Abstract
Diagnosis of Parkinson's disease (PD) is commonly based on medical observations and assessment of clinical signs, including the characterization of a variety of motor symptoms. However, traditional diagnostic approaches may suffer from subjectivity as they rely on the evaluation of movements that are sometimes subtle to human eyes and therefore difficult to classify, leading to possible misclassification. In the meantime, early non-motor symptoms of PD may be mild and can be caused by many other conditions. Therefore, these symptoms are often overlooked, making diagnosis of PD at an early stage challenging. To address these difficulties and to refine the diagnosis and assessment procedures of PD, machine learning methods have been implemented for the classification of PD and healthy controls or patients with similar clinical presentations (e.g., movement disorders or other Parkinsonian syndromes). To provide a comprehensive overview of data modalities and machine learning methods that have been used in the diagnosis and differential diagnosis of PD, in this study, we conducted a literature review of studies published until February 14, 2020, using the PubMed and IEEE Xplore databases. A total of 209 studies were included, extracted for relevant information and presented in this review, with an investigation of their aims, sources of data, types of data, machine learning methods and associated outcomes. These studies demonstrate a high potential for adaptation of machine learning methods and novel biomarkers in clinical decision making, leading to increasingly systematic, informed diagnosis of PD.
Keywords: Parkinson's disease, machine learning, deep learning, diagnosis, differential diagnosis
Introduction
Parkinson's disease (PD) is one of the most common neurodegenerative diseases with a prevalence rate of 1% in the population above 60 years old, affecting 1–2 people per 1,000 (Tysnes and Storstein, 2017). The estimated global population affected by PD has more than doubled from 1990 to 2016 (from 2.5 million to 6.1 million), which is a result of increased number of elderly people and age-standardized prevalence rates (Dorsey et al., 2018). PD is a progressive neurological disorder associated with motor and non-motor features (Jankovic, 2008) which comprises multiple aspects of movements, including planning, initiation and execution (Contreras-Vidal and Stelmach, 1995).
During its development, movement-related symptoms such as tremor, rigidity and difficulties in initiation can be observed, prior to cognitive and behavioral alterations including dementia (Opara et al., 2012). PD severely affects patients' quality of life (QoL), social functions and family relationships, and places heavy economic burdens at individual and society levels (Johnson et al., 2013; Kowal et al., 2013; Yang and Chen, 2017).
The diagnosis of PD is traditionally based on motor symptoms. Despite the establishment of cardinal signs of PD in clinical assessments, most of the rating scales used in the evaluation of disease severity have not been fully evaluated and validated (Jankovic, 2008). Although non-motor symptoms (e.g., cognitive changes such as problems with attention and planning, sleep disorders, sensory abnormalities such as olfactory dysfunction) are present in many patients prior to the onset of PD (Jankovic, 2008; Tremblay et al., 2017), they lack specificity, are complicated to assess and/or yield variability from patient to patient (Zesiewicz et al., 2006). Therefore, non-motor symptoms do not yet allow for diagnosis of PD independently (Braak et al., 2003), although some have been used as supportive diagnostic criteria (Postuma et al., 2015).
Machine learning techniques are being increasingly applied in the healthcare sector. As its name implies, machine learning allows for a computer program to learn and extract meaningful representation from data in a semi-automatic manner. For the diagnosis of PD, machine learning models have been applied to a multitude of data modalities, including handwritten patterns (Drotár et al., 2015; Pereira et al., 2018), movement (Yang et al., 2009; Wahid et al., 2015; Pham and Yan, 2018), neuroimaging (Cherubini et al., 2014a; Choi et al., 2017; Segovia et al., 2019), voice (Sakar et al., 2013; Ma et al., 2014), cerebrospinal fluid (CSF) (Lewitt et al., 2013; Maass et al., 2020), cardiac scintigraphy (Nuvoli et al., 2019), serum (Váradi et al., 2019), and optical coherence tomography (OCT) (Nunes et al., 2019). Machine learning also allows for combining different modalities, such as magnetic resonance imaging (MRI) and single-photon emission computed tomography (SPECT) data (Cherubini et al., 2014b; Wang et al., 2017), in the diagnosis of PD. By using machine learning approaches, we may therefore identify relevant features that are not traditionally used in the clinical diagnosis of PD and rely on these alternative measures to detect PD in preclinical stages or atypical forms.
In recent years, the number of publications on the application of machine learning to the diagnosis of PD has increased. Although previous studies have reviewed the use of machine learning in the diagnosis and assessment of PD, they were limited to the analysis of motor symptoms, kinematics, and wearable sensor data (Ahlrichs and Lawo, 2013; Ramdhani et al., 2018; Belić et al., 2019). Moreover, some of these reviews only included studies published between 2015 and 2016 (Pereira et al., 2019). In this study, we aim to (a) comprehensively summarize all published studies that applied machine learning models to the diagnosis of PD for an exhaustive overview of data sources, data types, machine learning models, and associated outcomes, (b) assess and compare the feasibility and efficiency of different machine learning methods in the diagnosis of PD, and (c) provide machine learning practitioners interested in the diagnosis of PD with an overview of previously used models and data modalities and the associated outcomes, and recommendations on how experimental protocols and results could be reported to facilitate reproduction. As a result, the application of machine learning to clinical and non-clinical data of different modalities has often led to high diagnostic accuracies in human participants, therefore may encourage the adaptation of machine learning algorithms and novel biomarkers in clinical settings to assist more accurate and informed decision making.
Methods
Search Strategy
A literature search was conducted on the PubMed (https://pubmed.ncbi.nlm.nih.gov) and IEEE Xplore (https://ieeexplore.ieee.org/search/advanced/command) databases on February 14, 2020 for all returned results. Boolean search strings used are shown in Table 1. No additional filters were applied in the literature search. All retrieved studies were systematically identified, screened and extracted for relevant information following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al., 2009).
Table 1.
Boolean search strings used for the retrieval of relevant publications on PubMed and IEEE Xplore databases.
| Database | Boolean search string |
|---|---|
| PubMed | (“Parkinson Disease”[Mesh] OR Parkinson*) AND (“Machine Learning”[Mesh] OR machine learn* OR machine-learn* OR deep learn* OR deep-learn*) AND (human OR patient) AND (“Diagnosis”[Mesh] OR diagnos* OR detect* OR classif* OR identif*) NOT review[Publication Type] |
| IEEE Xplore | (Parkinson*) AND (machine learn* OR machine-learn* OR deep learn* OR deep-learn*) AND (human OR patient) AND (diagnosis OR diagnose OR diagnosing OR detection OR detect OR detecting OR classification OR classify OR classifying OR identification OR identify OR identifying) |
Inclusion and Exclusion Criteria
Studies that satisfy one or more of the following criteria and used machine learning methods were included:
Classification of PD from healthy controls (HC),
Classification of PD from Parkinsonism (e.g., progressive supranuclear palsy (PSP) and multiple system atrophy (MSA)), and
Classification of PD from other movement disorders (e.g., essential tremor (ET)).
Studies falling into one or more of the following categories were excluded:
Studies related to Parkinsonism or/and diseases other than PD that did not involve classification or detection of PD (e.g., differential diagnosis of PSP, MSA, and other atypical Parkinsonian disorders),
Studies not related to the diagnosis of PD (e.g., subtyping or severity assessment, analysis of behavior, disease progression, treatment outcome prediction, identification, and localization of brain structures or parameter optimization during surgery),
Studies related to the diagnosis of PD, but performed analysis and assessed model performance at sample level (e.g., classification using individual MRI scans without aggregating scan-level performance to patient level),
Classification of PD from non-Parkinsonism (e.g., Alzheimer's disease),
Study did not use metrics that measure classification performance,
Study used organisms other than human (e.g., Caenorhabditis elegans, mice or rats),
Study did not provide sufficient or accurate descriptions of machine learning methods, datasets or subjects used (e.g., does not provide sample size, or incorrectly described the dataset(s) used),
Not original journal article or conference proceedings papers (e.g., review and viewpoint paper), and
In languages other than English.
Data Extraction
The following information is included in the data extraction table: (1) objectives, (2) type of diagnosis (diagnosis, differential diagnosis, sub-typing), (3) data source, (4) data type, (5) number of subjects, (6) machine learning method(s), splitting strategy and cross validation, (7) associated outcomes, (8) year, and (9) reference.
For studies published online first and archived in another year, “year of publication” was defined as the year during which the study was published online. If this information was unavailable, the year in which the article was copyrighted was regarded as the year of publication. For studies that introduced novel models and used existing models merely for comparison, information related to the novel models was extracted. Classification of PD and scans without evidence for dopaminergic deficit (SWEDD) was treated as subtyping (Erro et al., 2016).
Study Objectives
To outline the different goals and objectives of included studies, we have further categorized them based on the type of diagnosis and their general aim. From the perspective of diagnostics, these studies could be divided into (a) the diagnosis or detection of PD (which compares data collected from PD patients and healthy controls), (b) differential diagnosis (discrimination between patients with idiopathic PD and patients with atypical Parkinsonism), and (c) sub-typing (discrimination among sub-types of PD).
Included studies were also analyzed for their general aim: For studies with a focus on the development of novel technical approaches to be used in the diagnosis of Parkinson's disease, e.g., new machine learning and deep learning models and architectures, data acquisition devices, and feature extraction algorithms that haven't been previously presented and/or employed, we defined them as (a) “methodology” studies. Studies that validate and investigate (a) the application of previously published and validated machine learning and deep learning models, and/or (b) the feasibility of introducing data modalities that are not commonly used in the machine learning-based diagnosis of PD (e.g., CSF data), were defined as (b) “clinical application” studies.
Model Evaluation
In the present study, accuracy was used to compare performance of machine learning models. For each data type, we summarized the type of machine learning models that led to the per-study highest accuracy. However, in some studies, only one machine learning model was tested. Therefore, we define “model associated with the per-study highest accuracy” as (a) the only model implemented and used in a study or (b) the model that achieved the highest accuracy or that was highlighted in studies that used multiple models. Results are expressed as mean (SD).
For studies reporting both training and testing/validation accuracy, testing or validation accuracy was considered. For studies that reported both validation and test accuracy, test accuracy was considered. For studies with more than one dataset or classification problem (e.g., HC vs. PD and HC vs. idiopathic hyposmia vs. PD), accuracy was averaged across datasets or classification problems. For studies that reported classification accuracy for each group of subjects individually, accuracy was averaged across groups. For studies reporting a range of accuracies or accuracies given by different cross validation methods or feature combinations, the highest accuracies were considered. In studies that compared HC with diseases other than PD or PD with diseases other than Parkinsonism, diagnosis of diseases other than PD or Parkinsonism (e.g., amyotrophic lateral sclerosis) was not considered. Accuracy of severity assessment was not considered.
Results
Literature Review
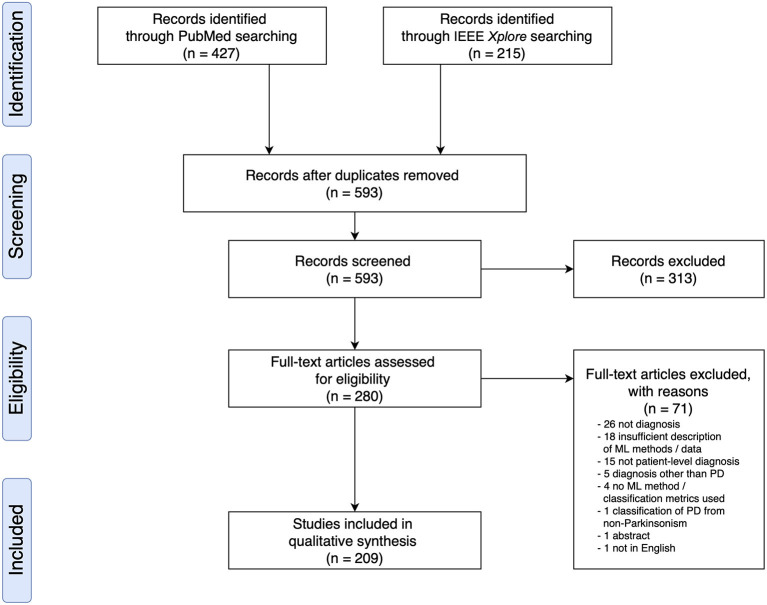
Based on the search criteria, we retrieved 427 (PubMed) and 215 (IEEEXplore) search results, leading to a total of 642 publications. After removing duplicates, we screened 593 publications for titles and abstracts, following which we excluded 313 based on the exclusion criteria and examined 280 full text articles. Overall, we included 209 research articles for data extraction (Figure 1 and see Supplementary Materials for a full list of included studies). All articles were published from the year 2009 onwards, and an increase in the number of papers published per year was observed (Supplementary Figure 1).
Figure 1.
PRISMA Flow Diagram of Literature Search and Selection Process showing the number of studies identified, screened, extracted, and included in the review.
Data Source and Sample Size
In 93 out of 209 studies (43.1%), original data were collected from human participants. In 108 studies (51.7%), data used were from public repositories and databases, including University of California at Irvine (UCI) Machine Learning Repository (Dua and Graff, 2018) (n = 44), Parkinson's Progression Markers Initiative (Marek et al., 2011) (PPMI; n = 33), PhysioNet (Goldberger et al., 2000) (n = 15), HandPD dataset (Pereira et al., 2015) (n = 6), mPower database (Bot et al., 2016) (n = 4), and 6 other databases (Mucha et al., 2018; Vlachostergiou et al., 2018; Bhati et al., 2019; Hsu et al., 2019; Taleb et al., 2019; Wodzinski et al., 2019; Table 2).
Table 2.
Source of data of the included studies.
| Data source/Database | Number of studies | Percentage |
|---|---|---|
| independent recruitment of human participants | 93 | 43.06% |
| UCI Machine Learning Repository | 44 | 20.37% |
| PPMI database | 33 | 15.28% |
| PhysioNet | 15 | 6.94% |
| HandPD dataset | 6 | 2.78% |
| mPower database | 4 | 1.85% |
| Other databases (1 PACS, 1 PaHaW, 1 PC-GITA database, 1 PDMultiMC database, 1 Neurovoz corpus, 1 The NTUA Parkinson Dataset) |
6 | 2.78% |
| Collected postmortem | 1 | 0.46% |
| Commercially sourced | 1 | 0.46% |
| Acquired at another institution | 1 | 0.46% |
| From another study | 1 | 0.46% |
| From the author's institutional database | 1 | 0.46% |
| Others (1 PPMI + Sheffield Teaching Hospitals NHS Foundation Trust; 1 PPMI + Seoul National University Hospital cohort; 1 UCI + collected from participants) |
3 | 1.39% |
PACS, Picture Archiving and Communication System; PaHaW, Parkinson's Disease Handwriting Database.
In 3 studies, data from public repositories were combined with data from local databases or participants (Agarwal et al., 2016; Choi et al., 2017; Taylor and Fenner, 2017). In the remaining studies, data were sourced (Wahid et al., 2015) from another study (Fernandez et al., 2013), collected at another institution (Segovia et al., 2019), obtained from the authors' institutional database (Nunes et al., 2019), collected postmortem (Lewitt et al., 2013), or commercially sourced (Váradi et al., 2019).
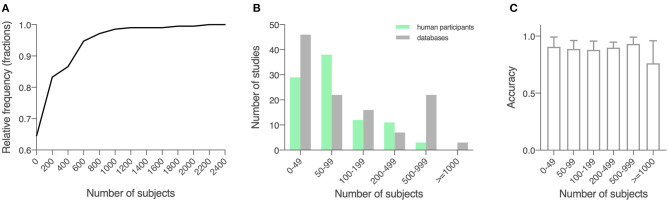
The 209 studies had an average sample size of 184.6 (289.3), with a smallest sample size of 10 (Kugler et al., 2013), and a largest sample size of 2,289 (Tracy et al., 2019; Figure 2A). For studies that recruited human participants (n = 93), data from an average of 118.0 (142.9) participants were collected (range: 10–920; Figure 2B). For other studies (n = 116), an average sample size of 238.1 (358.5) was reported (range: 30–2,289; Figure 2B). For a description of average accuracy reported in these studies in relation to sample size, see Figure 2C.
Figure 2.
Sample size of the included studies. (A) Cumulative relative frequency graph depicting the frequency of the sample sizes studied. (B) Histogram depicting the frequency of a sample size of 0–50, 50–100, 100–200, 200–500, 500–100, and over 1,000 for studies using locally recruited human participants and studies using previously published open databases. Green, studies using locally recruited human participants; gray, studies using data sourced from public databases. (C) Model performance as measured by accuracy in relation to sample size, shown in means (SD).
Study Objectives
In included studies, although “diagnosis of PD” was used as the search criteria, machine learning had been applied for diagnosis (PD vs. HC), differential diagnosis (idiopathic PD vs. atypical Parkinsonism) and sub-typing (differentiation of sub-types of PD) purposes. Most studies focused on diagnosis (n = 168, 80.4%) or differential diagnosis (n = 20, 9.6%). Fourteen studies performed both diagnosis and differential diagnosis (6.7%), 5 studies (2.4%) diagnosed and subtyped PD, 2 studies (1.0%) included diagnosis, differential diagnosis, and subtyping.
Among the included studies, a total of 132 studies (63.2%) implemented and tested a machine learning method, a model architecture, a diagnostic system, a feature extraction algorithm, or a device for non-invasive, low-cost data acquisition that hasn't been established for the detection and early diagnosis of PD (methodology studies). In 77 studies (36.8%), previously proposed and validated machine learning methods were tested in clinical settings for early detection of PD, identification of novel biomarkers or examination of uncommonly used data modalities for the diagnosis of PD (e.g., CSF; clinical application studies).
Comparing Studies With Different Objectives
Source of Data
In the 132 studies that proposed or tested novel machine learning methods (i.e., methodology studies), a majority used data from publicly available databases (n = 89, 67.4%). Data collected from human participants were used in 41 studies (31.1%) and the two remaining studies (1.5%) used commercially sourced data or data from both existing public databases and local participants specifically recruited for the study. Out of the 77 studies that used machine learning models in clinical settings (i.e., clinical application studies), 52 (67.5%) collected data from human participants, 22 (28.6%) used data from public databases. Two (2.6%) studies obtained data from a database and a local cohort, and 1 (1.3%) study collected data postmortem.
Data Modality
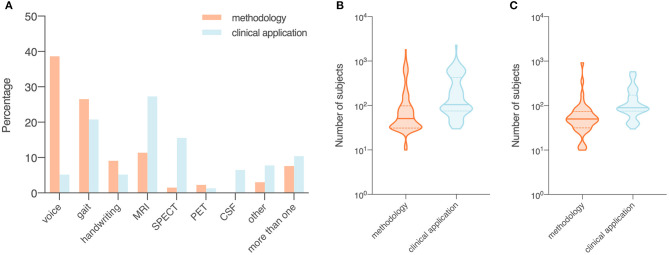
In methodology studies, the most commonly used data modalities were voice recordings (n = 51, 38.6%), movement (n = 35, 26.5%), and MRI data (n = 15, 11.4%). For studies on clinical applications, MRI data (n = 21, 27.3%), movement (n = 16, 20.8%), and SPECT imaging data (n = 12, 15.6%) were of high relevance. All studies using CSF features (n = 5) focused on validation of existing machine learning models in a clinical setting (Figure 3A).
Figure 3.
Data modality (A) and number of subjects (B,C) of included studies, summarized by objectives (i.e., methodology or clinical application). Orange, studies with a focus on the development of a novel technical approach to be used in the diagnosis of Parkinson's disease (i.e., methodology); blue, studies that investigate the use of published machine learning models or novel data modalities (i.e., clinical application). (A) Proportion of data modalities in included studies displayed as percentages. (B) Sample size in all included studies. (C) Sample size in studies that collected data from recruited human participants. Data shown are means (SD).
Number of Subjects
The average sample size was 137.1 for the 132 methodology studies (Figure 3B). For 41 out of the 132 studies that used data from recruited human participants, the average sample size was 81.7 (Figure 3C). In the 77 studies on clinical applications, the average sample size was 266.2 (Figure 3B). For 52 out of the 77 clinical studies that collected data from recruited participants, the average sample size was 145.9 (Figure 3C).
Machine Learning Methods Applied to the Diagnosis of PD
We divided 448 machine learning models from the 209 studies into 8 categories: (1) support vector machine (SVM) and variants (n = 132 from 130 studies), (2) neural networks (n = 76 from 62 studies), (3) ensemble learning (n = 82 from 57 studies), (4) nearest neighbor and variants (n = 33 from 33 studies), (5) regression (n = 31 from 31 studies), (6) decision tree (n = 28 from 27 studies), (7) naïve Bayes (n = 26, from 26 studies), and (8) discriminant analysis (n = 12 from 12 studies). A small percentage of models used did not fall into any of the categories (n = 28, used in 24 studies).
On average, 2.14 machine learning models per study were applied to the diagnosis of PD. One study may have used more than one category of models. For a full description of data types used to train each type of machine learning models and the associated outcomes, see Supplementary Materials and Supplementary Figure 2.
Performance Metrics
Various metrics have been used to assess the performance of machine learning models (Table 3). The most common metric was accuracy (n = 174, 83.3%), which was used individually (n = 55) or in combination with other metrics (n = 119) in model evaluation. Among the 174 studies that used accuracy, some have combined accuracy with sensitivity (i.e., recall) and specificity (n = 42), or with sensitivity, specificity and AUC (n = 16), or with recall (i.e., sensitivity), precision and F1 score (n = 7) for a more systematic understanding of model performance. A total of 35 studies (16.7%) used metrics other than accuracy. In these studies, the most used performance metrics were AUC (n = 19), sensitivity (n = 17), and specificity (n = 14), and the three were often applied together (n = 9) with or without other metrics.
Table 3.
Performance metrics used in the evaluation of machine learning models.
| Performance metric | Definition | Number of studies |
|---|---|---|
| Accuracy | 174 | |
| Sensitivity (recall) | 110 | |
| Specificity (TNR) | 94 | |
| AUC | The two-dimensional area under the Receiver Operating Characteristic (ROC) curve | 60 |
| MCC | 9 | |
| Precision (PPV) | 31 | |
| NPV | 8 | |
| F1 score | 25 | |
| Others (7 kappa; 4 error rate; 3 EER; 1 MSE; 1 LOR; 1 confusion matrix; 1 cross validation score; 1 YI; 1 FPR; 1 FNR; 1 G-mean; 1 PE; 5 combination of metrics) |
N/A | 28 |
TNR, true negative rate; AUC, Area under the ROC Curve; MCC, Matthews correlation coefficient; PPV, positive predictive value; NPV, negative predictive value; EER, equal error rate; MSE, mean squared error; LOR, log odds ratio; YI, Youden's Index; FPR, false positive rate; FNR, false negative rate; PE, probability excess.
Data Types and Associated Outcomes
Out of 209 studies, 122 (58.4%) applied machine learning methods to movement-related data, i.e., voice recordings (n = 55, 26.3%), movement data (n = 51, 24.4%), or handwritten patterns (n = 16, 7.7%). Imaging modalities analyzed including MRI (n = 36, 17.2%), SPECT (n = 14, 6.7%), and positron emission tomography (PET; n = 4, 1.9%). Five studies analyzed CSF samples (2.4%). In 18 studies (8.6%), a combination of different types of data was used.
Ten studies (4.8%) used data that do not belong to any categories mentioned above, such as single nucleotide polymorphisms (Cibulka et al., 2019) (SNPs), electromyography (EMG) (Kugler et al., 2013), OCT (Nunes et al., 2019), cardiac scintigraphy (Nuvoli et al., 2019), Patient Questionnaire of Movement Disorder Society Unified Parkinson's Disease Rating Scale (MDS-UPDRS) (Prashanth and Dutta Roy, 2018), whole-blood gene expression profiles (Shamir et al., 2017), transcranial sonography (Shi et al., 2018) (TCS), eye movements (Tseng et al., 2013), electroencephalography (EEG) (Vanegas et al., 2018), and serum samples (Váradi et al., 2019).
Given that studies used different data modalities and sources, and sometimes different samples of the same database, a summary of model performance, instead of direct comparison across studies, is provided.
