Abstract
The consequences of damage to the mitochondrial genome (mtDNA) are poorly understood, although mtDNA is more susceptible to damage resulting from some genotoxicants than nuclear DNA (nucDNA), and many environmental toxicants target the mitochondria. Reports from the toxicological literature suggest that exposure to early-life mitochondrial damage could lead to deleterious consequences later in life (the “Developmental Origins of Health and Disease” paradigm), but reports from other fields often report beneficial (“mitohormetic”) responses to such damage. Here, we tested the effects of low (causing no change in lifespan) levels of ultraviolet C (UVC)-induced, irreparable mtDNA damage during early development in Caenorhabditis elegans. This exposure led to life-long reductions in mtDNA copy number and steady-state ATP levels, accompanied by increased oxygen consumption and altered metabolite profiles, suggesting inefficient mitochondrial function. Exposed nematodes were also developmentally delayed, reached smaller adult size, and were rendered more susceptible to subsequent exposure to chemical mitotoxicants. Metabolomic and genetic analysis of key signaling and metabolic pathways supported redox and mitochondrial stress-response signaling during early development as a mechanism for establishing these persistent alterations. Our results highlight the importance of early-life exposures to environmental pollutants, especially in the context of exposure to chemicals that target mitochondria.
Keywords: Mitochondrial DNA damage, Bioenergetics, Redox signaling, Mitochondrial function, Environmental toxicants, Developmental exposures
Graphical abstract
Highlights
-
•
Early life mtDNA damage led to lifelong deficits in mitochondrial function.
-
•
C. elegans developed slowly and were sensitive to chemical exposures as adults.
-
•
Redox signaling is a mechanism that establishes these persistent alterations.
-
•
Data are consistent with the Developmental Origins of Health and Disease model.
1. Introduction
The mitochondrial genome (mtDNA) is small (16,569 bases in humans) and encodes a small number of genes: 13 proteins, 22 tRNAs and 2 rRNAs in humans, with similar numbers in most metazoans. Nonetheless, it is critical: the proteins are essential subunits of the mitochondrial respiratory chain (MRC) complexes, while the tRNAs and rRNAs are required for translation of those proteins. The health significance of mtDNA homeostasis is demonstrated by the fact that a large number of diseases are caused by mutations [1] or depletion [2] of mtDNA, which is normally present in thousands of copies per cell. In addition to these specific mtDNA-related diseases, mitochondrial dysfunction has been implicated in many more common human diseases including cancer, diabetes, metabolic syndrome and neurodegenerative conditions [3]. Further support for the importance of mitochondrial function to health is provided by evidence for toxicity associated with many pharmaceuticals that target mitochondria [4].
Globally, environmental pollutants are a major environmental driver of loss of years of life, conservatively estimated at three times greater than malaria, AIDS, and tuberculosis combined [5]. Developmental and low-level exposures are of particular concern [6,7]. The evidence that mitochondria are important targets of many environmental toxicants is growing, yet the consequences of such mitotoxicity are poorly understood [8]. mtDNA may be a particularly important target, because of the lack of some DNA repair pathways, in particular nucleotide excision repair (NER) [9]. Without NER, DNA lesions caused by common environmental stressors such as ultraviolet radiation, polycyclic aromatic hydrocarbons, and mycotoxins are irreparable in mtDNA. In vitro, such lesions can impair the progression of the mitochondrial DNA polymerase γ [10,11], potentially resulting in decreased mtDNA copy number or mutagenesis in vivo. However, despite the fact that many common environmental exposures cause irreparable mtDNA damage [12], the in vivo effects of such damage remain poorly understood.
We have previously employed ultraviolet C radiation (UVC) exposures to generate mtDNA damage that is irreparable due to the absence of NER (nucDNA damage is also caused, but is efficiently repaired). UVC is not environmentally relevant because stratospheric ozone absorbs UVC wavelengths; however, it provides a useful laboratory tool for near-exclusive production of damage (photodimers) that is repaired by NER, compared to UVB and UVA which also produce significant oxidative DNA damage. We reported that early-life (1st larval stage) exposures to such damage reduced levels of mtDNA, mtRNAs, total steady-state ATP levels, and oxygen consumption in developing C. elegans [13,14]. What might the long-term impact of such mtDNA damage be? Although irreparable, UVC- induced mtDNA damage is slowly removed via a process involving autophagic machinery in both adult C. elegans and human cells in culture [13,15]. We also found that the nematode mitochondrial DNA polymerase γ, polg-1, was dramatically upregulated by UVC exposure [14], suggesting the potential for an adaptive response. An adaptive response appears to be consistent with multiple examples of developmental mitochondrial dysfunction resulting in beneficial later-life outcomes in C. elegans and other species, including stress resistance and increased lifespan, sometimes termed “mitohormesis” [[16], [17], [18], [19], [20]]. On the other hand, a growing body of environmental health literature suggests that environmental exposures during development can lead to adverse outcomes later in life, a paradigm sometimes named the Developmental Origins of Health and Disease (DOHaD) [21,22]. A recent example identified long-term mitochondrial dysfunction associated with developmental mitochondrial toxicity resulting from exposure to the Complex I inhibitor rotenone [23].
Here, we asked if a low-level, in vivo, developmental environmental exposure that causes irreparable mtDNA damage would result in hormetic or adverse later life outcomes related to mitochondrial function, and tested the mechanisms mediating observed changes.
2. Results
We employed a previously developed [13,14] protocol that involves exposing growth-arrested L1 larvae to 7.5 J/m2 UVC three times (Fig. 1A), 24 h apart, in order to cause a high level of irreparable mtDNA damage while permitting nucDNA repair. Importantly, this is not a highly toxic level of UVC exposure, as demonstrated by the fact that there was no effect on lifespan (Fig. 1B). However, this protocol consistently leads to a significant reduction in N2 (wild-type strain) worm length that is maintained at least 72 h post-UVC exposure (Fig. 1C). We then examined mitochondrial and organismal phenotypes at later timepoints, and tested signaling pathways involved in the response to this damage. Many of these studies were conducted using strains (glp-1 and PE255, an ATP reporter strain created in a glp-4 background) in which germline proliferation fails at 25 °C [24], in order to exclude the confounding effects of the high level of mtDNA replication that occurs during gametogenesis [25].
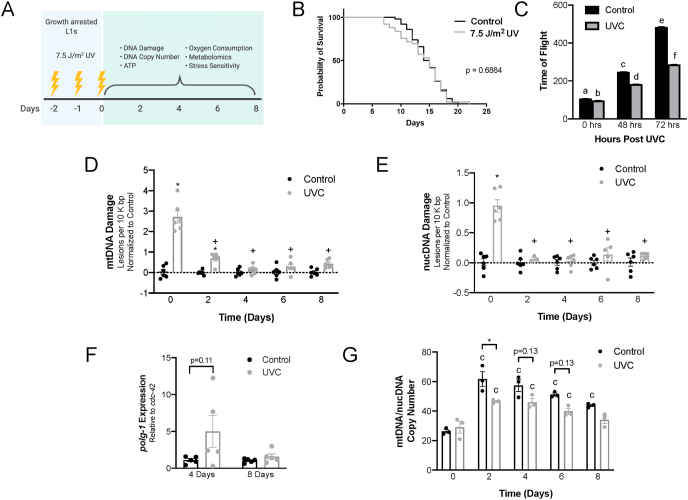
Fig. 1.
Early-life, irreparable mtDNA damage does not affect lifespan and is removed quickly A. Schematic depicting experimental design. Growth arrested L1s were exposed to UVC radiation three times over 48 h and transferred to normal culture conditions. Assays were performed at 0-, 2-, 4-, 6-, and 8-days post UVC exposure. Many studies were performed in germline proliferation-deficient strains. B. Lifespan is unchanged in N2 nematodes exposed to 7.5 J/m2 UVC radiation 3 times over 48 h as unfed, arrested L1 larvae (p = 0.688, Mantel-Cox test). Median lifespan was 15 days for both controls and UVC exposed, while maximal lifespans were 20 days in the controls and 22 days in UVC exposed (20 °C). Lifespan days were counted from hatch. n = 25–26 per group per experiment. Survival curve combines two independent experiments. C. UVC decreases time of flight (TOF; a surrogate for worm length) at 0-, 48-, and 72-h post-UVC exposure. Two-way ANOVA with Tukey's correction for multiple comparisons. n > 4000 per group. 2 independent experiments. D. UVC induces mtDNA damage that is not repaired but is removed in somatic cells (glp-1 strain raised at 25 °C) by four days post-UVC exposure. Data is normalized to control at each timepoint. Two-way ANOVA with Bonferroni's correction for multiple comparisons (9 comparisons). n = 3 per group per experiment (2 independent experiments). *p ≤ 0.05 compared to control at same timepoint. + p ≤ 0.05 compared to UVC at day 0. E. nucDNA damage does not accumulate and is quickly repaired. Two-way ANOVA with Bonferroni's correction for multiple comparisons (9 comparisons). n = 3 per group per experiment (2 independent experiments). *p ≤ 0.05 compared to control at same timepoint. + p ≤ 0.05 compared to UVC at day 0. F. Induction of transcription of the mtDNA polymerase γ (polg-1) is large but variable at 4 days post exposure in the UVC-exposed animals compared to control animals. Two-way ANOVA with Tukey's multiple comparisons correction. n = 5 per group. G. mtDNA/nucDNA copy number ratio in UVC exposed glp-1 nematodes is significantly lower than control animals at 2 days post-exposure and has a strong trend of remaining reduced for the length of the experiment. Two-way ANOVA with Bonferroni's correction for multiple comparisons (13 comparisons). n = 1 per group per experiment (3 independent experiments). *p ≤ 0.05. “c” indicates p ≤ 0.05 compared to control at day 0.
