Abstract
A specific aptameric sequence has been immobilized on short polyethyleneglycol (PEG) interface on gold nano-film deposited on a D-shaped plastic optical fiber (POFs) probe, and the protein binding has been monitored exploiting the very sensitive surface plasmon resonance (SPR) phenomenon. The receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein has been specifically used to develop an aptasensor. Surface analysis techniques coupled to fluorescence microscopy and plasmonic analysis have been utilized to characterize the biointerface. Spanning a wide protein range (25 ÷ 1000 nM), the SARS-Cov-2 spike protein was detected with a Limit of Detection (LoD) of about 37 nM. Different interferents (BSA, AH1N1 hemagglutinin protein and MERS spike protein) have been tested confirming the specificity of our aptasensor. Finally, a preliminary test in diluted human serum encouraged its application in a point-of-care device, since POF-based aptasensor represent a potentially low-cost compact biosensor, characterized by a rapid response, a small size and could be an ideal laboratory portable diagnostic tool.
Keywords: Self assembled monolayer (SAM), Aptamer, Sars-CoV-2, Plastic optical fiber (POF), Polyethyleneglycol (PEG), Surface plasmon resonance (SPR)
Graphical abstract
1. Introduction
At the end of 2019 in China a new coronavirus appeared, causing one of the main pandemic event of the last years [1]. This virus spread into humans infecting first the respiratory system and then other ones (hepatic, central nervous and gastrointestinal systems) [1] and due to the large genome similarity (almost 96%) with Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), it is named SARS-CoV-2 [2].
SARS-CoV-2 virus is a positive sense single-stranded RNA genome that encodes for 27 proteins and four structural proteins with specific roles: the spike surface glycoprotein (S) mediates receptor binding and membrane fusion, the matrix protein (M) and small envelope protein (E) are essential for virus assembly while the nucleocapsid protein (N) is responsible for transcription and replication of viral RNA, its packaging into virions and interaction with cell cycle [[1], [2], [3]]. Actually some compounds have been approved for curing this infection, many vaccines are under evaluation [1,4,5] and few of them are in these days under administration.
Other than using vaccination, in order to decrease the spreading of the virus infection we can act in two directions as suggested by Talebian and co-workers [6]: the first route is to develop efficient viral disinfectants and the second one is to develop highly sensitive, rapid and accurate detection systems. Reverse transcription polymerase chain reaction (RT-PCR) is at the present the recommended method for the identification of SARS-CoV-2 cases (approved by the World Health Organization), however a high rate of false-negative Results compromises an accurate test (mainly due to low genetic material) [7]: it is moreover complex and costly, requires specialized diagnostic laboratories and hours or days to provide a response [3]. Alternative approaches are under development: a nucleic acid isothermal amplification is under evaluation and viral protein antigen or antibodies created in response to SARS-CoV-2 infection can be used for diagnosing [8]. The cytochine levels was also found an important parameter not only to monitor disease progress but also to verify the effects of anti-inflammatory therapies [9]. Several point-of-care systems (POCs) based on antibodies detection are on the market [2,3,10,11], but the antibody levels necessary to become detectable in blood may require days after viral infection and their concentration is patient dependent [8,10]. It is therefore crucial to develop detection systems able to reveal the virus presence as early as possible. In this context, Qiu and co-workers report on a biosensor that combines a plasmonic photothermal effect (used to hybridize a specific DNA probe with the RNA viral sequence) and a localized surface plasmon resonance reaching a limit of detection (LoD) of 0.22 pM [12], while Moitra et al. hybridize a DNA antisense with N-gene of SARS-CoV-2 causing gold nanoparticles agglomeration (LoD of 0.18 ng/μL) [13]. Alternatively, graphene sheets have been decorated with specific antibody against SARS-CoV-2 spike protein and using a field-effect transistor (FET)-based device a LoD of 1 fg/ml has been achieved [14]. Despite the impressive LoD reached, FET-based POC are not so diffuse, mainly because important challenges has to performed before approaching the biosensor market [15].
An alternative biorecognition system is based on aptamers, biological molecules used in pharmacology since 2004 [16]. Thanks to their small size, functionality in non-physiological buffers, resistance to harsh conditions (in terms of temperature and pH), on-demand chemical modifications and high reproducibility, aptamers are ideal bioreceptor for POC devices. They have been already used in the past for virus detection [[17], [18], [19], [20], [21], [22], [23], [24], [25], [26]] and their use in the development of POC for the detection of SARS-CoV-2 has been already suggested [7,27,28]: Chen and coworkers [7] tested the ability of the DNA aptamer previously developed against SARS-CoV N protein [17] to recognize the N protein of SARS-CoV-2 thanks to the high homology between the two protein sequences. They found three sequences able to bind the protein with high affinity, while recently Zhang et al. [27] discovered four new aptameric sequences able to recognize N protein and used it in a sandwich configuration coupled with antibody to develop a lateral flow system, reaching a LoD of 1 ng/ml in buffer. An amplification system, using two previously reported aptamers against N protein, reached a LoD of 37.5 pg/ml in buffer (30.9 pg/ml in 100% human serum), coupling ligation DNA region and allowing an amplification via qPCR [28]. Other amplification systems can be applied, mainly based on antibodies [29] or antibody coupled with nanoparticles [30], or on a nanostructuration of the sensing platform [31], as reported on optical fiber based biosensors, to enhance the sensor performances.
Among biosensors, optical-based ones are in fact largely used [[32], [33], [34], [35]] and plastic optical fibers (POFs) present several advantages respect to glass fiber [36].
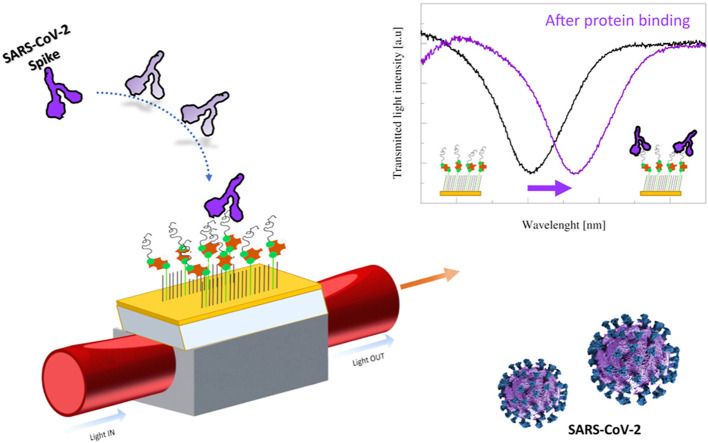
In this work, we immobilized an aptamer specific for the recognition of the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein, published by Song with an affinity constant (Kd) of 5.8 nM [37], on a SPR D-shaped POF probe, previously used in other bio-chemical applications [36]. The aptameric sequence was here immobilized on a short PEG interface [38] and a chemical-physical and functional characterization was performed by X-ray Photoelectron Spectroscopy (XPS), static contact angle (CA), atomic force microscopy (AFM), fluorescence microscopy, and plasmonic study. SARS-CoV-2 spike protein binding was optically measured without any amplification systems. Moreover, the specificity of the biosensor system was tested using both non-sense aptamer sequence and non-specific proteins (BSA, AH1N1 hemagglutinin protein and MERS spike protein), confirming the high specificity of our sensor. Our data pave the way for the development of an optical POC device useful in the early detection of SARS-CoV-2 virus.
