Abstract
Background
Immunocompromised patients show prolonged shedding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in nasopharyngeal swabs. We report a case of prolonged persistence of viable SARS-CoV-2 associated with clinical relapses of coronavirus disease 2019 (COVID-19) in a patient with mantle cell lymphoma who underwent treatment with rituximab, bendamustine, cytarabine with consequent lymphopenia and hypogammaglobulinemia.
Methods
Nasopharyngeal swabs and blood samples were tested for SARS-CoV-2 by real-time polymerase chain reaction (RT-PCR). On 5 positive nasopharyngeal swabs, we performed viral culture and next-generation sequencing. We analyzed the patient’s adaptive and innate immunity to characterize T- and NK-cell subsets.
Results
SARS-CoV-2 RT-PCR on nasopharyngeal swabs samples remained positive for 268 days. All 5 performed viral cultures were positive, and genomic analysis confirmed a persistent infection with the same strain. Viremia resulted positive in 3 out of 4 COVID-19 clinical relapses and cleared each time after remdesivir treatment. The T- and NK-cell dynamic was different in aviremic and viremic samples, and no SARS-CoV-2-specific antibodies were detected throughout the disease course.
Conclusions
In our patient, SARS-CoV-2 persisted with proven infectivity for >8 months. Viremia was associated with COVID-19 relapses, and remdesivir treatment was effective in viremia clearance and symptom remission, although it was unable to clear the virus from the upper respiratory airways. During the viremic phase, we observed a low frequency of terminal effector CD8+ T lymphocytes in peripheral blood; these are probably recruited in inflammatory tissue for viral eradication. In addition, we found a high level of NK-cell repertoire perturbation with relevant involvement during SARS-CoV-2 viremia.
Keywords: hematological, immunological response, SARS-CoV-2, viral shedding, viremia
Persistence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral shedding and recurrence of positive SARS-CoV-2 nasopharyngeal swabs after a negative sample and symptom remission have been documented both in asymptomatic and symptomatic patients [1]. Nevertheless, viral infectivity cannot be determined with real-time polymerase chain reaction (RT-PCR), which detects only viral RNA, and viral culture and genome sequencing to determine the infectivity and genomic asset of the virus cannot be routinely used in clinical practise, leading to uncertainty about definitions of re-infection, relapse, prolonged viral shedding, and infectiousness. Studies assessing the infectivity of these isolates have documented that viral infectivity declines within 10 days of infection, with a possible tail of another 10 days, both in asymptomatic and symptomatic individuals [2]. In immunocompromised patients, prolonged viral shedding with proven viral infectivity has increasingly been reported [3–7], and in 2 cases it has been associated with disease relapse [3, 4].
Adaptive and innate immunity play a vital role in viral clearance: During many acute infections, including SARS-CoV-2, the presence of CD8+ cytotoxic T cells capable of secreting an array of molecules such granzymes to eradicate virus from the host has been described [8]. At the same time, CD4+ helper T cells can assist cytotoxic T cells and enhance their ability to clear pathogens. However, persistent stimulation by the virus may induce T-cell exhaustion as a state of T-cell dysfunction, leading to loss of cytokine production and reduced function [9].
Our case represents the longest SARS-CoV-2 viral persistence with proven viral infectivity in an immunocompromised patient who experienced 4 clinically relevant coronavirus disease 2019 (COVID-19) relapses. Relapses were associated with positive viremia and responded to remdesivir treatment with viremia clearance and symptom remission but with persistence of viable virus in the upper respiratory airways. We observed a difference in T and NK cells’ distribution between the aviremic and viremic phases.
METHODS
RT-PCR on nasopharyngeal swabs was performed with the Allplex 2019 nCOV Seegene–Three Target assay (sensitivity, 98.2%; specificity, 100%) [10].
Viral isolation and culture from nasopharyngeal swab samples was performed on VERO E6 cells (Figure 1). We extracted RNA for confirmation of SARS-CoV-2 by RT-PCR and next-generation sequencing. We obtained full-genome sequences from the supernatant at different time points with Miseq Illumina (Illumina, Inc., San Diego, CA, USA) using 2×150 paired-end sequencing. We mapped and aligned the results to the reference genome obtained from Gisaid (https://www.gisaid.org/; accession ID: EPI_ISL_412973) [11] using Geneious software, version 9.1.5 (http://www.geneious.com) [12]. We used the Nextclade and Pangolin systems (freely available at https://clades.nextstrain.org/ and https://pangolin.cog-uk.io/, respectively) [13, 14] for strain classification.