Voice Recordings (n = 55)
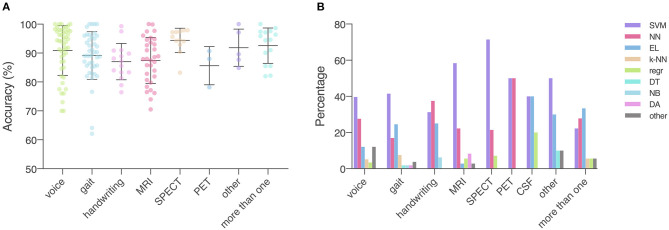
The 49 studies that used accuracy to evaluate machine learning models achieved an average accuracy of 90.9 (8.6) % (Figure 4A), ranging from 70.0% (Kraipeerapun and Amornsamankul, 2015; Ali et al., 2019a) to 100.0% (Hariharan et al., 2014; Abiyev and Abizade, 2016; Ali et al., 2019c; Dastjerd et al., 2019). In 3 studies, the highest accuracy was achieved by two types of machine learning models individually, namely regression or SVM (Ali et al., 2019a), neural network or SVM (Hariharan et al., 2014), and ensemble learning or SVM (Mandal and Sairam, 2013). The per-study highest accuracy was achieved with SVM in 23 studies (39.7%), with neural network in 16 studies (27.6%), with ensemble learning in 7 studies (12.1%), with nearest neighbor in 3 studies (5.2%), and with regression in 2 studies (3.4%). Models that do not belong to any given categories led to the per-study highest accuracy in 7 studies (12.1%; Figure 4B).
Figure 4.
Data type, machine learning models applied, and accuracy. (A) Accuracy achieved in individual studies and average accuracy for each data type. Error bar: standard deviation. (B) Distribution of machine learning models applied per data type. MRI, magnetic resonance imaging; SPECT, single-photon emission computed tomography; PET, positron emission tomography; CSF, cerebrospinal fluid; SVM, support vector machine; NN, neural network; EL, ensemble learning; k-NN, nearest neighbor; regr, regression; DT, decision tree; NB, naïve Bayes; DA, discriminant analysis; other: data/models that do not belong to any of the given categories.
Voice recordings from the UCI machine learning repository were used in 42 studies (Table 4). Among the 42 studies, 39 used accuracy to evaluate classification performance and the average accuracy was 92.0 (9.0) %. The lowest accuracy was 70.0% and the highest accuracy was 100.0%. Eight out of 9 studies that collected voice recordings from human participants used accuracy as the performance metric, and the average, lowest and highest accuracies were 87.7 (6.8) %, 77.5%, and 98.6%, respectively. The 4 remaining studies used data from the Neurovoz corpus (n = 1), mPower database (n = 1), PC-GITA database (n = 1), or data from both the UCI machine learning repository and human participants (n = 1). Two out of these 4 studies used accuracy to evaluate model performance and reported an accuracy of 81.6 and 91.7%.
Table 4.
Studies that applied machine learning models to voice recordings to diagnose PD (n = 55).
| Objectives | Type of diagnosis | Source of data | Number of subjects (n) | Machine learning method(s), splitting strategy and cross validation | Outcomes | Year | References |
|---|---|---|---|---|---|---|---|
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Fuzzy neural system with 10-fold cross validation | Testing accuracy = 100% | 2016 | Abiyev and Abizade, 2016 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | RPART, C4.5, PART, Bagging CART, random forest, Boosted C5.0, SVM | SVM: | 2019 | Aich et al., 2019 |
| Accuracy = 97.57% | |||||||
| Sensitivity = 0.9756 | |||||||
| Specificity = 0.9987 | |||||||
| NPV = 0.9995 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | DBN of 2 RBMs | Testing accuracy = 94% | 2016 | Al-Fatlawi et al., 2016 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | EFMM-OneR with 10-fold cross validation or 5-fold cross validation | Accuracy = 94.21% | 2019 | Sayaydeha and Mohammad, 2019 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | Linear regression, LDA, Gaussian naïve Bayes, decision tree, KNN, SVM-linear, SVM-RBF with leave-one-subject-out cross validation | Logistic regression or SVM-linear accuracy = 70% | 2019 | Ali et al., 2019a |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | LDA-NN-GA with leave-one-subject-out cross validation | Training: | 2019 | Ali et al., 2019c |
| Accuracy = 95% | |||||||
| Sensitivity = 95% | |||||||
| Test: | |||||||
| Accuracy = 100% | |||||||
| Sensitivity = 100% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | NNge with AdaBoost with 10-fold cross validation | Accuracy = 96.30% | 2018 | Alqahtani et al., 2018 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Logistic regression, KNN, naïve Bayes, SVM, decision tree, random forest, DNN with 10-fold cross validation | KNN accuracy = 95.513% | 2018 | Anand et al., 2018 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | MLP with a train-validation-test ratio of 50:20:30 | Training accuracy = 97.86% | 2012 | Bakar et al., 2012 |
| Test accuracy = 92.96% | |||||||
| MSE = 0.03552 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31 (8 HC + 23 PD) for dataset 1 and 68 (20 HC + 48 PD) for dataset 2 | FKNN, SVM, KELM with 10-fold cross validation | FKNN accuracy = 97.89% | 2018 | Cai et al., 2018 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | SVM, logistic regression, ET, gradient boosting, random forest with train-test split ratio = 80:20 | Logistic regression accuracy = 76.03% | 2019 | Celik and Omurca, 2019 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | MLP, GRNN with a training-test ratio of 50:50 | GRNN: | 2016 | Çimen and Bolat, 2016 |
| Error rate = 0.0995 (spread parameter = 195.1189) | |||||||
| Error rate = 0.0958 (spread parameter = 1.2) | |||||||
| Error rate = 0.0928 (spread parameter = 364.8) | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | ECFA-SVM with 10-fold cross validation | Accuracy = 97.95% | 2017 | Dash et al., 2017 |
| Sensitivity = 97.90% | |||||||
| Precision = 97.90% | |||||||
| F-measure = 97.90% | |||||||
| Specificity = 96.50% | |||||||
| AUC = 97.20% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | Fuzzy classifier with 10-fold cross validation, leave-one-out cross validation or a train-test ratio of 70:30 | Accuracy = 100% | 2019 | Dastjerd et al., 2019 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Averaged perceptron, BPM, boosted decision tree, decision forests, decision jungle, locally deep SVM, logistic regression, NN, SVM with 10-fold cross-validation | Boosted decision trees: | 2017 | Dinesh and He, 2017 |
| Accuracy = 0.912105 | |||||||
| Precision = 0.935714 | |||||||
| F-score = 0.942368 | |||||||
| AUC = 0.966293 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 50; 8 HC + 42 PD | KNN, SVM, ELM with a train-validation ratio of 70:30 | SVM: | 2017 | Erdogdu Sakar et al., 2017 |
| Accuracy = 96.43% | |||||||
| MCC = 0.77 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 252; 64 HC + 188 PD | CNN with leave-one-person-out cross validation | Accuracy = 0.869 | 2019 | Gunduz, 2019 |
| F-measure = 0.917 | |||||||
| MCC = 0.632 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | SVM, logistic regression, KNN, DNN with a train-test ratio of 70:30 | DNN: | 2018 | Haq et al., 2018 |
| Accuracy = 98% | |||||||
| Specificity = 95% | |||||||
| sensitivity = 99% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | SVM-RBF, SVM-linear with 10-fold cross validation | Accuracy = 99% | 2019 | Haq et al., 2019 |
| Specificity = 99% | |||||||
| Sensitivity = 100% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | LS-SVM, PNN, GRNN with conventional (train-test ratio of 50:50) and 10-fold cross validation | LS-SVM or PNN or GRNN: | 2014 | Hariharan et al., 2014 |
| Accuracy = 100% | |||||||
| Precision = 100% | |||||||
| Sensitivity = 100% | |||||||
| specificity = 100% | |||||||
| AUC = 100 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Random tree, SVM-linear, FBANN with 10-fold cross validation | FBANN: | 2014 | Islam et al., 2014 |
| Accuracy = 97.37% | |||||||
| Sensitivity = 98.60% | |||||||
| Specificity = 93.62% | |||||||
| FPR = 6.38% | |||||||
| Precision = 0.979 | |||||||
| MSE = 0.027 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | SVM-linear with 5-fold cross validation | Error rate ~0.13 | 2012 | Ji and Li, 2012 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | Decision tree, random forest, SVM, GBM, XGBoost | SVM-linear: | 2018 | Junior et al., 2018 |
| FNR = 10% | |||||||
| Accuracy = 0.725 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | CART, SVM, ANN | SVM accuracy = 93.84% | 2020 | Karapinar Senturk, 2020 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | Dataset 1: 31; 8 HC + 23 PD Dataset 2: 40; 20 HC + 20 PD |
EWNN with a train-test ratio of 90:10 and cross validation | Dataset 1: Accuracy = 92.9% |
2018 | Khan et al., 2018 |
| Ensemble classification accuracy = 100.0% | |||||||
| Sensitivity = 100.0% | |||||||
| MCC = 100.0% | |||||||
| Dataset 2: | |||||||
| Accuracy = 66.3% | |||||||
| Ensemble classification accuracy = 90.0% | |||||||
| Sensitivity = 93.0% | |||||||
| Specificity = 97.0% | |||||||
| MCC = 87.0% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | Stacked generalization with CMTNN with 10-fold cross validation | Accuracy = ~70% | 2015 | Kraipeerapun and Amornsamankul, 2015 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 40; 20 HC + 20 PD | HMM, SVM | HMM: | 2019 | Kuresan et al., 2019 |
| Accuracy = 95.16% | |||||||
| Sensitivity = 93.55% | |||||||
| Specificity = 91.67% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | IGWO-KELM with 10-fold cross validation | Iteration number = 100 | 2017 | Li et al., 2017 |
| Accuracy = 97.45% | |||||||
| Sensitivity = 99.38% | |||||||
| Specificity = 93.48% | |||||||
| Precision = 97.33% | |||||||
| G-mean = 96.38% | |||||||
| F-measure = 98.34% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | SCFW-KELM with 10-fold cross validation | Accuracy = 99.49% | 2014 | Ma et al., 2014 |
| Sensitivity = 100% | |||||||
| Specificity = 99.39% | |||||||
| AUC = 99.69% | |||||||
| F-measure = 0.9966 | |||||||
| Kappa = 0.9863 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | SVM-RBF with 10-fold cross validation | Accuracy = 96.29% | 2016 | Ma et al., 2016 |
| Sensitivity = 95.00% | |||||||
| Specificity = 97.50% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Logistic regression, NN, SVM, SMO, Pegasos, AdaBoost, ensemble selection, FURIA, rotation forest Bayesian network with 10-fold cross-validation | Average accuracy across all models = 97.06% SMO, Pegasos, or AdaBoost accuracy = 98.24% |
2013 | Mandal and Sairam, 2013 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Logistic regression, KNN, SVM, naïve Bayes, decision tree, random forest, ANN | ANN: | 2018 | Marar et al., 2018 |
| Accuracy = 94.87% | |||||||
| Specificity = 96.55% | |||||||
| Sensitivity = 90% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | Dataset 1: 31; 8 HC + 23 PD | KNN | Dataset 1 accuracy = 90% | 2017 | Moharkan et al., 2017 |
| Dataset 2: 40; 20 HC + 20 PD | Dataset 2 accuracy = 65% | ||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Rotation forest ensemble with 10-fold cross validation | Accuracy = 87.1% | 2011 | Ozcift and Gulten, 2011 |
| Kappa error = 0.63 | |||||||
| AUC = 0.860 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Rotation forest ensemble | Accuracy = 96.93% | 2012 | Ozcift, 2012 |
| Kappa = 0.92 | |||||||
| AUC = 0.97 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | SVM-RBF with 10-fold cross validation or a train-test ratio of 50:50 | 10-fold cross validation: | 2016 | Peker, 2016 |
| Accuracy = 98.95% | |||||||
| Sensitivity = 96.12% | |||||||
| Specificity = 100% | |||||||
| F-measure = 0.9795 | |||||||
| Kappa = 0.9735 | |||||||
| AUC = 0.9808 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | ELM with 10-fold cross validation | Accuracy = 88.72% | 2016 | Shahsavari et al., 2016 |
| Recall = 94.33% | |||||||
| Precision = 90.48% | |||||||
| F-score = 92.36% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Ensemble learning with 10-fold cross validation | Accuracy = 90.6% | 2019 | Sheibani et al., 2019 |
| Sensitivity = 95.8% | |||||||
| Specificity = 75% | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | GLRA, SVM, bagging ensemble with 5-fold cross validation | Bagging: | 2017 | Wu et al., 2017 |
| Sensitivity = 0.9796 | |||||||
| Specificity = 0.6875 | |||||||
| MCC = 0.6977 | |||||||
| AUC = 0.9558 | |||||||
| SVM: | |||||||
| Sensitivity = 0.9252 | |||||||
| specificity = 0.8542 | |||||||
| MCC = 0.7592 | |||||||
| AUC = 0.9349 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | Decision tree classifier, logistic regression, SVM with 10-fold cross validation | SVM: | 2011 | Yadav et al., 2011 |
| Accuracy = 0.76 | |||||||
| Sensitivity = 0.9745 | |||||||
| Specificity = 0.13 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 80; 40 HC + 40 PD | KNN, SVM with 10-fold cross validation | SVM: | 2019 | Yaman et al., 2020 |
| Accuracy = 91.25% | |||||||
| Precision = 0.9125 | |||||||
| Recall = 0.9125 | |||||||
| F-Measure = 0.9125 | |||||||
| Classification of PD from HC | Diagnosis | UCI machine learning repository | 31; 8 HC + 23 PD | MAP, SVM-RBF, FLDA with 5-fold cross validation | MAP: | 2014 | Yang et al., 2014 |
| Accuracy = 91.8% | |||||||
| Sensitivity = 0.986 | |||||||
| Specificity = 0.708 | |||||||
| AUC = 0.94 | |||||||
| Classification of PD from other disorders | Differential diagnosis | Collected from participants | 50; 30 PD + 9 MSA + 5 FND + 1 somatization + 1 dystonia + 2 CD + 1 ET + 1 GPD | SVM, KNN, DA, naïve Bayes, classification tree with LOSO | SVM-linear: | 2016 | Benba et al., 2016a |
| Accuracy = 90% | |||||||
| Sensitivity = 90% | |||||||
| Specificity = 90% | |||||||
| MCC = 0.794067 | |||||||
| PE = 0.788177 | |||||||
| Classification of PD from other disorders | Differential diagnosis | Collected from participants | 40; 20 PD + 9 MSA + 5 FND + 1 somatization + 1 dystonia + 2 CD + 1ET + 1 GPD | SVM (RBF, linear, polynomial, and MLP kernels) with LOSO | SVM-linear accuracy = 85% | 2016 | Benba et al., 2016b |
| Classification of PD from HC and assess the severity of PD | Diagnosis | Collected from participants | 52; 9 HC + 43 PD | SVM-RBF with cross validation | Accuracy = 81.8% | 2014 | Frid et al., 2014 |
| Classification of PD from HC | Diagnosis | Collected from participants | 54; 27 HC + 27 PD | SVM with stratified 10-fold cross validation or leave-one-out cross validation | Accuracy = 94.4% | 2018 | Montaña et al., 2018 |
| Specificity = 100% | |||||||
| Sensitivity = 88.9% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 40; 20 HC + 20 PD | KNN, SVM-linear, SVM-RBF with leave-one-subject-out or summarized leave-one-out | SVM-linear: | 2013 | Sakar et al., 2013 |
| Accuracy = 77.50% | |||||||
| MCC = 0.5507 | |||||||
| Sensitivity = 80.00% | |||||||
| Specificity = 75.00% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 78; 27 HC + 51 PD | KNN, SVM-linear, SVM-RBF, ANN, DNN with leave-one-out cross validation | SVM-RBF: | 2017 | Sztahó et al., 2017 |
| Accuracy = 84.62% | |||||||
| Precision = 88.04% | |||||||
| Recall = 78.65% | |||||||
| Classification of PD from HC and assess the severity of PD | Diagnosis | Collected from participants | 88; 33 HC + 55 PD | KNN, SVM-linear, SVM-RBF, ANN, DNN with leave-one-subject-out cross validation | SVM-RBF: | 2019 | Sztahó et al., 2019 |
| Accuracy = 89.3% | |||||||
| Sensitivity = 90.2% | |||||||
| Specificity = 87.9% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 43; 10 HC + 33 PD | Random forests, SVM with 10-fold cross validation and a train-test ratio of 90:10 | SVM accuracy = 98.6% | 2012 | Tsanas et al., 2012 |
| Classification of PD from HC | Diagnosis | Collected from participants | 99; 35 HC + 64 PD | Random forest with internal out-of-bag (OOB) validation | EER = 19.27% | 2017 | Vaiciukynas et al., 2017 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository and participants | 40 and 28; 20 HC + 20 PD and 28 PD, respectively | ELM | Training data: | 2016 | Agarwal et al., 2016 |
| Accuracy = 90.76% | |||||||
| MCC = 0.815 | |||||||
| Test data: | |||||||
| Accuracy = 81.55% | |||||||
| Classification of PD from HC | Diagnosis | The Neurovoz corpus | 108; 56 HC + 52 PD | Siamese LSTM-based NN with 10-fold cross- validation | EER = 1.9% | 2019 | Bhati et al., 2019 |
| Classification of PD from HC | Diagnosis | mPower database | 2,289; 2,023 HC + 246 PD | L2-regularized logistic regression, random forest, gradient boosted decision trees with 5-fold cross validation | Gradient boosted decision trees: | 2019 | Tracy et al., 2019 |
| Recall = 0.797 | |||||||
| Precision = 0.901 | |||||||
| F1-score = 0.836 | |||||||
| Classification of PD from HC | Diagnosis | PC-GITA database | 100; 50 HC + 50 PD | ResNet with train-validation ratio of 90:10 | Precision = 0.92 | 2019 | Wodzinski et al., 2019 |
| Recall = 0.92 | |||||||
| F1-score = 0.92 | |||||||
| Accuracy = 91.7% |
ANN, artificial neural network; AUC, area under the receiver operating characteristic (ROC) curve; CART, classification and regression trees; CD, cervical dystonia; CMTNN, complementary neural network; CNN, convolutional neural network; DA, discriminant analysis; DBN, deep belief network; DNN, deep neural network; ECFA, enhanced chaos-based firefly algorithm; EFMM-OneR, enhanced fuzzy min-max neural network with the OneR attribute evaluator; ELM, extreme Learning machine; ET, extra trees or essential tremor; EWNN, evolutionary wavelet neural network; FBANN, feedforward back-propagation based artificial neural network; FKNN, fuzzy k-nearest neighbor; FLDA, Fisher's linear discriminant analysis; FND, functional neurological disorder; FNR, false negative rate; FPR, false positive rate; FURIA, fuzzy unordered rule induction algorithm; GA, genetic algorithm; GBM, gradient boosting machine; GLRA, generalized logistic regression analysis; GPD, generalized paroxysmal dystonia; GRNN, general(ized) regression neural network; HC, healthy control; HMM, hidden Markov model; IGWO-KELM, improved gray wolf optimization and kernel(-based) extreme learning machine; KELM, kernel-based extreme learning machine; KNN, k-nearest neighbors; LDA, linear discriminant analysis; LOSO, leave-one-subject-out; LS-SVM, least-square support vector machine; LSTM, long short-term memory; MAP, maximum a posteriori decision rule; MCC, Matthews correlation coefficient; MLP, multilayer perceptron; MSA, multiple system atrophy; MSE, mean squared error; NN, neural network; NNge, non-nested generalized exemplars; NPV, negative predictive value; PD, Parkinson's disease; PNN, probabilistic neural network; RBM, restricted Boltzmann machine; ResNet, residual neural network; RPART, recursive partitioning and regression trees; SCFW-KELM, subtractive clustering features weighting and kernel-based extreme learning machine; SMO, sequential minimal optimization; SVM, support vector machine; SVM-linear, support vector machine with linear kernel; SVM-RBF, support vector machine with radial basis function kernel; XGBoost, extreme gradient boosting.
Movement Data (n = 51)
The 43 out of 51 studies using accuracy to assess model performance achieved an average accuracy of 89.1 (8.3) %, ranging from 62.1% (Prince and de Vos, 2018) to 100.0% (Surangsrirat et al., 2016; Joshi et al., 2017; Pham, 2018; Pham and Yan, 2018; Figure 4A). One study reported three machine learning methods (SVM, nearest neighbor and decision tree) achieving the highest accuracy individually (Félix et al., 2019). Out of the 51 studies, the per-study highest accuracy was achieved with SVM in 22 studies (41.5%), with ensemble learning in 13 studies (24.5%), with neural network in 9 studies (17.0%), with nearest neighbor in 4 studies (7.5%), with discriminant analysis in 1 study (1.9%), with naïve Bayes in 1 study (1.9%), and with decision tree in 1 study (1.9%). Models that do not belong to any given categories were associated with the highest per-study accuracy in two studies (3.8%; Figure 4B).
Among the 33 studies that collected movement data from recruited participants, 25 used accuracy in model evaluation, leading to an average accuracy of 87.0 (7.3) % (Table 5). The lowest and highest accuracies were 64.1% (Martínez et al., 2018) and 100.0% (Surangsrirat et al., 2016), respectively. Fifteen studies used data from the PhysioNet database (Table 5) and had an average accuracy of 94.4 (4.6) %, a lowest accuracy of 86.4% and a highest accuracy of 100%. Three studies used data from the mPower database (n = 2) or data sourced from another study (n = 1), and the average accuracy of these studies was 80.6 (16.2) %.
Table 5.
