Abstract
Hepatocellular carcinoma (HCC) is a common primary malignant tumor with a high mortality rate. Increasing evidence suggests that ribosomal protein LP1 (RPLP1) is involved in the progression of different types of cancer. Thus, the present study aimed to investigate the underlying molecular mechanism of RPLP1 in HCC progression. The cellular behaviors of Hep3b cells were assessed via Cell Counting Kit-8, colony formation, wound healing and Transwell assays. Western blot analysis was performed to detect protein expression levels, while reverse transcription-quantitative PCR analysis was performed to detect mRNA expression levels. The results demonstrated that RPLP1 was highly expressed in HCC tissues and cells, and the overexpression of RPLP1 was associated with a less favorable prognosis of patients with HCC. Notably, downregulation of RPLP1 significantly suppressed the proliferation, migration and invasion of Hep3b cells. Taken together, the results of the present study suggested that RPLP1 acts as an oncogene in HCC, and thus may be used to treat patients with HCC.
Keywords: hepatocellular carcinoma, ribosomal protein LP1, proliferation, invasion, migration
Introduction
Hepatocellular carcinoma (HCC) is estimated to be the sixth most common primary malignant tumor and the second leading cause of cancer-associated mortality worldwide (1-3). The most common etiological factor for HCC is the cirrhotic liver, which provides a pro-carcinogenic intrahepatic environment (4,5). The most notable risk factors, including chronic viral hepatitis caused by hepatitis B virus (HBV) or hepatitis C virus infection, alcohol abuse, non-alcoholic fatty liver disease and other abnormal metabolic conditions, contribute to the development of cirrhosis, particularly in East Asian countries where HBV infection is endemic (6-8). HCC is generally diagnosed in individuals at an advanced stage due to the lack of early symptoms (6). Despite advancements in the comprehensive treatments based on surgery, the postoperative recurrence of HCC seriously threatens the quality of life of ~30% those affected, with limited treatment options (9). Thus, it remains crucial to develop novel therapeutic strategies to improve the outcome of patients with HCC.
The onset and development of HCC involves multi-step biological processes, which are regulated by multiple factors and signaling molecules (10). Ribosomal protein LP1 (RPLP1) consists of 114 amino acids and is a member of the ribosomal protein L12P family (11). Increasing evidence suggests that RPLP1 plays a crucial role in the elongation step of protein synthesis (12,13). Upregulated RPLP1 expression facilitates tumorigenesis and immortalizes primary cells, contributing to cellular transformation (14). It has been reported that RPLP1 may act as a prognostic biomarker and anti-metastatic candidate therapeutic target in triple-negative breast cancer (14). In addition, RPLP1 expression has been demonstrated to be upregulated in biopsy specimens taken from patients with colon cancer (11). A previous study demonstrated that RPLP1 expression is significantly associated with the progression of gynecological tumors, including serous ovarian cancers and endometrial carcinomas (15). However, the role of RPLP1 in HCC progression remains unknown. Thus, the present study aimed to investigate the role of RPLP1 in HCC progression.
The present study aimed to investigate the role of RPLP1 in HCC progression and assess its effect on the cellular behaviors of human HCC cells. The present study provided novel insight into understanding the HCC pathogenesis and indicating RPLP1 may be of value as a promising therapeutic target for HCC treatment.
Materials and methods
Cell culture and transfection
Human liver HHL-5 cells and the HCC cell lines, SK-HEP-1, Hep3b and Huh-7, were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. All cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 µg/ml streptomycin and 100 U/ml penicillin (all Gibco; Thermo Fisher Scientific, Inc.), at 37˚C with 5% CO2.