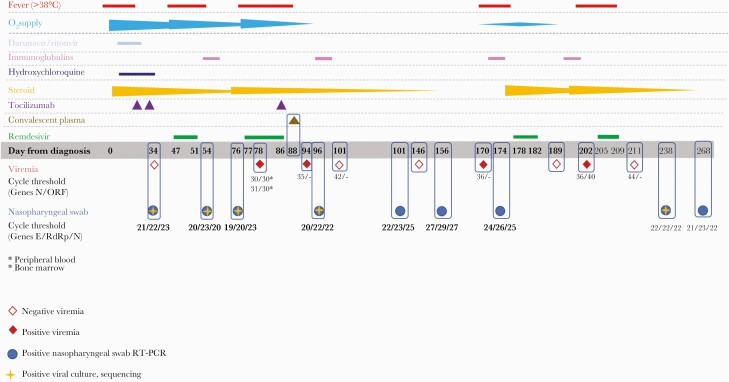
Figure 1.
Timeline of SARS-CoV-2 RT-PCR Ct values in nasopharyngeal swabs, SARS-CoV-2 RT-PCR Ct values in viremia (full rhombus = positive viremia, empty rhombus = negative viremia [Ct value for positive RT-PCR: 41 cycles]), administered treatments, O2 supply, and fever from March 2020 to December 2020. The full blue circle represents positive RT-PCR of nasopharyngeal swabs; the full blue circle with an orange cross indicates samples that underwent viral culture and sequencing. Abbreviation: RT-PCR, real-time polymerase chain reaction.
SARS-CoV-2 immunoglobulin G (IgG) and immunoglobulin M (IgM) antibody testing was performed using the Thermo Scientific COVID-19 Total Antibody ELISA test.
We assessed T cells and NK cells’ activity on 1 viremic (Day 84) blood sample and 1 aviremic (Day 146) blood sample by BD Fortessa X20 flow cytometer (BD Biosciences) using BD FACSDiva (version 8.0; BD Biosciences) and the Activation Induced Marker (AIM) assay [15], a cytokine-independent approach using specific peptide megapools (MP) identified by bioinformatic approaches [16]. We used multidimensional data reduction analysis through t-dependent Stochastic Neighbor Embedding (t-SNE) analysis to examine CD8+ T cells’ marker expression.
Patient Consent
The patient was aware of the conduct and dissemination of this case study and provided verbal and written informed consent allowing the dissemination of this case study. Unfortunately, the COVID-19-related emergency situation did not allow for safe storage of the written consent.
The study was carried out in accordance with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of Liguria Region (Comitato Etico Regione Liguria; N. CER Liguria 114/2020–ID 10420).
RESULTS
Case History
On March 24, 2020, during the first SARS-CoV-2 wave in Italy [17], a 70-year-old male was hospitalized in Genoa, Italy, for sudden onset of fever, dry cough, and respiratory distress. The patient had been recently diagnosed (January 2020) with non-Hodgkin lymphoma (mantle cell type, blastoid variant stage IV) and completed 2 cycles of rituximab, bendamustine, cytarabine chemo-immunotherapy in early March 2020. A follow-up computed tomography (CT) scan documented lymphoma remission. His other treated comorbidities included arterial hypertension, atrial flutter, benign prostatic hypertrophy, and hypercholesterolemia. He presented B-cell depletion (CD19+ count, 0; total lymphocyte count, 0.20 × 109/L) and hypo-IgG and -IgM (6.010 g/L; cutoff, 8.000–17.000 g/L; and 0.147; cutoff, 0.400–4.000, respectively).
His nasopharyngeal swab was positive by SARS-CoV-2 RT-PCR (Figure 1), and his CT scan showed bilateral interstitial pneumonia compatible with COVID-19. Blood tests showed anemia (Hb, 7.2 g/dL), absolute lymphopenia (0.20 × 109L), increased CRP (28.5 mg/L; cutoff, 0.5 mg/L), IL-6 (57.2 ng/L; cutoff, <3.4 ng/L), ferritin (875 mcg/L; cutoff, 400 mcg/L), and D-dimer (900 mcg/L; cutoff, 500 mcg/L). We define his initial day of presentation with severe respiratory failure and fever and SARS-CoV-2 RT-PCR-positive nasopharyngeal swab as day 0 of SARS-CoV-2 infection. The patient immediately required noninvasive mechanical ventilation with continuous positive airway pressure (CPAP). We started medical treatment with darunavir/ritonavir 800 mg/100 mg QD, hydroxychloroquine (400 mg twice daily [BID]), intravenous (IV) methylprednisolone (1 mg/kg/d), IV tocilizumab (8 mg/kg, 2 doses), and ceftaroline 600 mg BID according to the local COVID-19 treatment protocol in place at that time.
