Abstract
Secretory organs are critical for organismal survival. Yet, the transcriptional regulatory mechanisms governing their development and maintenance remain unclear for most model secretory organs. The Drosophila embryonic salivary gland (SG) remedies this deficiency as one of the few organs wherein direct connections from the expression of the early patterning genes to cell specification to organ architecture and functional specialization can be made. Few other models of secretion can be accorded this distinction. Studies from the past three decades have made enormous strides in parsing out the roles of distinct transcription factors (TFs) that direct major steps in furnishing this secretory organ. In the first step of specifying the salivary gland, the activity of the Hox factors Sex combs reduced, Extradenticle, and Homothorax activate expression of fork head (fkh), sage, and CrebA, which code for the major suite of TFs that carry forward the task of organ building and maintenance. Then, in the second key step of building the SG, the program for cell fate maintenance and morphogenesis is deployed. Fkh maintains the secretory cell fate by regulating its own expression and that of sage and CrebA. Fkh and Sage maintain secretory cell viability by actively blocking apoptotic cell death. Fkh, along with two other TFs, Hkb and Rib, also coordinates organ morphogenesis, transforming two plates of precursor cells on the embryo surface into elongated internalized epithelial tubes. Acquisition of functional specialization, the third key step, is mediated by CrebA and Fkh working in concert with Sage and yet another TF, Sens. CrebA directly upregulates expression of all of the components of the secretory machinery as well as other genes (e.g., Xbp1) necessary for managing the physiological stress that inexorably accompanies high secretory load. Secretory cargo specificity is controlled by Sage and Sens in collaboration with Fkh. Investigations have also uncovered roles for various signaling pathways, e.g., Dpp signaling, EGF signaling, GPCR signaling, and cytoskeletal signaling, and their interactions within the gene regulatory networks that specify, build, and specialize the SG. Collectively, studies of the SG have expanded our knowledge of secretory dynamics, cell polarity, and cytoskeletal mechanics in the context of organ development and function. Notably, the embryonic SG has made the singular contribution as a model system that revealed the core function of CrebA in scaling up secretory capacity, thus, serving as the pioneer system in which the conserved roles of the mammalian Creb3/3L-family orthologues were first discovered.
Keywords: Drosophila, morphogenesis, salivary gland, secretion
Introduction
Professional secretory organs go beyond their own requirements
Secretion is a fundamental facet of cell physiology. It impacts the survival and function of prokaryotes by enabling key biological processes that include microenvironment sculpting, e.g., extracellular polymeric substances in biofilms (Hall-Stoodley et al., 2004); cell-to-cell communication, e.g., autoinducer in quorum sensing (Ahmer, 2004); and pathogenic potential, e.g., lung pathogenesis by Legionella pneumophila (Hiller et al., 2018). In eukaryotes, secretion is the cornerstone of homeostatic function and survival not only for unicellular (e.g., yeast) and colonial organisms (e.g., Dictyostelium), but also for the multicellular phyla, e.g., plants and animals (Chung and Zeng, 2017; Goncalves et al., 2019; Guo et al., 2017; Huber and O’Day, 2017). It is the secretion of digestive enzymes that allows organisms to extract essential nutrition and vitamins from their consumption. Secretions also lubricate organs to prevent tissue damage in multi-organ systems while also protecting the organism from the pathogens it encounters. Accordingly, no organ system in higher metazoans is exempt from the profound impact of secretory dynamics, as they are integral to organismal physiology. The mechanisms that affect eukaryotic cell secretion, moreover, unfold at the interface of major membrane-bound organelles, the endoplasmic reticulum and the Golgi complex, thereby spotlighting these expansive organelles while also rendering an unparalleled context for discovering aspects of novel physiological and regulatory mechanisms underlying their coordinated function. Uncovering the cellular networks involved in secretory organ development is, therefore, expected to contribute to a better understanding of organismal development itself, from both an organellar and a cell physiological perspective.
Beyond its role as one of the facets of basal physiology in a variety of cell types (e.g., muscle), secretion attains a premier role in defining the function of select cell types (e.g., plasma cells). Indeed, a fully functional human plasma cell can perform the astonishing feat of secreting up to its own weight in antibody each day (Bromage et al., 2009). Specialized secretory organs, furthermore, produce quantitatively abundant outputs. For example, the mammary glands of high-producing dairy cows secrete up to forty-four liters of milk per day—an amount nearly twice their daily dry matter intake (Kolver and Muller, 1998). Specialized secretory organs, moreover, are attuned to producing qualitatively diverse products essential for organismal foraging and defense, e.g., spider silk and snake venom (Harmer et al., 2011; Villar-Briones and Aird, 2018). Specialized secretory organs are also critical from a pathophysiological standpoint due to their roles as reservoirs and transmitters of pathogens afflicting humans and, thus, as potential targets for intervention, e.g., salivary glands of ticks and mosquitos (Simo et al., 2017; Wells and Andrew, 2019).
Since the secretory physiology of higher organisms is typically governed by functional cellular networks in the form of specialized secretory organs, studies of the embryonic development of organs specialized for secretion have maximally contributed to improving our understanding of secretory organ biology (Byrnes et al., 2018; Huebner et al., 2014). Glandular organs are the hallmark of secretion, and have, hitherto, proved ideal models for studying its underlying molecular and cellular mechanisms (Byrnes et al., 2018; Romagnoli et al., 2020; Yang et al., 2019). Within the assortment of model systems available for such studies, however, the Drosophila embryonic salivary gland (SG) is unmatched. Its relative morphological simplicity coupled with a low redundancy of genes and the availability of powerful genetic and molecular biological tools in this organism have made the SG a salient model system in the efforts to understand the gene regulatory network(s) involved in the assembly and maintenance of specialized secretory organs. Here, we review the morphogenetic trajectory of the Drosophila embryonic SG by highlighting the key roles of transcription factors (TFs) in its construction and functional priming. Our discussion underscores the advantages of this model system for the identification and characterization of key factors involved in secretory organ specialization and maintenance (Table 1). In other words, the Drosophila SG, in its utility as a model system, exemplifies functional cellular networks specialized for secretion.
Table 1. The major transcription factors involved in SG specification, maintenance, morphogenesis, survival and secretion and their orthologues in humans and major model systems.
The genes involved in SG specification are among the most highly conserved, whereas the genes involved in regulating secretory content are among the least conserved. CrebA and it orthologues are among the few SG TF genes that have been demonstrated to have the same functions in increasing secretory capacity in mammals based on functional characterization of mutant phenotypes within each species (mice and humans) and/or their ability to regulate the same targets when expressed in Drosophila embryos.
| SG Specification | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drosophila melanogaster | Arabidopsis thaliana | Dictyostelium discoideum | Saccharomyces cervisaie | Caenorhabditis elegans | Anopheles gambiae | Aedes aegypti | Ciona intestinalis | Homo sapiens | ||||
| Scr | Hox 20 | G0284293 | Very weak homology to mating type regulators | lin-39 | AGAP004659 | AAEL009949 | Hox5 | HoxA5 | HoxB5 | HoxC5 | ||
| % query cover | 10% | 15% | 29% | 100% | 100% | 25% | 21% | 36% | 27% | |||
| % identity | 52% | 35% | 53 | 48% | 51% | 66% | 72% | 50% | 63% | |||
| E value | 2.00E-05 | 3.00E-05 | 1.00E-33 | 1.00E-111 | 7.00E-107 | 8.00E-44 | 5.00E-44 | 3.00E-43 | 8.00E-42 | |||
| exd | knotted-like/BEL1-like | G0272967 | Tos8P | Cup9P | ceh-20 | AGAP004696 | exd | TF protein | Pbx1 | Pbx2 | Pbx3 | Pbx4 |
| % query cover | 26%/16% | 15% | 21% | 14% | 87% | 100% | 100% | 81% | 92% | 81% | 92% | 75% |
| % identity | 32/42% | 41% | 38% | 47% | 59% | 85% | 85% | 72% | 76% | 79% | 74% | 75% |
| E value | 1.00E-08 | 1.00E-07 | 1.00E-09 | 8.00E-09 | 1.00E-132 | 0 | 0 | 9.00E-143 | 6.00E-172 | 2.00E-166 | 2.00E-166 | 2.00E-156 |
| hth | AT1G69890 | G0280473 | Tos8P | Cup9P | unc-62 | AGAP002178 | hth | TF protein | Meis1 | Meis2 | Meis3 | |
| % query cover | 13% | 13% | 17% | 13% | 63% | 100% | 49% | 78% | 95% | 95% | 58% | |
| % identity | 57% | 54% | 44% | 47% | 48% | 72% | 85% | 52% | 52% | 53% | 73% | |
| E value | 5.00E-17 | 5.00E-15 | 4.00E-15 | 4.00E-13 | 1.00E-49 | 0 | 2.00E-145 | 1.00E-109 | 6.00E-128 | 8.00E-127 | 4.00E-60 | |
| SG Morphogenesis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hkb | Zn finger C2H2 | Zn finger C2H2 | Azf1p | Crz1p | EGR homolog | AGAP004517 | Zn finger 623 | Zn fing.132 | Kr-like 10 | Sp5-like | TF SP5 | Other TF-SP | Kr-like TFs |
| % query cover | 32% | 32% | 27% | 37% | 28% | 100% | 100% | 30% | 57% | 49% | 37% | 28–41% | 26–42% |
| % identity | 40 % | 46% | 47% | 36% | 48% | 46% | 49% | 47% | 35% | 48% | 52% | 44–50% | 39–49% |
| E value | 5.00E-11 | 3.00E-14 | 2.00E-17 | 1.00E-16 | 4.00E-21 | 9.00E-76 | 5.00E-83 | 2.00E-22 | 3.00E-22 | 9.00E-22 | 4.00E-23 | E-21 - E-22 | E-21/E-22 |
| SG Morphogenesis, Maintenance of SG expression, Suppression of duct genes, SG cell survival, Secretome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| fkh | No clear orthologue | No clear orthologue | Hcm1p | Fkh2 | pha-4 | AGAP001671 | fkh | FoxA1-A | FOXA2 | FOXA1 | FOXA3 |
| % query cover | 17% | 15% | 39% | 89% | 91% | 28% | 24% | 36% | 22% | ||
| % identity | 49% | 46% | 51% | 65% | 64% | 72% | 85% | 65% | 81% | ||
| E value | 8.00E-18 | 8.00E-17 | 1.00E-58 | 2.00E-180 | 6.00E-179 | 1.00E-65 | 4.00E-75 | 1.00E-72 | 2.00E-63 | ||
| SG cell survival, Secretome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| sage | No clear orthologue | No clear orthologue | No clear orthologue | ndf-1 | hlh-6 | AGAP013335 | LOC5578695 | TF protein | Mesp2 | Ngn3 | Mesp1 |
| % query cover | 22% | 23% | 25% | 27% | 41% | 20% | 23% | 20% | |||
| % identity | 47% | 41% | 74% | 72% | 31% | 51% | 48% | 51% | |||
| E value | 3.00E0–8 | 6.00E-07 | 1.00E-29 | 7.00E-32 | 2.00E-05 | 5.00E-09 | 1.00E-08 | 1.00E-08 | |||
| SG cell survival, Secretome | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sens | Zn finger C2H2 | Zn finger C2H2 | Azf1p | F45B8.4 | AGAP010926 | sens | fez Zn finger 2-like | Gfi-1b | Gfi-1 |
| % query cover | 20% | 24% | 21% | 21% | 48% | 61% | 19% | 20% | 20% |
| % identity | 35% | 39% | 36% | 83% | 86% | 86% | 83% | 85% | 87% |
| E value | 6.00E-10 | 6.00E-19 | 4.00E-21 | 1.00E-64 | 9.00E-69 | 1.00E-69 | 1.00E-58 | 1.00E-63 | 7.00E-63 |
| Secretory capacity increases | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CrebA | bZIP TF | No clear orthologue | No clear orthologue | crh-2 | AGAP011038 | CrebA | TF protein | CREB3L1 | CREB3L2 | CREB3L3 | CREB3L4 | CREB3 |
| % query cover | 11% | 21% | 96% | 60% | 25% | 21% | 36% | 20% | 20% | 20% | ||
| % identity | 40% | 54% | 45% | 50% | 55% | 62% | 49% | 53% | 52% | 53% | ||
| E value | 1.00E-04 | 2.00E-29 | 4.00E-80 | 4.00E-62 | 4.00E-36 | 6.00E-34 | 5.00E-25 | 2.00E-25 | 5.00E-26 | 2.00E-22 | ||
The Drosophila SG—an historical overview of its research utility
Drosophila larval SGs are famous for their giant polytene chromosomes, which have undergone up to 10 rounds of division without physical separation (Li and Chen, 2015). Polytene chromosomes are the functional equivalent of interphase chromosomes with the activity state of different chromosomal regions reflected in intense dark bands, lighter “gray” bands and interbands (Kolesnikova, 2018; Zykova et al., 2018). The highly condensed chromosomal bands—visible with a range of DNA-binding chromophores—correspond to inactive, late-replicating “interstitial heterochromatic” regions harboring genes that are not expressed in SG cells (Zykova et al., 2018). The open, unstained interbands correspond to the regions of the transcription start sites of housekeeping genes with the lighter gray bands corresponding to their downstream protein coding regions (Demakova et al., 2020). The stereotypical banding patterns of SG polytene chromosomes have been extensively utilized to map chromosomal rearrangements, including deletions, duplications and translocations. As molecular tools were developed, DNA was directly mapped to specific chromosome positions by in situ hybridization (Schmidt and Kubli, 1980; Vaslet et al., 1980). In situ hybridization on chromosomal aberrations disrupting gene function expedited cloning in the 1970s through the 1990s (Baker et al., 1991; Scott et al., 1983). Over the past 30+ years, SG polytene chromosomes have been used to localize a variety of chromatin factors in vivo, including RNA polymerases, epigenetic modifiers, dosage compensation proteins, transcription factors, and even chromosome-specific RNAs, providing a genome-wide overview of chromosome organization and associations among chromatin factors (Boros, 2012; Fox et al., 2010; Franke and Baker, 1999; Jamrich et al., 1977; Kabisch and Bautz, 1983; Kuroda et al., 2016; Zink and Paro, 1989).