Hep3b cells were seeded into 96-well (2x103 cells/well) or six-well (1x105 cells/well) plates and transfected with short hairpin (sh) RNA-RPLP1 (100 nM) using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C for 12 h. Cells were cultured until they reached 60-70% confluence. RPLP1 shRNA-1 (5'-CATTAAAGCAGCCGGTGTAAATGTTGAGC-3') and RPLP1 shRNA-2 (5'-GAAGAAAGTGGAAGCATTCAAGAGATGCT-3') or the negative control RNA (non-targeting) were synthesized by Shanghai GeneChem Co., Ltd. Reverse transcription-quantitative (RT-q)PCR analysis was performed to assess transfection efficiency. Hep3b cells were used for subsequent experimentation 48 h post-transfection. RPLP1 expression was assessed by screening the Gene Expression Profiling Interactive Analysis database (http://gepia.cancer-pku.cn) to determine the association between RPLP1 expression and HCC progression. Moreover, the overall survival (OS) analyzed using the GEPIA database (http://gepia.cancer-pku.cn/) to investigate the role of RPLP1 in HCC progression and the UALCAN database (http://ualcan.path.uab.edu) was used to determine the association between RPLP1 expression and tumor grade of HCC.
RT-qPCR
Total RNA was extracted from transfected Hep3b cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The prepared reaction mixture was incubated at 37˚C for 60 min after brief centrifugation (14,000 x g for 5 min at 4˚C), followed by incubation at 85˚C for 5 min for RT. qPCR was subsequently performed using the SYBR Premix EX Taq™ II kit (Takara Bio, Inc.) on an ABI Prism 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Pre-denaturation at 95˚C for 5 min, followed by 40 cycles at 95˚C for 15 sec, 60˚C for 30 sec and 72˚C for 30 sec. The following primer sequences were used for qPCR: RPLP1, Forward: 5'-TGGCCTGGCTTGTTTGC-3' and reverse: 5'-CTCGGATTCTTCTTTCTTTGCTT-3'; and GAPDH, forward: 5'-CTCCTCCACCTTTGACGCTG-3' and reverse: 5'-TCCTCTTGTGCTCTTGCTGG-3'. Relative expression levels were calculated using the 2-ΔΔCq method (16) and normalized to the internal reference gene GAPDH.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 (Dojindo Molecular Technologies, Inc.) assay was performed to assess the viability of transfected Hep3b cells, according to the manufacturer's instructions. Briefly, Hep3b cells transfected with or without RPLP1 shRNA-1 were seeded into 96-well plates at a density of 2x103 cells/well and incubated for 0, 24, 48 and 72 h at 37˚C. Following incubation, 10 µl CCK-8 solution was added to each well for another 2 h and cell viability was analyzed at a wavelength of 450 nm using a microtiter plate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
Transfected Hep3b cells were seeded into six-well plates at a density of 1x103 cells/well and cultured for 4 weeks at 37˚C, without disturbing. Cell colonies were subsequently fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 10 min at room temperature. Stained cells were counted in five randomly selected fields of view by eye and images were captured.
Western blotting
Total lysates were extracted from transfected Hep3b cells using RIPA buffer with protease inhibitors cocktail (both Sigma-Aldrich; Merck KGaA). Protein concentration was measured using the BCA assay kit (Bio-Rad Laboratories, Inc.) and 25 µg protein/lane was separated using 10% SDS-PAGE gels. The separated proteins were subsequently transferred onto PVDF membranes (EMD Millipore) and blocked with 5% skimmed milk at room temperature for 2 h. The membranes were incubated with primary antibodies against: Minichromosome maintenance (MCM; cat. no. #3228), proliferating cell nuclear antigen (PCNA; cat. no. #13110), vimentin (cat. no. #5741,), snail (cat. no. #3895), slug (cat. no. #9585) (all 1:1,000), β-catenin (1:500; cat. no. #9582), N-cadherin (cat. no. #13116), E-cadherin (cat. no. #5296) (all 1:500), claudin-1 (1:1,000, #4933, Cell Signaling Technology, Inc.), matrix metalloproteinase (MMP)-2 (cat. no. #4022,), matrix metalloproteinase (MMP)-9 (cat. no. #3852), tissue inhibitor of MMP-1 (TIMP-1; cat. no. #8946) (all 1:1,000) and GAPDH (1:2,000; cat. no. #5174) (all Cell Signaling Technology, Inc.) overnight at 4˚C. After washing with PBS, the membranes were incubated with secondary HRP-conjugated goat anti-rabbit IgG (1:5,000; cat. no. #7074; Cell Signaling Technology, Inc.) at room temperature for 2 h, and then visualized using an ECL chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.). At last, the gray values of bands were detected using ImageJ software (version 1.48; National Institutes of Health) and normalized to GAPDH.
