Abstract
It is crucial to replicate or mimic the human digestive system conditions closely in model systems to have the food digestion-related data as accurate as possible. Thus, the data obtained could contribute to studies like those on the relationship between health and nutrition. This review aims to express the human digestion system’s role in food digestion and compare the capability of the models used in simulations, especially the dynamic in vitro models. Activities of the human digestive system governing food digestion and the food matrix’s disintegration mechanism in the digestive system were discussed. Dynamic in vitro models and their relevance to the human digestive system were described. Advancements in the last 20 years, as well as limitations of those artificial systems, with prospects, were discussed. Extensive use and improvement on these models will extend our knowledge of the food matrix and digestive system’s complex interaction. Thus, it will be possible to design next-generation foods with improved health benefits.
Keywords: In vitro, food digestion; Dynamic model; Food matrix
Graphical abstract
Higlights
-
•
Digestion is a combination of versatile and multiple scales physicochemical processes.
-
•
Food composition, structure, and processing affect food digestion.
-
•
There are many dynamic in vitro models used in food digestion studies.
-
•
Results from the models should be interpreted with care.
1. Introduction
Understanding the outcome of the ingested food components in the human digestive system is an area of interest for researchers because of its relation to nutrition and health. Foods contain components that can have either beneficial or adverse effects on human health. Consumed foods are combinations of diverse phases and structures depending on their sources, formulations, and processes used for their production. The digestive system must physically and chemically break down ingested food to release its components, which will be further metabolized to be used by the body.
The foods’ composition and structure have a significant role in their nutritional and functional performances during digestion (Bornhorst & Singh, 2012, 2013; Bornhorst et al., 2014; Bornhorst, 2017; Mackie, 2017; Dupont et al., 2018). The food matrix affects the release of nutrients and their journey to the body’s target sites during digestion. Therefore, it is essential to understand the underlying mechanisms affecting the release of the food components during digestion for health benefit.
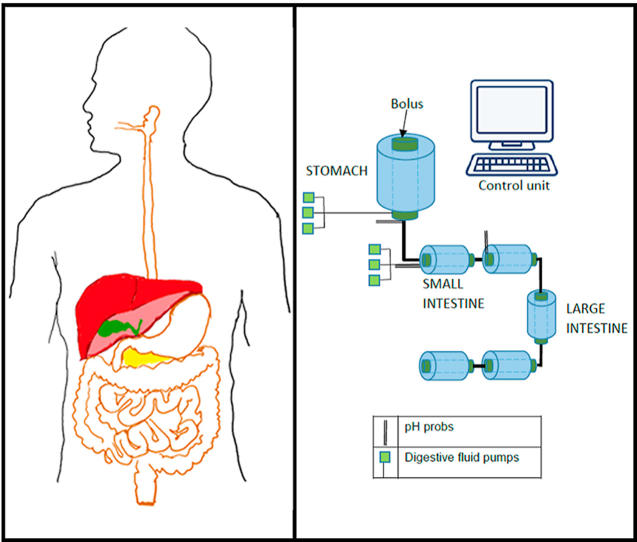
Digestion of food in the human digestive system is a complex combination of versatile and multiple-scale physicochemical processes that steer the food intake, disintegration to suitable forms, absorption of the basic units, transportation to related organs, and purging the remaining waste. The human digestive system consists of the digestive tract and the accessory organs controlled by the neural network and the hormones (Saladin, 2017). The digestive tract can be described as an open-ended tube with a total length of about 8–9 m, extending from mouth to anus, consisting of the pharynx, esophagus, stomach, and small and large intestines (Fig. 1). Accessory organs are the teeth, tongue, salivary glands, liver, gall bladder, and pancreas (Bender et al., 2005; Saladin, 2017). Each part of the digestive tract has a specific function, where altogether, they perform the extraction of the digested products and the disposal of wastes.
Fig. 1.
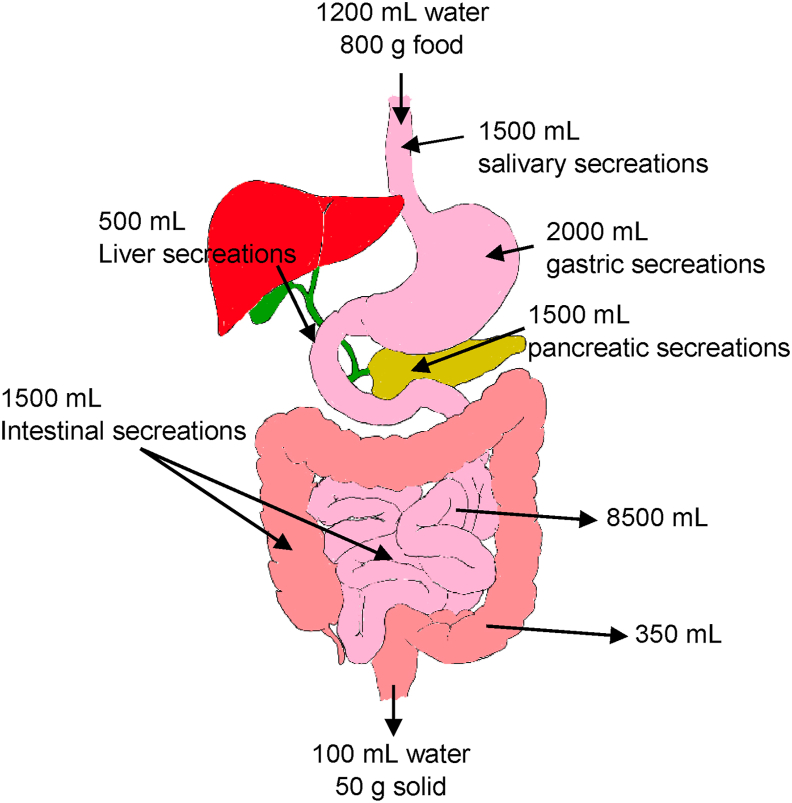
Digestive secretions and absorption of water (numeric values are from Smith and Morton (2010)).
Increased interest in modifying the matrix and structural characteristics of foods to optimize their digestion and absorption behavior for health benefit requires implementing many food digestion studies in the digestive tract. However, studying human digestion’s intricate process is complicated, costly, varies from person to person, and constrained by ethical considerations (Directive 2001/20/EU, 2001). The usage of animal models as an alternative should also be avoided as much as possible (Directive, 2010/63/EU, 2010). Therefore, these limitations and considerations have led researchers to design and use in vitro models to simulate the human digestive system for research.
The interaction between the food and human digestive systems controlling the disintegration of foods and releasing nutrients is quite complicated. This review describes the physiological processes of the human digestive system steering food digestion. The disintegration mechanism of the food matrix in the digestive system was discussed. Models used to simulate the digestion, and their relevance to the human digestive system was analyzed.
2. Effect of food matrix on digestion
The disintegration mechanism differs among foods and is related to the original food structure and how it changes during digestion. Although most of the breakdown process for the solid foods happens in the mouth, the rest of the disintegration occurs while transiting the digestive system, especially in the stomach (Bornhorst and Singh, 2013; Bornhorst et al., 2013; Drechsler and Ferrua, 2016). Forces that food particles experience in the stomach are less than those applied by the teeth in the mouth. The maximum force that the human jaw can apply is about 400 N, while the force that the walls of the stomach (antrum) can apply is about 0.2–2 N depending on the subjects (Ferrua et al., 2011; Lentle and Janssen, 2010, Lentle and Janssen, 2011; Lentle, 2018a).
Fragmentation, abrasion, and dissolution behaviors of the food in the digestive system determine their effect on human health. Fragmentation (cleavage into smaller pieces) and abrasion (erosion of the surface by shear stress) are the two primary disintegration mechanisms of the food (Brandstaeter et al., 2019). When the forces received by the food due to contractions and peristaltic movements in the stomach are higher than the critical value for the internal forces holding the food matrix together, fragmentation will be the dominant mechanism, and the food will be broken down into relatively larger pieces (Ferrua et al., 2011). When the applied stresses are smaller, surface abrasion will be dominant, and the smaller fragments leave the surface (Ferrua et al., 2011; Drechsler and Ferrua, 2016). The acidic environment and the digestive system’s enzymes also contribute to the leaching of solids to the digestion medium.
The kinetic of food disintegration is affected by the physiological conditions in the digestive system like mechanical forces, pH, temperature, and enzymes, as well as by the meal properties such as composition, amount, texture, structure, and viscosity (Kong and Singh, 2008a, Kong and Singh, 2008b; 2009; 2010; Ferrua and Singh, 2010, Ferrua and Singh, 2011; Ferrua et al., 2011; Bornhorst et al., 2015; Drechsler and Ferrua, 2016; Mulet-Cabero et al., 2019). Some foods are more susceptible to fragmentation than other foods under similar mechanical forces (Kong and Singh, 2008a; Ferrua et al., 2011). Water absorption, acid hydrolysis, and enzymatic reactions may cause softening of the ingested foods. Cohesive forces holding the food matrix together may attenuate, leading to a shift in the disintegration mode from erosion to fragmentation (Kong and Singh, 2008a; Ferrua et al., 2011). For example, cereal products like bread and snacks become very soft after mixing and absorption of saliva in the mouth, and then they fragment easily. On the other hand, for foods with a rigid and hard texture like carrots and nuts, erosion is the dominant disintegration mechanism (Kong and Singh, 2008a; Ferrua et al., 2011). Moreover, diffusion processes inside the chyme are affected by physiological conditions such as pH, temperature, viscosity, and degree of mixing as well as by the structure and content of the food. One of the studies attempting to explain and quantify the disintegration mechanisms, Bornhorst et al. (2016), suggested a classification system for the food breakdown similar to the one used by the pharmaceutical industry as a tool to be used for product development. Therefore, all modes of transport phenomena, mass, heat, and momentum, should be considered together in analyzing and simulating the digestive system.
3. Food digestion in the human digestive system
The digestive tract that begins at the mouth continues as the throat (pharynx), the esophagus, the stomach, the small intestine (duodenum, jejunum, and ileum), the large intestine (cecum, ascending colon, transverse colon, descending colon, and sigmoid colon) and rectum, ending at the anus where the wastes are excreted (Fig. 1). Digestive juices are secreted by the salivary glands, gastric glands, pancreas, and liver with its adjuncts (the gallbladder and bile ducts). Secretions from the digestive organs alter the physical properties of the digested meal by dilution (Fig. 1). On average, humans can produce 1.5 L saliva, 2 L stomach secretions, and 0.5 L bile solution.