Polyteny is not limited to the SG, and the degree of polytenization is linked to cell size and function (Hochstrasser, 1987; Lamb, 1982; Richards, 1980; Rudkin, 1972). Indeed, all Drosophila larval tissues typically cease normal mitotic divisions and resort to polytenization (Smith and Orr-Weaver, 1991). SG cells, however, undergo the highest level of polytenization in keeping with their role as the major secretory organ in the larva (Rudkin, 1972). The increased number of templates for transcription provided by polytenization, combined with increased rates of translation, and amplification of the secretory machinery (Abrams and Andrew, 2005; Fox et al., 2010; Johnson et al., 2020), allow these professional secretory cells to produce and secrete sufficient levels of the relevant proteins for the rapidly growing organism to survive and advance through subsequent life stages.
The utility of the Drosophila SG for scientific discovery goes well beyond its use in gene cloning and studies of chromatin factors. Over the past 30 years, the SG has become a premier model organ for understanding how cell types are initially specified in the early embryo and for how genetic programs controlling cell type specialization are organized and deployed. Indeed, studies of the SG have revealed how HOX proteins specify a wide range of distinct cell fates within their domains of expression (Andrew et al., 1994; Henderson et al., 1999; Panzer et al., 1992). The SG system has revealed an ancient and highly conserved family of TFs, the CrebA/Creb3-family (see below), that coordinately upregulates the proteins of the entire secretory machinery (Abrams and Andrew, 2005; Fox et al., 2010; Johnson et al., 2020). SG studies have also highlighted the capacity for TFs to multitask by exposing the roles for a single, highly conserved factor, Fork head (see below), in controlling cell fates within the organ, in cell survival, in chromosome polytenization, and in activation of its secretome (Fox et al., 2013; Haberman et al., 2003; Maruyama et al., 2011; Myat and Andrew, 2000a). Recent studies have linked this same TF to the coordinated cytoskeletal events required to transform the SG placodes into three-dimensional elongated tubes (Chung et al., 2017; Roper, 2012). High-resolution live imaging of late larval SGs has uncovered additional roles for the cytoskeleton in high-level bulk secretion at critical life stages (Rousso et al., 2016; Tran et al., 2015). Indeed, this remarkable insect organ has revealed secrets of secretion relevant to all higher animals.
Salivary gland specification
The first critical step in parsing out functions within a developing organ is to specify the fates of cells that carry out those functions (Fig. 1A). With the SG, and indeed all other organs in the Drosophila embryo, this process involves integration of patterning information along the anterior-posterior (AP) axis with that along the dorsal-ventral (DV) axis (Haberman et al., 2003; Henderson et al., 1999; Panzer et al., 1992). The organization—prior to the onset of organogenesis—of nearly all cells in a surface monolayer on the Drosophila embryo makes specification of cell fate a two-dimensional (2D) problem, which is solved by each cell expressing and/or being exposed to a unique combination of factors along the two major body axes corresponding to the position of that cell within this “2D” space.
Figure 1. Specification and development of SG cell types.
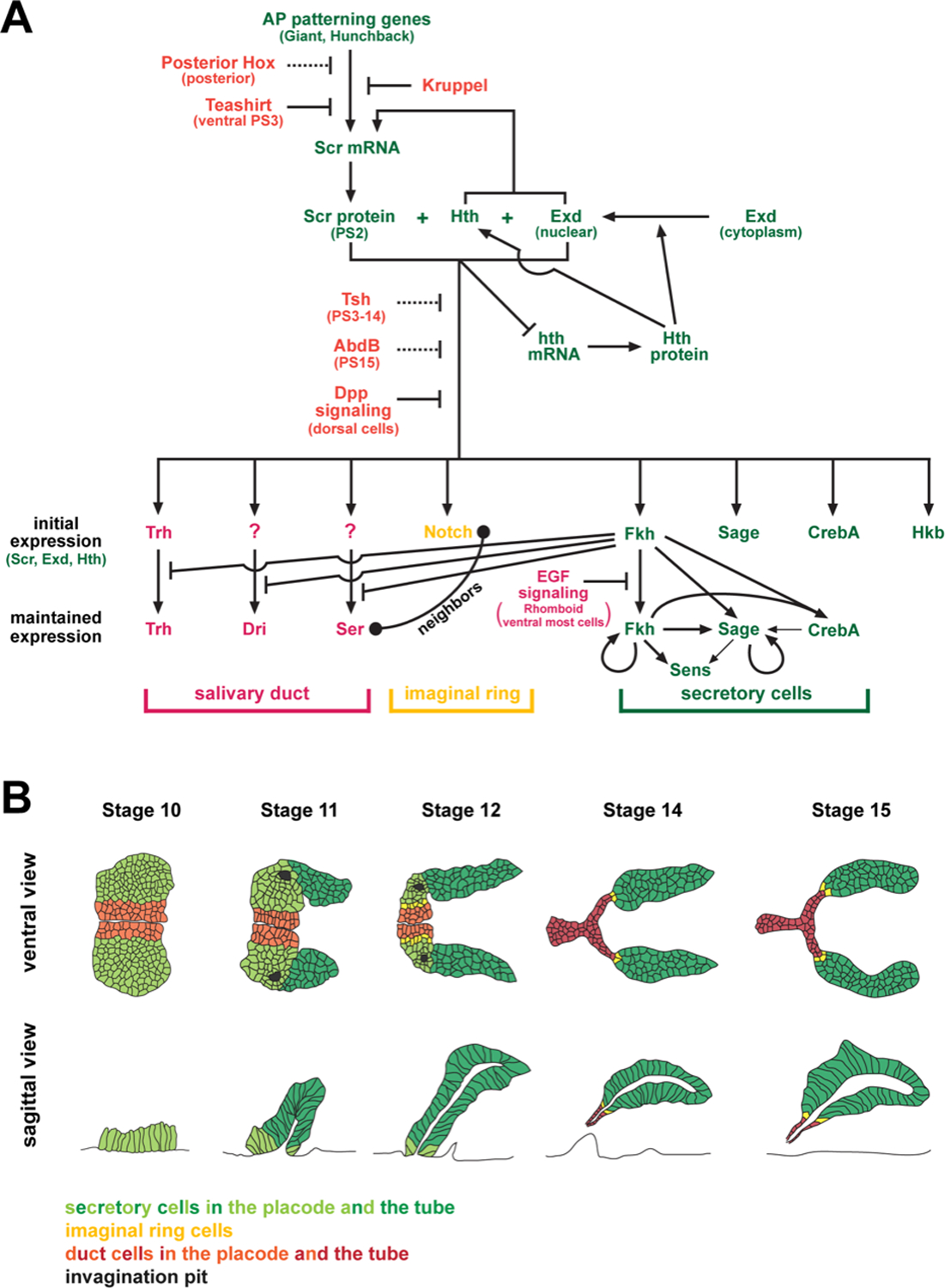
(A) Gene network involved in specifying SGs to form in the ventral domains of PS2 and in the activation and maintenance of key TFs and signaling pathways involved in cell fate choices within the SG and in maintaining those cell fates. Green indicates factors involved in positive regulation of SGs and genes expressed in secretory cells. Red indicates factors involved in negative regulation of SGs or expressed in salivary duct cells. Notch (yellow) signaling is required to specify imaginal ring cells. (B) Secretory, duct and imaginal ring cells of the SG arise from different dorsal ventral positions within the primordia. The more dorsal cells form the secretory tubes (green cells). The most ventral cells form the salivary duct (red cells). The cells in between become the imaginal ring (yellow cells).
Sex combs reduced specifies salivary glands
The highly conserved and well-characterized Hox transcription factors are expressed in limited domains along the AP axis of the embryo, and it is the Hox protein Sex combs reduced (Scr) that specifies SGs (Fig. 1A). Scr, the single fly orthologue of the HoxA5, HoxB5 and HoxC5 genes (Table 1), is initially expressed throughout parasegment 2 (PS2)— the “segment” in which SGs normally form (Panzer et al., 1992; Riley et al., 1987). At later stages, Scr is also expressed in PS3, but expression there is confined to only dorsal cells (Riley et al., 1987). In the absence of Scr function, SG-specific genes are not expressed and SGs fail to form (Andrew et al., 1994; Panzer et al., 1992). When Scr is expressed everywhere, cells outside of PS2 express a variety of SG genes (Andrew et al., 1994; Panzer et al., 1992) and cells in more anterior segments—PS0 and PS1—not only express all tested SG markers, but also form internalized SG tubes (Andrew et al., 1994).
Cofactors are required for the induction of salivary gland cell fate by Scr
Two more broadly-expressed and highly conserved Hox-domain containing TFs, Extradenticle (Exd)—the single fly version of the four Pbx proteins in mammals—and Homothorax (Hth)—the single fly version of the three Meis proteins in mammals (Fig. 1A; Table 1)—are also required to form SGs (Henderson and Andrew, 2000). With loss of either gene, SGs are absent. Exd and Hth form a complex with Scr, or with other AP Hox proteins, to provide specificity for target site recognition and DNA binding (Joshi et al., 2010; Lelli et al., 2011; Ryoo and Mann, 1999). Interestingly, with the loss of exd or hth, expression of Scr disappears prematurely, but only in the SG precursor cells of PS2 (Henderson and Andrew, 2000). Exd and Hth, thus, allow Scr expression to persist through the initiation of SG cell fate. Tight temporal control of Hox gene regulation limits their role to early stages of SG formation. Hence, as SGs invaginate, Scr prevents Hth-dependent nuclear localization of Exd by negatively regulating hth expression (Ryoo and Mann, 1999). This autoregulatory feedback loop, therefore, allows Scr to limit its own expression through only the early stages of SG formation. The disappearance of Scr, Hth and Exd from the SG as tube morphogenesis ensues indicates that although these proteins are essential for SG fates, they neither maintain nor directly implement this cell fate (Henderson and Andrew, 2000).
Posterior factors block SG specification
The limited formation of additional SGs in only PS0 and PS1 with global expression of Scr suggests that either other factors are also required or that negative factors block Scr induction of SG cell fates. Indeed, the zinc finger transcription factor Teashirt (Tsh) (Fig. 1A) prevents Scr from inducing SGs in the thorax and most abdominal segments (Andrew et al., 1994). Ubiquitous expression of Scr in tsh null mutants results in the formation of SGs in every segment from PS0 through PS14. Tsh functions at two levels to block SG formation: (i) Tsh prevents Scr from inducing SG fates in the trunk, and (ii) it blocks expression of Scr in the ventral cells of PS3. Other Hox genes also prevent Scr from inducing SG fates (Andrew et al., 1994). The Hox protein Abdominal B (AbdB) (Fig. 1A) blocks SG formation in the last segment of the embryo (PS15), where Tsh is not expressed (Mathies et al., 1994). Global expression of the Hox proteins Antennapedia (Antp) or Ultrabithorax (Ubx) (Fig. 1A), moreover, completely blocks SG formation, even in PS2 (Andrew et al., 1994). However, when either Antp or Ubx are expressed globally along with Scr, SGs form in PS2, and in PS0 and PS1. Indeed, Antp and Ubx block SG formation by preventing Scr expression. Together, these results reveal the requirement for an exquisite balance between both positive and negative regulation of gene expression by the Hox factors for SG specification.
Dpp signaling limits SG formation to ventral cells
SG formation is limited to the ventral cells of PS2, and ventral cells of other segments when Scr is expressed ectopically. Loss of the TGF-β pathway ligand ,Dpp (Fig. 1A), or of any of its downstream signaling components—receptors, effectors, or nuclear factors—results in most or all PS2 cells becoming SG (Henderson et al., 1999; Panzer et al., 1992). Correspondingly, expression of Dpp throughout the embryo blocks SG formation. Dpp not only disrupts the SG-inducing capabilities of Scr, but also stabilizes the latter’s expression. In the dorsal cells of PS2 and PS3, Scr transcript and protein levels are higher and persist much longer than in the SG precursors (Henderson et al., 1999). This finding suggests that Scr complexes with the TFs downstream of Dpp to both maintain its own expression and to redirect its activity for specification of dorsal cell fates.