Immunofluorescence
When Hep3b cells were cultured to the appropriate densities, cells in co-culture plates were fixed with 4% paraformaldehyde for 10 min at room temperature and subsequently permeabilized with 0.1% Triton X-100 for 10 min. Following blocking with 5% BSA and 10% horse serum (Sigma-aldrich; Merck KGaA) in PBST for 1 h at room temperature, Hep3b cells were incubated with primary antibody against Ki-67 (1:200; cat. no. #12075; Cell Signaling Technology, Inc.) overnight at 4˚C. Following the primary incubation, cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:200; cat. no. ab150077; Abcam) for 1 h at room temperature. Cell nuclei were stained with DAPI for 5 min at room temperature. Hep3b cells were washed three times with PBST prior to observation under a light microscope at x200 magnification and images were captured (Olympus Corporation).
Wound healing assay
Hep3b cells were seeded into six-well plates at a density of 1x105 cells/well and transfected with or without shRNA-RPLP1. Once cells reached 100% confluence, the monolayers were scratched using 200-µl sterile pipette tips to create straight linear wounds. Cells were cultured in serum-free medium for 24 h at 37˚C, with 5% CO2. Wounds were observed in five randomly selected fields using an inverted fluorescence microscope at 100x magnification and images were captured. The recovered wound area (%) at the indicated time point (24 h) was calculated according to the following formula: [(Wound width at 0 h)-(wound width at 24 h)]/wound width at 0 h.
Transwell assay
Following transfection, Hep3b cells were resuspended in serum-free medium and plated in the upper chambers of Transwell plates precoated with Matrigel at 37˚C for 30 min. Complete medium (600 µl) was plated in the lower chambers. Following incubation for 24 h at 37˚C with 5% CO2, cells in the upper chambers were gently removed using a wet cotton swab. Cells in the lower chambers were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min, both at room temperature. Stained cells were counted in five randomly selected fields using an inverted light microscope and images were captured (magnification, x200).
Statistical analysis
Statistical analysis was performed using SPSS 13.0 software (SPSS Inc.). All experiments were performed in triplicate and data are presented as the mean ± standard error of mean (unless otherwise shown). A two-tailed unpaired Student's t-test was used to compare differences between two groups, while one-way ANOVA followed by Dunnett's post hoc tests were used to compare differences between multiple groups. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
RPLP1 is highly expressed in HCC tissues and cells, and overexpression of RPLP1 is associated with the prognosis of patients with HCC
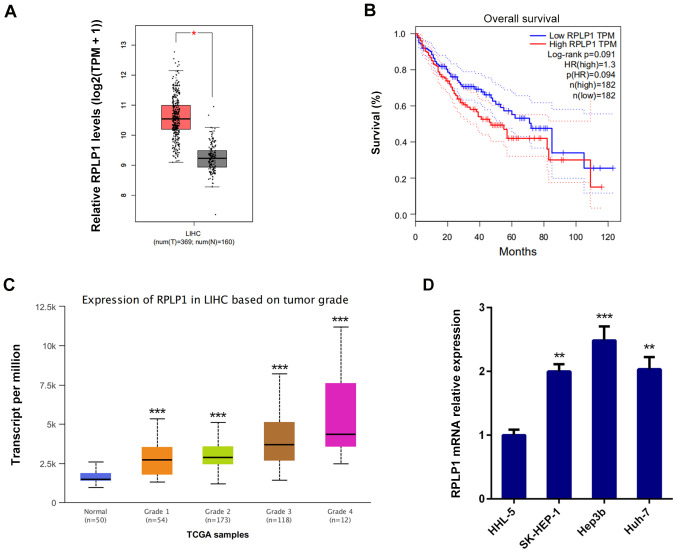
RPLP1 expression was assessed by screening the Gene Expression Profiling Interactive Analysis database (http://gepia.cancer-pku.cn) to determine the association between RPLP1 expression and HCC progression. The results demonstrated that RPLP1 was highly expressed in HCC tissues compared with adjacent liver tissues (Fig. 1A) (529 cases; P=1.6x10-12).