3.1. Mouth
Food enters the digestion system through the mouth and is broken down into small pieces while mixed with saliva to speed its progression through the digestive system. Textural and rheological properties of foods as well as age, gender, and eating ability of individuals affect the oral processing behavior (Chen, 2009; Ketel et al., 2019).
The mechanical digestion of foods starts by chewing. The teeth in the human mouth are responsible for different tasks like cutting, tearing, grinding, and shearing (Rogers, 2011; Mosca and Chen, 2016). Biting force can range approximately from 100 to 400 N depending on the individual (Mosca and Chen, 2016; Marze, 2017a, Marze, 2017b). Solid foods are more difficult to manipulate. Therefore, the amount of food taken in by the mouth decreases from liquid foods to solids (Chen, 2009; Bornhorst et al., 2016; Aguayo-mendoza et al., 2019; Ketel et al., 2019). Moreover, the bolus’ mean particle size depends on the physical properties of the food, smaller for hard and larger for softer foods (Chen, 2009).
Bolus formation in the mouth includes deformation, deterioration, and disintegration processes, which are accomplished by the coordinated effort of the teeth, tongue, and saliva secretion mechanism (Rogers, 2011; Mosca and Chen, 2016). Saliva, which is about 99% water, contains sodium, potassium, calcium, bicarbonate, mucin, and enzymes (amylase, lingual lipase). The saliva secreted by the salivary glands moistens and starts to dissolve the food with the amylase enzyme action that splits starch into disaccharide molecules of maltose (Table 1). The bolus is propelled from mouth to esophagus by the pharynx during the swallowing process. The pressure of 10 kPa can be generated on the tongue’s surface during swallowing (Chen, 2009).
Table 1.
Summary of the physical and chemical processes occurring in the human digestive system.
| Section | Physical Process | Chemical Process | Conditions (adult) | References |
|---|---|---|---|---|
| Mouth |
|
|
pH: 5-7 Transit time: 5s −2 min Saliva flow rate:
|
(Chen, 2009, Marze, 2017a, Marze, 2017b, Mosca and Chen, 2016, Rogers, 2011) |
| Esophagus |
|
– | Transit time: 8–10 s solid1-2 liquid | (Bender et al., 2005, Buettner et al., 2001, Rogers, 2011) |
| Stomach |
|
|
pH: 1-3 Transit time: 15 min - 4 h Gastric juice secretion: 1–3 L/day Contraction frequency: 3 cycles/min |
(Boland et al., 2014, Chen et al., 2011, Miftahof, 2017, Rogers, 2011, Sullivan, 2009) |
| Small Intestine |
|
|
pH: 6–7.5 Transit time: 1–5 h Pancreatic juice secretion: ~1.5 L/day |
(Rogers, 2011, Saladin, 2017, Smith and Morton, 2010, Sullivan, 2009) |
| Large Intestine |
|
|
pH: 5-7 Transit time: 12–24 h Microbiota: ~1011-1012 (>1000 different species) |
(Rogers, 2011, Saladin, 2017, Smith and Morton, 2010, Sullivan, 2009) |
Peristaltic contractions of the esophagus transfer the bolus to the stomach (Buettner et al., 2001; Rogers, 2011; Bornhorst et al., 2016). Transport of material through the esophagus (length: ~ 25 cm and diameter: 1.5–2 cm) takes approximately 8–10 s (Buettner et al., 2001; Bender et al., 2005; Rogers, 2011). When the bolus arrives at the stomach junction, the bolus can enter the stomach by the relaxed esophageal sphincter (a muscular cuff). Even though the transport of liquids through the esophagus may be faster than solids due to the gravity, they wait (8–10 s) for the arrival of the peristaltic relaxation before entering into the stomach (Rogers, 2011).
3.2. Stomach
In the stomach, which is a J-shaped muscular pouch, the digested food goes through a mechanical and chemical disintegration (Table 1). The stomach where the digested food is diluted with the gastric juice works like a container, mixer, and sieve (Bornhorst and Paul Singh, 2014, Brandstaeter et al., 2019). The stomach is the main digestion region. Even though digestion of the starch and triglycerides can start in the mouth due to amylase and lingual lipase in the saliva, protein digestion starts in the stomach (Sullivan, 2009).
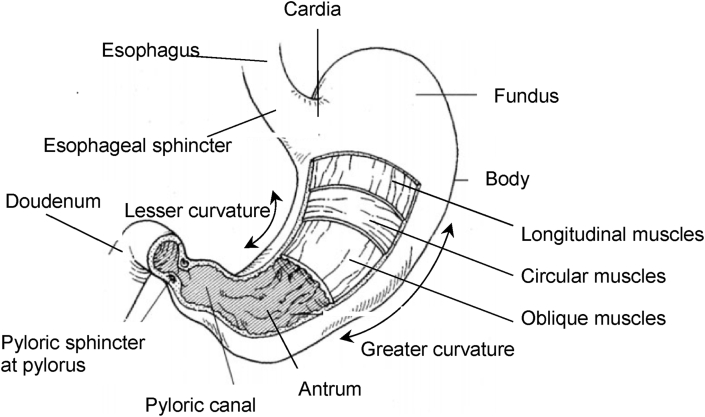
The concave border of the human stomach on the right is named as the lesser curvature, and the convex border to the left is named as the greater curvature (Fig. 2). The section that the stomach is connected to the esophagus is called the cardia (Miftahof, 2017). Fundus, body (corpus), and antrum (Fig. 2) are the main sections of the stomach (Brandstaeter et al., 2019). The stomach’s proximal region regulates the pressure for food storage, and the distal stomach functions as a gastric pump (Liu et al., 2019a). After a meal, the fundus, which sticks out like a dome, usually stores swallowed air. The largest section of the stomach, the body, serves as a reservoir for the ingested meal (Fig. 2). The antrum has a conical shape and is between the lower section of the body and the pyloric canal. The pylorus is the stomach’s slimmest portion and has enclosed by thick, smooth muscle layers (Miftahof, 2017, Rogers, 2011).
Fig. 2.
Anatomy of the stomach.
The sphincters, connecting the stomach to the esophagus and the duodenum (Fig. 2), are tightly controlled by the nervous system and responsible for the flow of chymus by rhythmic relaxation of the ring of smooth muscles from time to time, allowing partially digested food to pass through (Bender et al., 2005). The pyloric sphincter is more potent than the esophageal sphincter (Fig. 2).
Layers of the stomach wall (~ 3–4 mm thick) such as gastric mucosa and muscles are responsible for gastric secretions to dissolve the food and for contractions to grind and push the food through to the pyloric sphincter. In the stomach, the change of pH is controlled, and the bolus from the mouth is mixed with the digestive enzymes. Gastric contractions mix food particles and digestive juice by grinding action and reduce particle sizes (<1–2 mm) to form the fluidized mixture called chyme (Ferrua and Singh, 2011; Ferrua et al., 2011; Rogers, 2011; Bornhorst, 2017). Chyme progresses into the small intestine at small amounts and regular intervals.
The mucosal lining of the stomach forms countless wrinkles and folds, known as rugae when it is empty. These folds disappear when the stomach expands. The empty human stomach has a volume of about 25–50 mL, and the volume becomes 1–1.5 L after an average meal, while the geometry can vary depending on the individual, body position, surrounding organs, ingested meal, and digestion time (Ferrua and Singh, 2010; Saladin, 2017). Stomach volume can increase up to 4 L when full by unfolding and stretching (Saladin, 2017; Marieb and Keller, 2018; Brandstaeter et al., 2019).
The digestion process in the stomach involves physical and chemical processes (Table 1). Physical parameters like contractions affect particle sizes, while chemical parameters such as secretion of acid and digestive enzymes have an impact on the softening of the food and hydrolysis of nutrients (Kong and Singh, 2008a). The acidic environment activates the secreted pepsin and causes denaturation of food. Gastric pH in the fasted state is in the range of 1–3; after the ingestion of a meal, it will rise to 5.5–7, and after the half emptying time, it will go down to 4–5, and after the stomach emptied it will turn to its basal value (Boland et al., 2014).
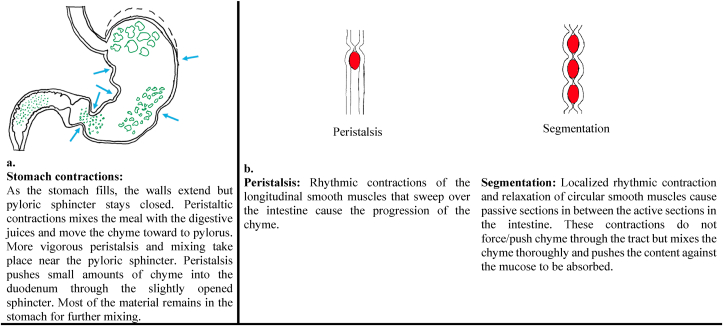
The fundus is the region where the food stays until some space becomes available for digestion. Some of the food that entered the stomach by relaxation of the esophageal sphincter stays in the fundus region, and the rest mixes with the gastric juice and moves toward the pylorus (Fig. 2). Vigorous mixing takes place near the pyloric sphincter. Because small amounts of chyme are allowed to pass through by the relaxed and opened pyloric sphincter, most of the material remains in the stomach for further mixing. As chyme is transferred to the small intestine, the food in the fundus region moves down for digestion (Fig. 2).
Gastric contractions are complex systems controlled by interrelated combinations of the body’s neural and hormonal systems and depend on the amount, composition, and physicochemical parameters of the ingested meal (Ferrua and Singh, 2010). In the antrum, generated compressive forces can be about 0.65 N for soft and 1.89 N for hard solids (Lentle, 2018). According to calculations based on magnetic resonance imaging (MRI) measurements, the maximum pressure experienced by the food particle surfaces is about 30–40 Pa (Lentle and Janssen, 2011; Lentle, 2018b).
The stomach wall consists of three layers of smooth muscle; longitudinal, circular, and oblique layers (Fig. 2). This combination of muscles lets the stomach not only push but mix and grind so the ingested meal can disintegrate into smaller particles. Gastric motility (Fig. 3a) can be defined as the contractions or relaxations of the smooth muscles in the stomach wall to coordinate the gastric accommodation, gastric mixing and emptying through antral contraction waves, and migrating motor complexes. (Brandstaeter et al., 2019). Gastric accommodation is the expansion of the stomach, especially in the proximal stomach, due to the reflex after ingestion of the meal to allow food storage. Antral peristalsis, or contraction waves, are ring-shaped muscular contractions propagating through the stomach wall and are in charge of mixing and emptying. Migrating motor complexes can be observed in the fasted state between meals (Brandstaeter et al., 2019).
