Abstract
Atherosclerosis is a primary cause of multiple types of cardiovascular disease, including myocardial infarction. In addition, injury of human umbilical vein endothelial cells (HUVECs) can lead to the development of atherosclerosis. Circular (circ)RNAs participate in atherosclerosis. It has previously been shown that circRNA cSMARCA5 (hsa_circ_0001445) expression is downregulated in atherosclerosis. However, the effects of hsa_circ_0001445 on the proliferation of HUVECs remain unclear. In order to mimic atherosclerosis in vitro, HUVECs were treated with oxidized low-density lipoprotein (oxLDL). The expression levels of specific genes and proteins were detected in HUVECs by reverse transcription-quantitative PCR and western blot analysis, respectively. Cell proliferation was assessed by Cell Counting Kit-8 and 5-Ethynyl-2′-deoxyuridine staining. Cell apoptosis and 5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine staining were examined by flow cytometry. In addition, the association between hsa_circ_0001445 and serine/arginine-rich splicing factor 1 (SRSF1) was investigated by RNA pull-down assay. hsa_circ_0001445 expression was downregulated in oxLDL-treated HUVECs. Moreover, oxLDL-induced inhibition of HUVEC proliferation was significantly reversed by overexpression of hsa_circ_0001445. oxLDL notably inhibited tube formation and mitochondrial membrane potential in HUVECs, while these effects were markedly reversed by hsa_circ_0001445 overexpression. Furthermore, overexpression of hsa_circ_0001445 reversed oxLDL-induced activation of β-catenin by binding to SRSF1. Collectively, these data demonstrated that overexpression of hsa_circ_0001445 reversed oxLDL-induced inhibition of HUVEC proliferation via activation of the SRSF1/β-catenin axis. These findings may provide novel targets for the treatment of atherosclerosis.
Keywords: atherosclerosis, hsa_circ_0001445, serine/arginine-rich splicing factor 1, β-catenin
Introduction
Atherosclerosis is a primary cause of myocardial infarction worldwide and atherosclerosis was present in ~60% of the people >60 years old in European and American countries in 2019 (1,2). Atherosclerosis is associated with several risk factors, including dysfunction of human umbilical vein endothelial cells (HUVECs) (3). Inappropriate proliferation, apoptosis, injury and migration of HUVECs lead to the progression of atherosclerosis (4,5). Therefore, the maintenance of the function of HUVECs is important for the treatment of atherosclerosis.
Circular (circ)RNAs are a class of endogenous RNA molecules with a stable closed loop structure (6). circRNAs serve important regulatory roles in cell biological functions, such as protein synthesis, gene expression and post-transcriptional modification (6). circRNAs have been shown to play a key role in the function of HUVECs during the progression of atherosclerosis. For example, Qin et al (7) demonstrated that knockdown of hsa_circ_0003645 expression alleviates inflammation and apoptosis in oxidized low-density lipoprotein (oxLDL)-induced HUVECs. Moreover, Wang et al (8) indicated that circ_0124644 exacerbates oxLDL-induced injury in HUVECs via sponging miR-149-5p in atherosclerosis. In addition, hsa_circ_0001445 expression is notably downregulated in atherosclerosis (9). However, the function of hsa_circ_0001445 in HUVECs remains unclear.
Serine/arginine-rich splicing factors (SRSFs) are well-characterized for their roles in atherosclerosis and are composed of one or two RNA recognition motifs and a serine/arginine-rich domain (10). Among these SRSFs, SRSF1 is a prototypical splicing factor that specifically binds to exonic enhancers and stimulates splicing (11). In addition, it has been reported that SRSF1 binds to hsa_circ_0001445 (12). However, the association between hsa_circ_0001445 and SRSF1 remains unclear.
Based on this evidence, the present study aimed to investigate the biological function of hsa_circ_0001445 in atherosclerosis in vitro.
Materials and methods
Cell culture and treatment
HUVECs (cat. no. CRL1730) were obtained from the American Type Culture Collection and cultured in RPMI-1640 medium, supplemented with 10% FBS and 2 mM glutamine (Sigma-Aldrich; Merck KGaA) at 37°C, 5% CO2. In order to establish an in vitro model of atherosclerosis, HUVECs were treated with oxLDL (50, 75 or 100 µg/ml) for 48 h at 37°C.
Cell transfection
The pcDNA3.1 vector (1 µg/µl), pDNA3.1-hsa_circ_0001445 (1 µg/µl) and pDNA3.1-SRSF1 (1 µg/µl) were purchased from Guangzhou RiboBio Co., Ltd., and transfected into HUVECs using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. After 24 h of transfection, transfected cells were used in subsequent experiments. The efficiency of hsa_circ_0001445 overexpression was detected by reverse transcription-quantitative (RT-q)PCR and the efficiency of SRSF1 overexpression was investigated by RT-qPCR and western blotting.
