Kumbhar et al. identify a new poly(ADP-ribose) interaction domain within the demethylase KDM5A that acts in concert with the histone variant macroH2A1.2 to localize this enzyme to DNA lesions, where it regulates damage-associated transcriptional responses and promotes DNA double-strand break repair.
Abstract
The histone demethylase KDM5A erases histone H3 lysine 4 methylation, which is involved in transcription and DNA damage responses (DDRs). While DDR functions of KDM5A have been identified, how KDM5A recognizes DNA lesion sites within chromatin is unknown. Here, we identify two factors that act upstream of KDM5A to promote its association with DNA damage sites. We have identified a noncanonical poly(ADP-ribose) (PAR)–binding region unique to KDM5A. Loss of the PAR-binding region or treatment with PAR polymerase (PARP) inhibitors (PARPi’s) blocks KDM5A–PAR interactions and DNA repair functions of KDM5A. The histone variant macroH2A1.2 is also specifically required for KDM5A recruitment and function at DNA damage sites, including homology-directed repair of DNA double-strand breaks and repression of transcription at DNA breaks. Overall, this work reveals the importance of PAR binding and macroH2A1.2 in KDM5A recognition of DNA lesion sites that drive transcriptional and repair activities at DNA breaks within chromatin that are essential for maintaining genome integrity.
Introduction
DNA is recurrently damaged by endogenous processes, including transcription and replication, as well as by dysregulated proteins and chemical reactions within cells (Chatterjee and Walker, 2017; Lindahl and Barnes, 2000; Tubbs and Nussenzweig, 2017; Xia et al., 2019). Exogenous agents, including UV light, chemicals, and cancer therapies (i.e., radiation), also damage DNA (Ciccia and Elledge, 2010; Jackson and Bartek, 2009). An inability to constrain and repair DNA damage can result in mutations and genome instability. Loss of genome integrity can have dire consequences for cellular and organismal homeostasis, including the development of cancer (Jeggo et al., 2016; Negrini et al., 2010). Cells employ an extensive network of proteins, referred to as the DNA damage response (DDR), that counteract DNA damage by sensing its presence in the genome, activating the appropriate response, and repairing the lesion (Ciccia and Elledge, 2010; Jackson and Bartek, 2009). Mutations within DDR-related genes are frequent in cancer (Knijnenburg et al., 2018), which helps explain the prevalence of genome instability in cancer. For example, DNA double-strand breaks (DSBs) are repaired mainly by homologous recombination (HR) and nonhomologous end joining (Chapman et al., 2012), with the HR factors breast cancer gene types 1 and 2 (BRCA1 and BRCA2) found recurrently mutated in breast and ovarian cancer (Stratton and Rahman, 2008). While mutations in DNA repair genes result in genome instability, these genetic alterations also provide vulnerabilities that can be targeted therapeutically in some genetic backgrounds (Ashworth and Lord, 2018; Jackson and Helleday, 2016; O’Connor, 2015; Pilié et al., 2019). This strategy has been successfully employed against BRCA-deficient cancers, which display synthetic lethality with poly(ADP-ribose; PAR) polymerase (PARP) inhibitors (PARPi’s), a treatment now used clinically (Bryant et al., 2005; Farmer et al., 2005; Fong et al., 2009; Lord and Ashworth, 2017).
Chromatin is an integral component of the DDR as DNA is organized by histones into chromatin, thereby controlling accessibility and DDR factor functions at DNA lesions that reside within the chromatin environment (Agarwal and Miller, 2016; Caron et al., 2019; Chiu et al., 2017; Clouaire and Legube, 2019; Dabin et al., 2016; Gong and Miller, 2019; Kim et al., 2019c; Tan and Huen, 2020). One such chromatin mechanism involved in DDR regulation is histone post-translational modification. Histone marks, including phosphorylation, methylation, acetylation, and ubiquitylation, act to regulate chromatin structure and function to coordinate the DDR with DNA-templated processes such as transcription, replication, and repair (reviewed in Kim et al., 2019c). Histone variants, including the histone H2A family variants H2AX, H2AZ, and macroH2A, also participate in the regulation of the DDR (Buschbeck and Hake, 2017; Corujo and Buschbeck, 2018; Kim et al., 2018; Turinetto and Giachino, 2015). Collectively, chromatin employs multiple mechanisms to engage DDR factors to facilitate DNA damage signaling and repair within the chromatin environment.
Histone methylation plays important roles in the DDR (Gong and Miller, 2019; Kim et al., 2019c). For example, lysine-specific demethylase 5 (KDM5) subfamily proteins, consisting of four jmjC proteins, KDM5A–D, target the transcription-associated marks H3K4me2/3 to regulate transcription and the DDR, including DSB repair, p53 regulation, and the response to oxidative stress (Batie et al., 2019; Bayo et al., 2018; Christensen et al., 2007; Gong et al., 2017; Hu et al., 2018; Klose et al., 2007; Li et al., 2014b; Xu et al., 2018). KDM5A (alias JARID1A) is recruited to DSBs, where it demethylates H3K4me3 to regulate the ZMYND8–nucleosome remodeling and histone deacetylation (NuRD) chromatin remodeling complex (Gong et al., 2017). ZMYND8 binds acetylation marks at DSBs through its acetyl-binding bromodomain, recruits NuRD to repress transcription, and promotes HR repair at DSBs (Gong et al., 2015; Gong et al., 2017; Gong and Miller, 2018; Savitsky et al., 2016; Spruijt et al., 2016). KDM5A is also found to be dysregulated in several cancers. For example, KDM5A is up-regulated in multiple cancers (Cao et al., 2014; Choi et al., 2018; Li et al., 2014a; Teng et al., 2013; Zeng et al., 2010) and promotes resistance to cancer treatments, including to DNA-damaging agents (Hou et al., 2012; Sharma et al., 2010; Vinogradova et al., 2016; Yang et al., 2019). KDM5B is also involved in cancer and radioresistance (Bayo et al., 2018; Xhabija and Kidder, 2019). These findings have highlighted KDM5A and other KDM5s as potential therapeutic targets in cancer. However, the potential connection between the DNA repair activities of KDM5A and its alterations in cancer have not been investigated. Although the function of histone demethylation by KDM5A in transcription and DNA damage repair have been described, the factors that regulate KDM5A in these processes are poorly understood.
Here, we have identified two mechanisms that regulate KDM5A localization to DNA damage sites. First, we identified a PAR interaction domain (PID) within KDM5A in its C terminus that is required to recruit KDM5A to DNA damage sites. This domain is unique to this KDM5 lysine demethylase and likely contains a coiled-coil domain, which is required for binding to PAR chains. KDM5A-PID by itself can localize to damage sites in a PARP-dependent manner, and its presence in KDM5A is required for its genome integrity–promoting functions, including DNA repair. Second, the histone H2A variant macroH2A1.2 promotes KDM5A recruitment to DNA damage. Functionally, loss of macroH2A1.2 results in defective HR and DSB-induced transcriptional repression, which phenocopies KDM5A deficiency. Thus, we propose that PAR binding and macroH2A1.2 recognition engage KDM5A to target this enzyme to DNA damage sites. Identification of KDM5A regulators in the DDR may provide valuable insights into their potential involvement in other biological processes dependent on this lysine demethylase, including transcription and cancer.
Results
KDM5A promotes genome integrity
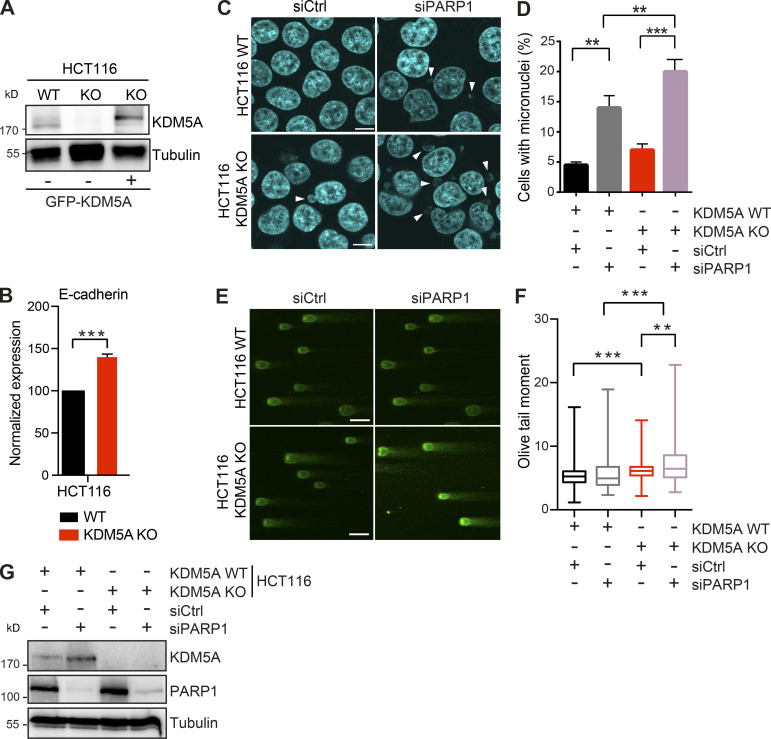
We previously demonstrated the involvement of KDM5A in regulating the ZMYND8-NuRD chromatin remodeling complex at DNA damage sites. The KDM5A–ZMYND8–NuRD axis functions to demethylate H3K4me3 at DNA damage sites, allowing ZMYND8-NuRD to localize to DNA lesions within transcriptionally active chromatin, where this pathway represses transcription and promotes HR repair (Gong et al., 2015; Gong et al., 2017; Gong and Miller, 2018; Savitsky et al., 2016; Spruijt et al., 2016). To gain further insight into the involvement of KDM5A in genome integrity pathways, we obtained colon carcinoma cell lines lacking KDM5A (see Materials and methods). Western blot (WB) analysis confirmed the loss of KDM5A protein in this HCT116 KDM5A-knockout (KDM5A-KO) cell line (Fig. S1 A). We also observed increased expression of the KDM5A target gene E-cadherin in HCT116 KDM5A-KO cells versus WT parental HCT116 cells, providing additional evidence for the functional loss of KDM5A in these cells (Fig. S1 B; Dabiri et al., 2019; Feng et al., 2017; Liang et al., 2015; Wang et al., 2013).
Figure S1.
KDM5A loss results in reduced genome integrity in siPARP1-depleted HCT116 cells. Related to Fig. 1. (A) WB analysis of cell lines from Fig. 1 A with the indicated antibodies. (B) Gene expression analysis of E-cadherin in WT and KDM5A-KO HCT116 cells. Total RNA was isolated for each cell type and analyzed using gene-specific primers. Data are normalized to B2M, ALAS1, and WT samples. Shown is mean ± SEM (n = 3). P values were calculated by unpaired Student’s t test (***, P < 0.001). (C) Micronuclei formation analysis in HCT116 WT and KDM5A-KO cells with or without siPARP1. PARP1 was depleted by siRNA for 48 h followed by DAPI staining to detect micronuclei. Scale bars, 10 µm. White arrowheads mark micronuclei. (D) Quantification of C. n = 2 with >100 cells quantified per condition per replicate. P values were calculated by unpaired Student’s t test (**, P < 0.01; ***, P < 0.001). (E) KDM5A loss results in increased DSBs as detected by neutral comet assay in cells deficient for PARP1. Cells were treated as in C and analyzed by comet assay as in Fig. 1 G. (F) Quantification of E. Olive tail moment for >100 cells quantified per condition per replicate and plotted as a box and whiskers; n = 2. P values were calculated by Tukey’s multiple comparison test (**, P < 0.01; ***, P < 0.001). (G) Validation of PARP1 protein depletion by siRNAs. WT and KDM5A-KO HCT116 cells were treated with siCtrl or siPARP1, and samples were evaluated by WB analysis with the indicated antibodies 48 h after transfection of the siRNAs. Error bars in B, D, and F represent SEM.
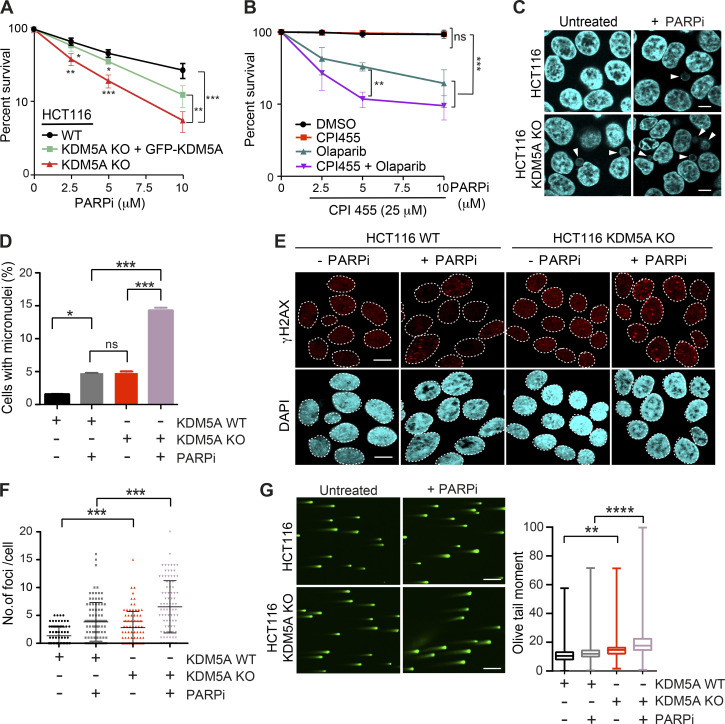
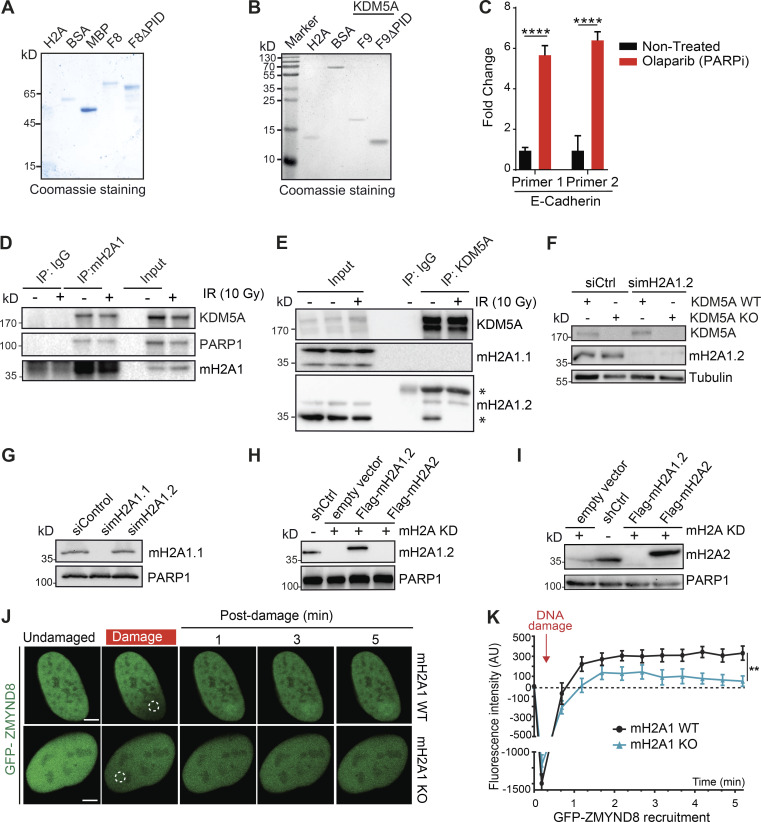
Consistent with the involvement of KDM5A in HR, cells deficient for KDM5A were sensitive to the PARPi olaparib (Fig. 1 A; KDM5A protein levels of cells analyzed in Fig. S1 A). PARPi sensitivity often correlates with HR deficiency because cells deficient for HR factors, such as BRCA1 or BRCA2, are sensitive to PARP inhibition (Bryant et al., 2005; Farmer et al., 2005; Lord and Ashworth, 2017; O’Connor, 2015). To validate that the observed effects were due to KDM5A loss, we reconstituted KDM5A-KO HCT116 cells with GFP-tagged full-length KDM5A (Fig. S1 A). The observed sensitivity of KDM5A-KO cells to PARPi’s was largely rescued by complementation with ectopically supplied KDM5A, confirming that these results were due to loss of KDM5A in these cells (Fig. 1 A). PARP1 activity is required for KDM5A localization to the DNA damage sites to allow efficient DSB repair by HR (Gong et al., 2017). Similar to KDM5A-KO cells, human osteosarcoma (U2OS) cells treated with the KDM5A inhibitor CPI455, which blocks the catalytic activity of this enzyme, displayed reduced cell survival after PARPi treatment compared with untreated cells or cells treated with either single inhibitor (Fig. 1 B). Thus, KDM5A deficiency results in loss of cell viability because of PARP inhibition.
Figure 1.
