Abstract
MYCN amplification is tightly associated with the poor prognosis of pediatric neuroblastoma (NB). The regulation of NB cell death by MYCN represents an important aspect, as it directly contributes to tumor progression and therapeutic resistance. However, the relationship between MYCN and cell death remains elusive. Ferroptosis is a newly identified cell death mode featured by lipid peroxide accumulation that can be attenuated by GPX4, yet whether and how MYCN regulates ferroptosis are not fully understood. Here, we report that MYCN-amplified NB cells are sensitive to GPX4-targeting ferroptosis inducers. Mechanically, MYCN expression reprograms the cellular iron metabolism by upregulating the expression of TFRC, which encodes transferrin receptor 1 as a key iron transporter on the cell membrane. Further, the increased iron uptake promotes the accumulation of labile iron pool, leading to enhanced lipid peroxide production. Consistently, TFRC overexpression in NB cells also induces selective sensitivity to GPX4 inhibition and ferroptosis. Moreover, we found that MYCN fails to alter the general lipid metabolism and the amount of cystine imported by System Xc(−) for glutathione synthesis, both of which contribute to ferroptosis in alternative contexts. In conclusion, NB cells harboring MYCN amplification are prone to undergo ferroptosis conferred by TFRC upregulation, suggesting that GPX4-targeting ferroptosis inducers or TFRC agonists can be potential strategies in treating MYCN-amplified NB.
Subject terms: Cancer metabolism, Cell death
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in human infants, with 90% of cases diagnosed by the age of 51. Genetically, frequent amplifications of the oncogene MYCN are identified in patients with NB, predicting poor prognosis independent of other factors2. Although MYCN inhibition leads to the suppression of NB in vitro, there are technical challenges in targeting MYCN clinically3,4. MYCN orchestrates multiple molecular pathways for cell growth, survival, metabolism, and death, dictating cancer cell fate and tumor progression5,6. In order to explore new treatments against MYCN-amplified NB, it is necessary to gain a deeper understanding of the biological functions mediated by MYCN.
Among the key cellular processes governed by MYCN, cell death is a puzzling phenomenon. On one hand, it is the cause of tumor growth under nutrient-deprived conditions whose adaptation promotes NB progression7,8; on other hand, abnormal expression of MYCN leads to the direct activation of cell death pathways9,10. This contradiction indicates that MYCN finely regulates the balance between cell survival and cell death. Recently, researchers discovered a new type of programmed cell death, namely ferroptosis, which is characterized by intracellular iron disorder and accumulation of lipid peroxides in the cell membrane11. Cell culture and animal experiments have shown that induction of ferroptosis can eliminate selective tumor cells that are resistant to apoptosis, such as in kidney cancer12, breast cancer13, lung cancer14, etc., which provides a new venue for overcoming drug resistance. System Xc(−) and GPX4 inhibition are the most common methods to induce ferroptosis. System Xc(−) imports cystine as a key substrate for synthesizing glutathione (GSH), which is the reducing equivalent used by GPX4 to antagonize ferroptosis15. Although MYCN has a role in regulating cellular redox balance16, the potential effects of MYCN on ferroptosis remain unclear.
In this study, we identified a new regulatory mechanism of ferroptosis by MYCN. We found that NB cells overexpressing MYCN are particularly sensitive to ferroptosis induced by GPX4 inhibition rather than system Xc(−) blockage. Moreover, MYCN amplification fails to cause significant changes in lipid species, suggesting that alternations of iron metabolism are mainly accounting for MYCN regulated ferroptosis. Therefore, we analyzed the expression of iron-related genes and revealed that TFRC, which encodes transferrin receptor 1 as an essential iron transporter on the cell membrane, plays a key role in the process of MYCN regulated ferroptosis. Similar to MYCN, TFRC significantly increases the intracellular iron load, and TFRC upregulation confers cell sensitivity to ferroptosis induced by GPX4 inhibition. Considering that MYCN and TFRC are both deregulated in a variety of tumor types, our finding points to a tumor vulnerability that can be therapeutically exploited.
Results
MYCN is a regulator of GPX4-dependent ferroptosis
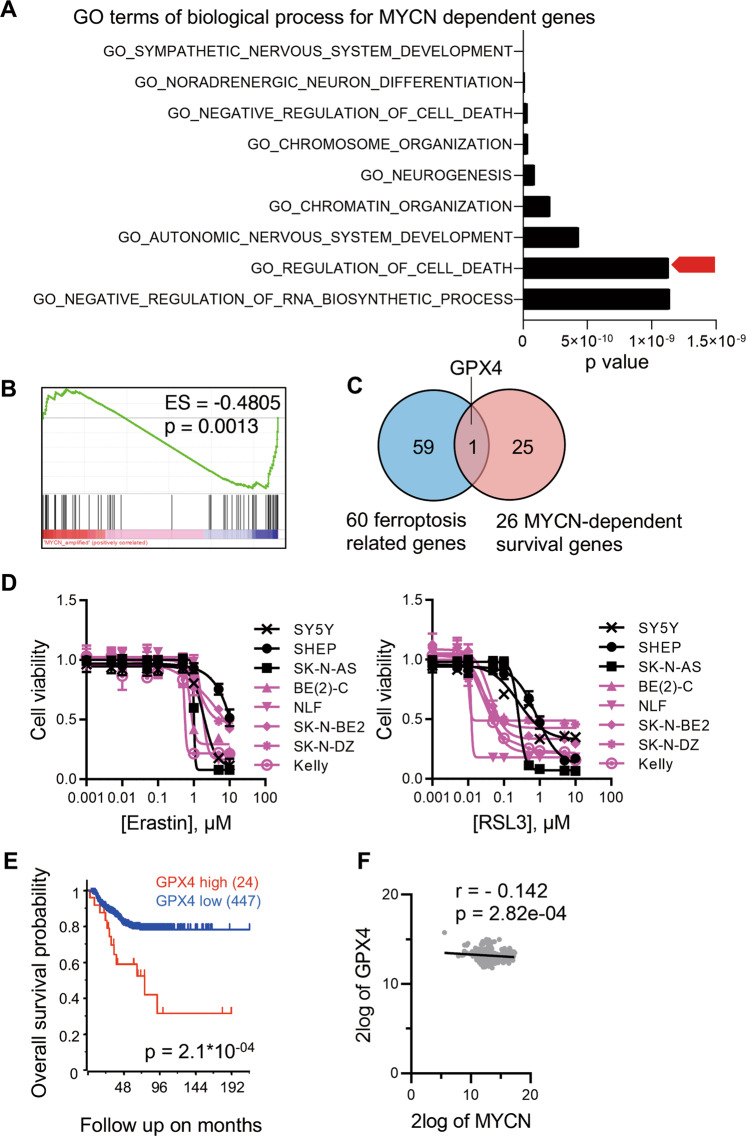
To study MYCN-dependent cellular processes in NB, we examined a collection of 147 genes essential for the survival of MYCN-amplified NB cells17 through pathway analysis using the Molecular Signatures Database v7.2. We discovered that genes regulating cell death are significantly enriched in this collection (Fig. 1A). Furthermore, we performed RNA-seq and gene set enrichment analysis (GSEA) of six human NB cell lines (MYCN amplified SK-N-BE2, BE(2)-C, NLF, SK-N-DZ, and MYCN non-amplified SHEP, SK-N-AS), and found that MYCN expression is closely associated with levels of ferroptosis-related genes as summarized by Liang et al.18 (Fig. 1B and Supplementary Table 1). Next, we compared the ferroptosis-related genes with those regulating MYCN-dependent cell survival (Supplementary Table 2), and identified GPX4 as the only overlapping gene (Fig. 1C). GPX4 is a well-established ferroptosis regulator neutralizing the oxidized lipid species19. To confirm the above results, multiple NB cell lines were treated respectively with two classic ferroptosis inducer, (1 S, 3 R)-RSL3 (hereafter RSL3 for short), a highly specific GPX4 inhibitor, and Erastin, a cystine-glutamate antiporter (system Xc(−)) inhibitor. Interestingly, MYCN amplification significantly increases the sensitivity of NB cells to RSL3, whereas all NB cells exhibit similar resistance to Erastin (Fig. 1D and Fig. S1A). Consistently, the effects of an alternative system Xc(−) inhibitor sulfasalazine (SAS)20 are weakly associated with MYCN amplification (Fig. S1B, C). These findings imply that MYCN is a key regulator of ferroptosis, and GPX4 might play an essential role in this process. Indeed, high expression of GPX4 in NB tumor tissues predicts poor prognosis (Fig. 1E). However, RNA-seq data reveal that there is no significant correlation between GPX4 and MYCN expression (Fig. 1F and Fig. S1D). Therefore, MYCN regulates GPX4-dependent ferroptosis likely through an unknown mechanism.