Fig. 3.
(a) Contractions of the stomach, (b) Peristalsis and segmentation in the digestive system.
Tonic or sustained contractions of all the stomach muscles coordinate the stomach’s accommodation to varying volumes of gastric content (Ferrua and Singh, 2010). During tonic contraction, the stomach wall contracts as a whole; thus, the lumen size decreases. The tonic contraction is independent of the mixing and peristaltic contractions; but, mixing and peristaltic contractions co-occur with the tonic contraction (Rogers, 2011).
A small contraction wave that produces slight indentations on the stomach wall starts in the upper part and slowly moves down toward the pyloric sphincter (Fig. 3a). Backward waves sweep back from the pyloric sphincter through the antrum. These contractions cause a pressure build-up and a retropulsive backflow in the antrum region (Chen et al., 2011). This combination of back and forth flow causes mixing, grinding, and erosion of food particles.
Another contracting wave, which is peristaltic in nature, originates in the corpus and slowly propagates toward the pyloric sphincter (Ferrua and Singh, 2010). This peristaltic contraction starts with small indentations, and indentations deepen as it goes down. When this peristaltic wave reaches the antrum, the indentation completely blocks and divides the lumen (Boland et al., 2014). This wave propelling the chymus moves over the antrum. These peristaltic antral contractions function as a pump for emptying the contents through the pyloric sphincter at a constant rate (Boland et al., 2014, Rogers, 2011).
Contractions in the antrum are more potent because the muscles are thicker than other regions of the stomach (Smith and Morton, 2010). The pyloric sphincter governs the passage of the stomach content to the duodenum and inhibits the content of the duodenum from going back to the stomach. Tiny food particles are pushed through a momentarily opened pyloric sphincter providing the duodenum to taste the gastric content. About 3 mL of chyme is purged to the duodenum at each contraction regularly (Saladin, 2017).
The mixing and the peristaltic contractions are produced at a regular rate of 2–3 cycles per minutes as a response to the ingested meal (Chen et al., 2011; Rogers, 2011; Bornhorst and Singh, 2014; Ferrua and Singh, 2015; Bornhorst et al., 2016; Lentle, 2018a). Migrating peristaltic waves move indigestible fractions from the stomach to the intestine every 120 min (Brandstaeter et al., 2019).
The inner surface of the stomach contains the gastric mucosa, which is a mucous membrane layer. The mucosa secretes 1.2–1.5 L gastric juice per day, which contains water, hydrochloric acid, electrolytes (sodium, potassium, calcium, phosphate, sulfate, and bicarbonate), mucus, enzymes (lipase and pepsinogen), hormones (gastrin, serotonin) and the intrinsic factor (Bender et al., 2005, Rogers, 2011). The stomach walls are protected from this highly acidic juice by the membrane adjacent to the stomach’s lumen. In the area bordering the epithelium, the acidity of the mucous layer is almost neutral (pH 7) because of the secreted bicarbonate from the mucosa, whereas the acidity at the lumen side is very high (pH 2).
The gastric mucosa is covered by mucus-secreting epithelial cells and other types of cells, such as mucoid cells, chief cells, gastrin cells, parietal cells, and other endocrine cells (Rogers, 2011). Mucoid cells secrete gastric mucus. Chief cells secrete gastric lipase and pepsinogen, which is converted to the active digestive enzyme pepsin by the stomach’s acidic conditions. Gastrin (G) cells secrete the acid-stimulating hormone gastrin as a response to meal intake, which reduces the pH and extends the stomach wall. Gastrin hormone, which binds the receptor sites of the parietal cells, triggers hydrogen ion production in the parietal cells. The secreted hydrogen ions form hydrochloric acid (HCl) by combining with chloride ions (Sullivan, 2009). Most of the water in the gastric juice is produced by the parietal cells, which produce an intrinsic factor (glycoprotein) that is essential for the absorption of B12 through the small intestine as well as the appetite-regulating hormone ghrelin. The serotonin hormone secreted from endocrine cells stimulates the contraction of stomach muscles (Bender et al., 2005).
The gastric secretion processes that consist of cephalic, gastric, and intestinal phases are complex coordination of neural and hormonal factors, which overlap (Rogers, 2011). The cephalic phase starts before food reaches the stomach; seeing, smelling, and tasting food stimulates the secretion of gastrin hormone (Bender et al., 2005). The gastric phase starts with the presence of food. The intestinal phase starts when food starts to leave the stomach after one to 3 h. With the help of receptors, the duodenal bulb (the juncture between the duodenum and the stomach) and the next section of the duodenum relax to allow fluidized chyme with sufficiently small particles to enter the small intestine. The intestinal phase involves complex simulatory and inhibitory processes. When acidic chyme coming from the stomach enters the duodenum, it triggers the secretion of hormones that slow or inhibit gastric secretion in order to prevent more acidic chyme from entering the small intestine (Bender et al., 2005).
3.3. Small intestine
Most of the processes that convert the food into suitable forms that the body can use happen in the small intestine, where the nutrients are absorbed (Lentle and Janssen, 2010, Lentle and Janssen, 2011; Tharakan, Norton, Fryer and Bakalis, 2010; Rogers, 2011). The small intestine is the longest (6–7 m) part of the gastrointestinal tract and folded to fit in the abdominal cavity (Fig. 2). It is joined to the stomach by the pylorus and to the colon by the ileocecal valve. The duodenum, jejunum, and ileum are the small intestine’s main segments, respectively (Fig. 1). The duodenum (23–28 cm long) is where the canals secrete pancreatic juice and bile open up. The jejunum (~ 2 m) is the first 40% of the small intestine beyond the duodenum. The remaining 60% is the ileum (~ 3 m), which joins the large intestine through the ileocecal valve.
Most of the secretions controlled by nerves and hormones occur in the duodenum; the secretions are minimal in the other parts of the small intestine. The pancreas is a large gland that produces enzymes and hormones. The pancreatic juice and the bile flow through the duodenal papilla (rounded projection into the duodenum) to enter the duodenum (Rogers, 2011).
When the chyme enters the duodenum, gastric secretions within the chyme continue their digestive processes for a short time in the small intestine. Incoming chyme stimulates the pancreas to release concentrated bicarbonate solution that neutralizes the highly acidic gastric juice (Sullivan, 2009). Other secretions from the pancreas, gallbladder, and glands in the intestinal wall increase the total volume of chyme. Pancreatic secretion includes many enzymes, proteases (trypsin and chymotrypsin), pancreatic lipase, and pancreatic amylase. Bile is an aqueous solution produced by the liver and stored in the gallbladder facilitates digestion and absorption of lipids, acting as an emulsifier. Bile consists of bile salts, phospholipids, cholesterol, bilirubin (a breakdown product of red blood cells), electrolytes, and water. Here, digestion enzymes break down proteins, carbohydrates, triglycerides, and nucleic acids to smaller sizes. Pancreatic amylase can split starch into disaccharides such as maltose, but, here, disaccharides such as sucrose (from sugar and fruits) and lactose (from milk) can not be broken down into their monomers yet.
The motor activities observed in the small intestine are segmenting and peristaltic contractions that provide mixing and transport of chyme (Fig. 3b). Segmenting contractions, which are the predominant motor action, mix and separate the intestinal chyme. A short segment of the intestinal wall (<1–2 cm) contracts and constricts the lumen to divide its contents. The number of segmenting contractions decreases gradually from 11 to 12 cycles per minute in the duodenum to 8–9 cycles per minute in the ileum (Boland et al., 2014, Rogers, 2011). Peristaltic contraction is an advancing wave of contraction that can propel the chyme at a rate of 2–2.5 cm/s (Tharakan et al., 2010; Bornhorst and Singh, 2014). These contractions provide the movement of the intestinal content by creating a pressure difference between the adjacent segments.
The small intestine’s diameter is 3–4 cm in diameter, but its total absorptive surface area is approximately 4500 square meters (Rogers, 2011, Sullivan, 2009; Capuano, 2017; Lambeau and McRorie, 2017; Saladin, 2017). Numerous concentric folds of the mucosa (plicae circulares) provide this large absorptive surface. The brush border, which is the specialized surface of the epithelial cells, is richer in proteins and lipids than the plasma membrane on the epithelial cells. The final breakdown of digestion products happens on the surface of the brush border. It consists of microvilli that secrete the enzymes (maltase-glucoamylase, sucrase-isomaltase, lactase, brush border peptidases, lipase) that hydrolyze disaccharides, peptides, and nucleotides to their basic units suitable for absorption, such as monosaccharides and amino acids. These monomers are absorbed by the intestinal wall and transported to the bloodstream. Solutes and water move through the surface epithelium of the mucosa. Most of the absorption happens in the proximal small intestine, but a few substances (vitamin B12 and bile salts) are absorbed in the ileum (Smith and Morton, 2010). The absorption rate of water and nutrients are different in the jejunum and the ileum because, in the proximal region, the villi are large, and the available brush border surface area increases.
3.4. Large intestine
The large intestine has a large diameter (~ 6 cm), but it is shorter (~ 150 cm) than the small intestine. The ileocecal valve joining the small and large intestine shields the opening of the ileum into the cecum. The large intestine absorbs water into the body, and the remaining thick waste is ejected to complete the digestion. Each day about 1.5–2 L of chyme passes through the ileocecal valve. The chyme volume becomes about 150 ml after the water absorption in the colon (Fig. 1b).
The large intestine has an anaerobic environment and is colonized by an involved community of microorganisms (~ 1011/g) formed mainly by obligate anaerobic bacteria. The microbiota produces a variety of enzymes that breakdown dietary fibers not digested in the small intestine. The bacteria also metabolize the bile salts and pancreatic enzymes that reach the colon. The colon also contains bacteria that synthesize some essential vitamins (niacin, vitamin B1, and vitamin K) (Rogers, 2011). The free fatty acids (by-products of fermentation) content increases from about 6 to 8 mM in the ileum to 32-29 mM in the cecum. Chyme content affects the composition of the microbiota and metabolites produced, which can impact health.