KDM5A deficiency results in genome instability and sensitivity to PARP inhibition. (A) Clonogenic survival assay of HCT116 WT, KDM5A-KO, and KDM5A-KO + GFP-KDM5A complemented cells. Cells were treated with the PARPi olaparib at the indicated doses for 24 h, and colonies were quantified after 2 wk. Error bars represent SD; n = 3. (B) Clonogenic survival assay of U2OS cells treated with DMSO, 25 µM CPI455, and olaparib at the indicated doses for 24 h. Colonies were analyzed as in A. Error bars represent SD; n = 3. (C) KDM5A suppresses micronuclei formation. HCT116 WT and KDM5A-KO cells were treated with 5 µM olaparib for 24 h and immunostained with DAPI to detect micronuclei. Scale bars, 10 µm. White arrowheads mark micronuclei. (D) Quantification of C. n = 2; >100 cells quantified per condition per replicate. (E and F) KDM5A deficiency leads to DNA damage that is exacerbated by PARP inhibition. HCT116 WT or KDM5A-KO cells were treated with or without 5 µM PARPi (olaparib) for 24 h and immunostained for the DNA break marker γH2AX. γH2AX foci are quantified and plotted in F. n = 2; >100 cells were quantified per condition per replicate. Error bars represent SEM. Scale bars, 20 µm. (G) Loss of KDM5A increases DSBs as detected by neutral comet assay. Cells were treated as in C. Scale bars, 100 µm. Right panel: quantification of left panel. Olive tail moment for >100 cells quantified per condition per replicate and plotted as a box and whiskers; n = 2. Error bars represent SEM. P values were calculated by Tukey’s multiple comparison test. P values for A, B, D, and E were calculated with unpaired Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
KDM5A-deficient cells treated with PARPi’s exhibited several phenotypes consistent with defective genome integrity pathways in these cells that were antagonized by PARPi treatment. This includes increased formation of micronuclei and persistence of DSBs as detected by the DNA damage marker histone variant H2AX phosphorylation (γH2AX; Fig. 1, C–F). These conditions also result in the appearance of persistent DSBs in KDM5A-KO cells as detected by neutral comet assays (Fig. 1 G). Depletion of PARP1 by siRNA gave similar phenotypes in KDM5A-KO cells, validating that PARP1 deficiency by either protein loss or inhibition resulted in increased DNA break formation and loss of genome integrity in cells lacking KDM5A (Fig. S1, C–F; efficiency of siRNA depletion of PARP1 was confirmed by WB analysis in Fig. S1 G).
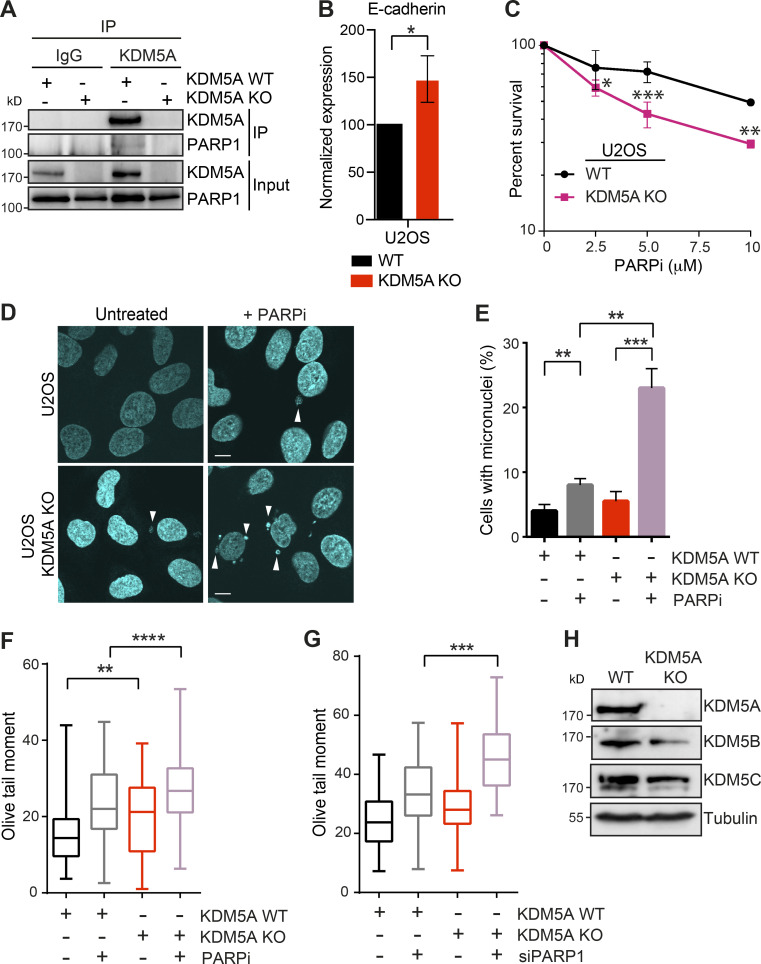
To further support the involvement of KDM5A in genome integrity, we performed the same experiments in U2OS cells lacking KDM5A, which we generated using CRISPR-Cas9 gene editing (Fig. S2 A). Like HCT116 cells, U2OS cells lacking KDM5A derepressed E-cadherin and were sensitive to PARPi treatment (Fig. S2, B and C). U2OS cells lacking KDM5A displayed increased micronuclei formation after PARPi treatment and an inability to clear DNA breaks following PARP1 inhibition by either olaparib or siRNA depletion of PARP1 (Fig. S2, F and G). Loss of KDM5A did not alter the expression of other JARID1 histone demethylases, KDM5B and KDM5C, suggesting these results are specifically due to KDM5A deficiency (Fig. S2 H). Collectively, these results validate our cell models for KDM5A loss and provide strong evidence for the role of KDM5A in genome integrity pathways, including those impacted by PARPi’s (i.e., DNA repair).
Figure S2.
KDM5A loss results in reduced genome integrity in siPARP1-depleted U2OS cells. Related to Fig. 1. (A) KDM5A co-IP and WB analysis in U2OS cells. KDM5A was immunoprecipitated from WT and KDM5A-KO cells, and precipitates were evaluated by WB analysis with the indicated antibodies. Analysis confirms loss of KDM5A protein in KDM5A-KO cells. (B) Gene expression analysis of E-cadherin in WT and KDM5A-KO U2OS cells. Experiments were performed as in Fig. S1 B. n = 3. Error bars represent SEM. P values were calculated by unpaired Student’s t test (*, P < 0.05). (C) U2OS cells lacking KDM5A are sensitive to PARPi. Experiments were performed as in Fig. 1 A. Error bars represent SD; n = 3. (D) Loss of KDM5A increases micronuclei in U2OS cells treated with PARPi. Experiments were performed as in Fig. 1 C. Scale bars, 10 µm. White arrowheads mark micronuclei. (E) Quantification of D. >100 cells were quantified per condition per replicate; n = 2. (F and G) KDM5A loss in U2OS cells results in persistent DSBs as detected by neutral comet assay in cells deficient for PARP1 by either inhibition (F) or siRNA depletion (G). Cells were treated and analyzed as in Fig. 1 G and Fig. S1 E. Olive tail moment for >100 cells quantified per condition per replicate; n = 2. Error bars represent SEM. P values were calculated by Tukey’s multiple comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (H) WB analysis of KDM5 proteins in WT and KDM5A-KO HCT116 cells. P values were calculated by unpaired Student’s t test for B, C, and E (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
KDM5A interacts with PARP1 and PAR chains in cells
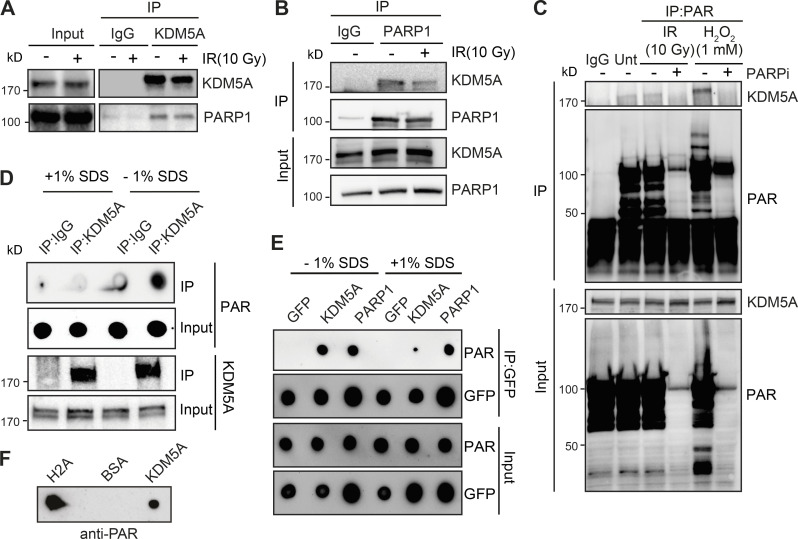
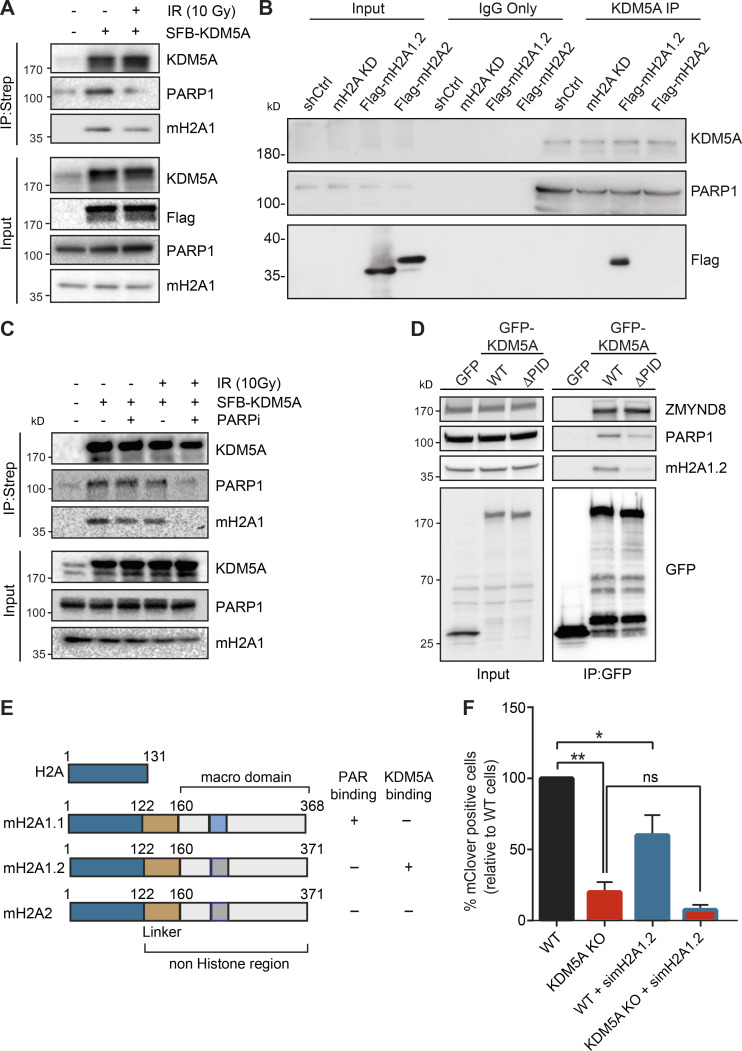
PARP1 recruitment and formation of PAR chains at sites of DNA damage are one of the earliest responses to DNA damage. PARylation promotes the rapid accumulation of DNA damage factors to DNA lesions, including DSBs (Ray Chaudhuri and Nussenzweig, 2017). To further delineate how PARP1 regulates KDM5A in the DDR, we tested whether KDM5A interacts with PARP1 directly by performing coimmunoprecipitation (co-IP) followed by WB analysis. IP of endogenous KDM5A with an anti-KDM5A antibody in human embryonic kidney (HEK293T) cells detected an interaction between endogenous PARP1 and KDM5A (Fig. 2 A). We obtained similar results in U2OS cells and further controlled for the specificity of the KDM5A antibody, as the immunoprecipitated signal and PARP1 interaction were abolished in samples from KDM5A-KO U2OS cells (Fig. S2 A). Reciprocal co-IP WB analysis using PARP1 antibodies in HEK293T cells corroborated the observed interaction between KDM5A and PARP1 (Fig. 2 B). The interaction between PARP1 and KDM5A was still detected and not increased in cells treated with ionizing radiation (IR) compared with cells that were undamaged (Fig. 2, A and B). Reduced PARP1–KDM5A interactions upon IR treatment could be due to increased binding of PARP1 to DNA or DNA damage–induced autoPARylation of PARP, behaviors of PARP1 that are known to disrupt protein–protein interactions (Thomas and Tulin, 2013).
Figure 2.
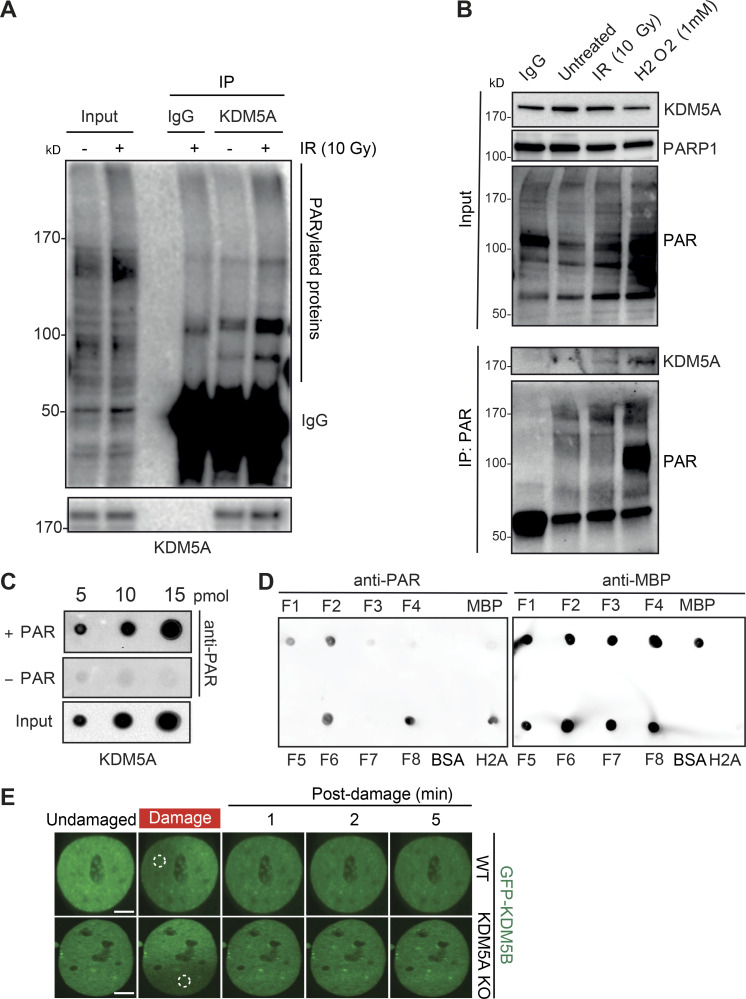
KDM5A interacts with PARP1 and PAR chains. (A) Endogenous KDM5A and PARP1 interact. KDM5A was immunoprecipitated from HEK293T cells with or without 10-Gy IR. Samples were evaluated by WB analysis with the indicated antibodies. (B) Reciprocal IP with endogenous PARP1 was performed as in A. (C) KDM5A binds to PAR in cells in a PARP-dependent manner. HEK293T cells were treated with olaparib (5 µM, 1 h) followed by 10-Gy IR or hydrogen peroxide (H2O2; 1 mM, 15 min) and examined by IP-WB analysis with an anti-PAR antibody. (D and E) KDM5A binds PAR noncovalently. U2OS cells were treated with 10-Gy IR for 2 min, and cell lysates were incubated with KDM5A or IgG antibodies with or without 1% SDS. Purified samples were either blotted on nitrocellulose membrane to detect PAR chains or resolved on SDS-PAGE gel and immunoblotted to detect KDM5A. In E, experiments were performed as in D in cells expressing GFP, GFP-KDM5A, or GFP-PARP1. Immunoprecipitated samples with or without SDS were blotted onto nitrocellulose membrane and detected with anti-PAR and anti-GFP for loading. (F) Purified KDM5A from U2OS cells binds PAR chains in vitro. Purified proteins were spotted onto nitrocellulose membrane and probed with purified PAR chains with binding detected with an anti-PAR antibody.