2.1. mtDNA damage accumulates over consecutive UVC exposures
We first tested the persistence of UVC-induced mtDNA damage during the worms’ lifespan. UVC-induced DNA damage largely comprises photodimers and 6,4-photoproducts, both of which consist of covalent bonds between adjacent bases; we use the term “photolesion” in this manuscript to describe this class of DNA damage. Although photolesions are not repaired in mtDNA, they are slowly and incompletely removed by autophagy in adult nematodes [13]; furthermore, damaged genomes may be diluted by the extensive mtDNA replication that occurs during development [25]. We measured significantly higher levels of mtDNA damage immediately after the third and final dose of UVC exposure (time 0 days) compared to the non-exposed controls at the same time point (Fig. 1D and Fig. S1A). mtDNA damage in UVC-exposed animals at day 2 was significantly higher than in their non-exposed controls. All other data points showed no distinguishable differences between amount of mtDNA damage in control and UVC-exposed animals at the same time point. This suggests that mtDNA damage is either removed rapidly via mechanisms of mitochondrial quality control, which seems unlikely given the slow kinetics of photodimer removal [13], or that the mtDNA damage is diluted due to rapid proliferation of mitochondria during development [14]. The apparent (though not significant) slight increase in mtDNA damage in UVC-exposed animals at day 8 could be a result of oxidative damage resulting from mitochondrial reactive oxygen species (ROS) production; however, this assay cannot distinguish between different types of DNA damage. A significant increase in nucDNA lesions in UVC-exposed compared to control animals was observed at day 0, but not at any other time point. (Fig.1E and Fig. S1B). Notably, there was approximately 3-fold higher mtDNA damage in UVC-exposed animals at day 0 compared to nucDNA damage, indicating that nucDNA photolesion damage was removed rapidly between arrested L1 UVC exposures. This replicates our previous observations [13,14] of much faster photolesion removal in nucDNA, presumably largely from NER.
2.2. Reduction in mtDNA copy number is observed throughout life following developmental UVC exposure
To further characterize DNA integrity following UVC exposure, we analyzed mitochondrial and nuclear DNA copy number. We previously found that UVC exposure induced an 8-fold increase in polg-1 (mitochondrial DNA polymerase γ) expression 48 h after UVC-exposure [14]. We confirmed that polg-1 expression shows a strong trend of remaining elevated up to 4 days post-exposure (p = 0.11) and returns to control levels of expression by 8 days post-exposure (Fig. 1F). We hypothesized that photolesions in mtDNA would block the progress of POLG-1 [11], resulting in reduced mtDNA content at early timepoints post-exposure. However, because mtDNA damage is not significantly different in control versus UVC-exposed worms in adulthood (Fig. 1D), we expected no difference in mtDNA copy number between groups in adulthood. Instead, mtDNA copy number per worm (Fig. S1C) and mtDNA/nucDNA ratio (Fig. 1G) was decreased throughout life in the UVC-exposed animals (p < 0.0001, main effect of UVC exposure). This was unexpected given the large proliferation of mtDNA copies during development and absence of detectable mtDNA damage in adults, and suggests that there could be other persistent effects of developmental exposure to UVC. There was no effect on nucDNA copy number (Fig. S1D). We observed similar results for the PE255 glp-4(bn2) strain (Fig. S1E), used to permit comparisons to ATP levels (below).
2.3. Developmental mtDNA damage did not result in detectable changes in mtRNA transcription in adults
We next asked whether this reduction in mtDNA copy number, and/or the presence of undetected photolesions that might block the mtRNA polymerase [26], would reduce the levels of RNAs for mitochondrial proteins, or affect the stoichiometry of MRC proteins. All MRC complexes except complex II contain proteins coded in both the mitochondrial and nuclear genomes, and proper stoichiometry depends on coordinated production of proteins from both genomes. Improper stoichiometry results in protective mitochondrial unfolded protein responses (UPRmt) [27], which can be triggered by reduced transcription of mtDNA-encoded MRC subunits [17]. To our surprise, mtDNA-encoded transcript levels in UVC-treated nematodes were unchanged at 4- and 8-days post-exposure (Fig. S2A). This lack of decreased transcription in the UVC-exposed animals might result from the multiplicity of mtDNAs buffering against transcriptional blocks present in a subset of mtDNAs, possibly in combination with compensatory increased transcription. However, a series of experiments designed to detect a compensatory transcriptional response in nucDNA-encoded MRC subunits and the TFAM homologue hmg-5 indicated that there were again no differences in transcription between control and UVC-exposed animals (Figs. S2B–C). Additionally, we tested for, but did not detect, transcriptional activation of the UPRmt (Fig. S2D). The atfs-1 mutant (which is deficient in mounting a UPRmt) was protected from larval growth inhibition (Fig. S2E), suggesting ATFS-1 mediated transcriptional response is important for inducing growth delay following UVC exposure. Finally, we asked whether exposure to chloramphenicol and doxycycline, two chemicals that inhibit mitochondrial protein translation and nematode larval development [28], would exacerbate UVC-mediated inhibition of larval growth [13]. We observed that both chloramphenicol and doxycycline further inhibited larval development after UVC exposure (Fig. S2F), indicating that, by itself, UVC-induced mtDNA damage does not entirely abrogate translation of proteins coded in mtDNA, even at the relatively high levels of mtDNA damage persisting in the larval stages [14]. Overall, this data suggests that mtDNA damage via UVC exposure does not result in overall shifts in transcriptional response of the mitochondrial or nuclear genomes, and that mitochondrial protein translation remains functional.
2.4. Mitochondrial respiration was altered in response to developmental mtDNA damage
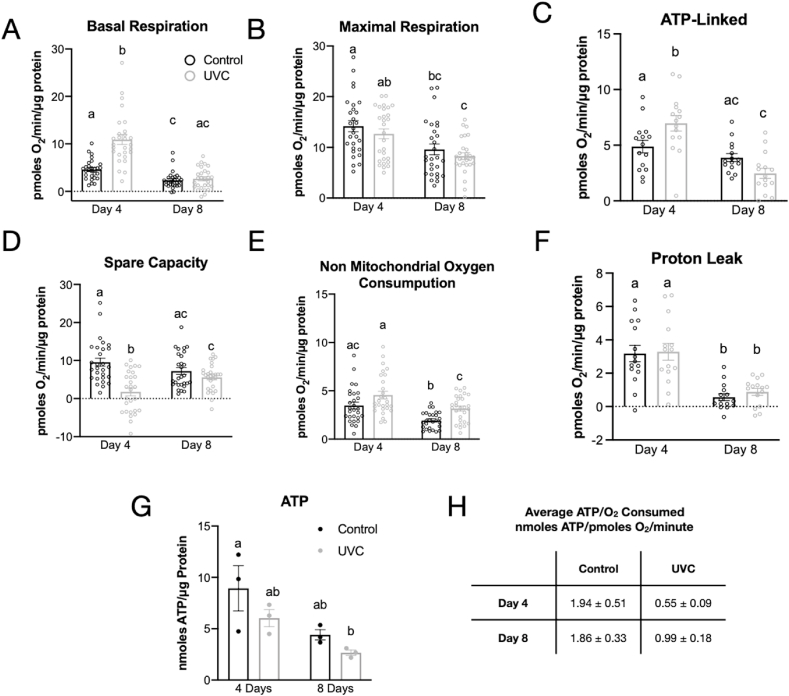
Given that mtDNA copy number was reduced, but RNAs of mitochondrial proteins were expressed normally after UVC exposure, we next asked if mitochondrial function in later life was impacted by an early life exposure to UVC. To directly assess mitochondrial function, we used the Seahorse XF Analyzer to measure basal, ATP-linked, maximal, and non-mitochondrial respiration rates at days 4- and 8- post-UVC in vivo [29]. Unexpectedly, basal oxygen consumption rates were significantly increased in UVC-exposed nematodes at 4 days post exposure, but not at 8 days post-exposure (Fig. 2A). Basal respiration at 8 days post-exposure was significantly reduced in both the control and UVC exposed groups compared to basal respiration at 4 days post-exposure, consistent with decreased basal respiration with increased age. At 4 days post-exposure, the mitochondrial uncoupler FCCP increased oxygen consumption in non-exposed animals 2.2-fold, but had no effect in UVC-exposed nematodes (Fig. 2B). Surprisingly, ATP-dependent OCR measured by inhibiting ATP synthase with DCCD was significantly increased in UVC-exposed animals, compared to non-exposed animals at 4 days post-exposure (Fig. 2C). Spare capacity (the difference of maximal and basal respiration) was significantly lower in UVC-exposed animals compared to non-exposed animals at 4 days post-exposure (Fig. 2D), indicating that UVC-exposed animals were operating at or near maximal respiration. Interestingly, non-mitochondrial respiration was similar in the exposed and non-exposed groups at 4 days post-exposure, but there was a significant increase in non-mitochondrial oxygen consumption in the UVC-exposed animals at 8 days post-exposure (Fig. 2E). While non-mitochondrial oxygen consumption was increased in the UVC-exposed animals, there was no detectable difference in proton leak (Fig. 2F), suggesting that mitochondrial membrane potential is likely unchanged between control and UVC-exposed animals.
Fig. 2.
Early-life mtDNA damage results in deficient mitochondrial respiration and decreased ATP per oxygen consumption. A. Basal respiratory rates were significantly increased in UVC-exposed nematodes at 4 days post-exposure, but not at 8 days. B. Maximal respiration was unchanged between control and UVC-treated groups but decreased with age. C. ATP-linked respiration rates were significantly increased in UVC-exposed nematodes at 4 days post-exposure. D. Spare capacity was significantly decreased in UVC-exposed animals 4 days after exposure and returned to control levels by 8 days post-exposure. E. Non mitochondrial oxygen consumption was significantly higher in UVC-exposed animals compared to control animals at 8 days post-exposure. F. No differences in proton leak were observed. For data shown in A–F: Data are presented as mean with error bars representing the standard error. Open circles are individual data points (2–4 independent experiments, n = 7–8 per group per experiment). Letters show which groups are significantly different (p ≤ 0.05). Two-way ANOVA with Tukey's correction for multiple comparisons. A UVC exposure of 2 J/m2 was used in panels C and F because these experiments were performed at a later date. This exposure level produced the same growth inhibition as characterized in the paper. G. UVC exposure leads to a trend of reduced steady-state ATP in glp-1 (q244). Two-way ANOVA with Tukey's correction for multiple comparisons. n = 3 per group. H. The ratio of ATP per oxygen consumed was calculated by dividing the average steady-state ATP value by the average basal respiration value for each group.