Contractions and reverse movements in the large intestine mix the chyme and push the contents toward the walls. Slow-wave contractions sweep at a frequency of 11 cycles per minute from the ascending colon to the descending colon, then decrease to 6 cycles per minute in the sigmoid colon and rectum. Local contractions migrate at a rate of 4 cm per second through the colon. Reverse movements are seen mainly in the upper (proximal) colon.
4. In vitro digestion models
Digestion models are tools that can be used to design novel food products for human health. The models can be instrumental in consciously designing food products by estimating the in vivo behavior after meals. In vitro digestion models have been developed since the 1990s to be used in food digestion studies (Ferrua and Singh, 2010).
It is possible to study food digestion by in vivo (human or animal) or in vitro methods. Both methods have their advantages and disadvantages. Even though in vivo studies can give direct results due to their high cost, the requirement of ethical evaluation and variation of the digestive system from person to person, in general, in vitro models are used. In vitro models are preferred in food, nutrition, and medical research because of their speed, cost, and reproducibility due to used standardized conditions compared to in vivo studies (Kong and Sing 2008a, 2008b, 2010; Ferrua et al., 2011; Hur, Lim, Decker and McClements, 2011; Vardakou et al., 2011; Lafond et al., 2015; Egger et al., 2016; Guerra et al., 2016). Bioaccessibility determinations can be conducted in vitro or ex vivo by use of tissues, cell cultures and artificial membranes to measure permeability or absorption of small molecules (Vega-Rojas et al., 2021; Caicedo-Lopez et al., 2019).
Many in vitro digestion systems are designed to work in static conditions using glass containers to mimic human digestion. These systems can not produce the mechanical forces and dynamic conditions that foods experience in the digestive system. Static models can be practical, inexpensive, and feasible choices to assess many experimental conditions and a large number of samples; however, dynamic models are the ones that can come closest to in vivo conditions.
In food digestion, substrate-enzyme ratio, pH profiles, and transport of digested products are important parameters. Therefore, in vitro, static models that lack dynamic and mechanic actions have limitations for accurately predicting accessible nutrients or food behavior during digestion. Therefore, different dynamic systems have been developed to mimic the human digestive system’s physiological conditions with reproducibility necessary for scientific studies. They can be either mono-compartmental or multi-compartmental (Guerra et al., 2012; Dupont et al., 2019; Liu et al., 2019a, 2019b).
These dynamic systems have the challenge of competition between technological complexity and biological significance (Guerra et al., 2012). Multi-compartmental systems usually have gastric and small intestinal compartments, but few include the colon. Dynamic models can simulate the change in pH, enzyme secretion, peristaltic forces, and microbial fermentation continuously (Liu et al., 2019b). A list of in vitro dynamic models developed, especially for food digestion studies, is presented below (Table 2). Not all dynamic in vitro models mimic mechanic, kinetic, and chemical physiological conditions of the digestive system altogether. Some only simulate chemical conditions, some primarily focus on mechanical conditions, and very few include all mechanical, dynamic, and chemical conditions (Table 2). In this review, models developed for pharmaceutical and medicinal products were not included due to different digestion mechanisms of food, pharmaceuticals, and drugs.
Table 2.
Characteristics of the selected in vitro dynamic models used in food digestion studies.
| SYSTEM |
Mouth |
Stomach |
Small Intestine |
Colon |
System Temperature Control |
References with images of the systems | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Mixing pH Saliva |
Secretions | Mixing | Emptying | Secretions | Mixing | Absorption |
Microbiota Mixing Absorption |
|||
| TIM | Prepared as a bolus | Syringe pumps | Peristaltic pumps | Peristaltic pumps | Syringe pumps | Peristaltic pumps | Dialysis (Hallow fibers) |
TIM-2 Feces Anaerobic conditions Peristaltic pumps Dialysate system |
Heating elements connected with temperature sensors for each compartment | (Bellmann et al., 2016, Lafond et al., 2015) |
| SHIME | Prepared as suspension | Fill and draw | Magnetic stirrer | Fill and draw | Fill and draw | Magnetic stirrer | – | Feces Anaerobic conditions Magnetic stirrer |
Heater and Thermostat | (Molly et al., 1993, Vissenaekens et al., 2017) |
| ESIN | Meal reservoir, progressive introduction of food for 20 min (1–8 mm) | Peristaltic pumps | Two inox pistons | Peristaltic pumps | Peristaltic pumps | Shaft stirrers with adjustable rotors | Dialysis (Hallow fibers) | – | Water bath and heating films | (Guerra et al., 2016) |
| DIDGI | Prepared as a bolus | Peristaltic pumps | Agitation with a rotating blade actuated by a motor | Peristaltic pumps | Peristaltic pumps | Agitation with a rotating blade actuated by a motor | – | – | Water bath | (de La Pomélie et al., 2019, Ménard et al., 2014) |
| SIMGI | Prepared as a bolus | Peristaltic pumps | Peristaltic pumps | Peristaltic pumps | Peristaltic pumps | Magnetic stirrer | – | Feces Anaerobic conditions Magnetic stirrer |
Water bath | (Barroso et al., 2015, Miralles et al., 2018) |
| DGM | Prepared as a bolus Can be loaded in real time |
Through perforated hoop Peristaltic pumps |
Piston and barrel up and down movement | Piston and cyclical movement | – | – | – | – | Water bath | (Vardakou et al., 2011, Wickham et al., 2012) |
| HGS | Prepared as a bolus | Peristaltic pumps | Rollers, belts, driving shafts, and pulley system | Peristaltic pump | – | – | – | – | 60 W light bulbs and thermostat | (Kong and Singh, 2010) |
| DGSM | Prepared as a bolus | Variable flow mini pump | A probe attached to a texture analyzer up and down movement | Variable flow mini pump | – | – | – | – | Water bath | (Tran Do et al., 2016) |
| DIVHS | Prepared as a bolus and feed at a rate of 72 mL/min | Peristaltic pumps | Eccentric wheels, rollers, motors, belts, and pulley system | Adjusting the tilting angles of the auxiliary emptying device | – | – | – | – | Electric lamp with a controller | (Wang et al., 2019) |
| c-GDS | Prepared as a bolus <5 mm cube particle size solids |
Diaphram pump | Plastic rollers | Syringe pump | – | – | – | – | Heater with a temperature controller | (Kobayashi et al., 2017, Kozu et al., 2017) |
| AGDS | As a suspension | Roller peristalsis | Driving wheels, belts, compression rollers | Flow pump | – | – | – | – | Computer controlled incubator | (Liu et al., 2019a, Liu et al., 2019b) |
| ARCOL | – | – | – | – | – | – | – | Feces Anaerobic conditions Magnetic stirrer Dialysate system |
Heater | (Cordonnier et al., 2015, Thévenot et al., 2013) |
In order to reproduce the conditions of the human digestive system, especially for food systems, the mechanic, dynamic and chemical conditions of each organ in the digestive system should be acquired. Usually, data obtained in vivo from human or animal subjects were used in simulations. These conditions can be outlined as transit times of ingested meal, pH profiles, temperature, contractions, and peristaltic movements, digestive secretion rates, and absorption of water and nutrients through the surface of the organs. Each model uses a protocol specific to their food sample and research interest. Protocols can be specialized to an individual (healthy adult, elderly, infant, or person with specific needs) as well as the properties of the meal (solid, liquid). In the protocols used for dynamic models, chyme transfer is controlled according to a power exponential curve given below. The curve allows the description of two-component emptying patterns for solid and liquid foods. The constants of the equation are determined by using observed in vivo data according to the properties of the meal (solid or liquid) and used in simulation models.
Where.
f: the fraction of remaining chyme
t1/2: half-time (time to empty 50% of the chyme)
β: coefficient describing the shape of the curve
t: time
Several dynamic in vitro models used in food digestion studies were described briefly below.
4.1. TNO gastrointestinal model (TIM)
The in vitro gastrointestinal model (TIM) was developed at TNO Nutrition and Food Research Center (Zeist, The Netherlands) in the early 1990s. TIM is a computer-controlled multi-compartmental dynamic system that simulates the gastrointestinal system (Table 2). The system aimed to simulate the main physiological conditions that change with time and location, such as contractions, transit time, pH, composition, and secretion rate of digestive fluids, absorption of nutrients, and water in a reproducible and controllable manner. In the system, computer simulations used the protocols that were prepared with valid in vivo data. Specialized protocols depending on age and health status, as well as on the type of food, can be generated.
TIM is one of the successful in vitro dynamic systems and has been widely used in food and pharmacology research to investigate the release and absorption behavior of nutrients and drug components (Ferrua et al., 2011). The TIM system consists of glass units for the stomach, duodenum, jejunum, and ileum. Each unit has a glass jacket with a flexible inner membrane to allow expansion and contraction of the walls. Pressures and the temperature of the water pumped through the glass jackets can be adjusted to simulate peristaltic contractions and body temperature. Peristaltic valves are used to connect each unit. Computer simulation controls the flow of the digested food through and between the compartments.
This group has TIM-1, TIM-2, Tiny TIM, and TIM-agc models (Bellmann et al., 2016). TIM-1 is the most used one with four compartments. TIM-2 is the system designed for the large intestine using fecal donations from volunteers (Liu et al., 2019b). TinyTIM is a simplified and higher capacity version of TIM-1. The stomach section is the same as TIM-1, whereas the small intestine has one compartment instead of three and has no ileum exit. TIMagc has an advanced gastric model (Bellmann et al., 2016). TIM, with its computer-controlled simulations of peristaltic contractions and membrane technologies, offers flexibility to be used for different groups (babies, adults, elderly, and animal species), including health and disease conditions. Even though TIM-1 and TIM-2 complement each other as the upper and lower sections of the gastrointestinal tract, they do not run jointly (Liu et al., 2019b).
4.2. Simulator of the human intestinal microbial ecosystem (SHIME®)
Simulator of the human intestinal microbial ecosystem (SHIME®, Ghent University-Prodigest, Belgium) has been developed to simulate the microbial ecosystem of the gastrointestinal tract (Molly et al., 1993). The system (Table 2) that contains five reactor stages simulates the stomach, the small intestine, and the regions of the large intestine together (Liu et al., 2019b). The first two reactors simulate the acidity and pepsin digestion of the stomach and digestive process of the small intestine by fill and draw principle, where the stomach was simulated by reactor 1, and the small intestine by reactor 2. Peristaltic pumps were used for the controlled transfer of the vessel contents and digestive juices. The last three vessels, which were stirred continuously with a magnetic stirrer, were used to simulate the sections of the large intestine, ascending, transverse, and descending colons. Transit time of the contents and pH values in the vessels can be controlled to mimic in vivo data (Molly et al., 1993; Douny et al., 2019). System temperature kept at 37 °C, and N2 is flushed every day for 15 min to secure anaerobic conditions (Possemiers et al., 2004). Fecal microbiota is used to simulate the metabolic fate of food.