Cell fate within the SG
Scr is required for all SG-specific cell types, including the large polytene secretory cells, the smaller, less polytenized, duct cells, and the diploid precursors to the adult SG—the “imaginal ring” cells (Fig. 1B). Precursors to each subtype derive from different spatial domains within SG-forming cells of PS2. The more laterally positioned ~144 cells become secretory, the most ventral ~50 cells become duct, and the ~10–12 cells in between become imaginal ring. The sequential deployment of two signaling pathways controls these cell fates. First, the most ventral cells of the SG primordia—in which the EGF ligand is processed and released by the Rhomboid protease—experience EGF signaling, and become duct (Kuo et al., 1996). As a consequence, key secretory-specific TFs are shut off in duct cells (Fig. 1A), most notably Fork head (Fkh)—the single Drosophila orthologue of the three vertebrate FoxA proteins (Table 1). In the non-duct cells, Fkh represses expression of a subset of genes, including trachealess (trh), dead ringer (dri) and Serrate (Ser), limiting their expression to the salivary duct (Haberman et al., 2003). Ser is a ligand for the receptor protein Notch, which is transiently upregulated in the remaining non-duct cells (Kidd et al., 1989). Salivary duct-expressed Ser then signals through Notch to its immediate neighbors to specify the imaginal ring cells, leaving the remaining, more lateral cells to become secretory (Haberman et al., 2003). The relative arrangement of the major cell types within the primordia is preserved in the differentiated SG tube: the laterally-positioned secretory cells populate the larger distal secretory tube, the duct cells populate the most proximal portions, and imaginal cells form a ring of cells separating the secretory cells from duct cells (Fig.1B).
Salivary gland fate maintenance and morphogenesis
Building in memory: Maintenance of cell fates
Since expression of the genes that specify the SG fate disappears shortly after SG development ensues, other factors must both “remember” and “carry out” the fate decision. Scr and its cofactors activate a small set of immediate early TFs (Fig. 1A; Table 1): The single Drosophila member of the winged-helix FoxA family protein—Fkh; the single orthologue to the five Creb3/3-like basic leucine zipper proteins of mammals—Cyclic-AMP response element binding protein A (CrebA); a zinc-finger protein related to a large family of SP-like/Kr-family TFs—Huckebein (Hkb); and a bHLH TF related to several mammalian TFs—Salivary gland-expressed bHLH (Sage). The genes encoding these proteins are likely directly activated by Scr/Exd/Hth (Abrams and Andrew, 2005; Abrams et al., 2006; Ryoo and Mann, 1999). Hkb (Fig. 1A) is only transiently expressed in the early SG, and is thus unlikely to maintain the SG fate decision (Myat and Andrew, 2000b, 2002). Fkh, however, maintains its own expression (Zhou et al., 2001) along with the expression of CrebA and Sage (Abrams and Andrew, 2005; Abrams et al., 2006), identifying it as a key component in cell fate maintenance. Together, Fkh and Sage activate expression of the C2H2-type zinc finger-containing TF Senseless (Sens) (Fox et al., 2013). Fkh, CrebA, Sage, and Sens continue to be expressed in the SG until the organ undergoes histolysis during pupation, when the larval SG is replaced with its adult equivalent (Fox et al., 2013). Imaginal ring cells actively divide in the late larval and early pupal stages and form the adult SG, which also expresses Fkh, CrebA, Sage, and Sens to very high levels [Flybase]. As will be discussed in detail below, CrebA increases SG cell secretory capacity, while Fkh along with Sage and Sens ensure cell survival and control the expression of SG-specific gene products (the secretome).
Combating death: Keeping the non-dividing SG cells alive until no longer needed
Like most tissues, the SG ceases normal mitotic divisions and begins polytenization almost as soon as it is specified (Smith and Orr-Weaver, 1991). Non-dividing cells must actively block cell death (O’Neill, 1991). In the SG, this task is one of many under the control of Fkh. In embryos lacking fkh, the SG undergoes massive apoptotic death at very early stages, prior to invagination (Myat and Andrew, 2000a). fkh mutant SG cells highly express the pro-apoptotic genes reaper (rpr) and head involution defective (hid) and exhibit features of programmed cell death (Myat and Andrew, 2000a). Correspondingly, deletion of the pro-apoptotic regulators in a fkh mutant rescues SG cells from death (Myat and Andrew, 2000a). Loss of either sage or sens also results in increased expression of rpr and hid resulting in massive apoptotic SG cell death, but at later developmental stages (Chandrasekaran and Beckendorf, 2003; Fox et al., 2013). This finding suggests that Fkh, Sage and Sens each function to keep death at bay. The dependence of Sage and Sens on Fkh function for their maintained expression means that loss of fkh results in the loss of all three proteins, whereas loss of sage has only minor effects on sens expression, and loss of sens has no effect on either Fkh or Sens (Fox et al., 2013). The relative effects of the loss of each of the three proteins and the consequent up-regulation of pro-apoptotic genes thus determine when death occurs. Importantly, this is the mechanism by which larval SG cells undergo histolysis during early pupation; Fkh expression is shut off in the early pupal SG by the hormones that control metamorphosis (Renault et al., 2001). RNAi induced loss of Fkh expression at earlier stages, moreover, results in the immediate demise of larval SG cells (Cao et al., 2007; Liu and Lehmann, 2008) and the continued forced expression of Fkh can forestall SG death (Liu and Lehmann, 2008).
Infusing form: Fkh and spatial transformations
fkh mutant SGs rescued from death by inactivation of the pro-apoptotic genes fail to internalize and form tubes, remaining on the surface through all of embryogenesis (Myat and Andrew, 2000a). fkh mutant SGs fail to undergo the coordinated apical constriction that has been shown to accompany invagination of a significant subset of secretory cells (Booth et al., 2014; Chung et al., 2017; Myat and Andrew, 2000a; Sanchez-Corrales et al., 2018) and they fail to form a supra-cellular myosin cable that surrounds the entire SG primordia and is thought to provide compressive forces to help fuel invagination (Chung et al., 2017; Roper, 2012; Sidor et al., 2020). To fully understand how the Fkh TF controls morphogenesis, genome-wide expression analyses were done to identify downstream targets, including both in situ- and microarray-based screens (Maruyama et al., 2011). Although the microarray-based screen was less successful in identifying SG targets, likely due to the broad expression of fkh in embryos and the relatively small number of cells in the SG, it did, however, lead to the discovery that Fkh is required for polytenization of SG chromosomes and the chromosomes of other larval tissues (Maruyama et al., 2011).
To date, two Fkh early-expressed transcriptional targets have been implicated in SG internalization (Fig. 2A,B): one encodes the secreted ligand Fog, which also functions in the invagination of the foregut, hindgut and mesoderm, and the other encodes the transmembrane protein Crumbs (Crb), best known for its role in specifying, maintaining, and expanding the apical domain of epithelial cells in a variety of organisms and tissue-specific contexts (Chung et al., 2017). In the mesoderm, Fog activates the G-protein coupled receptor (GPCR) known as Mist, which acts through the Galpha protein Concertina (Cta) to activate RhoGEF2 (reviewed by (Manning and Rogers, 2014). From RhoGEF2, the activation cascade involves the GTPase Rho1 and the Rho kinase (Rok), of which the latter phosphorylates and activates an apical medial pool of non-muscle myosin II (NMII) to drive apical constriction. The activities of some, but not all, of the same mesodermal components function to drive coordinated apical constriction in the SG (Fig 2A), including Fog (Chung et al., 2017), RhoGEF2 (Nikolaidou and Barrett, 2004), Rho1 (Xu et al., 2008), Rok, and NMII (Chung et al., 2017). The receptor Mist is not expressed in the SG; thus, it is likely that a related SG-expressed GPCR functions in this capacity (Chung et al., 2017). Likewise, the Galpha protein that activates RhoGEF2 in the SG is yet to be discovered. Localization of these Fog-dependent signaling events to the apical medial domain of SG cells is likely due to the apical secretion of Fog and the (likely) apical localization of its putative receptor. The putative localization of two RhoGAPs to the basal surface may also contribute (Kolesnikov and Beckendorf, 2007). Indeed, polarized vesicle trafficking is a key cellular event underlying downstream cytoskeletal changes; disruption of the microtubule network in early SGs results in a failure to accumulate the apical-medial pools of NMII driving apical constriction (Booth et al., 2014). The SG TF Hkb mediates apical trafficking of secretory vesicles along microtubules through its downstream target Klarsicht (Klar) (Myat and Andrew, 2002) and could target Fog and possibly its SG receptor to the apical domain (Fig 2A). hkb loss-of-function and overexpression phenotypes are consistent with a role in determining where in the primordia apical constriction occurs and invagination begins, as well as in expanding the apical surface post-invagination (Myat and Andrew, 2000b, 2002). Notably, the localized dynamic expression of Hkb within the SG primordia presages apical constriction (Myat and Andrew, 2002).
Figure 2. Fkh mediated morphogenetic events in SG tube formation and positioning.
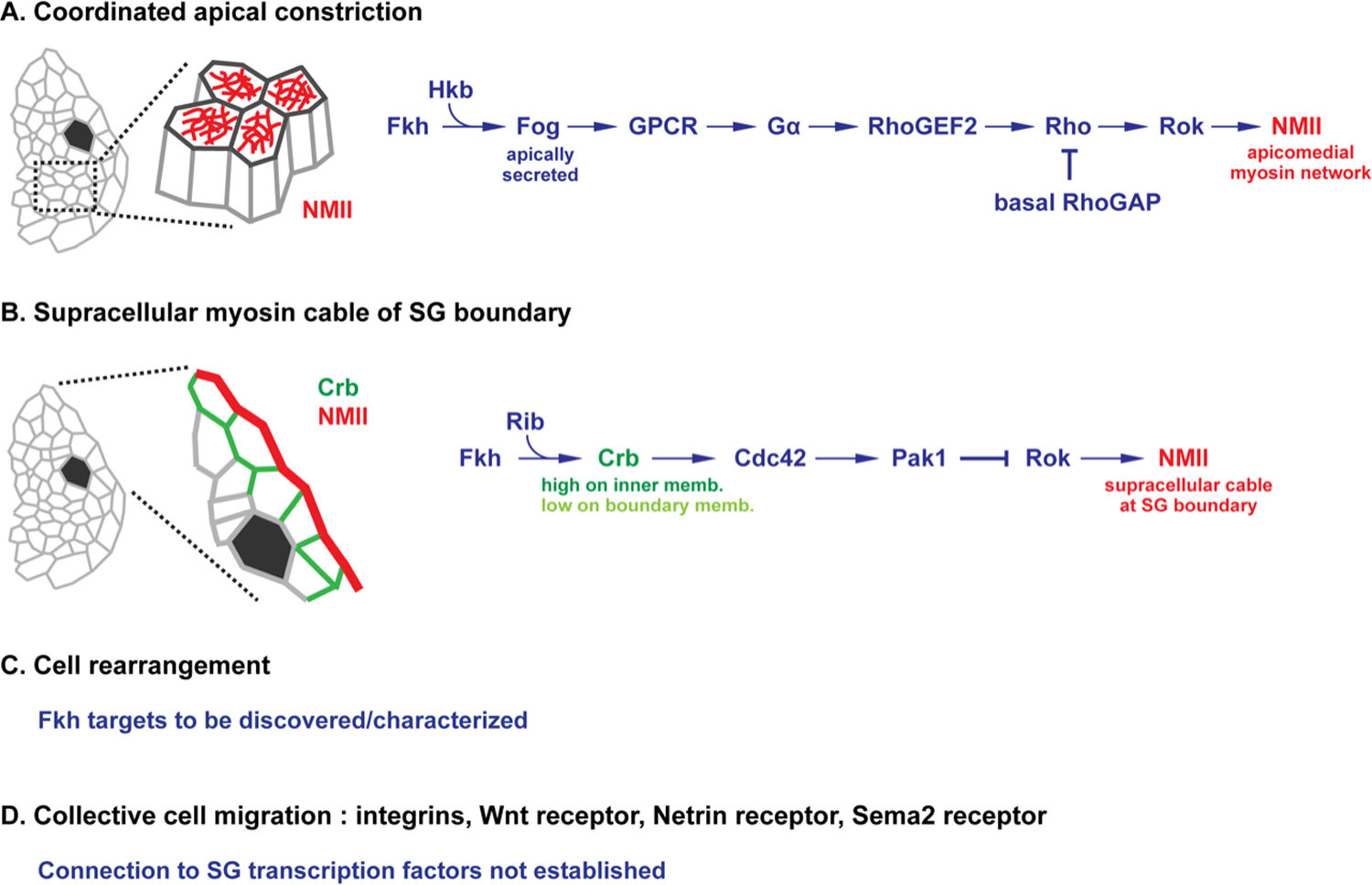
(A) Fkh activates expression of Fog to initiate the molecular/cellular events of apical constriction. Hkb is proposed to localize Fog secretion (and potentially the localization of its yet-to-be characterized GPCR receptor) to the apical surface. Basally-localized RhoGAPs may also help localize Rho activation to the apical domain. (B) Fkh, along with Rib, transcriptionally elevate Crb expression within the SG primordia. The high-levels of transmembrane Crb in membranes shared between SG cells versus membranes between SG cells and their non-SG neighbors preferentially localizes active Rok to the external boundary membrane of the SG, leading to the formation of a supracellular myosin cable surrounding the entire SG. (C) Based on the severity of SG morphological phenotypes associated with loss of fkh versus loss of Crb or of fog, it is proposed that Fkh is also involved in cell rearrangement. (D) Collective migration is required for the correct final position of the SG within the embryo. A number of signaling pathways are required for migration and navigation of the SG. How these pathways fit into the transcriptional network controlling morphogenesis awaits discovery.
