Abstract
Bacteria encase their cytoplasmic membrane with peptidoglycan (PG) to maintain the shape of the cell and protect it from bursting. The enlargement of the PG layer is facilitated by the coordinated activities of PG synthesising and -cleaving enzymes. In Escherichia coli, the cytoplasmic membrane-bound lytic transglycosylase MltG associates with PG synthases and was suggested to terminate the polymerisation of PG glycan strands. Using pull-down and surface plasmon resonance, we detected interactions between MltG from Bacillus subtilis and two PG synthases; the class A PBP1 and the class B PBP2B. Using in vitro PG synthesis assays with radio-labelled or fluorophore-labelled B. subtilis-type and/or E. coli-type lipid II, we showed that both, BsMltG and EcMltG, are lytic tranglycosylases and that their activity is higher during ongoing glycan strand polymerisation. MltG competed with the transpeptidase activity of class A PBPs, but had no effect on their glycosyltransferase activity, and produced glycan strands with a length of 7 disaccharide units from cleavage in the nascent strands. We hypothesize that MltG cleaves the nascent strands to produce short glycan strands that are used in the cell for a yet unknown process.
Abbreviations: PBP, penicillin-binding protein; PG, peptidoglycan; TPase, transpeptidase; GTase, glycosyltransferase; LT, lytic transglycosylase
Keywords: Peptidoglycan, Lytic transglycosylase, Penicillin-binding protein
Introduction
Most bacteria are engulfed by peptidoglycan (PG), a mesh-like molecule that maintains the shape of the cell and protects it from bursting due to the turgor (Vollmer et al., 2008). Growing cells continuously synthesise, modify and cleave PG to maintain cell integrity (Vollmer et al., 2008). PG is synthesised from the precursor lipid II. Glycosyltransferases (GTases) polymerize glycan strands and DD-transpeptidases (TPases) form peptide crosslinks (Barrett et al., 2007, Egan et al., 2020, Lovering et al., 2012, Van Heijenoort, 2001). PG synthases comprise bifunctional class A penicillin-binding proteins (PBPs) with GTase and TPase activity, SEDS and Mtg proteins with GTase activity, and monofunctional class B PBPs with TPase activity (Egan et al., 2015, Meeske et al., 2016, Taguchi et al., 2019).
In Bacillus subtilis, BsPBP1 is the most abundant class A PBP with roles in PG synthesis during length growth and cell division (Claessen et al., 2008, Pedersen et al., 1999). Escherichia coli has two semi-redundant class A PBPs, EcPBP1A and EcPBP1B, with preferential roles in elongation and cell division, respectively (Banzhaf et al., 2012, Bertsche et al., 2006, Yousif et al., 1985). EcPBP1A and EcPBP1B are activated by the outer membrane-anchored lipoproteins LpoA and LpoB, respectively (Paradis-Bleau et al., 2010, Typas et al., 2010). The SEDS proteins BsFtsW and EcFtsW interact with their cognate class B PBP, BsPBP2B and EcPBP3, respectively, and synthesise PG in the test tube (Fraipont et al., 2011, Rohs et al., 2018, Sjodt et al., 2018, Taguchi et al., 2019).
In B. subtilis and E. coli, glycan strands terminate with 1,6-anhydroMurNAc residues that harbours an intramolecular ring from C1 to C6 (Atrih et al., 1999, Glauner et al., 1988, Höltje et al., 1975). AnhydroMurNAc sugars are synthesised by lytic transglycosylases (LTs) that catalyse the non-hydrolytic cleavage of the glycosidic bond between the N-acetylmuramic acid and N-acetylglucosamine (Höltje et al., 1975). B. subtilis has five known or predicted LTs, YomI, SleB, CwlQ, YuiC and SleC, of which some may play roles in the lysis of the spore cortex for germination (Kumazawa et al., 2007, Quay et al., 2015, Sudiarta et al., 2010, Smith et al., 2000). E. coli has nine LTs, of which MltA, MltB, MltC, MltD, MltE, MltF and RlpA are lipoproteins anchored to the outer membrane (Jorgenson et al., 2014, van Heijenoort, 2011), Slt is a periplasmic enzyme (Höltje et al., 1975), and EcMltG an inner membrane protein (Yunck et al., 2016).
Bioinformatics analysis showed that EcMltG and its defining YceG domain are widely distributed in the bacterial domain (Yunck et al., 2016). EcMltG is not essential and cells lacking EcMltG have no growth defect but the overexpression of EcMltG in cells lacking the synthase EcPBP1B resulted in loss of rod shape and cell death (Yunck et al., 2016). Purified EcMltG has a weak activity against PG causing the release of glycan strands with anhydroMurNAc ends, and EcMltG associates with EcPBP1B and RodA but not with EcPBP1A, GTase inactive EcPBP1B E233Q or RodA D262A in bacterial two-hybrid experiments (Bohrhunter et al., 2020, Yunck et al., 2016), hence, EcMltG has been hypothesized to act as a terminase of glycan strand synthesis. In Streptococcus pneumoniae, SpMltG has an additional cytoplasmic domain of unknown function and the loss of SpMltG resulted in a growth defect and a more spherical cell shape (Tsui et al., 2016).
B. subtilis has two MltG homologues with a predicted YceG-like domain, the forespore germination-specific SleB (Moriyama et al., 1999), and YrrL. Here, we focused on YrrL which has 32% amino acid sequence identity and 50% similarity with EcMltG extending over 356 amino acid residues (Fig. S1). We renamed YrrL to BsMltG. We show that purified BsMltG interacts with several PBPs and that it has lytic transglycosylase activity on nascent glycan strands, but is inactive against crosslinked PG, producing short glycan strands with a length of 7 disaccharide residues.
Methods
Media and general methods
Bacterial strains and plasmids used in this work are listed in Tables S1 and S2, respectively, and primers are listed in Table S3. E. coli cells were cultivated in Luria Britani (LB). Fresh cultures were inoculated with overnight culture and grown at 30 or 37˚C with continuous shaking. For solid media, 1% agar (Bacteriological agar no. 1, Oxoid) was added in addition to the appropriate antibiotic concentration (50 µg/ml Kanamycin). Competent E. coli cells were produced according to Hanahan et al. (1991), and heat shock transformations were used for DH5α as described by Bergmans et al. (1981). DNA was purified using DNA purification kit (Quiagen) as per manufacturer's instructions.
Cloning of strains and plasmids
Plasmids were generated using a ligase-free cloning method, which requires 4 sets of primers and 2 DNA templates: plasmid DNA and genomic DNA. pET-28a(+) plasmid was amplified using JS128-JS129 primers, and inserts were amplified using the oligonucleotides in Table S3. Plasmid construction was performed as described in Richardson et al. (2016). For site directed mutagenesis, a PCR reaction was performed using NEB Q5 high fidelity polymerase, plasmid construct as a template, and the corresponding primers from Table S3. Plasmids were purified using Mini-prep kit (Quiagen) as per manufacturer’s instruction.