The group has other new models for different purposes. M-SHIME® (Mucus-SHIME) contains a mucosal compartment integrated to the colonic regions of SHIME® to evaluate the fraction of microorganisms that can selectively adhere to the mucous layer that covers the gut wall and play an essential role as a barrier against pathogens (AbbeeleVan Den et al., 2013). Evaluating the changes in the bacterial adhesion for several reasons is crucial in investigating host-microbiota interactions and resulting health effects. Another model developed by the group is TWINSHIME®. In this system, two identical SHIME® systems run in parallel under identical conditions to be able to have a direct comparison of two different samples at the same time (Vissenaekens et al., 2017). Diseased conditions such as inflammatory bowel disease have also been simulated with specific protocols (Dupont et al., 2019). This model’s strength is the incorporation of the human microbiota. However, it lacks the dialysis modules in the small and large intestines. Furthermore, using magnetic stirrers for mixing of the vessels is not as good as peristaltic movements (Liu et al., 2019b).
4.3. Engineered stomach and small intestinal (ESIN) system
Engineered stomach and small intestinal (ESIN) system have been developed at the University of Auvergne (Clermont-Ferrand, France). This dynamic system (Table 2) has been reported to aim to overcome some limitations of otherwise complex and useful models like TIM and SHIME (Guerra et al., 2012). Limitations were expressed as using ground food rather than a close imitation of real food bolus with realistic size particles at the stomach entrance and not representing the differential emptying of liquids and solids in the stomach seen in vivo (Dupont et al., 2019; Guerra et al., 2016). This dynamic model includes a patented (WO2009087314 A1) design for the stomach section (Dupont et al., 2019).
The ESIN model has six vessels, a meal reservoir, salivary container, the stomach, and the small intestine, duodenum, jejunum, and ileum segments. Food particles are advanced gradually into the stomach via the meal reservoir. Solid particles can pass the pylorus only if their sizes are reduced to 1–2 mm. The passage of the small particles (<2 mm) and liquids into the second vessel was achieved by an indentation inside the stomach vessel (Guerra et al., 2016). Solid particles larger than 2 mm stay in the gastric vessel for further digestion.
Liquids and solids particles are emptied by two peristaltic pumps that are programmed to follow an exponential model separately. Liquid fractions follow the exponential model without a lag phase period, while the solid factions have a 30 min lag phase (Guerra et al., 2016). In vivo data were used to simulate temperature, variations in pH values, flow rates of secretions, retention times, and absorption of nutrients in the digestive system.
4.4. In vitro dynamic system (DIDGI ®)
This dynamic digestion system (Table 2), which is developed at the French National Institute for Agricultural Research (INRA, Rennes, France) simulates the stomach and the small intestine with two glass jacketed vessels. The jackets are filled with water that is pumped from a temperature-controlled water bath. A Teflon membrane with 2 mm holes is placed before the transfer pump between the gastric and the intestinal compartment mimicking the sieving effect of the pylorus in humans. The temperature, pH, flow rate of the meal, digestive secretions and emptying rate for each compartment are controlled by computer simulation designed with the data obtained from in vivo observations (Ménard et al., 2014; de La Pomélie et al., 2019).
4.5. Simulator of the gastrointestinal tract (Simgi®)
This dynamic simulator (Table 2) has been developed to reproduce gastrointestinal digestion and colonic fermentation at the Institute of Food Science Research (Madrid, Spain). The simgi® model consists of five compartments simulating the stomach, small intestine, and the ascending, transverse, and descending colon (Barroso et al., 2015; Mackie et al., 2017; Miralles, del Barrio, Cueva, Recio & Amigo, 2018; Tamargo et al., 2018). The stomach section has two transparent and rigid plastic vessels that cover a flexible silicone container. Water at 37 °C pumped through the jacket between the plastic modules, and the flexible container is used to simulate peristaltic movements (Barroso et al., 2015). Flow rates of the digestive secretions, temperature, and pressure values are controlled through a computer program to follow the defined values. Emptying curves are controlled according to an exponential equation described by Elashoff et al.. The remaining four compartments, the small intestine, the ascending, transverse and descending colon are continuously stirred reactors operating under anaerobic conditions and controlled pH. The system has collecting points in each of the compartments to carry out the biochemical and microbiological analysis. This system combines peristaltic contractions and chyme transit behaviour for gastric and small intestine compartments (Liu et al., 2019b).
4.6. Dynamic gastric model (DGM)
The model (DGM) has been developed at the Institute of Food Research (Norwich, UK). This computer-controlled model (Table 2) simulating only the stomach can process real size chewed meals and simulates the physiological conditions such as mixing, the transit of meals, and forces observed in the stomach (Wickham et al., 2012; Dupont et al., 2019). This model simulates three stages of the in vivo conditions: 1. stage: the proximal stomach for ingestion and mixing; 2. stage: the antrum for higher shear rate; 3. stage: the duodenum, first part of the small intestine (Bornhorst and Singh, 2012). In the model, the disintegration of the food particles was obtained by a stationary outer and mobile inner tube (Wickham et al., 2012; Ferrua et al., 2011). Temperature, transit time, and flow rates of acid, salts, and enzymes are controlled and adjusted to desired physiological rates. The model produces a cyclical (0.05 Hz) mixing in the body of the stomach and preferential antral sieving (Wickham et al., 2012). In the system, with the collection of data obtained by the samples delivered from the antrum over time, emptying profiles, particle size reduction, and mass transfer data can be obtained but, peristaltic movement is not accurately simulated (Liu et al., 2019b).
4.7. Human gastric simulator (HGS)
Human gastric simulator (HGS) is designed at the University of California, Davis, USA (Kong and Singh, 2010). The model (Table 2) simulates peristaltic contractions, secretions and emptying rates seen in the stomach. The mechanical driving system composed of rollers, a latex reservoir, secretion, emptying and temperature control systems (Kong and Singh, 2010). The latex stomach vessel can be filled with food and digestive fluids, and peristaltic contractions were created by the 12 rollers steered by a motor assembly. The flow rates of secretions and temperature can be adjusted, and stomach contents can be emptied through pylorus simulating in vivo conditions (Kong and Singh, 2010). Two generations of the HGS model exists (Kong and Singh 2010; Dupont et al., 2019). The model called IHGS (improved human gastric simulator) has a better capability of simulating the shape, contour, contractions, and other physical parameters of the human stomach.
4.8. Dynamic gastric simulation model (DGSM)
This is a design based on the dynamic gastric simulation model proposed by Chen et al. (2011) at the University of Leeds (Leeds, UK). The model (Table 2) simulates compressive forces, secretions, and gastric emptying to remove digest to simulate digestion in the stomach. The model has an acrylic vessel insulated with water. A PVC probe attached to a texture analyzer is used to measure the forces observed. Up and down movements of the probe attached to a texture analyzer produces the gastric contractions. The probe moves down to the bottom of the vessel until leaving a 2 mm gap to compress and generate food particles <2 mm at the bottom, then cycles back up (at a rate of 3 cycles/min). Variable flow mini pumps are used for gastric secretions and emptying (Tran Do et al., 2016).
4.9. Near real dynamic in vitro human stomach (new DIVHS) system
This dynamic in vitro human stomach (new DIVHS) model is an advanced design based on the previous models DIVRS-I, DIVRS-II, and RD-IV-HSM at the Suzhou Key Laboratory of Green Chemical Engineering, Soochow University (Suzhou, China) (Wang et al., 2019). The new DIVHS (Table 2) includes a stomach and duodenum vessel, a driving instrument, a system for secreting and emptying, and a box with temperature control. J-shaped soft elastic silicone vessel is produced by 3D-printing technology to have a similar dimension and folded inner morphology of the human stomach. Contractions are simulated by a series of motors, rollers, and eccentric wheels. A separate roller-added system improves the disintegration and sieving ability. An auxiliary emptying device controls the gastric emptying rate by adjusting the tilting angles (Wang et al., 2019). An exponential model was used to generate gastric emptying rates for the solid and liquid fractions where the solid fractions had a lag phase, whereas the liquid fractions did not have a lag phase.
4.10. Human gastric digestion simulator (GDS)
This human gastric digestion simulator (GDS) was designed to simulate the antrum at the Food Research Institute, NARO (Ibaraki, Japan). The GDS (Table 2) consists of a gastric compartment, a roller system, and a temperature control system with a transparent parallel window to view the food disintegration mechanism (Kozu et al., 2014; Kobayashi et al., 2017). The simplified antrum geometry has a trapezoidal shape resembling the human antrum with a smaller diameter toward the pylorus. The deformable sidewalls allow the simulation of progressive antral contraction waves by pushing a pair of rollers. The rollers move down at a speed of several mm/s at a frequency of a few cycles/min. The digestion process, which can last for up to 180 min can be followed with a video camera to analyze the changes in the food particles. This group also developed a continuous GDS (c-GDS) equipped with gastric secretion and emptying systems (Kozu et al., 2017).
4.11. Artificial gastric digestive system (AGDS)
This artificial gastric digestive system (AGDS) is developed at the School of Food Science and Biotechnology, Zhejiang Gongshang University (Zhejiang, China). Size, shape, and the folds of the stomach wall are generated by using 3D printing technology in the silicone stomach model (Table 2) (Liu et al., 2019b). Peristaltic contractions of the stomach were produced by two sets of symmetrical rollers and one set of contrary rollers. A computer program controlled the forces, temperature, pH, gastric juice secretions, and gastric emptying. The mechanical forces exerted on the external wall of the silicone stomach were measured by a texture analyzer to estimate the forces experienced by the chyme (Liu et al., 2019b).
4.12. Artificial colon (ARCOL)
The artificial colon (ARCOL) is a fermenter (Applikon, Schiedam, The Netherlands) that simulates the human colonic environment (Cordonnier et al., 2015; Thévenot et al., 2013). The system (Table 2) simulates the pH, body temperature, supply of ileal effluents, retention time, anaerobic conditions, and absorption of water and fermentation metabolites (Blanquet-Diot et al., 2012; Cordonnier et al., 2015). Feces from healthy volunteers are used in the reactor. Fecal suspension prepared under anaerobic conditions flushed with O2-free N2 gas.