Fkh, along with another more broadly expressed TF known as Ribbon (Rib), increases Crb expression in the SG to levels much higher than in the surrounding, non-SG epithelial cells (Chung et al., 2017; Kerman et al., 2008). In the SG placode, high Crb levels mediate intercellular homotypic binding of Crb in neighboring SG cells through its extracellular domains (Fig 2B; (Roper, 2012)). As a consequence of the relatively lower levels of Crb in the surrounding non-SG epithelia, homotypic Crb binding is significantly lower in boundary membranes between SG cells and non-SG cells (Roper, 2012). High-level Crb through CDC42 recruits the kinase Pak1 to the inner cell membranes (Sidor et al., 2020). In turn, Pak1 phosphorylates Rok to increase the latter’s dissociation rate from the membrane. Since on-rates of Rok are proposed to be unaffected by Pak1-dependent phosphorylation, this leads to increased Rok at boundary membranes. In turn, Rok phosphorylates and activates NMII, ultimately leading to the formation of a supra-cellular Myosin cable around the boundary of the entire SG primordia (Sidor et al., 2020). Laser ablation experiments suggest that this Myosin cable is under increased tension, and, thus, could generate centripetal force to fuel invagination (Roper, 2012).
Another process concomitant with SG invagination is cell rearrangement (Fig. 2C). Indeed, it has been proposed as a major mechanism driving internalization of the SG cells that do not undergo apical constriction (Sanchez-Corrales et al., 2018). Since SGs still internalize in both fog and in crb mutants (Wodarz et al., 1993) and can internalize when apical constriction is blocked (Chung et al., 2017), the complete failure of SG cells to internalize in fkh mutants could be the consequent failure to activate multiple independent cellular events: Fog-dependent apical constriction, Crb-dependent centripetal force provided by the Myosin cable encircling the SG primordia, and the—as yet undescribed—Fkh-dependent SG factors that mediate cell rearrangement. Future studies are expected to reveal both missing pathway components and additional relevant Fkh targets, allowing this SG morphogenetic model to be tested directly.
To arrive at their final correct position in the embryo, SGs undergo collective cell migration, remaining fully polarized epithelia as the nascent tubes actively migrate on surrounding mesodermal tissues. This integrin-dependent process relies on signals emanating from a variety of tissues for navigation, including the CNS, fat body, and visceral mesoderm (Bradley et al., 2003; Harris and Beckendorf, 2007; Harris et al., 2007; Kolesnikov and Beckendorf, 2005; Vining et al., 2005). The transcriptional links between Fkh and the signaling components of navigation remain undiscovered (Fig 2D).
Salivary Gland Functional Specialization
What to produce? Sage and the regulation of the secretome
Cargo production in the SG is directed by multiple TFs acting in concert—Fkh, Sage, Sens and, to a lesser extent, CrebA (Fox et al., 2013; Johnson et al., 2020) (Fig. 3A). Cooperative or combinatorial regulation of cargo by these TFs occurs with a major contribution from Sage and Fkh. Sage and Fkh activate expression of genes encoding both transmembrane and SG-secreted cargo proteins and their modifying enzymes (Fox et al., 2013). Indeed, nearly 75% of the genes differentially expressed due to changes in sage expression code for proteins that traverse the secretory pathway or proteins that function within secretory organelles to modify cargo. The majority of genes down-regulated upon loss of sage encode secretory proteins; among them are secreted glutamate/aspartate-rich proteins, a tripartite gene cluster that encodes related mucins, and two SG-specific prolyl-4α-hydroxylases (Abrams et al., 2006; Fox et al., 2013). All transcriptional targets of Sage require Fkh for their SG expression. The combined activities of Sage and Fkh are, moreover, not only necessary but also sufficient for the transcriptional activation of secreted and transmembrane cargo genes (Fox et al., 2013). Additional layers of SG-cargo regulation are provided by Sens and CrebA. Sens, the loss of which resembles the loss of Sage with regard to SG phenotypes (Chandrasekaran and Beckendorf, 2003; Fox et al., 2013), appears to boost the expression of a subset of Fkh- and Sage-regulated SG cargo genes (Fox et al., 2013). Sens also colocalizes with Fkh and Sage on shared loci of the SG polytene chromosomes. CrebA, in addition to its primary role in the regulation of SG secretory capacity (see below), increases expression of SG cargo genes in two ways, by directly binding them for transcriptional up-regulation and/or by boosting the expression of sage (Johnson et al., 2020). Adding yet another layer to the regulatory architecture of SG cargos is the transcriptional control by CrebA of the gene encoding Tudor staphylococcal nuclease (Tudor-SN), which likely stabilizes the ER-associated mRNAs encoding secreted cargo proteins (Johnson et al., 2020).
Figure 3. Transcriptional regulation of secretory capacity and the secretome.
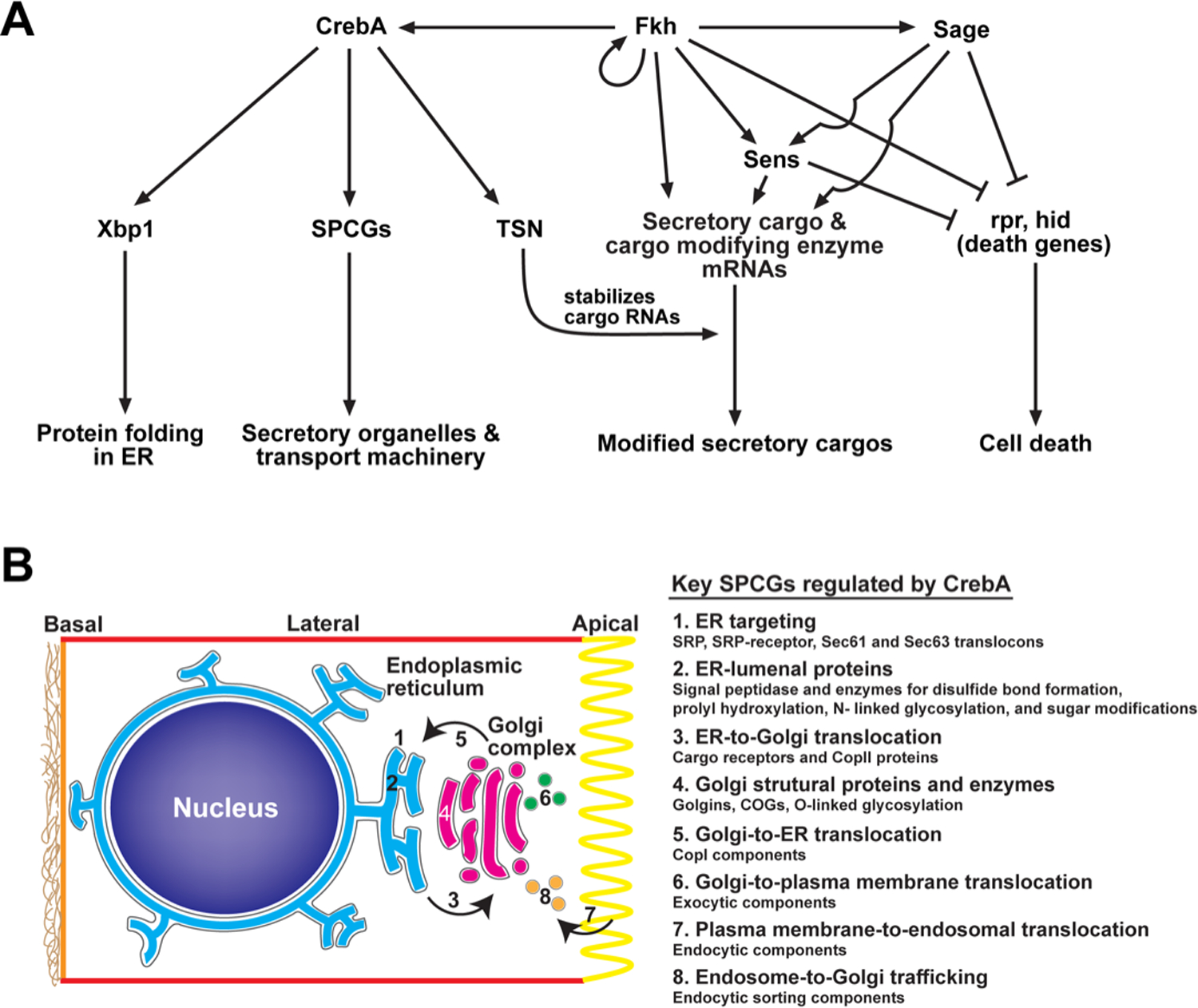
(A) CrebA increases secretory capacity by direct binding and activation of core secretory pathway component genes (SPCGs), Xbp1, and TSN. Fkh works with Sage and Sens to control what is made in and secreted from the SG (cargo) and how cargo is modified as it goes through the secretory pathway. Fkh, Sage and Sens also block apoptotic cell death by blocking expression of pro-apoptotic genes rpr and hid. B. Cartoon showing the organelles of the secretory system and what secretory processes are transcriptionally regulated by CrebA.
Since Fkh expression in the embryo is not confined to the SG, unlike its target (Sage), it is likely that Fkh gains SG cell specificity through its cooperation with Sage. Hence, the regulation of secretory cargo genes by Sage is fundamental to its role as a SG-cell specificity factor. As mentioned earlier, Fkh, Sage, and Sens are also critical for SG cell survival (Fig 3A). The collaboration between a FoxA protein (Fkh) and a tissue-specific bHLH protein (Sage) may be a conserved/generic paradigm for achieving cell specificity and survival by FoxA proteins, e.g., Neurogenin & FoxA1 and Neurogenin & FoxA2 in dopaminergic neurons (Kele et al., 2006; Lin et al., 2009), Ptf1& FoxA proteins in the pancreas (Gao et al., 2010; Krapp et al., 1996; Krapp et al., 1998), and HLH-6 & PHA-4 proteins in the C. elegans pharynx (Gaudet and Mango, 2002; Smit et al., 2008).
Fkh and Sage function, which had been observed in the Drosophila SG, is likely conserved in the SG- (organ) homologs of other insect orders (Table 1). The Fkh counterpart (Silk Gland Factor 1, SGF-1) has long been known to bind and regulate expression of silk protein encoding genes in the silk gland cells of the lepidopteran Bombyx mori (Julien et al., 2002; Mach et al., 1995). More recently, BmSage (Sage), in direct association with SGF-1, has been shown to bind and activate expression of the Fibroin heavy chain cargo—an essential ingredient for silk production (Zhao et al., 2014). Sage expression levels are, moreover, directly proportional to silk yields, with the high-yield strains showing higher levels of sage transcripts compared to the transcript levels in the low-yield strain, thus, indicating the primacy of Sage for cargo regulation. Similar to the Drosophila sage loss-of-function phenotype, which results in secretory defects concomitant with SG morphological defects during the early and intermediate stages of gland development in conjunction with secretory cell loss during late stages (Fox et al., 2013), Bmsage loss-of-function also leads to secretion deficiency concomitant with remarkable silk gland morphological defects and the loss of middle and posterior compartments of the gland (Xin et al., 2015). Taken together, these results suggest the conserved functional collaboration between Sage and Fkh across insect species in the regulation of SG cargo and secretory cell survival.
A question of scale: Boosting secretory capacity by CrebA
Organogenetic processes commence with cell specification and culminate with cell specialization. Functional specialization of the SG is evident not only in the ability of its constituent cells to produce appropriately modified tissue-specific cargo, but also in the remarkable increase of their secretory capacity in a process regulated by CrebA (Fig. 3), which is the single Creb3-family member in Drosophila (Table 1). CrebA is expressed in several embryonic, larval, and adult organs, with highest levels in the embryonic and larval SGs (Smolik et al., 1992). High expression of proteins required for ER-targeting and translocation, and of those mediating ER-Golgi transport is also observed in the embryonic SG; hence, in the search for early-expressed TFs involved in activating genes crucial for SG secretory capacity, CrebA emerged as the most likely candidate for their regulation (Abrams and Andrew, 2005). Expression of all early secretory pathway component genes (SPCGs) is lost in both fkh and CrebA mutant embryonic SGs. However, SPCG expression is lost in only late fkh mutants, whereas its loss occurs in both early and late CrebA mutants (Abrams and Andrew, 2005). Since Fkh is required to maintain the SG CrebA expression following the latter’s initiation by Scr (and its cofactors), the effects of Fkh on secretory capacity are likely to be entirely through CrebA, with the latter functioning as the direct regulator of SPCGs in the embryonic SG.
