Abstract
Background
Both severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection and persistent infection have been reported, but sequence characteristics in these scenarios have not been described. We assessed published cases of SARS-CoV-2 reinfection and persistence, characterizing the hallmarks of reinfecting sequences and the rate of viral evolution in persistent infection.
Methods
A systematic review of PubMed was conducted to identify cases of SARS-CoV-2 reinfection and persistence with available sequences. Nucleotide and amino acid changes in the reinfecting sequence were compared with both the initial and contemporaneous community variants. Time-measured phylogenetic reconstruction was performed to compare intrahost viral evolution in persistent SARS-CoV-2 to community-driven evolution.
Results
Twenty reinfection and 9 persistent infection cases were identified. Reports of reinfection cases spanned a broad distribution of ages, baseline health status, reinfection severity, and occurred as early as 1.5 months or >8 months after the initial infection. The reinfecting viral sequences had a median of 17.5 nucleotide changes with enrichment in the ORF8 and N genes. The number of changes did not differ by the severity of reinfection and reinfecting variants were similar to the contemporaneous sequences circulating in the community. Patients with persistent coronavirus disease 2019 (COVID-19) demonstrated more rapid accumulation of sequence changes than seen with community-driven evolution with continued evolution during convalescent plasma or monoclonal antibody treatment.
Conclusions
Reinfecting SARS-CoV-2 viral genomes largely mirror contemporaneous circulating sequences in that geographic region, while persistent COVID-19 has been largely described in immunosuppressed individuals and is associated with accelerated viral evolution.
Keywords: SARS-CoV-2, persistent COVID-19, reinfection, sequence analysis, immunosuppression
Reinfecting SARS-CoV-2 viral genomes largely mirror contemporaneous circulating sequences in that geographic region, while persistent COVID-19 has been largely described in immunosuppressed individuals and is associated with accelerated viral evolution.
After resolution of coronavirus disease 2019 (COVID-19) following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, antibodies against SARS-CoV-2 persist in the majority of patients for 6 months or more [1]. Despite this, there have now been a number of reports of COVID-19 reinfection, spanning a broad range of age groups, time frames, and disease severity [2–7]. There remains a great deal of uncertainty over the viral characteristics of reinfection cases, including the degree of sequence heterogeneity and the location of new mutations between the initial and reinfecting variants, if any. In addition, the diagnosis of COVID-19 reinfection has been complicated by the increasing reports of persistent COVID-19 infection, especially in immunosuppressed individuals. Like reinfection cases, persistent COVID-19 can also span the range of disease severity, from asymptomatic to severe disease, and recurrent symptoms can last for months [8–11]. Differentiating between persistence and reinfection can be challenging, and little is known about differences in the location and quantity of SARS-CoV-2 mutations in these scenarios. We performed an analysis of SARS-CoV-2 sequences from published cases of COVID-19 reinfection and persistence, characterizing the hallmarks of reinfecting sequences and the rate of viral evolution in persistent infection.
METHODS
Data Search and Selection Criteria
We conducted a systematic literature review in PubMed through 8 March 2021 for cases of persistent COVID-19 using the search term “((covid or sars-CoV-2) AND (persistent or persistence or prolonged)) AND (sequence or evolution).” A search for COVID-19 reinfection reports was made using the terms “(covid or sars-CoV-2) AND (reinfection).” Both peer-reviewed and preprint results were evaluated. We used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) for reviewing literature and for reporting search results. Additional preprints that appeared through Google search and that met our criteria were also included. For cases of reinfection, papers were included if the authors described it as a case of reinfection diagnosed more than 30 days after the initial infection and if whole-genome SARS-CoV-2 sequences or sites of mutations relative to a reference sequence (eg, Wuhan-Hu-1) from both infection time points were available. Of the 291 results from the search, 14 articles met the inclusion criteria and were included in the present report along with 2 additional preprints that were identified (Supplementary Figure 1A).
Persistent cases were included if the authors described it as a case of persistent COVID-19 infection and if longitudinal whole-genome SARS-CoV-2 sequences were available. The search returned 129 results, 7 of which met the inclusion criteria and were included in the present report along with 1 other preprint (Supplementary Figure 1B). Only sequences obtained directly from patient respiratory tract samples were included in our analysis to exclude the possibility of sequence changes during the ex vivo culture process. Three cases were excluded due to uncertainty in their classification as either reinfection or persistent infection cases (Supplementary Methods, Supplementary Table 1, Supplementary Figure 2).
Sequences were analyzed for mutations using Nextclade (https://clades.nextstrain.org/) and snp-sites (https://github.com/sanger-pathogens/snp-sites). The degree of reinfection severity, either more or less severe compared with the first infection, was classified based on an explicit determination by the authors of each article or by comparing symptoms, duration of illness, and hospitalization status between both episodes.
