Graphical abstract
Keywords: COVID-19, Neuropilin-1, SARS-CoV-2 S1 CendR, EG00229, EG01377
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for causing the current coronavirus 2019 (COVID-19) pandemic, uses its spike (S1) protein for host cell attachment and entry. Apart from angiotensin-converting enzyme 2, neuropilin-1 (NRP1) has been recently found to serve as another host factor for SARS-CoV-2 infection; thus, blocking S1–NRP1 interaction can be a potential treatment for COVID-19. Herein, molecular recognition between SARS-CoV-2 S1 C-end rule (CendR) heptapeptide including small-molecule antagonists (EG00229 and EG01377) and the NRP1 was investigated using molecular dynamics simulations and binding free energy calculations based on MM-PBSA method. The binding affinity and the number of hot-spot residues of EG01377/NRP1 complex were higher than those of CendR/NRP1 and EG00229/NRP1 systems, in line with the reported experimental data as well as with the lower water accessibility at the ligand-binding site. The (i) T316, P317, and D320 and (ii) S346, T349, and Y353 residues of NRP1 were confirmed to respectively form H-bonds with the positively charged guanidinium group and the negatively charged carboxyl moiety of all studied ligands. Moreover, Rosetta protein design was employed to improve the binding affinity between CendR peptide and NRP1. The newly designed peptides, especially R683G and A684M, exhibited higher binding efficiency than the native CendR heptapeptide as well as the small-molecule EG00229 by forming more H-bonds and hydrophobic interactions with NPR1, suggesting that these designed peptides could be promising NRP1 inhibitors to combat SARS-CoV-2 infection.
1. Introduction
The expanding of coronavirus disease 2019 (COVID-19) pandemic has had an unexpected impact on current society and humanity. In May 2021, the world health organization (WHO) declared over 167 million infections and 3.4 million deaths globally (https://covid19.who.int). Researchers have been attempted to develop an antiviral drug or a vaccine for controlling and reducing the COVID-19 cases, but now reliable pharmaceutical interventions against this disease have not been reported yet.
The COVID-19 is caused by a new coronavirus, namely severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], [2], [3]. Patients infected with SARS-CoV-2 may present mild to severe symptoms, including fever, cough, sore throat, tiredness, chest pain, and severe pneumonia [4], [5]. Although both SARS-CoV-1 and SARS-CoV-2 are originated from bats, the human-to-human transmission of SARS-CoV-2 is easier than that of SAR-CoV-1 [1], [6]. Structurally, SARS-CoV-2 consists of four major proteins, including the envelope (E), membrane (M), nucleocapsid (N), and spike (S) proteins [7], [8]. The S protein of SARS-CoV-2 comprises S1 and S2 subunits that are responsible for the host cell attachment and entry. The S1 subunit of SARS-CoV-2 S protein directly binds to the angiotensin-converting enzyme 2 (ACE2) receptor of human cells, facilitating the viral infection [9], [10], [11]. Till now, many studies have been attempted to discover and develop the molecules that specifically disrupt the protein–protein interaction between S1 and ACE2 for preventing SARS-CoV-2 infection [12], [13], [14].
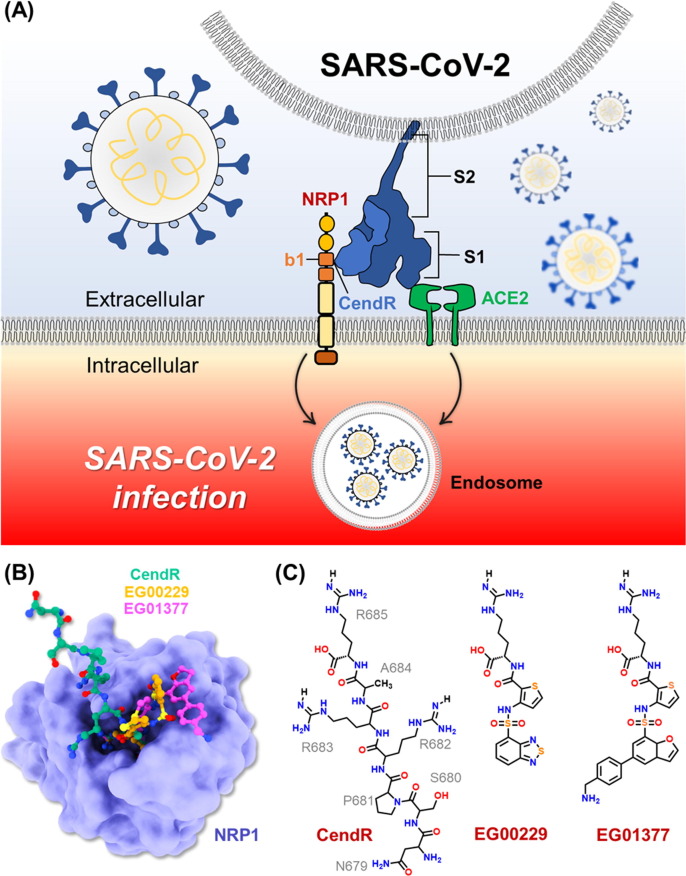
Recent studies have shown that SARS-CoV-2 S1 protein not only binds to the ACE2 but also the neuropilin-1 (NRP1) receptor (Fig. 1 A) [8], [15]. Interestingly, suppression of NRP1 expression in ACE2-expressing cells significantly reduced SARS-CoV-2 infection [16]. Based on X-ray crystal structure, the SARS-CoV-2 S1 protein binds to NRP1 receptor via its Arg-Arg-Ala-Arg C-terminal sequence (682RRAR685), called “C-end rule (CendR)” motif [16]. Site-directed mutagenesis indicated that C-terminal R685 of S1 protein is crucial for the binding of NRP1 [16]. Moreover, blocking this interaction using selective inhibitor, EG00229, can inhibit the viral infection, suggesting that S1–NRP1 interaction serves as a potential anti-SARS-CoV-2 target. Therefore, in