Abstract
BACKGROUND AND PURPOSE: The molecular characteristics of intracranial aneurysms are still poorly documented. A rabbit elastase aneurysm model has been helpful in the evaluation of devices and strategies involved in endovascular treatment of aneurysms. The goal of this project was to document the molecular changes, assessed by gene chip microarrays, associated with the creation of aneurysms in this model compared with the contralateral carotid artery.
MATERIALS AND METHODS: A microarray of rabbit genes of interest was constructed using rabbit nucleotide sequences from GenBank. Elastase-induced saccular aneurysms were created at the origin of the right common carotid artery in 4 rabbits. Twelve weeks after aneurysm creation, RNA was isolated from the aneurysm as well as the contralateral common carotid artery and used for microarray experiments. Reverse transcription-polymerase chain reaction (RT-PCR) was performed on 1 animal as a confirmatory test.
RESULTS: Ninety-six (46%) of 209 genes in the microarray were differentially expressed in the rabbit aneurysm compared with the contralateral common carotid artery. In general, differential gene expression followed specific molecular pathways. Similarities were found between rabbit aneurysms and human intracranial aneurysms, including increased metalloproteinase activity and decreased production of the extracellular matrix. RT-PCR results confirmed the differential expression found by the gene chip microarray.
CONCLUSIONS: The molecular characteristics of the rabbit elastase-induced saccular aneurysm are described. The rabbit aneurysm model shares some molecular features with human intracranial aneurysms. Future studies can use the rabbit model and the new rabbit gene chip microarray to study the molecular aspects of saccular aneurysms.
We are entering the genetic era of medicine, where ideas such as genetic screening and gene-directed therapy are becoming mainstream technologies. Genetic medicine may also extend to the field of intracranial aneurysms.1-5 However, genetic studies on intracranial aneurysms are limited because of the difficulty of obtaining tissue. Animal models with well-defined molecular characteristics may be of value in the study of the molecular pathophysiology of saccular aneurysms.
The rabbit elastase-induced saccular aneurysm model has been applied to the study of saccular aneurysms.6-9 In this model, saccular aneurysms are surgically created from the origin of the right common carotid artery using distal ligation of the artery and injection of elastase into the artery wall.9 The resultant aneurysms have a morphology and size similar to that of human intracranial aneurysms.8 These aneurysms are also histologically similar to human aneurysms in that they have lost their elastic membrane.8 Little is known about the molecular characteristics of these aneurysms.
The purpose of this study was to apply gene chip technology to delineate the molecular characteristics of saccular aneurysms in the rabbit model. Gene chip technology allows study of gene expression in hundreds of genes in many different molecular pathways. We initially created a rabbit gene chip microarray containing 209 genes that were each individually selected because of their relevance to vascular physiology or pathology. We applied this gene chip on 4 experimental aneurysms to compare the gene expression of tissue within the aneurysm cavity with that of the control, contralateral common carotid artery. We used reverse transcription-polymerase chain reaction (RT-PCR) as a confirmatory test to demonstrate the validity of the gene chip microarray results.
Materials and Methods
Selecting Genes for the Rabbit Gene Chip Microarray
We constructed a rabbit gene chip with the ability to detect differences in 209 genes (Table 1). To choose candidate genes, we initially searched the current literature on human intracranial aneurysms5,10-25 and abdominal aortic aneurysms.26-30 We identified approximately 400 genes of interest. These genes can be grouped into 7 molecular and physiologic themes, all relevant to vascular physiology and pathology: activation of the coagulation cascade,2,11,16 recruitment of inflammatory cells such as monocytes and macrophages,10,11 production of antioxidative genes,1-3,11,13,15,30-33 apoptosis of smooth muscle cells,12,34-37 production of proteolytic extracellular matrix enzymes,3,5,10,16,18,19,38,39 decreased production of the extracellular matrix,3,11,16,21-25,38 and activation of cell signaling cascades.10,12,26-29 We then searched GenBank to identify sequenced rabbit genes. Of the 400 genes of interest, we identified 209 rabbit genes that had been sequenced and posted on the GenBank data base. The microarray of oligonucleotides of interested genes was constructed commercially (Operon, Huntsville, Ala).