Figure 1.
RPLP1 is highly expressed in HCC tissues and cells, and overexpression of RPLP1 is associated with the prognosis of patients with HCC. (A) High RPLP1 expression in HCC tissue is associated with a poor OS rate. (B) OS analysis was performed using the log-rank, according to follow-up information of patients with HCC. (C) RPLP1 expression was associated with tumor grade of HCC. (D) Reverse transcription-quantitative PCR analysis was performed to detect RPLP1 mRNA expression in the human liver cell line and the HCC cell lines. *P<0.05, **P<0.01 and ***P<0.001 vs. respective control. RPLP1, ribosomal protein LP1; HCC, hepatocellular carcinoma; OS, overall survival; TPM, transcripts per million; TCGA, The Cancer Genome Atlas; LIHC, liver hepatocellular carcinoma.
To further investigate the role of RPLP1 in HCC progression, the overall survival (OS) datasets was analyzed by GEPIA database (http://gepia.cancer-pku.cn/). As presented in Fig. 1B, high RPLP1 expression was associated with poor prognosis and high mortality, and the OS rate (log-rank P=0.091; hazard ratio=1.3) were less favorable in patients with high RPLP1 expression levels.
The UALCAN database (http://ualcan.path.uab.edu) was used to determine the association between RPLP1 expression and tumor grade of HCC. As presented in Fig. 1C, RPLP1 expression was positively associated with tumor grade. Taken together, these results suggested that RPLP1 may promote HCC development. In addition, RT-qPCR analysis was performed to detect RPLP1 expression in human liver HHL-5 cells and the HCC cell lines, SK-HEP-1, Hep3b and Huh-7. The results demonstrated that RPLP1 was highly expressed in the HCC cell lines compared with normal liver cells, and the most significant overexpression was observed in Hep3b cells (Fig. 1D). Thus, Hep3b cells were selected for subsequent experiments.
Downregulation of RPLP1 inhibits the proliferation of Hep3b cells
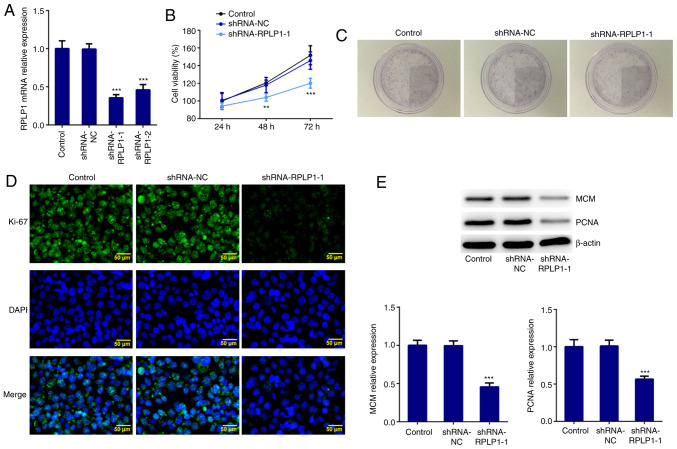
RPLP1 expression was downregulated using shRNA-RPLP1-1, shRNA-RPLP1-2 and shRNA-NC. Due to the low expression level induced by shRNA-RPLP1-1 (Fig. 2A), this shRNA was selected for subsequent experimentation. The results of the CCK-8 assay demonstrated that downregulation of RPLP1 significantly suppressed Hep3b cell viability (Fig. 2B). The results of the colony formation assay revealed that the proliferation rate of Hep3b cells transfected with shRNA-RPLP1-1 markedly decreased compared with the control and shRNA-NC groups (Fig. 2C). Furthermore, the expression levels of the standard markers of proliferation, Ki-67, PCNA and MCM (17) were assessed via western blotting and immunofluorescence. The results demonstrated that the expression levels of PCNA, MCM and Ki-67 decreased in the shRNA-RPLP1-1 group compared with the control and shRNA-NC groups (Fig. 2D and E). Taken together, these results suggested that downregulation of RPLP1 suppresses the proliferation of Hep3b cells.
Figure 2.
