Abstract
Background
BRAF mutant melanoma patients are commonly treated with anti-BRAF therapeutic strategies. However, many factors, including the percentage of BRAF-mutated cells, may contribute to the great variability in patient outcomes.
Patients and methods
The BRAF variant allele frequency (VAF; defined as the percentage of mutated alleles) of primary and secondary melanoma lesions, obtained from 327 patients with different disease stages, was assessed by pyrosequencing. The BRAF mutation rate and VAF were then correlated with melanoma pathological features and patients’ clinical characteristics. Kaplan–Meier curves were used to study the correlations between BRAF VAF, overall survival (OS), and progression-free survival (PFS) in a subset of 62 patients treated by anti-BRAF/anti-MEK therapy after metastatic progression.
Results
A highly heterogeneous BRAF VAF was identified (3%-90%). Besides being correlated with age, a higher BRAF VAF level was related to moderate lymphocytic infiltration (P = 0.017), to melanoma thickness according to Clark levels, (level V versus III, P = 0.004; level V versus IV, P = 0.04), to lymph node metastases rather than cutaneous (P = 0.04) or visceral (P = 0.03) secondary lesions. In particular, a BRAF VAF >25% was significantly associated with a favorable outcome in patients treated with the combination of anti-BRAF/anti-MEK drug (OS P = 0.04; PFS P = 0.019), retaining a significant value as an independent factor for the OS and the PFS in the multivariate analysis (P = 0.014 and P = 0.003, respectively).
Conclusion
These results definitively support the role of the BRAF VAF as a potential prognostic and predictive biomarker in melanoma patients in the context of BRAF inhibition.
Key words: melanoma, BRAF mutations, variant allele frequency, increased survival
Highlights
-
•
In melanoma the response to anti-BRAF targeted therapies is heterogeneous and influenced by several features.
-
•
The role of the BRAF VAF as provider of sensitivity to target therapies is debated.
-
•
We found that high BRAF VAFs are associated with patient age, melanoma thickness, non-brisk TILs and lymph node metastases.
-
•
We proved the independent prognostic value of high BRAF VAFs in melanoma patients treated with targeted therapies.
-
•
The quantitative evaluation of BRAF mutations allows stratifying melanoma patients to the BRAF/MEK targeted treatment.
Introduction
Melanoma is the most aggressive cancer among all skin tumors. The American Cancer Society reported about 100 000 new melanoma cases and 7300 estimated patient deaths in 2019 with an increasing trend of both incidence and death rates.1 About 50% of patients carry a somatic mutation of the BRAF gene, which encodes for a serine-threonine kinase involved in the control of the MAPK pathway (BRAF/MEK/ERK).2 The BRAF p.V600E (c. 1799T > A) pathogenic variant is detected in up to 90% of the BRAF-mutated melanomas along with other rarer variants in the same codon (p.V600K, p.V600D, p.V600R, and p.V600M) or in the surrounding amino acid residues.3 In addition to BRAF, driver mutations in the NRAS gene have been reported in 15%-25% of cutaneous melanomas.4 BRAF and NRAS mutations are almost always mutually exclusive.5
The molecular profiling of melanomas has become prominent following the approval of BRAF-mutated selective inhibitors as single agent4,5 or in combination with the BRAF downstream effector MEK inhibitors.6 Melanomas harboring a BRAF mutation are characterized by more aggressive clinical features, resulting in shorter patient survival.7,8 However, the approval of BRAF-targeting agents has noticeably improved progression-free survival (PFS) and overall survival (OS) in melanoma patients. Yet, as for many other forms of neoplasia,9, 10, 11 inter-patient heterogeneity and tumor sub-clonality may influence the response to treatments and result in variable clinical outcomes in BRAF-mutated melanomas.12
We previously reported a sun-exposure-dependent distribution of the BRAF p.V600E mutation in either nevi or melanocytic lesions, thus suggesting that this oncogene variant may not be clonally selected in melanoma progression.13 The relationship between the BRAF variant allele frequency (VAF) and the clinical outcome in patients treated with BRAF inhibitors was evaluated in previous studies with controversial results.14, 15, 16, 17 Satzger et al.18 reported that the allele frequency of the BRAF mutations does not impact on overall and disease-free survival of melanoma patients treated with BRAF-targeted therapy, whereas Stagni et al.19 observed that BRAF VAF is an independent predictor of PFS in melanoma patients treated with monotherapy or combined anti-BRAF and anti-MEK agents. However, both studies lacked correlations with the histopathological features of the lesions.
In order to assess whether BRAF VAF can affect pathological features, outcome, and treatment responsiveness to target therapies in melanoma patients, the mutation rate and the allele frequency of BRAF mutations were correlated with the clinical-pathological features of a large cohort of 345 primary and secondary melanocytic lesions.
Patients and methods
Patient cohort
Our cohort consisted of 345 melanoma lesions derived from 327 patients consecutively diagnosed at the Candiolo Cancer Institute, FPO-IRCCS, between 2010 and 2018. Familial melanoma was excluded according to their clinical history. All lesions were clustered into seven subsets according to their site of origin [trunk, limbs, facial/scalp, acral, ocular, visceral, and occult primary melanoma (OPM)] and into three groups according to metastatic spread (lymph nodal, cutaneous, and visceral). For 208 patients, NRAS mutational status was also available. Clinical and pathological data for the total cohort are reported in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100133 and in Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100133 for the NRAS cohort.
Follow-up data were obtained for 62 patients diagnosed or progressed to stage IV disease. The BRAF test was assessed on 18 primary melanomas and 44 metastatic lesions. After metastatic progression, patients were treated with the BRAF inhibitor alone (MONO, n = 21, dabrafenib or vemurafenib) or with a combination of anti-BRAF and anti-MEK drugs (COMBO n = 41, dabrafenib plus trametinib or vemurafenib plus cobimetinib). Tumor assessment was carried out at baseline by computed tomography (CT) scan or magnetic resonance imaging (MRI) scan of the head, chest, abdomen, and pelvis, and skin lesions were photographed. Scans were assessed every 8 weeks for a total of 56 weeks and were checked every 12 weeks thereafter until disease progression or death. Response to treatment was defined according to RECIST criteria. Considering the best response to disease, the patients were clustered according to disease progression (PD), stable disease (SD), and patients with response, either partial or complete [partial response (PR) or complete response (CR), respectively]. We estimated the overall survival from diagnosis (OSd) as the time from diagnosis to death or to the last follow-up, the overall survival from treatment onset (OSt) as the time from the beginning of therapy to death or last follow-up, and the PFS, as the interval between the beginning of the therapy to the time of progression. We also evaluated the lactate dehydrogenase (LDH) level in blood at the beginning of the anti-BRAF therapy and patients were clustered in high LDH (>450 U/l, 20 patients) and low LDH (<450 U/l, 42 patients) levels. The cohort diagrams are summarized in Supplementary Figure S1A and B, available at https://doi.org/10.1016/j.esmoop.2021.100133.