2. Materials and methods
Materials. SARS-CoV-2 (2019-nCoV) Spike S1 + S2 ECD-His Recombinant Protein, MERS-CoV spike antigen protein and Influenza AH1N1 (A/New Caledonia/20/1999) Hemagglutinin/HA-specific B cell probe (His Tag) were purchased from Sino Biological Inc. (China). DNA-aptamer sequences purchased from IDT Integrated DNA technologies (Leuven, Belgium) with HPLC purification were reported in Supporting Information file (Table S1). Purified Thrombin (THR), streptavidin from Streptomyces avidinii (SA), bovine serum albumin (BSA), Human Serum (from male AB clotted whole blood) and all powders for buffers were purchased from Sigma-Aldrich s.r.l. (Milan, Italy). The monoclonal anti-thrombin antibody produced in mouse was from Abcam (Cambridge, UK), the anti-mouse HRP-conjugated antibody was from Life Technologies and Cy3-Streptavidin (Cy3-SA) was from Zymed Laboratories (San Francisco, CA, USA). SuperSignal West Femto Chemiluminescent Substrate kit was purchased from Thermo Scientific (Rockford, IL USA) and 100 nm of gold evaporated on silicon wafer from Agar Scientific (United Kingdom) was utilized. M-dPEG®8-Thiol and Biotin-dPEG®3-Lipoamide for the self-assembled monolayer (SAM) were purchased from Stratech (United Kingdom).
2.1. SPR D-shaped POF platform and measurement setup
SPR-POF sensor architecture has been already reported [39]. A detailed description and a picture of the experimental setup were reported in the Supporting Information file (Fig. S1). The measurement protocol in buffer and diluted human serum was the following: 50 μL of binding buffer (PBS, pH = 7.4, including 136.8 mM NaCl, 10.1 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4, 0.55 mM MgCl2) or real solution (diluted human serum) were dropped over the sensing region; after an incubation of 10 min, the spectrum was acquired, then a washing step in the buffer was applied and the spectrum was acquired again dropping fresh buffer. Different protein solutions were deposited over the sensor (spanning a range between 12.5 and 1000 nM of protein) and the measurement protocol was repeated. The transmission spectra were recorded and normalized to air. This processing of the data was carried out by Matlab software, whereas the normalized transmission spectra were analyzed fitting the curve with a Voigt function to find the minimum value using OriginPro8.5, Origin Lab. Corp. (Northampton, MA, USA). The whole procedure took about 20 min for the entire measurement.
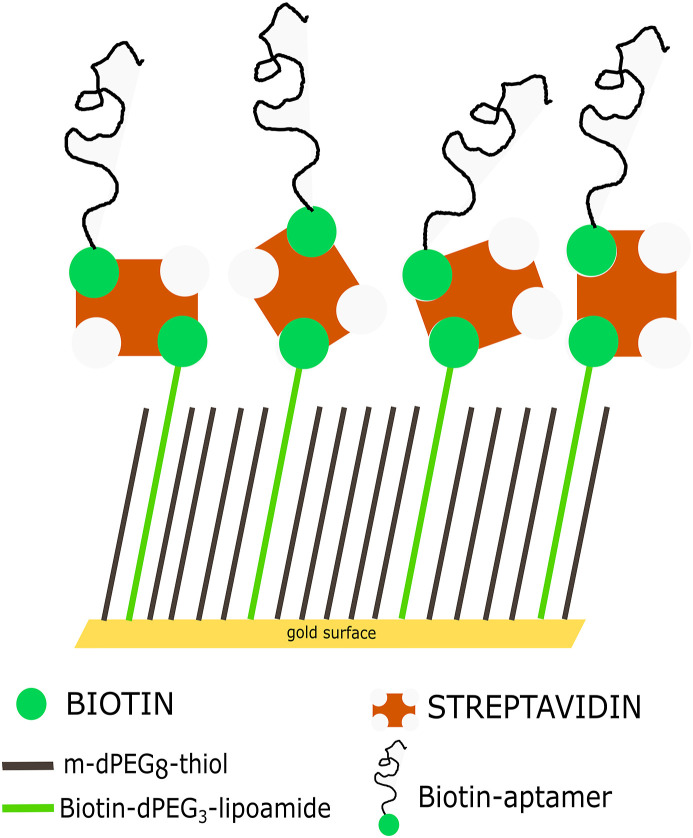
Gold derivatization. Gold derivatization (both flat samples and gold-coated POF) was achieved in three steps: SAM formation, SA immobilization and biotin-aptamer binding. SAM formation was obtained first cleaning the surface with an Argon plasma at 6.8 W of RF power for 2 min, then immersing it in a 0.2 mM of a PEGthiol and BiotinPEGlipo mixture at 8:2 molar ratio (in MilliQ water) at room temperature for an overnight incubation, followed by a washing step in ultrapure water and drying with nitrogen. For streptavidin coating, a 5 μg/ml solution (SA or Cy3-SA) in phosphate buffer (10 mM phosphate, 138 mM NaCl, 2.7 mM KCl, pH 7.4) for 1 h was applied. Finally, after washing in buffer, 10 μM of biotin-aptamer solution (previously heated at 95 °C for 1 min and quickly cooled on ice to unfold the sequence) for 3 h in the same buffer was applied, followed by a washing in buffer. The interface picture is reported in Scheme 1 .
Scheme 1.
Scheme of the surface derivatization based on aptamer. A mixed layer obtained from a 0.2 mM mixture of PEGthiol and BiotinPEGlipo in 8:2 molar ratio was prepared on gold and a streptavidin coating was next performed. Finally, a biotin-modified aptamer was immobilized on the streptavidin layer.
SAM and aptamer layer stability. SAM stability was assessed by aging the surfaces in different conditions and verifying their ability to bind Cy3-SA through fluorescence microscopy measurements. Freshly prepared SAMs were aged up to 33 days (at 22 °C or 4 °C) in three different conditions: i) in the incubation solution (SAM solution), or ii) in MilliQ water after a washing step (WAT) or iii) dried at room temperature after a washing step in MilliQ water (DRY). 5 μg/ml of Cy3-SA solution in phosphate buffer for 1 h was applied and after washing, the surfaces were analyzed with a fluorescence microscope. The effect of aging on the aptamer layer was instead tested using an aptamer specific for thrombin recognition immobilized with the same protocol evaluating its ability to bind thrombin by means of a chemiluminescence protocol [38]. The aptamer-treated surfaces were aged in PBS buffer (PBS) or PBS + 20% DMSO (PBS + DMSO) or dried (DRY) at 4 °C for several days. At selected time, 50 nM of thrombin was incubated in buffer (Tris 50 mM, EDTA 1 mM, MgCl21 mM, KCl 150 mM pH 7.4) for 1 h on a slow orbital shaker and then washed in the same buffer and passivated in 3% w/v BSA for 30 min. An anti-thrombin mouse antibody solution (2.5 μg/ml) in 3% w/v BSA for 30 min was applied and after three extensive washing steps, an anti-mouse HRP-conjugated secondary antibody (2 μg/ml) in 3% w/v BSA for 30 min was incubated. After three washing in buffer, the chemiluminescence signal was developed, acquired using a ChemDoc-It (Bio-Rad) as imaging system for 0.5 s and quantified using the ImageJ software [40].
