Abstract
Favipiravir is a broad-spectrum inhibitor of viral RNA polymerase. It is currently used as a possible treatment for coronavirus disease 2019 (COVID-19). Pre-clinical or clinical trials of favipiravir require robust, sensitive, and accurate bioanalytical methods for quantitation of favipiravir levels. Recently, several studies have been reported about developing a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method for measuring favipiravir levels. However, these methods were validated predominantly for plasma samples, electrospray ionization was operated only in negative or positive mode, and clinical application of these methods has not been applied for patients with COVID-19. This study aimed was to develop a validated LC-MS/MS method for the measurement of favipiravir levels in positive and negative electrospray ionization mode and to perform a pilot study in patients with COVID-19 receiving favipiravir to demonstrate the applicability of this method in biological samples. Simple protein precipitation was used for the extraction of favipiravir from the desired matrix. Favipiravir levels were quantitated using MS / MS with an electrospray ionization source in positive and negative multiple reaction monitoring (MRM) mode. The chromatographic detection was performed on a reverse-phase Phenomenex C18 column (50 mm × 4.6 mm, 5 µm, 100 Å) with gradient elution using 0.1% formic acid in water and 0.1% formic acid in methanol as mobile phase. The method was linear over the concentration ranges of 0.048–50 µg/mL (in negative ionization mode) and 0.062–50 µg/mL (in positive ionization mode) with a correlation coefficient (r2) better than 0.998. The total run time was 3.5 min. The intra-assay and inter-assay %CV values were less than 7.2% and 8.0%, respectively. A simple, rapid and robust LC-MS / MS method was developed for the measurement of favipiravir and validation studies were performed. The validated method was successfully applied for drug level measurement in COVID-19 patients receiving favipiravir.
Keywords: Favipiravir, COVID-19, Tandem mass spectrometry, Validation
Abbreviations: CLSI, The Clinical & Laboratory Standards Institute; COVID-19, coronavirus disease 2019; HQC, high quality control; LC-MS/MS, liquid chromatography-tandem mass spectrometry; LLOQ, lower limit of quantification; LOD, limit of detection; LQC, low-quality control; MRM, multiple reaction monitoring; MQC1, medium 1 quality control; MQC2, medium 2 quality control; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2
1. Introduction
Since December 2019, a new coronavirus (severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)) infection has spread rapidly, causing respiratory disease coronavirus disease 2019 (COVID-19), threatening the health of many people [1]. The disease was first reported in Wuhan, China, but has now evolved into a pandemic, and it is currently known greater than 156 million people globally in over 219 countries have been infected and more than 3.2 million people have died of COVID-19 [2], [3]. At the end of March 2021, the reported hospitalization rates were 12.5% for over 65 years old, 9.5% for the aged 50–64, and the case fatality rate was estimated to be around 2.3% globally [4], [5]. The symptoms of COVID-19 can range from asymptomatic to acute respiratory distress syndrome or multi-organ dysfunction. The common clinical symptoms in patients are fever, cough, fatigue, joint pain, sore throat, and shortness of breath [6].
COVID-19 has become a global health emergency, with the lack of effective medical treatment and incipient vaccines [7]. Various treatments and prophylactic approaches are used to treat or prevent COVID-19, including oxygen therapy, ventilation support, convalescent plasma therapy, cell therapy, vaccines and drugs [8]. To date, global coronavirus vaccine research and development has covered the categories of live virus and inactivated vaccines, subunit vaccines, vector vaccines, nucleic acid vaccines (mRNA vaccines and DNA vaccines) [9]. Some of the authorized vaccines available for COVID-19 include Comirnaty (BioNTech, Germany, mRNA-based vaccine, 95% efficacy), Moderna COVID-19 Vaccine (Moderna Inc., USA, mRNA-based vaccine, 94.5% efficacy), Sputnik V (Gamaleya, Russia, Vector vaccine (Adenovirus Ad5 and Ad26), 92% efficacy), CoronaVac (Sinovac, China, inactivated vaccine, 50–91% efficacy), Janssen COVID-19 (Janssen Biotech Inc, USA, Vector vaccine (Adenovirus Ad26), 76.7–85.4% efficacy), AstraZeneca (Vector vaccine (Adenovirus), AstraZeneca, University of Oxford, 70% efficacy), Covaxin (Bharat Biotech, India, inactivated vaccine, 81% efficacy) [10]. Antimalarial agents (hydroxychloroquine, chloroquine), nucleotide analogues (remdesivir), nucleoside analogues (favipiravir, ribavirin), protease inhibitors (lopinavir, ritonavir), immunotherapy agents (tocilizumab, sarilumab), corticosteroids (dexamethasone, methylprednisolone), non-steroidal anti-inflammatory agents (naproxen, aspirin), antibiotics (azithromycin, teicoplanin), anticoagulants (nafamostat) are commonly registered drugs in COVID-19 clinical trials [8], [11], [12], [13].
One of the agents investigated for the COVID-19 treatment is favipiravir (Avigan™), (T-705), (6-fluoro-3-hydroxy-2-pyrazinecarboxamide), an oral pyrazinecarboxamide derivative [14]. Favipiravir, a purine analog and a potent RNA-dependent RNA polymerase inhibitor [15]. It is effective against a large number of RNA viruses including influenza, arena-, bunya-, flavi- and filo-, noroviruses, West Nile virus, yellow fever virus, Ebola virus, Lassa virus [16]. Favipiravir is approved for influenza treatment and is currently considered in the treatment of COVID-19 [15]. It has a well-established safety profile and any serious adverse events related to favipiravir have not been reported. The most common side effects reported in clinical trials are increased serum uric acid levels, gastrointestinal side effects, increased transaminase levels and decreased neutrophil count [17]. Favipiravir reaches maximum blood concentration 2 h after oral administration and has a short elimination half-life of 2–5.5 h. It is 54% bound to plasma proteins (albumin and α1-acid glycoprotein) [18]. Favipiravir is a prodrug that is metabolized to its active metabolite, favipiravir-ribofuranosyl-5′-triphosphate, by phosphoribosylation in the cell. It has been reported that the human hypoxanthine–guanine phosphoribosyltransferase enzyme may play a role in this activation process [19]. Favipiravir is also metabolized in the liver to its inactive metabolite T-705 M1 mainly via aldehyde oxidase, partly xanthine oxidase. Favipiravir is extensively metabolized and only 1% is excreted a parent drug via the urinary.
