Abstract
We combined viral genome sequencing with contact tracing to investigate introduction and evolution of severe acute respiratory syndrome coronavirus 2 lineages in Santa Clara County, California, from 27 January to 21 March 2020. From 558 persons with coronavirus disease 2019, 101 genomes from 143 available clinical samples comprised 17 lineages, including SCC1 (n = 41), WA1 (n = 9; including the first 2 reported deaths in the United States, with postmortem diagnosis), D614G (n = 4), ancestral Wuhan Hu-1 (n = 21), and 13 others (n = 26). Public health intervention may have curtailed the persistence of lineages that appeared transiently during February and March. By August, only D614G lineages introduced after 21 March were circulating in Santa Clara County.
Keywords: SARS-CoV-2, COVID-19, viral whole-genome sequencing, public health surveillance, epidemiology, D614G lineage, viral evolution
We used viral whole-genome sequencing with contact tracing to investigate the introduction, transmission, and fate of 17 severe acute respiratory syndrome coronavirus 2 lineages into Santa Clara County, California, early in the pandemic in the United States (January–March 2020).
The coronavirus disease 2019 (COVID-19) pandemic from the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged from Wuhan, China, in December 2019 and rapidly spread throughout the world, causing approximately 163 million cases and 3.4 million deaths as of 16 May 2021 [1]. The first confirmed SARS-CoV-2 case in the United States was diagnosed in a resident of Washington State on 20 January 2020 [2]; since then, multiple introductions into the United States been reported [3–9], resulting in widespread community dissemination nationwide [9]. For outbreaks caused by SARS-CoV-2, health response and action play critical roles in the recognition and isolation of suspected infectious cases. Contact tracing is a classic epidemiologic tool to study outbreaks of infectious disease and track patterns of transmission that can inform public health interventions [10].
Genomic epidemiology using viral whole-genome sequencing (WGS) complements contact tracing during outbreak investigations and can track virus evolution and spread in an epidemic [11]. WGS of SARS-CoV-2 has been used to identify (1) undetected transmission of the WA1 lineage associated with the first reported SARS-CoV-2 case in the United States from Washington State in January 2020 [3], (2) multiple introductions of SARS-CoV-2 lineages into Northern California [4], coast-to-coast transmission [5], and (3) importation of a viral lineage containing a D614G mutation (A23403G single-nucleotide variant [SNV]) in the viral spike protein to New York from Europe [6, 8, 12], with subsequent dispersion throughout the United States [12]. However, few studies to date have included sampling and analysis of dynamic changes in SARS-CoV-2 genotypes within a single community over time. In the current study we sequenced a demographically representative sampling of SARS-CoV-2 strains circulating in Santa Clara County (SCC) from 27 January to 21 March 2020 and analyzed publicly available viral WGS data to mid-October 2020, to investigate the introduction, transmission, and persistence or disappearance of SARS-CoV-2 lineages in this community.
METHODS
Ethics
Nasopharyngeal and/or oropharyngeal swab specimens were collected for the purpose of diagnostic testing as part of public health practice during the pandemic response at the SCC Public Health Laboratory (SCCPHL), California Department of Public Health, and the US Centers for Disease Control and Prevention (CDC). Viral WGS was performed at the University of California, San Francisco (UCSF) genomics laboratory with the approval of UCSF’s Institutional Review Board (protocol no. 11-05519). Viral WGS studies of samples submitted to the SCCPHL were designated exempt by the Committee for the Protection of Human Subjects (project no. 2020–30; issued under the California Health and Human Services Agency’s Federal Wide Assurance no. 00000681 with the Office of Human Research Protections).
Sample Collection, Quantitative Reverse-Transcription Polymerase Chain Reaction Testing, and Contact Tracing Investigation
SARS-CoV-2 nasopharyngeal and/or oropharyngeal samples were collected in SCC from 27 January to 21 March 2020 (Supplementary Methods). For the autopsy cases, formalin-fixed paraffin-embedded tissue specimens from 2 persons who died from unknown causes on 6 and 17 February 2020 were submitted to the US CDC for analysis. Quantitative real-time reverse-transcription polymerase chain reaction testing for laboratory diagnosis of COVID-19 was performed initially by the CDC and subsequently by the SCCPHL [13]. Contact tracing was performed according to standardized protocols (Supplementary Methods).
Viral WGS, Assembly, and Phylogenetic Analysis
Viral WGS and Sanger sequencing confirmation of SNVs was performed as described (Supplementary Methods) [4, 14]. Complete, high-quality SARS-CoV-2 (n = 19 922) genomes from the global COVID-19 pandemic with collection date information, which had been sequenced from samples obtained from infected persons on or before 23 March 2020, were downloaded from the Global Initiative on Sharing All Influenza Data (GISAID) database (10 August 12020 build) [15, 16], expanded to include SARS-CoV-2 genomes, and processed using the Nextstrain bioinformatics pipeline Augur [17]. After addition of the 101 newly sequenced genomes in the current study to the data set, a total of 20 223 genomes were aligned using MAFFT v7.4 software [18] as implemented in Augur, and a maximum-likelihood phylogenetic tree was constructed using IQTREE v1.6 software [19]. Branch locations were estimated using a maximum-likelihood discrete traits model. The resulting tree was visualized in the Nextstrain Web application Auspice [17] and using Geneious v11.1.5 software [20]. Smaller subtrees consisting of viruses in the WA1, SCC1, and SCC3 lineages were also constructed using the Augur pipeline. Multiple sequence alignments of clusters were generated using MAFFT v7.388 software [18] and visualized using Geneious software (Supplementary Methods). Lineage and cluster information extracted from the phylogenetic analyses was merged with the information stored in the California Reportable Disease Information Exchange (CalREDIE) database.
