Summary
Optical mapping (OM) provides single-molecule readouts of fluorescently labeled sequence motifs on long fragments of DNA, resolved to nucleotide-level coordinates. With the advent of microfluidic technologies for analysis of DNA molecules, it is possible to inexpensively generate long OM data ( kbp) at high coverage. In addition to scaffolding for de novo assembly, OM data can be aligned to a reference genome for identification of genomic structural variants. We introduce FaNDOM (Fast Nested Distance Seeding of Optical Maps)—an optical map alignment tool that greatly reduces the search space of the alignment process. On four benchmark human datasets, FaNDOM was significantly (4–14×) faster than competing tools while maintaining comparable sensitivity and specificity. We used FaNDOM to map variants in three cancer cell lines and identified many biologically interesting structural variants, including deletions, duplications, gene fusions and gene-disrupting rearrangements. FaNDOM is publicly available at https://github.com/jluebeck/FaNDOM.
Data science maturity: DSML 3: Development/Pre-production: Data science output has been rolled out/validated across multiple domains/problems
Highlights
-
•
FaNDOM is a fast open-source aligner for OM data
-
•
It utilizes a novel filtering strategy to reduce the search space of alignment
-
•
The method enables discovery of large, complex genomic structural variants
-
•
Structural variants suggested by FaNDOM include gene fusions and gene disruptions
The bigger picture
Optical mapping (OM) is a rapidly maturing strategy for detecting large-scale rearrangements in genomes, leveraging ultra-long fragments of DNA imaged at very high depth of coverage (>100×). OM data reflect an orthogonal strategy to DNA sequencing, instead utilizing image-based detection of fluorescent tags associated with specific DNA motifs. The resulting data can be aligned back to the reference genome for discovery of genomic rearrangements and karyotypic abnormalities. Existing methods, however, are computationally demanding, making discovery harder. We present a novel method, FaNDOM, for alignment of OM data to the reference genome, and the additional discovery of structural variants. FaNDOM utilizes fast filtering algorithms based on constructing graph-based chains of seed matches, achieving orders of magnitude speedup, while maintaining high sensitivity, enabling a more comprehensive search of complex structural variations involving hundreds of kbp.
Optical mapping data is an orthogonal technique to DNA sequencing for the identification of genomic structural variants (SVs). We present a method, FaNDOM, which performs fast alignment of optical mapping data to the reference genome for identification of SVs. FaNDOM utilizes a novel filtering strategy, vastly reducing the search space of the alignment process, enabling rapid discovery of biologically interesting events.
Introduction
Optical mapping (OM) is a rapidly maturing genome-mapping technology whose historical antecedents are at least a few decades old.1 In the much older restriction-mapping technique, the use of sequence-specific restriction sites in a genome enabled unique “fingerprints” of the DNA. The initial restriction site maps were used to compare and position clones (genetic linkage maps) before sequencing.2 Now, OM provides single-molecule readouts of the locations of fluorescently labeled sequence motifs on long fragments of DNA, resolved to nucleotide-level coordinates.3 Despite the development of competing capillary sequencing and next-generation sequencing methods, optical maps continue to play an important role in scaffolding and assembly. With the advent of microfluidic technologies for high throughput of individual molecules and fluorescence-based visualization of covalently marked sites (labels), it is possible to generate high coverage (100× of the human genome) with long OM molecules (150 kbp) for $500–1,000. For instance, the OM datasets analyzed in this paper had a median length of 191 kbp.
As the optical mapping technology evolves, the error profiles found in OM data also change. Bionano optical mapping (Bionano Genomics, San Diego, CA) uses direct covalent labeling of fluorescent molecules onto DNA fragments, as opposed to previous generations of OM, which used nickases. Its sources of error are orthogonal to DNA sequencing technologies,4 and currently include incomplete labeling of donor sequences, false-positive labels, and imprecise resolution about exact locations of imaged labels. Other technology-specific phenomena, such as possible molecular chimerism or molecular stretching, also contribute to error. Computational methods, which handle OM data, must capture these various errors.
Given its uses for scaffold construction in de novo assembly projects,5, 6, 7 optical mapping has matured to becoming a routine part of assembly pipelines for complex and/or large genomes. As a first step of this process, the OM fragments themselves are assembled into much larger (and error-corrected) OM contigs. The samples considered by our study had a median OM contig N50 of 38.4 Mbp. To achieve this, a computationally challenging problem of identifying overlapping OM fragments must be addressed. Much of the previous work about that problem uses dynamic programming algorithms to compare and align restriction maps,8 and now extends to optical maps.9,10 Newer methods, such as Kohdista11 and MalignerIX,12 tackle the overlapping fragment identification problems. Indexing and alignment-based methods have also been developed to map a sequence contig to a reference optical map genome, a requirement for scaffolding.13,14
Here, we consider the slightly different problem of mapping optical maps to a reference human genome for the purposes of identifying structural variants (SVs).15,16 Such methods have been effective in identifying genomic abnormalities in Mendelian disease17,18 as well as cancer.19, 20, 21 Due to similar algorithmics, general methods for pairwise alignment or scaffolding, including Valouev,22 SOMA,23 TWIN,13 and MalignerDP,12 could be used in principle for mapping optical maps to an in-silico-digested reference genomic sequence. However, most of these methods do not repurpose well in practice, especially on data from the latest Bionano platform. Moreover, they do not call SVs. In contrast, OMBlast24 and RefAligner25 have previously demonstrated superior performance on Bionano data.24,26 RefAligner specifically has been configured to call SVs. A new software, OMSV,27 now combines RefAligner and OMBlast output to call SVs. Notably, RefAligner is a closed-source proprietary method, available only as pre-compiled binaries for specific hardware, and is very resource intensive, as described in the Results.
