Abstract
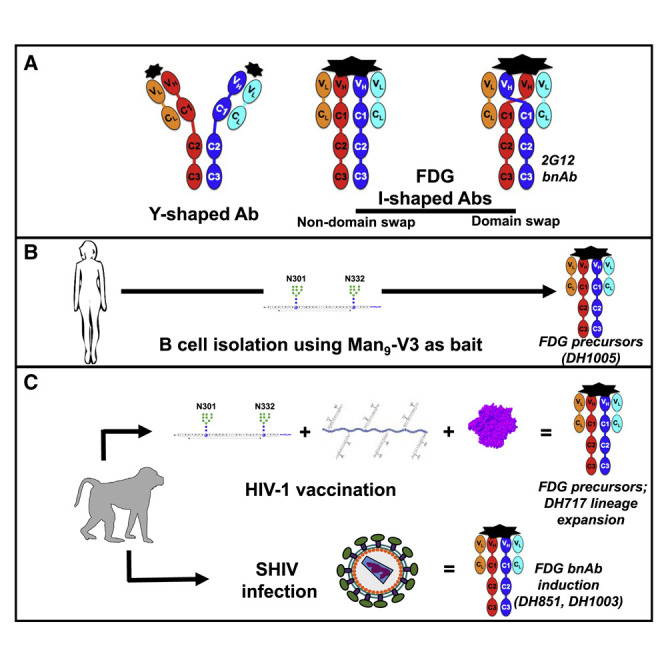
Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) as a model antigen. 2G12 is a broadly neutralizing Ab (bnAb) that targets a conserved glycan patch on Env of geographically diverse HIV-1 strains using a unique heavy-chain (VH) domain-swapped architecture that results in fragment antigen-binding (Fab) dimerization. Here, we describe HIV-1 Env Fab-dimerized glycan (FDG)-reactive bnAbs without VH-swapped domains from simian-human immunodeficiency virus (SHIV)-infected macaques. FDG Abs also recognized cell-surface glycans on diverse pathogens, including yeast and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike. FDG precursors were expanded by glycan-bearing immunogens in macaques and were abundant in HIV-1-naive humans. Moreover, FDG precursors were predominately mutated IgM+IgD+CD27+, thus suggesting that they originated from a pool of antigen-experienced IgM+ or marginal zone B cells.
Keywords: natural Abs, glycan-dependent Ab binding, HIV-1 Env glycans, SARS-CoV-2 spike glycans, Fab dimerization, FDG Abs, marginal zone B cells, IgM-memory B cells
Graphical abstract
Structural and functional analyses identify a category of glycan-reactive antibodies in macaques and humans that are marked by the dimerization of the antigen-binding fragment. These antibodies are involved in HIV neutralization and also recognize the S2 protein of SARS-CoV-2.
Introduction
Many pathogens, including human immunodeficiency virus (HIV) envelope (Env) (Doores et al., 2010a; Bonomelli et al., 2011; Cao et al., 2017), yeast (Masuoka, 2004; Hall and Gow, 2013), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike proteins (Watanabe et al., 2020) express surface glycans. Anti-glycan antibodies (Abs) constitute a subset of natural Abs that are present prior to stimulation by cognate antigen, can exist as different isotypes, and provide the first line of defense against bacterial, fungal, and viral infections (Holodick et al., 2017). In humans, natural Abs can be mutated either after antigen stimulation or by unknown processes as immunoglobulin M (IgM) Abs in the pre-immune B cell repertoire (Holodick et al., 2017; Klein et al., 1998) and have been postulated to be derived from marginal zone and transitional B cells (Casali et al., 1987; Griffin et al., 2011; Quách et al., 2016; Weller et al., 2004; Weill et al., 2009).
HIV-type 1 (HIV-1) Env is heavily glycosylated, thus Abs have limited access to epitopes targeted by broadly neutralizing Abs (bnAbs) that are effective against multiple geographically diverse HIV-1 strains (Doores et al., 2010a; Bonomelli et al., 2011; Cao et al., 2017; Pritchard et al., 2015). HIV-1 Env-reactive bnAbs have been described that used long complementary-determining region (CDR) loops to gain access to bnAb epitopes in the Env protein, while also making stabilizing contacts with surrounding glycans (Walker et al., 2009; Bonsignori et al., 2011, 2017; Pejchal et al., 2011; Garces et al., 2014, 2015; Andrabi et al., 2015; Doria-Rose et al., 2015; Gorman et al., 2016; Fera et al., 2018). Thus, investigators have long sought anti-glycan Abs as a means to turn an effective HIV-1 defense into a vulnerability (Scanlan et al., 2007; Doores, 2015; Crispin et al., 2018; Daniels and Saunders, 2019). However, glycans found on HIV-1 and other pathogens are also present on host molecules (Shackelford and Strominger, 1983; Yu et al., 2010; Blixt et al., 2004), suggesting that the induction of Abs targeting pathogen glycans may be controlled by immune tolerance mechanisms (Scanlan et al., 2007). The Ab 2G12 has been the only example to date of an HIV-1 bnAb that interacts solely with glycans on Env (Calarese et al., 2003). 2G12 has a relatively short variable heavy (VH) domain CDR3 loop and a unique domain-swapped architecture in which the VH domains of the two fragment antigen-binding (Fab) arms of the 2G12 IgG swap to create a Fab-dimerized multivalent surface that facilitates strong interaction with glycans on Env (Doores et al., 2010b; Murin et al., 2014; Scanlan et al., 2002; Calarese et al., 2003; Chuang et al., 2019). Fab dimerization by 2G12 has been shown to be essential for optimal HIV-1 biological function, thus distinguishing it from conventional polyreactive Abs with low-level glycan interactions (Doores et al., 2010b).
The bnAb 2G12 binds HIV-1 Env with nanomolar affinity, primarily due to its multivalent recognition of a unique glycan cluster (Scanlan et al., 2002). Structures of 2G12 determined using X-ray crystallography (Calarese et al., 2003; Wu et al., 2013), negative stain, and cryoelectron microscopy (cryo-EM) (Chuang et al., 2019; Murin et al., 2014; Seabright et al., 2020), have revealed details of the VH domain-swapped Fab-dimerized 2G12 architecture and its multiple glycan binding sites; both features are critical for high-affinity binding to HIV-1 Env. In addition to HIV-1 Env, 2G12 also bound Candida albicans yeast, which presents a similar Man(α1-2)Man motif as HIV-1 Env glycans (Doores et al., 2010b; Gemmill and Trimble, 1999; Scanlan et al., 2002). Thus, yeast glycans have been implicated in the induction of 2G12-like B cell responses (Scanlan et al., 2007).
Here, we report multiple Fab-dimerized glycan-reactive (FDG) Abs that target HIV-1 Env. FDG B cells were common in rhesus macaques (RMs) before virus infection or vaccination and resided in a mutated IgM+IgD+CD27+ B cell pool in HIV-1 naive humans. Fab dimerization occurred by several mechanisms, including Fab-Fab disulfide linkage, hydrophobic interactions, and hydrogen bonding—none of which required VH domain swapping for HIV-1 neutralization. FDG Abs, in addition to binding yeast and HIV-1 Env glycans, also recognized a glycan cluster in the S2 subunit of the SARS-CoV-2 spike. Thus, our study emphasized the specificity of FDG Abs for glycosylated proteins and suggested that this natural FDG Ab precursor pool can target diverse human pathogens.
Results
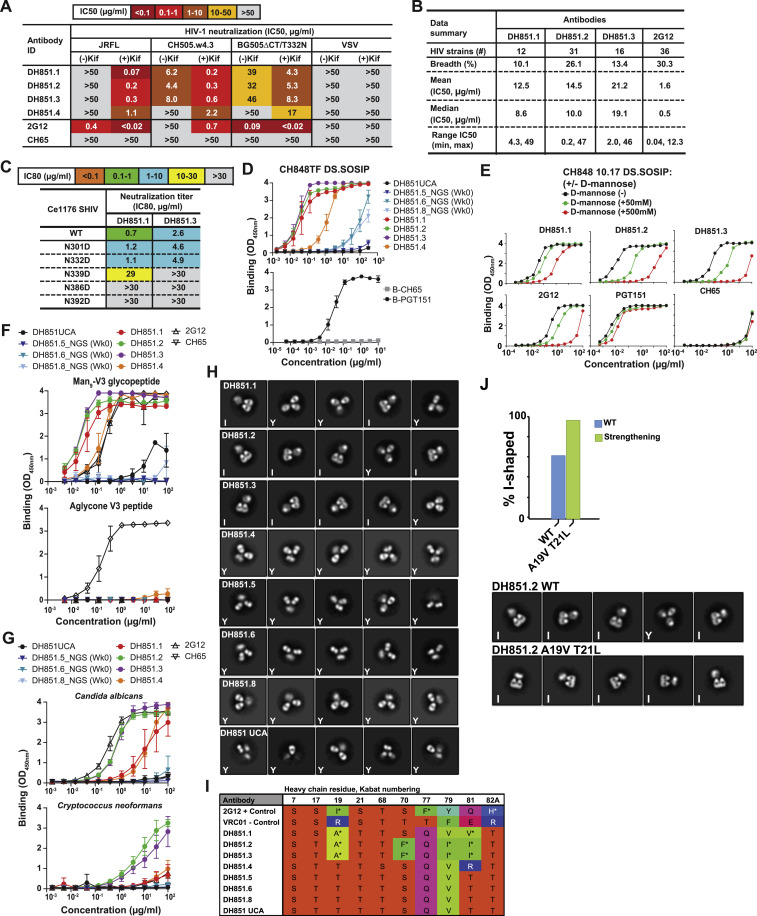
Vaccine-induced HIV-1 Env glycan-reactive Abs
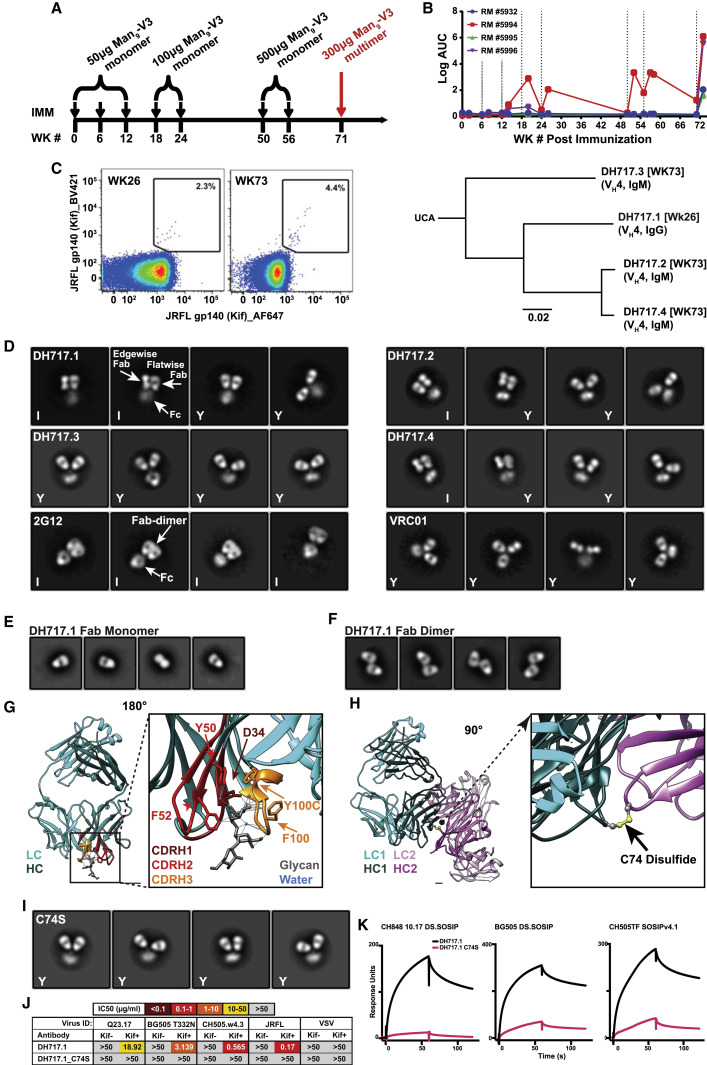
We previously reported a glycopeptide (Man9-V3) mimic of a bnAb epitope containing high-mannose glycoforms on native HIV-1 Env trimer (Alam et al., 2017; Fera et al., 2018). In an effort to induce bnAbs that target high-mannose glycans on HIV-1 Env, four RMs previously immunized with monomeric Man9-V3 (Alam et al., 2017) were subsequently immunized with a multimeric form of Man9-V3 (Figure 1 A). After repeated immunizations with monomeric Man9-V3, plasma Abs from two of the four RMs bound Man9-V3; whereas plasma from all four RMs bound Man9-V3 following a single immunization with multimeric Man9-V3 (Figure 1B). From a representative macaque, we isolated a 4-member DH717 Ab clonal lineage; DH717.1 IgG and DH717.2-DH717.4 IgMs were isolated before and after immunization with multimeric Man9-V3, respectively (Figure 1C; Data S3). Recombinant DH717 monoclonal Abs (mAbs) demonstrated glycan-dependent binding to Man9-V3 (Alam et al., 2017), and bound soluble recombinant HIV-1 Env trimers (Saunders et al., 2019), Candida albicans or Cryptococcus neoformans yeast glycans, and high mannose glycans including Man9GlcNAc2 (Figures 2 B–2D, S1 A, and S1B). DH717 mAbs neutralized env-pseudotyped HIV-1 bearing Envs with Man9-enriched glycans but did not neutralize corresponding wild-type HIV-1 bearing Envs with heterogeneous glycoforms (Figure S1C). Negative stain electron microscopy (NSEM) of DH717.1, DH717.2, and DH717.4 revealed a mixture of canonical Y-shaped Abs, as well as I-shaped Fab-dimerized Abs reminiscent of the I-shaped HIV-1 bnAb 2G12 (Figure 1D). In 2G12, the two Fabs lie side-by-side and in the same plane; thus, both Fabs are seen in a flatwise view via NSEM. In contrast, in DH717, one Fab is seen flatwise, whereas the other Fab is seen edgewise, suggesting that the two Fabs are turned relative to each other 90° about their long axes (Figure 1D). This arrangement was confirmed by 3D negative stain reconstruction of DH717.1 IgG (Figure S1D).
Figure 1.
Characteristics of HIV-1 vaccine-induced Env glycan-reactive neutralizing B cell lineage, DH717
(A) Schedule of Man9-V3 immunizations in RMs.
(B) Plasma IgG at weeks (WK) post infection from four immunized RMs were tested in ELISA for binding to Man9-V3. Data shown are representative from multiple assays and binding titer was reported as log area under the curve (AUC). Vertical dotted lines indicate immunization time points.
(C) Representative flow cytometry sort plot of HIV-1 Env-reactive memory B cells detected at WK 26 and 73. Phylogram of VH genes isolated from a clonal lineage of DH717 Abs found at WK 26 and 73.
(D) NSEM 2D class averages for DH717.1–DH717.4, and HIV-1 bnAbs 2G12 and VRC01, are labeled I or Y according to shape.
(E and F) Monomeric (E) and dimeric (F) Fabs are also shown.
(G) Crystal structure of DH717.1 Fab monomer (VH and Vλ in dark and light teal, respectively) bound to Man9-V3. The terminal glycan-moieties of Man9-V3 (gray sticks) bound a pocket formed by the CDRH1-3 loops (dark red, red and orange) of the DH717.1 Fab. Inset shows a zoomed-in view of glycan binding.
(H) Crystal structure of DH717.1 dimer with dark teal or pink VH, and light teal or pink Vλ. Inset shows a zoomed-in view of the inter-Fab disulfide (yellow spheres).
(I) NSEM of cysteine to serine DH717.1 mutant (C74S).
(J) DH717.1 wild-type and C74S mutant IgGs were tested for neutralization of env-pseudotyped HIV-1 bearing Envs with heterogeneous [−Kif] or Man9-enriched [+Kif] glycans in TZM-bl cells in a single assay. Neutralization titers were reported as IC50 in μg/mL. These Abs were also tested in a SPR assay for binding soluble stabilized forms of recombinant HIV-1 Env trimers (K); shown are SPR binding curves for each Ab.
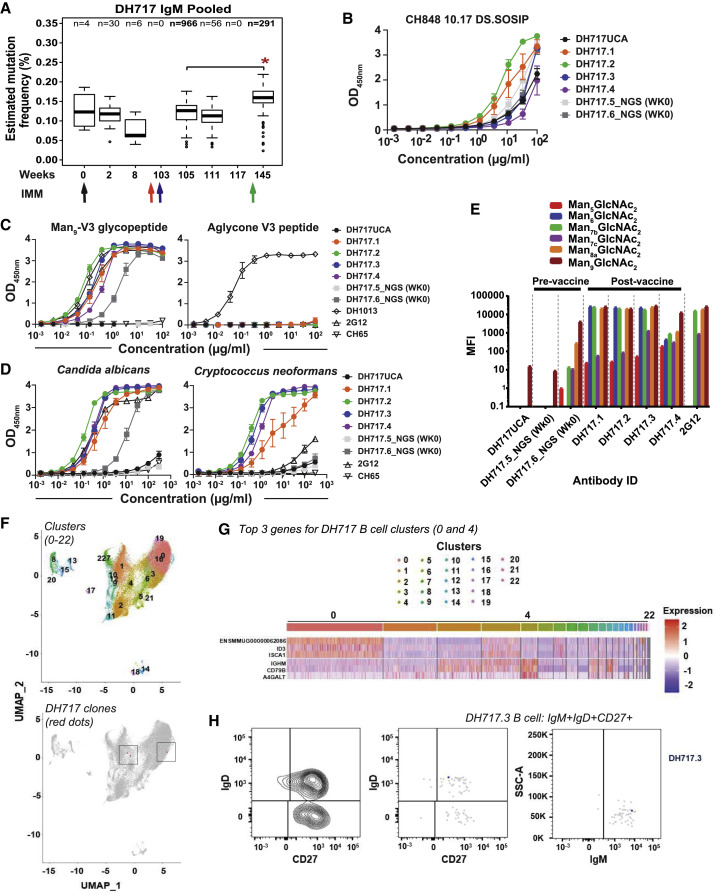
Figure 2.
The ontogeny and dynamics of HIV-1 vaccine-induced DH717 FDG Abs
(A) Absolute counts of unique VH reads for blood-derived DH717 lineage members at longitudinal time points (weeks). ∗p < 0.0001 (non-parametric Wilcoxon test) for increase in the distribution of estimated mutation frequencies at week 105 compared to week 145. The arrows indicate immunization time points with the initial Man9-V3 monomer (black arrow), Man9-V3 multimer (red arrow), nanoparticle (green arrow), and the first of five recombinant HIV-1 Env trimers (blue arrow).
(B–D) Recombinant mAbs bearing the week 0 (DH717.5_NGS and DH717.6_NGS), near-germline (DH717UCA), and post-vaccination (DH717.1-DH717.4) Ab sequences were tested in ELISA for binding soluble stabilized form of recombinant HIV-1 Env trimer (B), Man9-V3 (C), and heat-killed yeast antigens (D). CH65 was used as a negative control mAb, whereas biotinylated trimer-specific mAbs (see Figure S1), and peptide (DH1013)- and Env glycan (2G12)-specific mAbs were used as positive controls. ELISA data shown are averaged from 3–5 separate assays for each mAb tested; error bars represent SEM. Binding was measured at OD450nm.
(E) Binding of DH717 mAbs to individual glycans in a luminex binding assay, where glycan reactivity was reported as MFI after background subtraction.
(F) Top panel: UMAP visualization of 23 transcriptionally unique immune cell clusters from peripheral blood. Bottom panel: clonally related DH717 B cells (shown in red dots within the rectangle outlines) detected among B cell clusters.
(G) Heatmap summary of gene enrichment analysis across the 23 clusters; gene upregulation is shown in red compared to gene downregulation shown in blue. Shown are the top three genes identified for the DH717 B cell clusters 0 and 4.
(H) Representative sort plots demonstrating IgM+IgD+CD27+ phenotype of a DH717 lineage B cell (blue dot) among antigen-specific B cells that were sorted (gray dots). The IgM sort plot displayed only the number of events from the IgD+CD27+ population as well as DH717.3 B cell. The contour plot (left) was used to establish the analysis gates.
See also Data S5 and STAR Methods.
Figure S1.
Characterization of HIV-1 vaccine-induced FDG Ab lineage, DH717, related to Figure 1
(A) DH717 mAbs as well as biotinylated (b) control mAbs were tested in ELISA for binding to soluble recombinant HIV-1 Env trimer (CH848 strain) in a single ELISA. Binding titers were reported as Log AUC. Control mAbs targeted different epitopes on HIV-1 Env: both Env glycan and peptide – PGT121, PGT125 and PGT128; linear peptide (V3 loop) reactive – 19B; glycan-only reactive – 2G12; co-receptor binding site – 17B; and CD4 binding site – DH493). (B) Binding titers of DH717 and control mAbs with heat-killed yeast antigens Candida albicans or Cryptococcus neoformans. MAbs were tested in standard diluent or diluent spiked with 0.5M D-mannose. Binding data were representative of duplicate ELISAs. (C) Kifunensine [Kif] enriches Man9-glycans on recombinant HIV-1 Envs (Saunders et al., 2017a) (see STAR Methods). DH717 mAbs were tested for neutralization against a multi-clade panel of env-pseudotyped HIV-1 bearing Envs with heterogeneous [(-)Kif] or Man9-enriched glycans [(+)Kif]. Neutralization was tested in TZM-bl cells in a single experiment and titers are reported as IC50 in μg/ml. (D) Negative stain 3D reconstruction of DH717 I-shaped Ab. Map shown as transparent surface. Atomic model of two DH717.1 Fabs shown as ribbon diagrams, fit as rigid bodies into the 20-Å resolution NSEM map using UCSF Chimera’s automatic fitmap function. As fit, the VH cysteine at residue 74 pointed toward one another (arrow) and the terminal sulfurs were 3.5 Å apart. (E) View of DH717.1 monomer crystal structure showed contact with aspartate, VH residue 34 (D34), and the man-9 ligand (green); electron density shown as mesh. (F) VH sequence alignment of DH717.1-4 with the inferred near-germline unmutated common ancestor (UCA). DH717.1, 0.2, and 0.4 have cysteine at residue 74, whereas DH717.3 and the UCA do not. (G) Close up views of DH717.1 dimer crystal structure, labeled by region. (H) Size exclusion chromatography (SEC) profile of DH717.1 C74S IgG. The SEC runs were performed on a Superose 6 Increase 10/300 column in running buffer composed of 10mM HEPES, pH 7.3, 150mM NaCl, 5% Glycerol. (I) Immobilization of DH717.1 wild-type (black lines) and DH717.1 C74S (gray lines) on anti-Fc surface. At least three technical repeats are shown for each. The Abs were captured on a CM5 chip by flowing 200 nM of the Ab over a flow cell immobilized with ~9000 RU of anti-human Fc Ab. Following this immobilization, binding was measured by flowing over 200 nM solution of Env in the running buffer HBS-EP+ that is composed of 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20. Env binding was shown in Figure 1K.