Correlation of Epidemiologic and Genomic Data
To assess whether the COVID-19 cases diagnosed by the SCCPHL were representative of those diagnosed in SCC during the period of our study, we compared by sex, age, race/ethnicity, and home address the information from all cases reported to CalREDIE. For cases classified as travel associated, such as imported cases, we evaluated whether the identified genomic lineage was consistent with the reported travel history. For all other COVID-19 cases that were determined to be locally acquired, we used the genomic data to confirm all links involving ≥2 persons that had been identified by contact tracing and epidemiologic investigation.
Determining the Fate of Circulating SARS-CoV-2 Lineages
After identification of the 17 lineages represented in the study, all genomes from California collected after 23 March 2020, sequenced, and deposited in GISAID as of 18 October 2020 were downloaded from GISAID. Combined with the 101 study genomes and previously analyzed California genomes collected on or before 23 March 2020, this yielded a total of 3660 total genomes. These 3660 longitudinally collected genomes from 27 January to 18 October 2020 were then screened for the presence or absence of the key single-nucleotide polymorphisms defining each of the 16 study lineages, using in-house Linux shell scripts.
Statistical Methods for Group Comparisons
For comparison of individual characteristics between COVID-19–infected persons with sequenced genomes and those for whom samples were unavailable for genomic sequencing or recovered genomes had insufficient coverage, we calculated P values using the χ 2 goodness-of-fit test. Differences were considered statistically significant at P < .05.
RESULTS
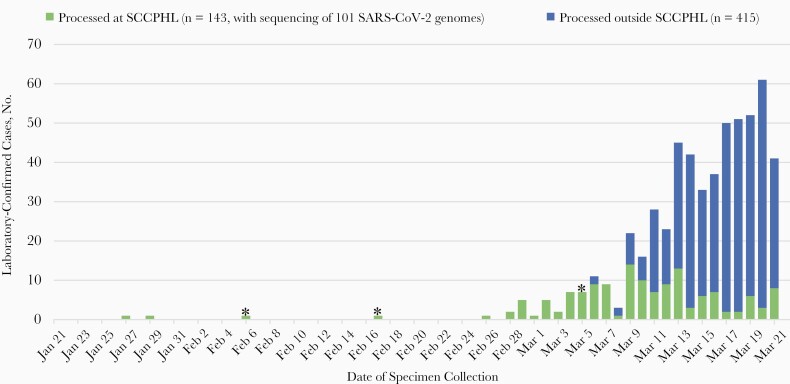
From 27 January to 21 March 2020, there were 558 SARS-CoV-2 positive cases diagnosed in SCC (all except 2 were in SCC residents) and reported to the statewide CalREDIE database (Figure 1). The SCCPHL and the CDC performed diagnostic testing on specimens from 143 of these 558 cases. Specimens from 101 of 143 cases (70.6%) had recoverable SARS-CoV-2 genomes with sufficient breadth of coverage (≥70%) across the genome for phylogenetic analysis. There were no statistically significant differences in sex or race/ethnicity between the 101 sequenced cases with viral WGS and 457 other cases in SCC (Table 1), but there were differences in age, with sequenced cases being older overall (P = .03). There was a higher proportion of deaths (P = .001) among the sequenced cases, a finding consistent with early criteria prioritizing testing of hospitalized persons with serious COVID-19 disease and with disproportionate sequencing of such cases during the January–March time frame of the study (Figure 1).
Figure 1.
Epidemic curve of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak in Santa Clara County, California, from 21 January to 21 March 2020. Laboratory-confirmed SARS-CoV-2 cases (n = 558) are shown by 2020 date. Asterisks denote collection dates for the 3 decedent medical examiner cases. Abbreviation: SCCPHL, Santa Clara County Public Health Laboratory.
Table 1.
Demographics of Coronavirus Disease 2019 Cases in Santa Clara County, California, January–March 2020
| Cases, No. (%) | |||||
|---|---|---|---|---|---|
| Characteristic | No Viral WGS (n = 457)a | Viral WGS (n = 101)b | P Valuec | ||
| Sex | |||||
| Female | 200 | 43.8 | 44 | 43.6 | .93 |
| Male | 254 | 55.6 | 57 | 56.4 | |
| Age group, y | |||||
| <20 | 13 | 2.8 | 7 | 6.9 | .03d |
| 21–30 | 49 | 10.7 | 6 | 5.9 | |
| 31–40 | 87 | 19 | 14 | 13.9 | |
| 41–50 | 98 | 21.4 | 18 | 17.8 | |
| 51–60 | 90 | 19.7 | 20 | 19.8 | |
| 61–70 | 52 | 11.4 | 15 | 14.9 | |
| >70 | 66 | 14.4 | 21 | 20.8 | |
| Race/ethnicity | |||||
| Asian/Pacific Islander | 111 | 24.3 | 39 | 38.6 |
.29 |
| Black or African American | 8 | 1.8 | 2 | 2 | |
| Latinx | 154 | 33.7 | 36 | 35.6 | |
| White | 99 | 21.7 | 18 | 17.8 | |
| Other | 22 | 4.8 | 6 | 5.9 | |
| Deceased | |||||
| No | 429 | 93.9 | 85 | 84.2 | .001d |
| Yes | 28 | 6.1 | 16 | 15.8 | |
Abbreviation: WGS, whole-genome sequencing.