this study, we aimed to investigate the binding pattern and susceptibility of SARS-CoV-2 S1 CendR heptapeptide as well as small-molecule inhibitors (EG00229 and EG01377; Fig. 1 C) [17], [18] against NRP1 receptor using all-atom molecular dynamics (MD) simulations and free energy calculations. Moreover, computational protein design with Rosetta was employed to improve the binding affinity between the SARS-CoV-2 S1 CendR and the NRP1 b1 domain. We hope that the information obtained from this study could be useful for the development of more effective drugs targeting NRP1 in the fight against COVID-19.
Fig. 1.
(A) Cartoon representation of SARS-CoV-2 infection. (B) Superimposed structures of NRP1 b1 domain in complex with CendR heptapeptide (PDB entry 7JJC) [19], EG00229 (PDB entry 3I97) [19], and EG01377 (PDB entry 6FMF) [20]. (C) Chemical structure of all studied ligands.
2. Computational methods
2.1. System preparation and molecular dynamics simulations
The crystal structures of human NRP1 b1 domain in complex with SARS-CoV-2 S1 CendR heptapeptide (PDB ID: 7JJC, chain C) [16], EG00229 (PDB ID: 3I97, chain A) [19], and EG01377 (PDB ID: 6FMF, chain A) [21] were obtained from the RSCB Protein Data Bank. The H++ server [22] was used to determine the protonation state of all amino acid residues at pH 7.4. The electrostatic potential (ESP) charges of EG00229 and EG01377 were calculated with the HF/6-31G* level of theory using Gaussian09 program [23]. The antechamber module of AMBER20 was employed to generate the restrained ESP (RESP) charges of the two small-molecule inhibitors. The AMBER ff14SB force field was applied for the protein, while other parameters were obtained from the generalized AMBER force field version 2 (GAFF2) [24]. Remaining missing parameters were generated by using parmchk2 modules. LEaP module in AMBER20 was used to add missing hydrogen atoms and solvate each complex in an isomeric truncated octahedral box of TIP3P water model [25] with a buffer distance of 13 Å. Chloride ions were added to neutralize the systems. The systems were then minimized to remove unfavorable interactions. Each procedure composed of steepest descent (SD) 2500 steps followed by conjugated gradient (CG) 2500 steps, in which the heavy atoms of proteins were first restrained with the force constant of 5.0 kcal mol−1 Å−2. Then, the protein backbones were restrained with the force constants of 10, 5, and 1 kcal mol−1 Å−2, respectively. At last, the systems were fully minimized without any restraint. After that, the systems were simulated under the periodic boundary condition [26], [27], [28] with a simulation time step of 0.002 ps using PMEMD module. The SHAKE algorithm [29] was used to constrain all bonds involving hydrogen atoms. The temperatures were controlled using Langevin dynamic technique [30] with the collision frequency of 1 ps−1. For the heating step, each system was gradually heated from 0 to 310 K for 200 ps, while the protein backbones were restrained with the force constant of 10 kcal mol−1 Å−2 in the NVT ensemble. In the equilibrating step, the systems were simulated at 310 K for 300 ps in the NVT ensemble. After that, the systems were simulated at 310 K and 1 atm in the NPT ensemble until reaching 100 ns.
2.2. Analysis
The root-mean-square displacement (RMSD) of complex and the number of hydrogen bonds (#H-bonds) between protein and ligand were calculated using cpptraj module [31] of AMBER20. The H-bond interactions were calculated using the following criteria: (i) distance between the H-bond donor (HD) and H-bond acceptor (HA) ≤ 3.0 Å and (ii) the angle of HD−H···HA ≥ 135°. The solvent-accessible surface area (SASA) was calculated using the NRP1 residues within 5 Å of small-molecule antagonists as solvent-exposed area. All energy calculations were performed using MMPBSA.py module [32] of AMBER20. The total binding free energy () and per-residue decomposition free energy () were calculated using molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) method [33] on 100 frames taken from the production phase.