Given that PARP1 catalytic activity promotes the recruitment of KDM5A to laser-induced DNA damage sites (Gong et al., 2017) and poly(ADP) ribosylation nucleates the localization of many proteins in the vicinity of DNA damage sites (Gibson and Kraus, 2012; Liu et al., 2017; Ray Chaudhuri and Nussenzweig, 2017), we next assessed if KDM5A was associated with PAR in cells. IP of KDM5A or anti-PAR antibodies revealed an association of PARylated proteins with KDM5A (Fig. 2 C and Fig. S3 A), suggesting that KDM5A or its associated proteins are PARylated and/or bind PAR chains. The increased binding between KDM5A and PAR was further enhanced by treatment with hydrogen peroxide, another DNA damage–inducing agent and potent activator of PARP1 (Fig. 2 C and Fig. S3 B). Treatment of cells with the PARPi olaparib inhibited KDM5A–PAR interactions (Fig. 2 C), further supporting that these interactions are PARP dependent. These results established a connection between KDM5A and PARylation in cells but do not distinguish between PAR binding or direct PARylation of KDM5A and/or an interacting protein. To address these various possibilities, KDM5A was immunoprecipitated from cells following DNA damage by IR under denaturing conditions using 1% SDS, and the presence of PAR chains was evaluated by WB analysis. KDM5A–PAR interactions were readily detected under native conditions (i.e., in the absence of SDS) and were abolished by the addition of SDS to denature the samples, which suggested KDM5A was not itself PARylated (Fig. 2 D). We further confirmed our assay conditions using PARP1, which is known to be directly PARylated upon DNA damage. Unlike KDM5A, anti-PAR dot-blot analysis of immunoprecipitated GFP-PARP1 after IR treatment revealed signals in both native and denaturing conditions (Fig. 2 E). Thus, these results are consistent with KDM5A binding PAR chains directly and not being itself a direct substrate for PARylation under these conditions. These interactions are also likely not mediated by nucleic acids, given that cell lysates were treated with a broad-spectrum DNA/RNA nuclease. Collectively, these results indicated that KDM5A interacts with PARP1 and PAR chains in cells in response to DNA damage.
Figure S3.
KDM5A interacts with PAR chains. Related to Figs. 2 and 4. (A) Endogenous KDM5A interacts with PAR chains. Untreated or IR-treated U2OS cells were immunoprecipitated with KDM5A antibody and analyzed with the indicated antibodies. (B) PAR IP identifies increased KDM5A interactions following DNA damage. Experiment was performed as in Fig. 2 C. (C) Immunoprecipitated KDM5A from U2OS cells binds PAR. Experiment was performed as in Fig. 2 E. (D) Analysis of MBP-KDM5A fragments binding to PAR. 10 pM of H2A, BSA, MBP, and MBP-KDM5A fragments F1–F8 were spotted onto nitrocellulose membrane, and PAR-binding analysis was performed as in C. (E) GFP-KDM5B is not recruited to laser-induced DNA damage in either HCT116 WT or KDM5A-KO cells. Scale bars, 5 µm. Dotted white circles specify damaged region.
KDM5A directly binds PAR chains in vitro
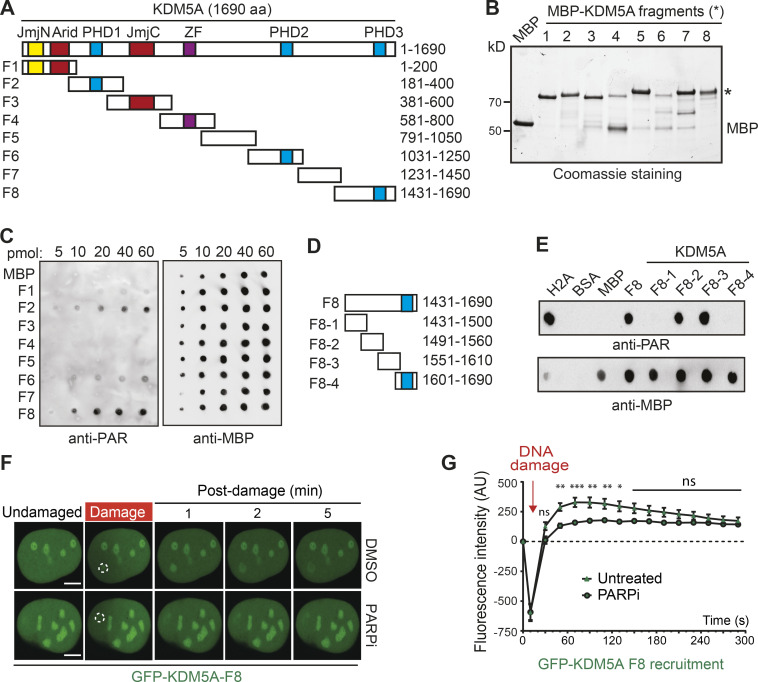
Our results could be explained by the direct binding of KDM5A to PAR chains as a mechanism to direct its association to DNA damage lesions. To test this hypothesis, we determined if KDM5A was capable of binding PAR chains directly using in vitro PAR-binding assays (Ahel et al., 2008; Kumbhar et al., 2018). Purified target proteins, including immunoprecipitated KDM5A, histone H2A (a strong PAR-binding protein), and BSA (negative control), were blotted onto nitrocellulose membrane and probed with purified PAR chains. Using purified KDM5A from U2OS cells, we observed KDM5A binding to PAR chains (Figs. 2 F and S3 C). These results were consistent with KDM5A binding directly to PAR chains and prompted us to identify the PAR-binding region within KDM5A. Several PAR-binding domains have been reported (Krietsch et al., 2013; Teloni and Altmeyer, 2016; Wei and Yu, 2016), but none of these are annotated within KDM5A in the Pfam database. Thus, to identify the PAR-binding region of KDM5A, we dissected full-length KDM5A into eight partially overlapping maltose-binding protein (MBP)-tagged fragments (F1–F8), ensuring that the main functional domains of KDM5A were preserved within the fragments (Fig. 3 A). These fragments were expressed and purified from Escherichia coli, then used in in vitro PAR-binding assays (Fig. 3 B). These experiments identified three putative PAR-binding regions, and the C-terminal fragment of KDM5A (F8), which contains aa 1431–1690, exhibited the strongest binding to PAR chains (Figs. 3 C and S3 D).
Figure 3.
Mapping of a PAR interaction domain within KDM5A. (A) Domain organization of KDM5A. Schematics for overlapping KDM5A fragments (F1–F8) are indicated. (B) Expression of affinity-purified, MBP-tagged KDM5A fragments. Purified fragments were analyzed by SDS-PAGE and Coomassie blue staining. (C) Purified MBP and MBP-KDM5A fragments F1–F8 were analyzed for PAR binding as in Fig. 2 F. (D) Schematics for KDM5A-F8 and derivatives. Blue box indicates PHD3 domain. (E) PAR binding resides in KDM5A-F8-2 and F8-3. PAR binding was performed as in C with indicated proteins. (F) GFP-KDM5A-F8 sufficient for PARP-dependent recruitment to laser damage. Laser microirradiation was performed in U2OS cells transfected with GFP-KDM5A-F8 with or without the PARPi olaparib (5 µM, 1 h). Damaged region is indicated by dotted white circle. Scale bars, 5 µm. (G) Quantification of F from two representative experiment where n ≥ 20 cells. Error bars represent SEM. P values were calculated with unpaired Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The KDM5A-F8 fragment contains the third plant homeodomain (PHD) of KDM5A, which interacts with its substrate H3K4me3 and is dispensable for supporting KDM5A localization to DNA damage sites (Gong et al., 2017; Wang et al., 2009). To further map the PID within KDM5A, we created four additional truncation mutations within F8, including one containing the PHD3 domain (Fig. 3 D). F8-2 (aa 1491–1560) and F8-3 (aa 1551–1610) displayed PAR binding, and, as expected, the fragment containing the PHD3 domain did not bind PAR (Fig. 3 E). These data show that the PAR-binding domain resides within aa 1491–1610 in the C terminus of KDM5A and, importantly, is independent from the PHD3 domain.
We next sought to determine if the KDM5A-F8 region was sufficient to support DNA damage localization. To this end, we generated GFP-tagged KDM5A-F8 (GFP-KDM5A-F8) and analyzed its ability to be recruited to laser-induced DNA damage in cells. We observed rapid recruitment of GFP-KDM5A-F8 to DNA damage sites after microirradiation (Fig. 3 F; quantified in Fig. 3 G). Retention of KDM5A-F8 was transient, with the maximum intensity of recruitment occurring within the first minute after damage induction followed by a steady decline in localization at damage sites. These recruitment dynamics were reminiscent of the short lifespan of PAR chains, which are rapidly removed by PAR hydrolase (Alvarez-Gonzalez and Althaus, 1989; Kassab et al., 2020; O’Sullivan et al., 2019). Consistent with the in vitro binding of PAR chains by KDM5A-F8, recruitment to DNA damage sites of this domain-containing fragment was highly reduced in cells treated with PARPi’s (Fig. 3 F; quantified in Fig. 3 G). In sum, these results map the PID of KDM5A within aa 1491–1610 and show that this region is sufficient for localization to damage sites in a PARP-dependent manner, supporting the identification of a PID within this region of KDM5A.
Characterization of the PAR-binding domain within KDM5A
We next considered whether other KDM5 proteins interact directly with PAR chains. To address this question, we performed DNA damage recruitment analyses of GFP-tagged KDMs and PAR-binding assays with purified GFP-KDM5A, GFP-KDM5B, and GFP-KDM5C from U2OS cells. We did not include the male-specific KDM5D protein for this analysis. KDM5 proteins share similar domain structures and are highly homologous (Fig. 4 A). While GFP-KDM5A is readily detected at laser-induced DNA damage sites, GFP-tagged KDM5B and KDM5C were not recruited to these lesions (Fig. 4, B and C). This result is consistent with another report that did not observe KDM5B recruitment to DNA damage after laser microirradiation (Li et al., 2014b). Since KDM5A and KDM5B are both reported to demethylate H3K4me3 at DNA damage sites, we also tested if KDM5B could be recruited to laser-induced DNA damage in cells lacking KDM5A. Even in cells lacking KDM5A, KDM5B was unable to associate with DNA damage sites to the extent detectable by this approach (Fig. S3 E), suggesting different mechanisms are responsible for regulating these two demethylases in the DDR. KDM5A, but not KDM5B and KDM5C, displayed PAR-binding activities (Fig. 4 D). Taken together, these data suggest that direct binding to PAR chains by KDM5A is a unique mechanism that regulates its damage localization compared with other KDM5 demethylases.
Figure 4.
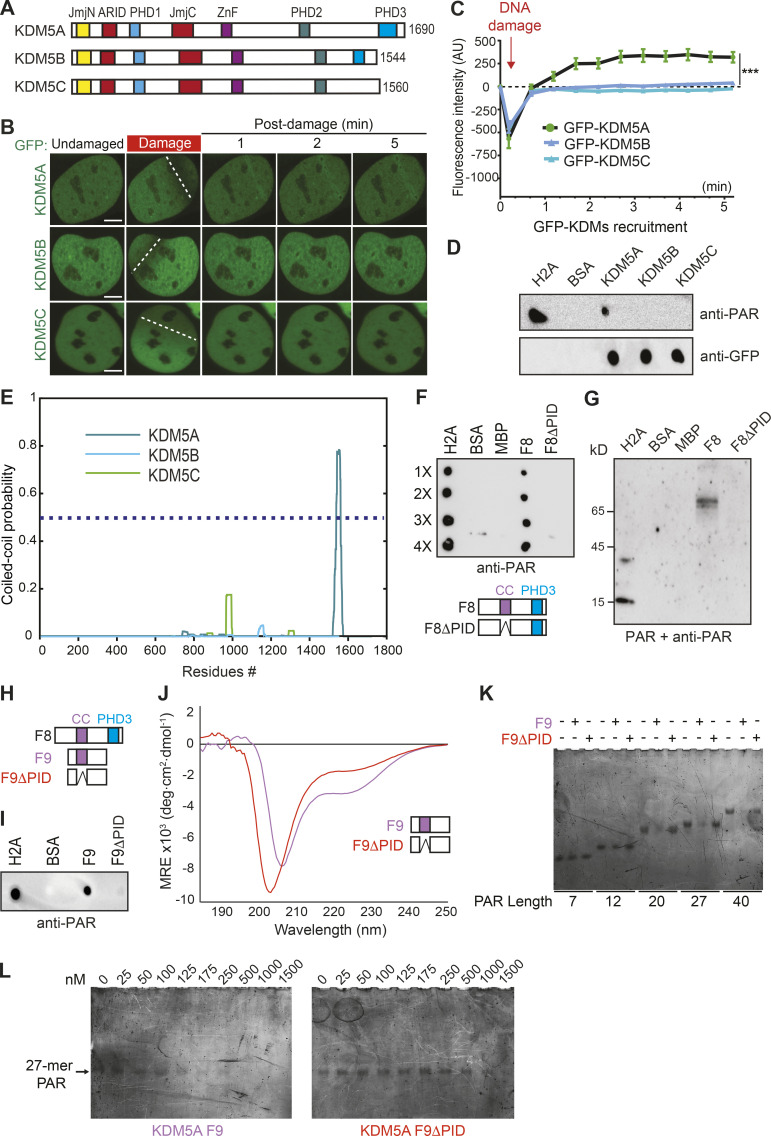
KDM5A contains a coiled-coil domain that facilitates preferential binding to longer PAR chains. (A) Schematic of human KDM5A, KDM5B, and KDM5C. (B) Recruitment of GFP-KDM5A, GFP-KDM5B, and GFP-KDM5C to laser-induced DNA damage sites using live-cell confocal microscopy and laser microirradiation. White dotted lines indicate path of laser damage. Scale bars, 5 µm. (C) Quantification of B from one representative experiment. n > 7 individual cells per condition. Two-way ANOVA with Sidak’s multiple comparison test was used for statistical analysis comparing KDM5A with KDM5B and KDM5C recruitment. Error bars represent SEM (***, P < 0.001). (D) KDM5A uniquely interacts with PAR chains. GFP-tagged KDM5A, KDM5B, and KDM5C were expressed and purified from U2OS cells. PAR binding was performed as in Fig. 2 F. (E) KDM5A contains a predicted coiled-coil region that is absent from KDM5B and KDM5C. Predicted per residue coiled-coil scores for dimers were calculated using MultiCoil (see Materials and methods). (F) KDM5A region predicted as a coiled-coil (residues 1501–1562) is required for PAR binding (PID). PAR-binding assay performed as in D. Diagram of KDM5A-F8 and KDM5A-F8ΔPID with putative coiled-coil (cc) and PHD3 domains indicated. (G) Far-WB analysis of KDM5A PAR binding. The indicated proteins were resolved by SDS-PAGE and blotted onto a nitrocellulose membrane. The membrane was incubated with PAR chains, and the indicated proteins were detected as in D. (H) Comparative diagram of KDM5A-F8 with KDM5A-F9 (1491–1610) and KDM5A-F9ΔPID (1491–1610Δ1501-1562). (I) PAR-binding analysis of KDM5A-F9 and KDM5A-F9ΔPID. PAR binding assay was performed as in D. (J) CD spectrum for the F9 region of KDM5A-F9 (purple) is consistent with the presence of α-helical protein elements, while the CD spectrum for F9ΔPID (1491–1610Δ1501-1562; red) exhibits an appreciable loss in this helicity, supporting the predictive model of a coiled-coil domain between residues 1501 and 1562. Notably, both KDM5A-F9 and F9ΔPID harbor significant intrinsically disordered protein regions. deg, degree; MRE, mean residue ellipticity. (K) KDM5A-F9, but not F9ΔPID, has preferential nM binding for medium and long PAR chains. (L) EMSA titrations demonstrate that KDM5A-F9 has nM binding for a medium-length 27-mer PAR chain, while PAR recognition by F9ΔPID is severely compromised.