Structural details of Env glycan recognition by DH717 Abs
We obtained a 2.6-Å crystal structure of monomeric DH717.1 Fab complexed with Man9-V3 (Figures 1E and 1G; Data S4). DH717.1 VH interacted with the Man9-glycan using three HCDR loops to form a binding pocket into which Man9-glycan was inserted (Figures 1G and S1E). No interactions were observed with the peptide itself, reminiscent of the glycan-only binding of 2G12 (Doores et al., 2010b; Murin et al., 2014; Stanfield et al., 2015; Calarese et al., 2003; Chuang et al., 2019). We also determined a 3.5-Å crystal structure of unliganded dimeric DH717.1 Fab (Figures 1F, 1H, and S1G; Data S4). The structure revealed two disulfide-linked Fab dimers in the asymmetric unit, with a disulfide bond linking two Fabs via VH gene residues cysteine 74 (Figure 1H). DH717.3, which only showed Y-shaped Abs, had a serine at this position (Figure S1F), and disrupting the disulfide in DH717.1 with a cysteine to serine (C74S) mutation resulted in loss of DH717.1 I-shape forms (Figure 1I) and reduction in binding and neutralizing activity (Figures 1J, 1K, and S1H). DH717.1 Fab monomer did not bind recombinant HIV-1 Env trimers, whereas the Fab dimer bound with micromolar affinity, and the DH717.1 IgG mAb bound with nanomolar affinity, thus demonstrating a role for avidity in Env glycan recognition of FDG Abs (Data S1). These data showed that an inter-Fab disulfide linkage was the mechanism of Fab-dimerization for DH717 Abs that conferred biological activity against HIV-1.
Ontogeny of vaccine-induced DH717 FDG Abs
We found VH genes prior to vaccination (week 0) with similar VHDJH rearrangements as the DH717 VH genes from Abs isolated post vaccination (Figure 2A), indicating that they are clonally related. Week 0 DH717 VH genes were mutated IgMs, thus suggesting a role for host or environmental antigens in their initiation. DH717 B cell lineage was expanded following immunizations with monomeric and multimeric Man9-V3 (Francica et al., 2019; Alexander et al., 2000; Alam et al., 2017) as well as a soluble recombinant HIV-1 Env trimer (Saunders et al., 2019) bearing near-native high mannose glycan patch (Figure 2A). Recombinant mAbs (DH717.5_NGS and DH717.6_NGS) bearing DH717 clonally related mutated IgM VH found at week 0 paired with post-vaccination DH717.1 Vλ had minimal or no binding to soluble recombinant Env trimers, Man9-V3, Candida albicans yeast antigens, and high mannose glycans (Figures 2B–2E). Additionally, the DH717 mAb bearing the near-germline unmutated common ancestor (UCA) genes weakly bound Candida albicans glycans but did not bind Man9-V3, whereas the mature DH717 lineage mAbs demonstrated affinity maturation after Man9-V3 immunization as shown by increased binding to Man9-V3, soluble recombinant HIV-1 Env trimer, and Candida albicans or Cryptococcus neoformans yeast glycans (Figures 2B–2E).
To define the nature and characteristics of circulating DH717 FDG B cells, we interrogated the transcriptome of blood DH717 B cells. We studied 23 transcriptionally unique B cell clusters in blood DH717 lineage B cells, 1 week after final immunization with the nanoparticle form of Man9-V3 (Figure 2F). The DH717 B cells were found in clusters 0 and 4 (Figure 2F) that had 315 enriched genes that achieved statistical significance with a 0.3–1.4 log fold change (Figure 2G; Data S5). B cell receptor signaling pathway was enriched in both clusters 0 and 4, and IgM was the top gene enriched in cluster 4 with statistical significance (log fold change = 0.9, p < 0.001) (Data S5). Interestingly, we found genes for IgM, phosphatases, and FcR-like proteins enriched in clusters 0 and 4, similar to transcripts previously found in human blood IgM memory and marginal zone (MZ) B cells (Weller et al., 2004). Moreover, we found that IgM DH717.3 displayed an IgM+IgD+CD27+ phenotype (Figure 2H) in agreement with previously identified human blood marginal zone B cells (Weill et al., 2009; Weller et al., 2004). Additional DH717 IgM Abs displayed IgM+ and IgD+CD27− or IgD−CD27+ phenotypes (not shown), thus implicating different differentiation pathways for DH717 lineage B cells.
Precursor FDG Abs in HIV-1-naive humans
We interrogated the B cell repertoire of nine HIV-1 naive individuals for candidate FDG Ab precursors that demonstrated Man9-V3 or yeast binding, and/or contained I-shaped Abs. Antigen-specific flow sorting revealed that candidate FDG Ab precursors were present at an average frequency of 1 per ∼340,000 B cells (Figure 3 A), which was considerably greater than the frequencies of bnAb precursors that targeted different HIV-1 Env epitopes (Steichen et al., 2019; Havenar-Daughton et al., 2018). From flow sorting, 37 glycan-reactive Abs used a variety of heavy and light chain genes, although 46% used Vκ2-24 for the light chain (Figure 3B) in contrast to a Vκ2-24 frequency of 0.3%–0.6% in HIV-1 naive individuals (DeKosky et al., 2015), demonstrating immunoglobulin repertoire skewing as previously reported for natural Abs (New et al., 2016). The majority of flow sorted candidate FDG precursor B cells (76.1%, 405/532) were IgM+IgD+CD27+ B cells (Figure 3C).
Figure 3.
Characteristics of HIV-1 Env glycan-reactive neutralizing Abs in the B cell repertoire of naive humans
(A) Repertoire analysis of blood-derived Env glycan-reactive B cells in nine HIV-1 naive individuals. Enriched B cells were studied via different sort strategies listed that used fluorophore-labeled Man9-V3 or soluble stabilized recombinant HIV-1 Env trimers.
(B) Immunogenetics of candidate Env glycan-reactive B cells (N = 37) that were isolated from individuals listed in (A).
(C) Repertoire of Env glycan-reactive B cells with IgM+IgD+CD27+ phenotype among six individuals. Flow cytometry data were analyzed from B cells sorted via strategies listed in (A); strategy “c” was used for LKP04.
(D–F) NSEM 2D class averages of DH1005 Ab with I-shaped Ab conformations denoted by “I” in image. 2D class averages of DH1005-SOSIP complex shows Fab-dimer binding to soluble stabilized recombinant HIV-1 Env trimer. Fc domain is not visible here because it lies outside the circular mask used during class averaging. Shown also are 2D class averages of DH1009 (E) and DH1010 (F) that displayed I-shaped Abs.
(G) Seven Env glycan-reactive mAbs were tested in ELISA for binding Man9-V3 and non-glycosylated aglycone V3 peptide and heat-killed yeast antigens. Data shown are from a representative ELISA. Reference mAbs included peptide-reactive DH1013, Env glycan-reactive DH717.2 and 2G12, trimer-reactive PGT151, and negative control CH65 Abs.
(H) Only DH1005 neutralized multi-clade env-pseudotyped HIV-1 bearing Envs with Man9-enriched glycans [Kif]. Neutralization titers are representative of two assays in TZM-bl cells, and neutralization titers were reported as IC50 in μg/mL. Control mAbs were 2G12 and CH65.
See also Data S3 and STAR Methods.
Seven FDG Abs were chosen, based on glycan binding activity, for expression as mAbs (Data S3). Three of these, IgG3 DH1005, IgM DH1009, and IgM DH1010, displayed a mix of Y- and I-shaped Abs (Figures 3D–3F), and the I-shaped form of DH1005 bound soluble recombinant HIV-1 Env trimer (Figure 3D). All seven mAbs showed glycan-dependent binding to Man9-V3 and bound Candida albicans and Cryptococcus neoformans yeast (Figure 3G). Only IgG3 DH1005 neutralized env-pseudotyped HIV-1 bearing Man9-enriched Envs (Figure 3H) but did not neutralize wild-type HIV-1 with Envs bearing heterogeneous glycoforms (not shown), thus exhibiting a neutralization profile similar to DH717 Abs that we classified as FDG precursors. These data demonstrated that FDG Abs were abundant in the B cell repertoire of HIV seronegative humans and may be found in the pool of IgM+IgD+CD27+ marginal zone B cells (Weller et al., 2004; Weill et al., 2009).
DH1010 Fab-dimerized IgM mAb isolated from an HIV-1 naive individual had a free cysteine at position 74 in the VH gene, similar to DH717 mAbs isolated from an HIV-1 vaccinated RM. Of 77,336 immunoglobulin heavy and light chain gene pairs studied in three HIV-1 naive individuals (not shown), none contained a cysteine at position 74. Additionally, none of 201 germline VH genes in the human Cloanalyst Ig gene database (Ramesh et al., 2017; see STAR Methods) had C74, and <1% of the Abs in the Abysis database (Swindells et al., 2017) had a VH gene encoding C74. These data demonstrated that human Abs with a cysteine in VH position 74 are rare but they may be selected by Man9-V3 and HIV-1 Env trimer as observed in RMs by selection of DH717 Abs.
Macaque SHIV-induced neutralizing FDG B cell lineages
Because the unique 2G12 bnAb arose in human HIV-1 infection (Buchacher et al., 1994), we sought to isolate FDG bnAbs from RMs infected with pathogenic SHIVs (Roark et al., 2021). Infection with HIV (Bonsignori et al., 2017) or a SHIV (Roark et al., 2021), with the same Env, elicited plasma bnAbs that targeted a high mannose glycan-containing epitope in humans and RMs, respectively. In one SHIV-infected RM (Roark et al., 2021), we interrogated the glycan-reactive memory B cell repertoires in blood and lymph node at week 52 post-infection, and blood at week 104 post-infection.
Autologous neutralizing FDG B cell lineage DH898
A seven-member IgG Ab clonal lineage, termed DH898 (Data S3), isolated from lymph node memory B cells at 52 weeks post SHIV infection, used macaque genes Vκ2 paired with VH1 encoding a 13-amino acid HCDR3 (Figure 4 A). DH898 mAbs mediated Env glycan-dependent neutralization of difficult-to-neutralize env-pseudotyped HIV-1 bearing Env from the autologous SHIV infection and neutralized heterologous env-pseudotyped HIV-1 bearing Envs with Man9-enriched glycans (Figure 4B) but neutralized none of 119 geographically diverse difficult-to-neutralize env-pseudotyped HIV-1 bearing heterologous Envs with heterogeneous glycoforms (Seaman et al., 2010; not shown). Like 2G12, DH898 mAbs demonstrated glycan-dependent binding to Man9-V3 (Figure 4C) and bound yeast glycans (Figure S2 A). DH898 mAbs also showed glycan-dependent binding to a soluble recombinant HIV-1 Env trimer (Saunders et al., 2019; Figures S2B and S2C). By NSEM, DH898 mAbs showed a mixture of canonical Y-shaped as well as I-shaped Abs (Calarese et al., 2003; Figures 4D and S2D).
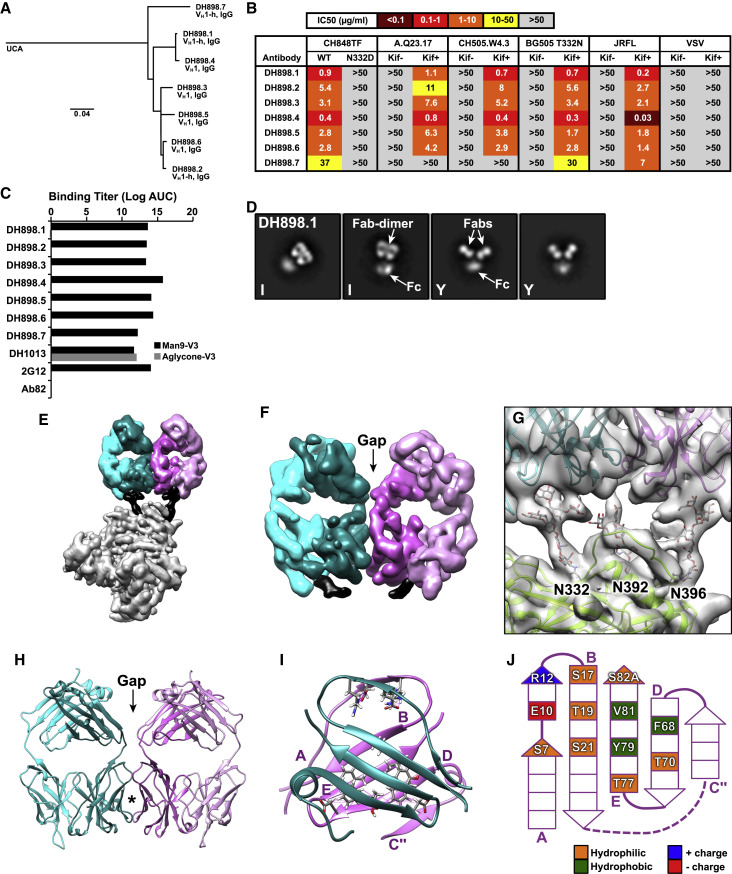
Figure 4.
Characteristics of HIV-1 Env glycan-reactive neutralizing B cell lineage, DH898, elicited by SHIV infection in RMs
(A) Phylogram and immunogenetics of DH898 neutralizing Abs isolated from a pathogenic SHIV-infected RM (6163).
(B) DH898 mAbs were tested for neutralization of env-pseudotyped HIV-1 bearing an autologous Env from SHIV infection. MAbs were tested for neutralizing HIV-1 bearing wild-type and glycan-deleted mutant Envs in TZM-bl cells. Data shown are from a representative assay and neutralization titers are IC50 in μg/mL.
(C) DH898 mAbs were tested for binding in ELISA to Man9-V3 (black bars) and non-glycosylated aglycone V3 peptide (gray bars). Binding titer was reported as log AUC. Control Abs tested included peptide-reactive DH1013, Env glycan-reactive 2G12, and negative-control anti-influenza CH65 mAbs. Data shown are representative of four assays.
(D) NSEM of DH898.1 IgG showing a mixture of I-shaped (I) and Y-shaped (Y) Abs.
(E) Segmented cryo-EM map of DH898.1 Fabs bound to a soluble stabilized HIV-1 Env trimer showing a single Fab-dimer bound to the trimer. Colors are as in Figure 1.
(F) Zoomed-in view of the Fab-dimer showing a gap between the two Fabs.
(G) A fitted model showing the Fab-dimer binding to high mannose glycans at Env positions 332, 392, and 396.
(H) Atomic model of the Fab-dimer showing a gap between the Fabs (arrow) and an interface between the Fv domains (star).
(I) The beta-sandwich view of the interface demonstrating that the beta- strands of the two VH domains are angled relative to one another.
(J) Schematic diagram of the Fab-dimer interface showing a patch of three hydrophobic or aromatic residues (green) surrounded by small hydrophilic residues (orange) and a complementary pair of charged residues (blue +, red −).
See also Figures S2 and S3, Data S3 and S4, and Videos S1 and S2.
Figure S2.
Characterization of neutralizing FDG Ab lineage (DH898) elicited by SHIV infection in RMs, related to Figure 4
DH898 mAbs were elicited by pathogenic SHIV (CH848TF) infection in RM6163 (see STAR Methods). (A) DH898 mAbs were tested in ELISA for binding to Man9-V3, and heat-killed yeast antigens Candida albicans or Cryptococcus neoformans, in the absence (black bars) or presence (white bars) of 1M D-mannose. Control mAbs are reactive to Env glycan (DH501 and 2G12), Env peptide (DH1013) or influenza (CH65). Binding titers were reported as Log AUC. Data shown were from a single ELISA that were in agreement with an independent experiment of DH898 mAbs binding to Man9-V3 and yeast antigens in the absence or presence of 0.5M D-mannose. (B) DH898 mAbs were tested in ELISA for binding soluble stabilized recombinant HIV-1 trimers, CH848 10.17 DS.SOSIP wild-type and mutant trimers, in a single experiment. Mutations in CH848 10.17 DS.SOSIP trimers included deletions of potential N-linked glycan sites in the V3 (N301A_N332A) or V1 (N133D_N138T) regions. Mutations also included amino acid insertions that filled glycan holes on the trimer (D230N_H289N). Binding levels were measured at OD450nm. CH848 10.17 SOSIP trimers were captured using PGT151. Biotinylated (B) 2G12, PGT128 (V3 glycan bnAb) and CH65 mAbs were tested as control Abs. (C) Binding levels of DH898 mAbs to CH848TF SOSIPv4.1 in one ELISA, representative of at least two independent experiments. CH848TF SOSIP trimer was captured using anti-AVI mAb. 2G12, PGT151 and CH65 were tested as control Abs. (D) NSEM class averages for DH898 mAbs with lineage member indicated at upper left of each panel. At the lower left corner, each class average was identified as Y-shaped (Y), I-shaped (I), or ambiguous (A). (E) Sequence analysis of key residues within the Fab-dimer interface shows three hydrophobic or aromatic residues in the interface, similar to 2G12. VH interface residues were numbered according to standard Kabat numbering (Wu and Kabat, 1970). Amino acids at each position were indicated by their one-letter code and colored according to Taylor (Taylor, 1997), with polar groups orange, hydrophobic and aromatic groups in shades of green to yellow, positively charged groups blue and negatively charged groups red. Residues that are rare for a particular position, i.e., ≤ 1% in the abYsis database (Swindells et al., 2017), are indicated with an asterisk. Bottom row indicates VRC01 as a non-Fab-dimerized negative control. (F) Bar graph indicating the fraction of I-shaped Abs for wild-type (blue bars), strengthening mutants (green bars), or disrupting mutants (red bars) in DH898 mAbs. Mutations were engineered in the Ab VH genes. The % I-shaped was estimated by the fraction of particle images that sorted into I-shaped classes as indicated in each of the NSEM 2D class averages shown in G. (G) NSEM 2D class averages from of DH898.4 wild-type (WT), strengthening double mutations T19I and T70F, and disrupting double mutations R64D and F68D. The class averages shown represented the average of ~8,000 to 20,000 individual particle images classified and averaged into ten classes, arranged from the most populated class at the top left to the least populated at the bottom right, and marked as I-shaped (I), Y-shaped (Y), or ambiguous (A). (H) DH898.4 wild-type and mutant mAbs were tested in ELISA for binding to HIV-1 CH848TF SOSIP trimer. MAbs were tested in technical replicates within a single ELISA and binding levels measured at OD450nm; error bars represent standard error of the mean. (I) DH898.4 wild-type and mutant mAbs weres tested for neutralization against autologous (CH848TF) HIV-1 strain bearing Env with heterogeneous glycoforms as well as heterologous HIV-1 isolates bearing Envs Man9-enriched glycans [Kif] in TZM-bl cells. These data were generated in a single neutralization assay and titers were reported as IC50 in μg/ml.