During the first month of hospitalization, the patient slowly recovered from acute respiratory failure; however, his follow-up SARS-CoV-2 RT-PCR nasopharyngeal swab results were consistently positive. On day 38, he underwent immunoglobulin administration (0.4 mg/kg/d for 3 days). On day 45, his fevers returned, and a repeat lung CT demonstrated increased interstitial lesions with a crazy-paving pattern. Bronchoalveolar fluid lavage results were as follows: SARS-CoV-2 RT-PCR positive; negative for bacterial, mycobacterial, and fungal growth with negative galactomannan; herpes simplex virus 1/2 DNA; positive for cytomegalovirus (CMV) DNA (3093 UI/mL). Serum CMV-DNA was 1181 UI/mL. Blood cultures, serum β-d-glucan, and serum galactomannan were negative.
Several days later, the patient developed acute respiratory failure requiring ventilation with helmet CPAP. Given his clinical symptoms and CT findings, we concluded that his symptoms were consistent with COVID-19 relapse and started remdesivir (200-mg loading dose, then 100 mg QD) for a total of 5 days. We monitored his plasma concentration of remdesivir: T0 concentration at day 3 of 29.35 ng/mL for remdesivir and 81.82 ng/µL for its metabolite GS-441524. Peak values (2 hours after the end of administration) were, respectively, 5364.31 ng/mL and 1310.86 ng/mL. After 6 days of helmet CPAP ventilation, on day 53, the patient was progressively weaned to low-flow oxygen support. He was afebrile but had a positive repeat nasopharyngeal swab for SARS-CoV-2, with RT-PCR cycle threshold values comparable to those of his initial swabs (Figure 1), suggestive of viral persistence.
On day 66, he presented with a new onset of high fever (38.4°C) and hypotension. Despite empiric broad-spectrum antibiotic therapy and successful treatment of an episode of CMV reactivation with ganciclovir, he remained persistently febrile. He developed severe respiratory failure again and required 4 days of ventilation with helmet CPAP. Repeat CT scan demonstrated persistent bilateral ground glass opacities and new bilateral pleural effusions. Repeat SARS-CoV-2 nasopharyngeal swab was positive, with RT-PCR cycle threshold (Ct) values comparable to previous assessments (Figure 1). SARS-CoV-2 viremia by RT-PCR was detected on day 78. A bone marrow aspirate and trephine biopsy were performed that were negative for lymphoma; however, SARS-CoV-2 RNA viremia was detected by RT-PCR in blood from bone marrow aspirate. Having excluded other infections and lymphoma relapse, we concluded that SARS-CoV-2 was responsible and attributed his signs and symptoms to a second COVID-19 relapse. We treated the patient with a 10-day course of remdesivir and added a single tocilizumab infusion (8 mg/kg). On day 88, the patient underwent COVID-19 convalescent plasma infusion. He slowly recovered, with progressive weaning from oxygen therapy and eventual clearance of SARS-CoV-2 viremia.
The third symptomatic COVID-19 relapse occurred in September 2020, with persistent fever and new onset of mild respiratory failure requiring low-flow oxygen supplementation, followed by newly detected SARS-CoV-2 viremia (Figure 1). We initiated another 5-day cycle of remdesivir with IV immunoglobulins (0.7 mg/kg/d for 3 days). Viral clearance in blood was observed 5 days after the last dose of remdesivir. The patient was weaned from oxygen, became afebrile, and was subsequently discharged. Repeat CT scan and positron emission tomography excluded lymphoma relapse.
The fourth COVID-19 relapse occurred in October 2020, when the patient was admitted for new fever in the absence of respiratory symptoms. SARS-CoV-2 viremia was detected (Figure 1), and we administered another 5-day cycle of remdesivir, which resulted in viremia clearance. The patient was eventually discharged to a long-term facility for SARS-CoV-2-positive patients. His last nasopharyngeal swab specimen was collected on day 268, and it was positive for SARS-CoV-2 (Figure 1). On day 271, the patient died following onset of hematuria, hypotension, and general wasting.