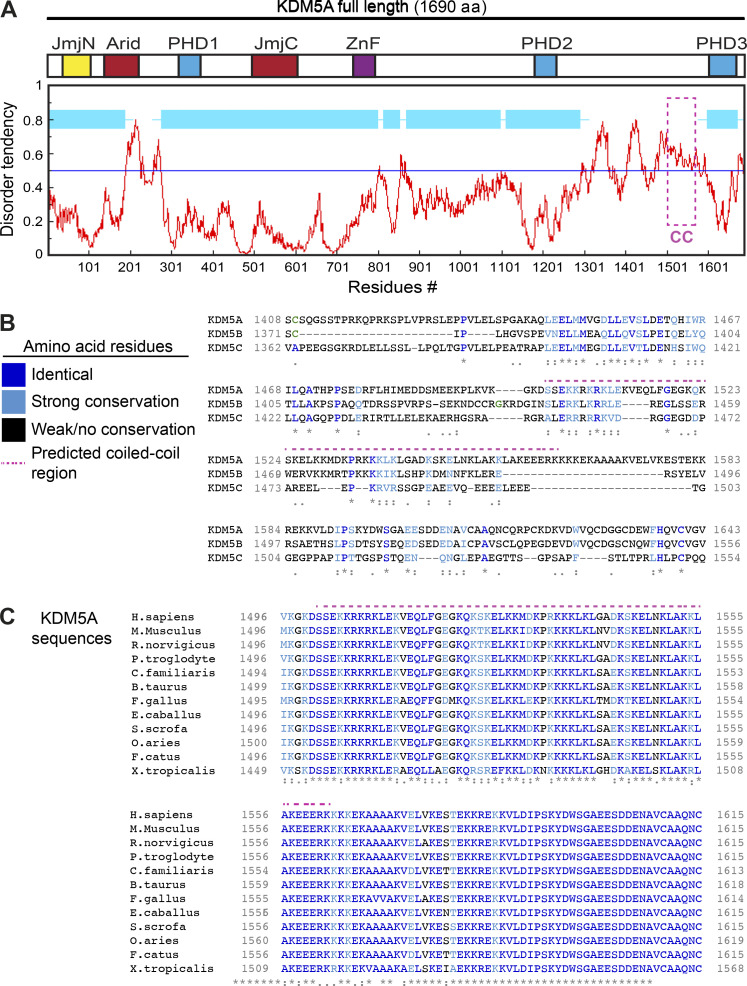
To further define the molecular properties of the PID in KDM5A, we performed a multicoil sequence analysis of KDM5A, KDM5B, and KDM5C. The MultiCoil algorithm predicts coiled-coil location and oligomerization in protein sequences (Wolf et al., 1997). This analysis revealed that the PAR-binding region of KDM5A contains a highly probable coiled-coil domain, which is absent in KDM5B and KDM5C (Fig. 4 E). The region consists of highly basic and hydrophobic amino acids and is predicted to be intrinsically disordered by IUPred, a prediction tool designed to identify protein regions with these biophysical properties that are known to be ideal for PAR binding (Fig. S4, A and B; Dosztányi et al., 2005). This putative coiled-coil PAR-binding domain of KDM5A is not conserved between KDM5B and KDM5C but is highly conserved across KDM5A homologues from higher eukaryotes (Fig. S4, B and C). Intrigued by these observations, we set out to further demonstrate that the putative coiled-coil region containing the PID (residues 1501–1562) is required for PAR binding. To this end, we deleted this region from MBP-tagged KDM5A-F8 to create KDM5A-F8ΔPID. KDM5A-F8 and KDM5A-F8ΔPID were recombinantly purified from E. coli and tested for PAR binding using an in vitro PAR-binding assay. This analysis confirmed that the KDM5A-PID contained within F8 was required for PAR binding because removal of the proposed PID abolished KDM5A-F8 interactions with PAR chains (Fig. 4, F and G; purified proteins shown in Fig. S5 A). We had previously identified two overlapping fragments within F8 of KDM5A that interacted with PAR (Fig. 3 E; fragments F8-2 and F8-3). We purified an untagged KDM5A 1491–1610 fragment (referred to as F9) containing these two domains along with an F9ΔPID (1491–1610Δ1501–1562) mutant of KDM5A and performed PAR-binding assays (schematic of KDM5A-F8 and -F9 constructs shown in Fig. 4 H). The results using these protein products were similar, showing PAR binding to be dependent on the PID of KDM5A (Fig. 4 I; purified proteins shown in Fig. S5 B). These analyses, along with our previous binding studies, support the presence of a PID within the C terminus of KDM5A that is specific to this KDM5 demethylase family member and that contains a putative coiled-coil domain.
Figure S4.
Identification of a conserved putative coiled-coil domain unique to KDM5A. Related to Fig. 4. (A) KDM5A PID is intrinsically disordered. Diagram of KDM5A aligned with a disorder tendency plot (red graph). The disorder tendency of KDM5A was calculated using IUPred. Globular, structured domains in KDM5A are shown in light blue boxes. Magenta box indicates PID. (B) Alignment of human KDM5A aa 1408–1623 and the corresponding regions of human KDM5B and KDM5C. Amino acids are color coordinated as indicated in the legend. PAR-binding–predicted coiled-coil region is indicated by the magenta dotted line. Multiple sequence alignment was performed using Clustal Omega. (C) A multiple sequence alignment of Homo sapiens KDM5A aa 1496–1615 with Mus musculus, Rattus norvegicus, Pan troglodytes, Canis familiaris, Bos taurus, Gallus gallus, Equus caballus, Sus scrofa, Ovis aries, Felis catus, and Xenopus tropicalis. KDM5A exhibits high conservation in this C-terminal region. Colored amino acids are as in B. For B and C, asterisks denote fully conserved residues; colon indicates conservation between strongly similar residues; period indicates conservation between weakly similar residues.
Figure S5.
DNA damage recruitment dynamics, dependencies, and interactions. Related to Figs. 4, 5, 6, and 7. (A and B) Purified proteins from Fig. 4, G and I, respectively, were analyzed by Coomassie blue staining after SDS-PAGE. (C) PARPi treatment increases E-cadherin expression. U2OS cells were treated with 25 µM PARPi for 16 h, and samples were analyzed by RT-qPCR. E-cadherin expression was normalized to untreated and GAPDH. Error bars are SEM; n = 3. P values were calculated by unpaired Student’s t test (****, P < 0.0001). (D) Co-IP with endogenous macroH2A1 was performed as in Fig. 2 A. (E) KDM5A interacts with macroH2A1.2 but not macroH2A1.1. Co-IP with endogenous KDM5A from HEK293T cells was performed as in Fig. 2 A. Asterisks mark bands that are not full-length/unmodified macroH2A1. (F) Validation of macroH2A1.2 antibody. KDM5A WT and KDM5A-KO HCT116 cells were treated with siCtrl or simacroH2A1.2 and evaluated by WB analysis with the indicated antibodies 48 h after transfection. (G) Validation of macroH2A1.1 antibody. HEK293T cells were treated with siCtrl, simacroH2A1.1, and simacroH2A1.2 and analyzed as in F. (H and I) WB analysis of HEPG2 macroH2A variant–expressing cells. Samples from Fig. 6 B were probed with macroH2A1.2 (H) and macroH2A2 (I) antibodies to confirm expression of these variants in these cells. (J) Damage recruitment of GFP-ZMYND8 is abolished in macroH2A1-KO U2OS cells. Dotted white circles indicate damaged region. Scale bars, 5 µm. (K) Quantification of J. n > 10 cells. Error bars represent SEM. P values were calculated by unpaired Student’s t test (**, P < 0.01).
To gain further insight into whether the KDM5A-F9 region contained characteristics of a coiled-coil domain, we performed circular dichroism (CD) spectroscopic analysis on both KDM5A-F9 and F9ΔPID. Consistent with the domain mapping and prediction analyses, KDM5A-F9 displayed an apparent α-helical component, as exemplified by the presence of CD spectral minima at both 222 nm and ∼208 nm (Fig. 4 J). Notably, however, the spectral region between 200 and 215 nm is more negative than would be expected for a predominantly helical protein and is indicative of the F9 region also containing intrinsically disordered protein elements, which is also consistent with our prediction models based on sequence (Fig. S4 A). Further CD analysis of F9ΔPID showed a significant reduction in helical signatures and resulted in a spectrum consistent with a predominantly disordered protein (Fig. 4 J). Together, these data suggest that an α-helical structural element resides within the region spanning KDM5A residues 1501–1562, which encompasses the predicted coiled-coil region and domain responsible for PAR binding.
To better quantify the ability of KDM5A-F9 to recognize PAR chains, electrophoretic gel mobility shift assay (EMSA) analysis was performed. To date, only a few studies have been directed toward quantitively characterizing the molecular recognition of PAR-binding proteins with their PAR ligands (Barkauskaite et al., 2013; Fahrer et al., 2007; Fahrer et al., 2010; Fischer et al., 2014; Krüger et al., 2019; Zhang et al., 2015). This has in part been due to limitations in obtaining sufficient quantities of PAR polymers of a defined length, an impediment that has been overcome (Tan et al., 2012). One intriguing trend that has emerged from these studies is that PAR-binding proteins appear to have PAR chain length–dependent binding preferences (Fahrer et al., 2007; Fahrer et al., 2010; Fischer et al., 2014). Consistent with these previous observations, KDM5A-F9 exhibits preferential recognition for medium- and long-chained PAR polymers, as evidenced by a significant depletion of the free PAR bands, while comparatively at this protein concentration (250 nM), KDM5A F9ΔPID was significantly compromised in recognizing any of the PAR chain lengths (Fig. 4 K). Notably, in the EMSA studies, a supershifted band representing the F9–PAR complex was not detectable even after an extensive effort to optimize the assay. The lack of smearing in the gel and absence of complex being trapped in the well suggests that the complex was not able to enter the gel for detection. Nevertheless, it is unequivocally clear from these studies that the F9 region of KDM5A is responsible for PAR chain recognition, while PAR binding by F9ΔPID is severely compromised.
While the lack of an observable F9–PAR complex band prevented quantification of binding affinities, it can nevertheless be seen from EMSA titration that under our assay conditions, KDM5A-F9 qualitatively binds a medium-sized 27-mer PAR chain with a relatively low nM affinity (∼100–175 nM), a range consistent with known PAR-binding domains (Fig. 4 L; Fahrer et al., 2007; Teloni and Altmeyer, 2016). A moderate capability for binding the 27-mer PAR chain by KDM5A F9ΔPID can only be seen at low μM concentrations (Fig. 4 L). The fact that KDM5A F9ΔPID can exhibit weak recognition of PAR chains at higher protein concentrations suggests that KDM5A-F9 may use additional protein elements outside of the coiled-coil domain to enhance PAR recognition. Taken together, these results are consistent with KDM5A containing a coiled-coil domain within residues 1501–1562 that binds PAR, with a preference for longer PAR chains.
KDM5A-PAR binding is essential for KDM5A recruitment and function at DNA damage sites
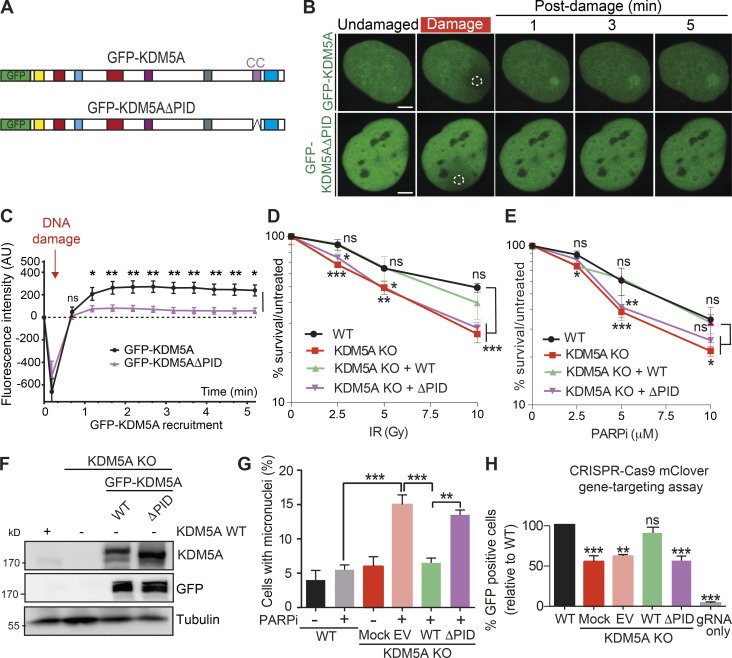
To test for the functional importance of the identified KDM5A PAR-binding region, we analyzed the ability of KDM5A protein lacking the PID to bind to sites of DNA damage and act in the DDR. Interestingly, GFP-KDM5A lacking the PAR-binding region (KDM5AΔPID) displayed reduced recruitment to DNA damage sites compared with full-length GFP-KDM5A (Fig. 5, A–C). To assess if PAR binding by KDM5A is involved in supporting its DDR activities, we reconstituted KDM5A-KO HCT116 cells with WT and ΔPID GFP-KDM5A, allowing us to directly address this question. While KDM5A-KO cells were sensitive to IR and PARPi’s, expression of WT KDM5A, but not KDM5AΔPID, was able to rescue the inability of KDM5A-KO cells to survive these treatments (Fig. 5, D and E; protein expression of KDM5A derivatives analyzed in Fig. 5 F). Similarly, KDM5A-KO cells displayed increased micronuclei formation upon PARPi treatment, which was rescued by WT KDM5A but not KDM5AΔPID (Fig. 5 G). Many of these phenotypes in KDM5A-KO cells could be explained by a defect in HR repair, which we previously reported in KDM5A siRNA-depleted U2OS cells (Gong et al., 2017). Using WT, KDM5A-KO, and KDM5A-KO reconstituted HCT116 cells, we measured HR efficiency using a CRISPR-Cas9/mClover gene-targeting assay (Kim et al., 2019b; Pinder et al., 2015). While KDM5A-KO cells displayed reduced HR compared with WT or KDM5A-KO rescued cells expressing WT KDM5A, expression of KDM5AΔPID was unable to rescue HR deficiencies in KDM5A-KO cells (Fig. 5 H). Interestingly, we observed that PARPi treatment in U2OS cells resulted in increased expression of the KDM5A repressed gene, E-cadherin, suggesting PAR binding by KDM5A may also participate in regulating the transcriptional repressive activities of this chromatin regulator (Fig. S5 C). Taken together, these data demonstrate that KDM5A utilizes a domain consisting of a coiled-coil region in its C terminus to bind PAR chains, which is required to localize this enzyme to damage sites, where it functions on chromatin to promote genome integrity and HR repair.
Figure 5.
The PID containing a coiled-coil region is required for damage localization and DDR functions of KDM5A. (A) Diagram of KDM5A constructs used in B. (B) KDM5A-PID is required for damage localization. Experiments were performed as in Fig. 3 F. Damaged regions are indicated by dotted white outlines. Scale bars, 5 µm. (C) Quantification of B from one representative experiment. Error bars represent SEM; n ≥ 10 cells. Damaged regions are indicated by a dotted white line. (D and E) KDM5A-PID is required for survival in response to IR and PARPi. Clonogenic survival assays were performed as in Fig. 1 A with the indicated ectopically expressed KDM5A genes. Error bars represent SD; n = 3. (F) WB analysis of WT and KDM5A-KO HCT116 cells, including GFP-KDM5A WT and ΔPID reconstituted cells. (G) KDM5A-PID suppresses micronuclei formation. Experiments were performed as in Fig. 1 C with the indicated KDM5A-KO cells and complemented cells as in D. Error bars represent SD; n = 2. P values were calculated by Tukey’s multiple comparison test (**, P < 0.01; ***, P < 0.001). (H) KDM5A-PID is required for efficient homology-directed repair (HDR). HDR efficiency in HCT116 WT and KDM5A-KO cells with or without SFB-KDM5A and SFB-KDM5AΔPID was determined using a CRISPR-mClover HR assay. GFP+ cells represent a repair event. The percentage of GFP+ cells was normalized to WT. Error bars represent SEM; n = 2. P values were calculated for C, D, E, and H with unpaired Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
KDM5A interacts with the histone variant macroH2A1.2
Given the importance of KDM5A in chromatin-based DNA repair activities, we sought to test if other histone pathways may regulate KDM5A activities. We focused on the histone variant macroH2A because this variant can bind and be regulated by PARP and is involved in HR (Khurana et al., 2014; Ruiz et al., 2019; Timinszky et al., 2009). macroH2A1 has two splice variants, macroH2A1.1 and macroH2A1.2, which differ in 32 amino acids within the C-terminal macrodomain, leading to the presence of a PAR-binding domain in macroH2A1.1 but not in macroH2A1.2 (Kozlowski et al., 2018; Timinszky et al., 2009). macroH2A2 is expressed from a second, independent gene and does not bind to PAR but is ∼80% identical with macroH2A1 (Posavec et al., 2013). IP of S protein–Flag-streptavidin–binding peptide (SFB)-tagged KDM5A from HEK293T cells revealed an interaction between KDM5A and macroH2A1, including after DNA damage induction by IR (Fig. 6 A). Reciprocal IP analysis using endogenous macroH2A1 antibodies corroborated these results (Fig. S5 D). To determine which macroH2A1 variant KDM5A interacted with, we performed endogenous KDM5A IP in HEK293T cells and immunoblotted with antibodies specific to macroH2A1.1 and macroH2A1.2. We observed that KDM5A interacted specifically with macroH2A1.2 (Fig. S5 E; antibody specificity for macroH2A1.1 and macroH2A1.2 is shown in Fig. S5, F and G). To further validate these results, we performed co-IP experiments with KDM5A and macroH2A using an HepG2 cell line in which all macroH2A variants are stably knocked down and complemented with stably expressed Flag-macroH2A1.2 or Flag-macroH2A2 (Douet et al., 2017). Co-IP WB analysis using HepG2 control and macroH2A-depleted derivatives identified an interaction with macroH2A1.2 but not macroH2A2 (Fig. 6 B; verification of macroH2A variant expression shown in Fig. S5, H and I). We next considered that the interaction between KDM5A, PARP1, and macroH2A1.2 may be regulated by DNA damage. To pursue this question, we performed co-IP experiments in cells untreated or treated with IR in the presence or absence of PARPi’s. As we previously demonstrated, KDM5A interacted with PARP1 and macroH2A1.2 in undamaged and damaged conditions (Fig. 6, A–C). However, specifically in IR-treated conditions, these interactions were dependent on PARP activity because the addition of PARPi’s reduced the interaction between KDM5A and both PARP1 and macroH2A (Fig. 6 C). KDM5A is known to interact with chromatin using its PHDs, with PHD1 interacting with unmodified histone H3 and PHD3 binding to H3K4me3 (Torres et al., 2015). Interestingly, KDM5A PHD1, but not PHD3, is required for DNA damage localization, and H3K4me3 is reduced following DNA damage (Gong et al., 2017). Thus, KDM5A engagement with chromatin, including macroH2A1.2, at damaged versus undamaged chromatin regions may rely on different binding modules within the protein that mediate different interactions that are themselves regulated by DNA damage, including PARylation. The requirement for PARP to support macroH2A1.2 and KDM5A interactions is also intriguing, given that macroH2A1.2 does not interact with PAR chains directly. We reasoned that PAR binding by KDM5A may mediate these interactions. To test this hypothesis, we evaluated whether KDM5A-PID is required for KDM5A interactions with PARP1 and macroH2A1.2. Indeed, KDM5AΔPID failed to associate with either macroH2A1.2 and PARP1 as efficiently as WT KDM5A (Fig. 6 D). These results identify KDM5A PAR binding as an important component of both PARP1 and mH2A1.2 interactions (summarized in Fig. 6 E).