aAmong 558 laboratory-confirmed cases in Santa Clara County, cases without viral WGS (n = 457) include cases with samples unavailable for sequencing (n = 420) and cases that were sequenced but yielded insufficient coverage of the viral genome (n = 37).
bComplete or near-complete viral genomes were recovered from 101 of 138 samples submitted to the Santa Clara County Public Health Laboratory.
c P values calculated using the χ 2 goodness-of-fit test.
dSignificant at the P = .05 level.
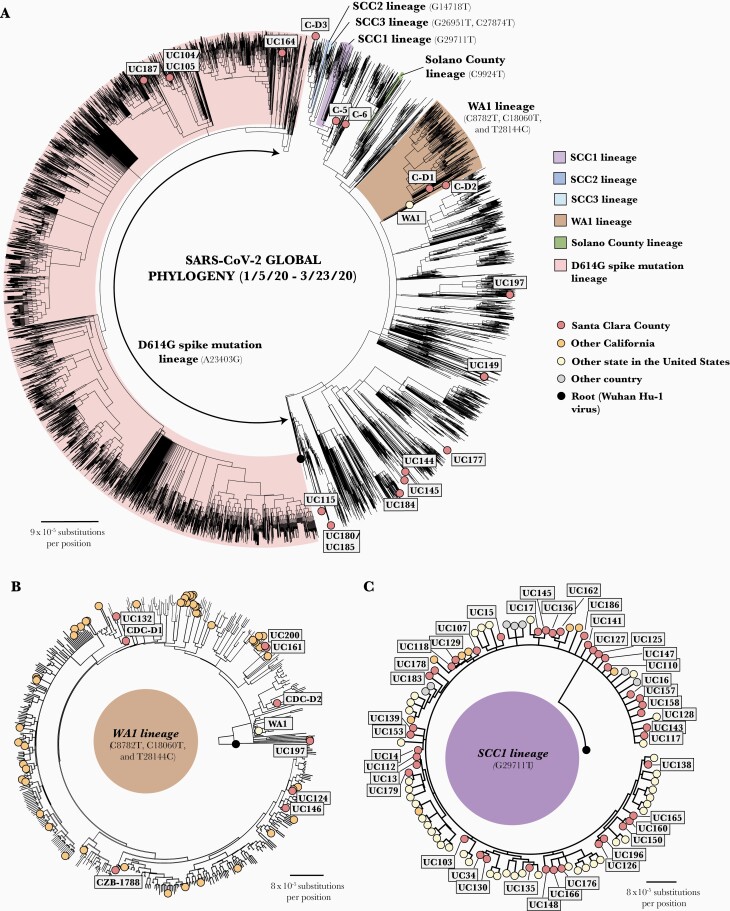
Phylogenetic analysis grouped the 101 SARS-CoV-2 genomic sequences into 17 different lineages, including the ancestral Wuhan Hu-1 lineage (n = 21), SCC1 (n = 41), SCC2 (n = 5), SCC3 (n = 9), WA1 (n = 9), a Solano County lineage associated with the first case of community transmission in the United States (n = 1), the D614G lineage associated with outbreaks in Europe and New York (n = 4), and 10 additional lineages defined by 1 or 4 SNVs identified in 11 persons (Figure 2A and Supplementary Table 1).
Figure 2.
Phylogenetic tree analysis of severe acute respiratory syndrome coronavirus (SARS-CoV-2) strains in Santa Clara county collected from 27 January to 21 March 2020. A, Radial view of Santa Clara SARS-CoV-2 viruses (n = 101) in the context of 19 922 global SARS-CoV-2 strains. Viral lineages are labeled on the tree and color coded. B, Radial view of WA1 lineage. The death case C-D2 (possessing 3 single-nucleotide variants [SNVs]) is situated on a side branch of the WA1 subtree, while the earlier death case C-D1 is positioned within the main WA1 cluster (with 5 common SNVs). Note that all genomes in the WA1 lineage are closely related to each other, and alignments can be artifactually driven by gaps (stretches of N’s) in the assembled genome. Thus, CZB-1788 is positioned away from C124 and UC146 despite all 3 individuals being passengers on the same cruise and having identical genomes. C, Overall view of SCC1 lineage sharing G29711T.