Table 1:
Genes that were included in the rabbit gene chip
| Proteolytic enzymes | a1-Antitrypsin,11,15,17 calpain,27 cathepsin B and D,10,27,29 collagenase,11,30 elastase,11 MMP-1,26 MMP-2,11,18 MMP-7,26 MMP-8,26,30 MMP-9,5,11,16,19,20,27-30 MMP-12,30 MMP-14,26 MT1-MMP,11,16 protease inhibitor,15,27 SPARC (also known as osteonectin),10 TIMP-2,11,19 TIMP-310 |
| Apoptosis of smooth muscle cells | Actinin,30 heat shock protein,26,27 c-Jun,12 caveolin,26 fas,26 myosin heavy chain,30 titin,27 tumor suppressor 53,26 |
| Inflammatory cell recruitment | ACE,11,14,15 albumin,27 angiotensin type 1 receptor,14 β-galactosidase-binding lectin,10 endothelial differentiation factor,26 HLA,10,15 IgG heavy chain,10 Ig-γ light chain,10 NFAT,27 vinculin,10 interferon γ,26,27 interleukins,26,27,29,30 MCP-1,26 neutrophil-activating protein,26 TNF26-28,30 |
| Coagulation cascade | Annexin III,26 factor XIII,11 haptoglobin, heme oxygenase,30 plasminogen activator inhibitor-1,11,26,30 PDGF-AA,14,26,27,29 platelet membrane glycoprotein IIIA,28 prothrombin, t-PA,16 thrombin receptor,26,27 thrombomodulin,26 thrombospondin 1,26 transferrin,27 u-PA,11,27 vitronectin15 |
| Extracellular matrix proteins | ADAM,27 bFGF,14 bone cartilage proteoglycan,26,29 BMP,26,27 collagen I,10,11,15,21,38 collagen III,10,11,15,21-24 collagen IV,10,11,15,21,25 collagen V,11,21 collagen VIII,11 fibronectin,10,11,38 fibromodulin,30 FK506 binding protein 12,27 insulin-like growth factor,26-28 laminin,11,15,28 lysyl oxidase,11 tenascin,11 TGF-β11,27 |
| Oxidative stress | Adenosine A3 receptor,27 alcohol dehydrogenase,26 apolipoprotein E,15,27,29 atrial natriuretic peptide,26 clusterin (apolipoprotein J),26 cytochrome P450,26 endothelin,26 GST,26 HIF-1, lipoxygenase,30 SOD,26 iNOS,11,13,27,30 VEGF,11 VEGF receptor11 |
| Cell signaling pathways | Acyl CoA binding protein,29 c-myc,26 calcineurin,27 calmodulin,27 CD86,27 elongation factor 1-δ,10 deoxyribonuclease,27 erythroblastic leukemia viral oncogene homolog 2,27 frizzled,27 glutamate receptor metabotropic,27 heparin-binding epidermal growth factor-like growth factor, insulin responsive glucose transporter 4,26 JNK MAPK,12,26 neuregulin,27 PLC,27 PKC,27,29 Serum- and glucocorticoid-inducible kinases ,27,29 protein phosphatase-1, Ras associated protein 2,27 RAL A GTP-binding protein,29 ras,14 retinoic acid receptor-β,26 rho/rac GEF,27,28 transcription factor egr1,26,27 tryptophan hydroxylase,27 tyrosine receptor kinase26 |
Note:—MMP indicates matrix metalloproteinase; MT1-MMP, membrane type metalloproteinase-1; SPARC, secreted protein acidic and rich in cysteine; TIMP, tissue inhibitor of metalloproteinase; ACE, angiotensin-converting enzyme; HLA, human leukocyte antigen; NFAT, nuclear factor of activated T cells; MCP, macrophage chemoattractant protein; TNF, tumor necrosis factor; ADAM, a disintegrin and metalloproteinase; PDGF, platelet-derived growth factor; u-PA, urokinase type plasminogen activator; bFGF, basic fibroblast growth factor; BMP, bone morphogenetic protein; TGF, transforming growth factor; SOD, superoxide dismutase; iNOS, inducible nitric-oxide synthase; VEGF, vascular endothelial growth factor; GST, glutathione S-transferase; HIF, hypoxia-inducible factor; JNK, c-Jun NH2-terminal kinase; MAPK, mitogen activated protein kinase; PLC, phospholipase C; PKC, protein kinase C; GEF, guanine nucleotide exchange factor.
Articles demonstrating the relevance of each gene to vascular pathophysiology are cited. The functions of these genes are grouped into 7 pathways that are relevant to intracranial aneurysms.
Rabbit Aneurysm Creation
Elastase-induced, saccular aneurysms were created in New Zealand white rabbits (body weight, 3–4 kg) using the rabbit elastase model. The Mayo Foundation Institutional Animal Care and Use Committee approved all procedures before initiation of the study. Detailed procedures for aneurysm creation have been described in depth elsewhere.9 In brief, anesthesia was induced with an intramuscular injection of ketamine, xylazine, and acepromazine (75, 5, and 1 mg/kg, respectively). Using sterile technique, the right common carotid artery (RCCA) was exposed and ligated distally. A 1–2-mm beveled arteriotomy was made, and a 5F vascular sheath was advanced retrograde in the RCCA to a point approximately 3 cm cephalad to the origin of RCCA. A 3F Fogarty balloon was advanced through the sheath to the level of the origin of the RCCA with fluoroscopic guidance and was inflated with iodinated contrast material. Porcine elastase (Worthington Biochemical Corporation, Lakewood, NJ) was incubated within the lumen of the common carotid above the inflated balloon for 20 minutes, after which the catheter, balloon, and sheath were removed. The RCCA was ligated below the sheath entry site, and the incision was closed. Subjects underwent intra-arterial digital subtraction angiography at the time of sacrifice with a 5F diagnostic catheter placed into the brachiocephalic artery from a femoral approach.
Tissue Harvest
Tissue was harvested 12 weeks after aneurysm creation. At the time of tissue harvest, the animals were deeply anesthetized and then euthanized by an overdose injection of pentobarbital. The entire aneurysm (approximately 1 cm long) and the contralateral common carotid artery were dissected free from the surrounding tissues and were then immediately frozen in liquid nitrogen. None of the aneurysms had clotted off or ruptured. The frozen tissues were stored at −70°C until the tissues were ready for RNA extraction.
RNA Extraction
RNA was isolated from frozen tissues by using the RNeasy fibrous tissue mini kit (QIAGEN, Valencia, Calif). The quantity of the RNA was measured using spectrophotometry, and the integrity of the RNA was confirmed by electrophoretic separation using the Agilent 2100 Bioanalyzer (Agilent, Palo Alto, Calif).