A more direct confirmation of CrebA as a major transcriptional regulator of SPCGs was obtained with the discovery that nearly 400 genes—a third of which were annotated as SPCGs—were downregulated in CrebA mutant embryos (Fox et al., 2010). Among these SPCG-targets of CrebA (Fig. 3B) are genes encoding the components of signal recognition particle and its receptor; Sec61 translocon complex; Sec63 complex; ER lumenal HSP70s; ER morphology proteins (e.g., Atlastin); signal peptide complex; factors implicated in N-linked glycosylation, disulfide bond formation, prolyl hydroxylation, sugar trimming and protein folding; ER cargo receptors; COPII components and their regulators; Golgi structural proteins (e.g., Grasp65); factors implicated in O-linked glycosylation; COPI components and factors involved in early Golgi retrograde traffic; Arf-1 GAPs; and, factors involved in ER retrieval from Golgi (e.g., KDEL-R). CrebA, moreover, is not only necessary, but is also sufficient for SPCG expression (Fox et al., 2010). More recent studies reveal that CrebA binds the enhancers of nearly every gene encoding a known component of the secretory machinery (Johnson et al., 2020). Thus, CrebA emerges as the singular Drosophila factor to mediate the augmented demands of increased secretory capacity required of the cells specialized for secretion and biosynthetic trafficking to support processes as diverse as membrane growth, dendritic arborization, and extracellular matrix (ECM) secretion (Rodrigues and Harris, 2019).
CrebA and the unfolded protein response (UPR)
Secretory cell specialization, i.e., functional priming, is inherently associated with high levels of protein folding and modification in the combined ER-Golgi machineries, consequently, stressing these organelles. This physiological-yet-undue stress on the system triggers a buffering mechanism by activating the UPR to restore ER homeostasis (Hetz, 2012). Three branches of the UPR are implicated in its regulation, and are initiated by the following mammalian stress sensors, all of which also have Drosophila orthologues: (i) stress sensors protein kinase RNA-like ER kinase (PERK), (ii) inositol-requiring enzyme 1α (IRE1α), and (iii) activating transcription factor 6 (ATF6). The UPR occurs as a biphasic process, the first phase recruiting the PERK-mediated pathway to curb protein influx into the ER by attenuating translation, i.e., the relaxation phase (Harding et al., 2000). The second phase utilizes both the IRE1α- and ATF6-mediated pathways, and triggers the expression of ERAD (ER-associated protein degradation) genes, e.g., X-box binding protein-1 (Xbp1), to restore ER function, i.e., the recovery phase.
The ER-membrane resident IRE1α is a kinase and endoribonuclease that, upon ER stress, dimerizes and autophosphorylates—triggering its RNase activity (cytosolic face)—to process the unspliced, inactive Xbp1. Spliced Xbp1 is an active TF that regulates the expression of proteins involved in folding, ERAD, protein quality control, and phospholipid synthesis (Acosta-Alvear et al., 2007; Sriburi et al., 2004; Symoens et al., 2013). Creb3/3L proteins, the orthologues of CrebA (Table 1; see below), and Xbp1, moreover, often collaborate to increase secretory capacity, and interestingly, share a consensus DNA-binding motif (Khetchoumian et al., 2019; Tanegashima et al., 2009). Indeed, active spliced Xbp1 is present in the Drosophila embryonic SG, where CrebA directly binds and activates its expression (Fig. 3A) (Johnson et al., 2020). Loss-of-function xbp1 phenotypes in the embryonic gland are relatively mild compared to loss of CrebA (Johnson et al., 2020), however, suggesting that Xbp1 supplements CrebA function only during peak secretory demands, or that Xbp1 plays a more significant role at later stages, consistent with its very high levels of expression in the larval, pupal and adult SG [Flybase].
CrebA orthologues—a window into secretory dynamics and the UPR
The investigation of CrebA’s role in SG development has offered profound insights into the mechanisms underlying the coordinate upregulation of SPCG expression in cells specialized for secretion in more complex organisms. All five mammalian orthologs (Table 1) of CrebA (Creb3/Luman, Creb3L1/OASIS, Creb3L2/BBF2H7, Creb3L3/CrebH, and Creb3L4/Creb4), contain a transmembrane domain and undergo regulated intramembrane proteolysis in the Golgi to release their cytosolic N-terminal domain for nuclear translocation and subsequent transcriptional regulation (Bailey et al., 2007). CrebA, unlike its mammalian orthologues, lacks a transmembrane domain and is constitutively nuclear [Fox et al., 2010]. Like all mammalian Creb3-family members, however, CrebA features the ATB (Adjacent To BZip) domain, a characteristic chain of ~30 amino acids adjacent to the highly conserved bZip DNA binding domain that distinguishes this class of bZIP TFs from its closest cousins (Barbosa et al., 2013). Studies conducted in myriad mammalian secretory systems that show tissue-specific expression of the Creb3-family members and case reports of humans with deficiency of Creb3-family members have also confirmed the conserved, core function of Creb3-family members as key regulators of SPCGs (Khetchoumian et al., 2019; Reiling et al., 2013; Sampieri et al., 2019).
Creb3/Luman is highly expressed in the liver and in the nervous system along with detectable expression in several other tissues (Ying et al., 2015). It is also implicated in the maturation of dendritic cells—a professional antigen-presenting cell-type of the myeloid lineage that primes the immune response (Sanecka et al., 2012). The functional maturation of dendritic cells occurs, in a remarkable parallel to the CrebA-directed specialization of SG secretory cells, via the Creb3-mediated upregulation of SPCGs (Eleveld-Trancikova et al., 2010).
Creb3L1/OASIS is expressed in several secretory tissues, i.e., astrocytes, skeletal tissues, salivary glands, intestine, prostate gland, and pancreas (Murakami et al., 2009; Nikaido et al., 2001; Omori et al., 2002). In osteoblasts, it is a major transcriptional activator of Col1a1, the type 1 collagen gene, and of ER stress response genes, with its loss causing severe osteopenia in mice (Murakami et al., 2009). Autosomal recessive Creb3L1-deficiency in humans causes oseteogenesis imperfecta (Cayami et al., 2019; Guillemyn et al., 2019; Keller et al., 2018; Symoens et al., 2013). The roles of Creb3L1 in the regulation of secretory components have also been confirmed in intestinal goblet cells (Asada et al., 2012), the pancreas (Vellanki et al., 2010), and thyroid cells (Garcia et al., 2017).
Creb3L2/BBF2H7 functions in various tissues, across the model organisms tested, in the context of secretory pathway regulation. Overexpression of the active form of Creb3L2 in Xenopus laevis embryonic explants leads to the activation of SPCGs (Tanegashima et al., 2009). Creb3L2 also regulates secretion in mouse chondrocytes. Its loss-of-function results in severely impaired cartilage formation, attributed to the loss of Creb3L2-dependent transcriptional activation of Sec23a—a component of COPII protein essential for anterograde (ER-to-Golgi) vesicle trafficking (Saito et al., 2009). Creb3L2-mediated activation of select COPII components is, furthermore, essential for ECM secretion during zebrafish craniofacial development (Melville et al., 2011). In the hormone-secreting cells of the mouse pituitary gland, Creb3L2 is regulated by the cell-differentiation factor Tpit. In these pituitary cells, Creb3L2 has been proposed as a factor scaling translation in addition to its generic role as a factor boosting secretion (Khetchoumian et al., 2019). Creb3L2 is also strongly induced to reprogram the secretory activity during the transition of B-cell to a plasma cell state (Al-Maskari et al., 2018).
Creb3L3/CrebH is highly expressed in hepatocytes and bridges ER-stress and the acute phase response. As part of the hepatocyte unfolded protein response (UPR), CrebH regulates the production of serum amyloid P-component and C-reactive protein—both, critical components of the acute phase response—during ER-stress (Zhang et al., 2006). Hepatocyte ER-stress also leads to the secretion of hepcidin, a peptide hormone involved in iron homeostasis, in yet another example of CrebH-mediated regulation of cellular response to mitigate aberrant protein quality control (Vecchi et al., 2009). CrebH also regulates the production of apolipoprotein cargos in hepatocytes (Barbosa et al., 2017; Barbosa et al., 2013; Xu et al., 2014), confers protection from bacterial endotoxin LPS by modulating lipid profiles (Dandekar et al., 2016), and it also regulates glucose and lipid metabolism in the liver and small intestine (Nakagawa and Shimano, 2018; Zeituni et al., 2016). CrebH, furthermore, plays a critical role in C. elegans egg shell integrity by its regulation of components involved in protein trafficking (Weicksel et al., 2016).
Creb3L4/Creb4 is expressed in several organs including the pancreas, liver, and gonads with enrichment in prostate epithelial cells (Ben Aicha et al., 2007). Its major regulatory targets include the KDEL receptor (KDELR3), chaperone proteins, GALNT3—the O-glycosylating enzyme, and a Golgi assembly protein. In yet another intriguing example for the role of Creb3-family members in terminal differentiation of specialized secretory cells, Creb3L4 regulation by SPDEF (SAM-pointed domain ETS factor) allows two highly secretory cell-types of the intestine—goblet cells and Paneth cells—to attain functional maturation (Gregorieff et al., 2009).
The functional conservation between Drosophila CrebA and the human Creb3-family members is, furthermore, evident from the upregulation of all SPCGs hitherto tested by ectopic expression of either CrebA or the active form of any of the human Creb3-family members in the drosophila embryo (Barbosa et al., 2013; Fox et al., 2010). Importantly, the ATB domain, unique to CrebA and its mammalian orthologues, is critical for this activity (Barbosa et al., 2013). Collectively, these studies demonstrate that CrebA and its Creb3-family orthologues act at the regulatory crossroads of SPCG upregulation and the associated physiological UPR, hence, affecting the regulatory core of what it means to be/become a cell or tissue specialized for high levels of secretion.
The big push: Regulated secretion and cytoskeletal dynamics
The functional priming and the subsequent maturation of the SG is realized in its final act as the regulated secretion of highly glycosylated adhesive proteins—known as salivary gland-secreted (Sgs) proteins or ‘glue’ proteins—at the onset of pupariation (Biyasheva et al., 2001). Glue proteins allow the pupa to attach to a solid substrate during metamorphosis (Beckendorf and Kafatos, 1976), wherein all of the larval structures are replaced by their adult equivalents. The glue proteins, whose synthesis is stimulated by a low dose of the hormone 20-hydroxyecdysone (20E), are packaged in small granules (∼1 μm diameter) that grow into larger secretory vesicles (3–8 μm diameter) through homotypic fusion (Tran et al., 2015). Upon a second stimulation with a higher titer of 20E, the secretory vesicles fuse with the apical plasma membrane (PM) of SG cells releasing the cargo glue proteins into the lumen (Biyasheva et al., 2001). Thus, glue protein secretion is a hormonally-regulated process in late larval SGs in contrast to the embryonic gland, where secretion appears to be largely constitutive (Myat and Andrew, 2002).
The large size of secretory vesicles, the availability of fluorescent markers to label cargo, membranes, and the cytoskeleton, along with the opportunity to experimentally manipulate its hormonally-regulated exocytosis make the Drosophila larval SG an excellent system for uncovering the molecular mechanisms underlying regulated secretion in living tissues (Tran and Ten Hagen, 2017). Recent studies have revealed the dynamic engagement of the actin cytoskeleton and a myosin motor protein in cargo secretion and have provided sufficient spatial and temporal resolution to reveal the molecular events underlying each step of secretion (Rousso et al., 2016; Tran et al., 2015). In SG cells, Sgs-GFP-containing vesicles become bigger and mature through homotypic fusion between smaller vesicles (Tran et al., 2015). Immediately after membrane fusion with the plasma membrane through fusion pore formation, secretory vesicles first expand, presumably by hydrostatic pressure from the lumen, and then collapse concomitant with cargo expulsion and vesicle membrane integration into the PM (Rousso et al., 2016; Tran et al., 2015).
Upon fusion pore formation, F-actin is actively disassembled along the apical plasma membrane at the site of fusion, suggesting that the apical F-actin network is a physical barrier preventing premature fusion. Subsequently, F-actin is assembled on the vesicle membrane starting in the vicinity of the PM and then expanding to generate an ‘actin coat’ surrounding the entire vesicle (Fig. 4B). Formation of this actin coat is preceded by membrane mixing between the vesicle and the PM as visualized by the presence of PI(4,5)P2 on the vesicle prior to F-actin assembly (Rousso et al., 2016; Tran et al., 2015). PI(4,5)P2, which is abundant on the apical PM, has been shown to facilitate actin polymerization by activating multiple actin nucleating proteins (Di Paolo and De Camilli, 2006). Diaphanous (Dia), a formin that nucleates and elongates linear actin filaments, is recruited to the secretory vesicles just prior to actin coat formation and the actin coat fails to form in dia mutant SGs. With this loss of the actin coat, Sgs-GFP secretion is reduced and results in the expansion of fused vesicles and compound exocytosis, i.e., exocytosis by the consecutive fusion of vesicles with other vesicles. The fusion pore still forms between the vesicle and the PM even in the absence of the actin coat, suggesting that the actin coat is required for efficient secretion, and to prevent vesicle expansion by hydrostatic pressure and homotypic fusion (Rousso et al., 2016).