To better understand possible functional consequences of increased basal oxygen consumption rates in the 4-day post-UVC exposure animals, we measured steady-state ATP levels using two methods. ATP levels in extracts from glp-1 nematodes at 4- and 8- days post-UVC exposure showed a strong trend of reduction compared to their relative controls (Fig. 2G). Consistent with previous results [30], ATP content decreased in an age-dependent fashion by roughly 50% between days 4 and 8 in both control and UVC-exposed animals. We also measured ATP using the in vivo ATP reporter strain PE255 and found similar results of decreased steady-state ATP in UVC-exposed animals compared to controls in later life stages (days 6–10 post-exposure) (Fig. S3A), with the largest reduction (44%) at day 8. As a complementary measure of energy availability, we measured movement, and found that even lower levels of developmental UVC exposure reduced spontaneous movement in adults 8 days post-exposure (Fig. S3B). Thus, surprisingly, we observed decreased steady-state ATP levels but increased ATP-linked respiration in the UVC-exposed animals 4 days post-exposure. This could result from an inefficient electron transport chain (ETC) that is leaking electrons to oxygen [31], or an increase in ATP utilization by pathways activated by the UVC exposure, such as stress response pathways. The lack of increased proton leak in the UVC-exposed animals appears inconsistent with a stress-induced adaptive response to mtROS generation. However, the increased non-mitochondrial OCR suggests that sources of non-mitochondrial oxygen consumption such as enzymatic ROS production could be responsible for production of ROS. Possibilities for non-mitochondrial oxygen consumption include NADPH oxidase [32,33], cytochrome P450s, and others.
The increased basal oxygen consumption and lack of spare respiratory capacity combined with reduced ATP levels in UVC-exposed nematodes suggested persistently inefficient mitochondrial function. The decrease in spare respiratory capacity might be partially explained by the decrease in mitochondrial content suggested by decreased mtDNA content, or by decreased availability of substrate. We also note that although at 8 days post exposure basal oxygen consumption rates were no longer elevated in UVC-treated worms, ATP levels still trended lower, suggesting that mitochondrial efficiency was also reduced, resulting in a reduction in ATP levels per unit oxygen consumed (Fig. 2H).
2.5. Metabolic flexibility is critical to tolerance of early-life UVC exposure
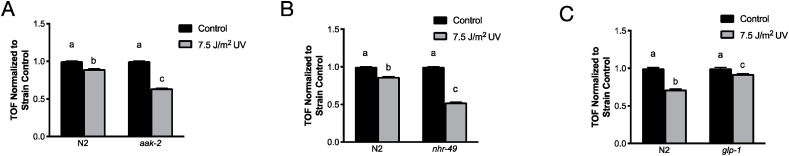
We hypothesized that in response to inefficient MRC function, UVC-exposed nematodes would shift metabolism towards alternate metabolic pathways. We tested whether strains carrying mutations in two major regulators of energy metabolism (aak-2, activated upon increase in the AMP:ATP ratio [34] and nhr-49, a nuclear hormone receptor that regulates energy metabolism in response to mitochondrial impairment [35] would be sensitized to UVC-induced inhibition of larval development [13]. aak-2 (Fig. 3A) and nhr-49 (Fig. 3B) mutant strains were both approximately two-fold more sensitive to early-life UVC exposure, supporting the importance of the nematodes’ capacity to regulate energy metabolism in response to early-life mtDNA damage. Importantly for extrapolating the results from mutant versus wild-type N2 studies to the outcomes observed in glp-1 animals used in other experiments in this manuscript, the growth decrease observed in the N2 strain was also observed, although to a lesser extent, in the glp-1 strain (Fig. 3C).
Fig. 3.
aak-2 and nrh-49 mutants are sensitive to UVC. A.aak-2 (36% decrease) and B.nhr-49 (48% decrease) are significantly growth delayed 72 h post UVC exposure compared to N2 animals exposed to UVC (10–13% decrease). C.glp-1 animals exposed to UVC had a mild growth delay (7.5% decrease) that was less than N2 animals exposed to UVC (30% decrease) in the same experiment. n > 500 in each group from 2 independent experiments. TOF values from UVC exposures are normalized to strain controls. Letters show which groups are significantly different (p ≤ 0.05). Two-way ANOVA with Tukey's correction for multiple comparisons.
Given this genetic evidence for protective metabolic restructuring, we next tested for alterations in transcript levels of key intermediary metabolic enzymes that regulate TCA cycle metabolism (pdp-1, pdhk-2), glycolysis (gpd-3), the glyoxylate cycle (gei-7), fatty acid B-oxidation (acs-2), and gluconeogenesis (pck-1), but detected no changes at either 4 or 8 days post-exposure (Fig. S4). However, the lack of altered transcriptional regulation of intermediary metabolic pathways does not rule out significant metabolic alterations, as there are many non-transcriptional mechanisms of enzyme activity regulation. To directly test for metabolic shifts, we next measured levels of organic acids, amino acids and acyl carnitines.
2.6. Targeted metabolomics supports NADPH stress and altered metabolic function in adults
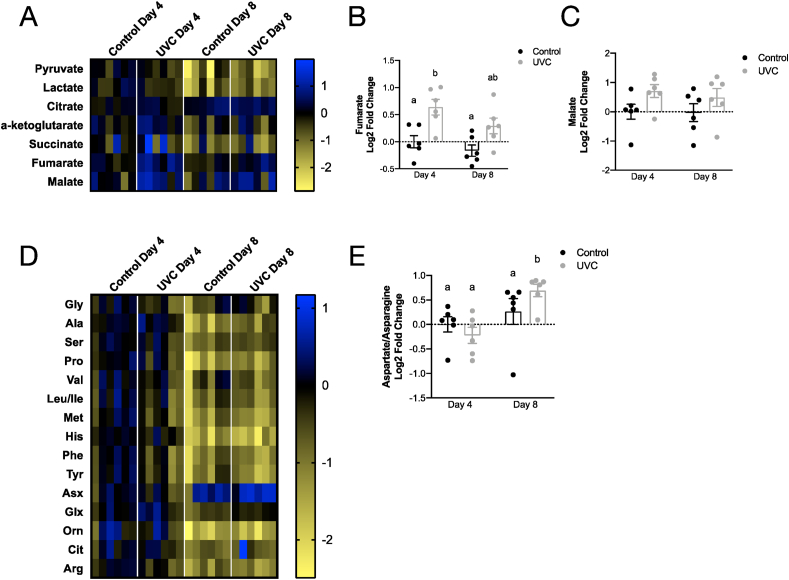
We observed age-related decreases in the levels of most organic acids between days 4 and 8 in both control and UVC-exposed animals (Fig. 4A). In contrast, fumarate and malate appeared to be elevated in the UVC-exposed animals compared to non-exposed controls and statistical analysis revealed that fumarate was significantly elevated in the UVC-exposed animals compared to controls at 4 days post-exposure (Fig. 4B). Additionally, there was a significant main effect of increased malate in the UVC-exposed animals compared to controls, though multiple comparisons testing did not reveal statistical significance at a specific time point (Fig. 4C). This increase suggests cataplerosis from the TCA cycle. Amino acid levels also generally declined with age, consistent with reduced overall metabolism in aging nematodes (Fig. 4D). Notably, Asx (aspartate/asparagine) was significantly increased in UVC-exposed animals compared to control animals at day 8 post-exposure (Fig. 4E). Together with increased cataplerosis from the TCA cycle, these differences in metabolites in the UVC-exposed animals could be indicative of activation of the malate-aspartate shuttle, potentially in response to redox stress [36]. However, given that metabolomics was performed in whole animal tissue, we cannot determine the impact of UVC exposure on redox status of specific cellular compartments. Lastly, a number of long chain acyl-carnitines (LCAC) had strong trends of being elevated in UVC samples at day 4 (Fig. S5A), which is often indicative of defects in, or altered regulation of, fatty acid beta-oxidation. When absolute LCAC levels were compared, UVC-related increases were seen in both C16:2-OH/C14:2-DC and C18–OH/C16-DC species (Fig. S5B). The potential defects in fatty acid oxidation and observations of cataplerosis from the TCA cycle, along with the UVC-exposed worms operating at near maximal rates of respiration, suggests that possibility that glycolysis in the UVC-exposed worms may be elevated in the UVC-exposed worms, leading to the generation of redox stress [37]. While the mechanism of increased redox stress cannot be fully elucidated from the metabolomics data, these data offer a few possibilities of how NADPH could be increased in the UVC-exposed animals and offers a potential explanation for the observed increase in non-mitochondrial oxygen consumption through an increase in NADPH oxidase activity in response to redox stress.
Fig. 4.
Metabolomics analysis suggests that the malate-aspartate shuttle is activated after UVC exposure. A. Heat map of organic acids at 4- and 8-days post-exposure in control and UVC-exposed animals. B. Fumarate is significantly increased in UVC-exposed animals at 4 days post-exposure. C. Malate trends toward increased in UVC-exposed compared to control animals at 4- and 8-days post-exposure. D. Heat map of amino acids at 4- and 8-days post-exposure in control and UVC animals. Generally, amino acid levels decline with age with no effect of UVC exposure. E. Asx (aspartic acid/asparagine) is significantly increased in UVC-exposed animals compared to controls at 8 days post-exposure. Error bars represent standard error. Letters show which groups are significantly different (p ≤ 0.05). Two-way ANOVA with Tukey's correction for multiple comparisons. n = 6 per group, 2 independent experiments.
2.7. Mitochondrial SOD mutants are more sensitive to UVC exposure
The data shown thus far provides evidence for NADPH stress as a mechanism of later life effects of early-life exposure to UVC. Specifically, we observed increased basal oxygen consumption (in the context of reduced ATP and movement) and evidence of increased flux through the malate-aspartate shuttle. Mitochondrial dysfunction, particularly in the context of increased overall oxygen consumption, can result in increased oxidative protein damage. We observed a significant increase in carbonyl groups of cytoplasmic (8%) and mitochondrial proteins (20%) 4 days post-UVC exposure (Fig. 5A and B), suggesting increased ROS production later in life in response to early-life mtDNA damage.