Protein purification
BsMltG, BsMltG E242A, BsPBP2B, EcMltG and EcMltG E218Q. BL21 (DE3) cells with the corresponding plasmids were grown in LB medium at 37˚C to OD600 0.5. Gene expression was induced with 1 mM IPTG for 3 h at 30˚C. Cells were pelleted by centrifugation (6371 × g / 4˚C/ 15 min) and resuspended in 40 ml buffer I (25 mM Tris/HCl, 1 M NaCl, pH 7.5) supplemented with protease inhibitor cocktail (PIC), phenylmethylsulfonyl fluoride (PMSF) and DNase (≈ 1 mg). Cells were sonicated for 3 × 20 s at 5, 16, 22, 33, 44 and 60 W. Membrane proteins were pelleted by ultracentrifuge at 133,907 × g at 4˚C, for 1 h, and the soluble fraction was discarded. The pellet was resuspended in running buffer (25 mM Tris/HCl, 1 M NaCl, 20 mM imidazole, 2% Triton X-100, pH 7.5). Solubilised proteins were mixed gently with equilibrated Ni-NTA beads for 24 h at 4˚C. The mixture was applied to a gravity column and bound proteins were eluted in elution buffer (25 mM Tris/HCl, 400 mM imidazole, 1 M NaCl, 0.2% Triton X-100, pH 7.5). Restriction grade thrombin was added to eluted proteins, and samples were dialysed against 2 × 2 l of dialysis buffer I (25 mM Tris/HCl, 500 mM NaCl, pH 6.5) and 2 × 2 l of dialysis buffer II (25 mM Tris/HCl, 100 mM NaCl, pH 6.5) for overnight at 4˚C. Ion exchange chromatography was performed using an Äkta Prime FPLC with a HiTrap SP HP column. Dialysed protein was injected onto the column equilibrated with buffer I (25 mM Tris/HCl, 100 mM NaCl, 0.2% Triton X-100, pH 6.5). Bound protein was eluted in a 50 ml gradient to buffer II (25 mM Tris/HCl, 1 M NaCl, 0.2% Triton X-100, pH 7.5).
PBP1 and PBP1 (S390A). The purification protocol was adopted from (Rismondo et al., 2016) and modified. BL21 (DE3) cells with corresponding plasmids were grown in 5 l of LB medium with 50 μg/ml kanamycin and 20 ml/l auto-induction medium (250 mg/l glycerol, 100 g/L α-lactose and 25 g/L glucose) for 18 h at 30˚C. Cells were harvested, and sonicated. Cell membranes were pelleted by ultracentrifuge at 133907 × g then resuspended in resuspension buffer (50 mM Hepes/NaOH, 500 mM NaCl, 3 mM MgCl2, 2% Triton X-100, 15% glycerol, 10 mM β-mercaptoethanol, pH 7.5). Membrane extracts were ultracentrifuged at 100000 × g and the supernatant was supplied with 20 mM imidazole. The first purification step was performed using a 5 ml HisTrap HP column attached to an ÄKTA Prime plus FPLC. Proteins were injected using running buffer (50 mM Hepes/NaOH, 500 mM NaCl, 3 mM MgCl2, 0.2% reduced Triton X-100, 15% glycerol, 20 mM imidazole, pH 7.5) and eluted with elution buffer (same as running buffer with 250 mM imidazole). Protein samples were mixed with restriction grade thrombin and dialysed overnight at 4˚C against 3 l dialysis buffer (25 mM Hepes/NaOH, 300 mM NaCl, 10% glycerol, 0.2% Triton X-100, pH 8.5). Next, ion exchange chromatography was performed using an Äkta Prime FPLC with a HiTrap SP HP column. Dialysed protein samples were injected into the column using buffer I (Same as the dialysis buffer with 0.2% Triton X-100) and eluted in a 200 ml gradient with buffer II (10 mM Hepes/NaOH, 1 M NaCl, 3 mM MgCl2, 0.2% Triton X-100, 12% glycerol, pH 7.5). The third purification step consisted of a size exclusion chromatography using a Superdex 75 column. Proteins were eluted with SEC buffer (10 mM Hepes/NaOH, 300 mM NaCl, 3 mM MgCl2, 0.2% Triton X-100, 12% glycerol, pH 7.5).
PBP1B, PBP1B S510A and LpoB were purified as described in (Born et al., 2006). PBP1A and LpoA were purified as described in (Born et al., 2006) and (Jean et al., 2014), respectively.
In vitro pulldown assay
This method was adapted from (Egan et al., 2015). Proteins (1 μM) were mixed in 200 μl binding buffer (10 mM Hepes/NaOH, 10 mM MgCl2, 150 mM NaCl, 0.05% Triton X-100, pH 7.5). Samples were applied to 100 μl of washed and equilibrated Ni-NTA superflow beads (Qiagen, The Netherlands) and incubated overnight at 4˚C with gentle mixing. The beads were then washed with wash buffer (10 mM Hepes/NaOH, 10 mM MgCl2, 500 mM NaCl, 50 mM imidazole, 0.05% Triton X-100, pH 7.5) and boiled in SDS–PAGE loading buffer. Beads were pelleted by centrifugation and samples analysed by SDS–PAGE. Gels were stained with Coomassie brilliant blue (Roth, Germany).
Surface Plasmon Resonance (SPR) assay
This method was adapted from (Vollmer et al., 1999). Binding assays were performed at 25˚C in running buffer (10 mM Tris/Maleate, 150 mM NaCl, 0.05% Triton X-100, pH 7.5). Proteins to be injected (analyte) were dialysed into 2 × 1 l of dialysis buffer (10 mM Tris/Maleate, 150 mM NaCl, pH 7.5) then centrifuged using a Beckman TLA120.2 rotor (90000 rpm, 30 min, 4˚C) to remove aggregates. The concentration of the protein was measured and the analytes were diluted in running buffer to 6 concentration ranges from 0 to 500 nM. It was important to make sure the Triton X-100 level in the analyte was as close to the running buffer as possible. SigmaPlot software (windows version 13.0) was used for kinetic calculations. Several repeats (at least 3) were required across a range of analyte concentrations. The KD (nM) of ligand binding was based on the assumption of a one site saturation with the use of the equation y = Bmax × x(KD + x) where y is the response (RU) for an analyte concentration in (nM), and Bmax is the maximum response recorded (RU).
In vitro glycosyltransferase activity assay
For assay using radio-labelled B. subtilis or E. coli lipid II, experiments were performed as published (Egan and Vollmer, 2016). For assay using ATTO-550 lipid II, experiments were performed as published in (Barrett et al., 2007, van’t Veer et al., 2016) with modifications according to (Egan et al., 2018). Glycan strands were separated by Tris-Tricine SDS-PAGE.
In vitro peptidoglycan synthesis assay
This method was performed as in (Cleverley et al., 2016) for BsPBP1, and as in (Bertsche et al., 2005) for EcPBP1B and EcPBP1A. For HPLC analysis, a linear gradient was used at 55˚C for 90 min, from 100% solvent A (50 mM sodium phosphate pH 4.31 + 0.0002% NaN3) to 100% solvent B (75 mM sodium phosphate, 15% methanol, pH 4.75) for BsPBP1 and EcPBP1B samples, and solvent B (75 mM sodium phosphate, 30% methanol, pH 4.75) for EcPBP1A samples.
Cell wall purification and muropeptide analysis
This method was adapted from (Atrih et al., 1999) and modified as per (Bisicchia et al., 2011).