Sequencing Dataset Compilation and Phylogenetic Tree Construction
The sequencing dataset contained a total of 262 globally representative SARS-CoV-2 genomes selected from GISAID (https://www.gisaid.org/) and sequences from the reinfection and persistence cases (Supplementary Methods, Supplementary Data 1). The sampled sequences were chosen to be representative of global sequence diversity throughout the time course of the pandemic. Sequences of variants of concern B.1.1.7 and B.1.351 were also included. Nucleotide sequence alignment was performed using MAFFT (Multiple Alignment using Fast Fourier Transform) [12]. Best-fit nucleotide substitution was calculated using model selection followed by maximum likelihood (ML) phylogenetic tree construction using IQ-Tree with 1000-bootstrap replicates [12].
Mutation Analysis
For reinfection cases, mutations were determined in 2 ways. First, nucleotide and amino acid changes were identified for the reinfection sequences relative to the first infection sequence. The frequency of nucleotide or amino acid changes within each gene was compared with the frequency of changes in the remainder of the genome by Fisher’s exact tests with a Bonferroni correction (for multiple comparisons). The relationship between disease severity and number of nucleotide or amino acid changes in the genome was assessed using a Mann-Whitney test. Second, to identify unique characteristics of reinfecting viruses, each of the first and reinfection sequences were compared with circulating sequences in the community as defined by the same Nextstrain clade sampled within 1 month obtained from the same geographic location uploaded to GISAID (Supplementary Table 2, Supplementary Methods, Supplementary Data 2). Rare mutations were determined as polymorphisms that were present only in the reinfecting sequence (not the initial variant) and found in less than 1% of contemporaneous community sequences. Mutation locations are graphically represented in Circos plots [13].
For persistent infections, sequence changes were assessed at 2 time intervals: before or after convalescent plasma or monoclonal antibody treatment. Sequences sampled before convalescent plasma or antibody treatment were compared with the first sequence sampled. For sequences sampled after convalescent plasma or antibody treatment, sequence changes (both nucleotide and amino acid) were determined relative to the last pretreatment sequence. Linear regression was used to estimate the rate of viral changes between 2 intervals. The slope of the trendline was compared with the latest global clock rate (29 March 2021) as estimated by NextStrain (https://nextstrain.org/ncov/global/).
Time-Measured Phylogenetic Analysis
The temporal signal of the ML tree was examined in TempEst [14] regressing on root-to-tip divergence, and outliers were inspected in the distribution of residuals. A high degree of clock-like behavior in the whole dataset was observed (R2 = 0.721) in root-to-tip regression analysis with the slope rate as 8.26E-4 and the rough ancestral time of the sample was calculated as 2019.84. This suggests that the whole dataset has a realistic temporal signal and is appropriate for an estimation of temporal parameters. No outliers were found in this sample. To further examine the temporal signal in the sequences from persistent patients (especially those with >2 sequences), separate root-to-tip regression analysis also supported temporal signal for a time-measured phylogeny. To compare the evolutionary rates between the reported persistent infections and the general population infections, time-measured phylogenetic reconstruction was conducted in Bayesian Evolutionary Analysis Sampling Trees (BEAST) version 1.10.4 [15]. Nine partitions, including 8 persistent patients and the global sequences, were used as separate groups of taxa, to estimate separate evolutionary rates. Due to large uncertainties with small samples, persistent patients with only 2 viral sequences were excluded from this analysis. A general time reversible (GTR) model was applied with gamma-distributed rate variations among sites. A log-normal relaxed molecular clock was used with an initial mean of 0.0008 and a uniform prior ranging from 0.0 to 1.0. A logistic growth tree prior was applied. Four independent Bayesian Markov Chain Monte Carlo (MCMC) chains of 100 million generations were performed with a sampling step every 10 000 generations to yield 10 000 trees per run. To ensure a sufficient effective sample size greater than 200, the convergence of 3 runs was diagnosed in Tracer version 1.7.1 (http://tree.bio.ed.ac.uk/software/tracer/) for all parameters. LogCombiner version 1.10.4 as part of the BEAST software package was used to combine the multiple runs to generate log and tree files after appropriate removal of the burn-in from each MCMC chain. The comparison of the evolutionary rates from the combined log file was analyzed and visualized in R version 4.0.2 (https://www.r-project.org/).
Statistical Analysis
Nonparametric Wilcoxon rank-sum or matched-pairs signed-rank tests were used to compare the number of amino acid changes between sequences. Statistical analyses were performed using GraphPad Prism 9 (GraphPad Software, San Diego, CA).