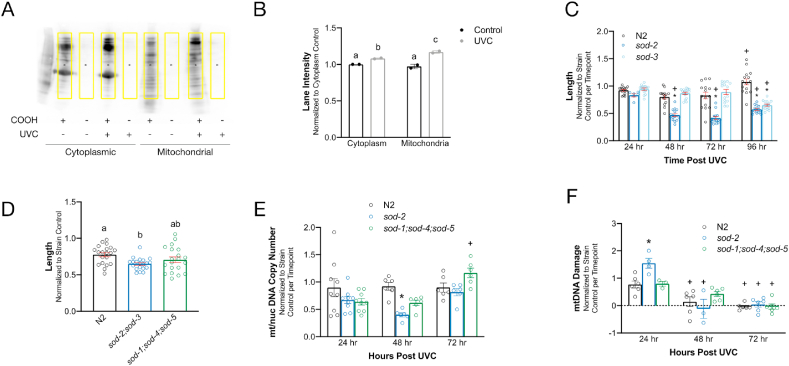
Fig. 5.
Mitochondrial SOD mutants are more sensitive to UVC exposure. A. Representative image of oxyblot at 4 days post-exposure demonstrates increased oxidative protein damage in UVC-exposed nematodes in both cytoplasmic and mitochondrial fractions. Yellow boxes indicate area of the blot that was quantified. COOH = derivatization reagent (required for detection), UVC = UVC exposure. B. Quantification of oxyblot. Error bars represent standard error. Letters show which groups are significantly different (p ≤ 0.05). Two-way ANOVA with Tukey's correction for multiple comparisons. n = 2 per group, 2 independent experiments. C.sod-2 and sod-3 mutants are more sensitive to UVC-induced growth delay 96 h post-exposure. Each UVC-exposed strain is normalized to its own non-exposed control at each time point. Two-way ANOVA with Bonferroni correction for multiple comparisons (17 comparisons). n = 6–15 per group. *p ≤ 0.05 compared to N2 UVC-exposed at same time point. + p ≤ 0.05 compared to 24 h UCV exposed of same strain. D. The mitochondrial SOD double mutant sod-2;sod-3, is more sensitive to UVC-induced growth delay 72 h post UVC exposure than wild type N2 though the triple cytosolic and extracellular combination knockout (sod-1;sod-4;sod-5) is not. One-way ANOVA with Tukey's correction for multiple comparisons. n = 20 per group. Different letters indicate a significant difference (p ≤ 0.05) between groups E. mtDNA:nucDNA ratio in SOD mutants over time. Each UVC-exposed strain is normalized to its own non-exposed control at each time point. Two-way ANOVA with Bonferroni correction for multiple comparisons (12 comparisons). n = 6–9 per group. *p ≤ 0.05 compared to N2 UVC-exposed at same time point. + p ≤ 0.05 compared to 24 h UVC exposed of same strain. F. mtDNA damage in SOD mutants. Each UVC-exposed strain is normalized to its own non-exposed control at each time point. mtDNA damage was increased by more than two-fold in UVC-treated sod-2 nematodes in comparison to both UVC-treated wild-type and sod-1;sod-4;sod-5 mutant nematodes at the 24 h timepoint. By 48 h and 72 h, damage levels had returned to our limit of detection in all strains. UVC-exposed sod-1;sod-4;sod-5 mutants were not statistically distinguishable from UVC-exposed N2s in terms of mtDNA damage at any timepoint. Two-way ANOVA with Bonferroni correction for multiple comparisons (12 comparisons). n = 3–6 per group. *p ≤ 0.05 compared to N2 UVC-exposed at same time point. + p ≤ 0.05 compared to 24 h UVC-exposed of same strain. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The ETC is a major source of intracellular ROS, and mitochondrial dysfunction can result in increased ROS production [38], including both superoxide anion (O2•−) and hydrogen peroxide (H2O2). Mitochondrial ROS may cause damage or serve as mitochondrial signaling molecules [39]. To investigate the potential functional importance of increased mitochondrial ROS generation, we tested the sensitivity to developmental UVC exposure of nematode strains carrying loss-of-function mutations in all 5 superoxide dismutase (SOD) enzymes (sod-1, sod-2, sod-3, sod-4, and sod-5), which convert O2•− to H2O2 [40]. We additionally looked at the effects of UVC exposure on the double sod-2;sod-3 mutant (mutations in both mitochondrial SOD proteins), and the triple sod-1;sod-4;sod-5 mutant (mutations in all cytosolic and extracellular SOD proteins).
UVC-induced larval delay was exacerbated in both mitochondrial SOD mutants (sod-2 and sod-3), with the delay occurring as soon as 48 h post-exposure in sod-2 nematodes and by 96 h post-exposure in sod-3 nematodes (Fig. 5C). Furthermore, the double sod-2;sod-3 mutant was significantly more growth delayed than wild-type 72 h after UVC exposure (p = 0.011), but the triple sod-1;sod-4;sod-5 mutant was not (p = 0.229) (Fig. 5D). The sod-2 mutant was more sensitive than the sod-2;sod-3 mutant, consistent with a previous report [41]. Based on this data, we used the sod-2 and sod-1;sod-4;sod-5 mutants to examine the role of mitochondrial and cytosolic superoxide dismutase proteins in the response to UVC exposure. We confirmed the growth delay at the molecular level by measuring mitochondrial and nuclear DNA content in wild-type and SOD mutants after UVC exposure. There was a significant delay in mtDNA:nucDNA copy number expansion in the sod-2 worms, but not the sod-1;sod-4;sod-5 worms at 48-h post-exposure (Fig. 5E). The reduction in this ratio was due to the reduction in mtDNA copy number expansion (Fig. S6A), as nucDNA copy number expansion was comparable in all three nematode strains post-UVC exposure (Fig. S6B). mtDNA damage at 24 h post-exposure was significantly increased in the sod-2 mutants compared to N2, but not in sod-1;sod-4;sod-5 mutants (Fig. 5F). Importantly, neither sod-2 mutant or sod-1;sod-4;sod-5 mutant non-exposed controls showed any change in mtDNA damage compared to N2 controls (Fig. S6C). We detected no statistically significant differences between strains in nucDNA damage (Fig. S6D).
Given the evidence of increased mitochondrial ROS, we looked at transcript levels of heat shock proteins in the N2, sod-2, and sod-1;sod-4;sod-5 strains to test for evidence of the heat shock response in different subcellular compartments, as well as heat shock contributions to ROS induction [42]. However, we did not detect any sod-2-specific increases in any tested mitochondrial (hsp-6, hsp-60a, and hsp-60b), cytosolic (hsp-16.12 and hsp-16.41), or endoplasmic reticular (hsp-4) heat shock protein (Figs. S6E–J). Thus, this experiment failed to support the hypothesis that an induction of the heat shock response in UVC-treated animals contributed to mitochondrial phenotypes or an increase in ROS production.
2.8. Skn-1 mediates response to UVC exposure
The delayed development and indications of increased mtDNA damage following UVC exposure in the mitochondrial, but not cytosolic SOD mutants could be explained in two ways: either mitochondrial O2•− causes the growth delay by causing oxidative damage; or conversion of mitochondrial O2•− to H2O2 is important in reducing the extent of the growth delay via adaptive signaling. Both H2O2 and O2•− can cause macromolecular damage, while only H2O2 can exit the mitochondria to act as a signaling molecule. To address the first hypothesis, we tested whether antioxidant treatment of 10 mM N-acetyl cysteine [43], 3 mM Trolox [[44], [45], [47]] or 1 and 5 μM MitoQ [47,48] (all previously shown to protect against various stressors in C. elegans) rescued UVC-induced growth delays. We observed no effect of antioxidant treatment on the UVC-induced growth delay (Fig. S7). To test the hypothesis that redox signaling is protective after UVC exposure, we examined the sensitivity of nematodes deficient in skn-1, the nematode homologue of the mammalian oxidant-responsive Nrf2 transcription factor. Nrf2 controls the expression of genes related to detoxification, antioxidant defenses, and intermediary metabolism [49], and nematodes lacking skn-1 are oxidative stress-sensitive [50]. Remarkably, skn-1 mutant nematodes showed only a 7% decrease in growth with UVC exposure (Fig. 6A), suggesting that the dramatic growth delay observed in wild-type nematodes after UVC exposure is partially dependent on the skn-1 mediated signaling response to ROS, not oxidative damage. We investigated the sensitivity of other transcription factors that mediate the ROS stress response. Nematodes with deletions in daf-16 (the nematode FOXO transcription factor [51,52]) and ced-4 (the nematode homologue of Apaf 1 [53]) showed a similar or more dramatic growth delay compared to the N2 nematodes, indicating that the response of these two transcription factors to UVC exposure does not contribute to the growth delay (Figs. S8A and B). Additionally, nematodes with genetically compromised ETC function and increased oxidative stress sensitivity [54] were not more resistant to UVC exposure: both gas-1 and mev-1 mutants were more growth delayed than wild type nematodes after UVC exposure (Figs. S8C and D). Together, these data suggest that skn-1 signaling, more than oxidative damage, is responsible for mediating the response to UVC exposure in wild-type nematodes. Lastly, we tested ATP levels in skn-1 mutants to determine if skn-1 signaling was responsible for the persistent phenotypes observed, and found that the skn-1 mutants were no more sensitive to reduction in ATP levels after UVC exposure than N2 (Fig. 6B), which is also inconsistent with an oxidative damage theory, since skn-1 mutants are oxidative stress-sensitive.
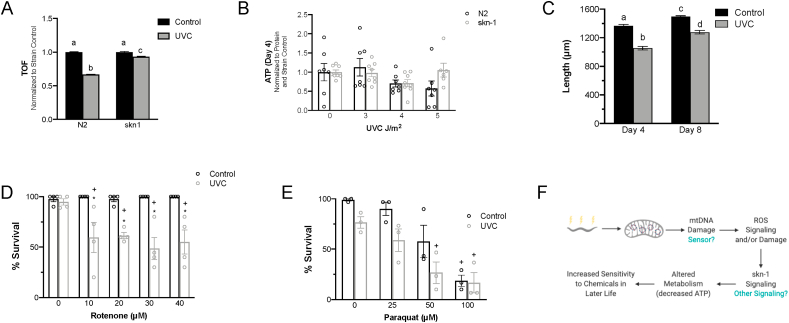
Fig. 6.