Chemical-physical analysis. The gold derivatization was analyzed via XPS, CA, AFM and fluorescence measurements.
A Kratos Axis UltraDLD instrument (Kratos Analytical Ltd, England) equipped with a hemispherical analyzer and a monochromatic AlKα (1486.6eV) X-ray source, in spectroscopy mode was used to perform XPS measurements. Photoelectrons were detected at two angles, 0° and 60° (sampling depth of about 10 and 3–4 nm respectively [41]). The main core lines acquired were O 1s, C 1s, N 1s, S 2p and Au 4f. The relative elemental percentage, that gives a semiquantitative analysis, was obtained by integrating the area under the core lines, by applying the Shirley background subtraction, and by correcting for the atomic sensitivity factors through a dedicated software [42]. Using the attenuation of the Au 4f7/2 signal between the different samples it was possible to calculate the overlayer thickness (dOL) through the following equation:
where I0 Au is the Au 4f7/2 peak intensity on the clean gold sample, IAu is the Au 4f7/2 peak intensity of the gold sample after treatment, and λOL is the attenuation length of Au 4f7/2 photoelectrons in the polymer overlayer (3.45 nm) [43].
The static contact angle was measured depositing 2 μl of deionized water droplets on the substrate (at least two droplets for sample), acquiring images with CMOS camera and analyzing it by Drop-Analysis, a plug-in of ImageJ software [40]. Results were reported as average value and respective standard deviations.
AFM measurements were performed using a Cypher AFM equipped with an environmental scanner (Oxford Instruments - Asylum Research, Germany). Samples were let dry in air and then inserted in the instrument. AC240TS (Olympus Corporation, Japan) cantilevers (nominal spring constant 2 N/m, nominal radius 7 nm, nominal resonance frequency 70 kHz) were used to acquire data in AC mode at least three different regions for every sample at a scanning speed of ~2 Hz. Images were flattened and rendered with ImageJ, using custom written routines to import AFM data.
The Cy3 fluorescence signal was monitored using a Leica DMLA fluorescence microscope equipped with a mercury lamp and a fluorescence filter N2.1 (Leica Microsystems, Germany). A 20× magnification objective was used to image all samples and fluorescence was measured with a cooled CCD camera (DFC420C, Leica Microsystems, Germany), analyzing the signal with the ImageJ software [40]. Calibration curves have been obtained as reported in the Supporting Information file (Fig. S2).
3. Results and discussion
Before proceeding with the optical measurements, the interface built as described in Scheme 1 has been characterized through chemiluminescence and fluorescence measurements, XPS, CA and AFM analysis.
3.1. Characterization of the aptamer layer
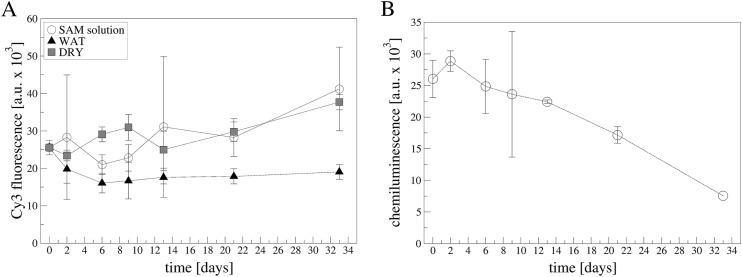
Besides the characterization of the aptamer layer on POF platform previously reported [38], here we focused on the evaluation of the stability of the platform over time, testing different conservation conditions both in terms of solutions and temperatures. First, the stability of the SAM layer was analyzed, testing different conditions, as reported in experimental section. A functional test was utilized to evaluate the goodness of the layer. A fluorescent streptavidin (Cy3-SA) was incubated on the different SAMs conserved for more than 30 days and the fluorescent signal was acquired at fixed intervals. As reported in Fig. 1 A and Fig. S3, the more stable condition seems to be the water conservation independently from the temperature. In the other conditions sometimes, an increase is recorded (especially at 4 °C, Fig. 1A) mainly due to the formation of aggregates on the surface (Fig. S4). Secondly, we tested the aging of the aptamer layer: for this purpose, an anti-THR aptamer was used applying an immune-chemiluminescence method previously used [38]. Fig. 1B showed that, up to about 12 days, the aptamer platform was able to recognize thrombin in an unaltered way (inside error bars) in PBS solution, but also in the other conditions namely PBS+20%DMSO and dried, as reported in Fig. S5. After 12 days a decrease in the ability to recognize thrombin has been observed in all conditions, suggesting a degradation in the aptamer functionality. However, other conditions are under exploitation to increase the time stability.
Fig. 1.
A) SAM aging in different conditions at 4 °C. Fluorescence microscopy data of SAMs incubated with Cy3-SA. SAMs aged in: SAM solution (open circles), MilliQ water (black triangles up) or dried (grey squares). Data are reported as mean value of 10 images acquired on two samples. Error bars represent the standard deviation. B) Apt-THR layer aging in PBS solution at 4 °C. Data are reported as mean value of two samples. Error bars represent the standard deviation. Exposition time:0.5 s.
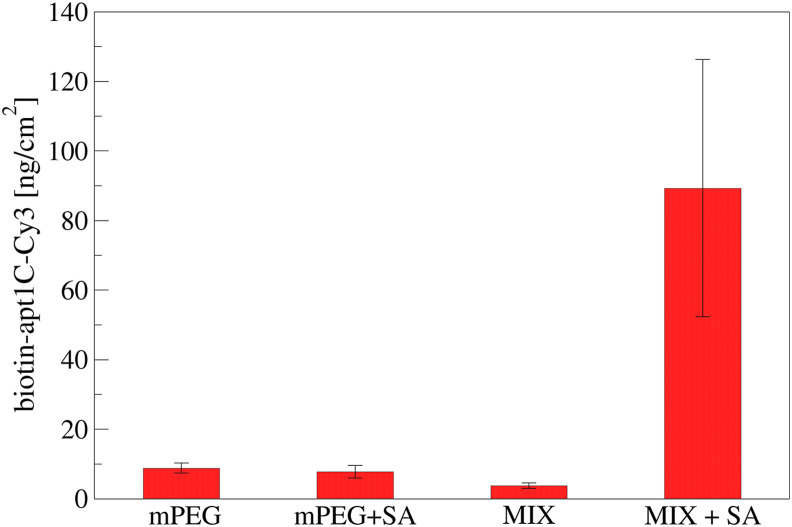
Using a fluorescent-labelled aptamer (apt-1C-Cy3) immobilized on SAM we obtained a quantification of the aptamer density. The incubation was performed on different surfaces: i) the specific surface (mPEG:biotinPEGlipoamide + SA), ii) a control surfaces with or without streptavidin (mPEG or mPEG + SA) and iii) the specific surface without streptavidin (mPEG:biotinPEGlipoamide). In this way it was possible to determine that no aspecific adsorption of the aptamer occurred on SAM made only of mPEG neither on mixed SAM without streptavidin (Fig. 2 ). Through an opportune calibration curve (reported in Fig. S2) the aptamer density resulted in 89.3 ± 36.9 ng/cm2, in good agreement with previous data [38]. Moreover, an additional experiment was performed varying the aptamer concentration from 1 up to 100 μM, confirming that 10 μM was enough to cover the surface (see Fig. S6).