RESULTS
Sequence Analysis of Reinfection Cases
A total of 20 cases from 16 reports were included in this analysis (Table 1) [2–7, 16–25]. A broad range of age groups were represented and 90% were under the age of 70 years. Most (80%) of the cases had no reported comorbidities, and while 1 patient had diabetes and end-stage renal disease, none had high-level immunosuppression. The interval between diagnosis of the first infection and the second infection ranged from 44 days to 282 days with a median of 113.5 days. Five patients had more severe illness during the second infection, while 6 had less severe symptoms on reinfection, including 2 who were asymptomatic on reinfection. Two cases were asymptomatic in both infections, 5 cases reported the same severity for both infections, and no information on infection severity was available for 2 cases (Table 1). Six cases reported reinfection with a virus from the same clade.
Table 1.
Reinfection Cases
| Patient | Authors | Publication/Preprint Server (Year) | Age | Sex | Comorbidities | Time Between Infections, Days | Second Infection Severity | First Infection Clade | Second Infection Clade |
|---|---|---|---|---|---|---|---|---|---|
| Re1 | Selhorst et al [5] | Clin Infect Dis (2020) | 39 | F | None | 185 | Less | 19A | 20A |
| Re2 | To et al [3] | Clin Infect Dis (2020) | 33 | M | None | 144 | Less | 19A | 20E |
| Re3 | Prado-Vivar et al [16] | Lancet Infect Dis (2021) | 46 | M | None | 63 | More | 20A | 19B |
| Re4 | Tillett et al [4] | Lancet Infect Dis (2020) | 25 | M | None | 48 | More | 20C | 20C |
| Re5 | Goldman et al [7] | medRxiv | 60–69 | N/A | Emphysema, hypertension | 139 | Less | 19B | 20A |
| Re6 | Resende et al [17] | Virological | 37 | F | None | 116 | Similar | 20B | 20B |
| Re7 | Harrington et al [6] | Clin Infect Dis (2021) | 78 | M | Diabetic nephropathy with hemodialysis, COPD, sleep apnea, ischemic heart disease | 250 | More | 19A | 20I (B.1.1.7) |
| Re8 | Van Elslande et al [2] | Clin Infect Dis (2021) | 51 | F | Asthma | 93 | Less | 20B | 19B |
| Re9 | Colson et al [18] | J Infect (2020) | 70 | M | None | 105 | Less | 20A | 20A.EU2 |
| Re10 | Nonaka et al [19] | Emerg Infect Dis (2021) | 45 | F | None | 147 | More | 20B | 20B |
| Re11-1 | Gupta et al [20] | Clin Infect Dis (2020) | 25 | M | None | 108 | Both asymptomatic | 19A | 20A |
| Re11-2 | 28 | F | None | 111 | Both asymptomatic | 20A | 20A | ||
| Re12-1 | Abu-Raddad et al [21] | Clin Infect Dis (2020) | 25–29 | M | N/A | 46 | N/A | 19A | 20A |
| Re12-2 | 40–44 | M | N/A | 71 | N/A | 19A | 20A | ||
| Re13-1 | Naveca et al [22 | Research Square | 29 | N/A | N/A | 281 | Similar | 20A | 20J (P.1) |
| Re13-2 | 50 | N/A | N/A | 153 | Similar | 20B | 20J (P.1) | ||
| Re13-3 | 40 | F | N/A | 282 | Similar | 20A | 20J (P.1) | ||
| Re14 | Vetter et al [23] | Clin Microbiol Infect (2021) | 36 | F | None | 205 | Similar | 20A | 20A.EU2 |
| Re15 | Kulkarni et al [24] | Clin Infect Dis (2021) | 61 | M | None | 44 | More | 20B | 20B |
| Re16 | Adrielle dos Santos et al [25] | J Infect (2021) | 40 | F | Systemic arterial hypertension, obesity | 53 | Less severe | 20A | 20A |
Abbreviations: Clin Infect Dis, Clinical Infectious Diseases; COPD, chronic obstructive pulmonary disease; F, female; M, male; Lancet Infect Dis, Lancet Infectious Diseases; J Infect, Journal of Infection; Emerg Infect Dis, Emerging Infectious Diseases; Clin Microbiol Infect, Clinical Microbiology and Infection; N/A, not available.