2.3. Peptide design
Rosetta 3.12 [34] was employed with the beta_nov16 energy function to design new mutants of SARS-CoV-2 S1 CendR peptide. The FastDesign protocol [35], [36] based on the FastRelax was used in this study. This protocol not only designed target residues but also attempted to optimize structure by allowing the movement of sidechains and backbones. The crystal structure of NRP1/SARS-CoV-2 S1 CendR complex was used as a template. To design peptides with improved binding affinity, positions 683 and 684 of CendR were selected as target residues due to their locations that were close to the arginine-binding cleft and could form potential interactions with the NRP1 protein. In each run, the residues within 5 Å of the target residue were allowed to move. The 50 independent runs [37] of design were performed on each position. The 50 independent runs without design were also performed and used as a reference for further analysis. The free energy () in Rosetta was calculated in Rosetta Energy Units (REU). Each of the designed peptides with the lowest free energy was selected. The binding free energy () of each complex was then evaluated using InterfaceAnalyzer module [38] of Rosetta. Finally, these complexes with designed peptides of CendR were subjected to MD simulation.
3. Results and discussion
3.1. System stability
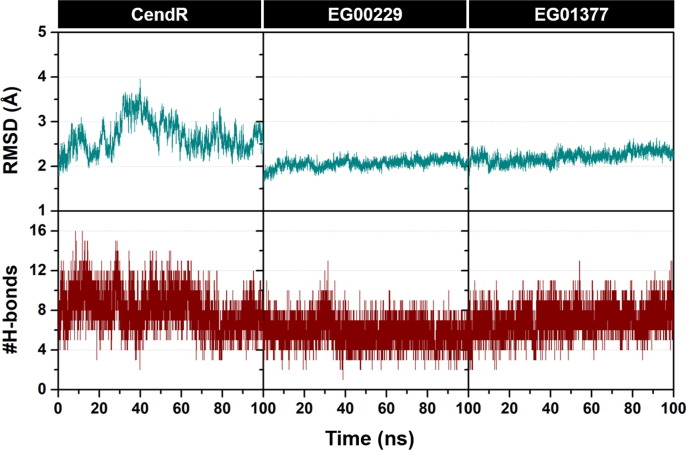
The stability of all simulated systems was determined using the calculations of root-mean-square displacement (RMSD) and the number of intermolecular H-bonds (#H-bonds) along the simulation time. The obtained results are plotted and summarized in Fig. 2 . We found that the structural flexibility of EG00229/NRP1 and EG01377/NRP1 complexes (RMSD of ~ 2.0 Å) was lower than that of CendR/NRP1 model (RMSD of ~ 2.0–3.0 Å), since the number of degrees of freedom of CendR heptapeptide is higher than that of EG00229 and EG01377. In the case of time evolution of #H-bonds, the CendR system exhibited higher fluctuation than the other two models, especially during the first 80 ns. Notably, #H-bonds of EG01377 system (7.32 ± 1.35 over the last 20 ns) was higher than that of the CendR (6.99 ± 1.31) and EG00229 (5.40 ± 1.14) models, suggesting that EG01377 exhibited the highest binding affinity with NRP1 (discussed in more detail later).
Fig. 2.
Time evolution of (top) all-atom RMSD and (bottom) #H-bonds of CendR heptapeptide (left), EG00229 (middle), and EG01377 (right) in complex with NRP1.
3.2. Inhibitory efficiency
The binding free energy (ΔG bind) of all studied ligands toward NRP1 was estimated using MM-PBSA method. As shown in Table 1 , the molecular mechanics energy (ΔE MM) in gas phase revealed that electrostatic interaction (ΔE ele) is the main force inducing protein–ligand complexation (ΔE ele of −328.35 ± 1.97, −203.22 ± 0.67, and −223.28 ± 0.70 kcal/mol for CendR, EG00229, and EG01377, respectively), which is ~ 7–14-fold stronger than the van der Waals (vdW) interaction (ΔE vdW of −23.81 ± 0.18, −25.60 ± 0.14, and 31.29 ± 0.15 kcal/mol for CendR, EG00229, and EG01377, respectively). This electrostatic-driven complexation is in good agreement with the reported binding of a prototypic CendR peptide, RPAR to NRP1 b1 domain [39] as well as the electrostatic surface of compound 3 (Lys(Har)-GlyΨ[Trl]GlyΨ[Trl]Arg) at the NRP1 b1 domain [40]. The electrostatic contribution of CendR system was higher than that of the two focused small-molecules antagonists, since the CendR has several polar moieties that can form H-bonds with NRP1 (discussed in more details later). Even though the CendR system exhibited the lowest ΔE MM value among the three systems, the high values of entropic term (TΔS) and solvation free energy (ΔG sol v) increased the total ΔG bind, leading to the low binding affinity with NRP1 b1 domain compared to the other two small-molecule inhibitors.