We introduce FaNDOM (Fast Nested Distance Seeding of Optical Maps)—an optical map alignment tool that introduces a novel method for seeding optical map alignments, greatly reducing the search space of the alignment process. FaNDOM is specifically optimized to handle data from the Bionano Saphyr optical mapping technology. The algorithmic and technology-specific improvements allow us to be significantly (4–14×) faster than competing tools while maintaining sensitivity and specificity. We used FaNDOM to map variants in three cancer cell lines and identified many structural variations, including deletion of tumor suppressor genes, duplications, gene fusions, and gene-disrupting rearrangements. FaNDOM is publicly available at https://github.com/jluebeck/FaNDOM.
Results
As OMBlast24,28 and RefAligner25 were the best-performing pre-existing methods for mapping Bionano optical maps to a reference genome, we compared the performance of FaNDOM against Bionano RefAligner (Solve3.5.1) and OMBlast (OMTools v.1.4a). We also attempted to benchmark TWIN and Kohdista, but they are not specifically designed for this problem and did not perform as well (Methods S8).
Saphyr optical map data are publicly available for samples NA12878, GM09888, GM08331, and GM24143. We collected 270,000 raw molecules from each sample, where more than 85% of each molecule aligned to reference, as reported by Bionano, using their own RefAligner tool. We then ran FaNDOM, OMBlast, and RefAligner on this testing set.
Running time
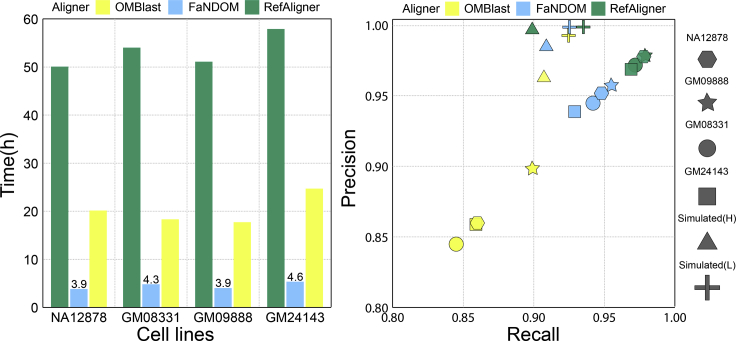
We note that RefAligner is already highly optimized for the Saphyr technology, and is only provided as pre-compiled binary code, which runs on specific machine architectures. All experiments were conducted on an Intel(R) Core(TM) i9-9900 CPU @3.10GHz with 32 GB of main memory running Ubuntu 18.04.3 LTS (Bionic Beaver), using 10 threads. The results (Figure 1A) showed that FaNDOM was 4–6× faster than OMBlast and 13–14× faster than RefAligner on all datasets, highlighting the speedups created by our filtering methods. FaNDOM required approximately 2–2.5 GB of RAM for each thread (Methods S6). While OMBlast required less memory, the memory usage increased with increase in molecule size, and did not scale well for Saphyr-assembled contigs. The OMBlast documentation suggests 200 Gb RAM for mapping assembled contigs.
Figure 1.
FaNDOM performance
(Left) Running time; (right) accuracy. The set of true positives (TP) were all mappings identified by at least two of the three methods. Recall = TP/(TP + FN), Precision = TP/(TP + FP).
Mapping accuracy
We compared the accuracy FaNDOM, RefAligner, and OMBlast reported mappings on simulated and real data. Unlike DNA sequencing read mapping, which has discrete character matches and mismatches, it is not trivial to designate an OM molecule alignment as correct or incorrect on real data. Instead, we treated a mapping as correct if it was supported by at least two of the three methods.
We simulated datasets with “high” and “low” error (Methods S7), where high (H) corresponded to a false-positive label rate of 4 per 100 kbp, and stretch factor with standard deviation 0.02, which matched the Saphyr technology (Methods S7). Low (L) corresponded to a false-positive label rate of 1 per 100 kbp and stretch factor with standard deviation 0.01. All tools performed well on low-error. On high-error data, the three methods had very similar recall, with FaNDOM marginally higher, while FaNDOM precision lay between RefAligner and OMBlast (Figure 1B). On the cell lines, RefAligner had the highest precision and recall followed by FaNDOM and OMBlast. We note that RefAligner is better positioned to incorporate specifics of the Saphyr technology. The lower recall for FaNDOM relative to RefAligner can be partially attributed to the occasional removal of true maps during the filtering step. The precision can be improved by post-alignment filtering, and will be part of future release of FaNDOM after more datasets have been analyzed.
FaNDOM was 5× and 15× faster that OMBlast and RefAligner on cell lines (Figure 1A) as well as simulations (Figure S7; note different scale). As expected, simulations show that the running time increases with higher error rate for all methods.