Fig. 1. MYCN is a regulator of GPX4-dependent ferroptosis.
A Genes selectively dependent on MYCN for NB cell survival are enriched within a variety of GO terms of biological processes, and the top nine processes are shown according to p values. B GSEA assays of ferroptosis-related genes showing enrichment with MYCN amplification based on RNA-seq data of six human NB cell lines. ES, enrichment score. C GPX4 is shared between ferroptosis-related and MYCN-dependent survival genes. D Cell viabilities when different concentrations of the GPX4 inhibitor (1S, 3R)-RSL3 (RSL3) and system Xc(−) inhibitor Erastin were used as ferroptosis inducers in multiple human NB cell lines (MYCN amplified SK-N-BE2, BE(2)-C, NLF, SK-N-DZ, and Kelly are represented in pink; and MYCN non-amplified SHEP, SK-N-AS, and SY5Y are shown in black; n = 4). E Kaplan–Meier survival curves for NB patients based on GPX4 mRNA expression. Gene expression and clinical data were retrieved from GSE45547. F Correlation analysis between GPX4 and MYCN expression based on the data retrieved from GSE45547. *p < 0.05, **p < 0.01.
MYCN confers cell sensitivity to ferroptosis upon GPX4 inhibition
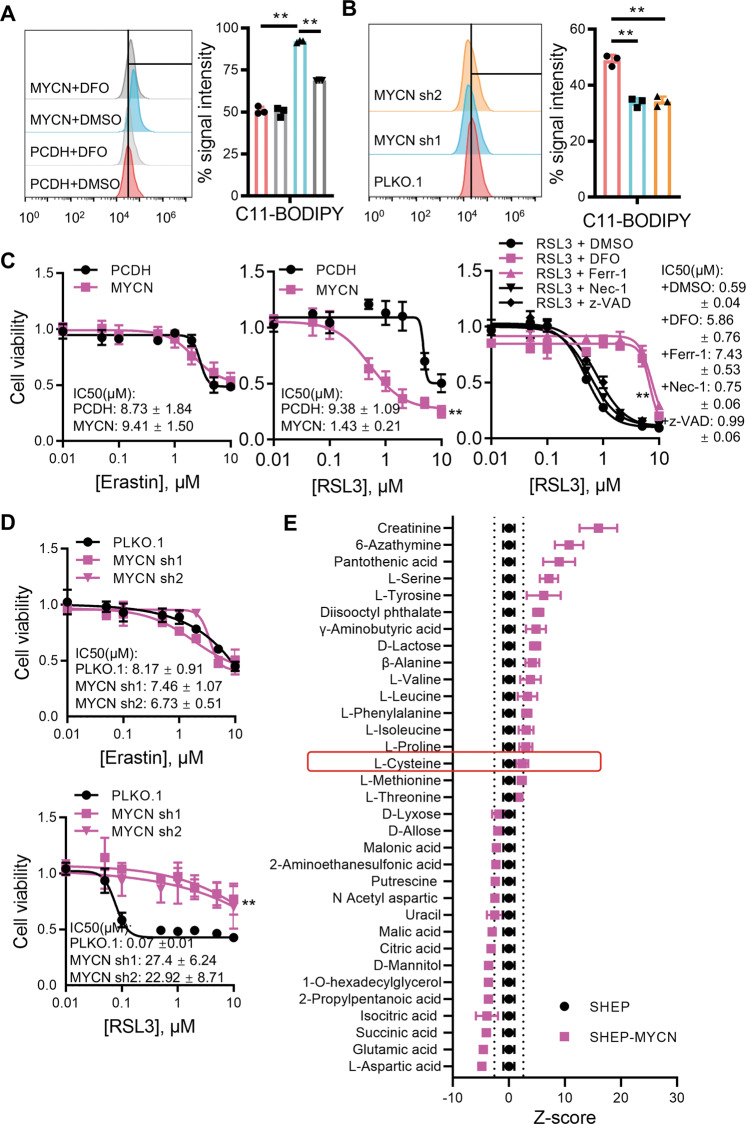
To confirm that MYCN is linked to ferroptosis, we ectopically expressed MYCN in SHEP, a MYCN non-amplified NB cell line, and found that MYCN expression significantly increases lipid peroxide contents. Conversely, lipid peroxides are reduced when MYCN is depleted in SK-N-BE2, a MYCN amplified NB cell line (Fig. 2A, B). Next, MYCN overexpression significantly increases the sensitivity of SHEP cells to RSL3, but not Erastin. Moreover, MYCN induced sensitivity towards RSL3 can only be rescued by treating cells with ferroptosis inhibitors, but not other cell death inhibitors (Fig. 2C). These results are further supported by the data in MYCN amplified SK-N-BE2 cells with shRNA-mediated MYCN depletion (Fig. 2D).
Fig. 2. MYCN confers cell sensitivity to ferroptosis upon GPX4 inhibition.
Lipid peroxides were detected by flow cytometry after incubation with C11-BODIPY in SHEP (A) or SK-N-BE2 (B) cells with or without MYCN. DFO, desferrioxamine, n = 3. C Cell viabilities of SHEP cells with or without MYCN overexpression incubated with Erastin (left), RSL3 (middle), or RSL3 combined with either ferroptosis inhibitor (10 μM DFO, 1 μM Ferr-1), apoptosis inhibitor (20 μM z-VAD), or necroptosis inhibitor (2 μM Necrostatin-1) (right). D Cell viabilities of SK-N-BE2 cells with or without MYCN knockdown incubated with Erastin or RSL3. E Metabolic analysis of SHEP cells with or without MYCN. Z-score ±2.75 is corresponding to p = 0.01 and depicted as a dotted line. Data are presented as mean ± SD of three replicates. Two-tailed unpaired t-tests were performed to calculate p values. **p < 0.01.