Table 1.
Average ΔGbind and its energy components (kcal/mol) of CendR, EG00229, and EG01377 in complex with NRP1 calculated with the MM-PBSA method. Data are shown as mean ± standard error of mean.
| Energy component (kcal/mol) |
CendR | EG00229 | EG01377 |
|---|---|---|---|
| Gas Term | |||
| −23.81 ± 0.18 | −25.60 ± 0.14 | −31.29 ± 0.15 | |
| −328.35 ± 1.97 | −203.22 ± 0.67 | −223.28 ± 0.70 | |
| −352.16 ± 2.00 | −228.82 ± 0.65 | −254.58 ± 0.70 | |
| 35.91 ± 1.07 | 27.71 ± 0.82 | 33.51 ± 0.57 | |
| Solvation Term | |||
| 314.75 ± 1.83 | 197.10 ± 0.55 | 213.56 ± 0.56 | |
| −5.91 ± 0.02 | −4.76 ± 0.01 | −5.72 ± 0.01 | |
| 308.84 ± 1.82 | 192.34 ± 0.54 | 207.84 ± 0.56 | |
| Binding Free Energy | |||
| −7.41 ± 0.34 | −8.76 ± 0.24 | −13.23 ± 0.28 | |
| Kd (µM) | 20.3 [41] | 5.1 [41] | 1.3 [20] |
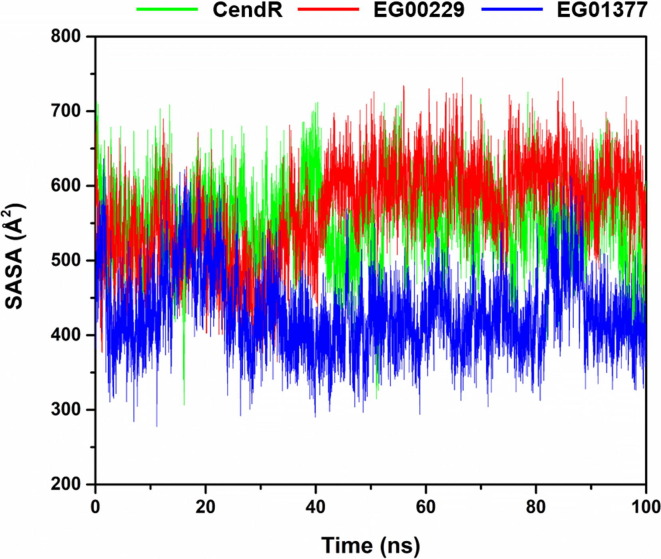
In the case of ΔG bind results, we found that EG01377 displayed the highest binding affinity with NRP1 (ΔG bind of −13.23 ± 0.28 kcal/mol) followed by EG00229 (−8.76 ± 0.24 kcal/mol) and CendR (−7.41 ± 0.34 kcal/mol), respectively. These results are in line with (i) the reported experimental data showing that the inhibitory activity against NRP1 of EG01377 (K d of 1.3 µM [20]) is higher than that of EG00229 (K d of 5.1 µM [41]) and CendR (K d of 20.3 µM [41]) and (ii) the solvation effect around the ligand-binding site (Fig. 3 ) demonstrating that the average SASA values of EG01377/NRP1 complex (430.62 ± 51.98 Å2) were lower than that of the other two systems (CendR/NRP1 complex (550.76 ± 0.53 Å2) and EG00229/NRP1 complex (566.28 ± 0.63 Å2)).
Fig. 3.
SASA plots along the simulation time of the three studied systems.
3.3. Key binding residues
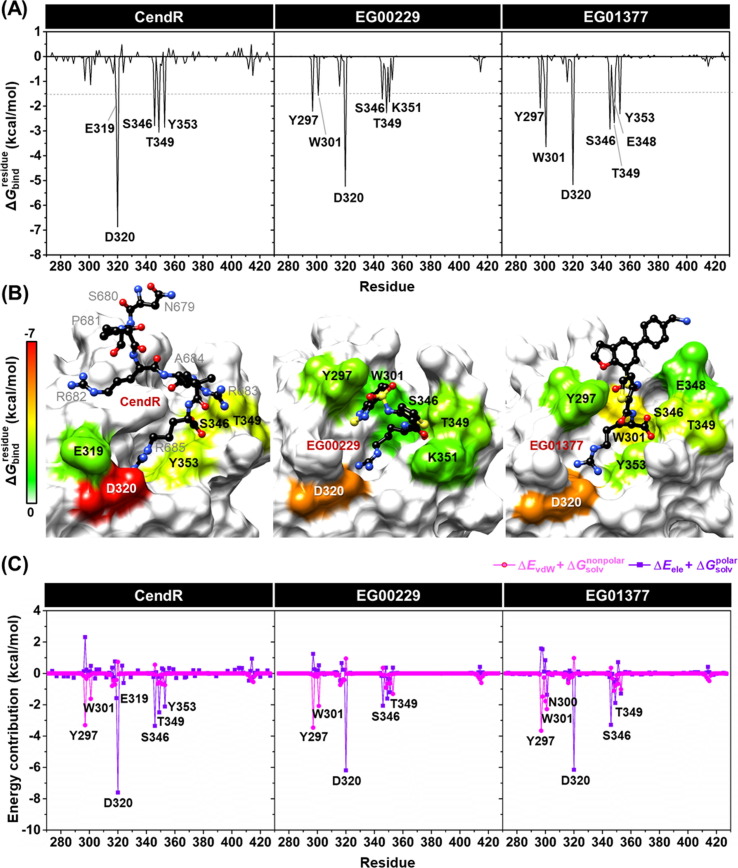
To investigate hot-spot residues involved in the binding of all studied ligands to NRP1, the calculation based on MM-PBSA method was conducted. The total energy contribution from each residue associated with ligand binding of all studied systems is shown in Fig. 4 , where the negative and positive values are energy stabilization and destabilization, respectively. It should be noted that only residues exhibiting the energy stabilization of ≤ −1.5 kcal/mol were taken into account.