SV detection
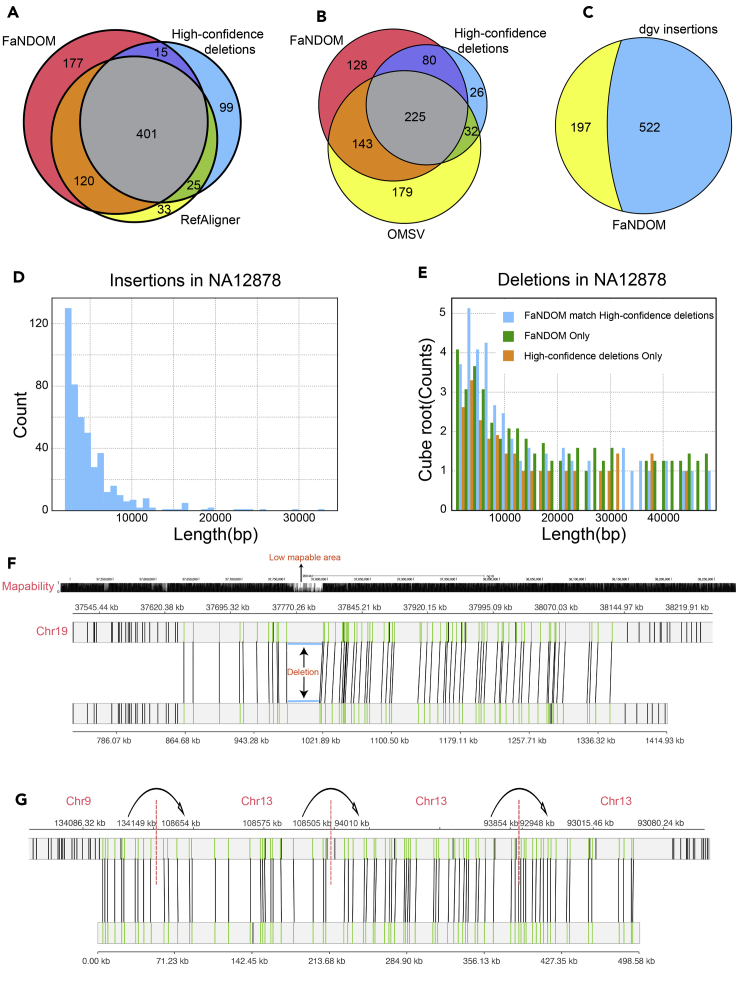
SV analysis continues to be a challenging problem requiring consensus from different methods and technologies. We compared the three methods using a benchmark of SV deletion calls of length >2,000bp on the genome NA12878. The benchmark was created previously using a multitude of technologies.29 Figure 2A compares the performance of FaNDOM and RefAligner using assembled OM contigs. FaNDOM and RefAligner had comparable recall identifying 77% and 79% of the high-confidence calls, respectively, despite FaNDOM using filtering strategies to make the runtime faster by an order of magnitude. FaNDOM was much more aggressive in calling deletions compared with RefAligner. Spot checking, many of the FaNDOM-specific deletion calls appeared to be accurate (e.g., see Figure 2F).
Figure 2.
SV calling performance
(A) Comparison of FaNDOM and RefAligner deletion calls on NA12878 against a benchmark dataset from Parikh et al.,29 using the hg19 reference.
(B) Comparison of FaNDOM and OMSV deletion calls on NA12878 against a benchmark created using multiple sequencing technologies published in Dixon et al.15 using the hg38 reference.
(C) Insertions identified by FaNDOM for NA12878. The blue region signifies insertion polymorphisms identified by FaNDOM also in the Database of Genomic Variants.
(D) Length distribution of FaNDOM insertion calls for NA12878.
(E) Length distribution of FaNDOM and benchmark deletion calls (Parikh et al.29).
(F) A FaNDOM deletion not in the Parikh et al.29 benchmark dataset likely due to its presence in a low mappability region.
(G) A FaNDOM alignment using assembled OM contigs that chains multiple breakpoints across 400 kbp on the K562 cell line. OM alignment visualizations were generated with MapOptics.30
OMSV27 is another recent method for detecting SVs with OM data. It is an integrative tool that combines the output of RefAligner and OMBlast together, and is therefore even more compute intensive. As we could not run OMBLast on Saphyr contig data, we compared FaNDOM calls against pre-computed OMSV calls on NA12878 mapped to the hg38 reference and compared the calls with a benchmark deletion call set15 on the hg38 reference (Figure 2B). The FaNDOM recall was 84% compared with the 70% recall of OMSV.
Detecting genomic insertions is one of the advantages of long-read technologies. FaNDOM predicted 719 insertions (Figure 2C). While there is no established call set of insertions for NA12878, 73% of the FaNDOM calls were previously reported as insertion polymorphisms in the Database of (human) Genomic Variants.31 FaNDOM also identified a few ultra-long insertions in OM contigs (Figure 2D) that would be challenging with any competing technology due to the insertion size.
We investigated the FaNDOM-specific SV calls for possible error. The high-confidence dataset29 has been collected by integrating a number of technologies, and is likely to be accurate. Nevertheless, many of its calls were discovered using short reads, while many of the FaNDOM-specific calls were kbp (e.g., see Figure 2E). In addition, some of the FaNDOM-specific calls are in regions of low mappability (typically low complexity or repetitive sequence). Those breakpoints typically cannot be captured by short reads, but can be captured by long OM contigs (e.g., chr19:37,760K–37,795K; Figure 2F), demonstrating the complementarity of OM data to sequencing technologies. Moreover, assembled optical map contigs enable the detection of multiple breakpoints in one contig. As an example, Figure 2G represents an assembled OM contig from the K562 cell line that covers translocation from chr9 to chr13 and multiple breakpoints in chr13 spanning 500 kbp.
SVs in cancer cell lines
We ran FaNDOM on assembled OM contigs as well as OM molecules for cancer cell lines K562, CAKI-2, and H460—all of which are known to carry extensive rearrangements. Table1 summarizes some of the rearrangements identified by FaNDOM on assembled OM contigs. The rearrangements identified by FaNDOM, which included 1,800 large ( kbp) indels, 133 interchromosomal translocations, 28 fold-back reads, and 223 breakpoints that disrupted an existing gene, among other rearrangements. In this study, we focused specifically on genes that were deleted, and on translocations that disrupted or fused two genes.
Table 1.
Rearrangements in cancer cell lines
| Cell lines | Indels | Interchromosomal translocations | Fold-back reads | Gene-disrupting breakpoints |
|---|---|---|---|---|
| CAKI-2 | 626 | 56 | 7 | 95 |
| H-460 | 571 | 26 | 4 | 62 |
| K562 | 603 | 21 | 17 | 66 |
The lung cancer cell line NCI-H460 has previously been documented to bear a focal amplification of the MYC/PVT1 region due to extrachromosomal DNA (ecDNA) and it has also been found to show evidence for intrachromosomal amplification in a homogeneously staining region (HSR).32 Previous reconstruction of the MYC amplified region revealed a complex duplicated structure, which suggested that the ecDNA element containing MYC/PVT1 had reintegrated as an HSR in a non-native location.20 The FaNDOM analysis identified a translocation from within the amplified ecDNA structure (chr8: 128,745, kbp) to a non-native location (chr12:7, 665k; Figure 3A) revealing chr12 to be the site of the HSR. Figure 3A also supports an inverted duplication at chromosome 8 as part of the amplified structure. In addition to recapitulating the breakpoints of the ecDNA, the FaNDOM analysis identified many partial or complete deletions of tumor suppressor genes, including LRP1B33 (chr2: 141,735K–142,155K), TUSC7A34 (non-coding; chr3: 116,295K–116,775K), FHIT35 (chr3: 60,405K–60,735K), LSAMP36 (chr3: 115,545K–116,145K). Notably, many of these deletions were on chr3. Many other rearrangements were identified providing a scenario of complex rearrangements in the cell line.