Figure 6.
KDM5A interacts with the histone variant macroH2A1.2. (A) KDM5A interacts with macroH2A1. Inducible SFB-KDM5A–expressing HEK293T cells were treated with or without 10-Gy IR. SFB-KDM5A was immunoprecipitated with streptavidin beads, and interactions were detected by WB analysis. Input shows expression and loading of proteins. (B) Endogenous KDM5A interacts with macroH2A1.2. Co-IPs were performed using HepG2 cells depleted with shRNA control (shCtrl) or shRNA macroH2A (mH2A knock down [KD]) and complemented shRNA-macroH2A with Flag-macroH2A1.2 or Flag-macroH2A2. (C) KDM5A and macroH2A1.2 interactions are PARP dependent. SFB-KDM5A–expressing HEK293T cells were treated with DMSO or olaparib (5 µM, 1 h) with or without subsequent 10-Gy IR treatments. Co-IPs were performed as in A. (D) The PID of KDM5A is required for macroH2A1.2 and PARP1 interactions. HEK293T cells expressing GFP, GFP-KDM5A, or GFP-KDM5AΔPID were analyzed by co-IP WB analysis with the indicated antibodies. (E) Comparison of domain structure of histone H2A and macroH2A variants with PAR- and KDM5A-binding summary. Dark blue represents a highly conserved region between canonical H2A and macroH2A variants; gold represents a linker region; light blue represents the PAR-binding region within the macrodomain; dark gray represents a region that has lost PAR binding. (F) KDM5A and macroH2A1.2 promote HDR. Measurement of HR efficiency in WT and KDM5A-KO cells with or without siRNA depletion of macroH2A1.2 was performed as in Fig. 5 H (n = 2). Error bars represent SEM. P values were calculated using an unpaired Student’s t test (*, P < 0.05; **, P < 0.01).
Histone variant macroH2A1.2 is required for KDM5A recruitment to DNA damage sites
Histone variant macroH2A1.2 but not macroH2A1.1 has been reported to be required for efficient DSB repair through HR by promoting BRCA1 retention at DSBs (Khurana et al., 2014). Since macroH2A1.2 and KDM5A are associated with DSB repair by HR, and because we identified an interaction between these proteins, we set out to determine if macroH2A1.2 and KDM5A collaborate to promote DNA repair. To begin to address this question, we measured HR efficiency using a CRISPR-Cas9/mClover assay in cells singly or doubly deficient for KDM5A and macroH2A1.2 (Kim et al., 2019b; Pinder et al., 2015). In agreement with our previous results and other studies, we observed that KDM5A or macroH2A1.2 deficiency resulted in decreased gene-targeting efficiency, which is a readout for HR, compared with WT control cells (Fig. 6 F). Depletion of H2A1.2 in KDM5A-KO cells did not further decrease HR efficiency significantly (Fig. 6 F; efficiency of H2A1.2 protein depletion by siRNA is shown in Fig. S5 F).
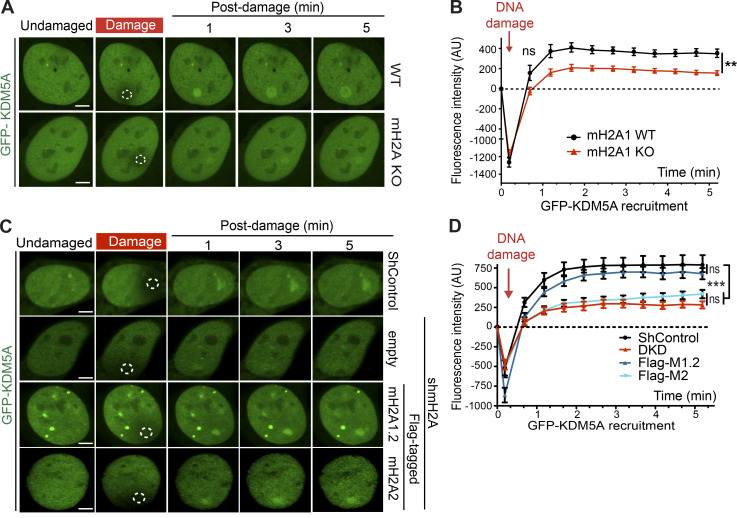
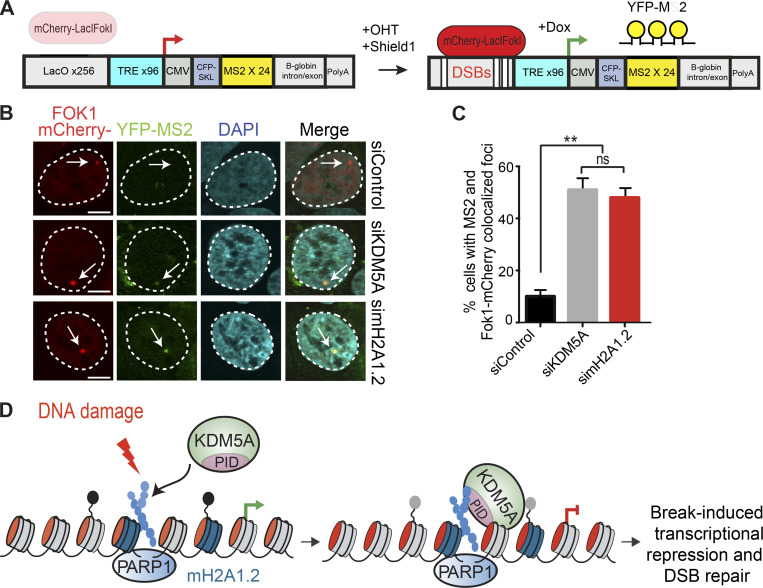
Considering these results, we speculated that macroH2A may regulate the recruitment of KDM5A to DNA damage sites. We previously generated CRISPR-Cas9 gene-edited U2OS cells that lack macroH2A (Leung et al., 2018). Using these cells, we sought to determine if macroH2A is required for KDM5A recruitment to laser-induced DNA damage sites. Consistent with this idea, we observed that GFP-KDM5A is recruited less efficiently to DNA damage sites in cells lacking macroH2A than in WT cells (Fig. 7, A and B). We extended these results by analyzing the recruitment of ZMYND8, a protein that relies on KDM5A to support its accumulation at DNA damage sites within transcriptionally active chromatin through H3K4me3 demethylation (Gong et al., 2017). Indeed, macroH2A also supported ZMYND8 damage recruitment (Fig. S5, J and K). Furthermore, similar and complementary results were obtained in HepG2 macroH2A-deficient cells, where reconstitution of these cells with macroH2A1.2 specifically led to a rescue of KDM5A localization at damage sites (Fig. 7, C and D). Finally, we reasoned that if macroH2A1.2 participated in the KDM5A–ZMYND8 DDR axis, this histone variant would also be required to suppress transcription after DSB induction, a pathway reliant on KDM5A and ZMYND8. To test this idea, we used a well-established cell-based system to analyze transcriptional repression following DSB formation. This system contains a lactose operator (LacO) array that is bound by an inducible Lac repressor protein (LacI)-FokI nuclease that creates DSBs upstream of a transcriptionally active gene (Fig. 8 A). U2OS cells containing this system also express YFP-tagged MS2, which binds to the hairpin structures that occur within the mRNA that is driven by the transgene downstream of the LacO array. Upon DSB induction, the downstream gene is suppressed, which halts production of the mRNA and abolishes the formation of colocalized mCherry-FokI and YFP-MS2 (Fig. 8, B and C). As expected, depletion of KDM5A resulted in defective transcriptional repression, causing an increased frequency of colocalized mCherry-FokI and YFP-MS2 foci. Depletion of macroH2A1.2 resulted in a loss of transcriptional repression, which phenocopied the loss of KDM5A (Fig. 8, B and C). These results demonstrate that macroH2A1.2 is required for break-induced transcriptional repression and suggest that macroH2A controls the KDM5A-ZMYND8 DDR pathway at least at the level of facilitating the recruitment of these factors to DNA damage sites.
Figure 7.
Histone variant macroH2A1.2 promotes KDM5A recruitment to DNA damage sites. (A) macroH2A1 promotes KDM5A damage localization. Analysis of GFP-KDM5A damage recruitment in macroH2A1-KO U2OS cells was performed as in Fig. 3 F. Scale bars, 5 µm. (B) Quantification of A. n ≥ 10 cells. Error bars represent SEM. (C) macroH2A1.2 promotes KDM5A damage recruitment. GFP-KDM5A damage recruitment was performed as in A. Dotted white circles indicate damaged regions. Scale bars, 5 µm. (D) Quantification of C as in B. n > 10 cells per condition. P values were calculated using an unpaired Student’s t test for B and D (**, P < 0.01; ***, P < 0.001).
Figure 8.
Histone variant macroH2A1.2 and KDM5A promote transcriptional repression at DNA breaks. (A) Schematic of FokI-inducible DSB reporter cell system (Tang et al., 2013). Upon 4-OHT and Shield1 treatment, mCherry-FokI endonuclease induces DSBs upstream of a Dox-inducible reporter gene. Nascent transcription is visualized by YFP-MS2 protein binding to stem-loop structures in the mRNA. DSBs are visualized by mCherry-FokI localization to the LacO loci. (B) macroH2A1.2 is required for transcriptional repression following DSBs. Nascent transcription was analyzed at FokI-induced DSBs in siControl-, siKDM5A-, and simacroH2A1.2-treated cells. The presence of MS2 foci indicates loss of DSB-induced transcriptional repression. Arrows indicate the location of the LacO array. Scale bars, 5 µm. (C) Quantification of B; n = 2 with >100 cells analyzed per condition per replicate. Error bars represent SEM. P values were calculated using an unpaired Student’s t test (**, P < 0.01). (D) Model for KDM5A regulation by PARP1 and macroH2A1.2. Following DNA damage, PARP1 promotes PARylation and recruitment of KDM5A to DNA damage sites. KDM5A engages PAR chains through its C-terminal coiled-coil–containing PID. macroH2A1.2 also promotes KDM5A accrual at DSBs to facilitate DSB-induced transcriptional repression and HR repair.
Discussion
Here, we report the identification of two molecular mechanisms that govern the DNA damage recruitment and function of KDM5A in the DDR through the employment of cellular, genetic, and biochemical approaches. Our results reveal that KDM5A directly binds to PAR chains at DNA damage sites through a newly identified PID that resides within the C terminus of KDM5A that is not found in other KDM5 demethylases. We also find that the histone variant macroH2A1.2 specifically interacts with KDM5A and promotes its recruitment to DNA damage sites. Our work thus identifies a PARP1/macroH2A1.2 pathway that acts upstream of KDM5A to control its interaction and functions at DNA break sites (Fig. 8 D).
Chromatin structure is known to be dynamically regulated upon DNA damage. Interestingly, PARP1 and macroH2A1.2 have opposing effects on chromatin structure. PARP1 and PARylation decompact chromatin, while macroH2A1 compacts chromatin, including through a mechanism involving PAR binding by the macrodomain of macroH2A1.1 (Khurana et al., 2014; Kozlowski et al., 2018; Krishnakumar and Kraus, 2010; Timinszky et al., 2009). If sufficiently expressed to high levels, macroH2A1.1 inhibits PARP1 activity in a manner dependent on its PAR-binding macrodomain (Kozlowski et al., 2018; Posavec Marjanović et al., 2017). MacroH2A1.2 does not bind PAR but is still able to compact chromatin through an activity located in the linker region (Kozlowski et al., 2018). This biphasic behavior of chromatin decompaction followed by compaction is observed at DNA damage sites and has been shown to correlate with HR through a macroH2A1.2-dependent mechanism (Khurana et al., 2014). Our data demonstrating PARP1-macroH2A1.2 regulation of KDM5A may shed light on how these previous observations relate to KDM5A.
KDM5A is rapidly recruited to damage sites in a PAR-binding–dependent manner (Fig. 5 B). While the PAR-binding region of KDM5A alone was rapidly recruited to damage sites, its association with breaks was very transient (Fig. 3, F and G). Transient PAR chains at breaks may be involved in the initial recruitment of KDM5A, but additional mechanisms may be needed for retention of this enzyme at DNA lesions. We posit that additional, multivalent interactions may act to channel PAR-binding proteins into a specific pathway versus others that are regulated also by PARylation, such as single-strand break repair, Okazaki fragment sensing, and transcription (Gibson and Kraus, 2012; Hanzlikova et al., 2018; Ray Chaudhuri and Nussenzweig, 2017). Our data may suggest that macroH2A1.2 likely serves this purpose for KDM5A at DSBs, given that they interact with each other and the requirement of this histone variant for KDM5A recruitment and function at DNA damage sites. In addition, we observed that KDM5A deficiency resulted in toxicity and genome instability as a result of PARPi treatment. While this may potentially be at odds with the finding that PARP1 promotes KDM5A recruitment to DNA damage sites, PARPi’s broadly impact genome integrity pathways in cells, including those that are HR proficient, during replication through PARP trapping and DSB formation, replication fork instability and Okazaki fragment processing, and other cellular functions, including chromatin remodeling and transcription (Azarm and Smith, 2020; Gupte et al., 2017). Indeed, while BRCA-deficient cells are exquisitely sensitive to PARPi’s through a concept known as “synthetic lethality,” HR-proficient cells also display cell toxicity to PARPi’s (Ashworth and Lord, 2018; Michelena et al., 2018). Thus, the sensitivity of KDM5A-deficient cells to PARP inhibition may not be entirely related to the involvement of PARylation in promoting KDM5A recruitment and involvement in HR repair.
Chromatin compaction at break sites may be related in part to our observation that macroH2A1.2 is required for transcriptional repression at DNA breaks. Indeed, KDM5A demethylates the transcription active mark H3K4me3 at DSBs and promotes the damage association of the ZMYND8–NuRD complex, which together act to repress transcription and promote HR repair of DSBs (Gong et al., 2015; Gong et al., 2017; Gong and Miller, 2018; Savitsky et al., 2016; Spruijt et al., 2016). Several other transcriptional repressive complexes, including PRC1 (polycomb repressive complex 1), PRC2, and NELF (negative elongation factor), as well as the histone variant H2AZ, are also required for DNA break–induced transcriptional repression (Caron et al., 2019; Puget et al., 2019; Tan and Huen, 2020). Given the identification of the histone variant macroH2A1.2 in this DDR pathway here, future studies are warranted to decipher other potential interactions between these DSB-associated transcriptional regulators.