Fig. 4.
(A) of CendR heptapeptide (left), EG00229 (middle), and EG01377 (right) in complex with NRP1 b1 coagulation factor domain. (B) Representative structures showing the ligand orientation in NRP1 b1 domain drawn from the last MD snapshot. The contributing residues of NRP1 involved in the binding of all studied ligands are colored according to their values, where the highest to lowest free energies are shaded from white to red, respectively. (C) Electrostatic (ΔEele + , purple) and vdW (ΔEvdW + , magenta) energy contributions from each residue of NRP1 b1 domain to the binding of CendR (left), EG00229 (middle), and EG01377 (right).
The obtained results demonstrated that there were five (E319, D320, S346, T349, and Y353), six (Y297, W301, D320, S346, T349, and K351), and seven (Y297, W301, D320, S346, T349, E348, and Y353) NRP1 residues associated with the binding of CendR, EG00229, and EG01377, respectively. These key binding residues are strongly supported by the alanine mutagenesis showing that Y297A, W301A, D320A, S346A, T349A, K351A, and Y353A result in a complete loss of biotinylated VEGF-A binding to NRP1 [19]. The highest contributing residues found in the EG01377/NRP1 complex was in good agreement with the ΔG bind results (Table 1) as mentioned earlier. Among nine hot-spot residues, D320 showed the lowest for all systems (−6.87, −5.24, and −5.17 kcal/mol for CendR, EG00229, and EG01377, respectively) via electrostatic attraction and H-bond formation with a positively charged guanidinium group of all studied ligands (Fig. 5 ), suggesting D320 as the most important key binding residue. This result is consistent with the mutational analysis mentioned above as well as the work of J. Daly et al. reported that the binding between the NRP1 and the alanine-substituted C-terminal R685 of CendR (R685A), which is the position that interacts with D320 residue of NRP1, was not observed [41].
Fig. 5.
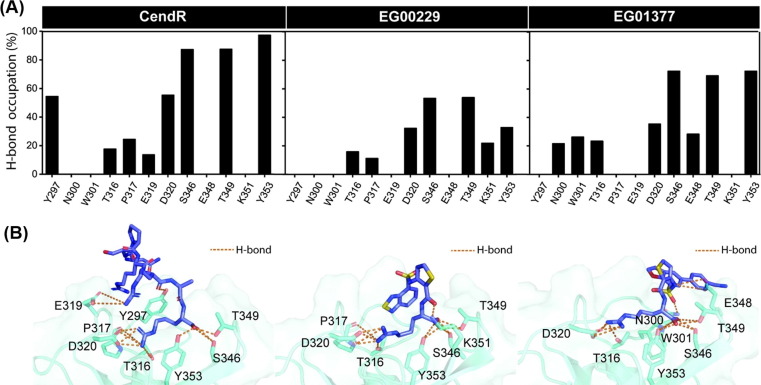
(A) Percentage of H-bond occupation of NRP1 contributing to the binding of (left) CendR heptapeptide, (middle) EG00229, and (right) EG01377. (B) Ligand orientation in the NRP1 b1 domain showing H-bond formations between protein and ligand(s). Orange dashed line indicates H-bond formation.
In terms of the energy contribution from electrostatic (ΔE ele + , purple line) and the vdW (ΔE vdW + , magenta line) energies from each NRP1 residue (Fig. 4 C), we found that the main energy contribution for stabilizing all studied ligands was the electrostatic energy (up to ~ −8.0 kcal/mol), especially for the D320 (electrostatically interacted with the positively charged guanidinium group of ligands) as well as the S346 and T349 (electrostatically interacted with the negatively charged carboxyl group of ligands), whereas the vdW contribution was observed in the range of ~ 1.0 to −3.5 kcal/mol as related to the ΔE MM results (Table 1).