Figure 3.
Examples of detected structural variants in cancer cell lines
(A) The chr8-chr12 translocation shows the integration of an Myc carrying ecDNA molecule onto chr12 in H460.
(B) A RACGAP1-AKAP6 fusion on CAKI-2.
(C) The BCR-ABL1 fusion on K562.
(D) Deletion of the genes ORC6 and MYLK3 with a partial inversion.
(E) A translocation that disrupts CDC25A and GRID1 but the direction is inconsistent with a fusion event.
(F) A “fold-back” inversion that duplicates and inverts GPC5 in K562.
In the renal cancer cell line CAKI-2, we observed deletions or disruptions involving tumor suppressor genes, including CFHR137 (chr1: 196,665K–197,295K), RNF217 (chr6: 125,265K–125,505K),38 RBFOX1 (chr16: 6,585K–7,155K),39 FBXL7 (chr5: 15,825K–15,945K).40 We also observed two fusions: TECRL1/GRIP1 (chr4: 65,205K, -, chr12: 66,975K, -) and RACGAP1/AKAP6 (Figure 3B, chr12: 50,385K, -, chr14: 33,255K, +). RACGAP1 displays tumor malignancy potential41 and is known to fuse with other genes, such as CERS5 and RAB34.42
K562 is a chronic myelogenous leukemia cell line with the Philadelphia chromosome. It was comprehensively analyzed recently using a multitude of technologies, including whole-genome sequencing and Hi-C.43 FaNDOM confirmed some of the rearrangements of the previous study, such as the BCR-ABL1 fusion (Figure 3C), between chr22 and chr9. Among other rearrangements, we also observed an atypical microdeletion in 22q11, almost identical to a deletion previously associated with a congenital syndrome,44 and a subset of a larger deletion reported for DiGeorge syndrome. The deletion encompasses the genes GSTT1, GSTT2, and GSTT2B, and deletions in these genes have previously been associated with esophageal cancer.45
While our results often matched the previously reported SVs,43 there were a few notable differences. For example, in contrast with the previous finding of an inversion involving ORC6, MYLK3 on chr16, we observed a deletion (16:46, 725K–46,845K; Figure 3D) that partially removed ORC6 as well as a microinversion involving MYLK3. In a second example, the Zhou et al. study also identified a fusion of CDC25A/GRID1.43 While we observe the same translocation, the directionality provided by the long reads suggests the disruption of the two genes, but not a fusion product (Figure 3E). We could confirm other chromosome 16 rearrangements, including an inverted duplication (88,605K–88,785 K), and another inverted duplication at chr13: 92,475K (Figure 3F).
Discussion
Improvements to the optical mapping technology in terms of accuracy and cost has made it competitive for SV detection. At the same time, the raw data are harder to interpret and motivate the development of public domain tools for interpretation. In this paper, we focus on speeding up the mapping by relying on a novel filtering strategy that greatly improved speed without a significant loss of accuracy. The filtering relies on two ideas: (1) for most high-quality optical maps, it is relatively easy to find seeds that locate the reference target region for a query, and (2), by merging distances, thousands of queries can identify their target seeds in a single search-and-merge strategy. The results demonstrate the viability of this trade-off, leading to high speedup over other tools with only a small loss of sensitivity.
We recognize that our proposed method uses many parameters and, for the most part, the parameters are empirically determined to work for Saphyr. The optimal parameter values will be determined only after a large number of datasets have been analyzed, and will need to be retrained for newer technologies. In addition, non-human genomes, such as plants, may also require some significant recalibration of parameters and low-complexity annotations, which we have not yet explored. Nevertheless, because we have used FaNDOM to analyze many tens of thousands of molecules, the current choice of parameters appears to be robust for the current technology. Taken together, our results point to the value of using OM as a complementary technology for structural variation identification.
The detection of SVs is a key benefit of the OM technology, but it is harder to benchmark given the lack of large-scale, robust truth datasets. Our results suggest that FaNDOM can identify discordant alignments and breakpoints with high sensitivity. As many of the calls are based on cutoffs that can be adjusted, the results do not reveal any fundamental limitation of the filtering, but indicate a lack of additional calibration against a true gold standard. Additional analysis will be needed to identify systemic sources of false-positive calls.
We note that calling the structural variation mechanism itself is a secondary process that will require integration with other information, including copy-number changes, and this will be a topic of ongoing research. For example, one possible improvement includes pruning deletion calls by limiting results to the regions with a decrease in copy number consistent with heterozygous or homozygous deletion. With further improvements and methods development, OM technologies could be used to replace cytogenetics as a method of choice for revealing large-scale genetic abnormalities in Mendelian diseases and cancer.17,20,21
Experimental procedures
Resource availability
Lead contact
Siavash Raeisi Dehkordi is the lead contact for this study and can be contacted by email at sraeisid@ucsd.edu.
Materials availability
This study did not generate any materials.
Data and code availability
The code for FaNDOM is available on GitHub at https://github.com/jluebeck/FaNDOM.
We used optical map data from the following individuals, and these data were obtained from the publicly available Bionano Saphyr datasets (https://bionanogenomics.com/library/datasets/)—NA12878, GM09888, GM08331, and GM24143. For cancer SV detection, we used previously published20 Bionano Saphyr data from cancer cell lines K562, CAKI-2, and NCI-H460.
