectra processing was performed on PC station using Topspin 2.0 software (Bruker BioSpin). TMAO methyl signals, located at around 1769 Hz in trichloroacetic acid/D2O mixtures, were identified and subsequently quantified using the TSP-d4 as internal reference. All TMAO quantifications were performed at least in triplicate with independent samples.
TMAO content was also quantified by LC-MS/MS, essentially as described (43). In short, 20 mg of frozen plant material was incubated overnight with 0.2 ml of 7% trichloroacetic acid (Sigma-Aldrich, catalog no. T6399) and clarified by centrifugation and filtration. The supernatant (10 μl) was analyzed by injection onto a silica column (ZORBAX RX-SIL, 110 Å, 5 μm, 150 mm by 2.1 mm, Agilent Technologies) at a flow rate of 0.3 ml min−1 interfaced with an LCMS-8030 Triple Quadrupole LC-MS/MS Mass Spectrometer (Shimadzu). A discontinuous gradient was generated to resolve the analytes by mixing solvent A (5 mM ammonium acetate) with solvent B (acetonitrile, Sigma-Aldrich, catalog no. 271004) at different ratios. TMAO was monitored using electrospray ionization in positive-ion mode (4.5 kV) with multiple reaction monitoring of precursor to product transition mass/charge ratio of 76.10 > 58.10 (quantifier) and 76.10 > 59 (qualifier), with a collision energy of −23 and −20, respectively. Different concentrations of TMAO were spiked into a mix of all samples to prepare the calibration curve. Quantifications were performed at least in triplicate with independent samples.
Expression, purification, and biochemical activity of FMOGS-OX5
The full-length FMOGS-OX5 cDNA was obtained by RT-PCR from total RNA of WT plants exposed 8 hours at 4°C, using specific primers (RCI5-ATG and RCI5-STOP; table S1). The resulting fragment was filled with Klenow and ligated into the Sma I site of pBluescript SK(+) (Stratagene) to yield the pBSFMOGS-OX5 plasmid. To obtain the GST::FMOGS-OX5 fusion, the pBSFMOGS-OX5 plasmid was digested with Bam HI and Sal I, and the fragment containing the FMOGS-OX5 cDNA filled with Klenow and ligated into the Sma I site of the pGEX-4T1 vector (GE Healthcare). To express the GST::FMOGS-OX5 fusion in E. coli, the pGEX-4T1 vector was transformed into the strain BL21(DE3) pLysS (Invitrogen). The purification of FMOGS-OX5 from E. coli, after inducing the expression culture with 1 mM IPTG during 24 hours at 28°C, was performed according to the manufacturer protocol. In summary, bacterial cells were collected by centrifugation, and the resulting pellet was lysed by incubation with lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 10% Triton X-100, and lysozyme (1 mg/ml)] during 30 min at 4°C and subsequent sonication. The extract, clarified by centrifugation, was incubated with Glutathione Sepharose 4B beads (GE Healthcare, catalog no. 17-0756-01) for 2 hours at room temperature, with gentle agitation. Then, beads were washed three times with washing buffer [50 mM tris-HCl (pH 7.5) and 150 mM NaCl], and FMOGS-OX5 protein was recovered by overnight incubation with Thrombin (40 U/ml; Cytiva, catalog no. 27-0846-01). The resulting purified FMOGS-OX5 protein extract was concentrated five times with Amicon Ultra 0.5 ml Centrifugal Filters (Merck, catalog no. UFC503008).
We determined the capacity of FMOGS-OX5 to catalyze the synthesis of TMAO from TMA essentially as described previously (44). Briefly, 2 or 8 nmol of FMOGS-OX5 purified from E. coli was added to a standard reaction mixture consisted of 2.5 μM flavin adenine dinucleotide, 300 μM reduced form of nicotinamide adenine dinucleotide phosphate, 3 mM MgCl2, and 20 mM potassium pyrophosphate buffer (pH 8.2) in a final volume of 0.5 ml. The reaction was started with the addition of 300 nmol of TMA (Sigma-Aldrich, catalog no. T7276). After incubating for 1 hour at 24°C, 55 μl of D2O was added to the reaction mix, and the TMAO generated was quantified by 1H-NMR spectrometry.