Studies that applied machine learning models to movement data to diagnose PD (n = 51).
| Objectives | Type of diagnosis | Source of data | Number of subjects (n) |
Machine learning method(s), splitting strategy and cross validation | Outcomes | Year | References |
|---|---|---|---|---|---|---|---|
| Classification of PD from HC | Diagnosis | Collected from participants | 103; 71 HC + 32 PD | Ensemble method of 8 models (SVM, MLP, logistic regression, random forest, NSVC, decision tree, KNN, QDA) | Sensitivity = 96% Specificity = 97% AUC = 0.98 |
2017 | Adams, 2017 |
| Classification of PD, HC and other neurological stance disorders | Diagnosis and differential diagnosis | Collected from participants | 293; 57 HC + 27 PD + 49 AVS + 12 PNP + 48 CA + 16 DN + 25 OT + 59 PPV | Ensemble method of 7 models (logistic regression, KNN, shallow and deep ANNs, SVM, random forest, extra-randomized trees) with 90% training and 10% testing data in stratified k-fold cross-validation | 8-class classification accuracy = 82.7% | 2019 | Ahmadi et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | 137; 38 HC + 99 PD | SVM with leave-one-out-cross validation | PD vs. HC accuracy = 92.3% | 2016 | Bernad-Elazari et al., 2016 |
| Mild vs. severe accuracy = 89.8% | |||||||
| Mild vs. HC accuracy = 85.9% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 30; 14 HC + 16 PD | SVM (linear, quadratic, cubic, Gaussian kernels), ANN, with 5-fold cross-validation | Classification with ANN: | 2019 | Buongiorno et al., 2019 |
| Accuracy = 89.4% | |||||||
| Sensitivity = 87.0% | |||||||
| Specificity = 91.8% | |||||||
| Severity assessment with ANN: | |||||||
| Accuracy = 95.0% | |||||||
| sensitivity = 90.0% | |||||||
| Specificity = 99.0% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 28; 12 HC + 16 PD | NN with a train-validation-test ratio of 70:15:15, SVM with leave-one-out cross-validation, logistic regression with 10-fold cross validation | SVM: Accuracy = 85.71% Sensitivity = 83.5% Specificity = 87.5% |
2017 | Butt et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | 28; 12 HC + 16 PD | Logistic regression, naïve Bayes, SVM with 10-fold cross validation | Naïve Bayes: | 2018 | Butt et al., 2018 |
| Accuracy = 81.45% | |||||||
| Sensitivity = 76% | |||||||
| Specificity = 86.5% | |||||||
| AUC = 0.811 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 54; 27 HC + 27 PD | Naïve Bayes, LDA, KNN, decision tree, SVM-linear, SVM-RBF, majority of votes with 5-fold cross validation | Majority of votes (weighted) accuracy = 96% | 2018 | Caramia et al., 2018 |
| Classification of PD, HC and PD, HC, IH | Diagnosis | Collected from participants | 90; 30 PD + 30 HC + 30 IH | SVM, random forest, naïve Bayes with 10-fold cross validation | Random forest: | 2019 | Cavallo et al., 2019 |
| HC vs. PD: | |||||||
| Accuracy = 0.950 | |||||||
| F-measure = 0.947 | |||||||
| HC + IH vs. PD: | |||||||
| Accuracy = 0.917 | |||||||
| F-measure = 0.912 | |||||||
| HC vs. IH vs. PD: | |||||||
| Accuracy = 0.789 | |||||||
| F-measure = 0.796 | |||||||
| Classification of PD from HC and classification of HC, MCI, PDNOMCI, and PDMCI | Diagnosis, differential diagnosis and subtyping | Collected from participants | PD vs. HC: | Decision tree, naïve Bayes, random forest, SVM, adaptive boosting (with decision tree or random forest) with 10-fold cross validation | Adaptive boosting with decision tree: | 2015 | Cook et al., 2015 |
| 75; 50 HC + 25 PD | PD vs. HC: | ||||||
| Accuracy = 0.79 | |||||||
| Subtyping: | AUC = 0.82 | ||||||
| 52; 18 HC + 16 PDNOMCI + 9 PDMCI + 9 MCI | Subtyping (HOA vs. MCI vs. PDNOMCI vs. PDMCI): | ||||||
| Accuracy = 0.85 | |||||||
| AUC = 0.96 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 580; 424 HC + 156 PD | Hidden Markov models with nearest neighbor classifier with cross validation and train-test ratio of 66.6:33.3 | Accuracy = 85.51% | 2017 | Cuzzolin et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | 80; 40 HC + 40 PD | Random forest, SVM with 10-fold cross validation | SVM-RBF: | 2017 | Djurić-Jovičić et al., 2017 |
| Accuracy = 85% | |||||||
| Sensitivity = 85% | |||||||
| Specificity = 82% | |||||||
| PPV = 86% | |||||||
| NPV = 83% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 13; 5 HC + 8 PD | SVM-RBF with leave-one-out cross validation | 100% HC and PD classified correctly (confusion matrix) | 2014 | Dror et al., 2014 |
| Classification of PD from HC | Diagnosis | Collected from participants | 75; 38 HC + 37 PD | SVM with leave-one-out cross validation | Accuracy = 85.61% | 2014 | Drotár et al., 2014 |
| Sensitivity = 85.95% | |||||||
| Specificity = 85.26% | |||||||
| Classification of PD from ET | Differential diagnosis | Collected from participants | 24; 13 PD + 11 ET | SVM-linear, SVM-RBF with leave-one-out cross validation | Accuracy = 83% | 2016 | Ghassemi et al., 2016 |
| Classification of PD from HC | Diagnosis | Collected from participants | 41; 22 HC + 19 PD | SVM, decision tree, random forest, linear regression with 10-fold and leave-one-individual out (L1O) cross validation | SVM accuracy = 0.89 | 2018 | Klein et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | 74; 33 young HC + 14 elderly HC + 27 PD | SVM with 10-fold cross validation | Sensitivity = ~90% | 2017 | Javed et al., 2018 |
| Classification of PD from HC and assess the severity of PD | Diagnosis | Collected from participants | 55; 20 HC + 35 PD | SVM with leave-one-out cross validation | PD diagnosis: | 2016 | Koçer and Oktay, 2016 |
| Accuracy = 89% | |||||||
| Precision = 0.91 | |||||||
| Recall = 0.94 | |||||||
| Severity assessment: | |||||||
| HYS 1 accuracy = 72% | |||||||
| HYS 2 accuracy = 77% | |||||||
| HYS 3 accuracy = 75% | |||||||
| HYS 4 accuracy = 33% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 45; 20 HC + 25 PD | Naïve Bayes, logistic regression, SVM, AdaBoost, C4.5, BagDT with 10-fold stratified cross-validation apart from BagDT | BagDT: Sensitivity = 82% Specificity = 90% AUC = 0.94 |
2015 | Kostikis et al., 2015 |
| Classification of PD from HC | Diagnosis | Collected from participants | 40; 26 HC + 14 PD | Random forest with leave-one-subject-out cross-validation | Accuracy = 94.6% Sensitivity = 91.5% Specificity = 97.2% |
2017 | Kuhner et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | 177; 70 HC + 107 PD | ESN with 10-fold cross validation | AUC = 0.852 | 2018 | Lacy et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | 39; 16 young HC + 12 elderly HC + 11 PD | LDA with leave-one-out cross validation | Multiclass classification (young HC vs. age-matched HC vs. PD): | 2018 | Martínez et al., 2018 |
| Accuracy = 64.1% | |||||||
| Sensitivity = 47.1% | |||||||
| Specificity = 77.3% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 38; 10 HC + 28 PD | SVM-Gaussian with leave-one-out cross validation | Training accuracy = 96.9% | 2018 | Oliveira H. M. et al., 2018 |
| Test accuracy = 76.6% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 30; 15 HC + 15 PD | SVM-RBF, PNN with 10-fold cross validation | SVM-RBF: | 2015 | Oung et al., 2015 |
| Accuracy = 88.80% | |||||||
| Sensitivity = 88.70% | |||||||
| Specificity = 88.15% | |||||||
| AUC = 88.48 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 45; 14 HC + 31 PD | Deep-MIL-CNN with LOSO or RkF | With LOSO: | 2019 | Papadopoulos et al., 2019 |
| Precision = 0.987 | |||||||
| Sensitivity = 0.9 | |||||||
| specificity = 0.993 | |||||||
| F1-score = 0.943 | |||||||
| With RkF: | |||||||
| Precision = 0.955 | |||||||
| Sensitivity = 0.828 | |||||||
| Specificity = 0.979 | |||||||
| F1-score = 0.897 | |||||||
| Classification of PD, HC and post-stroke | Diagnosis and differential diagnosis | Collected from participants | 11; 3 HC + 5 PD + 3 post-stroke | MTFL with 10-fold cross validation | PD vs. HC AUC = 0.983 | 2017 | Papavasileiou et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | 182; 94 HC + 88 PD | LSTM, CNN-1D, CNN-LSTM with 5-fold cross-validation and a training-test ratio of 90:10 | CNN-LSTM: | 2019 | Reyes et al., 2019 |
| Accuracy = 83.1% | |||||||
| Precision = 83.5% | |||||||
| Recall = 83.4% | |||||||
| F1-score = 81% | |||||||
| Kappa = 64% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 60; 30 HC + 30 PD | Naïve Bayes, KNN, SVM with leave-one-out cross validation | SVM: | 2019 | Ricci et al., 2020 |
| Accuracy = 95% | |||||||
| Precision = 0.951 | |||||||
| AUC = 0.950 | |||||||
| Classification of PD, HC and IH | Diagnosis and differential diagnosis | Collected from participants | 90; 30 HC + 30 PD + 30 IH | SVM-polynomial, random forest, naïve Bayes with 10-fold cross validation | HC vs. PD, naïve Bayes or random forest: | 2018 | Rovini et al., 2018 |
| Precision = 0.967 | |||||||
| Recall = 0.967 | |||||||
| Specificity = 0.967 | |||||||
| Accuracy = 0.967 | |||||||
| F-measure = 0.967 | |||||||
| HC + IH vs. PD, random forest: | |||||||
| Precision = 1.000 | |||||||
| Recall = 0.933 | |||||||
| Specificity = 1.000 | |||||||
| Accuracy = 0.978 | |||||||
| F-measure = 0.966 | |||||||
| Multiclass classification, random forest: | |||||||
| Precision = 0.784 | |||||||
| Recall = 0.778 | |||||||
| Specificity = 0.889 | |||||||
| Accuracy = 0.778 | |||||||
| F-measure = 0.781 | |||||||
| Classification of PD, HC and IH | Diagnosis and differential diagnosis | Collected from participants | 45; 15 HC + 15 PD + 15 IH | SVM-polynomial, random forest with 5-fold cross validation | HC vs. PD, random forest: | 2019 | Rovini et al., 2019 |
| Precision = 1.000 | |||||||
| Recall = 1.000 | |||||||
| Specificity = 1.000 | |||||||
| Accuracy = 1.000 | |||||||
| F-measure = 1.000 | |||||||
| Multiclass classification (HC vs. IH vs. PD), random forest: | |||||||
| Precision = 0.930 | |||||||
| Recall = 0.911 | |||||||
| Specificity = 0.956 | |||||||
| Accuracy = 0.911 | |||||||
| F-measure = 0.920 | |||||||
| Classification of PD from ET | Differential diagnosis | Collected from participants | 52; 32 PD + 20 ET | SVM-linear with 10-fold cross validation | Accuracy = 1 | 2016 | Surangsrirat et al., 2016 |
| Sensitivity = 1 | |||||||
| Specificity = 1 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 12; 10 HC + 2 PD | Naive Bayes, LogitBoost, random forest, SVM with 10-fold cross-validation | Random forest: | 2017 | Tahavori et al., 2017 |
| Accuracy = 92.29% | |||||||
| Precision = 0.99 | |||||||
| Recall = 0.99 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 39; 16 HC + 23 PD | SVM-RBF with 10-fold stratified cross validation | Sensitivity = 88.9% | 2010 | Tien et al., 2010 |
| Specificity = 100% | |||||||
| Precision = 100% | |||||||
| FPR = 0.0% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 60; 30 HC + 30 PD | Logistic regression, naïve Bayes, random forest, decision tree with 10-fold cross validation | Random forest: | 2018 | Urcuqui et al., 2018 |
| Accuracy = 82% | |||||||
| False negative rate = 23% | |||||||
| False positive rate = 12% | |||||||
| Classification of PD from HC | Diagnosis | PhysioNet | 47; 18 HC + 29 PD | SVM, KNN, random forest, decision tree | SVM with cubic kernel: | 2017 | Alam et al., 2017 |
| Accuracy = 93.6% | |||||||
| Sensitivity = 93.1% | |||||||
| Specificity = 94.1% | |||||||
| Classification of PD from HC | Diagnosis | PhysioNet | 34; 17 HC + 17 PD | MLP, SVM, decision tree | MLP: | 2018 | Alaskar and Hussain, 2018 |
| Accuracy = 91.18% | |||||||
| Sensitivity = 1 | |||||||
| Specificity = 0.83 | |||||||
| Error = 0.09 | |||||||
| AUC = 0.92 | |||||||
| Classification of PD from HC and assess the severity of PD | Diagnosis | PhysioNet | 166; 73 HC + 93 PD | 1D-CNN, 2D-CNN, LSTM, decision tree, logistic regression, SVM, MLP | 2D-CNN and LSTM accuracy = 96.0% | 2019 | Alharthi and Ozanyan, 2019 |
| Classification of PD from HC | Diagnosis | PhysioNet | 146; 60 HC + 86 PD | SVM-Gaussian with 3- or 5-fold cross validation | Accuracy = 100%, 88.88%, and 100% in three test groups | 2019 | Andrei et al., 2019 |
| Classification of PD from HC | Diagnosis | PhysioNet | 166; 73 HC + 93 PD | ANN, SVM, naïve Bayes with cross validation | ANN accuracy = 86.75% | 2017 | Baby et al., 2017 |
| Classification of PD from HC | Diagnosis | PhysioNet | 31; 16 HC + 15 PD | SVM-linear, KNN, naïve Bayes, LDA, decision tree with leave-one-out cross validation | SVM, KNN and decision tree accuracy = 96.8% | 2019 | Félix et al., 2019 |
| Classification of PD from HC | Diagnosis | PhysioNet | 31; 16 HC + 15 PD | SVM-linear with leave-one-out cross validation | Accuracy = 100% | 2017 | Joshi et al., 2017 |
| Classification of PD from HC | Diagnosis | PhysioNet | 165; 72 HC + 93 PD | KNN, CART, decision tree, random forest, naïve Bayes, SVM-polynomial, SVM-linear, K-means, GMM with leave-one-out cross validation | SVM: Accuracy = 90.32% Precision = 90.55% Recall = 90.21% F-measure = 90.38% |
2019 | Khoury et al., 2019 |
| Classification of ALS, HD, PD from HC | Diagnosis | PhysioNet | 64; 16 HC + 15 PD + 13 ALS + 20 HD | String grammar unsupervised possibilistic fuzzy C-medians with FKNN, with 4-fold cross validation | PD vs. HC accuracy = 96.43% | 2018 | Klomsae et al., 2018 |
| Classification of PD from HC | Diagnosis | PhysioNet | 166; 73 HC + 93 PD | Logistic regression, decision trees, random forest, SVM-Linear, SVM-RBF, SVM-Poly, KNN with cross validation | KNN: | 2018 | Mittra and Rustagi, 2018 |
| Accuracy = 93.08% | |||||||
| Precision = 89.58% | |||||||
| Recall = 84.31% | |||||||
| F1-score = 86.86% | |||||||
| Classification of PD from HC | Diagnosis | PhysioNet | 85; 43 HC + 42 PD | LS-SVM with leave-one-out, 2- or 10-fold cross validation | Leave-one-out cross validation: | 2018 | Pham, 2018 |
| AUC = 1 | |||||||
| Sensitivity = 100% | |||||||
| Specificity = 100% | |||||||
| Accuracy = 100% | |||||||
| 10-fold cross validation: | |||||||
| AUC = 0.89 | |||||||
| Sensitivity = 85.00% | |||||||
| Specificity = 73.21% | |||||||
| Accuracy = 79.31% | |||||||
| Classification of PD from HC | Diagnosis | PhysioNet | 165; 72 HC + 93 PD | LS-SVM with leave-one-out, 2- or 5- or 10-fold cross validation | Accuracy = 100% | 2018 | Pham and Yan, 2018 |
| Sensitivity = 100% | |||||||
| Specificity = 100% | |||||||
| AUC = 1 | |||||||
| Classification of PD from HC | Diagnosis | PhysioNet | 166; 73 HC + 93 PD | DCALSTM with stratified 5-fold cross validation | Sensitivity = 99.10% | 2019 | Xia et al., 2020 |
| Specificity = 99.01% | |||||||
| Accuracy = 99.07% | |||||||
| Classification of HC, PD, ALS and HD | Diagnosis and differential diagnosis | PhysioNet | 64; 16 HC + 15 PD + 13 ALS + 20 HD | SVM-RBF with 10-fold cross validation | PD vs. HC: | 2009 | Yang et al., 2009 |
| Accuracy = 86.43% | |||||||
| AUC = 0.92 | |||||||
| Classification of PD, HD, ALS and ND from HC | Diagnosis | PhysioNet | 64; 16 HC + 15 PD + 13 ALS + 20 HD | Adaptive neuro-fuzzy inference system with leave-one-out cross validation | PD vs. HC: | 2018 | Ye et al., 2018 |
| Accuracy = 90.32% | |||||||
| Sensitivity = 86.67% | |||||||
| Specificity = 93.75% | |||||||
| Classification of PD from HC and assess the severity of PD | Diagnosis | mPower database | 50; 22 HC + 28 PD | Random forest, bagged trees, SVM, KNN with 10-fold cross validation | Random forest: | 2017 | Abujrida et al., 2017 |
| PD vs. HC accuracy = 87.03% | |||||||
| PD severity assessment accuracy = 85.8% | |||||||
| Classification of PD from HC | Diagnosis | mPower database | 1,815; 866 HC + 949 PD | CNN with 10-fold cross validation | Accuracy = 62.1% | 2018 | Prince and de Vos, 2018 |
| F1 score = 63.4% | |||||||
| AUC = 63.5% | |||||||
| Classification of PD from HC | Diagnosis | Dataset from Fernandez et al., 2013 | 49; 26 HC + 23 PD | KFD-RBF, naïve Bayes, KNN, SVM-RBF, random forest with 10-fold cross validation | Random forest accuracy = 92.6% | 2015 | Wahid et al., 2015 |
ALS, amyotrophic lateral sclerosis; ANN, artificial neural network; AUC, area under the receiver operating characteristic (ROC) curve; AVS, acute unilateral vestibulopathy; BagDT, bootstrap aggregation for a random forest of decision trees; CA, anterior lobe cerebella atrophy; CART, classification and regression trees; DCALSTM, dual-modal with each branch has a convolutional network followed by an attention-enhanced bi-directional LSTM; DN, downbeat nystagmus syndrome; ESN, echo state network; FKNN, fuzzy k-nearest neighbor; GMM, Gaussian mixture model; HC, healthy control; HD, Huntington's disease; IH, idiopathic hyposmia; KFD, kernel Fisher discriminant; KNN, k-nearest neighbors; LDA, linear discriminant analysis; LOSO, leave-one-subject-out; LS-SVM, least-squares support vector machine; LSTM, long short-term memory; MCI, mild cognitive impairment; MIL, multiple-instance learning; MLP, multilayer perceptron; MTFL, multi-task feature learning; NN, neural network; NSVC, nu-support vector classification; OT, primary orthostatic tremor; PD, Parkinson's disease; PDMCI, PD participants who met criteria for mild cognitive impairment; PDNOMCI, PD participants with no indication of mild cognitive impairment; PNN, probabilistic neural network; PNP, sensory polyneuropathy; PPV, phobic postural vertigo; QDA, quadratic discriminant analysis; RkF, repeated k-fold; SVM, support vector machine; SVM-Poly, support vector machine with polynomial kernel; SVM-RBF, support vector machine with radial basis function kernel.
MRI (n = 36)
Average accuracy of the 32 studies that used accuracy to evaluate the performance of machine learning models was 87.5 (8.0) %. In these studies, the lowest accuracy was 70.5% (Liu L. et al., 2016) and the highest accuracy was 100.0% (Cigdem et al., 2019; Figure 4A). Out of the 36 studies, the per-study highest accuracy was obtained with SVM in 21 studies (58.3%), with neural network in 8 studies (22.2%), with discriminant analysis in 3 studies (8.3%), with regression in 2 studies (5.6%), and with ensemble learning in 1 study (2.8%). One study (2.8%) obtained the highest per-study accuracy using models that do not belong to any of the given categories (Figure 4B). In 8 of 36 studies, neural networks were directly applied to MRI data, while the remaining studies used machine learning models to learn from extracted features, e.g., cortical thickness and volume of brain regions, to diagnose PD.
Out of 17 studies that used MRI data from the PPMI database, 16 used accuracy to evaluate model performance and the average accuracy was 87.9 (8.0) %. The lowest and highest accuracies were 70.5 and 99.9%, respectively (Table 6). In 16 out of 19 studies that acquired MRI data from human participants, accuracy was used to evaluate classification performance and an average accuracy was 87.0 (8.1) % was achieved. The lowest reported accuracy was 76.2% and the highest reported accuracy was 100% (Table 6).
Table 6.
Studies that applied machine learning models to MRI data to diagnose PD (n = 36).