The function of GPX4 is to convert toxic lipid peroxides to nontoxic alcohol lipids using the reducing equivalent glutathione. The system Xc(−) imports cystine, the extracellular form of cysteine, for glutathione synthesis, therefore functioning upstream of GPX4. To test whether the intracellular cysteine level can be regulated by MYCN, we use mass spectrometry to identify altered metabolites upon MYCN overexpression, and found that MYCN does not significantly affect the intracellular cysteine level (Fig. 2E). Meanwhile, our metabolic analysis confirms that MYCN amplification elevates serine levels, which is consistent with previous findings such as by Xia et al.21. These results demonstrate that cellular cystine import is independent of MYCN. Together, we conclude that MYCN induces NB cell sensitivity to ferroptosis upon GPX4 inhibition, through a mechanism non-correlated with cysteine metabolism.
MYCN increases the intracellular iron load
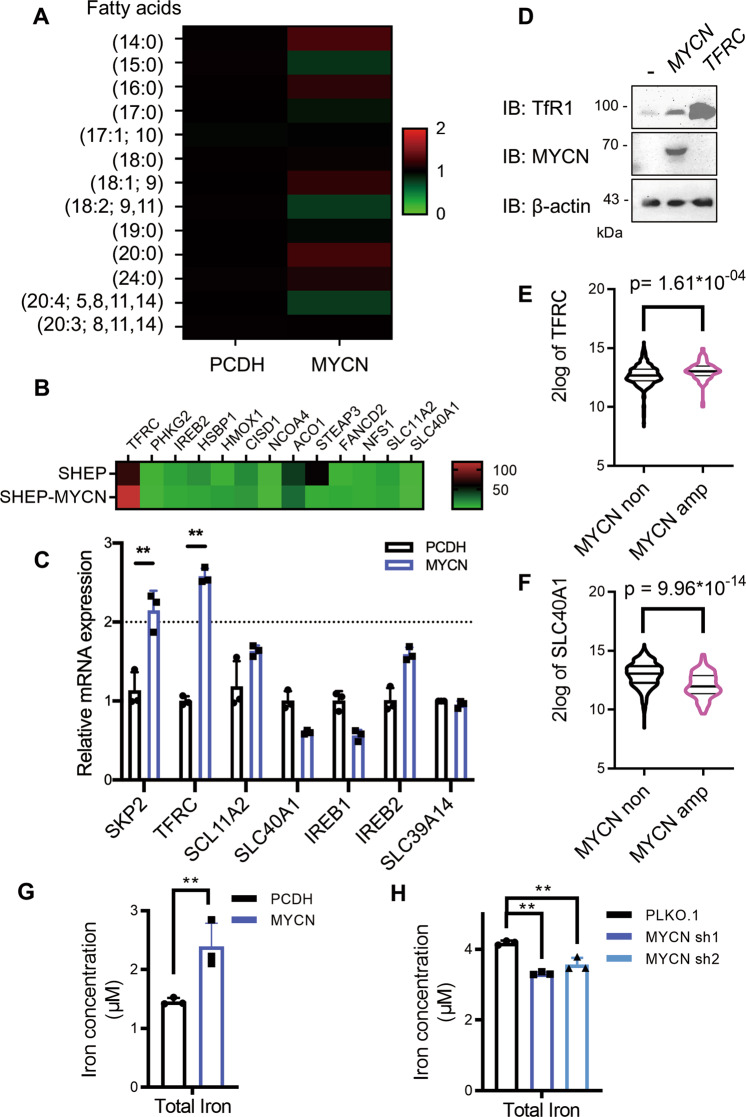
Deregulated iron metabolism and abnormal lipid synthesis are both required to induce ferroptosis22. To this end, we examined the fatty acid content upon MYCN overexpression by mass spectrometry and found that MYCN has no significant influence on cellular fatty acid pools (Fig. 3A). Next, we analyzed the expression of several ferroptosis-related genes as summarized by Hassannia et al.23, and identified TFRC as the most upregulated one both in RNA-seq data (Fig. 3B) and RT-PCR results (Fig. 3C). TFRC encodes transferrin receptor 1 (TfR1), which is the prevalent iron importer on the cell membrane. Consistently, MYCN upregulation enhances the protein level of TfR1 in NB cells (Fig. 3D), which parallels with TFRC levels in NB tumor tissues (Fig. 3E). Interestingly, we noticed that the expression of SLC40A1, which encodes the only known iron exporter ferroportin, is slightly but reproductively reduced upon MYCN overexpression (Fig. 3C), and RNA-seq analysis of NB tumor tissues also supports this argument (Fig. 3F). These results suggest that MYCN tends to increase the iron load in NB cells. To confirm this, we tested the level of the total iron pool (ferric plus ferrous ion) upon MYCN manipulation. As expected, MYCN overexpression increases the iron level in SHEP cells, whereas MYCN depletion reduces the iron level in SK-N-BE2 cells (Fig. 3G, H). These results indicate that MYCN mediates GPX4-dependent ferroptosis likely via regulating iron metabolism.
Fig. 3. MYCN increases the intracellular iron load.
A Heatmap showing changes in an array of fatty acids in SHEP cells with or without MYCN. Data are shown as relative changes in abundance compared to the PCDH vector. B Heatmap showing the expression profile of ferroptosis-associated metabolic genes. C Expression of MYCN induced iron metabolic genes analyzed by RT-qPCR, and SKP2 is positive control of MYCN targeted genes. D Western blot analysis of SHEP cells with or without MYCN expression using indicated antibodies. E, F TFRC and SLC40A1 expression in NB tumor tissues with or without MYCN amplification. Expression data were retrieved from GSE45547. Cellular iron load is detected with the colorimetric ferrozine-based assay in SHEP G and SK-N-BE2 H cells with or without MYCN, and values are normalized to protein concentration. Data are presented as mean ± SD of three replicates. Two-tailed unpaired t-tests were performed to calculate p values. **p < 0.01.
TFRC photocopies MYCN to increase the labile iron pool
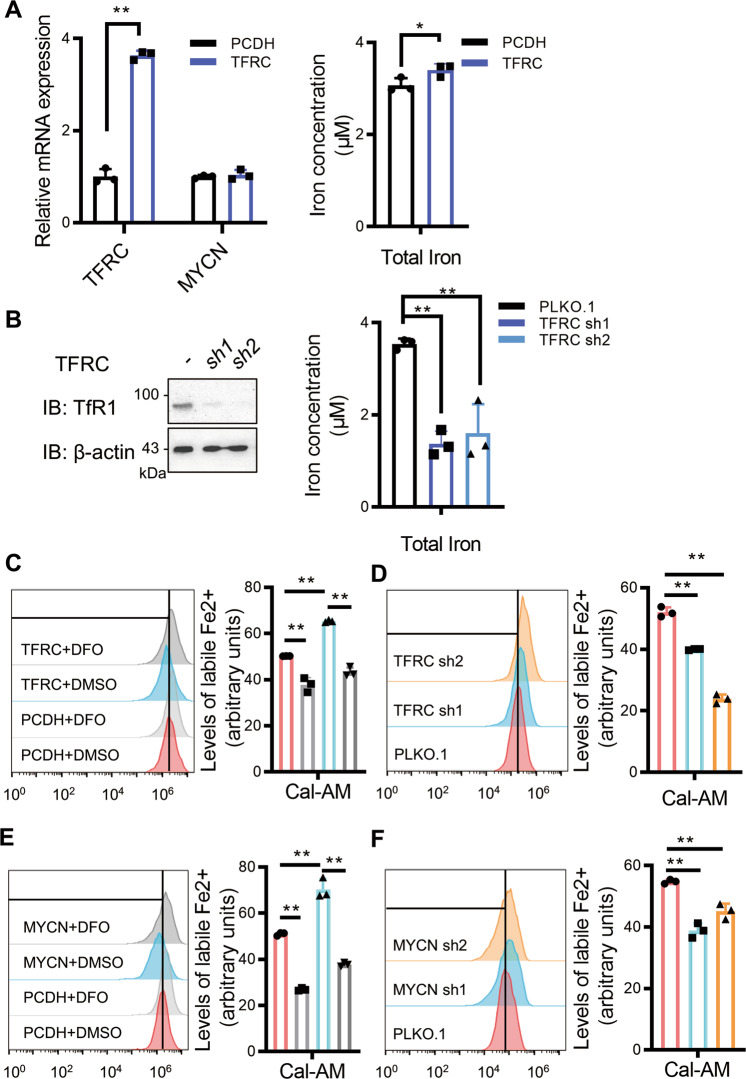
To confirm that MYCN-induced TFRC expression promotes iron uptake, we overexpressed TFRC in SHEP and depleted TFRC in SK-N-BE2 cells, respectively. Indeed, TFRC expression levels correlate with the cellular iron load (Fig. 4A, B). The Fe3+ ion transported by TfR1 undergoes endocytosis, reduction to Fe2+, and importation to the cytosol composing the labile iron pool (LIP). Fe2+ promotes the formation of lipid peroxides by Fenton reactions. Based on these knowledges, we reasoned that increased cellular iron load would result in LIP upregulation, thereby promoting lipid peroxide formation. To test this, we detected intracellular LIP levels with the calcein acetoxymethyl ester (Cal-AM) probe as previously described24. Indeed, the LIP levels are increased upon TFRC overexpression and deceased upon TFRC knockdown (Fig. 4C, D), which photocopy the effects of MYCN (Fig. 4E, F). Therefore, MYCN selectively sensitizes NB cells to ferroptosis by increasing the intracellular LIP.