Viral Sampling and Analysis
RT-PCR Ct values of repeat nasopharyngeal swab samples are displayed in Figure 1, together with viremia assessments. All nasopharyngeal swabs were positive for SARS-CoV-2, with Ct values persistently demonstrating acute infection. We assessed viremia 9 times during the disease course, and the patient tested positive 4 times: All positive samples were obtained within the first days after symptom onset in 3 of the patient’s 4 COVID-19 disease relapses with clinically relevant symptoms. Viremia clearance was observed all 3 times, always after remdesivir treatment (5, 7, and 2 days after the end of treatment, respectively).
We assessed viral infectivity by viral culture on 5 nasopharyngeal swab samples: Cultures were all positive for viral replication, with a cytopathic effect observed at 48 hours. Comparing the isolated and sequenced viral strains by genomic analysis, we observed an increasing number of in-host mutations from April 7 to November 20. We summarize the lineage assignment and observed mutations in Table 1. Patient sequences all belonged to clade 20B, corresponding to lineage B.1.1. According to data from the literature [18–25], no mutations related to possible resistance to remdesivir were observed, also analysing mutations present with a frequency of less than 1% (Supplementary Table 1).
Table 1.
Lineage Classification of Strains Included in the Study According to Nextclade and Pangolin Identified Mutations With Cutoff >90%
| Sequence Date | Clade | Pangolin Lineage | Total Amino Acid Changes | ORF1a | ORF1b | S | ORF3a | N | ORF14 |
|---|---|---|---|---|---|---|---|---|---|
| Day 34 | 20B | B.1.1 | 7 | P1640L | P314L | D614G | R203K, G204R | G50N, G50E | |
| Day 54 | 20B | B.1.1 | 13 | K291E, K291N, T1322R, T1638I, R3561K, A4396V | P314L | D614G | S74F | R203K, G204R | G50N, G50E |
| Day76 | 20B | B.1.1 | 15 | T1322R, T1638I, R3561K, A3705V, S4398L | P314L | D614G, W1214C, I1221K, G1223C | G44A | R203K, G204R | G50N, G50E |
| Day 96 | 20B | B.1.1 | 13 | T283P, K291T, K292T, V306I, T1322R, T1638I, S4398L | P314L | D614G | R203K, G204R | G50N, G50E | |
| Day 238 | 20B | B.1.1 | 20 | R24C, V86F, T1322R, V1570L, T1638I, T4087I, S4398L | P314L, C455Y | H69Y, H69P, V70G, D614G, S982A | T151I | R203K, G204R | G50N, G50E |
Immunological Analysis
We performed SARS-CoV-2 serology repeatedly throughout the disease course. In all cases, no specific antibodies (IgG or IgM) were detected in the patient’s serum, even after infusion of convalescent plasma (which had an unspecified anti-SARS-CoV-2 titer).
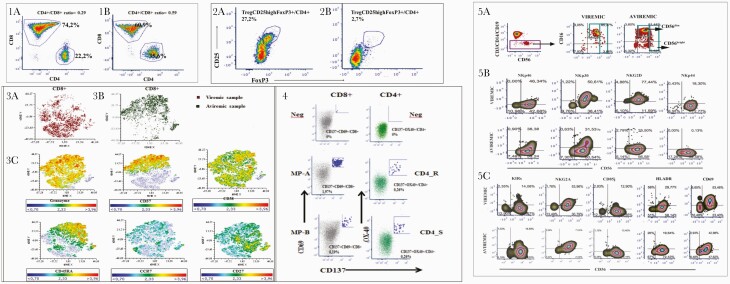
Detailed analysis by flow cytometry on peripheral blood mononuclear cells to characterize the T-cell dynamic in viremic and aviremic samples showed that CD4+ and CD8+ T cells in viremic and aviremic samples clustered differently, highlighting profound modifications in both the regulatory and effector arms of T-cell adaptive immunity. We found decreased CD4+/CD8+ ratios in both examined samples, with recovery of this ratio in the second (aviremic) sample (Figure 2, panels 1A and 1B). With regard to regulatory T cells (Treg), their proportions were particularly high in the viremic phase, when the proportion of total CD4+ cells was lower, as shown by a high frequency of CD4+FoxP3+CD25+ T cells, while their proportions dropped in the aviremic sample (Figure 2, panels 2A and 2B).