Figure 4. Regulation of the cytoskeleton is key to secretion both in early embryos and in larvae.
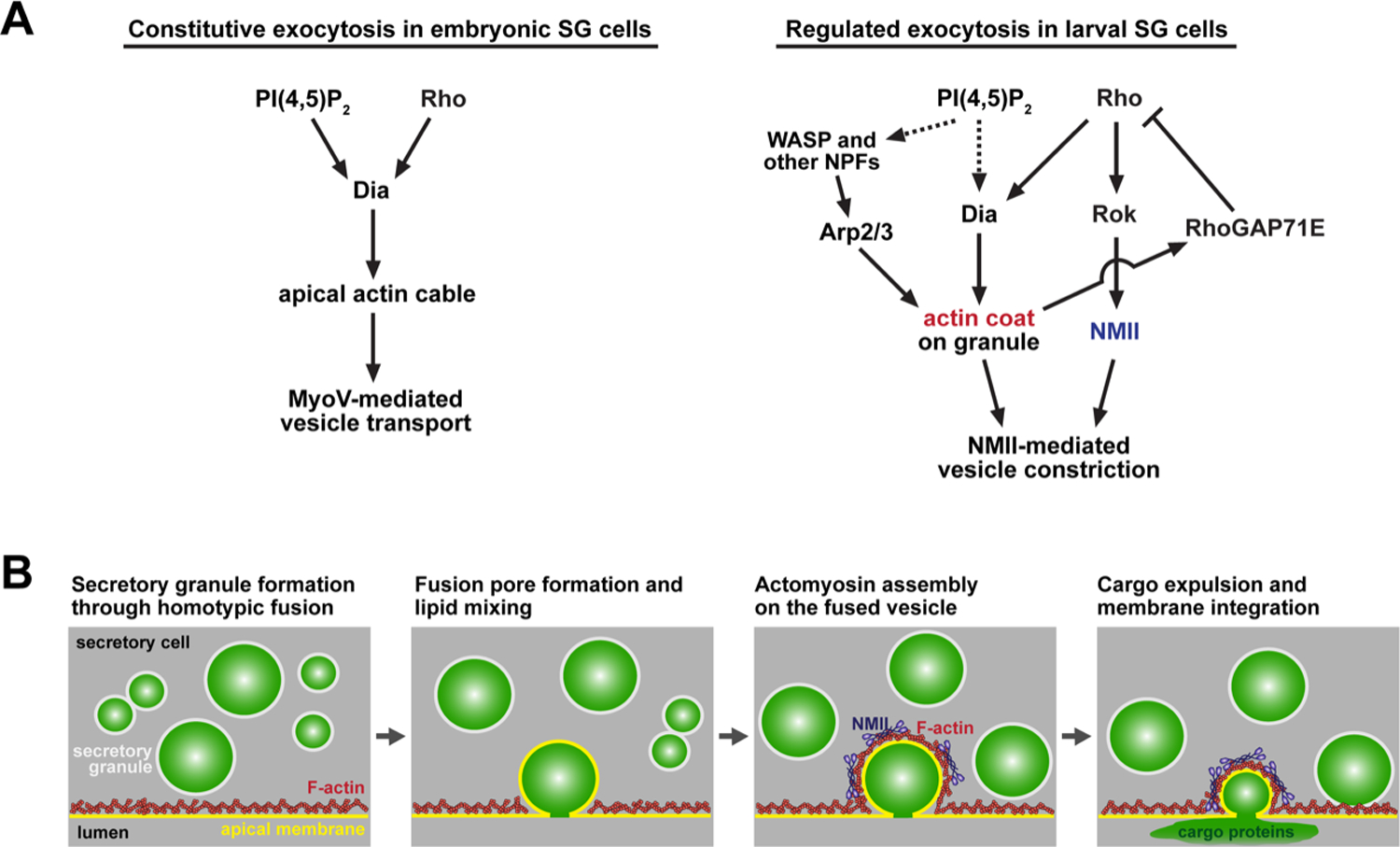
(A) Regulatory hierarchies controlling the actinomyosin network are required in embryos to target vesicles for apical secretion (left) and in larvae for efficient secretion of salivary glue proteins (right). (B) Sequence of cellular and molecular events governing efficient exocytosis of secretory vesicle content in late larval SGs.
Interestingly, Dia plays a different role in constitutive exocytosis in the Drosophila embryonic SG. Instead of coating the secretory vesicles with F-actin, Dia generates linear actin cables in the apical domain of embryonic SG cells (Fig. 4A, left panel) that function as a track for the motor protein MyoV to transport exocytic vesicles to the apical membrane (Massarwa et al., 2009). The apical localization of Dia is mediated by PI(4,5)P2 through its interaction with the N-terminal basic domain of Dia (Rousso et al., 2013). Dia-mediated F-actin cable formation is also required to coordinate apical transport of secretory vesicles in the mouse submandibular salivary gland and pancreatic cells, and in other Drosophila tubular organs, including the trachea and hindgut, implying a universal role for formin proteins in apical secretion (Geron et al., 2013; Massarwa et al., 2009). It is unclear if the large secretory vesicles in the larval SG require F-actin cables for their trafficking.
Once the secretory vesicle of the late larval SG is coated with linear actin filaments, the actin nucleator Arp2/3 complex is recruited and generates a branched F-actin network (Fig 4). The activators of the Arp2/3 complex WASp, Whamy and SCAR also localize on the secretory vesicle. Depletion of the Arp2/3 complex activity by knocking down Arp proteins or WASp did not cause compound exocytosis but interfered with cargo secretion and vesicle integration into the plasma membrane (Tran et al., 2015). This suggests that the branched actin network has a function distinct from that of linear F-actin on the secretory vesicle. Whereas the linear actin network prevents the mature vesicles from fusing with each other and provides a platform for the branched actin polymerization by the Arp2/3 complex, the branched actin network is required to generate the force necessary to complete cargo expulsion and membrane fusion. The temporal and physical separation between fusion pore formation and cargo expulsion attests to the requirement of force on the secretory vesicle to complete exocytosis.
Non-muscle myosin II (NMII) is a motor protein that generates contractile force on actin filaments (Vicente-Manzanares et al., 2009). NMII is recruited to the secretory vesicle later than actin (Tran et al., 2015). Reduction of NMII activity by knocking down Zipper (Zip, Drosophila NMII heavy chain) does not affect the size of secretory vesicles but it results in a decrease in the levels of glue protein secretion, similar to Arp2/3 complex depletion (Rousso et al., 2016). These outcomes suggest that NMII acts on the branched actin network and exerts contractile force to compress the vesicle, driving both efficient secretion of cargo proteins and the merging of the vesicle membrane with the PM. NMII activation requires regulatory light chain phosphorylation by either Rho-associated kinase (Rok) or myosin light chain kinase, MLCK. As in other contexts, Rok is activated by binding of Rho GTPase (Vicente-Manzanares et al., 2009). Depletion of Rok, or of Rho, results in similar defects in vesicle compression and cargo release as knockdown of NMII, indicating that the Rho-Rok-NMII signaling pathway functions in SG secretion (Rousso et al., 2016). As an upstream activator of Dia, Rho also regulates actin coat formation. A more recent study has revealed that negative feedback regulation of Rho activity by actin-dependent recruitment of a Rho GTPase inhibitor RhoGAP71E (Rho GTPase-activating protein 71E) to the fused vesicle promotes vesicular actin coat disassembly (Segal et al., 2018). This feedback inhibition on Rho activity induces oscillations of actin coat assembly and disassembly, which is necessary for efficient vesicle constriction. The mechanism by which this dynamic actin turnover affects actomyosin contractility on the vesicle remains unclear.
The active role of the actin cytoskeleton and NMII in secretion is not restricted to Drosophila larval SGs. Intravital microscopic imaging techniques reveal that F-actin and NMII are recruited to large secretory granules upon their fusion with the apical plasma membrane in rat submandibular SG cells. This recruitment provides the driving force that facilitates the collapse of the secretory granules into the apical membrane (Masedunskas et al., 2011). Interestingly, two isoforms of mammalian non-muscle myoII, NMIIA and NMIIB, are involved, and they control distinct steps in the secretory process (Milberg et al., 2017). Morphological changes of the secretory granules in either NMIIA- or NMIIB-deprived SG cells suggest that NMIIB stabilizes the granule membrane after fusion and pushes the membrane toward the plasma membrane, whereas NMIIA completes the membrane integration likely by controlling progressive expansion of the fusion pore (Ebrahim et al., 2018). Since Drosophila has only a single NMII, these two mechanical processes—vesicle membrane compression and fusion pore expansion—are likely to be mediated by the same myosin motor.
Thus, the actin cytoskeleton plays multiple critical roles in SG secretion. Before vesicle fusion, the apical actin network prevents premature fusion to the plasma membrane. The actin coat on the secretory vesicle maintains vesicle integrity and promotes the construction of the contractile machinery with branched actin filaments and NMII. Force generated by NMII drives cargo release and membrane integration. The conserved mechanical function of the actomyosin cytoskeleton provides a robust and well-regulated system for effective secretion of bulky cargos in specialized secretory organs.
Outlook
From its earliest utility for genetic mapping in the famous fly room at Columbia University, the SG cells of Drosophila have blazed the trail as an exceptional model system for the study of several key biological questions, e.g., tubulogenesis (Chung et al., 2017; Kolesnikov and Beckendorf, 2007; Myat and Andrew, 2000a; 2000b, 2002), cell signaling (Kupinski et al., 2013; Yavari et al., 2010; Vuong et al., 2018; Zhu et al., 2003), and tumor microenvironment (Mitchell et al., 2015; Yang et al., 2019). The highlight of this review is the major advantage offered by the embryonic SG cells for studies of the fundamental organizing principles of cellular secretion. With a relatively small set of TFs organized into the regulatory network(s), almost every aspect of secretory biology has become amenable for thorough investigations using the SG as the model system. The lack of gene redundancy in Drosophila has permitted us to gain complete “coverage” of the key TFs involved in secretory cell specification and maintenance (Scr, Exd, Hth, Fkh), survival (Fkh, Sage, Sens), tube formation and elongation (Fkh, Hkb, Rib), and functional specialization (Fkh, Sage, Sens, CrebA). These studies have also allowed us to connect different cell physiological activities through transcriptional regulation, e.g., the secretory pathway activity and the UPR. Studies of secretion in these embryonic and larval cells, furthermore, provide a developmental perspective to furnish profound insights on the evolutionarily conserved mechanisms of secretory organ developmental and functional regulation. Altogether, the results discussed in this review showcase the Drosophila embryonic SG as a paragon for the investigation of transcriptional programs, which orchestrate the entire spectrum of cell physiological processes required to produce a functional network of cells specialized for secretion. Hence, the Drosophila SG can be expected to continue to offer insights into the secrets of secretion.
Acknowledgements
We would like to thank former and current members of the Andrew, Beckendorf, Lehmann, Röper, Shilo, and Ten Hagen Labs for their major contributions to our understanding of Drosophila SG biology.