Fig. 2.
A fluorescent aptamer (apt-1C-Cy3) was immobilized on different surfaces: i) PEGthiol (mPEG), ii) PEGthiol + streptavidin (mPEG + SA), iii) PEGthiol:BiotinPEGlipo mixture in a 8:2 molar ratio (MIX) or iv) PEGlipo:BiotinPEGlipo mixture in a 8:2 molar ratio + streptavidin (MIX + SA). Three independent experiments have been performed. Five images have been acquired for each sample, reporting data as mean value and standard deviation.
3.2. Chemical-physical characterization
All the derivatization steps have been analyzed via surface analytical techniques. XPS determine the chemical-physical modification of the surface giving details on which chemical elements and in which chemical bonds are involved at the interface. CA analysis merges morphological and chemical modification on a large scale on the surface, while AFM gives morphological information at the nanometric scale. All these techniques together allowed to verify the changes at the gold interface due to the subsequent modification steps.
Table 1 reports the XPS quantification at 0° take-off angle with the relative thickness quantification computed by the integrated area. A clear surface modification is observed after SAM formation, with the decrease of the substrate signal (gold) and an increase in the carbon content in addition to the appearance of nitrogen and sulphur signals, typical markers of hetero-functional PEG chains. Also the increase in the contact angle measurement confirmed the surface modification, in good agreement with previous data [38]. The subsequent steps, i.e. streptavidin deposition and biotinylated aptamer immobilization did not change significantly the chemical composition and the hydrophilicity of the interface, while the spike protein binding, changed the chemistry in a significant way. XPS data revealed an increase in the typical markers of proteins (i.e. C, N, O) and a higher surface coating. A larger thickness is here computed and a higher contact angle is recorded, probably due to high heterogeneity of the surface. XPS data recorded at 60° take-off angle confirmed this behavior (Table S2 and Table S3) and a more detailed analysis was performed analyzing the core lines levels at that angle (Fig. S7). The presence of SAM on gold was confirmed, besides from an increase in C–C/C–H bonds at 285 eV, by the appearance of C–O/C–N bond at 286.5 eV and N–C O (amide) bonds at 288.3 eV present in the biotin-P3-lipo reagent, which are also the same group (peptide bond) indicative of proteins (SA and spike), in good agreement with literature [[43], [44], [45]]. Looking at the oxygen core line, a decrease in the component related to PEG chain is recorded (O2 at about 533 eV in Table S3), suggesting a coverage of the SAM layer. The other elements have been detected: nitrogen at 400 eV, sulphur doublet at 162 eV and gold at about 84 eV.
Table 1.
Chemical characterization determined by XPS, CA and AFM on different samples: argon plasma treated gold (Au), after SAM formation (SAM), after streptavidin immobilization (SAM + SA), after aptamer binding (SAM + SA + APT) and after 100 nM SARS-CoV-2 spike protein incubation (SAM + SA + APT + SPIKE). XPS data have been acquired at 0° take-off angle and standard error does not exceed the 1–2% of the reported value. AFM data have been acquired on areas from 200 × 200 nm2 to 800 × 800 nm2.
| Samples | O 1s (%) | C 1s (%) | N 1s (%) | S 2p (%) | Au 4f (%) | Thickness from XPS data [nm] | Contact angle [°] | Average roughness [nm] |
|---|---|---|---|---|---|---|---|---|
| Au | 16.4 | 14.6 | – | – | 68.9 | – | <5 | 1.38 ± 0.20 |
| SAM | 16.0 | 41.9 | 3.2 | 2.1 | 36.7 | 1.13 | 32.7 ± 1.1 | 1.86 ± 0.04 |
| SAM + SA | 15.4 | 42.0 | 3.7 | 2.3 | 36.7 | 1.31 | 34.1 ± 0.3 | 2.32 ± 0.07 |
| SAM + SA + APT | 16.0 | 41.8 | 3.3 | 2.3 | 36.7 | 1.42 | 32.7 ± 0.2 | 2.21 ± 0.16 |
| SAM + SA + APT + SPIKE | 17.2 | 51.2 | 7.7 | 1.7 | 22.2 | 2.79 | 74.5 ± 1.6 | 2.21 ± 0.17 |
The same samples have been analyzed by AFM (Fig. S8), finding a quite uniform morphology at the microscale. The roughness, reported in Table 1, has been found to increase up to the adhesion of SA, while no further significative variations were found following the further steps of functionalization.
Altogether these data clearly indicate that gold surface has been modified and spike protein was immobilized on it.
3.3. Optical measurements
In SPR probe the plasmonic wave penetration occurs in the thin bioreceptor dielectric film and in the bulk solution over it. Consequently, the resonance variation due to a functionalized step must always be measured in a bulk solution with the same refractive index. So, the immobilization of the aptamer sequence on the SPR-POF sensor surface can be confirmed by SPR measurements, using the proposed optical platform directly, exploiting the SPR spectra measured with the same bulk solution present on the bare surface and on the functionalized surface. The changes in SPR spectra before and after the gold derivatization were reported in Fig. S9. A red shift in the SPR normalized transmission spectrum was indeed detected when aptamers were immobilized on the gold sensor surface; in fact, when the bioreceptor is present on the gold and the bulk solution does not change, the refractive index increases with respect to the bare surface condition. The shift in the position of the transmission hole (Δλ) due to the derivatization process is 3.1 ± 1.2 nm (measured on five platforms).