Downregulation of RPLP1 inhibits the proliferation of Hep3b cells. (A) Reverse transcription-quantitative PCR analysis was performed to detect RPLP1 mRNA expression in Hep3b cells. (B) A Cell Counting Kit-8 assay was performed to assess cell viability. (C) A colony formation assay was performed to assess cell proliferation. (D) Immunofluorescence analysis was performed to detect Ki-67 expression. Scale bars, 50 µm. (E) Western blot analysis was performed to detect the protein expression levels of MCM and PCNA. **P<0.01, ***P<0.001 vs. control. RPLP1, ribosomal protein LP1; MCM, minichromosome maintenance; PCNA, proliferating cell nuclear antigen; sh, short hairpin; NC, negative control.
Downregulation of RPLP1 affects the expression of epithelial-to-mesenchymal (EMT)-associated proteins
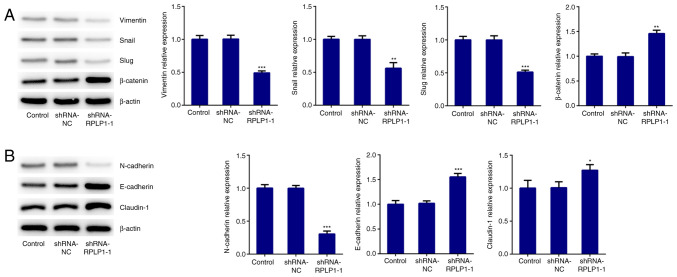
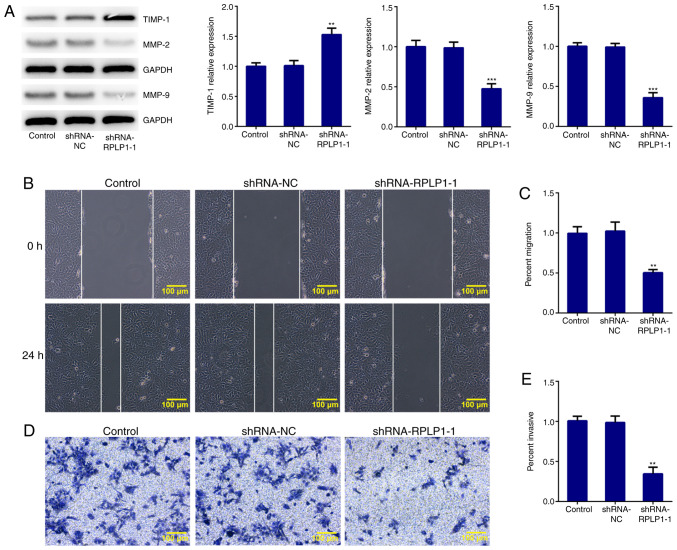
To further investigate the effect of RPLP1 on cellular biological behaviors, proteins related to EMT were detected via western blot analysis. The results demonstrated that downregulation of RPLP1 significantly decreased the expression levels of vimentin, snail and slug, and increased β-catenin expression (Fig. 3A). In addition, N-cadherin expression (mesenchymal cell marker) (18) notably decreased following downregulation of RPLP1 expression, while the expression levels of E-cadherin (epithelial cell marker) (18) and claudin-1 significantly increased following downregulation of RPLP1 (Fig. 3B). Downregulation of RPLP1 also suppressed the expression levels of MMP-2 and MMP-9, and notably increased TIMP-1 expression (Fig. 4A). These results implied that downregulation of RPLP1 inhibited the EMT process of Hep3b cells.
Figure 3.
Downregulation of RPLP1 suppresses the expression levels of EMT-associated proteins in Hep3b cells. (A and B) Western blot analysis was performed to detect the protein expression levels of vimentin, snail, slug, β-catenin, N-cadherin, E-cadherin and claudin-1. *P<0.05, **P<0.01 and ***P<0.001 vs. control. RPLP1, ribosomal protein LP1; EMT, epithelial-to-mesenchymal transition; sh, short hairpin; NC, negative control.
Figure 4.