Fluorescent cRNA Synthesis (In Vitro Transcription)
Total RNA (1 μg) was amplified and synthesized into 2 μg of complementary RNA (cRNA). The amplification technique was tested to confirm proportional amplification of each RNA strand. cRNA was then labeled with red and green fluorescent dyes for the purpose of the microarray experiment (Low RNA Input Fluorescent RNA Amplification kit; Agilent).
Fluorescent cRNA Hybridization Protocol
Two micrograms of the aneurysm cRNA (labeled with the fluorescent red tag) and 2 μg of the common carotid artery cRNA (labeled with the fluorescent green tag) were then mixed and hybridized to the microarray. The colors were switched and the hybridization was repeated in duplicate to control for the effect of the dye tag on hybridization.
Scanning
The microarray slides were then scanned, and the computer reported the intensity of the red and green fluorescent dyes for each spot on the microarray (GenePix 4000B; Molecular Devices, Sunnyvale, Calif). For example, a red spot would mean that the aneurysm messenger RNA (mRNA) dominates, a green spot would mean that the carotid artery mRNA dominates, and a mixed color spot would mean that the mRNA is equally represented.
RT-PCR
To validate the results obtained in the microarray, RT-PCR was performed on 1 of the rabbit tissue samples. RNA (100 ng) was reverse-transcribed to complementary DNA (cDNA) using Superscript III (Invitrogen, Carlsbad, Calif). PCR was performed with primers specific for caveolin, matrix metalloproteinase (MMP)-2, MMP-9, vascular cell adhesion molecule 1 (VCAM-1), endothelin-1, tissue inhibitor of metalloproteinase-2 (TIMP-2), and β-actin on a Thermocycler (Applied Biosystems, Foster City, Calif) by using PCR kit (Invitrogen).
Statistical Analyses
Analyses were done using the base-2 logarithm transform of the median signal intensity, and all analyses were conducted using SAS Version 9 statistical software (SAS Institute, Cary, NC). Normalization of the data were performed using 2-channel fastlo, a semiparametric approach that corrects for intensity-dependent effects developed by Eckel et al.40 The parametric component consisted of additive main effect for gene. The nonparametric component consisted of a set of nonparametric loess smoothers, one for each array, dye, and block combination. The normalized signal intensity for each observation was estimated by subtracting the predicted values obtained from the nonparametric component from the original base-2 logarithm transform of the median signal intensity.
To test for differential expression between aneurysm and the control arteries, a mixed-effects linear model was fit for each gene. The normalized expression values were the dependent variable in the mixed-effects linear models, array and dye were fit as fixed effect co-variates, and rabbit was included as a random effect. The “t” test statistics and corresponding P values, calculated from a linear contrast, were used as a measure of the mean change in expression between treatment groups relative to the variability. The genes were ranked according to their P values, and genes with a P value < .05 were used to identify pathways for further investigation.
Results
In the gene chip microarray experiment, 96 (46%) of 209 genes demonstrated statistically significant differential expression between the aneurysm tissue and the control contralateral carotid artery tissue (P < .05). Table 2 lists the 96 differentially expressed genes in decreasing order of statistical significance.
Table 2:
The 96 genes that had a statistically significant (P < .05) differential expression between the aneurysm and the control artery
| Rank | Gene | +/− |
|---|---|---|
| 1 | Fast skeletal muscle troponin C mRNA | − |
| 2 | Haptoglobin mRNA | + |
| 3 | Endothelial differentiation gene 1 protein | + |
| 4 | Muscle α-actinin subunit, N′-terminal region | − |
| 5 | Matrix metalloproteinase 9 | + |
| 6 | Caveolin | − |
| 7 | Alcohol dehydrogenase, class I | + |
| 8 | Endothelin-1 | − |
| 9 | Myosin light chain 2 | |
| 10 | Osteoglycin | − |
| 11 | Type VIII collagen α -1 chain | − |
| 12 | Osteopontin | + |