3.4. Protein-ligand hydrogen bonding
Since electrostatic attraction was the main force inducing protein–ligand complexations (Table 1), we further investigated the structural insights into the intermolecular H-bond formation between ligand(s) and NRP1 during the last 20 ns of simulation using the defining criteria described in material and method section. The percentage of H-bond occupation (%H-bond) is plotted in Fig. 5 A, whereas the ligand orientation showing H-bond formations is illustrated in Fig. 5 B. Note that only NRP1 residues that exhibited %H-bond of > 10% were taken into account.
As expected, all studied ligands formed H-bonds with several polar and charged residues in the NRP1 b1 coagulation factor domain. There were eight (Y297, T316, P317, E319, D320, S346, T349, and Y353), seven (T316, P317, D320, S346, T349, K351, and Y353), and eight (N300, W301, T316, D320, S346, E348, T349, and Y353) residues responsible for H-bond formations in CendR, EG00229, and EG01377 systems, respectively. The (i) T316, P317, and D320 and (ii) S346, T349, and Y353 residues were found to respectively form H-bonds with the positively charged guanidinium group and the negatively charged carboxyl moiety of all studied ligands, consistent with the crystalized form of NRP1 in complex with CendR heptapeptide [16], EG00229 [19], EG01377 [20], and tuftsin [42]. It should be noted that the number of strong H-bonds (>70% occupancy) of CendR (3 residues: S346, T349, and Y353) was higher than that of EG01377 (2 residues: S346 and Y353) and EG00229 (0 residue) models, respectively, in line with the ΔE ele results (Table 1).
3.5. Key binding residues in SARS-CoV-2 S1 CendR peptide
To evaluate the hot-spot residues of CendR peptide upon the binding of NRP1, the and RMSD calculations were conducted. As shown in Fig. 6 , the positively charged R685 residue of CendR showed the lowest (−5.44 kcal/mol) and the highest stability (RMSD of 1.98 ± 0.41 Å averaged over 100 ns). The guanidinium group of R685 electrostatically interacted with the negatively charged D320 (Fig. 6 B) and formed H-bonds with T316, P317, and D320 residues, while its C-terminal carboxyl group formed H-bonds with S346, T349, and Y353 of NRP1 (Fig. 6 C). The R682, R683, and A684 residues of CendR exhibited the of ~ −1.0 to −4.0 kcal/mol and the moderate stability (RMSD of 5.38 ± 1.97 Å, 5.05 ± 1.55 Å, and 2.57 ± 0.85 Å for R682, R683, and A684, respectively), whereas the other residues, including N679, S680, and P681 (located far away from the arginine-binding cleft of NRP1) showed the highest (~ 0 kcal/mol) and the lowest stability (RMSD of 10.52 ± 4.54, 7.65 ± 3.32, and 5.91 ± 2.3 Å for N679, S680, and P681, respectively). Altogether, the R685 residue of SARS-CoV-2 S1 CendR peptide is suggested as the most important hot-spot residue against NRP1.
Fig. 6.
(A) of SARS-CoV-2 S1 CendR toward the binding of NRP1 b1 domain. (B) Representative structures showing the ligand orientation in NRP1 b1 domain drawn from the last MD snapshot. The contributing residues involved in the binding of CendR are colored according to its values, where the highest to lowest free energies are shaded from white to red, respectively. (C) Percentage of H-bond occupation of NRP1 contributing to the binding of CendR peptide. (D) RMSD of each residue of CendR peptide along the simulation time.
3.6. Computational design of NRP1 peptide binders
To enhance the binding affinity between the SARS-CoV-2 S1 CendR and the NRP1 b1 domain, computational protein design with Rosetta was employed. The R683 and A684 residues of CendR were selected as target positions, since they located near the arginine-binding pocket of NRP1 and exhibited the moderate stability and binding efficiency against NRP1. The R685 was not selected for peptide design because (i) it showed strong binding affinity with NRP1 and (ii) its guanidinium group was a key functional group for the binding of NRP1 via electrostatic attraction (Fig. 4, Fig. 5, Fig. 6). Rosetta provided six designed peptides, including R683D, R683G, A684E, A684K, A684L, and A684M. Interestingly, the ΔG bind ( R osetta) of all designed peptides was lower than that of the wild-type CendR (ΔΔG bind ( R osetta) of < 0) (Fig. 7 A). To validate this, MD simulations and free energy calculations based on MM-PBSA method were then performed. The obtained results (Fig. 7 B-C) revealed that all six designed peptides showed higher binding efficiency than the wild-type CendR. Among them, R683G (ΔG bind of −11.94 ± 0.26 kcal/mol) and A684M (ΔG bind of −11.78 ± 0.29 kcal/mol) were the two most promising NRP1 inhibitors, since their predicted binding efficiency is better than the other designed peptides as well as the small-molecule antagonist EG00229. Replacement of arginine by glycine (R683G) may reduce the steric effect on the peptide structure, allowing its N-terminal residues (679NSPR682) to interact with NRP1 (Fig. 7 D-E), while replacement of alanine by methionine (A684M) increased the hydrophobic interaction between peptide and NRP1. Notably, both designed peptides also formed more H-bonds with NPR1 residues 680–682 compared to the parent CendR heptapeptide (Fig. 7 F). Altogether, these designed peptides are suggested as promising inhibitors of SARS-CoV-2 S1 CendR motif interaction with NRP1
Fig. 7.