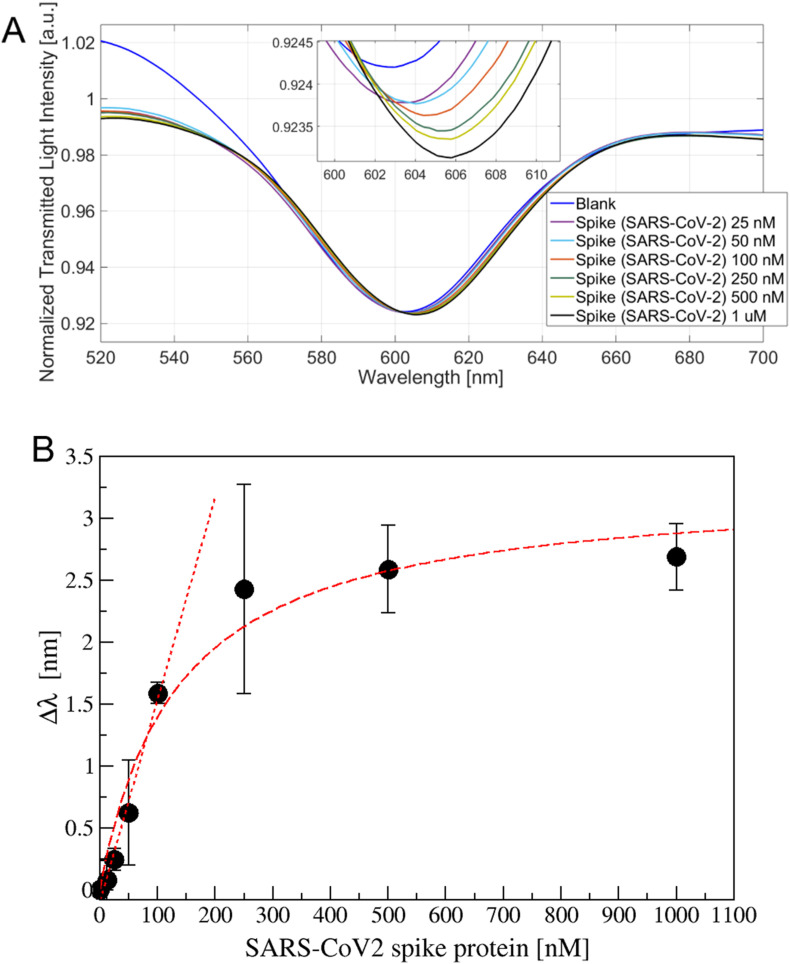
The aptamer-modified SPR-POF probe has been tested in different configurations: the apt-1C-aptamer specifically designed versus SARS-CoV-2 spike protein has been tested against its specific target and aspecific ones (BSA, AH1N1 hemagglutinin protein, MERS spike protein), while the apt-NS, an aspecific aptamer, has been tested against SARS-CoV-2 spike protein. To determine the optimal incubation time before acquiring the spectrum, different kinetics have been measured (Fig. S10). Several SARS-CoV-2 spike protein concentrations have been tested (12.5, 100, and 250 nM reported in Fig. S10) suggesting that 10 min was the sufficient time to wait before washing and acquiring the spectrum. Spectra acquired on apt-1C-functionalized platform after different SARS-CoV-2 spike protein incubation were reported in Fig. 3 A, observing a clear red shift. When the binding with the analyte occurs, the refractive index in contact with the gold surface increases shifting on the right the resonance wavelength. The experimental data (the resonance wavelengths) reported in Fig. 3B, were fitted by Langmuir equation as described in Supporting Information file (Table S4).
Fig. 3.
A) Transmission spectra for different SARS-CoV-2 spike protein concentration (25 ÷ 1000 nM) with a zoom of the resonance wavelengths region (inset). B) Resonance shift (Δλ) versus the spike protein concentration (nM), measured on at least three sensor for each value. Data are reported as mean value with standard deviation. Langmuir and linear fitting are also reported.
The Limit of Detection (LoD), has been calculated, considering the linear behavior at low protein concentration, as the ratio of the mean value of the blank plus three times its standard deviation and the slope in the linear range (linear slope, equal to 0.018 nm/nM), according to Refs. [46,47], resulting in 36.7 nM.
Our limit of detection is not so far from that published by Stanborough and coworkers, who using our same aptamer sequence on a standard SPR instrument, observed that 5 nM spike protein can be clearly distinguished from the baseline [48]; however, they used a conventional SPR instrument where instrument setup and microfluidic are well optimized and also a different chemistry based on thiols.
Recently, another publication reports the detection of SARS-CoV-2 spike protein using a specific molecular imprinted polymer (MIP) receptor combined with the same SPR-POF probe [49]. With respect to the MIP-SPR-POF system, our aptamer-based platform has a LoD seven times better (as reported in the supplementary file).
The choice of the biorecognition system is driven by different aspects: the kind of target analyte, the constrains of the detection platform, the immobilization chemistry, the stability on time of the biorecognition layer and also the overall costs of the system. Both systems present pro and cons on all these aspects and a careful evaluation has to be performed. Even if aptamers can result more “soft” when compared to MIP, an aptamer based surface is easy adaptable to a different application just changing the sequence. Moreover, its small size allows the employment on the optical platform, where the thickness of the bioreceptor layer is a crucial aspect.
We can however improve the performances of our device playing on two aspects: the biorecognition system or the optical platform. Concerning the improvement driven by the biorecognition system, a sandwich configuration can be applied. Tran and coworkers for instance, used an amplification system based on the binding of a polyclonal antibody to the complex aptamer-antigen followed by the binding of protein A coated gold nanoparticles for the detection of a peanut allergen [30]. In this way they gained almost one order of magnitude in buffer and were able to detect the allergen in food matrix samples. In a similar way, Loyez et al., improved of two orders of magnitude the glass-based optical fiber system performance using an antibody system [29]. From an optical point of view, our plasmonic sensor system can be improved if tapered POFs will be used instead of regular POFs [50]. Alternatively nanoparticles/nanostructures can be exploited, as also reported by Allsop and coworkers, who build gold nanoantennas on the fiber sensor to enhance the localized surface plasmon resonance effect and were able to detect bisphenol A in attomolar concentration [31].
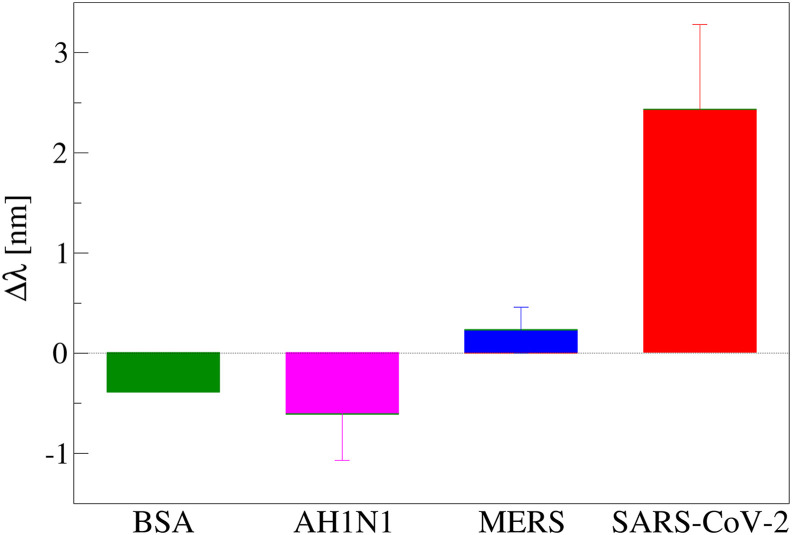
To verify the specificity of the aptasensor, two kind of experiments have been performed. The first one was done using aspecific proteins (BSA, AH1N1 hemagglutinin protein and MERS spike protein) on the apt-1C-modified POF sensor specific for SARS-CoV-2 spike protein, while the other one was performed using an aspecific aptamer layer (apt-NS). In Fig. 4 is reported the comparison incubating 250 nM of the different proteins on the apt-1C-modified POF platform and a positive shift was observed only when the specific SARS-CoV-2 spike protein was incubated. To confirm this behavior, a dose-response curve was performed with BSA and MERS spike protein (since albumin is the most abundant protein in human serum and MERS spike protein is a representative competitor for the virus), adding finally SARS-Cov-2 spike protein to be sure that aptasensor was responsive.