| 13 | Neutrophil attractant/activation protein-1 (NAP-1) | − |
| 14 | Interleukin 1 receptor antagonist | + |
| 15 | Apolipoprotein E | + |
| 16 | IL-8 receptor | − |
| 17 | Cytochrome P4B1 isoform | − |
| 18 | Monocyte chemoattractant protein-1 (MCP-1) | + |
| 19 | Tissue inhibitor of metalloproteinase-4 (TIMP-4) | + |
| 20 | Myosin heavy chain (MHC) | − |
| 21 | Transforming growth factor-β 2 | − |
| 22 | Heme oxygenase 1 (HO1) | + |
| 23 | Al adenosine receptor | + |
| 24 | Osteonectin | + |
| 25 | p53 protein | − |
| 26 | Peroxisome proliferator activated receptor (PPAR) gamma 3 | + |
| 27 | Cathepsin L | + |
| 28 | Apolipoprotein B | − |
| 29 | Elongation factor 1 β | + |
| 30 | Calmodulin-dependent protein kinase II-delta dash | − |
| 31 | Secreted frizzled-related protein 2 | + |
| 32 | Glutathione S-transferase | − |
| 33 | Vascular cell adhesion molecule (VCAM-1) | + |
| 34 | Endothelin converting enzyme (ECE1) | − |
| 35 | Platelet-derived growth factor-β | + |
| 36 | Matrix metalloproteinase-12 | + |
| 37 | Decorin=proteoglycan | − |
| 38 | Galectin-3 | + |
| 39 | Glycoprotein IIb (GPIIb) | + |
| 40 | Metallo-elastase, macrophage | + |
| 41 | Fertilin α subunit | − |
| 42 | Growth response 1 zinc-finger transcription factor | − |
| 43 | α2 type I collagen | − |
| 44 | α3 type IV collagen | + |
| 45 | Apolipoprotein D | + |
| 46 | Endothelin B receptor | − |
| 47 | Insulin-like growth factor 1 precursor (IGF-1) | + |
| 48 | Neuregulin 1 α isoform | − |
| 49 | Protein phosphatase-1 catalytic subunit (EC 3.1.3.-) | + |
| 50 | Heme oxygenase-2 | − |
| 51 | Class II alcohol dehydrogenase, isozyme 1 | + |
| 52 | Functional messenger ribonucleoprotein particle major protein p50 | − |
| 53 | factor XI | − |
| 54 | Rho-associated-Ser/Thr kinase | + |
| 55 | Acyl CoA binding protein | − |
| 56 | Protein kinase Cβ (PKC-β) | + |
| 57 | Bone morphogenetic protein 2 (BMP-2) | − |
| 58 | Heat shock protein 47 | + |
| 59 | Retinoic acid receptor RXR α mRNA | + |
| 60 | α 1 type X collagen | + |
| 61 | Coagulation factor VII | − |
| 62 | Interleukin-2 | − |
| 63 | Elongation factor 1 delta (Rabef1D) | + |
| 64 | fas antigen spliced variant | − |
| 65 | glucose transporter type 4 (GLUT4) | − |
| 66 | Cellular disintegrin ADAM 6e (ADAM 6e) | − |
| 67 | Atrial natriuretic polypeptides | + |
| 68 | Matrix metalloproteinase-2 | + |
| 69 | Major histocompatibility complex class II RLA-DR-α gene | + |
| 70 | Tissue inhibitor of metalloproteinase-2 (TIMP2) | + |
| 71 | Adenosine A3 receptor mRNA | − |
| 72 | Bone morphogenetic protein 7 | − |
| 73 | Forkhead transcription factor L2 (FoxL2) gene | − |
| 74 | Neuronal nitric oxide synthase NOS1 | + |
| 75 | Tissue-type plasminogen activator mRNA | + |
| 76 | Endothelin receptor type A | − |
| 77 | Phosphorylase kinase α subunit mRNA | − |
| 78 | α-1-antitrypsin | + |
| 79 | Thrombomodulin precursor (Thbd) | − |
| 80 | Neutrophil-activating peptide 78 (ENA-78) | − |
| 81 | Calcineurin A β | − |
| 82 | α-1 type V collagen (COL5A1) | − |
| 83 | Interferon γ | − |
| 84 | NADPH-cytochrome P450 reductase | − |
| 85 | Fibroblast growth factor | − |
| 86 | Thrombospondin 2-like protein (THBS2) | + |
| 87 | Tryptophan hydroxylase | − |
| 88 | Hyperoxia induced (H1) 1 gene encoding a TIMP | + |
| 89 | Inducible nitric oxide synthase (NOS2) | − |
| 90 | Glycoprotein IIIa (GPIIIa) | + |
| 91 | Elongation factor 1 γ | + |
| 92 | CD86 | − |
| 93 | Calmodulin-dependent protein kinase II-γ dash2 | − |
| 94 | Protein inhibitor of neuronal nitric oxide synthase | + |
| 95 | Vascular endothelial growth factor receptor 3 | + |
| 96 | Ksp-cadherin | − |
Note:—The genes are ranked according to the degree of significance of the differential expression. + indicates that the gene was overexpressed in the aneurysm; −, the gene was underexpressed in the aneurysm.
Table 3 categorizes the genes into their respective molecular pathways. A few trends in rabbit aneurysm gene expression are recognizable from these data. The proteolytic enzymes are activated (11 of 12 genes), many of the extracellular matrix (ECM) genes are down-regulated (9 of 13 genes), and the smooth muscle genes are down-regulated (4 of 4 genes).