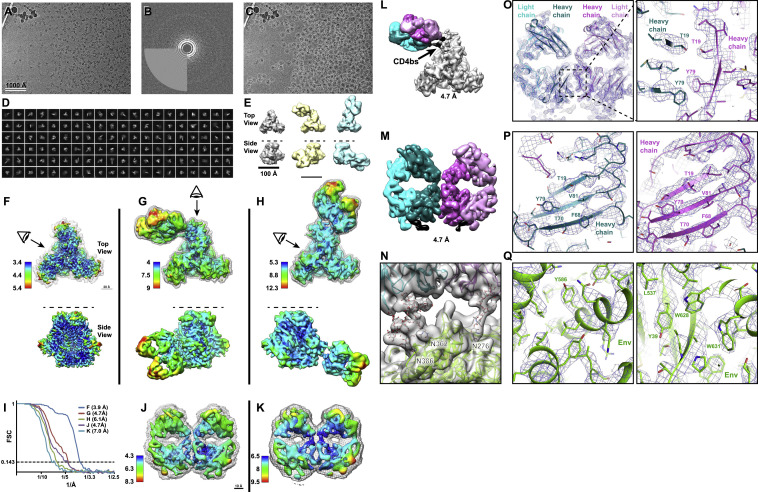
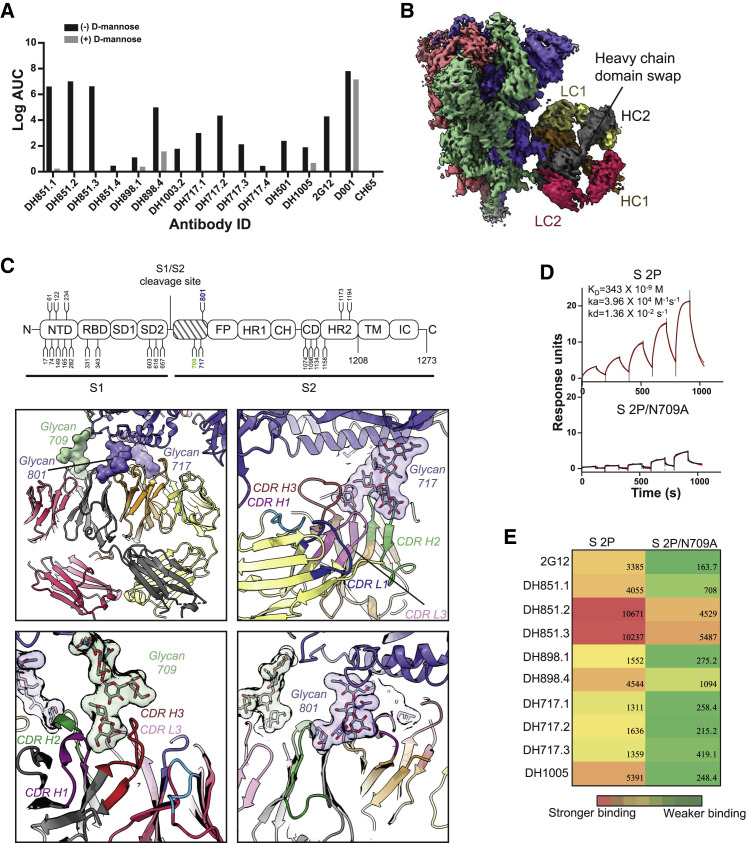
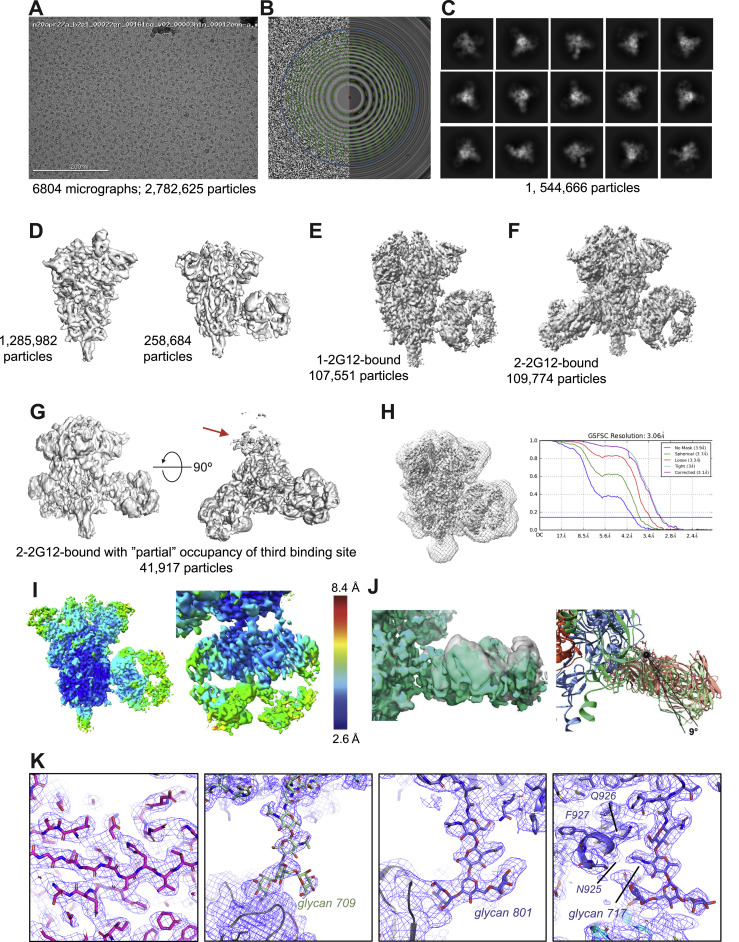
To visualize the DH898 epitope and Fab dimer interface, we determined cryo-EM structures of a soluble recombinant HIV-1 Env trimer bound to recombinant DH898.1 Fab (Figures 4E–4G and S3 ; Data S4; Videos S1 and S2). We identified three populations—an unliganded trimer, and two other populations, each with a single DH898 Fab dimer bound to two different epitopes. In one population, the Fab dimer bound multiple glycans at Env positions 332, 392, and 396, which included high mannose glycans targeted by 2G12 (Figures 4G and S3; Video S2), and was consistent with DH898 IgG mAbs dependence on glycan at Env position 332 for neutralization of the autologous strain (Figure 4B). In the second population, the Fab dimer bound glycans at Env positions 386, 362, and 276 around the Env CD4-binding site (Figures S3L–S3N; Video S2). In both complexes, the Fab dimer interfaces were well resolved and did not show VH domain-swapping (Figures 4F and S3M). Instead, the DH898 Fabs dimerized via a side-by-side association of their VH domains, mediated by interfacial aromatic and hydrophobic residues (Figures 4H, S2E, S3O, and S3P). The DH898 Fab dimer interface also involved a glutamate-arginine pair at residues E10 and R12 that were located such that they could form salt bridges between the two VH chains (Figure 4I-4J), analogous to a D72 and R57 aspartate-arginine pair seen in 2G12 (Calarese et al., 2003). Mutations predicted to disrupt the Fab dimer interface yielded a lower proportion of I-shaped Abs and loss of Ab binding activity. Conversely, mutations predicted to strengthen the Fab dimer interface resulted in higher proportion of I-shaped Abs, albeit without any enhancement of binding and neutralizing activity (Figures S2F–S2I), suggesting that the I-shaped conformation is necessary but not sufficient for enhanced function. These data demonstrated that DH898 were FDG Abs and suggested that they could be bnAb precursors.
Figure S3.
DH898.1 complex cryo-EM data processing summary and structural details, related to Figure 4
(A) Representative micrograph; (B) Power spectrum of micrograph, and fitted contrast transfer function; (C) Picked particles in white circles; and (D) Representative 2D class averages. (E) Ab initio volumes that showed three particle populations: free trimer, Fab-dimer bound to glycans near the CD4-binding site (bs), and Fab-dimer bound to glycans near the base of the V3 loop (left to right) seen in top and side views (top to bottom). Dashed line in side view indicated viral membrane location. (F) Top and side view of free soluble stabilized recombinant HIV-1 Env trimer (SOSIP) colored by local resolution. Mesh surface indicates mask used for FSC calculation. Eye indicates viewing direction in side view. (G) HIV-1 Env SOSIP with Fab-dimer bound to glycans near the CD4bs. (H) HIV-1 Env SOSIP with Fab-dimer bound to glycans near the base of the V3 loop. (I) Gold standard Fourier Shell Correlation (FSC) curves for F-H and J-K indicating global resolution ranging from 3.9 – 7.0 Å. (J) Local refinement of Fab-dimer from CD4bs particle set. (K) Local refinement of Fab-dimer only from V3-glycan bound particle set. (L) Segmented cryo-EM map showing Fab-dimer bound near the CD4bs. Gold-standard FSC resolution is indicated below each map. (M) Local refinement of the Fab-dimer bound near to the CD4bs of a HIV-1 Env SOSIP. (N) Close-up view of the epitope near the CD4bs with a HIV-1 Env SOSIP and Fab-dimer models shown as ribbons; glycans shown as sticks; and the cryo-EM map shown as a transparent surface. (O) (left) Local refined, density modified map of the DH898.1 Fab dimer with cryo-EM reconstruction shown as blue mesh and underlying fitted model in cartoon representation, and (right) Zoomed-in view of the Fab dimer interface with select interfacial residues shown as sticks. (P) Views of the Fab dimer interface rotated 90° clockwise (left) or counter-clockwise (right) relate to the view shown in panel O. (Q) Zoomed-in views of Env regions in the apo structure.
Broadly neutralizing FDG B cell lineage DH851
A second four-member clonal lineage, termed DH851 (Data S3), comprised of one IgM and three IgG Abs, was isolated from week 52 lymph node and blood (IgG DH851.1-DH851.3) and week 104 blood (IgM DH851.4) memory B cells; DH851 Abs used macaque genes Vλ2 paired with VH2 encoding an HCDR3 of 15 amino acids (Figure 5 A). DH851 IgG mAbs potently neutralized both autologous and heterologous HIV-1 isolates bearing Envs with Man9-glycans (Figure S4 A). Only those DH851 mAbs that were class-switched IgGs in vivo (DH851.1, DH851.2, and DH851.3) broadly neutralized autologous and heterologous difficult-to-neutralize HIV-1 isolates bearing Envs with heterogeneous glycoforms, whereas DH851.4 (an IgM) did not (Figure 5B). DH851.1, DH851.2, and DH851.3 mAbs showed 26% neutralization breadth against a panel of 119 geographically diverse HIV-1 strains (Figure S4B) and were dependent on glycans at Env position 339, 386, and 392 glycans for neutralization (Figure S4C), which included high mannose glycans targeted by 2G12 (Chuang et al., 2019). Like 2G12, DH851 mAbs bound soluble recombinant HIV-1 Env trimers and Man9-V3 in a glycan-dependent manner, and bound Candida albicans or Cryptococcus neoformans yeast glycans (Figures 5C and S4D–S4G). Both bnAbs 2G12 and IgG DH851.2 cross-blocked each other for binding recombinant HIV-1 Envs (Figures 5D and 5E).
Figure 5.
Characteristics of HIV-1 Env glycan-reactive bnAb B cell lineage, DH851, elicited by SHIV infection in RMs
(A) Phylogram and immunogenetics of DH851 bnAbs isolated from a pathogenic SHIV-infected RM (6163).
(B) DH851 mAbs were tested for neutralization of env-pseudotyped HIV-1 bearing Envs with difficult-to-neutralize autologous and heterologous strains. Data shown are representative of multiple neutralization assays tested in TZM-bl cells with different Ab lots, and neutralization titers were reported as IC50 in μg/mL.
(C) DH851 mAbs were tested in ELISA for binding to Man9-V3 (black bars) and non-glycosylated aglycone V3 peptide (gray bars). Reference Abs tested included peptide-reactive DH1013, Env glycan-reactive 2G12, and negative control anti-influenza CH65 mAbs. Data shown are representative of four assays and binding titers were reported as Log AUC.
(D and E) DH851 and 2G12 mAbs were tested for blocking each other via competition ELISA. Data show percent blocking of biotinylated (b) DH851 and 2G12 binding to soluble recombinant HIV-1 Env monomeric protein by competing non-biotinylated mAbs at varying concentrations. 2G12 blocking was representative of three experiments, but DH851 blocking data shown were from a single experiment.
(F) NSEM of DH851.3 showing a mix of Y-shaped (Y) and I-shaped (I) Abs.
(G) Segmented cryo-EM map of DH851.3 bound to soluble stabilized recombinant HIV-1 Env trimer showing six Fab domains arranged in three Fab-dimer pairs and binding to the trimer with branched glycans well resolved. Trimer, gray; glycans, black; Fab VH, teal or dark pink; Fab Vλ, cyan or light pink.
(H) View enlarged, turned 40°, and at a higher contour level shows a single Fab-dimer with a gap in the middle indicating that the two Fabs sit side-by-side and are not VH domain-swapped.
(I) Zoomed-in view of the map shown as transparent surface with the fitted model shown in cartoon representation and glycans shown as sticks.
(J) Atomic model of the Fab-dimer showing a gap between the Fabs (arrow) and an interface between the Fv domains (star).
(K) The beta-sandwich view of the interface (starred region in J, turned 90°) showed the beta-strands of the two VH as nearly parallel. Pink beta-strands were labeled A–E.
(L) Schematic diagram of the Fab-dimer interface showing a patch of four hydrophobic or aromatic residues (green) surrounded by small hydrophilic residues (orange). Beta strands in the schematic correspond to the pink strands in (K) and were labeled A– E accordingly.
See also Figures S4 and S5 and Data S2, S3, and S4.
Figure S4.
Characterization of FDG bnAb lineage (DH851) elicited by SHIV infection in RMs, related to Figure 5
DH851 mAbs were elicited by pathogenic SHIV (CH848TF) infection in RM6163 (see STAR Methods). (A) DH851 mAbs were tested for neutralization against env-pseudotyped HIV-1 bearing Envs with Man9-enriched glycans [(+) Kif] and heterogeneous glycoforms [(-) Kif]. Neutralization was tested in TZM-bl cells and titers reported as IC50 in μg/ml. (B) Summary of neutralization profile for DH851 and 2G12 bnAbs that were tested against 119 difficult-to-neutralize multi-clade HIV-1 strains in TZM-bl cells (Seaman et al., 2010). (C) DH851 neutralization epitopes mapped on a SHIV bearing wild-type or mutant HIV-1 Ce1176-strain Envs. Mutant Envs had deletion of potential N-linked glycosylation sites that constituted the Env glycan-containing bnAb epitope targeted by glycan-only Abs as well as Abs that bound both Env glycans and peptide (GDIR motif) (Kong et al., 2013). While DH851 mAbs also neutralized HIV-1 Ce1176 strain, a mutation in the GDIR motif (D325N) did not impact the neutralization sensitivity of HIV-1 Ce1176 (not shown). (D) DH851 mAbs bearing sequences for clonally-related VH genes from week 0 (DH851.5, DH851.6 and DH851.8), computationally-inferred near-germline unmutated common ancestor (DH851UCA), and post-infection (DH851.1-DH851.4) Abs were tested in ELISA for binding to soluble recombinant HIV-1 Env trimer (SOSIP) of autologous CH848TF strain in triplicate experiments. Biotinylated (B) PGT151 (Env trimer reactive) and CH65 (influenza reactive) mAbs were tested as controls. Binding levels were measured at OD450nm. (E) DH851 mAbs were tested in ELISA for binding to CH848 10.17 DS.SOSIP in the presence (+) or absence (-) of 0.05-0.5M D-mannose, compared with control mAbs 2G12, PGT151 and CH65. Data shown were from a single experiment that was in agreement with two additional experiments of DH851 mAbs binding to CH848 10.17 gp120 ± D-mannose. (F) DH851 mAbs were tested in ELISA for binding to Man9-V3 and non-glycosylated aglycone V3 peptide in triplicate experiments; error bars represent standard error of the mean. DH1013 (peptide reactive), 2G12 (Env glycan reactive) and CH65 mAbs were tested as control mAbs. (G) DH851 mAbs were tested in ELISA for binding to heat-killed yeast antigens, Candida albicans or Cryptococcus neoformans in triplicate experiments; error bars represent standard error of the mean. 2G12 and CH65 were tested as control mAbs. (H) NSEM 2D class averages of DH851 mAbs displaying I- and Y-shaped Abs. (I) Sequence analysis of key residues within the Fab-dimer interface. (I) Bar graph indicating the fraction of I-shaped Abs for wild-type (blue bars) and strengthening mutants (green bars) in DH851 mAbs. Mutations were engineered in the Ab VH genes. The % I-shaped was estimated by the fraction of particle images that sorted into I-shaped classes as indicated in each of the NSEM 2D class averages from DH851.2 wild-type (WT) and strengthening double mutations A19T and T21L. The class averages shown represented images classified and averaged into five classes, and marked as I-shaped (I) or Y-shaped (Y).
NSEM of DH851 IgGs (DH851.1-DH851.3) revealed a mixture of Y- and I-shaped Abs (Figures 5F and S4H). In contrast, neither the mAbs bearing the near-germline DH851UCA genes nor the non-bnAb DH851 lineage member, DH851.4, were I-shaped (Figure S4H). The lack of I-shaped configurations for DH851.4 Ab and its inability to neutralize wild-type HIV-1 isolates bearing Envs with heterogeneous glycoforms suggested that Fab dimerization was required for heterologous HIV-1 neutralization.
In a 5.6-Å cryo-EM reconstruction of DH851.3 bound to a soluble recombinant HIV Env trimer, we observed Env bound to three side-by-side Fab dimers (Figures 5G, 5H and S5 ; Data S4). Unlike 2G12, and like the I-shaped DH898 Abs, the DH851.3 Fab dimers did not show VH-domain-swapping, with a clear gap between the two Fabs in the cryo-EM reconstruction. The DH851.3 Fab dimers bound Env glycans at positions 241, 289, and 334 located within a previously described Env glycan-containing bnAb epitope near the V3 loop (Kong et al., 2013) and consistent with the observed cross-blocking with 2G12 (Figures 5D and 5E). DH851 interactions with Env were dominated by its VH domains (Figure 5I), which were rotated such that the beta-strands of both chains were nearly parallel (Figures 5J and 5K), as opposed to being angled as seen in DH898 (Figure 4I); nevertheless, the interface featured hydrophobic and aromatic residues similar to 2G12 and DH898 (Figures 5L, S3P, S4I, and S5). DH851.4 and the DH851UCA lacked key hydrophobic residues (Figure S4I), and NSEM did not show Fab-dimerization (Figure S4H). In particular, DH851.4 has a T81R mutation that would place a bulky charged residue in the dimer interface, analogous to the I19R mutation that disrupts VH domain-swapping and Fab dimerization in 2G12 (Doores et al., 2010b). In DH851, a dimer strengthening double mutant, A19V+T21L, increased the fraction of I-shaped Abs as expected (Figure S4J).
Figure S5.
DH851.3 complex cryo-EM data processing summary and structural details, related to Figure 5
(A) Representative micrograph; (B) Power spectrum of micrograph, and fitted contrast transfer function; (C) Picked particles in white circles; and (D) Representative 2D class averages. (E) Ab initio volume of DH851, bound to a soluble stabilized HIV-1 Env trimer (SOSIP) seen in top and side view. Dashed line in side view indicated viral membrane location. (F) Refined 3D map, without filtering or sharpening and colored by local resolution from 5.5 to 12.5 Å, blue to red. (G) Map after local filtering and B-factor sharpening. Fab constant domains are noisy and at this contour level mostly disappear and are only seen as small, disconnected blobs, but greater details of the complex can be seen. (H) Gold standard Fourier Shell Correlation (FSC) curves indicated global resolution ranging from 5.6 to 8.7 Å. (I) 2D class average shows view of Fab in complex with a HIV-1 Env SOSIP looking down the central axis. Red box shows region where the DH851.3 Fab dimer contacts the adjacent SOSIP protomer. (J) Fitted model shown as ribbon diagram with cryo-EM map shown as transparent volume. (K) Close up of contact seen in panel (I) showed that it may involve the framework region 3 of the light chain, and Env residues around 426-431 or 105-112. (L) View of the DH851.3 Fab dimer bound to Env glycans. Cryo-EM reconstruction was shown as a blue mesh, with underlying fitted coordinates in cartoon and stick representation. (M) and (N) Zoomed-in views of the Fab dimer interface shown within the dotted rectangle in the left panel.
The structures of DH898 and DH851 Fab dimers suggested a common signature for non-domain-swapped VH-VH Fab dimerization mediated by interfacial hydrophobic and aromatic residues. This sequence signature was also observed in DH501 (Data S2A), a glycan-reactive Ab previously isolated from a macaque following HIV-1 Env immunization (Saunders et al., 2017a). NSEM of DH501 IgG showed the presence of I-shaped Abs (Data S2B). Recombinant DH501 Fabs also showed Fab dimer populations (Data S2C), and the previously published DH501 crystal structure (PDB: 5IIE) revealed two Fabs within the unit cell, with their VH-domains in a side-by-side orientation reminiscent of the DH851 and DH898 Fab dimers (Data S2D). Mutations designed to strengthen or weaken the crystallographic dimer interface, increased or decreased the proportion of I-shaped DH501 IgG population observed, respectively (Data S2E–S2G), thus confirming that the crystallographic dimer was in fact a biologically relevant Fab dimer. When docked as a rigid body, the DH501 dimer showed good fit to the DH851 cryo-EM map (Data S2H). Analysis of the DH501 crystallographic dimer thus provided high-resolution insights into a non-domain-swapped Fab dimer interface (Data S2J–S2O). These data revealed common sequence and structural features of non-domain-swapped FDG Abs DH898, DH851, and DH501 (Figure S2, Figure S4E and S4I; Data S2I) that includes the same VH hydrophobic pattern seen in 2G12, involving rare framework mutations.
We isolated another FDG bnAb lineage (DH1003) from a RM infected with a pathogenic SHIV different from the one that elicited DH851 (Data S2P and S3). Sequence analysis of DH1003 lineage Abs showed numerous hydrophobic residues among the key interface residues implicated in Fab dimerization (Data S2Q). Representative DH1003 mAbs showed predominantly I-shaped Abs by NSEM, demonstrated glycan-dependent binding to Man9-V3, bound to Candida albicans, and neutralized autologous and heterologous difficult-to-neutralize HIV-1 isolates bearing Envs with heterogeneous glycoforms (deCamp et al., 2014; Data S2S–S2V). These data demonstrated the reproducibility of FDG bnAb generation in macaque SHIV infections.
Ontogeny of SHIV-induced FDG Abs
Class-switched and mutated DH898 and DH851 clonally related VH genes were found in peripheral blood cells collected prior to macaque SHIV infection (week 0), suggesting that host or environmental antigens may have initiated FDG Ab lineages. We found 596 DH898 and 235 DH851 clonally related sequences that were predominantly IgM. To determine the binding profile of week 0 clonally related DH851 VH sequences, recombinant mAbs bearing week 0 DH851 IgG (DH851.5) or IgM (DH851.6 and DH851.8) clonally related VHDJH were paired with DH851 Vλ genes. These pre-infection DH851 mAbs bound weakly to yeast antigens and soluble recombinant HIV-1 Env trimers, but did not bind Man9-V3; in contrast, post-SHIV-infection DH851 mAbs robustly bound yeast antigens, Man9-V3, and soluble recombinant HIV-1 Env trimers (Figures S4D, S4F, and S4G). Pre- and post-infection IgM DH851 lineage Abs showed only Y-shaped populations by NSEM, whereas post-SHIV infection IgG DH851 lineage Abs showed Fab-dimerized, I-shaped populations (Figure S4H). These data suggested that FDG precursors existed as mutated or class-switched non-Fab-dimerized Abs prior to SHIV infection, with lineage stimulation potentially triggered by glycan-bearing host or environmental antigens.