International Travel as a Risk Factor for COVID-19
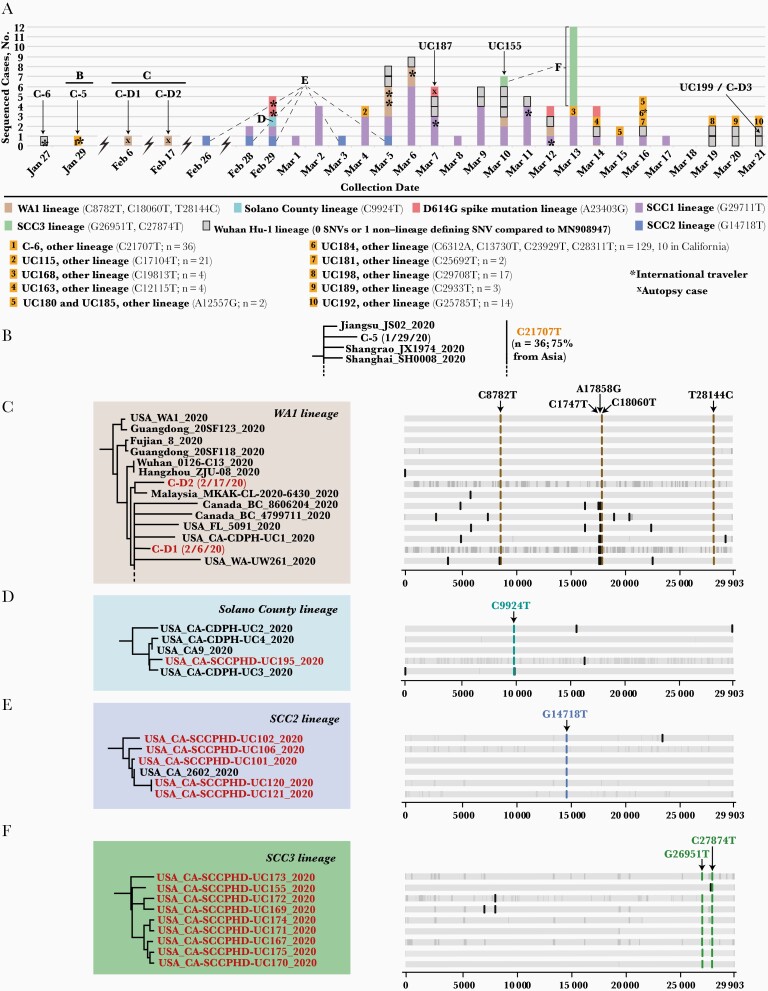
The first 2 cases in January 2020 were identified in international travelers (deposited in the GISAID database as US/CDC-5/2020 and USA/CDC-6/2020 and abbreviated as C-5 and C-6, with cases hereafter also referenced by their individual virus abbreviations) (Figures 2A and 3A and Supplementary Table 1). Consistent with their recent travel history to China, C-5 and C-6 were assigned to the ancestral Wuhan Hu-1 lineage (with 0 SNVs) and an Asian lineage defined by only 1 SNV relative to the Wuhan Hu-1 lineage (C21707T), respectively (Figure 3B). Of the other 8 persons in our series with a history of international travel, the viral genomes from 6 were positioned in clusters by phylogenetic analysis that included genomes from other cases sequenced from the geographic locations where they traveled. For instance, a couple (UC104 and UC105) were confirmed SARS-CoV-2 positive a few days after returning to California from a trip to the Middle East; their samples yielded genomes assigned to the D614G lineage and positioned within a cluster that included sequences from Egypt and Saudi Arabia (Figures 2A and 3A; Table 2, cluster N). Another couple (UC124 and CZB-1788) traveling aboard a cruise ship [21] became sick after disembarking and tested positive in early March. A third person UC146 also traveling on the cruise had COVID-19 diagnosed at approximately the same time.
Figure 3.
Single-nucleotide variant (SNV) analysis of Santa Clara severe acute respiratory syndrome coronavirus 2 strains collected between 27 January and 21 March 2020. A, Daily composition of viral lineages in Santa Clara County. The 7 main lineages are represented in different colors, while the remaining 10 lineages are given numerical designations. B, C-5 and related strains from China. C, Phylogenetic tree of WA1 strains, including 2 early death cases and their defining SNVs. Note that given the size of the WA1 subtree, not all of the 9 WA1 genomes recovered in this study are shown (dashed line). D, Solano County lineage and defining SNV. E, SCC2 lineage and the defining G14718T. F, SCC3 lineage and defining G26951T and C27874 SNVs. In B–F, highlighted red font denotes the genome as belonging to the 101 cases in the study. In C–F, the vertical gray lines represent regions of the genome with missing coverage, represented as nucleotide stretches of N’s in the assembled sequence.
Table 2.