Results
BsMltG interacts with BsPBP1 and BsPBP2B
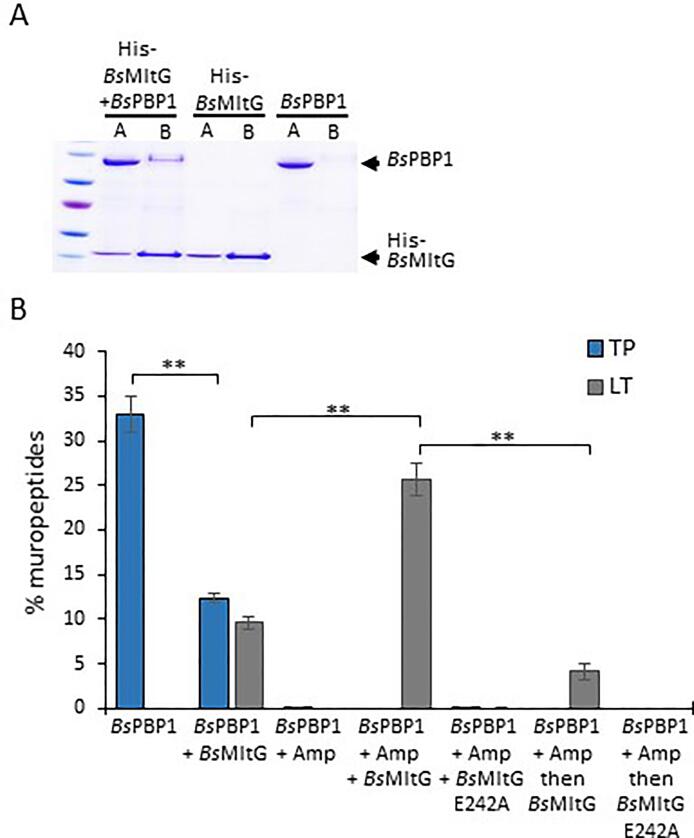
EcMltG presumably associates with EcPBP1B based on bacterial two-hybrid experiments (Yunck et al., 2016). To test if BsMltG interacts with a class A PBP, we purified hexahistidine tagged BsMltG (His-BsMltG) and tested its possible interaction with BsPBP1 in a pull-down experiment. SDS-PAGE analysis shows that His-BsMltG bound to Ni-NTA, and BsPBP1 was not retained by the beads in the absence of His-BsMltG (Fig. 1A). However, BsPBP1 was present in the bound fraction when applied together with His-BsMltG suggesting the two proteins interact. The interaction between BsMltG and BsPBP1 was also tested by surface plasmon resonance (SPR), but due to the high level of unspecific binding of proteins to the chip surface, we were not able to confirm the interaction by SPR.
Fig. 1.
BsMltG interacts with BsPBP1 and is active during ongoing glycan strand polymerisation. (A) Pull-down assays performed to test if BsMltG interacts BsPBP1. The Coomassie-stained SDS-PAGE analysis shows His-BsMltG and BsPBP1 in the applied and bound fractions suggesting that His-BsMltG pulled down BsPBP1. A, Applied; B, bound fractions. (B) Level of crosslinked and anhydro-N-acetylmuramic acid containing muropeptides generated in reactions with BsPBP1 or BsMltG versions, or combinations, in assays with the B. subtilis-type lipid II. The PG product was digested with cellosyl and the resulting muropeptides were reduced and separated by HPLC. Representative HPLC chromatograms are shown in Fig. S4. Values are the mean ± standard deviation of three independent experiments. BsMltG has lytic transglycosylase activity in the presence of BsPBP1. BsMltG reduces the TPase activity of BsPBP1. Blue columns: TPase products (TP); Grey columns: LT products (LT); MltG E242A, catalytically inactive version; Amp; Ampicillin. **, P < 0.01 (T-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also tested if His-BsMltG interacts with the monofunctional cell division-specific BsPBP2B in pull-down experiment. SDS-PAGE analysis shows that His-BsMltG bound to Ni-NTA and BsPBP2B was not pulled down by the beads in the absence of His-BsMltG (Fig. S2A). Interestingly, BsPBP2B in the presence of His-BsMltG appeared in both the pre-Ni-NTA sample (applied) and bound fractions indicating that His-BsMltG pulled down BsPBP2B and suggesting that BsPBP2B interacts with His-BsMltG.
Next, we used SPR to study the interaction between BsMltG and BsPBP2B and determine the dissociation constant. We first immobilized ampicillin (amp) on two traces of a sensor chip. On one trace, PBP2B was covalently bound to the immobilized ampicillin (via its TPase domain). The second trace received no BsPBP2B and served as control. We next incubated both surfaces with β-lactamase to digest ampicillin molecules not bound to PBP2B. Upon the injection of BsMltG, the surface with immobilized PBP2B showed a significant increase in response unit during association (from 0 to 300 s) compared to the control surface upon the injection of BsMltG (Fig. S2B). The response almost reached equilibrium towards the end of the association (from 200 to 300 s). Subsequently, running buffer was injected after 300 s causing the dissociation of the analyte. These results indicate an interaction between BsMltG and BsPBP2B. The binding curve was generated by plotting the response in (RU) during equilibrium against analyte concentration (Fig. S2C). The KD of the BsMltG and BsPBP2B interaction was 128 ± 28 nM, calculated based on a one site interaction model, from three independent experiments using Sigma Plot software. These results suggest that BsMltG interacts with both class A and B PBPs during PG synthesis in B. subtilis.
LT activity of BsMltG
EcMltG showed weak LT activity against purified EcPG, and the loss of EcMltG in cells resulted in longer glycan strands (Yunck et al., 2016). At first, we tested BsMltG LT activity against B. subtilis PG, however, BsMltG did not release muropeptides, suggesting that BsMltG was not active against PG (Fig. S3). Next, assuming that BsMltG processes glycan strands, and supported by the observed interaction of BsMltG with BsPBP1 and BsPBP2B, we hypothesised that BsMltG might be active during ongoing PG synthesis. Since SEDS proteins were shown to be active only in the presence of 20–30% DMSO, which is known to disrupt some protein–protein interactions (Egan et al., 2018), we characterised the activity of MltG in the presence of class A PBPs which are active in the absence of DMSO. Consequently, the activity of purified BsMltG and BsPBP1 were tested in vitro against the B. subtilis type lipid II (with amidated meso-Dap at position 3). A catalytically-inactive BsMltG variant possessing an Ala residue instead of the putative catalytic Glu242 was used in control experiments. BsPBP1 polymerised glycan strands and crosslinked peptide stems producing PG, which was digested by cellosyl and analysed by HPLC (Fig. S4). BsPBP1 alone produced a PG with 32.9% of the peptides participating in crosslinks, whereas, BsMltG alone had no activity against lipid II (Fig. 1B, S4). BsPBP1 in the presence of BsMltG produced a PG with significantly reduced level of peptides in crosslinks (12.3%), and we noticed an additional peak (9.6%) corresponding to GlcNAc-anhydroMurNAc-pentapeptide (PentaAnh), identified by LC-MS/MS with 991.4340 amu (neutral mass, theoretical: 991.4346), eluting at 64 min. The peak corresponding to PentaAnh was not present in samples with BsPBP1 and BsMltG E242A (Fig. S4), confirming that the E242 glutamate residue is essential for BsMltG LT activity. These results suggest that BsMltG has LT activity and reduces the TPase activity of BsPBP1.