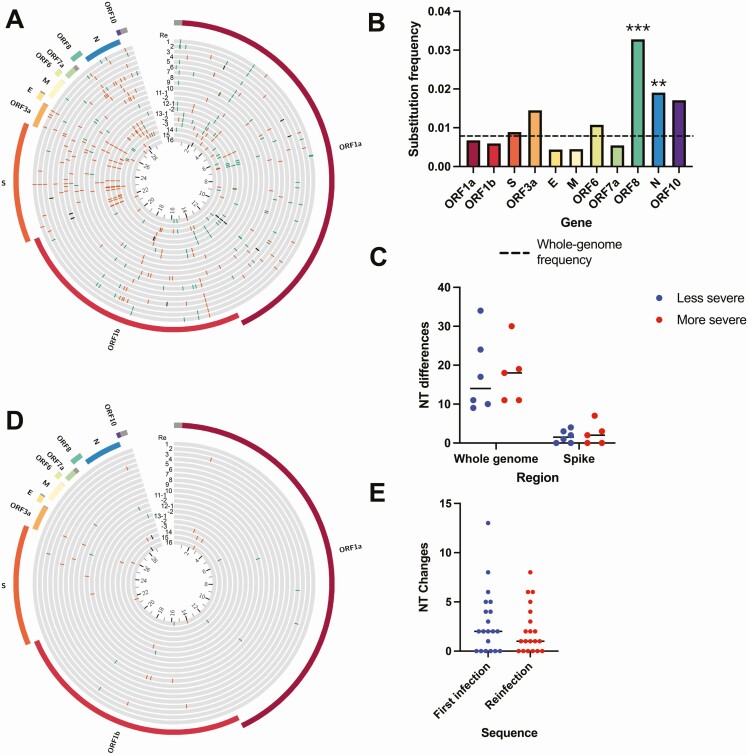
Phylogenetic analysis demonstrated distinct branching for the 2 sequences in each of the reinfection cases, corroborating results discussed in the original reports (Figure 1). We compared nucleotide and amino acid changes in the reinfecting viral sequence with the initial sequence and found a median of 17.5 nucleotide changes (range: 9–37) and 9 amino acid changes (range: 6–24) compared with the original sequence (Figure 2A). The nucleotide changes between the initial and reinfecting sequences were distributed across the SARS-CoV-2 genome, with significantly higher frequencies of changes in open reading frame (ORF) 8 (ORF8) (P < .001) and N (P = .001) (Figure 2B). A similar pattern was observed with amino acid changes (Supplementary Figure 3A). All but 2 reinfection cases had at least 1 substitution or deletion in the S gene (Supplementary Table 3). Next, we assessed whether reinfection with a more divergent second virus resulted in more severe disease. We found no significant differences in the number of nucleotide or amino acid changes in the reinfecting virus compared with the original viral variant when categorized by the severity of the reinfection (Figure 2C, Supplementary Figure 3B). Both the initial and reinfecting SARS-CoV-2 variants were similar to the sequences circulating in the community at the time of reinfection. The initial infecting variant harbored a median of only 2 rare nucleotide mutations compared with contemporaneous circulating variants in the community and the reinfecting variant contained a median of only 1 rare nucleotide mutation (Figure 2D–E, Supplemental Figure 3C).
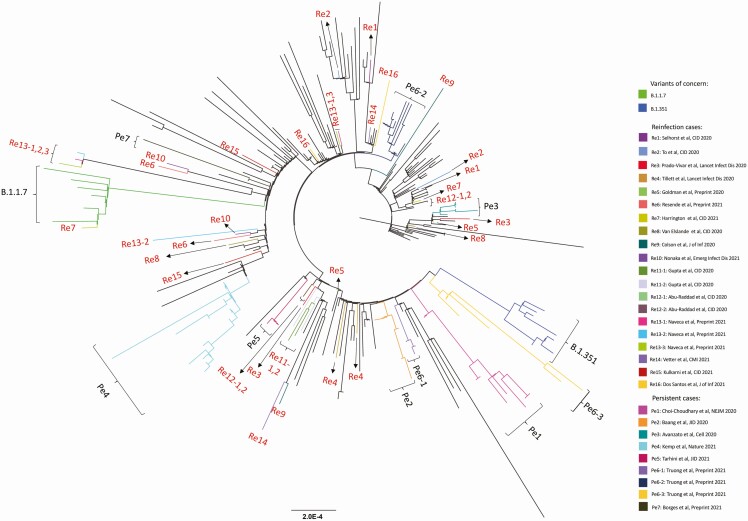
Figure 1.
Maximum-likelihood phylogenetic tree of sequences from persistent COVID-19 cases (Pe1–Pe7), COVID-19 reinfection cases (Re1–Re16), the variants of concern B.1.1.7 and B.1.351, and globally sampled sequences from GISAID. Abbreviations: CID, Clinical Infectious Diseases; COVID-19, coronavirus disease 2019; NEJM, New England Journal of Medicine; Lancet Infect Dis, Lancet Infectious Diseases; J of Inf, Journal of Infection; Emerg Infect Dis, Emerging Infectious Diseases; CMI, Clinical Microbiology and Infection; JID, Journal of Infectious Diseases .
Figure 2.