In COVID-19, a multi-drug regimen is applied to patients, especially if there are comorbidities. Therefore, the risk of drug-drug interaction increases. Studies have reported that coadministration of acetaminophen and favipiravir caused an increase in levels of acetaminophen and acetaminophen glucuronide by approximately 20% and 30%, respectively. Potential drug interactions should be carefully monitored when agents that inhibit aldehyde oxidase (raloxifene, tamoxifen, estradiol, cimetidine, felodipine, amlodipine, verapamil) or metabolize via aldehyde oxidase (citalopram, zaleplon, famciclovir, sulindac) are used with favipiravir [18], [20]. Therefore, monitoring of favipiravir levels is extremely important for an effective and safe treatment. Until now, several LC-MS/MS [21], [22], [23], [24], [25], [26] and high-performance liquid chromatography methods [27], [28], [29] have been reported for the quantitation of favipiravir levels. These HPLC methods have low sensitivity, require long analysis time and larger sample volumes. LC-MS/MS is the gold standard method for therapeutic drug level monitoring [30]. However, validation studies are lacking for most of these methods [21], [22], [23]. Recently, several studies have been reported about developing a validated LC-MS/MS method for measuring favipiravir levels [24], [25], [26]. However, these methods were validated for plasma samples, electrospray ionization was operated only in negative or positive mode, and clinical application of these methods has not been applied for patients with COVID-19. Therefore, we conducted validation studies by developing LC-MS/MS method in both negative and positive electrospray ionization mode for the measurement of favipiravir levels. The validated bioanalytical methods were successfully applied for drug level measurement in COVID-19 patients receiving favipiravir. The method developed differently from the others was validated for serum samples and favipiravir levels were compared in serum and plasma samples taken from COVID-19 patients.
2. Materials and methods
2.1. LC-MS/MS analysis
2.1.1. Chemicals and reagents
Atorvastatin, favipiravir, acetonitrile, methanol, HPLC grade water, formic acid were obtained from Sigma Aldrich (St. Louis, MO, USA).
2.1.2. Sample/Solution preparation
Individual stock solutions (1 mg / mL favipiravir and 0.5 mg / mL atorvastatin) were prepared separately in methanol for the preparation of quality control samples and calibration standards. Stock solutions were stable for up to 4 weeks at −20 °C. Calibration and quality control samples solutions were prepared fresh every time before analysis. Calibration and quality control (QC) samples were prepared by spiking 900 μL of drug free remnant human serum with 100 μL of favipiravir working standard solutions. The blank serum pool, which was used as a matrix throughout the study, was prepared by homogeneously mixing the serum samples obtained from 30 males and 30 females white blood donors (between the ages of 18–60 years) donating to the blood donation center of our hospital. Favipiravir calibration standards were prepared in drug-free remnant serum samples at final concentrations of 0.048, 0.097, 0.195, 0.390, 0.781, 1.56, 3.12, 6.25, 12.5, 25 and 50 μg/mL. The final serum concentrations of the lower limit of quantitation (LLOQC), low QC (LQC), medium QC 1 (MQC1), medium QC 2 (MQC2), high QC (HQC) were 0.048, 0.144, 25, 37.5 and 50 μg/mL for the negative mode, and 0.062, 0.186, 25, 37.5, and 50 μg/mL for the positive mode, respectively. Surrogate internal standard atorvastatin was prepared in methanol at 1250 ng/mL final concentration.
The sample preparation procedure of favipiravir was briefly, 100 μL internal standard (1250 ng/mL atorvastatin) and 600 μL acetonitrile were added to 250 μL sample or standard solution and vortexed for 30 s. Afterward, the reaction mixture was centrifuged at 2000 × g for 10 min and 30 μL of supernatant was injected into the LC-MS/MS system.
2.1.3. LC-mrm/ms
Shimadzu HPLC system (Kyoto, Japan) consisted of a pump (LC-20 AD), an automatic sampler (SIL-20 AC HT) and a unit for online degasser (DGU-20A3). Mass spectrometric analyses were performed using an API 3200 triple quadrupole mass spectrometer (Applied Biosystems/MDS Sciex) equipped with an electrospray ion source (ESI) operating in positive and negative mode. Chromatographic separation was performed using a Phenomenex C18 HPLC column (50 mm × 4.6 mm, 5 µm, 100 Å). The mobile phase A consisted of water containing 0.1% formic acid and mobile phase B consisted of methanol containing 0.1% formic acid. The percentage of mobile phase B was changed as follows: 0.0 min, 25%; 1.0 min, 50%; 2.0 min, 100; 2.8 min 25%, 3.5 min 25%. The HPLC column was re-equilibrated at 25% acetonitrile for 0.5 min before the next injection. The total run time including equilibration was 3.5 min. The flow rate was 1.0 mL/min, column and autosampler temperature were set at 25 °C and 4 °C, respectively.
MS/MS parameters were optimized with aqueous standard solutions of favipiravir infused at 15 µL/min into the MS/MS via a Hamilton syringe infusion pump. For ionization source optimization, the criteria for determining the best ionization parameters were spray stability (Total Ion Chromatogram (TIC) variation below 10%), signal intensity and quality. Fragment ions for favipiravir were present in the highest abundance at 158.2/85.1 (quantifier), 158.2/113.1 (qualifier) m/z and 156.2/85.1 (qualifier), 156.2/113.1 (quantifier) m/z for positive and negative modes, respectively. Fragment ions for atorvastatin were present in the highest abundance at 559.0/250.2 m/z in positive mode, and 557.0/278.2 m/z in negative mode. The main method optimization parameters for mass spectrometry were determined as ionspray voltage, 4500 V; ion source temperature, 400 °C; gas1, 40 psi; gas2, 40 psi; curtain gas, 20 psi, collision gas, 6 psi. Declustering entrance, collision cell exit potential, collision energy 40, 8, 31 V and 75, 6, 53 V, for favipiravir and atorvastatin, respectively for positive ionization. The main method optimization parameters for mass spectrometry were determined as ion spray voltage, −4000 V; ion source temperature, 450 °C; gas1, 60 psi; gas2, 60 psi; curtain gas, 25 psi, collision gas, 5 psi. Declustering entrance, collision cell exit potential, collision energy –32, −7, −25 V and −55, −7, −44 V, for favipiravir and atorvastatin for negative ionization.
2.1.4. Method validation
The method validation study was performed according to Clinical and Laboratory Standards Institute (CLSI) C62-A: Liquid Chromatography-Mass Spectrometry Methods guidelines and The Food and Drug Administration (FDA) guidelines [31], [32]. Linearity, precision, recovery, matrix effect, stability, carry-over, selectivity, and specificity parameters were evaluated in this study.
Each calibration curve was generated by analysis of two replicates of one blank sample, one zero sample (blank with internal standard added), and eleven non-zero standards. The calibration curves were generated by plotting the peak area ratios of analyte / internal standard versus nominal analyte concentrations. The calibration curve was established using the following criteria: (1) the mean value should be within ± 15% of the theoretical value, except at the LLOQ, where it should not deviate by more than ± 20%; (2) the correlation coefficients (r2) of all calibration curves should be more than 0.980.