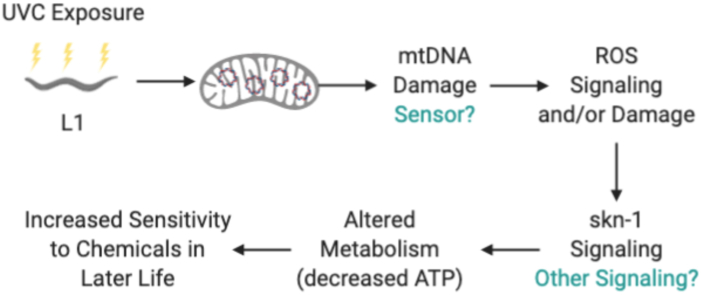
Developmental UVC exposure results in skn-1 signaling that mediates phenotypes in the adult nematode A.skn-1 mutants are protected from UVC-induced growth delay 72 h post UVC exposure. n > 500 in each group from 2 independent experiments. TOF values from UVC exposures are normalized to strain controls. Letters show which groups are significantly different (p ≤ 0.05). Two-way ANOVA with Tukey's correction for multiple comparisons. B.skn-1 mutants are protected from UVC-induced decrease in ATP levels 4 days post UVC exposure. Two-way ANOVA with Bonferroni correction for multiple comparisons (10 comparisons). C. Adult size was reduced in response to UVC at both day 4 and day 8. Letters show which groups are significantly different (p ≤ 0.05). Two-way ANOVA with Tukey's correction for multiple comparisons. n = 60–70 individuals in 2 experimental replicates. D. UVC-exposed nematodes are more sensitive to the complex I inhibitor rotenone, but not to E. paraquat. n = 4 per group. Two-way ANOVA with Bonferroni correction for multiple comparisons (13 in D and 10 in E). + p ≤ 0.05 compared to no drug treatment, no UVC exposure control. F. Model for ROS involvement in the development of lifelong effects as a result of early-life exposure to UVC. UVC exposure in early life leads to mtDNA damage that may be sensed by the cell. This damage, or unidentified sensor activity, leads to ROS generated from the ETC. skn-1 is activated by increased ROS generation, leading to decreased ATP and persistent phenotypes in adults.
2.9. Early life mtDNA damage results in decreased stress resistance in adulthood
As described earlier, there is evidence for both negative and positive effects of mitochondrial dysfunction and toxicity during development. Developmental mtDNA damage resulted in many of the same phenotypes as mitohormesis, suggesting the potential for a common mechanism. However, we found that although adult size was significantly (10–20%) reduced in response to developmental mtDNA damage (Fig. 6C) and ATP levels were reduced (Fig. 2 and S6), consistent with mitohormetic phenotypes, lifespan was statistically unchanged from that of control nematodes (Fig. 1B), whereas mitohormesis is associated with lifespan extension. Median lifespans were 15 days in both control and UVC treated animals, and maximal lifespans were 20 and 22 days, in control and UVC-treated animals, respectively. However, organisms outside of the laboratory are exposed to a variety of stressors, and there is evidence for “mitohormetic” responses resulting in increased stress resistance [55]. Instead of increased stress resistance, however, we found that UVC-treated nematodes were more sensitive to rotenone (Fig. 6D) than their untreated counterparts, though we detected no difference in sensitivity to paraquat (Fig. 6E).
3. Discussion
3.1. DOHaD or lifespan extension and stress resistance
Our results indicate that irreparable mtDNA damage incurred early in C. elegans development can lead to a lifelong reduction of mtDNA:nucDNA copy number, increased basal oxygen consumption and lack of spare respiratory capacity in the context of reduced ATP levels, and sensitivity to later-life chemical challenge, thus supporting the DOHaD hypothesis. These results are in contrast to many others in the literature that find hormetic responses, including increased lifespan and stress resistance, in response to knockdown of ETC genes [16,17,20,56,57] and some chemical exposures [18,57]. While there are several phenotypes shared between reported hormetic responses in the literature and the phenotypes we observed, including small adult size and reduced ATP levels, the lack of hormesis we observed is not entirely unexpected, as there were also differences in observed phenotypes. In particular, activation of the mitochondrial unfolded protein response, which triggers compensatory mechanisms that extend lifespan [17], was not observed in our experiments. We also did not observe a change in lifespan. We note that the majority of the treatments reported in the literature to trigger mitohormesis, particularly RNAi, result in major losses of protein or function. In our experiments, it appears that transcripts and proteins coded in the mtDNA were produced at sufficient levels despite the presence of transcription-blocking photolesions, presumably due to the multiplicity of mtDNAs. It may be that most environmental chemical exposures belong to a mechanistic category distinct from and possibly poorly modeled by gene knockdown, gene knockout, or specific pharmacological mitochondrial poisoning, because most environmental mitotoxicants are relatively non-specific [8].
More broadly, we suggest that observations of “mitohormesis” should be interpreted cautiously. RNAi experiments are different than what is seen in most human mitochondrial diseases, most of which result from mutations in mitochondrial gene, rather than reductions in transcript and protein levels. That mutations and RNAi knockdown are not identical is further underscored by the fact that in C. elegans, many strains that harbor mutations in genes that when knocked down with RNAi result in lifespan extension, actually have reduced lifespans [20]. Nonetheless, some mutations in ETC subunit genes do lead to lifespan extension. For example, an nduf-7 (complex 1) mutation increases ROS, activates the UPRmt and extends lifespan [58]. Another report indicates that relatively smaller perturbations of mitochondrial function can lead to later-life, mitohormetic effects by depressing the normally programmed repression of the heat shock response in adulthood [59]. Possible explanations for our differing results include: 1) different mechanisms of mitochondrial toxicity or different mutation-induced forms of protein dysfunction may have different long-term consequences; 2) examination of different health-related endpoints may reveal apparently hormetic vs deleterious consequences; 3) exposure level may be critical: i.e., it is possible that even lower levels of mtDNA damage than those we employed would cause a more classically hormetic response. Both toxicological [60] and gene knock-down [20] models of hormesis propose beneficial effects only at low levels of inhibition or dysfunction. Another caveat with interpreting model organism hormesis results is suggested by the fact that lifespan extension in C. elegans is not always directly relevant to humans, as mutations in C. elegans mitochondrial genes that increase lifespan can lead to shortened lifespan in humans [20]. Finally, there are tradeoffs associated with extending lifespan, bringing into question the apparently unambiguously positive connotation of the term “mitohormesis.” Reduced fertility, smaller adult size, and reduced movement are often seen in conjunction with lifespan extension. These results are consistent with data from recent toxicological studies which have provided evidence for later-life, adverse effects of mitochondrial toxicity. Perhaps most prominently among such reports, developmental exposure to nucleoside reverse transcriptase inhibitors, which interfere with mtDNA homeostasis, results in later-life mtDNA mutations, reduced mtDNA copy number, and reduced mitochondrial function in humans [61], rodents [62] and primates [63]. Other examples include latent mitochondrial myocardial toxicity seen in patients receiving doxorubicin as chemotherapy [64], latent liver carcinogenicity in mice after short term exposure to dichloroacetate [65], and increased rates of mitochondrial diseases later in life among survivors of childhood cancers [66].
3.2. A role for mitochondrial reactive oxygen species and mitochondrial retrograde signaling in metabolic programming
One of the most striking effects observed in response to early life mtDNA damage was the persistent reduction in steady state ATP levels, especially since this persisted even after mtDNA lesions were removed, and mtDNA-encoded ETC transcript levels were at or above those in untreated nematodes. mtDNA copy number was reduced at this time, but only approximately 20%, which by itself is not sufficient to reduce ATP levels in C. elegans [67]. Similarly, the lack of change in mtRNA levels or activation of UPRmt, suggest that the changes in ATP levels and other phenotypic effects result from signaling, rather than being direct functional deficits resulting from mtDNA damage or copy number changes.
Increased protein oxidation, increased oxygen consumption despite decreased ATP levels and movement, and the reduced sensitivity of skn-1 mutants to developmental mtDNA damage-induced growth delay, all support mtROS signaling as a mechanism for the regulation of growth delay (Fig. 6F). Increased oxidative stress is also consistent with metabolomic shifts suggesting activation of the malate-aspartate shuttle or increased glycolysis, which could explain some of the increases observed in non-mitochondrial oxygen consumption. In the context of oxidative stress resulting from UVC-induced mtDNA damage, much of the reducing equivalent pool in the mitochondria could be used for reducing oxidized glutathione, making less available for OXPHOS and ATP production, while at the same time reducing the mitochondrial membrane potential or limiting availability of reducing equivalents for the ETC, consistent with a reduced spare respiratory capacity.
Our data suggest that developmental UVC exposure results in a low level of mtROS that plays two roles, both causing damage and acting as signaling molecules, with a dominant long-term role for alterations in developmental ROS signaling. ROS play an important signaling role during development, and disruption of this signaling can alter differentiation and proliferation [68]. Short term exposure to H2O2 in young nematodes results in many similar outcomes to those seen here, including reduced movement and ATP levels, though these effects are transient [69]. We hypothesize that in our experiments, aberrant ROS generation during development led to skn-1-mediated metabolic alterations that program the developing nematode for life. In wild-type worms, the toxic effects of O2•− are ameliorated by the action of SODs, and thus insufficient to cause developmental delay, as indicated both by the sensitivity of the mitochondrial SOD mutants and by the inability of pharmacological antioxidants to rescue growth delay. mtROS, however, appears to be required for a SKN-1-mediated growth delay and metabolic remodeling, as indicated by the initially counterintuitive finding that skn-1 mutants are protected from developmental delay, and by the absence of a reduction in ATP levels in UVC-exposed skn-1 mutants. More broadly, given that ROS are both damaging and signaling molecules, it is not surprising that alterations in either direction of their tightly regulated developmental levels may have short-term (i.e., growth) and long-term (i.e., metabolic programming) consequences. Finally, the fact that atfs-1 mutants were also less sensitive to UVC-induced growth delay suggests a role for this transcription factor in mediating growth delay.