Glycan-dependent binding to SARS-CoV-2 spike protein by FDG Abs
Site-specific glycan analysis demonstrated that oligomannose-type glycans, including high mannose constitutes 28% of the glycans on the recombinant spike (S) protein of SARS-CoV-2, the causative agent of the current COVID-19 pandemic (Watanabe et al., 2020). Because FDG Abs recognized high mannose glycans, we tested them for binding SARS-CoV-2 S protein. FDG mAbs, including bnAbs 2G12 and DH851, bound the SARS-Cov-2 S protein to varying degrees in a glycan-dependent manner (Figures 6 A and S6 ). A 3.1-Å cryo-EM reconstruction revealed a 2G12 Fab dimer bound to the S2 subunit of the SARS-CoV-2 S protein, with high-mannose glycans 717 and 801 from one protomer and glycan 709 from the adjacent protomer clustered together to form the 2G12 epitope (Figures 6B, 6C, and S7 ). The VH domain-swapped configuration of the 2G12 Fab dimer and its binding interface with the SARS-CoV-2 S protein were well-resolved, with the bound glycans clearly visible in the cryo-EM map. Deletion of glycan 709 reduced or eliminated 2G12 binding to the panel of FDG Abs tested (Figures 6D, 6E, and S6), thus demonstrating a common epitope for the FDG mAbs on the SARS-CoV-2 S protein. Additionally, we found that 2G12 also bound the SARS-CoV-2 and MERS S ectodomains (Figure S6).
Figure 6.
Glycan-dependent binding of FDG Abs to recombinant SARS-CoV-2 spike
(A) FDG mAbs were tested for binding recombinant SARS-CoV-2 Spike (S) in ELISA. Ab binding was assessed in the absence (−) or presence (+) of 1M D-mannose. Binding Ab titers were reported as Log AUC. Controls were SARS-CoV-1 RBD (D001) and influenza HA (CH65) reactive mAbs. Data shown are from a representative assay.
(B) Cryo-EM reconstruction of 2G12 in complex with recombinant SARS-CoV-2 S. The cryo-EM map was colored by chain. SARS-CoV-2 S chains were colored salmon, green, and blue. The 2G12 chains are colored dark gray and orange for the heavy chains (HC), and yellow and dark pink for the light chains (LC).
(C) Top: schematic showing domain organization of SARS-CoV-2 S, with positions of N-linked glycosylation sequons numbered and shown as branches. Bottom: zoomed-in view of domain-swapped, dimerized 2G12 Fab interacting with SARS-CoV-2 S. The structure was colored by chain as in (B), with 2G12 and SARS-CoV-2 S shown in cartoon representation and the interacting glycans in surface representation.
(D) Binding of 2G12 Fab dimer to (top) unmutated and (bottom) N709A mutant recombinant S proteins, measured by SPR using single-cycle kinetics. The black lines show the data and the red lines show the fit of the data to a 1:1 Langmuir binding model.
(E) Binding of the unmutated and N709A mutant S proteins to a panel of FDG Abs measured by SPR; data shown as a heatmap for Log AUC binding.
See also Figures S6 and S7 and Data S4.
Figure S6.
Characterization of glycan-dependent binding of FDG Abs to recombinant SARS-CoV-2 spike protein, related to Figure 6
(A) FDG, SARS-CoV-1 RBD (D001), and influenza HA (CH65) mAbs were tested in ELISA for binding to recombinant SARS-CoV-2 Spike (S) protein. mAb binding was assessed in the absence (-) or presence (+) of D-mannose [1M] to determine if free high mannose can outcompete glycans on the S protein for binding. Binding Ab titers reported as Log AUC were shown in Figure 1. (B) FDG mAbs were tested in ELISA for binding to a set of commercially available constructs expressing the SARS-CoV-2 S1 and S2 extracellular domain (left), S2 domain (middle), and the receptor binding domain (right). Black and white bars show binding in the absence and presence of D-mannose [1M], respectively. Binding Ab titers were reported Log AUC. (C) We tested 2G12 mAb for binding to (from top to bottom) soluble stabilized recombinant HIV-1 Env trimer (CH505TF SOSIP), Man9-V3 and non-glycosylated aglycone V3 peptide, and recombinant SARS-CoV-2 spike ectodomain. HIV-1 CH505TF SOSIP was captured using mouse anti-AVI-tag mAb, whereas SARS-CoV-2 ectodomain and peptides (Man9-V3 and Aglycone) were captured using streptavidin. Blue and red symbols indicated binding in the absence and presence of D-mannose [1M], respectively. Binding was measured at OD450nm. All ELISAs (A-C) were done using BSA-based buffers (see STAR Methods). Data shown are from a representative assay. (D) Size-exclusion chromatogram of protein A affinity purified 2G12 IgG. (E) NSEM 2D class averages of (top) 2G12 IgG dimer, (bottom) 2G12 IgG monomer and (right) 2G12 Fab obtained by digesting 2G12 IgG monomer with papain. (F) SPR sensorgrams showing binding of 2G12 IgG dimer (red line) and 2G12 IgG monomer (black line) to the SARS-CoV-2 S protein. (G) We tested FDG mAbs for binding to the unmutated SARS-CoV-2 S and the N709-glycan deleted variant. Binding was assessed by SPR by capturing the unmutated spike and the N709-glycan deleted spike on flow cells 2 and 4 of a streptavidin coated (SA) chip, and flowing over a 200 nM solution of each Ab simultaneously over all four flow cells. Flow cells 1 and 3 were used as reference flow cells for flow cells 2 and 4, respectively. Buffer blanks were run in a similar manner and the sensorgrams were double-referenced by first subtracting the signal from the reference flow cell and then subtracting the reference-corrected buffer blank. CR3022 IgG and ACE-2 tagged with a mouse-Fc region were used as controls. (H) Binding of 2G12 to spike proteins of SARS-CoV-2 (red), SARS-CoV (blue) and MERS-CoV (green). The SARS-CoV-2/2G12 complex is shown with the spike in red and the bound 2G12 as a transparent pink surface, with the glycans contacting 2G12 shown as sticks and the respective Asn residues as spheres. Similar glycosylation was observed for the SARS-CoV and MERS-CoV S proteins (Asn residues shown as spheres). SPR binding data shown for SARS-CoV-2 (red), SARS-CoV (blue) and MERS-CoV (green) S proteins. The data shown are representative of three independent experiments, and for each dataset the graph showed three technical replicates. IgGs were captured on flow cells of a CM5 chip immobilized with human Anti-Fc antibody (8000RU). 200 nM solution of the SARS-CoV-2 spike was flowed over the flow cells. The surface was regenerated between injections by flowing over 3M MgCl2 solution for 10 s with flow rate of 100μl/min.
Figure S7.
Cryo-EM data processing details for SARS-CoV-2 S protein complex with 2G12, related to Figure 6
(A) Representative micrograph. (B) CTF fit (C) Representative 2D class averages. (D) Maps for (left) unliganded and (right) 2G12-bound S obtained after 3 D classification. (E-G) Refined maps for SARS-CoV-2 S protein bound to (E) 1-2G12, (F) 2-2G12-, and (G) 2-2G12 (with partial occupancy at the third binding site) Fab2 molecules. Red arrow in (G) points to disordered 2G12 Fab2 bound at the third binding site. (H) (Left) Map combining all particles and focusing refinement on the region within the masks that is shown as a gray mesh overlaid on the final refined map shown as a gray surface. (Right) Fourier shell correlation curves. (I) (Left) Cryo-EM reconstruction of 2G12 bound to the SARS-CoV-2 spike colored by local resolution. (Right) Zoomed-in view showing the cryo-EM reconstruction of the bound 2G12 Fab. (J) (Left) Two distinct states were resolved from the cryo-EM data by heterogeneous classification. Density for the two observed states were shown in green and gray. (Right) Cartoon representation of the SARS-CoV-2 S-protein (bright green, bright orange, blue) and the two 2G12 orientations. The axis of rotation hinged around glycan 709 is represented by a gray cylinder. (K) Zoomed-in view of (from left to right) a region in the S2 domain with map shown as blue mesh and fitted model shown as sticks; glycan 709; glycan 801; glycan 717 bound to 2G12. While not in direct contact with the bound antibody, the HR1 helix may play an indirect role in the binding by stabilizing glycan 717 via a stacking interaction with residues N925 and Q926.
Previous reports indicated that the 2G12 bnAb was not polyreactive for binding recombinant human autoantigens (Haynes et al., 2005; Liu et al., 2015), but a recent study demonstrated that glycan-dependent HIV-1 Env bnAbs also bound host cells from HIV-1 uninfected individuals in a glycan-dependent manner (Blazkova et al., 2019). Similarly, we found that 12 of 23 (52%) human or macaque FDG Abs and 2G12 bnAb bound HEp2 cells but were either weakly or non-reactive with a panel of human autoantigens (Figure 7 ). The specificity of FDG mAbs for glycans suggested that HEp2 cell-FDG reactivity was glycan-mediated.
Figure 7.
Polyreactivity profile of FDG Abs
(A) Indirect immunofluorescence assay testing reactivity of FDG Abs in HEp-2 cells. Each mAb was tested at 50 and 25 μg/mL in duplicate reactions; a representative of the replicate data was reported. Positivity scores were determined relative to RM positive (DH1037) and negative (DH570.30) control mAbs. Staining patterns were identified using the Zeus Scientific pattern guide.
(B) Images of representative mAbs staining of HEp-2 cells. Ab reactivity pattern in HEP-2 cells were described in (A).
(C) Nine autoantigens were tested for reactivity by FDG Abs using a commercially available AtheNA Multi-Lyte ANA kit (Zeus Scientific). Serially diluted mAbs were tested for binding and the data analyzed using an AtheNA software. The dash lines represented the positivity score (121 units), which was consistent across independent experiments. 4E10 and CH65 represented positive and negative control mAbs, respectively. Data shown were for representative FDG mAbs that were reactive with HEp-2 cells (A and B).
See also STAR Methods.
Discussion
In this study, we identified a new category of prevalent glycan-reactive Abs that target diverse pathogens including HIV-1, SARS-CoV-2, and yeast. Although the Fab dimerized architectures of these glycan-reactive Abs bear resemblance to the unique glycan-reactive, VH-domain-swapped HIV-1 bnAb 2G12, they differ from 2G12 in their diverse mechanisms of Fab dimerization that occur without VH domain swapping.
FDG Abs isolated prior to SHIV infection or HIV vaccination were mainly mutated IgM, thus raising the hypothesis for MZ or IgM-memory B cells being a source for these Abs. The B cell sources of natural Abs have been well characterized in the spleen and MZ of mice (Baumgarth, 2016; Lalor et al., 1989; Ichikawa et al., 2015), but the phenotype of the B cells that generate natural Abs in peripheral blood of humans are not fully established (Holodick et al., 2017; Griffin et al., 2011). Our data demonstrated that human and RM FDG B cells were predominately IgM+IgD+CD27+, in agreement with circulating MZ B cells described in humans (Weller et al., 2004). FDG and 2G12 bnAbs have relatively short HCDR3 segments, target only glycans, and are more common than HIV-1 bnAbs with longer HCDR3s (Haynes and Verkoczy, 2014; Steichen et al., 2019). FDG Abs also used varied immunogenetics, thus indicative of a category of Abs, unlike previous classes of bnAb lineages that use more restricted heavy and light chain genes (Zhou et al., 2013; Bonsignori et al., 2018).
We previously showed that in a clonal bnAb lineage (termed DH270), early lineage members only neutralized HIV-1 bearing Man9-enriched Envs, whereas mature bnAbs neutralized autologous and heterologous HIV-1 bearing Envs with heterogeneous glycoforms (Bonsignori et al., 2017), thus demonstrating evolution of Abs within a lineage to achieve bnAb status through recognition of Envs with heterogeneous glycoforms. Similarly, although FDG precursors generally neutralized only HIV-1 bearing Envs with high mannose glycans, FDG bnAbs neutralized HIV-1 bearing Envs with heterogeneous glycoforms.
Our data also demonstrated that FDG and 2G12 mAbs had limited polyreactivity. It was postulated that the recognition of a unique glycan patch on HIV-1 Env was key for 2G12 maturation and neutralization potency (Scanlan et al., 2002). FDG Abs can also recognize the HIV-1 Env glycan patch, thus supporting the hypothesis for affinity maturation of FDG B cell lineages via HIV Env selection. Moreover, we demonstrated that avidity was important for glycan recognition by FDG Abs as vaccine-induced DH717 Fab had minimal Env-binding compared to DH717 IgG. We reported two examples where the Y-shaped forms of the Abs were less optimal for HIV-1 Env glycan recognition. First, 2G12 mutations that disrupted its Fab dimer resulted in loss of glycan binding (Huber et al., 2010). Second, Y-shaped DH851.4 IgM showed decreased binding and limited neutralization capacity compared to the Fab-dimerized DH851.1-DH851.3 IgG Abs within the same lineage. Thus, through Fab dimerization, FDG Abs acquired improved affinity and/or avidity for binding Env glycans, including the high mannose patch targeted by 2G12.
FDG mAbs bound a glycan epitope in the S2 subunit of the recombinant SARS-CoV-2 S protein. Consistent with a quaternary epitope, 2G12 binding was sensitive to SARS-CoV-2 S conformation with cold denaturation of the 2P S ectodomain resulting in reduced 2G12 binding (Edwards et al., 2021). Although these HIV-1 Env-directed FDG mAbs did not neutralize SARS-CoV-2 (not shown), our results suggest that FDG bnAbs may be matured to glycans on SARS-CoV-2 S. It remains unknown whether SARS-CoV-2 S-directed FDG mAbs have non-neutralizing protective effects.
The RM from which the FDG bnAb DH851 clonal lineage was isolated had relatively modest heterologous neutralizing plasma Abs that mapped to glycans in the high mannose patch as well as the peptide motif (GDIR) (Roark et al., 2021). Thus, it is likely that FDG bnAbs that target only glycans were minor components of the B cell repertoire of this macaque.
Other modalities of Fab-dimerization have been recently described, including head-to-head dimerization mediated by the CDR loops and intermolecular Fab dimers (Plath et al., 2016). These Abs have been shown to target non-HIV antigens, including CD20 (Rougé et al., 2020) and malaria parasite (Imkeller et al., 2018), although the interactions they mediate differ substantially from the glycan interactions with the FDG Abs we describe in this study, where intramolecular Fab dimerization within the Ab is a mechanism for enhancing weak glycan interactions via avidity. Mouse B cell lines bearing mature 2G12 exhibited domain-swapped forms of the BCR (Doores et al., 2013), but it remains unknown if the membrane-bound BCR of FDG bnAbs is analogous to BCR cross-linking that results in B cell activation (Yang and Reth, 2010a, 2010b).
It is important to note that FDG bnAbs with the capacity to neutralize autologous and heterologous difficult-to-neutralize HIV-1 bearing heterogeneous glycoforms in this report were only isolated from SHIV-infected RMs. SHIV-induced DH851 and DH1003 FDG bnAbs were class-switched, suggesting B cell maturation events occurred in SHIV infection that did not occur during precursor expansion with vaccination. We have previously shown that HIV-1-infected individuals who make bnAbs have multiple immunoregulatory abnormalities that create a permissive immunological environment for bnAb development (Moody et al., 2016; Bradley et al., 2018; Roskin et al., 2020). It will be of interest to determine if SHIV-infected RMs that make bnAbs have similar immunoregulatory abnormalities and whether FDG bnAbs can be found in chronic HIV-1-infected humans. Our data showed that we expanded FDG B cell lineages via vaccination, thus implying that FDG B cells were not tightly controlled by immune tolerance. Finally, that FDG Abs can bind multiple glycan clusters and use multiple BCRs, combined with FDG precursors being well represented in human B cell repertoire make this new category of glycan-reactive B cells an attractive target to consider for induction by HIV-1 vaccines. Since the submission of this paper, Lee et al. have reported that the FDG Ab 2G12 can neutralize strains of influenza (Lee et al., 2021), further demonstrating the breadth of cross-binding activity of the 2G12 FDG Ab for glycosylated virus pathogens.
Limitations of the study
We evaluated glycan-reactivity of FDG Abs on recombinant HIV-1 Env and SARS-CoV-2 S proteins, which may exclude different glycoforms only found on these surface proteins on the intact viruses in vivo. The frequency of FDG precursors was estimated in peripheral blood cells of nine HIV-1 naive individuals, thus future studies in a larger cohort of individuals and in other immune tissues will be informative about the prevalence of these Abs. We defined blood-derived FDG B cells as MZ-like, thus future studies of spleen-derived MZ B cells will verify this pool of B cells as a source of FDG Abs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 | BD | Cat#552852; RRID:AB_394493 |
| CD14 | BioLegend | Cat#301832; RRID:AB_2563629 |
| CD16 | BD | Cat#557744; RRID:AB_396850 |
| IgD | Southern Biotech | Cat#2030-09; RRID:AB_2795630 |
| CD27 | BioLegend | Cat#302816; RRID:AB_571977 |
| CD20 | BioLegend | Cat#302336; RRID:AB_2563806 |
| CD38 | Beckman Coulter | Cat# IM2371U; RRID:AB_131330 |
| CD19 | BD | Cat#557791; RRID:AB_396873 |
| AbC-mAb | AVIDITY | AbC-mAb |
| Goat anti-human IgG-HRP | Jackson ImmunoResearch Laboratories | Cat#109-035-098; RRID:AB_2337586 |
| Mouse anti-monkey IgG-HRP | Southern Biotech | Cat#4700-05; RRID:AB_2796069 |
| Streptavidin-HRP | Thermo Scientific | Ref#21130 |
| Mouse-Human chimeric mAb D001 | Sino Biological Inc | Cat#40150-D001; RRID:AB_2827980 |
| Bacterial and virus strains | ||
| Simian-HIV bearing TF Envs | Li et al., 2016 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Glycopeptide | Alam et al., 2017 | Man9-V3 |
| Deglycosylated peptide | Alam et al., 2017 | Aglycone V3 |
| Nanoparticle glycopeptide | Francica et al., 2019 | Star-Man9-V3 |
| CH848.3.D0949.10.17CHIM.6R.SOSIP.664v4.1/293F | Bonsignori et al., 2017; Saunders et al., 2019 | CH848 10.17 |
| CH848.3.D0836.10.31CHIM.6R.SOSIP.664v4.1/293F | This paper; Bonsignori et al., 2017 | CH848 10.31 |
| CH848.3.D0358.80.06CHIM.6R.SOSIP.664v4.1/293F | This paper; Bonsignori et al., 2017 | CH848 80.06 |
| CH848.3.D1432.5.41CHIM.6R.SOSIP.664v4.1/293F | This paper; Bonsignori et al., 2017 | CH848 5.41 |
| CH848.3.D0526.25.02CHIM.6R.SOSIP.664v4.1/293F | This paper; Bonsignori et al., 2017 | CH848 25.02 |
| SARS-CoV-2 S1+S2 ECD | Sino Biological Inc | Cat #40589-V08B1 |
| SARS-CoV-2 S2 ECD | Sino Biological Inc | Cat #40590-V08B |
| SARS-CoV-2 RBD | Genescript | Cat#Z03483 |
| Critical commercial assays | ||
| EasySep Human Pan-B Cell Enrichment Kit | STEMCELL Technologies | Cat#19554 |
| ExpiFectamine 293 transfection reagents | Life Technologies, GIBCO | Cat#A14524 |
| BIOTIN-X-NHS | Cayman Chemicals | Cat#13316 |
| ZEUS IFA ANA HEp-2 Test System 25 × 12 wells | ZEUS Scientific | Cat #FA2400 |
| AtheNA ANA-II Plus Kit | ZEUS Scientific | Cat#A21101 |
| Chromium Single Cell V(D)J Enrichment Kit, Human B Cell | 10X Genomics | PN-1000016 |
| Chromium Single Cell 5′ Library Construction Kit | 10X Genomics | PN-1000020 |
| Chromium Next GEM Chip G Single Cell Kit | 10X Genomics | PN-1000127 |
| Chromium i7 Multiplex Kit | 10X Genomics | PN-120262 |
| Deposited data | ||
| DH717.1 Fab monomer in complex with man9 glycan | This paper | PDB: 6VTU |
| Crystal structure of the disulfide linked DH717.1 Fab dimer, derived from a macaque HIV-1 vaccine-induced Env glycan-reactive neutralizing antibody B cell lineage | This paper | PDB: 6XRJ |
| Cryo-EM structure of DH898.1 Fab-dimer from local refinement of the Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | This paper | PDB: 7L6M: EMDB: EMD-23145 |
| Cryo-EM structure of CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | This paper | PDB: 7L6O; EMDB: EMD-23124 |
| Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | This paper | PDB: 7LUA; EMDB: EMD-23519 |
| Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env | This paper | PDB: 7LU9; EMDB: EMD-23518 |
| Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from focused refinement | This paper | PDB: 7L09; EMDB: EMD-23097 |
| Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | This paper | PDB: 7L06; EMDB: EMD-23095 |
| Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12 | This paper | PDB: 7L02; EMDB: EMD-23094 |
| Cryo-electron microscopy reconstruction of antibody DH898.1 Fab-dimer bound to glycans 332, 392, and 396 of HIV Env CH848 10.17 SOSIP trimer | This paper | EMDB: EMD-23152 |
| Cryo-electron microscopy local refinement of antibody DH898.1 Fab-dimer bound to glycans 332, 392, and 396 of HIV Env CH848 10.17 SOSIP trimer | This paper | EMDB: EMD-23153 |
| Recombinant mAbs | This paper | MT470283–MT470354 |
| Gene expression data | This paper | PRJNA685659 |
| Experimental models: cell lines | ||
| HEK293T | ATCC | Cat #CRL3216 |
| Experimental models: organisms/strains | ||
| Candida albicans | This paper | SC5314 |
| Crytococcus neoformans | This paper | H99 |
| Software and algorithms | ||
| Cloanalyst Ig gene database | Ramesh et al., 2017 | https://www.bu.edu/computationalimmunology/research/software/ |
| Cell Ranger Single Cell Software Suite v3.1 | 10X Genomics; Zheng et al., 2017a | N/A |
| Surat r package v3.2.0 | Macosko et al., 2015; Satija et al., 2015; Stuart et al., 2019 | N/A |
| Unicorn 7.0.2 software | GE Healthcare | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Wilton B. Williams (wilton.williams@mc.duke.edu).
Materials availability
The data presented in this manuscript, and research materials used in this study are available from Duke University upon request and subsequent execution of an appropriate materials transfer agreement.