The precision and accuracy were evaluated using quality control samples prepared at LLOQ, low, medium 1, medium 2, and high QC levels of favipiravir. The inter-day precision studies were performed analyzing four replicates per level at the LLOQ, low, medium 1, medium 2 and high QC samples for 5 consecutive days. The intra-day precision study was carried out by analyzing a total of 40 replicates, 20 in the morning and 20 in the afternoon for each level. For quality control samples at low level, medium1, medium 2 level, and high level, the intra- and inter-assay precision should be equal to or less than 15% and the intra- and inter-assay accuracy should be between 85% and 115%. For LLOQ samples, the intra- and inter-assay precision should be no more than 20% and the intra- and inter-assay accuracy should be from 80% to 120%.
Matrix effects and recovery were determined using 6 different negative matrix sources at low, medium and high concentrations (n = 3) and were considered acceptable within ± 25% of the target area at the respective concentration. According to the CLSI guideline, the recovery values should be between 85% and 115% at the points where the concentration level is more than 3 times the LLOQ, while it should be between 80% and 120% at low concentration levels. Percent matrix bias (100-%ME) should be less than 15%.
In the carry-over study, low and high level analyte spiked remnant blank serum samples were sequentially analyzed For the acceptability of this study, the calculated carry-over results should be less than 3 × SDlow-low results.
The stability of favipiravir in serum samples following four freeze - thaw cycles (room temperature to − 20° C to room temperature), long-term sample storage (−20° C for 45 days) was evaluated by determining at LQC, MQC and HQC concentrations (n = 3). The acceptability criteria was that all calculated bias% values were less than 15% .
The selectivity was evaluated by comparing the chromatogram of six different blank serum samples with that of favipiravir and internal standard spiked serum samples. The acceptability criteria was that the blank and zero calibrators were free of interference in the retention times of the favipiravir and internal standard [31], [32].
2.1.5. Application to clinical samples
The validated LC-MS/MS method was applied to randomized remnant serum samples from patients with COVID-19 receiving favipiravir (n = 55). Analysis was performed in accordance with the Declaration of Helsinki and ethical approval was obtained from the Selcuk University local Ethics Committee (Number: 2021/151, Date: 10/03/2021). The patients were administered 1,600 mg of FPV twice daily on day 1, followed by 600 mg twice daily from day 2 to day 5. In less than 12 h from the last dosage, blood samples were taken immediately and transferred to the laboratory and serum samples were obtained from collection tubes without gel additive by centrifugation at 2000 × g for 10 min, plasma samples were obtained from EDTA containing tubes by centrifugation at 2000 × g at 4 °C for 10 min. Serum and plasma samples were separated and stored at 80◦C and analyzed within two weeks. 30 samples were used for the comparison of the different matrix (serum and plasma) in a total of 55 patient samples.
2.1.6. Data analysis
Statistical analysis was performed using EP Evaluator Release 8 version (Data Innovations, South Burlington, VT), Excel (2010) programs. Data analysis was processed by SCIEX Analyst® 1.6.2 Software. Analyte concentrations were calculated using the internal standard method. The standard curves were generated from the peak area ratios of analyte / internal standard and the nominal analyte concentrations using a linear regression analysis y = a + bx with a weighting factor (1 / x2).
The limit of blank (LOB) value was calculated by analysis of 20 replicates of the blank serum sample and the following formula: LOB = meanblank + 1.65 SDblank. The LOD value was calculated by the following formula after the analysis of 20 replicates of low-level analyte spiked serum samples: LOD = LOB + 1.645 (SDlow concentration sample). The recovery results are calculated as the average of “measured value/expected value” ratio (%). The matrix effect was calculated with the following formula: (ME% = (mean post-extracted peak area / mean un-extracted peak area) × 100). The bias% value was calculated as: Bias%= . The %CV values were calculated with the following formula: %CV = .
3. Results
3.1. Method development
Different pre-treatment steps such as protein precipitation, evaporation, liquid–liquid extraction (LLE) were tried to extract favipiravir from the biological matrix. Peak intensities and shapes were satisfactory in these methods, however protein precipitation was preferred due to its simple, fast, and economical nature. Various organic solvents such as acetonitrile, ethanol, and methanol were used for protein precipitation, the best peak shape and lowest matrix effect were achieved with acetonitrile. Favipiravir is used in high dose (2x8 loading on the first day, 2x3 maintenance dose on the other 4 days) in the treatment of COVID-19. The current method developed based on protein precipitation has sufficient sensitivity (LLOQ) and selectivity for quantitation of favipiravir in the patient population. Therefore, only protein precipitation was preferred due to the advantages mentioned above.
Different mobile phases including methanol–water, acetonitrile–water were evaluated by isocratic or gradient elution to improve chromatographic separation, and peak shape. To increase the ionization, additives such as formic acid, ammonium formate, acetic acid were added in different concentrations, either alone or together. The best separation, peak shape and sensitivity were obtained by gradient elution of mobile phase A: water containing 0.1% formic acid and mobile phase B: methanol containing 0.1% formic acid. The use of a short chromatography column Phenomenex C18 (50 mm × 4.6 mm, 5 µm, 100 Å) allowed separation of analyte and internal standard and elution in a very short time. Different flow rates and run times from 1 min to 15 min have been tried. The best peak shape and separation were achieved at 1.0 mL / min flow rate and 3.5 min run time. Multiple reaction monitoring (MRM) scans in both negative and positive ion mode of the molecular ion and the two most predominant fragments for each analyte were utilized. Although the analyte and internal standard responded better to negative ionization, satisfactory responses were obtained in both negative and positive ionization modes. Therefore, validation studies were performed using both ionization modes for the analysis of favipiravir levels.
3.2. Method validation of favipiravir
3.2.1. Linearity
Linearity study was performed according to CLSI EP06-A protocol [31]. Standard solutions of favipiravir were prepared at 0.048, 0.097, 0.195, 0.390, 0.781, 1.56, 3.12, 6.25, 12.5, 25 and 50 μg/mL concentration levels by spiking the drug into remnant human blank serum obtained from at least 6 different healthy subjects. The calibration curve with eleven concentration levels was established. Each standard was measured in duplicate, along with a blank sample without internal standard and a zero-sample with internal standard. The calibration curves were generated by plotting the peak area ratios of analyte / internal standard versus nominal analyte concentrations. The results were evaluated by linear regression analysis. The correlation coefficients of favipiravir calibration curves were found as 0.9991 and 0.9987 for positive and negative ionization modes, respectively. The negative and positive mass spectrometric method was linear at the 0.048–50 µg/mL for favipiravir.