Clinical-pathological data were recovered after obtaining informed consent from all the patients, approved by the medical ethical committee of the FPO-IRCCS, and carried out according to the principles of the Declaration of Helsinki.
Specimens, DNA extraction, and pyrosequencing analysis
DNA was purified from formalin-fixed, paraffin-embedded (FFPE) tissues. Before extraction, two independent pathologists reviewed the histological slides and recorded information regarding: (i) tumor-infiltrating lymphocytes (TILs) (absent, non-brisk, and brisk); (ii) Clark I-V levels and Breslow levels corresponding to thickness in mm; (iii) mitotic index for the primary lesions (number of mitoses/mm2); (iv) site of metastases (cutaneous, lymph nodal, or visceral); (v) eligibility for molecular assessment of the BRAF molecular test (>100 cells and >50% tumoral cells). Tumor areas with >50% of tumor cells were selected based on hematoxylin–eosin-stained slides and dissected from four different 6-μm FFPE sections. DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Germany) following the manufacturer’s protocol. All DNA samples were quantified using DeNovix Spectrophotometer (Wilmington, DE) and Qubit fluorometer (ThermoFisher Scientific, Waltham, MA).
The BRAF exon 15 analysis was carried out using the pyrosequencing method, which permits evaluating both the type and the allele fraction of the BRAF [Seq ID: Locus Reference Genomic (LRG) 299; https://www.lrg-sequence.org/]20 codons 599, 600, 601, and 602.13 The assay was carried out using the Anti-EGFR MoAb response® (BRAF status) kit (Diatech Pharmacogenetics, Jesi, Italy) following the manufacturer’s protocol. The same approach was applied to sequence NRAS [Seq ID: LRG 92; https://www.lrg-sequence.org/]20 codons 12, 13, 58, 59, 61, 117, and 146 using the Anti-EGFR MoAb response® (NRAS status) kit (Diatech Pharmacogenetics, Jesi, Italy). The mutation rate (MR) was defined as the number of mutated samples in the population and the VAF as the percentage of the mutated peak in the BRAF sequence
Statistical analyses
Data were analyzed by using the IBM SPSS Statistics, Version 20.0. (IBM Corp Armonk, NY). The parametric (t-student) or non-parametric (Mann–Whitney) tests were used to evaluate differences in the distribution of the BRAF MR, VAF, and the different histopathological variables of the melanomas. Pearson’s correlation coefficient was used to define the relationship between age and BRAF variants whereas Spearman’s correlation coefficient defined the relationship between the VAF and the normalized VAF. The chi-squared test was applied to infer proportions when assessing the presence of an association between BRAF MR and VAF with the type of metastasis. The same tests were applied to evaluate the association between NRAS mutations and pre-clinical features. For the survival analyses, OS and PFS were evaluated by the Kaplan–Meier method and analyzed with the log-rank test. Surviving or disease progression-free patients were censored at the date of the last follow-up. Multivariate Cox regression model (backward, conditional) considering all the clinical-pathological characteristics of the patients was applied to evaluate the strengths of the VAF values as OS or PFS independent factors. A P < 0.05 was considered statistically significant.
Results
BRAF MR/BRAF VAF and clinical data
Our cohort of 327 patients comprised 144 female and 183 male patients, with a mean age of 62 years (range 15-91). DNA samples were obtained from 141 primary lesions and 204 metastatic specimens. BRAF was mutated in 156/345 (45%) of the analyzed specimens with the BRAF p.V600E mutation being by far the most represented (89%). The rare mutations included 11 p.V600K and 6 different substitutions located between codon 599 and codon 602. BRAF VAF data followed a normal distribution with mean and median values of 33% and 31%, respectively (range 3%-90%). Considering only the BRAF p.V600E mutated samples, the mean and the median VAF values were both ∼30% (range 3.5%-90%). As for the rare mutations, their mean and median VAF values were higher but not statistically different (34% and 32%, respectively). Details regarding BRAF MR and VAF for all the mutations are reported in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100133. Differences were detected in terms of BRAF VAF distribution considering sex and clinical stage while an inverse correlation of the MR and the VAF was found (r Pearson = −0.96, P = 0.035, Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2021.100133) clustering the mutated patients into four age-based quartile groups (group 1: <51 years; group 2: 51-64 years; group 3: 65-74 years; group 4: >74 years) (Supplementary Figure S2B, available at https://doi.org/10.1016/j.esmoop.2021.100133). The metastatic lesions showed a slightly higher but not significant median MR and VAF compared with the primary melanomas (MR: 49% versus 40%; VAF: 34.1% versus 31.3%, P = 0.22).
BRAF MR/BRAF VAF, different melanoma sites and metastatic spreading
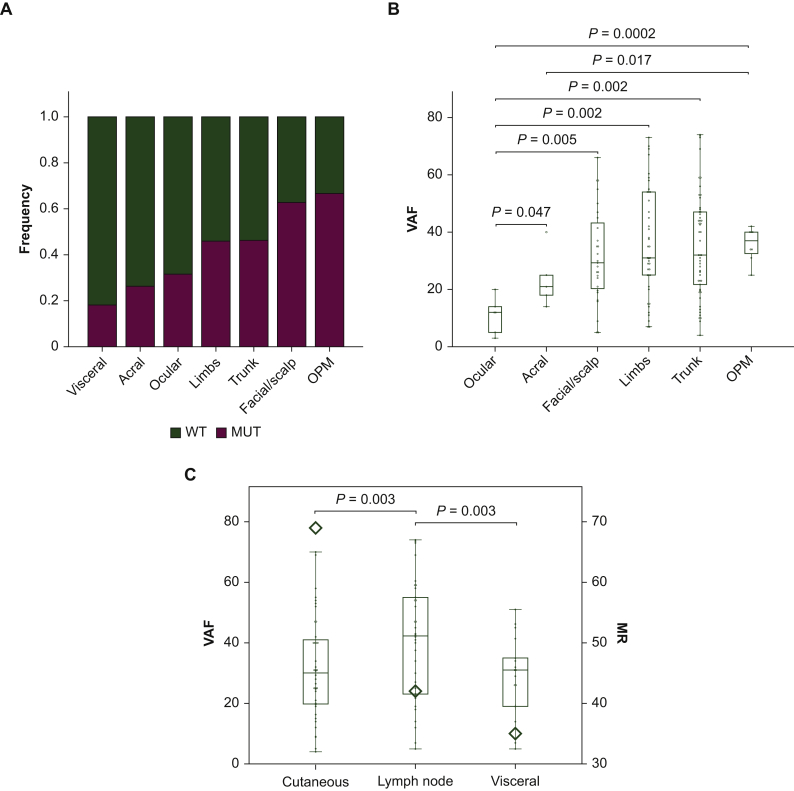
We checked for possible relationships of BRAF MR and BRAF VAF with the site of primary melanomas and the metastases (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100133). The highest MR was detected in specimens derived from primary facial/scalp melanomas (63%). Specimens from trunk and limb melanomas showed a similar MR (46%), whereas samples derived from acral and visceral neoplasia had the lowest MR levels (26% and 18%, respectively). Regarding metastatic lesions, the highest MR (67%) was found in the subset of patients with OPM (Figure 1A).