Coronavirus Disease 2019 Clusters Defined by Epidemiologic and/or a Common Genomic Lineage
| Clustera | Description | Known Cases, No. | Viral Genomes, No.b | Case Abbreviations | Lineage (SNVs) |
|---|---|---|---|---|---|
| A | Solano County clusterc | 4 | 1 | UC195 | Solano County lineage (C9924T) |
| B | Household and HCW-associated cluster of 5 | 5 | 5 | UC101, UC102, UC106, UC120, UC121 | SCC2 lineage (G14718T) |
| C | Household cluster of 9d | 9 | 8 | UC167, UC169, UC170, UC171, UC172, UC173, UC174, UC175 | SCC3 lineage (G26951T and/or C27874T) |
| D | Cluster at local churche | 7 | 3 | UC161, UC197, UC200 | WA1 lineage (C8782T, C18060T, T28144C, C17747T, A17858G) |
| E | Household cluster of 4, including a couple and 2 in-home caregiversf | 4 | 2 | UC149 and UC151 | Wuhan Hu-1 lineage (0 SNVs) |
| F | Parent and child attending a school-hosted gathering (from which there had been other reported cases) | 2 | 2 | UC137 and UC159 | Wuhan Hu-1 lineageg |
| G | SJC airport cluster | 11 | 9 | UC13, UC14, UC15, UC34 and UC143 (workplace); UC16, UC17 (household contacts of UC13); UC117,h UC138 (HCW for UC13 and UC14) | SCC1 lineage (G29711T) |
| H | Grocery store clusteri | 6 | 3 | UC141, UC179, UC196 | SCC1 lineage (G29711T) |
| I | Cluster of 4 includes a parent who traveled to Asia, his child, a tenant, and a friend/coworker of the childj | 4 | 4 | UC135, UC150, UC158, UC166 | SCC1 lineage (G29711T) |
| J | Household cluster of 4, including a couple with history of out-of-state travel and their child | 4 | 3 | UC130, UC139, UC153 | SCC1 lineage (G29711T) |
| K | Two couple-family with history of out-of-state travel | 4 | 2 | UC107 and UC118 | SCC1 lineage (G29711T) |
| L | Household cluster of 3; multigenerational family | 3 | 3 | UC160, UC176, UC178 | SCC1 lineage (G29711T) |
| M | Couple and sibling with history of travel within California | 3 | 3 | UC103, UC110, UC112 | SCC1 lineage (G29711T) |
| N | Couple returning from a trip to the Middle East | 2 | 2 | UC104, UC105 | D614G lineage (A23403G) |
| O | Cruise ship travelers, including a couple and an additional unrelated person [21] | ≥78 | 3 | UC124, UC146, and CZB-1788 | WA1 lineage (C8782T, C18060T, T28144C, C17747T, A17858G) |
Abbreviations: HCW, healthcare worker; SJC airport, San Jose International Airport; SNVs, single-nucleotide variants.
aCluster defined as ≥2 persons with epidemiologic links confirmed by viral whole-genome sequencing (WGS).
bViral genomes from 53 cases, each part of a defined cluster, are shown in this table (the remaining 48 of 101 total cases in this study were not part of a cluster); viral genomes are not available for some cases, as the corresponding clinical specimen was not available for viral WGS.
cThe Solano County cluster (Solano County C9924T lineage) includes Solano County case UC4 from a previous publication [4].
dAnother case (UC155) was assigned to the SCC3 lineage, but there is no known epidemiologic link with the household cluster of 9.
eViral genomic sequencing identified 1 additional case in the cluster, which was missed by contact tracing.
fSamples from the 2 caregivers were not available for sequencing.
gParent and child with 0 SNVs relative to ancestral Wuhan Hu-1 genome.
hReported as UC35 in a previous publication [4].
iGrocery store cluster (SCC1 G29711T lineage) includes Solano County case UC21 from a previous publication [4].
jThe last 3 persons listed are also HCWs.
These 3 virus strains were found to be of the WA1 lineage (Figures 2B and 3A; Table 2, cluster O), sharing 5 SNVs in common with sequenced genomes from passengers and crew aboard the cruise ship and the majority of WA1 lineage viruses circulating in northern California and Washington State in during February and March 2020 [3, 4, 21]. Another international travel–related case (UC184) occurred in a person who traveled to Asia in mid-March and died after returning home. UC184 was assigned to a lineage characterized by 4 distinct SNVs (C6312A, C13730T, C23929T, and C28311T) (Figures 2A and 3A) [22]. We found 129 cases of this 4-SNV lineage reported globally as of 21 March 2020, with major clusters in India [22], southeast Asia, and California (n = 10 cases). Two of 10 persons with international travel history (UC135, who traveled to Asia, and UC162, who returned from a trip to Central America but also attended a large party in the San Francisco Bay Area) were found to be infected with viruses of the SCC1 lineage (Figures 2C and 3A).
Retrospectively Identified COVID-19 Deaths
To assess whether there were cases and deaths associated with COVID-19 in California at a time when testing for COVID-19 was limited and widespread community transmission of COVID-19 had not yet been recognized, the California Department of Public Health provided recommendations to county medical examiners on 29 April 2020 that persons who died between 17 December 2019 and 16 March 2020 from suspected COVID-19 should have postmortem specimens collected and submitted to the CDC for analysis. CDC confirmation of SARS-CoV-2 infection in postmortem tissue specimens was obtained April 2020 from 2 persons who had died at home in February from an unknown respiratory illness [7]. The viral genomes associated with both cases, C-D1 and C-D2, were determined by the CDC to be part of the WA1 lineage, with 5 and 3 SNVs, respectively (Figures 2B and 3C) [4], suggesting that infection had likely been acquired locally. In a third medical examiner case in an elderly man who died at home (UC187), clinical samples were tested at the SCCPHL and found to be positive for SARS-CoV-2; the virus was subsequently shown by viral WGS to belong to the D614G lineage (Figures 2A and 3A).