3.2.2. LOD (Limit of detection) and LLOQ (Lower limit of quantitation)
LLOQ has been determined in according to FDA guidelines [32]. The Food and Drug Administration (FDA) guideline recommends the use of data from ≥ five replicates of a spiked sample from at least three different runs. The precision should be ± 20% CV and the accuracy should be ± 20% of nominal analyte concentration. Accordingly, 4 replicates of the low level analyte spiked sample were analyzed for five days by determining the concentration level at which the signal / noise ratio was approximately 10. Based on the measurement results, precision and accuracy results are calculated. The LLOQ value was determined as 0.062 µg/mL in the positive mode and 0.048 µg/mL in the negative mode. %CV values calculated from the inter-day precision study in LLOQ value of favipiravir were 7.9% and 7.4% for positive and negative modes, respectively. The accuracy values were determined as 105.2% and 98.7% in positive and negative modes, respectively. The limit of blank (LOB) value was calculated by analysis of 20 replicates of the blank serum sample and the following formula: LOB = meanblank + 1.65 SDblank. The LOD value was calculated by the following formula after the analysis of 20 replicates of low-level analyte spiked serum samples: LOD = LOB + 1.645 (SD low concentration sample). The LOD values were determined as 0.059 µg/mL and 0.045 µg/mL for positive and negative modes, respectively [33].
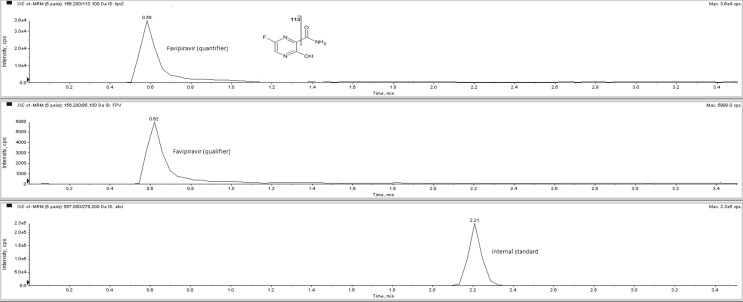
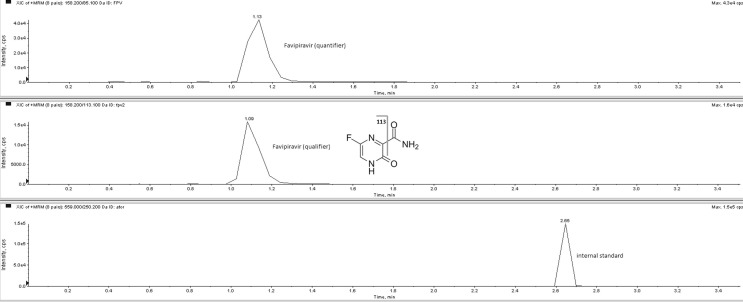
The chromatogram of a serum sample taken from a patient 8 h after tablet administration containing 200 mg favipiravir was shown in Fig. 1 (negative ionization mode) and Fig. 2 (positive ionization mode).
Fig. 1.
The chromatogram of a serum sample taken from a patient 8 h after tablet administration containing 200 mg favipiravir (in negative ionization mode). Serum favipiravir concentration calculated as 19.9 µg/mL.
Fig. 2.
The chromatogram of a serum sample taken from a patient 8 h after tablet administration containing 200 mg favipiravir (in positive ionization mode). Serum favipiravir concentration calculated as 21.2 µg/mL.
3.2.3. Intra- and inter-day precision
FDA guidelines recommend analyzing at least 5 replicates in at least 3 different independent runes for each of the five QC (LLOQ, low, medium 1, medium 2, and high QC) levels in the precision study. The inter-day precision studies were performed analyzing four replicates per level at the LLOQ, low, medium 1, medium 2, and high QC samples (Table 1 ) for 5 consecutive days. The intra-day precision study was carried out by analyzing a total of 40 replicates, 20 in the morning and 20 in the afternoon for each level [32].
Table 1.
The intra- and inter-day precision study results of favipiravir.
| Intra-day(Positive mode) | Inter-day(Positive mode) | ||||||
|---|---|---|---|---|---|---|---|
| QC | Concentration(μg/mL) | Mean(μg/mL) | Accuracy% | Precision(%CV) | Mean(μg/mL) | Accuracy% | Precision(%CV) |
| LLOQC | 0.062 | 0.064 | 103.6 | 7.1 | 0.065 | 105.2 | 7.9 |
| LQC | 0.186 | 0.188 | 101.1 | 5.9 | 0.194 | 104.4 | 6.6 |
| MQC1 | 25 | 26.4 | 105.4 | 4.4 | 25.1 | 100.5 | 5.5 |
| MQC2 | 37.5 | 36.7 | 97.9 | 4.1 | 36.3 | 96.7 | 4.9 |
| HQC | 50 | 49.9 | 99.8 | 3.1 | 49.9 | 99.8 | 3.8 |
| Intra-day(Negative mode) | Inter-day(Negative mode) | ||||||
| QC | Concentration(μg/mL) | Mean(μg/mL) | Accuracy% | Precision(%CV) | Mean(μg/mL) | Accuracy% | Precision(%CV) |
| LLOQC | 0.048 | 0.049 | 101.1 | 5.6 | 0.047 | 98.7 | 7.4 |
| LQC | 0.144 | 0.143 | 99.8 | 4.8 | 0.146 | 103.1 | 6.4 |
| MQC1 | 25 | 24.8 | 99.3 | 4.6 | 26.2 | 104.6 | 4.9 |
| MQC2 | 37.5 | 38.0 | 101.4 | 3.5 | 37.4 | 99.8 | 4.4 |
| HQC | 50 | 50.9 | 101.8 | 3.3 | 51.2 | 102.4 | 3.9 |
LLOQ: lower limit of quantification; LQC: low QC; MQC1: medium 1 QC; MQC2: medium 2 QC; HQC: high QC.
The results were summarized in Table 1.
3.2.4. Recovery and matrix effect study
The recovery study was performed according to the CLSI EP6-A protocol [31]. The three different sample sets consisting of low, medium and high-level analytes selected throughout the calibration curve were prepared by spiking into at least 6 different remnant blank serum samples (Table 2 ). The recovery study results were calculated as the average “measured value/expected value” ratio (%). The matrix effect is determined by two common methods. These are post-column infusion and post-extraction spike methods. We performed the matrix effect study with the second method described by Chambers et al [34]. In this method, the response of the analyte in neat solution is compared with the response of the spiked analyte to the pre-treated blank matrix. Accordingly, the analyte response in a mixture of water: acetonitrile (50:50, v/v%) containing three different levels of analyte selected along the calibration curve was compared with the analyte response obtained as a result of spiking to the pre-treated remnant blank serum sample, and the matrix effect was calculated with the following formula: (ME% = (mean post-extracted peak area / mean un-extracted peak area) × 100). The results were expressed in Table 2.
Table 2.