Fig. 4. TFRC photocopies MYCN to increase the labile iron pool.
A Iron load and RT-qPCR analysis of SHEP cells with or without TFRC overexpression. B Iron load and western blot analysis of SK-N-BE2 cells with or without TFRC depletion. Cellular labile iron pool (LIP) was detected using the probe calcein acetoxymethyl ester (Cal-AM) in SHEP (C) and SK-N-BE2 (D) cells with or without TFRC. LIP was similarly detected in SHEP (E) and SK-N-BE2 (F) cells with or without MYCN. Data are presented as mean ± SD of three replicates. Two-tailed unpaired t-tests were performed to calculate p values. *p < 0.05, **p < 0.01.
TFRC mediates MYCN induced, GPX4-dependent ferroptosis
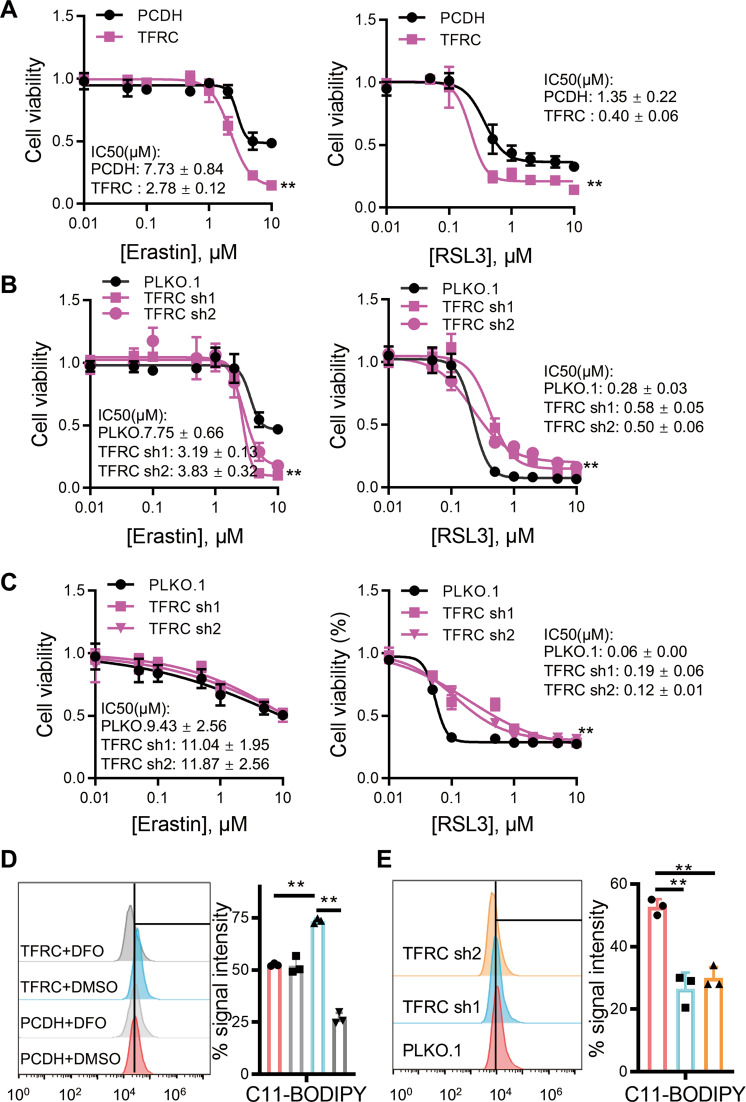
Our data have shown that MYCN upregulates TFRC expression, thereby regulating ferroptosis. Next, we explored whether TFRC mediates GPX4-dependent ferroptosis, in a similar way as MYCN. In MYCN non-amplified SHEP cells, we found that TFRC overexpression makes cells sensitive to ferroptosis induced by both System Xc(−) and GPX4 inhibition (Fig. 5A), whereas TFRC knockdown only rescues ferroptosis induced by GPX4 inhibition, but not system Xc(−) inhibition (Fig. 5B). In SK-N-BE2 cells with amplified MYCN, we also found that knockdown of TFRC rescues the GPX4-dependent but not Xc−-dependent ferroptosis, and the rescue effects are substantially enhanced (Fig. 5C). Moreover, we found that the system Xc(−) inhibitor SAS upregulates TFRC expression, and TFRC-depleted, MYCN-overexpressed NB cells exhibit high resistance to SAS (Fig. S1E–G). The above results suggest that TFRC indeed mediates GPX4-dependent ferroptosis, which can be further reinforced by MYCN amplification. Consistent with it, the lipid peroxide amount is significantly elevated with ectopic TFRC expression, and decreased upon TFRC knockdown (Fig. 5D, E).
Fig. 5. TFRC mediates MYCN–induced, GPX4-dependent ferroptosis.
A Cell viability of SHEP cells with or without TFRC overexpression incubated with Erastin (left) or RSL3 (right). Cell viabilities of SHEP (B) or SK-N-BE2 (C) cells with or without TFRC depletion incubated with Erastin (left) or RSL3 (right). Amounts of cellular lipid peroxides measured by C11-BODIPY in SHEP (D) or SK-N-BE2 (E) cells with or without TFRC. Data are presented as mean ± SD of three replicates. Two-tailed unpaired t-tests were performed to calculate p values. **p < 0.01.