The temperature of 37 °C, pH of 6.3, and mean retention time of 36 h were used in the model. Mixtures consisting of carbohydrate, protein, lipid, mineral, and vitamins consecutively introduced into the reactor to mimic the ileal effluents. After inoculation, the flushing with O2-free N2 gas is ceased to allow the fermentative process to maintain anaerobic conditions in the reactor. A dialysis system with hollow fiber membranes (cut-off 30 kDa) is used to simulate the electrolyte and metabolite concentrations in vivo.
Only a few (TIM, SHIME, ESIN, DIDGI, and SIMGI) of the above dynamic models (Table 2) simulate both the stomach and small intestine compartments as interconnected modules. SHIME and SIMGI models also simulate the fermentation of the large intestine connected to the small intestine. While TIM, ESIN, and DIDGI are highly sophisticated systems and focus on reproducing the mechanical, dynamic, and chemical physiological conditions in the stomach and small intestine close to reality, SHIME and SIMGI are systems that focus entirely on the large intestine fermentation and cannot accurately mimic the contractions of the stomach and small intestine (Table 2). The meal chamber in the ESIN model allows meals to be delivered to the stomach as mixtures of particles of realistic size, progressively. DIDGI does not have a dialysis system as TIM and ESIN to simulate the absorption of the digested nutrients in the small intestine.
The other group, DGM, HGS, DGSM, DIVHS, c-DGS, and AGDS, are models aiming to simulate the mechanical, dynamic, and chemical conditions of the stomach only (Table 2). These successful and sophisticated models simulating the stomach have differences. DGM model focused on the representation of the mechanical forces by using piston and barrel, but peristalsis and dynamic conditions of the stomach were not considered. Due to its windowed design, the GDS model allows the monitoring of stomach contents during digestion. In DIVHS and AGDS systems, stomach geometry is modeled very close to reality. Finally, ARCOL is a reactor that simulates fermentation in the large intestine only (Table 2). TIM-2 is also an independent module that simulates the large bowel fermentation only (Table 2).
The surge of articles published in the area indicates the requirement of engineering physiologically relevant in vitro models to use to understand the food digestion mechanism and use in the development of novel foods (Jin, Ou, Decker, McClements, 2011; Guerra et al., 2012; Alminger et al., 2014; Bornhorst et al., 2016; Marze, 2017a; Shani-Levi et al., 2017; Bohn et al., 2018; Lucas-González, Viuda-Martos, Perez-Alvarez and Fernandez-Lopez, 2018; Pimentel, Burton, Vergeres & Dupont, 2018; Bellmann et al., 2019; Brodkorb et al., 2019; Costa and Ahluwalia, 2019; Dupont et al., 2019; Keppler, O'Meara, Bakalis, Fryer and Bornhorst, 2020). Digestion is regulated by nerves and hormones that facilitate acid release, and increase muscular movements in the stomach (Rogers, 2011). There is no definitive model to use for the interactions of the digestive system’s simulatory and inhibitory effects with food intake. As we gain information on these interactions, it will be possible to fine-tune the simulations. As an example of such improvement, to improve the prediction behavior of the existing models, TIM group used TIMagc with an in silico artificial neural network to estimate the appetite parameters after consumption of different meals. They have achieved a reasonable prediction for appetite in humans. Other improvement areas would be related to simulation of the large intestine. Although many large intestine models use human feces bacteria, challenges still exist. Variations in the ratio of bacterial populations from the feces and inadequate simulation of the real shape and movements of the large intestine can limit the simulation studies (Liu et al., 2019b). The role of gut microbiota in digestion also needs further exploration. Even though the results obtained from these complex models should be analyzed with care and require confirmation with human studies, their contribution to the accumulation of scientific data is substantial.
5. Conclusions
Many in vitro simulation tools have been designed to reproduce the complexity of the human digestive system. Many of them are very complex systems with their sometimes in-house computer control and the mechanic or dynamic systems designs. They attempt to reproduce the physiological conditions of the human digestive system as close as possible by the use of observed in vivo data obtained from human subjects. They use complex computer control systems to adjust the dynamics of pH, transit time, and digestive secretions. Sophisticated roller or piston systems or application of regulated pressure waves by peristaltic pumps have been used to simulate the fluid dynamics and mechanical forces in the digestive system. Carrier-mediated transport and passive diffusion through the small intestine epithelial cells were simulated as passive diffusion by using a filter medium or by exposing the digested samples to cell lines. The systems sometimes used a texture analyzer to measure and control the applied forces in the stomach. Even individual organs can be modeled to close perfection in anatomy and physiology. There are examples of early models like a human duodenum model by Wright et al. (2016). Even though it was not possible to perfectly match the complex human digestive system, it was possible to simulate the digestive system’s critical mechanical, dynamic, and biochemical processes in a robust, repeatable manner. The models have been improved extensively over the years, and some have been used by the food, nutrition, and medical industry widely. Still, new models show up, as seen in a recent real-size stomach model developed by Li et al. (2019).
CRediT authorship contribution statement
Ilkay Sensoy: Conceptualization, Investigation, Writing - orginal draft, Writing – review & editing.
Declaration of competing interest
The author declares no conflicts of interest.
References
- AbbeeleVan Den P., Venema K., WieleVan T.De, Verstraete W., Possemiers S. Different human gut models reveal the distinct fermentation patterns of Arabinoxylan versus inulin. J Agric Food Chem. 2013 doi: 10.1021/jf4021784. [DOI] [PubMed] [Google Scholar]
- Aguayo-mendoza M.G., Ketel E.C., Linden E. Van Der, Forde C.G., Piqueras-fiszman B., Stieger M. Oral processing behavior of drinkable , spoonable and chewable foods is primarily determined by rheological and mechanical food properties. Food Qual. Prefer. 2019;71:87–95. doi: 10.1016/j.foodqual.2018.06.006. June 2018. [DOI] [Google Scholar]
- Alminger M., Aura A.M., Bohn T., Dufour C., El S.N., Gomes A. In vitro models for studying secondary plant metabolite digestion and bioaccessibility. Compr. Rev. Food Sci. Food Saf. 2014;13(4):413–436. doi: 10.1111/1541-4337.12081. [DOI] [PubMed] [Google Scholar]
- Barroso E., Cueva C., Peláez C., Martínez-Cuesta M.C., Requena T. Development of human colonic microbiota in the computer-controlled dynamic SIMulator of the GastroIntestinal tract SIMGI. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 2015;61(2):283–289. doi: 10.1016/j.lwt.2014.12.014. [DOI] [Google Scholar]
- Bellmann S., Lelieveld J., Gorissen T., Minekus M., Havenaar R. Development of an advanced in vitro model of the stomach and its evaluation versus human gastric physiology. FRIN. 2016;88:191–198. doi: 10.1016/j.foodres.2016.01.030. [DOI] [Google Scholar]
- Bellmann S., Krishnan S., de Graaf A., de Ligt R.A., Pasman W.J., Minekus M., Havenaar R. Appetite ratings of foods are predictable with an in vitro advanced gastrointestinal model in combination with an in silico artificial neural network. Food Res. Int. 2019;122:77–86. doi: 10.1016/j.foodres.2019.03.051. [DOI] [PubMed] [Google Scholar]
- Bender L., Harding D., Kennedy D., Lee G., Stokes&Taylor J.B., editors. The Facts On File Illustrated Guide to the Human Body: Digestive System. Facts on File, Inc; New York: 2005. [Google Scholar]
- Blanquet-Diot S., Denis S., Chalancon S., Chaira F., Cardot J.M., Alric M. Use of artificial digestive systems to investigate the biopharmaceutical factors influencing the survival of probiotic yeast during gastrointestinal transit in humans. Pharmaceut. Res. 2012;29(6):1444–1453. doi: 10.1007/s11095-011-0620-5. [DOI] [PubMed] [Google Scholar]
- Bohn T., Carriere F., Day L., Deglaire A., Egger L., Freitas D. Correlation between in vitro and in vivo data on food digestion. What can we predict with static in vitro digestion models? Crit. Rev. Food Sci. Nutr. 2018;58(13):2239–2261. doi: 10.1080/10408398.2017.1315362. [DOI] [PubMed] [Google Scholar]
- Boland M., Golding M., Singh H., editors. Food Structures, Digestion and Health. Elsevier Inc; New York: 2014. [DOI] [Google Scholar]
- Bornhorst G.M. Gastric mixing during food digestion : mechanisms and applications. Annu. Rev. Food Sci. Technol. 2017;8(1):523–542. doi: 10.1146/annurev-food-030216-025802. [DOI] [PubMed] [Google Scholar]
- Bornhorst G.M. Gastric mixing during food Digestion : mechanisms and applications. Annu. Rev. Food Sci. 2017;8(1):523–542. doi: 10.1146/annurev-food-030216-025802. [DOI] [PubMed] [Google Scholar]
- Bornhorst G.M., Ferrua M.J., Rutherfurd S.M., Heldman D.R., Singh R.P. Rheological properties and textural attributes of cooked Brown and white rice during gastric digestion in vivo. Food Biophys. 2013;8(2):137–150. doi: 10.1007/s11483-013-9288-1. [DOI] [Google Scholar]