Data and code availability
The variable heavy and light chain gene sequences for recombinant mAbs were deposited in GenBank and can be accessed using the following accession numbers; GenBank: MT470283–MT470354. The transcriptome sequencing data were deposited in NCBI Sequence Read Archive (SRA) under Bioproject: PRJNA685659.The atomic models and cryo-EM reconstructions and generated during this study are available at PDB and EMBD (https://www.rcsb.org; http://emsearch.rutgers.edu) under the accession codes PDB: 6VTU, 6XRJ, 7L6M, 7L6O, 7LUA, 7LU9, 7L09, 7L06, and 7L02 and EMDB: EMD-23145, EMD-23124, EMD-23519, EMD-23518, EMD-23097, EMD-23095, EMD-23094, EMD-23152, EMD-23153.
Experimental model and subject details
Rhesus macaques (RMs)
All RMs were maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animals. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
Immunization of Indian origin RMs and blood draws were performed at (Bioqual Inc., Rockville, MD). The four animals in this study were as follows: RM5932 (male, 3.5 years old), RM5994 (male, ∼4 years old), RM5995 (female, 5.25 years old), and RM5996 (female, 5.25 years old). DH717 lineage mAbs were isolated from RM5996. All vaccine doses were divided in to equal volume and injected in the bilateral quadriceps muscles. The RMs were first immunized with a dose-escalation of Man9-V3 monomer (50 μg, 100 μg, and 500 μg) each formulated in 25 μg GLA-SE adjuvant. 50 μg of monomeric Man9-V3 was administered at weeks 0, 6, and 12. 100 μg of monomeric Man9-V3 was administered at weeks 18 and 24. 500 μg of monomeric Man9-V3 was administered at weeks 50 and 56. Dose escalation of monomeric Man9-V3 was then followed by 300 μg of Man9-V3 multimer (∼6-mer) scaffolded onto N-(2-Hydroxypropyl) methacrylamide (HPMA) linear copolymer in 25 μg of GLA-SE given at week 71. Subsequently, RMs were immunized with a series of sequential soluble stabilized recombinant HIV-1 Env trimers (termed SOSIPs) (200 μg each) adjuvanted in 1 mg of Hiltonol Poly ICLC adjuvant (Oncovir). HIV-1 Env SOSIP trimers used for immunization were derived from CH848 Env strains previously identified as candidate immunogens for eliciting bnAbs that target glycans on the Env high mannose patch in conjunction with the Env peptide backbone (Bonsignori et al., 2017); 10.17 (week 103), 10.31 (week 109), 80.06 (week 115), 5.41 (week 127) and 25.02 (week 135). RMs were finally immunized with Star-Man9-V3 bearing the PADRE T cell helper epitope (300 μg) in 25 μg GLA-SE at week 143. Blood samples were collected two weeks post-immunization. The RM immunogenicity study in this paper was designed to evaluate the immunogenicity and dose of Man9-V3 monomer and to determine the effect of multimerization of Man9-V3 and later full envelope trimers, as boosts. The rational for the doses used were based on our previous experience with peptide immunogens (Zhang et al., 2016). Thus, we designed a dose escalation immunization study with the Man9-V3 monomer to study its immunogenicity and determine whether increased valency by multimerization improved the immunogenicity of the glycopeptide. Man9-V3 was previously used as a bait to isolate an HIV-1 Env bnAb that targeted N332 glycan in the high mannose patch in conjunction with the GDIR motif in the Env peptide backbone (Alam et al., 2017), both of which comprise the V3 glycan bnAb epitope. Thus, Man9-V3 was postulated to be a possible prime for bnAb precursors that target Env-glycan containing epitopes, including V3 glycan bnAbs; and the 5-valent soluble recombinant HIV-1 Env trimers were subsequently postulated to be candidate immunogens to boost these bnAb precursor B cells (Bonsignori et al., 2017). Hence, our approach to prime with Man9-V3 and boost with multimerized Man9-V3 and then the Env trimers. However, our studies showed that Man9-V3 and one of the trimers (CH848 10.17) were able to select FDG B cells rather than V3-glycan bnAbs, and are thus candidate immunogens for future vaccine strategies to elicit FDG Abs.
SHIV infection of Indian RMs were performed at (Bioqual Inc., Rockville, MD) and previously described (Roark et al., 2021). HIV-1 transmitted-founder (TF) Envs were derived from viruses that established infection in humans, thus TF Env-bearing SHIVs (Li et al., 2016) provided a novel model for studying HIV-1 infection as described (Roark et al., 2021). Blood and plasma samples were collected for binding and neutralization assays. HIV/SHIV bearing CH848TF Env was referred to as an autologous virus strain. DH851 and DH898 mAbs were isolated from a single macaque (RM6163; female, ∼5 years old) that was infected with a SHIV bearing HIV-1 CH848TF Env strain, and DH1003 mAbs were isolated from a different macaque (RM10N011; male, ∼8 years old) that was infected with a SHIV bearing HIV-1 BG505.T332N Env strain.
HIV-1 naive humans
Our cohort of nine HIV-1 seronegative adults aged 18-48 years (mix of both males and females) were bled for leukapheresis PBMCs in the summer of 2019 or earlier, thus it is unlikely that they were COVID-19 convalescent subjects. From our observation that FDG mAbs bound glycans on SARS-CoV-2 spike, we hypothesized that FDG Abs can be elicited by SARS-CoV-2 spike high mannose glycans and this is now currently being actively explored in COVID-19 convalescent subjects.
Candida albicans (SC5314) and Cryptococcus neoformans (H99) preparation
A single colony of yeast was inoculated into 250 mL YPD broth and grown for 2 days at 30°C on a shaking incubator set at 225 rpm. The cells were harvested and washed thrice with PBS. The pellets were resuspended in 11 mL PBS and the cell concentration was determined (C. albicans, 3.40x10e9 CFUs/ml and C. neoformans, 1.62x10e9 CFUs/ml, respectively). To heat-kill the yeast, the cells were incubated for 24h at 60°C in a water bath. Three hundred microliters of the heat-killed yeast were plated onto YPD agar, incubated for 7 days to ensure no viable yeast were present.
Method details
Synthesis of polymer-based glycopeptide (Man9-V3) constructs
The Man9-V3 monomer comprising a 30 amino acid V3-glcocopeptide with high-mannose (Man9GlcNAc2) glycans at position 301 and 332 based on the clade B JRFL mini-V3 construct (Pejchal et al., 2011) was synthesized as previously described (Alam et al., 2017). Man9-V3 multimer was synthesized by reacting multiple copies of a Man9-V3 monomer with a multivalent amino group-reactive HPMA-based co-polymer that was prepared as previously described (Lynn et al., 2015). The concept, design and synthesis of Man9 multimerized on star-shaped polymers (Star-Man9-V3), composed of HPMA-based polymer arms radially extending from a poly(amidoamine) (PAMAM) dendrimer core, with the universal PADRE T helper epitope (Alexander et al., 2000) has been described (Francica et al., 2019). In this study, Man9-V3 multimer and Star-Man9-V3 (∼50nm construct containing multiple Man9-V3 units) were used as macromolecular nanosized forms of Man9-V3 monomer for immunization of RMs.
Expression of recombinant HIV-1 Env proteins
Production of recombinant HIV-1 Envs, including gp120 monomers, and unstabilized gp140 and stabilized SOSIP trimers was previously described (Francica et al., 2019; Saunders et al., 2017a, 2017b, 2019). Recombinant Envs were used for binding assays and NHP immunizations. The main Env strains for soluble stabilized recombinant HIV-1 Env trimers (SOSIPs) used in this study were previously described (Bonsignori et al., 2017; Saunders et al., 2019) and the complete nomenclatures are shown below: CH848.3.TFchim.6R.SOSIP664v4.1; CH848.3.D0949.10.17chim.6R.DS.SOSIP664; CH848.3.D0949.10.17chim.6R.DS.SOSIP664_N301A; CH848.3.D0949.10.17chim.6R.DS.SOSIP664_ N301AN332A; CH848.3.D0949.10.17chim.6R.SOSIP664v4.1_N133DN138T; CH848.3.D0949.10.17chim.6R.DS.SOSIP664_N133DN138T; and CH848.3.D0949.10.17chim.6R.DS.SOSIP664v4.1degly4.
We tested wild-type and mutant (N301A, N301AN332A, N133DN138T and degly4) Env trimers listed above to evaluate mAb binding. For HIV-1 Env trimer immunizations in macaques, the following HIV-1 Env CH848 strains were generated as soluble stabilized recombinant trimers (SOSIPs); CH848 10.17., CH848 10.31, CH848 80.06, CH848 5.41 and CH848 25.02. These trimers constitute a sequential 5-valent series of CH848 Env stabilized trimers that had evolved during human CH848TF infection and were predicted to be involved in the induction of HIV-1 Env bnAbs that target glycans in the high mannose in conjunction with the GDIR motif in the Env peptide (Bonsignori et al., 2017). Since macaques were infected with a pathogenic SHIV bearing HIV-1 CH848TF Env, recombinant HIV-1 Env trimers bearing CH848TF were referred to as autologous Envs when testing mAbs isolated from SHIV-infected macaques. In instances where high mannose glycosylation was desired, Kifunensine (Sigma-Aldrich) was dissolved in phosphate-buffered saline (PBS) and added once to the cell culture media to a final concentration of 25 μM. The cells were cultured for 5 days and on the fifth day the cell culture media was cleared of cells by centrifugation and filtered with 0.8 um filter. The cell culture was concentrated with a Vivaflow 50 (Sartorius) with a 10 kd MWCO. The concentrated cell culture supernatant was rotated with lectin beads (Vistar Labs) overnight at 4°C. The beads were pelleted by centrifugation the next day and resuspended in MES wash buffer. The lectin beads were washed twice and the protein was eluted with methyl-α-pyranoside. The protein was buffer exchanged into PBS and stored at –80°C.
Expression of recombinant Coronavirus spikes
The SARS-CoV-2, SARS and MERS ectodomain constructs were produced and purified as previously described (Wrapp et al., 2020). Briefly, genes encoding residues 1−1208 of SARS-CoV-2 S (GenBank: MN908947), residues 1-1291 for MERS S (GenBank KJ650098.1) and residues 1-1190 for SARS S (UniProt P59594-1) with proline substitutions at residues 986 and 987 (SARS-CoV-2), 1060-1061 (MERS) and 968-969 (SARS) and a “GSAS” substitution at the furin cleavage site (residues 682–685) for SARS-CoV-2 was synthesized and cloned into the mammalian expression vector pαH. In all constructs, a C-terminal T4 fibritin trimerization motif, an HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag were also included. All other mutants were introduced in these backgrounds. Expression plasmids encoding the ectodomains sequences were used to transiently transfect FreeStyle293F cells using Turbo293 (SpeedBiosystems). Protein were purified on the sixth day post transfection from the filtered supernatant using StrepTactin resin (IBA). Affinity purified protein was run over a Superose 6/300 increase column. The purified proteins were flash-frozen in liquid nitrogen and stored at −80°C.
Isolation of Abs from single B cells
Antigen-specific memory B cells were isolated from RMs or humans using fluorophore-labeled antigens as hooks in various sort strategies outlined below. Antigens used for sorts were conjugated with Alexa Fluor 647 (AF647), Brilliant Violet 421 (BV421), or VioBright 515 (VB515) fluorophores. With the exception of peptides, immune cells were concurrently incubated with a cocktail of staining Abs and fluorophore-labeled antigens. Fluorophore-labeled peptides required an incubation with immune cells prior to addition of the staining antibody cocktail to improve binding efficiency. Fluorophore-labeled peptides were used at a ratio of 2.5μg per 10 million cells, recombinant gp120 and gp140 proteins at 10μg per 10 million cells, and SOSIP trimers at 90pmol per 10 million cells, to stain antigen-specific memory B cells. All incubations took place in approximately 1mL of buffer per 10 million cells. Flow cytometry sorts were performed using a BD FACSAria II (BD Biosciences, San Jose, CA), and the data were analyzed using FlowJo (Treestar, Ashland, OR). Single cell Env-reactive Abs were CD3 (BD Cat#552852; 5μl per test)/ CD14 (BioLegend Cat#301832; 2.5μl per test)/ CD16 (BD Cat#557744; 5μl per test), and/or surface IgD (Southern Biotech Cat#2030-09; 1μl per test) negative. Macaque B cells were also CD27+/− (BioLegend Cat#302816; 5μl per test) and CD20 (BioLegend Cat#302336; 1μl per test) positive, while human B cells were CD38+/− (Beckman Coulter Cat#IM2371U; 1μl per test) and CD19 (BD Cat#557791; 1μl per test) positive.
Single sorted B wells were obtained from RMs and humans using previously described flow cytometry-based sorts (Williams et al., 2017b; Alam et al., 2017; Zhang et al., 2016). Cells were individually sorted into 96 well plates containing lysis buffer and immediately stored at −80, as previously described (Liao et al., 2009). Rhesus macaque and human VHDJH and VLJL segments were isolated by single-cell PCR approaches that were previously described (Liao et al., 2009; Alam et al., 2017). Antibody sequences were analyzed using a custom-built bioinformatics pipeline for base-calling, contig assembly, quality trimming, immunogenetic annotation with Cloanalyst (https://www.bu.edu/computationalimmunology/research/software/), VDJ sequence quality filtering, functionality assessment, and isotyping. Antigen-specific single B cells were indexed sorted, thus allowing us to perform a backgate analysis to identify individual IgM+IgD+CD27+ cells overlayed against the total sorted B cell populations in FlowJo (Treestar, Ashland, OR). In vaccinated RMs, Env-reactive B cells were sorted using soluble unstabilized recombinant HIV-1 Env trimer (HIV-1 JRFL gp140) bearing Envs with Man9-enriched protein (Kifunensine-treated Env) conjugated to both AF647 and BV421 fluorophores. The antigens for sorts of SHIV-infected B cells used tetramerized Man9-V3 and HIV-1 Env SOSIPs. The following flow cytometry sort strategies were used to investigate the repertoire of SHIV-elicited Env-reactive B cells, including glycan-reactive B cells: LN wk52 [Man9-V3 peptide (+) and Aglycone V3 peptide (-); CH848TF gp120 (+) and CH848TF gp120 N332A (-); and CH848TF SOSIPv4.1 (+)]; PBMCs wk52 [CH848TF gp120 (+) and CH848TF gp120 N332A (-)]; and PBMCs wk104 [Man9-V3 peptide (+) and Aglycone V3 peptide (-);CH848TF SOSIPv4.1 (+); CH848.d949.10.17 DS.SOSIP (+) and CH848.d949.10.17 DS.SOSIP N332A (-); and CH848.d949.10.17 DS.SOSIP (+) and CH848TF SOSIPv4.1 (-)].
In HIV-1 naive humans, Env-reactive B cells were sorted using glycopeptides and HIV-1 Env SOSIPs in the following differential sort strategies: (#1) Man9-V3 (+) versus Aglycone V3 peptide (-) in a dual or triple color sort strategy; (#2) CH848 10.17 DS.SOSIP (+) versus CH848 10.17 DS.SOSIP_N332A (-); and (#3) CH848 10.17 DS.SOSIP_N133DN138T (+) versus CH848 10.17 DS.SOSIP_N133DN138T_N332A (-). In three individuals (LPK01-LPK03), we sorted 20M PBMCs using strategy #1 (dual color differential sort) or strategy #2 (total for 2 sorts = 40 million PBMCs). For dual color differential sort strategy #1, we sorted B cells that were reactive with Man9-V3 conjugated to one fluorophore, but not the aglycone V3 peptide conjugated to a different fluorophore. In one individual (LPK04), we studied B cells enriched from 100 million leukapheresis PBMCs per experiment using sort strategy #3 (total for 2 sorts = 200 million PBMCs). In five individuals (LKP05-LKP09), we studied B cells enriched from 100 million leukapheresis PBMCs using sort strategy #1 (triple color differential sort); here we used a more stringent approach to sort B cells that were reactive with Man9-V3 conjugated to two different fluorophores, but was not reactive with aglycone V3 peptide conjugated to a third fluorophore. For subjects LKP04-LKP09, we performed negative B cell enrichment from 100M PBMCs in each experiment using a commercially available B cell enrichment kit according to manufacturer’s protocol (StemCell Technologies; Vancouver, BC, Canada). We calculated the frequency of human FDG B cells as the percent of all sorted B cell-derived recombinant IgGs produced in small-scale transfection that bound Man9-V3 but not aglycone V3 peptide and/or yeast antigens in ELISA, relative to the total number of B cells analyzed via flow cytometry sort per individual. The estimated frequency of human FDG B cells was reported as the average frequency of FDG B cells across the nine individuals studied.
Expression of recombinant mAbs
Recombinant mAbs were first generated in human embryonic kidney epithelial cell lines (HEK293T, ATCC, Manassas, VA; Cat#CRL3216) in small scale transfections using linear cassettes encoding antibody heavy and light chain genes that were PCR amplified from single B cells (Liao et al., 2009). For generation of larger quantities commercially-obtained (GeneScript, Piscataway, NJ) plasmids with antibody heavy and light chain genes were used to transfect suspension Expi 293i cells using ExpiFectamine 293 transfection reagents (Life Technologies, GIBCO; Cat#A14524) as described (Bonsignori et al., 2017). Purified recombinant mAbs were dialyzed against PBS, analyzed, and stored at 4°C. All recombinant mAbs were expressed from plasmids encoding a human or macaque IgG constant region, and were QC’ed in western blot for appropriate heavy and light chain protein expression and/or via size exclusion chromatograpgy (SEC) for a single peak indicative of stable antibody expression.
Expression of DH717 Fabs for structural studies
To purify DH717.1 Fab monomer for crystallization studies, the heavy- and light-chain variable and constant domains were cloned into a modified pVRC-8400 expression vector using NheI and NotI restriction sites and the tissue plasminogen activation signal sequence. The C terminus of the heavy-chain constructs contained a non-cleavable 6x-histidine tag. Fabs were expressed using transient transfection of HEK293T cells using linear polyethylenimine (PEI) following manufacturer’s protocols. After 5 days of expression, supernatants were clarified by centrifugation. His-tagged Fabs were loaded onto Ni-NTA superflow resin (QIAGEN) preequilibrated with Buffer A (10 mM Tris, pH 7.5, 100 mM NaCl), washed with Buffer A + 350 mM imidazole, and eluted with Buffer A + 350 mM imidazole, Fabs were then purified by gel filtration chromatography in Buffer A using a superdex 200 analytical column. The fractions corresponding to monomer were pooled and concentrated. While intermolecular disulfide bonds can occur due to purification artifacts, intramolecular disulfide are likely to be formed in vivo because of the structural proximity of two unpaired cysteines, such as what we see in the DH717.1 Fab dimers.
HIV-1 Env and glycopeptide binding of mAbs via ELISA
Envelope binding to recombinant mAbs, and plasma or sera were tested in ELISA as previously described (Williams et al., 2017a; Saunders et al., 2019). Binding was assessed using synthetic peptides, recombinant HIV-1 Env gp120, gp140, and stabilized chimeric SOSIP trimers, and heat-killed yeast antigens. ELISA protocols had modifications that supported binding to different antigens. In general, antigens were directly coated to Nunc-absorb (ThermoFisher) plates overnight at 4°C or captured using streptavidin, AbC-mAb (AVIDITY, Colorado, USA) or PGT151 mAb that were coated to Nunc-absorp plates overnight at 4°C. Unbound proteins were washed away and the plates were blocked with either goat serum-based (SuperBlock) (Liao et al., 2009) or non-serum-based (Alam et al., 2017) blocking media for 1 hour. Serial dilution of serum and mAbs were added to the plate for 60 and 90 minutes, respectively. Binding Abs were detected with specie-specific HRP-labeled anti-IgG Fc Abs using 20μl per reaction with 1 hour incubation at room temperature. HRP detection was subsequently quantified with 3,3′,5,5′-tetramethylbenzidine (TMB). ELISA binding levels were measured at an optical density of 450 nm (OD450nm) and binding titers were analyzed as area-under-curve of the log-transformed concentrations (Log AUC).
Binding to biotin-Man9-V3 and biotin-aglycone-V3 was determined as previously described using streptavidin for capturing each peptide on Nunc-absorb ELISA plates (Alam et al., 2017; Bonsignori et al., 2017). In brief, we coated 30ng of streptavidin in 15μl at 2μg/ml per well of a 384-well Nunc-absorb ELISA plate, sealed and incubated overnight at 4°C. Synthetic peptides (10ng) were added to streptavidin in 10μl at 1μg/ml for one hour at room temperature to facilitate peptide capture. Binding to yeast antigens was tested at 1:400 (Cryptococcus neoformans) and 1:2000 (Candida albicans) dilutions made in sodium bicarbonate buffer; 15μl of each yeast solution was coated per well of a 384-well Nunc-absorb ELISA plate, sealed and incubated overnight at 4°C. Yeast antigen reactivity was performed using ELISA conditions for assessing glycan reactivity on glycopeptides (Alam et al., 2017). HRP-conjugated specie-specific secondary Abs were used to detect antibody binding to synthetic peptides and yeast antigens - Goat anti-human IgG-HRP, 1:15000 dilution (Jackson ImmunoResearch Laboratories, Cat# 109-035-098); and Mouse anti-monkey IgG-HRP, 1:20000 dilution (Southern Biotech, Cat# 4700-05). The yeast dilutions tested in ELISA were found to be the lowest dilutions at which we observed maximal binding by glycan-reactive Abs, such as 2G12 in optimization assays. DH1013 isolated from RM5996 was used as a positive control mAb for peptide binding to Man9-V3 and Aglycone V3 peptides, and 2G12 or DH501 (Saunders et al., 2017a) were used as positive control mAbs for binding Man9-V3, but not Aglycone V3 peptide. 2G12 bnAb was also used as a reference mAb for Env glycan and Candida albicans yeast antigen binding (Doores et al., 2010b).