RT-qPCR
Total RNA was extracted from cell lines with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc.) according to the manufacturer's instructions. Then, RT-qPCR was performed using a SYBR® Premix Ex Taq™ II kit (Takara Bio, Inc.) on a 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling conditions: 60°C for 1 min, 90°C for 15 min, followed by 40 cycles of 90°C for 15 sec and 55°C for 60 sec. β-actin was used as the internal control. The primer sequences used were as follows: hsa_circ_0001445 forward, 5′-GAGAAAAACAAAAGGGAGGCTT-3′ and reverse, 5′-TTGAAGCAAACACATGTGTTGC-3′; SRSF1 forward, 5′-GCGACGGCTATGATTACGATG-3′ and reverse, 5′-ACATACATCACCTGCTTCACGC-3′; and β-actin forward, 5′-GTCCACCGCAAATGCTTCTA-3′ and reverse, 5′-TGCTGTCACCTTCACCGTTC-3′. The 2−ΔΔCq method (13) was used to measure the relative expression levels of genes.
Cell Counting Kit (CCK)-8 assay
HUVECs were seeded in 96-well plates (5×103 per well) overnight. The cells were treated with 100 µg/ml oxLDL, hsa_circ_0001445 OE or oxLDL + hsa_circ_0001445 OE for 0, 24, 48 and 72 h. A total of 10 µl CCK-8 (Beyotime Institute of Biotechnology) was added to each well and the cells were incubated for 2 h at 37°C. Finally, the absorbance of HUVECs was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Oil red O staining of lipid accumulation in cells
Culture media was removed from HUVECs and cells were washed twice with PBS. The cells were then fixed with 4% paraformaldehyde at room temperature for 30 min. Oil Red O working solution was prepared fresh from 0.5% (w/v) stock solution, which was diluted with water at a ratio of 6:4 (Oil Red O:water). Cells were then incubated with Oil Red O for 30 min at room temperature, washed gently with PBS three times to remove excess non-specific staining and observed under a light microscope (magnification, ×200; Olympus Corporation).
RNA pull-down assay
RNA pull-down assay was performed using Biotin RNA Labeling Mix (Roche Diagnostics) to transcribe and label probe-control (ctrl) or probe-hsa_circ_0001445 in vitro. An RNA structure buffer (Thermo Fisher Scientific, Inc.) was used to induce secondary structure formation from the biotin-labeled RNAs. Streptavidin beads (Thermo Fisher Scientific, Inc.) were washed three times with 500 µl RNA immunoprecipitation washing buffer (Thermo Fisher Scientific, Inc.) and added to the biotinylated RNA samples at 4°C overnight. The mixture was separated by a magnetic field to obtain streptavidin bead-RNA complexes. Subsequently, lysates of HUVECs (60 µl) were added to the complexes and incubated on a rotating platform at room temperature for 1 h. The incubated mixture was separated with a magnetic field to obtain streptavidin bead-RNA-protein complexes.
In vitro angiogenesis assay
Angiogenesis assay was conducted using 6-well plates that were pre-coated with Matrigel (BD Biosciences) at 37°C for 24 h. Subsequently, 1×105 HUVECs were seeded into each well. Following 24 h cell culture at 37°C, the formation of capillary-like structures was photographed and the branchpoints were counted. The tube length was analyzed by the AxioVision Rel software version 4.8 (Carl Zeiss AG).
Transwell assay
Transwell chambers (8 mm pore; Corning, Inc.) were used to determine the rate of cell migration. The aforementioned cell culture medium containing 4×105 cells/200 µl was added into the upper chamber. Cell culture medium supplemented with 10% FBS was added to the lower chamber of the well. Following culture at 37°C for 24 h, the migrated cells were fixed with 4% paraformaldehyde for 20 min at room temperature, followed by staining with 4% crystal violet solution for 5 min at room temperature. The migrated cells were counted using a light microscope (200× magnification; Olympus Corporation).
Western blotting
Total protein was isolated from cell lysates using RIPA buffer (Shanghai GenePharma Co., Ltd.) and quantified by BCA protein assay kit (Beyotime Institute of Biotechnology). The proteins (40 µg per lane) were resolved on 10% SDS-PAGE and subsequently transferred onto PVDF (Bio-Rad Laboratories, Inc.) membranes. Following blocking with 5% skimmed milk in TBST (0.05% Tween-20) at room temperature for 1 h, the membranes were incubated with primary antibodies (all 1:1,000) at 4°C overnight. The following day, membranes were incubated with HRP-conjugated secondary anti-rabbit antibody (cat. no. ab7090; 1:5,000 Abcam) at room temperature for 1 h. The membranes were scanned using an Odyssey Imaging system and analyzed with Odyssey v2.0 software (LI-COR Biosciences). The primary antibodies used in the present study were as follows: Anti-β-catenin (cat. no. ab223075; Abcam), anti-SRSF1 (cat. no. 32-4500; Thermo Fisher Scientific, Inc.), anti-E-cadherin (cat. no. ab40772; Abcam), anti-Bcl-2 (cat. no. ab182858; Abcam), anti-Bax (cat. no. ab182733; Abcam) and anti-β-actin (cat. no. ab8226; Abcam). β-actin was used as an internal control. Enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.) was used to visualize the protein bands. ImageJ software (version 2.0; National Institutes of Health) was used to quantify the intensity of the bands.