Table 3:
The differentially expressed genes are grouped into their respective molecular pathways
| Proteolytic enzymes | Apoptosis of smooth muscle cells | ||
| α-1-Antitrypsin | + | Caveolin | − |
| Cathepsin L | + | Fas antigen spliced variant | − |
| Cellular disintegrin ADAM 6e | − | Fast skeletal muscle troponin C mRNA | − |
| Hyperoxia induced (HI) 1 gene encoding a TIMP | + | Galectin-3 | + |
| Matrix metalloproteinase-2 | + | Heat shock protein 47 | + |
| Matrix metalloproteinase-9 | + | Muscle α -actinin subunit, N′-terminal region | − |
| Matrix metalloproteinase-12 | + | Myosin heavy chain (MHC) | − |
| Metallo-elastase, macrophage | + | Myosin light chain 2 | − |
| Osteonectin = SPARC | + | PPAR γ 3 | + |
| Tissue inhibitor of metalloproteinase-2 (TIMP-2) | + | p53 protein | − |
| Tissue inhibitor of metalloproteinase-4 (TIMP-4) | + | Secreted frizzled-related protein 2 | + |
| Coagulation cascade | Inflammatory cell recruitment | ||
| Coagulation factor VII | − | Adhesion molecule VCAM-1 (VCAM-1) | + |
| Factor XI | − | Endothelial differentiation gene 1 protein | + |
| Glycoprotein IIb (GPIIb) | + | Interferon γ | − |
| Glycoprotein IIIa (GPIIIa) | + | Interleukin-1 receptor antagonist | + |
| Haptoglobin mRNA | + | Interleukin-2 | − |
| Heme oxygenase-1 (HO1) | + | Interleukin-8 receptor | − |
| Heme oxygenase-2 | − | MHC class II RLA-DR-α gene | + |
| Platelet-derived growth factor-β | + | Monocyte chemoattractant protein-1 (MCP-1) | + |
| Thrombomodulin precursor (inhibitor of coagulation) | − | Neutrophil-activating peptide 78 (ENA-78) | − |
| Thrombospondin 2-like protein (THBS2) | + | Neutrophil attractant/activation protein-1 (NAP-1) | − |
| Tissue-type plasminogen activator mRNA | + | ||
| ECM proteins | Oxidative stress | ||
| α-1 type V collagen (COL5A1) | − | A1 Adenosine receptor | + |
| α-1 type X collagen | + | A3 Adenosine receptor | − |
| α-2 type I collagen | − | Alcohol dehydrogenase, class I | + |
| α-3 type IV collagen | +* | Apolipoprotein B | − |
| Bone morphogenetic protein 2 (BMP-2) | − | Apolipoprotein E | + |
| Bone morphogenetic protein 7 (BMP-7) | − | Atrial natriuretic polypeptide | + |
| Decorin=proteoglycan | − | Class II alcohol dehydrogenase, isozyme 1 | + |
| Fibroblast growth factor | − | Clone pApoD-L) apolipoprotein D (apoD) | + |
| Insulin-like growth factor 1 precursor (IGF-1) | + | CYP4B1 isoform | − |
| Osteoglycin | − | Endothelin-1 | − |
| Osteopontin | + | Endothelin B receptor | − |
| Transforming growth factor-β 2 | − | Endothelin converting enzyme (ECE1) | − |
| Type VIII collagen α -1 chain | − | Endothelin receptor type A | − |
| Glutathione S-transferase | − | ||
| Inducible nitric oxide synthase (NOS2) | − | ||
| NADPH-cytochrome P450 reductase | − | ||
| Neuronal nitric oxide synthase NOS1 | + | ||
| Protein inhibitor of neuronal nitric oxide synthase | + | ||
| Vascular endothelial growth factor receptor 3 | + |
Note:—+ indicates that the gene was overexpressed in the aneurysm; −, the gene was underexpressed in the aneurysm.
The RT-PCR results confirmed that VCAM-1, MMP-2, MMP-9, and TIMP-2 were all overexpressed in the aneurysm, whereas endothelin-1 and caveolin were underexpressed in the aneurysm (Fig 1). The RT-PCR–derived expression profile of these 6 genes matches the results of the gene chip microarray data.
Fig 1.
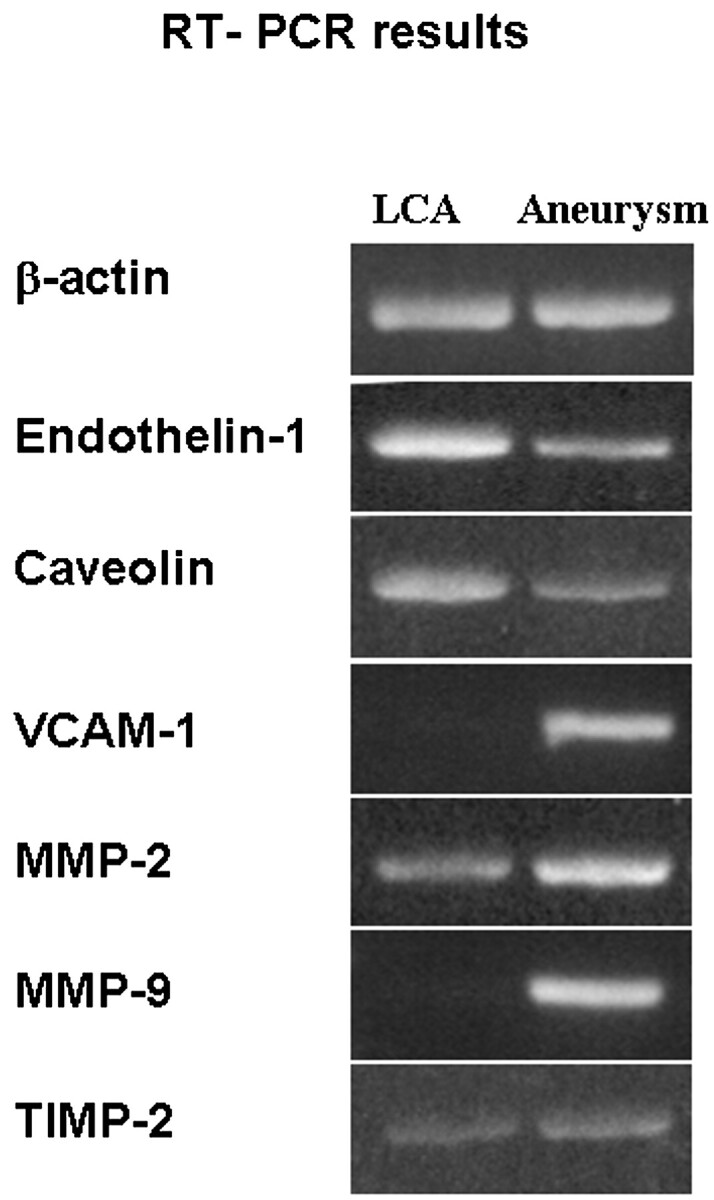
RT-PCR data. The bands reveal that endothelin and caveolin are underexpressed in aneurysms, whereas VCAM-1, MMP-2, MMP-9, and TIMP-2 are overexpressed in aneurysms. β-Actin is the control that is equally expressed in both the aneurysm and the control.