Downregulation of RPLP1 suppresses the migratory and invasive abilities of Hep3b cells. (A) Western blot analysis was performed to detect the protein expression levels of MMP-2, MMP-9 and TIMP-1. (B and C) Wound healing assay was performed to assess cell migration. Scale bars, 100 µm. (D and E) Transwell assay was performed to assess cell invasion. Scale bars, 100 µm. **P<0.01 and ***P<0.001 vs. control. RPLP1, ribosomal protein LP1; MMP, matrix metalloproteinase; TIMP-1, tissue inhibitor of matrix metalloproteinase-1; sh, short hairpin; NC, negative control.
Downregulation of RPLP1 suppresses the migration and invasion of Hep3b cells
The wound healing and Transwell assays were performed to confirm the effect of RPLP1 downregulation on the migratory and invasive abilities of Hep3b cells. As presented in Fig. 4B-E, the migratory and invasive abilities of Hep3b cells were significantly inhibited following downregulation of RPLP1. Taken together, these results suggested that downregulation of RPLP1 inhibits the potency of invasion and migration of Hep3b cells.
Discussion
HCC is the sixth most common primary malignant tumor, which poses a great threat to human health and life, with ~800,000 mortalities annually worldwide (19,20). Increasing evidence suggests that several biomarkers can be used to predict cancer progression and are associated with the prognosis of patients with different types of cancer (21,22). Notably, the results of the present study demonstrated that RPLP1 expression was upregulated in HCC tissues compared with adjacent liver tissues and was significantly associated with less favorable prognosis of patients with HCC. In addition, the results demonstrated an association between RPLP1 expression and HCC pathogenesis, which merits further investigation for use of RPLP1 as a potential target for HCC treatment.
High potency of invasion and migration of cancer cells promotes cancer progression and is associated with poor prognosis of patients with cancer (23,24). A previous study reported that overexpression of RPLP1 promoted the proliferation, migration and invasion of cervical cancer cells (25). The results of the present study demonstrated that RPLP1 was highly expressed in HCC cell lines, which is consistent with the previous study. The postoperative quality of life and prognosis of patients with HCC is restrained by the strong invasion and migration potential of tumor cell (26). Thus, the proliferative, migratory and invasive abilities of Hep3b cells were assessed in the present study. RPLP1 expression was downregulated following transfection of Hep3b cells with shRNA-RPLP1. Ki67, PCNA and MCM proteins are the standard proliferative markers that are commonly applied to analyze the proliferative activity of a cell population (27). The results of the present study demonstrated that downregulation of RPLP1 significantly decreased the expression levels of Ki67, PCNA and MCM, and suppressed the viability and proliferation of Hep3b cells. Collectively, these results suggested that downregulation of RPLP1 inhibits the proliferation of HCC cells. In addition, the expression levels of the EMT-related proteins, and the migratory and invasive abilities of Hep3b cells, were assessed by a series of functional experiments. The loss of epithelial marker, E-cadherin, accompanied by the gain of the key mesenchymal markers, N-cadherin and vimentin, suggests that the cells underwent EMT (28). As a component of the cadherin complex, β-catenin plays a key role in localization (29). During the process of EMT, β-catenin dissociates from the cadherin complex and is translocated into the nucleus where it functions as a transcription factor, which regulates the expression of several genes associated with cancer metastasis including Wnt and p53(29). In addition, MMP-2 and MMP-9 expression are associated with tumor invasion and metastasis of malignant tumors (30). In the present study, RPLP1 silencing decreased the expression levels of vimentin, snail, slug, N-cadherin, MMP-2 and MMP-9, and increased the expression levels of β-catenin, E-cadherin, claudin-1 and TIMP-1, suggesting that RPLP1 may contribute to the induction of EMT. Hence, increased RPLP1 expression may promote HCC metastasis via regulation of EMT and enhanced cell motility. Moreover, the results of the wound healing and Transwell assays demonstrated that RPLP1 silencing remarkably suppressed the migratory and invasive abilities of Hep3b cells. Taken together, these results suggested that downregulation of RPLP1 significantly inhibits the proliferation, migration and invasion of Hep3b cells. Thus, RPLP1 may play a key role in HCC occurrence, progression, invasion and metastasis and so may be used as a target for HCC treatment.