| Objectives | Type of diagnosis | Source of data | Number of subjects (n) |
Machine learning method(s), splitting strategy and cross validation | Outcomes | Year | References |
|---|---|---|---|---|---|---|---|
| Classification of PD from MSA | Differential diagnosis | Collected from participants | 150; 54 HC + 65 PD + 31 MSA | SVM with leave-one-out-cross validation | MSA vs. PD: | 2019 | Abos et al., 2019 |
| Accuracy = 0.79 | |||||||
| Sensitivity = 0.71 | |||||||
| Specificity = 0.86 | |||||||
| MSA vs. HC: | |||||||
| Accuracy = 0.79 | |||||||
| Sensitivity = 0.84 | |||||||
| Specificity = 0.74 | |||||||
| MSA vs. subsample of PD: | |||||||
| Accuracy = 0.84 | |||||||
| Sensitivity = 0.77 | |||||||
| Specificity = 0.90 | |||||||
| Classification of PD from MSA | Differential diagnosis | Collected from participants | 151; 59 HC + 62 PD + 30 MSA | SVM with leave-one-out-cross validation | Accuracy = 77.17% | 2019 | Baggio et al., 2019 |
| Sensitivity = 83.33% | |||||||
| Specificity = 74.19% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 94; 50 HC + 44 PD | CNN with 85 subjects for training and 9 for testing | Training accuracy = 95.24% | 2019 | Banerjee et al., 2019 |
| Testing accuracy = 88.88% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 47; 26 HC + 21 PD | SVM-linear with leave-one-out cross validation | Accuracy = 93.62% | 2015 | Chen et al., 2015 |
| Sensitivity = 90.47% | |||||||
| Specificity = 96.15% | |||||||
| Classification of PD from PSP | Differential diagnosis | Collected from participants | 78; 57 PD + 21 PSP | SVM with leave-one-out cross validation | Accuracy = 100% | 2013 | Cherubini et al., 2014a |
| Sensitivity = 1 | |||||||
| Specificity = 1 | |||||||
| Classification of PD, MSA, PSP and HC | Diagnosis and differential diagnosis | Collected from participants | 106; 36 HC + 35 PD + 16 MSA + 19 PSP | Elastic Net regularized logistic regression with nested 10-fold cross validation | HC vs. PD/MSA-P/PSP: | 2017 | Du et al., 2017 |
| AUC = 0.88 | |||||||
| Sensitivity = 0.80 | |||||||
| Specificity = 0.83 | |||||||
| PPV = 0.82 | |||||||
| NPV = 0.81 | |||||||
| HC vs. PD: | |||||||
| AUC = 0.91 | |||||||
| Sensitivity = 0.86 | |||||||
| Specificity = 0.80 | |||||||
| PPV = 0.82 | |||||||
| NPV = 0.89 | |||||||
| PD vs. MSA/PSP: | |||||||
| AUC = 0.94 | |||||||
| Sensitivity = 0.86 | |||||||
| Specificity = 0.87 | |||||||
| PPV = 0.88 | |||||||
| NPV = 0.84 | |||||||
| PD vs. MSA: | |||||||
| AUC = 0.99 | |||||||
| Sensitivity = 0.97 | |||||||
| Specificity = 1.00 | |||||||
| PPV = 1.00 | |||||||
| NPV = 0.93 | |||||||
| PD vs. PSP: | |||||||
| AUC = 0.99 | |||||||
| Sensitivity = 0.97 | |||||||
| Specificity = 1.00 | |||||||
| PPV = 1.00 | |||||||
| NPV = 0.94 | |||||||
| MSA vs. PSP: | |||||||
| AUC = 0.98 | |||||||
| Sensitivity = 0.94 | |||||||
| Specificity = 1.00 | |||||||
| PPV = 1.00 | |||||||
| NPV = 0.93 | |||||||
| Classification of HC, PD, MSA and PSP | Diagnosis and differential diagnosis | Collected from participants | 64; 22 HC + 21 PD + 11 MSA + 10 PSP | SVM-linear with leave-one-out cross validation | PD vs. HC: | 2011 | Focke et al., 2011 |
| Accuracy = 41.86% | |||||||
| Sensitivity = 38.10% | |||||||
| Specificity = 45.45% | |||||||
| PD vs. MSA: | |||||||
| Accuracy = 71.87% | |||||||
| Sensitivity = 36.36% | |||||||
| Specificity = 90.48% | |||||||
| PD vs. PSP: | |||||||
| Accuracy = 96.77% | |||||||
| Sensitivity = 90% | |||||||
| Specificity = 100% | |||||||
| MSA vs. PSP: | |||||||
| Accuracy = 76.19% | |||||||
| MSA vs. HC: | |||||||
| Accuracy = 78.78% | |||||||
| Sensitivity = 54.55% | |||||||
| Specificity = 90.91% | |||||||
| PSP vs. HC: | |||||||
| Accuracy = 93.75% | |||||||
| Sensitivity = 90.00% | |||||||
| Specificity = 95.45% | |||||||
| Classification of PD and atypical PD | Differential diagnosis | Collected from participants | 40; 17 PD + 23 atypical PD | SVM-RBF with 10-fold cross-validation | Accuracy = 97.50% | 2012 | Haller et al., 2012 |
| TPR = 0.94 | |||||||
| FPR = 0.00 | |||||||
| TNR = 1.00 | |||||||
| FNR = 0.06 | |||||||
| Classification of PD and other forms of Parkinsonism | Differential diagnosis | Collected from participants | 36; 16 PD + 20 other Parkinsonism | SVM-RBF with 10-fold cross validation | Accuracy = 86.92% | 2012 | Haller et al., 2013 |
| TP = 0.87 | |||||||
| FP = 0.14 | |||||||
| TN = 0.87 | |||||||
| FN = 0.13 | |||||||
| Classification of HC, PD, PSP, MSA-C and MSA-P | Diagnosis and differential diagnosis | Collected from participants | 464; 73 HC + 204 PD + 106 PSP + 21 MSA-C + 60 MSA-P | SVM-RBF with 10-fold cross validation | PD vs. HC: | 2016 | Huppertz et al., 2016 |
| Sensitivity = 65.2% | |||||||
| Specificity = 67.1% | |||||||
| Accuracy = 65.7% | |||||||
| PD vs. PSP: | |||||||
| Sensitivity = 82.5% | |||||||
| Specificity = 86.8% | |||||||
| Accuracy = 85.3% | |||||||
| PD vs. MSA-C: | |||||||
| Sensitivity = 76.2% | |||||||
| Specificity = 96.1% | |||||||
| Accuracy = 94.2% | |||||||
| PD vs. MSA-P: | |||||||
| Sensitivity = 86.7% | |||||||
| Specificity = 92.2% | |||||||
| Accuracy = 90.5% | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 42; 21 HC + 21 PD | SVM-linear with stratified 10-fold cross validation | Accuracy = 78.33% | 2017 | Kamagata et al., 2017 |
| Precision = 85.00% | |||||||
| Recall = 81.67% | |||||||
| AUC = 85.28% | |||||||
| Classification of PD, PSP, MSA-P and HC | Diagnosis and differential diagnosis | Collected from participants | 419; 142 HC + 125 PD + 98 PSP + 54 MSA-P | CNN with train-validation ratio of 85:15 | PD: | 2019 | Kiryu et al., 2019 |
| Sensitivity = 94.4% | |||||||
| Specificity = 97.8% | |||||||
| Accuracy = 96.8% | |||||||
| AUC = 0.995 | |||||||
| PSP: | |||||||
| Sensitivity = 84.6% | |||||||
| Specificity = 96.0% | |||||||
| Accuracy = 93.7% | |||||||
| AUC = 0.982 | |||||||
| MSA-P: | |||||||
| Sensitivity = 77.8% | |||||||
| Specificity = 98.1% | |||||||
| Accuracy = 95.2% | |||||||
| AUC = 0.990 | |||||||
| HC: | |||||||
| Sensitivity = 100.0% | |||||||
| Specificity = 97.5% | |||||||
| Accuracy = 98.4% | |||||||
| AUC = 1.000 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 65; 31 HC + 34 PD | FCP with 36 out of the 65 subjects as the training set | AUC = 0.997 | 2016 | Liu H. et al., 2016 |
| Classification of PD, PSP, MSA-C and MSA-P | Differential diagnosis | Collected from participants | 85; 47 PD + 22 PSP + 9 MSA-C + 7 MSA-P | SVM-linear with leave-one-out cross validation | 4-class classification (MSA-C vs. MSA-P vs. PSP vs. PD) accuracy = 88% | 2017 | Morisi et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | 89; 47 HC + 42 PD | Boosted logistic regression with nested cross-validation | Accuracy = 76.2% | 2019 | Rubbert et al., 2019 |
| Sensitivity = 81% | |||||||
| Specificity = 72.7% | |||||||
| Classification of PD, PSP and HC | Diagnosis and differential diagnosis | Collected from participants | 84; 28 HC + 28 PSP + 28 PD | SVM-linear with leave-one-out cross validation | PD vs. HC: | 2014 | Salvatore et al., 2014 |
| Accuracy = 85.8% | |||||||
| Specificity = 86.0% | |||||||
| Sensitivity = 86.0% | |||||||
| PSP vs. HC: | |||||||
| Accuracy = 89.1% | |||||||
| Specificity = 89.1% | |||||||
| Sensitivity = 89.5% | |||||||
| PSP vs. PD: | |||||||
| Accuracy = 88.9% | |||||||
| Specificity = 88.5% | |||||||
| Sensitivity = 89.5% | |||||||
| Classification of PD, APS (MSA, PSP) and HC | Diagnosis and differential diagnosis | Collected from participants | 100; 35 HC + 45 PD + 20 APS | CNN-DL, CR-ML, RA-ML with 5-fold cross-validation | PD vs. HC with CNN-DL: | 2019 | Shinde et al., 2019 |
| Test accuracy = 80.0% | |||||||
| Test sensitivity = 0.86 | |||||||
| Test specificity = 0.70 | |||||||
| Test AUC = 0.913 | |||||||
| PD vs. APS with CNN-DL: | |||||||
| Test accuracy = 85.7% | |||||||
| Test sensitivity = 1.00 | |||||||
| Test specificity = 0.50 | |||||||
| Test AUC = 0.911 | |||||||
| Classification of PD from HC | Diagnosis | Collected from participants | 101; 50 HC + 51 PD | SVM-RBF with leave-one-out cross validation | Sensitivity = 92% Specificity = 87% |
2017 | Tang et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | 85; 40 HC + 45 PD | SVM-linear with leave-one-out, 5-fold, 0.632-fold (1-1/e), 2-fold cross validation | Accuracy = 97.7% | 2016 | Zeng et al., 2017 |
| Classification of PD from HC | Diagnosis | PPMI database | 543; 169 HC + 374 PD | RLDA with JFSS with 10-fold cross validation | Accuracy = 81.9% | 2016 | Adeli et al., 2016 |
| Classification of PD from HC | Diagnosis | PPMI database | 543; 169 HC + 374 PD | RFS-LDA with 10-fold cross validation | Accuracy = 79.8% | 2019 | Adeli et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database | 543; 169 HC + 374 PD | Random forest (for feature selection and clinical score); SVM with 10-fold stratified cross validation | Accuracy = 0.93 | 2018 | Amoroso et al., 2018 |
| AUC = 0.97 | |||||||
| Sensitivity = 0.93 | |||||||
| Specificity = 0.92 | |||||||
| Classification of PD, HC and prodromal | Diagnosis | PPMI database | 906; 203 HC + 66 prodromal + 637 PD | MLP, XgBoost, random forest, SVM with 5-fold cross validation | MLP: | 2020 | Chakraborty et al., 2020 |
| Accuracy = 95.3% | |||||||
| Recall = 95.41% | |||||||
| Precision = 97.28% | |||||||
| F1-score = 94% | |||||||
| Classification of PD from HC | Diagnosis | PPMI database | Dataset 1: 15; 6 HC + 9 PD | SVM with leave-one-out cross validation | Dataset 1: | 2014 | Chen et al., 2014 |
| EER = 87% | |||||||
| Dataset 2: 39; 21 HC + 18 PD | Accuracy = 80% | ||||||
| AUC = 0.907 | |||||||
| Dataset 2: | |||||||
| EER = 73% | |||||||
| Accuracy = 68% | |||||||
| AUC = 0.780 | |||||||
| Classification of PD from HC | Diagnosis | PPMI database | 80; 40 HC + 40 PD | Naïve Bayes, SVM-RBF with 10-fold cross validation | SVM: | 2019 | Cigdem et al., 2019 |
| Accuracy = 87.50% | |||||||
| Sensitivity = 85.00% | |||||||
| Specificity = 90.00% | |||||||
| AUC = 90.00% | |||||||
| Classification of PD from HC | Diagnosis | PPMI database | 37; 18 HC + 19 PD | SVM-linear with leave-one-out cross validation | Accuracy = 94.59% | 2017 | Kazeminejad et al., 2017 |
| Classification of PD, HC and SWEDD | Diagnosis and subtyping | PPMI database | 238; 62 HC + 142 PD + 34 SWEDD | Joint learning with 10-fold cross validation | HC vs. PD: | 2018 | Lei et al., 2019 |
| Accuracy = 91.12% | |||||||
| AUC = 94.88% | |||||||
| HC vs. SWEDD: | |||||||
| Accuracy = 94.89% | |||||||
| AUC = 97.80% | |||||||
| PD vs. SWEDD: | |||||||
| accuracy = 92.12% | |||||||
| AUC = 93.82% | |||||||
| Classification of PD and SWEDD from HC | Diagnosis | PPMI database | Baseline: 238; 62 HC + 142 PD + 34 SWEDD12 months: 186; 54 HC + 123 PD + 9 SWEDD 24 months: 127; 7 HC + 88 PD + 22 SWEDD |
SSAE with 10-fold cross validation |
HC vs. PD: Accuracy = 85.24%, 88.14%, and 96.19% for baseline, 12 m, and 24 mHC vs. SWEDD: Accuracy = 89.67%, 95.24%, and 93.10% for baseline, 12 m, and 24 m |
2019 | Li et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database | 112; 56 HC + 56 PD | RLDA with 8-fold cross validation | Accuracy = 70.5% | 2016 | Liu L. et al., 2016 |
| AUC = 71.1 | |||||||
| Classification of PD from HC | Diagnosis | PPMI database | 60; 30 HC + 30 PD | SVM, ELM with train-test ratio of 80:20 | ELM: | 2016 | Pahuja and Nagabhushan, 2016 |
| Training accuracy = 94.87% | |||||||
| Testing accuracy = 90.97% | |||||||
| Sensitivity = 0.9245 | |||||||
| Specificity = 0.9730 | |||||||
| Classification of PD from HC | Diagnosis | PPMI database | 172; 103 HC + 69 PD | Multi-kernel SVM with 10-fold cross validation | 2017 | Peng et al., 2017 | |
| Accuracy = 85.78% | |||||||
| Specificity = 87.79% | |||||||
| Sensitivity = 87.64% | |||||||
| AUC = 0.8363 | |||||||
| Classification of PD from HC | Diagnosis and subtyping | PPMI database | 109; 32 HC + 77 PD (55 PD-NC + 22 PD-MCI) | SVM with 2-fold cross validation | PD vs. HC: | 2016 | Peng et al., 2016 |
| Accuracy = 92.35% | |||||||
| Sensitivity = 0.9035 | |||||||
| Specificity = 0.9431 | |||||||
| AUC = 0.9744 | |||||||
| PD-MCI vs. HC: | |||||||
| Accuracy = 83.91% | |||||||
| Sensitivity = 0.8355 | |||||||
| Specificity = 0.8587 | |||||||
| AUC = 0.9184 | |||||||
| PD-MCI vs. PD-NC: | |||||||
| Accuracy = 80.84% | |||||||
| Sensitivity = 0.7705 | |||||||
| Specificity = 0.8457 | |||||||
| AUC = 0.8677 | |||||||
| Classification of PD, HC and SWEDD | Diagnosis and subtyping | PPMI database | 831; 245 HC + 518 PD + 68 SWEDD | LSSVM-RBF with cross validation | Accuracy = 99.9% Specificity = 100% Sensitivity = 99.4% |
2015 | Singh and Samavedham, 2015 |
| Classification of PD, HC and SWEDD | Diagnosis and differential diagnosis | PPMI database | 741; 262 HC + 408 PD + 71 SWEDD | LSSVM-RBF with 10-fold cross validation | PD vs. HC accuracy = 95.37% | 2018 | Singh et al., 2018 |
| PD vs. SWEDD accuracy = 96.04% | |||||||
| SWEDD vs. HC accuracy = 93.03% | |||||||
| Classification of PD from HC | Diagnosis | PPMI database | 408; 204 HC + 204 PD | CNN (VGG and ResNet) | ResNet50 accuracy = 88.6% | 2019 | Yagis et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database | 754; 158 HC + 596 PD | FCN, GCN with 5-fold cross validation | AUC = 95.37% | 2018 | Zhang et al., 2018 |
APS, atypical parkinsonian syndromes; AUC, area under the receiver operating characteristic (ROC) curve; CNN, convolutional neural network; CNN-DL, convolutional neural network with discriminative localization; CR-ML, contrast ratio classifier; EER, equal error rate; ELM, extreme learning machine; FCN, fully connected network; FCP, folded concave penalized (learning); FN, false negative; FNR, false negative rate; FP, false positive; FPR, false positive rate; GCN, graph convolutional network; HC, healthy control; JFSS, joint feature-sample selection; LSSVM, least-squares support vector machine; MLP, multilayer perceptron; MSA, multiple system atrophy; MSA-C, multiple system atrophy with a cerebellar syndrome; MSA-P, multiple system atrophy with a parkinsonian type; PD, Parkinson's disease; PD-MCI, PD participants who met criteria for mild cognitive impairment; PD-NC, PD participants with no indication of mild cognitive impairment; PSP, progressive supranuclear palsy; RA-ML, radiomics based classifier; ResNet, residual neural network; RFS-LDA, robust feature-sample linear discriminant analysis; RLDA, robust linear discriminant analysis; SSAE, stacked sparse auto-encoder; SVM, support vector machine; SVM-RBF, support vector machine with radial basis function kernel; SWEDD, PD with scans without evidence of dopaminergic deficit; TN, true negative; TNR, true negative rate; TP, true positive; TPR, true positive rate; XgBoost, extreme gradient boosting.
Handwriting Patterns (n = 16)
Fifteen out of 16 studies used accuracy in model evaluation and the average accuracy was 87.0 (6.3) % (Table 7). Among these studies, the lowest accuracy was 76.44% (Ali et al., 2019b) and the highest accuracy was 99.3% (Pereira et al., 2018; Figure 4A). The highest accuracy per-study was obtained with neural network in 6 studies (37.5%), with SVM in 5 studies (31.3%), with ensemble learning in 4 studies (25.0%), and with naïve Bayes in 1 study (6.3%; Figure 4B).
Table 7.
Studies that applied machine learning models to handwritten patterns, SPECT, PET, CSF, other data types and combinations of data to diagnose PD (n = 67).
| Objectives | Type of diagnosis | Source of data | Type of data | Number of subjects (n) | Machine learning method(s), splitting strategy and cross validation | Outcomes | Year | References |
|---|---|---|---|---|---|---|---|---|
| Classification of PD from HC | Diagnosis | HandPD | Handwritten patterns | 92; 18 HC + 74 PD | LDA, KNN, Gaussian naïve Bayes, decision tree, Chi2 with Adaboost with 5- or 4-fold stratified cross validation | Chi-2 with Adaboost: Accuracy = 76.44% Sensitivity = 70.94% Specificity = 81.94% |
2019 | Ali et al., 2019b |
| Classification of PD (PD + SWEDD) from HC | Diagnosis | PPMI database | More than one | 388; 194 HC + 168 PD + 26 SWEDD | Ensemble method of several SVM with linear kernel with leave-one-out cross validation | Accuracy = 94.38% | 2018 | Castillo-Barnes et al., 2018 |
| Classification of PD from HC | Diagnosis | PPMI database | More than one | 586; 184 HC + 402 PD | MLP, BayesNet, random forest, boosted logistic regression with a train-test ratio of 70:30 | Boosted logistic regression: Accuracy = 97.159% AUC curve = 98.9% |
2016 | Challa et al., 2016 |
| Classification of tPD from rET | Differential diagnosis | Collected from participants | More than one | 30; 15 tPD + 15rET | Multi-kernel SVM with leave-one-out cross validation | Accuracy = 100% | 2014 | Cherubini et al., 2014b |
| Classfication of PD, HC and atypical PD | Diagnosis, differential diagnosis and subtyping | PPMI database and SNUH cohort | SPECT imaging data | PPMI: 701; 193 HC + 431 PD + 77 SWEDD snuh: 82 PD |
CNN with train-validation ratio of 90:10 | PPMI: Accuracy = 96.0% Sensitivity = 94.2% Specificity = 100% SNUH: Accuracy = 98.8% Sensitivity = 98.6% Specificity = 100% |
2017 | Choi et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | Other | 270; 120 HC + 150 PD | Random forest | Classification error = 49.6% (rs11240569) Classification error = 44.8% (rs708727) Classification error = 49.3% (rs823156) |
2019 | Cibulka et al., 2019 |
| Classification of PD from HC | Diagnosis | HandPD | Handwritten patterns | 92; 18 HC + 74 PD | Naïve Bayes, OPF, SVM with cross-validation | SVM-RBF accuracy = 85.54% | 2018 | de Souza et al., 2018 |
| Classification of PD from HC | Diagnosis | PPMI database | More than one | 1194; 816 HC + 378 PD | BoostPark | Accuracy = 0.901 AUC-ROC = 0.977 AUC-PR = 0.947 F1-score = 0.851 |
2017 | Dhami et al., 2017 |
| Classification of PD and HC, and PD + SWEDD and HC | Diagnosis | PPMI database | More than one | 430; 127 HC + 263 PD + 40 SWEDD | AdaBoost, SVM, naïve Bayes, decision tree, KNN, K-Means with 5-fold cross validation | PD vs. HC (adaboost): Accuracy = 0.98954704 Sensitivity = 0.97831978 Specificity = 0.99796748 PPV = 0.99723757 NPV = 0.98396794 LOR = 10.0058805 PD + SWEDD vs HC (adaboost): Accuracy = 0.9825784 Sensitivity = 0.97560976 Specificity = 0.98780488 PPV = 0.98360656 NPV = 0.98181818 LOR = 8.08332861 |
2016 | Dinov et al., 2016 |
| Classification of PD from HC | Diagnosis | Collected from participants | CSF | Cohort 1: 160; 80 HC + 80 PD Cohort 2: 60; 30 HC + 30 PD |
Elastic Net and gradient boosted regression with 10-fold cross validation | Ensemble of 60 decision trees identified with gradient boosted model: Sensitivity = 85% Specificity = 75% PPV = 77% NPV = 83% AUC = 0.77 |
2018 | Dos Santos et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 75; 38 HC + 37 PD | SVM-RBF with stratified 10-fold cross-validation | Accuracy = 88.13% Sensitivity = 89.47% Specificity = 91.89% |
2015 | Drotár et al., 2015 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 75; 38 HC + 37 PD | KNN, ensemble AdaBoost, SVM | SVM: Accuracy = 81.3% Sensitivity = 87.4% Specificity = 80.9% |
2016 | Drotár et al., 2016 |
| Classification of IPD, VaP and HC | Differential diagnosis | Collected from participants | More than one | 45; 15 HC + 15 IPD + 15 VaP | MLP, DBN with 10-fold cross validation | IPD + VaP vs HC with MLP: Accuracy = 95.68% Specificity = 98.08% Sensitivity = 92.44% VaP vs. IPD with DBN: Accuracy = 75.33% Specificity = 72.31% Sensitivity = 79.18% |
2018 | Fernandes et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | More than one | 75; 15 HC + 60 PD blood: 75; 15 HC + 60 PD FDOPA PET: 58; 14 HC + 44 PD FDG PET: 67; 16 HC + 51 PD |
SVM-linear, random forest with leave-one-out cross validation | SVM AUC for FDOPA + metabolomics: 0.98 SVM AUC for FDG + metabolomics: 0.91 |
2019 | Glaab et al., 2019 |
| Classification of PD, HC and SWEDD | Diagnosis and subtyping | PPMI database | More than one | 666; 415 HC + 189 PD + 62 SWEDD | EPNN, PNN, SVM, KNN, classification tree with train-test ratio of 90:10 | EPNN: PD vs SWEDD vs HC accuracy = 92.5% PD vs HC accuracy = 98.6% SWEDD vs HC accuracy = 92.0% PD vs. SWEDD accuracy = 95.3% |
2015 | Hirschauer et al., 2015 |
| Classification of PD from HC and assess the severity of PD | Diagnosis | Picture Archiving and Communication System (PACS) | SPECT imaging data | 202; 6 HC + 102 mild PD + 94 severe PD | Linear regression, SVM-RBF with a train-test ratio of 50:50 | SVM-RBF: Sensitivity = 0.828 Specificity = 1.000 PPV = 0.837 NPV = 0.667 Accuracy = 0.832 AUC = 0.845 Kappa = 0.680 |
2019 | Hsu et al., 2019 |
| Classification of PD from VP | Differential diagnosis | Collected from participants | SPECT imaging data | 244; 164 PD + 80 VP | Logistic regression, LDA, SVM with 10-fold cross-validation | SVM: Accuracy = 0.904 Sensitivity = 0.954 Specificity = 0.801 AUC = 0.954 |
2014 | Huertas-Fernández et al., 2015 |
| Classification of PD from HC | Diagnosis | Collected from participants | SPECT imaging data | 208; 108 HC + 100 PD | SVM, KNN, NM with 3-fold cross validation | SVM: Sensitivity = 89.02% Specificity = 93.21% AUC = 0.9681 |
2012 | Illan et al., 2012 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 72; 15 HC + 57 PD | CNN with 10-fold cross validation or leave-one-out cross validation | Accuracy = 88.89% | 2018 | Khatamino et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | Other | 10; 5 HC + 5 PD | SVM with leave-one-subject-out cross validation | Sensitivity = 0.90 Specificity = 0.90 |
2013 | Kugler et al., 2013 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | Handwritten patterns | 72; 15 HC + 57 PD | SVM-linear, SVM-RBF, KNN with leave-one-subject-out cross validation | SVM-linear: Accuracy = 97.52% MCC = 0.9150 F-score = 0.9828 |
2019 | İ et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected postmortem | CSF | 105; 57 HC + 48 PD | SVM with 10-fold cross validation | Sensitivity = 65% Specificity = 79% AUC = 0.79 |
2013 | Lewitt et al., 2013 |
| Classification of PD from HC | Diagnosis | Collected from participants | CSF | 78; 42 HC + 36 PD | Random forest and extreme gradient tree boosting with 10-fold cross validation | Extreme gradient tree boosting: Specificity = 78.6% Sensitivity = 83.3% AUC = 83.9% |
2018 | Maass et al., 2018 |
| Classification of PD from HC or NPH | Diagnosis and differential diagnosis | Collected from participants | CSF | 157; 68 HC + 82 PD + 7 NPH | SVM with 10-fold cross validation or leave-one-out cross validation | Cohort 1, PD vs HC: AUC = 0.76 Cohort 2, PD vs HC: AUC = 0.78 Cohort 3, PD vs HC: AUC = 0.31 Cohort 4, PD vs NPH: AUC = 0.88 |
2020 | Maass et al., 2020 |
| Classification of PD from HC | Diagnosis | PPMI database | More than one | 550; 157 HC + 342 PD + 51 SWEDD | SVM, random forest, MLP, logistic regression, KNN with nested cross-validation | Motor features, SVM: Accuracy = 78.4% AUC = 84.7% Non-motor features, KNN: Accuracy = 82.2% AUC = 88% |
2018 | Mabrouk et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database | SPECT imaging data | 642; 194 HC + 448 PD | CNN (LENET53D, ALEXNET3D) with 10-fold stratified cross-validation | ALEXNET3D: Accuracy = 94.1% AUC = 0.984 |
2018 | Martinez-Murcia et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 75; 10 HC + 65 PD | MLP, non-linear SVM, random forest, logistic regression with stratified 10-fold cross-validation | MLP: Accuracy = 84% Sensitivity = 75.7% Specificity = 88.9% Weighted Kappa = 0.65 AUC = 0.86 |
2015 | Memedi et al., 2015 |
| Classification of PD from HC | Diagnosis | Parkinson's Disease Handwriting Database (PaHaW) | Handwritten patterns | 69; 36 HC + 33 PD | Random forest with stratified 7-fold cross-validation | Accuracy = 89.81% Sensitivity = 88.63% Specificity = 90.87% MCC = 0.8039 |
2018 | Mucha et al., 2018 |
| Classification of PD, MSA, PSP, CBS and HC | Differential diagnosis | Collected from participants | SPECT imaging data | 578; 208 HC + 280 PD + 21 MSA + 41 PSP + 28 CBS | SVM with 5-fold cross-validation | Accuracy = 58.4–92.9% | 2019 | Nicastro et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 30; 15 HC + 15 PD | KNN, decision tree, random forest, SVM, AdaBoost with 3-fold cross validation | Random forest accuracy = 0.91 | 2018 | Nõmm et al., 2018 |
| Classification of HC, AD and PD | Diagnosis and differential diagnosis | The authors' institutional oct database | Other | 75; 27 HC + 28 PD + 20 AD | SVM-RBF with 2-, 5- and 10-fold cross validation | Accuracy = 87.7% HC sensitivity = 96.2% HC specificity = 88.2% PD sensitivity = 87.0% PD specificity = 100.0% |
2019 | Nunes et al., 2019 |
| Classification of idiopathic PD, atypical Parkinsonian and ET | Differential diagnosis | Collected from participants | Other | 85; 50 idiopathic PD + 26 atypical PD + 9 ET | SVM, random forest with leave-one-out cross validation | SVM accuracy = 100% Random forest accuracy = 98.5% |
2019 | Nuvoli et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database | SPECT imaging data | 654; 209 HC + 445 PD | SVM-linear with leave-one-out cross validation | Accuracy = 97.86% Sensitivity = 97.75% Specificity = 98.09% |
2015 | Oliveira and Castelo-Branco, 2015 |
| Classification of PD from HC | Diagnosis | PPMI database | SPECT imaging data | 652; 209 HC + 443 PD | SVM-linear, KNN, logistic regression with leave-one-out cross validation | SVM-linear: Accuracy = 97.9% Sensitivity = 98.0% Specificity = 97.6% |