PARP1 was one of the first factors identified that is required for break-induced transcriptional repression (Chou et al., 2010), and it was previously shown to regulate DNA damage recruitment of KDM5A (Gong et al., 2017). The identification of a PID in KDM5A was unexpected for several reasons. KDM5B, a related KDM5 demethylase, is also involved in DSB repair but is PARylated directly, which inhibits its activity at transcriptionally active sites (Bayo et al., 2018; Krishnakumar and Kraus, 2010; Li et al., 2014b). In addition, KDM5A does not share homology with any known PAR-binding domains (Krietsch et al., 2013; Teloni and Altmeyer, 2016; Wei and Yu, 2016). Our extensive mapping, functional, and biochemical analyses identified a putative coiled-coil domain near the C terminus of KDM5A able to bind PAR both in vitro and in cells, which is sufficient to localize to DNA damage sites in a PARP-dependent manner. Our data identifying α-helical properties within this region, which are features of coiled-coil domains (Truebestein and Leonard, 2016), further strengthen the likelihood that this region is a bona fide coiled-coil domain. Our evidence reveals that this coiled-coil domain can facilitate PAR binding, which is necessary for KDM5A recruitment and activities at DNA damage sites. These findings have several important implications. Coiled-coil domains are common in proteins, including several involved in the DDR, and play important roles in protein–protein interactions, protein multimerization, and conformational changes (Truebestein and Leonard, 2016). In the DDR, dimerization of CtIP, a BRCA1-interacting protein, requires an N-terminal coiled-coil region (Dubin et al., 2004), while the BRCA1–PALB2 interaction is mediated by coiled-coil regions present in both proteins (Song et al., 2018). It will be important to test additional coiled-coil domains in other proteins for PAR binding, which may reveal additional mechanisms by which PARP1 regulates not only the DDR but also other PARP-dependent processes, such as transcription. Finally, the KDM5A-PID is unique to KDM5A demethylase, as KDM5B and KDM5C do not contain an identifiable coiled-coil domain, nor do they readily bind PAR (Fig. 4). Given that KDM5A is overexpressed in cancer and is involved in therapy-associated drug resistance (Cao et al., 2014; Choi et al., 2018; Hou et al., 2012; Li et al., 2014a; Sharma et al., 2010; Teng et al., 2013; Vinogradova et al., 2016; Yang et al., 2019; Zeng et al., 2010), our data provide the rationale for testing the potential use of PARPi’s in these settings, which may selectively inhibit KDM5A function.
In sum, this work has identified two regulatory mechanisms, PAR binding and macroH2A, that control KDM5A interactions and function at DNA damage sites (Fig. 8 D). These findings exemplify how chromatin acts as a platform for DNA-templated processes, including DNA repair and transcription, that must be remodeled by chromatin-modifying enzymes to transition from one activity to another. In this example, upon DNA damage, PARP1 and macroH2A1.2 regulate the chromatin environment to trigger KDM5A damage association, resulting in histone demethylation and recruitment of the ZMND8–NuRD complex, which collectively act to repress transcription and promote DNA DSB repair to maintain genome and epigenome integrity.
Materials and methods
Cell lines and cell culture
U2OS and HEK293T cells were purchased from American Type Culture Collection. HCT116 WT and HCT116 KDM5A-KO cells were purchased from Horizon Discovery. To knock out KDM5A, a PGK promoter neomycin resistance cassette flanked by LoxP sites was integrated into both alleles of KDM5A at exon 11, which removes this exon, making both KDM5A alleles nonfunctional in this cell line. All cell lines were maintained in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C and 5% CO2. Inducible cell lines (SFB-tagged KDM5A) were established and maintained in medium with 0.2 mg/ml hygromycin B (Invitrogen). To establish the inducible SFB-tagged KDM5A-expressing cell lines, pcDNA5/flippase (Flp) recognition target (FRT)/pCMV-tetO2–containing, SFB-tagged constructs were transfected with pOG44 Flp recombinase expression vector into Flp/In T-REx HEK293T cells. After 48 h, cells were treated with 0.2 mg/ml hygromycin B (Invitrogen) for the selection of cells with integration of the construct at the FRT locus. All cell lines were routinely checked for mycoplasma contamination.
The U2OS DSB reporter cell line used for transcriptional analysis was a gift from Roger Greenberg (University of Pennsylvania, Philadelphia, PA). This cell line stably expresses an mCherry-LacI-Fok1 nuclease domain fusion protein that also contains a modified ER receptor and destabilization domain. Upon Shield1 and 4-hydroxytamoxifen (4-OHT) addition, the mCherry LacI-FokI protein is stabilized and induces DSBs within the LacO genomic loci, which are located upstream of a doxycycline (Dox)-inducible YFP-MS2 reporter gene (Tang et al., 2013). Cells were treated with 1 µM Shield1 and 1 mM 4-OHT for 3 h to induce site-specific DSBs and 1 µg/ml Dox for an additional 3 h to induce transcription.
The generation of HepG2 cells (American Type Culture Collection; HB-8065) with stable shRNA-mediated knockdown of both macroH2A1 and macroH2A2 proteins is described in Douet et al. (2017). Cell lines stably expressing Flag-tagged macroH2A1.2 and macroH2A2 proteins in the double-knockdown background were achieved by retroviral transduction. GP2-293 cells were used as packaging cells to produce retroviral particles. 4 × 106 GP2-293 cells were seeded in a 10-cm plate and cultured to 60–70% confluency, then transfected with 10 µg of pBabe.puro plasmids containing either Flag-tagged full-length mouse macroH2A1.2 or macroH2A2 sequences and 3 µg of pCMV-VSV-G mixed in a 1× Hepes-buffered saline solution (2× Hepes-buffered saline: 272 mM NaCl, 2.8 mM Na2HPO4, 55 mM Hepes, pH 7) containing 125 mM CaCl2. The supernatant containing viral particles produced by GP2-293 cells was collected at 24 h and 48 h after transfection, filtered using a 0.45-µm filter, supplemented with 8 µg/ml Polybrene (Sigma-Aldrich), and added to the target HepG2 cells cultured in 6-well plates at 60–70% confluency. Cells were then centrifuged for 45 min at 1,200 rpm at 37°C, incubated at 37°C for 45 min, and then cultured overnight in fresh media. The same process was repeated 24 h after the first infection. The cells were selected with 2 µg/ml puromycin. The necessary selection time was determined by using a negative control plasmid without resistance.
Cloning and plasmids
KDM5A was cloned into the pDONR201 Gateway vector. The pDONR201 clones were transferred into GFP- or SFB-tagged destination vectors using the Gateway LR cloning system (Invitrogen). KDM5AΔPID deletion mutant was generated by PCR in pDONR201 vector following a standard cloning method and then subcloned into a Gateway destination vector derived from pcDNA5/FRT/TO plasmids containing the NLS and SFB or GFP tagged in the N terminus. KDM5A fragments were amplified from KDM5A cDNA using Q5 High Fidelity polymerase (New England Biolabs) with primers containing SalI and NotI recognition sites. PCR products were then cloned into SalI and NotI sites of a pMAL-C5X vector (New England Biolabs). All constructs were validated by DNA Sanger sequencing.
Gene editing by CRISPR-Cas9
KDM5A gene KOs in U2OS cells were generated as previously described (Gong et al., 2017). The sequence of the single-guide RNA used to edit the gene for human KDM5A was 5′-CGGATGCGGCCGATAAAGCT-3′. Loss of KDM5A protein expression by gene editing in U2OS clones was validated by WB analysis using a mouse anti-KDM5A antibody (ab78322; Abcam).
siRNA depletion and overexpression
For knockdown experiments, 2.5 × 105 cells were seeded in 6-well plates. Cells were transfected with siRNA duplexes targeting the corresponding protein or scrambled mixes using Lipofectamine RNAiMAX (Invitrogen) reagents as per the manufacturer’s recommendations. Knockdown cells were cultured for 48–72 h before further processing. The siRNAs used were as follows: KDM5A, 5′-GCAAAUGAGACAACGGAAA-3′; macroH2A1.2, 5′-CUGAACCUUAUUCACAGUGAA-3′. For KDM5A rescue experiments or overexpression, cells were transfected with 2 µg of emGFP-KDM5A or emGFP-pcDNA 6.2 empty vector using either Lipofectamine 2000 or Lipofectamine 3000 (Invitrogen) and then allowed to recover for 24 h before proceeding to assay further. In the case of Dox-inducible SFB- or GFP-tagged pcDNA5/FRT/TO plasmids containing KDM5A or KDM5AΔPID, 1 µg/ml Dox was used for at least 24 h to induce expression of the indicated transgene.
IP
Cells (U2OS or HEK293T) were lysed in NETN buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40, protease inhibitor cocktail) containing TurboNuclease (Accelagen) at 4°C for 1 h. Cell lysates were centrifuged at 15,000 rpm at 4°C for 10 min. The lysate was then incubated with appropriate antibodies for 12 h and then conjugated with Dynabead Protein A or Protein G beads (Invitrogen) for an additional 1 h. SFB- or GFP-tagged proteins were immunoprecipitated with streptavidin Dynabeads (Invitrogen) or GFP-Trap (Chromotek). After three or four washes with NETN buffer, bead-bound proteins were eluted with 2× sample loading buffer (120 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 1% bromophenol blue) and resolved on 4–15% gradient SDS-PAGE gel. Proteins were identified by an appropriate antibody by WB analysis using anti-rabbit or anti-mouse IgG HRP-conjugated secondary antibodies (7074S and 7076S; Cell Signaling Technology).
Antibodies
The following antibodies were used in this study: mouse anti-KDM5A (ab78322; Abcam), rabbit anti-KDM5B (A301-813; Bethyl Laboratories, Inc.), rabbit anti-KDM5C (A301-034; Bethyl Laboratories, Inc.), rabbit anti–β-tubulin (ab6046; Abcam), mouse anti-γH2AX (05-636, clone JBW301; Sigma-Aldrich), rabbit anti-GFP (A11122; Invitrogen), rabbit anti-PARP1 (9542; Cell Signaling Technology), mouse anti-PAR (4335-MC-100; Trevigen), rabbit anti–pan-ADP-ribose (MABE1016; Sigma-Aldrich), rabbit anti-MBP (Ab9084; Abcam), mouse anti-Flag (F1804; Sigma-Aldrich), rabbit anti-macroH2A1 (Ab37264; Abcam), rabbit anti-macroH2A1.1 (12455S [D5F6N]; Cell Signaling Technology), mouse anti-macroH2A1.2 (MABE61 [14G7]; Sigma-Aldrich), rabbit IgG (NI01; Calbiochem), mouse IgG (NI03; Calbiochem), and rabbit anti-ZMYND8 (A302-089; Bethyl Laboratories, Inc.).
Immunofluorescence
For γH2AX foci and micronuclei analysis, cells were seeded onto glass coverslips and 24 h later pretreated with DMSO or 5 µM olaparib (10621; Cayman Chemical). After 24 h, cells were fixed with 2% (vol/vol) PFA for 15 min, permeabilized with 0.5% Triton X-100 in PBS for 15 min on ice, and blocked with 5% BSA/PBS for 1 h. Coverslips were then incubated with anti-γH2AX antibody (1:1,000) for 1 h at RT. After three washes in PBS, samples were incubated with Alexa Fluor 594 goat anti-mouse IgG (H+L) secondary antibody (A11032; Invitrogen) for 1 h at RT, and slides were mounted with VECTASHIELD mounting medium with DAPI (Vector Laboratories). Cells were imaged using a FluoView 3000 confocal microscope (Olympus) using TruSpectral high-efficiency gallium arsenide phosphide spectral detectors with a 60× plan apochromat oil objective lens with 1.42 NA. Image acquisition was performed with FV-10 ASW3.1 software (Olympus). Quantification of γH2AX foci and micronuclei was performed using ImageJ software.
Clonogenic cell survival assay
Clonogenic cell survival was analyzed using a colony-forming assay. Briefly, 500 U2OS or HCT116 WT and KDM5A-KO cells were plated in 6-well plates 24 h before treatment. Cells were incubated with increasing concentrations of PARPi (olaparib), and, 24 h later, PARPi-containing media were replaced with normal DMEM and incubated for 14 d in a tissue culture incubator (37°C, 5% CO2). DMSO was used as a control. Cells were pretreated with KDM5A inhibitor (CPI455, 2573; Axon Medchem) alone or in combination with olaparib wherever indicated. Colonies were fixed and stained with crystal violet solution (0.5% crystal violet in 20% ethanol). Results were normalized to WT untreated cells.
Neutral comet assay
HCT116 WT or KDM5A-KO cells were treated with 5 µM DMSO or olaparib for 24 h. After treatment, DNA breaks were analyzed using the CometAssay Reagent Kit (Trevigen) according to the manufacturer’s instructions. After electrophoresis, DNA was stained with SYBR Green (Invitrogen), and images were acquired with a FluoView 3000 confocal microscope (Olympus) using a 10× objective with 0.3 NA (Olympus). Comet tail moments (Olive moments) were calculated by using CometScore 2.0 software.
X-ray irradiation
Indicated doses of IR were delivered by an x-ray generator (Faxitron x-ray system; RX650).
Laser-induced live-cell imaging
Laser-induced live-cell imaging was performed as previously described (Kim et al., 2019a). In brief, cells were seeded onto glass-bottomed dishes (Pelco; Ted Pella Inc.) and were presensitized by adding 10 µM BrdU (B9285; Sigma-Aldrich) for 24 h before laser-induced damage. For PARPi treatment, cells were pretreated with 5 µM PARPi (olaparib) for 1 h before laser-induced DNA damage. Laser damage was induced in a defined region of interest (ROI) using a 405-nm, 50-mW laser (150 frames at 60% at a scan speed of 20.0 µs/pixel) mounted on a FluoView 3000 confocal microscope (Olympus). Cells were incubated during laser damage experimentation in a 37°C temperature-controlled WSKM series Tokai Hit top-stage incubator chamber with 5% CO2. Cell imaging was performed on the FluoView 3000 confocal microscope (Olympus) using TruSpectral high-efficiency gallium arsenide phosphide spectral detectors. Cells were incubated in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin during imaging. The relocalization intensity of GFP fused proteins at the damage sites were analyzed using FV-10 ASW3.1 software (Olympus). All images were captured using a 60× apochromatic oil objective lens with a 1.42 NA. Recruitment intensities of GFP fused proteins were quantified by FV-10 ASW3.1 software (Olympus). To calculate the fluorescence intensity at the DNA lesions, the intensity of the damaged ROI (ROID) and the undamaged ROI (ROIU) are first normalized to the fluorescence intensity at time point 0 (before damage). The relative fluorescence intensity for each time point is calculated as follows: after DNA damage (ROID/ROIU)/before DNA damage (ROID/ROIu) × 100. GraphPad Prism (version 6.0) software was used to calculate the SEM for the fluorescence intensity at damaged sites.
CRISPR-mClover HR repair assay
CRISPR-mClover HR assay was performed as previously described (Kim et al., 2019b). Briefly, mClover-HR donor plasmid (98970; Addgene) and Cas9-gRNA vector (98971, Addgene; single-guide RNA target sequence: 5′-GGTTGGCAGCGCTGCCCGCG-3′) were transfected into U2OS WT cells, U2OS KDM5A KO cells, and U2OS KDM5A KO cells expressing SFB-tagged WT or ΔPID KDM5A constructs. Cells were transfected 24 h before mClover-HR donor plasmid and Cas9-gRNA vector transfection. After transfection, cells were incubated for 48 h, and GFP-positive cells representing repair events by HR were analyzed using a BD Accuri flow cytometer. Quantification was performed using FlowJo software.
Recombinant protein purification
U2OS cells expressing full-length GFP-tagged KDM5A, KDM5B, and KDM5C were harvested and lysed in high-salt buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1% Triton X-100, and 1 mM DTT). The cell lysate was incubated with GFP-Trap beads for 1 h. Beads were extensively washed five times with high-salt buffer, and GFP fusion protein was eluted with 200 mM glycine, pH 2.5. Eluted proteins were neutralized with 1 M Tris, pH 10.4. Eluted proteins were used in a PAR-binding assay, as described below. MBP fused KDM5A fragments were expressed in BL21 codon plus RIPL competent E. coli cells (230280; Agilent Technologies) purified and eluted from amylose resin with maltose (E8021; New England Biolabs) as described previously (Kumbhar et al., 2018). KDM5A 1491–1610 (F9) and KDM5A F9ΔPID were cleaved from MBP with factor Xa protease (P8010; New England Biolabs) and further purified using a HiTrap SP HP column (GE29-0513-24; GE Healthcare) on an ÄKTA Pure fast protein liquid chromatograph (GE Healthcare).
PAR-binding assay
An in vitro PAR-binding assay was performed as previously described (Kumbhar et al., 2018). Briefly, proteins were spotted onto nitrocellulose membrane (10600007; GE Healthcare). The air-dried membrane was then blocked with 5% BSA in PBS and incubated membrane with 10 nM PAR chains for 1 h at RT. The membrane was washed extensively and incubated overnight with an anti-PAR antibody. Immunoblot analysis was performed to detect the PAR signal. To check for PAR binding of KDM5A fragments, an equal quantity of each fragment was spotted onto the nitrocellulose membrane, and the assay was performed as explained above.
Nascent transcript detection at DNA damage sites
U2OS reporter cells (from the Greenberg laboratory, described above) with siRNA transfections were seeded onto 12-mm round coverslips in 6-well plates. To induce site-specific DSBs, U2OS reporter cells were treated with 1 µM Shield1 ligand (632189; Takara) and 1 mM 4-OHT for 4 h for mCherry-FokI expression. Transcription was induced from an integrated reporter gene by treating cells with 1 µg/ml Dox for an additional 3 h. Cells on coverslips were fixed with 4% PFA, permeabilized with 0.5% Triton X-100 in PBS, washed, and mounted on glass slides with VECTASHIELD mounting medium with DAPI. Cells were visualized using a FluoView 3000 confocal microscope (Olympus) as described above.