Method details
Conceptually, define an optical map as a sorted list of numeric values, representing the relative positions of labels on a fragment of DNA (Figure S1A). These numeric lists can be generated for any collection of individual OM molecules, assembled OM molecules, or from in-silico-predicted label positions on the reference genome. FaNDOM utilizes standard optical map data formats (.bnx or .cmap), where each imaged DNA fragment has been pre-converted to label position lists specified in base pair coordinates. An overview of the structure of the FaNDOM software is available in Figure S1B.
Pre-processing
Query fragments with length kbp or containing less than 10 labels were filtered out from mapping. Similarly, queries containing consecutive labels with distance kbp were removed (Methods S2).
Scaling refers to a systematic translation of physical inter-label distances into nucleotide distances. The Saphyr instrument performs a calibration to scale distances, estimating the number of base pairs present per image pixel. The process can on occasion be erroneous.46 To recalibrate, FaNDOM randomly selects 250 molecules and estimates a corrected scaling factor using a grid search in a range of values between 0.96 and 1.2. The range was determined by experimenting from a set of 38 human samples (Methods S3). The rescaled molecules in each iteration are aligned to the reference. The scaling factor that achieves the highest total alignment score is selected for rescaling molecules before alignment.
Assembled OM contigs can be very large, often exceeding thousands of labels. As the alignment time grows quadratically with length, FaNDOM pre-processes assembled OM contigs by splitting them into smaller fragments, each containing 75 labels, with an overlap of 50 labels between endpoints of consecutive fragments. When alignment is completed, FaNDOM merges the alignments from overlapping fragments from assembled OM contigs to produce a complete alignment for the OM contig. In the case of conflicting alignments between overlapping contig fragments, FaNDOM maintains both partial alignments.
We convert the reference genome into a collection of expected label locations based on the in silico presence of the labeling motif throughout the reference. If the distance between two consecutive reference labels is less than 800 bp, they are replaced with the average of the two locations to account for the potential inability of resolving nearby OM labels (Methods S2). We also adapted a Bionano method25 to identify and mask low-complexity regions in the human genome. Formally, denote a low-complexity region as containing at least five consecutive labels where the distance between adjacent labels is identical within 10% tolerance. Those could result in spurious alignments and are masked out. Specifically, in reference genome build hg19, 1.5 Mbp, which (0.04% of total reference genome) was masked out, while in hg38, 2.8 Mbp (0.09% of the total reference genome) was masked out (see Table S1 for masked regions).
Optical map alignment
The crux of a mapping procedure is an alignment of an optical map query to an in silico optical map of a reference sequence interval. The alignment maps query labels to the reference labels so that the inter-label distances between the query and reference are preserved (Figure S1).
The alignment of optical maps is a well-studied problem.1,22 FaNDOM's scoring function follows previous methodologies, but diverges slightly. Consider reference R of length m and reference Q of length n labels. For and , define as the optimum score of aligning a subsequence (local alignment) ending at label j on R with a subsequence ending at label q on query Q. S can be computed using the following banded dynamic programming recurrence, where the band size is d:
| (Equation 1) |
where, Score_region scores a match after penalizing for discrepancies in the match. Specifically, for , let , denote the number of unmatched labels in the query and reference, respectively. Then,
We set to represent a perfect match score. Empirical tests (Methods S4) indicated that a wide range of showed identical performance. Increasing resulted in the same alignments but with tighter boundaries. We chose the distance scale parameter and false-label parameter (Figure S4). After computing initial alignments for molecules, FaNDOM then identifies molecules, which are candidates for local/partial alignment discovery, as a prelude to SV analysis. In this partial alignment mode (see computing partial alignments for SV detection section below), where split-molecule alignments are allowed, FaNDOM computes more stringent partial alignments (, ).
Alignment running time suggests the necessity of filtering.
The ungapped alignment algorithm has complexity . Despite algorithmic improvements and optimizations, our empirical results suggested that aligning a collection of two million OM fragments representing (100×) whole-genome coverage against every position on the human genome would take cpu-h. While assembly of OM fragments into contigs reduces the number of query sequences, the OM contigs are longer and the estimated time remains cpu-h. Therefore, similar to the Bionano RefAligner25 and OMBlast,24,28 we deploy a filtering strategy, where, for each query molecule, the goal is to identify a small collection of reference intervals to align the query with. The filter must be fast, sensitive (defined by the probability of the true reference location being included in the filtered reference intervals), and efficient (defined by the number of filtered regions per query—smaller being better). The filtered regions, or seeds are used to compute alignments and return the full or partial mappings of each query OM fragment or contig.
Search-and-merge filtering for optical maps
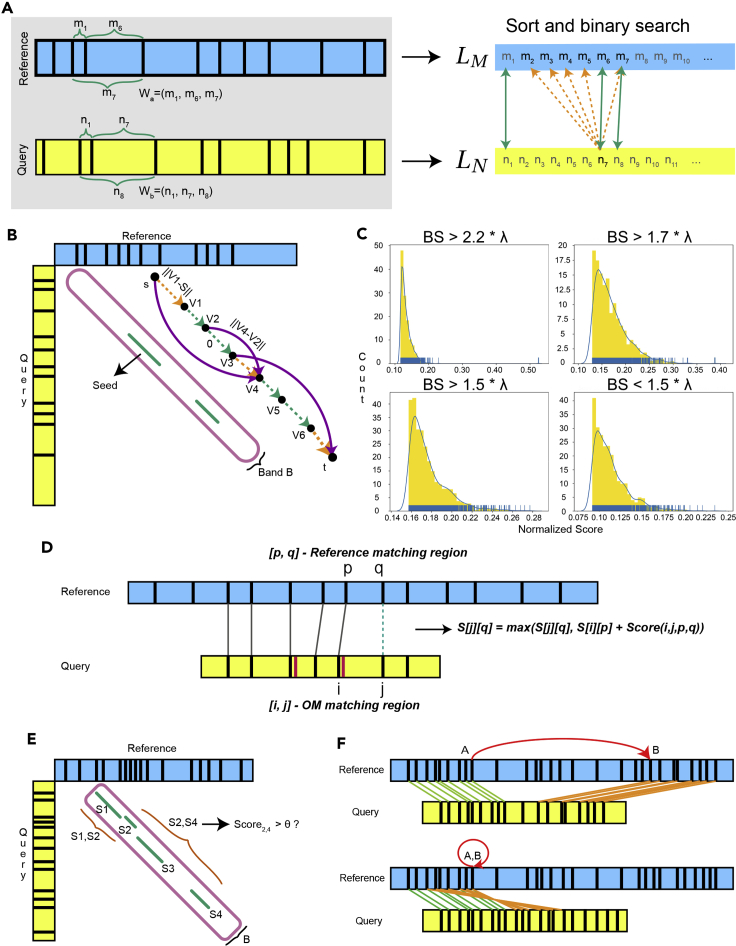
The key idea of filtering is that in a correct alignment there are some parts of query and reference, which are highly similar to each other, or that all inter-label distances in those regions are practically equivalent. Let (respectively, ), denote the genomic distance between labels in R (respectively, Q). Denote a window in the reference as a collection of distances for all . Windows , in the query OMs are defined similarly. Let
A default value of was chosen empirically (Methods S4). In the search-and-merge procedure, we sort all genomic distances from every window of the reference (typically a chromosome) to a list (Figure 4A). Similarly, for a collection of query OMs, we merge all sorted distances from all windows of each query in the collection into list . Each distance (respectively, ) is associated with all reference windows (respectively, query windows) containing distance x (respectively, y).