2017 | Oliveira F. et al., 2018 |
| Classification of PD and non-PD (ET, drug-induced Parkinsonism) | Differential diagnosis | Collected from participants | SPECT imaging data | 90; 56 PD + 34 non-PD | SVM-RBF with leave-one-out or 5-fold cross validation | Accuracy = 95.6% | 2014 | Palumbo et al., 2014 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 55; 18 HC + 37 PD | Naïve Bayes, OPF, SVM-RBF with 10-fold cross validation | Naïve Bayes accuracy = 78.9% | 2015 | Pereira et al., 2015 |
| Classification of PD from HC | Diagnosis | HandPD | Handwritten patterns | 92; 18 HC + 74 PD | Naïve Bayes, OPF, SVM-RBF with cross-validation | SVM-RBF recognition rate (sensitivity) = 66.72% | 2016 | Pereira et al., 2016a |
| Classification of PD from HC | Diagnosis | Extended handpd dataset with signals extracted from a smart pen | Handwritten patterns | 35; 21 HC + 14 PD | CNN with cross validation with a train:test ratio of 75:25 or 50:50 | Accuracy = 87.14% | 2016 | Pereira et al., 2016b |
| Classification of PD from HC | Diagnosis | HandPD | Handwritten patterns | 92; 18 HC + 74 PD | CNN, OPF, SVM, naïve Bayes with train-test split = 50:50 | CNN-Cifar10 accuracy = 99.30% Early stage accuracy with CNN-ImageNet = 96.35% or 94.01% for Exam 3 or Exam 4 |
2018 | Pereira et al., 2018 |
| Classification of PD from HC | Diagnosis | UCI machine learning repository | More than one | Dataset 1: 40; 20 HC + 20 PD dataset 2: 77; 15 HC + 62 PD |
Random forest, KNN, SVM-RBF, ensemble method with 5-fold cross validation | Ensemble method: Accuracy = 95.89% Specificity = 100% Sensitivity = 91.43% |
2019 | Pham et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database | More than one | 618; 195 HC + 423 PD | SVM-linear, SVM-RBF, classification tree with a train-test ratio of 70:30 | SVM-RBF, test set: Accuracy = 85.48% Sensitivity = 90.55% Specificity = 74.58% AUC = 88.22% |
2014 | Prashanth et al., 2014 |
| Classification of PD from HC | Diagnosis and subtyping | PPMI database | SPECT imaging data | 715; 208 HC + 427 PD + 80 SWEDD | SVM, naïve Bayes, boosted trees, random forest with 10-fold cross validation | SVM: Accuracy = 97.29% Sensitivity = 97.37% Specificity = 97.18% AUC = 99.26 |
2016 | Prashanth et al., 2017 |
| Classification of PD from HC | Diagnosis | PPMI database | More than one | 584; 183 HC + 401 PD | Naïve Bayes, SVM-RBF, boosted trees, random forest with 10-fold cross validation | SVM: Accuracy = 96.40% Sensitivity = 97.03% Specificity = 95.01% AUC = 98.88% |
2016 | Prashanth et al., 2016 |
| Classification of PD from HC | Diagnosis | PPMI database | Other | 626; 180 HC + 446 PD | Logistic regression, random forests, boosted trees, SVM with cross validation | Accuracy > 95% AUC > 95% Random forests: Accuracy = 96.20–97.14% (95% CI) |
2018 | Prashanth and Dutta Roy, 2018 |
| Classification of PD from HC | Diagnosis | mPower database | More than one | 133 out of 1,513 with complete source data; 46 HC + 87 PD | Logistic regression, random forests, DNN, CNN, Classifier Ensemble, Multi-Source Ensemble learning with stratified 10-fold cross validation | Ensemble learning: Accuracy = 82.0% F1-score = 87.1% |
2019 | Prince et al., 2019 |
| Classification of PD from HC | Diagnosis | HandPD | Handwritten patterns | 35; 21 HC + 14 PD | Bidirectional Gated Recurrent Units with a train-validation-test ratio of 40:10:50 or 65:10:25 | The Spiral dataset: Accuracy = 89.48% Precision = 0.848 Recall = 0.955 F1-score = 0.897 The Meander dataset: Accuracy = 92.24% Precision = 0.952 Recall = 0.883 F1-score = 0.924 |
2019 | Ribeiro et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | Handwritten patterns | 130; 39 elderly HC + 40 young HC + 39 PD + 6 PD (validation set) + 6 HC (validation set) | KNN, SVM-Gaussian, random forest with leave-one-out cross validation | SVM for PD vs young HC: Accuracy = 94.0% Sensitivity = 0.94 Specificity = 0.94 F1-score = 0.94 SVM for PD vs elderly HC: Accuracy = 89.3% Sensitivity = 0.89 Specificity = 0.89 F1-score = 0.89 Random forest for validation set: Accuracy = 83.3% Sensitivity = 0.92 Specificity = 0.93 F1-score = 0.92 |
2019 | Rios-Urrego et al., 2019 |
| Classification of IPD from non-IPD | Differential diagnosis | Collected from participants | PET imaging | 87; 39 IPD + 48 non-IPD (24 MSA + 24 PSP) | SVM with leave-one-out cross validation | Accuracy = 78.16% Sensitivity = 69.29% Specificity = 85.42% |
2015 | Segovia et al., 2015 |
| Classification of PD from HC | Diagnosis | Dataset from “Virgen de la Victoria” hospital | SPECT imaging data | 189; 94 HC + 95 PD | SVM with 10-fold cross validation | Accuracy = 94.25% Sensitivity = 91.26% Specificity = 96.17% |
2019 | Segovia et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | Other | 486; 233 HC + 205 PD + 48 NDD | SVM-linear with leave-batch-out cross validation | Validation AUC = 0.79 Test AUC = 0.74 |
2017 | Shamir et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | PET imaging | 350; 225 HC + 125 PD | GLS-DBN with a train-validation ratio of 80:20 | Test dataset 1: Accuracy = 90% Sensitivity = 0.96 Specificity = 0.84 AUC = 0.9120 Test dataset 2: Accuracy = 86% Sensitivity = 0.92 Specificity = 0.80 AUC = 0.8992 |
2019 | Shen et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | Other | 33; 18 HC + 15 PD | SMMKL-linear with leave-one-out cross validation | Accuracy = 84.85% Sensitivity = 80.00% Specificity = 88.89% YI = 68.89% PPV = 85.71% NPV = 84.21% F1 score = 82.76% |
2018 | Shi et al., 2018 |
| Classification of PD from HC | Diagnosis | Collected from participants | More than one | Plasma samples: 156; 76 HC + 80 PD; CSF samples: 77; 37 HC + 40 PD |
PLS, random forest with 10-fold cross validation with train-test ratio of 70:30 | PLS: AUC (plasma) = 0.77 AUC (CSF) = 0.90 |
2018 | Stoessel et al., 2018 |
| Classification of PD from HC | Diagnosis | PPMI database | SPECT imaging data | 658; 210 HC + 448 PD | Logistic Lasso with 10-fold cross validation | Test errors: FP = 2.83% FN = 3.78% Net error = 3.47% |
2017 | Tagare et al., 2017 |
| Classification of PD from HC | Diagnosis | PDMultiMC | handwritten patterns | 42; 21 HC + 21 PD | CNN, CNN-BLSTM with stratified 3-fold cross validation | CNN: Accuracy = 83.33% Sensitivity = 85.71% Specificity = 80.95% CNN-BLSTM: Accuracy = 83.33% Sensitivity = 71.43% Specificity = 95.24% |
2019 | Taleb et al., 2019 |
| Classification of PD from HC | Diagnosis | PPMI database and local database | SPECT imaging data | Local: 304; 113 Non-PDD + 191 PD PPMI: 657; 209 HC + 448 PD |
SVM with stratified, nested 10-fold cross-validation | Local data: Accuracy = 0.88 to 0.92 PPMI: Accuracy = 0.95 to 0.97 |
2017 | Taylor and Fenner, 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | CSF | 87; 43 HC + 44 PD | Logistic regression | Sensitivity = 0.797 specIFICITy = 0.800 AUC = 0.833 |
2017 | Trezzi et al., 2017 |
| Classification of PD from HC | Diagnosis | Collected from participants | Other | 38; 24 HC + 14 PD | SVM-RFE with repeated leave-one-out bootstrap validation | Accuracy = 89.6% | 2013 | Tseng et al., 2013 |
| Classification of MSA and PD | Differential diagnosis | Collected from participants | More than one | 85; 25 HC + 30 PD + 30 MSA-P | NN | AUC = 0.775 | 2019 | Tsuda et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | Other | 59; 30 HC + 29 PD | Logistic regression, decision tree, extra tree | Extra tree AUC = 0.99422 | 2018 | Vanegas et al., 2018 |
| Classification of PD from HC | Diagnosis | Commercially sourced | Other | 30; 15 HC + 15 PD | Decision tree | Cross validation score = 0.86 (male) Cross validation score = 0.63 (female) |
2019 | Váradi et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | More than one | 84; 40 HC + 44 PD | CNN with train-validation-test ratio of 80:10:10 | Accuracy = 97.6% AUC = 0.988 |
2018 | Vásquez-Correa et al., 2019 |
| Classification of PD and Parkinsonism | Differential diagnosis | The NTUA Parkinson Dataset | More than one | 78; 55 PD + 23 Parkinsonism | MTL with DNN | Accuracy = 0.91 Precision = 0.83 Sensitivity = 1.0 Specificity = 0.83 AUC = 0.92 |
2018 | Vlachostergiou et al., 2018 |
| Classification of PD from HC | Diagnosis | PPMI database | More than one | 534; 165 HC + 369 PD | pGTL with 10-fold cross validation | Accuracy = 97.4% | 2017 | Wang et al., 2017 |
| Classification of PD from HC | Diagnosis | PPMI database | SPECT imaging data | 645; 207 HC + 438 PD | CNN with train-validation-test ratio of 60:20:20 | Accuracy = 0.972 Sensitivity = 0.983 Specificity = 0.962 |
2019 | Wenzel et al., 2019 |
| Classification of PD from HC | Diagnosis | Collected from participants | PET imaging | Cohort 1: 182; 91 HC + 91 PD Cohort 2: 48; 26 HC + 22 PD |
SVM-linear, SVM-sigmoid, SVM-RBF with 5-fold cross validation | Cohort 1: Accuracy = 91.26% Sensitivity = 89.43% Specificity = 93.27% Cohort 2: Accuracy = 90.18% Sensitivity = 82.05% Specificity = 92.05% |
2019 | Wu et al., 2019 |
| Classification of PD, MSA and PSP | Differential diagnosis | Collected from participants | PET imaging | 920; 502 PD + 239 MSA + 179 PSP | 3D residual CNN with 6-fold cross validation | Classification of PD: Sensitivity = 97.7% Specificity = 94.1% PPV = 95.5% NPV = 97.0% Classification of MSA: Sensitivity = 96.8% Specificity = 99.5% PPV = 98.7% NPV = 98.7% Classification of PSP: Sensitivity = 83.3% Specificity = 98.3% PPV = 90.0% NPV = 97.8% |
2019 | Zhao et al., 2019 |
AD, Alzheimer's disease; AUC or AUC-ROC, area under the receiver operating characteristic (ROC) curve; AUC-PR, area under the precision-recall (PR) curve; BLSTM, bidirectional long short-term memory; CBS, corticobasal syndrome; CNN, convolutional neural network; CSF, cerebrospinal fluid; DBN, deep belief network; DNN, deep neural network; EPNN, enhanced probabilistic neural network; ET, essential tremor; FN, false negative; FP, false positive; GLS-DBN, group Lasso sparse deep belief network; HC, healthy control; IPD, idiopathic Parkinson's disease; KNN, k-nearest neighbors; LDA, linear discriminant analysis; LOR, log odds ratio; MCC, Matthews correlation coefficient; MLP, multilayer perceptron; MSA, multiple system atrophy; MSA-P, Parkinson's variant of multiple system atrophy; MTL, multi-task learning; NDD, neurodegenerative disease; NM, nearest mean; non-PDD, patients without pre-synaptic dopaminergic deficit; NPH, normal pressure hydrocephalus; NPV, negative predictive value; OPF, optimum-path forest; PD, Parkinson's disease; PET, positron emission tomography; pGTL, progressive graph-based transductive learning; PLS, partial least square; PNN, probabilistic neural network; PPV, positive predictive value; PSP, progressive supranuclear palsy; rET, essential tremor with rest tremor; SMMKL, soft margin multiple kernel learning; SPECT, single-photon emission computed tomography; SVM, support vector machine; SVM-RBF, support vector machine with radial basis function kernel; SVM-RFE, support vector machine-recursive feature elimination; SWEDD, PD with scans without evidence of dopaminergic deficit; tPD, tremor-dominant Parkinson's disease; VaP or VP, vascular Parkinsonism; YI, Youden's Index.
SPECT (n = 14)
Average accuracy of 12 out of 14 studies that used accuracy to measure the performance of machine learning models was 94.4 (4.2) % (Table 7). The lowest reported accuracy was 83.2% (Hsu et al., 2019) and 97.9% (Oliveira F. et al., 2018; Figure 4A). SVM led to the highest per-study accuracy in 10 out of 14 studies (71.4%). The highest per-study accuracy was obtained with neural networks in 3 studies (21.4%) and with regression in 1 study (7.1%; Figure 4B).
PET (n = 4)
All 4 studies used sensitivity and specificity (Table 7) in model evaluation while 3 used accuracy. Average accuracy of the 3 studies was 85.6 (6.6) %, with a lowest accuracy of 78.16% (Segovia et al., 2015) and a highest accuracy of 90.72% (Wu et al., 2019; Figure 4A). Half of the 4 studies (50.0%) obtained the highest per-study accuracy with SVM (Segovia et al., 2015; Wu et al., 2019) and the other half (50.0%) with neural networks (Figure 4B).
CSF (n = 5)
All 5 studies used AUC, instead of accuracy, to evaluate machine learning models (Table 7). The average AUC was 0.8 (0.1), the lowest AUC was 0.6825 (Maass et al., 2020) and the highest AUC was 0.839 (Maass et al., 2018), respectively. Two studies obtained the highest per-study AUC with ensemble learning, 2 studies with SVM and 1 study with regression (Figure 4B).
Other Types of Data (n = 10)
Only 5 studies used accuracy to measure the performance of machine learning models (Table 7). An average accuracy of 91.9 (6.4) % was obtained, with a lowest accuracy of 84.85% (Shi et al., 2018) and a highest accuracy of 100% (Nuvoli et al., 2019; Figure 4A). Out of the 10 studies, 5 (50%) used SVM to achieve the per-study highest accuracy, 3 (30%) used ensemble learning, 1 (10%) used decision trees and 1 (10%) used machine learning models that do not belong to any given categories (Figure 4B).
Combination of More Than One Data Type (n = 18)
Out of the 18 studies that used more than one type of data, 15 used accuracy in model evaluation (Table 7). An average accuracy of 92.6 (6.1) % was obtained, and the lowest and highest accuracy among the 15 studies was 82.0% (Prince et al., 2019) and 100.0% (Cherubini et al., 2014b), respectively (Figure 4A). The per-study highest accuracy was achieved with ensemble learning in 6 studies (33.3%), with neural network in 5 studies (27.8%), with SVM in 4 studies (22.2%), with regression in 1 (5.6%) study and with nearest neighbor (5.6%) in 1 study. One study (5.6%) used machine learning models that do not belong to any given categories to obtain the highest per-study accuracy (Figure 4B).
Discussion
Principal Findings
In this review, we present results from published studies that applied machine learning to the diagnosis and differential diagnosis of PD. Since the number of included papers was relatively large, we focused on a high-level summary rather than a detailed description of methodology and direct comparison of outcomes of individual studies. We also provide an overview of sample size, data source and data type, for a more in-depth understanding of methodological differences across studies and their outcomes. Furthermore, we assessed (a) how large the participant pool/dataset was, (b) to what extent new data (i.e., unpublished, raw data acquired from locally recruited human participants) were collected and used, (c) the feasibility of machine learning and the possibility of introducing new biomarkers in the diagnosis of PD. Overall, methodology studies that proposed and tested novel technical approaches (e.g., machine learning and deep learning models, data acquisition devices, and feature extraction algorithms) have repetitively shown that features extracted from data modalities including voice recordings and handwritten patterns could lead to high patient-level diagnostic performance, while facilitating accessible and non-invasive data acquisition. Nevertheless, only a small number of studies further validated these technical approaches in clinical settings using local human participants recruited specifically for these studies, indicating a gap between model development and their clinical applications.
A per-study diagnostic accuracy above chance levels was achieved in all studies that used accuracy in model evaluation (Figure 4A). Apart from studies using CSF data that measured model performance with AUC, classification accuracy associated with 8 other data types ranged between 85.6% (PET) and 94.4% (SPECT), with an average of 89.9 (3.0) %. Therefore, although the small number of studies of some data types may not allow for a generalizable prediction of how well these data types can help us differentiate PD from HC or atypical Parkinsonian disorders, the application of machine learning to a variety of data types led to high accuracy in the diagnosis of PD. In addition, an accuracy significantly above chance levels was achieved in all machine learning models (Supplementary Table 1), while SVM, neural networks and ensemble learning were among the most popular model choices, all yielding great applicability to a variety of data modalities. In the meantime, when compared with other models, they led to the per-study highest classification accuracy in >50% of all cases (50.7, 51.9, and 52.3%, respectively; Supplementary Table 1). Despite the high diagnostic accuracy and performance reported, in a number of studies, data splitting strategies and the use of cross validation were not specified. For data modalities such as 3D MRI scans, when 2D slices are extracted from 3D volumes, multiple slices could be generated for one subject. Having data from the same subject across training, validation and tests sets can lead to a biased data split (Wen et al., 2020), causing data leakage and overestimation of model performance, thus compromising reproducibility of published results.
As previously discussed (Belić et al., 2019), although satisfactory diagnostic outcomes could be achieved, sample size in few studies was extremely small (<15 subjects). The application of some machine learning models, especially neural networks, typically rely on a large dataset. Nevertheless, collecting data from a large pool of participants remains challenging in clinical studies, and data generated are commonly of high dimensionality and small sample size (Vabalas et al., 2019). To address this challenge, one solution is to combine data from a local cohort with public repositories including PPMI, UCI machine learning repository, PhysioNet and many others, depending on the type of data that have been collected from the local cohort. Furthermore, when a great difference in group size is observed (i.e., class imbalance problem), labeling all samples after the majority class may lead to an undesired high accuracy. In this case, evaluating machine learning models with other metrics including precision, recall and F-1 score is recommended (Jeni et al., 2013).
Even though high diagnostic accuracy of PD has been achieved in clinical settings, machine learning approaches have also reached high accuracy as shown in the present study, while models including SVM and neural networks are particularly useful in (a) diagnosis of PD using data modalities that have been overlooked in clinical decision making (e.g., voice), and (b) identification of features of high relevance from these data. For example, the use of machine learning models with feature selection techniques allows for assessing the relative importance of features of a large feature space in order to select the most differentiating ones, which is conventionally challenging using manual approaches. For the discovery of novel markers allowing for non-invasive diagnostic options with relatively high accuracy, e.g., handwritten patterns, a small number of studies have been conducted, mostly using data from published databases. Given that these databases generally included handwritten patterns from a small number of diagnosed PD patients, sometimes under 15, it would be of great importance to validate the use of handwritten patterns in early diagnosis of PD in clinical studies of a larger scale. In the meantime, diagnosing PD using more than one data modality has led to promising results. Accordingly, supplying clinicians with non-motor data and machine learning approaches may support clinical decision making in patients with ambiguous symptom presentations, and/or improve diagnosis at an earlier stage.
An issue observed in many included studies was the insufficient or inaccurate description of methods or results, and some failed to provide accurate information of the number and type of subjects used (for example, methodology studies on early diagnosis of PD missing a table summarizing the characteristics of subjects, therefore it was challenging to understand the stage of PD in recruited patients), or how machine learning models were implemented, trained and tested. Infrequently, authors skipped basic information such as number of subjects and their medical conditions and referred to another publication. Although we attempted to list model hyperparameters and cross-validation strategies in the data extraction table, many included studies did not make this information available in the main text, leading to potential difficulties in replicating the results. Apart from these, rounding errors or inconsistent reporting of results also exist. Furthermore, although we treated the differentiation of PD from SWEDD as subtyping, there is ongoing controversy regarding whether it should be considered as differential diagnosis or subtyping (Lee et al., 2014; Erro et al., 2016; Chou, 2017; Kwon et al., 2018). Given these limitations, clinicians interested in adapting machine learning models or implementing diagnostic systems based on novel biomarkers are advised to interpret published results with care. Further, in this context we would like to stress the need for uniform reporting standards in studies using machine learning.
In both machine learning research and clinical settings, appropriately interpreting published results and methodologies is a necessary step toward an understanding of state-of-the-art methods. Therefore, vagueness in reporting not only compromises the interpretation of results but makes further methodological developments based on published research unnecessarily challenging. Moreover, for medical doctors interested in learning how machine learning methods could be applied in their domains, insufficient description of methods may lead to incorrect model implementation and failure of replication.
To enable efficient replication of published results, detailed descriptions of (a) model and architecture (hyperparameters, number and type of layers, layer-specific parameter settings, regularization strategies, activation functions), (b) implementation (programming language, machine learning and deep learning libraries used, model training and testing, metrics and model evaluation, validation strategy, optimization), and (c) version numbers of software/libraries used for both preprocessing and model implementation, are often desirable, as newer software versions may lead to differences in pre-processing and model implementation stages (Chepkoech et al., 2016).
Due to the use of imbalanced datasets in medical sciences, reporting model performance with a confusion matrix may give rise to a more comprehensive understanding of the model's ability to discriminate between PD and healthy controls. In the meantime, due to costs associated with acquisition of patient data, researchers often need to expand data collected from a local cohort using data sourced from publicly available databases or published studies. Nevertheless, unclear description of data acquisition and pre-processing protocols in some published studies may lead to challenges in the integration of newly acquired data and previously published data. Taken together, to facilitate early, refined diagnosis of PD and efficient application of novel machine learning approaches in a clinical setting, and to allow for improved reproducibility of studies on machine learning-based diagnosis and assessment of PD, a higher transparency in reporting data collection, pre-processing protocols, model implementation, and study outcomes is required.
Limitations
In the present study, we have excluded research articles in languages other than English and results published in the form of conference abstracts, posters, and talks. Despite the ongoing discussion of advantages and importance of including conference abstracts in systematic reviews and reviews (Scherer and Saldanha, 2019), conference abstracts often do not report sufficient key information which is why we had to exclude them. However, this may lead to a publication and result bias. In addition, since the aim of the present review is to assess and summarize published studies on the detection and early diagnosis of PD, we noticed that few large-scale, multi-centric studies on subtyping or/and severity assessment of PD were therefore excluded. Given the current challenges in subtyping, severity assessment and prognosis of PD, a further step toward a more systematic understanding of the application of machine learning to neurodegenerative diseases would be to review these studies.
Moreover, due to the high inter-study variance in the data source and presentation of results, it was challenging to directly compare outcomes associated with each type of model across studies, as some studies failed to indicate whether model performance was evaluated using a test set, and/or results given by models that did not yield the best per-study performance. Results of published studies were discussed and summarized based on data and machine learning models used, and for data modalities such as PET (n = 4) or CSF (n = 5), the number of studies were too small despite the high total number of studies included. Therefore, it was improbable to assess the general performance of machine learning techniques when PET or CSF data are used.