RT quantitative PCR (RT-qPCR)
For analysis of E-cadherin mRNA levels upon PARPi treatment, total RNAs from untreated and PARPi-treated (0.25 µM olaparib overnight) U2OS cells were purified using the RNeasy Mini Kit (Qiagen) and treated with RNase-free DNase I (Qiagen) following the manufacturer’s protocol. 1 µg of total RNA for each sample was used to synthesize cDNA by the SuperScript III First-Strand Synthesis System (Invitrogen). For E-cadherin mRNA analysis in KDM5A-KO cells, RNA was purified using the RNA/Protein Purification Plus Kit (48200; Norgen) according to the manufacturer’s recommendations. 250 ng of total RNA was reverse transcribed into cDNA with the qScript cDNA SuperMix kit (95048; Quantabio). RT-qPCR was performed using the StepOnePlus Real-Time PCR system (Applied Biosystems) with SYBR Green Master Mix (Applied Biosystems) and the indicated gene-specific primers: ALAS1 forward: 5′-CCTTGGCCTTAGCAGTTTTG-3′; ALAS1 reverse: 5′-CCAAGATGATGGAAGTTGGG-3′; B2M forward: 5′-AATGTCGGATGGATGAAACC-3′; B2M reverse: 5′-TCTCTCTTTCTGGCCTGGAG-3′; GAPDH forward: 5′-CAATGACCCCTTCATTGACC-3′; GAPDH reverse: 5′-GATCTCGCTCCTGGAAGATG-3′; E-cadherin primer 1 forward: 5′-TGCCCAGAAAATGAAAAAGG-3′; E-cadherin primer 1 reverse: 5′-GTCACCTTCAGCCATCCTGT-3′; E-cadherin primer 2 forward: 5′-CAGGTCTCCTCTTGGCTCTG-3′; E-cadherin primer 2 reverse: 5′-ACTTTGAATCGGGTGTCGAG-3′.
CD spectroscopy
All CD spectra were measured at RT with a JASCO J-815 spectropolarimeter. Each protein spectrum was collected as an average of four scans using a quartz 0.1-cm cell on 50 µM samples in 20 mM Tris (pH 7.2), 100 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine. CD spectral data were acquired using a 1-nm bandwidth scanning from 185 to 250 nm at a speed of 50 nm/min, data pitch of 0.5 nm, and response time of 4 s. Each protein sample was autocorrected with a baseline spectrum of matching sample buffer that was collected under the same conditions. Raw ellipticities were converted to mean residue ellipticity to normalize the CD spectra between protein samples.
Synthesis, purification, and fractionation of PAR chains
Synthesis, purification, and fractionation of PAR chains were conducted following previously established protocols (Chen et al., 2018; Tan et al., 2012). Briefly, PAR polymers were enzymatically synthesized in a 10-ml reaction containing 50 mM Tris-HCl (pH 8), 4 mM MgCl2, 250 µM DTT, 20 mM NaCl, 5 mM NAD+, 10 µg/ml 8-mer oligonucleotide (5′-GGAATTCC-3′), and 150 nM human PARP1 at 37°C for 60–90 min. PAR chains were detached from modified PARP by treating the reaction with DNase I for 1 h and proteinase K for 2 h, respectively, and precipitated with 20% TCA (wt/vol). Further purification and fractionation of PAR chains (0.85 μmol) was performed by ion exchange HPLC using a DNAPac PA 100, 4 × 250–mm analytical column preequilibrated with buffer A (25 mM Tris, pH 9.0) and a previously published 152-min variable gradient (0 min [0% B], 6 min [0% B], 12 min [30% B], 18 min [40% B], 48 min [47% B], 78 min [49% B], 138 min [55% B], 144 min [63% B], 145 min [100% B], 148 min [100% B], 149 min [0% B], 152 min [0% B]) with buffer B (25 mM Tris, pH 9.0, and 1 M NaCl) at a flow rate of 1 ml/min and a detector setting of 260 nm (Tan et al., 2012). PAR chain HPLC fractions were manually collected, dialyzed against double-distilled water (ddH2O) for 24 h to desalt, and lyophilized. Dried powders were resuspended in a minimal volume (15–20 µl) of 20 mM Tris (pH 7.0), and fractionated PAR chain concentrations were determined by UV analysis at 258 nm using an extinction coefficient of 1,300 cm−1 M−1. The sizes of purified PAR chains in each fraction were identified by mass spectrometry at the University of Utah Department of Chemistry Mass Spectrometry Core Facility. In brief, 10–50 µM fractionated PAR chains in ddH2O were submitted for analysis on a Waters ACQUITY I-class ultra-high-pressure liquid chromatograph coupled to a Waters Xevo G2-S quadrupole time-of-flight mass spectrometer operating in negative ion mode (2.5-kV capillary voltage, 25-V sampling cone voltage, 150°C source temperature, 500°C desolvation temperature, nitrogen desolvation at 800 liters/h). An ACQUITY UPLC BEH C18 VanGuard precolumn (2.1 × 5 mm) preequilibrated in solvent A (1 mM ammonium formate) was used for filtering and further desalting samples before analysis using mass spectrometry. The samples were eluted from the column using a linear solvent gradient: {0–1 min [1% B (methanol)] or 1–5 min [90% B]} at a flow rate of 0.3 ml/min. The acquired mass spectra were processed and deconvoluted using MassLynx 4.1 software.
EMSA
For EMSAs, fractionated PAR chains (75 nM) were incubated with a single concentration (250 nM) or varying amounts of either KDM5A-F9 or KDM5A F9ΔPID in 10 µl binding buffer (20 mM Tris, pH 7.2, 100 mM NaCl, 1 mM tris[2-carboxyethyl]phosphine, 10% wt/vol sucrose). After incubation at RT for 30 min, reaction mixtures were separated by electrophoresis in 8% (wt/vol) polyacrylamide gels using 1× Tris-borate-EDTA (pH 8.3) for 50 min at 100 V. Gels were soaked in ddH2O supplemented with 1× SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific) and visualized on an Amersham Typhoon biomolecular imager (GE Healthcare).
Bioinformatics
The MultiCoil algorithm hosted at http://cb.csail.mit.edu/cb/multicoil/cgi-bin/multicoil.cgi was used for prediction of the coiled-coil region in KDM5A–C (Wolf et al., 1997). A coiled-coil probability cutoff score of 0.5 and window size 28 were used to obtain dimeric or trimeric coiled-coil predictions. KDM5A predicted intrinsic disorder was determined using IUPred (Dosztányi et al., 2005). Multiple sequence alignments were performed using Clustal Omega.
Quantification and statistical analyses
GraphPad Prism (version 6.0) software was used for statistical analysis. All results are presented as mean ± SEM unless otherwise specified. P values were calculated using unpaired t tests unless otherwise indicated. P values <0.05 were considered as statistically significant.
Primers
Primers used for KDM5A molecular cloning (with their restriction sites in primers underlined) are as follows: KDM5A F1: forward: 5′-TTGCGGCCGCATGGCGGGCGTGGGGCCGGGGGGCTA-3′, reverse: 5′-TTGTCGACATCAGTGCTGAGAACCTCAGGCTC-3′; KDM5A F2: forward: 5′-TTGCGGCCGCGTGCAGATGCCTAATTTAGATCTTA-3′, reverse: 5′-TTGTCGACGCTGCTTACCAGCCGCCAAAATTC-3′; KDM5A F3: forward: 5′-TTGCGGCCGCCCAGTCCATATGGTTCCCACAGAA-3′, reverse: 5′-TTGTCGACATTTACACATTGACGTCCAATGGG-3′; KDM5A F4: forward: 5′-TTGCGGCCGCGCTGAAGCTGTGAACTTCTGTACT-3′, reverse: 5′-TTGTCGACCAGCTGAGCCACAGAAGCACA-3′; KDM5A F5: forward: 5′-TTGCGGCCGCGCTGAGACCTGTGCTTCTGTGGCT-3′, reverse: 5′-TTGTCGACTGCTGCTGCTACCTGTGATTCCAC-3′; KDM5A F6: forward: 5′-TTGCGGCCGCCGCCCTATTCCTGTGCGTCTTGAA-3′, reverse: 5′-TTGTCGACTGTCAAACACTGCAGGGCCTCTCC-3′; KDM5A F7: forward: 5′-TTGCGGCCGCGTATCCCTTCAGAAGTTGCCCGTA-3′, reverse: 5′-TTGTCGACCATCATAAGTTCTTCCAGTTGTGC-3′; KDM5A F8: forward: 5′-TTGCGGCCGCGAACCTCCAGTGCTGGAGTTGTCA-3′, reverse: 5′-TTGTCGACCTAACTGGTCTCTTTAAGATCCTCCAT-3′; KDM5A F8-1: ]forward: 5′-TTGCGGCCGCGAACCTCCAGTGCTGGAGTTGTCA-3′, reverse: 5′-TTGTCGACGTCCTTTCCTTTCACTTTTAGTGG-3′; KDM5A F8-2: forward: 5′-TTGCGGCCGCGAGAAACCACTAAAAGTGAAAGGA-3′, reverse: 5′-TTGTCGACCTCTTCTTCTTTTGCTAGTTTCTT-3′; KDM5A F8-3: forward: 5′-TTGCGGCCGCCTGGCCAAGAAACTAGCAAAAGAA-3′, reverse: 5′-TTGTCGACGCACACAGCATTCTCATCATCAGA-3′; KDM5A F8-4: forward: 5′-TTGCGGCCGCGAGGAGTCTGATGATGAGAATGCT-3′, reverse: 5′-TTGTCGACCTAACTGGTCTCTTTAAGATCCTCCAT-3′; KDM5A F8ΔPID: forward: 5′-AAAAAGAAGGAGAAGGCTGCTGCAGCC-3′, reverse: 5′-GTCCTTTCCTTTCACTTTTAGTGGTTTCTC-3′; KDM5A F9: forward: 5′-TTGCGGCCGCGAGAAACCACTAAAAGTGAAAGGA-3′, reverse: 5′-TTGTCGGACGCACACAGCATTCTCATCATCAGA-3′.
Online supplemental material
Fig. S1 is related to Fig. 1 and shows validation of KDM5A KO in HCT116 cells and the defects in genome integrity pathways resulting from KDM5A deficiency in these cells. Fig. S2 is related to Fig. 1 and analyzes KDM5A loss in human U2OS cells and reports the associated deficiencies in genome integrity in these cells. Fig. S2 also shows that PARPi interacts with KDM5A and that other KDMs are expressed normally in KDM5A U2OS KO cells compared with WT U2OS cells. Fig. S3 is related to Fig. 2 and Fig. 4 and depicts the biochemical analysis of KDM5A interactions with PAR and PARylated proteins. Fig. S3 also shows that KDM5B is not recruited to laser damage in either WT or KDM5A-KO HCT116 cells. Fig. S4 is related to Fig. 4 and shows the intrinsically disordered regions of KDM5A, including in the predicted coiled-coil region, and compares sequence homology of this putative coiled-coil region of human KDM5A with human KDM5B and KDM5C, as well as KDM5A proteins from other species. Fig. S5 is related to Figs. 4, 5, 6, and 7 and depicts the purified protein fragments of KDM5A (Fig. 4) and the induction of E-cadherin expression by PARPi treatment (Fig. 5). Fig. S5 also shows that KDM5A interacts with macroH2A1.2, validates macroH2A variant expression and depletion by siRNAs (Fig. 6), and demonstrates that ZMYND8 recruitment is deficient in macroH2A1-KO U2OS cells (Fig. 7).
Supplementary Material
Acknowledgments
We thank Roger Greenberg (University of Pennsylvania) for providing the FokI DSB cell line and Hung-wen Liu (University of Texas at Austin, Austin, TX) for providing PAR chains and recombinant PARP1. We thank Dr. A.M. Fleming (University of Utah, Salt Lake City, UT) for his ion exchange HPLC expertise. We thank members of the Miller laboratory for helpful discussions and comments on the manuscript.
B. Xhemalce acknowledges support from the National Institutes of Health (R01 GM127802). Funding in A. Matouschek’s laboratory was provided by the National Institute of General Medical Sciences (R01GM124501) and the Welch Foundation (F-1817). Research support for work performed in M. Buschbeck’s laboratory was provided through the Fondo Europeo de Desarrollo Regional/Ministerio de Ciencia e Innovación—Agencia Estatal de Investigación (RTI2018-094005-B-I00 and ISCIII PIE16/00011). The work performed by B.A. Buck-Koehntop was supported by the National Science Foundation (MCB-1715379). Funding for this project from K.M. Miller’s laboratory was provided by the American Cancer Society (RSG-16-042-01-DMC) and the National Institutes of Health, National Cancer Institute (R01CA1982279 and R01CA201268).
The authors declare no competing financial interests.
Author contributions: R. Kumbhar performed the experiments unless otherwise indicated. R. Kumbhar and K.M. Miller designed and analyzed the experiments. A. Sanchez performed co-IPs in macroH2A variant cell lines and validation of macroH2A siRNA treatments and qPCR data for E-cadherin upon PARPi treatment. J. Perren performed KDM5A fragment cloning and laser microirradiation of KDM5A-F8 experiments. F. Gong generated U2OS KDM5A-KO and stable SFB-KDM5A HEK293T cells lines. F. Medina and A. Matouschek assisted with protein purifications. S.K. Devanathan and B. Xhemalce performed RT-qPCR analyses in KDM5A-KO cells. D. Corujo and M. Buschbeck generated and characterized macroH2A-deficient cells and rescued HepG2 cells. B.A. Buck-Koehntop performed and analyzed CD, PAR chain length purifications, and EMSA analyses. K.M. Miller conceived the study and supervised the project. R. Kumbhar and K.M. Miller wrote the manuscript with additional input from all other authors.