A previous study reported that the RPLP protein deficiency resulted in reactive oxygen species accumulation and MAPK1/ERK2 signaling pathway activation in colon cancer cells (11). However, the detailed mechanism underlying the role of RPLP remains unclear. Hence, future studies will be performed to further investigate the role of RPLP1 in HCC progression.
In conclusion, the results of the present study demonstrated that RPLP1 was highly expressed in HCC tissues and cells, and overexpression of RPLP1 was associated with the prognosis of patients with HCC. Notably, downregulation of RPLP1 suppressed the proliferation, migration and invasion of Hep3b cells. Collectively, the results of the present study suggested that RPLP1 acts as an oncogene in HCC, and thus may be used to treat patients with HCC.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CX and LQ designed the experiment and drafted the manuscript. CX, KC and DP performed the experiments and analyzed the data. CX and LQ reviewed the manuscript. All authors read and approved the final manuscript. CX and LQ confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Xiao JX, Xu W, Fei X, Hao F, Wang N, Chen Y, Wang J. Anillin facilitates cell proliferation and induces tumor growth of hepatocellular carcinoma via miR-138/SOX4 axis regulation. Transl Oncol. 2020;13(100815) doi: 10.1016/j.tranon.2020.100815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vilgrain V, Pereira H, Assenat E, Guiu B, Ilonca AD, Pageaux GP, Sibert A, Bouattour M, Lebtahi R, Allaham W, et al. Efficacy and safety of selective internal radiotherapy with yttrium-90 resin microspheres compared with sorafenib in locally advanced and inoperable hepatocellular carcinoma (SARAH): An open-label randomised controlled phase 3 trial. Lancet Oncol. 2017;18:1624–1636. doi: 10.1016/S1470-2045(17)30683-6. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Coskun M. Hepatocellular carcinoma in the cirrhotic liver: Evaluation using computed tomography and magnetic resonance imaging. Exp Clin Transplant. 2017;15 (Suppl 2):S36–S44. doi: 10.6002/ect.TOND16.L10. [DOI] [PubMed] [Google Scholar]
- 5.Granito A, Bolondi L. Non-transplant therapies for patients with hepatocellular carcinoma and Child-Pugh-Turcotte class B cirrhosis. Lancet Oncol. 2017;18:e101–e112. doi: 10.1016/S1470-2045(16)30569-1. [DOI] [PubMed] [Google Scholar]
- 6.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16(1) doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahtab MA, Chaudhury M, Uddin MH, Noor EASM, Rahim MA, Alam MA, Moben AL, Khondaker FA, Choudhury MF, Sarkar MJ, et al. Cost assessment of Hepatitis B virus-related hepatitis in Bangladesh. Euroasian J Hepatogastroenterol. 2016;6:163–166. doi: 10.5005/jp-journals-10018-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarma MP, Bhattacharjee M, Kar P, Medhi S. Detection of HBV genotype C in hepatocellular carcinoma patients from north east India: A brief report. Asian Pac J Cancer Prev. 2018;19:1741–1746. doi: 10.22034/APJCP.2018.19.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reig M, Mariño Z, Perelló C, Iñarrairaegui M, Ribeiro A, Lens S, Díaz A, Vilana R, Darnell A, Varela M, et al. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J Hepatol. 2016;65:719–726. doi: 10.1016/j.jhep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Dituri F, Mancarella S, Cigliano A, Chieti A, Giannelli G. TGF-β as multifaceted orchestrator in HCC progression: Signaling, EMT, immune microenvironment, and novel therapeutic perspectives. Semin Liver Dis. 2019;39:53–69. doi: 10.1055/s-0038-1676121. [DOI] [PubMed] [Google Scholar]