Comparison of viral sequences from reinfection cases. (A) Circos plot showing location of nucleotide changes in the reinfecting sequence relative to the initial infection sequence for each of the 20 cases. The inner ring indicates nucleotide position in kilobases. Synonymous changes are in green, nonsynonymous changes in orange, deletions in black. (B) Nucleotide substitution frequency pooled across all reinfection cases for each SARS-CoV-2 gene. The dashed line indicates global substitution frequency across the whole genome. Substitution frequency for each gene was compared with the substitution frequency in the rest of the genome using Fisher’s exact tests. P values were corrected for multiple comparisons using the Bonferroni correction. **P < .01, and ***P < .001. (C) Nucleotide changes in the second infection relative to the first infection by clinical disease severity. Mutations shown for the whole genome and S gene. P = .67, Mann-Whitney test. (D) Circos plot showing the location of nucleotide mutations from the second infection relative to other viruses circulating at the same time in the same geographic region. Only rare mutations present in <1% of contemporaneous community sequences are shown. (E) Number of rare nucleotide polymorphisms at each time point relative to circulating sequences in the community. P = .26, Wilcoxon matched-pairs signed-rank test. Abbreviations: E, envelope; M, membrane; NT, nucleotide; N, nucleocapsid; ORF, open reading frame; S, spike; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Sequence Analysis of Persistent COVID-19 Cases
A total of 9 cases from 7 reports describing persistent infection were retrieved from our literature search. Of these 9 cases, all but one had B-cell immunodeficiency [8–10, 11, 26–28]. Four were treated with B-cell–depleting therapy for lymphoma or autoimmune disorders, while 4 had B-cell lymphomas treated with chemotherapy (Table 2). One patient had advanced human immunodeficiency virus (HIV) infection with a CD4+ count of 0 cells/mm3 and diminished CD19+ cell counts. The median length of infection was 154 days and 33% of the cases ended in death. One patient had asymptomatic disease throughout [9]. Four patients were treated with convalescent plasma at least once during their illness [9, 10, 11, 27], and 1 patient was treated with the monoclonal antibodies casirivimab and imdevimab [8].
Table 2.
Persistent Cases
| Patient | Case | Publication (Year) | Age | Sex | Underlying Conditions | Immunosuppressants | Antiviral Treatment | Infection Length, Days | Fatal? |
|---|---|---|---|---|---|---|---|---|---|
| Pe1 | Choi et al [8] | New Engl J Med (2020) | 45 | M | Antiphospholipid antibody syndrome | Rituximab, eculizumab, cyclophosphamide, corticosteroids | Remdesivir, casirivimab and imdevimab | 154 | Yes |
| Pe2 | Baang et al [10] | J Infect Dis (2021) | 60 | M | Refractory mantle cell lymphoma | CD20 bispecific Ab, B-cell directed Ab, cyclophosphamide, doxorubicin, prednisone | Remdesivir, convalescent plasma | 156 | Yes |
| Pe3 | Avanzato et al [9] | Cell (2020) | 71 | F | CLL, acquired hypogammaglobulinemia | … | Convalescent plasma | 156 | No |
| Pe4 | Kemp et al [11] | Nature (2021) | … | … | Marginal B-cell lymphoma, hypogammaglobulinemia | B-cell–depleting therapy | Remdesivir, convalescent plasma | 102 | Yes |
| Pe5 | Tarhini et al [27] | J Infect Dis (2021) | 66 | M | HIV, multifocal leukoencephalopathy from JC virus | … | … | 124 | No |
| Pe6–1 | TTruong et al [28] | EBioMedicine (2021) | <5 | F | B-cell ALL | Dexamethasone, vincristine, peg- asparaginase, methotrexate, 6-MP, doxorubicin | … | 91 | No |
| Pe6–2 | 20–25 | M | B-cell ALL | CD-19–directed CAR-T cell therapy (tisagenlecleucel), cylcophosphamide, fludarabine | Remdesivir, convalescent plasma | 250+ | No | ||
| Pe6–3 | <5 | M | B-cell ALL | Cyclophosphamide, cytarabine, thioguanine, vincristine, dexamethasone, methotrexate, mercaptopurine | … | 196 | No | ||
| Pe7 | Borges et al [29] | Virological | 61 | F | Diffuse large B-cell lymphoma | Chemotherapy (unspecified), methotrexate, corticosteroids | Remdesivir | 197 | No |
Abbreviations: Ab, antibody; ALL, acute lymphoblastic leukemia; CAR, chimeric antigen receptor; CLL, chronic lymphocytic leukemia; F, female; HIV, human immunodeficiency virus; JC virus, John Cunningham virus; J Infect Dis, Journal of Infectious Diseases; M, male; New Engl J Med, New England Journal of Medicine.