Figure 1.
BRAF MR and VAF correlations with the primary site of melanomas.
(A) Bar stacked chart of the MR (frequency) for each sample, clustered for the primary site. (B) Box and whisker plot defining the VAF distribution for each primary site; the absence of visceral samples in the box plot is due to the too low number of mutated patients.2 (C) Double Y-axis box and whisker plot for the MR and the VAF (only for metastatic samples) grouped for the site of the metastasis; on the left Y-axis is reported the BRAF VAF, whereas on the right axis the MR; the MR was defined as the average MR by the rhombus symbol.
OPM, occult primary melanoma; MR, mutation rate; VAF, variant allele frequency; WT, wild type.
Facial/scalp, limb, and trunk melanomas displayed a heterogeneous distribution with comparable median VAF levels while samples from ocular, acral, and OPM lesions were more homogeneously grouped and exhibited the median lowest (ocular and acral) and highest (OPM) VAF value (Figure 1B). Overall, the BRAF VAF of the ocular-derived cases was statistically different from that of the other groups.
We also investigated whether the site of the metastatic spreading was related to a specific pattern of the BRAF MR/VAF. Cutaneous metastases were more frequently mutated than lymph node and visceral lesions (P = 0.003 for both) (Figure 1C). However, lymph node metastases displayed higher BRAF VAF in comparison with cutaneous and visceral metastases (P = 0.04 and P = 0.03, respectively) (Figure 1C).
BRAF MR/BRAF VAF and pathological features of primary melanomas
In order to investigate the potential effects of the tumor cellularity on the BRAF VAF, we normalized VAF considering tumor cellularity defined as the mean cell percentage calculated on five different sample fields provided by the detailed histological revision of 120 mutated samples. Tumor cellularity ranged between 55% and 90%. As reported in Supplementary Figure S3, available at https://doi.org/10.1016/j.esmoop.2021.100133, Spearman’s correlation revealed a linear trend between original and normalized VAF values (r = 0.96), hence implying that VAF values were not significantly influenced by tumor cell content.
We then assessed possible correlations between BRAF status and specific features of primary melanomas, such as TILs, melanoma thickness (Clark's level and Breslow classification), and mitotic count.
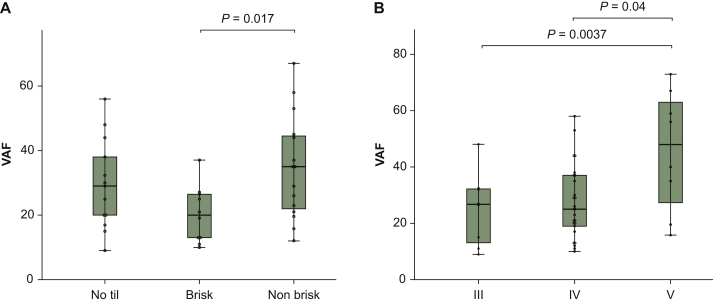
No differences in MR and VAF were found comparing TIL negative with TIL positive (brisk + non-brisk) melanomas; however, lesions with a moderate infiltrate (non-brisk) had a significantly higher VAF compared with those with severe TIL infiltration (brisk) (P = 0.017) (Figure 2A).
Figure 2.
BRAF MR and VAF correlations with both primary and metastatic features.
(A) Box and whisker plot reporting the VAF (only for primary melanoma cases) of the groups defined for the TIL. (B) Box and whisker plot reporting the VAF (only for primary melanoma cases) of the groups defined by Clark's level.
MR, mutation rate; VAF, variant allele frequency; TIL, tumor-infiltrating lymphocyte.
Regarding tumor thickness, there was a positive association between the VAF values and the depth of the lesions, as defined by the Breslow classification (r = 0.517, P = 0.001). Moreover, Clark V level melanomas showed a statistically significant higher VAF compared with those of level III and IV (P = 0.004 and P = 0.04, respectively) (Figure 2B). No correlation was observed with the mitotic count of primary tumors.
BRAF MR/BRAF VAF and NRAS MR/NRAS VAF
NRAS MR and VAF were evaluated by pyrosequencing and compared with BRAF MR and VAF in 208 lesions of the cohort. NRAS was mutated in 28 (13.4%) of the analyzed specimens, whereas 113 (54.4%) showed a BRAF mutation and 67 (32.2%) were wild type for both genes. Coexisting mutations were not identified. Most of the NRAS mutations were detected on the codon 61 (26/28), with a prevalence of p.Q61R variant (13/28) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100133). NRAS mutations showed higher VAFs than BRAF (P = 0.015), with mean and median values of 42% and 40%, respectively (range 11%-80%). Metastatic lesions (11 cases) had a slightly higher but not significant NRAS VAF compared with primary melanomas (17 cases) (median VAF: 39.5% versus 44%, P = 0.41). NRAS MR and VAF were not correlated with the site of primary lesions (10 trunk, 14 limbs, 3 facial, and 1 acral) or the type of metastases (4 cutaneous, 3 lymph node, and 4 visceral). Finally, no association was evidenced between NRAS VAF and the specific TIL subsets, whereas thicker lesions, according to Clark levels, displayed a higher but not significant NRAS VAF (P = 0.077) (Supplementary Figure S4A–E, available at https://doi.org/10.1016/j.esmoop.2021.100133).