Figure 2.
Panel 1, Comparison of CD4+/CD8+ ratio in CD3+ T cells derived from viremic (panel 1A) and aviremic (panel 1B) samples. Panel 2, Comparison of CD4+FoxP3+CD25+ Treg cells in CD4+ T cells derived from viremic (panel 2A) and aviremic (panel 2B) samples. Panel 3, t-SNE analysis to the multiparametric analyses performed on CD8+ T cells in both samples. Panel 3A, t-SNE color plot of CD8+ lymphocytes in viremic samples. Panel 3B, t-SNE plots of concatenated CD8+ lymphocytes in aviremic samples. Panel 3C, description of phenotypic distributions to maturation and senescence markers, such as CD45RA, CCR7, CD27, CD38, CD57, and granzyme expression. Panel 4, FACS plot analysis of SARS-CoV-2-specific CD8+ and CD4+ T-cell responses of aviremic sample. To measure SARS-CoV-2-specific CD4+ and CD8+ T cells, peripheral blood mononuclear cells were stimulated with a spike MP (MP_S) and the class II MP representing all the proteomes without spike (“nonspike,” MP CD4_R) and HLA A and HLA B peptide megapools (MP A, MP B). SARS-CoV-2-specific CD4+ T-cell responses have been expressed as frequency of CD137+OX40+ cells on total CD4+ T-cell population, whereas SARS-CoV-2-specific CD8+ T-cell responses have been expressed as frequency of CD137+CD69+ cells on total CD8+ T-cell population. Panel 5, Flow cytometric analysis of NK cells at viremic or aviremic time points. Panel 5A, Gating strategy to identify peripheral blood NK cells. Panel 5B, Flow cytometric analysis of the expression of activating (NKp46, NKp30, NKp44, NKG2D) NK cell receptors. Panel 5C, Flow cytometric analysis of inhibitory NK cell receptors (KIRs, NKG2A, CD85j) and activation markers (HLADR, CD69) expressed on the surface of NK cells circulating in peripheral blood in the presence or absence of viremia. To identify SARS-CoV-2-specific CD8+ T cells, 2 class I peptide MPs have been used, based on epitope predictions for the 12 most common HLA A and B alleles, which collectively encompass 628 predicted HLA class I CD8+ T-cell epitopes from the entire SARS-CoV-2 proteome (CD8 MP-A and MP-B).
The t-SNE analysis performed on CD8+ T cells further highlighted marker expression differences between the 2 samples, showing that CD8+ T cells clustered differently in viremic and aviremic samples, respectively (Figure 2, panels 3A and 3B). Accordingly, differences in phenotypic distributions were observed with regard to maturation and senescence markers, such as CD45RA, CCR7, CD27, CD38, CD57, and granzyme expression (Figure 2, panel 3C). In particular, the expression of CD57, a marker of terminally differentiated cells, was a highly discriminating factor when comparing the 2 samples. We observed a significant increase of CD57+ effector memory (CD45RA+CCR7-CD27-CD57+) in the aviremic sample that was associated with granzyme expression, confirming a relationship between cell differentiation and cytolytic enzymes in terminal effector CD8+ T lymphocytes. In fact, the dynamic picture of CD8+ T-cell maturation showed a larger fraction of CD45RA+CCR7- terminal effector cells in the aviremic sample (68% of total CD8+ T cells) than that analyzed in the viremic phase (18% of total CD8+ T cells).
In fact, results from the AIM assay exhibited a significant frequency of CD137+CD69+ CD8+ T cells specific to 2 MP peptides representative of structural proteins (HLA A and HLA B peptide MPs; 1.9% and 0.19% of total peripheral CD8+ T cells, respectively) and a high percentage of CD137+OX40+ CD4+ T cells specific to spike MP (CD4-S) and all nonspike protein MP (CD4-R; 0.26% of total peripheral CD4+ T cells) (Figure 2, panel 4).