Measurement of mitochondrial membrane potential (MMP)
The changes in MMP were assessed via 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine (JC-1) staining using a FACScan flow cytometer (BD Biosciences). The cells were plated into 6-well plates (3×105 cells/well) overnight, harvested, washed twice with PBS and finally resuspended in 0.5 ml complete medium containing 2 µM JC-1 at 37°C for 20 min.
Immunofluorescence analysis
HUVECs (5×103) were seeded in 24-well plates and incubated overnight. The following day, the cells were treated with oxLDL, hsa_circ_0001445 OE, oxLDL + hsa_circ_0001445 OE or oxLDL + hsa_circ_0001445 OE + SRSF1 OE at 37°C for 72 h, blocked with 10% goat serum (Invitrogen; Thermo Fisher Scientific, Inc.) for 30 min at room temperature and finally incubated with anti-5-ethynyl-2′-deoxyuridine (EdU) antibody (1:1,000; cat. no. ab222421; Abcam) at 4°C overnight, followed by incubation with HRP-conjugated goat anti-rabbit IgG (1:5,000; ab7090; Abcam) at 37°C for 1 h. Subsequently, the nuclei were stained with DAPI (5 µg/µl, Beyotime Institute of Biotechnology) for 5 min at room temperature. Finally, the cells were observed using a fluorescence microscope (magnification, ×200; Olympus CX23; Olympus Corporation). The images was analyzed by ImageJ software (version 2.0; National Institutes of Health).
Cell apoptosis analysis
HUVECs were trypsinized, washed with PBS and resuspended in Annexin V binding buffer. The cells were stained with 5 µl FITC (BD Biosciences) and 5 µl propidium iodide (BD Biosciences) at 4°C for 15 min. Finally, they were analyzed using flow cytometry (BD Biosciences) to assess the cell apoptotic rate. The data was quantified by FlowJo (v7.6.5; FlowJo LLC).
Statistical analysis
Each experiment was performed in three independent repeats. The data are presented as the mean ± SD. Comparisons between two groups were analyzed using unpaired Student's t-test. Comparisons between multiple groups were performed using one-way ANOVA followed by post hoc Tukey's test. The analysis was performed using GraphPad Prism 7 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
oxLDL-induced inhibition of HUVEC proliferation is reversed by hsa_circ_0001445 OE
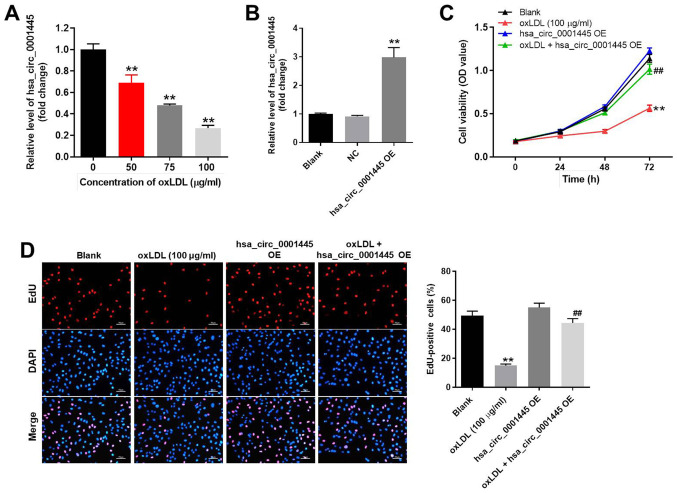
In order to mimic atherosclerosis in vitro, HUVECs were treated with different concentrations of oxLDL (50, 75 or 100 µg/ml). oxLDL significantly inhibited the expression levels of hsa_circ_0001445 in HUVECs in a dose-dependent manner (Fig. 1A). Based on this evidence, 100 µg/ml oxLDL was selected for use in subsequent experiments. Expression levels of hsa_circ_0001445 in HUVECs were notably upregulated by pcDNA3.1-hsa_circ_0001445 (Fig. 1B). HUVEC viability was significantly decreased by oxLDL; this effect was reversed in the presence of hsa_circ_0001445 OE (Fig. 1C). hsa_circ_0001445 OE reversed the antiproliferative effect of oxLDL on HUVECs (Fig. 1D). Taken together, the results indicated that oxLDL-induced inhibition of HUVEC proliferation was reversed by hsa_circ_0001445 OE.