Gene expression analyses and RNA-seq experiments
For gene expression analyses, total RNAs were obtained using Nzyol reagent (Nzytech, catalog no. MB18501) according to the manufacturer’s protocol. RNA samples were treated with deoxyribonuclease I (Roche, catalog no. 4716728001) and quantified with a Nanodrop spectrophotometer (Thermo Fisher Scientific). cDNAs were synthesized from each sample with an iScript cDNA synthesis kit (Bio-Rad, catalog no. 1708891), and RT-qPCRs were performed with SsoFast EvaGreen Supermix (Bio-Rad, catalog no. 1725201) in a Bio-Rad iQ2 thermocycler. In all cases, the relative expression values were calculated using the At4g24610 gene as a reference (45). Primers used are listed in table S1. All reactions were carried out in triplicate using three independent RNA samples.
For RNA-seq experiments, total RNAs were extracted from 12-day-old WT and FMO5-OE T1 plants growing in petri dishes under control conditions using TRIzol reagent (Thermo Fisher Scientific, catalog no. 15596026) and subsequently purified with the RNeasy Plant Mini Kit (Qiagen, catalog no. 74904). cDNA libraries were generated from three independent RNA preparations each. RNA quality determination, library preparation, and subsequent sequencing in an Illumina HiSeq 2000 platform were performed by the staff of the Beijing Genome Institute. Approximately, 20 million 50-bp single-end reads per sample were generated, and >90% of them were aligned to the TAIR10 Col-0 reference genome using SOAP2 (46), with default parameters. Gene expression levels were calculated using the RPKM (reads per kilobase per million reads) method (47). Differentially expressed genes (DEGs) were identified using the algorithm developed by Audic and Claverie (48) to obtain a P value for each gene between any pair of samples. Then, an FDR analysis was performed to determine the threshold of P values in multiple tests. We established a FDR ≤ 0.001 and a fold change ±2 as cutoffs for any given DEG.
The Expression Browser tool of the Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca) (49) was used to determine which genes, among those that were up-regulated in the FMO5-OE T1 plants in our RNA-seq data, had been described to be induced in response to abiotic stresses. Selected settings were “AtGenExpress-stress series” as dataset and “cold stress,” “drought stress,” and “salt stress” as research areas (23). All tissue types, growth stages, and time points were considered, output options were set to “average of replicate treatments relative to average of appropriate control,” and induction was only contemplated when fold change was equal to or higher than twofold.
Phylogenetic analysis
The phylogenetic tree was generated by using the neighbor-joining method with the Geneious Tree Builder program (Geneious 11.1.2., Biomatters Ltd) with default parameters. Protein sequences were downloaded from the National Center for Biotechnology Information. The corresponding accession numbers are as follows: FMOOX-GS1 (Q9SS04), FMOOX-GS2 (Q94K43), FMOOX-GS3 (Q9SXE1), FMOOX-GS4 (Q93Y23), FMOOX-GS5 (A8MRX0), FMOOX-GS6 (Q9FWW3), and FMOOX-GS7 (Q9FWW6).
Statistical analysis
All data reported about the experiments are presented as means ± SD of at least three independent biological replicates, consisting of pools of at least 20 plants each. Statistical analyses were performed with the Prism 6 software (GraphPad Software Inc.). Comparisons between two groups were made by one-sided t test. Comparisons between multiple groups were realized by one-way or two-way analysis of variance (ANOVA) followed by Dunnett’s post hoc test, depending on whether one or two variables were considered, respectively.