3.3. Unexpected mtDNA damage and copy number dynamics
The patterns of change in mtDNA copy number and mtDNA damage were also unexpected in a number of ways. With regards to mtDNA damage in the glp-1 germline-deficient strains (Fig. 1D), the intriguing reappearance of mtDNA lesions 8 days post-exposure suggests that mtDNA damage may accumulate more quickly with aging in the context of early life exposure than in control animals (in which we did not detect damage at 8 days), possibly due to persistently increased oxidative stress or reduced repair capacity due to reduced ATP or nucleotide availability.
The complete (at least within our limits of detection) removal of photolesions from mtDNA in developing larvae is in contrast to previous work from our lab in somatic cells of adult nematodes, which had shown just under 40% damage removal after 72 h in adult nematodes [13]. mtDNA replication appears to stop or at least fall below detection in adult nematodes, and copy number slowly declines with a half-life of approximately 10 days [70]. This half-life roughly corresponds to the rate at which damage is removed, suggesting that mtDNA damage removal in adult nematodes is tied to turnover of mitochondrial genomes. In contrast, mtDNA replication is essential and extensive during development. We hypothesize that increased mtDNA replication (leading to dilution of damaged genomes with new, undamaged copies) and, potentially, more frequent turnover leads to more complete reduction in per-nucleotide levels of damage during development. Supporting this hypothesis, the increase in mtDNA copy number between days 0 and 2 (3.88-fold) is proportionally similar to the reduction in lesion frequency over the same time period (3.98-fold). This would suggest that photodimers in mtDNA are not removed in any quantitatively important fashion during the first 2 days of C. elegans development.
With regard to mtDNA copy number, complete lesion removal and induction of polg-1 suggest that mtDNA replication should be unimpeded, yet reductions in mtDNA:nucDNA copy number persist throughout adulthood. However, mtDNA replication appears to cease between 3 and 6 days of age in untreated nematodes [70]. Therefore, it is possible that replication of some mtDNAs may be blocked by lesions until the point at which replication stops, resulting in fewer mtDNA copies per cell. Alternately, it is possible that a reduction in mtDNA copy number results from increased removal. Consistent with this theory, oxidative damage in cultured mammalian cells induced strand breaks in mtDNA, leading to degradation of mitochondrial genomes [71].
A limitation of our study is the inability to draw conclusions about cell-type specific effects of either mtDNA damage or copy number which may be important because mitochondria vary significantly between cell types [72]. Cell- and tissue-specific differences include turnover rates [73] and levels of mitophagy [74], such that damaged genomes or alterations in copy number may persist only in some cell types. This is an intriguing hypothesis, as mitochondrial dysfunction specifically in neurons results in whole organism effects in C. elegans [56], and is implicated in many human diseases [75].
4. Conclusions
Overall, these results identify life-long redox signaling-mediated alterations in mitochondrial function resulting from a low-level exposure that had no effect on lifespan. The outcomes later in life were largely deleterious, although we cannot exclude that the metabolic changes we observed may be advantageous under certain circumstances. These results are potentially important in the context of human health, because environmental exposures that result in irreparable mtDNA damage are common [12], there are increasing reports of pollutants affecting mitochondria [8,76], and we now understand that pollution is the major environmental driver of loss of life globally [5], despite the fact that we know very little about the toxic effects of the great majority of anthropogenically-produced chemicals [7].
5. Methods
5.1. C. elegans strains and culture conditions
Populations of C. elegans were maintained on K-agar plates seeded with E. coli OP50 bacteria, unless otherwise noted. N2 (wild-type), JK1107 glp-1(q224), sod-2 (gk257 I), sod-3 (gk235), gas-1 (fc21), mev-1(Kn1), glp-1 (e2141), daf-16 (mu86), skn-1 (zu670), SJ4100 zclS13 [hsp-6:GFP], SJ4058 zclS9 [hsp-60:GFP], and pkc-1 (nj1) were obtained from the Caenorhabditis Genetics Center (CGC), University of Minnesota. PE255 glp-4(bn2) were provided by Christina Lagido, University of Aberdeen (Aberdeen, UK). The sod-2;sod-3 [sod-2 (gk257) I; sod-3 (tm760) X] double mutant strain and sod-1;sod-4;sod-5 triple mutant strain [sod-1 (tm776);sod-5 (tm1146) II; sod-4 (gk101) III] were a kind gift from Bart Braeckman (Ghent University, Ghent, Belgium).
5.2. UVC exposure, DNA damage and genome copy number
UVC exposures were conducted in a custom-built exposure cabinet as previously described [13]. UVC output was measured with a UVX radiometer (UVP, Upland, CA). Synchronized (by bleach/NaOH egg isolation [77]). L1 nematodes were maintained on plates that contain no bacterial food, and therefore do not develop for UVC exposures, and were then transferred to seeded plates, as previously described [14]. Mitochondrial and nuclear DNA damage levels and mtDNA copy number were measured as previously described [78]. All PCR conditions and primer sequences can be found in Table S1.
5.3. ATP levels and oxygen consumption
ATP levels were measured in two different strains by two methods. First, using the JK1107 glp-1(q224) strain, ATP levels were measured as described in [79] using the Molecular Probes ATP determination Kit (Invitrogen/Life Technologies, Carlsbad, CA, USA). Second, the firefly luciferase expressing PE255 glp-4(bn2) strain was used to investigate relative, steady state ATP levels in vivo, in live nematodes at 2, 4, 6, 8, 10 and 12 days post final UVC dose, as previously described [80]. Details of both protocols are available in the supplemental methods section. Oxygen consumption was measured using a Seahorse Biosciences XFe24 extracellular flux analyzer as described [81].
5.4. Gene expression assays
RNA was isolated from between 1000 and 2000 nematodes according to the Qiagen RNeasy Min Kit protocol. cDNA was created from 100 ng of the isolated RNA using the High Capacity Reverse Transcription kit (Life Technologies) per the manufacturer's instructions. Gene expression was measured via real-time PCR using the Power SYBR Green PCR Master Mix (Life Technologies). Changes in expression levels were calculated based on the standard delta-delta-cT method, compared to housekeeping genes cdc-42 and pmp-3. Primer sequences and PCR conditions can be found in Table S2.
5.5. Targeted metabolomics
Nematodes (approximately 7500 for each sample) were washed, incubated in K-medium at 20 °C for 30 min to allow for gut clearance, and washed twice in ice-cold PBS. Worms were resuspended in 0.6% formic acid and stored at −80 °C. Samples were thawed on ice, lysed by sonication and aliquots were removed for total protein determination. 270 μL acetonitrile was added to each sample, and they were vortexed for 1 min and centrifuged at 15,000×g for 10 min to pellet proteins. Amino acids, acylcarnitines and organic acids were analyzed using stable isotope dilution technique. Amino acids and acylcarnitine measurements were made by flow injection tandem mass spectrometry using sample preparation methods described previously [82,83]. The data were acquired using a Waters TQD mass spectrometer equipped with AcquityTM UPLC system and controlled by MassLynx 4.1 operating system (Waters, Milford, MA). Organic acids were quantified using methods described previously [84] employing Trace Ultra GC coupled to ISQ MS operating under Xcalibur 2.2 (Thermo Fisher Scientific, Austin, TX). Metabolite levels were normalized to total protein as determined by BCA (Thermo Scientific, Rockford, IL) and fold changes compared to day 4 control samples were calculated.
5.6. Mitochondrial isolation and oxyblot
Mitochondria were isolated essentially as described [85]. Briefly, worms were washed off plates, rinsed twice in K-media and allowed to clear guts for 30 min in K-medium at room temperature. All steps from here were conducted on ice. Worms were washed once in K-medium and twice in 10 mL MSM-E buffer (MSM-E − 150 mL MSM (=40.08 g mannitol, 23.96 g sucrose, 1.047 g MOPS, 1L milliQ H2O, pH 7.4) plus 3 mL 0.1 M EDTA), resuspended in 1 mL MSM-E and lysed in a glass-Teflon potter homogenizer. Worms were microscopically monitored for lysis. Once sufficiently lysed, one volume MSM-EB (50 mL MSM-E + 0.2 g fatty acid-free BSA) was added and samples were centrifuged at 300 x G for 10 min. Supernatant was transferred to a fresh tube and kept, and pellet was re-extracted. Supernatants from the two extractions were combined and spun at 7000 x G for 10 min to yield the mitochondrial pellet, and supernatant samples were taken as the cytoplasmic fraction. Mitochondrial pellets were washed once in MSM-E and once in MSM. Oxyblot was performed following the manufacturer's instructions (MilliporeSigma, Burlington, MA).
5.7. Growth, antioxidant rescue, protein translation exacerbation, and lethality assays
Laval growth was assessed by measuring nematode size at 72 h post-exposure, using either a COPAS Biosorter (Union Biometrica, Holliston, MA), which measures extinction and time of flight of individual C. elegans [46], or microscopic imaging. For the latter, aliquots of nematodes were frozen, thawed and imaged at 10× magnification on a Zeis Axioskop. Nematode length was measured using NIS elements BR software (Nikon Inc. Melville, NY, USA). In many cases, growth is presented as percent control to permit comparisons between the methods and between strains that may develop at different rates under control conditions. In some experiments, larvae were staged (L1 through adult) at 24, 48, 72, and 96 h post-final exposure. For chemical exacerbation experiments, the mitochondrial translation inhibitors doxycycline and chloramphenicol were added after plates cooled, and nematodes were exposed to chemicals throughout development after UVC exposure. For trolox, MitoQ, and N-acetyl cysteine antioxidant rescue experiments, compounds were added to the medium prior to pouring plates, nematodes were exposed to antioxidants throughout development after UVC exposure, and size was measured on the COPAS Biosorter. Lethality assays were conducted 4 days after the 3rd UVC exposure. 10–15 nematodes were transferred to plates containing either rotenone or paraquat that were seeded with UVC-inactivated UvrA deficient OP50 E. coli to reduce bacterial metabolism of the chemicals [86]. Nematodes were scored for survival 24 h later and were counted as dead if they did not respond (move) to gentle prodding with a worm pick.
5.8. Lifespan analysis
Lifespan was determined on K agar plates with an OP50 lawn at 20 °C. Approximately 25 synchronized (as above) L1 nematodes were plated per condition were assayed in triplicate experiments. Beginning one day after reaching L4 and continuing until reproduction stopped, all adults were transferred daily onto a new plate, leaving offspring on the old plate. Nematodes were monitored daily by tapping the adults on the head. Animals were considered dead if no movement was observed following repeated probing. Individual lifespans were calculated from egg (day 0) until death.