BRAF MR/BRAF VAF and patient clinical outcomes
Finally, we analyzed the response and the outcome of the 62 patients with available clinical data treated with target therapy after metastatic progression.
Twelve patients experienced disease progression (PD) (19%) and 18 SD (29%). Of the 32 patients with response to treatments, 19 achieved PR and 13 a CR (31% and 21%, respectively). Overall, the median OSd was 46 months [95% confidence interval (CI) 30-60 months] while the median OSt was 15 months (95% CI 9-21 months). The median PFS was 8 months (95% CI 5-11 months). As for the whole cohort, the BRAF p.V600E mutation represented the most prevalent variant (90%). In addition, five cases carried the p.V600K substitution and one patient exhibited the very rare p.T599I variant. The mean and the median VAF were 28% and 25%, respectively.
BRAF inhibitor alone (MONO) was given to 21 patients and the combination of anti-BRAF and anti-MEK inhibitors (COMBO) to the remaining 41 patients. The ‘baseline’ mutational BRAF profile was assessed before starting the therapy for all the patients. As expected, the patients treated with COMBO showed an increased survival rate (OSt P = 0.007 and PFS P = 0.004) compared with the MONO therapeutic approach.
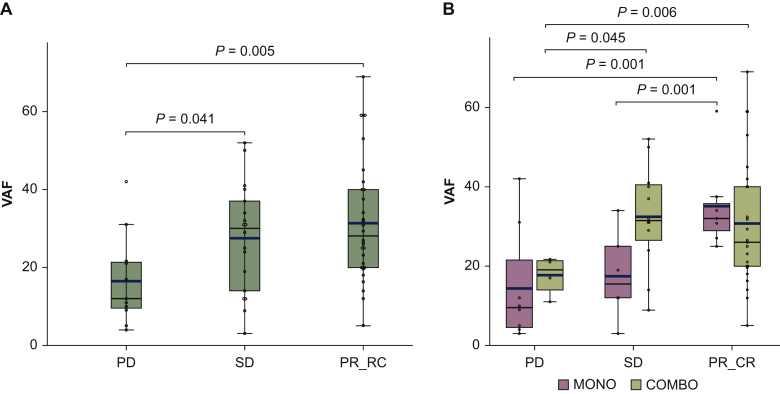
In particular, responsive (CR and PR) and SD patients exhibited significantly higher VAFs compared with the group with tumor progression (PD) (P = 0.005 and P = 0.041 respectively, Figure 3A and Table 1).
Figure 3.
Patient response to treatment and the VAF distribution.
(A) Box and whisker plot defining the BRAF VAF in the followed-up cohort grouped for the best response rate. The thick line represents the average value for each group. (B) Box and whisker plot defining the BRAF VAF in the followed-up cohort clustered for the response rate and for the therapy type; the thick line denotes the average VAF value for each group.
CR, complete response; PD, progression disease; PR, partial response; SD, stable disease.
Table 1.
Univariate and multivariate analyses
| OS diagnosis (OSd) |
|||||
|---|---|---|---|---|---|
| Median months (95% CI) | Univariate analysis |
Multivariate analysis |
|||
| HR (95% CI) | P | HR (95% CI) | P | ||
| Sex | |||||
| Female | 66 (41.08-90.91) | 1 | 1 | ||
| Male | 40 (31.94-48.01) | 1.28 (0.70-2.22) | 0.40 | 1.10 (0.71-2.29) | 0.50 |
| BRAF VAF | |||||
| HIGH | 68 (35.45-100.66) | 1 | 1 | ||
| LOW | 40 (33.51-46.80) | 1.88 (0.99-3.29) | 0.046 | 1.55 (0.85-2.85) | 0.15 |
| Therapy | |||||
| COMBO | 52 (28.65-75.35) | 1 | 1 | ||
| MONO | 45 (33.78-56.22) | 1.46 (0.80-2.66) | 0.20 | 1.40 (0.72-2.70) | 0.32 |
| Stage | |||||
| I-II | 75 (56.85-93.15) | 1 | 1 | ||
| III | 40 (36.70-43.3) | 1.43 (0.71-2.86) | 0.26 | 1.47 (0.75-2.90) | 0.23 |
| IV | 16 (5.35-33.53) | 4.77 (1.30-17.51) | 0.001 | 5.10 (2.19-11.78) | 0.0001 |
| LDH | |||||
| High | 15 (37.03-94.97) | 1 | 1 | ||
| Low | 8 (11.66-42.33) | 0.15 (0.06-0.34) | 0.001 | 0.21 (0.09-0.435) | 0.0001 |
| Therapy + VAF | |||||
| MONO low VAF | 40 (24.50-55.49) | 1 | |||
| MONO high VAF | 45 (5.36-108.85) | 1.54 (0.62-3.77) | 0.30 | ||
| COMBO low VAF | 39 (30.68-47.316) | 1.18 (0.51-2.681) | 0.69 | ||
| COMBO high VAF | 77 (33.68-120.37) | 2.22 (0.90-5.63) | 0.045 | ||
| OS from treatment onset (OSt) |
|||||
|---|---|---|---|---|---|
| Median months (95% CI) | Univariate analysis |
Multivariate analysis |
|||
| HR (95% CI) | P | HR (95% CI) | P | ||
| Sex | |||||
| Female | 19 (7.41-30.6) | 1 | 1 | ||
| Male | 13 (7.42-18.6) | 1.26 (0.72-2.22) | 0.70 | 1.062 (0.592-1.90) | 0.85 |
| BRAF VAF | |||||
| HIGH | 21 (5.28-36.71) | 1 | 1 | ||
| LOW | 10 (7.40-12.59) | 1.89 (1.01-3.47) | 0.024 | 2.14 (1.168-3.93) | 0.014 |
| Therapy | |||||
| COMBO | 21 (7.23-34.77) | 1 | |||
| MONO | 10 (7.04-12.69) | 2.39 (1.20-4.71) | 0.007 | 3.04 (1.59-5.81) | 0.001 |
| Stage | |||||
| I-II | 26 (7.64-44.36) | 1 | 1 | ||