Acknowledgments
We thank W. Ji (Northeast Agricultural University, Harbin, China) for the FMO6-OE transgenic line, and J. A. Jarillo, J. J. Sanchez-Serrano, and R. Solano for critical reading of the manuscript. We also thank J. Cañada and E. Calviño for the help with NMR measurements. Funding: Research in the Salinas laboratory was supported by grants BIO2016-79187-R from AEI/FEDER, UE, and PID2019-106987RB-100/AEI/10.13039/5011033. Author contributions: R.C., J.J.-B., and J.S. designed the research. R.C., R.L.-C., and M.Á.B. performed the research. R.C., R.L.-C., M.Á.B., J.J.-B., and J.S. analyzed the data. R.C. and J.S. wrote the paper. Competing interests: R.C., R.L.-C., and J.S. are inventors on a patent related to this work filed by Consejo Superior de Investigaciones Científicas and Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (no. WO2010034862A1, filed 16 September 2009, published 01 April 2010). R.C. and J.S. are inventors on two patents related to this work filed by Plant Response Biotech SL and Consejo Superior de Investigaciones Científicas (no. US_9085776_B2, filed 27 December 2013, published 21 July 2015, and no. US_9198416_B2, filed 19 February 2015, published 01 December 2015). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All primers used in this study are described in table S1. Sequence data from all genes mentioned in this manuscript can be found in the GenBank/EMBL data libraries under the accession numbers listed in table S4. The full names of these genes are also included in table S4. The RNA-seq data from this article have been submitted to the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) and assigned the identifier accession no. GSE144502.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/21/eabd9296/DC1
REFERENCES AND NOTES
- 1.Zhu J. K., Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y., Ding Y., Yang S., Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Chen T. H. H., Murata N., Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 5, 250–257 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Chen T. H. H., Murata N., Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ. 34, 1–20 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Sharma A., Shahzad B., Kumar V., Kohli S. K., Sidhu G. P. S., Bali A. S., Handa N., Kapoor D., Bhardwaj R., Zheng B., Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomolecules 9, 285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slama I., Abdelly C., Bouchereau A., Flowers T., Savouré A., Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 115, 433–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tognetti J. A., Pontis H. G., Martínez-Noël G. M. A., Sucrose signaling in plants: A world yet to be explored. Plant Signal. Behav. 8, e23316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashraf M., Foolad M. R., Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59, 206–216 (2007). [Google Scholar]
- 9.Szabados L., Savouré A., Proline: A multifunctional amino acid. Trends Plant Sci. 15, 89–97 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Zeisel S. H., Warrier M., Trimethylamine N-oxide, the microbiome, and heart and kidney disease. Annu. Rev. Nutr. 37, 157–181 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Liao Y.-T., Manson A. C., DeLyser M. R., Noid W. G., Cremer P. S., Trimethylamine N-oxide stabilizes proteins via a distinct mechanism compared with betaine and glycine. Proc. Natl. Acad. Sci. U.S.A. 114, 2479–2484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fennema D., Phillips I. R., Shephard E. A., Trimethylamine and trimethylamine N-oxide, a flavin-containing monooxygenase 3 (FMO3)-mediated host-microbiome metabolic axis implicated in health and disease. Drug Metab. Dispos. 44, 1839–1850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yancey P. H., Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Strambini G. B., Gonnelli M., Singular efficacy of trimethylamine N-oxide to counter protein destabilization in ice. Biochemistry 47, 3322–3331 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Jethva P. N., Udgaonkar J. B., The osmolyte TMAO modulates protein folding cooperativity by altering global protein stability. Biochemistry 57, 5851–5863 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Catalá R., Santos E., Alonso J. M., Ecker J. R., Martínez-Zapater J. M., Salinas J., Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15, 2940–2951 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J., Hansen B. G., Ober J. A., Kliebenstein D. J., Halkier B. A., Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 148, 1721–1733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tierens K. F. M. J., Thomma B. P. H. J., Brouwer M., Schmidt J., Kistner K., Porzel A., Mauch-Mani B., Cammue B. P. A., Broekaert W. F., Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125, 1688–1699 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beekwilder J., van Leeuwen W., van Dam N. M., Bertossi M., Grandi V., Mizzi L., Soloviev M., Szabados L., Molthoff J. W., Schipper B., Verbocht H., de Vos R. C. H., Morandini P., Aarts M. G. M., Bovy A., The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLOS ONE 3, e2068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Laan T., Kloots T., Beekman M., Kindt A., Dubbelman A.