Fig. 4.
Resonance wavelength variation (Δλ), with respect to the blank, obtained incubating 250 nM of different proteins (BSA, AH1H1 hemagglutinin protein, MERS spike protein and SARS spike protein) on apt-1C- modified platform (at least two platform for protein); error bars represent standard deviation.
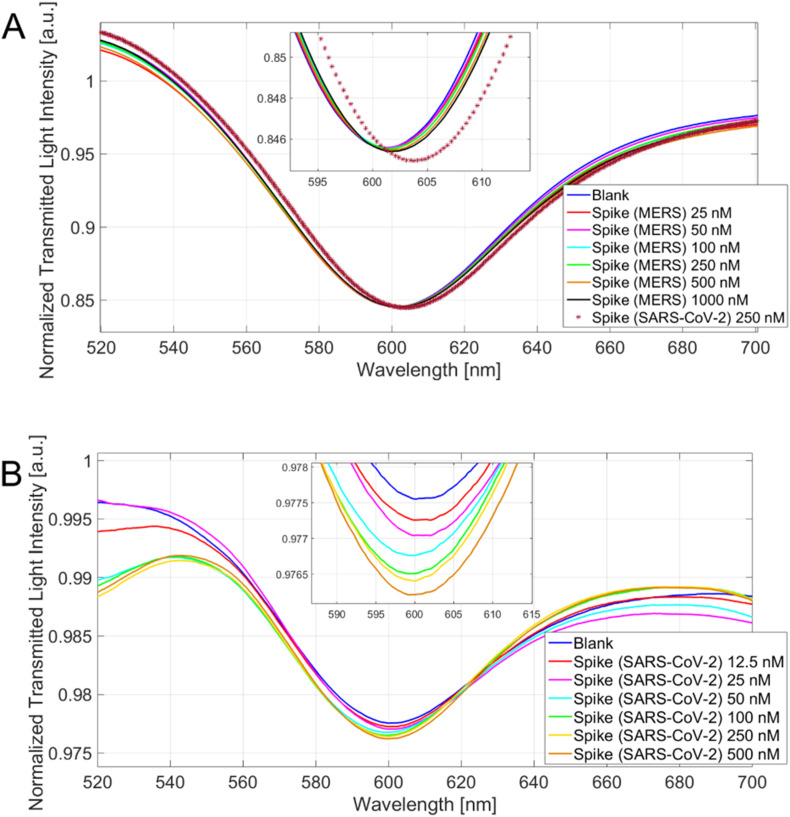
Fig. 5A reports the MERS spike protein addition in the same range (25 ÷ 1000 nM) without significant red shifts, until the final addition of 250 nM SARS-CoV-2 that causes a significant shift. A similar behavior for BSA was observed and reported in Fig. S11.
Fig. 5.
A) Spectra for different MERS spike protein concentrations (25 ÷ 1000 nM) and a SARS-CoV-2 spike protein concentration (250 nM) with a zoom of the resonance wavelengths region (inset). B) Spectra for different SARS-CoV-2 spike protein concentration (12.5 ÷ 500 nM) on the apt–NS–modified platform and a zoom of the resonance wavelengths region (inset).
Moreover, the specificity was proved testing the SARS-CoV-2 spike protein bonding on apt–NS–modified POF probe, an aspecific aptamer sensor. As reported in Fig. 5B, no red shift has been observed, confirming the absence of interaction with un-specific sequence. The dose-response curves on all tested interferents were reported in Fig. S12 and compared with SARS-CoV-2 curve, highlighting the specific response of our platform.
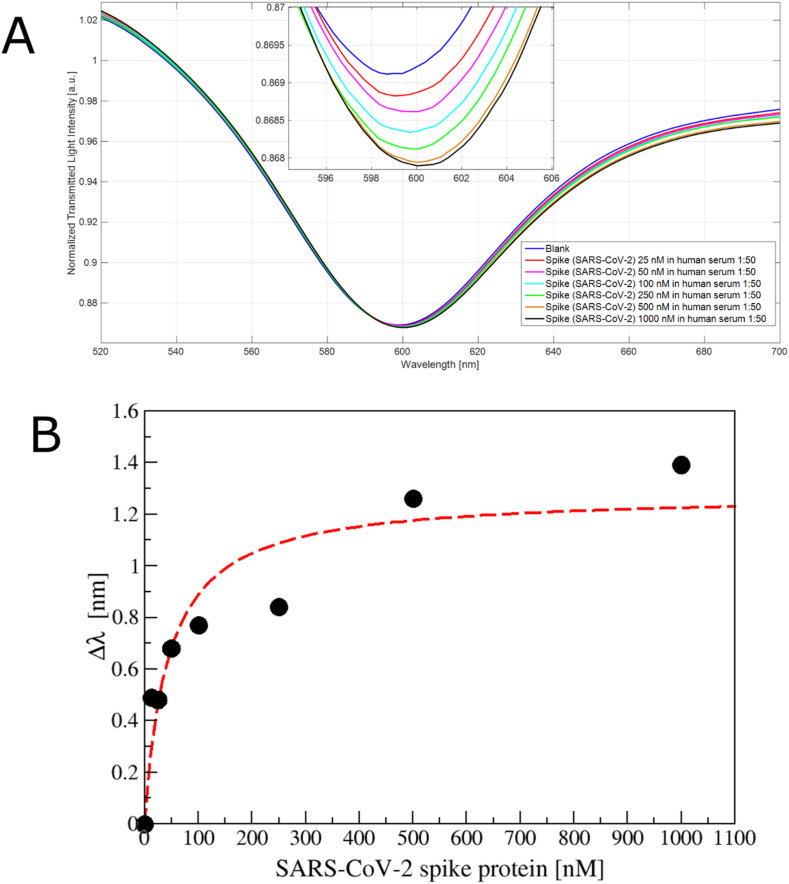
Finally, a preliminary test was performed in diluted human serum (1:50 v/v), observing a red shift (Fig. 6 A). These resonance variations are reported in Fig. 6B, suggesting that the spike protein could also be detected in 1:50 v/v diluted human serum. Analytical parameters of Langmuir fitting are reported in Table S5 and the computed LoD resulted 75.26 nM.
Fig. 6.
A) SPR transmission spectra, normalized to the spectrum in air, incubating different concentration of SARS-CoV-2 spike protein (25 ÷ 1000 nM) in human serum diluted 1:50 in phosphate buffer on the apt1C- aptamer functionalized POF. Inset: zoom of the resonance wavelengths region. B) Plot of the resonance shift (Δλ), with respect to the blank, versus the spike protein concentration (nM) diluted in human serum (1:50 v/v), reporting also the Langmuir fit.