References
- Agarwal, P., and Miller K.M.. 2016. The nucleosome: orchestrating DNA damage signaling and repair within chromatin. Biochem. Cell Biol. 94:381–395. 10.1139/bcb-2016-0017 [DOI] [PubMed] [Google Scholar]
- Ahel, I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., and West S.C.. 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 451:81–85. 10.1038/nature06420 [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez, R., and Althaus F.R.. 1989. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat. Res. 218:67–74. 10.1016/0921-8777(89)90012-8 [DOI] [PubMed] [Google Scholar]
- Ashworth, A., and Lord C.J.. 2018. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 15:564–576. 10.1038/s41571-018-0055-6 [DOI] [PubMed] [Google Scholar]
- Azarm, K., and Smith S.. 2020. Nuclear PARPs and genome integrity. Genes Dev. 34:285–301. 10.1101/gad.334730.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskaite, E., Brassington A., Tan E.S., Warwicker J., Dunstan M.S., Banos B., Lafite P., Ahel M., Mitchison T.J., Ahel I., et al. 2013. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat. Commun. 4:2164. 10.1038/ncomms3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batie, M., Frost J., Frost M., Wilson J.W., Schofield P., and Rocha S.. 2019. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science. 363:1222–1226. 10.1126/science.aau5870 [DOI] [PubMed] [Google Scholar]
- Bayo, J., Tran T.A., Wang L., Peña-Llopis S., Das A.K., and Martinez E.D.. 2018. Jumonji inhibitors overcome radioresistance in cancer through changes in H3K4 methylation at double-strand breaks. Cell Rep. 25:1040–1050.e5. 10.1016/j.celrep.2018.09.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., and Helleday T.. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 434:913–917. 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Buschbeck, M., and Hake S.B.. 2017. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 18:299–314. 10.1038/nrm.2016.166 [DOI] [PubMed] [Google Scholar]
- Cao, J., Liu Z., Cheung W.K., Zhao M., Chen S.Y., Chan S.W., Booth C.J., Nguyen D.X., and Yan Q.. 2014. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Rep. 6:868–877. 10.1016/j.celrep.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, P., van der Linden J., and van Attikum H.. 2019. Bon voyage: A transcriptional journey around DNA breaks. DNA Repair (Amst.). 82:102686. 10.1016/j.dnarep.2019.102686 [DOI] [PubMed] [Google Scholar]
- Chapman, J.R., Taylor M.R., and Boulton S.J.. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 47:497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Chatterjee, N., and Walker G.C.. 2017. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58:235–263. 10.1002/em.22087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.K., Lin W.L., Chen Z., and Liu H.W.. 2018. PARP-1-dependent recruitment of cold-inducible RNA-binding protein promotes double-strand break repair and genome stability. Proc. Natl. Acad. Sci. USA. 115:E1759–E1768. 10.1073/pnas.1713912115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, L.Y., Gong F., and Miller K.M.. 2017. Bromodomain proteins: repairing DNA damage within chromatin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160286. 10.1098/rstb.2016.0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.J., Joo H.S., Won H.Y., Min K.W., Kim H.Y., Son T., Oh Y.H., Lee J.Y., and Kong G.. 2018. Role of RBP2-induced ER and IGF1R-ErbB signaling in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 110:400–410. 10.1093/jnci/djx207 [DOI] [PubMed] [Google Scholar]
- Chou, D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiácovo M.P., and Elledge S.J.. 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 107:18475–18480. 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., and Helin K.. 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 128:1063–1076. 10.1016/j.cell.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Ciccia, A., and Elledge S.J.. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire, T., and Legube G.. 2019. A snapshot on the cis chromatin response to DNA double-strand breaks. Trends Genet. 35:330–345. 10.1016/j.tig.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Corujo, D., and Buschbeck M.. 2018. Post-translational modifications of H2A histone variants and their role in cancer. Cancers (Basel). 10:59. 10.3390/cancers10030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabin, J., Fortuny A., and Polo S.E.. 2016. Epigenome maintenance in response to DNA damage. Mol. Cell. 62:712–727. 10.1016/j.molcel.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri, Y., Gama-Brambila R.A., Taškova K., Herold K., Reuter S., Adjaye J., Utikal J., Mrowka R., Wang J., Andrade-Navarro M.A., et al. 2019. Imidazopyridines as potent KDM5 demethylase inhibitors promoting reprogramming efficiency of human iPSCs. iScience. 12:168–181. 10.1016/j.isci.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi, Z., Csizmok V., Tompa P., and Simon I.. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 21:3433–3434. 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- Douet, J., Corujo D., Malinverni R., Renauld J., Sansoni V., Posavec Marjanović M., Cantariño N., Valero V., Mongelard F., Bouvet P., et al. 2017. MacroH2A histone variants maintain nuclear organization and heterochromatin architecture. J. Cell Sci. 130:1570–1582. 10.1242/jcs.199216 [DOI] [PubMed] [Google Scholar]
- Dubin, M.J., Stokes P.H., Sum E.Y., Williams R.S., Valova V.A., Robinson P.J., Lindeman G.J., Glover J.N., Visvader J.E., and Matthews J.M.. 2004. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J. Biol. Chem. 279:26932–26938. 10.1074/jbc.M313974200 [DOI] [PubMed] [Google Scholar]
- Fahrer, J., Kranaster R., Altmeyer M., Marx A., and Bürkle A.. 2007. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 35:e143. 10.1093/nar/gkm944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer, J., Popp O., Malanga M., Beneke S., Markovitz D.M., Ferrando-May E., Bürkle A., and Kappes F.. 2010. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 49:7119–7130. 10.1021/bi1004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434:917–921. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Feng, T., Wang Y., Lang Y., and Zhang Y.. 2017. KDM5A promotes proliferation and EMT in ovarian cancer and closely correlates with PTX resistance. Mol. Med. Rep. 16:3573–3580. 10.3892/mmr.2017.6960 [DOI] [PubMed] [Google Scholar]
- Fischer, J.M., Popp O., Gebhard D., Veith S., Fischbach A., Beneke S., Leitenstorfer A., Bergemann J., Scheffner M., Ferrando-May E., et al. 2014. Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function. FEBS J. 281:3625–3641. 10.1111/febs.12885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361:123–134. 10.1056/NEJMoa0900212 [DOI] [PubMed] [Google Scholar]
- Gibson, B.A., and Kraus W.L.. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13:411–424. 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- Gong, F., and Miller K.M.. 2018. Double duty: ZMYND8 in the DNA damage response and cancer. Cell Cycle. 17:414–420. 10.1080/15384101.2017.1376150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, F., and Miller K.M.. 2019. Histone methylation and the DNA damage response. Mutat. Res. 780:37–47. 10.1016/j.mrrev.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S., et al. 2015. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29:197–211. 10.1101/gad.252189.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, F., Clouaire T., Aguirrebengoa M., Legube G., and Miller K.M.. 2017. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 216:1959–1974. 10.1083/jcb.201611135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte, R., Liu Z., and Kraus W.L.. 2017. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31:101–126. 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzlikova, H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., and Caldecott K.W.. 2018. The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell. 71:319–331.e3. 10.1016/j.molcel.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, J., Wu J., Dombkowski A., Zhang K., Holowatyj A., Boerner J.L., and Yang Z.Q.. 2012. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am. J. Transl. Res. 4:247–256. [PMC free article] [PubMed] [Google Scholar]
- Hu, D., Jablonowski C., Cheng P.H., AlTahan A., Li C., Wang Y., Palmer L., Lan C., Sun B., Abu-Zaid A., et al. 2018. KDM5A regulates a translational program that controls p53 protein expression. iScience. 9:84–100. 10.1016/j.isci.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.P., and Helleday T.. 2016. Drugging DNA repair. Science. 352:1178–1179. 10.1126/science.aab0958 [DOI] [PubMed] [Google Scholar]
- Jeggo, P.A., Pearl L.H., and Carr A.M.. 2016. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 16:35–42. 10.1038/nrc.2015.4 [DOI] [PubMed] [Google Scholar]
- Kassab, M.A., Yu L.L., and Yu X.. 2020. Targeting dePARylation for cancer therapy. Cell Biosci. 10:7. 10.1186/s13578-020-0375-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana, S., Kruhlak M.J., Kim J., Tran A.D., Liu J., Nyswaner K., Shi L., Jailwala P., Sung M.H., Hakim O., et al. 2014. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 8:1049–1062. 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Oberdoerffer P., and Khurana S.. 2018. The histone variant macroH2A1 is a splicing-modulated caretaker of genome integrity and tumor growth. Mol. Cell. Oncol. 5:e1441629. 10.1080/23723556.2018.1441629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.J., Kumbhar R., Gong F., and Miller K.M.. 2019a. In time and space: laser microirradiation and the DNA damage response. Methods Mol. Biol. 1999:61–74. 10.1007/978-1-4939-9500-4_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.J., Lee S.Y., Gong F., Battenhouse A.M., Boutz D.R., Bashyal A., Refvik S.T., Chiang C.M., Xhemalce B., Paull T.T., et al. 2019b. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 33:1751–1774. 10.1101/gad.331231.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.J., Lee S.Y., and Miller K.M.. 2019c. Preserving genome integrity and function: the DNA damage response and histone modifications. Crit. Rev. Biochem. Mol. Biol. 54:208–241. 10.1080/10409238.2019.1620676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, R.J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D.G., Zhang Y., and Kaelin W.G. Jr. 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 128:889–900. 10.1016/j.cell.2007.02.013 [DOI] [PubMed] [Google Scholar]
- Knijnenburg, T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D., Fan H., Shen H., Way G.P., Greene C.S., et al. The Cancer Genome Atlas Research Network . 2018. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 23:239–254.e6. 10.1016/j.celrep.2018.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski, M., Corujo D., Hothorn M., Guberovic I., Mandemaker I.K., Blessing C., Sporn J., Gutierrez-Triana A., Smith R., Portmann T., et al. 2018. MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 19:e44445. 10.15252/embr.201744445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietsch, J., Rouleau M., Pic É., Ethier C., Dawson T.M., Dawson V.L., Masson J.Y., Poirier G.G., and Gagné J.P.. 2013. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 34:1066–1087. 10.1016/j.mam.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar, R., and Kraus W.L.. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell. 39:736–749. 10.1016/j.molcel.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger, A., Stier A., Fischbach A., Bürkle A., Hauser K., and Mangerich A.. 2019. Interactions of p53 with poly(ADP-ribose) and DNA induce distinct changes in protein structure as revealed by ATR-FTIR spectroscopy. Nucleic Acids Res. 47:4843–4858. 10.1093/nar/gkz175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhar, R., Vidal-Eychenié S., Kontopoulos D.G., Larroque M., Larroque C., Basbous J., Kossida S., Ribeyre C., and Constantinou A.. 2018. Recruitment of ubiquitin-activating enzyme UBA1 to DNA by poly(ADP-ribose) promotes ATR signalling. Life Sci. Alliance. 1:e201800096. 10.26508/lsa.201800096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J.W.C., Emery L.E., and Miller K.M.. 2018. CRISPR/Cas9 gene editing of human histone H2A variant H2AX and MacroH2A. Methods Mol. Biol. 1832:255–269. 10.1007/978-1-4939-8663-7_14 [DOI] [PubMed] [Google Scholar]
- Li, L., Wang L., Song P., Geng X., Liang X., Zhou M., Wang Y., Chen C., Jia J., and Zeng J.. 2014a. Critical role of histone demethylase RBP2 in human gastric cancer angiogenesis. Mol. Cancer. 13:81. 10.1186/1476-4598-13-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Liu L., Yang S., Song N., Zhou X., Gao J., Yu N., Shan L., Wang Q., Liang J., et al. 2014b. Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. USA. 111:7096–7101. 10.1073/pnas.1324036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X., Zeng J., Wang L., Shen L., Ma X., Li S., Wu Y., Ma L., Ci X., Guo Q., et al. 2015. Histone demethylase RBP2 promotes malignant progression of gastric cancer through TGF-β1-(p-Smad3)-RBP2-E-cadherin-Smad3 feedback circuit. Oncotarget. 6:17661–17674. 10.18632/oncotarget.3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, T., and Barnes D.E.. 2000. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 65:127–133. 10.1101/sqb.2000.65.127 [DOI] [PubMed] [Google Scholar]
- Liu, C., Vyas A., Kassab M.A., Singh A.K., and Yu X.. 2017. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45:8129–8141. 10.1093/nar/gkx565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.J., and Ashworth A.. 2017. PARP inhibitors: synthetic lethality in the clinic. Science. 355:1152–1158. 10.1126/science.aam7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelena, J., Lezaja A., Teloni F., Schmid T., Imhof R., and Altmeyer M.. 2018. Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance. Nat. Commun. 9:2678. 10.1038/s41467-018-05031-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini, S., Gorgoulis V.G., and Halazonetis T.D.. 2010. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11:220–228. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- O’Connor, M.J. 2015. Targeting the DNA damage response in cancer. Mol. Cell. 60:547–560. 10.1016/j.molcel.2015.10.040 [DOI] [PubMed] [Google Scholar]
- O’Sullivan, J., Tedim Ferreira M., Gagné J.P., Sharma A.K., Hendzel M.J., Masson J.Y., and Poirier G.G.. 2019. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 10:1182. 10.1038/s41467-019-08859-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilié, P.G., Tang C., Mills G.B., and Yap T.A.. 2019. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 16:81–104. 10.1038/s41571-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder, J., Salsman J., and Dellaire G.. 2015. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43:9379–9392. 10.1093/nar/gkv993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posavec, M., Timinszky G., and Buschbeck M.. 2013. Macro domains as metabolite sensors on chromatin. Cell. Mol. Life Sci. 70:1509–1524. 10.1007/s00018-013-1294-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posavec Marjanović, M., Hurtado-Bagès S., Lassi M., Valero V., Malinverni R., Delage H., Navarro M., Corujo D., Guberovic I., Douet J., et al. 2017. MacroH2A1.1 regulates mitochondrial respiration by limiting nuclear NAD+ consumption. Nat. Struct. Mol. Biol. 24:902–910. 10.1038/nsmb.3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puget, N., Miller K.M., and Legube G.. 2019. Non-canonical DNA/RNA structures during transcription-coupled double-strand break repair: roadblocks or bona fide repair intermediates? DNA Repair (Amst.). 81:102661. 10.1016/j.dnarep.2019.102661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri, A., and Nussenzweig A.. 2017. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18:610–621. 10.1038/nrm.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, P.D., Hamilton G.A., Park J.W., and Gamble M.J.. 2019. MacroH2A1 regulation of poly(ADP-ribose) synthesis and stability prevents necrosis and promotes DNA repair. Mol. Cell. Biol. 40:e00230-19. 10.1128/MCB.00230-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky, P., Krojer T., Fujisawa T., Lambert J.P., Picaud S., Wang C.Y., Shanle E.K., Krajewski K., Friedrichsen H., Kanapin A., et al. 2016. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 17:2724–2737. 10.1016/j.celrep.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 141:69–80. 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, F., Li M., Liu G., Swapna G.V.T., Daigham N.S., Xia B., Montelione G.T., and Bunting S.F.. 2018. Antiparallel coiled-coil interactions mediate the homodimerization of the DNA damage-repair protein PALB2. Biochemistry. 57:6581–6591. 10.1021/acs.biochem.8b00789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt, C.G., Luijsterburg M.S., Menafra R., Lindeboom R.G., Jansen P.W., Edupuganti R.R., Baltissen M.P., Wiegant W.W., Voelker-Albert M.C., Matarese F., et al. 2016. ZMYND8 co-localizes with NuRD on target genes and regulates poly(ADP-ribose)-dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep. 17:783–798. 10.1016/j.celrep.2016.09.037 [DOI] [PubMed] [Google Scholar]
- Stratton, M.R., and Rahman N.. 2008. The emerging landscape of breast cancer susceptibility. Nat. Genet. 40:17–22. 10.1038/ng.2007.53 [DOI] [PubMed] [Google Scholar]
- Tan, X.Y., and Huen M.S.Y.. 2020. Perfecting DNA double-strand break repair on transcribed chromatin. Essays Biochem. 64:705–719. 10.1042/EBC20190094 [DOI] [PubMed] [Google Scholar]
- Tan, E.S., Krukenberg K.A., and Mitchison T.J.. 2012. Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers. Anal. Biochem. 428:126–136. 10.1016/j.ab.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., and Greenberg R.A.. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20:317–325. 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teloni, F., and Altmeyer M.. 2016. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 44:993–1006. 10.1093/nar/gkv1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Y.C., Lee C.F., Li Y.S., Chen Y.R., Hsiao P.W., Chan M.Y., Lin F.M., Huang H.D., Chen Y.T., Jeng Y.M., et al. 2013. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 73:4711–4721. 10.1158/0008-5472.CAN-12-3165 [DOI] [PubMed] [Google Scholar]
- Thomas, C., and Tulin A.V.. 2013. Poly-ADP-ribose polymerase: machinery for nuclear processes. Mol. Aspects Med. 34:1124–1137. 10.1016/j.mam.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky, G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K., et al. 2009. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 16:923–929. 10.1038/nsmb.1664 [DOI] [PubMed] [Google Scholar]
- Torres, I.O., Kuchenbecker K.M., Nnadi C.I., Fletterick R.J., Kelly M.J., and Fujimori D.G.. 2015. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 6:6204. 10.1038/ncomms7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truebestein, L., and Leonard T.A.. 2016. Coiled-coils: the long and short of it. BioEssays. 38:903–916. 10.1002/bies.201600062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs, A., and Nussenzweig A.. 2017. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 168:644–656. 10.1016/j.cell.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto, V., and Giachino C.. 2015. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 43:2489–2498. 10.1093/nar/gkv061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova, M., Gehling V.S., Gustafson A., Arora S., Tindell C.A., Wilson C., Williamson K.E., Guler G.D., Gangurde P., Manieri W., et al. 2016. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12:531–538. 10.1038/nchembio.2085 [DOI] [PubMed] [Google Scholar]
- Wang, G.G., Song J., Wang Z., Dormann H.L., Casadio F., Li H., Luo J.L., Patel D.J., and Allis C.D.. 2009. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 459:847–851. 10.1038/nature08036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Wang Y., Wu H., and Hu L.. 2013. RBP2 induces epithelial-mesenchymal transition in non-small cell lung cancer. PLoS One. 8:e84735. 10.1371/journal.pone.0084735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, H., and Yu X.. 2016. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics. 14:131–139. 10.1016/j.gpb.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, E., Kim P.S., and Berger B.. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179–1189. 10.1002/pro.5560060606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhabija, B., and Kidder B.L.. 2019. KDM5B is a master regulator of the H3K4-methylome in stem cells, development and cancer. Semin. Cancer Biol. 57:79–85. 10.1016/j.semcancer.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J., Chiu L.Y., Nehring R.B., Bravo Núñez M.A., Mei Q., Perez M., Zhai Y., Fitzgerald D.M., Pribis J.P., Wang Y., et al. 2019. Bacteria-to-human protein networks reveal origins of endogenous DNA damage. Cell. 176:127–143.e24. 10.1016/j.cell.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Zhou B., Zhao X., Zhu L., Xu J., Jiang Z., Chen D., Wei Q., Han M., Feng L., et al. 2018. KDM5B demethylates H3K4 to recruit XRCC1 and promote chemoresistance. Int. J. Biol. Sci. 14:1122–1132. 10.7150/ijbs.25881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G.J., Ko C.N., Zhong H.J., Leung C.H., and Ma D.L.. 2019. Structure-based discovery of a selective KDM5A inhibitor that exhibits anti-cancer activity via inducing cell cycle arrest and senescence in breast cancer cell lines. Cancers (Basel). 11:92. 10.3390/cancers11010092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, J., Ge Z., Wang L., Li Q., Wang N., Björkholm M., Jia J., and Xu D.. 2010. The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 138:981–992. 10.1053/j.gastro.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Zhang, F., Shi J., Bian C., and Yu X.. 2015. Poly(ADP-ribose) mediates the BRCA2-dependent early DNA damage response. Cell Rep. 13:678–689. 10.1016/j.celrep.2015.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.