-C., Harms A., van Duijn C. M., Slagboom P. E., Hankemeier T., Fast LC-ESI-MS/MS analysis and influence of sampling conditions for gut metabolites in plasma and serum. Sci. Rep. 9, 12370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong W., Li J., Yu Q., Cang W., Xu R., Wang Y., Ji W., Two novel flavin-containing monooxygenases involved in biosynthesis of aliphatic glucosinolates. Front. Plant Sci. 7, 1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawkar G. M., Kang C. H., Maibam P., Park J. H., Jung Y. J., Chae H. B., Chi Y. H., Jung I. J., Kim W. Y., Yun D.-J., Lee S. Y., HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 2084–2089 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K., The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y., Deyholos M. K., Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S., Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLOS Genet. 8, e1002767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubois M., Skirycz A., Claeys H., Maleux K., Dhondt S., De Bodt S., Vanden Bossche R., De Milde L., Yoshizumi T., Matsui M., Inzé D., ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catala R., Lopez-Cobollo R., Mar Castellano M., Angosto T., Alonso J. M., Ecker J. R., Salinas J., The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittler R., Kim Y. S., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J. K., Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J., Jiang H., Xu Y., Li H., Wu X., Xie Q., Li C., The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48, 1148–1158 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Zhao L., Wang C., Zhu F., Li Y., Mild osmotic stress promotes 4-methoxy indolyl-3-methyl glucosinolate biosynthesis mediated by the MKK9–MPK3/MPK6 cascade in Arabidopsis. Plant Cell Rep. 36, 543–555 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Fu Y., Ma H., Chen S., Gu T., Gong J., Control of proline accumulation under drought via a novel pathway comprising the histone methylase CAU1 and the transcription factor ANAC055. J. Exp. Bot. 69, 579–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishizawa A., Yabuta Y., Shigeoka S., Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147, 1251–1263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park E.-J., Jeknic Z., Chen T. H. H., Exogenous application of glycinebetaine increases chilling tolerance in tomato plants. Plant Cell Physiol. 47, 706–714 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Teft W. A., Morse B. L., Leake B. F., Wilson A., Mansell S. E., Hegele R. A., Ho R. H., Kim R. B., Identification and characterization of trimethylamine-N-oxide uptake and efflux transporters. Mol. Pharm. 14, 310–318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodes D., Hanson A. D., Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 44, 357–384 (1993). [Google Scholar]
- 36.Park E.-J., Jeknić Z., Pino M.-T., Murata N., Chen T. H.-H., Glycinebetaine accumulation is more effective in chloroplasts than in the cytosol for protecting transgenic tomato plants against abiotic stress. Plant Cell Environ. 30, 994–1005 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Einset J., Nielsen E., Connolly E. L., Bones A., Sparstad T., Winge P., Zhu J.-K., Membrane-trafficking RabA4c involved in the effect of glycine betaine on recovery from chilling stress in Arabidopsis. Physiol. Plant. 130, 511–518 (2007). [Google Scholar]
- 38.Hwang G., Kim S., Cho J.-Y., Paik I., Kim J.-I., Oh E., Trehalose-6-phosphate signaling regulates thermoresponsive hypocotyl growth in Arabidopsis thaliana. EMBO Rep. 20, e47828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baulcombe D. C., Saunders G. R., Bevan M. W., Mayo M. A., Harrison B. D., Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature 321, 446–449 (1986). [Google Scholar]
- 40.Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H., Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Purdy S. J., Bussell J. D., Nunn C. P., Smith S. M., Leaves of the Arabidopsis maltose exporter1 mutant exhibit a metabolic profile with features of cold acclimation in the warm. PLOS ONE 8, e79412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shumilina E., Slizyte R., Mozuraityte R., Dykyy A., Stein T. A., Dikiy A., Quality changes of salmon by-products during storage: Assessment and quantification by NMR. Food Chem. 211, 803–811 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wang Z., Levison B. S., Hazen J. E., Donahue L., Li X.-M., Hazen S. L., Measurement of trimethylamine-N-oxide by stable isotope dilution liquid chromatography tandem mass spectrometry. Anal. Biochem. 455, 35–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agústsson I., Strøm A. R., Biosynthesis and turnover of trimethylamine oxide in the teleost cod, Gadus morhua. J. Biol. Chem. 256, 8045–8049 (1981). [PubMed] [Google Scholar]
- 45.Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W.-R., Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J., SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Audic S., Claverie J. M., The significance of digital gene expression profiles. Genome Res. 7, 986–995 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Toufighi K., Brady S. M., Austin R., Ly E., Provart N. J., The botany array resource: e-Northerns, expression Angling, and promoter analyses. Plant J. 43, 153–163 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/21/eabd9296/DC1