5.9. Analysis of movement
Synchronized glp-1 L1 larvae were obtained by bleach-sodium hydroxide isolation of eggs (described above), transferred to unseeded no-peptone K-agar, and exposed to 3 sequential doses, 24 h apart, of 0, 3, or 4 J/m2 UVC as described above. Next, nematodes were transferred to K-agar plates seeded with OP50 and were grown at 25° for either 4 or 8 days. For each treatment group, six 60-s videos of nematode locomotion were recorded on a Nikon SMZ1500 stereomicroscope using NIS-Elements BR software. Analysis was completed using Fiji/ImageJ. Nematode pathways were constructed using the first 10 s of each video and quantified using the Analyze Particles function. Pathway area was normalized to mean nematode size for each treatment group.
5.10. Measurement of mitochondrial unfolded protein response
UPRmt activation was measured in hsp-6 (strain SJ4100) and hsp-60 (strain SJ4058) GFP fusion reporter strain 4 days post 2.5 or 5 J/m2 UVC exposure. Chloramphenicol exposure was included as a positive control. Worms were washed with K-medium, and approximately 100 worms in 100 μL K-medium were aliquoted into wells of a white 96 well plate (70 worms per well for SW4100 chloramphenicol exposure). GFP fluorescence was measured from 4 wells on a FLUOstar Optima microplate reader and adjusted for number of worms per well.
5.11. Statistical analysis
Specific analyses are indicated in the manuscript text of figure legends. In Figures, error bars indicate standard errors of the mean. In any case where asterisks are employed, p ≤ 0.05. Correction for multiple comparisons is indicated in the figure legends.
Author contributions
Kathleen A. Hershberger: formal analysis, writing - original draft preparation, writing - review and editing, visualization. John P. Rooney: conceptualization, methodology, formal analysis, investigation, writing - original draft preparation. Elena A. Turner: investigation. Lauren J. Donoghue: investigation. Rakesh Bodhicharla: investigation. Laura L. Maurer: investigation. Ian T. Ryde: investigation. Jina J Kim: investigation. Rashmi Joglekar: investigation. Jonathan D. Hibshman: investigation, writing - review and editing. Latasha L. Smith: investigation. Dhaval P. Bhatt: investigation. Olga R. Ilkayeva: methodology, investigation, resources. Matthew D. Hirschey: supervision. Joel N. Meyer: conceptualization, writing - review and editing, supervision, project administration, funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the National Institute of Environmental Health Sciences (R01ES017540, T32ES021432, and P42ES010356). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Tracey Crocker for assistance with the doxycycline and chloramphenicol experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102000.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chinnery P.F. Mitochondrial disease in adults: what's old and what's new? EMBO Mol. Med. 2015;7(12):1503–1512. doi: 10.15252/emmm.201505079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suomalainen A., Isohanni P. Mitochondrial DNA depletion syndromes--many genes, common mechanisms. Neuromuscul. Disord. : NMD. 2010;20(7):429–437. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dykens J.A., Will Y. The significance of mitochondrial toxicity testing in drug development. Drug Discov. Today. 2007;12(17–18):777–785. doi: 10.1016/j.drudis.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Landrigan P.J., Fuller R., Acosta N.J.R., Adeyi O., Arnold R., Basu N.N.…Zhong M. The Lancet Commission on pollution and health. Lancet. 2018;391(10119):462–512. doi: 10.1016/S0140-6736(17)32345-0. [DOI] [PubMed] [Google Scholar]
- 6.Braun J.M., Gray K. Challenges to studying the health effects of early life environmental chemical exposures on children's health. PLoS Biol. 2017;15(12) doi: 10.1371/journal.pbio.2002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross L., Birnbaum L.S. Regulating toxic chemicals for public and environmental health. PLoS Biol. 2017;15(12) doi: 10.1371/journal.pbio.2004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer J.N., Hartman J.H., Mello D.F. Mitochondrial toxicity. Toxicol. Sci. 2018;162(1):15–23. doi: 10.1093/toxsci/kfy008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roubicek D.A., de Souza-Pinto N.C. Mitochondria and mitochondrial DNA as relevant targets for environmental contaminants. Toxicology. 2017;391:100–108. doi: 10.1016/j.tox.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Graziewicz M.A., Sayer J.M., Jerina D.M., Copeland W.C. Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 2004;32(1):397–405. doi: 10.1093/nar/gkh213. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14729924 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasiviswanathan R., Gustafson M.A., Copeland W.C., Meyer J.N. Human mitochondrial DNA polymerase gamma exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 2012;287(12):9222–9229. doi: 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer J.N., Leung M.C., Rooney J.P., Sendoel A., Hengartner M.O., Kisby G.E., Bess A.S. Mitochondria as a target of environmental toxicants. Toxicol. Sci. 2013;134(1):1–17. doi: 10.1093/toxsci/kft102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bess A.S., Crocker T.L., Ryde I.T., Meyer J.N. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung M.C., Rooney J.P., Ryde I.T., Bernal A.J., Bess A.S., Crocker T.L.…Meyer J.N. Effects of early life exposure to ultraviolet C radiation on mitochondrial DNA content, transcription, ATP production, and oxygen consumption in developing Caenorhabditis elegans. BMC Pharmacol Toxicol. 2013;14:9. doi: 10.1186/2050-6511-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bess A.S., Ryde I.T., Hinton D.E., Meyer J.N. UVC-induced mitochondrial degradation via autophagy correlates with mtDNA damage removal in primary human fibroblasts. J. Biochem. Mol. Toxicol. 2013;27(1):28–41. doi: 10.1002/jbt.21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G.…Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–2401. doi: 10.1126/science.1077780. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12471266 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 17.Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G.…Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Canto C.…Auwerx J. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng L.F., Ng L.T., van Breugel M., Halliwell B., Gruber J. Mitochondrial DNA damage does not determine C. elegans lifespan. Front. Genet. 2019;10:311. doi: 10.3389/fgene.2019.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rea S.L., Ventura N., Johnson T.E. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5(10):e259. doi: 10.1371/journal.pbio.0050259. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17914900 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barouki R., Gluckman P.D., Grandjean P., Hanson M., Heindel J.J. Developmental origins of non-communicable disease: implications for research and public health. Environ. Health. 2012;11:42. doi: 10.1186/1476-069X-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandjean P., Barouki R., Bellinger D.C., Casteleyn L., Chadwick L.H., Cordier S.…Heindel J.J. Life-long implications of developmental exposure to environmental stressors: new perspectives. Endocrinology. 2015;156(10):3408–3415. doi: 10.1210/EN.2015-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozoya O.A., Xu F., Grenet D., Wang T., Grimm S.A., Godfrey V.…Santos J.H. Single nucleotide resolution analysis reveals pervasive, long-lasting DNA methylation changes by developmental exposure to a mitochondrial toxicant. Cell Rep. 2020;32(11):108131. doi: 10.1016/j.celrep.2020.108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimble J., Crittenden S.L. vols. 1–14. WormBook; 2005. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18050413 (Germline Proliferation and its Control). Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang W.Y., Lemire B.D. Mitochondrial genome content is regulated during nematode development. Biochem. Biophys. Res. Commun. 2002;291(1):8–16. doi: 10.1006/bbrc.2002.6394. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11829454 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 26.Cline S.D. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim. Biophys. Acta. 2012;1819(9–10):979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shpilka T., Haynes C.M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018;19(2):109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 28.Tsang W.Y., Sayles L.C., Grad L.I., Pilgrim D.B., Lemire B.D. Mitochondrial respiratory chain deficiency in Caenorhabditis elegans results in developmental arrest and increased life span. J. Biol. Chem. 2001;276(34):32240–32246. doi: 10.1074/jbc.M103999200. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11410594 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 29.Luz A.L., Smith L.L., Rooney J.P., Meyer J.N. Seahorse Extracellular Flux-based analysis of cellular respiration in Caenorhabditis elegans. Current Protocols in Toxicology. 2015;66 doi: 10.1002/0471140856.tx2507s66. 25.27.21-25.27.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braeckman B.P., Houthoofd K., De Vreese A., Vanfleteren J.R. Assaying metabolic activity in ageing Caenorhabditis elegans. Mech. Ageing Dev. 2002;123(2–3):105–119. doi: 10.1016/s0047-6374(01)00331-1. Retrieved from <Go to ISI>://000177152000008. [DOI] [PubMed] [Google Scholar]
- 31.Jastroch M., Divakaruni A.S., Mookerjee S., Treberg J.R., Brand M.D. Mitochondrial proton and electron leaks. Essays Biochem. 2010;47:53–67. doi: 10.1042/bse0470053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller B., Lewis N., Adeniyi T., Leese H.J., Brison D.R., Sturmey R.G. Application of extracellular flux analysis for determining mitochondrial function in mammalian oocytes and early embryos. Sci Rep-Uk. 2019;9:16778. doi: 10.1038/s41598-019-53066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apfeld J., O'Connor G., McDonagh T., DiStefano P.S., Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18(24):3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Gilst M.R., Hadjivassiliou H., Jolly A., Yamamoto K.R. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3(2):e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C., Chen H., Zhang J., Hong Y., Ding X., Ying W. Malate-aspartate shuttle mediates the intracellular ATP levels, antioxidation capacity and survival of differentiated PC12 cells. Int J Physiology Pathophysiol Pharmacol. 2014;6:109–114. [PMC free article] [PubMed] [Google Scholar]
- 37.Liemburg-Apers D.C., Willems P.H.G.M., Koopman W.J.H., Grefte S. Interactions between mitochondrial reactive oxygen species and cellular glucose metabolism. Arch. Toxicol. 2015;89:1209–1226. doi: 10.1007/s00204-015-1520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blajszczak C., Bonini M.G. Mitochondria targeting by environmental stressors: implications for redox cellular signaling. Toxicology. 2017;391:84–89. doi: 10.1016/j.tox.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244(22):6049–6055. https://www.ncbi.nlm.nih.gov/pubmed/5389100 Retrieved from. [PubMed] [Google Scholar]