| III | 16 (8.22-23.78) | 1.33 (0.69-2.56) | 0.38 | 1.01 (0.442-1.76) | 0.72 |
| IV | 7 (6.08-11.92) | 2.30 (0.96-6.14) | 0.03 | 1.94 (0.91-3.77) | 0.045 |
| LDH | |||||
| High | 21 (11.02-30.98) | 1 | 1 | ||
| Low | 6 (1.62-10.38) | 0.11 (0.04-0.23) | <0.0001 | 0.25 (0.13-0.48) | 0.0001 |
| Therapy + VAF | |||||
| MONO low VAF | 9 (6.93-11.06) | 1 | |||
| MONO high VAF | 12 (3.90-20.09) | 1.74 (0.70-4.32) | 0.17 | ||
| COMBO low VAF | 12 (9.22-14.77) | 2.09 (0.84-5.24) | 0.06 | ||
| COMBO high VAF | 34 (8.83-21.17) | 3.55 (1.21-10.33) | 0.0006 | ||
| PFS diagnosis |
|||||
|---|---|---|---|---|---|
| Median months (95% CI) | Univariate analysis |
Multivariate analysis |
|||
| HR (95% CI) | P | HR (95% CI) | P | ||
| Sex | |||||
| Female | 9 (3.53-14.47) | 1 | 1 | ||
| Male | 8 (3.78-12.21) | 1.01 (0.56-1.78) | 0.74 | 1.12 (0.63-2.00) | 0.70 |
| BRAF VAF | |||||
| HIGH | 16 (4.6-30.80) | 1 | 1 | ||
| LOW | 7 (5.11-8.91) | 2.00 (1.09-3.66) | 0.019 | 2.79 (1.35-4.58) | 0.001 |
| Therapy | |||||
| COMBO | 13 (4.37-21.63) | 1 | 1 | ||
| MONO | 5 (3.52-6.48) | 2.158 (1.10-4.21) | 0.004 | 2.92 (1.56-5.74) | 0.006 |
| Stage | |||||
| I-II | 12 (4.57-19.43) | 1 | 1 | ||
| III | 8 (1.88-14.12) | 1.15 (0.60-2.18) | 0.64 | 1.01 (0.39-1.81) | 0.87 |
| IV | 6 (5.07-10.92) | 1.99 (0.66-3.4) | 0.038 | 1.59 (0.60-2.96) | 0.24 |
| LDH | |||||
| High | 12 (2.703-21.297) | 1 | 1 | ||
| Low | 6 (4.998-7.002) | 0. | 0.004 | 0.41 (0.22-0.77) | 0.0001 |
| Therapy + VAF | |||||
| MONO low VAF | 3 (1.48-4.51) | 1 | |||
| MONO high VAF | 9 (3.85-16.55) | 3.05 (1.10-8.43) | 0.0037 | ||
| COMBO low VAF | 10 (3.18-16.24) | 4.38 (1.37-13.97) | 0.0001 | ||
| COMBO high VAF | 26 (10.54-41.46) | 5.10 (1.48-17.42) | 0.0001 | ||
P < 0.05 (in bold) are considered significant.
CI, confidence interval; HR, hazard ratio; LDH, lactate dehydrogenase; OS, overall survival; VAF, variant allele frequency.
Only patients with low VAF levels did not respond to the COMBO treatment, whereas patients with the highest VAF levels were also responders for the MONO treatment (PC/CR) (Figure 3B, and Table 1)
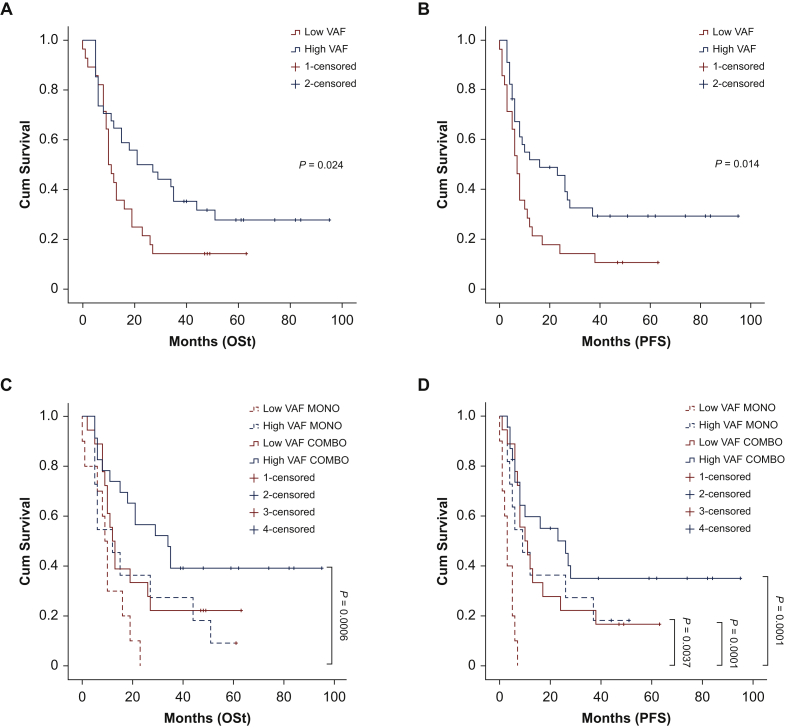
To estimate the influence of BRAF VAF on patient survival, we used the cut-off of 25% median VAF to dichotomize cases in low VAF and high VAF. Longer OSd, OSt, and PFS were observed in patients with high VAF (P = 0.046, P = 0.04, and P = 0.019, respectively) (Figure 4A and B and Supplementary Figure S5A, available at https://doi.org/10.1016/j.esmoop.2021.100133, Table 1). When patients were clustered according to VAF values (high versus low) and type of therapeutic approach (MONO versus COMBO), the COMBO-treated, high VAF patients showed significantly longer OSt and PFS compared with the other groups. However, the survival curves of COMBO-treated/low VAF patients and MONO-treated/high VAF patients tended to overlap. The worse OS and PFS rates were observed for MONO-treated/low VAF patients (Figure 4C and D and Supplementary Figure S5B available at https://doi.org/10.1016/j.esmoop.2021.100133).
Figure 4.
Survival data and the BRAF VAF.
(A) OSt survival curve for low and high VAF patients (cut-off 25%). High VAF curve shows a statistically higher survival compared with low VAF curve. (B) PFS survival curve for low and high VAF patients (cut-off 25%). High VAF curve indicates a statistically higher time to progression compared with the low VAF curve. (C) OSt survival curve for low VAF (<25%), high VAF (>25%), MONO- and COMBO-treated patients; high VAF-COMBO curve shows a statistically higher survival compared with the low VAF-COMBO curve, high VAF-COMBO dotted curve, and low VAF-MONO dotted curve. (D) PFS survival curve for low VAF (<25%), high VAF (>25%), MONO- and COMBO-treated patients. High VAF-COMBO solid curve shows a statistically higher survival compared with the low VAF-COMBO solid curve, high VAF-COMBO dotted curve, and low VAF-MONO dotted curve.