The recovery and matrix effect studies results of favipiravir.
| Positive mode | Negative mode | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recovery | Matrix effect | Recovery | Matrix effect | |||||||||
| Concentration(μg/mL) | 0.062 | 25 | 50 | 0.062 | 25 | 50 | 0.048 | 25 | 50 | 0.048 | 25 | 50 |
| Results(%) | 110.1 | 101.7 | 99.8 | 7.1 | 5.1 | 2.9 | 108.2 | 99.8 | 102.4 | 9.7 | 7.1 | 5.5 |
3.2.5. Stability study
The stability study was carried out according to the CLSI EP25-A protocol [31]. The stability study was performed using remnant blank serum samples containing spiked analyte at three different concentration levels, low, medium and high, selected along the calibration curve (Table 3 ). In this context, firstly, the effect of the freeze–thaw process on sample stability was investigated. After determining the concentration of analyte on the day of preparation for each level, the remaining 1000 µl of the sample were aliquoted. The concentration of favipiravir was measured in each thaw cycle by performing 4 freeze–thaw cycles (room temperature to − 20° C to room temperature). Secondly, after measuring favipiravir levels on the preparation day, three samples containing 250 μL of each sample were stored at −20 °C. Favipiravir levels were measured by thawing each eppendorf at 15, 30 and 45 days. After each analysis, the bias% value was calculated compared to the day of collection (expected value) using the following formula:
Table 3.
The stability study results of favipiravir (bias%).
| Concentration(μg/mL) | Positive mode | Negative mode | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frozen (-20 °C) for 45 day | Freeze-thaw stability | Frozen (-20 °C) for 45 day | Freeze-thaw stability | |||||||||
| 0.062 25 50 |
15. day(%) | 30. day(%) | 45. day(%) | 2.(%) | 3.(%) | 4.(%) | 15. day(%) | 30. day(%) | 45. day(%) | 2.(%) | 3.(%) | 4.(%) |
| −3.7 | −6.3 | −7.9 | −4.1 | −7.7 | −9.2 | −3.9 | −6.5 | −8.9 | −4.2 | −8.1 | −10.9 | |
| −2.5 | −6.1 | −7.1 | −3.7 | −7.5 | −8.1 | −3.1 | −5.6 | −7.5 | −3.7 | −6.2 | −7.9 | |
| −2.9 | −5.4 | −7.4 | −4.2 | −6.5 | −7.7 | −3.8 | −5.7 | −8.1 | −4.1 | −7.6 | −7.6 | |
The results were expressed in Table 3.
3.2.6. Carry-over study
The carryover study was performed according to the CLSI EP10-A protocol [31]. In this study, low and high level analyte spiked remnant blank serum samples were sequentially analyzed according to the order in the EP Evaluator Release program. The mean of high-low results, mean of low-low results, 3 × SDlow-low results and carry-over values were calculated through the program. In the program, the carry-over value was calculated with the following formula: Carry-over = (mean of high-low results) - (mean of low-low results). For the acceptability of this study, the calculated carry-over results should be less than 3 × SDlow-low results. In our study, the mean of high-low results were calculated as 0.110 µg/mL in positive mode and 0.099 µg/mL in negative mode. The mean of low-low results are calculated as 0.100 µg/mL in the method applied in positive mode and 0.088 µg/mL in the method applied in negative mode. Accordingly, the carry-over values were calculated as 0.010 µg/mL in the positive method and 0.011 µg/mL in the negative method, respectively. SDlow-low results were calculated as 0.010 and 0.084 µg/mL, respectively. Therefore, the allowable carry-over values should be less than 0.030 and 0.252 µg/mL, respectively (less than 3 × SDlow-low results). Therefore, our carry-over results were at acceptable levels in both methods.
3.2.7. Selectivity and specificity study
The selectivity and specificity studies were carried out in according to FDA guidelines [32]. In the selectivity study, the remnant blank serum sample was analyzed and no interfering peak was found in retention times corresponding to favipiravir and internal standard (atorvastatin) in the chromatogram of the blank. In addition, the internal standard response in the blank remnant serum did not exceed 5% of the response in the other calibration and working solutions.
As part of the specificity study, an interference study was conducted with allopurinol according to CLSI EP7-A [31]. As part of the specificity study, an interference study was conducted with allopurinol. 0%, 25%, 50%, 75%, and 100% of the remnant spiked serum pool containing 2000 ng/mL allopurinol was added to the remnant serum sample spiked low, medium and high-level favipiravir. The results are expressed as bias% in Table 4 .
Table 4.
The interference study results of favipiravir (bias%).
| Positive mode | |||||
|---|---|---|---|---|---|
| Concentration(μg/mL) | D1 4.1 3.9 3.5 |
D2 4.2 4.3 3.9 |
D3 4.8 3.8 4.2 |
D4 6.5 5.7 4.9 |
D5 |
| 0.062 (bias%) | 7.8 | ||||
| 25 (bias%) | 5.9 | ||||
| 50 (bias%) | 5.1 | ||||
| Concentration(μg/mL) | Negative mode | ||||
| 0.048 (bias%) | D1 4.3 3.7 3.6 |
D2 4.5 4.4 4.4 |
D3 7.1 4.9 3.8 |
D4 7.4 4.5 4.8 |
D5 |
| 7.9 | |||||
| 25 (bias%) | 5.8 | ||||
| 50 (bias%) | 5.1 | ||||
D1: 0%, D2: 25%, D3: 50%, D4: 75%, D5:100% interferant (allopurinol) level.
3.3. Application to clinical samples
The serum concentration of favipiravir was 11.06 (1.73–32.88) μg/mL in Day-1 (n = 18; dose: 3,200 mg), 7.19 (0.70–34.30) μg/mL in Day-2 to Day-5 (n = 30; dose: 1,200 mg) and 1.78 (0.65–3.57) μg/mL in Day-6 to Day-8 (n = 7) in negative method. The serum concentration of favipiravir was 11.78 (n = 18; 1.66–31.0) μg/mL in Day-1 (n = 18; 3,200 mg), 6.93 (0.62–35.62) μg/mL in Day-2 to Day-5 (n = 30; 1,200 mg) and 1.83 (0.63–3.77) μg/mL in Day-6 to Day-8 (n = 7) in positive method. There was no statistically significant difference (p = 0.795) between serum favipiravir levels measured in both methods ((6.64 (0.65–34.30) for negative mode; 6.85 (0.62–35.62) for positive mode). According to serum-plasma comparison in negative mode, serum favipiravir levels [3.17 (0.64–29.9) μg/mL] were slightly higher compared to plasma favipiravir levels [3.10 (0.63–27.4) μg/mL] (p = 0.912). Also, there was no significant difference for serum favipiravir levels [3.50 (0.62–28.0) μg/mL] were compared to plasma favipiravir levels [3.76 (0.63–29.8) μg/mL] in positive mode (p = 0.871).