Figure 4.
The FaNDOM workflow
(A) Search-and-merge filtering step in which genomic distances extracted from windows (, ) and added to lists and . The lists and are merged and seeds are identified.
(B) Packing seeds into bands step, in which for each band B seeds inside it are formed into a directed acyclic graph, G, and the band is scored by finding the shortest path from s to t.
(C) Different score threshold possibilities for the band score distribution of bands for a single query. The best score is denoted as “BS.”
(D) Dynamic programming for the alignment module in FaNDOM.
(E) Seed selection for partial alignment, which scores bands based on the shortest path between each pair of seeds inside the band B.
(F) SV detection module, which finds breakpoints based on multiple partial alignments. The alignment on top shows a breakpoint from A to B, the lower alignment visualizes an inversion, or “fold-back.”
Next, the sorted lists are “search-merged” (Figure 4A). For each element we perform two binary searches to identify the smallest and largest distances such that . For all “matches” () where , we increment the match score of all window pairs associated with x and y. Finally, for all reference labels a, query labels b, such that , a seed , is generated, with representing direction of match.
Packing seeds into bands
For each reference label a, and each query OM, FaNDOM explores a diagonal band around a of width (default value ; Methods S4). Label a is filtered out if contains fewer than seeds (Methods S4). For retained bands, an edge-weighted directed acyclic graph G is constructed as follows: each node u in G corresponds to a pair of (query, reference) labels , where (respectively, ) represents the nucleotide distance of the query label (respectively, reference label) from the first query (reference) label. Also, add nodes and corresponding to the start and end of band . For each seed u in the band, designate nodes corresponding to start, middle, and end of the seed. With few exceptions, we use Euclidean distances for edge weights so that . Specifically,
-
1.
For each seed u, add edges and with weights 0 each; edge with weight , and edge with weight .
-
2.
For each pair of seeds such that and , add edge with weight .
-
3.
For each pair of seeds such that and , add edge with weight .
We use dynamic programming to compute the weight of the shortest (least-weight) path from s to t in G. The score of band is given by
A similar process is used for seeds in the reverse direction, with , . For each query OM, we save the highest scoring 150 bands.
As a first idea, we could align the query map with the reference region for each of the 150 bands, and still achieve high speed and sensitivity. However, we observed that, in some cases, the top-scoring bands were significantly more likely to yield true alignments than other high-scoring bands, and that the correct region was near the tail of the band score distribution and could be identified without aligning every candidate. We empirically fit the band scores to an exponential distribution with parameter λ and used the following empirical guidelines for scoring (Methods S4). For each query
A band that is selected for alignment is converted to reference alignment boundaries by using the reference coordinate of the source node s, and the query molecule Q of length . Specifically, for a padding factor p (default ), the region to on the reference is used to align to the query molecule.
Computing partial alignments for SV detection
We identify SVs in two steps. First, queries that are either (1) unaligned, (2) have a mean alignment score less than 5,000/label, (3) the alignment does not cover 80% of the query length, or (4) has a total alignment length ⩽25 kbp, are targeted for partial alignments. The banding procedure is identical. For partial alignments, we compute local shortest paths between all pairs of seeds as long as kbp and the path contains at least four labels. If the corresponding band score
then the region gets a score of , and the top 300 candidate regions, each designated by a pair of nodes, are selected for alignment and re-ranking. A gapped alignment module is used and, if the score exceeds a threshold, the partial or gapped alignment is reported.
FaNDOM currently identifies discordant alignments (defined below) and breakpoints, which form the core of any SV discovery strategy, and defers the calling of actual SVs to a subsequent script that can be customized by the user. Recall that an alignment is a chain of matches . For alignments below a threshold score, if there exists such that (1) , (2) , and (3) , then a discordant alignment is called. Discordant alignments typically represent insertions/deletions, but may also represent small inversions flanked by high-quality alignments on both sides.
Breakpoints refer to a pair of coordinates that are non-adjacent on the reference, but are together on the query. Consider two partial alignments that involve the same query molecule, described by and . Note that could potentially be on a different chromosome than . Define using , and . FaNDOM calls a breakpoint () if there is no partial alignment involving the labels between and . Breakpoints are clustered if their endpoints are within 30 kbp, and each breakpoint is listed along with its “support,” or the number of alignments consistent with the breakpoint. Subsequent scripts are used to describe the rearrangement that creates the breakpoint. For example, describes a homozygous (respectively, heterozygous) deletion if and are on the same chromosome and the fragment coverage in the interval is 0 (respectively, half of normal coverage).