CR, complete response; Cum survival, cumulative survival; OSt, overall survival treatment onset; PFS, progression-free survival; PR, partial response; SD, stable disease; VAF, variant allele frequency.
BRAF VAF still remained an independent factor for longer OSt and PFS in multivariate analysis, including the VAF levels, the stage at diagnosis (I-II, III, IV), the sex, the age at diagnosis, the type of therapy, and the LDH level. Data of the analyses are reported in Table 1.
Discussion
Based on the improved clinical outcomes achieved by targeted therapies in melanoma patients, the assessment of BRAF mutational status has been recognized as a fundamental diagnostic tool for the choice of treatment. However, not all patients experience comparable disease control and survival rate. Using a quantitative method for BRAF mutation evaluation, we report a detailed analysis of BRAF mutations from a large cohort of different stage melanoma patients.
In our cohort, approximately half of the patients carried a BRAF hotspot mutation. The BRAF mutation prevalence results are in line with those previously reported.21, 22, 23 Heinzerling and colleagues observed a BRAF MR of ∼45%, using pyrosequencing to assess BRAF status in 187 melanoma patients.24 However, only a limited number of studies have been focused on the BRAF VAF.14, 15, 16, 17 In our analyses, the VAF of BRAF mutations followed a Gaussian distribution, with the average and the median level at 33% and 30%, respectively. Similarly, a previous report regarding a detailed BRAF analysis of lesions with >80% of tumor cells described a wide heterogeneity of VAF, ranging between 10% and 90%.25 Stagni and co-workers applying a correction for the copy number variation of the BRAF locus on lesions with >80% of tumor cells reported a BRAF VAF of ∼54%.19 This frequency, which is higher compared with our findings, can possibly be explained by the smaller cohort they analyzed (42 patients) and the larger number of high-grade melanomas.
The age of melanoma onset correlated with specific features of BRAF mutations. The MR was higher in tumors of younger patients, whereas older patients showed increased VAF levels. The documented correlation of sun-exposed body regions with the BRAF mutation incidence26 and VAF13 suggests a lifetime of ultraviolet rays exposure in elderly patients. Ultraviolet incidence may also explain the different body distribution of the BRAF VAF of melanomas. As a matter of fact, lesions with primary location in the trunk, limbs, and face/scalp showed a higher level of BRAF VAF compared with those with ocular and acral origin. The rarity of the BRAF mutation in ocular27 and acral melanomas28 can further support this difference. We also found a high BRAF MR and VAF in melanomas derived from unknown primary lesions. Gos et al. evidenced that the high incidence of BRAF and NRAS mutations, in the absence of KIT alterations, suggested a cutaneous origin from sun-damaged skin.29
We demonstrated a highly heterogeneous distribution of the BRAF mutations in the different metastatic subsets. Although the cutaneous secondary lesions were the most frequently mutated, the lymph node metastases displayed the highest BRAF VAF, thus supporting different pathways of anatomic and/or biological selections on BRAF-mutated cells. Accordingly, Adler et al. reported the lymph nodes as the main site of secondary lesions of BRAF-mutated primary melanomas.30 In visceral metastases, we found a reduced MR in agreement with other studies28 and VAF was not directly correlated with that of the primary site.31
Only limited data report the correlation between BRAF mutations and TIL infiltration. We showed a higher VAF in the non-brisk melanoma compared with the brisk lesions.32 A statistical association between the presence of TILs and the BRAF mutation has been previously reported.33 Similarly, half of primary BRAF-mutated melanomas fell into the non-brisk phenotype and ∼30% into the brisk group.34 Although the significance of TILs in primary melanomas is still debated, in vitro data have shown the importance of the immunomodulation associated with the BRAF-targeted therapy.35 Immunotherapy agents allow for durable responses only in a limited fraction of patients; therefore the combination with BRAF-targeted inhibitors can be a successful approach. Results from the IMspire150 trial showed a substantial increase of PFS in patients treated with combined anti-BRAF, anti-MEK, and atezolizumab immune checkpoint therapy, even if with inter-patient heterogeneity.36 Therefore, studying VAF and TILs could be of interest when selecting patients for therapeutic options, including immunotherapies.
Tumor thickness, evaluated as a continuous variable with the Breslow depth or as a discrete class using the Clark level, was reported as a clear negative prognostic factor for melanoma patients.33,37 However, no correlation between the BRAF MR and melanoma thickness has been previously described.38,39 Here we showed that thicker melanomas carry a higher BRAF VAF.
As shown earlier,40 no coexisting BRAF and NRAS activating mutations were detected in our cohort. Moreover, in line with a previous report,33 the NRAS VAF values were generally higher than those of BRAF, but did not correlate with sites of primary lesions, type of metastatic spreading, and lymphocytic infiltration. This evidence demonstrates that BRAF and NRAS mutations act differently and independently in melanoma progression.
With respect to the correlation with BRAF VAF and clinical outcomes, our findings agree with some of the previously reported data. Stagni and colleagues19 showed a strong correlation of BRAF VAF with PFS, but not with the response rate. Lebbe et al.16 found that the BRAF p.V600E mutation was able to predict responses to vemurafenib treatment in metastatic melanomas. However, Mesbah Ardakani and co-workers, studying only 33 metastatic patients treated with BRAF inhibitors, proved that OS and PFS were independent of the BRAF VAF.17 Similarly, no differences between low and high VAF patients were reported by Satzger and colleagues, who scored 75 melanomas using a BRAF VAF cut-off of 18%.18
In our experience, analyzing 62 metastatic patients with available follow-up data and using the median level of BRAF VAF (25%), we demonstrated that a lower BRAF VAF identified patients with a faster progression and a lower response rate. Patients with high VAF showed prolonged PFS and OSt in multivariate analyses, proving that BRAF VAF can be used to select patients who might benefit from anti-BRAF strategies. The impact of the stage at diagnosis, and possibly the multiple treatments received during the patients’ oncological history, led to a lesser VAF effect on the OSd in the Cox model. Nevertheless, when patients were classified according to a composed model (type of therapy and the BRAF VAF discrete level), high VAF identified patients with a better outcome for all the survival metrics. Patients with high BRAF VAF treated with MONO therapy displayed comparable survival rates compared with low VAF melanomas treated with the COMBO, suggesting that low BRAF VAF might induce only partial activation of the downstream MEK with a consequent reduced responsiveness to the COMBO treatment.