Previously, EcMltG was shown to release only uncrosslinked anhydroMurNAc muropeptides from purified PG (Yunck et al., 2016) and it might therefore favour uncrosslinked glycan strands for activity. Consequently, we added ampicillin to samples to block the TPase activity of BsPBP1 (Fig. 1B and S4). BsMltG produced significantly more (25.7%) LT product in the presence of ampicillin-inhibited BsPBP1, suggesting that BsMltG shows enhanced activity against uncrosslinked, nascent PG strands. Next, to test if BsMltG requires ongoing PG synthesis by BsPBP1 for activity, glycan strands were first synthesised by BsPBP1 in the presence of ampicillin followed by digest with BsMltG (BsPBP1 + Amp then BsMltG sample, Fig. 1B, and S4). In this sample there was approximately 5 times fewer LT products present as in the sample in which BsMltG was present during glycan strand synthesis, suggesting that BsMltG is most active during ongoing glycan strand polymerisation.
To test if BsMltG relies on both, ongoing PG synthesis and its interaction with BsPBP1, for higher activity, its activity was tested against nascent PG strands polymerized from B. subtilis lipid II by EcPBP1B (instead of BsPBP1). BsMltG was less active in the sample with EcPBP1B compared to BsPBP1, producing 4.5% PentaAnh (Fig. S5). Additionally, BsMltG showed high activity against glycan strands produced by BsPBP1 from E. coli type lipid II (with non-amidated meso-Dap), suggesting that the amidation of the meso-DAP has no effect on the activity of BsMltG (Fig. S5).
To test if BsMltG has an endo-LT activity, PG material synthesised by PBP1 in the presence of ampicillin and BsMltG was split into two aliquots, one was digested with the muramidase cellosyl and the other not, followed by HPLC analysis. An exo-LT activity of BsMltG should generate PentaAnh, which elutes in 1 peak at 64 min. However, PentaAnh was not present in the sample with BsMltG that was not digested with cellosyl. The sample digested first with BsMltG and then with cellosyl contained PentaAnh (from the MurNAcAnh termini of the glycan strands) and Penta (from within the glycan strands and the GlcNAc termini) at a ratio of 1/5.2 (Fig. S5), showing that BsMltG was an endo-specific lytic transglycosylase that released glycan strands with an average length of 5–6 disaccharide units from nascent PG strands synthesized by PBP1.
EcMltG reduces the TPase activity of EcPBP1A and EcPBP1B
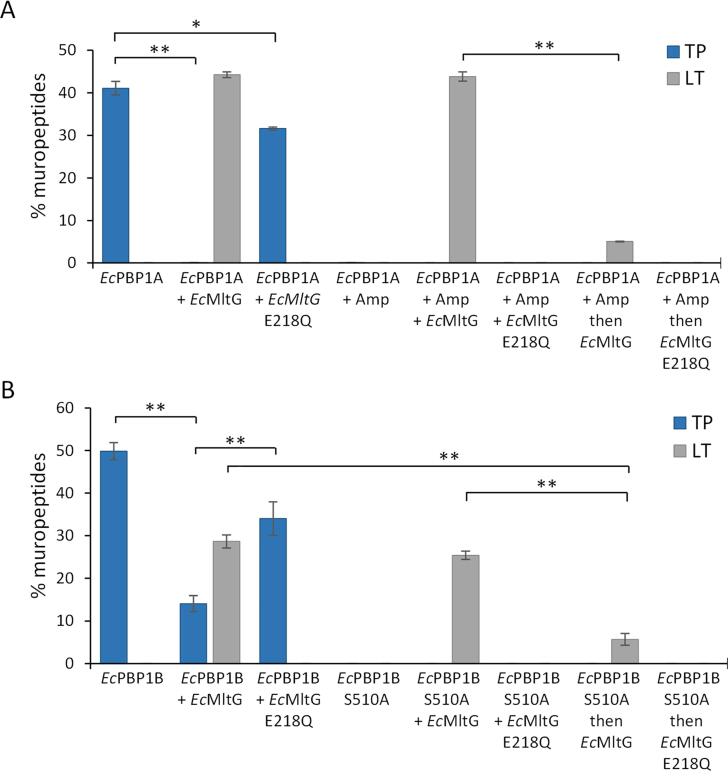
BsMltG was more active in the presence of ongoing glycan strand synthesis by BsPBP1, and reduced the TPase activity of the latter. Since EcMltG showed weak LT activity against E. coli PG and interacts with EcPBP1B, we hypothesised that EcMltG is active during PG synthesis by EcPBP1A or EcPBP1B. To test this hypothesis, we performed PG synthesis experiments using EcPBPs and E. coli-type lipid II. EcPBP1A alone produced PG with 41.1% of the peptides present in crosslinks (Fig. 2 and S6), however, EcPBP1A produced no crosslinks in the presence of EcMltG and the sample contained 44.3% PentaAnh (Fig. 2 and S6), suggesting that EcMltG inhibits the TPase activity of EcPBP1A. EcPBP1A produced fewer crosslinks (31.6%) also in the presence of catalytically inactive EcMltG E218Q, as expected this sample did not contain PentaAnh, suggesting that inactive EcMltG is also able to reduce the TPase activity of EcPBP1A. Additionally, the sample prepared with EcPBP1A (+Amp) and then digested with EcMltG contained significantly lower LT activity (5.0%) (Fig. 2 and S6), suggesting that, similar to BsMltG, EcMltG is most active during ongoing PG synthesis.
Fig. 2.
EcMltG favours ongoing glycan strand polymerisation for LT activity. Levels of crosslinked muropeptides (TP: blue columns) and LT product (LT: grey columns) obtained after cellosyl-digestion of the PG synthesised by EcPBP1A, EcPBP1B and/or EcMltG, from E. coli-type lipid II. Values are the mean ± variation of two independent experiments. EcMltG has a lytic transglycosylase activity in the presence of EcPBP1A or EcPBP1B. EcMltG reduces the TPase activity of EcPBP1A and EcPBP1B. EcPBP1B S510A and BsMltG E218Q, catalytically inactive versions (controls); Amp, Ampicillin. **, P < 0.01; *, P < 0.05 (T-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
EcPBP1B alone produced PG with 49.9% peptides in crosslinks but in the presence of EcMltG approximately 3.5 × fewer peptides were present in crosslinks (14.1%), and 28.7% PentaAnh was produced by EcMltG (Fig. 2 and S7). The presence of EcMltG E218Q resulted also in a significant decrease in TPase products generated by EcPBP1B (34.0%). EcMltG produced 25.4% PentaAnh in the presence of ongoing PG synthesis by a TPase inactive EcPBP1B S510A, close to the levels observed in the presence of active EcPBP1B with EcMltG (Fig. 2 and S7). However, EcMltG produced only 5.7% PentaAnh when it was added after PG synthesis by EcPBP1B S510A, confirming that EcMltG is more active during ongoing glycan strand polymerisation by either EcPBP1A or EcPBP1B.