- 41.Gruber J., Ng L.F., Fong S., Wong Y.T., Koh S.A., Chen C.B., Halliwell B. Mitochondrial changes in ageing Caenorhabditis elegans--what do we learn from superoxide dismutase knockouts? PloS One. 2011;6(5) doi: 10.1371/journal.pone.0019444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal S., Ganesh S. Perinuclear mitochondrial clustering, increased ROS levels, and HIF1 are required for the activation of HSF1 by heat stress. J. Cell Sci. 2020;133(13):jcs245589. doi: 10.1242/jcs.245589. [DOI] [PubMed] [Google Scholar]
- 43.Yang W., Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8(12) doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn J.-M., Eom H.-J., Yang X., Meyer J.N., Choi J. Comparative toxicity of silver nanoparticles on oxidative stress and DNA damage in the nematode, Caenorhabditis elegans. Chemosphere. 2014;108:343–352. doi: 10.1016/j.chemosphere.2014.01.078. [DOI] [PubMed] [Google Scholar]
- 45.Benedetti M.G., Foster A.L., Vantipalli M.C., White M.P., Sampayo J.N., Gill M.S.…Lithgow G.J. Compounds that confer thermal stress resistance and extended lifespan. Exp. Gerontol. 2008;43(10):882–891. doi: 10.1016/j.exger.2008.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X., Gondikas A.P., Marinakos S.M., Auffan M., Liu J., Hsu-Kim H., Meyer J.N. Mechanism of silver nanoparticle toxicity is dependent on dissolved silver and surface coating in Caenorhabditis elegans. Environ. Sci. Technol. 2012;46(2):1119–1127. doi: 10.1021/es202417t. [DOI] [PubMed] [Google Scholar]
- 47.Luz A.L., Godebo T.R., Smith L.L., Leuthner T.C., Maurer L.L., Meyer J.N. Deficiencies in mitochondrial dynamics sensitize Caenorhabditis elegans to arsenite and other mitochondrial toxicants by reducing mitochondrial adaptability. Toxicology. 2017;387:81–94. doi: 10.1016/j.tox.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng L.F., Gruber J., Cheah I.K., Goo C.K., Cheong W.F., Shui G., Sit K.P., Wenk M.R., Halliwell B. The mitochondria-targeted antioxidant MitoQ extends lifespan and improves healthspan of a transgenic Caenorhabditis elegans model of Alzheimer disease. Free Radical Biol. Med. 2014;71:390–401. doi: 10.1016/j.freeradbiomed.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 50.An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17(15):1882–1893. doi: 10.1101/gad.1107803. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12869585 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin K., Dorman J.B., Rodan A., Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–1322. doi: 10.1126/science.278.5341.1319. https://www.ncbi.nlm.nih.gov/pubmed/9360933 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 52.Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J.…Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424(6946):277–283. doi: 10.1038/nature01789. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12845331 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 53.Yee C., Yang W., Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157(4):897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dingley S., Polyak E., Lightfoot R., Ostrovsky J., Rao M., Greco T.…Falk M.J. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 2010;10(2):125–136. doi: 10.1016/j.mito.2009.11.003. S1567-7249(09)00172-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yun J., Finkel T. Mitohormesis. Cell Metab. 2014;19(5):757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durieux J., Wolff S., Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zubovych I.O., Straud S., Roth M.G. Mitochondrial dysfunction confers resistance to multiple drugs in Caenorhabditis elegans. Mol. Biol. Cell. 2010;21(6):956–969. doi: 10.1091/mbc.E09-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rauthan M., Pilon M. A chemical screen to identify inducers of the mitochondrial unfolded protein response in C. elegans. Worm. 2015;4(4) doi: 10.1080/21624054.2015.1096490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labbadia J., Brielmann R.M., Neto M.F., Lin Y.F., Haynes C.M., Morimoto R.I. Mitochondrial stress restores the heat shock response and prevents proteostasis collapse during aging. Cell Rep. 2017;21(6):1481–1494. doi: 10.1016/j.celrep.2017.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calabrese E.J. Hormetic mechanisms. Crit. Rev. Toxicol. 2013;43(7):580–606. doi: 10.3109/10408444.2013.808172. [DOI] [PubMed] [Google Scholar]
- 61.Poirier M.C., Gibbons A.T., Rugeles M.T., Andre-Schmutz I., Blanche S. Fetal consequences of maternal antiretroviral nucleoside reverse transcriptase inhibitor use in human and nonhuman primate pregnancy. Curr. Opin. Pediatr. 2015;27(2):233–239. doi: 10.1097/MOP.0000000000000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Nguyen P., Baris T.Z., Poirier M.C. Molecular analysis of mitochondrial compromise in rodent cardiomyocytes exposed long term to nucleoside reverse transcriptase inhibitors (NRTIs) Cardiovasc. Toxicol. 2012;12(2):123–134. doi: 10.1007/s12012-011-9148-5. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y., Shim Park E., Gibbons A.T., Shide E.D., Divi R.L., Woodward R.A., Poirier M.C. Mitochondrial compromise in 3-year old patas monkeys exposed in utero to human-equivalent antiretroviral therapies. Environ. Mol. Mutagen. 2016;57(7):526–534. doi: 10.1002/em.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berthiaume J.M., Wallace K.B. Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol. Toxicol. 2007;23(1):15–25. doi: 10.1007/s10565-006-0140-y. [DOI] [PubMed] [Google Scholar]
- 65.Wood C.E., Hester S.D., Chorley B.N., Carswell G., George M.H., Ward W.…Deangelo A.B. Latent carcinogenicity of early-life exposure to dichloroacetic acid in mice. Carcinogenesis. 2015;36(7):782–791. doi: 10.1093/carcin/bgv057. [DOI] [PubMed] [Google Scholar]
- 66.Hudson M.M., Ness K.K., Gurney J.G., Mulrooney D.A., Chemaitilly W., Krull K.R.…Robison L.L. Clinical ascertainment of health outcomes among adults treated for childhood cancer. J. Am. Med. Assoc. 2013;309(22):2371–2381. doi: 10.1001/jama.2013.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luz A.L., Meyer J.N. Effects of reduced mitochondrial DNA content on secondary mitochondrial toxicant exposure in Caenorhabditis elegans. Mitochondrion. 2016;30:255–264. doi: 10.1016/j.mito.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hernandez-Garcia D., Wood C.D., Castro-Obregon S., Covarrubias L. Reactive oxygen species: a radical role in development? Free Radic. Biol. Med. 2010;49(2):130–143. doi: 10.1016/j.freeradbiomed.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 69.Kumsta C., Thamsen M., Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxidants Redox Signal. 2011;14(6):1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rooney J.P., Luz A.L., Gonzalez-Hunt C.P., Bodhicharla R., Ryde I.T., Anbalagan C., Meyer J.N. Effects of 5 '-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Exp. Gerontol. 2014;56:69–76. doi: 10.1016/j.exger.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shokolenko I., Venediktova N., Bochkareva A., Wilson G.L., Alexeyev M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37(8):2539–2548. doi: 10.1093/Nar/Gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vafai S.B., Mootha V.K. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491(7424):374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 73.Menzies R.A., Gold P.H. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 1971;246(8):2425–2429. https://www.ncbi.nlm.nih.gov/pubmed/5553400 Retrieved from. [PubMed] [Google Scholar]
- 74.Van Laar V.S., Arnold B., Cassady S.J., Chu C.T., Burton E.A., Berman S.B. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 2011;20(5):927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coskun P., Wyrembak J., Schriner S.E., Chen H.W., Marciniack C., Laferla F., Wallace D.C. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim. Biophys. Acta. 2012;1820(5):553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer J.N., Chan S.S.L. Sources, mechanisms, and consequences of chemical-induced mitochondrial toxicity. Toxicology. 2017 doi: 10.1016/j.tox.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewis J.A., Fleming J.T. Basic culture methods. In: Epstein H.F., Shakes D.C., editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press; San Digo, CA: 1995. pp. 3–29. [Google Scholar]
- 78.Gonzalez-Hunt C.P., Rooney J.P., Ryde I.T., Anbalagan C., Joglekar R., Meyer J.N. PCR-based analysis of mitochondrial DNA copy number, mitochondrial DNA damage, and nuclear DNA damage. Curr Protoc Toxicol. 2016;67 doi: 10.1002/0471140856.tx2011s67. 20 11 21-20 11 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brys K., Castelein N., Matthijssens F., Vanfleteren J.R., Braeckman B.P. Disruption of insulin signalling preserves bioenergetic competence of mitochondria in ageing Caenorhabditis elegans. BMC Biol. 2010;8:91. doi: 10.1186/1741-7007-8-91. 1741-7007-8-91 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagido C., Pettitt J., Flett A., Glover L.A. Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 2008;8:7. doi: 10.1186/1472-6793-8-7. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18384668 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luz A.L., Rooney J.P., Kubik L.L., Gonzalez C.P., Song D.H., Meyer J.N. Mitochondrial morphology and fundamental parameters of the mitochondrial respiratory chain are altered in Caenorhabditis elegans strains deficient in mitochondrial dynamics and homeostasis processes. PloS One. 2015;10(6) doi: 10.1371/journal.pone.0130940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.An J., Muoio D.M., Shiota M., Fujimoto Y., Cline G.W., Shulman G.I.…Newgard C.B. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med. 2004;10(3):268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 83.Ferrara C.T., Wang P., Neto E.C., Stevens R.D., Bain J.R., Wenner B.R.…Attie A.D. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4(3) doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jensen M.V., Joseph J.W., Ilkayeva O., Burgess S., Lu D., Ronnebaum S.M.…Newgard C.B. Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J. Biol. Chem. 2006;281(31):22342–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- 85.Falk M.J., Kayser E.B., Morgan P.G., Sedensky M.M. Mitochondrial complex I function modulates volatile anesthetic sensitivity in C. elegans. Curr. Biol. 2006;16(16):1641–1645. doi: 10.1016/j.cub.2006.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maurer L.L., Ryde I.T., Yang X., Meyer J.N. Caenorhabditis elegans as a model for toxic effects of nanoparticles: lethality, growth, and reproduction. Curr Protoc Toxicol. 2015;66 doi: 10.1002/0471140856.tx2010s66. 20 10 21-20 10 25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.