Even though our study has produced innovative results, we are aware they still present some limitations. Mostly, the strong heterogeneity in the VAF distribution of the responder group suggests the need for further investigation to increase the significance of the BRAF VAF as a molecular marker. Secondly, it is a retrospective study on melanoma lesions from different origins, which is a confounding factor that we aimed to reduce by applying the Cox multivariate model. Moreover, we were able to obtain follow-up data for only 62 patients (40%) of the 153 mutated patients. In addition, molecular profiling of melanomas carried out by next-generation sequencing (NGS) has recently demonstrated that low-frequency pathogenic alterations affecting genes involved in several cancer-related pathways coexist differently with BRAF mutations.41,42 Thus, we cannot rule out that any of these rare mutations might modulate the correlation of BRAF VAF with the clinical and pathological features of melanomas.
Pyrosequencing represents a well-assessed technique to evaluate BRAF VAF. Nevertheless, alternative methods could be validated in the future for a wider clinical application of this analysis. Immunohistochemical detection of BRAF V600E by VE1 antibody could be a fast and cost-effective approach.43, 44, 45, 46 However, at present, there are important limitations due to a lack of VE1 immunostaining standardization and information concerning the relationship between BRAF gene quantitative allelic variations and its protein expression level.
In conclusion, we confirmed the prevalence of BRAF alterations and we demonstrated that high BRAF VAF was associated with patient age, melanoma thickness, non-brisk TILs, and lymph node metastases compared with the distant ones (cutaneous and visceral). More importantly, our results strongly demonstrate the significant prognostic value of high BRAF VAF (>25%) in patients treated with a combination of anti-BRAF and anti-MEK treatment.
To corroborate these data, the evaluation of BRAF VAF in other prospective, independent, and larger cohorts is warranted. However, we envision that along with qualitative assays, BRAF VAF could be used as a biomarker for the selection of BRAF-targeting agents.
Acknowledgements
EB is the recipient of a PhD fellowship from the Department of Medical Sciences, University of Torino (‘Dipartimenti di Eccellenza 2018-2022’, Project No. D15D18000410001), thanks to tutor Prof. Caterina Marchiò. The authors also thank the technical support of Dr Simona De Rosa.
Funding
This work was supported by FPRC 5x1000 MIUR 2015 ‘Futuro’ to AS (no grant number).
Disclosure
The authors have declared no conflicts of interest.
Data sharing
All data relevant to the study are included in the article or uploaded as online supplemental information. Individual patient data cannot be shared.
Disclaimer
The views expressed are those of the author(s) and not necessarily those of the Candiolo Cancer Institute or the University of Turin.
Ethics approval
Clinical-pathological data were recovered after obtaining the informed consent from all the patients, approved by the medical ethical committee of the FPO-IRCCS, and carried out according to the principles of the Declaration of Helsinki.
Contributor Information
A. Sapino, Email: anna.sapino@ircc.it.
T. Venesio, Email: tiziana.venesio@ircc.it.
Supplementary data
References
- 1.Society AC A.C. Cancer Facts & Figures 2019. 2019. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2019/cancer-facts-and-figures-2019.pdf Cited 2018. Available at.
- 2.Davies H., Bignell G.R., Cox C. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Batus M., Waheed S., Ruby C. Optimal management of metastatic melanoma: current strategies and future directions. Am J Clin Dermatol. 2013;14(3):179–194. doi: 10.1007/s40257-013-0025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas N Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omholt K., Platz A., Kanter L. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9(17):6483–6488. [PubMed] [Google Scholar]
- 6.Chapman P.B., Hauschild A., Robert C. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert C., Karaszewska B., Schachter J. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 8.Larkin J., Ascierto P.A., Dreno B. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371(20):1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 9.Cheng L., Lopez-Beltran A., Massari F. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod Pathol. 2018;31(1):24–38. doi: 10.1038/modpathol.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugdahl E., Kalvenes M.B., Puntervoll H.E. BRAF-V600E expression in primary nodular melanoma is associated with aggressive tumour features and reduced survival. Br J Cancer. 2016;114(7):801–808. doi: 10.1038/bjc.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Sousa V.M.L., Carvalho L. Heterogeneity in lung cancer. Pathobiology. 2018;85(1-2):96–107. doi: 10.1159/000487440. [DOI] [PubMed] [Google Scholar]
- 12.Molinari C., Marisi G., Passardi A. Heterogeneity in colorectal cancer: a challenge for personalized medicine? Int J Mol Sci. 2018;19(12):3733. doi: 10.3390/ijms19123733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turashvili G., Brogi E. Tumor heterogeneity in breast cancer. Front Med (Lausanne) 2017;4:227. doi: 10.3389/fmed.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yancovitz M., Litterman A., Yoon J. Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations in primary and metastatic melanoma. PLoS One. 2012;7(1):e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venesio T., Chiorino G., Balsamo A. In melanocytic lesions the fraction of BRAF V600E alleles is associated with sun exposure but unrelated to ERK phosphorylation. Mod Pathol. 2008;21(6):716–726. doi: 10.1038/modpathol.2008.41. [DOI] [PubMed] [Google Scholar]
- 16.Lebbe C., How-Kit A., Battistella M. BRAF(V600) mutation levels predict response to vemurafenib in metastatic melanoma. Melanoma Res. 2014;24(4):415–418. doi: 10.1097/CMR.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 17.Mesbah Ardakani N., Leslie C., Grieu-Iacopetta F. Clinical and therapeutic implications of BRAF mutation heterogeneity in metastatic melanoma. Pigment Cell Melanoma Res. 2017;30(2):233–242. doi: 10.1111/pcmr.12569. [DOI] [PubMed] [Google Scholar]
- 18.Satzger I., Marks L., Kerick M. Allele frequencies of BRAFV600 mutations in primary melanomas and matched metastases and their relevance for BRAF inhibitor therapy in metastatic melanoma. Oncotarget. 2015;6(35):37895–37905. doi: 10.18632/oncotarget.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stagni C., Zamuner C., Elefanti L. BRAF gene copy number and mutant allele frequency correlate with time to progression in metastatic melanoma patients treated with MAPK inhibitors. Mol Cancer Ther. 2018;17(6):1332–1340. doi: 10.1158/1535-7163.MCT-17-1124. [DOI] [PubMed] [Google Scholar]