Introduction of New SARS-CoV-2 Lineages
On 26 February 2020, the first case of community transmission of SARS-CoV-2 in California (UC4) was reported [4], and the SCC Public Health Department was notified. One extended family member (UC195) had a positive SARS-CoV-2 test in late February during the 14-day quarantine period (Figure 3A and D; Table 2, cluster A). The genome of his SARS-CoV-2 strain had the same C9924T SNV as UC4 that defines the Solano County lineage (Figure 3D) [4]. Concomitant with this intercounty transmission event, an elderly woman (UC101) was hospitalized with SARS-CoV-2 infection, and contact tracing eventually identified an additional 4 infections in 2 family members (UC102 and UC106), a healthcare worker (HCW) at the hospital (UC121), and a close contact of the HCW (UC120). All 5 strains were assigned to a previously undescribed lineage containing a G14178 SNV, named SCC2 (Figure 3A and D; Table 2, cluster B).
A notable example of how the genomic surveillance directly informed contact tracing efforts involved cases UC200, UC197, and UC161. Genomic analysis of samples collected from these 3 individuals in mid-March revealed that all 3 viruses were of the WA1 lineage and shared 5 SNVs (Figures 2B and 3A; Table 2, cluster D). The genomic linkage guided further contact investigation interviews showing that UC161, previously classified as a community transmission case with unknown source, attended the same church as UC200 and UC197. Another local cluster was identified when a member of a large household became ill in late February, followed by SARS-CoV-2 infection of an additional 8 household members (UC167, UC169, and UC170–UC175). All viral genomes sequenced from this cluster were assigned to a single lineage containing G26591T and C27874T SNV, named SCC3 (Figures 2A, 3A, and 3F; Table 2, cluster C). Phylogenetic analysis revealed an additional SCC3 lineage genome (UC155) containing the C27874T SNV and corresponding to a COVID-19 case diagnosed in an unrelated SCC resident (Figure 3A and 3F). No epidemiologic link between this person and the large household cluster was identified by contact tracing.
On 29 February, the SCC Public Health Department initiated an investigation of a COVID-19 outbreak among workers at San Jose International Airport. Of 11 confirmed cases, all 9 with available viral genomes, sequenced from 5 workers, 2 household contacts, and 2 HCWs, were of the SCC1 lineage that shares the G29711T SNV (Figures 2A, 2C, and 3A; Table 2, cluster G). Overall, 41 genomes of 101 in the current study were assigned to the SCC1 lineage. Epidemiologic links were known a priori in 27 cases (69.5%), grouped into 7 clusters, including the aforementioned San Jose airport cluster [4] that includes a household transmission event and 2 HCWs, a cluster associated with a grocery store that also involved a resident from Solano County [4], and 5 other household transmission events, of which 2 had a history of domestic travel and 2 a history of international travel (Table 2, clusters G–M). No epidemiologic links were found among the remaining 14 SCC1 lineage genomes, indicating cryptic transmission of this lineage in California beginning in late February 2020 (Figure 3A and Supplementary Table 1).
Wuhan Hu-1 and D614G SARS-CoV-2 Lineages
Twenty-one of 101 SARS-CoV-2 genomes in this study (21.8%) differed from the ancestral Wuhan Hu-1 lineage by 0 SNVs or 1 non–lineage-defining SNV (Figure 3A). One of these genomes was sequenced from a young man who died at home in March (UC199; nasal swab/C-D3; postmortem formalin-fixed paraffin-embedded lung tissue). Paired household cases harboring the Wuhan Hu-1 lineage were also identified in an elderly couple (UC149 and UC151) and in a parent and child (UC137 and UC159) who attended a school-hosted gathering from which there had been other reported cases (Table 2, clusters E and F).
Phylogenetic analysis identified only 4 of 101 virus genomes (4.0%) containing the D614G (A23403G) spike mutation (Figure 2A). These included the aforementioned couple (UC104 and UC105) who traveled to the Middle East in February (Table 2, cluster N), an aforementioned death case (UC187; early March), and a middle-aged man (UC164; mid-March) without a known exposure risk factor.
Introduction of 10 Other SARS-CoV-2 Lineages
In addition to Wuhan Hu-1, WA1, SSC1, SCC2, SSC3, Solano County, and D614G, 10 other lineages were identified among cases in our series, including the aforementioned 4-SNV lineage in returning traveler UC184 (Figures 2A and 3A). For the majority of these lineages (9 of 10 [90%]), only 1 person from SCC was identified as being infected by a virus from each lineage, and these singleton cases were attributed to unknown community exposure. The only exceptions were UC180 and UC185; both male adults were infected with the A12557G lineage, although an epidemiologic link between the 2 cases was not determined.
Dynamic Changes of SARS-CoV-2 Genotypes in SCC Over Time
We performed genotype analysis of all 3660 full-length sequenced genomes from California deposited in the GISAID database that had been collected from 27 January to 30 September 2020. In January 2020, sequenced genomes from SCC corresponded mostly to Asian lineages, with 0–1 SNVs compared with the ancestral Wuhan Hu-1 lineage. The WA1, SCC1, and D614G lineages emerged in February, and SCC1 expanded to become the single dominant lineage in the county in March (accounting for approximately 25% of the sequenced genomes during that month and 40.6% of the complete sample set) (Figure 4A). The SCC1 and WA1 lineages declined in number and disappeared in March and June, respectively, while the proportion of genomes from the D614G lineage rapidly increased, becoming the single predominant genotype in SCC by June (Figure 4A and B). The A12557G and C25692T lineages were common in April and May, the latter lineage in part owing to its association with a large skilled nursing facility outbreak (unpublished data) but disappeared afterward (Figure 4A and B).
Figure 4.