EcMltG has lower activity in the presence of activated EcPBP1A and EcPBP1B.
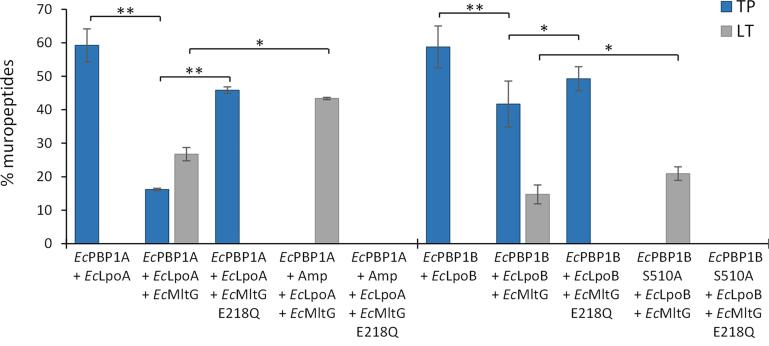
The outer membrane-anchored lipoprotein LpoA activates EcPBP1A by direct interaction and stimulates its TPase activity, whereas, LpoB interacts with EcPBP1B and stimulates its GTase and TPase activities (Egan et al., 2014, Lupoli et al., 2014, Typas et al., 2010). To test the effect of MltG on glycan strand synthesis by Lpo-activated PBPs, we assayed the GTase and TPase activity of EcPBPs, and the LT activity of EcMltG, in the presence of the PBP activators. As expected, the activation of EcPBP1A by LpoA resulted in increased levels of peptides present in crosslinks (59.2%) compared to EcPBP1A alone (41.1%) (Fig. 2, Fig. 3). The addition of EcMltG resulted in a significant decrease in peptides present in crosslinks to 16.2% (Fig. 3 and S8A) which, however, was above the 0% peptides in crosslinks without LpoA (Fig. 2). These results suggest that the activation of EcPBP1A rescued some of the TPase activity of PBP1A despite the presence of EcMltG. On the other hand, EcMltG synthesised 26.7% PentaAnh in the presence of EcPBP1A and LpoA (Fig. 3 and S8A), which was significantly lower than in the presence of EcPBP1A without LpoA (44.3%) (Fig. 2). Blocking the TPase activity of EcPBP1A with ampicillin (EcPBP1A, Amp, LpoA, EcMltG sample) resulted in a significant increase in PentaAnh (43.4%). These results show that the increase in TPase activity is matched by a decrease in LT activity, and vice versa, and suggest that EcPBP1A and EcMltG compete with each other for the glycan strand substrates.
Fig. 3.
EcMltG is active in the presence of activated EcPBP1A and EcPBP1B. Level of crosslinked muropeptides (TP: blue columns) and LT products (grey columns) obtained after cellosyl-digestion of PG synthesised by Lpo-activated EcPBP1A and EcPBP1B in the presence or absence of EcMltG versions, in assays with E. coli-type lipid II. Values are the mean ± variation of two independent experiments. EcMltG reduces the TPase activity of both, activated EcPBP1A and EcPBP1B. EcMltG had lower LT activity in the presence of Lpo-activated EcPBP1A or EcPBP1B, compared to EcPBP1A or EcPBP1B alone. EcPBP1B S510A and EcMltG E218Q, catalytically inactive versions (controls); Amp, Ampicillin. **, P < 0.01; *, P < 0.05 (T-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
EcPBP1B produced PG with 58.8% peptides in crosslinks in the presence of LpoB (Fig. 3 and S8B), however, the addition of EcMltG resulted in a significant decrease in TPase products to 41.7% and the production of 14.7% PentaAnh. The sample of EcPBP1B with LpoB and EcMltG E218Q also showed lower levels of TPase products (49.3%) compared to EcPBP1B with LpoB (Fig. 3 and S8B), suggesting that catalytically active or inactive EcMltG compromises the TPase activity of LpoB-activated EcPBP1B. EcMltG produced 20.9% PentaAnh in the presence of EcPBP1B S510A and LpoB (Fig. 3) compared to 25.4% in the presence of EcPBP1B S510A alone (Fig. 2B), suggesting that increased glycan synthesis rate resulted in a small decrease in EcMltG LT activity. Taken together, these results suggest that EcMltG processes growing PG strands and this LT activity decreases the TPase activity of class A PBPs, presumably by competing for the same PG strands, despite the activation by the Lpo protein.
MltG has no effect on the GTase activity of class A PBPs
The effect of MltG on the GTase activity of PBPs was tested using fluorescent-labelled Dansyl-lipid II. The polymerisation of glycan strands from Dansyl-lipid II and the digestion of these by a muramidase results in a decrease in fluorescence over time. As predicted, EcMltG or BsMltG alone did not cause a decrease in fluorescence over time demonstrating that both enzymes have no GTase activity (Fig. S9A and B). BsPBP1 alone, or BsPBP1 with BsMltG showed similar decrease in fluorescence showing that BsMltG does not affect the GTase activity of BsPBP1 (Fig. S9A). Similarly, EcPBP1A showed a decrease in fluorescence alone or in the presence of EcMltG and/or LpoA, suggesting that neither EcMltG nor LpoA affects the GTase activity of EcPBP1A (Fig. S9B). Confirming previous data (Egan et al., 2014), the addition of LpoB resulted in a faster decrease in fluorescence by EcPBP1B, but EcMltG had no effect on EcPBP1B alone or EcPBP1B/LpoB (Fig. S9C). These data show that EcMltG has no effect on the GTase activity of PBP1B, or its activation by LpoB.
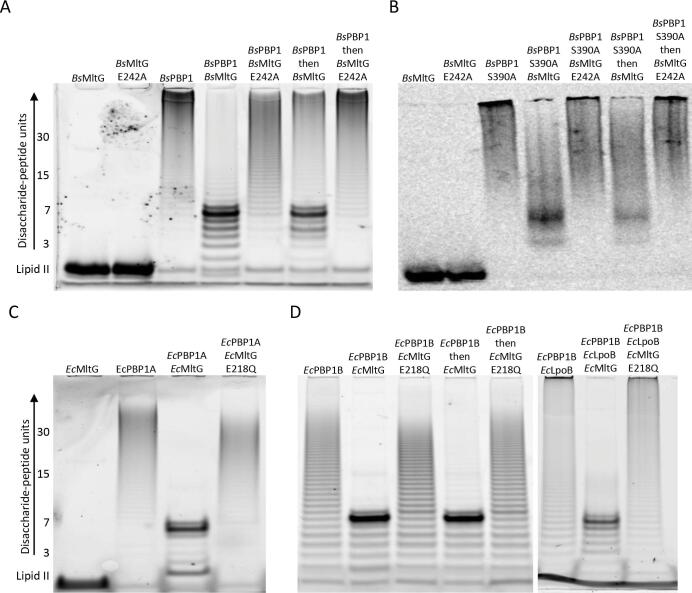
EcMltG and BsMltG process glycan strands after the seventh disaccharide repeat
EcMltG and BsMltG cleaved newly synthesised glycan strands, suggesting a role in glycan length determination. To test this hypothesis, the activity of PBPs and MltG was tested against radioactive or fluorescent (ATTO-550) lipid II in the presence of ampicillin, and the resulting glycan strands were analysed by SDS-PAGE. BsPBP1 alone produced long glycan strands that migrated at the top of the gel (Fig. 4A). The presence of BsMltG and not BsMltG E242A caused the formation of shorter glycan strands, with a distinctive increase in strands with 7 disaccharide units. Adding BsMltG after the strands were synthesized, also generated glycan strands with 7 disaccharide units, however, longer glycan strands were also detected suggesting that BsMltG was less active, consistent with the previous experiments (Fig. 1B). Testing the activity of BsMltG and/or BsPBP1 against radioactive lipid II without the fluorophore produced similar results with a significant increase in short strands, albeit the glycan strand separation in the gel was poorer in this case (Fig. 4B). EcMltG was also tested using the fluorescent lipid II in the presence of EcPBP1A and EcPBP1B (Fig. 4C and D). EcPBP1A or EcPBP1B alone produced long glycan strands, however, the presence of EcMltG decreased the glycan strand length and, again, there was a significant increase in strands with 7 disaccharide units (Fig. 4C–D). This suggests that MltG from both species produces glycan strands with 7 disaccharide units when it cleaves the nascent strands during GTase reactions. Next, we tested if a faster rate of glycan synthesis would affect PG cleavage by EcMltG by testing EcPBP1B with LpoB and EcMltG against lipid II (Fig. 4D). EcPBP1B and LpoB produced glycan strands with a broader length distribution (Egan et al., 2018). The addition of EcMltG resulted in a similar processing of the glycan strands and a clear increase in the strands with 7 disaccharide repeats, suggesting that the length of strands generated by EcMltG is independent from the rate of glycan polymerisation (Fig. 4D).