Acknowledgments
The research was supported by a grant from the NIH (GM114362). We would like to thank Andy Pang of Bionano Genomics, Inc. for his feedback and assistance with data interpretation and explanations of the Bionano pipelines.
Author contributions
S.R.D., J.L., and V.B. designed the study, developed the algorithms, conducted analysis, and wrote the paper. S.R.D. and J.L. developed the code for FaNDOM.
Declaration of interests
V.B. is a co-founder, consultant, and SAB member of and has equity interest in Boundless Bio, Inc. (BB) and Digital Proteomics, LLC (DP) and also receives income from DP. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Published: May 3, 2020
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2021.100248.
Contributor Information
Siavash Raeisi Dehkordi, Email: sraeisid@ucsd.edu.
Vineet Bafna, Email: vbafna@ucsd.edu.
Supplemental information
References
- 1.Schwartz D.C., Li X., Hernandez L.I., Ramnarain S.P., Huff E.J., Wang Y.K. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 2.Botstein D., White R.L., Skolnick M., Davis R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 3.Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D., Deshpande P., Cao H., Nagarajan N., Xiao M., Kwok P.Y. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P., Jing X., Ren J., Cao H., Hao P., Li X. Modelling BioNano optical data and simulation study of genome map assembly. Bioinformatics. 2018;34:3966–3974. doi: 10.1093/bioinformatics/bty456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S., Wei F., Nguyen J., Bechner M., Potamousis K., Goldstein S., Pape L., Mehan M.R., Churas C., Pasternak S. A single molecule scaffold for the maize genome. PLoS Genet. 2009;5:e1000711. doi: 10.1371/journal.pgen.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teague B., Waterman M.S., Goldstein S., Potamousis K., Zhou S., Reslewic S., Sarkar D., Valouev A., Churas C., Kidd J.M. High-resolution human genome structure by single-molecule analysis. Proc. Natl. Acad. Sci. U S A. 2010;107:10848–10853. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan W., Jiang T., Lonardi S. OMGS: optical map-based genome scaffolding. J. Comput. Biol. 2020;27:519–533. doi: 10.1089/cmb.2019.0310. [DOI] [PubMed] [Google Scholar]
- 8.Huang X., Waterman M.S., Oct Dynamic programming algorithms for restriction map comparison. Comput. Appl. Biosci. 1992;8:511–520. doi: 10.1093/bioinformatics/8.5.511. [DOI] [PubMed] [Google Scholar]
- 9.Anantharaman T.S., Mishra B., Schwartz D.C. Genomics via optical mapping. II: ordered restriction maps. J. Comput. Biol. 1997;4:91–118. doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- 10.Valouev A., Li L., Liu Y.-C., Schwartz D.C., Yang Y., Zhang Y., Waterman M.S. Alignment of optical maps. In: Miyano S., Mesirov J., Kasif S., Istrail S., Pevzner P.A., Waterman M., editors. Research in Computational Molecular Biology. Springer Berlin Heidelberg; 2005. pp. 489–504. [Google Scholar]
- 11.Muggli M.D., Puglisi S.J., Boucher C. Kohdista: an efficient method to index and query possible Rmap alignments. Algorithms Mol. Biol. 2019;14:25. doi: 10.1186/s13015-019-0160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendelowitz L.M., Schwartz D.C., Pop M. Maligner: a fast ordered restriction map aligner. Bioinformatics. 2016;32:1016–1022. doi: 10.1093/bioinformatics/btv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muggli, M., Puglisi, S.J., Boucher, C., 2014. Efficient indexed alignment of contigs to optical maps, 68–81.
- 14.Leinonen M., Salmela L. Optical map guided genome assembly. BMC Bioinformatics. 2020;21:285. doi: 10.1186/s12859-020-03623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon J.R., Xu J., Dileep V., Zhan Y., Song F., Le V.T., Yardimci G.G., Chakraborty A., Bann D.V., Wang Y. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 2018;50:1388–1398. doi: 10.1038/s41588-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barseghyan H., Tang W., Wang R.T., Almalvez M., Segura E., Bramble M.S., Lipson A., Douine E.D., Lee H., Delot E.C. Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 2017;9:90. doi: 10.1186/s13073-017-0479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai Y., Li P., Wang Z., Liang F., Yang F., Fang L., Huang Y., Huang S., Zhou J., Wang D. Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD) J. Med. Genet. 2020;57:109–120. doi: 10.1136/jmedgenet-2019-106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan E.K.F., Cameron D.L., Petersen D.C., Lyons R.J., Baldi B.F., Papenfuss A.T., Thomas D.M., Hayes V.M. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018;28:726–738. doi: 10.1101/gr.227975.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luebeck J., Coruh C., Dehkordi S.R., Lange J.T., Turner K.M., Deshpande V., Pai D.A., Zhang C., Rajkumar U., Law J.A. AmpliconReconstructor integrates NGS and optical mapping to resolve the complex structures of focal amplifications. Nat. Commun. 2020;11:4374. doi: 10.1038/s41467-020-18099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neveling K., Mantere T., Vermeulen S., Oorsprong M., van Beek R., Kater-Baats E., Pauper M., van der Zande G., Smeets D., Weghuis D.O. Next generation cytogenetics: comprehensive assessment of 48 leukemia genomes by genome imaging. bioRxiv. 2020 doi: 10.1101/2020.02.06.935742. 2020.02.06.935742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valouev A., Schwartz D.C., Zhou S., Waterman M.S. An algorithm for assembly of ordered restriction maps from single DNA molecules. Proc. Natl. Acad. Sci. U S A. 2006;103:15770–15775. doi: 10.1073/pnas.0604040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagarajan N., Read T.D., Pop M. Scaffolding and validation of bacterial genome assemblies using optical restriction maps. Bioinformatics. 2008;24:1229–1235. doi: 10.1093/bioinformatics/btn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung A.K.-Y., Kwok T.-P., Wan R., Xiao M., Kwok P.