For ELISAs with HIV-1 Env SOSIPs bearing an Avi-tag, the trimer was captured by the AbC- mAb antibody against C terminus Avi-tag, while non Avi-tagged SOSIP trimers were captured by PGT151 bnAb expressed in 293 cells as described (Saunders et al., 2019; Williams et al., 2017a). In brief, Avi-tag and PGT151 mAbs were coated at 30ng per well of a 384-well plate in 15μl at 2μg/ml. HIV-1 Env SOSIPs (20ng) were added to Avi-tag or PGT mAbs in 10μl at 2μg/ml for one hour at room temperature to facilitate SOSIP capture. PGT151 mAb was used as a positive control for binding Avi-tagged SOSIP, while influenza-specific antibody CH65 (Whittle et al., 2011) was used as a negative control. HRP-conjugated specie-specific secondary Abs were used to detect binding Abs (see above). For non-Avi-tagged SOSIPs that were captured by PGT151 mAb, biotinylated mAbs were used as positive (B-2G12 and B-PGT128) and negative (B-CH65) controls - biotinylated-mAbs were tested at 10μg/ml and 3-fold dilutions. This latter approach was used to test only rhesus mAbs and b-mAbs, since the secondary Abs would not detect human PGT151 mAb used for SOSP capture; Mouse anti-monkey IgG-HRP (see above) and Streptavidin-HRP, 1:30000 dilution (Thermo Scientific, Ref# 21130). FDG mAbs, DH1005-DH1011, bound Man9-V3 but not aglycone V3 peptide in ELISA (Figure 3). Additionally, DH1005-DH1011 had varied binding levels to Env SOSIP trimers (not shown). Competitive ELISA to assess cross-blocking of recombinant mAbs were previously described (Alam et al., 2013; Saunders et al., 2017a; Bonsignori et al., 2017). We biotinylated the Abs using the following product: BIOTIN-X-NHS, Cayman Chemicals, Cat# 13316. Here, we studied biotinylated 2G12 and DH851 mAbs in competition ELISAs using CH848TF or CH848 10.17 gp120 Envs.
Competitive inhibition of biotiniylated-mAbs was measured as a percent of binding in the presence of a competing non-biotinylated mAb relative to binding in the absence of this competing mAb. A successful assay had positive control blocking ≥ 40%. DH851 blocking on CH848TF gp120 was from a single experiment (Figure 5), but the results were similarly observed for blocking on CH848 10.17 gp120 in three separate experiments (not shown).
Glycan-dependent binding of FDG mAbs to SARS-CoV-2 spike protein
Recombinant FDG mAbs were tested for binding SARS-CoV-2 trimer (lot 62KJ) in ELISA in the absence or presence of single monomer D-mannose sugar. SARS-CoV-2 trimeric spike protein was captured by streptavidin to individual wells of a 384-well plate, and serially diluted mAbs were tested for binding. ELISA binding assays were optimized to use SARS-CoV-2 spike protein that was aliquotted and frozen upon production; desired aliquots of frozen spike protein were thawed once at 37° and stored overnight at room temperature. SARS-CoV-2 spike protein, and Man9-V3 and Aglycone V3 peptides were captured via streptavidin on Nunc-absorb ELISA plates using PBS-based buffers and assay conditions as previously described (Alam et al., 2017; Bonsignori et al., 2017). HIV-1 CH505TF SOSIP trimer and commercially-obtained constructs of SARS-CoV-2 spike ectodomain (S1+S2 ECD, S2 ECD and RBD) (Sino Biological Inc Cat#40589-V08B1 and 40590-V08B respectively and RBD from Genescript Cat#Z03483) were captured using mouse anti-AVI-tag mAb (Avidity LLC, Aurora, CO). In brief, we coated 30ng of streptavidin in 15μl at 2μg/ml or 30 ng of mouse anti-AVI-tag mAb in 15μl at 2μg/ml per well of a 384-well Nunc-absorb ELISA plate, sealed and incubated overnight at 4°C. SARS-CoV-2 spike protein (2μg/ml) were added to streptavidin in 10μl per well of a 384-well plate for one hour at room temperature to facilitate protein capture. HIV-1 CH505TF SOSIP, and recombinant SARS-CoV-2 S1+S2 ECD, S2 ECD and RBD spike proteins, at final concentration of 2 μg/ml were added to anti-AVI mAb in 10μl per well of a 384-well plate for one hour at room temperature to facilitate protein capture. Mouse anti-monkey IgG-HRP (Southern Biotech, CAT# 4700-05) or Goat anti-human IgG-HRP (Jackson ImmunoResearch Laboratories, CAT# 109-035-098) secondary Abs were used to detect mAb bound to the SARS-CoV-2 spike protein. HRP detection was subsequently quantified with 3,3′,5,5′-tetramethylbenzidine (TMB) by measuring binding levels at an absorbance of 450nm; binding titers were reported as Log area under the curve (AUC).
Commercially obtained D-mannose (Sigma, St. Louis, MO) was used to outcompete mAb binding to glycans on SARS-CoV-2; D-mannose solutions were produced in ELISA buffers. Competition ELISAs with D-mannose were performed in two separate assays; each with [0.5M] (data not shown) or [1.0M] D-mannose. Mouse-Human chimeric mAb D001 (SARS-CoV RBD mAb; Sino Biological Inc Cat#40150-D001) was used as a control mAb; D001 was tested at 2μg/ml and 2-fold serial dilutions (10x), in contrast to all other mAbs tested at 100μg/ml and 3-fold serial dilutions (10x). Anti-influenza CH65 mAb was used a negative control mAb. We found that goat serum containing buffer (Superblock) – PBS, 4% (w/v) whey protein (BiPro USA), 15% normal goat serum (Invitrogen), 0.5% Tween 20, and 0.05% sodium azide (Sigma-Aldrich) – inhibited binding of glycan-dependent FDG mAbs to SARS-CoV-2 spike protein, whereas positive control RBD-binding mAb D001 bound the spike protein in the presence of superblock. ELISAs using superblock were previously described (Williams et al., 2017a). Moreover, whereas the 2G12 from Polymun bound to S protein in surface plasmon reasonance, it was less potent than the recombinantly expressed 2G12 Abs, and did not bind well in ELISA; commercial 2G12 from Polymun was provided in a maltose (mannose) buffer. D-mannose more competitively inhibited binding to fungal glycans by some FDG Abs. For example, SHIV-elicited DH898 mAbs were not outcompeted for binding as much as vaccine-induced DH717 mAbs. These data suggested that FDG Abs have differences in affinities for free-glycans in solution compared to glycan clusters on HIV-1 Env or the promiscuity in binding different glycoforms. In fact, structural analysis of DH898 mAbs in complex with recombinant HIV-1 Env trimers showed that DH898 mAbs bound glycans at two different epitopes on HIV-1 Env (Figures 4 and S3). These data suggested that DH898 mAbs have a higher propensity to bind different glycan arrangements on HIV-1 Env and not only the high mannose patch where the vaccine-induced DH717 Abs, SHIV-induced DH851 bnAbs and 2G12 have been shown to bind.
Neutralization assays
mAb neutralizing activity was assessed in TZM-bl cells as described (Montefiori et al., 2012; LaBranche et al., 2019; Montefiori, 2009). Env-pseudotyped HIV-1 bearing Envs used in SHIVs for infection were referred to as autologous viruses, whereas viruses bearing Envs from global panels of geographically diverse multi-clade strains were referred to as heterologous viruses. Difficult-to-neutralize or tier 2 autologous or heterologous HIV-1 isolates are generally used to assess neutralization potency and breadth of HIV-1 Abs; global panel of HIV-1 strains (deCamp et al., 2014) and119 heterologous tier 2 isolates (Seaman et al., 2010). Env-pseudotyped HIV-1 bearing Envs with Man9-enriched glycans (termed Kif-treated HIV-1 strains) were grown in cells treated with 25μM Kifunensine to facilitate high mannose glycan expression on HIV-1 Envs and Kif-treated HIV-1 strains were tested for neutralization in TZM-bl cells as previously described (Saunders et al., 2017a). Env-pseudotyped HIV bearing Envs with Man9-enriched glycans express Env expressed in cell lines, but it remains unknown how additional glycoforms present on the Env generated in vivo might impact the binding specificities and function of our FDG mAbs that preferentially target Man9-glycans. For neutralization assays, a mixture of CH01+CH31 bnAbs is used as a positive control for neutralization of all HIV-1 strains, and murine leukemia virus (MLv) or vesicular stomatitis virus (VSV) were used as negative retrovirus controls. For neutralization assays, a positive for neutralizing antibody activity in a sample is based on the criterion of > 3X the observed background against the MLV negative control pseudovirus. Note that DH898 mAbs were tested for neutralization of all tier 2 HIV-1 strains in the global panel, except CNE8. Additionally, we found neutralization titer variabilities in DH851 mAbs across different batches of mAbs, in agreement with the unstable dimerization of Fabs for these Abs. Only DH1005 neutralized [Kif]-treated HIV strains (Figure 3), but none of the human FDG mAbs (DH1005-DH1010) neutralized non-[Kif]-treated HIV-1 strains (not shown).
Glycan binding of Abs oligomannose bead immunoassay
Recombinant mAbs were tested for binding glycans in an oligomannose bead immunoassay as described (Saunders et al., 2017a). Custom glycan microspheres were generated with different individual glycans to evaluate mAb binding. Binding of mAb to glycan was determined with A Bio-Plex 200 HTS (Bio-Rad) machine with the Bioplex manager software (Bio-Rad) was used to quantify binding of mAb to glycan, and binding was measured as background-subtracted fluorescence.
Anti-nuclear Ab (ANA) reactivity
mAb reactivity to nine autoantigens was measured using the AtheNA Multi-Lyte ANA kit (Zeus scientific, Cat#A21101). MAbs were 2-fold serially diluted starting at 50 mcg/mL. The secondary antibody provided with the kit was Phycoerythrin-conjugated goat anti-human IgG. The kit SOP was followed for the remainder of the assay. Samples were analyzed using AtheNA software. Positive (+) specimens received a score > 120, and negative (-) specimens received a score < 100. Samples that score 100-120 were considered indeterminate. FDG mAbs that had ANA reactivity were shown in Figure 7. The following mAbs were negative for autoantigen binding (not shown): DH851.2, DH717.1, DH717.3, DH1005-DH1008, and DH1010-DH1011.
HEp-2 cell staining
Indirect immunofluorescence binding of mAbs to human epithelial (HEp-2) cells (ZEUS IFA ANA HEp-2 Test System, Zeus Scientific, Somerville, NJ) was performed per manufacturer’s instructions. Briefly, 20μL of diluted mAbs (50μg/ml, and 25μg/ml) were added to the appropriate wells on the antinuclear antibody (ANA) slide. Slides were incubated for 20 minutes at room temperature in a humid chamber, and then washed with 1X PBS. Goat Anti-Rhesus Ig(H+L) FITC secondary was added at a concentration of 30μg/ml to each well. The slides were incubated for 20 minutes, then washed twice, dried, fixed with 33% glycerol and coverslipped. Slides were imaged using an Olympus AX70 microscope with a SpotFlex FX1520 camera. Images were acquired on a 40X objective using the FITC fluorescence channel with different acquisition time as indicated on each image. Positivity was determined by comparing Abs of interest to positive and negative non-human primate antibody controls, DH1037 and DH570.30 respectively. Staining patterns were identified using the Zeus Scientific pattern guide found on the website and previously reported (Buchner et al., 2014; Huang et al., 2012; Haynes et al., 2005).
Next-generation sequencing (NGS) and analysis of Ab genes
Illumina MiSeq sequencing of Ab heavy chain VDJ sequences was performed on peripheral blood cells as previously described (Zhang et al., 2016; Williams et al., 2017b). Briefly, for each time point, RNA from peripheral cells was divided equally into two separate portions that were used to generate independent cDNA aliquots for sequencing to confirm the absence or presence of Ab sequences of interest. Paired-end sequences were merged using FLASH (Magoč and Salzberg, 2011), quality filtered (Q score > 30 for > 95% of sequence) and deduplicated. V, D, and J gene segment, clonal relatedness testing and reconstruction of clonal lineage trees were performed using the Cloanalyst software package (Kepler, 2013). Immunogenetics information of rhesus and human antibody sequences were assigned by Cloanalyst using Cloanalyst’s default libraries of rhesus and human immunoglobulin genes, respectively (https://www.bu.edu/computationalimmunology/research/software/). B cell clonality was determined based on similar heavy chain rearrangements and CDR3 length as described (Kepler et al., 2014). For Abs that were inferred as being clonally-related by Cloanalyst, and had the same VH sequence but different alleles, we manually inspected the VH sequence for similar residues in the HCDR3, including the non-templated nucleotides.
The near-germline or unmutated common ancestor (UCA) of Ab lineages were inferred using the Cloanalyst software program with the macaque immunoglobulin gene library. DH851UCA was initially inferred based on the isolation of DH851.1-DH851.3 lineage members. DH851UCA.2 was later inferred using DH851.1-DH851.4. We found that DH851UCA and DH851UCA.2 had one amino acid difference in a framework region of both the heavy and light chain genes; the UCA sequences were deposited in GenBank. Data shown in this study described the characteristics of DH851UCA. For recombinant Ab expression, week 0 mAbs had clonally-related heavy chain genes detected via NGS paired with DH851.1 light chain, while the DH851UCA had the Cloanalyst-inferred heavy and light chain genes.
To exclude the possibility of generating unmutated common ancestor Abs that do not account for the allelic variability observed in the RM model (Ramesh et al., 2017; Corcoran et al., 2016), we generated a NGS dataset as described above that comprised only of VH1 (DH898 lineage), VH2 (DH851 lineage) and VH4 (DH717 lineage) IgM+ repertoires at baseline or week 0 of the RMs. Here, we sought to find unmutated or least mutated FDG Ab clonal lineage members indicated from macaque baseline PBMCs. Week 0 clonally-related mAbs were generated by pairing the week 0 NGS-derived heavy chain genes with the light chain of the single B cell-derived Ab isolated following infection or vaccination as described (Williams et al., 2015). Clonally-related DH717 VH genes identified via VH gene deep sequencing post the Star-Man9-V3 boost had higher average mutation frequencies compared to DH717 VH genes identified after the initial trimer boost. However, our attempts to express recombinant mAbs bearing the mutated DH717 VH genes paired with the DH717.1 Vλ failed, so it remains unknown whether these mutations improved antibody function.
Week 0 DH898 and DH851 FDG lineage precursors that existed prior to SHIV CH848TF infection in macaque RM6163 were evaluated using NGS in two separate experiments. cDNA of isotype-specific VH1 and VH2 genes were amplified from peripheral blood-extracted RNA and split into separate samples for cDNA library preparation and NGS (Run A and B). Sequences were quality filtered and computationally analyzed, and DH898 and DH851 clonal lineages assessed using Cloanalyst software program. We found 2 IgG and 594 IgM DH898 clonal lineage members among ∼3.5 million unique VH1 sequences at week 0; IgA (151760), IgG (860757), and IgM (2457675). We found 21 IgA, 66 IgG and 148 IgM DH851 clonal lineage members among ∼2.1 million unique VH2 sequences at week 0; IgA (516938), IgG (1282669), and IgM (759778).
High-resolution single cell VDJ and transcriptome sequencing and analysis
RM VDJ libraries were generated following the manufacturers protocol (Chromium Next GEM Single Cell V(D)J protocol (v1.1, Rev E)) with three modifications. First, 15 μl of cDNA was used as input for the first BCR enrichment (step 4.1a). Second, RM-specific reverse primers were used to amplify BCR transcripts (step 4.1b and 4.3a). Third, we used 9 PCR cycles in the first BCR enrichment (step 4.1) and 11 PCR cycles for the second BCR enrichment (step 4.3). Primers were designed to amplify the BCR sequences from RMs using the previously published macaque Ig libraries (Ramesh et al., 2017) and Cloanalyst default rhesus Ig library (https://www.bu.edu/computationalimmunology/research/software/). Reverse primers were designed to sit in the constant region for all isotypes (IgA, IgG, IgE, IgM, IgD) and both light chains (IgK, IgL). First round primers (5′ to 3′) were as follows for IgA, IgG, IgE, IgM, IgD, IgK, and IgL are [IgA] GGCGGGAAGTTTATGACGGTC, [IgG] TTGTCCACCTTGGTGTTGCT, [IgE] ATGGAGTGTGCACCACAT, [IgM] CCTGCATGACATCCTTGGAAG, [IgD] AGGTGCCAGGTGACAGTC, [IgK] TGTCCTGCTCTGTGACACT, and [IgL] TGACCTGGCAGCTGTAGCT. Second round primers (5′ to 3′) were as follows [IgA] CTTTTGCTCCAGGTCACGT, [IgG] CACGCTGCTGAGGGAGT, [IgE] GTCACCATCACCGGCT, [IgM] GCCAACGGCCACTTCGTT, [IgD] GGTTGTACCCAGTTATCAAGCAT, [IgK] TCCACCTTCCACTTTACGCT, and [IgL] TAGCTGCTGGCYGC.
Illumina NGS-generated raw data files were processed using the Cell Ranger single cell gene expression software provided by 10X Genomics. Sequences were quality filtered, followed by assembly and annotation using Cell Ranger adapted to contain RM Ig genes from three reference libraries (the Cloanalyst default Ig library – see link above, IMGT, and Ramesh et al. [2017]). In order to adapt Cell Ranger to contain rhesus Ig libraries, we manually compiled all three RM Ig reference libraries and reformatted them to be compatible with Cell Ranger as described (https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/advanced/references#segment). Next, an updated text file specifying the sequences of the RM inner enrichment primers (second round primers shown above) was compiled and formatted for compatibility with Cell Ranger as described in the documentation for running customized libraries in Cell Ranger that is provided by 10X Genomics. The customized RM Ig reference library described above and the file containing the inner enrichment primers were then used in Cell Ranger to build and annotate contigs. Contigs assembled by Cell Ranger were reannotated and analyzed using only the Cloanalyst software with default macaque Ig library to determine B cell immunogenetics and clonality.
For transcriptome analyses of macaque single B cells, the Cell Ranger Single Cell Software Suite (version 3.1) was used to generate sequencing fastq files and to perform sample de-multiplexing, barcode processing, reference alignment and single-cell 3′ gene counting (Zheng et al., 2017a). Reads were aligned to the RM genome (Mmul_10). Matrices of cell barcodes and gene counts generated by Cell Ranger were loaded into Seurat R package (v3.2.0) for graph-based cell clustering, dimensionality reduction and data visualization (Macosko et al., 2015; Satija et al., 2015; Stuart et al., 2019). Differentially expressed genes between cell clusters or groups were determined using Seurat by the Likelihood-ratio test (McDavid et al., 2013). Graphs and plots were generated using the Seurat and ggplot2 R packages and Graphpad Prism version 8.
Surface plasmon resonance (SPR)
The binding of DH717.1 and DH717.1 C76S mAbs to 5 different CH848 Env’s was assessed by surface plasmon resonance on Biacore T-200 (GE-Healthcare) at 25°C with HBS-EP+ (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20) as the running buffer. The Abs were captured on a CM5 chip by flowing 200 nM of the mAb over a flow cell immobilized with ∼9000 RU of anti-human Fc antibody. Binding was measured by flowing over 200 nM solution of Env’s in running buffer. 2G12 was used as a control and it was immobilized on one of the flow cells. The surface was regenerated between injections by flowing over 3M MgCl2 solution for 10 s with flow rate of 100 μl/min. Blank sensorgrams were obtained by injection of same volume of HBS-EP+ buffer in place of trimer solutions. Sensorgrams of the concentration series were corrected with corresponding blank curves.
The SPR binding curves of 2G12 and DH717.1 mAbs and Fabs against CH848 10.17 DS.SOSIP and CH848 10.17 DS.SOSIP_N133DN138T trimers were obtained using the Biacore S200 instrument (GE Healthcare) in HBS-N 1X running buffer. Biotinylated CH848 DS.SOSIP trimers were immobilized onto a CM3 sensor via streptavidin to a level of 300-350RU. The 2G12 and DH717.2 mAbs and Fabs were diluted down to 50μg/mL and injected over the SOSIP trimer at 30μL/min for 180 s. A blank streptavidin surface was used for reference subtraction to account for non-specific binding. SPR single cycle kinetic and affinity measurements of the 2G12 and DH717.1 mAbs and Fabs were also obtained using the Biacore S200 instrument with the CH848 DS.SOSIP trimers directly immobilized to a level of 300-350RU via streptavidin. Five sequential injections of DH717.1 mAb and 2G12 mAb diluted from 5nM to 50nM were injected over the immobilized CH848 DS.SOSIP trimers at a flow rate of 50uL/min for 120 s per injection. 2G12 Fab2 was injected from 50nM to 500nM and DH717.1 Fab dimer was injected from 250nM to 2000nM over the CH848 DS.SOSIP trimers. The dissociation length of the single cycle injections was 600 s followed by regeneration with a 20 s pulse of 25mM NaOH. Results were analyzed using the Biacore S200 Evaluation Software (GE Healthcare). A blank streptavidin surface as well as buffer binding were used for double reference subtraction to account for non-specific antibody binding and signal drift. Subsequent curve fitting analysis was performed using the 1:1 Langmuir model with a local Rmax for both 2G12 and DH717.1 mAb and the heterogeneous ligand model for 2G12 Fab2. The reported kinetic binding curves are representative of 2 datasets.
For affinity measurements, the DH717.1 Fab was purified using size exclusion chromatography (SEC-FPLC) in PBS 1x. Approximately 10mg of DH717.1 Fab was loaded onto a Superdex 200 increase 10/300 column using a 500 μL loop and run at 0.5 mL/min using an Äkta Pure system (GE Healthcare). DH717.1 Fab peaks were collected via fractionation using a 96-well plate and were analyzed using the Unicorn 7.0.2 software (GE Healthcare). Using a linear regression derived from running protein standards (GE Healthcare) of known molecular weight, the molecular weights of the Fab monomer and dimer peaks were estimated. The monomer and dimer peak fractions were then pooled and concentrated using 10k, 0.5mL centrifugal filters (Amicon) and concentrations were measured using a NanoDrop UV-VIS spectrophotometer. Following SEC purification, DH717.1 Fab monomer and dimer stability were assessed using SDS-PAGE gel electrophoresis and re-analysis by SEC-FPLC. SDS-PAGE gel electrophoresis was performed using the BioRad system and 4%–15% TGX stain free gels. 7μg of each DH717.1 Fab fragment was loaded onto the gel under both reducing and non-reducing conditions. 10μg of a Precision Plus Protein standard (BioRad) was also loaded to help verify the size of the fragments. The gel was run at 200V for 35-40 minutes in Tris/Glycine/SDS running buffer. The Fab bands were analyzed via the Gel Doc EZ imager (BioRad). For SEC-FPLC re-analysis, 40μg of each DH717.1 Fab monomer and dimer were loaded onto a Superdex 200 increase 10/300 column using a 100 μL loop and run at 0.5 mL/min using the aforementioned Äkta Pure system.