The effect of complex biological matrix reduced by a factor of 2 the LoD of our system, maintaining a similar affinity constant (k value), encouraging its application in biomedical field. Moreover, even if our LoD is higher than that obtained using lateral flow system [27] or ELISA assay [14], considering that the virion has a molecular weight three order of magnitude higher with respect to the spike protein [51], we expect to be able to measure the virions binding.
4. Conclusions
The development of highly sensitive, specific, and fast sensor system for an early detection of biomarkers and virus is crucial. In this context, we developed a highly sensitive and specific optical aptasensor for the detection of receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein. The biorecognition element (an aptamer-based interface) is immobilized on a short PEG SAM that has been characterized via analytical techniques (XPS, CA and AFM), fluorescence and chemiluminescence methods, and via plasmonic analysis to assess their stability and their goodness. The specific biosensor has then been built on an SPR in POFs device and optical measurements confirmed the specific protein binding in a nanomolar range. The obtained ability to detect the SARS-CoV-2 Spike protein in the nanomolar range is similar to that recently achieved exploiting a similar bio-receptor on an SPR conventional probe [48] and a similar SPR platform in POFs combined with a specific MIP receptor layer [49].
Due to the easy-to-implement fabrication procedure of the POF-SPR device, our work will lead to a platform that will be potentially industrialized in a straightforward way.
Author contributions
Nunzio Cennamo, Conceptualization, Data curation, Formal analysis: binding measurements, Funding acquisition, Methodology, Writing – review & editing. Laura Pasquardini, Conceptualization, Data curation, Formal analysis: binding measurements, surface functionalization, Methodology, Writing – original draft, Writing – review & editing. Francesco Arcadio, Data curation, Formal analysis: binding measurements, Lorenzo Lunelli, AFM Formal analysis, Writing – review & editing. Lia Vanzetti, XPS measurements, Writing – review & editing. Vincenzo Carafa, Conceptualization, Data curation, Formal analysis: binding measurements, Funding acquisition, Methodology, Writing – review & editing. Lucia Altucci, Conceptualization, Data curation, Supervision, Writing – review & editing. Luigi Zeni, Conceptualization, Data curation, Funding acquisition, Supervision, Writing – review & editing. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the VALERE program of the University of Campania “L. Vanvitelli” (Italy), CAMPANIA project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122532.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Gupta A., Kumar S., Kumar R., Choudhary A.K., Kumari K., Singh P., Kumar V. COVID-19: emergence of infectious diseases, nanotechnology aspects, challenges, and future perspectives. ChemistrySelect. 2020;5:7521–7533. doi: 10.1002/slct.202001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 3.Kubina R., Dziedzic A. Molecular and serological tests for COVID-19. A comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10 doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik Y.S., Kumar N., Sircar S., Kaushik R., Bhat S., Dhama K., Gupta P., Goyal K., Singh M.P., Ghoshal U., El Zowalaty M.E., Vinodhkumar O.R., Yatoo M.I., Tiwari R., Pathak M., Patel S.K., Sah R., Rodriguez-Morales A.J., Ganesh B., Kumar P., Singh R.K. Coronavirus disease pandemic (Covid-19): challenges and a global perspective. Pathogens. 2020;9:1–31. doi: 10.3390/pathogens9070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piyush R., Rajarshi K., Chatterjee A., Khan R., Ray S. Nucleic acid-based therapy for coronavirus disease 2019. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talebian S., Wallace G.G., Schroeder A., Stellacci F., Conde J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020;15:618–621. doi: 10.1038/s41565-020-0751-0. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z., Wu Q., Chen J., Ni X., Dai J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol. Sin. 2020;35:351–354. doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Younes N., Al-Sadeq D.W., AL-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12:1–27. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell S.M., Alba-Patiño A., Barón E., Borges M., Gonzalez-Freire M., De La Rica R. Biosensors for managing the COVID-19 cytokine storm: challenges ahead. ACS Sens. 2020;5:1506–1513. doi: 10.1021/acssensors.0c00979. [DOI] [PubMed] [Google Scholar]
- 10.Ghaffari A., Meurant R., Ardakani A. COVID-19 serological tests: how well do they actually perform? Diagnostics. 2020;10:1–14. doi: 10.3390/diagnostics10070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riccò M., Ferraro P., Gualerzi G., Ranzieri S., Henry B.M., Ben Said Y., Pyatigorskaya N.V., Nevolina E., Wu J., Bragazzi N.L., Signorelli C. Point-of-Care diagnostic tests for detecting SARS-CoV-2 antibodies: a systematic review and meta-analysis of real-world data. J. Clin. Med. 2020;9:1515. doi: 10.3390/jcm9051515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate Severe Acute respiratory Syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 13.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 15.Sadighbayan D., Hasanzadeh M., Ghafar-Zadeh E. Biosensing based on field-effect transistors (FET): recent progress and challenges. TrAC Trends Anal. Chem. 2020;133:116067. doi: 10.1016/j.trac.2020.116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berzal-Herranz A., Romero-López C. Two examples of RNA aptamers with antiviral activity. Are aptamers the wished antiviral drugs? Pharmaceuticals. 2020;13:1–10. doi: 10.3390/ph13080157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho S.J., Woo H.M., Kim K.S., Oh J.W., Jeong Y.J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wandtke T., Woźniak J., Kopiński P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7:751–780. doi: 10.3390/v7020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González V.M., Elena Martín M., Fernández G., García-Sacristán A. Use of aptamers as diagnostics tools and antiviral agents for human viruses. Pharmaceuticals. 2016;9:1–34. doi: 10.3390/ph9040078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Percze K., Szakács Z., Scholz É., András J., Szeitner Z., Den Kieboom C.H.V., Ferwerda G., Jonge M.I.D., Gyurcsányi R.E., Mészáros T. Aptamers for respiratory syncytial virus detection. Sci. Rep. 2017;7:1–11. doi: 10.1038/srep42794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou X., Wu J., Gu J., Shen L., Mao L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. https://www.frontiersin.org/article/10.3389/fmicb.2019.01462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H.Y., Jia W.N., Li X.Y., Zhang L., Liu C., Wu J. Advances in detection of infectious agents by aptamer-based technologies. Emerg. Microb. Infect. 2020:1–38. doi: 10.1080/22221751.2020.1792352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee I., Kim S.E., Lee J., Woo D.H., Lee S., Pyo H., Song C.S., Lee J. A self-calibrating electrochemical aptasensing platform: correcting external interference errors for the reliable and stable detection of avian influenza viruses. Biosens. Bioelectron. 2020;152:112010. doi: 10.1016/j.bios.2020.112010. [DOI] [PubMed] [Google Scholar]