TFRC is a direct downstream target of MYCN
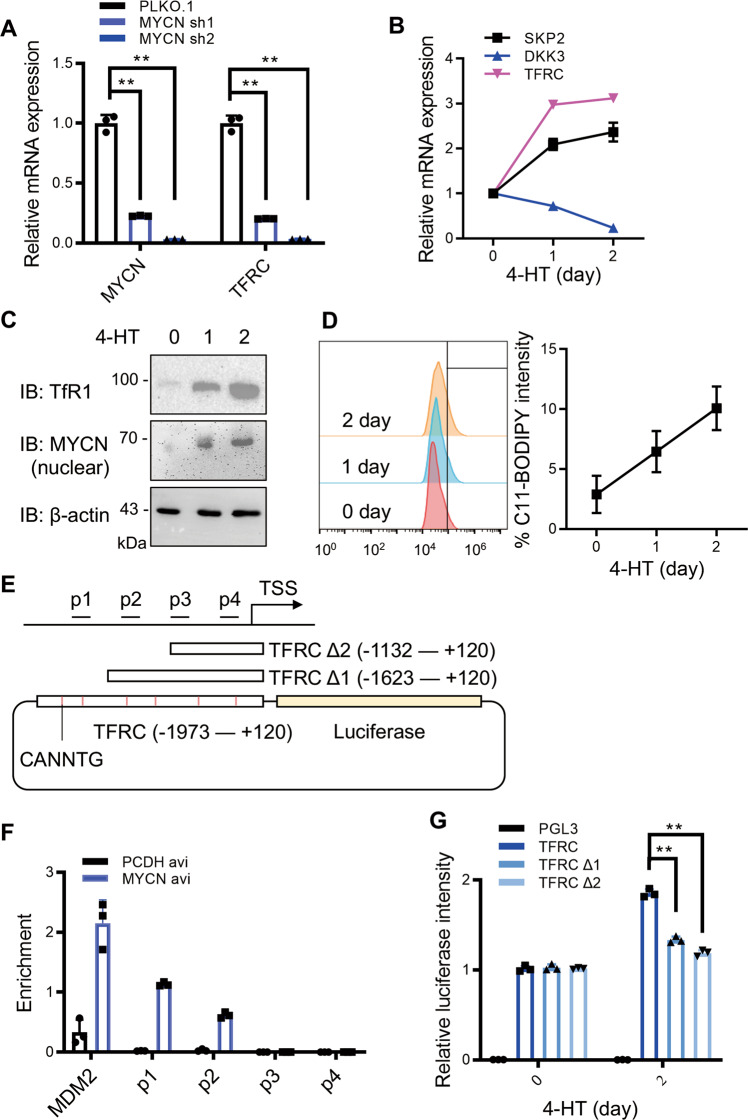
Given that MYCN is a well-established transcription factor, we speculated that MYCN drives TFRC expression through direct transcriptional regulation. Indeed, we confirmed that TFRC expression is responsive to either up or down-regulation of MYCN (Figs. 3C and 6A). Then, we established an inducible MYCN–ER chimeric protein expression system in SHEP cells, in which nuclear translocation and activity of MYCN can be induced by 4-hydroxytamoxifen (4-HT) treatments. Upon 4-HT stimulation, TFRC expression is elevated at both mRNA and protein levels (Fig. 6B, C), paralleled with lipid peroxide formation in a time-dependent manner (Fig. 6D). To further dissect exactly how MYCN regulates TFRC expression, we performed ChIP-qPCR assays and found that MYCN mainly binds to the P1 and P2 regions, which are located ~1.5 kb away from the transcription starting site (TSS) within the TFRC promoter (Fig. 6E, F). In addition, we cloned the TFRC promoter region from −1973 to +120 bp surrounding the TSS as shown in Fig. 6E and generated corresponding truncations. We found that MYCN-promoted luciferase signals are strongly attenuated when the region of −1973 to −1623 is eliminated (Fig. 6G), demonstrating that MYCN regulates TFRC expression by binding to this region. Therefore, we conclude that MYCN induces GPX4-dependent ferroptosis by upregulating TFRC as a direct downstream target.
Fig. 6. TFRC is a direct downstream target of MYCN.
A RT-qPCR analysis of SK-N-BE2 cells with or without MYCN depletion. MYCN activation induced by 4-hydroxytamoxifen (4-HT) treatment promotes TFRC expression at both mRNA B and protein C levels. SKP2, DKK3 were used as positive and negative controls, respectively. D Amounts of cellular lipid peroxides measured by C11-BODIPY upon MYCN activation. E Primers designed for ChIP-qPCR analysis, and sequence variants for luciferase assays based on the TFRC promoter region. F ChIP-qPCR analysis of different binding sites of MYCN on the TFRC promoter. MDM2 is a positive control for MYCN binding. G Dual-luciferase assays based on (E). Signal values were normalized to no 4-HT treatment (day 0). Data are presented as mean ± SD of three replicates. Two-tailed unpaired t-tests were performed to calculate p values. **p < 0.01.
Discussion
The MYC family member MYCN plays important role in the development of the central nervous system25,26, and its abnormal expression correlates with the poor prognosis of NB patients. Meanwhile, elevated activity of c-MYC, the predominant MYC isoform, also accelerates the progression of low-grade NB27, indicating that MYC family members share similar functions during NB development. Ferroptosis is a novel type of programmed cell death characterized by iron-dependent, unleashed lipid peroxidation, which has been identified in recent years. Cystine deprivation rendered by system Xc(−) blockage, or redox imbalance by GPX4 inhibition are two classic approaches to induce ferroptosis15. Our study revealed that MYCN sensitizes NB cells to ferroptosis, which is largely dependent on GPX4 inhibition rather than cystine deprivation. This is mainly because the abnormal level of MYCN stimulates the expression of TFRC, increases the intracellular levels of iron load and LIP, ultimately leads to enhanced lipid peroxidation and a tendency towards ferroptosis.
One important function of MYCN in NB is upregulating glutamine transporters, such as ASCT2, thereby promoting glutamine addiction28,29, a hallmark of tumor metabolic reprogramming. In NB cells, the glutamic acid level is decreased upon MYCN amplification (Fig. 2E), confirming that MYCN promotes glutamine consumption. It was reported that glutaminolysis and iron are both essential for Erastin-induced ferroptosis in an ischemia/reperfusion model30, implying that MYCN might induce ferroptosis through system Xc(−) in that context. However, our results reveal that MYCN induces ferroptosis in NB cells independent of system Xc(−), suggesting that the molecular mechanism underlying MYCN-induced ferroptosis is highly context-dependent.
Notably, Floros et al. recently reported that MYCN-amplified NB cells are more sensitive to the system Xc(−) inhibitor sulfasalazine (SAS)31. Indeed, MYCN-amplified NB cells are slightly sensitive to SAS as reflected by their smaller IC50 values compared to non-amplified cells, although no statistical significance was achieved in our hands (Fig. S1C). This trend is seemingly contradictory to what we observed with Erastin. However, SAS equally inhibits other targets such as NFκB and reduced folate carrier32,33 in addition to System Xc(−), which may help explain this discrepancy. Interestingly, Floros et al. noticed that SAS treatment of MYCN-amplified NB in vivo upregulate TFRC expression through an uncharacterized mechanism, which can be reproduced by us in vitro (Fig. S1E). Since we demonstrated that enhanced TFRC expression is responsible for increased cellular sensitivity to ferroptosis, we reasoned that SAS may exert its effect through a similar mechanism. Indeed, TFRC-depleted, MYCN-overexpressed NB cells exhibit high resistance to SAS (Fig. S1F, G), demonstrating that SAS sensitizes MYCN-amplified NB cells to ferroptosis at least partially through TFRC upregulation. This argument can be further supported by the study from Floros et al. For instance, SAS is capable of inducing ~70% cell death in MYCN-amplified NB cells. However, knockdown of two System Xc(−) components SLC7A11 and SLC3A2 induce only ~25% cell death in the same cell line31, confirming that SAS harbors considerable off-target effects. In conclusion, the complex effects of SAS treatments leading to ferroptosis cannot be solely linked to the sensitivity of NB cells to System Xc(−) inhibition.