Figure 1.
hsa_circ_0001445 OE promotes proliferation of oxLDL-treated HUVECs. (A) HUVECs were treated with 0, 50, 75 or 100 µg/ml oxLDL for 48 h. Then, the expression of hsa_circ_0001445 in HUVECs was detected by RT-qPCR. (B) HUVECs were transfected with hsa_circ_0001445 OE for 24 h. Then, the expression of hsa_circ_0001445 in HUVECs was measured by RT-qPCR. (C) HUVECs were treated with oxLDL (100 µg/ml), hsa_circ_0001445 OE or oxLDL + hsa_circ_0001445 OE for 0, 24, 48 or 72 h. The viability of HUVECs was detected by Cell Counting Kit-8 assay. (D) Proliferation of HUVECs was detected by EdU staining (red). The number of EdU-positive cells was calculated. Blue immunofluorescence indicates DAPI. **P<0.01 vs. blank, ##P<0.01 vs. oxLDL (100 µg/ml). OE, overexpression; oxLDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; RT-q, reverse transcription-quantitative; EdU, 5-ethynyl-2′-deoxyuridine; OD, optical density; NC, negative control.
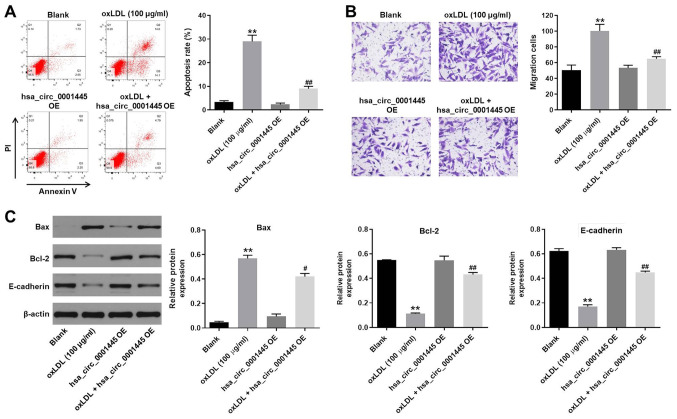
hsa_circ_0001445 OE reverses oxLDL-induced apoptosis and migration of HUVECs
In order to detect cell apoptosis, flow cytometry was performed. oxLDL induced apoptosis of HUVECs, while its apoptotic effect was significantly inhibited by hsa_circ_0001445 OE (Fig. 2A). oxLDL-induced increase in cell migration was inhibited by hsa_circ_0001445 OE (Fig. 2B). In addition, the expression levels of Bax in HUVECs were upregulated by oxLDL, while this effect was reversed by hsa_circ_0001445 OE (Fig. 2C). By contrast, the oxLDL-induced decrease in Bcl-2 and E-cadherin expression levels was partially rescued in the presence of hsa_circ_0001445 OE (Fig. 2C). Taken together, the data demonstrated that hsa_circ_0001445 OE reversed oxLDL-induced apoptosis and migration of HUVECs.
Figure 2.
hsa_circ_0001445 OE reverses oxLDL-induced apoptosis and migration of HUVECs. (A) Apoptosis of HUVECs was tested by flow cytometry. (B) Migration of HUVECs was measured by Transwell assay. (C) Protein expression levels of Bax, Bcl-2 and E-cadherin in HUVECs were detected by western blot analysis. The relative expression levels were normalized to β-actin. **P<0.01 vs. blank, #P<0.05, ##P<0.01 vs. oxLDL (100 µg/ml). OE, overexpression; oxLDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; E-cadherin, epithelial cadherin.
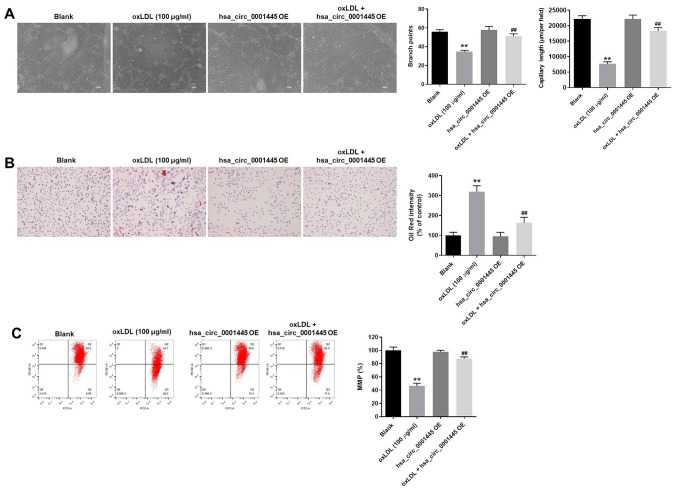
hsa_circ_0001445 OE inhibits progression of angiogenesis in vitro
The role of hsa_circ_0001445 in atherosclerosis was investigated by angiogenesis assay. The branch points of capillary-like structures in HUVECs were significantly decreased by oxLDL treatment, while this effect was improved in the presence of hsa_circ_0001445 OE (Fig. 3A). Similarly, oxLDL-induced decrease in tube length of HUVECs was reversed by hsa_circ_0001445 OE (Fig. 3A). In addition, hsa_circ_0001445 OE relieved oxLDL-induced lipid accumulation and mitochondrial injury in HUVECs (Fig. 3B and C) and significantly inhibited the progression of angiogenesis in vitro.