Fig. 4.
BsMltG and EcMltG cleave PG glycan strands after the seventh disaccharide unit. Glycan strand length assays for products of BsPBP1, EcPBP1A and EcPBP1B formed in the presence of MltG from B. subtilis or E. coli. B. subtilis Proteins were incubated with fluorescent ATTO-550 lipid II (A) or [14C]-GlcNAc-labelled lipid II (B), and ampicillin to block cross-linking. (C) E. coli proteins incubated with fluorescent lipid and ampicillin. PG synthesised by the enzymes was resolved by SDS-PAGE and visualised by fluorescence imaging. The position of the substrates is marked by ‘Lipid II’, the length of the glycan strand is given in disaccharide units.
Discussion
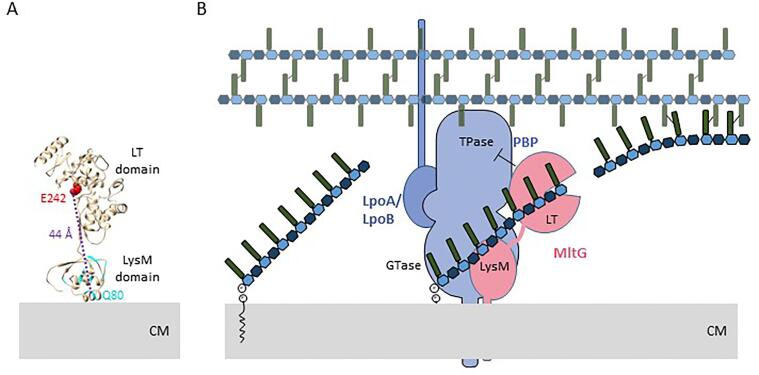
BsMltG showed 67% sequence similarity and 53% identity to Lmo1499, a membrane bound lytic transglycosylase from Listeria monocytogenes (LmMltG). LmMltG has a transmembrane region followed by a LysM domain, involved in PG binding (Buist et al., 2008), and a catalytic domain close to the C-terminus of the protein (PDB: 4IIW), and this domain organization is conserved in BsMltG (Fig. 5A and S1). The LysM domain of MltG could mediate the binding to newly synthesised glycan strand in close proximity to the cytoplasmic membrane (Fig. 5). Interestingly, MltG produced glycan strands with 7 disaccharide residues, in the presence or absence of ongoing PG synthesis, suggesting that MltG binds a nascent glycan strand protruding from the membrane and cleaves it after 7 disaccharide units. In the model of BsMltG the distance between the LT catalytic residue E242 and one of the distant and conserved LysM domain residues Q80 is ~ 44 Å (Fig. 5A and S1), which corresponds to the length of 4–5 disaccharide units, given the length of one disaccharide unit of ~ 10 Å (Fig. 5A) (Carlström, 1962, Carlström, 1957). Presumably, the length of the glycan strand produced by MltG depends also on the distance of the LT active site to the terminal disaccharide unit. Future work will explore whether MltG can be used to produce a sufficient amount of glycan strands with a desired length for biochemical or structural studies.
Fig. 5.
Model of glycan strand cleavage by MltG. (A) BsMltG residues were modelled using the crystal structure of Lmo1499 (PDB: 4IIW) as template (Yang et al., 2020). Highly conserved residues in the LysM domain are in cyan (Fig. S1), and the catalytic E242 residue is in red. The distance from the LT catalytic residue E242 to one of the distant and conserved LysM domain residues, Q80, is ~ 44 Å. (B) MltG is recruited to PG synthesis sites, presumably by interacting with PBPs, and competes with their TPase activity, cleaving in the nascent glycan strands to produce strands with 7 disaccharide units. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Maintaining the integrity of the PG layer is crucial to protect the cell from bursting due to its turgor. In this manuscript, we showed that MltG cleaves in nascent PG strands produced by class A PBPs and competes with their TPase activity even in the presence of the Lpo activators. These results are consistent with published data showing that the loss of MltG function suppressed a lethal deficiency in class A PBP activity (Bohrhunter et al., 2020), presumably by alleviating the reduction of TPase activity exerted by MltG on PBPs, and the decrease in PG cleavage. Interestingly, the absence or overexpression of EcMltG has no effect on the fitness of wild type E. coli cells (Yunck et al., 2016).
What is the cellular function of MltG? It was previously hypothesized that MltG is a 'terminase' for glycan strand polymerization (Yunck et al., 2016). Prior to Yunck et al., 2016, the term 'terminase' was only used for an enzyme involved in bacteriophage lambda genome packaging (Catalano, 2000) and not for enzymes cleaving in nascent polymers. MltG could generate short glycan strands either by terminating the GTase activity of the class A PBP after the nascent glycan strand reaches a certain length, causing glycan strand release. Alternatively, MltG cleaves within the nascent glycan strand during its synthesis, without affecting the GTase reaction. Our data support the latter hypothesis since the addition of MltG did not show an effect on the GTase activity of class A PBPs, which continued in the presence of MltG (Fig. S9). Additionally, MltG might be needed to detach a nascent glycan strand from the membrane after it has been incorporated into the PG layer (Yunck et al., 2016; Fig. 5B). Indeed, pulse chase experiments showed the formation of MurNAcAnh-containing LT products shortly after glycan strand polymerization suggesting that LT enzymes may act in close proximity to PG synthases or as part of the PG synthesis machinery (Burman and Park, 1983, Glauner and Höltje, 1990). Consistent with this hypothesis, BsMltG interacts with BsPBP1 and BsPBP2B, and EcMltG associates with GTase active EcPBP1B and RodA (Bohrhunter et al., 2020, Yunck et al., 2016), which presumably targets MltG to the newly synthesised PG strands (Fig. 5B). However, this function does not appear to be essential and it remained unclear whether the detachment of glycan strands from the membrane occurs in the mltG mutant and, if so, if it is important. Our discoveries that MltG produces 7-disaccharide long glycan strands, which was corroborated by work done in the group of Suzanne Walker (preprint on BioRxiv, Taguchi and Walker, 2021) and that it competes with the TPase function of class A PBPs over the use of glycan strands, could point to another function. We hypothesize that MltG utilizes some of the nascent PG made by PG synthases to produce short glycan strands that are used for a yet unknown process. These glycan strands could be used to initiate the polymerisation of a new glycan strand, or they could be used to repair defects in the PG layer. For example, the GTase function of PBP1B is needed together with the LD-transpeptidase LtdD and the DD-carboxypeptidase PBP6A for cell survival upon the inhibition of LPS export to the outer membrane (Morè et al., 2019). It remains to be seen whether MltG participates in the repair of defective PG or has other cellular functions.