References
- Abrams EW, Andrew DJ, 2005. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development 132, 2743–2758. [DOI] [PubMed] [Google Scholar]
- Abrams EW, Mihoulides WK, Andrew DJ, 2006. Fork head and Sage maintain a uniform and patent salivary gland lumen through regulation of two downstream target genes, PH4alphaSG1 and PH4alphaSG2. Development 133, 3517–3527. [DOI] [PubMed] [Google Scholar]
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD, 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27, 53–66. [DOI] [PubMed] [Google Scholar]
- Ahmer BM, 2004. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol 52, 933–945. [DOI] [PubMed] [Google Scholar]
- Al-Maskari M, Care MA, Robinson E, Cocco M, Tooze RM, Doody GM, 2018. Site-1 protease function is essential for the generation of antibody secreting cells and reprogramming for secretory activity. Sci Rep 8, 14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DJ, Horner MA, Petitt MG, Smolik SM, Scott MP, 1994. Setting limits on homeotic gene function: restraint of Sex combs reduced activity by teashirt and other homeotic genes. EMBO J 13, 1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada R, Saito A, Kawasaki N, Kanemoto S, Iwamoto H, Oki M, Miyagi H, Izumi S, Imaizumi K, 2012. The endoplasmic reticulum stress transducer OASIS is involved in the terminal differentiation of goblet cells in the large intestine. J Biol Chem 287, 8144–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, Barreca C, O’Hare P, 2007. Trafficking of the bZIP transmembrane transcription factor CREB-H into alternate pathways of ERAD and stress-regulated intramembrane proteolysis. Traffic 8, 1796–1814. [DOI] [PubMed] [Google Scholar]
- Baker BS, Hoff G, Kaufman TC, Wolfner MF, Hazelrigg T, 1991. The doublesex locus of Drosophila melanogaster and its flanking regions: a cytogenetic analysis. Genetics 127, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa S, Carreira S, O’Hare P, 2017. GSK-3-mediated phosphorylation couples ER-Golgi transport and nuclear stabilization of the CREB-H transcription factor to mediate apolipoprotein secretion. Mol Biol Cell 28, 1565–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa S, Fasanella G, Carreira S, Llarena M, Fox R, Barreca C, Andrew D, O’Hare P, 2013. An orchestrated program regulating secretory pathway genes and cargos by the transmembrane transcription factor CREB-H. Traffic 14, 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckendorf SK, Kafatos FC, 1976. Differentiation in the salivary glands of Drosophila melanogaster: characterization of the glue proteins and their developmental appearance. Cell 9, 365–373. [DOI] [PubMed] [Google Scholar]
- Ben Aicha S, Lessard J, Pelletier M, Fournier A, Calvo E, Labrie C, 2007. Transcriptional profiling of genes that are regulated by the endoplasmic reticulum-bound transcription factor AIbZIP/CREB3L4 in prostate cells. Physiol Genomics 31, 295–305. [DOI] [PubMed] [Google Scholar]
- Biyasheva A, Do TV, Lu Y, Vaskova M, Andres AJ, 2001. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev Biol 231, 234–251. [DOI] [PubMed] [Google Scholar]
- Booth AJR, Blanchard GB, Adams RJ, Roper K, 2014. A dynamic microtubule cytoskeleton directs medial actomyosin function during tube formation. Dev Cell 29, 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros IM, 2012. Histone modification in Drosophila. Brief Funct Genomics 11, 319–331. [DOI] [PubMed] [Google Scholar]
- Bradley PL, Myat MM, Comeaux CA, Andrew DJ, 2003. Posterior migration of the salivary gland requires an intact visceral mesoderm and integrin function. Dev Biol 257, 249–262. [DOI] [PubMed] [Google Scholar]
- Bromage E, Stephens R, Hassoun L, 2009. The third dimension of ELISPOTs: quantifying antibody secretion from individual plasma cells. J Immunol Methods 346, 75–79. [DOI] [PubMed] [Google Scholar]
- Byrnes LE, Wong DM, Subramaniam M, Meyer NP, Gilchrist CL, Knox SM, Tward AD, Ye CJ, Sneddon JB, 2018. Lineage dynamics of murine pancreatic development at single-cell resolution. Nat Commun 9, 3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Liu Y, Lehmann M, 2007. Fork head controls the timing and tissue selectivity of steroid-induced developmental cell death. J Cell Biol 176, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayami FK, Maugeri A, Treurniet S, Setijowati ED, Teunissen BP, Eekhoff EMW, Pals G, Faradz SM, Micha D, 2019. The first family with adult osteogenesis imperfecta caused by a novel homozygous mutation in CREB3L1. Mol Genet Genomic Med 7, e823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran V, Beckendorf SK, 2003. senseless is necessary for the survival of embryonic salivary glands in Drosophila. Development 130, 4719–4728. [DOI] [PubMed] [Google Scholar]
- Chung KP, Zeng Y, 2017. An Overview of Protein Secretion in Plant Cells. Methods Mol Biol 1662, 19–32. [DOI] [PubMed] [Google Scholar]
- Chung S, Kim S, Andrew DJ, 2017. Uncoupling apical constriction from tissue invagination. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar A, Qiu Y, Kim H, Wang J, Hou X, Zhang X, Zheng Z, Mendez R, Yu FS, Kumar A, Fang D, Sun F, Zhang K, 2016. Toll-like Receptor (TLR) Signaling Interacts with CREBH to Modulate High-density Lipoprotein (HDL) in Response to Bacterial Endotoxin. J Biol Chem 291, 23149–23158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demakova OV, Demakov SA, Boldyreva LV, Zykova TY, Levitsky VG, Semeshin VF, Pokholkova GV, Sidorenko DS, Goncharov FP, Belyaeva ES, Zhimulev IF, 2020. Faint gray bands in Drosophila melanogaster polytene chromosomes are formed by coding sequences of housekeeping genes. Chromosoma 129, 25–44. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P, 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657. [DOI] [PubMed] [Google Scholar]
- Ebrahim S, Liu J, Weigert R, 2018. The Actomyosin Cytoskeleton Drives Micron-Scale Membrane Remodeling In Vivo Via the Generation of Mechanical Forces to Balance Membrane Tension Gradients. Bioessays 40, e1800032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleveld-Trancikova D, Sanecka A, van Hout-Kuijer MA, Looman MW, Hendriks IA, Jansen BJ, Adema GJ, 2010. DC-STAMP interacts with ER-resident transcription factor LUMAN which becomes activated during DC maturation. Mol Immunol 47, 1963–1973. [DOI] [PubMed] [Google Scholar]
- Fox RM, Hanlon CD, Andrew DJ, 2010. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol 191, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Vaishnavi A, Maruyama R, Andrew DJ, 2013. Organ-specific gene expression: the bHLH protein Sage provides tissue specificity to Drosophila FoxA. Development 140, 2160–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Baker BS, 1999. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell 4, 117–122. [DOI] [PubMed] [Google Scholar]
- Gao N, Le Lay J, Qin W, Doliba N, Schug J, Fox AJ, Smirnova O, Matschinsky FM, Kaestner KH, 2010. Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature beta-cell. Mol Endocrinol 24, 1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia IA, Torres Demichelis V, Viale DL, Di Giusto P, Ezhova Y, Polishchuk RS, Sampieri L, Martinez H, Sztul E, Alvarez C, 2017. CREB3L1-mediated functional and structural adaptation of the secretory pathway in hormone-stimulated thyroid cells. J Cell Sci 130, 4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE, 2002. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295, 821–825. [DOI] [PubMed] [Google Scholar]
- Geron E, Schejter ED, Shilo BZ, 2013. Directing exocrine secretory vesicles to the apical membrane by actin cables generated by the formin mDia1. Proc Natl Acad Sci U S A 110, 10652–10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves DS, Ferreira MDS, Guimaraes AJ, 2019. Extracellular Vesicles from the Protozoa Acanthamoeba castellanii: Their Role in Pathogenesis, Environmental Adaptation and Potential Applications. Bioengineering (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Stange DE, Kujala P, Begthel H, van den Born M, Korving J, Peters PJ, Clevers H, 2009. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology 137, 1333–1345 e1331–1333. [DOI] [PubMed] [Google Scholar]
- Guillemyn B, Kayserili H, Demuynck L, Sips P, De Paepe A, Syx D, Coucke PJ, Malfait F, Symoens S, 2019. A homozygous pathogenic missense variant broadens the phenotypic and mutational spectrum of CREB3L1-related osteogenesis imperfecta. Hum Mol Genet 28, 1801–1809. [DOI] [PubMed] [Google Scholar]
- Guo Y, Yang F, Tang X, 2017. An Overview of Protein Secretion in Yeast and Animal Cells. Methods Mol Biol 1662, 1–17. [DOI] [PubMed] [Google Scholar]
- Haberman AS, Isaac DD, Andrew DJ, 2003. Specification of cell fates within the salivary gland primordium. Dev Biol 258, 443–453. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P, 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2, 95–108. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D, 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harmer AM, Blackledge TA, Madin JS, Herberstein ME, 2011. High-performance spider webs: integrating biomechanics, ecology and behaviour. J R Soc Interface 8, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KE, Beckendorf SK, 2007. Different Wnt signals act through the Frizzled and RYK receptors during Drosophila salivary gland migration. Development 134, 2017–2025. [DOI] [PubMed] [Google Scholar]
- Harris KE, Schnittke N, Beckendorf SK, 2007. Two ligands signal through the Drosophila PDGF/VEGF receptor to ensure proper salivary gland positioning. Mech Dev 124, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KD, Andrew DJ, 2000. Regulation and function of Scr, exd, and hth in the Drosophila salivary gland. Dev Biol 217, 362–374. [DOI] [PubMed] [Google Scholar]
- Henderson KD, Isaac DD, Andrew DJ, 1999. Cell fate specification in the Drosophila salivary gland: the integration of homeotic gene function with the DPP signaling cascade. Dev Biol 205, 10–21. [DOI] [PubMed] [Google Scholar]
- Hetz C, 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13, 89–102. [DOI] [PubMed] [Google Scholar]
- Hiller M, Lang C, Michel W, Flieger A, 2018. Secreted phospholipases of the lung pathogen Legionella pneumophila. Int J Med Microbiol 308, 168–175. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, 1987. Chromosome structure in four wild-type polytene tissues of Drosophila melanogaster. The 87A and 87C heat shock loci are induced unequally in the midgut in a manner dependent on growth temperature. Chromosoma 95, 197–208. [DOI] [PubMed] [Google Scholar]
- Huber RJ, O’Day DH, 2017. Extracellular matrix dynamics and functions in the social amoeba Dictyostelium: A critical review. Biochim Biophys Acta Gen Subj 1861, 2971–2980. [DOI] [PubMed] [Google Scholar]
- Huebner RJ, Lechler T, Ewald AJ, 2014. Developmental stratification of the mammary epithelium occurs through symmetry-breaking vertical divisions of apically positioned luminal cells. Development 141, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrich M, Greenleaf AL, Bautz EK, 1977. Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster. Proc Natl Acad Sci U S A 74, 2079–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DM, Wells MB, Loganathan R, Lee JS, Levings D, Bastien A, Slattery M, Andrew DJ, 2020. CrebA increases secretory capacity through direct transcriptional regulation of the secretory machinery, a subset of secretory cargo, and other key regulators Traffic 21, 560–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Sun L, Mann R, 2010. Dissecting the functional specificities of two Hox proteins. Genes Dev 24, 1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Bordeaux MC, Garel A, Couble P, 2002. Fork head alternative binding drives stage-specific gene expression in the silk gland of Bombyx mori. Insect Biochem Mol Biol 32, 377–387. [DOI] [PubMed] [Google Scholar]
- Kabisch R, Bautz EK, 1983. Differential distribution of RNA polymerase B and nonhistone chromosomal proteins in polytene chromosomes of Drosophila melanogaster. EMBO J 2, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL, 2006. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133, 495–505. [DOI] [PubMed] [Google Scholar]
- Keller RB, Tran TT, Pyott SM, Pepin MG, Savarirayan R, McGillivray G, Nickerson DA, Bamshad MJ, Byers PH, 2018. Monoallelic and biallelic CREB3L1 variant causes mild and severe osteogenesis imperfecta, respectively. Genet Med 20, 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman BE, Cheshire AM, Myat MM, Andrew DJ, 2008. Ribbon modulates apical membrane during tube elongation through Crumbs and Moesin. Dev Biol 320, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetchoumian K, Balsalobre A, Mayran A, Christian H, Chenard V, St-Pierre J, Drouin J, 2019. Pituitary cell translation and secretory capacities are enhanced cell autonomously by the transcription factor Creb3l2. Nat Commun 10, 3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Baylies MK, Gasic GP, Young MW, 1989. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev 3, 1113–1129. [DOI] [PubMed] [Google Scholar]
- Kolesnikov T, Beckendorf SK, 2005. NETRIN and SLIT guide salivary gland migration. Dev Biol 284, 102–111. [DOI] [PubMed] [Google Scholar]
- Kolesnikov T, Beckendorf SK, 2007. 18 wheeler regulates apical constriction of salivary gland cells via the Rho-GTPase-signaling pathway. Dev Biol 307, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova TD, 2018. Banding Pattern of Polytene Chromosomes as a Representation of Universal Principles of Chromatin Organization into Topological Domains. Biochemistry (Mosc) 83, 338–349. [DOI] [PubMed] [Google Scholar]
- Kolver ES, Muller LD, 1998. Performance and nutrient intake of high producing Holstein cows consuming pasture or a total mixed ration. J Dairy Sci 81, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK, 1996. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J 15, 4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK, 1998. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev 12, 3752–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YM, Jones N, Zhou B, Panzer S, Larson V, Beckendorf SK, 1996. Salivary duct determination in Drosophila: roles of the EGF receptor signalling pathway and the transcription factors fork head and trachealess. Development 122, 1909–1917. [DOI] [PubMed] [Google Scholar]