Figure 3.
hsa_circ_0001445 OE inhibits progression of angiogenesis in vitro. (A) Formation of capillary-like structures was photographed in HUVECs. The branch points were calculated. The capillary length was tested. (B) Lipid accumulation in HUVECs was tested by Oil Red O staining. (C) Changes in MMP were measured by 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine staining using a FACSan flow cytometer. The ratio of MMP was calculated. **P<0.01 vs. blank, ##P<0.01 vs. oxLDL (100 µg/ml). OE, overexpression; HUVECs, human umbilical vein endothelial cells; MMP, mitochondrial membrane potential.
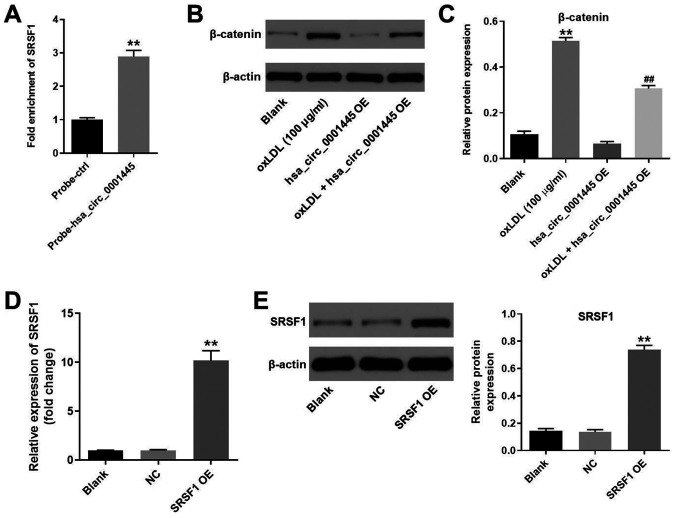
hsa_circ_0001445 binds to SRSF1 and reverses oxLDL-induced activation of β-catenin
It has been shown that hsa_circ_0001445 binds to SRSF1 (12,14). Therefore, the present study investigated the association between hsa_circ_0001445 expression levels and SRSF1 in HUVECs by RNA pull-down assay. As expected, the fold enrichment of SRSF1 in HUVECs was increased in probe-hsa_circ_0001445 compared with that in probe-ctrl cells (Fig. 4A). Moreover, the protein levels of β-catenin in HUVECs were upregulated by oxLDL (Fig. 4B and C). However, the effects of oxLDL on β-catenin expression were partially inhibited in the presence of hsa_circ_0001445 OE (Fig. 4B and C). HUVECs were transfected with pcDNA3.1-SRSF1, and the efficiency of cell transfection was tested. Expression of SRSF1 in HUVECs was significantly upregulated by pcDNA3.1-SRSF1 (Fig. 4D and E). In summary, the data demonstrated that hsa_circ_0001445 reversed oxLDL-induced activation of β-catenin via binding to SRSF1.
Figure 4.
hsa_circ_0001445 binds to SRSF1 and reverses oxLDL-induced activation of β-catenin. (A) RNA pull-down assay was performed to investigate the association between hsa_circ_0001445 and SRSF1. (B) Protein levels of SRSF1 in HUVECs were tested by western blot analysis. (C) Relative expression levels were normalized to β-actin. (D) HUVECs were transfected with pcDNA3.1 or pcDNA3.1-SRSF1. (E) Efficiency of cell transfection was tested by reverse transcription-quantitative PCR and western blot analysis. The relative expression levels were y normalized to β-actin. **P<0.01 vs. blank, ##P<0.01 vs. oxLDL (100 µg/ml). SRSF1, serine/arginine-rich splicing factor 1; oxLDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; ctrl, control; NC, negative control; OE, overexpression.
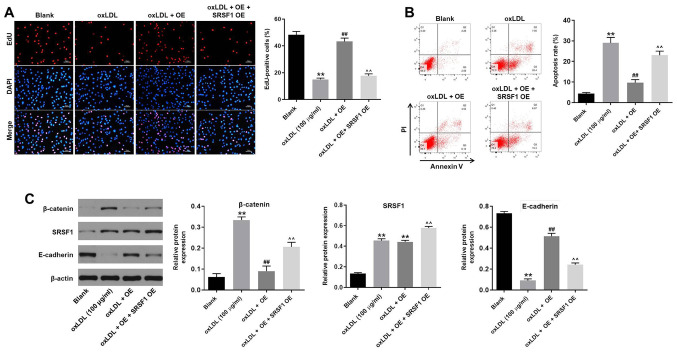
hsa_circ_0001445 OE reverses oxLDL-induced HUVEC proliferation inhibition by binding to SRSF1
In order to assess the association between hsa_circ_0001445 and SRSF1, EdU staining and flow cytometry were performed. hsa_circ_0001445 OE reversed the antiproliferative effect of oxLDL on HUVECs, while SRSF1 OE reversed this effect (Fig. 5A). Moreover, SRSF1 OE partially rescued the antiapoptotic effects of hsa_circ_0001445 on oxLDL-treated HUVECs (Fig. 5B), whereas oxLDL-induced activation of β-catenin was significantly inhibited in the presence of hsa_circ_0001445 OE. hsa_circ_0001445 OE exerted limited effects on the expression of SRSF1 in oxLDL-treated HUVECs (Fig. 5C). By contrast, protein levels of E-cadherin in HUVECs were inhibited in the presence of oxLDL, while this effect was significantly reversed by hsa_circ_0001445 OE (Fig. 5C). Furthermore, the effects of hsa_circ_0001445 on SRSF1, E-cadherin and β-catenin were significantly inhibited by SRSF1 OE (Fig. 5C). Taken together, the results indicated that hsa_circ_0001445 OE reversed oxLDL-induced inhibition of HUVEC proliferation via binding to SRSF1.