There are multiple major differences between our studies and the work by Floros et al. First, they were focused on targeting the redox balance of MYCN-amplified NB cells to induce ferroptosis, using different small molecule inhibitors such as buthionine-(S,R)-sulfoximine (BSO), sulfasalazine (SAS), and auranofin. All these inhibitors function to antagonize the ROS-scavenging activities in MYCN-amplified NB cells. In contrast, our work highlights TFRC as the major downstream target of MYCN to confer cell sensitivities to ferroptosis inhibitors. We particularly demonstrated that TFRC depletion greatly suppresses MYCN-mediated ferroptosis, as companied by the reduction of the intracellular iron pool. Moreover, TFRC overexpression phenocopies MYCN amplification to increase the intracellular iron pool and sensitize these tumor cells to ferroptosis. Given that ferroptosis requires both elevated iron levels and dampened ROS-neutralizing activities, these two studies compensate with each other in understanding the vulnerability of MYCN-amplified NB cells towards ferroptosis. Second, work by Floros et al. suggested that MYCN binds to the TFRC promoter and upregulates TFRC expression by analyzing relevant data in a public ChIP-seq database. Conversely, we performed ChIP-PCR analyses to identify the MYCN-binding region within the TFRC promoter, and further confirmed this result by luciferase reporter assays. Third, they found that MYCN-amplified NB cells are sensitive to the System Xc(−) inhibitor SAS, whereas our study suggested that these cells are resistant to the System Xc(−) inhibitor Erastin.
Iron is an essential element for cell growth and survival, the deregulation of which determines cell fate. In the process of ferroptosis, iron acts as a catalyst for lipid peroxidation through Fenton reactions23. TfR1 encoded by TFRC is the main iron transporter in cells, and it has been reported that SLC39A14 is an additional iron transporter34, although our experiment shows that MYCN fails to regulate SLC39A14 expression (Fig. 3C). In addition, several classical MYCN binding motifs (CANNTG) are present in the TFRC promoter, and our ChIP-qPCR analysis reveals that MYCN precisely binds to a motif-containing region distal to TSS. The specific recognition and regulation of TFRC by MYCN is consistent with previous findings35. Recently, the Stockwell group reported that TfR1 clusters on the cell membrane can be used as a detection marker for ferroptosis36. They observed that TfR1 expression is increased during ferroptosis induction, but the mechanism is unknown. Our study supports the MYC signaling as an important candidate. Moreover, it has been reported that TfR1 depletion confers cell resistance to Erastin-induced ferroptosis under abnormal RAS activities37. However, we found that TFRC knockdown has no significant impact on Erastin-induced ferroptosis in MYCN-amplified cells. These results suggest that Erastin-induced ferroptosis harbors a regulatory mechanism independent of TFRC, which is likely decided by different oncogenic signals. Although MYC is one of the first identified oncogenes38,39, it can function to induce p53-dependent cell death, similar to the case that oncogenic RAS can initiate cell senescence40,41. Whether and how different oncogenes cooperate to determine cancer cell fate is pivotal for developing new cancer therapeutics.
Materials and methods
Cell culture
Human NB cell lines with MYCN amplification (SK-N-BE2, BE(2)-C, NLF, SK-N-DZ, Kelly) and without MYCN amplification (SHEP, SK-N-AS, SY5Y) were grown in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin, under 5% CO2 at 37 °C. Cell authentication was confirmed by the short tandem repeat analysis, and mycoplasma tests were weekly performed. Cell stocks were created within five passages, and experiments were completed within ten passages.
Antibodies and reagents
Antibodies and reagents were obtained from commercial sources. Specifically, anti-transferrin receptor, rabbit antibody (ab84036) was from Abcam; N-Myc (D1V2A) rabbit mAb (84406), beta-actin (8H10D10) mouse mAb, HRP-linked mouse IgG (7076 S), and rabbit IgG (7074S) were from Cell Signaling Technology (Massachusetts, USA). Erastin (HY-15763), (1 S,3 R)-RSL3 (HY-100218A), deferoxamine mesylate (DFO), Z-VAD(OMe)-FMK(HY-16658), Necrostatin-1 (HY-15760) were from MedChemExpress (New Jersey, USA); (Z)-4-hydroxytamoxifen (T4420) and sulfasalazine (SAS, T0907) from Target Molecule Corp. (Massachusetts, USA); BODIPY™ 581/591 C11 (D3861), Cal-AM (C3099), Ferrozine (160601), and Neocuproine (N1501) from Invitrogen, Thermofisher (California, USA). Polyethylenimine (PEI) Linear, MW 25000 (24769-2) was from Polysciences Inc (Pennsylvania, USA). CCK-8 reagent is from Beyotime Inst Biotech (C0040, Shanghai, China).
Constructs
MYCN and TFRC expression plasmids were constructed by cloning the open reading frame of the corresponding cDNA into the multiple cloning sites of the PCDH vector. shRNA sequences were cloned into PLKO.1 vector according to Addgene’s protocol. For the luciferase assay, promoter sequences were cloned into pGL3 Luciferase Reporter Vectors (E1741, Promega, Wisconsin, USA).
RNA-seq analysis
Total RNA was extracted from each sample using the standard TRIzol protocol. The Agilent 2100 Bioanalyzer was employed to verify RNA quality. cDNA libraries were generated and processed for sequencing following the Illumina TrueSeq version 2 library preparation kit’s protocol. The cDNA libraries were then sequenced by Illumina HiSeq 2500 with paired-end 2 × 75 bp reads using the HiSeq Control Software (version 2.0.10). The quality of raw reads was evaluated by FastQC, and reads alignment was performed using STAR (version 2.3.0). We used SAMTools to sort the aligned reads, and performed gene-level read quantifications with HTSeq (version 0.10.0).
Bioinformatic analyses
Gene expression correlation and survival analyses for NB patients (retrieved from GSE4554742 and GSE4971043) were conducted using the R2 Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi), and the resulting data and p values were calculated according to online instructions. Gene Ontology (GO) analysis of MYCN-dependent genes17 was performed using the Molecular Signatures Database v7.2 online tool (http://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp). For GSEA analysis, the expression data of six NB cells by RNA-seq were used, and cell lines were divided into two groups based on the conventional MYCN status (MYCN amplified: SK-N-BE2, BE(2)-C, NLF, SK-N-DZ v.s. MYCN non-amplified: SHEP, SK-N-AS). Lists and expression data of ferroptosis-related and MYCN-dependent survival genes are provided in Supplementary Tables 1 and 2.
Viral production and transduction
HEK-293T cells were seeded on a 10-cm culture dish with 50% confluence. After attachment, cells were co-transfected with the lentiviral expression plasmid (10 μg), viral packaging plasmid (psPAX2, Addgene 12260, 5 μg), and envelope plasmid (pMD2.G, Addgene 12259, 2 μg) using PEI following the manufacturer’s protocols. Viral supernatants were collected and filtered after transfection. For viral transduction, target cells were seeded and incubated with a medium supplemented with viral particles and 8 mg/mL polybrene for 6–8 h. Virus-infected cells were selected with corresponding antibiotics.