(A) ΔΔGbind(Rosetta) (REU) of all designed peptides obtained from Rosetta. Note that the ΔΔGbind was calculated using the following equation: ΔΔGbind = ΔGbind (mutant) – ΔGbind (wild-type). (B) ΔGbind (kcal/mol) of all designed peptides obtained from MM-PBSA method. (C) Representative structures of the two most promising peptides CendRR683G and CendRA684M showing the ligand orientation in NRP1 b1 domain drawn from the last MD snapshot. (D) of CendRWT, CendRR683G, and CendRA684M contributing to the binding of NRP1. (E) of NRP1 contributing to the binding of CendRWT, CendRR683G, and CendRA684M. (F) Percentage of H-bond occupation of NRP1 contributing to the binding of (left) CendRWT, (middle) CendRR683G, and (right) CendRA684M. %H-bond occupation (%H-bondoc) was classified into four levels: (i) strong H-bond (%H-bondoc of > 75%), (ii) medium H-bond (75% ≥ %H-bondoc > 50%), and (iii) weak H-bond (50% ≥ %H-bondoc > 10%).
4. Conclusion
In this work, the binding mechanism and susceptibility of the SARS-CoV-2 S1 CendR peptide as well as the small-molecule antagonists EG00229 and EG01377 in complex with the NRP1 b1 domain were fully revealed by all-atom MD simulations and free energy calculations based on MM-PBSA method. From ΔG bind calculation, the susceptibility against the NRP1 of all studied ligands was ranked in the order of EG01377 > EG00229 > CendR, in line with the reported experimental data. The molecular complexation of all studied systems was driven mainly by electrostatic attraction rather than vdW interaction. Among nine hot-spot residues, D320 showed the lowest via electrostatic attraction and H-bond formation with the positively charged guanidinium group of all studied ligands. According to computational protein design with Rosetta, all six designed CendR peptides (R683D, R683G, A684E, A684K, A684L, and A684M) exhibited higher binding efficiency with NRP1 than the wild-type CendR heptapeptide, and, among them, R683G and A684M are suggested as promising peptide inhibitors of SARS-CoV-2 S1 CendR motif interaction with NRP1. Altogether, the obtained structural and energetic information at the atomic level from this work could be helpful for further design and development of more specific NRP1 inhibitors in the fight against COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank the Department of Chemistry, Faculty of Science, Silpakorn University for computing resources.
Funding
This research was supported by the Invitation Research Fund from the Faculty of Medicine, Khon Kaen University (IN64227) to P.M.
References
- 1.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S. The Lancet. 2020;395:514. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. The Lancet. 2020;395:565. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nutho B., Mahalapbutr P., Hengphasatporn K., Pattaranggoon N.C., Simanon N., Shigeta Y., Hannongbua S., Rungrotmongkol T. Biochemistry. 2020;59:1769. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. The Lancet. 2020;395:507. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. The lancet. 2020;395:497. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X. J. Med.Virol. 2020;92:424. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kielian M. Science. 2020;370:765. doi: 10.1126/science.abf0732. [DOI] [PubMed] [Google Scholar]
- 9.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Nature. 2020;581:215. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Nature. 2020:1. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadi-Alijanvand H., Rouhani M. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00348. [DOI] [PubMed] [Google Scholar]
- 12.Prajapat M., Shekhar N., Sarma P., Avti P., Singh S., Kaur H., Bhattacharyya A., Kumar S., Sharma S., Prakash A. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benítez-Cardoza C.G., Vique-Sánchez J.L. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y., Král P. ACS nano. 2020;14:5143. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M. Science. 2020;370:856. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Science. 2020;370:861. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarvis A., Allerston C.K., Jia H., Herzog B., Garza-Garcia A., Winfield N., Ellard K., Aqil R., Lynch R., Chapman C. J. Med. Chem. 2010;53:2215. doi: 10.1021/jm901755g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell J., Mota F., Steadman D., Soudy C., Miyauchi J.T., Crosby S., Jarvis A., Reisinger T., Winfield N., Evans G. J. Med. Chem. 2018;61:4135. doi: 10.1021/acs.jmedchem.8b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis A., Allerston C.K., Jia H., Herzog B., Garza-Garcia A., Winfield N., Ellard K., Aqil R., Lynch R., Chapman C., Hartzoulakis B., Nally J., Stewart M., Cheng L., Menon M., Tickner M., Djordjevic S., Driscoll P.C., Zachary I., Selwood D.L. J. Med. Chem. 2010;53:2215. doi: 10.1021/jm901755g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell J., Mota F., Steadman D., Soudy C., Miyauchi J.T., Crosby S., Jarvis A., Reisinger T., Winfield N., Evans G., Finniear A., Yelland T., Chou Y.-T., Chan A.W.E., O'Leary A., Cheng L., Liu D., Fotinou C., Milagre C., Martin J.F., Jia H., Frankel P., Djordjevic S., Tsirka S.E., Zachary I.C., Selwood D.L. J. Med. Chem. 2018;61:4135. doi: 10.1021/acs.jmedchem.8b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell J., Mota F., Steadman D., Soudy C., Miyauchi J.T., Crosby S., Jarvis A., Reisinger T., Winfield N., Evans G., Finniear A., Yelland T., Chou Y.-T., Chan A.W.E., O’Leary A., Cheng L., Liu D., Fotinou C., Milagre C., Martin J.F., Jia H., Frankel P., Djordjevic S., Tsirka S.E., Zachary I.C., Selwood D.L. J. Med. Chem. 2018;61:4135. doi: 10.1021/acs.jmedchem.8b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anandakrishnan R., Aguilar B., Onufriev A.V. Nucleic Acids Res. 2012;40:W537. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. Frisch, G.W. Trucks, H.B. Schlegel, G.E. Scuseria, M.A. Robb, J.R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. Petersson, Gaussian, Inc., Wallingford CT, 2009.
- 24.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. J. Comput. Chem. 2004;25:1157. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. The Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- 26.Mahalapbutr P., Sangkhawasi M., Kammarabutr J., Chamni S., Rungrotmongkol T. Curr Top Med Chem. 2020;20:2046. doi: 10.2174/1568026619666191118110155. [DOI] [PubMed] [Google Scholar]
- 27.Mahalapbutr P., Lee V.S., Rungrotmongkol T. J. Agric. Food Chem. 2020;68:7974. doi: 10.1021/acs.jafc.0c02580. [DOI] [PubMed] [Google Scholar]
- 28.Mahalapbutr P., Kongtaworn N., Rungrotmongkol T. Comput. Struct. Biotechnol. J. 2020;18:2757. doi: 10.1016/j.csbj.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York D.M., Darden T.A., Pedersen L.G. The Journal of Chemical Physics. 1993;99:8345. [Google Scholar]
- 30.Wu X., Brooks B.R. Chem. Phys.Lett. 2003;381:512. [Google Scholar]
- 31.Roe D.R., Cheatham T.E. J. Chem. Theory Comput. 2013;9:3084. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 32.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. J. Chem. Theory Comput. 2012;8:3314. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 33.Genheden S., Ryde U. Expert Opin Drug Discov. 2015;10:449. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., Davis I.W., Cooper S., Treuille A., Mandell D.J., Richter F., Ban Y.-E.A., Fleishman S.J., Corn J.E., Kim D.E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J.J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J.J., Kuhlman B., Baker D., Bradley P. MethodsEnzymol. 2011;487:545. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.G. Bhardwaj, V.K. Mulligan, C.D. Bahl, J.M. Gilmore, P.J. Harvey, O. Cheneval, G.W. Buchko, S.V.S.R.K. Pulavarti, Q. Kaas, A. Eletsky, P.-S. Huang, W.A. Johnsen, P. Greisen, Jr., G.J. Rocklin, Y. Song, T.W. Linsky, A. Watkins, S.A. Rettie, X. Xu, L.P. Carter, R. Bonneau, J.M. Olson, E. Coutsias, C.E. Correnti, T. Szyperski, D.J. Craik, D. Baker, Nature 538 (2016) 329. [DOI] [PMC free article] [PubMed]
- 36.Loshbaugh A.L., Kortemme T. Proteins. 2020;88:206. doi: 10.1002/prot.25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poultney C.S., Butterfoss G.L., Gutwein M.R., Drew K., Gresham D., Gunsalus K.C., Shasha D.E., Bonneau R. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0023947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stranges P.B., Kuhlman B. Protein Sci. 2013;22:74. doi: 10.1002/pro.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haspel N., Zanuy D., Nussinov R., Teesalu T., Ruoslahti E., Aleman C. Biochemistry. 2011;50:1755. doi: 10.1021/bi101662j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedorczyk B., Lipiński P.F.J., Puszko A.K., Tymecka D., Wilenska B., Dudka W., Perret G.Y., Wieczorek R. A. Misicka. 2019;24:1756. doi: 10.3390/molecules24091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J. Y. Yamauchi. 2020;370:861. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starzec A., Miteva M.A., Ladam P., Villoutreix B.O., Perret G.Y. Bioorg. Med. Chem. 2014;22:4042. doi: 10.1016/j.bmc.2014.05.068. [DOI] [PubMed] [Google Scholar]