Discussion
This study gives a gross picture of the molecular characteristics of the elastase-induced rabbit aneurysm. Proteolytic activity is markedly elevated above control tissue, with activation of metalloproteinases, while the production of the extracellular matrix and smooth muscle genes are inhibited. Variables that could produce these molecular changes include exposure to elastase, altered shear stress due to the new morphology of the arterial wall, and the surgical operation itself.
Human aneurysms display molecular changes similar to those seen in this animal model. In humans, aneurysm tissue metalloproteinases are activated,19,20,39 production of the extracellular matrix is inhibited,21-25 and smooth muscle cell production is significantly decreased as a result of apoptosis.34-36 Because of these molecular similarities between the experimental rabbit aneurysms and human intracranial aneurysms, it is possible that the rabbit aneurysm model could be used to study the molecular pathophysiology of saccular aneurysms.
To our knowledge, this is the first report ever to use a gene chip microarray that was specifically made from rabbit oligonucleotide sequences and the first to study the genetic profile of rabbit aneurysms. The RT-PCR results are consistent with the gene chip microarray data, suggesting that the differential expression found by the gene chip microarrays is reliable. Gene chip microarray technology is so sensitive that the slight difference in homology across species can lead to significant changes in microarray binding.41 Consequently, we believe that the use of human gene chip microarrays on animals such as rabbits should be avoided.
Other animal models for intracranial aneurysms have been used to study the molecular nature of aneurysms. Most of these studies have focused on individual genes.13,35,42-45 However, in a recent study similar to our own, human gene chip microarray technology was used on the swine model to classify the molecular character of the swine aneurysm.46 Although their study was innovative, their data were collected using human gene chips and thus have the potential of bias because of the imperfect homology of genes across species. Furthermore, in our opinion, the swine model does not resemble human aneurysms as well as the rabbit model does.8 For example, the aneurysm in the swine model contains an intact internal elastic lamina and tunica media and the venous pouch that forms the swine aneurysm thromboses spontaneously.
This study has several limitations. First, the rabbit genome is not yet fully elucidated, and some of the genes of interest in intracranial aneurysm pathology are not yet sequenced. Consequently, we were not able to construct a gene chip that contained every gene of known significance to intracranial aneurysm pathology. However, if a specific gene was not in the data base, there was usually a closely related gene that we could include in its place. Second, this study was performed on only 4 aneurysms. Fortunately, even this small number of subjects yielded results that showed statistically significant differential expression in 96 genes. This high yield of differential expression reveals the care that was taken in the selection of candidate genes for the gene chip. Third, the contralateral left carotid artery is imperfect as a control because the flow in this artery is increased after right common carotid artery occlusion. The advantages of the contralateral carotid as a control are that it is from the same animal and it is the same type of vessel as the vessel used to create the aneurysm (they are both from common carotid arteries). Fourth, these data reveal genetic expression at the RNA level. Because the cell regulates protein expression at the levels of both transcription and translation, RNA levels do not necessarily correlate with protein levels. Future proteomic studies are necessary to confirm the results of this study.
The rabbit, as a molecular model, will be a valuable tool in future studies. We can do controlled experiments on rabbit aneurysms that would not be feasible in humans. For example, we can modulate blood pressure in rabbits, modify the shape of the aneurysm in rabbits, or use various medications in rabbits and then determine the molecular effects of these changes by using the rabbit gene chip microarray. Future experiments can also focus on many of the new genes of interest revealed by this microarray study. For example, differentially expressed genes such as atrial natriuretic polypeptide, monocyte chemoattractant protein, endothelin, and peroxisome proliferator-activated receptors34,36,47 will be particularly interesting given their known function and their logical connection to intracranial aneurysms. In summary, by combining the advantages of the rabbit model and the gene chip microarray, we will be a step closer to attaining an understanding of the molecular biology of intracranial aneurysms.
Conclusions
The molecular characteristics of the rabbit elastase-induced saccular aneurysm are described. The rabbit aneurysm model shares some molecular features with human intracranial aneurysms, including increased metalloproteinase activity, decreased production of the extracellular matrix, and decreased production of smooth muscle associated genes. This knowledge will be helpful in future studies that use the rabbit model and the new rabbit gene chip microarray to study the molecular effects of controlled experiments.
Footnotes
This work was presented at the 44th Annual Meeting of the American Society of Neuroradiology; April 29–May 5, 2006; San Diego, Calif.