Cell viability assay
Cell viability was detected with CCK-8 according to the manufacturer’s protocol. Briefly, 5 × 103 cells were seeded in 96-well plates per well and treated with different compounds. Twenty-four hours later, cells are incubated with CCK-8 for 2 h at 37 °C. Viability values were obtained by a plate reader at 450 nm.
Total iron analysis
Total iron concentrations of cells were measured by a colorimetric ferrozine-based assay as previously reported44. Briefly, 5 × 105 cells were seeded in 6 cm dishes for 24 h, collected with 50 mM NaOH, and incubate at room temperature (RT) for 2 h. Iron ion was extracted with iron-releasing buffer (1.4 M HCl with 4.5% KMnO4, 1:1) at 60 °C for 2 h, followed by incubation with 30 μL detecting buffer (6.5 mM Ferrozine, 6.5 mM Neocuproine, 2.5 M ammonium acetate, 1 M ascorbic acid) at RT for 30 min. Absorbance was measured with a 96-well plates reader at 550 nm. The iron contents were calculated based on FeCl3 standard curves and normalized to protein concentration.
Labile iron pool (LIP) analysis
Calcein acetoxymethyl ester (Cal-AM) is a low-toxic and non-fluorescence dye that can easily pass through the cell membrane. Cal-AM emits fluorescence after being digested by esterase in living cells and quenched in the presence of Fe2+, which can be used as a LIP probe as previously described24. In this study, 2 × 105 cells were seeded on six-well plates for 24 h, after washing with PBS three times, and incubated with 20 nM Cal-AM in HBSS at 37 °C for 30 min. Then, Cells were trypsinized, suspended, and analyzed using a BD Accuri C6 flow cytometer with a 488 nm laser on an FL1 detector.
Measurement of lipid peroxides
Cells were incubated with 200 nM C11-BODIPY in HBSS at 37 °C for 30 min, and analyzed using a BD Accuri C6 flow cytometer with a 488 nm laser on an FL1 detector.
ChIP-qPCR
Assays were performed by following a modified Abcam’s Chromatin immunoprecipitation protocol. Briefly, cells overexpressing biotinylated-tagged proteins (avi tag) were fixed in 1% formaldehyde at RT for 10 min and stopped by the addition of 0.125 M glycine. Cells were then harvested in ChIP Lysis Buffer (50 mM HEPES-KOH pH = 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1X protease inhibitor cocktail). Lysates were sonicated and DNA sheared to an average length of 200–500 bp. Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase A, Proteinase K, and 65 °C heat for de-crosslinking, followed by ethanol precipitation. The resuspended pellet was incubated with magnetic streptavidin beads (Z5482, Promega, Wisconsin, USA) for 4 h rocking at 4 °C, washed four times, and eluted from beads with SDS elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH = 8). Crosslinks were reversed by high salt incubation for 4 h at 65 °C. ChIP DNA was purified by phenol–chloroform extraction, followed by quantitative real-time PCR analysis. Primers used for ChIP-qPCR:
TFRC p1: fw: TGAGGTCAGGAATTCGAGACAAG; rev: GAGGAGTGACGGGATTTTATC;
TFRC p2: fw: GCAGCAATGCTCCTGCTCA; rev: GCGACAGAGCAAGATTCCATC;
TFRC p3: fw: GGGAAGAGTAAAAGCCCAAGG; rev: GAACAGCCCTTTAAGAAGCAAATC;
TFRC p4: fw: GGCTGCAAAATACATCTTCACAAG; rev: GCCTTGAAATGTACGTGCAGG;
MDM2 promoter fw: AGCCTTTGTGCGGTTCGTG; rev: CCCCCGTGACCTTTACCCTG.
Quantitative real-time PCR analysis
Total RNA was extracted from cells using the RNA Midiprep kit (AP-MN-MS- RNA-250, Axygen, California, USA) according to the manufacturer’s protocol. Then, cDNAs were generated using a Reverse Transcription kit (RR036A, Takara, Beijing, China). Quantitative real-time PCR was performed using the TB Green Premix Ex Taq qPCR kit (AQ101, Transgen Biotech, Beijing, China). Primers used for RT-qPCR:
18S: fw: CTACCACATCCAAGGAAGGCAG; rev: TTTTTCGTCACTACCTCCCCG;
MYCN:fw: CCACAAGGCCCTCAGTACC; rev: TCTTCCTCTTCATCATCTTCATCA;
GPX4:fw: TGGGAAATGCCATCAAGTG; rev: GGGGCAGGTCCTTCTCTATC;
SLC11A2: fw: CTGCACCATGAGGAAGAAGC; rev: TGGATACCTGAGTGGCTGAGT;
SLC40A1: fw: CCCCAGCTCTAGCTGTGAAA; rev: CAGGGGTTTTGGCTCAGTAT;
IREB1: fw: CCTCAGCCCCTGTCAAAA; rev: GATTACTGATGGCCACGTGTT;
IREB2:fw: AGAAATATGGTTCAGGAAACTCCA; rev: GCCAAAACAGCTTTCACACC;
SLC39A14: fw: AAGGCCCTACTCAACCACCT; rev: CGACTGCTCGCTGAAATTGTG
TFRC: fw: ACCTGTCCAGACAATCTCCAG; rev: TGTTTTCCAGTCAGAGGGACA;
DKK3: fw: GAAGGAGCCACGAGTGCAT; rev: CCTCATGCTGTCAAGCCAGA;
SKP2: fw: GACGCTATGCACAGGAAGCA; rev: CCTTTAGCAGCTCAGGGAGG.
Luciferase reporter assay
Dual-Luciferase Reporter Assay System (E1910, Promega, Wisconsin, USA) was used. Briefly, pGL3 vectors carrying TFRC promoter sequences were transfected into SHEP cells. Cells were then transferred to six-well plates with 2 × 105 per well. MYCN activity was induced with 40 nM 4-HT treatments. Luciferase signals were measured 48 h later. Firefly luciferase signals were divided by Renilla luciferase signals, and rescaled to set the control signals equal to 1.
Primers for cloning the TFRC promoter: fw: CCTGCAAATACCAGCATTGTTTG; TFRCΔ1: fw GAAGGAACACCACAGGGGAGCA; TFRCΔ2 fw: GGCGGTGAGAATCCCAAGTACT; The reverse primer for all above: GACACGAGGGTCGGTGTAGTTC.
Western blot
Briefly, cells were lysed with RIPA buffer (20 mM Tris-HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton-X) supplemented with the protease inhibitor cocktail. Lysates were resolved on SDS-PAGE followed by immunoblotting. Primary antibodies were used following instructions in the antibody dilution buffer (1% BSA, 0.05% sodium azide in 0.05 M TBS, pH = 7.6). ChemiDocTM Touch Imaging System (Bio-Rad, California, USA) was used to develop the blot on the 0.2 μm diameter PVDF membrane.
Metabolic analysis
For GC-MS assays, cells were washed with pre-cold physiological saline buffer and harvested in HPLC grade methanol containing internal standards (Ribitol). After sonication and 20,000 g centrifugation at 4 °C, the supernatants were transferred to new 1.5 mL tubes. The supernatants were then transferred into the rotary evaporator and evaporate completely. After treatment with derivatization reagent, samples were analyzed with GC-MS according to the manufacturer protocol. For LC-MS assays, cells were washed with pre-cold physiological saline buffer and harvested in HPLC grade methanol. Then, lipids were extracted with chloroform after 12,000 g centrifugation at 4 °C. The lower phase was evaporated with nitrogen and esterified in 5% sulfuric acid in methanol. The extracts were analyzed by capillary electrophoresis time-of-flight mass spectrometry. Peak areas were normalized to standards and cell numbers.