- Bornhorst G.M., Ferrua M.J., Singh R.P. A proposed food breakdown classification system to predict food behavior during gastric digestion. J. Food Sci. 2015;80(5):R924–R934. doi: 10.1111/1750-3841.12846. [DOI] [PubMed] [Google Scholar]
- Bornhorst G.M., Gouseti O., Wickham M.S.J., Bakalis S. Eng. Digest: Multiscale Processes of Food Digestion. 2016;81(3):534–543. doi: 10.1111/1750-3841.13216. [DOI] [PubMed] [Google Scholar]
- Bornhorst G.M., Paul Singh R. Gastric digestion in vivo and in vitro: how the structural aspects of food influence the digestion process. Annu. Rev. Food Sci. Technol. 2014;5:111–132. doi: 10.1146/annurev-food-030713-092346. [DOI] [PubMed] [Google Scholar]
- Bornhorst G.M., Rutherfurd S.M., Roman M.J., Burri B.J., Moughan P.J., Singh R.P. Gastric pH distribution and mixing of soft and rigid food particles in the stomach using a dual-marker technique. 2014. 292-300. [DOI]
- Bornhorst G.M., Singh R.P. Kinetics of in Vitro bread bolus digestion with varying oral and gastric digestion parameters. Food Biophy. 2013;8(1):50–59. doi: 10.1007/s11483-013-9283-6. [DOI] [Google Scholar]
- Bornhorst G.M., Singh R.P. Bolus formation and disintegration during digestion of food carbohydrates. Compr. Rev. Food Sci. Food Saf. 2012;11(2):101–118. doi: 10.1111/j.1541-4337.2011.00172.x. [DOI] [Google Scholar]
- Brandstaeter S., Fuchs S.L., Aydin R.C., Cyron C.J. Mechanics of the stomach: a review of an emerging field of biomechanics. GAMM-Mitteilungen. 2019;42(3):1–17. doi: 10.1002/gamm.201900001. [DOI] [Google Scholar]
- Brodkorb A., Egger L., Alminger M., Alvito P., Assunção R., Ballance S. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019;14(4):991–1014. doi: 10.1038/s41596-018-0119-1. [DOI] [PubMed] [Google Scholar]
- Buettner A., Beer A., Hannig C., Settles M. Observation of the swallowing process by application of videofluoroscopy and real-time magnetic resonance imaging - consequences for retronasal aroma stimulation. Chem. Senses. 2001;26(9):1211–1219. doi: 10.1093/chemse/26.9.1211. [DOI] [PubMed] [Google Scholar]
- Caicedo-Lopez L.H., Luzardo-Ocampo I., Cuellar-Nuñez M.L., Campos-Vega R., Mendoza S., Loarca-Piña G. Effect of the in vitro gastrointestinal digestion on free-phenolic compounds and mono/oligosaccharides from Moringa oleifera leaves: bioaccessibility, intestinal permeability and antioxidant capacity. Food Res. Int. 2019;120:631–642. doi: 10.1016/j.foodres.2018.11.017. July 2018. [DOI] [PubMed] [Google Scholar]
- Capuano E. The behavior of dietary fi ber in the gastrointestinal tract determines its physiological effect. Crit. Rev. Food Sci. Nutr. 2017;57(16):3543–3564. doi: 10.1080/10408398.2016.1180501. [DOI] [PubMed] [Google Scholar]
- Chen J. Food oral processing-A review. Food Hydrocolloids. 2009;23(1):1–25. doi: 10.1016/j.foodhyd.2007.11.013. [DOI] [Google Scholar]
- Chen J., Gaikwad V., Holmes M., Murray B., Povey M., Wang Y., Zhang Y. Development of a simple model device for in vitro gastric digestion investigation. Food and Function. 2011;2(3–4):174–182. doi: 10.1039/c0fo00159g. [DOI] [PubMed] [Google Scholar]
- Costa J., Ahluwalia A. Advances and current challenges in intestinal in vitro model engineering: a digest. Front. Bioeng. Biotechnol. 2019;7(144):1–14. doi: 10.3389/fbioe.2019.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordonnier C., Thévenot J., Etienne-Mesmin L., Denis S., Alric M., Livrelli V., Blanquet-Diot S. Dynamic in vitro models of the human gastrointestinal tract as relevant tools to assess the survival of probiotic strains and their interactions with gut microbiota. Microorganisms. 2015;3(4):725–745. doi: 10.3390/microorganisms3040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Pomélie D., Santé-Lhoutellier V., Sayd T., Théron L., Gatellier P. Using a dynamic artificial digestive system to investigate heme iron nitrosylation during gastro-intestinal transit. Food Chem. 2019;281(September 2018):231–235. doi: 10.1016/j.foodchem.2018.12.094. [DOI] [PubMed] [Google Scholar]
- Directive 2001/20/EU No 2010/63/EU of the European Parliament and of the Council of 4 April 2001 on the approximation of the laws, regulations and administrative provisions of the Member States relating to the implementation of good clinical practice in the conduct of clinical trials on medicinal products for human use. Off. J. Eur. Communities. 2001:L121/34–44. http://data.europa.eu/eli/dir/2001/20/oj [PubMed] [Google Scholar]
- Directive 2010/63/Eu No 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Euro. Union, L. 2010;276:33–79. http://data.europa.eu/eli/dir/2010/63/oj [Google Scholar]
- Douny C., Dufourny S., Brose F., Verachtert P., Rondia P., Lebrun S., Marzorati M., Everaert N., Delcenserie V., Scippo M. Development of an analytical method to detect short-chain fatty acids by SPME-GC – MS in samples coming from an in vitro gastrointestinal model. J. Chromatogr. B. 2019;1124(June):188–196. doi: 10.1016/j.jchromb.2019.06.013. [DOI] [PubMed] [Google Scholar]
- Drechsler K.C., Ferrua M.J. Modelling the breakdown mechanics of solid foods during gastric digestion. FRIN. 2016;88:181–190. doi: 10.1016/j.foodres.2016.02.019. [DOI] [Google Scholar]
- Dupont D., Alric M., Bornhorst G., Cueva C., Deglaire A., Denis S., Ferrua M., Havenaar R., Lelieveld J., Mackie A.R., Marzorati M., Minekus M., Miralles B., Recio I., Abbeele P. Van Den. Can dynamic in vitro digestion systems mimic the physiological reality ? Crit. Rev. Food Sci. Nutr. 2019:1–17. doi: 10.1080/10408398.2017.1421900. 0(0) [DOI] [PubMed] [Google Scholar]
- Dupont Didier, Le S., Marze S., Souchon I. Structuring food to control its disintegration in the gastrointestinal tract and optimize nutrient bioavailability. Innovat. Food Sci. Emerg. Technol. 2018;46:83–90. doi: 10.1016/j.ifset.2017.10.005. March 2017. [DOI] [Google Scholar]
- Egger L., Menard O., Delgado-Andrade C., Alvito P., Assunçao R., Balance S. The harmonized INFOGEST in vitro digestion method: from knowledge to action. Food Res. Int. 2016;88:217–225. doi: 10.1016/j.foodres.2015.12.006. [DOI] [Google Scholar]
- Ferrua Maria J., Kong F., Singh R.P. Computational modeling of gastric digestion and the role of food material properties. Trends Food Sci. Technol. 2011;22(9):480–491. doi: 10.1016/j.tifs.2011.04.007. [DOI] [Google Scholar]
- Ferrua M.J., Singh R.P. Modeling the fluid dynamics in a human stomach to gain insight of food digestion. J. Food Sci. 2010;75(7):R151–R162. doi: 10.1111/j.1750-3841.2010.01748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrua M.J., Singh R.P. Computational modelling of gastric digestion: current challenges and future directions. Curr. Opin. Food Sci. 2015;4:116–123. doi: 10.1016/j.cofs.2015.06.005. [DOI] [Google Scholar]
- Ferrua Maria J., Singh R.P. Understanding the fluid dynamics of gastric digestion using computational modeling. Procedia Food Science. 2011;1:1465–1472. doi: 10.1016/j.profoo.2011.09.217. [DOI] [Google Scholar]
- Guerra A., Denis S., le Goff O., Sicardi V., François O., Yao A.F., Garrait G., Manzi A.P., Beyssac E., Alric M., Blanquet-Diot S. Development and validation of a new dynamic computer-controlled model of the human stomach and small intestine. Biotechnol. Bioeng. 2016;113(6):1325–1335. doi: 10.1002/bit.25890. [DOI] [PubMed] [Google Scholar]
- Guerra A., Etienne-Mesmin L., Livrelli V., Denis S., Blanquet-Diot S., Alric M. Relevance and challenges in modeling human gastric and small intestinal digestion. Trends Biotechnol. 2012;30(11):591–600. doi: 10.1016/j.tibtech.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Hur S.J., Lim B.O., Decker E.A., McClements D.J. In vitro human digestion models for food applications. Food Chem. 2011;125(1):1–12. doi: 10.1016/j.foodchem.2010.08.036. [DOI] [Google Scholar]
- Jin S., Ou B., Decker E.A., McClements D.J. In vitro human digestion models for food applications. Food Chem. 2011;125(1):1–12. doi: 10.1016/j.foodchem.2010.08.036. [DOI] [Google Scholar]
- Keppler S., O'Meara S., Bakalis S., Fryer P.J., Bornhorst G.M. Characterization of individual particle movement during in vitro gastric digestion in the Human Gastric Simulator (HGS) J. Food Eng. 2020;264:109674. doi: 10.1016/j.jfoodeng.2019.07.021. [DOI] [Google Scholar]
- Ketel E.C., Aguayo-mendoza M.G., Wijk R. A De, Graaf C.De, Piqueras-fiszman B., Stieger M. Age , gender , ethnicity and eating capability in fl uence oral processing behaviour of liquid , semi-solid and solid foods di ff erently. Food Res. Int. 2019;119(September 2018):143–151. doi: 10.1016/j.foodres.2019.01.048. [DOI] [PubMed] [Google Scholar]
- Kobayashi I., Kozu H., Wang Z., Isoda H., Ichikawa S. Development and fundamental characteristics of a human gastric digestion simulator for analysis of food disintegration. Jpn. Agric. Res. Q. 2017;51(1):17–25. doi: 10.6090/jarq.51.17. [DOI] [Google Scholar]
- Kong F., Singh R.P. Disintegration of solid foods in human stomach. J. Food Sci. 2008;73(5):R67–R80. doi: 10.1111/j.1750-3841.2008.00766.x. [DOI] [PubMed] [Google Scholar]
- Kong F., Singh R.P. A model stomach system to investigate disintegration kinetics of solid foods during gastric digestion. J. Food Sci. 2008;73(5):E202–E210. doi: 10.1111/j.1750-3841.2008.00745.x. [DOI] [PubMed] [Google Scholar]
- Kong Fanbin, Singh R.P. Modes of disintegration of solid foods in simulated gastric environment. Food Biophys. 2009;4(3):180–190. doi: 10.1007/s11483-009-9116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Fanbin, Singh R.P. A human gastric simulator (HGS) to study food digestion in human stomach. J. Food Sci. 2010;75(9) doi: 10.1111/j.1750-3841.2010.01856.x. [DOI] [PubMed] [Google Scholar]