Figure 5.
hsa_circ_0001445 OE reverses oxLDL-induced inhibition of HUVEC growth via binding to SRSF1. (A) HUVECs were treated with oxLDL (100 µg/ml), hsa_circ_0001445 OE, oxLDL + hsa_circ_0001445 OE or oxLDL + hsa_circ_0001445 OE + SRSF1 OE. Then, the proliferation of HUVECs was tested by EdU staining. (B) Apoptosis of HUVECs was tested by flow cytometry. (C) Protein expression levels of β-catenin, SRSF1 and E-cadherin in HUVECs were detected by western blot analysis. The relative expression levels were normalized to β-actin. **P<0.01 vs. blank, ##P<0.01 vs. oxLDL (100 µg/ml), ^^P<0.01 vs. oxLDL + OE. OE, overexpression; oxLDL, oxidized low-density lipoprotein; HUVEC, human umbilical vein endothelial cells; SRSF1, serine/arginine-rich splicing factor 1; EdU, 5-ethynyl-2′-deoxyuridine; E-cadherin, epithelial cadherin.
Discussion
Atherosclerosis is a complex multicellular process involving vascular injury and atheroma formation, which is associated with vascular inflammation, leading to increased expression levels of pro-inflammatory cytokines, pentraxin-3, monocyte chemotactic protein, vascular cell adhesion molecule-1, intercellular adhesion molecule-1 and E-selectin in monocytes and endothelial cells (15,16). During the progression of atherosclerosis, oxLDL induces lipid accumulation in macrophages, thus leading to the formation of foam cells (17). In the present study, it was demonstrated that hsa_circ_0001445 expression was significantly decreased in oxLDL-treated HUVECs. It has been previously shown that hsa_circ_0001445 participates in the development of certain diseases. For example, hsa_circ_0001445 serves as a key biomarker for osteoporosis in postmenopausal women (18) and can modulate VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme by binding to SRSF1 (12). The present study investigated the biological function of hsa_circ_0001445 in atherosclerosis, revealing that hsa_circ_0001445 may serve as an inhibitor during the progression of this disease.
The present study investigated the mechanism by which hsa_circ_0001445 OE mediates the progression of atherosclerosis. In this study, SRSF1 could bind to hsa_circ_0001445. SRSF1 is involved in several cellular processes. For instance, LncRNA GASAL1 can interact with SRSF1 to regulate trophoblast cell proliferation, invasion, and apoptosis via the mTOR signaling (19); Sun and Hu (20) found that SRSF1 upregulation could induce the apoptosis of cardiomyocytes. It has been reported that SRSF1 is a risk factor involved in coronary disease, including atherosclerosis (21). In addition, previous studies have shown that hsa_circ_0001445 binds to SRSF1 in glioblastoma multiforme (12,14). The present study demonstrated the association between hsa_circ_0001445 and SRSF1 in HUVECs, confirming that hsa_circ_0001445 reversed oxLDL-induced inhibition of HUVEC proliferation by binding to SRRSF1.
The association between SRSF1 and β-catenin has been previously reported (22,23). SRSF1 recruit β-catenin and promote its expression (23,24). In the present study, hsa_circ_0001445 OE inhibited the expression of β-catenin in oxLDL-treated HUVECs. Therefore, the present study revealed the association between SRSF1 and β-catenin in oxLDL-treated HUVECs, suggesting that hsa_circ_0001445 OE decreased expression levels of β-catenin via binding to SRSF1. Moreover, Wnt/β-catenin signaling has been reported to serve an important role in the development of coronary artery disease, including atherosclerosis (25,26). Wu et al (27) demonstrated that inactivation of the Wnt/β-catenin signaling pathway reverses oxLDL-induced inhibition of HUVEC proliferation. Our research indicates that hsa_circ_0001445 reverses oxLDL-induced inhibition of HUVEC proliferation via inactivation of β-catenin. It has also been shown that activation of Wnt/β-catenin reverses oxLDL-induced foam cell formation in macrophages during the development of atherosclerosis (28). Therefore, more studies are required to investigate the association between β-catenin and atherosclerosis. E-cadherin is a crucial mediator in the epithelial-mesenchymal transition (EMT) process (29,30) and it has been shown that β-catenin negatively regulates expression of E-cadherin, thereby promoting EMT (31,32). The findings of the present study indicated that E-cadherin expression in oxLDL-HUVECs was upregulated by hsa_circ_0001445 OE. In summary, the data suggested that hsa_circ_0001445 OE reversed oxLDL-induced inhibition of HUVEC proliferation via activation of the SRSF1/β-catenin/EMT axis.