The binding FDG Abs to CoV spikes was assessed by surface plasmon resonance on Biacore T-200 (GE-Healthcare) at 25°C with HBS-EP+ (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20) as the running buffer. For the data shown in Figure 6E, S proteins were captured via their C-terminal Twin Streptactin tags at ∼1000 RU levels on two alternate flow cells of a streptavidin coated chip, and 200 nM of each FDG antibody was flowed over all four flow cells; the binding curves were double-reference subtracted. The surface was regenerated between injections by flowing over 1M NaCl in 50mM NaOH solution for 10 s with flow rate of 100μl/min.
For determining affinity and kinetics of 2G12 interaction with SARS-CoV-2 spike, single cycle kinetics analysis was performed on the SARS-CoV-2 S protein immobilized on a streptavidin (SA) chip with five injections of 2G12 Fab, prepared by digestion with papain as described previously (Murin et al., 2014), at 3.125 nM, 6.25 nM, 12.5 nM, 25 nM and 50 nM concentrations. The data were fit to a 1:1 Langmuir binding model.
Structural analyses
Crystallography
DH717.1 IgG was expressed and purified as above. IgG samples were digested with papain and further purified to produce Fab fragment. DH717.1 Fab was purified via size exclusion chromatography to separate the Fab monomers and dimers. The DH717.1 Fab monomer was mixed with 3 molar excess of Man9-V3 at a final complex concentration of 15 mg/mL for crystallization trials. The complex crystallized over a 100 μl reservoir of 100mM Na Citrate, pH 5.0, 30% PEG 4000 in 1-2 days at room temperature. The DH717.1 Fab dimer, concentrated to 10 mg/ml in 0.1M HEPES buffer with 0.15M NaCl, was crystallized over a 60 μl reservoir of 0.1M citric acid pH 3.5, 14% PEG 1,000 in a drop composed of 0.5 μl protein plus 0.5 μl reservoir. The crystals were cryoprotected by brief immersion in reservoir solution supplemented with 25% glycerol for the DH717.1 Fab monomer-glycopeptide complex, and 30% ethylene glycol for the DH717.1 Fab dimer, before being flash frozen in liquid nitrogen. Diffraction data of the monomer-glycopeptide complex were collected at NE-CAT beamline 24-ID-E and data of dimer were collected at SER-CAT with an incident beam of 1 Å in wavelength. Data were processed using HKL-2000 (Otwinowski and Minor, 1997). Matthews analysis of the monomer-glycopeptide complex and dimer data suggested 1 Fab and four Fabs present in the unit cell, respectively (Matthews, 1968). Molecular replacement calculations for the monomer-glycopeptide complex were carried out with PHASER, using published DH270.5 (Protein Data Bank (PDB) ID 5TTP) as the starting model. The Fab model was separated into its variable and constant domains for molecular replacement. A molecular replacement solution for the dimeric DH717.1 with four Fabs was found using Phaser with search models DH717.1 Fv (above) and the constant region from DH522 UCA (Terwilliger et al., 2008; Williams et al., 2017b). The solution was improved through alternating rounds of manual rebuilding in Coot and reciprocal space refinement in PHENIX, and geometry optimization using Rosetta-Phenix refinement (phenix.rosetta_refine) (Emsley et al., 2010; Adams et al., 2010). The crystal structure of the monomeric DH717.1 Fab complex showed well-defined electron density for the three protein-proximal mannose residues, while the rest of the Man9 glycan and the V3 peptide were disordered.
Negative-stain electron microscopy (NSEM)
MAbs were diluted to 100 μg/ml final concentration with buffer containing 10 mM NaCl, 20 mM HEPES buffer, pH 7.4, 5% glycerol and 7.5 mM glutaraldehyde After 5-minute incubation, excess glutaraldehyude was quenched by adding sufficient 1 M Tris stock for a final Tris 75 mM for 5 minutes; then samples were stained with 2% uranyl formate. Images were obtained with a Philips 420 electron microscope operated at 120 kV, at 82,000 × magnification and a 4.02 Å pixel size. The RELION program was used for CTF correction, automatic particle picking and 2D class averaging of the single-particle images. For human FDG precursor recombinant mAbs, only DH1005, DH1009 and DH1010 showed a mixture of I- and Y- shaped conformations (Figure 3); the remaining mAbs only showed Y-shaped Abs (not shown).
Cryo-electron microscopy (cryo-EM) of HIV-1 Env complexes
For the DH898.1-bound complex, purified HIV-1 Env was mixed with 6-fold molar excess of recombinantly expressed and purified DH898.1 Fab to obtain a final concentration of 1.5 mg/ml Env, and incubated for 2 hours at room temperature. 2.5 uL of protein was deposited on a Quantifoil-1.2/1.3 holey carbon grid (Electron Microscopy Sciences, PA) that had been glow discharged for 30 s in a PELCO easiGlow Glow Discharge Cleaning System. After a 30 s incubation in > 95% humidity, excess protein was blotted away for 2.5 s before being plunge frozen into liquid ethane using a Leica EM GP2 plunge freezer (Leica Microsystems). For the DH851 Abs, attempts to obtain a structure of the complex by mixing with Fabs that were either recombinantly produced or obtained by proteolytic digestion of the IgG were unsuccessful, yielding mostly unliganded Env trimer populations and very small fractions of Env bound to Fab dimer (not shown). We next prepared a complex of DH851.3 bound to HIV-1 Env amenable to structural analysis by first making a complex of the Env with DH851.3 IgG, which was purified by size exclusion chromatography. The purified complex was then subjected to proteolytic digestion with HRV3c protease that cleaved at a site engineered between the Fab and Fc regions. Following purification over a protein A column to remove the Fc, the complex was purified by SEC, and concentrated to 1.5 mg/ml. Cryo-EM grids were prepared in a manner similar to the DH898.1 complex.
Cryo-EM imaging was performed on a FEI Titan Krios microscope (Thermo Fisher Scientific) operated at 300 kV, aligned for parallel illumination. Data collection images were acquired with a Gatan K3 detector operated in counting mode with a calibrated physical pixel size of 1.066 Å with a defocus range between −1.0 and −3.5 μm using the Latitude S software (Gatan Inc.). The dose rate used was ∼1.0 e-/Å2·s to ensure operation in the linear range of the detector. The total exposure time was 4 s, and intermediate frames were recorded every 0.067 s giving an accumulated dose of ∼60 e-/Å2 and a total of 60 frames per image. A total of 6058 and 3230 images were collected for the DH851.3- and DH898.1-bound complexes, respectively. Cryo-EM image quality was monitored on-the-fly during data collection using automated processing routines.
Data processing was performed within cryoSPARC (Punjani et al., 2017) including particle picking, multiple rounds of 2D classification, ab initio reconstruction, heterogeneous and homogeneous map refinements, and non-uniform map refinements.
For DH898.1 reconstructions, micrographs were imported into cryoSPARC, CTF-corrected, and manually curated to eliminate astigmatism or defocus outliers and micrographs with excessive ice contamination, yielding 2978 final micrographs. Particles were picked using Blob Picker on 500 micrographs with a minimum and maximum particle diameter of 150 and 250 Å for circular blobs, yielding 259,587 particles, which were extracted with a 320-pixel box size and subjected to 2D class averaging with 100 classes. From these, seven classes were chosen for template picking applied to the full final micrograph set, yielding 1,655,400 particles that were extracted with a 256-pixel box size and subjected to multiple rounds of 2D classification and manual selection, multi-class ab initio refinements and multi-class heterogeneous refinements to eliminate junk particles. Early rounds of analysis quickly identified three particle populations consisting of unliganded Env SOSIP trimer, SOSIP trimer with a single Fab dimer bound near the CD4-binding site (CD4bs), and Fab dimers either free or bound to a poorly resolved blob of the approximate size of an Env gp120 subunit. An intermediate stage hetero-refinement with 91,638 particles used eight starting models that included four CD4bs SOSIP-Fab dimer complexes, two unliganded SOSIP trimers, and two Fab-blob complexes. This refinement yielded two junk classes (4,440 particles total), two Fab-blob classes (13,160 particles total), one unliganded SOSIP class (18,058), two CD4bs classes (40,350 total particles), and a new class with a single Fab dimer bound near the V3 glycan supersite of the SOSIP trimer (15,630 particles). We then returned to an earlier, larger stack with 324,010 particles, and subjected this to a hetero-refinement using the unliganded SOSIP, the CD4bs complex and the V3 complex as starting models, which yielded unliganded SOSIP classes with 82,025 particles, and CD4bs-complex classes with 72,951 particles, and a V3-complex class with 21,502 particles. These particle stacks for the unliganded SOSIP and the CD4bs-complex were further cleaned to remove junk particles and yielded final reconstructions with 48,127 and 59,012 particles and achieving final global resolutions of 3.9 and 4.7 Å, respectively, as measured by cryoSPARC’s auto-masked, phase-randomized-corrected gold standard FSC calculation at the FSC 0.143 level. The map of the V3-complex class was used to generate ten 2D templates that were used to re-pick the micrographs, yielding 1,827,101 particles extracted with 320-pixel box size. This stack was subjected to 2D classification and manual selection, multi-class ab initio refinements and multi-class heterogeneous refinements to yield a final reconstruction at 6.1-Å resolution with 14,873 particles. Using cryoSPARC’s 3D Variability tool we noted substantial movement of the fab dimer relative to the SOSIP trimer in the CD4bs complex; therefore we performed particle subtraction and local refinements focused on the fab dimer of the CD4bs-complex and the V3-complex. The local refinements reported the same or slightly worse resolution compared to the homogeneous refinements; however, the density for the Fab dimer was better defined as judged by eye, particularly the appearance of beta-strands within the Fab dimer interface. Local resolution at the FSC 0.5 level was estimated for all final reconstructions and maps were locally filtered and sharpened at 1.5x the reported B-value from the final refinements. Images were created in UCSF Chimera.
For the movies of the DH898.1-bound complex, a homogeneous set of ∼122k particles were identified and re-extracted from the original dose-weighted micrographs using a binning factor 2. 3D classification in RELION (Scheres, 2012) was performed imposing C1 symmetry and using different models generated by cryoSPARC as references for alignment. All models were low-pass filtered to 60 Å before refinement. For the binding site located around a CD4 binding site glycan cluster, we identified one class with 42,745 particles that was consistent with the cryoSPARC reference model and subjected them to further 3D auto refinement. After convergence, the refinement was continued with a soft shape mask and resulted in an overall structure at 6.4-Å resolution. To obtain higher resolution, multibody refinement (Nakane et al., 2018) was performed on the refined particles using two individual soft masks (one for the Fab dimer, another for the Env trimer) created with UCSF Chimera (Pettersen et al., 2004) and RELION. The overall resolution for the gp120 trimer portion reached 4.7 Å and for the Fab dimer the estimated resolution was 6.7 Å. At this point we switched to un-binned particles and repeated the same processing strategy, but without significant improvements. We then carried out focused refinement on the Env trimer component using the re-extracted particles and imposing C3 symmetry, which lead to an improved map at 4.3-Å resolution. Based on these refinement parameters, CTF-refine and Bayesian-polishing were performed on the particles. Another round of 3D auto refine was performed using the polished particles and the reference model of the entire complex (with a soft shape mask applied), resulting in an overall resolution of 4.8 Å. Multibody refinement was performed once again based on these updated refinements. The final resolution for the gp120 trimer after post-processing was 4.65 Å and 5.8 Å for the Fab dimer. For the V3-binding site, 17,510 particles were classified out using the V3-binding reference model generated by cryoSPARC. The first round of 3D auto refinement yielded an 8.14 Å resolution map, and multibody refinement resulted in a 7.6 Å map for the gp120 portion and 8.4 Å for the Fab dimer. Given the lower resolution of this refinement and the fewer number of particles compared with binding-site1, CTF-refine and Bayesian polishing were not used for this class. In order to visualize the principal motions present in the data identified by the multibody refinement procedure, we reconstructed intermediate volumes using different relative orientations of each body and produced movies using the ‘Volume Series’ tool from ChimeraX (Goddard et al., 2018). Additional editing of the videos was performed with Movavi Video Editor (Movavi) to label the different components and provide visual aids.
For DH851.3 reconstruction, micrographs were imported into cryoSPARC and CTF-corrected. Particles were picked using Blob Picker on 500 micrographs with a minimum and maximum particle diameter of 125 and 250 Å for elliptical blobs, yielding 99,285 particles, which were extracted with a 384-pixel box size, down-sampled to 128 pixel box size, and subjected to 2D class averaging with 100 classes. From these classes, five were chosen for template picking applied to the full micrograph set, yielding 1,276,458 particles that were extracted with a 384-pixel box size and subjected to multiple rounds of 2D classification and manual selection, multi-class ab initio refinements and multi-class heterogeneous refinements to eliminate junk particles. Refinements that imposed C3 symmetry yielded ∼4.8-Å overall resolution, but at the expense of losing density in the constant domains, making it difficult to trace the elbow region to determine whether the Fab dimer was or was not domain-swapped. A 3D variability analysis confirmed that there was considerable flexibility within the constant domains, so the C3 refinement was not pursued further. The final non-uniform C1 reconstruction with 52,398 particles yielded a 5.6-Å overall resolution. Particle subtraction and local refinements focused on the Fab dimer did not improve resolution or the definition of the Fab dimer map, as judged by the lack of visible beta-strands.
Cryo-EM model fitting
Starting model for the CH848 SOSIP used for the DH898 complex was from PDB 6UM6. Starting model for the CH5050 SOSIP used for the DH851 complex was a homology model built using PDB 5VN8 as a template by submitting the DH898 sequence to SWISS-MODEL (Waterhouse et al., 2018). Homology models for DH898 and DH851 Fabs were built with SWISS-MODEL using the top reported template. Intermediate models were generated from the starting models using Rosetta (Song et al., 2013) to rebuild 500-1000 new models into the cryo-EM maps and selecting the model with the best combination of Rosetta energy and fit to the map. The cryo-EM half maps of CH848 SOSIP trimer and DH898.1 Fab dimer were used to improve the maps using a density modification program ResolveCryoEM (Terwilliger et al., 2020). The models were then fit manually in Coot (Emsley et al., 2010) following iterative refinement using Isolde (Croll, 2018) and subsequent manual coordinate fitting in Coot. These models were rigid fit into DH898.1 fab dimer bound to CH848 SOSIP trimer complex map, and the interacting glycans were fit manually in Coot followed by refinement in Isolde. DH851 structure was fit manually in Coot followed by refinement in Isolde as well. Structure and map analysis was performed using PyMol and ChimeraX (Goddard et al., 2018).
Cryo-EM of SARS-CoV-2 complexes
Purified SARS-CoV-2 spike preparations were diluted to a final concentration of ∼1 mg/mL in 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3, were mixed with 6-fold molar excess of 2G12 Fab and incubated for 2 hours at room tempearture. 2.5 uL of protein was deposited on a CF-1.2/1.3 grid that had been glow discharged for 30 s in a PELCO easiGlow Glow Discharge Cleaning System. After a 30 s incubation in > 95% humidity, excess protein was blotted away for 2.5 s before being plunge frozen into liquid ethane using a Leica EM GP2 plunge freezer (Leica Microsystems).
Cryo-EM imaging was performed on a FEI Titan Krios microscope (Thermo Fisher Scientific) operated at 300 kV. Data were acquired with a Gatan K3 detector operated in counting mode using the Leginon system (Suloway et al., 2005). The K3 experimental parameters are 2.5 exposure with 50 frames at 50 ms framerate with a total dose of 66.43 e-/A2 and a pixel size of ∼1.058 Å/px. This system was also energy-filtered with a slit width of 20 eV. A total of 6804 images were collected.
Cryo-EM image quality was monitored on-the-fly during data collection using automated processing routines. Individual frames were aligned and dose-weighted using MotionCor2 (Zheng et al., 2017b) implemented within the Appion pipeline (Lander et al., 2009). Data processing was performed within cryoSPARC (Punjani et al., 2017) including particle picking, multiple rounds of 2D classification, ab initio reconstruction, heterogeneous and homogeneous map refinements, and non-uniform map refinements. Heterogenous classifications were performed within RELION (Scheres, 2012) using masks produced in Chimera.
Structure of the all ‘down’ state (PDB ID 6VXX) previously published SARS-CoV-2 ectodomain, and a structure of the 2G12 Fab bound to Man1-2 (PDB ID 6N35) were used to fit the cryo-EM maps in ChimeraX. Coordinates were then fit manually in Coot (Emsley et al., 2010) following iterative refinement using Isolde (Croll, 2018) and subsequent manual coordinate fitting in Coot. Structure and map analysis was performed using PyMol and ChimeraX (Goddard et al., 2018).
Dimerization sequence analysis
For sequence analysis of DH851 Abs (see Figure S4), 2G12 was used as a positive control, which has four hydrophobic residues participating in contacts within the Fab dimer interface (Calarese et al., 2003); heavy chain residues 19, 77, 79 and 81. Residue 81 in the heavy chain of 2G12 was considered hydrophobic, because the amphipathic glutamine (Q) side chain was participating in a π-N bond with the aromatic phenylalanine (F) residue 77 of the adjacent heavy chain (Calarese et al., 2003). The isoleucine (I) at position 19 is especially important, because a single mutation to arginine (R) at this position is sufficient to completely disrupt Fab-dimerization and domain-swapping in 2G12 (Doores et al., 2010b). The 2G12 interface contains three rare interface residues (asterisks), which were not present in the germline sequence (Huber et al., 2010) and thus represent mutations acquired during antibody affinity maturation. In contrast to the I-shaped 2G12, the Y-shaped antibody VRC01 has only a single hydrophobic residue, no rare residues, and three charged residues in these positions, including an arginine at position 19. The three DH851 Abs that showed I-shaped, Fab-dimerized Abs, DH851.1 to DH851.3, have three or four hydrophobic interface residues, most of them rare, and no charged residues. In contrast, DH851.4 to DH851.6, DH851.8 and the DH851UCA have only a single hydrophobic interface residue and these Abs are all Y-shaped, similar to the negative control VRC01. For DH1003 lineage analysis (see Data S2Q), the specific pattern of hydrophobic residues within the interface correlated with the phylogram branch points, as indicated in the last column, suggesting that evolution of the lineage correlated with Fab-dimerization; this hypothesis will be explored further in subsequent studies.
Quantification and statistical analysis
We used a statistical test to look for distribution shifts between two time points (week 105 and 145) and the DH717 lineage cells within the RMs were considered the unit of analysis (Figure 2A). The asterisk indicated significance at p < 0.0001, using a non-parametirc Wilcoxon test performed in SAS 9.4 (SAS Institute, Cary, NC). In ELISA (Figures 2, S2, and S4), error bars represented the standard error of the mean (GraphPad Prism) for mAb binding levels that was generated from multiple independent assay.
Acknowledgments
We thank the DHVI teams from Program Management, Computational Biology, and the Biostatistics Center for their support. Thomas B. Kepler provided computational support, and Charles Giamberardino and Jennifer Tenor generated heat-killed yeast antigens. The following individuals provided technical assistance: Andy (Shi) Huang, Shi-Mao Xia, Melissa Cooper, Kara Anasti, Maggie Barr, Callie Vivian, Giovanna Hernandez, Esther Lee, Aja Sanzone, and Paige Rawls. Cryo-EM data were collected at the Shared Materials Instrumentation Facility at Duke University and National Center for CryoEM Access and Training and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247). The adjuvant GLA-SE was provided by Infectious Disease Research Institute, Seattle, WA 98102, USA; Christopher Fox and Steve Reed. The use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (contract W-31-109-Eng-38). This study utilized the computational resources offered by Duke Research Computing (https://rc.duke.edu). This work was funded by NIH, NIAID, DAIDS grants Center for HIV/Vaccine Immunology-Immunogen Discovery (CHAVI-ID) (UM1-AI100645) and Consortia for HIV/AIDS Vaccine Development (CHAVD) (UM-1AI44371) to B.F.H.; NIH (R01-AI145687 to P.A., R01-AI140897 to W.B.W., R01-AI128832 to G.K., and R01-AI120801 to K.O.S.); Translating Duke Health Initiative (to P.A.); MSTP (T32GM007171 to R.R.M.); NIAID (F30-AI22982-0 to R.R.M. and P01-AI131251 to G.M.S.); DARPA (N66001-09-C-2082 to G.D.S.); NIH/NIAID (UC6AI058607 to G.D.S. and HHSN272201800004C to C.C.L.); Duke CFAR; Intramural Research Program of the VRC, NIAID, NIH (to G.B.E.S.-J. and P.D.K.); and amfAR and Swarthmore College Startup and Faculty Research Funds (109502-61-RKVA to D.F.).
Author contributions
B.F.H. conceived and designed the study, evaluated all data, and wrote the paper. P.A. (SARS-CoV-2-related and structural studies), W.B.W. (macaque and human B cell repertoire studies), R.R.M. (vaccination study workup), and R.J.E. (structural studies) designed and performed different aspects of the study, co-wrote, and edited the paper. D.F., K. Manne, N.I.N., R.H., A.L.H., M.J.B., Y.Z., A.B., K. Mansouri, M.S.L., V.S., M.K., N.B., G.B.E.S.-J., and P.D.K. performed or contributed to structural studies. J.P. (fungal antigens) and K.O.S. (recombinant proteins and glycan array) provided key reagents and tools. M.A.M. and A.Y.A. performed FACS and data analyses. T.B., K.W., M. Berry, T.E., and B.H. generated and analyzed BCR repertoires and transcriptome gene sequences. S.M.A., K.J., S.G., R.J.P., M.D., A.F., J.S., D.C.M., C.C.L., M. Bonsignori, M.S., T.O., and G.D.S. performed functional characterization of mAbs. S.S. performed NHP immunizations and care. B.A., W.E.W., J.R.F., G.M.L., R.L., and R.A.S. designed or constructed synthetic peptides. G.K. contributed to B cell lineage studies. G.M.S. and H.L. designed SHIV infection of RMs.