Flow cytometric analysis of NK cells in peripheral blood mononuclear cells, as defined by cells selected as CD3-CD14-CD19- by a negative gate, showed that during the clinical viremic phase associated with SARS-CoV-2 symptoms, CD56brightCD16+/- NK cells were decreased compared with the subsequent clinical condition (Figure 2, panel 5A), characterized by absent symptoms following remdesivir treatment. In addition, the viremic phase was associated with a 3-fold increase in NKG2D+CD56+ and a 140-fold increase in NKp44+CD56+ NK cells, while minor or no increases were seen for the other natural cytotoxicity receptors (NKp30, NKp46) on circulating NK cells (Figure 2, panel 5B). Increased NKp44+ NK cell frequency during viremia was associated with increased NK cell activation, as indicated by HLA-DR expression (Figure 2, panel C), accounting for 33% of CD56+ NK cells. On the other hand, the circulation of NKp44+CD56+ NK cells was shut off when viremia was absent, although some level of NK-cell activation (21% of CD56+ NK cells) persisted.
DISCUSSION
In this case study, we describe a case of 8-month persistence of SARS-CoV-2 RT-PCR-positive nasopharyngeal swab in an immunosuppressed patient with non-Hodgkin lymphoma, with 4 clinically symptomatic relapses of COVID-19. During each relapse, the patient received a cycle of remdesivir that resulted in viremia clearance; however, viable virus remained detectable on nasopharyngeal swabs. To the best our knowledge, this is the longest persistence of SARS-CoV-2 RT-PCR-positive nasopharyngeal swabs reported. Our report suggests that this persistence may be due to viable SARS-CoV-2 virus that can determine a well-defined innate and adaptive cellular immune response compatible with disappearance of viremia, whose presence correlated with symptomatic relapses, as previously reported [26, 27].
Remdesivir was effective in clearing blood viremia and was associated with clinical recovery every time it was administered but was unable to clear SARS-CoV-2 from the upper airways, as inferred by the persistence of viable virus on repeat nasopharyngeal swabs. A 10-day treatment course with remdesivir was performed on 1 occasion and was associated with viremia clearance and a prolonged (70 days) disease-free period. Subsequent treatment courses were 5 days long based on data demonstrating noninferiority of 5-day courses vs 10-day courses [28]. As other authors have done, we can speculate that longer courses of remdesivir might be beneficial in immunocompromised patients [29, 30]; however, at the time of writing, no definite evidence in this regard has been established, and 5 days were sufficient for viremia clearance and clinical improvement in our case. Furthermore, prolonged antiviral administration, as a prophylaxis of clinical relapse, is an unexplored yet interesting option.
Convalescent plasma therapy has proven efficacious in viremia clearance and has resulted in resolution of COVID-19 symptoms in specific circumstances, such as in patients with B-cell depletion after anti-CD20 treatment [31]. In our case, a single treatment course was administered because of an apparent lack of benefit inferred from negative data from randomized trials [32, 33] and because of difficulties obtaining convalescent plasma with documented high neutralizing antibody levels. Unfortunately, treatment with monoclonal antibodies was unavailable at the time. The use of monoclonal antibodies for chronic SARS-CoV-2 infection with COVID-19 recurrences in immunocompromised patients who are unlikely to develop a natural immune response to SARS-CoV-2 due to B-cell depletion could offer a promising management strategy, particularly after anti-CD20-containing chemotherapy.
As the disease course of this immunosuppressed patient was characterized by persistent viral shedding with intermittent viremia, we set out to study whether an adaptive and innate immune response could be characterized in the different viremic phases. Taken together, our analyses highlighted profound differences in both the regulatory and effector arms of T-cell adaptive immunity in viremic and aviremic samples with an activated cytotoxic phenotype displayed by acute-phase SARS-CoV-2-specific T cells, as previously reported [34, 35].
During the viremic phase, we observed a high frequency of CD4+FoxP3+CD25+ T cells (Treg cells, 10-fold higher than in the aviremic phase) and a reduced frequency of terminal effector CD8+ T lymphocytes (18% vs 68%), which is attributable to CD8+ T cells’ ability to migrate into tissues affected by inflammation to control the virus. This finding is in line with the dual function performed by Treg during SARS-CoV-2 infection, as it may indicate that in patients with COVID-19 CD4+ Treg cells are generated/recruited in the attempt to counteract the overwhelming inflammatory response, but also that Treg expansion may hamper virus-specific effector responses.
Moreover, the analysis of exhaustion marker expression confirmed the different dynamics in the viremic and aviremic samples. In fact, terminally differentiated memory cells were more present in the aviremic sample with respect to the viremic sample, and consequently the frequency of CD8+ PD-1+ and CD8+EOMES+ cells decreased in the aviremic sample (CD8+PD-1+/CD8+ and CD8+EOMES+/CD8+ were 53% and 49% in the viremic sample, respectively, while CD8+PD-1+/CD8+ and CD8+EOMES+/CD8+ were 15.7% and 22.3% in the aviremic sample, respectively). These data seem to suggest the elimination of a brake on the immune response. The analysis of specific T-cell responses in the aviremic sample confirms this hypothesis, and a fraction of specific CD4+ and CD8+ T cells were described in the aviremic sample.