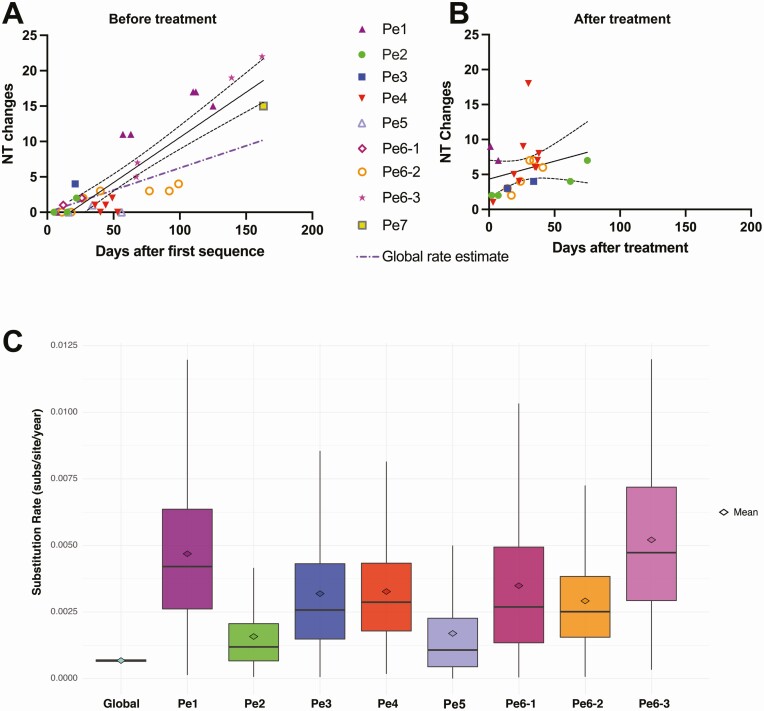
Phylogenetic analysis revealed that, for each of the 9 patients, sequences formed a distinct cluster, confirming what was found in the original reports (Figure 1). New mutations emerging over time were detected in all of the patients with persistent COVID-19, with further changes identified after treatment with convalescent plasma or monoclonal antibodies (Supplementary Figure 4). Mutations occurred with significantly higher frequency in S (P < .001) and ORF7a (P = .02) and lower frequency in ORF1a (P = .02) (Figure 3A, Supplementary Figure 5A). The rate of viral evolution was plotted for each patient both for the interval before and after convalescent plasma/antibody treatment. Before antiviral treatment, the rate of sequence changes over time appeared faster than the Nextstrain estimate for the global rate of SARS-CoV-2 evolution (dotted purple line in Figure 3B, Supplementary Figure 5B). Treatment with convalescent plasma or antibody cocktail was insufficient to halt intrahost viral evolution (Figure 3C, Supplementary Figure 5C).
Figure 3.
SARS-CoV-2 mutation location and evolutionary rate in the persistent COVID-19 cases. (A) Nucleotide substitution frequency pooled across all persistent cases for each SARS-CoV-2 gene. The dashed line indicates global substitution frequency across the whole genome. Substitution frequency for each gene was compared with the substitution frequency in the rest of the genome using Fisher’s exact tests. P values were corrected for multiple comparisons using the Bonferroni correction. *P < .05, and ***P < .001. (B) Nucleotide changes in samples taken prior to convalescent plasma or monoclonal antibody treatment relative to first sampled sequence in each persistently infected patient. Regression line and 95% confidence bands are shown. The purple dash-dotted line is the global rate estimate obtained from Nextstrain. (C) Nucleotide changes in samples taken after convalescent plasma or monoclonal antibody treatment relative to the last sample taken prior to treatment in each persistently infected patient. Regression line and 95% confidence bands are shown. (D) Substitution rate (nucleotide substitutions per site per year) of sampled global SARS-CoV-2 sequences relative to persistent patients based on Markov chain Monte Carlo time-measured phylogenetic reconstruction. Box plots show median and interquartile ranges of estimated substitution rates. The mean, median, and 95% HPD interval can be found in Supplementary Table 4. Abbreviations: COVID-19, coronavirus disease 2019; HPD, highest posterior density; Nt, nucleotide; Pe, persistent; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
We also performed time-measured phylogenetic reconstruction with the pretreatment persistent sequences to compare the rate of intrahost viral evolution in persistent COVID-19 with the rate of community-driven evolution. This analysis provided further evidence that SARS-CoV-2 evolution appeared faster in these persistent-infection individuals compared with the rate in the general public population, although substantial uncertainties are shown in these estimates given the limited sequence sampling in each patient (Figure 3D, Supplementary Table 4).