4. Discussion
Since COVID-19 has become a rapidly spreading pandemic that causes many deaths worldwide, various treatment options have been tried to reduce or prevent the clinical symptoms of the disease. Currently, there is no specific treatment for COVID-19, however various antivirals are authorized by different national organizations. Most of these drugs are being investigated in preclinical and clinical studies. However, to achieve the desired effect on SARS-CoV-2, determination of the most appropriate dose and pharmacokinetic studies are required. Therefore, to accurately quantify these drugs in patient samples, it is necessary to develop sensitive, selective, new measurement methods [35].
Favipiravir was approved against influenza in Japan (2014), and currently, with the increase of the COVID-19 epidemic, its use was approved against SARS-CoV-2 in Europe, Turkey, Egypt, Bangladesh, Ukraine, Japan, Uzbekistan, Russia, Moldova, Kazakhstan, Dubai, and Saudi Arabia [36]. Favipiravir is generally well tolerated by patients [17]. One of the most important disadvantages of favipiravir is its use in high doses (it is used at a daily dose of 1200 mg for 2–14 days following a 3200 mg loading dose) [37]. In COVID-19, a multi-drug regimen is applied to patients, especially if there are comorbidities. Therefore, the risk of drug-drug interaction increases [20]. Considering the broad spectrum of favipiravir, its potential role in COVID-19 treatment and the important of drug-related pharmacokinetic studies, it is clear that a reliable, practical and robust measurement method is required for the measurement of favipiravir levels. Therefore, in our study, we aimed to develop an LC-MS / MS method for the measurement of favipiravir levels and to carry out validation studies.
As a result of our literature reviews, several LC-MS/MS method reported for the measurement of favipiravir levels was reached. Some of these studies investigate favipiravir levels in an in vitro model (cell culture medium) of Zika infection, serum/plasma favipiravir levels in Ebola-infected patients, and favipiravir contamination in river samples, however these studies lack validation studies [21], [22], [23]. Recently, several validated LC-MS/MS methods for measuring favipiravir levels have been reported [24], [25], [26]. Habler et al. [25] reported a validated two-dimensional isotope dilution LC–MS/MS method for multiplex analysis of the seven repurposed drug COVID-19 (remdesivir (plus metabolite GS-441524), chloroquine, hydroxychloroquine, lopinavir, ritonavir, favipiravir and azithromycin). In the method, online solid-phase extraction was applied, and there is no data for the measurement of favipiravir levels in patient samples. The reported %CV values for this method for favipiravir ranged from 1.27% to 5.57%, with a mean recovery of 99.45%. Rezk et al. [26] conducted a bioequivalence study in healthy volunteers (n = 30, after a single oral dose 200 mg) using the validated UPLC–MS/MS method developed for the measurement of plasma favipiravir levels. The reported LLOQ value of this method for favipiravir was 0.25 μg/mL, the recovery value varied between 81.95 and 92.56% and the precision of the method varied between 9.79 and 19.50%. The total analysis time was 4.5 min. The accuracy and precision of the tandem mass spectrometric method developed by Curley et al. [24] ranged between 89% and 110%, 101% and 106%, respectively.
Only three methods were reached for the development of a validated HPLC method for the quantitation of favipiravir levels [27], [28], [29]. However, these methods were for the measurement of favipiravir levels in pharmaceutical dosage forms and had disadvantages in terms of long analysis time (from 15 min to 60 min) and low sensitivity. In contrast, our method required a sample volume of 250 µl and a simple pretreatment step based on protein precipitation for extraction of favipiravir from the matrix. The total run time was 3.5 min.
The negative and positive mass spectrometric method was linear at the 0.048–50 µg/mL for favipiravir. The correlation coefficients of favipiravir calibration curves were found as 0.9991 and 0.9987 for positive and negative ionization mode methods, respectively. The LLOQ value was determined 0.062 µg/mL in the positive mode and 0.048 µg/mL in the negative mode. The calibration range established for favipiravir was suitable to quantify clinical samples.
The intra-assay %CV values were less than 7.2% in positive mode and less than 5.7% in negative mode. The inter-assay %CV values were less than 8.0% in positive mode and less than 7.5% in negative mode. %CV values are acceptable in both methods. According to FDA guidelines, the imprecision should be ± 20% CV at the LLOQ level, and ± 15% CV except for the LLOQ and our results were within these limits.
In the validation study performed in positive ionization mode, the mean recovery was 103.8% (99.8%-110.1%), while the matrix effect varied between 2.9% and 7.1%. In the validation study performed in negative ionization mode, the mean recovery was 103.5% (102.4%-108.2%), while the matrix effect varied between 5.5% and 9.7%. According to the CLSI guideline, in order for the recovery results to be acceptable, the recovery values should be between 85% and 115% at the points where the concentration level is more than 3 times the LLOQ, while it should be between 80% and 120% at low concentration levels. The extraction method used recovered almost all favipiravir from the serum and resulted in consistent and negligible matrix effects.
The remnant blank serum sample was analyzed and no interfering peak was found in retention times corresponding to favipiravir and internal standard (atorvastatin) in the chromatogram of the blank. Furthermore, the internal standard response in the blank remnant serum did not exceed 5% of the response in the other calibration and working solutions. These findings demonstrate that the method is selective for the quantification of favipiravir in human serum.
In the interference studies carried out within the scope of the specificity study, no interference effect was found on favipiravir analysis of allopurinol. It was determined that the calculated bias% values varied between 3.5% and 7.8% in the positive ionization mode in the interference study, while it varied between 3.6% and 7.9% in the negative mode. In the interference study, the bias% value should be less than 15%. Therefore, allopurinol doesn’t interfere with favipiravir analysis in the present method.
The carry-over value was calculated with the following formula: Carry-over = (mean of high-low results) - (mean of low-low results). For the acceptability of this study, the calculated carry-over results should be less than 3 × SDlow-low results. In our study, the mean of high-low results were calculated as 0.110 µg/mL in positive mode and 0.099 µg/mL in negative mode. The mean of low-low results are calculated as 0.100 µg/mL in the method applied in positive mode and 0.088 µg/mL in the method applied in negative mode. Accordingly, the carry-over values were calculated as 0.010 µg/mL in the positive method and 0.011 µg/mL in the negative method, respectively. SDlow-low results were calculated as 0.010 and 0.084 µg/mL, respectively. The allowable carry-over values should be less than 0.030 and 0.252 µg/mL, respectively (less than 3 × SDlow-low results). The carry-over between the high and low levels is negligible.