Conclusions
To the best of our knowledge, the present study is the first review which included results from all studies that applied machine learning methods to the diagnosis of PD. Here, we presented included studies in a high-level summary, providing access to information including (a) machine learning methods that have been used in the diagnosis of PD and associated outcomes, (b) types of clinical, behavioral and biometric data that could be used for rendering more accurate diagnoses, (c) potential biomarkers for assisting clinical decision making, and (d) other highly relevant information, including databases that could be used to enlarge and enrich smaller datasets. In summary, realization of machine learning-assisted diagnosis of PD yields high potential for a more systematic clinical decision-making system, while adaptation of novel biomarkers may give rise to easier access to PD diagnosis at an earlier stage. Machine learning approaches therefore have the potential to provide clinicians with additional tools to screen, detect or diagnose PD.
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JM conceived and designed the study, collected the data, performed the analysis, and wrote the paper. CD and JF supervised the research. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Antje Haehner for her comments on the manuscript. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and Québec Bio-Imaging Network.
Footnotes
Funding. JM was supported by the Québec Bio-Imaging Network Postdoctoral Fellowship (FRSQ—Réseaux de recherche thématiques; Dossier: 35450). JF was supported by FRQS (#283144), Parkinson Québec, Parkinson Canada (PPG-2020-0000000061), and CIHR (#PJT173514).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2021.633752/full#supplementary-material
References
- Abiyev R. H., Abizade S. (2016). Diagnosing Parkinson's diseases using fuzzy neural system. Comput. Mathe. Methods Med. 2016:1267919. 10.1155/2016/1267919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abos A., Baggio H. C., Segura B., Campabadal A., Uribe C., Giraldo D. M., et al. (2019). Differentiation of multiple system atrophy from Parkinson's disease by structural connectivity derived from probabilistic tractography. Sci. Rep. 9:16488. 10.1038/s41598-019-52829-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abujrida H., Agu E., Pahlavan K. (2017). Smartphone-based gait assessment to infer Parkinson's disease severity using crowdsourced data, in 2017 IEEE Healthcare Innovations and Point of Care Technologies (HI-POCT) (Bethesda, MD: ), 208–211. 10.1109/HIC.2017.8227621 [DOI] [Google Scholar]
- Adams W. R. (2017). High-accuracy detection of early Parkinson's Disease using multiple characteristics of finger movement while typing. PLoS ONE 12:e0188226. 10.1371/journal.pone.0188226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeli E., Shi F., An L., Wee C.-Y., Wu G., Wang T., et al. (2016). Joint feature-sample selection and robust diagnosis of Parkinson's disease from MRI data. NeuroImage 141, 206–219. 10.1016/j.neuroimage.2016.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeli E., Thung K.-H., An L., Wu G., Shi F., Wang T., et al. (2019). Semi-supervised discriminative classification robust to sample-outliers and feature-noises. IEEE Trans. Pattern Anal. Mach. Intell. 41, 515–522. 10.1109/TPAMI.2018.2794470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Chandrayan S., Sahu S. S. (2016). Prediction of Parkinson's disease using speech signal with Extreme Learning Machine, in 2016 International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT) (Chennai: ), 3776–3779. 10.1109/ICEEOT.2016.7755419 [DOI] [Google Scholar]
- Ahlrichs C., Lawo M. (2013). Parkinson's disease motor symptoms in machine learning: a review. arXiv preprint arXiv:1312.3825. 10.5121/hiij.2013.2401 [DOI] [Google Scholar]
- Ahmadi S. A., Vivar G., Frei J., Nowoshilow S., Bardins S., Brandt T., et al. (2019). Towards computerized diagnosis of neurological stance disorders: data mining and machine learning of posturography and sway. J. Neurol. 266(Suppl 1), 108–117. 10.1007/s00415-019-09458-y [DOI] [PubMed] [Google Scholar]
- Aich S., Kim H., younga K., Hui K. L., Al-Absi A. A., Sain M. (2019). A supervised machine learning approach using different feature selection techniques on voice datasets for prediction of Parkinson's disease, in 2019 21st International Conference on Advanced Communication Technology (ICACT) (PyeongChang: ), 1116–1121. 10.23919/ICACT.2019.8701961 [DOI] [Google Scholar]
- Alam M. N., Garg A., Munia T. T. K., Fazel-Rezai R., Tavakolian K. (2017). Vertical ground reaction force marker for Parkinson's disease. PLoS ONE 12:e0175951. 10.1371/journal.pone.0175951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaskar H., Hussain A. (2018). Prediction of Parkinson disease using gait signals, in 2018 11th International Conference on Developments in eSystems Engineering (DeSE) (Cambridge: ), 23–26. 10.1109/DeSE.2018.00011 [DOI] [Google Scholar]
- Al-Fatlawi A. H., Jabardi M. H., Ling S. H. (2016). Efficient diagnosis system for Parkinson's disease using deep belief network, in 2016 IEEE Congress on Evolutionary Computation (CEC) (Vancouver, BC: ), 1324–1330. 10.1109/CEC.2016.7743941 [DOI] [Google Scholar]
- Alharthi A. S., Ozanyan K. B. (2019). Deep learning for ground reaction force data analysis: application to wide-area floor sensing, in 2019 IEEE 28th International Symposium on Industrial Electronics (ISIE) (Vancouver, BC: )„ 1401–1406. 10.1109/ISIE.2019.8781511 [DOI] [Google Scholar]
- Ali L., Khan S. U., Arshad M., Ali S., Anwar M. (2019a). A multi-model framework for evaluating type of speech samples having complementary information about Parkinson's disease, in 2019 International Conference on Electrical, Communication, and Computer Engineering (ICECCE) (Swat: ), 1–5. 10.1109/ICECCE47252.2019.8940696 [DOI] [Google Scholar]
- Ali L., Zhu C., Golilarz N. A., Javeed A., Zhou M., Liu Y. (2019b). Reliable Parkinson's disease detection by analyzing handwritten drawings: construction of an unbiased cascaded learning system based on feature selection and adaptive boosting model. IEEE Access 7, 116480–116489. 10.1109/ACCESS.2019.2932037 [DOI] [Google Scholar]
- Ali L., Zhu C., Zhang Z., Liu Y. (2019c). Automated detection of Parkinson's disease based on multiple types of sustained phonations using linear discriminant analysis and genetically optimized neural network. IEEE J. Transl. Eng. Health Med. 7, 1–10. 10.1109/JTEHM.2019.2940900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani E. J., Alshamrani F. H., Syed H. F., Olatunji S. O. (2018). Classification of Parkinson's disease using NNge classification algorithm, in 2018 21st Saudi Computer Society National Computer Conference (NCC) (Riyadh: ), 1–7. 10.1109/NCG.2018.8592989 [DOI] [Google Scholar]
- Amoroso N., La Rocca M., Monaco A., Bellotti R., Tangaro S. (2018). Complex networks reveal early MRI markers of Parkinson's disease. Med. Image Anal. 48, 12–24. 10.1016/j.media.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Anand A., Haque M. A., Alex J. S. R., Venkatesan N. (2018). Evaluation of machine learning and deep learning algorithms combined with dimentionality reduction techniques for classification of Parkinson's disease, in 2018 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT) (Louisville, KY: ), 342–347. 10.1109/ISSPIT.2018.8642776 [DOI] [Google Scholar]
- Andrei A., Tău?an A., Ionescu B. (2019). Parkinson's disease detection from gait patterns, in 2019 E-Health and Bioengineering Conference (EHB) (Iasi: ), 1–4. 10.1109/EHB47216.2019.8969942 [DOI] [Google Scholar]
- Baby M. S., Saji A. J., Kumar C. S. (2017). Parkinsons disease classification using wavelet transform based feature extraction of gait data, in 2017 International Conference on Circuit, Power and Computing Technologies (ICCPCT) (Kollam: ), 1–6. 10.1109/ICCPCT.2017.8074230 [DOI] [Google Scholar]
- Baggio H. C., Abos A., Segura B., Campabadal A., Uribe C., Giraldo D. M., et al. (2019). Cerebellar resting-state functional connectivity in Parkinson's disease and multiple system atrophy: characterization of abnormalities and potential for differential diagnosis at the single-patient level. NeuroImage. Clin. 22:101720. 10.1016/j.nicl.2019.101720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakar Z. A., Ispawi D. I., Ibrahim N. F., Tahir N. M. (2012). Classification of Parkinson's disease based on Multilayer Perceptrons (MLPs) neural network and ANOVA as a feature extraction, in 2012 IEEE 8th International Colloquium on Signal Processing and its Applications) (Malacca: ), 63–67. 10.1109/CSPA.2012.6194692 [DOI] [Google Scholar]
- Banerjee M., Chakraborty R., Archer D., Vaillancourt D., Vemuri B. C. (2019). DMR-CNN: a CNN tailored For DMR scans with applications to PD classification, in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) (Venice: ), 388–391. 10.1109/ISBI.2019.8759558 [DOI] [Google Scholar]
- Belić M., Bobić V., BadŽa M., Šolaja N., Ã?urić-Jovičić M., Kostić V. S. (2019). Artificial intelligence for assisting diagnostics and assessment of Parkinson's disease–a review. Clin. Neurol. Neurosurg. 184:105442. 10.1016/j.clineuro.2019.105442 [DOI] [PubMed] [Google Scholar]
- Benba A., Jilbab A., Hammouch A. (2016a). Discriminating between patients with Parkinson's and neurological diseases using cepstral analysis. IEEE Trans. Neural Syst. Rehab. Eng. 24, 1100–1108. 10.1109/TNSRE.2016.2533582 [DOI] [PubMed] [Google Scholar]
- Benba A., Jilbab A., Hammouch A., Sandabad S. (2016b). Using RASTA-PLP for discriminating between different neurological diseases, in 2016 International Conference on Electrical and Information Technologies (ICEIT) (Tangiers: ), 406–409. 10.1109/EITech.2016.7519630 [DOI] [Google Scholar]
- Bernad-Elazari H., Herman T., Mirelman A., Gazit E., Giladi N., Hausdorff J. M. (2016). Objective characterization of daily living transitions in patients with Parkinson's disease using a single body-fixed sensor. J. Neurol. 263, 1544–1551. 10.1007/s00415-016-8164-6 [DOI] [PubMed] [Google Scholar]
- Bhati S., Velazquez L. M., Villalba J., Dehak N. (2019). LSTM siamese network for Parkinson's disease detection from speech, in 2019 IEEE Global Conference on Signal and Information Processing (GlobalSIP) (Ottawa, ON: ), 1–5. 10.1109/GlobalSIP45357.2019.8969430 [DOI] [Google Scholar]
- Bot B. M., Suver C., Neto E. C., Kellen M., Klein A., Bare C., et al. (2016). The mPower study, Parkinson disease mobile data collected using ResearchKit. Sci. Data 3, 1–9. 10.1038/sdata.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Del Tredici K., Rüb U., De Vos R. A., Steur E. N. J., Braak E. (2003). Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging 24, 197–211. 10.1016/S0197-4580(02)00065-9 [DOI] [PubMed] [Google Scholar]
- Buongiorno D., Bortone I., Cascarano G. D., Trotta G. F., Brunetti A., Bevilacqua V. (2019). A low-cost vision system based on the analysis of motor features for recognition and severity rating of Parkinson's Disease. BMC Med. Inform. Decision Making 19(Suppl 9):243. 10.1186/s12911-019-0987-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A. H., Rovini E., Dolciotti C., Bongioanni P., De Petris G., Cavallo F. (2017). Leap motion evaluation for assessment of upper limb motor skills in Parkinson's disease. IEEE Int. Conf. Rehabil. Robot. 2017, 116–121. 10.1109/ICORR.2017.8009232 [DOI] [PubMed] [Google Scholar]
- Butt A. H., Rovini E., Dolciotti C., De Petris G., Bongioanni P., Carboncini M. C., et al. (2018). Objective and automatic classification of Parkinson disease with Leap Motion controller. Biomed. Eng. Online 17:168. 10.1186/s12938-018-0600-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Gu J., Wen C., Zhao D., Huang C., Huang H., et al. (2018). An intelligent Parkinson's disease diagnostic system based on a chaotic bacterial foraging optimization enhanced fuzzy KNN approach. Comp. Math. Methods Med. 2018:2396952. 10.1155/2018/2396952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramia C., Torricelli D., Schmid M., Munoz-Gonzalez A., Gonzalez-Vargas J., Grandas F., et al. (2018). IMU-based classification of Parkinson's disease from gait: a sensitivity analysis on sensor location and feature selection. IEEE J. Biomed. Health Inf. 22, 1765–1774. 10.1109/JBHI.2018.2865218 [DOI] [PubMed] [Google Scholar]
- Castillo-Barnes D., Ramírez J., Segovia F., Martínez-Murcia F. J., Salas-Gonzalez D., Górriz J. M. (2018). Robust ensemble classification methodology for I123-Ioflupane SPECT images and multiple heterogeneous biomarkers in the diagnosis of Parkinson's disease. Front. Neuroinf. 12:53. 10.3389/fninf.2018.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo F., Moschetti A., Esposito D., Maremmani C., Rovini E. (2019). Upper limb motor pre-clinical assessment in Parkinson's disease using machine learning. Parkinsonism Relat. Disord. 63, 111–116. 10.1016/j.parkreldis.2019.02.028 [DOI] [PubMed] [Google Scholar]
- Celik E., Omurca S. I. (2019). Improving Parkinson's disease diagnosis with machine learning methods, in 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT) (Istanbul: ), 1–4. 10.1109/EBBT.2019.8742057 [DOI] [Google Scholar]
- Chakraborty S., Aich S., Kim H.-C. (2020). 3D textural, morphological and statistical analysis of voxel of interests in 3T MRI scans for the detection of Parkinson's disease using artificial neural networks. Healthcare 8:E34. 10.3390/healthcare8010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa K. N. R., Pagolu V. S., Panda G., Majhi B. (2016). An improved approach for prediction of Parkinson's disease using machine learning techniques, in 2016 International Conference on Signal Processing, Communication, Power and Embedded System (SCOPES) (Paralakhemundi: ), 1446–1451. 10.1109/SCOPES.2016.7955679 [DOI] [Google Scholar]
- Chen Y., Storrs J., Tan L., Mazlack L. J., Lee J.-H., Lu L. J. (2014). Detecting brain structural changes as biomarker from magnetic resonance images using a local feature based SVM approach. J. Neurosci. Methods 221, 22–31. 10.1016/j.jneumeth.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Chen Y., Yang W., Long J., Zhang Y., Feng J., Li Y., et al. (2015). Discriminative analysis of Parkinson's disease based on whole-brain functional connectivity. PLoS ONE 10:e0124153. 10.1371/journal.pone.0124153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepkoech J. L., Walhovd K. B., Grydeland H., Fjell A. M., Alzheimer's Disease Neuroimaging Initiative (2016). Effects of change in FreeSurfer version on classification accuracy of patients with Alzheimer's disease and mild cognitive impairment. Hum. Brain Mapp. 37, 1831–1841. 10.1002/hbm.23139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini A., Morelli M., Nisticó R., Salsone M., Arabia G., Vasta R., et al. (2014a). Magnetic resonance support vector machine discriminates between Parkinson disease and progressive supranuclear palsy. Move. Disord. 29, 266–269. 10.1002/mds.25737 [DOI] [PubMed] [Google Scholar]
- Cherubini A., Nisticó R., Novellino F., Salsone M., Nigro S., Donzuso G., et al. (2014b). Magnetic resonance support vector machine discriminates essential tremor with rest tremor from tremor-dominant Parkinson disease. Move. Disord. 29, 1216–1219. 10.1002/mds.25869 [DOI] [PubMed] [Google Scholar]
- Choi H., Ha S., Im H. J., Paek S. H., Lee D. S. (2017). Refining diagnosis of Parkinson's disease with deep learning-based interpretation of dopamine transporter imaging. NeuroImage Clin. 16, 586–594. 10.1016/j.nicl.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. L. (2017). Diagnosis and Differential Diagnosis of Parkinson Disease. Waltham, MA: UpToDate. [Google Scholar]
- Cibulka M., Brodnanova M., Grendar M., Grofik M., Kurca E., Pilchova I., et al. (2019). SNPs rs11240569, rs708727, and rs823156 in SLC41A1 do not discriminate between slovak patients with idiopathic parkinson's disease and healthy controls: statistics and machine-learning evidence. Int. J. Mol. Sci. 20:4688. 10.3390/ijms20194688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigdem O., Demirel H., Unay D. (2019). The performance of local-learning based clustering feature selection method on the diagnosis of Parkinson's disease using structural MRI, in 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC) (Bari: ), 1286–1291. 10.1109/SMC.2019.8914611 [DOI] [Google Scholar]
- Çimen S., Bolat B. (2016). Diagnosis of Parkinson's disease by using ANN, in 2016 International Conference on Global Trends in Signal Processing, Information Computing and Communication (ICGTSPICC) (Jalgaon: ), 119–121. 10.1109/ICGTSPICC.2016.7955281 [DOI] [Google Scholar]
- Contreras-Vidal J., Stelmach G. E. (1995). Effects of Parkinsonism on motor control. Life Sci. 58, 165–176. 10.1016/0024-3205(95)02237-6 [DOI] [PubMed] [Google Scholar]
- Cook D. J., Schmitter-Edgecombe M., Dawadi P. (2015). Analyzing activity behavior and movement in a naturalistic environment using smart home techniques. IEEE J. Biomed. Health Inf. 19, 1882–1892. 10.1109/JBHI.2015.2461659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzzolin F., Sapienza M., Esser P., Saha S., Franssen M. M., Collett J., et al. (2017). Metric learning for Parkinsonian identification from IMU gait measurements. Gait Posture 54, 127–132. 10.1016/j.gaitpost.2017.02.012 [DOI] [PubMed] [Google Scholar]
- Dash S., Thulasiram R., Thulasiraman P. (2017). An enhanced chaos-based firefly model for Parkinson's disease diagnosis and classification, in 2017 International Conference on Information Technology (ICIT) (Bhubaneswar: ), 159–164. 10.1109/ICIT.2017.43 [DOI] [Google Scholar]
- Dastjerd N. K., Sert O. C., Ozyer T., Alhajj R. (2019). Fuzzy classification methods based diagnosis of Parkinson's disease from speech test cases. Curr. Aging Sci. 12, 100–120. 10.2174/1874609812666190625140311 [DOI] [PubMed] [Google Scholar]
- de Souza J. W. M., Alves S. S. A., Rebouças E. d,.S, Almeida J. S., Rebouças Filho P.P. (2018). A new approach to diagnose Parkinson's disease using a structural cooccurrence matrix for a similarity analysis. Comput. Intell. Neurosci. 2018:7613282. 10.1155/2018/7613282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhami D. S., Soni A., Page D., Natarajan S. (2017). Identifying Parkinson's patients: a functional gradient boosting approach. Artif. Intell. Med. Conf. Artif. Intell. Med. 10259, 332–337. 10.1007/978-3-319-59758-4_39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh A., He J. (2017). Using machine learning to diagnose Parkinson's disease from voice recordings, in 2017 IEEE MIT Undergraduate Research Technology Conference (URTC) (Cambridge, MA: ), 1–4. 10.1109/URTC.2017.8284216 [DOI] [Google Scholar]
- Dinov I. D., Heavner B., Tang M., Glusman G., Chard K., Darcy M., et al. (2016). Predictive big data analytics: a study of parkinson's disease using large, complex, heterogeneous, incongruent, multi-source and incomplete observations. PLoS ONE 11:e0157077. 10.1371/journal.pone.0157077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurić-Jovičić M., Belić M., Stanković I., Radovanović S., Kostić V. S. (2017). Selection of gait parameters for differential diagnostics of patients with de novo Parkinson's disease. Neurol. Res. 39, 853–861. 10.1080/01616412.2017.1348690 [DOI] [PubMed] [Google Scholar]
- Dorsey E. R., Elbaz A., Nichols E., Abd-Allah F., Abdelalim A., Adsuar J. C., et al. (2018). Global, regional, and national burden of Parkinson's disease, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 17, 939–953. 10.1016/S1474-4422(18)30295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos M. C. T., Scheller D., Schulte C., Mesa I. R., Colman P., Bujac S. R., et al. (2018). Evaluation of cerebrospinal fluid proteins as potential biomarkers for early stage Parkinson's disease diagnosis. PLoS ONE 13:e0206536. 10.1371/journal.pone.0206536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror B., Yanai E., Frid A., Peleg N., Goldenthal N., Schlesinger I., et al. (2014). Automatic assessment of Parkinson's disease from natural hands movements using 3D depth sensor, in 2014 IEEE 28th Convention of Electrical & Electronics Engineers in Israel (IEEEI) (Eilat: ), 1–5. 10.1109/EEEI.2014.7005763 [DOI] [Google Scholar]
- Drotár P., Mekyska J., Rektorová I., Masarová L., Smékal Z., Faundez-Zanuy M. (2014). Analysis of in-air movement in handwriting: a novel marker for Parkinson's disease. Comp. Methods Progr. Biomed. 117, 405–411. 10.1016/j.cmpb.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Drotár P., Mekyska J., Rektorová I., Masarová L., Smékal Z., Faundez-Zanuy M. (2015). Decision support framework for Parkinson's disease based on novel handwriting markers. IEEE Trans. Neural Syst. Rehabil. Eng. 23, 508–516. 10.1109/TNSRE.2014.2359997 [DOI] [PubMed] [Google Scholar]
- Drotár P., Mekyska J., Rektorová I., Masarová L., Smékal Z., Faundez-Zanuy M. (2016). Evaluation of handwriting kinematics and pressure for differential diagnosis of Parkinson's disease. Artif. Intell. Med. 67, 39–46. 10.1016/j.artmed.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Du G., Lewis M. M., Kanekar S., Sterling N. W., He L., Kong L., et al. (2017). Combined Diffusion tensor imaging and apparent transverse relaxation rate differentiate Parkinson disease and atypical Parkinsonism. Am. J. Neuroradiol. 38, 966–972. 10.3174/ajnr.A5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua D., Graff C. (2018). UCI Machine Learning Repository. Irvine, CA: University of California, School of Information and Computer Science. [Google Scholar]
- Erdogdu Sakar B., Serbes G., Sakar C. O. (2017). Analyzing the effectiveness of vocal features in early telediagnosis of Parkinson's disease. PLoS ONE 12:e0182428. 10.1371/journal.pone.0182428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erro R., Schneider S. A., Stamelou M., Quinn N. P., Bhatia K. P. (2016). What do patients with scans without evidence of dopaminergic deficit (SWEDD) have? New evidence and continuing controversies. J. Neurol. Neurosurg. Psychiatry 87, 319–323. 10.1136/jnnp-2014-310256 [DOI] [PubMed] [Google Scholar]
- Félix J. P., Vieira F. H. T., Cardoso Á. A., Ferreira M. V. G., Franco R. A. P., Ribeiro M. A., et al. (2019). A Parkinson's disease classification method: an approach using gait dynamics and detrended fluctuation analysis, in 2019 IEEE Canadian Conference of Electrical and Computer Engineering (CCECE) (Edmonton, AB: ), 1–4. 10.1109/CCECE.2019.8861759 [DOI] [Google Scholar]
- Fernandes C., Fonseca L., Ferreira F., Gago M., Costa L., Sousa N., et al. (2018). Artificial neural networks classification of patients with Parkinsonism based on gait, in 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (Madrid: ), 2024–2030. 10.1109/BIBM.2018.8621466 [DOI] [Google Scholar]