Conclusion
Our results suggest that inner-membrane bound lytic transglycosylases like MltG can modulate PG synthesis not only through their catalytic activities but also by interacting with and regulating the activities of PG synthases, and ultimately determining the structure of the PG.
CRediT authorship contribution statement
Jad Sassine: Conceptualization, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization. Manuel Pazos: Investigation, Writing - review & editing. Eefjan Breukink: Resources, Writing - review & editing. Waldemar Vollmer: Conceptualization, Methodology, Validation, Data curation, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the European Union, Marie Curie ITN AMBER, 317338 (WV) and the UK Medical Research Council (MR/N002679/1) (WV). We thank Dr Joe Gray, Newcastle University, for MS analysis of muropeptides.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tcsw.2021.100053.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Atrih, A., Bacher, G., Williamson, M.P., Foster, S.J., 1999. Analysis of peptidoglycan Structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181, 3956–3966. https://jb.asm.org/content/181/13/3956.long. [DOI] [PMC free article] [PubMed]
- Banzhaf M., van den Berg van Saparoea B., Terrak M., Fraipont C., Egan A., Philippe J., Zapun A., Breukink E., Nguyen-Distèche M., den Blaauwen T., Vollmer W. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 2012;85:179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- Barrett D., Andrew Wang T.S., Yuan Y., Zhang Y., Kahne D., Walker S. Analysis of glycan polymers produced by peptidoglycan glycosyltransferases. J. Biol. Chem. 2007;282:31964–31971. doi: 10.1074/jbc.M705440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmans H.E.N., Van Die I.M., Hoekstra W.P.M. Transformation in Escherichia coli: stages in the process. J. Bacteriol. 1981;146:564–570. doi: 10.1128/jb.146.2.564-570.1981. https://jb.asm.org/content/146/2/564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U., Breukink E., Kast T., Vollmer W. In vitro murein (Peptidoglycan) synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J. Biol. Chem. 2005;280:38096–38101. doi: 10.1074/jbc.M508646200. [DOI] [PubMed] [Google Scholar]
- Bertsche U., Kast T., Wolf B., Fraipont C., Aarsman M.E.G., Kannenberg K., Von Rechenberg M., Nguyen-Distèche M., Den Blaauwen T., Höltje J.-V., Vollmer W. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 2006;61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- Bisicchia P., Bui N.K., Aldridge C., Vollmer W., Devine K.M. Acquisition of VanB-type vancomycin resistance by Bacillus subtilis: the impact on gene expression, cell wall composition and morphology. Mol. Microbiol. 2011;81:157–178. doi: 10.1111/j.1365-2958.2011.07684.x. [DOI] [PubMed] [Google Scholar]
- Bohrhunter J.L., Rohs P.D.A., Torres G., Yunck R., Bernhardt T.G. MltG activity antagonizes cell wall synthesis by both types of peptidoglycan polymerases in Escherichia coli. Mol. Microbiol. 2020;mmi.14660 doi: 10.1111/mmi.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born P., Breukink E., Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J. Biol. Chem. 2006;281:26985–26993. doi: 10.1074/jbc.M604083200. [DOI] [PubMed] [Google Scholar]
- Buist G., Steen A., Kok J., Kuipers O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Burman L.G., Park J.T. Changes in the composition of Escherichia coli murein as it ages during exponential growth. J. Bacteriol. 1983;155:447–453. doi: 10.1128/jb.155.2.447-453.1983. https://jb.asm.org/content/155/2/447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlström D. The polysaccharide chain of chitin. Biochim. Biophys. Acta. 1962;59:361–364. doi: 10.1016/0006-3002(62)90185-3. [DOI] [PubMed] [Google Scholar]
- Carlström D. The crystal structure of α-chitin (poly-N-acetyl-D-glucosamine) J. Biophys. Biochem. Cytol. 1957;3:669–683. doi: 10.1083/jcb.3.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano C.E., 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life. Sci. 57, 128–148. https://link.springer.com/article/10.1007/s000180050503. [DOI] [PMC free article] [PubMed]
- Claessen D., Emmins R., Hamoen L.W., Daniel R.A., Errington J., Edwards D.H. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- Cleverley R.M., Rismondo J., Lockhart-Cairns M.P., Van Bentum P.T., Egan A.J.F., Vollmer W., Halbedel S., Baldock C., Breukink E., Lewis R.J. Subunit arrangement in GpsB, a regulator of cell wall biosynthesis. Microb. Drug Resist. 2016;22:446–460. doi: 10.1089/mdr.2016.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A.J.F., Biboy J., van’t Veer I., Breukink E., Vollmer W. Activities and regulation of peptidoglycan synthases. Philos. Trans. R. Soc. B Biol. Sci. 2015;370:20150031. doi: 10.1098/rstb.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A.J.F., Errington J., Vollmer W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020;18:446–460. doi: 10.1038/s41579-020-0366-3. [DOI] [PubMed] [Google Scholar]
- Egan, A.J.F., Jean, N.L., Koumoutsi, A., Bougault, C.M., Biboy, J., Sassine, J., Solovyova, A.S., Breukink, E., Typas, A., Vollmer, W., Simorre, J.P., 2014. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc. Natl. Acad. Sci. U. S. A. 111, 8197–8202. https://doi.org/10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed]
- Egan A.J.F., Maya-Martinez R., Ayala I., Bougault C.M., Banzhaf M., Breukink E., Vollmer W., Simorre J.-P. Induced conformational changes activate the peptidoglycan synthase PBP1B. Mol. Microbiol. 2018;110:335–356. doi: 10.1111/mmi.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A.J.F., Vollmer W. Continuous fluorescence assay for peptidoglycan glycosyltransferases. Methods Mol. Biol. 2016;1440:171–184. doi: 10.1007/978-1-4939-3676-2_13. [DOI] [PubMed] [Google Scholar]
- Fraipont C., Alexeeva S., Wolf B., van der Ploeg R., Schloesser M., den Blaauwen T., Nguyen-Disteche M. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology. 2011;157:251–259. doi: 10.1099/mic.0.040071-0. [DOI] [PubMed] [Google Scholar]
- Glauner B., Höltje J.V. Growth pattern of the murein sacculus of Escherichia coli. J. Biol. Chem. 1990;265:18988–18996. doi: 10.1016/S0021-9258(17)30613-0. [DOI] [PubMed] [Google Scholar]
- Glauner, B., Höltje, J.V., Schwarz, U., 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088–10095. https://www.pnas.org/content/111/22/8197. [PubMed]
- Hanahan D., Jessee J., Bloom F.R. Plasmid transformation of Escherichia coli and other bacteria. Meth. Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-A. [DOI] [PubMed] [Google Scholar]