Furthermore, we observed a high level of NK-cell repertoire perturbation with relevant involvement of NKp44+ and NKG2D+ NK cells during SARS-CoV-2 viremia. A considerable degree of activation of NK cells was observed, with increased HLA-DR expression. This level of activation is comparable to previous findings observed in people with HIV with either detectable [36] or undetectable viremia due to suppressive antiretroviral therapy [37] or in elite controllers and long-term nonprogressors [38]. A level of NK-cell activation comparable to our case was also found in immunocompetent, symptomatic patients with COVID-19 pneumonia requiring oxygen supplementation at the time of admission as inpatients [39]. The change in frequency of NKp44- and NKG2D-expressing circulating NK cells observed in viremic and aviremic disease phases contributes to the notion of the presence and role of NK cells and the innate reactivity underlying adaptive T-cell responses and to their association to transient control of virus replication. In our experience, in immunocompetent patients with severe COVID-19, concerning the cytotoxic CD8+ T-cell compartment, we observed a decrease of naïve cells and increased stem central memory and terminal effector memory cells.
In conclusion, this case study adds to the existing literature on COVID-19 relapses and prolonged SARS-CoV-2 shedding in immunocompromised patients [3–7], highlighting the need for special attention in this particular population both in terms of clinical management comprising isolation measures and immunosuppressive treatment [5, 40], as well as the need for possible immunotherapeutic interventions, including monoclonal antibodies and vaccination.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Thanks to B.C. and his family for enduring the hardest times with dignity and determination and for always trusting us.
Thanks to Dr. Rosanna Vagge for her tireless work and for being always available.
A special thanks to Dr. Justin Koh, who provided a fast and accurate English revision.
Financial support. The authors received no specific funding for this work.
Potential conflicts of interest. None of the authors reported any conflict of interest relevant to the present work. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. C.S., C.D., and M.M. drafted the manuscript and collected the data; B.B., A.O., A.L., A.B., and G.Z. performed the virological analysis; A.D.M., F.B., D.F., A.P., T.A., R.D.P., and C.D. carried out the immunological analysis and data interpretation; S.B. performed the therapeutic drug monitoring analysis; A.D.B., M.B., G.S., E.D., F.B., and G.B. revised and provided conceptual insight to the manuscript.
References
- 1. Cevik M, Tate M, Lloyd O, et al. . SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021; 2:e13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrne AW, McEvoy D, Collins AB, et al. . Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open 2020; 10:e039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi B, Choudhary MC, Regan J, et al. . Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baang JH, Smith C, Mirabelli C, et al. . Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aydillo T, Gonzalez-Reiche AS, Aslam S, et al. . Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 2020; 383:2586–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakajima Y, Ogai A, Furukawa K, et al. . Prolonged viral shedding of SARS-CoV-2 in an immunocompromised patient. J Infect Chemother 2021; . 27:387–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avanzato VA, Matson MJ, Seifert SN, et al. . Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–12.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diao B, Wang C, Tan Y, et al. . Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 2020; 11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilk AJ, Rustagi A, Zhao NQ, et al. . A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 2020; 26:1070–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liotti FM, Menchinelli G, Marchetti S, et al. . Evaluation of three commercial assays for SARS-CoV-2 molecular detection in upper respiratory tract samples. Eur J Clin Microbiol Infect Dis 2021; 40:269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Global Initiative on Sharing All Influenza Data (GISAID). Clade and Lineage Nomenclature Aids in Genomic Epidemiology Studies of Active hCoV-19 Viruses. Munich: GISAID; 2020. Available at: https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/. Accessed 20 November 2020. [Google Scholar]
- 12. Kearse M, Moir R, Wilson A, et al. . Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nextclade. Available at: https://clades.nextstrain.org/. Accessed 17 December 2020.
- 14.Pangolin COVID-19 Lineage Assigner. Available at: https://pangolin.cog-uk.io/. Accessed 17 December 2020.