There were certain limitations to the present study. The sponging of specific miRNAs by hsa_circ_0001445 in HUVECs was not investigated. Only in vitro data were presented and the analysis lacked in vivo results. The effect of oxLDL + hsa_circ_0001445 OE + SRSF1 OE on angiogenesis and migration of cells needs to be further investigated. Further in vivo assays using multiple cell types are also required to validate the results of the present study. Therefore, additional investigations are needed in future.
Taken together, the data from the present study demonstrated that hsa_circ_0001445 OE reversed oxLDL-induced inhibition of HUVEC proliferation via activation of the SRSF1/β-catenin/EMT axis. The findings may provide information for the identification of potential treatment options for atherosclerosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LD conceived and supervised the study. GL designed the study. GL, SC and SX performed the experiments and analyzed the data. LD and GL confirmed the authenticity of all the raw data. All authors reviewed the results and read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liang H, Chen M, Qi F, Shi L, Duan Z, Yang R, He J, Lou B, Li Y, Yang Q. The proatherosclerotic function of indoleamine 2, 3-dioxygenase 1 in the developmental stage of atherosclerosis. Signal Transduct Target Ther. 2019;4:23. doi: 10.1038/s41392-019-0058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W, Yin R, Zhu X, Yang S, Wang J, Zhou Z, Pan X, Ma A. Mesenchymal stem-cell-derived exosomal miR-145 inhibits atherosclerosis by targeting JAM-A. Mol Ther Nucleic Acids. 2020;23:119–131. doi: 10.1016/j.omtn.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo X, Fu H, Xu C, Dong Y, Wu Z, Li D, Sun Y, Shen M, Wang L, Li Z, Duan Y. Efficient treatment of atherosclerosis by dexamethasone acetate and rapamycin Co-Loaded mPEG-DSPE calcium phosphate nanoparticles. J Biomed Nanotechnol. 2020;16:810–826. doi: 10.1166/jbn.2020.2936. [DOI] [PubMed] [Google Scholar]
- 4.Yin J, Hou X, Yang S. MicroRNA-338-3p promotes ox-LDL-induced endothelial cell injury through targeting BAMBI and activating TGF-β/Smad pathway. J Cell Physiol. 2019;234:11577–11586. doi: 10.1002/jcp.27814. [DOI] [PubMed] [Google Scholar]
- 5.Dang H, Song B, Dong R, Zhang H. Atorvastatin reverses the dysfunction of human umbilical vein endothelial cells induced by angiotensin II. Exp Ther Med. 2018;16:5286–5297. doi: 10.3892/etm.2018.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han BH, Song CH, Yoon JJ, Kim HY, Seo CS, Kang DG, Lee YJ, Lee HS. Anti-vascular inflammatory effect of ethanol extract from securinega suffruticosa in human umbilical vein endothelial cells. Nutrients. 2020;12:3448. doi: 10.3390/nu12113448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin M, Wang W, Zhou H, Wang X, Wang F, Wang H. Circular RNA circ_0003645 silencing alleviates inflammation and apoptosis via the NF-κB pathway in endothelial cells induced by oxLDL. Gene. 2020;755:144900. doi: 10.1016/j.gene.2020.144900. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Li Y, Liu Z, Ma X, Li M, Lu Q, Li Y, Lu Z, Niu L, Fan Z, Lei Z. Circular RNA circ_0124644 exacerbates the ox-LDL-induced endothelial injury in human vascular endothelial cells through regulating PAPP-A by acting as a sponge of miR-149-5p. Mol Cell Biochem. 2020;471:51–61. doi: 10.1007/s11010-020-03764-0. [DOI] [PubMed] [Google Scholar]
- 9.Vilades D, Martinez-Camblor P, Ferrero-Gregori A, Bar C, Lu D, Xiao K, Vea A, Nasarre L, Sanchez Vega J, Leta R, et al. Plasma circular RNA hsa_circ_0001445 and coronary artery disease: Performance as a biomarker. FASEB J. 2020;34:4403–4414. doi: 10.1096/fj.201902507R. [DOI] [PubMed] [Google Scholar]
- 10.Long JC, Caceres JF. The SR protein family of splicing factors: Master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 11.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/S1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 12.Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi G, Caltabiano R, et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (Basel) 2019;11:194. doi: 10.3390/cancers11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F, Morrone A, Caltabiano R, Barbagallo GM, Ragusa M, et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19:480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang JW, Jiang X, Li ZL, Jiang CR. MicroRNA-328-5p alleviates macrophage lipid accumulation through the histone deacetylase 3/ATP-binding cassette transporter A1 pathway. Lipids. 2021 Mar 4; doi: 10.1002/lipd.12297. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 16.Christophersen DV, Moller P, Thomsen MB, Lykkesfeldt J, Loft S, Wallin H, Vogel U, Jacobsen NR. Accelerated atherosclerosis caused by serum amyloid A response in lungs of ApoE(−/-) mice. FASEB J. 2021;35:e21307. doi: 10.1096/fj.202002017R. [DOI] [PubMed] [Google Scholar]