DISCUSSION
We conducted a systematic review and pooled analysis of sequences from reports of COVID-19 reinfection and persistent infection. Reports of reinfection cases demonstrate a wide range of situations, spanning a broad distribution of ages, baseline health status, and reinfection severity compared with the initial infection. Reinfection occurred as early as 1.5 months or more than 8 months after the initial infection. Common explanations for the presence of reinfection involves either waning SARS-CoV-2 antibodies or the presence of viral escape mutations [29, 30]. While most cases of SARS-CoV-2 reinfection did involve infection with a different clade (including the variants of concern B.1.1.7 and P.1), it is noteworthy that mutations were identified throughout the genomes and the frequency of mutations within the S gene was not elevated relative to the rest of the genome. In addition, individuals with more severe reinfections did not have significantly greater frequency of S gene mutations. Interestingly, the genes with the highest frequency of mutations were ORF8 and N. ORF8 is a rapidly evolving accessory protein that may antagonize host immune function [31], while the nucleocapsid is a vital structural protein that also serves as a target for both humoral and cell-mediated immune responses [32]. Finally, the presence of rare mutations was uncommon in the reinfecting virus, which largely mirrored the contemporaneously circulating variants in the region of infection. However, the reinfecting variants generally contained a substantial number of mutations compared with the initial variant, including frequent changes in the S gene, and additional studies are needed to assess whether these changes may have contributed to the risk of repeat infection.
While the number of immunosuppressed individuals with available sequences remains limited, the results suggest that the rate of viral evolution (measuring both synonymous and nonsynonymous changes) is accelerated within immunosuppressed individuals. In addition, treatment with convalescent plasma or monoclonal antibody cocktails was insufficient to fully halt viral evolution and the emergence of viral escape with treatment has been documented [11, 33]. Mutations associated with immune escape and/or more efficient replication kinetics, including E484K, S494P, N501Y, and N-terminal spike deletions, have been observed in both immunosuppressed individuals and the novel variants of concern [34, 35]. The results raise the possibility that novel variants, including those harboring escape mutations against current treatments, could arise from immunosuppressed individuals and suggest that immunosuppressed individuals should be a focus of public health efforts. Among the current reports of persistent COVID-19, B-cell dysfunction appears to be a common thread, including in reports that were not included in this analysis due to a lack of available full-length sequences [36–40]. It is important to note, however, that T-cell function may also play a role in protection against SARS-CoV-2 [41] and a subset of these patients also included concurrent suppression of other aspects of the immune response. Additional studies are needed to fully define the type and intensity of immunosuppression that would place patients at greatest risk of persistent COVID-19.
Two factors generally differentiated between reinfection and persistent infection scenarios: first, reinfections have so far been largely described in immunocompetent individuals while the majority of persistent COVID cases have been in immunosuppressed patients. Second, phylogenetic analysis can generally differentiate between reinfection and persistent infection, especially in cases where persistent infection allowed the longitudinal collection of more than 2 sequences. However, given the slow rate of SARS-CoV-2 evolution and limited viral diversity [42], it can be challenging to differentiate between reinfection and persistent infection, especially in situations with limited sampling and/or duration between samples.
A limitation of this work is that it relies on case reports, which can be influenced by publication bias and limits our statistical power. However, to date, there have been no systematic, large-scale, sequence-based studies of COVID-19 reinfection or persistent infections. This is partly due to the rarity of these types of cases and that initial infecting sequences are frequently unavailable for comparison with reinfecting or persistently infecting variants. Overall, our results demonstrate the need to further explore factors that increase the risk of breakthrough reinfections and persistent COVID-19. This line of investigation will have important implications on the durability of currently available vaccines and for preventing the rise of novel variants.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Jeremy Luban and Ronald Bosch for their feedback and discussion.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported in part by National Institutes of Health (NIH) grant 106701. X. Q. was supported by the NIH Models of Infectious Disease Agent Study (MIDAS) through the National Institute of General Medical Sciences under award number U54GM088558.