Statistical analysis
Data were presented as mean ± SD. Statistical significance was analyzed using a two-tailed unpaired t-test and assessed by p values as reported in figure legends. Kaplan–Meier srvival analyses employed the log-rank test.
Supplementary information
Acknowledgements
We thank the Metabolic Innovation Centre at Sun Yat-sen University for assistance in metabolic and fatty acid assays.
Author contributions
Conceived the study and analyzed the data: Z.L., J-X.B., C.P., X.G. and B.L. Designed and performed the experiments: Y.L., Q.Y., Y.S., Y.J., G.L., X.Y., L.X., J.D. and Y.W., Wrote the manuscript: C.P., X.G., and B.L.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFA0502600 to B.L.); the National Natural Science Foundation of China (82073074 to B.L., 42076095 to Y.S., and 82003194 to C.P.); Guangdong Natural Science Fund, China (2021A1515011207 to X.G.); the National Program for Support of Top-Notch Young Professionals (to J-X.B.); Chang Jiang Scholars Program (to J-X.B.); Special Support Program of Guangdong (to J-X.B.); Sun Yat-sen University Young Teacher Key Cultivate Project (17ykjc24); Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S638); the postdoctoral research initiation fund by Guangzhou Institute of Pediatrics, Guangzhou Women and Children’s Medical Center.
Data availability
Our raw RNA seq deposited in NCBI SRA database, and the accession ID is PRJNA720775.
Ethics statement
We do not process samples of human participants in this study. Analyzed gene expressions of human NB tissues were obtained from a public database, which was approved by the Ethics Committee of the Medical Faculty of University of Cologne or local universities of collaborators according to related reference. The study was performed in accordance with the Declaration of Helsinki.
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by B. Zhivotovsky
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuxiong Lu, Qing Yang, Yubin Su, Yin Ji
Contributor Information
Xiaoqiong Gu, Email: guxiaoqiong@gwcmc.org.
Bo Li, Email: libo47@mail.sysu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-03790-w.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Matthay KK, et al. Neuroblastoma. Nat. Rev. Dis. Prim. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 3.Diskin SJ, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rickman DS, Schulte JH, Eilers M. The expanding world of N-MYC-driven tumors. Cancer Discov. 2018;8:150–163. doi: 10.1158/2159-8290.CD-17-0273. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Perez M. V., Henley A. B. Arsenian-Henriksson M. The MYCN protein in health and disease. Genes8, 113 (2017). [DOI] [PMC free article] [PubMed]
- 6.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013;3:a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Y, et al. N-Myc promotes therapeutic resistance development of neuroendocrine prostate cancer by differentially regulating miR-421/ATM pathway. Mol. Cancer. 2019;18:11. doi: 10.1186/s12943-019-0941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen RR, et al. MYCN induces neuroblastoma in primary neural crest cells. Oncogene. 2017;36:5075–5082. doi: 10.1038/onc.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon SB. MYC and the control of apoptosis. Cold Spring Harb. Perspect. Med. 2014;4:a014407. doi: 10.1101/cshperspect.a014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King D, et al. MYCN expression induces replication stress and sensitivity to PARP inhibition in neuroblastoma. Oncotarget. 2020;11:2141–2159. doi: 10.18632/oncotarget.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng H, Stockwell BR. Unsolved mysteries: how does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16:e2006203. doi: 10.1371/journal.pbio.2006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miess H, et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene. 2018;37:5435–5450. doi: 10.1038/s41388-018-0315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chekhun VF, et al. Iron metabolism disturbances in the MCF-7 human breast cancer cells with acquired resistance to doxorubicin and cisplatin. Int. J. Oncol. 2013;43:1481–1486. doi: 10.3892/ijo.2013.2063. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez SW, et al. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017;551:639–643. doi: 10.1038/nature24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stockwell BR, Jiang X. The chemistry and biology of ferroptosis. Cell Chem. Biol. 2020;27:365–375. doi: 10.1016/j.chembiol.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tjaden B, et al. N-Myc-induced metabolic rewiring creates novel therapeutic vulnerabilities in neuroblastoma. Sci. Rep. 2020;10:7157. doi: 10.1038/s41598-020-64040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durbin AD, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 2018;50:1240–1246. doi: 10.1038/s41588-018-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang JY, et al. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int. J. Biol. Sci. 2020;16:2430–2441. doi: 10.7150/ijbs.45050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadian K, Stockwell BR. SnapShot: ferroptosis. Cell. 2020;181:1188–1188.e1. doi: 10.1016/j.cell.2020.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla K, et al. Inhibition of xc− transporter-mediated cystine uptake by sulfasalazine analogs. Bioorg. Med. Chem. Lett. 2011;21:6184–6187. doi: 10.1016/j.bmcl.2011.07.081. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, et al. Metabolic reprogramming by MYCN confers dependence on the serine-glycine-one-carbon biosynthetic pathway. Cancer Res. 2019;79:3837–3850. doi: 10.1158/0008-5472.CAN-18-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon SJ, Stockwell BR. The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 2019;3:35–54. doi: 10.1146/annurev-cancerbio-030518-055844. [DOI] [Google Scholar]
- 23.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 2020;22:476–486. doi: 10.1038/s41556-020-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes. Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawai S, et al. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 27.Fredlund E, Ringner M, Maris JM, Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc. Natl Acad. Sci. USA. 2008;105:14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren P, et al. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J. Pathol. 2015;235:90–100. doi: 10.1002/path.4429. [DOI] [PubMed] [Google Scholar]
- 29.Qing G, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22:631–644. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floros K. V., et al. MYCN-amplified neuroblastoma is addicted to iron and vulnerable to inhibition of the system Xc−/glutathione axis. Cancer Res.10.1158/0008-5472.CAN-20-1641 (2021). [DOI] [PMC free article] [PubMed]
- 32.Weber CK, Liptay S, Wirth T, Adler G, Schmid RM. Suppression of NF-kappaB activity by sulfasalazine is mediated by direct inhibition of IkappaB kinases alpha and beta. Gastroenterology. 2000;119:1209–1218. doi: 10.1053/gast.2000.19458. [DOI] [PubMed] [Google Scholar]
- 33.Jansen G, et al. Sulfasalazine is a potent inhibitor of the reduced folate carrier: implications for combination therapies with methotrexate in rheumatoid arthritis. Arthritis Rheum. 2004;50:2130–2139. doi: 10.1002/art.20375. [DOI] [PubMed] [Google Scholar]
- 34.Jenkitkasemwong S, et al. SLC39A14 is required for the development of hepatocellular iron overload in murine models of hereditary hemochromatosis. Cell Metab. 2015;22:138–150. doi: 10.1016/j.cmet.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T, et al. MYCN drives glutaminolysis in neuroblastoma and confers sensitivity to an ROS augmenting agent. Cell Death Dis. 2018;9:220. doi: 10.1038/s41419-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng H, et al. Transferrin receptor is a specific ferroptosis marker. Cell Rep. 2020;30:3411–3423.e7. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 39.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 40.Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, et al. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- 42.Kocak H, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal. Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our raw RNA seq deposited in NCBI SRA database, and the accession ID is PRJNA720775.