The developed method was applied to randomized patient samples. The measured favipiravir levels in the patient samples were within the calibration range of the analysis (0.048–50 µg / mL). Seven critically ill patients with COVID-19 who were admitted to the intensive care unit and required mechanical ventilation patients were enrolled in the study conducted by Irie et al. [38] 49 blood samples were collected from patients who were eligible to assess favipiravir levels. In most of these samples, favipiravir trough levels (after 8–12 h) were lower than the lower limit of quantification (1 μg/mL) and half-maximal effective concentration (9.7 μg/mL) previously tested in vitro against SARS-CoV-2. According to the pharmacokinetic study performed with healthy subjects in the AVIGAN package insert, (day 1: 1,600 mg b.i.d.,2–5: 600 mg b.i.d.), favipiravir trough levels have been reported to be 20–60 μg/mL [39], [40]. Lou et al. [41] measured plasma favipiravir levels on days 1 (n = 7, within 1 h following last drug dose), 4 (n = 8, within 7–8 h) and 7 (n = 5, within 7–8 h) in patients with COVID-19 who received favipiravir (day 1: 1,600 mg or 2200 mg b.i.d., 2--14: 600 mg t.i.d). Plasma favipiravir levels were reported as 32.2 ± 18.7 μg / mL, 11.8 ± 11.6 μg / mL and 21.8 ± 28.7 μg / mL, respectively. In the pharmacokinetic study performed by Nguyen et al. [22] in Ebola patients, favipiravir was administered in doublet dosage (day 0: 6,000 mg (2,400 mg, 2,400 mg, 1,200 mg q8h), day 1–9: 1,200 mg b.i.d.). The median (min–max) plasma favipiravir trough concentrations were 46.1 (2.3–106.9) μg/mL on day 2 and 25.9 (0–173.2) μg / mL on day 4.
In our study, favipiravir levels were measured in both modes in serum samples collected less than 12 h after the last dosage. The serum concentration of favipiravir was measured using negative and positive modes in our study, they were found as 11.06 (1.73–32.88) μg/mL 11.78 (n = 18; 1.66–31.0) μg/mL in Day-1 (n = 18; dose: 3,200 mg), 7.19 (0.70–34.30) μg/mL and 6.93 (0.62–35.62) μg/mL in Day-2 to Day-5 (n = 30; dose: 1,200 mg) and 1.78 (0.65–3.57) μg/mL and 1.83 (0.63–3.77) μg/mL in Day-6 to Day-8 (n = 7), respectively. The favipiravir levels of the patients whose blood samples were taken on day 1 were close to the in vitro EC50 value (9.7 μg / mL) reported by Wang et al. [42], but lower than the EC50 value (greater than15.7 μg / mL) reported by Lou et al. [41] Serum favipiravir levels of patients with blood drawn on the 2nd and 5th days of treatment were below 7.2 μg/mL, while it was less than 2 μg/mL in the post-treatment group. Mass spectrometry was operated in negative ionization mode and positive ionization mode for measurement of serum favipiravir levels, and serum favipiravir levels were found consistent in both modes (p = 0.795). In addition, plasma and serum favipiravir levels were largely similar when measured by the validated method for serum samples (p = 0.912).
5. Conclusion
The LC-MS/MS method was developed for the measurement of favipiravir and validation studies were performed. The validated method was successfully applied for drug level measurement in COVID-19 patients receiving favipiravir. Simple sample preparation, cost-effectiveness, rapid quantification, satisfactory precision, accuracy and sensitivity are the main advantages of the method. The assay can be used for routine favipiravir therapeutic drug monitoring in clinical laboratories. However, the limitations of the study are the lack of a multiplexed drug method, the use of the surrogate internal standard, the measurement of favipiravir levels in a limited number of patients, and the fact that favipiravir levels were not measured for each patient on consecutive days. To elucidate the pharmacokinetic and pharmacodynamic characteristics of favipiravir in COVID-19 patients, further studies are required to measure drug blood levels in well-defined conditions and to investigate the relationship between adverse effects and blood drug levels in a larger population.
CRediT authorship contribution statement
Duygu Eryavuz Onmaz: Conceptualization, Methodology, Software, Validation, Investigation, Writing - original draft. Sedat Abusoglu: Conceptualization, Methodology, Writing - review & editing. Mustafa Onmaz: Conceptualization, Methodology, Validation, Data curation. Fatma Humeyra Yerlikaya: Conceptualization, Methodology, Visualization. Ali Unlu: Software, Writing - original draft, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to thank the Selcuk University for this study.
References
- 1.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worldometer, coronavirus. https://www.worldometers.info/coronavirus/, 2021 (accessed 07 May 2021).
- 4.Centers for Disease Control And Prevention (CDC), Coronavirus disease 2019, Cases&Data, COVID-NET Laboratory - confirmed COVID-19 hospitalizations. https://gis.cdc.gov/grasp/covidnet/covid19_3.html /, 2021 (accessed 11 April 2021).
- 5.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baj J., Karakuła-Juchnowicz H., Teresiński G., Buszewicz G., Ciesielka M., Sitarz E., Forma A., Karakuła K., Flieger W., Portincasa P., Maciejewski R. COVID-19: Specific and Non-Specific Clinical Manifestations and Symptoms: The Current State of Knowledge. J. Clin. Med. 2020;9:1753. doi: 10.3390/jcm9061753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezzi G., Piga E.J., Binolfi A., Armas P. CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. Int. J. Mol. Sci. 2021;22:2614. doi: 10.3390/ijms22052614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.F. Babaei, M. Mirzababaei, M. Nassiri-Asl, H. Hosseinzadeh, Review of registered clinical trials for the treatment of COVID-19, Drug. Dev. Res. https://doi.org/10.1002/ddr.21762(2020). [DOI] [PMC free article] [PubMed]
- 9.Silveira M.M., Moreira G.M.S.G., Mendonça M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021;267:118919. doi: 10.1016/j.lfs.2020.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Y., Pang Y., Lyu Z., Wang R., Wu X., You C., Zhao H., Manickam S., Lester E., Wu T., Pang C.H. The COVID-19 Vaccines: Recent Development. Challenges and Prospects, Vaccines. 2021;9:349. doi: 10.3390/vaccines9040349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R. Cannalire, C. Cerchia, A.R. Beccari, F.S. Di Leva, V. Summa, Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities, Journal of Medicinal Chemistry, Nat. Rev. Immunol. https://doi.org/10.1021/acs.jmedchem.0c01140(2020). [DOI] [PMC free article] [PubMed]