- 15. Grifoni A, Weiskopf D, Ramirez SI, et al. . Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181:1489–501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martini S, Nielsen M, Peters B, Sette A. The Immune Epitope Database and Analysis Resource Program 2003-2018: reflections and outlook. Immunogenetics 2020; 72:57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Istituto Superiore di Sanità, Epidemiology for Public Health. COVID-19 integrated surveillance data in Italy. Available at:https://www.epicentro.iss.it/en/coronavirus/sars-cov-2-dashboard. Accessed 19 November 2020.
- 18. Lin CH, Yang CY, Ou SC, et al. . The impacts of antivirals on the coronavirus genome structure and subsequent pathogenicity, virus fitness and antiviral design. Biomedicines 2020; 8:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Padhi AK, Shukla R, Tripathi T. Rational design of the remdesivir binding site in the RNA-dependent RNA polymerase of SARS-CoV-2: implications for potential resistance. iScience 2021; 24. [DOI] [PMC free article] [PubMed]
- 20. Shannon A, Le NT, Selisko B, et al. . Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res 2020; 178:104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agostini ML, Andres EL, Sims AC, et al. . Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 2018; 9:e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo MK, Albariño CG, Perry JK, et al. . Remdesivir targets a structurally analogous region of the Ebola virus and SARS-CoV-2 polymerases. Proc Natl Acad Sci U S A 2020; 117:26946–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martinot M, Jary A, Fafi-Kremer S, et al. . Remdesivir failure with SARS-CoV-2 RNA-dependent RNA-polymerase mutation in a B-cell immunodeficient patient with protracted Covid-19. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tchesnokov EP, Gordon CJ, Woolner E, et al. . Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem 2020; 295:16156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Artese A, Svicher V, Costa G, et al. . Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses. Drug Resist Updat 2020; 53:100721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hagman K, Hedenstierna M, Gille-Johnson P, et al. . SARS-CoV-2 RNA in serum as predictor of severe outcome in COVID-19: a retrospective cohort study. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bermejo-Martin JF, González-Rivera M, Almansa R, et al. . Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care 2020; 24:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldman JD, Lye DCB, Hui DS, et al. ; GS-US-540-5773 Investigators . Remdesivir for 5 or 10 days in patients with severe Covid-19. N Engl J Med 2020; 383: 1827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Helleberg M, Niemann CU, Moestrup KS, et al. . Persistent COVID-19 in an immunocompromised patient temporarily responsive to two courses of remdesivir therapy. J Infect Dis 2020; 222:1103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Camprubí D, Gaya A, Marcos MA, et al. . Persistent replication of SARS-CoV-2 in a severely immunocompromised patient treated with several courses of remdesivir. Int J Infect Dis 2021; 104:379–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hueso T, Pouderoux C, Péré H, et al. . Convalescent plasma therapy for B-cell-depleted patients with protracted COVID-19. Blood 2020; 136:2290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li L, Zhang W, Hu Y, et al. . Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial [published correction appears in JAMA 2020; 324:519]. JAMA 2020; 324:460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agarwal A, Mukherjee A, Kumar G, et al. ; PLACID Trial Collaborators . Convalescent plasma in the management of moderate COVID-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) [published correction appears in BMJ 2020; 371:m4232]. BMJ 2020; 371:m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. ; Karolinska COVID-19 Study Group . Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020; 183:158–68.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thevarajan I, Nguyen THO, Koutsakos M, et al. . Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med 2020; 26:453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fogli M, Costa P, Murdaca G, et al. . Significant NK cell activation associated with decreased cytolytic function in peripheral blood of HIV-1-infected patients. Eur J Immunol 2004; 34:2313–21. [DOI] [PubMed] [Google Scholar]
- 37. Lichtfuss GF, Cheng WJ, Farsakoglu Y, et al. . Virologically suppressed HIV patients show activation of NK cells and persistent innate immune activation. J Immunol 2012; 189:1491–9. [DOI] [PubMed] [Google Scholar]
- 38. Marras F, Nicco E, Bozzano F, et al. . Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci U S A 2013; 110:11970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bozzano F, Dentone C, Perrone C, et al. ; GECOVID study group . Extensive activation, tissue trafficking, turnover and functional impairment of NK cells in COVID-19 patients at disease onset associates with subsequent disease severity. PLoS Pathog 2021; 17:e1009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lamure S, Duléry R, Di Blasi R, et al. . Determinants of outcome in Covid-19 hospitalized patients with lymphoma: a retrospective multicentric cohort study. EClinicalMedicine 2020; 27:100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.