Potential conflicts of interest. J. Z. L. has consulted for Abbvie. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Dan JM, Mateus J, Kato Y, et al. . Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021; 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Elslande J, Vermeersch P, Vandervoort K, et al. . Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis 2021; 73:354–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. To KK, Hung IF, Ip JD, et al. . COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis 2021; 73:e2946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tillett RL, Sevinsky JR, Hartley PD, et al. . Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis 2021; 21:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Selhorst P, Van Ierssel S, Michiels J, et al. . Symptomatic SARS-CoV-2 reinfection of a health care worker in a Belgian nosocomial outbreak despite primary neutralizing antibody response. Clin Infect Dis 2021; 73:e2985–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrington D, Kele B, Pereira S, et al. . Confirmed reinfection with SARS-CoV-2 variant VOC-202012/01. Clin Infect Dis 2021; 73:1946–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldman JD, Wang K, Roltgen K, et al. . Reinfection with SARS-CoV-2 and failure of humoral immunity: a case report. medRxiv [Preprint]. 2020. [Google Scholar]
- 8. Choi B, Choudhary MC, Regan J, et al. . Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Avanzato VA, Matson MJ, Seifert SN, et al. . Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–12, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baang JH, Smith C, Mirabelli C, et al. . Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kemp SA, Collier DA, Datir RP, et al. . SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021; 592:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 2016; 44:W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krzywinski M, Schein J, Birol I, et al. . Circos: an information aesthetic for comparative genom ics. Genome Res 2009; 19:1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2016; 2:vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012; 29:1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, et al. . A case of SARS-CoV-2 reinfection in Ecuador. Lancet Infect Dis 2020. doi: 10.1016/S1473-3099(20)30910-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Resende PC, de Vasconcelos RHT, Arantes I, et al. . Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil. Virological 2021. Available at: https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584. Accessed 11 January 2021. [Google Scholar]
- 18. Colson P, Finaud M, Levy N, Lagier JC, Raoult D. Evidence of SARS-CoV-2 re-infection with a different genotype. J Infect 2020. doi: 10.1016/j.jinf.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nonaka CKV, Franco MM, Gräf T, et al. . Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil . Emerg Infect Dis 2021; 27:1522–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gupta V, Bhoyar RC, Jain A, et al. . Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis 2021; 73:e2823–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abu-Raddad LJ, Chemaitelly H, Malek JA, et al. . Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. Clin Infect Dis 2021; 73:e1830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naveca FG, da Costa C, Nascimento V, et al. . Three SARS-CoV-2 reinfection cases by the new variant of concern (VOC) P.1/501Y.V3. Res Square 2021. Available at: https://www.researchsquare.com/article/rs-318392/v1. Accessed 20 March 2021. [Google Scholar]
- 23. Vetter P, Cordey S, Schibler M, et al. . Clinical, virologic and immunologic features of a mild case of SARS-CoV-2 reinfection. Clin Microbiol Infect 2021. doi: 10.1016/j.cmi.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kulkarni O, Narreddy S, Zaveri L, Kalal IG, Tallapaka KB, Sowpati DT. Evidence of SARS-CoV-2 reinfection without mutations in Spike protein. Clin Infect Dis 2021; 73:e1239–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adrielle Dos Santos L, Filho PGG, Silva AMF, et al. . Recurrent COVID-19 including evidence of reinfection and enhanced severity in thirty Brazilian healthcare workers. J Infect 2021; 82:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tarhini H, Recoing A, Bridier-Nahmias A, et al. . Long term SARS-CoV-2 infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis 2021. doi: 10.1093/infdis/jiab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Truong TT, Ryutov A, Pandey U, et al. . Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: A consecutive case series. EBioMedicine 2021; 67:103355 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borges V, Isidro J, Cunha M, et al. . Long-term evolution of SARS-CoV-2 in an immunocompromised patient with non-Hodgkin lymphoma. Virological 2021. Available at: https://virological.org/t/long-term-evolution-of-sars-cov-2-in-an-immunocompromised-patient-with-non-hodgkin-lymphoma/621. Accessed 8 March 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wibmer CK, Ayres F, Hermanus T, et al. . SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 2021; 27:622–5. [DOI] [PubMed] [Google Scholar]
- 30. Weisblum Y, Schmidt F, Zhang F, et al. . Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. 2021; 118(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelde A, Bilich T, Heitmann JS, et al. . SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol 2021; 22:74–85. [DOI] [PubMed] [Google Scholar]
- 33. Starr TN, Greaney AJ, Addetia A, et al. . Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021; 371:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Volz E, Mishra S, Chand M, et al. . Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 35. Tegally H, Wilkinson E, Giovanetti M, et al. . Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592:438–43. [DOI] [PubMed] [Google Scholar]
- 36. CaCamprubí D, Gaya A, Marcos MA, et al. . Persistent replication of SARS-CoV-2 in a severely immunocompromised patient treated with several courses of remdesivir. Int J Infect Dis 2021; 104:379–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hensley MK, Bain WG, Jacobs J, et al. . Intractable COVID-19 and prolonged SARS-CoV-2 replication in a CAR-T-cell therapy recipient: a case study. Clin Infect Dis 2021; 73:e815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinot M, Jary A, Fafi-Kremer S, et al. . Remdesivir failure with SARS-CoV-2 RNA-dependent RNA-polymerase mutation in a B-cell immunodeficient patient with protracted COVID-19. Clin Infect Dis 2021; 73:e1762–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Helleberg M, Niemann CU, Moestrup KS, et al. . Persistent COVID-19 in an immunocompromised patient temporarily responsive to two courses of remdesivir therapy. J Infect Dis 2020; 222:1103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sepulcri C, Dentone C, Mikulska M, et al. . The longest persistence of viable SARS-CoV-2 with recurrence of viremia and relapsing symptomatic COVID-19 in an immunocompromised patient – a case study. medRxiv [Preprint]. 2021. doi: 10.1093/ofid/ofab217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McMahan K, Yu J, Mercado NB, et al. . Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021; 590(7847):630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rausch JW, Capoferri AA, Katusiime MG, Patro SC, Kearney MF. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc Natl Acad Sci USA 2020; 117:24614–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.