- 24.Le T.T., Chang P., Benton D.J., McCauley J.W., Iqbal M., Cass A.E.G. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017;89:6781–6786. doi: 10.1021/acs.analchem.7b01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K.H., Zeng H. Aptamer-based ELISA assay for highly specific and sensitive detection of zika NS1 protein. Anal. Chem. 2017;89:12743–12748. doi: 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- 26.Kang J., Yeom G., Ha S.J., Kim M.G. Development of a DNA aptamer selection method based on the heterogeneous sandwich form and its application in a colorimetric assay for influenza A virus detection. New J. Chem. 2019;43:6883–6889. doi: 10.1039/c8nj06458j. [DOI] [Google Scholar]
- 27.Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., Li Q., Cheng H., Zhang W., Luo Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020;56:10235–10238. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
- 28.Liu R., He L., Hu Y., Luo Z., Zhang J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020;11:12157–12164. doi: 10.1039/d0sc03920a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loyez M., Lobry M., Hassan E.M., DeRosa M.C., Caucheteur C., Wattiez R. HER2 breast cancer biomarker detection using a sandwich optical fiber assay. Talanta. 2021;221 doi: 10.1016/j.talanta.2020.121452. [DOI] [PubMed] [Google Scholar]
- 30.Tran D.T., Knez K., Janssen K.P., Pollet J., Spasic D., Lammertyn J. Selection of aptamers against Ara h 1 protein for FO-SPR biosensing of peanut allergens in food matrices. Biosens. Bioelectron. 2013;43:245–251. doi: 10.1016/j.bios.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 31.Allsop T.D.P., Neal R., Wang C., Nagel D.A., Hine A.V., Culverhouse P., Ania Castañón J.D., Webb D.J., Scarano S., Minunni M. An ultra-sensitive aptasensor on optical fibre for the direct detection of bisphenol A. Biosens. Bioelectron. 2019;135:102–110. doi: 10.1016/j.bios.2019.02.043. [DOI] [PubMed] [Google Scholar]
- 32.Caucheteur C., Guo T., Albert J. Review of plasmonic fiber optic biochemical sensors: improving the limit of detection. Anal. Bioanal. Chem. 2015;407:3883–3897. doi: 10.1007/s00216-014-8411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S., Singh R. Recent optical sensing technologies for the detection of various biomolecules: Review. Opt Laser. Technol. 2021;134:106620. doi: 10.1016/j.optlastec.2020.106620. [DOI] [Google Scholar]
- 34.Li Y., Xin H., Zhang Y., Li B. Optical fiber technologies for nanomanipulation and biodetection: a review. J. Lightwave Technol. 2021;39:251–262. doi: 10.1109/JLT.2020.3023456. [DOI] [Google Scholar]
- 35.Soler M., Estévez M.C., Cardenosa-Rubio M., Astua A., Lechuga L.M. How nanophotonic label-free biosensors can contribute to rapid and massive diagnostics of respiratory virus infections: COVID-19 case. ACS Sens. 2020 doi: 10.1021/acssensors.0c01180. [DOI] [PubMed] [Google Scholar]
- 36.Cennamo N., Pesavento M., Zeni L. Sensors and Actuators : B . Chemical A review on simple and highly sensitive plastic optical fiber probes for bio-chemical sensing. Sensor. Actuator. B Chem. 2021;331:129393. doi: 10.1016/j.snb.2020.129393. [DOI] [Google Scholar]
- 37.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 38.Cennamo N., Pasquardini L., Arcadio F., Vanzetti L.E., Bossi A.M., Zeni L. D-shaped plastic optical fibre aptasensor for fast thrombin detection in nanomolar range. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cennamo N., Massarotti D., Conte L., Zeni L. Low cost sensors based on SPR in a plastic optical fiber for biosensor implementation. Sensors. 2011;11:11752–11760. doi: 10.3390/s111211752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z., Han W., Kozodaev D., Brokken-Zijp J.C.M., de With G., Thüne P.C. Surface properties of poly(dimethylsiloxane)-based inorganic/organic hybrid materials. Polymer. 2006;47:1150–1158. doi: 10.1016/j.polymer.2005.12.057. [DOI] [Google Scholar]
- 42.Speranza G., Canteri R. RxpsG a new open project for Photoelectron and Electron Spectroscopy data processing. SoftwareX. 2019;10:100282. doi: 10.1016/j.softx.2019.100282. [DOI] [Google Scholar]
- 43.Al-Ani A., Boden A., Al Kobaisi M., Pingle H., Wang P.-Y., Kingshott P. The influence of PEG-thiol derivatives on controlling cellular and bacterial interactions with gold surfaces. Appl. Surf. Sci. 2018;462:980–990. doi: 10.1016/j.apsusc.2018.08.136. [DOI] [Google Scholar]
- 44.Briand E., Humblot V., Landoulsi J., Petronis S., Pradier C.M., Kasemo B., Svedhem S. Chemical modifications of Au/SiO 2 template substrates for patterned biofunctional surfaces. Langmuir. 2011;27:678–685. doi: 10.1021/la101858y. [DOI] [PubMed] [Google Scholar]
- 45.Aydoğdu Tığ G., Pekyardımcı Ş. An electrochemical sandwich-type aptasensor for determination of lipocalin-2 based on graphene oxide/polymer composite and gold nanoparticles. Talanta. 2020;210:120666. doi: 10.1016/j.talanta.2019.120666. [DOI] [PubMed] [Google Scholar]
- 46.Chiavaioli F., Gouveia C.A.J., Jorge P.A.S., Baldini F. Towards a uniform metrological assessment of grating-based optical fiber sensors: from refractometers to biosensors. Biosensors. 2017;7 doi: 10.3390/bios7020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiavaioli F., Baldini F., Tombelli S., Trono C., Giannetti A. Biosensing with optical fiber gratings. Nanophotonics. 2017;6:663–679. doi: 10.1515/nanoph-2016-0178. [DOI] [Google Scholar]
- 48.Stanborough T., Given F.M., Koch B., Sheen C.R., Stowers-Hull A.B., Waterland M.R., Crittenden D.L. Optical detection of CoV-SARS-2 viral proteins to sub-picomolar concentrations. ACS Omega. 2021;6:6404–6413. doi: 10.1021/acsomega.1c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cennamo N., D’agostino G., Perri C., Arcadio F., Chiaretti G., Parisio E.M., Camarlinghi G., Vettori C., Di Marzo F., Cennamo R., Porto G., Zeni L. Proof of concept for a quick and highly sensitive on-site detection of sars-cov-2 by plasmonic optical fibers and molecularly imprinted polymers. Sensors. 2021;21:1–17. doi: 10.3390/s21051681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cennamo N., D'Agostino G., Pesavento M., Zeni L. High selectivity and sensitivity sensor based on MIP and SPR in tapered plastic optical fibers for the detection of l-nicotine. Sensor. Actuator. B Chem. 2014;191:529–536. doi: 10.1016/j.snb.2013.10.067. [DOI] [Google Scholar]
- 51.Popovic M., Minceva M. Thermodynamic insight into viral infections 2: empirical formulas, molecular compositions and thermodynamic properties of SARS, MERS and SARS-CoV-2 (COVID-19) viruses. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.