- Kuroda MI, Hilfiker A, Lucchesi JC, 2016. Dosage Compensation in Drosophila-a Model for the Coordinate Regulation of Transcription. Genetics 204, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb MJ, 1982. The DNA content of polytene nuclei in midgut and Malpighian tubule cells of adultDrosophila melanogaster. Wilehm Roux Arch Dev Biol 191, 381–384. [DOI] [PubMed] [Google Scholar]
- Lelli KM, Noro B, Mann RS, 2011. Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity. Proc Natl Acad Sci U S A 108, 21122–21127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Chen FG, 2015. [Advances in understanding Drosophila salivary gland polytene chromosome and its applications in genetics teaching]. Yi Chuan 37, 605–612. [DOI] [PubMed] [Google Scholar]
- Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M, Kaestner KH, Ang SL, 2009. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol 333, 386–396. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lehmann M, 2008. Genes and biological processes controlled by the Drosophila FOXA orthologue Fork head. Insect Mol Biol 17, 91–101. [DOI] [PubMed] [Google Scholar]
- Mach V, Takiya S, Ohno K, Handa H, Imai T, Suzuki Y, 1995. Silk gland factor-1 involved in the regulation of Bombyx sericin-1 gene contains fork head motif. J Biol Chem 270, 9340–9346. [DOI] [PubMed] [Google Scholar]
- Manning AJ, Rogers SL, 2014. The Fog signaling pathway: insights into signaling in morphogenesis. Dev Biol 394, 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Grevengoed E, Stempniewicz P, Andrew DJ, 2011. Genome-wide analysis reveals a major role in cell fate maintenance and an unexpected role in endoreduplication for the Drosophila FoxA gene Fork head. PLoS One 6, e20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masedunskas A, Sramkova M, Parente L, Sales KU, Amornphimoltham P, Bugge TH, Weigert R, 2011. Role for the actomyosin complex in regulated exocytosis revealed by intravital microscopy. Proc Natl Acad Sci U S A 108, 13552–13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R, Schejter ED, Shilo BZ, 2009. Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Dev Cell 16, 877–888. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Kerridge S, Scott MP, 1994. Role of the teashirt gene in Drosophila midgut morphogenesis: secreted proteins mediate the action of homeotic genes. Development 120, 2799–2809. [DOI] [PubMed] [Google Scholar]
- Melville DB, Montero-Balaguer M, Levic DS, Bradley K, Smith JR, Hatzopoulos AK, Knapik EW, 2011. The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis. Dis Model Mech 4, 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milberg O, Shitara A, Ebrahim S, Masedunskas A, Tora M, Tran DT, Chen Y, Conti MA, Adelstein RS, Ten Hagen KG, Weigert R, 2017. Concerted actions of distinct nonmuscle myosin II isoforms drive intracellular membrane remodeling in live animals. J Cell Biol 216, 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K, 2009. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 11, 1205–1211. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ, 2000a. Fork head prevents apoptosis and promotes cell shape change during formation of the Drosophila salivary glands. Development 127, 4217–4226. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ, 2000b. Organ shape in the Drosophila salivary gland is controlled by regulated, sequential internalization of the primordia. Development 127, 679–691. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ, 2002. Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell 111, 879–891. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Shimano H, 2018. CREBH Regulates Systemic Glucose and Lipid Metabolism. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido T, Yokoya S, Mori T, Hagino S, Iseki K, Zhang Y, Takeuchi M, Takaki H, Kikuchi S, Wanaka A, 2001. Expression of the novel transcription factor OASIS, which belongs to the CREB/ATF family, in mouse embryo with special reference to bone development. Histochem Cell Biol 116, 141–148. [DOI] [PubMed] [Google Scholar]
- Nikolaidou KK, Barrett K, 2004. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr Biol 14, 1822–1826. [DOI] [PubMed] [Google Scholar]
- O’Neill MC, 1991. A general method for modeling cell populations undergoing G1----G0 transitions during development. J Theor Biol 153, 499–518. [DOI] [PubMed] [Google Scholar]
- Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S, 2002. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun 293, 470–477. [DOI] [PubMed] [Google Scholar]
- Panzer S, Weigel D, Beckendorf SK, 1992. Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development 114, 49–57. [DOI] [PubMed] [Google Scholar]
- Reiling JH, Olive AJ, Sanyal S, Carette JE, Brummelkamp TR, Ploegh HL, Starnbach MN, Sabatini DM, 2013. A CREB3-ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat Cell Biol 15, 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault N, King-Jones K, Lehmann M, 2001. Downregulation of the tissue-specific transcription factor Fork head by Broad-Complex mediates a stage-specific hormone response. Development 128, 3729–3737. [DOI] [PubMed] [Google Scholar]
- Richards G, 1980. The polytene chromosomes in the fat body nuclei of Drosophila melanogaster. Chromosoma 79, 241–250. [DOI] [PubMed] [Google Scholar]
- Riley PD, Carroll SB, Scott MP, 1987. The expression and regulation of Sex combs reduced protein in Drosophila embryos. Genes Dev 1, 716–730. [DOI] [PubMed] [Google Scholar]
- Rodrigues FF, Harris TJC, 2019. Key roles of Arf small G proteins and biosynthetic trafficking for animal development. Small GTPases 10, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli M, Bresson L, Di-Cicco A, Perez-Lanzon M, Legoix P, Baulande S, de la Grange P, De Arcangelis A, Georges-Labouesse E, Sonnenberg A, Deugnier MA, Glukhova MA, Faraldo MM, 2020. Laminin-binding integrins are essential for the maintenance of functional mammary secretory epithelium in lactation. Development 147. [DOI] [PubMed] [Google Scholar]
- Roper K, 2012. Anisotropy of Crumbs and aPKC drives myosin cable assembly during tube formation. Dev Cell 23, 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso T, Schejter ED, Shilo BZ, 2016. Orchestrated content release from Drosophila glue-protein vesicles by a contractile actomyosin network. Nat Cell Biol 18, 181–190. [DOI] [PubMed] [Google Scholar]
- Rousso T, Shewan AM, Mostov KE, Schejter ED, Shilo BZ, 2013. Apical targeting of the formin Diaphanous in Drosophila tubular epithelia. Elife 2, e00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin GT, 1972. Replication in polytene chromosomes. Results Probl Cell Differ 4, 59–85. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Mann RS, 1999. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev 13, 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Imaizumi K, 2009. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol 11, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Sampieri L, Di Giusto P, Alvarez C, 2019. CREB3 Transcription Factors: ER-Golgi Stress Transducers as Hubs for Cellular Homeostasis. Front Cell Dev Biol 7, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Corrales YE, Blanchard GB, Roper K, 2018. Radially patterned cell behaviours during tube budding from an epithelium. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanecka A, Ansems M, van Hout-Kuijer MA, Looman MW, Prosser AC, Welten S, Gilissen C, Sama IE, Huynen MA, Veltman JA, Jansen BJ, Eleveld-Trancikova D, Adema GJ, 2012. Analysis of genes regulated by the transcription factor LUMAN identifies ApoA4 as a target gene in dendritic cells. Mol Immunol 50, 66–73. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Kubli E, 1980. The localization of tRNA5Asn, tRNAHis, and tRNAAla genes from drosophila melanogaster by in situ hybridization to polytene salivary gland chromosomes. Chromosoma 80, 277–287. [DOI] [PubMed] [Google Scholar]
- Scott MP, Weiner AJ, Hazelrigg TI, Polisky BA, Pirrotta V, Scalenghe F, Kaufman TC, 1983. The molecular organization of the Antennapedia locus of Drosophila. Cell 35, 763–776. [DOI] [PubMed] [Google Scholar]
- Segal D, Zaritsky A, Schejter ED, Shilo BZ, 2018. Feedback inhibition of actin on Rho mediates content release from large secretory vesicles. J Cell Biol 217, 1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidor C, Stevens TJ, Jin L, Boulanger J, Roper K, 2020. Rho-Kinase Planar Polarization at Tissue Boundaries Depends on Phospho-regulation of Membrane Residence Time. Dev Cell 52, 364–378 e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo L, Kazimirova M, Richardson J, Bonnet SI, 2017. The Essential Role of Tick Salivary Glands and Saliva in Tick Feeding and Pathogen Transmission. Front Cell Infect Microbiol 7, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit RB, Schnabel R, Gaudet J, 2008. The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function. PLoS Genet 4, e1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Orr-Weaver TL, 1991. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112, 997–1008. [DOI] [PubMed] [Google Scholar]
- Smolik SM, Rose RE, Goodman RH, 1992. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol 12, 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW, 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symoens S, Malfait F, D’Hondt S, Callewaert B, Dheedene A, Steyaert W, Bachinger HP, De Paepe A, Kayserili H, Coucke PJ, 2013. Deficiency for the ER-stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet J Rare Dis 8, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanegashima K, Zhao H, Rebbert ML, Dawid IB, 2009. Coordinated activation of the secretory pathway during notochord formation in the Xenopus embryo. Development 136, 3543–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DT, Masedunskas A, Weigert R, Ten Hagen KG, 2015. Arp2/3-mediated F-actin formation controls regulated exocytosis in vivo. Nat Commun 6, 10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DT, Ten Hagen KG, 2017. Real-time insights into regulated exocytosis. J Cell Sci 130, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaslet CA, O’Connell P, Izquierdo M, Rosbash M, 1980. Isolation and mapping of a cloned ribosomal protein gene of Drosophila melanogaster. Nature 285, 674–676. [DOI] [PubMed] [Google Scholar]
- Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A, 2009. ER stress controls iron metabolism through induction of hepcidin. Science 325, 877–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanki RN, Zhang L, Guney MA, Rocheleau JV, Gannon M, Volchuk A, 2010. OASIS/CREB3L1 induces expression of genes involved in extracellular matrix production but not classical endoplasmic reticulum stress response genes in pancreatic beta-cells. Endocrinology 151, 4146–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR, 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Briones A, Aird SD, 2018. Organic and Peptidyl Constituents of Snake Venoms: The Picture Is Vastly More Complex Than We Imagined. Toxins (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining MS, Bradley PL, Comeaux CA, Andrew DJ, 2005. Organ positioning in Drosophila requires complex tissue-tissue interactions. Dev Biol 287, 19–34. [DOI] [PubMed] [Google Scholar]
- Weicksel SE, Mahadav A, Moyle M, Cipriani PG, Kudron M, Pincus Z, Bahmanyar S, Abriola L, Merkel J, Gutwein M, Fernandez AG, Piano F, Gunsalus KC, Reinke V, 2016. A novel small molecule that disrupts a key event during the oocyte-to-embryo transition in C. elegans. Development 143, 3540–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MB, Andrew DJ, 2019. Anopheles Salivary Gland Architecture Shapes Plasmodium Sporozoite Availability for Transmission. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Grawe F, Knust E, 1993. CRUMBS is involved in the control of apical protein targeting during Drosophila epithelial development. Mech Dev 44, 175–187. [DOI] [PubMed] [Google Scholar]
- Xin HH, Zhang DP, Chen RT, Cai ZZ, Lu Y, Liang S, Miao YG, 2015. TRANSCRIPTION FACTOR Bmsage PLAYS A CRUCIAL ROLE IN SILK GLAND GENERATION IN SILKWORM, Bombyx mori. Arch Insect Biochem Physiol 90, 59–69. [DOI] [PubMed] [Google Scholar]
- Xu N, Keung B, Myat MM, 2008. Rho GTPase controls invagination and cohesive migration of the Drosophila salivary gland through Crumbs and Rho-kinase. Dev Biol 321, 88–100. [DOI] [PubMed] [Google Scholar]
- Xu X, Park JG, So JS, Hur KY, Lee AH, 2014. Transcriptional regulation of apolipoprotein A-IV by the transcription factor CREBH. J Lipid Res 55, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li LC, Lamaoqiezhong, Wang X, Wang WH, Wang YC, Xu CR, 2019. The contributions of mesoderm-derived cells in liver development. Semin Cell Dev Biol 92, 63–76. [DOI] [PubMed] [Google Scholar]
- Ying Z, Zhang R, Verge VM, Misra V, 2015. Cloning and characterization of rat Luman/CREB3, a transcription factor highly expressed in nervous system tissue. J Mol Neurosci 55, 347–354. [DOI] [PubMed] [Google Scholar]
- Zeituni EM, Wilson MH, Zheng X, Iglesias PA, Sepanski MA, Siddiqi MA, Anderson JL, Zheng Y, Farber SA, 2016. Endoplasmic Reticulum Lipid Flux Influences Enterocyte Nuclear Morphology and Lipid-dependent Transcriptional Responses. J Biol Chem 291, 23804–23816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ, 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599. [DOI] [PubMed] [Google Scholar]
- Zhao XM, Liu C, Li QY, Hu WB, Zhou MT, Nie HY, Zhang YX, Peng ZC, Zhao P, Xia QY, 2014. Basic helix-loop-helix transcription factor Bmsage is involved in regulation of fibroin H-chain gene via interaction with SGF1 in Bombyx mori. PLoS One 9, e94091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Bagri A, Beckendorf SK, 2001. Salivary gland determination in Drosophila: a salivary-specific, fork head enhancer integrates spatial pattern and allows fork head autoregulation. Dev Biol 237, 54–67. [DOI] [PubMed] [Google Scholar]
- Zhu AJ, Zheng L, Suyama K, Scott MP, 2003. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev 17, 1240–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink B, Paro R, 1989. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature 337, 468–471. [DOI] [PubMed] [Google Scholar]
- Zykova TY, Levitsky VG, Belyaeva ES, Zhimulev IF, 2018. Polytene Chromosomes - A Portrait of Functional Organization of the Drosophila Genome. Curr Genomics 19, 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]