- 20.MacArthur J.A., Morales J., Tully R.E. Locus Reference Genomic: reference sequences for the reporting of clinically relevant sequence variants. Nucleic Acids Res. 2014;42:D873–D878. doi: 10.1093/nar/gkt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer J., Buttner P., Murali R. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res. 2011;24(2):345–351. doi: 10.1111/j.1755-148X.2011.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heppt M.V., Siepmann T., Engel J. Prognostic significance of BRAF and NRAS mutations in melanoma: a German study from routine care. BMC Cancer. 2017;17(1):536. doi: 10.1186/s12885-017-3529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pracht M., Mogha A., Lespagnol A. Prognostic and predictive values of oncogenic BRAF, NRAS, c-KIT and MITF in cutaneous and mucous melanoma. J Eur Acad Dermatol Venereol. 2015;29(8):1530–1538. doi: 10.1111/jdv.12910. [DOI] [PubMed] [Google Scholar]
- 24.Heinzerling L., Baiter M., Kuhnapfel S. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br J Cancer. 2013;109(11):2833–2841. doi: 10.1038/bjc.2013.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helias-Rodzewicz Z., Funck-Brentano E., Baudoux L. Variations of BRAF mutant allele percentage in melanomas. BMC Cancer. 2015;15:497. doi: 10.1186/s12885-015-1515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas N.E., Edmiston S.N., Alexander A. Number of nevi and early-life ambient UV exposure are associated with BRAF-mutant melanoma. Cancer Epidemiol Biomarkers Prev. 2007;16(5):991–997. doi: 10.1158/1055-9965.EPI-06-1038. [DOI] [PubMed] [Google Scholar]
- 27.Malaponte G., Libra M., Gangemi P. Detection of BRAF gene mutation in primary choroidal melanoma tissue. Cancer Biol Ther. 2006;5(2):225–227. doi: 10.4161/cbt.5.2.2429. [DOI] [PubMed] [Google Scholar]
- 28.Saldanha G., Potter L., Daforno P. Cutaneous melanoma subtypes show different BRAF and NRAS mutation frequencies. Clin Cancer Res. 2006;12(15):4499–4505. doi: 10.1158/1078-0432.CCR-05-2447. [DOI] [PubMed] [Google Scholar]
- 29.Gos A., Jurkowska M., van Akkooi A. Molecular characterization and patient outcome of melanoma nodal metastases and an unknown primary site. Ann Surg Oncol. 2014;21(13):4317–4323. doi: 10.1245/s10434-014-3799-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler N.R., Wolfe R., Kelly J.W. Tumour mutation status and sites of metastasis in patients with cutaneous melanoma. Br J Cancer. 2017;117(7):1026–1035. doi: 10.1038/bjc.2017.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doma V., Karpati S., Raso E. Dynamic and unpredictable changes in mutant allele fractions of BRAF and NRAS during visceral progression of cutaneous malignant melanoma. BMC Cancer. 2019;19(1):786. doi: 10.1186/s12885-019-5990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnes T.A., Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117(4):451–460. doi: 10.1038/bjc.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edlundh-Rose E., Egyhazi S., Omholt K. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16(6):471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 34.Leslie C., Bowyer S.E., White A. FOXP3+ T regulatory lymphocytes in primary melanoma are associated with BRAF mutation but not with response to BRAF inhibitor. Pathology. 2015;47(6):557–563. doi: 10.1097/PAT.0000000000000314. [DOI] [PubMed] [Google Scholar]
- 35.Boni A., Cogdill A.P., Dang P. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010;70(13):5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- 36.Gutzmer R., Stroyakovskiy D., Gogas H. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;395(10240):1835–1844. doi: 10.1016/S0140-6736(20)30934-X. [DOI] [PubMed] [Google Scholar]
- 37.Bhatia P., Friedlander P., Zakaria E.A. Impact of BRAF mutation status in the prognosis of cutaneous melanoma: an area of ongoing research. Ann Transl Med. 2015;3(2):24. doi: 10.3978/j.issn.2305-5839.2014.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S.Y., Kim S.N., Hahn H.J. Metaanalysis of BRAF mutations and clinicopathologic characteristics in primary melanoma. J Am Acad Dermatol. 2015;72(6):1036–1046.e2. doi: 10.1016/j.jaad.2015.02.1113. [DOI] [PubMed] [Google Scholar]
- 39.Lee J.H., Choi J.W., Kim Y.S. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164(4):776–784. doi: 10.1111/j.1365-2133.2010.10185.x. [DOI] [PubMed] [Google Scholar]
- 40.Lokhandwala P.M., Tseng L.H., Rodriguez E. Clinical mutational profiling and categorization of BRAF mutations in melanomas using next generation sequencing. BMC Cancer. 2019;19(1):665. doi: 10.1186/s12885-019-5864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byeon S., Cho H.J., Jang K.T. Molecular profiling of Asian patients with advanced melanoma receiving check-point inhibitor treatment. ESMO Open. 2020;6(1):100002. doi: 10.1016/j.esmoop.2020.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ticha I., Hojny J., Michalkova R. A comprehensive evaluation of pathogenic mutations in primary cutaneous melanomas, including the identification of novel loss-of-function variants. Sci Rep. 2019;9(1):17050. doi: 10.1038/s41598-019-53636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo M.C., Paterson A., Maraka J. A UK feasibility and validation study of the VE1 monoclonal antibody immunohistochemistry stain for BRAF-V600E mutations in metastatic melanoma. Br J Cancer. 2016;115(2):223–227. doi: 10.1038/bjc.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kakavand H., Walker E., Lum T. BRAF(V600E) and NRAS(Q61L/Q61R) mutation analysis in metastatic melanoma using immunohistochemistry: a study of 754 cases highlighting potential pitfalls and guidelines for interpretation and reporting. Histopathology. 2016;69(4):680–686. doi: 10.1111/his.12992. [DOI] [PubMed] [Google Scholar]
- 45.Orchard G.E., Wojcik K., Rickaby W. Immunohistochemical detection of V600E BRAF mutation is a useful primary screening tool for malignant melanoma. Br J Biomed Sci. 2019;76(2):77–82. doi: 10.1080/09674845.2019.1592885. [DOI] [PubMed] [Google Scholar]
- 46.Vallee A., Denis-Musquer M., Herbreteau G. Prospective evaluation of two screening methods for molecular testing of metastatic melanoma: diagnostic performance of BRAF V600E immunohistochemistry and of a NRAS-BRAF fully automated real-time PCR-based assay. PLoS One. 2019;14(8):e0221123. doi: 10.1371/journal.pone.0221123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.