-Y., Yip K.Y., Chan T.-F. OMBlast: alignment tool for optical mapping using a seed-and-extend approach. Bioinformatics. 2016;33:311–319. doi: 10.1093/bioinformatics/btw620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelton J.M., Coleman M.C., Herndon N., Lu N., Lam E.T., Anantharaman T., Sheth P., Brown S.J. Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics. 2015;16:734. doi: 10.1186/s12864-015-1911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Y., Chung C.Y., Chan T.F. Advances in optical mapping for genomic research. Comput. Struct. Biotechnol. J. 2020;18:2051–2062. doi: 10.1016/j.csbj.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Leung A.K., Kwok T.P., Lai Y.Y.Y., Pang I.K., Chung G.T., Mak A.C.Y., Poon A., Chu C., Li M. OMSV enables accurate and comprehensive identification of large structural variations from nanochannel-based single-molecule optical maps. Genome Biol. 2017;18:230. doi: 10.1186/s13059-017-1356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung A.K.-Y., Jin N., Yip K.Y., Chan T.-F. OMTools: a software package for visualizing and processing optical mapping data. Bioinformatics. 2017;33:2933–2935. doi: 10.1093/bioinformatics/btx317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parikh H., Mohiyuddin M., Lam H.Y., Iyer H., Chen D., Pratt M., Bartha G., Spies N., Losert W., Zook J.M. svclassify: a method to establish benchmark structural variant calls. BMC Genomics. 2016;17:64. doi: 10.1186/s12864-016-2366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgin J., Molitor C., Mohareb F. MapOptics: a light-weight, cross-platform visualization tool for optical mapping alignment. Bioinformatics. 2019;35:2671–2673. doi: 10.1093/bioinformatics/bty1013. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner K.M., Deshpande V., Beyter D., Koga T., Rusert J., Lee C., Li B., Arden K., Ren B., Nathanson D.A. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C.X., Li Y., Obermoeller-McCormick L.M., Schwartz A.L., Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J. Biol. Chem. 2001;276:28889–28896. doi: 10.1074/jbc.M102727200. [DOI] [PubMed] [Google Scholar]
- 34.Li N., Shi K., Li W. TUSC7: a novel tumor suppressor long non-coding RNA in human cancers. J. Cell Physiol. 2018;233:6401–6407. doi: 10.1002/jcp.26544. [DOI] [PubMed] [Google Scholar]
- 35.Waters C.E., Saldivar J.C., Hosseini S.A., Huebner K. The FHIT gene product: tumor suppressor and genome ”caretaker”. Cell Mol. Life Sci. 2014;71:4577–4587. doi: 10.1007/s00018-014-1722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kresse S.H., Ohnstad H.O., Paulsen E.B., Bjerkehagen B., Szuhai K., Serra M., Schaefer K.L., Myklebost O., Meza-Zepeda L.A. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer. Aug 2009;48:679–693. doi: 10.1002/gcc.20675. [DOI] [PubMed] [Google Scholar]
- 37.Wu G., Yan Y., Wang X., Ren X., Chen X., Zeng S., Wei J., Qian L., Yang X., Ou C. CFHR1 is a potentially downregulated gene in lung adenocarcinoma. Mol. Med. Rep. 2019;20:3642–3648. doi: 10.3892/mmr.2019.10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontanari Krause L.M., Japp A.S., Krause A., Mooster J., Chopra M., Muschen M., Bohlander S.K. Identification and characterization of OSTL (RNF217) encoding a RING-IBR-RING protein adjacent to a translocation breakpoint involving ETV6 in childhood ALL. Sci. Rep. 2014;4:6565. doi: 10.1038/srep06565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengupta N., Yau C., Sakthianandeswaren A., Mouradov D., Gibbs P., Suraweera N., Cazier J.B., Polanco-Echeverry G., Ghosh A., Thaha M. Analysis of colorectal cancers in British Bangladeshi identifies early onset, frequent mucinous histotype and a high prevalence of RBFOX1 deletion. Mol. Cancer. 2013;12:1. doi: 10.1186/1476-4598-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong J., Zhou Y., Liu D., Huo J., Jun F-box proteins involved in cancer-associated drug resistance. Oncol. Lett. 2018;15:8891–8900. doi: 10.3892/ol.2018.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imaoka H., Toiyama Y., Saigusa S., Kawamura M., Kawamoto A., Okugawa Y., Hiro J., Tanaka K., Inoue Y., Mohri Y. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 2015;36:346–354. doi: 10.1093/carcin/bgu327. [DOI] [PubMed] [Google Scholar]
- 42.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou B., Ho S.S., Greer S.U., Zhu X., Bell J.M., Arthur J.G., Spies N., Zhang X., Byeon S., Pattni R. Comprehensive, integrated, and phased whole-genome analysis of the primary ENCODE cell line K562. Genome Res. 2019;29:472–484. doi: 10.1101/gr.234948.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi H., Wang Z. Atypical microdeletion in 22q11 deletion syndrome reveals new candidate causative genes: a case report and literature review. Medicine (Baltimore) 2018;97:e9936. doi: 10.1097/MD.0000000000009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matejcic M., Li D., Prescott N.J., Lewis C.M., Mathew C.G., Parker M.I. Association of a deletion of GSTT2B with an altered risk of oesophageal squamous cell carcinoma in a South African population: a case-control study. PLoS One. 2011;6:e29366. doi: 10.1371/journal.pone.0029366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinhart W.F., Reifenberger J.G., Gupta D., Muralidhar A., Sheats J., Cao H., Dorfman K.D. Distribution of distances between DNA barcode labels in nanochannels close to the persistence length. J. Chem. Phys. 2015;142:064902. doi: 10.1063/1.4907552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for FaNDOM is available on GitHub at https://github.com/jluebeck/FaNDOM.
We used optical map data from the following individuals, and these data were obtained from the publicly available Bionano Saphyr datasets (https://bionanogenomics.com/library/datasets/)—NA12878, GM09888, GM08331, and GM24143. For cancer SV detection, we used previously published20 Bionano Saphyr data from cancer cell lines K562, CAKI-2, and NCI-H460.