- Höltje, J.V., Mirelman, D., Sharon, N., Schwarz, U., 1975. Novel type of murein transglycosylase in Escherichia coli. J. Bacteriol. 124, 1067–1076. https://jb.asm.org/content/124/3/1067. [DOI] [PMC free article] [PubMed]
- Jean N.L., Bougault C.M., Lodge A., Derouaux A., Callens G., Egan A.J.F., Ayala I., Lewis R.J., Vollmer W., Simorre J.-P. Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: implications for PBP1A stimulation. Structure. 2014;22:1047–1054. doi: 10.1016/j.str.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson M.A., Chen Y., Yahashiri A., Popham D.L., Weiss D.S. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol. Microbiol. 2014;124:1067-1076. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa T., Masayama A., Fukuoka S., Makino S., Yoshimura T., Moriyama R. Mode of action of a germination-specific cortex-lytic enzyme, SleC, of Clostridium perfringens S40. Biosci. Biotechnol. Biochem. 2007;71:884–892. doi: 10.1271/bbb.60511. [DOI] [PubMed] [Google Scholar]
- Lovering A.L., Safadi S.S., Strynadka N.C.J. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- Lupoli T.J., Lebar M.D., Markovski M., Bernhardt T., Kahne D., Walker S. Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J. Am. Chem. Soc. 2014;136:52–55. doi: 10.1021/ja410813j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske A.J., Riley E.P., Robins W.P., Uehara T., Mekalanos J.J., Kahne D., Walker S., Kruse A.C., Bernhardt T.G., Rudner D.Z. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morè N., Martorana A.M., Biboy J., Otten C., Winkle M., Serrano C.K.G., Montón Silva A., Atkinson L., Yau H., Breukink E., den Blaauwen T., Vollmer W., Polissi A. Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. MBio. 2019;10:e02729–e2818. doi: 10.1128/mBio.02729-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama R., Fukuoka H., Miyata S., Kudoh S., Hattori A., Kozuka S., Yasuda Y., Tochikubo K., Makino S. Expression of a germination-specific amidase, sleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J. Bacteriol. 1999;181:2373–2378. doi: 10.1128/jb.181.8.2373-2378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau, C., Markovski, M., Uehara, T., Lupoli, T.J., Walker, S., Kahne, D.E., Bernhardt, T.G., 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 143, 1110–1120. https://doi.org/10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed]
- Pedersen, L.B., Angert, E.R., Setlow, P., 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181, 3201–3211. https://jb.asm.org/content/181/10/3201. [DOI] [PMC free article] [PubMed]
- Quay D.H.X., Cole A.R., Cryar A., Thalassinos K., Williams M., Bhakta S., Keep N.H. Structure of the stationary phase survival protein YuiC from B. subtilis. BMC Struct. Biol. 2015;15:1–14. doi: 10.1186/s12900-015-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson T.T., Harran O., Murray H. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature. 2016;534:412–416. doi: 10.1038/nature17962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismondo J., Cleverley R.M., Lane H.V., Großhennig S., Steglich A., Möller L., Mannala G.K., Hain T., Lewis R.J., Halbedel S. Structure of the bacterial cell division determinant GpsB and its interaction with penicillin-binding proteins. Mol. Microbiol. 2016;99:978–998. doi: 10.1111/mmi.13279. [DOI] [PubMed] [Google Scholar]
- Rohs P.D.A., Buss J., Sim S.I., Squyres G.R., Srisuknimit V., Smith M., Cho H., Sjodt M., Kruse A.C., Garner E.C., Walker S., Kahne D.E., Bernhardt T.G. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 2018;14:e1007726. doi: 10.1371/journal.pgen.1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodt M., Brock K., Dobihal G., Rohs P.D.A., Green A.G., Hopf T.A., Meeske A.J., Srisuknimit V., Kahne D., Walker S., Marks D.S., Bernhardt T.G., Rudner D.Z., Kruse A.C. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature. 2018;556:118–121. doi: 10.1038/nature25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.J., Blackman S.A., Foster S.J. Autolysins of Bacillus subtilis: Multiple enzymes with multiple functions. Microbiology. 2000;146:249–262. doi: 10.1115/1.4026364. [DOI] [PubMed] [Google Scholar]
- Sudiarta I.P., Fukushima T., Sekiguchi J. Bacillus subtilis CwlQ (previous YjbJ) is a bifunctional enzyme exhibiting muramidase and soluble-lytic transglycosylase activities. Biochem. Biophys. Res. Commun. 2010;398:606–612. doi: 10.1016/j.bbrc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Taguchi, A., Walker, S., 2021. Biochemical reconstitution defines new functions for membrane-bound glycosidases in assembly of the bacterial cell wall. bioRxiv 2021.03.06.434200; doi: https://doi.org/10.1101/2021.03.06.434200. [DOI] [PMC free article] [PubMed]
- Taguchi A., Welsh M.A., Marmont L.S., Lee W., Sjodt M., Kruse A.C., Kahne D., Bernhardt T.G., Walker S. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 2019;4:587–594. doi: 10.1038/s41564-018-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.-C.T., Zheng J.J., Magallon A.N., Ryan J.D., Yunck R., Rued B.E., Bernhardt T.G., Winkler M.E. Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol. Microbiol. 2016;100:1039–1065. doi: 10.1111/mmi.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A., Banzhaf M., Den Berg Van, Van Saparoea B., Verheul J., Biboy J., Nichols R.J., Zietek M., Beilharz K., Kannenberg K., Von Rechenberg M., Breukink E., Den Blaauwen T., Gross C.A., Vollmer W. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Veer I.L., Leloup N.O.L., Egan A.J.F., Janssen B.J.C., Martin N.I., Vollmer W., Breukink E. Site-specific immobilization of the peptidoglycan synthase PBP1B on a surface plasmon resonance chip surface. ChemBioChem. 2016;17:2250–2256. doi: 10.1002/cbic.201600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heijenoort, J., 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11, 25–36. https://academic.oup.com/glycob/article/11/3/25R/677256. [DOI] [PubMed]
- Vollmer W., Blanot D., de Pedro M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W., von Rechenberg M., Höltje J.V. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- Yang J., Anishchenko I., Park H., Peng Z., Ovchinnikov S., Baker D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. 2020;117:1496–1503. doi: 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousif S.Y., Broome-Smith J.K., Spratt B.G., Beveridge T., Tropini C., Salick M., Crone W.C., Gopinathan A., Huang K.C., Weibel D.B. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 1985;131:2839–2845. doi: 10.1016/j.cels.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Yunck R., Cho H., Bernhardt T.G. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol. Microbiol. 2016;99:700–718. doi: 10.1111/mmi.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.