Dynamic change of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genotypes in Santa Clara County (SCC) over time. A, Proportion of main viral lineages every month between January and September. B, Dynamic change curve of major viral lineages over time in the SCC community from January to September. The y-axis represents the number of SARS-CoV-2 cases. C, Bar graph of other viral lineages between January and September. D, Fate of the SARS-CoV-2 lineages identified in this study. The dates that the lineage was last identified in SCC and in California by genomic sequencing are provided in red text, with red X’s denoting disappearance of the lineage from the county by that date.
Similarly, additional lineages that were introduced to SCC from January to March 2020, including those associated with discrete household clusters (Solano County, G14718T, and G26591T), disappeared by August 2020 (Figure 4A and 4C). Overall, similar longitudinal changes in lineage frequency were observed across the state of California (Figure 4A and B). In September 2020, all sequenced genomes in SCC and California were of the D614G lineage (Figure 4A–4C), However, an analysis of additional SNVs in the 4 D614G genomes sequenced from SCC from January to March 2020 revealed that these sublineages disappeared from SCC and California by 5 August (Figure 4C, left), indicating that continuation of the D614G lineage in SCC was most likely due to ongoing introduction into the county after March 2020 rather than persistent community transmission.
DISCUSSION
In the current study, we combined the power of genomic epidemiology with public health surveillance using contact tracing to monitor the introduction and community transmission of at least 17 SARS-CoV-2 lineages circulating in SCC, California, from 27 January to 21 March 2020. We identified 2 cases in which the infection was initially thought to have been associated with international travel by contact tracing, but viral genome analysis suggested that the individual had likely been infected by a locally circulating strain (SCC1). Viral WGS also identified a new epidemiologic link at a local church between seemingly unrelated cases. Finally, we were able to elucidate the cause of death in 3 previously unexplained cases as unrecognized SARS-CoV-2 infections, and to determine their phylogenetic placement in the WA1 lineage. Genomic epidemiology has rapidly emerged as an indispensable tool for investigating and monitoring spread of outbreaks such as COVID-19.
The 3 decedent cases in our study highlight the unmet need for expanded SARS-CoV-2 testing during the early stages of the pandemic in the United States, which would have likely revealed cases of cryptic viral transmission not linked to ostensible travel history. They also underscore the value of performing autopsies and postmortem testing early as an additional system for identifying the spread and shortening the time to assess the threat of the virus in a community. A robust public health genomic surveillance system of sufficient scope and scale to address pandemic threats such as SARS-CoV-2 needs access to many different types of samples for testing [23].
The D614G lineage containing a spike protein coding mutation is thought to have arisen in Germany from China in late January 2020 [24], and rapidly spread via travel through Europe, and from there, to the United States, associated with a large outbreak in New York City [6, 8, 12]. Epidemiologic, in vitro cell culture, and rodent model data to date [12, 25, 26] support the notion that D614G lineage viruses achieve higher viral loads and are more infectious than other strains, although, notably, there is no evidence of increased pathogenicity. Thus, a potential fitness advantage may explain the persistence and predominance of the D614G lineage in SCC, the United States, and globally [12, 27], although some have attributed the rise of D614G lineage to random founder effects [28]. The disappearance of the sublineages from the 4 sequenced D614G viruses in the study indicate that the surge in D614G cases in the county during the summer was mainly fueled by ongoing exogenous introduction.
Our results confirm that SARS-CoV-2 community transmission was already occurring by late January 2020, when available testing was extremely limited and earlier than the first officially reported case in SCC on 27 February [29]. Given the diversity of viral lineages uncovered in this study, it is likely that no local intervention, short of shutting down all travel into and out of the region, could have prevented these repeated introductions into SCC. Thus, given that “stay-in place” mandates were not enacted in SCC until 16 March and statewide until 19 March, community transmission may have been inevitable, although earlier public health interventions, such as social distancing and masking, would likely have reduced the size of the outbreak in SCC. Nevertheless, the disappearance of all of 17 introduced lineages suggest that these public health mandates may have supported local eradication. However, given ongoing introductions of SARS-CoV-2, local control of SARS-CoV-2 transmission becomes impractical without concurrent containment at the state and national levels.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the US Centers for Disease Control and Prevention (CDC) and the California Department of Public Health teams that joined the Santa Clara County (SCC) Public Health Department and conducted many of the epidemiologic and contact tracing investigations, the staff from the University of California, San Francisco, Center for Advanced Technology core facility for sequencing samples, and the staff from the China Basin clinical laboratory for the Illumina NextSeq sequencing efforts. We thank the staff of the CDC Infectious Diseases Pathology Branch for analysis of autopsy tissues and facilitating the submission of tissue specimens. We also thank all the authors and research groups who have contributed genome data to the Global Initiative on Sharing All Influenza Data (GISAID) database (author credits for specific GISAID contributions may be found at https://www.gisaid.org).