- 17.Dubland JA. Role of inflammatory cytokines in genesis and treatment of atherosclerosis. Looking at foam cells through a different lens. Trends Cardiovasc Med. 2021 Mar 3; doi: 10.1016/j.tcm.2021.02.008. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 18.Xiang S, Wu Y, Shi H, Xue L, Luo K, Ding Y. Circular RNA hsa_circ_0001445 in plasma as a novel biomarker for osteoporosis in postmenopausal women. Biomark Med. 2020;14:1599–1607. doi: 10.2217/bmm-2020-0447. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Zhang Q, Ma N. LncRNA GASAL1 interacts with SRSF1 to regulate trophoblast cell proliferation, invasion, and apoptosis via the mTOR signaling pathway. Cell Transplant. 2020;29:963689720965182. doi: 10.1177/0963689720965182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Hu ZQ. LncRNA HOTAIR aggravates myocardial ischemia-reperfusion injury by sponging microRNA-126 to upregulate SRSF1. Eur Rev Med Pharmacol Sci. 2020;24:9046–9054. doi: 10.26355/eurrev_202009_22850. [DOI] [PubMed] [Google Scholar]
- 21.Tejedor JR, Tilgner H, Iannone C, Guigo R, Valcarcel J. Role of six single nucleotide polymorphisms, risk factors in coronary disease, in OLR1 alternative splicing. RNA. 2015;21:1187–1202. doi: 10.1261/rna.049890.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, Chen L, Lv Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J Cell Mol Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Huang B, Shi Z, Han J, Wang Y, Huangfu J, Wu W. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. EMBO Mol Med. 2013;5:737–750. doi: 10.1002/emmm.201202218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorsen K, Mansilla F, Schepeler T, Øster B, Rasmussen MH, Dyrskjøt L, Karni R, Akerman M, Krainer AR, Laurberg S, et al. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002998. M110.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Neogi A, Mani A. The role of Wnt signalling in development of coronary artery disease and its risk factors. Open Biol. 2020;10:200128. doi: 10.1098/rsob.200128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singla B, Lin HP, Chen A, Ahn W, Ghoshal P, Cherian-Shaw M, White J, Stansfield BK, Csanyi G. Role of R-spondin 2 in arterial lymphangiogenesis and atherosclerosis. Cardiovasc Res. 2020 Aug 4; doi: 10.1093/cvr/cvaa244. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Liu T, Hou H. Knockdown of LINC00657 inhibits ox-LDL-induced endothelial cell injury by regulating miR-30c-5p/Wnt7b/β-catenin. Mol Cell Biochem. 2020;472:145–155. doi: 10.1007/s11010-020-03793-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Wei M, Liu G, Song C, Yang M, Cao Z, Zheng M. Silencing protease-activated Receptor-2 alleviates ox-LDL-induced lipid accumulation, inflammation and apoptosis via activation of Wnt/β-catenin signaling. Gen Physiol Biophys. 2020;39:437–448. doi: 10.4149/gpb_2020014. [DOI] [PubMed] [Google Scholar]
- 29.Hu C, Li M, Guo T, Wang S, Huang W, Yang K, Liao Z, Wang J, Zhang F, Wang H. Anti-metastasis activity of curcumin against breast cancer via the inhibition of stem cell-like properties and EMT. Phytomedicine. 2019;58:152740. doi: 10.1016/j.phymed.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L, Li J, Liu Y, Zhou W, Shan Y, Fan X, Zhou X, Shan B, Song Y, Zhan Q. Flotillin1 promotes EMT of human small cell lung cancer via TGF-β signaling pathway. Cancer Biol Med. 2018;15:400–414. doi: 10.20892/j.issn.2095-3941.2018.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, He B. MiR-760 inhibits the progression of non-small cell lung cancer through blocking ROS1/Ras/Raf/MEK/ERK pathway. Biosci Rep. 2020 Apr 29; doi: 10.1042/BSR20182483. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 32.Yang D, Li Q, Shang R, Yao L, Wu L, Zhang M, Zhang L, Xu M, Lu Z, Zhou J, et al. WNT4 secreted by tumor tissues promotes tumor progression in colorectal cancer by activation of the Wnt/β-catenin signalling pathway. J Exp Clin Cancer Res. 2020;39:251. doi: 10.1186/s13046-020-01774-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.