- 13.Mehta N., Mazer-Amirshahi M., Alkindi N., Pourmand A. Pharmacotherapy in COVID-19; A narrative review for emergency providers. Am. J. Emerg. Med. 2020;38:1488–1493. doi: 10.1016/j.ajem.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghasemnejad-Berenji M., Pashapour S. Favipiravir and COVID-19: A Simplified Summary. Drug. Res (Stuttg) 2021;71:166–170. doi: 10.1055/a-1296-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabbous H.M., Abd-Elsalam S., El-Sayed M.H., Sherief A.F., Ebeid F.F.S., El Ghafar M.S.A., Soliman S., Elbahnasawy M., Badawi R., Tageldin M.A. Efficacy of favipiravir in COVID-19 treatment: a multi-center randomized study. Arch. Virol. 2021;166:949–954. doi: 10.1007/s00705-021-04956-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur R.J., Charan J., Dutta S., Sharma P., Bhardwaj P., Sharma P., Lugova H., Krishnapillai A., Islam S., Haque M., Misra S. Favipiravir Use in COVID-19: Analysis of Suspected Adverse Drug Events Reported in the WHO Database. Infect. Drug Resist. 2020;13:4427–4438. doi: 10.2147/IDR.S287934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Y.-X., Chen X.-P. Favipiravir: Pharmacokinetics and Concerns About Clinical Trials for 2019-nCoV Infection. Clin. Pharmacol. Ther. 2020;108 doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 19.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishima E., Anzai N., Miyazaki M., Abe T. Uric Acid Elevation by Favipiravir, an Antiviral Drug. Tohoku J. Exp. Med. 2020;251:87–90. doi: 10.1620/tjem.251.87. [DOI] [PubMed] [Google Scholar]
- 21.Pires de Mello C.P., Tao X., Kim T.H., Vicchiarelli M., Bulitta J.B., Kaushik A., Brown A.N. Clinical Regimens of Favipiravir Inhibit Zika Virus Replication in the Hollow-Fiber Infection Model. Antimicrob. Agents Chemother. 2018;62:e00967–00918. doi: 10.1128/AAC.00967-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen T.H., Guedj J., Anglaret X., Laouénan C., Madelain V., Taburet A.M., Baize S., Sissoko D., Pastorino B., Rodallec A., Piorkowski G., Carazo S., Conde M.N., Gala J.L., Bore J.A., Carbonnelle C., Jacquot F., Raoul H., Malvy D., de Lamballerie X., Mentré F. Favipiravir pharmacokinetics in Ebola-Infected patients of the JIKI trial reveals concentrations lower than targeted. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azuma T., Ishida M., Hisamatsu K., Yunoki A., Otomo K., Kunitou M., Shimizu M., Hosomaru K., Mikata S., Mino Y. Fate of new three anti-influenza drugs and one prodrug in the water environment. Chemosphere. 2017;169:550–557. doi: 10.1016/j.chemosphere.2016.11.102. [DOI] [PubMed] [Google Scholar]
- 24.Curley P., Neary M., Arshad U., Tatham L., Pertinez H., Box H., Rajoli R., Valentijn A., Sharp J., Rannard S., Owen A. Development of a highly sensitive bioanalytical assay for the quantification of favipiravir. 2021 doi: 10.1101/2021.02.03.429628. [DOI] [Google Scholar]
- 25.Habler K., Brügel M., Teupser D., Liebchen U., Scharf C., Schönermarck U., Vogeser M., Paal M. Simultaneous quantification of seven repurposed COVID-19 drugs remdesivir (plus metabolite GS-441524), chloroquine, hydroxychloroquine, lopinavir, ritonavir, favipiravir and azithromycin by a two-dimensional isotope dilution LC-MS/MS method in human serum. J. Pharm. Biomed. Anal. 2021;196 doi: 10.1016/j.jpba.2021.113935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M.R. Rezk, K.A. Badr, N.S. Abdel-Naby, M.M. Ayyad, A novel, rapid and simple UPLC–MS/MS method for quantification of favipiravir in human plasma: Application to a bioequivalence study, Biomed. Chromatogr. n/a e5098. [DOI] [PubMed]
- 27.I. Bulduk, HPLC-UV method for quantification of favipiravir in pharmaceutical formulations, Acta Chromatogr. https://doi.org/10.1556/1326.2020.00828(2020).
- 28.China patent (CN104914185B). A kind of Favipiravir has the HPLC assay method of related substance. 21.09.2016.
- 29.Favipiravir. 2015;16:09. [Google Scholar]
- 30.Koal T., Römling R., Svoboda M., Resch K., Kaever V. LC-MS/MS As Gold-standard For TDM Of Antiretroviral Drugs: 76. Ther. Drug Monit. 2005;27 [Google Scholar]
- 31.Wayne P.A. Liquid Chromatography-Mass Spectrometry Methods; Approved Guidelines, Pennsylvania, USA; Clinical and Laboratory Standards Institute: 2014. CLSI document C62-A. [Google Scholar]
- 32.Guidance for Industry, Bionanalytical Method Validation, US Department of Health and Human Services/Food and Drug Administration Centre for Drug Evaluation and Research (CDER)/Centre for Veterinary Medicine (CVM) (2018) May.
- 33.J.O. Westgard, Basic method validation, fourth ed., Method validation: the detection limit experiment, Madison, 2008, pp.169-173.
- 34.Chambers E., Wagrowski-Diehl D.M., Lu Z., Mazzeo J.R. Systematic and comprehensive strategy for reducing matrix effects in LC/MS/MS analyses. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;852(1–2):22–34. doi: 10.1016/j.jchromb.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Acquavia M.A., Foti L., Pascale R., Nicolò A., Brancaleone V., Cataldi T.R.I., Martelli G., Scrano L., Bianco G. Detection and quantification of Covid-19 antiviral drugs in biological fluids and tissues. Talanta. 2021;224 doi: 10.1016/j.talanta.2020.121862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrawal U., Raju R., Udwadia Z. Favipiravir: A new and emerging antiviral option in COVID-19. Med. J. Armed. Forces India. 2020;76 doi: 10.1016/j.mjafi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Łagocka R., Dziedziejko V., Kłos P., Pawlik A. Favipiravir in Therapy of Viral Infections. J. Clin. Med. 2021;10 doi: 10.3390/jcm10020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irie K., Nakagawa A., Fujita H., Tamura R., Eto M., Ikesue H., Muroi N., Tomii K., Hashida T. Pharmacokinetics of Favipiravir in Critically Ill Patients With COVID-19. Clin. Transl. Sci. 2020;13:880–885. doi: 10.1111/cts.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Package insert Avigan Tablet 200 mg (English translation) by Toyama Chemical Co., Ltd., November 2017. <https://www.cdc.gov.tw/File/Get/ht8jU iB_MI-aKnlwstwzvw>. Accessed April 10, 2021.
- 40.Report on the Deliberation Results Avigan Tablet 200 mg by Pharmaceuticals and Medical Devices Agency (PMDA), March 4, 2014. <https://www.pmda.go.jp/files/00021 0319.pdf>. Accessed April 10, 2021.
- 41.Lou Y., Liu L., Yao H., Hu X., Su J., Xu K., Luo R., Yang X., He L., Lu X., Zhao Q., Liang T., Qiu Y. Clinical Outcomes and Plasma Concentrations of Baloxavir Marboxil and Favipiravir in COVID-19 Patients: An Exploratory Randomized, Controlled Trial. Eur. J. Pharm. Sci. 2021;157:105631. doi: 10.1016/j.ejps.2020.105631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]