- 11.Artero-Castro A, Kondoh H, Fernandez-Marcos PJ, Serrano M, Ramon y Cajal S, Lleonart ME. Rplp1 bypasses replicative senescence and contributes to transformation. Exp Cell Res. 2009;315:1372–1383. doi: 10.1016/j.yexcr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Artero-Castro A, Perez-Alea M, Feliciano A, Leal JA, Genestar M, Castellvi J, Peg V, Ramon y Cajal S, Lleonart ME. Disruption of the ribosomal P complex leads to stress-induced autophagy. Autophagy. 2015;11:1499–1519. doi: 10.1080/15548627.2015.1063764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos RK, Wong B, Xie X, Lu YF, Shi PY, Pompon J, Garcia-Blanco MA, Bradrick SS. RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J Virol. 2017;91(e01706) doi: 10.1128/JVI.01706-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Z, Xu Q, Wang X, Wang J, Mu X, Cai Y, Qian Y, Shao W, Shao Z. RPLP1 promotes tumor metastasis and is associated with a poor prognosis in triple-negative breast cancer patients. Cancer Cell Int. 2018;18(170) doi: 10.1186/s12935-018-0658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artero-Castro A, Castellvi J, Garcia A, Hernandez J, Ramon y Cajal S, Lleonart ME. Expression of the ribosomal proteins Rplp0, Rplp1, and Rplp2 in gynecologic tumors. Hum Pathol. 2011;42:194–203. doi: 10.1016/j.humpath.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Roels S, Tilmant K, Ducatelle R. PCNA and Ki67 proliferation markers as criteria for prediction of clinical behaviour of melanocytic tumours in cats and dogs. J Comp Pathol. 1999;121:13–24. doi: 10.1053/jcpa.1998.0291. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Wang Y, Ding H. doi: 10.1007/s12094-021-02611-y. COL4A1, negatively regulated by XPD and miR-29a-3p, promotes cell proliferation, migration, invasion and epithelial-mesenchymal transition in liver cancer cells. Clin Transl Oncol: Apr 23, 2021 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 19.Beudeker BJB, Boonstra A. Circulating biomarkers for early detection of hepatocellular carcinoma. Therap Adv Gastroenterol. 2020;13(1756284820931734) doi: 10.1177/1756284820931734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leiphrakpam PD, Lazenby AJ, Chowdhury S, Smith LM, Mathiesen M, Brattain MG, Wang J, Black JD, Are C. Prognostic and therapeutic implications of NHERF1 expression and regulation in colorectal cancer. J Surg Oncol. 2020;121:545–560. doi: 10.1002/jso.25805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang XJ, Yu Q, Chi P, Lin HM, Lu XR, Huang Y, Xu ZB, Huang SH, Sun YW, Ye DX. Identification of gene biomarkers to predict responses to neoadjuvant chemoradiotherapy in patients with rectal cancer and pathways enrichment analysis. Zhonghua Wei Chang Wai Ke Za Zhi. 2019;22:1183–1187. doi: 10.3760/cma.j.issn.1671-0274.2019.12.015. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 23.Liu YP, Chen WD, Li WN, Zhang M. Overexpression of FNDC1 relates to poor prognosis and its knockdown impairs cell invasion and migration in gastric cancer. Technol Cancer Res Treat. 2019;18(1533033819869928) doi: 10.1177/1533033819869928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu ZH, Wang YM, Jiang YZ, Ma SJ, Zhong Q, Wan YY, Wang XW. NID2 can serve as a potential prognosis prediction biomarker and promotes the invasion and migration of gastric cancer. Pathol Res Pract. 2019;215(152553) doi: 10.1016/j.prp.2019.152553. [DOI] [PubMed] [Google Scholar]
- 25.Xia L, Yue Y, Li M, Zhang YN, Zhao L, Lu W, Wang X, Xie X. CNN3 acts as a potential oncogene in cervical cancer by affecting RPLP1 mRNA expression. Sci Rep. 2020;10(2427) doi: 10.1038/s41598-020-58947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang G, Zhang G. Upregulation of FoxP4 in HCC promotes migration and invasion through regulation of EMT. Oncol Lett. 2019;17:3944–3951. doi: 10.3892/ol.2019.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juríková M, Danihel Ľ, Polák S, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15(18) doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tafrihi M, Nakhaei Sistani R. E-Cadherin/β-catenin complex: A target for anticancer and antimetastasis plants/plant-derived compounds. Nutr Cancer. 2017;69:702–722. doi: 10.1080/01635581.2017.1320415. [DOI] [PubMed] [Google Scholar]
- 30.Bjorklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta. 2005;1755:37–69. doi: 10.1016/j.bbcan.2005.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.