- Kozu H., Kobayashi I., Nakajima M., Neves M.A., Uemura K., Isoda H., Ichikawa S. Mixing characterization of liquid contents in human gastric digestion simulator equipped with gastric secretion and emptying. Biochem. Eng. J. 2017;122:85–90. doi: 10.1016/j.bej.2016.10.013. [DOI] [Google Scholar]
- Kozu H., Nakata Y., Nakajima M., Neves M.A., Uemura K., Sato S., Kobayashi I., Ichikawa S. Development of a human gastric digestion simulator equipped with peristalsis function for the direct observation and analysis of the food digestion process. Food Sci. Technol. Res. 2014;20(2):225–233. doi: 10.3136/fstr.20.225. [DOI] [Google Scholar]
- Lambeau K.V., McRorie J.W. Fiber supplements and clinically proven health benefits: how to recognize and recommend an effective fiber therapy. J. Am. Assoc. Nurse Pract. 2017;29(4):216–223. doi: 10.1002/2327-6924.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafond M., Bouza B., Eyrichine S., Rouffineau F., Saulnier L., Giardina T., Bonnin E., Preynat A. In vitro gastrointestinal digestion study of two wheat cultivars and evaluation of xylanase supplementation. J. Anim. Sci. Biotechnol. 2015;6(1):11–14. doi: 10.1186/s40104-015-0002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentle R.G., Janssen P.W.M. Manipulating digestion with foods designed to change the physical characteristics of digesta. Crit. Rev. Food Sci. Nutr. 2010;50(2):130–145. doi: 10.1080/10408390802248726. [DOI] [PubMed] [Google Scholar]
- Lentle R.G. Deconstructing the physical processes of digestion: reductionist approaches may provide greater understanding. Food and Function. 2018;9(8):4069–4084. doi: 10.1039/c8fo00722e. [DOI] [PubMed] [Google Scholar]
- Lentle R.G. 2018. Function digestion : reductionist approaches may; pp. 4069–4084. [DOI] [PubMed] [Google Scholar]
- Lentle R.G., Janssen P.W.M. Springer; New York: 2011. The Physical Process of Digestion. [Google Scholar]
- Li Y., Fortner L., Kong F. Development of a Gastric Simulation Model (GSM) incorporating gastric geometry and peristalsis for food digestion study. Food Res. Int. 2019;125:108598. doi: 10.1016/j.foodres.2019.108598. [DOI] [PubMed] [Google Scholar]
- Liu W., Fu D., Zhang X., Chai J., Tian S., Han J. Development and validation of a new artificial gastric digestive system. Food Res. Int. 2019;122(April):183–190. doi: 10.1016/j.foodres.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Liu W., Ye A., Han F., Han J. Advances and challenges in liposome digestion: surface interaction, biological fate, and GIT modeling. Adv. Colloid Interface Sci. 2019;263:52–67. doi: 10.1016/j.cis.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Lucas-González R., Viuda-Martos M., Pérez-Alvarez J.A., Fernández-López J. vol. 107. 2018. pp. 423–436. (Vitro Digestion Models Suitable for Foods: Opportunities for New Fields of Application and Challenges). [DOI] [PubMed] [Google Scholar]
- Mackie A. Food: more than the sum of its parts. Current Opinion in Food Science. 2017;16:120–124. doi: 10.1016/j.cofs.2017.07.004. [DOI] [Google Scholar]
- Mackie A.R., Lesmes U., Day L., Le Feunteun S., Deglaire A., Menard O., Macierzanka A., Dupont D., Recio I., Miralles B., Golding M., Santé-Lhoutelier V., Bohn T., Freitas D., Portmann R., Egger L., Moscovici A., Rémond D., Wooster T.J., Carriere F. Correlation between in vitro and in vivo data on food digestion. What can we predict with static in vitro digestion models? Crit. Rev. Food Sci. Nutr. 2017;58(13):2239–2261. doi: 10.1080/10408398.2017.1315362. [DOI] [PubMed] [Google Scholar]
- Marieb E.N., Keller S.M. 12th. Pearson Education Limited; England: 2018. Essentials of Human Anatomy and Physiology. [Google Scholar]
- Marze S. Bioavailability of Nutrients and Micronutrients: Advances in Modeling and In Vitro Approaches. Ann. Rev. Food Sci. Technol. 2017;8(1):35–55. doi: 10.1146/annurev-food-030216-030055. [DOI] [PubMed] [Google Scholar]
- Marze S. Ch. 14 Modeling of Food Digestion. In: Becker S.M., editor. Modeling of Microscale Transport in Biological Processes. Academic Press, Elsevier Inc; New York: 2017. pp. 353–374. [DOI] [Google Scholar]
- Ménard O., Cattenoz T., Guillemin H., Souchon I., Deglaire A., Dupont D., Picque D. Validation of a new in vitro dynamic system to simulate infant digestion. Food Chem. 2014;145:1039–1045. doi: 10.1016/j.foodchem.2013.09.036. [DOI] [PubMed] [Google Scholar]
- Miftahof R.N. Biomechanics of the Human Stomach. Springer International; New York: 2017. [DOI] [Google Scholar]
- Miralles B., del Barrio R., Cueva C., Recio I., Amigo L. Dynamic gastric digestion of a commercial whey protein concentrate†. J. Sci. Food Agric. 2018;98(5):1873–1879. doi: 10.1002/jsfa.8668. [DOI] [PubMed] [Google Scholar]
- Molly K., Vande Woestyne M., Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993;39(2):254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- Mosca A.C., Chen J. Food oral management : physiology and objective assessment. Curr. Opin. Food Sci. 2016;9:11–20. doi: 10.1016/j.cofs.2016.03.003. [DOI] [Google Scholar]
- Mulet-Cabero A.I., Mackie A.R., Wilde P.J., Fenelon M.A., Brodkorb A. Structural mechanism and kinetics of in vitro gastric digestion are affected by process-induced changes in bovine milk. Food Hydrocolloids. 2019;86:172–183. doi: 10.1016/j.foodhyd.2018.03.035. [DOI] [Google Scholar]
- Pimentel G., Burton K.J., Vergères G., Dupont D. The role of foodomics to understand the digestion/bioactivity relationship of food. Curr. Opin. Food Sci. 2018;22:67–73. doi: 10.1016/j.cofs.2018.02.002. [DOI] [Google Scholar]
- Possemiers S., Verth K., Uyttendaele S., Verstraete W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 2004;49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Rogers K (ed) The Digestive System. Britannica Educational Publishing; New York: 2011. [Google Scholar]
- Saladin K.S. Human Anatomy. 5th Edition. McGraw-Hill Education; New York: 2017. [Google Scholar]
- Shani-Levi C., Alvito P., Andrés A., Assunção R., Barberá R., Blanquet-Diot S. Extending in vitro digestion models to specific human populations: perspectives, practical tools and bio-relevant information. Trends Food Sci. Technol. 2017;60:52–63. doi: 10.1016/j.tifs.2016.10.017. [DOI] [Google Scholar]
- Smith M.E., Morton D.G. Systems of The Body. The Digestive System. Basic Science and Clinical Conditions. 2nd Edition. Elsevier Limited; New York: 2010. [Google Scholar]
- Sullivan R. The Human Body, How It Works, Digestion and Nutrition. Chelsea House Publications; 2009. [Google Scholar]
- Tamargo A., Cueva C., Laguna L., Moreno-Arribas M.V., Muñoz L.A. Understanding the impact of chia seed mucilage on human gut microbiota by using the dynamic gastrointestinal model simgi. Journal of Functional Foods. 2018;50(September):104–111. doi: 10.1016/j.jff.2018.09.028. [DOI] [Google Scholar]
- Thévenot J., Etienne-Mesmin L., Denis S., Chalancon S., Alric M., Livrelli V., Blanquet-Diot S. Enterohemorrhagic Escherichia coli O157: H7 survival in an in vitro model of the human large intestine and interactions with probiotic yeasts and resident microbiota. Appl. Environ. Microbiol. 2013;79(3):1058–1064. doi: 10.1128/AEM.03303-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakan A., Norton I.T., Fryer P.J., Bakalis S. Mass transfer and nutrient absorption in a simulated model of small intestine. J. Food Sci. 2010;75(6):E339–E346. doi: 10.1111/j.1750-3841.2010.01659.x. [DOI] [PubMed] [Google Scholar]
- Tran Do D.H., Kong F., Penet C., Winetzky D., Gregory K. Using a dynamic stomach model to study efficacy of supplemental enzymes during simulated digestion. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 2016;65:580–588. doi: 10.1016/j.lwt.2015.08.054. [DOI] [Google Scholar]
- Vardakou M., Mercuri A., Barker S.A., Craig D.Q.M., Faulks R.M., Wickham M.S.J. Achieving antral grinding forces in biorelevant in Vitro models: comparing the USP dissolution apparatus II and the dynamic gastric model with human in Vivo data. AAPS PharmSciTech. 2011;12(2):620–626. doi: 10.1208/s12249-011-9616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rojas L.J., Luzardo-Ocampo I., Mosqueda J., Palmerín-Carreño D.M., Escobedo-Reyes A., Blanco-Labra A., Escobar-García K., García-Gasca T. Bioaccessibility and in vitro intestinal permeability of a recombinant lectin from tepary bean (Phaseolus acutifolius) using the everted intestine assay. Int. J. Mol. Sci. 2021;22(3):1–22. doi: 10.3390/ijms22031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenaekens H., Pitart J., Romo-Vaquero M., Esp J.C., Grootaert C., Selma V., Tomas-barberan F.A. Gastrointestinal simulation model TWIN-SHIME shows differences between human urolithin-metabotypes in gut microbiota composition, pomegranate polyphenol metabolism, and transport along the intestinal tract. J. Agric. Food Chem. 2017;65:5480–5493. doi: 10.1021/acs.jafc.7b02049. [DOI] [PubMed] [Google Scholar]
- Wang J., Wu P., Liu M., Liao Z., Wang Y., Dong Z., Chen X.D. An advanced near real dynamic: in vitro human stomach system to study gastric digestion and emptying of beef stew and cooked rice. Food and Function. 2019;10(5):2914–2925. doi: 10.1039/c8fo02586j. [DOI] [PubMed] [Google Scholar]
- Wickham M.J.S., Faulks R.M., Mann J., Mandalari G. The design, operation, and application of a dynamic gastric model. Dissolution Technol. 2012;19(3):15–22. doi: 10.14227/DT190312P15. [DOI] [Google Scholar]
- Wright N.D., Kong F., Williams B.S., Fortner L. A human duodenum model ( HDM ) to study transport and digestion of intestinal contents. J. Food Eng. 2016;171:129–136. doi: 10.1016/j.jfoodeng.2015.10.013. [DOI] [Google Scholar]