References
- 1.Khurana VG, Smith LA, Baker TA, et al. Protective vasomotor effects of in vivo recombinant endothelial nitric oxide synthase gene expression in a canine model of cerebral vasospasm. Stroke 2002;33:782–89 [DOI] [PubMed] [Google Scholar]
- 2.Khurana VG, Sohni YR, Mangrum WI, et al. Section on cerebrovascular surgery: Galbraith Award: Endothelial nitric oxide synthase (eNOS) and heme oxygenase-1 (HO-1) gene polymorphisms predict susceptibility to aneurysmal subarachnoid hemorrhage (SAH) and post-SAH cerebral vasospasm. Clin Neurosurg 2004;51:343–50 [PubMed] [Google Scholar]
- 3.Ribourtout E, Raymond J. Gene therapy and endovascular treatment of intracranial aneurysms. Stroke 2004;35:786–93 [DOI] [PubMed] [Google Scholar]
- 4.Abrahams JM, Forman MS, Grady MS, et al. Delivery of human vascular endothelial growth factor with platinum coils enhances wall thickening and coil impregnation in a rat aneurysm model. AJNR Am J Neuroradiol 2001;22:1410–17 [PMC free article] [PubMed] [Google Scholar]
- 5.Peters DG, Kassam A, St Jean PL, et al. Functional polymorphism in the matrix metalloproteinase-9 promoter as a potential risk factor for intracranial aneurysm. Stroke 1999;30:2612–16 [DOI] [PubMed] [Google Scholar]
- 6.Stehbens WE. The ultrastructure of experimental aneurysms in rabbits. Pathology 1985;17:87–95 [DOI] [PubMed] [Google Scholar]
- 7.Stehbens WE. In re: histological and morphologic comparison of experimental aneurysms with human intracranial aneurysms. AJNR Am J Neuroradiol 2000;21:1769–73 [PMC free article] [PubMed] [Google Scholar]
- 8.Abruzzo T, Shengelaia GG, Dawson RC 3rd, et al. Histologic and morphologic comparison of experimental aneurysms with human intracranial aneurysms. AJNR Am J Neuroradiol 1998;19:1309–14 [PMC free article] [PubMed] [Google Scholar]
- 9.Altes TA, Cloft HJ, Short JG, et al. 1999 ARRS Executive Council Award. Creation of saccular aneurysms in the rabbit: a model suitable for testing endovascular devices. American Roentgen Ray Society. AJR Am J Roentgenol 2000;174:349–54 [DOI] [PubMed] [Google Scholar]
- 10.Peters DG, Kassam AB, Feingold E, et al. Molecular anatomy of an intracranial aneurysm: coordinated expression of genes involved in wound healing and tissue remodeling. Stroke 2001;32:1036–42 [DOI] [PubMed] [Google Scholar]
- 11.Krex D, Schackert HK, Schackert G. Genesis of cerebral aneurysms—an update. Acta Neurochirurgica 2001;143:429–48; discussion 448–49 [DOI] [PubMed] [Google Scholar]
- 12.Takagi Y, Ishikawa M, Nozaki K, et al. Increased expression of phosphorylated c-Jun amino-terminal kinase and phosphorylated c-Jun in human cerebral aneurysms: role of the c-Jun amino-terminal kinase/c-Jun pathway in apoptosis of vascular walls. Neurosurgery 2002;51:997–1002; discussion 1002–04 [DOI] [PubMed] [Google Scholar]
- 13.Fukuda, S., et al. Prevention of rat cerebral aneurysm formation by inhibition of nitric oxide synthase. Circulation 2000;101:2532–38 [DOI] [PubMed] [Google Scholar]
- 14.Ohkuma, H., et al. Role of a decreased expression of the local renin-angiotensin system in the etiology of cerebral aneurysms. Circulation 2003;108:785–87 [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Fugleholm K, Day LB, et al. Molecular pathogenesis of subarachnoid haemorrhage. Int J Biochem Cell Biol 2003;35:1341–60 [DOI] [PubMed] [Google Scholar]
- 16.Bruno G, Todor R, Lewis I, et al. Vascular extracellular matrix remodeling in cerebral aneurysms. J Neurosurg 1998;89:431–40 [DOI] [PubMed] [Google Scholar]
- 17.St Jean P, Hart B, Webster M, et al. Alpha-1-antitrypsin deficiency in aneurysmal disease. Hum Hered 1996;46:92–97 [DOI] [PubMed] [Google Scholar]
- 18.Todor DR, Lewis I, Bruno G, et al. Identification of a serum gelatinase associated with the occurrence of cerebral aneurysms as pro-matrix metalloproteinase-2. Stroke 1998;29:1580–83 [DOI] [PubMed] [Google Scholar]
- 19.Kim SC, Singh M, Huang J, et al. Matrix metalloproteinase-9 in cerebral aneurysms. Neurosurgery 1997;41:642–66; discussion 646–47 [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Han L, Wang S. In situ localization of mRNA gelatinases in cerebral aneurysms. Chung-Hua i Hsueh Tsa Chih 1999;79:822–24 [PubMed] [Google Scholar]
- 21.Mimata C, Kitaoka M, Nagahiro S, et al. Differential distribution and expressions of collagens in the cerebral aneurysmal wall. Acta Neuropathologica 1997;94:197–206 [DOI] [PubMed] [Google Scholar]
- 22.Ostergaard JR, Reske-Nielsen E, Buhl J. Deficiency of reticular fibers in cerebral arteries. On the etiology of saccular aneurysms in childhood. Br J Neurosurg 1989;3:113–15 [DOI] [PubMed] [Google Scholar]
- 23.Neil-Dwyer G, Bartlett JR, Nicholls AC, et al. Collagen deficiency and ruptured cerebral aneurysms. A clinical and biochemical study. J Neurosurg 1983;59:16–20 [DOI] [PubMed] [Google Scholar]