Author contributions. E. V. and C. Y. C. conceived and designed the study. E. V., C. A. K., M. A. J., B. Bonin, S. L. R., G. S. H., W. L., and T. M. performed case intake, contact tracing, and epidemiologic and medical examiner’s investigations and analyses. E. V., B. Bonin, and D. A. W. coordinated quantitative real-time reverse-transcription polymerase chain reaction testing of SCC cases. X. D. and C. Y. C. oversaw and coordinated the sequencing efforts. X. D., G. Y., and C. W. performed viral WGS and Sanger sequencing confirmation. B. Bushnell and S. F. assembled and curated the viral genomes. W. L., Y. T., C. R. P, S. T., and J. Bhatnagar performed quantitative real-time reverse-transcription polymerase chain reaction and sequenced viral genomes from COVID-19 cases submitted to the CDC, including the 2 initial COVID-19–associated decedent cases in SCC and the United States. The CZB CovidTracker Consortium and J. Batson sequenced and assembled viral genomes from SCC and other California counties from April onward. X. D., T. M., and C. Y. C. performed the genomic, phylogenetic, and comparative SNV analyses. E. V., X. D., C. A. K., and C. Y. C. wrote the manuscript. C. Y. C. designed the figures. E. V., X. D., C. A. K., M. A. J., S. L. R., D. A. W., J. Batson, T. M., and C. Y. C. edited and provided feedback on the manuscript. All authors read the manuscript and agree to its contents.
Disclaimer. The opinions expressed by the do not necessarily reflect the opinions of the CDC or the institutions with which the authors are affiliated.
Financial support. This work was supported by the Innovative Genomics Institute (C. Y. C.), the New Frontiers in Research Fund provided by the Canadian Institutes of Health Research (C. Y. C.), and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R33-AI129455 to C. Y. C.).
Potential conflicts of interest. X. D. and C. Y. C. are inventors on a patent application on the metagenomic sequencing with spiked primer enrichment method, titled “Spiked Primer Design for Targeted Enrichment of Metagenomic Libraries” (US application 62/667,344; filed 4 May 2018 by the University of California, San Francisco). All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data, materials, and software availability. Assembled severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomes in this study were uploaded to GISAID as FASTA files (accession nos. provided in Supplementary Table 1) and can be visualized on a continually updated phylogenetic tree using Nextstrain. The bioinformatics pipeline for assembly of SARS-CoV-2 genomes from raw FASTA files is open source and available for download at https://sourceforge.net/projects/bbmap/.
References
- 1. Dong E, Du H, Gardner L. An interactive Web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20:533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holshue ML, DeBolt C, Lindquist S, et al. . First case of 2019 novel coronavirus in the United States. N Engl J Med 2020; 382:929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bedford T, Greninger AL, Roychoudhury P, et al. ; Seattle Flu Study Investigators . Cryptic transmission of SARS-CoV-2 in Washington State. Science 2020; 370:571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deng X, Gu W, Federman S, et al. . Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science 2020; 369:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fauver JR, Petrone ME, Hodcroft EB, et al. . Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell 2020; 181:990–6 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, et al. . Introductions and early spread of SARS-CoV-2 in the New York City area. Science 2020; 369:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jorden MA, Rudman SL, Villarino E, et al. . Evidence for limited early spread of COVID-19 within the United States, January-February 2020. MMWR Morb Mortal Wkly Rep 2020; 69:680–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maurano MT, Ramaswami S, Zappile P, et al. . Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City region. Genome Res 2020; 30:1781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuchat A; CDC COVID-19 Response Team . Public health response to the initiation and spread of pandemic COVID-19 in the United States, February 24-April 21, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keeling MJ, Hollingsworth TD, Read JM. Efficacy of contact tracing for the containment of the 2019 novel coronavirus (COVID-19). J Epidemiol Community Health 2020; 74:861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet 2018; 19:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Korber B, Fischer WM, Gnanakaran S, et al. . Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182:812–27 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Centers for Disease Control and Prevention. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2020. https://www.fda.gov/media/134922/download. Accessed 8 November 2020.
- 14. Paden CR, Tao Y, Queen K, et al. . Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020; 26:2401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 2017; 1:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill 2017; 22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hadfield J, Megill C, Bell SM, et al. . Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kearse M, Moir R, Wilson A, et al. . Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moriarty LF, Plucinski MM, Marston BJ, et al. ; CDC Cruise Ship Response Team; California Department of Public Health COVID-19 Team; Solano County COVID-19 Team . Public health responses to COVID-19 outbreaks on cruise ships—worldwide, February-March 2020. MMWR Morb Mortal Wkly Rep 2020; 69:347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banu S, Jolly B, Mukherjee P, et al. . A distinct phylogenetic cluster of Indian severe acute respiratory syndrome coronavirus 2 isolates. Open Forum Infect Dis 2020; 7:ofaa434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rockefeller Foundation. Implementation framework: toward a national genomic surveillance network. New York, NY: Rockefeller Foundation, 2021. [Google Scholar]
- 24. Isabel S, Graña-Miraglia L, Gutierrez JM, et al. . Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci Rep 2020; 10:14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou B, Thao TTN, Hoffmann D, et al. . SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021; 592:122–27. [DOI] [PubMed] [Google Scholar]
- 26. Plante JA, Liu Y, Liu J, et al. . Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021; 592:116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Volz E, Hill V, McCrone JT, et al. . Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021; 184:64–75 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Dorp L, Richard D, Tan CCS, Shaw LP, Acman M, Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun 2020; 11:5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zwald ML, Lin W, Sondermeyer Cooksey GL, et al. . Rapid sentinel surveillance for COVID-19—Santa Clara County, California, March 2020. MMWR Morb Mortal Wkly Rep 2020; 69:419–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.