- Fernandez K. M., Roemmich R. T., Stegemöller E. L., Amano S., Thompson A., Okun M. S., et al. (2013). Gait initiation impairments in both Essential Tremor and Parkinson's disease. Gait Posture 38, 956–961. 10.1016/j.gaitpost.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focke N. K., Helms G., Scheewe S., Pantel P. M., Bachmann C. G., Dechent P., et al. (2011). Individual voxel-based subtype prediction can differentiate progressive supranuclear palsy from idiopathic Parkinson syndrome and healthy controls. Human Brain Mapp. 32, 1905–1915. 10.1002/hbm.21161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frid A., Safra E. J., Hazan H., Lokey L. L., Hilu D., Manevitz L., et al. (2014). Computational diagnosis of Parkinson's disease directly from natural speech using machine learning techniques, in 2014 IEEE International Conference on Software Science, Technology and Engineering (Ramat Gan: ), 50–53. 10.1109/SWSTE.2014.17 [DOI] [Google Scholar]
- Ghassemi N. H., Marxreiter F., Pasluosta C. F., Kugler P., Schlachetzki J., Schramm A., et al. (2016). Combined accelerometer and EMG analysis to differentiate essential tremor from Parkinson's disease. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 672–675. 10.1109/EMBC.2016.7590791 [DOI] [PubMed] [Google Scholar]
- Glaab E., Trezzi J.-P., Greuel A., Jäger C., Hodak Z., Drzezga A., et al. (2019). Integrative analysis of blood metabolomics and PET brain neuroimaging data for Parkinson's disease. Neurobiol. Dis. 124, 555–562. 10.1016/j.nbd.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Goldberger A. L., Amaral L. A., Glass L., Hausdorff J. M., Ivanov P. C., Mark R. G., et al. (2000). PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101, e215–e220. 10.1161/01.CIR.101.23.e215 [DOI] [PubMed] [Google Scholar]
- Gunduz H. (2019). Deep learning-based parkinson's disease classification using vocal feature sets. IEEE Access 7, 115540–115551. 10.1109/ACCESS.2019.2936564 [DOI] [Google Scholar]
- Haller S., Badoud S., Nguyen D., Barnaure I., Montandon M. L., Lovblad K. O., et al. (2013). Differentiation between Parkinson disease and other forms of Parkinsonism using support vector machine analysis of susceptibility-weighted imaging (SWI): initial results. Eur. Radiol. 23, 12–19. 10.1007/s00330-012-2579-y [DOI] [PubMed] [Google Scholar]
- Haller S., Badoud S., Nguyen D., Garibotto V., Lovblad K. O., Burkhard P. R. (2012). Individual detection of patients with Parkinson disease using support vector machine analysis of diffusion tensor imaging data: initial results. Am. J. Neuroradiol. 33, 2123–2128. 10.3174/ajnr.A3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq A. U., Li J., Memon M. H., Khan J., Din S. U., Ahad I., et al. (2018). Comparative analysis of the classification performance of machine learning classifiers and deep neural network classifier for prediction of Parkinson disease, in 2018 15th International Computer Conference on Wavelet Active Media Technology and Information Processing (ICCWAMTIP) (Chengdu: ), 101–106. [Google Scholar]
- Haq A. U., Li J. P., Memon M. H., khan J., Malik A., Ahmad T., et al. (2019). Feature selection based on L1-Norm support vector machine and effective recognition system for Parkinson's disease using voice recordings. IEEE Access 7, 37718–37734. 10.1109/ACCESS.2019.2906350 [DOI] [Google Scholar]
- Hariharan M., Polat K., Sindhu R. (2014). A new hybrid intelligent system for accurate detection of Parkinson's disease. Comput. Methods Programs Biomed. 113, 904–913. 10.1016/j.cmpb.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Hirschauer T. J., Adeli H., Buford J. A. (2015). Computer-aided diagnosis of Parkinson's disease using enhanced probabilistic neural network. J. Med. Syst. 39:179. 10.1007/s10916-015-0353-9 [DOI] [PubMed] [Google Scholar]
- Hsu S.-Y., Lin H.-C., Chen T.-B., Du W.-C., Hsu Y.-H., Wu Y.-C., et al. (2019). Feasible classified models for Parkinson disease from (99m)Tc-TRODAT-1 SPECT imaging. Sensors 19:1740. 10.3390/s19071740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas-Fernández I., García-Gómez F. J., García-Solís D., Benítez-Rivero S., Marín-Oyaga V. A., Jesús S., et al. (2015). Machine learning models for the differential diagnosis of vascular parkinsonism and Parkinson's disease using [(123)I]FP-CIT SPECT. Eur. J. Nucl. Med. Mol. Imag. 42, 112–119. 10.1007/s00259-014-2882-8 [DOI] [PubMed] [Google Scholar]
- Huppertz H.-J., Möller L., Südmeyer M., Hilker R., Hattingen E., Egger K., et al. (2016). Differentiation of neurodegenerative parkinsonian syndromes by volumetric magnetic resonance imaging analysis and support vector machine classification. Mov. Disord. 31, 1506–1517. 10.1002/mds.26715 [DOI] [PubMed] [Google Scholar]
- I K., Ulukaya S., Erdem O. (2019). Classification of Parkinson's disease using dynamic time warping, in 2019 27th Telecommunications Forum (TELFOR) (Belgrade: ), 1–4 [Google Scholar]
- Illan I. A., Gorrz J. M., Ramirez J., Segovia F., Jimenez-Hoyuela J. M., Ortega Lozano S. J. (2012). Automatic assistance to Parkinson's disease diagnosis in DaTSCAN SPECT imaging. Med. Phys. 39, 5971–5980. 10.1118/1.4742055 [DOI] [PubMed] [Google Scholar]
- Islam M. S., Parvez I., Hai D., Goswami P. (2014). Performance comparison of heterogeneous classifiers for detection of Parkinson's disease using voice disorder (dysphonia), in 2014 International Conference on Informatics, Electronics & Vision (ICIEV) (Dhaka: ), 1–7. 10.1109/ICIEV.2014.6850849 [DOI] [Google Scholar]
- Jankovic J. (2008). Parkinson's disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 79, 368–376. 10.1136/jnnp.2007.131045 [DOI] [PubMed] [Google Scholar]
- Javed F., Thomas I., Memedi M. (2018). A comparison of feature selection methods when using motion sensors data: a case study in Parkinson's disease. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 5426–5429. 10.1109/EMBC.2018.8513683 [DOI] [PubMed] [Google Scholar]
- Jeni L. A., Cohn J. F., De La Torre F. (2013). Facing imbalanced data–recommendations for the use of performance metrics, in 2013 Humaine Association Conference on Affective Computing and Intelligent Interaction (Geneva: IEEE; ), 245–251. 10.1109/ACII.2013.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Li Y. (2012). Energy-based feature ranking for assessing the dysphonia measurements in Parkinson detection. IET Signal Proc. 6, 300–305. 10.1049/iet-spr.2011.0186 [DOI] [Google Scholar]
- Johnson S. J., Diener M. D., Kaltenboeck A., Birnbaum H. G., Siderowf A. D. (2013). An economic model of P arkinson's disease: implications for slowing progression in the United States. Move. Disord. 28, 319–326. 10.1002/mds.25328 [DOI] [PubMed] [Google Scholar]
- Joshi D., Khajuria A., Joshi P. (2017). An automatic non-invasive method for Parkinson's disease classification. Comput. Methods Programs Biomed. 145, 135–145. 10.1016/j.cmpb.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Junior S. B., Costa V. G. T., Chen S., Guido R. C. (2018). U-healthcare system for pre-diagnosis of Parkinson's disease from voice signal, in 2018 IEEE International Symposium on Multimedia (ISM) (Taichung: ), 271–274. [Google Scholar]
- Kamagata K., Zalesky A., Hatano T., Di Biase M. A., El Samad O., Saiki S., et al. (2017). Connectome analysis with diffusion MRI in idiopathic Parkinson's disease: evaluation using multi-shell, multi-tissue, constrained spherical deconvolution. NeuroImage. Clin. 17, 518–529. 10.1016/j.nicl.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapinar Senturk Z. (2020). Early diagnosis of Parkinson's disease using machine learning algorithms. Med. Hypoth. 138:109603. 10.1016/j.mehy.2020.109603 [DOI] [PubMed] [Google Scholar]
- Kazeminejad A., Golbabaei S., Soltanian-Zadeh H. (2017). Graph theoretical metrics and machine learning for diagnosis of Parkinson's disease using rs-fMRI, in 2017 Artificial Intelligence and Signal Processing Conference (AISP) (Shiraz: ), 134–139. 10.1109/AISP.2017.8324124 [DOI] [Google Scholar]
- Khan M. M., Mendes A., Chalup S. K. (2018). Evolutionary Wavelet Neural Network ensembles for breast cancer and Parkinson's disease prediction. PLoS ONE 13:e0192192. 10.1371/journal.pone.0192192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatamino P., I C., Özyilmaz L. (2018). A deep learning-CNN based system for medical diagnosis: an application on Parkinson's disease handwriting drawings, in 2018 6th International Conference on Control Engineering & Information Technology (CEIT) (Istanbul: ), 1–6. 10.1109/CEIT.2018.8751879 [DOI] [Google Scholar]
- Khoury N., Attal F., Amirat Y., Oukhellou L., Mohammed S. (2019). Data-driven based approach to aid Parkinson's disease diagnosis. Sensors 19:242. 10.3390/s19020242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu S., Yasaka K., Akai H., Nakata Y., Sugomori Y., Hara S., et al. (2019). Deep learning to differentiate parkinsonian disorders separately using single midsagittal MR imaging: a proof of concept study. Eur. Radiol. 29, 6891–6899. 10.1007/s00330-019-06327-0 [DOI] [PubMed] [Google Scholar]
- Klein Y., Djaldetti R., Keller Y., Bachelet I. (2017). Motor dysfunction and touch-slang in user interface data. Scientific reports 7:4702. 10.1038/s41598-017-04893-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomsae A., Auephanwiriyakul S., Theera-Umpon N. (2018). String grammar unsupervised possibilistic fuzzy C-medians for gait pattern classification in patients with neurodegenerative diseases. Comput. Intell. Neurosci. 2018:1869565. 10.1155/2018/1869565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koçer A., Oktay A. B. (2016). Nintendo Wii assessment of Hoehn and Yahr score with Parkinson's disease tremor. Technol. Health Care 24, 185–191. 10.3233/THC-151124 [DOI] [PubMed] [Google Scholar]
- Kostikis N., Hristu-Varsakelis D., Arnaoutoglou M., Kotsavasiloglou C. (2015). A smartphone-based tool for assessing Parkinsonian hand tremor. IEEE J. Biomed. Health Inf. 19, 1835–1842. 10.1109/JBHI.2015.2471093 [DOI] [PubMed] [Google Scholar]
- Kowal S. L., Dall T. M., Chakrabarti R., Storm M. V., Jain A. (2013). The current and projected economic burden of Parkinson's disease in the United States. Move. Disord. 28, 311–318. 10.1002/mds.25292 [DOI] [PubMed] [Google Scholar]
- Kraipeerapun P., Amornsamankul S. (2015). Using stacked generalization and complementary neural networks to predict Parkinson's disease, in 2015 11th International Conference on Natural Computation (ICNC) (Zhangjiajie: ), 1290–1294. 10.1109/ICNC.2015.7378178 [DOI] [Google Scholar]
- Kugler P., Jaremenko C., Schlachetzki J., Winkler J., Klucken J., Eskofier B. (2013). Automatic recognition of Parkinson's disease using surface electromyography during standardized gait tests. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2013, 5781–5784. 10.1109/EMBC.2013.6610865 [DOI] [PubMed] [Google Scholar]
- Kuhner A., Schubert T., Cenciarini M., Wiesmeier I. K., Coenen V. A., Burgard W., et al. (2017). Correlations between motor symptoms across different motor tasks, quantified via random forest feature classification in Parkinson's disease. Front. Neurol. 8:607. 10.3389/fneur.2017.00607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuresan H., Samiappan D., Masunda S. (2019). Fusion of WPT and MFCC feature extraction in Parkinson's disease diagnosis. Technol. Health Care 27, 363–372. 10.3233/THC-181306 [DOI] [PubMed] [Google Scholar]
- Kwon D.-Y., Kwon Y., Kim J.-W. (2018). Quantitative analysis of finger and forearm movements in patients with off state early stage Parkinson's disease and scans without evidence of dopaminergic deficit (SWEDD). Parkinsonism Related Disord. 57, 33–38. 10.1016/j.parkreldis.2018.07.012 [DOI] [PubMed] [Google Scholar]
- Lacy S. E., Smith S. L., Lones M. A. (2018). Using echo state networks for classification: A case study in Parkinson's disease diagnosis. Artif. Intell. Med. 86, 53–59. 10.1016/j.artmed.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Lee M. J., Kim S. L., Lyoo C. H., Lee M. S. (2014). Kinematic analysis in patients with Parkinson's disease and SWEDD. J. Parkinsons Dis. 4, 421–430. 10.3233/JPD-130233 [DOI] [PubMed] [Google Scholar]
- Lei H., Huang Z., Zhou F., Elazab A., Tan E.-L., Li H., et al. (2019). Parkinson's disease diagnosis via joint learning from multiple modalities and relations. IEEE J. Biomed. Health Inf. 23, 1437–1449. 10.1109/JBHI.2018.2868420 [DOI] [PubMed] [Google Scholar]
- Lewitt P. A., Li J., Lu M., Beach T. G., Adler C. H., Guo L., et al. (2013). 3-hydroxykynurenine and other Parkinson's disease biomarkers discovered by metabolomic analysis. Move. Disord. 28, 1653–1660. 10.1002/mds.25555 [DOI] [PubMed] [Google Scholar]
- Li Q., Chen H., Huang H., Zhao X., Cai Z., Tong C., et al. (2017). An enhanced grey wolf optimization based feature selection wrapped kernel extreme learning machine for medical diagnosis. Comput. Math. Methods Med. 2017:9512741. 10.1155/2017/9512741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lei H., Zhou F., Gardezi J., Lei B. (2019). Longitudinal and Multi-modal Data Learning for Parkinson's Disease Diagnosis via Stacked Sparse Auto-encoder, in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) (Venice: ), 384–387. 10.1109/ISBI.2019.8759385 [DOI] [Google Scholar]
- Liu H., Du G., Zhang L., Lewis M. M., Wang X., Yao T., et al. (2016). Folded concave penalized learning in identifying multimodal MRI marker for Parkinson's disease. J. Neurosci. Methods 268, 1–6. 10.1016/j.jneumeth.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang Q., Adeli E., Zhang L., Zhang H., Shen D. (2016). Feature selection based on iterative canonical correlation analysis for automatic diagnosis of Parkinson's disease. Med. Image Comput. Computer Assist. Interv. 9901, 1–8. 10.1007/978-3-319-46723-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Ouyang J., Chen H.-L., Zhao X.-H. (2014). An efficient diagnosis system for Parkinson's disease using kernel-based extreme learning machine with subtractive clustering features weighting approach. Computat. Math. Methods Med. 2014:985789. 10.1155/2014/985789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Tan T., Zhou H., Gao T. (2016). Support Vector Machine-recursive feature elimination for the diagnosis of Parkinson disease based on speech analysis, in 2016 Seventh International Conference on Intelligent Control and Information Processing (ICICIP) (Siem Reap: ), 34–40. 10.1109/ICICIP.2016.7885912 [DOI] [Google Scholar]
- Maass F., Michalke B., Leha A., Boerger M., Zerr I., Koch J.-C., et al. (2018). Elemental fingerprint as a cerebrospinal fluid biomarker for the diagnosis of Parkinson's disease. J. Neurochemistry 145, 342–351. 10.1111/jnc.14316 [DOI] [PubMed] [Google Scholar]
- Maass F., Michalke B., Willkommen D., Leha A., Schulte C., Tönges L., et al. (2020). Elemental fingerprint: reassessment of a cerebrospinal fluid biomarker for Parkinson's disease. Neurobiol. Dis. 134:104677. 10.1016/j.nbd.2019.104677 [DOI] [PubMed] [Google Scholar]
- Mabrouk R., Chikhaoui B., Bentabet L. (2019). Machine learning based classification using clinical and DaTSCAN SPECT imaging features: a study on Parkinson's disease and SWEDD. IEEE Trans. Rad. Plasma Med. Sci. 3, 170–177. 10.1109/TRPMS.2018.2877754 [DOI] [Google Scholar]
- Mandal I., Sairam N. (2013). Accurate telemonitoring of Parkinson's disease diagnosis using robust inference system. Int. J. Med. Informatics 82, 359–377. 10.1016/j.ijmedinf.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Marar S., Swain D., Hiwarkar V., Motwani N., Awari A. (2018). Predicting the occurrence of Parkinson's disease using various classification models, in 2018 International Conference on Advanced Computation and Telecommunication (ICACAT) (Bhopal: ), 1–5. 10.1109/ICACAT.2018.8933579 [DOI] [Google Scholar]
- Marek K., Jennings D., Lasch S., Siderowf A., Tanner C., Simuni T., et al. (2011). The parkinson progression marker initiative (PPMI). Progress Neurobiol. 95, 629–635. 10.1016/j.pneurobio.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez M., Villagra F., Castellote J. M., Pastor M. A. (2018). Kinematic and kinetic patterns related to free-walking in Parkinson's disease. Sensors 18:4224. 10.3390/s18124224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Murcia F. J., Górriz J. M., Ramírez J., Ortiz A. (2018). Convolutional neural networks for neuroimaging in Parkinson's disease: is preprocessing needed? Int. J. Neural Syst. 28:1850035. 10.1142/S0129065718500351 [DOI] [PubMed] [Google Scholar]
- Memedi M., Sadikov A., Groznik V., Žabkar J., Možina M., Bergquist F., et al. (2015). Automatic spiral analysis for objective assessment of motor symptoms in Parkinson's disease. Sensors 15, 23727–23744. 10.3390/s150923727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittra Y., Rustagi V. (2018). Classification of subjects with Parkinson's disease using gait data analysis, in 2018 International Conference on Automation and Computational Engineering (ICACE) (Greater Noida: ), 84–89. 10.1109/ICACE.2018.8687022 [DOI] [Google Scholar]
- Moharkan Z. A., Garg H., Chodhury T., Kumar P. (2017). A classification based Parkinson detection system, in 2017 International Conference On Smart Technologies For Smart Nation (SmartTechCon) (Bengaluru: ), 1509–1513. 10.1109/SmartTechCon.2017.8358616 [DOI] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Inter. Med. 151, 264–269. 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- Montaña D., Campos-Roca Y., Pérez C. J. (2018). A Diadochokinesis-based expert system considering articulatory features of plosive consonants for early detection of Parkinson's disease. Comp. Methods Programs Biomed. 154, 89–97. 10.1016/j.cmpb.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Morisi R., Manners D. N., Gnecco G., Lanconelli N., Testa C., Evangelisti S., et al. (2018). Multi-class parkinsonian disorders classification with quantitative MR markers and graph-based features using support vector machines. Parkinsonism Relat. Disord. 47, 64–70. 10.1016/j.parkreldis.2017.11.343 [DOI] [PubMed] [Google Scholar]
- Mucha J., Mekyska J., Faundez-Zanuy M., Lopez-De-Ipina K., Zvoncak V., Galaz Z., et al. (2018). Advanced Parkinson's disease dysgraphia analysis based on fractional derivatives of online handwriting, in 2018 10th International Congress on Ultra Modern Telecommunications and Control Systems and Workshops (ICUMT) (Moscow: ), 1–6. 10.1109/ICUMT.2018.8631265 [DOI] [Google Scholar]
- Nicastro N., Wegrzyk J., Preti M. G., Fleury V., Van de Ville D., Garibotto V., et al. (2019). Classification of degenerative parkinsonism subtypes by support-vector-machine analysis and striatal (123)I-FP-CIT indices. J. Neurol. 266, 1771–1781. 10.1007/s00415-019-09330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nõmm S., Bard,õš K., Toomela A., Medijainen K., Taba P. (2018). Detailed analysis of the Luria's alternating seriestests for Parkinson's disease diagnostics, in 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA) (Orlando, FL: ), 1347–1352. 10.1109/ICMLA.2018.00219 [DOI] [Google Scholar]
- Nunes A., Silva G., Duque C., Januário C., Santana I., Ambrósio A. F., et al. (2019). Retinal texture biomarkers may help to discriminate between Alzheimer's, Parkinson's, and healthy controls. PLoS One 14:e0218826. 10.1371/journal.pone.0218826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuvoli S., Spanu A., Fravolini M. L., Bianconi F., Cascianelli S., Madeddu G., et al. (2019). [(123)I]Metaiodobenzylguanidine (MIBG) cardiac scintigraphy and automated classification techniques in Parkinsonian disorders. Mol. Imaging Biol. 22, 703–710. 10.1007/s11307-019-01406-6 [DOI] [PubMed] [Google Scholar]
- Oliveira F. P. M., Castelo-Branco M. (2015). Computer-aided diagnosis of Parkinson's disease based on [(123)I]FP-CIT SPECT binding potential images, using the voxels-as-features approach and support vector machines. J. Neural Eng. 12:026008. 10.1088/1741-2560/12/2/026008 [DOI] [PubMed] [Google Scholar]
- Oliveira F. P. M., Faria D. B., Costa D. C., Castelo-Branco M., Tavares J. M. R. S. (2018). Extraction, selection and comparison of features for an effective automated computer-aided diagnosis of Parkinson's disease based on [(123)I]FP-CIT SPECT images. Eur. J. Nucl. Med. Mol. Imaging 45, 1052–1062. 10.1007/s00259-017-3918-7 [DOI] [PubMed] [Google Scholar]
- Oliveira H. M., Machado A. R. P., Andrade A. O. (2018a). On the use of t-distributed stochastic neighbor embedding for data visualization and classification of individuals with Parkinson's disease. Comput. Math. Methods Med. 2018:8019232. 10.1155/2018/8019232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opara J., Brola W., Leonardi M., Błaszczyk B. (2012). Quality of life in Parkinsons disease. J. Med. Life 5:375. [PMC free article] [PubMed] [Google Scholar]
- Oung Q. W., Hariharan M., Lee H. L., Basah S. N., Sarillee M., Lee C. H. (2015). Wearable multimodal sensors for evaluation of patients with Parkinson disease, in 2015 IEEE International Conference on Control System, Computing and Engineering (ICCSCE) (Penang: ), 269–274. 10.1109/ICCSCE.2015.7482196 [DOI] [Google Scholar]
- Ozcift A. (2012). SVM feature selection based rotation forest ensemble classifiers to improve computer-aided diagnosis of Parkinson disease. J. Med. Syst. 36, 2141–2147. 10.1007/s10916-011-9678-1 [DOI] [PubMed] [Google Scholar]
- Ozcift A., Gulten A. (2011). Classifier ensemble construction with rotation forest to improve medical diagnosis performance of machine learning algorithms. Comp. Methods Progr. Biomed. 104, 443–451. 10.1016/j.cmpb.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Pahuja G., Nagabhushan T. N. (2016). A novel GA-ELM approach for Parkinson's disease detection using brain structural T1-weighted MRI data, in 2016 Second International Conference on Cognitive Computing and Information Processing (CCIP) (Mysuru: ), 1–6. 10.1109/CCIP.2016.7802848 [DOI] [Google Scholar]
- Palumbo B., Fravolini M. L., Buresta T., Pompili F., Forini N., Nigro P., et al. (2014). Diagnostic accuracy of Parkinson disease by support vector machine (SVM) analysis of 123I-FP-CIT brain SPECT data: implications of putaminal findings and age. Medicine 93:e228. 10.1097/MD.0000000000000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos A., Kyritsis K., Klingelhoefer L., Bostanjopoulou S., Chaudhuri K. R., Delopoulos A. (2019). Detecting Parkinsonian tremor from IMU data collected in-the-wild using deep multiple-instance learning. IEEE J. Biomed. Health Inform. 24, 2559–2569. 10.1109/JBHI.2019.2961748 [DOI] [PubMed] [Google Scholar]
- Papavasileiou I., Zhang W., Wang X., Bi J., Zhang L., Han S. (2017). Classification of neurological gait disorders using multi-task feature learning, in 2017 IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE) (Philadelphia, PA: ), 195–204. 10.1109/CHASE.2017.78 [DOI] [Google Scholar]
- Peker M. (2016). A decision support system to improve medical diagnosis using a combination of k-medoids clustering based attribute weighting and SVM. J. Med. Syst. 40:116. 10.1007/s10916-016-0477-6 [DOI] [PubMed] [Google Scholar]
- Peng B., Wang S., Zhou Z., Liu Y., Tong B., Zhang T., et al. (2017). A multilevel-ROI-features-based machine learning method for detection of morphometric biomarkers in Parkinson's disease. Neurosci. Lett. 651, 88–94. 10.1016/j.neulet.2017.04.034 [DOI] [PubMed] [Google Scholar]