- 24.Pope FM, Nicholls AC, Narcisi P, et al. Some patients with cerebral aneurysms are deficient in type III collagen. Lancet 1981;1:973–75 [DOI] [PubMed] [Google Scholar]
- 25.Majamaa K, Savolainen ER, Myllyla VV. Synthesis of structurally unstable type III procollagen in patients with cerebral artery aneurysm. Biochim Biophys Acta 1992;1138:191–96 [DOI] [PubMed] [Google Scholar]
- 26.Brooks AR, Lelkes PI, Rubanyi GM. Gene expression profiling of human aortic endothelial cells exposed to disturbed flow and steady laminar flow. Physiol Genomics 2002;9:27–41 [DOI] [PubMed] [Google Scholar]
- 27.Absi TS, Sundt TM 3rd, Tung WS, et al. Altered patterns of gene expression distinguishing ascending aortic aneurysms from abdominal aortic aneurysms: complementary DNA expression profiling in the molecular characterization of aortic disease. J Thoracic Cardiovasc Surg 2003;126:344–57; discussion 357 [DOI] [PubMed] [Google Scholar]
- 28.Armstrong PJ, Johanning JM, Calton WC Jr, et al. Differential gene expression in human abdominal aorta: aneurysmal versus occlusive disease. J Vasc Surg 2002;35:346–55 [DOI] [PubMed] [Google Scholar]
- 29.Tung WS, Lee JK, Thompson RW. Simultaneous analysis of 1176 gene products in normal human aorta and abdominal aortic aneurysms using a membrane-based complementary DNA expression array. J Vasc Surg 2001;34:143–50 [DOI] [PubMed] [Google Scholar]
- 30.Yajima N, Masuda M, Miyazaki M, et al. Oxidative stress is involved in the development of experimental abdominal aortic aneurysm: a study of the transcription profile with complementary DNA microarray. J Vasc Surg 2002;36:379–85 [DOI] [PubMed] [Google Scholar]
- 31.Dunn LT, Stewart E, Murray GD, et al. The influence of apolipoprotein E genotype on outcome after spontaneous subarachnoid hemorrhage: a preliminary study. Neurosurgery 1006;48:1006–10 [DOI] [PubMed] [Google Scholar]
- 32.Khurana VG, Sohni YR, Mangrum WI, et al. Endothelial nitric oxide synthase T-786C single nucleotide polymorphism: a putative genetic marker differentiating small versus large ruptured intracranial aneurysms. Stroke 2003;34:2555–59 [DOI] [PubMed] [Google Scholar]
- 33.Suzuki Y, et al. Nitric oxide metabolites in the cisternal cerebral spinal fluid of patients with subarachnoid hemorrhage. Neurosurgery 1997;41:807–11; discussion 811–12 [DOI] [PubMed] [Google Scholar]
- 34.Hara A, Yoshimi N, Mori H. Evidence for apoptosis in human intracranial aneurysms. Neurol Res 1998;20:127–30 [DOI] [PubMed] [Google Scholar]
- 35.Kondo S, Hashimoto N, Kikuchi H, et al. Apoptosis of medial smooth muscle cells in the development of saccular cerebral aneurysms in rats. Stroke 1998;29:181–88; discussion 189 [DOI] [PubMed] [Google Scholar]
- 36.Sakaki T, Kohmura E, Kishiguchi T, et al. Loss and apoptosis of smooth muscle cells in intracranial aneurysms. Studies with in situ DNA end labeling and antibody against single-stranded DNA. Acta Neurochir (Wien) 1997;139:469–74; discussion 474–75 [DOI] [PubMed] [Google Scholar]
- 37.Stehbens WE. Apoptosis and matrix vesicles in the genesis of arterial aneurysms of cerebral arteries. Stroke 1998;29:1478–80 [DOI] [PubMed] [Google Scholar]
- 38.Austin G, Fisher S, Dickson D, et al. The significance of the extracellular matrix in intracranial aneurysms. Ann Clin Lab Sci 1993;23:97–105 [PubMed] [Google Scholar]
- 39.Gaetani P, Rodriguez y Baena R, Tartara F, et al. Metalloproteases and intracranial vascular lesions. Neurol Res 1999;21:385–90 [DOI] [PubMed] [Google Scholar]
- 40.Eckel JE, Gennings C, Therneau TM, et al. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics 2005;21:1078–83 [DOI] [PubMed] [Google Scholar]
- 41.Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 1996;14:1675–80 [DOI] [PubMed] [Google Scholar]
- 42.Sadamasa N, Nozaki K, Hashimoto N. Disruption of gene for inducible nitric oxide synthase reduces progression of cerebral aneurysms. Stroke 2003;34:2980–84 [DOI] [PubMed] [Google Scholar]
- 43.Coutard M. Experimental cerebral aneurysms in the female heterozygous Blotchy mouse. Int J Exp Pathol 1999;80:357–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres VE, Cai Y, Chen X, et al. Vascular expression of polycystin-2. J Am Soc Nephrol 2001;12:1–9 [DOI] [PubMed] [Google Scholar]
- 45.Arnaout MA. The vasculopathy of autosomal dominant polycystic kidney disease: insights from animal models. Kidney Int 2000;58:2599–610 [DOI] [PubMed] [Google Scholar]
- 46.Lee DW, Yuki I, Murayama Y, et al. A histologic and molecular analysis of thrombus organization and healing in the swine experimental aneurysm model. Presentation at the American Society of Neuroradiology, San Diego, Calif; May 4,2006. .
- 47.Sakomura Y, Nagashima H, Aoka Y, et al, Expression of peroxisome proliferator-activated receptor-gamma in vascular smooth muscle cells is upregulated in cystic medial degeneration of annuloaortic ectasia in Marfan syndrome. Circulation 2002;106(12 Suppl 1):24. [PubMed] [Google Scholar]