Declaration of interests
B.A. and W.E.W. are co-founders of Chemitope Technologies, and G.M.L. is a founder of Avidea Technologies that now commercially produce peptides used in our HIV-1 vaccination regimen. The remaining authors declare no competing interests.
Published: May 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.04.042.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., Liao H.X., Tomaras G.D., Bonsignori M., Tsao C.Y., Hwang K.K., Chen H., Lloyd K.E., Bowman C., Sutherland L., et al. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J. Virol. 2013;87:1554–1568. doi: 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., Aussedat B., Vohra Y., Meyerhoff R.R., Cale E.M., Walkowicz W.E., Radakovich N.A., Anasti K., Armand L., Parks R., et al. Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci. Transl. Med. 2017;9:eaai7521. doi: 10.1126/scitranslmed.aai7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J., del Guercio M.F., Maewal A., Qiao L., Fikes J., Chesnut R.W., Paulson J., Bundle D.R., DeFrees S., Sette A. Linear PADRE T helper epitope and carbohydrate B cell epitope conjugates induce specific high titer IgG antibody responses. J. Immunol. 2000;164:1625–1633. doi: 10.4049/jimmunol.164.3.1625. [DOI] [PubMed] [Google Scholar]
- Andrabi R., Voss J.E., Liang C.H., Briney B., McCoy L.E., Wu C.Y., Wong C.H., Poignard P., Burton D.R. Identification of Common Features in Prototype Broadly Neutralizing Antibodies to HIV Envelope V2 Apex to Facilitate Vaccine Design. Immunity. 2015;43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N. B-1 Cell Heterogeneity and the Regulation of Natural and Antigen-Induced IgM Production. Front. Immunol. 2016;7:324. doi: 10.3389/fimmu.2016.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazkova J., Refsland E.W., Clarridge K.E., Shi V., Justement J.S., Huiting E.D., Gittens K.R., Chen X., Schmidt S.D., Liu C., et al. Glycan-dependent HIV-specific neutralizing antibodies bind to cells of uninfected individuals. J. Clin. Invest. 2019;129:4832–4837. doi: 10.1172/JCI125955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O., Head S., Mondala T., Scanlan C., Huflejt M.E., Alvarez R., Bryan M.C., Fazio F., Calarese D., Stevens J., et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C., Doores K.J., Dunlop D.C., Thaney V., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS ONE. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Hwang K.K., Chen X., Tsao C.Y., Morris L., Gray E., Marshall D.J., Crump J.A., Kapiga S.H., Sam N.E., et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Kreider E.F., Fera D., Meyerhoff R.R., Bradley T., Wiehe K., Alam S.M., Aussedat B., Walkowicz W.E., Hwang K.K., et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 2017;9:eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Scott E., Wiehe K., Easterhoff D., Alam S.M., Hwang K.K., Cooper M., Xia S.M., Zhang R., Montefiori D.C., et al. Inference of the HIV-1 VRC01 Antibody Lineage Unmutated Common Ancestor Reveals Alternative Pathways to Overcome a Key Glycan Barrier. Immunity. 2018;49:1162–1174.e8. doi: 10.1016/j.immuni.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T., Peppa D., Pedroza-Pacheco I., Li D., Cain D.W., Henao R., Venkat V., Hora B., Chen Y., Vandergrift N.A., et al. RAB11FIP5 Expression and Altered Natural Killer Cell Function Are Associated with Induction of HIV Broadly Neutralizing Antibody Responses. Cell. 2018;175:387–399.e17. doi: 10.1016/j.cell.2018.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A., Predl R., Strutzenberger K., Steinfellner W., Trkola A., Purtscher M., Gruber G., Tauer C., Steindl F., Jungbauer A., et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Buchner C., Bryant C., Eslami A., Lakos G. Anti-nuclear antibody screening using HEp-2 cells. J. Vis. Exp. 2014;(88):e51211. doi: 10.3791/51211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese D.A., Scanlan C.N., Zwick M.B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M.R., Stanfield R.L., Roux K.H., et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Cao L., Diedrich J.K., Kulp D.W., Pauthner M., He L., Park S.R., Sok D., Su C.Y., Delahunty C.M., Menis S., et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali P., Burastero S.E., Nakamura M., Inghirami G., Notkins A.L. Human lymphocytes making rheumatoid factor and antibody to ssDNA belong to Leu-1+ B-cell subset. Science. 1987;236:77–81. doi: 10.1126/science.3105056. [DOI] [PubMed] [Google Scholar]
- Chuang G.Y., Zhou J., Acharya P., Rawi R., Shen C.H., Sheng Z., Zhang B., Zhou T., Bailer R.T., Dandey V.P., et al. Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition. Structure. 2019;27:196–206.e6. doi: 10.1016/j.str.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran M.M., Phad G.E., Vázquez Bernat N., Stahl-Hennig C., Sumida N., Persson M.A., Martin M., Karlsson Hedestam G.B. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun. 2016;7:13642. doi: 10.1038/ncomms13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M., Ward A.B., Wilson I.A. Structure and Immune Recognition of the HIV Glycan Shield. Annu. Rev. Biophys. 2018;47:499–523. doi: 10.1146/annurev-biophys-060414-034156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll T.I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C.N., Saunders K.O. Antibody responses to the HIV-1 envelope high mannose patch. Adv. Immunol. 2019;143:11–73. doi: 10.1016/bs.ai.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCamp A., Hraber P., Bailer R.T., Seaman M.S., Ochsenbauer C., Kappes J., Gottardo R., Edlefsen P., Self S., Tang H., et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2014;88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky B.J., Kojima T., Rodin A., Charab W., Ippolito G.C., Ellington A.D., Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- Doores K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015;282:4679–4691. doi: 10.1111/febs.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J., Bonomelli C., Harvey D.J., Vasiljevic S., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. USA. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J., Fulton Z., Huber M., Wilson I.A., Burton D.R. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J. Virol. 2010;84:10690–10699. doi: 10.1128/JVI.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J., Huber M., Le K.M., Wang S.K., Doyle-Cooper C., Cooper A., Pantophlet R., Wong C.H., Nemazee D., Burton D.R. 2G12-expressing B cell lines may aid in HIV carbohydrate vaccine design strategies. J. Virol. 2013;87:2234–2241. doi: 10.1128/JVI.02820-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Bhiman J.N., Roark R.S., Schramm C.A., Gorman J., Chuang G.Y., Pancera M., Cale E.M., Ernandes M.J., Louder M.K., et al. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J. Virol. 2015;90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.J., Mansouri K., Stalls V., Manne K., Watts B., Parks R., Janowska K., Gobeil S.M.C., Kopp M., Li D., et al. Cold sensitivity of the SARS-CoV-2 spike ectodomain. Nat. Struct. Mol. Biol. 2021;28:128–131. doi: 10.1038/s41594-020-00547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera D., Lee M.S., Wiehe K., Meyerhoff R.R., Piai A., Bonsignori M., Aussedat B., Walkowicz W.E., Ton T., Zhou J.O., et al. HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope. Nat. Commun. 2018;9:1111. doi: 10.1038/s41467-018-03565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francica J.R., Laga R., Lynn G.M., Mužíková G., Androvič L., Aussedat B., Walkowicz W.E., Padhan K., Ramirez-Valdez R.A., Parks R., et al. Star nanoparticles delivering HIV-1 peptide minimal immunogens elicit near-native envelope antibody responses in nonhuman primates. PLoS Biol. 2019;17:e3000328. doi: 10.1371/journal.pbio.3000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F., Sok D., Kong L., McBride R., Kim H.J., Saye-Francisco K.F., Julien J.P., Hua Y., Cupo A., Moore J.P., et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F., Lee J.H., de Val N., de la Pena A.T., Kong L., Puchades C., Hua Y., Stanfield R.L., Burton D.R., Moore J.P., et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill T.R., Trimble R.B. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta. 1999;1426:227–237. doi: 10.1016/s0304-4165(98)00126-3. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J., Soto C., Yang M.M., Davenport T.M., Guttman M., Bailer R.T., Chambers M., Chuang G.Y., DeKosky B.J., Doria-Rose N.A., et al. NISC Comparative Sequencing Program Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat. Struct. Mol. Biol. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.O., Holodick N.E., Rothstein T.L. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70- J. Exp. Med. 2011;208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Gow N.A. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 2013;90:1147–1161. doi: 10.1111/mmi.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Sarkar A., Kulp D.W., Toy L., Hu X., Deresa I., Kalyuzhniy O., Kaushik K., Upadhyay A.A., Menis S., et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 2018;10:eaat0381. doi: 10.1126/scitranslmed.aat0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344:588–589. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Fleming J., St Clair E.W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R.M., Plonk K., Staats H.F., et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Holodick N.E., Rodríguez-Zhurbenko N., Hernández A.M. Defining Natural Antibodies. Front. Immunol. 2017;8:872. doi: 10.3389/fimmu.2017.00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M., Le K.M., Doores K.J., Fulton Z., Stanfield R.L., Wilson I.A., Burton D.R. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J. Virol. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa D., Asano M., Shinton S.A., Brill-Dashoff J., Formica A.M., Velcich A., Hardy R.R., Hayakawa K. Natural anti-intestinal goblet cell autoantibody production from marginal zone B cells. J. Immunol. 2015;194:606–614. doi: 10.4049/jimmunol.1402383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkeller K., Scally S.W., Bosch A., Martí G.P., Costa G., Triller G., Murugan R., Renna V., Jumaa H., Kremsner P.G., et al. Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope. Science. 2018;360:1358–1362. doi: 10.1126/science.aar5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler T.B. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler T.B., Munshaw S., Wiehe K., Zhang R., Yu J.S., Woods C.W., Denny T.N., Tomaras G.D., Alam S.M., Moody M.A., et al. Reconstructing a B-Cell Clonal Lineage. II. Mutation, Selection, and Affinity Maturation. Front. Immunol. 2014;5:170. doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Rajewsky K., Küppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Lee J.H., Doores K.J., Murin C.D., Julien J.P., McBride R., Liu Y., Marozsan A., Cupo A., Klasse P.J., et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche C.C., Henderson R., Hsu A., Behrens S., Chen X., Zhou T., Wiehe K., Saunders K.O., Alam S.M., Bonsignori M., et al. Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLoS Pathog. 2019;15:e1008026. doi: 10.1371/journal.ppat.1008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalor P.A., Herzenberg L.A., Adams S., Stall A.M. Feedback regulation of murine Ly-1 B cell development. Eur. J. Immunol. 1989;19:507–513. doi: 10.1002/eji.1830190315. [DOI] [PubMed] [Google Scholar]
- Lander G.C., Stagg S.M., Voss N.R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P.W., et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-C.D., Watanabe Y., Wu N.C., Han J., Kumar S., Pholcharee T., Seabright G.E., Allen J.D., Lin C.-W., Yang J.-R., et al. A cross-neutralizing antibody between HIV-1 and influenza virus. PLOS PATHOGENS. 2021;17 doi: 10.1371/journal.ppat.1009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang S., Kong R., Ding W., Lee F.H., Parker Z., Kim E., Learn G.H., Hahn P., Policicchio B., et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2016;113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K., et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Yang G., Wiehe K., Nicely N.I., Vandergrift N.A., Rountree W., Bonsignori M., Alam S.M., Gao J., Haynes B.F., Kelsoe G. Polyreactivity and autoreactivity among HIV-1 antibodies. J. Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn G.M., Laga R., Darrah P.A., Ishizuka A.S., Balaci A.J., Dulcey A.E., Pechar M., Pola R., Gerner M.Y., Yamamoto A., et al. In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat. Biotechnol. 2015;33:1201–1210. doi: 10.1038/nbt.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka J. Surface glycans of Candida albicans and other pathogenic fungi: physiological roles, clinical uses, and experimental challenges. Clin. Microbiol. Rev. 2004;17:281–310. doi: 10.1128/CMR.17.2.281-310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McDavid A., Finak G., Chattopadyay P.K., Dominguez M., Lamoreaux L., Ma S.S., Roederer M., Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- Montefiori D.C., Karnasuta C., Huang Y., Ahmed H., Gilbert P., de Souza M.S., McLinden R., Tovanabutra S., Laurence-Chenine A., Sanders-Buell E., et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J. Infect. Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M.A., Pedroza-Pacheco I., Vandergrift N.A., Chui C., Lloyd K.E., Parks R., Soderberg K.A., Ogbe A.T., Cohen M.S., Liao H.X., et al. Immune perturbations in HIV-1-infected individuals who make broadly neutralizing antibodies. Sci. Immunol. 2016;1:aag0851. doi: 10.1126/sciimmunol.aag0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin C.D., Julien J.P., Sok D., Stanfield R.L., Khayat R., Cupo A., Moore J.P., Burton D.R., Wilson I.A., Ward A.B. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J. Virol. 2014;88:10177–10188. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T., Kimanius D., Lindahl E., Scheres S.H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife. 2018;7:e36861. doi: 10.7554/eLife.36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J.S., King R.G., Kearney J.F. Manipulation of the glycan-specific natural antibody repertoire for immunotherapy. Immunol. Rev. 2016;270:32–50. doi: 10.1111/imr.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M., et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plath F., Ringler P., Graff-Meyer A., Stahlberg H., Lauer M.E., Rufer A.C., Graewert M.A., Svergun D., Gellermann G., Finkler C., et al. Characterization of mAb dimers reveals predominant dimer forms common in therapeutic mAbs. MAbs. 2016;8:928–940. doi: 10.1080/19420862.2016.1168960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L.K., Vasiljevic S., Ozorowski G., Seabright G.E., Cupo A., Ringe R., Kim H.J., Sanders R.W., Doores K.J., Burton D.R., et al. Structural Constraints Determine the Glycosylation of HIV-1 Envelope Trimers. Cell Rep. 2015;11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Quách T.D., Rodríguez-Zhurbenko N., Hopkins T.J., Guo X., Hernández A.M., Li W., Rothstein T.L. Distinctions among Circulating Antibody-Secreting Cell Populations, Including B-1 Cells, in Human Adult Peripheral Blood. J. Immunol. 2016;196:1060–1069. doi: 10.4049/jimmunol.1501843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A., Darko S., Hua A., Overman G., Ransier A., Francica J.R., Trama A., Tomaras G.D., Haynes B.F., Douek D.C., Kepler T.B. Structure and Diversity of the Rhesus Macaque Immunoglobulin Loci through Multiple De Novo Genome Assemblies. Front. Immunol. 2017;8:1407. doi: 10.3389/fimmu.2017.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roark R.S., Li H., Williams W.B., Chug H., Mason R.D., Gorman J., Wang S., Lee F.H., Rando J., Bonsignori M., et al. Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth. Science. 2021;371:eabd2638. doi: 10.1126/science.abd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskin K.M., Jackson K.J.L., Lee J.Y., Hoh R.A., Joshi S.A., Hwang K.K., Bonsignori M., Pedroza-Pacheco I., Liao H.X., Moody M.A., et al. Aberrant B cell repertoire selection associated with HIV neutralizing antibody breadth. Nat. Immunol. 2020;21:199–209. doi: 10.1038/s41590-019-0581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougé L., Chiang N., Steffek M., Kugel C., Croll T.I., Tam C., Estevez A., Arthur C.P., Koth C.M., Ciferri C., et al. Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab. Science. 2020;367:1224–1230. doi: 10.1126/science.aaz9356. [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Nicely N.I., Wiehe K., Bonsignori M., Meyerhoff R.R., Parks R., Walkowicz W.E., Aussedat B., Wu N.R., Cai F., et al. Vaccine Elicitation of High Mannose-Dependent Neutralizing Antibodies against the V3-Glycan Broadly Neutralizing Epitope in Nonhuman Primates. Cell Rep. 2017;18:2175–2188. doi: 10.1016/j.celrep.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Verkoczy L.K., Jiang C., Zhang J., Parks R., Chen H., Housman M., Bouton-Verville H., Shen X., Trama A.M., et al. Vaccine Induction of Heterologous Tier 2 HIV-1 Neutralizing Antibodies in Animal Models. Cell Rep. 2017;21:3681–3690. doi: 10.1016/j.celrep.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Wiehe K., Tian M., Acharya P., Bradley T., Alam S.M., Go E.P., Scearce R., Sutherland L., Henderson R., et al. Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science. 2019;366:eaay7199. doi: 10.1126/science.aay7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan C.N., Pantophlet R., Wormald M.R., Ollmann Saphire E., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Rudd P.M., Burton D.R. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan C.N., Offer J., Zitzmann N., Dwek R.A. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright G.E., Cottrell C.A., van Gils M.J., D’addabbo A., Harvey D.J., Behrens A.J., Allen J.D., Watanabe Y., Scaringi N., Polveroni T.M., et al. Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions. Structure. 2020;28:897–909.e6. doi: 10.1016/j.str.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.S., Janes H., Hawkins N., Grandpre L.E., Devoy C., Giri A., Coffey R.T., Harris L., Wood B., Daniels M.G., et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford D.A., Strominger J.L. Analysis of the oligosaccharides on the HLA-DR and DC1 B cell antigens. J. Immunol. 1983;130:274–282. [PubMed] [Google Scholar]
- Song Y., DiMaio F., Wang R.Y., Kim D., Miles C., Brunette T., Thompson J., Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield R.L., De Castro C., Marzaioli A.M., Wilson I.A., Pantophlet R. Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose. Glycobiology. 2015;25:412–419. doi: 10.1093/glycob/cwu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen J.M., Lin Y.C., Havenar-Daughton C., Pecetta S., Ozorowski G., Willis J.R., Toy L., Sok D., Liguori A., Kratochvil S., et al. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science. 2019;366:eaax4380. doi: 10.1126/science.aax4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Swindells M.B., Porter C.T., Couch M., Hurst J., Abhinandan K.R., Nielsen J.H., Macindoe G., Hetherington J., Martin A.C. abYsis: Integrated Antibody Sequence and Structure-Management, Analysis, and Prediction. J. Mol. Biol. 2017;429:356–364. doi: 10.1016/j.jmb.2016.08.019. [DOI] [PubMed] [Google Scholar]
- Taylor W.R. Residual colours: a proposal for aminochromography. Protein Eng. 1997;10:743–746. doi: 10.1093/protein/10.7.743. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Ludtke S.J., Read R.J., Adams P.D., Afonine P.V. Improvement of cryo-EM maps by density modification. Nat. Methods. 2020;17:923–927. doi: 10.1038/s41592-020-0914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill J.C., Weller S., Reynaud C.A. Human marginal zone B cells. Annu. Rev. Immunol. 2009;27:267–285. doi: 10.1146/annurev.immunol.021908.132607. [DOI] [PubMed] [Google Scholar]
- Weller S., Braun M.C., Tan B.K., Rosenwald A., Cordier C., Conley M.E., Plebani A., Kumararatne D.S., Bonnet D., Tournilhac O., et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J.R., Zhang R., Khurana S., King L.R., Manischewitz J., Golding H., Dormitzer P.R., Haynes B.F., Walter E.B., Moody M.A., et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.B., Liao H.X., Moody M.A., Kepler T.B., Alam S.M., Gao F., Wiehe K., Trama A.M., Jones K., Zhang R., et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.D., Ofek G., Schätzle S., McDaniel J.R., Lu X., Nicely N.I., Wu L., Lougheed C.S., Bradley T., Louder M.K., et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol. 2017;2:eaal2200. doi: 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.B., Zhang J., Jiang C., Nicely N.I., Fera D., Luo K., Moody M.A., Liao H.X., Alam S.M., Kepler T.B., et al. Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat. Commun. 2017;8:1732. doi: 10.1038/s41467-017-01336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.T., Kabat E.A. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J. Exp. Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., West A.P., Jr., Kim H.J., Thornton M.E., Ward A.B., Bjorkman P.J. Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12. Cell Rep. 2013;5:1443–1455. doi: 10.1016/j.celrep.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Reth M. Oligomeric organization of the B-cell antigen receptor on resting cells. Nature. 2010;467:465–469. doi: 10.1038/nature09357. [DOI] [PubMed] [Google Scholar]
- Yang J., Reth M. The dissociation activation model of B cell antigen receptor triggering. FEBS Lett. 2010;584:4872–4877. doi: 10.1016/j.febslet.2010.09.045. [DOI] [PubMed] [Google Scholar]
- Yu J.S., Ma B.J., Scearce R.M., Liao H.X., Haynes B.F. Anti-Ebola MAb 17A3 reacts with bovine and human alpha-2-macroglobulin proteins. J. Virol. Methods. 2010;168:248–250. doi: 10.1016/j.jviromet.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Verkoczy L., Wiehe K., Munir Alam S., Nicely N.I., Santra S., Bradley T., Pemble C.W., 4th, Zhang J., Gao F., et al. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci. Transl. Med. 2016;8:336ra62. doi: 10.1126/scitranslmed.aaf0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I.S., Altae-Tran H.R., Chuang G.Y., Joyce M.G., et al. NISC Comparative Sequencing Program Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variable heavy and light chain gene sequences for recombinant mAbs were deposited in GenBank and can be accessed using the following accession numbers; GenBank: MT470283–MT470354. The transcriptome sequencing data were deposited in NCBI Sequence Read Archive (SRA) under Bioproject: PRJNA685659.The atomic models and cryo-EM reconstructions and generated during this study are available at PDB and EMBD (https://www.rcsb.org; http://emsearch.rutgers.edu) under the accession codes PDB: 6VTU, 6XRJ, 7L6M, 7L6O, 7LUA, 7LU9, 7L09, 7L06, and 7L02 and EMDB: EMD-23145, EMD-23124, EMD-23519, EMD-23518, EMD-23097, EMD-23095, EMD-23094, EMD-23152, EMD-23153.