- Peng B., Zhou Z., Geng C., Tong B., Zhou Z., Zhang T., et al. (2016). Computer aided analysis of cognitive disorder in patients with Parkinsonism using machine learning method with multilevel ROI-based features, in 2016 9th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI) (Datong: ), 1792–1796. 10.1109/CISP-BMEI.2016.7853008 [DOI] [Google Scholar]
- Pereira C. R., Pereira D. R., da Silva F. A., Hook C., Weber S. A., Pereira L. A., et al. (2015). A step towards the automated diagnosis of parkinson's disease: Analyzing handwriting movements, in 2015 IEEE 28th International Symposium on Computer-Based Medical Systems (Sao Carlos: IEEE; ), 171–176. 10.1109/CBMS.2015.34 [DOI] [Google Scholar]
- Pereira C. R., Pereira D. R., Rosa G. H., Albuquerque V. H. C., Weber S. A. T., Hook C., et al. (2018). Handwritten dynamics assessment through convolutional neural networks: An application to Parkinson's disease identification. Artif. Intell. Med. 87, 67–77. 10.1016/j.artmed.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Pereira C. R., Pereira D. R., Silva F. A., Masieiro J. P., Weber S. A. T., Hook C., et al. (2016a). A new computer vision-based approach to aid the diagnosis of Parkinson's disease. Comp. Methods Progr. Biomed. 136, 79–88. 10.1016/j.cmpb.2016.08.005 [DOI] [PubMed] [Google Scholar]
- Pereira C. R., Pereira D. R., Weber S. A., Hook C., de Albuquerque V. H. C., Papa J. P. (2019). A survey on computer-assisted Parkinson's disease diagnosis. Artif. Intell. Med. 95, 48–63. 10.1016/j.artmed.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Pereira C. R., Weber S. A. T., Hook C., Rosa G. H., Papa J. P. (2016b). Deep learning-aided Parkinson's disease diagnosis from handwritten dynamics, in 2016 29th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI) (São Paulo: ), 340–346. 10.1109/SIBGRAPI.2016.054 [DOI] [Google Scholar]
- Pham H. N., Do T. T. T., Chan K. Y. J., Sen G., Han A. Y. K., Lim P., et al. (2019). Multimodal detection of Parkinson disease based on vocal and improved spiral test, in 2019 International Conference on System Science and Engineering (ICSSE) (Dong Hoi: ), 279–284. 10.1109/ICSSE.2019.8823309 [DOI] [Google Scholar]
- Pham T. D. (2018). Pattern analysis of computer keystroke time series in healthy control and early-stage Parkinson's disease subjects using fuzzy recurrence and scalable recurrence network features. J. Neurosci. Methods 307, 194–202. 10.1016/j.jneumeth.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Pham T. D., Yan H. (2018). Tensor decomposition of gait dynamics in Parkinson's disease. IEEE Trans. Bio-Med. Eng. 65, 1820–1827. 10.1109/TBME.2017.2779884 [DOI] [PubMed] [Google Scholar]
- Postuma R. B., Berg D., Stern M., Poewe W., Olanow C. W., Oertel W., et al. (2015). MDS clinical diagnostic criteria for Parkinson's disease. Move. Disord. 30, 1591–1601. 10.1002/mds.26424 [DOI] [PubMed] [Google Scholar]
- Prashanth R., Dutta Roy S. (2018). Early detection of Parkinson's disease through patient questionnaire and predictive modelling. In. J. Med. Inform. 119, 75–87. 10.1016/j.ijmedinf.2018.09.008 [DOI] [PubMed] [Google Scholar]
- Prashanth R., Dutta Roy S., Mandal P. K., Ghosh S. (2016). High-accuracy detection of early Parkinson's disease through multimodal features and machine learning. Int. J. Med. Inform. 90, 13–21. 10.1016/j.ijmedinf.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Prashanth R., Roy S. D., Mandal P. K., Ghosh S. (2014). Parkinson's disease detection using olfactory loss and REM sleep disorder features, in Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2014, 5764–5767. 10.1109/EMBC.2014.6944937 [DOI] [PubMed] [Google Scholar]
- Prashanth R., Roy S. D., Mandal P. K., Ghosh S. (2017). High-accuracy classification of Parkinson's disease through shape analysis and surface fitting in 123I-Ioflupane SPECT imaging. IEEE J. Biomed. Health Inform. 21, 794–802. 10.1109/JBHI.2016.2547901 [DOI] [PubMed] [Google Scholar]
- Prince J., Andreotti F., Vos M. D. (2019). Multi-source ensemble learning for the remote prediction of Parkinson's disease in the presence of source-wise missing data. IEEE Trans. Biomed. Eng. 66, 1402–1411. 10.1109/TBME.2018.2873252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince J., de Vos M. (2018). A deep learning framework for the remote detection of Parkinson's disease using smart-phone sensor data, in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (Honolulu, HI: ), 3144–3147. 10.1109/EMBC.2018.8512972 [DOI] [PubMed] [Google Scholar]
- Ramdhani R. A., Khojandi A., Shylo O., Kopell B. H. (2018). Optimizing clinical assessments in Parkinson's disease through the use of wearable sensors and data driven modeling. Front. Comput. Neurosci. 12:72. 10.3389/fncom.2018.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J. F., Montealegre J. S., Castano Y. J., Urcuqui C., Navarro A. (2019). LSTM and convolution networks exploration for Parkinson's diagnosis, in 2019 IEEE Colombian Conference on Communications and Computing (COLCOM) (Barranquilla: ), 1–4. 10.1109/ColComCon.2019.8809160 [DOI] [Google Scholar]
- Ribeiro L. C. F., Afonso L. C. S., Papa J. P. (2019). Bag of samplings for computer-assisted Parkinson's disease diagnosis based on recurrent neural networks. Comp. Biol. Med. 115:103477. 10.1016/j.compbiomed.2019.103477 [DOI] [PubMed] [Google Scholar]
- Ricci M., Lazzaro G. D., Pisani A., Mercuri N. B., Giannini F., Saggio G. (2020). Assessment of motor impairments in early untreated parkinson's disease patients: the wearable electronics impact. IEEE J. Biomed. Health Inform. 24, 120–130. 10.1109/JBHI.2019.2903627 [DOI] [PubMed] [Google Scholar]
- Rios-Urrego C. D., Vásquez-Correa J. C., Vargas-Bonilla J. F., Nöth E., Lopera F., Orozco-Arroyave J. R. (2019). Analysis and evaluation of handwriting in patients with Parkinson's disease using kinematic, geometrical, and non-linear features. Comput. Methods Progr. Biomed. 173, 43–52. 10.1016/j.cmpb.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Rovini E., Maremmani C., Moschetti A., Esposito D., Cavallo F. (2018). Comparative motor pre-clinical assessment in parkinson's disease using supervised machine learning approaches. Annals Biomed. Eng. 46, 2057–2068. 10.1007/s10439-018-2104-9 [DOI] [PubMed] [Google Scholar]
- Rovini E., Moschetti A., Fiorini L., Esposito D., Maremmani C., Cavallo F. (2019). Wearable sensors for prodromal motor assessment of parkinson's disease using supervised learning, in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 4318–4321. 10.1109/EMBC.2019.8856804 [DOI] [PubMed] [Google Scholar]
- Rubbert C., Mathys C., Jockwitz C., Hartmann C. J., Eickhoff S. B., Hoffstaedter F., et al. (2019). Machine-learning identifies Parkinson's disease patients based on resting-state between-network functional connectivity. Br. J. Radiol. 92:20180886. 10.1259/bjr.20180886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakar B. E., Isenkul M. E., Sakar C. O., Sertbas A., Gurgen F., Delil S., et al. (2013). Collection and analysis of a Parkinson speech dataset with multiple types of sound recordings. IEEE J. Biomed. Health Inform. 17, 828–834. 10.1109/JBHI.2013.2245674 [DOI] [PubMed] [Google Scholar]
- Salvatore C., Cerasa A., Castiglioni I., Gallivanone F., Augimeri A., Lopez M., et al. (2014). Machine learning on brain MRI data for differential diagnosis of Parkinson's disease and progressive supranuclear palsy. J. Neurosci. Methods 222, 230–237. 10.1016/j.jneumeth.2013.11.016 [DOI] [PubMed] [Google Scholar]
- Sayaydeha O. N. A., Mohammad M. F. (2019). Diagnosis of the Parkinson disease using enhanced fuzzy min-max neural network and OneR attribute evaluation method, in 2019 International Conference on Advanced Science and Engineering (ICOASE) (Zakho-Duhok: ), 64–69. 10.1109/ICOASE.2019.8723870 [DOI] [Google Scholar]
- Scherer R. W., Saldanha I. J. (2019). How should systematic reviewers handle conference abstracts? A view from the trenches. Syst. Rev. 8:264. 10.1186/s13643-019-1188-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia F., Górriz J. M., Ramírez J., Levin J., Schuberth M., Brendel M., et al. (2015). Analysis of 18F-DMFP PET data using multikernel classification in order to assist the diagnosis of Parkinsonism, in 2015 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) (San Diego, CA: ), 1–4. 10.1109/NSSMIC.2015.7582227 [DOI] [Google Scholar]
- Segovia F., Górriz J. M., Ramírez J., Martínez-Murcia F. J., Castillo-Barnes D. (2019). Assisted diagnosis of Parkinsonism based on the striatal morphology. Int. J. Neural Syst. 29:1950011. 10.1142/S0129065719500114 [DOI] [PubMed] [Google Scholar]
- Shahsavari M. K., Rashidi H., Bakhsh H. R. (2016). Efficient classification of Parkinson's disease using extreme learning machine and hybrid particle swarm optimization, in 2016 4th International Conference on Control, Instrumentation, and Automation (ICCIA) (Qazvin: ), 148–154. 10.1109/ICCIAutom.2016.7483152 [DOI] [Google Scholar]
- Shamir R., Klein C., Amar D., Vollstedt E.-J., Bonin M., Usenovic M., et al. (2017). Analysis of blood-based gene expression in idiopathic Parkinson disease. Neurology 89, 1676–1683. 10.1212/WNL.0000000000004516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheibani R., Nikookar E., Alavi S. E. (2019). An ensemble method for diagnosis of Parkinson's disease based on voice measurements. J. Med. Signals Sens. 9, 221–226. 10.4103/jmss.JMSS_57_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T., Jiang J., Lin W., Ge J., Wu P., Zhou Y., et al. (2019). Use of overlapping group LASSO sparse deep belief network to discriminate Parkinson's disease and normal control. Front. Neurosci. 13:396. 10.3389/fnins.2019.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Yan M., Dong Y., Zheng X., Zhang Q., An H. (2018). Multiple kernel learning based classification of Parkinson's disease with multi-modal transcranial sonography, in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (Honolulu, HI: ), 61–64. 10.1109/EMBC.2018.8512194 [DOI] [PubMed] [Google Scholar]
- Shinde S., Prasad S., Saboo Y., Kaushick R., Saini J., Pal P. K., et al. (2019). Predictive markers for Parkinson's disease using deep neural nets on neuromelanin sensitive MRI. NeuroImage. Clin. 22:101748. 10.1016/j.nicl.2019.101748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Samavedham L. (2015). Unsupervised learning based feature extraction for differential diagnosis of neurodegenerative diseases: a case study on early-stage diagnosis of Parkinson disease. J. Neurosci. Methods 256, 30–40. 10.1016/j.jneumeth.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Singh G., Samavedham L., Lim E. C.-H., Alzheimer's Disease Neuroimaging I., Parkinson Progression Marker I. (2018). Determination of imaging biomarkers to decipher disease trajectories and differential diagnosis of neurodegenerative diseases (DIsease TreND). J. Neurosci. Methods 305, 105–116. 10.1016/j.jneumeth.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Stoessel D., Schulte C., Teixeira Dos Santos M. C., Scheller D., Rebollo-Mesa I., Deuschle C., et al. (2018). Promising metabolite profiles in the plasma and CSF of early clinical Parkinson's disease. Front. Aging Neurosci. 10:51. 10.3389/fnagi.2018.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surangsrirat D., Thanawattano C., Pongthornseri R., Dumnin S., Anan C., Bhidayasiri R. (2016). Support vector machine classification of Parkinson's disease and essential tremor subjects based on temporal fluctuation. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 6389–6392. 10.1109/EMBC.2016.7592190 [DOI] [PubMed] [Google Scholar]
- Sztahó D., Tulics M. G., Vicsi K., Valálik I. (2017). Automatic estimation of severity of Parkinson's disease based on speech rhythm related features, in 2017 8th IEEE International Conference on Cognitive Infocommunications (CogInfoCom) (Debrecen: ), 000011–000016. 10.1109/CogInfoCom.2017.8268208 [DOI] [Google Scholar]
- Sztahó D., Valálik I., Vicsi K. (2019). Parkinson's disease severity estimation on Hungarian speech using various speech tasks, in 2019 International Conference on Speech Technology and Human-Computer Dialogue (SpeD) (Timisoara: ), 1–6. 10.1109/SPED.2019.8906277 [DOI] [Google Scholar]
- Tagare H. D., DeLorenzo C., Chelikani S., Saperstein L., Fulbright R. K. (2017). Voxel-based logistic analysis of PPMI control and Parkinson's disease DaTscans. NeuroImage 152, 299–311. 10.1016/j.neuroimage.2017.02.067 [DOI] [PubMed] [Google Scholar]
- Tahavori F., Stack E., Agarwal V., Burnett M., Ashburn A., Hoseinitabatabaei S. A., et al. (2017). Physical activity recognition of elderly people and people with parkinson's (PwP) during standard mobility tests using wearable sensors, in 2017 International Smart Cities Conference (ISC2) (Wuxi: ), 1–4. 10.1109/ISC2.2017.8090858 [DOI] [Google Scholar]
- Taleb C., Khachab M., Mokbel C., Likforman-Sulem L. (2019). Visual representation of online handwriting time series for deep learning Parkinson's disease detection, in 2019 International Conference on Document Analysis and Recognition Workshops (ICDARW) (Sydney, NSW: ), 25–30. 10.1109/ICDARW.2019.50111 [DOI] [Google Scholar]
- Tang Y., Meng L., Wan C. M., Liu Z. H., Liao W. H., Yan X. X., et al. (2017). Identifying the presence of Parkinson's disease using low-frequency fluctuations in BOLD signals. Neurosci. Lett. 645, 1–6. 10.1016/j.neulet.2017.02.056 [DOI] [PubMed] [Google Scholar]
- Taylor J. C., Fenner J. W. (2017). Comparison of machine learning and semi-quantification algorithms for (I123)FP-CIT classification: the beginning of the end for semi-quantification? EJNMMI Phys. 4:29. 10.1186/s40658-017-0196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien I., Glaser S. D., Aminoff M. J. (2010). Characterization of gait abnormalities in Parkinson's disease using a wireless inertial sensor system, in Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2010, 3353–3356. 10.1109/IEMBS.2010.5627904 [DOI] [PubMed] [Google Scholar]
- Tracy J. M., Özkanca Y., Atkins D. C., Hosseini Ghomi R. (2019). Investigating voice as a biomarker: deep phenotyping methods for early detection of Parkinson's disease. J. Biomed. Inform. 104:103362. 10.1016/j.jbi.2019.103362 [DOI] [PubMed] [Google Scholar]
- Tremblay C., Martel P. D., Frasnelli J. (2017). Trigeminal system in Parkinson's disease: a potential avenue to detect Parkinson-specific olfactory dysfunction. Parkinsonism Relat. Dis. 44, 85–90. 10.1016/j.parkreldis.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Trezzi J.-P., Galozzi S., Jaeger C., Barkovits K., Brockmann K., Maetzler W., et al. (2017). Distinct metabolomic signature in cerebrospinal fluid in early parkinson's disease. Mov. Disord. 32, 1401–1408. 10.1002/mds.27132 [DOI] [PubMed] [Google Scholar]
- Tsanas A., Little M. A., McSharry P. E., Spielman J., Ramig L. O. (2012). Novel speech signal processing algorithms for high-accuracy classification of Parkinson's disease. IEEE Trans. Bio. Med. Eng. 59, 1264–1271. 10.1109/TBME.2012.2183367 [DOI] [PubMed] [Google Scholar]
- Tseng P.-H., Cameron I. G. M., Pari G., Reynolds J. N., Munoz D. P., Itti L. (2013). High-throughput classification of clinical populations from natural viewing eye movements. J. Neurol. 260, 275–284. 10.1007/s00415-012-6631-2 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Asano S., Kato Y., Murai K., Miyazaki M. (2019). Differential diagnosis of multiple system atrophy with predominant parkinsonism and Parkinson's disease using neural networks. J. Neurol. Sci. 401, 19–26. 10.1016/j.jns.2019.04.014 [DOI] [PubMed] [Google Scholar]
- Tysnes O.-B., Storstein A. (2017). Epidemiology of Parkinson's disease. J. Neural Trans. 124, 901–905. 10.1007/s00702-017-1686-y [DOI] [PubMed] [Google Scholar]
- Urcuqui C., Castaño Y., Delgado J., Navarro A., Diaz J., Muñoz B., et al. (2018). Exploring Machine Learning to Analyze Parkinson's Disease Patients, in 2018 14th International Conference on Semantics, Knowledge and Grids (SKG) (Guangzhou: ), 160–166. 10.1109/SKG.2018.00029 [DOI] [Google Scholar]
- Vabalas A., Gowen E., Poliakoff E., Casson A. J. (2019). Machine learning algorithm validation with a limited sample size. PLoS ONE 14:e0224365. 10.1371/journal.pone.0224365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiciukynas E., Verikas A., Gelzinis A., Bacauskiene M. (2017). Detecting Parkinson's disease from sustained phonation and speech signals. PLoS ONE 12:e0185613. 10.1371/journal.pone.0185613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas M. I., Ghilardi M. F., Kelly S. P., Blangero A. (2018). Machine learning for EEG-based biomarkers in Parkinson's disease, in 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (Madrid: ), 2661–2665. 10.1109/BIBM.2018.8621498 [DOI] [Google Scholar]
- Váradi C., Nehéz K., Hornyák O., Viskolcz B., Bones J. (2019). Serum N-glycosylation in Parkinson's disease: a novel approach for potential alterations. Molecules 24:2220. 10.3390/molecules24122220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez-Correa J. C., Arias-Vergara T., Orozco-Arroyave J. R., Eskofier B., Klucken J., Nöth E. (2019). Multimodal assessment of parkinson's disease: a deep learning approach. IEEE J. Biomed. Health Inform. 23, 1618–1630. 10.1109/JBHI.2018.2866873 [DOI] [PubMed] [Google Scholar]
- Vlachostergiou A., Tagaris A., Stafylopatis A., Kollias S. (2018). Multi-task learning for predicting Parkinson's disease based on medical imaging information, in 2018 25th IEEE International Conference on Image Processing (ICIP) (Athens: ), 2052–2056. 10.1109/ICIP.2018.8451398 [DOI] [Google Scholar]
- Wahid F., Begg R. K., Hass C. J., Halgamuge S., Ackland D. C. (2015). Classification of Parkinson's disease gait using spatial-temporal gait features. IEEE J. Biomed. Health Inform. 19, 1794–1802. 10.1109/JBHI.2015.2450232 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhu X., Adeli E., Zhu Y., Nie F., Munsell B., et al. (2017). Multi-modal classification of neurodegenerative disease by progressive graph-based transductive learning. Med Image Anal. 39, 218–230. 10.1016/j.media.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Thibeau-Sutre E., Diaz-Melo M., Samper-González J., Routier A., Bottani S., et al. (2020). Convolutional neural networks for classification of Alzheimer's disease: overview and reproducible evaluation. Med. Image Anal. 63:101694. 10.1016/j.media.2020.101694 [DOI] [PubMed] [Google Scholar]
- Wenzel M., Milletari F., Krüger J., Lange C., Schenk M., Apostolova I., et al. (2019). Automatic classification of dopamine transporter SPECT: deep convolutional neural networks can be trained to be robust with respect to variable image characteristics. Eur. J. Nuclear Med. Mol. Imaging 46, 2800–2811. 10.1007/s00259-019-04502-5 [DOI] [PubMed] [Google Scholar]
- Wodzinski M., Skalski A., Hemmerling D., Orozco-Arroyave J. R., Nöth E. (2019). Deep learning approach to Parkinson's disease detection using voice recordings and convolutional neural network dedicated to image classification, in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (Berlin: ), 717–720. 10.1109/EMBC.2019.8856972 [DOI] [PubMed] [Google Scholar]
- Wu Y., Chen P., Yao Y., Ye X., Xiao Y., Liao L., et al. (2017). Dysphonic voice pattern analysis of patients in Parkinson's disease using minimum interclass probability risk feature selection and bagging ensemble learning methods. Comput. Math. Methods Med. 2017:4201984. 10.1155/2017/4201984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Jiang J.-H., Chen L., Lu J.-Y., Ge J.-J., Liu F.-T., et al. (2019). Use of radiomic features and support vector machine to distinguish Parkinson's disease cases from normal controls. Ann. Transl. Med. 7:773. 10.21037/atm.2019.11.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Yao Z., Ye Q., Cheng N. (2020). A dual-modal attention-enhanced deep learning network for quantification of Parkinson's disease characteristics. IEEE Trans. Neural Syst. Rehab. Eng. 28, 42–51. 10.1109/TNSRE.2019.2946194 [DOI] [PubMed] [Google Scholar]
- Yadav G., Kumar Y., Sahoo G. (2011). Predication of Parkinson's disease using data mining methods: a comparative analysis of tree, statistical, and support vector machine classifiers. Ind. J. Med. Sci. 65, 231–242. 10.4103/0019-5359.107023 [DOI] [PubMed] [Google Scholar]
- Yagis E., Herrera A. G. S. D., Citi L. (2019). Generalization Performance of Deep Learning Models in Neurodegenerative Disease Classification, in 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (San Diego, CA: ), 1692–1698. 10.1109/BIBM47256.2019.8983088 [DOI] [Google Scholar]
- Yaman O., Ertam F., Tuncer T. (2020). Automated Parkinson's disease recognition based on statistical pooling method using acoustic features. Med. Hypoth. 135:109483. 10.1016/j.mehy.2019.109483 [DOI] [PubMed] [Google Scholar]
- Yang J.-X., Chen L. (2017). Economic burden analysis of Parkinson's disease patients in China. Parkinson's Dis. 2017:8762939. 10.1155/2017/8762939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Zheng H., Wang H., McClean S. (2009). Feature selection and construction for the discrimination of neurodegenerative diseases based on gait analysis, in 2009 3rd International Conference on Pervasive Computing Technologies for Healthcare (London: ), 1–7. 10.4108/ICST.PERVASIVEHEALTH2009.6053 [DOI] [Google Scholar]
- Yang S., Zheng F., Luo X., Cai S., Wu Y., Liu K., et al. (2014). Effective dysphonia detection using feature dimension reduction and kernel density estimation for patients with Parkinson's disease. PLoS ONE 9:e88825. 10.1371/journal.pone.0088825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Xia Y., Yao Z. (2018). Classification of gait patterns in patients with neurodegenerative disease using adaptive neuro-fuzzy inference system. Computat. Math. Methods Med. 2018:9831252. 10.1155/2018/9831252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.-L., Xie L., Shen H., Luo Z., Fang P., Hou Y., et al. (2017). Differentiating patients with Parkinson's disease from normal controls using gray matter in the cerebellum. Cerebellum 16, 151–157. 10.1007/s12311-016-0781-1 [DOI] [PubMed] [Google Scholar]
- Zesiewicz T. A., Sullivan K. L., Hauser R. A. (2006). Nonmotor symptoms of Parkinson's disease. Expert Rev. Neurother. 6, 1811–1822. 10.1586/14737175.6.12.1811 [DOI] [PubMed] [Google Scholar]
- Zhang X., He L., Chen K., Luo Y., Zhou J., Wang F. (2018). Multi-view graph convolutional network and its applications on neuroimage analysis for Parkinson's disease. Annu. Symp. Proc. 2018, 1147–1156. [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wu P., Wang J., Li H., Navab N., Yakushev I., et al. (2019). A 3D deep residual convolutional neural network for differential diagnosis of Parkinsonian syndromes on 18F-FDG PET images, in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 3531–3534. 10.1109/EMBC.2019.8856747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.